
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 08 June 2023
Sec. Mucosal Immunity
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1177504
This article is part of the Research TopicMucosal Vaccines for the Induction of Antimicrobial Immunoglobulin AView all 6 articles
The human gastrointestinal tract harbors trillions of commensal microorganisms. Emerging evidence points to a possible link between intestinal fungal dysbiosis and antifungal mucosal immunity in inflammatory bowel disease, especially in Crohn’s disease (CD). As a protective factor for the gut mucosa, secretory immunoglobulin A (SIgA) prevents bacteria from invading the intestinal epithelium and maintains a healthy microbiota community. In recent years, the roles of antifungal SIgA antibodies in mucosal immunity, including the regulation of intestinal immunity binding to hyphae-associated virulence factors, are becoming increasingly recognized. Here we review the current knowledge on intestinal fungal dysbiosis and antifungal mucosal immunity in healthy individuals and in patients with CD, discuss the factors governing antifungal SIgA responses in the intestinal mucosa in the latter group, and highlight potential antifungal vaccines targeting SIgA to prevent CD.
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis, is a chronic, relapsing and incurable inflammatory disease of the intestine, with growing global prevalence in the 21st century (1, 2). IBD pathogenesis involves several factors, including genetic susceptibility, environmental exposure, gut microbiota alterations, and both innate and adaptive immune responses (3, 4). Recent studies have revealed intestinal fungi as a key factor contributing to intestinal mucosal immunity and IBD development, which has raised questions regarding the specificity, functions, and mechanisms of induction of antifungal mucosal antibody responses in the gut (4–7).
In patients with IBD, structural and functional changes in the gut microbiota, including bacteria, viruses, fungi, and protozoa, disrupt the gut mucosal homeostasis, resulting in persistent and excessive immune system activation (5, 7). Many studies have focused on the role of bacteria in IBD, however, ample evidence from gut microbiota studies and studies on the immune responses to intestinal fungi suggests a potential link between fungi and IBD (7). Although fungi account for only approximately 0.1% of the total microorganisms in the gut, they have recently attracted attention for their role in IBD pathogenesis (6, 8, 9). SIgA is the most abundant antibody on the intestinal mucosal surfaces in humans and many other mammals, with approximately 3 g per day secreted into the human gut lumen (10–13). Acting as the first line of defense against pathogens, SIgA defends the intestinal epithelium by coating a substantial fraction of gut bacteria and maintaining homeostasis of the commensal microbiota (12, 14–18). Compared to healthy controls, mice and humans with reduced IgA secretion levels show an altered gut microbiota composition and increased susceptibility to IBD and other inflammatory diseases (15, 19, 20). The percentage of IgA-coated bacteria is dramatically increased in the feces of patients with IBD, particularly in patients with CD aged 17–40 years (21–23). In addition, in patients with CD, IgA-bound fecal bacterial frequencies are positively correlated with disease activity (23).
Notably, interactions between SIgA and gut bacteria may play a critical role in IBD severity (12, 23–25). However, the potential involvement of antifungal SIgA responses in CD remains largely unknown. In this review, we focus our discussion on recent advances in the understanding of intestinal fungal homeostasis and antifungal mucosal immunity in healthy and diseased (inflammatory) states. Further, we explore the roles and mechanisms of antifungal SIgA responses in the intestinal mucosa in patients with CD.
A study by Iliev et al. revealed that the presence of fungi, as measured by quantitative (q)PCR of fungal ribosomal DNA in the mucosa, increased from the ileum to the colon, with the highest frequencies in the distal colon (26). In the gastrointestinal tract (GIT) of humans, there are three major fungal phyla: Ascomycota, Basidiomycota, and Chytridiomycota; some research studies indicate that the colonic mucosa-associated fungal microbiota is dominated by Basidiomycota and Ascomycota (27–31), whereas other have indicated that Ascomycota are significantly more common than Basidiomycota in the human intestinal fungal microbial community (29, 30, 32–34). Using internal transcribed spacer (ITS) 2 sequencing, one study reported Saccharomyces cerevisiae and Candida albicans as the most commonly detected fungal species in stool samples of healthy subjects (35).
Numerous studies have revealed alterations in the fungal microbiota in patients with IBD. However, consensus is lacking because of the high variability of microbiomes among individuals from different areas and with different dietary preferences, and the use of different sample types and methodologies among studies (Table 1). Researchers have observed increased fungal diversity in colonic biopsy tissue samples or fecal samples from patients with CD in comparison to controls (36, 37). However, some recent studies of mucosa-associated microbes or stool samples reported little or no change in fungal diversity (alpha diversity) in patients with CD, but a significant increase in the global fungus load in both inflamed and non-inflamed mucosa during CD flare (29, 30). Moreover, decreased intestinal fungal diversity in stool samples from pediatric or adult patients with IBD compared to that in healthy subjects has also been reported (38, 39) (Table 1).
Some studies have revealed an increased abundance of Basidiomycota and a decreased abundance of Ascomycota, resulting in a higher Basidiomycota-to-Ascomycota ratio, in patients with CD compared to healthy controls (32, 38, 42, 43). However, other studies have indicated that the Basidiomycota-to-Ascomycota ratio is decreased in patients with CD (29, 30, 37, 40). Moreover, patients with CD have a decreased proportion of S. cerevisiae and an increased proportion of C. albicans in their gut compared to healthy subjects (30, 38). C. albicans can increase the production of pro-inflammatory cytokines, such as interferon (IFN)-γ, interleukin (IL)-17, and tumor necrosis factor (TNF)-α, and disrupt the gut microbiota composition to exacerbate gut inflammation (44–46). Besides C. albicans, Candida tropicalis and Candida glabrata are also significantly more abundant in patients with CD (29, 33). Recent studies have uncovered evidence for a strong association of Malassezia restricta and Malassezia globosa with CD, and M. restricta exacerbated colitis in mice in a CARD9-dependent manner (29, 32). Debaryomyces, particularly Debaryomyces hansenii, has been found to be enriched in inflamed intestinal tissues of patients with CD and to prevent colonic healing in the absence of altered Schaedler flora bacteria via the myeloid cell–IFN-γ–CCL5 axis (41) (Table 1).
With the rise of high-throughput sequencing, recent years have seen a steady increase in studies on the human intestinal fungal microbiota. Nevertheless, it is difficult to pinpoint the intestinal fungal alterations in patients with CD versus healthy people without differentiating the living areas and dietary preferences of study participants, the specific tissues sampled and the sampling methods used, and the methodology used to analyze the gut mycobiome. Clearly, there remain many challenges to characterizing the mycobiome. However, study findings have clearly revealed a strong association between the gut fungal microbiota and mucosal inflammation or disease activity of CD, which has drawn attention to the gut fungal microbiota composition and its possible roles in CD.
Several genome-wide association studies have identified >200 loci that contribute to the risk of IBD development (47–49). These studies revealed a common polymorphism in the gene encoding caspase recruitment domain-containing protein 9 (CARD9), as one of the strongest genetic risk factors linked to CD (4, 50, 51). The CARD9 gene is located on chromosome 9q34.3 and encodes a protein with 536 amino acids, containing an N-terminal CARD domain and a C-terminal coiled-coil domain (52–54). Expressed by myeloid cells, CARD9 is a signaling adapter protein that plays a key role in antifungal innate immunity in mice and humans, suggesting the potential links between IBD and host antifungal immunity.
As many researches reported, some CARD9 variants are closely associated with increased risk for IBD, while others are shown to be protective for IBD (55–57). The single-nucleotide polymorphism (SNP) rs4077515 in CARD9, substituting asparagine for serine at position 12 (S12N) in the protein CARD9 (CARD9S12N), is associated with increased expression of CARD9 mRNA and the development of CD (58, 59). A recent study reported that M. restricta, one of the intestinal mucosa-associated fungi significantly more abundant in patients with CD than in healthy controls, was particularly present in patients with CD carrying the CARD9S12N risk allele, and exacerbated colitis via CARD9 in a dextran sulfate sodium (DSS)-induced colitis mouse model (32). Conversely, CARD9 S12N Δ11 (with deletion of exon 11) fails to recruit TRIM62 to mediate CARD9 ubiquitination due to the lack of a functional C-terminal domain (57, 60). As a protective variant, it leads to negative activities of CARD9-dependent cytokine signaling including decreased TNF-α and IL-6 production (60).
