- 1Department of Clinical Laboratory, Sun Yat-sen Memorial Hospital of Sun Yat-sen University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-sen Memorial Hospital of Sun Yat-sen University, Guangzhou, China
Background: Inflammation serves as an essential driver of liver cirrhosis (LC) incidence. Accordingly, a meta-analysis was carried out to explore the association between specific polymorphisms in the interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α) genes and the incidence of LC based on comparisons of genotype and allele frequencies.
Objectives: To study the relationship between TNF-α rs361525 and IFN-γ rs2430561 polymorphisms and the risk of LC.
Methods: A database search was performed for all studies published as of September 10, 2022. The strength of risk relationships was assessed based on odds ratios (ORs) with 95% confidence intervals (CIs).
Results: Pooled analyses were conducted for one common TNF-α polymorphism (rs361525) as well as one common IFN-γ polymorphism (rs2430561). Both of these SNPs were identified as LC-related risk factors. Specifically, rs361525 was related to LC incidence in both alcoholic liver cirrhosis (OR: 1.86, 95%CI: 1.03-3.34) and hepatitis B virus (HBV)-related cirrhosis cases (OR: 1.44, 95%CI: 1.00-2.06) when using an allelic contrast model. Moreover, rs2430561 was significantly related to LC in an Asian population (OR: 1.45, 95%CI: 1.13-1.86) and in the context of HBV-related cirrhosis (OR: 1.48, 95%CI: 1.13-1.93) when using an allelic contrast model.
Conclusion: These findings indicate that rs361525 and rs2430561 represent LC-related risk factors, although additional large-scale clinical and case-control studies will be vital to confirm these results.
Introduction
Liver cirrhosis (LC) was responsible for an estimated 2.4% of global mortality in 2017 (1), more than 45% of all deaths in Western nations and over 1.32 million deaths throughout the globe (2).
Many different clinical trials have been developed and implemented in recent years with the goal of defining more effective anti-fibrotic therapies, with phase II and III trials having been performed for a range of agents including obethicolic acid, selonsertib, and elafibranor. While some of these studies have yielded promising findings, in general the benefits have been modest and the outlook for anti-fibrotic treatment remains poor. Accordingly, the most effective strategies currently available to protect against hepatic fibrosis center on its prevention and on alleviating known risk factors associated with this condition (3, 4).
The risk factors known to be most closely related to LC incidence include chronic infections with hepatitis B or C virus (HBV and HCV), which are respectively associated with 39.64 million and 30.36 million cases, as well as nonalcoholic fatty liver disease (NAFLD), which is linked to 10.26 million cases, and alcoholic liver disease (ALD), which is associated with 26.04 million cases. In addition, roughly 16.62 million cases are linked with other underlying causes (5, 6).
Both host genetic factors and immune activity can shape the progression of chronic viral hepatitis and associated infections, making them important determinants of LC risk. Single nucleotide polymorphisms (SNPs) are the most prevalent and extensively explored form of genetic variation (7, 8), and SNPs in genes such as PON1(9), GSTM1(10), and CTLA4(11) have been linked with LC.
Growth factors and cytokines such as members of the interferon (IFN) and tumor necrosis factor (TNF) family are vital components of host immune activity that can shape viral infection-related processes and the subsequent development of fibrosis (12, 13). In a prior study, our group conducted a meta-analysis examining the relationship between mutations in the IL-6 and IL-10 genes (rs1800871, rs1800872, rs1800795, rs1800796, rs1800797, rs1800896) and LC risk, ultimately identifying the IL-10 -592 and -1082 polymorphisms as being closely linked to LC susceptibility(14). Other reports have specifically focused on links between IFN-γ and TNF-α genes polymorphisms and cirrhotic disease (15).
TNF-α is a potent inflammatory and immunomodulatory cytokine, and the gene that encodes it is associated with the class III MHC region (16). Signaling through the TNF-α/NF-kB axis has been linked to damage, fibrotic activity, and cirrhosis in the liver (17, 18). IFN-γ is another pro-inflammatory cytokine that can influence hematopoietic stem cell development at baseline and serves as a major pathogenic factor associated with various forms of hematopoiesis-related disease stats such as hemophagocytic lymphohistiocytosis, aplastic anemia, and LC (19).
In one recent meta-analysis of 22 studies incorporating 2,638 LC patients and 2,905 control individuals, the TNF-α -308A/G polymorphism was found to be unrelated to the risk of developing LC (20). No similar analyses, however, have been performed for the common rs361525 (-238 A>G) polymorphism in this gene or for the common IFN-γ rs2430561 polymorphism (+874T/A).
To date, no studies have performed any comprehensive assessments of the TNF-α rs361525(21–33) and IFN-γ rs2430561(8, 29, 34–40) polymorphisms. Given the above findings and in an effort to overcome limitations associated with small study sample sizes or ethnic/regional variation among study cohorts, the present meta-analysis was thus performed to attempt to more fully clarify the link between these two SNPs in inflammatory cytokine genes and LC risk in an effort to provide a foundation for future clinical reference.
Materials and methods
Study selection
In the current study, relevant papers published as of September 10, 2022, were retrieved from PubMed and other databases using the following search terms: “tumor necrosis factor-α or TNF-α”, “interferon-γ or IFN-γ”, “polymorphism or variation or mutation”, “rs361525”, “rs2430561” and “liver cirrhosis or nonalcoholic fatty liver disease or primary biliary cirrhosis”. This analysis included the research with the largest sample size when several studies used the same set of clinical data.
Inclusion criteria
Research studies were only considered if they fulfilled the following requirements: (a) studies in which the criteria used to diagnose LC were clearly defined, including liver biopsy, CT, MRI, B-ultrasound, or endoscopic retrograde cholangiopancreatography, (b) studies of correlations between the designated TNF-α and IFN-γ gene polymorphisms (rs361525 or rs2430561) and LC risk, (c) studies using cohort or case-control designs, (d) studies with enough data to allow for the calculation of 95% confidence interval (95%CI) values, odds ratio (OR), and (e) studies were not duplicated.
Data extraction
Data extracted from individual studies included first author, country, subject ethnicity, number of cases, year of publication, number of controls, source of control subjects, HWE in the control group, genotyping approach, and LC type. Two investigators (Minghui Zheng, Hong Luo) independently extracted data, with any discrepancies being resolved through discussion and consensus.
Statistical analysis
Relationships between these IFN-γ and TNF-α polymorphisms of interest and LC risk were analyzed based on ORs and 95% CIs. Analyzing the significance of ORs was performed using the Z-test (41), while heterogeneity was assessed with the Q-test and I2 statistic (42, 43). I2 values < 25%, ~50%, and >75% were respectively associated with low, moderate, and high degrees of heterogeneity (44). When heterogeneity was absent, a fixed-effect Mantel-Haenszel model was used (Pheterogeneity > 0.1), whereas a random-effects DerSimonian-Laird model was instead employed in the presence of heterogeneity (45, 46). Sensitivity analyses were used to assess result stability through a leave-one-out approach (47). Pearson’s chi-square test was used to assess whether polymorphism frequencies diverged from the expected HWE (48). The potential for publication bias was examined through a combination of Begg’s funnel plots and Egger’s regression test (49). Stata 11.0 (StataCorp LP, TX, USA) was used for all statistical analyses.
Protein-protein interaction network analysis
To better explore links between TNF-α and IFN-γ and LC pathogenesis, Protein-protein interaction networks involving these two genes were generated using the STRING database (http://string-db.org/) (50).
Results
Study selection
Initial literature searches of PubMed and other databases led to the identification of 659 studies (Figure 1), including 445 pertaining to the TNF-α rs361525 polymorphism and 214 pertaining to the IFN-γ rs2430561 polymorphism. Of these studies, 503 were excluded following title and abstract review (no relationship between above two polymorphisms and liver disease), 156 articles were left after reviewing full-text, additional 135 articles were excluded due to following reasons: duplicates (including 36 articles), meta-analyses or systematic reviews (including 11 articles), articles focused on other polymorphisms (including 38 articles), review (including 44 articles), or clinical trials (including 6 articles). The remaining 21 studies (8, 21–40) met with inclusion criteria for this analysis and were thus incorporated into this study, including 13 pertaining to the TNF-α rs361525 polymorphism (5, 5, and 2 case-control studies related to PBC, HBV, and ALC, respectively) and 9 pertaining to the IFN-γ rs2430561 polymorphism. In all studies, DNA was isolated from samples of patient blood (Table 1). Minor allele frequency (MAF) reports for these two SNPs were assessed in 6 different global populations with the 1000 Genomes Browser (https://www.ncbi.nlm.nih.gov/snp/) (Figure 2). The data of Figure 2 is from the online website (https://www.ncbi.nlm.nih.gov/snp/), which is objective and based on statistic data from several genome-wide association studies and published articles. In addition, the difference among different ethnicity may be from ethnicity themselves or other polymorphisms, including other IFN‐γ and TNF‐α polymorphisms.

Figure 1 A flowchart outlining the search strategy utilized for the identification of association studies related to the relationship between the TNF-α rs361525/IFN-γ rs2430561 polymorphisms and the risk of LC.
