- Department of General Practice, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
Background: Lung cancer is a common comorbidity of heart failure (HF). The early identification of the risk factors for lung cancer in patients with HF is crucial to early diagnosis and prognosis. Furthermore, oxidative stress and immune responses are the two critical biological processes shared by HF and lung cancer. Therefore, our study aimed to select the core genes in HF and then investigate the potential mechanisms underlying HF and lung cancer, including oxidative stress and immune responses through the selected genes.
Methods: Differentially expressed genes (DEGs) were analyzed for HF using datasets extracted from the Gene Expression Omnibus database. Functional enrichment analysis was subsequently performed. Next, weighted gene co-expression network analysis was performed to select the core gene modules. Support vector machine models, the random forest method, and the least absolute shrinkage and selection operator (LASSO) algorithm were applied to construct a multigene signature. The diagnostic values of the signature genes were measured using receiver operating characteristic curves. Functional analysis of the signature genes and immune landscape was performed using single-sample gene set enrichment analysis. Finally, the oxidative stress–related genes in these signature genes were identified and validated in vitro in lung cancer cell lines.
Results: The DEGs in the GSE57338 dataset were screened, and this dataset was then clustered into six modules using weighted gene co-expression network analysis; MEblue was significantly associated with HF (cor = −0.72, p < 0.001). Signature genes including extracellular matrix protein 2 (ECM2), methyltransferase-like 7B (METTL7B), meiosis-specific nuclear structural 1 (MNS1), and secreted frizzled-related protein 4 (SFRP4) were selected using support vector machine models, the LASSO algorithm, and the random forest method. The respective areas under the curve of the receiver operating characteristic curves of ECM2, METTL7B, MNS1, and SFRP4 were 0.939, 0.854, 0.941, and 0.926, respectively. Single-sample gene set enrichment analysis revealed significant differences in the immune landscape of the patients with HF and healthy subjects. Functional analysis also suggested that these signature genes may be involved in oxidative stress. In particular, METTL7B was highly expressed in lung cancer cell lines. Meanwhile, the correlation between METTL7B and oxidative stress was further verified using flow cytometry.
Conclusion: We identified that ECM2, METTL7B, MNS1, and SFRP4 exhibit remarkable diagnostic performance in patients with HF. Of note, METTL7B may be involved in the co-occurrence of HF and lung cancer by affecting the oxidative stress immune responses.
1 Introduction
Heart failure (HF) is a devastating clinical syndrome characterized by symptoms and/or signs caused by structural and/or functional impairments in ventricular filling or blood ejection (1). The global prevalence of HF across patients of all ages is 1.3% but can be as high as 8.3% in people aged >50 years; moreover, the incidence of HF increases with age (2). HF can be classified on the basis of the onset type (acute vs. chronic), the affected ventricle (left vs. right), and the type of circulation affected (systolic vs. diastolic) (3). Hypertension, obesity, prediabetes, diabetes, and atherosclerotic cardiovascular disease are the main risk factors for HF (4). The leading causes of HF include ischemic heart disease and myocardial infarction, hypertension, and valvular heart disease (1). The pathophysiological mechanisms of HF involve comprehensive biological processes, including ischemia (5), mechanical stress (6, 7), autoimmune disease (8), microbial infections (9), and genetic diseases.
In western countries, patients with HF have an average of five different diseases simultaneously, with cancer being one of the main causes of non-cardiovascular death in patients with HF (10). In addition, patients with HF have a significantly increased risk of cancer (approximately 76%) as compared with those without HF (10–12). Banke et al. reported that lung cancer was one of the most common malignant diagnoses in the HF cohort, with a proportion of approximately 15.7% (13). The prevalence of HF and cancer has increased with the aging population and advancing medical technology. The co-existence of cancer in patients with HF is becoming more common owing to the similar pathogenesis of and risk factors for the two diseases, including inflammation, angiogenesis, clonal hematopoiesis, metabolic remodeling and extracellular matrix (ECM), and stromal cell infiltration (14, 15). In particular, the risk of respiratory cancer is increased by 91% among patients with HF (10). The causes of the co-existence of HF and lung cancer are not entirely clear; however, research has suggested that the two diseases share some risk factors, such as smoking, advanced age, high blood pressure, diabetes, and obesity (16–19). Moreover, some lung cancer treatments, such as radiation and chemotherapy, can also damage the heart, thereby increasing the risk of HF (20).
To investigate the correlation between HF and lung cancer, first, we aimed to use multiple bioinformatic approaches to identify the signature genes in HF. Next, we performed enrichment pathway analyses to investigate the correlations in these genes and the roles that they play in lung cancer. Our results will provide clinicians with new insights into the diagnosis and treatment of lung cancer.
2 Method and materials
2.1 Data acquisition and the identification of differentially expressed gene
The GSE57338 dataset related to HF was downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). The differentially expressed genes (DEGs) between patients with HF and healthy subjects were selected using the R software’s limma package (21) (criteria: p < 0.05 and |log fold change (FC)| > 1), and the results were visualized as volcano plots and heatmaps.
2.2 Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analysis
The enrichment of the functions of the selected DEGs was performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis with the cluster profiler package in R (22).
2.3 Weighted gene co-expression network analysis
A weighted gene co-expression network analysis (WGCNA) (23) was performed using the gene expression profiles from the GSE57338 dataset to explore the hub genes based on the soft threshold power chosen by the pickSoftThreshold function. The dynamic tree-cutting method was then used to identify the co-expressed gene modules. The Pearson correlation coefficients between the module eigengenes and HF were evaluated to acquire the hub module.
2.4 Signature gene identification
First, the candidate hub genes were identified through the intersection of the DEGs and key module genes. Second, the signature genes were selected according to calculations conducted using support vector machine (SVM) models, the least absolute shrinkage and selection operator (LASSO) algorithm (24), and the random forest (RF) model (25). Finally, the area under the receiver operating characteristic (AUC-ROC) curve was used to evaluate the diagnostic efficiency of each selected signature gene. An AUC > 0.7 indicated a favorable diagnostic performance.
2.5 The immune landscape and functional analysis
The single-sample gene set enrichment analysis (ssGSEA) algorithm was used with R packages (GSVA and GSEABase) (26) to comprehensively assess the immunologic characteristics between the patients with HF and healthy subjects. The marker genes of different immune cells were derived from previous studies (27) and are listed in Table S1. Next, functional analysis of the signature genes was performed using the ssGSEA algorithm.
2.6 Identification of oxidative stress–related genes
The oxidative stress–related genes were selected on the basis of the background information obtained from GeneGards (https://www.genecards.org/). A total of 10,022 oxidative stress–related genes were screened.
2.7 Cell culture
The human lung cancer cell lines A549 and PC9 and the normal human lung epithelial cell line BEAS2B were obtained from the American Type Culture Collection (Manassas, VA, USA). A549 was cultured in Roswell Park Memorial Institute-1640 medium, whereas PC9 and BEAS2B were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum at 37°C in an incubator with 5% CO2.
2.8 Quantitative polymerase chain reaction and Western blotting
Total RNA was isolated from the cells using the RNA-Quick Purification Kit (ESscience, RN001, China). A reverse transcription kit (Vazyme, R333-01, China) was used to reverse-transcribe RNA into cDNA, and real-time fluorescence quantitative PCR (Vazyme, Q711-02, China) was performed using Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the control. Finally, the relative mRNA expression of METTL7B was calculated using the 2−ΔΔCt technique to assess METTL7B expression in lung cancer cell lines and the normal human lung epithelial cell line. The sequences of the PCR primers used in this study are listed in the Supplementary Information (Table S2).
