
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 23 January 2023
Sec. Microbial Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1072655
This article is part of the Research Topic Targeting the microbiota to attenuate chronic inflammation View all 16 articles
Although the microbiota has largely been associated with the pathogenesis of viral infections, most studies using omics techniques are correlational and hypothesis-generating. The mechanisms affecting the immune responses to viral infections are still being fully understood. Here we focus on the two most important sexually transmitted persistent viruses, HPV and HIV. Sophisticated omics techniques are boosting our ability to understand microbiota-pathogen-host interactions from a functional perspective by surveying the host and bacterial protein and metabolite production using systems biology approaches. However, while these strategies have allowed describing interaction networks to identify potential novel microbiota-associated biomarkers or therapeutic targets to prevent or treat infectious diseases, the analyses are typically based on highly dimensional datasets —thousands of features in small cohorts of patients—. As a result, we are far from getting to their clinical use. Here we provide a broad overview of how the microbiota influences the immune responses to HIV and HPV disease. Furthermore, we highlight experimental approaches to understand better the microbiota-host-virus interactions that might increase our potential to identify biomarkers and therapeutic agents with clinical applications.
Evolutionary and ecological mechanisms have favored the cooperation of microorganisms that ensure critical functions for host fitness, such as the response against viral infections. The largest fraction of the microbiota resides in close interaction with the mucosa-associated lymphoid tissue (MALT) (1). Therefore, the expectations that the microbiota could exert a clinically relevant impact on viral infections, such as HPV and HIV, are high. The pathogenesis of HPV and HIV infection is intimately associated with the MALT, from the early establishment of infection to their persistence or progression (2–4). For example, HIV infection causes chronic defects in mucosal immunity (5, 6) and translocation of microbial products from the gut to the blood. These changes promote T cell activation, monocyte activation, and proinflammatory cytokine release (7–10). In HPV, the gut microbiota appears to influence viral persistence, immune responses, the host-mucosal environment, and HPV-related cancer progression (11, 12).
Although omics technologies have allowed us to map the functional alterations produced by viral infections, many studies show correlations, and we lack a granular understanding of the underlying mechanisms for the functional alterations. Omics techniques have allowed linking specific microbiome profiles to certain disease phenotypes (13, 14). The influence of bacterial proteins and metabolites on disease is gaining interest and being more deeply studied (15–21). However, most studies in the field are still correlational and hypothesis-generating. Furthermore, although proteomics and metabolomics are helpful tools to infer pathways and generate hypotheses and they have become increasingly efficient, their results still have limitations and biases and warrant experimental validation. Thus, following the enthusiasm of omics-based studies, the classical approach of designing hypothesis-driven studies focused on digging deeper into particular questions after interrogating highly dimensional datasets, is gaining attention. Here, we review specifically the current concepts on the reciprocal interactions between the microbiota and two persistent viral infections, HIV and HPV. We discuss the opportunities for omics techniques and their limitations in the field, highlight examples of studies aimed at understanding the consequences of the microbiota in HIV and HPV infections, and summarize the experimental approaches that have improved our mechanistic insight.
Correlations between changes in gut mucosa leading to “dysbiosis” (i.e., alterations in the intestinal microbiota) and viral infections are commonly studied. The commensal microbiota appears to be a significant determinant of the acquisition and replication of some pathogenic viruses. This may include mechanisms not well understood yet, including pathogen growth regulation, competitive metabolic interactions, localization in intestinal niches, and host-immune response induction (22–26). However, as we will review below, there is a clear connection between the microbiota status and the clinical course of HIV and HPV.
Acute HIV infection exerts dramatic and perhaps irreversible MALT damage (27, 28). The fact that HIV replication has been associated with a loss of anti-inflammatory bacteria (20, 29) has spurred research into the hypothesis that HIV infection affects the microbiota and that this altered microbiota may contribute to persistent inflammation, increasing the risk of comorbidities. Mechanistically, HIV infection could affect the microbiota by inducing depletion of Th17 cells in MALT, enteropathy, mucosal inflammation, aberrant cytokine production, and intestinal epithelial cell damage (25, 30–34). In the context of HIV infection, microbial-induced immune activation occurs and correlates with markers of intestinal damage, suggesting that the microbiota is a relevant driver of systemic inflammation (35–41). Even the changes appreciated in the oral microbiota of people living with HIV (PLWH), who exhibit an increased prevalence of dental caries and periodontal inflammation, seem to be connected to shifts in systemic immune responses (reviewed in (42)). Specific Lactobacillus species-rich vaginal microbiota have been associated to protection from HIV infection (last reviewed in (43)).
It is now widely accepted that impairment of intestinal integrity and dysbiosis lead to translocation of bacterial derivatives from the gut to the bloodstream, resulting in chronic inflammation. This may occur by immunosuppressive or immunostimulatory mechanisms and via various non-mutually exclusive processes, including augmented antigenicity, adjuvanticity, or bystander T-cell activation (44, 45). This fact has been studied before for HIV-associated inflammation, which has been associated with an increase of active microorganisms leading to different pathways related to immune modification (46), These pathways include (i) decreased amino acid catabolism, leading to nutritional deficits (47). (ii) induction of indolamine-2,3-dioxygenase-1 (IDO1) leading to an increased transformation of tryptophan into the immunosuppressive kynurenine derivatives, bacterial translocation, and systemic inflammation, which has been linked with excess mortality risk ,45). (iii) increased butyrate synthesis, which, among other functions, tempers intestinal inflammation (48). and (iv) accumulation of inflammatory molecules, such as arachidonic acid and leukotriene-B4 (49).
Inflammatory biomarkers levels remain increased in PLWH even when ART is started early (50). Chronic inflammation has consistently been associated with an excess risk of comorbidities during treated HIV infection and is suggested as a contributing risk factor (51, 52). Thus, the HIV field has pursued whether the microbiota affects inflammation during treated infection. For example, microbiota metabolic profiles affect HIV inflammation by promoting changes in glutathione metabolism and zeatin biosynthesis, butyrate production, or tryptophan catabolism (46, 49, 50, 53). Furthermore, a well-defined deleterious consequence of HIV infection is bacterial translocation triggering immune activation (54–56). A few sequence-based and ultramicroscopic studies have uncovered a blood bacterial DNA profile in HIV. Following acute SIV infection in macaques, analysis of bacterial DNA isolated from the colon, liver, and mesenteric lymph nodes demonstrated a preference for the phylum Proteobacteria to translocate to these compartments and an increased metabolic activity of Proteobacteria within the colonic lumen (57). In a study in PLWH diagnosed with advanced disease and starting ART, we also found that Proteobacteria was the predominant phylum in the blood, indicating commonalities in the mechanisms by which bacterial translocate from the gut into the bloodstream between SIV and HIV infections. The same study showed that ART initiation in late-presenters attenuated the bacterial signature of untreated HIV infection, characterized by the presence of DNA from commensal bacteria with pathogenic potential (58). Relationships between the translocated microbiome, systemic inflammation, and clinical outcomes were described in a different study showing increased CD4 T cell counts following one year of ART that were associated with high Serratia abundance, innate proinflammatory cytokines and metabolites driving Th17 gene expression signatures, and restoration of mucosal immunity (59).
Current evidence supports investigating therapeutic strategies for immune modulation in HIV. However, so far, no intervention targeting the microbiota of PLWH either by using prebiotics (16, 16, 60), probiotics (61–63), synbiotics (64), rifaximin (65, 66), or even fecal microbiota transplants (67, 68) have convincingly proved to effectively temper inflammation or enhance boost immune recovery following ART initiation. In general, there was lack of standardization in the outcomes assessed (ranging from studies designed to assess T cell changes (60, 64) to exploratory studies evaluating multiple markers of T cell activation (69) or soluble markers of inflammation or bacterial translocation), the duration of the intervention (from weeks (16, 61, 62, 69) up to one year), the disease status (from ART naive patients followed without ART (60) or patients presenting at advanced stages of the disease (64) starting ART to patients under ART-mediated HIV RNA suppression (61–63), and even the dosage and components of the prebiotic or probiotic mixtures (16, 62–64, 69). Therefore, this still represents a field of active research, and has been extensively reviewed elsewhere (70).
We know less about the impact of HPV infection on the microbiota epithelial surface integrity, mucosal state, and immune regulation, all factors related to HPV persistence and progression to cancer (46, 71–74). For example, metabolites associated with the vaginal microbiome, including biogenic amines, glutathione, and lipids, have been implicated in HPV persistence (75). It has been described that the microbiota composition can affect all these factors in the context of HPV infection (76–80). A meta-analysis found that Lactobacillus iners and non-Lactobacilli species dominance in the vaginal microbiota is associated with a higher risk of persistent HPV infection and dysplasia (22) compared to the dominance of L. iners and L. crispatus (81, 82).
