- 1Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, China
- 2International Research Center for Marine Biosciences, Ministry of Science and Technology, Shanghai Ocean University, Shanghai, China
- 3College of Animal Science and Technology, Guangxi University, Nanning, China
- 4SinoNorway Fish Gastrointestinal Microbiota Joint Lab, Feed Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
P2X receptors, including seven subtypes, i.e., P2X1-7, are the ligand-gated ion channels activated by the extracellular ATP playing the critical roles in inflammation and immune response. Even though the immune functions of P2X receptors have been characterized extensively in mammals, their functions in fish remain largely unknown. In this study, four P2X receptor homologues were characterized in spotted sea bass (Lateolabrax maculatus), which were named LmP2X2, LmP2X4, LmP2X5, and LmP2X7. Their tissue distributions and expression patterns were then investigated by real-time quantitative PCR (qPCR). Furthermore, their functions in regulating the expressions of inflammation-associated genes and possible signaling pathway were examined by qPCR and luciferase assay. The results showed that they share similar topological structures, conserved genomic organization, and gene synteny with their counterparts in other species previously investigated. And the four P2X receptors were expressed constitutively in the tested tissues. In addition, the expression of each of the four receptor genes was significantly induced by stimulation of Edwardsiella tarda and/or pathogen-associated molecular patterns (PAMPs) in vivo. Also, in primary head kidney leukocytes of spotted sea bass, LmP2X2 and LmP2X5 were induced by using PAMPs and/or ATP. Notably, the expressions of CCL2, IL-8, and TNF-α recognized as the pro-inflammatory cytokines, and of the four apoptosis-related genes, i.e., caspase3, caspase6, caspase7, and P53, were differentially upregulated in the HEK 293T cells with over-expressed LmP2X2 and/or LmP2X7 following ATP stimulation. Also, the over-expression of LmP2X4 can upregulate the expressions of IL-8, caspase6, caspase7, and P53, and LmP2X5 upregulates of IL-8, TNF-α, caspase7, and P53. Then in the present study it was demonstrated that the activation of any one of the four receptors significantly upregulated the activity of NF-κB promoter, suggesting that the activated LmP2Xs may regulate the expressions of pro-inflammatory cytokines via the NF-κB pathway. Taken together, the four P2X receptors were identified firstly from fish species in Perciformes, and they participate in innate immune response of spotted sea bass possibly by regulating the expressions of the inflammation-related genes. Our study provides the new evidences for the P2X receptors’ involvement in fish immunity.
1 Introduction
Adenosine triphosphate (ATP) is well known as the universal energy currency in cellular metabolism (1). However, there are growing evidences that ATP is released from activated or stressed cells due to inflammation, body injury, and/or apoptosis to perform its immune functions (1–3). Extracellular ATP is a kind of damage-associated molecular patterns (DAMPs), involved in activating the innate immune defense mechanisms, inflammasome activation, phagocytosis, etc. (1, 4, 5). Importantly, these functions are performed by extracellular ATP via the activation of P2 receptors on the surface of cell (5, 6). P2 receptors can be classified into two subfamilies, i.e. P2X as ligand-gated ion channels and P2Y being G protein-coupled receptors (1, 7). P2X receptors consist of 7 members (P2X1-7), forming either homo- or hetero-trimeric channels (8, 9). All the P2Xs share similar topological structures, including an extracellular (ECL) region, two transmembrane (TM) domains, and both N- and C-terminal regions in the cytosol (7, 10). Differently, although their natural agonist is extracellular ATP, each P2X receptor has a different affinity for ATP from the micromolar to the millimolar levels (7).
In mammals, P2Xs are widely expressed in immune cells, such as macrophages, T cells, and B cells, to participate in various immune responses, including the release of cytokines, apoptosis, NF-κB activation, etc. (1, 4, 5, 11). Among the P2X subtypes, as the essential moderator of both inflammation and immunity, P2X7 has received the most attention (12), which has been shown to extensively engage in innate and adaptive immunity (6, 12–17). Besides P2X7, P2X2 and P2X4 were also involved in the release of pro-inflammatory cytokines, T cell activation, and the differentiation of CD4+ T cells (4, 7, 18–23). P2X2/3 or P2X3 activation is associated with inflammatory hyperalgesia (24), and P2X5 is required for Listeria monocytogenes-induced inflammasome activation and IL-1β production (25). In summary, A large number of studies have indicated that P2X receptors play important roles in inflammation and immune response in mammals.
Compared to mammals, evidences for the involvement of P2X receptors in fish inflammation and immunity are still limited. To date, six P2X receptors except P2X6 have been cloned in zebrafish (Danio rerio) (26, 27) and some P2X receptors have also been identified in other fish species, such as P2X7 in gilthead seabream (Sparus aurata L.) (28) and ayu (Plecoglossus altivelis) (29), and P2X2, P2X4, and P2X7 in Japanese flounder (Paralichthys olivaceus) (30–32). Similarly, P2X7 of fish was relatively more studied, which was proved to play a critical role in innate immunity (28, 33). For instances, P2X7 of Japanese flounder was involved in innate immune response by moderating the expressions of pro-inflammatory cytokines (30), and P2X7 in ayu macrophages could mediate cell death, phagocytosis, and bacterial killing (29). In addition, P2X2 and P2X4 in Japanese flounder have been proved to take part in the innate immunity (31, 32). To our knowledge, so for there has been no report on the immunological function of P2X5 in fish. Therefore, the immune functions of P2X receptors in fish need to be extensively studied. Our previous study has indicated that ATP can be released from cells through pannexin1, connexin32, and connexin43 channels in inflammatory situation in spotted sea bass (Lateolabrax maculatus) (34). Therefore, it is needed to explore further the functions of extracellular ATP in spotted sea bass. In this study, we characterized four P2X receptors in spotted sea bass, which was their first identification in Perciformes. Their tissue distributions and the regulation of expressions by Edwardsiella tarda infection and PAMPs stimulation were then investigated. Furthermore, their functions, particularly for P2X5 unnoticed previously in fish, in regulating the expressions of inflammation-associated genes and possible signaling pathway were examined. Undoubtedly, this study will be helpful in enriching and further understanding the innate immune response mediated by extracellular ATP in fish.
2 Materials and methods
2.1 Experimental fish
Spotted sea bass (100 ± 10 g) were sourced from a fish farm in Hangzhou city, Zhejiang province, China. Fish were farmed in a fish aquaculture system at 26 ± 2 °C and fed with commercial feed of sea bass once a day for more than two weeks before experiments. Healthy fish without any pathological signs were then selected for the experiments.
2.2 Cloning of P2Xs from spotted sea bass
According to previously described methods, the first strand cDNA was obtained and the full-length cDNA sequences were then cloned and verified as the method described previously (35). In details, total RNA was extracted from head kidney and liver of spotted sea bass for gene cloning using the TRIzol reagent (Ambion, USA). The first strand cDNA was reverse-synthesized from total RNA using the Hifair® II 1st Strand cDNA Synthesis Kit (gDNA digester plus) (YEASEN, China). For the 5´- and 3´-RACE, nested PCR was performed with Premix Ex Taq™ Hot Start Mix (TaKaRa, Japan). The complete coding sequences were verified by sequencing the PCR products amplified using one pair of primers located at the 5´- and 3´- untranslated region. All primers were summarized in Supplementary Table 1.
2.3 Bioinformatics analysis
Programs on NCBI (https://www.ncbi.nlm.nih.gov/) and Expasy (http://www.expasy.org/tools) website were used to analyze nucleotide & protein sequences and the Ensembl (http://www.ensembl.org/) and NCBI genome database were analyzed to infer the genomic organization & syntenic relationships. Phylogenetic tree and multiple sequence alignment were analyzed by the same methods (35) and then the phylogenetic tree was embellished using the iTOL website (https://itol.embl.de/) (36). Concretely, multiple sequence alignments were performed using the ClustalW program and processed using the GeneDoc program. Phylogenetic tree was constructed by the Neighbour-Joining method using the MEGA X program and the tree were bootstrapped for 10,000 replications.
2.4 Tissue expression analysis
Following the same method, 8 tissue samples including liver, spleen, head kidney, intestine, skin, gill, muscle, and brain of healthy spotted sea bass were obtained for the preparation of assay templates (35, 37). The first strand cDNA was reverse-synthesized from 5 μg total RNA using Hifair® II 1st Strand cDNA Synthesis SuperMix (gDNA digester plus) (YEASEN, China). The real-time quantitative PCR (qPCR) was performed in triplicate using Hieff® qPCR SYBR Green Master Mix (No Rox) (YEASEN, China), and all qPCR primers were shown in Supplementary Table 1. The relative expression levels of target genes were normalized to that of EF1α.
2.5 Expression analysis after in vivo stimulation
For the challenge experiment, fish were i.p. injected with 500 μL Lipopolysaccharides (LPS, 1 mg/mL), Polyinosinic-polycytidylic acid (Poly (I:C), 1 mg/mL), Edwardsiella tarda (1×105 CFU/mL) or phosphate buffered saline (PBS, pH 7.4, Gibco). Tissues were sampled at 6, 12, 24, and 48 h after injection, homogenized in 1 mL TRIzol, and reverse transcribed into cDNA for qPCR analysis as described above (37). E. tarda was cultured in tryptic soy agar medium or tryptic soy broth medium at 28°C and prepared as previously described (37). LPS and Poly (I:C) were purchased from Sigma-Aldrich (USA).
2.6 Expression analysis after in vitro stimulation
Primary head kidney leukocytes of healthy spotted sea bass were isolated by a discontinuous 51% Percoll gradient as previously described (38). The leukocytes (1×107/well) were cultured in a 6-well plate (Corning, USA) with DMEM-F12 medium (Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco) and 1% Pen/Strep (Gibco, USA) in a CO2 incubator at 28 °C, and then treated with 50 μg/mL peptidoglycan (PGN, Sigma-Aldrich), 100 μg/mL LPS, 50 μg/mL Poly (I:C), 100 μM or 1mM ATP (Sigma-Aldrich, USA), an equal volume of PBS was used as the control. Each condition was done in Quadruplicate. Cell samples were collected for qPCR at 6, 12, 24, and 48 h after stimulation. Total RNA was extracted using TRIzol reagent and reverse transcribed into cDNA for qPCR analysis as described above.
