
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 09 March 2023
Sec. Microbial Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1131447
 Wiwat Chancharoenthana1,2*
Wiwat Chancharoenthana1,2* Supitcha Kamolratanakul1,2
Supitcha Kamolratanakul1,2 Peerapat Visitchanakun3
Peerapat Visitchanakun3 Supistha Sontidejkul3
Supistha Sontidejkul3 Thanya Cheibchalard4,5
Thanya Cheibchalard4,5 Naraporn Somboonna5,6
Naraporn Somboonna5,6 Sarn Settachaimongkon7
Sarn Settachaimongkon7 Asada Leelahavanichkul3*
Asada Leelahavanichkul3*The impacts of metabolomic changes (reduced short-chain-fatty acids; SCFAs) in uremic condition is not fully understood. Once daily Candida gavage with or without probiotics (different times of administration) for 1 week prior to bilateral nephrectomy (Bil Nep) in 8-week-old C57BL6 mice as the possible models more resemble human conditions were performed. Candida-administered Bil Nep mice demonstrated more severe conditions than Bil Nep alone as indicated by mortality (n = 10/group) and other 48 h parameters (n = 6-8/group), including serum cytokines, leaky gut (FITC-dextran assay, endotoxemia, serum beta-glucan, and loss of Zona-occludens-1), and dysbiosis (increased Enterobacteriaceae with decreased diversity in microbiome analysis) (n = 3/group for fecal microbiome) without the difference in uremia (serum creatinine). With nuclear magnetic resonance metabolome analysis (n = 3-5/group), Bil Nep reduced fecal butyric (and propionic) acid and blood 3-hydroxy butyrate compared with sham and Candida-Bil Nep altered metabolomic patterns compared with Bil Nep alone. Then, Lacticaseibacillus rhamnosus dfa1 (SCFA-producing Lacticaseibacilli) (n = 8/group) attenuated the model severity (mortality, leaky gut, serum cytokines, and increased fecal butyrate) of Bil Nep mice (n = 6/group) (regardless of Candida). In enterocytes (Caco-2 cells), butyrate attenuated injury induced by indoxyl sulfate (a gut-derived uremic toxin) as indicated by transepithelial electrical resistance, supernatant IL-8, NFκB expression, and cell energy status (mitochondria and glycolysis activities by extracellular flux analysis). In conclusion, the reduced butyrate by uremia was not enhanced by Candida administration; however, the presence of Candida in the gut induced a leaky gut that was attenuated by SCFA-producing probiotics. Our data support the use of probiotics in uremia.
Uremia is an accumulation of toxins in the blood due to the loss of renal function from both chronic and acute kidney injury that are common health-care problems worldwide (1). With an inadequate excretion of uremic toxins through the urine, several water-soluble toxins are excreted into the intestine as an alternative route causing an alteration in the intestinal environments that select the growth of some organisms (gut dysbiosis) (2). As such, the hydrolysis of urea (a major uremic toxin) by urease-producing bacteria and the digestion of other toxins facilitates the survival of some microbes (3–5). Indeed, uremia from both chronic kidney disease (CKD) and acute kidney injury (AKI) induced gut dysbiosis (6–8) and the dysbiosis also worsens uremic complications through several mechanisms (9). Accordingly, gut bacteria during uremia-induced dysbiosis facilitated the production of gut-derived uremic toxins, such as p-cresol, indoxyl sulfate, and trimethylamine N-oxide, which enhanced damage to endothelium, kidney, heart, and intestine through chronic inflammation, oxidative stress, and atherosclerosis (10, 11). Additionally, uremic toxins have a direct impact on the enterocytes resulting in enterocytic cell death (2) and also reduce short-chain-fatty-acid (SCFA)-producing bacteria (12). All of these factors from uremia induce defects of gut permeability (or gut barrier) allowing the translocation of pathogen molecules from the gut into the blood circulation (gut leakage or leaky gut). Indeed, the presence of endotoxin, a major cell wall component of Gram-negative bacteria (the most abundance gut organisms), and (1➔3)-β-D-glucan (BG), the main molecule in the cell wall of fungi (the second most abundance gut microbes), in the blood of patients with uremia are demonstrated as a proof of concept for uremia-induced leaky gut (13, 14). During uremia, the presence of these pathogen molecules in blood further enhances uremic toxin-induced chronic inflammatory responses, especially with the induction of innate immunity against microbial molecules that foreign to the host (15). Hence, uremia induces gut dysbiosis and chronic inflammation through impacts of uremia on gut microbes (selection of some bacteria) and host cells (cell injury by oxidative stress from the toxins), then the dysbiosis causes a more severe systemic inflammation with renal function worsening through the leaky gut. However, impacts of uremia on gut dysbiosis in the current literature are mostly focusing on gut bacteria, although gut fungi, especially Candida albicans in human intestines, are the second most abundance microbes in the gut that demonstrate some interaction with gut bacteria and enterocytes (16). Despite the larger size of fungi (10–12 µm of Candida in a yeast form) than bacteria (0.5–2 µm), the fungal abundance in feces by gene copies using 18S rRNA is 1,000-fold lower than 16S rRNA of bacteria with approximately 267 fungal species compared with more than 3,500 bacterial species in the gut (17). The bacterial community varies in quantity and composition from the stomach to the colon (102 versus 1011 cells/gram feces in the stomach and colon, respectively), whereas fungi seem to be localized mostly in the colon, with an average of 106 fungal cells per gram of colon content (18). Interestingly, the presence of gut fungi selectively induces the growth of some gut bacteria (dysbiosis), partly due to i) a digestion ability toward BG of fungal cell wall (glucanase enzymes) as mixing BG into the culture medium enhances the growth of some bacteria (19, 20), and ii) the bacterial tolerance against Candida toxins (21). Then, fungi in the gut can alter bacterial compositions in the gut and contribute to leaky gut-induced systemic inflammation in uremia through glucanemia and endotoxemia (22) and the abundance of gut organisms might be correlated with the level of pathogen molecules in blood during uremia-induced leaky gut (23). Due to the difference in the abundance of fungi in rodents versus humans, Candida administration might make the models more resemble humans (19, 24–26). Indeed, the abundance of Candida spp. in mouse feces is not high enough to be detectable by stool culture (27), which is different from cultures of human feces (28). Unfortunately, impacts of gut fungi, especially C. albicans, in several conditions with leaky gut is not properly considered different from gut bacteria, partly because gut fungi do not seem to cause illness directly. Although we previously demonstrated an impact of oral Candida administration in acute and chronic uremia through bilateral nephrectomy and 5/6 nephrectomy models (2, 12, 29, 30), respectively, the impact of gut fungi on metabolome analysis has never been described. As such, SCFAs (acetate, propionate, and butyrate) are metabolic products of anaerobic bacterial fermentation, especially on the complex carbohydrates-rich diets, in the intestine that is important for the maintenance of intestinal homeostasis (31–33). Due to the possible depletion of SCFAs by uremic toxin-induced dysbiosis, the administration of probiotics, the health beneficial organisms used in several situations, might attenuate leaky gut and systemic inflammation in several conditions, including uremia (34, 35). Although the benefits of probiotics in uremic conditions (acute and chronic kidney injury) are mentioned (7), the exploration of SCFAs and metabolome in uremia is still less. Hence, our objective was to explore the impact of gut Candida on the metabolome changes, especially SCFAs, in feces and in blood during acute uremia and also tested the effectiveness of Lacticaseibacillus rhamnosus dfa1 which are the recently isolated SCFA-producing Lacticaseibacilli from the Thai healthy volunteers from our previous study (36). We hypothesized that SCFAs from probiotics themselves or from other probiotic-promoted bacteria might attenuate uremia-induced intestinal damage and tested the hypothesis in vivo and in vitro.
Male 8-week-old C57BL/6 mice from Nomura Siam International (Pathumwan, Bangkok, Thailand) were used according to the approval by the Institutional Animal Care and Use Committee of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand, following the animal care and use protocol U.S. National Institutes of Health (NIH). Candida-administered bilateral nephrectomy (Bil Nep) or Bil Nep alone, as previously described, were conducted to test the possible impact of gut fungi. Briefly, Candida albicans from the American Type Culture Collection (ATCC 90028) (Fisher Scientific, Waltham, MA, USA) prepared in Sabouraud dextrose broth (SDB) (Oxoid, Hampshire, UK) at 1 × 106 CFU in a 0.3-mL phosphate buffer solution (PBS), or PBS alone, following by Lacticaseibacillus rhamnosus dfa1 that isolated from Thai population (Chulalongkorn University, Bangkok, Thailand) (36) at 1 × 108 CFU in 0.3 mL PBS, or PBS alone, were orally administered at 8 and 12 a.m., respectively, for 7 days prior to Bil Nep surgery. Subsequently, Bil Nep was performed 6 h after the last oral gavage (approximately at 6 p.m.) through abdominal incision according to previous publications (37–39). In the sham group, renal vessels and ureters were only identified before closing the abdominal incision, and fentanyl at 0.03 mg/kg of body weight in 0.5 mL normal saline solution (NSS) was subcutaneously injected after the operation for both analgesia and fluid replacement. Notably, the L. rhamnosus were cultured on de Man-Rogosa-Sharpe (MRS) agar (Oxoid) under anaerobic conditions with gas generation sachets (AnaeroPack-Anaero; Mitsubishi Gas Chemical Co., Inc., Japan) at 37°C for 48 h before quantitative preparation by the determination of the optical density at 600 nm (OD600) as previously described (2). There were 6 mice per group in sham and 8 mice per group for Bil Nep alone and Candida-administered Bil Nep. For probiotics experiments, there were 6 mice per group for sham, Bil Nep, and probiotic-administered Bil Nep, while there were 8 mice per group for Candida-administered Bil Nep with and without probiotics. In the non-survival experiments, mice were sacrificed at 48 h post-surgery under isoflurane anesthesia (with blood and colon collection). Survival analysis was performed using other groups of mice with 10 mice per group and the moribund mice were humanly sacrificed. Colons, 2 cm distal to caecum, were put in tissue frozen in optimal cutting temperature (OCT) compound (Tissue-Tek OCT compound; Sakura Finetek USA, Inc., Torrance, CA, USA) for fluorescent microscopic evaluation. Feces from all parts of the colon were combined and collected for fecal microbiome analysis. Of note, the data at 0 h were collected 3 days before the operation and were used as the baseline values.
Serum creatinine (renal injury) and serum cytokines were evaluated by QuantiChrom creatinine colorimetric assay (DICT-500) (Bioassay, Hayward, CA, USA) and enzyme-linked immunosorbent assay (ELISA) (Invitrogen, Carlsbad, CA, USA), respectively. Gut leakage was determined by i) the detection of fluorescein isothiocyanate-dextran (FITC-dextran), an intestinal nonabsorbable molecule in serum, after an oral administration, ii) serum endotoxin (LPS, a major cell wall components of Gram-negative bacteria), iii) bacteremia, and iv) staining of Zona occludens-1 (ZO-1, an enterocyte tight junction molecule) as previously described (2). For FITC-dextran assays, 0.5 mL of FITC-dextran (molecular weight, 4.4 kDa) (Sigma-Aldrich) at 25 mg/mL was orally administered 3 h prior to blood collection at sacrifice before analysis by fluorescence spectroscopy (Varioskan Flash; Thermo Scientific) at excitation and emission wavelengths of 485 and 528 nm, respectively, with a standard curve of FITC-dextran. Serum LPS was determined by HEK-Blue LPS detection (InvivoGen, San Diego, CA, USA) and bacterial burdens were evaluated by adding 25 µL of blood samples into blood agar for 24 h incubation at 37°C before colony enumeration. For ZO-1 determination, the colons in the 5-µm-thick-frozen sections were stained with a primary antibody against ZO-1 (61-7300) (Thermo Fisher Scientific) (1:200) followed by the secondary antibody Alexa Fluor 546 goat anti-rabbit IgG (A-11035) (Life Technologies, USA) (1:200) and 4’,6-diamidino-2-phenylindole (DAPI; BioLegend, USA) (1:1,000) (nucleus staining color) before visualization and analyzed with a Zeiss LSM 800 confocal microscope (Carl Zeiss, USA) following a previous publication (2).
