
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 28 March 2023
Sec. Microbial Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1140627
Introduction: Upon infection, insect hosts simultaneously express a cocktail of antimicrobial peptides (AMPs) which can impede pathogen colonization and increase host fitness. It has been proposed that such a cocktail might be adaptive if the effects of co-expressed AMPs are greater than the sum of individual activities. This could potentially prevent the evolution of bacterial resistance. However, in vivo studies on AMPs in combination are scarce. Attacins are one of the relatively large AMP families, which show anti-Gram-negative activity in vitro.
Material and methods: Here, we used RNA interference (RNAi) to silence three members of the Attacin family genes in the mealworm beetle, Tenebrio molitor: (TmAttacin1a (TmAtt1a), TmAttacin1b (TmAtt1b), and TmAttacin2 (TmAtt2) both individually and in combination. We then infected T. molitor with the Gram negative entomopathogen Pseudomonas entomophila.
Results: We found that survival of the beetles was only affected by the knockdown of TmAttacin1b, TmAttacin2 and the knockdown of all three Attacins together. Triple knockdown, rather than individual or double knockdowns of AMPs, changes the temporal dynamics of their efficiency in controlling the colonization of P. entomophila in the insect body.
Discussion: More precisely, AMP gene expression influences P. entomophila load early in the infection process, resulting in differences in host survival. Our results highlight the importance of studying AMP-interactions in vivo.
Hosts ranging from insects to mammals use conserved innate immune pathways for effective protection from pathogens. The downstream effectors of NF-κB signaling pathways, the antimicrobial peptides (AMPs) or host defense peptides, are inducible short proteins that play versatile roles in host physiology such as antimicrobial and anti-tumor activities and wound healing (1–4). Prior in vitro studies have found that AMP expression is long-lasting in several insect species and, remarkably, the expressed cocktails contain some AMPs that are not effective against the infective agent in isolation (5–8). Insects have been reported to simultaneously synthesize an effective repertoire of AMP combinations, either AMP members of the same family or different families (9–11). In addition, AMPs exhibit a greater effect in combination with other AMPs, which suggests that the immune system produces synergistic combinations to enhance their activities upon infections (3, 12).
Until recently, little was known about the in vivo AMP activities of single AMPs or AMPs in combination. Zanchi et al. (3) reported the importance of AMP cocktails in determining survival in persistent experimental Staphylococcus aureus infections, using RNAi-based single and double knockdown of three highly induced Tenebrio molitor AMPs (Tenecin 1, Tenecin 2, and Tenecin 4). In another study using CRISPR/Cas9-Induced loss-of-function, Hanson et al. (13) knocked out 10 out of 14 AMPs individually and in selected combinations and generated three group of flies including Defensin mutants, Drosocin/Attacins/Diptericins mutants, and Metchnikowin/Drosomycin mutants. They found that there are either additive or synergistic combinations of AMPs with either anti-Gram-negative or anti-fungal activities in Drosophila melanogaster, as well as in vivo specific AMP-pathogen interactions (3, 13). Moreover, insects express several AMPs of the same family simultaneously, including within the Attacin gene family.
After the first identification of Attacins in Hyalophora cecropia in 1983 (14), different Attacins from other insect orders including Diptera (D. melanogaster, Glossina morsitans, and Hermetia illucens) (15–18), Lepidoptera (Hyphantria cunea, Spodoptera exigua) (19–21), and Coleoptera (T. molitor) (22) have been described, and their antimicrobial activities have been investigated. The widespread presence of Attacin peptides in numerous insect orders suggest that Attacins are evolutionarily conserved (23, 24). Attacins as relatively large proteins have antimicrobial activities against Gram-negative bacteria (e.g., Escherichia coli) (15, 16, 19, 22). Moreover, earlier studies underlined potential pharmacological applications of Attacins. For example, in vitro inhibitory activity of the parasite, Trypanosoma brucei by the purified recombinant Attacin from G. m. morsitans (15), and also antibacterial activity against methicillin-resistant S. aureus (MRSA) by a recombinant Attacin from H. illucens (17). However, these studies either characterized the function of the Attacins in vivo or investigated the in vitro antimicrobial activity individually. The extent, however, to which in vitro activities of AMPs reflect the effects of their expression in an organism during an infection remains unclear. More studies are needed to better understand how AMPs act singly or in a mixture with other AMPs in vivo. For example, if the interactions between AMPs modify their spectrum of action. Alternatively, it could be proposed that the simultaneous expression of AMPs of the same family offers a certain degree of redundancy in the AMP cocktail. Such a redundancy could be adaptive if it could make the insect host resilient to the impairment of the activity of one or several AMPs during an interaction with a pathogen. Here we use gene knockdowns to study the in vivo effects of combinations of T. molitor Attacins in response to P. entomophila infection.
We use the mealworm beetle T. molitor as a model host to gain insight into the host-pathogen interaction upon bacterial infection and to uncover the importance combinations of Attacins. Previous studies have highlighted that antimicrobial activities of T. molitor AMPs in vitro do not reflect activities in vivo. There are several possible reasons for this such as synergisms among AMPs and local pH and salt concentration (3, 25). To date, there are five identified families of AMPs in T. molitor including the Tenecin (25, 26), Attacin (22), Defensin (27), Coleoptericin (28), Cecropin (29), and Thaumatin-like protein families (30). Some members of these families are named Tenecins for historical reasons.
P. entomophila is an entomopathogenic, Gram-negative bacterium that stimulates the induction of the immune deficiency (Imd) pathway in adult and larval D. melanogaster both systemically and locally in the gut following digestion. This results in the production of AMPs (31). P. entomophila has been shown to be highly pathogenic in a number of insect species, but is benign towards plants, suggesting that P. entomophila is a useful model to study host-pathogen interactions and a potential candidate for biocontrol agents (32). Previous studies of local and systemic innate immune responses in D. melanogaster and Drosophila suzukii after P. entomophila infection further support this conclusion (32–34):
We here knockdown members of one AMP family in vivo for the first time, investigating the effect of knocking down individual AMPs and their combinations on host response to infection with P. entomophila. We address whether depletion of several members of the Attacin gene family in T. molitor (TmAttacins) influences host survival and bacterial load, taking advantage of RNAi to provide single-, double-, and triple-knockdown of the Attacin family genes.
Early instar mealworms were purchased from a commercial supplier (Reptile Food Handels-u. Zucht GmbH, Berlin, Germany). A group of 500 larvae were maintained in plastic containers (20 cm × 20 cm × 9 cm). They were fed wheat bran ad libitum and kept in the dark at 25 ± 3°C. Larvae were supplemented with a piece of fresh apple every two days as a source of moisture and nutrition. Every 48 h pupae were collected and sexed using a dissecting microscope (ZEISS stereo microscope STEMI 305). Previous studies have shown sexual dimorphism in immune function in T. molitor. Therefore, to reduce variation in survival and/or immune traits’ expression due to sex, only females were retained for experiments. These females were kept under the controlled conditions from eclosion onwards (35). All experimental individuals were 9-11 days-old and weighed 0.11-0.14 g.
To examine whether P. entomophila infection influences the expression levels of three TmAttacin genes, T. molitor females were exposed to either PBS or P. entomophila in PBS and expression changes in the Attacin genes confirmed via qRT-PCR. Following this confirmation, single- (TmAtt1a, TmAtt1b, and TmAtt2), double- (TmAtt1a-Att2 and TmAtt1b-Att2), and triple-knockdowns (TmAtt1a-Att1b-Att2) were performed using RNAi. Double-stranded enhanced green fluorescent protein (dsEGFP) was injected as a negative control for the nonspecific effects of dsRNA. In each combination or single knockdown (KD) experiment, we needed to determine the time point at which the depletion in transcripts of our gene (s) of interest compared to dsEGFP was significant in T. molitor females. Therefore, efficiency of RNAi target gene suppression was evaluated in adult females collected from dsRNA- and dsEGFP-treated beetles. Based on these data, we selected the time points for P. entomophila infection. No infections were carried out before wounds from the knockdown injection had sufficiently healed. The immune tissues (haemolymph, fat body, gut, and Malpighian tubules) were dissected from infected beetles of all treatments for the first three days following infection and the survival of a second set of beetles was monitored daily for 35 days in parallel. A previous study on S. aureus-infected T. molitor females suggests that this time window captures most of the mortality caused by the infection without being confounded by senescence (3).
The beetles used in the survival assays were assigned to one of the following treatments: eitheruntreated control (designated as full control) or one of the three dsRNA-treated groups, dsEGFP (negative control) or gene (s) of interest-dsRNA (single-, double-, and triple-knockdown) groups. Each of these groups were divided into two subgroups, injected with PBS (control) or P. entomophila (Figure S1).
For bacterial load assays, colony forming units (CFU) were recovered from dsRNA-treated beetles only, namely dsEGFP and dsRNA (target gene (s) following P. entomophila infection (Figure S1).
P. entomophila L48 (provided by Alexandro Rodríguez-Rojas) was cultured from 50% glycerol frozen stock at 28 ± 2°C in ten mL of Luria-Bertani (LB) broth overnight. 100 μL of overnight culture was harvested and reincubated in ten mL LB broth for a further 2 h under the same conditions. Cultures were interrupted when an optical density of 0.5 at 600 nm (OD600) was reached, measured by spectrophotometer (Ultrospec 10, Amersham Biosciences). The subcultured bacteria were concentrated by centrifugation and the cell pellet was washed three times with phosphate buffered saline (PBS) and then was resuspended. Based on the OD600, the culture was adjusted to 1 × 104 cells/µL.
Infections were performed by injecting dsRNA-treated adults in the intersegmental membrane, between the fourth and fifth abdominal sternites, parallel to the anterior-posterior axis with a capillary needle filled with 5 μL of 1 × 104 cells/µL of P. entomophila. Based on preliminary experiments, this infection concentration was likely to yield progressive mortality over time (Figure S2), allowing us to detect the effect of the various knockdowns of TmAttacin genes. After treatment, beetles were kept in cohorts of 15 individuals.
In T. molitor, it was shown that last-instar larvae induce TmAtt1a and -1b in response to E. coli, suggesting the anti-Gram-negative activity of TmAttacin family (22). Thus, to elucidate whether this effect is also elicited by P. entomophila, we examined changes in the transcriptional abundance of TmAttacin1a, TmAttacin1b, and TmAttacin2 following infection. To do so, we injected P. entomophila into female beetles (9-11 days-old) and measured changes in the expression patterns in our genes of interest using relative quantitative PCR (qRT-PCR). Four individual beetles were pooled together and homogenized with a pestle in liquid nitrogen before being transferred to 2 mL microcentrifuge tubes (Safe-Lock tubes, Eppendorf). Total mRNA was extracted using the Quick-RNA Tissue/Insect Microprep Kit (ZYMO Research Europe GmbH), following the manufacturer’s instructions. The resulting mRNA was stored at -80°C until being used for qRT-PCR. qRT-PCR was performed using Power SYBR™ Green RNA-to-CT™ 1-Step Kit (Applied Biosystems TM) with extracted mRNA and specific primers as listed in Table S1. The PCR amplification conditions were as follows: 95 °C for 5 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 30 s. Two technical replicates were carried out for each gene of interest along with the housekeeping gene T. molitor 60S ribosomal protein L27a (TmL27a) and averaged for each gene. The mean Ct of the AMP gene of interest was normalized using the mean Ct of TmL27a by calculating the mean Ct TmL27a - mean Ct of the gene of interest (delta Ct). Gene expression levels were expressed as 2^ (delta-delta Ct) (36).
