- 1State Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, Engineering Research Center of Green Development for Conventional Aquatic Biological Industry in the Yangtze River Economic Belt, Ministry of Education, College of Fisheries, Huazhong Agricultural University, Wuhan, China
- 2Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
Antimicrobial peptides are important components of the host innate immune system, forming the first line of defense against infectious microorganisms. Among them, liver-expressed antimicrobial peptides (LEAPs) are a family of antimicrobial peptides that widely exist in vertebrates. LEAPs include two types, named LEAP-1 and LEAP-2, and many teleost fish have two or more LEAP-2s. In this study, LEAP-2C from rainbow trout and grass carp were discovered, both of which are composed of 3 exons and 2 introns. The antibacterial functions of the multiple LEAPs were systematically compared in rainbow trout and grass carp. The gene expression pattern revealed that rainbow trout and grass carp LEAP-1, LEAP-2A, LEAP-2B and/or LEAP-2C were differentially expressed in various tissues/organs, mainly in liver. After bacterial infection, the expression levels of LEAP-1, LEAP-2A, LEAP-2B and/or LEAP-2C in the liver and gut of rainbow trout and grass carp increased to varying degrees. Moreover, the antibacterial assay and bacterial membrane permeability assay showed that rainbow trout and grass carp LEAP-1, LEAP-2A, LEAP-2B and LEAP-2C all have antibacterial activities against a variety of Gram-positive and Gram-negative bacteria with varying levels through membrane rupture. Furthermore, cell transfection assay showed that only rainbow trout LEAP-1, but not LEAP-2, can lead to the internalization of ferroportin, the only iron exporter on cell surface, indicating that only LEAP-1 possess iron metabolism regulation activity in teleost fish. Taken together, this study systematically compared the antibacterial function of LEAPs in teleost fish and the results suggest that multiple LEAPs can enhance the immunity of teleost fish through different expression patterns and different antibacterial activities to various bacteria.
Introduction
Antimicrobial peptides (AMPs) are widely found in organisms and are considered to be the first line of host defense against pathogens (1, 2). With the increasing resistance of bacteria to traditional antibiotics, AMPs have gradually become one of the important alternatives to antibiotics (3, 4). There are two main antibacterial mechanisms of AMPs: direct sterilization and immune regulation, and the sterilization mechanism can be divided into membrane targeting and non-membrane targeting (5). In addition to their broad-spectrum antibacterial activities, AMPs can also inhibit fungi, viruses and parasites (6). Compared with other species, some AMPs derived from fish, shrimp, crabs and other aquatic animals have unique structures and functions (7, 8).
Liver-expressed antimicrobial peptide (LEAP) is a class of antibacterial peptides expressed and secreted by liver, including LEAP-1 and LEAP-2. Both LEAP-1 and LEAP-2 can be separated and purified from human plasma (9, 10). LEAP-1 exists as a single gene in the vast majority of mammals, which is highly expressed in the liver, followed by heart and brain (9). Because of its dual functions of antibacterial and iron regulation, LEAP-1 is also known as hepcidin. LEAP-1 mainly binds to ferroportin (Fpn), the only known iron exporter on the surface of cells (11), to facilitate its internalization, thus affecting the absorption and release of iron ion in iron storage cells (12–14). Fish have two hepcidin isoforms, hamp1 and hamp2; however, hamp1 is found to exist in both ray-finned and lobe-finned fish, while hamp2 is only found to exist in ray-finned fish, and the tissue distribution of hepcidin in most teleost fish is polymorphic (15–17).
Mammalian LEAP-2 is a single gene, which is mainly expressed by the liver, and to a certain extent by other tissues (18). For example, mouse LEAP-2 is mainly expressed in the liver and small intestine (18). Studies have shown that the mature peptide 38-77 of human LEAP-2 has different killing effects on both Gram-positive and Gram-negative bacteria, while its cleaved form 44-77 loses antibacterial activity but participates in the blood circulation of the body (19). Therefore, LEAP-2 may have other physiological functions besides sterilization. However, unlike mammals with a single LEAP-2, three LEAP-2s have been discovered in fish species, including LEAP-2A, LEAP-2B and LEAP-2C (20–23), and the tissue expression patterns in different fish species are also different (22, 24). Teleost LEAP-2 can enhance the bacterial killing efficiency of monocytes/macrophages (25, 26) and has a synergistic effect with antibiotics on the killing of drug-resistant bacteria (27).
So far, systematic comparative studies on the antibacterial functions of LEAPs are still blank. Therefore, this study was devoted to analyzing the antibacterial functions of LEAPs and conducting a systematic comparative study to lay a theoretical foundation for their use as feed additives to prevent bacterial diseases in fish.
Materials and methods
Experimental fish and cells
Rainbow trout (Oncorhynchus mykiss) (30 ± 5 g) and grass carp (Ctenopharyngodon idella) (200 ± 20 g) were purchased from Dujiang Dam Rainbow Trout Farm (Chengdu, China) and Xiantao Hatchery (Xiantao, China) respectively. They were maintained and acclimated to the laboratory conditions for at least two weeks before experiments. All animal experiments in this study were approved by the Committee on the Ethics of Animal Experiments at Huazhong Agricultural University.
Human embryonic kidney 293T (HEK293T) cells were cultured in 5% CO2 at 37°C. Dulbecco’s modified Eagle’s medium (DMEM) (HyClone) was supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 g/mL penicillin (Sigma-Aldrich), and 100 g/mL streptomycin (Sigma-Aldrich).
Searching, identification, and localization of LEAPs in rainbow trout and grass carp genome
The chromosome-level genome of grass carp was assembled by our laboratory and deposited in the NCBI BioProjects with the accession number PRJNA745929 (28). The published fish LEAPs, especially zebrafish LEAPs, were used to search against the grass carp genome using the Basic Local Alignment Search Tool (BLAST). Interestingly, in addition to LEAP-2A and LEAP-2B, LEAP-2C was found in grass carp. The grass carp LEAP-2C was cloned and submitted to the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under the accession number OQ026323. Similarly, the rainbow trout LEAP-2C was cloned and submitted to the GenBank database under the accession number GQ870279.1.
Sequence alignment and phylogenetic analysis
The signal peptide of the deduced amino acid sequences was predicted using the SignalP 5.0 Server (https://services.healthtech.dtu.dk/service.php?SignalP-5.0). The LEAPs gene organizations, including exon, intron and UTR were determined by aligning the cDNA sequences with the gene sequences. The protein sequence identity was calculated using the BioEdit software (version 7.0.9). Multiple sequence alignment was conducted with the ClustalX program (version 3.0), and phylogenetic tree was constructed based on the alignments using the neighbour-joining method with 1000 bootstrap times using the MEGA program (version 4.1). All the sequences used for multiple sequence alignment and phylogenetic analysis were listed in Table 1.
The mRNA expression of LEAPs in rainbow trout and grass carp tissues
Four healthy rainbow trout and grass carp were anesthetized with MS222 (1:10000), then the blood was removed from the body by cardiac perfusion using phosphate-buffered saline (PBS; pH 7.4; Gibco). The head kidney, spleen, gut, gill, skin, liver, heart, and muscle were collected, and the total RNA was extracted using the TRIzol Reagent (Takara). The cDNA was synthesized using the PrimeScript™ RT Reagent Kit contains gDNA Eraser (Takara). The mRNA expression levels of LEAPs were detected by quantitative real-time PCR (qPCR) using the CFX Connect™ Real-Time System (Bio-Rad). The primers used are listed in Table 2. The reaction mixture (20 μl) contained 1 μl cDNA, 10 μl SsoAdvanced™ SYBR Green Supermix (Bio-Rad), 1 μl forward primer (10 μM each) and 1 μl reverse primer (10 μM each). The amplification program was as follows: 95°C for 5 min, 45 cycles of amplification (95°C for 5 s and 60°C for 30 s), and then 65°C for 5 s. The tissue expression levels of LEAPs were determined using 2−ΔCt method with β-actin as the internal reference.
