
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 14 September 2023
Sec. Vaccines and Molecular Therapeutics
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1167965
 Weiming Wang1†
Weiming Wang1† Khanghy Truong2†
Khanghy Truong2† Chaobaihui Ye1
Chaobaihui Ye1 Suman Sharma2
Suman Sharma2 Huan He1
Huan He1 Lihong Liu1
Lihong Liu1 Michael Wen1
Michael Wen1 Anisha Misra2
Anisha Misra2 Paul Zhou1*
Paul Zhou1* Jason T. Kimata2*
Jason T. Kimata2*HIV-1 infection of target cells can occur through either cell-free virions or cell-cell transmission in a virological synapse, with the latter mechanism of infection reported to be 100- to 1,000-fold more efficient. Neutralizing antibodies and entry inhibitors effectively block cell-free HIV-1, but with few exceptions, they display much less inhibitory activity against cell-mediated HIV-1 transmission. Previously, we showed that engineering HIV-1 target cells by genetically linking single-chain variable fragments (scFvs) of antibodies to glycosyl phosphatidylinositol (GPI) potently blocks infection by cell-free virions and cell-mediated infection by immature dendritic cell (iDC)-captured HIV-1. Expression of scFvs on CD4+ cell lines by transduction with X5 derived anti-HIV-1 Env antibody linked to a GPI attachment signal directs GPI-anchored scFvs into lipid rafts of the plasma membrane. In this study, we further characterize the effect of GPI-scFv X5 on cell-cell HIV-1 transmission from DCs to target cells. We report that expression of GPI-scFv X5 in transduced CD4+ cell lines and human primary CD4+ T cells potently restricts viral replication in iDC- or mDC-captured HIV-1 in trans. Using live-cell imaging, we observed that when GPI-GFP or GPI-scFv X5 transduced T cells are co-cultured with iDCs, GPI-anchored proteins enrich in contact zones and subsequently migrate from T cells into DCs, suggesting that transferred GPI-scFv X5 interferes with HIV-1 infection of iDCs. We conclude that GPI-scFv X5 on the surface of transduced CD4+ T cells not only potently blocks cell-mediated infection by DCs, but it transfers from transduced cells to the surface of iDCs and neutralizes HIV-1 replication in iDCs. Our findings have important implications for HIV-1 antibody-based immunotherapies as they demonstrate a viral inhibitory effect that extends beyond the transduced CD4+ T cells to iDCs which can enhance HIV-1 replication.
Upon crossing the epithelial barrier, HIV-1 encounters and infects a wide range of cells in the underlying mucosal tissues including professional antigen presenting cells such as dendritic cells through CCR5 mediated viral entry. Immature DCs (iDCs) play a significant role in promoting the dissemination and amplification of HIV-1 infection. iDCs capture HIV-1 and facilitate infection of nearby CD4+ T cells by trafficking captured virions to draining lymphoid tissues where, upon transition to mature DCs (mDCs), they transmit the virus to CD4+ T cells (1–3). In vitro the transmission of HIV-1 from DCs to CD4+ T cells occurs via two mechanisms: 1) trans-infection, i.e. cell-cell transmission by capture and transfer of HIV-1 from iDCs or mDCs to CD4+ T cells which can occur within a few hours post infection with or without virus internalization (4–9); and 2) cis-infection, i.e. cell-free transmission where nascent virions from infected iDCs bud and infect CD4+ T cells which can occur within a few days and is sensitive to antiviral therapy (ART) (8–12).
iDCs or mDCs can mediate cell-cell transmission of captured HIV-1 to CD4+ T cells via virological synapses (13). Within the viral synapse, captured virions are liberated from their invaginated holding membrane compartment of DCs and engage CD4 and co-receptors (CCR5 or CXCR4) on CD4+ T cells. This high local concentration of virions enhances the efficiency of HIV-1 transmission (6, 7). Indeed, cell-cell trans-infection appears to be 100- to 1,000-fold more efficient for the spread of HIV-1 in vitro (7, 14, 15). While the relative impact of cell-free and cell-cell transmission in vivo remains to be defined, in a bone marrow-liver-thymus (BLT) humanized mouse model, HIV-1-infected CD4+ T cells in lymph nodes were found to be mobile, capable of establishing virological synapses, and form syncytia. Of note, blocking egress of migratory T cells from lymph nodes into efferent lymph vessels and interrupting T cell recirculation by the use of a S1PR1 (sphingosine 1-phosphate receptor 1) antagonist FTY720 at early onset of HIV-1 infection resulted in limited HIV-1 dissemination and reduction of plasma viremia (16), indicating that cell-cell transmission of HIV-1 may be important in establishment of systemic HIV-1 infection.
Neutralizing antibodies and pharmacologic entry inhibitors effectively block cell-free transmission of HIV-1. However, with few exceptions, they are less capable of blocking cell-cell viral transmission (13–15, 17–26). In T cell-T cell co-culture where HIV-infected donor T cells were added to uninfected T cells, viral neutralization was achieved only when virus-infected donor T cells were pretreated with antibodies before being added to target T cells (14, 15, 18, 19, 24–26). Additionally, Reh et al. tested a panel of 16 broadly neutralizing antibodies against 11 HIV-1 strains using cell-free virus and cell-cell viral transmission assays and concluded that capacity of broadly neutralizing antibodies to inhibit cell-cell viral transmission of HIV-1 is not only strain- and epitope-dependent, but also dependent on the window of action during the entry process in early infection (19). Further, the reports on trans-infection of DC-CD4+ T cell in co-cultures have been variable due to variations in assay systems used by different research groups (17, 20–23, 27, 28). For example, Su et al. showed that targeting HIV-1 entry receptor gp120 with antibodies 2G12, b12, VRC01, VRC03 and targeting HIV-1 fusion receptor gp41 with antibodies 2F5 and 4E10 blocked HIV-1 trans-infection (17). Conversely, Sagar et al. showed that only anti-gp41 antibodies 2F5 and 4E10, but not anti-gp120 antibodies 2G12, b12, VRC01, and PG16 could block trans-infection (23). Moreover, van Montfort et al. showed that HIV-1 bound to antibodies 2F5, 4E10 and 10E8, but not bound to b12, NIH45-46, and VRC01, could still be captured by DCs and subsequently infect CD4+ T cells (21, 22).
Lipid rafts in the plasma membrane are critical for the formation of virological synapses that mediate cell-cell transmission (29). Lipid rafts are specialized dynamic micro-domains of plasma membrane enriched for cholesterol, sphingolipids, and glycerophospholipids (30). They have been shown to function as doorways of HIV-1 budding as well as for HIV-1 entry into CD4+ T cells and macrophages (31–34). Importantly, the CD4 receptor has been found to be located within lipid rafts of plasma membrane (35, 36). Thus, directing HIV-1 neutralizing antibodies to lipid rafts within the virological synapse would be expected to enhance restriction of cell-cell viral transmission.
Previously, we showed that engineering CD4+ T cells to express membrane anchored viral inhibitors (MAVIs) protects modified cells from depletion and potently restricts HIV-1 (37). We genetically linked single chain variable fragments (scFv) or heavy chain third complementarity determining region (HCDR3) of human monoclonal antibodies specific for the HIV-1 envelope with a glycosyl phosphatidylinositol (GPI) attachment signal derived from decay accelerating factor (DAF) (38). Using these constructs, we demonstrated that the GPI anchor targeted the scFvs or HCDR3 molecules to lipid rafts of the plasma membrane of cells. While inhibition primarily occurred by blocking virus-receptor interactions, we also showed that GPI-anchored scFvs targeting the HIV Env can block viral replication from within cells, inhibiting Env incorporation into virions and reducing viral infectivity (39). Interestingly, GPI-scFvs based on antibodies X5 and 48d as well as GPI-HCDR3 PG9 and PG16 exhibited potent and broad neutralizing activity against diverse cell-free and iDC-captured HIV-1 infection (37, 40). In this study, we investigated the effect of GPI-scFv X5 expression on CD4+ cell lines and primary human CD4+ T cells with regards to their ability to block trans-infection of HIV-1 captured by iDCs or mDCs. Utilizing virological and live-cell imaging methods, we found that GPI-scFv X5 not only significantly inhibits cell-mediated infection by DCs, but also transfers to the surface of iDCs, leading to neutralization of HIV-1 replication within these cells. Taken together, our results suggest that using GPI-scFv-based MAVIs that can effectively restrict both HIV-1 cis- and trans-infection may be a promising approach to achieve a functional cure for HIV.
To generate monocyte-derived iDCs and mDCs, human monocytes were isolated from peripheral blood mononuclear cells (PBMCs) using anti-CD14 antibody-coated beads and stimulated with GM-CSF and IL-4 as previously described (37). Figure 1A shows that human monocyte-derived iDCs expressed high levels of CD209 (DC-SIGN) and CD11c, reduced level of CD14, but not CD3, indicating the absence of contaminating T cells in the cultures of iDCs. The iDCs were then maturated into mDCs with LPS stimulation. Figure 1B shows that mDCs expressed higher levels of CD86, CD83 and HLA-DR, but lower levels of CD209, as compared to those in iDCs.
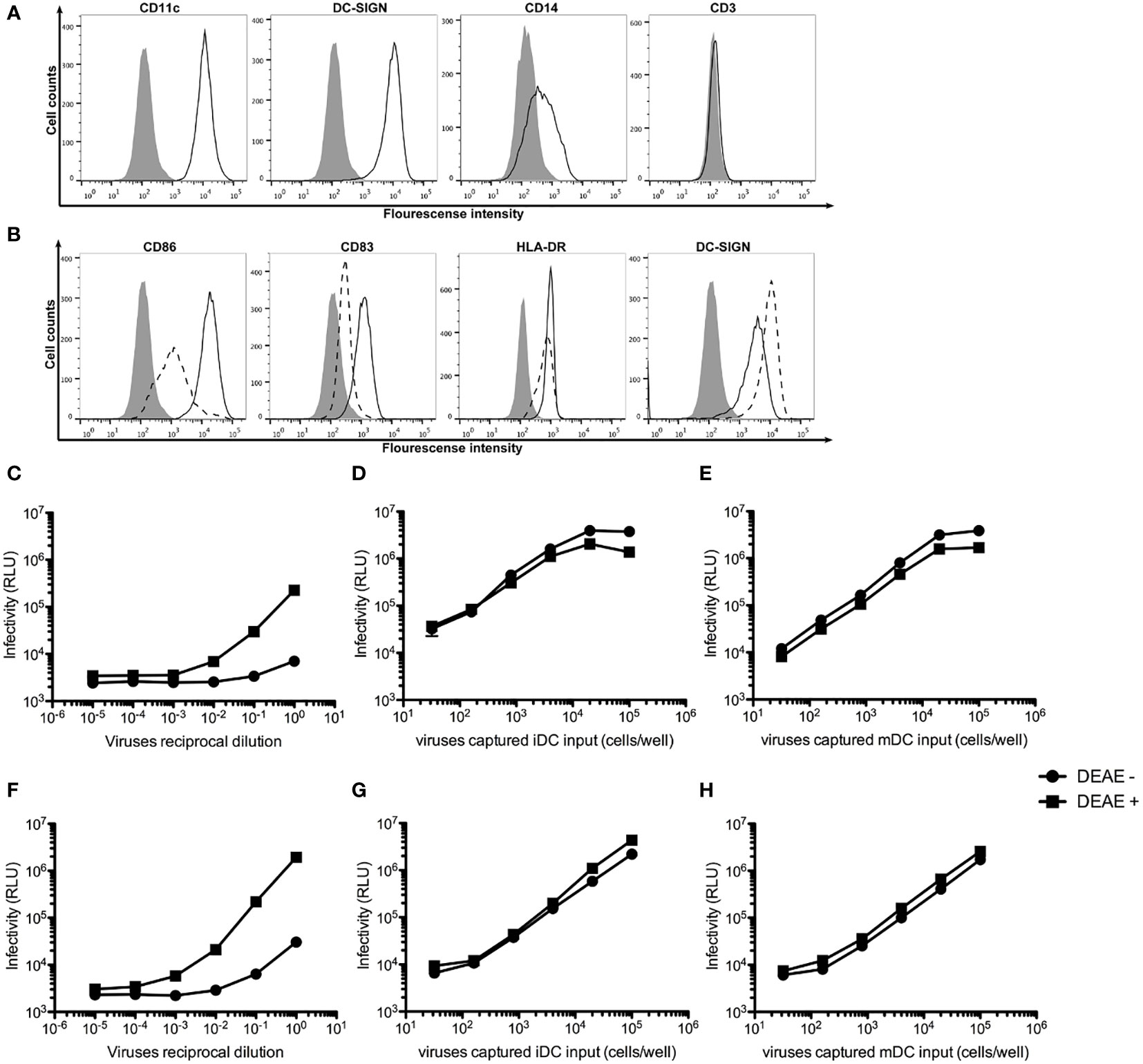
Figure 1 DEAE-dextran increases cell-free HIV-1 infection, but not trans-infection of HIV-1. (A) Histogram analysis of the expression of CD11c, CD209 (DC-SIGN), CD14 and CD3 on the surface of human monocyte-derived iDCs (open area) as compared unstained cells (solid area). (B) Histogram analysis of the expression of the maturation markers CD86, CD83, CD209 and HLA-DR on the surface of mDCs (continuous lines) as compared with iDCs (dash line). (C, D) Infectivity as measured by Relative Light Units (RLU) to TZM.bl cells by various reciprocal dilutions of HIV-1 AD8 (C) or Bru-3 (D) stocks in the presence or absence of DEAE-Dextran. (E, F) Infectivity as measured by RLU to TZM.bl cells by various amount of HIV-1 AD8 (E) or Bru-3 (F) captured by iDCs in the presence or absence of DEAE-Dextran. (G, H) Infectivity as measured by RLU to TZM.bl cells by various amount of HIV-1 AD8 (G) or Bru-3 (H) captured by mDCs in the presence or absence of DEAE-Dextran.
