
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Immunol., 25 January 2023
Sec. Molecular Innate Immunity
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1125635
This article is part of the Research TopicMicrobiota in Skin Inflammatory DiseasesView all 14 articles
Atopic dermatitis (AD) is a common chronic inflammatory skin disease that significantly affects the patient’s quality of life. A disrupted skin barrier, type 2 cytokine-dominated inflammation, and microbial dysbiosis with increased Staphylococcus aureus colonization are critical components of AD pathogenesis. Patients with AD exhibit decreased expression of antimicrobial peptides (AMPs) which is linked to increased colonization by Staphylococcus aureus. The skin microbiome itself is a source of several AMPs. These host- and microbiome-derived AMPs define the microbial landscape of the skin based on their differential antimicrobial activity against a range of skin microbes or their quorum sensing inhibitory properties. These are particularly important in preventing and limiting dysbiotic colonization with Staphylococcus aureus. In addition, AMPs are critical for immune homeostasis. In this article, we share our perspectives about the implications of microbial derived AMPs in AD patients and their potential effects on overlapping factors involved in AD. We argue and discuss the potential of bacterial AMPs as therapeutics in AD.
Atopic dermatitis (AD), also called atopic eczema, is one of the most common skin diseases in humans (1). The prevalence of AD ranges from 15% to 25% in children and from 7% to 10% in adults (2). AD shows remarkable heterogeneity in clinical presentation, but in most patients, the disease manifests as a condition with extensive eczematous lesions with red, dry, scaly patches that are intensely itchy. Patients with AD often suffer from sleep deprivation and depression, which significantly affects their quality of life (3, 4). The prevalence of AD varies by geographic location and has nearly doubled to tripled in developed countries in recent decades. One explanation for it is provided by the “biodiversity hypothesis”, i.e., that a reduced contact to natural environments in early life does lead to a failure of enrichment in the human microbiome, disturbs immune balance and leads in turn to allergy and inflammatory disorders (5–7).
Although the pathogenesis of AD is complex and not fully understood, three key features are involved in its development. These include (i) disruption of epidermal barrier function, (ii) an excessive immune response mediated by type 2 cells, including Th2, and innate lymphoid cells (ILCs) cells, and (iii) skin microbial dysbiosis with excessive growth of Staphylococcus aureus (S. aureus). AD patients exhibit altered stratum corneum lipids composition, decreased moisture content, and increased permeability to environmental molecules in both lesional and nonlesional skin (8, 9). Defects in structural barrier proteins (e.g., filaggrin) and the itch-scratch cycle are among the major causes of the observed barrier deficiencies in AD (10, 11). Epidermal barrier disruption leads to excessive secretion of damage-associated molecular patterns (DAMPs) and alarmins such as thymic stromal lymphopoietin (TSLP), IL-1ß, IL-25, and IL-33. Alarmins promote the secretion of cytokines IL-13 and IL-5 by immune cells such as dendritic cells and ILCs, which further promote a type 2 cell-mediated immune response (12). Increased allergen permeation, along with a Th2-cell mediated immune response lead IgE isotype switching (13); a hallmark of chronic recurring AD. Skin microbiome, the third feature involved in the pathogenesis of AD will be discussed in detail in the next sections.
The skin harbours millions of bacteria, fungi, viruses, archaea, and skin mites that together form the skin microbiome. Bacteria constitute the most abundant kingdom in the skin microbiome with the major genera being Cutibacterium, Corynebacterium, and Staphylococcus (14, 15). The composition and abundance of these genera depend upon the skin site, physiology, and microenvironments (sebaceous, dry, and moist). Skin microbiome is now known to be an imperative component of skin homeostasis maintenance, which includes development of skin’s barrier functions (16), immune system, breakdown of natural products and protection against invading pathogens (14, 17).
The implications of microbial shifts correlate with several dermatological diseases most notably, AD (18). The prevalence of S. aureus in patients with AD is approximately 20 times higher than the skin of healthy controls. The lesional skin of AD patients shows higher prevalence of S.aureus (up to 70%) than nonlesional skin of the same patients (39%) (19). A positive correlation was found between S. aureus density on lesional and nonlesional skin and disease severity (20). Moreover, the higher abundance of S. aureus in AD patients is independent of age group, ethnicity, and geographic location (21, 22).
S. aureus secretes several metabolites, also known as virulence factors, which are responsible in part for the proinflammatory activity and barrier destruction in AD lesions (Figure 1). The enzyme ceramidase secreted by S. aureus lowers lipid and fatty acid levels and makes the skin permeable to allergens (23). Lower fatty acid levels also lead to decreased formation of phospholipid hydrolysis products in sebum and sweat, which increase skin surface pH and further promote S. aureus growth. Cell surface proteins of S. aureus such as clumping factors A and B and fibronectin-binding proteins support attachment to the uppermost skin, the stratum corneum (24, 25). S. aureus is known to secrete several proteases and is also able to induce protease secretion from host (26). Staphophain, inactivates host defence peptides, thus promoting S. aureus colonization (27). Proteases also promote the permeation of allergens through the stratum corneum by acting on barrier proteins and tight junctions (28). Alpha toxin secreted by S. aureus is cytotoxic to keratinocytes and alters the integrity of E-cadherin, compromising barrier function (29). Staphylococcal enterotoxins (SE) (SEA, SEB, and SEC), phenol-soluble modulins, and lipoproteins target key immune pathways and create a pro-inflammatory environment characteristic of patients with AD (29). The mechanisms of virulence factors secreted by S. aureus that play a role in the pathogenesis of AD have been described in detail elsewhere (30–32).
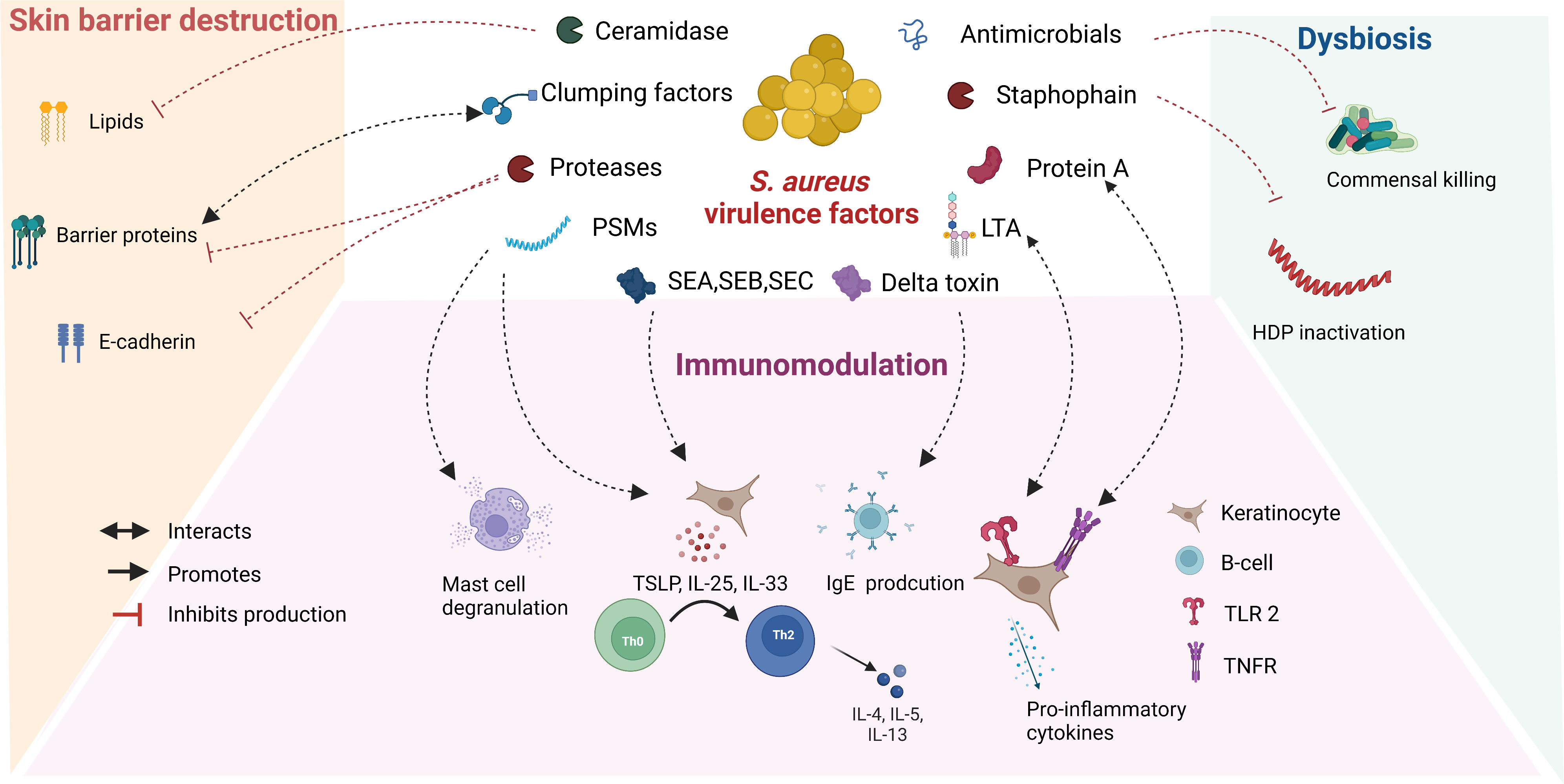
Figure 1 Role of S. aureus secreted virulence factors in the pathogenesis of atopic dermatitis: S. aureus secretes several virulence factors, which interact with overlapping features involved in the pathogenesis of AD. Abbreviations: VF: virulence factors, PSMs: Phenol soluble modulins, SE: staphylococcal enterotoxins, LTA: Lipoteichoic acid, HDP: Host defence peptide, TSLP: Thymic stromal lymphopoietin, TLR: Toll like receptor, TNFR: Tumor necrosis factor receptor.
So far, there appears to be no evidence linking the presence of a single virulence factor to AD severity. S. aureus strains isolated from AD skin differ from those isolated from healthy individuals (33). Fleury et al. showed that the frequency of isolation of S. aureus strains belonging to clonal complex (CC) 1 is higher than that of strains isolated from the nasal cavity of healthy children, which have a higher frequency of isolation of CC30 (24). Byrd et al. performed shotgun metagenome sequence analysis of the skin microbiome at the strain level in patients with AD throughout the disease course. They found that the severity of AD is strain-specific, with isolated strains from severe AD lesions being phylogenetically similar. This suggests that specific combinations of virulence factors expressed by certain strains may be responsible for the pathogenicity of S. aureus observed in AD (34).
