- 1State Key Laboratory of Marine Resource Utilization in South China Sea, Hainan University, Haikou, China
- 2Collaborative Innovation Center of Marine Science and Technology, Hainan University, Haikou, China
- 3Hainan Provincial Key Laboratory for Tropical Hydrobiology and Biotechnology, College of Marine Science, Hainan University, Haikou, China
Introduction: The transcription factor interferon regulatory factor 3 (IRF3) plays an important role in host defence against viral infections. However, its role during bacterial infection in teleosts remains unclear. In the present study, we evaluated the antibacterial effects of Trachinotus ovatus IRF3 (TroIRF3) and how it regulates type I interferon (IFN).
Methods: Subcellular localisation experiments, overexpression, and quantitative real-time PCR (qRT-PCR) were performed to examine the nuclear localisation signal (NLS) of TroIRF3 and its role in the antibacterial regulatory function of TroIRF3. We assessed the binding activity of TroIRF3 to the IFNa3 promoter by luciferase reporter assay.
Results and Discussion: The results showed that TroIRF3 was constitutively expressed at high levels in the gill and liver. TroIRF3 was significantly upregulated and transferred from the cytoplasm to the nucleus after Vibrio harveyi infection. By overexpressing TroIRF3, the fish were able to inhibit the replication of V. harveyi, whereas knocking it down increased bacterial replication. Moreover, the overexpression of TroIRF3 increased type I interferon (IFNa3) production and the IFN signalling molecules. The NLS, which is from the 64–127 amino acids of TroIRF3, contains the basic amino acids KR74/75 and RK82/84. The results proved that NLS is required for the efficient nuclear import of TroIRF3 and that the NLS domain of TroIRF3 consists of the key amino acids KR74/75 and RK82/84. The findings also showed that NLS plays a key role in the antibacterial immunity and upregulation of TroIFNa3 induced by TroIRF3. Moreover, TroIRF3 induces TroIFNa3 promoter activity, whereas these effects are inhibited when the NLS domain is deficient. Overall, our results suggested that TroIRF3 is involved in the antibacterial immunity and regulation of type I IFN in T. ovatus and that the NLS of TroIRF3 is vital for IRF3-mediated antibacterial responses, which will aid in understanding the immune role of fish IRF3.
1 Introduction
Innate immunity is the first line of defence against invading viruses and bacteria. The innate antiviral mechanism in mammals involves interferon (IFN), which induces the expression of IFN-relative genes (1–3). IFN can be divided into two major categories based on its cysteine residues. In all teleost fish lineages, type I IFNs contain two cysteine residues; however, type II IFNs contain four cysteine residues in a few species (4–7). In both vertebrates and invertebrates, interferon regulatory factors (IRFs) regulate type I and II IFN transcriptional activation (8).
Thus far, nine IRF family members have been found in mammals, 10 in birds and 11 in fish (9). The N-terminal DNA binding domain (DBD), consisting of five tryptophan repeats, is highly homologous between IRF members. The DBD forms a helix-turn-helix structure that binds specifically to IFN-stimulated response element (ISRE) sequences in the promoters of IFNβ and IFN-stimulated genes (ISGs) (10). Except for IRF1 and IRF2, the C-terminal of each IRF has an IRF association domain (IAD), which is used to interact with other IRFs and other factors (11–13). IRF3 and IRF7 contain a serine-rich region (SRD) at the C-terminus that controls their transcriptional activities when phosphorylated by viruses (11–14). Two transcription factors, IRF3 and IRF7 control the expression of type I IFNs (15, 16). The antiviral effects of several innate immune receptors, including retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), toll-like receptors (TLRs) and DNA sensors, are mediated by IRF3 (17–19). All TLRs in mammals (except TLR3) transduce signals through the MyD88-dependent pathway using molecules such as the TNFR-associated factor 6 (TRAF6) (17, 20–22). In general, pathogen invasion initiates an immune response by activating the immune signalling pathways. As a member of the IRF family, IRF3 plays an important role in immune signalling. Following bacterial infection, IRF3 is responsible for controlling the expression of IFNs and ISGs (23). ISGs include ISG15, MAX interactor I (MXI) and Viperin1 (24).
Accurate cellular localization plays a crucial role in the effective function of most signaling proteins, the ability of IRF3 to translocate into the nucleus is essential for its biological activity. Nuclear localization signals (NLSs) and nuclear export signals (NESs) are intrinsic targeting signals required to facilitate the shuttle between the nucleus and the cytoplasm for proteins larger than 45 kDa (25). IRF3 accumulates in the cytoplasm under resting conditions and adopts an auto-inhibited conformation, which is mediated by the NES. When infected, IRF3 enters the nucleus via NLS after being phosphorylated and dimerized at its C-terminus. IRF3 and the coactivator CBP/p300 form a complex and then bind the IFN-α/β promoters and the ISRE sequences of the targeted genes (14, 26, 27). Several studies have found that bacterial products, such as lipopolysaccharide (LPS), can activate toll-like receptors to induce type I IFN expression through IRF3 signalling (28–31). However, the role of IRF3 in bacterial infection is less understood.
The characteristics and functions of IRF3s have been reported in several species of teleost fish, such as Epinephelus coioides, Cynoglossus semilaevis, Carassius auratus L., Salmo salar, Larimichthys crocea, and Paralichthys olivaceus (32–37). In a previous study, red-spotted grouper nervous necrosis virus replication was significantly decreased in E. coioides when IRF3 was overexpressed (32). For C. auratus L., fish IFN or polyinosinic-polycytidylic acid (poly I:C) induced the phosphorylation of IRF3 and translocation from the cytoplasm to the nucleus. In addition, C. auratus L. IRF3 overexpression induced IFN production, which subsequently triggered ISG transcription via signal transducer and activator of transcription 1 (STAT1) (33). Moreover, viral and bacterial stimulation significantly induced C. semilaevis IRF3 (34). Although some studies have investigated IRF3 in fish, its function, particularly its role in antibacterial immunity, remains largely unknown.
Vibrio harveyi is a gram-negative opportunistic pathogenic bacterium of many commercially farmed fish species (38, 39). Trachinotus ovatus (golden pompano) is one of the most economically important aquaculture species in China that suffers from V. harveyi, resulting in serious economic losses (40). IFNa3 is a member of type I IFNs in T. ovatus. Previous study demonstrated that IRF1, IRF2, IRF5, and IRF7 positively regulate IFNa3 expression in T. ovatus (41, 42). The purpose of this study was to assess the antibacterial immune effects of IRF3 in T. ovatus (TroIRF3) and its role in the regulation of IFNa3. Based on these investigations, we further examine the NLS in TroIRF3 and its role in the host antibacterial immune response induced by TroIRF3.
2 Materials and methods
2.1 Fish, cell lines and bacteria
T. ovatus were purchased from a commercial fish farm in Hainan, China. Before the experiment, the fish were acclimated at 26°C in a flow-through water system for one week. All animal experiments conducted during this study were approved by the Animal Care and Use Committee at Hainan University. Human embryonic kidney (HEK293T) cells were cultured at ambient CO2 and 37°C in Dulbecco’s modified Eagle’s medium (DMEM, USA), the DMEM contained 1% penicillin and streptomycin (Pen Strept) and 10% foetal bovine serum (FBS). Golden pompano snout (GPS) cells were generously donated by the South China Sea Institute of Oceanology Chinese Academy of Sciences. The GPS cells were grown at 26°C with Leibovitz’s L-15 medium containing 10% FBS (Gibco, USA) (42, 43). V. harveyi was grown at 28°C in Luria-Bertani broth (38).
2.2 TroIRF3 cloning
Based on the transcriptome data, the primers TroIRF3-F/TroIRF3-R were used to clone the open reading frame (ORF) of TroIRF3 by polymerase chain reaction (PCR) amplification (Table S1). Sequencing was performed following gel purification of the PCR products into the pEASY®-T1Simple Vector (Transgen, Beijing, China). A homology search of the TroIRF3 protein sequence was conducted using BLAST programs on NCBI (http://www.ncbi.nlm.nih.gov/blast). Signal peptide prediction was performed using SignalP 5.1. Thereafter, DNAMAN (Lynnon Biosoft, USA) was used to align the amino acid sequences. SWISS-MODEL was used to predict the TroIRF3’s tertiary structure. A phylogenetic analysis was performed using neighbour-joining methods through MEGA 6.0.
2.3 Expression profiles of TroIRF3 in normal tissues and when challenged with V. harveyi
A total of 11 tissues were obtained from 15 healthy T. ovatus (blood, head kidney, liver, intestine, spleen, gill, muscle, skin, brain, stomach and heart). To avoid the affection of the individual difference of fish, the fish tissues were mixed aseptically into a sample with five identical samples. The tissue samples were stored at −80°C until RNA extraction was performed. The T. ovatus were divided into two groups: control and bacterial challenge. The challenge group received 0.1 ml of V. harveyi (2 × 106 CFU/ml), while the control group received the same volume of PBS. At 6, 9, 12 and 24 hours post-injection (hpi), 15 individuals in each group were randomly sacrificed, and the liver, spleen and head-kidney were sampled. Five identical tissues from each group were mixed aseptically to form a sample. The total RNA was extracted from the harvested samples using the Total RNA Extraction Kit (Promega, China). The cDNA was synthesised using the Reverse Transcription System (Promega, China). An SYBR ExScript qRT–PCR kit (LS2062, Promega, China) was used to perform quantitative real-time PCR (qRT-PCR) on a qTOWER3 Real-Time PCR (qPCR) system (Analytik Jena, Germany). The internal reference gene was β-2 microglobulin (B2M), and the relative expression levels of TroIRF3 were calculated using the 2^-ΔΔCT method (44). The primers are summarised in Table S1.
2.4 TroIRF3 overexpression and V. harveyi infection in vivo
The eukaryotic expression vector pTroIRF3 was constructed for TroIRF3 overexpression in vivo. TroIRF3 was cloned into the pCN3’s Sma I site and transformed into Escherichia coli, DH5α. pTroIRF3 was extracted using the Endo Free Plasmid Kit (Tiangen, China). The fish (weight 15.2 ± 3.1 g) were randomised into three groups (N=12), with each receiving a 0.1 ml injection of pTroIRF3 (150 μg/ml), pCN3 (empty vector, 150 μg/ml) or PBS (control). To detect the expression level of TroIRF3, the liver, spleen and head-kidney of three fish from each group were collected after five days of intramuscular injection of plasmids. After the isolation of total RNA and cDNA synthesis, TroIRF3 expression was detected by qRT-PCR with primer TroIRF3-RT-F/R (Table S1). B2M was used as the internal reference gene. V. harveyi (105 CFU/fish) was injected into the remaining fish. The liver, spleen and head-kidney of three fish in each group were homogenised at 6, 9 and 12 hpi for bacterial count analysis.
