
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 02 February 2023
Sec. Cancer Immunity and Immunotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1067520
This article is part of the Research TopicTherapeutic Targeting Of MDSC In The Tumor And Immune MicroenvironmentView all 10 articles
Lung tumours are widespread pathological conditions that attract much attention due to their high incidence of death. The immune system contributes to the progression of these diseases, especially non-small cell lung cancer, resulting in the fast evolution of immune-targeted therapy. Myeloid-derived suppressor cells (MDSCs) have been suggested to promote the progression of cancer in the lungs by suppressing the immune response through various mechanisms. Herein, we summarized the clinical studies on lung cancer related to MDSCs. However, it is noteworthy to mention the discovery of long non-coding RNAs (lncRNAs) that had different phenotypes and could regulate MDSCs in lung cancer. Therefore, by reviewing the different phenotypes of lncRNAs and their regulation on MDSCs, we summarized the lncRNAs’ impact on the progression of lung tumours. Data highlight LncRNAs as anti-cancer agents. Hence, we aim to discuss their possibilities to inhibit tumour growth and trigger the development of immunosuppressive factors such as MDSCs in lung cancer through the regulation of lncRNAs. The ultimate purpose is to propose novel and efficient therapy methods for curing patients with lung tumours.
Concerning cancer deaths, lung tumours are recognized as one of the leading causes globally. They are further classified in the category as conditions whose rates of morbidity and mortality, as well as the degrees of malignancy, are the highest. Along with the industrialization progress and environmental changes, the aetiology of lung cancer has become even more complex (1). According to sources, in 2018, many patients diagnosed with non-small cell lung cancer (NSCLC) in stage III were recorded, corresponding to 80% of all cases (2). Despite the ongoing efforts to develop efficient anti-cancer therapies, NSCLC has become among the most lethal cancers worldwide (3). In recent years, tumour-related immunosuppression has become the focal point in targeted therapy for lung tumours. In this respect, bone marrow-derived suppressor cells (MDSCs) have attracted much attention. As early as 1970, studies have pointed out the MDSCs relation to tumour development (4). Its role in the tumour microenvironment has been pivotal and turned into a new target for cancer therapy (5). In a study by Gabrilovich and Nagaraj, the authors confirmed that MDSCs are heterogeneous cell populations derivative from the bone marrow precursor and immature cells (IMC). Under standard conditions, IMC can quickly specialize into dendritic cells (DC), mature granulocytes, and macrophages. On the occasions of cancer, infection, inflammation or other illnesses, the authors have reported an increase in the number of MDSCs and inhibition of IMC differentiation into developed cells of the bone marrow (6).
Moreover, MDSCs are known for their potent immunosuppressive function. Immunotherapy with immune checkpoint inhibitors has been reported to control the long-term effects of tumours to a certain extent. However, due to the potential repercussions of MDSCs’ massive expansion, the MDSCs-induced immunosuppression has been considered the mechanism that effectively hinders the immune checkpoint blockade (7) Regarding lncRNAs, they have been implicated in the progression of tumours and various roles depending on the different expression types. Importantly, lncRNAs have multiple functions that are not limited to regulating the MDSCs generation, recruitment and immunosuppression. They also target multiple pathways simultaneously. The latter allows lncRNAs to act as markers in diagnosing tumours and are highly valued for targeted therapy (8). The modulatory effect of lncRNAs on MDSCs and the regulation of lung tumour tissue may open new horizons in treating individuals with lung tumours. The current paper is mainly based on the targeted therapy of MDSCs that lncRNAs have regulated to change the survival rate of patients with lung tumours. First, to elaborate on the importance of MDSCs in patients diagnosed with lung tumours, we have analyzed the MDSCs mechanism and investigated the clinical studies focused on the matter. Secondly, we studied the role of differential lncRNA expression in these cells and the impact on tumour progression. Finally, we have included clinical studies in which lncRNAs regulated various tumours, justifying their potential therapeutic value in tumours.
Cell heterogeneity is evident in MDSCs as they include two major subpopulations, those of granulocytes (G-MDSCs) and monocytes (M-MDSCs) in both human and animal (mice) models. They are derived from granulocytes or monocytes and represent relatively stable forms of pathologic activation of these blood cell populations (9). In mice, CD11b+ Ly6G-Ly6Chigh and CD11b+Ly6G+Ly6Clow stand for the phenotype of M-MDSCs and G-MDSCs, respectively. Related studies have confirmed CD11b+ GR-1lowcells’ ability as the most efficient in suppressing the immune system in contrast to CD11b+ GR-1high cells, regarded as the least effective. In humans, the MDSCs’ complexity is even greater and constitutes CD11b+CD14+HLA-DRlow/neg M-MDSCs and CD11b+CD14-CD15+ G-MDSCs populations. The MDSCs in cancer patients express granulocyte markers and bone marrow cell markers like CD11b and CD33, which are the most common. However, there is still a need for a profound exploration of the surface markers of M-MDSCs and G-MDSCs due to the differentiation of the MDSCs’ phenotype in various diseases (10, 11).Besides, a small group of bone marrow progenitors with MDSCs characteristics is identified only in humans and is named the “early MDSC” group. The group is mainly composed of bone marrow progenitors and precursors that account for less than 5% of the total number of MDSCs (12). However, many ongoing efforts have been made to report the surface markers of MDSCs. With fluorescence-activated cell sorting (FACS) for evaluating the multicolour immunofluorescence staining, several phenotypes of MDSCs in lung cancer have been reported (13).
The potent immunosuppressive effects of MDSCs, are present in most cancers (14). For example, the tumour microenvironment (TME) comprises different cell populations in a complex matrix. Cellular components of TME are markers that can regulate cancer processes like tumour proliferation, angiogenesis, invasion and metastasis, chemotherapy resistance, etc. Therefore, TME has become a new target for cancer treatment (15, 16) Furthermore, during tumour progression, the cells undergo alternations in their metabolism to satisfy their energy needs, with the ultimate goal of achieving proliferation and differentiation of the tumour—the last results in nutrition competition between the immune cells and immune modulators in the TME of MDSCs. Thus, increased glycolysis, fatty acid metabolism, and up-regulation of enzymes that are essential metabolites of catabolism are observed in the MDSCs tumour microenvironment. The last grants MDSCs immunosuppressive function (17). Other investigations have demonstrated that the MDSCs’ immunomodulatory role is mainly based on the inhibition of T cells (18).This happens by various mechanisms. The first one is described by the production of reactive oxygen species (ROS), and reactive nitrogen species (RNS) can result in blocking T cells activation and function by MDSCs. As reported by Wang et al., G-MDSCs are mainly responsible for producing ROS and arginase-1 (ARG-1).
In contrast, M-MDSCs mainly produce ARG-1 and nitric oxide (NO) to exert immunosuppressive effects (19). The half-life of the produced NO is more extended, while it requires cell entry. Nonetheless, there is no requirement for close contact between M-MDSCs and T cells, enabling M-MDSCs effectively inhibit non-specific T cell responses (20). One prominent feature of MDSCs is the up-regulation of ROS produced by G-MDSCs in mice and individuals diagnosed with cancer. The immunosuppressive effect of MDSCs can be significantly enhanced by the expression of ROS in cancer patients and mice (21). Concretely, superoxide anion (O2-), hydrogen peroxide (H2O2) and peroxynitrite (PNT) represent the abovementioned family of ROS. However, NO reacts with O2- to form PNT, which then prevents the recognition of the antigen/major histocompatibility complex (MHC) peptide by nitrification of the MHC class I molecules and the T cell receptors (TCRs). They reduce the TCR affinity for antigen-MHC complex and block T cell migration by nitrification of T cell-specific chemokines (22, 23). The second mechanism providing MDSCs with immunomodulatory roles includes the depletion of L-cysteine and arginine by MDSCs, which are required for the proliferation and activation of T cell (24). The ARG-1 produced by G-MDSCs and M-MDSCs metabolizes L-arginine to L-ornithine in the urea cycle, making L-arginine induce its exhaustion in the TME. Other studies have revealed that PEGylated forms of Arg I (PEG-ARG I) in mice promote tumour growth. This is accompanied by augmented amounts of MDSCs (25).On the other hand, cysteine’s role in T cell activation is indispensable. Factually, macrophages and DCs deliver to T cells the obtained cysteine, which the DCs and macrophages metabolise to methionine during the standard antigen processing and presentation. Moreover, in TME is observed a reduction in the activation of T cells and cysteine production by DCs as a result of a deprivation of macrophages of cysteine and DCs. The above is due to a large number of cysteine depletion by MDSCs (26).
The third mechanism includes the interaction between MDSCs with T cells that migrate to the lymph glands and T-cell activity via the expression of ADAM17 (metalloprotease structural domain 17 and a disintegrin) on their cell membrane. The MDSCs interfere. It has been reported that the latter leads to the downregulation of the homing receptor CD62L (L-selectin) on T cells (24).When it comes to effective anti-cancer immunity, and for purposes of action, it requires the transportation to the tumour of activated T cells and the activation of tumour-reactive T cells. Meanwhile, CD62L has been regarded as an essential molecule in this process, directing primitive lymphocytes to lymph glands in the periphery.
Furthermore, CD62L has proven necessary for housing the primitive T cells in the lymph glands, activating molecules on and in the cell membrane. Conversely, the suppressed activity of CD62L on primitive T cells by MDSCs decreases the chance of primitive T cells being delivered to the activation site. The last functions as MDSCs’ suppressive anti-tumour immune response (27).
