- 1School of Pharmacy, Nanchang University, Nanchang, China
- 22nd Abdominal Surgery Department, Jiangxi Cancer Hospital, Nanchang, China
Arecoline is an alkaloid extracted from betel nut, which has various pharmacological effects. In the present study, we showed that arecoline aggravated experimental acute ulcerative colitis (UC) induced by dextran sodium sulfate (DSS) in mice. We measured body weight and colon length, evaluated disease activity index, colon pathology sections, and levels of colonic inflammatory factors. Arecoline exacerbated the clinical signs of UC and the colonic inflammatory response in mice. The results of 16S rRNA sequencing of fecal samples showed a significant decrease in the percentage of probiotic bacteria Ligilactobacillus, Limosilactobacillus and Lactobacillus and a significant increase in the percentage of conditionally pathogenic bacteria Odoribacter and Bacteroides after arecoline treatment. Serum untargeted metabolomics showed that arecoline intervention reduced the levels of ergothioneine, pentostatin, diadenosine tetraphosphate and other metabolites and modulated nicotinate and nicotinamide metabolism, metabolic pathways, glyoxylate and dicarboxylate metabolism, and other metabolic pathways of intestinal microorganisms. According to the combined microbial and metabolite analysis, arecoline influences metabolite levels by modulating the intestinal microbiota. In summary, it was found that arecoline treatment exacerbated colonic injury and intestinal inflammatory responses in UC mice, disrupted the host’s intestinal flora, and affected changes in flora metabolites, thereby exacerbating the development of colonic inflammation. Therefore, the consumption of betel nut can be associated with the risk of aggravating UC.
Introduction
Ulcerative colitis (UC) is a form of inflammatory bowel disease (IBD) that has become a globally widespread and disease that poses a serious health risk given its increasing incidence (1). UC is a chronic idiopathic inflammatory disease with common symptoms including blood in the stool and diarrhea and mainly involves the colon and rectum. Its pathogenesis includes environmental factors, an unhealthy lifestyle, immune imbalance and dysbiosis of the gut microbiota (2, 3).Meanwhile, UC is also closely related to colorectal cancer, and studies have shown that long-term UC increases the risk of developing colorectal cancer (4).
Gut microbiota are closely related to the development of UC, and dysbiosis of the gut flora can increase the severity of UC (5). Studies have shown that a significant increase in the abundance of Bacteroidota and a relative decrease in the abundance of Firmicutes in the intestines of UC patients and reduced ratio of Firmicutes to Bacteroides (F/B) contribute to the development of UC (6).5-aminosalicylate sodium (5-ASA), cyclosporine, and glucocorticoids are the traditional drugs used to treat UC. However, these drugs are associated with serious side effects. Probiotics have fewer side effects than traditional drugs and can treat and alleviate the occurrence of UC (7). The probiotic Lactobacillus plantarum HNU082 (Lp082) was found to protect the mucosal barrier of the intestine, modulate the gut microbiota and reduce the symptoms of intestinal inflammation for the purpose of treating UC (8). Furthermore, live and pasteurized Akkermansia muciniphila effectively alleviated the symptoms of colitis in mice by promoting the proliferation of beneficial intestinal bacteria, increasing the production of short-chain fatty acids and suppressing intestinal inflammation (9).
Herbal extracts have also been extensively studied for their ability to inhibit the development of UC in mice. For example, the polyphenol extract of Thymus vulgaris L. could protect the intestinal epithelial barrier, modulate the intestinal microbiota, and inhibit the TLR4/NF-κB-NLRP3 inflammatory vesicle pathway to improve UC in mice (10). Intestinal flora metabolites are closely related to the maintenance of the intestinal barrier and the balance of the intestinal immune microenvironment. Short-chain fatty acids (SCFAs) have been shown to have immunomodulatory effects by reducing the production of pro-inflammatory factors to reduce the inflammatory response and mitigate the development of UC (11, 12).
Arecoline is the active ingredient of betel nut, and its pharmacological action has important effects on the nervous, cardiovascular, endocrine, and digestive systems (13–15). Some studies have shown that arecoline can increase inflammatory cell infiltration in the oral epithelium and affect the synthesis of various inflammatory mediators, thus leading to the development of oral epithelial fibrosis (16, 17). More importantly, long-term stimulation by arecoline may lead to further development of oral squamous cell carcinoma (18, 19). One study found that consumption of betel nut induces the peripheral blood mononuclear cells (PBMC) to secrete inflammatory cytokines such as IL-1β, IL-8, TNF-α, and IL-6, leading to their increased circulation (20). In long-term studies, arecoline has great research significance for its role in the body (21). However, it remains unexplored whether the development of UC is affected by arecoline.
In this study, the effect of arecoline on dextran sodium sulfate (DSS)-induced UC was investigated. We assessed the clinical signs by observing the colonic length, weight change, and disease activity index of mice, and the inflammatory response based on their colonic pathological changes and the concentrations of colonic tissue inflammatory factors. We also performed 16S rRNA sequencing on fecal samples and untargeted metabolomics analysis on serum samples to explore the specific mechanism of arecoline action on UC. The results suggest that arecoline may exacerbate DSS-induced UC by affecting the intestinal microbiota and its metabolites.
Materials and methods
Animal experimental design
We purchased 18 C57BL/6 male mice (age: 6–8 weeks) from Jiangsu Jicuiyaokang Biotechnology Co (Nanjing, China). The mice were housed at the Jiangxi Academy of Sciences, Royo Biotech Co., Ltd. (Nanchang, China). They were housed at a temperature of 25°C with a 24-h light-dark cycle. Mice had ad libitum access to food and water. After 7 days of acclimation, the mice were divided into three groups (n=6 per group): (1) Control group that was allowed ad libitum access to food and water; (2) DSS group: ad libitum diet was allowed for 1–7 days and 3% DSS (Meilunbio, Dalian) was added to the drinking water for 8–14 days; (3) Arec group: ad libitum diet was allowed for 1–7 days, 3% DSS was added to the drinking water for 8–14 days, and 5 mg/kg arecoline (Yuanye, Shanghai) was administered daily by tube feeding.
Colonic length and histological analysis
Mice were killed after anesthesia, and their colon was dissected out and its length measured. Colon samples were preserved and fixed in 4% paraformaldehyde solution and then embedded in paraffin. The colon tissue was cut into 4-µm–thick sections, and then the colon sections were stained using hematoxylin and eosin (H&E). The stained slides were examined under an inverted microscope and photographed.
Assessment of disease activity index
Throughout the experiment, the mice were weighed daily and fecal conditions were recorded. The stool properties and body weight were scored together, and the scores were summed and recorded as the DAI (22). Assessment was performed according to the following scoring criteria:
weight change: 0, no weight loss; 1, weight loss of 1–5%; 2, weight loss of 5–10%; 3, weight loss of 10–15%; 4: weight loss of >15%.
stool consistency: 0, normal; 2, dilute stool; 4, diarrhea.
blood in the stool: 0, no blood; 2, visual pellet bleeding; 4, severe bloody stool and blood around the anus.
Enzyme-linked immunosorbent assay
Colon tissue stored at -80°C was thawed and mixed with phosphate-buffered saline (PBS) at a volume of 1:9 by weight and placed in a tissue grinder for homogenization, followed by centrifugation at 3000×g for 4 min at 5°C. We quantified the supernatant after centrifugation for quantitative ELISA (Shenkebio, Wuhan) of TNF-α, IL-1β, and IL-6 according to the manufacturer’s instructions.
16S rRNA sequencing
Mice fecal samples were collected after excretion and stored immediately in liquid nitrogen. We extracted genomic DNA by the Cetyltrimethylammonium Bromide (CTAB) method. The purity and concentration of DNA was assessed using 2% agarose gel electrophoresis; the DNA samples were diluted in sterile water to 1 ng/μl. After electrophoretic detection, the DNA was purified using magnetic beads and quantified by enzyme marker. Electrophoretic detection was again performed using 2% agarose gels, followed by recovery of target bands using Qiagen’s gel recovery kit (Qiagen, Germany). We used TruSeq® DNA PCR-Free sample preparation kit to construct libraries. Libraries were constructed and quantified by Qubit and Q-PCR, and the qualified libraries were sequenced using NovaSeq6000. 16S rRNA sequencing was provided by Metware Biotechnology Co., Ltd. (Wuhan, China).