A Lactobacillus-depleted microbiome has been associated with a proinflammatory environment that may increase malignant cell proliferation and HPV E6 and E7 oncogene expression (22, 83, 84) and promote coinfections by other pathogens such as Chlamydia trachomatis (80). Specifically, it has been shown that HPV down-regulates some innate molecules, such as SLPI, S100A7, elafin, HβD1, and TNFα/LPS that are used by some Lactobacillus species as an amino acid source sustaining their growth, in keeping with their decreased abundance in microbiome analyses of HPV infected individuals (80). Even virome alterations are associated with features of the vaginal microbiota and genital inflammation changes related to HPV infection (85).
Some authors have connected the expression of proinflammatory and chemotactic cytokines related to HPV-induced carcinogenesis with an increased presence of Sneathia or Gardnerella in the vaginal microenvironment (86–90). Furthermore, even though a certain level of inflammation has been described as potentially beneficial to decrease HPV dissemination, several studies have shown specific inflammation markers as related to the progression to a carcinogenic status that could be used as clinical markers to prevent high-grade squamous intraepithelial lesions (46, 86, 90–95) and specific metabolic profiles (96–98). However, most studies in the field are cross-sectional, so it is hard to assess whether the microbiota influences HPV infection or vice versa.
Viruses infecting epithelial cells can profoundly affect the mucosal immune system—the central habitat of the mucosal microbiota—altering the immunological signals required to orchestrate commensal colonization and possibly affecting systemic immune responses and other processes. While from an applied perspective, the gut microbiota functionalities are more relevant for health, most studies have focused on the compositional level, and only fewer studies have focused on the functional consequences. Some microbiota-associated mechanisms possibly influencing the clinical course have been characterized for HIV and HPV (Table 1).
Studying the interactions between host factors and pathogens is complex, especially when a third term—a virus— is added to the multifaceted dichotomy of host and microbiota. However, multi-omic techniques have allowed applying systems biology approaches and ecological concepts to analyze host-microbiota interactions during viral infections (130, 131). These approaches have boosted our ability to understand viral infection to the level in which we are starting to appreciate the importance of the commensal bacterial communities on the pathogenesis of diseases that not so long ago were assumed to depend only on the interactions between viruses and human cells. Nevertheless, current omic techniques have several limitations, such as the scarcity of standardized methods to integrate the different omic levels (132). More importantly, the research potential and fascination with the increasingly efficient omics approaches have often relegated hypothesis-driven research to a second position. We believe that, while the microbiome research primarily relying on 16S rRNA gene studies has been crucial to generate hypotheses, the field needs to move towards more mechanistic, hypothesis-driven studies and applied research.
Technologies such as Next Generation Sequencing (NGS), RNA sequencing (RNA-seq), and mass spectrometry (MS/MS) and all their different variations have already been used to describe the global landscape of viral-host interactions. Typically, gain and loss-of-function studies are performed to study the differential expression of DNA or RNA, either of human or bacterial origin, after viral infections. However, since this cannot capture the whole picture of the complex interactions between the virus and the host, mass spectrometry started being used to study the complete proteome, secretome, and metabolome, and even for bacterial identification in clinical microbiology (133). In addition, some studies have used these technologies, even performing integration of some of them (134), to study the role of the microbiome in the inflammation state produced by HIV infection (20) and reviewed in (24, 46, 135) and in pathogenesis and progression to cancer after HPV infection (74, 93, 136).
Improvements in meta-omic techniques have mainly been used to study the totality of the aimed compounds (genes, proteins, and metabolites) in a set of commensal organisms (metagenomics, metaproteomics, and metametabolomics). Currently, sophisticated versions of these methodologies are becoming more commonly used. For example, shallow metagenomics sequencing is being used to obtain strain-level resolution (137). This, together with the development of advanced computational methods (138), is increasing our resolution allowing the identification of novel strains with probiotic potential, an unmet need by previous studies with prebiotics or probiotics in PLWH (39, 64). Other thriving methods include single-cell technologies, which allow isolating, culturing, and characterizing the genomes and transcriptomes of individual microbes in complex communities (139), or tridimensional mapping of the host microbiota interactions within the mucosa, which is advancing our understanding of the microbiota-immune response interactions to the next level (140).
Inside and outside the HIV and HPV fields, the lack of methodological standardization is one of the main limitations in the study of the microbiome and challenges reproducibility (141). For example, a comparison of the clinical impacts of the use of probiotic showed very different results (142) (see Table 2). Although, as discussed before, technologies are improving, and now is possible to perform whole genome shotgun sequencing to enhance the detection of diversity, prediction of genes, and accuracy of bacterial species detection (166). However, it is also important to complement the studies by using omics other than genomics to obtain information at the functional level, although there are also challenges regarding the standardization of these methodologies (136, 167, 168). One of these challenges is the integration of datasets (169), which has led to the proposal of the use of machine learning and artificial intelligence for this task, which also have intrinsic limitations (170).
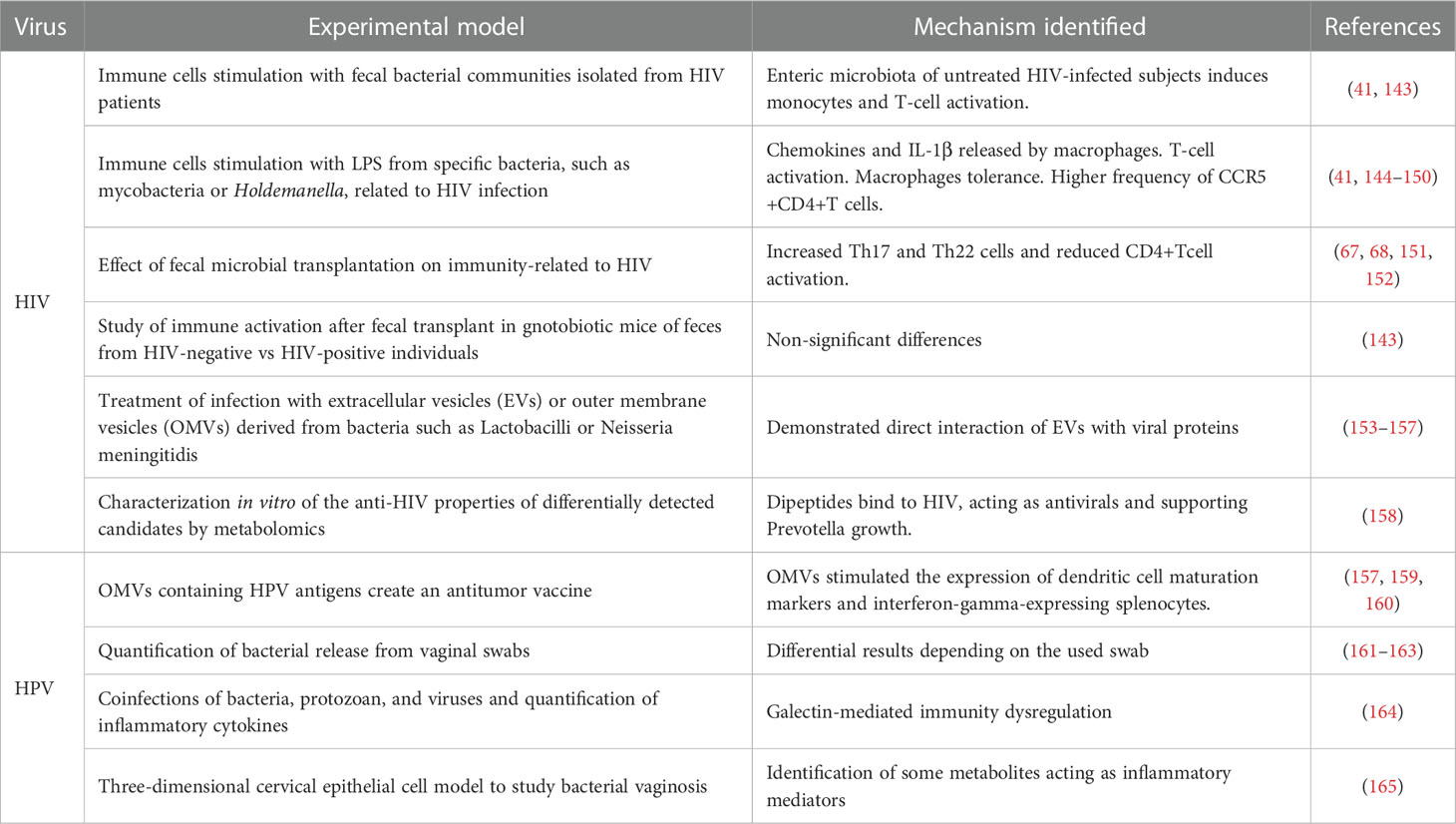
Table 2 Summary of the experimental models used to study the effects of microbiota on HIV and HPV infection.
