- 1School of Life Science and Bio-Pharmaceutics, Shenyang Pharmaceutical University, Shenyang, China
- 2School of Medical Devices, Shenyang Pharmaceutical University, Shenyang, China
Pattern recognition receptors (PRRs), as the “sensors” in the immune response, play a prominent role in recognizing pathogen-associated molecular patterns (PAMPs) and initiating an effective defense response to pathogens in Lepidoptera. It is becoming increasingly clear that damage-associated molecular patterns (DAMPs) normally play a physiological role within cells; however, when exposed to extracellular, they may become “part-time” critical signals of the immune response. Based on research in recent years, we review herein typical PRRs of Lepidoptera, including peptidoglycan recognition protein (PGRP), gram-negative binding protein (GNBP), β-1,3-glucan recognition protein (βGRP), C-type lectin (CTL), and scavenger receptor (SR). We also outline the ways in which DAMPs participate in the immune response and the correlation between PRRs and immune escape. Taken together, these findings suggest that the role of PRRs in insect innate immunity may be much greater than expected and that it is possible to recognize a broader range of signaling molecules.
1 Introduction
Lepidoptera species are the largest phytophagous insects (1), comprising the second largest order of Insecta after Coleoptera (2). Some of the species are economic insects, as represented by Bombyx mori, Antheraea pernyi, and Thitarodes xiaojinensis, whereas others are typical pests in agriculture, such as Helicoverpa armigera, Spodoptera exigua, Manduca sexta, and Galleria mellonella. It is noteworthy that there are 472 species of edible insects in sub-Saharan Africa, of which 31% are members of Lepidoptera (3). Meanwhile, Lepidopterans were first applied in the study of innate immunity (4), and emerging data suggest that they also meet the expectations of researchers for using the “3R” principle (replacement, reduction and refinement) in animal experimentation (5). During the long process of evolution, insects have relied on their innate immune system, without adaptive immunity, to resist invasion of exogenous pathogens, and this is the natural advantage of insects as attractive models for studying innate immunity. There are also certain vital reasons, as follows. (i) The virulence factors of human pathogenic microorganisms are similar between insects and mammals, and their virulence is equivalent (6). (ii) Pathogens infect insects and mammals via identical mechanisms, including adhesion, invasion, systemic transmission and evasion of the immune response (7). (iii) The physical barrier and innate immune system of insects and mammals show a high degree of homology in function (8, 9). (iv) Insects are easy to breed, convenient to manipulate and economical. For a long time, Drosophila has been the chief insect model in gene level research; as a mini-host, it is advantageous for use for forward and reverse genetics (6). However, it also has some shortcomings, such as the inability to be propagated at 37°C, a small size and hemolymph volume (7); in addition, a wealth of operating experience and special laboratory equipment are needed (e.g., microsyringes) (10). Lepidopterans arguably have more advantages in the study of protein levels, with consideration of physiological and immune characteristics. First, it is easier to extract tissues and collect more hemolymph due to the larger size of Lepidopteran larvae. In addition, it is known that the larvae of G. mellonella can be propagated at 37°C, which is equivalent to the body temperature of the mammalian host (5). As temperature has been proven to play a significant role in expression of virulence factors, this feature is extremely important in analyzing the innate immune response to pathogens (11).
The innate immune system in insects is composed of two arms: humoral and cellular mechanisms. Humoral immunity entails conversion of prophenoloxidase (PPO) to active phenoloxidase (PO) (12, 13) and expression of genes encoding antimicrobial peptides (AMPs) (14–17) through the Toll and IMD pathways. The above two pathways share many similar features with the Toll-like receptor (TLR) and tumor necrosis factor-α (TNF-α) receptor signaling cascades in mammals (18, 19). Active PO catalyzes conversion of phenols to quinones and promotes formation of melanin, which then participates in cellular immunity (20, 21). AMP directly kills invading microorganisms by interacting with microbial membranes, destroying membrane structures and interfering with internal mechanisms (14, 22). Cellular immunity is mainly mediated via hemocytes. If the pathogen is able to breach the physical barriers of the host, hemocytes will be recruited to the site of infection, phagocytosing or killing the pathogens at the membrane or intracellular level (14). In addition, hemocytes and pathogens form microaggregates (21, 23), which accumulate into nodules that are directly encapsulated and eliminated by melanin synthesized by active PO. Pattern recognition receptors (PRRs) are the “sensor” in the immune response that initiates the above two aspects; that is, they are a class of proteins expressed by innate immune cells that recognize invasive pathogens. This recognition involves a process of distinguishing “self” and “nonself” together with the conservative structure of pathogens (bacteria, fungi, viruses and other pathogenic microorganisms have specific structural components completely different from the host body), which are ordinarily referred to as pathogen-related molecular patterns (PAMPs) (24, 25). The role of this recognition process is also the basis of the classical concept of innate immunity (26). In general, PAMPs are mostly located on the cell surface of pathogens, with a few located inside cells. PAMPs can be divided according to their chemical nature into the following categories: (i) polysaccharide compounds, such as peptidoglycan (PGN), β-1,3-glucan, zymosan, lipopolysaccharide (LPS), and capsular polysaccharide (27–29); (ii) lipid compounds, such as LPS, lipoteichoic acid (LTA), and lipoarabinomannan (LAM) (29–31); (iii) proteins and polypeptides, such as flagellin and capsid protein (32); and (iv) nucleic acids, such as dsRNA, ssRNA of viruses and unmethylated CpG of bacteria (33).
Increasing evidence shows that PRRs play crucial roles in identifying endogenous molecules released by damaged cells, which are called damage-associated molecular patterns (DAMPs) (24). A cornerstone concept of DAMPs is that the molecules that act in response to various stresses and damage are host derived rather than pathogen or environment derived (34). DAMPs are regarded as endogenous danger signals because they induce aseptic inflammation without infection and induce an innate immune response that is similar to the response caused by PAMPs (35–37). Although the theory of DAMPs was advanced earlier, in 1994, Polly Matzinger proposed the “danger” theory under the assumption that injured tissues release intracellular molecules to activate the immune system (38). It was not until the high mobility group box 1 (HMGB1) (39), uric acid crystals (40), and Hsp family (41) were successively considered to be DAMPs that this theory began to be widely applied. In the past 30 years, more than 30 types of DAMPs have been confirmed in mammals, located outside the cell (e.g., lmw hyaluronic acid; fibrinogen fibrinogen) or intracellularly, including particles represented by defensins and substances distributed in various structures, such as the cytosol (e.g., S100 proteins, heat shock proteins), nucleus (e.g., HMGB1, DNA, RNA), mitochondria (e.g., mtDNA, formyl peptide), ER (e.g., calreticulin), and plasma membrane (e.g., Syndecans, Glypicans) (37). Dorsal switch protein 1 (DSP1) is a novel kind of DAMP in insects and an ortholog of mammalian HMGB1. Although there are currently few confirmed insect DAMPs, it was recently reported that DSP1 participates in activating the immune response in S. exigua after Bacillus thuringiensis (Bt) infection (42), providing concrete evidence for the study of insect DAMPs.
PAMPs and DAMPs, either exogenous or endogenous, are danger signals for the body. Detecting them rapidly and responding in time is the key process for insects to initiate the innate immune response and maintain homeostasis. PRRs are the “sensors” that initiate the immunoreaction. Here, we review new findings on pattern recognition receptors in Lepidoptera. In addition, we summarize the research progress of insect DAMPs in recent years. Our purpose is to deepen understanding of the mechanism of innate immunity of insects and reveal possible problems (Table 1).
2 The recognition effect of PRRs on PAMPs
What kind of special structure does a pathogen have? Considering this problem may help us to understand why PRRs can quickly identify pathogens and mediate the host’s immune response. A huge portion of PAMPs, mostly polysaccharides, lipids or the proteins or nucleic acids of viruses, are frequently the basic building blocks of pathogens. Such characteristic molecules render PRRs more efficient and accurate in recognizing PAMPs (Figure 1). In this section, Lepidopteran insects are taken as an example to briefly describe new PRRs and the characteristics of their involvement in the immune response, for instance, the type of pathogen they recognize and the type of immune response they mediate.
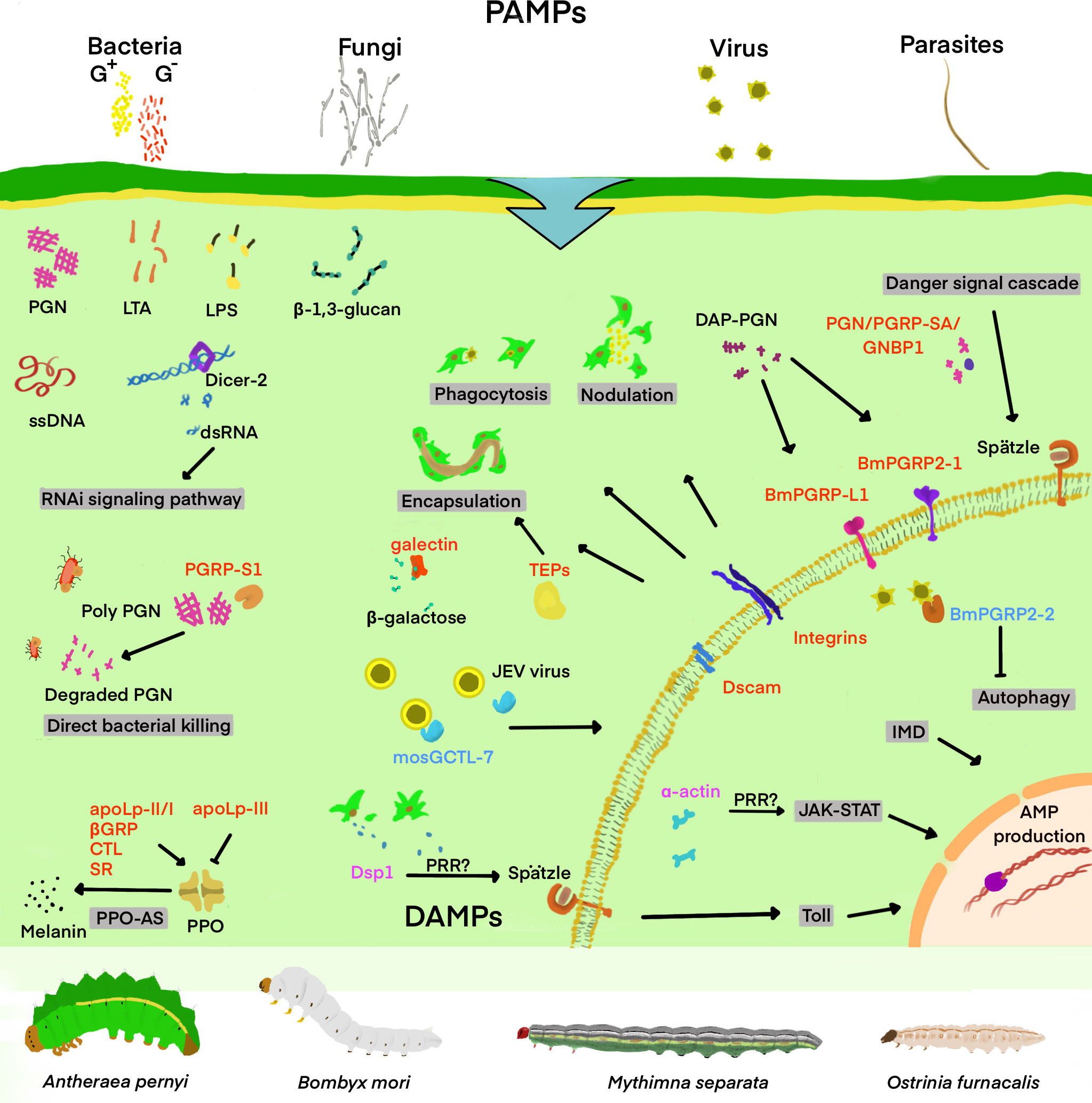
Figure 1 Schematic diagram of important insect’s PRRs involved in the recognition of “danger signal” and the regulation of downstream immune signaling pathways. Infectious invaders include viruses, fungi, gram-positive bacteria (G+), gram-negative bacteria (G-), and other parasites. DAMPs and PAMPs refer to injury-related and pathogen-related molecular patterns, respectively. Some of the PAMPs and DAMPs shown in the figure include double-stranded RNA (dsRNA), single-stranded DNA (ssDNA), β-1,3-glucan, lipopolysaccharide (LPS), lipoteichoic acid (LTA), and peptidoglycan (PGN), Dorsal switch protein 1(Dsp1), and α-actin. The pattern recognition receptors (PRRs) are classified into the secretory and the transmembrane type, and are marked in red in the figure. The secretory PRRs described here consist of short-type peptidoglycan recognition protein (PGRP-Sx); Gram-negative bacteria-binding protein (GNBP); β-1,3-glucan recognition protein (βGRP); C-type lectin (CTL); Scavenger receptor (SR); Apolipoprotein I-III (ApoLp-II/I, III);and thioester-containing protein (TEP), while transmembrane PRRs mainly include Toll, long-type PGRP (PGRP-Lx), integrins and Down syndrome cell adhesion molecule (Dscam). In addition, TEP is also identified as an opsonin of various immune responses, with the PRRs-triggered or regulated immune responses and signaling pathways mentioned in the review as shaded. Damage-associated mode molecule (DAMP) is labeled as magenta, which transmits signals in cells and regulates downstream immune pathways. The molecules labeled with blue are mosquito galactose-specific C-type lectin 7(mosGCTL-7) and B. mori peptidoglycan recognition protein 2-2 (BmPGRP2-2), which are involved in the immune escape of the virus, helping to infect the virus and inhibiting the downstream immune response.