2.7 Expression analysis of inflammation-associated genes in LmP2Xs-overexpressing HEK 293T cells after ATP stimulation
Based on the obtained CDS sequences, the recombinant pcDNA3.1 expression plasmid for LmP2Xs were constructed, which were named as pcDNA3.1-LmP2X2, pcDNA3.1-LmP2X4, pcDNA3.1-LmP2X5 and pcDNA3.1-LmP2X7. HEK 293T cells were then transfected with pcDNA3.1-LmP2Xs or pcDNA3.1 using the PEI 40K Transfection Reagent (Servicebio, China) according to the manufacturer’s instruction. The transfected cells were cultured in DMEM basic medium (Gibco, USA) containing 10% FBS and 1% Pen/Strep in a CO2 incubator at 37°C and the medium was replaced with fresh medium after 24 h. The cells were then stimulated with different concentrations of ATP (100 μM, 1 mM, or 5 mM), and were collected for qPCR after 6 h. The empty plasmid transfected cells by PBS stimulation served as the negative control. Each condition was done in triplicate. Total RNA was extracted using TRIzol reagent and reverse transcribed into cDNA for qPCR analysis as described above. All qPCR primers were shown in Supplementary Table 1.
2.8 Luciferase assay
The pcDNA3.1-LmP2Xs plasmid or pcDNA3.1 plasmid was co-transfected with the pGL4.32 (luc2P/NF-κB-RE/Hygro, NF-κB reporter plasmid, Promega) and pRL-TK (renilla luciferase control plasmid, Promega) into HEK 293T cells. The transfected cells were cultured for 24 h, and then were stimulated with ATP (100 μM, 1 mM, or 5 mM) or PBS (control). After 6 h, the cells were collected and then lysed for measuring the luciferase activity by using the dual-luciferase reporter assay system (Promega, USA) on a LumiPro (YPHBIO, China). Each condition was done in triplicate.
2.9 Subcellular localization
The pEGFP-N1 expression plasmid containing the coding sequence (CDS) of LmP2X2, LmP2X4, LmP2X5, or LmP2X7 were constructed, i.e. pEGFP-N1-LmP2X2, pEGFP-N1-LmP2X4, pEGFP-N1-LmP2X5, and pEGFP-N1-LmP2X7. We then transfected the recombinant pEGFP-N1 plasmids into HEK 293T cells by the same methods as before. As described by the method described previously (39), the transfected cells were cultured in coverslips pretreated with poly-L-lysine (Solarbio, China) at 24 h. Next, cells were stained with DAPI (Solarbio, China) and observed using a laser confocal microscope (Leica TCS SP8, Germany).
2.10 Statistical analysis of date
Data were analyzed using the SPSS package 20.0 by one-way ANOVA, “p<0.05” or “p<0.01” is considered significant between treatment groups and control groups (35).
3 Results
3.1 Sequence features of the four P2X receptors in spotted sea bass
The sequences obtained in this study have been deposited in the NCBI database with the following accession numbers: OP651016 (LmP2X2), OP651019 (LmP2X4), OP651020 (LmP2X5), and OP651021 (LmP2X7). Meanwhile, the sequence features of LmP2Xs, including the length of cDNA, 5´-untranslated region (UTR), 3´-UTR, and open reading frame (ORF) were summarized in Table 1. In addition, the cDNA and deduced amino acid sequence information of LmP2Xs was shown in Supplementary Figure 1.
Like other species, multiple sequence alignment revealed that LmP2Xs retain two highly conserved transmembrane domains (TM1 and TM2), a P2X receptor signature motif, and 10 conserved cysteine residues in the extracellular domain; At the same time, they contain the conserved protein kinase C (PKC) site and the YXXXK motif which located in the intracellular N-terminal region and C-terminal region, respectively (Figure 1, Supplementary Figure 2). Compared with other LmP2X members, LmP2X7 has a longer C-terminal region and contains an additional LPS/lipid-binding domain (Figure 1). In addition, the phylogenetic tree was constructed to infer the homologous relationships among P2Xs from various species. As shown in Figure 2, LmP2Xs were clustered with their homologues from other species, well reflecting the phylogeny of the chosen species.
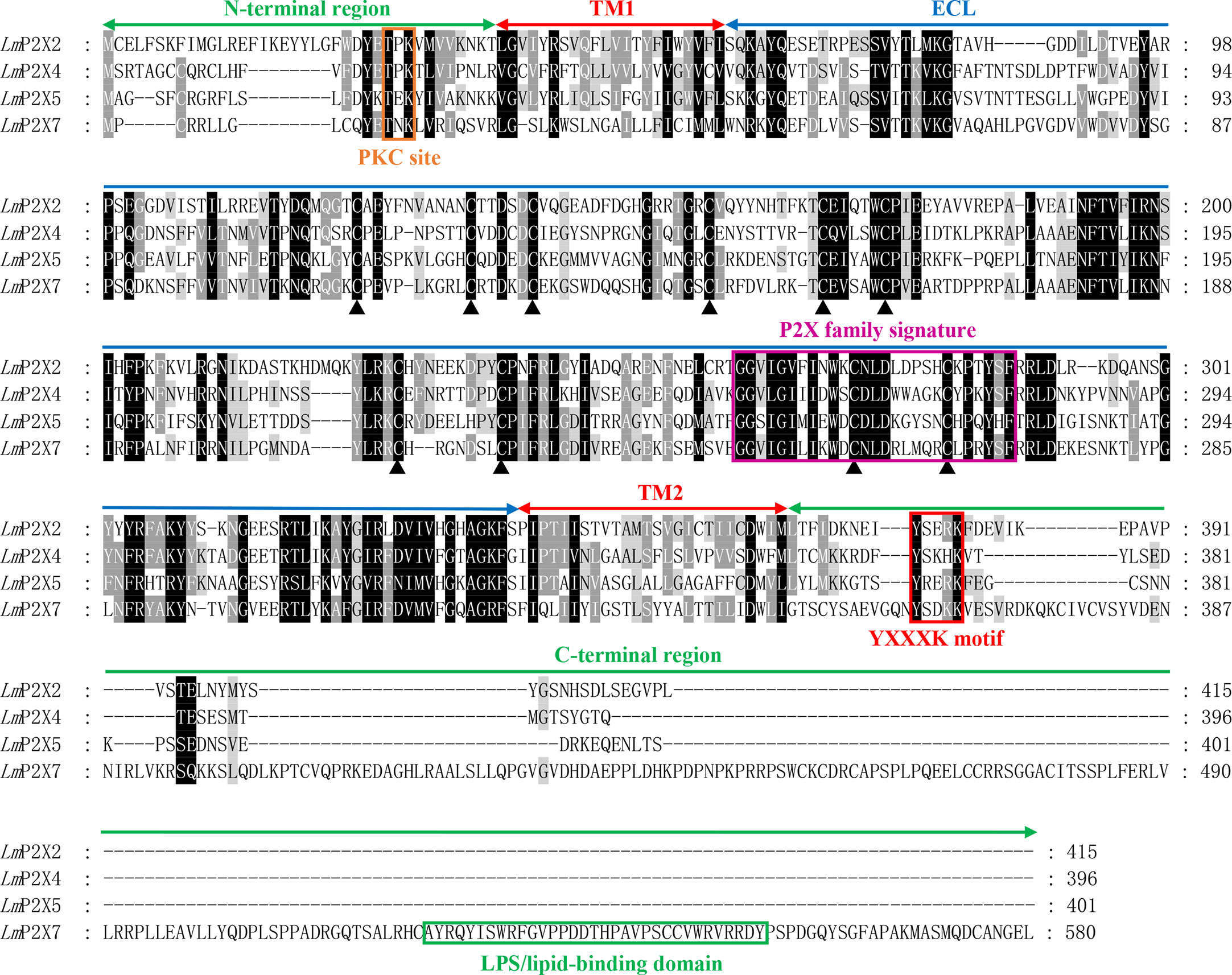
Figure 1 Multiple sequence alignment analysis of P2Xs. The N-terminal region, two transmembrane domains (TM1-2), the extracellular loop (ECL), and the C-terminal region of LmP2Xs are marked above the alignment. Symbol (▴) indicates the conserved cysteine residues. The PKC site, P2X family signature motif, YXXXK motif, and LPS/lipid-binding domain are boxed in orange, pink, red, and green, respectively.
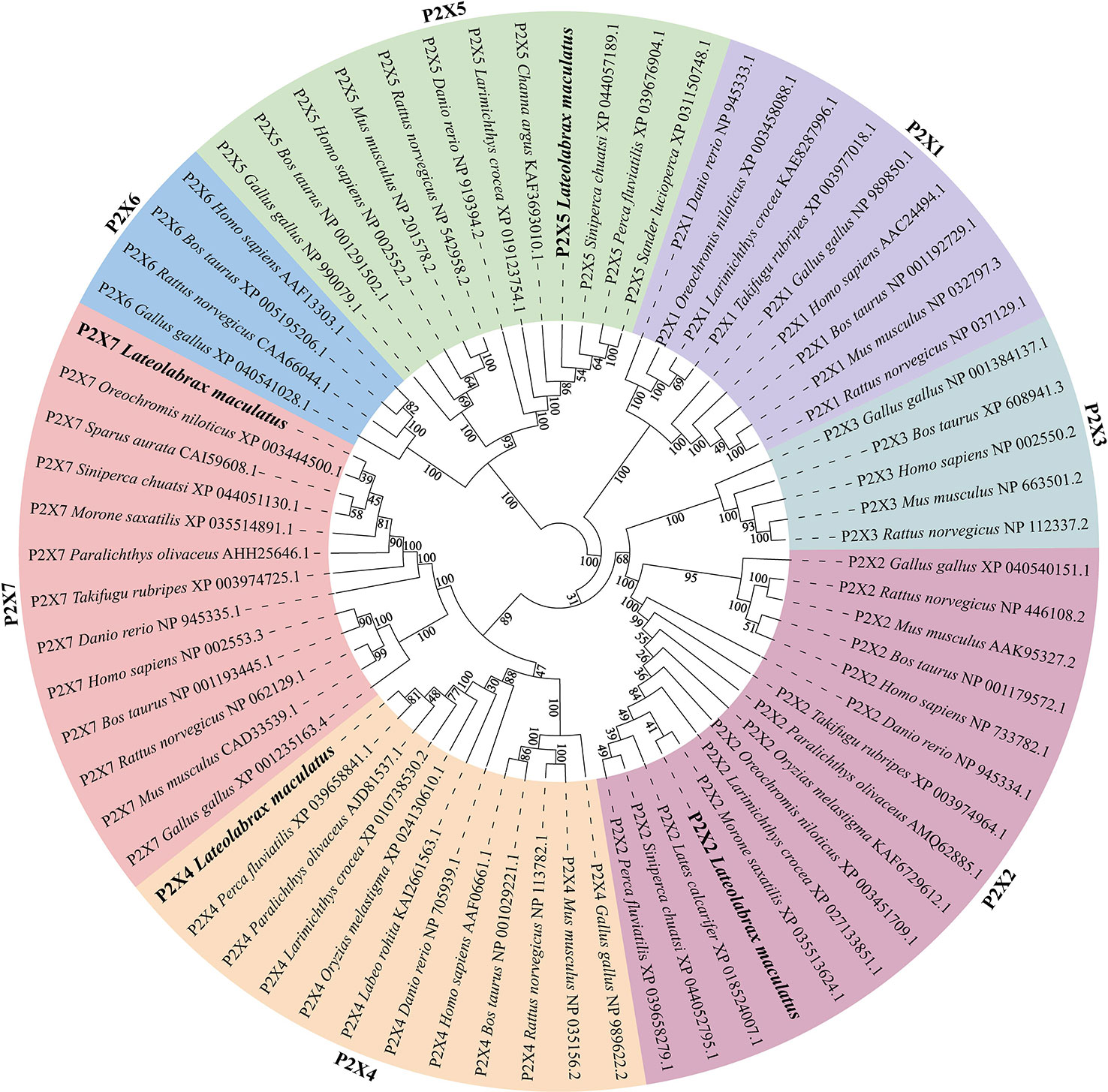
Figure 2 Phylogenetic tree analysis of LmP2Xs with the selected P2X family members. Phylogenetic tree was constructed using the NJ method within the MEGAX program and run for 10,000 replications. The percentages of bootstrap values for branches are indicated and the LmP2Xs are shown in bold.