Omic technologies result in compositional profiles and large taxonomic lists for which we lack culture methods in most cases. ‘Culturomics’—a high-throughput culture method— and MALDI-TOF mass spectrometry allow the growth of fastidious bacteria together with the identification of several bacterial species and longer incubation periods. However, these techniques have only allowed us to partially overcome the previously mentioned limitations (171). Furthermore, validation of results obtained from the omic techniques is challenging since, in most of the cases, if validation is performed, only a few of the most statistically significant hits are selected for validation. Even when results are validated, any assumption made or reductionist approach used in the experimental design need to be revisited in order to ensure that the results are physiologically relevant and translation to their clinical use can be performed.
In the case of microbiota studies, omic techniques may often result in compositional profiles and large taxonomic lists for which we lack culture methods in most cases. ‘Culturomics’—a high-throughput culture method—allows the growth of fastidious bacteria and more extended incubation periods, and MALDI-TOF mass spectrometry allows the identification of several bacterial species. However, these techniques have only allowed us to partially overcome the previously mentioned limitations (171).
Thus, the previously described shortcomings pose an enormous challenge to unleashing the clinical potential of microbiota role in medicine. If we want to assess the causal-effect relationship better and move towards applied microbiome research, it will be necessary to start with a clinical question and use the most consistent methodology to perform hypothesis-driven research that identifies convincing interactions and confounders. For this, we will need to define first the best hypothesis inspired by a clinical question. Then, from the hypothesis, we should carefully design the experimental approaches (e.g. different omics) and analysis (e.g. network models) and further perform experimental validation, including controls and complementary data sets (e.g., qPCR to confirm sequencing, immunoblot to confirm proteomic, fluorescence resonance energy transfer to confirm AP-MS data, infection kinetics, results validation in external cohorts, etc).
For example, we recently sought to solve a clinical need using applied microbiome research. We asked whether the microbiome could be harnessed to improve the prevention of anal precancer—a leading neoplasia in PWLH— for which we need better screening tools. After investigating a discovery and a validation cohort of at-risk patients, we discovered twelve proteins, previously reported to be associated with cancer progression, that were overexpressed in the anal bacteria from subjects with precancerous lesions. Since these proteins contribute to succinyl-CoA and cobalamin production, we measured the intracellular bacterial concentrations of these metabolites. We discovered that cobalamin and succinyl-CoA were increased in the anal microbiome of patients with anal precancer and overperformed the reference test—anal cytology—. Furthermore, we validated the findings in an external validation cohort, and we demonstrated greater in vitro production of succinyl-CoA and cobalamin in bacteria associated with HSIL or cancer vs. those presumably protective (172). Therefore, starting from a clinical question and integrating data from different omic levels we were able to define a new microbiome-based tool that could help in the prevention of a common cancer in PLWH by discovering two powerful biomarkers of anal precancer that could improve anal cancer prevention.
To overcome the limitations mentioned before and demonstrate the mechanisms driving the effects of the microbiota on HIV and HPV infection, hypothesis-driven experimental designs based on the information generated from the omics technologies should be encouraged. Some leading studies using this approach have been performed in HIV and HPV fields and are summarized in Table 2.
Although some improvements are being established in the experimental designs to demonstrate mechanisms led by microbiota components, there are still several limitations. These include a lack of standardization of the methods for obtaining the samples; understanding of differences on the effect of microbiota compartments (such as feces, tissues or EVs) and finding their correct origin (173); or extrapolation of findings in other model organisms, such as rodents (174), to human diseases, that are unrealistic. Furthermore, the in vivo models have been helpful in the past in proving the functional consequences of the microbiota. However, the differences between the animal models and the human anatomy, immune system, and genetic background are significant, and the type and mechanisms of interactions of the host with the pathogens are hard to reproduce, even in humanized mice models. Even more, if we only look at studies based on the human model, we still find difficulties in setting up proper validations and standardizations. For example, a significant challenge for human studies is controlling for confounding factors beyond age, sex, and sexual preferences (175), such as host genetic, diet, life style or presence of other pathologies or infections.
Although in the last decade, we have witnessed remarkable advances in the field of the microbiota in HIV and HPV infections, we still need to improve our understanding of the specific mechanisms by which the microbiota influences HIV and HPV pathogenesis and how effectively modulate the relevant microbiota-host interactions through targeted interventions. The current state-of-the-art suggests that the microbiota could offer relevant clinical applications for HIV and HPV diseases that might prove suitable to stratify the risk of HIV acquisition (reviewed in (176)), helping to the diagnosis of comorbidities (e.g., tuberculosis or anal dysplasia). This field might also advance the therapeutic options for HIV and HPV, including the development of new treatments or adjuvants through probiotics or postbiotics that could lead to more personalized medicine approaches, including targeting chronic inflammation (67), enhancing immune recovery (60), or facilitating HPV clearance (177). However, if we want to translate our current knowledge into clinical applications, we will have to overcome several methodological challenges, such as standardization of the methods to assess the species level and identify unknown microorganisms that represent today a significant fraction of the microbiota. Advancing culturomic approaches, microbiome-imaging techniques, multiomic integration, and validating the findings in hypothesis-driven experimental designs will also help the field to move forward. Finally, we will need to validate the conclusions from translational research in observational or interventional studies designed ad hoc to test previously generated hypotheses. While one decade of research has paved the road for investigating clinical applications of the microbiome in HIV and HPV infections, we face the challenge of learning how to harness the microbiome in medicine in the next years.
EM: writing first draft. All authors: revision and writing of manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the Instituto de Salud Carlos III and Fondos FEDER, Acción Estratégica en Salud (PI18/00154, ICI20/00058 and PI21/00041).
We thank all the participants and funders who support research in this field.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Comstock LE, Kasper DL. Bacterial glycans: Key mediators of diverse host immune responses. Cell (2006) 126:847–50. doi: 10.1016/j.cell.2006.08.021
2. Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A (2006) 103:12115–20. doi: 10.1073/PNAS.0605127103
3. Saw JHW. Characterizing the uncultivated microbial minority: towards understanding the roles of the rare biosphere in microbial communities. mSystems (2021) 6:6. doi: 10.1128/mSystems.00773-21
4. Dhar D, Mohanty A. Gut microbiota and covid-19- possible link and implications. Virus Res (2020) 285:198018. doi: 10.1016/j.virusres.2020.198018
5. Brenchley JM, Douek DC. The mucosal barrier and immune activation in HIV pathogenesis. Curr Opin HIV AIDS (2008) 3:356–61. doi: 10.1097/COH.0b013e3282f9ae9c
6. Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity (2013) 39:633–45. doi: 10.1016/j.immuni.2013.10.001
7. Ancuta P, Kamat A, Kunstman KJ, Kim E-Y, Autissier P, Wurcel A, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS One (2008) 3:e2516. doi: 10.1371/journal.pone.0002516
8. Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis (2015) 211:19–27. doi: 10.1093/infdis/jiu409
9. Ericsen AJ, Lauck M, Mohns MS, DiNapoli SR, Mutschler JP, Greene JM, et al. Microbial translocation and inflammation occur in hyperacute immunodeficiency virus infection and compromise host control of virus replication. PLoS Pathog (2016) 12:e1006048. doi: 10.1371/journal.ppat.1006048
10. Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med (2006) 12:1365–71. doi: 10.1038/nm1511
11. Santella B, Schettino MT, Franci G, De Franciscis P, Colacurci N, Schiattarella A, et al. Microbiota and HPV: The role of viral infection on vaginal microbiota. J Med Virol (2022) 94:4478–84. doi: 10.1002/jmv.27837
12. Mortaki D, Gkegkes ID, Psomiadou V, Blontzos N, Prodromidou A, Lefkopoulos F, et al. Vaginal microbiota and human papillomavirus: a systematic review. J Turk Ger Gynecol Assoc (2020) 21:193–200. doi: 10.4274/jtgga.galenos.2019.2019.0051
13. Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet (2017) 18:690–9. doi: 10.1038/nrg.2017.63
14. Goodrich JK, Davenport ER, Clark AG, Ley RE. The relationship between the human genome and microbiome comes into view. Annu Rev Genet (2017) 51:413–33. doi: 10.1146/annurev-genet-110711-155532
15. Vázquez-castellanos JF, Jiménez-hernández SSN, Dolores M, Sara R, David G, Manuel R, et al. Interplay between gut microbiota metabolism and in fl ammation in HIV infection ISME J. (2018) 12, 1964–76. doi: 10.1038/s41396-018-0151-8
16. Serrano-Villar S, Vázquez-Castellanos JF, Vallejo A, Latorre A, Sainz T, Ferrando-Martínez S, et al. The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol (2017) 10:1279–93. doi: 10.1038/mi.2016.122
17. Karu N, Deng L, Slae M, Guo AC, Sajed T, Huynh H, et al. A review on human fecal metabolomics: Methods, applications and the human fecal metabolome database. Analytica Chimica Acta (2018) 1030:1–24. doi: 10.1016/j.aca.2018.05.031
18. Colosimo DA, Kohn JA, Luo PM, Piscotta FJ, Han SM, Pickard AJ, et al. Mapping interactions of microbial metabolites with human G-Protein-Coupled receptors. Cell Host Microbe (2019) 26:273–82.e7. doi: 10.1016/j.chom.2019.07.002
19. Sperk M, Ambikan AT, Ray S, Singh K, Mikaeloff F, Diez RC, et al. Fecal metabolome signature in the HIV-1 elite control phenotype: Enrichment of dipeptides acts as an HIV-1 antagonist but a prevotella agonist. J Virol (2021) 95:1–13. doi: 10.1128/jvi.00479-21
20. Serrano-Villar S, Rojo D, Martínez-Martínez M, Deusch S, Vázquez-Castellanos JF, Bargiela R, et al. Gut bacteria metabolism impacts immune recovery in HIV-infected individuals. EBioMedicine (2016) 8:203–16. doi: 10.1016/j.ebiom.2016.04.033
21. Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol (2011) 19:349–59. doi: 10.1016/j.tim.2011.05.006
22. Norenhag J, Du J, Olovsson M, Verstraelen H, Engstrand L, Brusselaers N. The vaginal microbiota, human papillomavirus and cervical dysplasia: a systematic review and network meta-analysis. BJOG : an Int J Obstetrics Gynaecol (2020) 127:171–80. doi: 10.1111/1471-0528.15854
23. Sehgal R, Bedi O, Trehanpati N. Role of microbiota in pathogenesis and management of viral hepatitis. Front Cell Infect Microbiol (2020) 10:341. doi: 10.3389/fcimb.2020.00341
24. Zevin AS, McKinnon L, Burgener A, Klatt NR. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS (2016) 11:182–90. doi: 10.1097/COH.0000000000000234
25. Ullrich R, Zeitz M, Riecken EO. Enteric immunologic abnormalities in human immunodeficiency virus infection. Semin Liver Dis (1992) 12:167–74. doi: 10.1055/s-2007-1007388
26. Robinson CM, Jesudhasan PR, Pfeiffer JK. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe (2014) 15:36–46. doi: 10.1016/j.chom.2013.12.004
27. Franzen C, Salzberger B, Fätkenheuer G, Eidt S, Schrappe M. [Mucosa-associated immune system in HIV-1 infection. T-cell subpopulations compared in different segments of the gastrointestinal tract]. Med Klin (Munich) (1992) 87:510–512, 549.
28. Gary EN, Kutzler MA. Defensive driving: Directing HIV-1 vaccine-induced humoral immunity to the mucosa with chemokine adjuvants. J Immunol Res (2018) 2018:1–14. doi: 10.1155/2018/3734207
29. McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome (2013) 1:26. doi: 10.1186/2049-2618-1-26
30. Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med (2004) 200:749–59. doi: 10.1084/jem.20040874
31. Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, et al. Severe CD4 + T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol (2003) 77:11708–17. doi: 10.1128/JVI.77.21.11708-11717.2003
32. Mehandru S, Poles MA, Tenner-Racz K, Manuelli V, Jean-Pierre P, Lopez P, et al. Mechanisms of gastrointestinal CD4 + T-cell depletion during acuteand early human immunodeficiency virus type 1 infection. J Virol (2007) 81:599–612. doi: 10.1128/JVI.01739-06
33. Sankaran S, George MD, Reay E, Guadalupe M, Flamm J, Prindiville T, et al. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J Virol (2008) 82:538–45. doi: 10.1128/JVI.01449-07
34. Epple H-J, Schneider T, Troeger H, Kunkel D, Allers K, Moos V, et al. Impairment of the intestinal barrier is evident in untreated but absent in suppressively treated HIV-infected patients. Gut (2009) 58:220 LP – 227. doi: 10.1136/gut.2008.150425
35. Perkins MR, Bartha I, Timmer JK, Liebner JC, Wolinsky D, Wollinsky D, et al. The interplay between host genetic variation, viral replication, and microbial translocation in untreated HIV-infected individuals. J Infect Dis (2015) 212:578–84. doi: 10.1093/infdis/jiv089
36. Somsouk M, Estes JD, Deleage C, Dunham RM, Albright R, Inadomi JM, et al. Gut epithelial barrier and systemic inflammation during chronic HIV infection. AIDS (2015) 29:43–51. doi: 10.1097/QAD.0000000000000511
37. Hunt PW, Sinclair E, Rodriguez B, Shive C, Clagett B, Funderburg N, et al. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis (2014) 210:1228–38. doi: 10.1093/infdis/jiu238
38. Chamoun MN, Blumenthal A, Sullivan MJ, Schembri MA, Ulett GC. Bacterial pathogenesis and interleukin-17: interconnecting mechanisms of immune regulation, host genetics, and microbial virulence that influence severity of infection. Crit Rev Microbiol (2018) 44:465–86. doi: 10.1080/1040841X.2018.1426556
39. Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol (2014) 7:983–94. doi: 10.1038/mi.2013.116
40. Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe (2013) 14:329–39. doi: 10.1016/j.chom.2013.08.006
41. Neff CP, Krueger O, Xiong K, Arif S, Nusbacher N, Schneider JM, et al. Fecal microbiota composition drives immune activation in HIV-infected individuals. EBioMedicine (2018) 30:192–202. doi: 10.1016/j.ebiom.2018.03.024
42. Coker MO, Cairo C, Garzino-Demo A. HIV-Associated interactions between oral microbiota and mucosal immune cells: Knowledge gaps and future directions. Front Immunol (2021) 12:676669. doi: 10.3389/fimmu.2021.676669
