- 1Medical Genetics, Department of Medical Sciences and Public Health, University of Cagliari, Cagliari, Italy
- 2AART-ODV (Association for the Advancement of Research on Transplantation), Cagliari, Italy
- 3Medical Genetics, R. Binaghi Hospital, Local Public Health and Social Care Unit (ASSL) of Cagliari, Cagliari, Italy
- 4Department of Medical Sciences and Public Health, University of Cagliari, Cagliari, Italy
- 5Liver Unit, University Hospital, Cagliari, Italy
- 6Anesthesia and Intensive Care Unit, R. Binaghi Hospital, Local Public Health and Social Care Unit (ASSL) of Cagliari, Cagliari, Italy
- 7Neurogenomics Division, Translational Genomics Research Institute (TGen), Phoenix, AZ, United States
- 8Section of Pathology, Oncology and Molecular Pathology Unit, Department of Biomedical Sciences, University of Cagliari, Cagliari, Italy
- 9GeneMos-APS (Association for Social Advancement), Reggio Calabria, Italy
- 10Centre for Research University Services (CeSAR, Centro Servizi di Ateneo per la Ricerca), University of Cagliari, Monserrato, Italy
Introduction: A large number of risk and protective factors have been identified during the SARS-CoV-2 pandemic which may influence the outcome of COVID-19. Among these, recent studies have explored the role of HLA-G molecules and their immunomodulatory effects in COVID-19, but there are very few reports exploring the genetic basis of these manifestations. The present study aims to investigate how host genetic factors, including HLA-G gene polymorphisms and sHLA-G, can affect SARS-CoV-2 infection.
Materials and Methods: We compared the immune-genetic and phenotypic characteristics between COVID-19 patients (n = 381) with varying degrees of severity of the disease and 420 healthy controls from Sardinia (Italy).
Results: HLA-G locus analysis showed that the extended haplotype HLA-G*01:01:01:01/UTR-1 was more prevalent in both COVID-19 patients and controls. In particular, this extended haplotype was more common among patients with mild symptoms than those with severe symptoms [22.7% vs 15.7%, OR = 0.634 (95% CI 0.440 – 0.913); P = 0.016]. Furthermore, the most significant HLA-G 3’UTR polymorphism (rs371194629) shows that the HLA-G 3’UTR Del/Del genotype frequency decreases gradually from 27.6% in paucisymptomatic patients to 15.9% in patients with severe symptoms (X2 = 7.095, P = 0.029), reaching the lowest frequency (7.0%) in ICU patients (X2 = 11.257, P = 0.004). However, no significant differences were observed for the soluble HLA-G levels in patients and controls. Finally, we showed that SARS-CoV-2 infection in the Sardinian population is also influenced by other genetic factors such as β-thalassemia trait (rs11549407C>T in the HBB gene), KIR2DS2/HLA-C C1+ group combination and the HLA-B*58:01, C*07:01, DRB1*03:01 haplotype which exert a protective effect [P = 0.005, P = 0.001 and P = 0.026 respectively]. Conversely, the Neanderthal LZTFL1 gene variant (rs35044562A>G) shows a detrimental consequence on the disease course [P = 0.001]. However, by using a logistic regression model, HLA-G 3’UTR Del/Del genotype was independent from the other significant variables [ORM = 0.4 (95% CI 0.2 – 0.7), PM = 6.5 x 10-4].
Conclusion: Our results reveal novel genetic variants which could potentially serve as biomarkers for disease prognosis and treatment, highlighting the importance of considering genetic factors in the management of COVID-19 patients.
1 Introduction
Over the past 20 years, human coronaviruses (HCoVs), such as SARS-CoV, MERS-CoV, and SARS-CoV-2, have resulted in outbreaks of serious respiratory illness (1–6). The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the ongoing coronavirus disease-19 (COVID-19) pandemic (7–10), which has led to more than 6.3 million deaths globally as of June 2022 (11–13). COVID-19 can result in severe respiratory distress syndrome, coagulopathies, septic shock, and multiple organ injuries (14–17). Studies have revealed differences in COVID-19 incidence and lethality based on gender and age, with a higher incidence in women and higher lethality in men (18). Additionally, young individuals (aged 0-24) have a lower COVID-19 incidence rate than a group of people over 65 years of age (19, 20). Vaccine development has appeared to be the most effective approach in slowing the spread of COVID-19 (21–26), but the emergence of new variants of concern (VOC) has challenged vaccine efficacy and durability (27–30).
Various clinical outcomes have been described among COVID-19 patients since the outbreak. While some patients remain asymptomatic, others may develop respiratory or multiorgan failure with potentially lethal outcomes (31, 32).
Genetic factors may influence the individual’s susceptibility or resistance to viral infections by regulating the immune response (33–38). In particular, recent studies have also highlighted the role of specific genetic variants associated with asymptomatic COVID-19, such as genes of the lectin pathway (39).
However, the majority of the studies have indicated that COVID-19 development and/or severity are associated with polymorphisms in innate and adaptive immune genes, including killer-cell immunoglobulin-like receptors (KIR) and human leukocyte antigens (HLA) class I and II as well as genes involved in viral response pathways (LZTFL1, OAS gene family) (40–43).
The HLA-G, a non-classical HLA class I molecule, has been shown to play a critical role in immune response modulation and has been implicated in various pathological processes including response to viral infections (44). It is physiologically expressed in extravillous cytotrophoblast cells and is an essential factor in maternal-fetal tolerance (45, 46). Given its central role in immunotolerance, it is involved in several pathological conditions such as carcinogenesis, acute and chronic inflammation, autoimmune diseases, organ transplantation, allergies, parasitic diseases and response to viral infections (47, 48). HLA-G molecules interact primarily with ILT-2/LILRB1, ILT-4/LILRB2 and KIR2DL4 receptors, the immune checkpoint, to exert its inhibitory effects on immune cells (45). The interaction of HLA-G molecules with these specific receptors inhibits the proliferation and maturation of dendritic cells, cytotoxic NK cells (CD56dim, CD16+), and induces apoptosis in CD8+ T cells, while reducing the proliferation of CD4+ T cells and B cells (49). Thus, HLA-G can interfere in many different immunological processes of both the innate and adaptive immune system leading to a reduced immune response. Because of the extensive number of alleles and their associated regulatory regions (IPD-IMGT/HLA database, version 3.24.0.1), HLA-G expression levels between individuals differ widely (50). Moreover, the complexity of this system is increased by regulation at the transcriptional and post-translational levels, resulting in the production of seven alternative transcripts, four of which are membrane-bound (HLA-G1-G4), and 3 are soluble (HLA-G5-G7) (51, 52). To date, there were found several single nucleotide polymorphisms (SNPs) and an HLA-G insertion/deletion (Ins/Del; rs3711944629) of 14-base pair on the 3’UTR, in which the Del-Del genotype has been associated with high expression of HLA-G mRNA, whereas the Ins-Ins genotype has been associated with lower mRNA production (45, 49, 53). Several haplotypes have been described in the 3’UTR (UTR-1, UTR-2, UTR-3, UTR-4, UTR-5, UTR-6/-18, and UTR-7) of this gene, suggesting that HLA-G may influence immune responses to different stimuli, including in viral infections (45). Although the exact mechanisms by which the immunomodulatory molecule HLA-G influences disease presentation and progression are still not fully understood, research has shown that viral infections can lead to an increase in both the cell surface membrane-bound and soluble peripheral expression of HLA-G (53).
The association between HLA-G expression and COVID-19 severity and progression has been studied in some studies with contradictory results. Two hypotheses have been proposed to explain this discrepancy: the first is that the immunosuppressive action of HLA-G enhances the virus’s ability to escape the immune system, while the second is that HLA-G expression and secretion are a robust response to inflammation during the viral infection (53, 54). This suggests that high levels of HLA-G molecules may inhibit neutrophil adhesion to endothelial cells, resulting in a negative association between elevated levels of HLA-G and disease progression (55, 56).
In most of the studies conducted to date, the attention has focused mainly on the expression of HLA-G and serum levels of the molecule in patients with severe COVID-19, rather than the genetic basis from which these manifestations result. Starting from this consideration, this study aims to investigate the genetic basis of the HLA-G and its role in the manifestation of SARS-CoV-2 infections. To this aim, we used the Sardinian population as a model of study, which is noteworthy for its high degree of genetic homogeneity. This makes it ideal for studying genetic and immunogenetic features, including the role of innate and adaptive immunity in viral infections.
In previous studies, carried out during the outbreak of the SARS-CoV-2- B.1.1.7 variant in Italy, we observed how specific genetic factors in the Sardinian population significantly impact the outcome of COVID-19 infections (36, 38, 43). We have found that certain genetic factors, including the HLA extended haplotype HLA-B58:01, C07:01, DRB1*03:01, β°39 C>T variant at the HBB gene and KIR2DS2 gene/HLA C1 group ligand combination, can positively influence the course of the disease and result in an asymptomatic outcome (36, 38, 43). While the Neanderthal-inherited haplotype (rs35044562, rs73064425, rs34326463, rs67959919) at the leucine zipper transcription factor-like 1 (LZTFL1) has been associated with an increased risk of serious symptoms (36).
In this study, we evaluated a new and larger group of unvaccinated individuals affected by COVID-19, who were enrolled during the spread of the B.1.617.1 (Delta) variant (13). This allowed us to investigate whether the previously identified risk and protective factors still played a role in the infection caused by this different SARS-CoV-2 variant. Moreover, we explored other genetic traits, such as OAS3 protective haplotype and G6PDH enzyme deficiencies, which have not been found critical in our past studies (36, 38).
This study aimed to confirm the robustness of previous findings with a new pool of individuals and investigate the role of HLA-G in COVID-19. In particular, we evaluated the role of HLA-G, both as a single factor and in correlation with other factors, to determine its strength in affecting the outcome of the disease. Throughout this study, the goal was to contribute to the development of a broader spectrum of prognostic factors that could be used in the future.
2 Materials and methods
2.1 Patients and controls selection
In this study, we analyzed data from 381 unvaccinated patients, recruited between 1 August and 30 October 2021 at the Covid Unit of the SS.Trinità Hospital in Cagliari (Italy). In this period, the predominant variant circulating worldwide was the SARS-CoV-2 Delta (B.1.617.2) (25). All the recruited patients were diagnosed with SARS-CoV-2 by RT-PCR from a nasopharyngeal swab. Following the WHO’s guidelines, patients were divided into two groups: 207 patients had been admitted to the Covid Unit of the SS.Trinità Hospital in Cagliari with moderate or severe disease, including 57 intensive care unit (ICU) patients, (Group S) and 174 asymptomatic or paucisymptomatic patients (Group A) were confined to home quarantine. Four hundred twenty unrelated healthy individuals, from the Sardinian Voluntary Bone Marrow Donor Registry, were enrolled as control group. According to three-generation family trees, both groups (patients and controls) were from South Sardinia.
2.2 Ethics statement
The research protocol was conducted at the Department of Medical Sciences and Public Health of the University of Cagliari, the University Hospital of Cagliari (AOUCA), and the SS. Trinità Hospital of the Sardinian Regional Company for the Protection of Health (ATS Sardegna) where patient recruitment took place. In accordance with the local human research committee’s national and institutional ethical standards, all patients and controls provided informed consent. The informed consent procedures in the study protocol are in accordance with the ethical guidelines outlined in the Declaration of Helsinki and have been approved by the responsible ethics committee (Ethics Committee of the Cagliari University Hospital; date of approval: May 27th, 2020; protocol number GT/2020/10894). Documents containing written informed consent are kept on file and included in each patient’s clinical records.
