- 1Department of Basic Veterinary, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, Sichuan, China
- 2Department of Aquaculture, College of Animal Science and Technology, Sichuan Agricultural University, Chengdu, Sichuan, China
Koi sleepy disease (KSD) is a high mortality and infection viral disease caused by carp edema virus (CEV), which was a serious threat to aquaculture of common carp and export trade of Koi worldwide. Asymptomatic infection is an important cause of the difficulty in preventing KSD and its worldwide spread, because asymptomatic infection can be activated under appropriate condition. However, the understanding of the molecular correlates of these infections is still unknown. The purpose of this study was to compare the pathology change, enzyme activity, immunoglobulin activity, host and viral gene expression differences in acutely infected and cohabiting asymptomatic Koi infected with CEV. Healthy Koi were used as a control. The gross pathology, histopathology and ultrastructural pathology showed the difference and characteristics damage to the tissues of Koi under different infection conditions. Periodic Acid-Schiff stain (PAS), enzyme activity and immunoglobulin activity revealed changes in the immune response of gill tissue between acutely infected, asymptomatic infected and healthy Koi. A total of 111 and 2484 upregulated genes and 257 and 4940 downregulated genes were founded in healthy Koi vs asymptomatic infected Koi and healthy Koi vs acutely infected Koi, respectively. Additionally, 878 upregulated genes and 1089 downregulated genes were identified in asymptomatic vs. acutely infected Koi. Immune gene categories and their corresponding genes in different comparison groups were revealed. A total of 3, 59 and 28 immune-related genes were identified in the group of healthy Koi vs asymptomatic infected Koi, healthy Koi vs acutely infected Koi and asymptomatic infected Koi vs acutely infected Koi, respectively. Nineteen immune-related genes have the same expression manner both in healthy Koi vs acutely infected Koi and asymptomatic Koi vs acutely infected Koi, while 9 immune-related genes were differentially expressed only in asymptomatic Koi vs acutely infected Koi, which may play a role in viral reactivation. In addition, 8 differentially expressed genes (DEGs) were validated by quantitative reverse transcription PCR (RT-qPCR), and the results were consistent with the RNA-Seq results. In conclusion, the data obtained in this study provide new evidence for further elucidating CEV-host interactions and the CEV infection mechanism and will facilitate the implementation of integrated strategies for controlling CEV infection and spread.
1 Introduction
Common carp is one of the most economically valuable freshwater fish in global aquaculture according to annual statistics of FAO (Food and Agriculture Organization of the United Nations). The majority of global production of common carp is destined for human consumption, but its ornamental variety, Koi carp, were also considered among most popular ornamental species of fish (1). Intensive culture and international trade of common carp have led to the global spread of virus pathogens of carp, including Koi herpesvirus (KHV), carp edema virus (CEV) and Spring Viraemia of Carp Virus (SVCV) (2–4). Because of the great threat and economic loss to aquaculture caused by these viral pathogens, SVCV and KHV were identified as a disease that were required to be declared by the World Organization for Animal Health (WOAH) (5), and CEV was also included in the Asia-Pacific Aquatic Animal Disease Surveillance List in 2017 (6).
Koi sleepy disease (KSD) is a fatal viral infection caused by Carp edema virus (CEV), which is a DNA double-stranded virus belonging to the poxvirus family (7). This virus had been associated with high morbidity and mortality rates of up to 70–100% (8). CEV was first discovered in Japan in the 1970s (9). Before 2012, it was mainly reported in Japan, and then gradually appeared around the worldwide. KSD has been reported in many countries, including Britain, France, Germany, Italy, India, Korea and China (10–15). It has brought great challenges to carp farming and trade all over the world. The main clinical signs of Koi infected with CEV are lethargy, difficulty in swimming, enophthalmia and swollen and rotten gills, which mainly occur in spring and autumn (4, 16). This virus usually occurs outbreaks at temperatures ranging from 15 to 25°C, but it can also occur when the temperature is below 10°C (8, 17, 18). The mortality rate is relatively lower and the duration is longer during the outbreak of low temperature (4, 19).
The occurrence of CEV had been reported in many places and caused huge economic losses. However, there is no suitable cell line for the culture of CEV at present, which made the isolation and identification of viruses extremely difficult and further hinders the study of pathogenic mechanisms (18). Currently, most of the research on CEV was based on samples from clinical outbreaks. Gills have been confirmed as the main target organs of CEV infection by immunohistochemistry, in situ hybridization and real-time fluorescence quantification (20, 21). Gills are not only respiratory organs of fish, but also play an important role in osmotic pressure balance, metabolic function and immune function (22). Metabolomic analysis and blood biochemical indicators showed that the disease was accompanied by a severe disturbance of osmotic pressure balance, which was largely associated with impaired gill respiratory and excretory functions (23). In addition, the significantly down-regulation of CD4, TCRα2 and IgM in gill tissue suggests that complex host-pathogen interactions within the gills can have immunosuppressive consequences (24). However, another study reported that significant upregulation of nine immune-related genes, including IFN-γ, TNF-α, and TGF-β, during CEV infection may induce an antigen-adaptive immune response to inhibits viral replication (25). Although current studies had showed that CEV infection can induce host immune response, there was still little about the mechanism of CEV infection and immune response. Moreover, knowledge of the expression profile and the differences in host genes before and after CEV infection was unknown.
In this study, clinical phenotype, nested PCR, RT-qPCR, histopathology and ultra-pathology were combined to analyze the CEV-infected Koi characteristic pathological injury. The transcriptome was used for the first time to analyze the gene expression in the gill tissue of the Koi under different CEV infection conditions (healthy, asymptomatic infection and acutely infected with obvious symptoms). The common and unique gene expression patterns of acutely infected, asymptomatic infected and clinically healthy Koi indicate the involvement of the modification of host immunity. This will also be beneficial to better understand the pathogenic mechanism of CEV infection and re-activation.
2 Materials and methods
2.1 Acquisition of Koi samples
In April 2022, persistent deaths in Koi fish farms have been discovered in Chengdu, Sichuan Province. The Koi had lethargy, floating heads, sunken eyeballs, and gill necrosis in the pond. Thirty Koi were randomly selected for parasite, bacteria and virus detection. The livers, spleens, kidneys, gills and other tissues of the Koi were quickly collected and stored in 10% neutral formalin and 2.5% glutaraldehyde for light microscopic and electron microscopic observation. Some tissues were stored at - 80°C for transcriptome analysis, DNA and RNA extraction.
2.2 DNA extraction and nested PCR detection
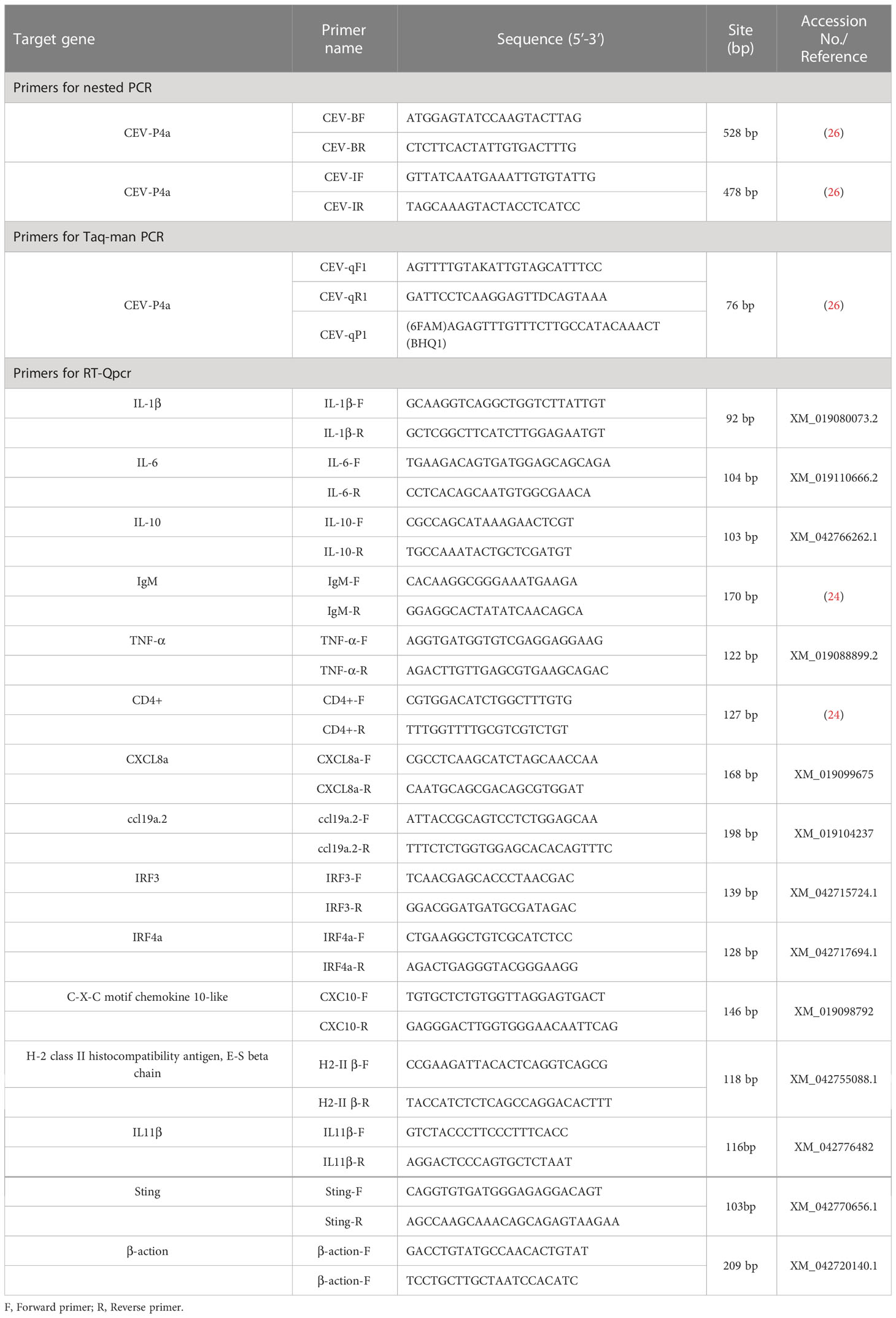
The tissues stored at -80°C were made into tissue homogenate according to the ratio of 1:10 (weight to volume ratio) of normal saline. Viral DNA was extracted from the tissues according to the instructions of TIANamp Genomic DNA Kit (Tiangen Biological Company, China). The purified DNA was stored at -20°C and used for Nested PCR detection (SC/T7229-2019).The primers used in this study were present in Table 1. PCR amplifications were performed using ProFlex PCR System (Thermo Fisher Scientific) with total volume of 25 μL containing 12.5 μL 2×PCR Master (Nanjing Vazyme Biotech Co., Ltd), 1 μL of DNA, 1 μL of the forward primer, 1 μL of reverse primer and 9.5 μL of double distilled water. The reaction conditions were used as follows: denaturation at 95°C for 3 min, followed by 30 cycles of 95°C for 15 s, 45°C for 15 s and 72°C for 15 s and elongation at 72°C for 5 min.