2.1 PGRP
Peptidoglycan recognition protein (PGRP), ubiquitous in vertebrates, mollusks, echinoderms and insects, is a pivotal PRR conserved from insects to mammals (54, 69). It plays a role in recognition, bacteriostasis or sterilization, agglutination, and amidase activity and participates in the immune response as a regulator. PGRPs mainly recognize the characteristic bacterial molecule PGN. PGRPs may also recognize LPS and β-1,3-glucan. PGRP can be divided into two types according to the length of the amino acid sequence: a long type (PGRP-L) and a short type (PGRP-S). The long type is an intracellular and transmembrane protein, and the short type is an extracellular protein (70). Furthermore, PGRPs can be divided into catalytic PGRPs and noncatalytic PGRPs according to the catalytic activity of amidase (71). All four mammalian PGRPs, PGLYRP1, PGLYRP2, PGLYRP 3 and PGLYRP4, have the ability to recognize bacterial PGN (72, 73). Among them, PGLYRP1, PGLYRP3 and PGLYRP4 have a direct bactericidal effect on gram-positive and gram-negative bacteria, but with no enzyme activity; PGLYRP2 exhibits amide enzyme activity to hydrolyze bacterial cell wall PGN (74, 75). There are some reports on the function of PGRP in reptiles. C-turtle-PGRP-S in the aquatic reptiles Chinese soft-shelled turtle Pelodiscus sinensis has been found to have PGN-binding and antibacterial activities (76). Research on fish PGRP has also gradually increased. Among the currently known 3 kinds of PGRPs in the orange-spotted grouper Epinephelus coioides, PGRP-S has direct antibacterial activity against two pathogens, Vibrio harveyi and Edwardsiella tarda, and it also significantly induces activation of NF-κB as a regulatory factor (77). PGRP-L1 and PGRP-L2 recognize and bind to PGN, play a role in activating NF-κB luciferase activity, and significantly inhibit growth of E. tarda (78). AjPGRP-S in the echinoderm Apostichopus japonicus has agglutination ability, strong antibacterial activity against Vibrio splendidus, V. harveyi, Vibrio parahaemolyticus, Staphylococcus aureus as well as Micrococcus luteus, and high amidase activity in the presence of Zn2+ (79). In the mollusk Crassostrea gigas, CgPGRPS2 and CgPGRPS4 recognize a variety of microorganisms and PAMPs, playing a pattern recognition role in the innate immune response of this oyster (80).
Compared with other species, the immune function of PGRP in insects has been studied in depth. In addition to the above functions, PGRP, which is an indispensable part of innate immunity, also participates in regulating synthesis of AMPs and activation of the PO system in insects. Insect PGRP mainly plays a recognition role upstream of the Toll versus Imd pathways (71). DAP-PGN in gram-negative bacteria activates the IMD pathway to produce AMPs, whereas Lys-PGN in gram-positive bacteria plays a role in the Toll pathway (81). In Drosophila melanogaster, extracellular PGRP-SA and PGRP-SD stimulate the Toll pathway after infection by gram-positive bacteria, and PGRP-SD promote repositioning of peptidoglycan on the cell surface to upregulate the Imd pathway (82–84). Membrane protein PGRP-LC (85–87) versus extracellular PGRP-LE acts upstream of the Imd pathway. Extracellular PGRP-LE participates in the PO pathway (88, 89), and intracellular PGRP-LE is involved in autophagy to defend against pathogens (e.g., Listeria monocytogenes) (90). Recent observations have shown that the affinity of PGRP-SC in Musca domestica for various intestinal polysaccharides, including LPS and β-1,3-glucan, is beneficial for maintaining intestinal equilibrium (91). Multiple lines of evidence indicate that PGRPs are closely related to regulation of AMPs in Lepidoptera. BmPGRP2-1 and BmPGRP-L1 in B. mori recognize DAP-PGN and then trigger the Imd pathway (43, 44). PxPGRP-S1 in Plutella xylostella and rPGRP6 in Ostrinia furnacalis appear to play a prominent role in synthesis of AMPs (48, 49). Amidase activity (71) is also an effective means for involvement of some PGRPs in the immune response, which is beneficial for destroying pathogens and maintaining immune homeostasis in the host. PGRP-S1 is capable of recognizing DAP-PGN and Lys-PGN and exerting amidase activity to degrade PGN, disrupting the bacterial surface (51). PGRP-S5 in B. mori is associated with downregulation of the IMD pathway induced by bacteria in the late stage, which is beneficial to prevent overactivation of the immune response, which might have an adverse effect on the host (92).
Recognition between PGRP and PGN also contributes to the PO pathway and plays multiple roles in cellular immunity. PGRP-S1, which was purified from the hemolymph of B. mori, was the first PGRP described. After PGN recognition, the PO pathway is activated (54), and PGRP-S4/S5 in B. mori has been proven to recognize PGN and trigger stimulation of the PO pathway (93, 94). PGRP-SA in A. pernyi has been shown to be a broad-spectrum pattern recognition receptor involved in activation of the PPO system and production of AMPs (95). PGRP-S1 of Diaphania pyloalis (Walker) recognizes two kinds of PGN and causes strong agglutination of Escherichia coli, M. luteus or S. aureus in the presence of Zn2+ (96).
Presently, most research exploring the role of PGRPs in innate immunity focuses on bacteria and fungi, and only a few studies have shown that PGRPs have the ability to respond to virus invasion. B. mori bidensovirus (BmBDV) infection significantly increases expression of BmPGRP-LB and BmPGRP-LE in B. mori and activates the Imd pathway (97). Induction of B. mori cytoplasmic polyhedrosis virus (BmCPV) increases expression of BmPGRPS2. Furthermore, overexpression of BmPGRPS2 activates the Imd pathway, elevates AMPs, and enhances the ability to resist infection by the virus (98). SePGRP-LB in S. exigua plays a similar role in S. exigua multiple nucleopolyhedrovirus (SeMNPV) infection and induces a significant increase in expression of Relish, a key gene in the IMD immune signaling pathway (53).
2.2 βGRP/GNBP
The β-1,3-glucan recognition protein family is one of the most characteristic pattern recognition receptor families in invertebrates and mainly includes βGRP and GNBP (99, 100). At present, this family has been identified mostly from insects and crustaceans; it mainly contributes to the innate immune response by participating in activation of the PO system. In crustaceans, this family is also called β-1,3-glucanase-related protein (BGRP) (101). PcBGRP in Procambarus clarkii exhibits strong binding to LPS and β-1,3-glucan and enhances PO activation in vitro and in vivo (102). The first βGRP isolated and purified was from B. mori, and it recognizes β-1,3-glucan in the fungal cell wall and initiates activation of the PPO cascade (103). In O. furnacalis, after induction by LPS and laminarin (soluble β-1,3-glucan), the mRNA content of OfβGRP3 increases, with the LPS-challenged group showing higher levels than the laminarin-challenged group. OfβGRP3 also activates the PO pathway after bacterial infection (56). βGRP1 in T. xiaojinensis is an essential receptor for activation of PO induced by Cordyceps militaris. However, failure of βGRP1 to recognize Ophiocordyceps sinensis is the main reason why the host does not undergo melanization after infection. Immunofluorescence detection has revealed a protective layer that prevents βGRP1 from recognizing fungi. βGRPs not only play a recognition role in Lepidopterans to trigger the innate immune response but also act in synergy with other immunity to promote antifungal defense (104). βGRP1 in T. xiaojinensis interacts with immulectin-8 (a kind of lectin) to promote encapsulation of pathogens (105). Although the role of βGRPs in viral infection needs further research, βGRPs are considered to have antiviral potential. This view is substantiated by the observation that overexpression of βGRP4 inhibits proliferation of B. mori nucleopolyhedrovirus (BmNPV) in B. mori ovary N (BmN) cells (57).
GNBPs also may be involved in resisting pathogens. The first member of the GNBP family was isolated and purified from B. mori, and it was named because of its strong binding ability to gram-negative bacteria (E. coli) but was later classified as a member of the β-1,3-glucan recognition protein family (106), also known as βGRP2 (107). The homologs GNBP1 and GNBP2 found in D. melanogaster are involved in defense against gram-positive bacteria (108–111), and GNBP3 correlates with defense against fungal infection (112, 113). GNBP6 in S. exigua significantly upregulates PO activity in vitro (114). Current research has shown that the immune response of S. exigua to fungal infection requires three PRRs (βGRP-1, βGRP-2 and GNBP3) to activate the Toll signaling pathway (115). In addition, PxβGBP is a β-1,3-glucan-binding protein identified in P. xylostella, which is similar to βGRP and participates in host immunity to fungal infection. Moreover, interference of dsPxβGBP sensitizes P. xylostella to Isaria cicadae infection (116).
