
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 17 May 2023
Sec. Vaccines and Molecular Therapeutics
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1166261
This article is part of the Research TopicCOVID-19 booster vaccination: increasing immunity against life-threatening infection.View all 29 articles
 Sina Hosseinian1
Sina Hosseinian1 Rafael de Assis2
Rafael de Assis2 Ghali Khalil1
Ghali Khalil1 Madeleine K. Luu3
Madeleine K. Luu3 Aarti Jain2
Aarti Jain2 Peter Horvath4
Peter Horvath4 Rie Nakajima2
Rie Nakajima2 Anton M. Palma4
Anton M. Palma4 Anthony Hoang3
Anthony Hoang3 Eisa Razzak3
Eisa Razzak3 Nicholas Garcia1
Nicholas Garcia1 Joshua Alger4
Joshua Alger4 Mina Kalantari5
Mina Kalantari5 Emily K. Silzel2
Emily K. Silzel2 Algis Jasinskas2
Algis Jasinskas2 Frank Zaldivar2,6
Frank Zaldivar2,6 Sebastian D. Schubl7
Sebastian D. Schubl7 Philip L. Felgner2*
Philip L. Felgner2* Saahir Khan8*
Saahir Khan8*Introduction: In the context of recurrent surges of SARS-CoV-2 infections, a detailed characterization of antibody persistence over a 6-month period following vaccine booster dose is necessary to crafting effective public health policies on repeat vaccination.
Methods: To characterize the SARS-CoV-2 antibody profile of a healthcare worker population over a 6-month period following mRNA vaccination and booster dose. 323 healthcare workers at an academic medical center in Orange County, California who had completed primary vaccination and booster dose against SARS-CoV-2 were recruited for the study. A total of 690 blood specimens over a 6-month period were collected via finger-stick blood and analyzed for the presence of antibodies against 9 SARS-CoV-2 antigens using a coronavirus antigen microarray.
Results: The primary outcome of this study was the average SARS-CoV-2 antibody level as measured using a novel coronavirus antigen microarray. Additional outcomes measured include levels of antibodies specific to SARS-CoV-2 variants including Delta, Omicron BA.1, and BA.2. We also measured SARS-CoV-2 neutralization capacity for a subset of the population to confirm correlation with antibody levels. Although antibodies against SARS-CoV-2 wane throughout the 6-month period following a booster dose, antibody levels remain higher than pre-boost levels. However, a booster dose of vaccine based on the original Wuhan strain generates approximately 3-fold lower antibody reactivity against Omicron variants BA.1 and BA.2 as compared to the vaccine strain. Despite waning antibody levels, neutralization activity against the vaccine strain is maintained throughout the 6-month period.
Discussion: In the context of recurrent surges of SARS-CoV-2 infections, our data indicate that breakthrough infections are likely driven by novel variants with different antibody specificity and not by time since last dose of vaccination, indicating that development of vaccinations specific to these novel variants is necessary to prevent future surges of SARS-CoV-2 infections.
Since the initial 2019 outbreak of the novel beta coronavirus SARS-CoV-2, rapid international spread of the COVID-19 disease has resulted in a global pandemic. In efforts to contain the spread and severity of COVID-19, the FDA approved the emergency distribution of mRNA vaccines BNT162b2 and mRNA-1273 in December of 2020. Both vaccines provide high rates of protective efficacy of up to 95% against the targeted virus strain following two doses administered at least 3-4 weeks apart (1, 2). There has been a rapid global increase in SARS-CoV-2 cases since then, mainly due to the high infectivity and antibody escape mutations of the new Omicron variants, as well as waning immunity from the BNT162b2 and mRNA-1273 vaccines (3). The FDA has since approved the administration of additional “booster” doses of mRNA vaccines. A booster vaccine dose has previously been effective at protecting against severe COVID-19-related outcomes (4) and has been shown to substantially increase neutralizing antibodies (5, 6). The neutralizing ability of the antibodies has also differed among SARS-CoV-2 variants (7). Therefore, it is of great importance to elucidate the effectiveness of the booster vaccine in maintaining a persistent antibody response in a population consistently exposed to novel variants of SARS-CoV-2.
Here, we seek to analyze the initial rise and waning of SARS-CoV-2 antibody responses induced by the third-dose mRNA vaccine booster in a healthcare worker population over a 6-month period using a coronavirus antigen microarray, with direct comparison of antibodies against multiple variants of concern. Binding antibodies against SARS-CoV-2 antigens have been shown to correlate strongly with neutralizing antibodies, which are a critical component of clinical immunity (8–11). We confirm the correlation of measured antibody responses with SARS-CoV-2 neutralizing capacity for a subset of 30 healthcare workers utilizing an FDA-authorized surrogate neutralization assay (12, 13).