2.3 Quantification of virus genome copies in tissues by real-time TaqMan qPCR
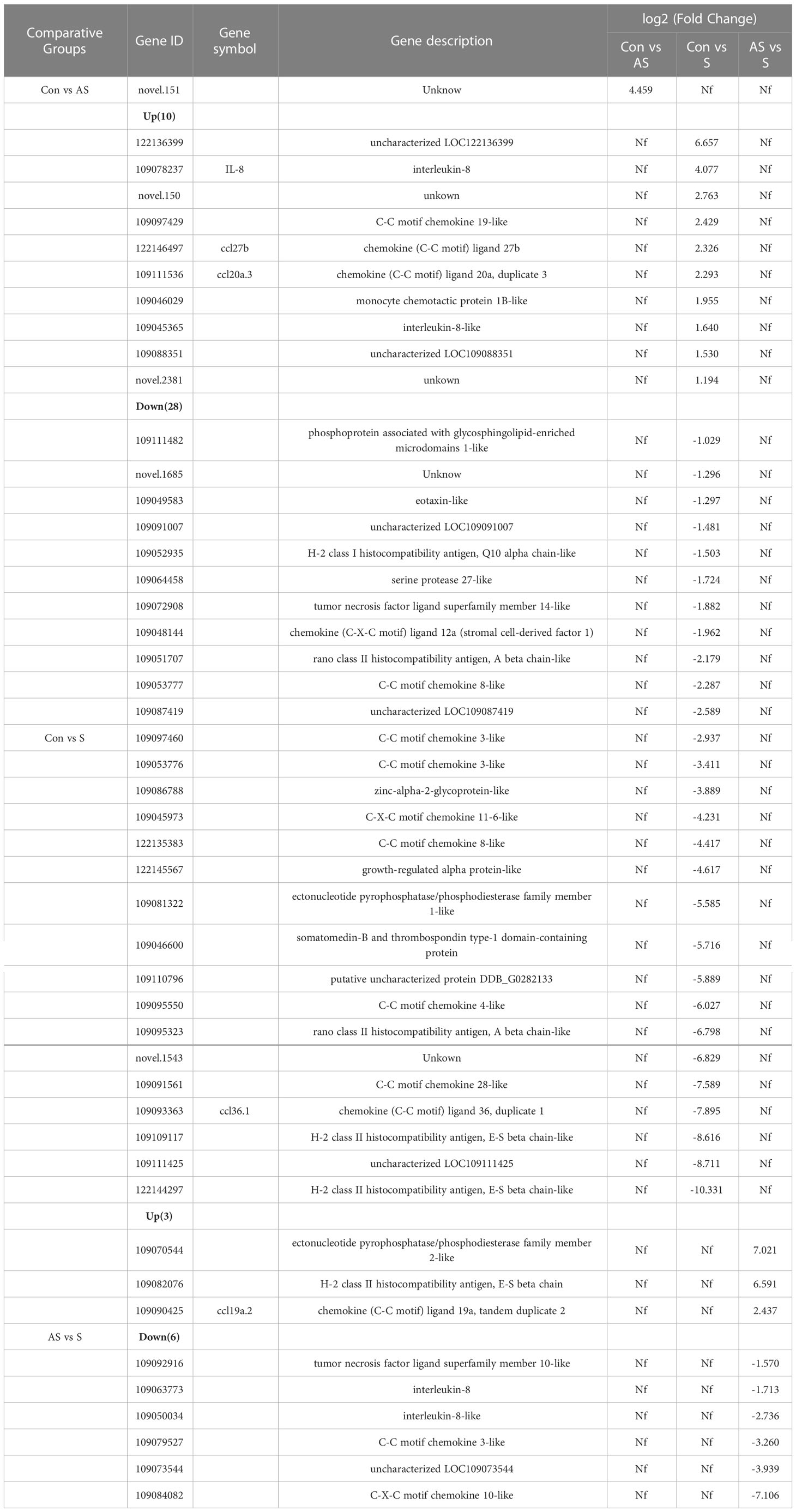
The primers and probes used for real-time TaqMan qPCR were synthesized according to the SC/T7229-2019 (26). The primers were used to amplify fragments of CEV partial 4a gene sequence. The amplicons were cloned into pMD19-T vector (Takara) and propagated in DH5α (Takara). The cloning procedure was following the manufacturer’s directions. The plasmids that successfully expressed the target gene were extracted according to TIANprep Mini Plasmid Kit (Tiangen Biological Company, China). The plasmid bearing the insert was diluted in DNase-free water in a 10-fold dilution series and used to generate a standard curve. The final reaction mixtures contained 2× Master Mix (ChamQ Universal SYBR qPCR Master Mix, Vazyme), 500 nM of each primer, 200 nM of the fluorescent probe and 250 ng of template DNA. The qPCR was performed using a real-time fluorescence quantitative PCR System (Bio-Rad, USA) including an initial denaturation step at 95°C for 5 min, followed by 45 cycles at 95°C for 10 s and 60°C for 30 s. Fluorescence data were analyzed using the real-time fluorescence quantitative PCR System (Bio-Rad). The results were presented as the total number of virus copies per 250 ng of DNA. From a standard curve constructed from 101 to 108 copies of recDNA plasmids of CEV and from aserially (10-fold) diluted CEV-positive sample as a template, we concluded that the qPCR gave the amplification curve y = - 3.32 x + 41.577 with a correlation R2 = 0.999 and efficiency of 100.1% (Range 99.3−105. 1%, Figure 2A).

Figure 1 Clinical signs and symptoms of Koi. (A, B) Koi had no clinical symptoms and pathology changes (C) The gill of sick Koi was necrotic, swollen, mucus increased significantly (C-1), slight bleeding in mandible and caudal fin (△). (D) The sick Koi has sunken eyes and slight redness around the eyeball, severe Gill necrosis, mainly characterized by Gill whitening and swelling (D-3), slight abdominal swelling (D-2), caudal fin bleeding (△).
2.4 Histopathology and ultramicropathology
The fixed tissue by 10% neutral formalin was washed overnight in running water, dehydrated in a graded ethanol series (50%, 70%, 80%, 90%, 100%× 2), cleaned in xylene, embed in paraffin, subsequently sectioned at approximately 5 μm thickness (LeicaRM2125RTS, Lycra, Germany), and stained with classical hematoxylin and eosin (H&E). The Gill tissue sections were scanned and analyzed by digital pathological scanner (VS120-S6-W (BX61VS)). The length, width and interlamellar area of gill lamella were measured by ImageJ. The Gill tissue samples were fixed with 2.5% glutaraldehyde and 1% osmium tetroxide, dehydrated using graded ethanol (30%→50%→70%→80%→90%→95%→100%×3), thick ultra-thin sections of 50 nm were prepared by ultra-thin section mechanism after infiltration and embedding, stained with uranium acetate and lead citrate, observation and image acquisition using transmission electron microscope (JEM-1400PLUS).
2.5 Periodic acid–Schiff staining
The prepared paraffin sections were oxidized 5min with 1% periodate, washed by current water for several times, stained with Schiff reagent, washed by running water, stained with hematoxylin, and sealed with neutral resin after dehydration. The gill tissue sections were scanned by digital pathological scanner (VS120-S6-W (BX61VS)). The number of mucous cells in the same visual field was recorded (0.024 mm2).
2.6 Enzyme activity analysis
The gill tissue samples were prepared into 10% tissue homogenate according to the ratio of gill weight to normal saline volume of 1:9. The lysozyme (LZM), alkaline phosphatase (AKP), acid phosphatase (ACP), catalase (CAT), glutathione strand hydrogenase (GSH-PX), glutathione (GSH), and superoxide dismutase (SOD) were determined following manufacturer’s instructions (Jian Cheng Bioengineering Institute, Nanjing, China).
2.7 ELISA analysis
In order to study the immune response in gill tissue of Koi infected by Carp edema virus at different infection conditions. The contents of related immunoglobulin (IgT and IgM) in gill of Koi was determined by fish immunoglobulin (IgM and IgT) enzyme-linked immunosorbent assay (ELISA) kit (MLBIO Biotechnology Co. Ltd, Shanghai, China) following manufacturer’s instructions.
2.8 RNA extraction, cDNA library construction, sequencing and analyses
The total RNA of gill tissue was extracted by Trizol method. Total amounts and integrity of RNA were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The mRNA with polyA tail was enriched by Oligo (dT) magnetic beads, and then the mRNA was randomly interrupted by divalent cations in Fragmentation Buffer, sifted out small fragments of about 370~420 bp. In order to select cDNA fragments of preferentially 370~420 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then PCR amplification, the PCR product was purified by AMPure XP beads, and the library was finally obtained.The clean reads obtained by removing connectors, empty reading, and low quality sequences. The Clean reads were aligned to the reference genome (https://www.ncbi.nlm.nih.gov/genome/?term=Cyprinus+carpio ) using Hisat2 (v2.0.5). DESeq2 software (1.20.0) was used to analyze the differential expression between groups. The unigenes were compared and predicted with known transcripts from six databases to obtain annotated information, including Gene Ontology (GO), Clusters of Orthologous Groups of proteins (COG), Kyoto of Encyclopedia of Genes and Genomes (KEGG), National Center for Biotechnology Information (NCBI) non-redundant protein sequences (NR), Swiss-Prot, and The Pfam protein families (Pfam). The expression level of the transcript was quantitatively analyzed by RSEM software (http://deweylab.github.io/RSEM/ ). Differential expression analysis of two conditions/groups (two biological replicates per condition) was performed using the DESeq2 algorithms. DESeq2 provide statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with adjusted P-value <=0.05 and |log2FoldChange|>=1.0 were assigned as differentially expressed.
2.9 Validation of differentially expressed transcripts by quantitative real-time PCR
To verify the high-throughput sequencing results from the RNA-Seq data, the upregulated or downregulated genes were detected by RT-qPCR. The primers (Table 1) used for RT-qPCR were designed with the Primer Premier 6.0 software. Quantitative real-time PCR assays were performed using a real-time fluorescence quantitative PCR System (Bio-Rad, USA) including an initial denaturation step at 95°C for 5 min, followed by 40 cycles at 95°C for 10 s and 60°C for 30 s. Fluorescence data were analyzed using the real-time fluorescence quantitative PCR System (Bio-Rad). All samples were analyzed in triplicate. The RNA expression of each target gene was normalized to β-actin expression, and the dates was estimated by the 2-△△CT method.
2.10 Data analysis
In this study, significance analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, La Jolla, CA). The gene expression were normalized by β-action and calculated by 2-△△CT method. All experiments were repeated three times at least and the results presented as mean ± SD. The significant level was set as P < 0.05 (*), P < 0.01 (**) , P < 0.001 (***), P < 0.0001 (****).
3 Results
3.1 Clinical sample analysis by nested PCR and quantitative real-time PCR
Thirty Koi were randomly collected for clinical observation and PCR detection, seven Koi showed obvious clinical signs, and the other showed no clinical symptoms. The main clinical signs were exhibited including loss of appetite, lethargy, loss of balance, floating heads, and other external signs such as swollen gills, enophthalmia, and congestion of the caudal fin (Figure 1). The fish were negative for parasite and bacterial infection (data not shown). However, nested PCR results showed that some Koi without clinical symptoms were also positive for CEV in tissue mixture, and the proportion was 13/23, although this PCR result was positive only in the second round of nested PCR. In order to further determine whether the asymptomatic Koi carried CEV virus, the viral load of per 250ng DNA in the mixed tissues of liver, spleen, kidney and gill was detected by Quantitative real-time PCR (qPCR). It was found that the Koi without obvious clinical symptoms also carried the virus, and the proportion was as high as 56.5%. However, the viral load of tissues mixtures of asymptomatic Koi was relatively lower than symptomatic Koi (Table 2, Figure 2B). Through clinical observation, nested PCR and qPCR detection, the Koi without clinical symptoms and negative result of nested PCR were defined as control, and those Koi without clinical symptoms and positive result of Nested PCR for second round were defined as asymptomatic infection, and the Koi with clinical symptoms and positive result of Nested PCR for both round were defined as symptomatic infection. The fish were free of other common viral diseases infection (such as SVCV, Cyprinid herpesvirus 1 (CyHV-1), Cyprinid herpesvirus 2 (CyHV-2) and KHV) were excluded by PCR (data not shown).

Table 2 The infection stages of Koi were identified by clinical phenotype, Nested PCR and Taq-man qPCR.
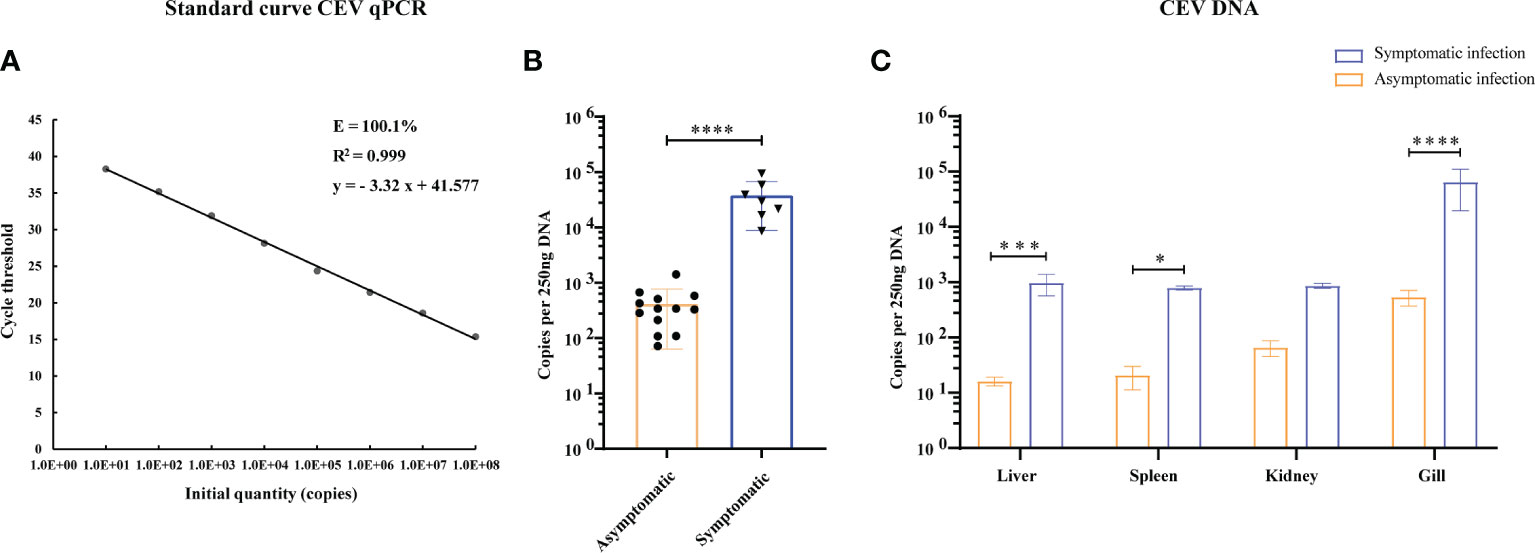
Figure 2 Viral loads in tissues of CEV were analyzed by TaqMan qPCR. (A) Standard curve of supercoiled recombinant plasmid DNA from 101 to 108 copies used for real time Taq-man PCR. Standard curve represents mean ± SD from three replicates. Ct (dRn): cycle threshold based on delta (change) of controlized fluorescence; R2: Correlation coefficient; E: efficiency. (B) Viral load of CEV in tissues mixture with or without clinical symptoms. (C) Viral loads in liver, spleen, kidney and gill tissues of Koi. The results were expressed as the means ± SD of the data observed for the 6 Koi analyzed per 250ng DNA. The significant level was set as P < 0.05 (*), P < 0.001 (***), P < 0.0001 (****).
