
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 22 May 2023
Sec. Viral Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1180959
 Shridhar Bale1‡
Shridhar Bale1‡ Lifei Yang1†‡
Lifei Yang1†‡ Mehrdad Alirezaei1
Mehrdad Alirezaei1 Richard Wilson1
Richard Wilson1 Takayuki Ota1
Takayuki Ota1 Esmeralda D. Doyle1
Esmeralda D. Doyle1 Christopher A. Cottrell1
Christopher A. Cottrell1 Javier Guenaga2
Javier Guenaga2 Karen Tran2
Karen Tran2 Wenjuan Li1
Wenjuan Li1 Leonidas Stamatatos3,4
Leonidas Stamatatos3,4 David Nemazee1
David Nemazee1 Andrew B. Ward2,5,6
Andrew B. Ward2,5,6 Richard T. Wyatt1,2,6*
Richard T. Wyatt1,2,6*An effective HIV vaccine likely requires the elicitation of neutralizing antibodies (NAbs) against multiple HIV-1 clades. The recently developed cleavage-independent native flexibly linked (NFL) envelope (Env) trimers exhibit well-ordered conformation and elicit autologous tier 2 NAbs in multiple animal models. Here, we investigated whether the fusion of molecular adjuvant C3d to the Env trimers can improve B- cell germinal center (GC) formation and antibody responses. To generate Env-C3d trimers, we performed a glycine-serine- based (G4S) flexible peptide linker screening and identified a linker range that allowed native folding. A 30–60- amino- acid- long linker facilitates Env-to-C3d association and achieves the secretion of well-ordered trimers and the structural integrity and functional integrity of Env and C3d. The fusion of C3d did not dramatically affect the antigenicity of the Env trimers and enhanced the ability of the Env trimers to engage and activate B cells in vitro. In mice, the fusion of C3d enhanced germinal center formation, the magnitude of Env-specific binding antibodies, and the avidity of the antibodies in the presence of an adjuvant. The Sigma Adjuvant System (SAS) did not affect the trimer integrity in vitro but contributed to altered immunogenicity in vivo, resulting in increased tier 1 neutralization, likely by increased exposure of variable region 3 (V3). Taken together, the results indicate that the fusion of the molecular adjuvant, C3d, to the Env trimers improves antibody responses and could be useful for Env-based vaccines against HIV.
Despite many attempts both preclinically and in the clinic, a broadly effective vaccine preventing HIV-1 infection remains elusive. Such a vaccine likely requires the elicitation of cross-neutralizing antibodies (NAbs) directed toward the surface-exposed trimeric envelope glycoproteins (Env) (1). NAbs are the correlate of protection for most licensed vaccines and are a major focus of HIV-1 vaccine research efforts. In recent years, developments in soluble HIV trimer design has resulted in the generation of multiple soluble native-like mimics, including SOSIPs, NFLs, and UFOs that exhibit a well-ordered, native-like Env conformation (2–6). Antigenic evaluation of these trimers, and their more stabilized variants, demonstrates that they preferentially present broadly neutralizing antibody (bNAb) epitopes while occluding non-neutralizing determinants. Despite advances in design and improvement of the trimer mimics, the antibodies elicited by these recombinant glycoproteins are often limited to neutralizing “tier 1” (lab-adapted) viruses and autologous tier 2 viruses depending upon the viral strain from which the trimers derive or the animal model used (7–13). These data indicate that challenges remain to develop an effective HIV-1 vaccine (14–16). However, recently we and others have elicited antibodies with the capacity to neutralize a set of heterologous tier 2 clinical isolates in animal models, demonstrating a proof of principle that this is possible following sequential Env trimer immunization (17, 18).
HIV-1 Env immunogens predominantly induce short-lived memory B- cell-dependent plasma Abs in the settings of both infection and vaccination (19). The magnitude and duration of antibody responses, affinity maturation, and the induction of B- cell memory are also important parameters to consider for development of an effective HIV-1 vaccine. Therefore, the means to improving the quality of the antibody responses is of great interest to the field and to basic B- cell biology, especially in the well-utilized mouse immune system. The use of molecular adjuvants, with specific and molecularly defined modes of action, has increased over the years (20). One relatively well-studied molecular adjuvant is a component of the complement pathway known as C3d (21). C3d is a cleavage product of complement component 3 and plays a key role in foreign antigen recognition, linking innate immunity to adaptive immune responses via binding to complement receptors (CD21, previously called CR2) (22). Following activation of the complement cascade by foreign proteins, C3d becomes covalently attached to the activating antigen. In turn, C3d can bind to CR2 present on B cells and follicular dendritic cells (FDCs), resulting in B- cell activation and initiation of an adaptive immune response (23) (Figures 1A, B, adapted from Carroll et al., Immunity 2012).
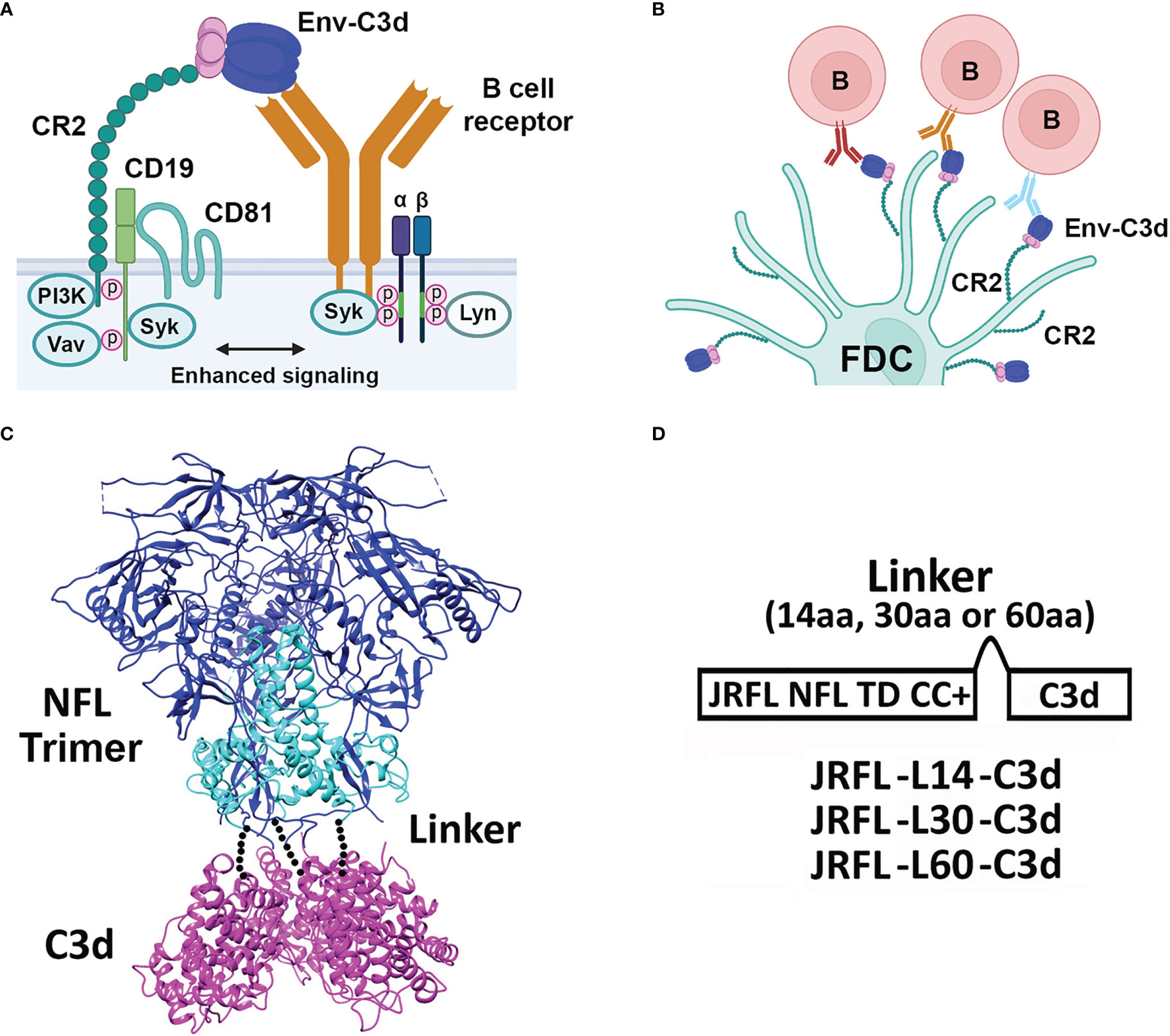
Figure 1 Activation by complement system, design, and schematic representation of HIV-1 Env-C3d fusion Trimers. (A) Cartoon model depicting enhanced activation of the innate immune system by simultaneous engagement of HIV-1 Env-C3d trimers with naïve B- cell receptors (BCRs) and CR2, CD19, and CD81 receptors of the complement system, respectively. (B) Further activation of the adaptive immune system in germinal centers by the engagement of CD2 receptors on follicular dendritic cells (FDCs) with BCR- attached Env-C3d trimers. (C) Model of Env-C3d (16055 NFL TD CC T569G, PDB 5UM8; human C3d, PDB: 3OED). The three Env gp120 subunits are shown in blue; the gp41 ectodomain subunits in cyan to form the gp140 trimers; mouse C3d in magenta. The three C3d subunits were manually fitted to the C-termini of the NFL trimer by visual inspection in PyMOL. The dotted lines represent the linker connecting the C-termini of NFL Env gp140 to the N-termini of C3d. (D) Linear schematic diagram of JRFL-C3d trimers. Mouse C3d was fused to the C-termini of NFL gp140 trimers by a flexible linker (G4S) of varying lengths (14, 30, and 60 amino acids to yield JRFL-L14-C3d, etc.). JRFL NFL TD CC+ (namely, JRFL) was used for the linker-length screening. The native signal peptide sequence of JRFL (or 426c NFL trimers) was replaced by the CD5 leader sequence to increase secretion in mammalian cells, as previously performed.
Dempsey et al. demonstrated previously that C3d can act as a molecular adjuvant when covalently fused to hen egg lysozyme (HEL), quantitatively lowering the amount of antigen needed to activate B cells 10,000- fold (21). We and others have used C3d fused to HIV-1 Env (gp120 or gp140) and confirmed the adjuvant effect of C3d in the absence of classical adjuvants but demonstrating less of an effect in the presence of a relatively robust “classical” adjuvant (24, 25). Very recently, it was shown that the lymph node follicles may be privileged sites with decreased proteolytic activity that may better maintain native antigen conformation (26). Thereby, targeting follicular FDCs may improve the elicitation of antibodies to native-like determinants. Accordingly, here we have sought to examine whether the fusion of C3d could improve the immunogenicity of the relatively recently developed conformationally near-native NFL Env trimers.
In this study, we genetically fused mouse C3d to the C-terminus of stabilized NFL Env trimers, previously developed and improved in our laboratory (4, 27). To accomplish this fusion, we initially performed a glycine-serine based (G4S) flexible peptide linker screening to identify a linker length that facilitated native-like trimer formation. We subsequently evaluated the Env-C3d fusion trimers regarding their biochemical, biophysical, and antigenic properties, as well as their conformational and functional integrity in vitro. Finally, we assessed the impact of C3d on the Env trimers regarding their immunogenicity in vivo, in the absence and presence of the Sigma Adjuvant System (SAS) adjuvant, in C57BL/6 mice and VRC01gHL knock-in mice.
We fused mouse C3d to the well-ordered NFL gp140 Env to assess whether this immunostimulatory molecule can function as an effective adjuvant in vivo, to better target these native-like trimers to appropriate immune cells and more efficiently elicit antibody responses to these HIV vaccine candidates. We began this study by genetically fusing C3d sequences to the 3′-end of sequences encoding NFL gp140 trimers. The basics for the NFL trimer design are previously reported, along with stepwise improvements that included stabilizing “trimer- derived” (TD) mutations inferred from the crystal structure of the clade A BG505 SOSIP and trimer and inspection of the primary sequence of each stabilized Env trimer (JR-FL, 16055, etc.). The NFL construct also included a critical internal 201–433- cysteine bridging sheet linkage, now termed “CC1” that renders the trimer resistant to CD4-induced conformational changes (4, 5, 27). A cartoon representation depicting the details of the HIV-1 Env-C3d trimer design is shown in Figure 1C.