DEAE-dextran has been shown to significantly increase cell-free HIV-1 infection, but to have little effect on cell-cell transmission of HIV-1 (14). Therefore, we tested the effect of DEAE-dextran on cell-free HIV-1 infection versus iDC- or mDC-mediated trans-infection of HIV-1. Figures 1C–H show that indeed, DEAE-dextran significantly increased cell-free HIV-1 infection (C and D), but has little effect on iDC- or mDC-mediated trans-infection (E to H). In the absence of DEAE-dextran, iDC- or mDC-mediated trans-infection was at least 100-fold more effective than free HIV-1 infection. Thus, all our subsequent DC-mediated trans-infection experiments were carried out in the absence of DEAE-dextran so that under this condition, the contribution of free HIV-1 infection is negligible.
To test the effect of GPI-scFv X5 on trans-infection by iDC-captured HIV-1, iDCs from healthy donors were incubated with CCR5-tropic HIV-1 AD8 or CXCR4-tropic HIV-1 Bru-3 for 2 hours. After the infection, cells were washed extensively to remove cell-free virions. TZM.bl reporter cells were used to evaluate trans-infection by iDC-captured HIV-1. iDCs with or without captured HIV-1 were co-cultured with TZM.bl, TZM.bl expressing GPI-scFv X5 or the GPI-scFv AB65 negative control construct. Figure 2 shows that while TZM.bl, TZM.bl-GPI-scFv X5 or AB65 cells alone yielded background levels of Relative Light Units (RLU), TZM.bl and TZM.bl-GPI-scFv AB65 cells co-cultured with iDCs harboring captured HIV-1 AD8 or Bru-3 exhibited significantly increased levels of RLU. In contrast, TZM.bl-GPI-scFv X5 cells co-cultured with iDCs containing captured HIV-1 AD8 or Bru-3 exhibited close to background levels of RLU (Figures 2A, B), indicating that in transduced TZM.bl cells GPI-scFv X5 significantly blocked trans-infection of HIV-1 by iDCs. Similar results were observed when mDCs with or without captured HIV-1 AD8 or Bru-3 were co-cultured with TZM.bl, TZM.bl-GPI-scFv X5 or AB65 cells (Figures 2C, D). These results have been observed in two other experiments using iDCs and mDCs derived from additional donors (Figure S1).
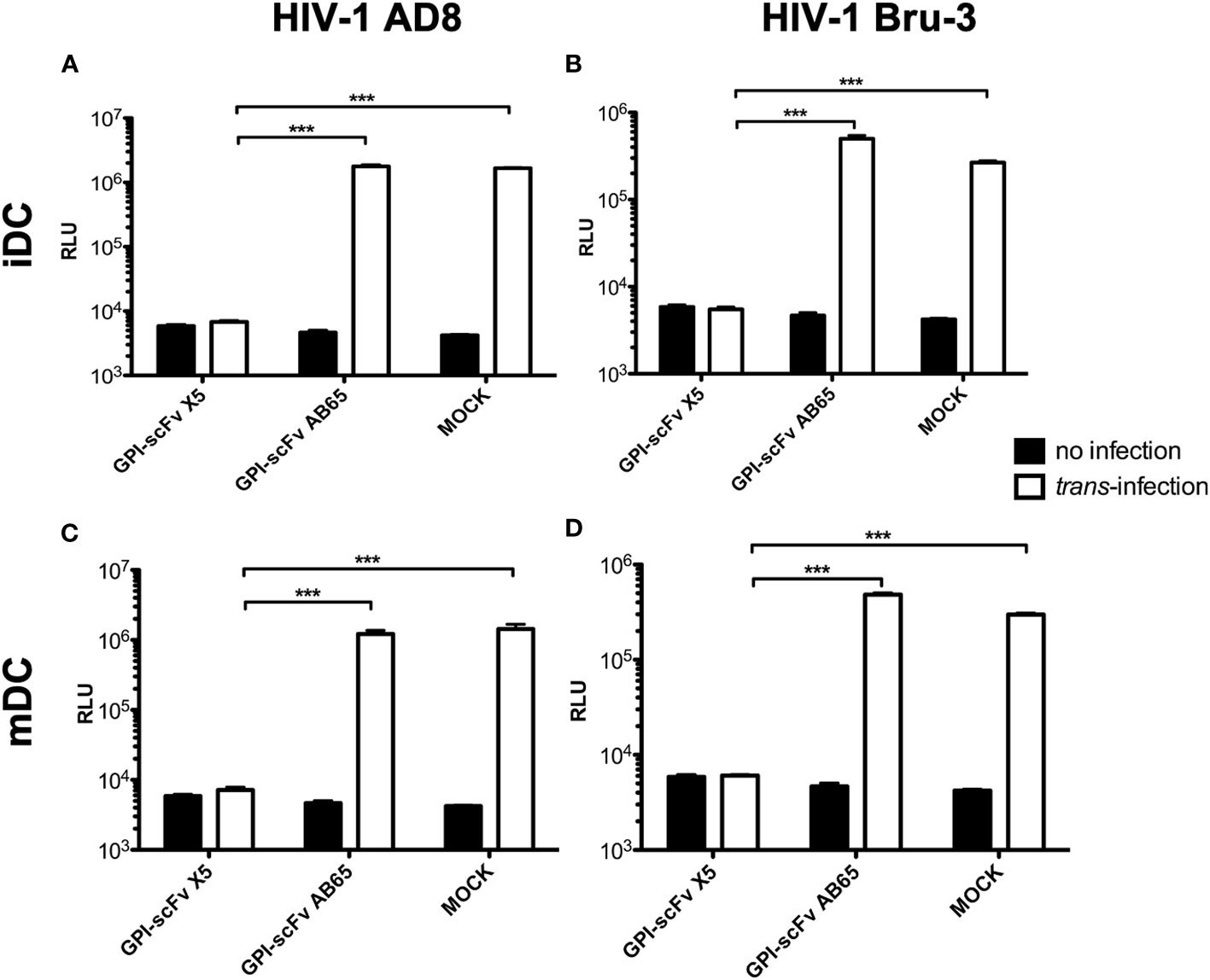
Figure 2 GPI-scFv X5-transduced TZM.bl cells are resistant to iDC- or mDC-mediated trans-infection of HIV-1. (A) RLU in mock-, GPI-scFv X5 or AB65-transduced TZM.bl cells with or without trans-infection of iDC-captured HIV-1 AD8. *** stands for P values = or < 0.001. (B) RLU in mock-, GPI-scFv X5 or AB65-transduced TZM.bl cells with or without trans-infection of iDC-captured HIV-1 Bru-3. *** stands for P values = or < 0.001. (C) RLU in mock-, GPI-scFv X5 or AB65-transduced TZM.bl cells with or without trans-infection of mDC-captured HIV-1 AD8. Infections were performed in triplicate. The average values ± standard deviation (SD) are shown. *** stands for P values = or < 0.001. (D) RLU in mock-, GPI-scFv X5 or AB65-transduced TZM.bl cells with or without trans-infection of mDC-captured HIV-1 Bru-3. *** stands for P values = or < 0.001.
While TZM.bl cells are a powerful tool for evaluating HIV-1 infection, they are of epithelial origin. We next tested the effect of GPI-X5 on trans-infection using CEMss T cells modified to express CCR5 as target cells. We found that depending on the coreceptor usage of the captured virus, distinctly different results were observed with iDCs. Interestingly, co-culture of the iDCs with captured R5-tropic HIV-1 AD8 and CEMss-CCR5-GPI-scFv X5 resulted in decreasing virus production compared to iDCs with HIV-1 AD8 alone, which increased over time. High levels of HIV-1 production were observed in co-cultures of iDCs with captured HIV-1 AD8 and CEMss-CCR5-GPI-scFv AB65 (Figure 3A). Co-culture of iDCs with captured X4-tropic HIV-1 Bru-3 and CEMss-CCR5-GPI-scFv X5 resulted in a constant but very low level of HIV-1 production similar to iDCs with captured HIV-1 Bru-3 alone, suggesting limited infection and spread. In contrast, co-culture of iDCs with captured HIV-1 Bru-3 and CEMss-CCR5-GPI-scFv AB65 resulted in a significantly increased level of HIV-1 replication (Figure 3B).
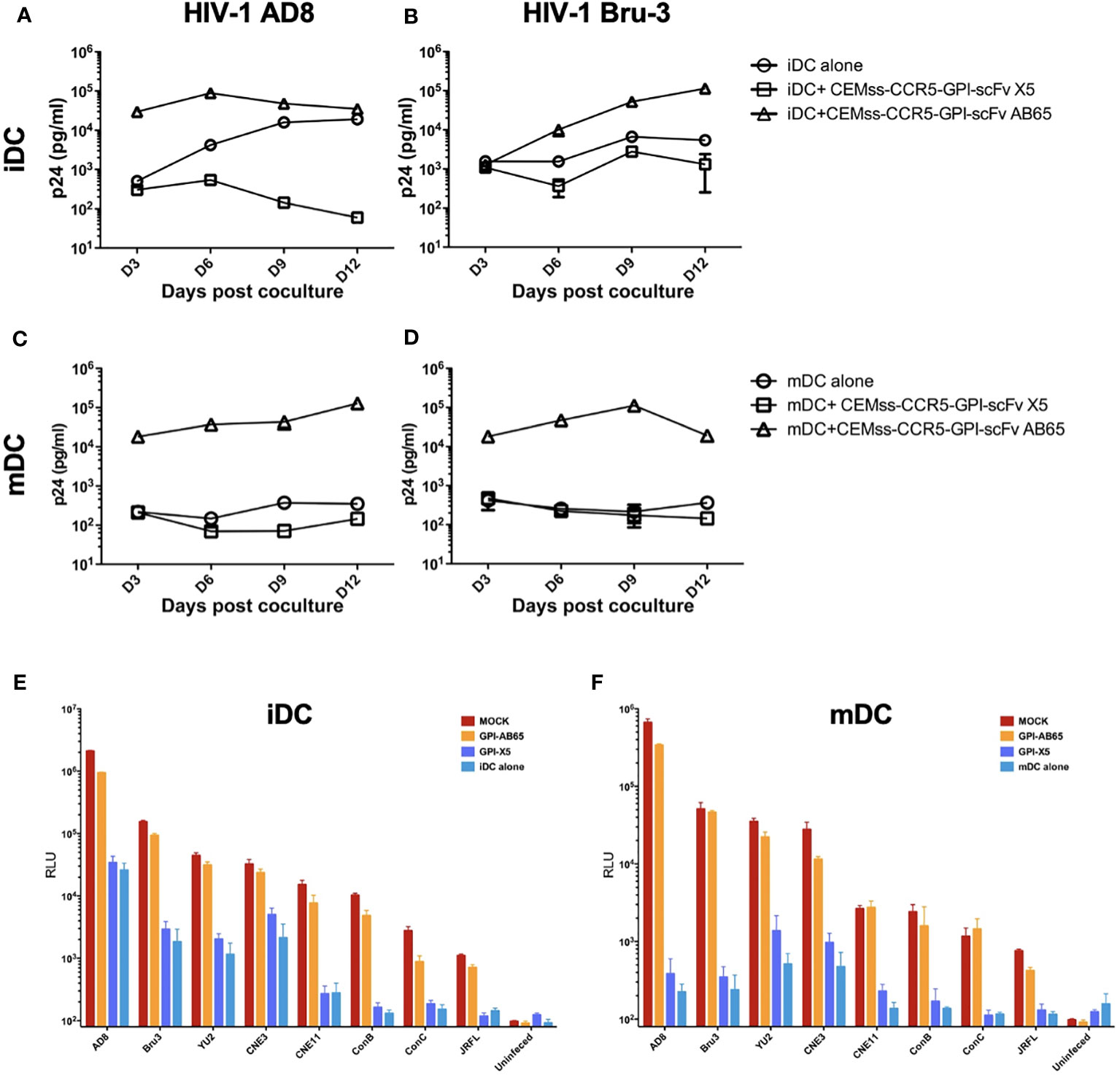
Figure 3 GPI-scFv X5-transduced CEMss-CCR5 cells are resistant to iDC- or mDC-mediated trans-infection of HIV-1. (A) HIV-1 gag p24 in the culture supernatants collected at days 3, 6, 9 and 12 post co-culture between mock-, GPI-scFv X5 or AB65-transduced CEMss-CCR5 cells and HIV-1 AD8-captured iDCs. (B) HIV-1 gag p24 in the culture supernatants collected at days 3, 6, 9 and 12 post co-culture between mock-, GPI-scFv X5 or AB65-transduced CEMss-CCR5 cells and HIV-1 Bru-3-captured iDCs. (C) HIV-1 gag p24 in the culture supernatants collected at days 3, 6, 9 and 12 post co-culture between mock-, GPI-scFv X5 or AB65-transduced CEMss-CCR5 cells and HIV-1 AD8-captured mDCs. (D) HIV-1 gag p24 in the culture supernatants collected at days 3, 6, 9 and 12 post co-culture between mock-, GPI-scFv X5 or AB65-transduced CEMss-CCR5 cells and HIV-1 Bru-3-captured mDCs. (E) RLU detected in mock-, GPI-scFv X5 or AB65-transduced CEMss-CCR5 cells trans-infected with or without HIV-1 pseudotyped AD8, Bru-3, Yu2, CNE3, CNE11, Con B, Con C or JRFL-captured iDCs at 48 hours post co-culture. RLU in HIV-1 pseudotyped AD8, Bru-3, Yu2, CNE3, CNE11, Con B, Con C or JRFL-captured iDCs alone were included for the comparison. (F) RLU detected in mock-, GPI-scFv X5 or AB65-transduced CEMss-CCR5 cells trans-infected with or without HIV-1 pseudotyped AD8, Bru-3, Yu2, CNE3, CNE11, Con B, Con C or JRFL-captured mDCs at 48 hours post co-culture. RLU in HIV-1 pseudotyped AD8, Bru-3, Yu2, CNE3, CNE11, Con B, Con C or JRFL-captured mDCs alone were included for the comparison. Infections were performed in triplicate. The average values ± standard deviation (SD) are shown.