To silence target gene expression by RNAi, total RNA was extracted (Direct-zol RNA Miniprep Plus Kits, ZYMO Research) from the late instar larvae showing the highest enrichment of TmAttacin1a (GenBank accession No. MF754109), TmAttacin1b (MF754110), TmAttacin2 (MF754108) (22) (Figures S3–S5), then cDNA fragments corresponding to each gene were produced using RevertAid™ Premium First Strand-cDNA-Synthese kit according to the manufacturer’s instructions. The cDNA was used as a template to amplify the fragments by PCR (KAPA2G Fast ReadyMix PCR Kit, KAPA Biosystems), following gene-specific primers tagged with a T7-promotor sequence in both 3’ and 5’ ends (Table S1). The PCR conditions were as follows: 95 °C for 2 min, followed by 30 cycles of denaturation at 95 °C for 20 s, annealing at 56 °C for 30 s, and extension at 72 °C for 5 min. A 508 bp PCR product of the Enhanced Green Fluorescent Protein (EGFP) gene derived from the plasmid (pGEM T-easy-GFP, Promega) was similarly amplified and used as a control for dsRNA (Table S1). After checking the length of the amplicons by running them on a 2% agarose gel, the PCR products were cleaned up using a kit (PCR/DNA Clean-Up DNA Kit, Roboklon). Using the resulting amplicons as template, in vitro transcription was carried out using a kit (HighYield T7 RNA Synthesis Kit, Jena Bioscience) as per the manufacturer’s recommendations. Next, the synthesized dsRNA was washed, and the RNA pellet was resuspended in nuclease-free water and kept at -20°C until further use.
To address the individual and combined functional importance of TmAttacin family genes in T. molitor immunity in vivo, we selectively silenced one, two or three representative genes by RNAi. In the case of single-knockdowns (TmAtt1a, TmAtt1b, and TmAtt2), adult females were injected with 1000 ± 100 ng of dsRNA (500 μg/μL in 2 μL), for double-knockdowns 2000 ± 100 ng (1000 μg/μL in 2 μL) (TmAtt1a-Att2 and TmAtt1b-Att2), and for triple-knockdown (TmAtt1a-Att1b-Att2) 3000 ± 100 ng of dsRNA in 6 µl total volume of nuclease-free water (i.e., circa 1000 ng for each representative genes). The same concentration of dsEGFP was injected for control insects, giving three control groups for single-dose, double-dose, and triple-dose controls, respectively.
To confirm RNAi efficiency of target genes, total RNA was extracted (n=4, pools of four adults per day) for single- (at first week and 14-day), double- (3-, 4- or 5-day), and triple-knockdown (4- and 5-day) at different time points post-exposure and then qRT-PCR carried out as described in section 2.4.
Next, to evaluate the viability of T. molitor females exposed to bacteria after silencing the target genes, there were six treatments in each set (single-, double-, and triple-knockdown) namely, full control - PBS, full control - P. entomophila, dsEGFP - PBS, dsEGFP - P. entomophila, dsRNA - PBS, dsRNA - P. entomophila. Survival assays were replicated twice in a separate set of beetles and bacterial cultures.
To address whether P. entomophila successfully survive in the T. molitor tissues or are killed by the host immune system, the immune tissues of TmAttacin knockdown and dsEGFP beetles were dissected on days 1, 2, and 3 post-infections. Sampling was stopped at this time point due to a high number of dead beetles, which led to a reduced sample size. We collected haemolymph, fat body, gut, and Malpighian tubules of each T. molitor female before pooling and homogenizing them in a 2 mL microcentrifuge tube (Safe-Lock tubes, Eppendorf) containing 250 μL of LB broth and two stainless steel beads (Ø 3 mm, Retsch) on ice. Collected tissues were homogenized at a frequency of 30 Hz for 20 seconds using a tissue homogenizer (Mill MM400, Retsch) before being centrifuged at 420 × g for 1 minute at 4 °C. Homogenates from each beetle were added into 180 μL of PBS and then serially diluted 1:10 to 1:105 (Table S2). Isolated P. entomophila were cultured on LB medium containing 1000 μg/mL of ampicillin for 24 h at 28°C (37). Four drops (5 μL) per insect were counted as replicates and averaged. We injected a subset of KD and control beetles with PBS to check whether we could retrieve colonies from their bodies: no colonies were recovered from uninfected dsRNA-treated females (n =15 beetles each per treatment).
All data were analyzed using the R software version 4.1.2 (38).
The survival of T. molitor females was analyzed with a Cox model for proportional hazards (checked with ‘coxzph’ function), with host survival as a response variable, and with knockdown treatments and infection treatments as explanatory variables along with experimental replicate as a random factor (package ‘coxme’) (39). When relevant, post-hoc comparisons were performed by looking at contrasts between treatment levels in the summary of the optimal model.
We analyzed the P. entomophila bacterial load retrieved from the beetles, measured as the number of CFU obtained on agar plates, with a generalized linear model fitted for a negative binomial distribution (‘glm.nb’ function of the ‘MASS’ package (40). CFU counts were the response variable, whereas time (days 1, 2 or 3) and knockdown treatments were the explanatory variables. Post hoc comparisons are performed by comparing the overlap of 95% confidence intervals (95%CI) around the estimates of the optimal model. The differences were considered significant if two 95%CI error bars overlap by no more than about half of their length (41). The effect plots for post hoc comparisons are presented in Figure S17.
In all analyses, we built the most complex model including all explanatory variables and interactions between them, as well as all the possible nested models and the null model. We then selected the optimal model based on the comparison of Akaike’s Information Criterion (AIC). Models with the lowest AIC - or lowest degree of complexity for an equivalent AIC - were retained as the optimal model. Models comprised within a delta AIC of two were considered equivalent (42).
Since different experiments on single-, double-, or triple-knockdowns had a different amount of dsEGFP as a negative control, the results yielded by different knockdown treatments were not directly comparable. However, we are able to circumvent this experimental feature by comparing the effect sizes of the knockdowns relative to their respective control treatments, both for bacterial load and host survival. This approach allows for a robust comparison of the effects of single, double, and triple knockdowns of the TmAttacin genes expression between experiments, as shown in Zanchi et al. (2017) (3). Therefore, to compare the effects of various knockdown treatments, we analyzed the effect size differences between each knockdown treatment and its respective control (dsEGFP) which is shown in the last section of the results.
In the case of the survival analysis, we used the Hazard Ratio (HR), obtained from the summary of the optimal model of each experiment, as a measure of effect size. The HR consists of the ratio of the hazard rates of the knockdown treatment to the respective dsEGFP control within each experiment, and represents the relative risk of dying of the knockdown versus dsEGFP (43).
In the case of the CFU counts, we chose Hedge’s g as an effect size, which like Cohen’s d describes the standardized mean difference of an effect but is further corrected for small sample sizes. It is calculated as the difference between the mean CFU counts of the knockdown treatment and the respective dsEGFP control, divided by the weighted pooled standard deviation for the two treatments (44).
We then plotted these parameters and the 95% confidence intervals around the effect for each knockdown experiment. An overlap of the confidence intervals around the HR with 1 indicates no significant difference between knockdown treatment and control in the risk of dying. A HR greater than 1 indicates a higher risk of dying, whereas a HR lower than 1 indicates a lower risk of dying in the knockdown treatment compared to control. Similarly, an overlap of the confidence intervals around Hedge’s g with 0 indicates no effect of the knockdown treatment compared to its respective dsEGFP control, a value greater than 0 indicates a positive effect, whereas a value smaller than 0 indicates a negative effect of the knockdown on CFU count compared to control. Both the HR and Hedge’s g were calculated using the package “effectsize” (45).
The result of the induction experiment was consistent with previous study that shows that bacterial challenge led to a significant increase in transcription of TmAttacin1a, -1b, and -2 compared to PBS-injected controls (Figure S6) (22). Based on this we carried out three sets of experiments with P. entomophila infection, in which single (TmAtt1a, TmAtt1b, and TmAtt2), pairs of (TmAtt1a-Att2 and TmAtt1b-Att2), and all three (TmAtt1a-Att1b-Att2) AMP genes were silenced.
In dsTmAtt1a-treated females, we observed a consistent decrease of TmAtt1a mRNA transcript, ranging from 4-day to 14-day post-exposure. Accordingly, the 4-day RNAi treatment was selected as a time point for bacterial challenge (Figure S7). For the series of experiments on TmAtt1b-silenced females were challenged 3-days post-RNAi treatment, at which point relative expression of TmAtt1b transcript was successfully suppressed (Figure S8). Similarly, we challenged TmAtt2-silenced females at 3-days post knockdown treatment (Figure S9). In dsTmAtt1a-Att2-treated beetles, females were infected at 4-days post dsRNA injection (Figure S10), while they were challenged at 3-day in dsTmAtt1b-Att2-treated beetles (Figure S11). Downregulation of the relative expression of TmAtt1a in dsTmAtt1a-Att1b-treated beetles was not successful (Figure S12).
Next, to tackle the effect of representative genes on each other, the transcription levels of all TmAttacin in dsTmAtt1a- and dsTmAtt1b-treated beetles were separately assessed. As can be seen in Figures S13, 14, silencing TmAtt1a (4-day) had no effect on expression level of either TmAtt1b or TmAtt2, similarly, TmAtt1b (3-day) knockdown did not influence transcript levels of TmAtt1a or TmAtt2. Based on the double-knockdown results, we altered the volume of representative genes to successfully silence all three genes. In triple-knockdown beetles we accounted for the reduced expression of TmAtt1a, TmAtt1b, and TmAtt2 after RNAi treatment of each gene separately. Therefore, we expected that 4-day post-exposure females would be optimal for challenging, which was confirmed via qRT-PCR (Figures S15, S16).
We tested whether depleting TmAtt1a transcripts could increase mortality of T. molitor females following challenge with P. entomophila. To do so, we infected beetles at 4-days post-RNAi treatment and monitored survival daily for 35 days. We observed that PBS-injected insects (dsTmAtt1a-PBS, dsEGFP-PBS, and full control-PBS) showed no death over 35 days, therefore we did not include them in the analysis, but presented them in the figures. We did not find an effect of knockdown treatment in P. entomophila injected beetles, since the survival of infected-dsTmAtt1a (dsTmAtt1a-P. entomophila, dsEGFP-P. entomophila, and full control-P. entomophila) was not significantly different from the control treatments (X²3.177 = 1.2, p = 0.55) (Figure 1A).