In this study, we began with the JR-FL NFL E160K TD CC+ trimer (“JRFL” in short for this work), as proof of principle to generate well-ordered Env trimers possessing genetically fused C3d from a single open reading frame. We covalently linked the N-terminus of C3d to the C-terminus of JRFL with a flexible linker (G4S) of varying lengths (Figure 1D). We determined previously that for cysteine linkage of the NFL trimers to maleimide-displaying liposomes, a cytoplasmic linker length greater than 13 residues is beneficial for efficient covalent linkage (28). Using this information, we first tested a 14- residue G4S linker (L14) between JRFL and C3d, designated as JRFL-L14-C3d. Following transient transfection in 293F cells, lectin purification, and size exclusion chromatography (SEC), we observed a broad protein peak instead of the expected distinct trimer peak in the SEC profile normally observed with the stabilized JR-FL trimers (5). Blue Native PAGE (BN-PAGE) revealed a broad protein “smear,” presumably non-native aggregates, with a small fraction of trimers (Figure S1). The non-native conformation of the presumed aggregates was confirmed by antigenic profiling characterized by poor recognition by the trimer-specific broadly neutralizing antibody (bNAb), PGT145. Although this was an unexpected outcome, we reasoned that this was likely due to a suboptimal linker length to permit proper trimer folding when tethered to the C3d adduct. The overall globular structure of human C3d, as well as primary sequence, is highly conserved between humans and mice (29, 30). Accordingly, we increased the linker length to both 30 and 60 residues (designated as JRFL-L30-C3d and JRFL-L60-C3d, respectively) to potentially restore native-like trimer folding.
The JRFL-L30-C3d and JRFL-L60-C3d variants were expressed in 293F cells and purified by lectin affinity purification followed by F105 negative-selection chromatography, as previously described (31). The two JRFL-C3d variants possessing the longer linkers appeared to form well-ordered trimers, each resolving as a single trimer peak by SEC (Figure 2A). For comparison, the SEC profile of the JRFL trimer is shown in Figure S1C. The yield of JRFL-L60-C3d trimers was approximately 0.8 mg/L, higher than that achieved by the JRFL-L30-C3d trimers (0.4 mg/L). The purified fusion proteins were resolved on BN-PAGE, revealing a migration pattern consistent with trimeric Env that generated a single band ~720 kDa by PAGE analysis (Figure 2B). The JRFL-L30-C3d and JRFL-L60-C3d fusions both migrated slightly more slowly in the gel than their counterpart JRFL trimers, consistent with the fusion of C3d adding ~90 kDa additional mass to the trimer (C3d is ~30 kDa). The successful fusion of C3d to JRFL was confirmed by Western blot analysis using Env-specific antibodies (2G12 and VRC01) and a mouse C3d-specific antibody, respectively (Figure 2C). Homogeneous trimer formation of JRFL-L30-C3d and JRFL-L60-C3d was also confirmed by negative stain-EM (NS-EM). NS-EM 2-D average analysis showed that JRFL-L30-C3d and JRFL-L60-C3d trimers were in closed, native-like conformation (Figure 2D). Then, we assessed the thermostability of JRFL-L30-C3d and JRFL-L60-C3d trimers by differential scanning calorimetry (DSC). JRFL-L30-C3d and JRFL-L60-C3d displayed a single thermal transition curve, confirming the homogeneity of trimers. The Tm of JRFL-L30-C3d and JRFL-L60-C3d were 70.38°C and 70.43°C, respectively (Figure 2E), values comparable with the Tm of JRFL (70°C) as previously described (27). These data indicated that the fusion of C3d to JRFL Env did not alter the thermostability or homogeneity of the trimer component.
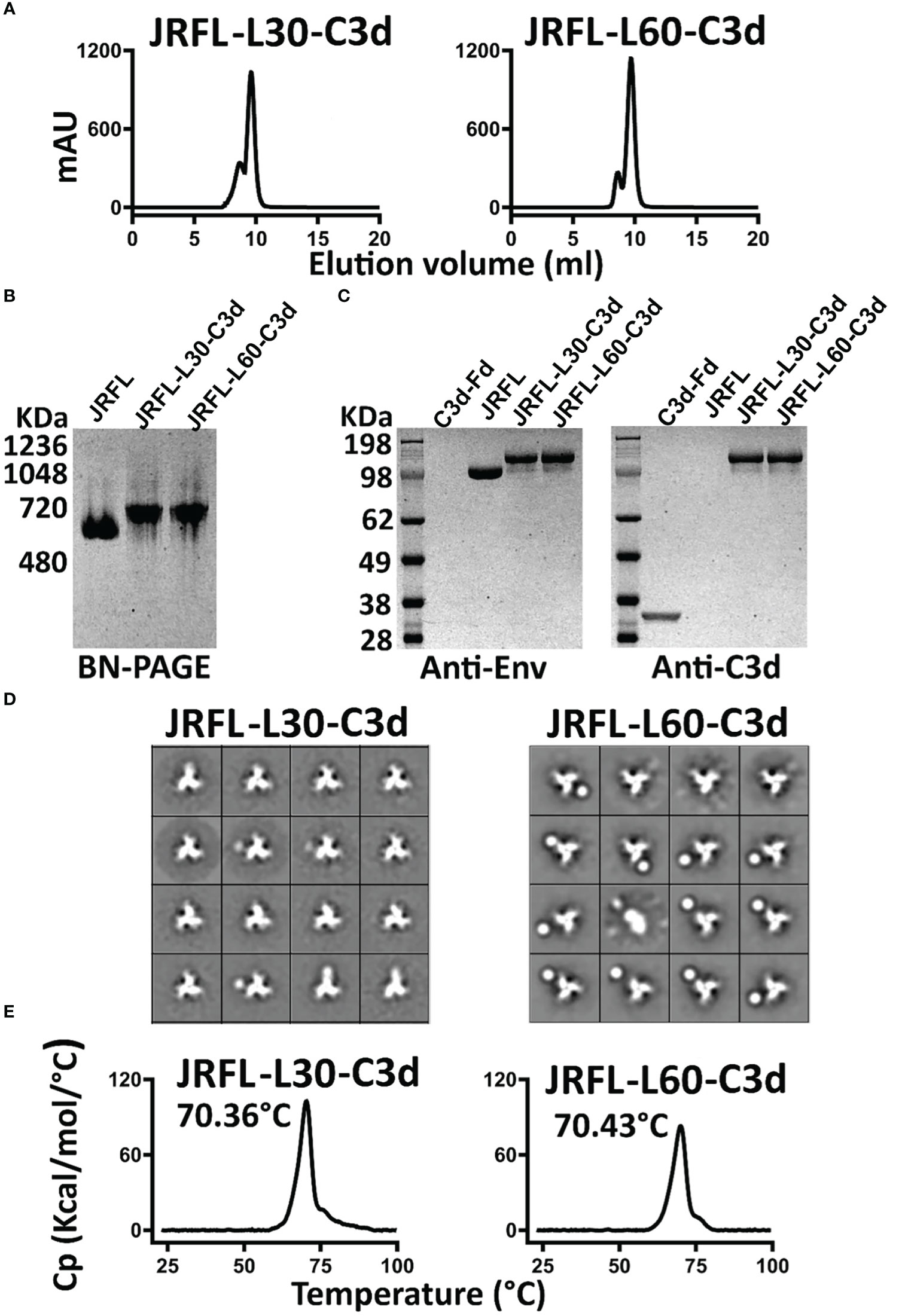
Figure 2 Biochemical and biophysical characterization of JRFL native-like-C3d fusion trimers. (A) SEC profile of JRFL-C3d trimers following lectin affinity purification and F105 negative selection. (B) Blue-native PAGE (BN-PAGE) analysis of JRFL-C3d trimers. (C) Western blot analysis of JRFL-C3d trimers. Protein identification was determined by anti-Env antibodies (2G12 and VRC01) or mouse C3d-specific antibody. Purified JRFL NFL native-like trimers and mouse C3d-foldon proteins were used as controls. (D) 2D averages from NS- EM of JRFL-C3d trimers revealed a native-like well-ordered structure. (E) DSC measurements of JRFL-C3d trimers. The Tm values are shown on top of each temperature peak.
To assess the potential impact of adding C3d to the JRFL Env trimers, we used a panel of mAbs to evaluate their level of binding to Env-C3d trimers and isogenic non-conjugated Env trimers. First, we tested recognition of JRFL-C3d trimers by C3d-specific Ab by Biolayer Interferometry. JRFL-L30-C3d and JRFL-L60-C3d showed similar recognition by the C3d-specific Ab, whereas there was no binding of their counterpart JRFL trimers, as expected (Figure 3A). Next, we tested recognition of the JRFL-C3d trimers by Env-specific mAbs. The JRFL-L30-C3d and JRFL-L60-C3d fusions were recognized similarly compared with unmodified JRFL trimers by bNAbs targeting the N-glycan shield (2G12), the glycan-dependent V3 332N region (PGT128), and slightly lower binding to bNAbs targeting the CD4 binding site (VRC01, VRC03), V2-apex (PGT145), and fusion peptide (VRC34) (Figure 3B). As expected, there was little or no binding detected by the non-neutralizing Abs F105 and 17b (Figure 3B). Taken together, Env-C3d fusion trimers with L30 and L60 linkers exhibit similar antigenicity profiles. These data demonstrate that the fusion of C3d to the NFL Env trimers did not largely affect the antigenicity of the Env trimers but that the longer linker appeared to be more favorable for the Env-C3d trimers to maintain functional integrity.
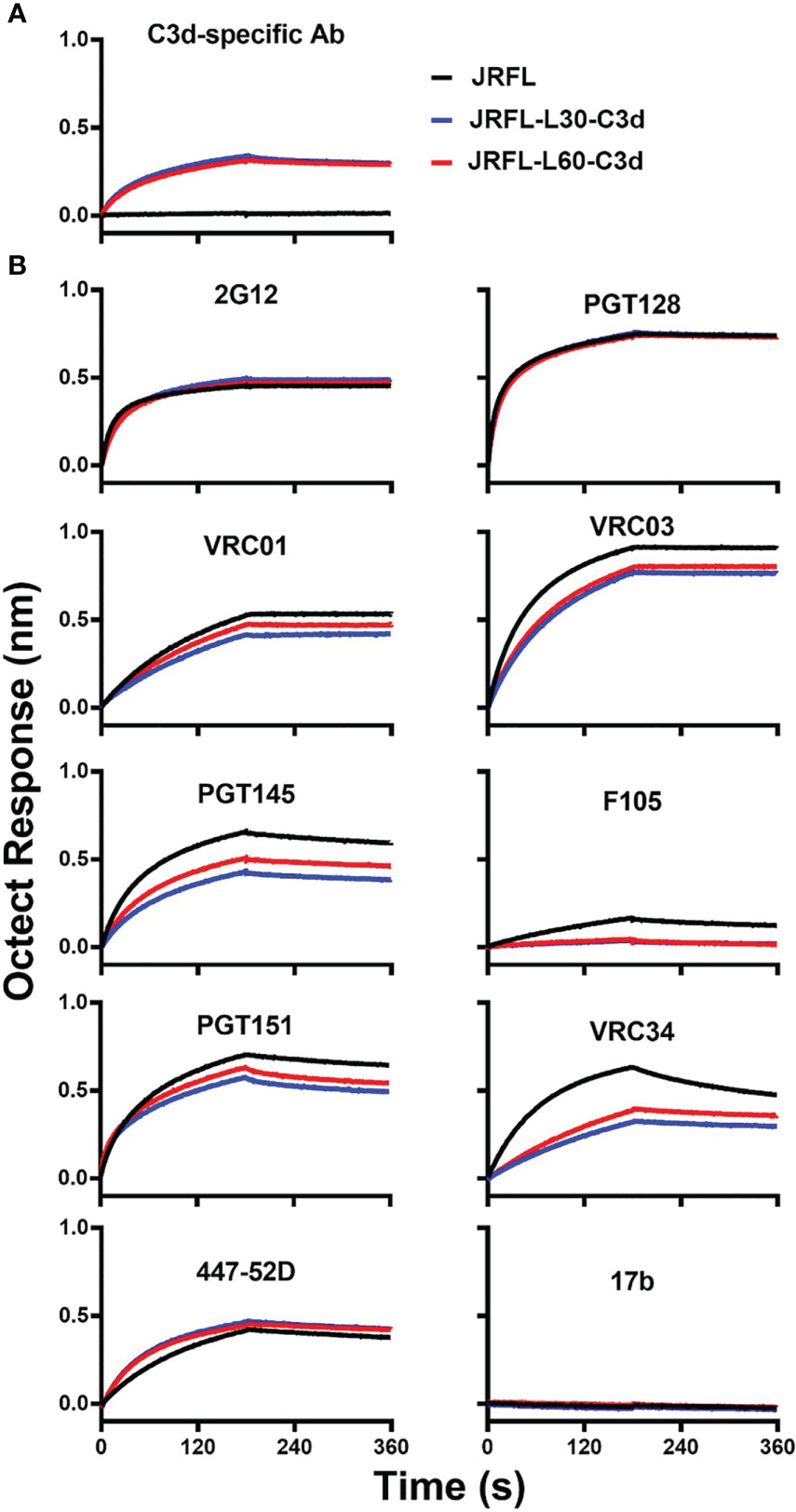
Figure 3 The JRFL-C3d trimers display a favorable antigenic profile as determined by Bio-layer interferometry (BLI). (A) JRFL-C3d trimers bind to the mouse C3d-specific antibody, which was immobilized on streptavidin sensors. Binding was not detected for JRFL trimer lacking the C3d domain. (B) JRFL and JRFL-C3d trimers are recognized by bNAbs and are not recognized by non-NAbs. Data are representative of at least two independent experiments.