3.2 Tissue viral load and histopathology injury
In order to study the relationship between the clinical signs, histopathological injury and viral load in the process of CEV infection in Koi, four tissues was analyzed by qPCR. The results showed that the viral load in the tissue of Koi with clinical symptoms was significantly higher than Koi without clinical symptoms (Figure 2B). The highest levels of viral DNA were found in the gills in both acutely infected and asymptomatic infection, which indicated that gill was the major target organ of CEV infection. There were also had significant differences of viral load in liver and spleen between acutely infected and asymptomatic infected Koi (Figure 2C).
Histopathology results showed that the lesion of gill tissue was the most serious injury organ in both acutely infected and asymptomatic infection Koi (Figure 3). The Koi in symptomatic infection group were showed pathology injury in liver, spleen and kidney as well (Figures 3I-K). However, the Koi in asymptomatic infection group were showed mild or no pathology injury in liver, spleen and kidney (Figures 3E-H). Pathological injure was consistent with the viral load in tissues from acutely infected and asymptomatic infected Koi. The gill lamellae of control Koi were arranged neatly, without swelling, exfoliation and necrosis of epithelial cells (Figures 4A, B). Asymptomatic infection Koi had slight proliferation at the base of gill lamellae, accounting for about 1/3 of the whole gill lamellae, epithelial cells swollen and diffusely attached to the gill lamellae and without exfoliation (Figures 4C, D). The gill lamellae of Koi with symptomatic infection proliferated obviously, hypertrophy, exfoliation and necrosis of branchial epithelial cells, occlusion of the branchial intralamellar spaces and fusion of secondary lamellae, and even the clav-shaped gill was presented (Figures 4E, F). The length, width and area between gill lamella were measured by Image J software. The length of gill lamellae did not change significantly, the width of gill lamellae was significantly different only between control and acutely infected Koi, and the area of branchial intralamellar spaces showed significant difference between each group (Figures 4G-I).

Figure 3 Histopathology change of Koi. (A-D) Histological changes in different tissues of control group Koi, (E-H) Histological changes in different tissues of asymptomatic infected Koi, (I-L) Histological changes in different tissues of symptomatic infected Koi.
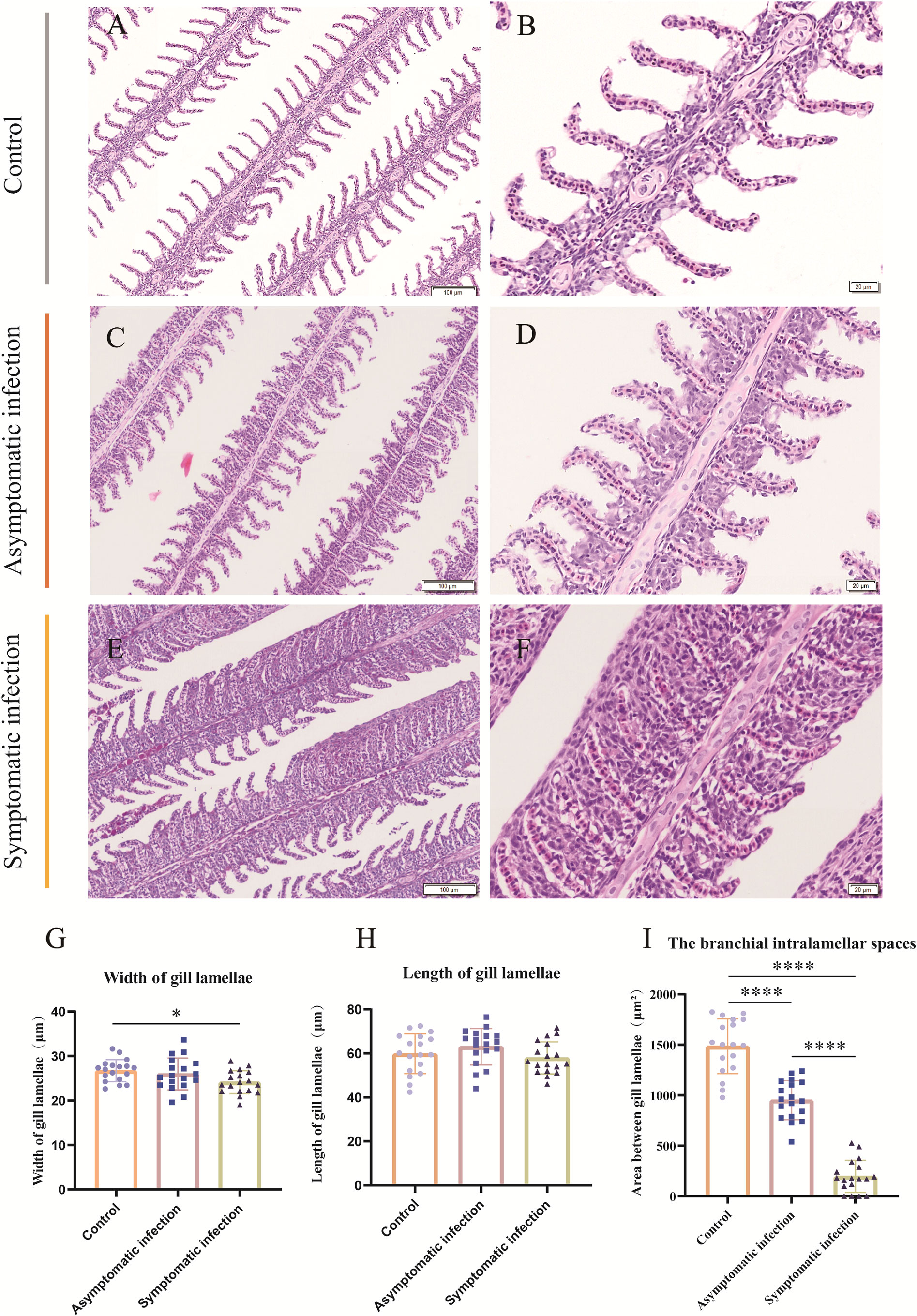
Figure 4 H&E staining, gill lamellae length, gill lamellae width and area between gill lamellae statistics of Koi gill tissues. (A, B) The gill of control Koi used as a control. (C, D) Gill of asymptomatic Koi with low viral load. (E, F) Gill of symptomatic Koi with high viral load. (G-I) The length, width of gill lamellae and area between gill lamellae. The significant level was set as: P < 0.05 (*), P < 0.0001 (****), (n =18).
3.3 Changes of mucous cells in gill tissue
The fish gill was a multipurpose organ, in addition to providing for aquatic gas exchange, plays dominant roles in osmotic and ionic regulation, acid-base regulation, and excretion of nitrogenous wastes (27). Importantly, it was one of peripheral mucosal immune organs of fish. Mucus cells play a crucial role in the immune regulation of gill tissue (22, 28). In order to detect the changes of mucous cells in gill tissue during CEV infection, PAS staining was performed on gill tissue. PAS staining showed that mucus cells of control Koi were attached to the base of gill lamella, and there were one or two mucus cells between each gill lamella (Figures 5A, B). Mucous cells of asymptomatic infected Koi migrated laterally from the base of gill lamella (Figures 5C, D). The mucous cells were scattered in the branchial intralamellar spaces (Figures 5E, F). The number of mucous cells in the gill tissue (0.024 mm2) was counted. The result showed that mucous cells increased significantly in the process of CEV infection and the number of mucous cells were the largest in Koi of symptomatic infection (Figure 5G).
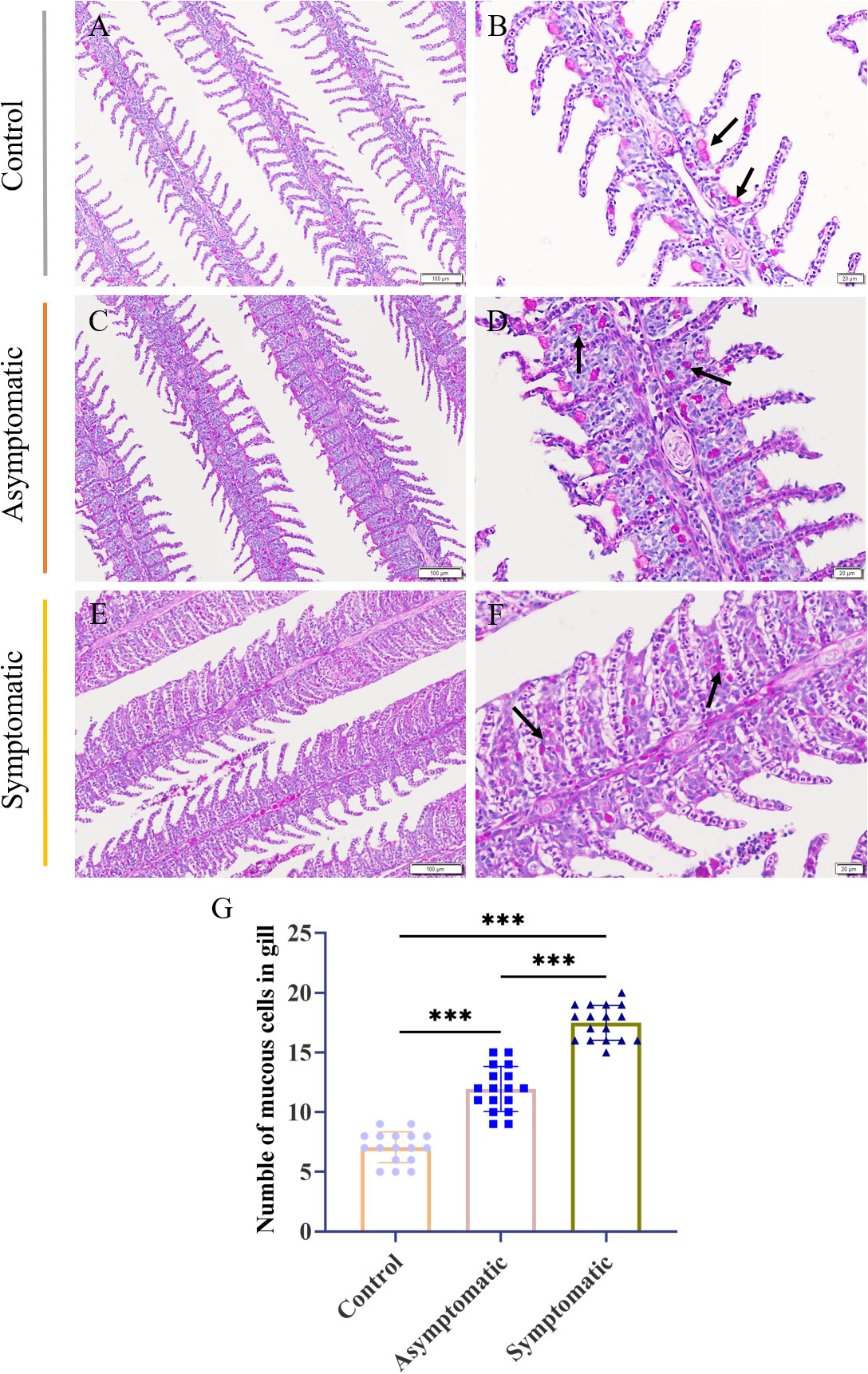
Figure 5 PAS staining and number of mucus cells of Koi gill tissues. (A, B) Gill of Koi in the control group. (C, D) Gill of asymptomatic Koi with low viral load. (E, F) Gill of symptomatic Koi with high viral load. (G) The number mucus cells of gill tissues. Significance: P < 0.001 (***) (n =18).