3.2 Genomic organization and synteny of the four P2X receptors in spotted sea bass
By comparing cDNA sequences with genomic sequences, the genomic organizations of P2Xs were confirmed (Figure 3). The CDS region of all LmP2Xs except LmP2X7 contained 12 exons and 11 introns, and the size of the exon at the same position was comparable. Corresponding to the longer C-terminal region, LmP2X7 contains an additional exon in the C-terminal region. Separately, the number and size of exons of LmP2Xs were highly conserved with their homologous of other species. Overall, the genomic organization of P2Xs was highly conserved in different species. Furthermore, gene synteny showed that the P2X2 and P2X5 locus have been well conserved during evolution, with P2X2 linked to FOXN4 and the P2X5 linked to TRPV1 (Figures 4A, C). On the contrary, the P2X4 and P2X7 locus have been relatively unconserved. Specifically, P2X4 was closely linked to P2X7 in human, mouse, chicken, zebrafish, and spotted gar, but not in fugu, large yellow croaker and spotted sea bass (Figure 4B). However, it is worth noting that P2X4 was linked to GUCD1 and ADORA2AA, and P2X7 was linked to GSTT1A and MMP11A in fish (Figure 4B).
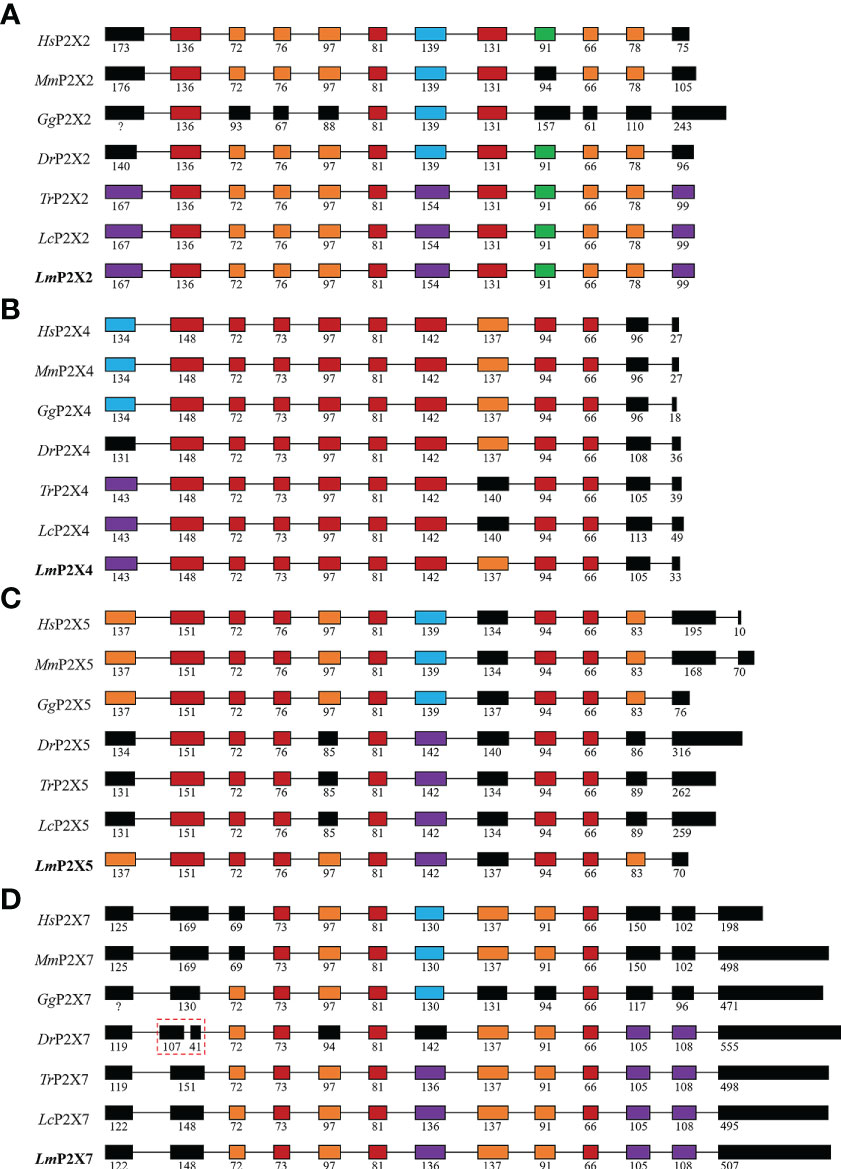
Figure 3 Genomic organization of P2X2 (A), P2X4 (B), P2X5 (C), and P2X7 (D). The solid boxes indicate coding exon. The numbers indicate the size (bp) of exons. Note that the size of exons and introns is disproportionate.
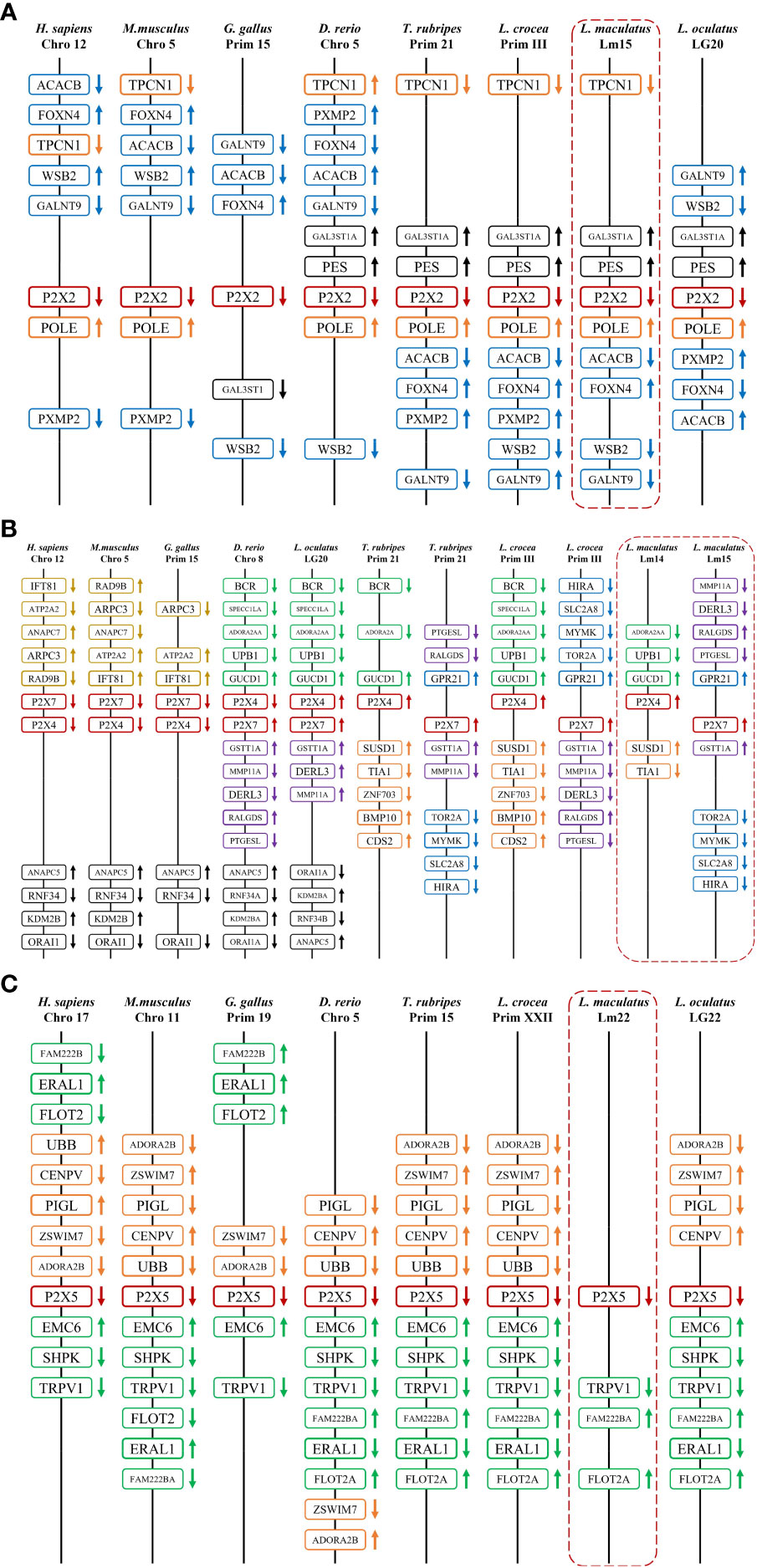
Figure 4 Gene synteny of P2X2 (A), P2X4/7 (B), and P2X5 (C). The LmP2Xs genome sequence data were obtained from the spotted sea bass genome database (https://www.ncbi.nlm.nih.gov/-genome/43909). Synteny information for other vertebrates was retrieved from the Ensembl database (http://www.ensembl.org/index.html). Arrows indicate transcription orientations.