Bacterial infection
To detect the immune responses of LEAPs during infection, rainbow trout and grass carp were intraperitoneally injected with 100 µL Aeromonas salmonicida BG1 (29) suspension culture (1×107 CFU/mL) and 200 µL Aeromonas hydrophila XS91-4-1 (30) suspension culture (8×106 CFU/mL) respectively, while the control fish were intraperitoneally injected with PBS instead after anesthetized with MS222 (1:10000). At 12 h, 1 d, 3 d, 5 d, and 7 d post-injection, the liver and gut of four individuals were sampled from each group. After the RNA extraction and cDNA synthesis, the expression levels of LEAPs in infected and control fish were determined by qPCR. The expression changes of LEAPs after infection were calculated using the 2−ΔΔCt method, with β-actin as the internal reference.
Peptide synthesis
The mature peptides of rainbow trout and grass carp LEAPs were synthesized by GL Biochem Ltd. The purity was confirmed to be higher than 95% by HPLC and MALDI-TOF mass spectroscopy. All peptides were stored at -80°C for later use.
Antibacterial activity assay
Six Gram-negative bacterial strains (Escherichia coli ATCC25922, A. hydrophila XS91-4-1, A. salmonicida BG1, A. sobria CR79-1-1, Edwardsiella ictaluri HSN-1, and Vibrio fluvialis WY91-24-3) and two Gram-positive bacterial strains (Micrococcus luteus ATCC10240 and Streptococcus agalactiae ATCC13813) were used in the antibacterial activity assay. Radial-diffusion assay (RDA) was conducted as previously described (31, 32). Briefly, 5 mL underlay agarose gel containing 0.03% (wt/vol) TSB, 1% (wt/vol) low electroendosmosis (EEO) agarose (Aladdin), 0.02% (vol/vol) Tween 20 (Amresco), and 4×106 CFU bacteria was added into each sterile petri dish (90 mm diameter). After the solidification of the agarose, 4 mm diameter wells were punched in the agar using a sterile steel borer. Then 6 μL peptide (125 μM) was added to each well, and sterile water was added as control. Agar plates were incubated for 3 h at the optimum bacterial growth temperature (28°C for A. hydrophila, A. salmonicida, A. sobria and E. ictaluri or 37°C for E. coli, V. fluvialis, M. luteus and S. agalactiae) to diffuse the peptides. Each plate was added 5 mL sterilized overlay agarose gel (containing 6% TSB and 1% low EEO agarose) and incubated for 24 h at the optimum bacterial growth temperature. The diameter of the clearance zone around each well was measured.
Bacterial membrane permeability assay
The membrane permeability of bacteria after the treatment of peptides was assessed by flow cytometry as previously described (33) with minor modifications. In brief, V. fluvialis WY91-24-3 were cultured to mid-logarithmic phase, then washed twice and resuspended to 106 CFU/ml with 10 mM NaPB (pH 7.4). Thereafter, 40 μl of peptides diluted in NaPB was added to 160 μl of V. fluvialis to give a final concentration of 8 μM. LL37 and NaPB was used as the positive and negative control, respectively. After being incubated at 37°C for 1 h, propidium iodide (PI; Sigma-Aldrich) was added to the bacterial suspension at a final concentration of 9 μM. The PI positive bacterial cells were detected using the flow cytometer FACSVerse (BD Biosciences) at 3,000 events and the data were analyzed using the FlowJo software v10 (Tree Star).
Plasmid construction and transfection
The Fpn of rainbow trout (GenBank: XM_021579209.2) was amplified using the cDNAs reverse-transcribed from liver RNA using the primers listed in Table 2. The PCR product was digested with restriction enzymes (Kpn 1and Xma 1), followed by ligation into pEGFP-N1 to construct pEGFP-Fpn. The plasmid pEGFP-Fpn was extracted from the Trans5α cells using an E.Z.N.A. Plasmid Maxi Kit (Omega). The extracted plasmid was transfected into the HEK293T cells using the TransIntro EL transfection reagents (TransGen Biotech) according to the manufacturer’s instructions. At 24 h after transfection, cycloheximide (Aladdin) was added to the HEK293T cells to give a final concentration of 75 μg/mL. After incubation for 3 h, the synthetic rainbow trout LEAPs were added to give a final concentration of 1 μM and incubated for 24 h. The changes in the position of Fpn-EGFP in the cells were investigated using a live cell station (Leica AF6000).
Statistical analysis
The statistic p value was calculated using the SPSS Statistics (version 19, IBM) by one-way ANOVA with a Dunnett post hoc test. A p value < 0.05 was considered statistically significant while a p value < 0.01 was considered highly significant.
Results
Homology comparison of LEAPs
Homologous sequence alignment showed that all LEAPs consist of a signal peptide, a propeptide and a mature peptide. The sequences of the mature peptides are more conserved than those of the signal peptides and propeptides. The mature peptide of LEAP-1 and LEAP-2 possessed eight and four conserved cysteines, respectively. Interestingly, from fish to mammals, the signature of the cleavage site (RXXR) between the propeptide and the mature peptide of LEAPs are conserved in evolution (Figure 1).

Figure 1 Amino acid sequence and domain organization of LEAP-1 (A) and LEAP-2 (B) from mammals, birds, amphibians, reptiles, and fish. The signal peptide, propeptide and mature peptide are denoted above the alignment. GenBank accession numbers of the selected sequences are listed in Table 1.
Gene structure and phylogenetic analyses of LEAPs
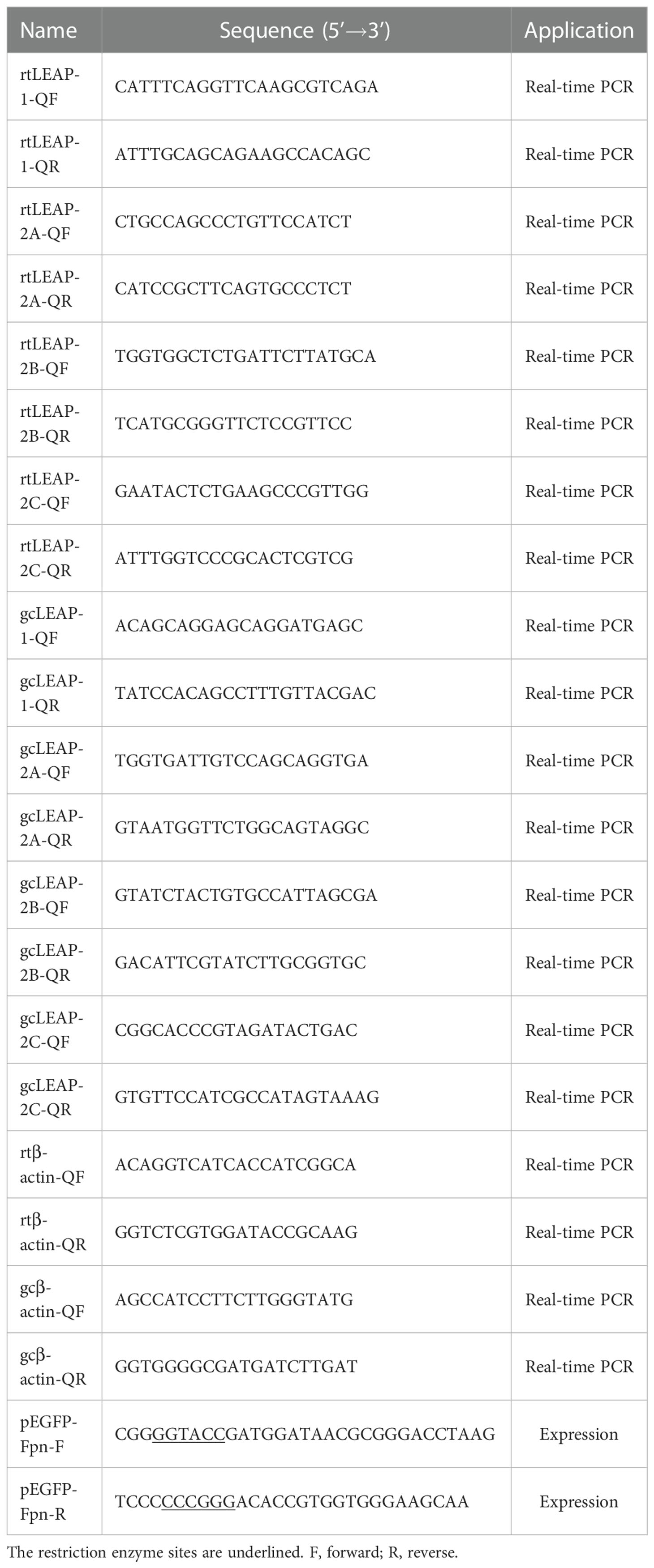
LEAP-1 and LEAP-2 have the same gene structure, which is composed of 3 exons and 2 introns. The 5’ and 3’ UTR of rainbow trout and grass carp LEAP-1 are longer than those of LEAP-1s from other species analyzed. The two introns of grass carp LEAP-2C are longer than those of LEAP-2Cs from other species analyzed (Figure 2A).
