
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 03 November 2023
Sec. Autoimmune and Autoinflammatory Disorders : Autoimmune Disorders
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1199896
 Weidong Xie1
Weidong Xie1 Haojie Jiang1
Haojie Jiang1 Yao Chen2
Yao Chen2 Huanhao Zhang3
Huanhao Zhang3 Yaoyu Song3
Yaoyu Song3 Zhaojie Yu3
Zhaojie Yu3 Huayan Gu4
Huayan Gu4 Hongkai Xu1
Hongkai Xu1 Saiyi Han5
Saiyi Han5 Sen Li6
Sen Li6 Naxin Liu1*
Naxin Liu1* Shaoliang Han1*
Shaoliang Han1*Background: Previous studies have shown a coexistence phenomenon between systemic lupus erythematosus (SLE) and inflammatory bowel disease (IBD), but the causal relationship between them is still unclear. Therefore, we conducted a two-sample Mendelian randomization (MR) analysis using publicly available summary statistics data to evaluate whether there was a causal relationship between the two diseases.
Methods: Summary statistics for SLE and IBD were downloaded from the Open Genome-Wide Association Study and the International Inflammatory Bowel Disease Genetics Consortium. European and East Asian populations were included in this MR work. We adopted a series of methods to select instrumental variables that are closely related to SLE and IBD. To make the conclusion more reliable, we applied a variety of different analysis methods, among which the inverse variance–weighted (IVW) method was the main method. In addition, heterogeneity, pleiotropy, and sensitivity were assessed to make the conclusions more convincing.
Results: In the European population, a negative causal relationship was observed between SLE and overall IBD (OR = 0.94; 95% CI = 0.90, 0.98; P < 0.004) and ulcerative colitis (UC) (OR = 0.93; 95% CI = 0.88, 0.98; P = 0.006). After removing outliers with Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO), the results remained consistent with IVW. However, there was no causal relationship between SLE and Crohn’s disease. In the East Asian population, no causal relationship was found between SLE and IBD.
Conclusion: Our results found that genetic susceptibility to SLE was associated with lower overall IBD risk and UC risk in European populations. In contrast, no association between SLE and IBD was found in East Asian populations. This work might enrich the previous research results, and it may provide some references for research in the future.
Inflammatory bowel disease (IBD) is a chronic idiopathic gastrointestinal disorder that includes Crohn’s disease (CD) and ulcerative colitis (UC) (1). Although a great deal of research has been done on IBD over a long period of time, the relationship between genetic susceptibility, environmental and other factors, and IBD is still controversial (2). Although IBD was previously considered a Western disease, the incidence of the disease has increased rapidly in Asia and other parts of the world in recent years (3). Over the past decade, IBD has become a huge burden on global public health (4). As a result, attempts have been made to clarify the risk and protective factors of IBD to better understand and prevent the disease.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that can involve multiple organs and tissues throughout the body, such as the skin and kidneys (5). The pathogenesis of SLE is complex, and the interplay of genetic and environmental factors is pointed out to be the cause of SLE (6). Although SLE and IBD seem to be two unrelated diseases, it has been found that some patients with SLE have IBD in combination (7), which seems to hint that there may be a link between the two diseases. Therefore, it is necessary to explore the relationship between the SLE and IBD further.
In this study, we conducted a Mendelian randomization (MR) study to further confirm the causal relationship between SLE and IBD. MR is an epidemiological analysis method that assesses the causal relationship between exposure and outcome by using single-nucleotide polymorphisms (SNPs) as instrumental variables (IVs) for exposure (8). The method avoids irrelevant confounders, such as environmental exposures, and it reduces the effect of reverse causality, thus making the results more convincing (9).
MR analyses need to fulfill the following three assumptions: (1) the IV is strongly associated with exposure; (2) the IV is not associated with any confounders affecting the exposure–outcome association; and (3) the IV affects the outcome only through exposure (Figure 1). It is important to note that the data used in this study are publicly available and free of charge, so there is no need to provide further ethical review and informed consent again.
