- 1Department of Respiratory Medicine, The Second Hospital of Jilin University, Changchun, Jilin, China
- 2Department of Immunology, College of Basic Medical Sciences, Jilin University, Changchun, China
Asthma is a heterogeneous airway disease characterized by airway inflammation and hyperresponsiveness. Autophagy is a self-degrading process that helps maintain cellular homeostasis. Dysregulation of autophagy is involved in the pathogenesis of many diseases. In the context of asthma, autophagy has been shown to be associated with inflammation, airway remodeling, and responsiveness to drug therapy. In-depth characterization of the role of autophagy in asthma can enhance the understanding of the pathogenesis, and provide a theoretical basis for the development of new biomarkers and targeted therapy for asthma. In this article, we focus on the relationship of autophagy and asthma, and discuss its implications for asthma pathogenesis and treatment.
1 Introduction
Asthma is a chronic airway inflammatory disease characterized by airway remodeling, airway hyperresponsiveness (AHR), and increased mucus secretion (1). It is a heterogeneous disease with multiple phenotypes and complex pathogenetic mechanisms. Various inflammatory cells and inflammatory mediators have been implicated in the pathophysiology of asthma. Adaptive and innate immunity play an indispensable role in the pathogenesis of asthma (2, 3). Corticosteroids are still the first-line treatment for asthma. In recent years, significant advances have been made in the identification of high Th2-type asthma-specific biomarkers and the development of new biologics (4, 5). However, treatment of low Th2 asthma and severe asthma infiltration remains a challenge. Asthma is classified as eosinophilic asthma (EA) and neutrophilic asthma (NA), paucigranulocytic asthma and mixed granulocytic asthma based on the proportion of inflammatory cells in the induced sputum. Clinically, EA shows good response to glucocorticoid therapy and newer biologic agents, while there is a paucity of very effective therapeutic agents for NA. This is likely attributable to the different pathogenesis of these asthma subtypes. Therefore, better elucidation of the molecular biology of different asthma inflammatory subtypes is a key research imperative to help identify novel biomarkers and more effective therapeutic targets.
Autophagy is an evolutionarily conserved catabolic process for digesting organelles and cytoplasmic components in lysosomes. Autophagy includes macroautophagy (hereinafter referred to as autophagy), microautophagy, and chaperone-mediated autophagy. Generally speaking, autophagy is macroautophagy, which is a metabolic process of eukaryotic cells and is essential for the maintenance of cellular homeostasis (6). Autophagy involves the following cellular processes: initiation, vesicle nucleation, elongation, membrane elongation, closure, maturation, and degradation. A stable ULK-inducible complex induces autophagosome formation during yeast initiation, and mTORC1 regulates its activity in different nutritional states (6). The nucleation stage mainly involves the formation of the PIK3C3 complex, and BECN1 is involved in the regulation of this process. Subsequently, autophagosomes are formed through the extension of the phagocyte membrane through the Atg12-Atg5-Atg16 complex and the Atg8/LC3 ubiquitination system. Autophagosomes combine with lysosomes to form autophagolysosomes, which are degraded by hydrolases (6). Autophagy contributes to the identification and clearance of infectious pathogens (7), and participates in the occurrence and development of various diseases by regulating inflammation, including cancer, cardiovascular diseases, and neurodegenerative diseases (8). In addition, LC3-associated phagocytosis (LAP) is a novel and unique form of LC3-dependent process of non-canonical autophagy that accelerates the degradation of phagosomes by promoting phagosomal and lysosomal fusion, regulates immunity and inflammation, and clears dead cells (9–11). Selective mitochondrial autophagy refers to mitochondria-specific autophagy that occurs in defective mitochondria following injury or stress (12). Research suggests that mitochondrial autophagy is a factor in asthma airway inflammation and a potential therapeutic target (13, 14). In recent years, several studies have shown the significance of regulation of autophagy in the context of asthma.
2 Autophagy and asthma
Aberrant expression of autophagy plays an important role in asthma. ATG5 single nucleotide polymorphism is closely associated with the development of childhood asthma (15). Autophagy-related genes are important in the immune microenvironment of asthma and may modulate the treatment outcomes. Autophagy-related genes have good classification function, as patients in the low autophagy subtype group were found to have more severe, glucocorticoid-resistant, and poorly controlled asthma (16). In a house dust mite (HDM)-induced asthma model, chloroquine was shown to attenuate airway inflammation, AHR, mucus and collagen production (17). In addition, autophagy also plays a role in severe asthma, as patients with severe asthma were found to have elevated levels of autophagy in peripheral blood and sputum granulocytes compared to those with non-severe asthma and healthy controls (18). Mice with severe asthma were found to have higher levels of autophagy compared to mice with relatively mild ovalbumin (OVA)-induced asthma (19). These findings suggest an important value of autophagy in the development and prognosis of asthma, opening up new directions for asthma treatment.
3 Autophagy and immune and non-immune cells in asthma
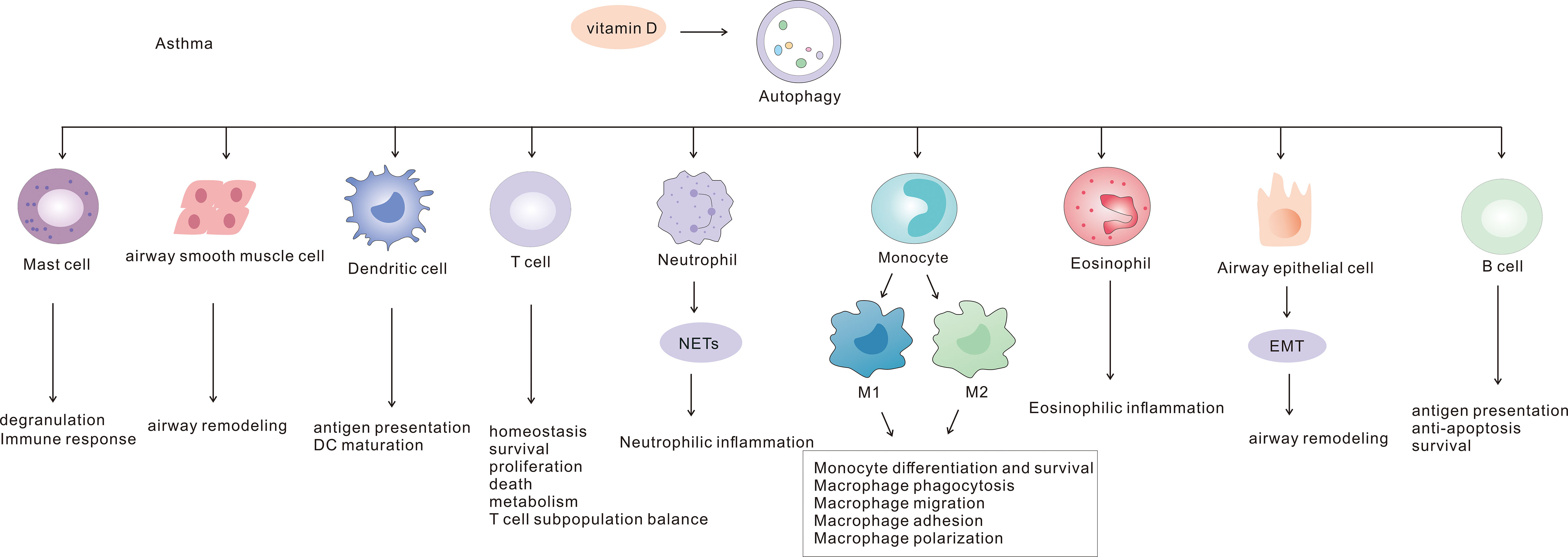
Autophagy is involved in the pathogenesis of asthma through interactions with a variety of immune and non-immune cells (Figure 1). In this paper we summarize the molecular mechanisms of the role of autophagy in asthma and its potential as a therapeutic target.