This study was approved by the institutional review board (IRB) of the University of California Irvine (UCI) prior to initiation of the study (protocol HS-20205818). Widespread mRNA vaccination of healthcare workers (HCWs) at UC Irvine Health began in December 2020, administering over 16,000 doses of mRNA-1273 (Moderna-NIAID) or the BNT162b2 (Pfizer-BioNTech) vaccines within the first 4 months. In September of 2021, the FDA approved and UCI secured and administered booster shots to all HCWs who wished to be additionally vaccinated. All HCWs working at the UCI Medical Center, located in Orange County, CA, who participated in our previous study (14) were invited to receive serological testing by providing serum blood samples via a fingerstick directly before vaccination, 1-2 weeks after vaccination, 2 months, 4 months, and 6 months after booster vaccination. All blood samples were brought to the Institute for Clinical and Translational Science Core Laboratory at the UCI Medical Center. Serum samples were centrifuged using the Eppendorf 5415R and spun at 3000xg for 5 minutes. Serum was quickly transferred into a clean sterile tube and frozen at -80°C until analyzed for IgGs. Reports of their serological test results were returned within 4 weeks of receiving the test. At each assessment, demographic and work-related characteristics, testing frequency, exposure risk, and symptom history were collected via surveys administered prior to serum sample collection. Longitudinal participation was encouraged through an aggressive email campaign as well as ensuring that participants received a report of their antibody levels, but not every subject participated at every time point. Subjects who participated provided written consent in the form of an electronic document that was physically signed using an iPad or cellular device.
953 independent finger stick blood serum samples were collected over the 6-month period for analysis. This analysis was restricted to 690 samples to adhere to the pre-specified guidelines (Table 1). Specimens were probed and analyzed on a coronavirus antigen microarray (CoVAM) for IgG and IgM antibodies against 37 antigens from SARS-CoV-2, other coronaviruses, and other respiratory viruses using a coronavirus antigen microarray. The CoVAM contained 10 SARS-CoV-2 antigens including nucleocapsid protein (NP) and several varying fragments of the spike (S) protein, as well as 4 SARS, 3 MERS, 12 Common CoV, and 8 influenza antigens. A full list of antigens used in the assay can be found in Supplementary Figure 1. Samples were tested in triplicate. For more detailed information regarding the coronavirus antigen microarray, analysis, refer to our previous work (14).
In summary, the raw data were checked for quality and normalized using a composite of methods described by Duarte (2013) and Bolstad (2003). Then, using a collection of known samples (here referred as “training set”, known to be positive or negative for SARS-CoV-2), a prediction model was constructed using a logistic regression, in which a combination of SARS-CoV-2 antigens were used as well as a Random Forest model using the full SARS-CoV-2 antigen collection. For single antigen reactivity prediction, a logistic regression model was built for each individual antigen, also using the training set. As a surrogate marker for exposure (in contrast to mRNA vaccination), reactivity to the nucleoprotein, as determined by the Logistic regression model, was used.
This model was found to be 93% sensitive and 98% specific in correctly classifying 91 PCR-positive cases and 88 pre-pandemic negative control (15). The model was then used to generate a weighted composite measure of IgG reactivity across antigens, with weights corresponding to each antigen’s relative importance in the model. This composite IgG reactivity measure was scaled up to represent the weighted mean fluorescence intensity (MFI) of all antigens assayed in the CoVAM. Here, we utilized a model containing all SARS-CoV-2 antigens as above with the exception of NP, as this antigen was used to classify prior exposure to SARS-CoV-2 in a subgroup analysis.
In order to characterize SARS-CoV-2 antibody response over time, we fit a third-degree polynomial model of the average IgG reactivity measure using all available data from n=323 HCWs. Due to the variability in the timing of the tests across individuals, we report the model-estimated average IgG reactivity means and standard error of the mean (SEM) error bars at pre-boost, 1-2 weeks, 2 months, 4 months and 6 months post-boost dose, and compared the changes over time. We then explored differences in long-term antibody response by individual characteristics hypothesized to influence the magnitude and durability of the vaccine-induced antibody response: history of PCR positivity [yes or no], presence of systemic side effects (side effects used in this study include fever, chills, cough, difficulty breathing, congestion, loss of smell or taste, new onset fatigue, myalgia other than site of injection, and headache [none, mild, moderate, or severe] exposure in community [yes or no], HCW role [patient care vs. non-patient care role, gender [male or female, by self-report], age [≥55 vs. <55 years, by self-report]. We tested each potential moderator individually by fitting the same linear mixed effect model with the inclusion of an interaction term between that variable and time (e.g., time * age≥55 vs. <55 years).
In order to determine the correlation between reactivity of SARS-CoV-2 antigens and reactivity of antigens from the other viruses in the array, a correlation matrix using the function rcorr() from the Hmisc (version 5.0-1) package. The correlation matrix was visualized using the corrplot (version 0.92) package. PCA analysis was performed using the tidyverse (version 1.3.2) and factoextra (version 1.0.7) packages. All analyses were conducted using R programing environment v4.1.1.
