- 1School of Pharmacy, Southwest Medical University, Luzhou, China
- 2Sichuan Institute for Translational Chinese Medicine, Sichuan Academy of Chinese Medical Sciences, State Key Laboratory of Quality Evaluation of Traditional Chinese Medicine, Sichuan Engineering Technology Research Center of Genuine Regional Drug, Sichuan Provincial Engineering Research Center of Formation Principle and Quality Evaluation of Genuine Medicinal Materials, Translational Chinese Medicine Key Laboratory of Sichuan Province, Chengdu, China
- 3Department of Radiation Oncology, Radiation Oncology Key Laboratory of Sichuan Province, Sichuan Clinical Research Center for Cancer, Sichuan Cancer Hospital and Institute, Sichuan Cancer Center, Affiliated Cancer Hospital of University of Electronic Science and Technology of China, Chengdu, China
- 4College of Food and Biological Engineering, Chengdu University, Chengdu, China
- 5Traditional Chinese Medicine Hospital Affiliated to Southwest Medical University, Classical Chinese Medicine Diagnosis and Treatment Center, Luzhou, China
The gut microbiota is not just a simple nutritional symbiosis that parasitizes the host; it is a complex and dynamic ecosystem that coevolves actively with the host and is involved in a variety of biological activities such as circadian rhythm regulation, energy metabolism, and immune response. The development of the immune system and immunological functions are significantly influenced by the interaction between the host and the microbiota. The interactions between gut microbiota and cancer are of a complex nature. The critical role that the gut microbiota plays in tumor occurrence, progression, and treatment is not clear despite the already done research. The development of precision medicine and cancer immunotherapy further emphasizes the importance and significance of the question of how the microbiota takes part in cancer development, progression, and treatment. This review summarizes recent literature on the relationship between the gut microbiome and cancer immunology. The findings suggest the existence of a “symbiotic microecosystem” formed by gut microbiota, metabolome, and host immunome that is fundamental for the pathogenesis analysis and the development of therapeutic strategies for cancer.
1 Introduction
Life, in the form of prokaryotes, first appeared on Earth approximately 3.8 billion years ago, while the earliest eukaryotic single-celled organisms emerged approximately 1.8 billion years ago. Evidence suggests that eukaryotes originated from the fusion and aggregation of prokaryotes into multicellular complexes, initially utilizing the genetic information of prokaryotes. This process led to the differentiation into animals and plants, with microorganisms playing a crucial role throughout the entire process. Hundreds of different types of microorganisms colonize the vertebrate intestine in a rather mutually beneficial interaction with the host (1–3). Among the gut microbiota, the members that play a dominant role are Firmicutes (Gram-positive bacteria without a true outer membrane) and Bacteroidetes (Gram-negative bacteria with an outer membrane) phyla (4–7). In healthy individuals, the gut microbiota maintains a dynamic balance between beneficial and opportunistic pathogenic bacteria (8). Moreover, these microorganisms actively contribute to the production of neurotransmitters, enzymes, and vitamins. For example, vitamins B and K produced by bacteria are involved in immune and metabolic functions (9–11). A dynamic equilibrium of the gut microbiota with beneficial bacteria predominating is the optimal condition for the gut microbiota. Disruptions to the dynamic equilibrium of the gut microbiota can lead to changes in the composition, amount, and activity of the microbial community (12, 13). These disruptions may occur due to factors such as age, dietary preferences, and illness. Consequently, the mucosal barrier may become impaired, leading to changes in cytokines and cell signaling, suppression of commensal bacteria and probiotic colonization, and increased proliferation of intestinal pathogens. These alterations can compromise both local and systemic immune responses (14). Damage to the mucosal barrier can result in the transfer of gut microbes to mesenteric lymph nodes (MLNs) and peripheral circulation, inducing Th17 and effector T-cell activation, promoting neutrophil infiltration, and activating local and systemic inflammatory responses (15).
The intestinal microbiota plays a complex and important role in the development and progression of tumors (16). The gut microbiota exhibits a bidirectional role in tumor development and progression. On one hand, certain bacteria can promote cancer by producing carcinogenic metabolites, inducing inflammation, and impairing immune responses. On the other hand, specific interventions such as probiotics and fecal microbiota transplantation (FMT) have demonstrated potential antitumor effects. Microbiota-driven carcinogenic mechanisms exhibit significant heterogeneity across different organs. For instance, specific bacteria have been implicated as pathogenic factors in gastric cancer, while changes in the intestinal microbiota and metabolites induced by dietary cholesterol drive NAFLD-HCC (non-alcoholic fatty liver disease-liver cancer) (17). However, the specific mechanisms involved in these processes remain largely unclear (18) (Figure 1).
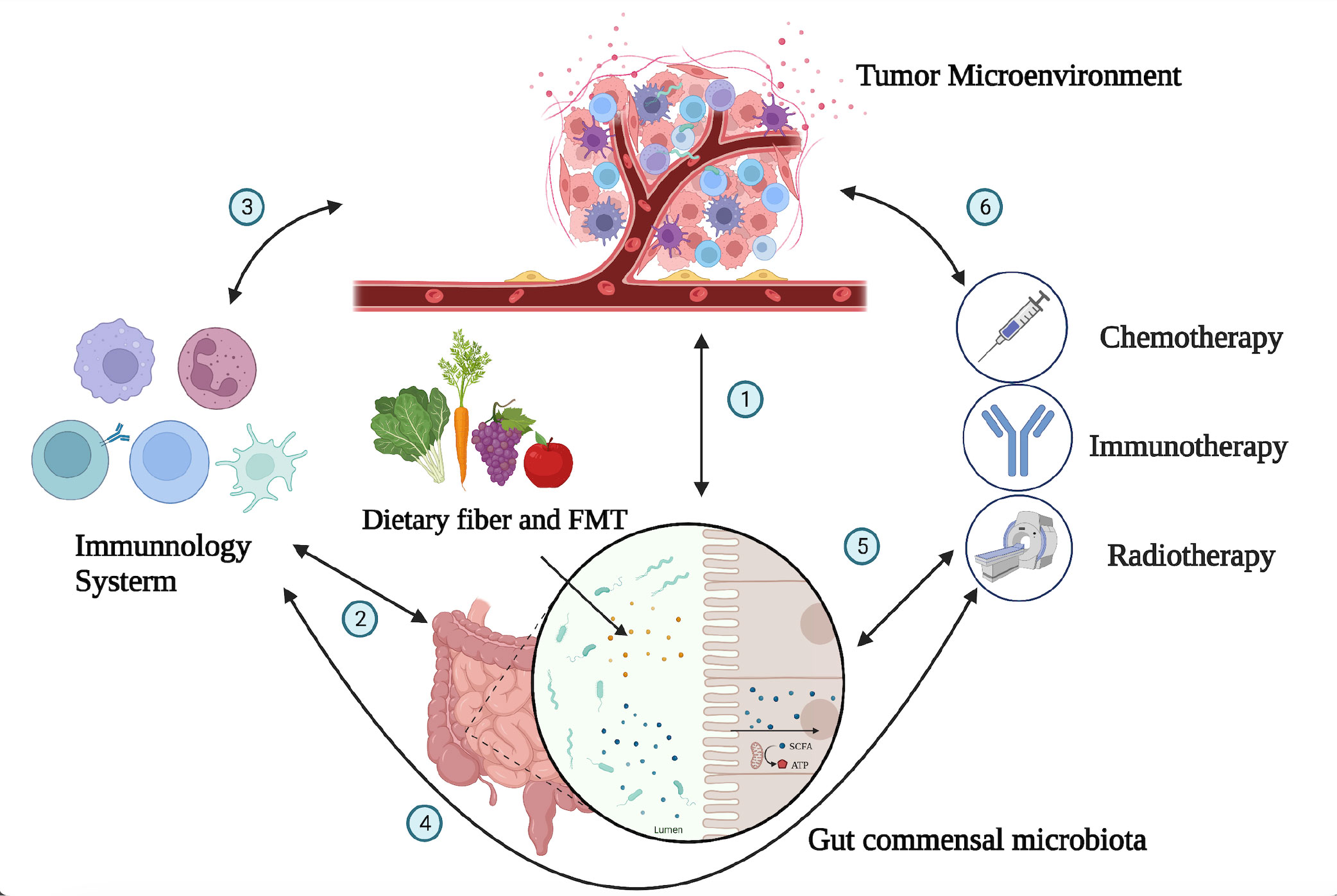
Figure 1 Interactions between the tumor microenvironment, gut microbiota, immune system, and immunotherapy. The gut microbiota can affect the occurrence and progression of tumors through a variety of mechanisms, including directly participating in the occurrence and progression of tumors; influencing the development and recognition of immune cells in the immune system, affecting the ability of immune cells to exert antitumor immunity; and collaborating with tumor treatment to improve treatment effectiveness. The numbers (1–6) in the figure demonstrate the dynamic and intricate connections among the tumor microenvironment, immune system, gut microbiota, and tumor treatment, with mutual influences on each other (1). The gut microbiota can influence tumor development through mechanisms such as modulating immune responses, affecting the growth and apoptosis of tumor cells, and regulating the expression of tumor-related genes. Conversely, changes in the tumor microenvironment can also impact the composition and function of the gut microbiota. The interplay between the tumor microenvironment and gut microbiota is a complex bidirectional relationship that holds significant implications for tumor development and treatment (2). The immune system can maintain immune balance in the gut by regulating the composition and function of the gut microbiota, and the gut microbiota can also influence the development and function of the immune system (5). The composition of the gut microbiota is related to individual responses to chemotherapy, radiation therapy, and immunotherapy. By modulating the composition and function of the gut microbiota, it may be possible to improve the effectiveness of tumor treatment and enhance patient survival rates and quality of life (3, 4, 6). The tumor microenvironment is closely related to the immune system, as cellular molecules and factors such as blood vessels within the tumor microenvironment can regulate immune system functions. Various treatment methods can impact the tumor microenvironment and immune system in different ways. Thus, there exists a complex interaction network among the immune system, tumor microenvironment, and tumor treatment.
2 The complicated bidirectional role of gut microbiome in cancer
2.1 Certain bacteria promote gastrointestinal cancer by inducing local inflammation and impairing the host immune response
If we consider the gut as the outer surface of the body (similar to the intestinal and skin surfaces), the gut microbiota belongs to the external symbiotic community. Clearly, it is not desirable to have such a situation inside cells or in peripheral blood and tissues (except in the case of individuals who develop symptoms after viral infections). In other words, the tolerance of the host immune system toward gut microbiota is mostly limited to the outer surface of tissues. To combat microbes that invade the internal tissues, the immune system needs to be activated or in a state of readiness (such as immune cells that are exposed to a large number of microbial antigens or immune receptor cells with diversity that can respond to microbial and mutated cell stimuli). This phenomenon is an important component of immune function because microbes from the outer surface of tissues can enter through various pathways. Without immune receptor cells in a state of readiness, we would struggle to defend against different types of microbial invasions. This also suggests that infants should be exposed to as many environmental microbes as possible soon after birth to stimulate the immune system and generate greater diversity, thereby maintaining a state of readiness. The presence of gut microbiota is crucial for maintaining the immune system’s readiness, and under normal circumstances, the two exist in a dynamic balance. However, if the quantity of a particular microbe or several microbes suddenly increases or remains at a consistently high level for a prolonged period, it can significantly impact the immune system. Apart from its pathogenicity (although current gut symbiotic microorganisms have evolved to have low virulence, a sharp increase in pathogenicity can occur when a large amount of the same strain becomes more virulent), it can greatly affect immune receptor diversity and immune spatial layout and directly contribute to the development of diseases. A classic example of a bacterial species that causes gastric cancer is Helicobacter pylori (19, 20). H. pylori overcomes the natural defense of the stomach by producing urease to neutralize the local acidic environment and using flagella, which have the ability to penetrate the mucous layer and interact with epithelial cells. Other characteristics contributing to the survival of H. pylori include its ability to adhere to the epithelium, produce catalase to neutralize hydrogen peroxide, and obtain nutrients (21). H. pylori is a cell toxin-associated antigen A (CagA) gene-positive bacterium that adheres to gastric epithelial cells by binding to the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) through the outer membrane adhesin HopQ (22) and then delivers the effector protein CagA, peptidoglycan metabolites, and DNA directly to epithelial cells through a type IV secretion system (23). Translocated CagA protein is localized to the cell membrane and then undergoes tyrosine phosphorylation at the Epiya site, mediated by the SRC family tyrosine kinase. Phosphorylated CagA interacts with intracellular signaling molecules through the Src Homology 2 (SH2) domain, releases its activity regulation, and triggers pathological effects on gastric cancer (24–26) (Figure 2, Table 1).
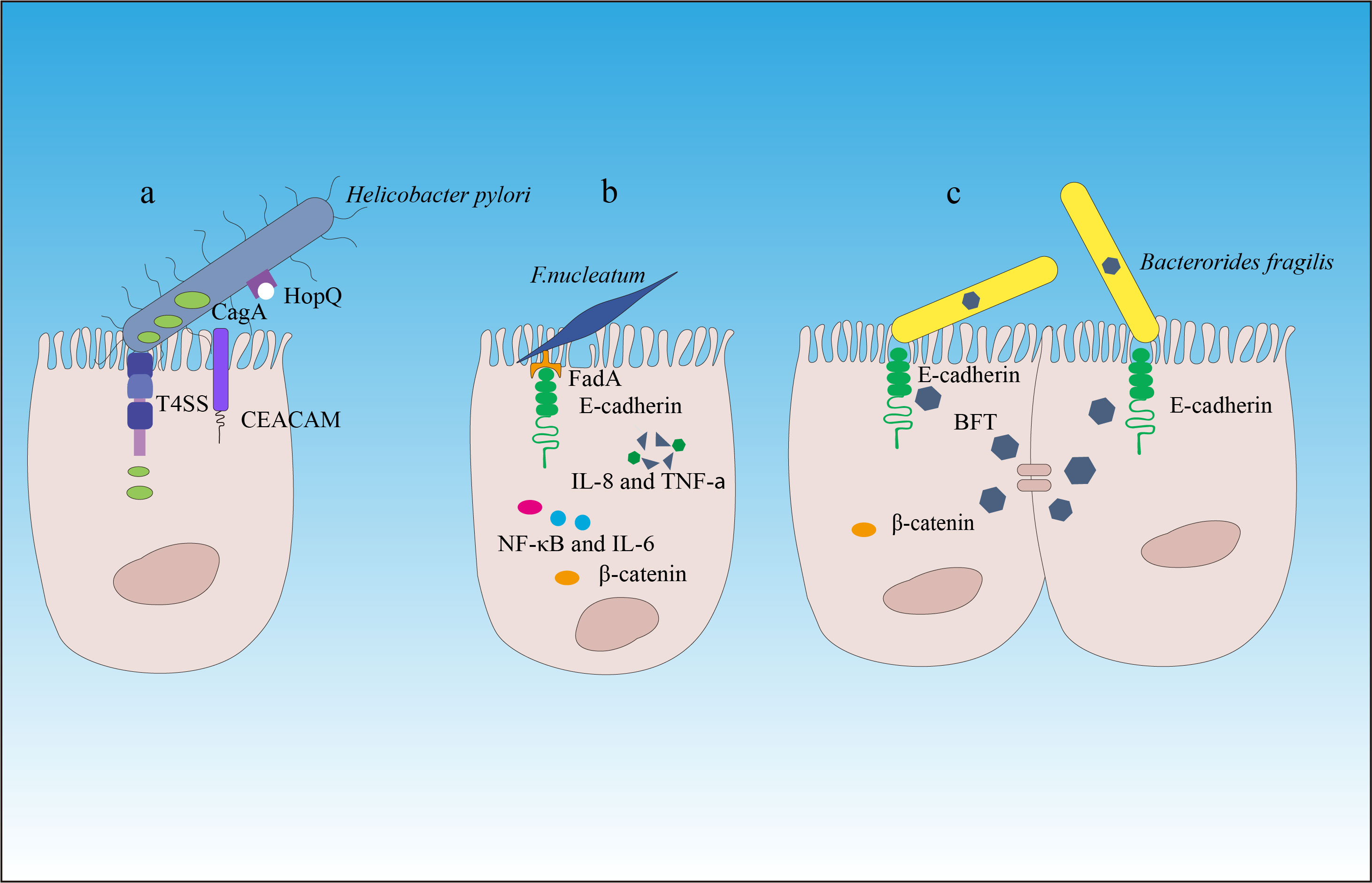
Figure 2 The carcinogenic mechanisms of Helicobacter pylori, Fusobacterium nucleatum, and Bacteroides fragilis. (A) Helicobacter pylori attaches to the gastric epithelial cells by binding to the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) through the outer membrane adhesin HopQ (22). Then, through the type IV secretion system of CagA, peptidoglycan metabolites, and DNA, the effector protein CagA is directly delivered into the epithelial cells to exert carcinogenic effects. (B) Through its mechanism of action, Fusobacterium nucleatum contributes to the development and progression of colorectal cancer. (C) Enterotoxigenic Bacteroides fragilis (ETBF) produces toxins that target the tight junctions of intestinal epithelial cells, cleaving E-cadherin and promoting inflammation and destruction of the intestinal mucosal barrier. This induces chronic intestinal inflammation and tissue damage in colorectal cancer. β-catenin signaling alterations are a frequent target of cancer-associated microbes. Some microbes bind E-cadherin on colonic epithelial cells, with altered polarity or within a disrupted barrier, and trigger β-catenin activation. Other microbes inject effectors (e.g., CagA) that activate β-catenin signaling, resulting in dysregulated cell growth, acquisition of stem cell–like qualities, and loss of cell polarity.