2.3 CTL
C-type lectins (CTLs), which are widely distributed in both vertebrates and invertebrates, are the most abundant and diverse superfamily of animal lectins (117). CTLs have been studied extensively in mammals. They are often referred to as C-type lectin receptors (CLRs), which play an important role in identifying and resisting pathogens and maintaining the homeostasis of the intestinal flora. For example, several CLRs, including Dectin-1, Dectin-2, Mincle, and Clecsf8, have been shown to be involved in mouse responses to mycobacteria via activation of the spleen tyrosine kinase (Syk)/caspase recruitment domain family member 9 (CARD9) signaling pathway (118, 119). Deletion of Dectin-1 and Dectin-2 affect the environment of the bacterial intestinal microbiota of mice and increase susceptibility to colitis (120). Dectin-1 specifically recognizes fungal cell wall β-glucan and then participates in activating the immune response of mice (121). In addition, CLRs in mammals play diverse roles in the development of inflammation and tumors. For example, Mincle in mice is able to inhibit clearance of dead cells and increase production of proinflammatory cytokines; it is involved in the persistent inflammation induced by cell death after acute kidney injury (122). In vitro results have shown that downregulation of DC-SIGN significantly inhibits the proliferation, cell cycle progression, migration and invasion of gastric cancer cells (123), suggesting that the expression level of DC-SIGN correlates positively with the occurrence and development of gastric cancer. However, mouse Dectin-2 and Dectin-3 enhances the phagocytic activity of Kupffer cells in the liver and promotes their phagocytosis and clearance of tumor cells, thereby suppressing liver metastasis of tumor cells (124).
Research on CTLs of other vertebrates mainly focuses on their recognition, agglutination or other functions in the process of pathogenic microorganism infection. In poultry, gene expression of cLL in chickens is significantly upregulated after induction by Avian Pathogenic Escherichia coli (APEC), suggesting that as a receptor, cLL may play an important role in innate defense against early pulmonary APEC infection in the host (125, 126). In fish, the expression level of OmLec1 in Onychostoma macrolepis is significantly increased after induction by Aeromonas hydrophila, showing agglutination activity against bacteria such as E. coli and S. aureus in vitro (127). SsCTL4 in black rockfish recognizes and agglutinates E. tarda and Vibrio anguillarum and inhibits E. tarda infection while promoting invasion of infectious spleen and kidney necrosis virus (ISKNV) through recognition (128). In addition, SmLec1 in Scophthalmus maximus stimulates renal lymphocyte proliferation and enhances the killing effect of macrophages on bacterial pathogens (129).
In invertebrates, CTLs have attracted much attention due to their abilities to recognize and bind sugar ligands, promote agglutination, participate in cellular immunity, mediate synthesis of AMPs and regulate the PO system (130–132). Hp-Lec in Hemifusus pugilinus displays a broad spectrum of bacterial agglutination activity and agglutination activity against vertebrate blood cells, as well as antifungal activity against Aspergillus niger and Aspergillus flavus (133). Cnlec-1 in Chlamys nobilis is upregulated after induction by three immunostimulants, V. parahaemolyticus, LPS and PolyI: C, and plays a role in the body’s immune response (134). LvLec in Litopenaeus vannamei is able to enhance V. harveyi-induced phagocytosis of blood cells, increase PO activity, and possibly regulate the immune response of blood cells through the cGMP-PKA pathway (135).
The role of some CTLs derived from Lepidopteran insects in innate immunity against exogenous pathogens has also been fully studied and characterized. The bracoviral-derived C-type lectin BLL2 from S. exigua is highly sensitive to a variety of bacteria and agglutinates a broad range of bacterial species (136). IML-10 in O. furnacalis binds to the surface of hemocytes and promotes their aggregation, thereby enhancing hemocytic encapsulation (63). 20E-HaEcR-HaUSP, a complex formed by the steroid hormone 20-hydroxyecdysone (20E), ecdysone receptor (HaEcR) and ultraspiracle (HaUSP), stimulates expression of CTL1 in H. armigera. Subsequently, HaCTL1 participates in cellular immunity (encapsulation and phagocytosis) to resist invading pathogens (nematodes and bacteria) (137). In addition, HaCTL3 promotes phagocytosis of hemocytes by bacteria (such as E. mundtii), and AMPs regulated by HaCTL3 exert a bactericidal effect. Both of the above strategies may lead to elimination and inhibition of bacteria in the hemolymph of H. armigera (64). CTL-14 in H. armigera is capable of recognizing fungal surface polysaccharides, forming aggregates with yeast polysaccharides and Beauveria bassiana conidia, and interacting with six melanin-related proteins to produce melanin (138). CTL-S6 in B. mori binds bacterial PGN strongly but has weak LPS binding ability. In addition, CTL-S6 is involved in encapsulation, activation of the PO pathway and melanogenesis (130). CTL-5 in B. mori has been proven to be an important PRR of the JAK/STAT signaling pathway, which mediates nodule melanization during fungal infection (139). Another CTL in B. mori, BmCTL10, binds to a variety of PAMPS and plays a role in enhancing encapsulation, nodulation and phagocytosis. It can also bind to blood cells to upregulate synthesis of AMPs. BmCTL10 even downregulates PPO expression as a modulator to protect the body from certain toxic compounds, maintaining immune homeostasis (140). Although little is known about the antiviral effects of CTLs in Lepidoptera, there is increasing evidence of antiviral potential. Expression of Ha-lectin in H. armigera is upregulated after H. armigera nuclear polyhedrosis virus (HaNPV) induction (141). Two bracovirus-associated lectins, Se-BLL2 and Se-BLL3, have been successively confirmed in recent years to exert antiviral activity during the immune process of S. exigua larvae against baculovirus (142) and Spodoptera frugiperda larvae against Junonia coenia densovirus (JcDV) (143). IML-2 in B. mori inhibits proliferation of BmNPV by promoting apoptosis (65).
2.4 Scavenger receptor
Scavenger receptors (SRs) are a class of transmembrane glycoproteins on the cell surface act as PRRs that directly recognize PAMPs as well as DAMPs and participates in the identification, phagocytosis and elimination of pathogens. SRs are generally divided into 12 categories: SR-A~J. Here, we focus on two categories, SR-B and SR-C. SR-B is a kind of PRR that exists in both vertebrates and invertebrates and mediates an immune response (144, 145). It has been reported that scavenger receptor class B1 (SCARB1), which is a typical SR-B, plays a complex biological function. In humans, SCARB1 serves as an important mediator of cholesterol homeostasis, and mutations in SCARB1 are associated with accelerated development of coronary artery disease (146). In mice, SRB1 regulates lymphocyte proliferation and cytokine production (147). SCARB1 in Serinus canaria has been shown to be a mediator of carotenoid uptake (148). SmSRB1 in S. maximus recognizes Streptococcus iniae, V. anguillarum, iridovirus, and multiple PAMPs. In addition, it may act as a coreceptor for TLRs and NLRs to regulate immune responses to pathogens (149). RpSR-BI in Ruditapes Philippinarum was confirmed to be a PRR with broad-spectrum recognition that may be used as an opsonin to participate in the innate immune response and enhance phagocytosis and chemotaxis of blood cells (150). EsSR-B2 in Eriocheir sinensis recognizes gram-positive and gram-negative bacteria, thus enhancing phagocytosis and stimulating expression of AMPs (151). AjSR-B in A. japonicus binds a variety of PAMPs and exhibits agglutinative activity against gram-positive and gram-negative bacteria (152). SR-B is also of interest in Lepidopteran insects. BmSCRB8 in B. mori enhances the bacterial clearance rate and promotes production of AMPs in vivo (66). MmSR-B1, an SR-B family member from Micropilits mediator (the natural enemy of many Lepidopteran agricultural pests), correlates significantly with synthesis of AMPs and phagocytosis by hemocytes (153). Interestingly, SR-C is only found in invertebrates and not in vertebrates (154). MjSRC in Marsupenaeus japonicus plays an important role in the antibacterial immunity of the host by enhancing phagocytosis and expression of AMPs (155). SR-C in D. melanogaster is a PRR associated with recognition and phagocytosis of invading bacteria (E. coli versus S. aureus) (156). SR-C in Tenebrio molitor is involved in phagocytosis of microorganisms such as bacteria (E. coli versus S. aureus) and fungi (Canidia albicans) (157). In recent years, the immune function of SR-C in silkworm was reported for the first time. As a pattern recognition receptor against bacteria, SR-C in B. mori recognizes and binds diverse types of PAMPs, especially Lys-type PGN, and initiates the immune response. Moreover, SR-C regulates expression of AMPs by activating Toll signaling (67).
2.5 Other PRRs
The understanding of innate immunity prompted the search for PRRs. In addition to the above PRRs, a portion of PRRs exhibit pattern recognition characteristics. Galectins are a type of β-galactoside-binding protein that act as a recognition and effector factor in innate immunity by recognizing polysaccharides on the surface of pathogenic microorganisms (158). It has been reported that Galectin-4 plays a recognition role in the fertilized eggs of the silkworm B. mori and can induce bacterial agglutination in vitro (159). The complement system is involved in mediating elimination of pathogens at an early stage of mammalian infection (160). Thioester (TE)-containing proteins (TEPs) are highly similar to mammalian C3 (161). Macroglobulin complement-related factor (MCR) in Aedes aegypti belongs to the TEP family, which functions with SR to regulate expression of AMPs, thus exerting anti-dengue virus (DENV) activity (162). Down syndrome cell adhesion molecule (Dscam) may be involved in various pathways, such as pathogen-specific recognition, phagocytosis, transduction of immune signals, and regulation of AMPs (163). Dscam in Anopheles gambiae mediates phagocytosis after bacterial infection (164).
3 The potential identification function of DAMPs by PRRs
In general, understanding of DAMPs is constantly being updated with deep investigation. Initially, DAMPs were expected to verify cell death. Subsequently, DAMPs were thought to be secreted or exposed by living cells that experience life-threatening stress. Recently, DAMPs have been found to be crucial to tissue healing after inflammation (165). DAMPs play a role in autoimmune diseases, osteoarthritis, cardiovascular diseases, neurodegenerative diseases and cancer in mammals. Hsp, ATP, HMGB1 and other typical DAMPs provide reference targets for disease diagnosis and treatment (37). Moreover, HMGB1 and ATP have been identified as prominent molecules for promoting regeneration (165).
HMGB1, which is a highly conserved nuclear protein expressed in mammals, normally functions as a DNA chaperone within cells (165, 166). During stress, HMGB1 is released, which can be used as a DAMP to activate the innate immune response. Toll-like receptor (167) and SR (168) in mammals have a recognition role in this process. DSP1, an ortholog of vertebrate HMGB1 (169, 170), is a type of known insect DAMP first discovered in D. melanogaster (169, 171). According to current research on insect DAMPs, we speculate that DSP1, as a DAMP triggering the insect signaling pathway, plays a role in amplifying signals during pathogen infection. In S. exigua, SeDSP1 is released into the circulatory system after infection by bacteria, and the Toll pathway is activated by triggering Spätzle. Then, AMPs and phospholipase A2 (PLA2) are produced. PLA2 catalyzes synthesis of Eicosanoids, which mediate both the cellular and humoral arms in insects. Xenorhabdus hominickii inhibits activation of the Toll signaling pathway by DSP1 (including activation of PLA2), which leads to significant immunosuppression in S. exigua (171). SeDSP1 also regulates production of reactive oxygen species (ROS) through the DSP1/PLA2/Ca2+/dual oxidation (Duox) signaling pathway and participates in the intestinal immune response (172). A similar mechanism has been found in mosquitoes (173).
Actin is another typical DAMP in mammals. Filamentous (F-) actin, an important protein involved in cell movement and contraction, is released by dead cells and recognized by an innate immune receptor called DNGR-1 (also known as CLEC9A, a member of the C-type lectin family) (174–176). Recent evidence has shown that α-actin, a cytoskeletal protein closely related to F-actin, is the key trigger for STAT activation in the JAK/STAT pathway in D. melanogaster (177).
We speculate that a portion of DAMPs may be conserved between vertebrates and invertebrates, with similar functions and recognized mechanisms. This review provides a basis for follow-up studies on insect DAMPs and shows the possibility of their complementation. Notably, although DAMPs have begun to be discovered in some insects, their paired receptors have not been clarified. What PRRs does DSP1 encounter before triggering Spätzle? In what way is α-actin involved in STAT activation? Are there any proteins in insects that are similar to the PRR that recognizes DAMPs in mammals? These questions are likely to be answered in the future.