The other mechanisms for MDSCs’ reduced immunity are explained in findings demonstrating that the regulatory T cells (Tregs) impact tumour immunosuppression. Tregs are considered to be immunosuppressive cells that promote cancer growth, and they are significantly increased in the peripheral blood of NSCLC patients (28). Although there are no proofs to relate MDSCs with the induction of Tregs, they produce a series of cytokines that allow the differentiation of Tregs (29, 30). MDSCs promote the amplification of natural Tregs through the production of IL-10, TGFβ, IFNγ and CD40-CD40L interactions and drive the development of the induced Tregs (24). Sinha et al. have reported that downregulation of the macrophage IL-12 and the IL-10 production is the outcome of T cell polarization to a type 2 pro-tumour phenotype (31). According to sources, the immune cells, specifically the B cells, NK cells, macrophages (Mø) and Treg cells, interact and can be regulated by MDSCs (13). The literature shows evidence for the interaction between DCs, tumour-associated macrophages (TAMS) and MDSCs in the TME. The outcome of this interaction is the enhancement of each cell population’s immunosuppressive activity (24).Thus, in addition to T cell inhibition, MDSCs interfere with the innate immune response by affecting various cells like the NKT, NK, and macrophages (Figure 1). Factually, the MDSCs’ impact on NK cells is intricate. One part can inhibit the death of the NK cell by hindering the production of IFN-γ. Importantly, due to the interaction of RAE-1 with NKG2D that takes place on the NK cells’ surface, the other part via the expression of RAE-1 can result in NK cells’ activation and provoke their death (24, 32). However, MDSCs’ profile is multi-dimensional. Indeed, they promote tumour cell invasion, drug resistance, tumour angiogenesis, pre-metastatic niche formation, and tumour metastasis while participating in tumour immunosuppressive response (33, 34) .It has been suggested by Yang et al. that the MDSCs’ non-immunosuppressive effects occur mainly via the promotion of tumour angiogenesis (35). Epithelial-mesenchymal transition (EMT) has been shown to play a key role in the process of tumor initiation and even metastasis, which can enhance the ability of cancer cells to enter the circulatory system to promote the metastasis of tumor cells. In the process of EMT, tumor cells lose polarity and cell-cell connection, and then enter a state of low proliferation, migration and invasion are enhanced (36). Studies have shown that EMT is related to the number of MDSCs, and EMT transcription factors can attract immunosuppressive cells MDSCs, leading to tumor immunosuppressive microenvironment. In turn, immunosuppressive factors induce EMT in tumor cells (37).The main mechanism is due to the fact that MDSCs and TAM further enhance chronic inflammation in the inflammatory microenvironment, which leads to EMT and enrichment of cancer stem cell-like cells (CSCs) and immune suppression may be the cause of drug resistance and metastasis (38), It is this feedback loop between EMT and immune suppression that promotes tumor progression. Therefore, the combination of immunotherapies targeting immunosuppressive cells such as MDSCs may be a promising treatment for EMT.
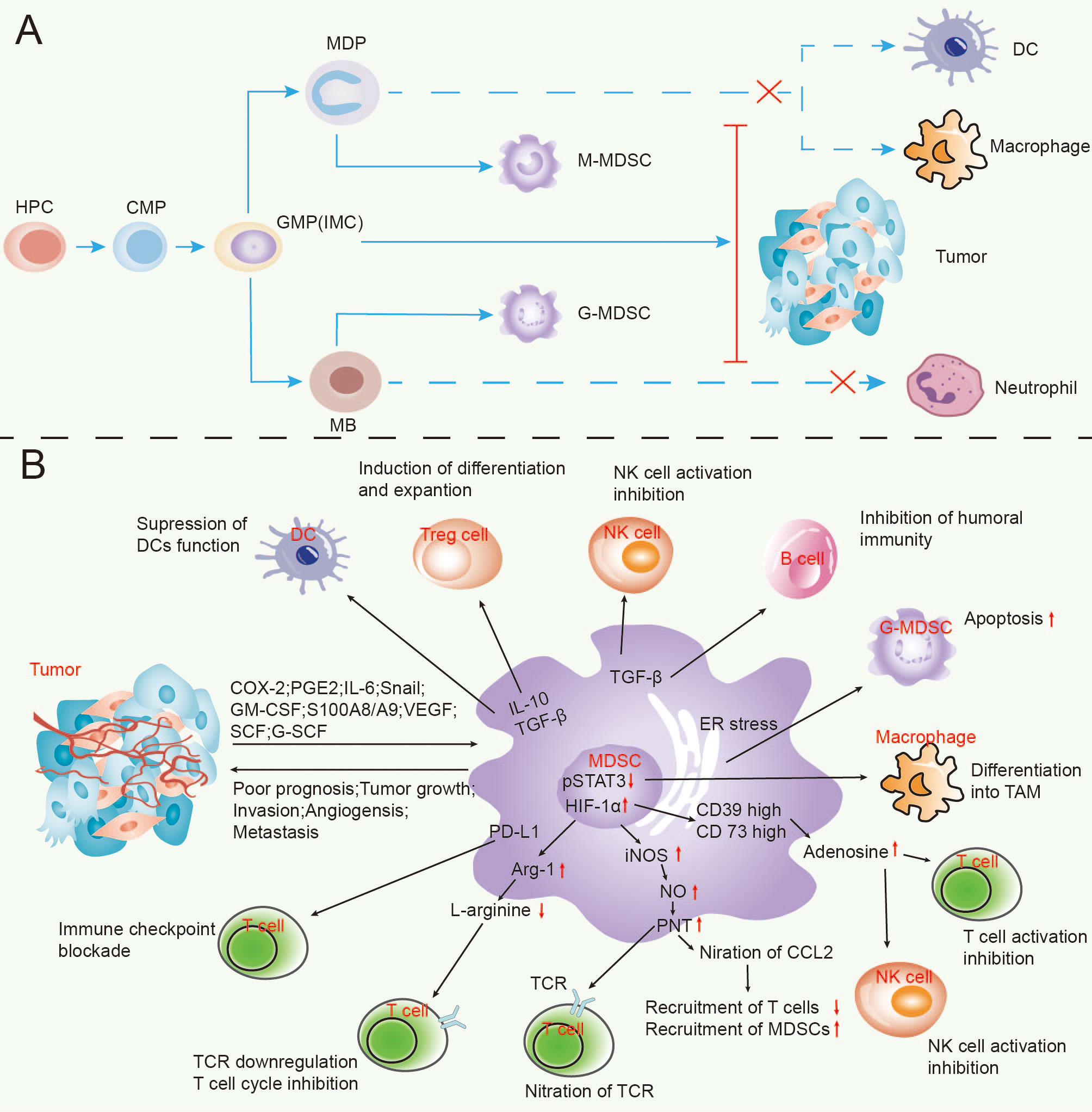
Figure 1 The process of MDSCs’ formation in lung cancer (A) and the appliance of MDSCs exert tumour immunosuppressive effects (B).
MDSCs represent the most significant immunosuppressive cellular population in individuals with lung tumours. In the stroma of the tumour, these cells limit the healing efficacy of anti-cancer curing approaches through their metabolic pathways and other modalities in response to complex TME (39).There are many clinical studies on MDSCs in lung tumours, and therapeutic strategies targeting MDSCs are also gradually emerging. We have summarized these clinical studies and presented them in Table 1.
Some data show a significant accumulation of M-MDSCs in NSCLC patients, especially in stage IV patients, compared to stage III, with the expected lower survival rate of patients with a higher accumulation of M-MDSCs (40).Other authors found that individuals with NSCLC exhibited an advanced proportion of G-MDSCs, linked with improved survival (41). At the same time, other work has reported on a new human MDSCs subpopulation, CD14 (+) HLA-DR (-/low), in another cohort of individuals with NSCLC. Among those individuals, the incidence and total number of CD14 (+) HLA-DR (-/low) cells in the peripheral blood were considerably augmented compared with those in healthy NSCLC patients (42). The authors hypothesized that this was related to tumour metastasis, adverse reactions to chemotherapy, and tumour immunosuppression. Other work reported the results of different clinical trials with patients with small cell lung cancer (SCLC), in which the depletion of MDSCs improved the immune response to vaccination and further enhanced the effect of immune interventions on cancer (43). It was shown that M-MDSCs from NSCLC patients demonstrated a worse prognosis (44).Seyed Sajjad Zadian et al. demonstrated that the frequency of M-MDSCs in NSCLC individuals was significantly more advanced than that in SCLC and healthy people, which impacted the differential diagnosis of NSCLC (45). Chien-Ying Liu et al. provided evidence of increased CD11b +/CD14 ϫ/CD15 +/CD33 + MDSC in the peripheral blood of NSCLC individuals, which played an essential role in mediating immunosuppression of NSCLC (46).Other authors recognized a unique subgroup of MDSCs in NSCLC patients, namely CD14 (+) S100A9 (+), which inhibited T cells through arginase, iNOS, and IL-13/IL-4Rα axis and were closely associated with adverse effects of chemotherapy (47). Some of the reported studies in Table 1 measured the amount of MDSCs in the peripheral blood mononuclear cells (PBMCS in untreated NSCLC individuals and made a comparison with the healthy ones (48). Their studies proved that the number of G-MDSC increased in NSCLC individuals, whereas further studies showed that G-MDSCs blocked T cell proliferation in vitro. The report by Tian Tian et al. who applied FACS sorting to analyze the peripheral blood of SCLC patients proved that the amount of CD14 (+) HLA-DR (-/low) MDSCs in the peripheral blood of SCLC individuals were considerably augmented. The authors further provided a clue that the amount of these MDSCs were predictors of poor prognosis of SCLC (49).