Fecal microbiota analysis was performed according to methods reported in previous publications (26, 40) using the total DNA from feces of individual mice. Mouse feces were collected by placing mice in metabolic cages (Hatteras Instruments, Cary, NC, USA) for a few hours before the collection of feces (0.25 g) from each mouse in different cages for microbiome analysis to avoid the influence of allocoprophagy (a habit of mice that ingest feces from other mice). The fecal microbiota analysis was performed in 3 fecal samples from 3 mice per group of sham, Bil Nep alone, and Bil Nep with Candida. Briefly, a power DNA isolation kit (MoBio, Carlsbad, CA, USA), metagenomic DNA quality determination (agarose gel electrophoresis and nanodrop spectrophotometry), universal prokaryotic primers; forward 515F (5’-GTGCCAGCMGCCGCGGTAA-3’) and reverse primer 806R (5’-GGACTACHVGGGTWTCTAAT-3’), and 16S rRNA V4 library (appended 50 Illumina adapter and 30 Golay barcode sequences) were used. Each sample (240 ng) was applied to the MiSeq300 sequencing platform (Illumina, San Diego, CA, USA) with Mothur’s standard quality screening operating procedures in MiSeq platform with aligned and assigned taxa (operational taxonomic units [OTUs]) based on default parameters were used (2).
Sample preparation and metabolome analysis by nuclear magnetic resonance (NMR) spectroscopy following previous publications was conducted (41, 42). Briefly, plasma (400 µL) or feces (0.2 g) at pH of 7.5 in 2.4 mL ultrapure water was vortexed for 10 minutes, centrifuged at 14,000 ×g for 10 minutes at 4°C before transferring the supernatant (500 μL) into an Eppendorf tube for filtering through a Pall Nanosep® (3 kDa molecular weight) (Pall life science, Ann Arbor, MI, USA). Then, the filtrate was mixed 1:1 (vol./vol.) with the buffer, which consisted of 300 mM KH2PO4, 10% (w/w) deuterium oxide (D2O), and 1 mM 3‐(Trimethylsilyl) propionic‐2, 2, 3, 3‐d4 acid sodium salt (TSP) at pH 7.5, as the internal standard (41). Additionally, NOESY (nuclear overhauser enhancement spectroscopy) 1D‐1H‐NMR measurements were performed in a 500 MHz NMR spectrometer (Bruker, Rheinstetten, Germany); the 1H NMR spectra were aligned and calibrated based on the internal standard (TSP) peak. For each spectrum, chemical shift (δ) across a range of 0.00-10.00 ppm was segmented (binning) with an interval of 0.02 ppm and the signal intensity in each bin was integrated using Topspin (V 4.0.7, Bruker Biospin) to derive a quantity of each spectrum. The identification of each spectrum (metabolite) was assigned according to the ChenomxNMR suite 8.5 library (Chenomx Inc., Alberta, Canada). The MetaboAnalyst 5.0 (http://www.metaboanalyst.ca/) was used for metabolome data normalization (i.e., by sample median and auto-scaled by mean-centering and dividing by the standard deviation of each variable), clustering algorithm by Ward’s method, and statistical analyses (43). Data visualizations were performed using GraphPad Prism version 8.0 software (GraphPad, La Jolla, CA, USA) and MetaboAnalyst 5.0. There were 5 samples (from 5 mice) in sham and 3 samples (from 3 mice) in Bil Nep and Candida-administered Bil Nep for fecal metabolome analysis. Meanwhile, there were 4 samples (from 4 mice) in sham, 3 samples (from 3 mice) in Bil Nep, and 4 samples (from 4 mice) of Candida-administered Bil Nep for blood metabolome analysis.
Both L. rhamnosus dfa1 and Enterococcus faecium dfa1 were prepared in MRS media as mentioned above, while Bifidobacterium longum dfa1 was cultured in the brain heart infusion (BHI) broth (Oxoid) according to the previous publications (36, 44). Then, short chain fatty acids (SCFAs) in the condition media were analyzed by gas chromatography–mass spectrometry (GC-MS) using the headspace solid-phase microextraction method with an Agilent 6890 GC equipped with an Agilent 5973 mass selective detector (Agilent Technologies) according to a previous publication (45). Briefly, the dimension of the column was 0.25 mm×30 m×0.25 μm with Helium carrier gas at 13.7 ml/min. The temperature program was 10 min isothermal at 50°C, 10 min rising to 240°C with 15°C/min. The injection port temperature is 200°C while the detector port temperature is 250°C. The mass spectrometer was operated in the electron impact mode at 70 eV with a scan range was 40–200 amu. A standard curve was obtained for the calculation of each SCFA concentration. For fecal SCFA, the fecal fatty acids were extracted before the determination by GC-MS according to a previous publication (46). In brief, feces (20 mg in 500 µL of NSS) were added with 10% H2SO4 before fatty acids separation by anhydrous ether (800 μL) and centrifuged (18,000 g for 15 min). Then, the upper ether phase was mixed with 0.25 g of anhydrous Na2SO4 for 30 min, centrifuged (18,000 g for 5 min), and SCFAs in the upper diethyl ether phase were determined by GC-MS as mentioned above.
The influence of indoxyl sulfate, a gut-derived uremia toxin, and butyrate, a well-known SCFA, in the enterocytes was examined using the Caco-2 cell line as previously described (12). As such, the Caco‐2 (ATCCHTB-37) (American Type Culture Collection, Manassas, VA, USA) at 2 × 106 cells/well in Dulbecco’s Modified Eagle Medium (DMEM) were incubated with the different concentrations of indoxyl sulfate (Sigma‐Aldrich, St. Louis, MO, USA) for 24 h before the determination of cell viability with the 2 h incubation (at 37°C in the dark) by 0.5 mg/mL of tetrazolium dye 3‐(4,5‐dimethylthiazol‐2‐yl) ‐2,5‐diphenyltetrazolium (MTT) solution (Thermo Fisher Scientific). The MTT assay is a colorimetric assay for measuring cell metabolic activity based on the ability of nicotinamide adenine dinucleotide phosphate (NADPH)-dependent cellular oxidoreductase enzymes to reduce the MTT tetrazolium dye into the insoluble purple color formazan. After the incubation, the MTT solution was removed and diluted with dimethyl sulfoxide (DMSO) (Thermo Fisher Scientific) before measurement with a Varioskan Flash microplate reader at an absorbance of optical density at 570 nm. On the other hand, Caco‐2 cells at 5 × 104 cells per well were seeded onto the upper compartment of 24‐well Boyden chamber trans wells (Sigma‐Aldrich), using high glucose DMEM supplemented with 20% Fetal Bovine Serum (FBS), 1% HEPES, 1% sodium pyruvate, and 1.3% Penicillin/Streptomycin for 15 days to establish the monolayer of the cells before 24 h incubation with indoxyl sulfate (Sigma‐Aldrich) (0.5 mM) alone or with butyrate (Sigma‐Aldrich) at the indicated concentrations. After that, the transepithelial electrical resistance (TEER) was measured as previously described (47) as demonstrate in ohm (Ω) × cm2 using the epithelial volt‐ohm meter (EVOM2™, World precision instruments, Sarasota, FL, USA) by placing electrodes in the supernatant at the basolateral chamber and in the apical chamber. The TEER values in media culture without Caco‐2 cells were used as a blank and were subtracted from all other measurements. In parallel, the supernatant cytokines were measured by ELISA (Invitrogen).
To explore the impact of uremic toxin and butyrate on cell energy status, the extracellular flux analysis using the Seahorse XFp Analyzers (Agilent, Santa Clara, CA, USA) for the determination of mitochondrial activity and glycolysis through the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), respectively (25, 48–50). In the Seahorse XFp Analyzers, mitochondrial ATP production rates are determined by the rate of oxygen consumption (OCR) in the oxidative phosphorylation pathway that is needed by mitochondria and the ECAR is a result of lactate production in the glycolysis pathway that is used as a representative for glycolysis activity. Both OCR and ECAR are simultaneously measured in real-time in culture well plates using fluorescent sensors in the machine analyzer. As such, Caco-2 cells (1 × 104 cells/well) were grown in modified DMEM, with 0.5 mM of indoxyl sulfate (a gut-derived uremic toxin) with or without 1 mM butyrate (a representative SCFA) for 24 h in the Seahorse cell culture plate before replacing by Seahorse substrates (glucose, pyruvate, and L-glutamine) (Agilent, 103575–100) in pH 7.4 at 37˚C for 1 h prior to the challenge with different metabolic interference compounds for mitochondrial reactions, including oligomycin, carbonyl cyanide-4-(trifluoromethoxy)-phenylhydrazone (FCCP), and rotenone/antimycin A, and for glycolysis intervention, including glucose, oligomycin, and 2-Deoxy-d-glucose (2-DG), according to the manufacturer’s instructions. Data from Seahorse Wave 2.6 software were also conducted based on the following equations: maximal respiration = (OCR between FCCP and rotenone/antimycin A) – (OCR after rotenone/antimycin A); respiratory reserve = (OCR between FCCP and rotenone/antimycin A) – (OCR before oligomycin); and glycolysis = ECAR between glucose and oligomycin.
Mean ± standard error (SE) was used for data presentation. The differences between groups were examined for statistical significance by one-way analysis of variance (ANOVA) followed by Tukey’s analysis or Student’s t-test for comparisons of multiple groups or 2 groups, respectively. All statistical analyses were performed with SPSS 11.5 software (SPSS, IL, USA) and Graph Pad Prism version 7.0 software (La Jolla, CA, USA). A p-value of < 0.05 was considered statistically significant.
To resemble human conditions, C. albicans was orally administered before bilateral nephrectomy (Bil Nep) surgery. As such, Candida worsened Bil Nep mice as indicated by mortality, liver injury (alanine transaminase), serum cytokines (TNF-α, IL-6, and IL-10), leaky gut by FITC-dextran assay, endotoxemia, serum (1➔3)-β-D-glucan (BG), and enterocyte tight junction protein (Zona occludens-1; ZO-1), but not renal injury (blood urea nitrogen and serum creatinine), and bacteremia, when compared with the control Bil Nep using only normal saline solution (NSS) gavage (Figures 1A–O). In sham mice, Candida did not alter any parameters when compared with sham control (data not shown). The increased endotoxin (LPS) and BG in serum (Figures 1K, L) along with enterocytes damage (ZO-1) (Figures 1N, O) of Bil Nep mice compared with sham mice indicated uremia-induced leaky gut which was more prominent in Candida-administered Bil Nep compared with Bil Nep alone.

Figure 1 Characteristics of bilateral nephrectomy (Bil Nep) mice orally administered by normal saline solution (NSS) or Candida albicans (Bil Nep+Cand) or control sham surgery (Sham) as determined by survival analysis (A), blood urea nitrogen (B), serum creatinine (C), serum cytokines (TNF-α, IL-6, and IL-10) (D–G), and parameters of gut barrier defect, including FITC-dextran assay (H), endotoxemia (I), (1→3-β-D-glucan) (J), bacteremia (K), and the abundance of tight junction molecule (Zona occluden-1; ZO-1) (L) in the colon (percentage of the green fluorescent color) with the representative fluorescent-stained pictures (original magnification 630x) (M–O) are demonstrated (n = 10/group for A and 6-8/group for others).
In parallel, Candida also induced gut dysbiosis as indicated by fecal microbiome analysis at 48 h of experiments (Figures 2A–D). With the collection of fecal samples from 3 mice in each experimental group, there was an elevation in Proteobacteria (a major phylum of Gram-negative bacteria including pathogenic microbes), Enterobacteriaceae (a group of pathogenic Gram-negative bacilli), and Muribaculaceae (Gram-negative anaerobes in Bacteroides group) in Candida-administered Bil Nep when compared with Bil Nep alone (Figures 2E–G). Additionally, Candida-induced dysbiosis was also indicated by the reduction in total bacterial abundance and the diversity (Chao-1 and Shannon scores) when compared with Bil Nep alone (Figures 2H–J). Proteobacteria of Bil Nep mice were also more prominent than sham mice despite the non-difference in total bacteria abundance and diversity (Figures 2A–J), supporting uremia-induced gut dysbiosis. Hence, Bil Nep without Candida demonstrated uremia-induced leaky gut through an enhanced pathogenic bacteria (Proteobacteria) resulting in endotoxemia and glucanemia with systemic inflammation-induced liver injury and nearly all of these parameters (fecal Proteobacteria, leaky gut, endotoxemia, glucanemia, serum TNF-a, and liver injury) was worsened by Candida administration in Bil Nep compared with Bil Nep alone.