When mDCs were used to mediate transmission of HIV-1, similar results were observed with R5- and X4-tropic viruses. Co-cultures of mDCs with captured HIV-1 AD8 or Bru-3 and CEMss-CCR5-GPI-scFv X5 resulted in very low level of HIV-1 replication like mDCs with HIV-1 AD8 or Bru-3. In contrast, co-culture of mDCs with captured HIV-1 AD8 or Bru-3 and CEMss-CCR5 expressing the negative control GPI-scFv AB65 construct resulted in a significantly higher level of HIV-1 production (Figures 3C, D). Similar results were observed in two additional experiments using iDCs and mDCs derived from different donors (Figure S2).
To specifically test the initial blocking of trans-infection by GPI-scFv X5, we performed single round infections with a panel of iDC- or mDC-captured HIV-luc pseudotyped with different Env proteins. Figures 3E, F show that inhibition of trans-infection by GPI-scFv X5 occurred when CEMss-CCR5-GPI-scFv X5 cells, but not CEMs-CCR5 or CEMss-CCR5-GPI-scFv AB65 cells, were co-cultured with iDCs or mDCs containing captured HIV-1 virions. These results were confirmed in two other independent experiments using iDCs and mDCs derived different donors (Figure S3).
Taken together, these data indicate that GPI-scFv X5 significantly blocks iDC or mDC-mediated trans-infection of CD4+ target cells by HIV-1. These results verify and extend our previously findings with iDCs (37). Furthermore, as the DCs were derived from different donors for Figures 2, 3A–F, the trans-inhibitory activity of GPI-X5 appears to be conserved.
Having demonstrated that GPI-scFv X5 in transduced CD4+ CEMss cell lines effectively blocked HIV-1 infection in trans from iDC or mDC, we next evaluated whether GPI-scFv X5 transduced human primary CD4+ T cells would be protected from DC-mediated HIV-1 infection. To facilitate monitoring transduced human primary CD4+ T cells, we inserted genes encoding GPI-HA-scFv X5 or AB65 into the pRRL-2A-eGFP vector, which expresses an enhanced green fluorescent protein (eGFP) as a marker. The resulting pRRL-GPI-HA-scFv X5- or AB65- 2A-eGFP transfer vectors (Figure 4A) were used to produce recombinant pseudotypes for transduction of human primary CD4+ T cells. Figure 4B shows that after a single round of transduction, over 80% of human primary CD4+ T cells became GFP+/GPI-HA-scFv+ double positive cells.

Figure 4 GPI-scFv X5-transduced human primary CD4+ T cells are resistant to trans-infection of iDC- or mDC-captured HIV-1. (A) Schematic diagram of lentiviral transfer vector pRRL-GPI-HA-scFv X5 or AB65-2A-GFP. RRE: rev response element; cPPT: central polypurine tract; EF1α: elongating factor 1α promoter; GFP: green fluorescent protein. (B) Expression of GFP and GPI-HA-scFv X5 (left panel) or AB65 (right panel) in human primary CD4+ T cells transduced with recombinant lentiviral vectors containing pRRL-GPI-HA-scFv X5 or AB65-2A-GFP, respectively, after a single round transduction. (C) A representative gating of p24+/GFP+ double positive cells in human primary CD4+ T cells transduced with recombinant lentiviral vectors containing pRRL-GPI-scFv X5 or AB65 trans-infected with or without iDCs containing captured HIV-1 AD8 at 2 days post co-culture. Human CD4+ T cells transduced with pRRL-GPI-scFv X5, but without iDCs-HIV-1 AD8 (upper-left panel). Human CD4+ T cells transduced with pRRL-GPI-scFv AB65, but without trans-infection with iDC-HIV-1 AD8 (lower left panel). Human CD4+ T cells transduced with pRRL-GPI-scFv X5 and cocultured with iDC- HIV-1 AD8 (upper right panel). Human CD4+ T cells transduced with pRRL-GPI-scFv AB65 and cocultured with iDC-HIV-1 AD8 (lower right panel). D to (G) Summary of percentages of Gag p24+/GFP+ cells at 3, 6, 9 and 12 days post co-culture from experiments using cells derived from three different human donors. (D) CD4+ T cells transduced with pRRL-GPI-HA-scFv X5 or AB65 and co-cultured with iDCs-HIV-1 AD8; (E) CD4+ T cells transduced with pRRL-GPI-scFv X5 or AB65 and co-cultured with iDCs-HIV-1 Bru-3; (F) CD4+ T cells transduced with pRRL-GPI-scFv X5 or AB65 and co-cultured with mDCs-HIV-1 AD8; (G) CD4+ T cells transduced with pRRL-GPI-scFv X5 or AB65 and co-cultured with mDCs-HIV-1 Bru-3. *stands for P<0.05; **stands for P<0.01; ***stands for P<0.001.
To determine if GPI-scFv X5 could effectively block trans-infection of autologous primary CD4+ T cells, iDCs or mDCs with captured HIV-1 were co-cultured with GPI-scFv X5 or AB65-transduced human primary CD4+ T cells. After co-culturing, HIV-1 infection was measured by intracellular HIV-1 Gag p24 staining. Figure 4C shows the gating of p24+/GFP+ double positive cells in GPI-scFv X5 or AB65-transduced human primary CD4+ T cells co-cultured with or without iDCs with captured HIV-1 AD8. Figures 4D–G summarize the percentage of Gag p24+/GFP+ cells at 3, 6, 9 and 12 days post co-culture of iDCs or mDCs with captured HIV-1 AD8 or Bru-3 and GPI-scFv X5 or AB65-transduced human primary CD4+ T cells from three individual donors and independent experiments. Co-culture of iDCs containing HIV-1 AD8 and GPI-scFv AB65-transduced human primary CD4+ T cells resulted in significant increases in the percentages of p24+/GFP+ cells with an average 35.6% at the peak (3 days) post co-culture. In contrast, co-culture of iDCs-HIV-1 AD8 and GPI-scFv X5-transduced human primary CD4+ T cells resulted in little or no p24+/GFP+ cells throughout the experiments (Figure 4D). Similarly, co-culture of iDCs-HIV-1 Bru-3 and GPI-scFv AB65-transduced human primary CD4+ T cells resulted in significant increase in the percentages of p24+/GFP+ cells with the average 51.7% at the peak (3 days) post co-culture. In contrast, co-culture of iDCs-HIV-1 Bru-3 and GPI-scFv X5-transduced human primary CD4+ T cells resulted in little or no HIV-1 p24+/GFP+ cells throughout the experiments (Figure 4E). Similar results were also obtained when co-cultures were carried out between HIV-1 mDCs with captured HIV-1 AD8 or Bru-3 and GPI-scFv X5 or AB65-transduced human primary CD4+ T cells (Figures 4F, G). Thus, in transduced human primary CD4+ T cells, GPI-scFv X5 significantly blocks trans-infection of iDC- or mDC-captured HIV-1, further supporting the findings in Figures 2, 3.
The co-culture experiments in Figure 3A suggested that expression of GPI-scFv X5 on T-cells may inhibit replication of R5-tropic HIV-1 AD8 in iDCs. To test whether replication of other HIV-1 strains in iDCs could be inhibited by GPI-scFv X5 expressing CD4+ T cells, we infected iDCs with two other R5-tropic HIV-1 clones, Mj4 and Yu-2. The iDCs were then co-cultured with or without CEMss-CCR5-GPI-scFv X5 or AB65 transduced cells. Figures 5A, B show that co-culture of iDCs containing either HIV-1 Mj4 or Yu-2 and CEMss-CCR5-GPI-scFv AB65 control resulted in high levels of HIV-1 production. Both viruses also replicated in the iDCs alone. In contrast, co-culturing iDCs infected with either HIV-1 Mj4 or Yu-2 and CEMss-CCR5-GPI-scFv X5 resulted in decreasing HIV-1 production. This experiment has been conducted two times with similar results. Thus, intercellular interactions may enable GPI-scFv X5 when expressed on CEMss-CCR5 cells to block replication of R5-tropic HIV-1 in neighboring iDCs.
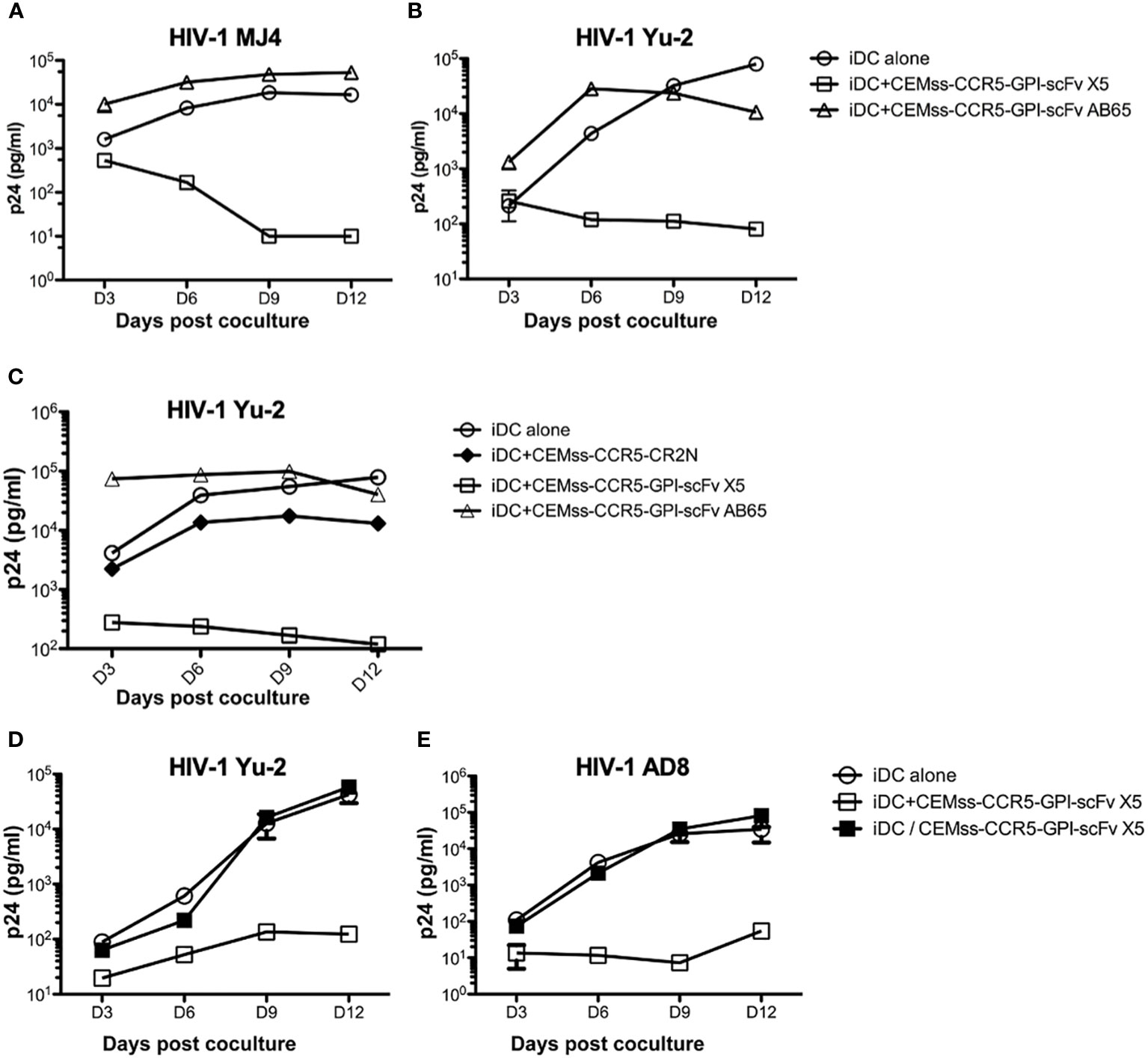
Figure 5 Trans-blockage of R5-tropic HIV-1 in iDCs by GPI-scFv X5. (A) HIV-1 Gag p24 in the culture supernatants collected at days 3, 6, 9 and 12 post co-culture between GPI-scFv X5 or AB65-transduced CEMss-CCR5 cells and iDCs with captured HIV-1 MJ4 or iDCs with HIV-1 MJ4 alone. (B) HIV-1 Gag p24 in the culture supernatants collected at days 3, 6, 9 and 12 post co-culture between GPI-scFv X5 or AB65-transduced CEMss-CCR5 cells and iDCs with captured HIV-1 Yu-2 or iDCs with HIV-1 Yu-2 alone. (C) HIV-1 Gag p24 in the culture supernatants collected at days 3, 6, 9 and 12 post co-culture between CEMss-CCR5-GPI-scFv X5 or AB65 or CEMss-CCR5-CR2N cells and iDCs with captured HIV-1 Yu-2 or iDCs with HIV-1 Yu-2 alone. D and (E) HIV-1 Gag p24 in the culture supernatants collected at days 3, 6, 9 and 12 post co-culture between CEMss-CCR5-GPI-scFv X5 cells and iDCs with captured HIV-1 Yu-2 (D) or AD8 (E) in a regular plate (open squares) versus a transwell plate (closed squares) or iDCs with HIV-1 Yu-2 (D) or AD8 (E) alone in a regular plate. Infections were performed in triplicate. The average values ± standard deviation (SD) are shown.