2.3 DNA extraction and genetic analysis
The genomic DNA from peripheral blood mononuclear cells was extracted following the standard methods (57). All 801 samples from patients and controls were genotyped at high resolution for the alleles at HLA-A, -B, -C, -DR and -G loci using Next-generation sequencing (NGS) AlloSeq Tx17 (CareDx) method based on Hybrid Capture Technology and performed on the Illumina platform. The data was analyzed using the AlloSeq Assign® software (v.1.0.2). The full-length HLA-G gene was sequenced through long-range PCR, including the 3’UTR non-coding region. Primers were designed using Primer3web (version 4.1.0), based on HLA-G RefSeqGene version NG_029039.1 (NCBI database), as previously described (58).
Starting with 1 ng of PCR product, the libraries were prepared using the Nextera XT DNA Library Preparation Kit. On MiSeq Illumina Sequencer, a pool of normalized libraries (4 nM) was loaded onto V3 flow cells for 600 cycles of paired-end sequencing. Alignment and variant calling of the FASTQ files were processed by MiSeq Reporter v2.6, and variant classification was performed using VariantStudio Software v3.0 (Illumina, Netherlands). Each variant was validated individually and then entered into appropriate spreadsheets for statistical analysis as reported later on.
The 3’UTR haplotypes of HLA-G were determined based on variations in their nucleotide sequences between +2945 and +3259 nucleotides of the 3’UTR using the methodology and nomenclature described elsewhere (47, 59–61).
Moreover, we performed the KIR typing in order to detect the presence of the 14 KIR genes KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5, KIR3DL1, KIR3DL2, KIR3DL3, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5 and KIR3DS1 using PCR-SSP with primers specific for each locus according to a previously reported method (62, 63).
We explored the Neanderthal haplotype in the LZTFL1 gene, which consists of the variants rs35044562, rs73064425, rs34326463, rs67959919, and is most strongly associated with the risk of developing a severe form of COVID-19. We considered rs35044562 as the index risk variant for severe infection (36).
Additionally, we examined another Neanderthal inherited variant located in the OAS3 gene (rs1156361) which has been associated with protection against severe COVID-19 (64). Our final step was to sequence the rs11549407 (C>T) variant at codon 39 of the hemoglobin subunit beta gene (HBB), the predominant mutation responsible for beta-thalassemia in Sardinia (65). Primer pairs for each region of interest were designed using Primer3web (version 4.1.0), as we previously reported (36).
The PCR reaction was performed according to the protocol supplied with AmpliTaq Gold™ DNA Polymerase (Applied Biosystems/Thermo Fisher Scientific, Waltham, MA) for each region containing the three SNPs: rs35044562, rs1156361 and rs11549407 respectively located within the LZTFL1, OAS3 and the HBB gene.
Sequencing was performed using the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA), with the same primers described previously followed by cleanup with CleanSEQ Dye-Terminator Removal Kit (Beckman Coulter, Inc.). Capillary electrophoresis was run on the ABI 3500 Genetic Analyzer (Applied Biosystems) and sequences were analyzed with Sequencher 5.3 (© 2017 Gene Codes Corporation).
2.4 Soluble HLA-G and G6PD activity quantification
Plasma samples were collected from all 381 convalescent COVID-19 patients, from one to six months after recovery, and 420 controls were recruited for this study. The levels of sHLA-G were determined using the sHLA-G ELISA assay kit (Exbio, Prague, Czech Republic), which detects both shedding HLA-G-1 and soluble HLA-G-5 molecules. The assay was conducted on the plasma samples immediately frozen after separation and stored at -80°C until use. Fifty µl of each sample were diluted 1:80 in the plasma-specific buffer. A six-point calibration curve was obtained using the human native HLA-G protein included in the kit. At the end of the reaction, optical density was measured using a microplate reader with a 450 nm filter. The limit of sensitivity was 0.6 U/ml. For all samples a technical replicate was included.
The activity of the G6PD enzyme was quantified by measuring an increase in absorbance of NADPH at 340 nm during the enzyme-catalyzed reaction (66). The assay was conducted using the Randox G6PD assay kit (Randox Laboratories Ltd., Crumlin, UK) as described in other studies (67).
2.5 Statistical analysis
Clinical and biochemical parameters of COVID-19 patients were reported using mean values and standard deviations (SD) or percentages, as appropriate. The Student’s t-test was used to compare continuous variables between patients and controls. The Fisher’s exact test was used to determine P values and odds ratios (ORs), along with their 95% confidence intervals (CIs), when we compared the categorical data between patients and controls or subgroups of patients. To account for multiple testing based on the number of HLA-G alleles or 3’UTR haplotypes analyzed, the P values (Pc) were adjusted. The results were considered statistically significant only when the adjusted P value (Pc) was lower than 0.05. Statistical analysis was performed with the R programming language (R version 4.2.2) [R core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/]. We examined the Hardy-Weinberg equilibrium (HWE) of the HLA-G 14bp Ins/Del polymorphism by computing and P values. Deviation from HWE was assessed using Haploview 4.0 software (Broad Institute, Cambridge, MA, USA) (68).
All tests were two-sided and only values of P < 0.05 were accepted as being statistically significant.
Soluble HLA-G plasma levels in controls, patients and subgroups of patients were represented by boxplots. We used the non-parametric Wilcoxon rank sum test (69) for comparisons between two groups and the Kruskal-Wallis rank sum test (70) for comparisons between three groups.
A multivariate logistic regression analysis was conducted to determine the independence of clinical and genetic variables with respect to age and gender, based on the results of the univariate analysis (PU< 0.05) that showed statistically significant differences between the groups (A and S patients). The univariate P values and ORs have adjusted accordingly to age and gender using a logistic regression model. The multivariate P values (PM) were corrected for multiple comparisons (PMC).
3 Results
3.1 Clinical characteristics of patients with COVID-19
Clinical and genetic characteristics of SARS-CoV-2 positive patients are shown in Table 1. The mean age at diagnosis was 56 years (mean ± SD: 56.0 ± 17.4). According to the results, 28.4% (n = 108) of the patients were under the age of 50, and 41.5% were over 65 years of age.
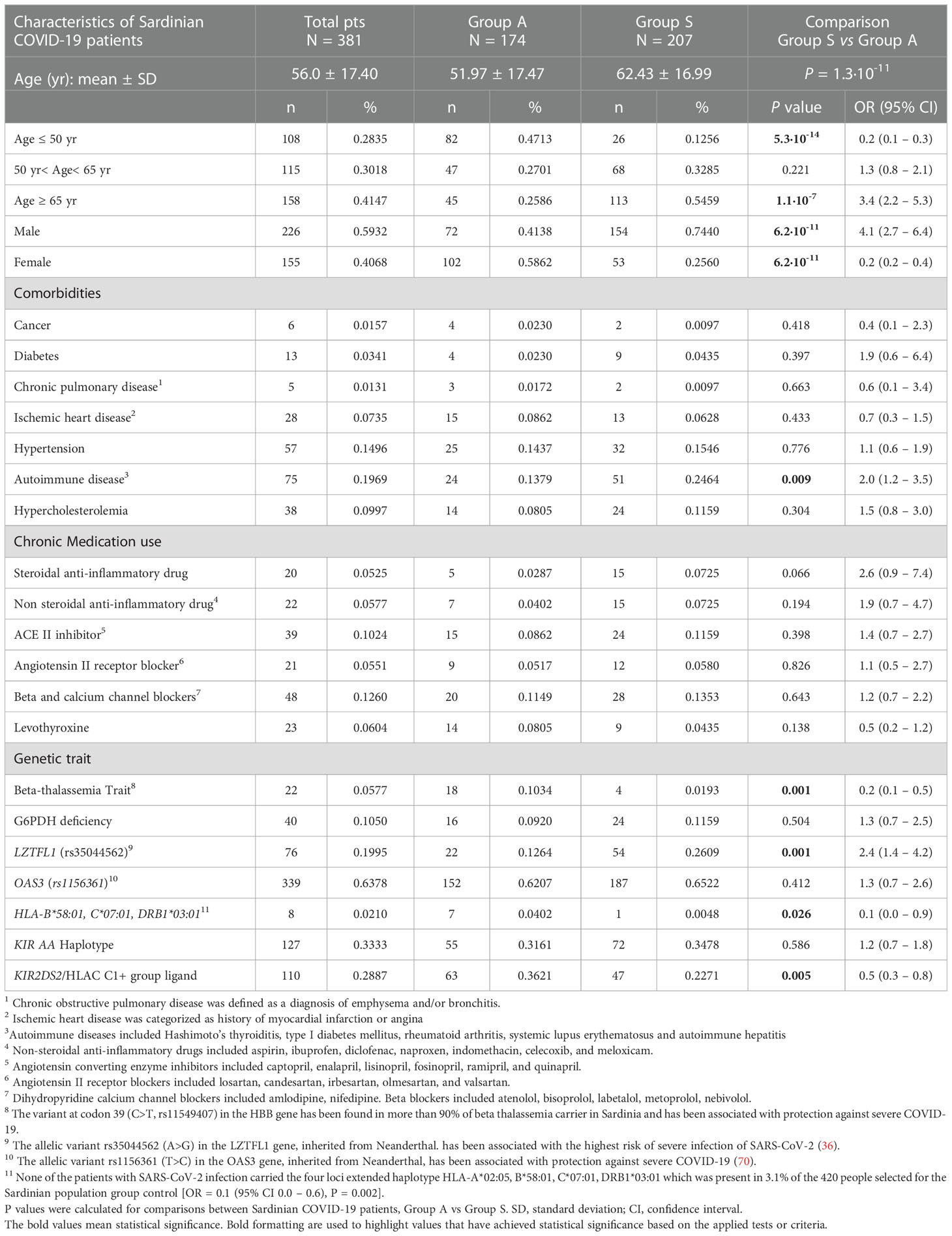
Table 1 Comparisons of baseline clinical and genetic parameters between COVID-19 patients with either asymptomatic/pauci-symptomatic (Group A) or moderate/severe (Group S) disease.
A significant proportion of patients over 65 years of age had more severe symptoms and clinical manifestations than adults less than 65 years old [OR = 3.4 (95% CI 2.2–5.3), P = 1.1 x 10−7].
On the other hand, a significant percentage of patients under the age of 50 seemed to have less severe symptoms than patients over 50 years old [OR = 0.2 (95% CI 0.1–0.3), P = 5.3 x 10−8].
COVID-19 occurred at the same rate in males and females, with males slightly more likely to contract the disease (59.3%). In addition, the results suggest that female patients are less susceptible to COVID-19 severe effects [74.4% males vs. 25.6% females, OR = 4.1 (95% CI 2.7–6.4), P = 6.2 x 10-11].
Our analysis also included several other comorbidities that may be involved in the disease course of COVID-19, in addition to sex and age. In our cohort, most patients with comorbidity had autoimmune diseases (19.7%). The remaining comorbidities were: 15% arterial hypertension, 10% hypercholesterolemia, 7.36% ischemic heart disease, 3.5% type I diabetes, 1.3% chronic pulmonary disease and 1.1% cancer. For each comorbidity present in the total cohort of patients, we compared group S (severe) and group A (asymptomatic) patients. There was no significant difference regarding comorbidities except for a higher prevalence of autoimmune disorders among patients with severe disease [24.6% A vs. 13.8% S, OR = 2.0 (95% CI 1.2–3.5), P = 0.009]. Moreover, there was no difference in chronic drug intake between the two groups of patients.