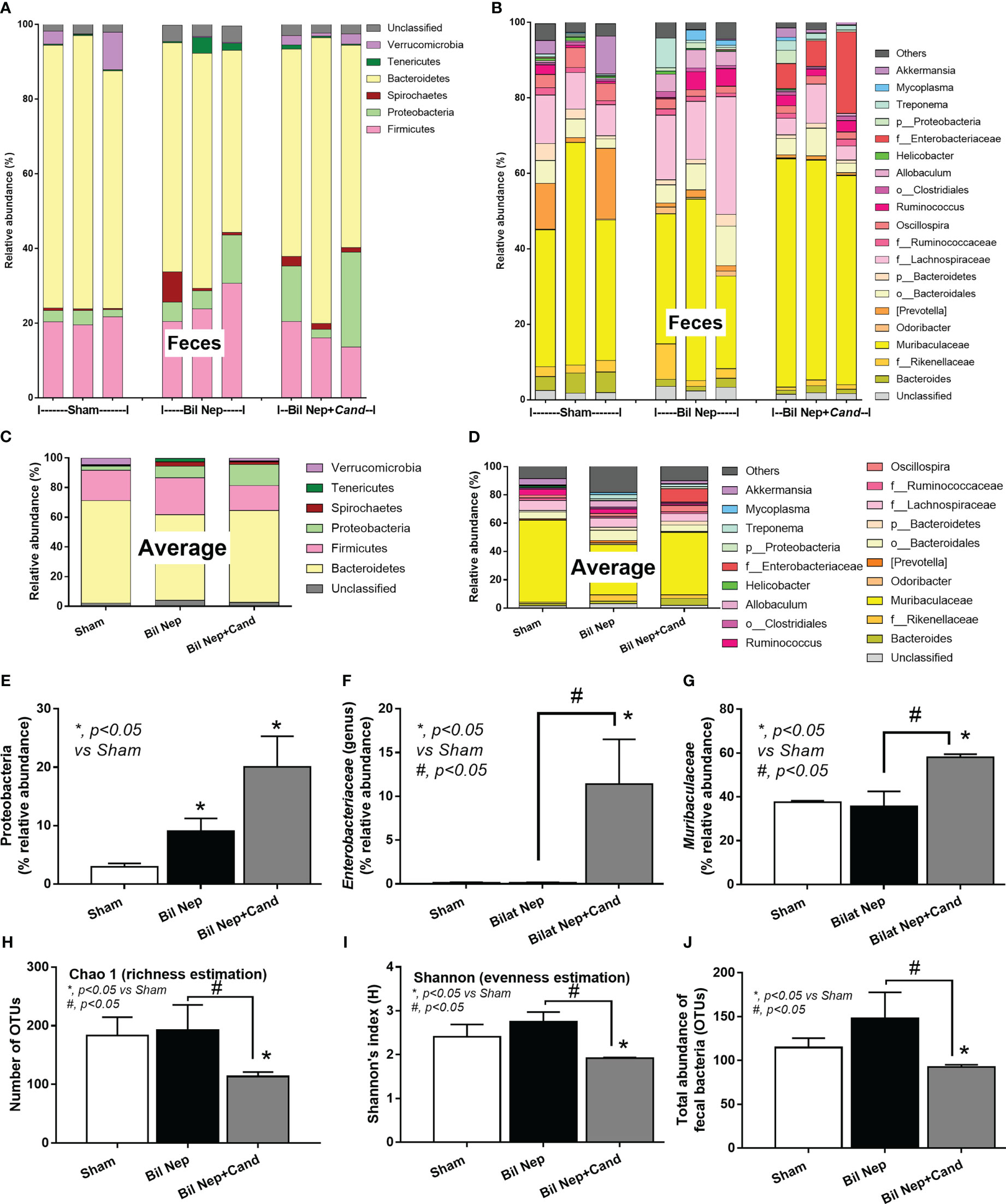
Figure 2 Fecal microbiota analysis of mice with sham, bilateral nephrectomy (Bil Nep), and Bil Nep with Candida administration at 48 h post-operation as indicated by the relative abundance of bacteria at the phylum and genus levels with the average value (A–D), the abundances of some bacteria in graph presentation (E–G), and the alpha diversity (Chao 1 and Shannon index) with a total abundance of bacteria in operational taxonomic units (OTUs) (H–J) are demonstrated.
The excretion of uremic toxins into the gut was not only inducing fecal dysbiosis (Figures 2A–J) but also altered the fecal metabolome (Figures 3A, B). In comparison with sham mice, Bil Nep mice demonstrated a significant increase in i) nitrogenous bases and derivatives (hypoxanthine, xanthine, and uracil), ii) amino acids (threonine, phenylalanine, lysine, valine, isoleucine, tyrosine, glycine, taurine, alanine, and leucine), iii) energy-related compounds (lactate, glucose, fumarate and ethanol) (Figure 3A). Meanwhile, Bil Nep induced a significant decrease in i) amino acids (lysine, aspartate, methionine, glutamine), ii) SCFAs (propionate and butyrate), and iii) an energy-related metabolite (pyruvate) when compared with sham feces (Figure 3A). The fecal metabolome of Bil Nep versus Candida-Bil Nep mice was similar except for the higher formate and acetate in the latter group (Figures 3B–D). Additionally, the separation between sham control versus Bil Nep feces and Bil Nep versus Candida-Bil Nep feces was demonstrated by several plot analyses, including principal component analysis (PCA; the data simplification into fewer summary dimensions while retaining trends and patterns) and partial least squares-discriminant analysis (PLS-DA; the data projection into the non-direct observed structure to find the fundamental relations between two matrices) with modified analyses, including orthogonal PLS-DA (OPLS-DA), and sparse PLS-DA (sPLS-DA) (Figures 4A–D). In parallel, blood metabolome in Bil Nep compared with sham demonstrated an increase in i) some amino acids (phenylalanine, lysine, and histidine), ii) uremic toxins (urea and creatinine), and iii) pyruvate, with a decrease in i) some amino acids (taurine, alanine, leucine, isoleucine, and valine), ii) energy-related compounds (glucose, lactate, and ethanol), and iii) SCFAs (3-hydroxybutyrate and acetate) (Figures 5A–D), supported by all plot analyses (Figures 6A–D). Meanwhile, the difference between Bil Nep versus Candida-Bil Nep was subtle (Figures 5A, B) as could be demonstrated only by PLS-DA and the modifications (OPLS-DA and sPLS-DA) (Figures 6B–D). Due to the importance of SCFAs in enterocyte homeostasis (18), the reduced butyrate (and propionate) in feces (Figure 3A) and 3-hydroxy butyrate in the blood (Figure 5A) of uremic mice, perhaps due to uremia-induced gut dysbiosis (Figures 2A–J), might partly be responsible for uremia-induced leaky gut (Figures 1J–O). In short, the uremia-induced metabolome alteration in feces and in blood of Bil Nep compared with sham mice from was clearly demonstrated, especially through the reduced butyrate (an important SCFA) in feces and in the blood; however, Candida administration did not worsen the decreased butyrate. These data implied that the worsened conditions of Candida-Bil Nep over Bil Nep alone (Figures 1J–O) were not due to reduced SCFA but possibly because of other adverse effects of the fungi, such as Candida-induced gut dysbiosis (Figures 2A–J) that causing a more severe leaky gut-induced systemic inflammation.
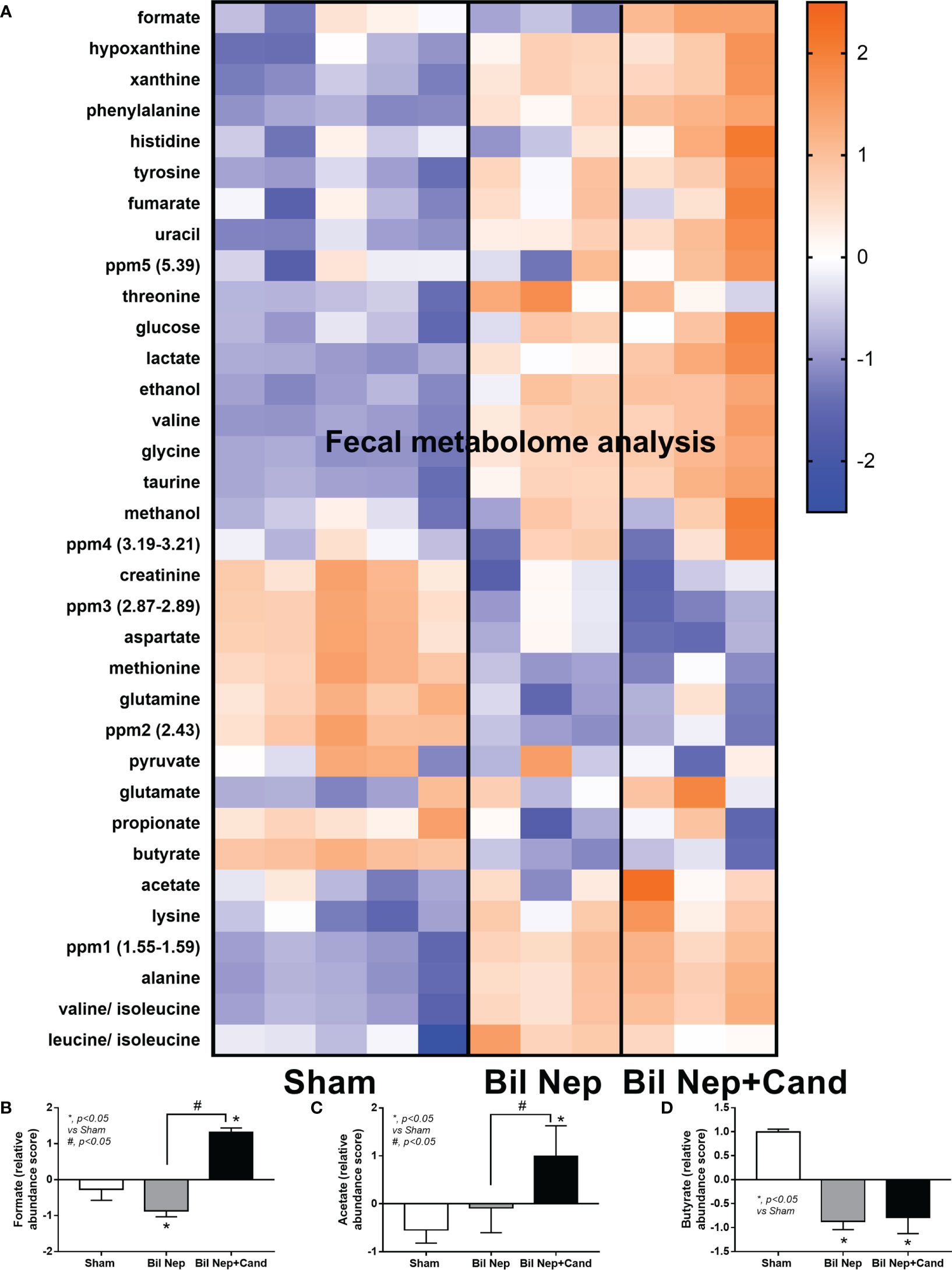
Figure 3 Fecal metabolome analysis of mice with sham, bilateral nephrectomy (Bil Nep), and Bil Nep with Candida administration at 48 h post-operation as indicated by the heat-map of the metabolites (A) with the graphs of some substances (formate, acetate, and butyrate) (B–D) are demonstrated. The color scale bars are demonstrated by log2 fold change. The average values from sham (5 samples) and Bil Nep with or without Candida (3 samples per group) are presented in the column graph for visualization of the comparison.