Interestingly, other authors analyzed tumour resections from NSCLC patients. The results showed that the occurrence of M-MDSCs in individuals with lung tumours was higher and that the accumulation of M-MDSCs and G-MDScs in the tumour site was higher than in peripheral blood. The authors further showed that the MDSCs amount in the peripheral blood could predict recurrence after surgery (50).The effect of the first-line treatment on peripheral blood MDSCs in NSCLC patients was analyzed by other authors. The obtained data presented that chemotherapy with Bevacizumab significantly decreased the levels of MDSCs in the peripheral blood of NSCLC individuals (51). Other authors proposed that the novel targets for immunotherapeutic drug combinations and the treatment of NSCLC through the galectin-9/TIM-3 pathway and mMDSCs were key to anti-PD-1 primary or secondary resistance (52). In patients with Nivolumab treatment, the reported data in another study showed that G-MDSCs could be used as potential immune biomarkers in NSCLC treated with Nivolumab and other second-line therapies (53). In another study by Pauline L de Goeje et al., it was demonstrated that immunoglobulin-like transcript 3 (ILT3) was expressed on MDSCs in fresh peripheral blood mononuclear cells (PBMCS) from individuals with NSCLC. The authors demonstrated that ILT3 cooperated with ligands on the T cells to inhibit T cells, thus augmenting the amount of MDSCs and decreasing survival (54).
Through the above clinical studies, it is not difficult to find that MDSCs are closely related to the prognosis of patients with lung cancer. Compared with normal people, patients with cancer have higher levels of MDSCs, and most of them achieve immunosuppression by inhibiting T cells. When the level of MDSCs in patients is higher, the OS of patients is lower. Lourdes Barrera et al. showed that the OS of patients with low G-MDSCs is better than that of patients with high G-MDSCs through a series of data studies, and the level of G-MDSCs is a potential prognosis of NSCLC disease progression (41). More MDSCs are found in patients with advanced disease, which is associated with poor prognosis. At the same time, primary drug resistance occurs during immunotherapy due to the presence of MDSCs. There are not many studies on the clinical relevance of MDSCs in human cancer, which mainly focus on the correlation between high levels of MDSCS and shorter OS or PFS in different cancers.
Indeed, it is interesting to note that the incidence of M-MDSC and G-MDSC is increased not only in the peripheral blood of patients but also in the neoplastic lesions. Both tumor-infiltrating MDSCs subsets were significantly elevated compared with circulating subsets, confirming that the tumor site had the strongest immunosuppressive effect. In particular, the frequency of tumor infiltration and circulating G-MDSCs correlated with tumor progression (55). In the study by Yoshikane Yamauchi et al., it was noted that the frequency of MDSCs in tumors was higher than that in peripheral blood of the same patients, and this accumulation was associated with increased concentrations of inflammatory mediators involved in MDSC migration to the tumor microenvironment and activation. Moreover, tumor G-MDSCs showed higher expression level of programmed death ligand 1 than the same cells in peripheral blood (50). Xinyu Tian et al. isolated MDSCs from tumor tissues of lung cancer patients by FCM and showed that RUNXOR was significantly associated with MDSCs-induced immunosuppression in lung cancer patients and may be a target for anti-tumor therapy (56). It has been suggested that in a mouse model, monocytic MDSCs can further mature into Tams in the tumor microenvironment, thereby allowing Tams to induce chemotherapy resistance through various mechanisms. Tumor-infiltrating CD68 Tams were analyzed and compared with blood S100A9 MDSCs from the same patients. Indicating their origin from S100A9 MDSCs, it was also found that the percentage of blood S100A9 MDSCs was closely correlated with the counts of S100A9 cells and CD68 TAM in tumor tissue, and patients with higher S100A9 and CD68 cell numbers also showed worse PFS (57).
In addition to clinical studies, several animal trials of MDSCs on lung cancer progression also exist, through which the key role of MDSCs in lung cancer can be supported. In the study of Mi So Park et al., it was found that the main mechanism by which the polypeptide N-acetyl-galactosaminyltransferase (GALNT3) inhibited the development and progression of lung cancer in xenograft and syngeneic mouse models was the ability of MDSCs to infiltrate the tumor site and subsequent angiogenesis, thereby inhibiting the development of lung cancer (58). Although treatment with immune checkpoint inhibitors (ICIs) improves overall survival in a subset of patients with NSCLC, co-occurring KRAS/LKB1 mutations can drive primary resistance to ICIs. Rui Li et al. therefore targeted G-MDSC enrichment as a potential mediator of immunosuppression in LKB1-deficient NSCLC and sensitized tumors to immunotherapy by overcoming MDSCs accumulation with all-trans retinoic acid in a LKB1-deficient NSCLC mouse model (59). Dickson Adah implanted tumors in mice, obtained whole tumors and tumor-derived sorted cells of tumor-bearing mice, and found that malaria infection significantly reduced the proportion of MDSCs and Treg in the lung tumor tissues of treated mice, and inhibited the expansion and activation of MDSCs and Treg in the tumor microenvironment (60). Xiaosan Su et al. also demonstrated that dexmedetomidine (DEX) induced the proliferation of M-MDSCs during the postoperative period in lung cancer patients by inducing spontaneous tumor metastasis in C57BL/6 mice and had a significant pro-angiogenic ability (61). Liran Levy et al. evaluated the effect of splenectomy in several mouse lung cancer models and found that the effect of splenectomy on tumor growth is essentially cell-mediated by MDSCs, which can be used to inhibit the growth of non-small cell lung cancer by depleting MDSCs (62).
Traditional therapeutic approaches for lung tumours are surgery, radio- and chemotherapy. Although some of these treatments are used as first-line therapy, the clinical studies mentioned above have shown that MDSCs can not only exert be immunosuppressive in the TME but could directly promote tumour advancement but also interfere with the prognosis of conventional treatments, thus making it more critical to treat lung cancer by targeting MDSCs. MDSCs were proposed as potential targets for the advance of anti-cancer lung treatment based on the following five aspects (1): promotion of myeloid differentiation (2); blockage of MDSC propagation (3); removal of MDSCs (4); functional decay of MDSCs, and (5) blockade of immune checkpoints (63).Further research on MDSCs showed that miRNAs/lncRNAs could regulate the specialization, propagation, and immunosuppressive roles of MDSCs in TME (64). Therefore, targeting miRNAs and lncRNAs to stop the development and expansion of MDSCs suppressor cells in the tumour environment appears more promising.
In the review’s subsequent chapters, we will focus on lncRNAs and their regulation on the generation, recruitment and immunosuppression of MDSCs.
LncRNAs are a diverse family of non-coding RNAs (ncRNAs). They encompass different ncRNAs like miRNAs, LncRNAs, snRNAs and CircRNAs (65).LncRNAs’ transcripts are longer than 200 nucleotides and are involved in the pathophysiology of many diseases (66). Relevant studies have pointed out that the number of human lncRNAs exceeds the number of protein-coding genes (67). The ENCODE project projected that the human genome contained over 28,000 different lncRNAs, most yet undiscovered (68).Some studies show that lncRNA categories have a high degree of diversity (69), ranging in number from a few hundred to several thousand nucleotides (70).RNA polymerase II transcribes lncRNAs, and according to their genomic localization, mode of action and function can be classified into intronic lncRNAs, intergenic lncRNAs (lincRNAs), enhancer lncRNAs (ELNcRNAs), bidirectional lncRNAs, and sense overlapping lncRNAs (64, 71). Some authors have proven that lncRNAs are mRNA precursors, and compared with mRNAs, ncRNAs show moderate sequence conservation, while lncRNA-pre-mRNA has a significant part in alternative splicing (72, 73). The expression of a variety of lncRNAs is abnormal in various diseases, especially malignant tumours. Some data show that lncRNAs regulate the bone marrow and immune cells. Their regulatory mechanisms are complex and diverse, and they have become vital regulators mediating cell activation, proliferation, differentiation, apoptosis and autophagy (74). Therefore, lncRNAs may have potential diagnostic, prognostic or therapeutic importance. Table 2 demonstrates the expression of lncRNAs and their regulatory effect on MDSCs.
The mouse plasmacytoma variant 1(Pvt1) gene represents a long non-coding RNA located on chromosome 15 (Ch 15) that was reported for the first time in 1985 (82). It is a candidate oncogene coding for a homologous lncRNA to the human Pvt1 gene, localized on Ch 8, specifically near the c-Myc locus on 8Q24. It encodes 52 ncRNA variants, including 26 linear and 26 circular isoforms and six microRNAs and is long 1.9 KB (83, 84). LncRNA PVT1 is recognized as an oncogene in many tumours. Its overexpression is associated with hepatocellular carcinoma, gastric, oesophagal, cervical, and bladder cancer and acute myeloid leukaemia (85–87), Yu Zheng et al. proves that Pvt1 is highly expressed in tumour-expanding G-MDSCs. The results show that lncRNA Pvt1 downregulation considerably blocked the immunosuppressive function of G-MDSC in vitro, reducing tumour development and suppressing anti-tumour immune responses. Since Pvt1 expression is augmented in tumour-infiltrated G-MDSCs more than in splenic G-MDSCs, the hypoxic conditions in TME are considered to trigger such a phenomenon. Therefore exposure of splenic G-MDSCs to hypoxic environments reveals an upregulation of both mRNA and protein levels of HIF-1α in these cells. HIF-1α role in the process is clarified by blocking its upregulation by its specific inhibitor YC-1. The results show restored upregulation of Pvt1 and c-myc in hypoxic environments, thus indicating that HIF-1α augmented Pvt1 expression in G-MDSCs cells under hypoxia (75).
RUNX1 overlapping RNA (RUNXOR) is a lncRNA transcribed by an upstream promoter and overlapping with RUNX1. It significantly controls bone marrow cells’ growth by targeting RUNT-associated transcription factor 1 (RUNX1) (88). LncRNA RUNXOR is about 260 KB long (89). As less research has been done on RUNXOR, dating back as far as 2014, RUNX1 is located on chromosome 21 and is usually disrupted by chromosomal translocations in haematopoietic malignancies. The most common observed translocation is t (8, 21), which is common in acute myeloid leukaemia (90). RUNXOR can regulate RUNX1 expression by recruiting the RUNX1 protein at its 3’ end, and upon binding to promoters and enhancers, it makes it participate in chromosomal translocations in malignant cells tumours (90).