S. aureus modulates the production of virulence factors necessary for its survival by sensing environmental factors such as cell density. This phenomenon is known as quorum sensing. The accessory gene regulator (agr) quorum sensing system is one of the best studied quorum sensing systems in S. aureus. The agr quorum sensing system recognizes cognate autoinducing peptides which are short peptides of 7-12 amino acids containing a cyclic thiolactone at the C- terminus (35). Interestingly, several virulence factors such as superantigens, lipases, and proteases involved in the pathogenesis of AD are also controlled by the agr quorum-sensing system (35, 36). Nakamura et al. demonstrated that agr virulence is essential for the epithelial degradation observed in AD. Moreover, the probability of developing AD is higher when S. aureus with a functional agr virulence system is present in childhood (37). These studies suggest that inhibition of agr quorum-sensing in addition to inhibition of S. aureus growth may be a promising therapeutic target in AD.
AD skin has lower microbial diversity compared to healthy individuals. This is associated with a lower abundance of the genera Streptococcus, Corynebacterium, and Cutibacterium, as well as members of the commensal Staphylococci with anti-S. aureus activity (38, 39). These bacterial communities play a critical role in the immune response to pathogens. Ridaura et al. showed that the genus Corynebacterium can promote IL-23 signalling and induce IL-17A γδT cells in the dermis, which recruit immune cells (40). At the species level, Cutibacterium acnes associated with healthy skin were shown to activate the release of extracellular traps from specialized Th17 subsets (41). Coagulase negative Staphylococci (CoNS) such as Staphylococcus cohnii activate the host steroid pathway and promote immunosuppression (42), while application of Staphylococcus epidermidis to mouse skin showed enhanced innate protection against Candida albicans by upregulating Th17 immune mediators such as S100A8 and S100A9 (43). Commensal microbes also play a critical role in epidermal barrier development and surface pH regulation (16, 44). For example, Roseomonas mucosa secretes glycerophospholipids which induce host epithelial repair by enhancing the cholinergic activation via TNFR2 signalling and Staphylococcus epidermidis increases the production of skin ceramides (45, 46), while Cutibacterium acnes secretes a lipase that converts triacylglycerols contained in sebum to propionic acid, which contributes to the acidification of the skin surface; a factor that limits the growth of S. aureus (47). Notably, commensals directly provide colonization resistance to pathogenic bacteria including S. aureus by secreting certain metabolites that inhibit their growth or virulence factor production. The most promising of these metabolites are bacterial-derived AMPs and are discussed in detail in later sections. Additionally, the host-derived AMPs also play a significant role in defining the skin microbiome composition and are discussed briefly as well.
Host-derived AMPs, also known as host defence peptides (HDPs), are mostly cationic peptides with a molecular weight of less than 10 kDa (48–51) but also include a few classes with larger molecules than 10 kDa (52, 53). These peptides are present on epithelial surfaces such as the oral mucosa, vaginal epithelia, skin etc. HDPs are either constitutively produced by keratinocytes and immune cells or induced in response to stimuli such as pathogen-associated molecular patterns (PAMPs) or inflammatory cytokines. Some important classes of HDPs include RNases, defensins, cathelicidins, dermcidin, and S100 class peptides (51, 54, 55).
HDPs show broad-spectrum but variable antimicrobial activity against different pathogens and members of the commensal microbiome. For example, RNAse7 showed higher inhibitory activity against E. Coli and Cutibacterium acnes compared to S. aureus (56). Interestingly, in the context of the vaginal microbiome, certain commensal bacteria have been shown to use constitutively expressed peptides (S100A7 and Elafin) as amino acid sources to ensure their survival (57). This phenomenon is most likely also found in skin epithelia but has not yet been studied. HDPs are well-known for their immunomodulatory and barrier-improving properties and play a critical role in the pathogenesis of diseases such as psoriasis and polymorphic light eruption underscoring the versatile role of HDPs in skin physiology (58–60).
Atopic skin exhibits lower levels of certain HDPs such as dermcidin, human beta-defensin-2 (hBD-2), human beta-defensin-3 (hBD-3), and cathelicidin, as well as increased expression of RNase7 and S100A7 (61–64). Moreover, a higher abundance of S. aureus in atopic skin and during disease flare-ups has also been linked to defective HDP expression (18, 65). Th2 cytokine activity and lower vitamin D levels are thought to be responsible for the altered HDP expression observed in AD (66–68). Interestingly, AD treatments such as UVB phototherapy improve expression of certain HDPs which has led researchers to question whether HDPs could potentially be used as a therapy in AD (69–71). This potential was recently indicated by Peng et.al, who showed that subcutaneous injection of hBD-3 in mice with AD alleviated inflammation through its barrier-improving properties (72). Several detailed reviews on host defence peptides are published during the last five years (58, 71, 73, 74).
Bacterial AMPs are small peptide substances usually restricted to short peptide bacteriocins that inhibit microorganisms by physically disrupting the cell membrane or interact with the intracellular components crucial for bacterial survival (75). Recently, bacteriocins and peptide molecules that inhibit the quorum-sensing activity of competing bacteria have gained increasing attention. The inhibitory effect on the quorum-sensing activity of effector bacteria results in reduced virulence, giving the producer a competitive advantage. These peptides are known as heterologous autoinducing peptides (AIPs) and are considered a class of bacterial AMPs in this review (Figure 2).
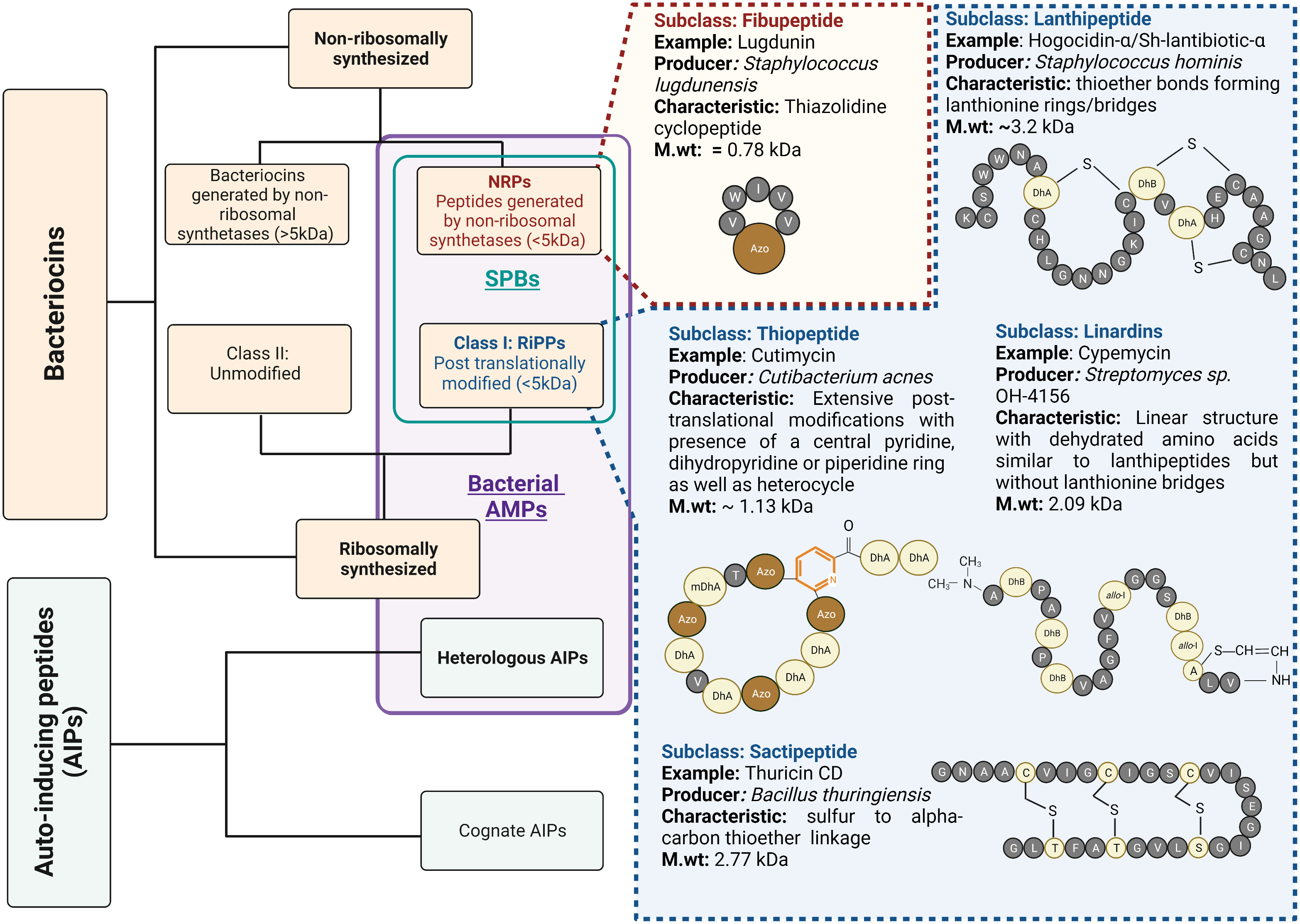
Figure 2 Bacterial AMPs and structural variety of short peptide bacteriocins: Bacterial AMPs include Short Peptide Bacteriocins (SPBs) with molecular weight less than 5kDa and Quorum quenching peptides, which are heterologous autoinducing peptides. The subcategory SPBs also includes the class I bacteriocins which undergo post-translational modifications of amino acids, also known as RiPPs, and the peptides synthesized by non-ribosomal synthetases (NRPs) with molecular weight less than 5kDa. SPBs constitute several subclasses based on their structural characteristics. The figure enlists some important classes with examples. Standard amino acids are described as single letter code with grey colour balls. The colour light yellow represents post-translationally modified amino acids. The prefix “Dh” stands for Dehydro and prefix “allo” represents stereo isomer of amino acid. The pyridine ring is coloured in orange. The azole moieties are indicated by abbreviation “Azo” and brown colour balls.
Bacteriocins are structurally diverse, and there are several classification systems that make them difficult to follow. Many authors restrict the term bacteriocin to ribosomally synthesized peptide antimicrobials with a molecular weight of less than 10 kDa (76, 77). They further divide bacteriocins into class I bacteriocins, which are post-translationally modified, and class II bacteriocins, which are unmodified. These class I bacteriocins are also referred to as ribosomally synthesized and post translationally modified peptides (RiPPs). Recently, however, the term bacteriocin has also been used to refer those AMPs generated by non-ribosomal synthetases (78–80). In the following, we will only discuss about the peptides with a molecular weight of less than 5 kDa, belonging to the group of RiPPs, and those peptides with a molecular weight of less than 5 kDa generated by non-ribosomal synthetases (NRPs). They are collectively known/described as short peptide bacteriocins (SPBs) (Figure 2).