The cell samples were collected, and total protein was extracted according to the manufacturer’s instructions. The BCA Protein Assay Kit (Beyotime Biotechnology, China) was used to measure the protein concentration. The proteins were separated by electrophoresis and then transferred from the gel onto polyvinylidene difluoride membranes (Merck Millipore, USA, cat. no. IPVH00010). Blocking was performed using 5% non-fat dry milk powder (room temperature, 1 h). Incubation with the following primary antibodies was performed overnight at 4°C: anti-METTL7B (1:1,000 dilution; Proteintech, cat. no. 17001-1-AP) and anti–β-actin (1:1,000 dilution; Huabio, cat. no. ET1701-80). The membranes were then incubated with horseradish peroxidase–conjugated secondary antibodies (room temperature, 1 h), and the binding was detected using an enhanced chemiluminescence reagent (Sangon Biotech, China, cat. no. C510045-0100).
2.9 Plasmid siRNA transfection
For plasmid transfection, lipofectamine 3000 (Invitrogen, cat. no. L3000008) was used for the A549 and PC9 cells. For small interfering RNA (siRNA) transfection, lipofectamine RNAiMAX (Invitrogen, cat. no. 13778075) was used for the corresponding cells.
2.10 Oxidative stress measurement
The intracellular reactive oxygen species (ROS) levels in the cell samples were measured using the Reactive Oxygen Species Assay Kit (Beyotime Biotechnology, China), according to the manufacturer’s instructions. Cells with METTL7B overexpression or knockdown were incubated with 2′,7′-dichlorodihydrofluorescein diacetate for 30 min at 37°C and measured using CytoFLEX S (Beckman Coulter, CA, USA).
2.11 Statistical analysis
All statistical analyses were performed using R software (version 4.1.2). p < 0.05 was considered statistically significant. Unless specifically stated, all p-values were two-tailed. The research flow chart is shown in Supplementary Figure S1.
3 Results
3.1 DEGs and functional analysis of heart failure–related genes
DEGs from the patients with HF and healthy controls were analyzed using the “limma” package. A total of 48 DEGs were finally screened, of which 26 were upregulated and 22 were downregulated (Figure 1A). The heatmap showed the DEGs between the patients with HF and healthy subjects (Figure 1B). The GO terms of the DEGs were enriched in the chord diagram (Figure 1C). As shown in the KEGG analysis, the top three enriched pathways were African trypanosomiasis, malaria, and complement and coagulation cascades (Figure 1D).

Figure 1 Identification of the DEGs in HF and functional enrichment analysis of the DEGs. (A) The green dots in the volcano map indicate the downregulated genes, the red dots indicate the upregulated genes, and the black dots indicate the genes with no significant differences. (B) The blue in the heat map represents the healthy samples, and the pink represents the HF samples. (C) The GO terms of the DEGs were enriched in the chord diagram. (D) KEGG analysis showing the functional enrichment pathways.
3.2 Construction of the WGCN
A co-expression network was established between the patients with HF and healthy subjects using the R package “WGCNA”. The soft threshold power was equivalent to 6 (Figure 2A). A cluster dendrogram was constructed (Figure 2B), and the data were clustered into six modules (Figure 2C). The relationship between the different modules and the patients with HF was assessed. The results suggested that MEblue, including 265 genes, was a pivotal module unrelated to the patients with HF (cor = 0.72, p < 0.001). The area with a total of 27 overlapping key DEGs was verified from the total DEGs and MEblue module–containing genes (Figure 2D).
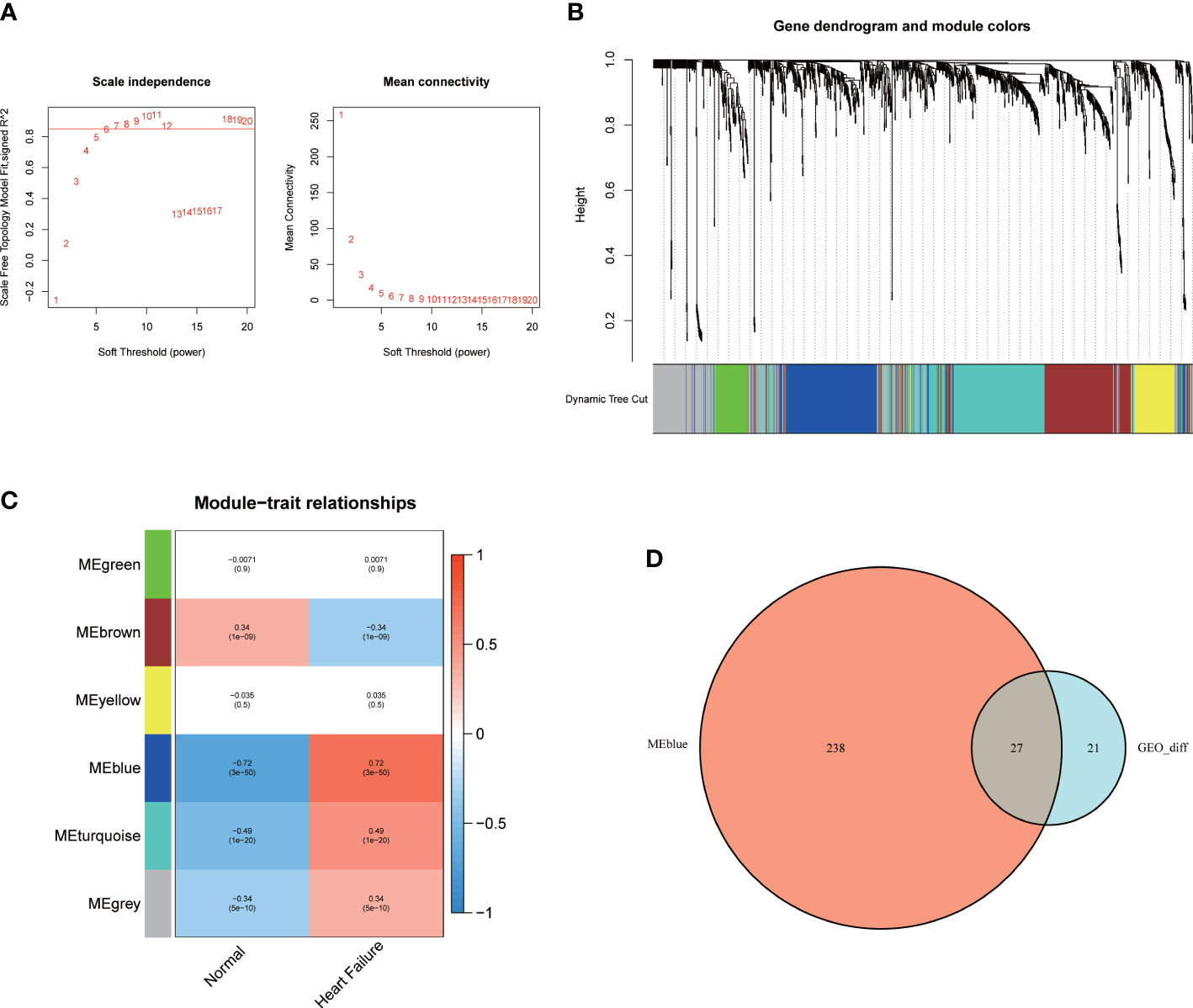
Figure 2 The WGCNA analysis and identification of candidate hub genes. (A) The soft threshold power and the mean connectivity of WGCNA. (B) The cluster dendrogram of WGCNA. (C) The clustered modules of WGCNA. (D) The venn diagram showed the interactions between the DEGs and genes in the MEblue module.