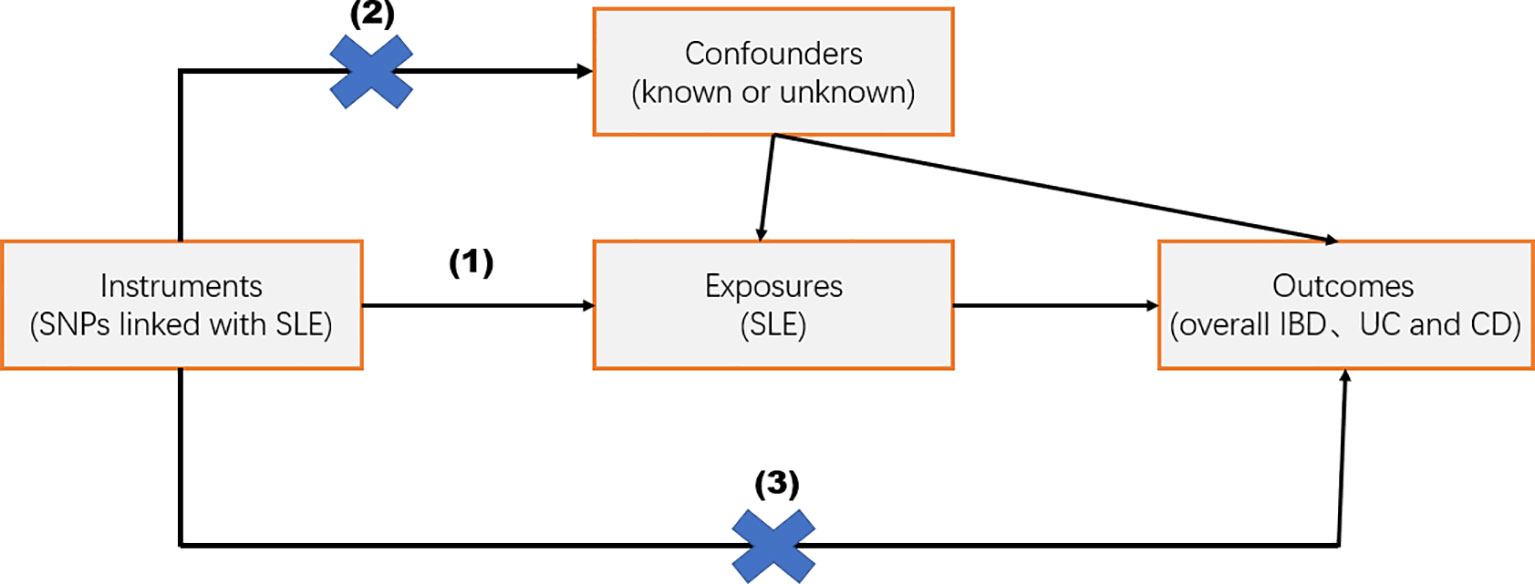
Figure 1 Overview of the study design. Mendelian randomization studies were based on three assumptions: (1) the instrumental variable (IV) was strongly related to exposure; (2) IV was independent of known or unknown confounding factors; and (3) IV affected the outcome only through exposure factors.
SLE-associated SNPs in European populations were extracted from a large-scale Genome-Wide Association Study (GWAS) study that included 5,201 cases and 9,066 controls (10). The IBD-associated GWAS data for the European population were obtained from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC), which was the world’s largest genetic database for IBD. The overall IBD-associated GWAS data for the European population contains 31,665 cases and 33,977 controls, UC for the European population contains 13,768 cases and 33,977 controls, and CD for the European population contains 17,897 cases and 33,977 controls (11). In addition, the SLE-associated SNPs in the East Asian population were derived from a large GWAS study containing 4,222 cases and 8,431 controls (12). Genetic data related to IBD in East Asian populations were also obtained from IIBDGC, with overall IBD in East Asian populations containing 2,824 cases and 3,719 controls, UC in East Asian populations containing 1,134 cases and 3,719 controls, and CD in East Asian populations containing 1,690 cases and 3,719 controls (11). The status of each data is listed in Table 1. These data are publicly available and can be accessed at https://gwas.mrcieu.ac.uk/ and www.ibdgenetics.org.