To test whether restriction of R5-tropic HIV-1 in the iDC-T cell co-cultures is due to blocking of cell-cell transmission to CD4+ T cells expressing GPI-scFv X5, we co-cultured iDCs-HIV-1 Yu-2 with or without CEMss-CCR5-GPI-scFv X5 or AB65 or CEMss-CCR5-CR2N cells. In CEMss-CCR5-CR2N cells, the ccr5 gene was disrupted using CRISPR/Cas9 system. Consequently, CEMss-CCR5-CR2N cells are rendered completely resistant to CCR5-tropic HIV-1 Yu-2 infection (41). Consistent with the experiment in Figure 5B, co-culture of iDCs-HIV-1 Yu-2 and CEMss-CCR5-GPI-scFv AB65 resulted in significantly higher levels of HIV-1 while co-culture of iDCs-HIV-1 Yu-2 with CEMss-CCR5-GPI-scFv X5 resulted in significant suppression of HIV-1 production than iDCs with virus alone (Figure 5C). Interestingly, co-culture of iDCs-HIV-1 Yu-2 and CEMss-CCR5-CR2N cells resulted in a delayed increase in virus production like iDCs with virus alone. While there is a small difference between iDC alone and iDC co-culture with CEMss-CCR5-CR2N, this was attributed to an over-growth of HIV-1 resistant CEMss-CCR5-CR2N cells, which reduced viability of the cells in the culture. These data indicate that CEMss-CCR5-CR2N cells resist trans-infection by iDC-captured HIV-1 Yu-2. However, without GPI-scFv X5 modification, these cells do not block viral replication in iDCs. Moreover, they suggest that the suppression of R5-tropic HIV-1 infection of iDCs observed in the co-cultures of iDCs-R5-tropic HIV-1 and GPI-scFv X5-expressing CEMss-CCR5 cells is not due to inhibition of trans-infection of the T cells by GPI-scFv X5.
To test whether the intercellular inhibition of R5-tropic HIV-1 replication in iDCs by GPI-scFv X5 transduced CD4+ T cells requires cell-cell contact, iDCs infected with CCR5 tropic HIV-1 AD8 or Yu-2 were co-cultured with or without CEMss-CCR5-GPI-scFv X5 separated by a trans-well barrier. Figures 5D, E show that co-culture of HIV-1 AD8 or Yu-2 infected iDCs and CEMss-CCR5-GPI-scFv X5 cells yields significantly lower levels of HIV-1 Gag p24 in the culture supernatants than co-culture of iDCs with only the viruses. Interestingly, separation of the iDCs and CEMss-CCR5-GPI-scFv X5 cells by a trans-well membrane abrogated the inhibitory effect, indicating that the intercellular inhibition of R5-tropic HIV-1 replication in iDCs by GPI-scFv X5 expressing CEMss-CCR5 cells requires close contact with iDCs. Taken together, these results suggest that expression of GPI-scFv X5 in neighboring T cells has the capacity to block HIV-1 replication in trans in iDCs.
Having shown that expression of GPI-scFv X5 on CEMss trans-restricts HIV-1 in iDCs, we next evaluated if GPI-scFv X5 expression on T cells would also block infection of neighboring untransduced CD4+ T cells. Human primary CD4+ T cells and CD8+ T cells were transduced with the pRRL-GPI-scFv X5-2A GFP vector as described above (Figure 4A). Figure 6A shows that after a single round of transduction, over 70% of CD4+ T cells and 50% of CD8+ T cells became GFP+ positive.
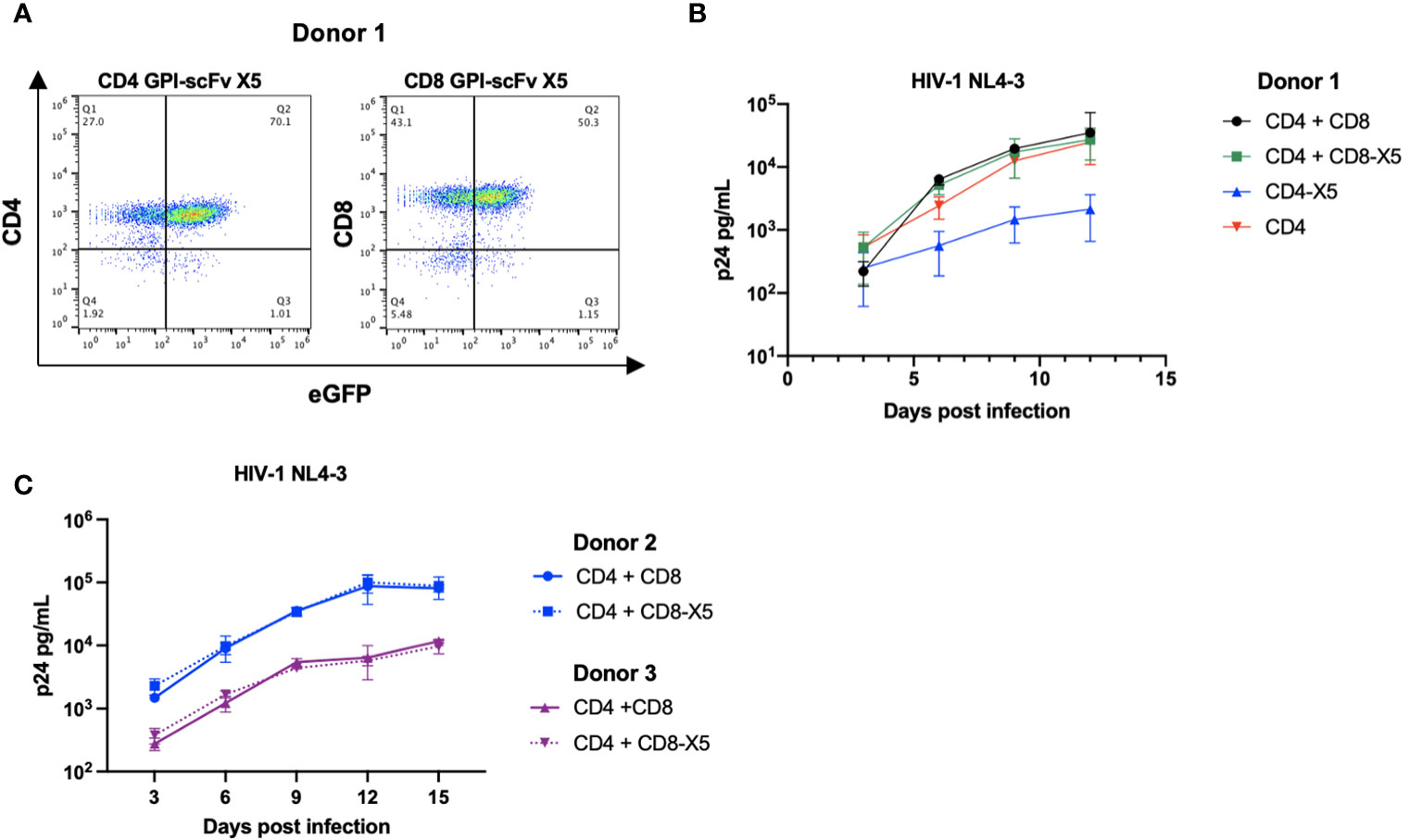
Figure 6 GPI-scFv X5 protects CD4 T cells in cis but not in trans when displayed on CD8 T cells. (A) Expression of CD4+ (left panel) or CD8+ (right panel) and GFP in human primary CD4+ T cells and CD8+ T cells transduced with recombinant lentiviral vectors containing pRRL-GPI-scFv X5-2A-GFP, respectively, after a single round transduction. (B) HIV-1 Gag p24 in the culture supernatants collected at days 3, 6, 9, and 12 post culture of CD4+, GPI-scFv X5 CD4+ or co-cultured with CD8+, or GPI-scFv X5 CD8+ infected with HIV-1 NL4-3. (C) Two additional donors, Donor 2 (blue) and Donor 3 (purple), were evaluated for GPI-scFv X5 modified CD8 T cell mediated trans protection of CD4 T cells. HIV-1 Gag p24 in the culture supernatants collected at days 3, 6, 9, 12, and 15 post co-culture of CD4+ with unmodified CD8+ (solid line) or GPI-scFv X5 CD8+ (dotted line) infected with HIV-1 NL4-3. Infections were performed in triplicate. The average values ± standard deviation (SD) are shown.
Human primary CD4+ T cells or primary GPI-scFv X5 transduced CD4+ T cells were cultured alone or co-cultured with autologous human primary CD8+ T cells or primary GPI-scFv X5 transduced CD8+ T cells, and then infected with HIV-1 NL4-3. Expression of GPI-scFv X5 on CD4+ T cells resulted in significant suppression of HIV-1 p24 production than CD4+ T cells that do not express GPI-scFv X5 (Figure 6B). In contrast, co-culture of CD4+ T cells with CD8+ T cells or GPI-scFv X5 transduced CD8+ T cells resulted in a similar level of HIV-1 production, suggesting that expression of GPI-scFv X5 on CD8+ T cells does not enhance control of HIV-1 in cocultures with CD4+ T cells (Figure 6B). Similar results were observed in additional experiments using human primary CD4+ and CD8+ T cells derived from different donors (Figure 6C).
To determine potential mechanisms of inhibition of HIV replication in iDCs by GPI-scFv X5 expressing CD4+ T cells, we first examined if recombinant GPI-anchored proteins can be transferred from transduced CEMss cells to iDCs. To evaluate transfer of GPI-anchored proteins, CEMss cells were transduced with a GPI-green fluorescent protein (GFP). iDCs were marked using a fluorescent DiD membrane dye and incubated with CEMss GPI-GFP for 2 hours. After co-culture, the movement of GPI-GFP were evaluated by Amnis ImageStream live-cell imaging. Figure 7A shows that after co-culture, GPI-GFP is significantly enriched on DiD dyed iDCs. Further, iDC and CEMss GPI-GFP cell-cell synapse can be seen where GPI-GFP is enriched on iDCs suggesting that transfer of GPI-GFP can occur from GPI-GFP transduced CEMss to iDCs after 2 hours of co-culture and may occur through cell-cell synapse engagement (Figure 7A). Cell populations were gated and analyzed by DiD and eGFP expression showing 50% of cells were double positive, indicating that the transfer of GPI anchored GFP occurs in half of cells when cocultured (Figures 7B, C). Next, we wanted to further confirm colocalization of GPI-GFP with the membrane dye DiD. Transfer of GPI-GFP from transduced CEMss cells to iDCs were evaluated after 2 hours of co-culture by Airyscan confocal microscopy. A similar pattern of enrichment of GPI-GFP in DiD dyed iDCs was also observed (Figures 7D, E). GPI-GFP is enriched on iDCs and co-localization of GPI-GFP and DiD dye on iDCs forming yellow punctae on iDCs were observed (Figure 7E). Taken together, GFP when anchored to GPI and expressed on CEMss cells by a single round of transduction migrates from CEMss transduced cells to iDCs after 2 hours of co-culture.