3.3 Tissue distributions of the four P2X receptors in spotted sea bass
Expressions of LmP2X2, LmP2X4, LmP2X5, and LmP2X7 were analyzed in various tissues by qPCR analysis (Figure 5). The results showed that all four LmP2Xs were constitutively expressed in the tissues examined. The highest expressions of LmP2Xs were observed in intestine, gill, and muscle, respectively. In addition, their expressions were lower in spleen and head kidney. Interestingly, the highest expression of LmP2X4 and the lower expressions of other LmP2Xs were both found in gill. Taken together, the differences in the distribution trends of LmP2Xs indicated that they were differentially regulated in eight tissues.
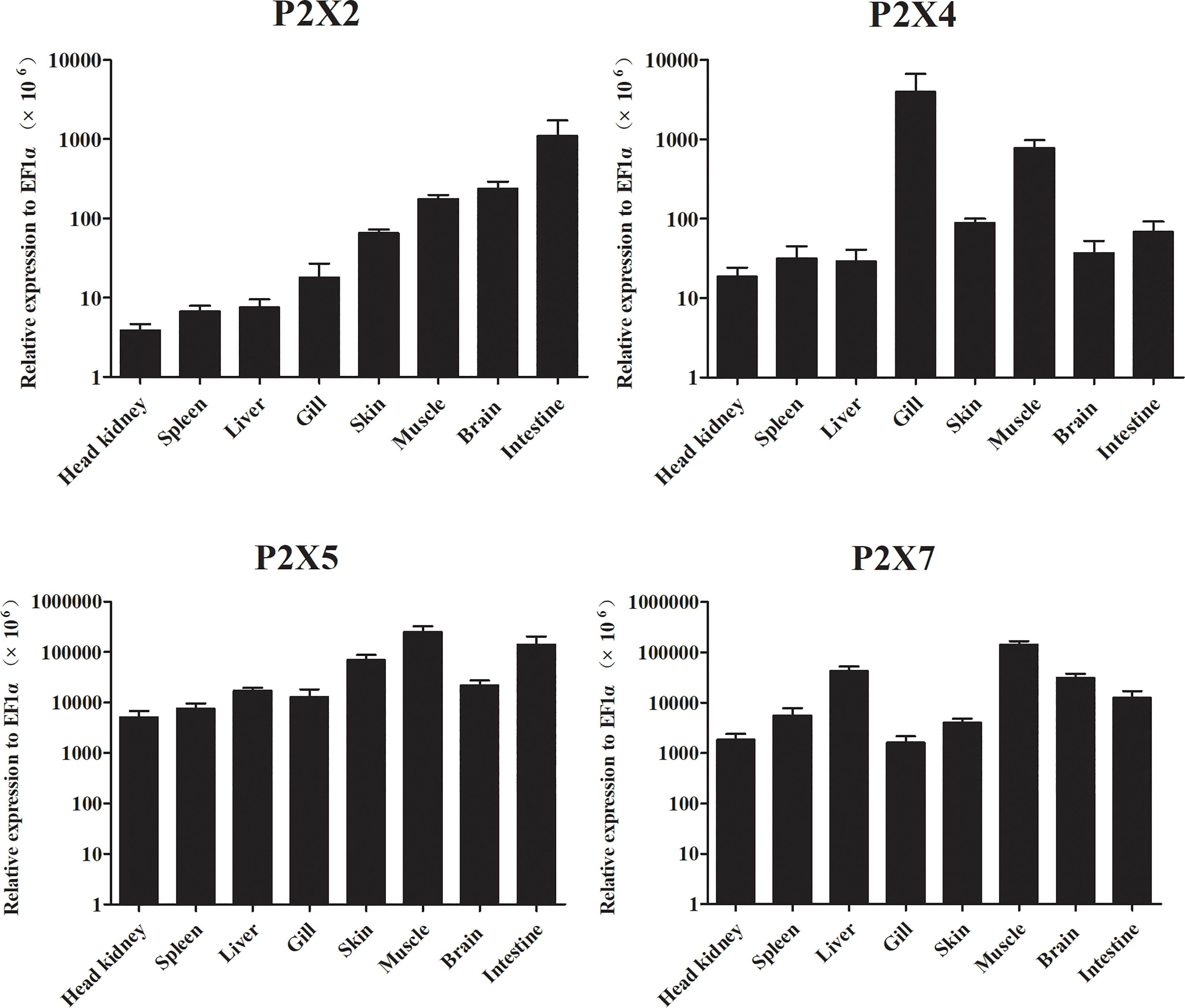
Figure 5 Expression analysis of LmP2Xs in tissues. The expression levels of each gene were normalized to that of EF1α. Data are presented as mean + SEM (N= 4).
3.4 Expressions of the four P2X receptors in spotted sea bass following injection with Poly (I:C), LPS, or Edwardsiella tarda
The mRNA expressions of the four P2X receptors in response to Poly (I:C), LPS, and E. tarda challenge were investigated in intestine, head kidney, spleen, and gill (Figure 6). In intestine, head kidney, and spleen, the expression of LmP2X2 was up-regulated to different degrees after Poly (I:C) and E. tarda stimulation, but down-regulated in intestine after LPS stimulation; In gill, the expression of LmP2X2 was down-regulated after three kinds of stimulation at 24 h and only up-regulated at 6 h after E. tarda stimulation (Figure 6A). As shown in Figure 6B, LmP2X4 were also up-regulated in intestine [after Poly (I:C) and LPS stimulation], gill [only at 24 h after Poly (I:C) stimulation], head kidney, and spleen. Noteworthy, LmP2X5 was induced in all examined tissues after the three kinds of stimulation, except in gill after LPS stimulation where no significant changes were observed (Figure 6C). Like LmP2X5, the expression of LmP2X7 was up-regulated to different degrees in all tissues after Poly (I:C) stimulation, furthermore, LPS and E. tarda stimulation also induced LmP2X7 in head kidney and gill (Figure 6D). Collectively, all the genes were up-regulated to different degrees after stimulation, but their expression patterns differed in different tissues.
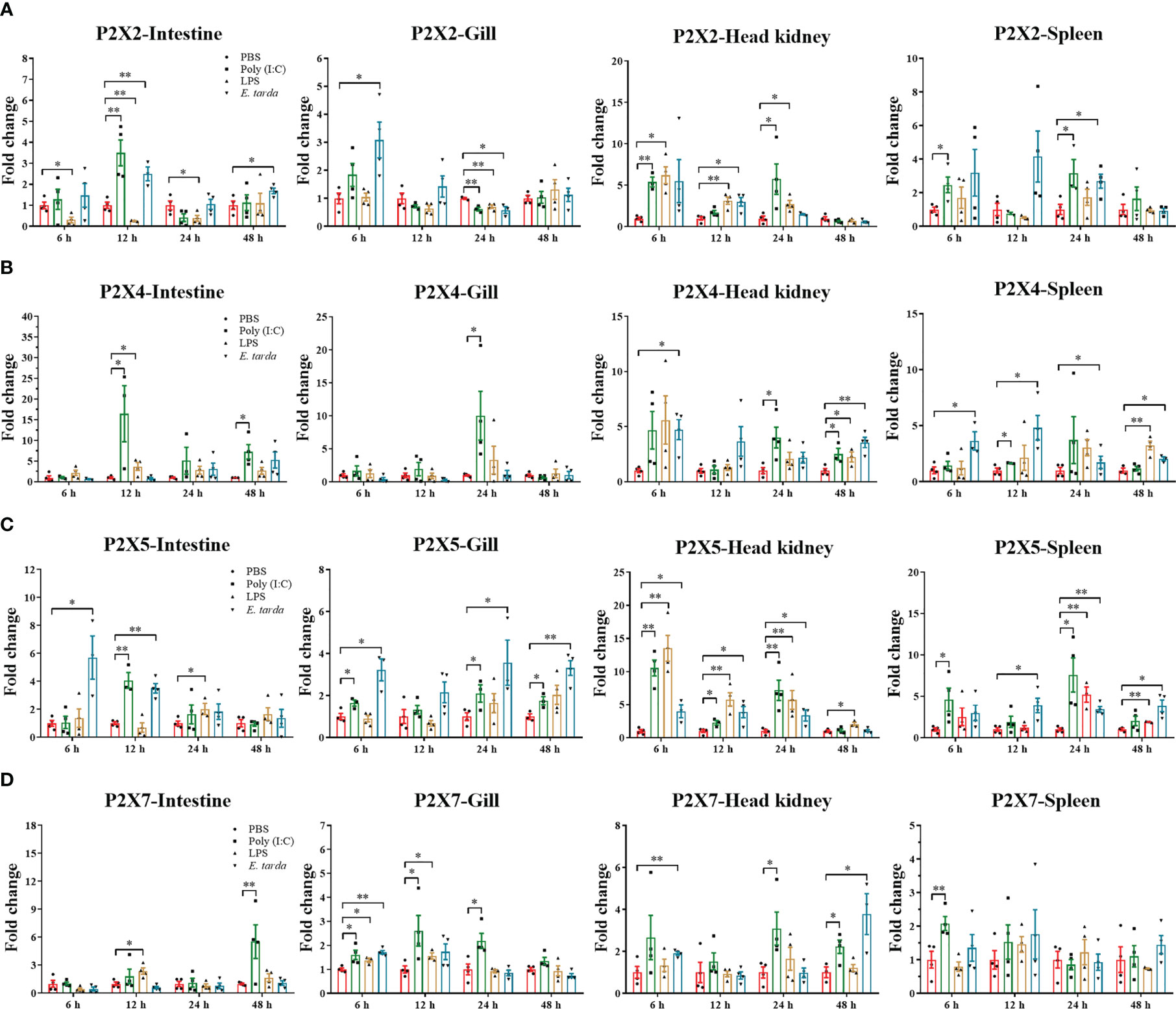
Figure 6 Expressions of LmP2X2 (A), LmP2X4 (B), LmP2X5 (C), and LmP2X7 (D) after LPS, Poly (I:C) or Edwardsiella tarda challenge. Spotted sea bass was i.p. injected with 500 μL LPS (1 mg/mL), Poly (I:C) (1 mg/mL), E tarda (1×105 CFU/mL), or PBS (control) and tissues were sampled at 6, 12, 24 and 48 h for qPCR analysis. The relative expression levels of target genes were normalized to that of EF1α and fold change was obtained by comparing the relative gene expression level of the treated group and control group (defined as 1). Data are shown as mean + SEM (N=3). *P<0.05 and **P<0.01 are considered significant difference.