3.4 Electron microscope observation
To determine the replication process of CEV virions in gill tissue, electron microscope observation was carried out, and the results showed that CEV virions replicate in the cytoplasm and range in size from 200-350nm (Figure 6), which was consistent with the results of other studies (16). Healty and asymptomatic Koi had no obvious pathological damage to their gill tissue, the mitochondrial structure was normal and the nucleus was uniformly colored (Figures 6A, B). A large number of cell necrosis and obvious mitochondrial swelling were clearly observed in the gill tissue of Koi with clinical symptoms by electron microscope (Figures 6C, D).

Figure 6 Transmission electron microscope observation of Koi gill tissues. (A) Gill of Koi in the control group. (B) Gill of Koi in asymptomatic group (C-D) Gill of Koi in symptomatic group. N, nucleus; M, mitochondria; Blue arrow, Necrotic cell; Red arrow, virus particles.
3.5 Enzyme activity and immunoglobulin concentration in gill tissue
In order to explore the effects of CEV infection on the gill tissue of Koi, enzyme activity and immunoglobulin level in gill tissue were detected in three groups. MDA, GSH-PX, GSH and CAT were detected as oxidative stress indicators, while enzyme activity (ACP, AKP and LZM) and immunoglobulin (IgM and IgT) were detected as immune-related indicators. According to the chemical analysis results, there was significant difference in oxidative stress enzymes activity of SOD, GSH and CAT in gill tissue between control group and symptomatic group Koi, but there was no significant difference between control and asymptomatic Koi (Figures 7A-D). The activity of ACP increased significantly than control Koi between asymptomatic infected Koi and acutely infected Koi (Figure 7E). The AKP activity increased significantly only between control Koi and symptomatic infected Koi (Figure 7F). Although the activity of LZM was higher after CEV infection, there was no significant difference (Figure 7G). Conversely, IgM concentrations were significantly decreased in both asymptomatic and symptomatic infected Koi, but IgT concentrations were significantly decreased in symptomatic infected Koi (Figures 7H, I).
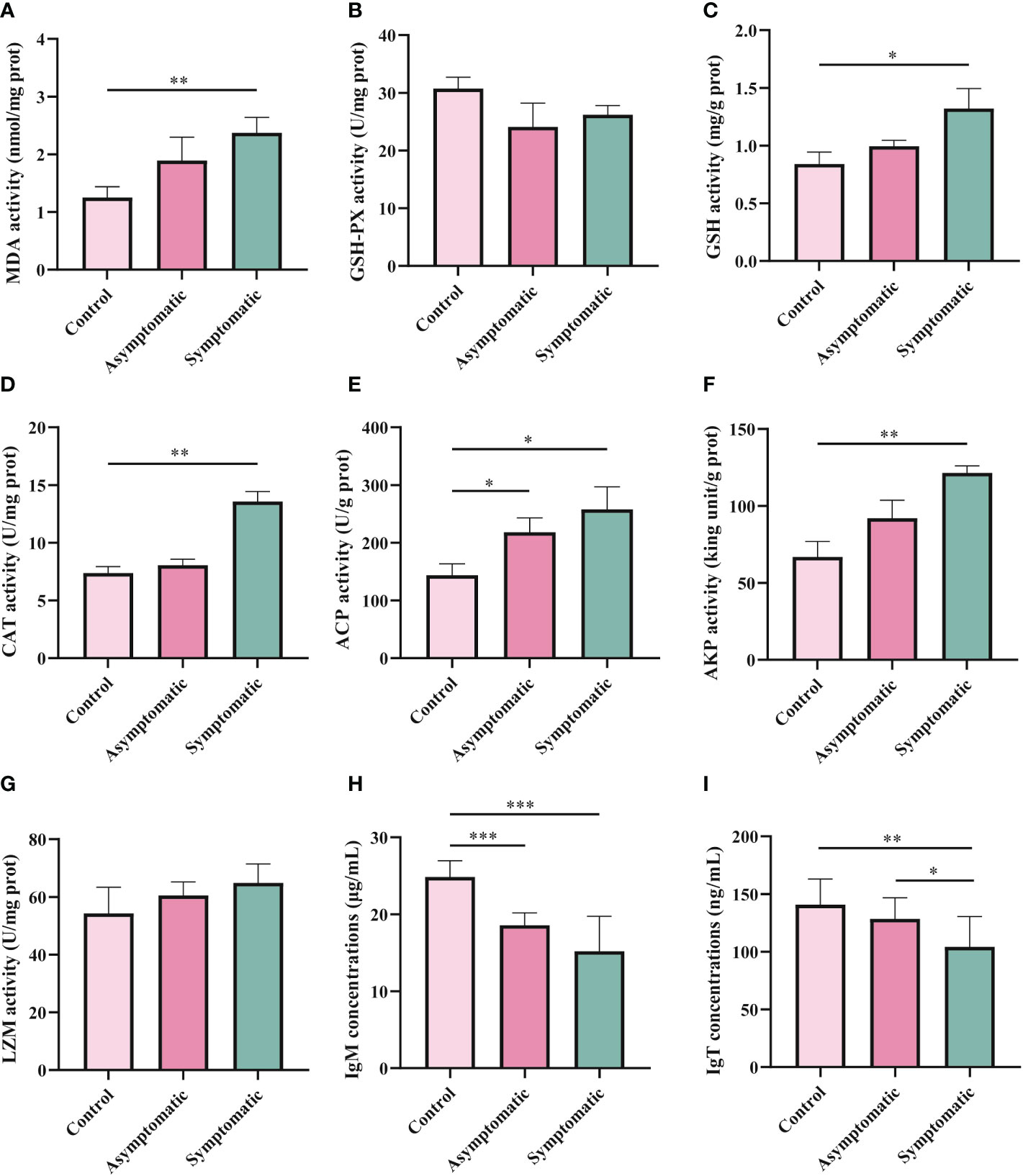
Figure 7 Enzyme activity and activity of immune-related substances were detection in Koi gill tissues. (A-D) Oxidative stress activity include catalase (CAT), glutathione strand hydrogenase (GSH-PX), glutathione (GSH), and superoxide dismutase (SOD). (E-I) Activity of immune-related substances were detected, include lysozyme, alkaline phosphatase, acid phosphatase, IgM and IgT,respectively. The significant level was set as P < 0.05 (*), P < 0.01 (**) , P < 0.001 (***).
3.6 The expression of immune-related genes in gill tissue
As the first line of defense against pathogens, gills play an important role in the removal of pathogens (29). In order to investigate the changes of immune-related functions in the gill tissue of Koi during CEV infection, immune-related genes (IL-1β, IL-6, TNF-α, IgM, CD4+ and IL-10) were detected by RT-qPCR. According to the results of RT-qPCR, the expression of TNF-α gene was significantly up-regulated after CEV infection (Figure 8C). A significant upregulation was observed in the expression of IL-1β,IL-6 and IL-10 in symptomatic infected Koi compared with control Koi and asymptomatic infected Koi (Figures 8A, B, F). On the contrary, the expressions of IgM and CD4+ genes were significantly down-regulated after CEV infection, which consist with the ELISA results of IgM (Figures 8D, E).

Figure 8 Expression of immune-related gene was measured in gill tissues of Koi at different stages during CEV infection. The RNA expression of each target gene was normalized to β-actin expression. The asterisks (*) indicate significant differences from two groups comparing each other. * indicate P < 0.05, ** indicate P <0.01, ***indicate P <0.001. Data are shown as mean ± SD of six fish.
3.7 Transcriptome analysis and gene expression statistics
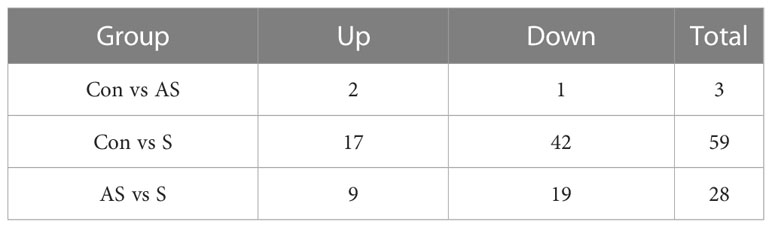
To investigate the mechanism of gill tissue damage during CEV infection, transcriptome analysis was performed on gill tissue between symptomatic infection, asymptomatic infection and healthy Koi. Nine samples were sequenced by Illumina NovaSeq 6000 sequencing platform, and a total of 42.76 GB of data was obtained, with an average of 4.75GB of data per sample. The clean readings of each sample were compared with the reference genome (https://www.ncbi.nlm.nih.gov/genome/?term=Cyprinus+carpio ), with a comparison rate ranging from 80.73% to 90.25%. Based on analysis, the total number of detected expressed genes was 51290, including 47696 known genes and 3594 predicted novel genes. The differentially expressed genes were analyzed by using variance analysis software DESeq2 (screening threshold for: |log2FC| ≥1, padjust<0.05). The statistical results showed that there were 878 and 2484 upregulated genes and 1089 and 4940 downregulated genes between control Koi vs asymptomatic infected Koi and control group Koi vs symptomatic infected Koi. In addition, the comparisons between asymptomatic infected Koi and symptomatic infected Koi revealed 111 upregulated genes and 257 downregulated genes (Figures 9A–C). Furthermore, a total of 21 genes were significantly different in the three comparison groups (Figure 9D), 13 genes were significantly up-regulated and 5 genes were significantly down-regulated in all comparison groups (Figures 9E, F). (In the co-expressed genes of all the differential genes, there will be cases where the same gene is up-regulated in one comparison group, but down-regulated in the other comparison group, and that gene will be counted in all the Venn diagram co-expressed genes of the differential genes, so the sum of up-regulated and down-regulated genes is different from that of total genes). The RNA-Seq data have been successfully deposited in the SRA database (https://www.ncbi.nlm.nih.gov/sra/PRJNA925573) with accession number PRJNA925573.
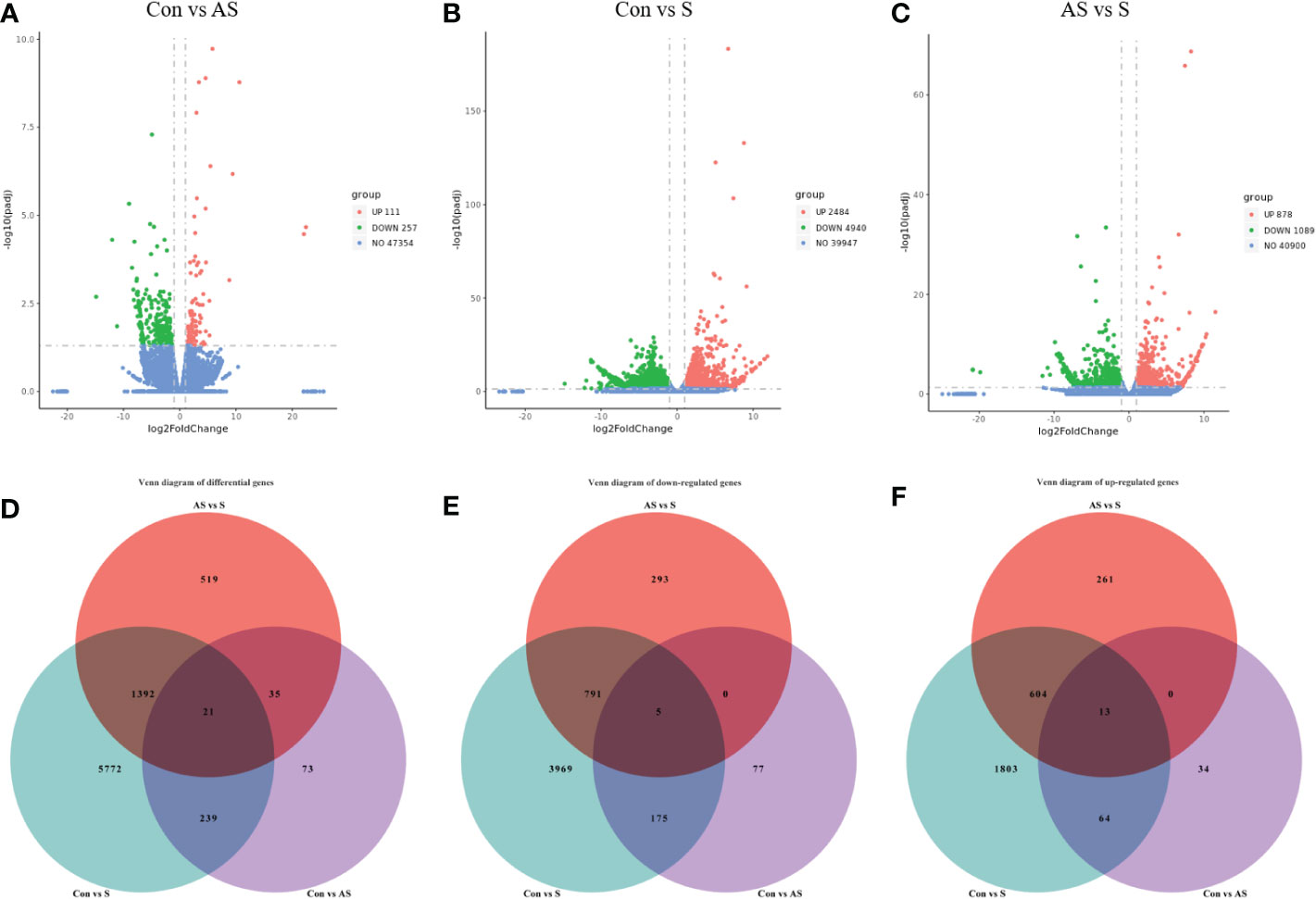
Figure 9 Differentially expressed genes (DEGs) analysis. (A) Control group Koi vs. asymptomatic infected Koi volcano plot. (B) Control group Koi vs. acutely infected Koi volcano plot. (C) Asymptomatic infected Koi vs. acutely infected Koi volcano plot. (D) Venn diagram of differentially expressed genes in control group Koi vs asymptomatic Koi, control group Koi vs. acutely infected Koi and asymptomatic infected Koi vs. acutely infected Koi; (E) Venn diagram of significantly downregulated genes in control vs asymptomatic Koi, Control group Koi vs. acutely infected Koi and asymptomatic infected Koi vs. acutely infected Koi; (F) Venn diagram of significantly upregulated genes in control vs asymptomatic Koi, Control group Koi vs. acutely infected Koi and asymptomatic infected Koi vs. acutely infected Koi.