To evaluate whether the fusion of C3d increases the ability of Env trimers to engage mouse B cells through C3d and CR2 interaction, we labeled JRFL and fusion JRFL-C3d trimers with Alexa Fluor 680 dye (AF680) and performed binding analysis by flow cytometry. The results show that JRFL-L30-C3d and JRFL-L60-C3d trimers bound well to PBMC-derived mouse B cells presumably through a trimer-C3d to CR2 interaction on the B cells. As expected, there was no detectable binding of JRFL trimers to B cells because there are no Env-specific B- cell receptors (BCRs) expressed on these normal mouse B cells (Figure 4A).
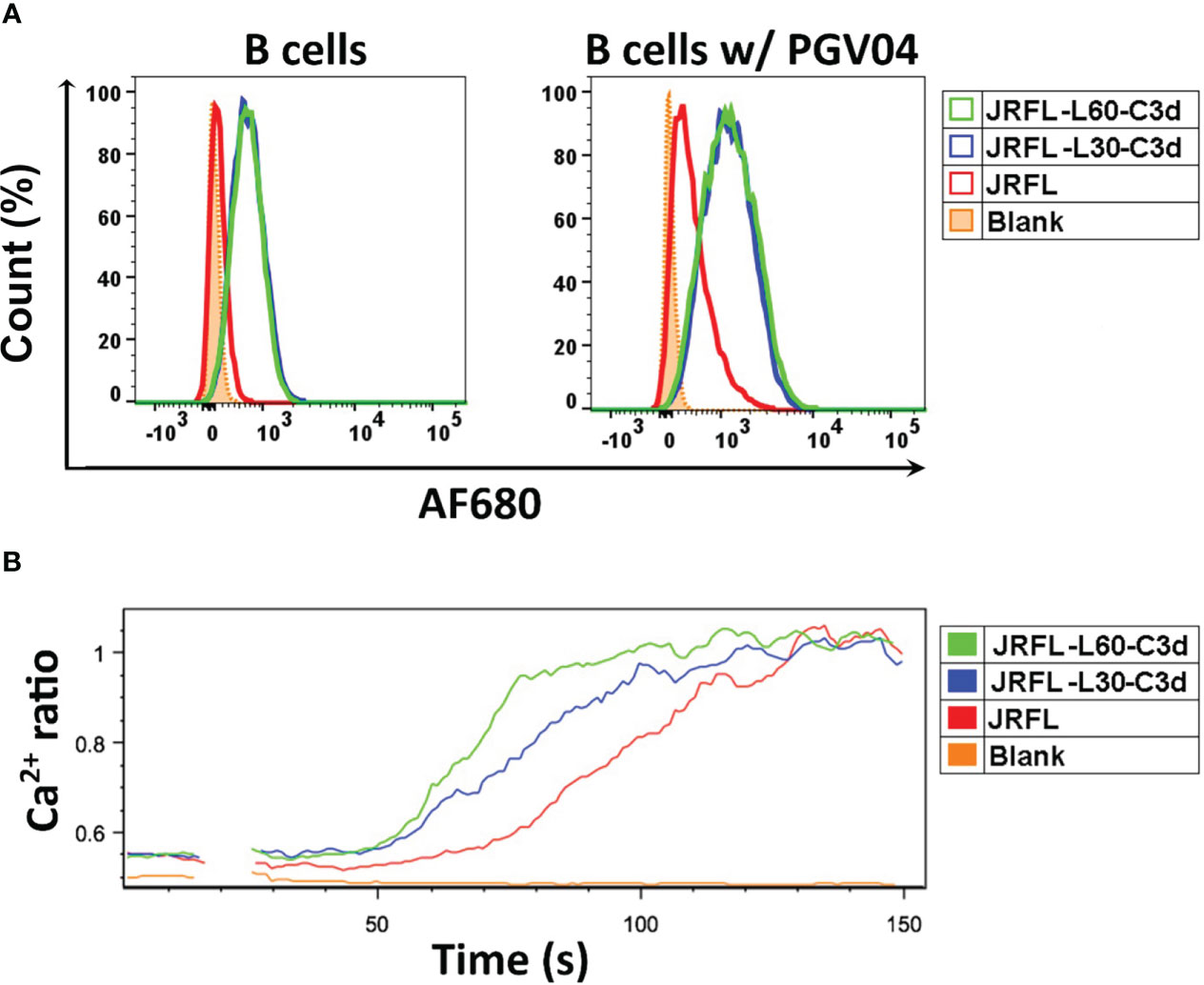
Figure 4 JRFL-C3d trimers bind and activate mouse B cells in vitro. (A) Flowcytometry analysis of JRFL-C3d trimers binding to mouse B cells with or without PGV04 expression on their surface. Percentage of maximum cell count is shown on the Y-axis, and relative fluorescence of Alexa Flour 680 (AF680) dye- conjugated trimers is shown on the X-axis. (B) Calcium flux analysis of B- cell activation by JRFL-C3d trimers. Mouse B cells expressing PGV04 on their surface were used. The resulting Ca2+ flux was measured over a period of 120 s. Representative data from two independent experiments is shown. JRFL trimers served as control for both the assays.
JRFL trimers showed modest binding reactivity to PBMC-derived normal mouse B cells, which display CR2 (CD21) and CD4bs-directed bNAb, PGV04, on their surface. However, the binding reactivities of JRFL-L30-C3d and JRFL-L60-C3d trimers were substantially increased with the addition of PGV04 expression, indicating that C3d enhanced the engagement of JRFL-C3d trimers to the mouse B cells through both C3d–CR2 interaction and the Env-specific BCRs (Figure 4A).
To further evaluate whether the fusion of C3d enhances the capability of the Env trimers to activate mouse B cells in vitro, we performed calcium flux analysis of B cells expressing the bNAb PGV04 using a flow cytometric assay (28, 32). B cells were stimulated with 50 μg/ml of soluble JRFL or JRFL-C3d trimers. The resulting Ca2+ flux (measurement of a 405/485-nm emission ratio of Indo-1 fluorescence upon excitation by UV) was analyzed over 120 s. Under the conditions tested, Ca2+ flux responses followed the trend JRFL-L60-C3d > JRFL-L30-C3d > JRFL NFL (Figure 4B). The results indicated that JRFL-C3d trimers activated the B cells more efficiently than the JRFL trimers by cross-linking CR2 and the BCRs (C3d-CR2 and Env-PGV04 in this case). In addition, JRFL-L60-C3d trimers generated much stronger B- cell activation than JRFL-L30-C3d, suggesting that the longer linker is more favorable for JRFL-C3d fusion trimers to achieve C3d-mediated functional integrity by engaging CR2s present on the B- cell plasma membrane. Finally, JRFL-L60-C3d trimers showed strong binding to soluble human CR2 protein by biolayer interferometry (BLI) (Figure S2) that was not detected using JRFL trimers lacking C3d fusion.
Taken together, the fusion of C3d improved the ability of Env trimers to engage and activate B cells in vitro. The fusion Env-C3d trimers preserved the functional integrity of both Env and C3d to engage Env-specific BCR and CR2, to activate B cells. The longer linker L60 was more favorable for Env-C3d fusion trimers to preserve the functional integrity of both the Env trimers and C3d. Accordingly, we selected the Env-L60-C3d trimers for subsequent immunogenicity studies in vivo.
We next tested the ability of JRFL-L60-C3d trimers to activate the germinal centers in vivo compared with the JRFL NFL trimer. Three groups of mice were immunized with JRFL NFL (13 mice), JRFL-L60-C3d (13 mice), and adjuvant SAS only (5 mice); 2 weeks after the immunization, six mice from the Env- immunized group and two mice from the adjuvant SAS only group were analyzed for GL7+ B cells (a surface marker specific for mouse germinal center B cells) in popliteal and inguinal lymph nodes (LNs) and spleens by flow cytometry. The fraction of GL7+ B cells in the population of CD45+/CD19+ B cells was found to be popliteal LNs > inguinal LNs > spleen. Although there was a trend, there was no statistically significant difference in the fraction of activated GL7+ B cells between the groups of mice immunized with JRFL or JRFL-L60-C3d trimers 2 weeks post priming immunization (Figure 5A). The remaining mice were boosted at week 4 and analyzed at week 6 for GL7+ B cells in the draining popliteal LNs as they had the highest signal. Activated GL7+ B cells in the mice immunized with JRFL-L60-C3d trimers were higher than those of mice immunized with JRFL NFL trimers (p = 0.0032) as well as for the adjuvant- only group (p = 0.0001) (Figure 5B).
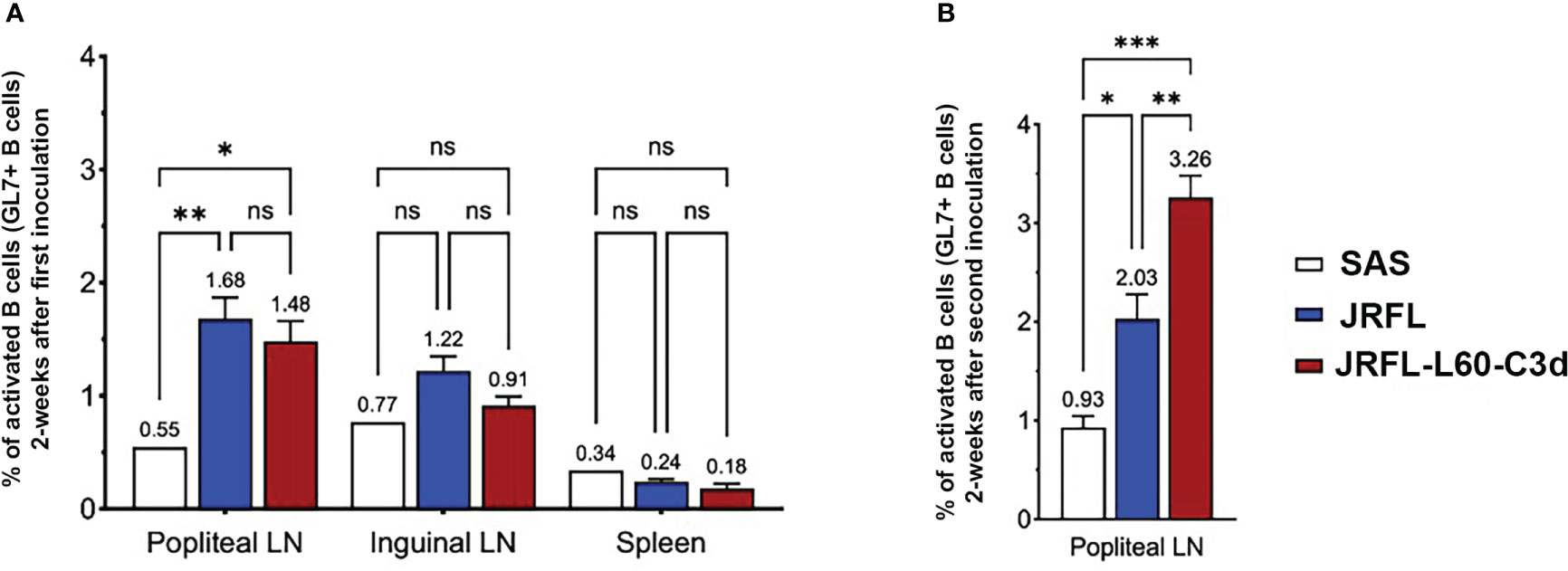
Figure 5 B cells from germinal centers in mice are better activated by JRFL-C3d trimers compared with those lacking C3d. (A) Comparison of activated GL7+ B cells measured by FACS after immunization with adjuvant- only, JRFL NFL, and JRFL-C3d trimers. Percent of activated GL7+ B cells were measured in draining popliteal and inguinal LNs and spleen 2 weeks after priming immunization. (B) Percent of activated GL7+ B cells in popliteal LN 6 weeks after two immunizations. The second immunization was performed at week 4. ns indicates non-significant. *, indicate P ≤ 0.05; **, indicate P ≤ 0.01; ***, indicate P ≤0.001.