In order to accurately assess neutralization capacity of SARS-CoV-2 antibodies, we utilized a FDA-authorized research use only surrogate virus neutralization test provided by GeneScript and carried out through a local commercial lab, Innovative Health Design lab located in Irvine, California. Samples and controls are pre-incubated with HRP bound RBD to allow for interaction and binding of neutralization antibodies to RBD-HRP. The mixture is subsequently added to a pre-coated plate of hACE2 protein. Unbound RBD-HRP or RBD-HRP bound to non-neutralizing antibody will be captured by the plate. Neutralization antibodies complexed to RBD-HRP remains in the supernatant and is removed through washout steps. The reaction is quenched and samples are read at 450 nm in a microtiter plate reader. Absorbance of the sample is inversely dependent on the titer of the neutralizing antibodies, numbers are shown as a percentage of controls. Sera of 30 individuals who participated at every timepoint were tested using this surrogate neutralization assay to assess neutralizing capacity for binding of spike RBD fragments from the original Wuhan strain to human ACE2 receptors.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
A total of 690 tests were analyzed from 323 HCWs throughout a 6-month period following booster vaccine dose (Table 1). Composite IgG antibody levels as measured by mean fluorescence intensity significantly increased in the 2 weeks following the booster vaccination (mean 23,303 vs 48,384, p <0.01) (Figure 1). Antibody levels continued to increase from 2 weeks post-boost until 2 months post-boost (mean 48,384 vs. 55,613, p < 0.01) but decreased significantly between months 2 and 4 (mean 55,613 vs. 50,105, p < 0.01) with a non-significant trend towards decreasing antibodies between months 4 and 6 (mean 50,105 vs 46,946, p = 0.14) Despite waning, antibody levels at 6 months post booster dose remain significantly higher than pre-boost levels.
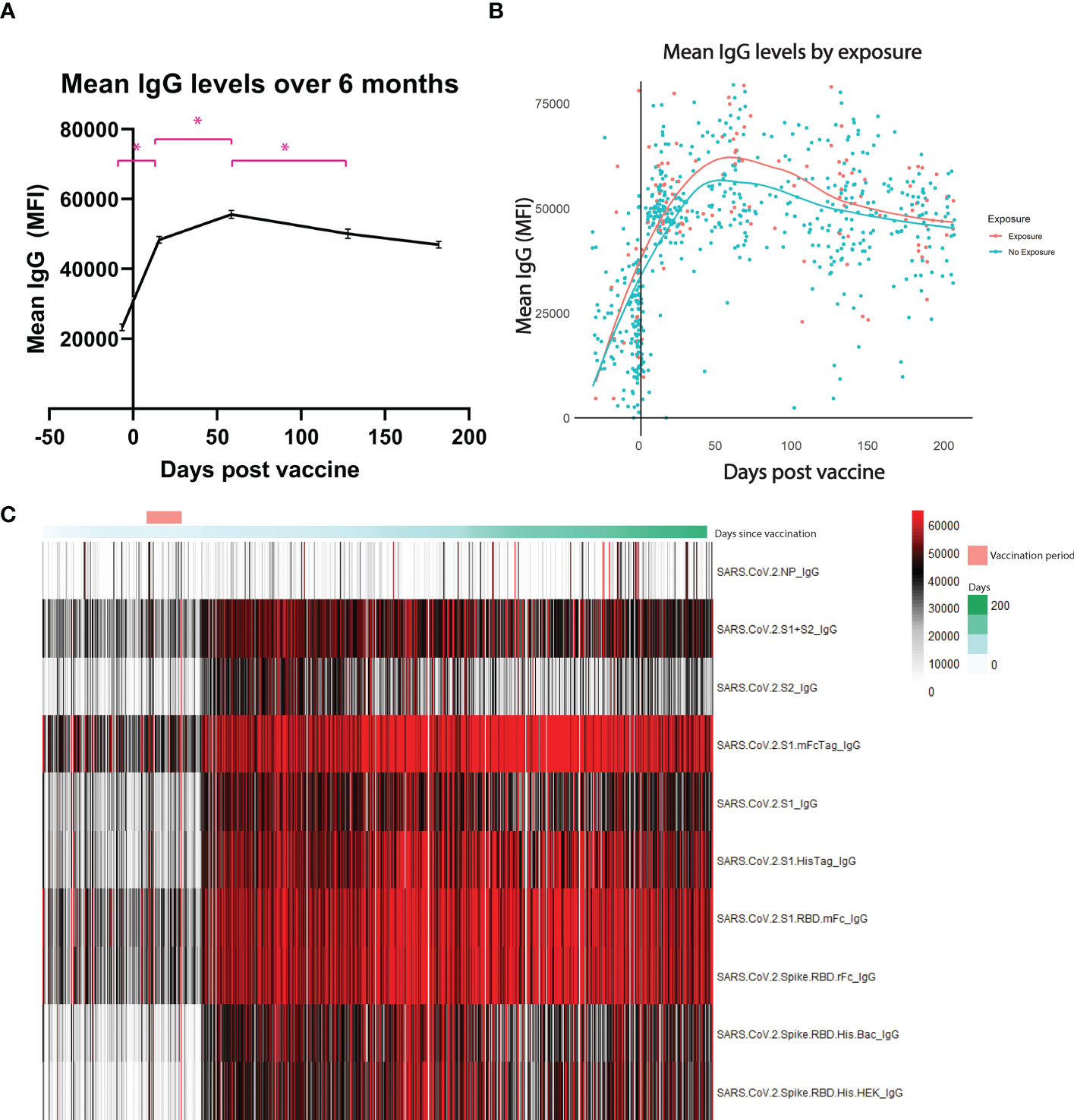
Figure 1 Antibody levels over 6 months. Mean IgG antibody levels measured by mean fluorescence intensity (MFI). (A) shows each individual sample collected over the 6-month period. (B) shows the difference between those who were exposed to the coronavirus at any point before the booster versus those who had no exposure. Exposure is deemed through presence of antibodies against the nucleocapsid protein, a protein only found in the virus and not in the vaccine. (C) is a heatmap of each antigen measured throughout the 6 months. * represents statistically significance, p < 0.05.
