- 1Department of Infectious Diseases, Istituto Superiore di Sanità, Rome, Italy
- 2National Center for Global Health, Istituto Superiore di Sanità, Rome, Italy
- 3Department of Oncology and Molecular Medicine, Istituto Superiore di Sanità, Rome, Italy
- 4Confocal Microscopy Unit, Core Facilities, Istituto Superiore di Sanità, Rome, Italy
- 5Center for Gender Medicine, Istituto Superiore di Sanità, Rome, Italy
- 6Department of Veterinary Public Health & Food Safety, Istituto Superiore di Sanità, Rome, Italy
- 7Center for Animal Research and Welfare, Istituto Superiore di Sanità, Rome, Italy
- 8Department of Medical Microbiology and Infection Prevention, Amsterdam UMC, University of Amsterdam, Amsterdam Institute for Infection and Immunity, Amsterdam, Netherlands
Several COVID-19 vaccine strategies utilizing new formulations for the induction of neutralizing antibodies (nAbs) and T cell immunity are still under evaluation in preclinical and clinical studies. Here we used Simian Immunodeficiency Virus (SIV)-based integrase defective lentiviral vector (IDLV) delivering different conformations of membrane-tethered Spike protein in the mouse immunogenicity model, with the aim of inducing persistent nAbs against multiple SARS-CoV-2 variants of concern (VoC). Spike modifications included prefusion-stabilizing double proline (2P) substitutions, mutations at the furin cleavage site (FCS), D614G mutation and truncation of the cytoplasmic tail (delta21) of ancestral and Beta (B.1.351) Spike, the latter mutation to markedly improve IDLV membrane-tethering. BALB/c mice were injected once with IDLV delivering the different forms of Spike or the recombinant trimeric Spike protein with 2P substitutions and FCS mutations in association with a squalene-based adjuvant. Anti-receptor binding domain (RBD) binding Abs, nAbs and T cell responses were detected up to six months from a single immunization with escalating doses of vaccines in all mice, but with different levels and kinetics. Results indicated that IDLV delivering the Spike protein with all the combined modifications, outperformed the other candidates in terms of T cell immunity and level of both binding Abs and nAbs soon after the single immunization and persistence over time, showing the best capacity to neutralize all formerly circulating VoC Alpha, Beta, Gamma and Delta. Although present, the lowest response was detected against Omicron variants (BA.1, BA.2 and BA.4/5), suggesting that the magnitude of immune evasion may be related to the higher genetic distance of Omicron as indicated by increased number of amino acid substitutions in Spike acquired during virus evolution.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the Betacoronavirus genus and is the agent causative of coronavirus disease 2019 (COVID-19). COVID-19 outbreak has been declared a pandemic by the World Health Organization (WHO) in March 2020 and soon after has become a global health priority, with over 757 million confirmed infections and more than 6.8 million deaths as of February 2023 (1). As a consequence, a global effort started to develop effective preventative interventions against SARS-CoV-2.
SARS-CoV-2 utilizes the transmembrane homotrimeric Spike glycoprotein to enter into target cells via the angiotensin-converting enzyme 2 (ACE2) receptor (2). As a consequence, neutralizing antibodies (nAbs) against the viral Spike protein are an essential component of the protective immune response against SARS-CoV-2 (3). Several effective vaccines delivering Spike, including mRNA, protein subunit, adenoviral vector, and whole-cell inactivated virus, showed efficacy in phase III trials and have received approval for use in many countries (4), and more are under consideration (5). While these vaccination approaches have proved remarkably successful in limiting viral spread and disease, mutations that affect transmission and disease severity have occurred throughout the pandemic. Indeed, the high infection rates and the immune selection pressure induced by the vaccines at a population level have accelerated the development of escape mutants, as demonstrated by the insurgence and spreading of several variants of interest and of concern (VoI and VoC), thus posing a threat to the long-term effectiveness of these vaccines. In particular, VoC have specific mutations in their Spike proteins that have been associated with breakthrough infections, increased transmissibility (6–11) and decreased sensitivity against neutralization by monoclonal Abs (mAbs), sera from vaccinated individuals and convalescent plasma (12–24). Therefore, vaccines against COVID-19 need continuous optimization and updating as a matter of urgency. In addition, the lack of sterilizing immunity and the modest durability of the protective immune responses induced by currently approved vaccines require additional boosters in a relatively short interval of time, decreasing the overall vaccine compliance. As a consequence, persistent and cross-reactive vaccination approaches should be sought to prevent close vaccination cycles, especially in developing countries with a low-resource setting, where the cost and logistics of vaccine campaigns are difficult (25–28).
Integrase-defective lentiviral vectors (IDLVs) offer a safe alternative vaccination approach with features similar to live attenuated virus including sustained antigen expression from the episomal forms of the vector, but in the absence of integration- and replication-competent virus (29–31). IDLVs showed their efficacy to induce high magnitude and long-lasting antigen-specific cellular and humoral immunity in mice, non-human primates (NHPs) and humans (32–36). Importantly, recent reports showed efficacy of non-integrating lentiviral vector against SARS-CoV-2 in mice immunogenicity studies (37, 38). In order to improve immunogenicity, we demonstrated that IDLV can be exploited for delivering immunogens also after pseudotyping of heterologous viral envelope glycoproteins on the vector’s particles, in addition to VSV.G. In particular, pseudotyping of IDLV with influenza virus hemagglutinin (HA) resulted in the induction of an anti-HA nAb response in mice, which was persistent for up to 24 weeks after a single immunization (39). More recently, we demonstrated that truncation of cytoplasmic tail of HIV-Env and SARS-CoV-2 Spike glycoproteins greatly improved the pseudotyping of lentiviral particles (34, 40), leading to persistent immunogenicity in the NHP model of immunization using HIV-Env (34).
Although IDLVs provide strong and durable immune responses, the encoded antigen also dictates the quality and magnitude of those responses, and different design elements of the Spike protein may influence the performance of the immune response after immunization. In this report, we have developed and used Simian Immunodeficiency Virus (SIV)-based IDLV for the delivery of different conformations of Spike (IDLV-CoV2) in the mouse immunogenicity model. In particular, we evaluated the introduction of substitutions to stabilize Spike in the prefusion conformation, including mutations at the furin cleavage site (FCS) and of two consecutive prolines (2P) in the hinge region of S2 portion (2, 41), truncation of the Spike protein cytoplasmic tail to favour Spike membrane-tethering on IDLV particles (40), and the inclusion of the D614G mutation to enhance the exposure of the receptor binding motif (RBM) (42). When combined together, these mutations increased the magnitude of nAbs against the autologous Spike compared to IDLV delivering wild-type Spike and to the subunit vaccine and elicited higher cross-reactive nAbs against all VoC, including Omicron. Importantly, neutralizing activity persisted up to 6 months from a single immunization even at the lowest dose of the IDLV expressing the fully modified Spike, confirming the ability of IDLV to elicit cross-reactive long term functional immunity.
Materials and methods
Plasmids construction
A schematic representation of the plasmids, transfer vectors and coding sequences used in this study is shown in Supplementary Figure 1. All Spike sequences expressed from lentiviral transfer vectors were obtained after codon optimization and cloning into pUC57 vector (GenScript Biotech, Rijswijk, Netherlands). Spike sequences were removed from pUC57 plasmids with AgeI/SalI restriction sites and cloned into the corresponding restriction sites of pGAE-GFP self-inactivating lentiviral transfer vector plasmid by substituting the GFP coding sequence (43) (Supplementary Figure 1A). Plasmid pGAE-Spike encodes the codon optimized full-length wild type SARS-CoV-2 Spike protein open reading frame (ORF) (Wuhan-Hu-1, GenBank: NC_045512.2); plasmid pGAE-S2PF encodes the Spike ORF, stabilized by the introduction of 2 prolines (2P, K986P and V987P) and by functional mutation of RRAR into GSAS at the furin cleavage site (FCS); plasmid pGAE-S2PGC encodes the Spike ORF and contains a 21 amino acid (aa) deletion in the cytoplasmic tail (delta21) with 2P and D614G mutations; plasmid pGAE-betaS2PGC encodes the delta21 codon optimized B.1.351 (beta) Spike ORF with the 2P stabilizing mutation; plasmid pGAE-S2PFGC encodes the delta21 Spike ORF with the 2P, D614G and FCS mutations; plasmids pGAE-JR, pGAE-Luc and pGAE-GFP encode the HIV-1JR-FL gp120 envelope, the luciferase and the GFP ORFs, respectively (44). Plasmids pAdSIVD64V and pAdSIV3+ and are the Integrase defective and Integrase competent packaging vectors, respectively (43). Plasmid phCMV-VSV.G encodes the vesicular stomatitis virus envelope glycoprotein G (VSV.G), used for pseudotyping of lentiviral vector (43).
Expression plasmids encoding wild type and variants of concern (VoC) Spike ORFs used in the pseudovirus neutralization assay were previously described (45, 46) and are depicted in Supplementary Figure 1B. Briefly, plasmid pSpike-C3 encodes the wild type (Wuhan-Hu-1) codon optimized SARS-CoV-2 delta21 Spike protein open reading frame (ORF) (40); plasmids pSpike-UKC3, pSpike-SAC3 and pSpike-BRC3 encode the delta21 codon optimized B.1.1.7 (Alpha), B.1.351 (Beta) and P.1 (Gamma) Spike ORFs, respectively, while plasmid pSpike-INC3 encodes the B.1.617.2 (Delta) Spike ORF with a 19 aa deletion in the cytoplasmic tail (45, 46). For construction of plasmid, expressing the delta21 codon optimized B.1.1.529 (Omicron) BA.1, BA.2 and BA.4/5 Spike ORF VoC, a NheI/BamHI fragment of DNA was removed from pUC57-BA.1, pUC57-BA.2 and pUC57-BA.4/5 plasmids (GenScript) and inserted into the pSpike-C3 plasmid at the corresponding restriction sites, to obtain pSpike-BA1C3, pSpike-BA2C3 and pSpike-BA4C3 plasmids. The B.1.1.7 Spike utilized for these studies contains the mutations: del69-70, del144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H. The B.1.351 Spike utilized for these studies contains the mutations: L18F, D80A, D215G, del242-244, R246I, K417N, E484K, N501Y, D614G, A701V. The P.1 Spike utilized for these studies contains the mutations: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I. The B.1.617.2 Spike utilized for these studies contains the following mutations: T19R, del157-158, L452R, T478K, D614G, P681R, D950N. The BA.1 Spike utilized for these studies contains the mutations: A67V, del69/70, T95I, G142D, del143-145, N211I, del212, ins214EPE, G339D, S371L, S373P, S375F, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, D796Y, N856K, Q954H, N969K, L981F. The BA.2 Spike utilized for these studies contains the following mutations: T19I, L24S, del25/27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K. The BA.4/5 Spike utilized for these studies contains the following mutations: T19I, L24S, del25/27, del69/70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K.
Production of integrase defective lentiviral vectors for immunization
293T Lenti-X cells (Clontech, Mountain View, CA, USA) were utilized for the production of SIV-based IDLV by transient transfection, as described (34). Cells were kept in Dulbecco’s modified Eagles medium, with high glucose 4.5 g/L (Gibco, Life Technologies Italia, Monza, Italy) and were supplemented with 100 units/ml penicillin/streptomycin (Gibco) and 10% fetal calf serum (Corning, Mediatech, Manassas, VA, USA). Cells (3.5x106 cells) were plated on 10 cm Petri dishes (Corning Incorporated-Life Sciences, Oneonta, NY, USA) and transfected with each self-inactivating lentiviral transfer vector plasmid pGAE expressing the different Spike(s), RBD or HIV-1JR-FL gp120 envelope, the packaging plasmid integrase-defective pAdSIVD64V and the VSV.G pseudotyping plasmid phCMV-VSV.G, to allow entry into target cells in vitro and in vivo, with the CalPhos™ Mammalian Transfection Kit (Clontech Laboratories, Inc, Mountain View, CA, USA) by following the recommendations of the manufacture using a plasmid ratio of 1:2:1 (transfer vector: packaging plasmid: VSV.G plasmid). Forty-eight hours post transfections, the supernatants of the cultures containing the IDLV pseudotyped with different conformation of Spike (IDLV-CoV2) (IDLV-Spike, IDLV-S-2PF, IDLV-S-2PGC, IDLV-betaS-2PGC and IDLV-S-2PFGC) and IDLV-JR were collected, filtered with a 0.45 μm pore size filter (Millipore Corporation, Billerica, MA, USA) and concentrated by ultracentrifugation for 2.5 h at 65,000 × g using a 20% sucrose cushion. Vector particles were dissolved in 1× phosphate buffered saline (PBS, Gibco) and stored at −80°C. Each stock of IDLV was titered using the reverse transcriptase (RT) activity assay (34).
Production of lentiviral vectors expressing Luciferase and pseudotyped with Spike variants
Spike variants pseudotyped lentiviral vectors expressing Luciferase (LV-Luc) were generated by transient transfection of 293T Lenti-X cells as previously described (40, 45, 46). In brief, 293T Lenti-X cells (3.5x106 cells) were plated on 10 cm Petri dishes (Corning) and transfected with the lentiviral transfer vector plasmid pGAE-Luc expressing the luciferase coding sequence (44), the packaging plasmid pAd-SIV3+ and each of the pseudotyping plasmid expressing Spike protein described above (Supplementary Figure 1B) or control VSV.G (phCMV-VSV.G) utilizing the JetPrime transfection kit (Polyplus Transfection, Illkirch, France) using a plasmid ratio of 1:2:1 (transfer vector: packaging plasmid: Spike/VSV.G plasmid). Forty-eight hours post transfection, the supernatants containing the LV-Luc pseudoviruses were collected, filtered with a 0.45 μm pore size filter (Millipore) and stored in 0.5 mL aliquotes at -80°C.