3.3 Selection of the signature genes using machine algorithms
The abovementioned 27 key genes were further screened using machine algorithms. Consequentially, four genes were selected by the SVM model (Figures 3A, B), 17 by the LASSO algorithm (Figures 3C, D), and 19 by the RF method, with an importance of >3 (Figures 3E, F). Finally, their intersection was considered so as to obtain four genes: extracellular matrix protein 2 (ECM2), methyltransferase-like 7B (METTL7B), meiosis-specific nuclear structural 1 (MNS1), and secreted frizzled-related protein 4 (SFRP4) (Figures 3E, F).
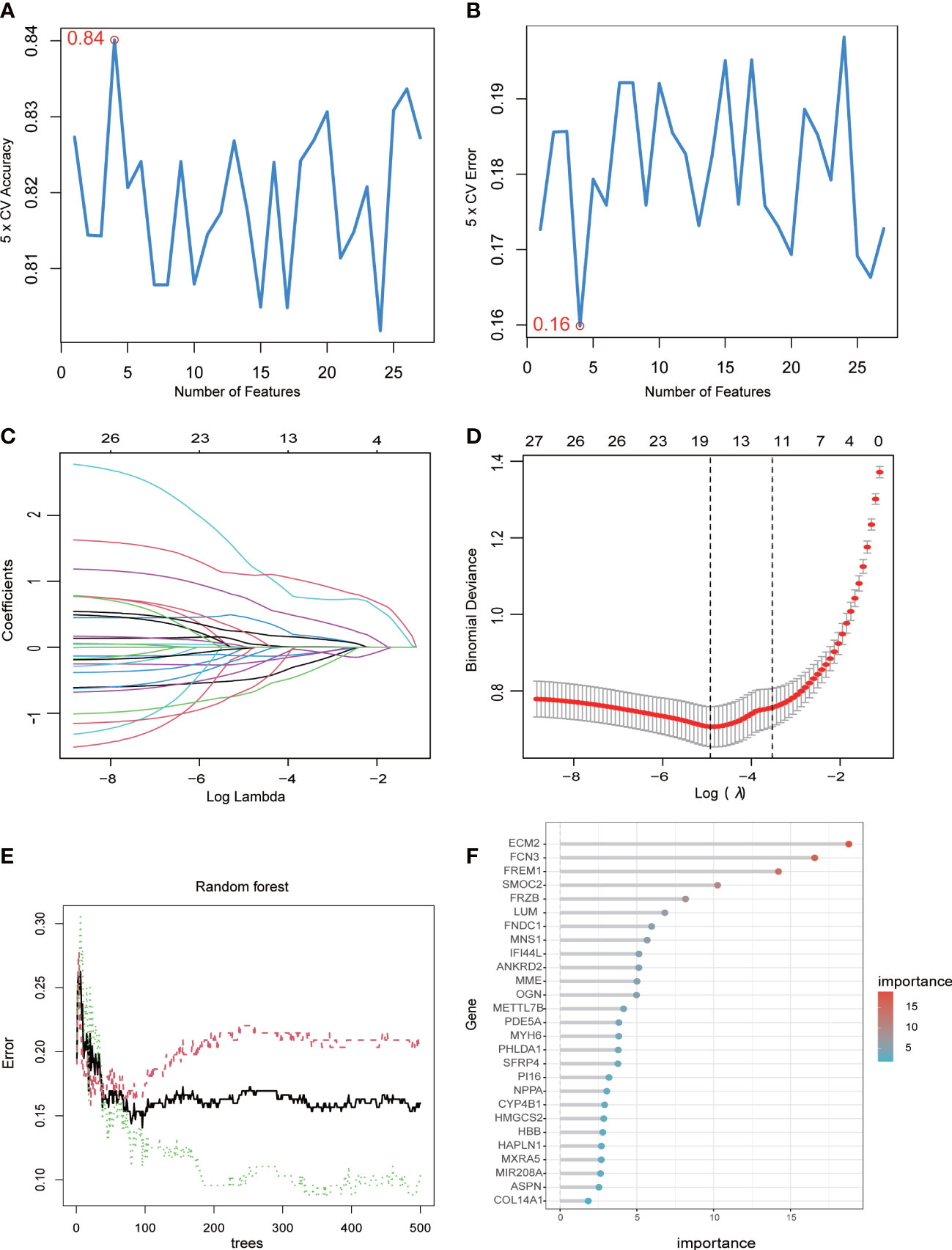
Figure 3 Machine learning algorithms determine signature genes. (A) The cross-validation accuracy of the SVM model. (B) The cross-validation error of the SVM model. (C) The LASSO algorithm showing variations in the size of coefficients for the parameters that shrank as the value of k penalty increased. (D) Penalty plot of the LASSO algorithm with error bars denoting standard errors. (E) The error rate confidence intervals of the RF model. (F) The relative importance of the genes in the RF model.
3.4 Diagnostic efficacy of the signature genes in predicting HF
An AUC-ROC of 0.939 for ECM2, 0.854 for METTL7B, 0.941 for MNS1, and 0.926 for SFRP4 was obtained (Figures 4A–D). Furthermore, the expression levels of these signature genes were significantly higher in the patients with HF than in healthy subjects, indicating the potential roles of these signature genes in HF (Figures 4E–H).
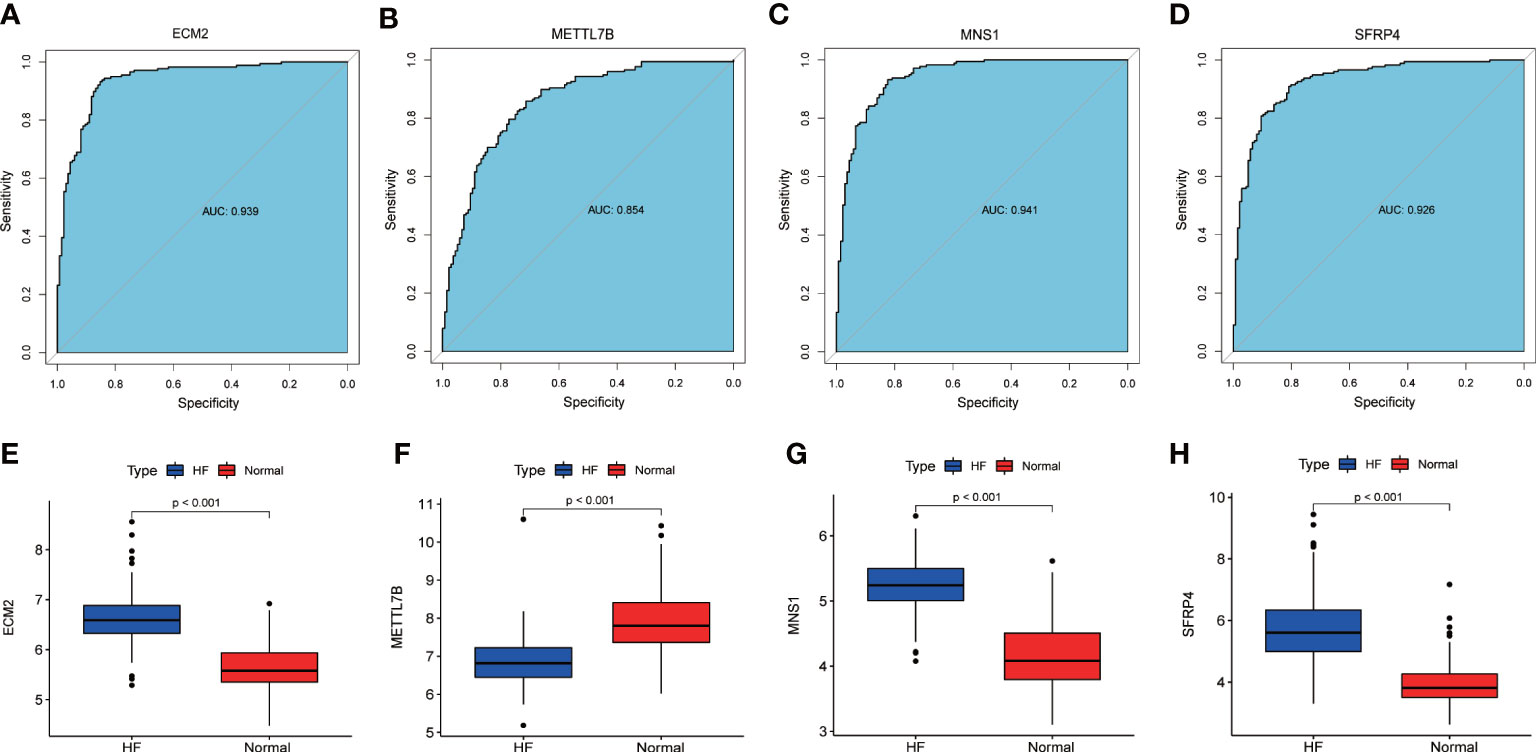
Figure 4 The ROC curve and expression of signature genes between the patients with HF and healthy subjects. (A–D) ROC curves of ECM2, METTL7B, MNS1, and SFRP4 between the patients with HF and healthy subjects. (E–H) Gene expression levels of ECM2, METTL7B, MNS1, and SFRP4 between the patients with HF and healthy subjects.
