
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 15 May 2023
Sec. Immunological Tolerance and Regulation
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1127743
This article is part of the Research TopicGlobal Excellence in Translational Immunology: Asia and Australasia 2023View all 7 articles
Bile acids (BAs) as cholesterol-derived molecules play an essential role in some physiological processes such as nutrient absorption, glucose homeostasis and regulation of energy expenditure. They are synthesized in the liver as primary BAs such as cholic acid (CA), chenodeoxycholic acid (CDCA) and conjugated forms. A variety of secondary BAs such as deoxycholic acid (DCA) and lithocholic acid (LCA) and their derivatives is synthesized in the intestine through the involvement of various microorganisms. In addition to essential physiological functions, BAs and their metabolites are also involved in the differentiation and functions of innate and adaptive immune cells such as macrophages (Macs), dendritic cells (DCs), myeloid derived suppressive cells (MDSCs), regulatory T cells (Treg), Breg cells, T helper (Th)17 cells, CD4 Th1 and Th2 cells, CD8 cells, B cells and NKT cells. Dysregulation of the BAs and their metabolites also affects development of some diseases such as inflammatory bowel diseases. We here summarize recent advances in how BAs and their metabolites maintain gut and systemic homeostasis, including the metabolism of the BAs and their derivatives, the role of BAs and their metabolites in the differentiation and function of immune cells, and the effects of BAs and their metabolites on immune-associated disorders.
Bile acid (BAs) are cholesterol-derived molecules involved in essential physiological processes including nutrient absorption, glucose homeostasis and regulation of energy expenditure (1). There are two main sites of BA biosynthesis, hepatocytes and gut microbiota. BAs are synthesized in the liver as primary BAs such as cholic acid (CA), chenodeoxycholic acid (CDCA) and their conjugated forms. A variety of secondary BAs such as deoxycholic acid (DCA) and lithocholic acid (LCA) and their derivatives, a large pool of bioactive molecules is synthesized in the intestine where they undergo bacteria-mediated transformation (2). BAs and their metabolites are abundant in the mammalian gut, and potentially distributed into other tissues and organs.
There exists a perfect immune system in different individuals, including innate immune cells such as macrophages (Macs), dendritic cells (DCs) and nature killer (NK) cells, and adaptive immune cells such as T cells and B cells. In addition to these cells, there also has a large amount of immune regulatory cells such as regulatory T cells (Treg cells), Breg cells, and innate immune lymphocytes (ILCs) to maintain local and systemic immune homeostasis. The differentiation and functions of these immune cells can be regulated by gut microbiota metabolites such as short-chain fatty acids (SCFAs) (3–5), tryptophan derived metabolites (6–8), and BA derivatives (9–12). BAs and their derivatives bind to multiple nuclear and cell surface receptors, which are expressed in the different immune cells such as Macs, DCs, MDSCs, Treg cells, Breg cells, ILCs, Th17 cells, CD4 Th1 cells, Th2 cells, CD8 cells, B cells and NKT cells. Each of BA and their derivatives has a different affinity for the receptor to which it can bind. While these receptors are bound and activated by different BA derivatives, they can affect the differentiation and function of different immune cells respectively. Understanding the effects of BAs and their metabolites on immune cells may elucidate a variety of disease states such as inflammatory bowel diseases, metabolic diseases, obesity, and other chronic inflammatory conditions. We here summarize recent advances in understanding the metabolism of BAs, the role of BAs and their derivatives in the differentiation and function of different immune cells, and the effects of BAs and their derivatives on immune-associated disorders.
BAs are the end-product of cholesterol metabolism (13, 14). The liver generates two primary BAs, i.e., CA and CDCA. The final products in the liver are mainly 3α-7α di-hydroxylated cholesterol derivatives, i.e., CDCA, and 3α-7α-12α-tri-hydroxylated derivatives, i.e., CA (14). These primary BAs in hepatocytes and/or in gut microbiota (15) are conjugated with glycine, taurine or other amino acids (15, 16). Then, the conjugated BAs are secreted into the intestine, becoming the substrate of an array of bacterial enzymes. This causes the generation of secondary BAs, i.e., LCA and DCA.
Secondary BAs DCA and LCA can be further modified into different derivatives by microbes (17). A range of oxo-, epi- and iso-derivatives of BAs is formed in the colon due to various dehydrogenation and epimerisation reactions in gut bacteria (18), such as 7-oxoCA, 7-oxoCDCA, 12-oxoCA and 12-oxoDCA (14). There also exist multiple forms of LCA derivatives such as allo-LCA, iso-LCA, isoalloLCA, 3-oxo-LCA, 3-oxoallo-LCA, and 3-ketoLCA (19, 20). In addition, the derivatives such as ursoDCA (UDCA) (21) and iso-DCA (9, 22) are also produced by 7α-hydroxysteroid dehydrogenase (7α-HSDH) and 7β-HSDH.
Gut microbiota is not only involved in the generation of conjugated BAs but also plays a critical role in the transformation of BAs into other metabolites (Figure 1). In human, there have four distinct ways to transform BAs, including deconjugation, dehydroxylation, oxidation, and epimerization, which have been well reviewed (18).
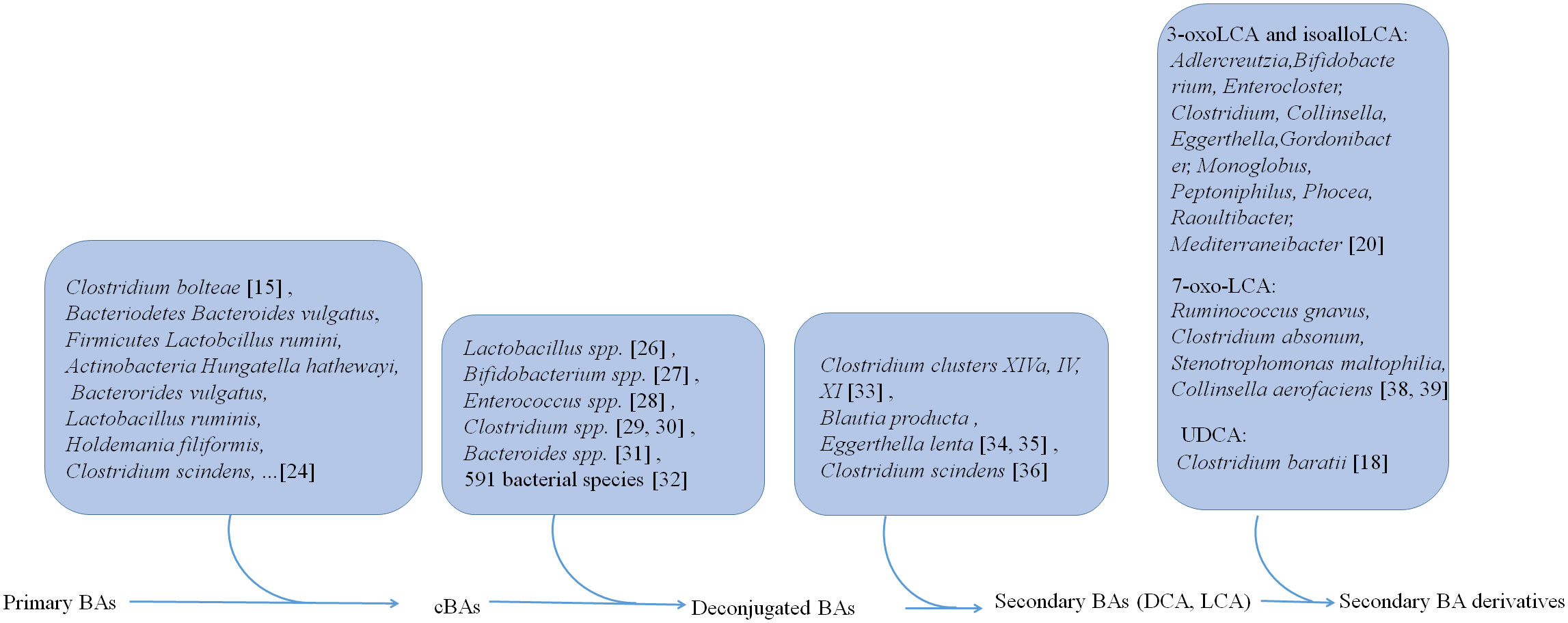
Figure 1 Gut microbiota is not only involved in the generation of conjugated BAs, but also plays a critical role in the transformation of BAs from conjugated BAs to deconjugated BAs and the generation of secondary BAs DCA and LCA and their derivatives.
Primary BAs can be conjugated with glycine and taurine in the liver. Then these conjugated BAs are released into the intestine via gallbladder (23). However, recent studies also show that gut microbiota such as Clostridium bolteae possesses an ability to conjugate BAs with phenylalanine, leucine, and tyrosine (15). 25 strains such as Bacteriodetes Bacteroides vulgatus, Firmicutes Lactobcillus ruminis and Actinobacteria Hungatella hathewayi, representing 24 species in the gut microbiota, can conjugate glycine to DCA, CDCA, or CA in vitro (24). 28 strains such as Bacterorides vulgatus, Lactobacillus ruminis, Holdemania filiformis, and Clostridium scindens, representing 27 species in the gut microbiota are capable of conjugating CDCA, DCA or CA to one or more other amino acids such as alanine, arginine and aspartate (24).
Liver derived conjugated BAs can be deconjugated in the small intestine by bile salt hydrolases (BSHs). These BSHs can be detected in gut microbiota (25) such as Lactobacillus spp (26)., Bifidobacterium spp (27)., Enterococcus spp (28)., Clostridium spp (29, 30)., and Bacteroides spp (31).. More recent studies show that BSHs can be found in 591 intestinal bacterial strains within 117 genera in human gut microbiota. Notably, 27.52% of these bacterial strains contains only BSH paralogs (32). These different phenotypes of BSHs exhibit different activity in the gut bacteria. BSH-T3, which is found in Lactobaclillus, shows the highest enzyme activity, whereas BSH-T5 and BSH-T6 mainly from Bacteroides, which have high percentage of paralogs, exhibit different enzyme and deconjugation activity (32).
After deconjugation, BAs can be converted into secondary BAs, i.e., DCA and LCA, and their derivatives by dehydroxylation, oxidation and epimerization.
Three distinct microbial 3α-, 7α-, and 12α- HSDHs, which result in oxidization and epimerization of specific hydroxyl groups on BAs can be found in gut microbes (33), such as Clostridium clusters XIVa, IV and XI. The bacteria such as C. scindens, C. hylemonae and C. perfringens are shown to produce enzymes capable of 3α-dehydrogenation. 3α-dehydrogenation also occurs in Blautia producta and Eggerthella lenta (34, 35). The BA transformations can also be carried out by 7-dehydroxylation in Clostridium scindens in vitro and in vivo (36). Recently, Funabashi et al. (37) showed that a set of six enzymes, which was necessary for conversion of CA to DCA, was engineered into a nonproducing bacteria, conferring production of DCA and LCA (37).