In this study, we selected IVs on the basis of the following criteria (13): (1) SNPs were strongly associated with SLE, so P < 5 × 10−8 was used as the primary screening condition; (2) SNPs were independent of confounding factors affecting SLE and IBD, and, to ensure that exposure-related IVs were independent, we excluded SNPs with linkage disequilibrium (R2 < 0.001, clumped = 10,000 kb). (3) SNPs were not directly associated with IBD and could only act on IBD through SLE.
In addition, we calculated the F-statistic to eliminate the bias in the results caused by weak IVs. The F-statistic was calculated as F = β2/se2, and the F-statistic was required to be >10 (14).
In this by study, MR analyses were performed with the TwoSampleMR software package (version 0.5.6) and R software (version 4.2.1) (15). In this study, we used three analysis methods, namely, MR Egger, weighted median, and inverse variance–weighted (IVW), to determine whether there was a causal relationship between SLE and IBD. Of these, we used IVW as the primary method for MR analyses, which used weighted regression of SNP-specific Wald ratios to assess the causal effect of exposure on outcomes (16). MR Egger and weighted median were used as complementary analyses to test the robustness of the results: (1) weighted median (17): this method was allowed for consistent estimates of causal effects to be provided even when up to 50% of the IVs were invalidated. (2) MR Egger (18): the MR-Egger method was able to assess whether genetic variants have pleiotropic effects on the outcome and provides consistent estimates of causal effects under weaker assumptions, but the method might increase the type 1 error rate. Moreover, we used the MR-PRESSO method to identify anomalous outliers with horizontal pleiotropy (19).
Several sensitivity analyses were used in this study to assess the robustness of the results. (1) Cochran’s Q test (20): the Cochran’s Q test assesses the heterogeneity among individual SNPs; if the P-value is greater than 0.05, then it is considered that there is no heterogeneity, in which case a fixed-effects IVW approach is used. If the P-value is less than 0.05, then the random effects IVW model is used. (2) MR Egger (21): this method was used to detect horizontal pleiotropy. (3) In addition, we used the leave-one-out method to determine whether the results of MR were significantly affected by any SNP.
We selected IVs according to the selection criteria described above. Consequently, 45 and 40 SNPs met the criteria as the IVs estimation of SLE in both Europe and East Asia, respectively (Supplementary Tables). The F-statistics were all greater than 10, and there was no evidence of weak instrumental bias (Supplementary Tables).
The present study showed that SLE exhibited negative associations for all three outcomes (IBD, UC, and CD). Among them, there was a statistically significant association between SLE and overall IBD (OR = 0.94; 95% CI = 0.90, 0.98; P < 0.004) and UC (OR = 0.93; 95% CI = 0.88, 0.98; P = 0.006), respectively, and the results of the MR-PRESSO after removing the outliers were still in agreement with the IVW results. However, there was no statistically significant relationship between SLE and CD (OR = 0.95; 95% CI = 0.88, 1.02; P = 0.14) (Figures 2, 3, Table 2).

Figure 3 Scatter plot of the relationship between SLE and IBD. (A) SLE and overall IBD (Europeans); (B) SLE and UC (Europeans); (C) SLE and CD (Europeans); (D) SLE and overall IBD (East Asians); (E) SLE and UC (East Asians); (F) SLE and CD (East Asians).
