
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 30 January 2023
Sec. Microbial Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1091165
This article is part of the Research Topic Ecology, Environment, and Human Microbiome Interaction with Infection View all 10 articles
Lung cancer is the leading cause of cancer-related deaths worldwide, and insights into its underlying mechanisms as well as potential therapeutic strategies are urgently needed. The microbiome plays an important role in human health, and is also responsible for the initiation and progression of lung cancer through its induction of inflammatory responses and participation in immune regulation, as well as for its role in the generation of metabolic disorders and genotoxicity. Here, the distribution of human microflora along with its biological functions, the relationship between the microbiome and clinical characteristics, and the role of the microbiome in clinical treatment of lung cancer were comprehensively reviewed. This review provides a basis for the current understanding of lung cancer mechanisms with a focus on the microbiome, and contributes to future decisions on treatment management.
Lung cancer has the highest morbidity and mortality worldwide, with approximately 2 million new cases and 1.76 million deaths in 2021 (1, 2). In recent years, researchers have found that more than 16% of cancer cases are related to infections, and most infections are caused by microorganisms (3). The relationship between microbes and cancer has attracted considerable attention in academia. Bacteria were first discovered in tumors over a hundred years ago, and the existence of microorganisms in various tumors has been successively reported (4, 5). Healthy lungs are traditionally thought to be sterile, but recent studies have found that they also harbor microbial communities, including Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria (6, 7). In addition, early epidemiological data have suggested that bacterial infections are common in lung cancer patients, especially as the disease progresses, and it is almost 50% to 70%. The pathogenic bacteria initially colonizing the lung might persist in patients with lung cancer as the disease progresses (8). Furthermore, the microflora residing outside the lung, such as the oral cavity, airways and gut, can also affect the occurrence and development of lung cancer, suggesting that the human microflora may play a direct or indirect role in lung cancer onset and progression. This article reviews the role of the human microbiome in lung cancer as well as providing a basis for a potential role of the microbiome in therapeutic methods and drug discovery of lung cancer.
Humans coexist with and host a variety of microbes, such as bacteria, fungi, and viruses. All these microorganisms inhabiting specific areas of the human body constitute the human microbiota, which plays an important role in physiological activities such as nutrient absorption, substance metabolism, and immune regulation, and is also closely related to the occurrence of diseases such as infectious diseases, metabolic disorders, and different cancer types.
The oral cavity contains more than 700 species of bacteria. Oral microorganisms reside in biofilms throughout the mouth and form an ecosystem that helps to maintain a healthy microenvironment. The oral microbiota was composed of Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Spirrochaetes, and Fusobacteria, accounting for 94% of the total classification. The remaining phyla, such as Saccharibacteria, Synergistetes, SR1, Gracilibacteria, Chlamydia, Chloroflexi, Tenericutes, and Chlorobi, account for 6% of the taxa. The oral microbiome is impressive in its breadth and depth: one milliliter of saliva contains 1.0×108 microbial cells and 700 different prokaryotic taxa. Among these, it contains bacteria, fungi, viruses, archaea, and protozoa, of which approximately 54% are culturable and have been identified, 14% are culturable and not identified, and 32% are unculturable (9).
When the human microbial group plan was launched in 2007, the lungs were not included among the sampled organs, in part as they were thought to be sterile (10). With the increasing development and popularity of high-throughput sequencing and sequence assembly technology, together with databases of sequenced organisms (11, 12), the identification and quantification of organisms from mixed metagenomic samples has been possible through high-throughput metagenomic sequencing, a convenient, and so far the fastest strategy for the study of lung microbes (13). Respiratory microbes grow rapidly in early life of the host and are influenced by the environment, age, and immune status of the host (14). Indeed, it has been proven that birth, the first postnatal hour, and the first 3 to 4 months of exposure to the living environment are important stages for a stable development of respiratory flora (15).
In healthy lungs, two phyla are mainly present, Bacteroidetes and Firmicutes, which constitute the pulmonary microbiota, whereas Prevotella and Veillonella spp. are dominant (16–18). Compared to the upper respiratory tract, the microbiota of the lung mucosa is phylogenetically diverse. In addition, the lower respiratory system is mainly composed of Pseudomonas, Streptococcus, Fusobacterium, Megacoccus and Sphingosphingomonas (18, 19). Some studies have shown that the lungs are susceptible to oropharyngeal bacterial colonies (16, 17, 20). For example, Bassis et al. compared the microbial composition in the oral and nasal cavities, lungs, and stomach of healthy adults and found that the microbial communities in gastric juices and alveolar lavage fluid (BAL) were mainly derived from the inhalation and colonization of oropharyngeal flora (21).
The gut provides a convenient habitat for all kinds of microorganisms, with comprise an estimated total of 1.0×1013 ˜ 1.0×1014. The human gut microbiota is composed of at least 1000 - 1200 species of bacteria, mainly Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia, and others. Among these, Firmicutes (64%) and Bacteroidetes (28%) were the main components in most individuals. Actinobacteria, Proteobacteria, and Verrucomicrobia were minor components. The human gut microbiome is extremely large and scientists have not been able to determine the number of gut microbes that people may carry. It has been estimated that a 70 kg adult (3.0×1013 cells) carries approximately 3.8×1013 bacteria (0.2 kg) (22, 23).
The composition of the human gut microbiota varies among populations, and the difference in individual composition is mainly reflected in the proportion of bacteria of each phylum. The diversity of species of gut microbiota in humans increases with time, mostly during the first three years (approximately 100 species in the first few weeks of life, 700 between six months and three years of age, and 1,000 in adulthood). Agedness is another stage at which the gut microbiota changes dramatically. At this stage, the number of facultative bacteria increases, the ratio of Bacteroidetes to Firmicutes increases, and that of Bifidobacterium decreases. Claesson et al. reported that, compared with young people, the differences in gut microbiota composition especially in Ruminococcaceae family (comprised of Ruminococcus, Sporobacter, and Faecalibacterium species), among individuals was significantly higher in the elderly, and the Bifidobacterium proportions, the Clostridium cluster IV, as well as the species diversity within each individual was significantly reduced, which is likely related to diet, health status, and immune system decay (24, 25).
Several decades ago, concentrations of intestinal bile acid were found to be much higher in breast cyst fluid than in serum in women with fibrocystic breast disease (26–28). Although the mechanisms for maintaining high bile concentrations within breast cysts remain to be studied, these studies suggest that breast tissue, like other parts of the body, is composed of an unique microbiome. Bacteria can also be detected in breast milk, possibly because microbes can travel from the surface of the skin into ducts and breast tissue. The most common genera in milk are Staphylococcus, Streptococcus, Lactobacillus, Pseudomonas, Bifidobacterium, Corynebacterium, Enterococcus, Acinetobacter, Rothia, Cutibacterium, Veillonella and Bacteroides (29). Urbaniak et al. studied the differences in microbial communities in 81 pairs of patients with and without breast cancer, and found that Proteobacteria composition was different between them, together with regional divergence (30).
The stomach has generally been considered to have fewer symbiotic bacteria because of its highly acidic environment and high protein hydrolase content (31, 32). However, recent studies have shown that a wide variety of bacteria can be found in the human stomach. Firmicutes and Proteobacteria are the major phyla, and Streptococcus and Prevotella are the major species in the stomach of individuals without Helicobacter pylori (HB) (33, 34) (Figure 1). Infection with HB can disturb the microbial community in the stomach (35).
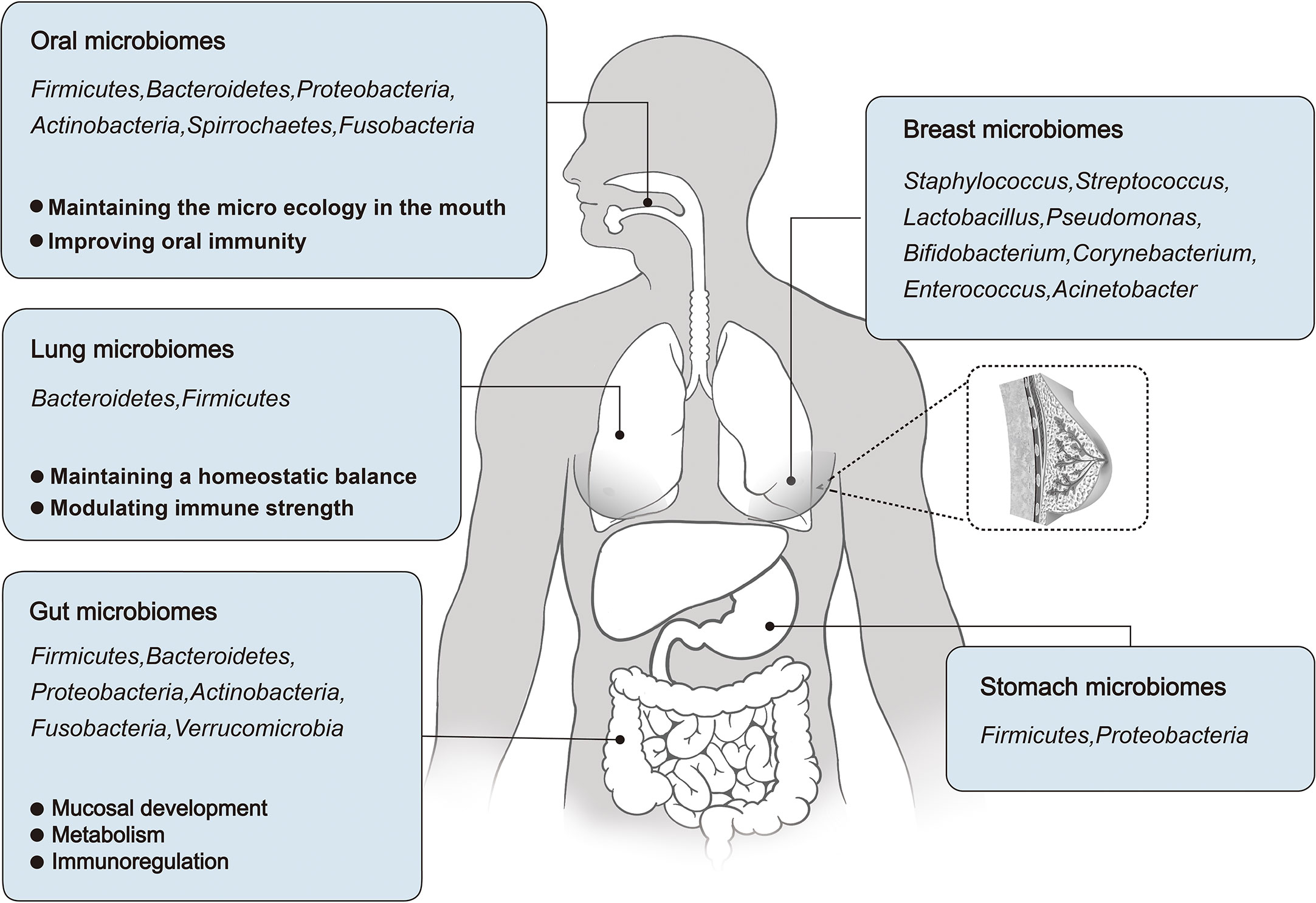
Figure 1 An overview of the microbial distribution in human body, and the roles of Oral, Lung, and Gut microbiomes in human development and physiological function.