3.5 Expressions of the four P2X receptors in primary head kidney leucocytes of spotted sea bass after PGN, LPS, Poly (I:C), or ATP stimulation
The expressions of LmP2Xs were investigated in primary head kidney leucocytes in response to PGN, LPS, Poly (I:C), and ATP (Figure 7). PGN, LPS, and Poly (I:C) are the major component of bacterial cell walls, bacterial endotoxin, and viral mimetic respectively, which are used to mimic bacterial and viral-mediated immune responses (37, 40). Here, we found that LmP2X2 and LmP2X5 were induced after all five kinds of stimulation and were most highly up-regulated by ATP stimulation. For LmP2X7, Poly (I:C) stimulation induced its expression at 6 h and 48 h, whereas ATP stimulation inhibited its expression at 6 h. In addition, there were no significant changes in LmP2X4 expression after all the stimulation.
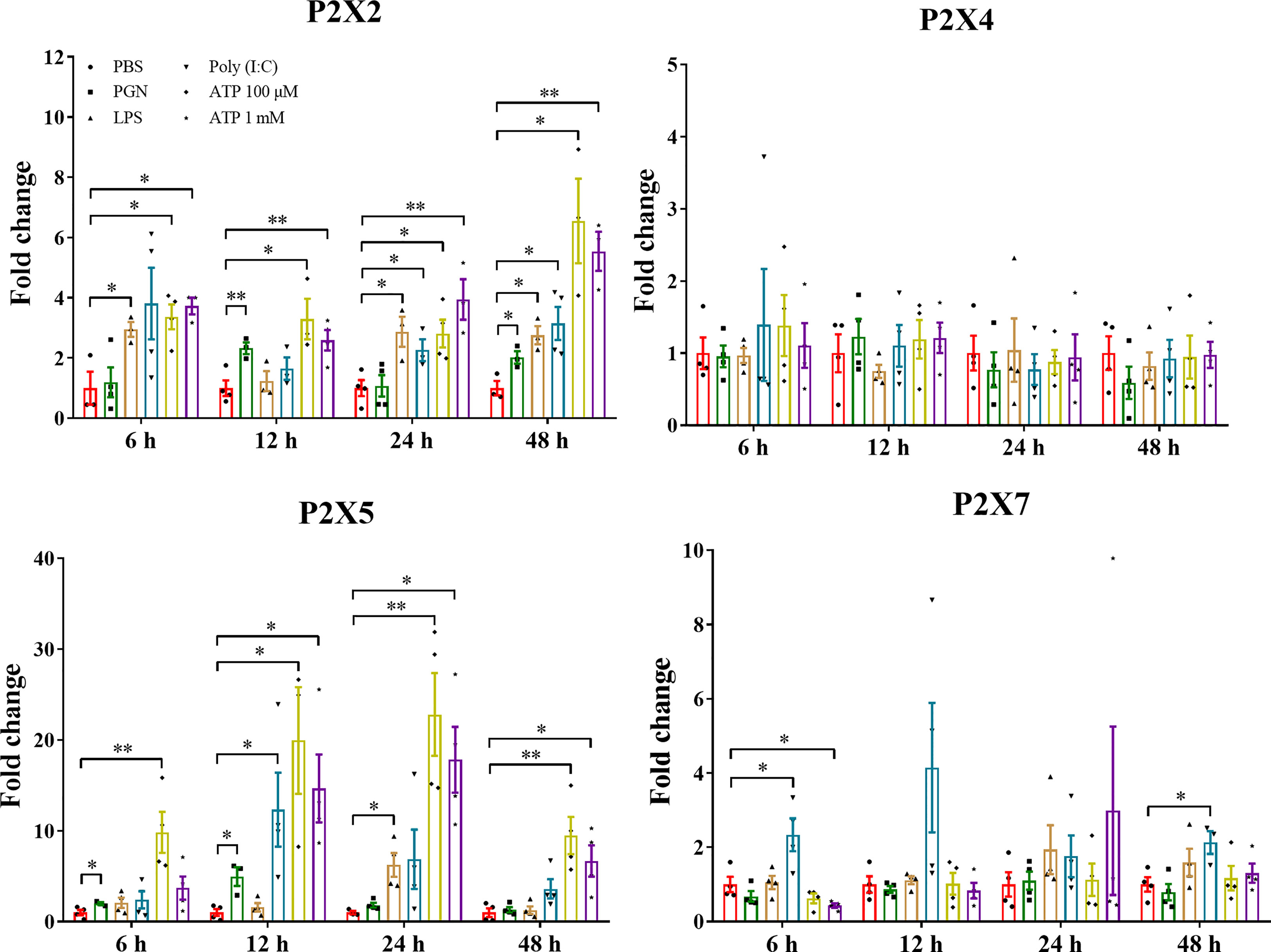
Figure 7 Expressions of LmP2Xs in primary head kidney leucocytes after stimulation with PGN, LPS, poly (I:C) or ATP. Primary head kidney leukocytes were isolated from the spotted sea bass head kidney and stimulated with PGN (50 μg/mL) LPS (100 μg/mL), LPS (100 μg/mL), Poly (I:C) (50 μg/mL), 100 μM ATP, 1 mM ATP, or PBS (control). The total RNA was extracted for qPCR analysis. The relative expression levels of target genes were normalized to that of EF1α and fold change was obtained by comparing the relative gene expression level of the treated group and control group (defined as 1). The data are shown as mean +SEM (N=3). *P<0.05 and **P<0.01 are considered significant difference.
3.6 Subcellular localization of the four P2X receptors
To determine the subcellular localization of LmP2Xs, plasmids pEGFP-N1-LmP2Xs were constructed to express the GFP-tagged LmP2X2, LmP2X4, LmP2X5, and LmP2X7 fusion protein in HEK 293T cells, respectively. As shown in Supplementary Figure 3, in HEK 293T cells transfected with expression plasmids, GFP fluorescence were mainly located on the cell membrane, while in HEK 293T cells transfected with empty plasmids, GFP fluorescence was mainly located in the intracellular area.
3.7 Effect of activated LmP2Xs on the expressions of inflammation-associated genes
The mRNA expressions of pro-inflammatory cytokines (CCL2, IL-8, and TNF-α) and apoptosis-related genes (caspase3, caspase6, caspase7, and P53) were analyzed in LmP2Xs-overexpressing HEK 293T cells after ATP stimulation (Figure 8). Compared to cells transfected with empty plasmids, the mRNA expressions of all 7 genes were significantly upregulated in cells overexpressing LmP2X7, and the mRNA expressions of all genes except caspase6 were also upregulated in cells overexpressing LmP2X2; However, cells overexpressing LmP2X2 were more sensitive to low concentration of ATP, whereas cells overexpressing LmP2X7 were sensitive to high concentration of ATP (Figure 8). Meanwhile, overexpression of LmP2X5 also upregulated the mRNA expressions of IL-8 and TNF-α by different concentrations of ATP stimulation, but overexpression of LmP2X4 only up-regulated the IL-8 mRNA expression following 0.1mM ATP stimulation (Figure 8A). In addition, overexpression of LmP2X4 or LmP2X5 up-regulated the mRNA expressions of caspase7 and P53 upon ATP stimulation, but had no effect on caspase3 (Figure 8B). Taken together, these results demonstrated that activated LmP2Xs can upregulate the expressions of inflammation-associated genes to various degrees.
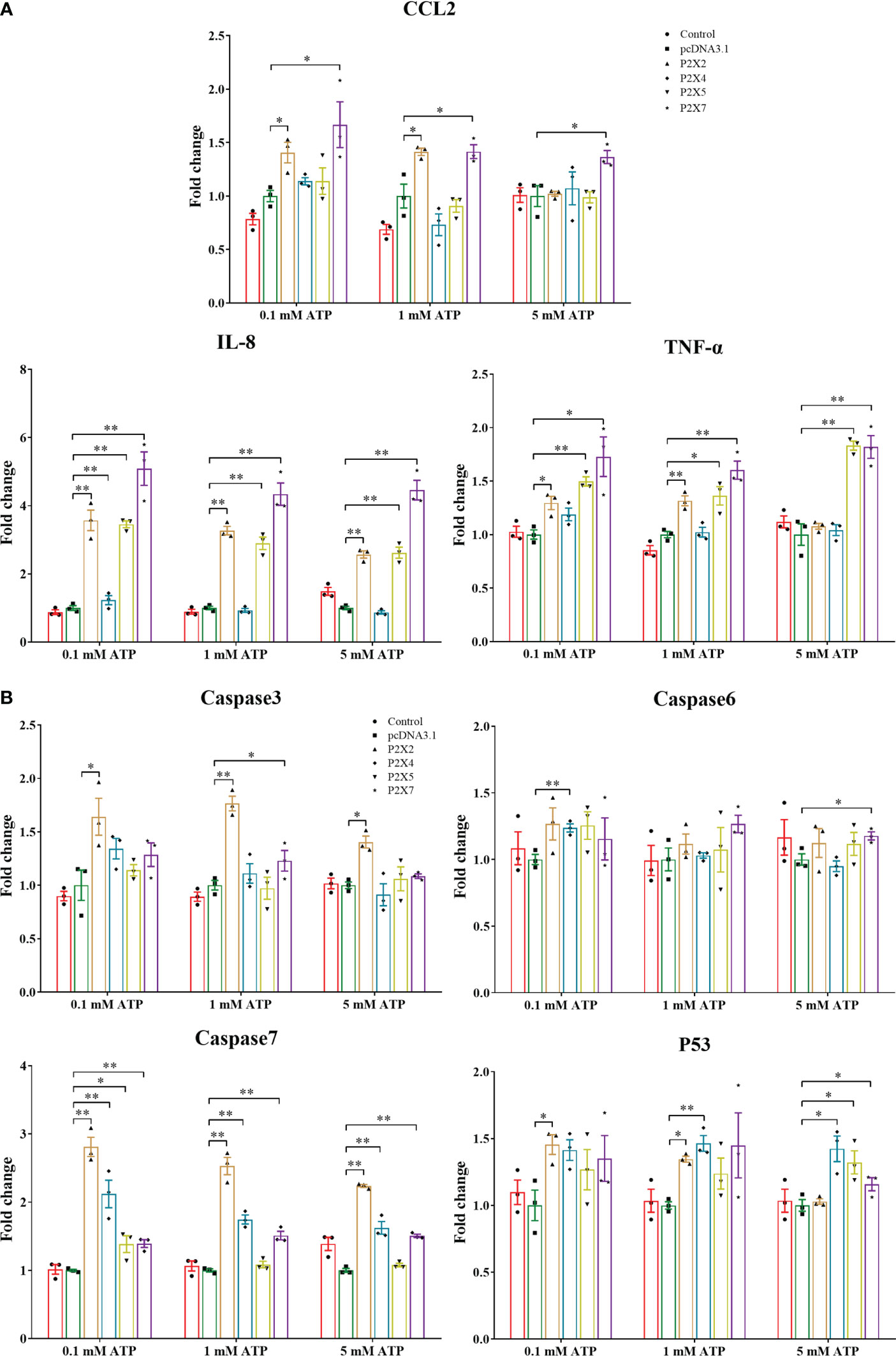
Figure 8 Effect of activated LmP2Xs on expressions of pro-inflammatory cytokines (A) and apoptosis-related genes (B) in the HEK 293T cells. HEK 293T cells were transfected with pcDNA3.1-LmP2Xs or pcDNA3.1. After 24 h, the cells were stimulated with (0.1 mM, 1 mM, or 5 mM) and were collected after 6 h. The empty plasmid transfected cells by PBS stimulation were served as control. The relative expression levels of target genes were normalized to that of βactin and fold change was obtained by comparing the relative gene expression level of the treated group and control group (defined as 1). The data are shown as mean +SEM (N=3). *P<0.05 and **P<0.01 are considered significant difference.
