- 1Division of Infectious Diseases, Department of Medicine, Washington University School of Medicine, St. Louis, MO, United States
- 2Infectious Diseases, Medicine Service, Veterans Affairs Saint Louis Health Care System, Saint Louis, MO, United States
The pathogenic Escherichia coli can be parsed into specific variants (pathovars) depending on their phenotypic behavior and/or expression of specific virulence factors. These pathogens are built around chromosomally-encoded core attributes and through acquisition of specific virulence genes that direct their interaction with the host. Engagement of E. coli pathovars with CEACAMs is determined both by core elements common to all E. coli as well as extrachromosomally-encoded pathovar-specific virulence traits, which target amino terminal immunoglobulin variable-like (IgV) regions of CEACAMs. Emerging data suggests that engagement of CEACAMs does not unilaterally benefit the pathogen and that these interactions may also provide an avenue for pathogen elimination.
Introduction
Pathogenic Escherichia coli exhibit extraordinary adaptation to multiple host niches, due in part to their remarkable genetic plasticity that has permitted the acquisition of a diverse array of virulence molecules. These pathogens are incredibly diverse as illustrated by whole genome sequencing studies which highlight the open nature of the E. coli pangenome (1). In essence, no two isolates of E. coli from different sources are likely to be “identical”. Despite this diversity, an important element of niche adaptation among pathogenic E. coli is the interaction of their respective adhesins with the family of eukaryotic molecules known as the Carcinoembryonic Antigen -related Cell Adhesion Molecules or CEACAMs. These molecules, encoded on human chromosome 19q13 (2), are cell surface proteins that participate in homotypic intercellular adhesion interactions. CEACAMs are present on many cell types including those lining intestinal and urogenital mucosae where E. coli pathogenic variants (pathovars) establish a niche, as well as immune cells which defend these tissues.
CEACAMs share a common architecture in which the amino terminal region of the protein forms a domain resembling the immunoglobulin variable region (IgV-like), while the remainder of extracellular portion of each protein is formed by a variable number of domains similar to immunoglobulin constant regions (3). Homotypic dimerization is primarily directed by the interaction of the N-terminal domains (4). The proteins are also heavily glycosylated along their lengths. Among the intestinal CEACAMs, the C terminal end of CEACAM1 is comprised of a transmembrane region and a cytoplasmic tail in which individual isoforms may contain Immunoreceptor Tyrosine-based Inhibitory Motifs (ITIM), while CEACAMs 5,6, and 7 each end in glycosylphosphotidyl inositol (GPI) anchors.
Biology of E. coli adhesin/CEACAM interactions
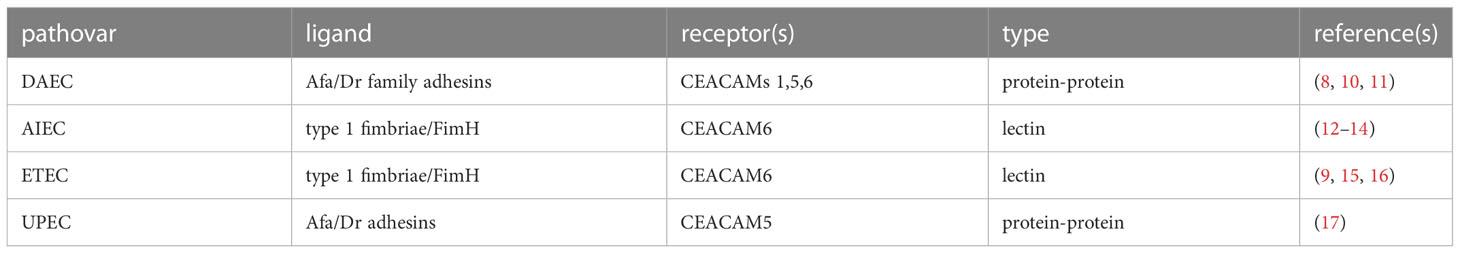
CEACAMs are among the most rapidly evolving proteins in humans. As the extracellular regions of these proteins serve as receptors for a number of important human pathogens, they appear to be under considerable adaptive pressure (5–7) driving CEACAM polymorphisms as well as species-specific diversity (5). Pathogenic E. coli are among the pathogens known to target CEACAM domains exposed on mucosal surfaces. Interestingly, the amino terminal IgV like domains of CEACAMs (Figure 1A) are defined targets for several adhesins expressed by the E. coli pathovars. To date, two major groups of structures- the Afa/Dr adhesins and type 1 pili have been shown to engage the CEACAM N-terminal domains (Table 1).
Figure 1 N-terminal IgV-like domains of CEACAMs (A) MUSCLE alignment of N-terminal CEACAM domains. Highlighted residues Phe 29 and Gln44 indicate amino acids shown to be critical for Afa/Dr interaction (8); residues at Asn71 and Asn78 indicate glycosylation sites important for interaction with type 1 pili (9) and the FimH adhesin. (B) Dimeric CEACAM5 N-terminal domain (PDB 2QSQ) (8) depicting location of Afa/Dr adhesin interactions. (C) Dimeric CEACAM6 N-terminal domain (PDB 4Y8A) (4) depicting location of sites critical for interaction with type 1 pili (9).