Members of SPBs possess potent narrow spectrum antibacterial activity at nanomolar concentrations, and have lower chances of resistance development (81). The important classes belonging to SPB group, and their structural details are shown in Figure 2. SPBs inhibit bacterial growth by several mechanisms. For example, the lantibitoic nisin binds to the lipid II, which is responsible for peptidoglycan synthesis, thus interfering with cell wall biosynthesis of target bacteria. In addition, its C-terminal region leads to the formation of pores that result in membrane disruption and efflux of bacterial metabolites necessary for growth (82, 83). Bottoromycins and Thiopeptides such as thiostrepton and micrococcin inhibit bacterial protein synthesis by binding to the 50s ribosomal subunit. Glycocins (e.g., sublancin 168) bind to and inhibit the function of the glucose phosphotransferase system and mechanosensitive channel (MscL) while Sactipeptides e.g., Ruminococcin C inhibits RNA polymerase (84).
The antimicrobial activity of SPBs is mainly directed against closely related bacteria (85). Moreover, not all susceptible members of the microbiome are equally targeted by SPBs; rather, certain species are more sensitive than others. For example, Subtilosin A, a RiPP produced by Bacillus subtilis, had a minimum inhibitory concentration (MIC) of 1.25 μg/ml against Streptococcus pyogens, but a MIC of 83.25 µg/ml against Streptococcus gordonii, a member of the same genus (86). This property makes SPBs an interesting therapeutic candidate for AD, in which dysbiotic colonization with single bacterial species is present.
SPBs are widely used in food and veterinary medicine. Medical applications in humans have experienced low growth due to insufficient investment. Today, however, there is growing interest in the potential of SPBs (76). A suitable example is Clostridium difficile-associated diarrhea (CDAD). It is well known that broad-spectrum antibiotic treatment for Clostridium difficile intestinal infections provides acute relief to patients but disrupts the gut microbiome with long-term use by depleting commensal bacteria necessary to control Clostridium difficile growth. This altered environment contributes to the thriving of Clostridium difficile and the secretion of toxins that cause diarrheal disease (87). In this case, the search for narrow spectrum bacteriocins from the commensal microbiome led to the discovery of a SPB of class RiPP- sactibitoic called thuricin CD, which showed potent inhibition of Clostridium difficile without affecting the commensal gut microbiome (88). In addition, the semisynthetic thiopeptide LEF571 has been tested in clinical trials for the treatment of CDAD but showed lower narrow spectrum activity compared with thuricin CD (89). Interestingly, NAI003, a derivative of thiopeptide GE2270A, showed selective activity against Cutibacterium acnes over the skin commensals. This thiopeptide has already completed a phase 1 clinical trial for the topical treatment of acne and provides evidence of use of SPBs as topical treatments for skin conditions (90). Lactocillin, a thiopeptide SPB isolated from a vaginal commensal lactobacillus was shown to inhibit several pathogens colonising skin or vagina and showed no antimicrobial activity against other lactobacilli species (91). Despite increasing research into how SPBs work, the exact reason for their selectivity is still not clear (92). However, this can be attributed to a combination of their properties such as amphipathicity, conformation, charge, hydrophilicity, and secondary structure.
Metagenomic analyses revealed that biosynthetic gene clusters encoding bacteriocins are ubiquitous in microbes associated with humans (91). O’Sullivan et al. further demonstrated that the human skin microbiome provides colonization resistance to pathogens by secreting a variety of novel bacteriocins (93). Within the skin microbiome, an inter-genera competition exists (Figure 3); for example, Cutibacterium acnes secretes a thiopeptide RiPP called cutimycin that inhibits members of Staphylococcus but does not affect the members of the genera Corynebacterium and Cutibacterium (94). However, the mechanism of action remains unknown. Similarly, RiPPs from lactobacilli have shown to inhibit members of Staphylococci, Cutibacterium and Corynebacterium (91, 95).
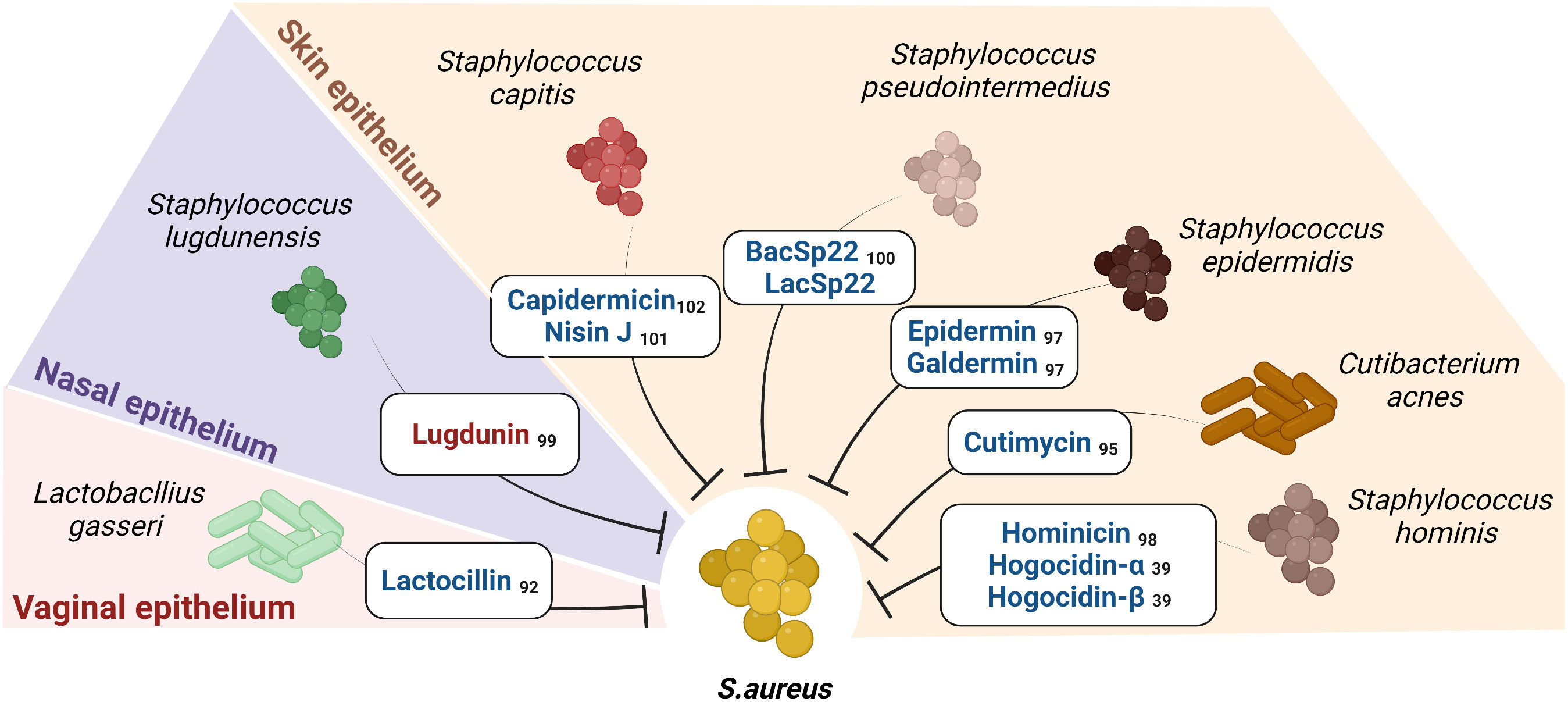
Figure 3 Short Peptide Bacteriocins (SPBs) secreted by certain commensals on human epithelia inhibit S.aureus: The figure enlists the SPBs isolated from human epithelial residing commensal bacteria known to possess narrow spectrum activity against S. aureus and sparing certain commensals (the color red represents NRPs, and color blue represents RiPPs). Subscripts denote references.
The chances of obtaining bacteriocins with a narrow spectrum of activity are greater if isolated from a phylogenetically similar species or a species that cohabits with the target species. Several staphylococcal-derived bacteriocins exhibit antimicrobial activity against S. aureus (96). Known SPBs are epidermin from Staphylococcus epidermidis (97), hominicin from Staphylococcus hominis (98), lugdunin from Staphylococcus lugdunensis (99), BacSp22 from Staphylococcus Pseudintermedius (100) and capidermicin and nisin J from Staphylococcus capitis (101, 102). An approach used by Nakatsuji et al. showed that several CoNS isolated from healthy skin inhibited S. aureus. In addition, they isolated and identified the S. hominis A9-derived RiPPs Sh-lantibiotic-alpha (Hogocidin-α) and Sh-lantibiotic-beta (Hogocidin-β), which inhibited S. aureus but showed no antimicrobial activity against commensal bacteria namely Staphylococcus epidermidis and Staphylococcus hominis (39).
Reports indicate that several SPBs modulate the host immune response (103). In bovine gut epithelium, oral administration of nisin for a short period resulted in an increased accumulation of CD4+ and CD8+ T lymphocytes (LT) and a decrease in B lymphocytes (104). However, it remains to be determined whether Nisin has as direct effect on epithelial or immune cells or an indirect effect mediated by gut microbiome changes. Interestingly, a higher concentration of nisin was shown to activate extracellular trap release (NETs) and increased intracellular superoxide levels in human neutrophils in-vitro (105). In contrast, in another study nisin showed high biological compatibility with explant cultures of rabbit vaginal tissue and did not exhibit immunomodulatory effects (106). This suggests that the immunomodulatory activities of SPBs may be dependent on the epithelia or the tissue under consideration. Lactobacilli are endogenous inhabitants of healthy skin. Hemert et al. investigated the immunomodulatory effects of Lactobacillus Plantarum (L. plantarum) by evaluating its ability to stimulate cytokine production in PBMCs. They found that L. plantarum strains stimulated the secretion of the anti-inflammatory cytokines IL -10 and IL -12 more than 10-fold. They moreover identified genetic loci responsible for immunomodulatory capabilities involving components of the bacteriocin biosynthesis and transport pathways (107), suggesting an anti-inflammatory effect of bacteriocins. Interestingly, Thiostrepton an SPB belonging to the thiopeptide RiPP class, was able to inhibit psoriasis-like inflammation induced by TLR7, TLR8, and TLR9 (108). Additionally, Lugdunin, an SPB belonging to the NRP class and produced by the nasal commensal Staphylococcus lugdunensis, provided multilevel protection against S. aureus. In addition to directly inhibiting S. aureus, it also enhanced the innate immune response by recruiting neutrophil granulocytes and monocytes in a mouse model of S. aureus infection. Similarly, BacSp222, a RiPP produced by a common skin colonizer, Staphylococcus pseudintermedius, showed immunomodulatory and cytotoxic properties apart from its antimicrobial activity (101).