Paik et al. identified 12 human gut bacterial genera including Adlercreutzia, Bifidobacterium, Enterocloster, Clostridium, Collinsella, Eggerthella, Gordonibacter, Monoglobus, Peptoniphilus, Phocea, Raoultibacter, and Mediterraneibacter, which could convert LCA to 3-oxoLCA and isoLCA (20). In addition, both metabolites 3-oxoLCA and iso-alloLCA were absent in germ-free (GF) mouse models, also suggesting that these derivations were from microbiota (12). Ruminococcus gnavus, Clostridium absonum, Stenotrophomonas maltophilia, and Collinsella aerofaciens contribute to the ursoDCA pool via conversion of 7-oxo-LCA in an nicotinamide adenine dinucleotide (NADH) or nicotinamide-adenine dinucleotide phosphate (NADPH)-dependent fashion (38, 39). 7α-epimerization to UDCA also occurs in the gut bacterium members such as Clostridium baratii (18).
BAs and their metabolites can act on the receptors expressed in Macs, DCs, MDSCs, Tregs, Th17 cells, ILCs, CD4 cells, CD8 cells, B cells and NKT cells to modulate their differentiation and function for gut and systemic homeostasis (12, 14, 40–42) (Figure 2). These receptors include a range of nuclear receptors such as farnesoid X receptor (FXR), liver-X-receptor (LXR), pregnane X receptor (PXR), vitamin D receptor (VDR), retinoid related orphan receptor (RORγt), constitutive androstane receptor (CAR), and membrane receptors such as G-protein BA receptor 1 (GPBAR1) (Takeda G protein-coupled receptor 5 (TGR5)), sphingosine-1-phosphate receptor 2 (S1PR2), cholinergic receptor muscarinic 2 and 3 (CHRM2 and 3), and MAS related GPR (G-protein coupled receptor) family member X4 (MRGPRX4) (43), which have been reviewed by Biagioli et al. (44).
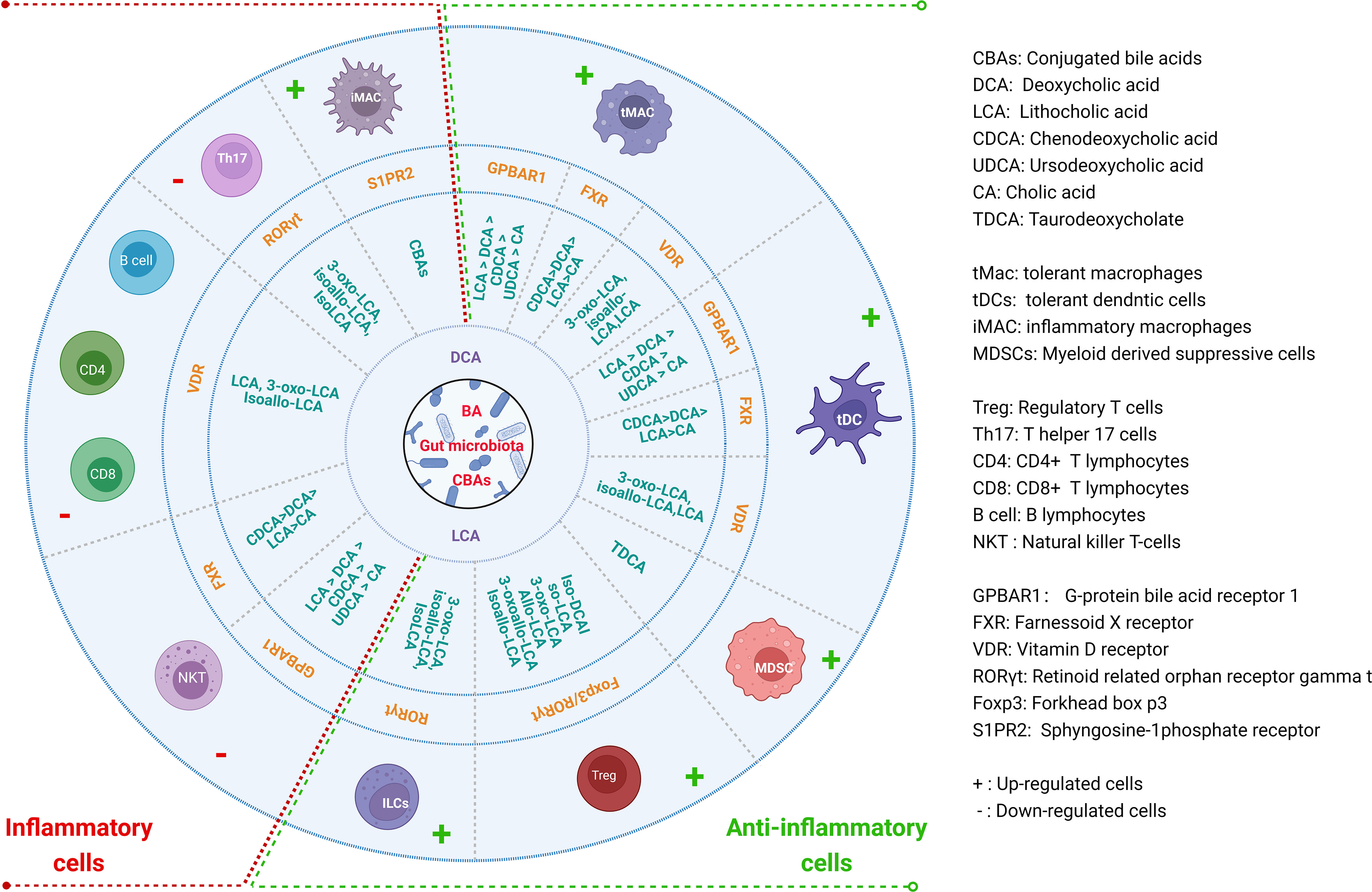
Figure 2 Regulations of BAs and their derivatives on the differentiation and function of immune cells for gut and systemic homeostasis. BAs and their derivatives not only promote the generation and function of anti-inflammatory cells, but also inhibit inflammatory cells through different receptors in the immune cells.
Macrophages (Macs) can be mainly divided two subpopulations, inflammatory macrophages (iMacs) and immune tolerogenic macrophages (tMacs). IMacs (M1) are mainly involved in pro-inflammatory responses, whereas tMacs (M2) are mainly involved in immune suppressive responses. Intestinal Macs reside either within the lamina propria (LP) or the muscle layer. Muller et al. (45) have discussed recent advances in gut Macs. In the resting intestine, mature resident (immune tolerogenic) ly6clow/-CX3CR1hiMHC IIhi Macs from inflammatory Ly6chigh monocytes/Macs can express IL-10 and maintain intestinal homeostasis (46). Studies found that BAs and their metabolites can induce immune tolerogenic Macs. However, BAs, especially cBAs also cause inflammatory Macs. The contradicts in the effects of the BAs on the Macs are derived from different receptors expressed in the Macs. The majority of BAs-activated receptors such as FXR, TGR5, VDR, LXRs, PXR and S1PR2 have been detected in myeloid cells (10).
TGR5 is essential to maintain a tolerogenic phenotype of the Macs (10, 47, 48). Its activation can promote Mac polarization from the M1 (pro-inflammatory phenotype) to the M2 (immune tolerogenic phenotype) Macs, and reduce pro-inflammatory cytokines (49). TGR5 activation also blocks NLRP3-dependent inflammation such as lipopolysaccharide-induced systemic inflammation, type-2 diabetes-related inflammation and alum-induced peritoneal inflammation (50, 51). Secondary BAs DCA or LCA can function as endogenous inhibitors of NLRP3 inflammasome activation by activating TGR5 (52), which can cause a TGR5-cAMP-PKA-dependent ubiquitination of NLRP3 to inhibit its activation (53). The knockout of TGR5 in mice can accelerate LPS-induced inflammation in the liver and abolish the suppressive effects of TGR5 agonist on inflammatory cytokines (54). TGR5 natural ligands are LCA > DCA > CDCA > UDCA > CA (55). FXR is also essential to maintain a tolerogenic phenotype of the Macs as demonstrated in FXR KO mice (10). FXR can activate SOCS3, CYP450 and fibroblasts growth factor 19 (FGF19) to inhibit inflammation. In addition, FXR also activates SHP to inhibit NF-κB, AP-1 and NLRP3 (56–59), and is recruited to the iNOS and IL-1β promoters to stabilize the NCoR1 complexes, which can make these genes in the basal state (60). The assembly of NLRP3 inflammasomes is also suppressed by FXR, which physically interacts with NLRP3 and caspase-1 (52, 61, 62). In addition, PXR as a nuclear receptor also binds to LCA (55). PXR activation decreases the expression of IL6, TNFα, and IL8 mRNAs (42).
Notably, high cellular concentrations (≈100–500 μM) of BAs, particularly the hydrophilic secondary BAs, might function as danger-associated molecular pattern molecules (DAMPs) to cause a calcium-dependent activation of NLRP3 inflammasome (52, 62). However, this happens only while Macs are preactivated after exposure to endotoxin (52, 62). The hydrophobic primary BA such as ChenoDCA (CDCA) can induce NLRP3 activation and secretion of IL-1β by promoting ROS production and K+ efflux in Macs (63). Hao H et al. also found that BAs synergistically with ATP induced a prolonged calcium influx and activated NLRP3 (53). In addition, the conjugated BAs such as tauroCA (TCA) can activate S1PR2, which is shown to promote immune cell infiltration and inflammation in mouse models (64). Activating S1PR2 promotes caspase-11-dependent Mac pyroptosis and worsens E. coli sepsis (65). S1PR2 is also activated by the conjugated BAs to result in proinflammatory effects that can increase liver damage (66). S1PR2 deficiency significantly reduces cholangiocyte proliferation and cholestatic injury (64). Blockade of S1PR2 inhibits S1P-induced NLRP3 priming and inflammatory cytokine secretion (67).
Dendritic cells (DCs) play a critical role in inducing protective adaptive immunity. However, DCs are also emerging as critical regulators of the immune responses (68). Secondary BA DCA suppresses LPS-induced expression of pro-inflammatory IL-1, IL-6, and TNFα in DCs (69), which can be rescued through DCA receptor TGR5 deficiency. The inhibitory effects of TGR5 are mediated through suppressing NF-κB by TGR5–cAMP–PKA signaling (69). BA-dependent TGR5 activation also induces the differentiation of human monocytes into IL-12 and TNF-α hypo-producing DCs via the TGR5-cAMP pathway (70). In addition, isoDCA can limit FXR activity in DCs and confer upon them an anti-inflammatory phenotype (9). The exposure of INT-747/obetiCA, which can activate FXR (10), greatly attenuates the differentiation CD14+ monocytes into mature DCs (71). A reduced number of activated DCs in the colon of mice administered with INT-747/obetiCA was also observed. In addition, VDR activation also inhibits the production of inflammatory cytokines, and the differentiation and maturation of DCs (72).
