- 1Key Laboratory of Mariculture & Stock Enhancement in North China’s Sea, Ministry of Agriculture and Rural Affairs, Dalian Ocean University, Dalian, China
- 2College of Life Science, Liaoning Normal University, Dalian, China
Nuclear factor-kappa B (NF-κB) pathways have a close relationship with many diseases, especially in terms of the regulation of inflammation and the immune response. Non-coding RNAs (ncRNAs) are a heterogeneous subset of endogenous RNAs that directly affect cellular function in the absence of proteins or peptide products; these include microRNAs (miRNAs), long noncoding RNAs (lncRNAs), circular RNAs (circRNAs), etc. Studies on the roles of ncRNAs in targeting the NF-κB pathways in aquatic animals are scarce. A few research studies have confirmed detailed regulatory mechanisms among ncRNAs and the NF-κB pathways in aquatic animals. This comprehensive review is presented concerning ncRNAs targeting the NF-κB pathway in aquatic animals and provides new insights into NF-κB pathways regulatory mechanisms of aquatic animals. The review discusses new possibilities for developing non-coding-RNA-based antiviral applications in fisheries.
1 Introduction
The nuclear factor-kappa B (NF-κB) pathways are well-known prototypical signaling pathways associated with inflammation, immune response, physiological stress, and disease occurrence in metazoans (1). According to differences in components and activation mechanisms, the NF-κB pathways can be further classified into canonical and non-canonical pathways (2). The canonical NF-κB pathway is currently recognized as a rapid and transient pathway closely related to pathogenesis of inflammatory diseases (1). This pathway includes NFκB1, p65 (also known as RelA), c-Rel, and activation of the signal-induced phosphorylation of IκB molecules by IκB kinases (IKKs) (3, 4). IKKs consist of two homologous catalytic subunits, IKKα and IKKβ (also known as IKK1 and IKK2), as well as a regulatory subunit IKKγ (also known as NF-κB essential modulator, NEMO) lacking catalytic capability. It has been demonstrated that IKKβ is essential for regulating canonical NF-κB activation with the support of the IKKγ subunit rather than the IKKα (4–6). In contrast to the canonical NF-κB pathway, the non-canonical NF-κB pathway is characterized as a slow and persistent pathway specifically associated with immune response and inflammatory diseases. The non-canonical NF-κB pathway includes NF-κB inducing kinase (NIK, also known as MAP3K14), IKKα, and RelB/p52 heterodimers (acting as transcription factors). The central event of non-canonical NF-κB pathway activation is NIK-IKKα axis-induced activation of RelB/p52 heterodimers.
Starting from the exploration of XrelA in developing embryos of the African clawed frog (Xenopus laevis) in 1994 (7), over the past 28 years both the canonical and the non-canonical NF-κB pathways have been extensively studied in various animals (7–11). In terms of aquatic animals, the immune regulatory functions of NF-κB pathways have been studied in amphibians, crustacean species, and teleost fishes, including in species such as the African clawed frog (7), Pacific white shrimp (Litopenaeus vannamei) (8), zebrafish (Danio rerio) (9, 10), and orange-spotted grouper (Epinephelus coioides) (11). However, most of the studies on NF-κB pathways in aquatic animals have focused on how the pathways regulate host immunity, while there is a lack of systematic summaries and analyses of the factors regulating the NF-κB pathways.
Non-coding RNAs (ncRNAs), a class of endogenous RNAs that are transcribed from DNA and affect cellular function through themselves rather than proteins or peptide products (12). ncRNAs can be classified into two main categories of small non-coding RNAs and long non-coding RNAs (lncRNAs) depending on their lengths. Small non-coding RNAs include small nuclear RNAs (snRNAs), microRNAs (miRNAs) and piwi-interacting RNAs (piRNAs), etc.; lncRNAs mainly include linear lncRNAs and circular RNAs (circRNAs) (12, 13). miRNAs are typically about 20–25 nucleotides (nt) in length and are generated from primary miRNAs (pri-miRNAs) (14). As regulatory molecules, miRNAs achieve their post-transcriptional regulation function through silencing or suppressing complexes leading to cleavage or translational downregulation of their target genes (14). lncRNAs are defined as transcripts with length greater than 200 nt and without evident protein coding function transcribed by RNA polymerase I, II or III (15). circular RNAs, as their name implies, have a circular structure which imparts additional stability to them against exonuclease cleavage. circRNAs can serve as transcriptional regulators, miRNA sponges, and as protein templates, decoys, scaffolds, and recruiters (13, 15). In recent years, increasing evidence has demonstrated that ncRNAs can regulate multiple physiological and pathological processes in aquatic animals such as teleost fishes (16, 17).
In this review, we focus on research advances concerning ncRNAs (mainly on miRNAs, linear lncRNAs and circRNAs) targeting the NF-κB pathways in aquatic animals such as teleost fishes and echinoderms (Table 1). Moreover, we will discuss the regulatory mechanisms of these ncRNAs involved in the NF-κB pathways and their potential as biomarkers or targets for early warning, control, and therapy of pathogen-induced inflammatory diseases in several aquatic animals.
2 miRNAs targeting NF-κB pathways
2.1 miRNAs targeting the canonical NF-κB pathways
The canonical NF-κB pathway can be activated rapidly in both innate and adaptive immune cells by numerous signals through various receptors, including pattern-recognition receptors (PRRs), T-cell receptors (TCRs), B-cell receptors (BCRs), and proinflammatory cytokine receptors (18–20). In addition, the activation of the canonical NF-κB pathway also requires the involvement of specific adaptor molecules, ubiquitin ligases, and protein kinases (21, 22). In aquatic animals, the role of the PRR family in the canonical NF-κB pathway has been extensively investigated.
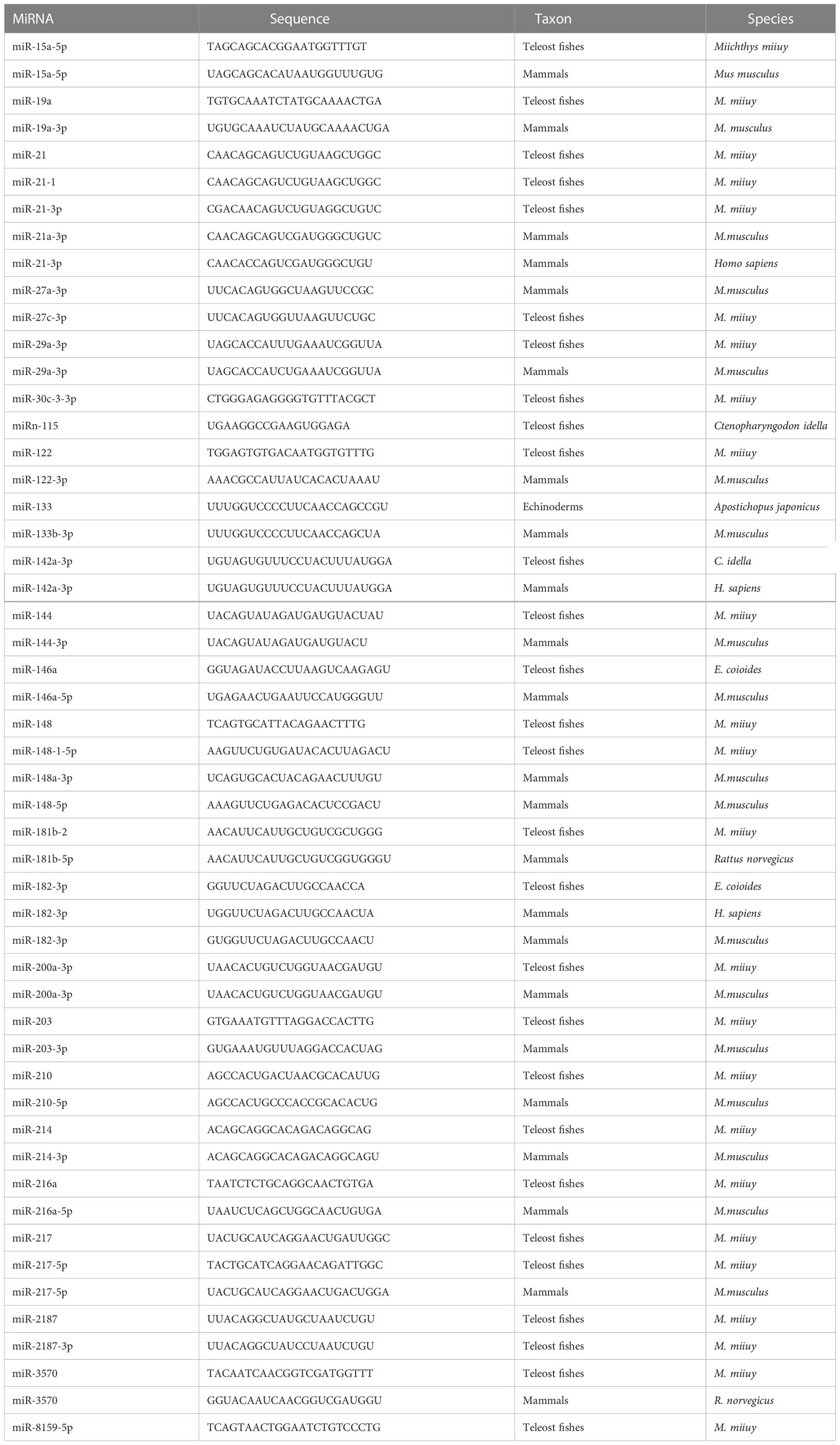
It is accepted that most aquatic animals primarily rely on their innate immunity modulated by PRRs involved in signaling pathways due to their poorly developed adaptive immune systems (23, 24). In aquatic animals, the toll-like receptor (TLR), NOD-like receptor (NLR), and RIG-I-like receptor (RLR) signaling pathways are the most well characterized (23–28) (Figure 1). Moreover, key members of those signaling pathways play a variety of roles in organism growth and development (Table 2). Therefore, we will discuss miRNAs targeting TLR, NLR, and RLR signaling pathways in this section in detail.
Figure 1 Schematic image of microRNA-mRNA interactions involved in nuclear factor-kappa B (NF-κB) pathways in aquatic animals.
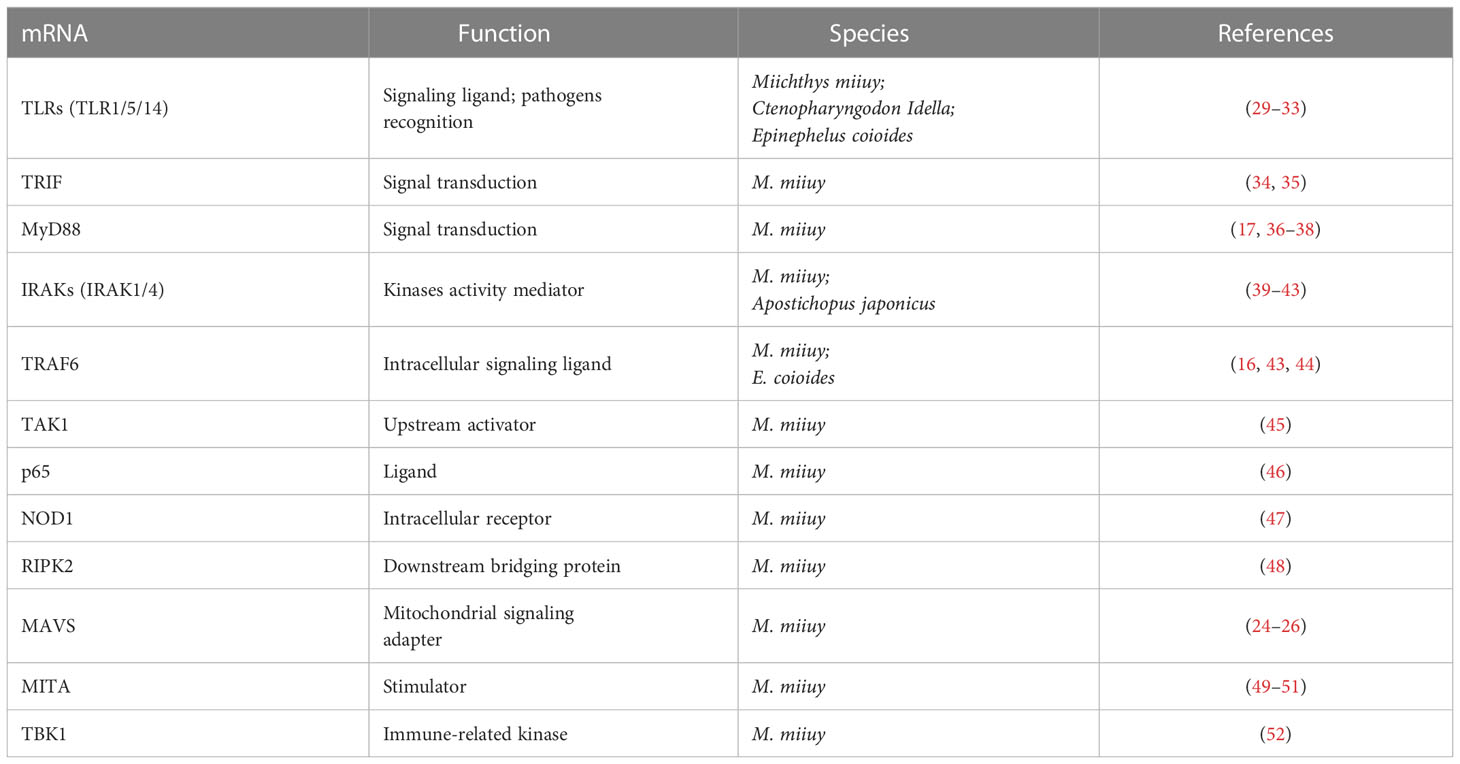
Table 2 Summary of identified mRNAs associated with the nuclear factor-kappa B (NF-κB) pathway in aquatic animals.
2.1.1 miRNAs targeting the TLR signaling pathways
The TLR pathways play a key role in the activation of innate immunity in eukaryotes by recognizing specific components of pathogens to detect pathogen invasion, which is known as a pathogen associated molecular pattern (PAMP) (53). The TLR pathways contain two axes, the “TLRs/TRIF-related adaptor molecule (TRAM)/Toll-IL-1 receptor domain-containing adaptor inducing interferon beta (TRIF)/TNFR-associated factor 6 (TRAF6)” axis and the “TLRs/Toll-IL-1 receptor adaptor protein (TIRAP)/Myeloid differentiation primary response gene 88 (MyD88)/IL-1 receptor-associated kinases (IRAKs, including IRAK1 and IRAK4)/TRAF6” axis. In aquatic animals, several key members of the TLR pathways have been identified, including cytoplasmic TLRs (TLR1, TLR5, and TLR14), TRIF, TRAF6, MyD88, IRAKs (IRAK1 and IRAK4), transforming growth factor beta-activated kinase 1 (TAK1), and p65 (Figure 1).