4 Immune escape
Occasionally, PRRs may act not as an “adversary” but an “accomplice” for pathogens. In mammals, viruses promote invasion by binding to pattern recognition receptors and escape from the innate immune response by interfering with signal transduction and cellular immunity (178). Immune escape by pathogens may lead to persistent infection and even cancer. For instance, Epstein-Barr virus (EBV), which was the first identified human virus associated with tumors, is closely related to development of nasopharyngeal carcinoma (NPC), gastric carcinoma (GC), and several lymphomas. Human papillomavirus (HPV) is associated with cervical, anal, penile, and head and neck squamous cell carcinomas. Furthermore, infection by these viruses is involved in tumor immune escape (47, 179–181). Cancer immune surveillance involves three basic processes of elimination, balance and escape, which is called the theory of cancer immunoediting (182, 183). According to the suggestion of this hypothesis, it is necessary to study the mechanism of immune escape for tumor treatment. Current data indicate that immune escape by pathogens, especially viruses, is closely related to PRRs and that this mechanism exists not only in mammals but also in insects. BmPGRP2-2 in B. mori, induced by BmNPV, negatively regulates phosphatase and tensin homolog (PTEN)-phosphoinositide 3-kinase (PI3K)/Akt signaling to inhibit apoptosis to promote replication of the virus (43). As a “bridge”, some CTLs in mosquitoes promote Flavivirus infection, such as West Nile virus (WNV) and Japanese encephalitis virus (JEV). The mosGCTL-1/WNV complex formed by mosGCTL-1 in A. aegypti and the invading WNV may be captured by the membrane protein mosPTP-1, which is a mosquito homolog of human CD45, at the surface of the cell membrane, promoting invasion by and spread of the virus (184). Studies have confirmed that mosGCTL-7 in mosquitoes is able to bind to the JEV envelope protein via an N-glycan at N154 in a calcium-dependent manner and promote viral infection through the mosGCTL-7/mosPTP-1 pathway (185). SRB1 in human hepatocytes and an SRB1-like receptor in mosquito cells acts as cell-binding proteins that bind to and facilitate internalization of DENV and Zika virus (ZIKV) NS1, resulting in high permeability of endothelial cells and downregulation of the innate immune response (186). In addition, knocking out GNBP in A. aegypti promotes clearance of DENV-2 (187).
5 Discussion
In the long process of evolution, organisms of various taxa have evolved immune systems with multiple structures and functions. Among them, the innate immune system is a common defense mechanism in animals, is especially important in invertebrates, and plays an important role in vertebrates such as zebrafish (188) (Table 2). Therefore, as a key link in innate immunity, PRRs have received extensive attention. PRRs of different organisms have both common and surprising characteristics, and they are closely related to each other. Some of the above-described Lepidopteran PRRs are homologs found in higher organisms, and the Toll-like receptors (TLRs) that have been well studied in vertebrates are conserved from Drosophila to human (197, 198). Hence, it is important to explore the immune function of PRRs and their association with PRRs of other organisms.
Based on the discussion herein about PRRs of Lepidoptera, the recognition role played by PRRs in insect innate immunity is essential for activation of signaling pathways. Partial PRRs can also directly interact with pathogens, for example, participating in agglutination, cleavage and even elimination. We have noticed that identification of hazardous signals by PRRs is not a single route of transmission but seems to comprise a large closely related network. Furthermore, PRRs form “attack complexes” with other proteins to enhance signal transduction. All of these factors are conducive to the efficient immune response of insects.
As more attention has been given to the role of PRRs in immunity, functional studies are becoming more comprehensive. Some research on PRRs has also been used in breeding of economic insects or formulating pesticides to kill agricultural pests. Here, we illustrate recognition of PRRs for PAMPs and DAMPs but do not discuss their symbiotic relationship or promotion of growth and development. Based on the above, we propose several possible future research directions.
i) Improving understanding of the antiviral pathway of Lepidoptera. Viruses do not have cellular structures, though nucleic acids are currently considered typical of PAMPs, and multiple studies have found that dsRNA, ssRNA, and DNA containing single-stranded unmethylated CpG motifs are recognized by TLR3, TLR7, and TLR9, respectively, in antiviral or antibacterial pathways in mammals (195, 199–202). However, the mechanism of virus recognition in insects is not very clear. For example, BmPGRP-S3 levels are increased after induction by the RNA virus BmCPV, but the specific mechanism needs to be further studied (203). A protein of the viral shell may also play a special role in the recognition process.
ii) Exploring PRRs that bind newly defined PAMPs. In investigations carried out on α-1,3-glucan based on the model of G. mellonella, it was validated that A. niger α-1,3-glucan, a virulence factor, can play a role in humoral and cellular immunity. Multiple PRRs were also shown to interact with α-1,3-glucan, but Apolipoprotein-III (ApoLp-III) was unable to recognize it. These data provide evidence that α-1,3-glucan is a PAMP that can be recognized by insects and a new direction for enriching understanding of PRRs (204–206).
iii) Searching for Lepidopteran DAMPs and their receptors. The DAMPs reported in Lepidoptera are limited, and PRRs capable of recognizing DAMPs are not clear and need further research.
These issues remind us that continuous attention to risk signaling molecules and pattern recognition receptors capable of recognizing them is indispensable for understanding the innate immunity of the host.
Author contributions
XW and RZ conceived the concepts, directed the writing and critically revised the manuscript. LZ carried out the manuscript drafting. JN and DF contributed to the design of the tables and figure. All authors have read and approved the content of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant numbers: 31970485); the Natural Science Foundation of Liaoning (Grant number: 2022-MS-249); the Project of Liaoning Provincial Department of Education (Grant number: LJKFZ20220259) and the Project of Shenyang Pharmaceutical University for Outstanding Young Teachers (Grant number: YQ202115).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mitter C, Davis DR, Cummings MP. Phylogeny and evolution of Lepidoptera. Annu Rev Entomol (2017) 62:265–83. doi: 10.1146/annurev-ento-031616-035125
2. Kristensen NP, Scoble M, Karsholt O. Lepidoptera Phylogeny and systematics: the state of inventorying moth and butterfly diversity. Zootaxa (2007) 1668:699–747. doi: 10.11646/ZOOTAXA.1668.1.30
3. Numbi Muya GM, Mutiaka BK, Bindelle J, Francis F, Caparros Megido R. Human consumption of insects in Sub-Saharan Africa: Lepidoptera and potential species for breeding. Insects (2022) 13(10):886. doi: 10.3390/insects13100886
4. Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol Rev (2004) 198:83–96. doi: 10.1111/j.0105-2896.2004.0120.x
5. Pereira MF, Rossi CC, da Silva GC, Rosa JN, Bazzolli DMS. Galleria mellonella as an infection model: an in-depth look at why it works and practical considerations for successful application. Pathog Dis (2020) 78(8). doi: 10.1093/femspd/ftaa056
6. Wang Y, Li DD, Jiang YY, Mylonakis E. Utility of insects for studying human pathogens and evaluating new antimicrobial agents. Adv Biochem Eng Biotechnol (2013) 135:1–25. doi: 10.1007/10_2013_194
7. Kemp MW, Massey RC. The use of insect models to study human pathogens. Drug Discovery Today: Dis Models (2007) 4(3):105–10. doi: 10.1016/j.ddmod.2007.06.007
8. Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science (1999) 284(5418):1313–8. doi: 10.1126/science.284.5418.1313
9. Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev (2001) 15(18):2321–42. doi: 10.1101/gad.909001
10. Wand ME, McCowen JWI, Nugent PG, Sutton JM. Complex interactions of klebsiella pneumoniae with the host immune system in a galleria mellonella infection model. J Med Microbiol (2013) 62(Pt 12):1790–8. doi: 10.1099/jmm.0.063032-0
11. Twittenhoff C, Heroven AK, Muhlen S, Dersch P, Narberhaus F. An RNA thermometer dictates production of a secreted bacterial toxin. PloS Pathog (2020) 16(1):e1008184. doi: 10.1371/journal.ppat.1008184
12. Wang Y, Jiang H, Cheng Y, An C, Chu Y, Raikhel AS, et al. Activation of aedes aegypti prophenoloxidase-3 and its role in the immune response against entomopathogenic fungi. Insect Mol Biol (2017) 26(5):552–63. doi: 10.1111/imb.12318
13. Xu L, Ma L, Wang W, Li L, Lu Z. Phenoloxidases are required for the pea aphid’s defence against bacterial and fungal infection. Insect Mol Biol (2019) 28(2):176–86. doi: 10.1111/imb.12536
14. Stanley D, Haas E, Kim Y. Beyond cellular immunity: on the biological significance of insect hemocytes. Cells (2023) 12(4). doi: 10.3390/cells12040599
15. Zhang QY, Yan ZB, Meng YM, Hong XY, Shao G, Ma JJ, et al. Antimicrobial peptides: mechanism of action, activity and clinical potential. Mil Med Res (2021) 8(1):48. doi: 10.1186/s40779-021-00343-2
16. Hetru C, Hoffmann JA. NF-kappaB in the immune response of drosophila. Cold Spring Harb Perspect Biol (2009) 1(6):a000232. doi: 10.1101/cshperspect.a000232
17. Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, et al. The drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol (2003) 4(5):478–84. doi: 10.1038/ni922
18. Hultmark D. Drosophila immunity: paths and patterns. Curr Opin Immunol (2003) 15(1):12–9. doi: 10.1016/s0952-7915(02)00005-5
19. Khush RS, Leulier F, Lemaitre B. Drosophila immunity: two paths to NF-kappaB. Trends Immunol (2001) 22(5):260–4. doi: 10.1016/s1471-4906(01)01887-7