In normal prostate, estimations concerning the microorganism number and composition are difficult since access to non-diseased prostate tissue is restricted. However, a number of previous studies have characterized the microbial composition in prostate cancer and normal surgically resected specimens, and found that no bacteria were present in normal prostate tissue (36–38). On the contrary, one study detected a positive result for bacteria in tissue specimens of benign prostatic hyperplasia (BPH) (39). However, it cannot be discarded that the positive result may owe to contamination (40). In addition, normal prostatic fluid may prevent microbial growth because of its highly antibacterial properties. Microbial invasion occurs only in the prostate upon prostatitis or other pathological occurrences (41).
The microbiome plays an important role in human development and physiology. In this context, changes in the oral microbiome may cause oral and systemic diseases (42) and an imbalance in the respiratory microbiome may affect the occurrence of lung diseases (43). The gut microbiome accounts for a relatively high proportion of the human body, and its functions have been fully studied, including nutrient metabolism and immune regulation. The following sections focus on the role of oral, respiratory, and intestinal microbiota in human development and physiological function (Figure 1).
The human oral microecosystem contains a large diversity of microorganisms, including bacteria, fungi, viruses, mycoplasma and protozoa. Of these, bacteria (about 700 species) make up the majority of the healthy oral microbiome and are mainly composed of six phyla, including Firmicutes, Actinobacteria, Proteobacteria, Fusobacteria, Bacteroides and Spirochaetas (44). In addition to bacteria, about 100 fungi also make up an important part of the oral microbiome, of which Candida is the most common. In the oral microecosystem, microbes such as bacteria and fungi attach to the surface of teeth and form a biofilm called plaque with the surrounding extracellular matrix in order to protect themselves from fluctuations in the oral environment and external drug stimuli and evade host defense mechanisms (45).
The balance of oral microecosystem not only contributes to the maintenance of oral health, but also has a potential impact on the overall health. Microorganisms in oral microecosystems achieve dynamic balance between each other and the host through complex interspecific interactions such as symbiosis, competition and confrontation (46). This paper summarizes the physiological function of normal microbial flora in oral cavity.
The normal microflora in oral cavity can maintain the microecological balance well. When pathogenic bacteria such as P. seudomonas aeruginosa invade, the oral flora inhibits their growth in saliva by producing lactic acid (47). Therefore, the normal oral flora plays an important role in preventing the invasion of pathogenic bacteria. However, disturbances in oral microecology such as oral flora imbalance or reduction of oral symbiotic bacteria provide opportunities for the invasion and colonization of respiratory pathogens such as Staphylococcus aureus, Pseudococcus aeruginosa, Enterococcus faecalis and Acinetobacter (48–51).
Natural aging, hypoplasia of parotid and submandibular glands, and medications (antihypertensive drugs, anticholinergics) can alter saliva composition or affect saliva secretion or flow rate, leading to dry mouth and poor oral hygiene (52). This may lead to the transfer of normal oral flora to communities containing more pathogens (53).
The respiratory tract is a complex organ system whose main function is the exchange of oxygen and carbon dioxide. It is divided into the upper respiratory tract, which includes the nasal passages, pharynx, larynx, and lower respiratory tract, which includes the conducting airways (trachea and bronchi), small airways (bronchioles), and respiratory areas (alveoli). Because the respiratory tract is connected to the outside world, a large number of airborne microorganisms and particles, including viruses, bacteria and fungi, continue to migrate or be removed from the respiratory tract. The bacterial burden of the upper respiratory tract is about 100-10000 times than that of the lower respiratory tract, and the nasal cavity is dominated by Propionibacterium, Corynebacterium, Staphylococcus and Moraxella. Prevotella, Vermicelli, Streptococcus, Haemophilus, Fusobacterium, Neisseria and Corynebacterium were predominant in oral cavity (54, 55). Prevotella, Vermicelli, and Streptococcus colonize in the lower respiratory tract, and these microbial compositions differ from those observed in the oral and nasal cavities (56). As mentioned earlier, the gut microbiome of young children stabilizes at about 3 years of age, similar to that of adults, and this pattern of community maturation is reproduced in the upper respiratory tract microbiome (14, 57, 58). The following is a comprehensive summary of the physiological function of respiratory microorganisms in human body.
The respiratory tract is the main site of continuous contact with exogenous microorganisms. Airway epithelium acts as a sensor for the presence of microorganisms, and its epithelial cells are in constant contact with the environment. This interaction is a key factor in maintaining stable homeostasis. The environmental conditions necessary for microbial growth in the respiratory tract (such as PH, temperature, nutrition, oxygen tension, and activation of inflammatory cells in the host) are heterogeneous, so considerable regional variation can be observed in a single healthy lung (59).
In health conditions, the microbiome can also regulate immune strength. Symbiotic fungi have been shown to influence the immune system and regulate the bacterial community, thus contributing to the recovery of bacterial flora after antibiotic treatment (60, 61).
Gut microorganisms can affect intestinal mucosal development and homeostasis. Comparative studies of conventional and germ-free animals have shown that the gut microbiota is essential for the formation and functional realization of the intestinal mucosal immune system during infancy (62). A poor development of villous capillaries in the infancy of sterile mice and a consequential still dysplasia in adulthood confirmed that the gut microbiota contributes to the formation of the intestinal immune ultrastructure (63). The gut microbiome also contributes to the development of intestinal intraepithelial lymphocytes (IILs). Compared with conventionally grown animals, the production of intestinal mucosal-associated lymphoid tissue and antibodies was strongly reduced, and the original center, cell lamina propria, and cell lymphoid follicles of the mesenteric lymph node were significantly decreased in germ-free animals. Meanwhile, gut microbiota plays an important immunomodulatory role in intestinal mucosal homeostasis, with direct consequences in human health (64, 65).
Gut microbiota improves nutrient metabolism. The gut is an important site of digestion and absorption in the human body. Here, gut microbiota can contribute in foor digestion and decomposition, also could promote intestinal peristalsis and inhibit the proliferation of pathogenic bacteria. Gut microbiota can also provide various substrates, enzymes, and energy necessary for human metabolism, and participate in metabolic processes. Among them, Firmicutes, Bacteroidetes, and some anaerobic microorganisms can decompose complex carbohydrates in the gut to produce short-chain fatty acids (SCFAs), such as acetic acid, propionic acid, and butyric acid (65–68). SCFAs are not only the energy source of gut microorganisms themselves and the intestinal epithelial cells of the host, participating in adipogenesis and gluconeogenesis, but can also regulate the intestinal immunity of the host, reducing the pH of the colonic environment and inhibiting harmful bacterial growth and colonic inflammation (69).
Most pathogens cannot compete with the resident microbiome for carbohydrate food sources and are therefore effectively excluded from the gut under normal circumstances. Thus, disruption of the gut ecosystem appears to play an important role in the establishment of pathogenic bacteria. For example, antibiotic treatment disrupts the cross-feeding network between mucinous and non-mucinous degradants and allows for pathogenic bacteria such as Salmonella typhimurium and Clostridioides difficile (70).