3.1 Autophagy and T cells
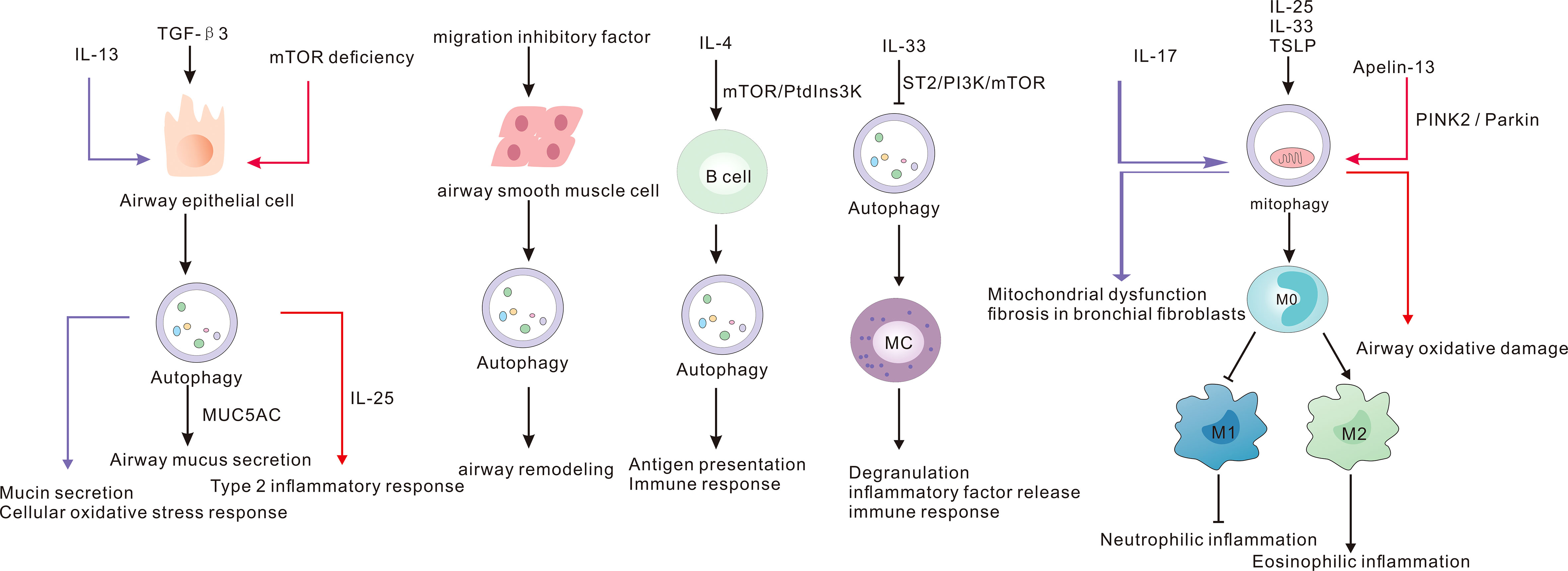
Differentiation of various T-cell subsets and the cytokines produced by them play an important role in different asthma inflammatory subtypes. The pathogenesis of EA is dominated by Th2 cells, whereas Th1 and Th17 cells play a dominant role in NA. Autophagy plays a vital role in T cell metabolism, and PIK3C3 deficiency impairs T cell metabolism and inhibits differentiation to Th1. PIK3C3 has a positive effect on aerobic glycolysis and maintenance of mitochondrial respiration in T cells (20). Autophagy is an essential regulatory mediator in T cell homeostasis, survival, proliferation, death, and functional roles (21–27). Autophagy controls the differentiation and activity of CD8+ T cells, and defective autophagy was found to result in a diminished immune response to secondary influenza virus infection (28). In addition autophagy is activated in activated CD4+ T cells and the level of autophagy is higher in Th2 than in Th1 (29). mTOR is a serine/threonine kinase that plays an important role in regulating cell growth, proliferation, aging and longevity (30, 31). mTOR participates in the pathogenesis of various diseases such as liver disease, breast cancer, acute lung injury, Alzheimer’s disease, and asthma by regulating autophagy (32–36). mTOR was shown to be associated with asthma status. Serum mTOR levels were higher in asthmatic patients than in healthy controls during acute exacerbation, but not during remission. In addition, mTOR showed a positive correlation with Th1/Th2, Th17/Tregs imbalance (37). Moreover, the mTOR inhibitor rapamycin was found to increase the proportion of Treg cells in mesenchymal stem cells and decrease the proportion of Th1 cells, while there was no significant change in Th17 cells (38). PI3K/AKT is an upstream pathway of mTOR and is closely related to airway inflammation. IL-27 was shown to attenuate airway remodeling in OVA asthmatic mice through the PI3K/AKT pathway (39). In HDM-stimulated asthmatic mice, brahma-related gene 1 inhibition of PI3K/AKT was found to exacerbate airway inflammation, whereas PI3K inhibitor attenuated this effect (40). In mice with OVA-induced allergic airway inflammation, TLR2 was found to induce autophagy through PI3K/AKT to promote airway inflammation. Moreover, the autophagy inhibitor 3 methyladenine alleviated inflammation (41). Luteolin and pingchuanning decoction improves asthma inflammation by inhibiting autophagy via activation of PI3K/AKT/mTOR pathway (42, 43). In addition, LAP supports the presentation of major histocompatibility complex (MHC)-II class antigen to helper CD4+ T cells which are key cells in the pathogenesis of asthma (44–46). In addition, LAP dysregulation can lead to autoimmune disease and inflammation (10). LAP was shown to stimulate macrophage dysregulation in chronic obstructive pulmonary disease (COPD) and cigarette smoke extracts, which may be involved in the pathogenesis of COPD (47). However, there is a need for further studies on its role in the pathogenesis of asthma.
Type 2 innate lymphoid cells (ILC2) are involved in the pathogenesis of Th2 asthma, and autophagy is involved in ILC2 activation and immune homeostasis. Lack of autophagy in ILC2 promotes glycolysis, inhibits fatty acid oxidation and TCA cycle, suppresses Th2 cytokine production, and attenuates asthma AHR (48). Sepsis caused by Methicillin-resistant Staphylococcus aureus (MRSA) infection was associated with elevated levels of autophagic protein and elevated serum levels of interferon-gamma (IFN-γ) and interleukin (IL)-17; in addition, rapamycin enhanced autophagy to reduce the ratio of Th1 and Th17 cells and promote survival in septic mice (49). The complex interplay between autophagy and T cell subsets and cytokines has given rise to an in-depth consideration of their role in asthma.
3.2 Autophagy and monocytes/macrophages
Macrophages are the main immune cells in asthma and are involved in phagocytosis, secretion of inflammatory factors, antigen presentation, and regulation of the immune response (50, 51). Studies have shown impaired phagocytosis of macrophages in children with non-eosinophilic asthma and poorly controlled asthma (52, 53). Macrophages can be classified into two phenotypes, i.e., classically activated M1 and substitution-activated M2, and these two macrophage states reflect Th1-Th2 polarization. M1 is stimulated by IFN-γ cytokines to produce Th1 cytokines (IL-6, IL-1β) that promote neutrophil inflammation, while M2 is stimulated by IL-4 and IL-13 to produce eosinophil-promoting cytokines (IL-10, TGF-β) (54). This has also been verified in animal models of asthma, where M1 polarisation was predominant in farm dust extracts-induced non-allergic asthma and where Th1 and Th17 cells and neutrophil infiltration were predominant. In contrast, in house dust mite-induced allergic asthma, M2 polarisation was predominant and Th2 cells and eosinophils were increased (55).
Paucigranulocytic asthma is a milder form of asthma characterized by macrophages as the predominant cell type. The molecular mechanisms underlying this type are not well understood. Macrophage polarization induces Th1 and Th2 cytokines involved in the pathogenetic mechanism of EA and NA. In contrast, autophagy has regulatory effects on the differentiation and survival of monocytes (56), phagocytosis, migration and adhesion functions of macrophages, macrophage polarization and bacterial killing. High glucose conditions can inhibit autophagy and promote macrophage migration and adhesion, providing a new strategy for the treatment of diabetic nephropathy (57). Autophagy is impaired in obese mice, and inhibition of macrophage autophagy promotes their polarization to M1 phenotype (58). In addition, lactobacillus induces autophagy by modulating Th1/Th2 cytokine balance to enhance mononuclear phagocyte response to Mycobacterium tuberculosis infection (59). Inhibition of autophagy enhances phagocytosis of MRSA in MRSA-infected mice (60). Pseudomonas aeruginosa infection promotes macrophage autophagy and limits macrophage intracellular killing, probably by activating NLRP3 vesicles and reducing the production of reactive oxygen species (ROS) and nitric oxide (NO); thus, inhibition of autophagy can promote clearance of bacteria (61, 62). In contrast, in Mycobacterium tuberculosis infection, autophagy inhibits the survival of Mycobacterium tuberculosis in a PI3K-dependent manner and contributes to an enhanced immune response (63). The different roles of autophagy may depend on the microenvironment and therefore specific studies are needed for different diseases.
3.3 Autophagy and dendritic cells
Dendritic cells play an important role in adaptive immunity because of their antigen-presenting role. Autophagy has multiple effects on the functional regulation of DCs. Ho et al. demonstrated that autophagy regulates DC antigen presentation (64). Autophagy promotes the survival of lipopolysaccharide-induced DCs under hypoxic conditions (65), and defective autophagy protein VPS34 increases MHC-I and class II antigen presentation by DC cells (66). In contrast, in a DC study of ATG5 and ATG7 deficiency, autophagy was shown to inhibit MHC-I and promote MHC-II class antigen presentation (67). DC deficiency of ATG5 promotes antiviral immune responses involving CD8+ cells (68). Autophagy in RSV-infected DCs was shown to facilitate CD4 cytokine production and DC maturation, promoting antiviral responses (69, 70).
3.4 Autophagy and B cells
The necessity of autophagy for B-cell polarization has yet to be investigated. ATG5 is involved in antigen presentation and B-cell polarization (71). Basal level autophagy is required for B-cell survival (72, 73). Dendritic cells and B-cell autophagy deficiency in toll-like receptor (TLR) 7-deficient mice was shown to promote activation of inflammatory vesicles and organ damage (74). While TLR7 agonists are beneficial in asthma, TLR7 agonists are effective airway diaphoresis substances (75, 76). In OVA-induced asthmatic mice, TLR7 was shown to attenuate airway inflammation by mediating the Nrf2 pathway (77). IL-4 induces enhanced B-cell autophagy in asthmatic mice via IAK, which contributes to antigen presentation and anti-apoptosis and is involved in asthma pathogenesis (78). B cells are important in the adaptive immune response and autophagy and its role in asthma deserves to be explored.