2.5 TroIRF3 knockdown and V. harveyi infection in vivo
The small interfering RNA (siRNA) of TroIRF3 was synthesised using the T7 RiboMAX™ Express RNAi System (Promega, USA). The primers used for siRNA synthesis are listed in Table S1. A total of 36 fish were randomly divided into three groups of 12 each and injected with 0.1 ml of PBS (control), siRNA-C (control, 15 μg/fish) or siRNA-TroIRF3 (15 μg/fish). At 12 h after intramuscular injection of siRNA, three fish were sacrificed for liver, spleen and head-kidney collection. Then, qRT–PCR was performed to detect the TroIRF3 expression levels. The rest of the fish were infected with V. harveyi (105 CFU/fish). The liver, spleen and head-kidney of three fish in each group were homogenised at 6, 9, and 12 hpi for bacterial count analysis.
2.6 Subcellular localisation
The eukaryotic expression vector pTroIRF3-N3 was created by cloning TroIRF3’s ORF into the Bgl II and Sal I sites of pEGFP-N3. Subcellular localisation experiments were performed to examine TroIRF3 localisation in the cell with or without V. harveyi infection. The cells were seeded on 24-well plates and grown overnight, and then Lipofectamine 2000 (Invitrogen, USA) was used to transfect pEGFP-N3 or pTroIRF3-N3 into the GPS cells. At 48 h post-transfection, the cells transfected with pTroIRF3-N3 were treated with 0.1 ml PBS (control) or V. harveyi (1×105 CFU/ml) for 6 h. The GPS cells were washed, stained and visualised with fluorescence microscopy (Leica, Wetzlar, Germany) after fixing in 4% paraformaldehyde.
Different PCR fragments of TroIRF3 were inserted into Bgl II and Sal I sites of pEGFP-N3 vector to constructe TroIRF3-GFP truncation mutants: pTroIRF3-(△NES)-N3, pTroIRF3-1-165-N3, pTroIRF3-1-417-(△NES)-N3, pTroIRF3-64-466-(△NES), or pTroIRF3-127-466-(△NES). Various plasmids were transfected into the GPS cells to detect the NLSs of TroIRF3. The cells were fixed, washed, stained and visualised under fluorescence microscopy at 48 h post-transfection.
Two TroIRF3 variants (KR74/75NG and RK82/84LQ) were designed to further detect the NLSs of IRF3 within the aa 64–127 region. The PCR fragments of KR74/75NG and RK82/84LQ were inserted into Bgl II and Sal I sites of pEGFP-N3 vector to construct two recombinant plasmids: pTroIRF3-KR74/75NG-N3 and pTroIRF3-RK82/84LQ-N3. The primers used are listed in Table S1. The GPS cells were transfected with plasmids (pTroIRF3-KR74/75NG-N3 or pTroIRF3-RK82/84LQ-N3). After 48 h, the cells were treated with 0.1 ml V. harveyi (1×105 CFU/ml) or PBS (control) for 6 h. Afterwards, the cells were fixed, washed, stained and visualised under fluorescence microscopy.
2.7 Role of NLSs sites in immune response in vivo
2.7.1 Role of NLSs in the antibacterial regulatory function of TroIRF3
To further investigate the role of these basic amino acids in TroIRF3’s antibacterial regulation, two recombinant plasmids (pTroIRF3-KR74/75NG and pTroIRF3-RK82/84LQ) were constructed by inserting the variants into the Sma I sites of the pCN3 vectors. The fish were randomised into four groups (12 fish per group) and injected with PBS (control), pTroIRF3 (15 μg/fish), pTroIRF3-KR74/75NG (15 μg/fish), or pTroIRF3- RK82/84LQ (15 μg/fish), respectively. After five days, the liver, spleen and head-kidney of three fish from each group were collected to detect the expression level of TroIRF3 or it mutants. The remaining fish were intraperitoneally infected with V. harveyi (2×106 CFU/fish). The liver, spleen and head-kidney of three fish in each group were homogenised at 6, 9 and 12 hpi for bacterial count analysis.
2.7.2 Role of NLSs in immune gene expression induced by TroIRF3
Twelve fish were randomised into four groups and injected with PBS (control), pTroIRF3, pTroIRF3-KR74/75NG or pTroIRF3-RK82/84LQ, respectively. After five days of injection, the spleens were collected, followed by RNA isolation and cDNA synthesisation. The relative transcription level of IFNa3, MXI, TRAF6, Viperin1 and mitochondrial antiviral signalling (MAVS) was detected by qRT-PCR. The primers are listed in Table S1.
2.8 Luciferase reporter assay
The TroIRF3 binding site in the promoter of TroIFNa3 was evaluated by luciferase reporter assay. Three truncation mutants of the TroIFNa3 promoter (−1649 to +1, –896 to +1 and −722 to +1) were amplified and inserted into the Kpn I and Xho I restriction sites of the pGL4-basic luciferase reporter plasmid (Promega, USA) to obtain the recombinant plasmid pGL4-TroIFNa3-p1, pGL4-TroIFNa3-p2 and pGL4-TroIFNa3-p3. The primers used are summarised in Table S1. Lipofectamine 2000 (Invitrogen, USA) was used to cotransfect pRL-TK (0.025 μg), pGL4-TroIFNa3-p1/p2/p3 (0.25 μg) and pTroIRF3 (0.25 μg) into the HEK293T cells. pRL-TK was used as an internal control. After 36 h of transfection, the firefly and renilla luciferase activities were measured using a dual-luciferase reporter assay system (Promega, USA). The activity ratio between the firefly luciferase and the renilla luciferase was used to calculate the luciferase activity. The effect of V. harveyi stimulation on the TroIFNa3 promoter activation mediated by TroIRF3 was detected. After 24 h of transfection, the transfected HEK293T cells were treated with PBS or V. harveyi (1×102 CFU/ml) for 12 h. Luciferase activity was measured in the lysed cells as described above.
Luciferase reporter assay was performed to detect whether the activation of the TroIFNa3 promoter was dependent on the pTroIRF3 concentration. The HEK293T cells were cotransfected with pRL-TK (0.025 μg), pGL4-TroIFNa3-p2 (0.25 μg) and different concentrations of pTroIRF3 (0, 0.01, 0.05, 0.1 or 0.2 μg). After 24 h, the transfected HEK293T cells were treated with V. harveyi (1×102 CFU/ml) for 12 h. Luciferase activity was measured in the lysed cells as described above.
Luciferase reporter assay was performed to detect the role of the basic amino acids (KR74/75 and RK82/84) in the TroIRF3-triggered TroIFNa3-p2 activation. The HEK293T cells were cotransfected with pGL4-TroIFNa3-p2, wild-type TroIRF3 or its variants (pTroIRF3-KR74/75NG or pTroIRF3-RK82/84LQ) and pRL-CMV. After 24 h, the cells were treated with V. harveyi (1×102 CFU/ml) for 12 h. Afterwards, the relative luciferase activity was tested as described above.
2.9 Statistical analysis
All data were analysed by SPSS 16.0 (SPSS, IL, USA) and GraphPad Prism 5 (GraphPad, CA, USA). The statistical significance level of this study was set at 0.05.
3 Results
3.1 Cloning and sequence analysis of TroIRF3
The full length of TroIRF3 cDNA contains a 1398 bp ORF encoding 465 amino acids (GenBank Accession No. AWY04222.1). In theory, it has a molecular mass of 52 kDa and an isoelectric point of 4.89. According to the amino acid alignments, TroIRF3 contains three conserved domains, namely, DBD, IAD and SRD (Figure 1A). Among the selected IRF3 sequences, TroIRF3 was closest to that of Seriola lalandi dorsalis (85.95%), followed by those of Siniperca chuatsi and Lateolabrax japonicus (81.94% and 82.37%, respectively) (Figure 1A). According to the SWISS-MODEL online software, TroIRF3 has a similar three-dimensional structure to human IRF3 (Figure 1B). In addition, the IRF3 from fish and mammals were located in two separate branches, while TroIRF3 was inserted in the fish cluster. TroIRF3 shows a close evolutionary relationship with S. lalandi dorsalis IRF3 (Figure 1C).
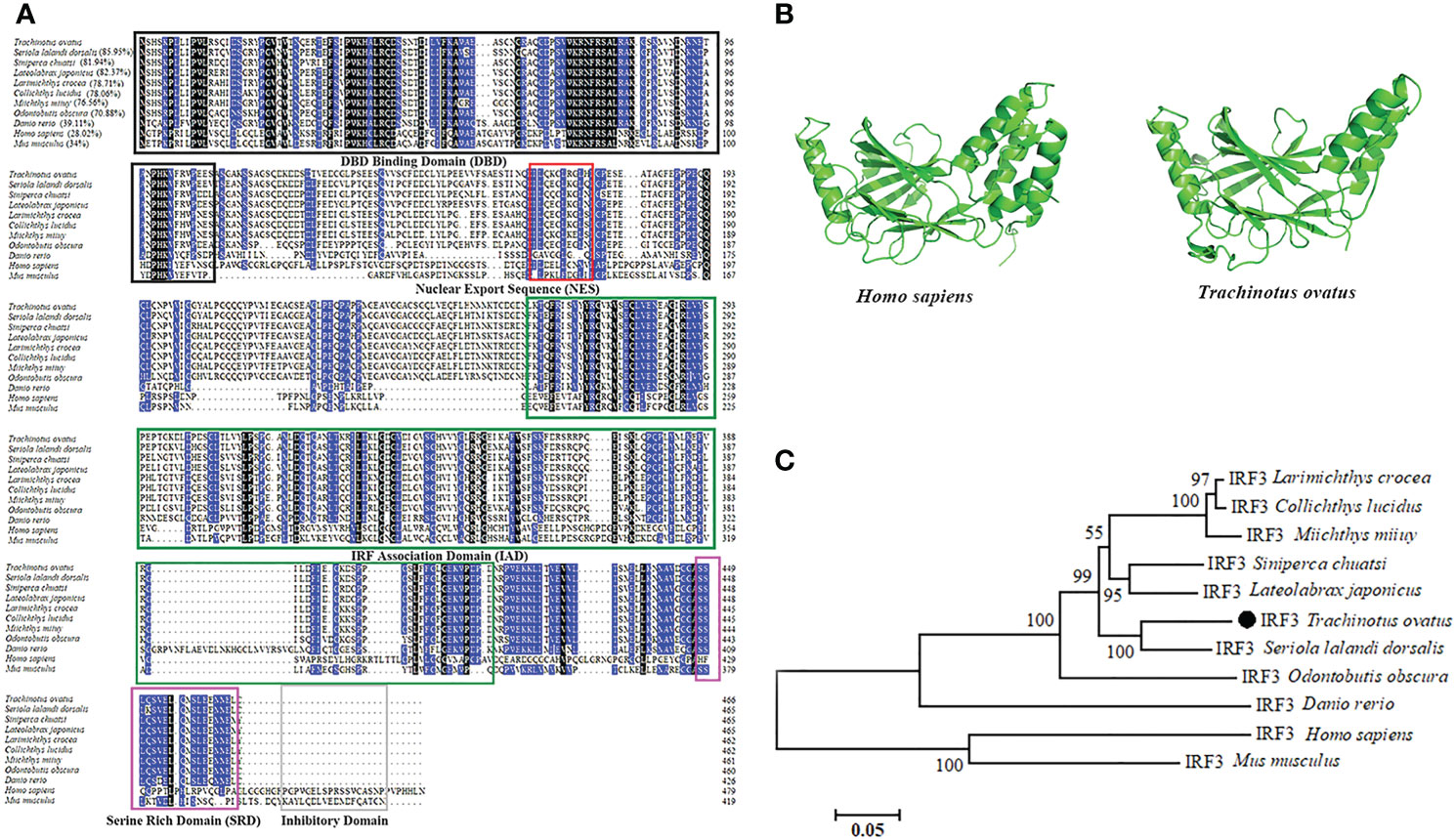
Figure 1 Sequence characteristics of TroIRF3. (A) Alignment of amino acid sequences from TroIRF3 and other IRF3 homologs. Identities between TroIRF3 and the compared sequences are shown in brackets. The background color of amino acid residues corresponds to the conservation degree (blue > 75%, black = 100%). The conserved DBD domain is boxed in black, the conserved IAD domain is boxed in green, the conserved SRD domain is boxed in purple, and the conserved inhibitory domain is boxed in gray. The nuclear export sequences (NES) are marked with the red box. (B) Analysis of the SWISS-MODEL prediction for TroIRF3 (SWISS-MODEL Template ID: 5jek.2) compared to human IRF3. (C) Phylogenetic analysis of TroIRF3. MEGA 6.0 was used to construct a phylogenetic tree based on neighbor-joining (NJ). The sequences of TroIRF3 genes used in this study were obtained from GenBank, a list of GenBank accession numbers can be found in Table S2.
