- 1Experimental Therapeutics Unit, Department of Medical Oncology, Hospital Clinico San Carlos and Health Research Institute of the Hospital Clinico San Carlos (IdISSC), Madrid, Spain
- 2Division of Medical Oncology and Hematology, Department of Medicine, Princess Margaret Cancer Centre and University of Toronto, Toronto, ON, Canada
- 3Department of Clinical Oncology, Velindre Cancer Centre, Cardiff, United Kingdom
- 4Department of Medical Oncology, Clinica Universidad de Navarra, Madrid, Spain
Introduction: Identification of modulators of the immune response with inhibitory properties that could be susceptible for therapeutic intervention is a key goal in cancer research. An example is the human leukocyte antigen G (HLA-G), a nonclassical major histocompatibility complex (MHC) class I molecule, involved in cancer progression.
Methods: In this article we performed a systematic review and meta-analysis on the association between HLA-G expression and outcome in solid tumors. This study was performed in accordance with PRISMA guidelines and registered in PROSPERO.
Results: A total of 25 studies met the inclusion criteria. These studies comprised data from 4871 patients reporting overall survival (OS), and 961 patients, reporting disease free survival (DFS). HLA-G expression was associated with worse OS (HR 2.09, 95% CI = 1.67 to 2.63; P < .001), that was higher in gastric (HR = 3.40; 95% CI = 1.64 to 7.03), pancreatic (HR = 1.72; 95% CI = 0.79 to 3.74) and colorectal (HR = 1.55; 95% CI = 1.16 to 2.07) cancer. No significant differences were observed between the most commonly utilized antibody (4H84) and other methods of detection. HLA-G expression was associated with DFS which approached but did not meet statistical significance.
Discussion: In summary, we describe the first meta-analysis associating HLA-G expression and worse survival in a variety of solid tumors.
Systematic Review Registration: https://www.crd.york.ac.uk/PROSPERO/, identifier CRD42022311973.
Introduction
Identification of modulators of the immune response with inhibitory properties that could be susceptible for therapeutic intervention is a key goal in cancer research. The human leukocyte antigen G (HLA-G) is a nonclassical major histocompatibility complex (MHC) class I molecule that belongs to the group of HLA-class Ib (1). HLA-G binds to the leukocyte Ig-like receptor subfamily B member 1 (LILRB1) and member 2 (LILRB2), and the killer immunoglobulin-like receptor 2DL4 (KIR2DL4) (2, 3). This molecule induces tolerance towards both innate and acquired immune cells. HLA-G has been associated with critical functions in maternal tolerance of the fetus during pregnancy, and its presence at this immunologically privileged site was proposed as a mechanism used by the fetus to avoid rejection by the mother’s immune system (1–5).
In recent years, the distribution of HLA-G in normal tissues has been found to be broader than initially thought. HLA-G molecules have been detected in embryonic (amniotic cells and fluid, endothelial cells from the chorionic villi and erythroid cells) and in some adult tissues (thymic epithelial cells or the bone marrow in cells of the erythropoietic lineage) (4).
This immune-suppressive function supports a role of HLA-G in tumor development and progression. In malignancy, pathologic HLA-G expression was first found in melanoma samples with no expression in adjacent normal tissues (6). Since this report, HLA-G expression has been associated with various malignancies such as breast cancer, colorectal cancer, cervical cancer, endometrial carcinoma, oesophageal cancer, Ewing sarcoma, gastric cancer, and lung cancer, among others (7). HLA-G expression in solid tumors has been associated with advanced disease stage, tumor metastasis, poor prognosis or poorer disease-free survival (7–10).
Recently agents targeting HLA-G or their receptors, such as the LILRB family of proteins have shown preclinical activity, and some of these agents have transitioned to clinical testing (11, 12). These drugs predominantly act on myeloid derived suppressor cells (13). Given the differential immune profile at different stages of cancer progression we aimed to understand the association between HLA-G expression and clinical parameters and with patient outcome.
In this systematic review, we aimed to explore the prognostic association between HLA-G in solid tumors with a particular focus on early-stage disease where the immune microenvironment plays a central role. We hypothesised that the expression of HLA-G would be associated with worse outcomes due to its inhibitory effects on the immune response.
Methods
Study design
This systematic review and meta-analysis were performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) and was conducted following the Cochrane Handbook for Systematic Reviews of Interventions recommendations. This study was registered in PROSPERO (registration number: CRD42022311973).
Search strategy and selection criteria
We searched MEDLINE (host: PubMed) and EMBASE to identify studies published from inception to April 24, 2022, and which evaluated: 1) HLA-G expression in solid tumors; 2) reporting hazard ratio (HR) and 95% confidence interval (CI) or a P value for overall survival (OS). In a secondary analysis, studies providing a HR for disease-free survival (DFS) were included. Only studies reporting on multivariable Cox proportional-hazards regression analyses were comprised. Those reporting univariable analyses were excluded. The titles identified by the initial search were evaluated, and potentially relevant publications were retrieved in full. Three authors (JBA, JDBF and GFH) reviewed the full articles for eligibility independently. Discrepancies were resolved by consensus. The following MeSH terms were used for the search: “neoplasm”, “malignancy”, “tumor”, “carcinoma”, “cancer” “human leukocyte antigen G” and “HLA-G”. We focused our search on studies performed in adults and reported in English. Non-human studies or preclinical studies, non-malignant pathology, haematological malignancies, case reports, literature reviews, letters to editors and studies with no survival data as specified in the eligibility criteria were excluded. With the emphasis being the association between HLA-G expression and outcomes in early-stage/curable malignancies, studies with more than 50% of patients in the metastatic setting were also excluded.
Data extraction
Data were collected by one reviewer (CMV) and quality assessment was conducted by a second author (EA). Discrepancies were resolved by consensus. All data were extracted from primary publications and their associated online appendices. Data were collected into an electronic database, which included the following data: study summary characteristics such as tumor type, setting (curative vs palliative), number of patients, proportion of patients with advanced and early stage disease, mean age, gender, median follow-up (months), type of HLA-G detection (tissue vs plasma), detection technique (Immunohistochemistry [IHC], Enzyme-Linked Immunosorbent Assay [ELISA] or mRNA), antibody used for detection (when appropriate) and, proportion of patients with HLA-G expression. We also extracted outcome data such as OS and DFS from multivariable Cox proportional-hazards regression analyses.