The Afa/Dr family of adhesins mediate mannose-resistant hemagglutination and adhesion. Members of this group of adhesion molecules share an operon organization that includes genes encoding a chaperone, an outer membrane protein (usher), and the adhesin. Afa operons encode an AfaB chaperone and an AfaC outer membrane protein, while AfaE molecules serve as afimbrial adhesins. Similarly, Dr systems are comprised of a DraB chaperone, a DraC usher, and a DraE protein which assemble to form thin homopolymeric fimbriae (18–20). Both AfaE as well as DraE can mediate interactions with CEACAMs as receptors (21).
Type 1 pili are ~1 µm long projections from the surface of E. coli that are also assembled through a chaperone usher pilus (CUP) pathway which directs the biogenesis of pili comprised of ~1000 copies of the major pilin subunit (FimA), and single copies of FimF and FimG adapter proteins that present the terminal FimH mannose-binding tip adhesin (22). All E. coli express chromosomally-encoded type 1 pili which mediate adhesion to mannosylated glycoproteins including CEACAMs. Early studies by Leusch et al, indicated that E. coli engage mannosylated residues on CEACAM5 and CEACAM6 (23–25).
Type 1 pili, long known to direct mannose-sensitive adhesion, bind to specific glycosylated residues which are projected on the exposed surface of the CEACAM6 (previously referred to as nonspecific cross-reacting antigen or NCA) (Figure 1) (9, 15). In contrast, the Afa/Dr adhesins are thought to engage residues on the inner dimerization face of CEACAMs (10) and are capable of disrupting CEACAM multimers (8).
Importance of CEACAM-E. coli interactions
Adherent-invasive E. coli and the pathogenesis of inflammatory bowel disease
Although the pathogenesis of inflammatory bowel disease is still being dissected, a prevailing concept is that aberrant interactions between intestinal microbes and the immune system are operative. Among the potential contributors to the pathogenesis of Crohn’s disease are the interactions between the host and E. coli known as adherent/invasive E. coli (AIEC). Although AIEC appear to lack canonical virulence factors found in other gastrointestinal E. coli pathovars (26), whole genome sequencing of isolates from patients with Crohn’s disease suggest that they are phylogenetically distinct from commensal E. coli (27). AIEC have the ability to engage CEACAM6 via FimH (28), with some AIEC having acquired pathoadaptive mutations within the fimH gene that augment interactions between FimH and CEACAM6, and which enhance the propensity of AIEC to induce intestinal inflammation (12). Enhanced expression of CEACAM6 in individuals with inflammatory bowel disease promoted by inflammatory mediators accelerates binding of AIEC (13, 29, 30), and AIEC infection of epithelial cells in vitro induces expression of multiple CEACAMs (31). Dumych, et al. also proposed that CEACAMs expressed on early apoptotic cells exhibit high mannose glycosylation at specific sites (Asn197, and Asn 224) and that FimH can induce the formation of membrane blebs that present these immature high-mannose glycans on their surface (32). Conversely, mannoside compounds which antagonize FimH -CEACAM6 interactions may interrupt AIEC colonization and mitigate subsequent inflammatory changes (14).
Altogether, however the precise role played by AIEC in the pathogenesis of Crohn’s disease is uncertain (33). Indeed, the pathogenesis of inflammatory bowel disease is complex, potentially involving not only AIEC, but human susceptibility genes including a loss of function variant of the protein tyrosine phosphatase non-receptor type 2 (PTPN2) gene (34). Notably, PTPN2 appears to modulate the gut microbiome (35) and loss of PTPN2 is associated with enhanced CEACAM expression and enhanced AIEC uptake and intracellular survival (36), suggesting that Crohn’s disease may reflect the interplay of pathogenic E. coli and distinctly susceptible hosts.
Diffusely adherent E. coli
The diffusely adhering E. coli (DAEC) are a diverse pathovar that have been associated with gastrointestinal (37–39) as well as urinary tract (40, 41) infections (42, 43). From the small number of whole genome sequences of these strains presently available, DAEC appear to be phylogenetically close to enteroaggregative E. coli, but are distinguished in their complement of adhesins as well as iron acquisition systems (44). Although DAEC have been isolated from patients with ulcerative colitis (UC) (45), their role in the molecular pathogenesis of UC remains unclear (33). These strains are defined phenotypically by their diffuse adherence pattern on cultured epithelial cells, and genetically by the expression of a group of virulence molecules known collectively as Afa/Dr adhesins which may constitute either fimbrial or afimbrial (Afa) structures which bind to the human decay accelerating factor (hDAF, also known as CD55) a glycosylphosphatidylinositol (GPI)-anchored glycoprotein present on many cell types including epithelial cells. A subset of this family of adhesins engage CEACAMs including CEACAM1, CEACAM5 (10, 46), and CEACAM6, which are recruited to the sites of bacterial attachment (46), along with lipid raft markers (47). CEACAM1-4L, a splice variant of CEACAM1, has an extended cytoplasmic L domain that can modulate cellular signaling following DAEC engagement (44). Engagement of either of the GPI-anchored CEACAM5 or CEACAM6 molecules can drive efficient internalization of Dr-fimbriated DAEC (21, 47). Interestingly, increased expression of Afa/Dr fimbriae has recently been associated with the emergence of the multidrug-resistant ST131 uropathogenic (UPEC) clones (48). Although UPEC expressing Afa/Dr fimbriae represent less than 10% of UTI isolates, engagement of CEACAMs in the urogenital epithelia suppresses exfoliation of the epithelial cells and enhances colonization (17). Notably, more recent studies demonstrate that CEACAM engagement by pathogenic bacteria results in delivery of bacterial nitric oxide that activates eukaryotic cGMP-dependent signaling pathways to enhance expression of CD105 (endoglin) (49). Increases in CD105 expression in turn abrogate detachment of cells targeted by bacteria by increasing their affinity for extracellular matrix (50).Therefore, engagement of CEACAMs could benefit the pathogen by facilitating colonization while suppressing host mechanisms for elimination.