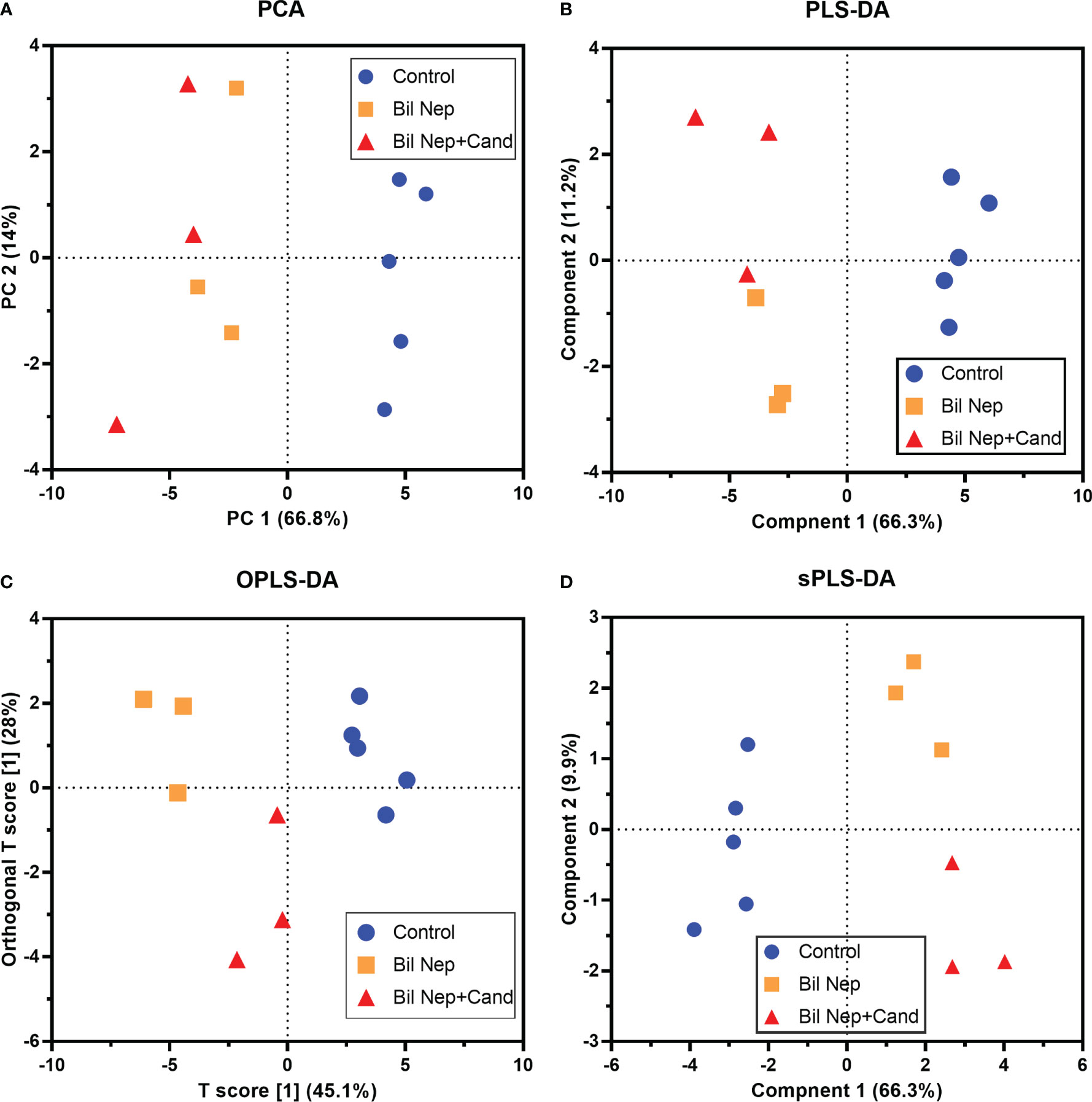
Figure 4 Fecal metabolome analysis of mice with sham, bilateral nephrectomy (Bil Nep), and Bil Nep with Candida administration at 48 h post-operation as indicated by several score plot analyses, including the relationships among plasma metabolome profiles by principal component analysis (PCA) (A), orthogonal partial least squares-discriminant analysis (OPLS-DA) (B), partial least squares-discriminant analysis (PLS-DA) (C), and sparse partial least squares - discriminant analysis (sPLS-DA) (D) are demonstrated.
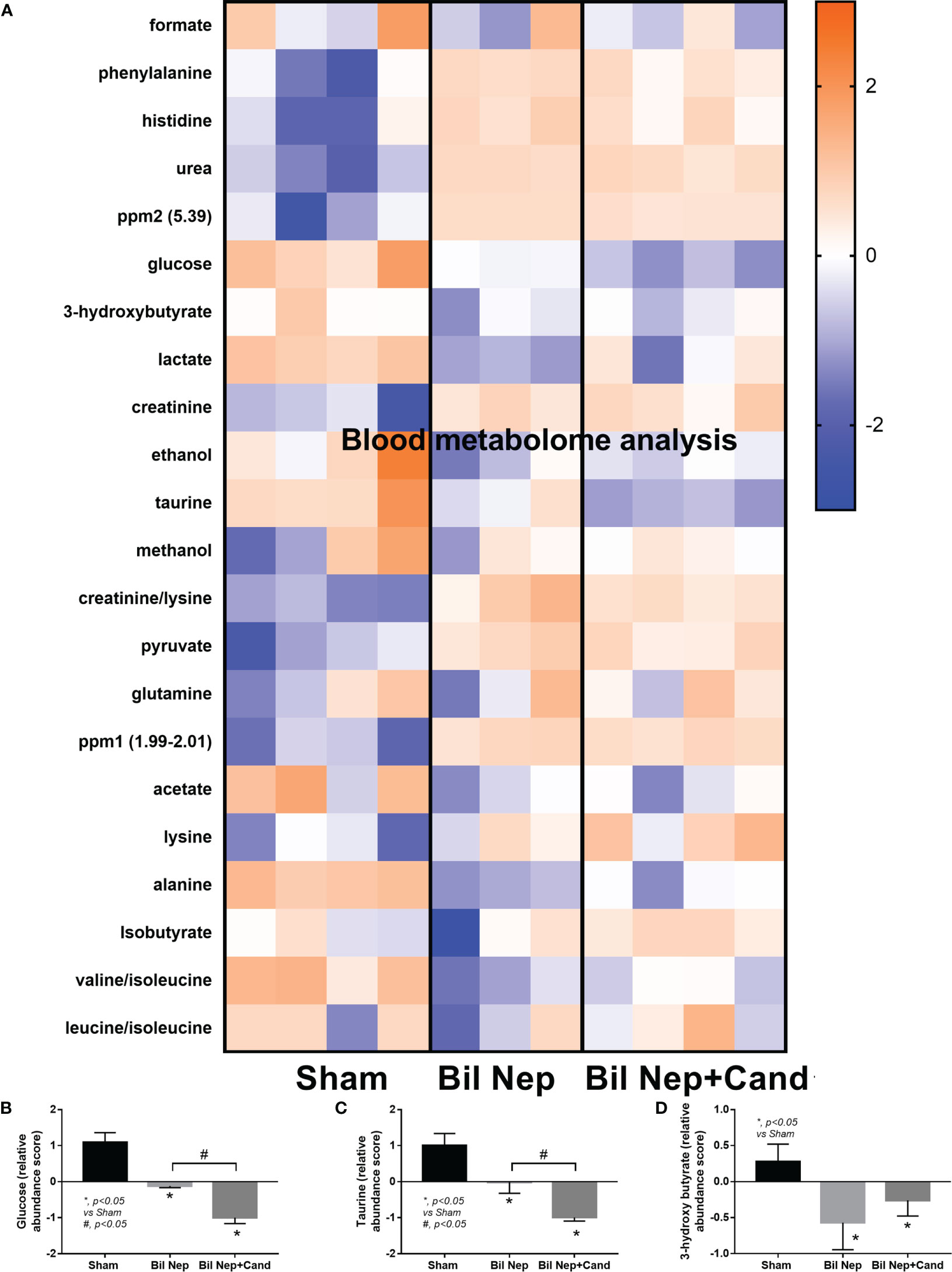
Figure 5 Blood metabolome analysis of mice with sham, bilateral nephrectomy (Bil Nep), and Bil Nep with Candida administration at 48 h post-operation as indicated by the heat-map of the metabolites (A) with the graphs of some substances (formate, acetate, and butyrate) (B–D) are demonstrated. The color scale bars are demonstrated by log2 fold change. The average values from sham (4 samples), Bil Nep (3 samples), and Bil Nep with Candida (3 samples) are presented in the column graph for visualization of the comparison.
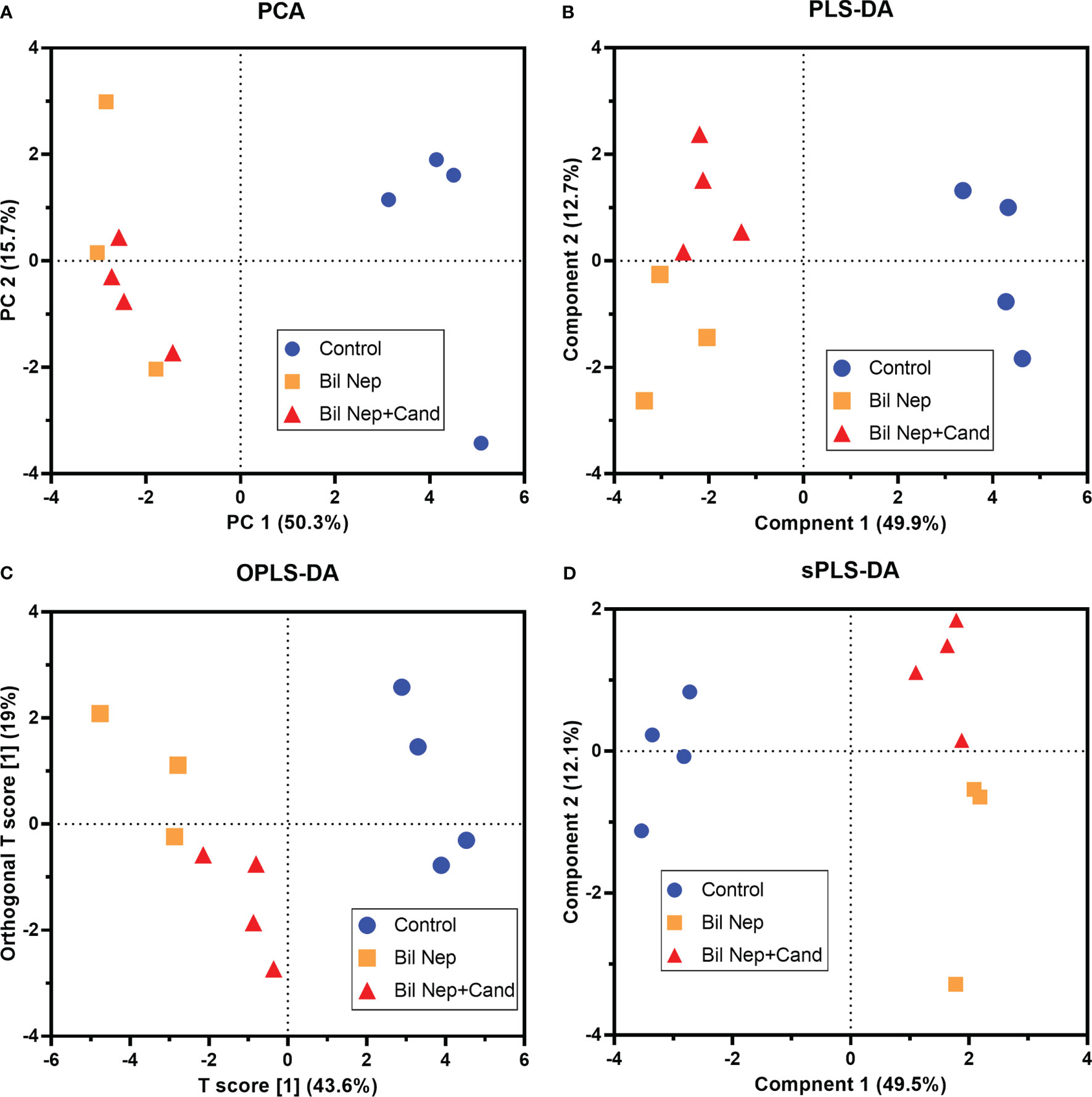
Figure 6 Blood metabolome analysis of mice with sham, bilateral nephrectomy (Bil Nep), and Bil Nep with Candida administration at 48 h post-operation as indicated by several score plot analyses, including the relationships among plasma metabolome profiles by principal component analysis (PCA) (A), orthogonal partial least squares-discriminant analysis (OPLS-DA) (B), partial least squares-discriminant analysis (PLS-DA) (C), and sparse partial least squares - discriminant analysis (sPLS-DA) (D) are demonstrated.
Although the increased SCFAs after probiotics administration might be produced from probiotics or the probiotics-promoted bacteria, SCFAs-producing probiotics ensure the enhanced production of SCFAs. As such, several SCFA-related compounds (acetic, butyric, and propionic acid) were detectable in the condition media of several probiotics from our library, including Lacticaseibacilli, Enterococci, and Bifidobacterium, without the significant differences among probiotics (Figures 7A-C). Due to the less difficult preparation processes, Lacticaseibacilli were selected to use in the mice. Although all mice were dead within 64 h post-Bil Nep, Candida-administered Bil Nep (without probiotics) demonstrated the earliest death as all mice died within 48 h (Figure 7D) implying an adverse effect of Candida gavage. In comparison with Bil Nep without Candida, Candida-Bil Nep demonstrated more prominent systemic inflammation (serum cytokines), and leaky gut (FITC-dextran assay and bacteremia but not endotoxemia), despite the similar uremic severity (serum creatinine) and the decreased fecal SCFAs (acetic, butyric, and propionic acid) (Figures 7E–N). In parallel, probiotics attenuated leaky gut severity in Bil Nep mice as indicated by FITC-dextran assay and endotoxemia (but not bacteremia) with enhanced fecal butyric acid without an alteration in other parameters (Figures 7D–N). In Candida-Bil Nep, probiotics improved survival rate, serum IL-6 (not TNF-α and IL-10), endotoxemia, and bacteremia with increased fecal butyric acid (Figures 7D–N).