Statistical analysis
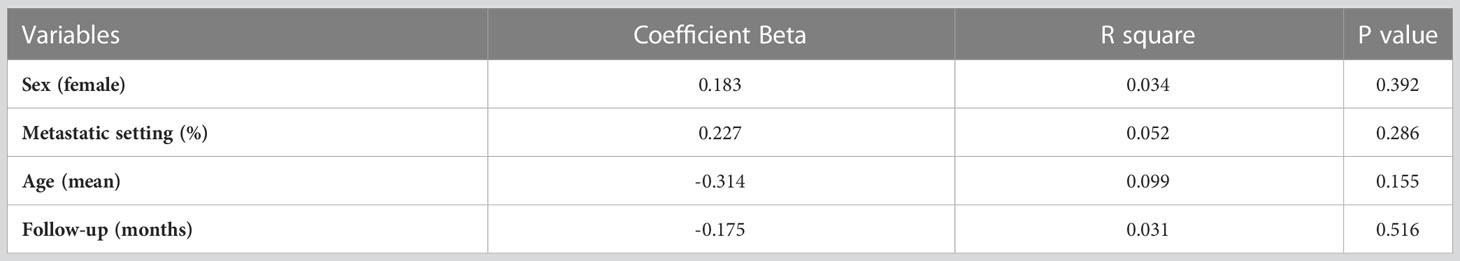
Extracted data were combined into a meta-analysis using Review Manager v5.4 analysis software (Cochrane Collaboration, Copenhagen, Denmark). In light of the expectation for substantial clinical heterogeneity, estimates of HRs were weighted and pooled using the generic inverse-variance and random-effect model (14, 15) irrespective of statistical heterogeneity. A post-hoc sensitivity analysis was also performed to exclude any studies in which HRs were not adjusted for age in multivariable analyses. Meta-regression was performed using SPSS version 28.0 (IBM Corp, Armonk, NY, USA) and comprised linear regression weighted by inverse-variance using the weighted least squares (mixed effect) function (16). The following variables were regressed against the natural logarithm for the HR for OS: mean age, proportion of females, proportion of patients with advanced stage, median follow-up (months) and proportion of HLA-G positive. For meta-analysis, statistical significance was defined as P <0.05. For meta-regression, in light of the small number of studies resulting in low statistical power, significance was defined quantitatively using thresholds defined by Burnand et al. (17) (quantitative significance defined as ß coefficient >0.28). Publication bias was explored by visual inspection of the funnel plots. No corrections were applied for multiple significance testing.
Results
Study characteristics
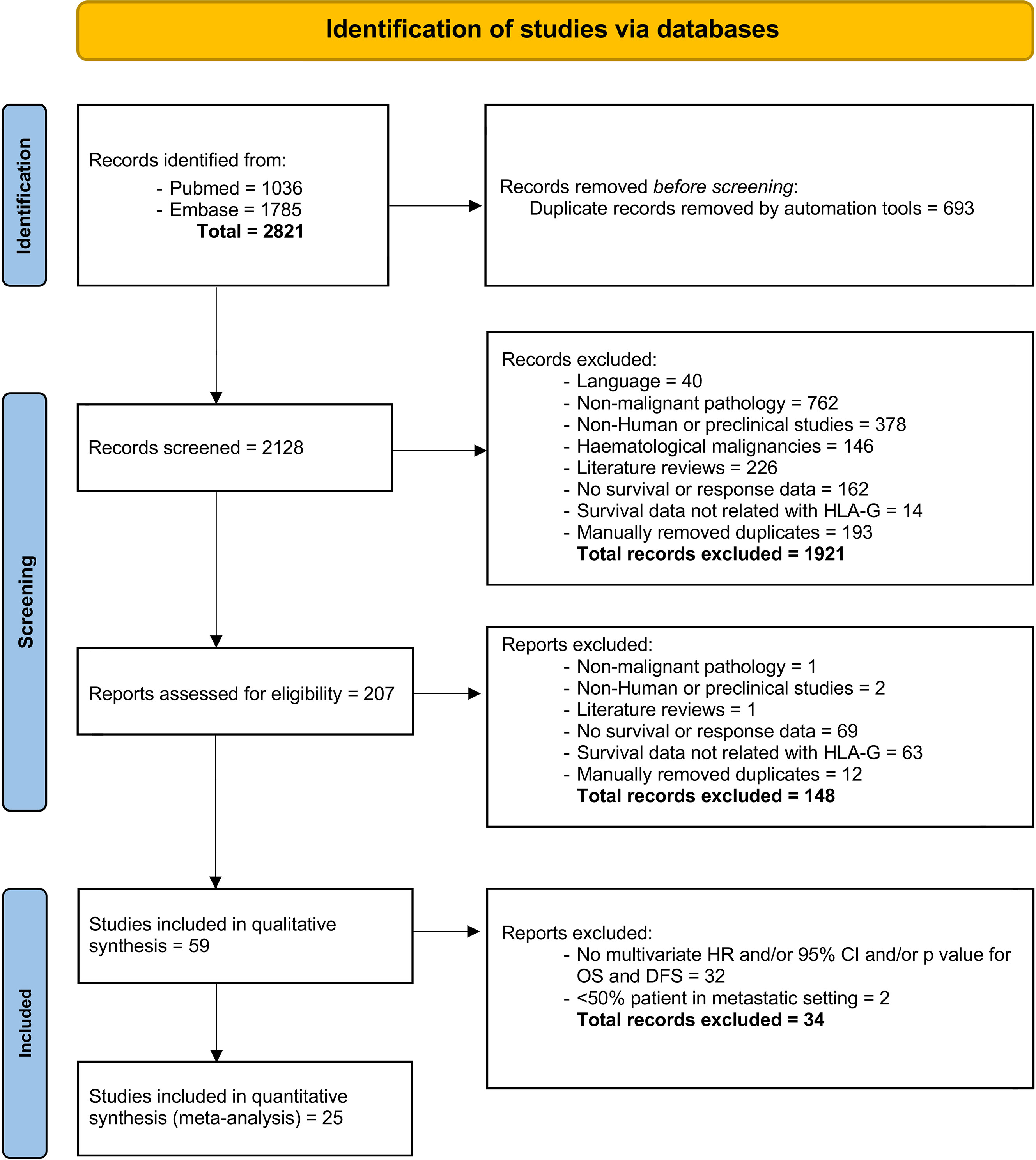
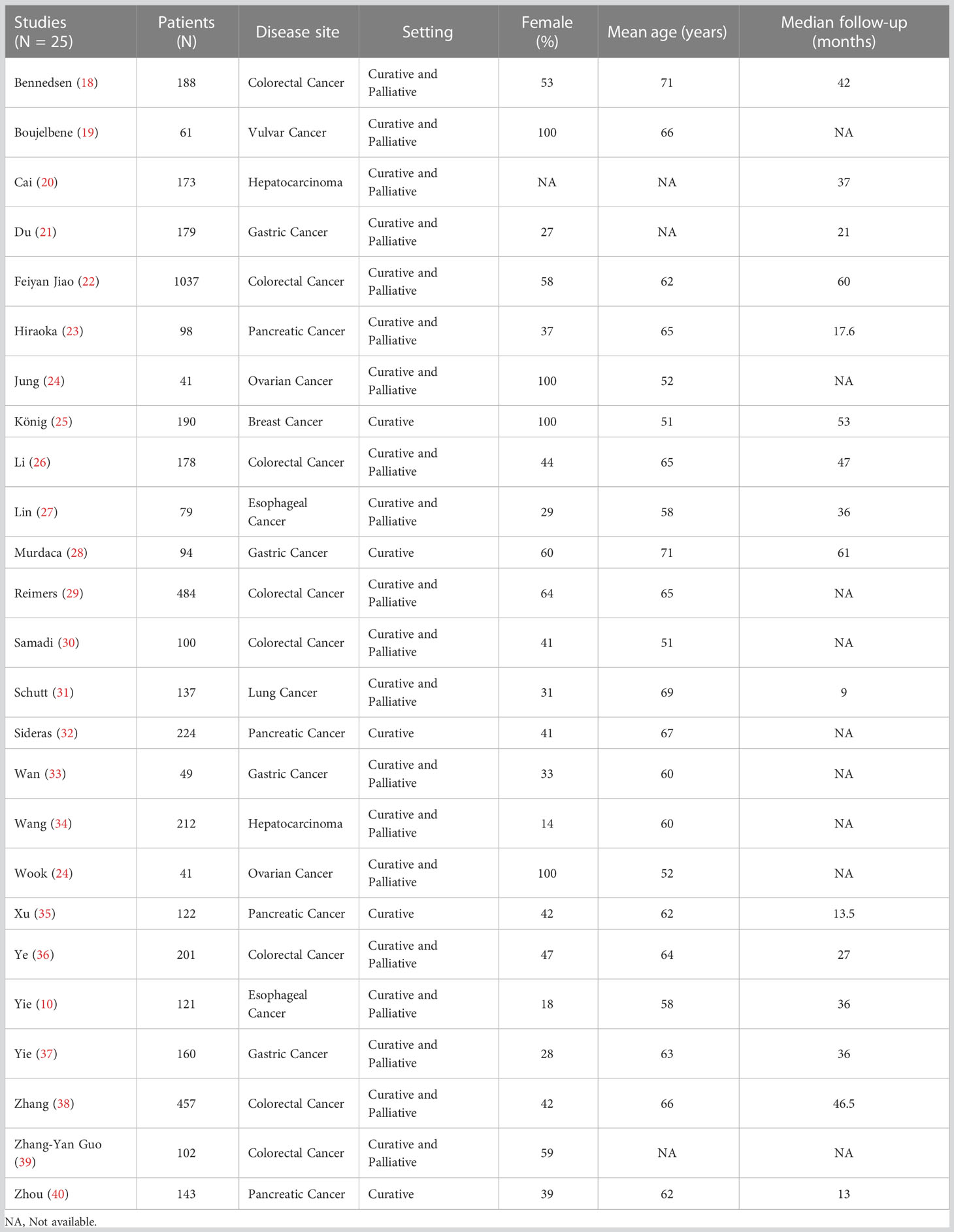
