
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 11 May 2023
Sec. Cancer Immunity and Immunotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1174138
This article is part of the Research Topic Emerging Talents in Cancer Immunity and Immunotherapy: 2022 View all 7 articles
 Pooya Farhangnia1,2,3
Pooya Farhangnia1,2,3 Shamim Mollazadeh Ghomi3
Shamim Mollazadeh Ghomi3 Shabnam Mollazadehghomi3
Shabnam Mollazadehghomi3 Hamid Nickho1,2
Hamid Nickho1,2 Mahzad Akbarpour3,4*
Mahzad Akbarpour3,4* Ali-Akbar Delbandi1,2*
Ali-Akbar Delbandi1,2*The signaling lymphocytic activation molecule (SLAM) family receptors were discovered in immune cells for the first time. The SLAM-family receptors are a significant player in cytotoxicity, humoral immune responses, autoimmune diseases, lymphocyte development, cell survival, and cell adhesion. There is growing evidence that SLAM-family receptors have been involved in cancer progression and heralded as a novel immune checkpoint on T cells. Previous studies have reported the role of SLAMs in tumor immunity in various cancers, including chronic lymphocytic leukemia, lymphoma, multiple myeloma, acute myeloid leukemia, hepatocellular carcinoma, head and neck squamous cell carcinoma, pancreas, lung, and melanoma. Evidence has deciphered that the SLAM-family receptors may be targeted for cancer immunotherapy. However, our understanding in this regard is not complete. This review will discuss the role of SLAM-family receptors in cancer immunotherapy. It will also provide an update on recent advances in SLAM-based targeted immunotherapies.
Cancer is the second most common cause of death before the age of 70, according to data from the World Health Organization (WHO). There are several cancer therapies available. They are selected for each patient depending on the kind of cancer and the stage of the disease at diagnosis. Chemotherapy, radiation therapy, stem cell transplantation, surgery, and immunotherapy are some treatments (1).
Cancer immunotherapy approaches may induce a considerable anti-tumor response. These may be achieved via various strategies, including immune checkpoint inhibitors (ICIs) and adoptive cellular therapies. Clinical trials treating patients with various cancers between 2010 and the present have shown impressive results employing various immunotherapeutic approaches. Immunotherapy-induced anti-tumor responses can persist for a considerable time after the treatment has ceased, unlike responses from therapies that target the tumor directly (1). Programmed cell death-1 (PD-1), cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), and programmed cell death ligand 1 (PD-L1) are examples of inhibitory immune checkpoint molecules (ICMs) that suppress immune responses and allow tumor immune escape. These ICMs are expressed by immune cells and tumor cells. Several ICIs, including ipilimumab, which targets CTLA-4, and nivolumab, which targets PD-1, have been authorized by the U.S. Food and Drug Administration (FDA) in the past ten years to treat a variety of cancers (2).
Signaling lymphocytic activation molecule (SLAM) family receptors were discovered in immune cells for the first time. In humans and mice, the genes that code for SLAM-family receptors are clustered on chromosome 1 in a 400 kilobase (kb) cluster (3). Recently, some members of this family have been identified as inhibitory immune checkpoints (4, 5). These receptors engage both activating and inhibitory SH2 domain-containing proteins via their immunoreceptor tyrosine-based switch motifs (ITSMs), which are found on both the activating and inhibitory SH2 domain-containing proteins. Evidence indicates that this family’s members are engaged in various physiological and pathological processes, including the regulation of immunological responses and the route of viral entry (6). The discovery of a genetic abnormality responsible for X-linked lymphoproliferative (XLP) disease highlights the functional relevance of SLAM molecules in immune response (7)—the XLP gene mutation codes for SLAM-associated protein (SAP) or SH2D1A, a small adapter-like protein. SAP comprises the Src homology 2 (SH2) domain and binds to tyrosines in the intracellular domain of SLAM-related receptors with great specificity and affinity (8). SLAM receptors in humans form a receptor complex when they bind with SAP and Ewing’s sarcoma-activated transcript 2 (EAT-2). Also, the EAT-2-related transducer (ERT; SH2D1W) is a member of the SAP family (9, 10).
Previous studies have reported the role of SLAMs in tumor immunity in a variety of cancers, including chronic lymphocytic leukemia (CLL) (11), lymphoma (12), multiple myeloma (MM) (13), acute myeloid leukemia (AML) (14), hepatocellular carcinoma (HCC) (15), head and neck squamous cell carcinoma (HNSCC) (16), pancreas (17), lung (17), and melanoma (18). Anti-CTLA-4 and anti-PD-1 mAbs have been incredibly effective. However, not all patients benefit from them. Therefore, the discovery of novel ICMs would facilitate the development of novel targeted immunotherapies, new drugs, enhanced therapeutic response, and improved patients survival. Thus, this review will focus on the current knowledge of the role of the SLAM family of receptors in regulating immune functions during tumor progression and recent findings describing how these molecules can be exploited as a novel target in cancer immunotherapy. As described in this review, several anti-SLAM mAbs are now under exploration in various cancer clinical trials.
The SLAM family of receptors is a group of type I transmembrane receptors that includes SLAMF1 (CD150; SLAM), SLAMF2 (CD48), SLAMF3 (Ly-9; CD229), SLAMF4 (CD244; 2B4), SLAMF5 (CD84), SLAMF6 (Ly108; NTB-A; CD352), SLAMF7 (CRACC; CS1; CD319), SLAMF8 (BLAME; CD353), and SLAMF9 (CD84-H1; SF2001; CD2F10) (Table 1) (6). All molecules of the SLAM family are members of the Immunoglobulin (Ig) superfamily. Each SLAM molecule has an extracellular segment consisting of two Ig-like domains (V-like variable and C2-like constant), a transmembrane segment, and a cytoplasmic tail bearing multiple tyrosine-based motifs TxYxxI/V (ITSM), (T is threonine, I is isoleucine, V is valine and X is any amino acid). The one exception to this structural organization is SLAMF3, which has four Ig-like domains in its extracellular domain. Additionally, SLAMF2, SLAMF8, and SLAMF9 lacks ITSM domains (Figure 1) (6). In immune cells that contain SLAM-family receptors containing ITSMs, SLAM/SAP and SLAM/EAT-2 complexes interact directly with Src family kinase Fyn and phospholipase Cγ, respectively, and stimulate an activating signal within these immune cells (76). On the one hand, groundbreaking studies revealed that SLAM molecules acted as activating receptors in immune cells that expressed adaptors belonging to the SAP family. However, those same receptors acted as inhibiting receptors in cells devoid of these adaptors. On the other hand, it has been revealed that SLAM-family receptors, particularly SLAMF4, might be either inhibitory or activating in cells carrying SAP family adaptors, depending on the circumstances (Figure 2) (7).
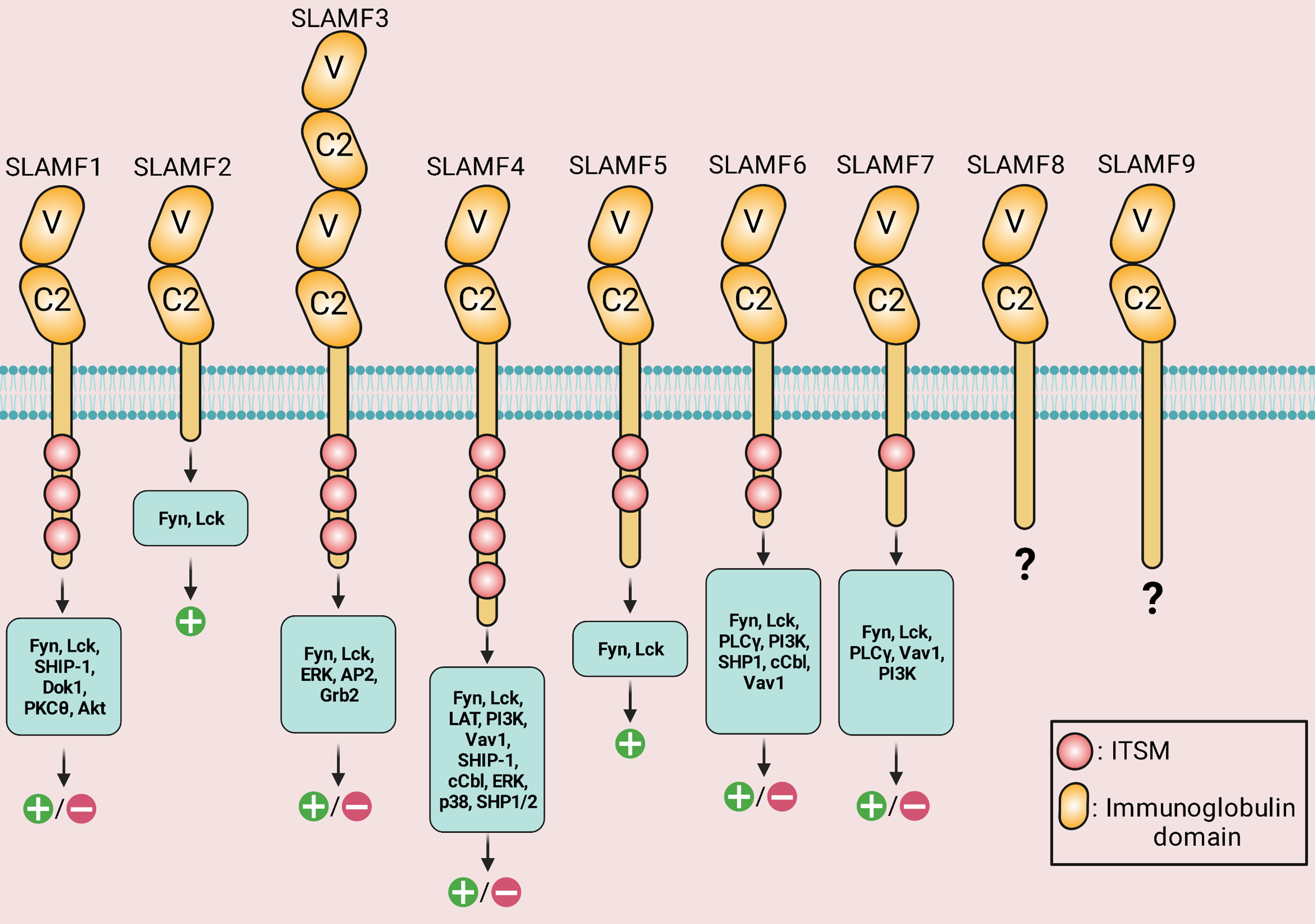
Figure 1 Structure, downstream signaling molecules, and the known signaling outcomes of signaling lymphocytic activation molecule (SLAM)-family receptors. IgV (V)- and IgC2 (C2)-like domains are found in the extracellular areas. Via the IgV-like domain, the receptors attach to self-ligands with great affinity. The cytoplasmic tail of every family member, except for SLAMF2, SLAMF8, and SLAMF9, has an immunoreceptor tyrosine-based switch motif (ITSM). SLAMF4 has the highest number of ITSM motifs. SLAMF3 has four Ig-like domains in its extracellular domain. Positive symbols and negative symbols indicate activatory outcome and inhibitory outcome, respectively.
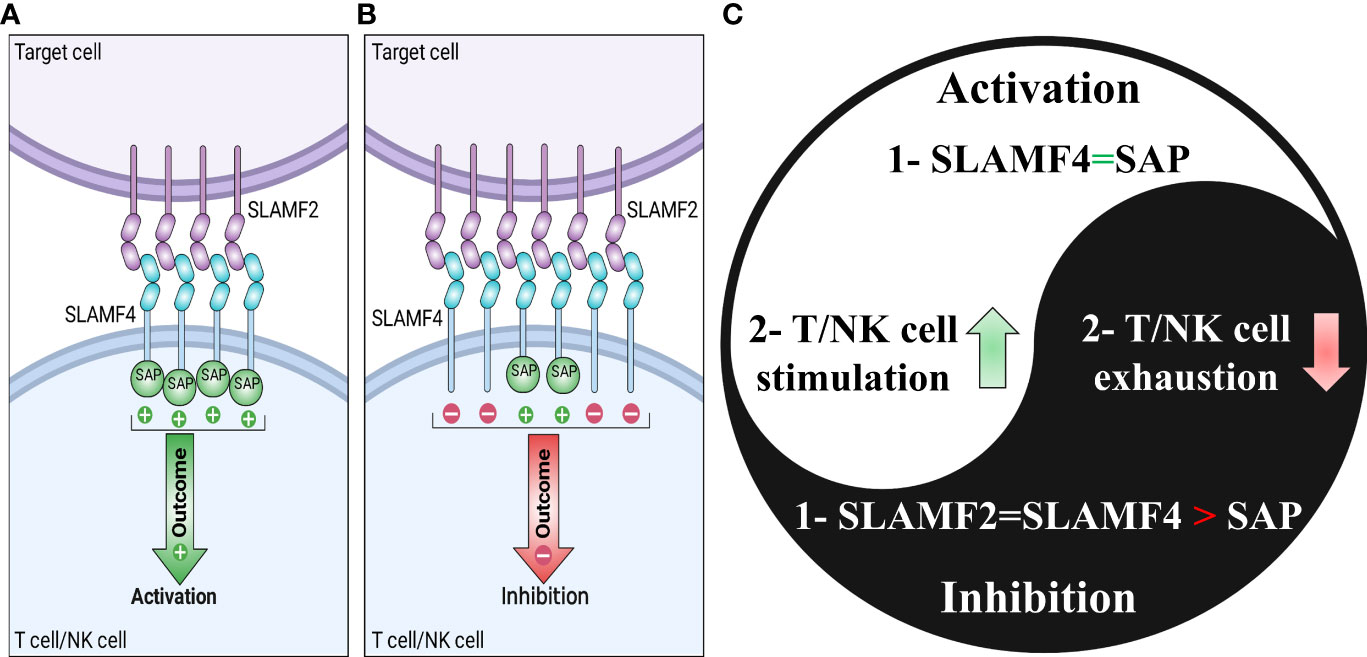
Figure 2 Signaling lymphocytic activation molecule-family receptor (SLAMF) 4-SLAMF2 interaction and dichotomous function of SLAMF4. SLAMF4 can affect T cells and natural killer (NK) cells in an activating or inhibiting manner. In the presence of adequate quantities of the adaptor protein SAP on the target cell, the SLAMF4-SLAMF2 interaction promotes NK or T cell activation (A). However, it results in an inhibitory signal in the absence of SAP on the target cell. (C) The Yin-yang of the SLAMF4 dual functions summarizes the parts of (A, B).