3.5 Autophagy and mast cells
Mast cells are key effector cells involved in the inflammation, airway hyperresponsiveness, and response to steroid treatment in asthma (79–81). Asthmatics have increased mast cells in airways (82). Mast cell activation correlates with asthma phenotype, and IL-33 and IgE stimulated activation of mast cells correlates with severe neutrophilic and eosinophilic asthma phenotypes, respectively (83). Mast cells were also shown to be associated with the response to corticosteroid treatment; in a study, the inhaled corticosteroid group showed reduction in mast cells in the epithelium and smooth muscle compared to the non-inhaled corticosteroid group (84). Autophagy is involved in mast cell degranulation and ATG7-deficient MC cells show severely impaired degranulation (85, 86). IL-33 expression is elevated in nasal epithelial cells in allergic rhinitis, inhibits autophagy and regulates degranulation and inflammatory factor release in mast cells via IL-33/ST2 (87). Mast cell autophagy contributes to defence against pathogenic bacteria. Pseudomonas aeruginosa infection promotes autophagy of airway epithelial cells and mast cells and facilitates bacterial clearance (88). Orosomucoid-like-3 (ORMDL3) is an asthma susceptibility gene. Studies have shown that ORMDL3 inhibits mast cell activation and degranulation and production of cytokines and chemokines through the activating transcription factor 6-related autophagic pathway, thereby suppressing the immune response (89). Mast cell autophagy is a potential therapeutic target in asthma worthy of future research.
3.6 Autophagy and eosinophils
EA is a classic allergic asthma that shows good response to glucocorticoids. The novel immunologic agents (e.g., anti-IL-4 monoclonal antibody, anti-IL-5 monoclonal antibody, anti-IgE monoclonal antibody) are also useful in this type of asthma. ATG5 is the main protein of the autophagic process. In vivo and in vitro experimental studies have shown that ATG5 deficiency leads to inhibition of eosinophil proliferation and differentiation, and enhancement of eosinophil extracellular traps formation and degranulation, promoting bacterial clearance (90). In addition, autophagy inhibits the release of the content of mature eosinophils, which may help reduce inflammation in EA (91). Impaired autophagy in obese HDM-sensitized asthmatic mice was found to aggravate airway inflammation and increase eosinophilic airway inflammation in ATG5-deficient mice with resistance to dexamethasone (92). Paradoxically, autophagy protein expression is increased in the peripheral blood of patients with severe asthma and IL-5 induces autophagy to promote the production of eosinophil cationic protein and eosinophil airway inflammation (18). Studies in asthmatic animals show that autophagy is associated with a severe eosinophil phenotype and that anti-IL-5 has a beneficial effect on asthma treatment by reducing autophagy (19). Moreover, the mTOR inhibitor rapamycin was shown to inhibit eosinophil differentiation in OVA-induced allergic asthma mice (93). The complex relationship between autophagy and eosinophils is inconclusive and may be related to the different pathogenesis of different asthma phenotypes, which remains to be elucidated.
3.7 Autophagy and neutrophils
Neutrophilic asthma is typically difficult to treat owing to resistance to glucocorticoid therapy and lack of other effective drugs. Moreover, biologic agents are mainly effective in EA. Autophagy has a multifaceted role in regulating neutrophil function. A study showed that genetic polymorphisms in the autophagy genes ATG5 and ATG7 are associated with NA inflammation (94). Autophagy is essential for neutrophil maturation and function. Defective autophagy leads to accumulation of immature neutrophils and impaired function, and autophagy can facilitate the differentiation process by inhibiting glycolysis and promoting energy metabolic shifts during lipolysis (95). In contrast, in immature neutrophils, autophagy has an inhibitory effect on neutrophil differentiation, possibly due to regulation of the p38-mTORC1 signaling pathway. The regulation of neutrophils by autophagy may be due to the regulation of multiple molecular mechanisms, and further experimental studies are needed (96). In addition, autophagy promotes neutrophil degranulation and is an important mediator in neutrophil inflammation (97). ATG5-deficient mice exacerbate neutrophil airway inflammation and cause exacerbation of steroid-resistant asthma involving IL-17A (98). Rapamycin can inhibit IL-17A production and suppress neutrophil inflammation (99).
Neutrophil extracellular traps (NETs) are trap networks consisting of DNA and various enzymatic proteins that form after neutrophils are stimulated to form cytoplasmic extracellular traps. It has an important anti-infective function and has a crucial role in asthma. The relationship between autophagy and NET is still controversial. Autophagy has been shown to be necessary for NET formation and to promote the formation of NETs (100). In an autophagy gene ATG5-deficient mouse model of senescence, TLR2 ligands were found to reduce NETs formation in neutrophils (101). Moreover, NETs formation was shown to be associated with autophagy and autophagy inducers promoted spontaneous release of NETs, further confirming that this relationship also exists in the OVA-induced asthma model (102). Increased levels of peripheral blood neutrophil autophagy protein were observed in patients with severe asthma relative to those in patients with non-severe asthma; in addition, a positive correlation between NET and autophagy was observed which may be involved in the pathogenesis of severe asthma by enhancing the eosinophil inflammatory response and epithelial cell damage (103). NET formation induced by phorbol myristate acetate (PMA) requires the involvement of autophagy and superoxide (100). Studies have also suggested a key role of autophagy in anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis, and autophagy was found to promote anti-LAMP-2 antibody and ANCA antibody-induced NET production (104, 105). However, PMA-stimulated neutrophil studies in ATG5-deficient mice showed that NET formation does not require the involvement of autophagy (106). Interestingly, it has been shown that autophagy has no significant effect on NETs generation. Autophagy gene ATG5 is not necessary for extracellular traps of neutrophils and eosinophils (97). Autophagy and NETs are associated with airway inflammation in severe asthma, and further studies are required for in-depth characterization of the relationship of autophagy and NETs with asthma. Mice lacking autophagy showed increased inflammation in IL-17-mediated bands of lung neutrophils. Absence of autophagy in lung CD11c cells induces neutrophil airway inflammation and airway hyperresponsiveness leading to exacerbation of asthma (98). Targeting autophagy-dependent NETs is a novel therapeutic strategy for asthma.
3.8 Autophagy and airway epithelial cells
Autophagy plays an important role in airway epithelial regeneration, lung development and morphogenesis (107, 108). IL-13 expression is elevated in EA, which can activate airway epithelial cell autophagy to promote mucus secretion and production of ROS (109). In addition, experiments in OVA-induced asthma mouse models and airway epithelial cells demonstrated that TGF-β3 promotes MUC5AC production for mucus secretion by regulating autophagy (110). Interestingly, specific knockout of mTOR in the airway epithelium induces autophagy and promotes allergic airway inflammation in mice (36). Epithelial mesenchymal transition (EMT) plays a key role in promoting asthma airway remodeling. In OVA-induced asthmatic mice, azithromycin was found to inhibit EMT and improve asthma airway remodeling (111). The link between autophagy and EMT has been widely studied in the context of cancer treatment and is gaining attention in asthma (112, 113). Autophagy is a key link in mesenchymal-to-epithelial transition (114). In vivo and in vitro studies have shown that FSTL1 activates autophagy and promotes EMT, thereby facilitating asthma airway remodeling (115). Further studies are required to demonstrate the mechanisms linking autophagy and EMT in asthma and to provide new strategies for asthma treatment.
3.9 Autophagy and airway smooth muscle cells
Airway smooth muscle is an important cell for airway remodeling in asthma. Azithromycin is one of the promising drugs for the treatment of NA; however, its exact mechanism of action remains to be elucidated. In addition, azithromycin was shown to reduce the proliferation of airway smooth muscle cells and induce autophagy (116). Knockdown of p62 in chronic asthma model inhibited the proliferation and migration of airway smooth muscle cells (117). Autophagy has important implications for the pathogenesis of cells and cytokines involved in asthma (Figure 2). Further studies are required for in-depth elucidation of its role.
3.10 Autophagy and vitamin D
Several studies have suggested that vitamin D may be an important mediator in different asthma phenotypes. However, there is no clear consensus regarding its specific mechanism of action. In a clinical trial including 54 adults with asthma, elevated serum vitamin D levels were associated with reduced AHR, improved lung function and increased sensitivity to glucocorticoids and exercise-induced bronchoconstriction (118, 119). In addition, vitamin D may be involved in the pathogenesis of asthma associated with obesity, and Vitamin D deficiency was shown to impair lung function in these patients (120). Vitamin D deficiency was also implicated as a factor in worsening of asthma control (121, 122). In asthmatic mice, administration of vitamin D was found to inhibit AHR and suppress inflammation (123). Moreover, a study showed that vitamin D and vitamin D receptor can regulate autophagy (124), although the mechanism of action in the context of asthma remains to be elucidated. Infection is a common trigger for worsening of asthma, and vitamin D plays a critical role in lung infection. In Aspergillus fumigatus-infected mice, the survival rate of vitamin D-sufficient mice was higher than that of the deficient group (125, 126). Vitamin D may protect against damage caused by Aspergillus fumigatus infection by reducing the expression of inflammatory factors such as NF-kB and IL-1β, IL-6 and inhibiting lysosome formation to reduce autophagy (125, 126). Autophagy-related proteins ATG5 and Becilin1 were increased in the OVA-induced asthma mouse model in the vitamin D-deficient group, compared with the vitamin D-sufficient group (127). In respiratory syncytial virus-induced asthmatic mice, vitamin D has the same effect of reducing inflammation, and it may act through the Notch1-HIF-1α signaling pathway to promote autophagy (128). Vitamin D has an important role in asthma, and the link between it and autophagy is worth exploring to provide new ideas for targeted asthma therapy.