The data was efficiently processed by FLASH (v1.2.11) and Qiime (v1.9.1). The valid data were clustered using the Uparse algorithm (USEARCH v7) to assign sequences with a similarity ≥97% to the same operational taxonomic units (OTUs). Taxonomic information was annotated using the Mothur algorithm of the SILVA v138.1 (http://www.arb-silva.de/) database. The Shannon index, Simpson index, ACE index, and Chao1 index were calculated using R software (v4.1.2) and QIIME (v1.9.1) to assess alpha diversity. We also performed principal co-ordinate analysis (PCoA) and analysis of similarities (ANOSIM) to assess beta diversity using QIIME and R software. Last, LEfSe analysis using LEfSe software was used to screen for LDA score ≥4.
Untargeted metabolomics
After thawing and vortexing the samples, 50 μL sample and 300 μL extract containing the internal standard (acetonitrile:methanol=1:4, V/V) were added to the centrifuge tube. The sample was again vortexed for 3 min and centrifuged at 12,000 rpm for 10 min at 4°C. Then, the 200 μL supernatant was collected and placed at -20°C for 30 min, followed by re-centrifuging at 12,000 rpm for 3 min at 4°C. This supernatant was collected for LC-MS analysis (Metware Biotechnology Co., Ltd., Wuhan, China). Samples were collected and analyzed according to the LC-MS system machine instructions. Meanwhile, we annotated the identified metabolites using the Kyoto Encyclopedia of Genes and Genomes (KEGG) compound database (http://www.kegg.jp/kegg/compound/) and then mapped the annotated metabolites to the KEGG pathway database (http://www.kegg.jp/kegg/pathway.html). Orthogonal partial least squares discriminant analysis of the three groups of differential serum metabolites was performed by R software. VIP (VIP>1) and P-value (P<0.05, Student’s t-test) were used for differential metabolite screening in both groups. For multiple group analysis, VIP (VIP>1) and P-value (P-value<0.05, ANOVA) were used for differential metabolite screening.
Statistical analysis
Data are expressed as arithmetic mean ± standard error of the mean (SEM). Data were analyzed and counted using GraphPad Prism 8.0. Statistically significant differences between the groups were assessed by one-way analysis of variance (ANOVA). Bivariate correlations were calculated using Pearson’s r coefficient. Heat maps were constructed using R software (v4.1.2). We investigated the correlation between gut microbiota and serum metabolites using Spearman’s correlation analysis. P<0.05 was considered to indicate statistically significant differences.
Results
Effect of arecoline on clinical symptoms of colitis in mice
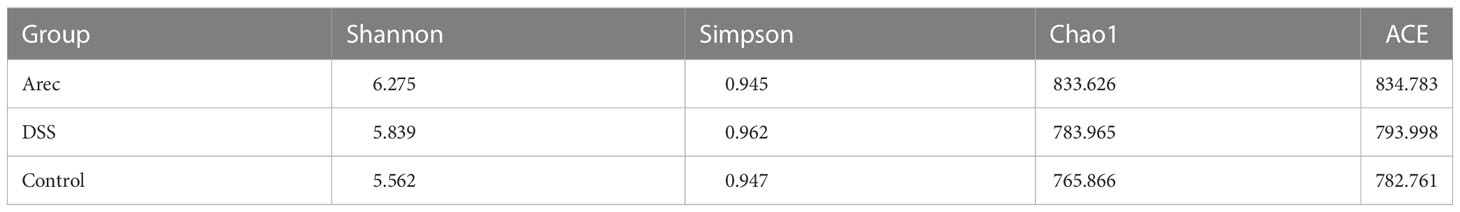
We used 3% DSS solution to induce colitis in mice to establish the UC model, which was used to study the effect of arecoline on colitis. Details of the study groups are presented in Figure 1A. By measuring the colon length of the three groups of mice, we found that mice in the DSS group had a shorter colon length than those in the control group (P<0.01); however, the shortening of the colon was more severe in the Arec group (P<0.001) (Figures 1B, C). With respect to weight loss, there was a significant difference in weight between the arecoline and control groups at day 14 (P<0.01); furthermore, arecoline exacerbated the weight loss in the DSS group mice (Figure 1D). We used the DAI scores to analyze body weight change and fecal properties (blood in stool and stool consistency) in all mice, and arecoline aggravated the clinical signs of DSS-induced acute ulcerative colitis in mice (Figure 1E).
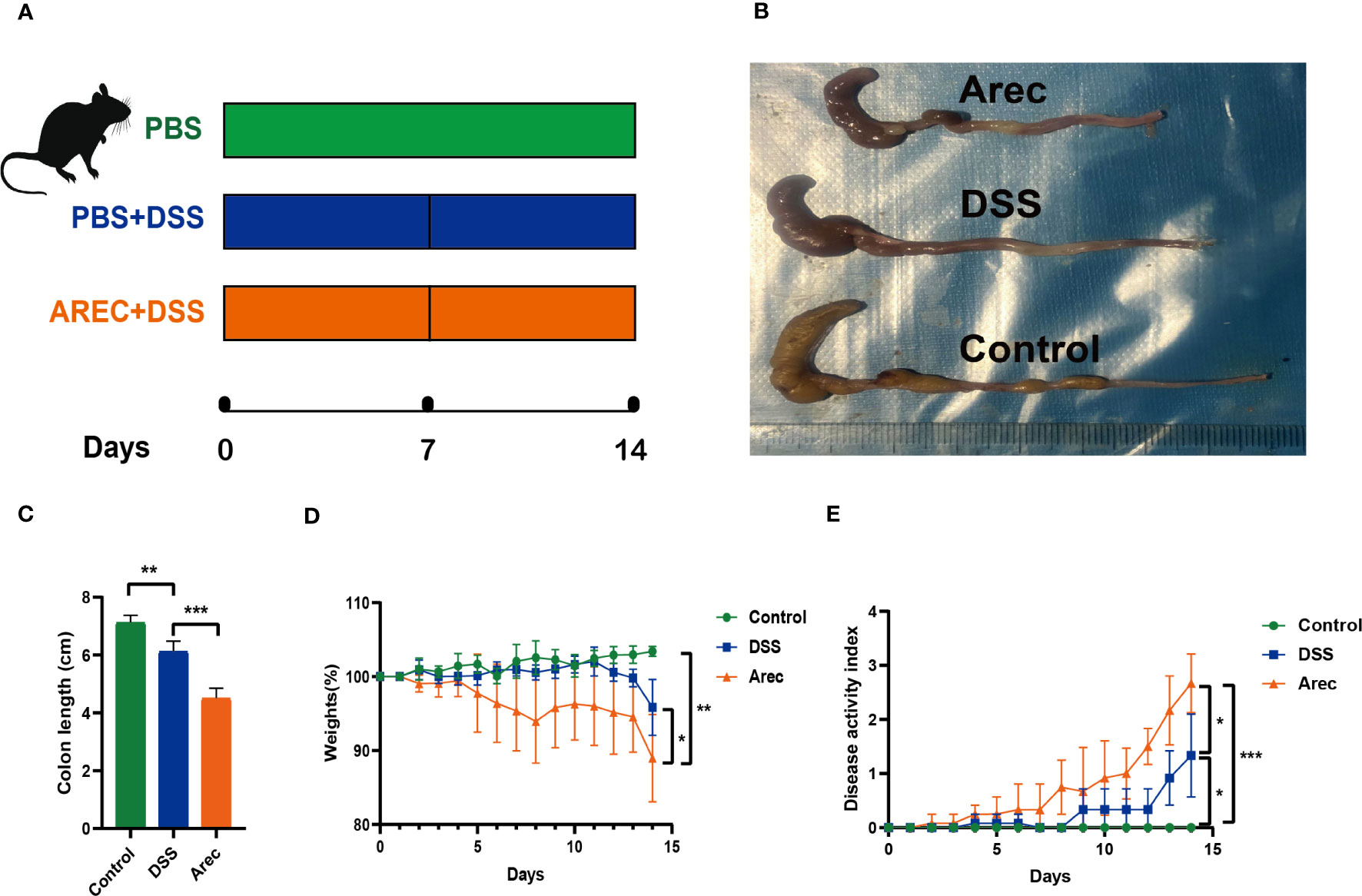
Figure 1 The effect of arecoline on clinical symptoms in mice with DSS-induced UC. (A) Establishment of a mouse UC model and treatment of arecoline. (B) Representative images of the colon in the Arec, DSS, and control groups of mice. (C) Colonic length in mice. (D) Body weight changes of the three groups of mice during the experiment. (E) Disease activity index of the three groups of mice. P<0.05 was statistically significant. *P<0.05, **P<0.01, and ***P<0.001.