Western blot
Concentrated preparations of IDLV were lysed in SDS loading buffer and resolved on SDS-PAGE (10% polyacrylamide) under reducing conditions. Nitrocellulose membrane was used to transfer gels with a Trans-Blot Turbo System (Bio-Rad Laboratories, Hercules, CA, USA). Filters were saturated with 5% nonfat dry milk dissolved in TBST (TBS with 0.1% Tween 20) for 1 hour and further incubated with anti-S2 polyclonal antibody (dilution 1:2000. Cat: 40590-T62, Sino Biological, Beijing, China), anti-S1 polyclonal antibody (dilution 1:1000. Cat: 40150-T62-COV2, Sino Biological, Beijing, China) or anti-HIV-1 SF2 p24 polyclonal Ab (dilution 1:4000. ARP-4250, NIH HIV Reagent Program, Manassas, VA, USA), followed by anti-rabbit horseradish peroxidase (HRP)-conjugated Ab (dilution 1:3000; Bio-Rad Laboratories, Hercules, CA, USA). Recombinant trimeric Spike with 2P and FCS mutations (47), RBD (45) and SIVmac239 p27 (ARP-13446, NIH HIV Reagent Program) proteins were used as positive controls, whereas preparations from unrelated LV or IDLVs were used as negative controls. WesternBright ECL detection system (Advansta, San Jose, CA, USA) was used as chemiluminescent substrate. Images were acquired and elaborated by ChemiDoc XRS+ System (Bio-Rad Laboratories, Hercules, CA, USA).
Flow cytometry
293T Lenti-X cells (3x105/well) were plated in 6-well plate and transfected with 1 µg of Spike-expressing plasmids or phCMV-VSV.G as a negative control using the CalPhos™ Mammalian Transfection Kit (Clontech). Forty-eight hours after transfection, cells were detached, counted and stained either with anti-S2 commercial antibody (Cat: 40590-T62, Sino Biological; 1:3000) followed by donkey anti-rabbit PE (Biolegend, San Diego, CA, USA; 4 µg/ml) or human monoclonal antibodies COVA2-15, COVA1-16, COVA1-18, COVA1-21 (47) (1 µg/ml) or CR3022 (Cat: ab273073, Abcam, Cambridge, UK; 5 µg/ml), followed by Goat anti-human IgG secondary AlexaFluor647 (Cat: 109-605-003, Jackson ImmunoResearch, 5 μg/ml). The expression of Spike was analyzed by flow cytometry utilizing a FACSCalibur (BD Biosciences, Milan, Italy), and the results were analyzed with Kaluza software (Beckman Coulter, Fullerton, CA, USA).
Transmission electron microscopy analysis
293T Lenti-X cells (3.5x106 cells) were transfected on 10 cm Petri dishes (Corning) to produce each IDLV-CoV2 as described above. At 48 hrs post transfection, cells were stained with anti-Spike COVA2-15 mAb (47), with the exception of IDLV producing beta Spike, which was stained with COVA1-16 mAb (47), followed by Goat Anti-Human IgG H and L (10 nm Gold) used as a secondary Ab (Abcam, Cambridge, UK). After staining, cells were fixed with 2.5% glutaraldehyde in cacodylate buffer 0.1 M, pH 7.2. Fixed cells were washed and post-fixed in 1% OsO4 using the same glutaraldehyde/cacodylate buffer. Fixed specimens were dehydrated by using a graded series of ethanol solutions and then embedded using an Agar 100 resin (Agar Scientific, Essex, UK). Ultrathin sections were placed on 200-mesh copper grids and then stained with lead citrate and uranyl acetate. Sections were analysed by using a Philips 208S transmission electron microscopy (TEM) at 100 kV.
Confocal laser scanner microscopy
293T Lenti-X cells (2.5x104/well) were plated onto L-polylysine (Sigma) treated 12-mm cover glasses inserted in 24-well microplates and then transduced with 5 MOI of each LV expressing Spikes used for immunization. At 48 hours post-transduction, the cells were washed and stained with anti-Spike COVA2-15 or COVA1-16 (47) mAbs followed by AlexaFluor 488 Goat anti-human IgG as a secondary Ab (Jackson ImmunoResearch, 0.4 µg/sample). The coverslips were extensively rinsed, fixed with cold methanol and then placed on the microscope slides using Vectashield antifade mounting medium, containing DAPI (Vector Labs, Burlingame, CA, USA).
Observations by confocal laser scanner mycroscopy (CLSM) were performed using a Zeiss LSM980 apparatus (Zeiss, Oberkochen, Germany), fitted with a Plan-Apochromat 63x/1.4 NA using the appropriate spectral laser lines. Acquisition and processing of images was performed by using Adobe Photoshop CS5 software programs (Adobe Systems, San Jose, CA, USA) and Zen Blue edition 3.3 (Zeiss). Cells which were stained with the secondary antibody were used to set up parameters of acquisition. Several fields (including >200 cells) were evaluated for each labeling condition, and shown are representative results.
Mouse immunization protocol
BALB/c mice, obtained from Charles River (Charles River, Calco, Como, Italy), were housed in the animal facility at the Istituto Superiore di Sanità (ISS, Rome, Italy) under specific pathogen-free conditions. All procedures have been performed in accordance with Italian legislation and European Union guidelines for animal care. All studies have been reviewed by the Service for Animal Welfare at ISS and authorized by the Italian Ministry of Healthy (Authorization n. 731/2020-PR, 21/7/20, prot. D9997.107). Mice (six animals per group) were immunized once intramuscularly (i.m.) with escalating doses (1.56x106 - 6.25x106 - 25x106 RT units/mice) of IDLV-CoV2 or Spike protein (10 and 1 µg/mouse) adjuvanted with Addavax (In vivogen Europe). Naïve and mice injected with the highest dose of IDLV-JR (mock), expressing the HIV-1JR-FL gp120 envelope (32), were used as negative controls. Retro orbital sampling of blood was carried out prior to immunization and at monthly intervals with glass Pasteur pipettes and sera were collected and stored at -80°C. Six months after the immunization, all mice were sacrificed and spleen harvested and processed for the analysis of T cell responses, as previously described (32). Briefly, single-cell suspensions of splenocytes were obtained after mechanical disruption of spleens in the presence of 3 mL of ACK followed by passage through cell strainers (Corning, Merk Life Science S.r.l., Italy). Splenocytes were then washed using RPMI 1640 medium supplemented with 1% penicillin/streptomycin, 2 mM L-glutamine, 10% FBS, and 50 mM 2-mercaptoethanol (complete medium). Splenocytes were centrifuged at 1500 rpm for 10 min at 4°C and then resuspended in complete medium, counted and stored in liquid Nitrogen.
ELISA
SARS-CoV-2 recombinant RBD (rRBD) protein produced in HEK293T cells (45) was used for coating 96 well plates (Greiner bio-one, Frickenhausen, Germany) using 0.1 µg/well of rRBD protein overnight at 4°C. Plates were washed and blocking was performed for 2 h by using 1% BSA (Sigma Chemicals) in 200 µL of PBS. Plasma from individual mice were added to wells in duplicate of serial dilutions and further incubated for 2 h at room temperature. After washing, the plates were incubated with biotin-conjugated goat anti-mouse IgG (Southern Biotech, Birmingham, AL, USA) for 2 h at room temperature. The plates were washed and streptavidin-conjugated horse radish peroxidase (HRP) (AnaSpec, Fremont, CA, USA) was added to the plates for 30 min at room temperature. The reaction was incubated with 3.3,5.5-tetramethylbenzidine substrate (SurModics BioFX, Edina, MN, USA) and then blocked using of H2SO4 1 M (50 µL). Endpoint titers were calculated as the reciprocal of the highest dilution with an absorbance value equal at least three times the values from naïve mice. Results were expressed as mean titer ± standard error of the mean (SEM) for each group.
Pseudovirus titration and neutralization assay
Preparations of LV-Luc pseudoviruses (LV-Luc/Spike-C3, LV-Luc/Spike-SAC3, LV-Luc/Spike-UKC3, LV-Luc/SpikeBRC3, LV-Luc/SpikeINC3, LV-Luc/SpikeOMC3 (BA.1, BA.2, BA.4/5) and LV-Luc/VSV.G) were titered in Vero E6 cells (Cercopithecus aethiops derived epithelial kidney, ATCC C1008). Cells (2.2x104 cells/well) were plated in 96-well plates (Viewplate, PerkinElmer). After forty-eight hours, luciferase expression was measured with a Varioskan luminometer (TermoFisher) by using the britelite plus Reporter Gene Assay System (PerkinElmer). In the neutralization assay were used dilutions providing 2x105 relative light units (RLU). In brief, serum serial 2-fold dilutions starting from 1:80 were incubated in duplicate with the LV-Luc pseudotypes at 37°C for 30 min in 96-deep well plates (Resnova, Roma, Italy). The mixture was then added to Vero E6 cells seeded in a 96-well Isoplate (Perkin Elmer, Groningen, Netherlands) at a density of 2.2x104 cells/well. Cell only and virus -only controls were included. After forty-eight hours, luciferase expression was measured as above by using the britelite plus Reporter Gene Assay System (Perkin Elmer). RLU numbers were transformed into percentage neutralization values, and relative to virus-only controls. Results were expressed as the inhibitory concentration (ID) 50, which corresponds to the dilution of serum providing 50% inhibition of the infection (corresponding to neutralization), compared to the virus-only control wells. ID50 was calculated with a linear interpolation method (40).
SARS-CoV-2 microneutralization assay
The SARS-CoV-2 isolate (GenBank: MT066156.1; Cat: NR-52284; SARS-Related Coronavirus 2, Isolate Italy-INMI1; BEI Resources, Manassas, VA, USA) was propagated by inoculation of 70% confluent Vero E6 cells in 175 cm2 cell culture flasks. The cells were kept in Dulbecco’s modified Eagles medium, high glucose 4.5 g/L (Gibco) supplemented with 2% fetal calf serum (Corning), 100 units/ml penicillin/streptomycin (Gibco), 1 mM sodium pyruvate (Gibco), and 1x non-essential amino acids (Gibco) (48). Briefly, cells were maintained at 37°C and supernatant collected at 72 hrs post inoculation, when a strong cytopathic effect (CPE) was observed. Supernatants were collected, clarified to remove cellular debris, aliquoted, and stored at -80°C. The tissue culture infectious dose 50 (TCID50) was determined on flat-bottom 96‐well culture plates of Vero E6 cells (2.2x103 cells/well) by end-point titration of serial 1 log dilutions (from 1 log to 11 log) of viral stocks. Cells were further incubated for 5 days and monitored daily for CPE. TCID50/ml of the SARS-CoV-2 was estimated as 7.5x106 TCID50/ml employing the Spearman-Karber method (49). Biosafety level 3 facilities were used for all viral manipulations.
The virus neutralization (VN) assay was executed as reported (50) using heat-inactivated samples (30 min at 56°C). Sera were mixed with an equal volume of 100 TCID50 SARS-CoV-2 and incubated for 1 hr at room temperature. A 100 µL of virus–serum mixture was subsequently added to 96-well plates containing a Vero E6 cell monolayer. After 4 days of incubation at 37°C and 5% CO2 in humidified atmosphere, the plates were inspected by an inverted optical microscope for presence/absence of CPE. The highest serum dilution that protected from CPE more than 50% of the cells was used to calculate the VN titer, which was expressed as the reciprocal of the highest serum dilution protecting from CPE.
IFNγ/IL-5 FluoroSpot assay
The assay was performed using Mabtech reagents and protocol (FluoroSpot Plus, Mabtech AB, Sweden). Briefly, 96-well plates were coated with anti-IFNγ and anti-IL-5 antibodies and then blocked with complete medium. Splenocytes were seeded at a density of 3x105/well and stimulated overnight with 1 µg/mL of SARS-CoV-2 Wuhan Spike peptide pool of 15-mer sequences with 11 amino acids overlap (PepTivator® SARS-CoV-2 Prot_S complete, Miltenyi Biotec, Bologna, Italy), as specific stimulation. Positive control included Concanavalin A (5 µg/mL, Sigma Chemicals), while the complete medium was utilized as a negative control. Wells were washed and incubated with anti-IFNγ and anti-IL-5 detection antibodies. After washes, wells were incubated with fluorophore-conjugates for 60 min and after washes a fluorescence enhancer was added. Spot Forming Cells (SFC) were counted using a FluoroSpot reader (AID iSpot, AID GmbH, Strassberg, Germany) and results were expressed as SFC/106 cells. The number of SFC counted in the wells treated with the medium (background) was subtracted from the number of SFC counted in the wells treated with the specific peptides. Samples were recorded positive when the number was equivalent to at least 50 specific SFC/106 cells and was two-fold higher than the background values.
Phylogenetic analysis
A total of 59 SARS-CoV-2 sequences were used for this study, 51 of which were downloaded from the GISAID database (51) and 8 were SARS-CoV-2 Spike protein sequences above described (Supplementary Table 1). The sequences were cut respect to the Spike protein positions of the Wuhan-Hu-1 Accession Number: NC_045512. The translation from nucleotide sequences to protein sequences was performed through Expansy tool (52). The sequence alignments were done using MAFFT v7 (53) with the Galaxy platform (54) and edited manually by utilizing BioEdit v. 7.2.6.1 (55). The maximum likelihood phylogenetic trees were estimated with the software PhyML version 3.0 (56). The statistical significance in the tree has been evaluated by using the Fast likelihood-based method, aLRT-SH like branch support. The Smart Model Selection software (57) was used to find the best-fit substitution models for protein alignments. In addition to the spike protein, three further protein alignments corresponding to the S1, S2, RBD portions were generated.
Genetic distance analyses
SARS-CoV-2 protein sequences of the S1, S2, RBD alignments were grouped according to the variant to which they belong and their genetic distances were calculated. The mean genetic distances between groups and the standard errors of the means were calculated using the MEGA v. 6 program by the bootstrap method with 1000 replicates (58). The amino acid distances were evaluated with the Equal Input Model (59).