As mentioned earlier, the agr quorum-sensing system of S. aureus plays a significant role in the secretion of virulence factors observed in AD (Figure 4). The system kicks in when cognate autoinducing peptides bind to a kinase receptor called AgrC. AgrC activates the downstream regulator AgrA, which triggers transcription of the agrBDCA operon and regulatory small RNA called RNAIII by binding to promoter regions P2 and P3. While agrBDCA is responsible for the production of quorum sensing machinery the RNAIII induces transcription of several virulence factors associated to the pathogenesis of AD (35).
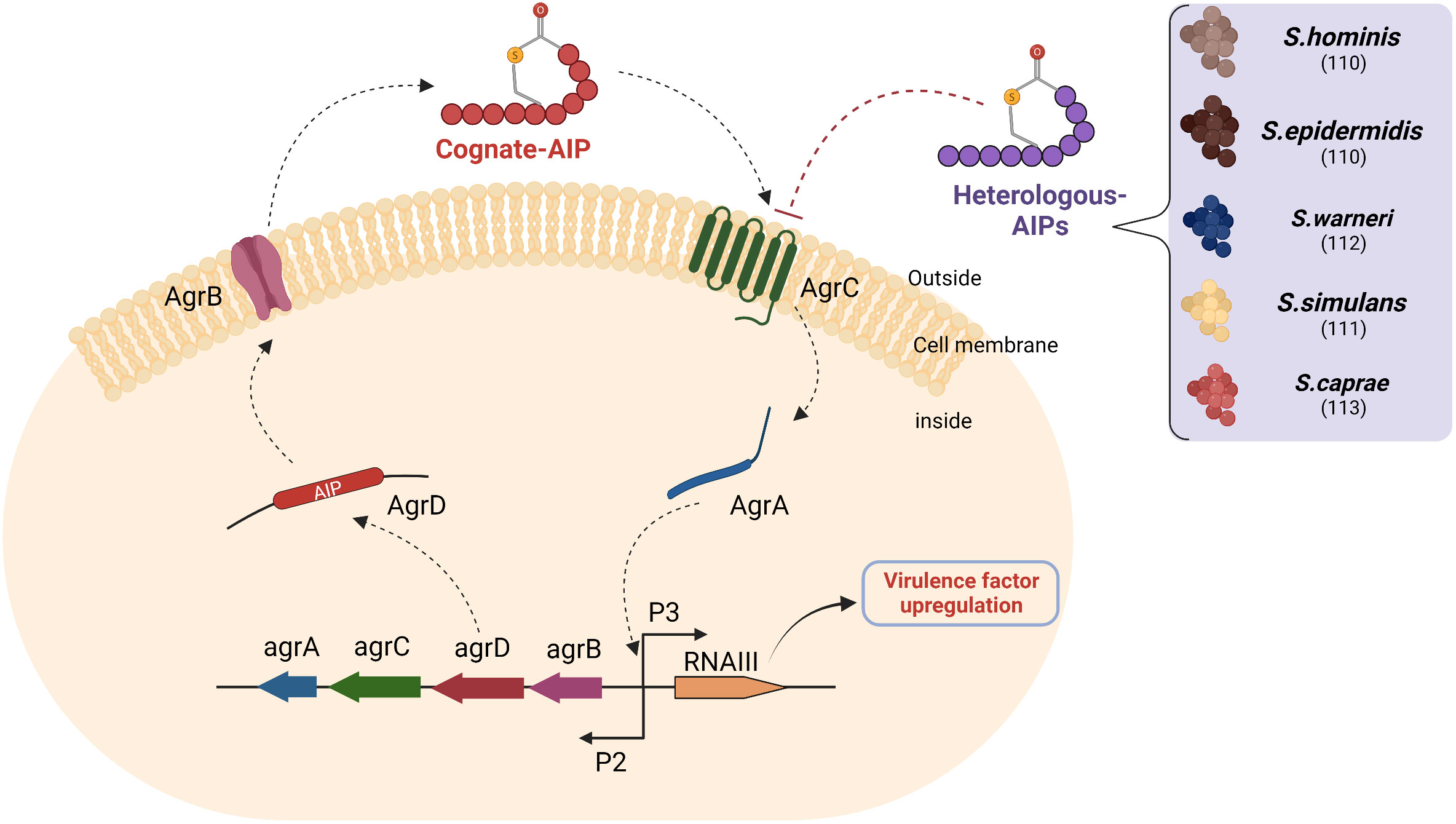
Figure 4 Heterologous AIPs inhibit agr quorum sensing of S. aureus: AIPs secreted by S. aureus (Cognate AIPs) and recognized via the AgrC receptor of the same species activate AgrA-mediated transcription of two divergent transcripts under the control of promoters (P), namely P2 and P3. The P2 promoter encodes the quorum-sensing machinery, while the P3 promoter encodes RNAIII. The RNAIII transcript is responsible for the expression of exoprotein virulence factors. AgrD is a precursor of autoinducing peptide, which is processed by AgrB and then exported to the extracellular space. Several heterologous AIPs secreted by CoNS compete with cognate AIP for binding to AgrC and inhibit downstream Agr signalling. The numbers in brackets denote references. Abbreviations: agr- accessory gene regulation, P2: Promoter region 2, P3: Promoter region 3.
The ability of certain bacterial supernatants to modulate the S. aureus agr system led to an interest in discovering the metabolites responsible for this phenomenon, which later became known as heterologous AIPs. These heterologous AIPs had similar structures to cognate AIPs (cyclic 7-12 amino acid long with a thiolactone group). This phenomenon is also referred to as quorum quenching and can be used as a therapeutic target in AD (109). Williams et al. identified and isolated an AIP from S. hominins with a potent inhibitory effect on the S. aureus quorum sensing (110). This AIP successfully inhibited S. aureus-mediated epidermal proteolysis and inflammation in mouse skin. Another study showed that synthetic heterologous AIPs identified from Staphylococcus simulans isolated from humans and cattle were able to reduce dermonecrotic and epicutaneous skin lesions in mouse models of methicillin-resistant S. aureus (MRSA) skin infections by inhibiting all agr quorum sensing signalling subtypes (111). Similarly, several peptide quorum quenchers, including those from Staphylococcus warneri, Staphylococcus capitis, and Staphylococcus epidermidis, have potential as therapeutic agents in AD (110, 112, 113). Interestingly, apicidin, a cyclic fungal tetrapeptide, also inhibited all agr QS systems in methicillin-resistant S. aureus (MRSA), suggesting that there is competition within kingdoms that can be exploited in the discovery of new quorum quenching AMPs (114).
The therapeutic potential of quorum quenching molecules is attributed in part to indirect immunomodulatory effects by quorum quenching, i.e., inhibition of Agr-associated virulence factors that interact with immune cells. However, in a recent publication by Pundir et al, many Gram-positive bacterial AIPs were shown to be recognized by the mast cell-specific receptor in humans and mice (Mrgprb2 and MRGPRX2). Among these AIPs, they found that the competence-stimulating peptide secreted by Streptococcus pneumoniae (CSP)-1 strongly activated Mrgprb2 and MRGPRX2 and induced effective mast cell degranulation that inhibited bacterial growth and biofilm formation (115).
Despite intensive research, a cure for AD is not yet possible. Currently, there are several treatment options for AD but due to the heterogeneous course of the disease, not all patients respond well, therefore there is an urgent need to develop novel treatment strategies. Topical antibiotics have shown little promise as the sole treatment for AD unless secondary infections are involved (116). In addition, treatment guidelines discourage the use of topical antibiotics (117). This is due to the broad-spectrum activity of marketed antibiotics, which not only inhibit S. aureus but also kill commensal bacteria, which, as mentioned earlier, are critical for homeostasis and resistance to S.aureus colonization. Recently, ATx201 (Niclosamide), a small molecule, was introduced as a promising AD therapy (118). The therapeutic potential was attributed to ATx201’s narrow spectrum activity against S. aureus without causing damage to the commensal microbiome (119). Interestingly, inhibition of S. aureus growth by live bacteriotherapy in AD has also shown promising results (120, 121). The mediators responsible for this effect were bacterial AMPs of class SPBs with a narrow spectrum of activity. Bacteriotherapy, while promising, also presents some challenges, e.g., we currently lack an understanding of the metabolism and interactions of individual bacteria when exposed to a complex microbial environment, as well as their long-term safety. This is especially true for the changes that result from interactions with mobile genetic elements. Long-term culture of bacteria can lead to spontaneous mutations that result in the loss or gain of undesirable functions. In addition, it is difficult to assess the purity and composition of live bacterial products compared to chemical components.
An alternative strategy is to use well-characterized bacterial metabolites, such as bacterial AMPs, to treat AD. Many SPBs have shown that they can be used in clinical practice (76). To date, however, most of them have been limited to use in animals. Certain patented SPBs, namely lugdunin, Hogocidin-α, and Hogocidin-ß, have already shown narrow-spectrum activity against S. aureus, but their therapeutic potential in AD remains uninvestigated. Further research is needed to explore and characterize the vast pool of undiscovered SPBs from the skin microbiome. Inadequate investment and lengthy identification and isolation procedures have severely hindered the development of SPBs as therapeutics. Technological advances in genomics have recently enabled the use of metagenome databases and the identification of novel bacteriocins from biosynthetic gene clusters using genome mining tools (122, 123). Mass spectrometry-assisted peptidomics is another popular technology commonly used to identify novel biomarkers from clinical blood samples for a variety of diseases. Recently, more robust techniques have enabled the application of this approach to a range of biological tissues (124). For example, Azkargorta et al. performed a differential peptidomic analysis of the natural peptide content of the endometrium and demonstrated the presence and activity of antimicrobial peptides in situ (125). Whether a similar approach can be used to identify novel bacterial AMPs from skin remains to be investigated but sounds promising.