Glycosylphosphatidylinositol (GPI) anchor is used by SLAMF2 (6, 7, 77). Unlike most receptors expressed on immune cells, SLAM molecules are routinely self-ligands and have homotypic cell-cell interaction. However, there is an exception. For instance, SLAMF2 interacts with SLAMF4, a heterophilic interaction (78). Overall, the SLAM family of receptors can be stimulated via homotypic or heterotypic cell-cell interactions.
The glycoprotein SLAMF1 or CD150 is found in T, B, natural killer (NK), and dendritic cells (DCs; Table 1). SLAMF1 extracellular domain is a measles virus receptor, and SLAMF1 functions as a co-stimulator on T and B cells. Although type 1 T helper (Th1) cells have significant levels of SLAMF1, Th2 cells have only trace quantities (19). SLAMF1 membrane proteins are found in plasma cells (79). Findings have indicated that normal B cells and B lymphoblastoid cell lines express SLAMF1 at equivalent mRNA and protein levels (80). SLAMF1 is involved in T cell and B cell activation and differentiation, germinal center formation, and antibody production (81). It is also expressed on thymocytes, where it is involved in thymocyte development and positive selection (20, 82).
In normal immune cells, SLAMF1 is known to recruit and activate intracellular signaling molecules, such as SH2-containing tyrosine phosphatase 2 (SHP-2), phosphatidylinositol 3-kinase (PI3K), and the mitogen-activated protein kinases (MAPKs) p38, upon binding to its ligands (77, 83). While, in tumor cells, SLAMF1 has been found to be up-regulated, and it regulates cell growth and survival (84). SLAMF1 is known to mediate the survival and growth of tumor cells by activating a PI3K/Akt/mTOR signaling pathway, which is involved in regulating cell metabolism, proliferation, and survival (85–88).
SLAMF2 (CD48) is a glycoprotein related to glycophosphatidylinositol expressed on the surface of immune cells such as mature lymphocytes and monocytes and functions as a ligand for SLAMF4 (Table 1). SLAMF2 is abundantly expressed on T cells, B cells, and plasma cells and is expressed at a lesser level on monocytes, neutrophils, and CD34+ cells but not on platelets or red blood cells (13, 78). SLAMF2 is expressed by human innate lymphoid cell (ILC) progenitors, which regulate the differentiation of these cells (89).
SLAMF4, also known as 2B4 or CD244, is a member of the immunoglobulin (Ig) superfamily, which belongs to the CD2 subgroup. SLAMF4 is expressed on the cell surfaces of NK cells, T cells, monocytes, eosinophils, myeloid-derived suppressor cells (MDSCs), and DCs (Table 1). SLAMF4 interacts with the ligand SLAMF2 on target cells to regulate immune activity and transmits stimulatory or inhibitory signals (Figure 2) (90). In the intestinal mucosa, SLAMF4 regulates the fine balance between tolerance maintenance and immune surveillance (91). SLAMF4 is a negative regulator of cytotoxic intraepithelial CD8+ T cell expansion in the small intestine, maintaining homeostasis (92).
In normal immune cells, the interaction between SLAMF2 and SLAMF4 involves the recruitment of various signaling molecules, including SAP and Src-family kinases such as Fyn and Lyn. Once activated, SLAMF4 signaling leads to downstream activation of MAPK and PI3K/Akt signaling pathways, which are essential for T cell activation and proliferation (76). In hematopoietic tumors, SLAMF4 hinders signals activating mTOR and Syk through the recruitment of SHP-2 (4). Furthermore, the interaction between SLAMF2 and SLAMF4 involves the recruitment of kinases such as LCK. Once activated, SLAMF2 signaling leads to downstream activation of MAPK and PI3K/Akt signaling pathways, which are essential for T cell activation and proliferation (76). Furthermore, the presence of SLAMF2 in DCs stimulates the activation of NK cells that express SLAMF4 through the activation of PLCγ, the induction of flux and the activation of ERK mediated by SAP or EAT-2 (93). If SAP and EAT-2 are not present, the interaction between SLAMF2 and SLAMF4 stimulates a signal that restrains NK cells with the help of proteins SHP1, SHP2, and SHIP-1 (7, 94–97). When the protein SAP is present in sufficient amounts, the interaction between SLAMF2 and SLAMF4 can help activate T cells or NK cells. As stated, the interaction between SLAMF2 and SLAMF4 has both co-stimulatory and co-inhibitory roles in immune cells. However, the specific function depends on whether SAP or EAT-2 expression is present (Figure 2) (76).
SLAMF3 (also known as Ly-9 or CD229) is expressed in T and B cells, NK cells, and thymocytes (Table 1) (40). SLAMF3 plays a role in diabetes in such a way that palmitic acid activates SLAMF3 on human T cells through the STAT5-PI3K/Akt pathway, exhibiting the chronic inflammatory characteristics of type 2 diabetes (98). The lack of the SLAMF3 revealed that this receptor negatively impacts B cell homeostasis by modifying the activation thresholds of B cell subsets in mice (99).
The immune synapse is formed by SLAM receptors clustering, including SLAMF1/3/5/6, which brings about the activation of Fyn through SAP binding to the cytoplasmic tails. This leads to the activation of PKCθ and Bcl-10, which in turn induces nuclear factor kappa B (NF-κB) activation and the production of Th2 cytokines (77, 100). Consequently, this plays a role in developing natural killer T (NKT) cells and CD4+ T cells. The cytoplasmic tails of SLAM receptors containing phosphorylated tyrosines associate with SHIP, docking protein 1 (DOK1), DOK2, and RAS-GAP, which can potentially take part in these processes (77). In hematopoietic tumors, SLAMF3 is categorized as a “don’t eat me” receptor present on macrophages. Its primary function is to hinder or inhibit “eat me” signals that activate mTOR and Syk, which are typically initiated by LRP1, through SHP-2 (4). In MM, SLAMF3 directly binds to the SHP-2 and growth factor receptor bound 2 (GRB2), which eventually activates downstream pathways, including MAPK/ERK signals (101).
SLAMF5 (CD84) is expressed on CD34+ hematopoietic progenitor cells, monocytes, macrophages, and B/T lymphocytes (Table 1). SLAMF5 expression reduces the proliferative ability of CD34+ cells, indicating that it is a marker for hematopoietic progenitor cells (102, 103). SLAMF5 is a negative moderator required to maintain and function IL-10+ regulatory B cells (104). In monocyte-derived DCs, SLAMF5 regulates autophagy and stabilizes interferon regulatory factor 8 (IRF8) (105). SLAMF5 acts as a negative regulator of the development of innate CD8+ T cells (106) and humoral immune response (107).
SLAMF5 through SAP binding helps regulate the intensity of T cell receptor (TCR) signals in primary T cell responses, which contributes to p73-mediated apoptosis. This process involves the recruitment of Fyn, which fine-tunes the TCR signal strength. Activated Fyn can impede Itch, a ubiquitin ligase, thereby preventing the degradation of p73 (77, 108). However, in tumor cells, SLAMF5-mediated signaling promotes cell survival (109). Thus, further research is needed to shed light on this discrepancy of SLAMF5 signaling in different contexts.
SLAMF6 (sometimes referred to as Ly108, NTB-A, or CD352) is an immunoglobulin superfamily homophilic cell-surface receptor and a type I transmembrane protein. SLAMF6 has two immunoglobin (Ig)-like extracellular domains and two ITSMs (61, 62, 110). The SLAMF6 receptor is found on the surface of a wide range of hematopoietic cells, including T cells, B cells, and NK cells (expression limited to humans; Table 1), and interactions between these cell types allow for a variety of immunomodulatory functions, including adhesion, innate T-lymphocyte development, neutrophil function, and NK and CD8+ T cell-mediated cytotoxicity (20, 111, 112).
On the one hand, in the T cell activation phase, SLAMF6 upon binding to its ligand can recruit Fyn, which in turn impedes the Itch molecule, resulting in p73-mediated apoptosis. On the other hand, when T cell enters their re-activation phase, SLAMF6 could prompt apoptosis. This apoptosis-inducing effect occurs via a mechanism that is dependent on SAP, but not on Fyn. According to this mechanism, SAP helps remove SHP1 from the cytoplasmic tail of SLAMF6. This, in turn, leads to stronger signaling from TCR, which eventually results in the expression of genes such as FasL and Bim that mediate the process of restimulation-induced cell death (RICD) (77, 108). Furthermore, as mentioned earlier, SLAMF6 can activate PKCθ and Bcl-10, which in turn induces NF-κB activation and the production of Th2 cytokines (77, 100). Furthermore, to enhance T cell function, SLAMF6 activates the small GTPase Rap1, thereby increasing T cell adhesiveness (113). In the cancer context, a study found that there is no difference at the level of phosphorylated ribosomal protein S6 regulating signaling pathways, including PI3K/AKT/mTOR and RAS-ERK between control T cells and SLAMF6-/- T cells (114), suggesting SLAMF6 might be associated with these signaling pathways. Overall, the exact mechanism of SLAMF6 in cancer cell signaling remains to be elucidated.
SLAMF7 is primarily expressed by NK cells, T lymphocytes, activated B cells, and macrophages (Table 1). One ITSM is found in the cytoplasmic domain of SLAMF7 and is involved in the interaction with SAP. The homophilic association between SLAMF7 and NK cells governs their cytolytic activity (65).
In cells lacking EAT-2 expression, following the homotypic interaction SLAMF7 between immune cells and myeloma cells, SHIP-1 recruitment is occurred, which requires tyrosine residue (Y-284) in the ITSM motif of SLAMF7. Thus, the immune cell is inhibited. In contrast, in cells with EAT-2 expression, following the interaction EAT-2 is mostly recruited to phosphorylated tyrosine residue (pY-304) within the ITSM of SLAMF7, and then PLCγ-1 and PLCγ-2 are recruited to phosphorylated tyrosine (pY-127) on EAT-2, which eventually leads to activate the ERK signaling pathway in immune cells. However, due to the absence of EAT-2 and CD45 expression in myeloma cells, these signaling pathways are revoked (115–117).
SLAMF8 (CD353 or BLAME; B lymphocyte activator macrophage expressed) includes an extracellular and transmembrane domain. However, unlike most other SLAM members, it contains no ITSM motifs, resulting in the prevention of SAP-dependent function (69). SLAMF8 is found in monocytes, DCs, and neutrophils (Table 1). It plays a critical role in regulating inflammation by acting as a negative regulator of forming reactive oxygen species (ROS) and migration (70). A study has demonstrated that T cells and NK cells express SLAMF8, which is weakly expressed in B cells. In contrast, SLAMF8 expression was restricted in plasmablasts and plasma cells (71).
SLAMF9 (also known as CD2F10) is found on myeloid cells (72), plasmacytoid DCs (73), tumor-associated macrophages (74), peritoneal B1 cells (75), and peritoneal macrophages (Table 1) (75).
Several studies have expanded current knowledge on SLAMF1 in the pathophysiology of CLL and lymphoma (12, 85, 87, 118–120). A study has demonstrated that SLAMF1 acts as a negative regulator of IL-10 expression and secretion on the surface of B cell CLL and is associated with favorable clinical outcomes (12). Another study revealed that through selective suppression of Akt and MAPK signaling, the interaction of SLAMF1 and CD180 receptor pathways contributes to the pathobiology of B cell CLL (87). In Epstein-Barr virus (EBV)-positive B cell lymphoma cell lines, SLAMF1 has been associated with drug resistance and promoted cancer cell survival (121, 122). In contrast, a study reported no association between SLAMF1 levels and chemosensitivity in B cell CLL (123). However, in B cell CLL, a high level of mRNA expression of the mSLAMF1 isoform is related to fludarabine sensitivity and cyclophosphamide resistance. In contrast, a high level of nSLAMF1 mRNA expression indicates fludarabine resistance (123).
SLAMF1 and CD180 are considered positive CD20 expression regulators that may promote the responsiveness of SLAMF1+CD180+ B cell CLL to cancer immunotherapies based on CD20 targeted therapy (123).
It has been demonstrated that the up-regulation of SLAMF1 and SLAMF7 is a favorable prognostic marker. Patients with CLL that do not have SLAMF1 and SLAMF7 down-regulation have more effective NK cell-mediated killing and, as a result, may have improved immunological control (124). Consistent with these results, an investigation have reported down-regulation of SLAMF1 as an unfavorable prognostic marker on time-to-first treatment and overall survival (125). An empirical investigation indicated that loss of SLAMF1 expression in CLL alters genetic pathways that orchestrate chemotaxis and autophagy and treatment responses, raising the possibility that these alterations are responsible for the worse clinical outcomes observed by patients with SLAMF1 low expression (11). A research has indicated that B lymphoma cells susceptibility to oncolytic virotherapy with measles increases with the up-regulation of SLAMF1 expression (126).
In choriocarcinoma cell lines, SLAMF1 may increase methotrexate resistance by triggering protective autophagy (127). A paper has reported that the nSLAMF1 isoform is the main SLAMF1 isoform in glioma cells and suggested that SLAMF1 expression in the central nervous system (CNS) tumors can serve as a new diagnostic marker as well as a potential target for innovative therapeutic approaches in the future (128). An in silico study based on The Cancer Genome Atlas (TCGA) database reported that the three-gene signature, including SLAMF1/CD27/SELL, could be used as a biomarker to predict the prognosis and personalized immunotherapy for cervical cancer (129). A study reveals no expression of SLAMF1 on the cell surface membrane in breast cancer cell lines, while the prostate cancer cell lines expressed SLAMF1 both in the cytoplasm and on the cell surface (130).
A therapeutic approach against lymphoid leukemia, lymphoma, and MM is a mAb directed against SLAMF2 because SLAMF2 is expressed in these malignancies (13). In vitro, a developed in-house anti-SLAMF2 mAb elicits modest antibody-dependent cell-mediated cytotoxicity and significant complement-dependent cytotoxicity against MM cell lines and primary MM plasma cells. In mouse models injected subcutaneously with MM cells, administration of the anti-SLAMF2 mAb dramatically slows tumor development. It hinders the development of MM cells injected directly into mice’s bone marrow (BM). Notably, the anti-SLAMF2 mAb does not impair normal CD34+ hematopoietic stem cells (13). In patients with AML, SLAMF2 is a favorable prognostic marker that is down-regulated. AML may evade NK cell immunosurveillance by down-regulation of SLAMF2 expression (131). Methylation regulates SLAMF2 expression, and a hypomethylating drug may up-regulate SLAMF2 expression, increasing NK cell-mediated cytotoxicity in vitro. Furthermore, increased SLAMF2 expression in vivo may reverse AML immune evasion and boost NK cell activity (132). Using the NK-92MI cell line and SLAMF2-knockdown leukemia cells, an analysis asserted that NK-92MI-targeted knockdown cells show a phenotype of reduced NK susceptibility (133).