Statistical analysis
Data were prepared using GraphPad Prism 9.4.1 (GraphPad Software Inc., San Diego CA, USA) and were expressed as the mean ± standard error of the mean (SEM) or median. Data were analyzed by ANOVA, Wilcoxon matched-pairs signed rank test, or Mann-Whitney to compare three or more groups. Pvalues < 0.05 were used as the threshold for statistical significance. The correlation between neutralization assays was evaluated by Spearman and Pearson correlation analyses.
Results
SARS-CoV-2 Spike immunogen design
In previous work we and other have shown that truncation of 13 to 21 amino acids (aa) at the cytoplasmic tail led to efficient incorporation of Spike (S) protein on lentiviral vectors (LV) (40, 60) and that functional mutation of FCS led to incorporation of Spike which was uncleaved at the S1/S2 junction (61, 62). We hypothesized that presence of different Spike conformations on lentiviral particles impacted on ensuing immune response after immunization, similarly to what has been shown with Ad26 vector (63). For immunogenicity studies in the mouse model, we engineered a series of lentiviral transfer vectors (pGAE) (44) expressing different membrane-tethered configurations derived from Wuhan-Hu-1 SARS-CoV-2 Spike protein coding sequence (GenBank: NC_045512.2) (Table 1 and Supplementary Figure 1A), including native full-length wild-type Spike (aa 1-1273, pGAE-Spike), full-length Spike with mutated FCS (RRAR to GSAS) and 2P substitutions (pGAE-S2PF), delta21 wild-type Spike with D614G mutation and 2P substitutions (aa 1-1252, pGAE-S2PGC) and delta21 wild-type Spike with D614G mutation, 2P substitutions and mutated FCS (pGAE-S2PFGC). The Beta (B.1.351) Spike sequence with delta21 and 2P substitutions (pGAE-betaS2PGC) was also generated to assess cross-neutralizing responses compared to the related Wuhan-based vaccine. The schematic representation of the plasmids and the characteristics of the vaccine candidates used in this study are shown in Table 1 and in Supplementary Figure 1A.
Expression of different Spike proteins was evaluated by using flow cytometry on 293T Lenti-X cells transfected with each transfer vectors using a commercial rabbit anti-Spike S2 polyclonal Ab (pAb), three human neutralizing monoclonal Abs (mAbs) (COVA2-15, COVA1-16 and COVA1-18) that recognize different epitopes of RBD (47), one non-RBD-binding neutralizing mAb (COVA1-21) and the SARS-CoV-1 neutralizing mAb CR3022 (64), which recognizes a conserved weakly neutralizing cryptic epitope in SARS-CoV-2 Spike, accessible in the open conformation of the protein with at least 2 RBD in the up-conformation, not overlapping with the ACE2 binding site within SARS-CoV-2 RBD (65–67). All membrane-bound Spikes were recognized by anti-S2 antibody, indicating their correct expression on the membrane of transfected cells (Supplementary Figure 2). The human mAbs COVA2-15, COVA1-16, COVA1-18 and COVA1-21 recognized all Wuhan-Hu-1 derived Spikes, whereas the beta variant was not recognized by COVA1-18 mAb, and poorly recognized by COVA2-15 and COVA1-21 mAbs. Interestingly, mAb CR3022 was overtly reactive only against Spike with mutated FCS, suggesting that this mAb recognizes an epitope that is accessible only in this Spike configuration.
IDLV-CoV2 are decorated with membrane tethered Spike proteins
IDLV expressing the different conformations of Spike (IDLV-CoV2) were produced to evaluate incorporation on pseudovirions. Transmission electron microscopy (TEM) images of 293T Lenti-X cells producing the IDLV-CoV2 showed presence of S on the released IDLVs and plasma membranes of the producing cells after staining with COVA2-15 or COVA1-16 mAbs, the latter used to stain cells producing IDLV-betaS-2PGC (Figure 1A). To further assess incorporation of the different configurations of Spike on pseudovirus, recovered and concentrated preparations of IDLV-CoV2 to be used for immunization were normalized for amount of p27 Gag and analyzed by Western blot (WB) using anti-S2 (S2 fragment) and anti-p27 Gag polyclonal antibodies (Figure 1B). Gag protein was detected similarly in all IDLV preparations and bands corresponding to the Spike protein were detected in all IDLV-CoV2 concentrated preparations, except for IDLV-GFP and IDLV-JR, encoding the codon optimized HIV-1JR-FL gp120 (32), as expected. In particular, the highest amount of Spike was incorporated in the IDLVs delivering the delta21 C-truncated version of the protein (IDLV-S-2PGC, IDLV-betaS-2PGC and IDLV-S-2PFGC), compared to IDLV-S-wt and IDLV-S-2PF. Since the removal of the FCS results in less S1 shedding of the S protein, IDLV-S-2PF and IDLV-S-2PFGC showed mainly presence of the stabilized uncleaved full-length S protein, while in both IDLV-S-2PGC (Wuhan-Hu-1 and Beta) and IDLV-S-wt the majority of the protein was cleaved at the FCS and showing on WB the S2 portion with the anti-S2 polyclonal Ab (Figure 1B). Interestingly, the presence of S2’ fragment in IDLV-CoV2 preparations incorporating high levels of S might be due to Spike engagement of low levels ACE2 expression on 293T cells (Supplementary Figure 3) during IDLV production, which primes S2′ site cleavage (68). These results demonstrate that different configurations of Spike are incorporated differently and according to their design in the vector particles used to immunize mice. To appreciate the membrane location of Spike expressed by the vector, 293T Lenti-X cells were transduced with LV pseudotyped with VSV.G protein and delivering the different Spikes, stained with anti-RBD human COVA2-15 or COVA1-16 mAbs and examined by confocal microscopy (CLSM) (Figure 2). The membrane expression of Spike was evident in the cells transduced with any vector, confirming their ability to express membrane bound antigens.
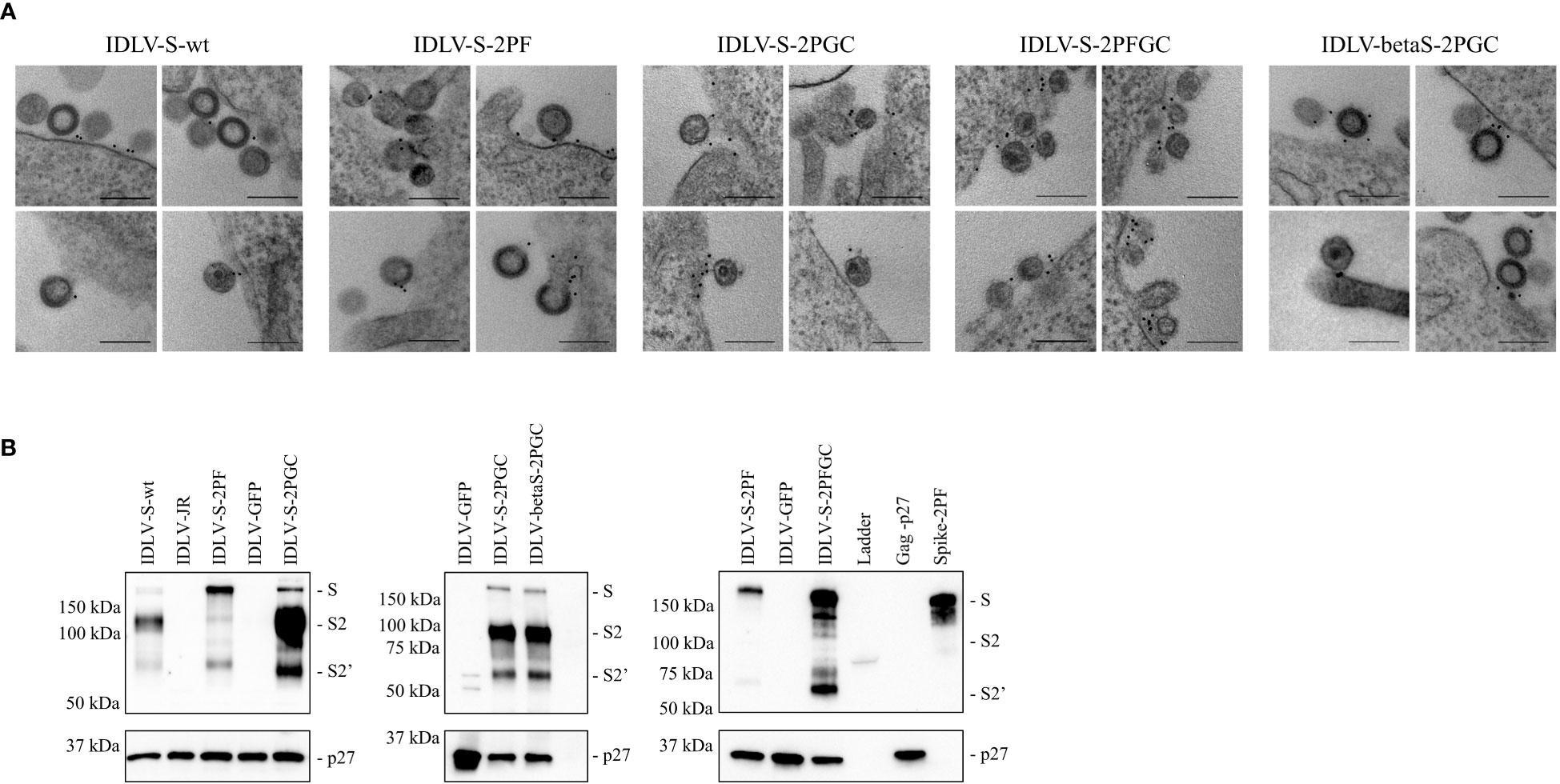
Figure 1 (A) IDLVs are pseudotyped with membrane-tethered Spike proteins. 293T Lenti-X cells producing the different IDLV-CoV2 were probed with anti-RBD COVA2-15 or COVA1-16 human mAbs and observed by TEM. Four representative images are shown for each sample. Bars, 0.2 μm. Shown are results from one representative of n = 2 experiments. (B) Spike incorporation on IDLV-CoV2 particles used for immunization. Western blot of lysates from concentrated preparation of IDLV-CoV2 pseudotyped with the different Spike conformations. IDLV-GFP and IDLV-JR are included as mock controls and SIV-Gag p27 protein (50 ng) as positive control. Filters were probed with polyclonal anti-S2 and anti-p27 Gag Abs. Molecular weights are indicated on the left. Bands corresponding to S, S2 and S2’ representing full-length and S2 or S2’ portion, are indicated. Blots derive from the same experiment and were processed in parallel. Shown are results from one representative of n = 3 experiments.
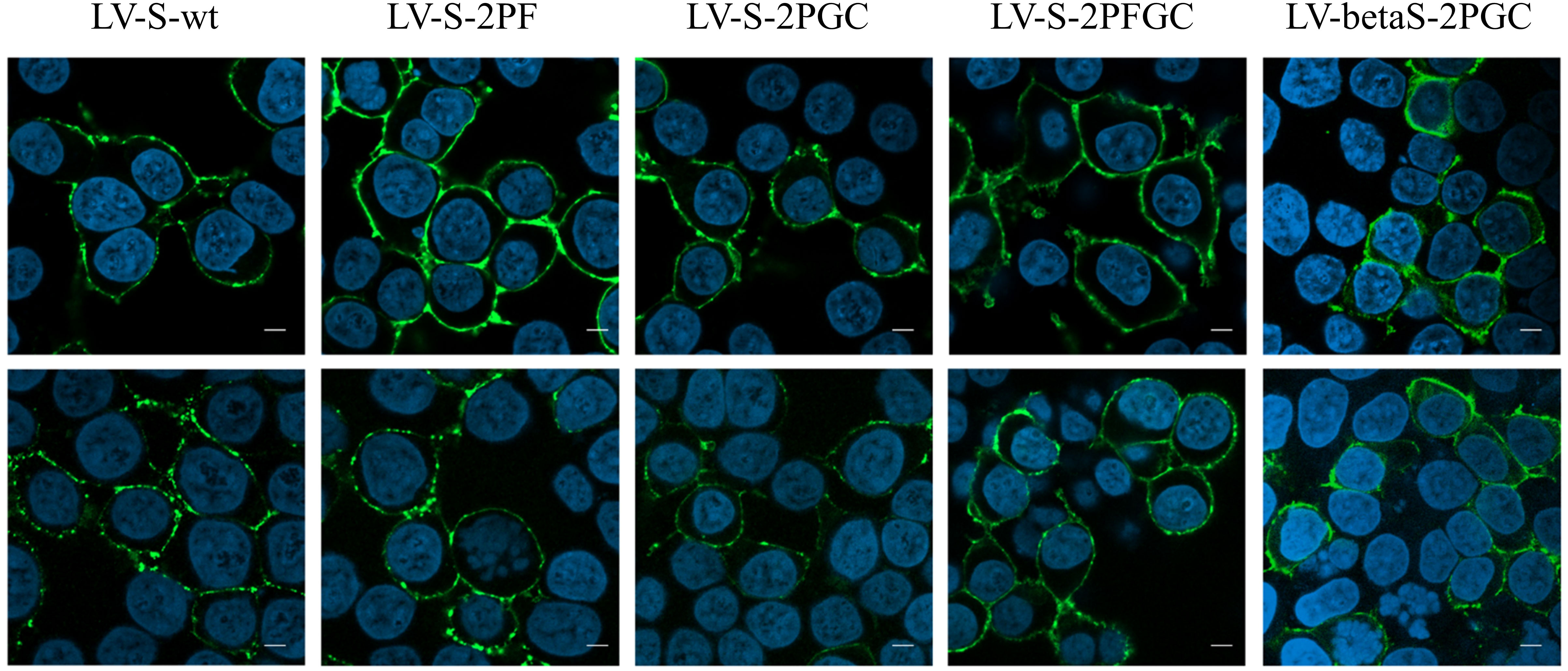
Figure 2 Spike expression from lentiviral vectors. Analysis by CLSM of 293T Lenti-X cells transduced with the indicated lentiviral vectors expressing the different Spike conformations. Cells were stained with anti-RBD COVA2-15 or COVA1-16 human mAbs followed by anti-human IgG Alexa Fluor 488 as secondary Ab (green). Nuclei were stained in blue with DAPI. Two images representing single central optical sections are shown for each sample. Shown are results from one representative of n = 3 experiments. Scale bar is 10 μm.