The dysbiosis of the gut microbiota is also closely related to the occurrence of colorectal cancer (CRC) (27, 65, 66). Previous research has revealed that the prevalence of several bacterial groups, such as Bacteroides fragilis and Fusobacterium nucleatum, in the fecal microbiota of CRC patients was higher than that of normal individuals (67–69). F. nucleatum has been shown to bind to host epithelial and endothelial cells through FadA adhesin and induce a series of inflammatory reactions mediated by Nuclear Factor-kappa B (NF-κB) and interleukin (IL)-6 (32–34). Hong et al. (70) have found that F. nucleatum abundance correlated with high glucose metabolism in patients with CRC. F. nucleatum induces a dramatic decline of m6A modifications in CRC cells and patient-derived xenograft (PDX) tissues by downregulating an m6A methyltransferase, METTL3, contributing to the induction of CRC aggressiveness (71). In addition, F. nucleatum can also inhibit the cytotoxic functions of tumor-infiltrating lymphocytes and natural killer (NK) cells by binding to the inhibitory immune receptor TIGIT through another adhesin, Fap2, thereby suppressing immune surveillance (33, 72). In addition, F. nucleatum may contribute to epithelial–mesenchymal transition (EMT), so it is tightly associated with cancer cell invasion, suppression of antitumor immune responses, stemness, and treatment resistance (73).
Enterotoxigenic Bacteroides fragilis (ETBF) that produces the metalloprotease Bacteroides fragilis toxin (BFT) promotes inflammation and disrupts the intestinal barrier function by targeting the tight junctions of intestinal epithelial cells (IECs), which is associated with acute diarrhea and inflammatory bowel disease (74), thereby inducing chronic inflammation and tissue damage in CRC (27, 28, 75–77). In addition, ETBF has been found to be enriched in the gut microbiota of CRC patients, and its enrichment is associated with poor prognosis of CRC (29, 78). ETBF, which is enriched in some human CRCs, can stimulate E-cadherin cleavage via BFT, leading to β-catenin activation (79). Studies have also found that ETBF plays an important role in promoting CRC through the Toll-like receptor 4 (TLR4)-Nuclear Factor of Activated T-cells 5 (NFAT5)-dependent upregulation of Jumonji domain-containing protein 2B (JMJD2B) levels in stem cell regulation (80, 81).
2.2 Gut microbiota also promotes extraintestinal cancers through bacterial translocation and production of bioactive molecules into circulation
Organs outside of the gastrointestinal tract are also remotely affected by the gut microbiota’s carcinogenic effects. A study found that certain gut microbiota, such as the Bacteroides and Ruminococcaceae, can contribute to the development of hepatocellular carcinoma (HCC) by exacerbating hepatic inflammation, accumulating toxic compounds, and causing liver steatosis (64). Obese and lean people have substantially distinct gut microbiota compositions, especially in terms of the proportion of bacteria that produce pro-inflammatory lipopolysaccharides (LPSs). In accordance with thorough experimental investigations, transplanting the microbiota of healthy individuals into obese mice can reduce steatosis while causing hepatic steatosis in mice that are fed normally (82).
Since symbiotic microbiota are frequently stable in the gastrointestinal system, researchers posed an essential question: where do tumor-related bacteria in remote organs come from? Geller et al. proposed that pancreatic ductal adenocarcinoma (PDAC)-related bacteria can retrogradely originate from the gastrointestinal tract (48, 83). Pushalkar et al. (49) provided evidence of bacterial migration from the gut to the pancreas, as well as a time-dependent association between gut dysbiosis and Kras activation in PDAC. Vitiello et al. (83) also found that gut dysbiosis can directly promote oncogenic signaling in the pancreas. Accordingly, dysbiosis and mislocalization of the gut microbiota have been associated with the onset of pancreatic and liver cancers, which is in line with the theory that the intestine, liver, and pancreas maintain continuous interactions. Microbiota can also indirectly affect tumor progression through the production and metabolism of bioactive molecules, which may reach tumors and metastatic sites via systemic circulation, such as bacterial LPS, which can enter the bloodstream and affect tumor formation in tissues far from the gastrointestinal tract (84). Deoxycholic acid (DCA) and lithocholic acid (LCA) can cause DNA damage by increasing the production of reactive oxygen species (ROS), leading to cell senescence, chronic inflammation, and tumorigenesis (85–87).
2.3 Colonization of certain strains of probiotics in gut may have an antitumor effect through modulating the immune system and reducing inflammation
Maintaining a healthy and balanced gut microbiota can help suppress tumor development. The symbiotic microbiota benefits from the nutrient-rich environment in the gut, where the microbiota will produce hundreds of proteins and metabolites that regulate key host functions, including nutrient processing, energy balance maintenance, and immune system development. The gut microbiota has complex impacts on the growth of tumors, and probiotics that colonize the gut can influence these processes by enhancing immune responses to antigens and antibodies, inhibiting monocyte proliferation, and upregulating anti-inflammatory cytokines such as IL-10 and IL-12. The gut microbiota contributes to reducing pro-inflammatory cytokines such as IL-1β and IL-6, exhibiting effective anti-inflammatory activity. Goldin et al. have found that probiotics play an important role in preventing CRC (88, 89).
Lactic acid bacteria and Bifidobacterium are involved in regulating pH and bile acid processes (90). Inhibiting the activity of glucoside and nitrite reductase and decreasing the synthesis of carcinogenic chemicals are two effects that Lactobacillus acidophilus can have on the gut’s putrefactive bacteria. Lactic acid bacteria and Bifidobacterium can physically degrade potential carcinogens and their metabolites, such as heterocyclic amines (91–93), nitrosamines, and aflatoxins, thereby inhibiting the development of various cancers such as gastric, esophageal, liver, colon, and bladder cancers (94). In female mice receiving subcutaneous injection of breast cancer cells (4T1), giving them plant-derived Lactobacillus rich in selenium nanoparticles (SeNPs) was shown to induce effective immune responses by increasing levels of pro-inflammatory cytokines interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), and IL-2 and increasing NK cell activity, significantly inhibiting tumor development and increasing survival rates compared to mice receiving only plant-derived Lactobacillus or control model mice (95). Interestingly, whether through preventative use of milk fermented with the Lactobacillus CRL431 strain of probiotics or starting use of milk fermented with the CRL431 strain of probiotics after injection of breast cancer cells (4T1), giving probiotics delayed or prevented tumor development compared to mice injected with tumor cells (96).
The most common treatment for non-muscle-invasive bladder cancer is transurethral resection of bladder tumor (TURBT) (97), followed by single-dose intravesical immunotherapy with Bacillus Calmette-Guérin (BCG), which is an effective method for preventing bladder cancer recurrence and progression after bladder surgery (98–100) (Table 2). BCG works by inducing nonspecific immune reactions and mediating antitumor effects through the activation of inflammatory responses. CD4+ and CD8+ lymphocytes, NK cells, granulosa cells, giant cells, and dendritic cells (DCs) may be involved (118–121). In a mouse model of subcutaneously implanted CT26 colon cancer cells, it was found that pretreatment with Lactobacillus plantarum (KC836552.1) significantly reduced tumor growth, prolonged survival time, activated innate immunity, and increased the intratumor levels of CD8+ T and NK cells (122). Although the antitumor effect of Lactobacillus rhamnosus (JF414108.1) on colon cancer is not clear, it has been shown to be more effective than BCG in reducing bladder cancer recurrence rates. L. rhamnosus GG (LGG) can recruit large numbers of neutrophils and macrophages to the tumor site, thereby promoting tumor regression (104).
In addition, certain specific types of bacteria can alter immune responses by promoting the development of certain subtypes of lymphocytes. For example, segmented filamentous bacteria (SFB) can induce the production of IL-17 and IL-22, which is beneficial for the production of Th17 cells in mice (123–125). When using a recombinant germ-free (GF) mouse model with the gut microbiota of gas-producing Clostridia and fragile bacteria, the fragile bacteria promote the production of regulatory T cells (Tregs) and IL-10-secreting T cells by binding surface polysaccharide A (PSA) to TLRs on Treg surfaces (126, 127). Sivan et al. compared melanoma growth in mice harboring different commensal microbiota and observed differences in spontaneous antitumor immunity that were abolished after mouse cohousing or FMT. The 16SrRNA sequencing of mouse feces identified Bifidobacterium that may cause differences in tumor growth and antitumor immunity in mice; studies have also found that the Bifidobacterium can enhance DC function, leading to the initiation and clustering of CD8+ T cells in the tumor microenvironment (TME), thereby improving the antitumor immune function (115, 128, 129).
2.4 Metabolites produced by gut microbiota could influence the function of the intestinal epithelial barrier as well as the immune response
Intestinal symbiotic microbiota can also affect host immunity through metabolites. Many metabolites produced by microbes come from undigested or partially digested dietary fibers in the body. The metabolites that play a major role in gut and human health are short-chain fatty acids (SCFAs), which link host nutrition to gut homeostasis. Through a variety of mechanisms, SCFAs regulate IECs’ functions, including proliferation and differentiation, and the subpopulations of intestinal endocrine cells that affect gut motility, intestinal barrier performance, and host metabolism. Recent studies have demonstrated improved intestinal barrier and immunological regulatory activities of SCFAs, particularly butyrate (130–132). Butyrate not only promotes the production of iTregs as a Histone Deacetylase (HDAC) inhibitor, which is crucial for intestinal balance, but also promotes iTreg differentiation by enhancing fatty acid oxidation (FAO) (133, 134). In CRC patients, there is a significant reduction in butyrate-producing bacteria, and intestinal tumor cells treated with butyrate-producing Clostridia show reduced proliferation and increased apoptosis. Butyrate-producing bacteria can suppress the development of intestinal tumors by regulating Wnt signaling and gut microbiota, indicating potential therapeutic efficacy against CRC (135, 136). In addition to SCFAs, other metabolites produced by microbes, such as lipoteichoic acid (LTA) and secondary bile acids, have a dual effect on tumor development, while lysophosphatidic acid and secondary bile acids promote tumor development (137).
3 The impact of gut microbiota on tumor immunity
3.1 The influence of gut microbiota on immunity
Gut microbiota plays a crucial role in the development and maintenance of the host immune system (Figure 3) (138, 139), and the types and distribution of gut microbiota can directly and indirectly affect the immune system and are closely related to the occurrence of various diseases (140). By modifying the gut metabolome, the variety and location of gut microbiota might affect the repertory of gut or peripheral blood immune cells, impairing the immune system’s ability to recognize microorganisms or tumor cells. In the early stages of life, gut microbiota shapes the immune system, and changes in gut microbiota can affect the development and maturation of the immune system later in life (22). Therefore, the diversity of the gut microbiota is crucial for the establishment of the immune regulatory network.
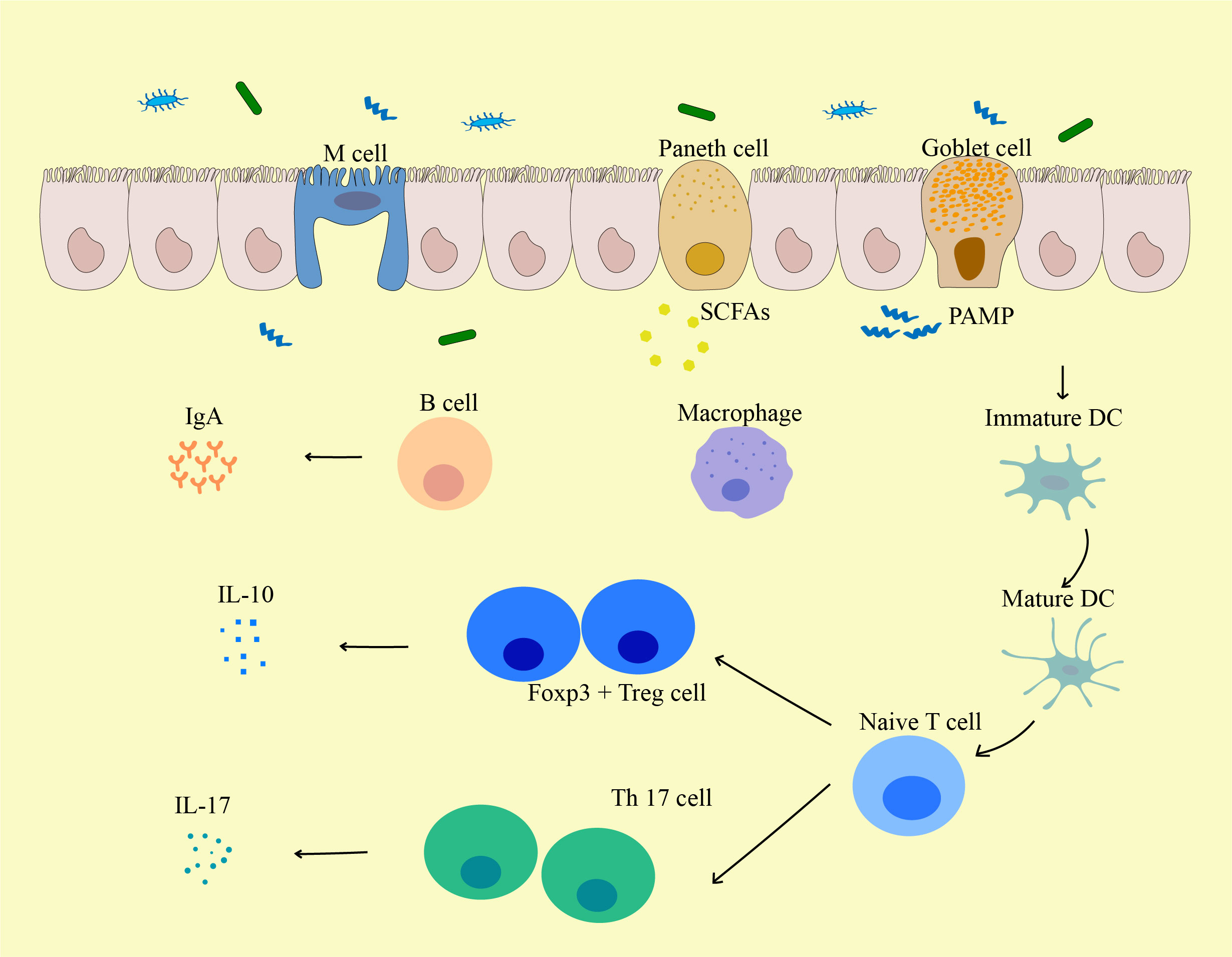
Figure 3 The influence of gut microbiota on the biological function of immune cells. Pathogen-Associated Molecular Pattern (PAMPs) from the gut induce the activation and maturation of antigen-presenting cells (APCs) including DCs. These APCs may then traffic to mesenteric lymph nodes to mediate the maturation of lymphocytes, and local DCs may be activated by bacterial metabolites (such as Short-chain fatty acids (SCFAs)) or bacteria themselves to migrate to mesenteric lymph nodes. Matured DCs further activate naive T cells to differentiate into effector T cells, Tregs, or Th17 cells, which can migrate back to the gut mucosa or systemic circulation. For local immune responses, Tregs secrete IL-10 and act on producing a local anti-inflammatory cytokine environment. Th17 cells secrete cytokines including IL-17 that induce IECs to form tight junctions and secrete antimicrobial proteins, and IL-17 can further lead to the release of other pro-inflammatory cytokines. PAMP, pathogen-associated molecular pattern; M cell, Microfold cell; SCFA, Short-chain fatty acids; DC, Dendritic cells.
