- 1National Clinical Research Center for Infectious Disease, Shenzhen Third People’s Hospital, Shenzhen, Guangdong, China
- 2Center for Cellular and Molecular Diagnostics, Tulane University School of Medicine, New Orleans, LA, United States
- 3Department of Biochemistry and Molecular Biology, Tulane University School of Medicine, New Orleans, LA, United States
Tuberculosis (TB) remains a major underdiagnosed public health threat worldwide, being responsible for more than 10 million cases and one million deaths annually. TB diagnosis has become more rapid with the development and adoption of molecular tests, but remains challenging with traditional TB diagnosis, but there has not been a critical review of this area. Here, we systematically review these approaches to assess their diagnostic potential and issues with the development and clinical evaluation of proposed CRISPR-based TB assays. Based on these observations, we propose constructive suggestions to improve sample pretreatment, method development, clinical validation, and accessibility of these assays to streamline future assay development and validation studies.
1 Introduction
It is estimated that one-quarter of the world population is infected with Mycobacterium tuberculosis (Mtb), and about 10 millions of these individuals develop tuberculosis (TB) annually, with more than 1 million TB-related deaths per year (1). Further, the global burden of TB and drug-resistant TB (DR-TB) has increased by 4.5% and 3% over the past year (1), deviating from the anticipated reduction rates required to meet the current schedule of the “End TB” strategy (2).
Early and accurate diagnosis of TB is critical for TB eradication efforts (3), but TB diagnosis remains challenging, and >35% of the estimated global TB cases are undiagnosed by current efforts (1, 4). This includes cases missed by insensitive sputum microbiology assays and immunoassays (5, 6), individuals who have difficulty producing diagnostic sputum samples (children, people living with HIV, extrapulmonary TB cases, or certain neurological impairments, including Dementia and Parkinson’s disease) (7–11), people living in remote high TB burden areas (4), and individuals infected with DR strain who have not undergone DR screening (1). Invasive (e.g., bronchoalveolar lavage or gastric aspirate) (12) and non-invasive (e.g., stool) (13) samples can be used as additional complementary specimens to improve pulmonary TB diagnosis in Patients that have difficulty producing expectorated sputum, while additional invasive biopsies are often required to diagnose extrapulmonary TB. However, these samples versus sputum may exhibit reduced diagnostic sensitivity and be more variable and difficult to obtain.
Research is ongoing to develop new TB biomarkers and detection technologies to enhance TB diagnosis. The application of new tools has accelerated the discovery of TB biomarkers, revealing many pathogen-derived biomarkers such as nucleic acids (Mtb DNA and RNA) and antigens (whole bacilli, cell components, or metabolites), as well as host-derived markers and signatures including antibodies, cytokines and chemokines, transcriptomic, proteomic and metabolic markers, and hematological effectors (14). However, only a small fraction (4%, 44/1008) of the biomarker candidates screened to date have shown diagnostic value in validation studies, and only a sputum-based PCR test for Mtb DNA (e.g., GeneXpert MTB/RIF, Xpert) is endorsed and promoted worldwide by the WHO for TB diagnosis (14). Xpert can rapidly diagnose TB with high sensitivity, and identify the most common form of initial drug resistance, when used to analyze sputum with high Mtb levels (15), but has poor diagnostic accuracy with low Mtb concentration (paucibacillary) samples (16) and for extrapulmonary TB (17). Its high cost also limits its accessibility in remote areas, which may explain why Xpert has not increased global TB detection rates (18). There is a pressing need for more efficient TB diagnostic tests, as described in the WHO target product profile (14, 19), which should employ easy-to-use techniques and sensitively detect or quantify TB-specific biomarkers in non-sputum samples to rapidly diagnose TB and respond to treatment.
Clustered regularly interspaced short palindromic repeats (CRISPR) sequence-specific cleavage activity provides a useful means to overcome challenges associated with TB diagnosis. CRISPR/Cas complexes utilize a short guide RNA to bind a specific target sequence, which activates their cis-cleavage activity to cut this target sequence and can also induce a trans-cleavage activity that cuts non-specific sequences while bound to its target sequence. This trans-cleavage activity can be used to repeatedly cleavage an abundant reporter oligonucleotide in proportion to the abundance of the target sequence for signal amplification (20, 21). CRISPR/Cas activity can thus be used to detect low copy number targets that differ by single-nucleotide polymorphisms (SNPs) (22, 23) to accurately detect trace nucleic acid (NA) or non-NA targets (24, 25) in complex clinical samples, including SNPs associated with microbial DR (26). CRISPR systems call also be easily integrated into portable platforms suitable for point-of-care (POC) testing (27–29). These features provide the opportunity to create CRISPR diagnostic platforms for cross-over the barriers of TB finding.
Several groups have realized the potential of CRISPR-based assays to overcome weaknesses associated with current tests employed for TB diagnosis (30–43). However, their studies largely ignore the drawbacks and challenges of CRISPR-based TB (CRISPR-TB) assays for methodology development and clinical validation studies, as these applications are still in their infancy. Here, we systematically reviewed current CRISPR-TB assays to identify their weakness, describe potential barriers to their future adoption, and propose steps that should be taken to enhance the development and translation of these assays.
2 Summary of current CRISPR-TB assay research
Fourteen studies have employed CRISPR to diagnose TB, identify DR-Mtb strains, and distinguish Mtb from nontuberculous mycobacteria (NTM) species that may produce similar symptoms but require different treatments (Figures 1A, S1). Most of these studies were published after 2019, indicating the recent nature of most of the interest in using CRISPR assays for TB diagnosis.