Effect of arecoline on the inflammatory response in mice colon
We observed by HE staining of colonic sections that the colonic structure was intact in the control group and severely disrupted in the DSS group, with destruction of the crypt and disappearance of glands, more severe disruption of the intestinal structure, and intestinal inflammation. The Arec group showed more aggregation of neutrophils than the other two groups (Figures 2A). The results of ELISA showed that the levels of inflammatory factors IL-6 (P<0.001), TNF-α (P<0.001), and IL-1β (P<0.001) were significantly higher in the colonic tissue of the Arec group than the DSS group (Figures 2B–D).
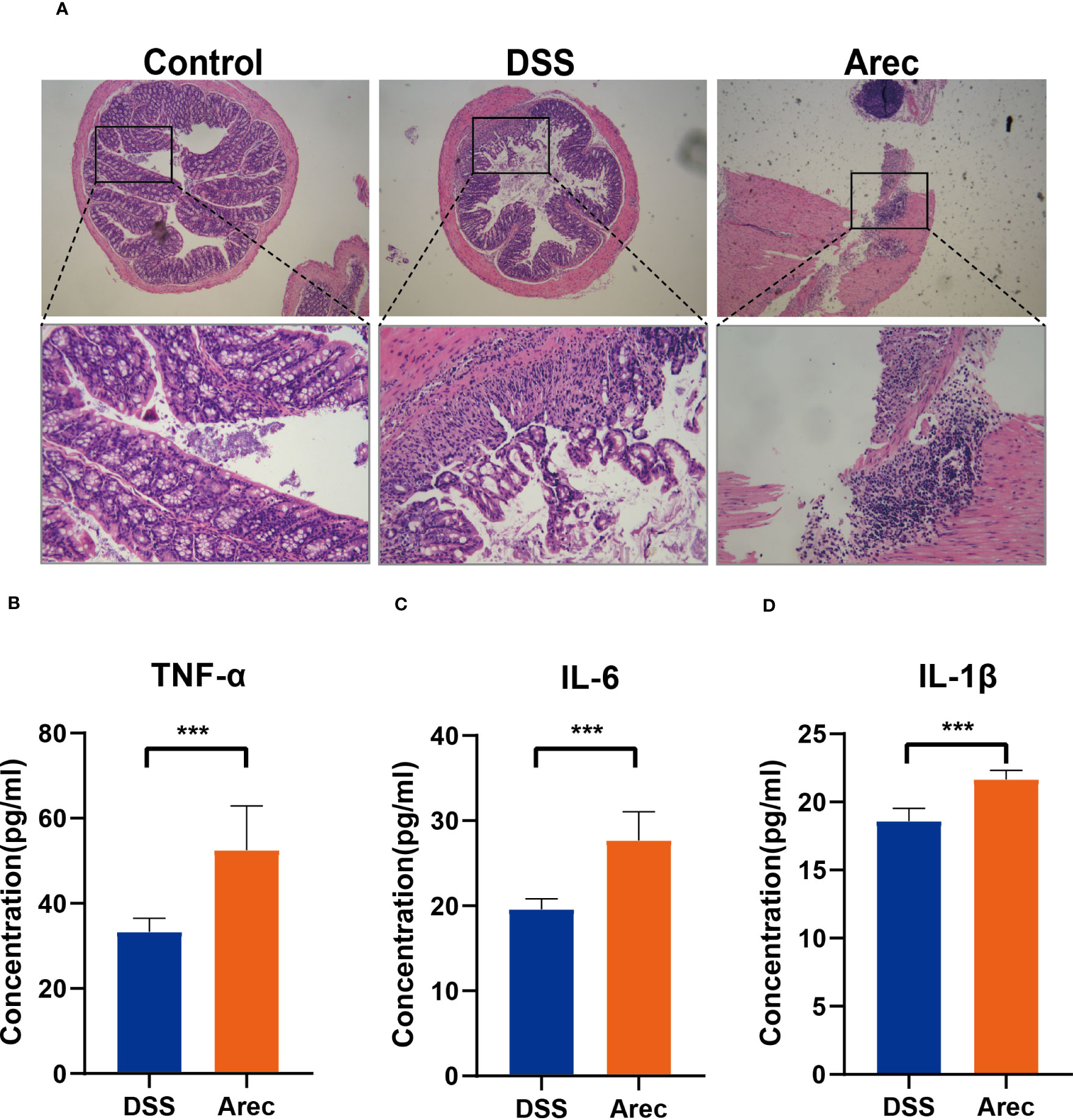
Figure 2 Histopathological sections of mouse colon with changes in inflammatory factors. (A) H&E stained images of mouse colon tissue (magnification 40×, 200×). (B–D) The concentration of inflammatory factors including TNF-α, IL-6, and IL-1β in the colonic tissue of mice in the DSS and Arec groups. P<0.05 was statistically significant. ***P<0.001.
Effect of arecoline on intestinal microbiota
It has been shown that the intestinal microbiota is related to digestion, absorption, metabolism, immunity, and other functions, and that dysbiosis of the intestinal flora is closely related to the occurrence of many diseases such as tumors, diarrhea, obesity, and cardiovascular and cerebrovascular diseases (23–25). Disruptions in the balance of the gut microbiota have also been reported to have many links to the development of UC (26, 27). Therefore, to investigate the effects of arecoline and DSS on intestinal flora, we sequenced 16S rRNA in fecal samples from all three groups and analyzed the alpha and beta diversity indices of the mouse intestinal microbiota. The Arec group had increased gut microbiota diversity as compared to the DSS and control groups according to the Shannon, Simpson, and ACE indices. According to the Chao1 index, the DSS group showed increased diversity of gut microbiota and the Arec group showed decreased diversity of gut microbiota compared to the control group (Table 1). OTU-based weighted UniFrac distance PCoA analysis showed significant differences in the gut microbiota composition of the three groups of mice (P<0.001) (Figure 3A). ANOSIM showed the same results, with significant differences between the control, DSS, and Arec groups (Figures 3B–D). At the phylum level, the Arec group showed an increase in the relative abundance of Bacteroidota and Firmicutes and a decrease in the relative abundance of Proteobacteria and Actinobacteria compared to the DSS group. The relative abundance of Proteobacteria and Bacteroidota increased in the DSS group compared to the control group, while the relative abundance of Firmicutes decreased significantly (Figure 3E). At the genus level, the relative abundance of Bacteroides and Odoribacter increased in the Arec group compared to the DSS group, while the abundance of Parasutterella, Allobaculum, Ligilactobacillus, Lactobacillus, and Limosilactobacillus decreased significantly (Figure 3F).