3.2 Expression pattern of TroIRF3 with or without bacterial challenge
The expression level of TroIRF3 was detected in 11 normal tissues (spleen, head-kidney, skin, muscle, gill, heart, liver, brain, stomach, blood and intestine) by qRT-PCR. TroIRF3 was constitutively distributed in all the tissues examined. TroIRF3 expression was high in the gill and liver but low in the spleen and blood (Figure 2A).
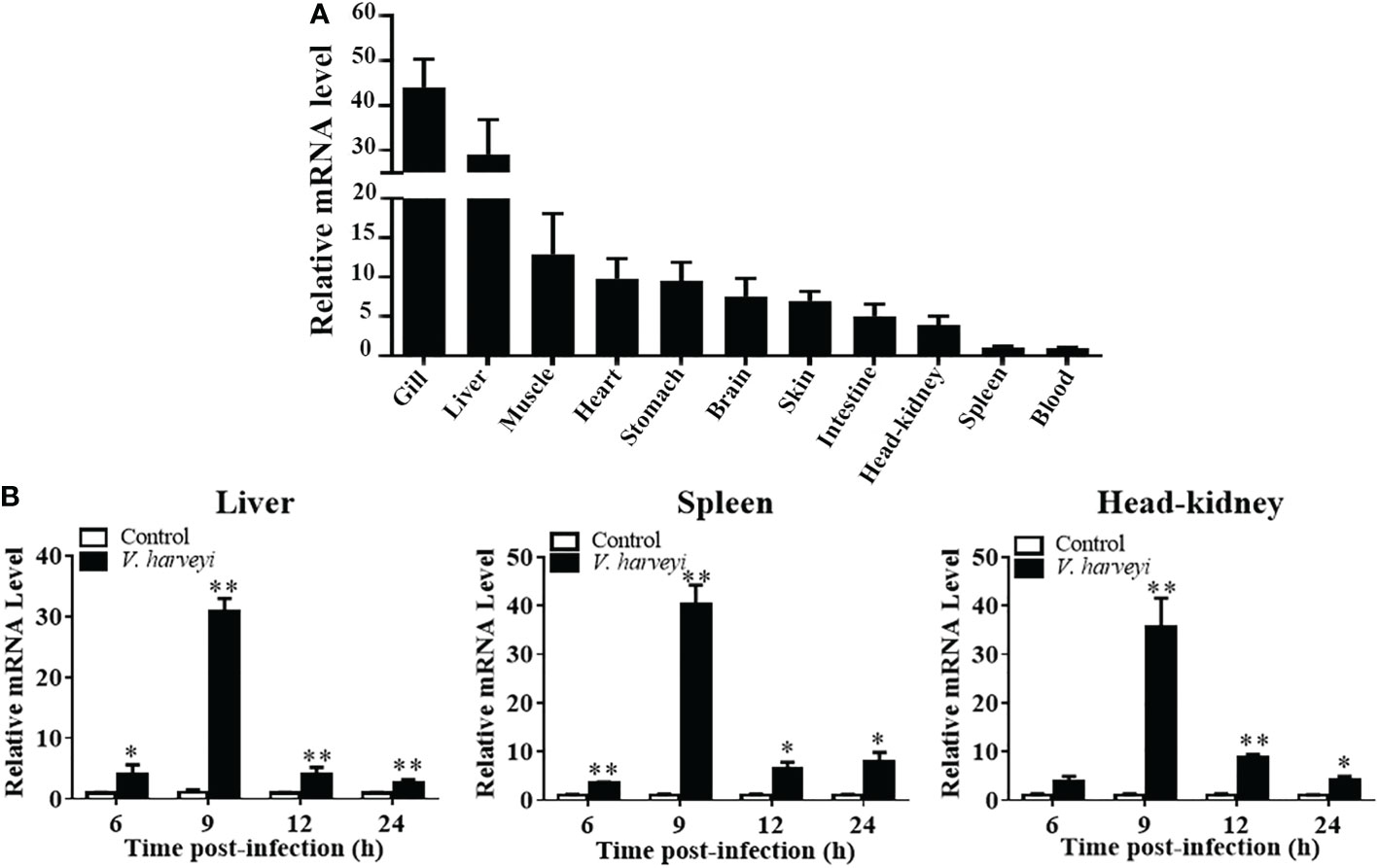
Figure 2 Expression profiles of TroIRF3 in normal tissues and when challenged with V. harveyi. (A) TroIRF3 expression levels in different tissues. (B) Expression of TroIRF3 in the liver, spleen, and head-kidney in response to the V. harveyi challenge. At every time point, control fish were set to have an average expression level of 1. Error bars display means ± SD (N = 3). N, indicates parallel experiments. *P < 0.05, **P < 0.01.
The level of TroIRF3 expression following V. harveyi infection was detected using qRT-PCR (Figure 2B). The level of TroIRF3 expression in the liver was increased from 6 hpi to 24 hpi (4.1-, 30.9-, 4.1-, and 2.7-folds at 6, 9, 12, and 24 hpi, respectively), the expression level of TroIRF3 in the spleen was increased from 6 hpi to 24 hpi (3.5-, 40.2-, 6.5-, and 7.8-folds at 6, 9, 12, and 24 hpi, respectively), and the expressions of TroIRF3 in the head-kidney were increased from 9 hpi to 24 hpi but manifested no difference at 6 hpi (35.5-, 8.8-, and 4.0-folds at 9, 12, and 24 hpi, respectively). Based on these results, TroIRF3 may contribute to antibacterial immunity.
3.3 Overexpression of TroIRF3 decreases V. harveyi replication in vivo
To clarify TroIRF3’s role in antibacterial immunity, we assessed the effects of TroIRF3 overexpression on the fish’s antibacterial ability. An increase in TroIRF3 expression was observed in fish treated with pTroIRF3, indicating the overexpression of TroIRF3 (Figure S1A). Compared with the control group, the number of bacteria in the tissues decreased in the TroIRF3-overexpressing fish (Figure 3A). The bacterial load in the liver was significantly decreased in the TroIRF3-overexpressing fish compared with the control fish by 7.1- and 8.3-fold at 6 and 9 hpi, respectively. The pattern of bacterial load in the spleen of the pTroIRF3-overexpressing fish was similar to that in the liver, which decreased significantly with 5.0- and 10.9-fold at 6 and 9 hpi, respectively. Compared with the control fish, TroIRF3 overexpression significantly reduced the number of bacterial colonies in the head-kidney by 6.8- and 9.6-fold at 9 and 12 hpi, respectively. No significant difference in bacterial loads was observed between the empty vector-injected fish and the control fish, which showed that the vector had no effect on the fish’s antibacterial immune responses.
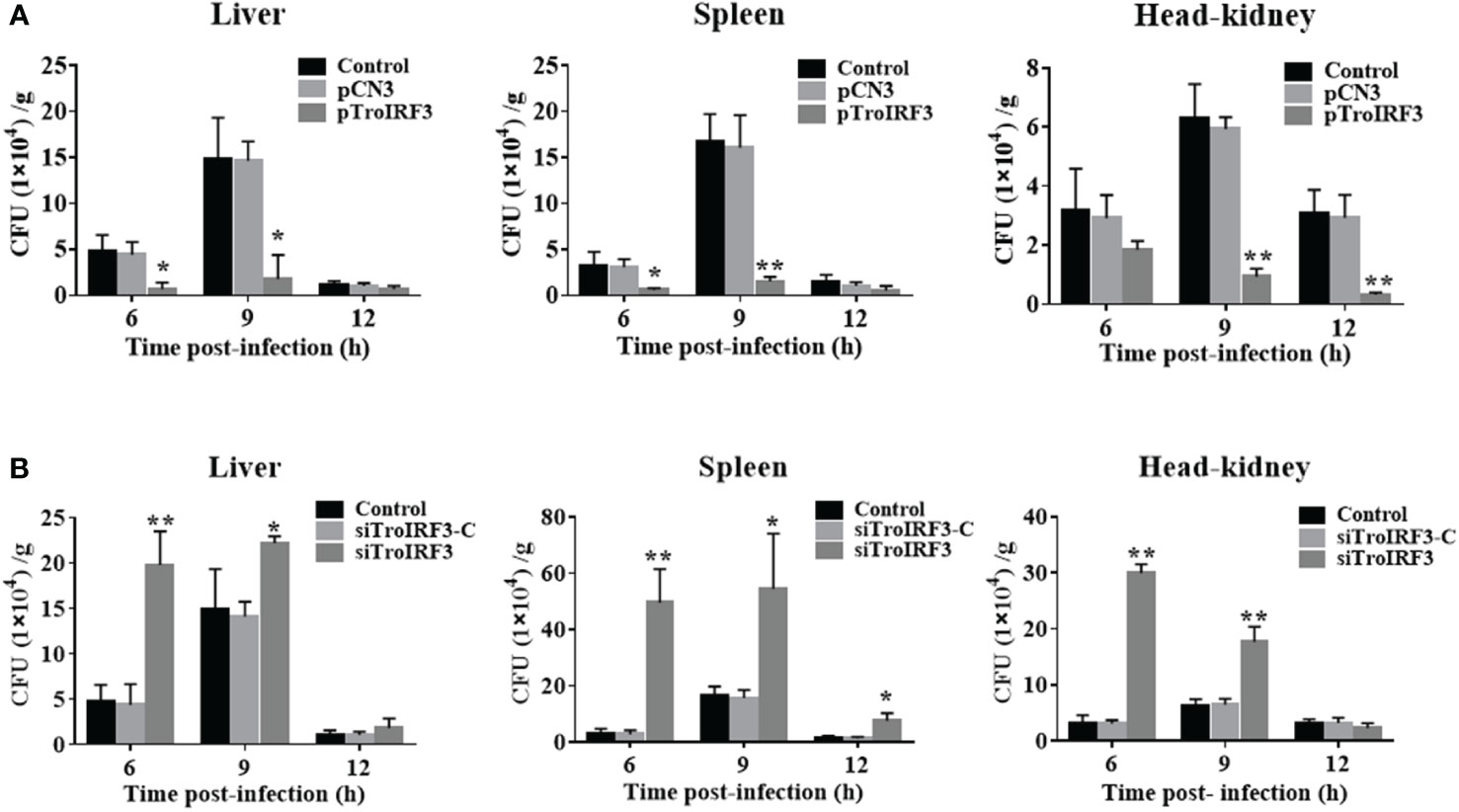
Figure 3 Effects of TroIRF3 on bacterial infection. (A) At five days after injection with PBS (Control), pCN3, and pTroIRF3, T. ovatus was infected with V. harveyi, and their liver, spleen, and head-kidney bacteria were determined at 6, 12, and 24 hpi. (B) T. ovatus was injected with siTroIRF3, siTroIRF3-C, or PBS (Control) for 12 h, then infected with V. harveyi, and their liver, spleen, and head-kidney bacteria were determined at 6, 12, and 24 hpi. The data are shown as the means ± SD (N =3), N, the number of fish used at each time point per group. *P < 0.05, **P < 0.01.