Several SPBs are active against multidrug-resistant S. aureus and are also less likely to develop resistance compared with conventional antibiotics (81). For example, Oyama et al. identified two antimicrobial peptides from the rumen microbiome metagenome data set that are active against multidrug-resistant S. aureus. When they examined the likelihood of resistance development after exposure to sub-MIC levels of these peptides, they found that the AMPs did not generate resistant mutants for 20 days. Moreover, the MIC remained within a 1-2-fold increase compared with mupirocin, a marketed topical antibiotic that had a 32-fold MIC increase (126). Nevertheless, there are reports of the development of resistance to the antimicrobial activity of HDPs (127, 128), so the possibility of resistance developing in SPBs cannot be excluded. Therefore, it is important to study the long-term development of resistance of S. aureus to SPBs before considering them for therapeutic use. It is known that the addition of SPBs to conventional antibiotics can have a synergistic effect against multidrug-resistant pathogens and also reduces the likelihood of resistance development (129, 130). This raises the likelihood that the use of multiple narrow-spectrum SPBs targeting different mechanisms could have synergistic and favorable microbial killing profiles and reduce the likelihood of resistance development. Some recent studies suggest this phenomenon (131, 132). Alternatively, a combination of bacterial AMPs with HDPs could also show synergistic activities and provide a favorable microbial killing profile, as recently shown by Bitschar et al. (99).
Certain bacterial AMPs have the potential to modulate skin immunity and possess cytotoxic activity (99, 100, 108, 115). On the one hand, this property makes bacterial AMPs of interest to AD, as there is evidence that an inadequate immune response due to a type 2 inflammatory environment and the absence of immune-enhancing cues from the commensal microbiome is a feature of AD (38, 133). On the other hand, it also raises the question of a possible cytotoxic effect of bacterial AMPs on the host. However, it should be kept in mind that bacterial AMPs derived from the human skin microbiome are ubiquitous on the skin and therefore may be better tolerated (91). NAI003, a derivative of a thiopeptide SPB, has completed a phase 1 clinical trial for acne treatment, providing evidence that SPBs can be safely used as topical agents (90). However, it will be extremely important to investigate the chronic effects of such bacterial AMPs at therapeutic concentrations on the skin and their potential to trigger inflammation.
Antimicrobial screening campaigns from the microbiome have so far focused only on the inhibition of pathogens; however, further research should also highlight the ability of bacteriocins to protect commensals. Targeted delivery of antimicrobials and the use of bacteriophages are other promising tools that can be used to kill specific microbes without harming commensals (134, 135).
Another way to render S. aureus ineffective is to affect its quorum sensing by bacterial AMPs. Studies targeting quorum sensing of S. aureus with synthetic AIPs have shown improved outcomes in AD mouse models and S. aureus-associated infections following the reduction of virulence factors. Inhibition of quorum sensing could reduce the production of proteases by S. aureus that are responsible for inactivating bacteriocins. This leads us to an interesting question: can a combination of SPBs and a heterologous AIP result in a better therapeutic outcome and a lower risk of resistance development in AD?
It is also important to note that S. aureus associated with AD, is non-communicable. This is because the conditions for S. aureus colonization and establishment of AD requires (I) altered barrier function (24, 136) or (II) loss of the commensal microbiota necessary to limit S. aureus (39), or (III) an inadequate immune response (66, 137) or their combination; all of which are normally intact in healthy individuals.
Several authors have put forward the idea that skin microbiome manipulation with targeted control of S. aureus colonization is a therapeutic aim in AD (30, 32, 38), and recent findings strongly support this idea (39, 110, 118, 121). However, it should be noted that not all patients with AD are colonized with S. aureus. This suggests that the disease is caused by multiple factors and their interaction with each other. It appears that certain commensal microbes which tend to act as pathobionts can take over the role of S. aureus in its absence. For example, Staphylococcus epidermidis, a skin commensal, has been shown to colonize the skin of AD and produce proteases that damage the host and induce expression of AD-associated proinflammatory cytokines in human primary keratinocytes (138, 139). However, further research is required to elucidate the fate and role of pathobionts like Staphylococcus epidermidis in AD. In addition, host gene defects, e.g., filaggrin, lower HDP expression owing to type 2 cytokine action (66, 140), and environmental factors may play an exacerbating role in AD. Whether therapies targeting S. aureus have any effect when S. aureus is not involved in AD remains to be determined but seems unrealistic. In such cases, it is important that the microbial AMPs used also target pathogenic Staphylococcus epidermidis, possess additional immunomodulatory activities and/or be supplemented with therapeutics that improve the skin barrier or immune system, or both.
In addition, studies using biologics targeting the immune system in AD patients show decreased S. aureus abundance after treatment (137, 141), suggesting the bidirectional nature of the disease and supporting the proposition that S. aureus is not the initiator of AD but rather a mediator that exacerbates the disease. This also leads to the question if microbial AMPs alone can lead to therapeutic efficacy equal to baseline level, which remains to be investigated.
Selective killing or virulence inhibition of S. aureus without damage to the commensal microbiome is an important therapeutic approach in AD. Evidence suggests that certain bacterial AMPs, which include short peptide bacteriocins and heterologous autoinducing peptides isolated from commensal microbiome, have the potential to treat AD by selectively inhibiting S. aureus or its virulence and/or by immunomodulation. This warrants the discovery of novel bacterial AMPs from members of the commensal microbiome of skin surface for the treatment of AD.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
VP and AJ conceived the ideas. AJ drafted the manuscript and figures. All authors critically revised the manuscript to provide important intellectual content. All authors contributed to the article and approved the submitted version.
This work was supported by Sanofi Regeneron Type2 innovation grant and funding from City of Graz to VP; FWF (I 4939-B) -ANR (ANR-20-CE91-0002) International joint project to PW, J-FN, MV and VP. PhD student AJ was supported by Medical University of Graz, and PhD program MolMed. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The authors would like to thank Dr. Thomas Eichmann, from CF mass spectrometry, Medical University of Graz, 8010, Graz, Austria for his feedback on the peptide structures in Figure 2. Figures created using Biorender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AD, atopic dermatitis; AMP, antimicrobial peptides; S.aureus, Staphylococcus aureus; CC, clonal complex; MIC, minimum inhibitory concentration; agr, accessory gene regulator; HDP, host defence peptides; PLE, Polymorphous light eruption; SPB, short peptide bacteriocins; RiPPs, Ribosomally synthesized and post translationally modified peptides; NRPs, Peptide bacteriocins synthesized by non-ribosomal synthetases; QS, quorum sensing; AIP, autoinducing peptide; Mrgprb2, mas-related G-protein coupled receptor member B2; MRGPRX2, Mas-related G-protein coupled receptor member X2.
1. Langan SM, Irvine AD, Weidinger S. Atopic dermatitis. Lancet (2020) 396(10247):345–60. doi: 10.1016/s0140-6736(20)31286-1
2. Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nat Rev Dis Primers (2018) 4(1):1. doi: 10.1038/s41572-018-0001-z
3. Fishbein AB, Mueller K, Kruse L, Boor P, Sheldon S, Zee P, et al. Sleep disturbance in children with Moderate/Severe atopic dermatitis: A case-control study. J Am Acad Dermatol (2018) 78(2):336–41. doi: 10.1016/j.jaad.2017.08.043
4. Chatrath S, Lei D, Yousaf M, Chavda R, Gabriel S, Silverberg JI. Longitudinal course and predictors of depressive symptoms in atopic dermatitis. J Am Acad Dermatol (2022) 87(3):582–91. doi: 10.1016/j.jaad.2022.04.061
5. Narla S, Silverberg JI. The role of environmental exposures in atopic dermatitis. Curr Allergy Asthma Rep (2020) 20(12):74. doi: 10.1007/s11882-020-00971-z
6. Haahtela T, Holgate S, Pawankar R, Akdis CA, Benjaponpitak S, Caraballo L, et al. The biodiversity hypothesis and allergic disease: World allergy organization position statement. World Allergy Organ J (2013) 6(1):3. doi: 10.1186/1939-4551-6-3
8. Janssens M, van Smeden J, Gooris GS, Bras W, Portale G, Caspers PJ, et al. Increase in short-chain ceramides correlates with an altered lipid organization and decreased barrier function in atopic eczema patients. J Lipid Res (2012) 53(12):2755–66. doi: 10.1194/jlr.P030338
9. Celebi Sozener Z, Ozdel Ozturk B, Cerci P, Turk M, Gorgulu Akin B, Akdis M, et al. Epithelial barrier hypothesis: Effect of the external exposome on the microbiome and epithelial barriers in allergic disease. Allergy (2022) 77(5):1418–49. doi: 10.1111/all.15240