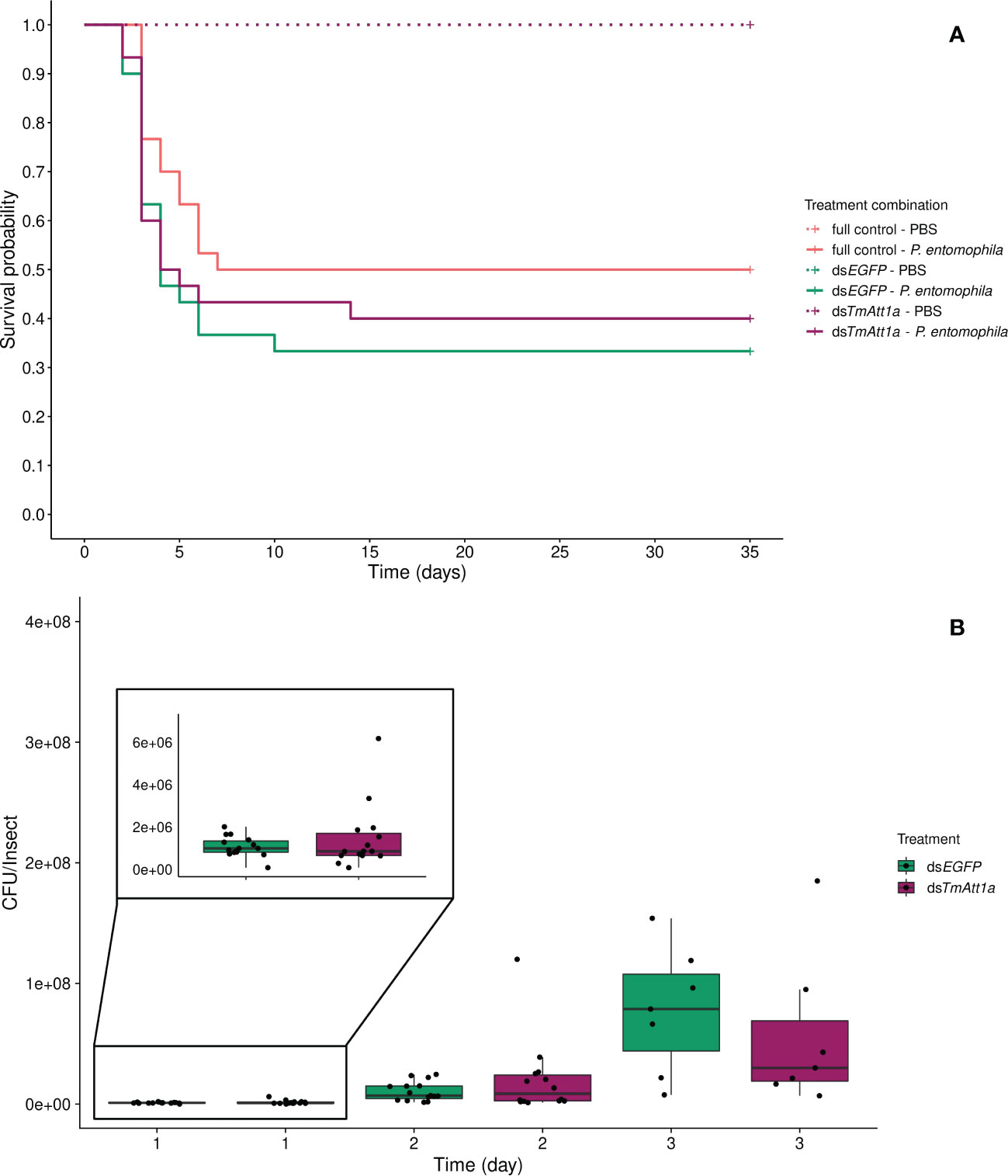
Figure 1 (A) Effect of TmAttacin1a (TmAtt1a) knockdown on survival of Pseudomonas entomophila infected Tenebrio molitor females over 35 days. Two independent experiments each performed on 15 females per treatment group. Each dsRNA-treated or dsRNA-untreated group was infected with P. entomophila on day 4 after dsRNA treatment based on the data on knockdown efficiency (Figure S7). Pink dashed line: Full control-PBS; Turquoise dashed line: dsEGFP - PBS; Purple dashed line: dsTmAtt1a - PBS; Pink line: Full control - P. entomophila; Turquoise line: dsEGFP - P. entomophila; Purple line: dsTmAtt1a - P. entomophila. (B) Bacterial load in dsTmAtt1a-silenced T. molitor challenged with P. entomophila at 1-, 2-, and 3-day post-infections. The colony-forming units (CFU) recovered from tissues of T. molitor females from dsTmAtt1a (purple) and dsEGFP (control, turquoise). In the box plots, the lower (first) quartile is the closest boundary to zero, the line within the box marks the median (second quartile), and the upper (third) quartile. Each dot represents the CFU count in an individual beetle. The bars represent the 1.5 interquartile. Sample sizes are given in Table S2.
Consistent with this observation, the single knockdown of TmAtt1a did not influence P. entomophila load in the bodies of the beetles, neither in interaction with time (time × KD treatment: X²2.72 = 2.667, p = 0.26) nor as a simple effect (X²1.72 = 1.17e-05, p = 1). CFU counts increased over 3 days (time: X²2.72 = 173.38, p < 0.0001) (Figure 1B, S17A).
The optimal model did not include an effect of the interaction between knockdown (KD) and infection treatments on beetle survival (KD. treatment × inf. treatment: X²5.175 = 3.41, p = 0.18). However, it is important to note that the mortality in the PBS injected beetles is negligible across all knockdown treatments (only 3 dead beetles) (Figure 2A).
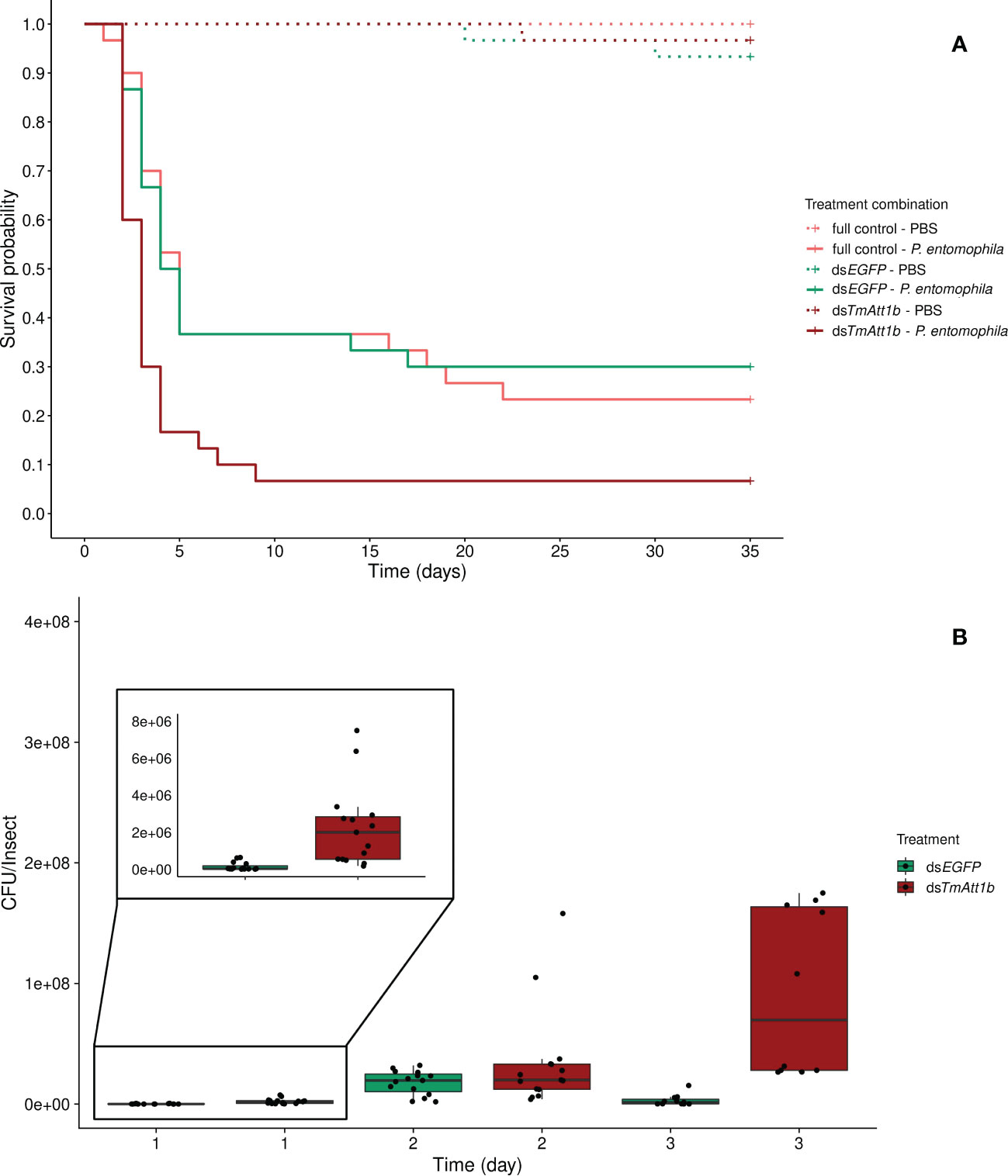
Figure 2 (A) Effect of TmAttacin1b (TmAtt1b) knockdown on survival of Pseudomonas entomophila infected Tenebrio molitor females over 35 days. dsEGFP was injected as a negative control for the nonspecific effects of dsRNA and the third group includes dsRNA untreated control. Two independent experiments each performed on 15 females per treatment group. Each dsRNA-treated or dsRNA-untreated groups were infected with P. entomophila on day 3 after dsRNA treatment based on the data on knockdown efficiency (Figure S8). Pink dashed line: Full control-PBS; Turquoise dashed line: dsEGFP - PBS; Burgundy dashed line: dsTmAtt1b - PBS; Pink line: Full control - P. entomophila; Turquoise line: dsEGFP - P. entomophila; Burgundy line: dsTmAtt1b - P. entomophila. (B) Bacterial load in dsTmAtt1b-silenced T. molitor challenged with P. entomophila at 1-, 2-, and 3-day post-infections. The colony-forming units (CFU) recovered from tissues of T. molitor females from dsTmAtt1b (burgundy) and dsEGFP (control, turquoise). In the box plots, the lower (first) quartile is the closest boundary to zero, the line within the box marks the median (second quartile), and the upper (third) quartile. Each dot represents the CFU count in an individual beetle. The bars represent the 1.5 interquartile. Sample sizes are given in Table S2.
Both knockdown and infection treatment influenced beetle survival as simple effects (KD. treatment: X²2.178 = 13.22, p = 0.0013; infection treatment: X²2.178 = 46.85, p < 0.0001). We observed a strong decrease in survival of beetles after P. entomophila challenge (Figure 2A). There was a decrease in survival when beetles that were treated with dsTmAtt1b and subsequently infected were compared to those treated with dsEGFP and infected (dsTmAtt1b/dsEGFP: z = 3.07, p = 0.0021) while no difference in survival of infected full control and infected dsEGFP was found (Full control/dsEGFP: z = -0.07, p = 0.95).
The dynamics of the bacterial load were significantly different in dsTmAtt1b-treated beetles compared to dsEGFP beetles over time (time × KD treatment: X²2.80 = 23.35, p = 8.512e-06). While the density of P. entomophila in dsEGFP-treated females increased between 1-day and 2-day post- infection, bacterial load of dsTmAtt1b-treated beetles showed higher densities starting from 1-day post infection. CFU counts of P. entomophila were similar between TmAtt1b knockdown treatment and control at 2-days post-infection. Interestingly, bacterial load of dsEGFP-treated females significantly decreased between 2-days and 3-days post-infection, whereas it remained similarly high between 2- and 3-days in dsTmAtt1b-treated beetles (Figures 2B, S17B).