Although SLE showed negative causal associations with overall IBD and UC in European populations, what is interesting was that, in this study, there was no statistically significant association between SLE and overall IBD (OR = 0.92; 95% CI = 0.84, 1.00; P = 0.052), UC (OR = 0.91; 95% CI = 0.79, 1.06; P = 0.22), and CD (OR = 0.92; 95% CI = 0.82, 1.03; P = 0.15) in the East Asian populations (Figures 2, 3, Table 2).
First, in the heterogeneity test, the P-value of Cochran’s Q test was less than 0.05, indicating heterogeneity among SNPs (Table 3). Therefore, in this MR analysis, we used the random-effects IVW method as the main analysis method. MR Egger regression intercepts showed limited evidence of horizontal pleiotropy for SLE-related IVs. In addition, the leave-one-out method showed that the potential causal link between SLE and IBD in the European population was not driven by a SNP (Supplementary Figure 4). Moreover, forest and volcano plots provided a more visual display of heterogeneity (Supplementary Figures 5, 6).
In the present study, two-sample MR study demonstrated that genetic susceptibility to SLE was causally associated with lower overall IBD risk and UC risk in European populations; however, no significant association between SLE and CD was found. Furthermore, interestingly, no association was found between SLE and IBD in East Asian populations. To the best of our knowledge, this is the first MR study to explore the causal relationship between SLE and IBD.
It is well known that SLE is considered an autoimmune disease, whereas previously, Mackay thought that IBD was not a strictly autoimmune disease because of the lack of specific serologic markers for IBD (22). However, in recent years, it has been found that the pathogenesis of IBD is inextricably linked to autoimmune factors (23). Recent studies have shown that the CXCL13/CXCR5 axis is activated in both SLE and IBD (24) and that its pathway is associated with the regulation of T cells. So far, the literature on the association between SLE and IBD consists mainly of a few case reports and case series (25), which seems to imply that the association between these two diseases is uncommon. It is worth noting, however, that the differentiation between SLE and IBD is sometimes difficult to make because SLE can also involve the gastrointestinal tract (25–27), and, thus, the association between the two diseases seems to be more than coincidental. In addition, it is noteworthy that patients with a combination of SLE and IBD do not have the perceived worse prognosis, and, on the contrary, patients tend to respond favorably to treatment (25, 28). To date, a relatively large database study has shown that the prevalence of CD is higher in patients with SLE (OR = 2.23; 95% CI = 1.46–3.4; P < 0.001) compared with controls, whereas SLE is not associated with UC (OR = 1.67; 95% CI = 0.99–2.815; P < 0.052) (29). This is contrary to our findings, which may be due to some confounding bias that is difficult to avoid, because people’s exposure to a risk factor may be related to a variety of factors, such as self-selection and occupation, and confounding bias exists when the given exposure is also closely related to another exposure that is associated with the outcome; studies based on management data may sometimes fail to accurately characterize the given exposures in each group, that is, population selection bias (30). In addition, the fact that the above study was conducted in a Middle Eastern population (Israelis), whereas the present study targeted European and East Asian populations, may also be a reason for the different conclusions, because the prevalence and pathogenesis of IBD may vary among different races and ethnic groups (31).
In this MR study, we confirmed a causal link between SLE and IBD in a European population. Previous studies have found that the Homo sapiens interferon regulatory factor 5 (IRF5) gene is strongly associated with the development of SLE (32), and a study from the United States suggests that polymorphisms in the IRF5 gene may be associated with protection against IBD (33); this is also consistent with our analysis (rs35000415, IRF5). In addition, further stratified analysis revealed an association between SLE and UC, but not between SLE and CD. Although there is an overlap in the pathogenesis of UC and CD, many recent studies have shown that they differ in genetics (34), pathogenesis (35, 36), cellular immunity (37), and response to probiotic therapies (38), which may explain the absence of a causal link between SLE and CD. However, further studies are needed to investigate the similarities and differences in genetics, intestinal flora, and pathogenesis between the two diseases. Talking about East Asian populations, no association was found between SLE and IBD, which might be due to the differences in the genes associated with SLE (39) and IBD (40) in East Asian populations compared with European populations. For example, Asian patients with IBD are less likely to carry mutants in the nucleotide-binding oligomerization domain 2 (NOD2) gene compared with Europeans (11), and studies have shown that NOD2 is strongly associated with the development of SLE (41). However, although there was no statistically significant difference between SLE and IBD in the East Asian population (P = 0.052), the P-value was very close to 0.05, which might be related to the small sample volume of the East Asian population, and, thus, a larger sample size is needed to further investigate the relationship between the two.