Besides playing a role in carbohydrate metabolism, gut microorganisms also participate in bile acid metabolism, tryptophan metabolism and other processes. Bile acids are produced in the liver and metabolized by enzymes produced by gut bacteria and are essential for maintaining a healthy gut microbiome, balancing lipid and carbohydrate metabolism, as well as innate immunity. The ability of intestinal flora to convert intestinal bile acid organisms into their unbound forms is critical to gastrointestinal .metabolic homeostasis, and these unbound bile acids activate bile acid signaling receptors (71, 72). The main bacterial genera involved in bile acid metabolism are Bacteroides, Clostridium, Lactobacillus, bifidobacterium and listeria (73, 74). Clinical patients with hepatoenteric diseases often present with intestinal ecological disorders characterized by reduced microbial diversity and a reduced abundance of firmicutes, leading to lower levels of intestinal secondary bile acids and higher levels of conjugated bile acids (75–77). Therefore, bile acid metabolism and intestinal flora interact, and when this balance is disrupted, a variety of clinical disease phenotypes can result.
Tryptophan metabolism is another important function of intestinal microorganisms to promote nutrient metabolism. As a nutrient enhancer, tryptophan plays a crucial role in the balance between intestinal immune tolerance and intestinal flora maintenance. Tryptophan is absorbed in the small intestine, but when it reaches the colon it can be broken down by gut bacteria such as Clostridium sporogenes, Escherichia coli and Lactobacillus to produce various indole derivatives that play an important role in key aspects of bacterial ecological balance (78–80).
Gut microorganisms regulate the human immune system through immune cells and their metabolites. Recent studies have shown that gut microorganisms can over-activate CD8+T cells, which can promote chronic inflammation and T-cell failure (81, 82). Signals from gut microbes also provide appropriate conditions for dendritic cell generation (83). Gut microorganisms can also participate in immune regulation through metabolites, which further guide or influence immune cells. For example, lactic acid and pyruvate, metabolites derived from gut microorganisms, can promote immune responses by inducing G-protein coupled receptor (GPR)-31 to mediate the production of intestinal C-X3-C Motif Chemokine Receptor (CX3CR)-1-positive dendritic cells (84). Furthermore, Odoribacter splanchnicus and Bilophila genus were negatively correlated with tumor necrosis factor (TNF)-α production following lipopolysaccharide (LPS) and C. albicans stimulation. Barnesiella was negatively associated with LPS-and B. fragilis-induced interferon (IFN)-γ production. This included common gut commensals, such as Dorea longicatena and Dorea formicigenerans, where higher species abundance was associated with higher IFN-γ levels in response to C. albicans hyphae. Both species of Dorea can metabolize sialic acids, which are usually found at the end of mucins; and the release of these acids is associated with mucin degradation, and may increase gut permeability. Both Streptococcus parasanguinis and Streptococcus australis were associated with IFN-γ production whereas other species, such as Streptococcus mitis/oralis/pneumoniae, were associated with IL-1β production. Also the correlation of Bifidobacterium pseudocatenulatum and IFN-γ was positive. In contrast, the correlation of Bifidobacterium adolescentis and TNF-α was negatively. In addition, P. distasonis was negatively associated with TNF-α and IL-1β after stimulation with C. albicans hyphae (85–88).
The microbiota may be specifically related to the pathological types of lung cancer tissues (Details in Table 1). Based on histological features, lung cancer can be divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), which can be further divided into adenocarcinoma (AC), squamous cell carcinoma (SCC), and large cell carcinoma (LCC). Klebsiella, Acidovorax, Polarmonas, and Rhodoferax have found to be more frequent in SCLC (89). This was later confirmed by Greathouse et al. (90). Xylobacter, Eufluobacter, and Clostridium were also positively correlated with SCLC occurrence. However, Prevotella and Pseudobutyrivibrio ruminis may be negatively correlated with SCLC (91).
The microbiome can be also used as a biomarker for NSCLC screening. Five bacterial genera showed abnormal abundance in the sputum of patients with NSCLC compared to that of controls (92). Also, contents of Prevotella, Lactobacillus, Rikenellaceae, Treptococcus, Enterobacteriaceae, Oscillospira, and Bacteroides plebeius were significantly higher in the feces of patients with NSCLC than in healthy controls (93). However, Leptum, Faecalibacterium prausnitzii, Ruminococcus, and Clostridia contents were found decreased in patients with NSCLC (94).
Furthermore, there were differences between the microbiomes of patients with SCC and AC. Acidovorax is enriched in SCC with TP53 mutations, but not in AC (90). Significant changes were observed in Capnocytophaga, Selenomonas, Veillonella, and Neisseria in SCC and AC saliva samples, whereas the microbiome of patients with SCC seemed to be more diverse than that of those with AC. Therefore, Acidovorax and Veillonella can be used as sputum biomarkers for SCC diagnosis (92) (95). SCC is specifically associated with Enterobacteriaceae microorganisms (96) (97). Levels of Capnocytophaga and Rothia were also higher in SCC than in AC. However, increases in Capnocytophaga, Selenomonas, Veillonella, and Neisseria were associated with AC (95). Capnocytophaga can be used as a diagnostic biomarker for AC sputum with 72% sensitivity and 85% specificity (92). In addition, Yu et al. observed an increased abundance of Thermus sp. and a decrease in the abundance of Ralstonia sp. In AC (98), whereas Greathouse et al. confirmed that Pseudomonas is specifically present in AC (89). In addition, John Cunningham (JC) virus was observed in tumor tissues and metastatic lymph nodes of patients with AC, suggesting that this virus may be involved in the occurrence of AC (99). Last, Huang et al. found that the number of Veillonella, Megacoccus, Actinomyces and Arthrobacter was significantly higher in AC than in SCC.
The microbiome features are closely associated with the progression of lung cancer (Table 1). In this line, Guo et al. found that Legionella was more abundant in patients with metastatic lung cancer (98). Also, Phascolarctobacterium has been found to be enriched in patients with clinical benefit and has been related to an extension of progression-free survival (PFS), whereas Dialister is more common in patients with progressive disease, and its higher abundance is related to reduction of progression-free and overall survival (OS) (100).
Huang et al. (101) sequenced 33 cases of broncholavage fluid (14 cases of squamous cell carcinoma and 19 cases of adenocarcinoma) and 52 cases of sputum samples (15 cases of squamous cell carcinoma and 37 cases of adenocarcinoma). The results showed that the number of Veillonell, Megasphaera, Actinomyces and Arthrobacter in lung adenocarcinoma without metastasis was significantly higher than that in lung squamous cell carcinoma without metastasis. The contents of Capnocytophaga and Rothia in metastatic lung adenocarcinoma were significantly lower than those in metastatic lung squamous cell carcinoma. Streptococcus content was significantly lower in lung adenocarcinoma with metastasis than in lung adenocarcinoma without metastasis. The contents of Veillonella and Rothia in lung squamous cell carcinoma with metastasis were significantly higher than those in lung squamous cell carcinoma without metastasis. Jungnickel et al. (102) found that the number and volume of metastatic cancer nodules in the lung of mice exposed to Haemophilus paraininfluenzae increased significantly. It is speculated that Haemophilus paraininfluenzae may promote the upregulation of TLR2 or TLR4, induce the high expression of cytokine IL-17C, aggravate the inflammatory response of neutrophils and thus play a role in promoting cancer. In basic experiments (102, 103), it was found that Hemophilus paraininfluenzae in the lung and the imbalance of lung flora promoted the metastasis of mouse cancer cells to the lung, indicating that lung flora was involved in the metastasis of lung cancer. Besides, the lung and gut microbiota may affect the prognosis of patients with lung cancer (104). The potential relationship between the lung microbiome and prognosis of lung cancer has been first demonstrated by Peters. Specifically, the abundance of Koribacteraceae in lung tissue is associated with an increase in relapse-free survival (RFS) and disease-free survival (DFS) in patients with lung cancer. On the contrary, the abundance of Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae was correlated with a decrease in RFS or DFS of lung cancer (105). These further indicated that the dynamic changes of some microflora might be related to the progression of lung cancer.
The human microbiome significantly affects the occurrence and development of lung cancer by regulating tumor cells and the microenvironment (106–108).
Proliferation, invasion, and metastasis are the core biological characteristics of tumor cells (109). The human microbiome can directly or indirectly affect lung cancer cell proliferation, invasion, metastasis, genomic instability, and mutations (Figure 2A).