3.2 Genetic traits influencing COVID-19 outcome
As a further step, we investigated genetic traits likely to influence the outcome of COVID-19 caused by the B.1.617.2 SARS-CoV-2 variant in the Sardinian population. The results of each genetic trait analyzed are presented in detail in Supplementary Tables (S1: Allele and Genotype distribution of rs35044562, rs1156361 and rs11549407 in SARS-CoV-2 patients; S2: HLA alleles and Haplotypes frequencies compared between Group A and S; S3: KIR genes and genotype frequencies compared between Group A and S; S4: Comparisons of KIR genes and their cognate HLA ligands between COVID-19 patients between Group A and S).
None of the patients with SARS-CoV-2 infection in group S carried the extended haplotype HLA-A*02:05, B*58:01, C*07:01, DRB1*03:01 whose frequency was 3.2% in the control group [OR 0.1 (95% CI 0.1 – 0.6), P = 0.002].
The three-locus HLA haplotype HLA-B*58:01, C*07:01, DRB1*03:01 was almost absent in group S [2.0% A vs 0.48% S, OR = 0.1 (95% CI 0.0 – 0.9), P = 0.026], as well as the KIR2DS2/HLA-C C1+ group ligand, which was more prevalent in group A [36.2% A vs 22.7% S, OR = 0.5 (95% CI 0.3 – 0.8), P = 0.005], suggesting their protective effect.
Similar results were obtained for the beta-thalassemia trait (rs11549407 C>T) that was found in 5.7% of patients, with higher prevalence in group A than in group S [10.3% A vs. 1.9% S, OR = 0.2 (95% CI 0.1 – 0.5), P = 0.001].
Additionally, the Neanderthal variant rs35044562 (A>G), reported to be associated with a severe form of COVID-19, was clearly associated with group S [12.6% A vs. 26.1% S, OR = 2.4 (95% CI 1.4 - 4.2), P = 0.001]. Finally, there was no significant difference between the two groups of patients in terms of frequencies in the KIR AA haplotype, G6PDH enzyme deficiency and OAS3 (rs1156361) polymorphism [62% A vs 65% S, OR = 1.1 (95% CI 0.8 – 1.7), P = 0.593].
3.2.1 Analysis of the locus HLA-G
3.2.1.1 A comparison of HLA-G alleles and 3’UTR haplotype frequencies between the population group and COVID-19 patients
HLA-G alleles and 3’UTR haplotype frequencies were compared in 381 COVID-19 patients and 420 healthy controls (Table 2). The analysis of extended haplotypes (HLA-G alleles and 3’UTR haplotype) showed there were few substantive differences in frequency between patients and healthy controls. In both groups, the most prevalent extended haplotypes were HLA-G*01:01:01:01/UTR-1 (27.5% controls, 18.9% patients), HLA-G*01:03:01:02/UTR-5 (15.6% controls, 9.8% patients), and HLA-G*01:01:02:01/UTR-2 (11.8% controls, 13.8% patients).
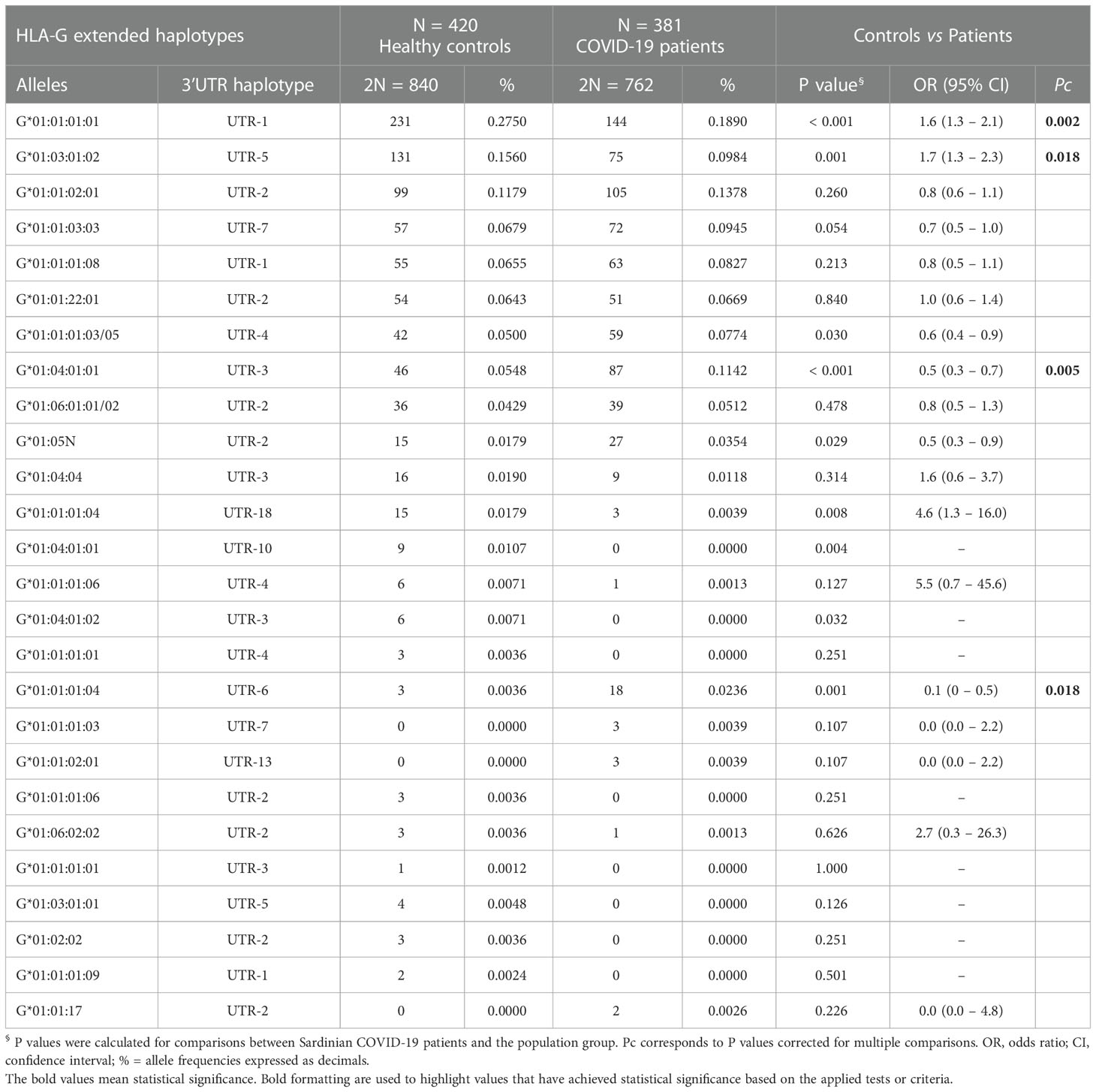
Table 2 Extended haplotypes (HLA-G alleles and 3’UTR haplotypes) frequencies in healthy controls and COVID-19 patients.
The HLA-G*01:01:01:01/UTR-1 and HLA-G*01:03:01:02/UTR-5 showed a lower and significantly different frequency in patients [(27.5% vs 18.9%, OR = 1.6 (95% CI 1.3 – 2.1); P< 0.001, Pc = 0.002) and [15.6% vs 9.8%, OR = 1.7 (95% CI 1.3 – 2.3); P = 0.001, Pc = 0.018)].
Conversely, the HLA-G*01:04:01:01/UTR-3 and HLA-G*01:01:01:04/UTR-6 were more prevalent in the patient’s group (5.5% vs 11.4% and 0.36% vs 2.4% respectively), with a significant difference only for the first of the two haplotypes: HLA-G*01:04:01:01/UTR-3 [OR = 0.5 (95% CI 0.3 – 0.7); P< 0.001, Pc = 0.005].
Additionally, we evaluated the frequencies of HLA-G 3’UTR haplotypes (Table 3). In both groups, UTR-1 was the most prevalent haplotype in both groups. Additionally, only UTR-1 and UTR-5 showed a significant difference in frequency between controls and patients [34.3% vs 27.2%, OR = 1.4 (95% CI 1.1 – 1.7); P = 0.002, Pc = 0.02 and 16.1% vs 9.8%, OR = 1.8 (95% CI 1.3 – 2.4); P< 0.001, Pc = 0.001 respectively]. UTR-6 and UTR-10 showed marginal significance in their P-values due to their low frequencies.
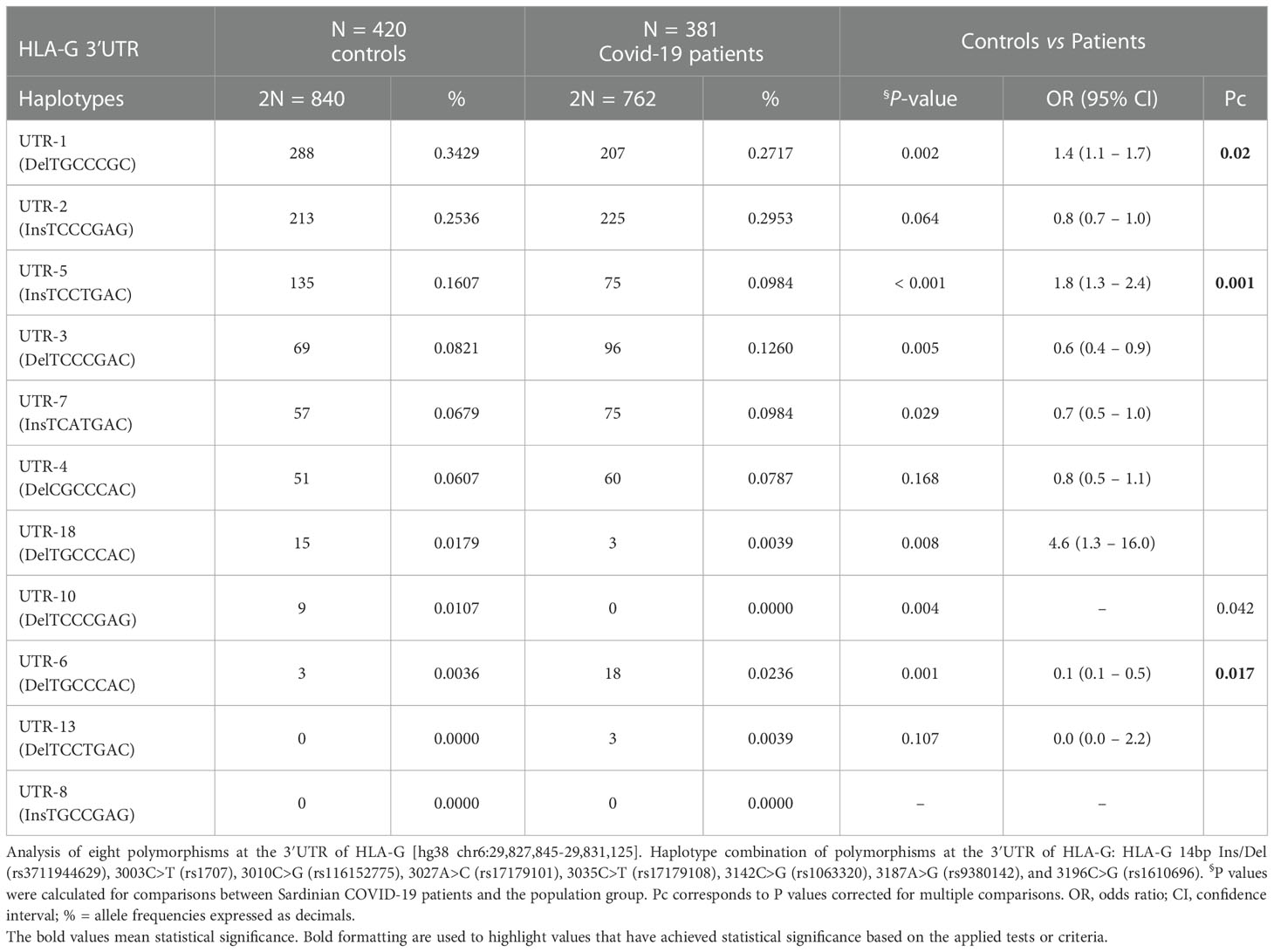
Table 3 Haplotype frequencies observed at the HLA-G 3’UTR polymorphic sites in population group controls and COVID-19 patients.