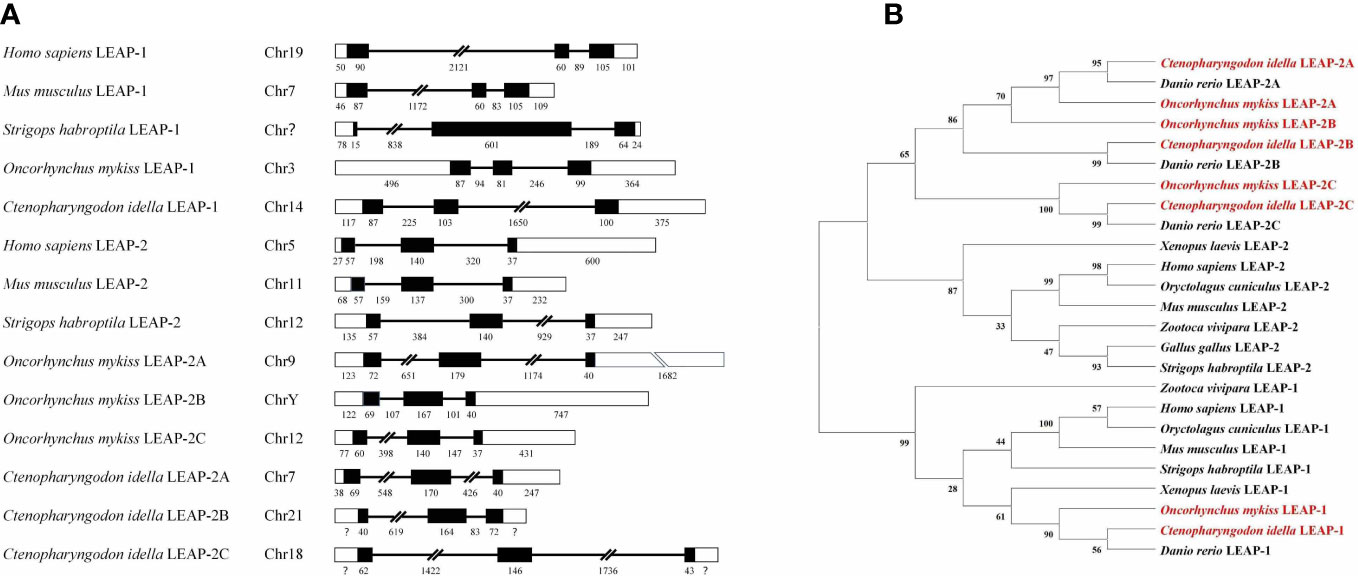
Figure 2 Gene structure and phylogenetic relationship of vertebrate LEAPs. (A) Gene structure of LEAPs from Homo sapiens, Mus musculus, Strigops habroptila, Oncorhynchus mykiss, and Ctenopharyngodon idella. The rectangles represent the exons, and the lines between them indicate the introns. Black and white areas indicate the coding regions and untranslated regions, respectively. The sizes of the exons and introns are shown by the numbers below. GenBank accession numbers of the selected sequences are listed in Table 1. The question mark means the length is unknown. (B) Phylogenetic relationship of LEAPs. A neighbour-joining phylogenetic tree was generated using the MEGA program with 1000 bootstrap replications. Rainbow trout and grass carp LEAPs are in red. GenBank accession numbers of the selected sequences are listed in Table 1.
In the phylogenetic tree constructed using the amino acid sequences of the LEAP-1 and LEAP-2 precursors (Figure 2B), the LEAP-1 sequences branched apart from the LEAP-2 sequences. Teleost LEAPs were distinguished from those of other vertebrates. LEAP-2 of grass carp, rainbow trout and zebrafish contain three members, named LEAP-2A, LEAP-2B and LEAP-2C.
Expression of LEAPs in rainbow trout and grass carp tissues
The mRNA expression levels of the LEAPs were evaluated in several healthy rainbow trout (Figure 3A) and grass carp (Figure 3B) tissues. Rainbow trout (rt) and grass carp (gc) LEAPs were differentially expressed in various tissues and organs, with an overall predominance of LEAP-1. In general, the LEAPs were highly expressed in the liver and lowly expressed in the gill, heart, and muscle. LEAP-2A was significantly expressed in the gut and skin of rainbow trout and grass carp, and the expression of LEAP-2C in the gut of rainbow trout was higher than that in the liver.