Myeloid derived suppressor cells (MDSCs) play a key role in the immune suppression in some diseases, especially in cancer, and also have prominent role in tumor angiogenesis, drug resistance, and promotion of tumor metastases (73). Many pathogens, ranging from viruses to multicellular parasites, can promote the expansion of MDSCs (74). These MDSCs can be divided into monocytic and granulocytic MDSCs. The BA derivative TDCA can increase the number of granulocytic MDSCs in the spleen of septic mice (75).
There have multiple CD4 T helper (Th) cell subsets such as FoxP3+ regulatory T cells, RORγt+IL17+ Th17 cells, T-bet+IFNγ+ Th1 cells and Gata3+IL4+IL13+ Th2 cells. These CD4 Th cells play a critical role in maintaining the immune homeostasis of individuals. Studies have found that the differentiation and function of these cells can be regulated by BAs and their derivatives.
1) FoxP3+T regulatory cells. FoxP3+T regulatory (Treg) cells express transcription factor Foxp3 (76, 77), and differentiate in the thymus or the periphery (78). BAs and their metabolites can affect the differentiation and function of Treg cells, which help protect against extracellular pathogens and maintain host immune tolerance, respectively (79). Indeed, secondary BAs such as isoalloLCA and isoDCA can promote the differentiation of Treg cells (9, 11, 12, 80, 81) (Figure 3). The isoalloLCA may be through the production of mitochondrial reactive oxygen species (mitoROS) to promote the expression of Foxp3 (11). Nuclear hormone receptor NR4A1 is also required for the regulation of isoalloLCA in Treg cells (80). Whereas the secondary BA derivatives isoDCA mediated Treg cells is through diminishing DC immunostimulatory properties (9). A distinct Treg population expressing the transcriptional factor RORγ can also be induced in the colonic LP by colonization with gut symbionts (81, 82). These RORγ+ Treg cells have a distinct phenotype (Helios– and Nrp1–). Their accumulation is influenced by enteric factors derived from diet or commensal colonization (12, 81). In addition, VDR also drives T cell maturation facilitating the induction of T regulatory cells (83) and reduces Th17 cell formation (84).
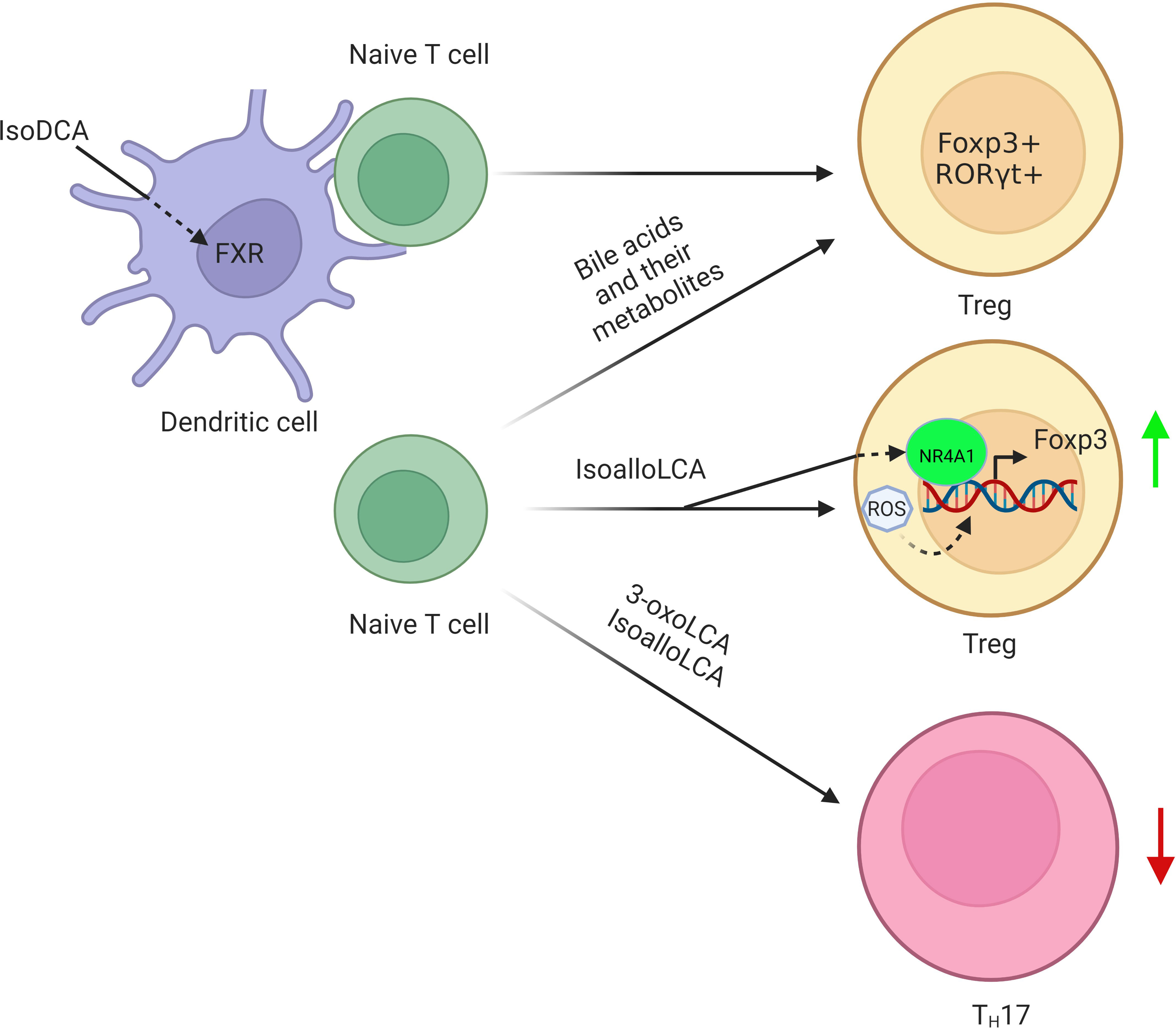
Figure 3 Regulation of BAs and their derivatives on the differentiation of Tregs. BA derivative isoDCA increases Foxp3 induction by diminishing DC immunostimulatory properties. IsoalloLCA promotes the differentiation of Treg cells through the production of mitochondrial reactive oxygen species, which lead to increased expression of Foxp3. Nuclear hormone receptor NR4A1 is also required for the effect of isoalloLCA on Treg cells. In addition, a distinct Treg population expressing the transcriptional factor RORγ is also induced in the colonic lamina propria by BAs and their metabolites.
2) RORγt+IL17+ Th17 cells. RORγt+IL17+ Th17 (Th17) cells cause autoimmunity and inflammation (85). The nuclear hormone retinoid-related orphan receptor γ (RORγ) is selectively expressed by Th17 cells, acting as a critical transcription factor for Th17 cell differentiation in chronic inflammation and autoimmune diseases (86). The BA metabolite 3-oxoLCA, which can directly bind to RORγ (11), inhibits Th17 cell differentiation by blocking the function of RORγ (11, 87). Similar to 3-oxoLCA, isoLCA also suppresses Th17 cell differentiation by inhibiting RORγ (20). A sulfated product of LCA, lithocholic acid 3-sulfate (CA-3-S) can also selectively inhibit Th17 cell differentiation by targeting RORγ (88). Thus, the inhibition of RORγ provides therapeutic benefits in the intestinal inflammation and reduces the frequencies of Th17 cells (89).
3) T-bet+IFNγ+ Th1 and Gata3+IL4+IL13+ Th2 cells. T-bet+IFNγ+ Th1 (Th1) and Gata3+IL4+IL13+ Th2 (Th2) cells can regulate appropriate cellular and humoral immune responses to pathogens and be involved in the progress of many diseases. Both IL-12 and IFN-γ make naive CD4+ T cells highly express T-bet and signal transducer and activator of transcription (STAT) 4 to differentiate to Th1 cells, while IL-4 makes naive CD4+ T cells highly express STAT6 and Gata3 to differentiate to Th2 cells. Through a VDR-dependent mechanism, the unconjugated LCA in physiological concentrations can inhibit the activation of human and mouse CD4+ Th1 cells, resulting in decreased TNFα and INF-γ production (90). VDR activation also promotes a shift from the Th1 to the Th2 phenotype through increased production of the transcription factors c-maf and Gata-3 (91). VDR can be activated by LCA and its metabolites such as 3-oxoLCA, 3-ketoLCA, LCA acetate, LCA propionate and iso-alloLCA. In addition, PXR activation also inhibit CD4 T cell proliferation in vitro. Liver puncture biopsy specimens from 34 matched patients before and after UDCA treatment showed the relationship between the infiltration of CD4 T cells and UDCA (92).
4) Innate lymphoid cells. Innate lymphoid cells (ILCs) are the importance in tissue homeostasis, morphogenesis, metabolism, repair, and regeneration. These cells can be divided into 3 groups, ILC1, ILC2 and ILC3 (93). In terms of function, ILC1s, ILC2s, and ILC3s mirror CD4+ Th1, Th2, and Th17 cells respectively (94). ILC3s can highly express BA receptors such as TGR5, FXR, and RORγt. The receptor RORγt is required for the generation of ILC3 (95).
BA derivatives 24-NorursoDCA (NorUDCA) can reshape immunometabolism in CD8+ T cells and alleviate hepatic inflammation (96). TCA inhibits the response to IFNα therapy in the patients with chronic hepatitis B through suppressing CD8+ T and NK cell function (97). Cholestatic mice are featured with dysfunctional T cells response, as indicated by decreased sub-population of CD4+ and CD8+ cells and increased CTLA-4+CD4+ and CD8+ subsets (98). Transcription factor VDR activation also reduces the ongoing proliferation of CD8+ cells (99). PXR is expressed in human CD8+ T lymphocytes. PXR activation also inhibits CD8+ cell proliferation in vitro.
B cells play an important role for immune response not only in antibody production but also in antigen presentation and cytokine production. BA receptor VDR activation reduces the proliferation of B lymphocytes (99), induces apoptosis of activated B cell apoptosis (100) and inhibits Ig production by B cells (101). Indeed, recent studies show that BAs can impair vaccine response, possibly via inhibiting post-class-switched memory B cell responses (102).