20. Wang Z, Luo J, Feng K, Zhou Y, Tang F. Prophenoloxidase of odontotermes formosanus (Shiraki) (Blattodea: termitidae) is a key gene in melanization and has a defensive role during bacterial infection. Int J Mol Sci (2022) 24(1). doi: 10.3390/ijms24010406
21. Prabu S, Jing D, Shabbir MZ, Yuan W, Wang Z, He K. Contribution of phenoloxidase activation mechanism to bt insecticidal protein resistance in Asian corn borer. Int J Biol Macromol (2020) 153:88–99. doi: 10.1016/j.ijbiomac.2020.03.003
22. Wang J, Dou X, Song J, Lyu Y, Zhu X, Xu L, et al. Antimicrobial peptides: promising alternatives in the post feeding antibiotic era. Med Res Rev (2019) 39(3):831–59. doi: 10.1002/med.21542
23. Cerenius L, Lee BL, Soderhall K. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol (2008) 29(6):263–71. doi: 10.1016/j.it.2008.02.009
24. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell (2010) 140(6):805–20. doi: 10.1016/j.cell.2010.01.022
25. Patin EC, Thompson A, Orr SJ. Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol (2019) 89:24–33. doi: 10.1016/j.semcdb.2018.03.003
26. Zhang W, Tettamanti G, Bassal T, Heryanto C, Eleftherianos I, Mohamed A. Regulators and signalling in insect antimicrobial innate immunity: functional molecules and cellular pathways. Cell Signal (2021) 83:110003. doi: 10.1016/j.cellsig.2021.110003
27. Calzas C, Lemire P, Auray G, Gerdts V, Gottschalk M, Segura M. Antibody response specific to the capsular polysaccharide is impaired in streptococcus suis serotype 2-infected animals. Infect Immun (2015) 83(1):441–53. doi: 10.1128/IAI.02427-14
28. Swaminathan CP, Brown PH, Roychowdhury A, Wang Q, Guan R, Silverman N, et al. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs). Proc Natl Acad Sci U.S.A. (2006) 103(3):684–9. doi: 10.1073/pnas.0507656103
29. Jiang H, Ma C, Lu ZQ, Kanost MR. Beta-1,3-glucan recognition protein-2 (betaGRP-2)from manduca sexta; an acute-phase protein that binds beta-1,3-glucan and lipoteichoic acid to aggregate fungi and bacteria and stimulate prophenoloxidase activation. Insect Biochem Mol Biol (2004) 34(1):89–100. doi: 10.1016/j.ibmb.2003.09.006
30. Ariki S, Koori K, Osaki T, Motoyama K, Inamori K, Kawabata S. A serine protease zymogen functions as a pattern-recognition receptor for lipopolysaccharides. Proc Natl Acad Sci U.S.A. (2004) 101(4):953–8. doi: 10.1073/pnas.0306904101
31. Geijtenbeek TB, Van Vliet SJ, Koppel EA, Sanchez-Hernandez M, Vandenbroucke-Grauls CM, Appelmelk B, et al. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med (2003) 197(1):7–17. doi: 10.1084/jem.20021229
32. Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by toll-like receptor 5. Nature (2001) 410(6832):1099–103. doi: 10.1038/35074106
33. Oliveira SC. Innate immune sensing of nucleic acids from pathogens. Microbes Infect (2014) 16(12):977–8. doi: 10.1016/j.micinf.2014.10.003
34. Jentho E, Weis S. DAMPs and innate immune training. Front Immunol (2021) 12:699563. doi: 10.3389/fimmu.2021.699563
35. Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol (2010) 10(12):826–37. doi: 10.1038/nri2873
36. Gong T, Liu L, Jiang W, Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat Rev Immunol (2020) 20(2):95–112. doi: 10.1038/s41577-019-0215-7
37. Roh JS, Sohn DH. Damage-associated molecular patterns in inflammatory diseases. Immune Netw (2018) 18(4):e27. doi: 10.4110/in.2018.18.e27
38. Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol (1994) 12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015
39. Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature (2002) 418(6894):191–5. doi: 10.1038/nature00858
40. Shi Y, Evans JE, Rock KL. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature (2003) 425(6957):516–21. doi: 10.1038/nature01991
41. Walter L. Allograft injury mediated by reactive oxygen species: from conserved proteins of drosophila to acute and chronic rejection of human transplants. part II: role of reactive oxygen species in the induction of the heat shock response as a regulator of innate. Transplant Rev (2003) 17(2):31–44. doi: 10.1053/trre.2003.2
42. Hrithik MTH, Ahmed S, Kim Y. Damage signal induced by bacillus thuringiensis infection triggers immune responses via a DAMP molecule in lepidopteran insect, spodoptera exigua. Dev Comp Immunol (2023) 139:104559. doi: 10.1016/j.dci.2022.104559
43. Jiang L, Liu W, Guo H, Dang Y, Cheng T, Yang W, et al. Distinct functions of bombyx mori peptidoglycan recognition protein 2 in immune responses to bacteria and viruses. Front Immunol (2019) 10:776. doi: 10.3389/fimmu.2019.00776
44. Zhan MY, Yang PJ, Rao XJ. Molecular cloning and analysis of PGRP-L1 and IMD from silkworm bombyx mori. Comp Biochem Physiol B Biochem Mol Biol (2018) 215:19–30. doi: 10.1016/j.cbpb.2017.10.002
45. Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol (2010) 708:181–204. doi: 10.1007/978-1-4419-8059-5_10
46. Wu C. Cloning of the peptidoglycan recognition protein PGRP-S2 from plutella xylostella and analysis of its microbial binding function. Fujian Normal Univ (2018).
47. Sun Q, Xu XX, Freed S, Huang WJ, Zheng Z, Wang S, et al. Molecular characterization of a short peptidoglycan recognition protein (PGRP-s) from Asian corn borer (Ostrinia furnacalis) and its role in triggering proPO activity. World J Microbiol Biotechnol (2014) 30(1):263–70. doi: 10.1007/s11274-013-1449-3
48. Zhang Z, Kong J, De Mandal S, Li S, Zheng Z, Jin F, et al. An immune-responsive PGRP-S1 regulates the expression of antibacterial peptide genes in diamondback moth, plutella xylostella (L.). Int J Biol Macromol (2020) 142:114–24. doi: 10.1016/j.ijbiomac.2019.09.081
49. Mei X, Peng P, Li C, Qiao P, He E, Qiu Z, et al. Peptidoglycan recognition protein 6 (PGRP6) from Asian corn borer, ostrinia furnacalis (Guenee) serve as a pattern recognition receptor in innate immune response. Arch Insect Biochem Physiol (2022) 111(4):e21955. doi: 10.1002/arch.21955
50. Wang X-W, Liu Y, Shen D, Zhou F, Wang G, An C. Identification of immunity-related genes in ostrinia furnacalis against entomopathogenic fungi by RNA-seq analysis. PloS One (2014) 9(1). doi: 10.1371/journal.pone.0086436
51. Yang PJ, Zhan MY, Yang LL, Liu QQ, Xu Y, Pan YM, et al. Characterization of PGRP-S1 from the oriental armyworm, mythimna separata. Dev Comp Immunol (2019) 90:121–9. doi: 10.1016/j.dci.2018.09.009
52. Du SX, Wu C, Su YH, Yang M. Cloning, expression and functional analysis of PGRP-S2 gene in plutella xylostella. J Enviro Entomol (2020) 42(02):410–8. doi: 10.3969/j.issn.1674-0858.2020.02.21
53. Li J, Li J, Jing Z, Yu Q, Zheng G, Zhang B, et al. Antiviral function of peptidoglycan recognition protein in spodoptera exigua (Lepidoptera: noctuidae). Insect Sci (2022). doi: 10.1111/1744-7917.13158
54. Yoshida H, Kinoshita K, Ashida M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, bombyx mori. J Biol Chem (1996) 271(23):13854–60. doi: 10.1074/jbc.271.23.13854
55. He YC, Luo CS, You HJ. Molecular characteristics and bioinformatics analysis of β-1,3-Glucan recognition protein βGRP1 gene of hepialus xiaojinensis larvae. Anhui Agric Sci (2021) 49(19):95–8 + 101. doi: 10.3969/j.issn.0517-6611.2021.19.023
56. Wu T, Zhao Y, Wang Z, Song Q, Wang Z, Xu Q, et al. Beta-1,3-Glucan recognition protein 3 activates the prophenoloxidase system in response to bacterial infection in ostrinia furnacalis guenee. Dev Comp Immunol (2018) 79:31–43. doi: 10.1016/j.dci.2017.10.004
57. Wang J, Zhu LB, Ma Y, Liu YX, Cao HH, Wang YL, et al. Bombyx mori β-1,3-Glucan recognition protein 4 (BmβGRP4) could inhibit the proliferation of b. mori Nucleopolyhedrovirus through Promoting Apoptosis. Insects (2021) 12(8):743. doi: 10.3390/insects12080743
58. Yu J, Xu XX, Gao YF, Jin FL. Molecular cloning of the c-type lectin gene PxCTL5 and its mRNA level changes under bacterial stimulation in plutella xylostella (Lepidoptera: plutellidae). Acta Entomologica (2017) 60(08):876–90. doi: 10.16380/j.kcxb.2017.08.004
59. Geng T, Lv DD, Huang YX, Hou CX, Qin GX, Guo XJ. JAK/STAT signaling pathway-mediated immune response in silkworm (Bombyx mori) challenged by beauveria bassiana. Gene (2016) 595(1):69–76. doi: 10.1016/j.gene.2016.09.043
60. Takase H, Watanabe A, Yoshizawa Y, Kitami M, Sato R. Identification and comparative analysis of three novel c-type lectins from the silkworm with functional implications in pathogen recognition. Dev Comp Immunol (2009) 33(6):789–800. doi: 10.1016/j.dci.2009.01.005
61. Rao XJ, Cao X, He Y, Hu Y, Zhang X, Chen YR, et al. Structural features, evolutionary relationships, and transcriptional regulation of c-type lectin-domain proteins in manduca sexta. Insect Biochem Mol Biol (2015) 62:75–85. doi: 10.1016/j.ibmb.2014.12.006
62. Shen DX, Chu Y, Hong F, Liu Y, Wang GR, An CJ. Molecular cloning and functional analysis of c-type lectin 6 (CTL6) in ostrinia furnacalis (Lepidoptera: crambidae). Acta Entomologica (2015) 58(11):1177–85. doi: 10.16380/j.kcxb.2015.11.004
63. Song ZK, Tian ML, Dong YP, Ren CB, Du Y, Hu J. The c-type lectin IML-10 promotes hemocytic encapsulation by enhancing aggregation of hemocytes in the Asian corn borer ostrinia furnacalis. Insect Biochem Mol Biol (2020) 118:103314. doi: 10.1016/j.ibmb.2020.103314
64. Wang W, Wang G, Zhuo X, Liu Y, Tang L, Liu X, et al. C-type lectin-mediated microbial homeostasis is critical for helicoverpa armigera larval growth and development. PloS Pathog (2020) 16(9):e1008901. doi: 10.1371/journal.ppat.1008901
65. Mei X, Li C, Peng P, Wang J, He E, Qiu Z, et al. Bombyx mori c-type lectin (BmIML-2) inhibits the proliferation of b. mori nucleopolyhedrovirus (BmNPV) through involvement in apoptosis. Int J Mol Sci (2022) 23(15). doi: 10.3390/ijms23158369
66. Zhang K, Hu X, Zhao Y, Pan G, Li C, Ji H, et al. Scavenger receptor B8 improves survivability by mediating innate immunity in silkworm, bombyx mori. Dev Comp Immunol (2021) 116:103917. doi: 10.1016/j.dci.2020.103917
67. Zhang K, Shen L, Wang X, Yang H, Zhang X, Pan G, et al. Scavenger receptor c regulates antimicrobial peptide expression by activating toll signaling in silkworm, bombyx mori. Int J Biol Macromol (2021) 191:396–404. doi: 10.1016/j.ijbiomac.2021.09.084