Of 2821 studies identified in the initial search, after removal duplications, 2128 studies were screened by title and abstract, with 207 reports were assessed for eligibility, including full text. Finally, a total of 25 studies (10, 18–40) met the inclusion criteria (see Figure 1 for study selection schema). Of these studies, 25 studies comprising data from 4871 patients reported OS outcomes and among them, only 5 studies (961 patients) reported DFS data in addition to OS. The characteristics of the included studies are shown in Tables 1, 2. Overall, 20 (80%) comprised studies with a mixed group of patients treated in the curative and palliative setting, 21 (84%) used tissue for HLA-G detection and 14 (56%) used anti-HLA-G antibody, clone 4H84. Supplementary Table 1 provides detailed information about the histological type, stage and the definition utilized for HLA-G expression positivity.
Overall survival
Twenty-five studies comprising 4871 patients reported HRs for OS. Seven of the eligible 25 studies (28%) reported a non-statistically significant HR (i.e., the 95% confidence intervals crossed 1); a forest plot of all studies is presented as Figure 2. Overall, HLA-G expression was associated with worse OS (HR 2.09, 95% CI = 1.67 to 2.63; P <.001). The funnel plot of HR for OS for HLA-G expression is shown in Figure 3. Visual inspection suggested potential publication bias with fewer studies with higher error (typically smaller sample size) showing a protective effect from HLA-G expression.
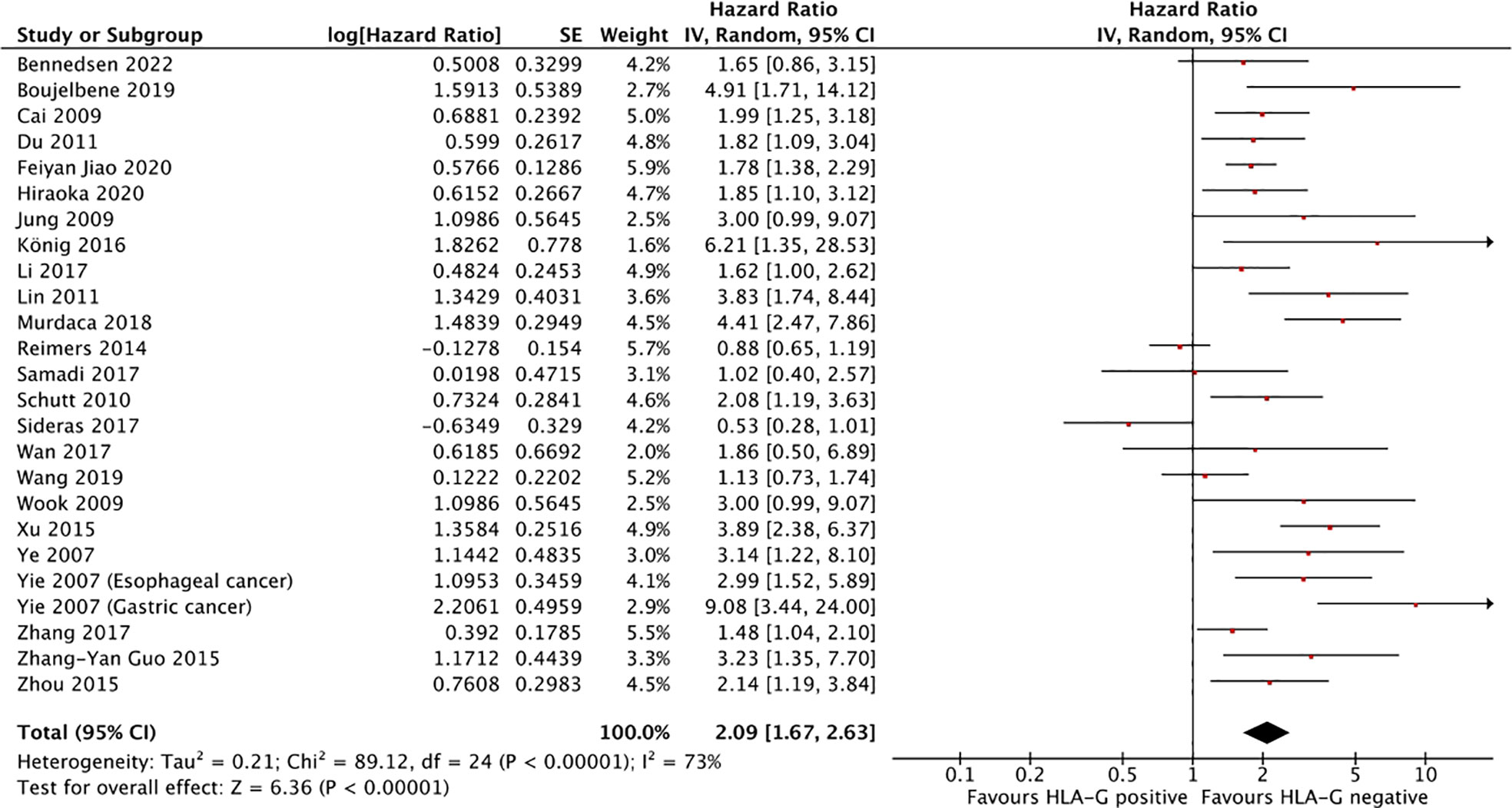
Figure 2 Forest plot showing hazard ratio for OS for HLA-G positive. Hazard ratios for each study are represented by the squares, the size of the square represents the weight of the study in the meta-analysis, and the horizontal line crossing the square represents the 95% confidence interval (CI). All statistical tests were two-sided.