NKT cells are an unusual population of T cells, which can recognize lipids presented by CD1d, a non-classical class I like molecule. These cells include two subtype, type I and II NKT cells, which pay a critical role in tumor immunity. Type I NKT cells generally promote tumor immunity; whereas type II NKT cells suppress it. But, type I NKT cells can also induce immunosuppressive cells such as Treg. BA receptor FXR activation in NKT cells results in a profound inhibition to produce osteopontin, a potent pro-inflammatory mediator along with IL-1β and IFN-γ (60). TGR5 agonists induce NKT cells polarization toward IL-10 secreting type I NKT cells and significantly expand the subset of IL-10 secreting type II NKT cells (103). Recent studies show that gut microbiota-mediated BA metabolism can regulate liver antitumor immunity via controlling an accumulation of NKT cells (104).
BAs and their metabolites play an important role in maintaining the homeostasis of local and system immunes. Damages of the homeostasis are related to the occurrence and development of immune-associated disorders such as gut diseases, metabolic diseases, tumors, neurodegenerative diseases, allergic diseases, autoimmune diseases and infectious diseases (Figure 4) (105).
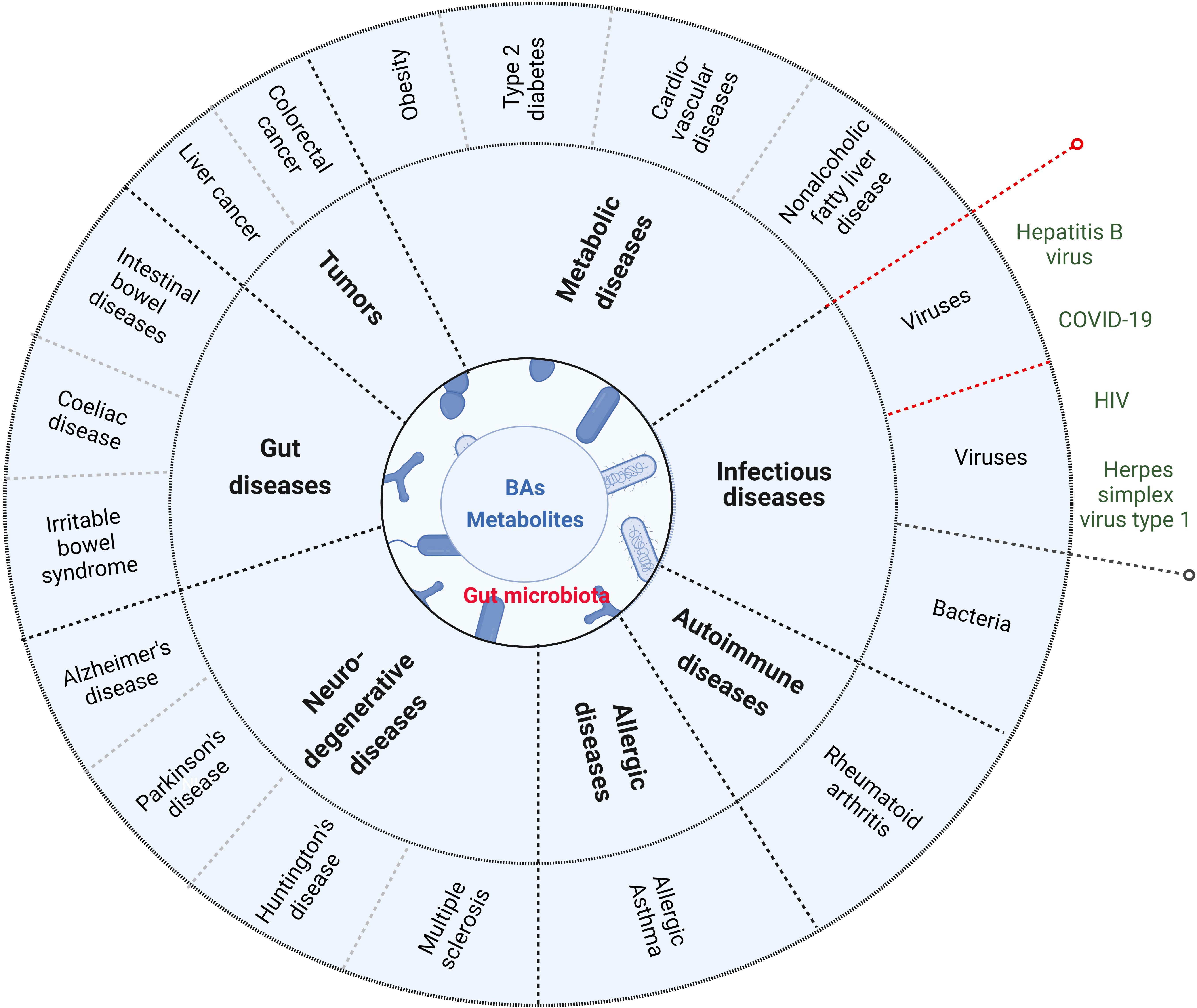
Figure 4 BAs and their metabolites are related to the occurrence and development of immune-associated disorders such as gut diseases (inflammatory bowel disease, coeliac disease and irritable bowel syndrome), metabolic diseases (obesity, type 2 disbetes, cardiovascular disease and nonalcoholic fatty liver disease), tumor (such as colorectal cancer and liver cancer), neurodegenerative disease (Alzheimer’s disease, Parkinson’s disease, Huntington’s disease and Multiple sclerosis), allergic diseases, autoimmune diseases and infections (such as bacterium infection). Red dotted lines indicate that BAs and their metabolites in virus infection can also promote innate immunes.
Gut diseases such as inflammatory bowel diseases (IBDs), coeliac disease and irritable bowel syndrome are gut inflammation associated diseases. Intestinal bowel diseases (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are chronic relapsing disorders (14). The effects of BAs on IBD have been reviewed (10, 44, 106). Accumulating evidences have shown that the gut microbiota plays a pivotal role in maintaining intestinal homeostasis. There exist decreased microbial diversity and abnormal microbial composition in the patients with IBD (99), which are characterized by increased phyla Proteobacteria and Fusobacteria (mainly Fusobacteria varium in UC and Fusobacteriaceae in CD patients), and reduced phyla Firmicutes (107–111). Since the majority of BSH expressing bacteria is members of Firmicutes phylum (112), these changes might impact on BA metabolism, which is related to the occurrence and development of IBD (113).
Recent metabolomics has revealed a consistent defect in the BA metabolism, which is companied with an increase in primary BAs and a reduction in secondary BAs in the patients with IBD (114, 115). Vantrappen et al. first demonstrated that the decrease in the BA pool size is inversely correlated with the Crohn’s disease activity index (116). There also exhibit a severe reduction in fecal secondary BAs such as DCA and LCA, and an increased abundance of primary bile acids such as CA and CDCA in active patients with IBD (115). The levels of 3-oxoLCA and isoLCA are also significantly reduced in the patients with IBD (20). Similar findings with increased primary BAs and their conjugated forms, and reduced secondary and unconjugated BAs (117) are also observed in paediatric patients with IBD (118). In addition, a reduction of LCA also impacts on activation of VDR, that is an anti-inflammatory receptor in macrophages. The patients with IBD are also characterized by an increased 3-sulfated DCA and LCA in the feces (119), suggesting that in addition to BSH-dependent deamidation, other biotransformations such as sulfatation might also be impaired in patients with IBD (119). Thus, the supplementation of secondary BAs may be a potential strategy for the therapy of the patients with IBD.
Primary and secondary BAs are identified as signaling molecules acting on a family of cell membrane and nuclear receptors such as TGR5, FXR, PXR and VDR, which are highly expressed in the gastrointestinal tract. Studies have demonstrated that both BA receptors FXR and TGR5 are essential to maintain a tolerogenic phenotype of intestine immune. Ablating these receptors can promote the polarization of intestinal T cells toward a pro-inflammatory phenotype (14). FXR or TGR5 KO mice are prone to develop an exaggerated inflammatory response upon exposure to dextran sodium sulfate (DSS) or trinitrobenzene sulfonate (TNBS) (120). Upon BA activation, FXR controls expression of genes, which can limit the inflammatory responses. FXR KO naïve mice are characterized by intestinal inflammation with increased expression of pro-inflammatory cytokines as compared to wild type (WT) mice (120). In addition to FXR, PXR is also involved in IBD. Human PXR activation represses intestinal immune response in a NF-κB dependent manner (121). Compared with WT mice, DSS induced colitis was more severe in PXR KO mice (122), which could be protected by pregnenolone 16α-carbonitrile (PCN, a human PXR agonist) (122). Notably, several studies showed that PXR polymorphisms had no markedly effects on the risk of IBD (123). Another nuclear receptor VDR can also be activated by the secondary BA LCA and/or its metabolites 3-oxoLCA and iso-alloLCA (12). Studies found that VDR plays a beneficial role in patients with IBD (124). Its polymorphisms are related with susceptibility to IBD (125). In mouse model of colitis, VDR KO can exacerbate the symptoms in IL-10 KO mice, whereas vitamin D supplementation improves the symptoms. Intestinal epithelial cells-specific VDR KO mice showed a more severe colitis than WT mice (126). Taken together, these receptors may provide new perspectives on the treatment of intestine diseases such as IBD.
Metabolic diseases such as obesity, type 2 diabetes (T2D), cardiovascular disease and non-alcoholic fatty liver disease (NAFLD), are generally considered a chronic inflammatory disease (127, 128). Altered bile acid metabolism can contribute to these chronic inflammatory diseases (129). BAs and their derivatives are valuable therapeutic agents for treating these inflammatory metabolic diseases (129).
Obesity is mainly induced by the disequilibrium of energy intake and energy expenditure, which results in metabolic disorders and chronic low-grade inflammation. UDCA supplementation can control diet-induced obesity in prenatally malnourished mice (130). Dietary acetic acid suppress high-fat diet-induced obesity in mice by altering taurine conjugated bile acids metabolism (131). Watanabe et al. reported that the administration of BAs to mice increased energy expenditure in brown adipose tissue (BAT), preventing obesity and resistance to insulin (132). The proportion of non-12-OH bile acids, including HCA, HDCA, glycohyodeoxycholic acid (GHDCA), UDCA, GUDCA, and CDCA in total bile acid is significantly lower in people with high body mass index (BMI), indicating that non-12-OH bile acids may contribute to the process of obesity (133). In the individuals with obesity, T2D and NAFLD, which are characterized by recruitment of immune cells, abnormal production of cellular inflammatory cytokines and acute phase reactants, and activation of inflammasomes, are associated with dysregulation of BA homeostasis (127, 134). Recent studies suggested that size and/or composition of BA pool changed in patients with T2D, and found that BAs and their derivatives improved T2D by reducing the levels of inflammatory cytokines (135). BA metabolism is also altered in patients with hepatic steatosis and glucose and lipid dysmetabolism (136). Dysregulation of BA metabolism was linked to steatosis, inflammation, and fibrosis in patients with NAFLD (137). Intervention of BAs could effectively control and prevent obesity and NAFLD (132, 138). Studies from animal models and human patients have found that NAFLD disease progression is closely associated with BA dysregulation (139–143).