3.8 GO Analysis of DEGs
To analyze the potential biological functions of the DEGs in the three comparative groups, GO annotation of the DEGs was performed according to Molecular Function (MF), Cellular Components (CC) and Biological Process (BP). According to the GO annotation results, the GO terms between control Koi vs asymptomatic infected Koi, control group Koi vs symptomatic infected Koi and asymptomatic infected Koi vs symptomatic infected Koi were 494, 1349 and 956. Ten GO terms with the most significant differences between the comparative groups were listed (Figures 10A–C). In this study, the identified MF terms mainly included catalysis, binding activity and molecular biological function, CC terms mainly included cell homeostasis, metabolic process and signal transduction, BF terms mainly included cell and organelle structure and composition.

Figure 10 GO classification of DEGs. Biological process, cellular component and molecular function. (A) GO analysis of control group vs asymptomatic group. (B) GO analysis of asymptomatic group vs symptomatic group (C) GO analysis of control group vs symptomatic group.
3.9 KEGG analysis of DEGs
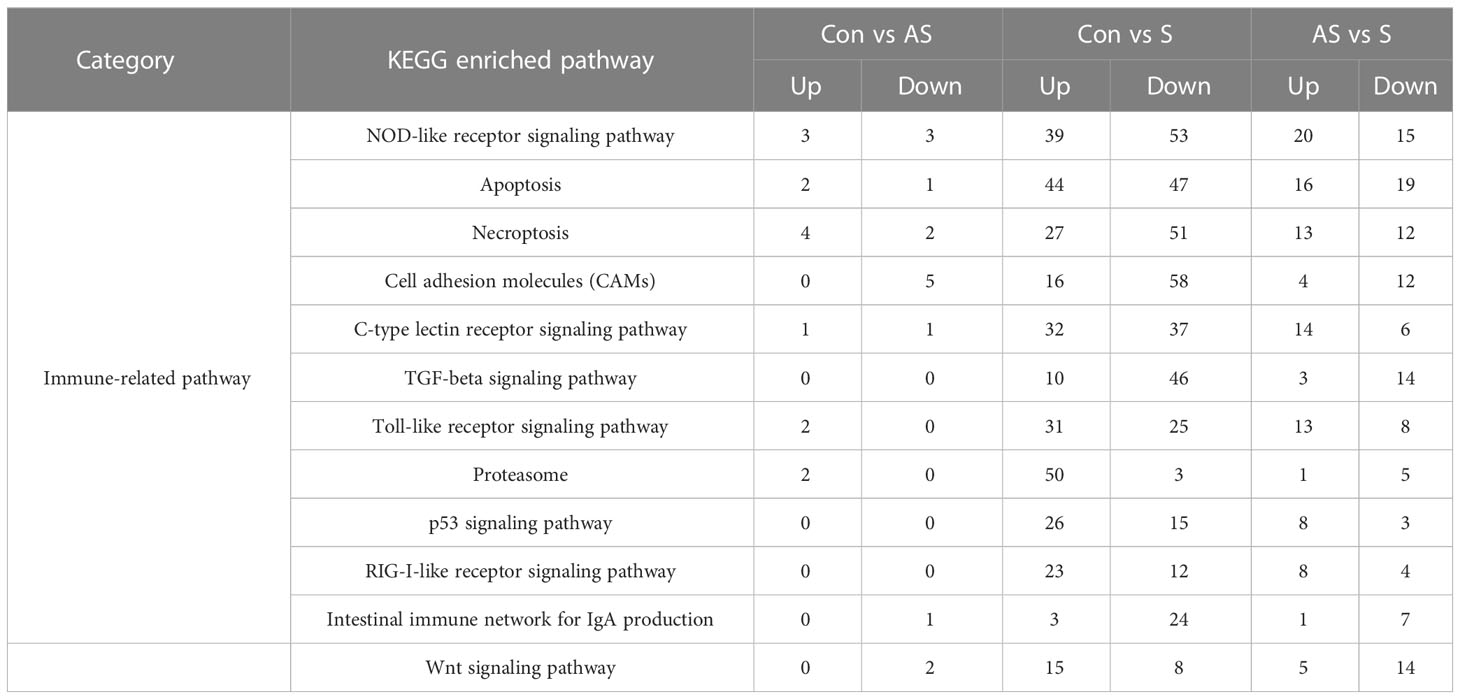
To explore which signaling pathways were involved in differentially expressed genes, the function of all differentially expressed genes in the KEGG database was annotated. A total of 88 signaling pathways were enriched between control group Koi and asymptomatic infected Koi, the pathways involved in the highest number of differentially expressed genes were mainly Ribosome (ccar03010), Lysine degradation (ccar00310), FoxO signaling pathway (ccar04068), and the differences of the pathways of Ribosome (ccar03010), Lysine degradation (ccar00310) and Glycosphingolipid biosynthesis-ganglio series (ccar00604) were the most significant. A total of 150 signaling pathways were enriched between control group and symptomatic infected group, the pathways involved in the highest number of differentially expressed genes were mainly Cellular senescence (ccar04218), NOD-like receptor signaling pathway (ccar04621), Apoptosis (ccar04210), and the differences of the pathways of Proteasome (ccar03050), Ribosome (ccar03010), ECM-receptor interaction (ccar04512) were the most significant. A total of 144 signaling pathways were enriched between asymptomatic infected Koi and symptomatic infected Koi, the pathways involved in the highest number of differentially expressed genes were mainly Cytokine-cytokine receptor interaction (ccar04060), Apoptosis (ccar04210), NOD-like receptor signaling pathway (ccar04621), and the differences of the pathways of Cytokine-cytokine receptor interaction (ccar04060), Arachidonic acid metabolism (ccar00590) and Apoptosis (ccar04210) were the most significant. The top 20 pathways with the smallest Q values were selected for KEGG enrichment analysis, and most of the signaling pathways were related to immunity (Figures 11A-F).
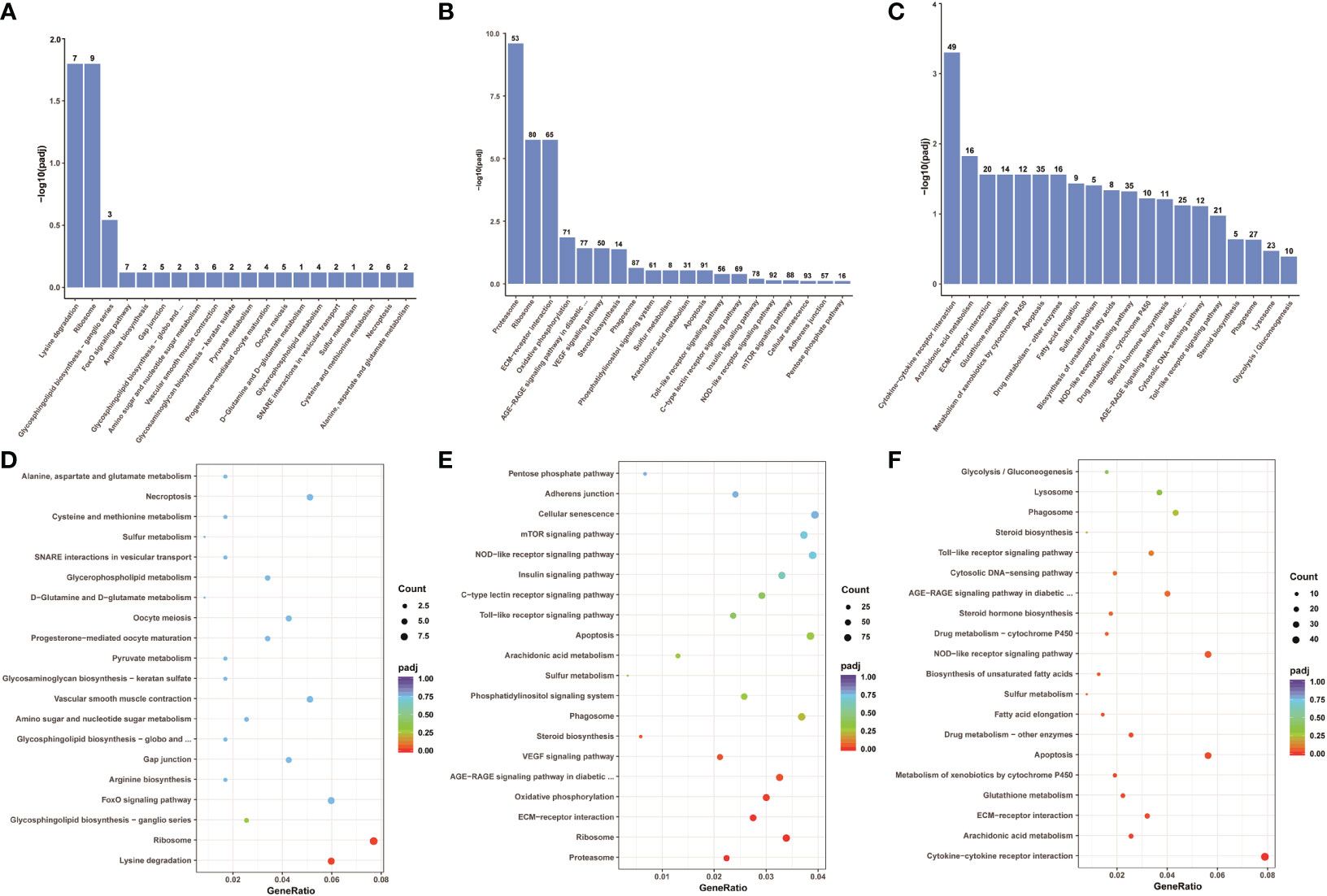
Figure 11 KEGG pathway enrichment analysis of the Koi transcriptome for all comparison groups. (A, B) The TOP 20 enriched pathway of DEGs between control group vs asymptomatic group. (C, D) The TOP 20 enriched pathway of DEGs between control group vs symptomatic group. (E, F) The top 20 enriched pathway of DEGs between asymptomatic group vs symptomatic group.