To infer natural exposure, we assessed reactivity to the SARS-CoV-2 nucleocapsid protein, as this antigen is not present in currently approved SARS-CoV-2 vaccines. Exposure threshold was determined by logistic regression using the training set as reference. As seen on Figure 1B, no significant difference was observed between the groups at any of the timepoints tested.
The cohort of HCWs were divided into subgroups based on their gender, age, presence of exposure in their community, worker role, presence of comorbidities, and history of PCR-confirmed SARS-CoV-2 infection. Individuals with a history of PCR-confirmed infection after their booster were found to have higher antibody levels at the 4 month timepoint (57,262 vs 40,846, p < 0.01) and at the 6 month timepoint (47,222 vs 42,451, p = 0.04) (Figure 2A). Vaccination systemic side effects were assessed through a 45-point subjective scale of severity encompassing 9 systemic side effects with subjective, self-reported severity from 0 to 5 points. Baseline antibody levels were equivalent between those with no systemic side effects and those with severe systemic side effects (21,639 vs 20,872, p = 0.85), but those with severe systemic side effects had significantly higher antibody levels at 2 months after booster vaccination (51,313 vs 69,849, p < 0.01), with a continued non-significant trend towards higher antibody levels compared to those without systemic side effects at later time points (Figure 2B). Stratification by community exposure, healthcare worker role, gender, and age did not yield any significant differences between groups (Figures 2C–F).
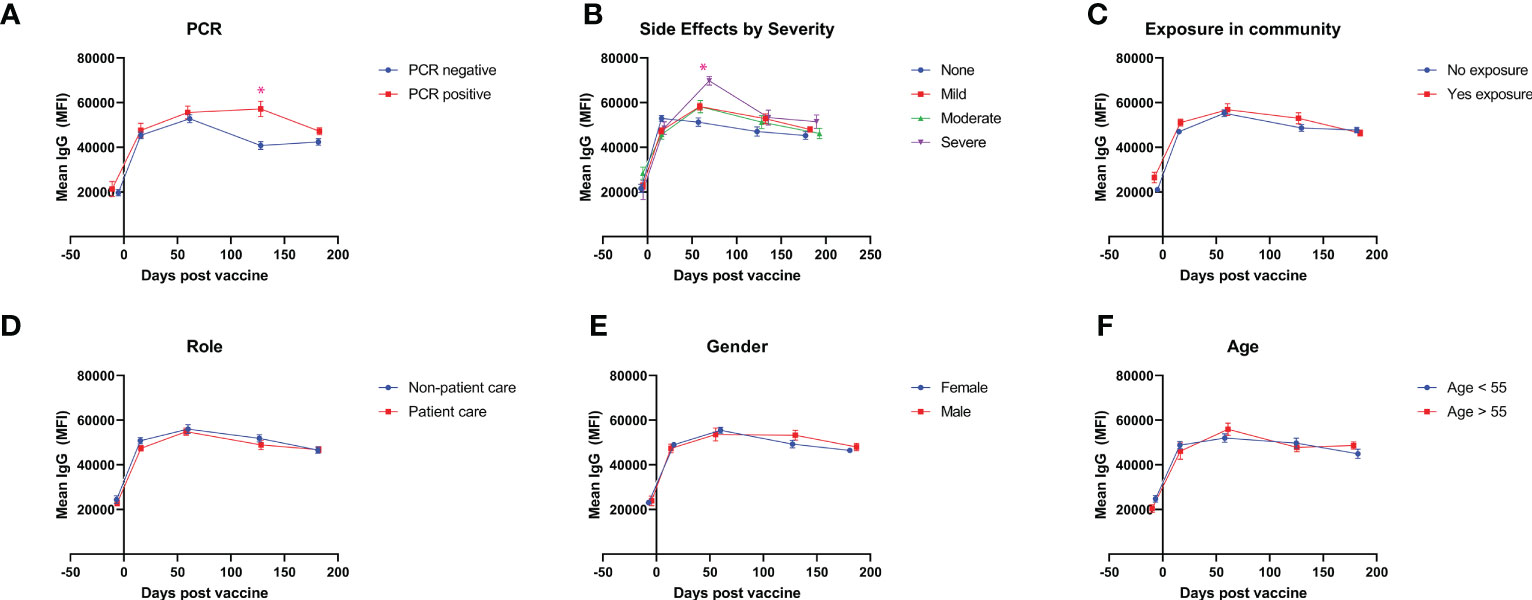
Figure 2 Antibody levels after vaccination for demographic subgroups. Mean IgG antibody levels measured throughout 6 months are compared for self-identified subgroups divided by (A) history of PCR-confirmed SARS-CoV-2 infection after booster vaccination, (B) severity of vaccine side-effects (C) exposure in community after booster vaccination, (D) healthcare worker role, (E) gender, and (F) age. * represents statistically significance, p < 0.05.