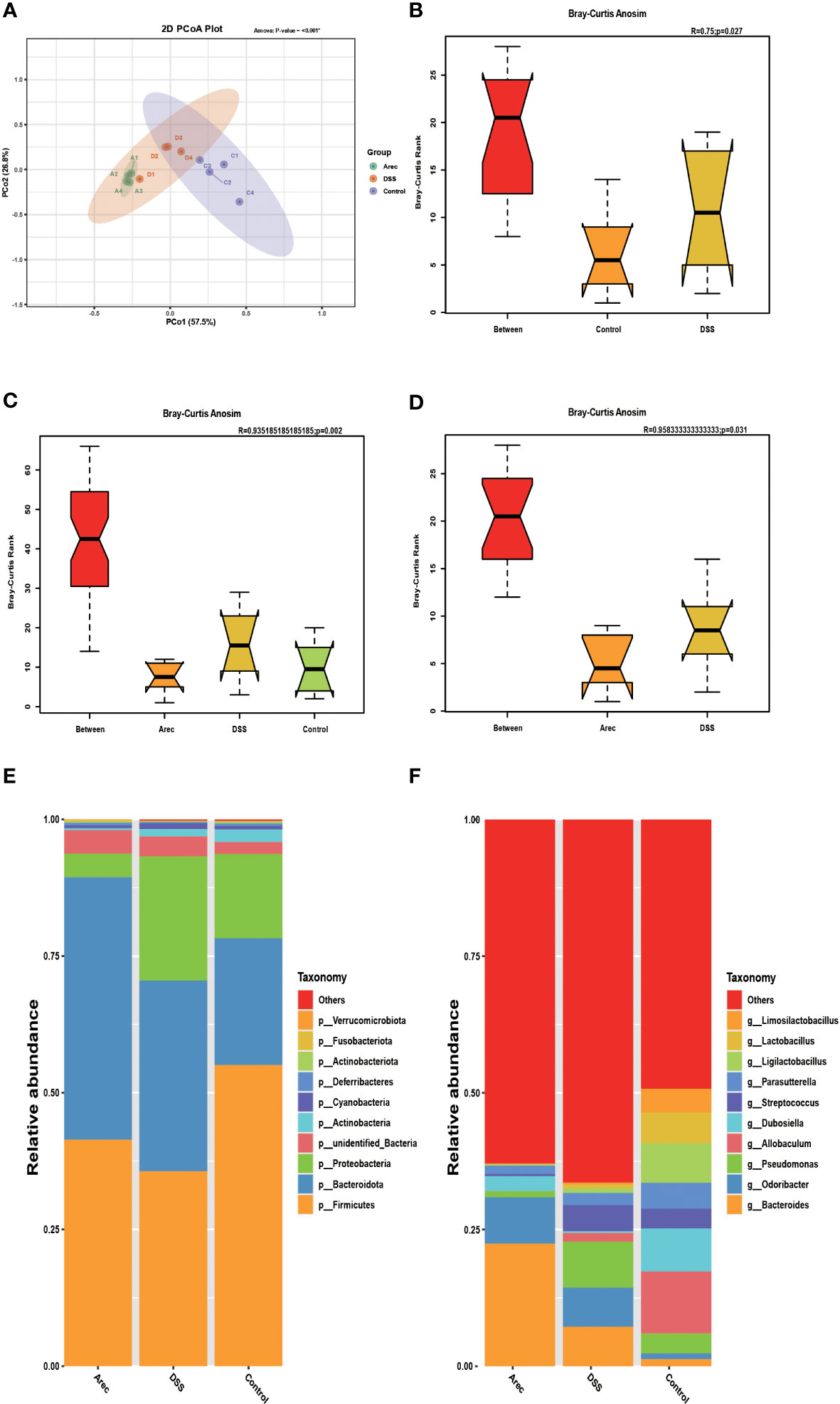
Figure 3 Arecoline and DSS interventions affect changes in intestinal flora. (A) PCoA analysis of OTU-based weighted UniFrac distances showed microbial differences between the groups. (B–D) OTU-based ANOSIM inter-group variation analysis. (E) Gut microbial composition of the three groups of mice at the portal level. (F) Intestinal microbial composition of the three groups of mice at the genus level. P<0.05 was statistically significant.
In the analysis of LEfSe results, the taxonomic phylogram showed that the main composition of the intestinal microbiota in the three groups of mice was Bacteroidota, Firmicutes, and Proteobacteria (Figure 4A). The LDA scores for the abundance of taxonomic units indicated that Bacteroides in the Arec group had higher LDA scores than in the DSS and control groups. By contrast, the control group was dominated by Lactobacillus (Figure 4B). As shown in Figures 4C–F, the population of Bacteroides and Bacteroides acidifaciens was significantly increased in the Arec group compared to the other two groups, while the population of Ligilactobacillus and Lactobacillus almost disappeared. Therefore, elevated abundance of Bacteroides and reduced abundance of Lactobacillus may be the reason for arecoline treatment leading to more severe colonic inflammation.
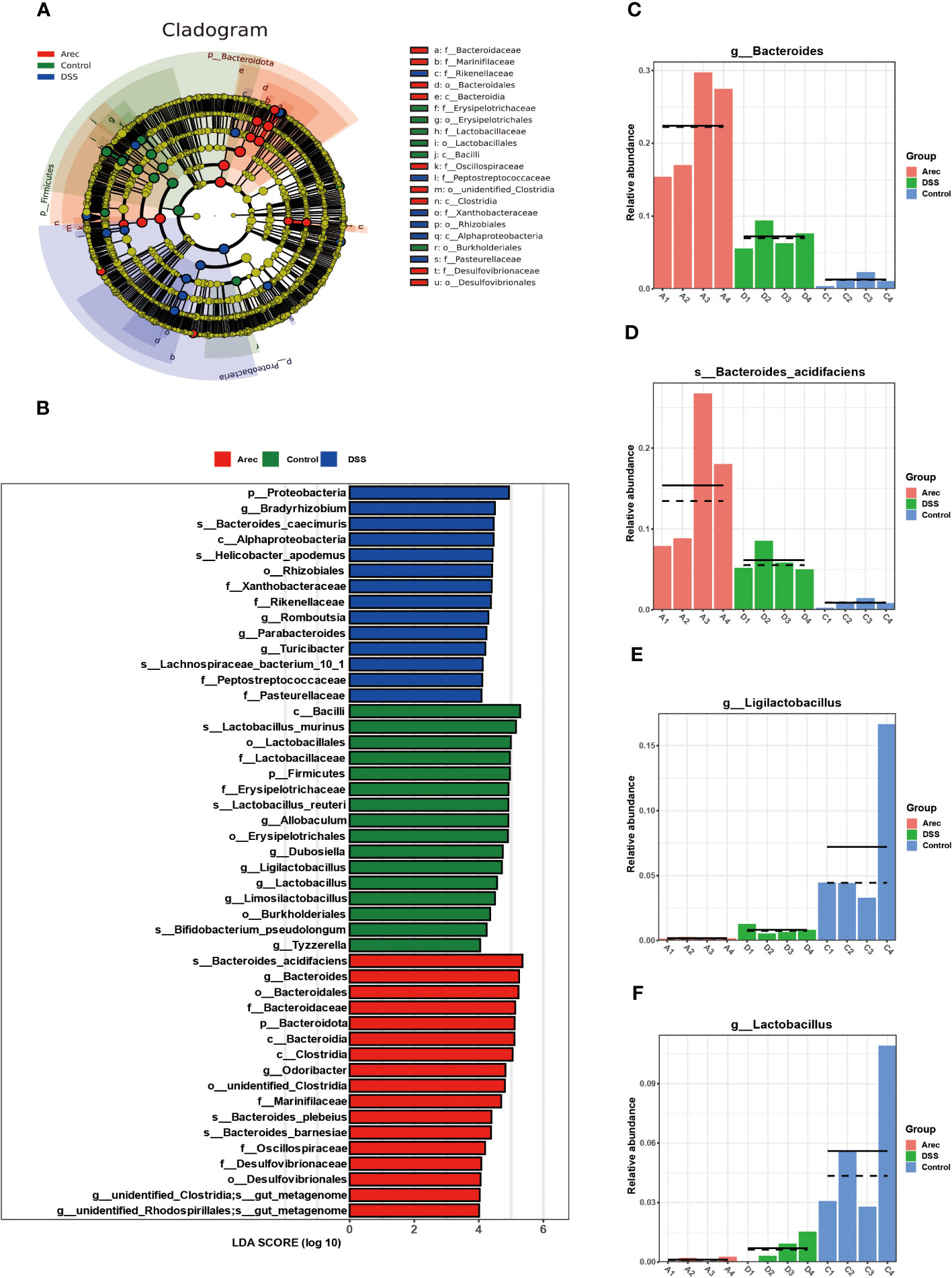
Figure 4 Results of LEfSe analysis of gut microbiota in the Arec, DSS, and control groups. (A) Taxonomic cladogram showing the main constituent taxa of the three groups of mouse intestinal microorganisms. (B) The distribution of LDA values in the histogram shows the species with LDA score greater than 4. (C–F) Difference maps of representative intestinal microorganisms three groups of mice.