A single immunization of IDLV-CoV2 elicits strong and persistent antibody responses
To evaluate the magnitude, quality and persistence of anti-Spike immunity, BALB/c mice were injected once intramuscularly (i.m.) with escalating doses of IDLV-CoV2 vaccines (1.56x106, 6.25x106 and 25x106 RT Units/mouse, hereafter indicated as Low Dose-LD, Medium Dose-MD and High Dose-HD) and compared to mice immunized with a subunit vaccine based on the purified S-2PF trimeric protein, at two different doses (10 and 1 µg/mouse) together with Addavax as an adjuvant. The kinetics of anti-RBD IgG titers in serum was evaluated by ELISA at 0, 1, 2, 3 and 6 months after immunization, using the Wuhan RBD monomer as coating protein (45). The kinetics of serum Ab response of each group and at each dose are depicted in Figures 3A, B. All vaccine candidates induced anti-RBD IgG Abs, according to the injected dose, starting from 1 month (the first point post-immunization evaluated) and increasing over time, mostly reaching a plateau at 3 months. All groups maintained the response up to 24 weeks, the experimental endpoint, at different levels. Overall, IDLV-S-wt induced the lowest and IDLV-S-2PFGC the highest anti-RBD Ab response at all injected doses (ANOVA, p<0.05). IDLV-S-2PF, IDLV-S-2PGC and the subunit vaccine induced similar Ab responses (ANOVA, p>0.05). IDLV-S-wt showed a peculiar kinetics of Ab response, increasing over time up to 6 months, reaching levels of Abs similar to the other vaccine candidates (Figures 3A, B).
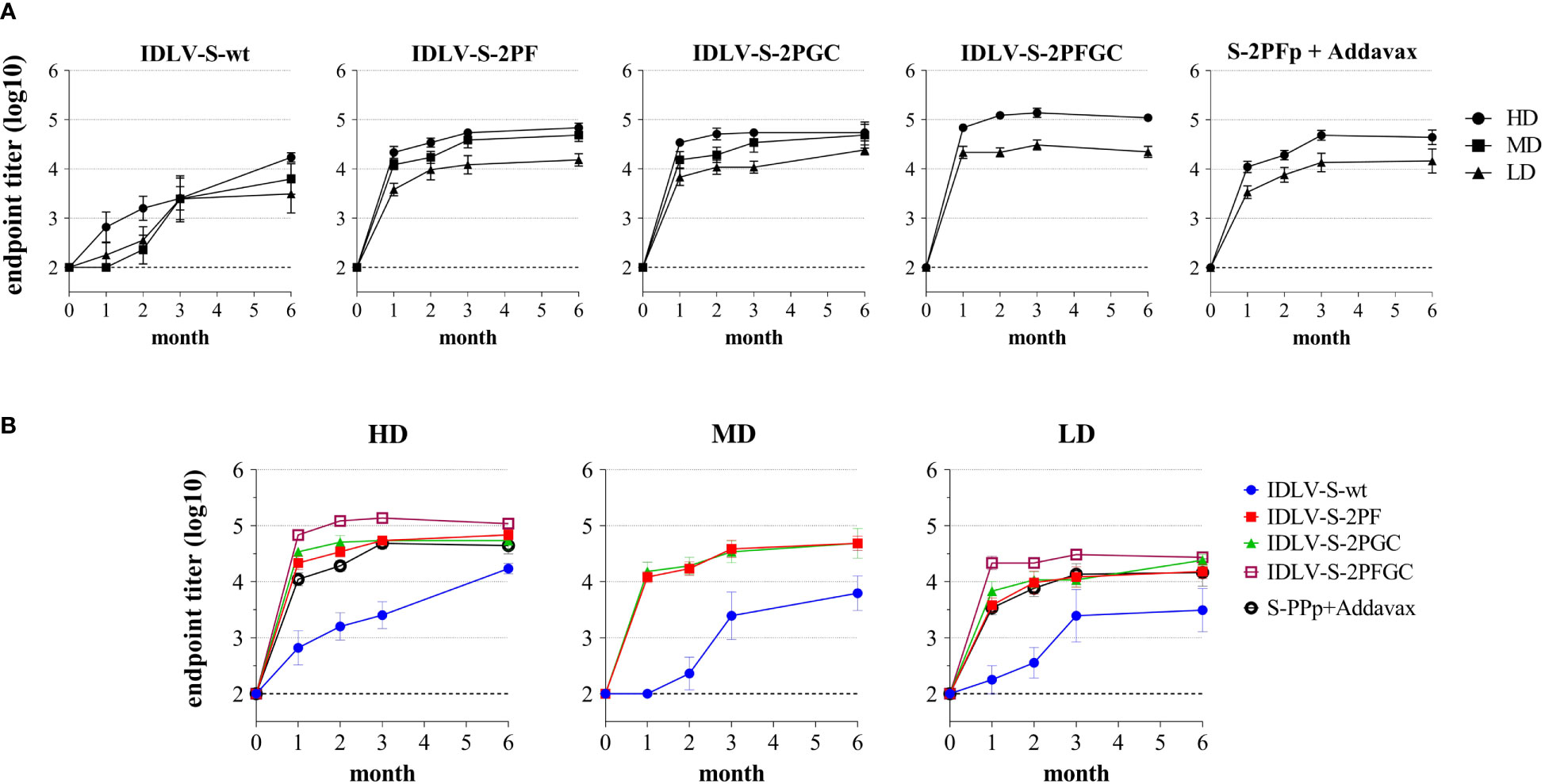
Figure 3 Kinetics of anti-RBD binding antibodies in immunized mice. BALB/c mice (N=5-6 animals per group) were vaccinated once with escalating doses (High dose, HD: 25x106; Medium dose, MD: 6.25x106; Low Dose, LD: 1.56x106 RT units/mouse) of SIV-based IDLVs expressing Spike or HD and LD of the purified Spike protein (10 and 1 µg/mouse) adjuvanted with Addavax. Anti-RBD IgG antibodies were evaluated at different time points (month) from immunization in sera of mice immunized with the indicated vaccines. Results are expressed as log10 mean endpoint titer ± SEM. The dotted line indicates the assay cut off (minimum serum dilution tested 1:100). (A) Each graph shows the results from mice of the same vaccine group at HD, MD and LD. (B) Each graph shows different vaccine groups at the same dose.
Level, kinetics and cross-neutralization of neutralizing antibodies are dependent on the immunogen configuration
Analysis of neutralizing Abs (nAbs) in serum of immunized mice was performed utilizing the pseudovirus assay, which is based on LV expressing luciferase (LV-Luc) pseudotyped with the truncated form of Spike derived from Wuhan-Hu-1 and VoC, as we previously described (40, 45, 46). A virus neutralization (VN) assay using the original infectious SARS-CoV-2 (isolate SARS-CoV-2/human/ITA/INMI1/2020) was initially performed using 31 serum samples to compare the assays and correlate the titers (ID50). The neutralization titers calculated using the two different assays correlated well with each other, as indicated by Spearmen and Pearson r values (Supplementary Figure 4). We observed a statistically significant difference between ID50 values, being the ID50 calculated using the pseudovirus neutralization assay higher than ID50 derived from infectious virus based test (median ID50: 1589 and 1280 in pseudovirus and infectious virus assays, respectively) probably due to higher sensitivity of the Luc-based assay.
The pseudovirus-based assay was used for the evaluation of presence and magnitude of nAbs over time in all immunized mice (Figure 4). All IDLV-CoV2 vaccines induced different levels of nAbs according to the injected dose within the same vaccine formulation (Figure 4A). Interestingly, the level of nAbs increased over time up to 6 months, especially in the HD treated animals, including mice immunized with the subunit vaccine at HD. Overall, IDLV-S-2PFGC induced the highest nAb response, while IDLV-S-wt induced the lowest response compared to all the other IDLV groups and at any dose, particularly evident at early time points (Figure 4B), thus in part paralleling the results obtained after evaluation of the binding Abs (Figure 3), but with a more evident difference among groups. In particular, IDLV-S-2PF and IDLV-S-2PGC consistently induced a higher and more rapid nAb response compared to that in mice immunized with S-2PFp+Addavax (ANOVA, p<0.05). The nAb response was barely detectable at 6 months in the sera of mice immunized with the subunit vaccine at LD.
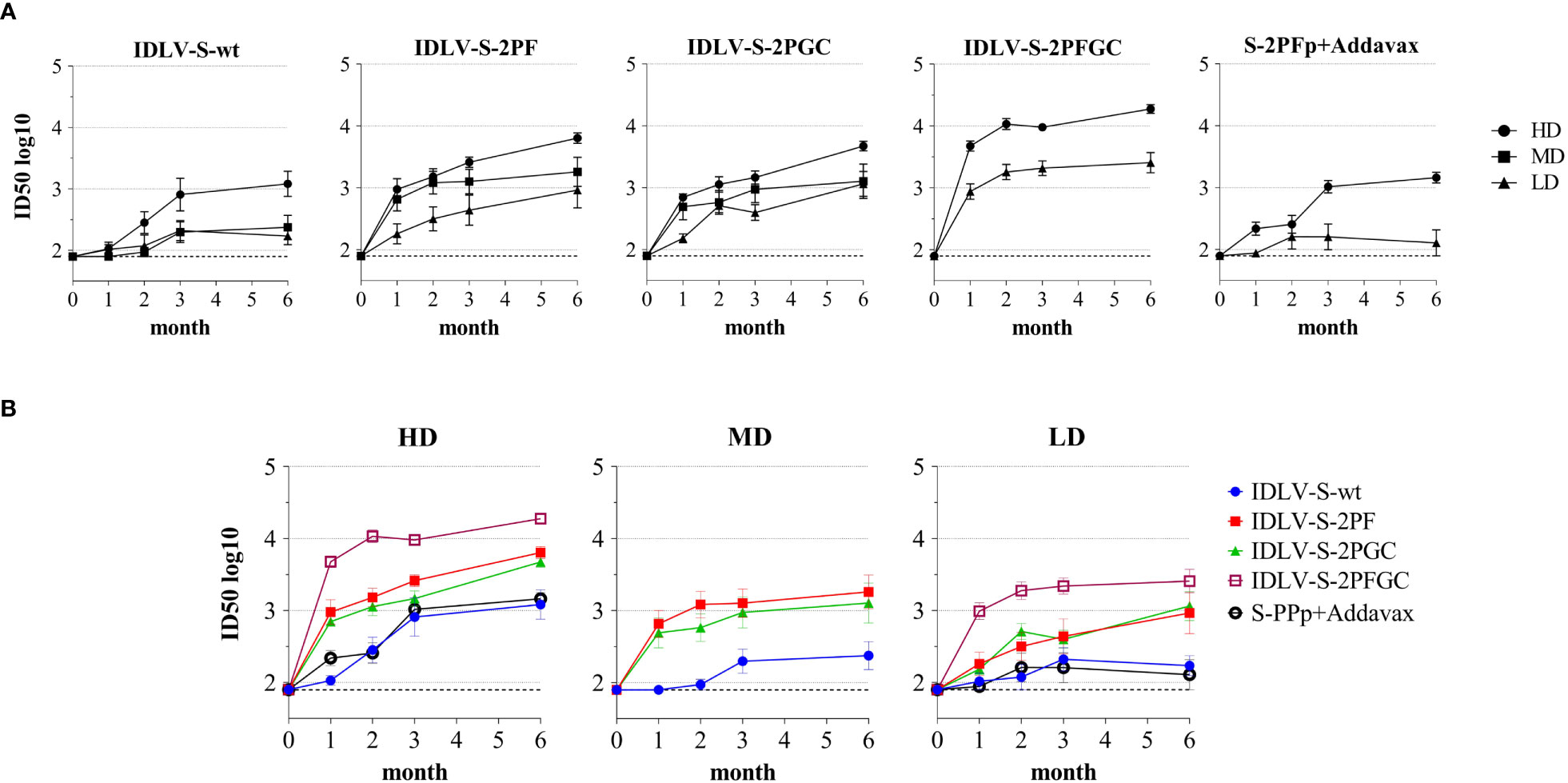
Figure 4 Kinetics of neutralizing antibodies in immunized mice. Anti-Wuhan neutralizing antibodies (nAbs) were evaluated at different time points (month) from immunization in sera of mice immunized with the indicated vaccines. Results are expressed as mean ID50 ± SEM with N=5-6 animals per group. HD, MD and LD: high, medium and low dose of vaccine, respectively. The dotted line indicates the assay cut off (minimum serum dilution tested 1:80 dilution). (A) Each graph shows results from mice of the same vaccine group at HD, MD and LD. (B) Each graph shows different vaccine groups at the same dose.