3.5 Immune landscape
Regarding the relationships between the different immune cell subtypes, tumor-infiltrating lymphocytes and neutrophils had the strongest positive correlation (r = 0.87), and T follicular helper (Tfh) and natural killer cells had the strongest negative correlation (r = −0.23) (Figure 5A). The ssGSEA analysis showed that the patients with HF were significantly prone to cytolytic activity, human leukocyte antigen response, inflammation promotion, T-cell co-stimulation, and type I interferon response, whereas the healthy subjects showed antigen-presenting cell co-inhibition, cytokine receptor interaction, checkpoint, and T-cell co-inhibition (Figure 5B). The patients with HF presented low infiltration of B cells, macrophages, plasmacytoid dendritic cells, and Tfh and T regulatory (Treg) cells but high infiltration of aDCs, CD8+ T cells, immature dendritic cells mast cells, natural killer cells, and Th1 cells (Figure 5C).
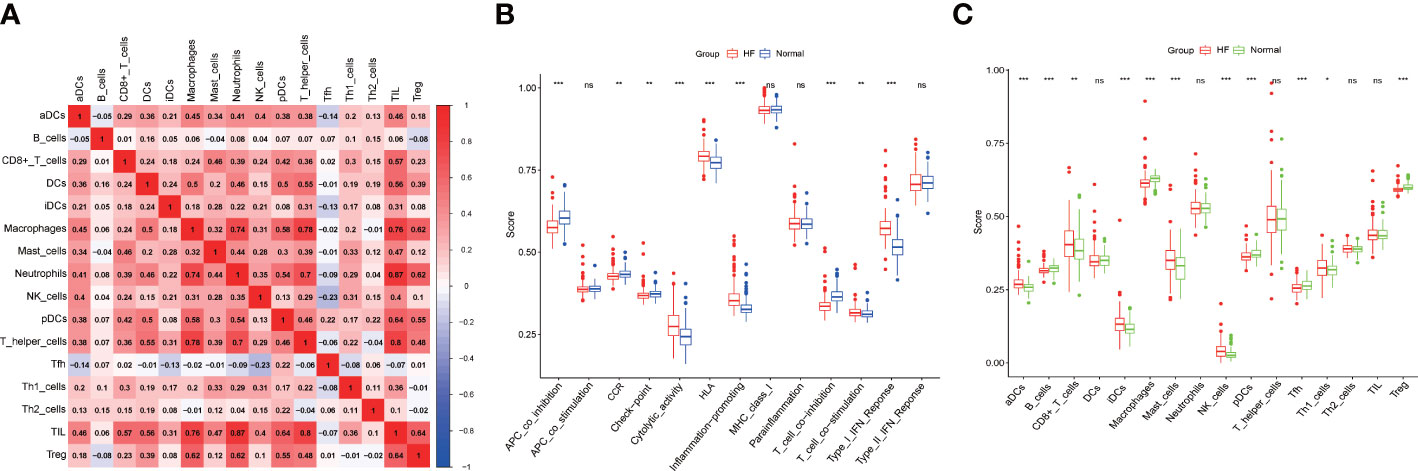
Figure 5 Immune landscape. (A) Immunological features. (B) Differences in the immune function between the patients with HF and healthy subjects. (C) Differences in the infiltration of immune cells between the patients with HF and healthy subjects. * p<0.05, ** p<0.01, *** p<0.001, “ns” means not significantly.
3.6 ssGSEA analysis and the identification of oxidative stress–related genes
The signaling pathways associated with the signature genes were analyzed using ssGSEA. ECM2, METTL7B, MNS1, and SFRP4 were significantly correlated with ROS generation (Figure 6). Subsequently, these four genes were intersected with the oxidative stress–related genes screened in the GeneGard database, and METTL7B was finally chosen for further analysis.
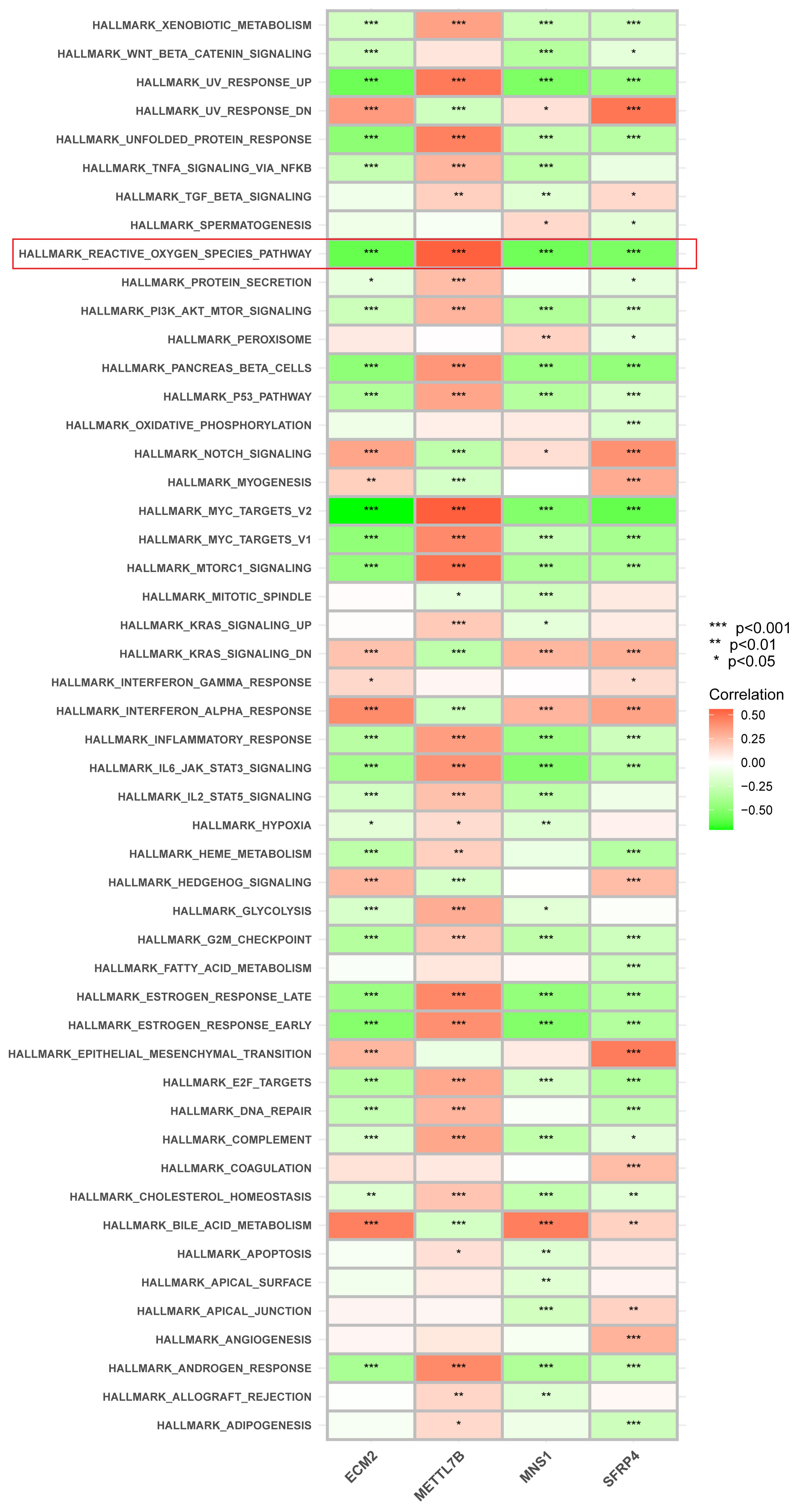
Figure 6 Correlation between each gene signature and pathway. ECM2, METTL7B, MNS1, and SFRP4 were significantly correlated with ROS generation. *p < 0.05, **p < 0.01, and ***P < 0.001.