Effect of arecoline on the metabolism of intestinal flora
The metabolites of the intestinal microbiota regulate all bodily functions. We analyzed the differential metabolites of the three groups of mice sera by performing untargeted metabolomics assays on their sera. In the positive ion mode, OPLS-DA showed a significant separation of metabolites in the three groups (Figure 5A). In addition, the metabolites were significantly separated between the DSS and control groups (Figure 5B) and between the Arec and DSS groups (Figure 5C). As shown in Figure 5D, the clustering analysis of different metabolites between the Arec group and DSS group in positive-ion mode showed that there were 114 differential metabolites between the two groups, with 25 up-regulated and 89 down-regulated ones. As shown in Figure 5E, the top 20 metabolic pathways including Nicotinate and nicotinamide metabolism, Metabolic pathways, and Glyoxylate and dicarboxylate metabolism were obtained using KEGG pathway enrichment analysis based on the different metabolite results between the Arec and DSS groups.
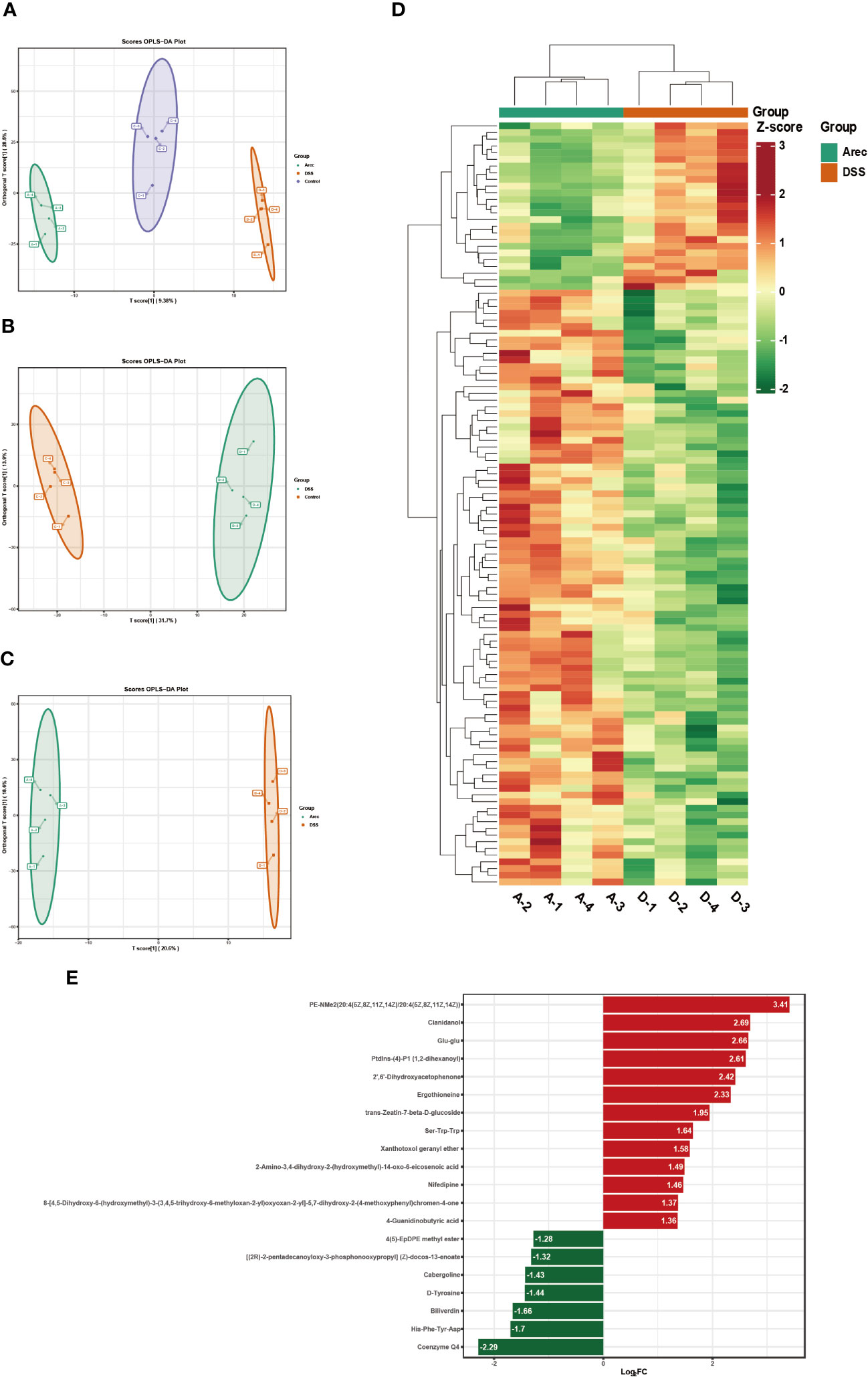
Figure 5 Arecoline regulates serum metabolism in positive-ion mode. (A–C) OPLS-DA based analysis showing differences among the Arec, DSS, and control groups; differences between the DSS and control groups, and differences between the Arec and DSS groups. (D) Heat map reflects the differential metabolites between the Arec and DSS groups (high levels in red, low levels in green). (E) KEGG enrichment maps of differential metabolites in the Arec and DSS groups.
We performed a joint analysis for intestinal flora and differential metabolites. Pentostatin, nifedipine, LPC (20:1/0:0), ergothioneine, and other metabolites increased with increasing abundance of Proteobacteria and Firmicutes and decreased with increasing abundance of Bacteroidota and Actinobacteria (Figure 6).
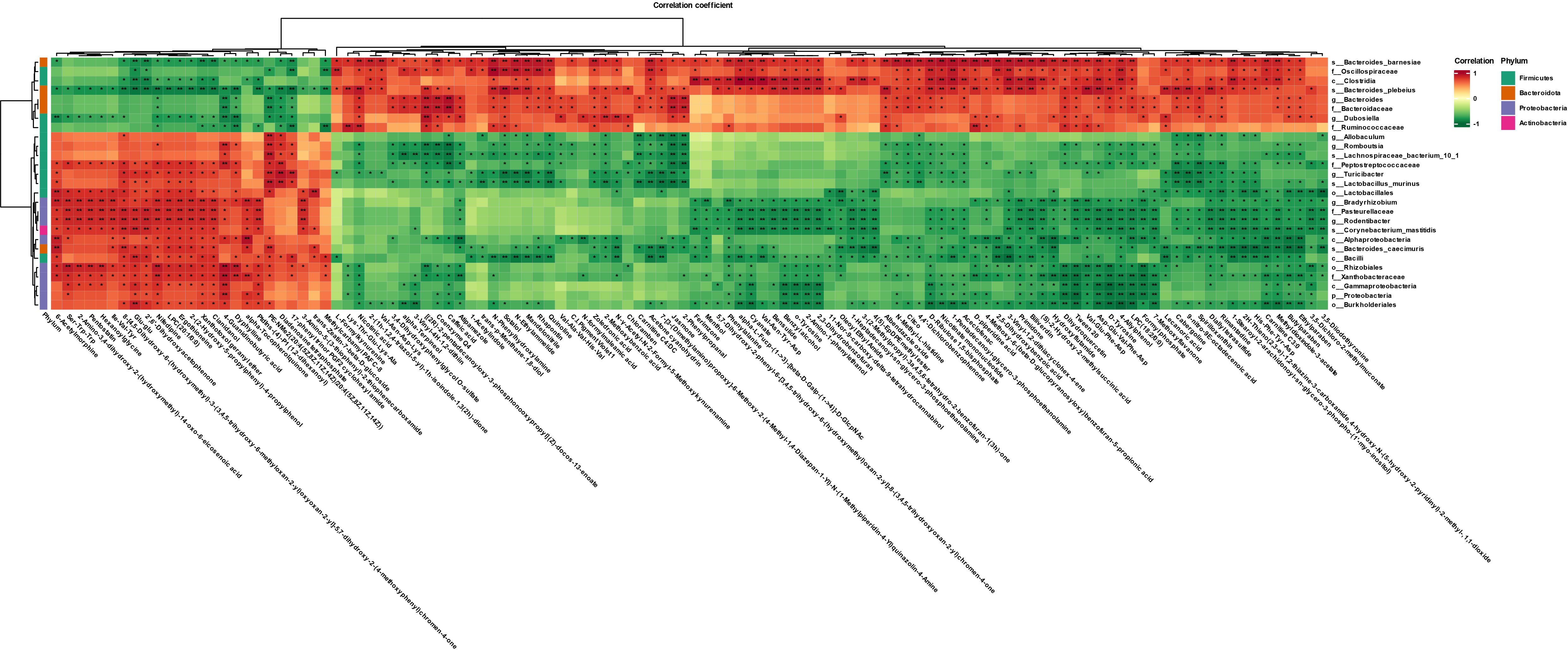
Figure 6 Heat map showing gut microbes associated with differential metabolites in the Arec and DSS groups (red indicates high levels, green indicates low levels). *P<0.05 and **P<0.01.