Similarly, to the KD of TmAtt1b, we found no interaction between knockdown and infection treatment on beetle survival (KD. treatment × infection treatment: X²5.120 = 4.98, p = 0.083). Instead, both the knockdown and the infection treatments affect survival of beetles as simple effects. Again, TmAtt2-silenced treatments showed a decreased survival compared to the dsEGFP control (KD. treatment: X²3.115 = 7.11, p = 0.029; dsTmAtt2/dsEGFP: z = 2.17; p = 0.03), whereas the survival of dsEGFP beetles does not differ compared to full control (Full control vs. dsEGFP: z = -0.07, p = 0.95) (Figure 3A). Moreover, as expected, infection by P. entomophila led to high mortality in beetles (infection treatment: X²3.115 = 111.02, p < 0.0001).
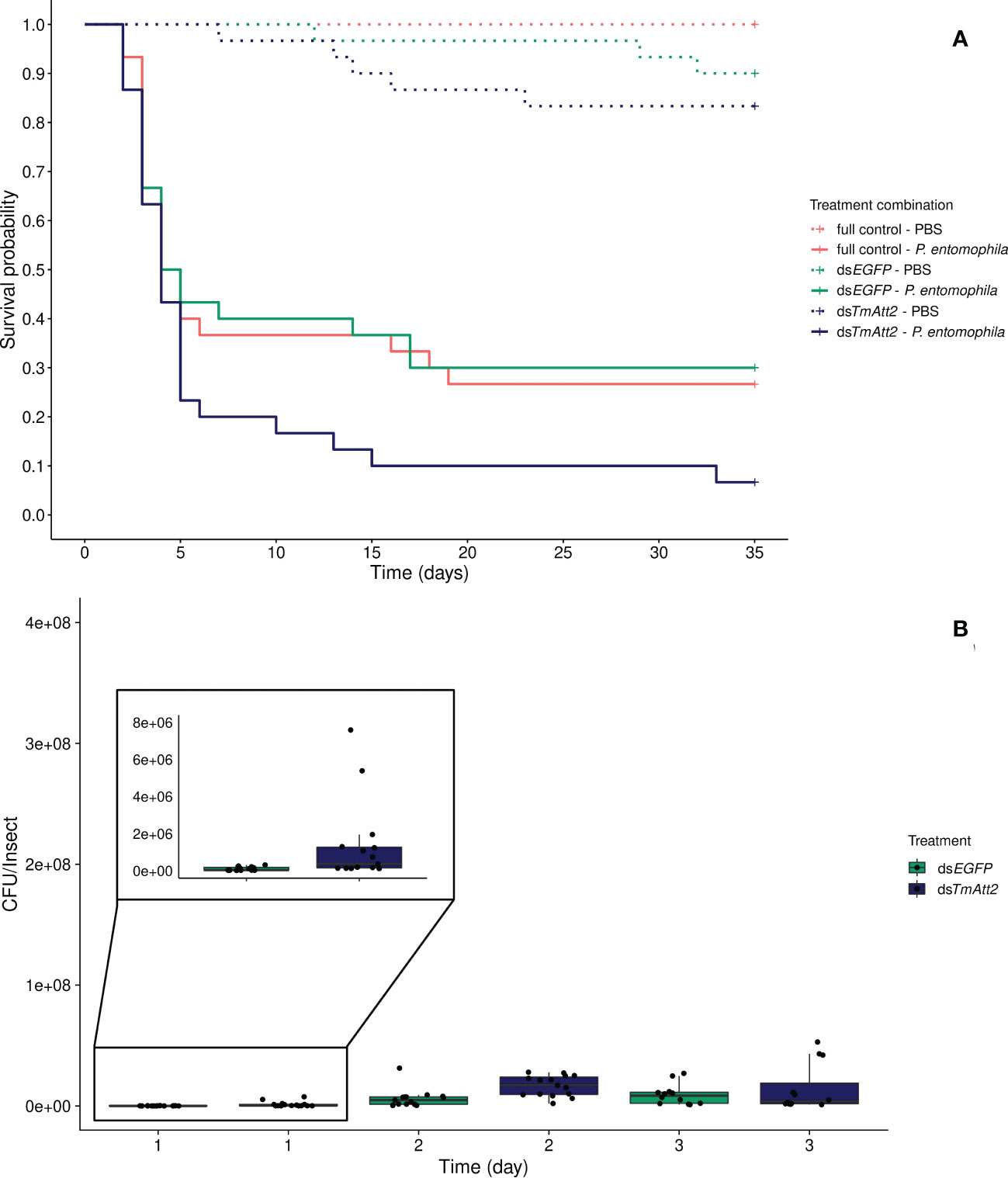
Figure 3 (A) Effect of TmAttacin2 (TmAtt2) knockdown on survival of Pseudomonas entomophila infected Tenebrio molitor females over 35 days. dsEGFP was injected as a negative control for the nonspecific effects of dsRNA and the third group includes dsRNA untreated control. Two independent experiments each performed on 15 females per treatment group. Each dsRNA-treated or dsRNA-untreated groups were infected with P. entomophila on day 3 after dsRNA treatment based on the data on knockdown efficiency (Figure S9). Pink dashed line: Full control-PBS; Turquoise dashed line: dsEGFP - PBS; Navy blue dashed line: dsTmAtt2 - PBS; Pink line: Full control - P. entomophila; Turquoise line: dsEGFP - P. entomophila; Navy blue line: dsTmAtt2 - P. entomophila. (B) Bacterial load in dsTmAtt2-silenced T. molitor challenged with P. entomophila at 1-, 2-, and 3-day post-infections. The colony-forming units (CFU) recovered from tissues of T. molitor females from dsTmAtt2 (navy blue) and dsEGFP (control, turquoise). In the box plots, the lower (first) quartile is the closest boundary to zero, the line within the box marks the median (second quartile), and the upper (third) quartile. Each dot represents the CFU count in an individual beetle. The bars represent the 1.5 interquartile. Sample sizes are given in Table S2.
The development of the bacterial load over time is different in TmAtt2-silenced beetles compared to the control (time × KD treatment: X²5.83 = 15.47, p = 0.00042). While CFU counts increased over 3 days in the control, as seen in the previous knockdowns, they already reached higher counts 1-day post-injection in dsTmAtt2-treated group compared to dsEGFP control. TmAtt2-silenced beetles increased and remained stable over the first two days post infection, whereas the control reached similar CFU counts at 3-days post challenge (Figures 3B, S17C).
The double KD of TmAtt1a and TmAtt2 treatment had no effect on survival, neither in interaction with the infection treatment (KD. treatment × infection treatment: X²5.175 = 1.23; p = 0.54) nor as a simple effect (KD. treatment: X²2.173 = 2.04, p = 0.36). Only the infection treatment was retained in the best model, where the survival of adult females infected with P. entomophila was significantly lower (X²1.174 = 32.35, p < 0.0001) (Figure 4A).
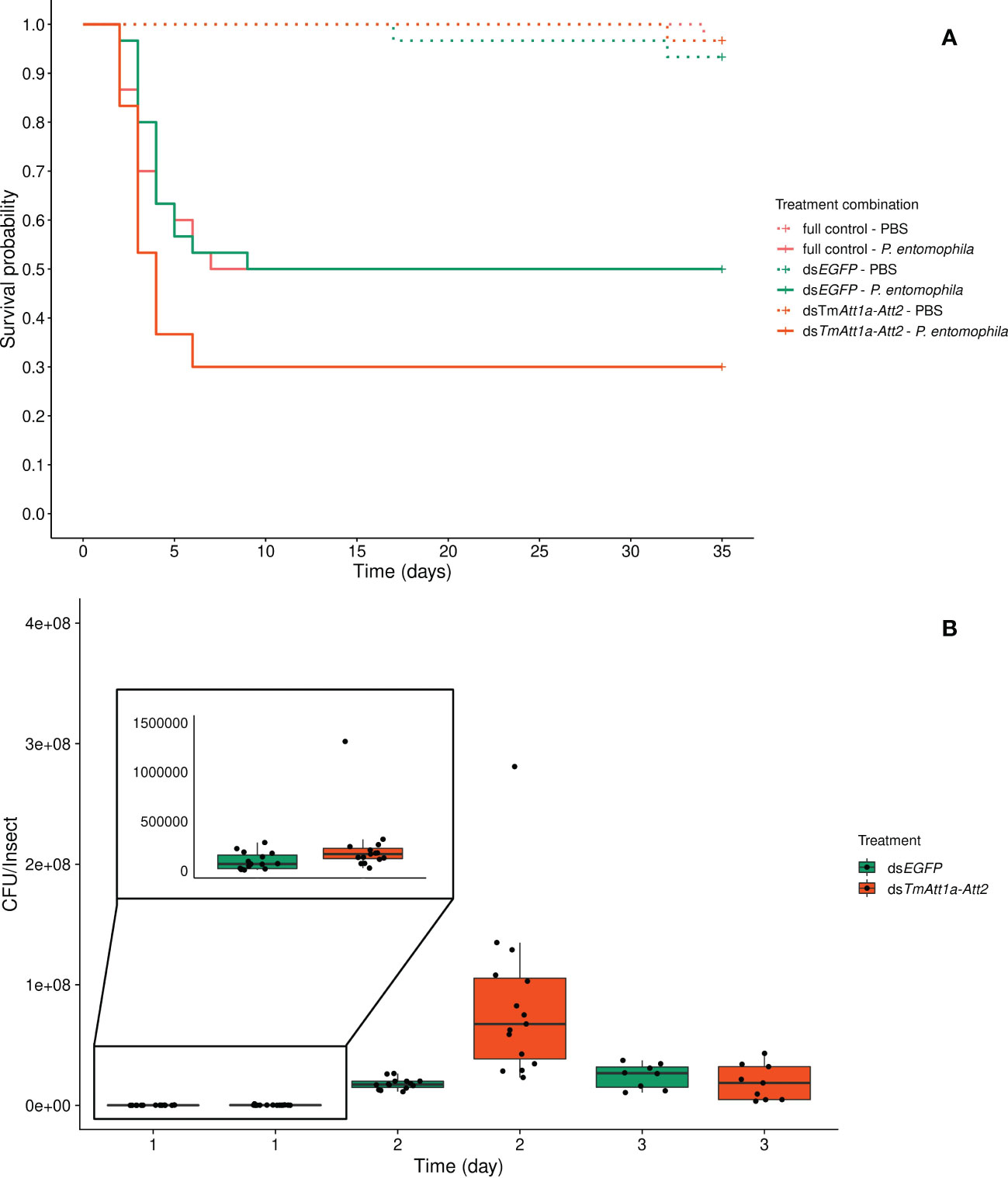
Figure 4 (A) Effect of TmAttacin1a-2 (TmAtt1a-Att2) double-knockdown on survival of Pseudomonas entomophila infected Tenebrio molitor females over 35 days. dsEGFP was injected as a negative control for the nonspecific effects of dsRNA and the third group includes dsRNA untreated control. Two independent experiments each performed on 15 females per treatment group. Each dsRNA-treated or dsRNA-untreated groups were infected with P. entomophila on day 4 after dsRNA treatment based on the data on knockdown efficiency (Figure S10). Pink dashed line: Full control-PBS; Turquoise dashed line: dsEGFP - PBS; Orange dashed line: dsTmAtt1a-Att2 - PBS; Pink line: Full control - P. entomophila; Turquoise line: dsEGFP - P. entomophila; Orange line: dsTmAtt1a-Att2 - P. entomophila. (B) Bacterial load in dsTmAtt1a-Att2-silenced T. molitor challenged with P. entomophila at 1-, 2-, and 3-day post-infection. The colony-forming units (CFU) recovered from tissues of T. molitor females from dsTmAtt1a-Att2 (orange) and dsEGFP (control, turquoise). In the box plots, the lower (first) quartile is the closest boundary to zero, the line within the box marks the median (second quartile), and the upper (third) quartile. Each dot represents the CFU count in an individual beetle. The bars represent the 1.5 interquartile. Sample sizes are given in Table S2.
