- 1Department of Biomedical Data Science and Bioinformatics, The Emmes Company, LLC, Rockville, MD, United States
- 2Division of Microbiology & Immunology, Emory National Primate Research Center, Emory University, Atlanta, GA, United States
- 3Department of Pathology & Laboratory Medicine, School of Medicine, Emory University, Atlanta, GA, United States
- 4Emory NPRC Genomics Core, Emory National Primate Research Center, Emory University, Atlanta, GA, United States
- 5Emory Vaccine Center, Emory University School of Medicine, Atlanta, GA, United States
- 6Hope Clinic of the Emory Vaccine Center, Emory University, Atlanta, GA, United States
- 7Division of Infectious Diseases, Allergy, and Immunology, Department of Internal Medicine, Saint Louis University School of Medicine, St. Louis, MO, United States
- 8McDonnell Genome Institute, Washington University, St. Louis, MO, United States
- 9Office of Biodefense, Research Resources and Translational Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD, United States
- 10Department of Medicine, Division of Infectious Diseases, Emory University School of Medicine, Emory University, Atlanta, GA, United States
- 11Center for Childhood Infections and Vaccines (CCIV) of Children’s Healthcare of Atlanta and Department of Pediatrics, Emory University School of Medicine, Atlanta, GA, United States
- 12New York University Vaccine Center, New York, NY, United States
- 13Department of Molecular Microbiology & Immunology, Saint Louis University, St. Louis, MO, United States
A corrigendum on
The Vacc-SeqQC project: Benchmarking RNA-Seq for clinical vaccine studies
by Goll JB, Bosinger SE, Jensen TL, Walum H, Grimes T, Tharp GK, Natrajan MS, Blazevic A, Head RD, Gelber CE, Steenbergen KJ, Patel NB, Sanz P, Rouphael NG, Anderson EJ, Mulligan MJ and Hoft DF (2023) Front. Immunol. 13:1093242. doi: 10.3389/fimmu.2022.1093242