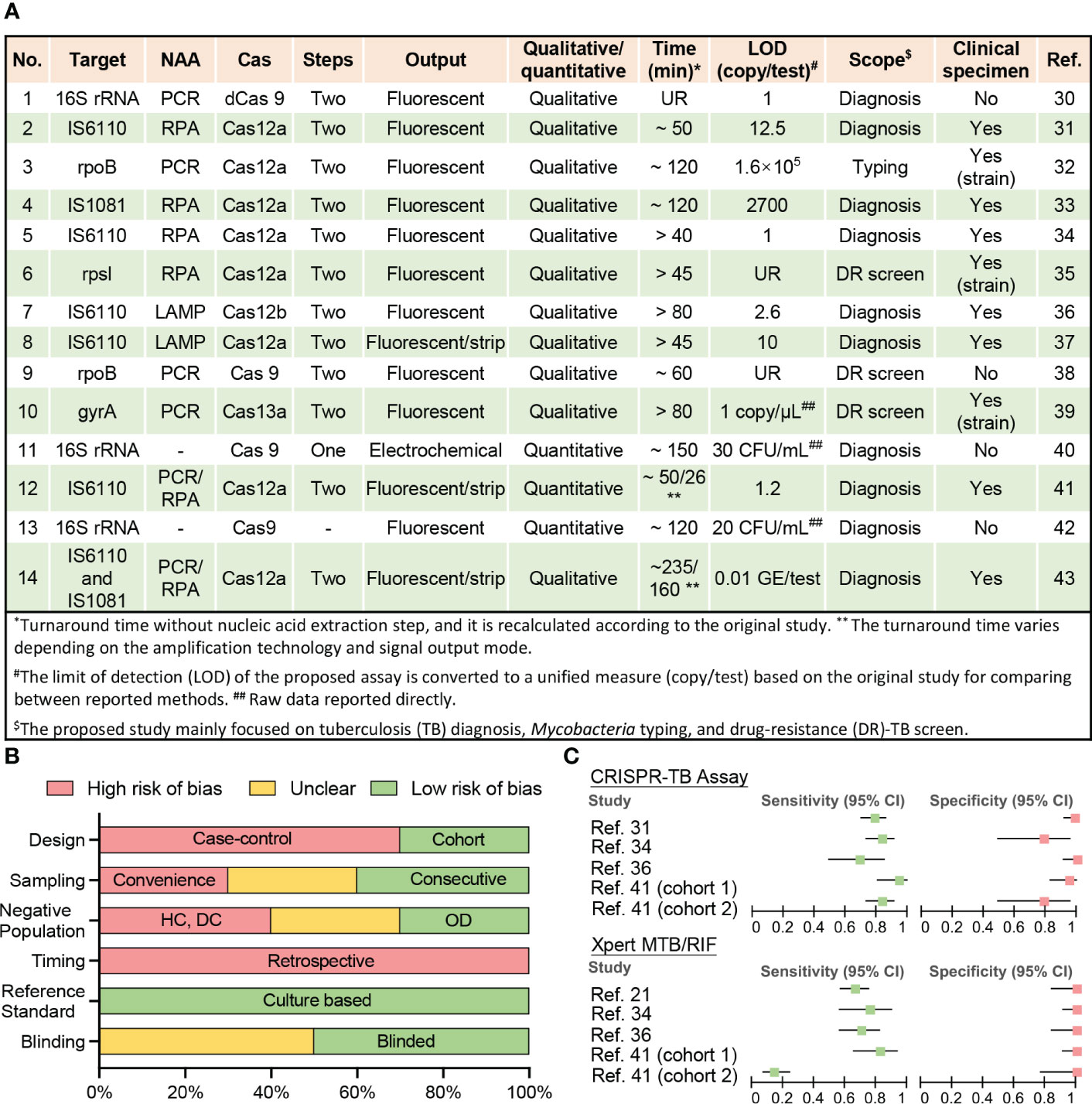
Figure 1 Current CRISPR-TB assay parameters and performance. (A) Methodological details of 12 studies employing CRISPR-TB assays. (B) Summary of QUADAS assessment results for the risk of bias in assessing data quality. Nine studies performed clinical sample evaluations (answers to QUADAS questions are listed in Table S1). The bar length reflects the frequency of an answer for each question. DC, disease contacts; DR, drug-resistant; HC, healthy control; LAMP, loop-mediated isothermal amplification; LOD, the limit of detection; NAA, nucleic acid amplification; OD, other diseases; PCR, polymerase chain reaction; RPA, recombinase polymerase amplification; UR, unreported. (C) Forest plot for the sensitivity and specificity of the CRISPR-TB assay and GeneXpert MTB/RIF (Xpert) results when compared with a composite reference standard. Details of the ten clinical specimen or strain validation studies, including control group information and diagnostic performance, are summarized in Table S2.
2.1 CRISPR-TB assay methodologies
Most reported assays still adhere to the original design paradigm of CRISPR assays, where CRISPR-based signal amplification is performed after an exponential nucleic acid amplification (NAA) step (21, 44, 45). This assay design detects trace levels of target NA sequences in clinical specimens to yield high analytical sensitivity (30, 34, 36, 41), but can prolong run times (> 1 h) (32, 33, 36, 39, 40, 43) (Figure 1A) and increase the risk of cross-contamination if amplified target NA sequences are transferred to separate CRISPR reactions. Most of these CRISPR assays employ recombinase polymerase amplification (RPA) or loop-mediated isothermal amplification (LAMP)-based isothermal amplification reactions for NAA to avoid the need for a thermocycler, which can facilitate the development of POC applications. However, many of these assays also generate fluorescent signals that require additional equipment to read and are thus less suitable for use in remote and resource-limited areas without the development of simple readout devices, although lateral flow strip-based visual readout approaches can present a good alternative for qualitative assays (37, 41, 43).
Proposed assays tend to employ Mtb-complex specific multi-copy genes (IS6110 and IS1081) as diagnostic targets as it provides higher diagnostic sensitivity (31, 33, 34, 36, 37, 41, 43) and several also detects changes in the rifampicin resistance (RR)-determining region (RRDR) of the rpoB gene (32, 38) to screen for drug resistance, since this region is altered in 95% of RR-TB cases, and most (>78%) multi-DR TB (MDR-TB) cases (1, 46). However, other Mtb-complex specific sequences in single or multi-copy genes (16s RNA) can also be used for TB diagnosis, while DR-related mutations in other genes (rpsl and gyrB) can be employed to predict resistance to other drugs used for TB treatment. It is worth noting that the sequence conservation among Mtb complex species (>99%) (47) poses a challenge when attempting to distinguish individual Mtb complex species. CRISPR assays can have single base specificity, but many of the current TB assays use IS6110 as a target and this sequence has also been detected in all Mtb complex species analyzed by these assays (M. bovis, M. bovis Bacillus Calmette–Guérin, M. africanum, and M. microti) (31, 37) Further work is therefore required to identify targets that can distinguish distinct Mtb complex species where this information would influence treatment decisions.
2.2 CRISPR-TB assay study quality
Well-designed clinical studies are required to evaluate the diagnostic performance of newly developed assays but have yet to be performed for most CRISPR-TB assays (Figure 1B and Table S1). Only ten studies have analyzed clinical samples, including three studies that used clinically isolated strains instead of patient samples (Figure 1A). All of these seven studies exhibit high bias using a modified Quality Assessment of Diagnostic Accuracy Studies (QUADAS) evaluation (14) (Table S1) primarily due to their retrospective and case-control designs, lack of consecutive sampling, and the use of controls that can inflate accuracy estimates (Figure 1B). These studies included 1219 individuals, most of whom (74%) were from China, and a substantial fraction (41%) of these individuals lacked reported demographic information and/or clinical characteristics, preventing an accurate assessment of the potential impact of population heterogeneities and comorbidities.
2.3 Diagnostic performance of CRISPR-TB assays
Nine of the ten studies provided at least one microbiological test result from Mtb culture and Xpert or had a clear clinical diagnosis to permit accurate assessment of the diagnostic performance of the proposed CRISPR-TB assay, but only four studies (involving five cohorts) were able to provide definitive Xpert results for methodological comparisons (Table S2 and Figure 1C). Unsurprisingly, CRISPR-TB assays tended to have higher diagnostic sensitivity than Xpert, with comparable or slightly decreased specificity, in most of these studies (Figure 1C), although these differences did not achieve significance, likely due to the limited number of individuals in these studies. However, CRISPR-TB assay sensitivity was significantly higher than Xpert (80.5% vs. 57.1%, p<0.05) for clinical TB cases with smear-negative sputum results (36). Further, CRISPR-TB assays have significant advantages over conventional tests when employed to analyze specimens that have low Mtb concentration (30), and thus be particularly useful in populations where this is a known problem (e.g., young children, patients living with HIV, etc.). For example, conventional TB assays exhibit very poor diagnostic performance (48, 49) in children living with HIV, representing a worst-case scenario for these assays. However, a CRISPR-based blood test for cell-free Mtb DNA diagnosed 83.3% of the children diagnosed with TB by microbiological finding or clinical algorithm, while Xpert sputum results identified only 14.5% of these children (Figure 1C) (41).