68. Huang X. Functional study on the silkworm class b scavenger receptor BmSRB3 in cellular lipid transport and immune response. Southwest Univ (2022). doi: 10.27684/d.cnki.gxndx.2022.000266
69. Kang D, Liu G, Lundström A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc Natl Acad Sci U.S.A. (1998) 95(17):10078–82. doi: 10.1073/pnas.95.17.10078
70. Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly drosophila melanogaster. Proc Natl Acad Sci U.S.A. (2000) 97(25):13772–7. doi: 10.1073/pnas.97.25.13772
71. Wang Q, Ren M, Liu X, Xia H, Chen K. Peptidoglycan recognition proteins in insect immunity. Mol Immunol (2019) 106:69–76. doi: 10.1016/j.molimm.2018.12.021
72. Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J Biol Chem (2001) 276(37):34686–94. doi: 10.1074/jbc.M105566200
73. Slonova D, Posvyatenko A, Kibardin A, Sysolyatina E, Lyssuk E, Ermolaeva S, et al. Human short peptidoglycan recognition protein PGLYRP1/Tag-7/PGRP-S inhibits listeria monocytogenes intracellular survival in macrophages. Front Cell Infect Microbiol (2020) 10:582803. doi: 10.3389/fcimb.2020.582803
74. Dziarski R, Gupta D. Review: mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun (2010) 16(3):168–74. doi: 10.1177/1753425910366059
75. Wang ZM, Li X, Cocklin RR, Wang M, Wang M, Fukase K, et al. Human peptidoglycan recognition protein-l is an n-acetylmuramoyl-L-alanine amidase. J Biol Chem (2003) 278(49):49044–52. doi: 10.1074/jbc.M307758200
76. Huang L, Chen SN, Gan Z, Nie P. Molecular and functional identification of a short-type peptidoglycan recognition protein, PGRP-s, in the Chinese soft-shelled turtle pelodiscus sinensis. Dev Comp Immunol (2021) 117:103965. doi: 10.1016/j.dci.2020.103965
77. Hou J, Gan Z, Chen S, Cheng J, Lu Y, Huang B, et al. Identification and functional characterization of a short-type peptidoglycan recognition protein, PGRP-s in the orange-spotted grouper, epinephelus coioides. Aquaculture Rep (2021) 20. doi: 10.1016/j.aqrep.2021.100739
78. Hou J, Hao W, Chang Li M, Gan Z, Chen SN, Lu YS, et al. Identification and characterization of two long-type peptidoglycan recognition proteins, PGRP-L1 and PGRP-L2, in the orange-spotted grouper, epinephelus coioides. Fish Shellfish Immunol (2023) 134:108580. doi: 10.1016/j.fsi.2023.108580
79. Hu Z, Cao X, Guo M, Li C. Identification and characterization of a novel short-type peptidoglycan recognition protein in apostichopus japonicus. Fish Shellfish Immunol (2020) 99:257–66. doi: 10.1016/j.fsi.2020.02.013
80. Yang C, Wang L, Jia Z, Yi Q, Xu Q, Wang W, et al. Two short peptidoglycan recognition proteins from crassostrea gigas with similar structure exhibited different PAMP binding activity. Dev Comp Immunol (2017) 70:9–18. doi: 10.1016/j.dci.2016.12.009
81. Valanne S, Wang JH, Ramet M. The drosophila toll signaling pathway. J Immunol (2011) 186(2):649–56. doi: 10.4049/jimmunol.1002302
82. Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila toll is activated by gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature (2001) 414(6865):756–9. doi: 10.1038/414756a
83. Monahan A, Kleino A, Silverman N. ReaDAPting the role of PGRP-SD in bacterial sensing and immune activation. Immunity (2016) 45(5):951–3. doi: 10.1016/j.immuni.2016.11.002
84. Iatsenko I, Kondo S, Mengin-Lecreulx D, Lemaitre B. PGRP-SD, an extracellular pattern-recognition receptor, enhances peptidoglycan-mediated activation of the drosophila imd pathway. Immunity (2016) 45(5):1013–23. doi: 10.1016/j.immuni.2016.10.029
85. Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in relish activation and antibacterial immune responses in drosophila. Science (2002) 296(5566):359–62. doi: 10.1126/science.1070216
86. Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, et al. The drosophila immune response against gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature (2002) 416(6881):640–4. doi: 10.1038/nature734
87. Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a drosophila receptor for e. coli. Nat (2002) 416(6881):644–8. doi: 10.1038/nature735
88. Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in drosophila larvae. Proc Natl Acad Sci U.S.A. (2002) 99(21):13705–10. doi: 10.1073/pnas.212301199
89. Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in drosophila immunity. EMBO J (2004) 23(23):4690–700. doi: 10.1038/sj.emboj.7600466
90. Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol (2008) 9(8):908–16. doi: 10.1038/ni.1634
91. Liang Y, Yang L, Wang Y, Tang T, Liu F, Zhang F. Peptidoglycan recognition protein SC (PGRP-SC) shapes gut microbiota richness, diversity and composition by modulating immunity in the house fly musca domestica. Insect Mol Biol (2023) 32(2):200–12. doi: 10.1111/imb.12824
92. Chen K, Zhou L, Chen F, Peng Y, Lu Z. Peptidoglycan recognition protein-S5 functions as a negative regulator of the antimicrobial peptide pathway in the silkworm, bombyx mori. Dev Comp Immunol (2016) 61:126–35. doi: 10.1016/j.dci.2016.03.023
93. Yang PJ, Zhan MY, Ye C, Yu XQ, Rao XJ. Molecular cloning and characterization of a short peptidoglycan recognition protein from silkworm bombyx mori. Insect Mol Biol (2017) 26(6):665–76. doi: 10.1111/imb.12330
94. Chen K, Liu C, He Y, Jiang H, Lu Z. A short-type peptidoglycan recognition protein from the silkworm: expression, characterization and involvement in the prophenoloxidase activation pathway. Dev Comp Immunol (2014) 45(1):1–9. doi: 10.1016/j.dci.2014.01.017
95. Zhao S, Wang X, Cai S, Zhang S, Luo H, Wu C, et al. A novel peptidoglycan recognition protein involved in the prophenoloxidase activation system and antimicrobial peptide production in antheraea pernyi. Dev Comp Immunol (2018) 86:78–85. doi: 10.1016/j.dci.2018.04.009
96. Shen D, Mei X, Guo J, Tong M, Xia D, Qiu Z, et al. Peptidoglycan recognition protein-S1 (PGRP-S1) from diaphania pyloalis (Walker) is involved in the agglutination and prophenoloxidase activation pathway. Gene (2022) 809:146004. doi: 10.1016/j.gene.2021.146004
97. Kumar D, Sun Z, Cao G, Xue R, Hu X, Gong C. Bombyx mori bidensovirus infection alters the intestinal microflora of fifth instar silkworm (Bombyx mori) larvae. J Invertebr Pathol (2019) 163:48–63. doi: 10.1016/j.jip.2019.03.004
98. Zhao P, Xia F, Jiang L, Guo H, Xu G, Sun Q, et al. Enhanced antiviral immunity against bombyx mori cytoplasmic polyhedrosis virus via overexpression of peptidoglycan recognition protein S2 in transgenic silkworms. Dev Comp Immunol (2018) 87:84–9. doi: 10.1016/j.dci.2018.05.021
99. Rao XJ, Zhong X, Lin XY, Huang XH, Yu XQ. Characterization of a novel manduca sexta beta-1, 3-glucan recognition protein (betaGRP3) with multiple functions. Insect Biochem Mol Biol (2014) 52:13–22. doi: 10.1016/j.ibmb.2014.06.003
100. Brown GD, Gordon S. Immune recognition of fungal beta-glucans. Cell Microbiol (2005) 7(4):471–9. doi: 10.1111/j.1462-5822.2005.00505.x
101. Huang Y, Ren Q. Research progress in innate immunity of freshwater crustaceans. Dev Comp Immunol (2020) 104:103569. doi: 10.1016/j.dci.2019.103569
102. Chai LQ, Meng JH, Gao J, Xu YH, Wang XW. Identification of a crustacean beta-1,3-glucanase related protein as a pattern recognition protein in antibacterial response. Fish Shellfish Immunol (2018) 80:155–64. doi: 10.1016/j.fsi.2018.06.004
103. Ochiai M, Ashida M. Purification of a beta-1,3-glucan recognition protein in the prophenoloxidase activating system from hemolymph of the silkworm, bombyx mori. J Biol Chem (1988) 263(24):12056–62. doi: 10.1016/s0021-9258(18)37892-x
104. Rao XJ, Zhan MY, Pan YM, Liu S, Yang PJ, Yang LL, et al. Immune functions of insect betaGRPs and their potential application. Dev Comp Immunol (2018) 83:80–8. doi: 10.1016/j.dci.2017.12.007
105. Meng Q, Wu PP, Li MM, Shu RH, Zhou GL, Zhang JH, et al. Distinct responses of thitarodes xiaojinensis beta-1,3-Glucan recognition protein-1 and immulectin-8 to ophiocordyceps sinensis and cordyceps militaris infection. J Immunol (2021) 207(1):200–9. doi: 10.4049/jimmunol.2000447
106. Lee WJ, Lee JD, Kravchenko VV, Ulevitch RJ, Brey PT. Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm, bombyx mori. Proc Natl Acad Sci U.S.A. (1996) 93(15):7888–93. doi: 10.1073/pnas.93.15.7888
107. Hughes AL. Evolution of the betaGRP/GNBP/beta-1,3-glucanase family of insects. Immunogenetics (2012) 64(7):549–58. doi: 10.1007/s00251-012-0610-8
108. Filipe SR, Tomasz A, Ligoxygakis P. Requirements of peptidoglycan structure that allow detection by the drosophila toll pathway. EMBO Rep (2005) 6(4):327–33. doi: 10.1038/sj.embor.7400371
109. Wang L, Weber AN, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. Sensing of gram-positive bacteria in drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J (2006) 25(20):5005–14. doi: 10.1038/sj.emboj.7601363
110. Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, et al. Dual activation of the drosophila toll pathway by two pattern recognition receptors. Science (2003) 302(5653):2126–30. doi: 10.1126/science.1085432
111. Pili-Floury S, Leulier F, Takahashi K, Saigo K, Samain E, Ueda R, et al. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against gram-positive bacterial infection in drosophila adults. J Biol Chem (2004) 279(13):12848–53. doi: 10.1074/jbc.M313324200
112. Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, et al. Dual detection of fungal infections in drosophila via recognition of glucans and sensing of virulence factors. Cell (2006) 127(7):1425–37. doi: 10.1016/j.cell.2006.10.046
113. Matskevich AA, Quintin J, Ferrandon D. The drosophila PRR GNBP3 assembles effector complexes involved in antifungal defenses independently of its toll-pathway activation function. Eur J Immunol (2010) 40(5):1244–54. doi: 10.1002/eji.200940164
114. Zhang Z, Zhu S, De Mandal S, Gao Y, Yu J, Zeng L, et al. Combined transcriptomic and proteomic analysis of developmental features in the immune system of plutella xylostella during larva-to-adult metamorphosis. Genomics (2022) 114(4):110381. doi: 10.1016/j.ygeno.2022.110381