Blood specimens from 30 healthcare workers were retrospectively chosen, based on specimen volume and availability at multiple time points and representativeness compared to the study population, to undergo further analysis using a more advanced microarray that included antigens against the original Wuhan strain and 3 variants of concern: Delta, Omicron BA.1, and BA.2. This analysis was restricted to analyze only the receptor-binding domain (RBD) for unbiased comparison between variants and included a total of 213 samples over the 6-month period, with a minimum of 27 samples per time point (Figure 3). Prior to booster vaccine dose, antibody levels against the Wuhan strain were not significantly different from the Delta strain (mean 3473 vs. 2449, p = 0.24) but were significantly lower against Omicron strains BA.1 and BA.2 (mean 3473 vs. 300, p < 0.01 and 3473 vs. 542, p < 0.01). At 2 weeks post booster vaccination, all variants had significantly lower antibody levels directed against them as compared to the Wuhan strain (Delta: 1.3 fold-reduction, p <0.01 Omicron BA.1 6.0 fold-reduction, p < 0.01; BA.2 3.4 fold reduction, p = <0.01). This trend persisted over the 6-month period.
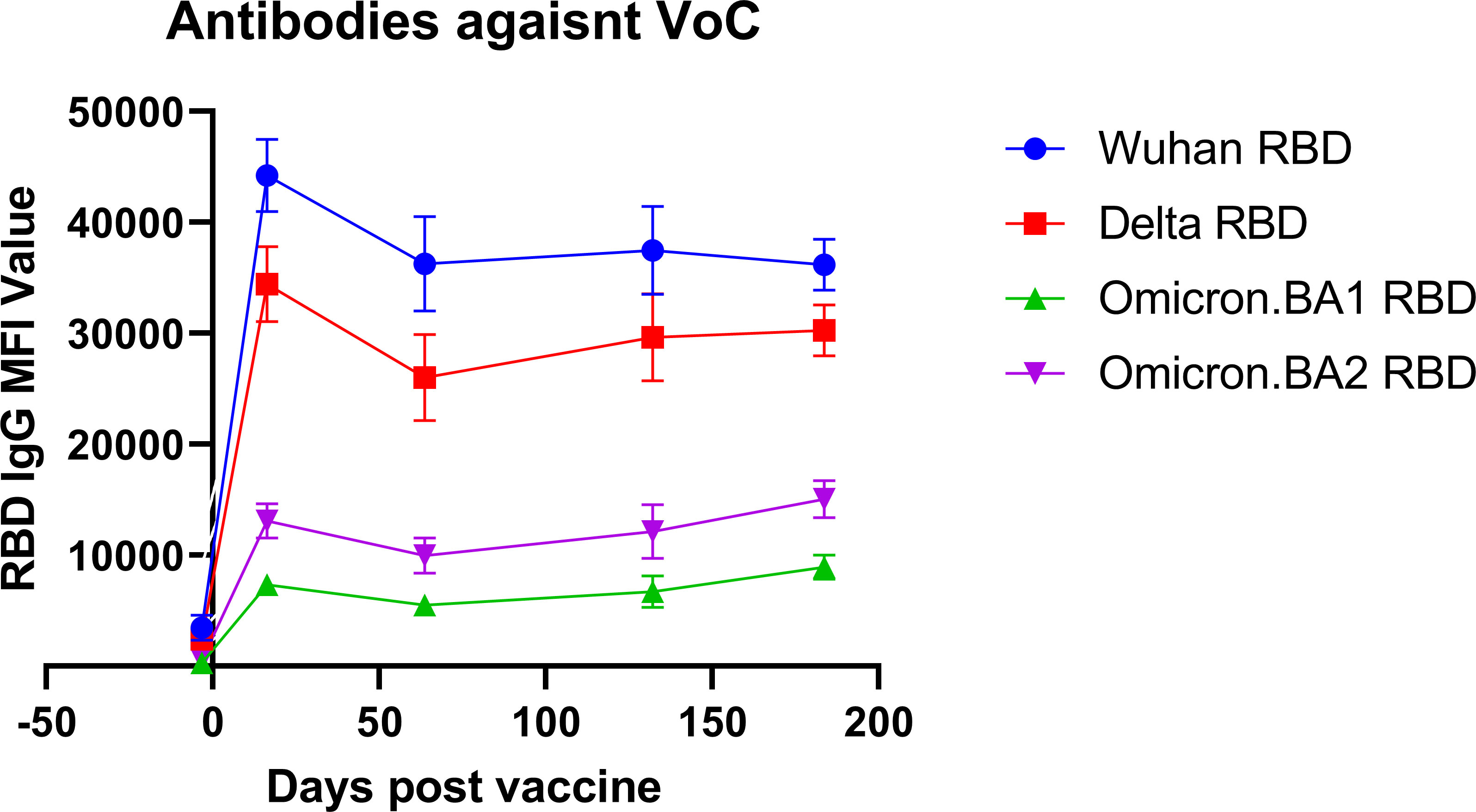
Figure 3 Antibody reactivity against variant RBD antigens. The antibody reactivity as measured by mean fluorescence intensity (MFI) against variant-specific receptor-binding domain (RBD) antigens is shown for different variants of concern (VoC) at different time points after vaccination.