As one of the major classes of the pattern-recognition receptors (PRRs), C-type lectin receptors (CLRs) can recognize the fungal pathogens and activate the subsequent antifungal immune responses in the gut (61). CLRs including Dectin-1, Dectin-2, Dectin-3 and Mincle can respectively recognize the fungal cell wall components such as β-glucan, α-mannose, glucosyl and mannosyl glycolipids (26, 50, 62–64). Upon recognizing the fungi, C-type lectin receptors recruit spleen tyrosine kinase (Syk), inducing CARD9 to couple to B cell leukemia-lymphoma 10 (BCL10) and mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1), ultimately leading to the activation of downstream nuclear factor kappa B (NF-κB) and production of cytokines through the CARD9-BCL10-MALT1 complex (65–68) (Figure 1).
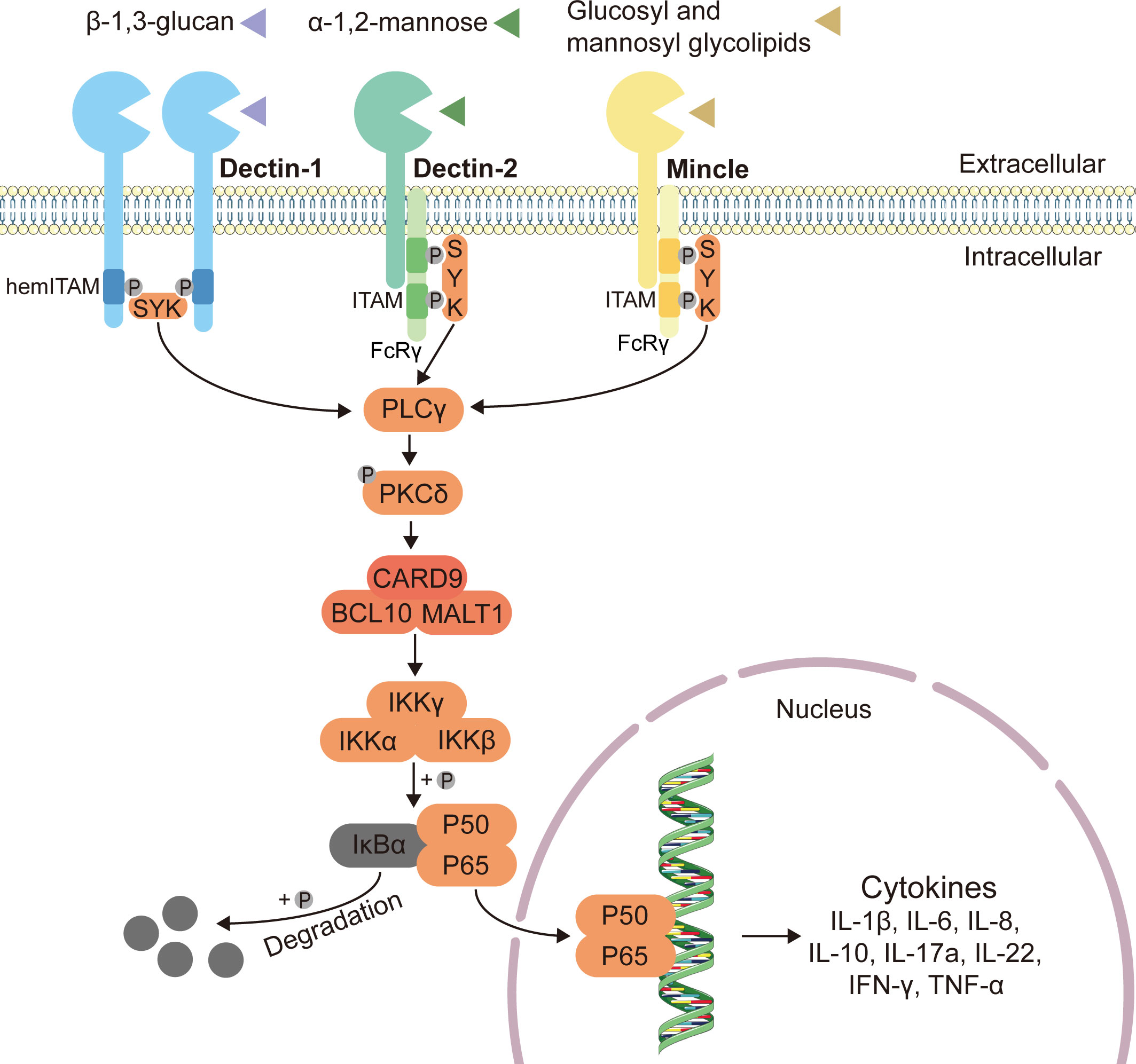
Figure 1 Classical innate antifungal signaling pathway through CLRs-Syk-CARD9. Expressed on myeloid cells, C-type lectin receptors (CLRs) including Dectin-1, Dectin-2 and Mincle can respectively recognize the fungal cell wall components such as β-1,3-glucan, α-1,2-mannose, glucosyl and mannosyl glycolipids, and induce intracellular antifungal signaling (26, 50, 62–64). The immunoreceptor tyrosine-based activating motif (ITAM)-coupled and the hemITAM-bearing CLRs, often considered as “activating” CLRs. Dectin-1, one of the hemITAM-bearing CLRs, can recruit spleen tyrosine kinase (Syk) directly, while the examples of ITAM-coupled CLRs, Dectin-2 and Mincle, indirectly activate Syk via the fragment crystallizable (Fc) receptor γ (FcRγ) adaptor chain (69). Syk activates phospholipase C (PLCγ), and then enhances the function of protein-kinase C (PKCδ), inducing the assembly of a CARD9–BCL10–MALT1 (caspase recruitment domain-containing protein 9–B cell leukemia-lymphoma 10–mucosa-associated lymphoid tissue lymphoma translocation protein 1) complex (65–68). The CARD9–BCL10–MALT1 complex serving as scaffolds mediates the activation of the canonical nuclear factor-κB (NF-κB) pathway (70). This mechanism activates the inhibitor of kappa B (IκB) kinase (IKK) complex, which phosphorylates NF-κB inhibitor-α (IκBα), leading to its degradation and the nuclear translocation of p65-p50 complexes (71, 72). Subsequently, the release of NF-κB dimers to the nucleus can activate gene transcription, and finally stimulate the production of related inflammatory cytokines.
Furthermore, Doron et al. presented evidence that C. albicans can induce the growth of germinal center (GC)-dependent IgG+ B cells by activating the Syk-CARD9 pathway in fractalkine/CX3C motif chemokine receptor 1 (CX3CR1)+ mononuclear phagocytes (MNPs), thus inducing the development of IgG antibodies to prevent systemic fungal infection (73). Moreover, intestinal fungi such as Candida may be recognized and phagocytosed by gut-resident CX3CR1+ MNPs in vivo. In patients with CD, there is a high correlation between a loss-of-function mutation (T280M) in the CX3CR1 gene and a reduction in antifungal IgG responses and anti-S. cerevisiae antibodies (ASCA), demonstrating a connection between CX3CR1+ MNPs and antifungal antibodies (74, 75). Several studies have found that high serum titers of ASCA IgG and IgA, which are directed against yeast cell wall-associated mannan, are a clinical biomarker for CD (76–79). With mannoproteins expressed by cell wall, C. albicans was shown to express ASCA epitopes and the abundance of C. tropicalis was also positively correlated with the level of ASCA (4, 33, 80). Since the genus Candida have been described as immunogens for ASCA as the CD biomarkers, it may be responsible for an abnormal immune response in CD.
In summary, ample genetic and serologic evidence suggests a possible association between intestinal fungi, antifungal antibodies, and CD.