3.4 Knockdown of TroIRF3 increases V. harveyi replication in vivo
The effects of TroIRF3 on antibacterial immune responses were further investigated, and TroIRF3 was knocked down using siRNA administration. The TroIRF3 gene was decreased in vivo using siRNA (Figure S1B). Compared with the control fish, the number of bacteria in the tissues obviously increased in the siTroIRF3-treated fish (Figure 3B). Compared with the control fish, the TroIRF3 knockdown fish exhibited an increased number of bacteria in the liver by 4.2- and 1.5-fold at 6 and 9 hpi, respectively. At 6, 9 and 12 hpi, the siTroIRF3-treated fish had higher bacterial colony counts in the spleen (15.4-, 3.3- and 4.9-fold). Compared with the control group, the siTroIRF3-treated fish had 9.5- and 2.8-fold higher bacterial loads in the head-kidneys at 6 and 9 hpi, respectively. The PBS-treated group and siTroIRF3-C-treated group (siRNA-C) showed similar bacterial loads, which indicated that the siRNA-C had no effect on the fish’s antibacterial immune responses. Together, TroIRF3 was involved in the antibacterial response against V. harveyi in vivo.
3.5 Mapping of the NLS domain of IRF3
The subcellular localisation of TroIRF3 with or without V. harveyi stimulation was studied using a fluorescence microscope. EGFP is detected in both cytoplasm and nucleus, TroIRF3 was located exclusively in the cytoplasm in the uninfected cells and translocated to the nucleus after infection with V. harveyi (Figure 4A). Given the presence of both NLS and NES in IRF3, its predominant cytoplasmic localisation suggested constitutive activities of NES before virus infection, nuclear exports dominated nuclear imports (45). The regulation function of IRF3 was dependent on its nuclear location. To define the TroIRF3’s NLS domain, the NES was inactivated to prevent interference. The protein of TroIRF3 (△NES) was located in the nucleus and cytoplasm of the GPS cells, suggesting that TroIRF3 is capable of entering and staying in the nucleus without NES (Figure 4C). To produce different mutants, TroIRF3’s N-terminal and C-terminal regions were truncated (Figure 4B). The C-terminal truncations (aa 1–417 and 1–165) and N-terminal truncations (aa 64–466) fused with GFP retained translocation into the nuclei, but the aa 127-466 did not (Figure 4C). According to these experiments, the TroIRF3’s NLS domain is located at the N-terminus of aa 64–127.
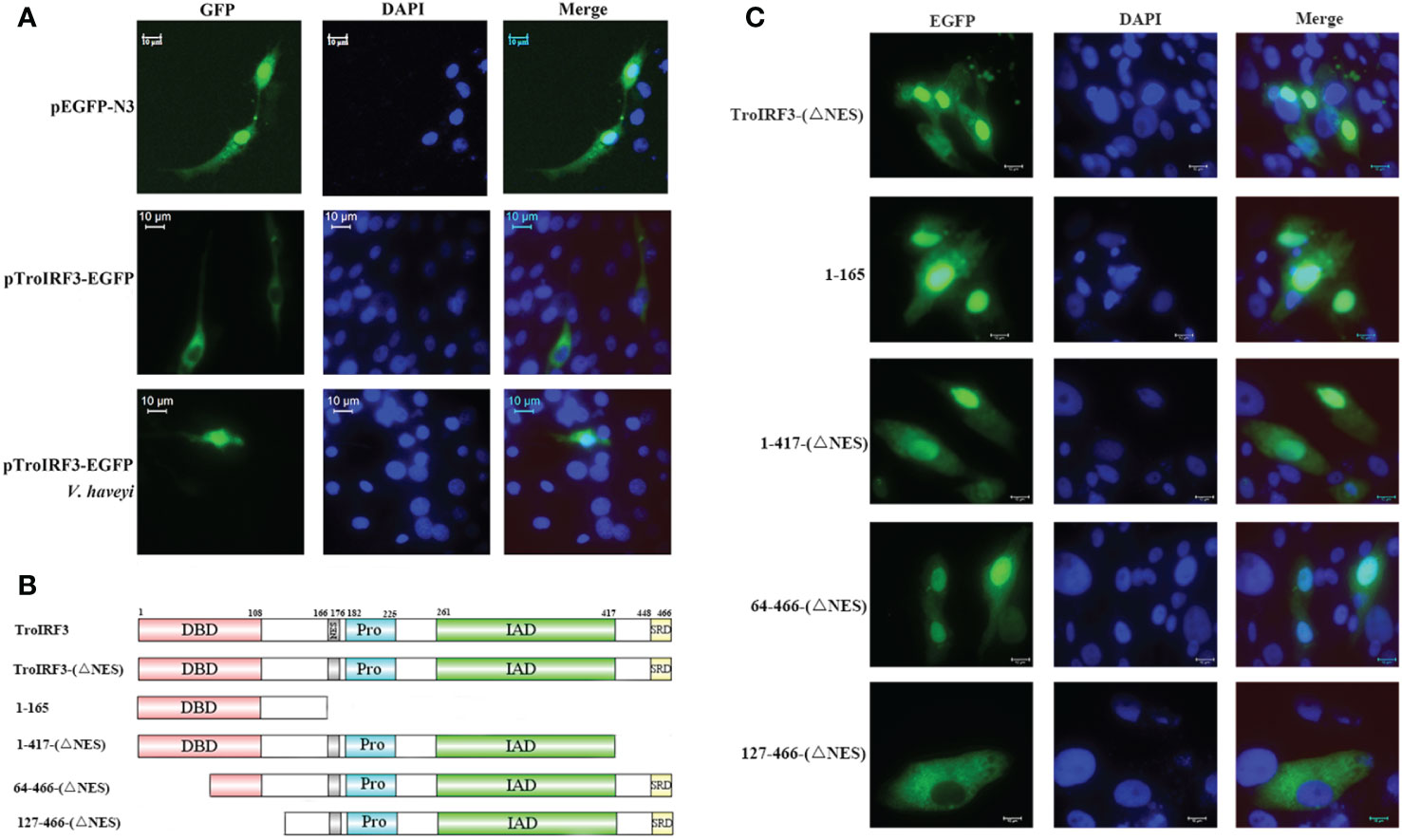
Figure 4 Mapping of the TroIRF3 NLS domain. (A) GPS cells were transfected with pEGFP-N3 or pTroIRF3-N3 for 48 h, and the cell transfected with pTroIRF3-N3 were incubated with PBS or V. haveyi for six hours, followed by inverted fluorescence microscopy imaging. DAPI was used to stain the nuclei of cells. (B) Schematic presentation of TroIRF3-GFP truncation mutants. (C) GPS cells were transfected with TroIRF3-GFP truncation mutant plasmids for 48 h before examination by an inverted fluorescence microscope.
3.6 Mutational analysis of IRF3 NLS
According to a previous study, mice IRF3 nuclear import requires both basic residue clusters (KR77/78 and RK86/87) (45). The TroIRF3’s NLS domain lies between amino acids 64 and 127. Site-directed mutagenesis was then constructed to determine whether they contribute to the nuclear accumulation of TroIRF3. In TroIRF3 mutants (KR74/75NG and RAK82/84LAQ), basic amino acids (KR74/75 and RK82/84) were replaced with uncharged amino acids (Figure 5A). The results of the subcellular localisation assays showed that V. harveyi infection caused TroIRF3, but not KR74/75NG and RK82/84LQ, to be localized in the nucleus (Figures 4A, 5B). Therefore, the NLS of TroIRF3 is composed of amino acid residues KR74/75 and RK82/84, which are critical in TroIRF3 nuclear accumulation.
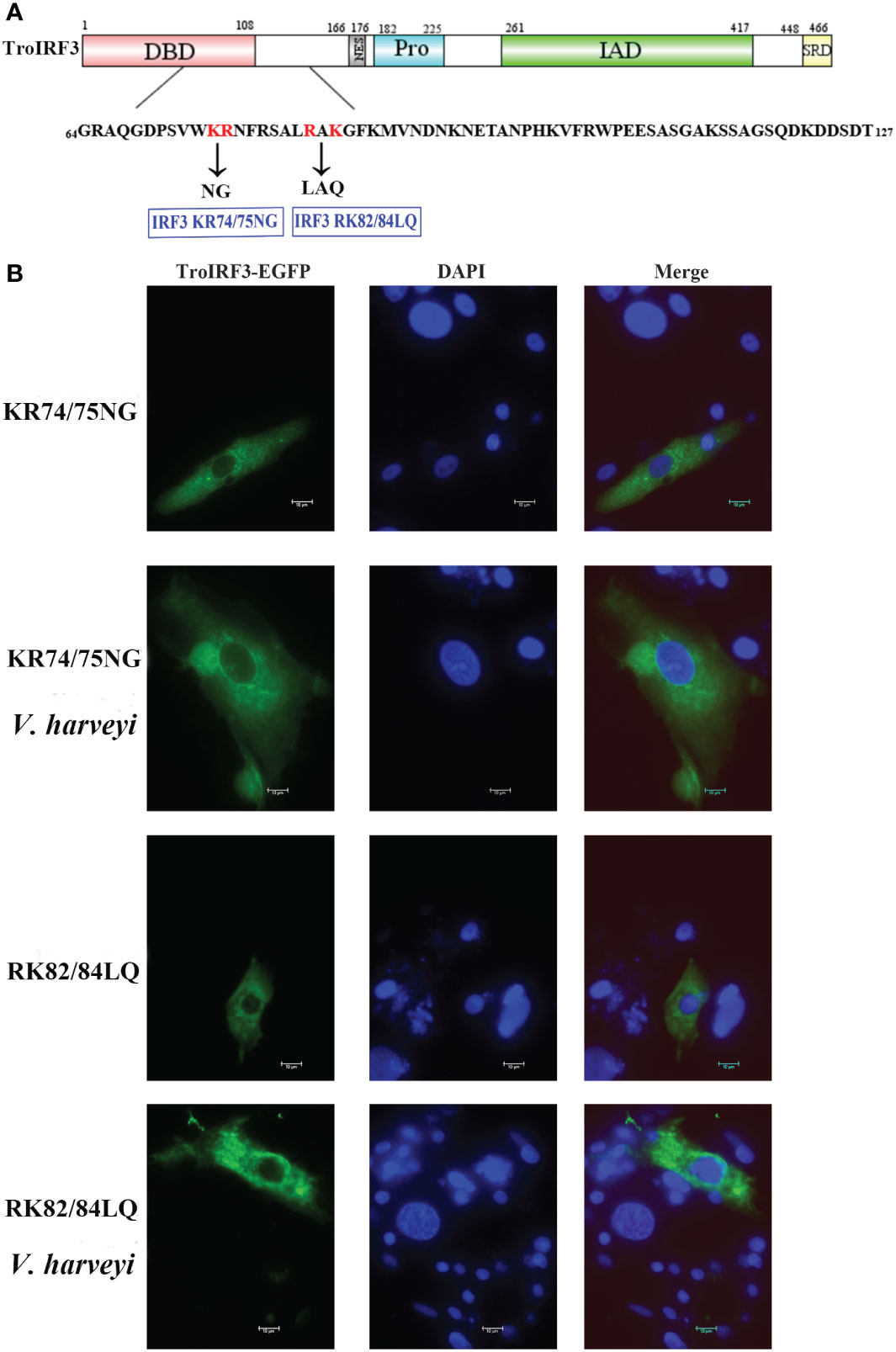
Figure 5 Subcellular distribution of nuclear import of TroIRF3 variants. (A) Schematic presentation of different TroIRF3-GFP variants. (B) The subcellular distribution of TroIRF3 variants with or without V. haveyi infection. GPS cells seeded overnight on a microscope in 24-well plates were transiently transfected with TroIRF3 variants for 48 h. The cells were stimulated by V. haveyi for 6 h, followed by inverted fluorescence microscopy imaging. DAPI was used to stain the nuclei of cells.