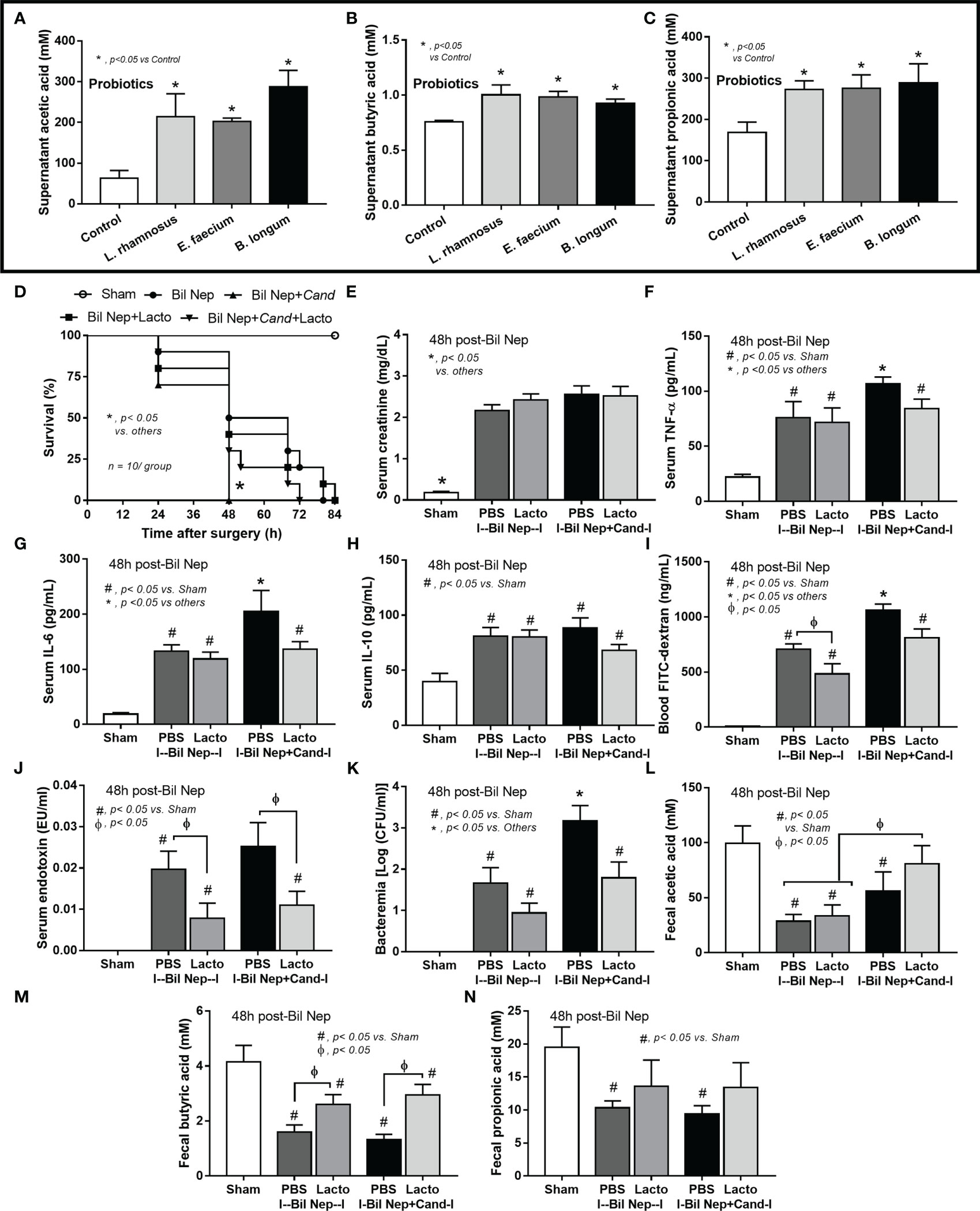
Figure 7 The short chain fatty acid (SCFA) (acetic, butyric, and propionic acids) in the condition media of several probiotics (Lacticaseibacillus rhamnosus, Enterococcus faecium, and Bifidobacterium longum) compared with the control media (control) (A–C) are demonstrated (independent triplicate experiments were performed). Characteristics of bilateral nephrectomy (Bil Nep) mice with or without Candida administration (Cand) after treatment by Lacticaseibacillus rhamnosus (Lacto) or normal saline solution (NSS) control versus sham mice as indicated by survival analysis (D), serum creatinine (E), serum cytokines (TNF-α, IL-6, and IL-10) (F–H), and parameters of gut barrier defect (FITC-dextran assay, endotoxemia, bacteremia) (I–K), and fecal SCFA (L–N) are demonstrated (n = 10/group for D and 6-8/group for E–N.
Because i) intestinal secretion of indoxyl sulfate (IS), a gut-derived uremic toxin transforming from indole (tryptophan derivatives of gut-dysbiosis bacteria) at the liver, in uremic conditions is mentioned (51), ii) the enterocyte toxicity of IS (52), and iii) the impact of SCFAs in enterocyte homeostasis (32) and the probiotics-increased fecal butyrate (Figure 7M), IS and butyrate were tested in enterocytes. Indeed, enterocyte toxicity of IS was demonstrated by MTT assay with a 50% reduction in cell viability from 2 mM of IS (Figure 8A). Despite the non-alteration of cell viability by 0.5 mM IS, this concentration induced enterocyte injury, as indicated by epithelial integrity (TEER), supernatant IL-8, and the upregulated inflammatory genes (IL-8 and NFκB), possibly from the reduction of cell energy (mitochondrial and glycolysis activities) (Figures 8B–J). Interestingly, butyrate (at 1 and 4 mM) similarly attenuated enterocyte injury (TEER, supernatant IL-8, IL-8, and NFκB) with an improvement of cell energy status (Figures 8B–J), perhaps through butyrate-related energy supplement (53, 54). In brief, Lacticaseibacilli were selected from several SCFA-producing probiotics to use in mice due to the simpler laboratory preparation and the probiotic administration enhanced fecal butyrate (Figure 7M) and attenuated the model severity, especially leaky gut, endotoxemia, and bacteremia (Figure 7I–K), similarly between Candida-Bil Nep and Bil Nep alone. The increased fecal butyrate, either from the administered probiotics or other probiotics-induced beneficial bacteria, partly due to the influence of butyrate on uremic toxin-induced enterocyte damage through enhanced cell integrity (TEER), anti-inflammation, and improved cell energy status (mitochondria and glycolysis activities) (Figure 8A–J).
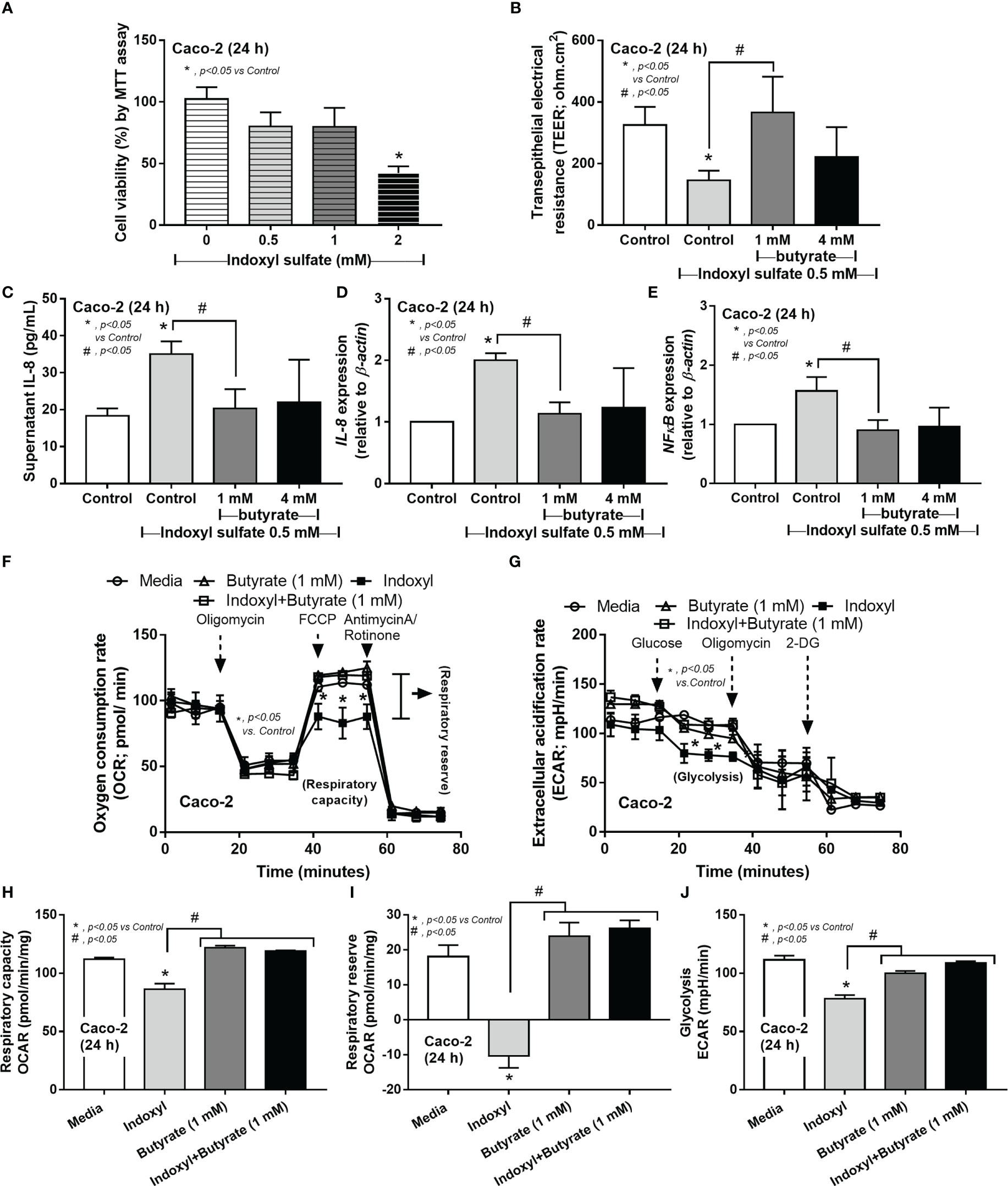
Figure 8 The cell viability (MTT assay) of enterocytes (Caco‐2 cells) after incubation by the different concentrations of indoxyl sulfate (A) is demonstrated. Characteristics of Caco-2 cells after incubation by 0.5 mM indoxyl sulfate with butyrate (1 and 4 mM) or control media (Control) as evaluated by transepithelial electrical resistance (TEER) (B) and supernatant IL-8 (C), expression of IL-8 and NF-κB (D, E) are shown. Additionally, the cell energy status of Caco-2 cells after being activated by 0.5 mM indoxyl sulfate with or without 1 mM butyrate or media control as indicated by indicators for mitochondrial function; oxygen consumption rate (OCR), and glycolysis activity; extracellular acidification rate (ECAR) (F, G) with some energy phenotype profile (maximal respiration, respiratory reserve, and glycolysis) (H–J) are also demonstrated (independent triplicate experiments were performed for all experiments).
Probiotics attenuated uremia-induced leaky gut in Bil Nep mice with or without Candida partly through the short-chain fatty acids (SCFAs)-improved cell energy of enterocytes.
The excretion of uremic toxins through the intestine during renal impairment is one of the obvious causes of uremia-induced leaky gut (55), as the direct injury from uremic toxins (such as urea, p-cresol, and indoxyl sulfate) against enterocytes is demonstrated in vitro (52, 56, 57), that is possibly an underlying mechanism of enterocyte apoptosis and leaky gut in uremia (2). The toxins (water-soluble and protein-bound compounds) contact intestinal epitheliums through the intestinal lumens (luminal side) and from blood delivery as some toxins (such as indoxyl sulfate) are produced in the blood (through livers) before the delivery to other organs, including kidney, brain, bone, and gut (58), while some toxins (such as p-cresol) are produced within the gut (59). Despite the different production sites, gut-derived uremic toxins (60) are partly responsible for uremia-induced gut dysbiosis, possibly due to the selection of bacteria by the toxins (enhanced bacteria that can utilize the toxins) (61–63) and the direct impacts of uremic toxins on enterocyte damages that also causing dysbiosis (gut inflammation-induced dysbiosis) (64). Here, the presence of Candida in the gut caused more severe dysbiosis than Bil Nep alone, as indicated by the increased Proteobacteria (especially Enterobacteria) with the possible direct epithelial invasion by Candida spp. (65), which might be correlated with the more severe leaky gut in Candida-Bil Nep compared with Bil Nep alone. Indeed, the selection of some bacterial groups by Candida, including Enterobacteria and some Enterobacter spp., is possibly due to the ability of bacteria to digest the Candida cell wall by β-glucanase and cellulolytic enzymes (19, 66). Additionally, the possibly bactericidal activity of some fungal toxins, such as candidalysin (a cytolytic pore-forming peptide), and fungal by-products, such as acetaldehyde, alcohol, and ammonia, also possibly alter some groups of bacteria more than other groups, leading to Candida-induced gut dysbiosis (67, 68). In a previous publication, Candida administration in sham control mice does not induce gut dysbiosis and systemic inflammation, while Candida administration in Bil Nep mice worsens gut dysbiosis and systemic inflammation (2) similar to our current results. Not only in acute uremia, Candida administration in chronic kidney disease (CKD) in 5/6 nephrectomy mice also induces gut dysbiosis and worsens systemic inflammation through leaky gut-induced endotoxemia and glucanemia that are severe enough to induce fibrosis in several internal organs (kidney, liver, and heart) approximately at 20-week post-surgery (12, 29). In this aspect, the presence of gut fungi enhances the severity of uremia-induced systemic inflammation partly through leaky gut-induced endotoxemia and glucanemia which might depend on the abundance of Gram-negative bacteria and fungi, respectively, in the gut. The monitoring of leaky gut and the abundance of endotoxin and beta-glucan in gut content might be interesting as biomarkers for monitoring of the disease severity in patients with acute or chronic uremia might be beneficial. More studies on these topics are interesting.
