- 1State Key Laboratory of Respiratory Diseases, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, Department of Critical Care Medicine, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- 2The First Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
A Corrigendum on:
Construction and validation of a robust prognostic model based on immune features in sepsis.
By Zheng Y, Liu B, Deng X, Chen Y, Huang Y, Zhang Y, Xu Y, Sang L, Liu X and Li Y (2022) Front. Immunol. 13:994295. doi: 10.3389/fimmu.2022.994295
In the published article, there was an error. The ‘pheatmap’ R package should be corrected to ‘ggplot2’ R package; false discovery rate (FDR) should be corrected to P-value.
A correction has been made to Materials and Methods, Differential expression analysis in sepsis. This sentence previously stated:
“All the genes in GSE65682 were differentially analyzed by using limma R packages (http://www.bioconductor.org/packages/release/bioc/html/limma.html) (11). The parameter for DEGs screened was│Log2Foldchange│≥0.5 and false discovery rate (FDR) < 0.05. The Volcano plots were drawn by ‘pheatmap’ R package. Then, the IRGs that were overlapping with DEGs were identified as DEIRGs. Similarly, DETFs were obtained by matching TFs with DEGs.”
The corrected sentence appears below:
“All the genes in GSE65682 were differentially analyzed by using limma R packages (http://www.bioconductor.org/packages/release/bioc/html/limma.html) (11). The parameter for DEGs screened was│Log2Foldchange│≥0.5 and P-value <0.05. The Volcano plots were drawn by ‘ggplot2’ R package. Then, the IRGs that were overlapping with DEGs were identified as DEIRGs. Similarly, DETFs were obtained by matching TFs with DEGs.”
In the published article, there was an error. The ‘survival R’ package should be corrected to ‘survival’ R package.
A further correction has been made to Materials and Methods, Construction of the prognostic prediction model in sepsis and development of nomogram, paragraph 1. This section previously stated:
“Based on the univariate Cox regression analysis, prognostic DEIRGs were recognized as the biomarkers for multivariate Cox regression analysis. According to the median risk score value, conducted between low-risk and high-risk groups by using ‘survival R’ package. To evaluate the sensitivity and specificity of the prediction model, the receiver operating characteristics (ROC) curve was calculated using the ‘survivalROC’ package. The area under the ROC curve (AUC) was used to evaluate the prognostic model: 0.5-0.7 (moderate), 0.7-0.8 (better), and >0.9 (excellent).”
The corrected sentence appears below:
“Based on the univariate Cox regression analysis, prognostic DEIRGs were recognized as the biomarkers for multivariate Cox regression analysis. According to the median risk score value, conducted between low-risk and high-risk groups by using ‘survival’ R package. To evaluate the sensitivity and specificity of the prediction model, the receiver operating characteristics (ROC) curve was calculated using the ‘survivalROC’ package. The area under the ROC curve (AUC) was used to evaluate the prognostic model: 0.5-0.7 (moderate), 0.7-0.8 (better), and >0.9 (excellent).”
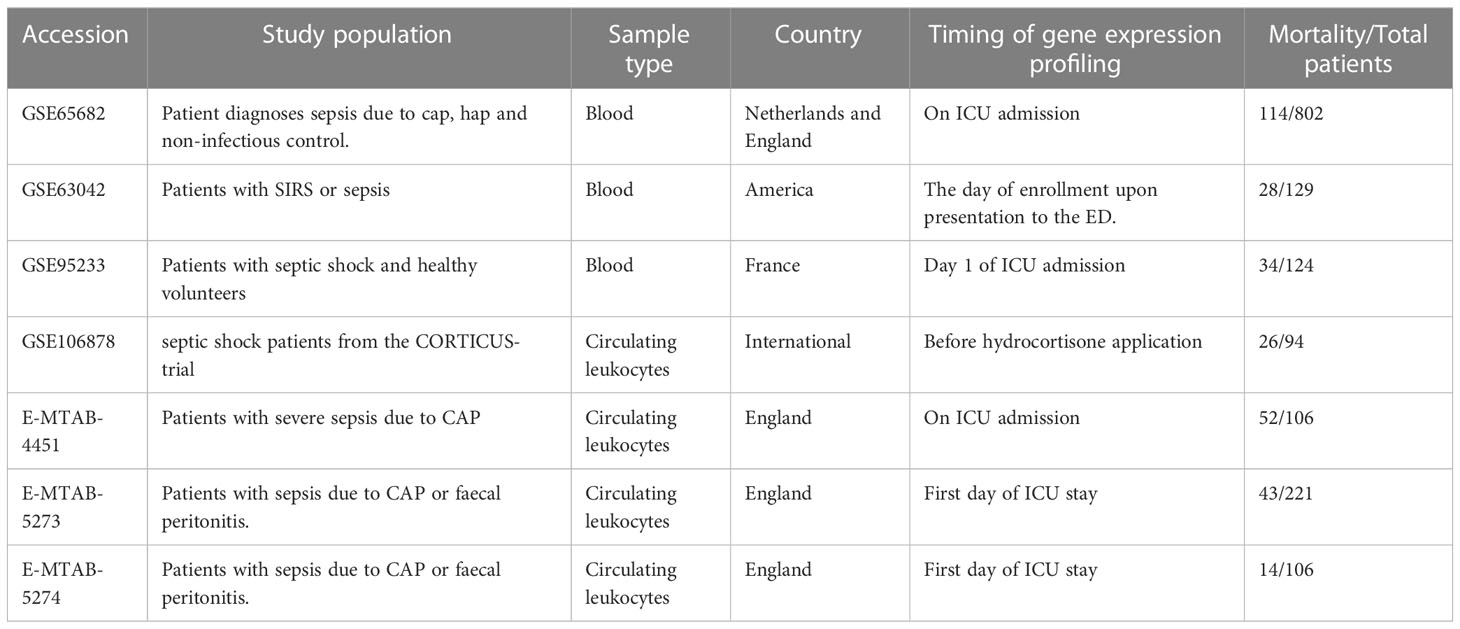
In the published article, there was an error in Table 1 as published. The name of dataset “GSE63062” was wrong, and it should be changed to “GSE63042”. The corrected Table 1 and its caption “Basic information of the datasets included in this study.” appear below.
The authors apologize for these errors and state that they do not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: sepsis, immune, prognostic model, 28-day mortality, immunosuppression
Citation: Zheng Y, Liu B, Deng X, Chen Y, Huang Y, Zhang Y, Xu Y, Sang L, Liu X and Li Y (2023) Corrigendum: Construction and validation of a robust prognostic model based on immune features in sepsis. Front. Immunol. 14:1146121. doi: 10.3389/fimmu.2023.1146121
Received: 17 January 2023; Accepted: 13 February 2023;
Published: 06 March 2023.
Edited and Reviewed by:
Pietro Ghezzi, University of Urbino Carlo Bo, ItalyCopyright © 2023 Zheng, Liu, Deng, Chen, Huang, Zhang, Xu, Sang, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yimin Li, ZHJ5aW1pbmxpQHZpcC4xNjMuY29t
†These authors have contributed equally to this work
 Yongxin Zheng
Yongxin Zheng Baiyun Liu1,2†
Baiyun Liu1,2† Xiumei Deng
Xiumei Deng Yongbo Huang
Yongbo Huang Yu Zhang
Yu Zhang Yonghao Xu
Yonghao Xu Ling Sang
Ling Sang Xiaoqing Liu
Xiaoqing Liu Yimin Li
Yimin Li