3.2.1.2 Correlation of HLA-G alleles and 3’UTR haplotype frequencies to the clinical manifestations of SARS-CoV-2 infection
The patients were then divided into two groups based on disease severity: Group A (asymptomatic and paucisymptomatic ill SARS-CoV-2 patients) and Group S (severely ill patients).
The results of the analysis of HLA-G alleles and 3’UTR haplotype frequencies in patients with SARS-CoV-2 infection appeared notable in relation to the severity of the clinical picture (Table 4). Among the twenty-six HLA-G extended haplotypes, only the HLA-G*01:01:01:01/UTR-1 and HLA-G*01:01:03:03/UTR-7 showed a significant association. In particular, the extended haplotype HLA-G*01:01:01:01/UTR-1 was more frequent in Group A [22.7% vs 15.7%, OR = 0.634 (95% CI 0.440 – 0.913); Pc = 0.016] (Table 5).
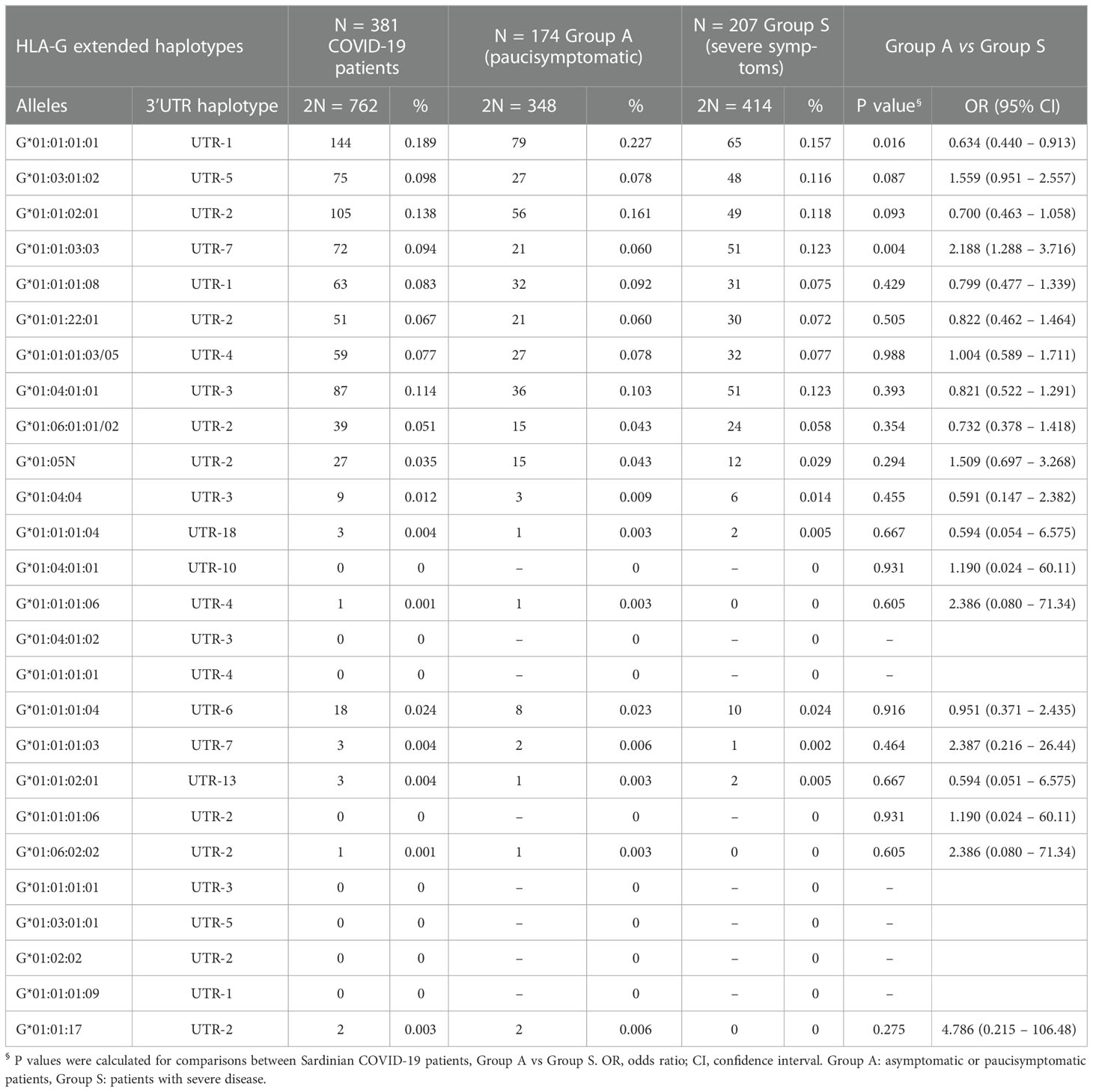
Table 4 Comparisons of HLA-G extended haplotypes among COVID-19 patients divided according to severity of the clinical manifestations.
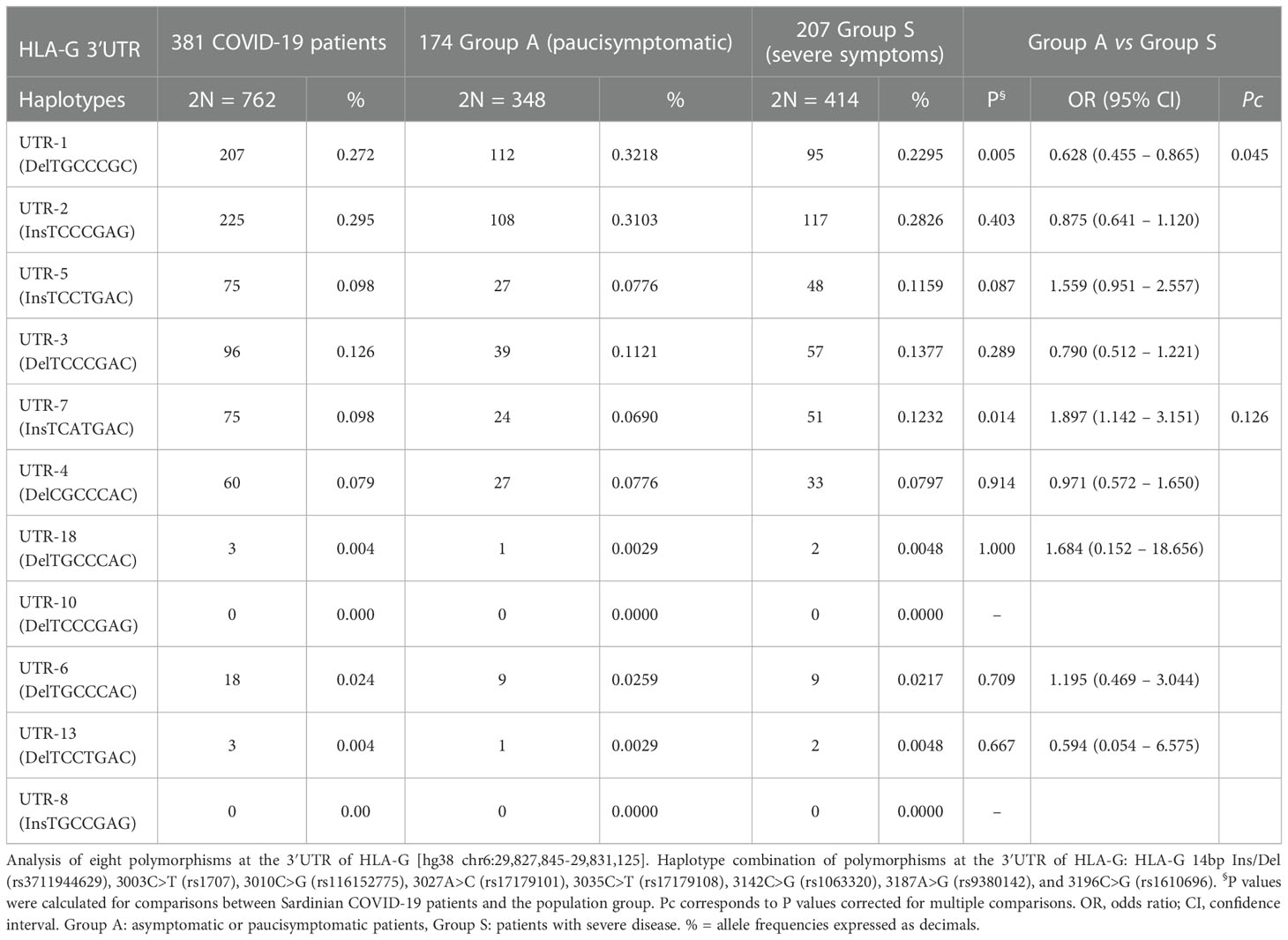
Table 5 Haplotype frequencies observed at the HLA-G 3’UTR polymorphic sites in COVID-19 patients divided according to severity of the clinical manifestations.
In addition, we examined the genotype and alleles frequencies distribution of the HLA-G 14bp Ins/Del polymorphism in both controls and patients (Table 6). Among the nine polymorphisms constituting the HLA-G UTR-1 haplotype, the Del variant (rs371194629, 14bp deletion) was found to be the most significant. The Ins/Del polymorphism variants revealed a distribution in Hardy–Weinberg equilibrium (HWE) both in group A patients and the control population. The P-values for the control population were not statistically significant ( = 0.789, P = 0.375 and = 1.031, P = 0.310, respectively). On the other hand, the HLA-G 14bp Ins/Del polymorphism in the total COVID-19 patient population and in Group S were not in HWE ( = 8.527, P = 0.003 and = 10.369, P = 0.001, respectively). Finally, the HWE for Intensive Care Unit (ICU) patients was close to being statistically significant ( = 3.079, P = 0.079).
The deviation from HWE is due to the reduced frequency of HLA-G Del variants among severe COVID-19 patients. The frequency of HLA-G Del/Del genotype decreases gradually from 27.6% in Group A to 15.9% in Group S (X2 = 7.095, P = 0.029), and reaches its lowest frequency of 7.0% among ICU patients (X2 = 11.257, P = 0.004).
The 14bp Ins/Del polymorphism at HLA-G 3’UTR implies an imbalance between Ins and Del variants, particularly in severe and ICU patients. As shown in Figure 1A Del allele was significantly less frequent in Group S and ICU patients compared to Group A and controls. Specifically, the Del variant was present in 35.1% (2N=40) of ICU patients and in 51.1% (2N= 429) of controls (ICU vs controls, P = 0.001) and in 54.3% (2N=189) of Group A (ICU vs Group A, P = 0.0004).