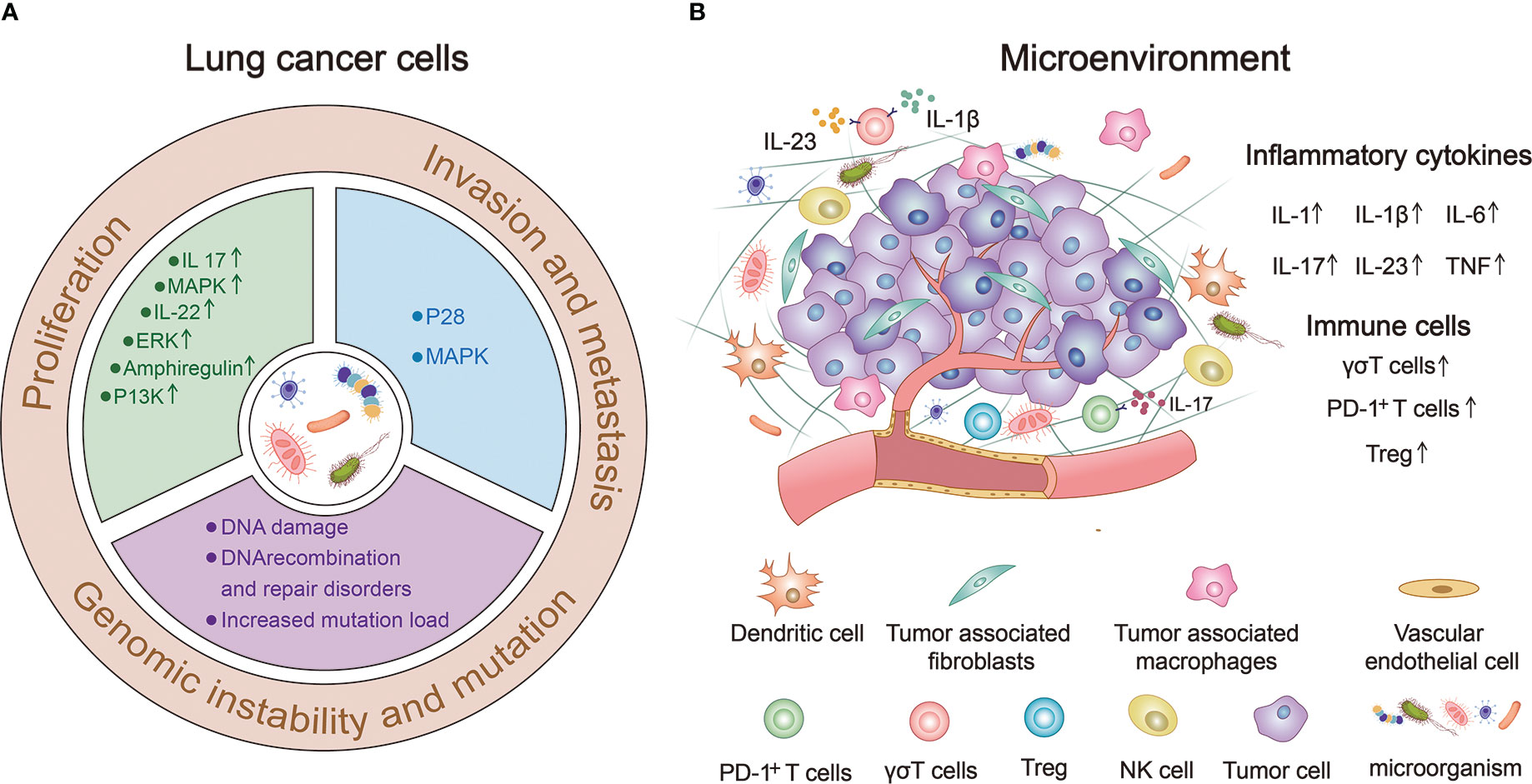
Figure 2 The human microbiome can significantly influence the occurrence and progression of lung cancer. (A) Human microbiota can directly or indirectly affect the proliferation, invasion, metastasis, genomic instability and mutation of lung cancer cells. (B) Human microorganisms participate in the composition of lung cancer microenvironment (TME) and regulate the occurrence and development of lung cancer by up-regulating the expression of immune cells and inflammatory factors.
Enrichment of lower airway microbiota and oral symbiotic bacteria frequently occurs in lung cancer, and these bacteria can trigger the host transcriptome associated with carcinogenesis. Compared with healthy people, extracellular signal-regulated kinase (ERK)- and phosphoinositide 3-kinase (PI3K)-signaling pathways of the lower airway transcriptome in patients with lung cancer are significantly upregulated, which is related to the enrichment of Streptococcus, Prevotella, and Veillonella oral groups in lower airways (110). Recent studies further found that tiny Vibrio is the most abundant microbe that drives the upregulation of interleukin (IL)-17, PI3K, mitogen-activated protein kinase (MAPK), and ERK pathways in the airway transcriptomes of patients with lung cancer and is associated with poor prognosis (111). In human lung cancer, not only is the pulmonary microflora changed, but the local adaptive immune gamma-delta (γδ)-T cells are also activated and directly promote the proliferation of tumor cells through effector molecules such as IL-22 and amphiregulin (103). In addition to the lung microbiota, other bacteria such as HP and its produced urease may also play an important role in lung mucosal proliferation and carcinogenesis. Recently, HP urease was found to enter the lung through gastroesophageal reflux and provide an antigenic trigger for pulmonary granuloma, which leads to subsequent lung mucosal proliferation and carcinogenesis (112).
Changes in the microbiota of patients with lung cancer may contribute to advancing disease progression. The “transition” of microorganisms to Firmicutes in the lower lobe of the lung may be a sign of increased pathogenicity and is associated with poorer prognosis (113). Such low airway microbiota is more common in stage IIIB - IV lung cancer with lymph node metastasis (111). In addition, the gut microbiota plays an important role in the invasion and metastasis of lung cancer. Toll-like receptors (TLRs) on the membrane surface of intestinal epithelial cells are pathogen-related recognition receptors that bind different microbial ligands, such as LPS, viral double-stranded RNA, and parasites and fungi-derived toxins (114). These enter the lungs and activate the adaptive immunity through TLRs, leading to T-cell differentiation and macrophage and dendritic cell activation. For example, TLR4 stimulation by heat-inactivated Escherichia coli increase the adhesion, migration and metastatic diffusion of NSCLC cells in vivo, mainly through p38 MAPK and ERK1/2 signaling pathways (115).
Microorganisms and their metabolites may produce tumorigenic effects by directly affecting epithelial cells or oncogenes (116). Pulmonary PAH-degrading bacteria, such as Massilia and Acidovorax, are more prevalent in smokers with lung cancer and TP53 mutations. The enrichment of these bacteria is combined with the trend of DNA recombination and repair pathway disorders, suggesting that contact of lung symbiotic microorganisms with tobacco may lead to mutations in host genes (117). An imbalance in the composition of microbial flora produces various toxins that lead to genotoxicity, promote the generation of free radicals, and cause DNA damage, thereby leading to a cycle arrest and apoptosis of cells without DNA repair systems (112). In addition, other microorganisms and their metabolites, such as HP, intestinal deoxycholic acid and shicholic acid, can cause DNA damage and increase the gene mutation load, thus inducing lung cancer (112, 118).
The tumor microenvironment (TME) is an environment composed of various physical and chemical factors surrounding tumor cells, including neighbor tumor cells, immune cells, stromal cells, extracellular matrix, and a variety of soluble molecules, and is an important aspect of the tumor. TME plays an important role in the occurrence and development of tumors (119). Figure 2B provides a good summary of the microbial involvement in the composition of lung TME and the mechanism of regulating the occurrence and development of lung cancer (120–122).
In a mouse model of KRAS-TP53 co-mutation (KP) lung cancer, airway microbiosis disorder caused by Tiny Vibrio led to the recruitment of Th17 cells, increased IL-17 production, increased PD-1+T cell-expression, and recruitment of neutrophils, which resulted in a reduced survival and increased the burden of lung tumors (111). Gut microbiota can also activate B cells, T cells, and other immune cells, which inflate the lungs through hemato-vascular or lymphatic pathways and activate the immune response to affect lung inflammation (114, 123–125). It has been reported that an imbalance in intestinal flora may regulate the TLR4/NF-KB signaling pathway of the lung immune system by modulating the intestinal barrier, activating pulmonary oxidative stress, and mediating the response to lung injury (126). Intestinal symbiotic bacteria and their metabolites, short-chain fatty acids (SCFAs), such as propionic acid and butyric acid in patients with NSCLC directly stimulate intestinal-epithelial cells to regulate the release of T-regulatory (Treg) cells (127). Treg cells can inhibit airway inflammation by stimulating SCFAs, suggesting that immune cells play an important role in microbial-mediated inflammation (125). In addition, HP produces some relevant adaptive immune effects on T cells, in addition to inducing extensive innate immune signal transduction effects in the lungs (128, 129).
Studies have shown correlations between lung cancer cell growth and unbalances in the airway microbial community. This locally dysregulated microbiome stimulates the production of IL-1β and IL-23 in myeloid cells, which in turn induce the proliferation and adaptive activation of lung-resident Vγ6+Vδ1+γδT-immune cells. Activated γδT cells produce IL-17, which promotes neutrophil infiltration and inflammation in the TME (103, 108). The theory of IL-17-mediated inflammatory pathway has also been confirmed in other studies and animal models (130, 131). Therefore, IL-17 produced by adaptive immune γδT cells plays a role in mediating the inflammatory pathways. In addition, increasing evidence suggests that HP contributes to inducing lung tumors. HP-derived LPS induces the production of pro-inflammatory factors, including IL-1, IL-6, and TNF. This inflammation can develop into chronic bronchitis which can be often accompanied by lung cancer (132).
Currently, the application of microbiomal knowledge to clinical research is a matter of extensive research. From the perspective of nutritional intervention, prebiotics and probiotics play indispensable roles. They can not only restore homeostasis of visceral organs or lower airways but also reduce microbial-induced inflammation, genotoxicity, and cell proliferation (133, 134) (Figure 3). Lee et al. found that Bifidobacterium was abundant in the intestinal tract of patients with NSCLC who responded to clinical treatment. Further, when a commercial Bifidobacterium strain was used to treat mice tumors with the same genotype the tumor load could be reduced by inducing the host immune response and cooperating with immunotherapeutic or chemotherapeutic drugs (135). Yusuke Tomita et al. used 588 strains of Clostridium butyricum (MIYAIRI 588 strain) to ameliorate symptoms associated with ecological disturbance caused by antibiotics (ATBs), suggesting that probiotic Clostridium butyricum therapy (CBT) has a positive effect on improving immune checkpoint blockade (ICB) in patients with cancer (136). On the other hand, oral administration of Lactobacillus acidophilus enhanced the antitumor effect of cisplatin, reduced tumor size, and improved the survival rate of mice (137). Therefore, prebiotics and probiotics can improve lung cancer treatment.