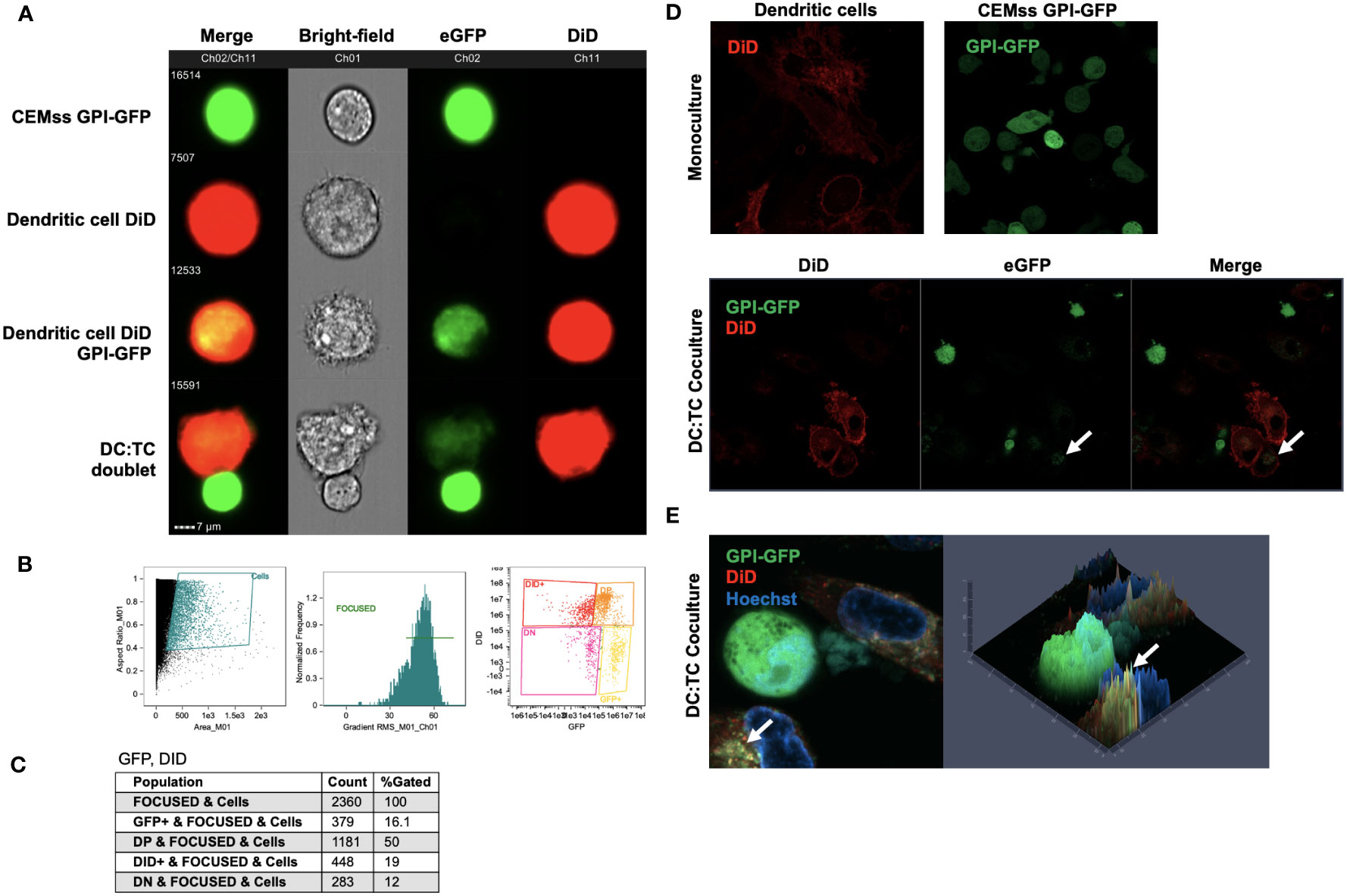
Figure 7 Migration of GPI-GFP from GPI-GFP transduced CEMss cells to iDCs. (A) Representative images by Amnis ImageStream Mark-II of single cell GPI-GFP CEMss cells, DiD dyed iDCs, GPI-GFP transfer to DiD dyed iDCs and cell-cell conjugates between DiD dyed iDCs and GPI-GFP CEMss cells. T cells are shown in Ch02 expressing GPI anchored GFP. DCs dyed with DiD are shown in Ch11. (B) Gating strategy for analysis of DC : TC cocultures. Cells were analyzed by aspect ratio vs area to identify single cells. RMS feature of Ch1 (Bright-field) were then used to plot gradient by normal frequency to select for focused events. Cells were finally selected for positivity in both Ch02 and Ch11 looking for expression of eGFP and DiD, respectively. (C) Population count of total cells sorted and counts of expression for GFP positive, double positive (DP), DiD positive, or double negative (DN) populations and relative percent. (D) AiryScan Confocal microscopy of monoculture DiD dyed DCs, CEMss GPI-GFP, or DC : TC coculture. The top left panel shows a representative image of DC monocultures stained with DiD dye. The top right panel shows CEMss cells that express GPI anchored GFP. The bottom three panels show DC : TC coculture at a 1:1 ratio. GPI-GFP was detected on DiD dyed dendritic cells when imaged after 2 hours of coculture as indicated by white arrows. (E) Left panel shows representative image by AiryScan confocal microscopy of DC : TC cocultures after 2 hours. DCs were pre-stained with DiD and cocultured with GPI-GFP CEMss cells. Cocultures were then stained with Hoechst nuclear blue dye. White arrows indicate detection of GPI-GFP on DCs. Right panel shows fluorescence signal intensity plot. Yellow punctae indicate GPI-GFP co-localization with DiD.
While we demonstrated that GPI-GFP could migrate from GPI-GFP transduced CEMss cells to iDCs, we next determined if scFvs anchored by GPI attachment were also transferred to iDCs. Further, it is possible that GPI-GFP may be taken up by iDCs internally and sequestered in vesicles but not displayed on the surface of the cell membrane. To evaluate this, we used CEMss cells transduced with the recombinant lentiviral vector pRRL-GPI-HA-scFv X5-2A-GFP. Cells transduced by pRRL-GPI-HA-scFv X5-2A-GFP result in internal GFP expression, marking transduced cells, while HA tagged GPI-scFvs are anchored to the outer leaflet of the plasma membrane. As previously shown, after a single round of transduction, cells are double positive for GPI-scFv and GFP (Figures 4A, B).
GPI-scFv X5 CEMss cells were cultured with DiD dyed iDCs for 2 hours and analyzed by Amnis ImageStream Mark-II. Surface expression of HA within the GPI-scFv X5 construct was then antibody-labeled with a PacBlue-conjugated anti-HA tag antibody. In focused cells, single versus doublet cells were gated by aspect ratio and subsequently analyzed for DiD and GFP expression (Figure 8A). Over 50,000 events were imaged with 16% of focused cells double positive for both GFP and DiD (Figure 8B). In Figure 8C, GPI-scFv X5 labeled with PacBlue-conjugated anti-HA are seen abundantly displayed on the surface of transduced CEMss GPI-scFv X5 cells that are marked by internal GFP expression. In DC : TC doublets, GPI-scFv X5 labeled by anti-HA PacBlue is seen enriched on the surfaces of iDCs dyed with DiD, suggesting that GPI-scFv X5 is transferred from transduced CEMss cells to iDCs after 2 hours of co-culture. Importantly, anti-HA PacBlue labeled GPI-scFv X5 is detected on the surface of DiD labeled iDCs, but not GFP, which is internally expressed in transduced CEMss cells, indicating that scFvs when anchored to GPI can be transferred from T cells to iDCs in co-culture (Figure 8C). When analyzing CEMss GPI-scFv X5 transduced cells, over 94% of cells are double positive for both GFP and PacBlue labeled HA tag after a single round of transduction (Figure 8C). Additionally, almost 50% of untransduced iDCs received GPI-scFv X5 from CEMss cells as detected by surface expression of HA tagged GPI-scFv X5 after just 2 hours of co-culture with CEMss GPI-scFv X5 transduced cells, (Figure 8E). Furthermore, GPI-scFv X5 CEMss cells when cocultured with primary CD4+ T cells were not found to pass GPI-scFv X5 from transduced CEMss cells to untransduced primary CD4+ T cells (Figures S4A–C). That is, in TC : TC co-cultures, GPI-X5 does not transfer from transduced CD4+ T cells to untransduced CD4+ T cells. Additionally, when DCs were added to TC : TC co-culture, transfer of GPI-scFv X5 from transduced CEMss cells to iDCs and subsequently from iDCs to naïve human primary CD4 T cells was not observed (Figures S5A, B).
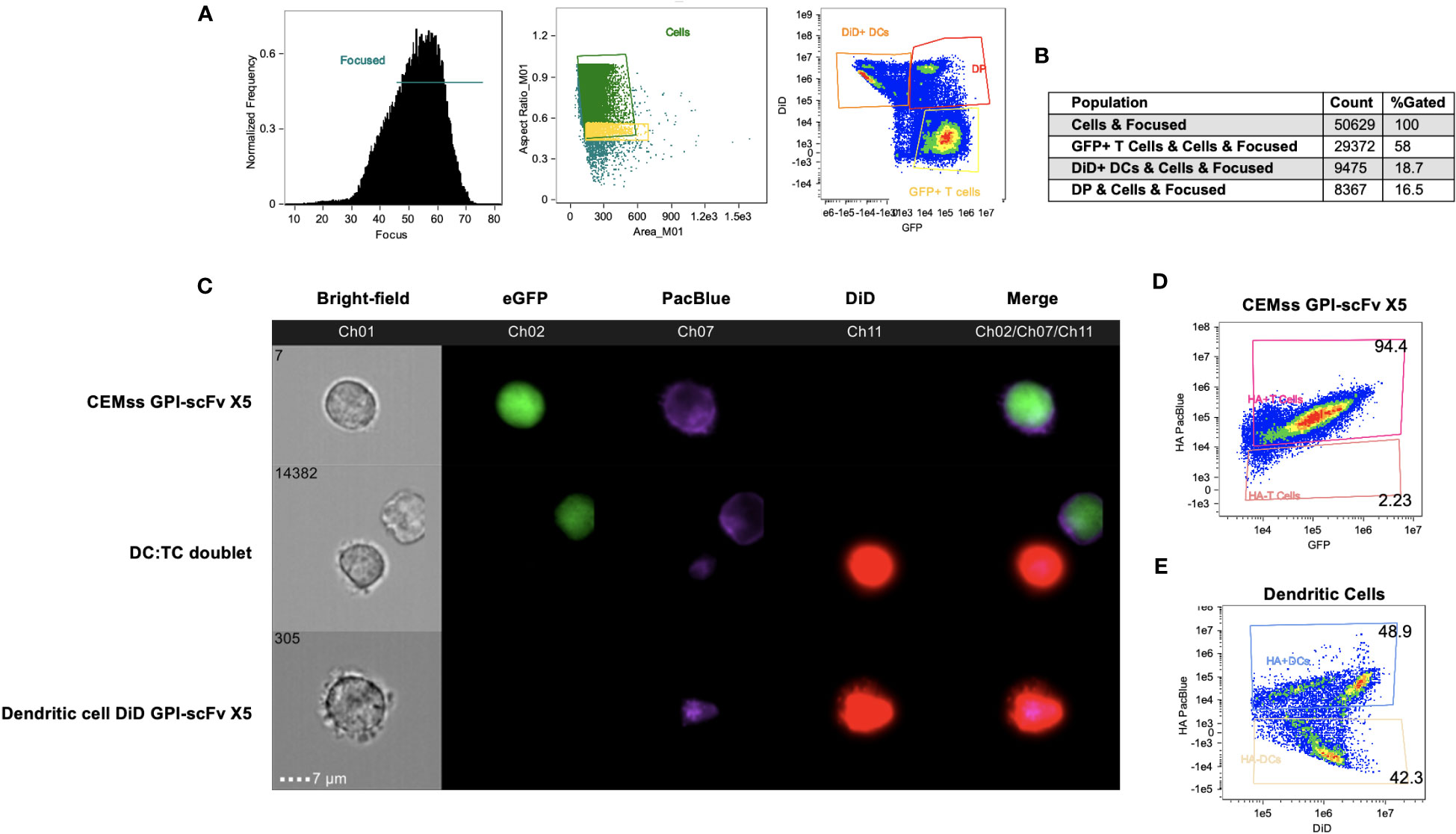
Figure 8 GPI-scFv X5 is transferred from CEMss cells to dendritic cells. GPI-scFv X5 CEMss cells were cocultured with DiD dyed DCs for 2 hours and analyzed by Amnis ImageStream Mark-II. (A) Gating strategy for analysis of DC : TC cocultures. RMS feature of Ch1 (Bright-field) were used to plot gradient by normal frequency to select for focused events. Cells were then analyzed by aspect ratio vs area to identify single cells and likely doublets. Cells were finally selected for positivity in both Ch02 and Ch11 looking for expression of eGFP and DiD, respectively. (B) Population count of total cells sorted and counts of expression for GFP positive, DiD positive, or double positive (DP) populations and relative percent. (C) Representative images of CEMss GPI-scFv X5 co-expressing GFP and GPI-HA-scFv X5 (top panel), DC : TC doublets show anti-HA PacBlue tagged GPI-HA-scFv X5 detected on DCs after DC : TC coculture (middle panel), and DiD dyed DC positive for GPI-HA-scFv X5 (bottom panel). (D) Coexpression of HA PacBlue and GFP on GPI-scFv X5 CEMss cells are 94.4% double positive. (E) DiD dyed DCs analyzed for coexpression of DiD and HA PacBlue after 2 hours of coculture with GPI-HA-scFv X5 CEMss reveals two distinct population of DiD+ HA PacBlue+ (48.9%) and DiD+ HA PacBlue- (42.3%).
Taken together, these data show that GPI-scFv X5 on transduced CEMss cells migrate to iDCs and are displayed on the outer membrane of iDCs after 2 hours of co-culture. This suggests that protection of iDCs in trans mediated by contact with CEMss GPI-scFv X5 cells may be due to transfer of GPI-scFv X5 from transduced CEMss to iDCs.
HIV-1 transmission occurs by both cell-free and cell-cell infection with cell-cell mechanisms found to be more efficient in spreading virus and less susceptible to inhibition by neutralizing antibodies and entry inhibitors than cell-free virus infection (13–15, 17–26). Furthermore, increasing evidence indicate that cell-cell transmission of HIV-1 may play an important role in the establishment of systemic infection as well as in virus spread within lymphoid tissues in vivo (16, 42). For example, Mooruka et al. showed that in a BLT humanized mouse model HIV-1-infected T cells in lymph nodes through virological synapses. Blocking the egress of T cells from lymph nodes into efferent lymph vessels at the onset of HIV-1 infection limited HIV-1 dissemination and reduced plasma viremia (16, 43). Sewald et al. showed that in secondary lymph tissues, HIV-1 and murine leukemia virus (MLV) are first captured by sinus-lining macrophages via CD169 that recognizes gangliosides. Virus-captured by macrophages then trans-infect B-1 cells. Infected B-1 cells then migrate into the lymph node to spread infection through virological synapses (42). Thus, development of an effective way to block cell-cell transmission of HIV-1 could have potential advantages for HIV-1 prevention and therapy.