Discussion
Ulcerative colitis is a chronic idiopathic inflammatory disease caused by an imbalance of immune mechanisms and environmental factors, which often manifests clinically as diarrhea, bleeding, and malabsorption, and is a significant threat to people’s physical and mental health (2, 3). The clinical signs and pathological changes in mice in the DSS group after 7 days of induction with 3% DSS indicated the success of our UC model. Our experiments revealed that 5 mg/kg arecoline aggravated the symptoms of colitis induced by DSS, and the Arec group showed higher DAI scores and showed more severe weight loss as well as shortening of the colon than the DSS and control groups. Pathological examination of the intestinal tract of mice showed that the DSS group had reduced crypts, mucosal breakdown, and inflammatory cell infiltration; moreover, the breakdown of the intestinal barrier and inflammatory cell infiltration were more severe with the addition of arecoline.
We also detected a significant increase in pro-inflammatory factors (IL-6, IL-1β, and TNF-α) in the Arec group by ELISA. Among them, the pro-inflammatory effect of IL-6 stems from its ability to prevent T-cell apoptosis, but it also affects the pro-ablative and repair functions of the epithelial barrier (28). TNF-α promotes T-cell proliferation and differentiation and aggravates intestinal inflammation (29). Studies have shown that elevated IL-1β correlates with the severity of acute inflammation and that inhibition of IL-1β expression alleviates the symptoms of colitis (30). These experimental results suggest that arecoline promotes the expression of inflammatory factors and aggravates UC; therefore, we investigated its mechanism of action in more depth.
As the study of UC has become more advanced, a large number of studies have shown that intestinal microorganisms play a central role in the development of UC (31, 32). Hence, we collected fecal samples from mice for 16S rRNA sequencing to examine their intestinal flora composition. The sequencing results showed significant differences in the gut microbial composition of the three groups. At the phylum level, the relative abundance of Proteobacteria was significantly increased after DSS treatment compared to the control group, consistent with previous studies. The study showed that the ratio of Firmicutes to Bacteroides (F/B) was reduced in the gut of UC patients (33), consistent with our experimental results, and the reduction of F/B was more significant with the addition of arecoline, which aggravated the dysbiosis of the intestinal flora caused by DSS.
At the genus level, arecoline exacerbated the effect of DSS on beneficial bacterial genera in the intestinal tract. Lactobacillus is a well-known beneficial genus in the intestine and has a protective effect on the gut in colitis (34, 35). The abundance of Lactobacillus was significantly reduced in the DSS group relative to the control group, and almost disappeared after the use of arecoline. Furthermore, the abundance of Limosilactobacillus (36, 37) and Ligilactobacillus (38, 39), which have antimicrobial and immune as well as intestinal barrier-enhancing abilities, were inhibited by DSS. Limosilactobacillus has been shown to influence the NF-κB signaling pathway to inhibit inflammation and prevent the development and progression of colitis (40). In addition, Limosilactobacillus also increases the induction and production of PD-1+ T follicular helper cell-dependent IgA, which alters the gut microbiome and prevents DSS-induced colitis and intestinal ecological dysbiosis (41). These results suggest that arecoline further exacerbates DSS-induced colitis by decreasing the abundance of beneficial bacterial genera in the mouse intestine, thereby exacerbating the development of colitis.
In addition, the relative abundance of Bacteroides in the intestine of DSS-treated mice was significantly increased in our assay, which is consistent with previous results of studies regarding the potential role of symbiotic Bacteroides in the induction of colonic inflammation in mice (42). It has also been reported that enterotoxigenic Bacteroides fragilis (ETBF) in Bacteroides increases the permeability of intestinal epithelial cells and disrupts epithelial barrier function by secreting enterotoxins, while ETBF colonization leads to acute or chronic inflammation of the intestine (43, 44). Therefore, based on our experimental results, it can be inferred that an increase in the relative abundance of Bacteroides may lead to the development of UC. The abundance of Odoribacter significantly increased with the use of DSS in our study, which was contrary to previous studies which reported that Odoribacter was more abundant in healthy individuals than in those with colitis (45). However, there are also many studies suggesting that Odoribacter may act as a conditional pathogenic agent and that its increased relative abundance may be positively associated with the occurrence of UC (46, 47). These results suggest that arecoline increases the relative abundance of conditionally pathogenic bacteria, thereby exacerbating DSS-induced colitis.
We analyzed the main components of intestinal microbiota of the three groups of mice by LEfSe analysis, in which the probiotic Lactobacillus obtained a higher score in the control group and the conditional pathogenic bacteria Bacteroides was higher in the Arec group. These results further corroborate our inference.
Intestinal microbiota can regulate immunity, maintain the intestinal environment, and provide energy to the body through its metabolites (48). For example, tryptophan and its metabolites can alleviate intestinal inflammation by reducing pro-inflammatory factors such as IL-22, and IL-17 (49). The production of SCFAs is essential for intestinal integrity, as they regulate intestinal pH and influence intestinal mucus production, thereby increasing mucosal immune function (50). In our study, metabolites such as ergothioneine, pentostatin, LPC (20:1/0:0), Glu-glu, and diadenosine tetraphosphate were reduced after arecoline treatment when compared with the DSS group. Ergothioneine is a common oxidant with strong antioxidant activity. It has been shown that oral administration of ergothioneine can increase the level of anti-inflammatory factors in the body and inhibit the TLR4/MyD88/NF-κB signaling pathway, thus protecting from colonic shortening and intestinal pathological damage in UC (51). Ergothioneine is thought to be a metabolite of the probiotic bacterium Limosilactobacillus, and the elevation of ergothioneine was positively correlated with Limosilactobacillus abundance (52). This is also consistent with the results of our combined gut microbial and metabolite analysis, wherein the relative population of Limosilactobacillus in the Arec group nearly disappeared, and arecoline may have reduced the amount of ergothioneine in mice by reducing the abundance of Limosilactobacillus. Pentostatin is a purine antimetabolite commonly used in the treatment of malignancies, which inhibits adenosine deaminase. Studies have shown that pentostatin can affect the release of pro-inflammatory factors and attenuate the effects of IL-10-/colitis (53). Our combined microbial and metabolite report showed that pentostatin levels increased with Proteobacteria and Firmicutes and decreased with Bacteroidota. Therefore, the decrease in pentostatin likely indicates that arecoline treatment increased the abundance of Bacteroidota and decreased the abundance of Proteobacteria. Diadenosine tetraphosphate is a metabolite produced under stressful conditions such as hypoxia and injury, and is widely found in prokaryotic and eukaryotic organisms for its role in regulating immune responses, gene expression repair, and DNA replication and synthesis (54). When DSS induced stress in mice, diadenosine tetraphosphate was secreted. However, the use of arecoline reduced the serum levels of diadenosine tetraphosphate, and according to our combined microbial and metabolite analysis report, it is possible that arecoline reduced the abundance of Allobaculum and Lactobacillus and increased the abundance of Bacteroides, resulting in a decrease of diadenosine tetraphosphate in mice. According to the metabolic pathway enrichment analysis based on the differential metabolite results, arecoline may aggravate UC in mice by modulating gut microbiota to regulate nicotinate and nicotinamide metabolism, metabolic pathways, and glyoxylate and dicarboxylate metabolism, which are important metabolic pathways. In summary, we suggest that arecoline regulates serum metabolite production by directly affecting the abundance of intestinal microbiota.