2.1.1.1 miRNAs targeting TLRs
TLRs are important signaling recruiters, and they can be classified into cell membrane surface receptors (e.g., TLR1, TLR2, TLR4, TLR5, and TLR6) and intracellular receptors (e.g., TLR3, TLR7, TLR8, and TLR9) depending on their location in the cell (29, 53). In aquatic animals, the identified TLRs include TLR1, TLR5, and TLR14 in fish (30–33, 54), TLR1, TLR2, and TLR4 in the Pacific oyster (Crassostrea gigas) (55), TLR3 in sea cucumbers (Apostichopus japonicus) (56), TLR2 in the green mud crab (Scylla paramamosain), and TLR6, TLR7, TLR8, and TLR9 in Pacific white shrimp. In recent years, studies on miRNAs targeting TLRs have been conducted in fish.
miR-200a-3p, miR-8159-5p, and miR-217-5p are three known miRNAs with lengths of 22 nt, 23 nt, and 23 nt, respectively, and seed regions of “-AACACUG-” (miR-200a-3p), “-CAGTAAC-” (miR-8159-5p), and “-CTGCATC-” (miR-217-5p), respectively. Several studies have reported that miR-200a-3p, miR-8159-5p, and miR-217-5p can bind to the 3’ untranslated region (UTR) of the TLR1 gene and exert negative regulation of post-transcriptional attenuation in the miiuy croaker (Miichthys miiuy) (Figure 2A). Pathogenic bacterial infection experiments indicated that the relative expression trends of miR-200a-3p, miR-8159-5p, and miR-217-5p were opposite to that of TLR1 in the spleen and leukocytes of miiuy croaker after infection with Vibrio anguillarum or stimulation with lipopolysaccharide (LPS), suggesting that miR-200a-3p, miR-8159-5p, and miR-217-5p may be involved in the immune response against pathogenic bacteria in fish by regulating the canonical NF-κB pathway via targeting TLR1 (30, 31).
Figure 2 Predicted microRNA (miRNA) binding sites in the 3’ untranslated regions (UTRs) of different genes of nuclear factor-kappa B (NF-κB) pathways in aquatic animals. (A) Predicted binding sites in the 3’ UTR of TLR1. (B) Predicted binding sites in the 3’ UTR of TLR5. (C) Predicted binding sites in the 3’ UTR of TLR5M (the membrane form of TLR5). (D) Predicted binding sites in the 3’ UTR of TLR14. (E) Predicted binding sites in the 3’ UTR of TRIF. (F) Predicted binding sites in the 3’ UTR of MyD88. (G) Predicted binding sites in the 3’ UTR of IRAK1. (H) Predicted binding sites in the 3’ UTR of IRAK4. (I) Predicted binding sites in the 3’ UTR of TRAF6. (J) Predicted binding sites in the 3’ UTR of TAK1. (K) Predicted binding sites in the 3’ UTR of p65. (L) Predicted binding sites in the 3’ UTR of NOD1. (M) Predicted binding sites in the 3’ UTR of RIPK2. (N) Predicted binding sites in the 3’ UTR of MAVS. (O) Predicted binding sites in the 3’ UTR of MITA. (P) Predicted binding sites in the 3’ UTR of TBK1. The red markings represent seed regions.
miR-142a-3p (32), miRn-115 (32), and miR-182-3p (33) have been identified as miRNAs targeting TLR5 homologous genes in aquatic animals. miR-142a-3p, miRn-115, and miR-182-3p are 23 nt, 18 nt and 20 nt in length, respectively, with seed regions of “-GUAGUGU-” (miR-142a-3p), “-GAAGGCC-” (miRn-115), and “-GUUCUAG-” (miR-182-3p), respectively (Figure 2B). Dual luciferase reporter gene assays showed that both miR-142a-3p and miRn-115 could bind to the 3’ UTR of grass carp (Ctenopharyngodon Idella) TLR5 and that they have a negative regulatory relationship (Figure 2B). Overexpression of miR-142a-3p or miRn-115 resulted in a significant decrease in the relative expression of the TLR5 gene in the kidneys of grass carp infected with Aeromonas hydrophila, and this in turn suppressed the expression of downstream genes (e.g., interleukin-1β (IL-1β), interleukin-8 (IL-8), and tumor necrosis factor-α (TNF-α)) to avoid an excessive inflammatory response, suggesting that miR-142a-3p or miRn-115 could indirectly suppress the inflammatory response mediated by the canonical NF-κB pathway via negatively regulating the expression of TLR5 homologous genes. miR-182-3p is another widely studied immunomodulatory-related miRNA in fish that has a length of 20 nt and a seed region of “-GUUCUAG-” (Figure 2C). As has been reported, miR-182-3p effectively inhibited the relative expression of TLR5M (the membrane form of TLR5) in the spleen cells of E. coioides during the immune response induced by flagellin of Staphylococcus parapsilosis (57) and consequently exerted organismal antibacterial immunity via inhibiting the activation of the TLR5M/MAPK/NF-κB pathway (32, 33).
miR-122 is a 22 nt miRNA that has a seed region of “-GGAGTG-”. miR-122 is involved in cell cycle regulation, cell differentiation, cell proliferation, and apoptosis processes in mammals (58–60). In aquatic animals, miR-122 is considered as a biomarker that contributes to the non-invasive diagnosis of liver injury in fish (61). TLR14 is a novel TLRs member that has been specifically identified in fish, including Japanese pufferfish (Takifugu rubripes) (GenBank accession No. AC156431.1), mandarin fish (Siniperca chuatsi) (GenBank accession No. MT594450.1), and miiuy croaker (M. miiuy) (GenBank accession No. KR709254.1) (54). A dual luciferase reporter gene assay indicated the presence of the seed region of miR-122 in the 3’ UTR of the TLR14 gene (which belongs to the TLR2 subfamily) in miiuy croaker (Figure 2D). It was reported that the relative expression levels of miR-122 and TLR14 exhibited opposite trends in the spleen and macrophages of miiuy croaker after V. anguillarum infection and LPS stimulation, suggesting that miR-122 may affect the TLR14/NF-κB signaling cascade response by negatively regulating the relative expression of TLR14 (54).
2.1.1.2 miRNAs targeting TRIF
TRIF, also known as TICAM-1 (62), is involved in immune response and disease regulation. Currently, TRIF homologs have been identified in a variety of aquatic organisms, including the Northeast Chinese lamprey (Lethenteron morii) (63), channel catfish (Ictalurus punctatus) (64), zebrafish (65), grass carp (66), orange-spotted grouper (67), and large yellow croaker (Larimichthys crocea) (34).
miRNAs targeting TRIF homologous genes in aquatic animals include miR-181b-2 (35), miR-21-1 (35), and miR-15a-5p (68) (Figure 2E). miR-181b-2, miR-21-1, and miR-15a-5p are 22 nt, 22 nt, and 21 nt in length, respectively, with highly similar seed regions of “-ACAUUCA-” (miR-181b-2), “-AACAGCA-” (miR-21-1), and “-AGCAGCA-” (miR-15a-5p), respectively. Dual luciferase reporter assays demonstrated that the seed regions of miR-181b-2, miR-21-1, and miR-15a-5p were all present in the 3’ UTR of TRIF in miiuy croaker (Figure 2E). The results of pathogenic infection experiments showed that the relative expression levels of miR-181b-2, miR-21-1, and miR-15a-5p in the macrophages and brain cells of miiuy croaker challenged by LPS stimulation and V. anguillarum or Siniperca chuatsi rhabdovirus (SCRV) infection showed opposite trends to those of TRIF (35). These observations suggested that miR-181b-2, miR-21-1, and miR-15a-5p could negatively regulate the expression of TRIF homologous genes and in turn regulate TRIF/NF-κB, ultimately inhibiting antibacterial and antiviral immune responses in fish (35).
2.1.1.3 miRNAs targeting MyD88
MyD88, a key node in the canonical NF-κB signaling pathway, is also an important target for several miRNAs (3). To date, MyD88 homologs have been identified in a variety of aquatic animals, including a marine gastropod (Littorina littorea) (69), a tropical sea cucumber (Holothuria leucospilota) (70), and a sea cucumber (A. japonicus) (36).
miR-148 (37), miR-214 (38), miR-19a (71), and miR-3570 (17) are known miRNAs that are 21 nt, 21 nt, 23 nt, and 22 nt in length, respectively, and contain seed regions “-CAGTGCA-” (miR-148), “-CAGCAGG-” (miR-214), “-GTGCAAA-” (miR-19a), and “-ACAATCA-” (miR-3570), respectively. These miRNAs have been validated to target and regulate the expression of MyD88 homologs in different aquatic animals (Figure 2F). For example, dual luciferase assays have confirmed that the seed regions of miR-148, miR-214, miR-19a, and miR-3570 were simultaneously present in the 3’ UTR of MyD88 in miiuy croaker (17, 37, 38, 71). Pathogenic bacterial infection experiments indicated that the relative expression trends of miR-148, miR-214, miR-19a, and miR-3570 were contrary to the relative expression levels of MyD88 in the spleen, macrophages, and kidney of miiuy croaker after Vibrio harveyi infection and LPS stimulation, suggesting that miR-148, miR-214, miR-19a, and miR-3570 can negatively regulate the expression of MyD88. These observations also indicated that miR-148, miR-214, miR-19a, and miR-3570 may be involved in pathogenic bacteria-induced immune responses in fish through inhibiting the canonical NF-κB pathway via targeting MyD88 homologs. Interestingly, it was reported that miR-19b (a member of the miR-19 family) with a seed region “-GUGCAAA-” significantly enhanced NF-κB activity in human HEK293 cells and mouse embryonic fibroblasts (72). Since there is no report concerning the role of miR-19b in aquatic animals, further study is necessary to clarify the regulation between miR-19b and the NF-κB pathways in aquatic animals.
2.1.1.4 miRNAs targeting IRAKs
IRAK1 and IRAK4 are the only two members of the IRAK family with kinase activity (73–75), and IRAK1 can mediate NF-κB signaling by being recruited by MyD88, in turn leading to the production of inflammatory factors such as IL-8 and TNF-α (76). IRAKs have been identified in many aquatic organisms; for example, IRAK1 homologs have been identified in red tailed shrimp (Fenneropenaeus penicillatus) (77) and Pacific white shrimp (78), and IRAK4 homologs have been identified in an abalone (Haliotis discus) (GenBank accession No. KU351646.1), a thick shell mussel (Mytilus coruscus) (79), and a sea cucumber (A. japonicus) (39). Dual luciferase reporter assay data showed that 3’ UTRs of both IRAK1 and IRAK4 in aquatic animals have multiple miRNA target binding sites (Figures 2G, H). miR-148-1-5p (40), miR-133 (41), miR-203 (42), miR-21 (43), and miR-27c-3p (80) are known miRNAs with lengths of 23 nt, 23 nt, 22 nt, 22 nt and 21 nt, respectively, and the seed regions “-AGUUCUG-” (miR-148-1-5p), “-UUGGUCC-” (miR-133), “-TGAAATG-” (miR-203), “-AACAGCA-” (miR-21), and “-UCAGACU-” (miR-27c-3p), respectively. miR-148-1-5p and miR-133 can regulate the expression of IRAK1, and miR-203, miR-21, and miR-27c-3p regulate the expression of IRAK4 (Figure 2H). Studies have shown that the expression levels of miR-148-1-5p in the brain cells of miiuy croaker and that of miR-133 in the coelomocytes of A. japonicus showed clear contradictory trends to those of IRAK1 homologous genes after pathogenic bacterial infection as well as to the expression trends of miRNAs (miR-203, miR-21, and miR-27c-3p) and IRAK4 in the liver, spleen, macrophages, kidney, and intestine cells of miiuy croaker. These observations indicated that miR-148-1-5p and miR-133 may act as negative regulators to inhibit the activation of the canonical NF-κB pathway by downregulating IRAK1 homologous gene expression and that miR-203, miR-21, and miR-27c-3p could ultimately inhibit the antibacterial and antiviral immune effects of aquatic organisms via downregulation of IRAK4 homologous gene expression.
2.1.1.5 miRNAs targeting TRAF6
TRAF6 is an important intracellular multifunctional signaling molecule, and it is one of the most widely studied members of the TRAF family (81, 82). It has been reported that TRAF6 activated the IKK complex, in turn leading to the activation of NF-κB and the expression of inflammatory cytokines (83). Based on current research, TRAF6 has been identified in a variety of aquatic organisms, including Pacific white shrimp (69), Zhikong scallop (Chlamys farreri) (84), and Hong Kong oyster (Crassostrea hongkongensis) (GenBank accession No. MK799968.1).
miR-2187 (16), miR-30c-3-3p (44), and miR-146a (85) are 21 nt, 22 nt, and 23 nt in length, respectively, with seed regions “-UACAGGC-” (miR-2187), “-TGGGAGA-” (miR-30c-3-3p), and “-GUAGAUA-” (miR-146a), respectively. Dual luciferase reporter assays showed that TRAF6 homologs in aquatic animals have binding sites (Figure 2I) on the 3’ UTR that exactly match the seed sequences of miR-2187, miR-30c-3-3p, and miR-146a. It has been reported that overexpression of miR-2187, miR-30c-3-3p, or miR-146a significantly suppressed the expression of TRAF6 in the liver and spleen cells of miiuy croaker infected with V. anguillarum and red-spotted grouper nervous necrosis virus (RGNNV), indicating a negative regulatory relationship between these miRNAs and TRAF6. In addition, overexpression of miR-2187, miR-30c-3-3p, or miR-146a also promoted the replication and occurrence of SCRV or RGNNV, suggesting that miR-2187, miR-30c-3-3p, or miR-146a could be induced and utilized by SCRV or RGNNV to be conducive to their infection and reproduction. Furthermore, it was also reported that miR-2187, miR-30c-3-3p, or miR-146a may possibly depress the expression of inflammatory factors (e.g., TNF-α, IL-8, or IL-1β) in an indirect way in the liver, kidney, and spleen cells of miiuy croaker or in the spleen of E. coioides after LPS stimulation or infection by RGNNV or SCRV (16, 44, 85). Combining all of the above observations, we conclude that miR-2187, miR-30c-3-3p, and miR-146a may enhance infection by SCRV and RGNNV through suppressing the NF-κB pathways and subsequently initiating inflammatory responses via targeting TRAF6 homologs in fish.