Furthermore, by binding straight to chromatin, RUNXOR orchestrates the long-range chromosomal inner loops. Xinyu Tian et al. proves that the RUNXOR and RUNX1 in MDSCs from the peripheral blood of lung cancer patients are differentially expressed in the tissues around the lung cancer compared to the normal tissues. The study results show that lncRNA RUNXOR is augmented in the lung cancer blood samples, while RUNX1 activity is reversely connected with immunosuppression in MDSCs. Moreover, the activity of RUNXOR is advanced in MDSCs in the lung tumour samples than in the adjacent tissues. The knockdown of RUNXOR also decreased arg1 activity in MDSCs. This suggest that RUNXOR expression is considerably related to MDSC-induced immunosuppression in lung tumours and may be a good aim for anti-tumour therapeutic approaches (56).
Lnc-chop is a novel lncRNA identified in MDSCs. It is positioned in the intronic region of the gene on Ch 11. Relevant data indicate that transcription factor C/EBPβ, C/EBP-homologous protein (CHOP) and phosphorylated STAT3 directly influence MDSCs growth (91).CHOP is coded by Ddit3 and takes part in the diminished production of significant factors associated with MDSCs functions, including ARG-1, PNT (peroxynitrite), and superoxide, thereby inhibiting MDSCs activity (92). In addition, lnc-CHOP is associated with lung, breast cancer, murine melanoma, and murine ovarian tumours. The results from the 2018 Yunhuan Gao’s trial notes that lnc-chop would be expressed in MDSCs mediated by factors involved in inflammation and tumour development, while lnc-chop potentiates MDSCs immunosuppressive activity in vivo and in vitro. These data indicates that lnc -chop regulated MDSCs specialization into M-MDSCs and that M-MDSCs have a more potent immunosuppressive effect. The mechanism behind this is that lnc-chop interacts with CHOP and the C/EBPβ isoform LIP to activate the C/EBPβ isoform LAP, thus lnc-chop triggers enhancement of H3K4me3 in the promoters of Arg-1, NOS2, NOX2 and COX2, which are implicated in MDSCs role in suppressing the immune system in TEM (76).
RNCR3, also known as LINC00599, is a lncRNA, which is highly conserved in mammals. In tumours, RNCR3 has oncogenic functions and promotes the progression of colorectal, prostate and brain cancers (93).RNCR3 is a crucial regulator of cell propagation, specialization, cell death, metastasis and atherosclerosis (94). Knockdown of RNCR3 leads to increased plasma amounts of TNF-α, CCL2 and IL-6, signifying its potential immunomodulatory functions. Wencong Shang’s trial confirms that RNCR3by acted as a competing endogenous RNA (ceRNA) during MDSC differentiation. Tying to miR-185-5p and releasing Chop stimulated their specialization and immunosuppressive activity. There is a close relationship between RNCR3, miR-185-5p and Chop, and the relationship between the three and how they regulated MDSCs is further elaborated in their subsequent experiments. As mentioned earlier, Chop triggers MDSCs’ specialization and activity in vivo, whereas its reduced activity blocks the activity of Arg-1 and iNOS. Therefore, the authors conclude that RNCR3 triggers MDSCs’ specialization by interacting with spongy miR-185-5p to free its target gene Chop. In addition, tumour microenvironmental molecules such as IL-6 triggered RNCR3 expression during MDSCs specialization and further promote their immunosuppressive activity (77).
Olfr29-ps1, a lncRNA distributes in the cytoplasm and nucleus, is a pseudogene, 963 bp in length, located on mouse Ch 4. Its sequence is preserved in vertebrates and is significantly overexpressed in peripheral blood mononuclear cells from individuals with colon and rectal cancer. It is linked with lung, breast, pancreatic, and uroepithelial cancer. Olfr29-ps1 is regulated by the pro-inflammatory cytokine IL6 and tumour-associated factors. IL6 up-regulates Olfr29-ps1 expression in MDSCs, while Olfr29-ps1 is considerably reduced in MDSCs in B16 tumour mice after IL6 knockdown. The Olfr29-ps1 knockdown results in lower NO, H2O2 and ROS in the cells, whereas increased NO, H2O2 and ROS are detected in MDSCs that overexpressed Olfr29-ps. Moreover, it is further confirmed that Olfr29-ps1 silencing diminished the protein levels of Arg-1, iNOS, Cox2 and Nox2, while the protein levels of Arg-1, iNOS, Cox2 and Nox2 are augmented in MDSCs that overexpressed Olfr29-ps1. These results signify that Olfr29-ps1 promoted Arg-1 production in M-MDSCs, and H2O2 in G-MDSCs, contributing to the MDSCs specialization and suppression of the immune system. MDSC overexpressing Olfr29-ps1 in murine models of melanoma resulted in larger, faster and heavier tumour growth. The mechanism by which Olfr29-ps1 affect MDSCs was further investigated. LncRNA pseudogene Olfr29-ps1 can directly sponge mir-214-3p and promote the differentiation and immunosuppressive function of M-MDSC in vitro and in vivo, which may be achieved by targeting MyD88. Data show that miR-214-3p diminished MyD88 mRNA and protein levels. Furthermore, the interaction between Olfr29-ps1 and miR-214-3p is reliant on the modification of m6A by Olfr29-ps1. Data show that lncRNA Olfr29-ps1 have seven conserved GGAC sequences, the most shared m6A sequences. Therefore, RIP-PCR Olfr29-ps1 is modified by m6A in IL6-induced MDSC. These results confirm that Olfr29-ps1 promoted MDSCs’ functions by forming miR-214-3p after mbA modification (78).
HOTAIR was discovered in 2007 by Rinn et al. (95). It is a 2.2-kilobase ncRNA located at the HOXC site, specifically in the intergenic place between HOXC11 and HOXC12 genes in the HOXC cluster on Ch 12. HOTAIR recruits a transcriptional corepressor polycombsin Complex 2 (PRC2) to repress the HOXD (homeobox gene cluster D) expression (96). In individuals diagnosed with lung tumours, HOTAIR activity is considerably augmented. Therefore, it is considered a new controller of lung tumours, which has great significance in the possible therapeutic approaches to lung cancer (97). HOTAIR is also overexpressed in nasopharyngeal carcinoma, breast, pancreatic, liver, gastric and non-small cell lung cancer (98).HOTAIR was proven to be linked with MDSCs functions in hepatocellular carcinoma lines. In HCC patients, MDSCs exerted their immunosuppressive functions by inducing regulatory T cells. Other authors demonstrated that HOTAIR induced the secretion of CCL2, which recruited TAM and MDSCs, in PBMCs co-cultured with HOTAIR overexpressing cells. It was also shown that HOTAIR played a crucial role in promoting macrophages and MDSCs by secreting cytokines and chemokines from HCC cells (79).
HOTAIRM1 (HOXA transcribed antisense RNA bone marrow specific 1) was discovered by (Xueqing Zhang et al., 2009). It is 1052bp in length and is located in the HOXA gene cluster between HOXA1 and HOXA2 on human Ch 7 (99). Initially, it was considered the most prominent intergenic transcript of granulocyte differentiation and up-regulation in NB4 promyelocytic leukaemia. Other authors proved that it was overexpressed in specific myeloid lines (100). Recent data show that HOTAIRM1 is a lncRNA that is abnormally active in different tumours and is related to hepatocellular carcinoma, colorectal, gastric, head and neck, ovarian, thyroid cancers, etc (101).Some authors detected that HOTAIRM1 expression was significantly reduced in tumour tissues. In addition, overexpression of HOTAIRM1 downregulated Arg1 expression levels and inhibited the effect of MDSCs. However, when HOTAIRM1 was overexpressed, the HOXA1, a target gene of HOTAIRM1, was significantly enhanced, suggesting that HOTAIRM1 could trigger HOXA1 expression in MDSCs. In contrast, its silencing or HOXA1 expression in MDSCs significantly reduced the frequency of MDSCs and their inhibitory function. It was further found that HOXA1 overexpression downregulated the activity of the immunosuppressive molecule Arg1 and ROS production in MDSCs and that HOXA1 overexpression enhanced CD4+ Th1 and CD8+ CTL cell initiation so that HOXA1 overexpression enhanced the anti-tumour T-cell response and suppressed the immunosuppressive effect of MDSCs, thereby delaying tumour progression (80).
Lnc-c/EBPβ is located on Ch 1 and 4. It is represented by three subtypes of C/EBPβ: liver-rich activator protein (LAP) and liver-rich repressor protein (LIP) (102). Studies on LNC-C/EBPβ are scarce. In 2018 some authors reported on the isolation of MDSCs from mice carrying melanoma, colon, ovarian and breast cancer. MDSCs subsets were classified to analyze the expression of RNC-C/EBPβ in different MDSC subsets. The results demonstrated that lnc-C/EBPβ was found in G-MDSC, M-MDSC and macrophages, and the amount of lnc-C/EBPβ was significantly augmented in G-MDSCs. It has been suggested that ARG-1, NOS2, NOX2 and COX2 activity was controlled by C/EBPβ, which lessened MDSCs’ immunosuppressive functions (103). The underlying mechanism included LNC-C/EBPβ knockdown, which changed the transcriptional activity of several genes, like interleukin4-induced gene-1 (IL4i1). However, LNC-C/EBPβ binding to C/EBPβ homotypes LIP and WDR5 was necessary for LNC-C/EBPβ-mediated IL4il silencing. The results showed that the expression of LNC-C/EBPβ was controlled by IL 6, while LNC-C/EBPβ potentially promoted the differentiation of PMN-MDSC. Furthermore, LNC-C/EBPβ hindered the specialization of MDSCs into M-MDSCs (81).
MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) is an extensively investigated lncRNA, especially in tumour biology. It is also recognized as NEAT2 (a nuclear-rich transcript 2), mainly localized in the nucleus and highly preserved among animals. In humans, it is found on chromosome 11q13. Its main transcript length is about 8kb in humans and 6.7kb in mice (104). It was one of the earliest lncRNA genes discovered, and some data linked it with metastasis in NSCLC individuals. It was overexpressed in NSCLC patients and appeared predictive of early-stage NSCLC in individuals with a high risk of metastasis. However, MALAT1 was not only highly expressed in lung cancer but participated in the progression and expansion of hepatocellular carcinoma, endometrial stromal sarcoma, cervical, breast cancer, osteosarcoma, colorectal cancer and others (105). Furthermore, MALAT1 regulated the molecular signalling pathways that drove cell division, apoptosis, cell cycle, metastasis, invasion, and immune response. It was linked to tumour site, size, salinization and cancer stage, so the MALAT1 abnormal expression in tumour tissues or body fluids could be used for diagnostic and prognostic purposes (106).Other authors proved that the average transcription activity of MALAT1 in PBMCs isolated from individuals with lung tumours was considerably reduced and was adversely linked with the amount MDSCs. These results proved that MDSCs and CTL were negatively correlated in PBMCs of individuals with lung tumours. However, according to Qinfeng Zhou et al., MALAT1 levels were not directly correlated with MDSCs and CTLs in PBMCs of patients with lung tumours. Therefore, it could be concluded that MALAT1 regulated the differentiation of MDSCs. This study also demonstrated that MALAT1 knockdown increased the number of MDSCs by regulating their differentiation, which provided a new understanding of the development of lung tumours and proposed a possible target for supplementary diagnosis and management of lung tumours (8).
Tumor microenvironment, such as hypoxia, the presence of IL-6 and other inflammatory factors, may promote the expression of lncRNA in MDSCs. For example, IL-6 or tumor-related factors can induce the overexpression of lnc C/EBPβ, LNC-CHOP, Olfr29-ps1, Pvt1 and RNCR3 in MDSCs, while chronic and low-dose stimulation of inflammation and tumor factors can also promote the down-regulation of lncRNAs. For example, HOTAIRM1 and MALAT1 are down-regulated in MDSCs from lung cancer patients. These microenvironmental factors produce different effects. From the above introduction, we can know that lnc-CHOP, Olfr29-ps1, Pvt1, RUNXOR, HOTAIR and RNCR3 can promote the immunosuppressive function and differentiation of MDSCs. lnc-C/EBPβ, HTOAIRM1 and MALAT1 blocked the immunosuppressive and differentiation functions of MDSCs, resulting in completely different effects. At the same time, some studies have shown that lncRNAs(such as Olfr29-ps1, lnc-CHOP, RNCR3 and RUNXOR) can participate in the first stage of MDSCs expansion and then participate in the second stage of MDSCS activation due to the identification of different targets, while Pvt1 can only participate in the second stage. In addition, lncrnas can also play cell-to-cell immunosuppressive and tumor-promoting roles through exosomes secreted by MDSCs.
LncRNAs control the immune cells’ functions by various mechanisms. The regulation of MDSCs in various tumour tissues by lncRNAs can promote their application in immunoregulatory anti-cancer therapy or as biomarkers. LncRNAs have dual roles in cancer. They can block or trigger its development. For example, the transmission of lncRNAs in exosomes triggers drug non-responsiveness. Data show that LncRNA H19 encapsulated into exosomes and unambiguously facilitated by hnRNPA2B1 is moved to non-resistant NSCLC cells, resulting in non-responsiveness to Gefitinib (36).In our study, we explored and summarized the direct regulation of lung cancer by lncRNAs and further investigated the indirect regulation of lncRNAs on lung cancer by regulating MDSCs (Figure 2).
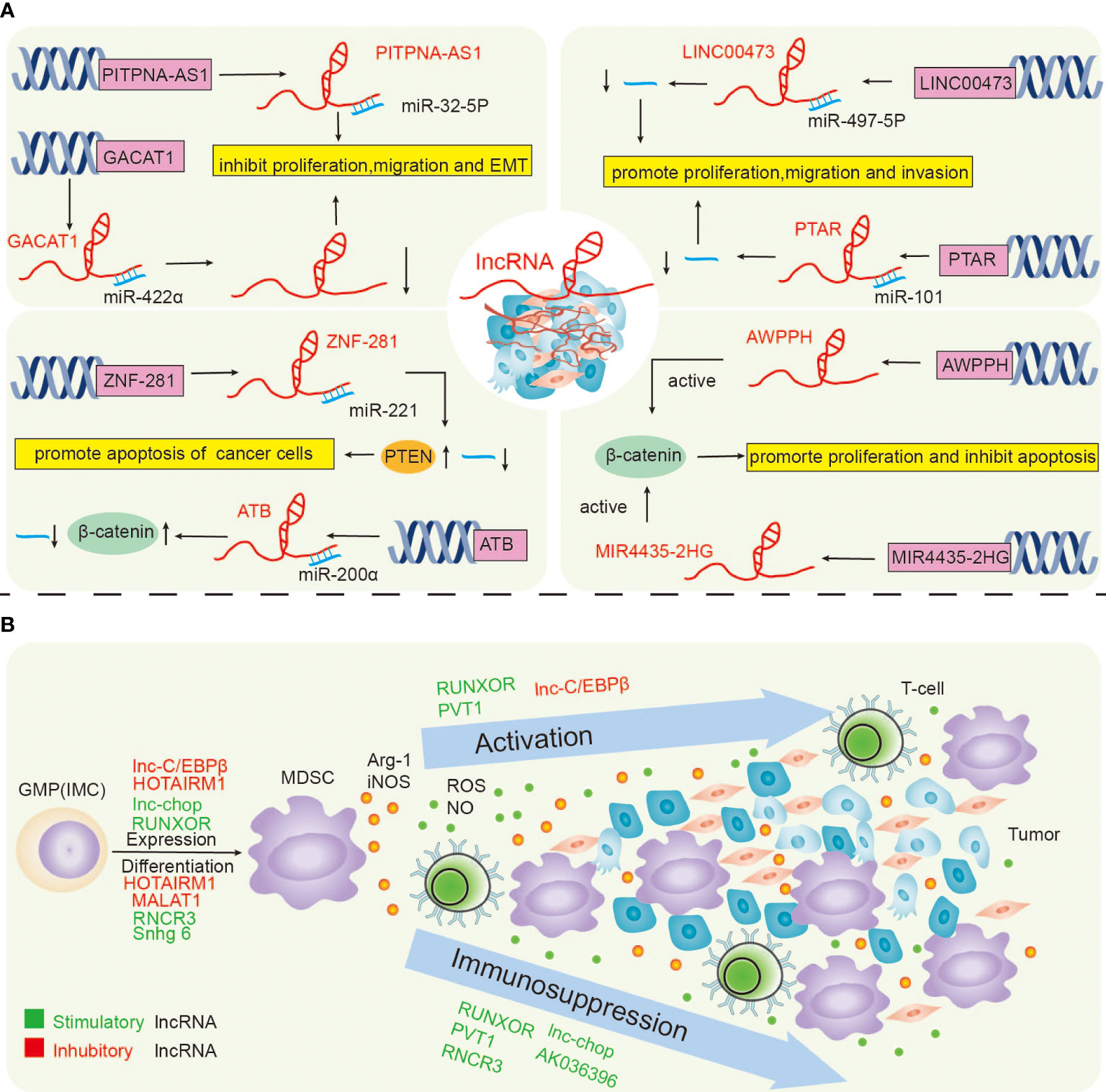
Figure 2 LncRNAs directly regulate lung cancer (A), and the lncRNAs regulate lung cancer by regulating MDSCs (B).
Taking lncRNAs as an entry point holds the promise of further improving the survival of individuals with lung tumours. Unfortunately, there is a lack of lncRNA-related lung cancer studies. Here, we summarize the available information on lncRNAs’ regulation of lung cancer and provide the findings in Table 3.
The roles of lncRNA PITPNA antisense RNA 1 (PITPNA-AS1) were studied in individuals with NSCLC. The results showed that the transcription levels of PitPNA-AS1 in NSCLC tissues were overexpressed in NSCLC tissues. However, its silencing blocked NSCLC propagation and metastasis. An interesting connection between PitPNA-AS1 and microRNA (miR)-32-5p was detected, proving that PITPNA-AS1 downregulation inhibited the progression of NSCLCs by directing Mir-32-5p. This suggested PITPNA-AS1 as a diagnostic and prognostic biomarker of NSCLC (107). Other authors proved that the lncRNA ZEB2-AS1 was overexpressed in NSCLC patients, which stimulated the epithelial-mesenchymal transition (EMT) in these patients, proving that lncRNA ZEB2-AS1 may become a new diagnostic, prognostic and therapeutic factor (108). Hongxia Wu et al. proposed that targeting the lncRNA NEAT1 could be a possible treatment for NSCLC (109). Some studies have pointed out that the zinc finger protein (ZNF) 281 could be a tumour suppressor lncRNA in glioma. Xin Lu et al. followed up on patients for 5 years to analyze the role of ZNF281 in NSCLC. They found that ZNF281 up-regulated the phosphatase and tensin homolog (PTEN) by down-regulating Mir-221 in NSCLC, thus constraining cancer cell propagation and death (110). The lncRNA LINC00473 was overexpressed in lung tumour tissues and NSCLC cells (A549 and H1299), resulting in a low 5-year patients’ OS (overall survival). Studies on lung tumour tissues demonstrated that LINC00473 interacted with Mir-497-5p and blocked its activity, thus promoting the propagation of NSCLC cells (111). To explore the relationship between lncRNA transforming associated RNA (PTAR) and Mir-101 in NSCLC, Wenjun Yu et al. conducted a series of studies during which they proved that lncRNA PTAR was up-regulated in NSCLC cells and could be combined with Mir-101 to inactivate it to stimulate the growth of NSCLC cells (112). GACAT1 (the lncRNA gastric cancer-associated transcript 1) plays a carcinogenic role in different types of cancer. It is overexpressed in NSCLC tissues, which may be associated with the adverse outcome of NSCLC patients. This finding delivers new knowledge for developing novel therapeutic approaches for NSCLC (113). AWPPH is a recently revealed lncRNA, which can be highly expressed in NSCLC tissues, thus stimulating the propagation of NSCLC cells, and considerably inhibiting the survival rate of patients with high AWPPH expression (114).LncRNA prostate cancer-associated transcript (PCAT) 19 also has a certain effect on the progression of NSCLC, PCAT19 was found overexpressed in NSCLC, which increased NSCLC cell proliferation and promoted the progression of NSCLC (115).Ting Wang et al. discovered the role of lncRNA-ATB in NSCLC using the in vitro cultured NSCLC NCI-H838 cell line. Their findings revealed that lncRNA-ATB promoted lung cancer progression by inhibiting miR-200a expression and reversed the promotion of β-linked protein expression to promote apoptosis in NSCLC cells (116).The newly discovered lncRNA SET binding factor 2 antisense RNA 1 (LncRNA SBF2-AS1) is involved in the progression of many cancers like lung, breast, hepatocellular carcinoma, thyroid, gastric, colorectal cancers and others (118). Interestingly, Weilong Ye et al. identified 13 lncRNAs related to Gefitinib metabolism and applied them to build the prognostic model of NSCLC patients (119). This proves that lncRNA studies are not limited to particular tumour tissue, and more and more studies have proposed new ideas for the diagnosis, treatment and prognosis of lncRNAs.