Due to the well-known association between gut bacteria and the alteration of small molecules (69), the metabolome analysis was evaluated. As such, the difference in the pattern of fecal metabolome between uremic mice and sham control was obvious, especially the reduction of amino acids, butyrate, and propionate (but not acetate), as indicated by principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) analyses. Perhaps, the enhanced fermentation of proteins in the gut during uremia-induced dysbiosis leads to an increase in amino acids in feces (62, 70) and the reduced normal microbiota from uremia decreases fecal SCFAs (71). Meanwhile, the reduced fecal creatinine in uremic mice compared with sham supported an increase in bacteria that can metabolite creatinine (62, 72). Here, the elevation of several amino acids, except for lysine, aspartate, methionine, and glutamine, supported the increased nonessential amino acids and reduced essential amino acids (histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine) in uremia (73). Indeed, the increase in glutamine and aspartate-associated proteins in uremia are previously mentioned (74, 75). Nevertheless, Candida gavage subtly altered uremic fecal metabolome with an increase in acetate and formate compared with uremia alone suggesting the promoted acetate- and formate-producing bacteria (perhaps through the digestion of Candida cell wall) (76, 77). In blood metabolome analysis, fewer compounds, possibly only with the high abundance, were detectable with a similar trend of the fecal metabolome, except for taurine and alanine (higher in feces but lower in the blood of uremia compared with sham), and pyruvate (lower in feces but higher in the blood of uremia compared with sham). This fecal-blood discordance of metabolome results indicates a complex correlation of the individual compounds such as the utilization by enterocytes and the transport mechanisms (78). In the blood of uremic mice compared with sham, the reduced 3-hydroxy butyrate (a main ketone body transforming from SCFAs by liver) (79) with the non-difference in acetate and isobutyrate (butyrate isomerization) (80) suggested the difference influences of different SCFAs in uremia. Despite the differences in PLS-DA analysis of blood metabolome between Candida-Bil Nep versus Bil Nep, the components in blood metabolome were not different between groups, while some alterations in fecal metabolome were detectable suggesting that Candida altered only the metabolites that were possibly too low to be detectable in blood. Despite a similarly low level of fecal SCFAs between Bil Nep versus Candida-Bil Nep, Candida-Bil Nep demonstrated more severe dysbiosis (the higher abundance of Proteobacteria with the lower bacterial diversity) suggesting that the presence of Candida in the gut and the selective gut bacteria from Candida had a less impact on fecal SCFAs but demonstrated a higher impact on leaky gut. However, Bil Nep mice with the presence of Candida in the gut might be more resemble to the patients due to the higher abundance of fungi in the human gut compared with mouse intestines (27, 28). Because of the higher disease severity of Candida-Bil Nep mice over Bil Nep alone, the probiotics that are effective in Bil Nep mice might be ineffective on the Candida-Bil Nep group and the test of probiotics on Candida-Bil nep mice might be more appropriate for further clinical translation.
Probiotics attenuate leaky gut from several causes, including acute and chronic uremia, through several mechanisms, including normalized gut dysbiosis, regulation of host immune responses, facilitated mucin production, and induced anti-inflammation by exopolysaccharide and SCFAs (81–83). Among these mechanisms, the production of SCFAs seems to be an interesting mechanism of probiotics as SCFAs, from probiotics themselves or from beneficial bacteria that are promoted by probiotics, are absorbed into enterocytes before using as an energy source (84) with induction of several anti-inflammatory signals (in enterocytes and immune cells) (85). Although the increased intestinal SCFAs after probiotic administration is well-known (86), data on probiotics-enhanced SCFAs in uremia is still less. In a healthy condition, normal gut microbiota enables the transformation of complex nutrients, including plant cell wall components, into simple sugars that are fermented to form SCFAs, mainly formate, acetate, propionate, and butyrate as contributed to more than 90% of SCFAs in the human gut (87, 88). With the reduced SCFAs, several probiotic bacteria can produce SCFAs, including Lacticaseibacillus rhamnosus GG, Bifidobacterium longum SP 07/3, and Enterococcus faecalis AG5 (83, 89). Although the non-SCFAs-producing probiotics can enhance intestinal SCFAs through the promotion of other beneficial bacteria in the gut, the use of SCFAs-producing bacteria possibly ensures SCFAs production. Despite several isolated probiotics with effectively produced SCFAs in our facility, Lacticaseibacillus rhamnosus dfa1 was selected to use due to the easier preparation process of the facultative anaerobes compared with the strictly anaerobic Bifidobacterium preparation. Indeed, the Lacticaseibacilli attenuated leaky gut and systemic inflammation with increased butyrate in uremia mice with or without Candida. Because of the low fecal butyrate both in feces and in blood of uremic mice here, the enhanced butyrate in feces might be a probiotic-activity biomarker as a previous report of low butyrate in uremia patients (90). However, uremia did not alter acetate (the most abundant SCFA) but reduced propionate only in feces (not in blood). Hence, patients with uremia with low fecal butyrate might be more benefit from the administration of SCFAs or probiotics-induced SCFAs and treatment monitoring of the probiotic effects in these patients through fecal metabolome changes (increased fecal butyrate when compared with the before treatment) might be useful. Notably, the treatment monitoring of probiotics effect by blood metabolome measurement might be less beneficial than fecal metabolome evaluation as a subtle difference in blood metabolome between Bil Nep mice with versus without probiotics.
During uremia, several uremic toxins induced damage to enterocytes as serum from patients with chronic kidney disease, urea, and some gut-derived uremic toxins reduced enterocyte integrity (transepithelial electrical resistance; TEER) (91, 92). Here, indoxyl sulfate, a gut-derived uremic toxin was used as a representative toxin for the in vitro test in enterocyte cell lines. As such, an impact of butyrate on uremic toxin (indoxyl sulfate)-stimulated enterocytes was demonstrated in vitro (enterocyte integrity and anti-inflammation) along with the increased enterocyte cell energy supporting a previous publication (93). The administration of butyrate as a chemical drug is interesting due to an easier production process than probiotic preparation; however, the proper dose adjustment will be necessary as butyrate toxicity is possible (94, 95). Although the more profound enhancement of mitochondrial functions over glycolysis by SCFAs (96, 97) and the increased cell status by butyrate as a source of acetyl coenzyme A (acetyl Co-A) in mitochondrial tricarboxylic acid (TCA) cycle (53) are previously reported, butyrate could not alter cell energy status in the control enterocytes here. On the other hand, indoxyl sulfate (IS) reduced both mitochondrial and glycolysis activity of enterocytes possibly through the enhanced oxidant species (98–100) that were toxic to both mitochondria (oxidant-induced mitochondrial injury) and glycolysis (blockage of glyceraldehyde-3-phosphate dehydrogenase by oxidants) (101). In enterocytes, butyrate normalized IS-induced mitochondrial defect possibly through the increased acetyl Co-A for mitochondrial TCA (102), while improved IS-reduced glycolysis through the improved cell conditions that facilitate glucose utilization (103). Notably, the probiotics were not administered in the sham control because there was no dysbiosis (the imbalance of gut microbiota correlating with the unhealthy condition) in sham mice with the well-known safety of L. rhamnosus probiotics (2, 7, 12, 25, 26, 47).
In conclusion, uremia altered metabolome characteristics in both feces, especially reduced butyrate, and the presence of Candida in the gut of Bil Nep mice worsened leaky gut-induced systemic inflammation. Despite the enhanced enterocyte damages by Candida, probiotics attenuated the model severity, as indicated by increased fecal butyrate, strengthened gut barrier, and reduced leaky gut-induced systemic inflammation. Additionally, the improved fecal butyrate levels after probiotic administration might be a monitoring biomarker for a proper probiotic effect. More studies on these topics are interesting.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA909950.
The animal study was reviewed and approved by The animal care and use protocol (SST 028/2564) was certified by the Institutional Animal Care and Use Committee of Chulalongkorn University’s Faculty of Medicine in Bangkok, Thailand, in compliance with the US National Institutes of Health criteria.
The followings are the authors’ contribution: conceptualization: WC and AL. Methodology: WC, SK, PV, SS, TC, NS, SS, and AL. Validation: SS, TC, SS, PV, and AL. Formal analysis: WC, SK, and AL. Investigation: NS, TC, PV, WC, and AL. Resources: WC, SK, and AL. Data curation: AL. Writing-original draft preparation: WC and AL. Writing review and editing: WC and AL. Supervision: AL. And funding acquisition: WC. All authors contributed to the article and approved the submitted version.