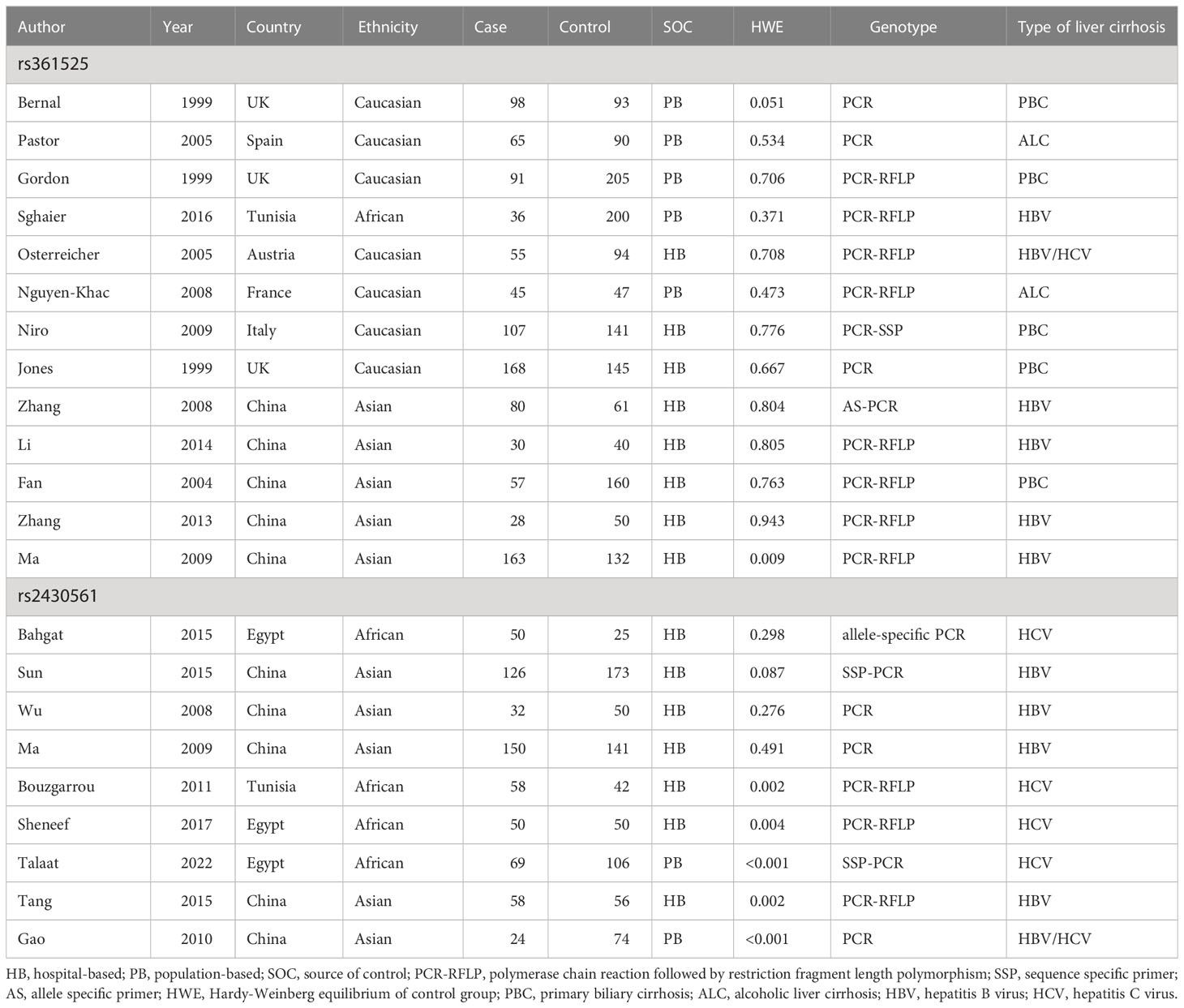
Table 1 Characteristics of included studies about polymorphisms in TNF-α and IFN-γ genes and cirrhosis of liver risk.
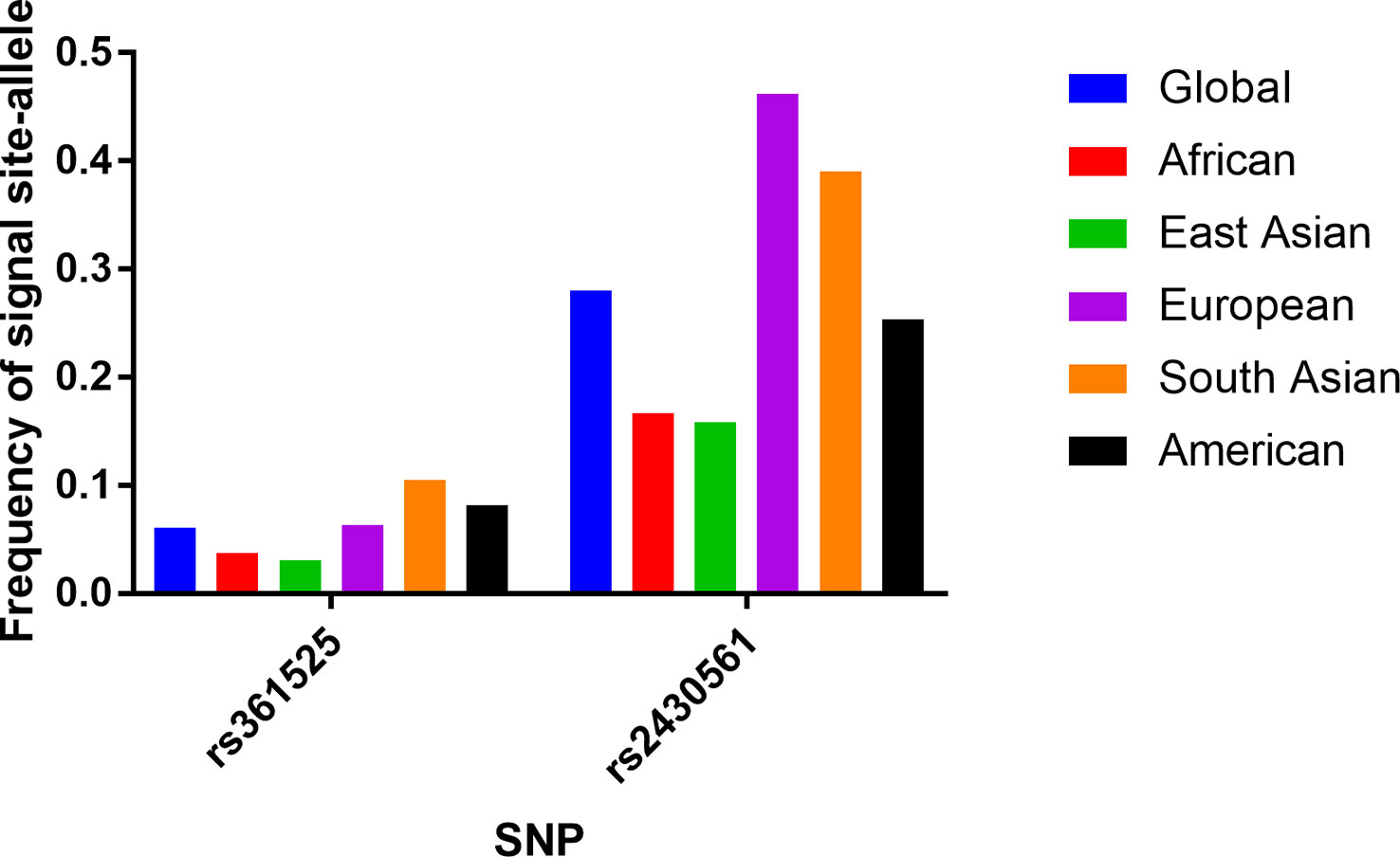
Figure 2 Minor (mutant) allele frequencies for rs361525 and rs2430561 polymorphisms based on data from the online 1000 Genomes database.
Quantitative result synthesis
TNF-α rs361525 polymorphism
In an overall analysis, no significant association with LC risk was detected when using an allelic contrast model (OR: 1.22, 95%CI:0.98-1.52, P = 0.233 for heterogeneity, P = 0.081, I2 = 20.9%). Subgroup analyses also failed to detect any significant association between this polymorphism and LC based on ethnicity or source of control subjects. However, positive relationships between rs361525 and LC risk were detected in the ALC (OR: 1.86, 95%CI:1.03-3.34, P = 0.420 for heterogeneity, P = 0.040, I2 = 7.1%) and HBV (OR: 1.44, 95%CI:1.00-2.06, P = 0.404 for heterogeneity, P = 0.047, I2 = 0.3%) subgroups using allelic contrast models (Figure 3; Table 2).