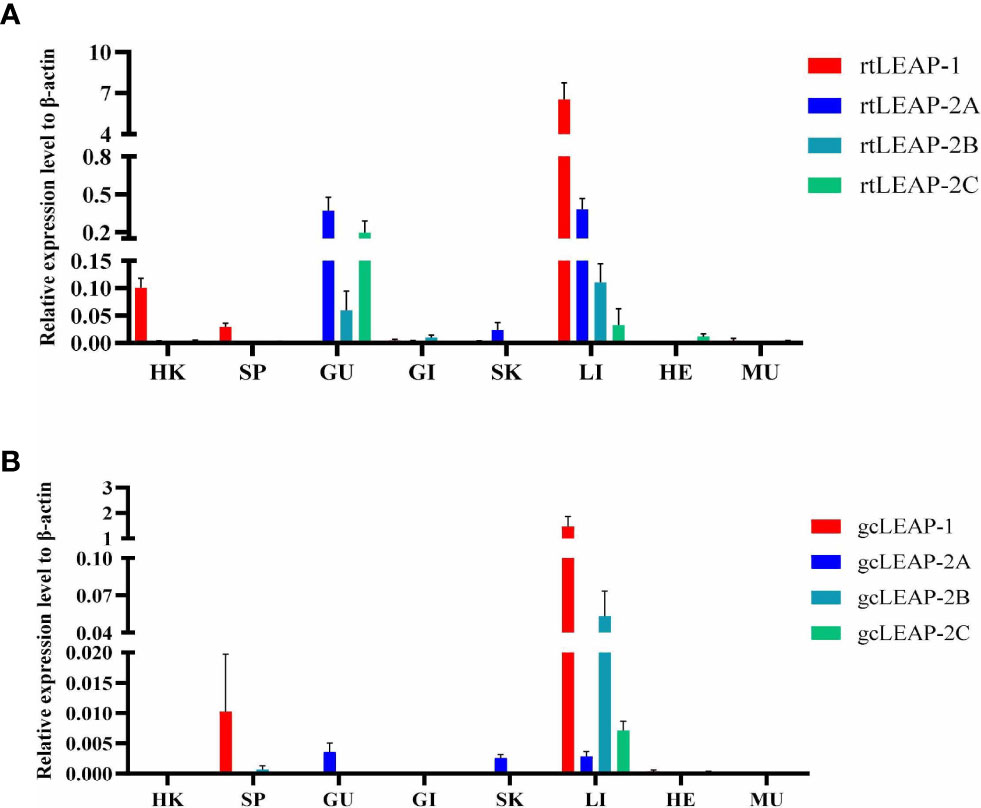
Figure 3 Expression patterns of rainbow trout (A) and grass carp (B) LEAPs in various tissues. The expression levels of LEAPs in lymphoid and non-lymphoid tissues were analyzed by qPCR and normalized against the expression of β-actin using the 2−ΔCt method. Abbreviations for tissues are as follows: HK, head kidney; SP, spleen; GU, gut; GI, gill; SK, skin; LI, liver; HE, heart; MU, muscle. Data for rainbow trout represent the mean ± SEM of four individuals, and data for grass carp represent the mean ± SEM of five individuals.
Induced expression of LEAPs by pathogenic bacteria
Expression of the total transcripts of rtLEAP-1 and rtLEAP-2 in the liver (Figure 4A) and gut (Figure 4B) of rainbow trout induced by A. salmonicida was studied. The expression level of rtLEAP-1 increased significantly in the liver from 12 h to 1 d, and in the gut from 12 h to 3 d after infection. The expression level of rtLEAP-2A increased significantly in the gut at 12 h after infection. The expression level of rtLEAP-2B increased significantly in the liver from 1 d to 5 d, and in the gut from 3 d to 7 d after infection. However, the expression level of rtLEAP-2C decreased significantly in the gut from 3 d to 7 d after infection.
Figure 4 Induced expressions of rtLEAPs (A, B) and gcLEAPs (C, D) in the liver and gut under the condition of infection. Fold changes were calculated by comparing the infected group with the control group (defined as 1) using the 2−ΔΔCt method. Data represent the mean ± SEM of four fish individuals. The p value was calculated by one-way ANOVA with a Dunnett post hoc test (*p < 0.05, **p < 0.01).
Expression of the total transcripts of gcLEAP-1 and gcLEAP-2 in the liver (Figure 4C) and gut (Figure 4D) of grass carp induced by A. hydrophila was studied. The expression levels of gcLEAP-1 and gcLEAP-2s increased from 12 h to 3 d after infection, and then gradually returned to normal level, with gcLEAP-2A increased most significantly in the liver and gcLEAP-1 increased most significantly in the gut.
Antibacterial activities of LEAPs
To investigate the antimicrobial properties of rainbow trout and grass carp LEAPs, we synthesized the mature peptides of rainbow trout and grass carp LEAPs and tested their antibacterial activities against a variety of bacterial strains. The results showed that all of the rainbow trout and grass carp LEAPs were antibacterial to Gram-negative (E. coli, A. hydrophila, A. salmonicida, A. sobria, E. ictaluri, and V. fluvialis) and Gram-positive bacteria (M. luteus and S. agalactiae) (Figures 5A, B). For the same bacterium, the antibacterial activities of different LEAPs are different. Interestingly, rainbow trout and grass carp LEAP-2C showed significant inhibitory effects on A. hydrophila, a common and harmful pathogen in fish farming.
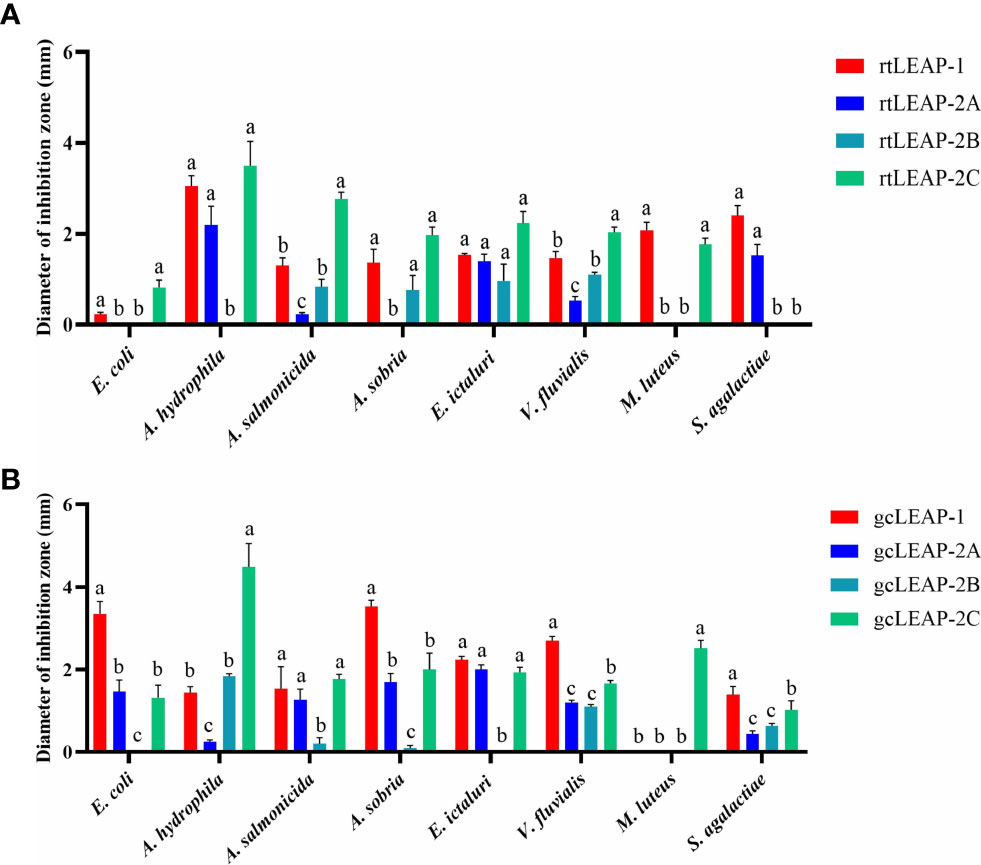
Figure 5 Antibacterial activities of rainbow trout (A) and grass carp (B) LEAPs against various bacteria. The underlay agarose gel containing E. coli ATCC25922, A. hydrophila XS91-4-1, A. salmonicida BG1, A. sobria CR79-1-1, E. ictaluri HSN-1, V. fluvialis WY91-24-3, M. luteus ATCC10240 and S. agalactiae ATCC13813 respectively was poured into sterile petri dishes. After the solidification, 4 mm diameter wells were punched in the agarose gel. Then 6 μL LEAP (125 μM) was added to each well, and sterile water was added as the control. Plates were incubated at the optimum bacterial growth temperature (28℃ for A. hydrophila, A. salmonicida, A. sobria and E. ictaluri or 37℃ for E. coli, V. fluvialis, M. luteus and S. agalactiae) for 3 h, and then the underlay agarose gel was covered with overlay agarose gel. The diameter of the inhibition zone around each well was measured after 24 h of incubation at the optimum bacterial growth temperature. Data represent the mean ± SEM of three independent experiments. Different letters indicate significant differences (p<0.05).