Enterotoxigenic E. coli (ETEC)
The enterotoxigenic E. coli (ETEC) are a diverse pathovar defined by the production of heat-labile toxin (LT) and/or heat-stable toxins (ST). These pathogens are a predominant cause of acute diarrheal illness as well as deaths due to diarrhea in developing countries among young children. Likewise, ETEC are perennially the major cause of traveler’s diarrhea (51, 52). ETEC have also been linked repeatedly to poorly understood sequelae among young children in LMICs including enteropathic changes to the small intestine and accompanying nutrient malabsorption and growth stunting (53–57).
The basic mechanisms by which ETEC enterotoxins cause diarrhea are known (58). Both toxins activate major cyclic nucleotide second messenger pathways in the cell. LT, like cholera toxin, stimulates adenylate cyclase resulting in increases in intracellular cAMP which in turn activates protein kinase A (PKA). PKA-mediated phosphorylation modulates the activity of cellular ion channels including the cystic fibrosis transmembrane regulator (CFTR) and the sodium-hydrogen ion exchanger (NHE3), resulting in the net export of salt and water into the intestinal lumen and watery diarrhea typical of ETEC. ST, similar to endogenous gastrointestinal peptides guanylin and uroguanylin, binds to guanylate cyclase C resulting in the increased production of cGMP. Increases in cGMP in turn activate Protein kinase G (PKG) which likewise phosphorylates and modulates ion channels resulting in diarrhea.
In contrast, the molecular basis of sequelae associated with ETEC, and the contribution of enterotoxins to enteropathic changes linked to ETEC remains enigmatic. Notably, cAMP and PKA are known to modulate hundreds of eukaryotic genes (59). Binding of cAMP to PKA, a heterotetramer comprised of two regulatory and two catalytic subunits, in the cytoplasm liberates PKA catalytic subunits to enter the nucleus (60) where they phosphorylate the cyclic AMP response element binding protein (CREB) at position S-133 (61, 62). The activated CREB transcription factor is then free to engage multiple cAMP-responsive elements (CRE, e.g., 5’-TGACGTCA-3’) in the regulatory regions of approximately 4000 genes within the human genome. Not surprisingly, recent studies of transcriptional modulation by LT have revealed that it impacts the expression of hundreds of genes in intestinal epithelial cells (63).
Among the many genes modulated by LT are those encoding CEACAMs expressed within the gastrointestinal tract. Although CEACAM expression is normally more robust in the colon, the expression of CEACAMs5, 6, and 7 in small intestinal epithelia are all substantially upregulated by LT as well as forskolin which also stimulates production of cAMP (16). Of note however, promoter regions of these genes appear to lack canonical CRE sites suggesting cAMP-mediated upregulation of their expression is indirect. This increased expression of CEACAM6 enhanced ETEC adhesion to target intestinal epithelial cells, with ETEC recruited specifically to regions of increased CEACAM expression. Conversely deletion of CEACAM6 by CRISPR-Cas9 resulted in a marked decrease in ETEC adhesion while restoration of CEACAM6 expression rescued the adhesion phenotype. Similarly, heterologous expression of CEACAM6 in HeLa cells resulted in marked increases in ETEC adhesion. Moreover, small intestinal biopsies of ETEC infected patients also demonstrated significant increases in CEACAM6 expression following infection. Together, these studies suggested that CEACAMs serve as important receptors for ETEC and that these pathogens exploit cAMP-dependent cellular pathways to alter CEACAM expression in establishing their niche in the small intestine.
Moreover, we demonstrated that FimH, the tip adhesin of type 1 pili, interacts specifically with CEACAM6 in a mannose-dependent fashion. Collectively, these data suggest that ETEC, via its plasmid-encoded heat-labile toxin stimulates the production CEACAMs to augment pathogen-host interactions mediated by chromosomally-encoded type 1 pili. In effect, ETEC alter the epithelial landscape, at least transiently, to suit the bacteria. Many important questions remain however, including whether these changes in CEACAM expression also impact other organisms including commensal E. coli and other E. coli pathovars that could also engage CEACAMs.