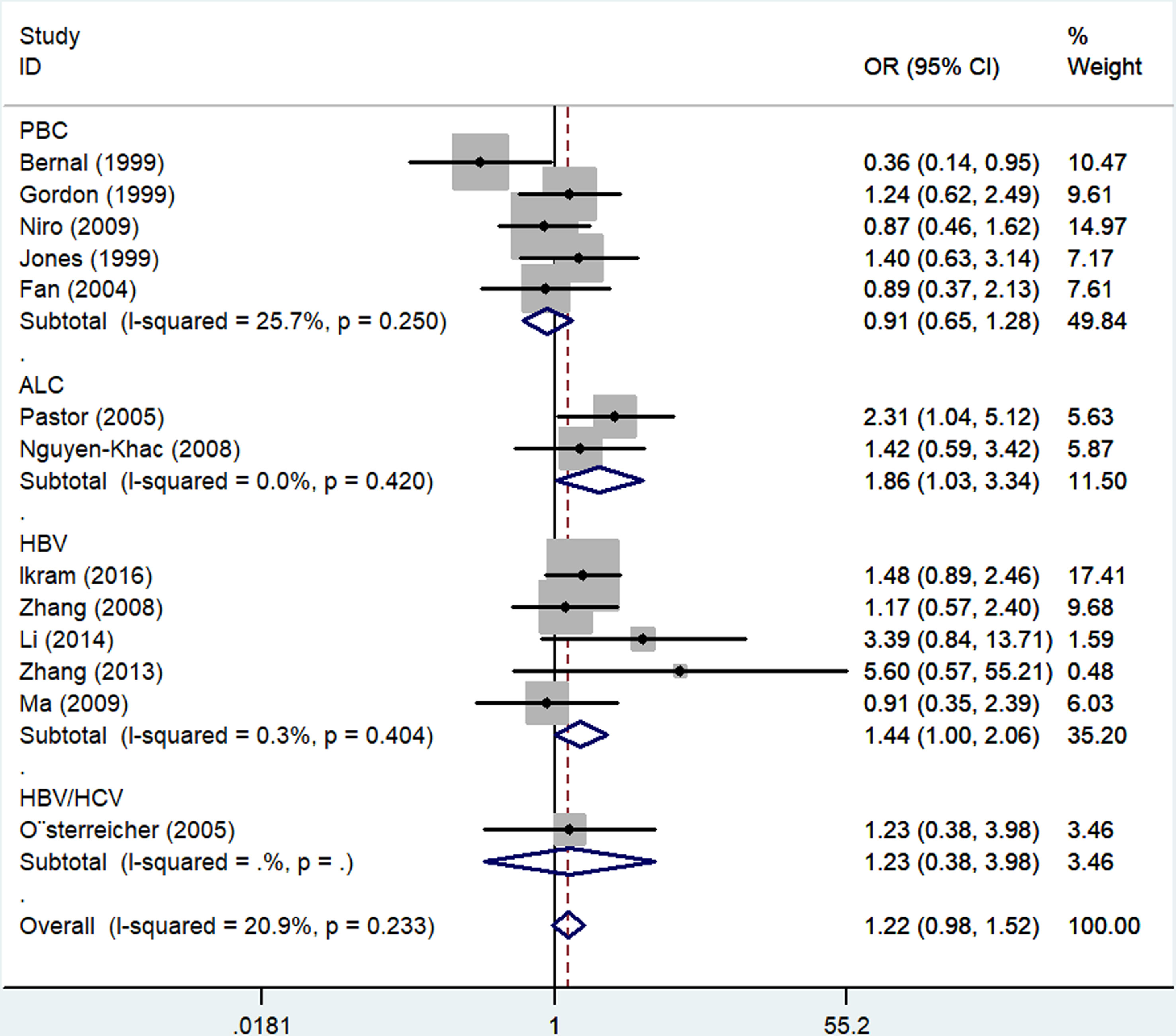
Figure 3 Forest plots depicting the association between rs361525 polymorphisms and LC risk in both the ALC and HBV LC subgroups generated using an allelic contrast model.
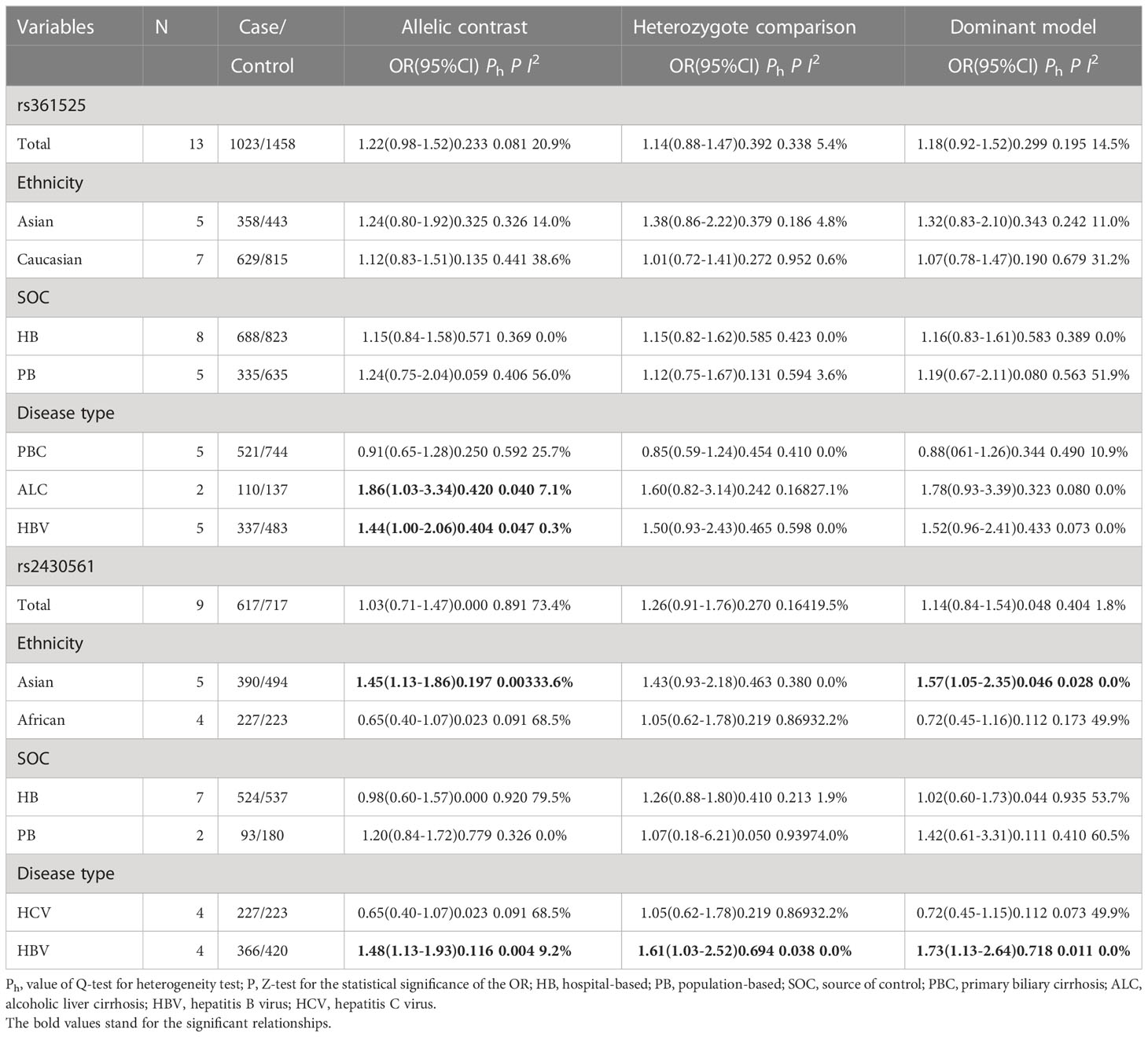
Table 2 Stratified analyses of TNF-α and IFN-γ genes’ common polymorphisms on cirrhosis of liver risk.
IFN-γ rs2430561 polymorphism
No overall association was detected between rs2430561 and risk of LC, and the same was true when assessing patients based on the source of control subjects. However, this polymorphism was linked to elevated LC risk in Asian individuals (allelic contrast: OR: 1.45, 95%CI:1.13-1.86, P = 0.197 for heterogeneity, P = 0.003, I2 = 33.6%, Figure 4; dominant model: OR: 1.57, 95%CI:1.05-2.35, P = 0.046 for heterogeneity, P = 0.028, I2 = 0.0%) and in patients with HBV (allelic contrast: OR: 1.48, 95%CI:1.13-1.93, P = 0.116 for heterogeneity, P = 0.004, I2 = 9.2%, Figure 5; heterozygote comparison: OR: 1.61, 95%CI:1.03-2.52, P = 0.694 for heterogeneity, P = 0.038, I2 = 0.0%; dominant model: OR: 1.73, 95%CI:1.13-2.64, P = 0.718 for heterogeneity, P = 0.011, I2 = 0.0%) (Table 2).
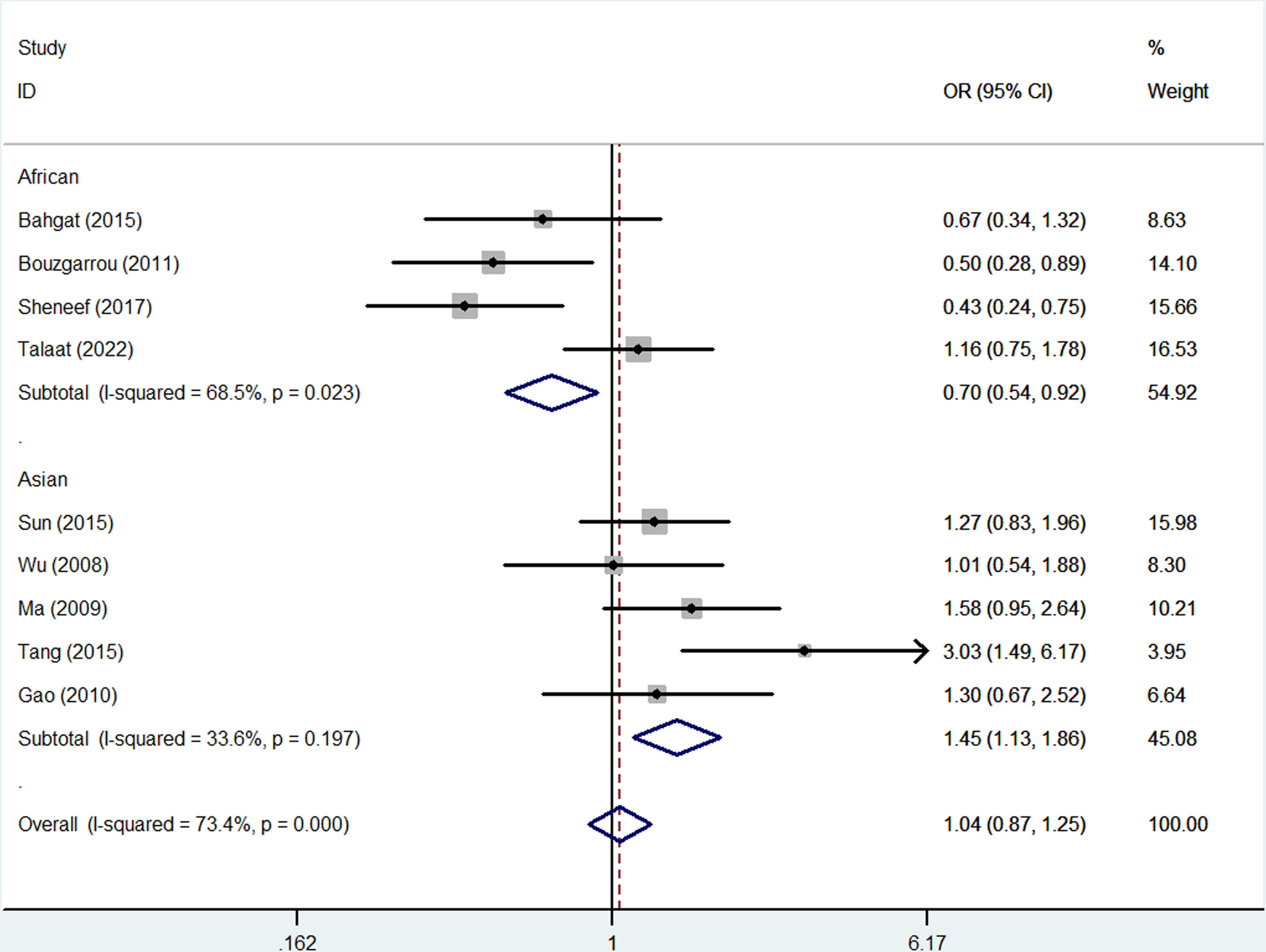
Figure 4 Forest plots corresponding to the association between LC risk and rs2430561 polymorphisms using an allelic contrast model in different ethnic subgroups.