115. Roy MC, Kim Y. Toll signal pathway activating eicosanoid biosynthesis shares its conserved upstream recognition components in a lepidopteran spodoptera exigua upon infection by metarhizium rileyi, an entomopathogenic fungus. J Invertebr Pathol (2022) 188:107707. doi: 10.1016/j.jip.2021.107707
116. Li S, Hao Z, Xu H, Gao Y, Zhang M, Liang J, et al. Silencing beta-1,3-glucan binding protein enhances the susceptibility of plutella xylostella to entomopathogenic fungus isaria cicadae. Pest Manag Sci (2022) 78(7):3117–27. doi: 10.1002/ps.6938
117. Alenton RR, Koiwai K, Miyaguchi K, Kondo H, Hirono I. Pathogen recognition of a novel c-type lectin from marsupenaeus japonicus reveals the divergent sugar-binding specificity of QAP motif. Sci Rep (2017) 7:45818. doi: 10.1038/srep45818
118. Marakalala MJ, Ndlovu H. Signaling c-type lectin receptors in antimycobacterial immunity. PloS Pathog (2017) 13(6):e1006333. doi: 10.1371/journal.ppat.1006333
119. Ishikawa E, Mori D, Yamasaki S. Recognition of mycobacterial lipids by immune receptors. Trends Immunol (2017) 38(1):66–76. doi: 10.1016/j.it.2016.10.009
120. Wang Y, Spatz M, Da Costa G, Michaudel C, Lapiere A, Danne C, et al. Deletion of both dectin-1 and dectin-2 affects the bacterial but not fungal gut microbiota and susceptibility to colitis in mice. Microbiome (2022) 10(1):91. doi: 10.1186/s40168-022-01273-4
121. Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol (2007) 8(1):31–8. doi: 10.1038/ni1408
122. Tanaka M, Saka-Tanaka M, Ochi K, Fujieda K, Sugiura Y, Miyamoto T, et al. C-type lectin mincle mediates cell death-triggered inflammation in acute kidney injury. J Exp Med (2020) 217(11). doi: 10.1084/jem.20192230
123. Li X, Na H, Xu L, Zhang X, Feng Z, Zhou X, et al. DC-SIGN mediates gastric cancer progression by regulating the JAK2/STAT3 signaling pathway and affecting LncRNA RP11-181G12.2 expression. BioMed Pharmacother (2020) 121:109644. doi: 10.1016/j.biopha.2019.109644
124. Kimura Y, Inoue A, Hangai S, Saijo S, Negishi H, Nishio J, et al. The innate immune receptor dectin-2 mediates the phagocytosis of cancer cells by kupffer cells for the suppression of liver metastasis. Proc Natl Acad Sci U.S.A. (2016) 113(49):14097–102. doi: 10.1073/pnas.1617903113
125. Liu H, Huang X, Xiong H, Liu M, Hu D, Wei C, et al. Co-Expression of surfactant protein a and chicken lung lectin in chicken respiratory system. Mol Immunol (2020) 122:49–53. doi: 10.1016/j.molimm.2020.03.018
126. Wu H, Xiong H, Huang X, Zhou Q, Hu D, Qi K, et al. Lung infection of avian pathogenic escherichia coli co-upregulates the expression of cSP-a and cLL in chickens. Res Vet Sci (2022) 152:99–106. doi: 10.1016/j.rvsc.2022.07.023
127. Shang-Guan XY, Cai YJ, Xu HZ, Cheng X, Zhang RF, Liu HX. A c-type lectin with a single CRD from onychostoma macrolepis mediates immune recognition against bacterial challenge. Fish Shellfish Immunol (2021) 115:160–70. doi: 10.1016/j.fsi.2021.06.007
128. Xue D, Guang-Hua W, Yan-Li S, Min Z, Yong-Hua H. Black rockfish c-type lectin, SsCTL4: a pattern recognition receptor that promotes bactericidal activity and virus escape from host immune defense. Fish Shellfish Immunol (2018) 79:340–50. doi: 10.1016/j.fsi.2018.05.033
129. Zhang M, Hu YH, Sun L. Identification and molecular analysis of a novel c-type lectin from scophthalmus maximus. Fish Shellfish Immunol (2010) 29(1):82–8. doi: 10.1016/j.fsi.2010.02.023
130. Shen D, Tong M, Guo J, Mei X, Xia D, Qiu Z, et al. A pattern recognition receptor c-type lectin-S6 (CTL-S6) is involved in the immune response in the silkworm (Lepidoptera: bombycidae). J Insect Sci (2021) 21(1). doi: 10.1093/jisesa/ieaa146
131. Lin Z, Wang JL, Cheng Y, Wang JX, Zou Z. Pattern recognition receptors from lepidopteran insects and their biological functions. Dev Comp Immunol (2020) 108:103688. doi: 10.1016/j.dci.2020.103688
132. Robinson MJ, Sancho D, Slack EC, LeibundGut-Landmann S, Reis e Sousa C. Myeloid c-type lectins in innate immunity. Nat Immunol (2006) 7(12):1258–65. doi: 10.1038/ni1417
133. Sivakamavalli J, Park K, Kwak IS, Vaseeharan B. Purification and partial characterization of carbohydrate-recognition protein c-type lectin from hemifusus pugilinus. Carbohydr Res (2021) 499:108224. doi: 10.1016/j.carres.2020.108224
134. Lu Y, Zhang H, Cheng D, Liu H, Li S, Ma H, et al. A multi-CRD c-type lectin gene cnlec-1 enhance the immunity response in noble scallop chlamys nobilis with higher carotenoids contents through up-regulating under different immunostimulants. Fish Shellfish Immunol (2018) 83:37–44. doi: 10.1016/j.fsi.2018.09.014
135. Li Y, Pan L, Yu J. The injection of one recombinant c-type lectin (LvLec) induced the immune response of hemocytes in litopenaeus vannamei. Fish Shellfish Immunol (2022) 124:324–31. doi: 10.1016/j.fsi.2022.04.017
136. Gasmi L, Ferre J, Herrero S. High bacterial agglutination activity in a single-CRD c-type lectin from spodoptera exigua (Lepidoptera: noctuidae). Biosensors (Basel) (2017) 7(1). doi: 10.3390/bios7010012
137. Wang GJ, Wang WW, Liu Y, Chai LQ, Wang GX, Liu XS, et al. Steroid hormone 20-hydroxyecdysone promotes CTL1-mediated cellular immunity in helicoverpa armigera. Insect Sci (2021) 28(5):1399–413. doi: 10.1111/1744-7917.12851
138. Cheng Y, Lin Z, Wang JM, Xing LS, Xiong GH, Zou Z. CTL14, a recognition receptor induced in late stage larvae, modulates anti-fungal immunity in cotton bollworm helicoverpa armigera. Dev Comp Immunol (2018) 84:142–52. doi: 10.1016/j.dci.2018.02.010
139. Geng T, Lu F, Wu H, Wang Y, Lou D, Tu N, et al. C-type lectin 5, a novel pattern recognition receptor for the JAK/STAT signaling pathway in bombyx mori. J Invertebr Pathol (2021) 179:107473. doi: 10.1016/j.jip.2020.107473
140. Liu FF, Liu Z, Li H, Zhang WT, Wang Q, Zhang BX, et al. CTL10 has multiple functions in the innate immune responses of the silkworm, bombyx mori. Dev Comp Immunol (2022) 127:104309. doi: 10.1016/j.dci.2021.104309
141. Chai LQ, Tian YY, Yang DT, Wang JX, Zhao XF. Molecular cloning and characterization of a c-type lectin from the cotton bollworm, helicoverpa armigera. Dev Comp Immunol (2008) 32(1):71–83. doi: 10.1016/j.dci.2007.04.006
142. Gasmi L, Boulain H, Gauthier J, Hua-Van A, Musset K, Jakubowska AK, et al. Recurrent domestication by Lepidoptera of genes from their parasites mediated by bracoviruses. PloS Genet (2015) 11(9):e1005470. doi: 10.1371/journal.pgen.1005470
143. Gasmi L, Jakubowska AK, Ferre J, Ogliastro M, Herrero S. Characterization of two groups of spodoptera exigua hubner (Lepidoptera: noctuidae) c-type lectins and insights into their role in defense against the densovirus JcDV. Arch Insect Biochem Physiol (2018) 97(1). doi: 10.1002/arch.21432
144. Prabhudas M, Bowdish D, Drickamer K, Febbraio M, Herz J, Kobzik L, et al. Standardizing scavenger receptor nomenclature. J Immunol (2014) 192(5):1997–2006. doi: 10.4049/jimmunol.1490003
145. Guillou A, Troha K, Wang H, Franc NC, Buchon N. The drosophila CD36 homologue croquemort is required to maintain immune and gut homeostasis during development and aging. PloS Pathog (2016) 12(10):e1005961. doi: 10.1371/journal.ppat.1005961
146. Zanoni P, Khetarpal SA, Larach DB, Hancock-Cerutti WF, Millar JS, Cuchel M, et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science (2016) 351(6278):1166–71. doi: 10.1126/science.aad3517
147. Feng H, Guo L, Wang D, Gao HQ, Hou GH, Zheng Z, et al. Deficiency of scavenger receptor BI leads to impaired lymphocyte homeostasis and autoimmune disorders in mice. Arterioscler Thromb Vasc Biol (2011) 31(11):2543–51. doi: 10.1161/ATVBAHA.111.234716
148. Toomey MB, Lopes RJ, Araujo PM, Johnson JD, Gazda MA, Afonso S, et al. High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc Natl Acad Sci U.S.A. (2017) 114(20):5219–24. doi: 10.1073/pnas.1700751114
149. Li C, Ge X, Su B, Fu Q, Wang B, Liu X, et al. Characterization of class b scavenger receptor type 1 (SRB1) in turbot (Scophthalmus maximus l.). Fish Shellfish Immunol (2020) 100:358–67. doi: 10.1016/j.fsi.2020.03.014
150. Yang D, Han Y, Chen L, Liu Y, Cao R, Wang Q, et al. Scavenger receptor class b type I (SR-BI) in ruditapes philippinarum: a versatile receptor with multiple functions. Fish Shellfish Immunol (2019) 88:328–34. doi: 10.1016/j.fsi.2019.03.009
151. Tang M, Li X, Yang L, Wang Q, Li W. A class b scavenger receptor mediates antimicrobial peptide secretion and phagocytosis in Chinese mitten crab (Eriocheir sinensis). Dev Comp Immunol (2020) 103:103496. doi: 10.1016/j.dci.2019.103496
152. Che Z, Shao Y, Zhang W, Zhao X, Guo M, Li C. Cloning and functional analysis of scavenger receptor b gene from the sea cucumber apostichopus japonicus. Dev Comp Immunol (2019) 99:103404. doi: 10.1016/j.dci.2019.103404
153. Zhou LZ, Wang RJ, Yan YY, Zeng S, Zou Z, Lu Z. Scavenger receptor B1 mediates phagocytosis and the antimicrobial peptide pathway in the endoparasitic wasp micropilits mediator. Dev Comp Immunol (2021) 119:104039. doi: 10.1016/j.dci.2021.104039
154. Alquraini A, El Khoury J. Scavenger receptors. Curr Biol (2020) 30(14):R790–R5. doi: 10.1016/j.cub.2020.05.051
155. Yang MC, Yang HT, Li J, Sun JJ, Bi WJ, Niu GJ, et al. Scavenger receptor c promotes bacterial clearance in kuruma shrimp marsupenaeus japonicus by enhancing hemocyte phagocytosis and AMP expression. Fish Shellfish Immunol (2017) 67:254–62. doi: 10.1016/j.fsi.2017.06.003
156. Rämet M, Pearson A, Manfruelli P, Li X KH, Göbel V, Chung E, et al. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity (2001) 15(6):1027–38. doi: 10.1016/s1074-7613(01)00249-7
157. Kim SG, Jo YH, Seong JH, Park KB, Noh MY, Cho JH, et al. TmSR-c, scavenger receptor class c, plays a pivotal role in antifungal and antibacterial immunity in the coleopteran insect tenebrio molitor. Insect Biochem Mol Biol (2017) 89:31–42. doi: 10.1016/j.ibmb.2017.08.007
158. Vasta GR. Galectins as pattern recognition receptors: structure, function, and evolution. Adv Exp Med Biol (2012) 946:21–36. doi: 10.1007/978-1-4614-0106-3_2
159. Rao XJ, Wu P, Shahzad T, Liu S, Chen L, Yang YF, et al. Characterization of a dual-CRD galectin in the silkworm bombyx mori. Dev Comp Immunol (2016) 60:149–59. doi: 10.1016/j.dci.2016.03.001