Different heavy and light chains assemble to create different immunoglobulins, which include five isotypes: IgG, IgA, IgM, IgE, and IgD. IgA is the most abundant immunoglobulin in humans, with a production rate of 66 mg/kg/day, and the most common antibody isotype at mucosal surfaces (81, 82). According to the amino acid composition in the hinge region and the number and locations of disulfide bonds in the heavy chain, IgA in humans is classified into two subclasses: IgA1 and IgA2. Serum IgA is comprised of 90% IgA1 and 10% IgA2, whereas IgA2 predominates in the colon (17, 83–87). The content ratio of IgA1 to IgA2 varies in different parts of the intestines, from 3:1 in the proximal small intestine to 1:3 in the colon (83, 85–88). With a shorter hinge region than IgA1, IgA2 seems more advantageous in the microbe-rich environment of the colon owing to its higher stability (86, 87, 89, 90). A recent study showed that the IgA1+ microbiota fraction in patients with CD was notably enriched in beneficial commensals and that local IgA2 selection of the microbiota correlated with disease activity in CD, suggesting that IgA1 has a pathogenic role in the lumen in CD, whereas IgA2 has a pathogenic role in tissues (91).
Further, IgA is expressed in different forms in different parts of the human body. Serum IgA is mostly present in the monomeric form and produced by plasma cells in the bone marrow, spleen, and lymph nodes (16, 17). In contrast, in mucosal secretions, SIgA mostly exists in a dimeric form, in which two SIgA copies are linked by a joining chain (83–85, 92). Besides stabilizing the dimeric IgA (dIgA) tail-to-tail, the joining chain also serves as a ligand for the polymeric immunoglobulin receptor (pIgR), which is an IgA and IgM transporter expressed on the basolateral surface of intestinal epithelial cells (85, 93). The resultant pIgR–dIgA complex is endocytosed and transferred across the intestinal epithelial cells, through a series of vesicles, to the gut lumen, where the extracellular portion of the pIgR is cleaved to yield a fragment termed secretory component, which then forms a complex with dIgA otherwise known as SIgA, which is released into the gut lumen (85, 94, 95).
SIgA, locally synthesized at the mucosal surfaces, is secreted mainly by plasma cells (PCs). The SIgA-producing PCs are primarily located within the gut‐associated lymphoid tissue (GALT) including Peyer’s patches (PPs), isolated lymphoid follicles (ILFs), and mesenteric lymph nodes (MLNs) (83, 84, 96–98). SIgA-producing plasma cell can be generated via both T cell-dependent (TD) and T cell-independent (TI) pathways (13, 15, 88, 99).
TD SIgA responses typically target protein antigens and involve iterative rounds of somatic hypermutation (SHM) and high affinity selection in PPs and MLNs, where GCs are constitutively active (15, 16). Furthermore, contact between CD40 on the surface of B cells and its ligand CD40L on T cells is necessary for TD responses (88, 100). Additionally, CD4+ T follicular helper cells, Foxp3+CD4+ T regulatory cells, and Th17 cells can release various cytokines, such as IL-4, IL-5, IL-6, IL-10, IL-13, IL-17, and IL-21, to promote IgA responses (84, 86, 101, 102).
T cell–deficient (TCRβδ−/−) mice possess a microbiota-reactive IgA repertoire coating non-overlapping commensal bacterial taxa when compared with that in T cell-sufficient mice, indicating that SIgA can be produced without T cells help (15, 100, 103). TI SIgA seems to occur mainly within the gut lamina propria (LP) and ILFs (17, 88, 92). The TI SIgA response to polyvalent antigens is primarily “natural” polyreactive and shows low-affinity for commensal bacteria with little SHM (16, 103). During the TI pathway, two tumor necrosis factor family members, B-cell activating factor of the TNF family (BAFF) and a proliferation‐inducing ligand (APRIL), are responsible for stimulating class switch recombination (CSR) to IgA (86, 100). However, the contribution and function of high-affinity TD SIgA and low-affinity TI SIgA in microbiota homeostasis and infection control remain controversial (92, 94).
The mucosal immune system can be principally divided into inductive and effector sites (104). GALT is the site of induction of intestinal immune responses, where antigens sampled from mucosal surfaces activate naive and memory T or B lymphocytes (105). By capturing particular antigens from microfold cells or dendritic cells, B cells can generate IgA via IgM-to-IgA CSR. To synthesize IgA, the Cµ exon in the immunoglobulin heavy-chain C-region (CH) gene must be exchanged with the downstream Cα exon by activation-induced cytidine deaminase (AID) and other factors in the intestinal microenvironment (84, 88, 98, 106–108).
IgA+ B cells differentiate during their migration to the effector sites in the lamina propria and act as effector cells after extravasation, retention, and differentiation (84, 104, 109, 110). After the terminal differentiation of B cells to PCs, at least 80% of all PCs are located in the small intestinal LP, mainly generating dIgA which binds to pIgR, and ultimately, the formed complex is secreted into the intestinal lumen as SIgA (111).
SIgA is involved in gut microbiota maintenance. It plays multiple protective roles by preventing the adhesion of commensal bacteria to epithelial cells, neutralizing toxins and pathogens, and suppressing (not killing) fast-growing bacterial species (112–114). IgA-bound pro-inflammatory bacteria have been found to be more prevalent in patients with IBD than in healthy subjects, whereas IgA-bound commensal bacteria and probiotics levels were decreased; notably, the percentage of IgA-coated bacteria was associated with disease severity (21, 24, 115). Importantly, in addition to bacteria, intestinal fungi can also be targeted by SIgA.
By establishing a multistep flow cytometry-based technique for multi-kingdom antibody profiling, researchers recently showed that a majority of intestinal fungi were bound to luminal SIgA rather than IgG or IgM in mouse feces, independent of serum supplementation, and systemic IgA made a small contribution to the antifungal IgA repertoire (73). These findings suggest that, unlike in the blood, antifungal IgA in the intestinal tract mainly exists in the form of SIgA. Recent studies have demonstrated that, under homeostatic conditions, intestinal colonization of C. albicans, a potentially pathogenic fungus, induces robust SIgA responses in the human gut and oral mucosa, which is associated with elevated IgA+ B cell frequencies in the PPs and in the LP (73, 116–118).
As the most prevalent dimorphic fungus in the human GIT, C. albicans colonizes the gut as a mixture of yeast and hypha cells with varying morphologies, modes of division, occurrences, and virulence (119, 120). In general, C. albicans yeast is more likely to colonize the human GIT, whereas the hyphal form tends to be more virulent and invasive; therefore, the transition from yeast to invasive hyphae generally plays a central role in C. albicans pathogenesis (120–122). The hyphae express diverse cell type-specific virulence factors, particularly, cell surface adhesins, such as agglutinin-like protein 3 precursor (Als3), secreted aspartyl proteinases (Sap), and the Ece1-derived cytolytic toxin candidalysin (Ece1-III), which contribute to fungal pathogenesis including tissue adhesion, invasion, and damage (119, 120, 123–125).
Recent work has shown that SIgA antibodies preferentially bind C. albicans hyphal morphotypes, targeting several hyphae-associated virulence factors (Als3, Sap6, and Ece1-III) (117, 118, 126). Consistent herewith, intervention with SIgA resulted in a reduction of C. albicans hyphae and promoted the growth of the yeast form (118). Additionally, studies have revealed a decrease in anti-C. albicans SIgA responses against hyphae-associated virulence factors and an increase in granular hyphal morphologies in the gut mucosa of patients with CD (117, 118). Together, these data suggest that antifungal SIgA, which prevents epithelial adhesion and invasion of C. albicans to reduce intestinal inflammation, has an important role in maintaining fungal commensalism in the gut and is dysregulated in patients with CD (Figure 2).