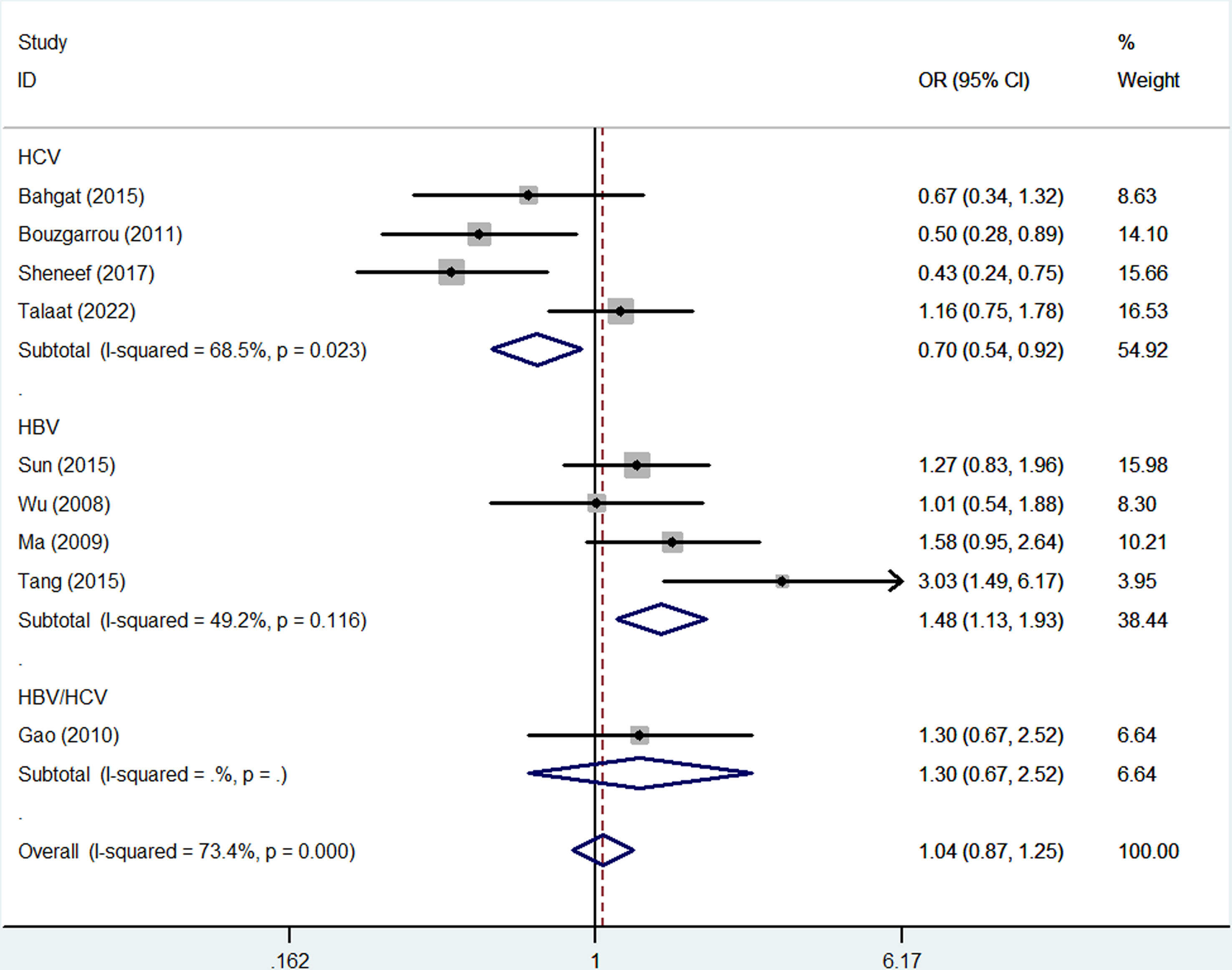
Figure 5 Forest plots corresponding to the association between LC risk and rs2430561 polymorphisms in different LC subgroups generated using an allelic contrast model.
Risk of bias and sensitivity analyses
Publication bias was evaluated using Begg’s funnel plots and Egger’s test. Allele-by-allelic comparisons in funnel plots showed asymmetry for these two polymorphisms, which is not attributable to publication bias. When using an allelic contrast model, Egger’s test also showed that there was no publication bias (rs361525: t = 0.6, P = 0.564 for Egger’s test; z = 0.18, P = 0.855 for Begg’s test; Figures 6A, B; rs2430561: t = -0.18, P = 0.861 for Egger’s test; z = 0.31, P = 0.754 for Begg’s test; Figures 7A, B) (Table 3).
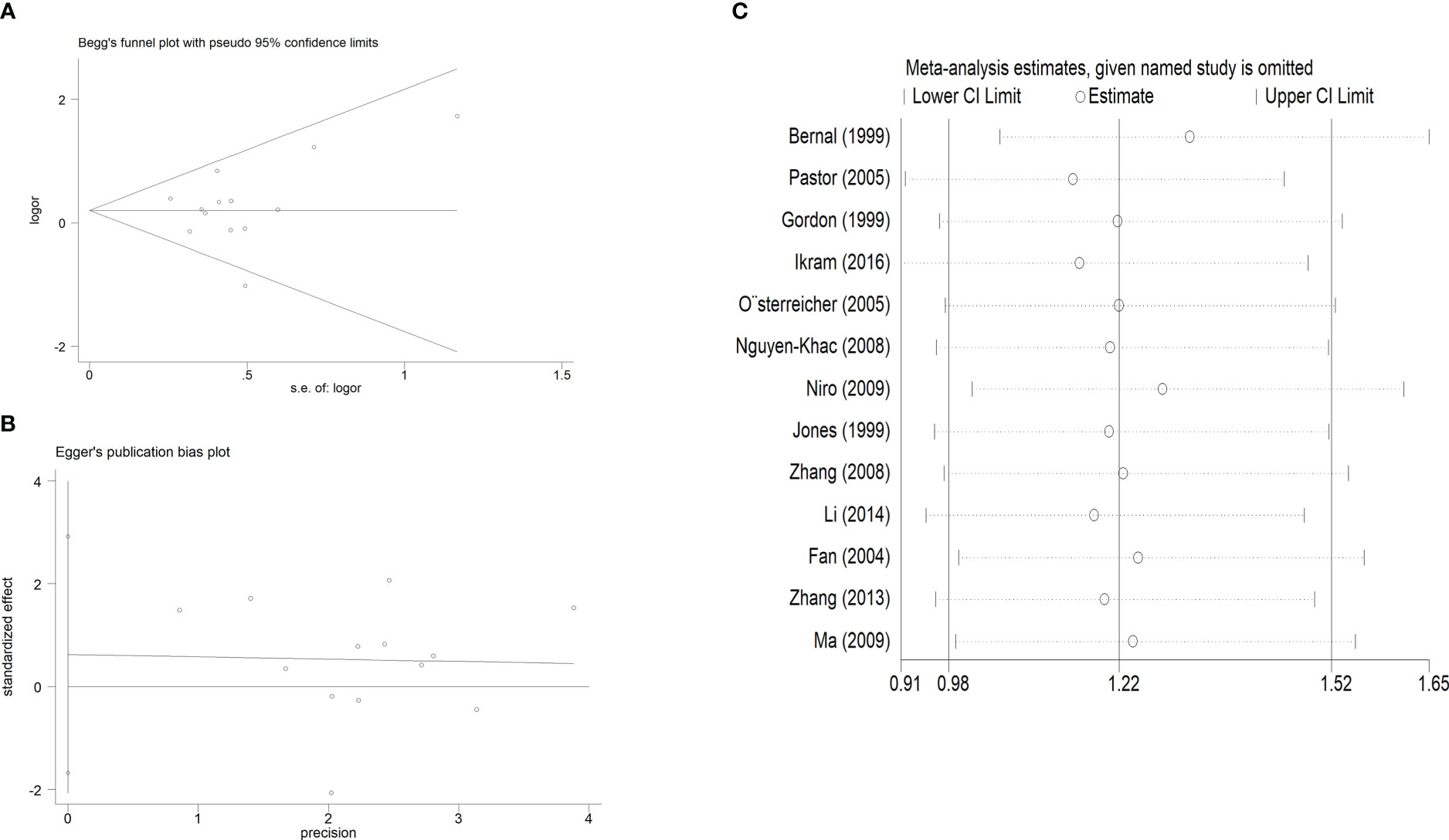
Figure 6 (A). Begg’s funnel plot test for publication bias, with individual points corresponding to specific studies, and the mean effect size being represented by the horizontal axis. Log [OR], natural logaritm of OR. (B). Egger’s publication bias plot. (C): Sensitivity analysis for the association between TNF-α rs361525 polymorphisms and LC risk.