The diversity of T-cell receptors (TCRs) in an immune repertoire with high levels of diversity is a key determinant of the host’s ability to resist various environmental pathogens (141). TCR diversity results from the random rearrangement of TCR gene segments and the fusion of TCRα and TCRβ chains during thymic T-cell maturation. As a response, the immunological repertoire can be created at different molecular levels from TCRs (142). The microbiota of a host is a complex community of microbial species that can form tissue-specific T-cell responses in mucosal tissues such as the respiratory tract, gastrointestinal tract, and urogenital tract and can induce CD4+ T cells to differentiate into various T helper cell subtypes, such as peripheral Foxp3+ Treg and Th17 cells in the gut (143). Early in life, the gut microbiota controls the location of innate lymphoid cells that express the transcription factor Promyelocytic Leukemia Zinc Finger (PLZF) in the thymus. The symbiotic microbiota’s extracellular signals that impact the gut immunoglobulin pool control the early B-cell lineage in the gut mucosa (144). The diversity of gut microbiota during early life colonization is critical for establishing an immune regulatory network that prevents the induction of mucosal immunoglobulin E (IgE), which is associated with allergy susceptibility (145). Though being limited to the neonatal stage in mice, TLR5-mediated negative selection of flagellated colonizing bacteria is an important process that determines the composition of the gut microbiota (146), and perturbations in this gut–thymus communication during early life can affect adult susceptibility to disease (136).
Microbes contribute to shaping the immune system (147), and GF mice, which lack a gut microbiota, are thought to have severe immune defects, including a lack of intestinal mucosal layer. Other defects include altered secretion of IgA and decreased size and function of Peyer’s patches and intestinal draining lymph nodes (mLNs) (148, 149). Intraepithelial lymphocytes (IELs) of type αβ and γδ in GF mice are substantially lower compared to typically colonized animals and can be significantly increased upon recolonization (150), and the development of thymic innate lymphoid cells in GF mice is impaired and lacks microbial ligands leading to defective TCR signaling (151). During colonization with the ubiquitous gut microbe B. fragilis, a bacterial polysaccharide (PSA) directs the cellular and physical maturation of the developing immune system. Compared with GF animals, PSA-mediated immune regulation during B. fragilis colonization includes the correction of systemic T-cell defects and Th1/Th2 imbalances and guidance of lymphoid organogenesis, and B. fragilis PSA mutants are unable to restore these immune functions. PSA expressed by intestinal DCs can activate CD4+ T cells and induce corresponding cytokine production (127). The thymic homeostasis of developing PLZF-expressing cells is likewise influenced by the gut microbiome. Specific developmental periods are crucial for the effect of gut–thymus communication on the thymus’s innate lymphoid cell development. Early-life antibiotic treatment can cause permanent damage to PLZF+ innate lymphoid cells in the thymus, while antibiotic treatment during adulthood does not result in damage (152). Colonization with a human commensal bacterium, segmented filamentous bacteria (SFB), but lacking PSA, can restore the thymic development of PLZF+ innate lymphoid cells in GF neonatal mice. Early in life, plasmacytoid DCs are influenced by the microbiota and migrate from the colon to the thymus to regulate the homeostasis of PLZF+ cells. More importantly, disturbance of thymic PLZF+ cells due to changes in the gut microbiota during early life affects susceptibility to diseases during adulthood, and this study identified a communication pathway between gut microbiota and thymic lymphoid cells during the neonatal period that regulates the host’s susceptibility to immunological diseases in later life (152). SFB is one of the few identified microbiota-specific TCRs (123). It induces polarization of Th17 cells, and these Th17 cells have a specific TCR for the SFB antigen (153, 154). Zegarra et al. found that early-life colonization of the gut commensal microbiota causes DCs in the intestine to transport microbial antigens to the thymus, followed by the induction of the expansion of microbiota-specific T cells. Once they enter the periphery, microbiota-specific T cells have the potential to become pathogenic or resist relevant pathogens. In this way, developing microbiota shapes and expands the thymic and peripheral T-cell repertoire, enhancing recognition of gut microbiota and pathogens (155). Stappenbeck et al. (156) examine the interactions between the gut microbiota, the small intestinal epithelium, and the villus’s mesenchymal microvascular network, and they show that the microbiota plays a key role in constructing this microvascular network, and that this regulation depends on a central component of the gut’s innate immune system: the Paneth cell. A conserved bacterial ligand produced by vitamin B synthesis activates the distinct innate T cells known as mucosal-associated invariant T (MAIT) cells, which link innate and adaptive immunity and are crucial in the body’s response to bacterial and viral infections. The development of thymic MAIT17 cells depends on 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU) produced by commensal bacteria on the mucosal surface (157, 158).
The gut mucosa consists of IECs and IELs, including Paneth cells that secrete antimicrobial peptides and goblet cells that produce mucus. The gut-associated lymphoid tissue (GALT) is the most significant component of the human immune system (159). The submucosa of the mucosa contains Peyer’s patches and various immune cells, such as antigen-presenting cells, innate lymphoid cells, CD4+ T cells, CD8+ T cells, and B cells (160). Plasma cells in the lamina propria secrete IgA into the intestinal lumen, which binds to various components of microorganisms, dietary and luminal antigens, preventing harmful antigens from directly interacting with the host immune system (161). Fermentation products of commensal microbiota such as butyrate can induce differentiation of colonic Tregs in mice (134). Bacterial metabolites or the bacteria themselves can activate local DCs (162), which migrate to draining lymph nodes and activate naive T cells to become effector T cells. These T cells can subsequently return to the intestinal mucosa or enter the systemic circulation as Treg or Th17 cells (163). Specific metabolites or bacterial by-products can shape DCs to favor a Treg or Th17 phenotype. Some bacterial metabolites can directly enter the bloodstream and further modulate the systemic immune system (164). To prevent infections and preserve immunological homeostasis, the gut microbiota and the host immune system continually interact and impact each other. Commensal bacteria can signal to immune cells in GALTs and MLNs via pattern recognition receptors (PRRs), such as TLRs that recognize pathogen-associated molecular patterns (PAMPs), such as bacterial LPS and flagellin, to stimulate downstream immune responses (164–167). Sensing the commensal microbiota through the TLR-MyD88 signaling pathway triggers several responses that are critical for maintaining host microbial homeostasis. MyD88-dependent bacterial signaling is required for the induction of epithelial antimicrobial proteins such as RegIIIγ (168, 169). Subsequently, the microbiota induces the repair of damaged IECs through a MyD88-dependent process (170).
Allied with the microbiota, the gut microbiome can affect the immune system by releasing different metabolites into the bloodstream, including SCFAs (171). SCFAs are ligands for HDAC inhibitors and G protein-coupled receptors (GPCRs); SCFA-driven HDAC inhibition tends to express tolerant and anti-inflammatory cell phenotypes, playing an important role in maintaining immune balance (172). SCFAs have been shown to enhance epithelial barrier function and immunological tolerance, as well as promote gut homeostasis through particular mechanisms such as increased mucin secretion in intestinal villi, inhibiting NF-κB, activating inflammatory vesicles and producing IL-18, and increasing the secretion of IgA from B cells. SCFAs can also reduce antigen-presenting cells and directly or indirectly act on local or resident antigen-presenting cells in other organs, such as in the brain and lungs, thereby reducing neuroinflammation and inflammation associated with allergic airway diseases (173, 174). SCFAs also play an important role in the functional development of microglia; microglial dysfunction in GF mice can be rescued by SCFA treatment (175). GPR109A (encoded by Niacr1) is a butyrate receptor in the colon and a receptor for niacin, which is produced by the gut microbiota and inhibits gut inflammation. Studies have found that GPR109A signaling promotes the anti-inflammatory properties of colonic macrophages and DCs, enabling them to induce the differentiation of Tregs and IL-10-producing T cells (176). The most effective anti-inflammatory property of SCFAs is probably their ability to promote the activity of Tregs, which suppress the activity of effector T cells. Acetate and propionate can stimulate the expansion of colonic Tregs (cTregs) that already exist, while butyrate and propionate can promote the de novo differentiation of naive T cells into Tregs (82, 177). Numerous additional metabolites created by the gut microbiota from dietary components also play a significant role in immunity. For instance, the gut microbiota can use arginine, another amino acid, to generate metabolites that regulate the immune system. Polyamines, such as putrescine (a diamine, N2), spermidine (N3), and spermine (N4), which are derived from arginine and produced and secreted by gut bacteria, are present in every living cell and play important roles in gene expression and proliferation. Oral polyamine intake can enhance the development and maintenance of intestinal mucosa and resident immune cells (178, 179).
3.2 The impact of gut microbiota on the tumor microenvironment
The TME is the environment in which tumors grow, and it consists of blood vessels surrounding tumor cells, immune cells, fibroblasts, bone marrow-derived inflammatory cells, different signaling chemicals, and extracellular matrix. The TME can regulate tumor growth, promote tumor invasion and metastasis, mediate tumor immune escape, and promote or weaken the carcinogenic process (180–183). The TME is a complex system that includes many different types of cells, abnormal blood vessels, and immune-suppressive cytokines and is one of the important reasons for tumor evasion of immune surveillance (184) (Figure 4).
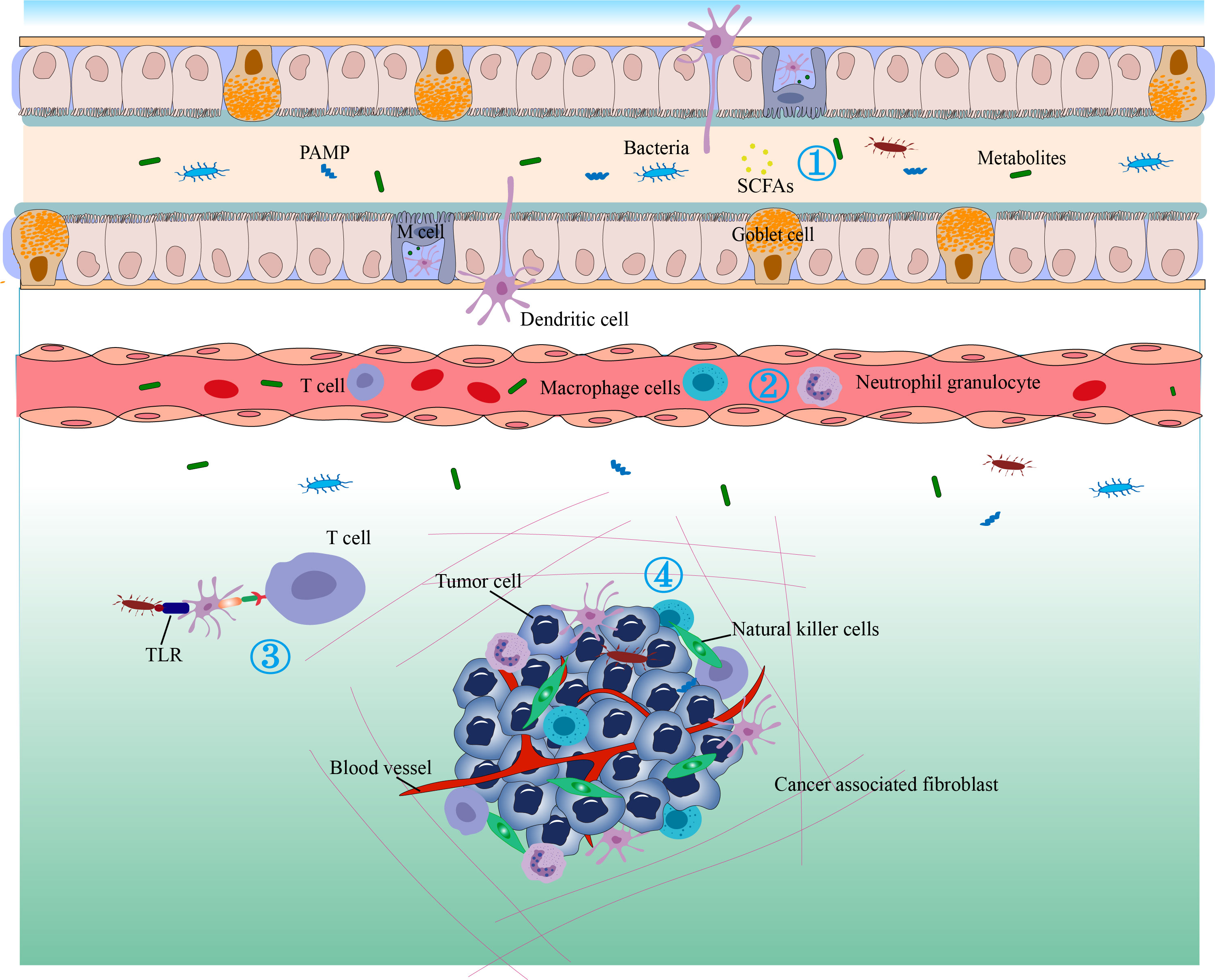
Figure 4 The antitumor immunity and immunotherapy effects of the gut microbiome. 1) Bacterial metabolites that enter the circulation can regulate gene expression in various cells. 2) Cytokines can be released in response to microbial stimulation in the GALT and may enter the circulation to regulate the immune function of downstream systems and stimulate the migration of immune cells. 3) The gut microbiota may translocate to distant tumor sites and alter their immune response or therapeutic efficacy. Immune cells in the gut-associated lymphoid tissue (GALT) can migrate to distant tumors under conditions of sensitive microbial signaling and perform immune-stimulating or inhibitory functions. 4) Bacterial metabolites, antimicrobial peptides, and bacteria can induce dendritic cells (DC) to migrate to lymph nodes to stimulate T cells and B cells. Microbe-associated molecular patterns (MAMPs) can regulate innate immunity by signaling through pattern recognition receptors such as TLRs.