In the present study, we demonstrated that after co-culturing iDCs or mDCs with captured HIV-1 and GPI-scFv-transduced CD4+ cell lines as well as human primary CD4+ T cells, the anti-HIV-1 GPI-scFv X5 inhibitor almost completely blocks trans-infection of the target cells (Figures 2–4). Importantly, CD4 is a lipid raft-associated protein and richly present in the contact zone (7, 35, 36). The epitope recognized by antibody X5 resides within a conserved region (amino acid residues 417 to 434) of the Env gp120 core, which is nearby the CD4 and co-receptor binding sites (44). We envision that when CD4 binds the native envelope spike on virions, GPI-scFv X5 residing in lipid rafts may quickly and efficiently engage the CD4-induced transiently exposed epitope in Env. This may block the conformational transition of Env at a pre-fusion intermediate step, and inhibit viral entry.
One surprising finding in this study is that GPI-scFv X5 not only effectively blocks HIV-1 cell-cell infection of CD4+ cell lines or human primary CD4+ T cells in trans from either iDC or mDC-captured HIV-1, but it also blocks HIV-1 infection of the iDCs (Figures 3, 5). The trans-inhibitory activity requires cell-cell contact with GPI-scfv X5 transduced CD4+ T cells. Inhibition was not observed when iDCs with captured HIV-1 were co-cultured with a R5-tropic HIV-1-resistant cell line, CEMss-CCR5-CR2N cells (Figure 5), indicating that the trans-infection neutralization of R5-tropic HIV-1 replication in iDCs is not due to the infection resistant phenotype of GPI-scFv X5-transduced CEMss-CCR5 cells. Additionally, GPI-scFv X5 expression on human primary CD8+ T cells does not enhance neutralization of HIV-1 in CD4+ T cells when co-cultured (Figure 6). Thus, the trans-inhibitory effect of GPI-scFv X5 may depend on stable synapses that occur between CD4+ T cells and iDCs, but not between T cells.
Using live cell microscopy and imaging flow cytometry, we demonstrated transfer of engineered GPI-anchored proteins from transduced T cells to iDCs using a GPI-GFP construct and GPI-scFv X5. The migration of GPI-GFP from transduced CEMss T cells to iDCs was observed when cells were co-cultured (Figure 7). Additionally, we found that GPI-scFv X5 migrates from CEMss transduced cells to iDCs in co-culture (Figure 8). These data provide further support that recombinant GPI-anchored proteins can be transferred from transduced T cells to untransduced iDCs. Importantly, we also showed that GPI-scFv X5 is displayed on the surface of iDCs where it would be available to neutralize infection. By contrast, GPI-scFv X5 was not observed to be transferred from transduced T cells to non-transduced T cells. Consequently, GPI-scFv X5 transduced T cells do not neutralize HIV-1 replication in non-transduced CD4+ T cells in coculture. Together, these data further support transfer of anti-HIV GPI-anchored scFv as a mechanism of inhibition of viral infection of iDCs.
Cell-cell transfer of GPI-anchored proteins has been shown in vivo, such as transfer of complement restriction factors CD55 and CD59 from erythrocytes to the vascular epithelium and from male genital tract epithelium to spermatozoa (45). The latter serves a physiological function during sperm maturation, sperm storage and fertilization (46). Thus, if cell-cell transfer of GPI-scFv X5 also occurs in vivo, this should have important implications for the utility of GPI-scFv inhibitor-based therapy against HIV-1.
Finally, anti-HIV GPI-anchored scFv inhibitors with such remarkable breadth of neutralization activity against both cell-free and cell-cell transmissions of HIV-1 should have potential either alone or in the combination with other anti-HIV-1 gene constructs, such as GPI-HCDR3 PG16 or GPI-C34 fusion inhibitor, to be developed into anti-viral agents for HIV-1 prevention and therapy (40, 47). For example, GPI-scFv-X5 and GPI-HCDR3 PG16, due to their different specificities, could be co-delivered into hematopoietic progenitor cells or human primary CD4+ T cells of HIV-1 patients ex vivo through lentiviral vector transduction. The GPI-anchored inhibitor modified hematopoietic progenitor cells or human primary CD4+ T cells could then be transfused to the patients as described by DiGiusto et al. and Tebas et al., respectively (48, 49). Alternatively, GPI-scFv X5 could be used to modify and protect HIV-specific T cells to improve their persistence and activity (50–53). HIV specific T cells are a reservoir that contributes to persistence and immune dysfunction (54). Protecting them from new rounds of infection may improve immune function and reduce the viral reservoir. However, many hurdles, such as efficient engraftment, self-renewal, and linage cell differentiation, sustainable transgene expression, avoidance of potential insertion mutagenesis, have to be worked out in HIV-1 infection humanized mouse models or SHIV infection macaque models before the clinical efficacy of GPI-scFv X5 or GPI-HCDR3 PG16-transduced hematopoietic progenitor cells or human primary CD4+ T cells can be tested in human patients. Taken together, we show that GPI-scFv X5 may be a promising approach for HIV-1 prevention and therapy by restricting HIV-1 cis- and trans-infection mechanisms of CD4+ cells.
The packaging cell line 293FT was purchased from Invitrogen Life Technologies and maintained in complete DMEM medium [i.e. high glucose DMEM supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml), streptomycin (100 μg/ml)] plus G418 (500 μg/ml) (Invitrogen Life Technologies). CEMss-CCR5 cells and stably transduced TZM.bl-GPI-scFv X5, TZM.bl-GPI-scFv AB65, CEMss-CCR5-GPI-scFv X5, CEMss-CCR5-GPI-scFv AB65 and CEMss-CCR5-CR2N were generated before and maintained in complete DMEM (41, 55).
The TZM.bl cell line was obtained from J. Kappes and X. Wu via the NIH AIDS Research and Reference Reagent Program (ARRRP; Germantown, MD). TZM.bl cells are a well described reporter cell line for their utility in HIV-1 studies. Briefly, TZM.bl cells are derived from HeLa cells that have been transduced to express HIV-1 receptors and co-receptors CD4, CCR5, and CXCR4, respectively (34). The cell line was additionally modified by lentiviral vectors to express an HIV-1 promoter controlling E. coli β-galactosidase and firefly luciferase reporters (56, 57). Thus, infection of TZM.bl cells by HIV-1 triggers expression of reporter constructs by HIV-1 Tat mediated mechanisms. Quantitation of luciferase by luminescence intensity and reported as Relative Light Units (RLU) were conducted utilizing the BrightGlo luciferase activity kit according to the manufacturer’s instructions (Promega).
Human peripheral blood mononuclear cells (PBMCs) of healthy donors were purchased from the Gulf Coast Regional Blood Center in Houston, Texas, Shanghai Blood Center or from the Blood Bank of the Shanghai Hospital, Shanghai, China. The iDCs were prepared as previously described (37). Briefly, human monocytes were isolated from buffy coats using a Ficoll gradient and a subsequent CD14 selection step using the MACS system according to manufacturer’s instructions (Miltenyi Biotec). Purified monocytes were differentiated into iDCs in the presence of IL-4 (50 ng/ml) and GM-CSF (50 ng/ml) (R&D System). On day 6, iDC phenotype was confirmed by flow cytometry using antibodies against CD3, CD11c, CD14, CD83, CD86 and CD209 (CD-SIGN) (see below).
To generate mDCs, iDCs were further cultured in the presence of LPS (100 ng/ml) for another 2 days. mDC phenotype was confirmed by flow cytometry using antibodies against CD83, CD86, CD209 and HLA-DR (see below).
Human primary CD4+ T cells were enriched from above CD14+ monocyte-depleted PBMCs by negative selecting magnetic beads according to the manufacturer’s instructions (Thermo Fisher Scientific) and resuspended in the complete RPMI 1640 medium (i.e. RPMI 1640 medium supplemented with 15% FBS, 2mM L-glutamine, 1 mM sodium pyruvate, penicillin [100 U/ml], and streptomycin [100 μg/ml]) supplemented with human rIL-2 (100 IU/ml, R&D System) before being activated and transduced with recombinant lentiviruses (see below).
Fusion genes encoding the GPI-scFv X5 or AB65 were amplified by PCR using pRRL-GPI-scFv X5 or AB65 as templates with a pair of primers (Forward: 5’-CCATGGGCTTGCTGCTGACTGGCAGCGGCGCCACCAACTTCA-3’, Reverse: 5’-AGTCGCCGTGAACGTTCTTTT-3’). GPI-scFv X5 or AB65 genes were transferred into pRRLsin-18.PPT.EF1a.GFP.Wpre (41). The resulting lentiviral transfer constructs were designated pRRL-GPI-scFv X5 or AB65-2A-GFP.
Recombinant lentiviral viruses were generated as described previously (37). Briefly, 4 X 106 293FT cells were seeded onto a P-100 dish in 10 ml complete DMEM. After culturing overnight, cells were co-transfected with 20 μg transfer construct pRRL-GPI-scFv X5 or AB65-2A-GFP, 10 μg packaging construct encoding HIV-1 Gag/Pol (pLP1), 7.5 μg plasmids encoding the vesicular stomatitis virus G protein envelope (pLP/VSV-G), and 7.5 μg HIV-1 Rev protein (pLP2) (Invitrogen) using a calcium phosphate precipitation method. Sixteen hours later, culture supernatants were removed and replaced with fresh complete DMEM plus 1 mM sodium butyrate (Sigma). After eight hours, supernatants were again removed and replaced with fresh DMEM plus 4% FBS. After another 20 hours, the culture supernatants were harvested and concentrated by ultracentrifugation. The recombinant lentivirus pellets were resuspended in a small volume of DMEM and stored in aliquots in a -80°C freezer. Recombinant lentivirus titers were determined as previously described (37).
To transduce human primary CD4+ T cells, human primary CD4+ T cells were enriched from CD14+ monocyte-depleted PBMCs by negative selecting magnetic beads (see above). 2.5 × 105 human CD4 T cells per well were activated by mixing with anti-CD3/CD28 antibody-coated beads (Thermo Fisher Scientific) at 1:1 ratio in 500 µl complete RPMI 1640 medium supplemented with human rIL-2 (100 IU/ml) in 48 well plates. After 24 hours, 5 x 106 TCID recombinant lentiviral viruses containing pRRL-GPI-scFv X5 or AB65-2A-GFP in complete RPMI 1640 supplemented with human rIL-2 (100 IU/ml) and 8 µg/ml polybrene were added into cell suspension at final volume of 750 µl. The plates were centrifuged at 1,500g and 37°C for 2 hours to facilitate transduction. After overnight incubation at 37°C, 500 µl supernatant was removed and 750 µl fresh complete RPMI 1640 medium supplemented with human rIL-2 (100 IU/ml) were added into cells. The anti-CD3/CD28 antibody-coated beads in human CD4+ T cells were removed by DynaMag magnet after 4 days activation. Transduced human CD4+ T cells were resuspended in 2 ml complete RPMI 1640 supplemented with human rIL-2 (100 IU/ml) and cultured in 24-well plates for additional 2 days. Transduction efficiency was estimated by GFP and GPI-scFv X5 or AB65 expression using FACS analysis. Untransduced human CD4+ T cells were used as a recipient cells in the trans-infection study (see below).
HIV-1 molecular clones AD8, Yu2, Mj4 (CCR5 tropic) and Bru-3 (CXCR4 tropic) were produced by transfecting proviral plasmids pAD8 (58), pYu2 (59), pMj4 (60) and pBru-3 (61) into 293 FT cells using a calcium phosphate precipitation as described before (37). The 50% tissue culture infection dose (TCID50) was determined by serial titration of viruses in TZM.bl cells (37).
To generate HIV-1 pseudotypes, 4 X 106 293FT packaging cells were co-transfected with 10 μg of an HIV-1-luciferase transfer vector and 1 μg of a DNA plasmid encoding one of several HIV-1 envelopes Bru-3, AD8, Yu2, JRFL, consensus B, consensus C, CNE3 and CNE11 using a calcium phosphate precipitation method (62). AD8, Yu2 and JRFL are derived from R5-tropic subtype B viruses (58, 63–66). CNE3 is derived from an R5-tropic CRF01_AE recombinant (66). CNE11 is derived from an R5-tropic subtype B’ virus. The pseudotype-containing supernatants were harvested and stored in aliquots at -80°C. Relative Light Units (RLU) of HIV-1 or 10A1 pseudotypes were determined as previously described (62).
To determine the phenotype of iDCs, monocyte-derived iDCs were incubated with PE-conjugated anti-CD83, CD86, DC-SIGN, or HLA-DR, and FITC-conjugated anti-CD14 and APC-conjugated anti-CD3, CD11c antibodies (BD BioSciences) for 45 min on ice. To determine the phenotype of mDCs, monocyte-derived mDCs were incubated with PE-conjugated anti-CD83, CD86, DC-SIGN or HLA-DR antibodies for 45 min on ice. Cells then were washed twice with FACS buffer (PBS containing 1% BSA and 0.02% NaN3) and fixed with 1% formaldehyde in 0.5 ml of FACS buffer. FACS analysis was performed on a FACScan LSR II (Becton Dickinson, Mountain View, CA). FACS data were analyzed with FlowJO 7.6.1.