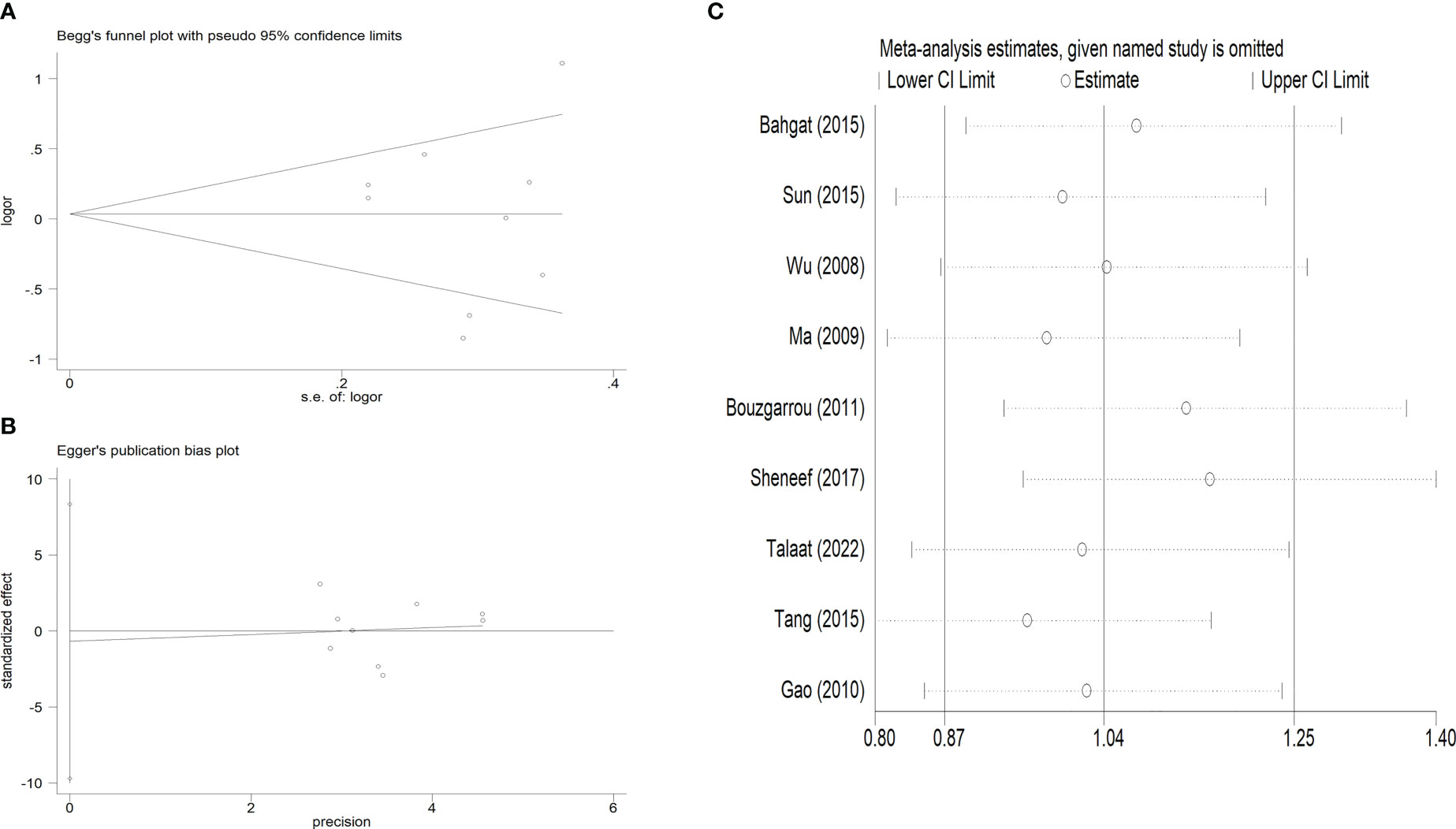
Figure 7 (A). Begg’s funnel plot test for publication bias. (B). Egger’s publication bias plot. (C): Sensitivity analysis for the association between IFN-γ rs2430561 polymorphisms and LC risk.
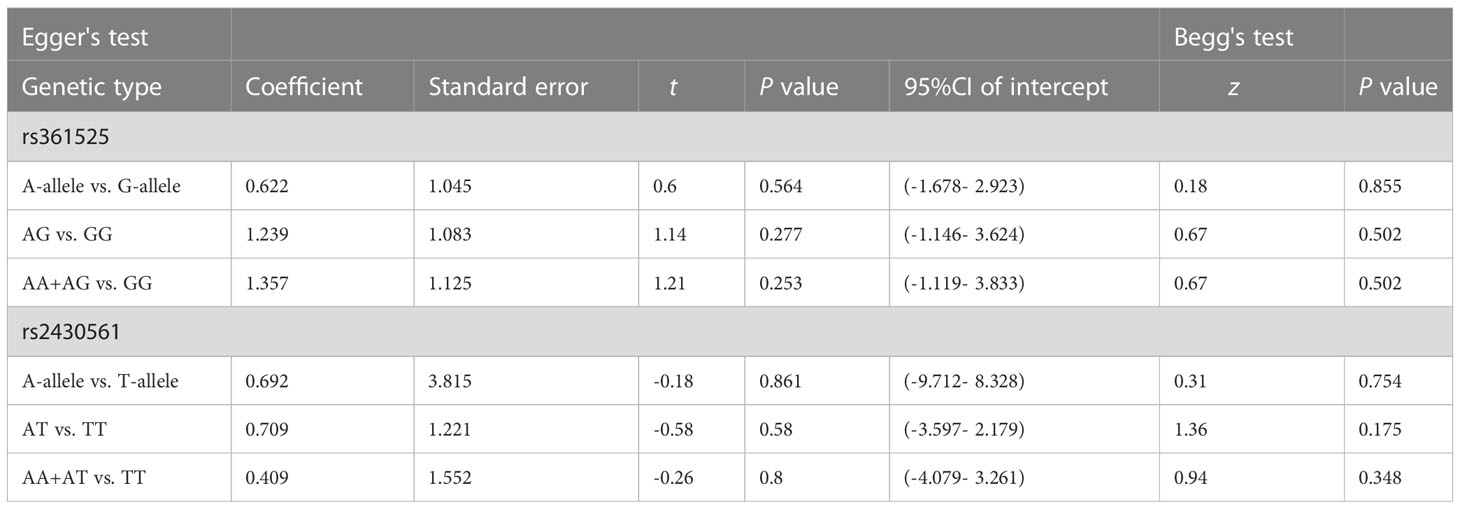
Table 3 Publication bias tests (Begg’s funnel plot and Egger’s test for publication bias test) for TNF-α and IFN-γ genes polymorphisms.
Additionally, sensitivity analysis was performed to explore factors with the potential to impact the stability of these overall analyses, and none of the included case-control studies were found to alter overall study conclusions under the three tested models for either of the analyzed polymorphisms (Figures 6C, 7C).
Protein-protein interaction network
Accordingly, the STRING database was used to identify other genes potentially associated with TNF-α or IFN-γ, with these genes then being used to establish putative protein-protein interaction networks (Figure 8). The majority of genes identified through this approach were associated with cytokines (TNFRSF1A, TNFRSF1B, TRADD, TRAF2, IL-10, TNFAIP3, IFNGR1, IFNGR2, IRF1, SOCS1), exhibiting interaction scores> 0.9 consistent with very close relationships.