3.7 Antibacterial function of TroIRF3 requires NLS
To determine whether NLS affects TroIRF3-mediated antibacterial defences, the number of V. harveyi in the fish treated with pTroIRF3 or the indicated TroIRF3 mutants (pTroIRF3-KR74/75NG and pTroIRF3-RK82/84LQ) was tested. TroIRF3 overexpression significantly suppressed V. harveyi replication in the fish, which was in line with our previous observations (Figure 6). However, the number of invaded bacteria in pTroIRF3-KR74/75NG- or pTroIRF3-RK82/84LQ-treated fish was enhanced compared with that in the wild-type pTroIRF3-treated fish (Figure 6). These results showed that basic amino acids KR74/75 and RK82/84 are required for the efficient antibacterial function of TroIRF3.
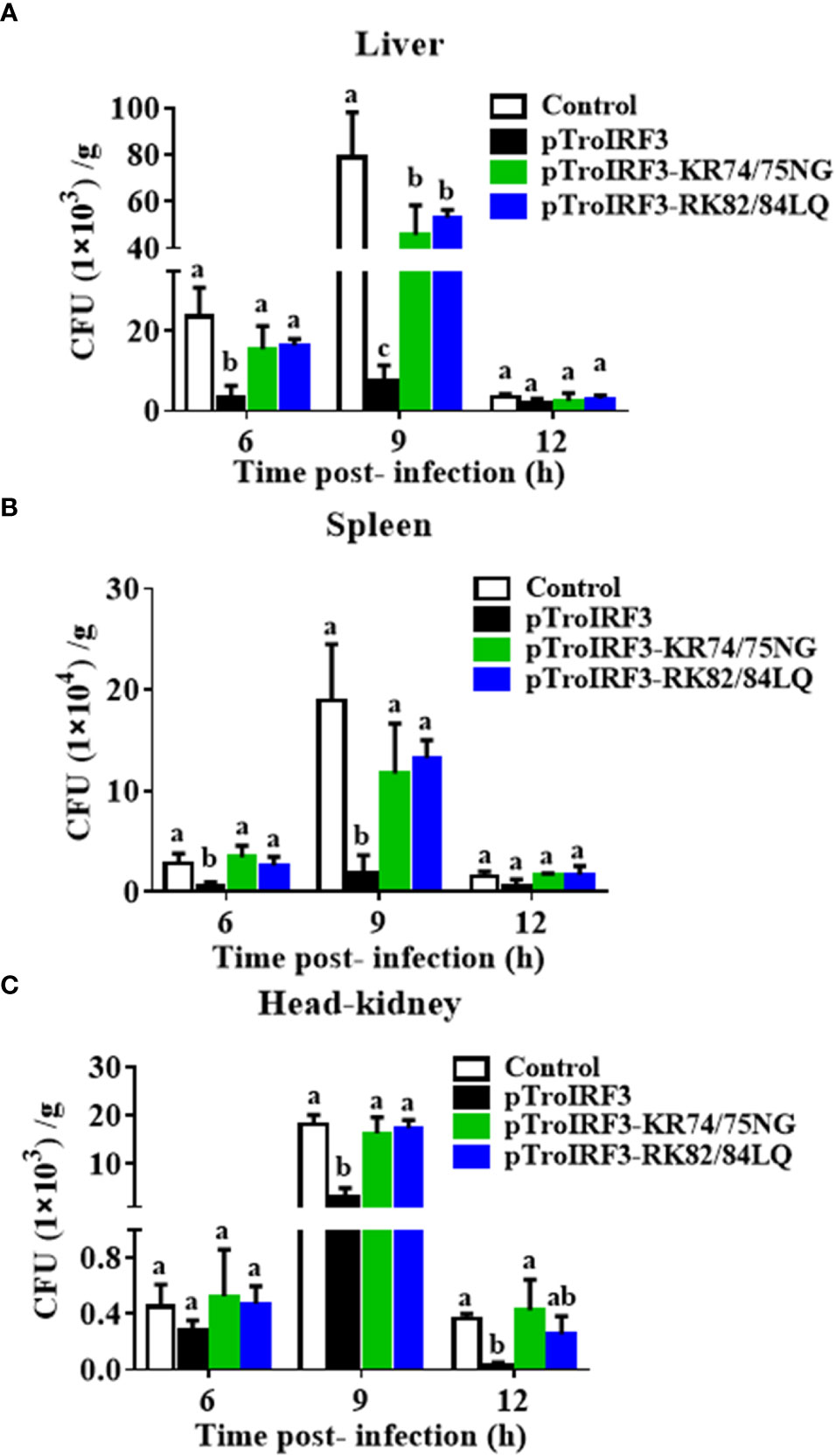
Figure 6 The function of NLS in antibacterial responses regulated by TroIRF3. T. ovatus were administered with PBS (control), pTroIRF3, pTroIRF3-KR74/75NG, or pTroIRF3-RK82/84LQ for 5 days, then injected intraperitoneally with V. harveyi, and the number of bacteria in the liver (A), spleen (B), and head-kidney (C) at different time points were quantified using the plate counting method. Data are expressed as means ± SD (N = 3). N, the number of fish used at each time point per group. Significant differences (P < 0.05) are indicated by different letters, and no significant differences are indicated by the same letters.
3.8 TroIRF3-mediated immune gene expression requires an intact NLS
To investigate the effects of NLS on the TroIRF3-mediated immune gene expression, the expression levels of immune genes (IFNa3, MXI, TRAF6, Viperin1 and MAVS) in the spleen of the fish treated with pTroIRF3 or the indicated TroIRF3 mutants (pTroIRF3-KR74/75NG and pTroIRF3-RK82/84LQ) were tested. In the pTroIRF3-, pTroIRF3-KR74/75NG- and pTroIRF3-RK82/84LQ-treated fish, the expression level of TroIRF3 and modified TroIRF3 mutants were significantly upregulated compared with the control fish (Figure S2). TroIRF3 remarkably enhanced the production of IFNa3, MXI, TRAF6, Viperin1 and MAVS. However, the expression level of the immune genes (IFNa3, MXI, TRAF6, Viperin1 and MAVS) in the pTroIRF3-KR74/75NG- or pTroIRF3-RK82/84LQ-treated fish was significantly reduced compared with that in the pTroIRF3-treated fish (Figure 7). Furthermore, the fish treated with pCN3, pTroIRF3- KR74/75NG or pTroIRF3-RK82/84LQ showed the same expression level of immune genes (Figure 7). These results showed that basic amino acids KR74/75 and RK82/84 are required for immune gene induction by TroIRF3.
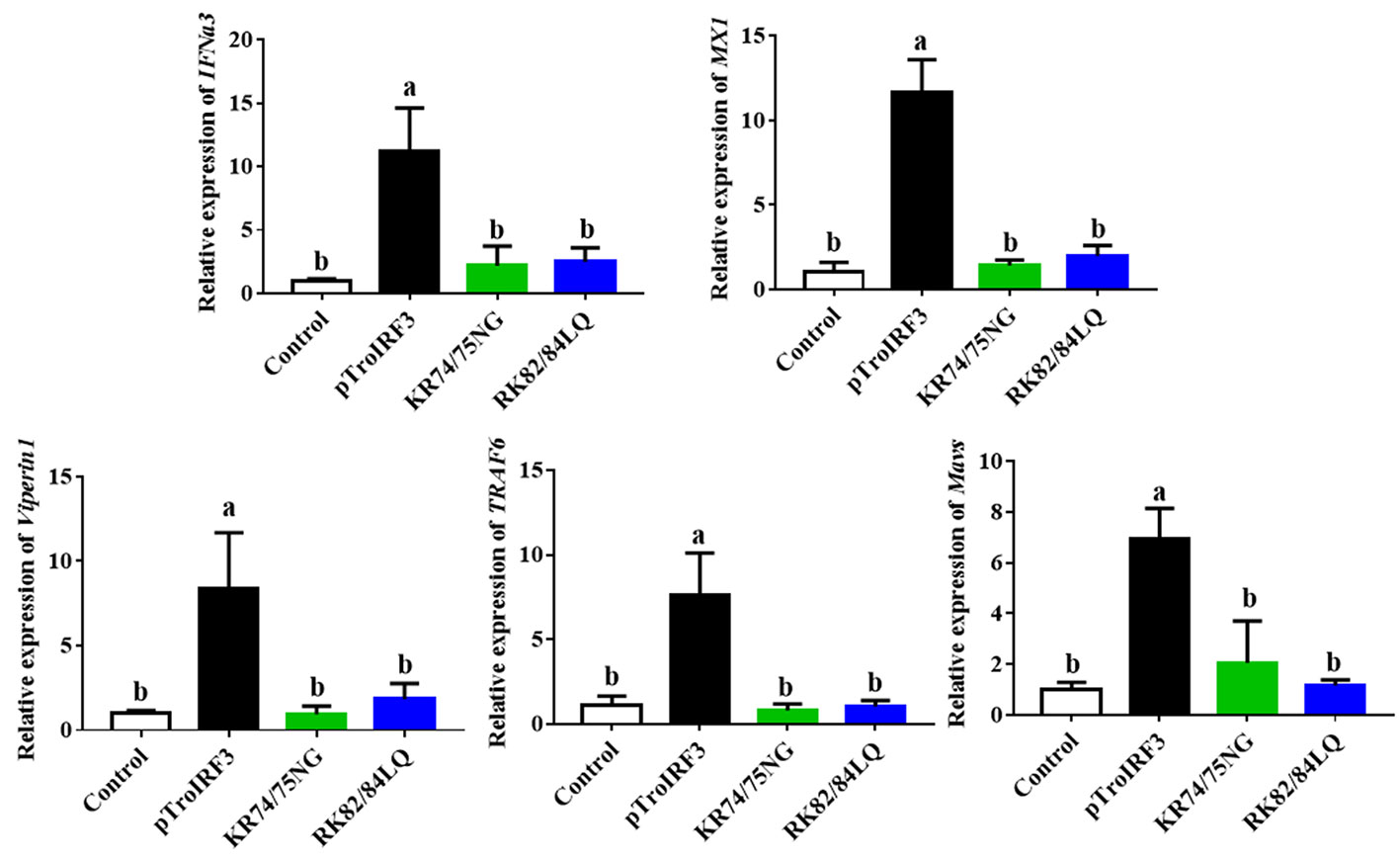
Figure 7 The function of NLS in the expression of interferon signaling molecules induced by TroIRF3. Golden pompano injected with pTroIRF3, pTroIRF3-KR74/75NG, pTroIRF3-RK82/84LQ, or PBS (control). After five days, the expression level of interferon signaling molecules in the spleen was detected. The results represent the mean ± SD (N=3), and N indicates the number of fish used per group. Significant differences (P < 0.05) are indicated by different letters, and no significant differences are indicated by the same letters.