To determine transgene expression, 3 X 105 mock-, pRRL-GPI-scFv X5 or AB65-2A-GFP-transduced human primary CD4 T cells were stained with rabbit anti-HA tag antibody at 40C for 40 min. Cells were washed twice with FACS buffer and stained with Alexa633 conjugated goat anti-rabbit IgG antibody on ice for 30 min. Cells then were washed twice with FACS buffer (PBS containing 1% BSA and 0.02% NaN3) and fixed with 1% formaldehyde in 0.5 ml of FACS buffer. FACS analysis was performed on a FACScan LSR II (Becton Dickinson, Mountain View, CA). FACS data were analyzed with FlowJO 7.6.1.
Intracellular HIV-1 Gag p24 detection was performed as described before (17). Briefly, at various indicated time intervals post co-culture between HIV-1-captured iDCs or mDCs and pRRL-GPI-scFv X5 or AB65-2A-GFP-transduced human primary CD4 T cells, small portion of cell mixture was harvested, washed with 1 ml FACS buffer, fixed and permeabilized by Cytofix/Cytoperm kit (Becton Dickinson). Cells were re-suspended in 50ul Perm/Wash buffer with 0.8 µl PE-conjugated anti-Gag p24 antibody (KC57, Beckman Coulter) and incubated for 30 min on ice. After washing twice with 1ml Perm/Wash buffer, the cells were re-suspended in 400 µl FACS buffer and analyzed by FACS Fortessa (Becton Dickinson) using software FlowJo and GraphPad Prism.
To examine DC-mediated HIV-1 infection of GPI-scFv transduced TZM.bl cells, human iDCs or mDCs were incubated with HIV-1 AD8 and Bru3 at MOI 5 for 2 hours in triplicate cultures. Cells were then washed 3 times with PBS to remove cell-free virions and seeded in triplicate in a 96 wells flat-bottom plate (2×104 per well) along with or without mock-, GPI-scFv X5- or AB65-transduced TZM.bl cells (1×104 per well). The cell-cell transmission of HIV-1, as measured by RLU in cell lysates, was determined after 48 hours by a BrightGlo Luciferase assay according to the manufacturer’s instructions (Promega).
To examine trans-infection of GPI-scFv transduced CEMss-CCR5 cells by DC-captured HIV-1, human iDCs or mDCs were incubated with HIV-1 AD8, Bru-Yu2, Mj4 and Bru3 at MOI 5 for 2 hours in triplicate cultures. Cells were then washed 3 times with PBS to remove cell-free virions and seeded in triplicate in a 24 wells flat-bottom plate (1×105 per well) along with or without GPI-scFv X5 or AB65-transduced CEMss-CCR5 or CEMss-CCR5-CR2N cells (2×105 per well). Cell mixtures were cultured in a total volume of 2 ml for 12 days. Every 3 days, 1.2 ml culture supernatants were collected and replaced with fresh complete RPMI 1640 medium. HIV-1 Gag p24 in the supernatant was measured by ELISA according to the manufacturer’s instructions (Zepto Metrix Co.).
In transwell experiments, 1 x 106 iDCs were incubated with HIV-1 AD8 or Bru-Yu2 at MOI of 5 at 37°C for 2 hours in triplicate cultures and washed extensively to remove cell-free virions. 2 x 105 CEMss-CCR5-GPI-scFv X5 or AB65 cells were seeded in 800 µl complete RPMI 1640 in the upper chamber of 24-well transwell plates (Corning) and 1 X 105 HIV-1-captured iDCs in 1ml RPMI1640 were added at the bottom chamber. Cells were incubated for 12 days. Every 3 days, 500 μl cells in upper layer were replaced by fresh complete RPMI 1640, while 670 µl supernatant in bottom layer were collected and replenished with fresh complete RPMI 1640 medium. The amount of HIV-1 Gag p24 in the culture supernatants was measured by ELISA according to the manufacturer’s instructions.
To test trans-infection of transduced human primary CD4+ T cells by DC-captured HIV-1, iDCs or mDCs were incubated with HIV-1 AD8 and Bru3 at MOI 5 for 2 hours in triplicate cultures. Cells were then washed 3 times with PBS to remove free viruses and seeded in triplicate in a 96 wells flat-bottom plate (2×105 per well) along with or without pRRL-GPI-scFv X5 or AB65-2A-GFP-transduced human primary CD4+ T cells (1×105 per well). Cell mixtures were co-cultured in a total volume of 200 µl for 12 days. at 6 hours, 1, 2, 3, 6, 9 and 12 days, small portions of cell suspension were harvested and replaced with fresh complete RPMI 1640 medium and human rIL-2 (100 IU/ml). Intracellular HIV-1 Gag p24 was analyzed by FACS (see above).
3×105 iDCs or mDCs were incubated with HIV-luc pseudotyped with either HIV-1 Env proteins or the 10A1 control in triplicate cultures. The amount of viral particles added to the DCs was equivalent to 2 ng HIV-1 Gag p24 at 37°C. After a 2 hour incubation, cells were washed extensively. 2×105 CEMss-CCR5-GPI-scFv X5 or AB65 cells were co-cultured with 1×105 iDCs or mDCs harboring captured virions, or iDCs or mDCs without virions, at 37°C for 48 hours in 1ml complete RPMI 1640 in 48wells plates. The cocultures were collected for assessing luciferase activity using BrightGlo luciferase activity kit according to the manufacturer’s instructions (Promega).
To evaluate if expression of GPI-scFv X5 on CD8 T cells enhances restriction of HIV-1 in trans of CD4 T cells, primary cells were isolated and transduced as previously described. Briefly, CD4 T cells were isolated from healthy human PBMCs and activated by PHA (5ug/ml) stimulation and expanded for one week. CD4 and CD8 T cells were transduced using lentiviral vectors as described above. Transduced or untransduced CD4 cells were plated in triplicate on a 96 well U-bottom plate (VWR). 2.5×105 cells CD4 and CD8 were co-cultured at a 1:1 ratio and suspended in 100 µl of RPMI media supplemented with IL-2 (100 IU/ml). Cell cultures in triplicate were infected for three hours with HIV-1 NL4-3 at an MOI of 0.01. Plates were spun down at 1200 rpm for 5 minutes and cells were washed two times with PBS. Co-cultures were collected and resuspended in 1 ml of RPMI supplemented with IL-2 (100 IU/mL) in 24 well plates. Supernatants were collected at three day intervals post infection and replenished with complete media. Experiments were independently performed with cells from three different donors.
To evaluate migration of GPI-GFP to iDCs, 1×106 iDCs were suspended in 250 µl of FluoroBrite DMEM (ThermoFisher) with 5 µl Vybrant DiD-Cell Labeling Solution (ThermoFisher) for 10 minutes at 37°C. iDCs were then washed three times and incubated in 500 µl FluoroBrite DMEM at a 1:1 ratio with CEMss-GPI GFP cells at 37°C for two hours in a 5mL polystyrene tube (Falcon). In iDC co-culture experiments with CEMss-GPI-HA-scFv X5 cells, co-cultures were stained with PacBlue conjugated anti-HA tag antibody (BioLegend) at 37°C for 40 min. Cells were washed twice with FACS buffer. Unfixed co-cultures were then resuspended in 50 µl FluoroBrite DMEM in micro-centrifuge tubes (Avantor VWR). FACS analysis and imaging was performed on a Amnis ImageStream Mk II Imaging Flow Cytometer (Luminex, Austin, TX). ImageStream data were analyzed with IDEAS 6.3.
To obtain confocal images of GPI-GFP transfer to iDCs, 5×105 iDCs were resuspended in 300 µl of complete DMEM and seeded in 8 well chamber slides (IBIDI) 24 hours before imaging. Media was removed and iDCs were washed once with PBS and replaced with FluoroBrite DMEM. 1.5 µl Vybrant DiD-Cell Labeling Solution (ThermoFisher) was added for 10 minutes at 37°C. Cells were then washed twice with PBS and 5×105 CEMss-GPI GFP cells in 300 µl of FluoroBrite DMEM were co-cultured with iDCs. Images were obtained after two hours by a LSM 980 with AiryScan 2 Confocal Microscope (Zeiss). Images were analyzed with ZEN 3.5.
The data in Figures 2, 4 were analyzed by two-way ANOVA and post-test Bonferroni comparison using GraphPad software. The data in Figures 5, 6 were compared by unpaired t-test using the same software. If 0.01≤p ≤ 0.05, it was labeled as *, while if 0.001≤p ≤ 0.01, it was labeled as **; if p ≤ 0.001, it was labeled as ***.
The original contributions presented in the study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding authors.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
KT, WW, PZ and JK conceived the project and designed the experiments. KT, WW, CY, SS, HH, LL, MW, and AM performed experiments and analyzed the data. KT, WW, JK, and PZ wrote the manuscript. The co-first authors, KT and WW, contributed equally and both earned the right list their name first in their C.V. All authors contributed to the article and approved the submitted version.
This work was jointly funded by the Chinese National Science Foundation and US National Institutes of Health (#81361120406 and AI106574) to PZ and JK). It was also supported by NIH grant AI116167 to JK, the Basic Science Core of the Texas Developmental CFAR (AI161943), instrumentation grant S10 OD028480 (PI: Dr. Mary K. Estes) for the LSM 980-Airyscan 2 microscope in the BCM BSL2 Microbiology Microscopy Core, and by the Cytometry and Cell Sorting Core at BCM with funding from the CPRIT Core Facility Support Award (CPRIT-RP180672) and the NIH (CA125123 and RR024574).