43. Armstrong E, Kaul R. Beyond bacterial vaginosis: vaginal lactobacilli and HIV risk. Microbiome (2021) 9:239. doi: 10.1186/s40168-021-01183-x
44. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell (2022) 185:8, 1356–72. doi: 10.1016/j.cell.2022.02.027
45. Park EM, Chelvanambi M, Bhutiani N, Kroemer G, Zitvogel L, Wargo JA. Targeting the gut and tumor microbiota in cancer. Nat Med (2022) 28:690–703. doi: 10.1038/s41591-022-01779-2
46. Serrano-Villar S, Moreno S, Ferrer M. The functional consequences of the microbiome in HIV: insights from metabolomic studies. Curr Opin HIV AIDS (2018) 13:88–94. doi: 10.1097/COH.0000000000000430
47. Serrano-Villar S, Rojo D, Martínez-Martínez M, Deusch S, Vázquez-Castellanos JF, Sainz T, et al. HIV Infection results in metabolic alterations in the gut microbiota different from those induced by other diseases. Sci Rep (2016) 6:26192. doi: 10.1038/srep26192
48. Quaranta MG, Vincentini O, Felli C, Spadaro F, Silano M, Moricoli D, et al. Exogenous HIV-1 nef upsets the IFN-γ-induced impairment of human intestinal epithelial integrity. PLoS One (2011) 6:e23442. doi: 10.1371/journal.pone.0023442
49. Benhar M, Shytaj IL, Stamler JS, Savarino A. Dual targeting of the thioredoxin and glutathione systems in cancer and HIV. J Clin Invest (2016) 126:1630–9. doi: 10.1172/JCI85339
50. Bhaskar A, Munshi M, Khan SZ, Fatima S, Arya R, Jameel S, et al. Measuring glutathione redox potential of HIV-1-infected macrophages. J Biol Chem (2015) 290:1020–38. doi: 10.1074/jbc.M114.588913
51. Grund B, Baker JV, Deeks SG, Wolfson J, Wentworth D, Cozzi-Lepri A, et al. Relevance of interleukin-6 and d-dimer for serious non-AIDS morbidity and death among HIV-positive adults on suppressive antiretroviral therapy. PloS One (2016) 11:e0155100. doi: 10.1371/journal.pone.0155100
52. Tenorio AR, Zheng Y, Bosch RJ, Deeks SG, Rodriguez B, Krishnan S, et al. Soluble markers of inflammation & coagulation , but not T-cell activation , predict non-AIDS- defining events during suppressive antiretroviral therapy (ART) J Infect Dis. (2014) 210:(8) 1248-59. doi: 10.1093/infdis/jiu254
53. Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Trans Med (2013) 5:193ra91. doi: 10.1126/scitranslmed.3006438
54. Giron LB, Tanes CE, Schleimann MH, Engen PA, Mattei LM, Anzurez A, et al. Sialylation and fucosylation modulate inflammasome-activating eIF2 signaling and microbial translocation during HIV infection. Mucosal Immunol (2020) 13:753–66. doi: 10.1038/s41385-020-0279-5
55. Oliva A, Aversano L, De Angelis M, Mascellino MT, Miele MC, Morelli S, et al. Persistent systemic microbial translocation, inflammation, and intestinal damage during clostridioides difficile infection. Open Forum Infect Dis (2020) 7:1. doi: 10.1093/ofid/ofz507
56. Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol (2012) 30:149–73. doi: 10.1146/annurev-immunol-020711-075001
57. Klase Z, Ortiz A, Deleage C, Mudd JC, Quiñones M, Schwartzman E, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol (2015) 8:1009–20. doi: 10.1038/mi.2014.128
58. Serrano-Villar S, Sanchez-Carrillo S, Talavera-Rodríguez A, Lelouvier B, Gutiérrez C, Vallejo A, et al. Blood bacterial profiles associated with human immunodeficiency virus infection and immune recovery. J Infect Dis (2021) 223:471–81. doi: 10.1093/infdis/jiaa379
59. Nganou-Makamdop K, Talla A, Sharma AA, Darko S, Ransier A, Laboune F, et al. Translocated microbiome composition determines immunological outcome in treated HIV infection. Cell (2021) 184:15, 3899–914. doi: 10.1016/j.cell.2021.05.023
60. Cahn P, Ruxrungtham K, Gazzard B, Diaz RS, Gori A, Kotler DP, et al. The immunomodulatory nutritional intervention NR100157 reduced CD4+ T-cell decline and immune activation: a 1-year multicenter randomized controlled double-blind trial in HIV-infected persons not receiving antiretroviral therapy (The BITE study). Clin Infect Dis (2013) 57:139–46. doi: 10.1093/cid/cit171
61. Villar-García J, Hernández JJ, Güerri-Fernández R, González A, Lerma E, Guelar A, et al. Effect of probiotics (Saccharomyces boulardii) on microbial translocation and inflammation in HIV-treated patients: a double-blind, randomized, placebo-controlled trial. J Acquir Immune Defic Syndr (2015) 68:256–63. doi: 10.1097/QAI.0000000000000468
62. Stiksrud B, Nowak P, Nwosu FC, Kvale D, Thalme A, Sonnerborg A, et al. Reduced levels of d-dimer and changes in gut microbiota composition after probiotic intervention in HIV-infected individuals on stable ART. JAIDS J Acquired Immune Deficiency Syndromes (2015) 70:329–37. doi: 10.1097/QAI.0000000000000784
63. Presti RM, Yeh E, Williams B, Landay A, Jacobson JM, Wilson C, et al. A randomized, placebo-controlled trial assessing the effect of VISBIOME ES probiotic in people with HIV on antiretroviral therapy. Open Forum Infect Dis (2021) 8:ofab550. doi: 10.1093/ofid/ofab550
64. Serrano-Villar S, de Lagarde M, Vázquez-Castellanos J, Vallejo A, Bernadino JI, Madrid N, et al. Effects of immunonutrition in advanced human immunodeficiency virus disease: A randomized placebo-controlled clinical trial (Promaltia study). Clin Infect Dis (2018) 68:120–30. doi: 10.1093/cid/ciy414
65. Williams BB, Green SJ, Bosch RJ, Chan ES, Jacobson JM, Margolis DM, et al. Four weeks of treatment with rifaximin fails to significantly alter microbial diversity in rectal samples of HIV-infected immune non-responders (ACTG A5286) which may be attributed to rectal swab use. PAI (2019) 4:235. doi: 10.20411/pai.v4i2.290
66. Tenorio AR, Chan ES, Bosch RJ, Macatangay BJC, Read SW, Yesmin S, et al. Rifaximin has a marginal impact on microbial translocation, T-cell activation and inflammation in HIV-positive immune non-responders to antiretroviral therapy – ACTG A5286. J Infect Dis (2015) 211:780–90. doi: 10.1093/infdis/jiu515
67. Serrano-Villar S, Talavera-Rodríguez A, Gosalbes MJ, Madrid N, Pérez-Molina JA, Elliott RJ, et al. Fecal microbiota transplantation in HIV: A pilot placebo-controlled study. Nat Commun (2021) 12:1139. doi: 10.1038/s41467-021-21472-1
68. Utay NS, Monczor AN, Somasunderam A, Lupo S, Jiang Z-D, Alexander AS, et al. Evaluation of six weekly oral fecal microbiota transplants in people with HIV. Pathog Immun (2020) 5:364–81. doi: 10.20411/pai.v5i1.388
69. Gori A, Rizzardini G, van’t Land B, Amor KB, van Schaik J, Torti C, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the “COPA” pilot randomized trial. Mucosal Immunol (2011) 4:554–63. doi: 10.1038/mi.2011.15
70. Kettelhut A, Bowman E, Funderburg NT. Immunomodulatory and anti-inflammatory strategies to reduce comorbidity risk in people with HIV. Curr HIV/AIDS Rep (2020) 17:394–404. doi: 10.1007/s11904-020-00509-y
71. Schiffman M, Doorbar J, Wentzensen N, de Sanjosé S, Fakhry C, Monk BJ, et al. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers (2016) 2:16086. doi: 10.1038/nrdp.2016.86
72. Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog (2009) 5:e1000318. doi: 10.1371/journal.ppat.1000318
73. Fernandes JV, Medeiros Fernandes TAA DE, Azevedo JCV DE, Cobucci RNO DE, Carvalho MGF, Andrade VS, et al. Link between chronic inflammation and human papillomavirus-induced carcinogenesis (Review). Oncol Lett (2015) 9:1015–26. doi: 10.3892/ol.2015.2884
74. Lin D, Kouzy R, Jaoude JA, Noticewala SS, Delgado Medrano AY, Klopp AH, et al. Microbiome factors in HPV-driven carcinogenesis and cancers. PloS Pathog (2020) 16:e1008524. doi: 10.1371/journal.ppat.1008524
75. Borgogna JC, Shardell MD, Santori EK, Nelson TM, Rath JM, Glover ED, et al. The vaginal metabolome and microbiota of cervical HPV-positive and HPV-negative women: a cross-sectional analysis. BJOG : an Int J Obstetrics Gynaecol (2020) 127:182–92. doi: 10.1111/1471-0528.15981