3.9 TroIFNa3 activation mediated by TroIRF3
Based on the predicted binding sites, consecutive truncated mutants of the TroIFNa3 promoter were constructed to investigate the TroIRF3-binding region in the TroIFNa3 promoter (Figure 8A). TroIFNa3-p2 had a higher response to TroIRF3 than the other mutants, suggesting that the region consists of –896 bp to +1 bp from the transcriptional start site, which contains the binding site for IRF3 (Figure 8A). The activation of the TroIFNa3 promoter by TroIRF3 was further enhanced by V. harveyi stimulation (Figure 8B). Furthermore, the activation of TroIFNa3-p2 is dependent on the TroIRF3 concentration (Figure 8C).
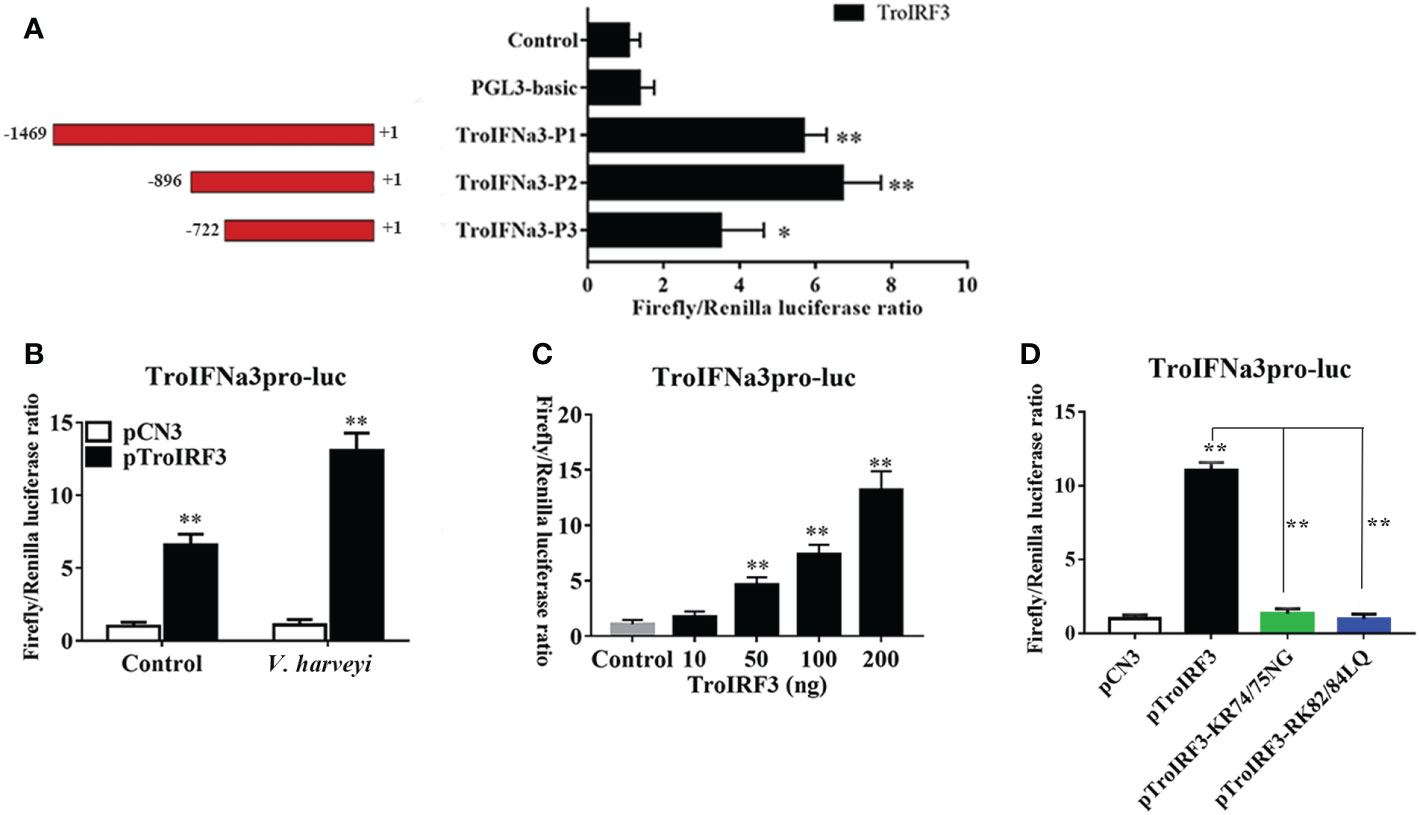
Figure 8 TroIRF3 regulated the activity of the TroIFNa3 promoter. (A) Three truncates of the TroIFNa3 promoter were constructed and transfected with pTroIRF3 into HEK 293T cells for 24 h, then harvested for detection of luciferase activity. (B) Activation of the TroIFNa3 promoter by TroIRF3. HEK293T cells seeded in 24-well plates were co-transfected with three plasmid sets: pCN3 (0.25 μg), pGL4-TroIFNa3-pro (0.25 μg), and pRL-TK; pTroIRF3 (0.25 μg), pGL4-TroIFNa3-pro (0.25 μg) and pRL-TK (0.025 μg). Cells were stimulated with or without V. harveyi after 24 h post-transfection, then harvested for detection of luciferase activity. (C) HEK293T cells seeded in 24-well plates were transfected with 0, 10, 50, 100, and 200 ng of pTroIRF3 along with pGL4-TroIFNa3-pro (0.25 μg) and pRL-TK (0.025 μg), then harvested for detection of luciferase activity. (D) The effect of NLS on TroIFNa3 promoter activation induced by TroIRF3. HEK293T cells seeded in 24-well plates were co-transfected with TroIFNa3 promoter (0.25 μg), as well as with pCN3, pTroIRF3, pTroIRF3-KR74/75NG, or pTroIRF3-RK82/84LQ (0.25 μg), plus 0.025 mg of pRL-CMV as an internal control. Cells were stimulated with V. harveyi for 12 h after 24 h post-transfection, then harvested for detection of luciferase activity. The values are represented as mean ± SD (N = 3). N, parallel experiments. *P < 0.05, **P < 0.01.
Subsequently, the potential effects of NLS on TroIRF3-mediated TroIFNa3-p2 activation were investigated using the luciferase reporter assay. TroIRF3 exhibited remarkably enhanced TroIFNa3-p2 activation following V. harveyi infection, whereas the other variants (pTroIRF3-KR74/75NG and pTroIRF3-RK82/84LQ) did not exhibit TroIFNa3-p2 activation under the same conditions (Figure 8D).
4 Discussion
IRF3 activates type I IFN and ISGs, which function as defences against viral and bacterial infections (46). TroIRF3 was identified and characterised in the present study. In addition, the functions of TroIRF3 were investigated, including the intracellular localisation of TroIRF3 and the ability of TroIRF3 to be an activator in antibacterial response and initiator of the transcription of TroIFNa3.
From fish to mammals, the DBD, IAD and SRD domains are conserved in vertebrate IRF3. The DBD domain consists of highly conserved tryptophan residues and forms a helix-turn-helix motif that binds to the ISRE in target promoters (33, 47, 48). After activation, the IRF3 molecules homodimerise through the IAD motif (49). SRD is critical for the phosphorylation IRF3 (12, 14). As with the other IRF3, TroIRF3 has a DBD on the N-terminus and IAD and SRD at the C-terminus. In the phylogenetic analysis, IRF3 from the Osteichthyes species and higher vertebrates cluster into two major groups. TroIRF3 is closely related to S. lalandi dorsalis IRF3, which belongs to Perciformes. Given its conserved structure and evolutionary history, TroIRF3 may play a conserved role in the immune response against pathogens.
Studies have found that IRF3 is predominantly expressed in the immune organs of several species, including Squaliobarbus curriculus and E. coioides (32, 50). In this study, TroIRF3 was most prominent in the gill and liver while relatively low in the spleen and blood. The gill and liver play a role in immunology or act as entry points for pathogens. TroIRF3 showed the highest expression level in gill, which may account for TroIRF3 could be involvement in the early immune response. The spleen and head-kidney from Odontobutis obscura also showed relatively low IRF3 expression levels (51). This observation may be attributed to the fact that the mRNA expression check was conducted in healthy fish, where IRF3 was not required for upregulation in the spleen and head-kidney (51). LPS treatment induces IRF3 transcripts in some fish species, such as L. crocea and Ctenopharyngodon idella (52, 53). In vivo, TroIRF3 transcript levels were also significantly upregulated following V. harveyi treatment. According to these results, T. ovatus’ innate immune response against V. harveyi may be mediated by TroIRF3.
In mammals, IRF3 plays a role in antiviral immunity. However, recent studies found that IRF3 also participates in bacterial immunity (54–56). In A549 cells, siRNA inhibiting IRF3 expression resulted in an overall increase in Legionella numbers (57). Chlamydophila pneumoniae replication was enhanced when IRF3 expression was inhibited by siRNA and attenuated by IFN-β treatment (58). Mice IRF3 deficiency leads to impaired clearance of P. aeruginosa from the lung (56). In this study, we showed that the bacterial load reduction in fish tissues was caused by TroIRF3 overexpression. Consistent with these findings, the inhibition of TroIRF3 expression by siTroIRF3 led to an increase in the number of bacteria. In addition, TroIRF3 upregulated key genes, such as IFNa3, MXI, TRAF6, Viperin1 and MAVS, which are involved in IFN/IRF-based signalling. IFN, MXI and Viperin have several functions that participate in the antibacterial and antiviral immune response (59–61). Moreover, TRAF6 and MAVS participate in the activation of IRF3 (62). These findings suggested that the overexpression of TroIRF3 upregulated the expression of IFN and IFN-related genes, leading to an antibacterial state to inhibit V. harveyi replication.