Mononuclear phagocytes (MPs), including monocytes (Mo), macrophages (Mac), and DCs, are the main innate immune cells and an important component of the TME. A recent study revealed the influence of the microbiota on MPs in the TME and innovatively proposed that MPs in the TME can be reshaped by microbiota to enhance the efficacy of immune checkpoint inhibitors (ICIs) (185). As sequencing technology advances, it is now known that various bacteria colonize human tumors, proliferate, and regulate immune function within tumors. The possible mechanism is that these bacteria can selectively settle in tumors that have a rich blood supply and relatively leaky vascular systems through the potential chemotactic gradient of necrotic cell debris. Once settled, they can selectively thrive in the relatively hypoxic TME (especially anaerobic or facultative anaerobic bacteria) (186).
Studies have shown that each cancer subtype has a unique microbiome with specific metabolic functions, and bacteria within tumors mainly exist in cancer cells and immune cells (187, 188). Therefore, the microbiome within tumors plays a crucial role in the development and treatment of tumors (69, 189–191).
The pancreas was once thought to be sterile, but new evidence shows that the microbiota within tumors can affect the progression and treatment of pancreatic cancer (48, 192). Researchers found that in 86 out of 113 tested human PDACs, representing 76% of the tumor tissues, there were bacteria present, mainly Gammaproteobacteria, which can metabolize the chemotherapeutic drug gemcitabine (2’, 2’-difluorodeoxycytidine) into its inactive form, 2’, 2’-difluorodeoxyuridine.
Resistance to gemcitabine is produced within tumors by Gammaproteobacteria, is dependent on bacterial cytidine deaminase (CDDL) expression, and can be removed by cotreatment with the antibiotic ciprofloxacin in a mouse model of CRC (48). Aykut et al. (50) found that Malassezia spp. were abundant in both genetically engineered mouse models and human pancreatic tumors, and that fungal or fecal bacterial transplantation selected from mice carrying PDAC accelerated tumor progression.
In addition, Riquelme et al. (193) found that the abundance of Pseudoxanthomonas, Saccharopolyspora, and Streptomyces spp. in tumors was highly predictive of long-term survival in pancreatic cancer patients, and a more diverse tumor microbiome composition was observed in long-term survivors, which may be attributed to the diverse microbiome promoting recruitment and activation of CD8+ T cells for an antitumor immune response.
According to research, the microbiota alters the immunological microenvironment of the lungs, promoting tumor formation. The resident immune cell network of the lung maintains lung tissue homeostasis while also providing immunological defense against invading infections (194). The development of lung cancer is closely associated with chronic inflammation, characterized by the infiltration of inflammatory cells and the accumulation of pro-inflammatory cytokines (such as cytokines, prostaglandins, and chemokines), which can stimulate various processes, including cell proliferation, angiogenesis, and metastasis (195, 196). Previous studies have shown that the local microbiome can activate lung-resident γδ T cells to cause lung adenocarcinoma-related inflammation. GF or antibiotic-treated mice have significant protective effects against KRAS mutation and p53 loss-induced lung cancer development. Mechanistically, commensal bacteria stimulate bone marrow cells to produce Myd88-dependent IL-1β and IL-23, inducing the proliferation and activation of Vγ6+Vδ1+γδ T cells that produce IL-17 and other effector molecules to promote inflammation and tumor cell proliferation (42).
Th2 cells and innate lymphoid cells 2 (ILC2) can stimulate tumor growth by secreting cytokines such as IL-4, IL-5, and IL-13. The oncogene Kras-G12D increases the expression of IL-33 in PDAC cells, and fungi in PDAC tissue can drive IL-33 secretion, further recruiting and activating Th2 cells and ILC2 in the tumor, ultimately inhibiting the antitumor immune response and promoting tumor progression (52).
In immune cells, neutrophils and Tregs are key cells for cancer growth and development (197–201). Neutrophils can activate the interaction between cancer cells and endothelial cells in the primary TME, thereby promoting tumor metastasis (202). Neutrophil cytokines, chemokines, growth factors, and serine proteases create a milieu that promotes tumor growth. Tumors control neutrophil differentiation in the early stages to produce multiple phenotypic and functionally polarized states that can affect tumor behavior (203). In melanoma, neutrophils recruited by TLR4 signaling can induce cancer cells to migrate to endothelial cells, promoting cancer metastasis (204). The number of neutrophils in peripheral blood and tumor tissue of individuals with various forms of cancer has increased. Importantly, these findings link neutrophils to worse clinical outcomes in cancer patients, implying that these cells may play a role in tumor promotion. In fact, many in vitro and in vivo functional studies have shown that tumor-stimulated neutrophils promote angiogenesis and immune suppression, as well as the migration, invasion, and metastasis of tumor cells (203, 205). However, the enrichment of gut microbiota reduces the number of neutrophils in circulation. One study reported that muscle atrophy in mice fed with Lactobacillus reuteri resulted in reduced systemic inflammation, better tumor suppression compared to the control group, and reduced numbers of neutrophils in the blood (206).
Tregs are essential for maintaining the balance of the immune system and balancing beneficial inflammatory responses during infection (207, 208). Tregs regulate host immune responses aggregated near the TME, inhibit antitumor inflammatory responses, and counteract antigen-specific effector T-cell responses (209). The TME promotes the differentiation and proliferation of Tregs and the secretion of immune-suppressive factors, thereby promoting immune suppression in tumor tissue (210). According to research by Arpaia et al., thymic-independent Treg production is stimulated by SCFAs called butyrate produced by symbiotic microbes during starch fermentation. Increased extrathymic differentiation of Tregs is the cause of the rise in Treg number following butyrate administration (82). In addition, based on animal model data, the Chinese herbal formula YYFZBJS decoction can regulate the natural gut flora, including B. fragilis and Lachnospiraceae, and prevent and inhibit the development of intestinal tumors by reducing the accumulation of CD4+ CD25+ Foxp3+ Tregs in the lymph nodes and MLNs of Apc Min/+ mice (211).
Based on the close interaction between the host microbiome and immune response in the TME, some scholars believe that regulating the gut microbiome to treat tumors is a feasible anticancer treatment strategy (164). The TME is impacted by the ongoing and positive interaction between the gut microbiome and microbial metabolites in the TME. This interaction affects IECs and host immunology, promoting or preventing the development of tumors (212). Ma et al. (213) analyzed the microbial composition of bacteria within prostate cancer tumors to determine the impact of the microbiome on prostate cancer metastatic growth. They identified microbial communities such as Listeria monocytogenes, Methylobacterium radiotolerans JCM 2831, Xanthomonas albilineans GPE PC73, and Bradyrhizobium japonicum that could effectively prevent prostate cancer growth (213).
In conclusion, the gut microbiome and tumor immunity are inextricably interconnected. The gut microbiome’s antitumor immune action requires additional investigation, even though the precise mechanism is still unclear in the modern era of tumor immunotherapy.
3.3 The impact of gut microbiota on tumor immunotherapy
Numerous studies have shown that antitumor therapies such as chemotherapy, radiation therapy, and immunotherapy can alter the gut microbiome of patients (214). Using the immune system for defense against cancer is a technique known as cancer immunotherapy. ICIs, oncolytic virus therapy, cancer vaccines, cytokine therapy, and adoptive cell transfer (ACT) are the main categories of current immunotherapies (215–223). Their common feature is to enhance the immune response, including innate immunity and/or adaptive immunity to clear cancer cells. Compared with other antitumor therapies, immunotherapy can significantly improve the survival rate of cancer patients. Nonetheless, there are still many patients who cannot benefit from immunotherapy because many factors, such as programmed cell death protein 1 ligand 1 (PD-L1) expression and tumor mutation burden, limit the response of many patients to immunotherapy (224).
Various gut microbiomes also play a regulatory role in cancer treatment. Microbial communities and their metabolites provide key signals for the development and function of the host immune system (174, 225–230). Using synthetic biology techniques, Canale et al. (231) created an engineered probiotic strain of Escherichia coli Nissle 1917. Its colonization in tumors raised the amount of L-arginine there, increased the number of tumor-infiltrating T cells, and significantly boosted tumor clearance when combined with PD-L1-blocking antibodies. The research discovered that these bacteria’s antitumor effects were mediated by L-arginine and were reliant on T cells (231).
The regulation of the gut microbiome on immune therapy response provides a new possibility for cancer treatment (Figure 3). Previous studies have shown that the gut microbiome may be a predictive biomarker of immunotherapy efficacy. The higher the diversity of the gut microbiome, the longer the objective response rate and survival rate of immunotherapy (232).
As early as 1813, it was reported that natural bacterial infections could be used as drugs against malignant tumors. Vautier reported that the tumor of a cancer patient with gas gangrene disappeared (224, 233). Fehleisen (1883) and later William B. Coley tested the live infection factor of dengue fever (later known as group A Streptococcus or pyogenic streptococcus) as a means of treating cancer (234–236).
Emerging evidence suggests that the gut microbiome can regulate antitumor immunity through various mechanisms (237). For example, certain antitumor microorganisms such as Bacteroides thetaiotaomicron and B. fragilis can activate DCs through TLR-4 signaling, promote Th1 and cytotoxic CD8+ T-cell responses, and help tumor immune surveillance and eradication (238). Bifidobacteria produce inosine, which enhances the cytotoxic activity of CD8+ T cells by agonizing adenosine 2A receptor signaling in T cells (239).
Although resident bacterial communities exist in the extraintestinal organs of healthy individuals, in cases of inflammation, the intestinal barrier permeability is further increased, enhancing the translocation of bacteria and bacterial components from the intestine to distant sites (240, 241). Bacterial translocation is not always associated with protumor inflammation but may also be associated with enhanced antitumor immunity. Cyclophosphamide induces immunogenic cell death in cancer cells and promotes the differentiation of Th17 and Th1 cells, thereby enhancing treatment efficacy (242, 243). Cyclophosphamide also causes increased intestinal permeability and bacterial translocation from the intestine to lymphoid organs. Surprisingly, bacterial translocation of Gram-positive bacterial species induced by cyclophosphamide results in the production of Th1 memory T cells and the differentiation of Th17 cells producing IFN-γ, which is crucial for antitumor immune responses during treatment (122, 244).
Radiation therapy can treat most tumors whether used alone or in combination. Increasing evidence shows an interaction between radiation exposure and the human intestinal microbiota. After radiation therapy, the structure and composition of the microbiota are directly altered, such as by the decrease in the relative abundance of beneficial microbiota such as Bifidobacterium and Faecalibacterium (245–247). When radiation therapy is used to treat tumors in the abdomen and pelvis, radiation may damage intestinal mucosal barrier function, affecting food absorption, and even causing immune changes (248–251). However, there is an interaction between radiation therapy and microbiota. Although Radiation therapy (RT) treatment leads to an imbalance in the intestinal microbiota, these changes in the microbiota may be an important determining factor for the effectiveness of radiation therapy against tumors (252).
Microbes have the potential to serve as biomarkers of response to ICIs (253). In melanoma, non-small cell lung cancer, urothelial carcinoma, and renal cell carcinoma, the impact of the gut microbiota on the efficacy and interactions of ICIs has been documented. By examining the oral and gut microbiome of 112 melanoma patients undergoing anti-Programmed Cell Death Protein 1 (PD-1) immunotherapy, significant differences in the diversity and composition of gut microbiomes were observed between responders and nonresponders, and when analyzing the patients’ fecal microbiomes, microbial α-diversity (P < 0.01) and the relative abundance of tumor Clostridiales (P < 0.01) were significantly higher. Transplanting fecal microbiota from patients with a favorable response to ICIs into GF mice enhanced their antitumor immunity (164). Mager et al. (239) found that Bifidobacterium pseudolongum, Lactobacillus johnsonii, and Olsenella can increase the efficacy of ICIs 4-fold in four cancer mouse models.
Resistance to ICIs targeting the PD-1/PD-L1 axis induces sustained clinical responses in a considerable proportion of cancer patients (254). We found that the main resistance to ICIs could be attributed to abnormal gut microbiota composition. Antibiotics inhibit the clinical benefits of ICIs in advanced cancer patients, and transplantation of fecal microbiota (FMT) from cancer patients who respond to ICIs into GF or antibiotic-treated mice can improve the antitumor effect of PD-1 blockade, whereas FMT from nonresponders cannot improve the efficacy of PD-1 (255).
Akkermansia muciniphila is a gut bacterium that has been shown to be associated with systemic effects on host metabolism and PD-1 checkpoint immune therapy and induces immunoglobulin G1 (IgG1) antibodies and antigen-specific T-cell responses in mice (256). The relative abundance of A. muciniphila was found to be correlated with the clinical response to ICIs in patients’ fecal samples as revealed by metagenomics at diagnosis. Oral supplementation of A. muciniphila restored the efficacy of PD-1 blockade in mice in a white blood cell-dependent manner by increasing the recruitment of CCR9+ CXCR3+ CD4+ T lymphocytes to the mouse tumor bed after FMT using nonresponder feces (229). Preliminary studies in mouse models have identified the role of the gut microbiota in supporting the efficacy of CpG oligonucleotide immunotherapy and immunostimulatory cyclophosphamide chemotherapy, and further research has demonstrated the immunostimulatory effects of specific bacteria such as Bifidobacterium and fragile Bifidobacteria that enhance the efficacy of ICIs in mouse models (115, 257). An experiment conducted at the University of Pittsburgh evaluated early data on the use of FMT combined with pembrolizumab to treat melanoma patients who had failed anti-PD-1 therapy. The report indicated that two out of three patients experienced stable disease or tumor regression (258). Another study evaluated FMT in patients with melanoma who had become resistant to PD-1 inhibitor therapy. One patient experienced illness regression after the initial scan, while another had a considerable reduction in disease load and lived for an additional 8 months. These patients’ tumor histology revealed increased immune cell infiltration, and sequencing of their gut microbiome revealed alterations in the bacterial population (259).