3 Challenges and outlook for CRISPR-TB assays
CRISPR assays are highly sensitive and specific, programmable, and easy-to-use. These features allow the ultra-sensitive detection of NA targets present at trace levels in complex samples, rapid target switching with different gRNA, and the development of streamlined assay platforms that can be operated in resource-limited settings. However, these properties, which have been extensively employed with assays for other diseases, appear to be underutilized in assays intended for TB diagnosis. As demonstrated in section-two, the methodologies of current CRISPR-TB assays are rudimentary, and high-quality clinical valuation studies are lacking.
3.1 Sample preparation for CRISPR analysis
Most CRISPR-TB assays employ column extraction protocols that involve multiple liquid transfers that confer a high risk for cross-contamination, and simple and efficient NA extraction procedures are not currently employed to avoid this issue. CRISPR assays are highly resistant to inhibitory components, and could, in theory, analyze specimens that have been subjected to chemical reduction or heating steps to inactivate nucleases and release target NAs from pathogens in these samples (31, 50). This would dramatically simplify sample handling and reduce contamination risks and facilitate the development of POC tests, although this approach could also reduce analytical sensitivity due since the approach would not concentrate sample NAs like conventional isolation procedures, and since inhibitory factors present in these lysates could attenuate target amplification in NAA-coupled CRISPR reactions.
Nano-/micro-technology may provide a means to balance assay sensitivity with streamlined sample processing approaches. For example, rapid procedures that enriched NAs using magnetic nanobeads (51) or fibrous materials (52, 53) can efficiently adsorb released SARS-COV-2 RNA for in situ target amplification without an elution step. Similarly, a microfluidics device that uses an electric field gradient to rapidly separate free SARS-COV-2 RNA (54) from other factors by isotachophoresis can significantly improve detection efficiency and diagnostic performance. However, while these approaches have been successful in SARS-COV-2 assays using sample lysates, significant optimization may be necessary to employ these methods for TB diagnosis, since Mtb lysis and nuclease deactivation steps may require more stringent conditions due to the structure and composition of the Mtb cell wall, and greater potential nuclease contributions from Mtb and its diagnostic clinical specimens. Further, optimized sample preparation procedures may need to be established for different specimen types (e.g., blood, urine, cerebrospinal fluid, stool, etc.) to permit their use in clinical applications. Incorporating the detection of non-sputum specimens into the scope of CRISPR diagnostics will maximize its ultra-sensitive properties and increase the microbiological confirmation rate of TB, which is a challenge for traditional NAA techniques.
CRISPR assays can also be used to sensitively detect non-NA targets using well-designed approaches where the binding of a functional NA reagent (e.g., aptamer/target NA complex) to a non-NA target releases a target NA sequence recognized by a CRISPR assay (24). CRISPR-TB assays could thus also potentially detect novel non-NA TB biomarkers in noninvasive or minimally invasive samples, such as Mtb-derived peptides or LAM in blood (55) or urine (56). Such approaches could provide additional opportunities to diagnose extrapulmonary TB, pediatric TB, and HIV-positive TB cases (57, 58) who are typically diagnosed with reduced sensitivity by standard methods and who are at increased risk for TB-related mortality. However, given the low and highly variable levels of valuable biomarkers in different samples, differentiated and efficient sample pre-treatment protocols may be required for different types of non-NA markers and different sample types. In addition, it may be critical to construct signal transduction systems with high matrix tolerance and compatibility with non-NA marker types to convert non-NA markers not identified by the CRISPR system into recognizable NA signals.
3.2 CRISPR assay workflow optimizations
Current CRISPR-TB assays typically utilize a separate NAA step to simplify assay development, but this increases the complexity, completion time, and contamination risk of the assay. The NAA and CRISPR reactions can be integrated into a single tube if an external force (e.g., centrifugation) is used to introduce CRISPR reagents after completion of the NAA step to avoid the potential for aerosol contamination during the addition of these reagents (59), although this still requires the use of consecutive NAA and CRISPR reactions that increase the sample-to-answer time of the assay.
Simultaneous NAA and CRISPR reactions can be performed in integrated NNA-CRISPR assays that use isothermal RPA or LAMP reactions for target amplification, which can simplify assay workflows to reduce assay performance times while avoiding the risk of contamination (51, 60, 61). However, such integrated NAA CRISPR reactions can also reduce detection sensitivity, since their buffer conditions may be suboptimal for both reactions and since these reactions are in direct competition (target amplification versus target cleavage), leading to assay designs that favor the NAA reaction to allow target accumulation over CRISPR cleavage and target detection.
It is also possible to eliminate the NAA step to simplify assay workflows and reduce reagent costs and the risk of cross-contamination, but it can significantly reduce assay sensitivity and thus necessitate the use of an ultrasensitive signal readout (62–65) or amplification system (66, 67) to detect weak signals produced in response to low concentration NA targets.
Thus, a CRISPR assay design must consider the workflow and diagnostic performance requirements for its intended application. For example, assays designed to have high diagnostic performance for paucibacillary TB cases (extrapulmonary TB, pediatric TB, and HIV-positive TB) may sacrifice procedure simplicity for sensitivity, while an assay intended as a POC test for TB diagnosis in the general population may prioritize a streamlined workflow over ultrasensitive detection.
3.3 Multiplex assays, target constraints, and quantitative assay readouts
Current CRISPR assays typically detect a single target and are non-quantitative. However, assays that detect a single target may produce false negatives due to strain-specific sequence variations. For example, a test that targets the multi-copy IS6110 insertion element should produce false negative results for Mtb strains that lack this insertion element (68, 69). Similarly, single target assays for DR-TB may miss alternate mutations in a gene associated with drug resistance and cannot detect mutations in other genes that confer resistance to other important drugs employed in anti-TB treatment regimens. For example, a test for DRTBRB targeting the RRDR fragment of rpoB to detect major mutations associated with rapamycin and rifabutin resistance could miss MDR-TB cases that lack these mutations (70).
Multiplex CRISPR assays could help address these issues, but are technically challenging to develop as single reaction tests since the trans cleavage activities of the CRISPR/Cas variants used for signal readout in the most popular and sensitive assays lack strong sequence specificity. Single-reaction multiplex CRISPR assays that employ multiple Cas proteins with distinct trans-cleavage substrate preferences have been proposed to address this issue, but no more than four targets can be detected in one reaction due to the limited number of Cas proteins with differential cleavage preferences (22), and even among these proteins there is the potential for significant off-target cleavage and the need to optimize an assay for all four activities.