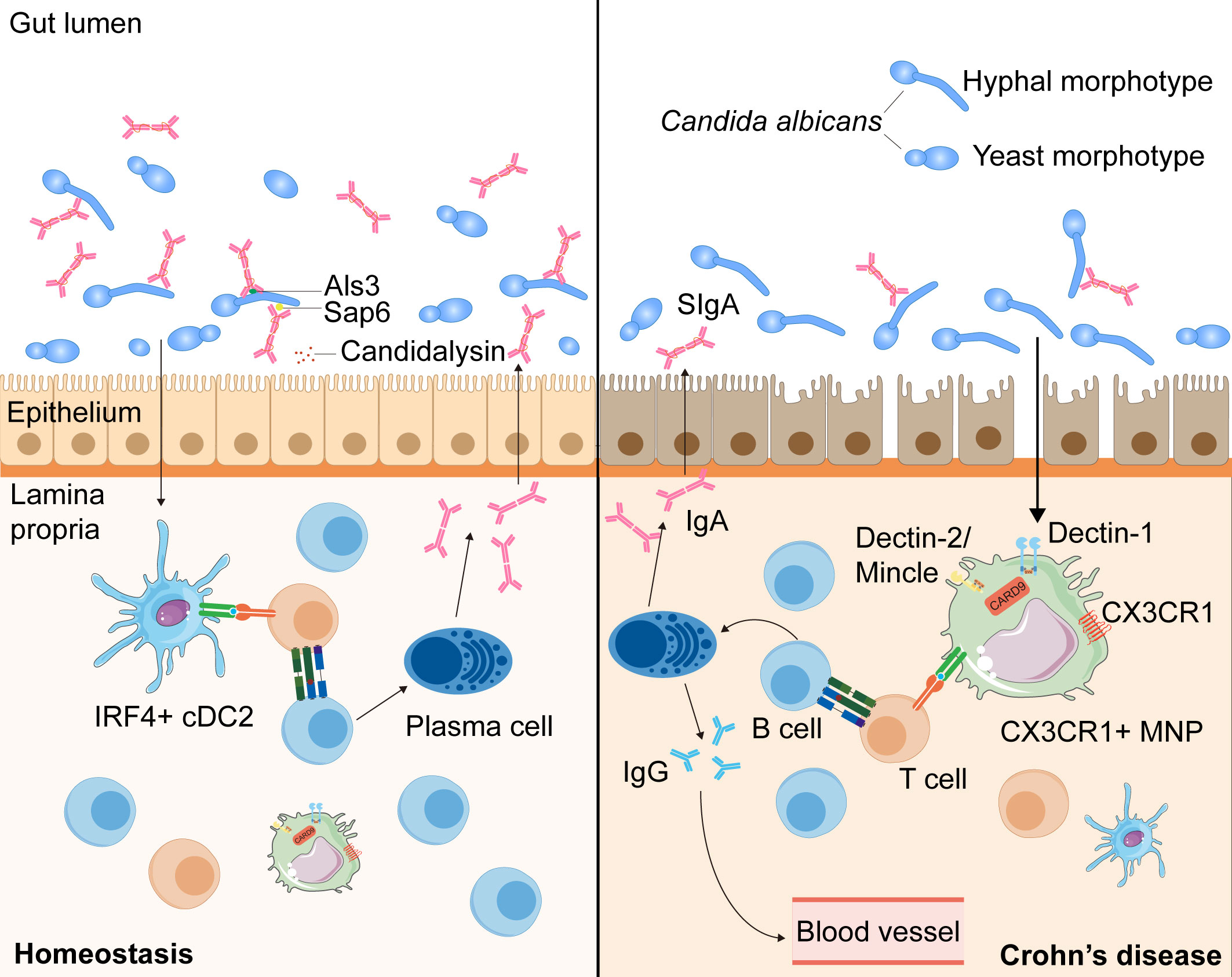
Figure 2 The intestinal SIgA responses targeting Candida albicans in the health and the patients with CD. During homeostasis, C. albicans, one of the commensal intestinal fungi, interacts to intestinal epithelial cells and can be recognized by IRF4+ cDC2s and CX3CR1+ MNPs, leading to the antibody CSR in mature B cells with the help of T cells (73, 116, 117). C. albicans can induce the growth of germinal center -dependent IgG+ B cells depending on the innate immunity regulator CARD9 and CARD9+CX3CR1+ MNPs, thus inducing the development of antifungal IgG antibodies to prevent systemic fungal infection (73). Moreover, through interaction with CX3CR1+ MNPs or cDC2s, the generation of SIgA targeting hyphae-associated fungal virulence factors (Als3, Sap6 and candidalysin) promotes a mutualistic relationship between the host and commensal fungi by suppressing the hyphal form of fungi (116, 117). In patients with CD, anti-C. albicans SIgA responses are decreased and hyphal morphotypes of C. albicans in the mucosa increased, which may alter the balance between the host and commensal fungi (118).
Unlike the production of systemic antifungal IgG, which depends solely on CX3CR1+ MNPs, anti-C. albicans SIgA responses are mediated by CX3CR1+ MNPs or CD11c+CD11b+CD103+ conventional dendritic cells (cDC2) via different pathways: one affecting IgA+ plasmablasts in the LP and the other affecting IgA+ B cells in the PPs (73, 74, 118, 127). Thus, intestinal fungi taken up by cDC2 and CX3CR1+ MNPs can induce antibody CSR in mature B cells towards the mycobiome (118). Through qPCR and flow-cytometric analysis, researchers have found that both cDC2 and CX3CR1+ MNPs express genes presenting antigens, but CX3CR1+ MNPs show higher expression of genes associated with fungal recognition, such as those encoding Dectin-1, Dectin-2, and Mincle (74). Furthermore, confocal microscopy examination has revealed that Candida was efficiently recognized by intestinal phagocytes in vivo, with >80% of all CX3CR1+ MNPs engulfing Candida (74). Moreover, a polymorphism in the coding region of the CX3CR1 gene is strongly associated with a decrease in antifungal antibody responses in patients with CD (74) (Figure 2).
Overall, these results suggest that, through interaction with CX3CR1+ MNP or cDC2, the generation of SIgA targeting hyphae-associated fungal virulence factors promotes a mutualistic relationship between the host and commensal fungi by suppressing the hyphal form of fungi (73, 117, 118). Furthermore, in patients with CD, anti-C. albicans SIgA responses are decreased and hyphal morphologies of fungi in the mucosa increased, which may alter the balance between the host and commensal fungi (118). However, the detailed mechanism by which SIgA antibodies are induced by intestinal fungi and bind to the fungi in order to limit intestinal tissue damage through B or T cells remains unclear. Therefore, further studies on the mechanisms of antifungal SIgA responses, especially in patients with CD, which may provide a deeper insight into mucosal immunity and the mycobiota in the pathogenesis of CD, are urgently needed.
Currently, only a few prophylactic or therapeutic vaccines against fungal infections are in clinical trials. According to the literature, anti-C. albicans vaccines can be designed to target specific virulence factors in order to reduce tissue damage. The NDV-3A vaccine, which contains the N-terminal region of the Als3 protein of C. albicans, is the most promising and advanced vaccine tested to date (128). This vaccine has been tested for safety and efficacy against recurrent vulvovaginal candidiasis in several clinical trials; it induced robust immunologic responses with significantly elevated serum anti-Als3 IgG and IgA1 titers, and stimulation of Als3-specific production of IFN-γ and IL-17A (126, 129). The NDV-3 vaccines also induced protective responses against Staphylococcus aureus and Candida auris infections in mice (130–132). Furthermore, NDV-3 vaccination prevented intestinal damage in C. albicans-monocolonized mice with colitis by inducing intestinal anti-Als3 IgA responses and reducing tissue-associated C. albicans in the colon (117).
As discussed above, the fungal microbiota is altered in patients with CD; fungal alpha diversity, fecal fungal load, and C. albicans abundance are increased in these patients (43). C. albicans can aggravate gut inflammation by driving Th17-mediated immune responses and inducing dysbiosis of the gut microbiome (31). Collectively, these findings exemplify that SIgA antibody responses against the C. albicans adhesin, Als3, can alleviate C. albicans-associated damage during colitis, which may be useful in relieving gut inflammation in patients with CD. Consequently, novel antifungal vaccines are worth developing through further exploring the pathways via which intestinal fungi induce SIgA antibody responses in order to partially protect the intestinal mucosa from inflammation or even prevent the occurrence and development of CD by maintaining fungal commensalism in the gut.