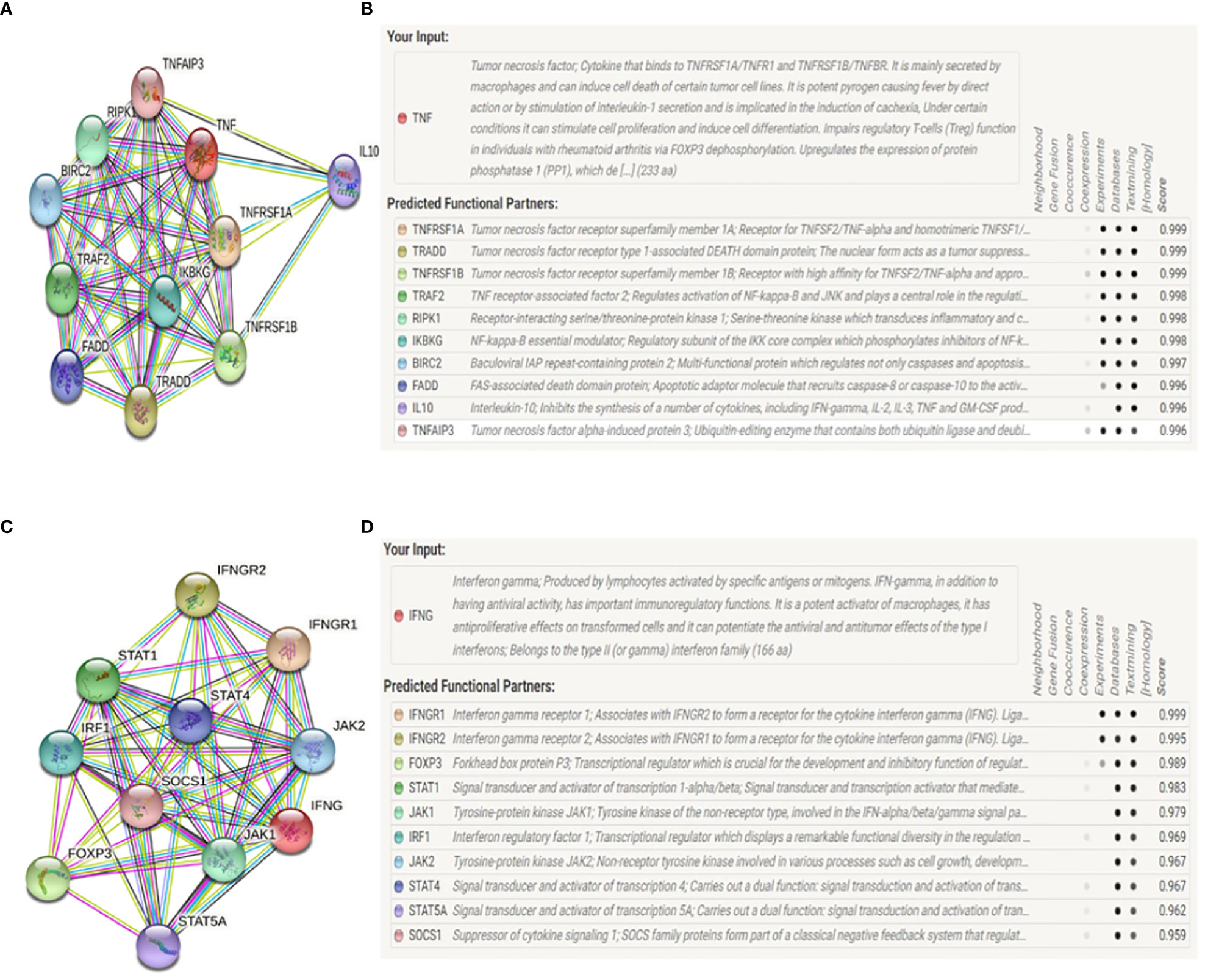
Figure 8 STRING network diagrams depicting interactions between human TNF-α or IFN-γ and other genes. (A, B). At least 10 genes were correlated with: TNFRSF1A: tumor necrosis factor receptor superfamily member 1A; TNFRSF1B: tumor necrosis factor receptor superfamily member 1B; TRADD: tumor necrosis factor receptor type 1-associated DEATH domain protein; TRAF2: TNF receptor-associated factor 2; RIPK1: receptor-interacting serine/threonine-protein kinase 1; FADD: FAS-associated death domine protein; IKBKG: NF-kappa-B essential modulate; BIRC2: baculoviral IAP repeat-containing protein; IL-10: interleukin-10; TNFAIP3: tumor necrosis factor alpha-induced protein 3. (C, D). At least 10 genes were correlated with: IFNGR1: IFN-γ receptor 1; IFNGR2: IFN-γ receptor 2; FOXP3: forkhead box protein P3; JAK1: tyrosine-protein kinase JAK1; JAK2: tyrosine-protein kinase JAK2; IRF1: interferon regulatory factor 1; STAT1: signal transducer and activator of transcription 1-alpha/beta; STAT4: signal transducer and activator of transcription 4; STAT5A: signal transducer and activator of transcription 5A; SOCS1: suppressor of cytokine signaling 1.
Discussion
Cirrhosis is the final progressive stage of hepatic fibrosis, and most cases of LC develop in patients with major risk factors such as chronic alcohol abuse or hepatitis caused by viral infections. Histologically, LC is characterized by extensive extracellular matrix material accumulating within the liver parenchyma including very high levels of fibrillar collagen, ultimately compromising the structural integrity and functional utility of this tissue (51).
LC is also closely associated with an elevated risk of hepatocellular carcinoma (HCC), with is the most prominent form of liver cancer in humans, resulting in high rates of morbidity and mortality owing to a lack of effective therapeutic interventions (52). The present study was developed to explore efforts to more effectively diagnose LC and reduce the medical burden associated with this disease by lowering patient mortality rates.
Short−chain fatty acids can influence the production of TNF-α and diverse other inflammatory mediators in response to signaling through the TLR4 pathway, playing important roles in inflammatory and fibrotic activity in the liver through the upregulation of TIMP-1 in activated hepatic stellate cells (HSCs) and through the inhibition of HSC apoptosis (53).
NK-derived IFN-γ can suppress the fibrotic differentiation of HSCs (54), in addition to suppressing proliferative activity, as well as the expression of α-SMA TGF-β (55). In certain contexts, IFN-γ can also promote activated HSC death through mechanisms mediated by the FasL-associated and TRIAL death domains (55). Prolonged HSC activation can contribute to LC incidence.
To date, a range of genes have been linked to the incidence of liver disease including CTLA-4, IL-18, TM6SF2, and GSTM1(10, 56, 57). Inflammation and fibrotic activity are particularly closely associated with LC risk, and various studies have accordingly highlighted possible links between TNF-α and IFN-γ SNPs and LC incidence. However, many of these studies were based on relatively small sample sizes such that their conclusions were not necessarily robust.
Pastor et al. (26) previously observed a link between the TNF-α rs361525 (-238) polymorphism and elevated alcoholic LC risk, while Bouzgarrou et al. (8) similarly observed a higher risk of LC incidence associated with the IFN-γ rs2430561 (+874) polymorphism. However, it is critical that these prior studies be combined and that analyses with larger sample sizes be performed to comprehensively examine the link between TNF-α and IFN-γ SNPs and LC risk.
In this study, in an effort to more fully solidify these relationships between TNF-α and IFN-γ SNPs and the odds of developing LC, a database search was conducted that ultimately identified 13 and 9 case-control studies respectively associated with the TNF-α rs361525 and IFN-γ rs2430561 polymorphisms. The pooled analysis of these results ultimately revealed that TNF-α rs361525 was linked to LC risk in the ALC and HBV subgroups, while IFN-γ rs2430561 was more strongly related to the risk of LC in HBV patients and Asian populations, potentially providing a valuable foundation for future efforts to prevent, diagnose, and treat LC in its earlier stages. This is a study that we need to pay attention to in the future, which is only aimed at determining the polymorphisms (rs361525, rs2430561) of two susceptible genes (TNF-α and IFN-γ) for LC. Based on this conclusion, physical examination of healthy people can be conducted to identify the population with gene mutations, and early intervention, observation, and follow-up can be conducted in such aspects as diet, behavioral habits, and emotional regulation to observe the risk of LC in such people, and further to verify the conclusion in this study, We wish to reduce the incidence rate and the mortality due to disease of LC through this exploration.
LC develops and progresses through a complex multi-factorial process, and the impact of any individual gene on the overall disease process may thus be limited. The STRING online database helps us to find potential interactive genes. The hub gene TNFRSF1A may offer value as a marker of PBC (58), and one GWAS study of 1,840 cases and 5,163 controls identified the TNFRSF1A rs1800693 SNP as a PBC-related risk factor (OR=1.23, 95%CI=1.14-1.33) (59). Peng et al. found TRADD expression to be associated with HBV-related LC and HCC incidence (60), while Lin et al. determined that LC model rats exhibited increased TRAF2 protein content in skeletal muscle samples (61). Moreover, Tan et al. found that RIPK1 acts as a promoter of inflammatory activity in the setting of HCC onset and liver fibrosis (62), and Higuchi et al. reported a link between deleterious TNFAIP3 alleles and autoimmune hepatitis with cirrhosis (63). While Falleti et al. identified two IFNGR1 gene polymorphisms, they failed to find them to be significantly associated with disease (64). In contrast, two IFNGR2 polymorphisms identified by Nalpas et al. that were in strong linkage disequilibrium were found to be closely related to the incidence of liver fibrosis (65). Ramadan et al. determined that reductions in CD4/FoxP3/CD25 levels were evident in patients treated with direct-acting antivirals, contributing to better immunological outcomes in the context of LC (66). There is a strong body of evidence in support of the role of cytokine-induced JAK/STAT signaling serving as a regulator of hepatic fibrosis and regenerative activity (67). Zhu et al. found sennoside A to be capable of inhibiting SOCS1 and decreasing HSC proliferation in liver fibrosis through the inhibition of inflammation (68). Accordingly, additional work should focus on the relationships between these TNF-α- and IFN-γ-associated genes, gene-gene interactions, and the pathogenesis of LC.
This study is subject to some limitations. For one, additional work focused on African and mixed populations is needed as these groups were underrepresented in studies published to date. In addition, LC is a complex multifactorial disease, and it is thus important that both gene-gene and gene-environment interactions be taken into consideration when examining the etiology of this disease. Particular lifestyle and environmental factors including sex, age, diet, smoking status, family history, history of parasite exposure, immunological factors, and viral infections may all shape the relationship between TNF-α or IFN-γ polymorphisms and LC. Further work is also necessary to assess whether LC patients face other complications including autoimmunity, hepatitis, abnormal liver function, and HCC incidence. These analyses also did not classify LC stage or pathogenic origin, highlighting important directions for additional study to more precisely guide the prevention and treatment of this disease. Furthermore, we only explored possible interacting genes through online databases, but through literature searches, we have not found that the interactions among 20 genes and the TNF-α or IFN-γ, which is also a topic for future research. Overall, this meta-analysis found some evidence for an association between TNF-α rs361525 and IFN-γ rs2430561 polymorphisms and the risk of LC suggesting that they may represent viable targets for clinical research aimed at predicting the future risk of severe liver disease. In the future research, we will also try to study a case-control study in our own cohort to confirm current conclusions.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
MZ, JL and WF: design research program, collect data, analyze statistics and write articles. LL, RD and HZ: collect data. HL: review the collected data. XL and CD: research design and paper review. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Guangdong Basic and Applied Basic Research Foundation, China (2023A1515012422, 2019A1515011646), National Natural Science Foundation of China (81401689), Guangzhou Science and Technology Plan Project (202102080298) and Yat-sen Sailing Research Funds of Sun Yatsen Memorial Hospital of Sun Yat-sen University, China (YXQH202004).