4 miRNA mediated autophagy in asthma
MicroRNAs (miRNAs) represent a new tool for targeted therapy for asthma. The link between miRNAs and autophagy in asthma has received increasing attention. Studies have demonstrated the involvement of miRNA-targeted autophagy in the pathogenesis of asthma. MiR-192-5p, miR-20a-5p, miR-34/miR-449, miRNA-335-5p, and miR-30a inhibit autophagy in asthma through different pathways. Overexpression of miR-192-5p was found to reduce the expression of MMP-16 and ATG7, attenuate airway remodeling and inhibit autophagy in asthmatic mice (129). Overexpressed miR-20a-5p acts as an autophagy inhibitor, targeting ATG7 to inhibit autophagy, apoptosis, fibrosis and airway inflammation in asthma (130). MiR-335-5p and miR-30a target ATG5 to suppress asthma inflammation (131, 132). MiR-34/miR-449 overexpression promotes Nur77 nuclear translocation and inhibits autophagy by downregulating IGFBP-3, reducing airway fibrosis and inflammation (133). Studies of the relation between miRNA and autophagy provide a basis for the elucidation of the mechanism of asthma. However, further studies are required to unravel the detailed process of miRNA regulation of autophagy to provide new ideas for the development of targeted drugs for asthma treatment.
5 Autophagy and asthma treatment
Corticosteroids are the drugs of choice for asthma treatment; however, they are not always effective, and some patients develop steroid resistance. Respiratory tract infections are a common cause of acute exacerbations of asthma. In recent years, several drugs targeting autophagy (inhibiting autophagy or activating autophagy) have been identified for their potential use in asthma (Table 1). However, there are some contradictory results, and there may be many mechanisms involved, which still needs further research. In vitro, budesonide and simvastatin inhibited mTOR-mediated autophagy in macrophages, decreased Beclin-1 and LC3 expression, increased p62 and IL-10 expression, inhibited autophagy and reduced asthma inflammation (135). In contrast, budesonide and simvastatin were found to activate autophagy through other pathways to reduce asthma inflammation. Budesonide can activate autophagy to exert anti-rhinovirus effect and has a protective effect against asthma attacks (138). Simvastatin activates autophagy in bronchial smooth muscle cells (BSMCs) to inhibit airway inflammation and airway remodeling for the treatment of asthma (137). In addition, a novel biologic anti-IL-5 antibody significantly reduced LC3 II expression in lung homogenates (19). High-dose luteolin, yeast-fermented prebiotic, ketamine and α1-antitrypsin inhibit autophagy in lung tissue ameliorating asthmatic mice (42, 134, 136, 144). Chinese herbal medicines can also regulate autophagy and can be potentially used for asthma treatment. Paeoniflorin modulates mitochondrial function by suppressing autophagy and reducing the expression of pro-inflammatory factors to alleviate asthmatic inflammation (145). Astragalin can alleviate ROS-promoted bronchial fibrosis by inhibiting the formation of airway autophagosomes (142). Acupuncture was shown to reduce airway inflammation and AHR by modulating endoplasmic reticulum stress and CD4+ T lymphocyte differentiation by inhibiting ATG5-mediated autophagy (146).
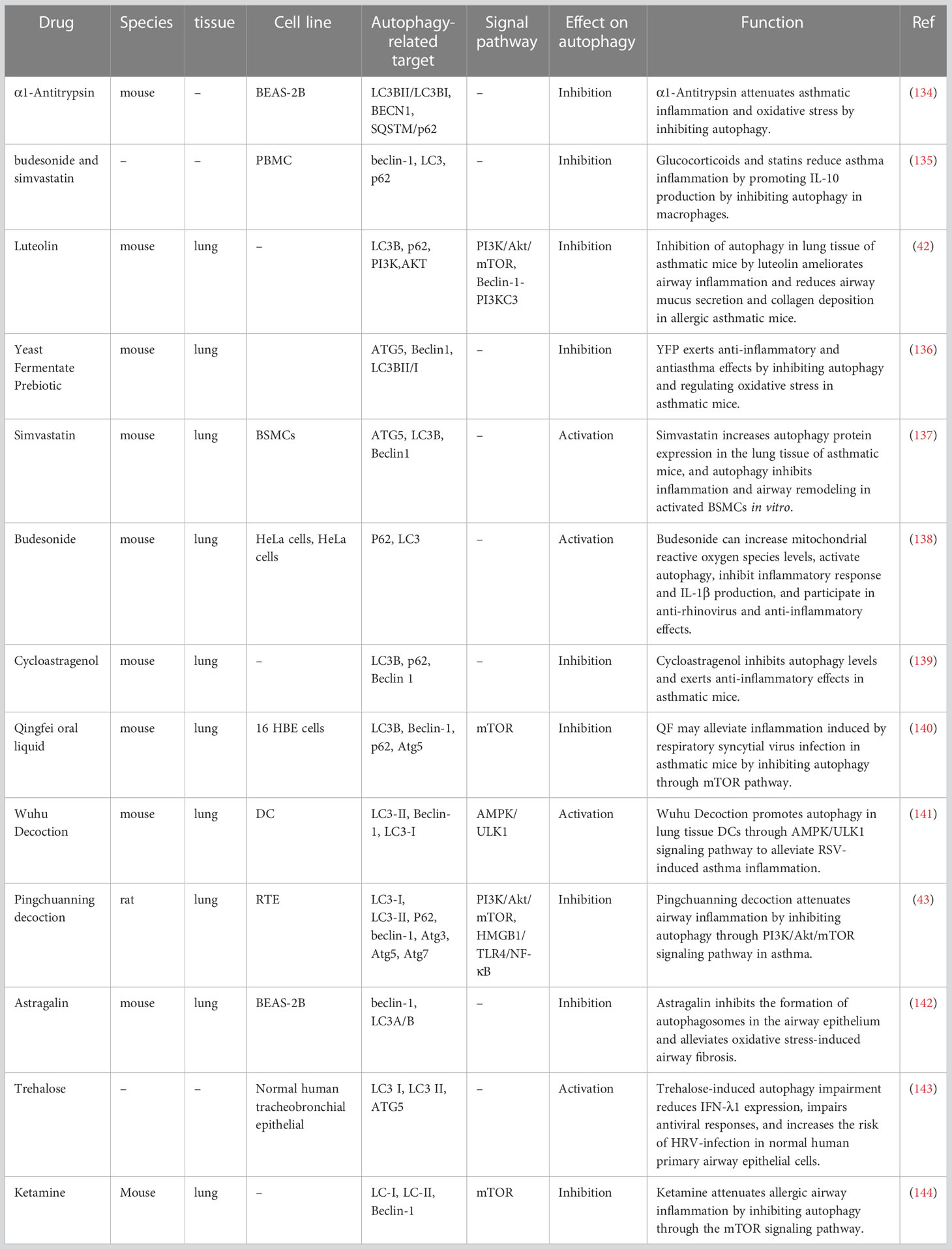
Table 1 Mechanisms of action of drug-mediated autophagy on allergic airway inflammation in vivo and in vitro.
6 Prospects and conclusion
Collectively, the available evidence suggests a key role of autophagy in the pathogenesis of asthma. Autophagy abnormality leads to the process of asthmatic airway inflammation and airway remodeling, indicating its promising role as a therapeutic target. This review also discusses the complex relationship between miRNAs and autophagy in asthma pathogenesis. However, there is a paucity of research on the underlying mechanism by which miRNA regulates autophagy in asthma. Most of the available evidence emanates from in vitro and in vivo preclinical studies. Development of targeted therapy for asthma is a key research imperative. The process of autophagy may be involved in regulating the immune response in different asthmatic inflammatory subtypes by influencing the antigen presentation, and secretion of cytokines by cells involved in asthma pathogenesis (e.g., B cells, T cells, macrophages, mast cells, neutrophils, and eosinophils). Based on the central role of autophagy in innate and adaptive immune responses, autophagy may play a beneficial or detrimental role in asthma, depending on the cellular microenvironment in asthmatic inflammatory subtypes. Further studies are required to unravel the specific mechanisms of the regulatory role of autophagy in asthma.
Author contributions
YH and PG designed and conceived the study, and reviewed the manuscript; WY and HD wrote and revised the manuscript; WL, SZ, and LZ collected the data and created the figures and tables. All authors read and approved the final manuscript for publication.
Funding
This study was supported by grant from the Natural Science Foundation of Jilin Province (YDZJ202201ZYTS052) and the National Science Foundation of China (82070037).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AHR, airway hyperresponsiveness; ANCA, anti-neutrophil cytoplasmic antibody; BSMCs, bronchial smooth muscle cells; COPD, chronic obstructive pulmonary disease; DC, Dendritic Cells; EA, eosinophilic asthma; EMT, Epithelial mesenchymal transition; HDM, house dust mite; IFN-γ, interferon-gamma; IL-, interleukin; ILC2, Type 2 innate lymphoid cells; LAP, LC3-associated phagocytosis; MC, Mast cell;MHC, major histocompatibility complex; miRNAs, MicroRNAs; MRSA, Methicillin-resistant Staphylococcus aureus; NA, neutrophilic asthma; NETs, Neutrophil extracellular traps; NO, nitric oxide; ORMDL3, orosomucoid-like-3; OVA, ovalbumin; PMA, phorbol myristate acetate; ROS, reactive oxygen species; TLR, toll-like receptor.