CEACAMs as innate defense molecules
The role of CEACAMs that are shed from intestinal epithelia and which could act as molecular decoys for pathogenic microbes remains largely unexplored. Nevertheless, these molecules have the potential to modulate pathogen-host interactions, particularly in the gastrointestinal tract. Large amounts of CEACAM5, estimated to be 50-70 mg, is normally shed in human feces (64–67) with the majority appearing in membrane-bound forms that can be released by cleavage of the GPI anchor with phosphatidylinositol-specific phospholipase C (PI_PLC). Studies of primary colonic epithelial cells in culture indicated that soluble CEACAMs may also be released by endogenous phospholipases (68). Although the precise molecular mechanisms by which CEACAMs are shed into the intestinal lumen and other mucosal spaces have not been definitely established, early studies of transformed intestinal epithelial cells indicated that CEACAMs are released in a directed fashion, specifically from the apical brush border surface (69). Subsequently it has been noted that HT-29 cells release significant amounts of extracellular vesicles (ECV) bearing CEACAMs on their surface, particularly when the cells are under stress (70). It has also been suggested that microvilli, which form the intestinal brush border, release ECV into the lumen (71, 72). Intriguingly, ECV (73) actively released from the tips of microvilli (74) comprising the intestinal brush border contain enzymatically active proteins, and can mitigate interactions of pathogenic E. coli with target epithelia (75). It has also been suggested that intestinal M cells express both CEACAM5 and CEACAM1 on their apical glycocalyx, but these cells differ from enterocytes in lacking the formation of CEACAM-laden vesicles (76). Others have indicated that CEACAM5 and CEACAM6 may be produced by goblet cells. (72, 77) and this appears to be supported by available human small intestine single cell RNAseq data (78).
It has also been suggested that innate responses to pathogen interaction can lead to induction of cytokines that alter expression of CEACAMs by intestinal epithelia, such as the induction of CEACAM5 and CEACAM6 by INF-γ (79). Conversely, others have argued that Gram-negative organisms might be able to impair release of CEA from intestinal epithelia (80).
Conclusion
The expression of a variety of CEACAMs by multiple cell types including those lining mucosal surfaces such as the gastrointestinal tract or on immune effector cells suggests that pathogenic E. coli are likely to encounter these molecules in their transit through human hosts. At present however, despite the substantial diversity of pathogenic E. coli our understanding of the contribution of these molecules in E. coli pathogen-host interactions is limited to a few select pathovars. Already, however, some important themes have emerged from studies of the molecular interactions between pathogen and host. E. coli appear to have adopted several strategies to engage CEACAMs. Ongoing studies are likely to further illuminate the critical nature of these interactions in directing the outcome of several important human infections.
Author contributions
Both authors contributed to the literature review. JF drafted the manuscript and figures. All authors contributed to the article and approved the submitted version.
Acknowledgments
JMF was supported by funding from the Department of Veterans Affairs (5I01BX001469-05), and the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) R01 AI170949, R01 AI089894, and support by the NIH Washington University DDRCC Grant NIDDK P30 DK052574. Research conducted by AS was also supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number T32AI007172. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, or the Department of Veterans Affairs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, et al. The pangenome structure of escherichia coli: comparative genomic analysis of e. coli commensal and pathogenic isolates. J Bacteriol (2008) 190:6881–93. doi: 10.1128/JB.00619-08
2. Teglund S, Olsen A, Khan WN, Frangsmyr L, Hammarstrom S. The pregnancy-specific glycoprotein (PSG) gene cluster on human chromosome 19: fine structure of the 11 PSG genes and identification of 6 new genes forming a third subgroup within the carcinoembryonic antigen (CEA) family. Genomics (1994) 23:669–84. doi: 10.1006/geno.1994.1556
3. Gray-Owen SD, Blumberg RS. CEACAM1: contact-dependent control of immunity. Nat Rev Immunol (2006) 6:433–46. doi: 10.1038/nri1864
4. Bonsor DA, Gunther S, Beadenkopf R, Beckett D, Sundberg EJ. Diverse oligomeric states of CEACAM IgV domains. Proc Natl Acad Sci U.S.A. (2015) 112:13561–6. doi: 10.1073/pnas.1509511112