Our study had several strengths. First, we used MR to assess the association between SLE and IBD, which was less susceptible to confounders, reverse causation, etc., than observational studies (42). Second, our exposed IVs were derived from large-scale GWAS, which provided robust and reliable gene-wide associated SNP associations, avoiding bias caused by weak instruments. In addition, we used the MR-PRESSO method to further confirm the reliability of this study.
However, there are some limitations to our study. First, although we used multiple methods to analyze horizontal pleiotropy and the results were consistent across multiple methods of analysis, there was still no guarantee that potential horizontal pleiotropy was ruled out completely. Second, the findings of this study could not be extrapolated to other races due to the racial limitations of this study. In addition, because of the limitations of sample volume and gene sequencing technology, we look forward to further exploring the association between SLE and IBD in larger-scale research in the future.
This MR work revealed a negative causal effect of SLE on overall IBD and UC in European populations, but not between SLE and CD. In contrast, there was no causal relationship between SLE and IBD in East Asian populations. Our results may enrich previous studies and may provide a reference for future animal experiments and clinical treatments.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
WX completed data analysis and most of the manuscript writing. NL and SH provided constructive comments and pertinent revisions to the manuscript. HZ, YS, ZY, GH, and YC completed the data collection. HJ, HX, and SH completed the data verification and checked the grammar of the article. SL provided constructive suggestions on the logic, grammar, and other issues of the article during the revision stage. In addition, he also provided meaningful suggestions on how to conduct further in-depth research in this field in the future. In addition, this article later received support from SL’s related funds: Natural Science Foundation of Zhejiang Province (LQ20H020002). All authors contributed to the article and approved the submitted version.
This project was sponsored by the Zhejiang Medical and Health Project (Key Talents) (2014RCA014), Wenzhou Municipal Science and Bureau (Y20210184), Natural Science Foundation of Zhejiang Province (LQ20H020002), and the Zhejiang Provincial Health Department Medical Support Discipline–Nutrition (11-ZC24).
We thank the above databases for making their data publicly available.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1199896/full#supplementary-material
1. Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet (2007) 369:1627–40. doi: 10.1016/S0140-6736(07)60750-8
2. Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology (2012) 142:46–54.e42. doi: 10.1053/j.gastro.2011.10.001
3. Aniwan S, Santiago P, Loftus EV Jr., Park SH. The epidemiology of inflammatory bowel disease in Asia and Asian immigrants to Western countries. United Eur Gastroenterol J (2022) 10:1063–76. doi: 10.1002/ueg2.12350
4. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol (2015) 12:720–7. doi: 10.1038/nrgastro.2015.150
5. Barber MRW, Drenkard C, Falasinnu T, Hoi A, Mak A, Kow NY, et al. Global epidemiology of systemic lupus erythematosus. Nat Rev Rheumatol (2021) 17:515–32. doi: 10.1038/s41584-021-00668-1
6. Barbhaiya M, Costenbader KH. Environmental exposures and the development of systemic lupus erythematosus. Curr Opin Rheumatol (2016) 28:497–505. doi: 10.1097/BOR.0000000000000318