References
1. Mims JW. Asthma: definitions and pathophysiology. Int Forum Allergy rhinology (2015) 5(Suppl 1):S2–6. doi: 10.1002/alr.21609
2. Kim HY, DeKruyff RH, Umetsu DT. The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol (2010) 11(7):577–84. doi: 10.1038/ni.1892
3. Boonpiyathad T, Sözener ZC, Satitsuksanoa P, Akdis CA. Immunologic mechanisms in asthma. Semin Immunol (2019) 46:101333. doi: 10.1016/j.smim.2019.101333
4. Reddel HK, Bacharier LB, Bateman ED, Brightling CE, Brusselle GG, Buhl R, et al. Global initiative for asthma strategy 2021: Executive summary and rationale for key changes. J Allergy Clin Immunol In Pract (2022) 10(1s):S1–s18. doi: 10.1111/resp.14174
5. Hynes G, Pavord ID. Targeted biologic therapy for asthma. Br Med Bulletin (2020) 133(1):16–35. doi: 10.1093/bmb/ldaa004
6. Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxidants Redox Signaling (2014) 20(3):460–73. doi: 10.1089/ars.2013.5371
7. Yano T, Kurata S. Intracellular recognition of pathogens and autophagy as an innate immune host defence. J Biochem (2011) 150(2):143–9. doi: 10.1093/jb/mvr083
8. Deretic V. Autophagy in inflammation, infection, and immunometabolism. Immunity (2021) 54(3):437–53. doi: 10.1016/j.immuni.2021.01.018
9. Romao S, Münz C. LC3-associated phagocytosis. Autophagy (2014) 10(3):526–8. doi: 10.4161/auto.27606
10. Heckmann BL, Boada-Romero E, Cunha LD, Magne J, Green DR. LC3-associated phagocytosis and inflammation. J Mol Biol (2017) 429(23):3561–76. doi: 10.1016/j.jmb.2017.08.012
11. Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA (2011) 108(42):17396–401. doi: 10.1073/pnas.1113421108
12. Montava-Garriga L, Ganley IG. Outstanding questions in mitophagy: What we do and do not know. J Mol Biol (2020) 432(1):206–30. doi: 10.1016/j.jmb.2019.06.032
13. Liu M, Zhang Y, Dong L, Guo Z. Apelin-13 facilitates mitochondria homeostasis via mitophagy to prevent against airway oxidative injury in asthma. Mol Immunol (2023) 153:1–9. doi: 10.1016/j.molimm.2022.11.012
14. Bai Q, Wang Z, Piao Y, Zhou X, Piao Q, Jiang J, et al. Sesamin alleviates asthma airway inflammation by regulating mitophagy and mitochondrial apoptosis. J Agric Food Chem (2022) 70(16):4921–33. doi: 10.1021/acs.jafc.1c07877
15. Martin LJ, Gupta J, Jyothula SS, Butsch Kovacic M, Biagini Myers JM, Patterson TL, et al. Functional variant in the autophagy-related 5 gene promotor is associated with childhood asthma. PLoS One (2012) 7(4):e33454. doi: 10.1371/journal.pone.0033454
16. Yang F, Kong J, Zong Y, Li Z, Lyu M, Li W, et al. Autophagy-related genes are involved in the progression and prognosis of asthma and regulate the immune microenvironment. Front Immunol (2022) 13:897835. doi: 10.3389/fimmu.2022.897835
17. McAlinden KD, Deshpande DA, Ghavami S, Xenaki D, Sohal SS, Oliver BG, et al. Autophagy activation in asthma airways remodeling. Am J Respir Cell Mol Biol (2019) 60(5):541–53. doi: 10.1165/rcmb.2018-0169OC
18. Ban GY, Pham DL, Trinh TH, Lee SI, Suh DH, Yang EM, et al. Autophagy mechanisms in sputum and peripheral blood cells of patients with severe asthma: a new therapeutic target. Clin Exp Allergy (2016) 46(1):48–59. doi: 10.1111/cea.12585
19. Liu JN, Suh DH, Trinh HK, Chwae YJ, Park HS, Shin YS. The role of autophagy in allergic inflammation: a new target for severe asthma. Exp Mol Med (2016) 48(7):e243. doi: 10.1038/emm.2016.38
20. Yang G, Song W, Postoak JL, Chen J, Martinez J, Zhang J, et al. Autophagy-related protein PIK3C3/VPS34 controls T cell metabolism and function. Autophagy (2021) 17(5):1193–204. doi: 10.1080/15548627.2020.1752979
21. Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci USA (2012) 109(22):8670–5. doi: 10.1073/pnas.1205305109
22. Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol (Baltimore Md: 1950) (2009) 182(7):4046–55. doi: 10.4049/jimmunol.0801143
23. Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med (2007) 204(1):25–31. doi: 10.1084/jem.20061303
24. Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol (Baltimore Md: 1950) (2011) 186(9):5313–22. doi: 10.4049/jimmunol.1002404
25. Pua HH, He YW. Maintaining T lymphocyte homeostasis: another duty of autophagy. Autophagy (2007) 3(3):266–7. doi: 10.4161/auto.3908
26. Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ (2012) 19(1):144–52. doi: 10.1038/cdd.2011.78
27. Raz Y, Guerrero-Ros I, Maier A, Slagboom PE, Atzmon G, Barzilai N, et al. Activation-induced autophagy is preserved in CD4+ T-cells in familial longevity. J Gerontol Ser A Biol Sci Med Sci (2017) 72(9):1201–6. doi: 10.1093/gerona/glx020
28. Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. eLife (2014) 3. doi: 10.7554/eLife.03706
29. Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, et al. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol (Baltimore Md: 1950) (2006) 177(8):5163–8. doi: 10.4049/jimmunol.177.8.5163
30. Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest (2015) 125(1):25–32. doi: 10.1172/JCI73939
31. Weichhart T. mTOR as regulator of lifespan, aging, and cellular senescence: A mini-review. Gerontology (2018) 64(2):127–34. doi: 10.1159/000484629
32. Wang H, Liu Y, Wang D, Xu Y, Dong R, Yang Y, et al. The upstream pathway of mTOR-mediated autophagy in liver diseases. Cells (2019) 8(12). doi: 10.3390/cells8121597
33. Liu T, Ye P, Ye Y, Han B. MicroRNA-216b targets HK2 to potentiate autophagy and apoptosis of breast cancer cells via the mTOR signaling pathway. Int J Biol Sci (2021) 17(11):2970–83. doi: 10.7150/ijbs.48933
34. Hu Y, Lou J, Mao YY, Lai TW, Liu LY, Zhu C, et al. Activation of MTOR in pulmonary epithelium promotes LPS-induced acute lung injury. Autophagy (2016) 12(12):2286–99. doi: 10.1080/15548627.2016.1230584
35. Li ZY, Chen LH, Zhao XY, Chen H, Sun YY, Lu MH, et al. Clemastine attenuates AD-like pathology in an AD model mouse via enhancing mTOR-mediated autophagy. Exp Neurology (2021) 342:113742. doi: 10.1016/j.expneurol.2021.113742
36. Li W, Wu Y, Zhao Y, Li Z, Chen H, Dong L, et al. MTOR suppresses autophagy-mediated production of IL25 in allergic airway inflammation. Thorax (2020) 75(12):1047–57. doi: 10.1136/thoraxjnl-2019-213771
37. Zhang Y, Jing Y, Qiao J, Luan B, Wang X, Wang L, et al. Activation of the mTOR signaling pathway is required for asthma onset. Sci Rep (2017) 7(1):4532. doi: 10.1038/s41598-017-04826-y
38. Cen S, Wang P, Xie Z, Yang R, Li J, Liu Z, et al. Autophagy enhances mesenchymal stem cell-mediated CD4(+) T cell migration and differentiation through CXCL8 and TGF-β1. Stem Cell Res Ther (2019) 10(1):265. doi: 10.1186/s13287-019-1380-0
39. Li X, Zhou L, Zhang Z, Liu Y, Liu J, Zhang C. IL-27 alleviates airway remodeling in a mouse model of asthma via PI3K/Akt pathway. Exp Lung Res (2020) 46(3-4):98–108. doi: 10.1080/01902148.2020.1740356
40. Zou W, Ding F, Niu C, Fu Z, Liu S. Brg1 aggravates airway inflammation in asthma via inhibition of the PI3K/Akt/mTOR pathway. Biochem Biophys Res Commun (2018) 503(4):3212–8. doi: 10.1016/j.bbrc.2018.08.127
41. Jiang X, Fang L, Wu H, Mei X, He F, Ding P, et al. TLR2 regulates allergic airway inflammation and autophagy through PI3K/Akt signaling pathway. Inflammation (2017) 40(4):1382–92. doi: 10.1007/s10753-017-0581-x
42. Wang S, Wuniqiemu T, Tang W, Teng F, Bian Q, Yi L, et al. Luteolin inhibits autophagy in allergic asthma by activating PI3K/Akt/mTOR signaling and inhibiting beclin-1-PI3KC3 complex. Int Immunopharmacol (2021) 94:107460. doi: 10.1016/j.intimp.2021.107460
43. Wang X, Gao Y, Yang Q, Fang X, Li Z. Pingchuanning decoction attenuates airway inflammation by suppressing autophagy via phosphatidylinositol 3-kinase/protein kinase b/mammalian target of rapamycin signaling pathway in rat models of asthma. J Cell Biochem (2019) 120(3):3833–44. doi: 10.1002/jcb.27665