2.1.1.6 miRNAs targeting TAK1
TAK1 is a member of the MAPK kinase family, and it can be activated by TNF, LPS, and Epstein-Barr virus latent membrane protein 1 (LMP1) (86–89). Additionally, one of the most important roles of TAK1 is acting as an upstream activator of the NF-κB pathways (90). In aquatic animals, TAK1 homologs have been identified in crustacean species and echinoderms such as the Pacific white shrimp (GenBank accession No. KU522004.1), a mud crab (Scylla paramamamosain) (GenBank accession No. MK319934.1), and a sea urchin (Paracentrotus lividus) (45).
miR-217 is the only known miRNA to date that may interact with TAK1 homologs in aquatic animals. This mRNA is 23 nt in length and has a “-CUGCAU-” seed region. Zhang et al. identified a binding site in the 3’ UTR of miiuy croaker TAK1 that exactly matched the seed sequence of miR-217 by using dual luciferase validation (91) (Figure 2J). In the Chinese mitten crab (Eriocheir sinensis), overexpression of miR-217 affects the replication of white spot syndrome virus (WSSV) and plays an active role in WSSV infection (92). Further in vivo and in vitro pathogenic infection experiments showed that the expression trends of miR-217 and TAK1 were contrary in the spleen and macrophages of miiuy croaker, indicating that miR-217 may be involved in the antibacterial and antiviral immune responses of fish through suppressing the NF-κB pathways via targeting TAK1 (91).
2.1.1.7 miRNAs targeting p65
The p65 protein is located at the end of the canonical NF-κB pathway. The transcription of inflammatory cytokines is modulated by binding to p50 and forming a “p65/p50” dimer. The “p65/p50” dimer enters the nucleus and binds to the κB site in the promoter or enhancer of the target gene (93–97). In aquatic animals, p65 homologous genes have been identified in teleost fishes such as zebrafish (9), olive flounder (Paralichthys olivaceus) (46), and common carp (Cyprinus carpio) (GenBank accession No. MN167531.1).
Currently, miR-216a is the only identified miRNA that can bind to the 3’ UTR of p65 homologs in fish (98) (Figure 2K). miR-216a is 22 nt in length and has the seed region “-AATCTC-”. A study by Xu et al. (98) showed that the expression of miR-216a was significantly upregulated in the spleen and macrophages of miiuy croaker. Overexpression of miR-216a could also affect the expression of inflammatory cytokines such as TNF-α, IL-1β, interleukin-6 (IL-6), and IL-8. In vivo and in vitro experiments demonstrated that miR-216a could downregulate the activation of the NF-κB pathways by negatively regulating the expression of miiuy croaker p65 at the post-transcription level. All of the above-mentioned studies indicate that miR-216a could suppress excessive and prolonged inflammatory responses in the organism through negatively regulating the NF-κB pathways via directly targeting p65 homologous genes in fish (98).
2.1.2 miRNAs targeting the NLR signaling pathways
NLRs are important PRRs that sense bacterial products in the cytoplasm, and they play crucial roles in the recognition of bacterial or viral invasion in eukaryotic cells (47, 99). Key members of the NLR pathways include the NLR family members nucleotide oligomerization domain 1 and 2 (NOD1 and NOD2), receptor interacting serine/threonine kinase 2 (RIPK2), NEMO, IKKs (IKKα and IKKβ), and p50/p65. In aquatic animals, several members of the NLR pathway have been identified, including NOD1 (48, 100) and RIPK2 (101).
2.1.2.1 miRNAs targeting NOD1
NOD1 is one of the most representative members of the NLR family, and it serves as an important intracellular receptor that effectively detects pathogenic components produced by various gram-negative bacteria in mammals (102, 103). NOD1 is involved in the antibacterial or antiviral-induced immune response by activating the NF-κB pathways and subsequent inflammatory responses (104, 105). In addition, NOD1 can also act as a receptor to enhance the immune response during viral infection (106). Currently, NOD1 has been identified in several aquatic organisms, including teleost fishes such as rainbow trout (Oncorhynchus mykiss) (GenBank accession No. KF484402.1), zebrafish (GenBank accession No. KC207831.1), mandarin fish (S. chuatsi) (GenBank accession No. KY974318.1), olive flounder (GenBank accession No. JF830013.1), and miiuy croaker (104).
miR-144 and miR-217 are 22 nt and 23 nt in length, respectively, with seed regions “-ACAGUAU-” (miR-144) and “-ACUGCAU-” (miR-217), respectively. Dual luciferase reporter assays have shown that NOD1 homologs in aquatic animals have binding sites in the 3’ UTR that exactly match the seed sequences of miR-144 and miR-217 (Figure 2L). Overexpression of miR-144 and miR-217 significantly inhibited the expression of miiuy croaker NOD1 at the post-transcription level in the spleen and macrophages of miiuy croaker infected with V. harveyi and stimulated by LPS, respectively, indicating a negative regulatory relationship between the above-mentioned miRNAs and NOD1 (100). Taken together, the above-mentioned studies suggest critical roles for miR-144 and miR-217 that may involve inhibiting organismal and antibacterial immunity via negatively regulating the NF-κB pathways and subsequent inflammatory responses by suppressing NOD1 expression (100).
2.1.2.2 miRNAs targeting RIPK2
RIPK2 is a key factor involved in the pathogen-induced immune response (107–109). As the downstream bridging protein in the NLR signaling pathway, RIPK2 can influence cellular signaling and cytokine production induced by NOD1 (110). Currently, RIPK2 homologs have been identified in a number of aquatic animals, including cyclostome and teleost fishes such as the Reissner lamprey (Lethenteron reissneri) (111), goldfish (Carassius auratus) (GenBank accession No. KJ636470.1), and cyprinid fish (Schizothorax prenanti) (GenBank accession No. MW113673.1).
miR-210 and miR-3570 are two known miRNAs having regulatory relationships with RIPK2 homologs (101). miR-210 and miR-3570 are both 22 nt in length, with seed regions “-CCACUG-” (miR-210) and “-CUGUU-” (miR-3570), respectively. Dual luciferase reporter assays revealed that both miR-210 and miR-3570 could precisely bind to the 3’ UTR of the RIPK2 gene in miiuy croaker (101) (Figure 2M). Pathogenic bacterial infection experiments showed that in the spleen and macrophages of miiuy croaker infected by V. harveyi and subjected to LPS stimulation, the relative expression trends of miR-210 and miR-3570 were significantly in contrast to those of RIPK2. Considering all of the above results, we can conclude that miR-210 and miR-3570 may participate in the immune response against pathogenic bacterial or LPS stimulation in fish by negatively regulating the NF-κB pathways via targeting RIPK2 (101).
2.1.3 miRNAs targeting the RLR signaling pathways
RLRs play a key role in virus recognition and subsequent induction of antiviral immune responses (49, 112–114). Studies have demonstrated that RLRs can activate a signaling cascade that subsequently leads to the production of type-I interferons (IFNs) after efficient recognition of viral infection in the organism’s cytoplasm. The main members of the RLR pathway are retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), mitochondrial antiviral signaling protein (MAVS), mediator of interferon regulatory factor 3 activation (MITA), and TANK-binding kinase 1 (TBK1). Key members of the RLR pathway identified in aquatic animals include MAVS (25–27), MITA (50–52), and TBK1 (115).
2.1.3.1 miRNAs targeting MAVS
MAVS acts as a mitochondrial antiviral signaling adapter that bridges the gap between RIG-I and MDA5 sensing of viral infection and downstream signaling. Activation of MAVS leads to the rapid production of antiviral cytokines. MAVS homologs have been identified in teleost fishes such as zebrafish (116) and Atlantic salmon (Salmo salar) (117).
miR-3570 (25), miR-122 (26), and miR-21-3p (27) have lengths of 22 nt, 22 nt, and 22 nt, respectively, with seed regions “-ACAATCA-” (miR-3570), “-GGAGUG- “ (miR-122), and “-GACAACA-” (miR-21-3p) (Figure 2N). Dual luciferase reporter gene assays showed that miR-3570, miR-122, and miR-21-3p can bind to the 3’ UTR of the miiuy croaker MAVS gene and exert a negative regulatory effect on post-transcriptional attenuation. Pathogenic infection and poly(I:C) challenge experiments indicated that in macrophages, intestine cells, and kidney cells of miiuy croaker after SCRV infection, the relative expression of levels miR-3570, miR-122, or miR-21-3p were completely opposite to that of MAVS. Hence, the expression level promoted viral replication by inhibiting the production of IFNs and antiviral genes (e.g., TNF-α, IL-1β, and Mx1), ultimately inhibiting the organism’s antiviral response and promoting the replication of SCRV (25–27). Overall, these studies indicated that miR-3570, miR-122, and miR-21-3p acted as negative regulators in suppressing organismal immune and antiviral responses through depressing the NF-κB pathways via directly targeting MAVS (25–27).
2.1.3.2 miRNAs targeting MITA
MITA is also known as a stimulator of interferon genes (STING) (118). As a crucial member of the RLR signaling pathway, MITA is involved in the regulation of signal transduction and the innate antiviral response in mammals (118). Compared to studies in mammals, there are relatively few studies related to MITA in aquatic animals.
miR-210, miR-21-3p, and miR-29a-3p are all 22 nt in length, with seed regions “-GCCACUG-” (miR-210), “-GACAACA-” (miR-21-3p), and “-AGCACCA-” (miR-29a-3p), respectively. Dual luciferase reporter assays demonstrated that MITA of miiuy croaker had binding sites (Figure 2O) in the 3’ UTR that precisely match the seed sequences of miR-210, miR-21-3p, and miR-29a-3p. Overexpression of miR-210, miR-21-3p, or miR-29a-3p suppressed the expression of MITA at both the mRNA and protein levels in the macrophages, intestinal cells and spleen cells of miiuy croaker after SCRV infection (50–52). Meanwhile, the expression of inflammatory cytokines (e.g., TNF-a, IL-6, and Mx1) was significantly inhibited. Taken together, these results suggest that miR-210, miR-21-3p, and miR-29a-3p may be involved in reducing cell proliferation, promoting viral replication, and inhibiting the organismal antiviral response by negatively regulating the NF-κB pathways via targeting MITA (50–52).
2.1.3.3 miRNAs targeting TBK1
TBK1 is a vital immune-related kinase involved in the production of IFNs to prevent invasion by pathogenic microorganisms (119, 120). To date, TBK1 homologous genes have been identified in several groups of aquatic animals, including mollusks and teleost fishes, in species such as marine gastropods (69), Pacific oyster (121), and zebrafish (122).
miR-2187-3p is the only miRNA that has a relationship with TBK1 homologs in aquatic animals (115). This miRNA is 21 nt in length, with a seed region “-UACAGGCU-”. Dual luciferase reporter gene assays demonstrated that TBK1 of miiuy croaker has a binding site in the 3’ UTR that completely matches the seed sequence of miR-2187-3p (Figure 2P) (115). In intestinal cells of miiuy croaker after SCRV infection, overexpression of miR-2187-3p downregulated the expression of TBK1 at both transcriptional and post-transcriptional levels. These observations suggested that miR-2187-3p may act as a negative regulator suppressing the organismal antiviral response through the NF-κB pathways via downregulating the relative expression of TBK1 (115).
As mentioned above, miRNAs involved in NF-κB pathways have now been identified in a variety of aquatic organisms, especially in teleost fishes (123–154) (Table 3). Notably, most of these identified miRNAs have corresponding homologous sequences in mammals. Phylogenetic analysis data indicated a high conservation of examined miRNA homologous sequences in both aquatic animals and mammals (Figure 3), suggesting that these miRNAs might have similar regulatory mechanisms in different species. Furthermore, some miRNAs (miR-133, miR-21 and miR-210) have been identified both in vertebrate and non-vertebrate aquatic animals, whereas some have only been identified in teleost fishes (Tables 3, 4). As the current relevant studies and evaluated aquatic species are still relatively few, whether there are differences in miRNAs regulating NF-κB pathways between vertebrate and non-vertebrate aquatic animals needs further exploration and justification.

Table 3 Summary of identified microRNAs (miRNAs) targeting genes of nuclear factor-kappa B (NF-κB) pathways in aquatic animals. “-”: negative regulation between miRNA and mRNA after experimental validation.