5. Kammerer R, Zimmermann W. Coevolution of activating and inhibitory receptors within mammalian carcinoembryonic antigen families. BMC Biol (2010) 8:12. doi: 10.1186/1741-7007-8-12
6. Adrian J, Bonsignore P, Hammer S, Frickey T, Hauck CR. Adaptation to host-specific bacterial pathogens drives rapid evolution of a human innate immune receptor. Curr Biol (2019) 29616-630:e615. doi: 10.1016/j.cub.2019.01.058
7. Baker EP, Sayegh R, Kohler KM, Borman W, Goodfellow CK, Brush ER, et al. Evolution of host-microbe cell adherence by receptor domain shuffling. Elife (2022) 11. doi: 10.7554/eLife.73330
8. Korotkova N, Yang Y, Le Trong I, Cota E, Demeler B, Marchant J, et al. Binding of Dr adhesins of escherichia coli to carcinoembryonic antigen triggers receptor dissociation. Mol Microbiol (2008) 67:420–34. doi: 10.1111/j.1365-2958.2007.06054.x
9. Sauter SL, Rutherfurd SM, Wagener C, Shively JE, Hefta SA. Identification of the specific oligosaccharide sites recognized by type 1 fimbriae from escherichia coli on nonspecific cross-reacting antigen, a CD66 cluster granulocyte glycoprotein. J Biol Chem (1993) 268:15510–6. doi: 10.1016/S0021-9258(18)82286-4
10. Korotkova N, Cota E, Lebedin Y, Monpouet S, Guignot J, Servin AL, et al. A subfamily of Dr adhesins of escherichia coli bind independently to decay-accelerating factor and the n-domain of carcinoembryonic antigen. J Biol Chem (2006) 281:29120–30. doi: 10.1074/jbc.M605681200
11. Berger CN, Billker O, Meyer TF, Servin AL, Kansau I. Differential recognition of members of the carcinoembryonic antigen family by Afa/Dr adhesins of diffusely adhering escherichia coli (Afa/Dr DAEC). Mol Microbiol (2004) 52:963–83. doi: 10.1111/j.1365-2958.2004.04033.x
12. Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, et al. Point mutations in FimH adhesin of crohn's disease-associated adherent-invasive escherichia coli enhance intestinal inflammatory response. PloS Pathog (2013) 9:e1003141. doi: 10.1371/journal.ppat.1003141
13. Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, et al. Crohn's disease adherent-invasive escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med (2009) 206:2179–89. doi: 10.1084/jem.20090741
14. Sivignon A, Yan X, Alvarez Dorta D, Bonnet R, Bouckaert J, Fleury E, et al. Development of heptylmannoside-based glycoconjugate antiadhesive compounds against adherent-invasive escherichia coli bacteria associated with crohn's disease. mBio (2015) 6:e01298–01215. doi: 10.1128/mBio.01298-15
15. Sauter SL, Rutherfurd SM, Wagener C, Shively JE, Hefta SA. Binding of nonspecific cross-reacting antigen, a granulocyte membrane glycoprotein, to escherichia coli expressing type 1 fimbriae. Infect Immun (1991) 59:2485–93. doi: 10.1128/iai.59.7.2485-2493.1991
16. Sheikh A, Tumala B, Vickers TJ, Alvarado D, Ciorba MA, Bhuiyan TR, et al. CEACAMs serve as toxin-stimulated receptors for enterotoxigenic escherichia coli. Proc Natl Acad Sci U.S.A. (2020) 117:29055–62. doi: 10.1073/pnas.2012480117
17. Muenzner P, Kengmo Tchoupa A, Klauser B, Brunner T, Putze J, Dobrindt U, et al. Uropathogenic e. coli exploit CEA to promote colonization of the urogenital tract mucosa. PloS Pathog (2016) 12:e1005608. doi: 10.1371/journal.ppat.1005608
18. Piatek R, Zalewska B, Bury K, Kur J. The chaperone-usher pathway of bacterial adhesin biogenesis – from molecular mechanism to strategies of anti-bacterial prevention and modern vaccine design. Acta Biochim Pol (2005) 52:639–46. doi: 10.18388/abp.2005_3424
19. Piatek R, Zalewska B, Kolaj O, Ferens M, Nowicki B, Kur J. Molecular aspects of biogenesis of escherichia coli Dr fimbriae: characterization of DraB-DraE complexes. Infect Immun (2005) 73:135–45. doi: 10.1128/IAI.73.1.135-145.2005
20. Servin AL. Pathogenesis of Afa/Dr diffusely adhering escherichia coli. Clin Microbiol Rev (2005) 18:264–92. doi: 10.1128/CMR.18.2.264-292.2005
21. Guignot J, Hudault S, Kansau I, Chau I, Servin AL. Human decay-accelerating factor and CEACAM receptor-mediated internalization and intracellular lifestyle of Afa/Dr diffusely adhering escherichia coli in epithelial cells. Infect Immun (2009) 77:517–31. doi: 10.1128/IAI.00695-08
22. Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol (2009) 7:765–74. doi: 10.1038/nrmicro2220
23. Leusch HG, Hefta SA, Drzeniek Z, Hummel K, Markos-Pusztai Z, Wagener C. Escherichia coli of human origin binds to carcinoembryonic antigen (CEA) and non-specific crossreacting antigen (NCA). FEBS Lett (1990) 261:405–9. doi: 10.1016/0014-5793(90)80603-G
24. Leusch HG, Drzeniek Z, Hefta SA, Markos-Pusztai Z, Wagener C. The putative role of members of the CEA-gene family (CEA, NCA an BGP) as ligands for the bacterial colonization of different human epithelial tissues. Zentralbl Bakteriol (1991) 275:118–22. doi: 10.1016/S0934-8840(11)80775-9