Conclusion
Our study shows that arecoline can aggravate the colonic damage of DSS-induced UC and increase the release of inflammatory factors. Based on our gut microbial sequencing results, we can conclude that UC in arecoline-treated aggravated mice may be mediated by modulation of the gut microbiota resulting in a decrease in the abundance of beneficial intestinal genera and an increase in the abundance of conditionally pathogenic bacteria. Moreover, the combined microbial and metabolite analysis showed that gut microbes were significantly associated with differential serum metabolites, and arecoline exacerbated UC in mice by affecting the abundance of intestinal flora that regulated serum metabolite concentrations. However, the exact mechanism of arecoline action needs to be further confirmed by targeted metabolomics. In addition, further studies are needed to investigate the effects of arecoline on patients with inflammatory bowel disease.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA977598.
Ethics statement
The animal study was reviewed and approved by The Institutional Animal Care and Use Committee of Nanchang Royo Biotech Co., Ltd.
Author contributions
DL and BY supervised the project and designed this study. HZ and TD performed the experiments and organized the manuscript. YC and WY conducted the data analysis. JR revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81760731, 81760587), the Jiangxi province Health Commission (202310886), the Outstanding Young Scientists Fund of Jiangxi Cancer Hospital (2021DYS03), and the Natural Science Foundation of the Jiangxi Province (No.20202BABL206093).
Acknowledgments
We thank Charlesworth (https://www.cwauthors.com.cn/) for editing this manuscript. We also thank Metware Biotechnology Co., Ltd. (Wuhan, China) for providing us with 16S rRNA sequencing and Untargeted Metabolomics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Segal JP, LeBlanc JF, Hart AL. Ulcerative colitis: an update. Clin Med (Lond) (2021) 21(2):135–9. doi: 10.7861/clinmed.2021-0080
2. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol (2015) 12(4):205–17. doi: 10.1038/nrgastro.2015.34
3. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet (2017) 389(10080):1756–70. doi: 10.1016/S0140-6736(16)32126-2
4. Yashiro M. Ulcerative colitis-associated colorectal cancer. World J Gastroenterol (2014) 20(44):16389–97. doi: 10.3748/wjg.v20.i44.16389
5. Shen ZH, Zhu CX, Quan YS, Yang ZY, Wu S, Luo WW, et al. Relationship between intestinal microbiota and ulcerative colitis: mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J Gastroenterol (2018) 24(1):5–14. doi: 10.3748/wjg.v24.i1.5
6. Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut (2018) 67(1):108–19. doi: 10.1136/gutjnl-2016-312135
7. Sang LX, Chang B, Zhang WL, Wu XM, Li XH, Jiang M. Remission induction and maintenance effect of probiotics on ulcerative colitis: a meta-analysis. World J Gastroenterol (2010) 16(15):1908–15. doi: 10.3748/wjg.v16.i15.1908
8. Wu Y, Jha R, Li A, Liu H, Zhang Z, Zhang C, et al. Probiotics (Lactobacillus plantarum HNU082) supplementation relieves ulcerative colitis by affecting intestinal barrier functions, immunity-related gene expression, gut microbiota, and metabolic pathways in mice. Microbiol Spectr (2022) 10(6):e0165122. doi: 10.1128/spectrum.01651-22
9. Xue L, Zhao Y, Wang H, Li Z, Wu T, Liu R, et al. The effects of live and pasteurized akkermansia muciniphila on DSS-induced ulcerative colitis, gut microbiota, and metabolomics in mice. Food Funct (2023) 14(10):4632–46. doi: 10.1039/d2fo03493j
10. Zhou Z, He W, Tian H, Zhan P, Liu J. Thyme (Thymus vulgaris l.) polyphenols ameliorate DSS-induced ulcerative colitis of mice by mitigating intestinal barrier damage, regulating gut microbiota, and suppressing TLR4/NF-κB-NLRP3 inflammasome pathways. Food Funct (2023) 14(2):1113–32. doi: 10.1039/d2fo02523j
11. Parada Venegas D, de la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
12. Rodríguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, et al. Differential intestinal anti-inflammatory effects of lactobacillus fermentum and lactobacillus salivarius in DSS mouse colitis: impact on microRNAs expression and microbiota composition. Mol Nutr Food Res (2017) 61(11):1700144. doi: 10.1002/mnfr.201700144
13. Bales A, Peterson MJ, Ojha S, Upadhaya K, Adhikari B, Barrett B. Associations between betel nut (areca catechu) and symptoms of schizophrenia among patients in Nepal: a longitudinal study. Psychiatry Res (2009) 169(3):203–11. doi: 10.1016/j.psychres.2008.06.006
14. Calogero AE, Kamilaris TC, Gomez MT, Johnson EO, Tartaglia ME, Gold PW, et al. The muscarinic cholinergic agonist arecoline stimulates the rat hypothalamic-pituitary-adrenal axis through a centrally-mediated corticotropin-releasing hormone-dependent mechanism. Endocrinology (1989) 125(5):2445–53. doi: 10.1210/endo-125-5-2445
15. Hsu HF, Tsou TC, Chao HR, Shy CG, Kuo YT, Tsai FY, et al. Effects of arecoline on adipogenesis, lipolysis, and glucose uptake of adipocytes-a possible role of betel-quid chewing in metabolic syndrome. Toxicol Appl Pharmacol (2010) 245(3):370–7. doi: 10.1016/j.taap.2010.04.008
16. Hosein M, Mohiuddin S, Fatima N. Association between grading of oral submucous fibrosis with frequency and consumption of areca nut and its derivatives in a wide age group: a multi-centric cross sectional study from Karachi, Pakistan. J Cancer Prev (2015) 20(3):216–22. doi: 10.15430/JCP.2015.20.3.216
17. Prabhu RV, Prabhu V, Chatra L, Shenai P, Suvarna N, Dandekeri S. Areca nut and its role in oral submucous fibrosis. J Clin Exp Dent (2014) 6(5):e569–75. doi: 10.4317/jced.51318
18. Chang NW, Pei RJ, Tseng HC, Yeh KT, Chan HC, Lee MR, et al. Co-Treating with arecoline and 4-nitroquinoline 1-oxide to establish a mouse model mimicking oral tumorigenesis. Chem Biol Interact (2010) 183(1):231–7. doi: 10.1016/j.cbi.2009.10.005
19. Cheng SJ, Ko HH, Cheng SL, Lee JJ, Chen HM, Chang HH, et al. Arecoline-stimulated placenta growth factor production in gingival epithelial cells: modulation by curcumin. Oral Dis (2013) 19(5):513–8. doi: 10.1111/odi.12034
20. Haque MF, Meghji S, Khitab U, Harris M. Oral submucous fibrosis patients have altered levels of cytokine production. J Oral Pathol Med (2000) 29(3):123–8. doi: 10.1034/j.1600-0714.2000.290304
21. Liu YJ, Peng W, Hu MB, Xu M, Wu CJ. The pharmacology, toxicology and potential applications of arecoline: a review. Pharm Biol (2016) 54(11):2753–60. doi: 10.3109/13880209.2016.1160251
22. Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nat Protoc (2007) 2(3):541–6. doi: 10.1038/nprot.2007.41
23. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell (2015) 161(2):264–76. doi: 10.1016/j.cell.2015.02.047