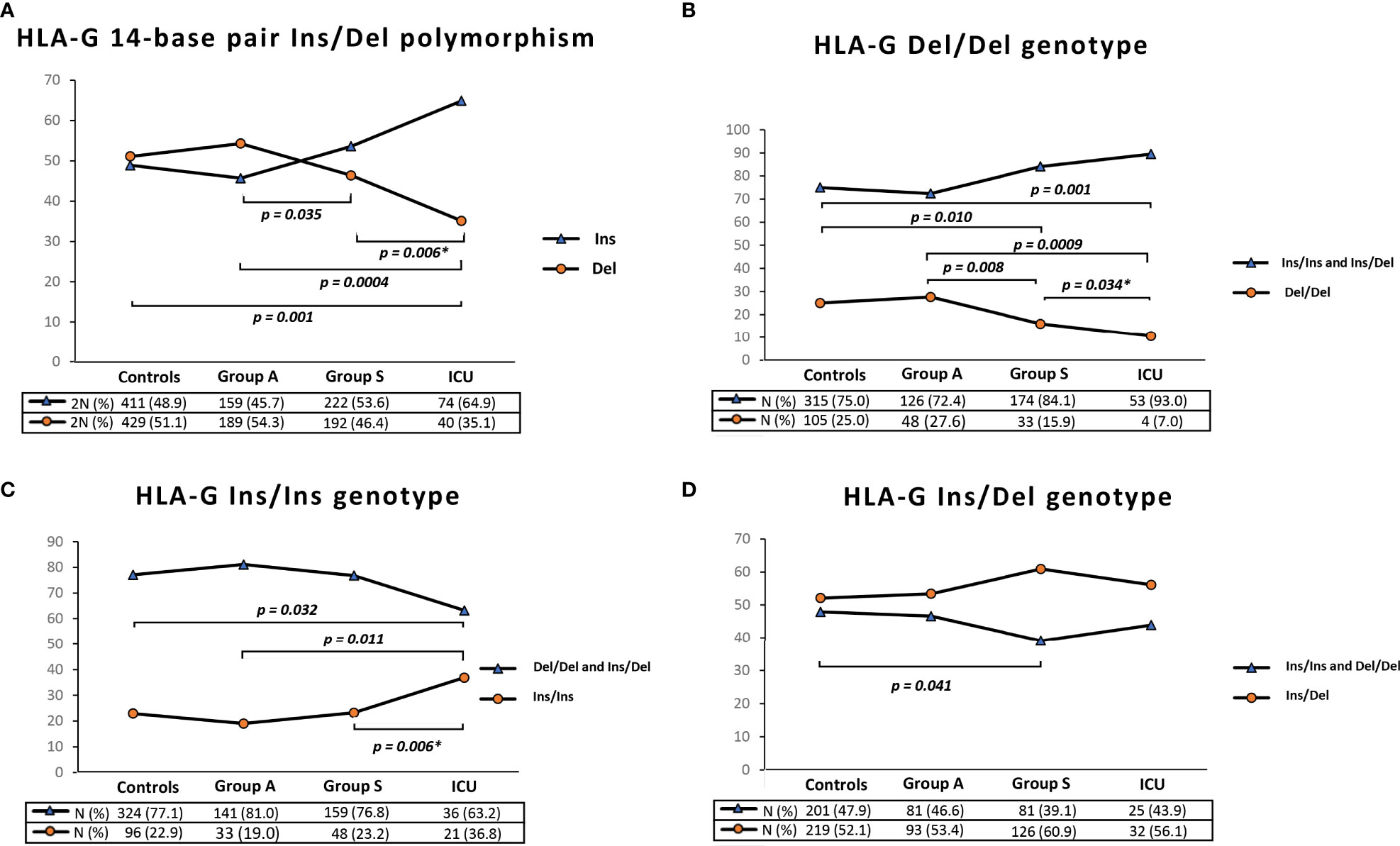
Figure 1 Graphical representation of HLA-G 3’UTR 14bp Ins/Del allele frequency (A), HLA-G 3’UTR 14bp Del/Del genotype frequency (B), HLA-G 3’UTR 14bp Ins/Ins genotype frequency (C) and HLA-G 3’UTR 14bp Ins/Del genotype frequency. Data extracted from controls, Group A (Paucisymptomatic patients), Group S (patients with severe symptoms) and ICU (critical patients admitted in Intensive Care Unit). P values were calculated by using the two-tailed Fisher’s exact test. Only P values less than 0.05 are reported in the figure corresponding to significant differences between the frequencies of the HLA-G 3’UTR 14-bp polymorphism (Ins or Del) and the HLA-G genotype (Del/Del or Ins/Ins and Ins/Del) in the control sample and in the groups of patients. Table S5 in the Supplementary Material reports the P values for all the possible comparisons between the HLA-G polymorphism and genotypes in the groups of controls and patients. *To calculate the P values between Group S and ICU, we excluded the patients in ICU from Group S.
Similarly, we observed a significant difference between Group S and Group A [46.4% (2N=192) vs 54.3% (2N=189), P = 0.035] and between Group S and ICU patients [35.1% (2N=40) vs 46.4% (2N=192), P = 0.006].
Based on genotype frequencies for the HLA-G 14-bp (Figure 1B), Del/Del genotype was significantly less common in Group S and in ICU patients [N= 33 (15.9%) and N=4 (7%), respectively] compared to Group A and controls [N= 48 (27.6%) and N=105 (25.0%), respectively] (Figure 1B).
On the other hand, we found the opposite situation for the Ins/Ins genotype. As shown in Figure 1C, this HLA-G 3’UTR genotype was significantly more frequent in ICU patients [N=21 (36.8%)] compared to Group S [N=48 (23.2%)], Group A [N=33 (19.0%)] and controls [N=96 (22.9%)].
Finally, the frequency of the Ins/Del genotype in Group S was slightly higher than in controls [N=126 (60.9%) vs N=219 (52.1%), P=0.041] (Figure 1D). Table S5 displays all the P values from the comparison of all groups.
3.2.1.3 Soluble HLA-G dosage
Soluble HLA-G (sHLA-G) levels were measured in healthy controls at the time of enrollment, and in patients at a time point between one to six months after complete recovery. The levels of sHLA-G were not measured in plasma obtained during the acute disease or during hospitalization because sHLA-G levels might be influenced by the pharmacologic treatment that was not standardized.
Results show that sHLA-G levels are similar between COVID-19 recovered patients and controls (Supplementary Figure S1A), with no statistically significant differences between the two groups [sHLA-G: Median (IQR) 5.9 (7.2) vs. 7.5 (8) U/mL P = 0.068]. We obtained the same results when we compared Group A vs. Group S (Supplementary Figure S1B), [sHLA-G: Median (IQR) 5.5 (4.8) vs. 7.3 (8.1) U/mL P = 0.960].
According to the literature, serum levels of HLA-G may be affected by the presence of Del allele. Patients were therefore divided into the genotype group: homozygous (Ins/Ins), heterozygous (Ins/Del) and homozygous (Del/Del). However, no significant differences were found between the three groups (Supplementary Figure S1C), [sHLA-G: Median (IQR) 5.3 (4.3) Ins/Ins vs 4.6 (7.2) Ins/Del vs 6.9 (8.2) Del/Del U/mL, P = 0.429].
Finally, we evaluated the effect of the Del/Ins genotype in Group A and Group S. No differences were found in this case either [Group A: Median (IQR) 4.6 (4.4) Ins/Ins vs 5.6 (5.4) Ins/Del vs. 5.7 (3.6) Del/Del U/mL P = 0.406] and [Group S: Median (IQR) 5.9 (2.4) Ins/Ins vs 4.3 (9.7) Ins/Del vs. 11.5 (4.6) Del/Del U/mL P = 0.368] (Supplementary Figures S1D, E respectively).
3.3 Multivariate analysis of clinical, immunological and genetic factors and the clinical manifestations of SARS-CoV-2 infection
As a further analysis, we performed a multivariate analysis based on a logistic regression model to calculate the independence of immunological and genetic variables from age and gender.
We included in the comparisons between the two groups of patients (A and S) the factors that were statistically significant in the univariate analysis (PU< 0.05): concomitant autoimmune diseases, beta-thalassemia trait, HLA-G 3’ UTR 14bp Del/Del, the KIR-ligand combination KIR2DS2/HLA-C C1+, the Neanderthal LZTFL1 polymorphism and the three loci HLA haplotype HLA-B*58:01, C*07:01, DRB1*03:01. The univariate P values and odds ratios of the statistically significant factors were adjusted according to age and gender (Table 7). The multivariate P values (PM) were corrected for multiple comparisons (PMC).
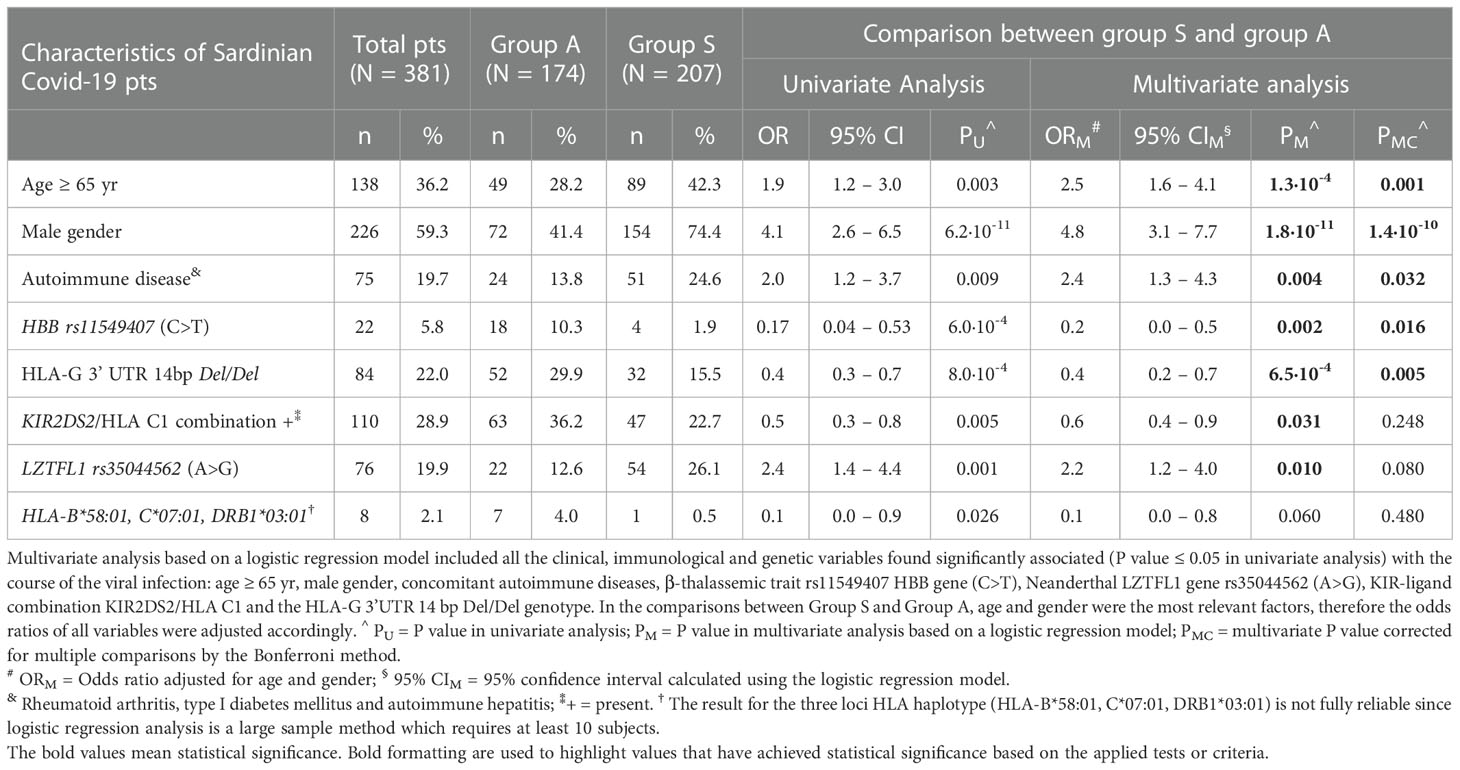
Table 7 Multivariate analysis of clinical, immunological and genetic factors associated with the course of the SARS-CoV-2 disease.