10. Irvine AD, McLean WH, Leung DY. Filaggrin mutations associated with skin and allergic diseases. N Engl J Med (2011) 365(14):1315–27. doi: 10.1056/NEJMra1011040
11. Mack MR, Kim BS. The itch-scratch cycle: A neuroimmune perspective. Trends Immunol (2018) 39(12):980–91. doi: 10.1016/j.it.2018.10.001
12. Hammad H, Lambrecht BN. Barrier epithelial cells and the control of type 2 immunity. Immunity (2015) 43(1):29–40. doi: 10.1016/j.immuni.2015.07.007
13. Geha RS, Jabara HH, Brodeur SR. The regulation of immunoglobulin e class-switch recombination. Nat Rev Immunol (2003) 3(9):721–32. doi: 10.1038/nri1181
14. Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol (2018) 16(3):143–55. doi: 10.1038/nrmicro.2017.157
15. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol (2011) 9(4):244–53. doi: 10.1038/nrmicro2537
16. Harris-Tryon TA, Grice EA. Microbiota and maintenance of skin barrier function. Science (2022) 376(6596):940–5. doi: 10.1126/science.abo0693
17. Schommer NN, Gallo RL. Structure and function of the human skin microbiome. Trends Microbiol (2013) 21(12):660–8. doi: 10.1016/j.tim.2013.10.001
18. Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res (2012) 22(5):850–9. doi: 10.1101/gr.131029.111
19. Totté JE, van der Feltz WT, Hennekam M, van Belkum A, van Zuuren EJ, Pasmans SG. Prevalence and odds of staphylococcus aureus carriage in atopic dermatitis: A systematic review and meta-analysis. Br J Dermatol (2016) 175(4):687–95. doi: 10.1111/bjd.14566
20. Tauber M, Balica S, Hsu CY, Jean-Decoster C, Lauze C, Redoules D, et al. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J Allergy Clin Immunol (2016) 137(4):1272–4.e3. doi: 10.1016/j.jaci.2015.07.052
21. Katsarou A, Armenaka M. Atopic dermatitis in older patients: Particular points. J Eur Acad Dermatol Venereol (2011) 25(1):12–8. doi: 10.1111/j.1468-3083.2010.03737.x
22. Ndhlovu GON, Abotsi RE, Shittu AO, Abdulgader SM, Jamrozy D, Dupont CL, et al. Molecular epidemiology of staphylococcus aureus in African children from rural and urban communities with atopic dermatitis. BMC Infect Dis (2021) 21(1):348. doi: 10.1186/s12879-021-06044-4
23. Ohnishi Y, Okino N, Ito M, Imayama S. Ceramidase activity in bacterial skin flora as a possible cause of ceramide deficiency in atopic dermatitis. Clin Diagn Lab Immunol (1999) 6(1):101–4. doi: 10.1128/cdli.6.1.101-104.1999
24. Fleury OM, McAleer MA, Feuillie C, Formosa-Dague C, Sansevere E, Bennett DE, et al. Clumping factor b promotes adherence of staphylococcus aureus to corneocytes in atopic dermatitis. Infect Immun (2017) 85(6):e00994-16. doi: 10.1128/iai.00994-16
25. Leung DY. New insights into atopic dermatitis: Role of skin barrier and immune dysregulation. Allergol Int (2013) 62(2):151–61. doi: 10.2332/allergolint.13-RAI-0564
26. Pietrocola G, Nobile G, Rindi S, Speziale P. Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Front Cell Infect Microbiol (2017) 7:166. doi: 10.3389/fcimb.2017.00166
27. Sonesson A, Przybyszewska K, Eriksson S, Mörgelin M, Kjellström S, Davies J, et al. Identification of bacterial biofilm and the staphylococcus aureus derived protease, staphopain, on the skin surface of patients with atopic dermatitis. Sci Rep (2017) 7(1):8689. doi: 10.1038/s41598-017-08046-2
28. Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, et al. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J Invest Dermatol (2016) 136(11):2192–200. doi: 10.1016/j.jid.2016.05.127
29. Seiti Yamada Yoshikawa F, Feitosa de Lima J, Notomi Sato M, Álefe Leuzzi Ramos Y, Aoki V, Leao Orfali R. Exploring the role of staphylococcus aureus toxins in atopic dermatitis. Toxins (Basel) (2019) 11(6):321. doi: 10.3390/toxins11060321
30. Geoghegan JA, Irvine AD, Foster TJ. Staphylococcus aureus and atopic dermatitis: A complex and evolving relationship. Trends Microbiol (2018) 26(6):484–97. doi: 10.1016/j.tim.2017.11.008
31. Kim J, Kim BE, Ahn K, Leung DYM. Interactions between atopic dermatitis and staphylococcus aureus infection: Clinical implications. Allergy Asthma Immunol Res (2019) 11(5):593–603. doi: 10.4168/aair.2019.11.5.593
32. Koh LF, Ong RY, Common JE. Skin microbiome of atopic dermatitis. Allergol Int (2022) 71(1):31–9. doi: 10.1016/j.alit.2021.11.001
33. Simpson EL, Villarreal M, Jepson B, Rafaels N, David G, Hanifin J, et al. Patients with atopic dermatitis colonized with staphylococcus aureus have a distinct phenotype and endotype. J Invest Dermatol (2018) 138(10):2224–33. doi: 10.1016/j.jid.2018.03.1517
34. Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng W-I, Conlan S, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Trans Med (2017) 9(397):eaal4651. doi: 10.1126/scitranslmed.aal4651
35. Jenul C, Horswill AR. Regulation of staphylococcus aureus virulence. Microbiol Spectr (2019) 7(2). doi: 10.1128/microbiolspec.GPP3-0031-2018
36. Lomholt H, Andersen KE, Kilian M. Staphylococcus aureus clonal dynamics and virulence factors in children with atopic dermatitis. J Invest Dermatol (2005) 125(5):977–82. doi: 10.1111/j.0022-202X.2005.23916.x
37. Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, et al. Staphylococcus agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med (2020) 12(551). doi: 10.1126/scitranslmed.aay4068
38. Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol (2019) 143(1):26–35. doi: 10.1016/j.jaci.2018.11.015
39. Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med (2017) 9(378). doi: 10.1126/scitranslmed.aah4680
40. Ridaura VK, Bouladoux N, Claesen J, Chen YE, Byrd AL, Constantinides MG, et al. Contextual control of skin immunity and inflammation by corynebacterium. J Exp Med (2018) 215(3):785–99. doi: 10.1084/jem.20171079
41. Thiboutot DM, Nelson AM. Keeping the peace: Commensal cutibacterium acnes trains Cd4+ Th17 cells to trap and kill. J Clin Invest (2021) 131(2). doi: 10.1172/jci145379
42. Ito Y, Sasaki T, Li Y, Tanoue T, Sugiura Y, Skelly AN, et al. Staphylococcus cohnii is a potentially biotherapeutic skin commensal alleviating skin inflammation. Cell Rep (2021) 35(4):109052. doi: 10.1016/j.celrep.2021.109052
43. Agak GW, Mouton A, Teles RM, Weston T, Morselli M, Andrade PR, et al. Extracellular traps released by antimicrobial Th17 cells contribute to host defense. J Clin Invest (2021) 131(2). doi: 10.1172/jci141594
44. Uberoi A, Bartow-McKenney C, Zheng Q, Flowers L, Campbell A, Knight SAB, et al. Commensal microbiota regulates skin barrier function and repair Via signaling through the aryl hydrocarbon receptor. Cell Host Microbe (2021) 29(8):1235–48.e8. doi: 10.1016/j.chom.2021.05.011
45. Zheng Y, Hunt RL, Villaruz AE, Fisher EL, Liu R, Liu Q, et al. Commensal staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host Microbe (2022) 30(3):301–13.e9. doi: 10.1016/j.chom.2022.01.004
46. Myles IA, Castillo CR, Barbian KD, Kanakabandi K, Virtaneva K, Fitzmeyer E, et al. Therapeutic responses to roseomonas mucosa in atopic dermatitis may involve lipid-mediated tnf-related epithelial repair. Sci Transl Med (2020) 12(560). doi: 10.1126/scitranslmed.aaz8631
47. Danby SG, Cork MJ. Ph in atopic dermatitis. Curr Probl Dermatol (2018) 54:95–107. doi: 10.1159/000489523
48. Hill CP, Yee J, Selsted ME, Eisenberg D. Crystal structure of defensin hnp-3, an amphiphilic dimer: Mechanisms of membrane permeabilization. Science (1991) 251(5000):1481–5. doi: 10.1126/science.2006422
49. Li X, Li Y, Han H, Miller DW, Wang G. Solution structures of human ll-37 fragments and nmr-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J Am Chem Soc (2006) 128(17):5776–85. doi: 10.1021/ja0584875
50. Nguyen VS, Tan KW, Ramesh K, Chew FT, Mok YK. Structural basis for the bacterial membrane insertion of dermcidin peptide, dcd-1l. Sci Rep (2017) 7(1):13923. doi: 10.1038/s41598-017-13600-z
51. Harder J, Gläser R, Schröder JM. Review: Human antimicrobial proteins — effectors of innate immunity. J Endotoxin Res (2007) 13:317 – 38. doi: 10.1177/0968051907088275
52. Singh P, Ali SA. Multifunctional role of S100 protein family in the immune system: An update. Cells (2022) 11(15). doi: 10.3390/cells11152274
53. Zasloff M. Antimicrobial rnases of human skin. J Invest Dermatol (2009) 129(9):2091–3. doi: 10.1038/jid.2009.216
54. Yamasaki K, Gallo RL. Antimicrobial peptides in human skin disease. Eur J Dermatol (2008) 18(1):11–21. doi: 10.1684/ejd.2008.0304
55. Oppenheim JJ, Biragyn A, Kwak LW, Yang D. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Ann Rheum Dis (2003) 62 Suppl 2(Suppl 2):ii17–21. doi: 10.1136/ard.62.suppl_2.ii17
56. Harder J, Schroder JM. Rnase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J Biol Chem (2002) 277(48):46779–84. doi: 10.1074/jbc.M207587200
57. Lebeau A, Bruyere D, Roncarati P, Peixoto P, Hervouet E, Cobraiville G, et al. Hpv infection alters vaginal microbiome through down-regulating host mucosal innate peptides used by lactobacilli as amino acid sources. Nat Commun (2022) 13(1):1076. doi: 10.1038/s41467-022-28724-8
58. Takahashi T, Gallo RL. The critical and multifunctional roles of antimicrobial peptides in dermatology. Dermatol Clin (2017) 35(1):39–50. doi: 10.1016/j.det.2016.07.006
59. Patra V, Mayer G, Gruber-Wackernagel A, Horn M, Lembo S, Wolf P. Unique profile of antimicrobial peptide expression in polymorphic light eruption lesions compared to healthy skin, atopic dermatitis, and psoriasis. Photodermatol Photoimmunol Photomed (2018) 34(2):137–44. doi: 10.1111/phpp.12355
60. Kiatsurayanon C, Niyonsaba F, Smithrithee R, Akiyama T, Ushio H, Hara M, et al. Host defense (Antimicrobial) peptide, human B-Defensin-3, improves the function of the epithelial tight-junction barrier in human keratinocytes. J Invest Dermatol (2014) 134(8):2163–73. doi: 10.1038/jid.2014.143