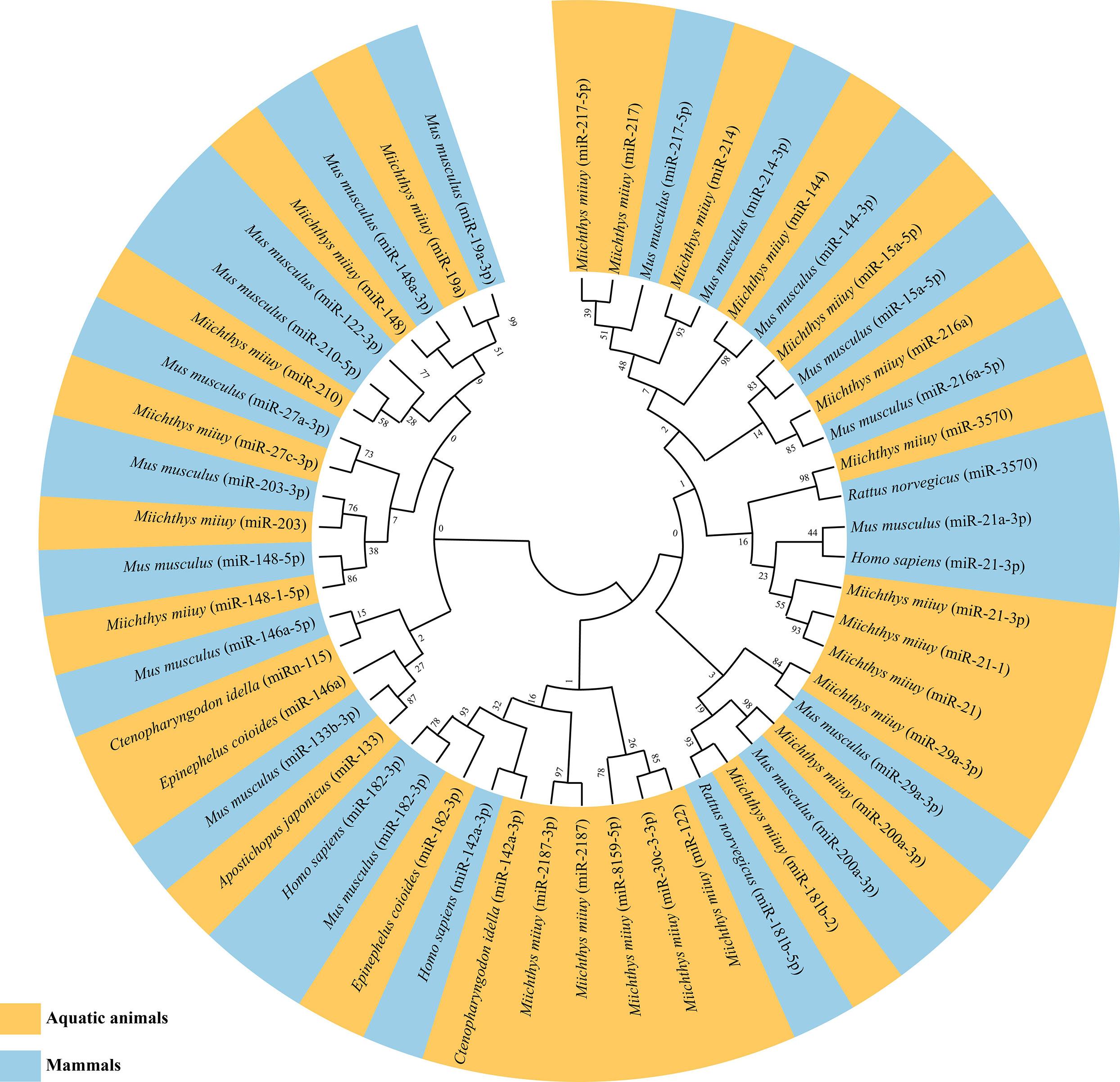
Figure 3 Maximum likelihood phylogenetic tree of identified microRNAs (miRNAs) regulating nuclear factor-kappa B (NF-κB) pathways in aquatic animals and mammals.
3 ceRNAs involved in the NF-κB pathways
Competing endogenous RNA (ceRNA) is defined as RNAs such as linear lncRNAs, circRNA and even mRNAs with the miRNA response element (MRE) that can bind competitively to miRNAs and make them nonfunctional (157). As a novel molecular mechanism in RNA interactions, the hypothesis of ceRNA action was first proposed in 2011 (157). It has been extensively documented that ceRNA can act as a sponge to attract and isolate miRNAs, thereby blocking the effects of miRNAs on their target genes (155–159). According to different ncRNA components, ceRNA networks can be classified into four types: “lncRNA-miRNA-mRNA”, “mRNA-miRNA-mRNA”, “circRNA-miRNA-mRNA”, and “pseudogene-miRNA-mRNA” (155, 159, 160). Several studies have reported that in aquatic animals, ceRNA regulatory networks formed by the participation of ncRNAs (lncRNAs, circRNAs, and miRNAs) play important roles in the innate immune response of the organism, especially in the NF-κB pathways (Figure 4, Table 5).
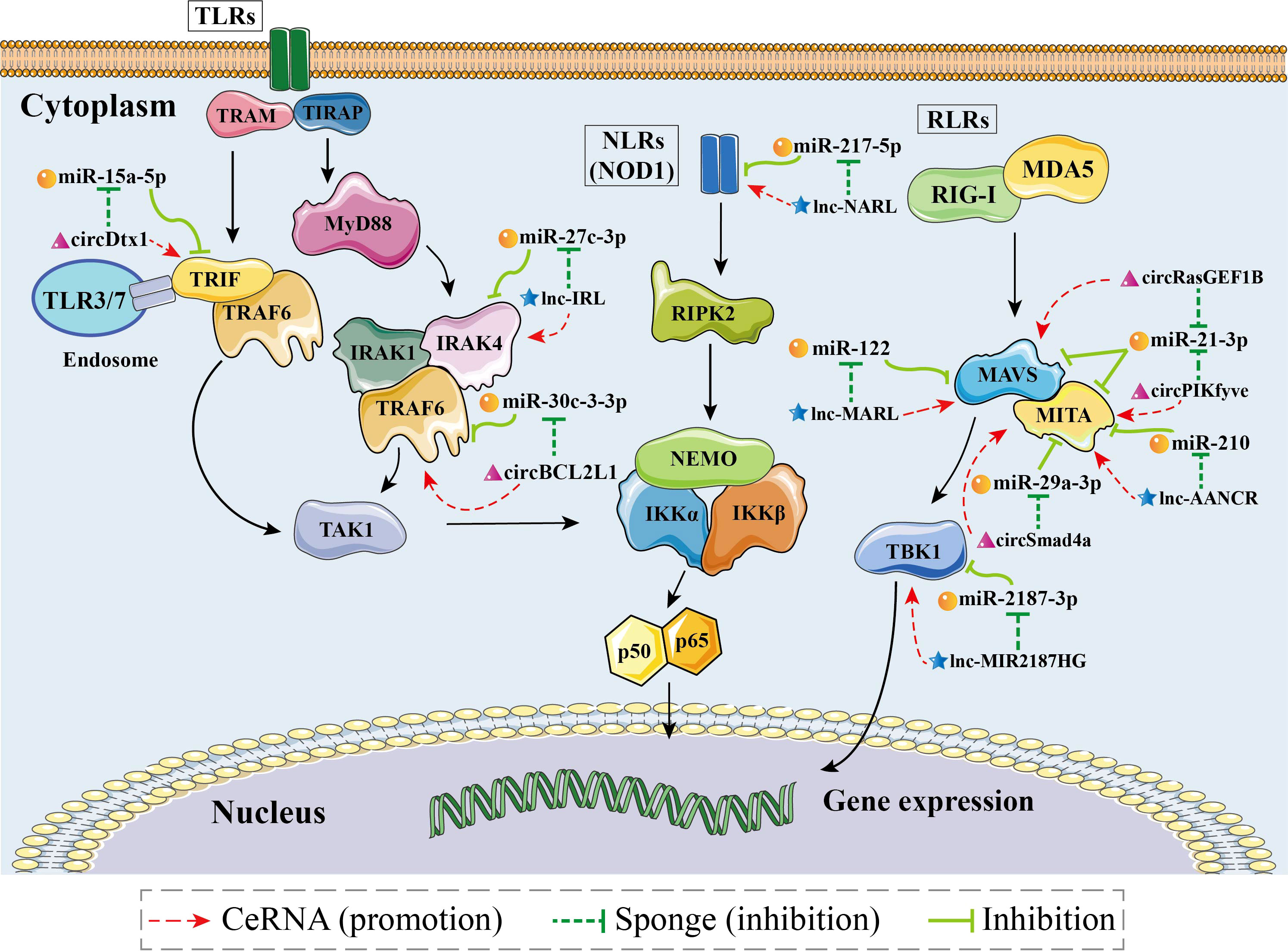
Figure 4 Schematic image of identified competing endogenous RNA (ceRNA) networks associated with nuclear factor-kappa B (NF-κB) pathways in aquatic animals.
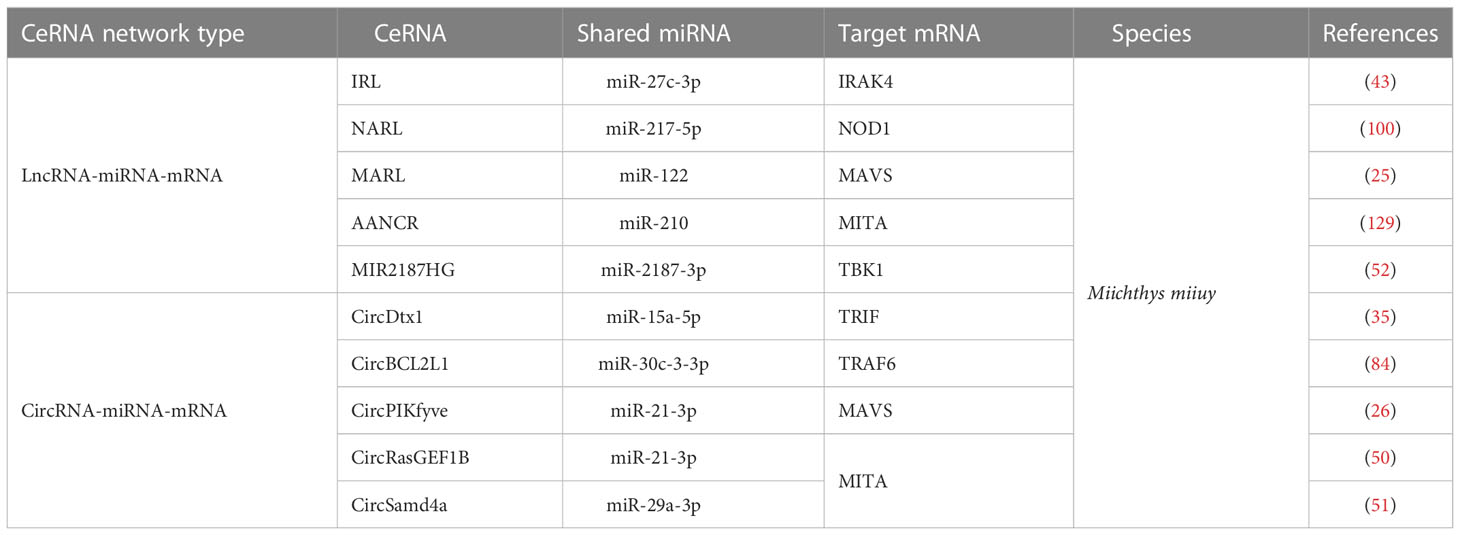
Table 5 Summary of identified competing endogenous RNAs (ceRNAs) associated with genes of the nuclear factor-kappa B (NF-κB) pathways in aquatic animals.
3.1 “lncRNA-miRNA-mRNA” networks regulating the NF-κB pathways
lncRNAs can achieve regulatory effects by interacting with other ncRNAs, mRNAs, proteins, and genomic DNA. lncRNAs play a crucial role in the regulation of gene expression during transcription and translation, as well as at the epigenetic level by performing different functions, including signaling, guidance, decoy, and sponge roles. Therefore, lncRNAs are considered to be pleiotropic and as “master regulators” of the genome (161). Among aquatic animals, lncRNAs have mostly been studied in teleost fishes. In this section, we discuss the “lncRNA-miRNA-mRNA” ceRNA networks that have been adequately identified in aquatic animals, including “IRAK4-related lncRNA (IRL)/miR-27c-3p/IRAK4” (80), “NOD1 antibacterial and antiviral-related lncRNA (NARL)/miR-217-5p/NOD1” (48), “MAVS antiviral-related lncRNA (MARL)/miR-122/MAVS” (26), “antiviral-associated lncRNA (AANCR)/miR-210/MITA” (162), and “MIR2187HG/miR-2187-3p/TBK1” (115).
3.1.1 “lncRNA-miRNA-mRNA” networks regulating the TLR signaling pathway
The “IRL/miR-27c-3p/IRAK4” network is a key ceRNA network related to immune defense in aquatic organisms (80). Dual luciferase reporter assays have demonstrated that IRL can bind to the seed sequence of miR-27c-3p (Figure 5A). Thus, IRL was able to activate and upregulate the expression of IRAK4 by functioning as a ceRNA, while miR-27c-3p acted as a repressor to IRAK4. Under the conditions of knockdown of IRL and overexpression of miR-27c-3p after LPS stimulation, the expression of IRAK4 and subsequent inflammatory factors was significantly suppressed, indicating that miR-27c-3p and IRL play important regulatory roles in the inflammatory response of miiuy croaker. IRAK4 has been repeatedly shown to be an important factor in the TLR-dependent immune response; it has a role in promoting cell proliferation, while inhibition of its expression promotes apoptosis (163–167). It has been reported that IRAK4 can induce innate antimicrobial responses through the NF-κB pathways, thereby activating inflammatory factors in response to external stimuli in fish (168–170). Collectively, IRL may promote organismal immune responses through upregulating the NF-κB pathways via sponging miR-27c-3p and acting as a ceRNA to IRAK4, while miR-27c-3p acts conversely (80). These studies suggested that the “IRL/miR-27-3p/IRAK4” network plays an important role in the inflammatory response, and the results contribute to further understanding of aquatic biological immunology, as well as to that of disease control mechanisms (80).
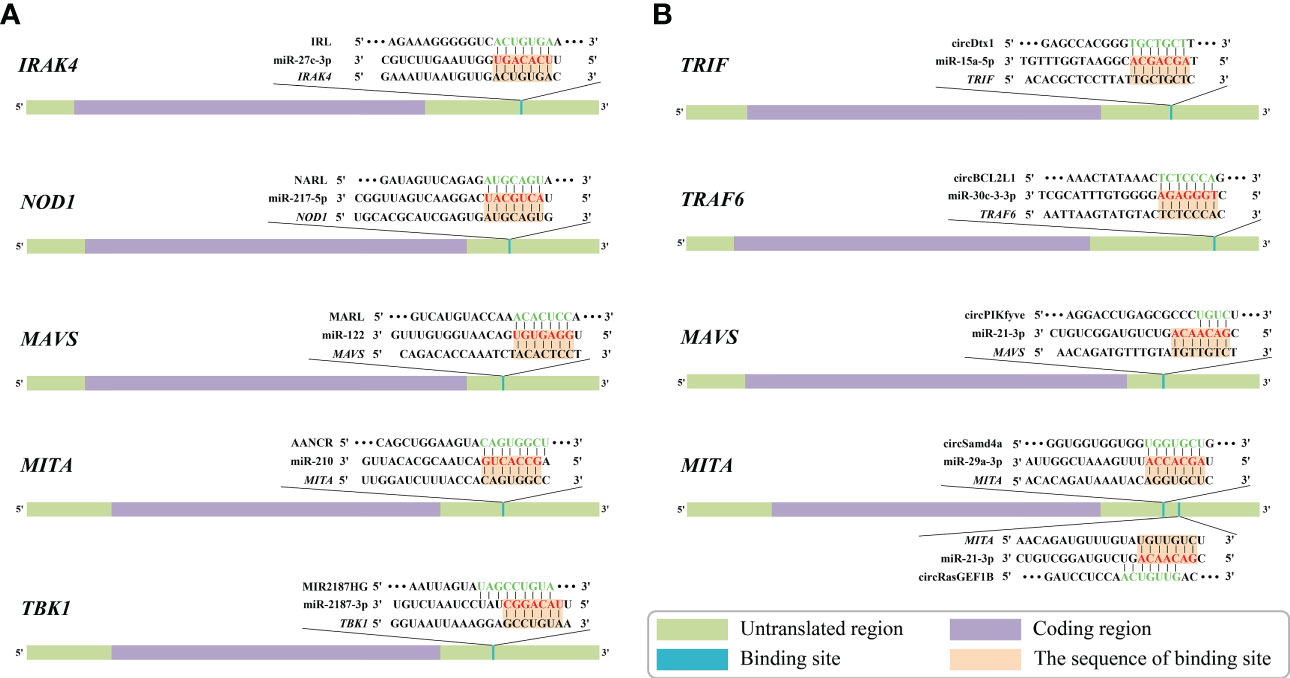
Figure 5 Predicted binding sites of identified competing endogenous RNAs (ceRNAs) associated with genes of nuclear factor-kappa B (NF-κB) pathways in aquatic animals. (A) Predicted binding sites of “long non-coding RNA (lncRNA)-microRNA (miRNA)-mRNA” ceRNA networks. (B) Predicted binding sites of “circular RNA (circRNA)-miRNA-mRNA” ceRNA networks. The red markings represent seed regions.