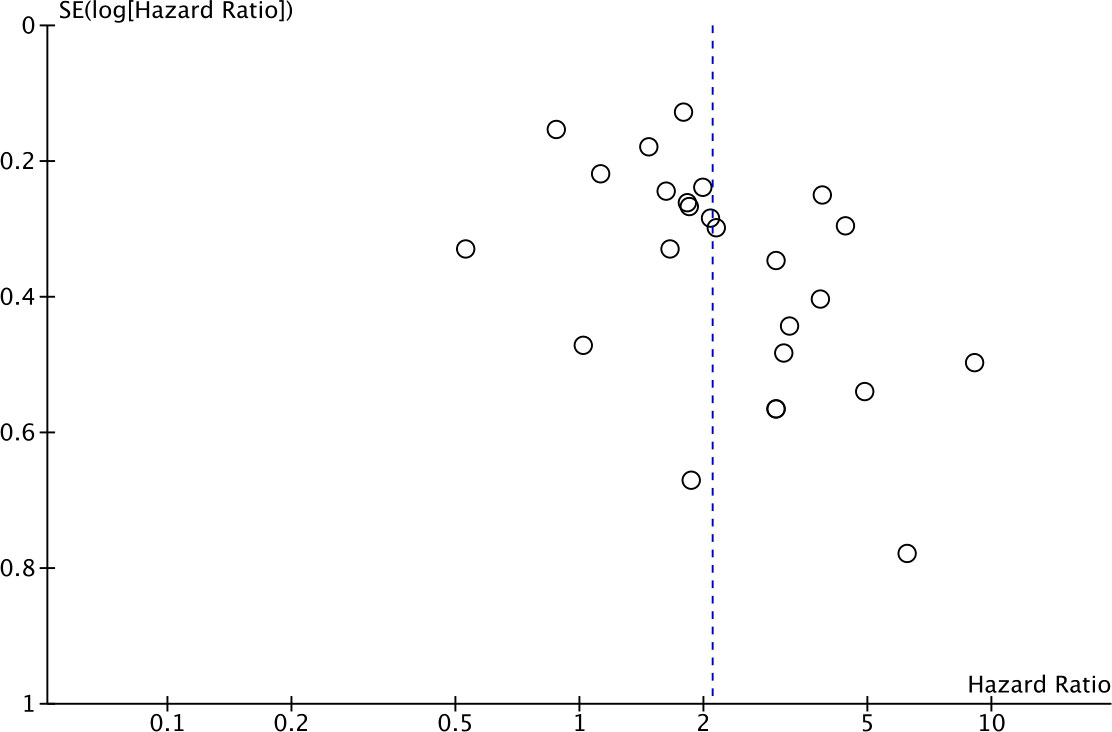
Figure 3 Funnel plot of hazard ratio for OS for HLA-G positive (horizontal axis) and the standard error (SE) for the hazard ratio (vertical axis). Each study is represented by one circle. The vertical line represents the pooled effect estimate.
The effect of HLA-G on OS among disease site subgroups is shown in Figure 4A. The prognostic effect of HLA-G was highest in gastric cancer (HR = 3.40; 95% CI = 1.64 to 7.03), followed by pancreatic cancer (HR = 1.72; 95% CI = 0.79 to 3.74) and colorectal cancer (HR = 1.55; 95% CI = 1.16 to 2.07). The hazard ratio for the subgroup of other unselected solid tumors was 2.45 (95% CI = 1.72 to 3.49). Differences between disease subgroups approached but did not meet statistical significance (P for subgroup difference = .10).