Antibacterial mechanism of LEAPs
To clarify the antibacterial mechanism of LEAPs, we detected the integrity of cell membrane by bacterial membrane permeability assay. The results showed that rainbow trout and grass carp LEAPs could significantly increase the permeability of the cell membrane of V. fluvialis within 1 h, and rtLEAP-1 had the most significant effect, which was similar to human AMP LL-37 (Figures 6A–K), suggesting that LEAPs kill bacteria through membrane permeabilizing action.
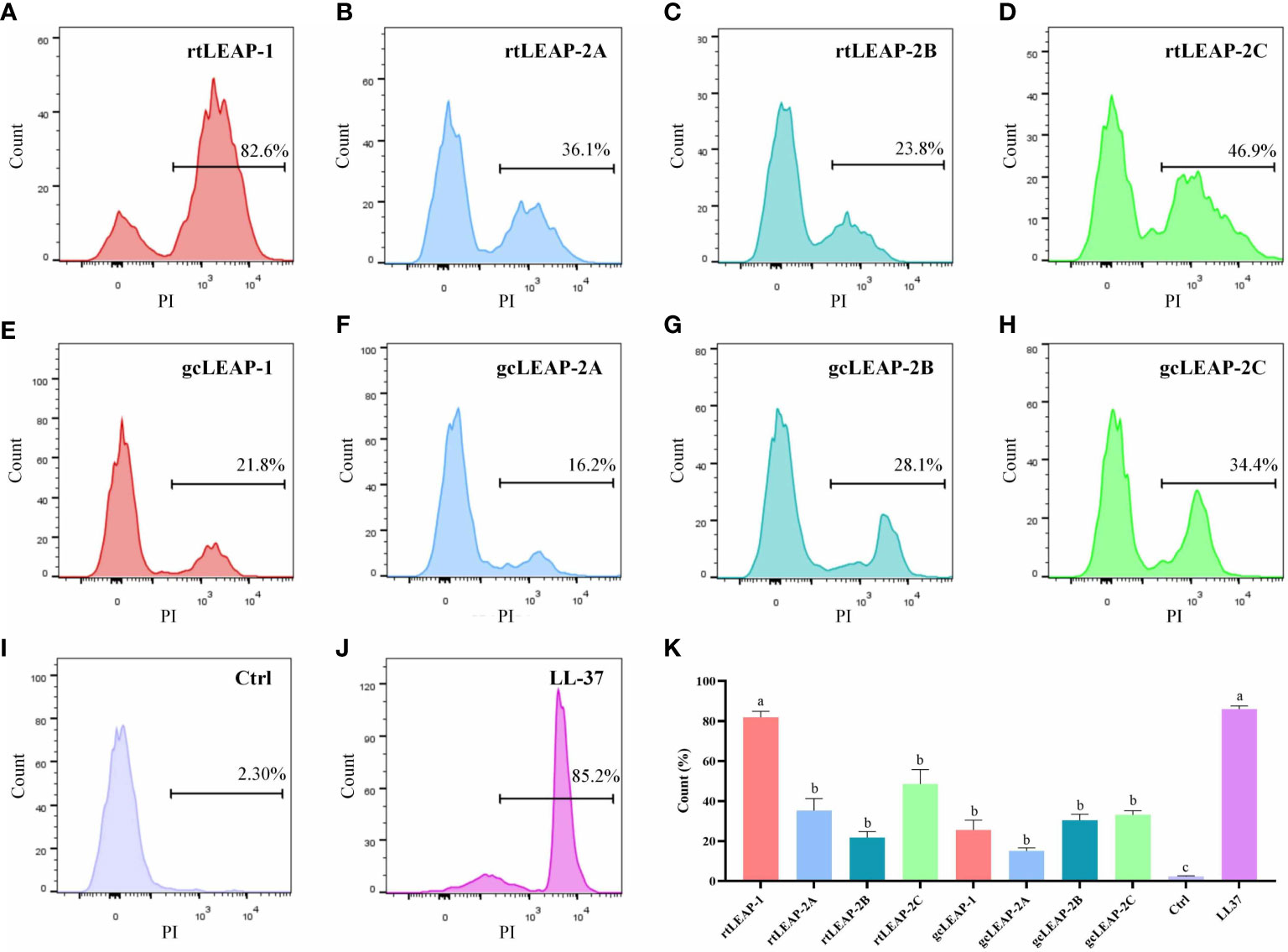
Figure 6 Antibacterial mechanisms of LEAPs. (A–J) Membrane permeability of V. fluvialis treated with LEAPs. The bacteria were incubated with LEAPs (8 μM) at 37°C for 1 h, and a bacterial suspension without peptide was included as a control (Ctrl). Then PI (9 μM) was added and the influx of PI was detected by flow cytometry. Data are representative results of three independent experiments. (K) The data in (A–J) were analyzed statistically. The p value was calculated by one-way ANOVA with a Dunnett post hoc test. Different letters indicate significant differences (p<0.05).
Comparison of the ability of LEAPs to internalize Fpn
Through the amino acid sequence alignment of the LEAP-1 mature peptides in rainbow trout, grass carp, zebrafish, European sea bass, Atlantic salmon, and turbot, the Q-S/I-H-L/I-S/A-L motif was found in the N-terminus of the mature peptides (Figure 7A). To compare the ability of LEAPs to internalize Fpn, rtLEAPs were added to the cells expressing Fpn-EGFP, and the changes in the position of Fpn-EGFP in the cells were investigated using a live cell station. The results showed that only rtLEAP-1 resulted in the loss of cell surface Fpn-EGFP and the presence of intracellular Fpn-EGFP (Figure 7B), indicating that only rtLEAP-1 can cause the internalization of Fpn, while rtLEAP-2A, rtLEAP-2B and rtLEAP-2C cannot.