Inflammation plays an important role in the development and progression of cardiovascular diseases (CVDs). Hypertension and hyperlipidemia, the key risk factors of CVDs, are related to inflammation in the heart and vessels (144). The signaling pathways mediated by immune and inflammatory mediators have been implicated within the atherosclerotic lesion (145). A growing number of studies have shown a strong relationship between gut microbiota and CVDs such as coronary atherosclerosis, hypertension and heart failure (146). High fiber diet significantly improved cardiac function through modulating the composition of intestinal flora and the production of metabolites production, including the biosynthesis of bile acids and linoleic acid metabolism (147). Dietary mannan oligosaccharides can increase fecal BA excretion and decrease atherosclerosis development (148).
BAs have been considered as pro-carcinogenic molecules (149, 150). Studies have also implied the involvement of BAs in colorectal, gastric, hepatocellular, pancreatic, breast, prostate and ovarian cancer (149). However, inflammation play a decisive role in inducing tumorigenesis, promoting tumor development, tumor invasion and migration (151). Human epidemiological evidence has confirmed the close relationship between chronic inflammation and tumorigenesis (152) such as that inflammation is a common medical complication in colorectal cancer (CRC) patients, which plays significant roles in tumor progression and immunosuppression (153). Some epidemiological studies have shown an association between fecal and serum BAs and CRC (154). TGR5 activation by UDCA and LCA can exert anti-inflammatory responses through TLR4 activation or by reducing pro-inflammatory cytokine production in the colon that can decrease the frequency of developing CRC (155). Altered BA metabolism also promoted helicobacter pylori-induced inflammation-driven gastric carcinogenesis (156). Remarkably decreasing percentages of serum conjugated DCA were closely associated with hepatocellular carcinoma (HCC) (157).
Various studies have shown the role of neuro-inflammation in the occurrence, diagnosis, and treatment of neurodegenerative diseases. Neuro-inflammation can trigger the formation of other factors responsible for causing several neuronal diseases including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), multiple sclerosis (MS), ischemia, and several others (158, 159). Parkinson disease (PD) is a progressive neurodegenerative disease that affects peripheral organs as well as the central nervous system and involves a fundamental role of neuro-inflammation in its pathophysiology. There is increasing evidence for inflammation as a determinant in the pathogenesis of Parkinson’s disease (160). UDCA and TUDCA have shown neuroprotective properties in these neurodegenerative diseases (161). TDCA is also as a potential therapeutic tool in neurodegenerative diseases (162).
Rheumatoid arthritis (RA) is an autoimmune disease characterized by joint destruction, synovitis, and pannus formation. Additional proinflammatory cytokines, such as IL-7, IL-17, IL-21, IL -23, GM-CSF, IL-1β, IL-18, IL-33 and IL-2 are involved in the pathogenesis of RA (163). Elevated levels of primary BAs have been found in the feces of some RA patients, which can be used to predict RA arthritis severity (164). Secondary BAs such as DCA and LCA can suppress macrophage cytokine production via FXR (165).
Obesity is a risk factor for the development of asthma and is associated with worsening symptoms and poor asthma control (166). Altered bile acid profiles have been reported in asthmatic patients. GCA, GDC, TCDC and taurocholate increased with asthma, compared to healthy individuals (167).
Bacterium and virus infection can cause inflammation. BAs are associated with infectious diseases such as Clostridioides difficile or Salmonella Typhimurium infection (105). BAs regulate immune responses upon ligation of these two receptors FXR and TGR5, which are located at the interface of the host immune system with the intestinal microbiota (10). However, studies have also reported that BAs activated several key innate signaling pathways to potentiate antiviral immunity (10). The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon (168, 169).
There are multiple forms of BAs such as conjugated and deconjugated primary BAs, secondary BAs DCA and LCA, and their derivatives. While primary BAs are generated in the liver, four distinct ways, including deconjugation, dehydroxylation, dehydrogenation and epimerization are used to transform primary BAs into secondary BAs and their derivatives by gut microbiota. These primary and secondary BAs and their derivatives can act on the receptors expressed in Macs, MDSCs, DCs, Tregs, Th17 cells, ILCs, CD4 cells, CD8 cells, B cells and NKT cells to modulate their differentiation and function, which can affect both the innate and adaptive immune responses for homeostasis. In addition, dysregulation of BA homeostasis is also found in the inflammation associated disorders such as IBD.
BAs have been used therapeutically in China for over 2500 years (170). Currently, the Food and Drug Administration (FDA) has approved a formulation of UDCA, Ursodiol, which has vast beneficial effects such as anti-inflammatory (171). It has been used to treat a variety of diseases such as cholesterol gallstones, primary biliary cirrhosis, primary sclerotic cholangitis, nonalcoholic fatty liver disease, chronic viral hepatitis C, recurrent colonic adenomas, cholestasis of pregnancy, and recurrent pancreatitis (171). With understanding of BAs and their metabolites on the local and systemic immunes, more precise therapy based on BA metabolites will be used in inflammation-associated diseases.
XS, YG made the figures and wrote the original manuscript. RY improved and wrote the final manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by NSFC grants 91842302, 81970488, 91029736, 9162910, 91442111 and 31570114.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab (2016) 24:41–50. doi: 10.1016/j.cmet.2016.05.005
2. Shapiro H, Kolodziejczyk AA, Halstuch D, Elinav E. Bile acids in glucose metabolism in health and disease. J Exp Med (2018) 215:383–96. doi: 10.1084/jem.20171965
3. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science (2013) 341:569–73. doi: 10.1126/science.1241165
4. Luu M, Visekruna A. Short-chain fatty acids: Bacterial messengers modulating the immunometabolism of T cells. Eur J Immunol (2019) 49:842–8. doi: 10.1002/eji.201848009
5. Parada Venegas D, de la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
6. Hezaveh K, Shinde RS, Klotgen A, Halaby MJ, Lamorte S, Ciudad MT, et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity (2022) 55:324–40.e328. doi: 10.1016/j.immuni.2022.01.006
7. Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory b cells. Cell Metab (2020) 31:837–51.e810. doi: 10.1016/j.cmet.2020.03.003
8. Su X, Gao Y, Yang R. Gut microbiota-derived tryptophan metabolites maintain gut and systemic homeostasis. Cells (2022) 11(15):2296. doi: 10.3390/cells11152296
9. Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature (2020) 581:475–9. doi: 10.1038/s41586-020-2193-0
10. Fiorucci S, Biagioli M, Zampella A, Distrutti E. Bile acids activated receptors regulate innate immunity. Front Immunol (2018) 9:1853. doi: 10.3389/fimmu.2018.01853
11. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control TH17 and treg cell differentiation. Nature (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
12. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature (2020) 577:410–5. doi: 10.1038/s41586-019-1865-0
13. Chiang JY. Bile acids: regulation of synthesis. J Lipid Res (2009) 50:1955–66. doi: 10.1194/jlr.R900010-JLR200
14. Fiorucci S, Carino A, Baldoni M, Santucci L, Costanzi E, Graziosi L, et al. Bile acid signaling in inflammatory bowel diseases. Dig Dis Sci (2021) 66:674–93. doi: 10.1007/s10620-020-06715-3
15. Quinn RA, Melnik AV, Vrbanac A, Fu T, Patras KA, Christy MP, et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature (2020) 579:123–9. doi: 10.1038/s41586-020-2047-9
16. Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem (2003) 72:137–74. doi: 10.1146/annurev.biochem.72.121801.161712
17. Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol (2015) 11:685–90. doi: 10.1038/nchembio.1864
18. Guzior DV, Quinn RA. Review: microbial transformations of human bile acids. Microbiome (2021) 9:140. doi: 10.1186/s40168-021-01101-1
19. Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature (2021) 599:458–64. doi: 10.1038/s41586-021-03832-5
20. Paik D, Yao L, Zhang Y, Bae S, D'Agostino GD, Zhang M, et al. Human gut bacteria produce TauEta17-modulating bile acid metabolites. Nature (2022) 603:907–12. doi: 10.1038/s41586-022-04480-z
21. Heinken A, Ravcheev DA, Baldini F, Heirendt L, Fleming RMT, Thiele I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome (2019) 7:75. doi: 10.1186/s40168-019-0689-3
22. Song I, Gotoh Y, Ogura Y, Hayashi T, Fukiya S, Yokota A. Comparative genomic and physiological analysis against clostridium scindens reveals eubacterium sp. c-25 as an atypical deoxycholic acid producer of the human gut microbiota. Microorganisms (2021) 9(11):2254. doi: 10.3390/microorganisms9112254
23. Kiriyama Y, Nochi H. Physiological role of bile acids modified by the gut microbiome. Microorganisms (2021) 10(1):68. doi: 10.3390/microorganisms10010068
24. Lucas LN, Barrett K, Kerby RL, Zhang Q, Cattaneo LE, Stevenson D, et al. Dominant bacterial phyla from the human gut show widespread ability to transform and conjugate bile acids. mSystems (2021) e0080521. doi: 10.1128/mSystems.00805-21