This research was supported by Specific League Funds from Mahidol University, fiscal year 2022 (WC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Ostermann M, Cerdá J. The burden of acute kidney injury and related financial issues. Contrib Nephrol (2018) 193:100–12. doi: 10.1159/000484967
2. Panpetch W, Kullapanich C, Dang CP, Visitchanakun P, Saisorn W, Wongphoom J, et al. Candida administration worsens uremia-induced gut leakage in bilateral nephrectomy mice, an impact of gut fungi and organismal molecules in uremia. mSystems (2021) 6:e01187–20. doi: 10.1128/mSystems.01187-20
3. Konieczna I, Zarnowiec P, Kwinkowski M, Kolesinska B, Fraczyk J, Kaminski Z, et al. Bacterial urease and its role in long-lasting human diseases. Curr Protein Pept Sci (2012) 13:789–806. doi: 10.2174/138920312804871094
4. Mitch WE. Effects of intestinal flora on nitrogen metabolism in patients with chronic renal failure. Am J Clin Nutr (1978) 31:1594–600. doi: 10.1093/ajcn/31.9.1594
5. Wong J, Piceno YM, DeSantis TZ, Pahl M, Andersen GL, Vaziri ND. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am J Nephrol (2014) 39:230–7. doi: 10.1159/000360010
6. Cao C, Zhu H, Yao Y, Zeng R. Gut dysbiosis and kidney diseases. Front Med (Lausanne) (2022) 9:829349. doi: 10.3389/fmed.2022.829349
7. Tungsanga S, Katavetin P, Panpetch W, Udompornpitak K, Saisorn W, Praditpornsilpa K, et al. Lacticaseibacillus rhamnosus L34 attenuates chronic kidney disease progression in a 5/6 nephrectomy mouse model through the excretion of anti-inflammatory molecules. Nephrol Dial Transplant (2022) 37:1429–42. doi: 10.1093/ndt/gfac032
8. Gong J, Noel S, Pluznick JL, Hamad ARA, Rabb H. Gut microbiota-kidney cross-talk in acute kidney injury. Semin Nephrol (2019) 39:107–16. doi: 10.1016/j.semnephrol.2018.10.009
9. Kim JE, Kim HE, Park JI, Cho H, Kwak MJ, Kim BY, et al. The association between gut microbiota and uremia of chronic kidney disease. Microorganisms (2020) 8:907. doi: 10.3390/microorganisms8060907
10. Graboski AL, Redinbo MR. Gut-derived protein-bound uremic toxins. Toxins (Basel) (2020) 12:590. doi: 10.3390/toxins12090590
11. Lau WL, Savoj J, Nakata MB, Vaziri ND. Altered microbiome in chronic kidney disease: systemic effects of gut-derived uremic toxins. Clin Sci (Lond) (2018) 132:509–22. doi: 10.1042/CS20171107
12. Tungsanga S, Panpetch W, Bhunyakarnjanarat T, Udompornpitak K, Katavetin P, Chancharoenthana W, et al. Uremia-induced gut barrier defect in 5/6 nephrectomized mice is worsened by candida administration through a synergy of uremic toxin, lipopolysaccharide, and (1➔3)-β-D-Glucan, but is attenuated by lacticaseibacillus rhamnosus L34. Int J Mol Sci (2022) 23:2511. doi: 10.3390/ijms23052511
13. Wong J, Zhang Y, Swift O, Finkelman M, Patidar A, Ramanarayanan S, et al. Beta-glucans in advanced CKD: role in endotoxaemia and inflammation. BMC Nephrol (2020) 21:118. doi: 10.1186/s12882-020-01779-9
14. Wong J, Vilar E, Farrington K. Endotoxemia in end-stage kidney disease. Semin Dial (2015) 28:59–67. doi: 10.1111/sdi.12280
15. Lau WL, Kalantar-Zadeh K, Vaziri ND. The gut as a source of inflammation in chronic kidney disease. Nephron (2015) 130:92–8. doi: 10.1159/000381990
16. Hiengrach P, Panpetch W, Chindamporn A, Leelahavanichkul A. Helicobacter pylori, protected from antibiotics and stresses inside Candida albicans vacuoles, cause gastritis in mice. Int J Mol Sci (2022) 23:8568. doi: 10.3390/ijms23158568
17. de Albuquerque GE, Moda BS, Serpa MS, Branco GP, Defelicibus A, Takenaka IKTM, et al. Evaluation of bacteria and fungi DNA abundance in human tissues. Genes (Basel) (2022) 13:237. doi: 10.3390/genes13020237
18. Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol (2019) 16:331–45. doi: 10.1038/s41575-019-0121-2
19. Hiengrach P, Panpetch W, Worasilchai N, Chindamporn A, Tumwasorn S, Jaroonwitchawan T, et al. Administration of candida albicans to dextran sulfate solution treated mice causes intestinal dysbiosis, emergence and dissemination of intestinal pseudomonas aeruginosa and lethal sepsis. Shock (2020) 53:189–98. doi: 10.1097/SHK.0000000000001339
20. Hiengrach P, Panpetch W, Chindamporn A, Leelahavanichkul A. Macrophage depletion alters bacterial gut microbiota partly through fungal overgrowth in feces that worsens cecal ligation and puncture sepsis mice. Sci Rep (2022) 12:9345. doi: 10.1038/s41598-022-13098-0
21. Nikou SA, Zhou C, Griffiths JS, Kotowicz NK, Coleman BM, Green MJ, et al. The candida albicans toxin candidalysin mediates distinct epithelial inflammatory responses through p38 and EGFR-ERK pathways. Sci Signal (2022) 15:eabj6915. doi: 10.1126/scisignal.abj6915
22. Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol (2017) 17:635–46. doi: 10.1038/nri.2017.55
23. Panpetch W, Somboonna N, Bulan DE, Issara-Amphorn J, Finkelman M, Worasilchai N, et al. Oral administration of live- or heat-killed candida albicans worsened cecal ligation and puncture sepsis in a murine model possibly due to an increased serum (1→3)-β-D-glucan. PloS One (2017) 12:e0181439. doi: 10.1371/journal.pone.0181439
24. Saithong S, Saisorn W, Dang CP, Visitchanakun P, Chiewchengchol D, Leelahavanichkul A. Candida administration worsens neutrophil extracellular traps in renal ischemia reperfusion injury mice: An impact of gut fungi on acute kidney injury. J Innate Immun (2022) 2022:502–17. doi: 10.1159/000521633
25. Panpetch W, Sawaswong V, Chanchaem P, Ondee T, Dang CP, Payungporn S, et al. Candida administration worsens cecal ligation and puncture-induced sepsis in obese mice through gut dysbiosis enhanced systemic inflammation, impact of pathogen-associated molecules from gut translocation and saturated fatty acid. Front Immunol (2020) 11:561652. doi: 10.3389/fimmu.2020.561652
26. Panpetch W, Hiengrach P, Nilgate S, Tumwasorn S, Somboonna N, Wilantho A, et al. Additional candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by lacticaseibacillus rhamnosus L34. Gut Microbes (2020) 11:465–80. doi: 10.1080/19490976.2019.1662712
27. Koh AY. Murine models of candida gastrointestinal colonization and dissemination. Eukaryot Cell (2013) 12:1416–22. doi: 10.1128/EC.00196-13
28. Borges FM, de Paula TO, Sarmiento MRA, de Oliveira MG, Pereira MLM, Toledo IV, et al. Fungal diversity of human gut microbiota among eutrophic, overweight, and obese individuals based on aerobic culture-dependent approach. Curr Microbiol (2018) 75:726–35. doi: 10.1007/s00284-018-1438-8
29. Tungsanga S, Udompornpitak K, Worasilchai J, Ratana-Aneckchai T, Wannigama DL, Katavetin P, et al. Candida administration in 5/6 nephrectomized mice enhanced fibrosis in internal organs: An impact of lipopolysaccharide and (1→3)-β-D-Glucan from leaky gut. Int J Mol Sci (2022) 23:15987. doi: 10.3390/ijms232415987
30. Issara-Amphorn J, Dang CP, Saisorn W, Limbutara K, Leelahavanichkul A. Candida administration in bilateral nephrectomy mice elevates serum (1→3)-β-D-glucan that enhances systemic inflammation through energy augmentation in macrophages. Int J Mol Sci (2021) 22:5031. doi: 10.3390/ijms22095031
31. Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne) (2020) 11:25. doi: 10.3389/fendo.2020.00025
32. Parada Venegas D, de la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
33. Zheng N, Gao Y, Zhu W, Meng D, Walker WA. Short chain fatty acids produced by colonizing intestinal commensal bacterial interaction with expressed breast milk are anti-inflammatory in human immature enterocytes. PloS One (2020) 15:e0229283. doi: 10.1371/journal.pone.0229283
34. Charoensappakit A, Sae-Khow K, Leelahavanichkul A. Gut barrier damage and gut translocation of pathogen molecules in lupus, an impact of innate immunity (Macrophages and neutrophils) in autoimmune disease. Int J Mol Sci (2022) 23:8223. doi: 10.3390/ijms23158223
35. Amornphimoltham P, Yuen PST, Star RA, Leelahavanichkul A. Gut leakage of fungal-derived inflammatory mediators: Part of a gut-Liver-Kidney axis in bacterial sepsis. Dig Dis Sci (2019) 64:2416–28. doi: 10.1007/s10620-019-05581-y
36. Ondee T, Pongpirul K, Janchot K, Kanacharoen S, Lertmongkolaksorn T, Wongsaroj L, et al. Lacticaseibacillus plantarum dfa1 outperforms enterococcus faecium dfa1 on anti-obesity in high fat-induced obesity mice possibly through the differences in gut dysbiosis attenuation, despite the similar anti-inflammatory properties. Nutrients (2021) 14:80. doi: 10.3390/nu14010080
37. Leelahavanichkul A, Somparn P, Panich T, Chancharoenthana W, Wongphom J, Pisitkun T, et al. Serum miRNA-122 in acute liver injury induced by kidney injury and sepsis in CD-1 mouse models. Hepatol Res (2015) 45:1341–52. doi: 10.1111/hepr.12501
38. Leelahavanichkul A, Somparn P, Issara-Amphorn J, Eiam-ong S, Avihingsanon Y, Hirankarn N, et al. Serum neutrophil gelatinase associated lipocalin (NGAL) outperforms serum creatinine in detecting sepsis-induced acute kidney injury, experiments on bilateral nephrectomy and bilateral ureter obstruction mouse models. Shock (2016) 45:570–6. doi: 10.1097/SHK.0000000000000530
39. Visitchanakun P, Tangtanatakul P, Trithiphen O, Soonthornchai W, Wongphoom J, Tachaboon S, et al. Plasma miR-370-3P as a biomarker of sepsis-associated encephalopathy, the transcriptomic profiling analysis of microrna-arrays from mouse brains. Shock (2020) 54:347–57. doi: 10.1097/SHK.0000000000001473
40. Panpetch W, Somboonna N, Bulan DE, Issara-Amphorn J, Worasilchai N, Finkelman M, et al. Gastrointestinal colonization of candida albicans increases serum (1→3)-β-D-Glucan, without candidemia, and worsens cecal ligation and puncture sepsis in murine model. Shock (2018) 49:62–70. doi: 10.1097/SHK.0000000000000896
41. Settachaimongkon S, Nout MJ, Antunes Fernandes EC, Hettinga KA, Vervoort JM, van Hooijdonk TC, et al. Influence of different proteolytic strains of streptococcus thermophilus in co-culture with lacticaseibacillus delbrueckii subsp. bulgaricus on the metabolite profile of set-yoghurt. . Int J Food Microbiol (2014) 177:29–36. doi: 10.1016/j.ijfoodmicro.2014.02.008
42. Lin Y, Ma C, Liu C, Wang Z, Yang J, Liu X, et al. NMR-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget (2016) 7:29454–64. doi: 10.18632/oncotarget.8762
43. Kaur SU, Oyeyemi BF, Shet A, Gopalan BP, Bhavesh NS, D H, et al. Plasma metabolomic study in perinatally HIV-infected children using 1H NMR spectroscopy reveals perturbed metabolites that sustain during therapy. PloS One (2020) 15(8):e0238316. doi: 10.1371/journal.pone.0238316
44. Panpetch W, Somboonna N, Palasuk M, Hiengrach P, Finkelman M, Tumwasorn S, et al. Oral candida administration in a clostridium difficile mouse model worsens disease severity but is attenuated by bifidobacterium. PloS One (2019) 14:e0210798. doi: 10.1371/journal.pone.0210798
45. Thitiratsakul B, Anprung P. Prebiotic activity score and bioactive compounds in longan (Dimocarpus longan lour.): influence of pectinase in enzyme-assisted extraction. J Food Sci Technol (2014) 51:1947–55. doi: 10.1007/s13197-014-1263-4
46. Wang G, Yang S, Sun S, Si Q, Wang L, Zhang Q, et al. Lacticaseibacillus rhamnosus strains relieve loperamide-induced constipation via different pathways independent of short-chain fatty acids. Front Cell Infect Microbiol (2020) 10:423. doi: 10.3389/fcimb.2020.00423
47. Panpetch W, Visitchanakun P, Saisorn W, Sawatpanich A, Chatthanathon P, Somboonna N, et al. Lacticaseibacillus rhamnosus attenuates Thai chili extracts induced gut inflammation and dysbiosis despite capsaicin bactericidal effect against the probiotics, a possible toxicity of high dose capsaicin. PloS One (2021) 16:e0261189. doi: 10.1371/journal.pone.0261189
48. Ondee T, Gillen J, Visitchanakun P, Somparn P, Issara-Amphorn J, Dang Phi C, et al. Lipocalin-2 (Lcn-2) attenuates polymicrobial sepsis with LPS preconditioning (LPS tolerance) in FcGRIIb deficient lupus mice. Cells (2019) 8:1064. doi: 10.3390/cells8091064