Reactivity to antigens from all viruses represented on the array was observed for multiple samples. However, no significant correlation between the reactivity to SARS-CoV-2 antigens and either endemic coronavirus (229E, NL63, OC43, HKU1) or influenza virus antigens was observed. In accordance with our previous observations and the literature, this can be explained by a low cross reactivity between SARS-CoV-2 S1 and RBD antigens and the same antigens on other coronaviruses. Nevertheless, as shown on Figure 4, the observed correlation between different fragments of SARS-CoV-2 spike antigen was high (averaging r > 0.8) except for the nucleoprotein (averaging a r < 0.3). This observation confirms that the microarray can distinguish between antibody responses induced by vaccines that include spike protein antigens and natural infection which includes exposure to both spike and nucleocapsid protein antigens.

Figure 4 Correlation of antibodies against other respiratory viruses. Using COVAM 5, we are able to measure antibodies against SARS, MERS, CoV, and influenza in addition to SARS-CoV-2. This figure represents the correlation coefficient for each antibody against antibodies from other respiratory viruses.
The same 30 healthcare workers who were selected to undergo variant testing were also selected for analysis of neutralization capacity. These 30 individuals expressed a similar trend to the overall cohort, with their average antibody levels initially increasing from pre-boost to 2 months post-boost, but significant waning between months 2 to 6. Signal inhibition of neutralizing titers increased from 77% before booster vaccination to 96% immediately after booster vaccination (p < 0.01). Despite waning antibody levels from months 2 to 6, signal inhibition did not wane significantly over this time (Supplementary Figure 2).
In summary, we report data on the boosting of both composite IgG antibody levels and antibody neutralizing activity by a third dose of mRNA vaccine against SARS-CoV-2. The strengths of our study include a 6-month follow-up period, the comparison of antibody responses against SARS-CoV-2 variants of concern, and inclusion of subgroup analyses with respect to demographics, vaccine side-effects, and breakthrough SARS-CoV-2 infection. The weaknesses of our study include limited power to detect differences in subgroups and limitations of the HCW population, which is a relatively young and healthy population with few individuals who are immunocompromised or over the age of 65.
Both IgG antibody levels and neutralization activity have been shown to wane over the course of months following vaccination (14, 16). The booster vaccine dose has been shown to significantly increase both IgG antibody levels and neutralization activity (5, 17). Similarly, the booster vaccine dose has been shown to reduce risk of SARS-CoV-2 breakthrough infections (6). However, previous studies have focused on short-term effect of booster vaccination on SARS-CoV-2 antibody responses (18), while we chose to observe the study population over a longer 6-month follow-up period.
Using our CoVAM assay, we observed a significant increase in antibody levels following the booster dose of the vaccine. Despite significant waning between months 2-6, the antibody levels at 6 months remain higher than pre-boost levels. We also show no significant difference between individuals who were exposed to SARS-CoV-2 (as determined through reactivity to the nucleocapsid protein) compared to those who had no previous exposure. This data is limited in the ability to detect a small effect of previous exposure by our small sample size of individuals with previous exposure. Others have shown longer-lasting immunity as exhibited by higher antibody levels and neutralization activity in those with previous infection 9 months after primary 2-dose COVID-19 mRNA vaccination (19). In the context of the high rate of breakthrough infections prior to 6 months observed in recent surges of SARS-CoV-2 infections, these data indicate that waning of antibody levels over time is not the primary driver of SARS-CoV-2 breakthrough infections.
We stratified study participants based on gender, age, community exposure, worker role, and breakthrough COVID-19 infection based on a history of PCR positivity. We did not find significant differences in antibody levels for subgroups stratified by gender, age, self-reported community exposure to COVID-19, or clinical role, although the study may have been underpowered to detect small differences within these subgroups. We had a small cohort of individuals who had breakthrough infections confirmed through PCR positivity, and these individuals showed less waning in antibody responses with significantly higher antibody levels at 4 and 6 months.
We, among others, hypothesized that the quantity and severity of systemic side effects may be correlated with a more robust immune response, resulting in higher antibody titers. We show that the presence of systemic side effects is associated with higher antibody titers up to 4 months following booster vaccination. Severe systemic side effects are associated with higher antibody levels in a longitudinal fashion throughout the 6-months period. These data can reassure patients that systemic side-effects associated with vaccination may represent a more robust immune response to the vaccine dose.
The neutralizing capacity of SARS-CoV-2 antibodies has been shown to increase quickly following the vaccine booster dose (20). While we focused on binding antibody levels in this study, we did confirm correlation between binding antibodies and neutralizing antibodies in a subset of patients using an FDA-authorized surrogate neutralization assay (12). We retroactively selected 30 individuals who participated at every time point available, including before booster vaccination, in order to get a complete timeline of neutralization throughout the 6 month period. We acknowledge that this may lead to some selection bias, as those who come to every collection offered may be more likely to be more compliant with masking and exposure guidelines. We report significant increase in neutralizing antibodies against the original Wuhan strain following booster vaccine dose with maintenance throughout the 6-month period in this cohort, despite waning binding antibody levels measured on the CoVAM. It is important to note that our neutralization data are limited in that our assay was likely saturated at post-boost time points, with our values being at 96% inhibition, which may not fully allow us to detect small differences between post-boost timepoints. However, based on these data, we hypothesize that the decrease in antibody levels over the course of the 6-month period is not sufficient to reduce neutralization capacity and therefore unlikely clinically significant given correlation between neutralization capacity and clinical immunity.