3.7 The potential roles of METTL7B in lung cancer
To investigate the roles of METTL7B in lung cancer, we first measured and compared the expression levels of METTL7B in lung cancer cell lines and normal human lung epithelial cells using qRT-PCR and Western blotting. Both the mRNA and protein expression levels of METTL7B were significantly higher in the two lung cell lines (A549 and PC9) than in the normal human lung epithelial cells (BEAS2B) (Figures 7A, B).
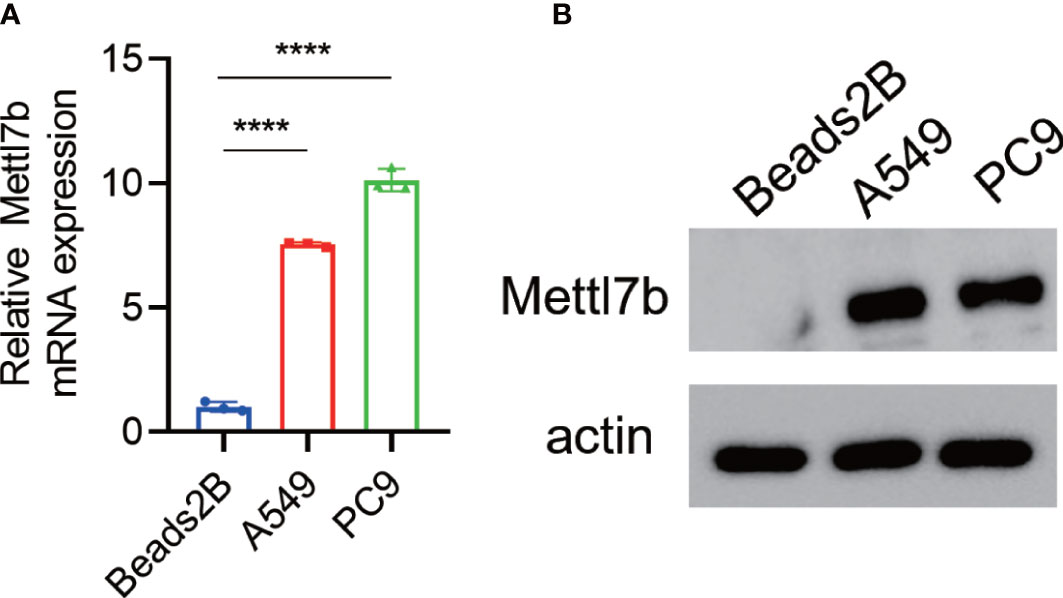
Figure 7 Overexpression of METTL7B in lung cancer cells. METTL7B mRNA (A) and protein levels (B) were increased in the A549 and PC9 cells. **** p<0.0001.
3.8 METTL7B is involved in ROS generation in lung cancer cells
The overexpression of METTL7B protein decreased the levels of intracellular ROS (Figures 8A–D). By contrast, METTL7B protein knockdown increased the levels of intracellular ROS (Figures 8E–H).
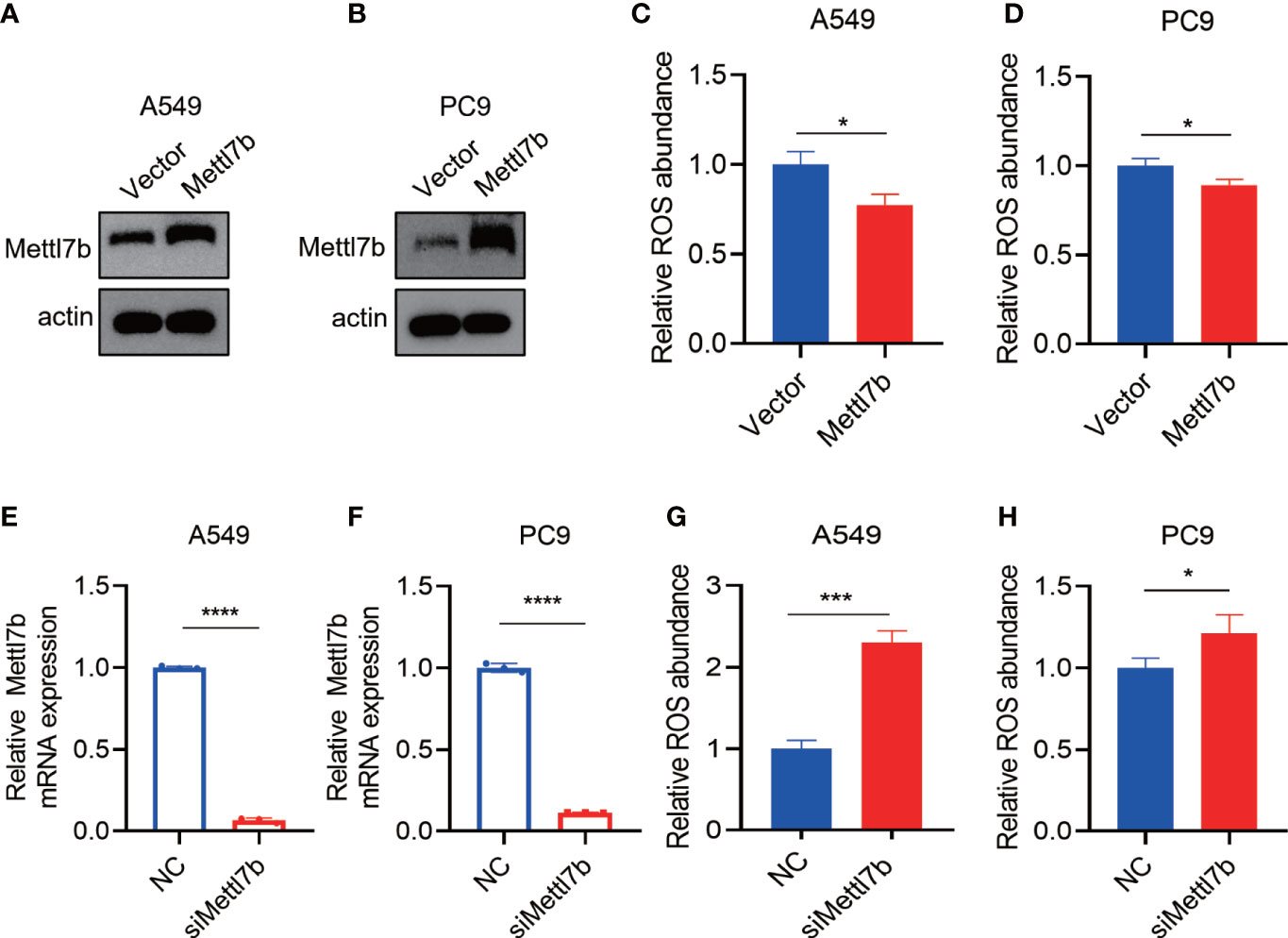
Figure 8 The METTL7B expression level is related to ROS generation. (A–D) After METTL7B was overexpressed in the A549 and PC9 cells, the relative ROS abundance was reduced. (E–H) mRNA was knocked down in the A549 and PC9 cells, and the relative ROS abundance was increased. * p<0.05, *** p<0.001, **** p<0.0001.