Microfluidic- or micro-droplet-based approaches may represent a better option for multiplex CRISPR assays as they can perform large numbers of distinct single-target tests in spatially separated regions to avoid target/reporter crosstalk difficulties while simultaneously detecting hundreds or thousands of distinct NA targets (71, 72).
Sequence considerations can also influence which specific target regions can be analyzed in a CRISPR assay, which can present a challenge when an assay must detect a specific sequence associated with a phenotype type of interest (e.g., a SNP associated with resistance to a specific drug). Most CRISPR/Cas systems used for sensitive NA detection require that their target NAs contain a protospacer adjacent motif (PAM) sequence, which is problematic when a sequence of interest does not contain this motif. This issue can be partially addressed by using NAA primers to introduce a PAM sequence into amplicons that contain the sequence of interest (61). However, there are limitations to this approach as this PAM sequence must be introduced in close proximity to the sequence of interest with minimal primer mismatch, and some PAM optimization may be required to obtain specificity for a SNP of interest. An alternate solution is to screen for or bioengineer Cas protein variants that exhibit fewer PAM constraints (73). Cas14 can recognize and cleave single-stranded DNA targets that lack PAM sequences (74), and could serve as a template for the design of new CAS proteins that lack a PAM sequence requirement.
Quantitative CRISPR assays are also necessary to rapidly determine Mtb burden and its real-time response to anti-TB therapy as a measure of disease severity and treatment efficacy (41). Standard curves can be used to quantify Mtb DNA levels in clinical specimens but can produce highly variable results when analyzing samples that contain only trace amounts of a target sequence (41). CRISPR assays that employ digital droplet technology to achieve absolute quantification can circumvent this problem (23, 75–80), but this approach requires additional equipment and resources. Smartphones have the signal acquisition and data processing properties required for portable quantitative assays suitable for use in resource-limited areas, and their network connectivity also provides a convenient means for data reporting for disease control efforts. Thus, the combination of CRISPR-based TB assays and smartphone-based readout devices, or other similar portable devices, has the potential to increase the capacity for TB screening and treatment monitoring.
3.4 Clinical validation studies
CRISPR-TB assays have exciting potential to improve TB diagnosis and management, but their translation as clinical applications require their validation in well-designed, adequately powered, and multicenter prospective clinical studies, which have not been conducted for any of the CRISPR-TB assays that have been reported to date. Such studies should ideally include cohorts of extrapulmonary TB, pediatric TB, and HIV-positive TB cases, as these individuals would most benefit from early diagnosis and treatment initiation to reduce their high mortality rates. These studies should also evaluate the relative utility of CRISPR-TB assay results obtained from several types of noninvasive or minimally invasive patient specimens (e.g., urine, stool, fingerstick blood samples) for TB diagnosis in different populations, and for their potential application as CRISPR-TB assays intended for use in resource-limited settings where obtaining sputum or invasive specimens can be difficult or infeasible. Moreover, given the high sensitivity CRISPR, it is recommended that these clinical evaluation studies employ a composite criterion to identify the TB-positive and TB-negative individuals, since the use of a single standard diagnostic method may misdiagnose TB cases, particularly in extrapulmonary, pediatric, or HIV-positive cohorts to skew CRISPR-TB assay sensitivity and specificity estimates.
3.5 Assay accessibility
Future studies should also focus on improving the accessibility of CRISPR-TB assays, as limited clinical laboratory resources or infrastructure may reduce access to or capacity for TB diagnostic tests in areas with high TB incidence and prevalence rates. Such assays should ideally integrate a rapid NA extraction method into the CRISPR-TB assay and employ a rapid and streamlined procedure that does not require significant additional equipment to perform. Ideally, such assays would integrate all their procedures into a single streamlined assay platform (e.g., a microfluidic chip or a test strip) in a direct sample-to-result assay format that would not require technical expertise or any equipment for sample processing or assay readout. Such integrated platforms could potentially be into wearable devices, such as masks (27), for streamlined real-time assessment TB assessment (81). Negative and positive controls should be incorporated into these platforms to permit immediate evaluation for adverse storage and contamination effects and other confounding factors that could decease the accuracy of assay results.
CRISPR-TB assays intended for use in remote and resource-limited areas should also account for the transport and storage conditions these assays will likely face, ideally during their initial development phase, since cold chains are often difficult to maintain in areas with high TB burden. Lyophilized CRISPR reagents can be stored for months at four degrees and weeks at room temperature without significant performance decreases (28, 82). However, assay developers should also determine the stability of a CRISPR-TB assay at the more variable ambient temperatures these assays might be likely to encounter in areas without temperature control. The efforts will also be required to promote the large-scale production of key reagents, such as Cas proteins, to reduce assay development and production costs and to shorten distribution distances, which will require a streamlined licensing procedure for the relevant patents, as has been done for NAA reagents.
4 Perspective on new CRISPR-TB assay development
In summary, CRISPR-TB assays have strong potential to improve the TB diagnosis of TB, but clinical validation studies are required to allow regulatory approval and commercialization for TB diagnosis and treatment evaluation. Substantial refinements are usually also required to translate an initial proof-of-concept CRISPR-TB assay suitable for use in a research laboratory to a clinical application that can be employed at a large-scale in clinical laboratories, clinics, or POC settings. We propose that future CRISPR-TB assays (Figure 2) should ideally employ an integrated platform for sample processing, NA enrichment, and coupled NAA and CRISPR detection that contains lyophilized reagents to minimize assay cold chain concerns. Such platforms should evaluate noninvasive or minimally invasive diagnostic specimens, employ streamlined workflows with rapid sample-to-result times, and employ a readout that can be quantified by a smart terminal that can report results to a central system to aid in TB control efforts or telemedicine interventions. We believe maturing CRISPR-TB assay approaches represent a powerful means of addressing current TB diagnosis and treatment evaluation challenges required to achieve the goals of current TB eradication efforts. Mature CRISPR-TB assays may also prove valuable in non-clinical applications, such as screening for active Mtb or Mtb complex infections in domestic livestock or wildlife populations (83, 84) as has been done with Xpert. This would be particularly valuable is these analyses could employ blood or fecal specimens to simplify sample collection or population level screening efforts.