We acknowledge that others have directly compared the neutralization capacity of vaccinee sera against SARS-CoV-2 variants of concern (20), however, estimates for the magnitude of antibody escape by the Omicron variant of concern vary widely from as low as 1.5-fold to as high as 32-fold (21) (22) (23, 24). Using an updated version of the CoVAM, we were able to directly compare antibodies elicited by booster vaccination against the spike protein receptor-binding domain antigens of multiple variants of concern. Our data show that antibodies elicited by booster vaccination directed against the original Wuhan strain show only a small non-significant decrease in binding to RBD antigen from the Delta variant but show significant decreases in binding to Omicron variants BA.1 (4-fold decrease) and BA.2 (3-fold decrease). Although the booster dose is effective in maintaining antibodies against Wuhan and Delta strain antigens, these data suggest that vaccines directed against Omicron strain antigens are needed to effectively prevent breakthrough COVID-19 infections.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by UCI IRB. The patients/participants provided their written informed consent to participate in this study.
SH, GK, ML, AP, SS, PF, and SK conceived and designed research. SH, GK, ML, AH, ES, and NG collected samples. PH, JA, and FZ prepared and stored samples. PF and SK designed the microarray. RA, AJ, RN, and AJ constructed the microarray and probed samples. RA analyzed data. SH, GK, SS, PF, and SK interpreted results of data. SH and RA prepared figures. SH, GK, ML, NG, and SK drafted the manuscript. SH and SK edited and revised the manuscript. SH, SS, PF, and SK obtained funding for the project. All authors contributed to the article and approved the submitted version.
This work was supported by two intramural research grants from the COVID-19 Basic, Translational, and Clinical Research Fund of the University of California Irvine and by the Emergency COVID-19 Research Seed Funding Opportunity from the University of California Office of the President [research grants R00RG2646, R01RG3745]. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position or policy of the University of California. This work was supported by a grant from the Surgical Infection Society Foundation. Research reported in this publication was supported by The Institute for Clinical and Translational Science of the National Institutes of Health under award number T35DK128788. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work utilized the Chao Family Comprehensive Cancer Center Experimental Tissue Shared Resource, supported by the National Cancer Institute of the National Institutes of Health [award number P30CA062203]. SK was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health [grant KL2 TR001416]. The project described was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health [grant UL1 TR001414]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The initial design and construction of the CoVAM was supported by the Prometheus-UMD contract sponsored by the Defense Advanced Research Projects Agency (DARPA) BTO under the auspices of Col. Matthew Hepburn [agreements N66001-17-2-4023, N66001-18-2-4015]. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position or policy of the funding agencies and no official endorsements should be inferred.
We thank Rodrigo Romo of UCI’s Institute for Clinical and Translational Science for their assistance in maintaining sample integrity. We thank Milind Vasudev, Marlene Jacobo, Dalia Breziner, and Joanne Mendoza. We thank Yannik Cadin for helping with the array’s quantification.
The coronavirus antigen microarray is intellectual property of the Regents of the University of California that is licensed for commercialization to Nanommune Inc. Irvine, CA, a private company for which PF is the largest shareholder and several co-authors RA, AaJ, RN, and SK also own shares. Nanommune Inc. has a business partnership with Sino Biological Inc. Beijing, China which expressed and purified the antigens used in this study.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1166261/full#supplementary-material
Supplementary Figure 1 | Antibodies tested through COVAM 5.
Supplementary Figure 2 | Neutralizing capacity of vaccinee sera. Neutralizing capacity of sera, measured as percent signal inhibition of binding of RBD antigen from the Wuhan strain to human ACE2 receptors compared to negative control sera, is shown for different time points after vaccination.
1. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
2. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
3. Goldberg Y, Mandel M, Bar-On YM, Bodenheimer O, Freedman L, Haas EJ, et al. Waning immunity after the BNT162b2 vaccine in Israel. N Engl J Med (2021) 385:e85. doi: 10.1056/NEJMoa2114228
4. Barda N, Dagan N, Cohen C, Hernan MA, Lipsitch M, Kohane IS, et al. Effectiveness of a third dose of the BNT162b2 mRNA COVID-19 vaccine for preventing severe outcomes in Israel: an observational study. Lancet (2021) 398:2093–100. doi: 10.1016/S0140-6736(21)02249-2