Acknowledgments
We are grateful to the authors of all the original studies included in the meta-analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Collaborators., N.B.o.D. Life expectancy and disease burden in the Nordic countries: results from the global burden of diseases, injuries, and risk factors study 2017. Lancet Public Health (2019) 4(12):e658–69. doi: 10.1016/s2468-2667(19)30224-5
2. Collaborators., G.C. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol Hepatol (2020) 5(3):245–66. doi: 10.1016/s2468-1253(19)30349-8
3. Damiris K, Tafesh ZH, Pyrsopoulos N. Efficacy and safety of anti-hepatic fibrosis drugs. World J Gastroenterol (2020) 26(41):6304–21. doi: 10.3748/wjg.v26.i41.6304
4. Guo YC, Lu LG. Antihepatic fibrosis drugs in clinical trials. J Clin Transl Hepatol (2020) 8(3):304–12. doi: 10.14218/jcth.2020.00023
5. Collaborators., G.D.a.I.I.a.P. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet (2018) 392(10159):1789–858. doi: 10.1016/s0140-6736(18)32279-7
6. Louissaint J, Deutsch-Link S, Tapper EB. Changing epidemiology of cirrhosis and hepatic encephalopathy. Clin Gastroenterol Hepatol (2022) 20(8s):S1–s8. doi: 10.1016/j.cgh.2022.04.036
7. Ganem D, Prince AM. Hepatitis b virus infection–natural history and clinical consequences. N Engl J Med (2004) 350(11):1118–29. doi: 10.1056/NEJMra031087
8. Bouzgarrou N, Hassen E, Farhat K, Bahri O, Gabbouj S, Maamouri N, et al. Combined analysis of interferon-gamma and interleukin-10 gene polymorphisms and chronic hepatitis c severity. Hum Immunol (2009) 70(4):230–6. doi: 10.1016/j.humimm.2009.01.019
9. Salari N, Kazeminia M, Mansouri K, Hosseinian-Far A, Mohammadi M. The activity and polymorphism of the PON1 in patients with chronic liver disease: a systematic review and meta-analysis. J Gastrointest Cancer (2022) 53(3):745–55. doi: 10.1007/s12029-021-00699-7
10. Gu Y, Zhao J, Ao L, Ma J, Bao K, Liu M, et al. The influence of polymorphic GSTM1 gene on the increased susceptibility of non-viral hepatic cirrhosis: evidence from observational studies. Eur J Med Res (2018) 23(1):34. doi: 10.1186/s40001-018-0331-z
11. Yang XC, Fujino M, Cai SJ, Li SW, Liu C, Li XK. Genetic polymorphisms of cytotoxic T-lymphocyte antigen 4 in primary biliary cholangitis: A meta-analysis. J Immunol Res (2017) 2017:5295164. doi: 10.1155/2017/5295164
12. Danis VA, Millington M, Hyland VJ, Grennan D. Cytokine production by normal human monocytes: inter-subject variation and relationship to an IL-1 receptor antagonist (IL-1Ra) gene polymorphism. Clin Exp Immunol (1995) 99(2):303–10. doi: 10.1111/j.1365-2249.1995.tb05549.x
13. Motavaf M, Safari S, Alavian SM. Interleukin 18 gene promoter polymorphisms and susceptibility to chronic hepatitis b infection: a review study. Hepat Mon (2014) 14(7):e19879. doi: 10.5812/hepatmon.19879
14. Zheng M, Fang W, Yu M, Ding R, Zeng H, Huang Y, et al. IL-6 and IL-10 gene polymorphisms and cirrhosis of liver risk from a comprehensive analysis. BMC Endocr Disord (2021) 21(1):242. doi: 10.1186/s12902-021-00906-3
15. Ferreira Sda C, Chachá SG, Souza FF, Teixeira AC, Santana Rde C, Deghaide NH, et al. IL-18, TNF, and IFN-γ alleles and genotypes are associated with susceptibility to chronic hepatitis b infection and severity of liver injury. J Med Virol (2015) 87(10):1689–96. doi: 10.1002/jmv.24225
16. Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U.S.A. (1997) 94(7):3195–9. doi: 10.1073/pnas.94.7.3195
17. Cai Q, Wang Z, Zhang R, Zhang L, Cui S, Lin H, et al. Huangjia ruangan granule inhibits inflammation in a rat model with liver fibrosis by regulating TNF/MAPK and NF-κB signaling pathways. Evid Based Complement Alternat Med (2022) 2022:8105306. doi: 10.1155/2022/8105306
18. Ebrahim HA, Kamar SS, Haidara MA, Latif NSA, Ellatif MA, ShamsEldeen AM, et al. Association of resveratrol with the suppression of TNF-α/NF-kB/iNOS/HIF-1α axis-mediated fibrosis and systemic hypertension in thioacetamide-induced liver injury. Naunyn Schmiedebergs Arch Pharmacol (2022) 395(9):1087–95. doi: 10.1007/s00210-022-02264-w
19. Morales-Mantilla DE, King KY. The role of interferon-gamma in hematopoietic stem cell development, homeostasis, and disease. Curr Stem Cell Rep (2018) 4(3):264–71. doi: 10.1007/s40778-018-0139-3
20. Liu C, Yang S. A meta-analysis of the influence of tumor necrosis factor-α-308 gene polymorphism on liver cirrhosis. J Healthc Eng (2022) 2022:9764770. doi: 10.1155/2022/9764770
21. Bernal W, Moloney M, Underhill J, Donaldson PT. Association of tumor necrosis factor polymorphism with primary sclerosing cholangitis. J Hepatol (1999) 30(2):237–41. doi: 10.1016/s0168-8278(99)80068-3
22. Gordon MA, Oppenheim E, Camp NJ, di Giovine FS, Duff GW, Gleeson D. Primary biliary cirrhosis shows association with genetic polymorphism of tumour necrosis factor alpha promoter region. J Hepatol (1999) 31(2):242–7. doi: 10.1016/s0168-8278(99)80220-7
23. Jones DEJ, Watt FE, Grove J, Newton JL, Daly AK. Tumour necrosis factor-a promoter polymorpl’dsms in primary biliary cirrhosis. J Hepatol (1999) 30:232–6.
24. Fan LY, Zhong RQ, Tu XQ, Thomas P, Ralph F. Genetic association of tumor necrosis factor (TNF)-alpha polymorphisms with primary biliary cirrhosis and autoimmune liver diseases in a Chinese population. Chin J Hepatol (2004) 12(3):160–2.
25. Osterreicher CH, Datz C, Stickel F, Hellerbrand C, Penz M, Hofer H, et al. TGF-beta1 codon 25 gene polymorphism is associated with cirrhosis in patients with hereditary hemochromatosis. Cytokine (2005) 31(2):142–8. doi: 10.1016/j.cyto.2005.03.005
26. Pastor IJ, Laso FJ, Romero A, González-Sarmiento R. -238 G>A polymorphism of tumor necrosis factor alpha gene (TNFA) is associated with alcoholic liver cirrhosis in alcoholic Spanish men. Alcohol Clin Exp Res (2005) 29(11):1928–31. doi: 10.1097/01.alc.0000187595.19324.ca
27. Nguyen-Khac E, Houchi H, Daoust M, Dupas JL, Naassila M. The -308 TNFalpha gene polymorphism in severe acute alcoholic hepatitis: identification of a new susceptibility marker. Alcohol Clin Exp Res (2008) 32(5):822–8. doi: 10.1111/j.1530-0277.2008.00629.x
28. Zhang JZ, Cui DL. Association of polymorphisms of TNF-a and IL-10 genes with outcomes of hepatitis b virus infection. Hebei Medical University (2008). Master’s Degree Thesis.
29. Ma J, Zhang KH. Association of cytokine gene polymorphisms with the susceptibility of HBV-related hepatic cirrhosis. Nanchang University (2009). Master’s Degree Thesis.
30. Niro GA, Poli F, Andriulli A, Bianchi I, Bernuzzi F, Caliari L, et al. TNF-alpha polymorphisms in primary biliary cirrhosis: a northern and southern Italian experience. Ann N Y Acad Sci (2009) 1173:557–63. doi: 10.1111/j.1749-6632.2009.04741.x
31. Zhang P, Wan PQ, Zhou CX, Tang F, Xuan WJ, Wu JZ, et al. Study of the correlation between polymorphism of tumor necrosis factor alpha gene promoter-238 and chronic hepatitis b virus infection. Guangxi Med J (2013) 35(10):1288–94. doi: 10.11675/j.issn.0253-4304.2013.10.05
32. Li H, Zhu JZ, Wang YL, Pan C. Relationship between polymorphism of tumor necrosis factor-alpha-238 gene promoter region and outcome of chronic hepatitis b virus infection. Fujian Med J (2014) 36(6):85–8.
33. Sghaier I, Zidi S, Mouelhi L, Dabbech R, Ghazouani E, Brochot E, et al. The relationship between TNF alpha gene polymorphisms (-238/-308), TNF RII VNTR (p75) and outcomes of hepatitis b virus infection in Tunisian population. Gene (2015) 568(2):140–5. doi: 10.1016/j.gene.2015.05.029
34. Wu JM, Sun H, Wu KW, HUang ZM, Wu JS, Chen J, et al. Relationship between IFN-γ gene polymorphism and clinical outcomes of hepatitis b virus infection. J Wenzhou Med Coll (2008) 38(3):241–4.