49. Dang CP, Leelahavanichkul A. Over-expression of miR-223 induces M2 macrophage through glycolysis alteration and attenuates LPS-induced sepsis mouse model, the cell-based therapy in sepsis. PloS One (2020) 15:e0236038. doi: 10.1371/journal.pone.0236038
50. Bhunyakarnjanarat T, Udompornpitak K, Saisorn W, Chantraprapawat B, Visitchanakun P, Dang CP, et al. Prominent indomethacin-induced enteropathy in fcgriib defi-cient lupus mice: An impact of macrophage responses and immune deposition in gut. Int J Mol Sci (2021) 22:1377. doi: 10.3390/ijms22031377
51. Morimoto K, Tominaga Y, Agatsuma Y, Miyamoto M, Kashiwagura S, Takahashi A, et al. Intestinal secretion of indoxyl sulfate as a possible compensatory excretion pathway in chronic kidney disease. Biopharm Drug Dispos (2018) 39:328–34. doi: 10.1002/bdd.2149
52. Adesso S, Ruocco M, Rapa SF, Piaz FD, Raffaele Di Iorio B, Popolo A, et al. Effect of indoxyl sulfate on the repair and intactness of intestinal epithelial cells: Role of reactive oxygen species' release. Int J Mol Sci (2019) 20:2280. doi: 10.3390/ijms20092280
53. Salvi PS, Cowles RA. Butyrate and the intestinal epithelium: Modulation of proliferation and inflammation in homeostasis and disease. Cells (2021) 10:1775. doi: 10.3390/cells10071775
54. Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab (2011) 13:517–26. doi: 10.1016/j.cmet.2011.02.018
55. Rysz J, Franczyk B, Ławiński J, Olszewski R, Ciałkowska-Rysz A, Gluba-Brzózka A. The impact of CKD on uremic toxins and gut microbiota. Toxins (Basel) (2021) 13:252. doi: 10.3390/toxins13040252
56. Vaziri ND, Yuan J, Norris K. Role of urea in intestinal barrier dysfunction and disruption of epithelial tight junction in chronic kidney disease. Am J Nephrol (2013) 37:1–6. doi: 10.1159/000345969
57. Al Hinai EA, Kullamethee P, Rowland IR, Swann J, Walton GE, Commane DM. Modelling the role of microbial p-cresol in colorectal genotoxicity. Gut Microbes (2019) 10:398–411. doi: 10.1080/19490976.2018.1534514
58. Leong SC, Sirich TL. Indoxyl sulfate-review of toxicity and therapeutic strategies. Toxins (Basel) (2016) 8:358. doi: 10.3390/toxins8120358
59. Vanholder R, De Smet R, Lesaffer G. P-cresol: a toxin revealing many neglected but relevant aspects of uraemic toxicity. Nephrol Dial Transplant (1999) 14:2813–5. doi: 10.1093/ndt/14.12.2813
60. Popkov VA, Zharikova AA, Demchenko EA, Andrianova NV, Zorov DB, Plotnikov EY. Gut microbiota as a source of uremic toxins. Int J Mol Sci (2022) 23:483. doi: 10.3390/ijms23010483
61. Non- H. Pylori bacteria with urease activity identified in the human stomach. Nat Clin Pract Gastroenterol Hepatol (2006) 3:605–5. doi: 10.1038/ncpgasthep0626
62. Ramezani A, Massy ZA, Meijers B, Evenepoel P, Vanholder R, Raj DS. Role of the gut microbiome in uremia: A potential therapeutic target. Am J Kidney Dis (2016) 67:483–98. doi: 10.1053/j.ajkd.2015.09.027
63. McDonald T, Drescher KM, Weber A, Tracy S. Creatinine inhibits bacterial replication. J Antibiot (Tokyo) (2012) 65:153–6. doi: 10.1038/ja.2011.131
64. Zeng MY, Inohara N, Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol (2017) 10:18–26. doi: 10.1038/mi.2016.75
65. Baldelli V, Scaldaferri F, Putignani L, Del Chierico F. The role of enterobacteriaceae in gut microbiota dysbiosis in inflammatory bowel diseases. Microorganisms (2021) 9:697. doi: 10.3390/microorganisms9040697
66. Lokapirnasari WP, Nazar DS, Nurhajati T, Supranianondo K, Yulianto AB. Production and assay of cellulolytic enzyme activity of enterobacter cloacae WPL 214 isolated from bovine rumen fluid waste of Surabaya abbatoir, Indonesia. Vet World (2015) 8:367–71. doi: 10.14202/vetworld.2015.367-371
67. Naglik JR, Gaffen SL, Hube B. Candidalysin: discovery and function in candida albicans infections. Curr Opin Microbiol (2019) 52:100–9. doi: 10.1016/j.mib.2019.06.002
68. Yajima D, Motani H, Kamei K, Sato Y, Hayakawa M, Iwase H. Ethanol production by candida albicans in postmortem human blood samples: effects of blood glucose level and dilution. Forensic Sci Int (2006) 164:116–21. doi: 10.1016/j.forsciint.2005.12.009
69. Ma Y, Liu X, Wang J. Small molecules in the big picture of gut microbiome-host cross-talk. EBioMedicine (2022) 81:104085. doi: 10.1016/j.ebiom.2022.104085
70. Kopple JD. Abnormal amino acid and protein metabolism in uremia. Kidney Int (1978) 14:340–8. doi: 10.1038/ki.1978.134
71. Magliocca G, Mone P, Di Iorio BR, Heidland A, Marzocco S. Short-chain fatty acids in chronic kidney disease: Focus on inflammation and oxidative stress regulation. Int J Mol Sci (2022) 23:5354. doi: 10.3390/ijms23105354
72. Harms C, Schleicher A, Collins MD, Andreesen JR. Tissierella creatinophila sp. nov., a gram-positive, anaerobic, non-spore-forming, creatinine-fermenting organism. Int J Syst Bacteriol (1998) 48 Pt 3:983–93. doi: 10.1099/00207713-48-3-983
73. Fürst P. Amino acid metabolism in uremia. J Am Coll Nutr (1989) 8:310–23. doi: 10.1080/07315724.1989.10720307
74. Fadel FI, Elshamaa MF, Essam RG, Elghoroury EA, El-Saeed GS, El-Toukhy SE, et al. Some amino acids levels: glutamine,glutamate, and homocysteine, in plasma of children with chronic kidney disease. Int J BioMed Sci (2014) 10:36–42.
75. Perna AF, Castaldo P, De Santo NG, di Carlo E, Cimmino A, Galletti P, et al. Plasma proteins containing damaged l-isoaspartyl residues are increased in uremia: implications for mechanism. Kidney Int (2001) 59:2299–308. doi: 10.1046/j.1523-1755.2001.00747.x
76. Zheng H, Xu P, Jiang Q, Xu Q, Zheng Y, Yan J, et al. Depletion of acetate-producing bacteria from the gut microbiota facilitates cognitive impairment through the gut-brain neural mechanism in diabetic mice. Microbiome (2021) 9:145. doi: 10.1186/s40168-021-01088-9
77. Laverde Gomez JA, Mukhopadhya I, Duncan SH, Louis P, Shaw S, Collie-Duguid E, et al. Formate cross-feeding and cooperative metabolic interactions revealed by transcriptomics in co-cultures of acetogenic and amylolytic human colonic bacteria. Environ Microbiol (2019) 21:259–71. doi: 10.1111/1462-2920.14454
78. Wilmanski T, Rappaport N, Earls JC, Magis AT, Manor O, Lovejoy J, et al. Blood metabolome predicts gut microbiome α-diversity in humans. Nat Biotechnol (2019) 37:1217–28. doi: 10.1038/s41587-019-0233-9
79. Chriett S, Dąbek A, Wojtala M, Vidal H, Balcerczyk A, Pirola L. Prominent action of butyrate over β-hydroxybutyrate as histone deacetylase inhibitor, transcriptional modulator and anti-inflammatory molecule. Sci Rep (2019) 9:742. doi: 10.1038/s41598-018-36941-9
80. Tholozan JL, Samain E, Grivet JP, Moletta R, Dubourguier HC, Albagnac G. Reductive carboxylation of propionate to butyrate in methanogenic ecosystems. Appl Environ Microbiol (1988) 54:441–5. doi: 10.1128/aem.54.2.441-445.1988
81. Ondee T, Pongpirul K, Visitchanakun P, Saisorn W, Kanacharoen S, Wongsaroj L, et al. Lactobacillus acidophilus LA5 improves saturated fat-induced obesity mouse model through the enhanced intestinal akkermansia muciniphila. Sci Rep (2021) 11:6367. doi: 10.1038/s41598-021-85449-2
82. Panpetch W, Chancharoenthana W, Bootdee K, Nilgate S, Finkelman M, Tumwasorn S, et al. Lactobacillus rhamnosus L34 attenuates gut translocation-induced bacterial sepsis in murine models of leaky gut. Infect Immun (2017) 86:e00700–17. doi: 10.1128/IAI.00700-17
83. LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact (2017) 16:79. doi: 10.1186/s12934-017-0691-z
84. Byrne CS, Chambers ES, Morrison DJ, Frost G. The role of short chain fatty acids in appetite regulation and energy homeostasis. Int J Obes (Lond) (2015) 39:1331–8. doi: 10.1038/ijo.2015.84
85. Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients (2011) 3:858–76. doi: 10.3390/nu3100858
86. Markowiak-Kopeć P. Śliżewska k. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients (2020) 12:1107. doi: 10.3390/nu12041107
87. van Zanten GC, Knudsen A, Röytiö H, Forssten S, Lawther M, Blennow A, et al. The effect of selected synbiotics on microbial composition and short-chain fatty acid production in a model system of the human colon. PloS One (2012) 7:e47212. doi: 10.1371/journal.pone.0047212
88. Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol (2016) 7:185. doi: 10.3389/fmicb.2016.00185
89. Mishra AK, Kumar SS, Ghosh AR. Probiotic enterococcus faecalis AG5 effectively assimilates cholesterol and produces fatty acids including propionate. FEMS Microbiol Lett (2019) 366:fnz039. doi: 10.1093/femsle/fnz039
90. Gao B, Jose A, Alonzo-Palma N, Malik T, Shankaranarayanan D, Regunathan-Shenk R, et al. Butyrate producing microbiota are reduced in chronic kidney diseases. Sci Rep (2021) 11:23530. doi: 10.1038/s41598-021-02865-0
91. Meijers B, Farré R, Dejongh S, Vicario M, Evenepoel P. Intestinal barrier function in chronic kidney disease. Toxins (Basel) (2018) 10:298. doi: 10.3390/toxins10070298
92. Huang Y, Zhou J, Wang S, Xiong J, Chen Y, Liu Y, et al. Indoxyl sulfate induces intestinal barrier injury through IRF1-DRP1 axis-mediated mitophagy impairment. Theranostics (2020) 10:7384–400. doi: 10.7150/thno.45455
93. Bekebrede AF, Deuren TV, Gerrits WJJ, Keijer J, Boer VCJ. Butyrate alters pyruvate flux and induces lipid accumulation in cultured colonocytes. Int J Mol Sci (2021) 22:10937. doi: 10.3390/ijms222010937
94. Peng L, He Z, Chen W, Holzman IR, Lin J. Effects of butyrate on intestinal barrier function in a caco-2 cell monolayer model of intestinal barrier. Pediatr Res (2007) 61:37–41. doi: 10.1203/01.pdr.0000250014.92242.f3
95. Kespohl M, Vachharajani N, Luu M, Harb H, Pautz S, Wolff S, et al. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4(+) T cells. Front Immunol (2017) 8:1036. doi: 10.3389/fimmu.2017.01036
96. Schönfeld P, Wojtczak L. Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J Lipid Res (2016) 57:943–54. doi: 10.1194/jlr.R067629
97. Geng HW, Yin FY, Zhang ZF, Gong X, Yang Y. Butyrate suppresses glucose metabolism of colorectal cancer cells via GPR109a-AKT signaling pathway and enhances chemotherapy. Front Mol Biosci (2021) 8:634874. doi: 10.3389/fmolb.2021.634874
98. Pieniazek A, Kopera M, Gwozdzinski L, Gwozdzinski K. Indoxyl sulfate induces oxidative changes in plasma and hemolysate. Molecules (2022) 27:3848. doi: 10.3390/molecules27123848
99. Dou L, Jourde-Chiche N, Faure V, Cerini C, Berland Y, Dignat-George F, et al. The uremic solute indoxyl sulfate induces oxidative stress in endothelial cells. J Thromb Haemost (2007) 5:1302–8. doi: 10.1111/j.1538-7836.2007.02540.x
100. Lu CL, Zheng CM, Lu KC, Liao MT, Wu KL, Ma MC. Indoxyl-Sulfate-Induced redox imbalance in chronic kidney disease. Antioxidants (Basel) (2021) 10:936. doi: 10.3390/antiox10060936
101. Quijano C, Trujillo M, Castro L, Trostchansky A. Interplay between oxidant species and energy metabolism. Redox Biol (2016) 8:28–42. doi: 10.1016/j.redox.2015.11.010
102. Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol (2015) 33:125–31. doi: 10.1016/j.ceb.2015.02.003
Keywords: uremia, microbiome, short-chain fatty acid, fungi, kidney
Citation: Chancharoenthana W, Kamolratanakul S, Visitchanakun P, Sontidejkul S, Cheibchalard T, Somboonna N, Settachaimongkon S and Leelahavanichkul A (2023) Lacticaseibacilli attenuated fecal dysbiosis and metabolome changes in Candida-administered bilateral nephrectomy mice. Front. Immunol. 14:1131447. doi: 10.3389/fimmu.2023.1131447
Received: 03 January 2023; Accepted: 27 February 2023;
Published: 09 March 2023.
Edited by:
Ahmed Sherif Attia, Cairo University, EgyptReviewed by:
Ali Mujtaba Shah, Shaheed Benazir Bhutto University of Veterinary & Animal Sciences, PakistanCopyright © 2023 Chancharoenthana, Kamolratanakul, Visitchanakun, Sontidejkul, Cheibchalard, Somboonna, Settachaimongkon and Leelahavanichkul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wiwat Chancharoenthana, d2l3YXQuY2hhQG1haGlkb2wuYWMudGg=; Asada Leelahavanichkul, YWxlZWxhaGF2YW5pdEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.