Next, we assessed whether the fusion of C3d can improve antibody responses elicited by the near-native HIV-1 Env NFL trimers in vivo. As outlined in Figure 6A, four groups of C57BL/6 (B6) mice were immunized with JRFL or JRFL-L60-C3d trimers in PBS (no adjuvant) or SAS adjuvant, which has supplanted the RIBI adjuvant system. This adjuvant contains MPLA and other immune-stimulating compounds in an emulsion formed by squalene, Tween 80, and water. Following the first immunization, all the animals in the JRFL-L60-C3d- immunized groups generated Env-specific antibodies; however, only one (in G1) and three (in G2) animals in JRFL- immunized groups generated Env-specific antibodies. Following the subsequent immunizations, all animals generated anti-Env antibodies and the magnitude of Env- binding antibodies increased significantly after the second immunization and plateau following the third and fourth immunizations (Figure 6B). The results showed that in the presence or absence of the SAS adjuvant, JRFL-L60-C3d trimers elicited higher-titer Env-specific IgG antibodies than JRFL (G3 vs. G1, G4 vs. G2), especially following the first immunization.
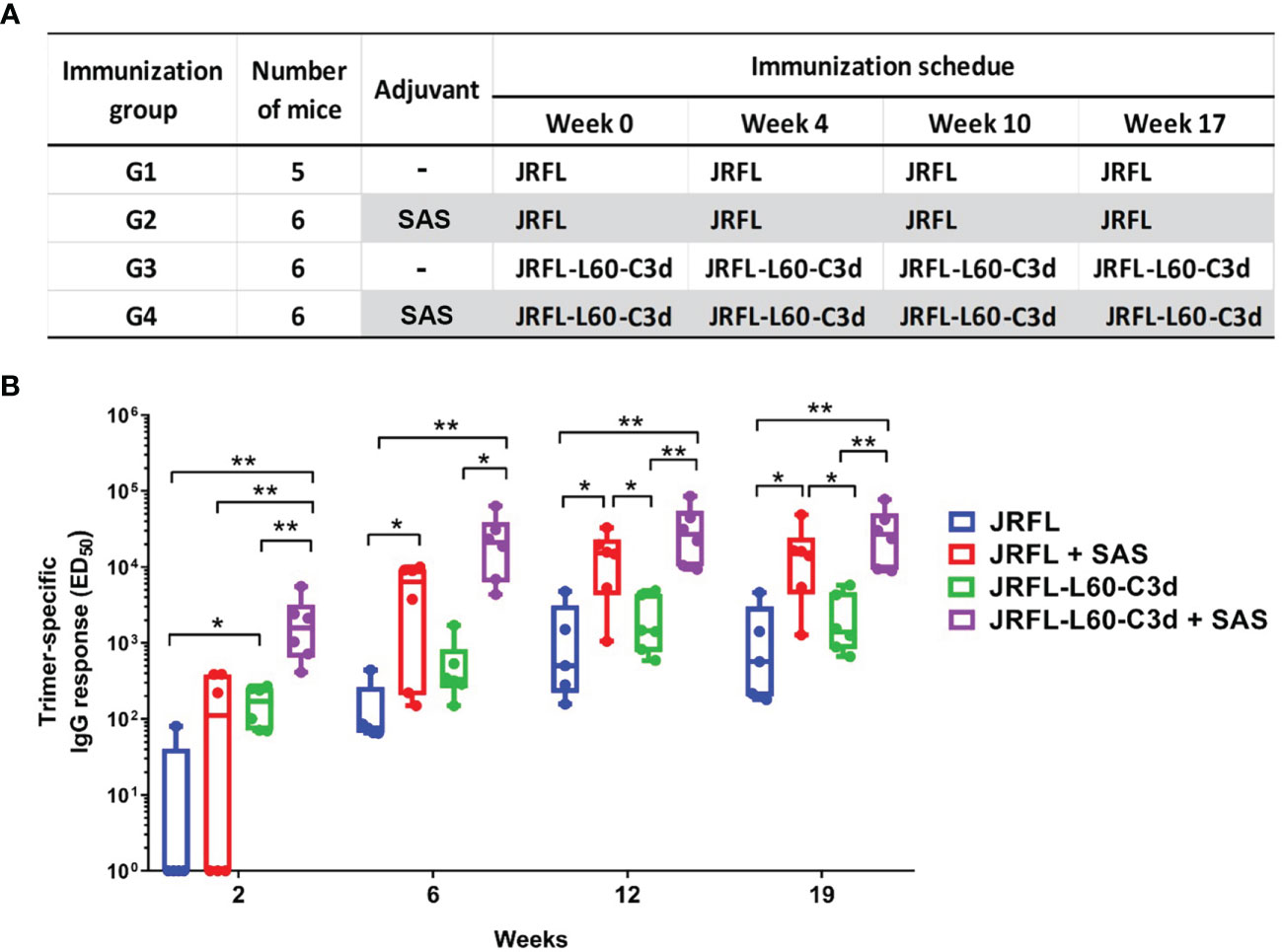
Figure 6 Immunization schedule and comparison of Env-specific binding antibody responses elicited in mice. (A) Immunization groups, antigens, and vaccine regimen. (B) JRFL trimer-specific IgG-binding titers were determined using 2G12-capture ELISA. JRFL-C3d fusion trimers elicit statistically significant higher binding antibodies than trimers lacking the C3d domain. The ED50 of each animal plotted, with geometric mean and min–max values shown. The box plots illustrate the following: horizontal line, median; plus, mean; box, interquartile range; whiskers, min/max. Statistical differences were evaluated by two-tailed Mann–Whitney U test. * indicates P ≤ 0.05; ** indicates P ≤ 0.01.
As shown in Figure 6B, the SAS adjuvant also enhanced the antibody responses for JRFL and JRFL-L60-C3d trimers in mice (G2 vs. G1, G4 vs. G3). Since adjuvant SAS contains MPLA, which is a TLR-4 agonist, we reasoned that the enhanced immunogenicity might be observed if both CD21 and TLR-4 pathways were targeted by the JRFL-C3d proteins in the presence of SAS. Indeed, the JRFL-C3d with SAS elicited significantly higher-titer Env-specific IgG antibodies than the JRFL without SAS after each immunization (G4 vs. G1). These data demonstrate that there is an additive effect of molecular adjuvant C3d and the SAS adjuvant to improve the immunogenicity of the Env trimers.
Overall, the results showed that molecular adjuvant C3d improves the ability of the Env trimers to elicit antibodies, to rapidly engage and activate naïve or germline B cells. In addition, there was an additive effect of C3d and the SAS adjuvant to improve the immunogenicity of the Env trimers.
To investigate if fusion of C3d can improve the avidity of Env-specific antibody responses, we performed sodium thiocyanate (NaSCN) displacement ELISAs. The avidity of Env-specific antibodies after the final immunization was measured by using the chaotropic agent NaSCN to potentially disrupt lower- affinity Env-antibody interactions. Under the stringent binding assay involving incubation in 1.5 M NaSCN, the avidity of the antibodies elicited by SAS- adjuvanted JRFL-L60-C3d trimers was higher and trending toward significance than that from SAS- adjuvanted JRFL trimer- vaccinated mice (G4 vs. G2, p = 0.0584) (Figure 7). However, in the absence of the SAS adjuvant, the JRFL-L60-C3d trimers did not drive antibody avidity compared with their counterpart JRFL trimers. Taken together, these results indicated that the fusion of C3d to Env may increase the affinity maturation of Env-specific antibodies in mice in the presence of the SAS adjuvant.

Figure 7 Avidity of the anti-Env IgG in mice after four immunizations. JRFL-specific NaSCN-displacement ELISA was used to measure the avidity index of anti-Env IgG in mouse sera at week 19 (2 weeks after the fourth immunization). The avidity index was calculated as the percentage of endpoint titers for NaSCN-treated samples to those of PBS-treated samples. The box plots illustrate the following: horizontal line, median; plus, mean; box, interquartile range; whiskers, min/max. Statistical differences were evaluated by two-tailed Mann–Whitney U test. * indicates P ≤ 0.05; ** indicates P ≤ 0.01. Data shown are representative from two independent experiments. JRFL-C3d trimers result in a statistically significant higher avidity index in the SAS adjuvant than in JRFL trimers alone in PBS and SAS adjuvant.
The serum samples (with ED50 values shown in Figure 6B) were further assessed for the capacity to neutralize pseudo-typed viruses by the standard TZM-bl assay (33). As shown in Table 1, substantial HIV-1- specific NAbs were elicited after the third immunization against tier 1 virus MN.3 in animals immunized with trimers in the SAS adjuvant. Following the fourth immunization, the neutralizing antibody response against tier 1 virus MN.3 was further increased. No reliably detected neutralization was observed against the antigen-matched tier 2 JRFL virus as expected in a mouse model, as seen previously (28, 34). Surprisingly, in the absence of the SAS adjuvant, the JRFL and JRFL-L60-C3d trimers did not elicit NAbs against MN.3 virus. MN.3 is a tier 1 clade B virus, which is particularly sensitive to V3-directed antibodies, whereas JRFL, from which our Env immunogens are derived, is a relatively resistant tier 2 clade B virus. The inability of the JRFL and JRFL-L60-C3d trimers to elicit NAbs against MN.3 likely reflects the maintenance of the native-like and desired closed native trimer conformation in aqueous solution in vitro. Low- neutralization titers against tier-1 viruses were also observed in rabbits and non-human primates after immunizations with native-like NFL trimers (10, 13, 35). However, when the immunogens were formulated in the SAS adjuvant, the JRFL and JRFL-L60-C3d trimers elicited a relatively strong neutralizing antibody response against the tier 1 MN.3 virus, indicating that the SAS adjuvant may be partially disruptive to the trimer integrity in vivo, resulting in an increased exposure of the V3 element. This protein determinant is often immunogenic and leads to the elicitation of antibodies that can neutralize lab-adapted isolates such as MN.3.