44. Ligeon LA, Pena-Francesch M, Münz C. Measuring oxidation within LC3-associated phagosomes that optimizes MHC class II restricted antigen presentation. Methods Cell Biol (2021) 164:187–200. doi: 10.1016/bs.mcb.2021.02.003
45. Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D, et al. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J Cell Biol (2013) 203(5):757–66. doi: 10.1083/jcb.201308173
46. Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem (2012) 287(41):34149–56. doi: 10.1074/jbc.M112.382812
47. Asare PF, Tran HB, Hurtado PR, Perkins GB, Nguyen P, Jersmann H, et al. Inhibition of LC3-associated phagocytosis in COPD and in response to cigarette smoke. Ther Adv Respir Disease (2021) 15:17534666211039769. doi: 10.1177/17534666211039769
48. Galle-Treger L, Hurrell BP, Lewis G, Howard E, Jahani PS, Banie H, et al. Autophagy is critical for group 2 innate lymphoid cell metabolic homeostasis and effector function. J Allergy Clin Immunol (2020) 145(2):502–17.e5. doi: 10.1016/j.jaci.2019.10.035
49. Zhang S, Huang X, Xiu H, Zhang Z, Zhang K, Cai J, et al. The attenuation of Th1 and Th17 responses via autophagy protects against methicillin-resistant staphylococcus aureus-induced sepsis. Microbes Infect (2021) 23(8):104833. doi: 10.1016/j.micinf.2021.104833
50. Fricker M, Gibson PG. Macrophage dysfunction in the pathogenesis and treatment of asthma. Eur Respir J (2017) 50(3). doi: 10.1183/13993003.00196-2017
51. VanHook AM. Macrophages present antigens in allergic asthma. Sci Signaling (2022) 15(755):adf2043. doi: 10.1126/scisignal.adf2043
52. Simpson JL, Gibson PG, Yang IA, Upham J, James A, Reynolds PN, et al. Impaired macrophage phagocytosis in non-eosinophilic asthma. Clin Exp Allergy (2013) 43(1):29–35. doi: 10.1111/j.1365-2222.2012.04075.x
53. Fitzpatrick AM, Holguin F, Teague WG, Brown LA. Alveolar macrophage phagocytosis is impaired in children with poorly controlled asthma. J Allergy Clin Immunol (2008) 121(6):1372–8, 8.e1-3. doi: 10.1016/j.jaci.2008.03.008
54. Saradna A, Do DC, Kumar S, Fu QL, Gao P. Macrophage polarization and allergic asthma. Trans research: J Lab Clin Med (2018) 191:1–14. doi: 10.1016/j.trsl.2017.09.002
55. Robbe P, Draijer C, Borg TR, Luinge M, Timens W, Wouters IM, et al. Distinct macrophage phenotypes in allergic and nonallergic lung inflammation. Am J Physiol Lung Cell Mol Physiol (2015) 308(4):L358–67. doi: 10.1152/ajplung.00341.2014
56. Zhang Y, Morgan MJ, Chen K, Choksi S, Liu ZG. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood (2012) 119(12):2895–905. doi: 10.1182/blood-2011-08-372383
57. Jiang Y, Zhao Y, Zhu X, Liu Y, Wu B, Guo Y, et al. Effects of autophagy on macrophage adhesion and migration in diabetic nephropathy. Renal Failure (2019) 41(1):682–90. doi: 10.1080/0886022X.2019.1632209
58. Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, et al. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy (2015) 11(2):271–84. doi: 10.1080/15548627.2015.1009787
59. Ghadimi D, de Vrese M, Heller KJ, Schrezenmeir J. Lactic acid bacteria enhance autophagic ability of mononuclear phagocytes by increasing Th1 autophagy-promoting cytokine (IFN-gamma) and nitric oxide (NO) levels and reducing Th2 autophagy-restraining cytokines (IL-4 and IL-13) in response to mycobacterium tuberculosis antigen. Int Immunopharmacol (2010) 10(6):694–706. doi: 10.1016/j.intimp.2010.03.014
60. Zhu Y, Li H, Ding S, Wang Y. Autophagy inhibition promotes phagocytosis of macrophage and protects mice from methicillin-resistant staphylococcus aureus pneumonia. J Cell Biochem (2018) 119(6):4808–14. doi: 10.1002/jcb.26677
61. Deng Q, Wang Y, Zhang Y, Li M, Li D, Huang X, et al. Pseudomonas aeruginosa triggers macrophage autophagy to escape intracellular killing by activation of the NLRP3 inflammasome. Infection Immunity (2016) 84(1):56–66. doi: 10.1128/IAI.00945-15
62. Wu Y, Li D, Wang Y, Chen K, Yang K, Huang X, et al. Pseudomonas aeruginosa promotes autophagy to suppress macrophage-mediated bacterial eradication. Int Immunopharmacol (2016) 38:214–22. doi: 10.1016/j.intimp.2016.04.044
63. Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and mycobacterium tuberculosis survival in infected macrophages. Cell (2004) 119(6):753–66. doi: 10.1016/j.cell.2004.11.038
64. Ho NI, Camps MGM, Verdoes M, Münz C, Ossendorp F. Autophagy regulates long-term cross-presentation by murine dendritic cells. Eur J Immunol (2021) 51(4):835–47. doi: 10.1002/eji.202048961
65. Monaci S, Aldinucci C, Rossi D, Giuntini G, Filippi I, Ulivieri C, et al. Hypoxia shapes autophagy in LPS-activated dendritic cells. Front Immunol (2020) 11:573646. doi: 10.3389/fimmu.2020.573646
66. Parekh VV, Pabbisetty SK, Wu L, Sebzda E, Martinez J, Zhang J, et al. Autophagy-related protein Vps34 controls the homeostasis and function of antigen cross-presenting CD8α(+) dendritic cells. Proc Natl Acad Sci USA (2017) 114(31):E6371–e80. doi: 10.1073/pnas.1706504114
67. Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity (2010) 32(2):227–39. doi: 10.1016/j.immuni.2009.12.006
68. Loi M, Müller A, Steinbach K, Niven J, Barreira da Silva R, Paul P, et al. Macroautophagy proteins control MHC class I levels on dendritic cells and shape anti-viral CD8(+) T cell responses. Cell Rep (2016) 15(5):1076–87. doi: 10.1016/j.celrep.2016.04.002
69. Morris S, Swanson MS, Lieberman A, Reed M, Yue Z, Lindell DM, et al. Autophagy-mediated dendritic cell activation is essential for innate cytokine production and APC function with respiratory syncytial virus responses. J Immunol (Baltimore Md: 1950) (2011) 187(8):3953–61. doi: 10.4049/jimmunol.1100524
70. Reed M, Morris SH, Jang S, Mukherjee S, Yue Z, Lukacs NW. Autophagy-inducing protein beclin-1 in dendritic cells regulates CD4 T cell responses and disease severity during respiratory syncytial virus infection. J Immunol (Baltimore Md: 1950) (2013) 191(5):2526–37. doi: 10.4049/jimmunol.1300477
71. Arbogast F, Arnold J, Hammann P, Kuhn L, Chicher J, Murera D, et al. ATG5 is required for b cell polarization and presentation of particulate antigens. Autophagy (2019) 15(2):280–94. doi: 10.1080/15548627.2018.1516327
72. Arnold J, Murera D, Arbogast F, Fauny JD, Muller S, Gros F. Autophagy is dispensable for b-cell development but essential for humoral autoimmune responses. Cell Death Differ (2016) 23(5):853–64. doi: 10.1038/cdd.2015.149
73. Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, et al. The autophagy gene ATG5 plays an essential role in b lymphocyte development. Autophagy (2008) 4(3):309–14. doi: 10.4161/auto.5474
74. Weindel CG, Richey LJ, Mehta AJ, Shah M, Huber BT. Autophagy in dendritic cells and b cells is critical for the inflammatory state of TLR7-mediated autoimmunity. J Immunol (Baltimore Md: 1950) (2017) 198(3):1081–92. doi: 10.4049/jimmunol.1601307
75. Kaufman EH, Fryer AD, Jacoby DB. Toll-like receptor 7 agonists are potent and rapid bronchodilators in guinea pigs. J Allergy Clin Immunol (2011) 127(2):462–9. doi: 10.1016/j.jaci.2010.10.029
76. Drake MG, Scott GD, Proskocil BJ, Fryer AD, Jacoby DB, Kaufman EH. Toll-like receptor 7 rapidly relaxes human airways. Am J Respir Crit Care Med (2013) 188(6):664–72. doi: 10.1164/rccm.201303-0442OC
77. Nadeem A, Siddiqui N, Al-Harbi NO, Al-Harbi MM, Ahmad SF. TLR-7 agonist attenuates airway reactivity and inflammation through Nrf2-mediated antioxidant protection in a murine model of allergic asthma. Int J Biochem Cell Biol (2016) 73:53–62. doi: 10.1016/j.biocel.2016.02.004
78. Xia F, Deng C, Jiang Y, Qu Y, Deng J, Cai Z, et al. IL4 (interleukin 4) induces autophagy in b cells leading to exacerbated asthma. Autophagy (2018) 14(3):450–64. doi: 10.1080/15548627.2017.1421884
79. Williams CM, Galli SJ. Mast cells can amplify airway reactivity and features of chronic inflammation in an asthma model in mice. J Exp Med (2000) 192(3):455–62. doi: 10.1084/jem.192.3.455