25. Bourgin M, Kriaa A, Mkaouar H, Mariaule V, Jablaoui A, Maguin E, et al. Bile salt hydrolases: At the crossroads of microbiota and human health. Microorganisms (2021) 9(6):1122. doi: 10.3390/microorganisms9061122
26. Foley MH, O'Flaherty S, Allen G, Rivera AJ, Stewart AK, Barrangou R, et al. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc Natl Acad Sci U S A (2021) 118(6):e2017709118. doi: 10.1073/pnas.2017709118
27. Xiao Y, Zhao J, Zhang H, Zhai Q, Chen W. Mining genome traits that determine the different gut colonization potential of lactobacillus and bifidobacterium species. Microb Genom (2021) 7(6):000581. doi: 10.1099/mgen.0.000581
28. Bhagwat A, Annapure US. In vitro assessment of metabolic profile of enterococcus strains of human origin. J Genet Eng Biotechnol (2019) 17:11. doi: 10.1186/s43141-019-0009-0
29. Coleman JP, Hudson LL. Cloning and characterization of a conjugated bile acid hydrolase gene from clostridium perfringens. Appl Environ Microbiol (1995) 61:2514–20. doi: 10.1128/aem.61.7.2514-2520.1995
30. Gopal-Srivastava R, Hylemon PB. Purification and characterization of bile salt hydrolase from clostridium perfringens. J Lipid Res (1988) 29:1079–85. doi: 10.1016/S0022-2275(20)38464-9
31. Adhikari AA, Seegar TCM, Ficarro SB, McCurry MD, Ramachandran D, Yao L, et al. Development of a covalent inhibitor of gut bacterial bile salt hydrolases. Nat Chem Biol (2020) 16:318–26. doi: 10.1038/s41589-020-0467-3
32. Song Z, Cai Y, Lao X, Wang X, Lin X, Cui Y, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome (2019) 7:9. doi: 10.1186/s40168-019-0628-3
33. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res (2006) 47:241–59. doi: 10.1194/jlr.R500013-JLR200
34. Harris SC, Devendran S, Mendez-Garcia C, Mythen SM, Wright CL, Fields CJ, et al. Bile acid oxidation by eggerthella lenta strains C592 and DSM 2243(T). Gut Microbes (2018) 9(6):523–39. doi: 10.1080/19490976.2018.1458180
35. Eggert T, Bakonyi D, Hummel W. Enzymatic routes for the synthesis of ursodeoxycholic acid. J Biotechnol (2014) 191:11–21. doi: 10.1016/j.jbiotec.2014.08.006
36. Marion S, Studer N, Desharnais L, Menin L, Escrig S, Meibom A, et al. In vitro and in vivo characterization of clostridium scindens bile acid transformations. Gut Microbes (2019) 10:481–503. doi: 10.1080/19490976.2018.1549420
37. Funabashi M, Grove TL, Wang M, Varma Y, McFadden ME, Brown LC, et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature (2020) 582:566–70. doi: 10.1038/s41586-020-2396-4
38. Lee JY, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A. Contribution of the 7beta-hydroxysteroid dehydrogenase from ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res (2013) 54:3062–9. doi: 10.1194/jlr.M039834
39. Ferrandi EE, Bertolesi GM, Polentini F, Negri A, Riva S, Monti D. In search of sustainable chemical processes: cloning, recombinant expression, and functional characterization of the 7alpha- and 7beta-hydroxysteroid dehydrogenases from clostridium absonum. Appl Microbiol Biotechnol (2012) 95:1221–33. doi: 10.1007/s00253-011-3798-x
40. Biagioli M, Carino A, Cipriani S, Francisci D, Marchiano S, Scarpelli P, et al. The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J Immunol (2017) 199:718–33. doi: 10.4049/jimmunol.1700183
41. Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, et al. Vitamin d receptor as an intestinal bile acid sensor. Science (2002) 296:1313–6. doi: 10.1126/science.1070477
42. Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A (2001) 98:3369–74. doi: 10.1073/pnas.051551698
43. Yu H, Zhao T, Liu S, Wu Q, Johnson O, Wu Z, et al. MRGPRX4 is a bile acid receptor for human cholestatic itch. Elife (2019) 8:e48431. doi: 10.7554/eLife.48431
44. Biagioli M, Marchiano S, Carino A, Di Giorgio C, Santucci L, Distrutti E, et al. Bile acids activated receptors in inflammatory bowel disease. Cells (2021) 10(6):1281. doi: 10.3390/cells10061281
45. Muller PA, Matheis F, Mucida D. Gut macrophages: key players in intestinal immunity and tissue physiology. Curr Opin Immunol (2020) 62:54–61. doi: 10.1016/j.coi.2019.11.011
46. Bain CC, Bravo-Blas A, Scott CL, Perdiguero EG, Geissmann F, Henri S, et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol (2014) 15:929–37. doi: 10.1038/ni.2967
47. Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem (2003) 278:9435–40. doi: 10.1074/jbc.M209706200
48. Haselow K, Bode JG, Wammers M, Ehlting C, Keitel V, Kleinebrecht L, et al. Bile acids PKA-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J Leukoc Biol (2013) 94:1253–64. doi: 10.1189/jlb.0812396
49. Shi Y, Su W, Zhang L, Shi C, Zhou J, Wang P, et al. TGR5 regulates macrophage inflammation in nonalcoholic steatohepatitis by modulating NLRP3 inflammasome activation. Front Immunol (2020) 11:609060. doi: 10.3389/fimmu.2020.609060
50. Pols TW, Nomura M, Harach T, Lo Sasso G, Oosterveer MH, Thomas C, et al. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab (2011) 14:747–57. doi: 10.1016/j.cmet.2011.11.006
51. Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity (2016) 45:944. doi: 10.1016/j.immuni.2016.10.009
52. Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity (2016) 45:802–16. doi: 10.1016/j.immuni.2016.09.008
53. Hao H, Cao L, Jiang C, Che Y, Zhang S, Takahashi S, et al. Farnesoid X receptor regulation of the NLRP3 inflammasome underlies cholestasis-associated sepsis. Cell Metab (2017) 25:856–867.e855. doi: 10.1016/j.cmet.2017.03.007
54. Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated b cells (NF-kappaB) in mice. Hepatology (2011) 54:1421–32. doi: 10.1002/hep.24525
55. Di Vincenzo F, Puca P, Lopetuso LR, Petito V, Masi L, Bartocci B, et al. Bile acid-related regulation of mucosal inflammation and intestinal motility: From pathogenesis to therapeutic application in IBD and microscopic colitis. Nutrients (2022) 14(13):2664. doi: 10.3390/nu14132664
56. Bertolini A, Fiorotto R, Strazzabosco M. Bile acids and their receptors: modulators and therapeutic targets in liver inflammation. Semin Immunopathol (2022) 44:547–64. doi: 10.1007/s00281-022-00935-7
57. Chanda D, Park JH, Choi HS. Molecular basis of endocrine regulation by orphan nuclear receptor small heterodimer partner. Endocr J (2008) 55:253–68. doi: 10.1507/endocrj.K07E-103
58. Fiorucci S, Distrutti E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol Med (2015) 21:702–14. doi: 10.1016/j.molmed.2015.09.001
59. Yang CS, Kim JJ, Kim TS, Lee PY, Kim SY, Lee HM, et al. Small heterodimer partner interacts with NLRP3 and negatively regulates activation of the NLRP3 inflammasome. Nat Commun (2015) 6:6115. doi: 10.1038/ncomms7115
60. Mencarelli A, Renga B, Migliorati M, Cipriani S, Distrutti E, Santucci L, et al. The bile acid sensor farnesoid X receptor is a modulator of liver immunity in a rodent model of acute hepatitis. J Immunol (2009) 183:6657–66. doi: 10.4049/jimmunol.0901347
61. Garcia-Irigoyen O, Moschetta AA. Novel protective role for FXR against inflammasome activation and endotoxemia. Cell Metab (2017) 25:763–4. doi: 10.1016/j.cmet.2017.03.014
62. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell (2014) 157:1013–22. doi: 10.1016/j.cell.2014.04.007
63. Gong Z, Zhou J, Zhao S, Tian C, Wang P, Xu C, et al. Chenodeoxycholic acid activates NLRP3 inflammasome and contributes to cholestatic liver fibrosis. Oncotarget (2016) 7:83951–63. doi: 10.18632/oncotarget.13796
64. Wang Y, Aoki H, Yang J, Peng K, Liu R, Li X, et al. The role of sphingosine 1-phosphate receptor 2 in bile-acid-induced cholangiocyte proliferation and cholestasis-induced liver injury in mice. Hepatology (2017) 65:2005–18. doi: 10.1002/hep.29076
65. Song F, Hou J, Chen Z, Cheng B, Lei R, Cui P, et al. Sphingosine-1-phosphate receptor 2 signaling promotes caspase-11-dependent macrophage pyroptosis and worsens escherichia coli sepsis outcome. Anesthesiology (2018) 129:311–20. doi: 10.1097/ALN.0000000000002196
66. Keitel V, Stindt J, Haussinger D. Bile acid-activated receptors: GPBAR1 (TGR5) and other G protein-coupled receptors. Handb Exp Pharmacol (2019) 256:19–49. doi: 10.1007/164_2019_230
67. Hou L, Yang L, Chang N, Zhao X, Zhou X, Dong C, et al. Macrophage sphingosine 1-phosphate receptor 2 blockade attenuates liver inflammation and fibrogenesis triggered by NLRP3 inflammasome. Front Immunol (2020) 11:1149. doi: 10.3389/fimmu.2020.01149
68. Gardner A, de Mingo Pulido A, Ruffell B. Dendritic cells and their role in immunotherapy. Front Immunol (2020) 11:924. doi: 10.3389/fimmu.2020.00924
69. Hu J, Wang C, Huang X, Yi S, Pan S, Zhang Y, et al. Gut microbiota-mediated secondary bile acids regulate dendritic cells to attenuate autoimmune uveitis through TGR5 signaling. Cell Rep (2021) 36:109726. doi: 10.1016/j.celrep.2021.109726
70. Ichikawa R, Takayama T, Yoneno K, Kamada N, Kitazume MT, Higuchi H, et al. Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells via a TGR5-dependent pathway. Immunology (2012) 136:153–62. doi: 10.1111/j.1365-2567.2012.03554.x
71. Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut (2011) 60:463–72. doi: 10.1136/gut.2010.212159
72. Szeles L, Keresztes G, Torocsik D, Balajthy Z, Krenacs L, Poliska S, et al. 1,25-dihydroxyvitamin D3 is an autonomous regulator of the transcriptional changes leading to a tolerogenic dendritic cell phenotype. J Immunol (2009) 182:2074–83. doi: 10.4049/jimmunol.0803345
73. Gabrilovich DI. Myeloid-derived suppressor cells. Cancer Immunol Res (2017) 5:3–8. doi: 10.1158/2326-6066.CIR-16-0297
74. Dorhoi A, Glaria E, Garcia-Tellez T, Nieuwenhuizen NE, Zelinskyy G, Favier B, et al. MDSCs in infectious diseases: regulation, roles, and readjustment. Cancer Immunol Immunother (2019) 68:673–85. doi: 10.1007/s00262-018-2277-y