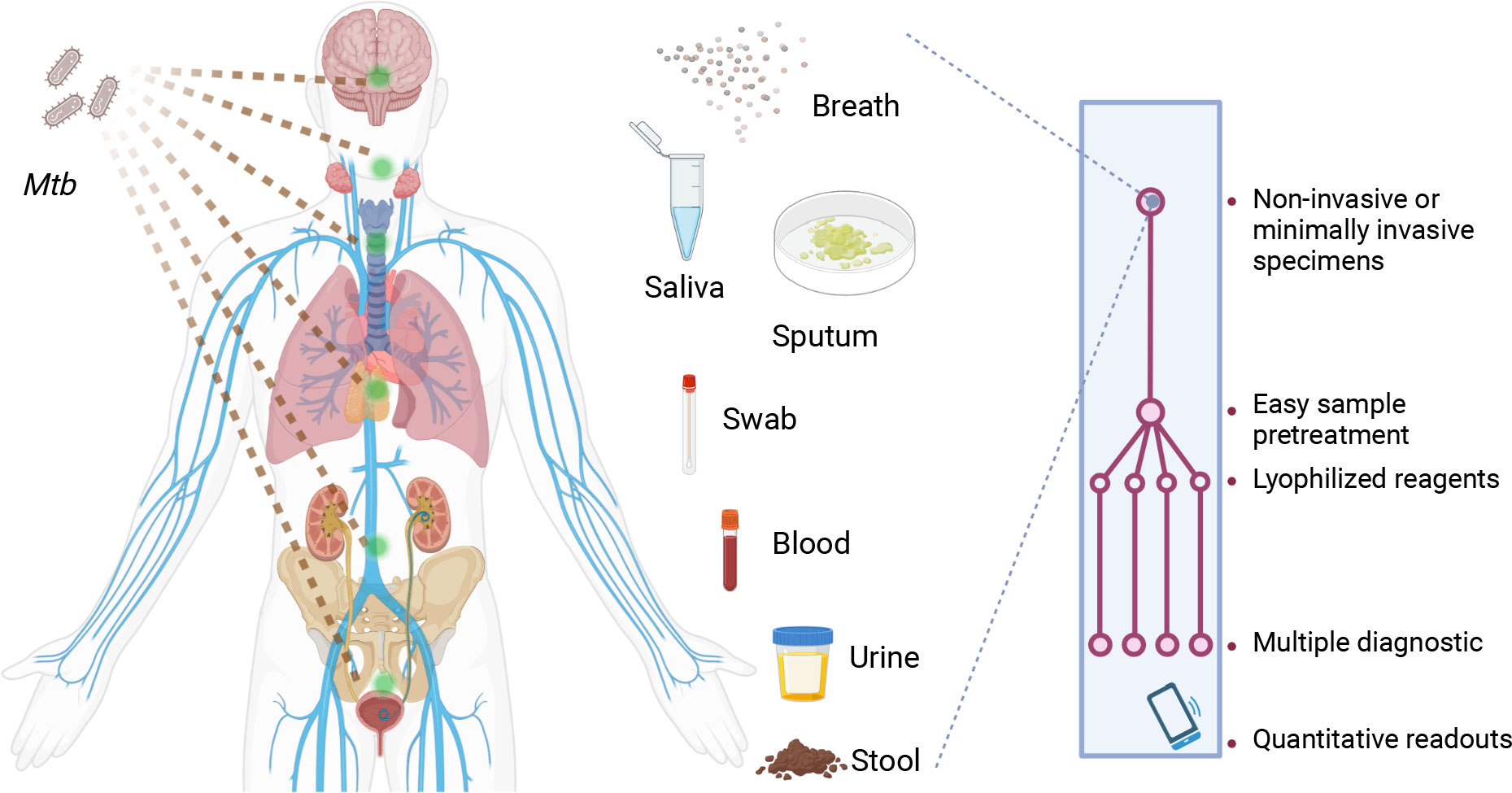
Figure 2 Characteristics desired for future CRISPR-TB assays. Future CRISPR-TB assays should ideally analyze noninvasive or minimally invasive diagnostic specimens (e.g., breath aerosol, saliva, sputum, swab, blood, urine and stool samples) and employ an integrated platform with lyophilized reagents to minimize cold chain concerns. These assays should integrate sample treatment and target detection reactions in a streamlined workflow to provide assay results within minutes. The assay readout should also provide quantitative results when read by a smart terminal, and this device should be able to report these results to a central system to facilitate TB control efforts or telemedicine interventions. Mtb, Mycobacterium tuberculosis.
Author contributions
ZH and SL conceived the design of this manuscript. ZH and GZ performed the dataset search, article screen, data extraction, and quality appraisal. ZH drafted the manuscript, and GZ, CL, and TH provided critical revision. All authors contributed to the article and approved the submitted version. SL was responsible for the decision to submit the manuscript.
Funding
The work was primarily supported by the Shenzhen Fund for Guangdong Provincial High-level Clinical Key Specialties (No. SZGSP010), the Special Fund for Science and Technology Innovation of Guangdong (No. 2020B1111170014), the Medical Scientific Research Foundation of Guangdong Province (No. A2023170), and Shenzhen Clinicla Research Center for Tuberculosis (202106171415099001).
Conflict of interest
ZH and TH are inventors on a provisional patent application related to this work US Patent US20230087018A1.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1172035/full#supplementary-material
Abbreviations
Mtb, mycobacterium tuberculosis; TB, tuberculosis; DR, drug-resistant; HIV, human immunodeficiency virus; PCR, polymerase chain reaction; Xpert, GeneXpert MTB/RIF; CRISPR, clustered regularly interspaced short palindromic repeats; NA, nucleic acid; SNP, single-nucleotide polymorphisms; POC, point-of-care; NTM, nontuberculous mycobacteria; NAA, nucleic acid amplification; RPA, recombinase polymerase amplification; LAMP, loop-mediated isothermal amplification; RR, rifampicin resistance; RRDR, rifampicin resistance-determining region; SARS−CoV−2, severe acute respiratory syndrome coronavirus 2; QUADAS, quality assessment of diagnostic accuracy studies; PAM, protospacer adjacent motif sequence.
