Immunogenomic Landscape in Breast Cancer Reveals Immunotherapeutically Relevant Gene Signatures
- 1College of Life and Health Sciences, Northeastern University, Shenyang, China
- 2Department of Immunology, Bengbu Medical College, Bengbu, China
- 3Department of Pathophysiology, Bengbu Medical College, Bengbu, China
- 4Department of Radiology, Tian Jin Fifth’s Central Hospital, Tianjin, China
- 5College of Pharmacy, Beihua University, Jilin, China
A Corrigendum on
Immunogenomic landscape in breast cancer reveals immunotherapeutically relevant gene signatures
by Wang T, Li T, Li B, Zhao J, Li Z, Sun M, Li Y, Zhao Y, Zhao S, He W, Guo X, Ge R, Wang L, Ding D, Liu S, Min S and Zhang X (2022) 13:805184. doi: 10.3389/fimmu.2022.805184
In the published article, there was an error in the IHC images of Figure 5B as published. Four repeat IHC images were displayed cursorily. The corrected Figure 5B and its caption “Construction of the immune biological signature. (A) DEGs were used to construct the immune biological signature. (B) Representative immunohistochemical images of infiltrated immune cells in Bengbu cohort between high- and low- ITBscore groups. (C) Kaplan-Meier curves for patients in the BRCA cohort divided into high and low ITBscore subgroups. (D) Alluvial diagram indicating immune subtypes in groups with different BRCA subtypes (basal, Her2, LumA, LumB, and normal), ITBscores, and survival outcomes. (E) Prognostic value of the ITBscore and classic clinicopathological covariates in the high/low ITBscore subgroups. (F) Distribution of the ITBscore among TCGA-BRCA molecular subtypes. “ appear below.
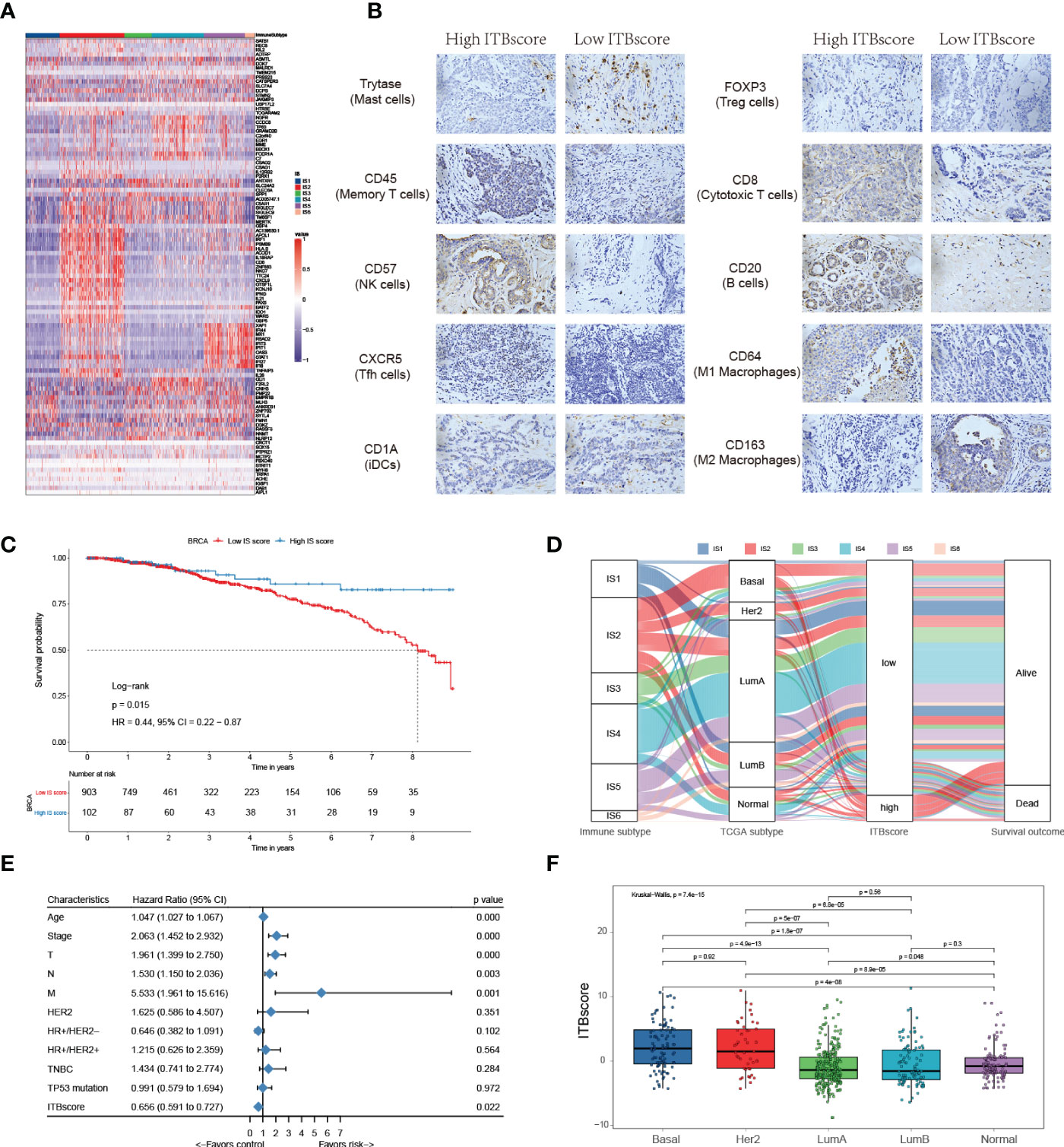
Figure 1 Construction of the immune biological signature. (A) DEGs were used to construct the immune biological signature. (B) Representative immunohistochemical images of infiltrated immune cells in Bengbu cohort between high- and low- ITBscore groups. (C) Kaplan-Meier curves for patients in the BRCA cohort divided into high and low ITBscore subgroups. (D) Alluvial diagram indicating immune subtypes in groups with different BRCA subtypes (basal, Her2, LumA, LumB, and normal), ITBscores, and survival outcomes. (E) Prognostic value of the ITBscore and classic clinicopathological covariates in the high/low ITBscore subgroups. (F) Distribution of the ITBscore among TCGA-BRCA molecular subtypes.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: immune subtype, tumor microenvironment, immune escape, immunotherapy, breast cancer
Citation: Wang T, Li T, Li B, Zhao J, Li Z, Sun M, Li Y, Zhao Y, Zhao S, He W, Guo X, Ge R, Wang L, Ding D, Liu S, Min S and Zhang X (2023) Corrigendum: Immunogenomic landscape in breast cancer reveals immunotherapeutically relevant gene signatures. Front. Immunol. 14:1134847. doi: 10.3389/fimmu.2023.1134847
Received: 31 December 2022; Accepted: 12 January 2023;
Published: 20 January 2023.
Edited and Reviewed by:
Riccardo Dolcetti, Peter MacCallum Cancer Centre, AustraliaCopyright © 2023 Wang, Li, Li, Zhao, Li, Sun, Li, Zhao, Zhao, He, Guo, Ge, Wang, Ding, Liu, Min and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaonan Zhang, emhhbmd4bkBiYm1jLmVkdS5jbg==
 Tao Wang
Tao Wang Tianye Li1
Tianye Li1 Baiqing Li
Baiqing Li Yan Li
Yan Li Xiao Guo
Xiao Guo Rongjing Ge
Rongjing Ge Xiaonan Zhang
Xiaonan Zhang