In patients with renal cell carcinoma (RCC), overexpression of SLAMF2, CD85, CD45, and PD-1 characterizes a predominantly inhibitory phenotype in circulating and tumor-infiltrating NK cells (134). In the HCC tumor microenvironment, growth differentiation factor 15 (GDF15) leads to anti-tumor immunosuppression via SLAMF2 expression on regulatory T cells (135). A study has concluded that SLAMF2 is a promising immunotherapeutic target in colorectal cancer (136).
Several studies indicate up-regulated expression of SLAMF3 on MM cells (Figure 3) (101, 137–139). It has been found that SLAMF3 is strongly and constitutively expressed in MM cells independent of the malignancy stage and that SLAMF3 knockdown/knockout reduces proliferative capacity and promotes drug-induced apoptosis in MM cells (101). In addition, high levels of serum-soluble SLAMF3 may be a valuable prognostic indicator for MM development (101). It has been suggested that immunotherapies targeting SLAMF3 eradicate the majority of tumor cells (Figure 3) and aid in eliminating chemotherapy-resistant cells that remain in the BM following therapy in patients with MM (139).
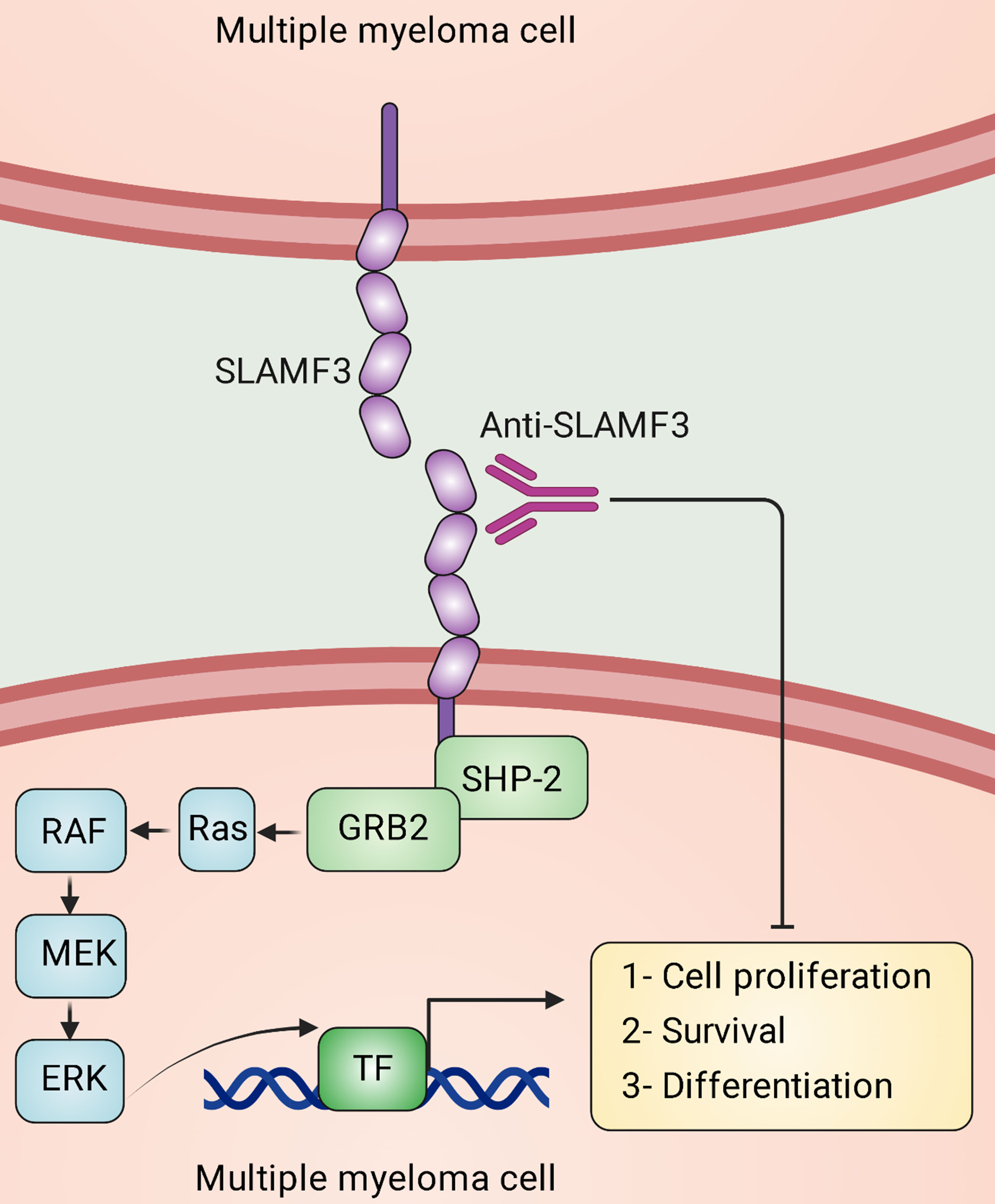
Figure 3 Signaling lymphocytic activation molecule family receptor (SLAMF) 3 signal transduction in multiple myeloma (MM) cells. In MM, homotypic interactions between SLAMF3 molecules are established, stimulating the ERK signaling pathway and specific transcription factors (TFs). This pathway eventually causes survival, proliferation, migration, differentiation, and anti-apoptotic responses in MM cells, which facilitates the progression of the malignancy. Anti-SLAMF3 antibodies might restrict the development of malignancy.
Hepatocyte proliferation and hepatocellular tumorigenesis are both controlled by SLAMF3. In human HCC samples and cell lines, SLAMF3 expression is considerably lower than in healthy primary human hepatocytes. High levels of SLAMF3 expression are restored in HCC cell lines, inhibiting cell proliferation, and migration while increasing apoptosis (15). A study has reported that SLAMF3 down-regulates multidrugs resistance protein-1 (MRP-1) expression, causing HCC cells sensitization to chemotherapy drugs (140). SLAMF3 also reduces STIM1 expression, which eventually decreases breast cancer cell migration. This molecule might be used to predict breast cancer development and aggressiveness (141). SLAMF3 is identified as inhibitory immune checkpoint and “don’t eat me” receptor on macrophages, causing the inhibition of hematopoietic tumor cells phagocytosis mediated by macrophages (4). Considering that SLAMF3 is a B and T lymphocyte cell surface receptor that is robustly expressed on MM cells, Radhakrishnan and colleagues have generated SLAMF3 chimeric antigen receptor (CAR) T cells that are highly active against MM plasma cells, memory B cells, and MM-propagating cells in vitro and in vivo (142).
In cancer studies, SLAMF4 plays a role in NK cells as an activating receptor and inhibitory receptor (Figure 2), CD8+ T cell exhaustion, and myeloid cells such as MDSCs (97). Additionally, SLAMF4 is identified as an inhibitory immune checkpoint and “don’t eat me” receptor on macrophages, causing the inhibition of tumor cells phagocytosis mediated by macrophages (4). Furthermore, SLAMF4 deletion improves phagocytosis of CD19-positive hematopoietic targets mediated by anti-CD19 CAR macrophages (4).
NK cells are essential for tumor cell surveillance. SLAMF4 is found in all NK cells, and it was in these cells that the signaling mechanisms of SLAMF4 were first discovered (97). Anti-SLAMF4 mAb therapy of mouse NK cells results in increased interferon-gamma (IFN-γ) production and non-major histocompatibility complex (MHC)-restricted tumor cell killing in vitro (46). Interactions between SLAMF4 and SLAMF2 are essential for optimum human NK cell proliferation in response to IL-2 and contribute to mouse NK cell proliferation, lytic capacity, and cytokine release (143, 144). Moreover, using anti-SLAMF4 mAb to crosslink SLAMF4 on human NK cells results in target cell lysis by NK cells (47). However, anti-SLAMF4 therapy inhibits IL2-stimulated proliferation in cultured human NK cells, showing that SLAMF4 might mediate activating or inhibitory signaling (47). Inlining with the inhibitory function of SLAMF4, murine NK cell effector activity is inhibited by SLAMF2+ target cells, inhibiting the SLAMF4-SLAMF2 interaction with specific mAb reverses this inhibition, resulting in increased target cell lysis (145). In vitro, SLAMF4 ligation reduces NK cell-mediated lysis of SLAMF2+ tumor cells and IFN-γ production by NK cells (48). In vivo, following anti-SLAMF4 mAb administration intravenously, the number of B16F10 syngeneic melanoma lung nodules in wild-type mice is considerably reduced (18). Tumor necrosis factor-alpha (TNF-α) and IFN-γ production are promoted when SLAMF4+ NK cells were co-cultured with tumor-infiltrating SLAMF2+/CD68+ monocytes/macrophages from patients with HCC. However, cytokine production and apoptosis decreases and increases, respectively (146). These findings establish an inhibitory role for SLAMF4 in mouse and human NK cells.
Tumor-infiltrating CD8+ T cells from HNSCC patients and mouse HNSCC-bearing models have significantly higher SLAMF4 expression linked with PD-1 molecule expression than healthy tissues. Additionally, SLAMF4 expression is higher on intratumoral DC and MDSC, and it is linked to PD-L1 expression and spontaneous production of immune-suppressive molecules. In vitro, SLAMF4 activation reduces the production of pro-inflammatory cytokines in human DCs. Notably, SLAMF4 knocked-out animal model dramatically reduces HNSCC tumor development. Interestingly, anti-SLAMF4 mAb therapy of mouse models reduces the growth of pre-existing HNSCC tumors and enhances tumor-infiltrating CD8+ T cells (16). SLAMF4 shows increased expression on exhausted CD8+ T cells in human cancers such as melanoma (147), MM (148), and AML (14). SLAMF4 is expressed on CD8+ T cells with an exhausted state in cancer animal models. T cells expressing co-inhibitory receptors SLAMF4, PD-1, and B- and T-lymphocyte attenuator (BTLA) are found in higher numbers in tumor-bearing animals compared to naive controls in syngeneic C57BL/6 mouse models of pancreatic adenocarcinoma and lung carcinoma (17).
SLAMF4 expression on MDSCs in tumor-bearing mice has been documented. A study found granulocytic myeloid-derived suppressor cells (Gr-MDSCs) expressing SLAMF4 in syngeneic tumor models. Additionally, significant variations in terms of function have been reported between SLAMF4+ Gr-MDSCs and SLAMF4– Gr-MDSCs. SLAMF4+ Gr-MDSCs inhibit antigen-specific CD8+ T cell responses substantially more than SLAMF4- Gr-MDSCs. Furthermore, SLAMF4 expression on Gr-MDSCs is linked to the generation of ROS and myeloperoxidase (149). Moreover, increased IL-2 and reduced IFN-γ production are linked to increased SLAMF4 expression on antigen-specific CD8+ T cells from the spleens of tumor-bearing mice (149).
A study has recently investigated that SLAMF5 is expressed on MM cells, candidating a novel therapeutic target in MM (132). This study indicates that cells producing macrophage migration inhibitory factor (MIF) induce SLAMF5 expression on cells in MM microenvironment and result in MDSCs accumulation and up-regulated PD-L1 expression on MDSCs (150). SLAMF5 inhibition diminishes MDSCs accumulation, increasing T cell activation and decreasing tumor burden (150). SLAMF5 acts as a regulator in favor of malignancy in CLL (109, 151–153). It has been suggested targeting SLAMF5 on BM-derived DCs as a novel therapeutic strategy, which may reduce CLL (151). SLAMF5 on CLL cells interacts with SLAMF5 on microenvironment cells, promoting cell survival on both sides. In vitro and in vivo, SLAMF5 inhibition causes CLL cell death (109). An investigation found that SLAMF5 promotes PD-L1 expression in CLL cells and their microenvironment and PD-1 expression in T cells (152).
SLAMF6/Ly108 facilitates macrophage M2 polarization, which aids in developing hepatocellular cancer (154). mAb directed against SLAMF6 considerably reduces the peritoneal cavity leukemic load and slows tumor development in B cell-related leukemias, lymphomas, and nonhematopoietic cancers such as melanoma. Moreover, anti-human SLAMF6 mAb restores exhausted CD8+ T cells in vitro (111).
According to the findings of a study, SLAMF6 is an inhibitory immune checkpoint whose absence allows potent CD8+ T cells to eliminate melanoma cells (114). TCR triggering of anti-melanoma CD8+ T cells in SLAMF6 knockout mice resulted in a robust effector phenotype, increased IFN-γ production, enhanced cytolysis, and improved outcomes in the adoptive transfer of SLAMF6-/- anti-melanoma CD8+ T cells to treat existing melanoma (114).
SLAMF7 is up-regulated by about 97% of myeloma cells, although its expression in normal cells is limited (155, 156). SLAMF7 was targeted in preclinical investigations by HuLuc63 (Elotuzumab; Elo) (155). HuLuc63 prevents MM cells from attaching to stromal cells in the BM and causes antibody-dependent cellular cytotoxicity (ADCC) against MM cells (Figure 4) (155, 157). Elo enhances ligation between NK and myeloma cells and induces NK cytotoxicity against myeloma cells in an ADCC-independent manner (158, 159). Moreover, Elo improves NK cell cytotoxicity against MM cells in a CD16-independent manner and promotes SLAMF7-SLAMF7 interactions (Figure 4) (160).