76. Chang AH, Parsonnet J. Role of bacteria in oncogenesis. Clin Microbiol Rev (2010) 23:837–57. doi: 10.1128/CMR.00012-10
77. Herrera S, Martínez-Sanz J, Serrano-Villar SHIV. Cancer, and the microbiota: Common pathways influencing different diseases. Front Immunol (2019) 10:1466. doi: 10.3389/fimmu.2019.01466
78. Piyathilake CJ, Ollberding NJ, Kumar R, Macaluso M, Alvarez RD, Morrow CD. Cervical microbiota associated with higher grade cervical intraepithelial neoplasia in women infected with high-risk human papillomaviruses. Cancer Prev Res (Phila) (2016) 9:357–66. doi: 10.1158/1940-6207.CAPR-15-0350
79. Mitra A, MacIntyre DA, Marchesi JR, Lee YS, Bennett PR, Kyrgiou M. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: what do we know and where are we going next? Microbiome (2016) 4:58. doi: 10.1186/s40168-016-0203-0
80. Tamarelle J, Thiébaut ACM, de Barbeyrac B, Bébéar C, Ravel J, Delarocque-Astagneau E, et al. The vaginal microbiota and its association with human papillomavirus, chlamydia trachomatis, neisseria gonorrhoeae and mycoplasma genitalium infections: a systematic review and meta-analysis. Clin Microbiol Infect (2019) 25:35–47. doi: 10.1016/j.cmi.2018.04.019
81. Usyk M, Zolnik CP, Castle PE, Porras C, Herrero R, Gradissimo A, et al. Cervicovaginal microbiome and natural history of HPV in a longitudinal study. PLoS Pathog (2020) 16:e1008376. doi: 10.1371/journal.ppat.1008376
82. Pierro F DI, Criscuolo AA, Dei Giudici A, Senatori R, Sesti F, Ciotti M, et al. Oral administration of lactobacillus crispatus M247 to papillomavirus-infected women: results of a preliminary, uncontrolled, open trial. Minerva Obstetrics Gynecol (2021) 73:621–31. doi: 10.23736/S2724-606X.21.04752-7
83. Kyrgiou M, Moscicki A-B. Vaginal microbiome and cervical cancer. Semin Cancer Biol (2022) 86:3, 189–98. doi: 10.1016/j.semcancer.2022.03.005
84. Alimena S, Davis J, Fichorova RN, Feldman S. The vaginal microbiome: A complex milieu affecting risk of human papillomavirus persistence and cervical cancer. Curr Problems Cancer (2022) 46:100877. doi: 10.1016/j.currproblcancer.2022.100877
85. Kaelin EA, Skidmore PT, Łaniewski P, Holland LA, Chase DM, Herbst-Kralovetz MM, et al. Cervicovaginal DNA virome alterations are associated with genital inflammation and microbiota composition. mSystems (2022) 7:e00064–22. doi: 10.1128/msystems.00064-22
86. Łaniewski P, Barnes D, Goulder A, Cui H, Roe DJ, Chase DM, et al. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep (2018) 8:7593. doi: 10.1038/s41598-018-25879-7
87. Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, Téllez-Sosa J, Martínez-Barnetche J, Cortina-Ceballos B, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study. PloS One (2016) 11:e0153274. doi: 10.1371/journal.pone.0153274
88. Peghini BC, Abdalla DR, Barcelos ACM, Teodoro L das GVL, Murta EFC, Michelin MA. Local cytokine profiles of patients with cervical intraepithelial and invasive neoplasia. Hum Immunol (2012) 73:920–6. doi: 10.1016/j.humimm.2012.06.003
89. Kemp TJ, Hildesheim A, García-Piñeres A, Williams MC, Shearer GM, Rodriguez AC, et al. Elevated systemic levels of inflammatory cytokines in older women with persistent cervical human papillomavirus infection. Cancer Epidemiol Biomarkers Prev (2010) 19:1954–9. doi: 10.1158/1055-9965.EPI-10-0184
90. Moscicki A-B, Shi B, Huang H, Barnard E, Li H. Cervical-vaginal microbiome and associated cytokine profiles in a prospective study of HPV 16 acquisition, persistence, and clearance. Front Cell Infect Microbiol (2020) 10:569022. doi: 10.3389/fcimb.2020.569022
91. Łaniewski P, Cui H, Roe DJ, Chase DM, Herbst-Kralovetz MM. Vaginal microbiota, genital inflammation, and neoplasia impact immune checkpoint protein profiles in the cervicovaginal microenvironment. NPJ Precis Onc (2020) 4:22. doi: 10.1038/s41698-020-0126-x
92. Łaniewski P, Cui H, Roe DJ, Barnes D, Goulder A, Monk BJ, et al. Features of the cervicovaginal microenvironment drive cancer biomarker signatures in patients across cervical carcinogenesis. Sci Rep (2019) 9:7333. doi: 10.1038/s41598-019-43849-5
93. Shannon B, Yi TJ, Perusini S, Gajer P, Ma B, Humphrys MS, et al. Association of HPV infection and clearance with cervicovaginal immunology and the vaginal microbiota. Mucosal Immunol (2017) 10:1310–9. doi: 10.1038/mi.2016.129
94. Gardella B, Pasquali MF, La Verde M, Cianci S, Torella M, Dominoni M. The complex interplay between vaginal microbiota, HPV infection, and immunological microenvironment in cervical intraepithelial neoplasia: A literature review. IJMS (2022) 23:7174. doi: 10.3390/ijms23137174
95. Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrús ML, Madrid N, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol (2015) 8:760–72. doi: 10.1038/mi.2014.107
96. Ilhan ZE, Łaniewski P, Thomas N, Roe DJ, Chase DM, Herbst-Kralovetz MM. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine (2019) 44:675–90. doi: 10.1016/j.ebiom.2019.04.028
97. Chorna N, Romaguera J, Godoy-Vitorino F. Cervicovaginal microbiome and urine metabolome paired analysis reveals niche partitioning of the microbiota in patients with human papilloma virus infections. Metabolites (2020) 10:36. doi: 10.3390/metabo10010036
98. Kamble A, Naik S, Talathi M, Jadhav D, Pingale S, Kaul-Ghanekar R. Cervicovaginal microbiota isolated from healthy women exhibit probiotic properties and antimicrobial activity against pathogens isolated from cervical cancer patients. Arch Microbiol (2022) 204:491. doi: 10.1007/s00203-022-03103-5
99. Masson L, Passmore J-AS, Liebenberg LJ, Werner L, Baxter C, Arnold KB, et al. Genital inflammation and the risk of HIV acquisition in women. Clin Infect Dis (2015) 61:260–9. doi: 10.1093/cid/civ298
100. Alisoltani A, Manhanzva MT, Potgieter M, Balle C, Bell L, Ross E, et al. Microbial function and genital inflammation in young south African women at high risk of HIV infection. Microbiome (2020) 8:165. doi: 10.1186/s40168-020-00932-8
101. Segal LN, Clemente JC, Li Y, Ruan C, Cao J, Danckers M, et al. Anaerobic bacterial fermentation products increase tuberculosis risk in antiretroviral-Drug-Treated HIV patients. Cell Host Microbe (2017) 21:530–7.e4. doi: 10.1016/j.chom.2017.03.003
102. Lebeau A, Bruyere D, Roncarati P, Peixoto P, Hervouet E, Cobraiville G, et al. HPV infection alters vaginal microbiome through down-regulating host mucosal innate peptides used by lactobacilli as amino acid sources. Nat Commun (2022) 13:1–20. doi: 10.1038/s41467-022-28724-8
103. Olmsted SS, Meyn LA, Rohan LC, Hillier SL. Glycosidase and proteinase activity of anaerobic gram-negative bacteria isolated from women with bacterial vaginosis. Sexually Transmitted Dis (2003) 30:257–61. doi: 10.1097/00007435-200303000-00016
104. Serrano-Villar S, Vásquez-Domínguez E, Pérez-Molina JA, Sainz T, de Benito A, Latorre A, et al. HPV, and microbiota: partners in crime? AIDS (2017) 31:591–4. doi: 10.1097/QAD.0000000000001352
105. Poropatich K, Paunesku T, Zander A, Wray B, Schipma M, Dalal P, et al. Elemental zn and its binding protein zinc-α2-Glycoprotein are elevated in HPV-positive oropharyngeal squamous cell carcinoma. Sci Rep (2019) 9:16965. doi: 10.1038/s41598-019-53268-1
106. Ino K, Yoshida N, Kajiyama H, Shibata K, Yamamoto E, Kidokoro K, et al. Indoleamine 2,3-dioxygenase is a novel prognostic indicator for endometrial cancer. Br J Cancer (2006) 95:1555–61. doi: 10.1038/sj.bjc.6603477
107. Schalper KA, Carvajal-Hausdorf D, McLaughlin J, Altan M, Velcheti V, Gaule P, et al. Differential expression and significance of PD-L1, IDO-1, and B7-H4 in human lung cancer. Clin Cancer Res (2017) 23:370–8. doi: 10.1158/1078-0432.CCR-16-0150
108. De Schutter T, Andrei G, Topalis D, Duraffour S, Mitera T, Naesens L, et al. Cidofovir treatment improves the pathology caused by the growth of human papillomavirus-positive cervical carcinoma xenografts in athymic nude mice. Cancer Lett (2013) 329:137–45. doi: 10.1016/j.canlet.2012.10.036