In recent studies, increasing evidence indicates that intestinal fungi and SIgA responses are essential part of mucosal immune regulation in CD. The evidence discussed in this review suggests that antifungal SIgA antibodies targeting hyphae-associated virulence factors through immune interaction are a critical regulator of fungal commensalism in the gut and are essential for protecting the intestinal mucosa (73, 74, 116–118). As anti-C. albicans SIgA responses are dysregulated in patients with CD, an imbalance in the gut fungal community can result in overgrowth by certain fungi, leading to inflammation and pathogenic consequences (118).
Despite the increasing interest in anti-C. albicans SIgA responses lately, the research about other fungal species interacting with SIgA is still worth exploring. Shapiro et al. identified increased levels of IgA coating of forty-three bacterial taxa in IBD compared with controls, combining bacterial fluorescence-activated cell sorting with 16S rRNA gene sequencing (133). Through a certain degree of improvement, this promising technology, IgA-SEQ, may also have potential applications for the analysis of IgA-coated fungi in the pathogenesis of CD to identify more fungal species targeted by SIgA (133, 134). In the future, much work is required to elucidate other species that induce antifungal SIgA responses, the functions of antifungal SIgA antibodies in intestinal mucosal immunity, and the detailed mechanisms governing the induction and regulation of antifungal SIgA in the GIT in CD. These explorations will likely lead to the development of novel strategies to protect the intestinal mucosa from inflammation and maintain intestinal fungal commensalism to prevent the occurrence and development of CD, including potential vaccines.
Although the understanding of mucosal immunity and SIgA in human diseases and homoeostasis has significantly improved over the past decades, the variety and complexity of the immune response processes, functions, and mechanisms of antifungal SIgA antibodies in the intestinal mucosa of healthy subjects and patients with CD remain to be fully unraveled.
MS conceived and designed the study, wrote the manuscript and finished the table and figures. MS and JJ reviewed the literature, wrote and edited the manuscript. HX analyzed articles and provided feedback on the content. YW supervised the study and reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Fund of China (81270447), the National Longitudinal Research of China (2012BAI06B03).
The authors would like to thank Editage (www.editage.cn) for English language editing. The Figures were partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CD, Crohn’s disease; SIgA, secretory immunoglobulin A; IBD, inflammatory bowel disease; qPCR, quantitative polymerase chain reaction; GIT, gastrointestinal tract; ITS, internal transcribed spacer; IFN, interferon; IL, interleukin; TNF, tumor necrosis factor; CARD9, caspase recruitment domain-containing protein 9; SNP, single-nucleotide polymorphism; PRRs, pattern-recognition receptors; CLRs, C-type lectin receptors; Syk, spleen tyrosine kinase; BCL10, B cell leukemia-lymphoma 10; MALT1, mucosa-associated lymphoid tissue lymphoma translocation protein 1; NF-κB, nuclear factor kappa B; DSS, dextran sulfate sodium; GC, germinal center; CX3CR1, CX3C motif chemokine receptor 1; MNPs, mononuclear phagocytes; ASCA, anti-Saccharomyces cerevisiae antibodies; dIgA, dimeric IgA; pIgR, polymeric immunoglobulin receptor; PCs, plasma cells; GALT, gut‐associated lymphoid tissue; PPs, Peyer’s patches; ILFs, isolated lymphoid follicles; MLNs, mesenteric lymph nodes; TD, T cell-dependent; TI, T cell-independent; SHM, somatic hypermutation; LP, lamina propria; TNF, tumor necrosis factor; BAFF, B-cell activating factor of the TNF family; APRIL, a proliferation‐inducing ligand; CSR, class switch recombination; DCs, dendritic cells; AID, activation-induced cytidine deaminase; Als3, agglutinin-like protein 3 precursor; Sap, secreted aspartyl proteinases; Ece1-III, Ece1-derived cytolytic toxin candidalysin; cDC2, CD11c+CD11b+CD103+ conventional DCs.
1. Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet (2017) 390:2769–78. doi: 10.1016/S0140-6736(17)32448-0
2. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol (2015) 12:720–7. doi: 10.1038/nrgastro.2015.150
3. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology (2014) 146:1489–99. doi: 10.1053/j.gastro.2014.02.009
4. Underhill DM, Braun J. Fungal microbiome in inflammatory bowel disease: a critical assessment. J Clin Invest (2022) 132:e155786. doi: 10.1172/JCI155786
5. Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology (2017) 152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012
6. Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol (2014) 14:405–16. doi: 10.1038/nri3684
7. Beheshti-Maal A, Shahrokh S, Ansari S, Mirsamadi ES, Yadegar A, Mirjalali H, et al. Gut mycobiome: the probable determinative role of fungi in IBD patients. Mycoses (2021) 64:468–76. doi: 10.1111/myc.13238
8. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalog established by metagenomic sequencing. Nature (2010) 464:59–65. doi: 10.1038/nature08821
9. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature (2011) 473:174–80. doi: 10.1038/nature09944
10. Rochereau N, Roblin X, Michaud E, Gayet R, Chanut B, Jospin F, et al. NOD2 deficiency increases retrograde transport of secretory IgA complexes in crohn’s disease. Nat Commun (2021) 12:261. doi: 10.1038/s41467-020-20348-0
11. Corthésy B. Multi-faceted functions of secretory IgA at mucosal surfaces. Front Immunol (2013) 4:185. doi: 10.3389/fimmu.2013.00185
12. Huus KE, Petersen C, Finlay BB. Diversity and dynamism of IgA–microbiota interactions. Nat Rev Immunol (2021) 21:514–25. doi: 10.1038/s41577-021-00506-1
13. Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol (2010) 28:243–73. doi: 10.1146/annurev-immunol-030409-101314
14. Caruso R, Lo BC, Núñez G. Host-microbiota interactions in inflammatory bowel disease. Nat Rev Immunol (2020) 20:411–26. doi: 10.1038/s41577-019-0268-7
15. Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin a. Immunity (2015) 43:541–53. doi: 10.1016/j.immuni.2015.08.007
16. Bunker JJ, Bendelac A. IgA responses to microbiota. Immunity (2018) 49:211–24. doi: 10.1016/j.immuni.2018.08.011
17. Pietrzak B, Tomela K, Olejnik-Schmidt A, Mackiewicz A, Schmidt M. Secretory IgA in intestinal mucosal secretions as an adaptive barrier against microbial cells. Int J Mol Sci (2020) 21:9254. doi: 10.3390/ijms21239254
18. Sutherland DB, Fagarasan S. IgA synthesis: a form of functional immune adaptation extending beyond gut. Curr Opin Immunol (2012) 24:261–8. doi: 10.1016/j.coi.2012.03.005
19. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature (2016) 535:75–84. doi: 10.1038/nature18848
20. Catanzaro JR, Strauss JD, Bielecka A, Porto AF, Lobo FM, Urban A, et al. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci Rep (2019) 9:13574. doi: 10.1038/s41598-019-49923-2
21. Lin R, Chen H, Shu W, Sun M, Fang L, Shi Y, et al. Clinical significance of soluble immunoglobulins a and G and their coated bacteria in feces of patients with inflammatory bowel disease. J Transl Med (2018) 16:359. doi: 10.1186/s12967-018-1723-0
22. Masu Y, Kanazawa Y, Kakuta Y, Shimoyama Y, Onodera M, Naito T, et al. Immunoglobulin subtype-coated bacteria are correlated with the disease activity of inflammatory bowel disease. Sci Rep (2021) 11:16672. doi: 10.1038/s41598-021-96289-5