3.1.2 "lncRNA-miRNA-mRNA" networks regulating the NLR signaling pathways
The “NARL/miR-217-5p/NOD1” network is the only known ceRNA network to date that regulates the NLR signaling pathway in aquatic animals (48). Zheng et al. demonstrated a binding site between NARL and miR-217-5p by using dual luciferase validation (Figure 5A). In this process, the expression levels of NARL and NOD1 increased, while those of miR-217-5p decreased; the functional attenuation of miR-217-5p thus resulted in the enhancement of the immune response and the NF-κB pathways in miiuy croaker. Specifically, NARL competitively binds to miR-217-5p in order to counteract its inhibition of NOD1 by exerting a sponge effect after LPS stimulation or SCRV infection, indicating that miR-217-5p and NARL have competitive relationships with MmiNOD1. It is generally acknowledged that NOD1 activates the NF-κB pathways to promote host production of multiple inflammatory cytokines to resist bacterial invasion in fish. Moreover, NOD1 can also act as an RNA virus receptor to enhance the immune response during viral infection (106). Considering all of this evidence, it seems that miR-217-5p/NOD1 can be further regulated by NARL to achieve more precise immune homeostasis (48). The above-mentioned studies revealed that miR-217-5p and NARL regulate NOD1 and subsequent immune defense processes of an organism through their mutually associated ceRNA activities. The “NARL/miR-217-5p/NOD1” network could be a key node for further understanding the molecular regulatory mechanisms of immune responses in aquatic organisms (48).
3.1.3 “lncRNA-miRNA-mRNA” networks regulating the RLR signaling pathways
Currently, three “lncRNA-miRNA-mRNA” ceRNA networks have been identified as regulating the RLR signaling pathway in aquatic animals, namely “MARL/miR-122/MAVS” (25), “AANCR/miR-210/MITA” (162), and “MIR2187HG/miR-2187-3p/TBK1” (115).
“MARL/miR-122/MAVS” was identified as a ceRNA network in aquatic animals. Dual luciferase reporter assays verified that MARL acts as a sponge by competitively binding to miR-122 (Figure 5A), thereby interacting with miR-122 and acting as a ceRNA of MAVS and in turn promoting the expression at both the mRNA level and the protein level, thereby enhancing the antiviral signaling pathway (26). Further functional experiments demonstrated that the expression of MAVS decreased after knockdown of MARL. In contrast, overexpression of miR-122 can suppress the activation of the NF-κB pathways and downstream inflammatory response, thereby helping the virus to evade the host antiviral response via inhibiting the expression of MAVS. The host signaling protein MAVS is critical in driving the antiviral innate immune response to RNA virus infection (171). MARL could counteract the upregulatory effect of miR-122 on SCRV replication, thus maintaining the stability of the antiviral response and ensuring an appropriate inflammatory response. Taken together, these observations indicated that the “MARL/miR-122/MAVS” network may play an important role in aquatic animals’ immune responses, especially inflammatory responses, as well as in the NF-κB signaling pathways (26).
The “AANCR/miR-210/MITA” network, a vital ceRNA network regulating the RLR signaling pathway related to MITA, was identified in miiuy croaker (162). Dual luciferase report assays demonstrated that AANCR has an intact binding site with miR-210 (Figure 5A). It has been reported that AANCR could act as a sponge for miR-210 that subsequently indirectly regulates the NF-κB pathways. This molecular regulatory mechanism of the ceRNA network contributes to the recognition and elimination of viruses by the host immune system (162).
The “MIR2187HG/miR-2187-3p/TBK1” network is a key ceRNA network related to antibacterial and antiviral responses in aquatic animals. TBK1 can promote canonical activation of NF-κB and interferon regulatory factor 3 (IRF3) to accelerate proinflammatory gene transcription (172). It has been reported that overexpression of MIR2187HG could inhibit the expression of miR-2187-3p and upregulate that of miiuy croaker TBK1 in a manner that competitively binds to miR-2187-3p (Figure 5A), consequently restoring the immune response mediated by the NF-κB pathways (115). MIR2187HG may promote organismal immune responses through upregulating the NF-κB pathways via sponging miR-2187-3p and being a ceRNA indirectly targeting TBK1 (115). Taken together, we can hypothesize that the “MIR2187HG/miR-2187-3p/TBK1” network could be a key entry point for further understanding the molecular regulatory mechanisms of immune responses in aquatic organisms (115).
3.2 “circRNA-miRNA-mRNA” networks regulating the NF-κB pathways
CircRNAs are novel ncRNAs with a stable structure of covalently closed continuous loops (173). Current data show that circRNAs can act as miRNA sponges and subsequently inactivate the post-transcriptional attenuation functions of corresponding miRNAs (174, 175). To date, most studies concerning circRNAs have focused on human diseases; studies on circRNAs’ targets (such as mRNA and miRNA) are largely lacking. In this section, we discuss five “circRNA-miRNA-mRNA” networks associated with NF-κB pathways, namely “circDtx1/miR-15a-5p/TRIF” (68), “circBCL2L1/miR-30c-3-3p/TRAF6” (44), “circPIKfyve/miR-21-3p/MAVS” (27), “circRasGEF1B/miR-21-3p/MITA” (51), and “circSamd4a/miR-29a-3p/MITA” (52).
3.2.1 "circRNA-miRNA-mRNA" networks regulating the TLR signaling pathways
The “CircDtx1/miR-15a-5p/TRIF” network (68) and the “circBCL2L1/miR-30c-3-3p/TRAF6” network (44) are two identified “circRNA/miRNA/mRNA” ceRNA networks regulating the TLR signaling pathway in aquatic animals.
The “circDtx1/miR-15a-5p/TRIF” network has been fully identified as being involved in the TLR signaling pathway in aquatic animals. Dual luciferase reporter assays have demonstrated that circDtx1 can bind to the seed sequence of miR-15a-5p that in turn inhibits the miR-15a-5p that had an intact binding site on the 3’ UTR of TRIF (Figure 5B). CircDtx1 acts as a ceRNA that upregulates the expression of TRIF at both the mRNA level and protein level by sponging miR-15a-5p, thereby promoting the organismal antiviral response via the NF-κB pathways (68). Overexpression of circDtx1 led to a decrease in the expression of miR-15a-5p, indicating a negative correlation. TRIF is a key member of the NF-κB pathways, and it has an important role in activating NF-κB signaling. These observations indicated that circDtx1 could function as a sponge of miR-15a-5p that forms the “circDtx1/miR-15a-5p/TRIF” ceRNA network to suppress viral replication and enhance immunological activity. These observations provide new insights into the role of circRNAs in host antiviral immunity (68).
Additionally, it was reported that circBCL2L1 could function as a molecular sponge of miR-30c-3-3p in the “circBCL2L1/miR-30c-3-3p/TRAF6” ceRNA network (44). As stated in previous studies, there is a strong correlation between TRAF6 and the innate immune response (83, 176). Dual luciferase reporter assays showed that circBCL2L1 had an intact binding site with miR-30c-3-3p (Figure 5B). Further experiments confirmed that overexpression of circBCL2L1 could promote organismal antibacterial and antiviral responses through enhancing the NF-κB pathways via competitively binding to miR-30c-3-3p and upregulating the relative expression of TRAF6. In addition, it was reported that circBCL2L1 could restore the attenuated immune response induced by miR-30c-3-3p and in turn maintain the stability of the immune response, thereby ensuring an appropriate inflammatory response.
3.2.2 “circRNA-miRNA-mRNA” networks regulating the RLR signaling pathways
Currently, there are three “circRNA-miRNA-mRNA” networks, the “circPIKfyve/miR-21-3p/MAVS” network (27), the “circRasGEF1B/miR-21-3p/MITA” network (51), and the “circSamd4a/miR-29a-3p/MITA” network (52), that have been identified as novel regulatory mechanisms of the RLR signaling pathway in aquatic animals.
Su et al. indicated that the “circPIKfyve/miR-21-3p/MAVS” network is a crucial ceRNA network associated with the RLR signaling pathway in aquatic organisms (27). Dual luciferase reporter assays demonstrated that circPIKfyve could target and bind to miR-21-3p (Figure 5B). Consequently, circPIKfyve could activate and upregulate the expression of MAVS by functioning as a ceRNA, while miR-21-3p acted as a repressor of MAVS. The expression levels of MAVS and inflammatory cytokines were remarkably reduced as a result of miR-21-3p overexpression and IRL knockdown after LPS stimulation. To date, studies have confirmed that MAVS is a key member of the RLR pathway-related innate antiviral immune response and NF-κB pathways whose activation leads to rapid production of antiviral cytokines (114, 177). miR-21-3p is a novel miRNA that directly targets MAVS, negatively regulates the relative expression of MAVS, and inhibits the antimicrobial response it mediates. Overall, the above observations suggest that circPIKfyve may promote the organismal immune response by upregulating the NF-κB pathways via sponging miR-21-3p and being a ceRNA to MAVS. This observation also highlights the finding that the “circPIKfyve/miR-21-3p/MAVS” network plays an important role in the inflammatory response as well as NF-κB pathways in aquatic animals.
It has been shown that the “circRasGEF1B/miR-21-3p/MITA” network (51) and the “circSamd4a/miR-29a-3p/MITA” network (52) are two ceRNA networks associated with MITA in aquatic animals. Dual luciferase reporter assays have demonstrated that miR-21-3p and miR-29a-3p are available to bind circRasGEF1B and circSamd4a, respectively (Figure 5B). circRasGEF1B and circSamd4a acted as sponges, competitively binding to their respective target miRNAs (miR-21-3p and miR-29a-3p, respectively). In this process, the expression of circRasGEF1B, circSamd4a, and MITA (mRNA and protein) increased, while the expression of miR-29a-3p decreased, indicating a negative regulatory relationship between the above-mentioned circRNAs and miRNAs. The above-mentioned miRNAs were able to reduce the expression of MITA and inhibit the antiviral response. In contrast, those circRNAs could counteract the promotive effect of the above-mentioned miRNAs on the replication of SCRV, thereby maintaining the stability of the antiviral response and ensuring an appropriate inflammatory response. The present findings indicate that the “circRasGEF1B/miR-21-3p/MITA” network and the “circSamd4a/miR-29a-3p/MITA” network may participate in the activation of the NF-κB pathways and subsequent production of inflammatory factors after RNA virus infection in aquatic animals (51, 52).
Through the summary in this section, it can be seen that research on ceRNAs involved in the NF-κB pathways in aquatic animals is currently far from adequate, as only one fish species (miiuy croaker) has been relatively comprehensively studied, and thus more comprehensive and exhaustive efforts should be made in the future.
4 Concluding remarks
The immune regulatory function of ncRNAs in both vertebrates and invertebrates has been an intense research topic for more than 10 years. Although not as extensively or as thoroughly studied as in mammals, many ncRNAs have been identified as transcriptional regulators of key genes in both the canonical and non-canonical NF-κB pathways in aquatic animals, especially in teleost fishes. There is no doubt that continuous mining of ncRNAs with immune regulatory potential would benefit the sustainable development of the rapidly expanding aquaculture industry worldwide.
Despite the above-mentioned advances, we still need to note that (1) we are just beginning to understand the immune regulatory function of ncRNAs; like looking at a leopard through a tube, the range of aquatic species should be further expanded to clarify the differences in ncRNAs regulating NF-κB pathways between vertebrate and non-vertebrate aquatic animals; in addition, there is also much more research needed to extensively identify ncRNAs (especially lncRNAs, circRNAs, piRNAs, and novel ncRNAs) associated with the NF-κB pathways or other immune related pathways in aquatic animals (2); further exploration is necessary to clarify the explicit mechanisms concerning how “ncRNA-mRNA” axes or ceRNA networks regulate the NF-κB pathways or other immune related pathways in aquatic animals; and (3) given the important links between the NF-κB pathways and immune capability of aquatic animals, further studies on breeding-valuable ncRNA markers targeting genes of the NF-κB pathways will facilitate the development of more accurate and effective molecular-assisted breeding strategies in aquaculture.