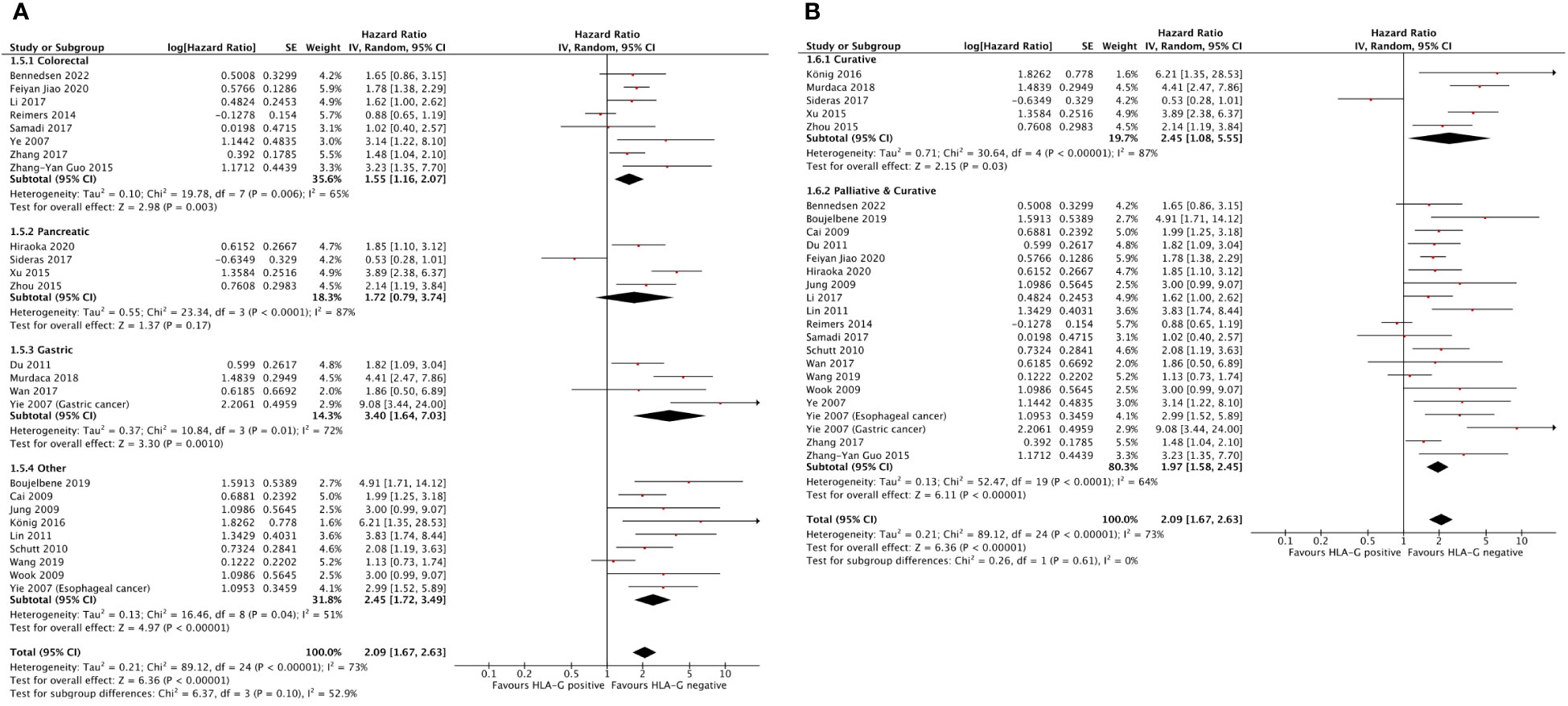
Figure 4 Forest plots showing hazard ratio for OS for HLA-G positive. (A) Hazard ratios by disease subgroups. (B) Hazard ratios by disease setting. Hazard ratios for each study are represented by the squares, the size of the square represents the weight of the study in the meta-analysis, and the horizontal line crossing the square represents the 95% confidence interval (CI). All statistical tests were two-sided.
The effect of HLA-G on OS among disease setting is shown in Figure 4B. The hazard ratios were 2.45 (95% CI = 1.08 to 5.55) for nonmetastatic disease and 1.97 (95% CI = 1.58 to 2.45) for a mixed group consisting of studies that included both metastatic and nonmetastatic patients. Although the effect of HLA-G expression in nonmetastatic disease was numerically higher value than in mixed metastatic and nonmetastatic disease, this difference was not statistically significant (P for subgroup difference = .61).
In the post-hoc sensitivity analysis, 3 studies (Cai et al, 2019; Du et al, 2011; Zhang-Yan Guo et al, 2015) (20, 21, 39) were excluded as they did not adjust for age in the multivariable model. Results showed an unchanged effect size (HR 2.10, 95% CI 1.62 to 2.71, p<.001).
The effect of HLA-G on OS among detection type, detection technique and antibody used are presented as Supplementary Figure 1 (available online). There was neither a magnitude of effect nor statistically significant difference between the most commonly utilized antibody (4H84) and other methods of detection of HLA-G expression.
Meta-regression analysis is presented in Table 3. Overall, there was a quantitatively significant, but non-statistically significant negative association between age and worse OS with HLA-G expression (β = 0.314; P = .155).
Disease-free survival
A total of 5 studies comprising 961 patients reported HRs for DFS. Two of these 5 studies (40%) reported a non-statistically significant HR (i.e., the 95% confidence intervals crossed 1). A forest plot of all studies is presented as Supplementary Figure 2 (available online). Overall, HLA-G expression was associated with DFS which approached but did not meet statistical significance (HR for DFS of 1.74 95% CI = 0.98 to 3.10; P <.06).
In the post-hoc sensitivity analysis, 1 study (Du et al, 2011) (21) was excluded as it did not adjust for age in the multivariable model. Again, results showed unchanged effect size (HR 1.69, 95% CI 0.83 to 3.47, p<.15), although with loss of power from a smaller cumulative sample size, confidence intervals were wider.
Discussion
In this article, we describe the association between expression of HLA-G and clinical outcome in several solid tumors. Given the success for antibody-mediated targeting of inhibitory immunologic signals on adaptive or innate immune cells, such as the targeting of programmed death receptor 1 (PD-1) and its ligand, and of CTLA4; focus has shifted to identify other receptors or ligands with immune inhibitory properties. Several of these targets are under evaluation and some have demonstrated benefit in patients (e.g. LAG3 antibodies in melanoma) (41).
HLA-G is another marker of interest in this setting. The presence of HLA-G is associated with physiological conditions where the immune system is inhibited, such as fetal tolerance during pregnancy (1–5). High expression of HLA-G in solid tumors has also been shown to play an immune-modulatory role in cancer (7, 8). It has been hypothesized that HLA-G works as an immune checkpoint inhibitor in cancer, allowing malignant cells to escape immune cell surveillance (3, 4). Furthermore, it has been shown that HLA-G expression in cancer cell lines can make them less susceptible to immune recognition and elimination (42). Consequently, HLA-G has been suggested as a potential target for therapeutic immune checkpoint inhibition (8, 43, 44).
In our analysis we confirmed data published in individual tumor sites and observed that higher expression of HLA-G is associated with worse survival in solid tumors. In line with this, a recent published meta-analysis covering only tumors in gastrointestinal indications, reported that elevated HLA-G expression was indicative of a poor prognosis and adverse clinicopathological parameters (45). Our study confirms these results adding additional tumor types, and a more represented population of early-stage/curable malignancies. Tumors with higher magnitude association with worse survival included gastric, pancreas and colorectal cancer. While this effect was observed predominantly in tumors with advanced disease, similar effects were observed on studies which included a mixed population of early-stage and advanced-stage patients. Of note the effect was stronger, although not statistically significant, in tumors with early-stage disease. This observation was likely the effect of lower statistical power. Although intra-heterogeneity is associated with a more immunosuppressive environment and less immunoreactivity, we did not observe differences in the prognostic role of HLA-G in early-stage tumors, which are usually more homogeneous, than in metastatic tumors (46, 47). Of note, in one study (Sideras et al., 2017) (32), associations were in the opposite direction of the pooled effect. In this study, positivity was considered as any case with positive staining. This threshold may not have been high enough to identify tumors in which HLA-G expression is meaningful. However, overall, data confirm the importance of this molecule favoring an immune suppressive environment from early stages of disease. Unfortunately, data for DFS were less robust as only 5 studies met the criteria for analysis. With the expectation of lower power in this group, the non-significant association with worse outcome was expected and most likely reflects low sampling in this cohort. Of interest, some studies not included in the current meta-analysis due to lack of reporting of outcome data suggest no association between HLA-G expression and a more aggressive features or adverse risk profile (48, 49).