80. Kobayashi T, Miura T, Haba T, Sato M, Serizawa I, Nagai H, et al. An essential role of mast cells in the development of airway hyperresponsiveness in a murine asthma model. J Immunol (Baltimore Md: 1950) (2000) 164(7):3855–61. doi: 10.4049/jimmunol.164.7.3855
81. Wang G, Baines KJ, Fu JJ, Wood LG, Simpson JL, McDonald VM, et al. Sputum mast cell subtypes relate to eosinophilia and corticosteroid response in asthma. Eur Respir J (2016) 47(4):1123–33. doi: 10.1183/13993003.01098-2015
82. Bradding P, Brightling C. Mast cell infiltration of airway smooth muscle in asthma. Respir Med (2007) 101(5):1045. doi: 10.1016/j.rmed.2007.01.004
83. Tiotiu A, Badi Y, Kermani NZ, Sanak M, Kolmert J, Wheelock CE, et al. Association of differential mast cell activation with granulocytic inflammation in severe asthma. Am J Respir Crit Care Med (2022) 205(4):397–411. doi: 10.1164/rccm.202102-0355OC
84. James A, Gyllfors P, Henriksson E, Dahlén SE, Adner M, Nilsson G, et al. Corticosteroid treatment selectively decreases mast cells in the smooth muscle and epithelium of asthmatic bronchi. Allergy (2012) 67(7):958–61. doi: 10.1111/j.1398-9995.2012.02836.x
85. Ushio H, Ueno T, Kojima Y, Komatsu M, Tanaka S, Yamamoto A, et al. Crucial role for autophagy in degranulation of mast cells. J Allergy Clin Immunol (2011) 127(5):1267–76.e6. doi: 10.1016/j.jaci.2010.12.1078
86. Nakano H, Ushio H. An unexpected role for autophagy in degranulation of mast cells. Autophagy (2011) 7(6):657–9. doi: 10.4161/auto.7.6.15384
87. Nian JB, Zeng M, Zheng J, Zeng LY, Fu Z, Huang QJ, et al. Epithelial cells expressed IL-33 to promote degranulation of mast cells through inhibition on ST2/PI3K/mTOR-mediated autophagy in allergic rhinitis. Cell Cycle (Georgetown Tex) (2020) 19(10):1132–42. doi: 10.1080/15384101.2020.1749402
88. Junkins RD, Shen A, Rosen K, McCormick C, Lin TJ. Autophagy enhances bacterial clearance during p. aeruginosa lung infection. PloS One (2013) 8(8):e72263. doi: 10.1371/journal.pone.0072263
89. Li J, Ullah MA, Jin H, Liang Y, Lin L, Wang J, et al. ORMDL3 functions as a negative regulator of antigen-mediated mast cell activation via an ATF6-UPR-Autophagy-Dependent pathway. Front Immunol (2021) 12:604974. doi: 10.3389/fimmu.2021.604974
90. Germic N, Hosseini A, Stojkov D, Oberson K, Claus M, Benarafa C, et al. ATG5 promotes eosinopoiesis but inhibits eosinophil effector functions. Blood (2021) 137(21):2958–69. doi: 10.1182/blood.2020010208
91. Silveira JS, Antunes GL, Kaiber DB, da Costa MS, Ferreira FS, Marques EP, et al. Autophagy induces eosinophil extracellular traps formation and allergic airway inflammation in a murine asthma model. J Cell Physiol (2020) 235(1):267–80. doi: 10.1002/jcp.28966
92. Suzuki Y, Aono Y, Akiyama N, Horiike Y, Naoi H, Horiguchi R, et al. Involvement of autophagy in exacerbation of eosinophilic airway inflammation in a murine model of obese asthma. Autophagy (2022) 18(9):2216–28. doi: 10.1080/15548627.2022.2025571
93. Hua W, Liu H, Xia LX, Tian BP, Huang HQ, Chen ZY, et al. Rapamycin inhibition of eosinophil differentiation attenuates allergic airway inflammation in mice. Respirology (Carlton Vic) (2015) 20(7):1055–65. doi: 10.1111/resp.12554
94. Pham DL, Kim SH, Losol P, Yang EM, Shin YS, Ye YM, et al. Association of autophagy related gene polymorphisms with neutrophilic airway inflammation in adult asthma. Korean J Internal Med (2016) 31(2):375–85. doi: 10.3904/kjim.2014.390
95. Riffelmacher T, Clarke A, Richter FC, Stranks A, Pandey S, Danielli S, et al. Autophagy-dependent generation of free fatty acids is critical for normal neutrophil differentiation. Immunity (2017) 47(3):466–80.e5. doi: 10.1016/j.immuni.2017.08.005
96. Rožman S, Yousefi S, Oberson K, Kaufmann T, Benarafa C, Simon HU. The generation of neutrophils in the bone marrow is controlled by autophagy. Cell Death Differ (2015) 22(3):445–56. doi: 10.1038/cdd.2014.169
97. Bhattacharya A, Wei Q, Shin JN, Abdel Fattah E, Bonilla DL, Xiang Q, et al. Autophagy is required for neutrophil-mediated inflammation. Cell Rep (2015) 12(11):1731–9. doi: 10.1016/j.celrep.2015.08.019
98. Suzuki Y, Maazi H, Sankaranarayanan I, Lam J, Khoo B, Soroosh P, et al. Lack of autophagy induces steroid-resistant airway inflammation. J Allergy Clin Immunol (2016) 137(5):1382–9.e9. doi: 10.1016/j.jaci.2015.09.033
99. Wu J, Zhong W, Zhang H, Yin Y. Mammalian target of rapamycin signaling enhances ovalbumin-induced neutrophilic airway inflammation by promoting Th17 cell polarization in murine noneosinophilic asthma model. Pediatr Allergy Immunol Pulmonol (2020) 33(1):25–32. doi: 10.1089/ped.2019.1088
100. Remijsen Q, Vanden Berghe T, Wirawan E, Asselbergh B, Parthoens E, De Rycke R, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res (2011) 21(2):290–304. doi: 10.1038/cr.2010.150
101. Xu F, Zhang C, Zou Z, Fan EKY, Chen L, Li Y, et al. Aging-related Atg5 defect impairs neutrophil extracellular traps formation. Immunology (2017) 151(4):417–32. doi: 10.1111/imm.12740
102. Guo Y, Gao F, Wang X, Pan Z, Wang Q, Xu S, et al. Spontaneous formation of neutrophil extracellular traps is associated with autophagy. Sci Rep (2021) 11(1):24005. doi: 10.1038/s41598-021-03520-4
103. Pham DL, Ban GY, Kim SH, Shin YS, Ye YM, Chwae YJ, et al. Neutrophil autophagy and extracellular DNA traps contribute to airway inflammation in severe asthma. Clin Exp Allergy (2017) 47(1):57–70. doi: 10.1111/cea.12859
104. Sha LL, Wang H, Wang C, Peng HY, Chen M, Zhao MH. Autophagy is induced by anti-neutrophil cytoplasmic abs and promotes neutrophil extracellular traps formation. Innate Immunity (2016) 22(8):658–65. doi: 10.1177/1753425916668981
105. Tang S, Zhang Y, Yin SW, Gao XJ, Shi WW, Wang Y, et al. Neutrophil extracellular trap formation is associated with autophagy-related signalling in ANCA-associated vasculitis. Clin Exp Immunol (2015) 180(3):408–18. doi: 10.1111/cei.12589
106. Germic N, Stojkov D, Oberson K, Yousefi S, Simon HU. Neither eosinophils nor neutrophils require ATG5-dependent autophagy for extracellular DNA trap formation. Immunology (2017) 152(3):517–25. doi: 10.1111/imm.12790
107. Li K, Li M, Li W, Yu H, Sun X, Zhang Q, et al. Airway epithelial regeneration requires autophagy and glucose metabolism. Cell Death Disease (2019) 10(12):875. doi: 10.1038/s41419-019-2111-2
108. Yeganeh B, Lee J, Ermini L, Lok I, Ackerley C, Post M. Autophagy is required for lung development and morphogenesis. J Clin Invest (2019) 129(7):2904–19. doi: 10.1172/JCI127307
109. Dickinson JD, Alevy Y, Malvin NP, Patel KK, Gunsten SP, Holtzman MJ, et al. IL13 activates autophagy to regulate secretion in airway epithelial cells. Autophagy (2016) 12(2):397–409. doi: 10.1080/15548627.2015.1056967
110. Zhang Y, Tang H, Yuan X, Ran Q, Wang X, Song Q, et al. TGF-β3 promotes MUC5AC hyper-expression by modulating autophagy pathway in airway epithelium. EBioMedicine (2018) 33:242–52. doi: 10.1016/j.ebiom.2018.06.032
111. Pu Y, Liu Y, Liao S, Miao S, Zhou L, Wan L. Azithromycin ameliorates OVA-induced airway remodeling in balb/c mice via suppression of epithelial-to-mesenchymal transition. Int Immunopharmacol (2018) 58:87–93. doi: 10.1016/j.intimp.2018.03.016
112. Chen HT, Liu H, Mao MJ, Tan Y, Mo XQ, Meng XJ, et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol Cancer (2019) 18(1):101. doi: 10.1186/s12943-019-1030-2
113. Erin N, Grahovac J, Brozovic A, Efferth T. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resistance Updates: Rev Commentaries Antimicrobial Anticancer Chemotherapy (2020) 53:100715. doi: 10.1016/j.drup.2020.100715
114. Zsiros V, Katz S, Dóczi N, Kiss AL. Autophagy is the key process in the re-establishment of the epitheloid phenotype during mesenchymal-epithelial transition (MET). Exp Cell Res (2017) 352(2):382–92. doi: 10.1016/j.yexcr.2017.02.031