References
1. WHO. Global tuberculosis report 2022. WHO (2022). https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022
2. Chakaya J, Petersen E, Nantanda R, Mungai BN, Migliori GB, Amanullah F, et al. The who global tuberculosis 2021 report - not so good news and turning the tide back to end TB. Int J Infect Dis (2022) 124(Suppl 1):S26–S9. doi: 10.1016/j.ijid.2022.03.011
3. Hamada Y, Cirillo DM, Matteelli A, Penn-Nicholson A, Rangaka MX, Ruhwald M. Tests for tuberculosis infection: landscape analysis. Eur Respir J (2021) 58(5):2100167. doi: 10.1183/13993003.00167-2021
4. Fleming KA, Horton S, Wilson ML, Atun R, DeStigter K, Flanigan J, et al. The lancet commission on diagnostics: transforming access to diagnostics. Lancet (2021) 398(10315):1997–2050. doi: 10.1016/S0140-6736(21)00673-5
5. Caulfield AJ, Wengenack NL. Diagnosis of active tuberculosis disease: from microscopy to molecular techniques. J Clin Tuberc Other Mycobact Dis (2016) 4:33–43. doi: 10.1016/j.jctube.2016.05.005
6. Sester M, Sotgiu G, Lange C, Giehl C, Girardi E, Migliori GB, et al. Interferon-γ release assays for the diagnosis of active tuberculosis: a systematic review and meta-analysis. Eur Respir J (2011) 37(1):100–11. doi: 10.1183/09031936.00114810
7. Connell TG, Zar HJ, Nicol MP. Advances in the diagnosis of pulmonary tuberculosis in HIV-infected and HIV-uninfected children. J Infect Dis (2011) 204(suppl_4):S1151–S8. doi: 10.1093/infdis/jir413
8. Gupta RK, Lucas SB, Fielding KL, Lawn SD. Prevalence of tuberculosis in post-mortem studies of HIV-infected adults and children in resource-limited settings: a systematic review and meta-analysis. AIDS (2015) 29(15):1987–2002. doi: 10.1097/QAD.0000000000000802
9. Lee JY. Diagnosis and treatment of extrapulmonary tuberculosis. Tuberc Respir Dis (2015) 78(2):47–55. doi: 10.4046/trd.2015.78.2.47
10. Peng YH, Chen CY, Su CH, Muo CH, Chen KF, Liao WC, et al. Increased risk of dementia among patients with pulmonary tuberculosis: a retrospective population-based cohort study. Am J Alzheimers Dis Other Demen (2015) 30(6):629–34. doi: 10.1177/1533317515577186
11. Shen CH, Chou CH, Liu FC, Lin TY, Huang WY, Wang YC, et al. Association between tuberculosis and Parkinson disease: a nationwide, population-based cohort study. Med (Baltimore) (2016) 95(8):e2883. doi: 10.1097/md.0000000000002883
12. Ahmad M, Ibrahim WH, Sarafandi SA, Shahzada KS, Ahmed S, Haq IU, et al. Diagnostic value of bronchoalveolar lavage in the subset of patients with negative sputum/smear and mycobacterial culture and a suspicion of pulmonary tuberculosis. Int J Infect Dis (2019) 82:96–101. doi: 10.1016/j.ijid.2019.03.021
13. MacLean E, Sulis G, Denkinger Claudia M, Johnston James C, Pai M, Ahmad Khan F. Diagnostic accuracy of stool Xpert MTB/RIF for detection of pulmonary tuberculosis in children: a systematic review and meta-analysis. J Clin Microbiol (2019) 57(6):10. doi: 10.1128/jcm.02057-18
14. MacLean E, Broger T, Yerlikaya S, Fernandez-Carballo BL, Pai M, Denkinger CM. A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol (2019) 4(5):748–58. doi: 10.1038/s41564-019-0380-2
15. Steingart KR, Sohn H, Schiller I, Kloda LA, Boehme CC, Pai M, et al. Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Db Syst Rev (2013) 1):CD009593. doi: 10.1002/14651858.CD009593.pub2
16. Ocheretina O, Brandao A, Pang Y, Rodrigues C, Banu S, Ssengooba W, et al. Impact of the bacillary load on the accuracy of rifampicin resistance results by Xpert® MTB/RIF. Int J Tuberc Lung Dis (2021) 25(11):881–5. doi: 10.5588/ijtld.21.0564
17. Allahyartorkaman M, Mirsaeidi M, Hamzehloo G, Amini S, Zakiloo M, Nasiri MJ. Low diagnostic accuracy of Xpert MTB/RIF assay for extrapulmonary tuberculosis: a multicenter surveillance. Sci Rep (2019) 9(1):18515. doi: 10.1038/s41598-019-55112-y
18. Walzl G, McNerney R, du Plessis N, Bates M, McHugh TD, Chegou NN, et al. Tuberculosis: advances and challenges in development of new diagnostics and biomarkers. Lancet Infect Dis (2018) 18(7):e199–210. doi: 10.1016/S1473-3099(18)30111-7
19. Gardiner JL, Karp CL. Transformative tools for tackling tuberculosis. J Exp Med (2015) 212(11):1759–69. doi: 10.1084/jem.20151468
20. Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature (2016) 532(7600):517–21. doi: 10.1038/nature17945
21. Gootenberg Jonathan S, Abudayyeh Omar O, Lee Jeong W, Essletzbichler P, Dy Aaron J, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science (2017) 356(6336):438–42. doi: 10.1126/science.aam9321
22. Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science (2018) 360(6387):439–44. doi: 10.1126/science.aaq0179
23. Yue H, Shu B, Tian T, Xiong E, Huang M, Zhu D, et al. Droplet Cas12a assay enables DNA quantification from unamplified samples at the single-molecule level. Nano Lett (2021) 21(11):4643–53. doi: 10.1021/acs.nanolett.1c00715
24. Xiong Y, Zhang J, Yang Z, Mou Q, Ma Y, Xiong Y, et al. Functional DNA regulated CRISPR-Cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets. J Am Chem Soc (2020) 142(1):207–13. doi: 10.1021/jacs.9b09211
25. Shen J, Zhou X, Shan Y, Yue H, Huang R, Hu J, et al. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat Commun (2020) 11(1):267. doi: 10.1038/s41467-019-14135-9
26. Wang S, Li H, Kou Z, Ren F, Jin Y, Yang L, et al. Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clin Microbiol Infect (2021) 27(3):443–50. doi: 10.1016/j.cmi.2020.04.018
27. Nguyen PQ, Soenksen LR, Donghia NM, Angenent-Mari NM, de Puig H, Huang A, et al. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat Biotechnol (2021) 39(11):1366–74. doi: 10.1038/s41587-021-00950-3
28. Arizti-Sanz J, Bradley AD, Zhang YB, Boehm CK, Freije CA, Grunberg ME, et al. Simplified Cas13-based assays for the fast identification of SARS-CoV-2 and its variants. Nat Biomed Eng (2022) 6(8):932–43. doi: 10.1038/s41551-022-00889-z
29. Chandrasekaran SS, Agrawal S, Fanton A, Jangid AR, Charrez B, Escajeda AM, et al. Rapid detection of SARS-CoV-2 Rna in saliva Via Cas13. Nat Biomed Eng (2022) 6(8):944–56. doi: 10.1038/s41551-022-00917-y
30. Zhang Y, Qian L, Wei W, Wang Y, Wang B, Lin P, et al. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synthetic Biol (2017) 6(2):211–6. doi: 10.1021/acssynbio.6b00215