For P. entomophila density inside the beetles, the best model retained an interaction between treatment and time (time × KD treatment: X²5.75 = 16.05, p = 0.00033). dsEGFP-treated beetles showed the same dynamics as described above, the CFU count increased between 1-day and 2-days post-infection and remained stable between 2- and 3-days post-infection. Double-knockdown of TmAtt1a and TmAtt2 resulted in a higher number of CFU recovered from beetles starting from 1-day after challenge. Moreover, it reached a higher CFU count than the control at 2-days post infection and at 3-days decreased to a level similar to controls (Figure 4B and S17D).
Despite a trend towards an interaction between KD and infection treatment (KD. treatment × infection treatment: X²3.177 = 5.185, p = 0.075), the best model is one in which beetle survival is explained by both simple effects of the infection (inf. treatment: X²2.179 = 50.46, p < 0.0001) and knockdown treatments (KD. treatment: X²2.179 = 9.859, p = 0.0072). As expected, P. entomophila infection decreased beetle survival. However, the effect of the KD treatment does not come from a lower survival of TmAtt1b-Att2 compared to dsEGFP (dsTmAtt1b-Att2/dsEGFP: z = 0.64, p = 0.52). It is caused by the fact that beetles with no injection of dsRNA outlived both dsEGFP (full control/dsEGFP: z = -2.50, p = 0.013) and dsTmAtt1b-Att2 (full control/dsTmAtt1b-Att2: z = -3.06, p = 0.002) (Figure 5A).
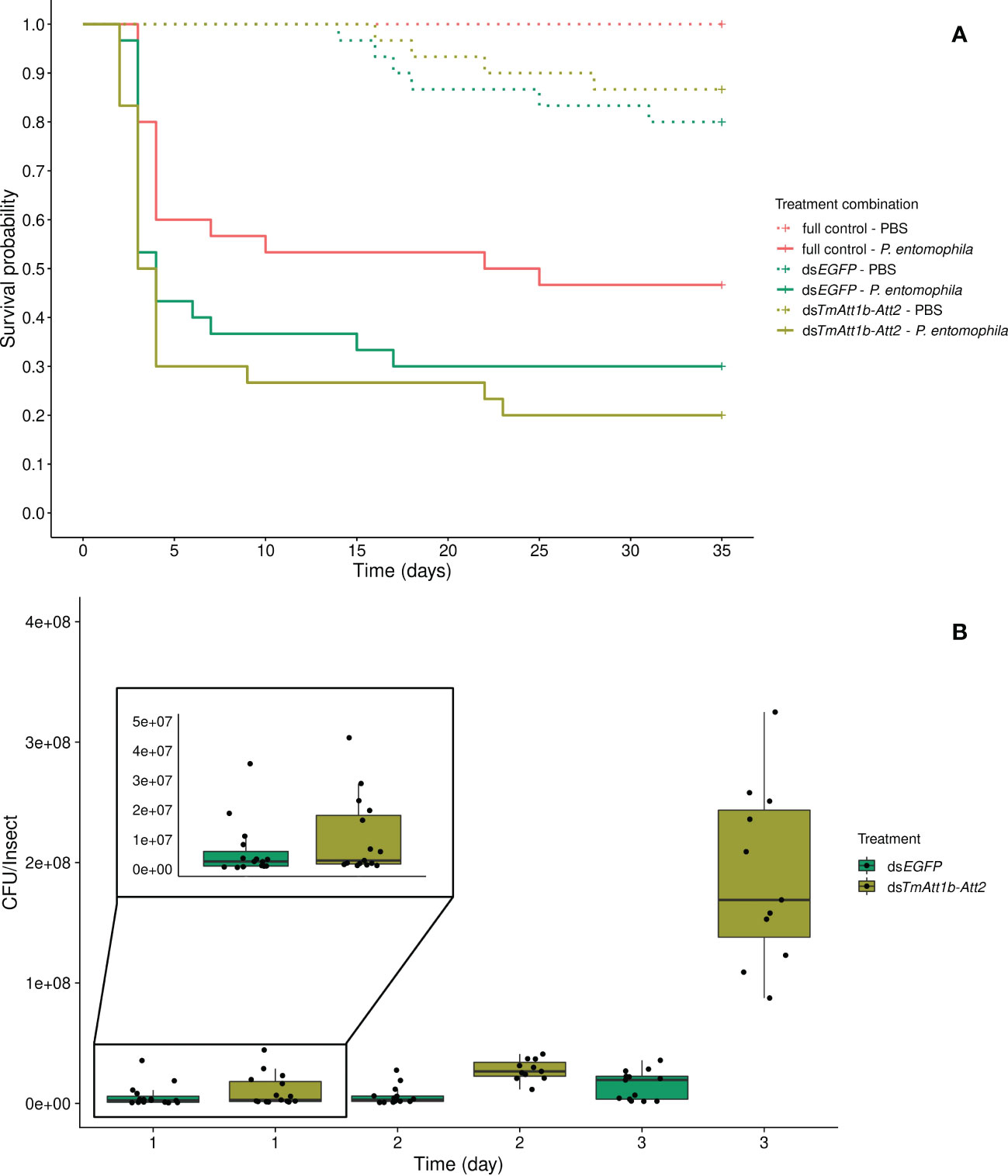
Figure 5 (A) Effect of TmAttacin1b-2 (TmAtt1b-Att2) double-knockdown on survival of Pseudomonas entomophila infected Tenebrio molitor females over 35 days. dsEGFP was injected as a negative control for the nonspecific effects of dsRNA and the third group includes dsRNA untreated control. Two independent experiments each performed on 15 females per treatment group. Each dsRNA-treated or dsRNA-untreated groups were infected with P. entomophila on day 3 after dsRNA treatment based on the data on knockdown efficiency (Figure S11). Pink dashed line: Full control-PBS; Turquoise dashed line: dsEGFP - PBS; Olive green dashed line: dsTmAtt1b-Att2 - PBS; Pink line: Full control - P. entomophila; Turquoise line: dsEGFP - P. entomophila; Olive green line: dsTmAtt1b-Att2 - P. entomophila. (B) Bacterial load in dsTmAtt1b-Att2-silenced T. molitor challenged with P. entomophila at 1-, 2-, and 3-day post-infection. The colony-forming units (CFU) recovered from tissues of T. molitor females from dsTmAtt1b-Att2 (olive green) and dsEGFP (control, turquoise). In the box plots, the lower (first) quartile is the closest boundary to zero, the line within the box marks the median (second quartile), and the upper (third) quartile. Each dot represents the CFU count in an individual beetle. The bars represent the 1.5 interquartile. Sample sizes are given in Table S2.
There was an interaction between KD treatment and time on the bacterial load recovered from the beetles (time × KD treatment: X²5.79 = 14.75, p = 0.00063). The CFU counts are similar at 1-day post-infection in both treatments but diverge at 2-days, where they reach higher concentrations in the KD. This effect persists at 3-days post-infection (Figures 5B, S17E). Therefore, the CFU counts observed higher in the double-knockdown treatment than in control at both 2-days and 3-days post injection.
The next question was how triple knockdown could influence survival of females upon P. entomophila infection, given that double knockdown did not reveal any significant difference in beetle survival (Figures 4, 5).
Beetles which were injected with PBS only (dsTmAtt1a-Att1b-Att2-PBS, dsEGFP-PBS, and full control-PBS) display no mortality over the time course of this experiment. We further carried out the survival analysis on P. entomophila injected beetles only, however PBS-injected treatments are displayed in the figures. We detected that knockdown treatment significantly affects the survival of P. entomophila injected beetles (KD. treatment:X²2.87 = 16.349, p = 0.00028). The triple knockdown of TmAtt1a, TmAtt1b, and TmAtt2 by RNAi reduced the survival of females following P. entomophila infection (dsTmAtt1a-Att1b-Att2/dsEGFP: z = 3.22, p = 0.0013) while survival of dsEGFP beetles does not differ compared to full control beetles (full control vs. dsEGFP: z = -0.42, p = 0.68) (Figure 6A). Since no effect of the KD is detectable among PBS-injected beetles, but a strong effect exists in P. entomophila-injected beetles, there is an interaction between KD and infection treatments on beetle survival.