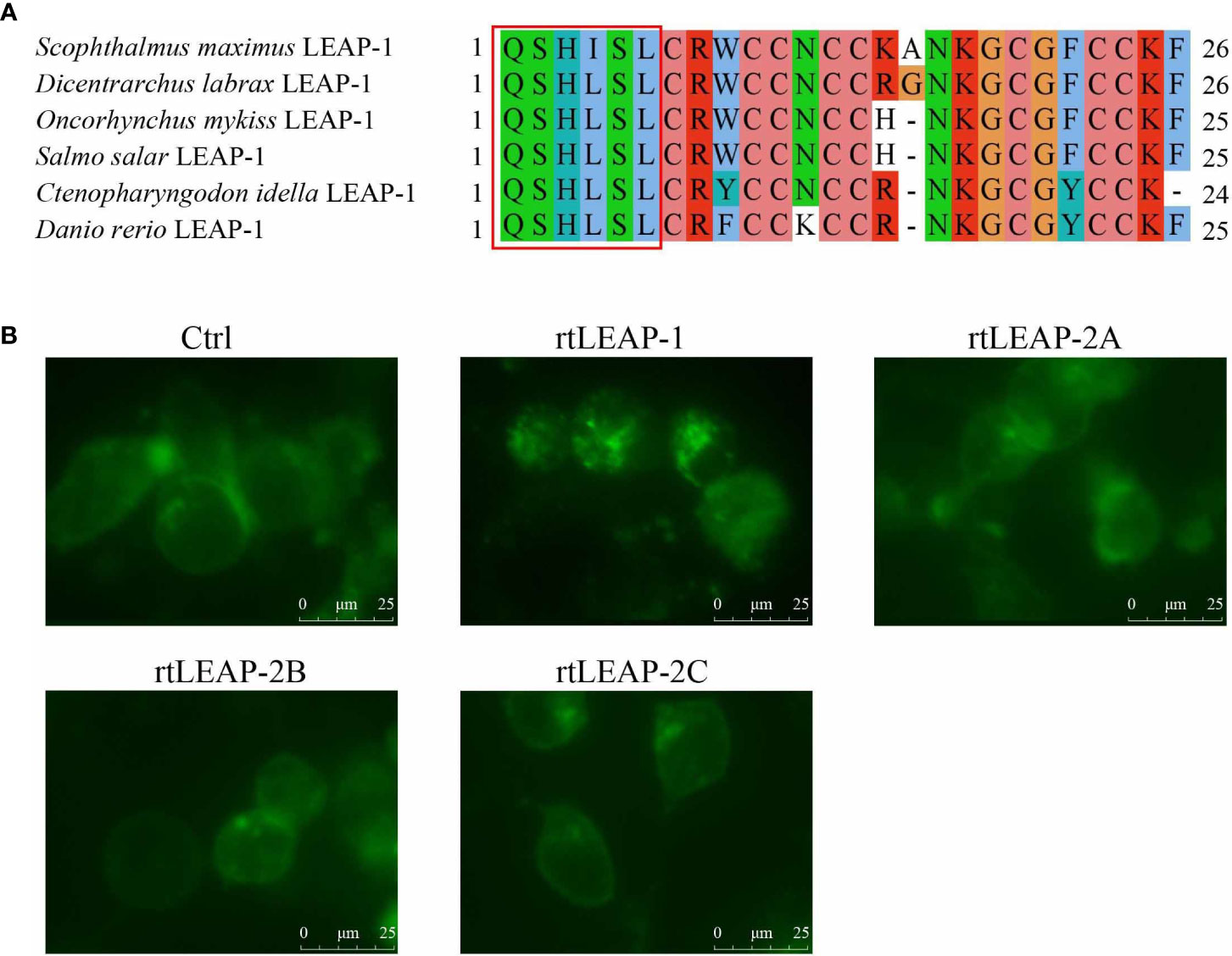
Figure 7 The effects of LEAPs on Fpn internalization. (A) Amino acid sequence alignment of LEAP-1 mature peptides in rainbow trout, grass carp, zebrafish, European sea bass, Atlantic salmon, and turbot. The Q-S/I-H-L/I-S/A-L motif at the N-terminus of the mature peptides are indicated with a red rectangle. GenBank accession numbers of the selected sequences are listed in Table 1. (B) The ability of LEAPs to internalize Fpn. HEK293T cells were seeded in 6-well plates and transfected with pEGFP-Fpn. After 24 h, cycloheximide (75 ug/mL) was added for 3 h, and then cells were incubated with rtLEAPs (1 μM) and PBS (blank control [Ctrl]) for 24 h. The images of cells were captured using a live cell station.
Discussion
Just as in tetrapods, there are also LEAP-1 and LEAP-2 in fish, but the difference is that fish LEAP-1 and LEAP-2 contain multiple genes. This is most likely due to the genome duplications and positive selection in fish, which suggests that different LEAPs may perform different functions. Multiple LEAPs existing in fish may be contributed to the immune defense of fish, which live in a complex aquatic environment.
Through amino acid sequence alignment, it was found that the LEAP-1 sequences of rainbow trout and grass carp were similar to other species, and the identity between rainbow trout and grass carp is 87.5%. Previous studies have shown that the LEAP-2 of rainbow trout (20) and grass carp (34) contains two genes, LEAP-2A and LEAP-2B, but in the current study we found that LEAP-2C also exists in these two species. Phylogenetic analysis indicated that LEAP-2 has evolved separately from LEAP-1 in vertebrates, resulting in LEAP-1 having iron regulation function, while LEAP-2 doing not.
As in many fish species and other vertebrates (19, 35–37), though predominantly expressed in the liver, both LEAP-1 and LEAP-2 were differentially expressed in other tissues and organs of rainbow trout and grass carp. A notable phenomenon was that LEAP-2A was significantly expressed in the gut and skin of rainbow trout and grass carp and LEAP-2B and LEAP-2C were significantly expressed in the gut of rainbow trout. Similar results were reported in blunt snout bream, whose LEAP-2 mRNA expression level was highest in the gut (38). Research has shown that common carp LEAP-2A mRNA expression level increased in the gut, gills and skin after infection (22). These suggest that LEAPs of teleost fish may play important roles in both systemic and mucosal immunity, respectively.
A significant induction of LEAP was observed after experimental bacterial challenge, with LEAP-1 being the most responsive gene in the liver and gut of both rainbow trout and grass carp. In rainbow trout, LEAP-1 expression increased significantly in the early stage of infection, and LEAP-2B expression increased significantly in the late stage of infection. In grass carp, the gene expression of all LEAPs increased after infection, and then gradually returned to normal levels. These results suggest that rtLEAPs may function for a longer period after pathogen infection to maintain robust immunity. Similar induced expression of LEAPs was reported in other fish species after bacterial infection. It is worth noting that the tissue expression patterns of LEAPs in different fish species have specificity, and different pathogens are likely to induce the expression of different LEAPs (35, 39–42). All of these studies, however, shared the same idea that multiple LEAPs in hosts perform important antimicrobial functions.
Rainbow trout and grass carp LEAPs have broad spectrum antibacterial activities against a various type of bacteria. Notably, gcLEAP-2A has antibacterial activity against several bacterial strains, including E. coli, which is different from the previous report that gcLEAP-2A has no antibacterial activity against E. coli (34). Our results suggest that the antibacterial immunity of fish was enhanced through LEAP expansion. Although LEAP-1 and LEAP-2 belong to different types, their bactericidal mechanisms are roughly the same. Rainbow trout and grass carp LEAPs kill bacteria by disrupt the cell membranes, which is similar to previous study on other species that LEAPs form multiple pairs of disulfide bonds through cysteine residues and form a stable β-sheet structure, which can destroy bacterial cell membrane, promote its permeability, and cause the leakage of cell contents and the death of bacterial cells, thus killing pathogenic bacteria (10, 43).
Previous study has shown that the expression of LEAP-1 is directly or indirectly regulated by iron storage anemia, hypoxia, inflammation, pathological conditions, cytokines, and other signals (44, 45). Under normal physiological conditions, the expression of LEAP-1 is negatively correlated with iron level (46–48). The conserved sequence Q-S/I-H-L/I-S/A-L, which is associated with iron regulation activity (49), is present at the N-terminus of the mature peptide of LEAP-1, but not LEAP-2, suggesting that LEAP-1 and LEAP-2 play different roles in iron regulation. Our results further confirm that only LEAP-1 has the ability to internalize Fpn. On the one hand, this reaction maintains the homeostasis of iron in teleost fish; on the other hand, it leads to the obstruction of iron outflow from cells and reduces the available iron content of extracellular pathogens (13, 50).
In conclusion, this study first reported the existence of the LEAP-2C in rainbow trout and grass carp and systematically compared the antibacterial function of LEAPs in these teleost fish, which suggested that multiple LEAPs can enhance the immunity of teleost fish through different expression patterns and different antibacterial activities to various bacteria. This enhanced our understanding of host innate immunity and provided new evidence and insights for systematically analyzing the antibacterial function of LEAPs in teleost fish.
Data availability statement
The data presented in the study are deposited in the GenBank repository (https://www.ncbi.nlm.nih.gov/genbank/), accession numbers GQ870279.1 and OQ026323.