4 Discussion
HF and cancer share many risk factors and epidemiological characteristics, such as high blood pressure, diabetes, obesity, smoking, unhealthy diet, and lack of physical activity. In addition, the two diseases may have some common triggering mechanisms, such as increased oxidative stress, low-level inflammatory responses, neurohormonal system activation, immune responses, and other pathological processes that can simultaneously promote the occurrence and development of HF and cancer. The relationship between the co-occurrence of HF and tumor and the mechanisms through which the two are related is not fully understood.
In this study, we tried to explore the signature genes involved in HF and investigate their potential roles in lung cancer using multiple bioinformatic techniques, including WGCNA, the LASSO algorithm, and the RF method. Four signature genes—ECM2, METTL7B, MNS1, and SFRP4—were selected and verified.
ECM2 protein is a member of the secreted protein acidic and rich in cysteine family; the proteins contained in this family are mainly related to various biological processes in the ECM (28). ECM2 gene is located in chromosome 5 (28), whereas ECM2 protein is expressed in various tissues, including the heart, brain, adrenal gland, epididymis, muscle, and lungs (29). The biological functions of ECM2 are unclear, and it is currently known to be associated with matrix assembly and cell adhesiveness (30, 31). ECM2 shares various similarities with the ECM protein. ECM2 remodeling is a key pathologic feature of HF; it is continuous and contributes to systolic and diastolic impairments (32).
METTL7B gene, also known as associated with lipid droplet protein 1, is located on chromosome 12q13.2. METTL7B protein belongs to the methyltransferase-like family (33) and contains a methyltransferase domain. METTL7B is involved in various diseases such as sepsis (34), lipid metabolism in non-alcoholic steatohepatitis (35, 36), and several tumors, including breast cancer (37), thyroid cancer (38), lung adenocarcinoma (39), and non–small cell lung cancer (40). Interestingly, of all these malignant diseases, the roles of METTL7B have been most widely reported in lung cancers. Liu et al. have reported that METTL7B is required for proliferation and tumorigenesis in non–small cell lung cancer (40). Li et al. have reported that METTL7B is a biomarker for prognosis and promotes the metastasis of lung adenocarcinoma cells (41). Moreover, METTL7B is mainly involved in regulating immunity and ROS generation, which are two important biological processes in both HF and lung cancer (42). In detail, METTL7B is not only a prognosis biomarker but is also involved in the tumorigenesis, proliferation (31) and metastasis of various lung cancers (32), making it a promising therapeutic target for different lung cancer subtypes (30). Therefore, we selected METTL7B for conducting an in-depth study of its potential role in the co-existence of HF and lung cancer. Our results showed that the METTL7B mRNA and protein levels were increased in two lung cancer cell lines (A549 and PC9 cells). Furthermore, after METTL7B was overexpressed in the A549 and PC9 cells, the relative ROS abundance was reduced. By contrast, METTL7B mRNA was knocked down in the A549 and PC9 cells, and the relative ROS abundance was increased. These results indicated that METTL7B may play a role in the co-occurrence of HF and lung cancer by affecting ROS-related pathways, which may act as an alternative target.
The MNS1 gene, located in chromosome 15, may play a role in the control of meiotic division and germ cell differentiation by regulating pairing and recombination during meiosis (43). MNS1 mutations are associated with the occurrence of situs inversus and male infertility (43, 44). Interestingly, recent research has shown that MNS1 may be used as a diagnostic variable when studying HF (26, 45). In 2022, Jiang et al. identified that the high expression of the MNS1 gene, together with fras1-related extracellular matrix 1 (FREM1), may affect the progression of HF by regulating bile acid, fatty acid, and heme metabolism (26). Later in the same year, by integrating three machine learning algorithms, Jiang et al. reported that FREM1 and MNS1 are diagnostic gene signatures for HF (45).
SFRP4 belongs to the SFRP family (46), which functions as soluble modulators of Wnt signaling (47). In detail, SFRP4 harbors a cysteine-rich domain homologous to the putative Wnt-binding site. The expression of SFRP4 in the ventricular myocardium is correlated with the expression of apoptosis-related genes. sFRP1-4 is expressed in cardiomyocytes, and the levels of sFRP3 and sFRP4 are elevated during HF (48).
Myocardial cells are supported by a matrix composed of vascular smooth muscle cells, endothelial cells, fibroblasts, immune cells, and their secreted cytokines and proteins. Changes in the microenvironment not only cause pathological changes such as myocardial cell hypertrophy and abnormal energy metabolism but also indirectly stimulate other organs such as tumor tissues through the action of paracrine or endocrine growth factors, cytokines, and chemokines through blood circulation (10, 49). ssGSEA analysis was conducted to analyze the immune cell infiltration between the HF and healthy groups and the correlation with the signature genes and related signaling pathways. Immune cells are involved in necrotic tissue clearance and infarct repair after myocardial infarction; they are also involved in the development of HF after myocardial infarction. In our study, the patients with HF were significantly prone to cytolytic activity, human leukocyte antigen activation, inflammation promotion, T-cell co-stimulation, and type I interferon response. Moreover, the patients with HF displayed low infiltration of B cells, macrophages, plasmacytoid dendritic cells, and Tfh and Treg cells. Treg cells are beneficial to the heart; they inhibit excessive inflammatory responses and promote stable scarring in the early stages of heart injury. However, the phenotype and function of Treg cells are altered in chronic HF (50). Moreover, different macrophage phenotypes have disparate roles in cardiovascular disease. Theoretically, manipulating macrophage phenotypes may be a means of regulating inflammation in the progression of HF (51).
Similarly, the tumor stroma is also composed of a large number of fibroblasts, lymphocytes, and macrophages and forms a complex molecular environment through the vigorous synthesis and secretion of a large number of protein components, microvesicles, exosomes, and miRNAs transported by them (52). Changes in the tumor microenvironment matrix composition provide the “soil” for the malignant growth of tumor cells; moreover, the rigid ECM (fibrosis) formed around and throughout the tumor creates a physical barrier that limits the spread of drugs to cancer cells and participates in the development of drug resistance (53). Bioinformatics and proteomics approaches have great potential for use in studying the composition of and dynamic changes in the microenvironment in HF and tumor tissues. The prevention and improvement of ECM remodeling and active tumor matrix formation are also key to the successful treatment of HF and tumors.
Our study has several limitations. First, small samples were obtained from public databases, which may lead to selection bias. Therefore, an external validation database should be used to improve the reliability of the results. Clinical trials with a larger sample size are necessary for further validation. Second, the molecular mechanisms underlying the roles of METTL7B in lung cancer and HF by regulating ROS and immune responses should be explored through more biological experiments.