In the published article, there was an error in the Figure 7 legend as published. The figure legend effect size values were incorrectly displayed as “>1.25, 51.5, 51.75, 52” instead of “>1.25, ≥1.5, ≥1.75, ≥2”. The corrected Figure 7 and its caption appear below.
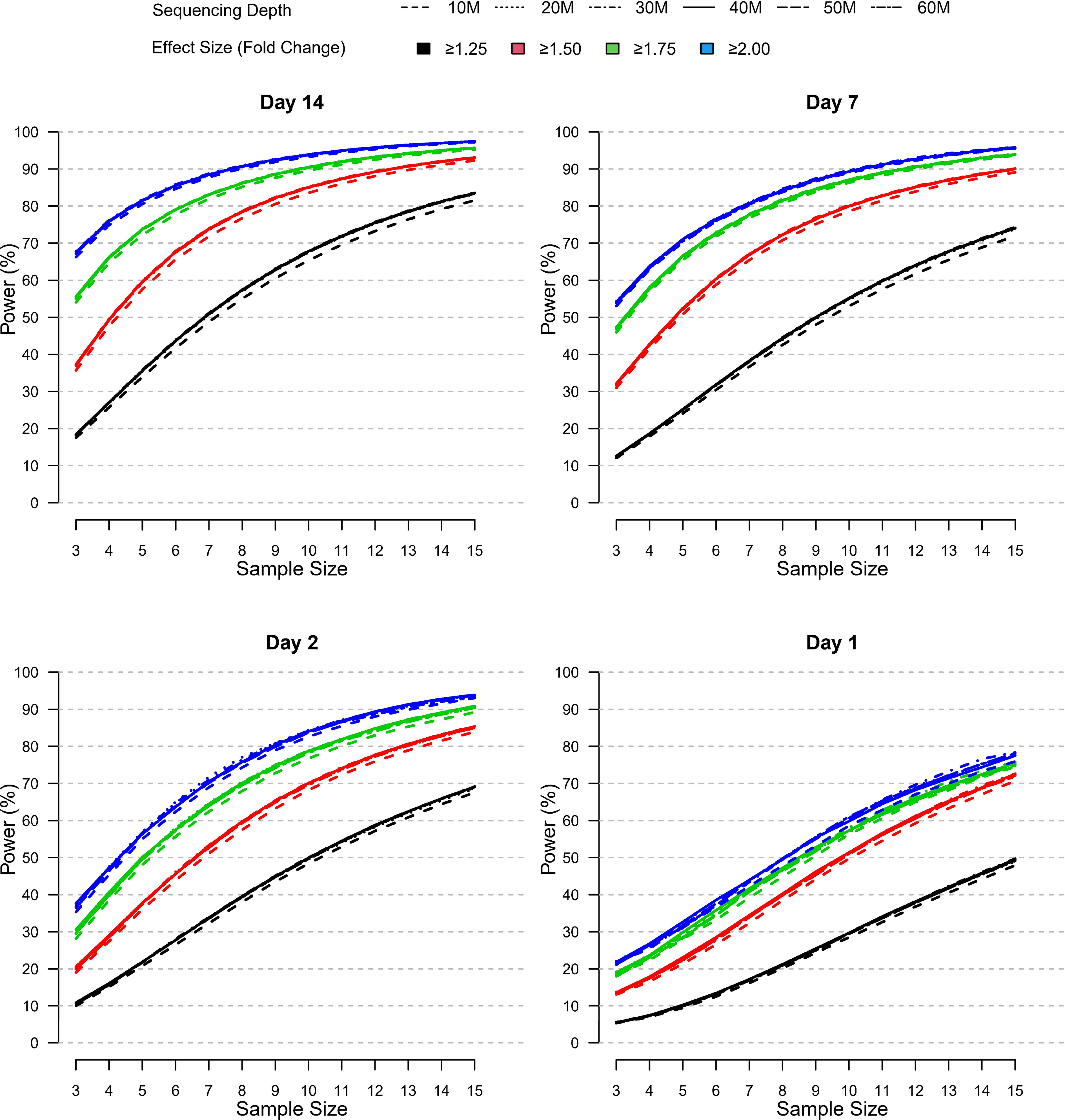
Figure 7 Relative power by sample size, effect size, and sequencing depth at each post-vaccination day as simulated using the modified PROPER R package. Days were sorted by decreasing vaccination effect based on overall fold changes and DEG responses observed for this study (see Figure 3A). Power was assessed for different fold-change cutoffs (indicated by color-coded lines), sequencing depth (as indicated by the line type), and sample size (x-axis).
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: RNA-Seq, statistical power, ERCC, tularemia vaccine (DVC-LVS), gene filtering, sequencing depth, read length, reproducibility
Citation: Goll JB, Bosinger SE, Jensen TL, Walum H, Grimes T, Tharp GK, Natrajan MS, Blazevic A, Head RD, Gelber CE, Steenbergen KJ, Patel NB, Sanz P, Rouphael NG, Anderson EJ, Mulligan MJ and Hoft DF (2023) Corrigendum: The Vacc-SeqQC project: Benchmarking RNA-Seq for clinical vaccine studies. Front. Immunol. 14:1163550. doi: 10.3389/fimmu.2023.1163550
Received: 10 February 2023; Accepted: 13 February 2023;
Published: 23 February 2023.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2023 Goll, Bosinger, Jensen, Walum, Grimes, Tharp, Natrajan, Blazevic, Head, Gelber, Steenbergen, Patel, Sanz, Rouphael, Anderson, Mulligan and Hoft. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel F. Hoft, RGFuaWVsLkhvZnRAaGVhbHRoLnNsdS5lZHU=; Mark J. Mulligan, bWFyay5tdWxsaWdhbkBueXVsYW5nb25lLm9yZw==
†These authors have contributed equally to this work
 Johannes B. Goll1†
Johannes B. Goll1† Steven E. Bosinger
Steven E. Bosinger Travis L. Jensen
Travis L. Jensen Hasse Walum
Hasse Walum Tyler Grimes
Tyler Grimes Gregory K. Tharp
Gregory K. Tharp Azra Blazevic
Azra Blazevic Kristen J. Steenbergen
Kristen J. Steenbergen Nadine G. Rouphael
Nadine G. Rouphael Daniel F. Hoft
Daniel F. Hoft