31. Ai JW, Zhou X, Xu T, Yang M, Chen Y, He GQ, et al. CRISPR-based rapid and ultra-sensitive diagnostic test for mycobacterium tuberculosis. Emerg Microbes Infect (2019) 8(1):1361–9. doi: 10.1080/22221751.2019.1664939
32. Xiao GH, He X, Zhang S, Liu YY, Liang ZH, Liu HM, et al. Cas12a/guide RNA-based platform for rapid and accurate identification of major mycobacterium species. J Clin Microbiol (2020) 58(2):e01368–19. doi: 10.1128/jcm.01368-19
33. Xu H, Zhang X, Cai Z, Dong X, Chen G, Li Z, et al. An isothermal method for sensitive detection of mycobacterium tuberculosis complex using clustered regularly interspaced short palindromic repeats/Cas12a cis and trans cleavage. J Mol Diagnost (2020) 22(8):1020–9. doi: 10.1016/j.jmoldx.2020.04.212
34. Li H, Cui X, Sun L, Deng X, Liu S, Zou X, et al. High concentration of Cas12a effector tolerates more mismatches on ssDNA. FASEB J (2021) 35(1):e21153. doi: 10.1096/fj.202001475R
35. Liu P, Wang X, Liang J, Dong Q, Zhang J, Liu D, et al. A recombinase polymerase amplification-coupled Cas12a mutant-based module for efficient detection of streptomycin-resistant mutations in mycobacterium tuberculosis. Front Microbiol (2021) 12:796916. doi: 10.3389/fmicb.2021.796916
36. Sam IK, Chen YY, Ma J, Li SY, Ying RY, Li LX, et al. TB-Quick: CRISPR-Cas12b-Assisted rapid and sensitive detection of mycobacterium tuberculosis. J Infect (2021) 83(1):54–60. doi: 10.1016/j.jinf.2021.04.032
37. Wang Y, Li JQ, Li SJ, Zhu X, Wang XX, Huang JF, et al. LAMP-CRISPR-Cas12-based diagnostic platform for detection of mycobacterium tuberculosis complex using real-time fluorescence or lateral flow test. Microchim Acta (2021) 188(10):347. doi: 10.1007/s00604-021-04985-w
38. Augustin L, Agarwal N. Designing a Cas9/Grna-assisted quantitative real-time PCR (CARP) assay for identification of point mutations leading to rifampicin resistance in the human pathogen mycobacterium tuberculosis. Gene (2023) 857:147173. doi: 10.1016/j.gene.2023.147173
39. Bai X, Gao P, Qian K, Yang J, Deng H, Fu T, et al. A highly sensitive and specific detection method for mycobacterium tuberculosis fluoroquinolone resistance mutations utilizing the CRISPR-Cas13a system. Front Microbiol (2022) 13:847373. doi: 10.3389/fmicb.2022.847373
40. Huang J, Liang Z, Liu Y, Zhou JD, He FJ. Development of an MSPQC nucleic acid sensor based on CRISPR/ Cas9 for the detection of mycobacterium tuberculosis. Anal Chem (2022) 94(32):11409–15. doi: 10.1021/acs.analchem.2c02538
41. Huang Z, LaCourse SM, Kay AW, Stern J, Escudero JN, Youngquist BM, et al. CRISPR detection of circulating cell-free mycobacterium tuberculosis DNA in adults and children, including children with HIV: a molecular diagnostics study. Lancet Microbe (2022) 3(7):e482–e92. doi: 10.1016/S2666-5247(22)00087-8
42. Zhou M, Li X, Wen H, Huang B, Ren J, Zhang J. The construction of CRISPR/Cas9-mediated FRET 16S rDNA sensor for detection of mycobacterium tuberculosis. Analyst (2023)148:2308–15. doi: 10.1039/D3AN00462G
43. Thakku SG, Lirette J, Murugesan K, Chen J, Theron G, Banaei N, et al. Genome-wide tiled detection of circulating mycobacterium tuberculosis cell-free DNA using Cas13. Nat Commun (2023) 14(1):1803. doi: 10.1038/s41467-023-37183-8
44. Chen Janice S, Ma E, Harrington Lucas B, Da Costa M, Tian X, Palefsky Joel M, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded dnase activity. Science (2018) 360(6387):436–9. doi: 10.1126/science.aar6245
45. Li SY, Cheng QX, Wang JM, Li XY, Zhang ZL, Gao S, et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discovery (2018) 4(1):20. doi: 10.1038/s41421-018-0028-z
46. Zaw MT, Emran NA, Lin Z. Mutations inside rifampicin-resistance determining region of rpoB gene associated with rifampicin-resistance in mycobacterium tuberculosis. J Infect Public Health (2018) 11(5):605–10. doi: 10.1016/j.jiph.2018.04.005
47. Brosch R, Gordon SV, Pym A, Eiglmeier K, Garnier T, Cole ST. Comparative genomics of the mycobacteria. Int J Med Microbiol (2000) 290(2):143–52. doi: 10.1016/S1438-4221(00)80083-1
48. Bell LCK, Noursadeghi M. Pathogenesis of HIV-1 and mycobacterium tuberculosis co-infection. Nat Rev Microbiol (2018) 16(2):80–90. doi: 10.1038/nrmicro.2017.128
49. Thomas TA. Tuberculosis in children. Pediatr Clin North Am (2017) 64(4):893–909. doi: 10.1016/j.pcl.2017.03.010
50. Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science (2018) 360(6387):444–8. doi: 10.1126/science.aas8836
51. Joung J, Ladha A, Saito M, Kim N-G, Woolley AE, Segel M, et al. Detection of SARS-CoV-2 with Sherlock one-pot testing. New Engl J Med (2020) 383(15):1492–4. doi: 10.1056/NEJMc2026172
52. de Puig H, Lee Rose A, Najjar D, Tan X, Soenksen Luis R, Angenent-Mari Nicolaas M, et al. Minimally instrumented Sherlock (Misherlock) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci Adv (2021) 7(32):eabh2944. doi: 10.1126/sciadv.abh2944
53. Mason MG, Botella JR. Rapid (30-second), equipment-free purification of nucleic acids using easy-to-make dipsticks. Nat Protoc (2020) 15(11):3663–77. doi: 10.1038/s41596-020-0392-7
54. Persat A, Marshall LA, Santiago JG. Purification of nucleic acids from whole blood using isotachophoresis. Anal Chem (2009) 81(22):9507–11. doi: 10.1021/ac901965v
55. Liu C, Zhao Z, Fan J, Lyon CJ, Wu HJ, Nedelkov D, et al. Quantification of circulating mycobacterium tuberculosis antigen peptides allows rapid diagnosis of active disease and treatment monitoring. Proc Natl Acad Sci (2017) 114(15):3969–74. doi: 10.1073/pnas.1621360114
56. Paris L, Magni R, Zaidi F, Araujo R, Saini N, Harpole M, et al. Urine lipoarabinomannan glycan in HIV-negative patients with pulmonary tuberculosis correlates with disease severity. Sci Trans Med (2017) 9(420):eaal2807. doi: 10.1126/scitranslmed.aal2807
57. Yu G, Shen Y, Ye B, Shi Y. Diagnostic accuracy of mycobacterium tuberculosis cell-free DNA for tuberculosis: a systematic review and meta-analysis. PloS One (2021) 16(6):e0253658. doi: 10.1371/journal.pone.0253658
58. Liu XH, Xia L, Song B, Wang H, Qian XQ, Wei JH, et al. Stool-based Xpert MTB/RIF ultra assay as a tool for detecting pulmonary tuberculosis in children with abnormal chest imaging: a prospective cohort study. J Infect (2021) 82(1):84–9. doi: 10.1016/j.jinf.2020.10.036