Ghada Mohamed Gamal El-din et al. verified the expression of serum markers RAB27A mRNA and RNA-RP11-510m2 in 20 individuals with lung tumours, 10 individuals with COPD and 10 controls in good physical shape. The results showed that the serum exosome RAB27A mRNA was positively correlated with lung cancer, whereas NRC-RNA-RP11-510m2 was negatively correlated, which could be developed as biomarkers for diagnosis and prognosis of lung tumours (120).LncRNAs HOTAIR has been shown to induce tumorigenesis of several cancer types. Chunlin Ke et al. explored the relationship between the four types of lncRNA HOTAIR and the susceptibility to lung cancer. One thousand seven hundred fifteen individuals with lung tumours and 2745 healthy subjects were recruited. The results proved that HOTAIR was important in lung cancer screening and prognosis prediction, especially for people with high-risk factors (121). Sik1-lnc is another lncRNA adjacent to salt-inducible kinase 1 (SIK1) and can be abnormally expressed in lung cancer. Other authors (Liu Yang et al.) studied the expression of SIK1 and SIK1-LNC in samples from lung cancer patients and established that the transcription activity of SIK1 and SIK1-LNC was decreased.
Moreover, SIK1-LNC considerably repressed the propagation of lung cancer cells, signifying that SIK1-LNC served as a novel biomarker and target for lung cancer therapeutic approaches (122).TSLC1, as a tumour suppressor gene in various cancers, is considerably repressed in NSCLC tissues and cell lines. The increased expression of TSLC1 inhibited the viability, migration and invasion of NSCLC cells. Other authors explored the biological mechanism of the antisense RNA of TSLC1, namely lncRNA RP11-713B9.1, in the development and progression of NSCLC. They analyzed the tumour tissues of 46 NSCLC patients. They established that the activity of RP11-713B9.1 was undoubtedly connected with TSLC1 (a tumour suppressor gene), while the overexpression of RP11-713B9.1 led to a substantial overexpression of TSLC1. In other words, the inhibition of RNA RP11-713B9.1 transcription backed NSCLC cell survival (123).
MDSCs are indispensable for the occurrence and progression of lung tumors. Therefore it is very logical to assume that lncRNAs could regulate lung tumors indirectly b modulating MDSCs’ activity. In a study by Qinfeng Zhou et al., the levels of MDSCs and ARG-1 were significantly increased in PBMCs of lung cancer patients. Simultaneously, the relative expression of lncRNA MALAT1 PBMCs of individuals with lung tumors was considerably reduced. The direct effect of MALAT1 on MDSCs was further confirmed by siRNA interference of MALAT1 expression, which resulted in the inhibition of MDSCs expansion (8). RUNXOR lncRNA is significantly related to MDSCS-induced immunosuppression in individuals with lung tumors and could serve as therapeutic targets. In another study, the expression of lncRNA RUNXOR in PBMCs isolated from individuals with lung tumors was studied by qRT-PCR. The level of RUNXOR in individuals with lung tumors was found to be augmented. Moreover, the authors detected a differential expression of RUNXOR in squamous cell lung and lung adenocarcinoma, thus suggesting that RUNXOR could be used to differentiate lung tumor types. Furthermore, the analysis of the peripheral blood of individuals with lung tumors showed that RUNXOR transcription was augmented with the increase of MDSCs percentage and Arg1 levels but was decreased with the increase of the Th1/CTL cell ratio. This suggested that the transcriptional activity of this lncRNA controlled the immunosuppressive function of MDSCs in these individuals (56). In another study, it was shown that lncRNA Snhg6 mainly exists in the cytoplasm.Furthermore, they proved that Snhg6 controlled MDSCs specialization by decreasing the stability of EZH2 without affecting their immunosuppressive functions (124). As mentioned above, the up-regulation of the lncRNA HOTAIRM1 down-regulated the inhibitory molecules in MDSCs. Other authors proved that HOTAIRM1 was conveyed in different types of lung cancer, especially in lung adenocarcinoma, and its transcription activity was considerably diminished in MDSCs from tumor tissues. Moreover, they showed that when HOTAIRM1 was overexpressed, Arg1 expression levels in MDSCs were down-regulated, thus inhibiting the propagation of MDSCs in lung tumors (125). In another study, the authors established that lncRNA F730016J06Rik (AK036396) was overexpressed in G-MDSCs, whereas its knockdown promoted the maturation of G-MDSCs and reduced their immunosuppression functions. Data show that the Fcnb protein can bind to some proteases to promote the production of ROS and Arg1 by MDSCs through the lectin pathway in granulocyte. The last accelerates the migration of MDSCs to tumor sites. Therefore, AK036396 can enhance the stability of Fcnb protein through the ubiquitin-proteasome system. Thus, the maturation and function of G-MDSCs can be regulated to accelerate immune suppression (126). Yu Zheng et al. implanted Lewis lung cancer (LLC) cells into mice, isolated MDSCs by microbeads and flow cytometry, and measured the transcriptional activity of Pvt1. The authors detected that Pvt1 was overexpressed in G-MDSCs derived from cancer samples. The immunosuppressive effect of tumor-infiltrating MDSCs was mainly due to increased Arg1 transcription and production of nitric oxide by HIF-1α. Coincidentally, Yu Zheng et al. first identified Pvt1 as a HIF-1α target in G-MDSCs of LLC mice cells under hypoxia. RNA interference of Arg1 diminished its activity, ROS production in G-MDSCs decreased, and the antitumor T-cell response was restored. From this perspective, the authors concluded that targeting Pvt1 attenuated G-MDSCS-mediated immune suppression (75). LncRNAs regulate tumors far beyond lung cancer. The proof comes from Zohreh Khodaii et al., who explored the LncRNA-Mir-mRNA complex to find new targets in a rectal tumor. The depletion of Lactobacillus acidophilus in individuals with rectal tumors induced the transcription of the lncRNA-Mir-mRNA network, which delivers new observing and treatment methods for rectal cancer patients (127). Other authors performed biopsies of tumor and non-tumor tissues from patients with gastric cancer. They showed that lncRNA PVT1 expression was increased, whereas the lncRNA ZFAS1 expression was decreased compared with non-tumor parts. PVT1 and ZFAS1 were biomarkers for detecting and treating gastric cancer cases (128). Furthermore, the lncRNA MEG3 was reported as a tumor suppressor in breast cancer. Battseren Bayarmaa et al. evaluated the impact of MEG3 polymorphism on neoadjuvant therapy in144 patients with breast cancer. The results showed that MEG3 polymorphism was associated with the chemotherapy response and toxicity of paclitaxel and cisplatin. This indicated that MEG3 polymorphism has the potential as a prognostic marker for breast cancer individuals (129). Xianmin Guan et al. obtained bone marrow samples from 146 pediatric patients with acute myeloid leukaemia (AML) and 73 patients with non-hematologic malignancies. They measured lncRNA-SOX6-1 expression to examine the association between lncRNA-SOX6-1 and AML. SOX6-1 transcription was augmented in the AML patients compared to the healthy volunteers, which promoted cell propagation while inhibiting cell death and was related to worse risk diversification and poorer treatment outcomes (130). Similarly, Zhenqing Tan et al. studied the relationship between INK4 expression and the clinical characteristics and prediction of AML in patients, they studied the transcription of ANRIL in bone marrow mononuclear cells (BMMCs) in 178 patients with initial AML and 30 healthy donors. Compared with healthy people, lncRNA ANRIL levels were increased in AML patients, and those with augmented ANRIL transcription had smaller event-free survival (EFS) and OS. Therefore, ANRIL was proposed as a biomarker for AML. Moreover, it has clinical relevance in assisting the diagnosis, treatment and prognosis prediction of AML and identifying potential drug targets of AML (131).
The development mechanisms of tumour cells are diverse, and the tumour microenvironment changes are also complex and miscellaneous. To successfully implement tumour immunotherapy, tumour suppressors must be removed. Recent data have shown that MDSCs are the chief controllers of cancer immune responses and inflammation in individuals with tumours as they intensely constrain the antitumor immune response of CD4+ T cells, CD8+ T cells and NK cells, thus triggering tumour growth. MDSCs play critical roles not only in lung cancer carcinogenesis but also in its progression and prognosis. Therefore, MDSCs are an attractive therapeutic target because they are carefully related to adverse effects. With the discovery of many novel lncRNAs and the general studies on their roles in different pathologies, particularly cancer, lncRNA research has become a new trend. Factors generated by the tumour and its accompanying hypoxia or inflammatory TME may sustain the expression of some non-coding RNAs. Based on lncRNAs, treatment strategies targeting MDSCs can recover our understanding of MDSCs and disclose new tumour propagation instruments.