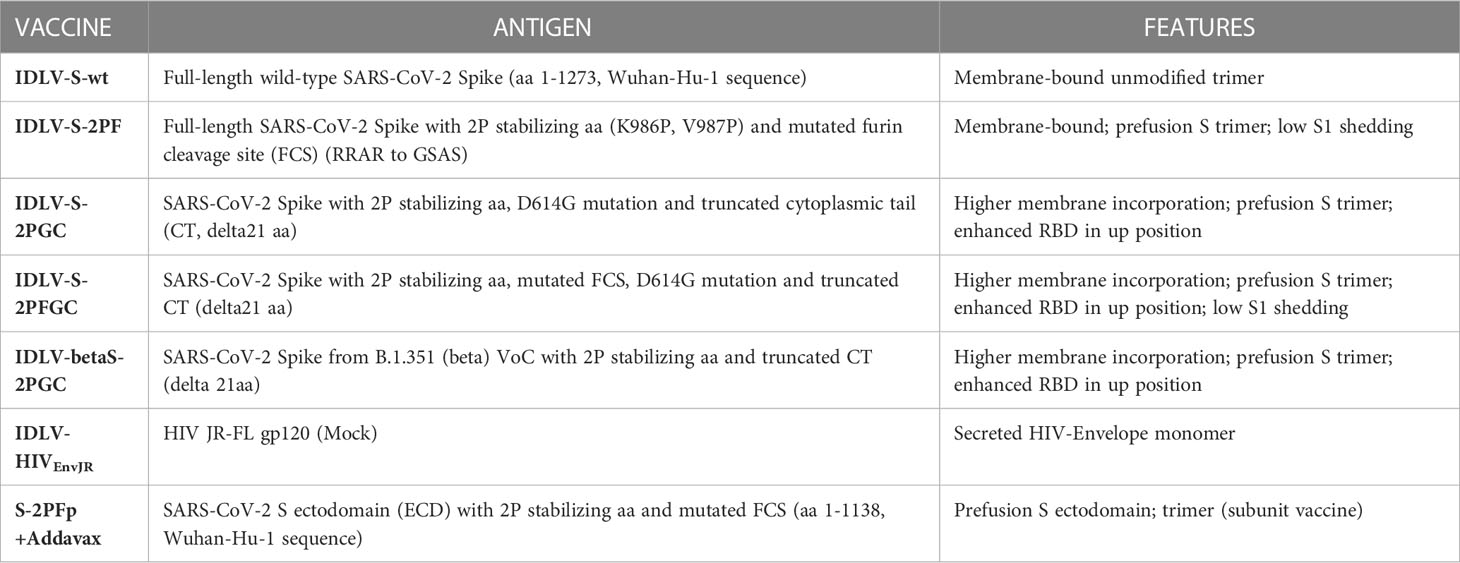
IDLV-betaS-2PGC vaccine, expressing the Beta Spike, was evaluated in the immunization protocol in order to assess and compare the cross-neutralizing responses using a Spike from the VoC able to better escape from the Wuhan-Hu-1 S vaccine induced nAbs, at the time when this study was conducted (69). A comparison of the anti-Wuhan Ab and nAb responses after immunization with IDLV-S-2PGC and the corresponding Beta variant IDLV-betaS-2PGC is shown in Supplementary Figure 5. The levels of both binding and nAbs were very similar in both vaccine groups at any dose and time point tested (Wilcoxon matched-pairs signed rank test, p>0.05).
Serum samples from all IDLV-CoV2 HD vaccinated mice, including IDLV-betaS-2PGC, were assayed for cross-neutralization ability using pseudoviruses pseudotyped with Spike from Alpha, Beta, Gamma, Delta and Omicron BA.1 VoC. Due to the small amount of mouse serum collected at each time point, 4 and 5 month-sera were dedicated to this analysis. As shown in Figure 5, the pattern of cross-neutralization is similar to that observed in human subjects vaccinated with Wuhan-based vaccines (46, 70, 71). Overall, compared to the Wuhan strain, homologous to the vaccine sequence, ID50 values of Alpha were >Gamma>Delta>Beta>Omicron. However, pattern and magnitude of neutralizing responses were different according to the Spike configuration delivered by each IDLV. In particular, we observed a high fold reduction of the ID50 against Delta compared to the vaccine strain in all vaccinated mice (Table 2), except for the IDLV-S-2PFGC group (2.41 fold reduction), where the vaccine induced nAbs were efficiently able to block the Delta pseudoviruses on target cells (Figure 5A and Table 2). Overall, IDLV-2PFGC, combining all modifications, outperformed the other candidates in terms of nAbs magnitude at early and later time points and breadth of neutralization against all VoC, including the Omicron BA.1 variant, although the latter at lower levels (Figure 5B). The IDLV-betaS-2PGC delivering the Beta Spike induced a better neutralization against Beta and Gamma pseudoviruses compared to the corresponding parental Wuhan-Hu-1 S immunogen (IDLV-S-2PGC), but neutralization against the Omicron BA.1 VoC was substantially reduced (Figure 5 and Table 2). Sera from IDLV-S-2PFGC immunized mice were also evaluated for neutralization of pseudoviruses enveloped with Spike protein from additional circulating Omicron BA.2 and BA.4/5 variants. As shown in Figure 5C, both at 4 and 24 weeks after immunization, BA.2 and BA.4/5 pseudoviruses were neutralized to levels lower than the parental BA.1.
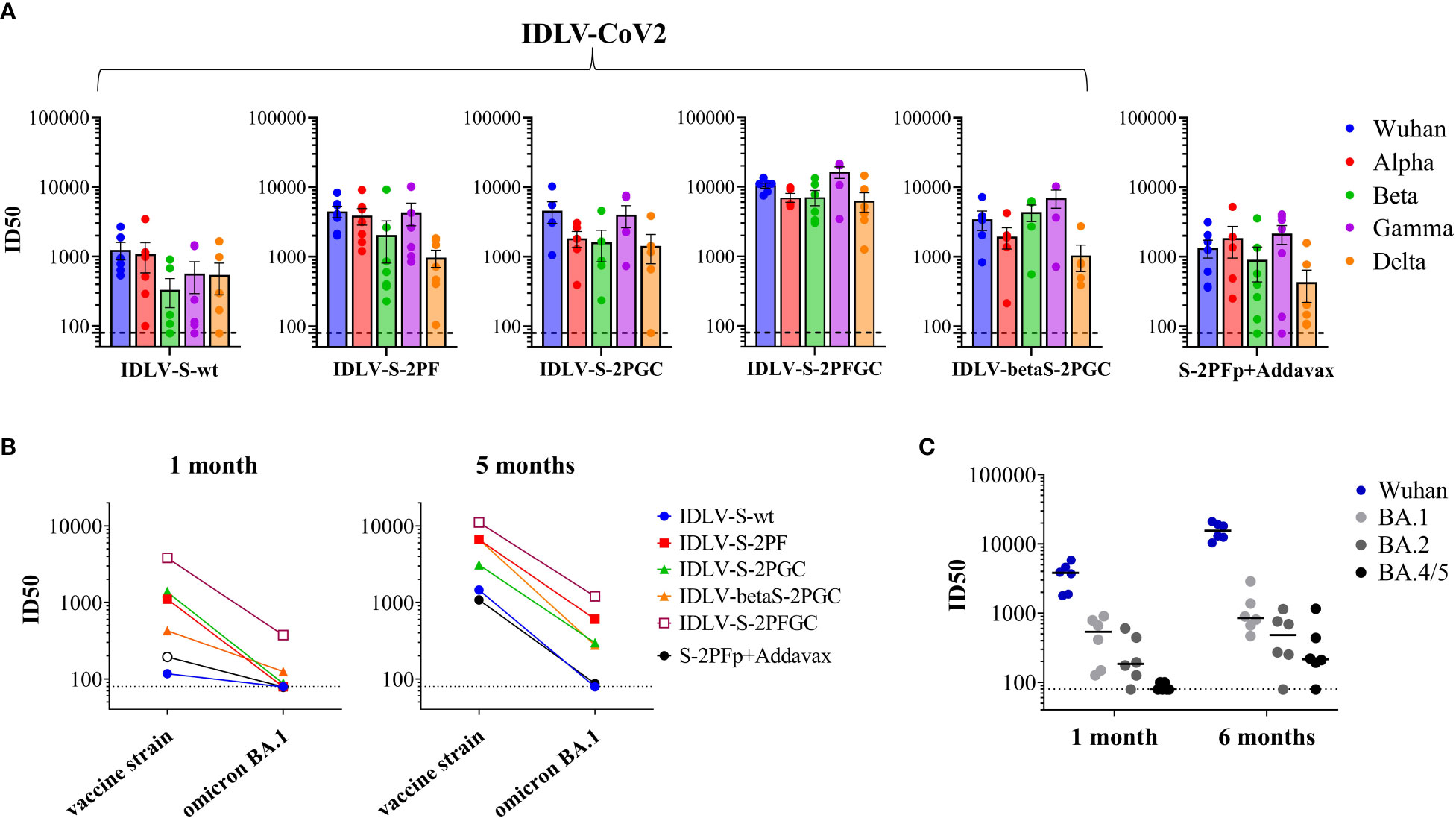
Figure 5 Cross-neutralization with VoC. (A) NAbs against each VoC were measured in serum samples collected at 4-5 months after immunization with HD vaccines, using LV-Luc pseudotyped with Spike from the indicated VoCs. Results are expressed as mean ID50 ± SEM. Each dot represents a single mouse. (B) Serum samples from early and late time points after immunization with HD vaccines were analyzed for cross-neutralization against the Omicron BA.1 variant. Results are expressed as median ID50. (C) Sera from IDLV-S-2PFGC were assayed for cross-neutralization against Omicron VoC at early and late time points after vaccination. Each dot represents a single mouse. The black line indicates the median ID50. The dotted lines indicate the assay cut off (minimum serum dilution tested 1:80 dilution).
Th-type T cell immunity persists up to six months from the immunization in IDLV-immunized mice
Since the S-specific T cell response may contribute to the vaccine efficacy, at least in terms of control of the clinical manifestation, IFNγ/IL-5 FluoroSpot was performed on splenocytes at 6 months after immunization in the HD groups using pools of peptides covering the entire Wuhan-Spike protein. As shown in Figure 6, Spike-specific IFNγ producing T cells were induced in all mice vaccinated with any IDLV, with IDLV-S-2PFGC showing the highest response compared to the other groups (Mann-Withney test p<0.001 vs S-2PF protein immunized animals; p<0.05 vs all other groups). Mice immunized with the protein subunit vaccine showed instead IL-5-producing T cells, as expected, since Addavax is a Th2-type adjuvant, which favours the induction of IL-5 (Figure 6) (72). Conversely, none of IDLV-vaccinated mice showed IL-5-producing T cells when stimulated with the Spike peptide pool, confirming the Th1-type profile of T cells induced by IDLV vaccination.
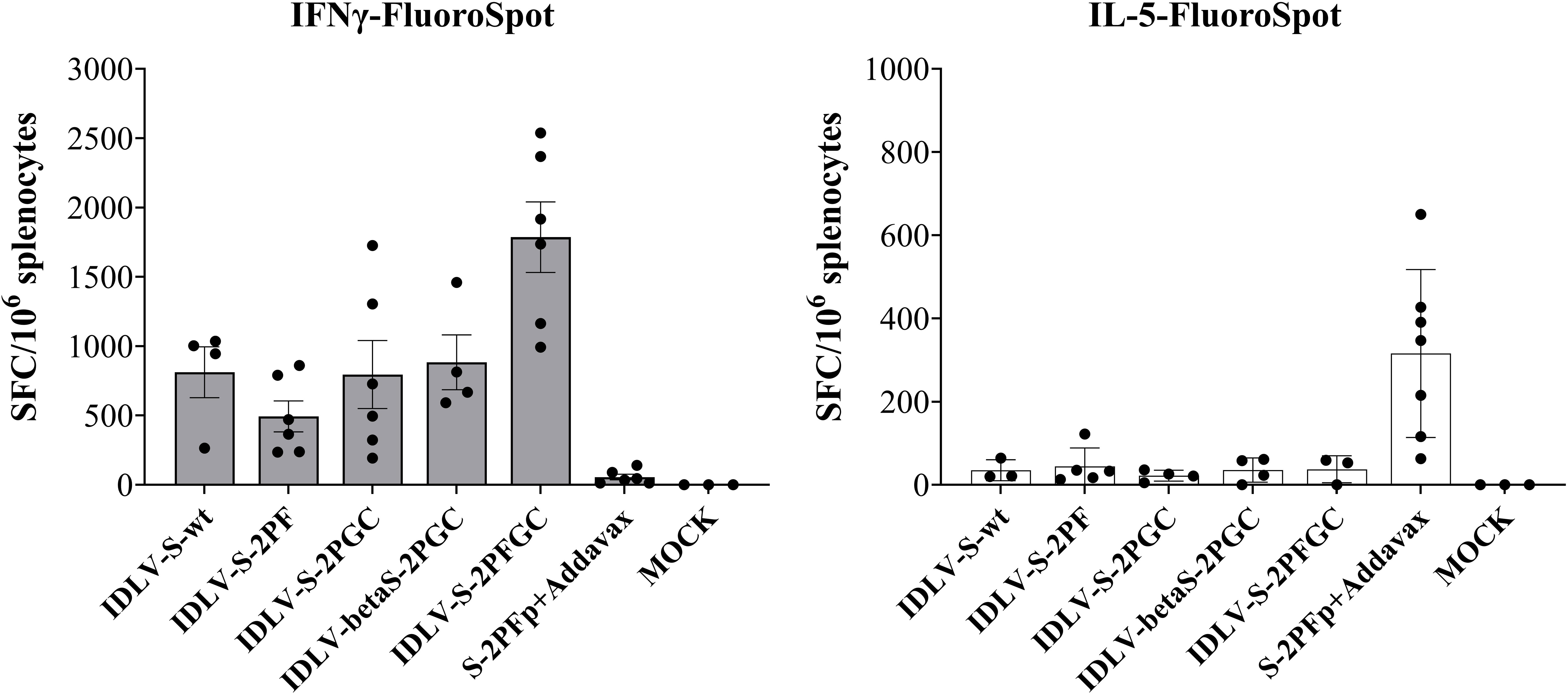
Figure 6 T cell immunity persists for six months after a single immunization. Splenocytes from HD groups recovered at 6 months after immunization were assayed for IFNγ (left panel) and IL-5 (right panel) producing T cells by IFNγ/IL-5-FluoroSpot. Cells were stimulated over night with Wuhan-Spike peptide pool. Data are expressed as mean ( ± SEM) specific Spot Forming Cells (SFC) per million splenocytes. Each dot represents a single mouse.