25. Leusch HG, Drzeniek Z, Markos-Pusztai Z, Wagener C. Binding of escherichia coli and salmonella strains to members of the carcinoembryonic antigen family: differential binding inhibition by aromatic alpha-glycosides of mannose. Infect Immun (1991) 59:2051–7. doi: 10.1128/iai.59.6.2051-2057.1991
26. Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an escherichia coli strain isolated from the ileal mucosa of a patient with crohn's disease. Infect Immun (1999) 67:4499–509. doi: 10.1128/IAI.67.9.4499-4509.1999
27. Desilets M, Deng X, Rao C, Ensminger AW, Krause DO, Sherman PM, et al. Genome-based definition of an inflammatory bowel disease-associated adherent-invasive escherichia coli pathovar. Inflammation Bowel Dis (2016) 22:1–12. doi: 10.1097/MIB.0000000000000574
28. Boudeau J, Barnich N, Darfeuille-Michaud A. Type 1 pili-mediated adherence of escherichia coli strain LF82 isolated from crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol Microbiol (2001) 39:1272–84. doi: 10.1111/j.1365-2958.2001.02315.x
29. Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive e. coli, supporting ileal mucosa colonization in crohn disease. J Clin Invest (2007) 117:1566–74. doi: 10.1172/JCI30504
30. Barnich N, Darfeuille-Michaud A. Abnormal CEACAM6 expression in crohn disease patients favors gut colonization and inflammation by adherent-invasive e. coli. Virulence (2010) 1:281–2. doi: 10.4161/viru.1.4.11510
31. Saiz-Gonzalo G, Hanrahan N, Rossini V, Singh R, Ahern M, Kelleher M, et al. Regulation of CEACAM family members by IBD-associated triggers in intestinal epithelial cells, their correlation to inflammation and relevance to IBD pathogenesis. Front Immunol (2021) 12:655960. doi: 10.3389/fimmu.2021.655960
32. Dumych T, Yamakawa N, Sivignon A, Garenaux E, Robakiewicz S, Coddeville B, et al. Oligomannose-rich membranes of dying intestinal epithelial cells promote host colonization by adherent-invasive e. coli. Front Microbiol (2018) 9:742. doi: 10.3389/fmicb.2018.00742
33. Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, Petersen AM. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin Microbiol Rev (2019) 32:e00060–18. doi: 10.1128/CMR.00060-18
34. Marchelletta RR, Krishnan M, Spalinger MR, Placone TW, Alvarez R, Sayoc-Becerra A, et al. T Cell protein tyrosine phosphatase protects intestinal barrier function by restricting epithelial tight junction remodeling. J Clin Invest (2021) 131:e138230. doi: 10.1172/JCI138230
35. Shawki A, Ramirez R, Spalinger MR, Ruegger PM, Sayoc-Becerra A, Santos AN, et al. The autoimmune susceptibility gene, PTPN2, restricts expansion of a novel mouse adherent-invasive e. coli. Gut Microbes (2020) 11:1547–66. doi: 10.1080/19490976.2020.1775538
36. Spalinger MR, Shawki A, Chatterjee P, Canale V, Santos A, Sayoc-Becerra A, et al. Autoimmune susceptibility gene PTPN2 is required for clearance of adherent-invasive escherichia coli by integrating bacterial uptake and lysosomal defence. Gut (2022) 71:89–99. doi: 10.1136/gutjnl-2020-323636
37. Bilge SS, Clausen CR, Lau W, Moseley SL. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated escherichia coli to HEp-2 cells. J Bacteriol (1989) 171:4281–9. doi: 10.1128/jb.171.8.4281-4289.1989
38. Giron JA, Jones T, Millan-Velasco F, Castro-Munoz E, Zarate L, Fry J, et al. Diffuse-adhering escherichia coli (DAEC) as a putative cause of diarrhea in Mayan children in Mexico. J Infect Dis (1991) 163:507–13. doi: 10.1093/infdis/163.3.507
39. Poitrineau P, Forestier C, Meyer M, Jallat C, Rich C, Malpuech G, et al. Retrospective case-control study of diffusely adhering escherichia coli and clinical features in children with diarrhea. J Clin Microbiol (1995) 33:1961–2. doi: 10.1128/jcm.33.7.1961-1962.1995
40. Labigne-Roussel A, Schmidt MA, Walz W, Falkow S. Genetic organization of the afimbrial adhesin operon and nucleotide sequence from a uropathogenic escherichia coli gene encoding an afimbrial adhesin. J Bacteriol (1985) 162:1285–92. doi: 10.1128/jb.162.3.1285-1292.1985
41. Nowicki B, Martens M, Hart A, Nowicki S. Gestational age-dependent distribution of escherichia coli fimbriae in pregnant patients with pyelonephritis. Ann N Y Acad Sci (1994) 730:290–1. doi: 10.1111/j.1749-6632.1994.tb44268.x
42. Nowicki B, Labigne A, Moseley S, Hull R, Hull S, Moulds J, et al. The Dr hemagglutinin, afimbrial adhesins And F1845 fimbriae of uropathogenic and diarrhea-associated escherichia coli belong to a family of hemagglutinins with Dr receptor recognition. Infect Immun (1990) 58:279–81. doi: 10.1128/iai.58.1.279-281.1990
43. Le Bouguenec C, Garcia MI, Ouin V, Desperrier JM, Gounon P, Labigne A. Characterization of plasmid-borne afa-3 gene clusters encoding afimbrial adhesins expressed by escherichia coli strains associated with intestinal or urinary tract infections. Infect Immun (1993) 61:5106–14. doi: 10.1128/iai.61.12.5106-5114.1993