Furthermore, the study of novel lncRNAs that regulate the activity of MDSCs is expected to enable their application in immunoregulatory therapy or as biomarkers. In particular, recent data proved that lncRNAs play a significant part in NSCLC development, which will provide a new direction for our subsequent research on lung cancer. However, the complex biological mechanism of MDSCs also poses new challenges for targeted therapy. In addition, the functional connection between lncRNAs and MDSCs does not seem strong enough, the investigations on the way the lncRNAs regulate MDSCs are yet in their infancy, and the clinical research on lncRNA-related lung cancer is very little. Nevertheless, it is hoped that with the development of social science and the progress of medical technology, more clinical studies in this field can be piloted to approve the possibility of regulating MDSCs in lung cancer by lncRNAs. The last will deliver novel ideas for lung cancer treatment and bring more benefits to lung cancer patients.
XP and JH designed and supervised the study. YL and YH reviewed the references. YL and XP wrote the manuscript. YL, YZ and TL contributed to tables and figures, XP and JH revised the manuscript. XP acquired funding. All authors contributed to the article and approved the submitted version.
This work was supported by National innovation and entrepreneurship training program for College Students (202010489017 to XP), Jingzhou Science and Technology Bureau Project (2022HC78 to XP), the College Students Innovative Entrepreneurial Training Program in Yangtze University (Yz2022297 to XP), the Scientific Research Project of Education Department of Yangtze university (JY2020134 to XP), Hubei Province Natural Science Foundation of China (2017CFB786 to XP).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Feng Q, Zhang H, Dong Z, Zhou Y, Ma J. Circulating 25-hydroxyvitamin d and lung cancer risk and survival: A dose-response meta-analysis of prospective cohort studies. Med (Baltimore) (2017) 96(45):e8613. doi: 10.1097/MD.0000000000008613
2. Feng QQ, Dong ZQ, Zhou Y, Zhang H, Long C. miR-16-1-3p targets TWIST1 to inhibit cell proliferation and invasion in NSCLC. Bratisl Lek Listy (2018) 119(1):60–5. doi: 10.4149/BLL_2018_012
3. Xie F, Li P, Gong J, Tan H, Ma J. Urinary cell-free DNA as a prognostic marker for KRAS-positive advanced-stage NSCLC. Clin Transl Oncol (2018) 20(5):591–8. doi: 10.1007/s12094-017-1754-7
4. Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer (2013) 13(10):739–52. doi: 10.1038/nrc3581
5. Wang T, Wang J, Jiang H, Ni M, Zou Y, Chen Y, et al. Targeted regulation of tumor microenvironment through the inhibition of MDSCs by curcumin loaded self-assembled nano-filaments. Mater Today Bio (2022) 15:100304. doi: 10.1016/j.mtbio.2022.100304
6. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol (2009) 9(3):162–74. doi: 10.1038/nri2506
7. Daveri E, Vergani E, Shahaj E, Bergamaschi L, La Magra S, Dosi M, et al. microRNAs shape myeloid cell-mediated resistance to cancer immunotherapy. Front Immunol (2020) 11:1214. doi: 10.3389/fimmu.2020.01214
8. Zhou Q, Tang X, Tian X, Tian J, Zhang Y, Ma J, et al. LncRNA MALAT1 negatively regulates MDSCs in patients with lung cancer. J Cancer (2018) 9(14):2436–42. doi: 10.7150/jca.24796
9. Grover A, Sanseviero E, Timosenko E, Gabrilovich DI. Myeloid-derived suppressor cells: A propitious road to clinic. Cancer Discov (2021) 11(11):2693–706. doi: 10.1158/2159-8290.CD-21-0764
10. Zhao Y, Wu T, Shao S, Shi and Y Zhao. Phenotype B. Development, and biological function of myeloid-derived suppressor cells. Oncoimmunology (2016) 5(2):e1004983. doi: 10.1080/2162402X.2015.1004983
11. Mandruzzato S, Brandau S, Britten CM, Bronte V, Damuzzo V, Gouttefangeas C, et al. Toward harmonized phenotyping of human myeloid-derived suppressor cells by flow cytometry: results from an interim study. Cancer Immunol Immunother (2016) 65(2):161–9. doi: 10.1007/s00262-015-1782-5
12. Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun (2016) 7:12150. doi: 10.1038/ncomms12150
13. Yang Z, Guo J, Weng L, Tang W, Jin S, Ma W. Myeloid-derived suppressor cells-new and exciting players in lung cancer. J Hematol Oncol (2020) 13(1):10. doi: 10.1186/s13045-020-0843-1
14. Sayyadioskoie SR, Schwacha MG. Myeloid-derived suppressor cells (MDSCs) and the immunoinflammatory response to injury (Mini review). Shock (2021) 56(5):658–66. doi: 10.1097/SHK.0000000000001795
15. Bader JE, Voss K, Rathmell JC. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell (2020) 78(6):1019–33. doi: 10.1016/j.molcel.2020.05.034
16. Lee SS, Cheah YK. The interplay between MicroRNAs and cellular components of tumour microenvironment (TME) on non-Small-Cell lung cancer (NSCLC) progression. J Immunol Res (2019) 2020:3046379. doi: 10.1155/2019/3046379
17. Hofer F, Di Sario G, Musiu C, Sartoris S, De Sanctis F, Ugel S. A complex metabolic network confers immunosuppressive functions to myeloid-derived suppressor cells (MDSCs) within the tumour microenvironment. Cells (2021) 10(10):2700. doi: 10.3390/cells10102700
18. Bizymi N, Papadaki HA. Myeloid-derived suppressor cells (MDSCs) in haematology. J Clin Med (2021) 11(1):1479–85. doi: 10.3390/jcm11010187
19. Wang Y, Tian J, Wang S. The potential therapeutic role of myeloid-derived suppressor cells in autoimmune arthritis. Semin Arthritis Rheum (2016) 45(4):490–5. doi: 10.1016/j.semarthrit.2015.07.003
20. Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol (2016) 37(3):208–20. doi: 10.1016/j.it.2016.01.004
21. Szuster-Ciesielska A, Hryciuk-Umer E, Stepulak A, Kupisz K, Kandefer-Szerszen M. Reactive oxygen species production by blood neutrophils of patients with laryngeal carcinoma and antioxidative enzyme activity in their blood. Acta Oncol (2004) 43(3):252–8. doi: 10.1080/02841860410029708
22. Xue Q, Yan Y, Zhang R, Xiong H. Regulation of iNOS on immune cells and its role in diseases. Int J Mol Sci (2018) 19(12):3805. doi: 10.3390/ijms19123805
23. Garcia-Ortiz A, Serrador JM. Nitric oxide signaling in T cell-mediated immunity. Trends Mol Med (2018) 24(4):412–27. doi: 10.1016/j.molmed.2018.02.002
24. Ostrand-Rosenberg S, Sinha P, Beury DW, Clements VK. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin Cancer Biol (2012) 22(4):275–81. doi: 10.1016/j.semcancer.2012.01.011
25. Fletcher M, Ramirez ME, Sierra RA, Raber P, Thevenot P, Al-Khami AA, et al. L-arginine depletion blunts antitumor T-cell responses by inducing myeloid-derived suppressor cells. Cancer Res (2015) 75(2):275–83. doi: 10.1158/0008-5472.CAN-14-1491
26. Srivastava MK, Sinha P, Clements VK, Rodriguez P, Ostrand-Rosenberg S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res (2010) 70(1):68–77. doi: 10.1158/0008-5472.CAN-09-2587
27. Hanson EM, Clements VK, Sinha P, Ilkovitch D, Ostrand-Rosenberg S. Myeloid-derived suppressor cells down-regulate l-selectin expression on CD4+ and CD8+ T cells. J Immunol (2009) 183(2):937–44. doi: 10.4049/jimmunol.0804253
28. Liu J, Wang H, Yu Q, Zheng S, Jiang Y, Liu Y, et al. RETRACTED: Aberrant frequency of IL-10-producing b cells and its association with treg and MDSC cells in non small cell lung carcinoma patients. Hum Immunol (2016) 77(1):84–9. doi: 10.1016/j.humimm.2015.10.015
29. Huang B, Pan PY, Li Q, Sato AI, Levy DE, Bromberg J, et al. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res (2006) 66(2):1123–31. doi: 10.1158/0008-5472.CAN-05-1299
30. Yang R, Cai Z, Zhang Y, t Yutzy WH, Roby KF, Roden RB. CD80 in immune suppression by mouse ovarian carcinoma-associated gr-1+CD11b+ myeloid cells. Cancer Res (2006) 66(13):6807–15. doi: 10.1158/0008-5472.CAN-05-3755
31. Sinha P, Clements VK, Bunt SK, Albelda SM, Ostrand-Rosenberg S. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J Immunol (2007) 179(2):977–83. doi: 10.4049/jimmunol.179.2.977
32. Liu C, Yu S, Kappes J, Wang J, Grizzle WE, Zinn KR, et al. Expansion of spleen myeloid suppressor cells represses NK cell cytotoxicity in tumor-bearing host. Blood (2007) 109(10):4336–42. doi: 10.1182/blood-2006-09-046201
33. Ortiz ML, Lu L, Ramachandran I, Gabrilovich DI. Myeloid-derived suppressor cells in the development of lung cancer. Cancer Immunol Res (2014) 2(1):50–8. doi: 10.1158/2326-6066.CIR-13-0129
34. Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol (2021) 21(8):485–98. doi: 10.1038/s41577-020-00490-y
35. Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, et al. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell (2004) 6(4):409–21. doi: 10.1016/j.ccr.2004.08.031
36. Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B (2021) 11(9):2783–97. doi: 10.1016/j.apsb.2021.01.001
37. Taki M, Abiko K, Ukita M, Murakami R, Yamanoi K, Yamaguchi K, et al. Tumor immune microenvironment during epithelial-mesenchymal transition. Clin Cancer Res (2021) 27(17):4669–79. doi: 10.1158/1078-0432.CCR-20-4459
38. Erin N, Grahovac J, Brozovic A, Efferth T. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resist Update (2020) 53:100715. doi: 10.1016/j.drup.2020.100715
39. Li Q, Xiang M. Metabolic reprograming of MDSCs within tumor microenvironment and targeting for cancer immunotherapy. Acta Pharmacol Sin (2022) 43(6):1337–48. doi: 10.1038/s41401-021-00776-4
40. Zahran AM, Hetta HF, Zahran ZAM, Rashad A, Rayan A, Mohamed DO, et al. Prognostic role of monocytic myeloid-derived suppressor cells in advanced non-Small-Cell lung cancer: Relation to different hematologic indices. J Immunol Res (2021) 2021:3241150. doi: 10.1155/2021/3241150
41. Barrera L, Montes-Servin E, Hernandez-Martinez JM, Orozco-Morales M, Montes-Servin E, Michel-Tello D, et al. Levels of peripheral blood polymorphonuclear myeloid-derived suppressor cells and selected cytokines are potentially prognostic of disease progression for patients with non-small cell lung cancer. Cancer Immunol Immunother (2018) 67(9):1393–406. doi: 10.1007/s00262-018-2196-y
42. Huang A, Zhang B, Wang B, Zhang F, Fan KX, Guo YJ. Increased CD14(+)HLA-DR (-/low) myeloid-derived suppressor cells correlate with extrathoracic metastasis and poor response to chemotherapy in non-small cell lung cancer patients. Cancer Immunol Immunother (2013) 62(9):1439–51. doi: 10.1007/s00262-013-1450-6
43. Iclozan C, Antonia S, Chiappori A, Chen DT, Gabrilovich D. Therapeutic regulation of myeloid-derived suppressor cells and immune response to cancer vaccine in patients with extensive stage small cell lung cancer. Cancer Immunol Immunother (2013) 62(5):909–18. doi: 10.1007/s00262-013-1396-8
44. Bronte G, Petracci E, De Matteis S, Canale M, Zampiva I, Priano I, et al. High levels of circulating monocytic myeloid-derived suppressive-like cells are associated with the primary resistance to immune checkpoint inhibitors in advanced non-small cell lung cancer: An exploratory analysis. Front Immunol (2022) 13:866561. doi: 10.3389/fimmu.2022.866561
45. Zadian SS, Adcock IM, Salimi B, Mortaz E. Circulating levels of monocytic myeloid-derived suppressor cells (M-MDSC) and CXCL-8 in non-small cell lung cancer (NSCLC). Tanaffos (2021) 20(1):15–21.