23. Rengarajan S, Vivio EE, Parkes M, Peterson DA, Roberson EDO, Newberry RD, et al. Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes (2020)11:405–20. doi: 10.1080/19490976.2019.1626683
24. Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin a coating identifies colitogenic bacteria in inflammatory bowel disease. Cell (2014) 158:1000–10. doi: 10.1016/j.cell.2014.08.006
25. Alipour M, Zaidi D, Valcheva R, Jovel J, Martínez I, Sergi C, et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohn’s Colitis (2016) 10:462–71. doi: 10.1093/ecco-jcc/jjv223
26. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the c-type lectin receptor dectin-1 influence colitis. Science (2012) 336:1314–7. doi: 10.1126/science.1221789
27. Li J, Chen D, Yu B, He J, Zheng P, Mao X, et al. Fungi in gastrointestinal tracts of human and mice: from community to functions. Microb Ecol (2018) 75:821–9. doi: 10.1007/s00248-017-1105-9
28. Scupham AJ, Presley LL, Wei B, Bent E, Griffith N, McPherson M, et al. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol (2006) 72:793–801. doi: 10.1128/AEM.72.1.793-801.2006
29. Liguori G, Lamas B, Richard ML, Brandi G, da Costa G, Hoffmann TW, et al. Fungal dysbiosis in mucosa-associated microbiota of crohn’s disease patients. J Crohns Colitis (2016) 10:296–305. doi: 10.1093/ecco-jcc/jjv209
30. Imai T, Inoue R, Kawada Y, Morita Y, Inatomi O, Nishida A, et al. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J Gastroenterol (2019) 54:149–59. doi: 10.1007/s00535-018-1530-7
31. Leonardi I, Gao IH, Lin W-Y, Allen M, Li XV, Fiers WD, et al. Mucosal fungi promote gut barrier function and social behavior via type 17 immunity. Cell (2022) 185:831–846.e14. doi: 10.1016/j.cell.2022.01.017
32. Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, et al. Malassezia is associated with crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe (2019) 25:377–388.e6. doi: 10.1016/j.chom.2019.01.007
33. Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial crohn’s disease. mBio (2016) 7:e01250–16. doi: 10.1128/mBio.01250-16
34. Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome (2017) 5:1–13. doi: 10.1186/s40168-017-0373-4
35. Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, et al. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere (2018) 3:e00092–18. doi: 10.1128/mSphere.00092-18
36. Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol (2008) 43:831–41. doi: 10.1080/00365520801935434
37. Nelson A, Stewart CJ, Kennedy NA, Lodge JK, Tremelling M, UK IBD Genetics Consortium, et al. The impact of NOD2 genetic variants on the gut mycobiota in crohn’s disease patients in remission and in individuals without gastrointestinal inflammation. J Crohn’s Colitis (2021) 15:800–12. doi: 10.1093/ecco-jcc/jjaa220
38. Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut (2017) 66:1039–48. doi: 10.1136/gutjnl-2015-310746
39. Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, et al. A fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflammation Bowel Dis (2015) 21:1948–56. doi: 10.1097/MIB.0000000000000454
40. Zeng L, Feng Z, Zhuo M, Wen Z, Zhu C, Tang C, et al. Fecal fungal microbiota alterations associated with clinical phenotypes in crohn’s disease in southwest China. PeerJ (2022) 10:e14260. doi: 10.7717/peerj.14260
41. Jain U, Ver Heul AM, Xiong S, Gregory MH, Demers EG, Kern JT, et al. Debaryomyces is enriched in crohn’s disease intestinal tissue and impairs healing in mice. Science (2021) 371:1154–9. doi: 10.1126/science.abd0919
42. Lam S, Zuo T, Ho M, Chan FKL, Chan PKS, Ng SC. Review article: fungal alterations in inflammatory bowel diseases. Aliment Pharmacol Ther (2019) 50:1159–71. doi: 10.1111/apt.15523
43. Zhang F, Aschenbrenner D, Yoo JY, Zuo T. The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe (2022) 3:e969–83. doi: 10.1016/S2666-5247(22)00203-8
44. Shao T-Y, Kakade P, Witchley JN, Frazer C, Murray KL, Ene IV, et al. Candida albicans oscillating UME6 expression during intestinal colonization primes systemic Th17 protective immunity. Cell Rep (2022) 39:110837. doi: 10.1016/j.celrep.2022.110837
45. Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, et al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against candida albicans. Cell (2019) 176:1340–1355.e15. doi: 10.1016/j.cell.2019.01.041
46. Yu M, Ding H, Gong S, Luo Y, Lin H, Mu Y, et al. Fungal dysbiosis facilitates inflammatory bowel disease by enhancing CD4+ T cell glutaminolysis. Front Cell Infect Microbiol (2023) 13:1140757. doi: 10.3389/fcimb.2023.1140757
47. Younis N, Zarif R, Mahfouz R. Inflammatory bowel disease: between genetics and microbiota. Mol Biol Rep (2020) 47:3053–63. doi: 10.1007/s11033-020-05318-5
48. de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet (2017) 49:256–61. doi: 10.1038/ng.3760
49. Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature (2020) 578:527–39. doi: 10.1038/s41586-020-2025-2
50. Wheeler ML, Limon JJ, Underhill DM. Immunity to commensal fungi: detente and disease. Annu Rev Pathol (2017) 12:359–85. doi: 10.1146/annurev-pathol-052016-100342
51. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature (2012) 491:119–24. doi: 10.1038/nature11582
52. Bertin J, Guo Y, Wang L, Srinivasula SM, Jacobson MD, Poyet JL, et al. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa b. J Biol Chem (2000) 275:41082–6. doi: 10.1074/jbc.C000726200
53. Ji C, Yang Z, Zhong X, Xia J. The role and mechanism of CARD9 gene polymorphism in diseases. Biomed J (2021) 44:560–6. doi: 10.1016/j.bj.2020.12.006
54. Goel S, Kuehn HS, Chinen J, Niemela J, Stoddard J, Yamanaka D, et al. CARD9 expression pattern, gene dosage, and immunodeficiency phenotype revisited. J Clin Immunol (2022) 42:336–49. doi: 10.1007/s10875-021-01173-6
55. Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, et al. Genetic analysis of innate immunity in crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet (2008) 82:1202–10. doi: 10.1016/j.ajhg.2008.03.016
56. Lee YH, Song GG. Pathway analysis of a genome-wide association study of ileal crohn’s disease. DNA Cell Biol (2012) 31:1549–54. doi: 10.1089/dna.2012.1605
57. Leshchiner ES, Rush JS, Durney MA, Cao Z, Dančík V, Chittick B, et al. Small-molecule inhibitors directly target CARD9 and mimic its protective variant in inflammatory bowel disease. Proc Natl Acad Sci U.S.A. (2017) 114:11392–7. doi: 10.1073/pnas.1705748114
58. Franke A, McGovern DPB, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Meta-analysis increases to 71 the tally of confirmed crohn’s disease susceptibility loci. Nat Genet (2010) 42:1118–25. doi: 10.1038/ng.717
59. Xu X, Xu J-F, Zheng G, Lu H-W, Duan J-L, Rui W, et al. CARD9S12N facilitates the production of IL-5 by alveolar macrophages for the induction of type 2 immune responses. Nat Immunol (2018) 19:547–60. doi: 10.1038/s41590-018-0112-4
60. Cao Z, Conway KL, Heath RJ, Rush JS, Leshchiner ES, Ramirez-Ortez ZG, et al. The ubiquitin ligase TRIM62 regulates CARD9-mediated anti-fungal immunity and intestinal inflammation. Immunity (2015) 43:715–26. doi: 10.1016/j.immuni.2015.10.005
61. Li T-H, Liu L, Hou Y-Y, Shen S-N, Wang T-T. C-type lectin receptor-mediated immune recognition and response of the microbiota in the gut. Gastroenterol Rep (2019) 7:312–21. doi: 10.1093/gastro/goz028
62. Wang Y, Zhang D, Hou Y, Shen S, Wang T. The adaptor protein CARD9, from fungal immunity to tumorigenesis. Am J Cancer Res (2020) 10:2203–25.