5 Conclusions and limitations
Taken together, we screened four signature genes–ECM2, METTL7B, MNS1, and SFRP4–that may assist in the early diagnosis of HF. Furthermore, immune cell infiltration in patients with HF and their association with the gene signatures were also investigated to find clues about immunity in patients with HF. Because METTL7B gene affects the development and progression of lung cancer, its role in lung cancer cell lines has been verified. Our study has great clinical significance. First, because of the stable incidence of cancer and the improved survival rate of patients with HF, cardiologists will more often encounter patients with HF with cancer symptoms and diagnosed cancers. Second, our study provides some clues about selecting patients, particularly elderly patients with HF, with a high risk of developing cancer.
Our study has several limitations. First, whether METTL7B is uniquely and aberrantly expressed in lung cancer or is universally abnormal in pan-cancers has not yet been explored. Thus, the expression pattern of METTL7B in cancer datasets should be further validated. Second, the score of each signaling pathway in the ssGSEA of lung cancers should be further investigated. Finally, the relationship between the expression levels of METTL7B and oxidative stress should be investigated in myocardial cells.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found within the article/Supplementary Materials.
Author contributions
RD, KY, and JR designed and analyzed the research study; RD, YL, YS, and JZ wrote and revised the manuscript; YS and JZ collected and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 72274169) and the 2022 Zhejiang Health and Science and Technology Program (grant number 2022RC132).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1167446/full#supplementary-material
Supplementary Figure 1 | The research flow chart.
Abbreviations
AUC, area under curve; DEGs, differentially expressed genes; ECM2, extracellular matrix protein 2; GEO, Gene Expression Omnibus; GO, Gene Ontology; HLA, human leukocyte antigen; KEGG, Kyoto Encyclopedia of Genes and Genomes; LASSO, least absolute shrinkage and selection operator; METTL7B, methyltransferase-like 7B; MNS1, meiosis-specific nuclear structural 1; ROC, receiver operating characteristic curve; ROS, reactive oxygen species; SFRP4, secreted frizzled-related protein 4; ssGSEA, single-sample gene set enrichment analysis; TIL, tumor-infiltrating lymphocytes; Tregs, regulatory T cells; WGCNA, weighted gene co-expression network analysis.
References
1. Heidenreich PA, Bozkurt B, Aguilar D, Allen LA, Byun JJ, Colvin MM, et al. 2022 AHA/ACC/HFSA guideline for the management of heart failure: a report of the American college of Cardiology/American heart association joint committee on clinical practice guidelines. Circulation (2022) 145(18):e895–e1032. doi: 10.1161/CIR.0000000000001063
2. Emmons-Bell S, Johnson C, Roth G. Prevalence, incidence and survival of heart failure: a systematic review. Heart (2022) 108(17):1351–60. doi: 10.1136/heartjnl-2021-320131
3. Lam CSP, Solomon SD. Classification of heart failure according to ejection fraction: JACC review topic of the week. J Am Coll Cardiol (2021) 77(25):3217–25. doi: 10.1016/j.jacc.2021.04.070
4. Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, et al. Heart disease and stroke statistics-2021 update: a report from the American heart association. Circulation (2021) 143(8):e254–743. doi: 10.1161/CIR.0000000000000950
5. Sun Y. Myocardial repair/remodelling following infarction: roles of local factors. Cardiovasc Res (2009) 81(3):482–90. doi: 10.1093/cvr/cvn333
6. Levine RA, Hagege AA, Judge DP, Padala M, Dal-Bianco JP, Aikawa E, et al. Mitral valve disease–morphology and mechanisms. Nat Rev Cardiol (2015) 12(12):689–710. doi: 10.1038/nrcardio.2015.161
7. Manning WJ. Asymptomatic aortic stenosis in the elderly: a clinical review. JAMA (2013) 310(14):1490–7. doi: 10.1001/jama.2013.279194
8. Eriksson U, Ricci R, Hunziker L, Kurrer MO, Oudit GY, Watts TH, et al. Dendritic cell-induced autoimmune heart failure requires cooperation between adaptive and innate immunity. Nat Med (2003) 9(12):1484–90. doi: 10.1038/nm960
10. O'Rourke K. Heart failure linked with an increased risk of cancer. Cancer (2022) 128(4):646. doi: 10.1002/cncr.34094
11. Warraich HJ, Allen LA, Mukamal KJ, Ship A, Kociol RD. Accuracy of physician prognosis in heart failure and lung cancer: comparison between physician estimates and model predicted survival. Palliat Med (2016) 30(7):684–9. doi: 10.1177/0269216315626048
12. Samejima Y, Iuchi A, Kanai T, Noda Y, Nasu S, Tanaka A, et al. Development of severe heart failure in a patient with squamous non-small-cell lung cancer during nivolumab treatment. Intern Med (2020) 59(16):2003–8. doi: 10.2169/internalmedicine.4550-20
13. Banke A, Schou M, Videbaek L, Moller JE, Torp-Pedersen C, Gustafsson F, et al. Incidence of cancer in patients with chronic heart failure: a long-term follow-up study. Eur J Heart Fail (2016) 18(3):260–6. doi: 10.1002/ejhf.472
14. Hasin T, Gerber Y, McNallan SM, Weston SA, Kushwaha SS, Nelson TJ, et al. Patients with heart failure have an increased risk of incident cancer. J Am Coll Cardiol (2013) 62(10):881–6. doi: 10.1016/j.jacc.2013.04.088
15. Aboumsallem JP, Moslehi J, de Boer RA. Reverse cardio-oncology: cancer development in patients with cardiovascular disease. J Am Heart Assoc (2020) 9(2):e013754. doi: 10.1161/JAHA.119.013754
16. Bozkurt B, Aguilar D, Deswal A, Dunbar SB, Francis GS, Horwich T, et al. Contributory risk and management of comorbidities of hypertension, obesity, diabetes mellitus, hyperlipidemia, and metabolic syndrome in chronic heart failure: a scientific statement from the American heart association. Circulation (2016) 134(23):e535–78. doi: 10.1161/CIR.0000000000000450
17. Zhou W, Liu G, Hung RJ, Haycock PC, Aldrich MC, Andrew AS, et al. Causal relationships between body mass index, smoking and lung cancer: univariable and multivariable mendelian randomization. Int J Cancer (2021) 148(5):1077–86. doi: 10.1002/ijc.33292
18. Raguraman R, Srivastava A, Munshi A, Ramesh R. Therapeutic approaches targeting molecular signaling pathways common to diabetes, lung diseases and cancer. Adv Drug Delivery Rev (2021) 178:113918. doi: 10.1016/j.addr.2021.113918
19. Lee SY, Kim MT, Jee SH, Im JS. Does hypertension increase mortality risk from lung cancer? a prospective cohort study on smoking, hypertension and lung cancer risk among Korean men. J Hypertens (2002) 20(4):617–22. doi: 10.1097/00004872-200204000-00017
20. Scott JM, Nilsen TS, Gupta D, Jones LW. Exercise therapy and cardiovascular toxicity in cancer. Circulation (2018) 137(11):1176–91. doi: 10.1161/CIRCULATIONAHA.117.024671
21. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res (2015) 43(7):e47. doi: 10.1093/nar/gkv007