3.8 Effect of activated LmP2Xs on the NF-κB pathway
Luciferase promoter assay was used to investigate the effect of activated LmP2Xs on the NF-κB pathway. As shown in Figure 9, in LmP2Xs-expressing cells, ATP stimulation resulted in significant increases of the luciferase activity compared to the control group (PBS). In detail, the NF-κB promoter activity was significantly up-regulated in LmP2X2 or LmP2X4-overexpressing HEK 293T cells after 1 mM and 5 mM ATP stimulation, in LmP2X5-overexpressing HEK 293T cells after 0.1 mM stimulation and in LmP2X7-overexpressing HEK 293T cells after 5 mM ATP stimulation. By contrast, ATP stimulation had no significant influence on the luciferase activity in pcDNA3.1-expressing cells. Taken together, these results suggested that ATP can activate the NF-κB pathway by interacting with LmP2Xs.
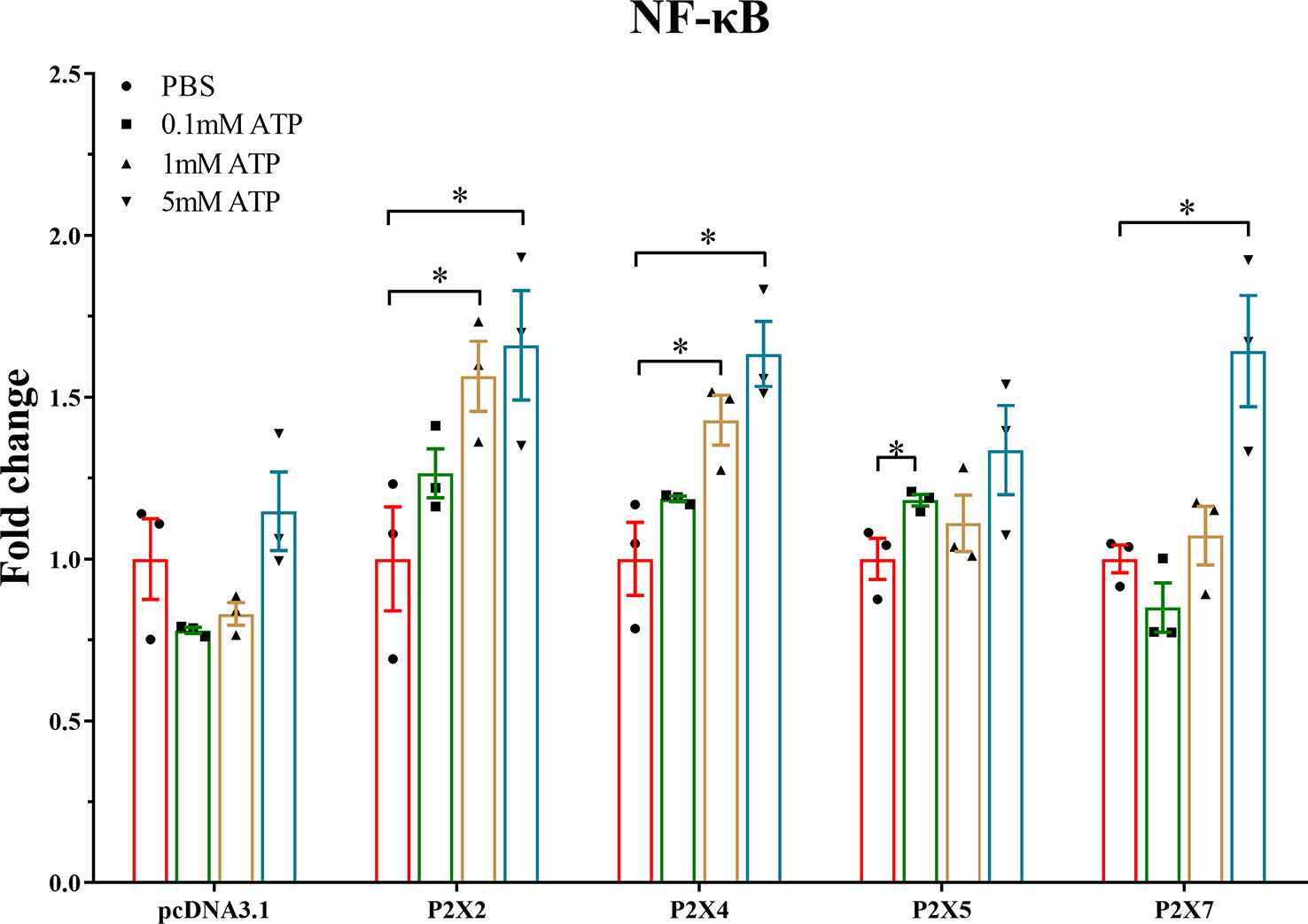
Figure 9 Effect of activated LmP2Xs on activity of NF-κB promoter in HEK 293T cells. The pGL4.32 and pRL-TK (negative control) were co-transfected with pcDNA3.1-LmP2Xs plasmid or pcDNA3.1 plasmid into HEK 293T cells for 24 h. Then the cells were stimulated with ATP (0.1 mM, 1 mM, or 5 mM) or PBS (control) and collected after 6 h. Fold change was obtained by comparing the relative Luciferase activity level of the stimulated group and control group (defined as 1). The data are shown as mean +SEM (N=3). *P<0.05 is considered significant difference.
4 Discussion
As important members of the inflammation and immune response, P2X receptors have been well-studied in mammals (4), however, their immunological functions in fish remain largely unclear. Therefore, their roles in fish inflammation and innate immunity need to be further investigation. In this study, four P2X receptors were characterized in spotted sea bass (Lateolabrax maculatus), which were named LmP2X2, LmP2X4, LmP2X5, and LmP2X7. They share conserved structures, including, a protein kinase C (PKC) site, ten conserved cysteine residues in the extracellular domain, and a YXXXK motif (Figure 1), which are critical for the expression and function of P2X receptors (41–43). Next, we found that they possess conserved genomic organization and gene synteny with their homologues in vertebrates (Figures 3, 4). Structural conservation of the four P2X receptors suggested that they may perform similar functions during evolution. As is known to all, teleost specific whole genome duplication (TSWGD) had resulted in the expansion of gene paralogues (44). Interestingly, although nine genes with high homology for different P2X receptors have been isolated in zebrafish (27, 45), no homologue of P2X6 has been identified in fish to date. Given the limited research on P2X receptors in fish, the presence or absence of P2X6 in teleost fish need to be clarified in future studies.
In healthy spotted sea bass, all four LmP2Xs were constitutively expressed in tissues examined, with lower expressions in head kidney and spleen, even though they are important immune tissues in fish. Similarly, the lower expressions of P2X2 and P2X4 were found in head kidney and spleen of Japanese flounder (31, 32). In addition, P2X7 was also relatively lower expressed in the two tissues of Japanese flounder (30) and gilthead seabream (28). These results suggest a similar expression pattern of P2X receptors in different fish. In addition, their expressions were higher in the mucosal immune tissue intestine or gill, suggesting that they may play an important immune function in defending against pathogenic invasion. On balance, we found that all LmP2X were co-expressed in various tissues, but with tissue preferences, demonstrating that multiple P2X receptors may function synergistically or complementarily, but in given tissues, different P2X receptors may contribute differently in the extracellular ATP-mediated immune response. However, these possibilities need to be further examined in the future. On the other hand, studies in mammals have shown that different P2X receptors are involved in regulating a variety of neural and physiological processes (46). In our study, different LmP2X were expressed at higher levels in brain, muscle or liver, indicating that they might be involved in physiological processes similar to those in mammals.
Previous studies have demonstrated that the expressions of mammalian P2X receptors can be up-regulated in tissues and immune cells after viral or bacterial infection or PAMP or ATP stimulation (4, 5). In fish, P2X2 and P2X4 have also been found significantly up-regulated after various TLR agonists (LPS, Poly (I:C) and zymosan) stimulation and bacterial infection in Japanese flounder (31, 32). In addition, P2X7 expression was up-regulated after bacterial infection and PAMP stimulation in Japanese flounder and ayu (29, 30). The expressions of P2X7-like receptor in the diploblastic coral and the invertebrate chordate amphioxus were also significantly challenged after LPS and ATP stimulation (47). Similarly, in that study, the expressions of all four LmP2Xs were changed significantly by stimulation with LPS, Poly (I:C) or E. tarda infection in vivo, with varying degrees in different tissues (Figure 6). Furthermore, LmP2X2 and LmP2X5 were also induced by stimulation with PAMP and ATP in primary head kidney leukocytes (Figure 7). Poly (I:C) is a synthetic double stranded RNA analog of viral nucleotide, and LPS as well as PGN are the main component of the outer membrane of bacteria. E. tarda is an intracellular bacterial pathogen and has been reported to infect vertebrates from fish to mammals (48). The severe disease and fatal cases by E. tarda have been reported in a wide range of fish species (37, 49). Thus, our results indicated that all four LmP2Xs were probably involved in both antiviral and antibacterial immune responses but with tissue specificity. Noteworthy, to our knowledge, this is the first report on the involvement of P2X5 in fish immunity.