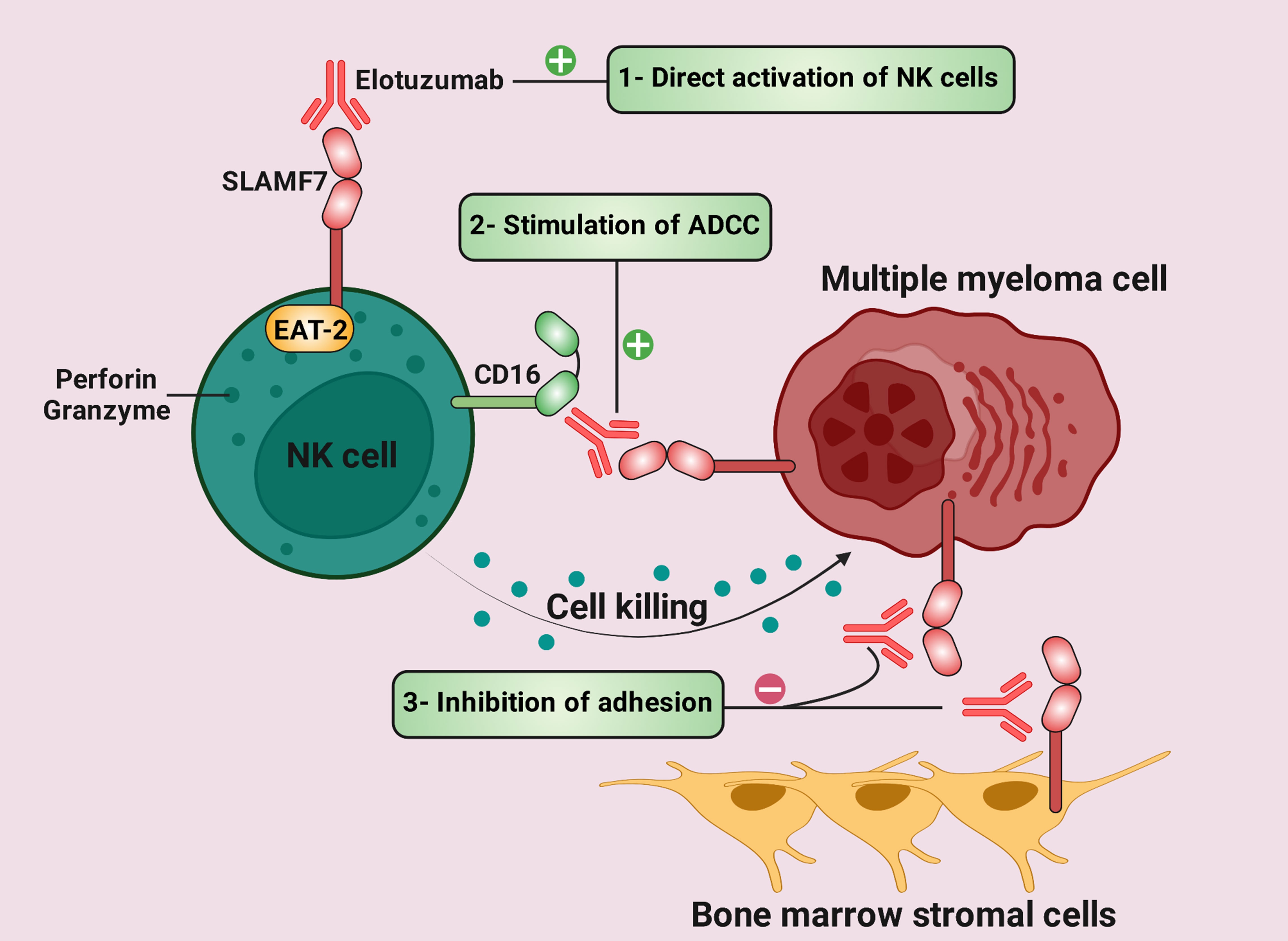
Figure 4 Therapeutic mechanisms of elotuzumab in multiple myeloma (MM) (1). Elotuzumab can bind to signaling lymphocytic activation molecule family receptor (SLAMF) 7, thus stimulating direct activation of natural killer (NK) cells via EAT-2. (2) While the Fc component of elotuzumab interacts with the activating Fc receptor CD16 on NK cells, the Fab portion binds to SLAMF7 on MM cells. This interaction results in antibody-dependent cellular cytotoxicity (ADCC). (3) Elotuzumab prevents MM cells from adhering to bone marrow stromal cells (BMSCs) because BMSCs and MM cells interact via SLAMF7-SLAMF7 homotypic interactions.
Elo can potentially activate ADCC in MM cells resistant to bortezomib (a proteasome inhibitor) and HSP90 inhibitors (6). A study found that combining Elo and bortezomib significantly exhibits in vivo anti-tumor activity in the MM xenograft model and enhances Elo-induced MM cell lysis (156). In phase 1 and the dose-escalation trial, Elo was generally well tolerated in patients with advanced MM, while it had no same efficiency as in the animal model (161). Elo increases the objective response rate (ORR) from 45% to 48% when given to patients on bortezomib therapy (162). In the phase II clinical trial, the combination of Elo, lenalidomide, and dexamethasone in 150 patients with MM improved progression-free survival from 6.9 to 9.7 months with a steady ORR from 66 to 63% (163). Elo has been shown to enhance ORR by 82% in combination with lenalidomide and low-dose dexamethasone (164). Table 2 provides more clinical trials regarding Elo therapy in patients with cancer.
Immunogenic peptides from the SLAMF7 antigen effectively activate specific cytotoxic T lymphocyte (CTL) clones. Accordingly, an immunogenic HLA-A2-specific SLAMF7239-247 peptide can induce antigen-specific CTL against MM cells, lysing tumor cells (172).
An empirical study highlighted that B16-F10 tumors grow more slowly in mice lacking SLAMF7 (179). Additionally, CD8+ T cells express lower levels of PD-1 and have a reduced capacity to manifest a T cell exhaustion phenotype in this animal model (179).
SLAMF7 is a decent target for CAR T cell therapy because of its high expression on myeloma cells and low expression on normal cells (173, 180–183). The distal variable (V) domain and the proximal constant 2 (C2) domain are two extracellular domains of the SLAMF7 protein. O’Neal and colleagues developed and evaluated SLAMF7 CAR T cells targeting the V domain of SLAMF7 (Luc90-SLAMF7 CAR T) and demonstrated anti-myeloma killing in vitro and in vivo using two myeloma mouse models (180). Another study corroborates the efficacy of SLAMF7 CAR T cells against MM cells. Adoptive transfer of SLAMF7 CAR T cells into MM-bearing mice shows potent anti-cancer efficacy. In vitro, adding lenalidomide to SLAMF7 CAR T cell improves immune functions such as cytotoxicity, memory maintenance, Th1 cytokine production, and immune synapse formation. Furthermore, the anti-cancer activity and durability of adoptively transferred SLAMF7 CAR T cells are also increased by lenalidomide in vivo (173).
One concern regarding SLAMF7 CAR T cell therapy may limit its use in the clinic. This limitation is that SLAMF7 is expressed in normal NK cells. Thus, incorporating a suicide gene with an anti-SLAMF7 CAR is reasonable. Accordingly, Amatya and colleagues invented a construct with a suicide gene and a CD28-containing anti-SLAMF7 CAR. The suicide gene encodes a dimerization domain linked to a caspase-9 domain. In vitro, T cells carrying the anti-SLAMF7 CAR with suicide-gene construct detect SLAMF7 and eradicate tumors in mice. Finally, the dimerizing chemical rimiducid is used to kill T cells that express this construct, preventing damage to normal immune cells (184).
A study has revealed that anaplastic large-cell lymphoma cell lines express SLAMF8 (185). Furthermore, SLAMF8 knockdown down-regulates SHP-2 activation and cell proliferation while increasing apoptosis in lymphoma cell lines (185). SLAMF8 orchestrates oncogenic KIT-mediated RAS-RAF-ERK signaling and SHP-2-mediated human neoplastic mast cell proliferation (186). Findings from analysis of the TCGA database revealed that high SLAMF8 expression is a prognostic factor in glioma, conferring reduced overall survival, chemotherapy resistance, and aggravated immunosuppression (187). In addition, high SLAMF8 expression is a favorable prognostic factor for better anti-PD1 immunotherapy efficacy in patients with gastrointestinal cancers (188).
SLAMF9 expression was discovered in malignant cell lines from the monocytic and lymphocytic origin using RT-PCR (189). In macrophages, SLAMF9 is a novel type-I transmembrane receptor with immunomodulatory characteristics. SLAMF9 affects pro-inflammatory cytokine production and migration in murine and human melanoma tumor-associated macrophages (74).
There are numerous potential hurdles in targeting SLAM family members, which deserve special attention. In the following, these challenges will be examined.
As discussed earlier, targeting SLAM molecules has been explored as a potential therapeutic strategy in cancer. However, as with any therapeutic target, there are potential toxicities associated with targeting SLAM family members. Some of the toxicities and AEs observed in preclinical and clinical studies include lymphopenia, fatigue, chest pain, gastroenteritis, neutropenia, thrombocytopenia, and hepatotoxicity (162, 164, 175, 176, 178, 190, 191). These AEs might due to the broad expression of SLAM family members on various immune cells and their complex regulatory functions within the immune system. Furthermore, targeting SLAM family members might also affect the normal immune response and increase the risk of infections, as some of these receptors are involved in controlling immune cell function (90), including the clearance of pathogens (21, 192). Therefore, careful consideration should be given to the potential toxicities associated with targeting SLAM family members, and appropriate safety measures should be taken to minimize the risk of AEs.
There is currently no clear evidence to suggest that targeting the SLAM receptors on cancer cells would lead to the up-regulation of other checkpoint molecules. The association between SLAM receptors and other checkpoint molecules such as PD-1, CTLA-4, and T cell immunoglobulin and mucin domain-containing protein 3 (TIM-3), is not well established. However, a study reported that SAP inhibits PD-1 signaling in T cells by interacting with CD28 (193). To prevent SHP-2 from dephosphorylating downstream phosphorylated CD28, SAP has indirect interactions with the PD-1 signaling complex (194). Additionally, in primary human T cells, suppressing SAP improves PD-1 ligation and function, which in turn suppresses IL-2 production (194, 195). Thus, it is postulated that targeting SLAM receptors could have downstream effects on other immune checkpoint molecules, but more research is needed to understand the potential interactions and outcomes of such therapies.
The function of SLAM receptors, including SLAMF1/3/4/6/7, is dichotomous (Figure 1). SLAMF3 function, for instance, is intricate and not well understood as it may have inhibitory (196) or stimulatory (197, 198) functions depending on the context of the immune response. SLAMF3 has been shown to have different effects on cancer depending on the cell types and the signaling pathways involved. In some cases, SLAMF3 has been reported to have an inhibitory effect on cancer. For instance, studies have shown that SLAMF3 can inhibit the growth and proliferation of certain types of cancer cells such as liver cancer and hematopoietic tumors (4, 15, 140). However, in other cases, SLAMF3 has been found to have a stimulatory effect on cancer. For instance, SLAMF3 has been shown to induce the proliferation of MM cells (101).
SLAMF7 appears to have both inhibitory and stimulatory effects in cancer, depending on the type of cancer and context in which it is expressed. In some types of cancer, such as MM and breast cancer, SLAMF7 has been found to be overexpressed (199), and its presence on cancer cells might aid them evade attack by the immune system. In this context, immunotherapies targeting SLAMF7, such as Elo and CAR T cell therapy, have been developed (173, 180–183, 200). On the other hand, in some other types of cancer, such as ovarian cancer and RCC, low expression of SLAMF7 has been associated with poor prognosis (179), suggesting that SLAMF7 may play a role in suppressing tumor growth in these contexts. Therefore, the impact of SLAMF3 and SLAMF7 on cancer is context-dependent and requires further investigation.
Overall, further study is required to identify if a given SLAM receptor acts as an activatory receptor or an inhibitory receptor in each cancer type. Functional assays using cancer cells and SLAM receptor-specific antibodies or CAR T cells might be one such strategy. This would aid in determining whether or not SLAM receptor activation enhances the development and survival of cancer cells. Instead, genetic knockdown or knockout approaches might be used to examine the impact of SLAM receptor deletion on the behavior of tumor cells. Eventually, investigating SLAM receptor expression in patient samples might help identify biomarkers and therapeutic targets for various cancers.
In the case of targeting SLAM molecules, the potential on-target off-tumor effects could include immune system dysfunction, as SLAM molecules play a crucial role in immune cells function. Not all SLAM receptors are expressed only on normal immune cells. While these receptors have a widespread expression on different immune cells such as T cells, B cells, and NK cells, also these molecules may be expressed in malignant cells. For instance, SLAMF7 is expressed in MM cells (201). Hence, immunotherapeutic strategies targeting SLAMF7 should be deployed with caution in patients with MM because of SLAMF7 expression on non-cancerous immune cells. One approach to minimize the risk of on-target off-tumor toxicities and optimize the benefits of targeting SLAM molecules might involve combining SLAM-targeted therapy with immune checkpoint inhibitors, which could help to mitigate these AEs and enhance the efficacy of the treatment.
Our knowledge of the role of SLAM-family receptors in cancer immunity has grown during the last several years. There is growing irrefutable proof that receptors belonging to the SLAM family mediate essential functions in immune cells. Studies have indicated that the SAP family of adaptors decides whether a SLAM receptor has a stimulatory or inhibitory function. Additionally, findings have established that SLAMF7 is a valid and safe drug target for the treatment of MM, and results statistically showed significant trends.
This article presented an outline of SLAM molecules with a focus on recent and previous findings on the role of SLAM family molecules in treating various cancers. Evidence indicates that some members of this family act as an inhibitory immune checkpoint. Thus, these molecules represent a novel therapeutic avenue to boost antitumoural immunity. Also, these molecules may be used as a predictor of cancer development and aggressiveness. Given recent breakthroughs in current knowledge and our understanding of the role of the SLAM-family receptors in tumor immunity, the receptors within this axis are potential candidates for hematological and solid tumors immunotherapy. However, a significant hurdle limiting the translation of ICIs to clinical therapeutic approaches is the corresponding AEs. For instance, serious AEs were reported in most patients with cancer receiving Elo (174). Overall, we tried to provide some powerful arguments which will shape our thinking about SLAM molecules in the context of cancer for the next years.
Several unresolved questions remain to be answered. 1 – Is the expression of SLAM molecules associated with the tumor stage? 2 – Which biomarkers predict the patient’s therapeutic response to SLAM inhibition? 3 – Considering the alterations in the elderly patients’ immune system, is the therapeutic response of this patient population to the inhibition of the SLAM molecule similar to other patients? 4 – Is there a possibility of any immune-related AEs associated with SLAM inhibition?