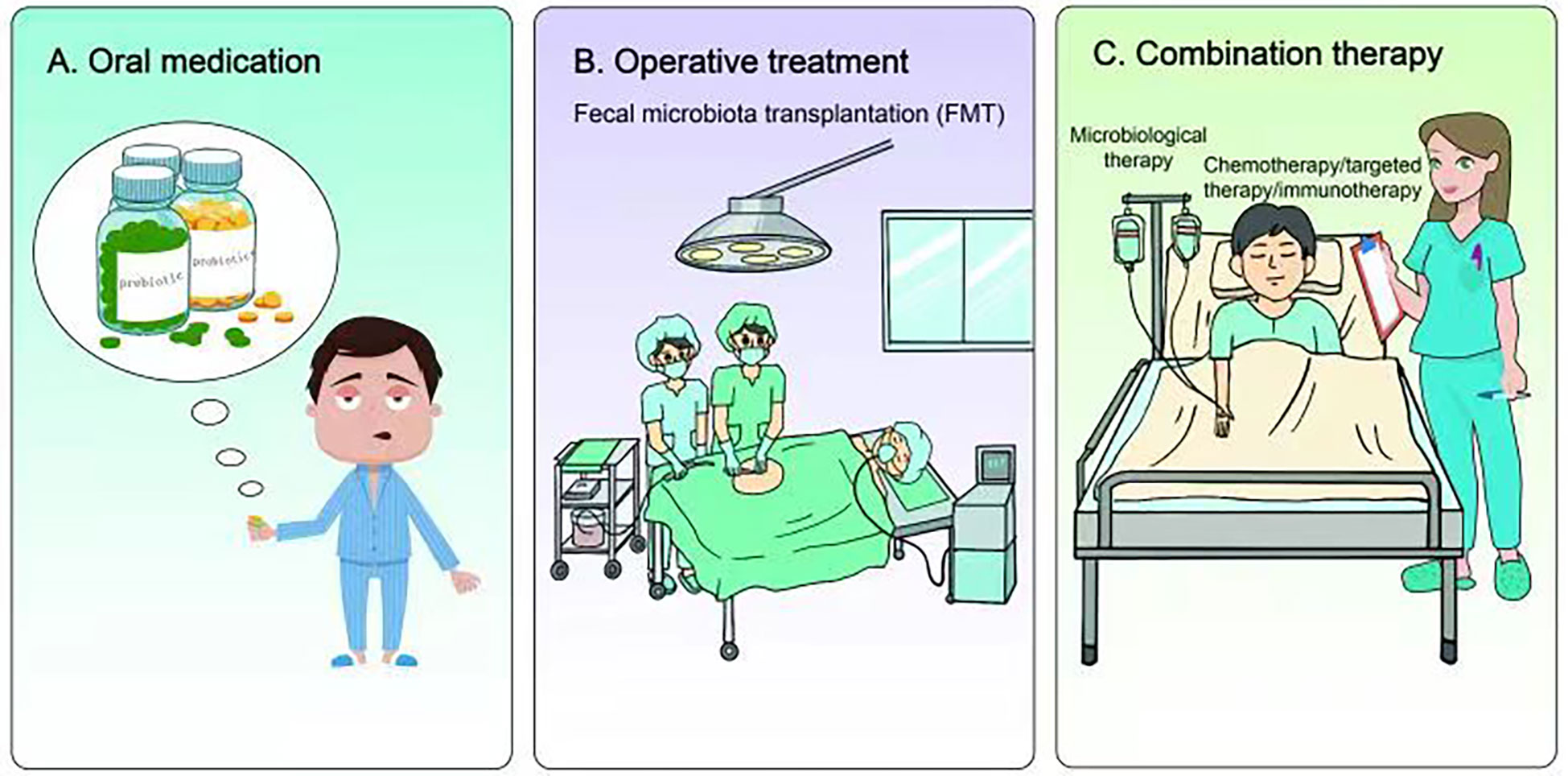
Figure 3 Research and application of microbiome in lung cancer treatment. (A) Nutritional intervention with prebiotics and probiotics can not only restore the homeostasis of internal organs or lower airway, but also reduce microbial-induced inflammation, genotoxicity and cell proliferation, thus improving the treatment of lung cancer. (B) Fecal microbiota transplantation (FMT) can also restore host homeostasis and reduce microbial-induced inflammation. Preclinical studies have shown that FMT therapy may have certain advantages in combating immunotherapy resistance in lung cancer. (C) Chemotherapy, targeted therapy or immunotherapy combined with microbial therapy can improve the clinical treatment effect of lung cancer patients.
In addition to nutritional intervention of prebiotics and probiotics, fecal microbiota transplantation (FMT) also restored host homeostasis and reduced microbial-induced inflammation (138, 139) (Figure 3). Although there is currently a lack of clinical application of FMT in lung cancer or other tumor types, previous preclinical studies have found that FMT could reverse the response to immunotherapy of drug-resistant patients by increasing the recruitment of CCR9+CXCR3+CD4+ T lymphocytes into tumor lesions in mice. These results indicate that FMT may have some advantages in battling resistance to lung cancer immunotherapy.
A number of studies have found that the gut microbiome of patients with lung cancer who respond to clinical treatment is significantly different from that of patients who do not respond, indicating that some favorable/unfavorable microorganisms are enriched in responders and non-responders respectively, thus implying a potentially predictive value for lung cancer clinical treatment (140–142) (Figure 3). Concerning chemotherapy, patients with advanced lung cancer treated with Enterococcus and Human Bariniella combined with immunochemotherapy showed longer PFS (143). In terms of targeted therapy, the role and therapeutic effects of the microbiota are very optimistic according to preclinical studies (144). In a mouse lung cancer model, Bacteroides ovatus and Bacteroides xylanisolvens were positively correlated with the treatment results. Oral or intragastric administration of these responsive bacteria could significantly improve the efficacy of Erlotinib and induce CXCL9 and IFN-γ expression (144). In immunotherapy, combined microbial therapy can improve the response to and effect of immune checkpoint inhibitors (ICIs). A recent study explored the role of gut microbes in the effectiveness of immunotherapy (145). The intestinal microbial community can affect the immune regulation mechanism by regulating T cell differentiation and significantly improve the therapeutic effect of ICI (140, 146–149). Mice using stool samples from patients who responded positively to immunotherapy, whereas mice using stool samples from patients who did not respond did not. A retrospective study reported that Clostridium butyricum treatment (CBT) before or after ICI treatment significantly extended patients’ progression-free survival (PFS)non-progressive survival and overall survival (OS) (136). Improved survival in these patients can be attributed to more efficient immunomodulatory effects.
The microbiome characteristics have significant effects in tumor development, however, how the microbiome responds to lung cancer, in particular, how lung cancer cells and TME shape the local microbial community of the lungs, is unknown. However, it has been shown that in colorectal cancer (CRC), loss of surface barrier function can cause tumor inflammation induced by symbiotic bacteria. In particular, the breakdown of tight connections between colon tumor cells allows bacterial degradation products such as LPS, to enter the tumor stroma, causing bone marrow-derived cells to be recruited to the TME. Therefore, understanding the interaction between the human microbiome and lung cancer cells, and identifying the cellular and molecular mediators involved in this interaction are relevant issues to be explored in order to find future potential targets for lung cancer treatment.
In addition, when considering the influence of microbiome on the efficacy of chemotherapy, targeted therapy, and immunotherapy for lung cancer, it is necessary to distinguish between the specific roles of the local lung microbiome, the distal gut microbiome, and oral bacteria in tumor growth and related immune responses (111). It is possible that selectively targeting one of these compartments may lead to different effects on lung cancer progression and treatment, thus providing new strategies for lung cancer treatments in the future.
Both authors contributed equally to the writing of the review.