61. Hata TR, Kotol P, Boguniewicz M, Taylor P, Paik A, Jackson M, et al. History of eczema herpeticum is associated with the inability to induce human B-defensin (Hbd)-2, hbd-3 and cathelicidin in the skin of patients with atopic dermatitis. Br J Dermatol (2010) 163(3):659–61. doi: 10.1111/j.1365-2133.2010.09892.x
62. Mallbris L, Carlén L, Wei T, Heilborn J, Nilsson MF, Granath F, et al. Injury downregulates the expression of the human cathelicidin protein Hcap18/Ll-37 in atopic dermatitis. Exp Dermatol (2010) 19(5):442–9. doi: 10.1111/j.1600-0625.2009.00918.x
63. Rieg S, Steffen H, Seeber S, Humeny A, Kalbacher H, Dietz K, et al. Deficiency of dermcidin-derived antimicrobial peptides in sweat of patients with atopic dermatitis correlates with an impaired innate defense of human skin in vivo. J Immunol (2005) 174(12):8003–10. doi: 10.4049/jimmunol.174.12.8003
64. Harder J, Dressel S, Wittersheim M, Cordes J, Meyer-Hoffert U, Mrowietz U, et al. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. J Invest Dermatol (2010) 130(5):1355–64. doi: 10.1038/jid.2009.432
65. Miller LS, Sørensen OE, Liu PT, Jalian HR, Eshtiaghpour D, Behmanesh BE, et al. Tgf-B regulates tlr expression and function on epidermal keratinocytes. J Immunol (2005) 174(10):6137–43. doi: 10.4049/jimmunol.174.10.6137
66. Hönzke S, Wallmeyer L, Ostrowski A, Radbruch M, Mundhenk L, Schäfer-Korting M, et al. Influence of Th2 cytokines on the cornified envelope, tight junction proteins, and ß-defensins in filaggrin-deficient skin equivalents. J Invest Dermatol (2016) 136(3):631–9. doi: 10.1016/j.jid.2015.11.007
67. Albenali LH, Danby S, Moustafa M, Brown K, Chittock J, Shackley F, et al. Vitamin d and antimicrobial peptide levels in patients with atopic dermatitis and atopic dermatitis complicated by eczema herpeticum: A pilot study. J Allergy Clin Immunol (2016) 138(6):1715–9.e4. doi: 10.1016/j.jaci.2016.05.039
68. Searing DA, Leung DY. Vitamin d in atopic dermatitis, asthma and allergic diseases. Immunol Allergy Clin North Am (2010) 30(3):397–409. doi: 10.1016/j.iac.2010.05.005
69. Vieyra-Garcia PA, Wolf P. A deep dive into uv-based phototherapy: Mechanisms of action and emerging molecular targets in inflammation and cancer. Pharmacol Ther (2021) 222:107784. doi: 10.1016/j.pharmthera.2020.107784
70. Suwanchote S, Waitayangkoon P, Chancheewa B, Inthanachai T, Niwetbowornchai N, Edwards SW, et al. Role of antimicrobial peptides in atopic dermatitis. Int J Dermatol (2021) 61(5):532–40. doi: 10.1111/ijd.15814
71. Mookherjee N, Anderson MA, Haagsman HP, Davidson DJ. Antimicrobial host defence peptides: Functions and clinical potential. Nat Rev Drug Discovery (2020) 19(5):311–32. doi: 10.1038/s41573-019-0058-8
72. Peng G, Tsukamoto S, Ikutama R, Le Thanh Nguyen H, Umehara Y, Trujillo-Paez JV, et al. Human-B-Defensin-3 attenuates atopic dermatitis-like inflammation through autophagy activation and the aryl hydrocarbon receptor signaling pathway. J Clin Invest (2022) 132(17). doi: 10.1172/jci156501
73. Hancock RE, Haney EF, Gill EE. The immunology of host defence peptides: Beyond antimicrobial activity. Nat Rev Immunol (2016) 16(5):321–34. doi: 10.1038/nri.2016.29
74. Nguyen HLT, Trujillo-Paez JV, Umehara Y, Yue H, Peng G, Kiatsurayanon C, et al. Role of antimicrobial peptides in skin barrier repair in individuals with atopic dermatitis. Int J Mol Sci (2020) 21(20):7607. doi: 10.3390/ijms21207607
75. Cotter PD, Hill C, Ross RP. Bacteriocins: Developing innate immunity for food. Nat Rev Microbiol (2005) 3(10):777–88. doi: 10.1038/nrmicro1273
76. Cotter PD, Ross RP, Hill C. Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol (2013) 11(2):95–105. doi: 10.1038/nrmicro2937
77. Soltani S, Hammami R, Cotter PD, Rebuffat S, Said LB, Gaudreau H, et al. Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiol Rev (2021) 45(1). doi: 10.1093/femsre/fuaa039
78. Lewis BB, Pamer EG. Microbiota-based therapies for clostridium difficile and antibiotic-resistant enteric infections. Annu Rev Microbiol (2017) 71(1):157–78. doi: 10.1146/annurev-micro-090816-093549
79. Wexler AG, Goodman AL. An insider's perspective: Bacteroides as a window into the microbiome. Nat Microbiol (2017) 2:17026. doi: 10.1038/nmicrobiol.2017.26
80. Otto M. Staphylococci in the human microbiome: The role of host and interbacterial interactions. Curr Opin Microbiol (2020) 53:71–7. doi: 10.1016/j.mib.2020.03.003
81. van Heel AJ, Montalban-Lopez M, Kuipers OP. Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert Opin Drug Metab Toxicol (2011) 7(6):675–80. doi: 10.1517/17425255.2011.573478
82. Breukink E, de Kruijff B. The lantibiotic nisin, a special case or not? Biochim Biophys Acta (1999) 1462(1-2):223–34. doi: 10.1016/s0005-2736(99)00208-4
83. Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chem Rev (2005) 105(2):633–84. doi: 10.1021/cr030105v
84. Acedo JZ, Chiorean S, Vederas JC, van Belkum MJ. The expanding structural variety among bacteriocins from gram-positive bacteria. FEMS Microbiol Rev (2018) 42(6):805–28. doi: 10.1093/femsre/fuy033
85. Hatakka K, Saxelin M. Probiotics in intestinal and non-intestinal infectious diseases–clinical evidence. Curr Pharm Des (2008) 14(14):1351–67. doi: 10.2174/138161208784480162
86. Shelburne CE, An FY, Dholpe V, Ramamoorthy A, Lopatin DE, Lantz MS. The spectrum of antimicrobial activity of the bacteriocin subtilosin a. J Antimicrob Chemother (2007) 59(2):297–300. doi: 10.1093/jac/dkl495
87. Nelson RL, Kelsey P, Leeman H, Meardon N, Patel H, Paul K, et al. Antibiotic treatment for clostridium difficile-associated diarrhea in adults. Cochrane Database Syst Rev (2011) 9):Cd004610. doi: 10.1002/14651858.CD004610.pub4
88. Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, et al. Thuricin cd, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium Difficile. Proc Natl Acad Sci (2010) 107(20):9352–7. doi: 10.1073/pnas.0913554107
89. Mullane K, Lee C, Bressler A, Buitrago M, Weiss K, Dabovic K, et al. Multicenter, randomized clinical trial to compare the safety and efficacy of Lff571 and vancomycin for clostridium difficile infections. Antimicrob Agents Chemother (2015) 59(3):1435–40. doi: 10.1128/aac.04251-14
90. Fabbretti A, He CG, Gaspari E, Maffioli S, Brandi L, Spurio R, et al. A derivative of the thiopeptide Ge2270a highly selective against propionibacterium acnes. Antimicrob Agents Chemother (2015) 59(8):4560–8. doi: 10.1128/aac.05155-14
91. Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell (2014) 158(6):1402–14. doi: 10.1016/j.cell.2014.08.032
92. Simons A, Alhanout K, Duval RE. Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms (2020) 8(5):639. doi: 10.3390/microorganisms8050639
93. O'Sullivan JN, Rea MC, O'Connor PM, Hill C, Ross RP. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol Ecol (2019) 95(2). doi: 10.1093/femsec/fiy241
94. Claesen J, Spagnolo JB, Ramos SF, Kurita KL, Byrd AL, Aksenov AA, et al. A cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci Transl Med (2020) 12(570). doi: 10.1126/scitranslmed.aay5445
95. Oh S, Kim S-H, Ko Y, Sim J-H, Kim KS, Lee S-H, et al. Effect of bacteriocin produced by lactococcus sp. hy 449 on skin-inflammatory bacteria. Food Chem Toxicol (2006) 44(8):1184–90. doi: 10.1016/j.fct.2005.08.008
96. Newstead LL, Varjonen K, Nuttall T, Paterson GK. Staphylococcal-produced bacteriocins and antimicrobial peptides: Their potential as alternative treatments for staphylococcus aureus infections. Antibiotics (Basel) (2020) 9(2):40. doi: 10.3390/antibiotics9020040
97. Panina I, Taldaev A, Efremov R, Chugunov A. Molecular dynamics insight into the lipid ii recognition by type a lantibiotics: Nisin, epidermin, and gallidermin. Micromachines (Basel) (2021) 12(10):1169. doi: 10.3390/mi12101169
98. Kim PI, Sohng JK, Sung C, Joo HS, Kim EM, Yamaguchi T, et al. Characterization and structure identification of an antimicrobial peptide, hominicin, produced by staphylococcus hominis mbbl 2-9. Biochem Biophys Res Commun (2010) 399(2):133–8. doi: 10.1016/j.bbrc.2010.07.024
99. Bitschar K, Sauer B, Focken J, Dehmer H, Moos S, Konnerth M, et al. Lugdunin amplifies innate immune responses in the skin in synergy with host- and microbiota-derived factors. Nat Commun (2019) 10(1):2730. doi: 10.1038/s41467-019-10646-7
100. Wladyka B, Piejko M, Bzowska M, Pieta P, Krzysik M, Mazurek L, et al. A peptide factor secreted by staphylococcus pseudintermedius exhibits properties of both bacteriocins and virulence factors. Sci Rep (2015) 5:14569. doi: 10.1038/srep14569
101. O'Sullivan JN, O'Connor PM, Rea MC, O'Sullivan O, Walsh CJ, Healy B, et al. A novel natural nisin variant, is produced by staphylococcus capitis sourced from the human skin microbiota. J Bacteriol (2020) 202(3). doi: 10.1128/jb.00639-19
102. Lynch D, O'Connor PM, Cotter PD, Hill C, Field D, Begley M. Identification and characterisation of capidermicin, a novel bacteriocin produced by staphylococcus capitis. PloS One (2019) 14(10):e0223541. doi: 10.1371/journal.pone.0223541
103. Hernández-González JC, Martínez-Tapia A, Lazcano-Hernández G, García-Pérez BE, Castrejón-Jiménez NS. Bacteriocins from lactic acid bacteria. a powerful alternative as antimicrobials, probiotics, and immunomodulators in veterinary medicine. Anim (Basel) (2021) 11(4):979. doi: 10.3390/toxins11060321
104. de Pablo MA, Gaforio JJ, Gallego AM, Ortega E, Gálvez AM, Alvarez de Cienfuegos López G. Evaluation of immunomodulatory effects of nisin-containing diets on mice. FEMS Immunol Med Microbiol (1999) 24(1):35–42. doi: 10.1111/j.1574-695X.1999.tb01262.x