Peng et al. performed 16SrRNA testing on fecal samples from 74 patients with advanced gastrointestinal tumors who received anti-PD-1/PD-L1 therapy. Before and during treatment, the patients who had higher proportions of Prevotella fungi bacteria in their fecal microbiota showed better PD-1/PD-L1 treatment responses and longer progression-free survival. Patients with higher relative abundances of Prevotella, Rumatococcaceae, and Trichophyton spp. had better treatment responses, and gut bacteria that produce SCFAs, including lactobacilli and streptococci, were positively correlated with the response to anti-PD-1/PD-L1 therapy in different types of gastrointestinal tumors. Microorganisms are potential response markers for ICIs (253). Bullman et al. found that human primary colon adenocarcinoma xenografts in mice retained living clostridial bacteria and their associated microbiota through consecutive passages. Treatment of mice carrying colon cancer xenografts with the antibiotic metronidazole reduced the clostridial load, cancer cell proliferation, and overall tumor growth. These findings support further investigation into antibacterial interventions as potential treatment methods for clostridial-related colon cancer patients (260). Tumor response rates and survival rates of Immune Checkpoint Blockade (ICB) cancer patients decreased when they received antibiotics during treatment (261). Routy et al. (229) found that if patients received antibiotics early, either before or after ICB treatment, their survival rates were significantly reduced. In a mouse model of melanoma, it was found that oral administration of Bifidobacteria alone can improve tumor control to the same extent as treatment with PD-L1-specific antibodies, and combined therapy almost eliminates tumor growth (230). The activation of antigen-presenting cells and increased cytotoxic T lymphocyte infiltration into the tumor may be the mechanisms underlying the improved antitumor response, although it is still unknown if microbiota-regulated CD4+ T cells may also stop tumor progression. According to studies, distinct species of B. thetaiotaomicron are required for Cytotoxic T-Lymphocyte-Associated Protein 4 (CTLA-4) inhibitors to exert antitumor effects. In mice and patients, specific T-cell responses to B. thetaiotaomicron or B. fragilis are correlated with the efficacy of CTLA-4 inhibitors, and antibiotic-treated or GF mice do not respond to CTLA inhibitors, which can be overcome by administering B. fragilis, immunizing with fragilis polysaccharides, or transferring fragilis-specific T cells (238). Recent studies have shown that the use of vancomycin can enhance the efficacy of Chimeric Antigen Receptor T-cell therapy (CAR-T) therapy in mouse models of cervical cancer. Mechanistically, vancomycin treatment induces an increase in systemic CD8α+ DCs, elevating IL-12 levels and maintaining the efficacy of systemically transferred antitumor T cells (262). The gut microbiota’s metabolites, like SCFAs, may also influence tumor immunotherapy, according to mounting data. One study showed that butyrate and propionate enhance the antitumor activity of Cytotoxic T Lymphocyte (CTL) and CAR-T cells through metabolic and epigenetic reprogramming, increasing the production of effector molecules such as CD25, IFN-γ, and TNF-α (263). Researchers revealed that they could dramatically improve the therapeutic impact of PD-1 mAb through providing mice oral doses of pectin, inulin, and other polysaccharide dietary fibers. Adding these prebiotics can increase the relative abundance of key symbiotic microbes, such as Akkermansia and Lactobacillus, and SCFAs, further promoting CD8+ T-cell infiltration into the tumor (264). In addition, SCFAs have been found to enhance the memory potential of antigen-induced CD8+ T cells and trigger their differentiation into stem-like Tcf1+PD-1+CD8+ T cells, which produce effective and long-lasting antitumor effects (265).
The exploration of how gut microbiota regulates the efficacy of immune therapy is proposed by Mager et al. (239) Gut microbiota may regulate the outcome of immune therapy by stimulating or inhibiting possible mechanisms of tumor immunity, such as bacterial metabolites entering the bloodstream and regulating the gene expression of various cells; regulation of innate immunity by pattern recognition receptors; complete live bacteria possibly transferring to distant tumors and affecting the immune response or drug activity; immune cells regulated by microbial signals in GALT can migrate and play immunostimulatory or immunosuppressive functions in distant tumors; and cytokines may be released through corresponding microbial stimulation in GALT, which could possibly enter the circulatory system and regulate the downstream immune function (173).
The effectiveness of immune therapy is significantly impacted by immunological resistance, and immune therapy susceptibility is correlated with gut microbiota. In addition to contributing to immune therapy for tumors, the gut microbiota may also affect immune resistance. Clostridium may play a role in patient resistance to chemotherapy, and a large-scale study showed that the abundance of Clostridium is associated with a decrease in overall survival (OS) (266). TLR4, which is expressed on CRC cells, is activated by Clostridia, making these tumor cells more resistant to oxaliplatin-induced cell death. This results in treatment failure and encourages chemotherapy resistance in CRC patients (72). Some strains of lactic acid bacteria, such as Lactobacillus fermentum, are also believed to weaken the response to immunotherapy (257). In addition to mediating therapeutic effects, the gut microbiota can also regulate the toxic effects of tumor treatment. The differences in gut microbiota composition are associated with graft-versus-host disease (GVHD) in different hematologic malignancies undergoing allogeneic stem cell transplantation (267–269).
Radiation therapy is an effective method for treating tumors. The interaction between gut microbiota and radiation therapy is bidirectional (246). Radiation therapy can disrupt the composition of the gut microbiota, which can have positive or negative effects on the efficacy of tumor treatment. Typically, this disruption manifests as a decrease in the abundance and diversity of the gut microbiota, an increase in harmful microbial populations (such as Proteobacteria and Clostridia), and a decrease in beneficial microbial populations (such as Firmicutes and Bacteroidetes) (245, 270).
Preclinical evidence suggests that patients exposed to broad-spectrum antibiotics may experience a reduction in the effectiveness of cancer radiotherapy. Changes in the gut microbiota caused by antibiotics may be a key factor contributing to this phenomenon (271). Another study found that whole-body irradiation enhanced the translocation of gut bacteria to the MLNs in a melanoma mouse model, resulting in a stronger anticancer response (272). Although increasing evidence suggests that the human gut microbiota has radioprotective effects, further research and exploration are needed to understand how the gut microbiota influences the response to radiation therapy.
4 Summary and outlook
The broad concept of microbiota extends beyond the gut microbiota and encompasses the microecology of various tissues within the body and even external ecological environments, forming a three-dimensional spatial microbiota system that profoundly influences the occurrence of diseases and the body’s resistance. There is an inseparable relationship between gut microbiota and tumors and tumor immunity. Imbalanced microbial ecology can induce tumor formation, and tumor formation can also cause microbial ecology disorders. The gut microbiota has undergone distinct modifications in various tumors, and these changes can be used as biomarkers for supplementary tumor diagnosis. The benefits brought about by utilizing probiotics in tumor immunotherapy are questionable, since the process by which microbial communities in the TME affect tumor progression is complex and mysterious. Some probiotics may hinder the effect of immunotherapy and even promote cancer progression. Although the specific mechanism is yet to be elucidated, the antitumor immune function of gut microbiota is worth further exploration in the current era of tumor immunotherapy. FMT has shown potential in tumor immunotherapy because of its effect on the microbiome (135, 273–276). Currently, FMT is being utilized more frequently beyond the treatment of metabolic syndrome, diabetes, Crohn’s disease, Parkinson’s disease, multiple sclerosis, psoriasis, anorexia nervosa, or Alzheimer’s disease (277–280). However, FMT still has many side effects, such as abdominal discomfort, cramps, bloating, diarrhea, or constipation, and its effect varies among individuals due to different microbial community compositions, as well as various factors such as age, diet, and medication (281). In addition, bacteriophages’ specific killing of gut microbiota provides direction for the specific elimination of microorganisms that promote tumor development and hinder microbial translocation for various reasons, as well as improving the efficacy of tumor treatment (282). Of course, the uncertainty introduced by these methods raises concerns about the role of the microbiota in tumor immunotherapy.
The interaction between microbial species, number, metabolites, and immune cells is one area that will require further investigation in order to fully understand the relationship between gut microbiota and immunotherapy. To create more individualized treatment plans, it is also essential to look into how different types of tumors are impacted by gut bacteria. In future studies, it is also important to consider the influence of an individual’s genetic background and lifestyle on gut microbiota to better understand the interaction between microbiota and tumor therapy. The findings in this paper will contribute to the development of novel tumor treatment plans, increased immunotherapy effectiveness, and decreased side effects.
In the future, the diagnosis and treatment of diseases will gradually transition from anti-disease, antibacterial, and antitumor drugs to comprehensive monitoring of the immune function. The evaluation of the comprehensive immune function is based on the balance of the overall and local dynamic microbiota, and the gut and skin are all components of the microbiota and places for maintaining normal immune function. Therefore, from the perspective of maintaining a normal comprehensive immune function, maintaining the balance of local microbiota is a prerequisite for maintaining overall balance, and the cross-dialog of different local microbiota forms a good symbiotic system. This kind of symbiosis is the core area for exploring the nature of health and the origin of diseases. Immunology’s advancements have embraced traditional Chinese and Western medicine in separate directions but ultimately to the same cause, creating a brand-new area of medicine. The evaluation of the comprehensive immune function must be carried out from the most fundamental perspective and presented in a digital immune force method, from mild regulation to dynamic detection of immunological normalization. This is also a trend in the evaluation of diagnosis and treatment and drug development and is one of the important trends in the development of precision immunology. It is also an important trend in the development of aging biology.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This research was supported by the Oncology Medical-Industrial Innovation Fund Project of University of Electronic Science and Technology, Sichuan Cancer Hospital (Grant No. ZYGX2021YGCX020), the Sichuan Cancer Hospital Youth Fund Project (Grant No. YB2021035, YB2021040), the National Natural Science Foundation of China (Grant No. 82204695, Grant No. 82104215), the Sichuan Provincial Research Institutes Basic Research Operations Fund Project (Grant No. A-2022N-Z-2), the Research Projects granted by Sichuan Administration of TCM (2021ZD004), and the Sichuan Academy of Traditional Chinese Medicine Research Project (Grant No. QNCJRSC2022-9).
Acknowledgments
In the process of composing this review, we extend our heartfelt gratitude to the following individuals for their support and assistance. It is their encouragement that has facilitated the smooth completion of this work. Firstly, I would like to express my sincerest appreciation to RL and RT. Thank you for providing invaluable advice and guidance throughout the entire research process. Your expertise and profound insights have played a pivotal role in driving my research forward, enabling me to delve more deeply into the issues and propose effective solutions. Secondly, I want to thank teachers JZ and YZ for their significant support on the project. I also want to express my gratitude to colleagues and friends such as ZC, GL, BL, SG, QY, SY, LX, and others for their assistance and valuable insights during the research process, as well as their contributions to the revision and proofreading of the article. Lastly, I want to acknowledge all those who have contributed directly or indirectly to this research endeavor. Your efforts and achievements have provided crucial references and foundations for my study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rakoff-Nahoum S, Medzhitov R. Role of the innate immune system and host-commensal mutualism. Curr Top Microbiol Immunol (2006) 308:1–18. doi: 10.1007/3-540-30657-9_1
2. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature (2011) 473(7346):174–80. doi: 10.1038/nature09944
3. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature (2012) 490(7418):55–60. doi: 10.1038/nature11450
4. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol (2004) 4(6):478–85. doi: 10.1038/nri1373
5. Adelman MW, Woodworth MH, Langelier C, Busch LM, Kempker JA, Kraft CS, et al. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486(7402):207–14. doi: 10.1038/nature11234
6. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (2005) 307(5717):1915–20. doi: 10.1126/science.1104816
7. Rea D, Coppola G, Palma G, Barbieri A, Luciano A, Del Prete P, et al. Microbiota effects on cancer: from risks to therapies. Oncotarget (2018) 9(25):17915–27. doi: 10.18632/oncotarget.24681
8. Shi N, Li N, Duan X, Niu H. Interaction between the gut microbiome and mucosal immune system. Mil Med Res (2017) 4:14. doi: 10.1186/s40779-017-0122-9
9. Lee KA, Luong MK, Shaw H, Nathan P, Bataille V, Spector TD. The gut microbiome: what the oncologist ought to know. Br J Cancer (2021) 125(9):1197–209. doi: 10.1038/s41416-021-01467-x
10. Keku TO, Dulal S, Deveaux A, Jovov B, Han X. The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol (2015) 308(5):G351–63. doi: 10.1152/ajpgi.00360.2012
11. Garrett WS. Cancer and the microbiota. Science (2015) 348(6230):80–6. doi: 10.1126/science.aaa4972
12. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and Nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med (2018) 379(22):2108–21. doi: 10.1056/NEJMoa1809615
13. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature (2014) 505(7484):559–63. doi: 10.1038/nature12820
14. Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol (2013) 14(7):646–53. doi: 10.1038/ni.2604
15. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol (2017) 17(4):219–32. doi: 10.1038/nri.2017.7
16. Wong-Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell (2021) 12(5):426–35. doi: 10.1007/s13238-020-00813-8
17. Zhang X, Coker OO, Chu ES, Fu K, Lau HCH, Wang YX, et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut (2021) 70(4):761–74. doi: 10.1136/gutjnl-2019-319664
18. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer (2013) 13(11):800–12. doi: 10.1038/nrc3610
19. Amieva M, Peek RM Jr. Pathobiology of helicobacter pylori-induced gastric cancer. Gastroenterology (2016) 150(1):64–78. doi: 10.1053/j.gastro.2015.09.004
20. Crowe SE. Helicobacter pylori infection. N Engl J Med (2019) 380(12):1158–65. doi: 10.1056/NEJMcp1710945
21. Chmiela M, Kupcinskas J. Review: Pathogenesis of Helicobacter pylori infection. Helicobacter (2019) 24(Suppl 1):e12638. doi: 10.1111/hel.12638
22. Hamway Y, Taxauer K, Moonens K, Neumeyer V, Fischer W, Schmitt V, et al. Cysteine residues in helicobacter pylori adhesin HopQ are required for CEACAM-HopQ interaction and subsequent CagA translocation. Microorganisms (2020) 8(4):465. doi: 10.3390/microorganisms8040465
23. Stein SC, Faber E, Bats SH, Murillo T, Speidel Y, Coombs N, et al. Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis. PloS Pathog (2017) 13(7):e1006514. doi: 10.1371/journal.ppat.1006514
24. Stein M, Bagnoli F, Halenbeck R, Rappuoli R, Fantl WJ, Covacci A. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol Microbiol (2002) 43(4):971–80. doi: 10.1046/j.1365-2958.2002.02781.x
25. Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J Biol Chem (2002) 277(9):6775–8. doi: 10.1074/jbc.C100754200
26. Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer (2004) 4(9):688–94. doi: 10.1038/nrc1433
27. Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest (2014) 124(10):4166–72. doi: 10.1172/JCI72334