5. Nemet I, Kliker L, Lustig Y, Zuckerman N, Erster O, Cohen C, et al. Third BNT162b2 vaccination neutralization of SARS-CoV-2 omicron infection. N Engl J Med (2022) 386:492–4. doi: 10.1056/NEJMc2119358
6. Amir O, Goldberg Y, Mandel M, Bar-On YM, Bodenheimer O, Ash N, et al. Protection following BNT162b2 booster in adolescents substantially exceeds that of a fresh 2-dose vaccine. Nat Commun (2022) 13:1971. doi: 10.1038/s41467-022-29578-w
7. Suryawanshi RK, Chen IP, Ma T, Syed AM, Brazer N, Saldhi P, et al. Limited cross-variant immunity from SARS-CoV-2 omicron without vaccination. Nature (2022). doi: 10.1038/s41586-022-04865-0
8. Rubio-Acero R, Castelletti N, Fingerle V, Olbrich L, Bakuli A, Wolfel R, et al. In search of the SARS-CoV-2 protection correlate: head-to-Head comparison of two quantitative S1 assays in pre-characterized oligo-/Asymptomatic patients. Infect Dis Ther (2021) 10:1505–18. doi: 10.1007/s40121-021-00475-x
9. Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet (2020) 396:1595–606. doi: 10.1016/S0140-6736(20)32137-1
10. Letizia AG, Ge Y, Vangeti S, Goforth C, Weir DL, Kuzmina NA, et al. SARS-CoV-2 seropositivity and subsequent infection risk in healthy young adults: a prospective cohort study. Lancet Respir Med (2021) 9:712–20. doi: 10.1016/S2213-2600(21)00158-2
11. Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med (2021) 27:1205–11. doi: 10.1038/s41591-021-01377-8
12. Valcourt EJ, Manguiat K, Robinson A, Chen JC, Dimitrova K, Philipson C, et al. Evaluation of a commercially-available surrogate virus neutralization test for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Diagn Microbiol Infect Dis (2021) 99:115294. doi: 10.1016/j.diagmicrobio.2020.115294
13. Tan CW, Manguiat K, Robinson A, Chen JC, Dimitrova K, Philipson C, et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol (2020) 38:1073–8. doi: 10.1038/s41587-020-0631-z
14. Hosseinian S, Powers K, Vasudev M, Palma AM, de Assis R, Jain A, et al. Persistence of SARS-CoV-2 antibodies in vaccinated health care workers analyzed by coronavirus antigen microarray. Front Immunol (2022) 13:817345. doi: 10.3389/fimmu.2022.817345
15. de Assis RR, Jain A, Nakajima R, Jasinskas A, Felgner J, Obiero JM, et al. Analysis of SARS-CoV-2 antibodies in COVID-19 convalescent blood using a coronavirus antigen microarray. Nat Commun (2021) 12:6. doi: 10.1038/s41467-020-20095-2
16. Levin EG, Lustig Y, Cohen C, Fluss R, Indenbaum V, Amit S, et al. Waning immune humoral response to BNT162b2 covid-19 vaccine over 6 months. N Engl J Med (2021) 385:e84. doi: 10.1056/NEJMoa2114583
17. Eliakim-Raz N, Leibovici-Weisman Y, Stemmer A, Ness A, Awwad M, Ghantous N, et al. Antibody titers before and after a third dose of the SARS-CoV-2 BNT162b2 vaccine in adults aged >/=60 years. JAMA (2021) 326:2203–4. doi: 10.1001/jama.2021.19885
18. Atmar RL, Lyke KE, Deming ME, Jackson LA, Branche A R, El Sahly HM, et al. Heterologous SARS-CoV-2 booster vaccinations - preliminary report. medRxiv (2021). doi: 10.1101/2021.10.10.21264827
19. Perico L, Todeschini M, Casiraghi F, Mister M, Pezzotta A, Peracchi T, et al. Long-term adaptive response in COVID-19 vaccine recipients and the effect of a booster dose. Front Immunol (2023) 14:1123158. doi: 10.3389/fimmu.2023.1123158
20. Basile K, Rockett RJ, McPhie K, Fennell M, Johnson-Mackinnon J, Agius JE, et al. Improved neutralization of the SARS-CoV-2 omicron variant after pfizer-BioNTech BNT162b2 COVID-19 vaccine boosting. bioRxiv (2021). doi: 10.1101/2021.12.12.472252
21. Cele S, Gazy I, Jackson L, Hwa SH, Tegally H, Lustig G, et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature (2021) 593:142–6. doi: 10.1038/s41586-021-03471-w
22. Yu X, Wei D, Xu W, Li Y, Li X, Zhang X, et al. Reduced sensitivity of SARS-CoV-2 omicron variant to antibody neutralization elicited by booster vaccination. Cell Discovery (2022) 8:4. doi: 10.1038/s41421-022-00375-5
23. Muik A, Lui BG, Wallisch AK, Bacher M, Muhl J, Reinholz J, et al. Neutralization of SARS-CoV-2 omicron by BNT162b2 mRNA vaccine-elicited human sera. Science (2022) 375:678–80. doi: 10.1126/science.abn7591
Keywords: serology, vaccine, mRNA, healthcare worker (HCW), SARS-CoV-2
Citation: Hosseinian S, de Assis R, Khalil G, Luu M K, Jain A, Horvath P, Nakajima R, Palma AM, Hoang A, Razzak E, Garcia N, Alger J, Kalantari M, Silzel EK, Jasinskas A, Zaldivar F, Schubl SD, Felgner PL and Khan S (2023) Analysis and comparison of SARS-CoV-2 variant antibodies and neutralizing activity for 6 months after a booster mRNA vaccine in a healthcare worker population. Front. Immunol. 14:1166261. doi: 10.3389/fimmu.2023.1166261
Received: 15 February 2023; Accepted: 02 May 2023;
Published: 17 May 2023.
Edited by:
Ritthideach Yorsaeng, Chulalongkorn University, ThailandCopyright © 2023 Hosseinian, de Assis, Khalil, Luu, Jain, Horvath, Nakajima, Palma, Hoang, Razzak, Garcia, Alger, Kalantari, Silzel, Jasinskas, Zaldivar, Schubl, Felgner and Khan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saahir Khan, c2FhaGlya2hAdXNjLmVkdQ==; Philip L. Felgner, cGZlbGduZXJAaHMudWNpLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.