44. Meza-Segura M, Zaidi MB, Vera-Ponce De Leon A, Moran-Garcia N, Martinez-Romero E, Nataro JP, et al. New insights into DAEC and EAEC pathogenesis and phylogeny. Front Cell Infect Microbiol (2020) 10:572951. doi: 10.3389/fcimb.2020.572951
45. Burke DA, Axon AT. Ulcerative colitis and escherichia coli with adhesive properties. J Clin Pathol (1987) 40:782–6. doi: 10.1136/jcp.40.7.782
46. Guignot J, Peiffer I, Bernet-Camard MF, Lublin DM, Carnoy C, Moseley SL, et al. Recruitment of CD55 and CD66e brush border-associated glycosylphosphatidylinositol-anchored proteins by members of the Afa/Dr diffusely adhering family of escherichia coli that infect the human polarized intestinal caco-2/TC7 cells. Infect Immun (2000) 68:3554–63. doi: 10.1128/IAI.68.6.3554-3563.2000
47. Korotkova N, Yarova-Yarovaya Y, Tchesnokova V, Yazvenko N, Carl MA, Stapleton AE, et al. Escherichia coli DraE adhesin-associated bacterial internalization by epithelial cells is promoted independently by decay-accelerating factor and carcinoembryonic antigen-related cell adhesion molecule binding and does not require the DraD invasin. Infect Immun (2008) 76:3869–80. doi: 10.1128/IAI.00427-08
48. Alvarez-Fraga L, Phan MD, Goh KGK, Nhu NTK, Hancock SJ, Allsopp LP, et al. Differential Afa/Dr fimbriae expression in the multidrug-resistant escherichia coli ST131 clone. mBio (2022) 13:e0351921. doi: 10.1128/mbio.03519-21
49. Muenzner P, Hauck CR. Neisseria gonorrhoeae blocks epithelial exfoliation by nitric-Oxide-Mediated metabolic cross talk to promote colonization in mice. Cell Host Microbe (2020) 27793-808:e795. doi: 10.1016/j.chom.2020.03.010
50. Muenzner P, Rohde M, Kneitz S, Hauck CR. CEACAM engagement by human pathogens enhances cell adhesion and counteracts bacteria-induced detachment of epithelial cells. J Cell Biol (2005) 170:825–36. doi: 10.1083/jcb.200412151
51. Steffen R, Hill DR, Dupont HL. Traveler's diarrhea: a clinical review. JAMA (2015) 313:71–80. doi: 10.1001/jama.2014.17006
52. Giddings SL, Stevens AM, Leung DT. Traveler's diarrhea. Med Clin North Am (2016) 100:317–30. doi: 10.1016/j.mcna.2015.08.017
53. Black RE, Brown KH, Becker S. Effects of diarrhea associated with specific enteropathogens on the growth of children in rural Bangladesh. Pediatrics (1984) 73:799–805. doi: 10.1542/peds.73.6.799
54. Qadri F, Saha A, Ahmed T, Al Tarique A, Begum YA, Svennerholm AM. Disease burden due to enterotoxigenic escherichia coli in the first 2 years of life in an urban community in Bangladesh. Infect Immun (2007) 75:3961–8. doi: 10.1128/IAI.00459-07
55. Kotloff KL, Nasrin D, Blackwelder WC, Wu Y, Farag T, Panchalingham S, et al. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the global enteric multicenter study (GEMS). Lancet Glob Health (2019) 7:e568–84. doi: 10.1016/S2214-109X(19)30076-2
56. Bagamian KH, Anderson JDT, Muhib F, Cumming O, Laytner LA, Wierzba TF, et al. Heterogeneity in enterotoxigenic escherichia coli and shigella infections in children under 5 years of age from 11 African countries: a subnational approach quantifying risk, mortality, morbidity, and stunting. Lancet Glob Health (2020) 8:e101–12. doi: 10.1016/S2214-109X(19)30456-5
57. Nasrin D, Blackwelder WC, Sommerfelt H, Wu Y, Farag TH, Panchalingam S, et al. Pathogens associated with linear growth faltering in children with diarrhea and impact of antibiotic treatment: The global enteric multicenter study. J Infect Dis (2021) 224(12 Suppl 2):S848–55. doi: 10.1093/infdis/jiab434
58. Fleckenstein JM, Sheikh A. Emerging themes in the molecular pathogenesis of enterotoxigenic escherichia coli. J Infect Dis (2021) 224(12 Suppl 2):S813–20. doi: 10.1093/infdis/jiab359
59. Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U.S.A. (2005) 102:4459–64. doi: 10.1073/pnas.0501076102
60. Hagiwara M, Brindle P, Harootunian A, Armstrong R, Rivier J, Vale W, et al. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase a. Mol Cell Biol (1993) 13:4852–9. doi: 10.1128/mcb.13.8.4852-4859.1993
61. Parker D, Ferreri K, Nakajima T, Lamorte VJ, Evans R, Koerber SC, et al. Phosphorylation of CREB at ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol (1996) 16:694–703. doi: 10.1128/MCB.16.2.694
62. Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol (2001) 2:599–609. doi: 10.1038/35085068
63. Sheikh A, Tumala B, Vickers TJ, Martin JC, Rosa BA, Sabui S, et al. Enterotoxigenic escherichia coli heat-labile toxin drives enteropathic changes in small intestinal epithelia. Nat Commun (2022) 13:6886. doi: 10.1038/s41467-022-34687-7
64. Freed DL, Taylor G. Carcinoembryonic antigen in faeces. Br Med J (1972) 1:85–7. doi: 10.1136/bmj.1.5792.85
65. Kuroki M, Koga Y, Matsuoka Y. Purification and characterization of carcinoembryonic antigen-related antigens in normal adult feces. Cancer Res (1981) 41:713–20.