28. Cheng WT, Kantilal HK, Davamani F. The mechanism of bacteroides fragilis toxin contributes to colon cancer formation. Malays J Med Sci (2020) 27(4):9–21. doi: 10.21315/mjms2020.27.4.2
29. Zamani S, Taslimi R, Sarabi A, Jasemi S, Sechi LA, Feizabadi MM. Enterotoxigenic bacteroides fragilis: A possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front Cell Infect Microbiol (2019) 9:449. doi: 10.3389/fcimb.2019.00449
30. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science (2018) 359(6375):592–7. doi: 10.1126/science.aah3648
31. Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carrá A, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science (2019) 363(6428):eaar7785. doi: 10.1126/science.aar7785
32. Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem (2007) 282(34):25000–9. doi: 10.1074/jbc.M611567200
33. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
34. Engevik MA, Danhof HA, Ruan W, Engevik AC, Chang-Graham AL, Engevik KA, et al. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. Host-Microbe Biol (2021) 12(2):e02706–20. doi: 10.1128/mBio.02706-20
35. Goedert JJ, Jones G, Hua X, Xu X, Yu G, Flores R, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J Natl Cancer Inst (2015) 107(8). doi: 10.1093/jnci/djv147
36. Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, et al. Distinct microbial signatures associated with different breast cancer types. Front Microbiol (2018) 9:951. doi: 10.3389/fmicb.2018.00951
37. Fernández MF, Reina-Pérez I, Astorga JM, Rodríguez-Carrillo A, Plaza-Díaz J, Fontana L. Breast cancer and its relationship with the microbiota. Int J Environ Res Public Health (2018) 15(8):1747. doi: 10.3390/ijerph15081747
38. Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun (2020) 11(1):3259. doi: 10.1038/s41467-020-16967-2
39. Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer (2016) 102:89–95. doi: 10.1016/j.lungcan.2016.10.016
40. Laroumagne S, Lepage B, Hermant C, Plat G, Phelippeau M, Bigay-Game L, et al. Bronchial colonisation in patients with lung cancer: a prospective study. Eur Respir J (2013) 42(1):220–9. doi: 10.1183/09031936.00062212
41. Liu HX, Tao LL, Zhang J, Zhu YG, Zheng Y, Liu D, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int J Cancer (2018) 142(4):769–78. doi: 10.1002/ijc.31098
42. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell (2019) 176(5):998–1013.e16. doi: 10.1016/j.cell.2018.12.040
43. Kullander J, Forslund O, Dillner J. Staphylococcus aureus and squamous cell carcinoma of the skin. Cancer Epidemiol Biomarkers Prev (2009) 18(2):472–8. doi: 10.1158/1055-9965.EPI-08-0905
44. Mizuhashi S, Kajihara I, Sawamura S, Kanemaru H, Makino K, Aoi J, et al. Skin microbiome in acral melanoma: Corynebacterium is associated with advanced melanoma. J Dermatol (2021) 48(1):e15–e6. doi: 10.1111/1346-8138.15633
45. Harkins CP, MacGibeny MA, Thompson K, Bubic B, Huang X, Brown I, et al. Cutaneous T-Cell Lymphoma Skin Microbiome Is Characterized by Shifts in Certain Commensal Bacteria but not Viruses when Compared with Healthy Controls. J Invest Dermatol (2021) 141(6):1604–8. doi: 10.1016/j.jid.2020.10.021
46. Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget (2015) 6(9):7209–20. doi: 10.18632/oncotarget.3109
47. Gaiser RA, Halimi A, Alkharaan H, Lu L, Davanian H, Healy K, et al. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut (2019) 68(12):2186–94. doi: 10.1136/gutjnl-2018-317458
48. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science (2017) 357(6356):1156–60. doi: 10.1126/science.aah5043
49. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discovery (2018) 8(4):403–16. doi: 10.1158/2159-8290.CD-17-1134
50. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature (2019) 574(7777):264–7. doi: 10.1038/s41586-019-1608-2
51. Bellotti R, Speth C, Adolph TE, Lass-Flörl C, Effenberger M, Öfner D, et al. Micro- and mycobiota dysbiosis in pancreatic ductal adenocarcinoma development. Cancers (Basel) (2021) 13(14):3431. doi: 10.3390/cancers13143431
52. Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, et al. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell (2022) 40(2):153–67.e11. doi: 10.1016/j.ccell.2022.01.003
53. Wang Z, Wang Q, Zhao J, Gong L, Zhang Y, Wang X, et al. Altered diversity and composition of the gut microbiome in patients with cervical cancer. AMB Express (2019) 9(1):40. doi: 10.1186/s13568-019-0763-z
54. Sims TT, Colbert LE, Zheng J, Delgado Medrano AY, Hoffman KL, Ramondetta L, et al. Gut microbial diversity and genus-level differences identified in cervical cancer patients versus healthy controls. Gynecol Oncol (2019) 155(2):237–44. doi: 10.1016/j.ygyno.2019.09.002
55. Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res (2016) 22(22):5574–81. doi: 10.1158/1078-0432.CCR-16-1786
56. Blackett KL, Siddhi SS, Cleary S, Steed H, Miller MH, Macfarlane S, et al. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett's and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther (2013) 37(11):1084–92. doi: 10.1111/apt.12317
57. Man SM. The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol (2011) 8(12):669–85. doi: 10.1038/nrgastro.2011.191
58. Sawada A, Fujiwara Y, Nagami Y, Tanaka F, Yamagami H, Tanigawa T, et al. Alteration of esophageal microbiome by antibiotic treatment does not affect incidence of rat esophageal adenocarcinoma. Dig Dis Sci (2016) 61(11):3161–8. doi: 10.1007/s10620-016-4263-6
59. Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature (2013) 499(7456):97–101. doi: 10.1038/nature12347
60. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature (2015) 517(7533):205–8. doi: 10.1038/nature13828
61. Dutta U, Garg PK, Kumar R, Tandon RK. Typhoid carriers among patients with gallstones are at increased risk for carcinoma of the gallbladder. Am J Gastroenterol (2000) 95(3):784–7. doi: 10.1111/j.1572-0241.2000.01860.x
62. Lazcano-Ponce EC, Miquel JF, Muñoz N, Herrero R, Ferrecio C, Wistuba II, et al. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin (2001) 51(6):349–64. doi: 10.3322/canjclin.51.6.349
63. Wistuba II, Gazdar AF. Gallbladder cancer: lessons from a rare tumour. Nat Rev Cancer (2004) 4(9):695–706. doi: 10.1038/nrc1429
64. Ponziani FR, Nicoletti A, Gasbarrini A, Pompili M. Diagnostic and therapeutic potential of the gut microbiota in patients with early hepatocellular carcinoma. Ther Adv Med Oncol (2019) 11:1758835919848184. doi: 10.1177/1758835919848184
65. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res (2012) 22(2):292–8. doi: 10.1101/gr.126573.111
66. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
67. Saus E, Iraola-Guzmán S, Willis JR, Brunet-Vega A, Gabaldón T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol Aspects Med (2019) 69:93–106. doi: 10.1016/j.mam.2019.05.001
68. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
69. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell (2017) 170(3):548–63.e16. doi: 10.1016/j.cell.2017.07.008
70. Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang X, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut (2021) 70(11):2123–37. doi: 10.1136/gutjnl-2020-322780
71. Chen S, Zhang L, Li M, Zhang Y, Sun M, Wang L, et al. Fusobacterium nucleatum reduces METTL3-mediated m(6)A modification and contributes to colorectal cancer metastasis. Nat Commun (2022) 13(1):1248. doi: 10.1038/s41467-022-28913-5
72. Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol (2019) 17(3):156–66. doi: 10.1038/s41579-018-0129-6
73. Zhang S, Li C, Liu J, Geng F, Shi X, Li Q, et al. Fusobacterium nucleatum promotes epithelial-mesenchymal transiton through regulation of the lncRNA MIR4435-2HG/miR-296-5p/Akt2/SNAI1 signaling pathway. FEBS J (2020) 287(18):4032–47. doi: 10.1111/febs.15233
74. Sears CL, Islam S, Saha A, Arjumand M, Alam NH, Faruque AS, et al. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin Infect Dis (2008) 47(6):797–803. doi: 10.1086/591130
75. Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis (2015) 60(2):208–15. doi: 10.1093/cid/ciu787
76. Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med (2009) 15(9):1016–22. doi: 10.1038/nm.2015
77. Ke Y, Bu S, Ma H, Gao L, Cai Y, Zhang Y, et al. Preventive and therapeutic effects of astaxanthin on depressive-like behaviors in high-fat diet and streptozotocin-treated rats. Front Pharmacol (2019) 10:1621. doi: 10.3389/fphar.2019.01621
78. Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A (2014) 111(51):18321–6. doi: 10.1073/pnas.1406199111
79. Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev (2009) 22(2):349–69. doi: 10.1128/CMR.00053-08
80. Liu QQ, Li CM, Fu LN, Wang HL, Tan J, Wang YQ, et al. Enterotoxigenic Bacteroides fragilis induces the stemness in colorectal cancer via upregulating histone demethylase JMJD2B. Gut Microbes (2020) 12(1):1788900. doi: 10.1080/19490976.2020.1788900
81. Ke Y, Weng M, Chhetri G, Usman M, Li Y, Yu Q, et al. Trappc9 deficiency in mice impairs learning and memory by causing imbalance of dopamine D1 and D2 neurons. Sci Adv (2020) 6(47):eabb7781. doi: 10.1126/sciadv.abb7781
82. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature (2013) 504(7480):451–5. doi: 10.1038/nature12726
83. Vitiello GA, Cohen DJ, Miller G. Harnessing the microbiome for pancreatic cancer immunotherapy. Trends Cancer (2019) 5(11):670–6. doi: 10.1016/j.trecan.2019.10.005
84. Shock T, Badang L, Ferguson B, Martinez-Guryn K. The interplay between diet, gut microbes, and host epigenetics in health and disease. J Nutr Biochem (2021) 95:108631. doi: 10.1016/j.jnutbio.2021.108631
85. Payne CM, Weber C, Crowley-Skillicorn C, Dvorak K, Bernstein H, Bernstein C, et al. Deoxycholate induces mitochondrial oxidative stress and activates NF-kappaB through multiple mechanisms in HCT-116 colon epithelial cells. Carcinogenesis (2007) 28(1):215–22. doi: 10.1093/carcin/bgl139
86. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol (2014) 12(10):661–72. doi: 10.1038/nrmicro3344
87. Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A (2011) 108(37):15354–9. doi: 10.1073/pnas.1010203108
88. Górska A, Przystupski D, Niemczura MJ, Kulbacka J. Probiotic bacteria: A promising tool in cancer prevention and therapy. Curr Microbiol (2019) 76(8):939–49. doi: 10.1007/s00284-019-01679-8
89. Goldin BR, Gorbach SL. Effect of Lactobacillus acidophilus dietary supplements on 1,2-dimethylhydrazine dihydrochloride-induced intestinal cancer in rats. J Natl Cancer Inst (1980) 64(2):263–5. doi: 10.1093/jnci/64.2.263
90. Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol (2018) 15(2):111–28. doi: 10.1038/nrgastro.2017.119
91. Nowak A, Kuberski S, Libudzisz Z. Probiotic lactic acid bacteria detoxify N-nitrosodimethylamine. Food Addit Contam Part A Chem Anal Control Expo Risk Assess (2014) 31(10):1678–87. doi: 10.1080/19440049.2014.943304
92. Nowak A, Libudzisz Z. Ability of probiotic Lactobacillus casei DN 114001 to bind or/and metabolise heterocyclic aromatic amines in vitro. Eur J Nutr (2009) 48(7):419–27. doi: 10.1007/s00394-009-0030-1
93. Nowak A, Czyżowska A, Stańczyk M. Protective activity of probiotic bacteria against 2-amino-3-methyl-3H-imidazo[4,5-f]quinoline (IQ) and 2-amino-1-methyl-6-phenyl-1H-imidazo[4,5-b]pyridine (PhIP) - an in vitro study. Food Addit Contam Part A Chem Anal Control Expo Risk Assess (2015) 32(11):1927–38. doi: 10.1080/19440049.2015.1084651
94. Lili Z, Junyan W, Hongfei Z, Baoqing Z, Bolin Z. Detoxification of cancerogenic compounds by lactic acid bacteria strains. Crit Rev Food Sci Nutr (2018) 58(16):2727–42. doi: 10.1080/10408398.2017.1339665
95. Yazdi MH, Mahdavi M, Kheradmand E, Shahverdi AR. The preventive oral supplementation of a selenium nanoparticle-enriched probiotic increases the immune response and lifespan of 4T1 breast cancer bearing mice. Arzneimittelforschung (2012) 62(11):525–31. doi: 10.1055/s-0032-1323700
96. Aragón F, Carino S, Perdigón G, de Moreno de LeBlanc A. The administration of milk fermented by the probiotic Lactobacillus casei CRL 431 exerts an immunomodulatory effect against a breast tumour in a mouse model. Immunobiology (2014) 219(6):457–64. doi: 10.1016/j.imbio.2014.02.005
97. Babjuk M, Burger M, Capoun O, Cohen D, Compérat EM, Dominguez Escrig JL, et al. European association of urology guidelines on non-muscle-invasive bladder cancer (Ta, T1, and carcinoma in situ). Eur Urol (2022) 81(1):75–94. doi: 10.1016/j.eururo.2021.08.010
98. Bellmunt J, Orsola A, Leow JJ, Wiegel T, De Santis M, Horwich A. Bladder cancer: ESMO Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol (2014) 25(Suppl 3):iii40–8. doi: 10.1093/annonc/mdu223
99. Moschini M, Gandaglia G, Dehò F, Salonia A, Briganti A, Montorsi F. Bladder cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol (2022) 33(5):561. doi: 10.1016/j.annonc.2022.01.075
100. Lenis AT, Lec PM, Chamie K, Mshs MD. Bladder cancer: A review. Jama (2020) 324(19):1980–91. doi: 10.1001/jama.2020.17598
101. Tsuda K, Yamanaka K, Linan W, Miyahara Y, Akeda T, Nakanishi T, et al. Intratumoral injection of Propionibacterium acnes suppresses malignant melanoma by enhancing Th1 immune responses. PloS One (2011) 6(12):e29020. doi: 10.1371/journal.pone.0029020
102. Nakatsuji T, Chen TH, Butcher AM, Trzoss LL, Nam SJ, Shirakawa KT, et al. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci Adv (2018) 4(2):eaao4502. doi: 10.1126/sciadv.aao4502
103. Weill FS, Cela EM, Paz ML, Ferrari A, Leoni J, González Maglio DH. Lipoteichoic acid from Lactobacillus rhamnosus GG as an oral photoprotective agent against UV-induced carcinogenesis. Br J Nutr (2013) 109(3):457–66. doi: 10.1017/S0007114512001225
104. Seow SW, Cai S, Rahmat JN, Bay BH, Lee YK, Chan YH, et al. Lactobacillus rhamnosus GG induces tumor regression in mice bearing orthotopic bladder tumors. Cancer Sci (2010) 101(3):751–8. doi: 10.1111/j.1349-7006.2009.01426.x
105. Yu Y, Dunaway S, Champer J, Kim J, Alikhan A. Changing our microbiome: probiotics in dermatology. Br J Dermatol (2020) 182(1):39–46. doi: 10.1111/bjd.18659
106. Orrhage K, Sillerström E, Gustafsson JA, Nord CE, Rafter J. Binding of mutagenic heterocyclic amines by intestinal and lactic acid bacteria. Mutat Res (1994) 311(2):239–48. doi: 10.1016/0027-5107(94)90182-1
107. Ismail B, Nampoothiri KM. Molecular characterization of an exopolysaccharide from a probiotic Lactobacillus plantarum MTCC 9510 and its efficacy to improve the texture of starchy food. J Food Sci Technol (2014) 51(12):4012–8. doi: 10.1007/s13197-013-0928-8
108. Wang Q, Wang K, Wu W, Lv L, Bian X, Yang L, et al. Administration of Bifidobacterium bifidum CGMCC 15068 modulates gut microbiota and metabolome in azoxymethane (AOM)/dextran sulphate sodium (DSS)-induced colitis-associated colon cancer (CAC) in mice. Appl Microbiol Biotechnol (2020) 104(13):5915–28. doi: 10.1007/s00253-020-10621-z
109. Kojima M, Wakai K, Tamakoshi K, Tokudome S, Toyoshima H, Watanabe Y, et al. Diet and colorectal cancer mortality: results from the Japan Collaborative Cohort Study. Nutr Cancer (2004) 50(1):23–32. doi: 10.1207/s15327914nc5001_4
110. Larsson SC, Bergkvist L, Rutegård J, Giovannucci E, Wolk A. Calcium and dairy food intakes are inversely associated with colorectal cancer risk in the Cohort of Swedish Men. Am J Clin Nutr (2006) 83(3):667–73; quiz 728-9. doi: 10.1093/ajcn.83.3.667
111. Deepak V, Ramachandran S, Balahmar RM, Pandian SR, Sivasubramaniam SD, Nellaiah H, et al. In vitro evaluation of anticancer properties of exopolysaccharides from Lactobacillus acidophilus in colon cancer cell lines. In Vitro Cell Dev Biol Anim (2016) 52(2):163–73. doi: 10.1007/s11626-015-9970-3
112. Wu J, Zhang Y, Ye L, Wang C. The anti-cancer effects and mechanisms of lactic acid bacteria exopolysaccharides in vitro: A review. Carbohydr Polym (2021) 253:117308. doi: 10.1016/j.carbpol.2020.117308
113. Eslami M, Yousefi B, Kokhaei P, Hemati M, Nejad ZR, Arabkari V, et al. Importance of probiotics in the prevention and treatment of colorectal cancer. J Cell Physiol (2019) 234(10):17127–43. doi: 10.1002/jcp.28473
114. Tomasi M, Dalsass M, Beghini F, Zanella I, Caproni E, Fantappiè L, et al. Commensal bifidobacterium strains enhance the efficacy of neo-epitope based cancer vaccines. Vaccines (Basel) (2021) 9(11):1356. doi: 10.3390/vaccines9111356
115. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
116. Lee SH, Cho SY, Yoon Y, Park C, Sohn J, Jeong JJ, et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat Microbiol (2021) 6(3):277–88. doi: 10.1038/s41564-020-00831-6
117. Li Q, Hu W, Liu WX, Zhao LY, Huang D, Liu XD, et al. Streptococcus thermophilus inhibits colorectal tumorigenesis through secreting β-galactosidase. Gastroenterology (2021) 160(4):1179–93.e14. doi: 10.1053/j.gastro.2020.09.003
118. Redelman-Sidi G, Glickman MS, Bochner BH. The mechanism of action of BCG therapy for bladder cancer–a current perspective. Nat Rev Urol (2014) 11(3):153–62. doi: 10.1038/nrurol.2014.15
119. Han J, Gu X, Li Y, Wu Q. Mechanisms of BCG in the treatment of bladder cancer-current understanding and the prospect. BioMed Pharmacother (2020) 129:110393. doi: 10.1016/j.biopha.2020.110393
120. Ingersoll MA, Albert ML. From infection to immunotherapy: host immune responses to bacteria at the bladder mucosa. Mucosal Immunol (2013) 6(6):1041–53. doi: 10.1038/mi.2013.72
121. Lee CW, Chen YJ, Wu HF, Chung YJ, Lee YC, Li CT, et al. Ketamine ameliorates severe traumatic event-induced antidepressant-resistant depression in a rat model through ERK activation. Prog Neuropsychopharmacol Biol Psychiatry (2019) 93:102–13. doi: 10.1016/j.pnpbp.2019.03.015
122. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science (2013) 342(6161):971–6. doi: 10.1126/science.1240537
123. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139(3):485–98. doi: 10.1016/j.cell.2009.09.033
124. Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe (2008) 4(4):337–49. doi: 10.1016/j.chom.2008.09.009
125. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell (2015) 163(2):367–80. doi: 10.1016/j.cell.2015.08.058
126. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science (2011) 332(6032):974–7. doi: 10.1126/science.1206095
127. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell (2005) 122(1):107–18. doi: 10.1016/j.cell.2005.05.007
128. Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature (2011) 469(7331):543–7. doi: 10.1038/nature09646
129. Chhetri G, Ke Y, Wang P, Usman M, Li Y, Sapp E, et al. Impaired XK recycling for importing manganese underlies striatal vulnerability in Huntington's disease. J Cell Biol (2022) 221(10):e202112073. doi: 10.1083/jcb.202112073
130. Martin-Gallausiaux C, Marinelli L, Blottière HM, Larraufie P, Lapaque N. SCFA: mechanisms and functional importance in the gut. Proc Nutr Soc (2021) 80(1):37–49. doi: 10.1017/S0029665120006916
131. Sun M, Wu W, Liu Z, Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol (2017) 52(1):1–8. doi: 10.1007/s00535-016-1242-9
132. Tomita Y, Ikeda T, Sakata S, Saruwatari K, Sato R, Iyama S, et al. Association of probiotic clostridium butyricum therapy with survival and response to immune checkpoint blockade in patients with lung cancer. Cancer Immunol Res (2020) 8(10):1236–42. doi: 10.1158/2326-6066.CIR-20-0051
133. Hao F, Tian M, Zhang X, Jin X, Jiang Y, Sun X, et al. Butyrate enhances CPT1A activity to promote fatty acid oxidation and iTreg differentiation. Proc Natl Acad Sci U.S.A. (2021) 118(22):e2014681118. doi: 10.1073/pnas.2014681118
134. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature (2013) 504(7480):446–50. doi: 10.1038/nature12721
135. Chen D, Jin D, Huang S, Wu J, Xu M, Liu T, et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett (2020) 469:456–67. doi: 10.1016/j.canlet.2019.11.019
136. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res (2020) 30(6):492–506. doi: 10.1038/s41422-020-0332-7
137. Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, et al. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab (2016) 4:11. doi: 10.1186/s40170-016-0151-y
138. Khan MAW, Ologun G, Arora R, McQuade JL, Wargo JA. Gut microbiome modulates response to cancer immunotherapy. Dig Dis Sci (2020) 65(3):885–96. doi: 10.1007/s10620-020-06111-x
139. McFall-Ngai M. Adaptive immunity: care for the community. Nature (2007) 445(7124):153. doi: 10.1038/445153a
140. Gomaa EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek (2020) 113(12):2019–40. doi: 10.1007/s10482-020-01474-7
141. Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol (2004) 4(2):123–32. doi: 10.1038/nri1292
142. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol (2014) 14(6):377–91. doi: 10.1038/nri3667
143. Kim KS. Regulation of T cell repertoires by commensal microbiota. Front Cell Infect Microbiol (2022) 12:1004339. doi: 10.3389/fcimb.2022.1004339
144. Wesemann DR, Portuguese AJ, Meyers RM, Gallagher MP, Cluff-Jones K, Magee JM, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature (2013) 501(7465):112–5. doi: 10.1038/nature12496
145. Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe (2013) 14(5):559–70. doi: 10.1016/j.chom.2013.10.004
146. Fulde M, Sommer F, Chassaing B, van Vorst K, Dupont A, Hensel M, et al. Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature (2018) 560(7719):489–93. doi: 10.1038/s41586-018-0395-5
147. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature (2016) 535(7610):75–84. doi: 10.1038/nature18848
148. Johansson ME, Jakobsson HE, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro AM, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe (2015) 18(5):582–92. doi: 10.1016/j.chom.2015.10.007
149. Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Stud Germfree Mice Am J Pathol (1963) 42(4):471–83.
150. Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology (1993) 79(1):32–7.
151. Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature (2003) 422(6928):164–9. doi: 10.1038/nature01433
152. Ennamorati M, Vasudevan C, Clerkin K, Halvorsen S, Verma S, Ibrahim S, et al. Intestinal microbes influence development of thymic lymphocytes in early life. Proc Natl Acad Sci U S A (2020) 117(5):2570–8. doi: 10.1073/pnas.1915047117
153. Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature (2014) 510(7503):152–6. doi: 10.1038/nature13279
154. Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity (2009) 31(4):677–89. doi: 10.1016/j.immuni.2009.08.020
155. Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu WH, Saldana-Morales FB, et al. Thymic development of gut-microbiota-specific T cells. Nature (2021) 594(7863):413–7. doi: 10.1038/s41586-021-03531-1
156. Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A (2002) 99(24):15451–5. doi: 10.1073/pnas.202604299
157. Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science (2019) 366(6464):494–9. doi: 10.1126/science.aaw2719
158. Toubal A, Nel I, Lotersztajn S, Lehuen A. Mucosal-associated invariant T cells and disease. Nat Rev Immunol (2019) 19(10):643–57. doi: 10.1038/s41577-019-0191-y
159. Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GM, Schütte A, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev (2014) 260(1):8–20. doi: 10.1111/imr.12182
160. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol (2014) 14(10):667–85. doi: 10.1038/nri3738
161. Roche AM, Richard AL, Rahkola JT, Janoff EN, Weiser JN. Antibody blocks acquisition of bacterial colonization through agglutination. Mucosal Immunol (2015) 8(1):176–85. doi: 10.1038/mi.2014.55
162. McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature (2012) 483(7389):345–9. doi: 10.1038/nature10863
163. Morton AM, Sefik E, Upadhyay R, Weissleder R, Benoist C, Mathis D. Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc Natl Acad Sci U S A (2014) 111(18):6696–701. doi: 10.1073/pnas.1405634111
164. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell (2018) 33(4):570–80. doi: 10.1016/j.ccell.2018.03.015
165. Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol (2014) 5:461. doi: 10.3389/fimmu.2014.00461
166. Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature (2011) 478(7368):250–4. doi: 10.1038/nature10434
167. Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol (2001) 2(4):361–7. doi: 10.1038/86373
168. Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med (2007) 204(8):1891–900. doi: 10.1084/jem.20070563
169. Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science (2011) 334(6053):255–8. doi: 10.1126/science.1209791
170. Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science (2007) 317(5834):124–7. doi: 10.1126/science.1140488
171. Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, et al. Gut-microbiota-targeted diets modulate human immune status. Cell (2021) 184(16):4137–53.e14. doi: 10.1016/j.cell.2021.06.019
172. Zhang Z, Tang H, Chen P, Xie H, Tao Y. Demystifying the manipulation of host immunity, metabolism, and extraintestinal tumors by the gut microbiome. Signal Transduct Target Ther (2019) 4:41. doi: 10.1038/s41392-019-0074-5
173. Matson V, Chervin CS, Gajewski TF. Cancer and the microbiome-influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology (2021) 160(2):600–13. doi: 10.1053/j.gastro.2020.11.041
174. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol (2016) 16(6):341–52. doi: 10.1038/nri.2016.42
175. Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci (2015) 18(7):965–77. doi: 10.1038/nn.4030
176. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity (2014) 40(1):128–39. doi: 10.1016/j.immuni.2013.12.007
177. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science (2013) 341(6145):569–73. doi: 10.1126/science.1241165
178. Michael AJ. Biosynthesis of polyamines and polyamine-containing molecules. Biochem J (2016) 473(15):2315–29. doi: 10.1042/BCJ20160185
179. Pérez-Cano FJ, González-Castro A, Castellote C, Franch A, Castell M. Influence of breast milk polyamines on suckling rat immune system maturation. Dev Comp Immunol (2010) 34(2):210–8. doi: 10.1016/j.dci.2009.10.001
180. Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity (2016) 44(3):647–58. doi: 10.1016/j.immuni.2016.02.006
181. Qiu Q, Lin Y, Ma Y, Li X, Liang J, Chen Z, et al. Exploring the emerging role of the gut microbiota and tumor microenvironment in cancer immunotherapy. Front Immunol (2020) 11:612202. doi: 10.3389/fimmu.2020.612202
182. Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med (2011) 17(3):320–9. doi: 10.1038/nm.2328
183. Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med (2018) 24(8):1178–91. doi: 10.1038/s41591-018-0085-8
184. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.CAN-18-3962
185. Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell (2021) 184(21):5338–56.e21. doi: 10.1016/j.cell.2021.09.019
186. Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med (2019) 25(3):377–88. doi: 10.1038/s41591-019-0377-7
187. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
188. Heymann CJF, Bard JM, Heymann MF, Heymann D, Bobin-Dubigeon C. The intratumoral microbiome: Characterization methods and functional impact. Cancer Lett (2021) 522:63–79. doi: 10.1016/j.canlet.2021.09.009
189. Meng S, Chen B, Yang J, Wang J, Zhu D, Meng Q, et al. Study of microbiomes in aseptically collected samples of human breast tissue using needle biopsy and the potential role of in situ tissue microbiomes for promoting malignancy. Front Oncol (2018) 8:318. doi: 10.3389/fonc.2018.00318
190. Boesch M, Horvath L, Baty F, Pircher A, Wolf D, Spahn S, et al. Compartmentalization of the host microbiome: how tumor microbiota shapes checkpoint immunotherapy outcome and offers therapeutic prospects. J Immunother Cancer (2022) 10(11):e005401. doi: 10.1136/jitc-2022-005401
191. Cogdill AP, Gaudreau PO, Arora R, Gopalakrishnan V, Wargo JA. The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol (2018) 39(11):900–20. doi: 10.1016/j.it.2018.09.007
192. Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature (2017) 551(7681):512–6. doi: 10.1038/nature24462
193. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell (2019) 178(4):795–806.e12. doi: 10.1016/j.cell.2019.07.008
194. Lloyd CM, Marsland BJ. Lung homeostasis: influence of age, microbes, and the immune system. Immunity (2017) 46(4):549–61. doi: 10.1016/j.immuni.2017.04.005
195. Budisan L, Zanoaga O, Braicu C, Pirlog R, Covaliu B, Esanu V, et al. Links between infections, lung cancer, and the immune system. Int J Mol Sci (2021) 22(17):9394. doi: 10.3390/ijms22179394
196. Palucka AK, Coussens LM. The basis of oncoimmunology. Cell (2016) 164(6):1233–47. doi: 10.1016/j.cell.2016.01.049
197. Erdman SE, Rao VP, Olipitz W, Taylor CL, Jackson EA, Levkovich T, et al. Unifying roles for regulatory T cells and inflammation in cancer. Int J Cancer (2010) 126(7):1651–65. doi: 10.1002/ijc.24923
198. Poutahidis T, Haigis KM, Rao VP, Nambiar PR, Taylor CL, Ge Z, et al. Rapid reversal of interleukin-6-dependent epithelial invasion in a mouse model of microbially induced colon carcinoma. Carcinogenesis (2007) 28(12):2614–23. doi: 10.1093/carcin/bgm180
199. Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer (2016) 16(7):431–46. doi: 10.1038/nrc.2016.52
200. Erdman SE, Rao VP, Poutahidis T, Rogers AB, Taylor CL, Jackson EA, et al. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc Natl Acad Sci U S A (2009) 106(4):1027–32. doi: 10.1073/pnas.0812347106
201. Shaul ME, Fridlender ZG. Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol (2019) 16(10):601–20. doi: 10.1038/s41571-019-0222-4
202. Hedrick CC, Malanchi I. Neutrophils in cancer: heterogeneous and multifaceted. Nat Rev Immunol (2022) 22(3):173–87. doi: 10.1038/s41577-021-00571-6
203. Dumitru CA, Lang S, Brandau S. Modulation of neutrophil granulocytes in the tumor microenvironment: mechanisms and consequences for tumor progression. Semin Cancer Biol (2013) 23(3):141–8. doi: 10.1016/j.semcancer.2013.02.005
204. Bald T, Quast T, Landsberg J, Rogava M, Glodde N, Lopez-Ramos D, et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature (2014) 507(7490):109–13. doi: 10.1038/nature13111
205. Jaillon S, Ponzetta A, Di Mitri D, Santoni A, Bonecchi R, Mantovani A. Neutrophil diversity and plasticity in tumour progression and therapy. Nat Rev Cancer (2020) 20(9):485–503. doi: 10.1038/s41568-020-0281-y
206. Varian BJ, Gourishetti S, Poutahidis T, Lakritz JR, Levkovich T, Kwok C, et al. Beneficial bacteria inhibit cachexia. Oncotarget (2016) 7(11):11803–16. doi: 10.18632/oncotarget.7730
207. Wing JB, Tanaka A, Sakaguchi S. Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity (2019) 50(2):302–16. doi: 10.1016/j.immuni.2019.01.020
208. Erdman SE, Poutahidis T. Cancer inflammation and regulatory T cells. Int J Cancer (2010) 127(4):768–79. doi: 10.1002/ijc.25430
209. Nishikawa H, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Curr Opin Immunol (2014) 27:1–7. doi: 10.1016/j.coi.2013.12.005
210. Wang YA, Li XL, Mo YZ, Fan CM, Tang L, Xiong F, et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol Cancer (2018) 17(1):168. doi: 10.1186/s12943-018-0913-y
211. Sui H, Zhang L, Gu K, Chai N, Ji Q, Zhou L, et al. YYFZBJS ameliorates colorectal cancer progression in Apc(Min/+) mice by remodeling gut microbiota and inhibiting regulatory T-cell generation. Cell Commun Signal (2020) 18(1):113. doi: 10.1186/s12964-020-00596-9
212. Sethi V, Kurtom S, Tarique M, Lavania S, Malchiodi Z, Hellmund L, et al. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology (2018) 155(1):33–7.e6. doi: 10.1053/j.gastro.2018.04.001
213. Ma J, Gnanasekar A, Lee A, Li WT, Haas M, Wang-Rodriguez J, et al. Influence of intratumor microbiome on clinical outcome and immune processes in prostate cancer. Cancers (Basel) (2020) 12(9):2524. doi: 10.3390/cancers12092524
214. Tan S, Li D, Zhu X. Cancer immunotherapy: Pros, cons and beyond. BioMed Pharmacother (2020) 124:109821. doi: 10.1016/j.biopha.2020.109821
215. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol (2020) 17(8):807–21. doi: 10.1038/s41423-020-0488-6
216. Kaeuferle T, Krauss R, Blaeschke F, Willier S, Feuchtinger T. Strategies of adoptive T -cell transfer to treat refractory viral infections post allogeneic stem cell transplantation. J Hematol Oncol (2019) 12(1):13. doi: 10.1186/s13045-019-0701-1
217. Fan J, Shang D, Han B, Song J, Chen H, Yang JM. Adoptive cell transfer: is it a promising immunotherapy for colorectal cancer? Theranostics (2018) 8(20):5784–800. doi: 10.7150/thno.29035
218. Sahin U, Türeci Ö. Personalized vaccines for cancer immunotherapy. Science (2018) 359(6382):1355–60. doi: 10.1126/science.aar7112
219. Miao L, Zhang Y, Huang L. mRNA vaccine for cancer immunotherapy. Mol Cancer (2021) 20(1):41. doi: 10.1186/s12943-021-01335-5
220. Harrington K, Freeman DJ, Kelly B, Harper J, Soria JC. Optimizing oncolytic virotherapy in cancer treatment. Nat Rev Drug Discovery (2019) 18(9):689–706. doi: 10.1038/s41573-019-0029-0
221. Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, et al. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell (2017) 170(6):1109–19.e10. doi: 10.1016/j.cell.2017.08.027
222. Briukhovetska D, Dörr J, Endres S, Libby P, Dinarello CA, Kobold S. Interleukins in cancer: from biology to therapy. Nat Rev Cancer (2021) 21(8):481–99. doi: 10.1038/s41568-021-00363-z
223. Propper DJ, Balkwill FR. Harnessing cytokines and chemokines for cancer therapy. Nat Rev Clin Oncol (2022) 19(4):237–53. doi: 10.1038/s41571-021-00588-9
224. Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature (2011) 480(7378):480–9. doi: 10.1038/nature10673
225. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity (2017) 46(4):562–76. doi: 10.1016/j.immuni.2017.04.008
226. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
227. Badgeley A, Anwar H, Modi K, Murphy P, Lakshmikuttyamma A. Effect of probiotics and gut microbiota on anti-cancer drugs: Mechanistic perspectives. Biochim Biophys Acta Rev Cancer (2021) 1875(1):188494. doi: 10.1016/j.bbcan.2020.188494
228. Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia (2017) 19(10):848–55. doi: 10.1016/j.neo.2017.08.004
229. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
230. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
231. Canale FP, Basso C, Antonini G, Perotti M, Li N, Sokolovska A, et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature (2021) 598(7882):662–6. doi: 10.1038/s41586-021-04003-2
232. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med (2019) 25(6):968–76. doi: 10.1038/s41591-019-0458-7
233. Mowday AM, Guise CP, Ackerley DF, Minton NP, Lambin P, Dubois LJ, et al. Advancing clostridia to clinical trial: past lessons and recent progress. Cancers (Basel) (2016) 8(7):63. doi: 10.3390/cancers8070063
234. Nauts HC, Swift WE, Coley BL. The treatment of malignant tumors by bacterial toxins as developed by the late William B. Coley, M.D., reviewed in the light of modern research. Cancer Res (1946) 6:205–16.