105. Begde D, Bundale S, Mashitha P, Rudra J, Nashikkar N, Upadhyay A. Immunomodulatory efficacy of nisin–a bacterial lantibiotic peptide. J Pept Sci (2011) 17(6):438–44. doi: 10.1002/psc.1341
106. Aranha CC, Gupta SM, Reddy KV. Assessment of cervicovaginal cytokine levels following exposure to microbicide nisin gel in rabbits. Cytokine (2008) 43(1):63–70. doi: 10.1016/j.cyto.2008.04.005
107. van Hemert S, Meijerink M, Molenaar D, Bron PA, de Vos P, Kleerebezem M, et al. Identification of lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol (2010) 10:293–. doi: 10.1186/1471-2180-10-293
108. Lai CY, Yeh DW, Lu CH, Liu YL, Huang LR, Kao CY, et al. Identification of thiostrepton as a novel inhibitor for psoriasis-like inflammation induced by Tlr7-9. J Immunol (2015) 195(8):3912–21. doi: 10.4049/jimmunol.1500194
109. Grandclément C, Tannières M, Moréra S, Dessaux Y, Faure D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol Rev (2016) 40(1):86–116. doi: 10.1093/femsre/fuv038
110. Williams MR, Costa SK, Zaramela LS, Khalil S, Todd DA, Winter HL, et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci Transl Med (2019) 11(490). doi: 10.1126/scitranslmed.aat8329
111. Brown MM, Kwiecinski JM, Cruz LM, Shahbandi A, Todd DA, Cech NB, et al. Novel peptide from commensal staphylococcus simulans blocks methicillin-resistant staphylococcus aureus quorum sensing and protects host skin from damage. Antimicrob Agents Chemother (2020) 64(6):e00172-20. doi: 10.1128/AAC.00172-20
112. Severn MM, Cho YK, Manzer HS, Bunch ZL, Shahbandi A, Todd DA, et al. The commensal staphylococcus warneri makes peptide inhibitors of mrsa quorum sensing that protect skin from atopic or necrotic damage. J Invest Dermatol (2022) 142(12):3349–52.e5. doi: 10.1016/j.jid.2022.05.1092
113. Paharik AE, Parlet CP, Chung N, Todd DA, Rodriguez EI, Van Dyke MJ, et al. Coagulase-negative staphylococcal strain prevents staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe (2017) 22(6):746–56.e5. doi: 10.1016/j.chom.2017.11.001
114. Parlet CP, Kavanaugh JS, Crosby HA, Raja HA, El-Elimat T, Todd DA, et al. Apicidin attenuates mrsa virulence through quorum-sensing inhibition and enhanced host defense. Cell Rep (2019) 27(1):187–98.e6. doi: 10.1016/j.celrep.2019.03.018
115. Pundir P, Liu R, Vasavda C, Serhan N, Limjunyawong N, Yee R, et al. A connective tissue mast-Cell-Specific receptor detects bacterial quorum-sensing molecules and mediates antibacterial immunity. Cell Host Microbe (2019) 26(1):114–22.e8. doi: 10.1016/j.chom.2019.06.003
116. Hwang J, Thompson A, Jaros J, Blackcloud P, Hsiao J, Shi VY. Updated understanding of staphylococcus aureus in atopic dermatitis: From virulence factors to commensals and clonal complexes. Exp Dermatol (2021) 30(10):1532–45. doi: 10.1111/exd.14435
117. Eichenfield LF, Tom WL, Berger TG, Krol A, Paller AS, Schwarzenberger K, et al. Guidelines of care for the management of atopic dermatitis: Section 2. management and treatment of atopic dermatitis with topical therapies. J Am Acad Dermatol (2014) 71(1):116–32. doi: 10.1016/j.jaad.2014.03.023
118. Weiss A, Delavenne E, Matias C, Lagler H, Simon D, Li P, et al. Topical niclosamide (Atx201) reduces staphylococcus aureus colonization and increases Shannon diversity of the skin microbiome in atopic dermatitis patients in a randomized, double-blind, placebo-controlled phase 2 trial. Clin Transl Med (2022) 12(5):e790. doi: 10.1002/ctm2.790
119. Chu CY. Targeting the cutaneous microbiota in atopic dermatitis: 'A new hope' or 'Attack of the cons'? Clin Transl Med (2022) 12(5):e865. doi: 10.1002/ctm2.865
120. Myles IA, Earland NJ, Anderson ED, Moore IN, Kieh MD, Williams KW, et al. First-in-Human topical microbiome transplantation with roseomonas mucosa for atopic dermatitis. JCI Insight (2018) 3(9). doi: 10.1172/jci.insight.120608
121. Nakatsuji T, Hata TR, Tong Y, Cheng JY, Shafiq F, Butcher AM, et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat Med (2021) 27(4):700–9. doi: 10.1038/s41591-021-01256-2
122. de Jong A, van Heel AJ, Kok J, Kuipers OP. Bagel2: Mining for bacteriocins in genomic data. Nucleic Acids Res (2010) 38(Web Server issue):W647–51. doi: 10.1093/nar/gkq365
123. Behsaz B, Bode E, Gurevich A, Shi Y-N, Grundmann F, Acharya D, et al. Integrating genomics and metabolomics for scalable non-ribosomal peptide discovery. Nat Commun (2021) 12(1):3225. doi: 10.1038/s41467-021-23502-4
124. Foreman RE, George AL, Reimann F, Gribble FM, Kay RG. Peptidomics: A review of clinical applications and methodologies. J Proteome Res (2021) 20(8):3782–97. doi: 10.1021/acs.jproteome.1c00295
125. Azkargorta M, Bregón-Villahoz M, Escobes I, Ibáñez-Pérez J, Iloro I, Iglesias M, et al. In-depth proteomics and natural peptidomics analyses reveal antibacterial peptides in human endometrial fluid. J Proteomics (2020) 216:103652. doi: 10.1016/j.jprot.2020.103652
126. Oyama LB, Olleik H, Teixeira ACN, Guidini MM, Pickup JA, Hui BYP, et al. In silico identification of two peptides with antibacterial activity against multidrug-resistant staphylococcus aureus. NPJ Biofilms Microbiomes (2022) 8(1):58. doi: 10.1038/s41522-022-00320-0
127. Cheung GYC, Fisher EL, McCausland JW, Choi J, Collins JWM, Dickey SW, et al. Antimicrobial peptide resistance mechanism contributes to staphylococcus aureus infection. J Infect Dis (2018) 217(7):1153–9. doi: 10.1093/infdis/jiy024
128. Kawada-Matsuo M, Le MN, Komatsuzawa H. Antibacterial peptides resistance in staphylococcus aureus: Various mechanisms and the association with pathogenicity. Genes (Basel) (2021) 12(10):1527. doi: 10.3390/genes12101527
129. Ovchinnikov KV, Kranjec C, Thorstensen T, Carlsen H, Diep DB. Bacteriocins revitalize non-effective penicillin G to overcome methicillin-resistant staphylococcus pseudintermedius. Antibiotics (Basel) (2022) 11(12):1691. doi: 10.3390/antibiotics11121691
130. Wolska KI, Grześ K, Kurek A. Synergy between novel antimicrobials and conventional antibiotics or bacteriocins. Pol J Microbiol (2012) 61(2):95–104. doi: 10.33073/pjm-2012-012
131. Soltani S, Biron E, Ben Said L, Subirade M, Fliss I. Bacteriocin-based synergetic consortia: A promising strategy to enhance antimicrobial activity and broaden the spectrum of inhibition. Microbiol Spectr (2022) 10(1):e0040621. doi: 10.1128/spectrum.00406-21
132. Mathur H, Field D, Rea MC, Cotter PD, Hill C, Ross RP. Bacteriocin-antimicrobial synergy: A medical and food perspective. Front Microbiol (2017) 8:1205. doi: 10.3389/fmicb.2017.01205
133. Kuo IH, Yoshida T, De Benedetto A, Beck LA. The cutaneous innate immune response in patients with atopic dermatitis. J Allergy Clin Immunol (2013) 131(2):266–78. doi: 10.1016/j.jaci.2012.12.1563
134. Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature (2019) 575(7783):505–11. doi: 10.1038/s41586-019-1742-x
135. Ting SY, Martínez-García E, Huang S, Bertolli SK, Kelly KA, Cutler KJ, et al. Targeted depletion of bacteria from mixed populations by programmable adhesion with antagonistic competitor cells. Cell Host Microbe (2020) 28(2):313–21.e6. doi: 10.1016/j.chom.2020.05.006
136. Riethmuller C, McAleer MA, Koppes SA, Abdayem R, Franz J, Haftek M, et al. Filaggrin breakdown products determine corneocyte conformation in patients with atopic dermatitis. J Allergy Clin Immunol (2015) 136(6):1573–80.e2. doi: 10.1016/j.jaci.2015.04.042
137. Callewaert C, Nakatsuji T, Knight R, Kosciolek T, Vrbanac A, Kotol P, et al. Il-4rα blockade by dupilumab decreases staphylococcus aureus colonization and increases microbial diversity in atopic dermatitis. J Invest Dermatol (2020) 140(1):191–202.e7. doi: 10.1016/j.jid.2019.05.024
138. Cau L, Williams MR, Butcher AM, Nakatsuji T, Kavanaugh JS, Cheng JY, et al. Staphylococcus epidermidis protease ecpa can be a deleterious component of the skin microbiome in atopic dermatitis. J Allergy Clin Immunol (2021) 147(3):955–66.e16. doi: 10.1016/j.jaci.2020.06.024
139. Ochlich D, Rademacher F, Drerup KA, Gläser R, Harder J. The influence of the commensal skin bacterium staphylococcus epidermidis on the epidermal barrier and inflammation: Implications for atopic dermatitis. Exp Dermatol (2022). doi: 10.1111/exd.14727
140. Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med (2002) 347(15):1151–60. doi: 10.1056/NEJMoa021481
141. Beck LA, Bieber T, Weidinger S, Tauber M, Saeki H, Irvine AD, et al. Tralokinumab treatment improves the skin microbiota by increasing the microbial diversity in adults with moderate-to-Severe atopic dermatitis: Analysis of microbial diversity in ecztra 1, a randomized controlled trial. J Am Acad Dermatol (2022). doi: 10.1016/j.jaad.2022.11.047
Keywords: antimicrobial peptides, atopic dermatitis, autoinducing peptides, bacteriocins, skin microbiome, Staphylococcus aureus
Citation: Joshi AA, Vocanson M, Nicolas J-F, Wolf P and Patra V (2023) Microbial derived antimicrobial peptides as potential therapeutics in atopic dermatitis. Front. Immunol. 14:1125635. doi: 10.3389/fimmu.2023.1125635
Received: 16 December 2022; Accepted: 09 January 2023;
Published: 25 January 2023.
Edited by:
Tej Pratap Singh, University of Pennsylvania, United StatesReviewed by:
Ian Antheni Myles, National Institutes of Health (NIH), United StatesCopyright © 2023 Joshi, Vocanson, Nicolas, Wolf and Patra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vijaykumar Patra, dmlqYXlrdW1hci5wYXRyYUBnbWFpbC5jb20=
†Present address: Vijaykumar Patra, L’Oréal Research and Innovation, Aulnay-sous-Bois, France
‡ORCID: Aaroh Anand Joshi, orcid.org/0000-0003-4385-1318
Marc Vocanson, orcid.org/0000-0002-0181-8862
Jean-Francois Nicolas, orcid.org/0000-0003-4204-803X
Peter Wolf, orcid.org/0000-0001-7777-9444
Vijaykumar Patra, orcid.org/0000-0002-7161-7767
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.