Several emerging therapies have been used in association with SLAM molecules. One of these novel paradigms is CAR T cells directed against SLAMF3 and SLAMF7. Clinical trials assessing the effects of these CAR T cells on progression-free survival and overall survival might be an intriguing research direction in solid and liquid tumors. mAb directed against SLAMF7, such as HuLuc63 or Elo, has emerged as a powerful tool in the armamentarium against MM (Figure 4). Many studies assessed the efficacy of Elo alone or in combination with therapeutic agents in MM. However, this mAb has not been applied in solid tumors and remains a conspicuous area of research with potential relevance to cancer patients. One of the main criticisms of targeting SLAM molecules is that they are also expressed in normal immune cells. This point limits the bench-to-bedside clinical success of SLAM-based targeted immunotherapies. Thus, preclinical and clinical investigations should examine whether SLAM molecule inhibitors have significant AEs. The definitive goal in combating tumors is to achieve the specific recognition and effective eradication of tumor cells with minimum and tolerable AEs. A significant hurdle limiting bench-to-bedside translation of ICIs directed against SLAM molecules is potential AEs. Overall, anti-SLAM mAbs-related toxicity might be a rare but severe side effect that deserves special attention. In order to effectively manage cancer patients who have toxicity due to SLAM-targeted immunotherapies, a better knowledge of the cause, early identification, and timely treatment are vital. Thus, further investigations underlying anti-SLAM mAbs-associated toxicity need to be undertaken.
PF and A-AD conceptualized the study. PF, ShamM, ShabM, and HN drafted the manuscript. MA and A-AD revised the article critically for important intellectual content. MA and A-AD supervised the study. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
mAb, Monoclonal antibody; PD-1, Programmed cell death-1; PD-L1, Programmed cell death ligand 1; CTLA-4, T-lymphocyte-associated protein 4; SLAM, Signaling lymphocytic activation molecule; ICI, Immune checkpoint inhibitor; ICM, Immune checkpoint molecule; MM, Multiple myeloma; CTL, Cytotoxic T lymphocyte; AE, Adverse effect; TCGA, The Cancer Genom Atlas; AML, Acute myeloid leukemia; CLL, Chronic lymphocytic leukemia; HCC, Hepatocellular carcinoma; HNSCC, Head and neck squamous cell carcinoma; ITSM, Immunoreceptor tyrosine-based switch motif; NK, Natural killer cell; CAR, Chimeric antigen receptor; ORR, Objective response rate; BM, Bone marrow; MDSC, Myeloid-derived suppressor cell; EAT-2, Ewing’s sarcoma-activated transcript 2; DC, Dendritic cell; ADCC, Antibody-dependent cellular cytotoxicity; TCR, T cell receptor; Th1, Type 1 T helper; IFN-γ, Interferon-gamma; Elo, Elotuzumab; ROS, Reactive oxygen species; Gr-MDSC, Granulocytic myeloid-derived suppressor cell; SHP-2, SH2-containing tyrosine phosphatase 2; PI3K, Phosphatidylinositol 3-kinase; MAPK, Mitogen-activated protein kinase.
1. Farhangnia P, Akbarpour M, Yazdanifar M, Aref AR, Delbandi A-A, Rezaei N. Advances in therapeutic targeting of immune checkpoints receptors within the CD96-TIGIT axis: clinical implications and future perspectives. Expert Rev Clin Immunol (2022). doi: 10.1080/1744666X.2022.2128107
2. Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov (2018) 8:1069–86. doi: 10.1158/2159-8290.CD-18-0367
3. Veillette A, Dong Z, Latour S. Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity (2007) 27:698–710. doi: 10.1016/j.immuni.2007.11.005
4. Dan L, Wei X, Yuande W, Jin F, Yuexi H, Juan D, et al. SLAMF3 and SLAMF4 are immune checkpoints that constrain macrophage phagocytosis of hematopoietic tumors. Sci Immunol (2022) 7:eabj5501. doi: 10.1126/sciimmunol.abj5501
5. Hajaj E, Zisman E, Tzaban S, Merims S, Cohen J, Klein S, et al. Alternative splicing of the inhibitory immune checkpoint receptor SLAMF6 generates a dominant positive form, boosting T-cell effector functions. Cancer Immunol Res (2021) 9:637–50. doi: 10.1158/2326-6066.CIR-20-0800
6. Fouquet G, Marcq I, Debuysscher V, Bayry J, Rabbind Singh A, Bengrine A, et al. Signaling lymphocytic activation molecules slam and cancers: friends or foes? Oncotarget (2018) 9:16248–62. doi: 10.18632/oncotarget.24575
7. Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol (2011) 29:665–705. doi: 10.1146/annurev-immunol-030409-101302
8. Morra M, Howie D, Grande MS, Sayos J, Wang N, Wu C, et al. X-Linked lymphoproliferative disease: a progressive immunodeficiency. Annu Rev Immunol (2001) 19:657–82. doi: 10.1146/annurev.immunol.19.1.657
9. Roncagalli R, Taylor JER, Zhang S, Shi X, Chen R, Cruz-Munoz M-E, et al. Negative regulation of natural killer cell function by EAT-2, a SAP-related adaptor. Nat Immunol (2005) 6:1002–10. doi: 10.1038/ni1242
10. Veillette A. SLAM-family receptors: immune regulators with or without SAP-family adaptors. Cold Spring Harb Perspect Biol (2010) 2:a002469. doi: 10.1101/cshperspect.a002469
11. Bologna C, Buonincontri R, Serra S, Vaisitti T, Audrito V, Brusa D, et al. SLAMF1 regulation of chemotaxis and autophagy determines CLL patient response. J Clin Invest (2016) 126:181–94. doi: 10.1172/JCI83013
12. Shcherbina V, Gordiienko I, Shlapatska L, Gluzman D, Sidorenko S. CD150 and CD180 are negative regulators of IL-10 expression and secretion in chronic lymphocytic leukemia b cells. Neoplasma (2021) 68:760–9. doi: 10.4149/neo_2021_210104N8
13. Hosen N, Ichihara H, Mugitani A, Aoyama Y, Fukuda Y, Kishida S, et al. CD48 as a novel molecular target for antibody therapy in multiple myeloma. Br J Haematol (2012) 156:213–24. doi: 10.1111/j.1365-2141.2011.08941.x
14. Lichtenegger FS, Schnorfeil FM, Emmerig K, Neitz JS, Beck B, Draenert R, et al. Pseudo-exhaustion of CD8+ T cells in AML. Blood (2013) 122:2615. doi: 10.1182/blood.V122.21.2615.2615
15. Marcq I, Nyga R, Cartier F, Amrathlal RS, Ossart C, Ouled-Haddou H, et al. Identification of SLAMF3 (CD229) as an inhibitor of hepatocellular carcinoma cell proliferation and tumour progression. PloS One (2013) 8:e82918. doi: 10.1371/journal.pone.0082918
16. Agresta L, Lehn M, Lampe K, Cantrell R, Hennies C, Szabo S, et al. CD244 represents a new therapeutic target in head and neck squamous cell carcinoma. J Immunother Cancer (2020) 8:e000245. doi: 10.1136/jitc-2019-000245
17. Mittal R, Wagener M, Breed ER, Liang Z, Yoseph BP, Burd EM, et al. Phenotypic T cell exhaustion in a murine model of bacterial infection in the setting of pre-existing malignancy. PloS One (2014) 9:e93523. doi: 10.1371/journal.pone.0093523
18. Johnson LA, Vaidya SV, Goldfarb RH, Mathew PA. 2B4(CD244)-mediated activation of NK cells reduces metastases of B16F10 melanoma in mice. Anticancer Res (2003) 23:3651–5.
19. Wang N, Morra M, Wu C, Gullo C, Howie D, Coyle T, et al. CD150 is a member of a family of genes that encode glycoproteins on the surface of hematopoietic cells. Immunogenetics (2001) 53:382–94. doi: 10.1007/s002510100337
20. Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity (2007) 27:751–62. doi: 10.1016/j.immuni.2007.08.020
21. Berger SB, Romero X, Ma C, Wang G, Faubion WA, Liao G, et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat Immunol (2010) 11:920–7. doi: 10.1038/ni.1931
22. Cannons JL, Qi H, Lu KT, Dutta M, Gomez-Rodriguez J, Cheng J, et al. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity (2010) 32:253–65. doi: 10.1016/j.immuni.2010.01.010
23. Sidorenko SP, Clark EA. Characterization of a cell surface glycoprotein IPO-3, expressed on activated human b and T lymphocytes. J Immunol (1993) 151:4614–24.
24. Mavaddat N, Mason DW, Atkinson PD, Evans EJ, Gilbert RJC, Stuart DI, et al. Signaling lymphocytic activation molecule (CDw150) is homophilic but self-associates with very low affinity*. J Biol Chem (2000) 275:28100–9. doi: 10.1074/jbc.M004117200
25. Castro AG, Hauser TM, Cocks BG, Abrams J, Zurawski S, Churakova T, et al. Molecular and functional characterization of mouse signaling lymphocytic activation molecule (SLAM): differential expression and responsiveness in Th1 and Th2 cells. J Immunol (1999) 163:5860–70.
26. Romero X, Benítez D, March S, Vilella R, Miralpeix M, Engel P. Differential expression of SAP and EAT-2-binding leukocyte cell-surface molecules CD84, CD150 (SLAM), CD229 (Ly9) and CD244 (2B4). Tissue Antigens (2004) 64:132–44. doi: 10.1111/j.1399-0039.2004.00247.x
27. Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell (2005) 121:1109–21. doi: 10.1016/j.cell.2005.05.026
28. Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature (2000) 406:893–7. doi: 10.1038/35022579
29. Bleharski JR, Niazi KR, Sieling PA, Cheng G, Modlin RL. Signaling lymphocytic activation molecule is expressed on CD40 ligand-activated dendritic cells and directly augments production of inflammatory cytokines. J Immunol (2001) 167:3174–81. doi: 10.4049/jimmunol.167.6.3174
30. Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, et al. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature (1998) 395:462–9. doi: 10.1038/26683
31. Erlenhoefer C, Wurzer WJ, Löffler S, Schneider-Schaulies S, ter Meulen V, Schneider-Schaulies J. CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J Virol (2001) 75:4499–505. doi: 10.1128/JVI.75.10.4499-4505.2001
32. Sidorenko SP, Clark EA. The dual-function CD150 receptor subfamily: the viral attraction. Nat Immunol (2003) 4:19–24. doi: 10.1038/ni0103-19
33. Kruse M, Meinl E, Henning G, Kuhnt C, Berchtold S, Berger T, et al. Signaling lymphocytic activation molecule is expressed on mature CD83+ dendritic cells and is up-regulated by IL-1 beta. J Immunol (2001) 167:1989–95. doi: 10.4049/jimmunol.167.4.1989
34. Nanda N, Andre P, Bao M, Clauser K, Deguzman F, Howie D, et al. Platelet aggregation induces platelet aggregate stability via SLAM family receptor signaling. Blood (2005) 106:3028–34. doi: 10.1182/blood-2005-01-0333
35. Cocks BG, Chang CC, Carballido JM, Yssel H, de Vries JE, Aversa G. A novel receptor involved in T-cell activation. Nature (1995) 376:260–3. doi: 10.1038/376260a0
36. Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, et al. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J Immunol (2010) 185:190–202. doi: 10.4049/jimmunol.0903505
37. Yokoyama S, Staunton D, Fisher R, Amiot M, Fortin JJ, Thorley-Lawson DA. Expression of the blast-1 activation/adhesion molecule and its identification as CD48. J Immunol (1991) 146:2192–200.
38. Baorto DM, Gao Z, Malaviya R, Dustin ML, van der Merwe A, Lublin DM, et al. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature (1997) 389:636–9. doi: 10.1038/39376
39. Kato K, Koyanagi M, Okada H, Takanashi T, Wong YW, Williams AF, et al. CD48 is a counter-receptor for mouse CD2 and is involved in T cell activation. J Exp Med (1992) 176:1241–9. doi: 10.1084/jem.176.5.1241
40. de la Fuente MA, Tovar V, Villamor N, Zapater N, Pizcueta P, Campo E, et al. Molecular characterization and expression of a novel human leukocyte cell-surface marker homologous to mouse ly-9. Blood (2001) 97:3513–20. doi: 10.1182/blood.v97.11.3513
41. Sandrin MS, Gumley TP, Henning MM, Vaughan HA, Gonez LJ, Trapani JA, et al. Isolation and characterization of cDNA clones for mouse ly-9. J Immunol (1992) 149:1636–41.