3.10 Analysis of immune-related genes and pathways in transcriptome data
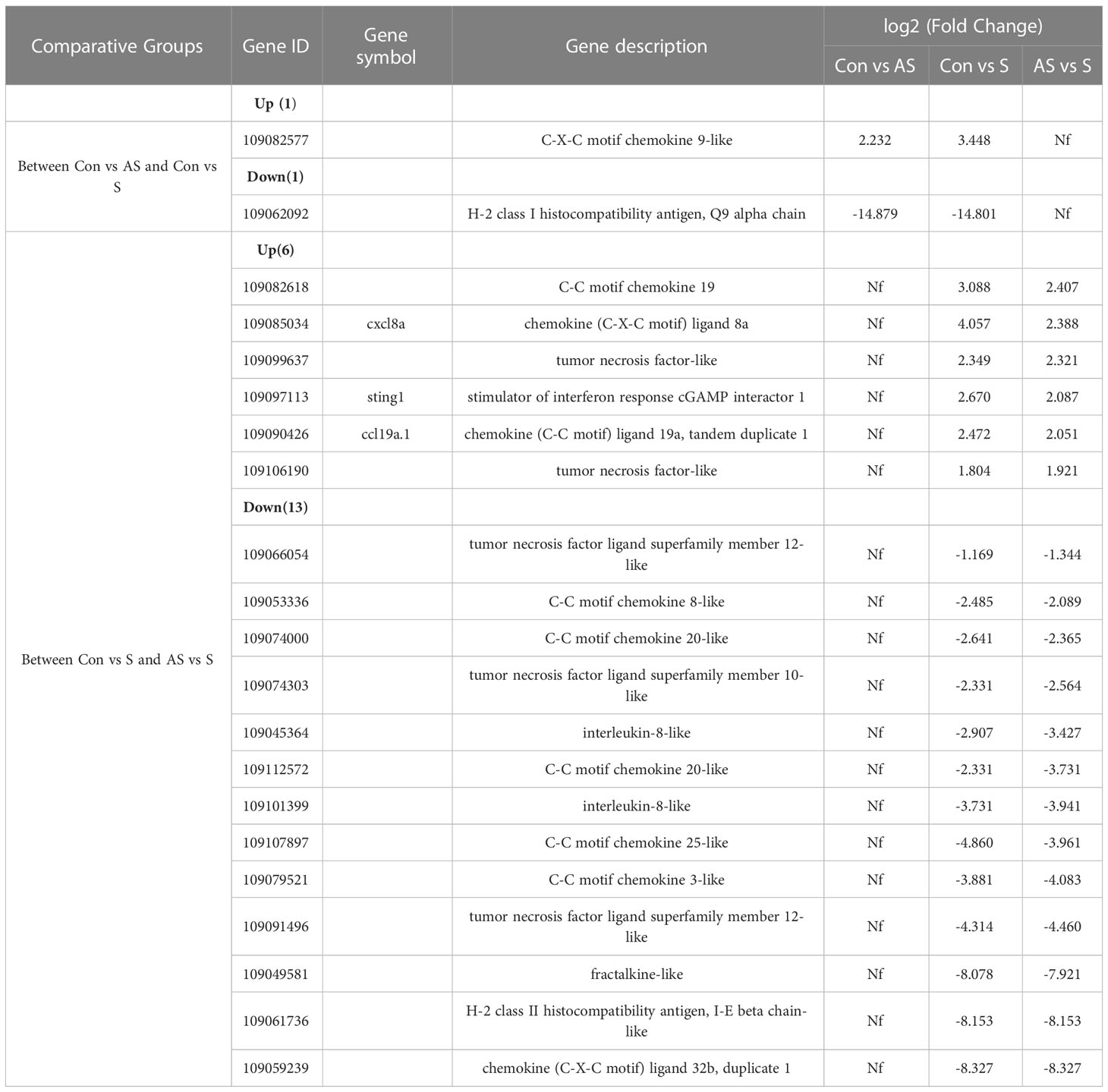
To analyze the mechanism of immunity during CEV infection, the immune-related pathways were screened (Table 3), and a bubble chart was used to clarify both the enrichment factors and gene numbers for revealed immune pathways (Figures 12A-C). The results showed that the three comparison groups mainly included NOD-like receptor signaling pathway, Toll-like receptor signaling pathway, C-type lectin receptor signaling pathway, Cell adhesion molecules (CAMs), Apoptosis and Necroptosis. In addition, in the comparison groups, among control group Koi vs asymptomatic infected Koi, control group Koi vs symptomatic infected Koi and asymptomatic infected Koi vs symptomatic infected Koi, the numbers of immune-related differential genes were 3, 59, 28, respectively (Table 4). The gene of CXC motif chemokine 9-like was significantly up-regulated during CEV infection, and the gene of H-2 class I histocompatibility antigen, Q9 alpha chain was significantly down-regulated. However, most of the genes showed significant difference expression only in the phase of symptomatic infection, the up-regulated genes mainly included cxcl8a, sting1, ccl19a.1, ccl27b and ccl20a.3, and the down-regulated genes mainly included fractalkine-like, CC motif chemokine 20-like, CC motif chemokine 25-like, CC motif chemokine 3-like, H-2 class II histocompatibility antigen, I-E beta chain-like and chemokine (CXC motif) ligand 32b, duplicate 1 (Table 5). To reveal the mechanism of immune system of gill tissue during the CEV infection from asymptomatic to symptomatic, the immune-related genes between asymptomatic and symptomatic Koi were compared (Table 6). The results showed that there were a total of nine genes upregulated, which mainly included cxcl8a, sting1, ccl19a.1 and ccl19a.2, and a total of 19 genes were down-regulated, including fractalkine-like, H-2 class II histocompatibility antigen, I-E beta chain-like, chemokine (CXC motif) ligand 32b, duplicate 1 and CXC motif chemokine 10-like, most of these genes belong to chemokines, which regulate immune responses by controlling chemotaxis of immune cells and play an important role in the process of CEV infection from asymptomatic to symptomatic.
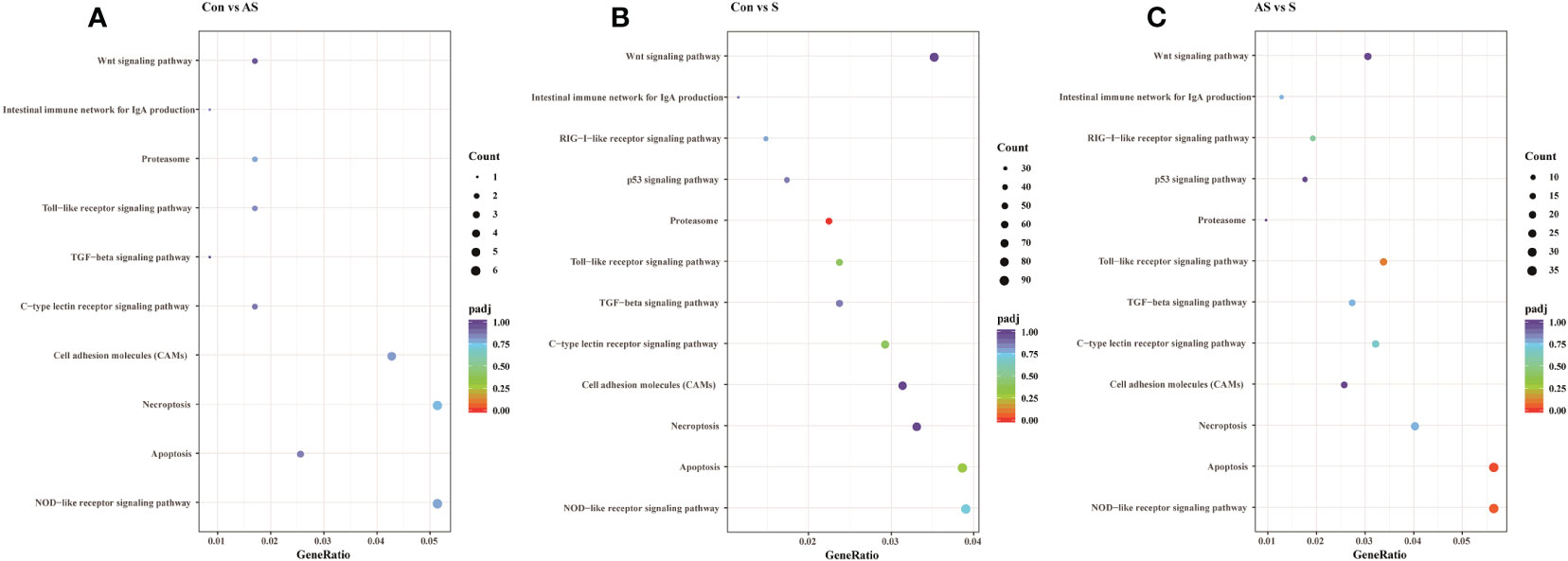
Figure 12 The pathways of immune-related DEG analyzed for the three groups. (A) The pathways of immune-related DEG for both comparison Control vs Asymptomatic group. (B) The pathways of immune-related DEG for both comparison Control vs Symptomatic group. (C) The pathways of immune-related DEG for both comparison group Asymptomatic vs Symptomatic group.
3.11 Validation of differentially expressed transcripts by RT-qPCR
To verify the validity and accuracy of transcriptome sequencing, significant DEGs were detected by RT-qPCR. CXCL8a, Sting, IRF3, IRF4a, IL11β C-X-C motif chemokine 10-like, H-2 class II histocompatibility antigen, E-S beta chain and ccl19a.2 were selected for RT-qPCR. CXCL8a, Sting, IRF3 and IL11β were up-regulated in the comparison groups between control and symptomatic Koi, and IRF4a was down-regulated in this comparison group. In addition, in the asymptomatic infected Koi vs symptomatic infected Koi, ccl19a.2 and H-2 class II histocompatibility antigen, E-S beta chain were up-regulated, and C-X-C motif chemokine 10-like was down-regulated. The results showed that the relative expression levels of DEGs were consistent with the RNA-seq data, demonstrating the validity of transcriptome analysis (Figure 13).
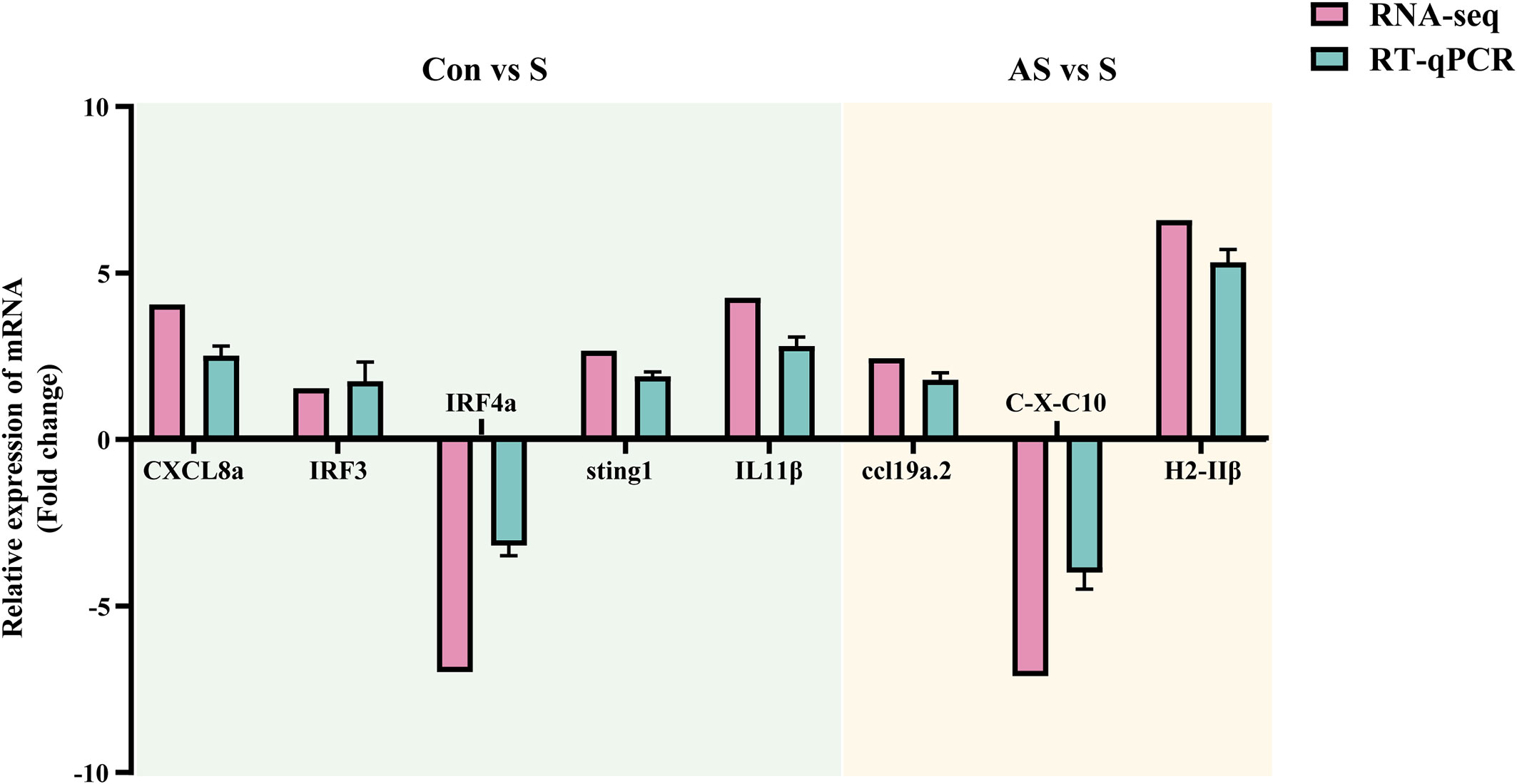
Figure 13 Validation of randomly selected DEGs by using RT-qPCR. The light blue area showed differentially expressed genes between control group Koi and symptomatic infected Koi. The light yellow area showed differentially expressed genes between asymptomatic infected and symptomatic infected Koi. Three biological replicates were set up for each sample.
4 Discussion
The mortality of KSD ranges from 5% to 100%, which may related to the virulence of CEV. The virulence of CEV strains ranges from highly pathogenic, causing death within a few days, to weakly pathogenic, causing subclinical or persistent infections with low levels of morbidity and mortality. Moreover, the subclinical or persistent infections can re-activated as acutely infection at some conditions, such as temperature changes according to seasonal changes. In this study, CEV was detected in tissues from acutely infected and asymptomatic Koi, while was not detected in control Koi by PCR. There were significant differences in pathology and viral load between asymptomatic and acutely infected Koi. The activity of immune-related enzymes, the difference changes of mucus cells and the expression of interleukins verified the differences in immune response of Koi under different infection conditions.