This study was funded by the National Natural Science Foundation of China (82002421), Young Talent Program of Tangdu Hospital, Health Research Fund of Shaanxi Province (2021B004) and Discipline Innovation Development Plan Project of Tangdu Hospital (2021LCYJ005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet (Lond Engl) (2021) 398(10299):535–54. doi: 10.1016/s0140-6736(21)00312-3
2. Rock CL, Thomson CA, Sullivan KR, Howe CL, Kushi LH, Caan BJ, et al. American Cancer society nutrition and physical activity guideline for cancer survivors. CA: Cancer J Clin (2022) 72(3):230–62. doi: 10.3322/caac.21719
3. de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol (2012) 13(6):607–15. doi: 10.1016/s1470-2045(12)70137-7
4. Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, et al. Distinct microbial signatures associated with different breast cancer types. Front Microbiol (2018) 9:951. doi: 10.3389/fmicb.2018.00951
5. Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature (2020) 579(7800):567–74. doi: 10.1038/s41586-020-2095-1
6. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med (2011) 184(8):957–63. doi: 10.1164/rccm.201104-0655OC
7. Apostolou P, Tsantsaridou A, Papasotiriou I, Toloudi M, Chatziioannou M, Giamouzis G. Bacterial and fungal microflora in surgically removed lung cancer samples. J Cardiothoracic Surg (2011) 6:137. doi: 10.1186/1749-8090-6-137
8. Akinosoglou KS, Karkoulias K, Marangos M. Infectious complications in patients with lung cancer. Eur Rev Med Pharmacol Sci (2013) 17(1):8–18. PMID: 23329518
9. Caselli E, Fabbri C, D'Accolti M, Soffritti I, Bassi C, Mazzacane S, et al. Defining the oral microbiome by whole-genome sequencing and resistome analysis: The complexity of the healthy picture. BMC Microbiol (2020) 20(1):120. doi: 10.1186/s12866-020-01801-y
10. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature (2007) 449(7164):804–10. doi: 10.1038/nature06244
11. Wood DE, Salzberg SL. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol (2014) 15(3):R46. doi: 10.1186/gb-2014-15-3-r46
12. Petersen TN, Lukjancenko O, Thomsen MCF, Maddalena Sperotto M, Lund O, Møller Aarestrup F, et al. Mgmapper: Reference based mapping and taxonomy annotation of metagenomics sequence reads. PloS One (2017) 12(5):e0176469. doi: 10.1371/journal.pone.0176469
13. Wooley JC, Ye Y. Metagenomics: Facts and artifacts, and computational challenges*. J Comput Sci Technol (2009) 25(1):71–81. doi: 10.1007/s11390-010-9306-4
14. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe (2015) 17(5):704–15. doi: 10.1016/j.chom.2015.03.008
15. Stiemsma LT, Turvey SE. Asthma and the microbiome: Defining the critical window in early life. Allergy Asthma Clin Immunol (2017) 13:3. doi: 10.1186/s13223-016-0173-6
16. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med (2013) 187(10):1067–75. doi: 10.1164/rccm.201210-1913OC
17. Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome (2013) 1(1):19. doi: 10.1186/2049-2618-1-19
18. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PloS One (2010) 5(1):e8578. doi: 10.1371/journal.pone.0008578
19. Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Trans Res (2012) 160(4):258–66. doi: 10.1016/j.trsl.2012.02.005
20. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc (2015) 12(6):821–30. doi: 10.1513/AnnalsATS.201501-029OC
21. Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio (2015) 6(2):e00037. doi: 10.1128/mBio.00037-15
22. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell (2016) 164(3):337–40. doi: 10.1016/j.cell.2016.01.013
23. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PloS Biol (2016) 14(8):e1002533. doi: 10.1371/journal.pbio.1002533
24. Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA (2011) 108 Suppl 1(Suppl 1):4586–91. doi: 10.1073/pnas.1000097107
25. Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature (2012) 488(7410):178–84. doi: 10.1038/nature11319
26. Raju U, Levitz M, Javitt NB. Bile acids in human breast cyst fluid: The identification of lithocholic acid. J Clin Endocrinol Metab (1990) 70(4):1030–4. doi: 10.1210/jcem-70-4-1030
27. Javitt NB, Budai K, Miller DG, Cahan AC, Raju U, Levitz M. Breast-gut connection: Origin of chenodeoxycholic acid in breast cyst fluid. Lancet (Lond Engl) (1994) 343(8898):633–5. doi: 10.1016/s0140-6736(94)92635-2
28. Swales KE, Korbonits M, Carpenter R, Walsh DT, Warner TD, Bishop-Bailey D. The farnesoid X receptor is expressed in breast cancer and regulates apoptosis and aromatase expression. Cancer Res (2006) 66(20):10120–6. doi: 10.1158/0008-5472.can-06-2399
29. Zimmermann P, Curtis N. Breast milk microbiota: A review of the factors that influence composition. J infection (2020) 81(1):17–47. doi: 10.1016/j.jinf.2020.01.023
30. Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB, Baban CK, et al. Microbiota of human breast tissue. Appl Environ Microbiol (2014) 80(10):3007–14. doi: 10.1128/aem.00242-14
31. Yang I, Nell S, Suerbaum S. Survival in hostile territory: The microbiota of the stomach. FEMS Microbiol Rev (2013) 37(5):736–61. doi: 10.1111/1574-6976.12027
32. Wu WM, Yang YS, Peng LH. Microbiota in the stomach: New insights. J Digestive Dis (2014) 15(2):54–61. doi: 10.1111/1751-2980.12116
33. Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA (2006) 103(3):732–7. doi: 10.1073/pnas.0506655103
34. Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, et al. Bacterial microbiota profiling in gastritis without helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PloS One (2009) 4(11):e7985. doi: 10.1371/journal.pone.0007985
35. Mailhe M, Ricaboni D, Vitton V, Gonzalez JM, Bachar D, Dubourg G, et al. Repertoire of the gut microbiota from stomach to colon using culturomics and next-generation sequencing. BMC Microbiol (2018) 18(1):157. doi: 10.1186/s12866-018-1304-7
36. Fassi Fehri L, Mak TN, Laube B, Brinkmann V, Ogilvie LA, Mollenkopf H, et al. Prevalence of propionibacterium acnes in diseased prostates and its inflammatory and transforming activity on prostate epithelial cells. Int J Med Microbiol IJMM (2011) 301(1):69–78. doi: 10.1016/j.ijmm.2010.08.014
37. Sfanos KS, Sauvageot J, Fedor HL, Dick JD, De Marzo AM, Isaacs WB. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate (2008) 68(3):306–20. doi: 10.1002/pros.20680
38. Cohen RJ, Shannon BA, McNeal JE, Shannon T, Garrett KL. Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: A possible link to cancer evolution? J Urol (2005) 173(6):1969–74. doi: 10.1097/01.ju.0000158161.15277.78
39. Hochreiter WW, Duncan JL, Schaeffer AJ. Evaluation of the bacterial flora of the prostate using a 16s rrna gene based polymerase chain reaction. J Urol (2000) 163(1):127–30.
40. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol (2014) 12:87. doi: 10.1186/s12915-014-0087-z
41. Fair WR, Parrish RF. Antibacterial substances in prostatic fluid. Prog Clin Biol Res (1981) 75a:247–64. PMID: 7041133
42. Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontology (2003) 30(7):644–54. doi: 10.1034/j.1600-051x.2003.00376.x
43. Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, et al. Application of a neutral community model to assess structuring of the human lung microbiome. mBio (2015) 6(1):e02284–14. doi: 10.1128/mBio.02284-14
44. Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol (2018) 200(4):525–40. doi: 10.1007/s00203-018-1505-3
45. Jhajharia K, Parolia A, Shetty KV, Mehta LK. Biofilm in endodontics: A review. J Int Soc Prev Community Dentistry (2015) 5(1):1–12. doi: 10.4103/2231-0762.151956
46. Hasan NA, Young BA, Minard-Smith AT, Saeed K, Li H, Heizer EM, et al. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PloS One (2014) 9(5):e97699. doi: 10.1371/journal.pone.0097699
47. He X, Hu W, He J, Guo L, Lux R, Shi W. Community-based interference against integration of pseudomonas aeruginosa into human salivary microbial biofilm. Mol Oral Microbiol (2011) 26(6):337–52. doi: 10.1111/j.2041-1014.2011.00622.x
48. Baena-Monroy T, Moreno-Maldonado V, Franco-Martínez F, Aldape-Barrios B, Quindós G, Sánchez-Vargas LO. Candida albicans, staphylococcus aureus and streptococcus mutans colonization in patients wearing dental prosthesis. Medicina oral patologia Oral y cirugia bucal (2005) 10 Suppl1:E27–39. PMID: 15800465
49. Passariello C, Puttini M, Iebba V, Pera P, Gigola P. Influence of oral conditions on colonization by highly toxigenic staphylococcus aureus strains. Oral Dis (2012) 18(4):402–9. doi: 10.1111/j.1601-0825.2011.01889.x
50. Colombo AV, Barbosa GM, Higashi D, di Micheli G, Rodrigues PH, Simionato MRL. Quantitative detection of staphylococcus aureus, enterococcus faecalis and pseudomonas aeruginosa in human oral epithelial cells from subjects with periodontitis and periodontal health. J Med Microbiol (2013) 62(Pt 10):1592–600. doi: 10.1099/jmm.0.055830-0
51. Souto R, Silva-Boghossian CM, Colombo AP. Prevalence of pseudomonas aeruginosa and acinetobacter spp. In: Subgingival biofilm and saliva of subjects with chronic periodontal infection. Brazilian journal of microbiology, vol. 45. (2014). p. 495–501. doi: 10.1590/s1517-83822014000200017