The results confirmed the strong association between the severe clinical manifestations of SARS-CoV-2 infection and five clinical and genetic factors after multiple testing correction, as shown in Table 7: I) age ≥ 65 years [ORM = 2.5 (95% CI 1.6 – 4.1), PM = 1.3 x 10-4, PMC = 0.001], II) male gender [ORM = 4.8 (95% CI 3.1 – 7.7), PM = 1.8·10-11, PMC = 1.4 x 10-10], III) autoimmune disease [ORM = 2.4 (95% CI 1.3 – 4.3), PM = 0.004, PMC = 0.032], IV) β-Thalassemia trait [ORM = 0.2 (95% CI 0.0 – 0.5), PM = 0.002, PMC = 0.016] and V) HLA-G 3’ UTR 14bp Del/Del [ORM = 0.4 (95% CI 0.2 – 0.7), PM = 6.5 x 10-4, PMC = 0.005]. The results confirmed that this specific HLA-G 3’UTR polymorphism plays a relevant role in protection against severe and life-threatening diseases.
4 Discussion
The clinical course of SARS-CoV-2 infection can be influenced by various clinical, genetic, and immunological factors, and may also depend on the specific variant of the virus, as observed in various studies. Since the first wave of the pandemic emerged, males and patients of advanced age have been more susceptible to severe clinical manifestations (71).
In the Sardinian population as well, these two clinical-demographic factors have been identified as the most relevant risk factors (43). Nevertheless, this study highlights additional clinical and genetic factors that may impact the progression of SARS-CoV-2 B.1.617.1 (Delta) variant infection among Sardinian population, beyond those previously identified during the spread of the B.1.1.7 variant (36, 43).
Among these factors, comorbidity with autoimmune diseases emerged as the most relevant clinical factor. The four most common autoimmune diseases associated with severe COVID-19 were Hashimoto’s thyroiditis, type I diabetes, rheumatoid arthritis, systemic lupus erythematosus, and autoimmune hepatitis.
According to the literature, patients with these immune-mediated diseases have an alteration of the cell-mediated immune response mechanism that facilitates a rapid release of proinflammatory cytokines and chemokines by T lymphocytes, resulting in the so-called “cytokine storm” that complicates COVID-19 course (9, 38, 43, 72).
Our study, in addition to these comorbidities, highlights the association of the Neanderthal haplotype at the LZTFL1 gene with severe COVID-19.
This haplotype, consisting of four SNPs (rs35044562, rs73064425, rs34326463, rs67959919) has exerted a negative influence on disease outcome. The findings are in line with previous studies in other populations showing that this Neanderthal haplotype is strongly linked to a severe form of COVID-19 (36, 73, 74).
The biological role of LZTFL1 in COVID-19 outcomes is still unclear. However, it should be noted that LZTFL1 gene expression is widely distributed in pulmonary epithelial cells, including those of the ciliated epithelium, which has been identified as a major target for SARS-CoV-2 infections. This gene encodes a cytosolic leucine zipper protein, which associates with the epithelial marker E-cadherin and participates in wound healing and immune response. According to previous studies, increased expression of LZTFL1 caused by a gain-of-function variant in an inducible enhancer may negatively affect the outcome in COVID-19 patients (75).
In contrast, according to previous studies in different populations, we confirmed that the other Neanderthal haplotype encompassing the OAS1, OAS2, and OAS3 genes (chr12: 113,350,796 to 113,425,679; hg19) does not provide any protection against severe SARS-CoV-2 infections in Sardinia population (65, 76, 77).
Another notable observation that emerged from this study was the low frequency of beta-thalassemia carriers in the group of patients with severe clinical manifestations. In particular, the most common mutation in Sardinia, the β°39 C>T (rs11549407), at the gene HBB, does not protect against infection but appears to enhance resistance in cases of severe disease.
Currently, the mechanism conferring resistance to severe COVID-19 infection remains to be elucidated. According to some studies, microRNAs produced by patients with hemoglobinopathies are involved in modulating the functions associated with several disease processes, including microbial defense (78).
Other researchers suggested that specific SARS-CoV-2 proteins may attack the heme on the 1-ß chain of hemoglobin, leading to iron dissociation, formation of porphyrin, and consequent oxidative damage (79).
This molecular process could be less frequent in patients with hemoglobinopathies. Nevertheless, this result needs to be further investigated by future studies since the literature does not provide unique data.
On one side, some studies have found the protective effect of the thalassemia trait and hemoglobinopathies (43, 78, 80). Other studies, however, reject this conclusion and suggest an increased mortality risk from COVID-19 in patients with hemoglobinopathies (81). This inconsistency is likely caused by the wide variety of mutations found in alpha and beta globin chains that are characteristic of different populations.
As a result of multivariate analysis, it was also confirmed that the KIR2DS2/HLA-C C1+ group-ligand combination can influence the outcome of COVID-19 disease independently of other genetic factors. During SARS-CoV-2 infection, the number of circulating NK cells decreases, and the cells express markers of exhaustion (TIM-3, PD-1, NKG2A), possibly due to high levels of IL-6 secreted from macrophages during the inflammatory process.
This combination results in a decreased secretion of IFN-γ and decreased degranulation (82, 83). As a result, NK cells may be less able to effectively fight infection when the KIR2DS2/HLA-C C1+ group ligand functional unit is not present.
The correct function of the NK cells requires a proper combination of activating and inhibiting KIR genes and their HLA ligands. Due to the defective combinations of KIR and HLA, the function of NK cells can be impaired, similar to what has been observed in COVID-19 patients from different ethnic backgrounds (43).
Our study aims to contribute to the understanding of SARS-CoV-2 infection by investigating the role of HLA-G polymorphism in the population.
So far, the research has concentrated on how this immunomodulatory molecule plays a role in virus immunopathogenesis in patients with SARS-CoV-2 infection. It has been shown that viruses can upregulate HLA-G molecules in both surface membrane-bound and peripheral soluble forms in cells infected with viruses (53). However, researchers have mainly focused on HLA-G expression and serum levels in COVID-19 patients, rather than focusing on the genetic basis of these manifestations.
The present study is the first to investigate the genetic basis of the HLA-G and how it contributes to symptoms associated with SARS-CoV-2 infection. Our first consideration is that the HLA-G genetic structure between COVID-19 patients and the controls does not differ significantly. Nevertheless, a significant reduction in the frequency of extended haplotype HLA-G*01:01:01:01/HLA-G/UTR-1 is observed in patients with severe illness (Group S) compared to controls and group A [P< 0.001, Pc =0.002]. On the contrary, our results show that the extended haplotypes HLA-G*01:03:01:02-HLA-G/UTR-5 and HLA-G*01:04:01/04-HLA-G/UTR-5 are more frequent in Group S than in Group A [P = 0.001, Pc = 0.018 and P< 0.001, Pc = 0.005 respectively].
A recent study has revealed that HLA-G*01:04 and HLA-G*01:03 products have a greater affinity for the heterodimer NKG2A/CD94 receptor than HLA-G*01:01. Therefore, this results in NKG2A to take on a much more potent immunosuppressive action, as it happens in severe COVID-19 infections (53, 84).
In addition, HLA-G UTR-1 is particularly rare in patients with severe clinical manifestations (Table 5). Among the nine polymorphisms that constitute the HLA-G UTR-1 haplotypes, 14bp Del (rs371194629), and HLA-G 3’ UTR +3187G (rs9380142) are the most significant ones. Indeed, about 30% of the Caucasian population has these two polymorphisms, which result in close linkage disequilibrium (60).
The “GenOMICC” study has previously conducted genome-wide association studies (GWAS) in 2,244 patients with a critical illness (85). The HLA-G 3’ UTR polymorphism +3187A (rs9380142) was reported as one of the genetic markers associated with severe COVID-19 disease, whereas the +3187G variant was not associated with severe disease (85). The study supports the hypothesis that the HLA-G/UTR-1 haplotype and its polymorphisms, HLA-G Del and HLA-G +3187G, might play a protective role against severe forms of the disease.
Furthermore, the protective effect is more pronounced in HLA-G Del/Del homozygous subjects, making it the most relevant genetic factor in the multivariate analysis (Table 7). This genotype, in fact, was found in only 7% of COVID-19 ICU patients (Figure 1B).
As several published studies have shown, the HLA-G Del polymorphism can result in increased expression of soluble HLA-G molecules by modifying mRNA stability or allowing post-transcriptional regulation (49, 86, 87).
Interestingly, where there was no infection, the levels of soluble HLA-G among patients who recovered from SARS-CoV-2 and those in the control group were not significantly different, even when grouped by their genotype (Ins/Ins, Ins/Del, and Del/Del). Similar results were described by Ali H Ad’hiah, where HLA-G genotypes did not significantly affect levels of the soluble molecule in 209 Iraqi patients (88).
However, SARS-CoV-2 infection, like other viral infections, can cause a marked variation in sHLA-G levels due to inflammation and upregulation of immune inhibitory receptors of which HLA-G is a ligand. In addition, a remarkable decrease in HLA-G+ immune cell numbers and exhaustion of host cellular immune responses are commonly observed in patients with severe COVID-19 illness (53).
It is possible that patients with the HLA-G Del/Del genotype may express more HLA-G molecules, leading to an increased number of immune-modulatory HLA-G+ cells in the host. In patients with COVID-19 disease, this could reduce the severity by limiting inflammation and cytokine storm responsible for critical cases of illness (89). Moreover, higher levels of sHLA-G are associated with increased expression of sICAM-1 and sE-selectin expression, which may contribute to improved clinical conditions in COVID-19 patients by reducing neutrophil adhesion to activated endothelium (56).
On the other hand, HLA-G Ins/Ins genotype has been associated in literature with lower surface expression of HLA-G. This reduction in surface expression may potentially worsen the immune response to viral infections, leading to increased tissue damage, which could explain why this genotype was more commonly observed in ICU patients.
In conclusion, this study is the first to thoroughly investigate the role of HLA-G genotypes in SARS-CoV2. Specifically, our results suggest that some HLA-G polymorphisms may positively impact the course of COVID-19 through their immunomodulatory effect.
The outcome of SARS-CoV-2 infection is dependent on a complex interaction between the virus and the host, which includes both virus-related factors like the variety of variants and viral load, as well as various genetic and immunological factors of the host. Host genetic factors, including some that have been shown to affect individual susceptibility to develop severe manifestations of COVID-19, should be taken into account along with well-established general risk factors like older age, male gender, and chronic comorbidities when predicting the severity and mortality associated with COVID-19.
It is essential to develop predictive algorithms based on these clinical, genetic, and immunological factors to identify categories of individuals at higher risk of severe short- and/or long-term clinical manifestations in case of SARS-CoV-2 infection. This need is urgent as novel pandemic waves caused by new COVID-19 variants could occur in the coming years (90).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA904643.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of the Cagliari University Hospital; date of approval: May 27th, 2020; protocol number GT/2020/10894. The patients/participants provided their written informed consent to participate in this study.
Author contributions
All authors contributed to the article and approved the submitted version.