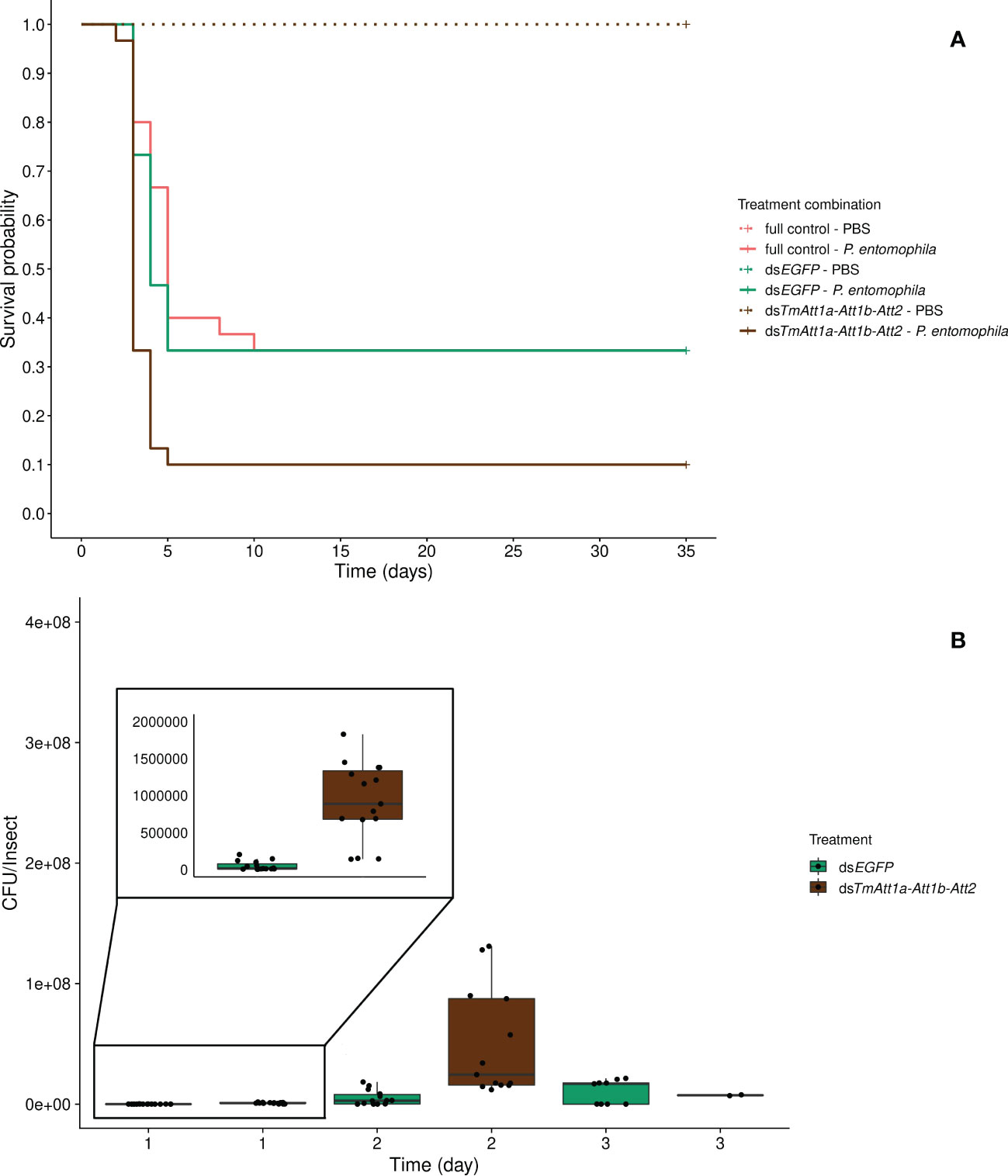
Figure 6 (A) Effect of TmAttacin1a-1b-2 (TmAtt1a-Att1b-Att2) triple-knockdown on survival of Pseudomonas entomophila infected Tenebrio molitor females over 35 days. dsEGFP was injected as a negative control for the nonspecific effects of dsRNA and the third group includes dsRNA untreated control. Two independent experiments each performed on 15 females per treatment group. Each dsRNA-treated or dsRNA-untreated groups were infected with P. entomophila on day 4 after dsRNA treatment based on the data on knockdown efficiency (Figure S15). Pink dashed line: Full control-PBS; Turquoise dashed line: dsEGFP - PBS; Brown dashed line: dsTmAtt1a-Att1b-Att2 - PBS; Pink line: Full control - P. entomophila; Turquoise line: dsEGFP - P. entomophila; Brown line: dsTmAtt1a-Att1b-Att2 - P. entomophila. (B) Bacterial load in dsTmAtt1a-Att1b-Att2-silenced T. molitor challenged with P. entomophila at 1-, 2-, and 3-day post-infection. The colony-forming units (CFU) recovered from tissues of T. molitor females from dsTmAtt1a-Att1b-Att2 (brown) and dsEGFP (control, turquoise). In the box plots, the lower (first) quartile is the closest boundary to zero, the line within the box marks the median (second quartile), and the upper (third) quartile. Each dot represents the CFU count in an individual beetle. The bars represent the 1.5 interquartile. Sample sizes are given in Table S2.
The optimal model does not include an interaction between time and knockdown treatment for bacterial loads (time × KD treatment: X²3.67 = 5.22, p = 0.074), however, the CFU counts are explained by both time (time: X²2.67 = 103.35, p <0.0001) and knockdown treatment (KD treatment: X²1.67 = 39.39, p <0.0001) as simple effects. P. entomophila load increased initially (1-day and 2-days post-infection) and remained stable at the last day in both treatments. CFU counts were overall higher in TmAtt1a-Att1b-Att2-silenced beetles than in the control (Figures 6B, S17F).
Since the different knockdown experiments have different controls, due to the different concentration of dsEGFP-injected, we normalized survival and pathogenic load of the KD beetles relative to their respective control, using effect size measures This approach can highlight potential interactions between AMPs, by showing that a multiple knockdown yields a higher effect than the sum of its single knockdowns, or on the contrary, by highlighting a lower effect of a multiple knockdown compared to single knockdowns. The following section summarizes the results of our experiments by calculating and presenting the effects of different knockdowns compared to their respective dsEGFP controls on the survival and the bacterial load of T. molitor females infected with P. entomophila.
The differential dynamics of the effects of KD on bacterial load was not reflected in the survival curves, as shown by the hazard ratios (HR) (Figure 7A). Instead, there is a remarkable correlation between the plot of the HR of the survival analyses and the plot of the Hedge’s g of the various treatments 1-day post injection (Figures 7A, B). Indeed, hazard ratios of the various treatments are explained by Hedge’s g on CFU count of the same treatments at 1-day post-infection (linear model: Hedge’s g: F1.4 = 20.41, p = 0.01). This is not the case at 2- (F1.4 = 0.073, p = 0.8) and 3-days (F1.4 = 0.1, p = 0.77) post-infection.
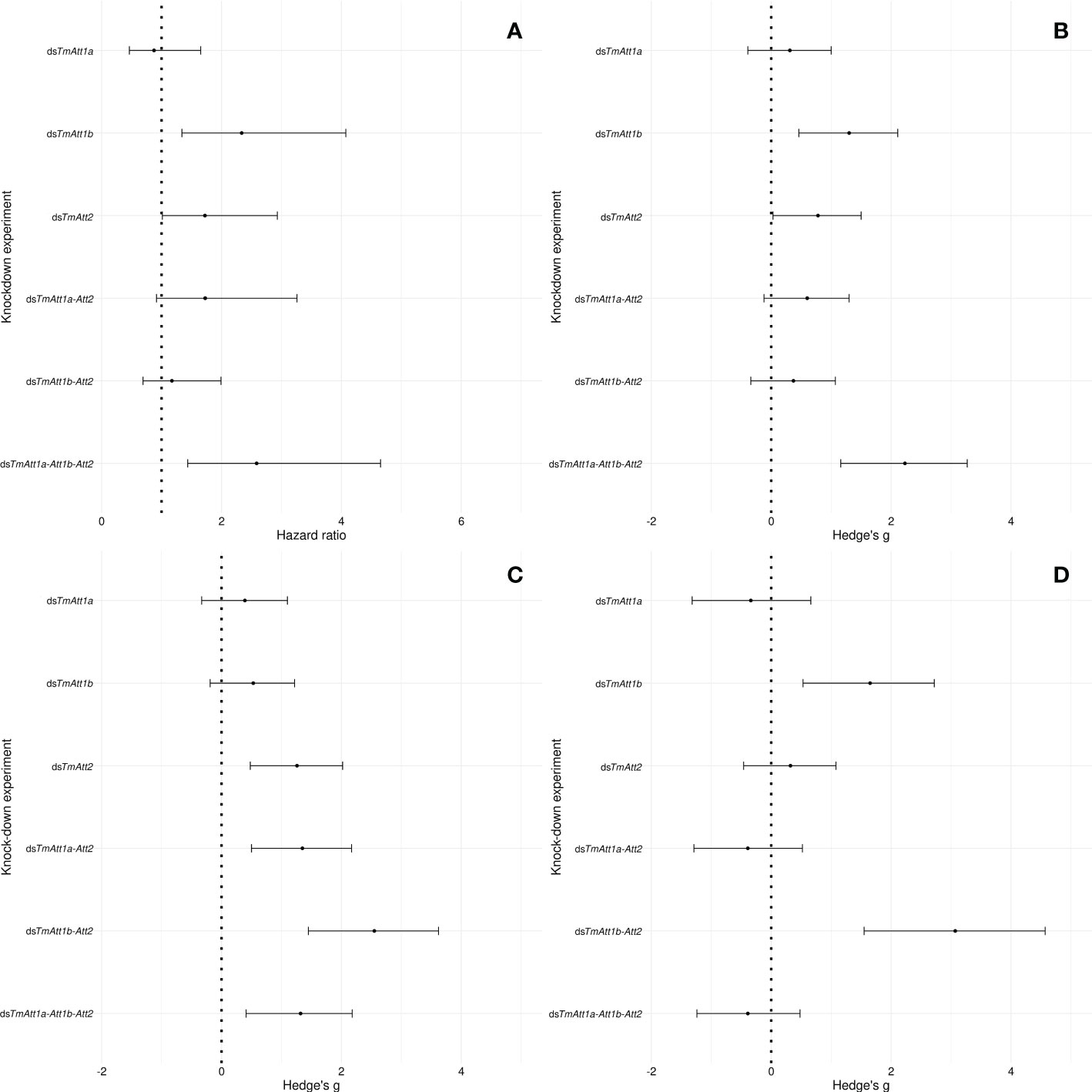
Figure 7 Forest plot representing the effects of the single, double, or triple knockdown of the AMP genes of interest on (A) survival of Tenebrio molitor females infected by Pseudomonas entomophila and (B) the number of P: entomophila CFU retrieved from females at 1-day, (C) 2-days, and (D) 3-days post-infection. In case of the survival, the effect size is calculated as the hazard ratio (HR) between knockdown and dsEGFP control groups (A). In case of CFU counts (B–D), it is calculated as the Hedge’s g between knockdown and dsEGFP control groups. Dots represent the HR and Hedge’s g, and the horizontal bars extend from the lower limit to the upper limit of the 95% confidence interval (95CI). An overlap of the 95CI with 1 (vertical line) means the knockdown has no effect on survival compared to control. A HR < 1 indicates a reduction in the hazard, whereas a HR > 1 indicates an increase in hazard In the knockdown compared to control, an overlap of the 95CI with 0 (vertical dotted line) indicates no significant effect of the knockdown treatments compared to control on CFU count, a positive value indicates a positive effect of the knockdown on CFU count, whereas a negative value indicates a negative effect of the knockdown on CFU count compared to control.
Looking more closely at the effect sizes of the various knockdown treatments on CFU counts over time, we notice that the effect of single knockdowns is relatively constant over time, i.e., the 95CI around the models’ estimates overlap within each single-knockdown treatment between time points.
Knockdown of TmAtt1a did not have an effect on the CFU count of P. entomophila over 3 days (95CI overlaps zero at each time points, Figures 7B–D). Knockdown of TmAtt2 had a larger effect at days 1 and 2 compared to the other single knockdowns, however, the effect disappeared at 3-days post-infection. Notably, a larger effect was observed in dsTmAtt1b- than in dsTmAtt2-treated females on day three. However, double-knockdown of TmAtt1b-Att2 yielded a slightly lower effect than both their single-knockdowns on day one. The effects of TmAtt2 and TmAtt1b knockdowns on CFU counts were dynamic over time.
The effect of double-knockdown of TmAtt1b-Att2 gradually increased over 3 days, from showing no effects on day one to reaching the highest effect than any other single and double knockdown at 3-days post-infection. Interestingly, this observation does not follow the dynamics of the effects of these genes when they were separately knocked down, since the single-knockdown of both genes had an effect on day one.
Finally, the effect of the triple-knockdown on P. entomophila load was higher at 1-day post infection compared to other knockdown experiments but decreased on day two. Of note, the sample size dropped at 3-day post-infection because all the highly infected individuals had died (Figures 7B–D).
AMPs as conserved molecules frequently combat pathogenic infections both synergistically and individually (12). Results of our study show that the knockdown of AMP genes in T. molitor either individually or in combinations of three AMP genes of the same family influences host survival following P. entomophila infection. T. molitor mortality is associated with bacterial load in host tissues, consistent with Drosophila studies (46, 47). Comparison of the survival and bacterial load in knockdown beetles with their corresponding controls in each single-, double-, or triple-knockdown experiment confirm this phenomenon. We find that silencing of three components of the AMP cocktail has stronger effects on host survival and bacterial load than knockdown of one or two components, and that the fate of the host-pathogen interaction in our system is determined relatively early in the infection process.