Under healthy conditions, mice IRF3 is mostly found in the cytoplasm of cell. After infection with P. aeruginosa, it is phosphorylated and transported to the nuclei (56). However, fish IRF3 localization varies. C. auratus L. IRF3 was located in the cytoplasm without stimuli but was observed in the nuclei after a poly I: C challenge (33). Oncorhynchus mykiss IRF3 is localized in both cytoplasm and nuclei with or without poly I: C and no apparent transfer from the cytoplasm to the nucleus was observed (63). When no bacterial infection is present, TroIRF3 initially resides in the GPS cells’ cytoplasm but undergoes nuclear translocation upon bacterial stimulation. The result suggests that TroIRF3 originally resided in the cytoplasm in an inactive form when no bacterial contamination was present, but when bacteria stimulated the cell, it was activated and translocated into the nucleus. Therefore, TroIRF3 is involved in antibacterial immunity.
IRF proteins contain both NLSs and NESs, so their subcellular distribution can be controlled to manage the IRF function (25). Before bacterial infection, TroIRF3 is predominantly localized in the cytoplasm, suggesting that nuclear export dominates nuclear import. As a result of bacterial infection, TroIRF3 resides mainly in the nucleus, which indicates that nuclear import dominates when TroIRF3 is activated. TroIRF3’s NLS was mapped to aa 64–127, which contains an α-helical pattern. The basic amino acids KR74/75 and RK82/84 are integral to TroIRF3’s nuclear import. KR74/75 and RK82/84 are 90° amino acids apart, which makes a spatial distance of one amino acid between these two clusters that interact with the binding sites in importin-α (Figure S3). Similarly, mice IRF3 localizes to the nucleus through a bipartite NLS located at the RK86/87 and KR77/78 sites (45). These results revealed that the amino acids that affected IRF3 nuclear localisation were conserved.
We further analysed the role of NLS in TroIRF3-mediated antibacterial responses and type I IFN expression. Our results showed that KR74/75 and RK82/84 in the NLS of TroIRF3 are involved in antibacterial immunity and immune gene (IFNa3, MXI, TRAF6, Viperin1 and MAVS) upregulation induced by TroIRF3. Similarly, mice IRF3 NLS plays an important role in the IFN response and antiviral immunity mediated by IRF3 (45). Our findings revealed that the NLSs enforced the nuclear localisation of TroIRF3 and were essential to antibacterial immunity in T. ovatus.
In DNA or RNA viral infection defence, vertebrate IRF3 is the key transcription factor in the signalling of type I interferon-dependent immune responses (18, 64, 65). Mammalian IRF3 has been shown to bind to the promoters of IFN and ISG and activate their transcription (66). As shown by the luciferase activity, the ectopic expression of TroIRF3 activated the TroIFNa3 promoter. The activation was dependent on the TroIRF3 concentration and further enhanced by V. harveyi stimulation. These results may reveal that TroIRF3 can activate TroIFNa3-mediated antibacterial responses. Similar results were also obtained in other species, such as Miichthys miiuy, Lateolabrax japonicus and Oreochromis niloticus. The IRF3 protein of O. niloticus enhances the activity of the IFN-β promoter, which plays a role in regulating the IFN response (67). M. miiuy IRF3 can activate the IFNα promoter, and the expressions increased with the increase in mmIRF3 quality (68). L. japonicus IRF3 overexpression significantly increased the promoter activity of zebrafish IFN1 (69). Furthermore, the amino acid residues KR74/75 and RK82/84 in TroIRF3’s NLS were essential for activating TroIFNa3-p2. These results showed that TroIRF3 acted as a transcription activator in immune responses and that the amino acid residues KR74/75 and RK82/84 are important to the immune regulation of TroIRF3.
In summary, the present study cloned and characterised TroIRF3. V. harveyi infection significantly increased TroIRF3 expression and shuttled it from the cytoplasm into the nucleus. TroIRF3 played an important role in the antibacterial immune response. Basic amino acids KR74/75 and RK82/84 were required for the efficient nuclear import of TroIRF3 and played a key role in the antibacterial immune response and upregulation of immune genes induced by TroIRF3. Furthermore, TroIRF3 activated the promoter of TroIFNa3, and the key amino acids KR74/75 and RK82/84 were essential for TroIFNa3-p2 activation induced by TroIRF3.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Animal Care and Use Committee of the Hainan University.
Author contributions
YS, ZC, YW, and YZ designed the research. PZ, CW, YS, and WY analyzed the data. YW and YZ supervised the study. YS, YW, ZC, and JL performed the research, analyzed the data, and wrote the manuscript with support from all authors. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (U22A20534, 42066007), and the Nanhai Famous Youth Project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1128196/full#supplementary-material
References
1. Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity (2006) 25:373–81. doi: 10.1016/j.immuni.2006.08.007
2. Gonzalez-Navajas JM, Lee J, David M and Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol (2012) 12:125–35. doi: 10.1038/nri3133
3. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol (2014) 14:36–49. doi: 10.1038/nri3581
4. Zou J, Tafalla C, Truckle J and Secombes CJ. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J Immunol (2007) 179:3859–71. doi: 10.4049/jimmunol.179.6.3859
5. Lopez-Munoz A, Roca FJ, Meseguer J and Mulero V. New insights into the evolution of IFNs: zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities. J Immunol (2009) 182:3440–9. doi: 10.4049/jimmunol.0802528
6. Zou J, Gorgoglione B, Taylor NGH, Summathed T, Lee PT, Panigrahi A, et al. Salmonids have an extraordinary complex type I IFN system: Characterization of the IFN oocus in rainbow trout Oncorhynchus mykiss reveals two novel IFN subgroups. J Of Immunol (2014) 193:2273–86. doi: 10.4049/jimmunol.1301796
7. Gan Z, Chen SN, Huang B, Zou J and Nie P. Fish type I and type II interferons: Composition, receptor usage, production and function. Rev In Aquacult (2020) 12:773–804. doi: 10.1111/raq.12349
8. Lu MM, Yang CY, Li MJ, Yi QL, Lu GX, Wu YC, et al. A conserved interferon regulation factor 1 (IRF-1) from pacific oyster Crassostrea gigas functioned as an activator of IFN pathway. Fish Shellfish Immunol (2018) 76:68–77. doi: 10.1016/j.fsi.2018.02.024
9. Stein C, Caccamo M, Laird G and Leptin M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol (2007) 8:R251. doi: 10.1186/gb-2007-8-11-r251
10. Panne D, Maniatis T and Harrison SC. An atomic model of the interferon-β enhanceosome. Cell (2007) 129:1111–23. doi: 10.1016/j.cell.2007.05.019
11. Panne D, McWhirter SM, Maniatis T and Harrison SC. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J Biol Chem (2007) 282:22816–22. doi: 10.1074/jbc.M703019200
12. Clement JF, Bibeau-Poirier A, Gravel SP, Grandvaux N, Bonneil E, Thibault P, et al. Phosphorylation of IRF-3 on ser 339 generates a hyperactive form of IRF-3 through regulation of dimerization and CBP association. J Virol (2008) 82:3984–96. doi: 10.1128/JVI.02526-07
13. Bergstroem B, Johnsen IB, Nguyen TT, Hagen L, Slupphaug G, Thommesen L, et al. Identification of a novel in vivo virus-targeted phosphorylation site in interferon regulatory factor-3 (IRF3). J Biol Chem (2010) 285:24904–14. doi: 10.1074/jbc.M109.084822
14. Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol (1998) 18:2986–96. doi: 10.1128/mcb.18.5.2986
15. Honda K, Takaoka A and Taniguchi T. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity (2006) 25:349–60. doi: 10.1016/j.immuni.2006.08.009
16. Ding Y, Ao JQ, Huang XH, Chen XH. Identification of two subgroups of type I IFNs in perciforme fish large yellow croaker Larimichthys crocea provides novel insights into function and regulation of fish type I IFNs. Front In Immunol (2016) 7:343. doi: 10.3389/Fimmu.2016.00343
17. Rajput A, Kovalenko A, Bogdanov K, Yang SH, Kang TB, Kim JC, et al. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8-Mediated cleavage of the RIP1 protein. Immunity (2011) 34:340–51. doi: 10.1016/j.immuni.2010.12.018
18. Honda K, Taniguchi T. IRFs: master regulators of signalling by toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol (2006) 6:644–58. doi: 10.1038/nri1900
19. Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature (2007) 448:501–5. doi: 10.1038/nature06013
20. Leulier F, Lemaitre B. Toll-like receptors–taking an evolutionary approach. Nat Rev Genet (2008) 9:165–78. doi: 10.1038/nrg2303
21. Suzuki N, Suzuki S and Yeh WC. IRAK-4 as the central TIR signaling mediator in innate immunity. Trends Immunol (2002) 23:503–6. doi: 10.1016/s1471-4906(02)02298-6
22. Janssens S, Beyaert R. Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol Cell (2003) 11:293–302. doi: 10.1016/s1097-2765(03)00053-4
23. Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PloS Pathog (2013) 9:e1003118. doi: 10.1371/journal.ppat.1003118
24. Tamura T, Yanai H, Savitsky D and Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol (2008) 26:535–84. doi: 10.1146/annurev.immunol.26.021607.090400
25. Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin α. J Biol Chem (2007) 282:5101–5. doi: 10.1074/jbc.R600026200
26. Servant MJ, Grandvaux N and Hiscott J. Multiple signaling pathways leading to the activation of interferon regulatory factor 3. Biochem Pharmacol (2002) 64:985–92. doi: 10.1016/s0006-2952(02)01165-6
27. Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E and Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J (1998) 17:1087–95. doi: 10.1093/emboj/17.4.1087
28. Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, et al. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity (2002) 17:251–63. doi: 10.1016/s1074-7613(02)00390-4