7. Jin X, Wang G, Xu X, Bai Y, An R, Jiang D. Coexistence of Crohn's disease and systemic lupus erythematosus: a case report and literature review. Eur J Gastroenterol Hepatol (2020) 32:1256–62. doi: 10.1097/MEG.0000000000001775
8. Ebrahim S, Davey Smith G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum Genet (2008) 123:15–33. doi: 10.1007/s00439-007-0448-6
9. Zoccali C, Testa A, Spoto B, Tripepi G, Mallamaci F. Mendelian randomization: a new approach to studying epidemiology in ESRD. Am J Kidney Dis (2006) 47:332–41. doi: 10.1053/j.ajkd.2005.10.027
10. Bentham J, Morris DL, Graham DSC, Pinder CL, Tombleson P, Behrens TW, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet (2015) 47:1457–64. doi: 10.1038/ng.3434
11. Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet (2015) 47:979–86. doi: 10.1038/ng.3359
12. Wang YF, Zhang Y, Lin Z, Zhang H, Wang TY, Cao Y, et al. Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity between ancestral groups. Nat Commun (2021) 12:772. doi: 10.1038/s41467-021-21049-y
13. Gagliano Taliun SA, Evans DM. Ten simple rules for conducting a mendelian randomization study. PloS Comput Biol (2021) 17:e1009238. doi: 10.1371/journal.pcbi.1009238
14. Wang B, Liu D, Song M, Wang W, Guo B, Wang Y. Immunoglobulin G N-glycan, inflammation and type 2 diabetes in East Asian and European populations: a Mendelian randomization study. Mol Med (2022) 28:114. doi: 10.1186/s10020-022-00543-z
15. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol (2017) 46:1734–9. doi: 10.1093/ije/dyx034
16. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol (2013) 37:658–65. doi: 10.1002/gepi.21758
17. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol (2016) 40:304–14. doi: 10.1002/gepi.21965
18. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
19. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
20. Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med (2015) 34:2926–40. doi: 10.1002/sim.6522
21. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol (2015) 44:512–25. doi: 10.1093/ije/dyv080
22. Mackay IR. Clustering and commonalities among autoimmune diseases. J Autoimmun (2009) 33:170–7. doi: 10.1016/j.jaut.2009.09.006
23. Guan Q. A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J Immunol Res (2019) 2019:7247238. doi: 10.1155/2019/7247238
24. Pan Z, Zhu T, Liu Y, Zhang N. Role of the CXCL13/CXCR5 axis in autoimmune diseases. Front Immunol (2022) 13:850998. doi: 10.3389/fimmu.2022.850998
25. Katsanos KH, Voulgari PV, Tsianos EV. Inflammatory bowel disease and lupus: a systematic review of the literature. J Crohns Colitis (2012) 6:735–42. doi: 10.1016/j.crohns.2012.03.005
26. Shimizu T, Nishinarita S, Son K, Tomita Y, Yoshihiro, Matsukawa, et al. [Crohn's disease with the onset resembling systemic lupus erythematosus]. Nihon Rinsho Meneki Gakkai Kaishi (1999) 22:164–9. doi: 10.2177/jsci.22.164
27. Buchman AL, Wilcox CM. Crohn's disease masquerading as systemic lupus erythematosus. South Med J (1995) 88:1081–3. doi: 10.1097/00007611-199510000-00018
28. Nitzan O, Elias M, Saliba WR. Systemic lupus erythematosus and inflammatory bowel disease. Eur J Intern Med (2006) 17:313–8. doi: 10.1016/j.ejim.2006.02.001
29. Shor DB, Dahan S, Comaneshter D, Cohen AD, Amital H. Does inflammatory bowel disease coexist with systemic lupus erythematosus? Autoimmun Rev (2016) 15:1034–7. doi: 10.1016/j.autrev.2016.07.027