59. Wang B, Wang R, Wang D, Wu J, Li J, Wang J, et al. Cas12aVDet: a CRISPR/Cas12a-based platform for rapid and visual nucleic acid detection. Anal Chem (2019) 91(19):12156–61. doi: 10.1021/acs.analchem.9b01526
60. Ding X, Yin K, Li Z, Lalla RV, Ballesteros E, Sfeir MM, et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun (2020) 11(1):4711. doi: 10.1038/s41467-020-18575-6
61. Li L, Li S, Wu N, Wu J, Wang G, Zhao G, et al. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synthetic Biol (2019) 8(10):2228–37. doi: 10.1021/acssynbio.9b00209
62. Yang W, Restrepo-Pérez L, Bengtson M, Heerema SJ, Birnie A, van der Torre J, et al. Detection of CRISPR-dCas9 on DNA with solid-state nanopores. Nano Lett (2018) 18(10):6469–74. doi: 10.1021/acs.nanolett.8b02968
63. Weckman NE, Ermann N, Gutierrez R, Chen K, Graham J, Tivony R, et al. Multiplexed DNA identification using site specific dCas9 barcodes and nanopore sensing. ACS Sens (2019) 4(8):2065–72. doi: 10.1021/acssensors.9b00686
64. Nouri R, Jiang Y, Lian XL, Guan W. Sequence-specific recognition of HIV-1 DNA with solid-state CRISPR-Cas12a-assisted nanopores (Scan). ACS Sens (2020) 5(5):1273–80. doi: 10.1021/acssensors.0c00497
65. Choi J-H, Lim J, Shin M, Paek S-H, Choi J-W. CRISPR-Cas12a-based nucleic acid amplification-free DNA biosensor Via Au nanoparticle-assisted metal-enhanced fluorescence and colorimetric analysis. Nano Lett (2021) 21(1):693–9. doi: 10.1021/acs.nanolett.0c04303
66. Shi K, Xie S, Tian R, Wang S, Lu Q, Gao D, et al. A CRISPR-Cas autocatalysis-driven feedback amplification network for supersensitive DNA diagnostics. Sci Adv (2021) 7(5):eabc7802. doi: 10.1126/sciadv.abc7802
67. Fozouni P, Son S, Díaz de León Derby M, Knott GJ, Gray CN, D’Ambrosio MV, et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell (2021) 184(2):323–33.e9. doi: 10.1016/j.cell.2020.12.001
68. Abraham PR, Chauhan A, Gangane R, Sharma VD, SHIVannavar C. Prevalence of mycobacterium tuberculosis with multiple copies of IS6110 elements in gulbarga, south India. Int J Mycobacteriol (2013) 2(4):237–9. doi: 10.1016/j.ijmyco.2013.09.003
69. Narayanan S, Das S, Garg R, Hari L, Rao VB, Frieden TR, et al. Molecular epidemiology of tuberculosis in a rural area of high prevalence in south India: implications for disease control and prevention. J Clin Microbiol (2002) 40(12):4785–8. doi: 10.1128/JCM.40.12.4785-4788.2002
70. Xu G, Liu H, Jia X, Wang X, Xu P. Mechanisms and detection methods of mycobacterium tuberculosis rifampicin resistance: the phenomenon of drug resistance is complex. Tuberculosis (2021) 128:102083. doi: 10.1016/j.tube.2021.102083
71. Ackerman CM, Myhrvold C, Thakku SG, Freije CA, Metsky HC, Yang DK, et al. Massively multiplexed nucleic acid detection with Cas13. Nature (2020) 582(7811):277–82. doi: 10.1038/s41586-020-2279-8
72. Welch NL, Zhu M, Hua C, Weller J, Mirhashemi ME, Nguyen TG, et al. Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants. Nat Med (2022) 28(5):1083–94. doi: 10.1038/s41591-022-01734-1
73. Collias D, Beisel CL. CRISPR technologies and the search for the PAM-free nuclease. Nat Commun (2021) 12(1):555. doi: 10.1038/s41467-020-20633-y
74. Wei Y, Yang Z, Zong C, Wang B, Ge X, Tan X, et al. Trans single-stranded DNA cleavage Via CRISPR/Cas14a1 activated by target RNA without destruction. Angewandte Chemie Int Edition (2021) 60(45):24241–7. doi: 10.1002/anie.202110384
75. Park JS, Hsieh K, Chen L, Kaushik A, Trick AY, Wang TH. Digital CRISPR/Cas-assisted assay for rapid and sensitive detection of SARS-CoV-2. Adv Sci (2021) 8(5):2003564. doi: 10.1002/advs.202003564
76. Tian T, Shu B, Jiang Y, Ye M, Liu L, Guo Z, et al. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano (2021) 15(1):1167–78. doi: 10.1021/acsnano.0c08165
77. Wu X, Tay JK, Goh CK, Chan C, Lee YH, Springs SL, et al. Digital CRISPR-based method for the rapid detection and absolute quantification of nucleic acids. Biomaterials (2021) 274:120876. doi: 10.1016/j.biomaterials.2021.120876
78. Luo X, Xue Y, Ju E, Tao Y, Li M, Zhou L, et al. Digital CRISPR/Cas12b-based platform enabled absolute quantification of viral RNA. Anal Chimica Acta (2022) 1192:339336. doi: 10.1016/j.aca.2021.339336
79. Ding X, Yin K, Li Z, Sfeir MM, Liu C. Sensitive quantitative detection of SARS-CoV-2 in clinical samples using digital warm-start CRISPR assay. Biosens Bioelectron (2021) 184:113218. doi: 10.1016/j.bios.2021.113218
80. Shinoda H, Taguchi Y, Nakagawa R, Makino A, Okazaki S, Nakano M, et al. Amplification-free RNA detection with CRISPR–Cas13. Commun Biol (2021) 4(1):476. doi: 10.1038/s42003-021-02001-8
81. Theron G, Limberis J, Venter R, Smith L, Pietersen E, Esmail A, et al. Bacterial and host determinants of cough aerosol culture positivity in patients with drug-resistant versus drug-susceptible tuberculosis. Nat Med (2020) 26(9):1435–43. doi: 10.1038/s41591-020-0940-2
82. Nguyen LT, Rananaware SR, Pizzano BLM, Stone BT, Jain PK. Clinical validation of engineered CRISPR/Cas12a for rapid SARS-CoV-2 detection. Commun Med (2022) 2(1):7. doi: 10.1038/s43856-021-00066-4
83. Goosen WJ, Kerr TJ, Kleynhans L, Warren RM, van Helden PD, Persing DH, et al. The Xpert MTB/RIF ultra assay detects mycobacterium tuberculosis complex DNA in white rhinoceros (Ceratotherium simum) and African elephants (Loxodonta africana). Sci Rep (2020) 10(1):14482. doi: 10.1038/s41598-020-71568-9
Keywords: tuberculosis, diagnosis, CRISPR, point-of-care, challenge and outlook
Citation: Huang Z, Zhang G, Lyon CJ, Hu TY and Lu S (2023) Outlook for CRISPR-based tuberculosis assays now in their infancy. Front. Immunol. 14:1172035. doi: 10.3389/fimmu.2023.1172035
Received: 23 February 2023; Accepted: 03 July 2023;
Published: 03 August 2023.
Edited by:
Xiao-Yong Fan, Fudan University, ChinaReviewed by:
Jin Wang, Shanghai Tolo Biotechnology Company Limited, ChinaWei Sha, Tongji University School of Medicine, China
Michel Drancourt, Aix-Marseille Université, France
Copyright © 2023 Huang, Zhang, Lyon, Hu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Huang, aHVhbmcuemhlbkBvdXRsb29rLmNvbQ==; Shuihua Lu, bHVzaHVpaHVhNjZAMTI2LmNvbQ==
 Zhen Huang
Zhen Huang Guoliang Zhang1
Guoliang Zhang1 Christopher J. Lyon
Christopher J. Lyon Shuihua Lu
Shuihua Lu