22. Zhuang Y, Qiao Z, Bi X, Han D, Jiang Q, Zhang Y, et al. Screening and bioinformatics analysis of crucial gene of heart failure and atrial fibrillation based on GEO database. Med (Kaunas) (2022) 58(10). doi: 10.3390/medicina58101319
23. Langfelder P, Horvath S. WGCNA: an r package for weighted correlation network analysis. BMC Bioinf (2008) 9:559. doi: 10.1186/1471-2105-9-559
24. Tibshirani R. The lasso method for variable selection in the cox model. Stat Med (1997) 16(4):385–95. doi: 10.1002/(SICI)1097-0258(19970228)16:4<385::AID-SIM380>3.0.CO;2-3
25. Izmirlian G. Application of the random forest classification algorithm to a SELDI-TOF proteomics study in the setting of a cancer prevention trial. Ann N Y Acad Sci (2004) 1020:154–74. doi: 10.1196/annals.1310.015
26. Jiang Y, Zhang Y, Zhao C. Integrated gene expression pro fi ling analysis reveals SERPINA3, FCN3, FREM1, MNS1 as candidate biomarkers in heart failure and their correlation with immune in fi ltration. J Thorac Dis (2022) 14(4):1106–19. doi: 10.21037/jtd-22-22
27. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity (2013) 39(4):782–95. doi: 10.1016/j.immuni.2013.10.003
28. Oritani K, Kanakura Y, Aoyama K, Yokota T, Copeland NG, Gilbert DJ, et al. Matrix glycoprotein SC1/ECM2 augments b lymphopoiesis. Blood (1997) 90(9):3404–13. doi: 10.1182/blood.V90.9.3404
29. Soderling JA, Reed MJ, Corsa A, Sage EH. Cloning and expression of murine SC1, a gene product homologous to SPARC. J Histochem Cytochem (1997) 45(6):823–35. doi: 10.1177/002215549704500607
30. Wang P, Yan F, Li Z, Yu Y, Parnell SE, Xiong Y. Impaired plasma membrane localization of ubiquitin ligase complex underlies 3-m syndrome development. J Clin Invest (2019) 129(10):4393–407. doi: 10.1172/JCI129107
31. Oritani K, Kincade PW. Lymphopoiesis and matrix glycoprotein SC1/ECM2. Leuk Lymp (1998) 32(1-2):1–7. doi: 10.3109/10428199809059241
32. Frangogiannis NG. The extracellular matrix in ischemic and nonischemic heart failure. Circ Res (2019) 125(1):117–46. doi: 10.1161/CIRCRESAHA.119.311148
33. Ignatova VV, Jansen P, Baltissen MP, Vermeulen M, Schneider R. The interactome of a family of potential methyltransferases in HeLa cells. Sci Rep (2019) 9(1):6584. doi: 10.1038/s41598-019-43010-2
34. Huang D, Yan H. Methyltransferase like 7B is upregulated in sepsis and modulates lipopolysaccharide-induced inflammatory response and macrophage polarization. Bioengineered (2022) 13(5):11753–66. doi: 10.1080/21655979.2022.2068892
35. Yang X, Yuan Y, Xie D. Low molecular pectin inhibited the lipid accumulation by upregulation of METTL7B. Appl Biochem Biotechnol (2021) 193(5):1469–81. doi: 10.1007/s12010-021-03486-z
36. Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, et al. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic (2006) 7(9):1254–69. doi: 10.1111/j.1600-0854.2006.00465.x
37. McKinnon CM, Mellor H. The tumor suppressor RhoBTB1 controls golgi integrity and breast cancer cell invasion through METTL7B. BMC Cancer (2017) 17(1):145. doi: 10.1186/s12885-017-3138-3
38. Ye D, Jiang Y, Sun Y, Li Y, Cai Y, Wang Q, et al. METTL7B promotes migration and invasion in thyroid cancer through epithelial-mesenchymal transition. J Mol Endocrinol (2019) 63(1):51–61. doi: 10.1530/JME-18-0261
39. Song H, Liu D, Wang L, Liu K, Chen C, Wang L, et al. Methyltransferase like 7B is a potential therapeutic target for reversing EGFR-TKIs resistance in lung adenocarcinoma. Mol Cancer (2022) 21(1):43. doi: 10.1186/s12943-022-01519-7
40. Liu D, Li W, Zhong F, Yin J, Zhou W, Li S, et al. METTL7B is required for cancer cell proliferation and tumorigenesis in non-small cell lung cancer. Front Pharmacol (2020) 11:178. doi: 10.3389/fphar.2020.00178
41. Li R, Mu C, Cao Y, Fan Y. METTL7B serves as a prognostic biomarker and promotes metastasis of lung adenocarcinoma cells. Ann Transl Med (2022) 10(16):895. doi: 10.21037/atm-22-3849
42. Xiong Y, Li M, Bai J, Sheng Y, Zhang Y. High level of METTL7B indicates poor prognosis of patients and is related to immunity in glioma. Front Oncol (2021) 11:650534. doi: 10.3389/fonc.2021.650534
43. Ta-Shma A, Hjeij R, Perles Z, Dougherty GW, Abu Zahira I, Letteboer SJF, et al. Homozygous loss-of-function mutations in MNS1 cause laterality defects and likely male infertility. PloS Genet (2018) 14(8):e1007602. doi: 10.1371/journal.pgen.1007602
44. Leslie JS, Rawlins LE, Chioza BA, Olubodun OR, Salter CG, Fasham J, et al. MNS1 variant associated with situs inversus and male infertility. Eur J Hum Genet (2020) 28(1):50–5. doi: 10.1038/s41431-019-0489-z
45. Jiang C, Jiang W. Integrated bioinformatics identifies FREM1 as a diagnostic gene signature for heart failure. Appl Bionics Biomech (2022) 2022:1425032. doi: 10.1155/2022/1425032
46. Jones SE, Jomary C. Secreted frizzled-related proteins: searching for relationships and patterns. Bioessays (2002) 24(9):811–20. doi: 10.1002/bies.10136
47. Pohl S, Scott R, Arfuso F, Perumal V, Dharmarajan A. Secreted frizzled-related protein 4 and its implications in cancer and apoptosis. Tumour Biol (2015) 36(1):143–52. doi: 10.1007/s13277-014-2956-z
48. Jin X, Guo B, Yan J, Yang R, Chang L, Wang Y, et al. Angiotensin II increases secreted frizzled-related protein 5 (sFRP5) expression through AT1 receptor/Rho/ROCK1/JNK signaling in cardiomyocytes. Mol Cell Biochem (2015) 408(1-2):215–22. doi: 10.1007/s11010-015-2497-9
49. Banerjee P, Kotla S, Reddy Velatooru L, Abe RJ, Davis EA, Cooke JP, et al. Senescence-associated secretory phenotype as a hinge between cardiovascular diseases and cancer. Front Cardiovasc Med (2021) 8:763930. doi: 10.3389/fcvm.2021.763930
50. Lu Y, Xia N, Cheng X. Regulatory T cells in chronic heart failure. Front Immunol (2021) 12:732794. doi: 10.3389/fimmu.2021.732794
51. Shirazi LF, Bissett J, Romeo F, Mehta JL. Role of inflammation in heart failure. Curr Atheroscler Rep (2017) 19(6):27. doi: 10.1007/s11883-017-0660-3
52. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.CAN-18-3962
Keywords: heart failure, lung cancer, comorbidity, immune infiltration, oxidative stress
Citation: Duan R, Ye K, Li Y, Sun Y, Zhu J and Ren J (2023) Heart failure–related genes associated with oxidative stress and the immune landscape in lung cancer. Front. Immunol. 14:1167446. doi: 10.3389/fimmu.2023.1167446
Received: 16 February 2023; Accepted: 02 May 2023;
Published: 18 May 2023.
Edited by:
Dongmei Zhang, Sichuan University, ChinaReviewed by:
Xiaofeng Qu, The Second Hospital of Dalian Medical University, ChinaJiaofang Shao, Nanjing Medical University, China
Zhe Zhen, First Affiliated Hospital of Sun Yat-sen University, China
Copyright © 2023 Duan, Ye, Li, Sun, Zhu and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingjing Ren, MzIwNDA5MkB6anUuZWR1LmNu
†These authors have contributed equally to this work
 Ruoshu Duan
Ruoshu Duan Kangli Ye†
Kangli Ye† Jiahong Zhu
Jiahong Zhu Jingjing Ren
Jingjing Ren