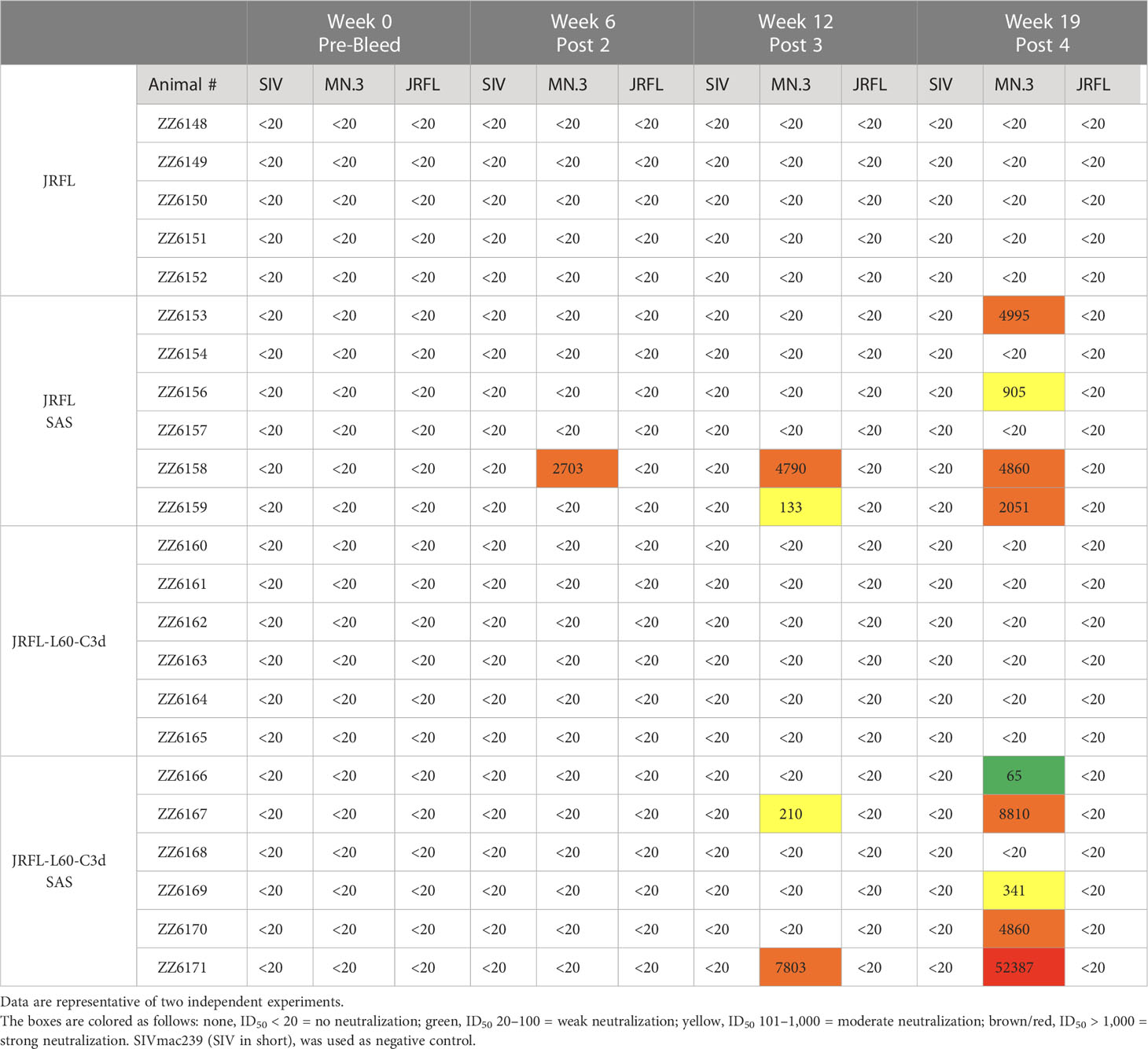
Table 1 Neutralization ID50 titers for sera from immunized mice against a panel of pseudo-typed viruses.
As described above, there is an additive effect of the molecular adjuvant C3d and the SAS adjuvant to improve the immunogenicity of the Env trimers. Interestingly, in the B6 mice immunization study, NAbs against MN.3 virus were only induced by immunogens in the presence of the SAS adjuvant, regardless of which antigen was inoculated into the mice. Neutralization against MN.3 is usually taken to indicate the elicitation of V3-directed antibodies to this “tier 1 virus” which exists in a more open Env spike conformation. As shown in Figure S3A, the SAS- adjuvanted immunogens elicited higher V3-directed antibodies than immunogens in PBS. It implies that the adjuvant SAS affects the trimer integrity in vivo. Moreover, there was a strong correlation of V3-specific binding antibody titers and the MN.3 neutralization titers (Figure S3B).
To further investigate whether the SAS adjuvant affects the Env trimer integrity in vitro, we tested the effect of SAS on the antigenicity of the Env trimers recognized by selected antibodies. We incubated His-tagged JRFL trimers in the presence or absence of the SAS adjuvant at room temperature or 37°C for 24 h. We captured the trimers on an ELISA plate via the His-tag and tested recognition of the trimers by a panel of mAbs. As shown in Figure S4, binding of the JRFL trimer to bNAbs or non-NAbs was quite similar in both the conditions tested, indicating that the SAS adjuvant did not affect the integrity of the trimer in vitro. Taken together, the SAS adjuvant likely alters the integrity of the JRFL trimers in vivo, as is evident by the elicitation of V3-directed binding and neutralizing antibodies even though SAS does not appreciably affect Env trimer integrity in vitro.
To investigate if we could extend the C3d fusion design to another trimer derived from a different HIV strain, we tested the C3d-fusion design to the 426c Env that is derived from a clade C virus. A stabilized 426c NFL engineered with three N-glycan-deletions (Δ276Δ460Δ463, termed ΔGly3) was used for this purpose (Figure 8A), as these trimers have been shown to engage the VRC01 class of precursor antibodies (36). We successfully expressed and purified the 426cΔGly3-C3d fusion trimers possessing both the L30 and L60 linkers (namely, 426cΔGly3-L30-C3d and 426c-ΔGly3-L60-C3d, respectively). Similar antigenic profiles were observed for both the –C3d fusion trimers, compared with their counterpart 426c-ΔGly3 trimers. Moreover, 426c-ΔGly3-L60-C3d trimers showed better recognition by trimer-specific bNAb PGT145 than 426c-ΔGly3-L30-C3d but a similar recognition by VRC01 and C3d-specific Ab (Figure 8B). We evaluated the effect of C3d fusion to 426c-ΔGly3 trimers in VRC01gHL mice to assess their capacity to activate antibody responses in this knock-in mouse model (the immunization schedule is shown in Figure 8C). After two immunizations in the SAS adjuvant, 426c-ΔGly3-L60-C3d trimers elicited significantly higher Env-specific IgG responses than that elicited by 426c-ΔGly3 trimers (Figure 8D). These data indicate that the fusion of C3d improves the ability of Env trimers to activate naïve or germline B cells in vivo.
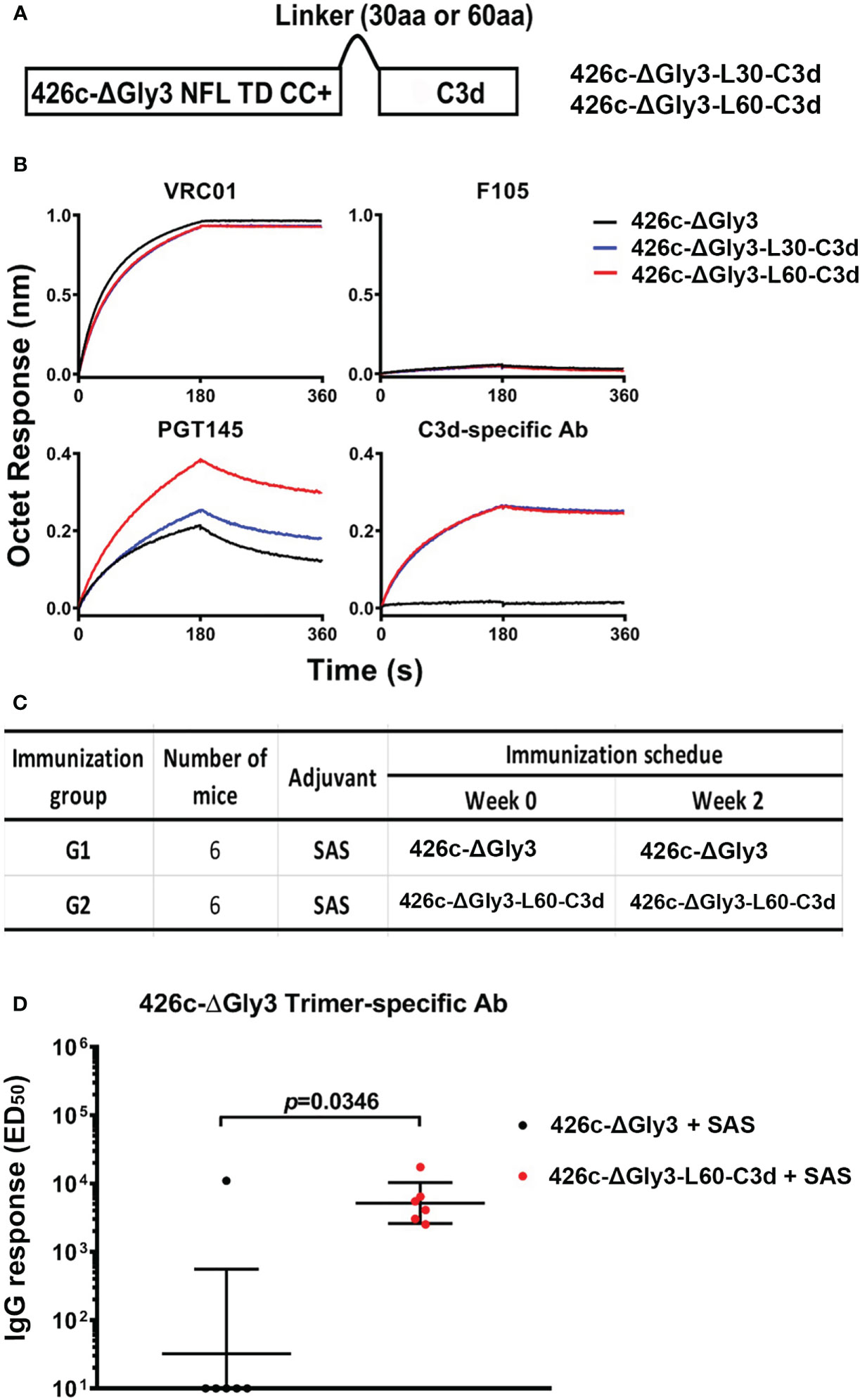
Figure 8 Design, antigenic characterization, and immunogenic analysis of 426c-ΔGly3-C3d trimers. (A) Schematic representation of 426c-ΔGly3-C3d fusion trimers. (B) 426c-ΔGly3-C3d trimers display favorable antigenic profile against Env-specific bNAbs, non-bNAbs, and mouse C3d-specific Ab. (C) Immunization groups and vaccine regimen. (D) ED50 binding Ab titers with geometric mean and geometric standard derivation against 426c-ΔGly3 trimers (captured by 2G12) in VRC01gHL mice after two immunizations. Data shown are an average of two independent measurements. Pre-immune sera from mice had no detectable specific IgG response and are not shown. Fusion of C3d domains to 426c-ΔGly3 trimers resulted in statistically significant improved binding titers in mice *, indicate P ≤ 0.05; **, indicate P ≤ 0.01; ***, indicate P ≤ 0.001. ns indicates non-significant..
The C3d domain naturally enhances activation of the innate and adaptive arms of immune system by interacting with the CR2 domain of the complement system. The goal of this study was to improve the humoral response to HIV-1 by fusing the molecular adjuvant, C3d, to the “newer generation” cleavage-independent, native-like NFL Env trimers. Accordingly, we generated fusion Env-C3d trimers from the clade B JRFL virus as well as from the clade C 426c virus. Env-C3d achieved a well-ordered trimeric structure and maintained the functional integrity of both the Env and C3d components, respectively. The fusion of C3d did not dramatically affect the antigenicity of the Env trimers; however, it enhanced the ability of the Env trimers to engage and activate B cells in vitro and in vivo. The SAS adjuvant does not affect the trimer integrity in vitro, but it is disruptive to the trimer integrity in vivo, resulting in an increased exposure of the V3 epitope. Nonetheless, in vivo, there is an additive effect of the SAS adjuvant and the molecular adjuvant C3d to improve the immunogenicity of the Env trimers. The fusion of C3d to the Env trimers improves the elicited antibody responses following vaccination, including Env-specific binding and neutralizing antibodies.
Initially, we screened the linker length between NFL Env and C3d. The 30- residue- (L30) and 60- residue- (L60) long linkers, rather than the 14- residue linker, generate well-ordered Env-C3d fusion trimers. Both Env-C3d variants exhibited similar biochemical, biophysical, structural, and antigenic properties, compared with their counterpart Env trimers. However, the yield of Env-L60-C3d was twofold higher than that of Env-L30-C3d. Env-L60-C3d showed slightly better binding reactivities to bNAbs and activation of B cells in vitro, compared with Env-L30-C3d, indicating that the L60 linker is likely better than the L30 linker for the fusion protein to achieve the structural and functional integrity of both the trimer and the C3d components. The 60-residue linker may not be the optimal linker length, as we did not assess intermediate linker lengths using stepwise increases of the G4S linker as we previously did to generate the original cleavage-independent NFL Env trimers (5). Nonetheless, the 60-residue linker was efficient to covalently fuse the NFL Env to the molecular adjuvant C3d, to form a well-ordered trimer, and to maintain their respective functional integrity.