63. Geijtenbeek TBH, Gringhuis SI. C-type lectin receptors in the control of T helper cell differentiation. Nat Rev Immunol (2016) 16:433–48. doi: 10.1038/nri.2016.55
64. Li M, Zhang R, Li J, Li J. The role of c-type lectin receptor signaling in the intestinal microbiota-Inflammation-Cancer axis. Front Immunol (2022) 13:894445. doi: 10.3389/fimmu.2022.894445
65. Malik A, Sharma D, Malireddi RKS, Guy CS, Chang T-C, Olsen SR, et al. SYK-CARD9 signaling axis promotes gut fungi-mediated inflammasome activation to restrict colitis and colon cancer. Immunity (2018) 49:515–530.e5. doi: 10.1016/j.immuni.2018.08.024
66. Ostrop J, Lang R. Contact, collaboration, and conflict: signal integration of syk-coupled c-type lectin receptors. J Immunol (2017) 198:1403–14. doi: 10.4049/jimmunol.1601665
67. Vornholz L, Ruland J. Physiological and pathological functions of CARD9 signaling in the innate immune system. In: Yamasaki S, editor. C-type lectins in immune homeostasis. Cham: Springer International Publishing (2020). p. 177–203. doi: 10.1007/82_2020_211
68. Liu X, Jiang B, Hao H, Liu Z. CARD9 signaling, inflammation, and diseases. Front Immunol (2022) 13:880879. doi: 10.3389/fimmu.2022.880879
69. Fischer S, Stegmann F, Gnanapragassam VS, Lepenies B. From structure to function – ligand recognition by myeloid c-type lectin receptors. Comput Struct Biotechnol J (2022) 20:5790–812. doi: 10.1016/j.csbj.2022.10.019
70. Roth S, Ruland J. Caspase recruitment domain-containing protein 9 signaling in innate immunity and inflammation. Trends Immunol (2013) 34:243–50. doi: 10.1016/j.it.2013.02.006
71. Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol (2009) 27:693–733. doi: 10.1146/annurev.immunol.021908.132641
72. Roth S, Bergmann H, Jaeger M, Yeroslaviz A, Neumann K, Koenig P-A, et al. Vav proteins are key regulators of Card9 signaling for innate antifungal immunity. Cell Rep (2016) 17:2572–83. doi: 10.1016/j.celrep.2016.11.018
73. Doron I, Leonardi I, Li XV, Fiers WD, Semon A, Bialt-DeCelie M, et al. Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell (2021) 184:1017–1031.e14. doi: 10.1016/j.cell.2021.01.016
74. Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, et al. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science (2018) 359:232–6. doi: 10.1126/science.aao1503
75. Iliev ID, Cadwell K. Effects of intestinal fungi and viruses on immune responses and inflammatory bowel diseases. Gastroenterology (2021) 160:1050–66. doi: 10.1053/j.gastro.2020.06.100
76. Frehn L, Jansen A, Bennek E, Mandic AD, Temizel I, Tischendorf S, et al. Distinct patterns of IgG and IgA against food and microbial antigens in serum and feces of patients with inflammatory bowel diseases. PloS One (2014) 9:e106750. doi: 10.1371/journal.pone.0106750
77. Joossens S, Reinisch W, Vermeire S, Sendid B, Poulain D, Peeters M, et al. The value of serologic markers in indeterminate colitis: a prospective follow-up study. Gastroenterology (2002) 122:1242–7. doi: 10.1053/gast.2002.32980
78. Torres J, Petralia F, Sato T, Wang P, Telesco SE, Choung RS, et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology (2020) 159:96–104. doi: 10.1053/j.gastro.2020.03.007
79. Bertha M, Vasantharoopan A, Kumar A, Bruce BB, Prince J, Hofmekler T, et al. IBD serology and disease outcomes in African americans with crohn’s disease. Inflammation Bowel Dis (2018) 24:209–16. doi: 10.1093/ibd/izx021
80. Standaert-Vitse A, Jouault T, Vandewalle P, Mille C, Seddik M, Sendid B, et al. Candida albicans is an immunogen for anti-saccharomyces cerevisiae antibody markers of crohn’s disease. Gastroenterology (2006) 130:1764–75. doi: 10.1053/j.gastro.2006.02.009
81. Fagarasan S, Honjo T. Intestinal IgA synthesis: regulation of front-line body defences. Nat Rev Immunol (2003) 3:63–72. doi: 10.1038/nri982
82. Yang C, Chen-Liaw A, Spindler MP, Tortorella D, Moran TM, Cerutti A, et al. Immunoglobulin a antibody composition is sculpted to bind the self gut microbiome. Sci Immunol (2022) 7:eabg3208. doi: 10.1126/sciimmunol.abg3208
83. Woof JM, Kerr MA. The function of immunoglobulin a in immunity. J Pathol (2006) 208:270–82. doi: 10.1002/path.1877
84. Li Y, Jin L, Chen T. The effects of secretory IgA in the mucosal immune system. BioMed Res Int (2020) 2020:2032057. doi: 10.1155/2020/2032057
85. Woof JM, Russell MW. Structure and function relationships in IgA. Mucosal Immunol (2011) 4:590–7. doi: 10.1038/mi.2011.39
86. Fleming A, Castro-Dopico T, Clatworthy MR. B cell class switching in intestinal immunity in health and disease. Scand J Immunol (2022) 95:e13139. doi: 10.1111/sji.13139
87. Lin M, Du L, Brandtzaeg P, Pan-Hammarström Q. IgA subclass switch recombination in human mucosal and systemic immune compartments. Mucosal Immunol (2014) 7:511–20. doi: 10.1038/mi.2013.68
88. Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol (2012) 12:821–32. doi: 10.1038/nri3322
89. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol (2014) 14:667–85. doi: 10.1038/nri3738
90. He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity (2007) 26:812–26. doi: 10.1016/j.immuni.2007.04.014
91. Michaud E, Waeckel L, Gayet R, Goguyer-Deschaumes R, Chanut B, Jospin F, et al. Alteration of microbiota antibody-mediated immune selection contributes to dysbiosis in inflammatory bowel diseases. EMBO Mol Med (2022) 14(8):e15386. doi: 10.15252/emmm.202115386
92. León ED, Francino MP. Roles of secretory immunoglobulin a in host-microbiota interactions in the gut ecosystem. Front Microbiol (2022) 13:880484. doi: 10.3389/fmicb.2022.880484
93. Brandtzaeg P, Johansen F-E. Mucosal b cells: phenotypic characteristics, transcriptional regulation, and homing properties. Immunol Rev (2005) 206:32–63. doi: 10.1111/j.0105-2896.2005.00283.x
94. Pabst O, Slack E. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol (2020) 13:12–21. doi: 10.1038/s41385-019-0227-4
95. Chen K, Magri G, Grasset EK, Cerutti A. Rethinking mucosal antibody responses: IgM, IgG and IgD join IgA. Nat Rev Immunol (2020) 20:427–41. doi: 10.1038/s41577-019-0261-1
96. Isho B, Florescu A, Wang AA, Gommerman JL. Fantastic IgA plasma cells and where to find them. Immunol Rev (2021) 303:119–37. doi: 10.1111/imr.12980
97. Mörbe UM, Jørgensen PB, Fenton TM, von Burg N, Riis LB, Spencer J, et al. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal Immunol (2021) 14:793–802. doi: 10.1038/s41385-021-00389-4
98. Fagarasan S, Kinoshita K, Muramatsu M, Ikuta K, Honjo T. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature (2001) 413:639–43. doi: 10.1038/35098100
99. Barone F, Patel P, Sanderson JD, Spencer J. Gut-associated lymphoid tissue contains the molecular machinery to support T-cell-dependent and T-cell-independent class switch recombination. Mucosal Immunol (2009) 2:495–503. doi: 10.1038/mi.2009.106
100. Hand TW, Reboldi A. Production and function of immunoglobulin a. Annu Rev Immunol (2021) 39:695–718. doi: 10.1146/annurev-immunol-102119-074236
101. Cao AT, Yao S, Gong B, Nurieva RI, Elson CO, Cong Y. Interleukin (IL)-21 promotes intestinal IgA response to microbiota. Mucosal Immunol (2015) 8:1072–82. doi: 10.1038/mi.2014.134