In mammalian, numerous studies indicated that extracellular ATP promotes the release of pro-inflammatory cytokines and chemokines, induces apoptosis, and activates MAPK and NF-κB through the activation of different P2X receptors (4–7). Similarly, extracellular ATP has been shown to induce the mRNA expressions of pro-inflammatory cytokines and immune-related genes, and to exert a pro-apoptotic effect by promoting the expressions and enzymatic activities of several caspases in fish immune cells (33, 50–53). Activated P2X7 induced the mRNA expressions of IL-1β and IL-6 in Japanese flounder head kidney primary cells (30) and mediated cell death of ayu macrophages (29). The binding of coral and amphioxus P2X7L and flounder P2X7 to extracellular ATP has been demonstrated to induce the both mRNA and protein expressions of IL-1β, IL-6, IL-8, and CCL2 in HEK 293T cells (47). In addition, some fish genes have been shown to modulate apoptosis in HEK 293T cells by regulating the mRNA expressions of apoptosis-related genes (39, 54). Here, we first demonstrated that LmP2X2, LmP2X4, LmP2X5 and LmP2X7 were localized on the cellular membrane and can be expressed in HEK 293T cells (Supplementary Figure 3). Then, we found that LmP2Xs activated by extracellular ATP differentially up-regulated the mRNA expressions of pro-inflammatory cytokines (CCL2, IL-8, and TNF-α) and apoptosis-related genes (caspase3, caspase6, caspase7, and P53) in HEK 293T cells (Figure 8), suggesting that activated LmP2Xs may be involved in inflammation and innate immune response by regulating the expressions of inflammation-associated genes. However, the direct immunological roles of P2X receptors, especially those other than P2X7, in fish cells needs to be further investigated.
We further investigated the effect of activated LmP2Xs on the NF-κB promoter, which is a critical signaling pathway controlling the expressions of inflammation-related genes (55). The results showed that ATP-activated LmP2Xs significantly increased the NF-κB promoter activity (Figure 9), implicating that ATP could activate the NF-κB pathway by interacting with LmP2Xs. Previous study also showed that extracellular ATP can induce NF-κB p65 subunit (p65) expression in Japanese flounder immune cells (53). As activation of the NF-κB pathway induces expressions of pro-inflammatory cytokines, these results provide strong evidence for activated P2X receptors up-regulate the expressions of inflammation-related genes via the NF-κB pathway in fish. However, their functional mechanisms need to be further clarified in the future.
P2X receptors have been demonstrated to have different affinities for extracellular ATP in mammals (7). All P2X receptors have high ATP-affinities (EC50 = 0.05-30 μM) except for P2X7, which has a low affinity for ATP (EC50 > 100 μM) (7, 56). However, in zebrafish, it was shown that P2X2 did not respond to low levels of ATP (30 μM) (27) and that P2X4 had a significantly lower affinity for ATP (EC50 = 274 ± 48 μM) than mammals (26). Japanese flounder P2X7 also exhibited a much lower ATP-affinity (EC50 = 790 ± 81 μM) than the P2X7 of mammalians (30). In addition, mouse P2X4 promoted ATP-dependent pore formation only at low ATP concentrations (≤ 1 mM) (57), while zebrafish P2X4 could be activated by higher ATP concentrations (≥ 3 mM) (26). Interestingly, in our study, LmP2X2 and LmP2X5 were induced by higher concentrations of ATP in head kidney primary leukocytes (Figure 7). At the same time, although LmP2X2, LmP2X4 and LmP2X5 were more sensitive to ATP than LmP2X7, all four LmP2Xs were activated by higher concentrations of ATP (≥ 1 mM) to perform their functions (Figures 8, 9). Taken together, the ATP-affinity of fish P2X receptors may be much lower than that of mammals. On the other hand, a number of key sites and structures of different mammalian P2X receptors to perform their functions have been reported (46). For example, it has been demonstrated that the TM2 structural domain of P2X7 has a key role in receptor function and the residues important for full P2X7 function have also been identified in mouse (57). However, these issues have not been well studied in fish. Therefore, the concentration range of ATP sensitivity and the key sites for the function of different fish P2X receptors need to be determined in the future to better explain these issues.
In summary, four P2X receptors (P2X2, P2X4, P2X5, and P2X7) were identified in spotted sea bass. They were constitutively expressed in the tissues and significantly upregulated by E. tarda infection and PAMP and ATP stimulation. Furthermore, activated LmP2Xs induced the expressions of inflammation-related genes, and significantly increased the NF-κB promoter activity. Thus, these four P2X receptors participated in innate immune response of spotted sea bass probably by regulating the expressions of inflammation-related genes. These results provide new evidence for the involvement of P2X receptors in fish immunity, which will be helpful in further understanding the innate immune response mediated by extracellular ATP in fish.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession numbers OP651016, OP651019, OP651020, and OP651021.
Ethics statement
The animal study was reviewed and approved by the ethics committee of laboratory animals of Shanghai Ocean University (SHOU-DW-2019-012).
Author contributions
ZS: investigation, methodology, data curation, and writing the original draft. QG: conceptualization, funding acquisition, project administration, supervision, and reviewing and editing. YW: supervision and reviewing and editing. ZZ: reviewing and editing. YC, CX, JG, and DL: investigation and methodology. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Key Research and Development Program of China (Grant Number: 2018YFD0900605).
Acknowledgments
We thank Dr. Haixia Xie, Institute of Hydrobiology, Chinese Academy of Sciences, for providing Edwardsiella tarda.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1181067/full#supplementary-material
References
1. Jacob F, Novo CP, Bachert C, Van Crombruggen K. Purinergic signaling in inflammatory cells: P2 receptor expression, functional effects, and modulation of inflammatory responses. Purinerg Signal (2013) 9(3):285–306. doi: 10.1007/s11302-013-9357-4
2. Faas MM, Saez T, de Vos P. Extracellular ATP and adenosine: the yin and yang in immune responses? Mol Aspects Med (2017) 55:9–19. doi: 10.1016/j.mam.2017.01.002
3. Trautmann A. Extracellular ATP in the immune system: more than just a "Danger signal". Sci Signal (2009) 2(56):pe6. doi: 10.1126/scisignal.256pe6
4. Alberto AVP, Ferreira NCD, Bonavita AGC, Nihei OK, de Farias FP, Bisaggio RD, et al. Physiologic roles of P2 receptors in leukocytes. J Leukocyte Biol (2022) 112:983–1012. doi: 10.1002/Jlb.2ru0421-226rr
5. Coutinho-Silva R, Savio LEB. Purinergic signalling in host innate immune defence against intracellular pathogens. Biochem Pharmacol (2021) 187:114405. doi: 10.1016/j.bcp.2021.114405
6. Adinolfi E, Giuliani AL, De Marchi E, Pegoraro A, Orioli E, Di Virgilio F. The P2X7 receptor: a main player in inflammation. Biochem Pharmacol (2018) 151:234–44. doi: 10.1016/j.bcp.2017.12.021
7. Vitiello L, Gorini S, Rosano G, la Sala A. Immunoregulation through extracellular nucleotides. Blood (2012) 120(3):511–8. doi: 10.1182/blood-2012-01-406496
8. Jarvis MF, Khakh BS. ATP-gated P2X cation-channels. Neuropharmacology (2009) 56(1):208–15. doi: 10.1016/j.neuropharm.2008.06.067
9. Coddou C, Yan ZH, Obsil T, Huidobro-Toro JP, Stojilkovic SS. Activation and regulation of purinergic P2X receptor channels. Pharmacol Rev (2011) 63(3):641–83. doi: 10.1124/pr.110.003129
10. Valera S, Hussy N, Evans RJ, Adami N, North RA, Surprenant A, et al. A new class of ligand-gated ion channel defined by P2X receptor for extracellular ATP. Nature (1994) 371(6497):516–9. doi: 10.1038/371516a0
11. North RA. Molecular physiology of P2X receptors. Physiol Rev (2002) 82(4):1013–67. doi: 10.1152/physrev.00015.2002
12. Cai XY, Yao Y, Teng F, Li YL, Wu LW, Yan W, et al. The role of P2X7 receptor in infection and metabolism: based on inflammation and immunity. Int Immunopharmacol (2021) 101:108297. doi: 10.1016/j.intimp.2021.108297
13. Rivas-Yanez E, Barrera-Avalos C, Parra-Tello B, Briceno P, Rosemblatt MV, Saavedra-Almarza J, et al. P2X7 receptor at the crossroads of T cell fate. Int J Mol Sci (2020) 21(14):4937. doi: 10.3390/ijms21144937
14. Chaves MM, Sinflorio DA, Thorstenberg ML, Martins MDA, Moreira-Souza ACA, Rangel TP, et al. Non-canonical NLRP3 inflammasome activation and IL-1 beta signaling are necessary to l. amazonensis control mediated by P2X7 receptor and leukotriene b-4. PloS Pathog (2019) 15(6):e1007887. doi: 10.1371/journal.ppat.1007887
15. Liu YH, Xiao YJ, Li ZY. P2X7 receptor positively regulates MYD88-dependent NF-kappa b activation. Cytokine (2011) 55(2):229–36. doi: 10.1016/j.cyto.2011.05.003
16. Deng Y, Zhou MT, Zhao XT, Xue XY, Liao L, Wang J, et al. Immune response studies based on P2X7 receptors: a mini-review. Curr Pharm Design (2022) 28(12):993–9. doi: 10.2174/1381612828666220131091325
17. Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S. The P2X7 receptor in infection and inflammation. Immunity (2017) 47(1):15–31. doi: 10.1016/j.immuni.2017.06.020
18. Vaccari JPD, Bastien D, Yurcisin G, Pineau I, Dietrich WD, De Koninck Y, et al. P2X(4) receptors influence inflammasome activation after spinal cord injury. J Neurosci (2012) 32(9):3058–66. doi: 10.1523/Jneurosci.4930-11.2012