24. Reinhardt C, Bergentall M, Greiner TU, Schaffner F, Ostergren-Lundén G, Petersen LC, et al. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature (2012) 483(7391):627–31. doi: 10.1038/nature10893
25. Gomaa EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek (2020) 113(12):2019–40. doi: 10.1007/s10482-020-01474-7
26. Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol (2020) 145(1):16–27. doi: 10.1016/j.jaci.2019.11.003
27. Ohkusa T, Koido S. Intestinal microbiota and ulcerative colitis. J Infect Chemother (2015) 21(11):761–8. doi: 10.1016/j.jiac.2015.07.010
28. Friedrich M, Pohin M, Powrie F. Cytokine networks in the pathophysiology of inflammatory bowel disease. Immunity (2019) 50(4):992–1006. doi: 10.1016/j.immuni.2019.03.017
29. Billmeier U, Dieterich W, Neurath MF, Atreya R. Molecular mechanism of action of anti-tumor necrosis factor antibodies in inflammatory bowel diseases. World J Gastroenterol (2016) 22(42):9300–13. doi: 10.3748/wjg.v22.i42.9300
30. Torretta S, Scagliola A, Ricci L, Mainini F, Di Marco S, Cuccovillo I, et al. D-mannose suppresses macrophage IL-1β production. Nat Commun (2020) 11(1):6343. doi: 10.1038/s41467-020-20164-6
31. Gonzalez CG, Mills RH, Zhu Q, Sauceda C, Knight R, Dulai PS, et al. Location-specific signatures of crohn’s disease at a multi-omics scale. Microbiome (2022) 10(1):133. doi: 10.1186/s40168-022-01331-x
32. Schirmer M, Denson L, Vlamakis H, Franzosa EA, Thomas S, Gotman NM, et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe (2018) 24(4):600–610.e4. doi: 10.1016/j.chom.2018.09.009
33. Kabeerdoss J, Jayakanthan P, Pugazhendhi S, Ramakrishna BS. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res (2015) 142(1):23–32. doi: 10.4103/0971-5916.162091
34. Wu J, Wei Z, Cheng P, Qian C, Xu F, Yang Y, et al. Rhein modulates host purine metabolism in intestine through gut microbiota and ameliorates experimental colitis. Theranostics (2020) 10(23):10665–79. doi: 10.7150/thno.43528
35. Matos RC, Leulier F. Lactobacilli-host mutualism: “learning on the fly”. Microb Cell Fact. (2014) 13 Suppl 1(Suppl 1):S6. doi: 10.1186/1475-2859-13-S1-S6
36. Walter J, Britton RA, Roos S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the lactobacillus reuteri paradigm. Proc Natl Acad Sci U S A. (2011) 108 Suppl 1(Suppl 1):4645–52. doi: 10.1073/pnas.1000099107
37. Piccioni A, Franza L, Vaccaro V, Saviano A, Zanza C, Candelli M, et al. Microbiota and probiotics: the role of limosilactobacillus reuteri in diverticulitis. Medicina (Kaunas) (2021) 57(8):802. doi: 10.3390/medicina57080802
38. Guerrero Sanchez M, Passot S, Campoy S, Olivares M, Fonseca F. Ligilactobacillus salivarius functionalities, applications, and manufacturing challenges. Appl Microbiol Biotechnol (2022) 106(1):57–80. doi: 10.1007/s00253-021-11694-0
39. Yakabe K, Higashi S, Akiyama M, Mori H, Murakami T, Toyoda A, et al. Dietary-protein sources modulate host susceptibility to clostridioides difficile infection through the gut microbiota. Cell Rep (2022) 40(11):111332. doi: 10.1016/j.celrep.2022.111332
40. Liu B, Yang L, Wu Y, Zhao X. Protective effect of Limosilactobacillus fermentum HFY06 on dextran sulfate sodium-induced colitis in mice. Front Microbiol (2022) 13:935792. doi: 10.3389/fmicb.2022.935792
41. Liu HY, Giraud A, Seignez C, Ahl D, Guo F, Sedin J, et al. Distinct b cell subsets in peyer’s patches convey probiotic effects by limosilactobacillus reuteri. Microbiome (2021) 9(1):198. doi: 10.1186/s40168-021-01128-4
42. Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, et al. Commensal bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe (2011) 9(5):390–403. doi: 10.1016/j.chom.2011.04.009
43. Zamani S, Hesam Shariati S, Zali MR, Asadzadeh Aghdaei H, Sarabi Asiabar A, Bokaie S, et al. Detection of enterotoxigenic Bacteroides fragilis in patients with ulcerative colitis. Gut Pathog (2017) 9:53. doi: 10.1186/s13099-017-0202-0
44. Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of bacteroides and prevotella spp. ulcerative colitis J Med Microbiol (2006) 55(Pt 5):617–24. doi: 10.1099/jmm.0.46198-0
45. Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SR, et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature (2019) 569(7758):663–71. doi: 10.1038/s41586-019-1236-x
46. Wei D, Xie L, Zhuang Z, Zhao N, Huang B, Tang Y, et al. Gut microbiota: a new strategy to study the mechanism of electroacupuncture and moxibustion in treating ulcerative colitis. Evid Based Complement Alternat Med (2019) 2019:9730176. doi: 10.1155/2019/9730176
47. Li W, Sun Y, Dai L, Chen H, Yi B, Niu J, et al. Ecological and network analyses identify four microbial species with potential significance for the diagnosis/treatment of ulcerative colitis (UC). BMC Microbiol (2021) 21(1):138. doi: 10.1186/s12866-021-02201-6
48. Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol (2014) 10(6):416–24. doi: 10.1038/nchembio.1535
49. Sun M, Ma N, He T, Johnston LJ, Ma X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit Rev Food Sci Nutr (2020) 60(10):1760–8. doi: 10.1080/10408398.2019.1598334
50. Blaak EE, Canfora EE, Theis S, Frost G, Groen AK, Mithieux G, et al. Short chain fatty acids in human gut and metabolic health. Benef Microbes (2020) 11(5):411–55. doi: 10.3920/BM2020.0057
51. Pang L, Wang T, Liao Q, Cheng Y, Wang D, Li J, et al. Protective role of ergothioneine isolated from pleurotus ostreatus against dextran sulfate sodium-induced ulcerative colitis in rat model. J Food Sci (2022) 87(1):415–26. doi: 10.1111/1750-3841.15982
52. Matsuda Y, Ozawa N, Shinozaki T, Wakabayashi KI, Suzuki K, Kawano Y, et al. Ergothioneine, a metabolite of the gut bacterium lactobacillus reuteri, protects against stress-induced sleep disturbances. Transl Psychiatry (2020) 10(1):170. doi: 10.1038/s41398-020-0855-1
53. Brown JB, Lee G, Grimm GR, Barrett TA. Therapeutic benefit of pentostatin in severe IL-10-/- colitis. Inflammation Bowel Dis (2008) 14(7):880–7. doi: 10.1002/ibd.20410
Keywords: arecoline, ulcerative colitis, gut microbiota, metabolomics, inflammatory responses
Citation: Zhao H, Ding T, Chen Y, Yang W, Rao J, Liu D and Yi B (2023) Arecoline aggravates acute ulcerative colitis in mice by affecting intestinal microbiota and serum metabolites. Front. Immunol. 14:1197922. doi: 10.3389/fimmu.2023.1197922
Received: 31 March 2023; Accepted: 23 June 2023;
Published: 10 July 2023.
Edited by:
Natividad Garrido Mesa, Kingston University, United KingdomReviewed by:
Kaijian Hou, Shantou University, ChinaKumaraswamy Naidu Chitrala, University of Houston, United States
Copyright © 2023 Zhao, Ding, Chen, Yang, Rao, Liu and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Liu, bGl1ZGFuMTIwMWp4QG5jdS5lZHUuY24=; Bo Yi, eWlibzc5MDUwOEAxNjMuY29t
 Hu Zhao
Hu Zhao Taohui Ding1,2
Taohui Ding1,2 Wenbin Yang
Wenbin Yang Jun Rao
Jun Rao Dan Liu
Dan Liu