109. Robles AI, Cooks T, Vega-Valle E, Vetizou M, Rose U, Miyanaga A, et al. Abstract PR07: Role of the microbiota in inflammation and lung cancer. Clin Cancer Res (2018) 24:PR07–7. doi: 10.1158/1557-3265.AACRIASLC18-PR07
110. McDermott AJ, Huffnagle GB. The microbiome and regulation of mucosal immunity. Immunology (2014) 142:24–31. doi: 10.1111/imm.12231
111. Rams TE, Andriolo M, Feik D, Abel SN, McGivern TM, Slots J. Microbiological study of HIV-related periodontitis. J Periodontol (1991) 62:74–81. doi: 10.1902/jop.1991.62.1.74
112. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature (2008) 453:620–5. doi: 10.1038/nature07008
113. Zheng J-J, Song J-H, Yu C-X, Wang F, Wang P-C, Meng J-W. Difference in vaginal microecology, local immunity and HPV infection among childbearing-age women with different degrees of cervical lesions in inner Mongolia. BMC Womens Health (2019) 19:109. doi: 10.1186/s12905-019-0806-2
114. Kehrmann J, Menzel J, Saeedghalati M, Obeid R, Schulze C, Holzendorf V, et al. Gut microbiota in human immunodeficiency virus–infected individuals linked to coronary heart disease. J Infect Dis (2019) 219:497–508. doi: 10.1093/infdis/jiy524
115. Haissman JM, Haugaard AK, Ostrowski SR, Berge RK, Hov JR, Trøseid M, et al. Microbiota-dependent metabolite and cardiovascular disease marker trimethylamine-n-oxide (TMAO) is associated with monocyte activation but not platelet function in untreated HIV infection. BMC Infect Dis (2017) 17:445. doi: 10.1186/s12879-017-2547-x
116. Missailidis C, Neogi U, Stenvinkel P, Trøseid M, Nowak P, Bergman P. The microbial metabolite trimethylamine-n-oxide in association with inflammation and microbial dysregulation in three HIV cohorts at various disease stages. AIDS (2018) 32:1589–98. doi: 10.1097/QAD.0000000000001813
117. Mostowska A, Myka M, Lianeri M, Roszak A, Jagodziński PP. Folate and choline metabolism gene variants and development of uterine cervical carcinoma. Clin Biochem (2011) 44:596–600. doi: 10.1016/j.clinbiochem.2011.02.007
118. Serrano-Villar S, Rojo D, Martínez-Martínez M, Deusch S, Vázquez-Castellanos JF, Bargiela R, et al. Gut bacteria metabolism impacts immune recovery in HIV-infected individuals. EBioMedicine (2016) 8:203–16. doi: 10.1016/j.ebiom.2016.04.033
119. Deusch S, Serrano-Villar S, Rojo D, Martínez-Martínez M, Bargiela R, Vázquez-Castellanos JF, et al. Effects of HIV, antiretroviral therapy and prebiotics on the active fraction of the gut microbiota. AIDS (2018) 32:1229–37. doi: 10.1097/QAD.0000000000001831
120. Torcia M. Interplay among vaginal microbiome, immune response and sexually transmitted viral infections. IJMS (2019) 20:266. doi: 10.3390/ijms20020266
121. Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PloS Pathog (2014) 10:e1003829. doi: 10.1371/journal.ppat.1003829
122. Rocafort M, Noguera-Julian M, Rivera J, Pastor L, Guillén Y, Langhorst J, et al. Evolution of the gut microbiome following acute HIV-1 infection. Microbiome (2019) 7:73. doi: 10.1186/s40168-019-0687-5
123. Lee SC, Chua LL, Yap SH, Khang TF, Leng CY, Raja Azwa RI, et al. Enrichment of gut-derived fusobacterium is associated with suboptimal immune recovery in HIV-infected individuals. Sci Rep (2018) 8:14277. doi: 10.1038/s41598-018-32585-x
124. Alizadehmohajer N, Shojaeifar S, Nedaeinia R, Esparvarinha M, Mohammadi F, Ferns GA, et al. Association between the microbiota and women’s cancers - cause or consequences? BioMed Pharmacother (2020) 127:110203. doi: 10.1016/j.biopha.2020.110203
125. Pérez-Santiago J, Gianella S, Massanella M, Spina CA, Karris MY, Var SR, et al. Gut lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS (2013) 27:1921–31. doi: 10.1097/qad.0b013e3283611816
126. Boccardo E, Lepique AP, Villa LL. The role of inflammation in HPV carcinogenesis. Carcinogenesis (2010) 31:1905–12. doi: 10.1093/carcin/bgq176
127. Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res (2014) 24:185–99. doi: 10.1101/gr.164806.113
128. Pett MR, Alazawi WOF, Roberts I, Dowen S, Smith DI, Stanley MA, et al. Acquisition of high-level chromosomal instability is associated with integration of human papillomavirus type 16 in cervical keratinocytes. Cancer Res (2004) 64:1359–68. doi: 10.1158/0008-5472.can-03-3214
129. Coussens LM, Werb Z. Inflammation and cancer. Nature (2002) 420:860–7. doi: 10.1038/nature01322
130. Albery GF, Becker DJ, Brierley L, Brook CE, Christofferson RC, Cohen LE, et al. The science of the host–virus network. Nat Microbiol (2021) 6:1483–92. doi: 10.1038/s41564-021-00999-5
131. Eckhardt M, Hultquist JF, Kaake RM, Hüttenhain R, Krogan NJ. A systems approach to infectious disease. Nat Rev Genet (2020) 21:339–54. doi: 10.1038/s41576-020-0212-5
132. Gonçalves RS, Musen MA. The variable quality of metadata about biological samples used in biomedical experiments. Sci Data (2019) 6:190021. doi: 10.1038/sdata.2019.21
133. Seng P, Abat C, Rolain JM, Colson P, Lagier J-C, Gouriet F, et al. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol (2013) 51:2182–94. doi: 10.1128/JCM.00492-13
134. Nyholm L, Koziol A, Marcos S, Botnen AB, Aizpurua O, Gopalakrishnan S, et al. Holo-omics: Integrated host-microbiota multi-omics for basic and applied biological research. iScience (2020) 23:101414. doi: 10.1016/j.isci.2020.101414
135. Dillon SM, Frank DN, Wilson CC. The gut microbiome and HIV-1 pathogenesis. AIDS (2016) 30:2737–51. doi: 10.1097/QAD.0000000000001289
136. Chorna N, Godoy-Vitorino F. A protocol for the multi-omic integration of cervical microbiota and urine metabolomics to understand human papillomavirus (HPV)-driven dysbiosis. Biomedicines (2020) 8:81. doi: 10.3390/biomedicines8040081
137. Snipen L, Angell I-L, Rognes T, Rudi K. Reduced metagenome sequencing for strain-resolution taxonomic profiles. Microbiome (2021) 9:79. doi: 10.1186/s40168-021-01019-8
138. Anyansi C, Straub TJ, Manson AL, Earl AM, Abeel T. Computational methods for strain-level microbial detection in colony and metagenome sequencing data. Front Microbiol (2020) 11:1925. doi: 10.3389/fmicb.2020.01925
139. Lloréns-Rico V, Simcock JA, Huys GRB, Raes J. Single-cell approaches in human microbiome research. Cell (2022) 185:2725–38. doi: 10.1016/j.cell.2022.06.040
140. Mondragón-Palomino O, Poceviciute R, Lignell A, Griffiths JA, Takko H, Ismagilov RF. Three-dimensional imaging for the quantification of spatial patterns in microbiota of the intestinal mucosa. Proc Natl Acad Sci USA (2022) 119:e2118483119. doi: 10.1073/pnas.2118483119
141. Mooser C, Gomez de Agüero M, Ganal-Vonarburg SC. Standardization in host–microbiota interaction studies: challenges, gnotobiology as a tool, and perspective. Curr Opin Microbiol (2018) 44:50–60. doi: 10.1016/j.mib.2018.07.007
142. Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ (2018) 361:k2179, k2179. doi: 10.1136/bmj.k2179
143. Li SX, Sen S, Schneider JM, Xiong K-N, Nusbacher NM, Moreno-Huizar N, et al. Gut microbiota from high-risk men who have sex with men drive immune activation in gnotobiotic mice and in vitro HIV infection. PloS Pathog (2019) 15:e1007611. doi: 10.1371/journal.ppat.1007611
144. Verani A, Scarlatti G, Comar M, Tresoldi E, Polo S, Giacca M, et al. C–c chemokines released by lipopolysaccharide (LPS)-stimulated human macrophages suppress HIV-1 infection in both macrophages and T cells. J Exp Med (1997) 185:805–16. doi: 10.1084/jem.185.5.805
145. Zhang L, Mosoian A, Schwartz ME, Florman SS, Gunasekaran G, Schiano T, et al. HIV Infection modulates IL-1β response to LPS stimulation through a TLR4-NLRP3 pathway in human liver macrophages. J Leukoc Biol (2019) 105:783–95. doi: 10.1002/JLB.4A1018-381R