Separate analysis of the single knockdown experiments shows that the effect of TmAtt1a knockdown neither increases bacterial load nor lowers host survival while bacterial load in the control increases gradually over three days post-infection. This was supported by the results of the double-knockdown of TmAtt1a in combination with TmAtt2, which does not seem to affect the efficiency of the latter. By contrast, both knockdowns of TmAtt1b and TmAtt2 dynamically affect bacterial load over time. Indeed, TmAtt1b knockdown increases bacterial load at both 1- and 3-days post-infection, while knockdown of TmAtt2 increases bacterial load only at 1-day post-infection. Interestingly, both these single knockdown treatments result in reduced beetle survival. Silencing TmAtt2 caused mortality within 15 days which was slower than in TmAtt1b knockdown females which died within 10 days (Figures 2, 3). However, overall mortality was similar. These results suggest the importance of the timing of AMP production in controlling of bacterial infection and highlight that the survival differences in knockdown Attacin females can be attributed to differences in their bacterial burden (46).
In the case of the double knockdowns, we observed that dsTmAtt1a-Att2 beetles showed an initial increased load of P. entomophila (1-day post-infection) but of lesser magnitude compared to the single knockdown of TmAtt2 and resulted in no detectable changes in host survival. In this regard, the increased bacterial burden might be responsible for the early host mortality, whereas a later decrease in bacterial load did not influence mortality. Similarly, double-knockdown of TmAtt1b-Att2 could be responsible for the drastic increase in P. entomophila colonization which on the contrary does not result in an increase in mortality rate. Therefore, both double knockdown treatments increased bacterial loads at later time points after infection but did not influence host survival.
P. entomophila infection resulted in the death of the majority of triple-knockdown females within 3 days (Figure 6A), leading to a low sample size in the bacterial load experiment at 3-days post-infection (Figure 6B), compared to single- and double-knockdown groups. Moreover, depletion of three Attacin gene transcripts led to increased bacterial colonization at 1- and 2-days post-infection. This is earlier in the infection process compared to other knockdown females. We only see an effect of TmAtt1a in our triple-knockdown experiments. However, due to the lack of information about the molecular mechanisms of Attacin interactions, we cannot decipher whether this effect comes from a synergistic action of TmAtt1a with TmAtt1b and/or TmAtt2 or an efficiency of TmAtt1a only when combined with TmAtt1b and TmAtt2.
It seems that the common denominator between the experiments in which host survival is affected is an increase in bacterial load on the first day after infection. Therefore, only effect size analysis allows for the correction of knockdown treatments by control treatment mortality and provides a meaningful comparison between experiments. Consistent with this, a striking finding of our dataset is that despite the fact that most mortality in these series of experiments occur within five days of infection (Figures 1–6), the outcome of the host-pathogen interaction appears to be fixed on the first day, as there is a concordance between bacterial load at this time point and mortality.
The relationship between pathogen load and host mortality is known to be affected by pathogen tolerance as well as pathogen resistance. The former has been defined as the tendency of the host to limit the effects of infection to a certain level, resulting in less loss of fitness so that the host lives longer. Resistance, on the other hand, actively reduces the growth of the pathogen (47, 48).The concordance we see indicates that the effects we observed on host survival are likely not due to changes in tolerance to the P. entomophila infection, but rather to changes in the ability of beetles to control P. entomophila load, or resistance. Our data also show that the number of CFUs, which appears to be critical for mortality is approximately 1 × 106 CFUs per beetle at 1-day post-infection, which is consistent with our preliminary data showing a clear dose-response (Figure S2).
This is consistent with the observations of Duneau et al. (46), who suggested that the faster the immune system is activated against infection, the higher the chance of D. melanogaster surviving. If immune responses are too late, flies will die when bacterial colonization reaches a certain level (46). Taken together, these findings are consistent with the fact that the risk of mortality increases markedly with increasing bacterial load early in the infection process, rather than just before host death (49). Interestingly, this observation was not made in Zanchi et al. (3), where T. molitor was exposed to an opportunistic pathogen. In this case differences in bacterial load were seen much later in the infection process. This co-occurrence between bacterial load and host death has also been observed in Drosophila (34, 46).
Following this reasoning, we focused on the effects of the different knockdown treatments on early bacterial load (1-day post-infection) and on host survival in response to P. entomophila infection to investigate the kind of interactions that might exist between our three focal genes. First, we note that while TmAtt1a alone has no effect on bacterial load and host survival, it is effective in combination with TmAtt1b and TmAtt2 (TmAtt1a-Att1b-Att2). Knockdown of TmAtt1a in combination with either TmAtt1b or TmAtt2 is inefficient (TmAtt1a-Att1b or TmAtt1a-Att2). This indicates an antagonistic effect of TmAtt1a on TmAtt1b and TmAtt2, however, the difference between the effect sizes of the knockdown treatments is not significant. Therefore, the datasets of the single- and double-knockdowns do not allow us to decide whether this pattern is the result of antagonism, redundancy, synergy, or an additive effect between the three TmAttacins in the context of a double-knockdown.
The effect of triple-knockdown on host survival and P.entomophila load is greatest at 1-day post-infection. This observation seems to rule out the possibility of an antagonistic interaction and suggests an additive effect of these three members of the TmAttacin family. The effect of the triple-knockdown, although larger, is not significantly different from the effect of the single knockdown of TmAtt1b, which could imply that the effects observed with the triple-knockdown treatment could be due exclusively to TmAtt1b. However, this seems unlikely because we do not observe a survival phenotype with the double-knockdown of TmAtt1b-Att2.
A closer look at the value of the effect sizes on the CFU counts at 1-day post-infection shows that the effect size of the triple-knockdown on the CFU counts is close to the effect size of the three single knockdowns combined (Hedge’s g TmAtt1a + TmAtt1b + TmAtt2 = 2.39 vs. 2.23 for the triple-knockdown). This is not the case for the double-knockdowns, where, on the contrary, the effect sizes of the double- knockdowns are close to the effect sizes of the corresponding single-knockdowns minus each other (Hedge’s g TmAtt2 – TmAtt1a = 0.47 vs. 0.60 for the double-knockdown; TmAtt1b – TmAtt2 = 0.52 vs. 0.37 for the double-knockdown). This suggests that the effects of single-, double-, and triple-knockdowns of our three AMPs alter the nature of the interactions between them.
These two possibilities were previously addressed by Hanson et al. (13), who knocked down a larger number of AMP genes across several gene families in D. melanogaster. This highlighted additive or synergistic anti-Gram-negative and antifungal activities between AMPs as well as in vivo specific AMP-pathogen interactions (3, 13). In a further study Carboni et al., 2021 used the flies from (13) to investigate the role of cecropins in fly immunity. While the knockout of all cecropins did not alter mortality of infected flies, the combined knockdown of the 4 cecropins with 10 other AMPs resulted in significant mortalities, further supporting the idea of higher-order interactions of AMPs (13, 50). Our data confirm that this is also the case in other insect species such as Tenebrio molitor. Our data also concur with in vitro experiments carried by Yu et al. (12), who reported strong synergistic effects of combinations of three or two AMPs from different families and host organisms, with synergy being stronger for combinations of three than for combination of two AMPs. This was also demonstrated in vivo between AMPs of different classes in Zanchi et al. (3), where the burden of an opportunistic pathogen in T. molitor is more efficiently controlled by a combination of AMPs than by single ones (3, 12). Overall, our knowledge on AMP interactions in vivo is still very limited, as studies have either knocked out groups of AMPs, for example (51) knocked out 10 Bomanins simultaneously, or the groups of AMPs knocked out by Hanson et al., 2019 (13, 51). The approach we presented here and in a previous publication (3), aims to knockdown all combinations of three different AMPs, here within one family. Future work needs to show what the nature of the interactions of AMPs in vivo is, as determining synergy for example is conceptually and empirically challenging (52).
To our knowledge, whether the depletion of transcripts of some AMP genes could be compensated by the upregulation of others has never been investigated, which would be an important direction for future research. We did not find evidence for compensation of the depletion of transcripts of TmAtt1a and TmAtt1b by overexpression of one other of our focal AMPs. Nevertheless, we cannot exclude that other members of the Attacin family or of other AMP families would be upregulated following the KD of one or several in vivo. This would add another level of interactions. In spite of this, our work demonstrates, that at least within the Attacin group, when AMPs are knocked down, the overall Attacin response in reduced, rendering beetles more susceptible to infection.
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Conceptualization: MK, CZ and JR. MK designed and performed the experiments. MK and CZ analyzed the data and wrote the manuscript. JR revised the manuscript and procured reagents and materials. All authors contributed to the article and approved the submitted version.
This research was supported by the Rising star junior fellowship awarded by the Department of Biology, Chemistry and Pharmacy at FU Berlin and Alexander von Humboldt Foundation to MK. JR and CZ were funded by Deutsche Forschungsgemeinschaft (DFG) (FOR5026).