Genetic distance among Wuhan and Omicron helps explain the escape from nAb responses
In order to better understand the limited antibody cross-reactivity to Omicron in sera of mice immunized with the optimized Wuhan-derived Spike, a phylogenetic analysis using amino acid sequences for the full-length Spike, RBD, S1 and S2 portions was performed. The maximum likelihood (ML) phylogenetic trees for the Spike, S1, S2 and RBD were shown in Figure 7A and Supplementary Figure 6. The ML tree performed on the Spike (Figure 7A) showed the SARS-CoV-2 variants located into two main supported clades. At one end of the tree the Wuhan strain and the Alpha, Beta, Gamma and Delta VoC were identified. On the other end, a supported clade including the Omicron variants (Omicron BA.1, BA.2, BA.4/5) was shown. In particular, the Omicron BA.2 appeared related and closer to BA.4/5 (Figure 7A), in respect to BA.1.
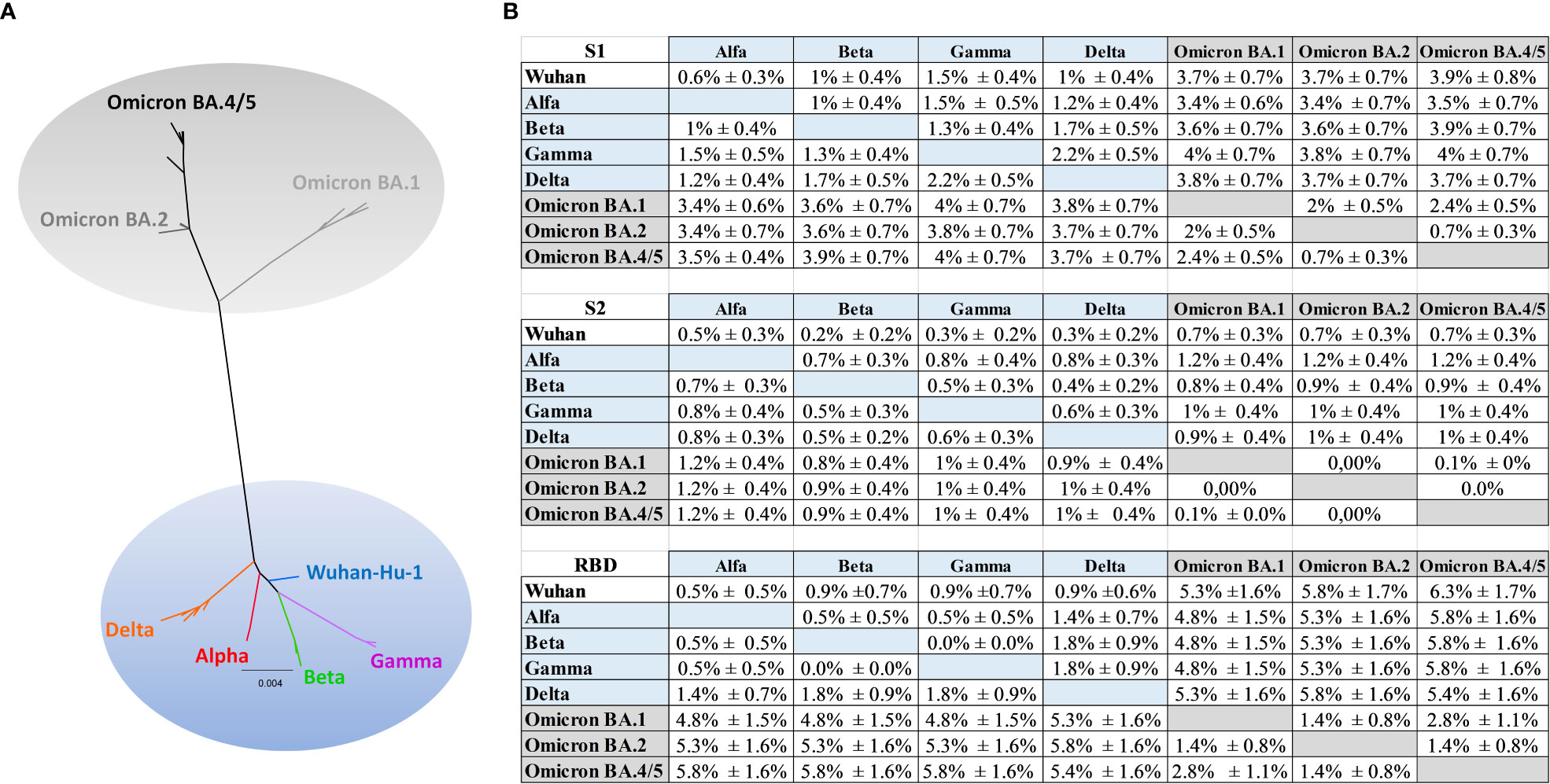
Figure 7 Phylogenetic analysis. (A) Maximum likelihood phylogenetic tree inferred from Spike amino acid sequences. The ancestral SARS-COV-2 sequence (Wuhan/Hu- 1/2019), the Alpha, Beta, Gamma, Delta and the Omicron (BA.1, BA.2, BA.4/5) variants are highlighted with different colors. The scale bar at the bottom of the tree corresponds to amino acid substitutions per site. The authors, originating and submitting laboratories of the sequences from GISAID used to generate the figures are listed in Supplementary Table 1. (B) Mean distances ± SEM at amino acid level for S1, S2 and RBD.
The phylogenetic analysis (Supplementary Figure 6) showed a higher distance (Figure 7B) between the group including Wuhan, Alpha, Beta, Gamma, Delta and the group including the Omicron (BA.1, BA.2, BA.4/5) in S1 respect to S2 portion, and a higher distance in RBD respect to S2 or to S1 (Figure 7B). In particular, the mean diversity at amino acid level between Wuhan and Omicron BA.1, BA.2, BA4/5 increased from 0.7% ± 0.3% (distance ± standard error) for S2 alignment to 3.7% ± 0.7% (Omicron BA.1, BA.2) and to 3.9% ± 0.8% (Omicron BA4/5) in the S1 alignment (Figure 7B), meanwhile increased to 5.3% ± 1.6% (Omicron BA.1) to 5.8% ± 1.7% (Omicron BA.2) and to 6.3 ± 1.7% (Omicron BA4/5) in RBD. Similarly, the mean genetic distance between Beta and Omicron increased from 0.8% ± 0.4%/0.9% ± 0.4% in S2 alignment to 3.6% ± 0.7% (Omicron BA.1, BA.2) and to 3.9% ± 0.7% (Omicron BA4/5) in the S1 alignment (Figure 7B); meanwhile the mean distance in RBD increased to 4.8% ± 1.5%, 5.3% ± 1.6%, 5.8% ± 1.6% respectively for Beta and Omicron BA.1, Beta and BA.2, Beta and BA.4/5.
Discussion
In this study we exploited SIV-based IDLVs delivering different configurations of SARS-CoV-2 Spike in order to induce a long-term, high-magnitude and cross-lineage neutralizing Ab response. We hypothesized that a strategy combining a rationally optimized trimeric Spike on the vector particles together with the sustained immunity provided by IDLV could enhance the quality of the humoral response after a single immunization.
In previous works, we and others showed that membrane-tethering of HIV-1 Env on the surface of a virus-like particles (VLP) or IDLV elicited broader immune responses compared to soluble HIV-Env (34, 73) and that lentiviral vectors can be successfully pseudotyped with SARS-CoV-2 Spike glycoprotein (40). Here, we show that IDLV-CoV2 can be pseudotyped with different configurations of membrane-tethered Spike, and that Spike protein design strongly affects incorporation on vector particles and the resulting immune responses elicited by IDLV. In particular, we evaluated the incorporation of different forms of Spike protein on lentiviral particles and investigated the importance of the immunogen design in the context of the IDLV, an enveloped viral vector which we have started exploiting for its ability to be pseudotyped with membrane-tethered glycoproteins. We reasoned that modifications aimed at increasing incorporation of full-length Spike on IDLV particles would have improved the immune response induced by immunization with IDLV expressing rationally designed immunogens. Our results confirmed that truncation of 21 aa at the C-terminus of the cytoplasmic tail greatly enhanced membrane incorporation of Spike on viral particles, as also observed in other settings (40, 69, 74, 75). Importantly, replacement of RRAR with GSAS at the FCS prevented S1/S2 cleavage, leading to incorporation of a higher amount of full-length Spike protein on IDLV. We hypothesized that this feature may favour the elicitation of nAbs compared to IDLV expressing Spike with an intact FCS, due to the higher amount of S1-associated neutralizing epitopes present on IDLV particles that can be sensed immediately by immune cells after the immunization. To further ameliorate the quality of the anti-Spike humoral response, we evaluated mutations aimed at improving exposure of neutralizing epitopes, including the introduction of 2P substitutions (K986P and V987P) for stabilizing Spike in the prefusion conformation (76, 77) and the inclusion of the D614G mutation, for enhancing RBM exposure (42) and Spike protein density (78) in the IDLV. These modifications have been already adopted, alone or in combination, using different Spike delivery systems, including adenoviral vectors (63, 79, 80), MVA (81) and mRNA vaccines (82, 83).
To assess and compare the immunogenicity of IDLV delivering different configurations of membrane-tethered Spike, mice were immunized once i.m. with scalar amounts of different IDLV-CoV2. Data clearly indicated that the prefusion stabilized Spike with 2P substitutions and mutated FCS, preventing S1/S2 cleavage, was a more effective immunogen in terms of level and persistence of autologous Ab and nAb response compared to the wild-type Spike which showed the poorest humoral immunity (IDLV-S-wt vs IDLV-S-2PF), in line with data from other studies (81, 84). We also investigated the role of the delivery system in the induction of humoral response, comparing IDLV delivering the purified prefusion stabilized Spike with 2P substitutions and mutated FCS (IDLV-S-2PF) with the same version of the Spike delivered as subunit vaccine administered with a squalene-based adjuvant. The level and quality of nAb responses following immunization with IDLV were significantly higher at any dose and time point analyzed.
Immunization with IDLV expressing membrane-tethered D614G Spike with truncated cytoplasmic tail induced higher IgG and autologous nAb levels compared to IDLV expressing full-length Spike (IDLV-S-wt vs IDLV-S-2PGC and IDLV-S-2PF vs IDLV-S-2PFGC), which was more evident at the early and late time points and with any dose of the injected IDLVs. Overall, IDLV-2PFGC, combining all the Spike modifications, outperformed the other candidates, in terms of level of both binding Abs and nAbs at early and later time points, showing the best ability to neutralize all tested VoC, including the circulating Omicron variants, although to a lesser extent. The high-density exposure of the full-length, stabilized and cytoplasmic tail truncated Spike trimers on the IDLV particles, avoiding S1 shedding, was likely responsible for the optimal induced immunity. Interestingly, the pattern of cross-neutralization was different among the tested Spike delivered by IDLVs, underlying the key role of the immunogen in driving the induction of functional nAbs. The Beta Spike variant delivered by IDLV (IDLV-betaS-2PGC) induced a better neutralization against autologous Beta and heterologous Gamma variants compared to the corresponding parental Wuhan-Hu-1 Spike immunogen (IDLV-S-2PGC), but did not improve the cross-neutralization of the other VoC, especially Omicron BA.1. Interestingly, recent data showed that the use of a vaccine adapted to the Omicron VoC may be beneficial to counteract the homologous variant (85), but can be detrimental for blocking the other known variants and potentially also some future new variants (86). This further underlines the importance of iterative testing and optimization of the antigen/vector combination to be included in the vaccine formulation. Indeed, in this study, immunization with IDLV-S-2PFGC, delivering fully optimized Wuhan Spike, led to high level of vaccine-induced immunity to SARS-CoV-2 against all early VoC including Alpha, Beta, Gamma and Delta. On the other hand, the same sera exhibited increased resistance to neutralization against Omicron VoC (BA.1>BA.2>BA.4/5). Evasion of immunity by the Omicron VoC may be attributable to the high number of amino acid substitutions which are present mostly in the S1 subunit of the Spike protein, as indicated by the higher genetic distance between Omicron and all the other VoC. Given the higher number of mutations in Spike, Omicron subvariants may be considered as belonging to a distinct SARS-CoV-2 group or serotype, as recently suggested (87). In addition to the number of mutations, also the effect on the conformation, structure and the combination of the effect of several mutations need to be considered. In this context, previous works have shown that the numerous mutations observed in the Omicron variants alter the conformation of antigenic sites (88, 89), leading to immunological escapes from neutralizing antibodies induced by the Wuhan-based vaccine. The procedure we followed for improving immunity against Spike may be successful against variants within a viral strain that contains a limited number of mutations, whereas a vaccine active against highly divergent strains should focus on portions of Spike that are more conserved, such as the S2 portion (90) where the genetic distance among VoC is much lower. Also, the vaccine should include more conserved viral proteins that induce effector CD8 responses in addition to humoral responses (91). Finally, since intranasal immunization with a different IDLV delivering the ancestral full-length unmodified Spike in ACE2+ rodent animal models showed evidence of protection after challenge (37, 38), a mucosal vaccine based on the optimized Spike delivered by IDLV may be the key for the induction of persistent and cross-neutralizing responses at the portal entry of the virus.