146. Tincati C, Bellistrì GM, Ancona G, Merlini E, d’Arminio Monforte A, Marchetti G. Role of In vitro stimulation with lipopolysaccharide on T-cell activation in HIV-infected antiretroviral-treated patients. Clin Dev Immunol (2012) 2012:1–9. doi: 10.1155/2012/935425
147. Equils O, Salehi KK, Cornataeanu R, Lu D, Singh S, Whittaker K, et al. Repeated lipopolysaccharide (LPS) exposure inhibits HIV replication in primary human macrophages☆. Microbes Infect (2006) 8:2469–76. doi: 10.1016/j.micinf.2006.06.002
148. Juffermans NP, Paxton WA, Dekkers PEP, Verbon A, de Jonge E, Speelman P, et al. Up-regulation of HIV coreceptors CXCR4 and CCR5 on CD4+ T cells during human endotoxemia and after stimulation with (myco)bacterial antigens: the role of cytokines. Blood (2000) 96:2649–54. doi: 10.1182/blood.V96.8.2649
149. Merlini E, Tincati C, Biasin M, Saulle I, Cazzaniga FA, d’Arminio Monforte A, et al. Stimulation of PBMC and monocyte-derived macrophages via toll-like receptor activates innate immune pathways in HIV-infected patients on virally suppressive combination antiretroviral therapy. Front Immunol (2016) 7:614. doi: 10.3389/fimmu.2016.00614
150. Yamada E, Martin CG, Moreno-Huizar N, Fouquier J, Neff CP, Coleman SL, et al. Intestinal microbial communities and Holdemanella isolated from HIV+/– men who have sex with men increase frequencies of lamina propria CCR5 + CD4 + T cells. Gut Microbes (2021) 13:1997292. doi: 10.1080/19490976.2021.1997292
151. Vujkovic-Cvijin I, Rutishauser RL, Pao M, Hunt PW, Lynch SV, McCune JM, et al. Limited engraftment of donor microbiome via one-time fecal microbial transplantation in treated HIV-infected individuals. Gut Microbes (2017) 8:440–50. doi: 10.1080/19490976.2017.1334034
152. Somsouk M, Vujkovic-Cvijin I, Pao M, Hunt P, McCune M. Safety of fecal microbial transplantation during treated HIV infection: 1325. Am J Gastroenterol (2015) 110:S578–9. doi: 10.14309/00000434-201510001-01325
153. Ñahui Palomino RA, Vanpouille C, Laghi L, Parolin C, Melikov K, Backlund P, et al. Extracellular vesicles from symbiotic vaginal lactobacilli inhibit HIV-1 infection of human tissues. Nat Commun (2019) 10:5656. doi: 10.1038/s41467-019-13468-9
154. Reza Aghasadeghi M, Sharifat Salmani A, Mehdi Sadat S, Javadi F, Memarnejadian A, Vahabpour R, et al. Application of outer membrane vesicle of neisseria meningitidis serogroup b as a new adjuvant to induce strongly Th1-oriented responses against HIV-1. CHR (2011) 9:630–5. doi: 10.2174/157016211798998772
155. Ñahui Palomino RA, Zicari S, Vanpouille C, Vitali B, Margolis L. Vaginal lactobacillus inhibits HIV-1 replication in human tissues ex vivo. Front Microbiol (2017) 8:906. doi: 10.3389/fmicb.2017.00906
156. Dong X-H, Ho M-H, Liu B, Hildreth J, Dash C, Goodwin JS, et al. Role of porphyromonas gingivalis outer membrane vesicles in oral mucosal transmission of HIV. Sci Rep (2018) 8:8812. doi: 10.1038/s41598-018-27284-6
157. Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol (2015) 15:375–87. doi: 10.1038/nri3837
158. Sperk M, Ambikan A, Ray S, Singh K, Mikaeloff F, Diez RC, et al. Novel mechanism of HIV elite control by enriching gut dipeptides as HIV-1 antagonist but prevotella agonist. Preprint (2020). 7(1):ofz507 doi: 10.21203/rs.3.rs-100746/v1
159. Wang S, Huang W, Li K, Yao Y, Yang X, Bai H, et al. Engineered outer membrane vesicle is potent to elicit HPV16E7-specific cellular immunity in a mouse model of TC-1 graft tumor. Int J Nanomedicine (2017) 12:6813–25. doi: 10.2147/IJN.S143264
160. Zhang Y, Fang Z, Li R, Huang X, Liu Q. Design of outer membrane vesicles as cancer vaccines: A new toolkit for cancer therapy. Cancers (2019) 11:1314. doi: 10.3390/cancers11091314
161. Warnke P, Warning L, Podbielski A. Some are more equal - a comparative study on swab uptake and release of bacterial suspensions. PLoS One (2014) 9:e102215. doi: 10.1371/journal.pone.0102215
162. Mitra A, MacIntyre DA, Mahajan V, Lee YS, Smith A, Marchesi JR, et al. Comparison of vaginal microbiota sampling techniques: cytobrush versus swab. Sci Rep (2017) 7:9802. doi: 10.1038/s41598-017-09844-4
163. Zasada AA, Zacharczuk K, Woźnica K, Główka M, Ziółkowski R, Malinowska E. The influence of a swab type on the results of point-of-care tests. AMB Expr (2020) 10:46. doi: 10.1186/s13568-020-00978-9
164. Fichorova RN, DeLong AK, Cu-Uvin S, King CC, Jamieson DJ, Klein RS, et al. Protozoan-Viral-Bacterial Co-infections alter galectin levels and associated immunity mediators in the female genital tract. Front Cell Infect Microbiol (2021) 11:649940. doi: 10.3389/fcimb.2021.649940
165. Łaniewski P, Herbst-Kralovetz MM. Bacterial vaginosis and health-associated bacteria modulate the immunometabolic landscape in 3D model of human cervix. NPJ Biofilms Microbiomes (2021) 7:88. doi: 10.1038/s41522-021-00259-8
166. Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun (2016) 469:967–77. doi: 10.1016/j.bbrc.2015.12.083
167. Fiori J, Turroni S, Candela M, Gotti R. Assessment of gut microbiota fecal metabolites by chromatographic targeted approaches. J Pharm Biomed Anal (2020) 177:112867. doi: 10.1016/j.jpba.2019.112867
168. Lin H, He Q-Y, Shi L, Sleeman M, Baker MS, Nice EC. Proteomics and the microbiome: pitfalls and potential. Expert Rev Proteomics (2019) 16:501–11. doi: 10.1080/14789450.2018.1523724
169. Morton JT, Aksenov AA, Nothias LF, Foulds JR, Quinn RA, Badri MH, et al. Learning representations of microbe–metabolite interactions. Nat Methods (2019) 16:1306–14. doi: 10.1038/s41592-019-0616-3
170. D’Adamo GL, Widdop JT, Giles EM. The future is now? clinical and translational aspects of “Omics” technologies. Immunol Cell Biol (2021) 99:168–76. doi: 10.1111/imcb.12404
171. Lagier J-C, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, et al. Culturing the human microbiota and culturomics. Nat Rev Microbiol (2018) 16:540–50. doi: 10.1038/s41579-018-0041-0
172. Serrano-Villar S, Saenz JS, Tincati C, Raju S, Moreno E, Bargiela R, et al. Microbiome-derived cobalamin and succinyl-CoA are powerful biomarkers for improved screening of anal cancer. (2022). doi: 10.21203/rs.3.rs-2326354/v1.
173. Ñahui Palomino RA, Vanpouille C, Costantini PE, Margolis L. Microbiota–host communications: Bacterial extracellular vesicles as a common language. PLoS Pathog (2021) 17:e1009508. doi: 10.1371/journal.ppat.1009508
174. Walter J, Armet AM, Finlay BB, Shanahan F. Establishing or exaggerating causality for the gut microbiome: Lessons from human microbiota-associated rodents. Cell (2020) 180:221–32. doi: 10.1016/j.cell.2019.12.025
175. Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y. Host variables confound gut microbiota studies of human disease. Nature (2020) 587:448–54. doi: 10.1038/s41586-020-2881-9
176. Abdool Karim SS, Baxter C, Passmore J-AS, McKinnon LR, Williams BL. The genital tract and rectal microbiomes: their role in HIV susceptibility and prevention in women. J Int AIDS Soc (2019) 22:e25300. doi: 10.1002/jia2.25300
Keywords: microbiota, HIV, HPV, inflammation, biomarkers, omics, personalized medicine
Citation: Moreno E, Ron R and Serrano-Villar S (2023) The microbiota as a modulator of mucosal inflammation and HIV/HPV pathogenesis: From association to causation. Front. Immunol. 14:1072655. doi: 10.3389/fimmu.2023.1072655
Received: 17 October 2022; Accepted: 06 January 2023;
Published: 23 January 2023.
Edited by:
Alfredo Garzino-Demo, University of Maryland, United StatesReviewed by:
Alasdair Leslie, Africa Health Research Institute (AHRI), South AfricaCopyright © 2023 Moreno, Ron and Serrano-Villar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elena Moreno, ZW1vbG1vQHNhbHVkLm1hZHJpZC5vcmc=; Sergio Serrano-Villar, c2VyZ2lvLnNlcnJhbm9Ac2FsdWQubWFkcmlkLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.