42. Sandrin MS, Henning MM, Lo MF, Baker E, Sutherland GR, McKenzie IF. Isolation and characterization of cDNA clones for Humly9: the human homologue of mouse Ly9. Immunogenetics (1996) 43:13–9. doi: 10.1007/BF00186599
43. Sayós J, Martín M, Chen A, Simarro M, Howie D, Morra M, et al. Cell surface receptors ly-9 and CD84 recruit the X-linked lymphoproliferative disease gene product SAP. Blood (2001) 97:3867–74. doi: 10.1182/blood.v97.12.3867
44. Sintes J, Vidal-Laliena M, Romero X, Tovar V, Engel P. Characterization of mouse CD229 (Ly9), a leukocyte cell surface molecule of the CD150 (SLAM) family. Tissue Antigens (2007) 70:355–62. doi: 10.1111/j.1399-0039.2007.00909.x
45. Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, et al. T Follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for b cells. J Immunol (2004) 173:68–78. doi: 10.4049/jimmunol.173.1.68
46. Garni-Wagner BA, Purohit A, Mathew PA, Bennett M, Kumar V. A novel function-associated molecule related to non-MHC-restricted cytotoxicity mediated by activated natural killer cells and T cells. J Immunol (1993) 151:60–70. doi: 10.4049/jimmunol.151.1.60
47. Valiante NM, Trinchieri G. Identification of a novel signal transduction surface molecule on human cytotoxic lymphocytes. J Exp Med (1993) 178:1397–406. doi: 10.1084/jem.178.4.1397
48. Lee K-M, McNerney ME, Stepp SE, Mathew PA, Schatzle JD, Bennett M, et al. 2B4 acts as a non-major histocompatibility complex binding inhibitory receptor on mouse natural killer cells. J Exp Med (2004) 199:1245–54. doi: 10.1084/jem.20031989
49. Sayós J, Nguyen KB, Wu C, Stepp SE, Howie D, Schatzle JD, et al. Potential pathways for regulation of NK and T cell responses: differential X-linked lymphoproliferative syndrome gene product SAP interactions with SLAM and 2B4. Int Immunol (2000) 12:1749–57. doi: 10.1093/intimm/12.12.1749
50. Munitz A, Bachelet I, Fraenkel S, Katz G, Mandelboim O, Simon H-U, et al. 2B4 (CD244) is expressed and functional on human eosinophils. J Immunol (2005) 174:110–8. doi: 10.4049/jimmunol.174.1.110
51. Tangye SG, Phillips JH, Lanier LL, Nichols KE. Functional requirement for SAP in 2B4-mediated activation of human natural killer cells as revealed by the X-linked lymphoproliferative syndrome. J Immunol (2000) 165:2932–6. doi: 10.4049/jimmunol.165.6.2932
52. Kambayashi T, Assarsson E, Chambers BJ, Ljunggren HG. Cutting edge: regulation of CD8(+) T cell proliferation by 2B4/CD48 interactions. J Immunol (2001) 167:6706–10. doi: 10.4049/jimmunol.167.12.6706
53. Brown MH, Boles K, Anton van der Merwe P, Kumar V, Mathew PA, Neil Barclay A. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J Exp Med (1998) 188:2083–90. doi: 10.1084/jem.188.11.2083
54. Latchman Y, McKay PF, Reiser H. Identification of the 2B4 molecule as a counter-receptor for CD48. J Immunol (1998) 161:5809–12. doi: 10.4049/jimmunol.161.11.5809
55. Speiser DE, Colonna M, Ayyoub M, Cella M, Pittet MJ, Batard P, et al. The activatory receptor 2B4 is expressed in vivo by human CD8+ effector alpha beta T cells. J Immunol (2001) 167:6165–70. doi: 10.4049/jimmunol.167.11.6165
56. Martin M, Romero X, de la Fuente MA, Tovar V, Zapater N, Esplugues E, et al. CD84 functions as a homophilic adhesion molecule and enhances IFN-gamma secretion: adhesion is mediated by ig-like domain 1. J Immunol (2001) 167:3668–76. doi: 10.4049/jimmunol.167.7.3668
57. Tangye SG, van de Weerdt BCM, Avery DT, Hodgkin PD. CD84 is up-regulated on a major population of human memory b cells and recruits the SH2 domain containing proteins SAP and EAT-2. Eur J Immunol (2002) 32:1640–9. doi: 10.1002/1521-4141(200206)32:6<1640::AID-IMMU1640>3.0.CO;2-S
58. Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature (2005) 435:452–8. doi: 10.1038/nature03555
59. de la Fuente MA, Pizcueta P, Nadal M, Bosch J, Engel P. CD84 leukocyte antigen is a new member of the ig superfamily. Blood (1997) 90:2398–405. doi: 10.1182/blood.V90.6.2398
60. de la Fuente MA, Tovar V, Pizcueta P, Nadal M, Bosch J, Engel P. Molecular cloning, characterization, and chromosomal localization of the mouse homologue of CD84, a member of the CD2 family of cell surface molecules. Immunogenetics (1999) 49:249–55. doi: 10.1007/s002510050490
61. Fraser CC, Howie D, Morra M, Qiu Y, Murphy C, Shen Q, et al. Identification and characterization of SF2000 and SF2001, two new members of the immune receptor SLAM/CD2 family. Immunogenetics (2002) 53:843–50. doi: 10.1007/s00251-001-0415-7
62. Peck SR, Ruley HE. Ly108: a new member of the mouse CD2 family of cell surface proteins. Immunogenetics (2000) 52:63–72. doi: 10.1007/s002510000252
63. Flaig RM, Stark S, Watzl C. Cutting edge: NTB-a activates NK cells via homophilic interaction. J Immunol (2004) 172:6524–7. doi: 10.4049/jimmunol.172.11.6524
64. Falco M, Marcenaro E, Romeo E, Bellora F, Marras D, Vély F, et al. Homophilic interaction of NTBA, a member of the CD2 molecular family: induction of cytotoxicity and cytokine release in human NK cells. Eur J Immunol (2004) 34:1663–72. doi: 10.1002/eji.200424886
65. Kumaresan PR, Lai WC, Chuang SS, Bennett M, Mathew PA. CS1, a novel member of the CD2 family, is homophilic and regulates NK cell function. Mol Immunol (2002) 39:1–8. doi: 10.1016/s0161-5890(02)00094-9
66. Bouchon A, Cella M, Grierson HL, Cohen JI, Colonna M. Activation of NK cell-mediated cytotoxicity by a SAP-independent receptor of the CD2 family. J Immunol (2001) 167:5517–21. doi: 10.4049/jimmunol.167.10.5517
67. Cruz-Munoz M-E, Dong Z, Shi X, Zhang S, Veillette A. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat Immunol (2009) 10:297–305. doi: 10.1038/ni.1693
68. Boles KS, Mathew PA. Molecular cloning of CS1, a novel human natural killer cell receptor belonging to the CD2 subset of the immunoglobulin superfamily. Immunogenetics (2001) 52:302–7. doi: 10.1007/s002510000274
69. Kingsbury GA, Feeney LA, Nong Y, Calandra SA, Murphy CJ, Corcoran JM, et al. Cloning, expression, and function of BLAME, a novel member of the CD2 family. J Immunol (2001) 166:5675–80. doi: 10.4049/jimmunol.166.9.5675
70. Wang G, van Driel BJ, Liao G, O’Keeffe MS, Halibozek PJ, Flipse J, et al. Migration of myeloid cells during inflammation is differentially regulated by the cell surface receptors Slamf1 and Slamf8. PloS One (2015) 10:e0121968. doi: 10.1371/journal.pone.0121968
71. Llinàs L, Lázaro A, de Salort J, Matesanz-Isabel J, Sintes J, Engel P. Expression profiles of novel cell surface molecules on b-cell subsets and plasma cells as analyzed by flow cytometry. Immunol Lett (2011) 134:113–21. doi: 10.1016/j.imlet.2010.10.009
72. Zeng X, Liu G, Peng W, He J, Cai C, Xiong W, et al. Combined deficiency of SLAMF8 and SLAMF9 prevents endotoxin-induced liver inflammation by downregulating TLR4 expression on macrophages. Cell Mol Immunol (2020) 17:153–62. doi: 10.1038/s41423-018-0191-z
73. Sever L, Radomir L, Stirm K, Wiener A, Schottlender N, Lewinsky H, et al. SLAMF9 regulates pDC homeostasis and function in health and disease. Proc Natl Acad Sci (2019) 116:16489–96. doi: 10.1073/pnas.1900079116
74. Dollt C, Michel J, Kloss L, Melchers S, Schledzewski K, Becker K, et al. The novel immunoglobulin super family receptor SLAMF9 identified in TAM of murine and human melanoma influences pro-inflammatory cytokine secretion and migration. Cell Death Dis (2018) 9:939. doi: 10.1038/s41419-018-1011-1
75. Wilson TJ, Clare S, Mikulin J, Johnson CM, Harcourt K, Lyons PA, et al. Signalling lymphocyte activation molecule family member 9 is found on select subsets of antigen-presenting cells and promotes resistance to salmonella infection. Immunology (2020) 159:393–403. doi: 10.1111/imm.13169
76. Ishibashi M, Morita R, Tamura H. Immune functions of signaling lymphocytic activation molecule family molecules in multiple myeloma. Cancers (Basel) (2021) 13. doi: 10.3390/cancers13020279
77. Detre C, Keszei M, Romero X, Tsokos GC, Terhorst C. SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin Immunopathol (2010) 32:157–71. doi: 10.1007/s00281-009-0193-0
78. Radhakrishnan SV, Bhardwaj N, Luetkens T, Atanackovic D. Novel anti-myeloma immunotherapies targeting the SLAM family of receptors. Oncoimmunology (2017) 6:e1308618. doi: 10.1080/2162402X.2017.1308618
79. Schoenhals M, Frecha C, Bruyer A, Caraux A, Veyrune JL, Jourdan M, et al. Efficient transduction of healthy and malignant plasma cells by lentiviral vectors pseudotyped with measles virus glycoproteins. Leukemia (2012) 26:1663–70. doi: 10.1038/leu.2012.36
80. Gordiienko IM, Shlapatska LM, Kovalevska LM, Sidorenko SP. Differential expression of CD150/SLAMF1 in normal and malignant b cells on the different stages of maturation. Exp Oncol (2016) 38:101–7. doi: 10.31768/2312-8852.2016.38(2):101-107
81. Karampetsou MP, Comte D, Suárez-Fueyo A, Katsuyama E, Yoshida N, Kono M, et al. Signaling lymphocytic activation molecule family member 1 engagement inhibits T cell-b cell interaction and diminishes interleukin-6 production and plasmablast differentiation in systemic lupus erythematosus. Arthritis Rheumatol (Hoboken NJ) (2019) 71:99–108. doi: 10.1002/art.40682
82. Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, et al. SAP couples fyn to SLAM immune receptors. Nat Cell Biol (2003) 5:155–60. doi: 10.1038/ncb920
83. Yurchenko M, Skjesol A, Ryan L, Richard GM, Kandasamy RK, Wang N, et al. SLAMF1 is required for TLR4-mediated TRAM-TRIF-dependent signaling in human macrophages. J Cell Biol (2018) 217:1411–29. doi: 10.1083/jcb.201707027
84. Gordiienko I, Shlapatska L, Kovalevska L, Sidorenko SP. SLAMF1/CD150 in hematologic malignancies: silent marker or active player? Clin Immunol (2019) 204:14–22. doi: 10.1016/j.clim.2018.10.015
85. Yurchenko MY, Kovalevska LM, Shlapatska LM, Berdova GG, Clark EA, Sidorenko SP. CD150 regulates JNK1/2 activation in normal and hodgkin’s lymphoma b cells. Immunol Cell Biol (2010) 88:565–74. doi: 10.1038/icb.2010.14
86. Mikhalap SV, Shlapatska LM, Yurchenko OV, Yurchenko MY, Berdova GG, Nichols KE, et al. The adaptor protein SH2D1A regulates signaling through CD150 (SLAM) in b cells. Blood (2004) 104:4063–70. doi: 10.1182/blood-2004-04-1273
87. Gordiienko I, Shlapatska L, Kholodniuk V, Sklyarenko L, Gluzman DF, Clark EA, et al. The interplay of CD150 and CD180 receptor pathways contribute to the pathobiology of chronic lymphocytic leukemia b cells by selective inhibition of akt and MAPK signaling. PloS One (2017) 12:e0185940. doi: 10.1371/journal.pone.0185940
88. Mikhalap SV, Shlapatska LM, Berdova AG, Law C-L, Clark EA, Sidorenko SP. CDw150 associates with src-homology 2-containing inositol phosphatase and modulates CD95-mediated Apoptosis1. J Immunol (1999) 162:5719–27. doi: 10.4049/jimmunol.162.10.5719
89. Tufa DM, Yingst AM, Trahan GD, Shank T, Jones D, Shim S, et al. Human innate lymphoid cell precursors express CD48 that modulates ILC differentiation through 2B4 signaling. Sci Immunol (2020) 5. doi: 10.1126/sciimmunol.aay4218
90. Sun L, Gang X, Li Z, Zhao X, Zhou T, Zhang S, et al. Advances in understanding the roles of CD244 (SLAMF4) in immune regulation and associated diseases. Front Immunol (2021) 12:648182. doi: 10.3389/fimmu.2021.648182
91. Liao G, Song J-H, O’Keeffe M, Calpe S, Castro W, Magelky E, et al. Regulation of expression of SLAMF4 on the surface of mucosal lymphocytes (120.13). J Immunol (2012) 188:120.13.