35. Gao QJ, Liu DW, Zhang SY, Jia M, Wu LH. Association between IFN-γ +874 polymorphisms and the clinical outcomes of hepatitis b and/or hepatitis c virus infection. Chin J Epidemiol (2010) 31(3):324–8. doi: 10.3760/cma.j.issn.0254-6450.2010.03.021
36. Bahgat NA, Kamal MM, Abdelaziz AO, Mohye MA, Shousha HI, Ahmed MM, et al. Interferon-γ and interleukin-10 gene polymorphisms are not predictors of chronic hepatitis c (Genotype-4) disease progression. Asian Pac J Cancer Prev (2015) 16(12):5025–30. doi: 10.7314/apjcp.2015.16.12.5025
37. Sun Y, Lu Y, Xie L, Deng Y, Li S, Qin X. Interferon gamma polymorphisms and hepatitis b virus-related liver cirrhosis risk in a Chinese population. Cancer Cell Int (2015) 15:35. doi: 10.1186/s12935-015-0184-2
38. Tang F, Zhou CX, Huang LY, Li QN, Xuan WJ. Relationship of polymorphisms of interleukin-10 -819T/C gene and interferon-γ +874T/A gene with prognosis of hepatitis b virus infection in guangxi han nationality. Guangxi Med J (2015) 37(3):324–8. doi: 10.11675/j.issn.0253-4304.2015.03.12
39. Sheneef A, Esmat MM, Mohammad AN, Mahmoud AA, Moghazy HM, Noureldin AK. Interleukin-10 and interferon gamma gene polymorphisms and hepatitis c virus-related liver cirrhosis risk. J Interferon Cytokine Res (2017) 37(4):175–80. doi: 10.1089/jir.2016.0106
40. Talaat RM, Elsharnoby S, Abdelkhalek MS, El-Shenawy SZ, Elmasry S. The impact of interferon-γ (IFN-γ) and IFN-γ-Inducible protein 10 (IP-10) genes’ polymorphism on risk of hepatitis c virus-related liver cirrhosis. Immunol Invest (2022) 51(3):688–704. doi: 10.1080/08820139.2020.1869251
41. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med (2002) 21(11):1539–58. doi: 10.1002/sim.1186
42. Cheng JW, Cheng SW, Ma XY, Cai JP, Li Y, Lu GC, et al. Myocilin polymorphisms and primary open-angle glaucoma: a systematic review and meta-analysis. PLoS One (2012) 7(9):e46632. doi: 10.1371/journal.pone.0046632
43. Zeng T, Guo FF, Zhang CL, Song FY, Zhao XL, Xie KQ. Roles of cytochrome P4502E1 gene polymorphisms and the risks of alcoholic liver disease: a meta-analysis. PLoS One (2013) 8(1):e54188. doi: 10.1371/journal.pone.0054188
44. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj (2003) 327(7414):557–60. doi: 10.1136/bmj.327.7414.557
45. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst (1959) 22(4):719–48.
46. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials (1986) 7(3):177–88. doi: 10.1016/0197-2456(86)90046-2
47. Mohammadi A, Azarnezhad A, Khanbabaei H, Izadpanah E, Abdollahzadeh R, Barreto GE, et al. Vitamin d receptor genetic polymorphisms and the risk of multiple sclerosis: A systematic review and meta-analysis. Steroids (2020) 158:108615. doi: 10.1016/j.steroids.2020.108615
48. Napolioni V. The relevance of checking population allele frequencies and hardy-Weinberg equilibrium in genetic association studies: the case of SLC6A4 5-HTTLPR polymorphism in a Chinese han irritable bowel syndrome association study. Immunol Lett (2014) 162(1 Pt A):276–8. doi: 10.1016/j.imlet.2014.08.009
49. Hayashino Y, Noguchi Y, Fukui T. Systematic evaluation and comparison of statistical tests for publication bias. J Epidemiol (2005) 15(6):235–43. doi: 10.2188/jea.15.235
50. Shao HB, Ren K, Gao SL, Zou JG, Mi YY, Zhang LF, et al. Human methionine synthase A2756G polymorphism increases susceptibility to prostate cancer. Aging (Albany NY) (2018) 10(7):1776–88. doi: 10.18632/aging.101509
51. Czaja AJ. Hepatic inflammation and progressive liver fibrosis in chronic liver disease. World J Gastroenterol (2014) 20(10):2515–32. doi: 10.3748/wjg.v20.i10.2515
52. Sánchez PS, Rigual MDM, Djouder N. Inflammatory and non-inflammatory mechanisms controlling cirrhosis development. Cancers (Basel) (2021) 13(20):5045. doi: 10.3390/cancers13205045
53. Stojsavljević S, Gomerčić Palčić M, Virović Jukić L, Smirčić Duvnjak L, Duvnjak M. Adipokines and proinflammatory cytokines, the key mediators in the pathogenesis of nonalcoholic fatty liver disease. World J Gastroenterol (2014) 20(48):18070–91. doi: 10.3748/wjg.v20.i48.18070
54. Shi J, Zhao J, Zhang X, Cheng Y, Hu J, Li Y, et al. Activated hepatic stellate cells impair NK cell anti-fibrosis capacity through a TGF-β-dependent emperipolesis in HBV cirrhotic patients. Sci Rep (2017) 7:44544. doi: 10.1038/srep44544
55. Yoon JH, Gores GJ. Death receptor-mediated apoptosis and the liver. J Hepatol (2002) 37(3):400–10. doi: 10.1016/s0168-8278(02)00209-x
56. Chen X, Zhou P, De L, Li B, Su S. The roles of transmembrane 6 superfamily member 2 rs58542926 polymorphism in chronic liver disease: A meta-analysis of 24,147 subjects. Mol Genet Genomic Med (2019) 7(8):e824. doi: 10.1002/mgg3.824
57. Zhang S, Yang X, Wang W. Associations of genetic polymorphisms in CTLA-4 and IL-18 with chronic liver diseases: Evidence from a meta-analysis. Genomics (2020) 112(2):1889–96. doi: 10.1016/j.ygeno.2019.11.001
58. Dong X, Yu X, Li H, Kang H. Identification of marker genes and pathways in patients with primary biliary cholangitis. J Comput Biol (2020) 27(6):923–33. doi: 10.1089/cmb.2019.0230
59. Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet (2011) 43(4):329–32. doi: 10.1038/ng.789
60. Peng Z, Zhang Y, Gu W, Wang Z, Li D, Zhang F, et al. Integration of the hepatitis b virus X fragment in hepatocellular carcinoma and its effects on the expression of multiple molecules: a key to the cell cycle and apoptosis. Int J Oncol (2005) 26(2):467–73. doi: org/10.3892/ijo.26.2.467
61. Lin SY, Wang YY, Chuang YH, Chen CJ. Skeletal muscle proteolysis is associated with sympathetic activation and TNF-α-ubiquitin-proteasome pathway in liver cirrhotic rats. J Gastroenterol Hepatol (2016) 31(4):890–6. doi: 10.1111/jgh.13159
62. Tan S, Zhao J, Sun Z, Cao S, Niu K, Zhong Y, et al. Hepatocyte-specific TAK1 deficiency drives RIPK1 kinase-dependent inflammation to promote liver fibrosis and hepatocellular carcinoma. Proc Natl Acad Sci U.S.A. (2020) 117(25):14231–42. doi: 10.1073/pnas.2005353117
63. Higuchi T, Oka S, Furukawa H, Nakamura M, Komori A, Abiru S, et al. Role of deleterious single nucleotide variants in the coding regions of TNFAIP3 for Japanese autoimmune hepatitis with cirrhosis. Sci Rep (2019) 9(1):7925. doi: 10.1038/s41598-019-44524-5
64. Falleti E, Fabris C, Toniutto P, Fontanini E, Cussigh A, Caldato M, et al. Genetic polymorphisms of inflammatory cytokines and liver fibrosis progression due to recurrent hepatitis c. J Interferon Cytokine Res (2007) 27(3):239–46. doi: 10.1089/jir.2006.0062
65. Nalpas B, Lavialle-Meziani R, Plancoulaine S, Jouanguy E, Nalpas A, Munteanu M, et al. Interferon gamma receptor 2 gene variants are associated with liver fibrosis in patients with chronic hepatitis c infection. Gut (2010) 59(8):1120–6. doi: 10.1136/gut.2009.202267
66. Ramadan HK, Badr G, Ramadan NK, Sayed A. Enhanced immune responses, PI3K/AKT and JAK/STAT signaling pathways following hepatitis c virus eradication by direct-acting antiviral therapy among Egyptian patients: a case control study. Pathog Dis (2021) 79(3):ftab008. doi: 10.1093/femspd/ftab008
67. El Sharkawy R, Thabet K, Lampertico P, Petta S, Mangia A, Berg T, et al. A STAT4 variant increases liver fibrosis risk in Caucasian patients with chronic hepatitis b. Aliment Pharmacol Ther (2018) 48(5):564–73. doi: 10.1111/apt.14866
Keywords: tumor necrosis factor-α, interferon-γ, liver cirrhosis, polymorphism, metaanalysis, risk
Citation: Zheng M, Li J, Fang W, Luo L, Ding R, Zeng H, Luo H, Lin X and Duan C (2023) The TNF-α rs361525 and IFN-γ rs2430561 polymorphisms are associated with liver cirrhosis risk: a comprehensive meta-analysis. Front. Immunol. 14:1129767. doi: 10.3389/fimmu.2023.1129767
Received: 30 December 2022; Accepted: 30 March 2023;
Published: 14 April 2023.
Edited by:
Thierry Thévenot, University Hospital of Besançon, FranceReviewed by:
Vladimir M. Pisarev, Federal Research and Clinical Center of Intensive Care Medicine and Rehabilitation, RussiaLivio Muccillo, University of Sannio, Italy
Copyright © 2023 Zheng, Li, Fang, Luo, Ding, Zeng, Luo, Lin and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianghua Lin, bHhpYW5naEBtYWlsLnN5c3UuZWR1LmNu; Chaohui Duan, ZHVhbmNoaEBtYWlsLnN5c3UuZWR1LmNu
†These authors have contributed equally to this work
 Minghui Zheng1,2†
Minghui Zheng1,2† Xianghua Lin
Xianghua Lin