In this study, after a single immunization in mice, nAbs increased over time, suggesting a maturation of B cell response allowing for the maintenance of B cell memory and the development of long lived plasmacells. This phenomenon is not observed in vaccinated humans, where nAb response wanes over time (46, 92) and suggests that some immunological aspects, including analysis of persistence of immunity and breadth of immune responses, should be verified in other animal models, such as non-human primates (93). Interestingly, a recent report showed that after three doses of mRNA vaccine in humans, the Spike-specific repertoire of human B cells was significantly increased, allowing for the development of high affinity and cross-neutralizing antibodies (94), suggesting that multiple antigen exposure is essential for B cell maturation. A recent report elegantly demonstrated that a slow-delivery (12 days) immunization approach and a long-prime in the non-human primate immunogenicity model induced a remarkable germinal center duration and B cell maturation resulting in an enhanced quality of antibody response (95). In this context, we previously demonstrated the persistence of antigen expression at the site of immunization and in draining lymph nodes after IDLV immunization (96) and the durability of IDLV-induced immunity in mice using IDLV delivering different viral antigens, including HBV-HBsAg (97), Influenza HA (39) and a membrane tethered HIV-1 Envelope sequence, the latest confirmed also in non-human primates (34). Persistent production of Spike protein upon immunization with IDLV may favour triggering of maturation of immune response, eventually resulting in higher antibody potency and breadth.
In addition to the humoral response, the T cell immunity represents a key element of the adaptive immune response to SARS-CoV-2 that contributes to reduce the virus replication and eventually the disease progression (91, 98). Previous work has shown that most of the SARS-CoV-2 T cell epitopes defined by different studies (99, 100) are conserved within VoC and consistently with this observation, the antigens containing the sequence variations are cross-recognized by infected and/or vaccinated individuals (101). Due to the high number of different epitopes and the epitope breadth on an individual level, the T cell escape appears unlikely. Those observations underline the importance of inducing T cell immunity following vaccination. In this study, Spike-specific IFNγ producing T cells were detected in all IDLV vaccinated mice at 6 months after immunization, confirming the ability of IDLV to elicit effective and durable T cell immunity (102).
The induction of long-term immunity provided by IDLV combined with rationally designed immunogens delivered both as transgene and pseudotyping glycoprotein on the lentiviral particles, resulted in a versatile and efficient platform to be exploited as a successful vaccine candidate against SARS-CoV-2. To confirm and validate these promising results, IDLV-S-2PFGC will be used in the non-human primate model of immunogenicity and infection, compared with the mRNA vaccines.
This study has some limitations. While IDLV pseudotyping and cross-lineage S-specific nAbs were demonstrated, the study was conducted using a small-animal model, without challenging with live virus. Although presence of nAbs correlates with protection (103), it is yet unknown the minimum level of nAbs which are required for providing protection after the infection, particularly in the mucosa of the respiratory tract. In this context, a study using an HIV-based IDLV expressing full-length wild type Wuhan-Hu-1 Spike in a prime-boost regimen in hamsters, showed strong reduction of viral copies after challenge with homologous SARS-CoV-2 (37), suggesting that an optimized Spike delivered by IDLV will be a promising vaccine strategy.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was authorized by the Italian Ministry of Healthy and reviewed by the Service for Animal Welfare at Istituto Superiore di Sanità (Authorization n. 731/2020-PR, 21/7/20, prot. D9997.107).
Author contributions
DN and ACara conceived and designed the study, analyzed the data, and wrote the manuscript. MB supervised animal experiments, performed ELISA and performed T-cell response experiments. AG and MP prepared the plasmids, the lentiviral vectors, performed flow cytometry experiments and neutralizing antibody assays. MLDA performed neutralizing antibody assays. ACan performed the Western blot assays. ZM performed ELISA and VN assay. SC performed confocal microscopy experiments. AT performed electron microscopy experiments. CF prepared plasmids. SM, ACap, MC, AI and RN produced the recombinant RBD protein. AV performed the animal experiments. MG and RS provided reagents and helped in study implementation. AP conceived and performed phylogenetic analyses. All authors contributed to the article and approved the submitted version.
Funding
This work was funded in part by NATO multi-year Project No. G5817 “New and Validated Tools for the Diagnosis and follow-up of SARS-CoV-2 Infected Individuals” and by Istituto Superiore di Sanità (ISS) intramural funds. This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 681137 (EAVI2020). This research was supported by EU funding within the NextGenerationEU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT).
Acknowledgments
We are grateful to Alice Zappitelli for technical support during vector production and FluoroSpot assays and to Tom Bijl and Jonne Snitselaar for producing Spike and monoclonal antibodies. We are grateful to Stefania Donnini for secretarial assistance and to Ferdinando Costa and Patrizia Cocco for technical support. We thank Fondation Dormeur, Vaduz for the donation of laboratory instruments relevant to this project to the ISS. The following reagents were obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: Polyclonal Anti-Human Immunodeficiency Virus Type 1 SF2 p24 (antiserum, Rabbit), ARP-4250, contributed by DAIDS/NIAID; produced by BioMelecular Technologies; SIVmac239 p27 Recombinant Protein from NIAID, DAIDS (cat# 13446). The following reagent was deposited by Dr. Maria R. Capobianchi for distribution through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate Italy-INMI1, NR-52284.
Conflict of interest
The authors declare that in this study the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1147953/full#supplementary-material
References
1. World Health Organization. WHO coronavirus (COVID-19) dashboard (2023). Available at: https://covid19.who.int/ (Accessed February 26, 2023).
2. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell (2020) 180:281–92.e6. doi: 10.1016/j.cell.2020.02.058
3. Gilbert PB, Montefiori DC, McDermott AB, Fong Y, Benkeser D, Deng W, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science (2022) 375:43–50. doi: 10.1126/science.abm3425
4. Sadarangani M, Marchant A, Kollmann TR. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat Rev Immunol (2021) 21:475–84. doi: 10.1038/s41577-021-00578-z
5. World Health Organization. Status of COVID-19 vaccines within WHO EUL/PQ evaluation process (2023). Available at: https://extranet.who.int/pqweb/sites/default/files/documents/Status_COVID_VAX_12January2023.pdf (Accessed February 26, 2023).
6. Rambaut A, Loman N, Pybus O, Barclay W, Barret J, Carabelli A, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological (2021). Available at: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
7. Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature (2021) 593:266–9. doi: 10.1038/s41586-021-03470-x
8. Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 variant of concern in south Africa. Nature (2021) 592:438–43. doi: 10.1038/s41586-021-03402-9
9. Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science (2021) 372:eabg3055. doi: 10.1126/science.abg3055
10. Sabino EC, Buss LF, Carvalho MPS, Prete CA, Crispim MAE, Fraiji NA, et al. Resurgence of COVID-19 in manaus, Brazil, despite high seroprevalence. Lancet (2021) 397:452–5. doi: 10.1016/S0140-6736(21)00183-5
11. Faria NR, Mellan TA, Whittaker C, Claro IM, Candido D da S, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in manaus, Brazil. Science (2021) 372:815–21. doi: 10.1126/science.abh2644
12. Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, et al. Evolution of antibody immunity to SARS-CoV-2. Nature (2021) 591:639–44. doi: 10.1038/s41586-021-03207-w
13. Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe (2021) 29:44–57.e9. doi: 10.1016/j.chom.2020.11.007
14. Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science (2021) 371:850–4. doi: 10.1126/science.abf9302
15. Liu Z, VanBlargan LA, Bloyet L-M, Rothlauf PW, Chen RE, Stumpf S, et al. Identification of SARS-CoV-2 mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe (2021) 29:477–88. doi: 10.1016/j.chom.2021.01.014
16. Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science (2020) 369:1014–8. doi: 10.1126/science.abd0831
17. Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell (2020) 182:1284–94.e9. doi: 10.1016/j.cell.2020.07.012
18. Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JCC, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife (2020) 9:1–31. doi: 10.7554/eLife.61312
19. Thomson EC, Rosen LE, Shepherd JG, Spreafico R, da Silva Filipe A, Wojcechowskyj JA, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell (2021) 184:1171–87.e20. doi: 10.1016/j.cell.2021.01.037
20. Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature (2021) 593:130–5. doi: 10.1038/s41586-021-03398-2
21. Graham C, Seow J, Huettner I, Khan H, Kouphou N, Acors S, et al. Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 spike is impacted by the B.1.1.7 variant. Immunity (2021) 54:1276–89. doi: 10.1016/j.immuni.2021.03.023
22. Shen X, Tang H, McDanal C, Wagh K, Fischer W, Theiler J, et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe (2021) 29:529–39.e3. doi: 10.1016/j.chom.2021.03.002
23. Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, et al. SARS-CoV-2 501Y.V2 escapes neutralization by south African COVID-19 donor plasma. Nat Med (2021) 27:622–5. doi: 10.1038/s41591-021-01285-x
24. Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature (2021) 592:616–22. doi: 10.1038/s41586-021-03324-6
25. Makoni M. The quest for more COVID-19 vaccinations in Africa. Lancet Respir Med (2022) 10:e70–1. doi: 10.1016/S2213-2600(22)00193-X
26. Wirsiy SF, Nkfusai NC, Ako-Arrey DE, Dongmo EK, Manjong FT, Cumber SN. Acceptability of COVID-19 vaccine in Africa. Int J Maternal Child Health AIDS (IJMA) (2021) 10:134–8. doi: 10.21106/ijma.482
27. Sulemane N, Armocida B, Valente M, Formenti B, Barigazzi S, Ussai S, et al. Vaccines hesitancy in Africa: How COVID-19 pandemic may affect malaria vaccination campaigns. J Prev Med Hyg (2022) 63:E1–3. doi: 10.15167/2421-4248/jpmh2022.63.1.2420
28. Ahiakpa JK, Cosmas NT, Anyiam FE, Enalume KO, Lawan I, Gabriel IB, et al. COVID-19 vaccines uptake: Public knowledge, awareness, perception and acceptance among adult africans. PLoS One (2022) 17:e0268230. doi: 10.1371/journal.pone.0268230
29. Cara A, Klotman ME. Retroviral e-DNA: Persistence and gene expression in nondividing immune cells. J Leukoc Biol (2006) 80:1013–7. doi: 10.1189/jlb.0306151
30. Wanisch K, Yáñez-Muñoz RJ. Integration-deficient lentiviral vectors: A slow coming of age. Mol Ther (2009) 17:1316–32. doi: 10.1038/mt.2009.122
31. Negri DRM, Michelini Z, Cara A. Toward integrase defective lentiviral vectors for genetic immunization. Curr HIV Res (2010) 8:274–81. doi: 10.2174/157016210791208622
32. Negri DRM, Michelini Z, Baroncelli S, Spada M, Vendetti S, Buffa V, et al. Successful immunization with a single injection of non-integrating lentiviral vector. Mol Ther (2007) 15:1716–23. doi: 10.1038/sj.mt.6300241
33. Coutant F, Frenkiel MP, Despres P, Charneau P. Protective antiviral immunity conferred by a nonintegrative lentiviral vector-based vaccine. PLoS One (2008) 3:e3973. doi: 10.1371/journal.pone.0003973
34. Gallinaro A, Pirillo MF, Aldon Y, Cecchetti S, Michelini Z, Tinari A, et al. Persistent immunogenicity of integrase defective lentiviral vectors delivering membrane-tethered native-like HIV-1 envelope trimers. NPJ Vaccines (2022) 7:44. doi: 10.1038/s41541-022-00465-1
35. Somaiah N, Block MS, Kim JW, Shapiro GI, Do KT, Hwu P, et al. First-in-class, first-in-human study evaluating LV305, a dendritic-cell tropic lentiviral vector, in sarcoma and other solid tumors expressing NY-ESO-1. Clin Cancer Res (2019) 25:5808–17. doi: 10.1158/1078-0432.CCR-19-1025
36. Pollack SM, Lu H, Gnjatic S, Somaiah N, O’Malley RB, Jones RL, et al. First-in-Human treatment with a dendritic cell-targeting lentiviral vector-expressing NY-ESO-1, LV305, induces deep, durable response in refractory metastatic synovial sarcoma patient. J Immunotherapy (2017) 40:302–6. doi: 10.1097/CJI.0000000000000183
37. Ku MW, Bourgine M, Authié P, Lopez J, Nemirov K, Moncoq F, et al. Intranasal vaccination with a lentiviral vector protects against SARS-CoV-2 in preclinical animal models. Cell Host Microbe (2021) 29:236–49.e6. doi: 10.1016/j.chom.2020.12.010
38. Ku M, Authié P, Bourgine M, Anna F, Noirat A, Moncoq F, et al. Brain cross-protection against SARS-CoV-2 variants by a lentiviral vaccine in new transgenic mice. EMBO Mol Med (2021) 13:e14459. doi: 10.15252/emmm.202114459
39. Gallinaro A, Borghi M, Bona R, Grasso F, Calzoletti L, Palladino L, et al. Integrase defective lentiviral vector as a vaccine platform for delivering influenza antigens. Front Immunol (2018) 9:171. doi: 10.3389/fimmu.2018.00171
40. Dispinseri S, Secchi M, Pirillo MF, Tolazzi M, Borghi M, Brigatti C, et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun (2021) 12(1):2670. doi: 10.1038/s41467-021-22958-8
41. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (2020) 367:1260–3. doi: 10.1126/science.abb2507
42. Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell (2020) 183:739–51.e8. doi: 10.1016/j.cell.2020.09.032
43. Michelini Z, Negri DRM, Baroncelli S, Spada M, Leone P, Bona R, et al. Development and use of SIV-based integrase defective lentiviral vector for immunization. Vaccine (2009) 27:4622–9. doi: 10.1016/j.vaccine.2009.05.070
44. Bona R, Michelini Z, Mazzei C, Gallinaro A, Canitano A, Borghi M, et al. Safety and efficiency modifications of SIV-based integrase-defective lentiviral vectors for immunization. Mol Ther Methods Clin Dev (2021) 23:263–75. doi: 10.1016/j.omtm.2021.09.011
45. Mariotti S, Capocefalo A, Chiantore MV, Iacobino A, Teloni R, de Angelis ML, et al. Isolation and characterization of mouse monoclonal antibodies that neutralize SARS-CoV-2 and its variants of concern alpha, beta, gamma and delta by binding conformational epitopes of glycosylated RBD with high potency. Front Immunol (2021) 12:750386. doi: 10.3389/fimmu.2021.750386
46. Dispinseri S, Marzinotto I, Brigatti C, Pirillo MF, Tolazzi M, Bazzigaluppi E, et al. Seasonal betacoronavirus antibodies’ expansion post-BNT161b2 vaccination associates with reduced SARS-CoV-2 VoC neutralization. J Clin Immunol (2022) 42:448–58. doi: 10.1007/s10875-021-01190-5
47. Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science (2020) 369:643–50. doi: 10.1126/science.abc5902
48. Michelini Z, Mazzei C, Magurano F, Baggieri M, Marchi A, Andreotti M, et al. Ultraviolet sanitizing system for sterilization of ambulances fleets and for real-time monitoring of their sterilization level. Int J Environ Res Public Health (2022) 19:331. doi: 10.3390/ijerph19010331
49. Hierholzer JC, Killington RA. Virus isolation and quantitation. In: Mahy BWJ, Kangro HO, editors. Virology methods manual. (Cambridge, MA: Academic Press) (1996). p. 25–46 p. doi: 10.1016/B978-012465330-6/50003-8
50. Manenti A, Maggetti M, Casa E, Martinuzzi D, Torelli A, Trombetta CM, et al. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J Med Virol (2020) 92:2096–104. doi: 10.1002/jmv.25986
51. GISAID. Official hCoV-19 reference sequence (2022). Available at: https://gisaid.org/wiv04/ (Accessed December 23, 2022).