Ethics statement
The animal study was reviewed and approved by the Committee on the Ethics of Animal Experiments at Huazhong Agricultural University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
XL performed most of the experiments, analyzed most of the data, and wrote the preliminary manuscript. Y-ZH searched the LEAP-2C in the grass carp genome and helped with the date analysis. Y-RP helped with the gene structure analysis. JL helped with the sampling of infection experiments. Y-BJ participated in the radial-diffusion assay. Y-AZ helped with the experiment design and revised the manuscript. X-JZ designed the research, analyzed some of the data, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFD0900505), the Laboratory of Lingnan Modern Agriculture Project (NT2021008), the China Agriculture Research System (CARS-46), and the Youth Talent Project of Engineering Research Center of Green Development for Conventional Aquatic Biological Industry in the Yangtze River Economic Belt (2662021SSSY004).
Acknowledgments
The authors are grateful to Zi-You Ma and Tao-Zhen Lu for their help in fish challenge experiments and sampling.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Kosciuczuk EM, Lisowski P, Jarczak J, Strzalkowska N, Jozwik A, Horbanczuk J, et al. Cathelicidins: Family of antimicrobial peptides. A Review. Mol Biol Rep (2012) 39(12):10957–70. doi: 10.1007/s11033-012-1997-x
2. Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev (2003) 55(1):27–55. doi: 10.1124/pr.55.1.2
3. Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Bba-Biomembranes (1999) 1462(1-2):11–28. doi: 10.1016/S0005-2736(99)00198-4
4. Zasloff M. Antimicrobial peptides of multicellular organisms. Nature (2002) 415(6870):389–95. doi: 10.1038/415389a
5. Won HS, Kang SJ, Lee BJ. Action mechanism and structural requirements of the antimicrobial peptides, gaegurins. Bba-Biomembranes (2009) 1788(8):1620–9. doi: 10.1016/j.bbamem.2008.10.021
6. Huan YC, Kong Q, Mou HJ, Yi HX. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front Microbiol (2020) 11:582779. doi: 10.3389/fmicb.2020.582779
7. Bachere E, Destoumieux D, Bulet P. Penaeidins, antimicrobial peptides of shrimp: A comparison with other effectors of innate immunity. Aquaculture (2000) 191(1-3):71–88. doi: 10.1016/S0044-8486(00)00419-1
8. Chen YC, Yang Y, Zhang C, Chen HY, Chen FY, Wang KJ. A novel antimicrobial peptide Sparamosin(26-54) from the mud crab Scylla paramamosain showing potent antifungal activity against Cryptococcus neoformans. Front Microbiol (2021) 12:746006. doi: 10.3389/fmicb.2021.746006
9. Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett (2000) 480(2-3):147–50. doi: 10.1016/S0014-5793(00)01920-7
10. Krause A, Sillard R, Kleemeier B, Kluver E, Maronde E, Conejo-Garcia JR, et al. Isolation and biochemical characterization of LEAP-2, a novel blood peptide expressed in the liver. Protein Sci (2003) 12(1):143–52. doi: 10.1110/ps.0213603
11. Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, et al. The iron exporter Ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab (2005) 1(3):191–200. doi: 10.1016/j.cmet.2005.01.003
12. Qiao B, Sugianto P, Fung E, del-Castillo-Rueda A, Moran-Jimenez MJ, Ganz T, et al. Hepcidin-induced endocytosis of ferroportin is dependent on ferroportin ubiquitination. Cell Metab (2012) 15(6):918–24. doi: 10.1016/j.cmet.2012.03.018
13. Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science (2004) 306(5704):2090–3. doi: 10.1126/science.1104742
14. Ramey G, Deschemin JC, Durel B, Canonne-Hergaux F, Nicolas G, Vaulont S. Hepcidin targets ferroportin for degradation in hepatocytes. Haematol-Hematol J (2010) 95(3):501–4. doi: 10.3324/haematol.2009.014399
15. Hilton KB, Lambert LA. Molecular evolution and characterization of hepcidin gene products in vertebrates. Gene (2008) 415(1-2):40–8. doi: 10.1016/j.gene.2008.02.016
16. Padhi A, Verghese B. Evidence for positive Darwinian selection on the hepcidin gene of perciform and pleuronectiform fishes. Mol Divers (2007) 11(3-4):119–30. doi: 10.1007/s11030-007-9066-4
17. Serna-Duque JA, Cuesta A, Esteban MA. Massive gene expansion of hepcidin, a host defense peptide, in gilthead seabream (Sparus aurata). Fish Shellfish Immunol (2022) 124:563–71. doi: 10.1016/j.fsi.2022.04.032
18. Mani BK, Puzziferri N, He ZY, Rodriguez JA, Osborne-Lawrence S, Metzger NP, et al. LEAP2 changes with body mass and food intake in humans and mice. J Clin Invest (2019) 129(9):3909–23. doi: 10.1172/Jci125332
19. Sang Y, Ramanathan B, Minton JE, Ross CR, Blecha F. Porcine liver-expressed antimicrobial peptides, hepcidin and LEAP-2: Cloning and induction by bacterial infection. Dev Comp Immunol (2006) 30(4):357–66. doi: 10.1016/j.dci.2005.06.004
20. Zhang YA, Zou J, Chang CI, Secombes CJ. Discovery and characterization of two types of liver-expressed antimicrobial peptide 2 (LEAP-2) genes in rainbow trout. Vet Immunol Immunopathol (2004) 101(3-4):259–69. doi: 10.1016/j.vetimm.2004.05.005
21. Li HX, Lu XJ, Li CH, Chen J. Molecular characterization and functional analysis of two distinct liver-expressed antimicrobial peptide 2 (LEAP-2) genes in Large yellow croaker (Larimichthys crocea). Fish Shellfish Immun (2014) 38(2):330–9. doi: 10.1016/j.fsi.2014.04.004
22. Yang GW, Guo HY, Li H, Shan SJ, Zhang XQ, Rombout JHWM, et al. Molecular characterization of LEAP-2 cDNA in common carp (Cyprinus carpio l.) and the differential expression upon a Vibrio anguillarum stimulus; indications for a significant immune role in skin. Fish Shellfish Immun (2014) 37(1):22–9. doi: 10.1016/j.fsi.2014.01.004
23. Zhang SH, Xu QQ, Du H, Qi ZT, Li YS, Huang J, et al. Evolution, expression, and characterisation of liver-expressed antimicrobial peptide genes in ancient chondrostean sturgeons. Fish Shellfish Immun (2018) 79:363–9. doi: 10.1016/j.fsi.2018.05.023
24. Li HX, Lu XJ, Li CH, Chen J. Molecular characterization of the liver-expressed antimicrobial peptide 2 (LEAP-2) in a teleost fish, plecoglossus altivelis: Antimicrobial activity and molecular mechanism. Mol Immunol (2015) 65(2):406–15. doi: 10.1016/j.molimm.2015.02.022
25. Chen J, Chen Q, Lu XJ, Chen J. The protection effect of LEAP-2 on the mudskipper (Boleophthalmus pectinirostris) against edwardsiella tarda infection is associated with its immunomodulatory activity on Monocytes/Macrophages. Fish Shellfish Immun (2016) 59:66–76. doi: 10.1016/j.fsi.2016.10.028