52. Mathews SA, Kurien BT, Scofield RH. Oral manifestations of sjögren's syndrome. J Dental Res (2008) 87(4):308–18. doi: 10.1177/154405910808700411
53. Scannapieco FA. Role of oral bacteria in respiratory infection. J periodontology (1999) 70(7):793–802. doi: 10.1902/jop.1999.70.7.793
54. Koch CD, Gladwin MT, Freeman BA, Lundberg JO, Weitzberg E, Morris A. Enterosalivary nitrate metabolism and the microbiome: Intersection of microbial metabolism, nitric oxide and diet in cardiac and pulmonary vascular health. Free Radical Biol Med (2017) 105:48–67. doi: 10.1016/j.freeradbiomed.2016.12.015
55. Huffnagle GB, Dickson RP, Lukacs NW. The respiratory tract microbiome and lung inflammation: A two-way Street. Mucosal Immunol (2017) 10(2):299–306. doi: 10.1038/mi.2016.108
56. Salisbury ML, Han MK, Dickson RP, Molyneaux PL. Microbiome in interstitial lung disease: From pathogenesis to treatment target. Curr Opin pulmonary Med (2017) 23(5):404–10. doi: 10.1097/mcp.0000000000000399
57. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature (2012) 486(7402):222–7. doi: 10.1038/nature11053
58. Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med (2014) 190(11):1283–92. doi: 10.1164/rccm.201407-1240OC
59. O'Dwyer DN, Dickson RP, Moore BB. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J Immunol (Baltimore Md 1950) (2016) 196(12):4839–47. doi: 10.4049/jimmunol.1600279
60. Erb Downward JR, Falkowski NR, Mason KL, Muraglia R, Huffnagle GB. Modulation of post-antibiotic bacterial community reassembly and host response by candida albicans. Sci Rep (2013) 3:2191. doi: 10.1038/srep02191
61. Marsland BJ, Gollwitzer ES. Host-microorganism interactions in lung diseases. Nat Rev Immunol (2014) 14(12):827–35. doi: 10.1038/nri3769
62. Allam-Ndoul B, Castonguay-Paradis S, Veilleux A. Gut microbiota and intestinal trans-epithelial permeability. Int J Mol Sci (2020) 21(17):6402. doi: 10.3390/ijms21176402
63. Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci CMLS (2017) 74(16):2959–77. doi: 10.1007/s00018-017-2509-x
64. Tytgat HLP, Nobrega FL, van der Oost J, de Vos WM. Bowel biofilms: Tipping points between a healthy and compromised gut? Trends Microbiol (2019) 27(1):17–25. doi: 10.1016/j.tim.2018.08.009
65. Tan S, Li D, Zhu X. Cancer immunotherapy: Pros, cons and beyond. Biomedicine pharmacotherapy = Biomedecine pharmacotherapie (2020) 124:109821. doi: 10.1016/j.biopha.2020.109821
66. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol (2017) 19(1):29–41. doi: 10.1111/1462-2920.13589
67. Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol (2017) 18(1):2. doi: 10.1186/s12865-016-0187-3
68. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J (2017) 474(11):1823–36. doi: 10.1042/bcj20160510
69. Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, et al. Acetate mediates a microbiome-Brain-β-Cell axis to promote metabolic syndrome. Nature (2016) 534(7606):213–7. doi: 10.1038/nature18309
70. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature (2013) 502(7469):96–9. doi: 10.1038/nature12503
71. Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov (2008) 7(8):678–93. doi: 10.1038/nrd2619
72. Zollner G, Wagner M, Trauner M. Nuclear receptors as drug targets in cholestasis and drug-induced hepatotoxicity. Pharmacol Ther (2010) 126(3):228–43. doi: 10.1016/j.pharmthera.2010.03.005
73. Chiang JY. Bile acids: Regulation of synthesis. J Lipid Res (2009) 50(10):1955–66. doi: 10.1194/jlr.R900010-JLR200
74. Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathog (Basel Switzerland) (2013) 3(1):14–24. doi: 10.3390/pathogens3010014
75. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci United States America (2007) 104(34):13780–5. doi: 10.1073/pnas.0706625104
76. Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of faecalibacterium prausnitzii in colitis microbiota. Inflammatory Bowel Dis (2009) 15(8):1183–9. doi: 10.1002/ibd.20903
77. Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflammatory Bowel Dis (2008) 14(2):147–61. doi: 10.1002/ibd.20330
78. Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev (2010) 34(4):426–44. doi: 10.1111/j.1574-6976.2009.00204.x
79. Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe (2014) 16(4):495–503. doi: 10.1016/j.chom.2014.09.001
80. Hubbard TD, Murray IA, Perdew GH. Indole and tryptophan metabolism: Endogenous and dietary routes to ah receptor activation. Drug Metab disposition: Biol fate chemicals (2015) 43(10):1522–35. doi: 10.1124/dmd.115.064246
81. Yu AI, Zhao L, Eaton KA, Ho S, Chen J, Poe S, et al. Gut microbiota modulate Cd8 t cell responses to influence colitis-associated tumorigenesis. Cell Rep (2020) 31(1):107471. doi: 10.1016/j.celrep.2020.03.035
82. Zou Z, Tao T, Li H, Zhu X. Mtor signaling pathway and mtor inhibitors in cancer: Progress and challenges. Cell bioscience (2020) 10:31. doi: 10.1186/s13578-020-00396-1
83. Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity (2012) 37(1):171–86. doi: 10.1016/j.immuni.2012.05.020
84. Morita N, Umemoto E, Fujita S, Hayashi A, Kikuta J, Kimura I, et al. Gpr31-dependent dendrite protrusion of intestinal Cx3cr1(+) cells by bacterial metabolites. Nature (2019) 566(7742):110–4. doi: 10.1038/s41586-019-0884-1
85. Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont ruminococcus gnavus is strain-dependent. PloS One (2013) 8(10):e76341. doi: 10.1371/journal.pone.0076341
86. López P, González-Rodríguez I, Gueimonde M, Margolles A, Suárez A. Immune response to bifidobacterium bifidum strains support Treg/Th17 plasticity. PloS One (2011) 6(9):e24776. doi: 10.1371/journal.pone.0024776
87. Ménard O, Butel MJ, Gaboriau-Routhiau V, Waligora-Dupriet AJ. Gnotobiotic mouse immune response induced by bifidobacterium sp. Strains Isolated Infants. Appl Environ Microbiol (2008) 74(3):660–6. doi: 10.1128/aem.01261-07
88. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell (2016) 167(4):1125–36.e8. doi: 10.1016/j.cell.2016.10.020
89. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the "Healthy" smoker and in copd. PloS One (2011) 6(2):e16384. doi: 10.1371/journal.pone.0016384
90. Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Interaction between the microbiome and Tp53 in human lung cancer. Genome Biol (2018) 19(1):123. doi: 10.1186/s13059-018-1501-6
91. Zhang L, Zhan H, Xu W, Yan S, Ng SC. The role of gut mycobiome in health and diseases. Ther Adv Gastroenterol (2021) 14:17562848211047130. doi: 10.1177/17562848211047130
92. Leng Q, Holden VK, Deepak J, Todd NW, Jiang F. Microbiota biomarkers for lung cancer. Diagnostics (Basel Switzerland) (2021) 11(3):407. doi: 10.3390/diagnostics11030407
93. Botticelli A, Vernocchi P, Marini F, Quagliariello A, Cerbelli B, Reddel S, et al. Gut metabolomics profiling of non-small cell lung cancer (Nsclc) patients under immunotherapy treatment. J Trans Med (2020) 18(1):49. doi: 10.1186/s12967-020-02231-0
94. Gui Q, Li H, Wang A, Zhao X, Tan Z, Chen L, et al. The association between gut butyrate-producing bacteria and non-Small-Cell lung cancer. J Clin Lab Anal (2020) 34(8):e23318. doi: 10.1002/jcla.23318
95. Yan X, Yang M, Liu J, Gao R, Hu J, Li J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res (2015) 5(10):3111–22. PMID: 26693063
96. Gomes S, Cavadas B, Ferreira JC, Marques PI, Monteiro C, Sucena M, et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci Rep (2019) 9(1):12838. doi: 10.1038/s41598-019-49195-w
97. Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Author correction: Interaction between the microbiome and Tp53 in human lung cancer. Genome Biol (2020) 21(1):41. doi: 10.1186/s13059-020-01961-0
98. Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol (2016) 17(1):163. doi: 10.1186/s13059-016-1021-1
99. Han AJ, Xiong M, Zong YS. Association of Epstein-Barr virus with lymphoepithelioma-like carcinoma of the lung in southern China. Am J Clin Pathol (2000) 114(2):220–6. doi: 10.1309/148k-nd54-6njx-na61
100. Zhang F, Ferrero M, Dong N, D'Auria G, Reyes-Prieto M, Herreros-Pomares A, et al. Analysis of the gut microbiota: An emerging source of biomarkers for immune checkpoint blockade therapy in non-small cell lung cancer. Cancers (2021) 13(11):2514. doi: 10.3390/cancers13112514
101. Huang D, Su X, Yuan M, Zhang S, He J, Deng Q, et al. The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am J Cancer Res (2019) 9(9):2047–63. PMID: 31598405
102. Jungnickel C, Schmidt LH, Bittigkoffer L, Wolf L, Wolf A, Ritzmann F, et al. Il-17c mediates the recruitment of tumor-associated neutrophils and lung tumor growth. Oncogene (2017) 36(29):4182–90. doi: 10.1038/onc.2017.28
103. Le Noci V, Guglielmetti S, Arioli S, Camisaschi C, Bianchi F, Sommariva M, et al. Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: A strategy to promote immunosurveillance against lung metastases. Cell Rep (2018) 24(13):3528–38. doi: 10.1016/j.celrep.2018.08.090
104. Dickson RP, Schultz MJ, van der Poll T, Schouten LR, Falkowski NR, Luth JE, et al. Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med (2020) 201(5):555–63. doi: 10.1164/rccm.201907-1487OC