Furthermore, the fusion of C3d enhanced the ability of the Env trimers to engage and activate B cells in vivo, reflected by the enhanced germinal center formation following two trimer inoculations. These results are consistent with recent work demonstrating that the follicular environment may be lower in proteolytic activity and that targeting to FDCs may benefit the elicited antibody to conformationally sensitive antigens such as the HIV Env trimer (26). In addition, Env-specific binding antibody response, especially in the presence of the SAS adjuvant, was detected with the C3d fusion to the NFL trimer. In the absence of the SAS adjuvant, the enhancement was evident after the first immunization, but not the following boosting, indicating that C3d alone was not sufficient to markedly improve the elicited antibody response. One potential reason is that following repeated immunization, the Env-C3d immunogens elicited de novo antibody responses instead of antibody affinity maturation. Furthermore, although mice are genetically inbred and are sufficient to generate Env binding titers, the mouse is not an ideal animal model to elicit neutralizing antibodies against the native HIV-1 Env trimer on the virus. In a previous study, BG505 SOSIP trimers failed to elicit autologous tier 2 NAb response to BG505.T332N in wild-type mice (34); however, more recently, He et al. reported that gp41ECTO-stabilized trimers were able to elicit tier 2 autologous NAbs (37). In this study, the Env-C3d trimers elicited tier 1 neutralizing antibodies only in the presence of the SAS adjuvant.
Elicitation of antibodies against non-neutralizing epitopes on Env may be an “immune distraction” to develop cross- neutralizing antibodies (7). Antibodies elicited against non-broadly neutralizing epitopes comprising the V3 epitope typically neutralize tier 1 MN.3 virus but fail to neutralize tier 2 viruses. A number of strategies have been employed to stabilize HIV-1 Env trimers with reduced exposure of non-neutralizing epitopes, including the V3 epitope (8, 11, 38–41). However, the reduced V3 responses do not enhance the elicitation of autologous or heterologous tier 2 neutralization. In our study, in the absence of the SAS adjuvant, the Env and Env-C3d trimers failed to elicit NAbs against the tier 1 MN.3 virus, which correlates with the low level of V3-specific binding antibody titers. However, in the presence of the SAS adjuvant, there are strong binding antibodies targeting the V3 epitope and neutralizing antibodies against the tier 1 MN.3 virus. The results indicate that, in vivo, the Env and Env-C3d trimers maintain a stable trimeric structure when formulated in physical conditions, but the SAS adjuvant is disruptive to the trimer integrity at the V3 epitope.
There have been few studies of how adjuvants affect HIV-1 Env proteins in vitro and in vivo (13, 42–46). For example, the complete Freund’s adjuvant impairs the conformational integrity of gp120 (45, 46). We previously demonstrated that BG505 NFL or SOSIP trimers retained the quaternary structure more effectively in ISCOMATRIX for up to 7 days than in Adjuplex (42). Adjuplex-induced conformational changes on BG505 NFL can affect the elicitation of tier 2 autologous neutralization and increased tier 1 neutralizing titers (13). Ozorowski et al. recently used a range of analytical techniques to investigate the effects of nine adjuvants on HIV-1 Env SOSIP trimers in vitro and found that most of the tested adjuvants have negligible impact on SOSIP trimers, including the SAS adjuvant (43). Similarly, in our study, the SAS adjuvant does not affect the trimers’ integrity in vitro, but it seems to affect the integrity of Env trimers in vivo by the indirect elicitation of V3-directed mAbs. Therefore, the effect of adjuvants on HIV-1 Env trimers in vitro may not always be reflected by their effects on Env trimers in vivo. The data indicate that the effects of adjuvant are far more complex and perhaps innate immune activation could also be involved (47, 48). If overall effects conferred by an adjuvant on the meta-stable HIV-1 trimer can only be determined in vivo, future selection of the appropriate adjuvant formulation(s) toward an effective HIV-1 vaccine should include both in vitro and in vivo testing.
In sum, we successfully engineered Env-C3d fusion trimers. The fusion of C3d to the NFL Env trimers improves B- cell antibody responses in animals, and the use of molecular adjuvant C3d could be useful for Env-based vaccines against HIV and other pathogens. The development of HIV-1 Env-C3d fusion immunogens is important as it might circumvent the need for a classical adjuvant, which can adversely affect spike conformation, as these trimers move into the clinic. These results have broader applications for other vaccines that may be extended to conformationally sensitive immunogens derived from other pathogens. Very recently, it was shown that the lymph node follicles may be privileged sites with decreased proteolytic activity that may better maintain native antigen conformation. Thereby, targeting follicular FDCs with Env-C3d fusion trimers may improve the elicitation of antibodies to native-like determinants (26).
Further studies are needed to increase and optimize the number of C3d domains attached per each protomer of the Env trimer. The newer constructs will be tested with different adjuvant formulations that maintain the conformational integrity of the antigens. Immunization studies in animal models other than mice (rabbits, guinea pigs, NHPs for example) with immunoglobulin repertoires that effectively recognize neutralizing determinants on the Env-C3d trimers that can result in tier 2 virus neutralization will provide valuable insights into further efficacy of C3d fusion to Env trimers.
To generate Env-C3d fusion trimers, we designed the constructs by fusing mouse C3d (21, 24) to the 3′-end of HIV-1 Env NFL TD CC+. The JRFL NFL TD CC+ (namely, JRFL) (5, 27) and C3d were linked by a flexible linker (G4S) of varying lengths (14aa, 30aa, 60aa), namely, JRFL-L14-C3d, JRFL-L30-C3d, and JRFL-L60-C3d, respectively.
426c Env with a three- N-glycan deletion (Δ276 Δ460 Δ463, namely ΔGly3) (36) was used as a template to generate 426c-ΔGly3 NFL TD CC+ (namely, 426c-ΔGly3), as previously described (27). Then, we fused the mouse C3d to the C-terminus of 426c-ΔGly3 through a flexible linker (G4S) of varying lengths (30aa, 60aa), namely, 426c-ΔGly3-L30-C3d and 426c-ΔGly3-L60-C3d, respectively. All native Env signal peptide sequences were replaced by the CD5 leader sequence to increase secretion. The gene constructs were codon optimized for mammalian expression, synthesized (GenScript), and inserted into the CMV/R vector, as previously described (27).
To generate the mouse C3d trimer, we fused the foldon trimeric motif to the C terminus of mouse C3d through two copies of the G4S linker. Foldon is a 27-residue trimerization domain at the C-terminal bacteriophage T4 fibritin (49–52). A His-tag was added to the C3d trimer for purification. The fusion gene named C3d-Fd was cloned into an inducible S2 expression vector pMT/BiP/V5-His (Invitrogen), as previously described (53, 54). The amino acid sequence of all the constructs used for this study is shown in Table S1.
The constructs expressing JRFL-C3d and 426c-ΔGly3-C3d fusion proteins with different linkers were transiently transfected into suspension 293F cells as previously described (55). Env proteins were harvested 4 days post transfection and purified by lectin affinity chromatography (Galanthus nivalis, Vector Labs) followed by size exclusion chromatography (SEC) on a Superdex 200 16/60 or Superdex 200 10/300 GL (GE Healthcare) columns. The trimer peak was subjected to negative selection by the non-neutralizing mAb F105 to remove disordered trimers (31). The flow-through from the F105 column, containing the well-ordered trimers, was resolved by a second SEC step.
C3d-Fd proteins were produced in stably transfected Drosophila S2 cells as previously described (53). Briefly, S2 cells were cotransfected with C3d-Fd and selection vector pCoBlast, which contains the blasticidin resistance gene (Invitrogen). After blasticidin selection, stable S2 transfectants were generated that were used for protein production. Briefly, when S2 cells reached 10 M/ml in 1 L of Express Five SFM medium without FBS, 5 μM CdCl2 was added into culture medium to induce protein expression; 3 days after the induction, the culture supernatants were harvested, clarified by centrifugation and filtration. Filtered supernatants were purified by Ni-NTA column followed by SEC.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blue-native PAGE (BN-PAGE), and western blot analyses were performed as described elsewhere (55). For western blot analysis, samples were separated through 3%–12% NuPAGE Gels and transferred onto polyvinylidene difluoride membranes (Invitrogen). The membranes were blocked in a solution of Tris-buffered saline containing 5% non-fat dry milk and 0.05% Tween 20 and subsequently probed with anti-Env antibodies (2G12 and VRC01) or biotinylated-goat anti-mouse C3d polyclonal antibody. Proteins were visualized with HRP-conjugated anti-human IgG antibody or streptavidin-peroxidase polymer according to the manufacturer’s instructions (Sigma).
The thermal transition points (Tm) of JRFL-C3d and 426c-ΔGly3-C3d fusion trimers were determined by differential scanning calorimetry (DSC) using a MicroCal VP- Capillary instrument (Malvern), as described previously (5, 27).
The purified Env-C3d fusion trimers were analyzed by negative stain electron microscopy (NS-EM). A 3- µl aliquot containing ~0.01 mg/ml of the sample was applied for 15 s onto a carbon-coated 400 Cu-mesh grid that had been glow- discharged at 30 mA for 30 s, then negatively stained with 0.7% uranyl formate for 45 s. Data were collected using an FEI T20 electron microscope operating at 200 kV, with an electron dose of ~45 e-/Å2 and a magnification of 80,000 × that resulted in a pixel size of 2.74 Å at the specimen plane. Images were acquired with an Eagle 2k × 2k CCD camera (FEI) using a nominal defocus of 1000 nm and the Serial EM software (56). Particles were selected from the micrographs and extracted, and a reference-free 2D class averages were obtained using RELION 2.1.0 (57).
Briefly, the BLI analysis was carried out on an Octet Red instrument (ForteBio) with IgGs immobilized on anti-human IgG Fc capture sensors (ForteBio). The Env-C3d trimers were assessed as free analytes in solution (PBS pH 7.4) at a final concentration of 200 nM. Association and dissociation were measured for 180 s. Data were analyzed using ForteBio version 7.1.
For mouse C3d detection, biotinylated-goat anti-mouse C3d polyclonal antibody was immobilized on streptavidin sensors. The fusion Env-C3d or C3d-Fd trimer proteins were assessed as free analytes in solution (PBS pH 7.4) at a final concentration of 200 nM.
For the binding of JRFL-C3d trimers to soluble CR2 protein, his-tagged human CR2 protein (Cat. #10811-H08H, Sino Biological Inc.) was immobilized on anti-HIS sensors. The fusion Env-C3d trimers were assessed as free analytes in solution (PBS pH 7.4) at a final concentration of 200 nM.
To test the capability of Env-C3d fusion trimers to bind CR2 on mouse B cells in vitro, we performed flowcytometry analysis. Briefly, we labeled trimers (JRFL, JRFL-L30-C3d, and JRFL-L60-C3d) with Alexa Fluor 680 dye (AF680) according to the manufacturer’s instructions (Thermo Scientific). Then, the PGV04 WEH-I231 cells were treated with 50 ng/μl doxycycline overnight for induction of PGV04 expression on the cell surface. PGV04 expression was confirmed with human constant chain kappa (hCk) expression by flowcytometry analysis. Non-treated PGV04 WEH-I231 cells, with no PGV04 expression on the cell surface, were included as control. WEH-I231 cells with or without PGV04 expression were incubated with AF680- labeled trimers (JRFL, JRFL-L30-C3d, JRFL-L60-C3d) or PBS on ice for 30 min. The cells were then washed twice with wash buffer (PBS containing 1% BSA and 0.02% NaN3) and fixed with 1% formaldehyde in 0.5 ml of wash buffer. Flowcytometry analyses were performed on a BD LSR II flow cytometer (BD Biosciences).
To test the capability of Env-C3d fusion proteins to activate mouse B cells in vitro, we performed calcium flux analysis (28, 32). Briefly, PGV04 WEH-I231 cells were treated with 50 ng/μl doxycycline overnight and confirmed for PGV04 expression. PGV04-expressing WEH-I231 cells were suspended at 4 million cells/ml in Hanks balanced salt solution (HBSS), labeled with 0.75 μM Indo-1 (Invitrogen) for 30 min at 37°C, and washed with 2 mM CaCl2-HBSS, followed by incubation for 30 min at 37°C. 300 μl of cells at 2 million cells/ml were then stimulated at 37°C with trimers (JRFL, JRFL-L30-C3d, and JRFL-L60-C3d) in solution at a final concentration of 50 μg/ml. Ca2+ signals were recorded for 120 s, measuring the 405/485-nm emission ratio of Indo-1 fluorescence upon UV excitation. Calcium flux analysis was performed on an LSR II cytometer (BD Biosciences). Kinetic analysis was performed using FlowJo (Tree Star).