When evaluating the effect of methods of analysis of HLA-G expression including the antibody used for IHC, or the analysis of the protein in human samples by ELISA, no differences were observed, and all the methods predicted for outcome to a similar magnitude. However, it is important to highlight (see Supplementary Table 1) that there was variability in the methods that each study used for analysis, including the cut-off used for consideration of increase HLA-G expression. Of note 56% of the studies utilized the 4H84 clone antibody which represents all α1 domain containing HLA-G or HLA-G5/6 isoforms (7). These data are of interest as they suggest the potential utility of serial liquid biopsies in the evaluation of this biomarker when evaluating therapeutic strategies against this pathway. In this regard, the use of ctDNA has recently demonstrated the potential to stratify patients and predict response to check point inhibitors (50).
Therapeutic strategies targeting HLA-G are currently in clinical development. For instance, MK4830 is an antibody in early phase clinical development targeting the HLA-G receptor, ITL4. In the early phase I study signs of activity were reported in PD-1 pretreated patients (51) Other studies evaluating antibodies targeting ITL2 such as BND22 are in preclinical and early clinical development (12, 52). Early signs of clinical activity have been reported in microsatellite instability-high (MSH-I) colorectal tumors and ovarian cancer, among other tumor types. These observations have supported further clinical exploration (53). In some of these studies, evaluation of HLA-G expression in human samples is explored to assess target inhibition as well as other pharmacodynamic markers.
Our study has limitations. First, this is a meta-analysis of published articles and therefore relies on summary data. Availability of individual patient data would have provided a more accurate estimate of effect. Second, we identified a potential for publication bias in the studies analyzed. Third, there was substantial heterogeneity between studies. While this was expected with the inclusion of diverse solid tumors, attempts to explore the impact of heterogeneity such as subgroup and sensitivity analyses may not have explained the effect of this heterogeneity in full. As such, there remains some uncertainty about effect size estimates. Some subgroups were small in size (e.g. early-stage disease) and therefore there was limited power to explore if magnitudes of effect observed were statistically significant or not. Finally, while all the hazard ratios which we extracted data were derived from multivariable analyses, the variables included in models of individual studies differed. It is for this reason that we elected to further explore variability in the magnitude of the hazard ratio utilizing meta-regression.
In summary, we describe the first meta-analysis evaluating HLA-G expression and its association with worse survival in a variety of solid tumors. This suggests a role of this ligand favoring an immune suppressed environment. Further prospective studies should explore the potential role of HLA-G as a therapeutic target, or as a biomarker of response to agents against the receptors LILRB1, LILRB2 or KIR2DL4.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1165813/full#supplementary-material
Supplementary Figure 1 | Forest plots showing hazard ratio for OS for HLA-G positive. (A) Hazard ratios by HLA-G detection type. (B) Hazard ratios by HLA-G detection technique. (C) Hazard ratios by antibody used. Hazard ratios for each study are represented by the squares, the size of the square represents the weight of the study in the meta-analysis, and the horizontal line crossing the square represents the 95% confidence interval (CI). All statistical tests were two-sided.
Supplementary Figure 2 | Forest plot showing hazard ratio for DFS for HLA-G positive. Hazard ratios for each study are represented by the squares, the size of the square represents the weight of the study in the meta-analysis, and the horizontal line crossing the square represents the 95% confidence interval (CI). All statistical tests were two-sided.
References
1. Xu X, Zhou Y, Wei H. Roles of HLA-G in the maternal-fetal immune microenvironment. Front Immunol (2020) 11:592010. doi: 10.3389/fimmu.2020.592010
2. Rouas-Freiss N, Khalil-Daher I, Riteau B, Menier C, Paul P, Dausset J, et al. The immunotolerance role of HLA-G. Semin Cancer Biol (1999) 9(1):3–12. doi: 10.1006/scbi.1998.0103
3. Carosella ED, Moreau P, Le Maoult J, Le Discorde M, Dausset J, Rouas-Freiss N. HLA-G molecules: from maternal-fetal tolerance to tissue acceptance. Adv Immunol (2003) 81:199–252. doi: 10.1016/S0065-2776(03)81006-4
4. Rouas-Freiss N, Moreau P, Ferrone S, Carosella ED. HLA-G proteins in cancer: do they provide tumor cells with an escape mechanism? Cancer Res (2005) 65(22):10139–44. doi: 10.1158/0008-5472.CAN-05-0097
5. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science (1990) 248(4952):220–3. doi: 10.1126/science.2326636
6. Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA. HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc Natl Acad Sci U S A. (1998) 95(8):4510–5. doi: 10.1073/pnas.95.8.4510
7. Lin A, Yan WH. HLA-G/ILTs targeted solid cancer immunotherapy: opportunities and challenges. Front Immunol (2021) 12:698677. doi: 10.3389/fimmu.2021.698677
8. Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: an immune checkpoint molecule. Adv Immunol (2015) 127:33–144. doi: 10.1016/bs.ai.2015.04.001
9. Li XJ, Zhang X, Lin A, Ruan YY, Yan WH. Human leukocyte antigen-G (HLA-G) expression in cervical cancer lesions is associated with disease progression. Hum Immunol (2012) 73(9):946–9. doi: 10.1016/j.humimm.2012.07.041
10. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of HLA-G is associated with prognosis in esophageal squamous cell carcinoma. Am J Clin Pathol (2007) 128(6):1002–9. doi: 10.1309/JNCW1QLDFB6AM9WE
11. Umiker B, Hashambhoy-Ramsay Y, Smith J, Rahman T, Mueller A, Davidson R. Inhibition of LILRB2 by a novel blocking antibody designed to reprogram immunosuppressive macrophages to drive T cell activation in tumors. Mol Cancer Ther (2023) 10:22–0351. doi: 10.1158/1535-7163.MCT-22-0351
12. Mandel I, Haves Ziv D, Goldshtein I, Peretz T, Alishekevitz D, Fridman Dror A, et al. BND-22, a first-in-class humanized ILT2-blocking antibody, promotes antitumor immunity and tumor regression. J Immunother Cancer (2022) 10(9):e004859. doi: 10.1136/jitc-2022-004859
13. Lin A, Yan WH. Human leukocyte antigen-G (HLA-G) expression in cancers: roles in immune evasion, metastasis and target for therapy. Mol Med (2015) 21(1):782–91. doi: 10.2119/molmed.2015.00083
14. Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, et al. Cochrane Handbook for Systematic Reviews of Interventions. 2nd Edition. Chichester (UK): John Wiley & Sons, (2019).
15. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials (1986) 7(3):177–88. doi: 10.1016/0197-2456(86)90046-2
16. Stanley TD, Doucouliagos H. Neither fixed nor random: weighted least squares meta-regression. Res Synth Methods (2017) 8(1):19–42. doi: 10.1002/jrsm.1211
17. Burnand B, Kernan WN, Feinstein AR. Indexes and boundaries for “quantitative significance” in statistical decisions. J Clin Epidemiol (1990) 43(12):1273–84. doi: 10.1016/0895-4356(90)90093-5
18. Bennedsen ALB, Cai L, Hasselager RP, Özcan AA, Mohamed KB, Eriksen JO, et al. An exploration of immunohistochemistry-based prognostic markers in patients undergoing curative resections for colon cancer. BMC Cancer (2022) 22(1):62. doi: 10.1186/s12885-022-09169-0
19. Boujelbene N, Ben Yahia H, Babay W, Gadria S, Zemni I, Azaiez H, et al. HLA-G, HLA-e, and IDO overexpression predicts a worse survival of Tunisian patients with vulvar squamous cell carcinoma. HLA (2019) 94(1):11–24. doi: 10.1111/tan.13536
20. Cai MY, Xu YF, Qiu SJ, Ju MJ, Gao Q, Li YW, et al. Human leukocyte antigen-G protein expression is an unfavorable prognostic predictor of hepatocellular carcinoma following curative resection. Clin Cancer Res (2009) 15(14):4686–93. doi: 10.1158/1078-0432.CCR-09-0463
21. Du L, Xiao X, Wang C, Zhang X, Zheng N, Wang L, et al. Human leukocyte antigen-G is closely associated with tumor immune escape in gastric cancer by increasing local regulatory T cells. Cancer Sci (2011) 102(7):1272–80. doi: 10.1111/j.1349-7006.2011.01951.x
22. Jiao F, Zhou J, Sun H, Song X, Song Y. Plasma soluble human leukocyte antigen G predicts the long-term prognosis in patients with colorectal cancer. Transl Cancer Res (2020) 9(6):4011–9. doi: 10.21037/tcr-20-2211
23. Hiraoka N, Ino Y, Hori S, Yamazaki-Itoh R, Naito C, Shimasaki M, et al. Expression of classical human leukocyte antigen class I antigens, HLA-e and HLA-G, is adversely prognostic in pancreatic cancer patients. Cancer Sci (2020) 111(8):3057–70. doi: 10.1111/cas.14514
24. Jung YW, Kim YT, Kim SW, Kim S, Kim JH, Cho NH, et al. Correlation of human leukocyte antigen-G (HLA-G) expression and disease progression in epithelial ovarian cancer. Reprod Sci (2009) 16(11):1103–11. doi: 10.1177/1933719109342131
25. König L, Kasimir-Bauer S, Hoffmann O, Bittner AK, Wagner B, Manvailer LF, et al. The prognostic impact of soluble and vesicular HLA-G and its relationship to circulating tumor cells in neoadjuvant treated breast cancer patients. Hum Immunol (2016) 77(9):791–9. doi: 10.1016/j.humimm.2016.01.002
26. Li JB, Ruan YY, Hu B, Dong SS, Bi TN, Lin A, et al. Importance of the plasma soluble HLA-G levels for prognostic stratification with traditional prognosticators in colorectal cancer. Oncotarget (2017) 8(30):48854–62. doi: 10.18632/oncotarget.16457
27. Lin A, Zhang X, Zhou WJ, Ruan YY, Xu DP, Wang Q, et al. Human leukocyte antigen-G expression is associated with a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Cancer (2011) 129(6):1382–90. doi: 10.1002/ijc.25807
28. Murdaca G, Calamaro P, Lantieri F, Pigozzi S, Mastracci L, Grillo F, et al. HLA-G expression in gastric carcinoma: clinicopathological correlations and prognostic impact. Virchows Arch (2018) 473(4):425–33. doi: 10.1007/s00428-018-2379-0
29. Reimers MS, Engels CC, Putter H, Morreau H, Liefers GJ, van de Velde CJ, et al. Prognostic value of HLA class I, HLA-e, HLA-G and tregs in rectal cancer: a retrospective cohort study. BMC Cancer (2014) 14:486. doi: 10.1186/1471-2407-14-486
30. Samadi R, Nazemalhosseini Mojarad E, Molaei M, Kazerouni F, Asadzadeh Aghdaei H, Navidini M, et al. Clinical value of human leucocyte antigen G (HLA-G) expression in the prognosis of colorectal cancer. Int J Cancer Manage (2017) 10(4):e9346. doi: 10.5812/ijcm.9346
31. Schütt P, Schütt B, Switala M, Bauer S, Stamatis G, Opalka B, et al. Prognostic relevance of soluble human leukocyte antigen-G and total human leukocyte antigen class I molecules in lung cancer patients. Hum Immunol (2010) 71(5):489–95. doi: 10.1016/j.humimm.2010.02.015
32. Sideras K, Biermann K, Yap K, Mancham S, Boor PPC, Hansen BE, et al. Tumor cell expression of immune inhibitory molecules and tumor-infiltrating lymphocyte count predict cancer-specific survival in pancreatic and ampullary cancer. Int J Cancer (2017) 141(3):572–82. doi: 10.1002/ijc.30760
33. Wan R, Wang ZW, Li H, Peng XD, Liu GY, Ou JM, et al. Human leukocyte antigen-G inhibits the anti-tumor effect of natural killer cells via immunoglobulin-like transcript 2 in gastric cancer. Cell Physiol Biochem (2017) 44(5):1828–41. doi: 10.1159/000485819
34. Wang XK, Liao XW, Yang CK, Yu TD, Liu ZQ, Gong YZ, et al. Diagnostic and prognostic biomarkers of human leukocyte antigen complex for hepatitis b virus-related hepatocellular carcinoma. J Cancer (2019) 10(21):5173–90. doi: 10.7150/jca.29655
35. Xu YF, Lu Y, Cheng H, Jiang J, Xu J, Long J, et al. High expression of human leukocyte antigen-G is associated with a poor prognosis in patients with PDAC. Curr Mol Med (2015) 15(4):360–7. doi: 10.2174/1566524015666150401102218
36. Ye SR, Yang H, Li K, Dong DD, Lin XM, Yie SM, et al. Human leukocyte antigen G expression: as a significant prognostic indicator for patients with colorectal cancer. Mod Pathol (2007) 20(3):375–83. doi: 10.1038/modpathol.3800751
37. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM, et al. Expression of human leukocyte antigen G (HLA-G) correlates with poor prognosis in gastric carcinoma. Ann Surg Oncol (2007) 14(10):2721–9. doi: 10.1245/s10434-007-9464-y
38. Zhang RL, Zhang X, Dong SS, Hu B, Han QY, Zhang JG, et al. Predictive value of different proportion of lesion HLA-G expression in colorectal cancer. Oncotarget (2017) 8(64):107441–51. doi: 10.18632/oncotarget.22487
39. Guo ZY, Lv YG, Wang L, Shi SJ, Yang F, Zheng GX, et al. Predictive value of HLA-G and HLA-e in the prognosis of colorectal cancer patients. Cell Immunol (2015) 293(1):10–6. doi: 10.1016/j.cellimm.2014.10.003
40. Zhou L, Niu ZY, Liang ZY, Zhou WX, You L, Wang MY, et al. HLA-G impairs host immune response and predicts poor prognosis in pancreatic cancer. Am J Transl Res (2015) 7(10):2036–44.