We would like to thank Elisa Bittermann and Diana Aldana Alvarez for technical support, Alexandro Rodríguez-Rojas for useful advice, and Brian Lazzaro, Sophie Armitage, and Charlotte Rafaluk-Mohrfor for comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1140627/full#supplementary-material
1. Lazzaro BP, Zasloff M, Rolff J. Antimicrobial peptides: Application informed by evolution. Science (2020) 368(6490). doi: 10.1126/science.aau5480
2. Oyama LB, Girdwood SE, Cookson AR, Fernandez-Fuentes N, Privé F, Vallin HE, et al. The rumen microbiome: an underexplored resource for novel antimicrobial discovery. NPJ Biofilms Microbiomes. (2017) 3(1):1–9. doi: 10.1038/s41522-017-0042-1
3. Zanchi C, Johnston PR, Rolff J. Evolution of defence cocktails: antimicrobial peptide combinations reduce mortality and persistent infection. Mol Ecology. (2017) 26(19):5334–43. doi: 10.1111/mec.14267
4. Parvy J-P, Yu Y, Dostalova A, Kondo S, Kurjan A, Bulet P, et al. The antimicrobial peptide defensin cooperates with tumour necrosis factor to drive tumour cell death in drosophila. Elife (2019) 8:e45061. doi: 10.7554/eLife.45061.019
5. Korner P, Schmid-Hempel P. In vivo dynamics of an immune response in the bumble bee bombus terrestris. J invertebrate pathology. (2004) 87(1):59–66. doi: 10.1016/j.jip.2004.07.004
6. Haine ER, Pollitt LC, Moret Y, Siva-Jothy MT, Rolff J. Temporal patterns in immune responses to a range of microbial insults (Tenebrio molitor). J Insect Physiol (2008) 54(6):1090–7. doi: 10.1016/j.jinsphys.2008.04.013
7. Johnston PR, Makarova O, Rolff J. Inducible defenses stay up late: temporal patterns of immune gene expression in tenebrio molitor. G3: Genes Genomes Genet (2014) 4(6):947–55. doi: 10.1534/g3.113.008516
8. Makarova O, Rodriguez-Rojas A, Eravci M, Weise C, Dobson A, Johnston P, et al. Antimicrobial defence and persistent infection in insects revisited. Philos Trans R Soc B: Biol Sci (2016) 371(1695):20150296. doi: 10.1098/rstb.2015.0296
9. Taniai K, Furukawa S, Shono T, Yamakawa M. Elicitors triggering the simultaneous gene expression of antibacterial proteins of the silkworm, bombyx mori. Biochem Biophys Res Commun (1996) 226(3):783–90. doi: 10.1006/bbrc.1996.1429
10. Pöppel A-K, Vogel H, Wiesner J, Vilcinskas A. Antimicrobial peptides expressed in medicinal maggots of the blow fly lucilia sericata show combinatorial activity against bacteria. Antimicrobial Agents chemotherapy. (2015) 59(5):2508–14. doi: 10.1128/AAC.05180-14
11. Rahnamaeian M, Cytryńska M, Zdybicka-Barabas A, Dobslaff K, Wiesner J, Twyman RM, et al. Insect antimicrobial peptides show potentiating functional interactions against gram-negative bacteria. Proc R Soc B: Biol Sci (2015) 282(1806):20150293. doi: 10.1098/rspb.2015.0293
12. Yu G, Baeder DY, Regoes RR, Rolff J. Combination effects of antimicrobial peptides. Antimicrobial Agents chemotherapy. (2016) 60(3):1717–24. doi: 10.1128/AAC.02434-15
13. Hanson MA, Dostálová A, Ceroni C, Poidevin M, Kondo S, Lemaitre B. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. Elife (2019) 8:511. doi: 10.7554/eLife.44341
14. Hultmark D, Engström A, Andersson K, Steiner H, Bennich H, Boman H. Insect immunity. attacins, a family of antibacterial proteins from hyalophora cecropia. EMBO J (1983) 2(4):571–6. doi: 10.1002/j.1460-2075.1983.tb01465.x
15. Hu Y, Aksoy S. An antimicrobial peptide with trypanocidal activity characterized from glossina morsitans morsitans. Insect Biochem Mol Biol (2005) 35(2):105–15. doi: 10.1016/j.ibmb.2004.10.007
16. Wang L-N, Yu B, Han G-Q, Chen D-W. Molecular cloning, expression in escherichia coli of attacin a gene from drosophila and detection of biological activity. Mol Biol Rep (2010) 37(5):2463–9. doi: 10.1007/s11033-009-9758-1
17. Shin HS, Park S-I. Novel attacin from hermetia illucens: cDNA cloning, characterization, and antibacterial properties. Preparative Biochem Biotechnol (2019) 49(3):279–85. doi: 10.1080/10826068.2018.1541807
18. Moretta A, Salvia R, Scieuzo C, Di Somma A, Vogel H, Pucci P, et al. A bioinformatic study of antimicrobial peptides identified in the black soldier fly (BSF) hermetia illucens (Diptera: Stratiomyidae). Sci Rep (2020) 10(1):16875. doi: 10.1038/s41598-020-74017-9
19. Kwon Y, Kim H, Kim Y, Kang Y, Lee I, Jin B, et al. Comparative analysis of two attacin genes from hyphantria cunea. Comp Biochem Physiol Part B: Biochem Mol Biol (2008) 151(2):213–20. doi: 10.1016/j.cbpb.2008.07.002
20. Bang K, Park S, Yoo JY, Cho S. Characterization and expression of attacin, an antibacterial protein-encoding gene, from the beet armyworm, spodoptera exigua (Hübner)(Insecta: Lepidoptera: Noctuidae). Mol Biol Rep (2012) 39(5):5151–9. doi: 10.1007/s11033-011-1311-3
21. Wang Z, Feng K, Tang F, Xu M. Activation of the host immune response in hyphantria cunea (Drury)(Lepidoptera: Noctuidae) induced by serratia marcescens bizio. Insects (2021) 12(11):983. doi: 10.3390/insects12110983
22. Jo YH, Park S, Park KB, Noh MY, Cho JH, Ko HJ, et al. In silico identification, characterization and expression analysis of attacin gene family in response to bacterial and fungal pathogens in tenebrio molitor. Entomological Res (2018) 48(1):45–54. doi: 10.1111/1748-5967.12287
23. Buonocore F, Fausto AM, Della Pelle G, Roncevic T, Gerdol M, Picchietti S. Attacins: a promising class of insect antimicrobial peptides. Antibiotics (2021) 10(2):212. doi: 10.3390/antibiotics10020212
24. Yi H-Y, Chowdhury M, Huang Y-D, Yu X-Q. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol (2014) 98(13):5807–22. doi: 10.1007/s00253-014-5792-6
25. Chae J-H, Kurokawa K, So Y-I, Hwang HO, Kim M-S, Park J-W, et al. Purification and characterization of tenecin 4, a new anti-gram-negative bacterial peptide, from the beetle tenebrio molitor. Dev Comp Immunol (2012) 36(3):540–6. doi: 10.1016/j.dci.2011.09.010
26. Lee YJ, Chung TJ, Park CW, Hahn Y, Chung JH, Lee BL, et al. Structure and expression of the tenecin 3 gene inTenebrio molitor. Biochem Biophys Res Commun (1996) 218(1):6–11. doi: 10.1006/bbrc.1996.0002
27. Jang HA, Park KB, Kim BB, Ali Mohammadie Kojour M, Bae YM, Baliarsingh S, et al. In silico identification and expression analyses of defensin genes in the mealworm beetle tenebrio molitor. Entomological Res (2020) 50(12):575–85. doi: 10.1111/1748-5967.12468
28. Jang HA, Park KB, Kim BB, Ali Mohammadie Kojour M, Bae YM, Baliarsingh S, et al. Bacterial but not fungal challenge up-regulates the transcription of coleoptericin genes in tenebrio molitor. Entomological Res (2020) 50(9):440–9. doi: 10.1111/1748-5967.12465
29. Ali Mohammadie Kojour M, Jang HA, Edosa TT, Keshavarz M, Kim BB, Bae YM, et al. Identification, in silico characterization, and expression analysis of tenebrio molitor cecropin-2. Entomological Res (2021) 51(2):74–82. doi: 10.1111/1748-5967.12476
30. Kim DH, Noh MY, Park KB, Jo YH. Expression profiles of two thaumatin-like protein (TmTLP) genes in responses to various micro-organisms from tenebrio molitor. Entomological Res (2017) 47(1):35–40. doi: 10.1111/1748-5967.12197
31. Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, Spellman P, et al. Drosophila host defense after oral infection by an entomopathogenic pseudomonas species. Proc Natl Acad Sci (2005) 102(32):11414–9. doi: 10.1073/pnas.0502240102
32. Dieppois G, Opota O, Lalucat J, Lemaitre B. Pseudomonas entomophila: a versatile bacterium with entomopathogenic properties. Pseudomonas: Springer; (2015) p:25–49. doi: 10.1007/978-94-017-9555-5_2
33. Carrau T, Thümecke S, Silva LM, Perez-Bravo D, Gärtner U, Taubert A, et al. The cellular innate immune response of the invasive pest insect drosophila suzukii against pseudomonas entomophila involves the release of extracellular traps. Cells (2021) 10(12):3320. doi: 10.3390/cells10123320
34. Hidalgo BA, Silva LM, Franz M, Regoes RR, Armitage SA. Decomposing virulence to understand bacterial clearance in persistent infections. Nat Commun (2022) 13(1):1–14. doi: 10.1038/s41467-022-32118-1
35. Rolff J, Armitage SA, Coltman DW. Genetic constraints and sexual dimorphism in immune defense. Evolution (2005) 59(8):1844–50. doi: 10.1111/j.00143820.2005.tb01831.x
36. Schmittgen T, Livak KJ. Analyzing real-time PC’R data hy the comparative (2 (“ i’) method. J 1 Nat Protoc (2008) 3(6):1101–8. doi: 10.1038/nprot.2008.73
37. Nikolouli K, Mossialos D. Functional characterization of TtgABC efflux pump of the RND family in the entomopathogenic bacterium pseudomonas entomophila. Ann Microbiol (2016) 66(1):499–503. doi: 10.1007/s13213-015-1119-9
38. Team RC. R: A language and environment for statistical computing Vol. 2021. . Vienna, Austria: R Foundation for Statistical Computing (2012).
40. Venables WN, Ripley BD. Random and mixed effects. Modern Appl Stat S: Springer; (2002), 271–300. doi: 10.1007/978-0-387-21706-2_10
41. Cumming G. Inference by eye: Reading the overlap of independent confidence intervals. Stat Med (2009) 28(2):205–20. doi: 10.1002/sim.3471
42. Akaike H. Canonical correlation analysis of time series and the use of an information criterion. Mathematics Sci engineering. 126: Elsevier; (1976) p:27–96. doi: 10.1016/S0076-5392(08)60869-3
43. Machin D, Cheung YB, Parmar M. Survival analysis: a practical approach. London: John Wiley & Sons (2006).
45. Ben-Shachar MS, Makowski D, Lüdecke D, Patil I, Wiernik B, Kelley K, et al. Effectsize: Indices of effect size and standardized parameters. J Open Source Softw (2021) 5. doi: 10.21105/joss.02815
46. Duneau D, Ferdy J-B, Revah J, Kondolf H, Ortiz GA, Lazzaro BP, et al. Stochastic variation in the initial phase of bacterial infection predicts the probability of survival in d. melanogaster. Elife (2017) 6. doi: 10.7554/eLife.28298
47. Nainu F, Asri RM, Arsyad A, Manggau MA, Amir MN. In vivo antibacterial activity of green algae ulva reticulata against staphylococcus aureus in drosophila model of infection. Pharmacognosy J (2018) 10(5):993–7. doi: 10.5530/pj.2018.5.169
48. Best A, White A, Boots M. Maintenance of host variation in tolerance to pathogens and parasites. Proc Natl Acad Sci (2008) 105(52):20786–91. doi: 10.1073/pnas.0809558105
49. Chambers MC, Jacobson E, Khalil S, Lazzaro BP. Consequences of chronic bacterial infection in drosophila melanogaster. PloS One (2019) 14(10):e0224440. doi: 10.1371/journal.pone.0224440
50. Carboni AL, Hanson MA, Lindsay SA, Wasserman SA, Lemaitre B. Cecropins contribute to drosophila host defense against a subset of fungal and gram-negative bacterial infection. Genetics (2022) 220(1):iyab188. doi: 10.1093/genetics/iyab188
51. Clemmons AW, Lindsay SA, Wasserman SA. An effector peptide family required for drosophila toll-mediated immunity. PloS Pathogens. (2015) 11(4):e1004876. doi: 10.1371/journal.ppat.1004876
Keywords: RNA interference, antimicrobial peptides (AMPs), host survival, Attacin, Pseudomonas entomophila, Tenebrio molitor
Citation: Keshavarz M, Zanchi C and Rolff J (2023) The effect of combined knockdowns of Attacins on survival and bacterial load in Tenebrio molitor. Front. Immunol. 14:1140627. doi: 10.3389/fimmu.2023.1140627
Received: 09 January 2023; Accepted: 14 March 2023;
Published: 28 March 2023.
Edited by:
Krishnendu Mukherjee, University Hospital Münster, GermanyReviewed by:
Heiko Vogel, Max Planck Institute for Chemical Ecology, GermanyCopyright © 2023 Keshavarz, Zanchi and Rolff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maryam Keshavarz, bWFyaWFrc2h2cnpAZ21haWwuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.