75. Chang S, Kim YH, Kim YJ, Kim YW, Moon S, Lee YY, et al. Taurodeoxycholate increases the number of myeloid-derived suppressor cells that ameliorate sepsis in mice. Front Immunol (2018) 9:1984. doi: 10.3389/fimmu.2018.01984
76. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science (2003) 299:1057–61. doi: 10.1126/science.1079490
77. Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for scurfin in CD4+CD25+ T regulatory cells. Nat Immunol (2003) 4:337–42. doi: 10.1038/ni909
78. Darrigues J, van Meerwijk JPM, Romagnoli P. Age-dependent changes in regulatory T lymphocyte development and function: A mini-review. Gerontology (2018) 64:28–35. doi: 10.1159/000478044
79. Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science (2006) 314:1461–3. doi: 10.1126/science.1135245
80. Li W, Hang S, Fang Y, Bae S, Zhang Y, Zhang M, et al. A bacterial bile acid metabolite modulates treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe (2021) 29:1366–77.e1369. doi: 10.1016/j.chom.2021.07.013
81. Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, et al. MUCOSAL IMMUNOLOGY. individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science (2015) 349:993–7. doi: 10.1126/science.aaa9420
82. Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG, et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science (2016) 351:858–63. doi: 10.1126/science.aac5560
83. Gregori S, Casorati M, Amuchastegui S, Smiroldo S, Davalli AM, Adorini L. Regulatory T cells induced by 1 alpha,25-dihydroxyvitamin D3 and mycophenolate mofetil treatment mediate transplantation tolerance. J Immunol (2001) 167:1945–53. doi: 10.4049/jimmunol.167.4.1945
84. Tang J, Zhou R, Luger D, Zhu W, Silver PB, Grajewski RS, et al. Calcitriol suppresses antiretinal autoimmunity through inhibitory effects on the Th17 effector response. J Immunol (2009) 182:4624–32. doi: 10.4049/jimmunol.0801543
85. Lee GR. The balance of Th17 versus treg cells in autoimmunity. Int J Mol Sci (2018) 19(3):730. doi: 10.3390/ijms19030730
86. Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol (2009) 27:485–517. doi: 10.1146/annurev.immunol.021908.132710
87. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell (2006) 126:1121–33. doi: 10.1016/j.cell.2006.07.035
88. Xiao R, Lei K, Kuok H, Deng W, Zhuang Y, Tang Y, et al. Synthesis and identification of lithocholic acid 3-sulfate as RORgammat ligand to inhibit Th17 cell differentiation. J Leukoc Biol (2022) 112:835–43. doi: 10.1002/JLB.1MA0122-513R
89. Withers DR, Hepworth MR, Wang X, Mackley EC, Halford EE, Dutton EE, et al. Transient inhibition of ROR-gammat therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med (2016) 22:319–23. doi: 10.1038/nm.4046
90. Pols TWH, Puchner T, Korkmaz HI, Vos M, Soeters MR, de Vries CJM. Lithocholic acid controls adaptive immune responses by inhibition of Th1 activation through the vitamin d receptor. PLoS One (2017) 12:e0176715. doi: 10.1371/journal.pone.0176715
91. Boonstra A, Barrat FJ, Crain C, Heath VL, Savelkoul HF, O'Garra A. 1alpha,25-dihydroxyvitamin d3 has a direct effect on naive CD4(+) T cells to enhance the development of Th2 cells. J Immunol (2001) 167:4974–80. doi: 10.4049/jimmunol.167.9.4974
92. Yu K, Li P, Xu T, Xu J, Wang K, Chai J, et al. Decreased infiltration of CD4(+) Th1 cells indicates a good response to ursodeoxycholic acid (UDCA) in primary biliary cholangitis. Pathol Res Pract (2021) 217:153291. doi: 10.1016/j.prp.2020.153291
93. Panda SK, Colonna M. Innate lymphoid cells in mucosal immunity. Front Immunol (2019) 10:861. doi: 10.3389/fimmu.2019.00861
94. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell (2018) 174:1054–66. doi: 10.1016/j.cell.2018.07.017
95. Lochner M, Ohnmacht C, Presley L, Bruhns P, Si-Tahar M, Sawa S, et al. Microbiota-induced tertiary lymphoid tissues aggravate inflammatory disease in the absence of RORgamma t and LTi cells. J Exp Med (2011) 208:125–34. doi: 10.1084/jem.20100052
96. Zhu C, Boucheron N, Muller AC, Majek P, Claudel T, Halilbasic E, et al. 24-norursodeoxycholic acid reshapes immunometabolism in CD8(+) T cells and alleviates hepatic inflammation. J Hepatol (2021) 75:1164–76. doi: 10.1016/j.jhep.2021.06.036
97. Xun Z, Lin J, Yu Q, Liu C, Huang J, Shang H, et al. Taurocholic acid inhibits the response to interferon-alpha therapy in patients with HBeAg-positive chronic hepatitis b by impairing CD8(+) T and NK cell function. Cell Mol Immunol (2021) 18:461–71. doi: 10.1038/s41423-020-00601-8
98. Ding C, Hong Y, Che Y, He T, Wang Y, Zhang S, et al. Bile acid restrained T cell activation explains cholestasis aggravated hepatitis b virus infection. FASEB J (2022) 36:e22468. doi: 10.1096/fj.202200332R
99. Bhalla AK, Amento EP, Krane SM. Differential effects of 1,25-dihydroxyvitamin D3 on human lymphocytes and monocyte/macrophages: inhibition of interleukin-2 and augmentation of interleukin-1 production. Cell Immunol (1986) 98:311–22. doi: 10.1016/0008-8749(86)90291-1
100. Chen S, Sims GP, Chen XX, Gu YY, Chen S, Lipsky PE. Modulatory effects of 1,25-dihydroxyvitamin D3 on human b cell differentiation. J Immunol (2007) 179:1634–47. doi: 10.4049/jimmunol.179.3.1634
101. Lemire JM, Adams JS, Sakai R, Jordan SC. 1 alpha,25-dihydroxyvitamin D3 suppresses proliferation and immunoglobulin production by normal human peripheral blood mononuclear cells. J Clin Invest (1984) 74:657–61. doi: 10.1172/JCI111465
102. Liu J, Fei Y, Zhou T, Ji H, Wu J, Gu X, et al. Bile acids impair vaccine response in children with biliary atresia. Front Immunol (2021) 12:642546. doi: 10.3389/fimmu.2021.642546
103. Biagioli M, Carino A, Fiorucci C, Marchiano S, Di Giorgio C, Roselli R, et al. GPBAR1 functions as gatekeeper for liver NKT cells and provides counterregulatory signals in mouse models of immune-mediated hepatitis. Cell Mol Gastroenterol Hepatol (2019) 8:447–73. doi: 10.1016/j.jcmgh.2019.06.003
104. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science (2018) 360(6391):eaan5931. doi: 10.1126/science.aan5931
105. Gruner N, Mattner J. Bile acids and microbiota: Multifaceted and versatile regulators of the liver-gut axis. Int J Mol Sci (2021) 22(3):1397. doi: 10.3390/ijms22031397
106. Kriaa A, Mariaule V, Jablaoui A, Rhimi S, Mkaouar H, Hernandez J, et al. Bile acids: Key players in inflammatory bowel diseases? Cells (2022) 11(5):901. doi: 10.3390/cells11050901
107. Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature (2019) 569:655–62. doi: 10.1038/s41586-019-1237-9
108. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA (2007) 104(34):13780–5. doi: 10.1073/pnas.0706625104
109. Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset crohn's disease. Cell Host Microbe (2014) 15:382–92. doi: 10.1016/j.chom.2014.02.005
110. Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell (2015) 160:447–60. doi: 10.1016/j.cell.2015.01.002
111. Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial crohn's disease. mBio (2016) 7(5):e01250–16. doi: 10.1128/mBio.01250-16
112. Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA (2008) 105(36):13580–5. doi: 10.1073/pnas.0804437105
113. Li N, Zhan S, Tian Z, Liu C, Xie Z, Zhang S, et al. Alterations in bile acid metabolism associated with inflammatory bowel disease. Inflammation Bowel Dis (2021) 27:1525–40. doi: 10.1093/ibd/izaa342
114. Thomas JP, Modos D, Rushbrook SM, Powell N, Korcsmaros T. The emerging role of bile acids in the pathogenesis of inflammatory bowel disease. Front Immunol (2022) 13:829525. doi: 10.3389/fimmu.2022.829525
115. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol (2019) 4:293–305. doi: 10.1038/s41564-018-0306-4
116. Vantrappen G, Ghoos Y, Rutgeerts P, Janssens J. Bile acid studies in uncomplicated crohn's disease. Gut (1977) 18:730–5. doi: 10.1136/gut.18.9.730
117. Wang Y, Gao X, Zhang X, Xiao F, Hu H, Li X, et al. Microbial and metabolic features associated with outcome of infliximab therapy in pediatric crohn's disease. Gut Microbes (2021) 13:1–18. doi: 10.1080/19490976.2021.1900996
118. Jacobs JP, Goudarzi M, Singh N, Tong M, McHardy IH, Ruegger P, et al. A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell Mol Gastroenterol Hepatol (2016) 2:750–66. doi: 10.1016/j.jcmgh.2016.06.004
119. Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut (2013) 62:531–9. doi: 10.1136/gutjnl-2012-302578
120. Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol (2009) 183:6251–61. doi: 10.4049/jimmunol.0803978
121. Mencarelli A, Migliorati M, Barbanti M, Cipriani S, Palladino G, Distrutti E, et al. Pregnane-x-receptor mediates the anti-inflammatory activities of rifaximin on detoxification pathways in intestinal epithelial cells. Biochem Pharmacol (2010) 80:1700–7. doi: 10.1016/j.bcp.2010.08.022
122. Okada K, Shoda J, Kano M, Suzuki S, Ohtake N, Yamamoto M, et al. Inchinkoto, a herbal medicine, and its ingredients dually exert Mrp2/MRP2-mediated choleresis and Nrf2-mediated antioxidative action in rat livers. Am J Physiol Gastrointest Liver Physiol (2007) 292:G1450–1463. doi: 10.1152/ajpgi.00302.2006
123. Guo X, Yan M. Pregnane X receptor polymorphisms and risk of inflammatory bowel disease: A meta-analysis. Immunol Invest (2017) 46:566–76. doi: 10.1080/08820139.2017.1322101
124. van der Post S, Jabbar KS, Birchenough G, Arike L, Akhtar N, Sjovall H, et al. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut (2019) 68:2142–51. doi: 10.1136/gutjnl-2018-317571
125. Xue LN, Xu KQ, Zhang W, Wang Q, Wu J, Wang XY. Associations between vitamin d receptor polymorphisms and susceptibility to ulcerative colitis and crohn's disease: a meta-analysis. Inflammation Bowel Dis (2013) 19:54–60. doi: 10.1002/ibd.22966
126. Kim JH, Yamaori S, Tanabe T, Johnson CH, Krausz KW, Kato S, et al. Implication of intestinal VDR deficiency in inflammatory bowel disease. Biochim Biophys Acta (2013) 1830:2118–28. doi: 10.1016/j.bbagen.2012.09.020