160. Shishido SN, Varahan S, Yuan K, Li X, Fleming SD. Humoral innate immune response and disease. Clin Immunol (2012) 144(2):142–58. doi: 10.1016/j.clim.2012.06.002
161. Dodds AW, Law SK. The phylogeny and evolution of the thioester bond-containing proteins C3, C4 and alpha 2-macroglobulin. Immunol Rev (1998) 166:15–26. doi: 10.1111/j.1600-065x.1998.tb01249.x
162. Xiao X, Liu Y, Zhang X, Wang J, Li Z, Pang X, et al. Complement-related proteins control the flavivirus infection of aedes aegypti by inducing antimicrobial peptides. PloS Pathog (2014) 10(4):e1004027. doi: 10.1371/journal.ppat.1004027
163. Ng TH, Kurtz J. Dscam in immunity: a question of diversity in insects and crustaceans. Dev Comp Immunol (2020) 105:103539. doi: 10.1016/j.dci.2019.103539
164. Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the anopheles gambiae innate immune system. PloS Biol (2006) 4(7):e229. doi: 10.1371/journal.pbio.0040229
165. Venereau E, Ceriotti C, Bianchi ME. DAMPs from cell death to new life. Front Immunol (2015) 6:422. doi: 10.3389/fimmu.2015.00422
167. Tsung A, Klune JR, Zhang X, Jeyabalan G, Cao Z, Peng X, et al. HMGB1 release induced by liver ischemia involves toll-like receptor 4–dependent reactive oxygen species production and calcium-mediated signaling. J Exp Med (2007) 204(12):2913–23. doi: 10.1084/jem.20070247
168. PrabhuDas MR, Baldwin CL, Bollyky PL, Bowdish DME, Drickamer K, Febbraio M, et al. A consensus definitive classification of scavenger receptors and their roles in health and disease. J Immunol (2017) 198(10):3775–89. doi: 10.4049/jimmunol.1700373
169. Mosrin-Huaman C, Canaple L, Locker D. DSP1 gene of drosophila melanogaster encodes an HMG-domain protein that plays multiple roles in development. Dev Genet (1998) 23(4):324–34. doi: 10.1002/(SICI)1520-6408(1998)23:4<324::AID-DVG7>3.0.CO;2-T
170. Decoville M, Giacomello E, Leng M, Locker D. DSP1, an HMG-like protein, is involved in the regulation of homeotic genes. Genetics (2001) 157(1):237–44. doi: 10.1093/genetics/157.1.237
171. Mollah MMI, Ahmed S, Kim Y. Immune mediation of HMG-like DSP1 via toll-spatzle pathway and its specific inhibition by salicylic acid analogs. PloS Pathog (2021) 17(3):e1009467. doi: 10.1371/journal.ppat.1009467
172. Roy MC, Ahmed S, Kim Y. Dorsal switch protein 1 as a damage signal in insect gut immunity to activate dual oxidase via an eicosanoid, PGE(2). Front Immunol (2022) 13:994626. doi: 10.3389/fimmu.2022.994626
173. Ahmed S, Sajjadian SM, Kim Y. HMGB1-like dorsal switch protein 1 triggers a damage signal in mosquito gut to activate dual oxidase via eicosanoids. J Innate Immun (2022) 14(6):657–72. doi: 10.1159/000524561
174. Ahrens S, Zelenay S, Sancho D, Hanc P, Kjaer S, Feest C, et al. F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity (2012) 36(4):635–45. doi: 10.1016/j.immuni.2012.03.008
175. Hanc P, Fujii T, Iborra S, Yamada Y, Huotari J, Schulz O, et al. Structure of the complex of f-actin and DNGR-1, a c-type lectin receptor involved in dendritic cell cross-presentation of dead cell-associated antigens. Immunity (2015) 42(5):839–49. doi: 10.1016/j.immuni.2015.04.009
176. Zhang JG, Czabotar PE, Policheni AN, Caminschi I, Wan SS, Kitsoulis S, et al. The dendritic cell receptor Clec9A binds damaged cells via exposed actin filaments. Immunity (2012) 36(4):646–57. doi: 10.1016/j.immuni.2012.03.009
177. Gordon O, Henry CM, Srinivasan N, Ahrens S, Franz A, Deddouche S, et al. α-actinin accounts for the bioactivity of actin preparations in inducing STAT target genes in drosophila melanogaster. Elife (2018) 7:e38636. doi: 10.7554/eLife.38636
178. Wang T, Xu L, Zhu B, Wang J, Zheng X. Immune escape mechanisms of severe fever with thrombocytopenia syndrome virus. Front Immunol (2022) 13:937684. doi: 10.3389/fimmu.2022.937684
179. Wu C, Li M, Meng H, Liu Y, Niu W, Zhou Y, et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci (2019) 62(5):640–7. doi: 10.1007/s11427-018-9461-5
180. Wang J, Ge J, Wang Y, Xiong F, Guo J, Jiang X, et al. EBV miRNAs BART11 and BART17-3p promote immune escape through the enhancer-mediated transcription of PD-L1. Nat Commun (2022) 13(1):866. doi: 10.1038/s41467-022-28479-2
181. Luo X, Donnelly CR, Gong W, Heath BR, Hao Y, Donnelly LA, et al. HPV16 drives cancer immune escape via NLRX1-mediated degradation of STING. J Clin Invest (2020) 130(4):1635–52. doi: 10.1172/JCI129497
182. Bates JP, Derakhshandeh R, Jones L, Webb TJ. Mechanisms of immune evasion in breast cancer. BMC Cancer (2018) 18(1):556. doi: 10.1186/s12885-018-4441-3
183. Luksza M, Sethna ZM, Rojas LA, Lihm J, Bravi B, Elhanati Y, et al. Neoantigen quality predicts immunoediting in survivors of pancreatic cancer. Nature (2022) 606(7913):389–95. doi: 10.1038/s41586-022-04735-9
184. Cheng G, Cox J, Wang P, Krishnan MN, Dai J, Qian F, et al. A c-type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes. Cell (2010) 142(5):714–25. doi: 10.1016/j.cell.2010.07.038
185. Liu K, Qian Y, Jung YS, Zhou B, Cao R, Shen T, et al. mosGCTL-7, a c-type lectin protein, mediates Japanese encephalitis virus infection in mosquitoes. J Virol (2017) 91(10):e01348-16. doi: 10.1128/JVI.01348-16
186. Alcalá AC, Maravillas JL, Meza D, Ramirez OT, Ludert JE, Palomares LA. Dengue virus NS1 uses scavenger receptor B1 as a cell receptor in cultured cells. J Virol (2022) 96(5):e0166421. doi: 10.1128/JVI.01664-21
187. Caicedo PA, Serrato IM, Sim S, Dimopoulos G, Coatsworth H, Lowenberger C, et al. Immune response-related genes associated to blocking midgut dengue virus infection in aedes aegypti strains that differ in susceptibility. Insect Sci (2019) 26(4):635–48. doi: 10.1111/1744-7917.12573
188. Li Y, Li Y, Cao X, Jin X, Jin T. Pattern recognition receptors in zebrafish provide functional and evolutionary insight into innate immune signaling pathways. Cell Mol Immunol (2017) 14(1):80–9. doi: 10.1038/cmi.2016.50
189. Dziarski R, Royet J, Gupta D. Peptidoglycan recognition proteins and lysozyme. Encyclopedia Immunobiology. (2016) . p:389–403. doi: 10.1016/B978-0-12-374279-7.02022-1
190. Wang M, Liu LH, Wang S, Li X, Lu X, Gupta D, et al. Human peptidoglycan recognition proteins require zinc to kill both gram-positive and gram-negative bacteria and are synergistic with antibacterial peptides. J Immunol (2007) 178(5):3116–25. doi: 10.4049/jimmunol.178.5.3116
191. Dziarski R, Gupta D. How innate immunity proteins kill bacteria and why they are not prone to resistance. Curr Genet (2018) 64(1):125–9. doi: 10.1007/s00294-017-0737-0
192. Zhou J, Valentini E, Boutros M. Microenvironmental innate immune signaling and cell mechanical responses promote tumor growth. Dev Cell (2021) 56(13):1884–99. doi: 10.1016/j.devcel.2021.06.007
193. Li X, Wang S, Qi J, Echtenkamp SF, Chatterjee R, Wang M, et al. Zebrafish peptidoglycan recognition proteins are bactericidal amidases essential for defense against bacterial infections. Immunity (2007) 27(3):518–29. doi: 10.1016/j.immuni.2007.07.020
194. Arroyo R, Echaide M, Wilmanowski R, Martin-Gonzalez A, Batllori E, Galindo A, et al. Structure and activity of human surfactant protein d from different natural sources. Am J Physiol Lung Cell Mol Physiol (2020) 319(1):L148–L58. doi: 10.1152/ajplung.00007.2020
195. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by toll-like receptor 3. Nature (2001) 413(6857):732–8. doi: 10.1038/35099560
196. Alesci A, Fumia A, Miller A, Calabrò C, Santini A, Cicero N, et al. Spirulina promotes macrophages aggregation in zebrafish (Danio rerio) liver. Nat Prod Res (2023) 37(5):743–9. doi: 10.1080/14786419.2022.2089883
197. Zhang J, Kong X, Zhou C, Li L, Nie G, Li X. Toll-like receptor recognition of bacteria in fish: ligand specificity and signal pathways. Fish Shellfish Immunol (2014) 41(2):380–8. doi: 10.1016/j.fsi.2014.09.022
198. Wang X, Smith C, Yin H. Targeting toll-like receptors with small molecule agents. Chem Soc Rev (2013) 42(12):4859–66. doi: 10.1039/c3cs60039d
199. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A toll-like receptor recognizes bacterial DNA. Nature (2000) 408(6813):740–5. doi: 10.1038/35047123
200. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science (2004) 303(5663):1529–31. doi: 10.1126/science.1093616
201. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science (2004) 303(5663):1526–9. doi: 10.1126/science.1093620
202. Ohto U, Shibata T, Tanji H, Ishida H, Krayukhina E, Uchiyama S, et al. Structural basis of CpG and inhibitory DNA recognition by toll-like receptor 9. Nature (2015) 520(7549):702–5. doi: 10.1038/nature14138
203. Gao K, Deng XY, Qian HY, Qin GX, Hou CX, Guo XJ. Cloning and expression analysis of a peptidoglycan recognition protein in silkworm related to virus infection. Gene (2014) 552(1):24–31. doi: 10.1016/j.gene.2014.09.008
204. Staczek S, Zdybicka-Barabas A, Pleszczynska M, Wiater A, Cytrynska M. Aspergillus niger alpha-1,3-glucan acts as a virulence factor by inhibiting the insect phenoloxidase system. J Invertebr Pathol (2020) 171:107341. doi: 10.1016/j.jip.2020.107341
205. Staczek S, Zdybicka-Barabas A, Wiater A, Pleszczynska M, Cytrynska M. Activation of cellular immune response in insect model host galleria mellonella by fungal alpha-1,3-glucan. Pathog Dis (2020) 78(9):ftaa062. doi: 10.1093/femspd/ftaa062
Keywords: pattern recognition receptor, Lepidoptera, pathogen-associated molecular pattern, damage-associated molecular pattern, immune escape
Citation: Zhao L, Niu J, Feng D, Wang X and Zhang R (2023) Immune functions of pattern recognition receptors in Lepidoptera. Front. Immunol. 14:1203061. doi: 10.3389/fimmu.2023.1203061
Received: 10 April 2023; Accepted: 05 June 2023;
Published: 16 June 2023.
Edited by:
Qiuning Liu, Yancheng Teachers University, ChinaReviewed by:
Muhammad Nadeem Abbas, Southwest University, ChinaErjun Ling, Shanghai Institutes for Biological Sciences (CAS), China
Copyright © 2023 Zhao, Niu, Feng, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Zhang, emhhbmdyb25nX3NwdUAxNjMuY29t; Xialu Wang, d2FuZ3hpYWx1MTk4NUAxNjMuY29t
 Lin Zhao
Lin Zhao Jinlan Niu
Jinlan Niu Disong Feng
Disong Feng Xialu Wang
Xialu Wang Rong Zhang
Rong Zhang