Author contributions
YYZ and YC conceived of the manuscript. The reference collection and data analysis were performed by TZ, YZ, and HY. The manuscript was written by YYZ, TZ, and YZ. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Key R & D Program of China (No. 2022YFD2400302) and the Liaoning Province Innovative Talent Support Program of Colleges and Universities (LR2020065).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study. Sig Transduct Target Ther (2020) 5(1):209. doi: 10.1038/s41392-020-00312-6
2. Sun SC. The non-canonical NF-κB pathway in immunity and inflammation. Nat Rev Immunol (2017) 17(9):545–58. doi: 10.1038/nri.2017.52
3. Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell (2008) 132(3):344–62. doi: 10.1016/j.cell.2008.01.020
4. Israel A. The IKK complex, a central regulator of NF-κB activation. Cold Spring Harbor Perspect Biol (2010) 2(3):a000158–a000158. doi: 10.1101/cshperspect.a000158
5. Ruland J. Return to homeostasis: downregulation of NF-κB responses. Nat Immunol (2011) 12(8):709–14. doi: 10.1038/ni.2055
6. Rao P, Hayden MS, Long M, Scott ML, West AP, Zhang D, et al. IκBβ acts to inhibit and activate gene expression during the inflammatory response. Nature (2010) 466(7310):1115–9. doi: 10.1038/nature09283
7. Richardson JC, Estrabot G, Woodland R. XrelA, a Xenopus maternal and zygotic homologue of the p65 subunit of NF-KB. characterisation of transcriptional properties in the developing embryo and identification of a negative interference mutant. Mech Dev (1994) 45(2):173–89. doi: 10.1016/0925-4773(94)90031-0
8. Wang P, Gu Z, Wan D, Zhang M, Weng S, Yu X, et al. The shrimp NF-κB pathway is activated by white spot syndrome virus (WSSV) 449 to facilitate the expression of WSSV069 (ie1), WSSV303 and WSSV371. PloS One (2011) 6(9):e24773. doi: 10.1371/journal.pone.0024773
9. Correa RG, Tergaonkar V, Ng JK, Dubova I, Izpisua-Belmonte JC, Verma IM. Characterization of NF-κB/IκB proteins in zebra fish and their involvement in notochord development. Mol Cell Biol (2004) 24(12):5257–68. doi: 10.1128/MCB.24.12.5257-5268.2004
10. Ouyang G, Liao Q, Zhang D, Rong F, Cai X, Fan S, et al. Zebrafish NF-κB/p65 is required for antiviral responses. J Immunol (2020) 204(11):3019–29. doi: 10.4049/jimmunol.1900309
11. He L, Zhao Y, Tang L, Yu X, Ye Z, Lin H, et al. Molecular characterization and functional analysis of IKKα in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol (2020) 101:159–67. doi: 10.1016/j.fsi.2020.03.029
12. Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet (2010) 11(8):559–71. doi: 10.1038/nrg2814
13. Xiao Z, Xiao Y, Lu X. Role of circRNA as biomarkers in the development and prognosis of colorectal cancer. Chin J Cancer Biother (2019) 26(1):116–20. doi: 10.3872/j.issn.1007-385x.2019.01.019
14. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J (2004) 23(20):4051–60. doi: 10.1038/sj.emboj.7600385
15. Zhou W, Cai Z, Liu J, Wang D, Ju H, Xu R. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer (2020) 19(1):172. doi: 10.1186/s12943-020-01286-3
16. Gao W, Chang R, Sun Y, Xu T. MicroRNA-2187 modulates the NF-κB and IRF3 pathway in teleost fish by targeting TRAF6. Front Immunol (2021) 12:647202. doi: 10.3389/fimmu.2021.647202
17. Chu Q, Sun Y, Cui J, Xu T. MicroRNA-3570 modulates the NF-κB pathway in teleost fish by targeting MyD88. J Immunol (2017) 198(8):3274–82. doi: 10.4049/jimmunol.1602064
18. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol (2010) 11(5):373–84. doi: 10.1038/ni.1863
19. Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol (2004) 5(5):392–401. doi: 10.1038/nrm1368
20. Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa b controls expression of inhibitor I kappa b alpha: evidence for an inducible autoregulatory pathway. Science (1993) 259(5103):1912–5. doi: 10.1126/science.8096091
21. Sun SC, Ley SC. New insights into NF-κB regulation and function. Trends Immunol (2008) 29(10):469–78. doi: 10.1016/j.it.2008.07.003
22. Häcker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci Stke (2006) 357:re13. doi: 10.1126/stke.3572006re13
23. Sahoo BR. Structure of fish toll-like receptors (TLR) and NOD-like receptors (NLR). Int J Biol Macromol (2020) 161:1602–17. doi: 10.1016/j.ijbiomac.2020.07.293
24. Canesi L, Auguste M, Balbi T, Prochazkova P. Soluble mediators of innate immunity in annelids and bivalve mollusks: A mini-review. Front Immunol (2022) 13:1051155. doi: 10.3389/fimmu.2022.1051155
25. Xu T, Chu Q, Cui J, Bi D. Inducible microRNA-3570 feedback inhibits the RIG-i-dependent innate immune response to rhabdovirus in teleost fish by targeting MAVS/IPS-1. J Virol (2018) 92(2):e01594–17. doi: 10.1128/JVI.01594-17
26. Chu Q, Xu T, Zheng W, Chang R, Zhang L. Long noncoding RNA MARL regulates antiviral responses through suppression miR-122-dependent MAVS downregulation in lower vertebrates. PloS Pathog (2020) 16(7):e1008670. doi: 10.1371/journal.ppat.1008670
27. Su H, Chu Q, Zheng W, Chang R, Gao W, Zhang L, et al. Circular RNA circPIKfyve acts as a sponge of miR-21-3p to enhance antiviral immunity through regulation of MAVS in teleost fish. J Virol (2021) 95(8):e02296–20. doi: 10.1128/JVI.02296-20
28. Takeda K, Akira S. TLR signaling pathways. Semin Immunol (2004) 16(1):3–9. doi: 10.1016/j.smim.2003.10.003
29. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity (2011) 34(5):637–50. doi: 10.1016/j.immuni.2011.05.006
30. Wang Y, Xu G, Han J, Xu T. miR-200a-3p regulates TLR1 expression in bacterial challenged miiuy croaker. Dev Comp Immunol (2016) 63:181–6. doi: 10.1016/j.dci.2016.06.004
31. Chu Q, Sun Y, Bi D, Cui J, Xu T. Up-regulated of miR-8159-5p and miR-217-5p by LPS stimulation negatively co-regulate TLR1 in miiuy croaker. Dev Comp Immunol (2017) 67:117–25. doi: 10.1016/j.dci.2016.11.004
32. Xu X, Shen Y, Fu J, Yu H, Huang W, Lu L, et al. MicroRNA-induced negative regulation of TLR-5 in grass carp. Ctenopharyngodon idella Sci Rep (2016) 6:18595. doi: 10.1038/srep18595
33. He L, Zhao M, Yu X, Zhao Y, Fu L, Qiao X, et al. MicroRNA-182-3p negatively regulates cytokines expression by targeting TLR5M in orange-spotted grouper. Epinephelus coioides Fish Shellfish Immunol (2019) 93:589–96. doi: 10.1016/j.fsi.2019.07.063
34. Zhu S, Xiang X, Xu X, Gao S, Mai K, Ai Q. TIR domain-containing adaptor-inducing interferon-β (TRIF) participates in antiviral immune responses and hepatic lipogenesis of large yellow croaker (Larimichthys crocea). Front Immunol (2019) 10:2506. doi: 10.3389/fimmu.2019.02506
35. Sun Y, Zhang L, Hong L, Zheng W, Cui J, Liu X, et al. MicroRNA-181b-2 and microRNA-21-1 negatively regulate NF-κB and IRF3-mediated innate immune responses via targeting TRIF in teleost. Front Immunol (2021) 12:734520. doi: 10.3389/fimmu.2021.734520
36. Lu Y, Li C, Zhang P, Shao Y, Su X, Li Y, et al. Two adaptor molecules of MyD88 and TRAF6 in Apostichopus japonicus toll signaling cascade: Molecular cloning and expression analysis. Dev Comp Immunol (2013) 41(4):498–504. doi: 10.1016/j.dci.2013.07.009
37. Chu Q, Gao Y, Bi D, Xu T. MicroRNA-148 as a negative regulator of the common TLR adaptor mediates inflammatory response in teleost fish. Sci Rep (2017) 7(1):4124. doi: 10.1038/s41598-017-04354-9
38. Chu Q, Sun Y, Cui J, Xu T. Inducible microRNA-214 contributes to the suppression of NF-κB-mediated inflammatory response via targeting myd88 gene in fish. J Biol Chem (2017) 292(13):5282–90. doi: 10.1074/jbc.M117.777078
39. Cui Y, Jiang L, Xing R, Wang Z, Wang Z, Shao Y, et al. Cloning, expression analysis and functional characterization of an interleukin-1 receptor-associated kinase 4 from Apostichopus japonicus. Mol Immunol (2018) 101:479–87. doi: 10.1016/j.molimm.2018.08.006
40. Chang R, Zheng W, Luo Q, Liu G, Xu T, Sun Y. miR-148-1-5p modulates NF-κB signaling pathway by targeting IRAK1 in miiuy croaker (Miichthys miiuy). Dev Comp Immunol (2021) 125:104229. doi: 10.1016/j.dci.2021.104229
41. Lu M, Zhang P, Li C, Lv Z, Zhang W, Jin C. miRNA-133 augments coelomocyte phagocytosis in bacteria-challenged Apostichopus japonicus via targeting the TLR component of IRAK-1 in vitro and in vivo. Sci Rep (2015) 5:12608. doi: 10.1038/srep12608
42. Xu T, Chu Q, Cui J, Zhao X. The inducible microRNA-203 in fish represses the inflammatory responses to gram-negative bacteria by targeting IL-1 receptor-associated kinase 4. J Biol Chem (2018) 293(4):1386–96. doi: 10.1074/jbc.RA117.000158
43. Chu Q, Yan X, Liu L, Xu T. The inducible microRNA-21 negatively modulates the inflammatory response in teleost fish via targeting IRAK4. Front Immunol (2019) 10:1623. doi: 10.3389/fimmu.2019.01623
44. Zheng W, Sun L, Yang L, Xu T. The circular RNA circBCL2L1 regulates innate immune responses via microRNA-mediated downregulation of TRAF6 in teleost fish. J Biol Chem (2021) 297(4):101199. doi: 10.1016/j.jbc.2021.101199
45. Röttinger E, Croce J, Lhomond G, Besnardeau L, Gache C, Lepage T. Nemo-like kinase (NLK) acts downstream of Notch/Delta signalling to downregulate TCF during mesoderm induction in the sea urchin embryo. Development (2006) 133(21):4341–53. doi: 10.1242/dev.02603
46. Kong HJ, Moon JH, Moon JY, Kim JM, Nam BH, Kim YO, et al. Cloning and functional characterization of the p65 subunit of NF-κB from olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol (2011) 30(1):406–11. doi: 10.1016/j.fsi.2010.11.024
47. Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to crohn’s disease. Nature (2001) 411(6837):603–6. doi: 10.1038/35079114
48. Zheng W, Chu Q, Xu T. The long noncoding RNA NARL regulates immune responses via microRNA-mediated NOD1 downregulation in teleost fish. J Biol Chem (2021) 296:100414. doi: 10.1016/j.jbc.2021.100414
49. Sun Q, Sun L, Liu H, Chen X, Seth RB, Forman J, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity (2006) 24(5):633–42. doi: 10.1016/j.immuni.2006.04.004
50. Xu T, Chu Q, Cui J. Rhabdovirus-inducible microRNA-210 modulates antiviral innate immune response via targeting STING/MITA in fish. J Immunol (2018) 201(3):982–94. doi: 10.4049/jimmunol.1800377
51. Chu Q, Zheng W, Su H, Zhang L, Chang R, Gao W, et al. A highly conserved circular RNA, circRasGEF1B, enhances antiviral immunity by regulating the miR-21-3p/MITA pathway in lower vertebrates. J Virol (2021) 95(7):e02145–20. doi: 10.1128/JVI.02145-20
52. Su H, Zheng W, Pan J, Lv X, Xin S, Xu T. Circular RNA circSamd4a regulates antiviral immunity in teleost fish by upregulating STING through sponging miR-29a-3p. J Immunol (2021) 207(11):2770–84. doi: 10.4049/jimmunol.2100469
53. Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity (2010) 32(3):305–15. doi: 10.1016/j.immuni.2010.03.012
54. Cui J, Chu Q, Xu T. miR-122 involved in the regulation of toll-like receptor signaling pathway after Vibrio anguillarum infection by targeting TLR14 in miiuy croaker. Fish Shellfish Immunol (2016) 58:67–72. doi: 10.1016/j.fsi.2016.09.027
55. Zhang Y, He X, Yu F, Xiang Z, Li J, Thorpe KL, et al. Characteristic and functional analysis of toll-like receptors (TLRs) in the lophotrocozoan, Crassostrea gigas, reveals ancient origin of TLR-mediated innate immunity. PloS One (2013) 8(10):e76464. doi: 10.1371/journal.pone.0076464
56. Sun H, Zhou Z, Dong Y, Yang A, Jiang B, Gao S, et al. Identification and expression analysis of two toll-like receptor genes from sea cucumber (Apostichopus japonicus). Fish Shellfish Immunol (2013) 34(1):147–58. doi: 10.1016/j.fsi.2012.10.014
57. He L, Liang Y, Yu X, Peng W, He J, Fu L, et al. Vibrio parahaemolyticus flagellin induces cytokines expression via toll-like receptor 5 pathway in orange-spotted grouper, Epinephelus coioides. Fish Shellfish Immunol (2019) 87:573–81. doi: 10.1016/j.fsi.2019.01.054
58. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol (2002) 12(9):735–9. doi: 10.1016/S0960-9822(02)00809-6
59. Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol (2004) 1(2):106–13. doi: 10.4161/rna.1.2.1066
60. Boutz DR, Collins PJ, Suresh U, Lu M, Ramírez CM, Fernández-Hernando C, et al. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J Biol Chem (2011) 286(20):18066–78. doi: 10.1074/jbc.M110.196451
61. Florczyk M, Brzuzan P, Krom J, Woźny M, Łakomiak A. miR-122-5p as a plasma biomarker of liver injury in fish exposed to microcystin-LR. J Fish Dis (2016) 39(6):741–51. doi: 10.1111/jfd.12406
62. Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in toll-like receptor 3–mediated interferon-β induction. Nat Immunol (2003) 4(2):161–7. doi: 10.1038/ni886