92. O’Keeffe MS, Song J-H, Liao G, De Calisto J, Halibozek PJ, Mora JR, et al. SLAMF4 is a negative regulator of expansion of cytotoxic intraepithelial CD8+ T cells that maintains homeostasis in the small intestine. Gastroenterology (2015) 148:991–1001.e4. doi: 10.1053/j.gastro.2015.02.003
93. Kis-Toth K, Tsokos GC. Engagement of SLAMF2/CD48 prolongs the time frame of effective T cell activation by supporting mature dendritic cell survival. J Immunol (2014) 192:4436–42. doi: 10.4049/jimmunol.1302909
94. Chen S, Dong Z. NK cell recognition of hematopoietic cells by SLAM-SAP families. Cell Mol Immunol (2019) 16:452–9. doi: 10.1038/s41423-019-0222-4
95. Meinke S, Watzl C. NK cell cytotoxicity mediated by 2B4 and NTB-a is dependent on SAP acting downstream of receptor phosphorylation. Front Immunol (2013) 4:3. doi: 10.3389/fimmu.2013.00003
96. Morandi B, Costa R, Falco M, Parolini S, De Maria A, Ratto G, et al. Distinctive lack of CD48 expression in subsets of human dendritic cells tunes NK cell activation. J Immunol (2005) 175:3690–7. doi: 10.4049/jimmunol.175.6.3690
97. Agresta L, Hoebe KHN, Janssen EM. The emerging role of CD244 signaling in immune cells of the tumor microenvironment. Front Immunol (2018) 9:2809. doi: 10.3389/fimmu.2018.02809
98. Zhou T, Wang G, Lyu Y, Wang L, Zuo S, Zou J, et al. Upregulation of SLAMF3 on human T cells is induced by palmitic acid through the STAT5-PI3K/Akt pathway and features the chronic inflammatory profiles of type 2 diabetes. Cell Death Dis (2019) 10:559. doi: 10.1038/s41419-019-1791-y
99. Wang N, Yigit B, van der Poel CE, Cuenca M, Carroll MC, Herzog RW, et al. The checkpoint regulator SLAMF3 preferentially prevents expansion of auto-reactive b cells generated by graft-vs.-Host disease. Front Immunol (2019) 10:831. doi: 10.3389/fimmu.2019.00831
100. Cannons JL, Yu LJ, Hill B, Mijares LA, Dombroski D, Nichols KE, et al. SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappaB1. Immunity (2004) 21:693–706. doi: 10.1016/j.immuni.2004.09.012
101. Ishibashi M, Takahashi R, Tsubota A, Sasaki M, Handa H, Imai Y, et al. SLAMF3-mediated signaling via ERK pathway activation promotes aggressive phenotypic behaviors in multiple myeloma. Mol Cancer Res (2020) 18:632–43. doi: 10.1158/1541-7786.MCR-19-0391
102. Zaiss M, Hirtreiter C, Rehli M, Rehm A, Kunz-Schughart LA, Andreesen R, et al. CD84 expression on human hematopoietic progenitor cells. Exp Hematol (2003) 31:798–805. doi: 10.1016/S0301-472X(03)00187-5
103. Sintes J, Romero X, Marin P, Terhorst C, Engel P. Differential expression of CD150 (SLAM) family receptors by human hematopoietic stem and progenitor cells. Exp Hematol (2008) 36:1199–204. doi: 10.1016/j.exphem.2008.03.015
104. Radomir L, Kramer MP, Perpinial M, Schottlender N, Rabani S, David K, et al. The survival and function of IL-10-producing regulatory b cells are negatively controlled by SLAMF5. Nat Commun (2021) 12:1893. doi: 10.1038/s41467-021-22230-z
105. Agod Z, Pazmandi K, Bencze D, Vereb G, Biro T, Szabo A, et al. Signaling lymphocyte activation molecule family 5 enhances autophagy and fine-tunes cytokine response in monocyte-derived dendritic cells via stabilization of interferon regulatory factor 8. Front Immunol (2018) 9:62. doi: 10.3389/fimmu.2018.00062
106. De Calisto J, Wang N, Wang G, Yigit B, Engel P, Terhorst C. SAP-dependent and -independent regulation of innate T cell development involving SLAMF receptors. Front Immunol (2014) 5:186. doi: 10.3389/fimmu.2014.00186
107. Wang N, Halibozek PJ, Yigit B, Zhao H, O’Keeffe MS, Sage P, et al. Negative regulation of humoral immunity due to interplay between the SLAMF1, SLAMF5, and SLAMF6 receptors. Front Immunol (2015) 6:158. doi: 10.3389/fimmu.2015.00158
108. Chen G, Tai AK, Lin M, Chang F, Terhorst C, Huber BT. Increased proliferation of CD8+ T cells in SAP-deficient mice is associated with impaired activation-induced cell death. Eur J Immunol (2007) 37:663–74. doi: 10.1002/eji.200636417
109. Marom A, Barak AF, Kramer MP, Lewinsky H, Binsky-Ehrenreich I, Cohen S, et al. CD84 mediates CLL-microenvironment interactions. Oncogene (2017) 36:628–38. doi: 10.1038/onc.2016.238
110. Cao E, Ramagopal UA, Fedorov A, Fedorov E, Yan Q, Lary JW, et al. NTB-a receptor crystal structure: insights into homophilic interactions in the signaling lymphocytic activation molecule receptor family. Immunity (2006) 25:559–70. doi: 10.1016/j.immuni.2006.06.020
111. Yigit B, Wang N, ten Hacken E, Chen S-S, Bhan AK, Suarez-Fueyo A, et al. SLAMF6 as a regulator of exhausted CD8+ T cells in cancer. Cancer Immunol Res (2019) 7:1485–96. doi: 10.1158/2326-6066.CIR-18-0664
112. Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, et al. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to b cells and NKT cell development. Immunity (2012) 36:986–1002. doi: 10.1016/j.immuni.2012.05.016
113. Dragovich MA, Adam K, Strazza M, Tocheva AS, Peled M, Mor A. SLAMF6 clustering is required to augment T cell activation. PloS One (2019) 14:e0218109. doi: 10.1371/journal.pone.0218109
114. Hajaj E, Eisenberg G, Klein S, Frankenburg S, Merims S, Ben David I, et al. SLAMF6 deficiency augments tumor killing and skews toward an effector phenotype revealing it as a novel T cell checkpoint. Elife (2020) 9. doi: 10.7554/eLife.52539
115. Campbell KS, Cohen AD, Pazina T. Mechanisms of NK cell activation and clinical activity of the therapeutic SLAMF7 antibody, elotuzumab in multiple myeloma. Front Immunol (2018) 9:2551. doi: 10.3389/fimmu.2018.02551
116. Wilson TJ, Garner LI, Metcalfe C, King E, Margraf S, Brown MH. Fine specificity and molecular competition in SLAM family receptor signalling. PloS One (2014) 9:e92184. doi: 10.1371/journal.pone.0092184
117. Pérez-Quintero L-A, Roncagalli R, Guo H, Latour S, Davidson D, Veillette A. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase cγ, ca++, and erk, leading to granule polarization. J Exp Med (2014) 211:727–42. doi: 10.1084/jem.20132038
118. Gordiienko I, Shlapatska L, Kholodniuk VM, Kovalevska L, Ivanivskaya TS, Sidorenko SP. CD150 and CD180 are involved in regulation of transcription factors expression in chronic lymphocytic leukemia cells. Exp Oncol (2017) 39:291–8. doi: 10.31768/2312-8852.2017.39(4):291-298
119. Yurchenko M, Shlapatska LM, Romanets OL, Ganshevskiy D, Kashuba E, Zamoshnikova A, et al. CD150-mediated akt signalling pathway in normal and malignant b cells. Exp Oncol (2011) 33:9–18.
120. Yurchenko MY, Kashuba EV, Shlapatska LM, Sivkovich SA, Sidorenko SP. The role of CD150-SH2D1A association in CD150 signaling in hodgkin’s lymphoma cell lines. Exp Oncol (2005) 27:24–30.
121. Yoon H, Kim EK, Ko YH. SLAMF1 contributes to cell survival through the AKT signaling pathway in farage cells. PloS One (2020) 15:e0238791. doi: 10.1371/journal.pone.0238791
122. Yoon H, Ko YH. LMP1+SLAMF1high cells are associated with drug resistance in Epstein-Barr virus-positive farage cells. Oncotarget (2017) 8:24621–34. doi: 10.18632/oncotarget.15600
123. Shcherbina V, Gordiienko I, Shlapatska L, Ivanivska T, Sidorenko S. Sensitivity of chronic lymphocytic leukemia cells to chemotherapeutic drugs ex vivo depends on expression status of cell surface receptors. Exp Oncol (2020) 42:16–24. doi: 10.32471/exp-oncology.2312-8852.vol-42-no-1.14093
124. von Wenserski L, Schultheiß C, Bolz S, Schliffke S, Simnica D, Willscher E, et al. SLAMF receptors negatively regulate b cell receptor signaling in chronic lymphocytic leukemia via recruitment of prohibitin-2. Leukemia (2021) 35:1073–86. doi: 10.1038/s41375-020-01025-z
125. Rigolin GM, Saccenti E, Melandri A, Cavallari M, Urso A, Rotondo F, et al. In chronic lymphocytic leukaemia, SLAMF1 deregulation is associated with genomic complexity and independently predicts a worse outcome. Br J Haematol (2021) 192:1068–72. doi: 10.1111/bjh.16865
126. Takeda S, Kanbayashi D, Kurata T, Yoshiyama H, Komano J. Enhanced susceptibility of b lymphoma cells to measles virus by Epstein–Barr virus type III latency that upregulates CD150/signaling lymphocytic activation molecule. Cancer Sci (2014) 105:211–8. doi: 10.1111/cas.12324
127. Shi D, Zhang Y, Tian Y. SLAMF1 promotes methotrexate resistance via activating autophagy in choriocarcinoma cells. Cancer Manag Res (2020) 12:13427–36. doi: 10.2147/CMAR.S278012
128. Romanets-Korbut O, Najakshin AM, Yurchenko M, Malysheva TA, Kovalevska L, Shlapatska LM, et al. Expression of CD150 in tumors of the central nervous system: identification of a novel isoform. PloS One (2015) 10:e0118302. doi: 10.1371/journal.pone.0118302
129. Chen Q, Qiu B, Zeng X, Hu L, Huang D, Chen K, et al. Identification of a tumor microenvironment-related gene signature to improve the prediction of cervical cancer prognosis. Cancer Cell Int (2021) 21:182. doi: 10.1186/s12935-021-01867-2
130. Gordiienko IM, Lykhova OO, Shcherbina VM, Shlapatska LM. SLAMF1/CD150 expression and topology in prostate and breast cancer cell lines. Exp Oncol (2021) 43:312–6.
131. Wang Z, Guan W, Wang M, Chen J, Zhang L, Xiao Y, et al. AML1-ETO inhibits acute myeloid leukemia immune escape by CD48. Leuk Lymphoma (2021) 62:937–43. doi: 10.1080/10428194.2020.1849680
132. Wang Z, Xiao Y, Guan W, Wang M, Chen J, Zhang L, et al. Acute myeloid leukemia immune escape by epigenetic CD48 silencing. Clin Sci (2020) 134:261–71. doi: 10.1042/CS20191170
133. Huang C-H, Liao Y-J, Chiou T-J, Huang H-T, Lin Y-H, Twu Y-C. TGF-β regulated leukemia cell susceptibility against NK targeting through the down-regulation of the CD48 expression. Immunobiology (2019) 224:649–58. doi: 10.1016/j.imbio.2019.07.002
134. Ziblat A, Iraolagoitia XLR, Nuñez SY, Torres NI, Secchiari F, Sierra JM, et al. Circulating and tumor-infiltrating NK cells from clear cell renal cell carcinoma patients exhibit a predominantly inhibitory phenotype characterized by overexpression of CD85j, CD45, CD48 and PD-1. Front Immunol (2021) 12:681615. doi: 10.3389/fimmu.2021.681615
135. Wang Z, He L, Li W, Xu C, Zhang J, Wang D, et al. GDF15 induces immunosuppression via CD48 on regulatory T cells in hepatocellular carcinoma. J Immunother Cancer (2021) 9:e002787. doi: 10.1136/jitc-2021-002787
136. Ma R, Qu X, Che X, Yang B, Li C, Hou K, et al. Comparative analysis and in vitro experiments of signatures and prognostic value of immune checkpoint genes in colorectal cancer. Onco Targets Ther (2021) 14:3517–34. doi: 10.2147/OTT.S304297
137. Atanackovic D, Panse J, Hildebrandt Y, Jadczak A, Kobold S, Cao Y, et al. Surface molecule CD229 as a novel target for the diagnosis and treatment of multiple myeloma. Haematologica (2011) 96:1512–20. doi: 10.3324/haematol.2010.036814
138. Ghogale S, Tauro W, Priyadarshini A, Subramanian PG, Patkar N, Yajamanam B, et al. SLAM family member “CD229”: a novel gating marker for plasma cells in flow cytometric immunophenotyping (FCI) of multiple myeloma (MM). Clin Lymphoma Myeloma Leuk (2017) 17:e19. doi: 10.1016/j.clml.2017.03.032
139. Yousef S, Kovacsovics-Bankowski M, Salama ME, Bhardwaj N, Steinbach M, Langemo A, et al. CD229 is expressed on the surface of plasma cells carrying an aberrant phenotype and chemotherapy-resistant precursor cells in multiple myeloma. Hum Vaccin Immunother (2015) 11:1606–11. doi: 10.1080/21645515.2015.1046658
140. Fouquet G, Debuysscher V, Ouled-Haddou H, Eugenio MS, Demey B, Singh AR, et al. Hepatocyte SLAMF3 reduced specifically the multidrugs resistance protein MRP-1 and increases HCC cells sensitization to anti-cancer drugs. Oncotarget (2016) 7:32493–503. doi: 10.18632/oncotarget.8679
141. Fouquet G, Marié C, Badaoui M, Demey B, Radoslavova S, Telliez M-S, et al. Mammary SLAMF3 regulates store-operated Ca2+ entry and migration through STIM1 in breast cancer cells and cell lines. J Cancer Sci Clin Ther (2020) 04. doi: 10.26502/jcsct.5079087
142. Radhakrishnan SV, Luetkens T, Scherer SD, Davis P, Vander Mause ER, Olson ML, et al. CD229 CAR T cells eliminate multiple myeloma and tumor propagating cells without fratricide. Nat Commun (2020) 11:798. doi: 10.1038/s41467-020-14619-z
143. Lee K-M, Forman JP, McNerney ME, Stepp S, Kuppireddi S, Guzior D, et al. Requirement of homotypic NK-cell interactions through 2B4(CD244)/CD48 in the generation of NK effector functions. Blood (2006) 107:3181–8. doi: 10.1182/blood-2005-01-0185
144. Kim E-O, Kim T-J, Kim N, Kim ST, Kumar V, Lee K-M. Homotypic cell to cell cross-talk among human natural killer cells reveals differential and overlapping roles of 2B4 and CD2. J Biol Chem (2010) 285:41755–64. doi: 10.1074/jbc.M110.137976
145. Mooney JM, Klem J, Wülfing C, Mijares LA, Schwartzberg PL, Bennett M, et al. The murine NK receptor 2B4 (CD244) exhibits inhibitory function independent of signaling lymphocytic activation molecule-associated protein expression. J Immunol (2004) 173:3953–61. doi: 10.4049/jimmunol.173.6.3953
146. Wu Y, Kuang D-M, Pan W-D, Wan Y-L, Lao X-M, Wang D, et al. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology (2013) 57:1107–16. doi: 10.1002/hep.26192
147. Baitsch L, Baumgaertner P, Devêvre E, Raghav SK, Legat A, Barba L, et al. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. J Clin Invest (2011) 121:2350–60. doi: 10.1172/JCI46102
148. Zelle-Rieser C, Thangavadivel S, Biedermann R, Brunner A, Stoitzner P, Willenbacher E, et al. T Cells in multiple myeloma display features of exhaustion and senescence at the tumor site. J Hematol Oncol (2016) 9:116. doi: 10.1186/s13045-016-0345-3
149. Youn J-I, Collazo M, Shalova IN, Biswas SK, Gabrilovich DI. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J Leukoc Biol (2012) 91:167–81. doi: 10.1189/jlb.0311177
150. Lewinsky H, Gunes EG, David K, Radomir L, Kramer MP, Pellegrino B, et al. CD84 is a regulator of the immunosuppressive microenvironment in multiple myeloma. JCI Insight (2021) 6. doi: 10.1172/jci.insight.141683
151. Barak AF, Lewinsky H, Perpinial M, Huber V, Radomir L, Kramer MP, et al. Bone marrow dendritic cells support the survival of chronic lymphocytic leukemia cells in a CD84 dependent manner. Oncogene (2020) 39:1997–2008. doi: 10.1038/s41388-019-1121-y
152. Lewinsky H, Barak AF, Huber V, Kramer MP, Radomir L, Sever L, et al. CD84 regulates PD-1/PD-L1 expression and function in chronic lymphocytic leukemia. J Clin Invest (2018) 128:5465–78. doi: 10.1172/JCI96610
153. Binsky-Ehrenreich I, Marom A, Sobotta MC, Shvidel L, Berrebi A, Hazan-Halevy I, et al. CD84 is a survival receptor for CLL cells. Oncogene (2014) 33:1006–16. doi: 10.1038/onc.2013.31
154. Meng Q, Duan X, Yang Q, Xue D, Liu Z, Li Y, et al. SLAMF6/Ly108 promotes the development of hepatocellular carcinoma via facilitating macrophage M2 polarization. Oncol Lett (2022) 23:83. doi: 10.3892/ol.2022.13203
155. Hsi ED, Steinle R, Balasa B, Szmania S, Draksharapu A, Shum BP, et al. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin Cancer Res (2008), 2775–84. doi: 10.1158/1078-0432.CCR-07-4246
156. van Rhee F, Szmania SM, Dillon M, van Abbema AM, Li X, Stone MK, et al. Combinatorial efficacy of anti-CS1 monoclonal antibody elotuzumab (HuLuc63) and bortezomib against multiple myeloma. Mol Cancer Ther (2009) 8:2616–24. doi: 10.1158/1535-7163.MCT-09-0483
157. Tai Y-T, Dillon M, Song W, Leiba M, Li X-F, Burger P, et al. Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces antibody-dependent cellular cytotoxicity in the bone marrow milieu. Blood (2008) 112:1329–37. doi: 10.1182/blood-2007-08-107292
158. Collins SM, Bakan CE, Swartzel GD, Hofmeister CC, Efebera YA, Kwon H, et al. Elotuzumab directly enhances NK cell cytotoxicity against myeloma via CS1 ligation: evidence for augmented NK cell function complementing ADCC. Cancer Immunol Immunother (2013) 62:1841–9. doi: 10.1007/s00262-013-1493-8
159. Guo H, Cruz-Munoz M-E, Wu N, Robbins M, Veillette A. Immune cell inhibition by SLAMF7 is mediated by a mechanism requiring src kinases, CD45, and SHIP-1 that is defective in multiple myeloma cells. Mol Cell Biol (2015) 35:41–51. doi: 10.1128/MCB.01107-14
160. Pazina T, James AM, Colby KB, Yang Y, Gale A, Jhatakia A, et al. Enhanced SLAMF7 homotypic interactions by elotuzumab improves NK cell killing of multiple myeloma. Cancer Immunol Res (2019) 7:1633–46. doi: 10.1158/2326-6066.CIR-18-0579
161. Zonder JA, Mohrbacher AF, Singhal S, van Rhee F, Bensinger WI, Ding H, et al. A phase 1, multicenter, open-label, dose escalation study of elotuzumab in patients with advanced multiple myeloma. Blood (2012) 120:552–9. doi: 10.1182/blood-2011-06-360552
162. Jakubowiak AJ, Benson DM, Bensinger W, Siegel DSD, Zimmerman TM, Mohrbacher A, et al. Phase I trial of anti-CS1 monoclonal antibody elotuzumab in combination with bortezomib in the treatment of relapsed/refractory multiple myeloma. J Clin Oncol (2012) 30:1960–5. doi: 10.1200/JCO.2011.37.7069
163. Jakubowiak A, Offidani M, Pégourie B, de la Rubia J, Garderet L, Laribi K, et al. Randomized phase 2 study: elotuzumab plus bortezomib/dexamethasone vs bortezomib/dexamethasone for relapsed/refractory MM. Blood (2016) 127:2833–40. doi: 10.1182/blood-2016-01-694604
164. Lonial S, Vij R, Harousseau J-L, Facon T, Moreau P, Mazumder A, et al. Elotuzumab in combination with lenalidomide and low-dose dexamethasone in relapsed or refractory multiple myeloma. J Clin Oncol (2012) 30:1953–9. doi: 10.1200/JCO.2011.37.2649
165. Miest TS, Frenzke M, Cattaneo R. Measles virus entry through the signaling lymphocyte activation molecule governs efficacy of mantle cell lymphoma radiovirotherapy. Mol Ther (2013) 21:2019–31. doi: 10.1038/mt.2013.171
166. Sun H, Norris BJ, Atkinson K, Biggs JC, Smith GM. Preclinical antitumor activity of an antibody against the leukocyte antigen CD48. Clin Cancer Res (1998) 4:895–900.