105. Peters BA, Hayes RB, Goparaju C, Reid C, Pass HI, Ahn J. The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol Biomarkers Prev (2019) 28(4):731–40. doi: 10.1158/1055-9965.epi-18-0966
106. Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol (2016) 1:16031. doi: 10.1038/nmicrobiol.2016.31
107. Palucka AK, Coussens LM. The basis of oncoimmunology. Cell (2016) 164(6):1233–47. doi: 10.1016/j.cell.2016.01.049
108. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development Via Γδ T cells. Cell (2019) 176(5):998–1013.e16. doi: 10.1016/j.cell.2018.12.040
109. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
110. Tsay JJ, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, et al. Airway microbiota is associated with upregulation of the Pi3k pathway in lung cancer. Am J Respir Crit Care Med (2018) 198(9):1188–98. doi: 10.1164/rccm.201710-2118OC
111. Tsay JJ, Wu BG, Sulaiman I, Gershner K, Schluger R, Li Y, et al. Lower airway dysbiosis affects lung cancer progression. Cancer Discov (2021) 11(2):293–307. doi: 10.1158/2159-8290.cd-20-0263
112. Alhinai EA, Walton GE, Commane DM. The role of the gut microbiota in colorectal cancer causation. Int J Mol Sci (2019) 20(21):5295. doi: 10.3390/ijms20215295
113. Bingula R, Filaire E, Molnar I, Delmas E, Berthon JY, Vasson MP, et al. Characterisation of microbiota in saliva, bronchoalveolar lavage fluid, non-malignant, peritumoural and tumour tissue in non-small cell lung cancer patients: A cross-sectional clinical trial. Respir Res (2020) 21(1):129. doi: 10.1186/s12931-020-01392-2
114. Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol (2015) 6:1085. doi: 10.3389/fmicb.2015.01085
115. Chow SC, Gowing SD, Cools-Lartigue JJ, Chen CB, Berube J, Yoon HW, et al. Gram negative bacteria increase non-small cell lung cancer metastasis Via toll-like receptor 4 activation and mitogen-activated protein kinase phosphorylation. Int J Cancer (2015) 136(6):1341–50. doi: 10.1002/ijc.29111
116. Ramírez-Labrada AG, Isla D, Artal A, Arias M, Rezusta A, Pardo J, et al. The influence of lung microbiota on lung carcinogenesis, immunity, and immunotherapy. Trends Cancer (2020) 6(2):86–97. doi: 10.1016/j.trecan.2019.12.007
117. Dong H, Tan Q, Xu Y, Zhu Y, Yao Y, Wang Y, et al. Convergent alteration of lung tissue microbiota and tumor cells in lung cancer. iScience (2022) 25(1):103638. doi: 10.1016/j.isci.2021.103638
118. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol (2014) 12(10):661–72. doi: 10.1038/nrmicro3344
119. Chen Q, Liu G, Liu S, Su H, Wang Y, Li J, et al. Remodeling the tumor microenvironment with emerging nanotherapeutics. Trends Pharmacol Sci (2018) 39(1):59–74. doi: 10.1016/j.tips.2017.10.009
120. Laroumagne S, Lepage B, Hermant C, Plat G, Phelippeau M, Bigay-Game L, et al. Bronchial colonisation in patients with lung cancer: A prospective study. Eur Respir J (2013) 42(1):220–9. doi: 10.1183/09031936.00062212
121. Liu HX, Tao LL, Zhang J, Zhu YG, Zheng Y, Liu D, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int J Cancer (2018) 142(4):769–78. doi: 10.1002/ijc.31098
122. Mao Q, Jiang F, Yin R, Wang J, Xia W, Dong G, et al. Interplay between the lung microbiome and lung cancer. Cancer Lett (2018) 415:40–8. doi: 10.1016/j.canlet.2017.11.036
123. Bingula R, Filaire M, Radosevic-Robin N, Bey M, Berthon JY, Bernalier-Donadille A, et al. Desired turbulence? gut-lung axis, immunity, and lung cancer. J Oncol (2017) 2017:5035371. doi: 10.1155/2017/5035371
124. Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol (2017) 15(1):55–63. doi: 10.1038/nrmicro.2016.142
125. Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut (2016) 65(4):575–83. doi: 10.1136/gutjnl-2015-309728
126. Tang J, Xu L, Zeng Y, Gong F. Effect of gut microbiota on lps-induced acute lung injury by regulating the Tlr4/Nf-kb signaling pathway. Int Immunopharmacol (2021) 91:107272. doi: 10.1016/j.intimp.2020.107272
127. Bingula R, Filaire M, Radosevic-Robin N, Berthon JY, Bernalier-Donadille A, Vasson MP, et al. Characterisation of gut, lung, and upper airways microbiota in patients with non-small cell lung carcinoma: Study protocol for case-control observational trial. Medicine (2018) 97(50):e13676. doi: 10.1097/md.0000000000013676
128. Altobelli A, Bauer M, Velez K, Cover TL, Müller A. Helicobacter pylori vaca targets myeloid cells in the gastric lamina propria to promote peripherally induced regulatory T-cell differentiation and persistent infection. mBio (2019) 10(2):e00261–19. doi: 10.1128/mBio.00261-19
129. van Wijck Y, John-Schuster G, van Schadewijk A, van den Oever RL, Obieglo K, Hiemstra PS, et al. Extract of helicobacter pylori ameliorates parameters of airway inflammation and goblet cell hyperplasia following repeated allergen exposure. Int Arch Allergy Immunol (2019) 180(1):1–9. doi: 10.1159/000500598
130. Zitvogel L, Kroemer G. Lower airway dysbiosis exacerbates lung cancer. Cancer Discovery (2021) 11(2):224–6. doi: 10.1158/2159-8290.cd-20-1641
131. Yadava K, Pattaroni C, Sichelstiel AK, Trompette A, Gollwitzer ES, Salami O, et al. Microbiota promotes chronic pulmonary inflammation by enhancing il-17a and autoantibodies. Am J Respir Crit Care Med (2016) 193(9):975–87. doi: 10.1164/rccm.201504-0779OC
132. Kanbay M, Kanbay A, Boyacioglu S. Helicobacter pylori infection as a possible risk factor for respiratory system disease: A review of the literature. Respir Med (2007) 101(2):203–9. doi: 10.1016/j.rmed.2006.04.022
133. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer (2013) 13(11):800–12. doi: 10.1038/nrc3610
134. Zmora N, Zeevi D, Korem T, Segal E, Elinav E. Taking it personally: Personalized utilization of the human microbiome in health and disease. Cell Host Microbe (2016) 19(1):12–20. doi: 10.1016/j.chom.2015.12.016
135. Lee SH, Cho SY, Yoon Y, Park C, Sohn J, Jeong JJ, et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat Microbiol (2021) 6(3):277–88. doi: 10.1038/s41564-020-00831-6
136. Tomita Y, Ikeda T, Sakata S, Saruwatari K, Sato R, Iyama S, et al. Association of probiotic clostridium butyricum therapy with survival and response to immune checkpoint blockade in patients with lung cancer. Cancer Immunol Res (2020) 8(10):1236–42. doi: 10.1158/2326-6066.cir-20-0051
137. Gui QF, Lu HF, Zhang CX, Xu ZR, Yang YH. Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genet Mol Res GMR (2015) 14(2):5642–51. doi: 10.4238/2015.May.25.16
138. Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: A randomised placebo-controlled trial. Lancet (Lond Engl) (2017) 389(10075):1218–28. doi: 10.1016/s0140-6736(17)30182-4
139. Ford AC. Stool as a treatment for ibs: More questions than answers? Lancet Gastroenterol Hepatol (2018) 3(1):2–3. doi: 10.1016/s2468-1253(17)30337-0
140. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of pd-1-Based immunotherapy against epithelial tumors. Sci (New York NY) (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
141. Song P, Yang D, Wang H, Cui X, Si X, Zhang X, et al. Relationship between intestinal flora structure and metabolite analysis and immunotherapy efficacy in Chinese nsclc patients. Thorac Cancer (2020) 11(6):1621–32. doi: 10.1111/1759-7714.13442
142. Zhang C, Wang J, Sun Z, Cao Y, Mu Z, Ji X. Commensal microbiota contributes to predicting the response to immune checkpoint inhibitors in non-Small-Cell lung cancer patients. Cancer Sci (2021) 112(8):3005–17. doi: 10.1111/cas.14979
143. Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, et al. Enterococcus hirae and barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity (2016) 45(4):931–43. doi: 10.1016/j.immuni.2016.09.009
144. Heshiki Y, Vazquez-Uribe R, Li J, Ni Y, Quainoo S, Imamovic L, et al. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome (2020) 8(1):28. doi: 10.1186/s40168-020-00811-2
145. Tartour E, Zitvogel L. Lung cancer: Potential targets for immunotherapy. Lancet Respir Med (2013) 1(7):551–63. doi: 10.1016/s2213-2600(13)70159-0
146. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-Pd-1 immunotherapy in melanoma patients. Sci (New York NY) (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
147. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-Pd-L1 efficacy. Sci (New York NY) (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
148. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Sci (New York NY) (2013) 342(6161):967–70. doi: 10.1126/science.1240527
Keywords: microbiome, pulmonary infection, immunology, clinical application, malignant mechanism, lung cancer
Citation: Sun Y, Wen M, Liu Y, Wang Y, Jing P, Gu Z, Jiang T and Wang W (2023) The human microbiome: A promising target for lung cancer treatment. Front. Immunol. 14:1091165. doi: 10.3389/fimmu.2023.1091165
Received: 06 November 2022; Accepted: 16 January 2023;
Published: 30 January 2023.
Edited by:
Matija Rijavec, University Clinic of Pulmonary and Allergic Diseases Golnik, SloveniaCopyright © 2023 Sun, Wen, Liu, Wang, Jing, Gu, Jiang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongping Gu, Z3UtemhvbmctcGluZ0AxNjMuY29t; Tao Jiang, amlhbmd0YW9jaGVzdEAxNjMuY29t; Wenchen Wang, MTgzOTIxODc5NzBAMTYzLmNvbQ==
†These authors contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.