29. Doyle SE, O'Connell R, Vaidya SA, Chow EK, Yee K and Cheng G. Toll-like receptor 3 mediates a more potent antiviral response than toll-like receptor 4. J Immunol (2003) 170:3565–71. doi: 10.4049/jimmunol.170.7.3565
30. Kawai T, Takeuchi O, Fujita T, Inoue J, Muhlradt PF, Sato S, et al. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J Immunol (2001) 167:5887–94. doi: 10.4049/jimmunol.167.10.5887
31. Sakaguchi S, Negishi H, Asagiri M, Nakajima C, Mizutani T, Takaoka A, et al. Essential role of IRF-3 in lipopolysaccharide-induced interferon-β gene expression and endotoxin shock. Biochem Biophys Res Commun (2003) 306:860–6. doi: 10.1016/s0006-291x(03)01049-0
32. Huang Y, Huang X, Cai J, OuYang Z, Wei S, Wei J, et al. Identification of orange-spotted grouper (Epinephelus coioides) interferon regulatory factor 3 involved in antiviral immune response against fish RNA virus. Fish Shellfish Immunol (2015) 42:345–52. doi: 10.1016/j.fsi.2014.11.025
33. Sun F, Zhang YB, Liu TK, Gan L, Yu FF, Liu Y, et al. Characterization of fish IRF3 as an IFN-inducible protein reveals evolving regulation of IFN response in vertebrates. J Of Immunol (2010) 185:7573–82. doi: 10.4049/jimmunol.1002401
34. Zhang J, Li YX, Hu YH. Molecular characterization and expression analysis of eleven interferon regulatory factors in half-smooth tongue sole, Cynoglossus semilaevis. Fish Shellfish Immunol (2015) 44:272–82. doi: 10.1016/j.fsi.2015.02.033
35. Iliev DB, Sobhkhez M. Fremmerlid K and Jorgensen J b, MyD88 interacts with interferon regulatory factor (IRF) 3 and IRF7 in Atlantic salmon (Salmo salar): Transgenic SsMyD88 modulates the IRF-induced type I interferon response and accumulates in aggresomes. J Biol Chem (2011) 286:42715–24. doi: 10.1074/jbc.M111.293969
36. Yao CL, Huang XN, Fan Z, Kong P and Wang ZY. Cloning and expression analysis of interferon regulatory factor (IRF) 3 and 7 in large yellow croaker. Larimichthys crocea Fish Shellfish Immunol (2012) 32:869–78. doi: 10.1016/j.fsi.2012.02.015
37. Ohtani M, Hikima J, Hwang SD, Morita T, Suzuki Y, Kato G, et al. Transcriptional regulation of type I interferon gene expression by interferon regulatory factor-3 in Japanese flounder. Paralichthys olivaceus Dev Comp Immunol (2012) 36:697–706. doi: 10.1016/j.dci.2011.10.008
38. Tu Z, Li H, Zhang X, Sun Y and Zhou Y. Complete genome sequence and comparative genomics of the golden pompano (Trachinotus ovatus) pathogen, Vibrio harveyi strain QT520. PeerJ (2017) 5:e4127. doi: 10.7717/peerj.4127
39. Chen Y, Zhou Y, Yang X, Cao Z, Chen X, Qin Q, et al. Insulin-like growth factor binding protein 3 gene of golden pompano (TroIGFBP3) promotes antimicrobial immune defense. Fish Shellfish Immunol (2020) 103:47–57. doi: 10.1016/j.fsi.2020.04.002
40. Wu Y, Zhou Y, Cao Z, Sun Y, Chen Y, Xiang Y, et al. Comparative analysis of the expression patterns of IL-1β, IL-11, and IL-34 in golden pompano (Trachinotus ovatus) following different pathogens challenge. Fish Shellfish Immunol (2019) 93:863–70. doi: 10.1016/j.fsi.2019.08.018
41. Zhu KC, Zhang N, Liu BS, Guo L, Guo HY, Jiang SG, et al. Functional analysis of IRF1 reveals its role in the activation of the type I IFN pathway in golden pompano, Trachinotus ovatus (Linnaeus 1758). Int J Of Mol Sci (2020) 21:2652. doi: 10.3390/Ijms21072652
42. Wu Y, Zhou Y, Cao Z, Chen X, Du H and Sun Y. Interferon regulatory factor 7 contributes to the host response during Vibrio harveyi infection in the golden pompano Trachinotus ovatus. Dev Comp Immunol (2021) 117:103959. doi: 10.1016/j.dci.2020.103959
43. Yu Y, Wei S, Wang Z, Huang X, Huang Y, Cai J, et al. Establishment of a new cell line from the snout tissue of golden pompano Trachinotus ovatus, and its application in virus susceptibility. J Fish Biol (2016) 88:2251–62. doi: 10.1111/jfb.12986
44. Chen XJ, Zhang XQ, Huang S, Cao ZJ, Qin QW, Hu WT, et al. Selection of reference genes for quantitative real-time RT-PCR on gene expression in golden pompano (Trachinotus ovatus). Polish J Of Veterinary Sci (2017) 20:583–94. doi: 10.1515/pjvs-2017-0071
45. Zhu M, Fang T, Li S, Meng K and Guo D. Bipartite nuclear localization signal controls nuclear import and DNA-binding activity of IFN regulatory factor 3. J Immunol (2015) 195:289–97. doi: 10.4049/jimmunol.1500232
46. Ysebrant de Lendonck L, Martinet V and Goriely S. Interferon regulatory factor 3 in adaptive immune responses. Cell Mol Life Sci (2014) 71:3873–83. doi: 10.1007/s00018-014-1653-9
47. Csumita M, Csermely A, Horvath A, Nagy G, Monori F, Goczi L, et al. Specific enhancer selection by IRF3, IRF5 and IRF9 is determined by ISRE half-sites, 5 ' and 3 ' flanking bases, collaborating transcription factors and the chromatin environment in a combinatorial fashion. Nucleic Acids Res (2020) 48:589–604. doi: 10.1093/nar/gkz1112
48. Petro TM. IFN regulatory factor 3 in health and disease. J Immunol (2020) 205:1981–89. doi: 10.4049/jimmunol.2000462
49. Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem (2007) 282:15325–9. doi: 10.1074/jbc.R700002200
50. Wang R, Li Y, Zhou Z, Liu Q, Zeng L and Xiao T. Involvement of interferon regulatory factor 3 from the barbel chub Squaliobarbus curriculus in the immune response against grass carp reovirus. Gene (2018) 648:5–11. doi: 10.1016/j.gene.2018.01.048
51. Li Z, Chen J, Li P, Li XY, Lu L and Li S. Functional characterization of dark sleeper (Odontobutis obscura) IRF3 in IFN regulation. Fish Shellfish Immunol (2019) 89:411–19. doi: 10.1016/j.fsi.2019.04.019
52. Yu N, Xu X, Qi G, Liu D, Chen X, Ran X, et al. Ctenopharyngodon idella TBK1 activates innate immune response via IRF7. Fish Shellfish Immunol (2018) 80:521–27. doi: 10.1016/j.fsi.2018.06.045
53. Zou PF, Shen JJ, Li Y, Zhang ZP, Wang YL. TRAF3 enhances TRIF-mediated signaling via NF-κB and IRF3 activation in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol (2020) 97:114–24. doi: 10.1016/j.fsi.2019.12.024
54. Juang YT, Lowther W, Kellum M, Au WC, Lin R, Hiscott J, et al. Primary activation of interferon a and interferon b gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci U.S.A. (1998) 95:9837–42. doi: 10.1073/pnas.95.17.9837
55. Yamashiro T, Sakamoto N, Kurosaki M, Kanazawa N, Tanabe Y, Nakagawa M, et al. Negative regulation of intracellular hepatitis c virus replication by interferon regulatory factor 3. J Gastroenterol (2006) 41:750–7. doi: 10.1007/s00535-006-1842-x
56. Carrigan SO, Junkins R, Yang YJ, Macneil A, Richardson C, Johnston B, et al. IFN regulatory factor 3 contributes to the host response during Pseudomonas aeruginosa lung infection in mice. J Immunol (2010) 185:3602–9. doi: 10.4049/jimmunol.0903429
57. Opitz B, Vinzing M, van Laak V, Schmeck B, Heine G, Gunther S, et al. Legionella pneumophila induces IFNβ in lung epithelial cells via IPS-1 and IRF3, which also control bacterial replication. J Biol Chem (2006) 281:36173–9. doi: 10.1074/jbc.M604638200
58. Buss C, Opitz B, Hocke AC, Lippmann J, van Laak V, Hippenstiel S, et al. Essential role of mitochondrial antiviral signaling, IFN regulatory factor (IRF)3, and IRF7 in Chlamydophila pneumoniae-mediated IFN-β response and control of bacterial replication in human endothelial cells. J Immunol (2010) 184:3072–8. doi: 10.4049/jimmunol.0902947
59. Helbig KJ, Teh MY, Crosse KM, Monson EA, Smith M, Tran EN, et al. The interferon stimulated gene viperin, restricts shigella. flexneri vitro Sci Rep (2019) 9:15598. doi: 10.1038/s41598-019-52130-8
60. Boxx GM, Cheng G. The roles of type I interferon in bacterial infection. Cell Host Microbe (2016) 19:760–9. doi: 10.1016/j.chom.2016.05.016
61. Teles RM, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science (2013) 339:1448–53. doi: 10.1126/science.1233665
62. Chen H, Jiang Z. The essential adaptors of innate immune signaling. Protein Cell (2013) 4:27–39. doi: 10.1007/s13238-012-2063-0
63. Holland JW, Bird S, Williamson B, Woudstra C, Mustafa A, Wang T, et al. Molecular characterization of IRF3 and IRF7 in rainbow trout, Oncorhynchus mykiss: Functional analysis and transcriptional modulation. Mol Immunol (2008) 46:269–85. doi: 10.1016/j.molimm.2008.08.265
64. Au WC, Moore PA, LaFleur DW, Tombal B, Pitha PM. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon a genes. J Biol Chem (1998) 273:29210–7. doi: 10.1074/jbc.273.44.29210
65. Taniguchi T, Takaoka A. A weak signal for strong responses: Interferon-α/β revisited. Nat Rev Mol Cell Biol (2001) 2:378–86. doi: 10.1038/35073080
66. Paun A, Pitha PM. The IRF family, revisited. Biochimie (2007) 89:744–53. doi: 10.1016/j.biochi.2007.01.014
67. Gu YF, Wei Q, Tang SJ, Chen XW, Zhao JL. Molecular characterization and functional analysis of IRF3 in tilapia (Oreochromis niloticus). Dev Comp Immunol (2016) 55:130–7. doi: 10.1016/j.dci.2015.10.011
68. Shu C, Chu Q, Bi D, Wang Y and Xu T. Identification and functional characterization of miiuy croaker IRF3 as an inducible protein involved regulation of IFN response. Fish Shellfish Immunol (2016) 54:499–506. doi: 10.1016/j.fsi.2016.04.136
Keywords: IRF3, Trachinotus ovatus, antimicrobial immunity, nuclear localization signal, IFNa3
Citation: Sun Y, Cao Z, Zhang P, Wei C, Li J, Wu Y and Zhou Y (2023) IFN regulatory factor 3 of golden pompano and its NLS domain are involved in antibacterial innate immunity and regulate the expression of type I interferon (IFNa3). Front. Immunol. 14:1128196. doi: 10.3389/fimmu.2023.1128196
Received: 20 December 2022; Accepted: 23 January 2023;
Published: 02 February 2023.
Edited by:
Mofei Li, Tianjin Normal University, ChinaReviewed by:
Weiwei Zhang, Ningbo University, ChinaJian Zhang, Yantai University, China
Min Zhang, Qingdao Agricultural University, China
Copyright © 2023 Sun, Cao, Zhang, Wei, Li, Wu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Wu, d3V5aW5nMDk0NzAwOUAxMjYuY29t; Yongcan Zhou, enljaG51QDE2My5jb20=
†These authors have contributed equally to this work
 Yun Sun1,2†
Yun Sun1,2† Jianlong Li
Jianlong Li Ying Wu
Ying Wu Yongcan Zhou
Yongcan Zhou