167. Greenaway S, Henniker AJ, Walsh M, Bradstock KF. A pilot clinical trial of two murine monoclonal antibodies fixing human complement in patients with chronic lymphatic leukaemia. Leuk Lymphoma (1994) 13:323–31. doi: 10.3109/10428199409056297
168. Lewis TS, Olson D, Gordon K, Sandall S, Quick M, Finn M, et al. SGN-CD48A: a novel humanized anti-CD48 antibody-drug conjugate for the treatment of multiple myeloma. Blood (2016) 128:4470. doi: 10.1182/blood.V128.22.4470.4470
169. Altvater B, Landmeier S, Pscherer S, Temme J, Schweer K, Kailayangiri S, et al. 2B4 (CD244) signaling by recombinant antigen-specific chimeric receptors costimulates natural killer cell activation to leukemia and neuroblastoma cells. Clin Cancer Res (2009) 15:4857–66. doi: 10.1158/1078-0432.CCR-08-2810
170. Korver W, Singh S, Liu S, Zhao X, Yonkovich S, Sweeney A, et al. The lymphoid cell surface receptor NTB-a: a novel monoclonal antibody target for leukaemia and lymphoma therapeutics. Br J Haematol (2007) 137:307–18. doi: 10.1111/j.1365-2141.2007.06569.x
171. Yigit B, Halibozek PJ, Chen S-S, O’Keeffe MS, Arnason J, Avigan D, et al. A combination of an anti-SLAMF6 antibody and ibrutinib efficiently abrogates expansion of chronic lymphocytic leukemia cells. Oncotarget (2016). doi: 10.18632/oncotarget.8378
172. Bae J, Song W, Smith R, Daley J, Tai Y-T, Anderson KC, et al. A novel immunogenic CS1-specific peptide inducing antigen-specific cytotoxic T lymphocytes targeting multiple myeloma. Br J Haematol (2012) 157:687–701. doi: 10.1111/j.1365-2141.2012.09111.x
173. Wang X, Walter M, Urak R, Weng L, Huynh C, Lim L, et al. Lenalidomide enhances the function of CS1 chimeric antigen receptor–redirected T cells against multiple myeloma. Clin Cancer Res (2018) 24:106–19. doi: 10.1158/1078-0432.CCR-17-0344
174. Dimopoulos MA, Lonial S, White D, Moreau P, Weisel K, San-Miguel J, et al. Elotuzumab, lenalidomide, and dexamethasone in RRMM: final overall survival results from the phase 3 randomized ELOQUENT-2 study. Blood Cancer J (2020) 10:91. doi: 10.1038/s41408-020-00357-4
175. Lonial S, Dimopoulos M, Palumbo A, White D, Grosicki S, Spicka I, et al. Elotuzumab therapy for relapsed or refractory multiple myeloma. N Engl J Med (2015) 373:621–31. doi: 10.1056/NEJMoa1505654
176. Mateos M-V, Granell M, Oriol A, Martinez-Lopez J, Blade J, Hernandez MT, et al. Elotuzumab in combination with thalidomide and low-dose dexamethasone: a phase 2 single-arm safety study in patients with relapsed/refractory multiple myeloma. Br J Haematol (2016) 175:448–56. doi: 10.1111/bjh.14263
177. Berdeja J, Jagannath S, Zonder J, Badros A, Kaufman JL, Manges R, et al. Pharmacokinetics and safety of elotuzumab combined with lenalidomide and dexamethasone in patients with multiple myeloma and various levels of renal impairment: results of a phase ib study. Clin Lymphoma Myeloma Leuk (2016) 16:129–38. doi: 10.1016/j.clml.2015.12.007
178. Richardson PG, Jagannath S, Moreau P, Jakubowiak AJ, Raab MS, Facon T, et al. Elotuzumab in combination with lenalidomide and dexamethasone in patients with relapsed multiple myeloma: final phase 2 results from the randomised, open-label, phase 1b-2 dose-escalation study. Lancet Haematol (2015) 2:e516–27. doi: 10.1016/S2352-3026(15)00197-0
179. O’Connell P, Hyslop S, Blake MK, Godbehere S, Amalfitano A, Aldhamen YA. SLAMF7 signaling reprograms T cells toward exhaustion in the tumor microenvironment. J Immunol (2021) 206:193–205. doi: 10.4049/jimmunol.2000300
180. O’Neal J, Ritchey JK, Cooper ML, Niswonger J, Sofía González L, Street E, et al. CS1 CAR-T targeting the distal domain of CS1 (SLAMF7) shows efficacy in high tumor burden myeloma model despite fratricide of CD8+CS1 expressing CAR-T cells. Leukemia (2022). doi: 10.1038/s41375-022-01559-4
181. Chu J, He S, Deng Y, Zhang J, Peng Y, Hughes T, et al. Genetic modification of T cells redirected toward CS1 enhances eradication of myeloma cells. Clin Cancer Res (2014) 20:3989–4000. doi: 10.1158/1078-0432.CCR-13-2510
182. Gogishvili T, Danhof S, Prommersberger S, Rydzek J, Schreder M, Brede C, et al. SLAMF7-CAR T cells eliminate myeloma and confer selective fratricide of SLAMF7(+) normal lymphocytes. Blood (2017) 130:2838–47. doi: 10.1182/blood-2017-04-778423
183. Danhof S, Rasche L, Mottok A, Steinmüller T, Zhou X, Schreder M, et al. Elotuzumab for the treatment of extramedullary myeloma: a retrospective analysis of clinical efficacy and SLAMF7 expression patterns. Ann Hematol (2021) 100:1537–46. doi: 10.1007/s00277-021-04447-6
184. Amatya C, Pegues MA, Lam N, Vanasse D, Geldres C, Choi S, et al. Development of CAR T cells expressing a suicide gene plus a chimeric antigen receptor targeting signaling lymphocytic-activation molecule F7. Mol Ther (2021) 29:702–17. doi: 10.1016/j.ymthe.2020.10.008
185. Sugimoto A, Kataoka TR, Ito H, Kitamura K, Saito N, Hirata M, et al. SLAM family member 8 is expressed in and enhances the growth of anaplastic large cell lymphoma. Sci Rep (2020) 10:2505. doi: 10.1038/s41598-020-59530-1
186. Sugimoto A, Kataoka TR, Ueshima C, Takei Y, Kitamura K, Hirata M, et al. SLAM family member 8 is involved in oncogenic KIT-mediated signalling in human mastocytosis. Exp Dermatol (2018) 27:641–6. doi: 10.1111/exd.13523
187. Zou C-Y, Guan G-F, Zhu C, Liu T-Q, Guo Q, Cheng W, et al. Costimulatory checkpoint SLAMF8 is an independent prognosis factor in glioma. CNS Neurosci Ther (2019) 25:333–42. doi: 10.1111/cns.13041
188. Zhang Q, Cheng L, Qin Y, Kong L, Shi X, Hu J, et al. SLAMF8 expression predicts the efficacy of anti-PD1 immunotherapy in gastrointestinal cancers. Clin Transl Immunol (2021) 10:e1347. doi: 10.1002/cti2.1347
189. Zhang W, Wan T, Li N, Yuan Z, He L, Zhu X, et al. Genetic approach to insight into the immunobiology of human dendritic cells and identification of CD84-H1, a novel CD84 homologue. Clin Cancer Res (2001) 7.
190. Gormley NJ, Ko C-W, Deisseroth A, Nie L, Kaminskas E, Kormanik N, et al. FDA Drug approval: elotuzumab in combination with lenalidomide and dexamethasone for the treatment of relapsed or refractory multiple myeloma. Clin Cancer Res (2017) 23:6759–63. doi: 10.1158/1078-0432.CCR-16-2870
191. Brioli A, Mügge L-O, Hochhaus A, Von Lilienfeld-Toal M. Safety issues and management of toxicities associated with new treatments for multiple myeloma. Expert Rev Hematol (2017) 10:193–205. doi: 10.1080/17474086.2017.1284584
192. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature (2011) 469:323–35. doi: 10.1038/nature09782
193. Sandigursky S, Philips MR, Mor A. SAP interacts with CD28 to inhibit PD-1 signaling in T lymphocytes. Clin Immunol (2020) 217:108485. doi: 10.1016/j.clim.2020.108485
194. Peled M, Tocheva AS, Sandigursky S, Nayak S, Philips EA, Nichols KE, et al. Affinity purification mass spectrometry analysis of PD-1 uncovers SAP as a new checkpoint inhibitor. Proc Natl Acad Sci USA (2018) 115:E468–77. doi: 10.1073/pnas.1710437115
195. Gartshteyn Y, Askanase AD, Mor A. SLAM associated protein signaling in T cells: tilting the balance toward autoimmunity. Front Immunol (2021) 12:654839. doi: 10.3389/fimmu.2021.654839
196. Dong Z, Davidson D, Pérez-Quintero LA, Kurosaki T, Swat W, Veillette A. The adaptor SAP controls NK cell activation by regulating the enzymes vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity (2012) 36:974–85. doi: 10.1016/j.immuni.2012.03.023
197. Chatterjee M, Hedrich CM, Rauen T, Ioannidis C, Terhorst C, Tsokos GC. CD3-T cell receptor co-stimulation through SLAMF3 and SLAMF6 receptors enhances RORγt recruitment to the IL17A promoter in human T lymphocytes. J Biol Chem (2012) 287:38168–77. doi: 10.1074/jbc.M112.415067
198. Chatterjee M, Rauen T, Kis-Toth K, Kyttaris VC, Hedrich CM, Terhorst C, et al. Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J Immunol (2012) 188:1206–12. doi: 10.4049/jimmunol.1102773
199. Assidi M. Strong prognostic value of SLAMF7 protein expression in patients with lymph node-positive breast cancer. Oncol Lett (2022) 24:433. doi: 10.3892/ol.2022.13553
200. Wang Y, Sanchez L, Siegel DS, Wang ML. Elotuzumab for the treatment of multiple myeloma. J Hematol Oncol (2016) 9:55. doi: 10.1186/s13045-016-0284-z
Keywords: SLAM, cancer immunotherapy, elotuzumab, CD244 (2B4), CD150, CD84, CD229, SLAMF7/CS1
Citation: Farhangnia P, Ghomi SM, Mollazadehghomi S, Nickho H, Akbarpour M and Delbandi A-A (2023) SLAM-family receptors come of age as a potential molecular target in cancer immunotherapy. Front. Immunol. 14:1174138. doi: 10.3389/fimmu.2023.1174138
Received: 25 February 2023; Accepted: 02 May 2023;
Published: 11 May 2023.
Edited by:
Cara Haymaker, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Srividya Swaminathan, City of Hope National Medical Center, United StatesCopyright © 2023 Farhangnia, Ghomi, Mollazadehghomi, Nickho, Akbarpour and Delbandi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali-Akbar Delbandi, RGVsYmFuZGkuYWtASXVtcy5hYy5pcg==; Mahzad Akbarpour, TWFrYmFycG91ckBtZWRpY2luZS5ic2QudWNoaWNhZ28uZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.