41. Tawbi HA, Schadendorf D, Lipson EJ, Ascierto PA, Matamala L, Castillo Gutiérrez E, et al. RELATIVITY-047 investigators. “Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma”. N Engl J Med (2022) 386(1):24–34. doi: 10.1056/NEJMoa2109970
42. Lin A, Yan WH. Heterogeneity of HLA-G expression in cancers: facing the challenges. Front Immunol (2018) 9:2164. doi: 10.3389/fimmu.2018.02164
43. Dumont C, Jacquier A, Verine J, Noel F, Goujon A, Wu CL, et al. CD8+PD-1-ILT2+ T cells are an intratumoral cytotoxic population selectively inhibited by the immune-checkpoint HLA-G. Cancer Immunol Res (2019) 7(10):1619–32. doi: 10.1158/2326-6066.CIR-18-0764
44. Tronik-Le Roux D, Sautreuil M, Bentriou M, Vérine J, Palma MB, Daouya M, et al. Comprehensive landscape of immune-checkpoints uncovered in clear cell renal cell carcinoma reveals new and emerging therapeutic targets. Cancer Immunol Immunother (2020) 69(7):1237–52. doi: 10.1007/s00262-020-02530-x
45. Peng Y, Xiao J, Li W, Li S, Xie B, He J, et al. Prognostic and clinicopathological value of human leukocyte antigen G in gastrointestinal cancers: a meta-analysis. Front Oncol (2021) 11:642902. doi: 10.3389/fonc.2021.642902
46. Johnson AL, Laterra J, Lopez-Bertoni H. Exploring glioblastoma stem cell heterogeneity: immune microenvironment modulation and therapeutic opportunities. Front Oncol (2022) 12:995498. doi: 10.3389/fonc.2022.995498
47. Espinet E, Klein L, Puré E, Singh SK. Mechanisms of PDAC subtype heterogeneity and therapy response. Trends Cancer (2022) 8(12):1060–71. doi: 10.1016/j.trecan.2022.08.005
48. Marletta S, Girolami I, Munari E, Pantanowitz L, Bernasconi R, Torresani E, et al. HLA-G expression in melanomas. Int Rev Immunol (2021) 40(5):330–43. doi: 10.1080/08830185.2020.1869732
49. Spurny C, Kailayangiri S, Altvater B, Jamitzky S, Hartmann W, Wardelmann E, et al. T Cell infiltration into Ewing sarcomas is associated with local expression of immune-inhibitory HLA-G. Oncotarget (2017) 9(5):6536–49. doi: 10.18632/oncotarget.23815
50. Powles T, Assaf ZJ, Davarpanah N, Banchereau R, Szabados BE, Yuen KC, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature (2021) 595(7867):432–7. doi: 10.1038/s41586-021-03642-9
51. Siu LL, Wang D, Hilton J, Geva R, Rasco D, Perets R, et al. First-in-Class anti-immunoglobulin-like transcript 4 myeloid-specific antibody MK-4830 abrogates a PD-1 resistance mechanism in patients with advanced solid tumors”. Clin Cancer Res (2022) 28(1):57–70. doi: 10.1158/1078-0432.CCR-21-2160
52. Lentz RW, Haves Ziv D, Goldshtein I, Peretz T, Alishekevitz D, Fridman Dror A, et al. Innate immune checkpoint inhibitors: the next breakthrough in medical oncology? Mol Cancer Ther (2021) 20(6):961–74. doi: 10.1158/1535-7163.MCT-21-0041
53. National Library of Medicine. Pembrolizumab with chemotherapy and MK-4830 for treating participants with ovarian cancer (MK-4830-002) (2022). Available at: https://www.clinicaltrials.gov/ct2/show/study/NCT05446870.
Keywords: tumors, HLA-G, human leukocyte antigen G, meta – analysis, cancer
Citation: Bartolome J, Molto C, Benitez-Fuentes JD, Fernandez-Hinojal G, Manzano A, Perez-Segura P, Mittal A, Tamimi F, Amir E and Ocana A (2023) Prognostic value of human leukocyte antigen G expression in solid tumors: a systematic review and meta-analysis. Front. Immunol. 14:1165813. doi: 10.3389/fimmu.2023.1165813
Received: 14 February 2023; Accepted: 02 May 2023;
Published: 18 May 2023.
Edited by:
Hai Fang, Shanghai Jiao Tong University, ChinaReviewed by:
Gunnar Wichmann, University Hospital Leipzig, GermanyStefano Marletta, University of Verona, Italy
Copyright © 2023 Bartolome, Molto, Benitez-Fuentes, Fernandez-Hinojal, Manzano, Perez-Segura, Mittal, Tamimi, Amir and Ocana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alberto Ocana, YWxiZXJ0by5vY2FuYUBzYWx1ZC5tYWRyaWQub3Jn; Eitan Amir, ZWl0YW5AdWhuLmNh
†These authors have contributed equally to this work and share first authorship
 Jorge Bartolome
Jorge Bartolome Consolacion Molto2†
Consolacion Molto2† Javier David Benitez-Fuentes
Javier David Benitez-Fuentes Gonzalo Fernandez-Hinojal
Gonzalo Fernandez-Hinojal Aranzazu Manzano
Aranzazu Manzano Faris Tamimi
Faris Tamimi Alberto Ocana
Alberto Ocana