19. Sakaki H, Fujiwaki T, Tsukimoto M, Kawaho A, Harada H, Kojima S. P2X4 receptor regulates P2X7 receptor-dependent IL-1 beta and IL-18 release in mouse bone marrow-derived dendritic cells. Biochem Bioph Res Co (2013) 432(3):406–11. doi: 10.1016/j.bbrc.2013.01.135
20. Huang P, Zou YJ, Zhong XZ, Cao Q, Zhao KX, Zhu MX, et al. P2X4 forms functional ATP-activated cation channels on lysosomal membranes regulated by luminal ph. J Biol Chem (2014) 289(25):17658–67. doi: 10.1074/jbc.M114.552158
21. Csoka B, Nemeth ZH, Szabo I, Davies DL, Varga ZV, Paloczi J, et al. Macrophage P2X4 receptors augment bacterial killing and protect against sepsis. JCI Insight (2018) 3(11):e99431. doi: 10.1172/jci.insight.99431
22. Woehrle T, Yip L, Elkhal A, Sumi Y, Chen Y, Yao YL, et al. Pannexin-1 hemichannel-mediated ATP release together with P2X1 and P2X4 receptors regulate T-cell activation at the immune synapse. Blood (2010) 116(18):3475–84. doi: 10.1182/blood-2010-04-277707
23. Kanellopoulos JM, Almeida-da-Silva CLC, Boudinot SR, Ojcius DM. Structural and functional features of the P2X4 receptor: an immunological perspective. Front Immunol (2021) 12:645834. doi: 10.3389/fimmu.2021.645834
24. Oliveira MCG, Pelegrini-Da-Silva A, Tambeli CH, Parada CA. Peripheral mechanisms underlying the essential role of P2X3,2/3 receptors in the development of inflammatory hyperalgesia. Pain (2009) 141(1-2):127–34. doi: 10.1016/j.pain.2008.10.024
25. Jeong YH, Walsh MC, Yu J, Shen H, Wherry EJ, Choi Y. Mice lacking the purinergic receptor P2X5 exhibit defective inflammasome activation and early susceptibility to listeria monocytogenes. J Immunol (2020) 205(3):760–6. doi: 10.4049/jimmunol.1901423
26. Diaz-Hernandez M, Cox JA, Migita K, Haines W, Egan TM, Voigt MM. Cloning and characterization of two novel zebrafish P2X receptor subunits. Biochem Bioph Res Co (2002) 295(4):849–53. doi: 10.1016/S0006-291x(02)00760-X
27. Kucenas S, Li Z, Cox JA, Egan TM, Voigt MM. Molecular characterization of the zebrafish P2X receptor subunit gene family. Neuroscience (2003) 121(4):935–45. doi: 10.1016/S0306-4522(03)00566-9
28. Lopez-Castejon G, Young MT, Meseguer J, Surprenant A, Mulero V. Characterization of ATP-gated P2X(7) receptors in fish provides new insights into the mechanism of release of the leaderless cytokine interleukin-1 beta. Mol Immunol (2007) 44(6):1286–99. doi: 10.1016/j.molimm.2006.05.015
29. He YQ, Chen J, Lu XJ, Shi YH. Characterization of P2X7R and its function in the macrophages of ayu, Plecoglossus altivelis. PloS One (2013) 8(2):e57505. doi: 10.1371/journal.pone.0057505
30. Li S, Li XJ, Coddou C, Geng XY, Wei JL, Sun JS. Molecular characterization and expression analysis of ATP-gated P2X7 receptor involved in Japanese flounder (Paralichthys olivaceus) innate immune response. PloS One (2014) 9(5):e96625. doi: 10.1371/journal.pone.0096625
31. Li S, Chen XL, Li XJ, Geng XY, Lin RX, Li M, et al. Molecular characterization of purinergic receptor P2X4 involved in Japanese flounder (Paralichthys olivaceus) innate immune response and its interaction with ATP release channel Pannexin1. Fish Shellfish Immun (2015) 47(1):100–9. doi: 10.1016/j.fsi.2015.08.029
32. Li S, Chen XL, Hao GX, Geng XY, Zhan WB, Sun JS. Identification and characterization of ATP-gated P2X2 receptor gene dominantly expressed in the Japanese flounder (Paralichthys olivaceus) head kidney macrophages. Fish Shellfish Immun (2016) 54:312–21. doi: 10.1016/j.fsi.2016.04.020
33. Li S, Zhang TT, Feng Y, Sun JS. Extracellular ATP-mediated purinergic immune signaling in teleost fish: a review. Aquaculture (2021) 537:736511. doi: 10.1016/j.aquaculture.2021.736511
34. Sun ZS, Xu C, Chen YX, Liu DJ, Wu P, Gao Q. Characterization of Pannexin1, Connexin32, and Connexin43 in spotted Sea bass (Lateolabrax maculatus): they are important neuro-related immune response genes involved in inflammation-induced ATP release. Front Immunol (2022) 13:870679. doi: 10.3389/fimmu.2022.870679
35. Sun ZS, Qin YT, Liu DJ, Wang BJ, Jia Z, Wang JY, et al. The evolution and functional characterization of CXC chemokines and receptors in lamprey. Dev Comp Immunol (2021) 116. doi: 10.1016/j.dci.2020.103905
36. Letunic I, Bork P. Interactive tree of life (ITOL) V5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res (2021) 49(W1):W293–W6. doi: 10.1093/nar/gkab301
37. Li X, Yuan SY, Sun ZS, Lei LN, Wan S, Wang JY, et al. Gene identification and functional analysis of peptidoglycan recognition protein from the spotted Sea bass (Lateolabrax maculatus). Fish Shellfish Immun (2020) 106:1014–24. doi: 10.1016/j.fsi.2020.08.041
38. Jiang XY, Gao JD, Xue YJ, Qin YT, Li X, Sun ZS, et al. Identification and expression analysis of il-4/13 receptors in grass carp Ctenopharyngodon idella. Fish Shellfish Immun (2019) 87:254–64. doi: 10.1016/j.fsi.2019.01.009
39. Chen KY, Tian JY, Wang JY, Jia Z, Zhang Q, Huang WJ, et al. Lipopolysaccharide-induced TNF alpha factor (LITAF) promotes inflammatory responses and activates apoptosis in zebrafish. Danio rerio. Gene (2021) 780:145487. doi: 10.1016/j.gene.2021.145487
40. Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol Rev (2004) 198:83–96. doi: 10.1111/j.0105-2896.2004.0120.x
41. Boue-Grabot E, Archambault V, Seguela P. A protein kinase c site highly conserved in P2X subunits controls the desensitization kinetics of P2X(2) ATP-gated channels. J Biol Chem (2000) 275(14):10190–5. doi: 10.1074/jbc.275.14.10190
42. Ennion SJ, Evans RJ. Conserved cysteine residues in the extracellular loop of the human P2X(1) receptor form disulfide bonds and are involved in receptor trafficking to the cell surface. Mol Pharmacol (2002) 61(2):303–11. doi: 10.1124/mol.61.2.303
43. Chaumont S, Jiang LH, Penna A, North RA, Rassendren F. Identification of a trafficking motif involved in the stabilization and polarization of P2X receptors. J Biol Chem (2004) 279(28):29628–38. doi: 10.1074/jbc.M403940200
44. Glasauer SMK, Neuhauss SCF. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics (2014) 289(6):1045–60. doi: 10.1007/s00438-014-0889-2
45. Appelbaum L, Skariah G, Mourrain P, Mignot E. Comparative expression of P2X receptors and ecto-nucleoside triphosphate diphosphohydrolase 3 in hypocretin and sensory neurons in zebrafish. Brain Res (2007) 1174:66–75. doi: 10.1016/j.brainres.2007.06.103
46. Khakh BS, North RA. P2X receptors as cell-surface ATP sensors in health and disease. Nature (2006) 442(7102):527–32. doi: 10.1038/nature04886
47. Duan H, Ni S, Yang S, Zhou Y, Zhang Y, Zhang S. Conservation of EATP perception throughout multicellular animal evolution: identification and functional characterization of coral and amphioxus P2X7-like receptors and flounder P2X7 receptor. Dev Comp Immunol (2020) 106:103641. doi: 10.1016/j.dci.2020.103641
48. Yi L, Nie P, Yu HB, Xie HX. Regulation of type iii secretion of translocon and effector proteins by the Esab/Esal/Esam complex in Edwardsiella tarda. Infect Immun (2017) 85(9):e00322–17. doi: 10.1128/IAI.00322-17
49. Mohanty BR, Sahoo PK. Edwardsiellosis in fish: a brief review. J Biosci (2007) 32(3):1331–44. doi: 10.1007/s12038-007-0143-8
50. Li S, Peng WJ, Li JF, Hao GX, Geng XY, Sun JS. Characterization of Japanese flounder (Paralichthys olivaceus) Caspase1 involved in extracellular ATP-mediated immune signaling in fish. Fish Shellfish Immun (2017) 67:536–45. doi: 10.1016/j.fsi.2017.06.043
51. Li S, Li JF, Peng WJ, Hao GX, Sun JS. Characterization of the responses of the caspase 2, 3, 6 and 8 genes to immune challenges and extracellular ATP stimulation in the Japanese flounder (Paralichthys olivaceus). BMC Vet Res (2019) 15:20. doi: 10.1186/s12917-018-1763-y
52. Zhang X, Liu ZT, Li C, Zhang Y, Wan LQ, Wei JG, et al. Characterization of orange-spotted grouper (Epinephelus coioides) asc and caspase-1 involved in extracellular ATP-mediated immune signaling in fish. Fish Shellfish Immun (2020) 97:58–71. doi: 10.1016/j.fsi.2019.12.023
53. Li S, Chen XL, Li JF, Li XJ, Zhang TX, Hao GX, et al. Extracellular ATP is a potent signaling molecule in the activation of the Japanese flounder (Paralichthys olivaceus) innate immune responses. Innate Immun-London (2020) 26(5):413–23. doi: 10.1177/1753425918804635
54. Zhao X, Huang WJ, Guo JH, Ji N, Feng JH, Chen KY, et al. Plaat1 promotes P53 degradation Via autophagy-lysosome pathway in zebrafish. Fish Shellfish Immun (2022) 125:48–53. doi: 10.1016/j.fsi.2022.05.001
55. Baeuerle PA. Pro-inflammatory signaling: last pieces in the NF-kappa b puzzle. Curr Biol (1998) 8(1):R19–22. doi: 10.1016/S0960-9822(98)70010-7
56. Zimmermann H. Extracellular ATP and other nucleotides-ubiquitous triggers of intercellular messenger release. Purinerg Signal (2016) 12(1):25–57. doi: 10.1007/s11302-015-9483-2
Keywords: P2X receptors, extracellular ATP, innate immunity, NF-κB pathway, Lateolabrax maculatus
Citation: Sun Z, Gao Q, Wei Y, Zhou Z, Chen Y, Xu C, Gao J and Liu D (2023) Activated P2X receptors can up-regulate the expressions of inflammation-related genes via NF-κB pathway in spotted sea bass (Lateolabrax maculatus). Front. Immunol. 14:1181067. doi: 10.3389/fimmu.2023.1181067
Received: 07 March 2023; Accepted: 18 April 2023;
Published: 04 May 2023.
Edited by:
Guan-Jun Yang, Ningbo University, ChinaReviewed by:
Francisco J. Morera, Universidad Austral de Chile, ChileJianfei Lu, Ningbo University, China
Copyright © 2023 Sun, Gao, Wei, Zhou, Chen, Xu, Gao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youchuan Wei, d2VpeWNAZ3h1LmVkdS5jbg==; Qian Gao, cWdhb0BzaG91LmVkdS5jbg==
 Zhaosheng Sun
Zhaosheng Sun Qian Gao
Qian Gao Youchuan Wei
Youchuan Wei Zhigang Zhou
Zhigang Zhou Yuxi Chen
Yuxi Chen Chong Xu
Chong Xu Jiaqi Gao
Jiaqi Gao Danjie Liu
Danjie Liu