The authors wish to thank Dr. L. Naldini at the University Torino Medical School, Torino, Italy for lentiviral transfer vector and Dr. B. H. Hahn at the University of Penn. for DNA plasmids encoding consensus B and C HIV-1 envelope proteins. Cell line TZM.bl and molecular clones of HIV-1 Bru-3, AD8, Mj4 and Yu2 as well as expression vectors pAD8, pBru-3, pYu-2, pJRFL and pNL4-3.luc.R-E- transfer vector were obtained through the AIDS Research and Reference Reagent Program, Division of the AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Germantown, MD. These reagents were originally developed and contributed by Drs. J. Kappes, X. Wu, N. Landau, R. Risser, J. Overbaugh, E. Freed, A. Adachi, M.A. Martin, I.R. Chen, G.W. Shaw and B.H. Hahn. The authors also thank Joel M. Sederstrom of the BCM Cytometry and Cell Sorting Core for assistance with the Amnis ImageStream imaging flow cytometer.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1167965/full#supplementary-material
1. Pope M, Haase AT. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nat Med (2003) 9(7):847–52. doi: 10.1038/nm0703-847
2. Shattock RJ, Moore JP. Inhibiting sexual transmission of HIV-1 infection. Nat Rev Microbiol (2003) 1(1):25–34. doi: 10.1038/nrmicro729
3. Steinman RM, Granelli-Piperno A, Pope M, Trumpfheller C, Ignatius R, Arrode G, et al. The interaction of immunodeficiency viruses with dendritic cells. Curr Top Microbiol Immunol (2003) 276:1–30. doi: 10.1007/978-3-662-06508-2_1
4. Arrighi JF, Pion M, Garcia E, Escola JM, van Kooyk Y, Geijtenbeek TB, et al. DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. J Exp Med (2004) 200(10):1279–88. doi: 10.1084/jem.20041356
5. Engering A, Van Vliet SJ, Geijtenbeek TB, Van Kooyk Y. Subset of DC-SIGN(+) dendritic cells in human blood transmits HIV-1 to T lymphocytes. Blood (2002) 100(5):1780–6. doi: 10.1182/blood-2001-12-0179
6. Izquierdo-Useros N, Blanco J, Erkizia I, Fernández-Figueras MT, Borràs FE, Naranjo-Gómez M, et al. Maturation of blood-derived dendritic cells enhances human immunodeficiency virus type 1 capture and transmission. J Virol (2007) 81(14):7559–70. doi: 10.1128/JVI.02572-06
7. McDonald D, Wu L, Bohks SM, KewalRamani VN, Unutmaz D, Hope TJ. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science (2003) 300(5623):1295–7. doi: 10.1126/science.1084238
8. Turville SG, Santos JJ, Frank I, Cameron PU, Wilkinson J, Miranda-Saksena M, et al. Immunodeficiency virus uptake, turnover, and 2-phase transfer in human dendritic cells. Blood (2004) 103(6):2170–9. doi: 10.1182/blood-2003-09-3129
9. Dong C, Janas AM, Wang JH, Olson WJ, Wu L. Characterization of human immunodeficiency virus type 1 replication in immature and mature dendritic cells reveals dissociable cis- and trans-infection. J Virol (2007) 81(20):11352–62. doi: 10.1128/JVI.01081-07
10. Burleigh L, Lozach PY, Schiffer C, Staropoli I, Pezo V, Porrot F, et al. Infection of dendritic cells (DCs), not DC-SIGN-mediated internalization of human immunodeficiency virus, is required for long-term transfer of virus to T cells. J Virol (2006) 80(6):2949–57. doi: 10.1128/JVI.80.6.2949-2957.2006
11. Nobile C, Petit C, Moris A, Skrabal K, Abastado JP, Mammano F, et al. Covert human immunodeficiency virus replication in dendritic cells and in DC-SIGN-expressing cells promotes long-term transmission to lymphocytes. J Virol (2005) 79(9):5386–99. doi: 10.1128/JVI.79.9.5386-5399.2005
12. Turville SG, Aravantinou M, Stössel H, Romani N, Robbiani M. Resolution of de novo HIV production and trafficking in immature dendritic cells. Nat Methods (2008) 5(1):75–85. doi: 10.1038/nmeth1137
13. Schiffner T, Sattentau QJ, Duncan CJ. Cell-to-cell spread of HIV-1 and evasion of neutralizing antibodies. Vaccine (2013) 31(49):5789–97. doi: 10.1016/j.vaccine.2013.10.020
14. Abela IA, Berlinger L, Schanz M, Reynell L, Günthard HF, Rusert P, et al. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PloS Pathog (2012) 8(4):e1002634. doi: 10.1371/journal.ppat.1002634
15. Chen P, Hübner W, Spinelli MA, Chen BK. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol (2007) 81(22):12582–95. doi: 10.1128/JVI.00381-07
16. Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, et al. HIV-infected T cells are migratory vehicles for viral dissemination. Nature (2012) 490(7419):283–7. doi: 10.1038/nature11398
17. Su B, Xu K, Lederle A, Peressin M, Biedma ME, Laumond G, et al. Neutralizing antibodies inhibit HIV-1 transfer from primary dendritic cells to autologous CD4 T lymphocytes. Blood (2012) 120(18):3708–17. doi: 10.1182/blood-2012-03-418913
18. Malbec M, Porrot F, Rua R, Horwitz J, Klein F, Halper-Stromberg A, et al. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J Exp Med (2013) 210(13):2813–21. doi: 10.1084/jem.20131244
19. Reh L, Magnus C, Schanz M, Weber J, Uhr T, Rusert P, et al. Capacity of broadly neutralizing antibodies to inhibit HIV-1 cell-cell transmission is strain- and epitope-dependent. PloS Pathog (2015) 11(7):e1004966. doi: 10.1371/journal.ppat.1004966
20. Frankel SS, Steinman RM, Michael NL, Kim SR, Bhardwaj N, Pope M, et al. Neutralizing monoclonal antibodies block human immunodeficiency virus type 1 infection of dendritic cells and transmission to T cells. J Virol (1998) 72(12):9788–94. doi: 10.1128/JVI.72.12.9788-9794.1998
21. van Montfort T, Nabatov AA, Geijtenbeek TB, Pollakis G, Paxton WA. Efficient capture of antibody neutralized HIV-1 by cells expressing DC-SIGN and transfer to CD4+ T lymphocytes. J Immunol (2007) 178(5):3177–85. doi: 10.4049/jimmunol.178.5.3177
22. van Montfort T, Thomas AA, Krawczyk PM, Berkhout B, Sanders RW, Paxton WA. Reactivation of neutralized HIV-1 by dendritic cells is dependent on the epitope bound by the antibody. J Immunol (2015) 195(8):3759–68. doi: 10.4049/jimmunol.1402344
23. Sagar M, Akiyama H, Etemad B, Ramirez N, Freitas I, Gummuluru S. Transmembrane domain membrane proximal external region but not surface unit-directed broadly neutralizing HIV-1 antibodies can restrict dendritic cell-mediated HIV-1 trans-infection. J Infect Dis (2012) 205(8):1248–57. doi: 10.1093/infdis/jis183
24. Durham ND, Yewdall AW, Chen P, Lee R, Zony C, Robinson JE, et al. Neutralization resistance of virological synapse-mediated HIV-1 Infection is regulated by the gp41 cytoplasmic tail. J Virol (2012) 86(14):7484–95. doi: 10.1128/JVI.00230-12
25. Martin N, Welsch S, Jolly C, Briggs JA, Vaux D, Sattentau QJ. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J Virol (2010) 84(7):3516–27. doi: 10.1128/JVI.02651-09
26. Massanella M, Puigdomènech I, Cabrera C, Fernandez-Figueras MT, Aucher A, Gaibelet G, et al. Antigp41 antibodies fail to block early events of virological synapses but inhibit HIV spread between T cells. AIDS (2009) 23(2):183–8. doi: 10.1097/QAD.0b013e32831ef1a3
27. Heath SL, Tew JG, Szakal AK, Burton GF. Follicular dendritic cells and human immunodeficiency virus infectivity. Nature (1995) 377(6551):740–4. doi: 10.1038/377740a0
28. Yu HJ, Reuter MA, McDonald D. HIV traffics through a specialized, surface-accessible intracellular compartment during trans-infection of T cells by mature dendritic cells. PloS Pathog (2008) 4(8):e1000134. doi: 10.1371/journal.ppat.1000134
29. Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 virological synapse formation in T cells requires lipid raft integrity. J Virol (2005) 79(18):12088–94. doi: 10.1128/JVI.79.18.12088-12094.2005
30. Simons K, Ikonen E. Functional rafts in cell membranes. Nature (1997) 387(6633):569–72. doi: 10.1038/42408
31. Liao Z, Cimakasky LM, Hampton R, Nguyen DH, Hildreth JE. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res Hum Retroviruses (2001) 17(11):1009–19. doi: 10.1089/088922201300343690
32. Chazal N, Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol Mol Biol Rev (2003) 67(2):226–37. doi: 10.1128/MMBR.67.2.226-237.2003
33. Popik W, Alce TM, Au WC. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4(+) T cells. J Virol (2002) 76(10):4709–22. doi: 10.1128/JVI.76.10.4709-4722.2002
34. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol (1998) 72(4):2855–64. doi: 10.1128/JVI.72.4.2855-2864.1998
35. Percherancier Y, Lagane B, Planchenault T, Staropoli I, Altmeyer R, Virelizier JL, et al. HIV-1 entry into T-cells is not dependent on CD4 and CCR5 localization to sphingolipid-enriched, detergent-resistant, raft membrane domains. J Biol Chem (2003) 278(5):3153–61. doi: 10.1074/jbc.M207371200
36. Nguyen DH, Giri B, Collins G, Taub DD. Dynamic reorganization of chemokine receptors, cholesterol, lipid rafts, and adhesion molecules to sites of CD4 engagement. Exp Cell Res (2005) 304(2):559–69. doi: 10.1016/j.yexcr.2004.11.022
37. Wen M, Arora R, Wang H, Liu L, Kimata JT, Zhou P. GPI-anchored single chain Fv–an effective way to capture transiently-exposed neutralization epitopes on HIV-1 envelope spike. Retrovirology (2010) 7:79. doi: 10.1186/1742-4690-7-79
38. Medof ME, Kinoshita T, Nussenzweig V. Inhibition of complement activation on the surface of cells after incorporation of decay-accelerating factor (DAF) into their membranes. J Exp Med (1984) 160(5):1558–78. doi: 10.1084/jem.160.5.1558
39. Misra A, Gleeson E, Wang W, Ye C, Zhou P, Kimata JT. Glycosyl-phosphatidylinositol-anchored anti-HIV env single-chain variable fragments interfere with HIV-1 env processing and viral infectivity. J Virol (2018) 92(7):e02080-17. doi: 10.1128/JVI.02080-17
40. Liu L, Wen M, Wang W, Wang S, Yang L, Liu Y, et al. Potent and broad anti-HIV-1 activity exhibited by a glycosyl-phosphatidylinositol-anchored peptide derived from the CDR H3 of broadly neutralizing antibody PG16. J Virol (2011) 85(17):8467–76. doi: 10.1128/JVI.00520-11
41. Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PloS One (2014) 9(12):e115987. doi: 10.1371/journal.pone.0115987
42. Sewald X, Ladinsky MS, Uchil PD, Beloor J, Pi R, Herrmann C, et al. Retroviruses use CD169-mediated trans-infection of permissive lymphocytes to establish infection. Science (2015) 350(6260):563–7. doi: 10.1126/science.aab2749
43. Lopez P, Koh WH, Hnatiuk R, Murooka TT. HIV infection stabilizes macrophage-T cell interactions to promote cell-cell HIV spread. J Virol (2019) 93(18):e00805-19. doi: 10.1128/JVI.00805-19
44. Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X, Binley JM, et al. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci USA (2002) 99(10):6913–8. doi: 10.1073/pnas.102562599
45. Kooyman DL, Byrne GW, McClellan S, Nielsen D, Tone M, Waldmann H, et al. In vivo transfer of GPI-linked complement restriction factors from erythrocytes to the endothelium. Science (1995) 269(5220):89–92. doi: 10.1126/science.7541557
46. Kirchhoff C, Hale G. Cell-to-cell transfer of glycosylphosphatidylinositol-anchored membrane proteins during sperm maturation. Mol Hum Reprod (1996) 2(3):177–84. doi: 10.1093/molehr/2.3.177
47. Liu L, Wen M, Zhu Q, Kimata JT, Zhou P. Glycosyl phosphatidylinositol-anchored C34 peptide derived from human immunodeficiency virus type 1 gp41 is a potent entry inhibitor. J Neuroimmune Pharmacol (2016) 11(3):601–10. doi: 10.1007/s11481-016-9681-x
48. Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med (2014) 370(10):901–10. doi: 10.1056/NEJMoa1300662
49. DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med (2010) 2(36):36ra43. doi: 10.1126/scitranslmed.3000931
50. Huang SH, Ren Y, Thomas AS, Chan D, Mueller S, Ward AR, et al. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest. (2018) 128(2):876–89. doi: 10.1172/JCI97555
51. Patel S, Chorvinsky E, Albihani S, Cruz CR, Jones RB, Shpall EJ, et al. HIV-specific T cells generated from naive T cells suppress HIV in vitro and recognize wide epitope breadths. Mol Ther (2018) 26(6):1435–46. doi: 10.1016/j.ymthe.2018.04.009
52. Sung JA, Patel S, Clohosey ML, Roesch L, Tripic T, Kuruc JD, et al. HIV-specific, ex vivo expanded T cell therapy: feasibility, safety, and efficacy in ART-suppressed HIV-infected individuals. Mol Ther (2018) 26(10):2496–506. doi: 10.1016/j.ymthe.2018.08.015
53. Lee PH, Keller MD, Hanley PJ, Bollard CM. Virus-specific T cell therapies for HIV: lessons learned from hematopoietic stem cell transplantation. Front Cell Infect Microbiol (2020) 10:298. doi: 10.3389/fcimb.2020.00298
54. Collora JA, Liu R, Pinto-Santini D, Ravindra N, Ganoza C, Lama JR, et al. Single-cell multiomics reveals persistence of HIV-1 in expanded cytotoxic T cell clones. Immunity (2022) 55(6):1013–31.e7. doi: 10.1016/j.immuni.2022.03.004
55. Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol (2009) 83(16):8289–92. doi: 10.1128/JVI.00709-09
56. Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother (2002) 46(6):1896–905. doi: 10.1128/AAC.46.6.1896-1905.2002
57. Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol (2009) 485:395–405. doi: 10.1007/978-1-59745-170-3_26
58. Theodore TS, Englund G, Buckler-White A, Buckler CE, Martin MA, Peden KW. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses (1996) 12(3):191–4. doi: 10.1089/aid.1996.12.191
59. Li Y, Hui H, Burgess CJ, Price RW, Sharp PM, Hahn BH, et al. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol (1992) 66(11):6587–600. doi: 10.1128/jvi.66.11.6587-6600.1992
60. Ndung'u T, Renjifo B, Essex M. Construction and analysis of an infectious human Immunodeficiency virus type 1 subtype C molecular clone. J Virol (2001) 75(11):4964–72. doi: 10.1128/JVI.75.11.4964-4972.2001
61. Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell (1985) 40(1):9–17. doi: 10.1016/0092-8674(85)90303-4
62. Tsai C, Caillet C, Hu H, Zhou F, Ding H, Zhang G, et al. Measurement of neutralizing antibody responses against H5N1 clades in immunized mice and ferrets using pseudotypes expressing influenza hemagglutinin and neuraminidase. Vaccine (2009) 27(48):6777–90. doi: 10.1016/j.vaccine.2009.08.056
63. Overbaugh J, Anderson RJ, Ndinya-Achola JO, Kreiss JK. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res Hum Retroviruses (1996) 12(2):107–15. doi: 10.1089/aid.1996.12.107
64. Koyanagi Y, Miles S, Mitsuyasu RT, Merrill JE, Vinters HV, Chen IS. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science (1987) 236(4803):819–22. doi: 10.1126/science.3646751
65. Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol (1991) 65(8):3973–85. doi: 10.1128/jvi.65.8.3973-3985.1991
Keywords: HIV, CD4 + T cells, dendritic cells, neutralization, single chain variable fragments, glycosyl phosphatidylinositol, immunotherapy
Citation: Wang W, Truong K, Ye C, Sharma S, He H, Liu L, Wen M, Misra A, Zhou P and Kimata JT (2023) Engineered CD4 T cells expressing a membrane anchored viral inhibitor restrict HIV-1 through cis and trans mechanisms. Front. Immunol. 14:1167965. doi: 10.3389/fimmu.2023.1167965
Received: 17 February 2023; Accepted: 31 August 2023;
Published: 14 September 2023.
Edited by:
Richard B. Markham, Johns Hopkins University, United StatesReviewed by:
Rebecca Veenhuis, Johns Hopkins University, United StatesCopyright © 2023 Wang, Truong, Ye, Sharma, He, Liu, Wen, Misra, Zhou and Kimata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul Zhou, cHphdmlndEBnbWFpbC5jb20=; Jason T. Kimata, amtpbWF0YUBiY20uZWR1
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.