30. Boyko EJ. Observational research–opportunities and limitations. J Diabetes Complications (2013) 27:642–8. doi: 10.1016/j.jdiacomp.2013.07.007
31. Barnes EL, Loftus EV Jr., Kappelman MD. Effects of race and ethnicity on diagnosis and management of inflammatory bowel diseases. Gastroenterology (2021) 160:677–89. doi: 10.1053/j.gastro.2020.08.064
32. Song S, De S, Nelson V, Chopra S, LaPan M, Kampta K, et al. Inhibition of IRF5 hyperactivation protects from lupus onset and severity. J Clin Invest (2020) 130:6700–17. doi: 10.1172/JCI120288
33. Gathungu G, Zhang CK, Zhang W, Cho JH. A two-marker haplotype in the IRF5 gene is associated with inflammatory bowel disease in a North American cohort. Genes Immun (2012) 13:351–5. doi: 10.1038/gene.2011.90
34. Waterman M, Xu W, Stempak JM, Milgrom R, Bernstein CN, Griffiths AM, et al. Distinct and overlapping genetic loci in Crohn's disease and ulcerative colitis: correlations with pathogenesis. Inflammation Bowel Dis (2011) 17:1936–42. doi: 10.1002/ibd.21579
35. Yang L, Tang S, Baker SS, Arijs I, Liu W, Alkhouri R, et al. Difference in pathomechanism between crohn's disease and ulcerative colitis revealed by colon transcriptome. Inflammation Bowel Dis (2019) 25:722–31. doi: 10.1093/ibd/izy359
36. Cordes F, Foell D, Ding JN, Varga G, Bettenworth D. Differential regulation of JAK/STAT-signaling in patients with ulcerative colitis and Crohn's disease. World J Gastroenterol (2020) 26:4055–75. doi: 10.3748/wjg.v26.i28.4055
37. Mitsialis V, Wall S, Liu P, Ordovas-Montanes J, Parmet T, Vukovic M, et al. Single-cell analyses of colon and blood reveal distinct immune cell signatures of ulcerative colitis and crohn's disease. Gastroenterology (2020) 159:591–608.e510. doi: 10.1053/j.gastro.2020.04.074
38. Bjarnason I, Sission G, Hayee B. A randomised, double-blind, placebo-controlled trial of a multi-strain probiotic in patients with asymptomatic ulcerative colitis and Crohn's disease. Inflammopharmacology (2019) 27:465–73. doi: 10.1007/s10787-019-00595-4
39. Morris DL, Sheng Y, Zhang Y, Wang YF, Zhu Z, Tombleson P, et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet (2016) 48:940–6. doi: 10.1038/ng.3603
40. McGovern DP, Kugathasan S, Cho JH. Genetics of inflammatory bowel diseases. Gastroenterology (2015) 149:1163–1176.e1162. doi: 10.1053/j.gastro.2015.08.001
41. De Jager PL, Graham R, Farwell L, Sawcer S, Richardson A, Behrens TW, et al. The role of inflammatory bowel disease susceptibility loci in multiple sclerosis and systemic lupus erythematosus. Genes Immun (2006) 7:327–34. doi: 10.1038/sj.gene.6364303
Keywords: Mendelian randomization (MR), systemic lupus erythematosus (SLE), inflammatory bowel disease (IBD), ulcerative colitis (UC), Crohn’s-disease (CD)
Citation: Xie W, Jiang H, Chen Y, Zhang H, Song Y, Yu Z, Gu H, Xu H, Han S, Li S, Liu N and Han S (2023) Association between systemic lupus erythematosus and inflammatory bowel disease in European and East Asian populations: a two-sample Mendelian randomization study. Front. Immunol. 14:1199896. doi: 10.3389/fimmu.2023.1199896
Received: 04 April 2023; Accepted: 18 October 2023;
Published: 03 November 2023.
Edited by:
Ryusuke Yoshimi, Yokohama City University, JapanReviewed by:
Juan C. Rueda, Faculty of Medicine and Engineering, ColombiaCopyright © 2023 Xie, Jiang, Chen, Zhang, Song, Yu, Gu, Xu, Han, Li, Liu and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naxin Liu, bGl1bmF4aW42OEB3emhvc3BpdGFsLmNu; Shaoliang Han, c2xoYW44OEAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.