52. SIB Swiss Institute of Bioinformatic. ExPASy - translate tool (2023). Available at: https://web.expasy.org/translate/ (Accessed February 26, 2023).
53. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol (2013) 30:772–80. doi: 10.1093/molbev/mst010
54. Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res (2016) 44:W3–W10. doi: 10.1093/nar/gkw343
55. Hall TA. BioEdit: A user friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium (1999) 41:95–8.
56. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol (2010) 59:307–21. doi: 10.1093/sysbio/syq010
57. Lefort V, Longueville JE, Gascuel O. SMS: Smart model selection in PhyML. Mol Biol Evol (2017) 34:2422–4. doi: 10.1093/MOLBEV/MSX149
58. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol (2013) 30:2725–9. doi: 10.1093/molbev/mst197
59. Megasoftware. Equal input model (Amino acids) (2023). Available at: https://www.megasoftware.net/webhelp/distancemethods_hc/hc_equal_input_model.htm (Accessed February 26, 2023).
60. Yu J, Li Z, He X, Gebre MS, Bondzie EA, Wan H, et al. Deletion of the SARS-CoV-2 spike cytoplasmic tail increases infectivity in pseudovirus neutralization assays. J Virol (2021) 95:e00044-21. doi: 10.1128/jvi.00044-21
61. Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol (2021) 6:899–909. doi: 10.1038/s41564-021-00908-w
62. Papa G, Mallery DL, Albecka A, Welch LG, Cattin-Ortolá J, Luptak J, et al. Furin cleavage of SARS-CoV-2 spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog (2021) 17(1):e1009246. doi: 10.1371/journal.ppat.1009246
63. Bos R, Rutten L, van der Lubbe JEM, Bakkers MJG, Hardenberg G, Wegmann F, et al. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines (2020) 91:5. doi: 10.1038/s41541-020-00243-x
64. Finkelstein MT, Mermelstein AG, Miller EP, Seth PC, Stancofski ESD, Fera D. Structural analysis of neutralizing epitopes of the SARS-CoV-2 spike to guide therapy and vaccine design strategies. Viruses (2021) 13:134. doi: 10.3390/v13010134
65. Yuan M, Wu NC, Zhu X, Lee C-CD, So RTY, Lv H, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science (2020) 368:630–3. doi: 10.1126/science.abb7269
66. Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect (2020) 9:382–5. doi: 10.1080/22221751.2020.1729069
67. ter Meulen J, van den Brink EN, Poon LLM, Marissen WE, Leung CSW, Cox F, et al. Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLoS Med (2006) 3:1071–9. doi: 10.1371/journal.pmed.0030237
68. Yu S, Zheng X, Zhou B, Li J, Chen M, Deng R, et al. SARS-CoV-2 spike engagement of ACE2 primes S2’ site cleavage and fusion initiation. PNAS (2022) 119:1–11. doi: 10.1073/pnas.2111199119/-/DCSupplemental
69. Garcia-Beltran WF, Lam EC, St. Denis K, Nitido AD, Garcia ZH, Hauser BM, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell (2021) 184:2372–83.e9. doi: 10.1016/j.cell.2021.03.013
70. Wall EC, Wu M, Harvey R, Kelly G, Warchal S, Sawyer C, et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet (2021) 397:2331–3. doi: 10.1016/S0140-6736(21)01290-3
71. Wang Q, Guo Y, Iketani S, Nair MS, Li Z, Mohri H, et al. Antibody evasion by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature (2022) 608:603–8. doi: 10.1038/s41586-022-05053-w
72. Knudsen NPH, Olsen A, Buonsanti C, Follmann F, Zhang Y, Coler RN, et al. Different human vaccine adjuvants promote distinct antigen-independent immunological signatures tailored to different pathogens. Sci Rep (2016) 6:19570. doi: 10.1038/srep19570
73. McBurney SP, Young KR, Ross TM. Membrane embedded HIV-1 envelope on the surface of a virus-like particle elicits broader immune responses than soluble envelopes. Virology (2007) 358:334–46. doi: 10.1016/j.virol.2006.08.032
74. Case JB, Rothlauf PW, Chen RE, Liu Z, Zhao H, Kim AS, et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe (2020) 28:475–85.e5. doi: 10.1016/j.chom.2020.06.021
75. Crawford KHD, Dingens AS, Eguia R, Wolf CR, Wilcox N, Logue JK, et al. Dynamics of neutralizing antibody titers in the months after severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis (2021) 223:197–205. doi: 10.1093/infdis/jiaa618
76. Bangaru S, Ozorowski G, Turner HL, Antanasijevic A, Huang D, Wang X, et al. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science (2020) 370:1089–94. doi: 10.1126/science.abe1502
77. Juraszek J, Rutten L, Blokland S, Bouchier P, Voorzaat R, Ritschel T, et al. Stabilizing the closed SARS-CoV-2 spike trimer. Nat Commun (2021) 12:244. doi: 10.1038/s41467-020-20321-x
78. Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun (2020) 11:6013. doi: 10.1038/s41467-020-19808-4
79. Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature (2020) 586:583–8. doi: 10.1038/s41586-020-2607-z
80. Capone S, Raggioli A, Gentile M, Battella S, Lahm A, Sommella A, et al. Immunogenicity of a new gorilla adenovirus vaccine candidate for COVID-19. Mol Ther (2021) 29:2412–23. doi: 10.1016/j.ymthe.2021.04.022
81. Meyer zu Natrup C, Tscherne A, Dahlke C, Ciurkiewicz M, Shin D-L, Fathi A, et al. Stabilized recombinant SARS-CoV-2 spike antigen enhances vaccine immunogenicity and protective capacity. J Clin Invest (2022) 132(24):e159895. doi: 10.1172/jci159895
82. Walsh EE, Frenck RW, Falsey AR, Kitchin N, Absalon J, Gurtman A, et al. Safety and immunogenicity of two RNA-based covid-19 vaccine candidates. New Engl J Med (2020) 383:2439–50. doi: 10.1056/nejmoa2027906
83. Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature (2020) 586:567–71. doi: 10.1038/s41586-020-2622-0
84. Graham SP, McLean RK, Spencer AJ, Belij-Rammerstorfer S, Wright D, Ulaszewska M, et al. Evaluation of the immunogenicity of prime-boost vaccination with the replication-deficient viral vectored COVID-19 vaccine candidate ChAdOx1 nCoV-19. NPJ Vaccines (2020) 5:69. doi: 10.1038/s41541-020-00221-3
85. Chalkias S, Harper C, Vrbicky K, Walsh SR, Essink B, Brosz A, et al. A bivalent omicron-containing booster vaccine against covid-19. New Engl J Med (2022) 387:1279–91. doi: 10.1056/nejmoa2208343
86. Wu Y, Shen Y, Wu N, Zhang X, Chen S, Yang C, et al. Omicron-specific mRNA vaccine elicits potent immune responses in mice, hamsters, and nonhuman primates. Cell Res (2022) 32:949–52. doi: 10.1038/s41422-022-00706-x
87. Simon-Loriere E, Schwartz O. Towards SARS-CoV-2 serotypes? Nat Rev Microbiol (2022) 20:187–8. doi: 10.1038/s41579-022-00708-x
88. Cui Z, Liu P, Wang N, Wang L, Fan K, Zhu Q, et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 omicron. Cell (2022) 185:860–71. doi: 10.1016/j.cell.2022.01.019
89. Iketani S, Liu L, Guo Y, Liu L, Chan JFW, Huang Y, et al. Antibody evasion properties of SARS-CoV-2 omicron sublineages. Nature (2022) 604:553–6. doi: 10.1038/s41586-022-04594-4
90. Ng KW, Faulkner N, Finsterbusch K, Wu M, Harvey R, Hussain S, et al. SARS-CoV-2 S2-targeted vaccination elicits broadly neutralizing antibodies. Sci Transl Med (2022) 14:eabn3715. doi: 10.1126/scitranslmed.abn3715
91. Ferrantelli F, Chiozzini C, Manfredi F, Leone P, Spada M, di Virgilio A, et al. Strong SARS-CoV-2 n-specific CD8+ T immunity induced by engineered extracellular vesicles associates with protection from lethal infection in mice. Viruses (2022) 14:329. doi: 10.3390/v14020329
92. Levin EG, Lustig Y, Cohen C, Fluss R, Indenbaum V, Amit S, et al. Waning immune humoral response to BNT162b2 covid-19 vaccine over 6 months. New Engl J Med (2021) 385:e84. doi: 10.1056/nejmoa2114583
93. Walls AC, VanBlargan LA, Wu K, Choi A, Navarro MJ, Lee D, et al. Distinct sensitivities to SARS-CoV-2 variants in vaccinated humans and mice. Cell Rep (2022) 40:111299. doi: 10.1016/j.celrep.2022.111299
94. Muecksch F, Wang Z, Cho A, Gaebler C, ben Tanfous T, DaSilva J, et al. Increased memory b cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature (2022) 607:128–34. doi: 10.1038/s41586-022-04778-y
95. Lee JH, Sutton HJ, Cottrell CA, Phung I, Ozorowski G, Sewall LM, et al. Long-primed germinal centres with enduring affinity maturation and clonal migration. Nature (2022) 609:998–1004. doi: 10.1038/s41586-022-05216-9
96. Lin YY, Belle I, Blasi M, Huang MN, Buckley AF, Rountree W, et al. Skeletal muscle is an antigen reservoir in integrase-defective lentiviral vector-induced long-term immunity. Mol Ther Methods Clin Dev (2020) 17:532–44. doi: 10.1016/j.omtm.2020.03.008
97. Karwacz K, Mukherjee S, Apolonia L, Blundell MP, Bouma G, Escors D, et al. Nonintegrating lentivector vaccines stimulate prolonged T-cell and antibody responses and are effective in tumor therapy. J Virol (2009) 83:3094–103. doi: 10.1128/jvi.02519-08
98. Sette A, Crotty S. Immunological memory to SARS-CoV-2 infection and COVID-19 vaccines. Immunol Rev (2022) 310:27–46. doi: 10.1111/imr.13089
99. Tarke A, Sidney J, Kidd CK, Dan JM, Ramirez SI, Yu ED, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med (2021) 2:100204. doi: 10.1016/j.xcrm.2021.100204
100. Kared H, Redd AD, Bloch EM, Bonny TS, Sumatoh H, Kairi F, et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J Clin Invest (2021) 131(5):e145476. doi: 10.1172/JCI145476
101. Tarke A, Sidney J, Methot N, Yu ED, Zhang Y, Dan JM, et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med (2021) 2:100355. doi: 10.1016/j.xcrm.2021.100355
102. Negri DRM, Michelini Z, Bona R, Blasi M, Filati P, Leone P, et al. Integrase-defective lentiviral-vector-based vaccine: A new vector for induction of T cell immunity. Expert Opin Biol Ther (2011) 11:739–50. doi: 10.1517/14712598.2011.571670
Keywords: lentiviral vector (LV), IDLV, vaccine, SARS-CoV-2, neutralizing Abs
Citation: Borghi M, Gallinaro A, Pirillo MF, Canitano A, Michelini Z, De Angelis ML, Cecchetti S, Tinari A, Falce C, Mariotti S, Capocefalo A, Chiantore MV, Iacobino A, Di Virgilio A, van Gils MJ, Sanders RW, Lo Presti A, Nisini R, Negri D and Cara A (2023) Different configurations of SARS-CoV-2 spike protein delivered by integrase-defective lentiviral vectors induce persistent functional immune responses, characterized by distinct immunogenicity profiles. Front. Immunol. 14:1147953. doi: 10.3389/fimmu.2023.1147953
Received: 19 January 2023; Accepted: 13 March 2023;
Published: 05 April 2023.
Edited by:
Ofer Levy, Division of Infectious Diseases, Boston Children’s Hospital and Harvard Medical School, United StatesReviewed by:
Domenico Tortorella, Icahn School of Medicine at Mount Sinai, United StatesDapeng Zhou, Tongji University, China
Copyright © 2023 Borghi, Gallinaro, Pirillo, Canitano, Michelini, De Angelis, Cecchetti, Tinari, Falce, Mariotti, Capocefalo, Chiantore, Iacobino, Di Virgilio, van Gils, Sanders, Lo Presti, Nisini, Negri and Cara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donatella Negri, donatella.negri@iss.it; Andrea Cara, andrea.cara@iss.it
†These authors share first authorship
‡These authors have contributed equally to this work and share last authorship
 Martina Borghi
Martina Borghi