66. Matsuoka Y, Kuroki M, Koga Y, Kuriyama H, Mori T, Kosaki G. Immunochemical differences among carcinoembryonic antigen in tumor tissues and related antigens in meconium and adult feces. Cancer Res (1982) 42:2012–8.
67. Matsuoka Y, Matsuo Y, Sugano K, Ohkura H, Kuroki M, Kuroki M. Characterization of carcinoembryonic antigen-related antigens in normal adult feces. Jpn J Cancer Res (1990) 81:514–9. doi: 10.1111/j.1349-7006.1990.tb02600.x
68. Kinugasa T, Kuroki M, Yamanaka T, Matsuo Y, Oikawa S, Nakazato H, et al. Non-proteolytic release of carcinoembryonic antigen from normal human colonic epithelial cells cultured in collagen gel. Int J Cancer (1994) 58:102–7. doi: 10.1002/ijc.2910580117
69. Fantini J, Rognoni JB, Culouscou JM, Pommier G, Marvaldi J, Tirard A. Induction of polarized apical expression and vectorial release of carcinoembryonic antigen (CEA) during the process of differentiation of HT29-D4 cells. J Cell Physiol (1989) 141:126–34. doi: 10.1002/jcp.1041410119
70. Muturi HT, Dreesen JD, Nilewski E, Jastrow H, Giebel B, Ergun S, et al. Tumor and endothelial cell-derived microvesicles carry distinct CEACAMs and influence T-cell behavior. PloS One (2013) 8:e74654. doi: 10.1371/journal.pone.0074654
71. Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol (1999) 9:67–81. doi: 10.1006/scbi.1998.0119
72. Hammarstrom S, Baranov V. Is there a role for CEA in innate immunity in the colon? Trends Microbiol (2001) 9:119–25. doi: 10.1016/S0966-842X(01)01952-7
73. Shifrin DA Jr., Demory Beckler M, Coffey RJ, Tyska MJ. Extracellular vesicles: communication, coercion, and conditioning. Mol Biol Cell (2013) 24:1253–9. doi: 10.1091/mbc.e12-08-0572
74. Mcconnell RE, Higginbotham JN, Shifrin DA Jr., Tabb DL, Coffey RJ, Tyska MJ. The enterocyte microvillus is a vesicle-generating organelle. J Cell Biol (2009) 185:1285–98. doi: 10.1083/jcb.200902147
75. Shifrin DA Jr., Mcconnell RE, Nambiar R, Higginbotham JN, Coffey RJ, Tyska MJ. Enterocyte microvillus-derived vesicles detoxify bacterial products and regulate epithelial-microbial interactions. Curr Biol (2012) 22:627–31. doi: 10.1016/j.cub.2012.02.022
76. Baranov V, Hammarstrom S. Carcinoembryonic antigen (CEA) and CEA-related cell adhesion molecule 1 (CEACAM1), apically expressed on human colonic m cells, are potential receptors for microbial adhesion. Histochem Cell Biol (2004) 121:83–9. doi: 10.1007/s00418-003-0613-5
77. Frangsmyr L, Baranov V, Hammarstrom S. Four carcinoembryonic antigen subfamily members, CEA, NCA, BGP and CGM2, selectively expressed in the normal human colonic epithelium, are integral components of the fuzzy coat. Tumour Biol (1999) 20:277–92. doi: 10.1159/000030075
78. Karlsson M, Zhang C, Mear L, Zhong W, Digre A, Katona B, et al. A single-cell type transcriptomics map of human tissues. Sci Adv (2021) 7:eabh2169. doi: 10.1126/sciadv.abh2169
79. Fahlgren A, Baranov V, Frangsmyr L, Zoubir F, Hammarstrom ML, Hammarstrom S. Interferon-gamma tempers the expression of carcinoembryonic antigen family molecules in human colon cells: a possible role in innate mucosal defence. Scand J Immunol (2003) 58:628–41. doi: 10.1111/j.1365-3083.2003.01342.x
Keywords: Escherichia coli, ETEC (enterotoxigenic Escherichia coli), CEACAM, Afa/dr family adhesins, type 1 fimbriae
Citation: Sheikh A and Fleckenstein JM (2023) Interactions of pathogenic Escherichia coli with CEACAMs. Front. Immunol. 14:1120331. doi: 10.3389/fimmu.2023.1120331
Received: 09 December 2022; Accepted: 30 January 2023;
Published: 14 February 2023.
Edited by:
Vu Ngo, Beckman Research Institute, City of Hope, United StatesReviewed by:
Cecilia Ambrosi, Università telematica San Raffaele, ItalyCopyright © 2023 Sheikh and Fleckenstein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James M. Fleckenstein, amZsZWNrZW5zdGVpbkB3dXN0bC5lZHU=
 Alaullah Sheikh
Alaullah Sheikh James M. Fleckenstein
James M. Fleckenstein