Funding
This study was partially funded by the Italian Ministry of University and Research, PNRR, mission 4, component 2, investment 1.3, (Partenariati estesi alle università, ai centri di ricerca, alle aziende per il finanziamento di progetti di ricerca di base), title HEAL ITALIA, project number PE00000019, CUP: F53C22000750006 (AP, University of Cagliari). This research was supported by “Fondazione di Sardegna”, grant n. 2023.0160. to the non-profit organization “Associazione per l’Avanzamento della Ricerca per i Trapianti (AART-ODV)”.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1138559/full#supplementary-material
References
1. Pene F, Merlat A, Vabret A, Rozenberg F, Buzyn A, Dreyfus F, et al. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin Infect Dis (2003) 37(7):929–32. doi: 10.1086/377612
2. Kirtipal N, Bharadwaj S, Kang SG. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect Genet Evol (2020) 85:104502. doi: 10.1016/j.meegid.2020.104502
3. Tian D, Sun Y, Zhou J, Ye Q. The global epidemic of SARS-CoV-2 variants and their mutational immune escape. J Med Virol (2022) 94(3):847–57. doi: 10.1002/jmv.27376
4. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature (2020) 579(7798):265–9. doi: 10.1038/s41586-020-2008-3
5. Menachery VD, Yount BL, Debbink K, Agnihothram S, Gralinski LE, Plante JA, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med (2015) 21(12):1508–13. doi: 10.1038/nm.3985
6. de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J Virol (2013) 87(14):7790–2. doi: 10.1128/JVI.01244-13
7. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (2020) 395(10224):565–74. doi: 10.1016/S0140-6736(20)30251-8
8. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature (2020) 579(7798):270–3. doi: 10.1038/s41586-020-2012-7
9. Ye Q, Wang B, Mao J. The pathogenesis and treatment of the `Cytokine storm' in COVID-19. J Infect (2020) 80(6):607–13. doi: 10.1016/j.jinf.2020.03.037
10. Ye Q, Wang B, Mao J, Fu J, Shang S, Shu Q, et al. Pidemiological analysis of COVID-19 and practical experience from China. J Med Virol (2020) 92(7):755–69. doi: 10.1002/jmv.25813
11. Tian D, Sun Y, Xu H, Ye Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 omicron variant. J Med Virol (2022) 94(6):2376–83. doi: 10.1002/jmv.27643
12. Drożdżal S, Rosik J, Lechowicz K, Machaj F, Szostak B, Przybyciński J, et al. An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist Update (2021) 59:100794. doi: 10.1016/j.drup.2021.100794
13. WHO coronavirus (COVID-19) dashboard. Available at: https://covid19.who.int/.
14. Ye Q, Lu D, Shang S, Fu J, Gong F, Shu Q, et al. Crosstalk between coronavirus disease 2019 and cardiovascular disease and its treatment. ESC Heart Fail (2020) 7(6):3464–72. doi: 10.1002/ehf2.12960
15. Han X, Ye Q. Kidney involvement in COVID-19 and its treatments. J Med Virol (2021) 93(3):1387–95. doi: 10.1002/jmv.26653
16. Tian D, Ye Q. Hepatic complications of COVID-19 and its treatment. J Med Virol (2020) 92(10):1818–24. doi: 10.1002/jmv.26036
17. Ye Q, Wang B, Zhang T, Xu J, Shang S. The mechanism and treatment of gastrointestinal symptoms in patients with COVID-19. Am J Physiol Gastrointest Liver Physiol (2020) 319(2):G245–52. doi: 10.1152/ajpgi.00148.2020
18. Hu D, Lou X, Meng N, Li Z, Teng Y, Zou Y, et al. Influence of age and gender on the epidemic of COVID-19 : evidence from 177 countries and territories-an exploratory, ecological study. Wien Klin Wochenschr (2021) 133(7-8):321–30. doi: 10.1007/s00508-021-01816-z
19. Mangia C, Russo A, Civitelli S, Gianicolo EAL. Sex/gender differences in COVID-19 lethality: what the data say, and do not say. Epidemiol Prev (2020) 44(5-6 Suppl 2):400–6. doi: 10.19191/EP20.5-6.S2.145
20. Hoffmann C, Wolf E. Older age groups and country-specific case fatality rates of COVID-19 in Europe, USA and Canada. Infection (2021) 49(1):111–6. doi: 10.1007/s15010-020-01538-w
21. Rauch S, Jasny E, Schmidt KE, Petsch B. New vaccine technologies to combat outbreak situations. Front Immunol (2018) 9:1963. doi: 10.3389/fimmu.2018.01963
22. Messina NL, Zimmermann P, Curtis N. The impact of vaccines on heterologous adaptive immunity. Clin Microbiol Infect (2019) 25(12):1484–93. doi: 10.1016/j.cmi.2019.02.016
23. Nohynek H, Wilder-Smith A. Does the world still need new covid-19 vaccines. N Engl J Med (2022) 386(22):2140–2. doi: 10.1056/NEJMe2204695
24. Fiolet T, Kherabi Y, MacDonald CJ, Ghosn J, Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clin Microbiol Infect (2022) 28(2):202–21. doi: 10.1016/j.cmi.2021.10.005
25. WHO (World Health Organization). Available at: https://www.who.int/ (Accessed 1 July 2022).
26. Chen X, Huang H, Ju J, Sun R, Zhang J. Impact of vaccination on the COVID-19 pandemic in U.S. states. Sci Rep (2022) 12(1):1554. doi: 10.1038/s41598-022-05498-z
27. Le Rutte EA, Shattock AJ, Chitnis N, Kelly SL, Penny MA. Modelling the impact of omicron and emerging variants on SARS-CoV-2 transmission and public health burden. Commun Med (Lond) (2022) 2:93. doi: 10.1038/s43856-022-00154-z
28. Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by omicron infection. Nature (2022) 608(7923):593–602. doi: 10.1038/s41586-022-04980-y
29. Singh DD, Parveen A, Yadav DK. SARS-CoV-2: emergence of new variants and effectiveness of vaccines. Front Cell Infect Microbiol (2021) 11:777212. doi: 10.3389/fcimb.2021.777212
30. Chenchula S, Karunakaran P, Sharma S, Chavan M. Current evidence on efficacy of COVID-19 booster dose vaccination against the omicron variant: a systematic review. J Med Virol (2022) 94(7):2969–76. doi: 10.1002/jmv.27697
31. D'Arena G, Penna A, Crocamo A, Sguazzo F, Viceconti R, Barlotti V, et al. Heterogeneity of clinical and radiological findings of COVID-19. Postgrad. Med J (2021) 97(1146):268–69. doi: 10.1136/postgradmedj-2020-137901
32. Luo X, Lv M, Zhang X, Estill J, Yang B, Lei R, et al. Clinical manifestations of COVID-19: an overview of 102 systematic reviews with evidence mapping. JEBM (2022) 15(3):201–15. doi: 10.1111/jebm.12483
33. Ji XS, Chen B, Ze B, Zhou WH. Human genetic basis of severe or critical illness in COVID-19. Front Cell Infect Microbiol (2022) 12:963239. doi: 10.3389/fcimb.2022.963239
34. Zhang Q, Bastard P, Cobat A, Casanova JL, Karbuz A, Gervais A, et al. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature (2022) 603(7902):587–98. doi: 10.1038/s41586-022-04447-0
35. Jesenak M, Brndiarova M, Urbancikova I, Rennerova Z, Vojtkova J, Bobcakova A, et al. Immune parameters and COVID-19 infection - associations with clinical severity and disease prognosis. Front Cell Infect Microbiol (2020) 10:364. doi: 10.3389/fcimb.2020.00364
36. Mocci S, Littera R, Tranquilli S, Provenzano A, Mascia A, Cannas F, et al. A protective HLA extended haplotype outweighs the major COVID-19 risk factor inherited from neanderthals in the sardinian population. Front Immunol (2022) 13:891147. doi: 10.3389/fimmu.2022.891147
37. Hernández-Doño S, Sánchez-González RA, Trujillo-Vizuet MG, Zamudio-Castellanos FY, García-Silva R, Bulos-Rodríguez P, et al. Protective HLA alleles against severe COVID-19: HLA-A*68 as an ancestral protection allele in tapachula-chiapas, Mexico. Clin Immunol (2022) 238:108990. doi: 10.1016/j.clim.2022.108990
38. Littera R, Campagna M, Deidda S, Angioni G, Cipri S, Melis M, et al. Human leukocyte antigen complex and other immunogenetic and clinical factors influence susceptibility or protection to SARS-CoV-2 infection and severity of the disease course. the sardinian experience. Front Immunol (2020) 11:605688. doi: 10.3389/fimmu.2020.605688
39. D'Alterio G, Lasorsa VA, Bonfiglio F, Cantalupo S, Rosato BE, Andolfo I, et al. Germline rare variants of lectin pathway genes predispose to asymptomatic SARS-CoV-2 infection in elderly individuals. Genet Med (2022) 24(8):1653–63. doi: 10.1016/j.gim.2022.04.007
40. Zeberg H. The major genetic risk factor for severe COVID-19 is associated with protection against HIV. Proc Natl Acad Sci U.S.A. (2022) 119(9):e2116435119. doi: 10.1073/pnas.2116435119
41. Banday AR, Stanifer ML, Florez-Vargas O, Onabajo OO, Papenberg BW, Zahoor MA, et al. Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries. Nat Genet (2022) 54(8):1103–16. doi: 10.1038/s41588-022-01113-z
42. Hajeer A, Jawdat D, Massadeh S, Aljawini N, Abedalthagafi MS, Arabi YM, et al. Association of KIR gene polymorphisms with COVID-19 disease. Clin Immunol (2022) 234:108911. doi: 10.1016/j.clim.2021.108911
43. Littera R, Chessa L, Deidda S, Angioni G, Campagna M, Lai S, et al. Natural killer-cell immunoglobulin-like receptors trigger differences in immune response to SARS-CoV-2 infection. PloS One (2021) 16(8):e0255608. doi: 10.1371/journal.pone.0255608
44. Attia JVD, Dessens CE, van de Water R, Houvast RD, Kuppen PJK, Krijgsman D. The molecular and functional characteristics of HLA-G and the interaction with its receptors: where to intervene for cancer immunotherapy. Int J Mol Sci (2020) 21(22):8678. doi: 10.3390/ijms21228678
45. Lv H, Lv H, Lin Z, Chen L, Zhu M, Hong D. Meta-analysis of correlationship between HLA-G 3'UTR 14-bp Ins/Del polymorphism and virus susceptibility. Med (Baltimore) (2018) . 97(38):e12262. doi: 10.1097/MD.0000000000012262
46. Poras I, Yaghi L, Martelli-Palomino G, Mendes-Junior CT, Muniz YC, Cagnin NF, et al. Haplotypes of the HLA-G 3' untranslated region respond to endogenous factors of HLA-g+ and HLA-g- cell lines differentially. PloS One (2017) 12(1):e0169032. doi: 10.1371/journal.pone.0169032
47. Castelli EC, Mendes-Junior CT, Deghaide NH, de Albuquerque RS, Muniz YC, Simões RT, et al. The genetic structure of 3'untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun (2010) 11(2):134–41. doi: 10.1038/gene.2009.74
48. Krijgsman D, Roelands J, Hendrickx W, Bedognetti D, Kuppen PJK. HLA-G: a new immune checkpoint in cancer. Int J Mol Sci (2020) 21(12):4528. doi: 10.3390/ijms21124528
49. Naji A, Menier C, Morandi F, Agaugué S, Maki G, Ferretti E, et al. Binding of HLA-G to ITIM-bearing ig-like transcript 2 receptor suppresses b cell responses. J Immunol (2014) 192(4):1536–46. doi: 10.4049/jimmunol.1300438
50. Martelli-Palomino G, Pancotto JA, Muniz YC, Mendes-Junior CT, Castelli EC, Massaro JD, et al. Polymorphic sites at the 3' untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PloS One (2013) 8(10):e71742. doi: 10.1371/journal.pone.0071742
51. Ishitani A, Geraghty DE. Alternative splicing of HLA-G transcripts yields proteins with primary structures resembling both class I and class II antigens. Proc Natl Acad Sci U.S.A. (1992) 89(9):3947–51. doi: 10.1073/pnas.89.9.3947
52. Kirszenbaum M, Moreau P, Gluckman E, Dausset J, Carosella E. An alternatively spliced form of HLA-G mRNA in human trophoblasts and evidence for the presence of HLA-G transcript in adult lymphocytes. Proc Natl Acad Sci U.S.A. (1994) 91(10):4209–13. doi: 10.1073/pnas.91.10.4209
53. Lin A, Yan WH. Perspective of HLA-G induced immunosuppression in SARS-CoV-2 infection. Front Immunol (2021) 12:788769. doi: 10.3389/fimmu.2021.788769
54. Al-Bayatee NT, Ad'hiah AH. Soluble HLA-G is upregulated in serum of patients with severe COVID-19. Hum Immunol (2021) 82(10):726–32. doi: 10.1016/j.humimm.2021.07.007
55. Jasinski-Bergner S, Schmiedel D, Mandelboim O, Seliger B. Role of HLA-G in viral infections. Front Immunol (2022) 13:826074. doi: 10.3389/fimmu.2022.826074
56. Bortolotti D, Gentili V, Rizzo S, Schiuma G, Beltrami S, Spadaro S, et al. Increased sHLA-G is associated with improved COVID-19 outcome and reduced neutrophil adhesion. Viruses (2021) 13(9):1855. doi: 10.3390/v13091855
57. QIAGEN. QIAamp DNA mini blood mini handbook (2022). Available at: https://www.qiagen.com/gb/shop/sample-technologies/dna/qiaamp-dna-mini-kit/#resources.