CEV is a double stranded DNA virus belonging to the family Poxviridae, and phylogenomic analysis has confirmed that CEV and the salmon gill poxvirus (SGPV) were sister species, and together form the deepest branch of the Chordopoxvirinae within the family Poxviridae (5). Although poxviruses were regarded as non-latent, experimental inoculation of poxviruses in mammalian cell lines has indicated that long-term latency presumably can occur (30, 31), and the poxviruses remained latent in the transfected immune cells for periods of several months to longer than 2 years (31, 32). Recently, CEV was detected in gill tissue of Koi imported from Asia to Germany, although the Koi showed no clinical symptoms (20). This might be a stress-related reactivation of a persistent CEV infection, showing a potential dispersal route of CEV, which also reported in fowl pox infections before (33). However, the current study did not pay close attention to the difference between asymptomatic and symptomatic CEV infection. Moreover, the understanding of the molecular correlates of the severity of these infections was still unknown. This study was distinguished between asymptomatic and symptomatic Koi by clinical signs, pathology changes, nested PCR and RT-qPCR. The results showed that the presence of CEV in asymptomatic Koi, and the proportion was as high as 56.5%, but the viral load of CEV was considerably low in tissues. This was basically consistent with the detection of CEV infection in asymptomatic Koi reported by Adamek (20, 34). Enzyme activity, activity of immune-related substances and expression of immune-related gene were compared at different stages during CEV infection. There were significant differences between acutely infected and asymptomatic Koi.
As a multifunctional organ, gills not only participate in the respiration of fish, but also play a significant role in metabolism, ammonia nitrogen and osmotic pressure balance (27). In addition, as one of the organs in direct contact with the external environment, gills were also the first line of defense against pathogens and play an important role in immunity (22). The main pathological changes during CEV infection were limited to the gills. The Koi infected with CEV exhibited floating heads, which is thought to be possibly due to hypoxia caused by damage to gill tissue. However, study had reported that the hemoglobin levels is rising during CEV infection (23), and fish have evolved this mechanism to cope with hypoxia (35). Severe impairment of osmoregulation was considered to be the potential cause of death of infected CyHV-3 (36). Since the Koi infected with CEV causes severe gill injury, whether severe impairment of osmoregulation was also considered to be the potential cause of death of infected CEV. The mortality rate of common carp of CEV infection can be decreased using a long-term curative 0.5% salt water bath (37), and the osmotic balance associated with hyponatremia and hyperammonemia during CEV infection have been demonstrated (24), suggesting that osmotic pressure imbalance may also be a potential cause of CEV pathogenesis (23, 24). Ammonia poisoning has a suppressive effect on innate immunity and affects the composition of immunoglobulin (38). There was also suggested that CEV could have direct immunosuppressive effect on host, but this need more investigations to be confirmed (1, 39).
The fish immune system is divided similarly to that of mammals into an innate (non-specific) and an adaptive (specific) part (40). Innate immunity is phylogenetically older than adaptive immunity and it is the first line of defence against spreading of the pathogen after its invasion into the body (40, 41). As the first line of defense against pathogen invasion, gills contain mucus cells and can secrete a large amount of mucus (28). The mucus contains a variety of active substances, such as mucopolysaccharides, glycoproteins, immunoglobulin and various enzymes, which play an important role in immune response (42). In this study, it was found that mucus cells migrated outward from the base of the branchial lamella during CEV infection. This migration was probably caused by the proliferation of the base cells of the branchial lamella, because the branchial lamella seriously proliferated during CEV infection. In addition, the number of mucus cells increased, especially when the pathological injury was more severe. This suggests that mucosal immunity plays an important role in the resistance to CEV invasion. Malondialdehyde (MDA) is a product of lipid peroxidation caused by free radicals in the body; it can polymerize with protein molecules in cells and impair control cell function by altering protein structure. MDA activity was elevated during CEV infection, and the difference was significant in Koi with clinical symptoms, suggesting that CEV infection can cause oxidative damage. The antioxidant enzymes CAT and GSH were elevated during CEV infection, glutathione peroxidase (GSH-Px) and catalase (CAT) can decompose toxic hydrogen peroxide (H2O2) into H2O and O2, which play an important role in enhancing the defense ability of phagocytes and the immune function of the body (43). AKP and ACP increased in the process of CEV infection, and the difference was significant in Koi with clinical symptoms. ACP and AKP are important phosphatases in the body, playing an important role in metabolism and immunity (44). Lysozyme can enhance the immune function of the body and play an important role in fighting against the invasion of various pathogens (45). Although lysozyme showed an increasing trend, there was no statistically significant difference. Higher expression of immune genes during the CEV infection may have inhibited viral replication and mount an antigenic adaptive response. Studies have shown that genes related to immune response were highly expressed after CEV infection, and these genes were basically involved in innate immune response (25). The results indicated that TNF-α and IL-10 were up-regulated in CEV infected koi in our study, which consistent with the previous report (25). In addition, immune-related enzyme activity also increased after CEV infection, such as lactic acid and alkaline phosphatase (23). This was consistent with our study, indicating that the enhanced innate immune response in the process of CEV infection can prevent the further spread of CEV in the body.
Besides innate immunity, fish immune system also has adaptive immunity, and immunoglobulin plays an important role as the main part of adaptive immunity (46). Unlike innate immunity, both immunoglobulin IgM and IgT were significantly decreased after CEV infection, and the down-regulation of immune-related genes IgM, CD4+ and up-regulation of IL10 further explained the inhibitory effect on adaptive immunity. In 2021, a paper reported that immune-related genes CD4, TCRα2 and IgM were significantly down-regulated in gill tissue after CEV infection, which may lead to immunosuppressive effects in the body (24). Transcriptome analysis showed that innate immunity was activated and acquired immunity was suppressed in early Vibrio alginolyticus infection of Larimichthys crocea. Innate immunity played an important role in resistance to early Vibrio alginolyticus infection of larimichthys crocea (47). Based on the above studies and analyses, adaptive immunosuppression may be induced during CEV infection, which may contribute to the immune escape of CEV and causing more serious damage to the body.
Transcriptome sequencing can effectively reflect the differential expression of different genes and play an important role in the study of pathogenic mechanism. Transcriptome analysis revealed the immune mechanism of CyHV-3 resistant carp strains, which laid a foundation for the screening of CyHV-3 resistant carp strains (48). Transcriptome analysis of the gill of Atlantic salmon (Salmo salar L.) affected by amoeba gill disease (AGD) reveals the role of tumor suppressor gene p53 in the pathogenesis of AGD, providing a good scientific basis for further prevention and treatment of AGD (49). Our studies have shown that CEV infection causes severe damage to gill tissue, in addition to dramatic changes in gill immune-related function, transcriptome analysis was performed on gill tissues of control, asymptomatic and clinically symptomatic Koi, and a large number of differentially expressed genes were annotated. GO analysis showed that in the comparison group of asymptomatic Koi vs symptomatic Koi, in biological function, the GO term enrichment of immune response and immune system response was the most significant and the number of differentially expressed genes was the largest, this suggests that immune function plays an important role in the progression from asymptomatic to symptomatic during CEV infection. In addition, the comparison group of control Koi vs symptomatic Koi showed a lot of differentially expressed immune-related genes. There was a total of 19 co-expressed genes between asymptomatic Koi vs symptomatic Koi and control Koi vs symptomatic Koi, including 6 up-regulated genes and 13 down-regulated genes. Up-regulated genes include cxcl8a, ccl19a.1, sting1 and tumor necrosis factor-like, and down-regulated genes include chemokines, such as C-C motif chemokine 8-like, C-C motif chemokine 20-like, C-C motif chemokine 25-like. CXCL8, also called interleukin-8, was a typical CXC chemokine that plays a key role in promoting inflammation (50). STING1 (also known as STING or TMEM173) was found to play a fundamental role in the production of type I interferons (IFNs) and pro-inflammatory cytokines in response to DNA derived from invading microbial pathogens or damaged hosts by activating multiple transcription factors (51). In mammals, the main target cells of CC subfamily chemokines are monocytes, macrophages and lymphocytes, but the specific chemokines of CC subfamily in fishes have not been determined (52). A total of nine genes were differentially expressed only in AS vs S group, including three up-regulated genes and six down-regulated genes. These differentially expressed genes were important in the process of CEV infection, especially the up-regulated genes H-2 class II histocompatibility antigen, E-S beta chain and ccl19a.2, those genes may the marker gene that makes Koi develop from asymptomatic to clinically symptomatic. KEGG enrichment analysis showed that many immune-related pathways were involved in the regulation of these genes, and NOD-like receptor signaling pathway, Cell adhesion molecules (CAMs), C-type lectin receptor signaling pathway, Toll-like receptor signaling pathway is the main enrichment pathway. NOD-like receptors, Toll-like receptors (TLRs) and C-Lectins are principal receptors of pattern-recognition receptors (PRRs), and the aim was to recognize pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) resulting in mobilization of downstream signaling responses leading to inflammation, microbial destruction and eventually activation of adaptive immune responses, playing an important role in resisting the invasion of pathogens (53, 54). These pathways may play an important role in immune response and show significant different in the course of CEV infection. How they regulate the changes in immune response, especially from asymptomatic to symptomatic infection, will be an interesting topic and worthy for further study.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJNA925573 (SRA).
Ethics statement
The animal study was reviewed and approved by Animal Care and Use Committee of Sichuan Agricultural University.
Author contributions
PO, YR, YZ, QL, DC and LY participated in material preparation and experimental investigation; QL, YG, HG, JF and HD performed research and analyzed results; YR and XH analyzed RNA-Seq data; WL prepared the experiment materials; ZC and GS developed methodology and discussed results; YR and LY designed research, wrote the manuscript and supervised the study. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Sichuan International Science and Technology Innovation Cooperation Foundation (grant number 2020YFH0156) and a funding from Chengdu Science and Technology Bureau (grant number 2022YF0501021SN).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Machat R, Pojezdal L, Piackova V, Faldyna M. Carp edema virus and immune response in carp (Cyprinus carpio): Current knowledge. J Fish Dis (2021) 44(4):371–8. doi: 10.1111/jfd.13335
2. Padhi SK, Tolo I, McEachran M, Primus A, Mor SK, Phelps NBD. Koi herpesvirus and carp oedema virus: Infections and coinfections during mortality events of wild common carp in the united states. J Fish Dis (2019) 42(11):1609–21. doi: 10.1111/jfd.13082
3. Bergmann SM, Jin Y, Franzke K, Grunow B, Wang Q, Klafack S. Koi herpesvirus (Khv) and khv disease (Khvd) - a recently updated overview. J Appl Microbiol (2020) 129(1):98–103. doi: 10.1111/jam.14616
4. Rehman T, Yin L, Latif MB, Zhou Y, Wang K, Geng Y, et al. Current findings on carp edema virus, control challenges, and future outlook. Aquacult Int (2020) 28(5):2015–26. doi: 10.1007/s10499-020-00573-6
5. Haenen O, Way K, Gorgoglione B, Ito T, Paley R, Bigarre L, et al. Novel viral infections threatening cyprinid fish. Bull OF THE Eur Assoc OF FISH PATHOL (2016) 36(1):11–23.
6. Lei Y. (2017). Important diseases of aquatic animals in the Asia-pacific region. Proceedings of the International Symposium on Health of Aquatic Animals and Nutrition Immunology 238-266+268-294, Guangzhou, Guangdong, China.
7. Lovy J, Friend SE, Al-Hussinee L, Waltzek TB. First report of carp edema virus in the mortality of wild common carp cyprinus carpio in north America. Dis Aquat Organ (2018) 131(3):177–86. doi: 10.3354/dao03296
8. Ouyang P, Zhou Y, Yang R, Yang Z, Wang K, Geng Y, et al. Outbreak of carp edema virus disease in cultured ornamental koi in a lower temperature in China. Aquacult Int (2019) 28(2):525–37. doi: 10.1007/s10499-019-00476-1
9. Hedrick RP, Antonio DB, Munn RJ. Poxvirus like agent associated with epizootic mortality in juvenile koi (Cyprinus carpio). FHS Newslett (1997) 25(3):1–2.