102. Cosovanu C, Neumann C. The many functions of Foxp3+ regulatory T cells in the intestine. Front Immunol (2020) 11:600973. doi: 10.3389/fimmu.2020.600973
103. Tezuka H, Abe Y, Asano J, Sato T, Liu J, Iwata M, et al. Prominent role for plasmacytoid dendritic cells in mucosal T cell-independent IgA induction. Immunity (2011) 34:247–57. doi: 10.1016/j.immuni.2011.02.002
104. Ahluwalia B, Magnusson MK, Öhman L. Mucosal immune system of the gastrointestinal tract: maintaining balance between the good and the bad. Scand J Gastroenterol (2017) 52:1185–93. doi: 10.1080/00365521.2017.1349173
105. Spencer J, Bemark M. Human intestinal b cells in inflammatory diseases. Nat Rev Gastroenterol Hepatol (2023) 20:254–65. doi: 10.1038/s41575-023-00755-6
106. Reboldi A, Arnon TI, Rodda LB, Atakilit A, Sheppard D, Cyster JG. IgA production requires b cell interaction with subepithelial dendritic cells in peyer’s patches. Science (2016) 352:aaf4822. doi: 10.1126/science.aaf4822
107. Suzuki K, Fagarasan S. Diverse regulatory pathways for IgA synthesis in the gut. Mucosal Immunol (2009) 2:468–71. doi: 10.1038/mi.2009.107
108. Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science (2002) 298:1424–7. doi: 10.1126/science.1077336
109. Brandtzaeg P, Kiyono H, Pabst R, Russell MW. Terminology: nomenclature of mucosa-associated lymphoid tissue. Mucosal Immunol (2008) 1:31–7. doi: 10.1038/mi.2007.9
110. Suzuki K, Fagarasan S. How host-bacterial interactions lead to IgA synthesis in the gut. Trends Immunol (2008) 29:523–31. doi: 10.1016/j.it.2008.08.001
111. van Egmond M, Damen CA, van Spriel AB, Vidarsson G, van Garderen E, van de Winkel JG. IgA and the IgA fc receptor. Trends Immunol (2001) 22:205–11. doi: 10.1016/s1471-4906(01)01873-7
112. Okai S, Usui F, Ohta M, Mori H, Kurokawa K, Matsumoto S, et al. Intestinal IgA as a modulator of the gut microbiota. Gut Microbes (2017) 8:486–92. doi: 10.1080/19490976.2017.1310357
113. Liu A, Wang X, Liang X, Wang W, Li C, Qian J, et al. Human umbilical cord mesenchymal stem cells regulate immunoglobulin a secretion and remodel the diversification of intestinal microbiota to improve colitis. Front Cell Infect Microbiol (2022) 12:960208. doi: 10.3389/fcimb.2022.960208
114. Huang M, Zhang M, Zhu H, Du X, Wang J. Mucosal vaccine delivery: a focus on the breakthrough of specific barriers. Acta Pharm Sin B (2022) 12:3456–74. doi: 10.1016/j.apsb.2022.07.002
115. Huang W, Huang H-L, Peng W, Liu Y-D, Zhou Y-L, Xu H-M, et al. Altered pattern of immunoglobulin a-targeted microbiota in inflammatory bowel disease after fecal transplantation. Front Microbiol (2022) 13:873018. doi: 10.3389/fmicb.2022.873018
116. Millet N, Solis NV, Swidergall M. Mucosal IgA prevents commensal candida albicans dysbiosis in the oral cavity. Front Immunol (2020) 11:555363. doi: 10.3389/fimmu.2020.555363
117. Ost KS, O’Meara TR, Stephens WZ, Chiaro T, Zhou H, Penman J, et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature (2021) 596:114–8. doi: 10.1038/s41586-021-03722-w
118. Doron I, Mesko M, Li XV, Kusakabe T, Leonardi I, Shaw DG, et al. Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in crohn’s disease. Nat Microbiol (2021) 6:1493–504. doi: 10.1038/s41564-021-00983-z
119. Noble SM, Gianetti BA, Witchley JN. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol (2017) 15:96–108. doi: 10.1038/nrmicro.2016.157
120. Witchley JN, Penumetcha P, Abon NV, Woolford CA, Mitchell AP, Noble SM. Candida albicans morphogenesis programs control the balance between gut commensalism and invasive infection. Cell Host Microbe (2019) 25:432–443.e6. doi: 10.1016/j.chom.2019.02.008
121. Gutierrez MW, van Tilburg Bernardes E, Changirwa D, McDonald B, Arrieta M-C. “Molding” immunity–modulation of mucosal and systemic immunity by the intestinal mycobiome in health and disease. Mucosal Immunol (2022) 15:573–83. doi: 10.1038/s41385-022-00515-w
122. Brand A. Hyphal growth in human fungal pathogens and its role in virulence. Int J Microbiol (2012) 2012:517529. doi: 10.1155/2012/517529
123. Liu Y, Filler SG. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell (2011) 10:168–73. doi: 10.1128/EC.00279-10
124. Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature (2016) 532:64–8. doi: 10.1038/nature17625
125. Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin A-P, et al. Transcription profiling of candida albicans cells undergoing the yeast-to-Hyphal transition. Mol Biol Cell (2002) 13:3452–65. doi: 10.1091/mbc.E02-05-0272
126. Edwards JE Jr, Schwartz MM, Schmidt CS, Sobel JD, Nyirjesy P, Schodel F, et al. A fungal immunotherapeutic vaccine (NDV-3A) for treatment of recurrent vulvovaginal candidiasis–a phase 2 randomized, double-blind, placebo-controlled trial. Clin Infect Dis (2018) 66:1928–36. doi: 10.1093/cid/ciy185
127. Kim Y-I, Song J-H, Ko H-J, Kweon M-N, Kang C-Y, Reinecker H-C, et al. CX3CR1+ macrophages and CD8+ T cells control intestinal IgA production. J Immunol (2018) 201:1287–94. doi: 10.4049/jimmunol.1701459
128. Swidergall M, LeibundGut-Landmann S. Immunosurveillance of candida albicans commensalism by the adaptive immune system. Mucosal Immunol (2022) 15:829–36. doi: 10.1038/s41385-022-00536-5
129. Schmidt CS, White CJ, Ibrahim AS, Filler SG, Fu Y, Yeaman MR, et al. NDV-3, a recombinant alum-adjuvanted vaccine for candida and staphylococcus aureus, is safe and immunogenic in healthy adults. Vaccine (2012) 30:7594–600. doi: 10.1016/j.vaccine.2012.10.038
130. Lin L, Ibrahim AS, Xu X, Farber JM, Avanesian V, Baquir B, et al. Th1-Th17 cells mediate protective adaptive immunity against staphylococcus aureus and candida albicans infection in mice. PloS Pathog (2009) 5:e1000703. doi: 10.1371/journal.ppat.1000703
131. Alqarihi A, Singh S, Edwards JE, Ibrahim AS, Uppuluri P. NDV-3A vaccination prevents c. albicans colonization of jugular vein catheters in mice. Sci Rep (2019) 9:6194. doi: 10.1038/s41598-019-42517-y
132. Singh S, Uppuluri P, Mamouei Z, Alqarihi A, Elhassan H, French S, et al. The NDV-3A vaccine protects mice from multidrug resistant candida auris infection. PloS Pathog (2019) 15:e1007460. doi: 10.1371/journal.ppat.1007460
133. Shapiro JM, de Zoete MR, Palm NW, Laenen Y, Bright R, Mallette M, et al. Immunoglobulin a targets a unique subset of the microbiota in inflammatory bowel disease. Cell Host Microbe (2021) 29:83–93.e3. doi: 10.1016/j.chom.2020.12.003
Keywords: intestinal fungi, secretory IgA (SIgA), Crohn’s disease, antifungal immunity, mucosal immunity
Citation: Sun M, Ju J, Xu H and Wang Y (2023) Intestinal fungi and antifungal secretory immunoglobulin A in Crohn’s disease. Front. Immunol. 14:1177504. doi: 10.3389/fimmu.2023.1177504
Received: 01 March 2023; Accepted: 30 May 2023;
Published: 08 June 2023.
Edited by:
Kathrin S. Michelsen, Cedars Sinai Medical Center, United StatesReviewed by:
Lael Werner, Sheba Medical Center, IsraelCopyright © 2023 Sun, Ju, Xu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yufang Wang, d2FuZ3l1ZmFuZzA0QDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.