For JRFL-C3d immunization studies, female C57BL/6 (B6) mice (7 to 8 weeks old), six animals per group, were used. B6 mice were inoculated subcutaneously at two sites with 20 μg of soluble trimers in PBS or formulated in 50% v/v Sigma Adjuvant System (SAS, containing 0.5 mg/ml MPLA) in a total volume of 200 μl. Inoculations were done at 0, 4, 10, and 17 weeks. Serum samples were collected 2 weeks before or after each inoculation (see the detailed immunization schedule in Figure 6A). One mouse (# ZZ6155) in group 1 died at week 4.
For 426c-ΔGly3-C3d immunization studies, germ-line VRC01 heavy- chain and light- chain (VRC01gHL) knock-in mice as described elsewhere (58) were used. Six animals per group were inoculated subcutaneously with 20 μg of soluble 426c-ΔGly3 or 426c-ΔGly3-L60-C3d trimers formulated in SAS in a total volume of 200 μl. Inoculations were done at 0 and 2 weeks. Bleeds were collected before inoculation and 2 weeks after second inoculation. The detailed immunization schedule is shown in Figure 8C.
JRFL and 426c trimer-specific IgG-binding titers were determined using 2G12-capture ELISA as previously described (10, 59, 60). Briefly, the 96 half-well ELISA plates were coated with 2G12 mAb (at 2 μg/ml) overnight at 4°C. After blocking and washing, the JRFL trimers at 2 μg/ml were captured onto the plates in blocking buffer for 1 h at 37°C. After washing, the plates were incubated with fivefold serial dilutions of sera starting at 1:20 dilution, for 1 h. The plates were further incubated with horseradish peroxidase (HRP)-coupled anti-mouse IgG at 1:5,000 dilution for 1 h and developed with HRP-3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution. The HRP-TMB reaction was stopped with 0.3 N sulfuric acid, and absorbance was measured at 450 nm. GraphPad Prism software was used to calculate half-maximal effective concentration of serum binding titers (ED50).
V3-specific IgG-binding titers were determined by coating 2 μg/ml of JRFL V3 peptide TRPNNNTRKSIHIGPGRAFYTTGEIIGDIRQAH (from amino acids 297–317 according to HxBc2 numbering) on the ELISA plates. The following steps were the same as described above without adding JRFL trimers. Endpoint titers for each sample were calculated based on their OD450 values, and OD450 values over threefold of the background were considered as positive.
To test the effect of the SAS adjuvant on the antigenicity of the Env trimer, we incubated the JRFL trimer containing a His-tag with or without the SAS adjuvant at the same ratio as used in immunization in vivo either at room temperature or at 37°C for 24 h. ELISA plates were coated with anti-His antibody. JRFL trimers with or without incubation in adjuvant were captured on the plates, and a panel of monoclonal antibodies were used to detect the integrity of the trimer.
To quantify 426c trimer- specific IgG-binding titers in VRC01gHL knock-in mice sera, we generated mouse VRC01 IgG1 Ab as a standard. Briefly, VRC01 heavy- and light- chain variable regions were cloned with mouse IgG1 Fc or mouse Kappa constant region into pCI-WPRE plasmid (modified pCI plasmid from Promega), respectively. Equal amounts of heavy- and light- chain plasmids were used to transfected 293F cells, and the supernatant was harvested at day 5. VRC01 mouse IgG1 in culture supernatants was purified with a HiTrap Protein G HP column and dialyzed against PBS. For quantification, ELISA plates coated with 2G12 (2 μg/ml) were incubated with the 426c-ΔGly3 trimer at 2 μg/ml. After washing, the plates were incubated with fivefold serially diluted mice sera starting at 1:500 dilution or serially diluted VRC01 mouse IgG1. Purified VRC01 mouse IgG1 was used as standard to calculate the IgG antibody concentration in immunized mouse sera. Non-linear fitting of absorbance data was performed in GraphPad Prism software.
Germinal center (GC) analysis was performed as previously reported with some adjustments (28). The intramuscular injections were at two sites (100 μl in each site) administered in the thigh muscles of the hind limb of 8-week-old female C57BL/6 mice (eight mice per group) at weeks 0 and 4 with the JRFL trimer (15 μg) or JRFL-L60-C3d trimer (20 μg) formulated in SAS Adjuvant System (Sigma-Aldrich). One group of mice (3 animals) received the SAS adjuvant as a negative control; 2 weeks post second immunization, the draining popliteal and inguinal lymph nodes and spleen were harvested and pressed through a 70-μm cell strainer to obtain a single-cell suspension. The cells were labeled with a LIVE/DEAD viability dye (fixable viability stain, FVS 450, 562247, BD, USA) to exclude dead cells and with PE rat anti-mouse CD19 (BD, 553786), PE Rat Anti-Mouse CD45R/B220 (Clone RA3-6B2, 561878), and fluorescein isothiocyanate (FITC) rat anti-mouse B-cell activation antigen (clone GL7 for germinal center B cells, BD, 553666) markers. Samples were then analyzed on a NovoCyte Flow Cytometer Systems (BD, USA). The post-acquisition analyses, including compensation, were performed using FlowJo software, version 10.8.1 (FlowJo LLC, USA).
Antibody avidity was evaluated by sodium thiocyanate (NaSCN) ELISA as described previously (61) with some modifications. ELISA plates were coated with JRFL Env trimers (at 2 μg/ml) overnight at 4°C. After blocking and washing, serially diluted mice sera were added onto the plates and incubated for 1 h at 37°C before exposure to 1.5 M NaSCN or PBS for 15 min at room temperature. Subsequent steps were performed similarly to 2G12 capture ELISA, as described in the previous section. Endpoint titers for each sample were calculated based on their OD450 values, and OD450 values over threefold of the background were considered as positive. The avidity index was calculated by dividing the endpoint titers for NaSCN-treated samples to those of PBS-treated samples and multiplying by 100.
Serum neutralization analysis was performed in TZM-bl cells, using a single round HIV-1 Env pseudovirus assay (33). To determine the sera dilution that resulted in a 50% reduction in relative luminescence units (RLU), serial dilutions of the sera were performed and the neutralization dose–response curves were fit by non-linear regression using a five-parameter hill slope equation. Diverse HIV-1 virus isolates were used in the neutralization assays. The neutralization titer was expressed as the reciprocal serum dilution that resulted in 50% reduction of RLU (ID50).
Analyses were performed with GraphPad Prism v8.0. Different groups in mouse immunizations were compared by two-tailed Mann–Whitney U test. Correlations between the NAb titers and the V3-specific binding antibody titers were assessed by using the non-parametric (two-tailed) Spearman rank correlation method. Data sets were considered statistically significant at a P value of ≤ 0.05.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
All mouse studies were performed at TSRI animal facilities under the approval of the TSRI Institutional Animal Care and Use Committee (IACUC) (study numbers - M1708, M1808, M1813, M1905). The committee uses the Guide for Care and Use of Laboratory Animals (National Academy Press), PHS policy, and the AWA as standards. All animal handling protocols were designed to minimize animal discomfort.
LY, SB, MA, JG, and RTW designed the research studies. LY, SB, RW, TO, MA, EDD, and CAC performed the experiments and data analysis. LY, SB, and RTW wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
The work was supported by HIVRAD grants P01 AI104722 and P01 AI157299 (SB, TO, MA, WL, EDD, RW, and RTW), grant P01 AI124337 (RTW), Scripps CHAVI-ID AI100663 (LY and RTW), grants R01 AI0988602 and R01 AI145055, Scripps CHAVD UMI AI144462 (RTW, CAC, and ABW), and funding from the International AIDS Vaccine Initiative (IAVI) (KT, JG, and RTW). The full list of donors for IAVI can be found on the website http://www.iavi.org. This study received funding from IAVI. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Authors JG, KT, and RTW are/were employed by IAVI.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1180959/full#supplementary-material
Supplementary Figure 1 | Characterization of JRFL-L14-C3d trimer. (A) SEC profile of JRFL-L14-C3d trimer following lectin affinity-purification. Fractions selected for further analysis are labeled. (B) Blue-native PAGE (BN-PAGE) analysis of fractions of JRFL-L14-C3d trimers from SEC purification. The band representing the trimer is marked in red. The major peak from SEC purification (fractions 8-10) is comprised of non-specific aggregates with a very small fraction of trimer. (C) SEC profile of JRFL NFL trimer following lectin affinity-purification and negative selection by F105.
Supplementary Figure 2 | Binding of JRFL-L60-C3d trimer to soluble human CR2 protein. Bio-layer interferometry (BLI) measurements show that JRFL-L60-C3d trimers bind to human CR2 protein. Binding was not observed with the control JRFL trimer lacking the C3d domain.
Supplementary Figure 3 | Neutralizing antibody titers correlate with V3-specific binding antibody titers. (A) Endpoint V3-specific binding antibody titers as determined by V3-peptide capture ELISA at week 19. (B) Midpoint neutralization titers (ID50) against tier 1 MN.3 pseudotyped virus are plotted against the V3-specific binding antibody titers (ED50). The spearman r-value and two-tailed P value are shown.
Supplementary Figure 4 | Effect of SAS adjuvant on the antigenicity of JRFL trimer. ELISA binding curves of selected bNAbs and non-bNAbs to JRFL trimers. The trimers were incubated +/- SAS adjuvant at room temperature (RT) or 37°C for 24 h prior to coating on the ELISA plate.
Supplementary Table 1 | Amino acid sequences of all antigens used for this study.
1. Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in hiv-1 infection. Nat Immunol (2015) 16(6):571–6. doi: 10.1038/ni.3158
2. Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, et al. A next-generation cleaved, soluble hiv-1 env trimer, Bg505 Sosip.664 Gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PloS Pathog (2013) 9(9):e1003618. doi: 10.1371/journal.ppat.1003618
3. Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, et al. Single-chain soluble Bg505.Sosip Gp140 trimers as structural and antigenic mimics of mature closed hiv-1 env. J Virol (2015) 89(10):5318–29. doi: 10.1128/JVI.03451-14
4. Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, et al. Structure-guided redesign increases the propensity of hiv env to generate highly stable soluble trimers. J Virol (2015) 90(6):2806–17. doi: 10.1128/JVI.02652-15
5. Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, et al. Cleavage-independent hiv-1 env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep (2015) 11(4):539–50. doi: 10.1016/j.celrep.2015.03.047S2211-1247(15)00326-5
6. Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, et al. Uncleaved prefusion-optimized Gp140 trimers derived from analysis of hiv-1 envelope metastability. Nat Commun (2016) 7:12040. doi: 10.1038/ncomms12040
7. Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, et al. Hiv-1 vaccines. hiv-1 neutralizing antibodies induced by native-like envelope trimers. Science (2015) 349(6244):aac4223. doi: 10.1126/science.aac4223
8. de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, et al. Immunogenicity of stabilized hiv-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell (2015) 163(7):1702–15. doi: 10.1016/j.cell.2015.11.056S0092-8674(15)01575-5
9. Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, et al. Sequential and simultaneous immunization of rabbits with hiv-1 envelope glycoprotein Sosip.664 trimers from clades a, b and c. PloS Pathog (2016) 12(9):e1005864. doi: 10.1371/journal.ppat.1005864PPATHOGENS-D-16-01438
10. Martinez-Murillo P, Tran K, Guenaga J, Lindgren G, Adori M, Feng Y, et al. Particulate array of well-ordered hiv clade c env trimers elicits neutralizing antibodies that display a unique V2 cap approach. Immunity (2017) 46(5):804–17.e7. doi: 10.1016/j.immuni.2017.04.021
11. Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by hiv envelope trimer immunization using optimized approaches. Immunity (2017) 46(6):1073–88.e6. doi: 10.1016/j.immuni.2017.05.007
12. McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, et al. Holes in the glycan shield of the native hiv envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep (2016) 16(9):2327–38. doi: 10.1016/j.celrep.2016.07.074S2211-1247(16)31020-8
13. Bale S, Martine A, Wilson R, Behrens AJ, Le Fourn V, de Val N, et al. Cleavage-independent hiv-1 trimers from cho cell lines elicit robust autologous tier 2 neutralizing antibodies. Front Immunol (2018) 9:1116. doi: 10.3389/fimmu.2018.01116
14. Ward AB, Wilson IA. The hiv-1 envelope glycoprotein structure: nailing down a moving target. Immunol Rev (2017) 275(1):21–32. doi: 10.1111/imr.12507
15. Sanders RW, Moore JP. Native-like env trimers as a platform for hiv-1 vaccine design. Immunol Rev (2017) 275(1):161–82. doi: 10.1111/imr.12481
16. Karlsson Hedestam GB, Guenaga J, Corcoran M, Wyatt RT. Evolution of b cell analysis and env trimer redesign. Immunol Rev (2017) 275(1):183–202. doi: 10.1111/imr.12515
17. Dubrovskaya V, Tran K, Ozorowski G, Guenaga J, Wilson R, Bale S, et al. Vaccination with glycan-modified hiv nfl envelope trimer-liposomes elicits broadly neutralizing antibodies to multiple sites of vulnerability. Immunity (2019) 51(5):915–29.e7. doi: 10.1016/j.immuni.2019.10.008
18. Kong R, Duan H, Sheng Z, Xu K, Acharya P, Chen X, et al. Antibody lineages with vaccine-induced antigen-binding hotspots develop broad hiv neutralization. Cell (2019) 178(3):567–84.e19. doi: 10.1016/j.cell.2019.06.030
19. Bonsignori M, Moody MA, Parks RJ, Holl TM, Kelsoe G, Hicks CB, et al. Hiv-1 envelope induces memory b cell responses that correlate with plasma antibody levels after envelope Gp120 protein vaccination or hiv-1 infection. J Immunol (2009) 183(4):2708–17. doi: 10.4049/jimmunol.0901068
20. Pulendran B, Arunachalam PS, O'Hagan DT. Emerging concepts in the science of vaccine adjuvants. Nat Rev Drug Discovery (2021) 20:454–75. doi: 10.1038/s41573-021-00163-y
21. Dempsey PW, Allison ME, Akkaraju S, Goodnow CC, Fearon DT. C3d of complement as a molecular adjuvant: bridging innate and acquired immunity. Science (1996) 271(5247):348–50. doi: 10.1126/science.271.5247.348
22. Gonzalez SF, Degn SE, Pitcher LA, Woodruff M, Heesters BA, Carroll MC. Trafficking of b cell antigen in lymph nodes. Annu Rev Immunol (2011) 29:215–33. doi: 10.1146/annurev-immunol-031210-101255
23. Carroll MC, Isenman DE. Regulation of humoral immunity by complement. Immunity (2012) 37(2):199–207. doi: 10.1016/j.immuni.2012.08.002
24. Koch M, Frazier J, Sodroski J, Wyatt R. Characterization of antibody responses to purified hiv-1 Gp120 glycoproteins fused with the molecular adjuvant C3d. Virology (2005) 340(2):277–84. doi: 10.1016/j.virol.2005.06.034
25. Ross TM, Xu Y, Green TD, Montefiori DC, Robinson HL. Enhanced avidity maturation of antibody to human immunodeficiency virus envelope: DNA vaccination with Gp120-C3d fusion proteins. AIDS Res Hum Retroviruses (2001) 17(9):829–35. doi: 10.1089/088922201750252025
26. Aung A, Cui A, Maiorino L, Amini AP, Gregory JR, Bukenya M, et al. Low protease activity in b cell follicles promotes retention of intact antigens after immunization. Science (2023) 379(6630):eabn8934. doi: 10.1126/science.abn8934
27. Guenaga J, Garces F, de Val N, Stanfield RL, Dubrovskaya V, Higgins B, et al. Glycine substitution at helix-to-Coil transitions facilitates the structural determination of a stabilized subtype c hiv envelope glycoprotein. Immunity (2017) 46(5):792–803.e3. doi: 10.1016/j.immuni.2017.04.014
28. Bale S, Goebrecht G, Stano A, Wilson R, Ota T, Tran K, et al. Covalent linkage of hiv-1 trimers to synthetic liposomes elicits improved b cell and antibody responses. J Virol (2017) 91(16). doi: 10.1128/JVI.00443-17
29. Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. X-Ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science (1998) 280(5367):1277–81. doi: 10.1126/science.280.5367.1277
30. van den Elsen JM, Isenman DE. A crystal structure of the complex between human complement receptor 2 and its ligand C3d. Science (2011) 332(6029):608–11. doi: 10.1126/science.1201954
31. Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, et al. Well-ordered trimeric hiv-1 subtype b and c soluble spike mimetics generated by negative selection display native-like properties. PloS Pathog (2015) 11(1):e1004570. doi: 10.1371/journal.ppat.1004570PPATHOGENS-D-14-01819
32. Ota T, Doyle-Cooper C, Cooper AB, Huber M, Falkowska E, Doores KJ, et al. Anti-hiv b cell lines as candidate vaccine biosensors. J Immunol (2012) 189(10):4816–24. doi: 10.4049/jimmunol.1202165
33. Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype b infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol (2005) 79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005
34. Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, et al. Murine antibody responses to cleaved soluble hiv-1 envelope trimers are highly restricted in specificity. J Virol (2015) 89(20):10383–98. doi: 10.1128/JVI.01653-15
35. Ingale J, Stano A, Guenaga J, Sharma SK, Nemazee D, Zwick MB, et al. High-density array of well-ordered hiv-1 spikes on synthetic liposomal nanoparticles efficiently activate b cells. Cell Rep (2016) 15(9):1986–99. doi: 10.1016/j.celrep.2016.04.078
36. McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, et al. Engineering hiv envelope protein to activate germline b cell receptors of broadly neutralizing anti-Cd4 binding site antibodies. J Exp Med (2013) 210(4):655–63. doi: 10.1084/jem.20122824
37. He L, Kumar S, Allen JD, Huang D, Lin X, Mann CJ, et al. Hiv-1 vaccine design through minimizing envelope metastability. Sci Adv (2018) 4(11):eaau6769. doi: 10.1126/sciadv.aau6769
38. Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, et al. Structure-based design of native-like hiv-1 envelope trimers to silence non-neutralizing epitopes and eliminate Cd4 binding. Nat Commun (2017) 8(1):1655. doi: 10.1038/s41467-017-01549-6
39. de Taeye SW, de la Pena AT, Vecchione A, Scutigliani E, Sliepen K, Burger JA, et al. Stabilization of the Gp120 V3 loop through hydrophobic interactions reduces the immunodominant V3-directed non-neutralizing response to hiv-1 envelope trimers. J Biol Chem (2018) 293(5):1688–701. doi: 10.1074/jbc.RA117.000709
40. Ringe RP, Ozorowski G, Rantalainen K, Struwe WB, Matthews K, Torres JL, et al. Reducing V3 antigenicity and immunogenicity on soluble, native-like hiv-1 env sosip trimers. J Virol (2017) 91(15). doi: 10.1128/JVI.00677-17
41. Cheng C, Pancera M, Bossert A, Schmidt SD, Chen RE, Chen X, et al. Immunogenicity of a prefusion hiv-1 envelope trimer in complex with a quaternary-Structure-Specific antibody. J Virol (2015) 90(6):2740–55. doi: 10.1128/JVI.02380-15
42. Feng Y, Tran K, Bale S, Kumar S, Guenaga J, Wilson R, et al. Thermostability of well-ordered hiv spikes correlates with the elicitation of autologous tier 2 neutralizing antibodies. PloS Pathog (2016) 12(8):e1005767. doi: 10.1371/journal.ppat.1005767PPATHOGENS-D-16-00497
43. Ozorowski G, Cupo A, Golabek M, LoPiccolo M, Ketas TA, Cavallary M, et al. Effects of adjuvants on hiv-1 envelope glycoprotein sosip trimers in vitro. J Virol (2018) 92(13). doi: 10.1128/JVI.00381-18
44. Fox CB, Kramer RM, Barnes VL, Dowling QM, Vedvick TS. Working together: interactions between vaccine antigens and adjuvants. Ther Adv Vaccines (2013) 1(1):7–20. doi: 10.1177/2051013613480144
45. Grundner C, Pancera M, Kang JM, Koch M, Sodroski J, Wyatt R. Factors limiting the immunogenicity of hiv-1 Gp120 envelope glycoproteins. Virology (2004) 330(1):233–48. doi: 10.1016/j.virol.2004.08.037
46. VanCott TC, Mascola JR, Kaminski RW, Kalyanaraman V, Hallberg PL, Burnett PR, et al. Antibodies with specificity to native Gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric Gp160. J Virol (1997) 71(6):4319–30. doi: 10.1128/jvi.71.6.4319-4330.1997
47. Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity (2010) 33(4):492–503. doi: 10.1016/j.immuni.2010.10.002
48. Liang F, Lindgren G, Sandgren KJ, Thompson EA, Francica JR, Seubert A, et al. Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci Transl Med (2017) 9(393). doi: 10.1126/scitranslmed.aal2094
49. Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, et al. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol (2000) 74(10):4746–54. doi: 10.1128/jvi.74.10.4746-4754.2000
50. Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol (2000) 74(12):5716–25. doi: 10.1128/JVI.74.12.5716-5725.2000
51. Tao Y, Strelkov SV, Mesyanzhinov VV, Rossmann MG. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the c-terminal domain. Structure (1997) 5(6):789–98. doi: 10.1016/S0969-2126(97)00233-5
52. Liu L, Wang W, Yang L, Ren H, Kimata JT, Zhou P. Trimeric glycosylphosphatidylinositol-anchored Hcdr3 of broadly neutralizing antibody Pg16 is a potent hiv-1 entry inhibitor. J Virol (2013) 87(3):1899–905. doi: 10.1128/JVI.01038-12
53. Yang L, Song Y, Li X, Huang X, Liu J, Ding H, et al. Hiv-1 virus-like particles produced by stably transfected drosophila S2 cells: a desirable vaccine component. J Virol (2012) 86(14):7662–76. doi: 10.1128/JVI.07164-11
54. Huang X, Zhu Q, Huang X, Yang L, Song Y, Zhu P, et al. In vivo electroporation in DNA-vlp prime-boost preferentially enhances hiv-1 envelope-specific Igg2a, neutralizing antibody and Cd8 T cell responses. Vaccine (2017) 35(16):2042–51. doi: 10.1016/j.vaccine.2017.03.006
55. Yang L, Sharma SK, Cottrell C, Guenaga J, Tran K, Wilson R, et al. Structure-guided redesign improves nfl hiv env trimer integrity and identifies an inter-protomer disulfide permitting post-expression cleavage. Front Immunol (2018) 9:1631. doi: 10.3389/fimmu.2018.01631
56. Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol (2005) 152(1):36–51. doi: 10.1016/j.jsb.2005.07.007
57. Fernandez-Leiro R, Scheres SHW. A pipeline approach to single-particle processing in relion. Acta Crystallogr D Struct Biol (2017) 73(Pt 6):496–502. doi: 10.1107/S2059798316019276
58. Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, et al. Precursor frequency and affinity determine b cell competitive fitness in germinal centers, tested with germline-targeting hiv vaccine immunogens. Immunity (2018) 48(1):133–46.e6. doi: 10.1016/j.immuni.2017.11.023
59. Hogan MJ, Conde-Motter A, Jordan APO, Yang L, Cleveland B, Guo W, et al. Increased surface expression of hiv-1 envelope is associated with improved antibody response in vaccinia Prime/Protein boost immunization. Virology (2017) 514:106–17. doi: 10.1016/j.virol.2017.10.013
60. Soldemo M, Adori M, Stark JM, Feng Y, Tran K, Wilson R, et al. Glutaraldehyde cross-linking of hiv-1 env trimers skews the antibody subclass response in mice. Front Immunol (2017) 8:1654. doi: 10.3389/fimmu.2017.01654
Keywords: HIV-1, C3d, immunogenicity, molecular adjuvant, vaccine
Citation: Bale S, Yang L, Alirezaei M, Wilson R, Ota T, Doyle ED, Cottrell C A, Guenaga J, Tran K, Li W, Stamatatos L, Nemazee D, Ward AB and Wyatt RT (2023) Fusion of the molecular adjuvant C3d to cleavage-independent native-like HIV-1 Env trimers improves the elicited antibody response. Front. Immunol. 14:1180959. doi: 10.3389/fimmu.2023.1180959
Received: 06 March 2023; Accepted: 27 April 2023;
Published: 22 May 2023.
Edited by:
Eric Hunter, Emory University, United StatesReviewed by:
Ann J Hessell, Oregon Health and Science University, United StatesCopyright © 2023 Bale, Yang, Alirezaei, Wilson, Ota, Doyle, Cottrell, Guenaga, Tran, Li, Stamatatos, Nemazee, Ward and Wyatt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard T. Wyatt, d3lhdHRAc2NyaXBwcy5lZHU=
†Present address: Lifei Yang, Lifei Yang, Department of Immunology, VIR Biotechnology, San Francisco, CA, United States
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.