63. Zhou Z, Zhang M, He Y, Bao S, Zhang X, Li W, et al. Identification and functional characterization of an immune adapter molecular TRIF in northeast Chinese lamprey (Lethenteron morii). Fish Shellfish Immunol (2022) 124:454–61. doi: 10.1016/j.fsi.2022.04.018
64. Baoprasertkul P, Peatman E, Somridhivej B, Liu Z. Toll-like receptor 3 and TICAM genes in catfish: species-specific expression profiles following infection with Edwardsiella ictaluri. Immunogenetics (2006) 58(10):817–30. doi: 10.1007/s00251-006-0144-z
65. Fan S, Chen S, Liu Y, Lin Y, Liu H, Guo L, et al. Zebrafish TRIF, a golgi-localized protein, participates in IFN induction and NF-κB activation. J Immunol (2008) 180(8):5373–83. doi: 10.4049/jimmunol.180.8.5373
66. Yang C, Li Q, Su J, Chen X, Wang Y, Peng L. Identification and functional characterizations of a novel TRIF gene from grass carp (Ctenopharyngodon idella). Dev Comp Immunol (2013) 41(2):222–9. doi: 10.1016/j.dci.2013.05.018
67. Wei J, Zhang X, Zang S, Qin Q. Expression and functional characterization of TRIF in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol (2017) 71:295–304. doi: 10.1016/j.fsi.2017.09.063
68. Zheng W, Chu Q, Yang L, Sun L, Xu T. Circular RNA circDtx1 regulates IRF3-mediated antiviral immune responses through suppression of miR-15a-5p-dependent TRIF downregulation in teleost fish. PloS Pathog (2021) 17(3):e1009438. doi: 10.1371/journal.ppat.1009438
69. Gorbushin AM. Toll-like signaling pathway in the transcriptome of Littorina littorea. Fish Shellfish Immunol (2020) 106:640–4. doi: 10.1016/j.fsi.2020.08.012
70. Zhao L, Jiang X, Chen T, Sun H, Ren C. Molecular characterization and functional analysis of MyD88 from the tropical sea cucumber, Holothuria leucospilota. Fish Shellfish Immunol (2018) 83:1–7. doi: 10.1016/j.fsi.2018.09.001
71. Zhao X, Chu Q, Cui J, Xu T. microRNA-19a as a negative regulator in TLR signaling pathway by direct targeting myeloid differentiation factor 88 in miiuy croaker. Dev Comp Immunol (2018) 87:171–5. doi: 10.1016/j.dci.2018.06.009
72. Gantier MP, Stunden HJ, McCoy CE, Behlke MA, Wang D, Kaparakis-Liaskos M, et al. A miR-19 regulon that controls NF-κB signaling. Nucleic Acids Res (2012) 40(16):8048–58. doi: 10.1093/nar/gks521
73. Cao Z, Henzel WJ, Gao X. IRAK: a kinase associated with the interleukin-1 receptor. Science (1996) 271(5252):1128–31. doi: 10.1126/science.271.5252.1128
74. Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci (2002) 99(8):5567–72. doi: 10.1073/pnas.082100399
75. Su L, Xu W, Huang A. IRAK family in inflammatory autoimmune diseases. Autoimmun Rev (2020) 19(3):102461. doi: 10.1016/j.autrev.2020.102461
76. Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity (1997) 7(6):837–47. doi: 10.1016/S1074-7613(00)80402-1
77. Xu Y, Huang Y, Cai S. Characterization and function analysis of interleukin-1 receptor-associated kinase-1 (IRAK-1) from Fenneropenaeus penicillatus. Fish Shellfish Immunol (2017) 61:111–9. doi: 10.1016/j.fsi.2016.12.030
78. Li C, Chen Y, Weng S, Li S, Zuo H, Yu X, et al. Presence of tube isoforms in Litopenaeus vannamei suggests various regulatory patterns of signal transduction in invertebrate NF-κB pathway. Dev Comp Immunol (2014) 42(2):174–85. doi: 10.1016/j.dci.2013.08.012
79. Qi P, Huang H, Guo B, Liao Z, Liu H, Tang Z, et al. A novel interleukin-1 receptor-associated kinase-4 from thick shell mussel Mytilus coruscus is involved in inflammatory response. Fish Shellfish Immunol (2019) 84:213–22. doi: 10.1016/j.fsi.2018.10.018
80. Zheng W, Chu Q, Xu T. Long noncoding RNA IRL regulates NF-κB-mediated immune responses through suppression of miR-27c-3p-dependent IRAK4 downregulation in teleost fish. J Bio Chem (2021) 296:100304. doi: 10.1016/j.jbc.2021.100304
81. Zhang X, Li C, Zhang L, Wu C, Han L, Jin G, et al. TRAF6 restricts p53 mitochondrial translocation, apoptosis, and tumor suppression. Mol Cell (2016) 64(4):803–14. doi: 10.1016/j.molcel.2016.10.002
82. Yoon K, Jung EJ, Lee SY. TRAF6-mediated regulation of the PI3 kinase (PI3K)–Akt–GSK3β cascade is required for TNF-induced cell survival. Biochem Biophy Res Commun (2008) 371(1):118–21. doi: 10.1016/j.bbrc.2008.04.007
83. Palsson-McDermott EM, Doyle SL, McGettrick AF, Hardy M, Husebye H, Banahan K, et al. TAG, a splice variant of the adaptor TRAM, negatively regulates the adaptor MyD88–independent TLR4 pathway. Nat Immunol (2009) 10(6):579–86. doi: 10.1038/ni.1727
84. Qiu L, Song L, Yu Y, Zhao J, Wang L, Zhang Q. Identification and expression of TRAF6 (TNF receptor-associated factor 6) gene in zhikong scallop. Chlamys farreri Fish Shellfish Immunol (2009) 26:359–67. doi: 10.1016/j.fsi.2008.10.010
85. Ni S, Yu Y, Wei J, Zhou L, Wei S, Yan Y, et al. MicroRNA-146a promotes red spotted grouper nervous necrosis virus (RGNNV) replication by targeting TRAF6 in orange spotted grouper, Epinephelus coioides. Fish Shellfish Immunol (2018) 72:9–13. doi: 10.1016/j.fsi.2017.10.020
86. Schenten D, Medzhitov R. The control of adaptive immune responses by the innate immune system. Adv Immunol (2011) . 109:87–124. doi: 10.1016/B978-0-12-387664-5.00003-0
87. Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol (2005) 6(11):1087–95. doi: 10.1038/ni1255
88. Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB. TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J Mol Biol (2003) 326(1):105–15. doi: 10.1016/S0022-2836(02)01404-3
89. Irie T, Muta T, Takeshige K. TAK1 mediates an activation signal from toll-like receptor(s) to nuclear factor-κB in lipopolysaccharide-stimulated macrophages. FEBS Lett (2000) 467(2-3):160–4. doi: 10.1016/S0014-5793(00)01146-7
90. Barton GM, Kagan JC. A cell biological view of toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol (2009) 9(8):535–42. doi: 10.1038/nri2587
91. Zhang L, Chu Q, Chang R, Xu T. Inducible microRNA-217 inhibits NF-κB– and IRF3-driven immune responses in lower vertebrates through targeting TAK1. J Immunol (2020) 205(6):1620–32. doi: 10.4049/jimmunol.2000341
92. Huang Y, Han K, Wang W, Ren Q. Host microRNA-217 promotes white spot syndrome virus infection by targeting tube in the Chinese mitten crab (Eriocheir sinensis). Front Cell Infect Microbiol (2017) 7:164. doi: 10.3389/fcimb.2017.00164
93. Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science (2003) 301(5633):640–3. doi: 10.1126/science.1087262
94. Wan J, Sun L, Mendoza JW, Chui YL, Huang DP, Chen ZJ, et al. Elucidation of the c-jun n-terminal kinase pathway mediated by Epstein-Barr virus-encoded latent membrane protein 1. Mol Cell Biol (2004) 24(1):192–9. doi: 10.1128/MCB.24.1.192-199.2004
95. Grilli M, Chiu JJ-S, Lenardo MJ. NF-κB and rel: participants in a multiform transcriptional regulatory system. Int Rev Cytol (1993) . 143:1–62. doi: 10.1016/S0074-7696(08)61873-2
96. Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa b family: intimate tales of association and dissociation. Genes Dev (1995) 9(22):2723–35. doi: 10.1101/gad.9.22.2723
97. Baker RG, Hayden MS, Ghosh S. NF-κB, inflammation, and metabolic disease. Cell Metab (2011) 13(1):11–22. doi: 10.1016/j.cmet.2010.12.008
98. Xu T, Chu Q, Cui J, Huo R. MicroRNA-216a inhibits NF-κB-mediated inflammatory cytokine production in teleost fish by modulating p65. Infect Immun (2018) 86(6):e00256–18. doi: 10.1128/IAI.00256-18
99. Girardin SE, Boneca IG, Carneiro LAM, Antignac A, Jéhanno M, Viala J, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science (2003) 300(5625):1584–7. doi: 10.1126/science.1084677
100. Chu Q, Bi D, Zheng W, Xu T. MicroRNA negatively regulates NF-κB-mediated immune responses by targeting NOD1 in the teleost fish Miichthys miiuy. Sci China Life Sci (2021) 64(5):803–15. doi: 10.1007/s11427-020-1777-y
101. Su H, Chang R, Zheng W, Sun Y, Xu T. microRNA-210 and microRNA-3570 negatively regulate NF-κB-mediated inflammatory responses by targeting RIPK2 in teleost fish. Front Immunol (2021) 12:617753. doi: 10.3389/fimmu.2021.617753
102. Bi D, Gao Y, Chu Q, Cui J, Xu T. NOD1 is the innate immune receptor for iE-DAP and can activate NF-κB pathway in teleost fish. Dev Comp Immunol (2017) 76:238–46. doi: 10.1016/j.dci.2017.06.012
103. Bi D, Wang Y, Gao Y, Li X, Chu Q, Cui J, et al. Recognition of lipopolysaccharide and activation of NF-κB by cytosolic sensor NOD1 in teleost fish. Front Immunol (2018) 9:1413. doi: 10.3389/fimmu.2018.01413
104. Li J, Gao Y, Xu T. Comparative genomic and evolution of vertebrate NOD1 and NOD2 genes and their immune response in miiuy croaker. Fish Shellfish Immunol (2015) 46:387–97. doi: 10.1016/j.fsi.2015.06.026
105. Swain B, Basu M, Samanta M. NOD1 and NOD2 receptors in mrigal (Cirrhinus mrigala): inductive expression and downstream signalling in ligand stimulation and bacterial infections. J Biosci (2013) 38(3):533–48. doi: 10.1007/s12038-013-9330-y
106. Wu X, Zhang J, Li P, Hu Y, Cao L, Ouyang S, et al. NOD1 promotes antiviral signaling by binding viral RNA and regulating the interaction of MDA5 and MAVS. J Immunol (2020) 204(8):2216–31. doi: 10.4049/jimmunol.1900667
107. Magalhaes JG, Lee J, Geddes K, Rubino S, Philpott DJ, Girardin SE. Essential role of Rip2 in the modulation of innate and adaptive immunity triggered by Nod1 and Nod2 ligands. Eur J Immunol (2011) 41(5):1445–55. doi: 10.1002/eji.201040827
108. Fang H, Wu X, Hu Y, Song Y, Zhang J, Chang M. NLRC3-like 1 inhibits NOD1-RIPK2 pathway via targeting RIPK2. Dev Comp Immunol (2020) 112:103769. doi: 10.1016/j.dci.2020.103769
109. Xie J, Belosevic M. Functional characterization of receptor-interacting serine/threonine kinase 2 (RIP2) of the goldfish (Carassius auratus l.). Dev Comp Immunol (2015) 48(1):76–85. doi: 10.1016/j.dci.2014.09.006
110. Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol (2007) 178(4):2380–6. doi: 10.4049/jimmunol.178.4.2380
111. Hou J, Pang Y, Li Q. Comprehensive evolutionary analysis of lamprey TNFR-associated factors (TRAFs) and receptor-interacting protein kinase (RIPKs) and insights into the functional characterization of TRAF3/6 and RIPK1. Front Immunol (2020) 11:663. doi: 10.3389/fimmu.2020.00663
112. Baccala R, Kono DH, Theofilopoulos AN. Interferons as pathogenic effectors in autoimmunity. Immunol Rev (2005) 204:9–26. doi: 10.1111/j.0105-2896.2005.00252.x
113. Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, et al. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem (2007) 282(10):7576–81. doi: 10.1074/jbc.M608618200
114. Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med (2006) 203(7):1795–803. doi: 10.1084/jem.20060792
115. Chang R, Zheng W, Sun Y, Geng S, Xu T. Long noncoding RNA MIR2187HG suppresses TBK1-mediated antiviral signaling by deriving miR-2187-3p in teleost fish. J Virol (2022) 96(1):e01484–21. doi: 10.1128/JVI.01484-21
116. Biacchesi S, LeBerre M, Lamoureux A, Louise Y, Lauret E, Boudinot P, et al. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J Virol (2009) 83(16):7815–27. doi: 10.1128/JVI.00404-09
117. Lauksund S, Svingerud T, Bergan V, Robertsen B. Atlantic Salmon IPS-1 mediates induction of IFNa1 and activation of NF-κB and localizes to mitochondria. Dev Comp Immunol (2009) 33(11):1196–204. doi: 10.1016/j.dci.2009.06.012
118. Ran Y, Shu H-B, Wang Y-Y. MITA/STING: A central and multifaceted mediator in innate immune response. Cytokine Growth Factor Rev (2014) 25(6):631–9. doi: 10.1016/j.cytogfr.2014.05.003
119. Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKϵ and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol (2003) 4(5):491–6. doi: 10.1038/ni921
120. Yoo JS, Kato H, Fujita T. Sensing viral invasion by RIG-I like receptors. Curr Opin Microbiol (2014) 20:131–8. doi: 10.1016/j.mib.2014.05.011
121. Tang X, Huang B, Zhang L, Li L, Zhang G. TANK-binding kinase-1 broadly affects oyster immune response to bacteria and viruses. Fish Shellfish Immunol (2016) 56:330–5. doi: 10.1016/j.fsi.2016.07.011
122. Zhang L, Chen W, Hu Y, Wu X, Nie P, Chang M. TBK1-like transcript negatively regulates the production of IFN and IFN-stimulated genes through RLRs-MAVS-TBK1 pathway. Fish Shellfish Immunol (2016) 54:135–43. doi: 10.1016/j.fsi.2016.04.002
123. Pasqualotto A, Ayres R, Longo L, Del Duca Lima D, Losch de Oliveira D, Alvares-da-Silva MR, et al. Chronic exposure to ethanol alters the expression of miR-155, miR-122 and miR-217 in alcoholic liver disease in an adult zebrafish model. Biomarkers (2021) 26(2):146–51. doi: 10.1080/1354750X.2021.1874051