235. Coley WB. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. 1893. Clin Orthop Relat Res (1991) 262):3–11.
236. Coley WB. II. Contribution to the knowledge of sarcoma. Ann Surg (1891) 14(3):199–220. doi: 10.1097/00000658-189112000-00015
237. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science (2021) 371(6536):eabc4552. doi: 10.1126/science.abc4552
238. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
239. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science (2020) 369(6510):1481–9. doi: 10.1126/science.abc3421
240. Hietbrink F, Besselink MG, Renooij W, de Smet MB, Draisma A, van der Hoeven H, et al. Systemic inflammation increases intestinal permeability during experimental human endotoxemia. Shock (2009) 32(4):374–8. doi: 10.1097/SHK.0b013e3181a2bcd6
241. Ha CWY, Martin A, Sepich-Poore GD, Shi B, Wang Y, Gouin K, et al. Translocation of viable gut microbiota to mesenteric adipose drives formation of creeping fat in humans. Cell (2020) 183(3):666–83.e17. doi: 10.1016/j.cell.2020.09.009
242. Sistigu A, Viaud S, Chaput N, Bracci L, Proietti E, Zitvogel L. Immunomodulatory effects of cyclophosphamide and implementations for vaccine design. Semin Immunopathol (2011) 33(4):369–83. doi: 10.1007/s00281-011-0245-0
243. Viaud S, Flament C, Zoubir M, Pautier P, LeCesne A, Ribrag V, et al. Cyclophosphamide induces differentiation of Th17 cells in cancer patients. Cancer Res (2011) 71(3):661–5. doi: 10.1158/0008-5472.CAN-10-1259
244. Buchta Rosean CM, Rutkowski MR. The influence of the commensal microbiota on distal tumor-promoting inflammation. Semin Immunol (2017) 32:62–73. doi: 10.1016/j.smim.2017.06.002
245. Fernandes A, Oliveira A, Soares R, Barata P. The effects of ionizing radiation on gut microbiota, a systematic review. Nutrients (2021) 13(9):3025. doi: 10.3390/nu13093025
246. Liu J, Liu C, Yue J. Radiotherapy and the gut microbiome: facts and fiction. Radiat Oncol (2021) 16(1):9. doi: 10.1186/s13014-020-01735-9
247. Deleu S, Machiels K, Raes J, Verbeke K, Vermeire S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? EBioMedicine (2021) 66:103293. doi: 10.1016/j.ebiom.2021.103293
248. Reis Ferreira M, Andreyev HJN, Mohammed K, Truelove L, Gowan SM, Li J, et al. Microbiota- and radiotherapy-induced gastrointestinal side-effects (MARS) study: A large pilot study of the microbiome in acute and late-radiation enteropathy. Clin Cancer Res (2019) 25(21):6487–500. doi: 10.1158/1078-0432.CCR-19-0960
249. Xavier JB, Young VB, Skufca J, Ginty F, Testerman T, Pearson AT, et al. The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends Cancer (2020) 6(3):192–204. doi: 10.1016/j.trecan.2020.01.004
250. Hollingsworth BA, Cassatt DR, DiCarlo AL, Rios CI, Satyamitra MM, Winters TA, et al. Acute radiation syndrome and the microbiome: impact and review. Front Pharmacol (2021) 12:643283. doi: 10.3389/fphar.2021.643283
251. Gerassy-Vainberg S, Blatt A, Danin-Poleg Y, Gershovich K, Sabo E, Nevelsky A, et al. Radiation induces proinflammatory dysbiosis: transmission of inflammatory susceptibility by host cytokine induction. Gut (2018) 67(1):97–107. doi: 10.1136/gutjnl-2017-313789
252. Uribe-Herranz M, Rafail S, Beghi S, Gil-de-Gómez L, Verginadis I, Bittinger K, et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J Clin Invest (2020) 130(1):466–79. doi: 10.1172/JCI124332
253. Peng Z, Cheng S, Kou Y, Wang Z, Jin R, Hu H, et al. The gut microbiome is associated with clinical response to anti-PD-1/PD-L1 immunotherapy in gastrointestinal cancer. Cancer Immunol Res (2020) 8(10):1251–61. doi: 10.1158/2326-6066.CIR-19-1014
254. Kono K, Nakajima S, Mimura K. Current status of immune checkpoint inhibitors for gastric cancer. Gastric Cancer (2020) 23(4):565–78. doi: 10.1007/s10120-020-01090-4
255. Derosa L, Zitvogel L. Antibiotics impair immunotherapy for urothelial cancer. Nat Rev Urol (2020) 17(11):605–6. doi: 10.1038/s41585-020-0373-1
256. Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science (2019) 364(6446):1179–84. doi: 10.1126/science.aaw7479
257. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (2013) 342(6161):967–70. doi: 10.1126/science.1240527
258. Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science (2021) 374(6575):1632–40. doi: 10.1126/science.aaz7015
259. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science (2021) 371(6529):602–9. doi: 10.1126/science.abb5920
260. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science (2017) 358(6369):1443–8. doi: 10.1126/science.aal5240
261. Huang XZ, Gao P, Song YX, Xu Y, Sun JX, Chen XW, et al. Antibiotic use and the efficacy of immune checkpoint inhibitors in cancer patients: a pooled analysis of 2740 cancer patients. Oncoimmunology (2019) 8(12):e1665973. doi: 10.1080/2162402X.2019.1665973
262. Uribe-Herranz M, Bittinger K, Rafail S, Guedan S, Pierini S, Tanes C, et al. Gut microbiota modulates adoptive cell therapy via CD8α dendritic cells and IL-12. JCI Insight (2018) 3(4):e94952. doi: 10.1172/jci.insight.94952
263. Luu M, Riester Z, Baldrich A, Reichardt N, Yuille S, Busetti A, et al. Microbial short-chain fatty acids modulate CD8(+) T cell responses and improve adoptive immunotherapy for cancer. Nat Commun (2021) 12(1):4077. doi: 10.1038/s41467-021-24331-1
264. Han K, Nam J, Xu J, Sun X, Huang X, Animasahun O, et al. Generation of systemic antitumour immunity via the in situ modulation of the gut microbiome by an orally administered inulin gel. Nat BioMed Eng (2021) 5(11):1377–88. doi: 10.1038/s41551-021-00749-2
265. Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8(+) T cells. Immunity (2019) 51(2):285–97.e5. doi: 10.1016/j.immuni.2019.06.002
266. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut (2016) 65(12):1973–80. doi: 10.1136/gutjnl-2015-310101
267. Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med (2012) 209(5):903–11. doi: 10.1084/jem.20112408
268. Weber D, Oefner PJ, Hiergeist A, Koestler J, Gessner A, Weber M, et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood (2015) 126(14):1723–8. doi: 10.1182/blood-2015-04-638858
269. Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant (2015) 21(8):1373–83. doi: 10.1016/j.bbmt.2015.04.016
270. Li Y, Zhang Y, Wei K, He J, Ding N, Hua J, et al. Review: effect of gut microbiota and its metabolite SCFAs on radiation-induced intestinal injury. Front Cell Infect Microbiol (2021) 11:577236. doi: 10.3389/fcimb.2021.577236
271. Nenclares P, Bhide SA, Sandoval-Insausti H, Pialat P, Gunn L, Melcher A, et al. Impact of antibiotic use during curative treatment of locally advanced head and neck cancers with chemotherapy and radiotherapy. Eur J Cancer (2020) 131:9–15. doi: 10.1016/j.ejca.2020.02.047
272. Paulos CM, Wrzesinski C, Kaiser A, Hinrichs CS, Chieppa M, Cassard L, et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest (2007) 117(8):2197–204. doi: 10.1172/JCI32205
273. Hanssen NMJ, de Vos WM, Nieuwdorp M. Fecal microbiota transplantation in human metabolic diseases: From a murky past to a bright future? Cell Metab (2021) 33(6):1098–110. doi: 10.1016/j.cmet.2021.05.005
274. Tian Y, Zhou Y, Huang S, Li J, Zhao K, Li X, et al. Fecal microbiota transplantation for ulcerative colitis: a prospective clinical study. BMC Gastroenterol (2019) 19(1):116. doi: 10.1186/s12876-019-1010-4
275. Cammarota G, Ianiro G. FMT for ulcerative colitis: closer to the turning point. Nat Rev Gastroenterol Hepatol (2019) 16(5):266–8. doi: 10.1038/s41575-019-0131-0
276. Allegretti JR, Kelly CR, Grinspan A, Mullish BH, Kassam Z, Fischer M. Outcomes of fecal microbiota transplantation in patients with inflammatory bowel diseases and recurrent clostridioides difficile infection. Gastroenterology (2020) 159(5):1982–4. doi: 10.1053/j.gastro.2020.07.045
277. Antushevich H. Fecal microbiota transplantation in disease therapy. Clin Chim Acta (2020) 503:90–8. doi: 10.1016/j.cca.2019.12.010
278. Aron-Wisnewsky J, Clément K, Nieuwdorp M. Fecal microbiota transplantation: a future therapeutic option for obesity/diabetes? Curr Diabetes Rep (2019) 19(8):51. doi: 10.1007/s11892-019-1180-z
279. Vendrik KEW, Ooijevaar RE, de Jong PRC, Laman JD, van Oosten BW, van Hilten JJ, et al. Fecal microbiota transplantation in neurological disorders. Front Cell Infect Microbiol (2020) 10:98. doi: 10.3389/fcimb.2020.00098
280. El-Salhy M, Hatlebakk JG, Gilja OH, Bråthen Kristoffersen A, Hausken T. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut (2020) 69(5):859–67. doi: 10.1136/gutjnl-2019-319630
281. Cresci GA, Bawden E. Gut microbiome: what we do and don't know. Nutr Clin Pract (2015) 30(6):734–46. doi: 10.1177/0884533615609899
Keywords: gut microbiome, cancer immunology, immunotherapy, tumor microenvironment, immunomodulation
Citation: Xue X, Li R, Chen Z, Li G, Liu B, Guo S, Yue Q, Yang S, Xie L, Zhang Y, Zhao J and Tan R (2023) The role of the symbiotic microecosystem in cancer: gut microbiota, metabolome, and host immunome. Front. Immunol. 14:1235827. doi: 10.3389/fimmu.2023.1235827
Received: 06 June 2023; Accepted: 12 July 2023;
Published: 24 August 2023.
Edited by:
Li Chai, Brigham and Women’s Hospital and Harvard Medical School, United StatesReviewed by:
Yuting Ke, Massachusetts Institute of Technology, United StatesHua Zhong, University of Hawaii at Manoa, United States
Adam Yongxin Ye, Boston Children’s Hospital and Harvard Medical School, United States
Jing Liu, University of Pennsylvania, United States
Copyright © 2023 Xue, Li, Chen, Li, Liu, Guo, Yue, Yang, Xie, Zhang, Zhao and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruirong Tan, cnVpcm9uZ3RhbmxhYkAxNjMuY29t; Junning Zhao, emFybXlAMTg5LmNu; Yiguan Zhang, eWlndWFuemhhbmdAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Xiaoyu Xue1,2†
Xiaoyu Xue1,2† Bisheng Liu
Bisheng Liu Yiguan Zhang
Yiguan Zhang Junning Zhao
Junning Zhao Ruirong Tan
Ruirong Tan