127. McGlone ER, Bloom SR. Bile acids and the metabolic syndrome. Ann Clin Biochem (2019) 56:326–37. doi: 10.1177/0004563218817798
128. Hachiya R, Tanaka M, Itoh M, Suganami T. Molecular mechanism of crosstalk between immune and metabolic systems in metabolic syndrome. Inflammation Regener (2022) 42:13. doi: 10.1186/s41232-022-00198-7
129. Chiang JYL, Ferrell JM. Bile acids as metabolic regulators and nutrient sensors. Annu Rev Nutr (2019) 39:175–200. doi: 10.1146/annurev-nutr-082018-124344
130. Ma H, Sales VM, Wolf AR, Subramanian S, Matthews TJ, Chen M, et al. Attenuated effects of bile acids on glucose metabolism and insulin sensitivity in a Male mouse model of prenatal undernutrition. Endocrinology (2017) 158:2441–52. doi: 10.1210/en.2017-00288
131. Wang R, Fan X, Lu Y, Chen D, Zhao Y, Qi K. Dietary acetic acid suppress high-fat diet-induced obesity in mice by altering taurine conjugated bile acids metabolism. Curr Res Food Sci (2022) 5:1976–84. doi: 10.1016/j.crfs.2022.10.021
132. Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature (2006) 439:484–9. doi: 10.1038/nature04330
133. Wei M, Huang F, Zhao L, Zhang Y, Yang W, Wang S, et al. A dysregulated bile acid-gut microbiota axis contributes to obesity susceptibility. EBioMedicine (2020) 55:102766. doi: 10.1016/j.ebiom.2020.102766
134. Zhang Q, Wu ZH, Zhao SS, Yang J, Chen L, Wang XY, et al. Identification and spatial visualization of dysregulated bile acid metabolism in high-fat diet-fed mice by mass spectral imaging. Front Nutr (2022) 9:858603. doi: 10.3389/fnut.2022.858603
135. Gao R, Meng X, Xue Y, Mao M, Liu Y, Tian X, et al. Bile acids-gut microbiota crosstalk contributes to the improvement of type 2 diabetes mellitus. Front Pharmacol (2022) 13:1027212. doi: 10.3389/fphar.2022.1027212
136. Agus A, Clement K, Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut (2021) 70:1174–82. doi: 10.1136/gutjnl-2020-323071
137. Jackson KG, Way GW, Zhou H. Bile acids and sphingolipids in non-alcoholic fatty liver disease. Chin Med J (Engl) (2022) 135:1163–71. doi: 10.1097/CM9.0000000000002156
138. Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut (2018) 67:1881–91. doi: 10.1136/gutjnl-2017-314307
139. Nimer N, Choucair I, Wang Z, Nemet I, Li L, Gukasyan J, et al. Bile acids profile, histopathological indices and genetic variants for non-alcoholic fatty liver disease progression. Metabolism (2021) 116:154457. doi: 10.1016/j.metabol.2020.154457
140. Wegermann K, Howe C, Henao R, Wang Y, Guy CD, Abdelmalek MF, et al. Serum bile acid, vitamin e, and serotonin metabolites are associated with future liver-related events in nonalcoholic fatty liver disease. Hepatol Commun (2021) 5:608–17. doi: 10.1002/hep4.1665
141. Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science (2009) 325:1254–7. doi: 10.1126/science.1176709
142. Hartmann P, Hochrath K, Horvath A, Chen P, Seebauer CT, Llorente C, et al. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology (2018) 67:2150–66. doi: 10.1002/hep.29676
143. Fang S, Suh JM, Reilly SM, Yu E, Osborn O, Lackey D, et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med (2015) 21:159–65. doi: 10.1038/nm.3760
144. Matsumori A. Targeting inflammation in the diagnosis, management, and prevention of cardiovascular diseases. Glob Heart (2022) 17:80. doi: 10.5334/gh.1156
145. Kong P, Cui ZY, Huang XF, Zhang DD, Guo RJ, Han M. Inflammation and atherosclerosis: signaling pathways and therapeutic intervention. Signal Transduct Target Ther (2022) 7:131. doi: 10.1038/s41392-022-00955-7
146. Qian B, Zhang K, Li Y, Sun K. Update on gut microbiota in cardiovascular diseases. Front Cell Infect Microbiol (2022) 12:1059349. doi: 10.3389/fcimb.2022.1059349
147. Zhao J, Cheng W, Lu H, Shan A, Zhang Q, Sun X, et al. High fiber diet attenuate the inflammation and adverse remodeling of myocardial infarction via modulation of gut microbiota and metabolites. Front Microbiol (2022) 13:1046912. doi: 10.3389/fmicb.2022.1046912
148. Hoving LR, Katiraei S, Heijink M, Pronk A, van der Wee-Pals L, Streefland T, et al. Dietary mannan oligosaccharides modulate gut microbiota, increase fecal bile acid excretion, and decrease plasma cholesterol and atherosclerosis development. Mol Nutr Food Res (2018) 62:e1700942. doi: 10.1002/mnfr.201700942
149. Rezen T, Rozman D, Kovacs T, Kovacs P, Sipos A, Bai P, et al. The role of bile acids in carcinogenesis. Cell Mol Life Sci (2022) 79:243. doi: 10.1007/s00018-022-04278-2
150. Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature (2013) 499:97–101. doi: 10.1038/nature12347
151. Tian Y, Cheng C, Wei Y, Yang F, Li G. The role of exosomes in inflammatory diseases and tumor-related inflammation. Cells (2022) 11(6):1005. doi: 10.3390/cells11061005
152. Yu W, Tu Y, Long Z, Liu J, Kong D, Peng J, et al. Reactive oxygen species bridge the gap between chronic inflammation and tumor development. Oxid Med Cell Longev (2022) 2022:2606928. doi: 10.1155/2022/2606928
153. Sui Q, Zhang X, Chen C, Tang J, Yu J, Li W, et al. Inflammation promotes resistance to immune checkpoint inhibitors in high microsatellite instability colorectal cancer. Nat Commun (2022) 13:7316. doi: 10.1038/s41467-022-35096-6
154. Kuhn T, Stepien M, Lopez-Nogueroles M, Damms-Machado A, Sookthai D, Johnson T, et al. Prediagnostic plasma bile acid levels and colon cancer risk: A prospective study. J Natl Cancer Inst (2020) 112:516–24. doi: 10.1093/jnci/djz166
155. Ward JBJ, Lajczak NK, Kelly OB, O'Dwyer AM, Giddam AK, Ni Gabhann J, et al. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am J Physiol Gastrointest Liver Physiol (2017) 312:G550–8. doi: 10.1152/ajpgi.00256.2016
156. Noto JM, Piazuelo MB, Shah SC, Romero-Gallo J, Hart JL, Di C, et al. Iron deficiency linked to altered bile acid metabolism promotes helicobacter pylori-induced inflammation-driven gastric carcinogenesis. J Clin Invest (2022) 132(10):e147822. doi: 10.1172/JCI147822
157. Shen R, Ke L, Li Q, Dang X, Shen S, Shen J, et al. Abnormal bile acid-microbiota crosstalk promotes the development of hepatocellular carcinoma. Hepatol Int (2022) 16:396–411. doi: 10.1007/s12072-022-10299-7
158. Rauf A, Badoni H, Abu-Izneid T, Olatunde A, Rahman MM, Painuli S, et al. Neuroinflammatory markers: Key indicators in the pathology of neurodegenerative diseases. Molecules (2022) 27(10):3194. doi: 10.3390/molecules27103194
159. Mou Y, Du Y, Zhou L, Yue J, Hu X, Liu Y, et al. Gut microbiota interact with the brain through systemic chronic inflammation: Implications on neuroinflammation, neurodegeneration, and aging. Front Immunol (2022) 13:796288. doi: 10.3389/fimmu.2022.796288
160. Bottigliengo D, Foco L, Seibler P, Klein C, Konig IR, Del Greco MFA. Mendelian randomization study investigating the causal role of inflammation on parkinson's disease. Brain (2022) 145:3444–53. doi: 10.1093/brain/awac193
161. Daruich A, Picard E, Guegan J, Jaworski T, Parenti L, Delaunay K, et al. Comparative analysis of urso- and tauroursodeoxycholic acid neuroprotective effects on retinal degeneration models. Pharm (Basel) (2022) 15(3):334. doi: 10.3390/ph15030334
162. Khalaf K, Tornese P, Cocco A, Albanese A. Tauroursodeoxycholic acid: a potential therapeutic tool in neurodegenerative diseases. Transl Neurodegener (2022) 11:33. doi: 10.1186/s40035-022-00307-z
163. Kondo N, Kuroda T, Kobayashi D. Cytokine networks in the pathogenesis of rheumatoid arthritis. Int J Mol Sci (2021) 22(20):10922. doi: 10.3390/ijms222010922
164. Fan Z, Ross RP, Stanton C, Hou B, Zhao J, Zhang H, et al. Lactobacillus casei CCFM1074 alleviates collagen-induced arthritis in rats via balancing Treg/Th17 and modulating the metabolites and gut microbiota. Front Immunol (2021) 12:680073. doi: 10.3389/fimmu.2021.680073
165. de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab (2013) 17:657–69. doi: 10.1016/j.cmet.2013.03.013
166. Holguin F, Bleecker ER, Busse WW, Calhoun WJ, Castro M, Erzurum SC, et al. Obesity and asthma: an association modified by age of asthma onset. J Allergy Clin Immunol (2011) 127:1486–93.e1482. doi: 10.1164/ajrccm-conference.2011.183.1_MeetingAbstracts.A2662
167. Crestani E, Harb H, Charbonnier LM, Leirer J, Motsinger-Reif A, Rachid R, et al. Untargeted metabolomic profiling identifies disease-specific signatures in food allergy and asthma. J Allergy Clin Immunol (2020) 145:897–906. doi: 10.1016/j.jaci.2019.10.014
168. Grau KR, Zhu S, Peterson ST, Helm EW, Philip D, Phillips M, et al. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat Microbiol (2020) 5:84–92. doi: 10.1038/s41564-019-0602-7
169. Hu MM, He WR, Gao P, Yang Q, He K, Cao LB, et al. Virus-induced accumulation of intracellular bile acids activates the TGR5-beta-arrestin-SRC axis to enable innate antiviral immunity. Cell Res (2019) 29:193–205. doi: 10.1038/s41422-018-0136-1
170. Wang DQ, Carey MC. Therapeutic uses of animal biles in traditional Chinese medicine: an ethnopharmacological, biophysical chemical and medicinal review. World J Gastroenterol (2014) 20:9952–75. doi: 10.3748/wjg.v20.i29.9952
Keywords: gut microbiota, bile acids, deoxycholic acid, lithocholic acid, tolerogenic macrophages, regulatory B cells, regulatory T cells
Citation: Su X, Gao Y and Yang R (2023) Gut microbiota derived bile acid metabolites maintain the homeostasis of gut and systemic immunity. Front. Immunol. 14:1127743. doi: 10.3389/fimmu.2023.1127743
Received: 19 December 2022; Accepted: 07 March 2023;
Published: 15 May 2023.
Edited by:
Antonio Riva, Foundation for Liver Research, United KingdomReviewed by:
Lindsey Edwards, King’s College London, United KingdomCopyright © 2023 Su, Gao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rongcun Yang, cnlhbmdAbmFua2FpLmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.