124. Qiang J, Tao YF, He J, Xu P, Bao JW, Sun YL. miR-122 promotes hepatic antioxidant defense of genetically improved farmed tilapia (GIFT, Oreochromis niloticus) exposed to cadmium by directly targeting a metallothionein gene. Aquat Toxicol (2017) 182:39–48. doi: 10.1016/j.aquatox.2016.11.009
125. Patra K, Rajaswini R, Murmu B, Rasal KD, Sahoo L, Saha A, et al. Identifying miRNAs in the modulation of gene regulation associated with ammonia toxicity in catfish, Clarias magur (Linnaeus, 1758). Mol Biol Rep (2022) 49(7):6249–59. doi: 10.1007/s11033-022-07424-y
126. Koganti PP, Wang J, Cleveland B, Ma H, Weber GM, Yao J. Estradiol regulates expression of miRNAs associated with myogenesis in rainbow trout. Mol Cell Endocrinol (2017) 443:1–14. doi: 10.1016/j.mce.2016.12.014
127. Zheng Z, Huang R, Tian R, Jiao Y, Du X. Pm-miR-133 hosting in one potential lncRNA regulates RhoA expression in pearl oyster Pinctada martensii. Gene (2016) 591(2):484–9. doi: 10.1016/j.gene.2016.06.051
128. Tao L, Pang Y, Wang A, Li L, Shen Y, Xu X, et al. Functional miR-142a-3p induces apoptosis and macrophage polarization by targeting tnfaip2 and glut3 in grass carp (Ctenopharyngodon idella). Front Immunol (2021) 12:633324. doi: 10.3389/fimmu.2021.633324
129. Lu X, Wei Y, Liu F. Direct regulation of p53 by miR-142a-3p mediates the survival of hematopoietic stem and progenitor cells in zebrafish. Cell Discovery (2015) 1:15027. doi: 10.1038/celldisc.2015.27
130. Wang X, Zheng Y, Ma Y, Du L, Chu F, Gu H, et al. Lipid metabolism disorder induced by up-regulation of miR-125b and miR-144 following β-diketone antibiotic exposure to F0-zebrafish (Danio rerio). Ecotoxicol Environ Saf (2018) 164:243–52. doi: 10.1016/j.ecoenv.2018.08.027
131. Chen C, Zhang J, Zhang M, You C, Liu Y, Wang S, et al. miR-146a is involved in the regulation of vertebrate LC-PUFA biosynthesis by targeting elovl5 as demonstrated in rabbitfish Siganus canaliculatus. Gene (2018) 676:306–14. doi: 10.1016/j.gene.2018.08.063
132. Zhu X, Li YL, Chen DX, Wu P, Yi T, Chen T, et al. Selection of reference genes for microRNA quantitative expression analysis in Chinese perch, Siniperca chuatsi. Int J Mol Sci (2015) 16(4):8310–23. doi: 10.3390/ijms16048310
133. Zhang D, Lu K, Dong Z, Jiang G, Xu W, Liu W. The effect of exposure to a high-fat diet on microRNA expression in the liver of blunt snout bream (Megalobrama amblycephala). PloS One (2014) 9(5):e96132. doi: 10.1371/journal.pone.0096132
134. Zhao C, Xie R, Qian Q, Yan J, Wang H, Wang X. Triclosan induced zebrafish immunotoxicity by targeting miR-19a and its gene socs3b to activate IL-6/STAT3 signaling pathway. Sci Total Environ (2022) 815:152916. doi: 10.1016/j.scitotenv.2022.152916
135. Zhong H, Zhou Y, Xu Q, Yan J, Zhang X, Zhang H, et al. Low expression of miR-19a-5p is associated with high mRNA expression of diacylglycerol O-acyltransferase 2 (DGAT2) in hybrid tilapia. Genomics (2021) 113(4):2392–9. doi: 10.1016/j.ygeno.2021.05.016
136. Zhang Q, Zheng S, Wang S, Wang W, Xing H, Xu S. Chlorpyrifos induced oxidative stress to promote apoptosis and autophagy through the regulation of miR-19a-AMPK axis in common carp. Fish Shellfish Immunol (2019) 93:1093–9. doi: 10.1016/j.fsi.2019.07.022
137. Bizuayehu TT, Furmanek T, Karlsen Ø, van der Meeren T, Edvardsen RB, Rønnestad I, et al. First feed affects the expressions of microRNA and their targets in Atlantic cod. Br J Nutr (2016) 115(7):1145–54. doi: 10.1017/S0007114516000155
138. Rajaram K, Harding RL, Hyde DR, Patton JG. miR-203 regulates progenitor cell proliferation during adult zebrafish retina regeneration. Dev Biol (2014) 392(2):393–403. doi: 10.1016/j.ydbio.2014.05.005
139. Wang W, Liu Q, Zhang T, Chen L, Li S, Xu S. Glyphosate induces lymphocyte cell dysfunction and apoptosis via regulation of miR-203 targeting of PIK3R1 in common carp (Cyprinus carpio l.). Fish Shellfish Immunol (2020) 101:51–7. doi: 10.1016/j.fsi.2020.03.047
140. Liu S, Yang Q, Chen Y, Liu Q, Wang W, Song J, et al. Integrated analysis of mRNA- and miRNA-seq in the ovary of rare minnow Gobiocypris rarus in response to 17α-methyltestosterone. Front Genet (2021) 12:695699. doi: 10.3389/fgene.2021.695699
141. Hoppe B, Pietsch S, Franke M, Engel S, Groth M, Platzer M, et al. MiR-21 is required for efficient kidney regeneration in fish. BMC Dev Biol (2015) 15:43. doi: 10.1186/s12861-015-0089-2
142. Zhao Y, Wu JW, Wang Y, Zhao JL. Role of miR-21 in alkalinity stress tolerance in tilapia. Biochem Biophys Res Commun (2016) 471(1):26–33. doi: 10.1016/j.bbrc.2016.02.007
143. Tao L, Xu X, Fang Y, Wang A, Zhou F, Shen Y, et al. miR-21 targets jnk and ccr7 to modulate the inflammatory response of grass carp following bacterial infection. Fish Shellfish Immunol (2019) 94:258–63. doi: 10.1016/j.fsi.2019.09.022
144. Wu Y, Lou QY, Ge F, Xiong Q. Quantitative proteomics analysis reveals novel targets of miR-21 in zebrafish embryos. Sci Rep (2017) 7(1):4022. doi: 10.1038/s41598-017-04166-x
145. Valenzuela-Muñoz V, Novoa B, Figueras A, Gallardo-Escárate C. Modulation of atlantic salmon miRNome response to sea louse infestation. Dev Comp Immunol (2017) 76:380–91. doi: 10.1016/j.dci.2017.07.009
146. Latimer M, Sabin N, Le Cam A, Seiliez I, Biga P, Gabillard JC. miR-210 expression is associated with methionine-induced differentiation of trout satellite cells. J Exp Biol (2017) 220:2932–8. doi: 10.1242/jeb.154484
147. Tse AC, Li JW, Chan TF, Wu RS, Lai KP. Hypoxia induces miR-210, leading to anti-apoptosis in ovarian follicular cells of marine medaka Oryzias melastigma. Aquat Toxicol (2015) 165:189–96. doi: 10.1016/j.aquatox.2015.06.002
148. Wang W, Zhong P, Yi JQ, Xu AX, Lin WY, Guo ZC, et al. Potential role for microRNA in facilitating physiological adaptation to hypoxia in the pacific whiteleg shrimp Litopenaeus vannamei. Fish Shellfish Immunol (2019) 84:361–9. doi: 10.1016/j.fsi.2018.09.079
149. Zhao Y, Lin Q, Li N, Babu VS, Fu X, Liu L, et al. MicroRNAs profiles of Chinese perch brain (CPB) cells infected with Siniperca chuatsi rhabdovirus (SCRV). Fish Shellfish Immunol (2019) 84:1075–82. doi: 10.1016/j.fsi.2018.11.020
150. Olena AF, Rao MB, Thatcher EJ, Wu SY, Patton JG. miR-216a regulates snx5, a novel notch signaling pathway component, during zebrafish retinal development. Dev Biol (2015) 400(1):72–81. doi: 10.1016/j.ydbio.2015.01.016
151. Zhang J, Zheng S, Wang S, Liu Q, Xu S. Cadmium-induced oxidative stress promotes apoptosis and necrosis through the regulation of the miR-216a-PI3K/AKT axis in common carp lymphocytes and antagonized by selenium. Chemosphere (2020) 258:127341. doi: 10.1016/j.chemosphere.2020.127341
152. Li H, Di G, Zhang Y, Liang J, Wang X, Xu Z, et al. miR-217 through SIRT1 regulates the immunotoxicity of cadmium in Cyprinus carpio. Comp Biochem Physiol C Toxicol Pharmacol (2021) 248:109086. doi: 10.1016/j.cbpc.2021.109086
153. Shwe A, Krasnov A, Visnovska T, Ramberg S, Østbye TK, Andreassen R. Differential expression of miRNAs and their predicted target genes indicates that gene expression in Atlantic salmon gill is post-transcriptionally regulated by miRNAs in the parr-smolt transformation and adaptation to sea water. Int J Mol Sci (2022) 23(15):8831. doi: 10.3390/ijms23158831
154. Cadonic IG, Ikert H, Craig PM. Acute air exposure modulates the microRNA abundance in stress responsive tissues and circulating extracellular vesicles in rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol Part D Genomics Proteomics (2020) 34:100661. doi: 10.1016/j.cbd.2020.100661
155. Bai Y, Long J, Liu Z, Lin J, Huang H, Wang D, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol (2019) 234(10):18837–48. doi: 10.1002/jcp.28522
156. Yan Y, Yu J, Liu H, Guo S, Zhang Y, Ye Y, et al. Construction of a long non-coding RNA-associated ceRNA network reveals potential prognostic lncRNA biomarkers in hepatocellular carcinoma. Pathol Res Pract (2018) 214(12):2031–8. doi: 10.1016/j.prp.2018.09.022
157. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell (2011) 146(3):353–8. doi: 10.1016/j.cell.2011.07.014
158. Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell (2014) 56(3):347–59. doi: 10.1016/j.molcel.2014.09.018
159. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol (2019) 234(7):10080–100. doi: 10.1002/jcp.27941
160. Arora S, Singh P, Dohare R, Jha R, Ali Syed M. Unravelling host-pathogen interactions: ceRNA network in SARS-CoV-2 infection (COVID-19). Gene (2020) 762:145057. doi: 10.1016/j.gene.2020.145057
161. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell (2011) 43(6):904–14. doi: 10.1016/j.molcel.2011.08.018
162. Chu Q, Xu T, Zheng W, Chang R, Zhang L. Long noncoding RNA AANCR modulates innate antiviral responses by blocking miR-210-dependent MITA downregulation in teleost fish, Miichthys miiuy. Sci China Life Sci (2021) 64(7):1131–48. doi: 10.1007/s11427-020-1789-5
163. Li X. IRAK4 in TLR/IL-1R signaling: Possible clinical applications. Eur J Immunol (2008) 38(3):614–8. doi: 10.1002/eji.200838161
164. Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, Sandercock AM, et al. An oligomeric signaling platform formed by the toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem (2009) 284(37):25404–11. doi: 10.1074/jbc.M109.022392
165. Lin S, Lo Y, Wu H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature (2010) 465(7300):885–90. doi: 10.1038/nature09121
166. Kollewe C, Mackensen A, Neumann D, Knop J, Cao P, Li S, et al. Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J Biol Chem (2004) 279(7):5227–36. doi: 10.1074/jbc.M309251200
167. Qian Y, Commane M, Ninomiya-Tsuji J, Matsumoto K, Li X. IRAK-mediated translocation of TRAF6 and TAB2 in the interleukin-1-induced activation of NFκB. J Biol Chem (2001) 276(45):41661–7. doi: 10.1074/jbc.M102262200
168. Zou P, Huang X, Yao C, Sun Q, Li Y, Zhu Q, et al. Cloning and functional characterization of IRAK4 in large yellow croaker (Larimichthys crocea) that associates with MyD88 but impairs NF-κB activation. Fish Shellf Immunol (2017) 63:452–64. doi: 10.1016/j.fsi.2016.12.019
169. Wu C, Xu X, Zhi X, Jiang Z, Li Y, Xie X, et al. Identification and functional characterization of IRAK-4 in grass carp (Ctenopharyngodon idellus). Fish Shellfish Immunol (2019) 87:438–48. doi: 10.1016/j.fsi.2019.01.031
170. Han X, Gao F, Lu M, Liu Z, Wang M, Ke X, et al. Molecular characterization, expression and functional analysis of IRAK1 and IRAK4 in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol (2020) 97:135–45. doi: 10.1016/j.fsi.2019.12.041
171. Vazquez C, Horner SM. MAVS coordination of antiviral innate immunity. J Virol (2015) 89(14):6974–7. doi: 10.1128/JVI.01918-14
172. Abe T, Barber GN. Cytosolic-DNA-Mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J Virol (2014) 88(10):5328–41. doi: 10.1128/JVI.00037-14
173. Suzuki H. Characterization of RNase r-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res (2006) 34(8):e63–3. doi: 10.1093/nar/gkl151
174. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature (2014) 505(7483):344–52. doi: 10.1038/nature12986
175. Zhao N, Deng Q, Zhu C, Zhang B. Application of extracellular vesicles in aquatic animals: a review of the latest decade. Rev Fish Sci Aquac (2022) 30(4):447–66. doi: 10.1080/23308249.2021.1985429
176. Becker CE, O’Neill LAJ. Inflammasomes in inflammatory disorders: the role of TLRs and their interactions with NLRs. Semin Immunopathol (2007) 29(3):239–48. doi: 10.1007/s00281-007-0081-4
Keywords: nuclear factor-κB, microRNA, long noncoding RNA, circular RNA, ceRNA network
Citation: Zhao T, Zou Y, Yan H, Chang Y and Zhan Y (2023) Non-coding RNAs targeting NF-κB pathways in aquatic animals: A review. Front. Immunol. 14:1091607. doi: 10.3389/fimmu.2023.1091607
Received: 07 November 2022; Accepted: 20 January 2023;
Published: 06 February 2023.
Edited by:
Saba Valadkhan, Case Western Reserve University, United StatesReviewed by:
Khalil Eslamloo, Memorial University of Newfoundland, CanadaYong Huang, Henan University of Science and Technology, China
Copyright © 2023 Zhao, Zou, Yan, Chang and Zhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoyao Zhan, enl5ZGxvdUBob3RtYWlsLmNvbQ==; Yaqing Chang, eXFrZXlsYWJAaG90bWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
 Tanjun Zhao
Tanjun Zhao Yang Zou
Yang Zou Hanyu Yan
Hanyu Yan Yaqing Chang
Yaqing Chang Yaoyao Zhan
Yaoyao Zhan