58. Nilsson LL, Funck T, Kjersgaard ND, Hviid TVF. Next-generation sequencing of HLA-G based on long-range polymerase chain reaction. HLA (2018) 92(3):144–53. doi: 10.1111/tan.13342
59. Castelli EC, Ramalho J, Porto IO, Lima TH, Felício LP, Sabbagh A, et al. Insights into HLA-G genetics provided by worldwide haplotype diversity. Front Immunol (2014) 5:476. doi: 10.3389/fimmu.2014.00476
60. Arnaiz-Villena A, Juarez I, Suarez-Trujillo F, López-Nares A, Vaquero C, Palacio-Gruber J, et al. HLA-G: function, polymorphisms and pathology. Int J Immunogenet (2021) 48(2):172–92. doi: 10.1111/iji.12513
61. de Almeida BS, Muniz YCN, Prompt AH, Castelli EC, Mendes-Junior CT, Donadi EA. Genetic association between HLA-G 14-bp polymorphism and diseases: a systematic review and meta-analysis. Hum Immunol (2018) 79(10):724–35. doi: 10.1016/j.humimm.2018.08.003
62. Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J, Moffatt MF, et al. Copy number variation leads to considerable diversity for b but not a haplotypes of the human KIR genes encoding NK cell receptors. Genome Res (2012) 22(10):1845–54. doi: 10.1101/gr.137976.112
63. Vilches C, Castaño J, Gómez-Lozano N, Estefanía E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens (2007) 70(5):415–22. doi: 10.1111/j.1399-0039.2007.00923.x
64. Zeberg H, Pääbo S. A genomic region associated with protection against severe COVID-19 is inherited from neandertals. Proc Natl Acad Sci U.S.A. (2021) 118(9):e2026309118. doi: 10.1073/pnas.2026309118
65. Rosatelli C, Leoni GB, Tuveri T, Scalas MT, Di Tucci A, Cao A. Beta thalassaemia mutations in sardinians: implications for prenatal diagnosis. J Med Genet (1987) 24(2):97–100. doi: 10.1136/jmg.24.2.97
66. Minucci A, Giardina B, Zuppi C, Capoluongo E. Glucose-6-phosphate dehydrogenase laboratory assay: how, when, and why. IUBMB Life (2009) 61(1):27–34. doi: 10.1002/iub.137
67. Okebe J, Amambua-Ngwa A, Parr J, Nishimura S, Daswani M, Takem EN, et al. The prevalence of glucose-6-phosphate dehydrogenase deficiency in Gambian school children. Malar J (2014) 13:148. doi: 10.1186/1475-2875-13-148
68. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (2005) 21(2):263–5. doi: 10.1093/bioinformatics/bth457
69. Kirkwood BR, Sterne JAC. Essential medical statistics. 2nd edition. Oxford UK: Blackwell Scientific Publications (2003).
70. William H, Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc (1952) 47(260):583–621. doi: 10.1080/01621459.1952.10483441
71. Lynch SM, Guo G, Gibson DS, Bjourson AJ, Rai TS. Role of senescence and aging in SARS-CoV-2 infection and COVID-19 disease. Cells (2021) 10(12):3367. doi: 10.3390/cells10123367
72. Liu Y, Sawalha AH, Lu Q. COVID-19 and autoimmune diseases. Curr Opin Rheumatol (2021) 33(2):155–62. doi: 10.1097/BOR.0000000000000776
73. Zeberg H, Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from neanderthals. Nature (2020) 587(7835):610–2. doi: 10.1038/s41586-020-2818-3
74. Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide association study of severe covid-19 with respiratory failure. N Engl J Med (2020) 383(16):1522–34. doi: 10.1056/NEJMoa2020283
75. Downes DJ, Cross AR, Hua P, Roberts N, Schwessinger R, Cutler AJ, et al. Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat Genet (2021) 53(11):1606–15. doi: 10.1038/s41588-021-00955-3
76. Kristiansen H, Scherer CA, McVean M, Iadonato SP, Vends S, Thavachelvam K, et al. Extracellular 2'-5' oligoadenylate synthetase stimulates RNase l-independent antiviral activity: a novel mechanism of virus-induced innate immunity. J Virol (2010) 84(22):11898–904. doi: 10.1128/JVI.01003-10
77. Huffman JE, Butler-Laporte G, Khan A, Pairo-Castineira E, Drivas TG, Peloso GM, et al. Multi-ancestry fine mapping implicates OAS1 splicing in risk of severe COVID-19. Nat Genet (2022) 54(2):125–7. doi: 10.1038/s41588-021-00996-8
78. Papadopoulos KI, Sutheesophon W, Manipalviratn S, Aw TC. A southeast Asian perspective on the COVID-19 pandemic: hemoglobin e (HbE)-trait confers resistance against COVID-19. Med Sci Monit Basic Res (2021) 27:e929207. doi: 10.12659/MSMBR.929207
79. Liu WZ, Li HL. COVID-19: attacks the 1-beta chain of hemoglobin to disrupt respiratory function and escape immunity . ChemRxiv. Cambridge: Cambridge Open Engage (2022).
80. Lansiaux E, Pébaÿ PP, Picard JL, Son-Forget J. COVID-19: beta-thalassemia subjects immunised. Med Hypotheses (2020) 142:109827. doi: 10.1016/j.mehy.2020.109827
81. Baba H, Shinkai A, Izuchi R, Azuma Y. Long-term results of short course chemotherapy of pulmonary tuberculosis. second study-b: results at 6 years after the end of 4-9 month chemotherapy of pulmonary tuberculosis. Kekkaku (1987) 62(10):511–20.
82. Gallardo-Zapata J, Maldonado-Bernal C. Natural killer cell exhaustion in SARS-CoV-2 infection. Innate Immun (2022) 28(6):189–98. doi: 10.1177/17534259221077750
83. Varchetta S, Mele D, Oliviero B, Mantovani S, Ludovisi S, Cerino A, et al. Unique immunological profile in patients with COVID-19. Cell Mol Immunol (2021) 18(3):604–12. doi: 10.1038/s41423-020-00557-9
84. Antonioli L, Fornai M, Pellegrini C, Blandizzi C. NKG2A and COVID-19: another brick in the wall. Cell Mol Immunol (2020) 17(6):672–4. doi: 10.1038/s41423-020-0450-7
85. Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in COVID-19. Nature (2021) 591(7848):92–8. doi: 10.1038/s41586-020-03065-y
86. Hviid TV, Rizzo R, Christiansen OB, Melchiorri L, Lindhard A, Baricordi OR. HLA-G and IL-10 in serum in relation to HLA-G genotype and polymorphisms. Immunogenetics (2004) 56(3):135–41. doi: 10.1007/s00251-004-0673-2
87. Amodio G, Gregori S. HLA-G Genotype/Expression/Disease association studies: success, hurdles, and perspectives. Front Immunol (2020) 11:1178. doi: 10.3389/fimmu.2020.01178
88. Ad'hiah AH, Al-Bayatee NT. HLA-G 14-bp insertion/deletion polymorphism and risk of coronavirus disease 2019 (COVID-19) among Iraqi patients. Hum Immunol (2022) 83(6):521–7. doi: 10.1016/j.humimm.2022.03.005
89. Ramzannezhad S, Tarighi M, Mohammadnia-Afrouzi M, Aghapour S, Bagherzadeh M, Ahmadnia Z, et al. The association of decreased HLA-g+ immune cell frequencies with critical COVID-19 patients. Microb Pathog (2022) 167:105550. doi: 10.1016/j.micpath.2022.105550
Keywords: COVID-19, Sardinian population, soluble HLA-G, HLA-G 3’UTR haplotypes, KIR2DS2 gene, neanderthal LZTFL1 variants
Citation: Mocci S, Littera R, Chessa L, Campagna M, Melis M, Ottelio CM, Piras IS, Lai S, Firinu D, Tranquilli S, Mascia A, Vacca M, Schirru D, Lecca LI, Rassu S, Cannas F, Sanna C, Carta MG, Sedda F, Giuressi E, Cipri S, Miglianti M, Perra A and Giglio S (2023) A review of the main genetic factors influencing the course of COVID-19 in Sardinia: the role of human leukocyte antigen-G. Front. Immunol. 14:1138559. doi: 10.3389/fimmu.2023.1138559
Received: 05 January 2023; Accepted: 23 May 2023;
Published: 05 June 2023.
Edited by:
Gajendra P. S. Raghava, Indraprastha Institute of Information Technology Delhi, IndiaReviewed by:
Mariza Gonçalves Morgado, Oswaldo Cruz Foundation (Fiocruz), BrazilMario Capasso, University of Naples Federico II, Italy
Annamaria Pasi, IRCCS Policlinico San Matteo- Immunogenetics Laboratory- Immunomematology and Transfusional Department, Italy
Copyright © 2023 Mocci, Littera, Chessa, Campagna, Melis, Ottelio, Piras, Lai, Firinu, Tranquilli, Mascia, Vacca, Schirru, Lecca, Rassu, Cannas, Sanna, Carta, Sedda, Giuressi, Cipri, Miglianti, Perra and Giglio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefano Mocci, c3RlZmFuby5tb2NjaS45QGdtYWlsLmNvbQ==; Roberto Littera, cm9ieS5saXR0ZXJAZ21haWwuY29t; Luchino Chessa, bHVjaGluby5jaGVzc2FAdW5pY2EuaXQ=; Andrea Perra, YW5kcmVhLnBlcnJhQHVuaWNhLml0
†These authors have contributed equally to this work
 Stefano Mocci
Stefano Mocci Roberto Littera
Roberto Littera Luchino Chessa
Luchino Chessa Marcello Campagna4
Marcello Campagna4 Maurizio Melis
Maurizio Melis Ignazio S. Piras
Ignazio S. Piras Davide Firinu
Davide Firinu Alessia Mascia
Alessia Mascia Daniele Schirru
Daniele Schirru Luigi Isaia Lecca
Luigi Isaia Lecca Federica Cannas
Federica Cannas Mauro Giovanni Carta
Mauro Giovanni Carta Francesca Sedda
Francesca Sedda Andrea Perra
Andrea Perra Sabrina Giglio
Sabrina Giglio