115. Liu T, Liu Y, Miller M, Cao L, Zhao J, Wu J, et al. Autophagy plays a role in FSTL1-induced epithelial mesenchymal transition and airway remodeling in asthma. Am J Physiol Lung Cell Mol Physiol (2017) 313(1):L27–l40. doi: 10.1152/ajplung.00510.2016
116. Stamatiou R, Paraskeva E, Boukas K, Gourgoulianis KI, Molyvdas PA, Hatziefthimiou AA. Azithromycin has an antiproliferative and autophagic effect on airway smooth muscle cells. Eur Respir J (2009) 34(3):721–30. doi: 10.1183/09031936.00089407
117. Yu H, Cheng Y, Zhang G, Wang X, Gu W, Guo X. p62-dependent autophagy in airway smooth muscle cells regulates metabolic reprogramming and promotes airway remodeling. Life Sci (2021) 266:118884. doi: 10.1016/j.lfs.2020.118884
118. Sutherland ER, Goleva E, Jackson LP, Stevens AD, Leung DY. Vitamin d levels, lung function, and steroid response in adult asthma. Am J Respir Crit Care Med (2010) 181(7):699–704. doi: 10.1164/rccm.200911-1710OC
119. Chinellato I, Piazza M, Sandri M, Peroni DG, Cardinale F, Piacentini GL, et al. Serum vitamin d levels and exercise-induced bronchoconstriction in children with asthma. Eur Respir J (2011) 37(6):1366–70. doi: 10.1183/09031936.00044710
120. Lautenbacher LA, Jariwala SP, Markowitz ME, Rastogi D. Vitamin d and pulmonary function in obese asthmatic children. Pediatr Pulmonol (2016) 51(12):1276–83. doi: 10.1002/ppul.23485
121. Kaaviyaa AT, Krishna V, Arunprasath TS, Ramanan PV. Vitamin d deficiency as a factor influencing asthma control in children. Indian Pediatrics (2018) 55(11):969–71. doi: 10.1007/s13312-018-1420-3
122. Confino-Cohen R, Brufman I, Goldberg A, Feldman BS. Asthma prevalence and asthma exacerbations: a large adult population-based study. Allergy (2014) 69(12):1673–80. doi: 10.1111/all.12508
123. Cho SW, Kim JH, Choi JH, Lim DH. Preventive and therapeutic effects of vitamin d in a mouse model of allergic asthma. Asian Pacific J Allergy Immunol (2019) 37(3):130–7. doi: 10.12932/AP-010218-0248
124. Sun J. VDR/vitamin d receptor regulates autophagic activity through ATG16L1. Autophagy (2016) 12(6):1057–8. doi: 10.1080/15548627.2015.1072670
125. Hu S, Dai J, Chen X. Vitamin d reduces autophagy by regulating NF-κB resistance to aspergillus fumigatus infection. Gene (2020) 753:144819. doi: 10.1016/j.gene.2020.144819
126. Dai J, Liang Y, Li H, Zhou W, Wang B, Gong A, et al. Vitamin d enhances resistance to aspergillus fumigatus in mice via inhibition of excessive autophagy. Am J Trans Res (2018) 10(2):381–91.
127. Zhou Y, Xue Y, Bao A, Han L, Bao W, Xia C, et al. Effect of vitamin d deficiency and supplementation in lactation and early life on allergic airway inflammation and the expression of autophagy-related genes in an ovalbumin mouse model. J Inflamm Res (2021) 14:4125–41. doi: 10.2147/JIR.S321642
128. Huang C, Peng M, Tong J, Zhong X, Xian J, Zhong L, et al. Vitamin d ameliorates asthma-induced lung injury by regulating HIF-1α/Notch1 signaling during autophagy. Food Sci Nutr (2022) 10(8):2773–85. doi: 10.1002/fsn3.2880
129. Lou L, Tian M, Chang J, Li F, Zhang G. MiRNA-192-5p attenuates airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7. Biomed Pharmacother (2020) 122:109692. doi: 10.1016/j.biopha.2019.109692
130. Yu Y, Men S, Zhang Y. miR-20a-5p ameliorates ovalbumin (OVA)-induced mouse model of allergic asthma through targeting ATG7-regulated cell death, fibrosis and inflammation. Int Immunopharmacol (2021) 95:107342. doi: 10.1016/j.intimp.2020.107342
131. Liang Q, He J, Yang Q, Zhang Q, Xu Y. MicroRNA-335-5p alleviates inflammatory response, airway fibrosis, and autophagy in childhood asthma through targeted regulation of autophagy related 5. Bioengineered (2022) 13(1):1791–801. doi: 10.1080/21655979.2021.1996315
132. Li BB, Chen YL, Pang F. MicroRNA-30a targets ATG5 and attenuates airway fibrosis in asthma by suppressing autophagy. Inflammation (2020) 43(1):44–53. doi: 10.1007/s10753-019-01076-0
133. Yin H, Zhang S, Sun Y, Li S, Ning Y, Dong Y, et al. MicroRNA-34/449 targets IGFBP-3 and attenuates airway remodeling by suppressing Nur77-mediated autophagy. Cell Death Disease (2017) 8(8):e2998. doi: 10.1038/cddis.2017.357
134. Huang CY, Hu RC, Li J, Chen BB, Dai AG. α1-antitrypsin alleviates inflammation and oxidative stress by suppressing autophagy in asthma. Cytokine (2021) 141:155454. doi: 10.1016/j.cyto.2021.155454
135. Maneechotesuwan K, Kasetsinsombat K, Wongkajornsilp A, Barnes PJ. Role of autophagy in regulating interleukin-10 and the responses to corticosteroids and statins in asthma. Clin Exp Allergy (2021) 51(12):1553–65. doi: 10.1111/cea.13825
136. Gong S, Ji X, Su J, Wang Y, Yan X, Wang G, et al. Yeast fermentate prebiotic ameliorates allergic asthma, associating with inhibiting inflammation and reducing oxidative stress level through suppressing autophagy. Mediators Inflamm (2021) 2021:4080935. doi: 10.1155/2021/4080935
137. Gu W, Cui R, Ding T, Li X, Peng J, Xu W, et al. Simvastatin alleviates airway inflammation and remodelling through up-regulation of autophagy in mouse models of asthma. Respirology (Carlton Vic) (2017) 22(3):533–41. doi: 10.1111/resp.12926
138. Kim SR, Song JH, Ahn JH, Lee GS, Ahn H, Yoon SI, et al. Antiviral and anti-inflammatory activity of budesonide against human rhinovirus infection mediated via autophagy activation. Antiviral Res (2018) 151:87–96. doi: 10.1016/j.antiviral.2018.01.012
139. Zhu X, Cao Y, Su M, Chen M, Li C, Yi L, et al. Cycloastragenol alleviates airway inflammation in asthmatic mice by inhibiting autophagy. Mol Med Rep (2021) 24(5). doi: 10.3892/mmr.2021.12445
140. Yu L, Wang J, Zou Y, Zeng H, Cheng W, Jing X. Qingfei oral liquid inhibited autophagy to alleviate inflammation via mTOR signaling pathway in RSV-infected asthmatic mice. Biomed Pharmacother (2021) 138:111449. doi: 10.1016/j.biopha.2021.111449
141. Chen X, Luo Y, Wang M, Sun L, Huang K, Li Y, et al. Wuhu decoction regulates dendritic cell autophagy in the treatment of respiratory syncytial virus (RSV)-induced mouse asthma by AMPK/ULK1 signaling pathway. Med Sci monitor: Int Med J Exp Clin Res (2019) 25:5389–400. doi: 10.12659/MSM.917692
142. Cho IH, Choi YJ, Gong JH, Shin D, Kang MK, Kang YH. Astragalin inhibits autophagy-associated airway epithelial fibrosis. Respir Res (2015) 16(1):51. doi: 10.1186/s12931-015-0211-9
143. Wu Q, Jiang D, Huang C, van Dyk LF, Li L, Chu HW. Trehalose-mediated autophagy impairs the anti-viral function of human primary airway epithelial cells. PLoS One (2015) 10(4):e0124524. doi: 10.1371/journal.pone.0124524
144. Zou H, Wang LX, Wang M, Cheng C, Li S, Shen Q, et al. MTOR-mediated autophagy is involved in the protective effect of ketamine on allergic airway inflammation. J Immunol Res (2019) 2019:5879714. doi: 10.1155/2019/5879714
145. Han X, Hu S, Yang Q, Sang X, Tang D, Cao G. Paeoniflorin ameliorates airway inflammation and immune response in ovalbumin induced asthmatic mice: From oxidative stress to autophagy. Phytomedicine: Int J Phytother Phytopharmacol (2022) 96:153835. doi: 10.1016/j.phymed.2021.153835
Keywords: asthma, autophagy, biomarker, therapeutic target, inflammation
Citation: Dong H, Yang W, Li W, Zhu S, Zhu L, Gao P and Hao Y (2023) New insights into autophagy in inflammatory subtypes of asthma. Front. Immunol. 14:1156086. doi: 10.3389/fimmu.2023.1156086
Received: 01 February 2023; Accepted: 27 March 2023;
Published: 06 April 2023.
Edited by:
Svetlana P. Chapoval, University of Maryland, United StatesReviewed by:
Achsah D. Keegan, University of Maryland, United StatesMohammad Asad, Albert Einstein College of Medicine, United States
Copyright © 2023 Dong, Yang, Li, Zhu, Zhu, Gao and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Gao, Z2FvcGVuZzEyMzRAc2luYS5jb20=; Yuqiu Hao, aGFvLnl1LnFpLnVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Hongna Dong1†
Hongna Dong1† Wei Yang
Wei Yang Peng Gao
Peng Gao