10. Adamek MA-O, Heling M, Bauer JA-O, Teitge F, Bergmann SM, Kleingeld DW, et al. It is everywhere-a survey on the presence of carp edema virus in carp populations in Germany. Transbound Emerg Dis. (2022) 69(4):2227–41. doi: 10.1111/tbed.14225
11. Marsella A, Pretto T, Abbadi M, Quartesan R, Cortinovis L, Fiocchi E, et al. Carp edema virus-related mortality in wild adult common carp (Cyprinus carpio) in Italy. J Fish Dis (2021) 44(7):939–47. doi: 10.1111/jfd.13353
12. Baud M, Pallandre L, Almeras F, Maillet L, Stone D, Bigarré L. Genetic diversity of the carp oedema virus in France. J Fish Dis (2021) 44(10):1531–42. doi: 10.1111/jfd.13474
13. Kim SW, Jun JW, Giri SS, Chi C, Yun S, Kim HJ, et al. First report of carp oedema virus infection of koi (Cyprinus carpio haematopterus) in the republic of Korea. Transbound Emerg Dis (2018) 65(2):315–20. doi: 10.1111/tbed.12782
14. Luo F, Lian Z, Niu Y, Lü A, Hu X, Xie X, et al. Molecular characterization of carp edema virus disease: An emerging threat to koi cyprinus carpio in China. Microb Pathog (2020) 149:104551. doi: 10.1016/j.micpath.2020.104551
15. Ouyang P, Yang R, Chen J, Wang K, Geng Y, Lai W, et al. First detection of carp edema virus in association with cyprinid herpesvirus 3 in cultured ornamental koi, cyprinus carpio l., in China. Aquaculture (2018) 490:162–8. doi: 10.1016/j.aquaculture.2018.02.037
16. Miyazaki T, Isshiki T, Katsuyuki H. Histopathological and electron microscopy studies on sleepy disease of koi cyprinus carpio koi in Japan. Dis Aquat Organ (2005) 65(3):197–207. doi: 10.3354/dao065197
17. Way K, Haenen O, Stone D, Adamek M, Bergmann SM, Bigarre L, et al. Emergence of carp edema virus (Cev) and its significance to European common carp and koi cyprinus carpio. Dis Aquat Organ (2017) 126(2):155–66. doi: 10.3354/dao03164
18. Pragyan D, Bajpai V, Suman K, Mohanty J, Sahoo PK. A review of current understanding on carp edema virus (CEV): A threatful entity in disguise. International Journal of Fisheries and Aquatic Studies (2019) 7(5):87–93.
19. Radosavljevic V, Adamek MA-O, Milicevic V, Maksimovic-Zoric J, Steinhagen DA-O. Occurrence of two novel viral pathogens (CEV and CyHV-2) affecting Serbian cyprinid aquaculture and ichthyofauna. J Fish Dis (2018) 41(5):851–4. doi: 10.1111/jfd.12789
20. Adamek M, Jung-Schroers V, Hellmann J, Teitge F, Bergmann SM, Runge M, et al. Concentration of carp edema virus (Cev) DNA in koi tissues affected by koi sleepy disease (Ksd). Dis Aquat Organ (2016) 119(3):245–51. doi: 10.3354/dao02994
21. lv X, Xu L, Zhang W, Wang N, Cao H, Wang X, et al. Research on histopathological changes and virus distribution situation of mirror carps infected with carp edema virus. Chineseanimal husbandry veterinary Med (2022) 49(03):1077–84. doi: 10.16431/j.cnki.1671-7236.2022.03.030
22. Koppang EO, Kvellestad A, Fischer U. Fish mucosal immunity: Gill. Mucosal Health Aquacult (2015) . p:93–133. doi: 10.1016/B978-0-12-417186-2.00005-4
23. Pikula J, Pojezdal L, Papezikova I, Minarova H, Mikulikova I, Bandouchova H, et al. Carp edema virus infection is associated with severe metabolic disturbance in fish. Front Vet Sci (2021) 8:679970. doi: 10.3389/fvets.2021.679970
24. Adamek M, Teitge F, Baumann I, Jung-Schroers V, El Rahman SA, Paley R, et al. Koi sleepy disease as a pathophysiological and immunological consequence of a branchial infection of common carp with carp edema virus. Virulence (2021) 12(1):1855–83. doi: 10.1080/21505594.2021.1948286
25. Kushala KB, Nithin MS, Girisha SK, Dheeraj SB, Sowndarya NS, Puneeth TG, et al. Fish immune responses to natural infection with carp edema virus (Koi sleepy disease): An emerging fish disease in India. Fish Shellfish Immunol (2022) 130:624–34. doi: 10.1016/j.fsi.2022.09.012
26. RURALAFFAIRS MOAA. Code of diagnosis for carp edema virus disease (Cevd) Sc/T7229- 2019. Beijing China Agriculture Press (2019).
27. Evans DH, Piermarini PM, Choe KP. The multifunctional fish gill: Dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev (2005) 85(1):97–177. doi: 10.1152/physrev.00050.2003
28. Salinas I, Fernandez-Montero A, Ding Y, Sunyer JO. Mucosal immunoglobulins of teleost fish: A decade of advances. Dev Comp Immunol (2021) 121:104079. doi: 10.1016/j.dci.2021.104079
29. Van Muiswinkel WB, Nakao M. A short history of research on immunity to infectious diseases in fish. Dev Comp Immunol (2014) 43(2):130–50. doi: 10.1016/j.dci.2013.08.016
30. Pogo BG, Friend C. Persistent infection of friend erythroleukemia cells with vaccinia virus. Proceedings of the National Academy of Sciences (1982) 79(15):4805–480. doi: 10.1073/pnas.79.15.4805
31. Buller RM, Palumbo GJ. Poxvirus pathogenesis. Microbiol Rev (1991) 55(1):80–122. doi: 10.1128/mr.55.1.80-122.1991
32. Abdelsalam EEE, Piačková V. Carp edema virus: Host selection and interaction, and potential factors affecting its introduction to the common carp population, distribution, and survival. Aquaculture (2023) 563:739009. doi: 10.1016/j.aquaculture.2022.739009
33. Lachish S, Bonsall MB, Lawson B, Cunningham AA, Sheldon BC. Individual and population-level impacts of an emerging poxvirus disease in a wild population of great tits. PloS One (2012) 7(11):e48545. doi: 10.1371/journal.pone.0048545
34. Adamek M, Oschilewski A, Wohlsein P, Jung-Schroers V, Teitge F, Dawson A, et al. Experimental infections of different carp strains with the carp edema virus (Cev) give insights into the infection biology of the virus and indicate possible solutions to problems caused by koi sleepy disease (Ksd) in carp aquaculture. Vet Res (2017) 48(1):12. doi: 10.1186/s13567-017-0416-7
35. Wells RMG. Chapter 6 blood-gas transport and hemoglobin function: Adaptations for functional and environmental hypoxia. Fish Physiol (2009) 27:255–99. doi: 10.1016/S1546-5098(08)00006-X
36. Negenborn J, van der Marel MC, Ganter M, Steinhagen D. Cyprinid herpesvirus-3 (Cyhv-3) disturbs osmotic balance in carp (Cyprinus carpio l.)–a potential cause of mortality. Vet Microbiol (2015) 177(3-4):280–8. doi: 10.1016/j.vetmic.2015.03.018
37. Séno R, Hata N, Oyamatsu T, Fukuda H. Curative effect of 0.5% salt water treatment on carp, cyprinus carpio, infected with carp edema virus (CEV) results mainly from reviving the physiological condition of the host. Aquac Sci (2003) 51(1):123–4. doi: 10.11233/AQUACULTURESCI1953.51.123
38. Wang L, Wang H, Shi W, Zhang Y, Zhu C, Pan Z, et al. Identification of genes and signaling pathways associated with immune response of hemibarbus maculatus (Bleeker, 1871) to ammonia stress. Aquaculture (2020) 524:735265. doi: 10.1016/j.aquaculture.2020.735265
39. Adamek M, Baska F, Vincze B, Steinhagen D. Carp edema virus from three genogroups is present in common carp in Hungary. Journal of fish diseases (2018) 41(3):463–68. doi: 10.1111/jfd.12744
40. Magnadottir B. Immunological control of fish diseases. Mar Biotechnol (NY) (2010) 12(4):361–79. doi: 10.1007/s10126-010-9279-x
41. Uribe C, Folch H, Enríquez R, Moran G. Innate and adaptive immunity in teleost fish: A review. Veterinarni Medicina (2011) 56:486–503. doi: 10.17221/3294-VETMED
42. Salinas I. The mucosal immune system of teleost fish. Biol (Basel) (2015) 4(3):525–39. doi: 10.3390/biology4030525
43. Gayashani Sandamalika WM, Kwon H, Lim C, Yang H, Lee J. The possible role of catalase in innate immunity and diminution of cellular oxidative stress: Insights into its molecular characteristics, antioxidant activity, DNA protection, and transcriptional regulation in response to immune stimuli in yellowtail clownfish (Amphiprion clarkii). Fish Shellfish Immunol (2021) 113:106–17. doi: 10.1016/j.fsi.2021.03.022
44. Ray GW, Liang D, Qh Y, Tan B, Xh D, Sy C, et al. Effects of replacing fishmeal with dietary soybean protein concentrate (Spc) on growth, serum biochemical indices, and antioxidative functions for juvenile shrimp litopenaeus vannamei. Aquaculture (2020) 516:734630. doi: 10.1016/j.aquaculture.2019.734630
45. Chen H, Chen X, Song TY, Ge JO. Expression profiles of zebrafish (Danio rerio) lysozymes and preparation of c-type lysozyme with high bacteriolytic activity against vibrio vulnificus. Antibiotics (Basel) (2022) 11(12):1803. doi: 10.3390/Antibiotics11121803
46. Huang X, Liu S, Zuo F, Luo L, Chen D, Ou Y, et al. Cmos enhanced the mucosal immune function of skin and gill of goldfish (Carassius auratus Linnaeus) to improve the resistance to ichthyophthirius multifiliis infection. Fish Shellfish Immunol (2022) 126:1–11. doi: 10.1016/j.fsi.2022.05.024
47. Chen A, Wang Y, Weng H, Chen X, Zhang W. Transcriptome analysis revealed the immune response characteristics of larimichthys crocea infected with vibrio alginolyticus. Acta Hydrobiol Sin (2022) 46(12):1845–54.
48. Jia Z, Wu N, Jiang X, Li H, Sun J, Shi M, et al. Integrative transcriptomic analysis reveals the immune mechanism for a cyhv-3-Resistant common carp strain. Front Immunol (2021) 12:687151. doi: 10.3389/fimmu.2021.687151
49. Morrison RN, Cooper GA, Koop BF, Rise ML, Bridle AR, Adams MB, et al. Transcriptome profiling the gills of amoebic gill disease (Agd)-affected Atlantic salmon (Salmo salar l.): A role for tumor suppressor P53 in agd pathogenesis? Physiol Genomics (2006) 26(1):15–34. doi: 10.1152/physiolgenomics.00320.2005
50. Zhou S, Mu Y, Ao J, Chen X. Molecular characterization and functional activity of Cxcl8_L3 in Large yellow croaker larimichthys crocea. Fish Shellfish Immunol (2018) 75:124–31. doi: 10.1016/j.fsi.2017.12.052
51. Zhang R, Kang R, Tang D. The Sting1 network regulates autophagy and cell death. Signal Transduct Target Ther (2021) 6(1):208. doi: 10.1038/s41392-021-00613-4
52. Ding Z, Cui H, Gu Z. Review on chemokines reseach in toleost fish. J Fishery Sci China (2021) 28(09):1227–37. doi: 10.12264/JFSC2021-0017
53. Ao J, Chen X. Research progress on pattern-recognition receptors in fish. Chin Bull Life Sci (2012) 24(09):1049–54. doi: 10.13376/j.cbls/2012.09.007
Keywords: Koi, carp edema virus (CEV), gill, immune response, RNA-seq
Citation: Ouyang P, Ren Y, Zhou Y, Li Q, Huang X, Chen D, Geng Y, Guo H, Fang J, Deng H, Lai W, Chen Z, Shu G and Yin L (2023) Characteristics of pathology and transcriptome profiling reveal features of immune response of acutely infected and asymptomatic infected of carp edema virus in Koi. Front. Immunol. 14:1142830. doi: 10.3389/fimmu.2023.1142830
Received: 12 January 2023; Accepted: 06 February 2023;
Published: 27 February 2023.
Edited by:
Joel Henrique Ellwanger, Federal University of Rio Grande do Sul, BrazilReviewed by:
Almas Jabeen, Jamia Millia Islamia, IndiaMarek Matras, National Veterinary Research Institute (NVRI), Poland
Copyright © 2023 Ouyang, Ren, Zhou, Li, Huang, Chen, Geng, Guo, Fang, Deng, Lai, Chen, Shu and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lizi Yin, eWlubGl6aUBzaWNhdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Ping Ouyang
Ping Ouyang Yongqiang Ren
Yongqiang Ren Yongheng Zhou1
Yongheng Zhou1 Yi Geng
Yi Geng Hongrui Guo
Hongrui Guo Jing Fang
Jing Fang Huidan Deng
Huidan Deng Zhengli Chen
Zhengli Chen Gang Shu
Gang Shu Lizi Yin
Lizi Yin