26. Chen J, Lv YP, Dai QM, Hu ZH, Liu ZM, Li JH. Host defense peptide LEAP-2 contributes to Monocyte/Macrophage polarization in barbel steed (Hemibarbus labeo). Fish Shellfish Immun (2019) 87:184–92. doi: 10.1016/j.fsi.2019.01.015
27. Chen Y, Wu J, Cheng HL, Dai Y, Wang YP, Yang HL, et al. Anti-infective effects of a fish-derived antimicrobial peptide against drug-resistant bacteria and its synergistic effects with antibiotic. Front Microbiol (2020) 11:602412. doi: 10.3389/fmicb.2020.602412
28. Wu CS, Ma ZY, Zheng GD, Zou SM, Zhang XJ, Zhang YA. Chromosome-level genome assembly of grass carp (Ctenopharyngodon idella) provides insights into its genome evolution. BMC Genomics (2022) 23(1):271. doi: 10.1186/s12864-022-08503-x
29. Long M, Zhao J, Li T, Tafalla C, Zhang Q, Wang X, et al. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. Salmonicida. J Proteomics (2015) 122:41–54. doi: 10.1016/j.jprot.2015.03.031
30. Lu T-Z, Liu X, Wu C-S, Ma Z-Y, Wang Y, Zhang Y-A, et al. Molecular and functional analyses of the primordial costimulatory molecule CD80/86 and its receptors CD28 and CD152 (CTLA-4) in a teleost fish. Front Immunol (2022) 13:885005. doi: 10.3389/fimmu.2022.885005
31. Nordahl EA, Rydengard V, Nyberg P, Nitsche DP, Morgelin M, Malmsten M, et al. Activation of the complement system generates antibacterial peptides. Proc Natl Acad Sci USA (2004) 101(48):16879–84. doi: 10.1073/pnas.0406678101
32. Pasupuleti M, Walse B, Nordahl EA, Morgelin M, Malmsten M, Schmidtchen A. Preservation of antimicrobial properties of complement peptide C3a, from invertebrates to humans. J Biol Chem (2007) 282(4):2520–8. doi: 10.1074/jbc.M607848200
33. Hu YZ, Ma ZY, Wu CS, Wang J, Zhang YA, Zhang XJ. LECT2 is a novel antibacterial protein in vertebrates. J Immunol (2022) 208(8):2037–53. doi: 10.4049/jimmunol.2100812
34. Liu F, Li JL, Yue GH, Fu JJ, Zhou ZF. Molecular cloning and expression analysis of the liver-expressed antimicrobial peptide 2 (LEAP-2) gene in grass carp. Veterinary Immunol Immunopathology (2010) 133(2-4):133–43. doi: 10.1016/j.vetimm.2009.07.014
35. Wang Y, Liu X, Ma L, Yu Y, Yu H, Mohammed S, et al. Identification and characterization of a hepcidin from half-smooth tongue sole Cynoglossus semilaevis. Fish Shellfish Immunol (2012) 33(2):213–9. doi: 10.1016/j.fsi.2012.04.011
36. Yang M, Chen B, Cai JJ, Peng H, Ling C, Yuan JJ, et al. Molecular characterization of hepcidin as-Hepc2 and as-Hepc6 in black porgy (Acanthopagrus schlegelii): Expression pattern responded to bacterial challenge and in vitro antimicrobial activity. Comp Biochem Physiol B Biochem Mol Biol (2011) 158(2):155–63. doi: 10.1016/j.cbpb.2010.11.003
37. Zhou JG, Wei JG, Xu D, Cui HC, Yan Y, Ou-Yang ZL, et al. Molecular cloning and characterization of two novel hepcidins from orange-spotted grouper. Epinephelus Coioides. Fish Shellfish Immunol (2011) 30(2):559–68. doi: 10.1016/j.fsi.2010.11.021
38. Liang T, Ji W, Zhang GR, Wei KJ, Feng K, Wang WM, et al. Molecular cloning and expression analysis of liver-expressed antimicrobial peptide 1 (LEAP-1) and LEAP-2 genes in the blunt snout bream (Megalobrama amblycephala). Fish Shellfish Immunol (2013) 35(2):553–63. doi: 10.1016/j.fsi.2013.05.021
39. Bao BL, Peatman E, Xu P, Li P, Zeng H, He CB, et al. The catfish liver-expressed antimicrobial peptide 2 (LEAP-2) gene is expressed in a wide range of tissues and developmentally regulated. Mol Immunol (2006) 43(4):367–77. doi: 10.1016/j.molimm.2005.02.014
40. Chen SL, Xu MY, Ji XS, Yu GC, Liu Y. Cloning, characterization, and expression analysis of hepcidin gene from red Sea bream (Chrysophrys major). Antimicrob Agents Ch (2005) 49(4):1608–12. doi: 10.1128/Aac.49.4.1608-1612.2005
41. Pereiro P, Figueras A, Novoa B. A novel hepcidin-like in turbot (Scophthalmus maximus l.) highly expressed after pathogen challenge but not after iron overload. Fish Shellfish Immunol (2012) 32(5):879–89. doi: 10.1016/j.fsi.2012.02.016
42. Kim YO, Park EM, Nam BH, Kong HJ, Kim WJ, Lee SJ. Identification and molecular characterization of two hepcidin genes from black rockfish (Sebastes schlegelii). Mol Cell Biochem (2008) 315(1-2):131–6. doi: 10.1007/s11010-008-9796-3
43. Rodrigues PNS, Vazquez-Dorado S, Neves JV, Wilson JM. Dual function of fish hepcidin: Response to experimental iron overload and bacterial infection in Sea bass (Dicentrarchus labrax). Dev Comp Immunol (2006) 30(12):1156–67. doi: 10.1016/j.dci.2006.02.005
44. Zhao N, Zhang AS, Enns CA. Iron regulation by hepcidin. J Clin Invest (2013) 123(6):2337–43. doi: 10.1172/JCI67225
45. Neves JV, Caldas C, Vieira I, Ramos MF, Rodrigues PN. Multiple hepcidins in a teleost fish, Dicentrarchus labrax: Different hepcidins for different roles. J Immunol (2015) 195(6):2696–709. doi: 10.4049/jimmunol.1501153
46. Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr (2006) 26:323–42. doi: 10.1146/annurev.nutr.26.061505.111303
47. Vyoral D, Jiri P. Therapeutic potential of hepcidin - the master regulator of iron metabolism. Pharmacol Res (2017) 115:242–54. doi: 10.1016/j.phrs.2016.11.010
48. Ward DM, Kaplan J. Ferroportin-mediated iron transport: Expression and regulation. Biochim Biophys Acta (2012) 1823(9):1426–33. doi: 10.1016/j.bbamcr.2012.03.004
49. Robertson LS, Iwanowicz LR, Marranca JM. Identification of centrarchid hepcidins and evidence that 17beta-estradiol disrupts constitutive expression of hepcidin-1 and inducible expression of hepcidin-2 in largemouth bass (Micropterus salmoides). Fish Shellfish Immunol (2009) 26(6):898–907. doi: 10.1016/j.fsi.2009.03.023
Keywords: rainbow trout, grass carp, LEAP-1, LEAP-2, expression pattern, antibacterial activity
Citation: Liu X, Hu Y-Z, Pan Y-R, Liu J, Jiang Y-B, Zhang Y-A and Zhang X-J (2023) Comparative study on antibacterial characteristics of the multiple liver expressed antimicrobial peptides (LEAPs) in teleost fish. Front. Immunol. 14:1128138. doi: 10.3389/fimmu.2023.1128138
Received: 20 December 2022; Accepted: 02 February 2023;
Published: 20 February 2023.
Edited by:
Sun Yun, Hainan University, ChinaReviewed by:
Xianghui Kong, Henan Normal University, ChinaZhi Liu, Huazhong University of Science and Technology, China
Copyright © 2023 Liu, Hu, Pan, Liu, Jiang, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-An Zhang, eW9uZ2FuemhhbmdAbWFpbC5oemF1LmVkdS5jbg==; Xu-Jie Zhang, eHVqaWV6aGFuZ0BtYWlsLmh6YXUuZWR1LmNu
 Xun Liu
Xun Liu Ya-Zhen Hu1
Ya-Zhen Hu1 Yong-An Zhang
Yong-An Zhang Xu-Jie Zhang
Xu-Jie Zhang