46. Liu CY, Wang YM, Wang CL, Feng PH, Ko HW, Liu YH, et al. Population alterations of l-arginase- and inducible nitric oxide synthase-expressed CD11b+/CD14(-)/CD15+/CD33+ myeloid-derived suppressor cells and CD8+ T lymphocytes in patients with advanced-stage non-small cell lung cancer. J Cancer Res Clin Oncol (2010) 136(1):35–45. doi: 10.1007/s00432-009-0634-0
47. Feng PH, Lee KY, Chang YL, Chan YF, Kuo LW, Lin TY, et al. CD14(+)S100A9(+) monocytic myeloid-derived suppressor cells and their clinical relevance in non-small cell lung cancer. Am J Respir Crit Care Med (2012) 186(10):1025–36. doi: 10.1164/rccm.201204-0636OC
48. Heuvers ME, Muskens F, Bezemer K, Lambers M, Dingemans AC, Groen HJM, et al. Arginase-1 mRNA expression correlates with myeloid-derived suppressor cell levels in peripheral blood of NSCLC patients. Lung Cancer (2013) 81(3):468–74. doi: 10.1016/j.lungcan.2013.06.005
49. Tian T, Gu X, Zhang B, Liu Y, Yuan C, Shao L, et al. Increased circulating CD14(+)HLA-DR-/low myeloid-derived suppressor cells are associated with poor prognosis in patients with small-cell lung cancer. Cancer biomark (2015) 15(4):425–32. doi: 10.3233/CBM-150473
50. Yamauchi Y, Safi S, Blattner C, Rathinasamy A, Umansky L, Juenger S, et al. Circulating and tumor myeloid-derived suppressor cells in resectable non-small cell lung cancer. Am J Respir Crit Care Med (2018) 198(6):777–87. doi: 10.1164/rccm.201708-1707OC
51. Koinis F, Vetsika EK, Aggouraki D, Skalidaki E, Koutoulaki A, Gkioulmpasani M, et al. Effect of first-line treatment on myeloid-derived suppressor cells' subpopulations in the peripheral blood of patients with non-small cell lung cancer. J Thorac Oncol (2016) 11(8):1263–72. doi: 10.1016/j.jtho.2016.04.026
52. Limagne E, Richard C, Thibaudin M, Fumet JD, Truntzer C, Lagrange A, et al. Tim-3/galectin-9 pathway and mMDSC control primary and secondary resistances to PD-1 blockade in lung cancer patients. Oncoimmunology (2019) 8(4):e1564505. doi: 10.1080/2162402X.2018.1564505
53. Passaro A, Mancuso P, Gandini S, Spitaleri G, Labanca V, Guerini-Rocco E, et al. Gr-MDSC-linked asset as a potential immune biomarker in pretreated NSCLC receiving nivolumab as second-line therapy. Clin Transl Oncol (2020) 22(4):603–11. doi: 10.1007/s12094-019-02166-z
54. de Goeje PL, Bezemer K, Heuvers ME, Dingemans AC, Groen HJ, Smit EF, et al. Immunoglobulin-like transcript 3 is expressed by myeloid-derived suppressor cells and correlates with survival in patients with non-small cell lung cancer. Oncoimmunology (2015) 4(7):e1014242. doi: 10.1080/2162402X.2015.1014242
55. De Cicco P, Ercolano G, Ianaro A. The new era of cancer immunotherapy: Targeting myeloid-derived suppressor cells to overcome immune evasion. Front Immunol (2020) 11:1680. doi: 10.3389/fimmu.2020.01680
56. Tian X, Ma J, Wang T, Tian J, Zheng Y, Peng R, et al. Long non-coding RNA RUNXOR accelerates MDSC-mediated immunosuppression in lung cancer. BMC Cancer (2018) 18(1):660. doi: 10.1186/s12885-018-4564-6
57. Feng PH, Yu CT, Chen KY, Luo CS, Wu SM, Liu CY, et al. S100A9(+) MDSC and TAM-mediated EGFR-TKI resistance in lung adenocarcinoma: the role of RELB. Oncotarget (2018) 9(7):7631–43. doi: 10.18632/oncotarget.24146
58. Park MS, Yang AY, Lee JE, Kim SK, Roe JS, Park MS, et al. GALNT3 suppresses lung cancer by inhibiting myeloid-derived suppressor cell infiltration and angiogenesis in a TNFR and c-MET pathway-dependent manner. Cancer Lett (2021) 521:294–307. doi: 10.1016/j.canlet.2021.08.015
59. Li R, Salehi-Rad R, Crosson W, Momcilovic M, Lim RJ, Ong SL, et al. Inhibition of granulocytic myeloid-derived suppressor cells overcomes resistance to immune checkpoint inhibition in LKB1-deficient non-small cell lung cancer. Cancer Res (2021) 81(12):3295–308. doi: 10.1158/0008-5472.CAN-20-3564
60. Adah D, Yang Y, Liu Q, Gadidasu K, Tao Z, Yu S, et al. Plasmodium infection inhibits the expansion and activation of MDSCs and tregs in the tumor microenvironment in a murine Lewis lung cancer model. Cell Commun Signal (2019) 17(1):32. doi: 10.1186/s12964-019-0342-6
61. Su X, Fan Y, Yang L, Huang J, Qiao F, Fang Y, et al. Dexmedetomidine expands monocytic myeloid-derived suppressor cells and promotes tumour metastasis after lung cancer surgery. J Transl Med (2018) 16(1):347. doi: 10.1186/s12967-018-1727-9
62. Levy L, Mishalian I, Bayuch R, Zolotarov L, Michaeli J, Fridlender ZG. Splenectomy inhibits non-small cell lung cancer growth by modulating anti-tumor adaptive and innate immune response. Oncoimmunology (2015) 4(4):e998469. doi: 10.1080/2162402X.2014.998469
63. Ma J, Xu H, Wang S. Immunosuppressive role of myeloid-derived suppressor cells and therapeutic targeting in lung cancer. J Immunol Res (2018) 2018:6319649. doi: 10.1155/2018/6319649
64. Liu X, Zhao S, Sui H, Liu H, Yao M, Su Y, et al. MicroRNAs/LncRNAs modulate MDSCs in tumor microenvironment. Front Oncol (2022) 12:772351. doi: 10.3389/fonc.2022.772351
65. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer (2018) 18(1):5–18. doi: 10.1038/nrc.2017.99
66. Yuan W, Xiong X, Du J, Fan Q, Wang R, Zhang X. LncRNA PVT1 accelerates LPS-induced septic acute kidney injury through targeting miR-17-5p and regulating NF-kappaB pathway. Int Urol Nephrol (2021) 53(11):2409–19. doi: 10.1007/s11255-021-02905-8
67. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet (2016) 17(1):47–62. doi: 10.1038/nrg.2015.10
68. Tragante V, Moore JH, Asselbergs FW. The ENCODE project and perspectives on pathways. Genet Epidemiol (2014) 38(4):275–80. doi: 10.1002/gepi.21802
69. Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet (2018) 19:9:535–48. doi: 10.1038/s41576-018-0017-y
70. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol (2021) 220(2):e202009045. doi: 10.1083/jcb.202009045
71. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res (2017) 77(15):3965–81. doi: 10.1158/0008-5472.CAN-16-2634
72. Ayupe AC, Tahira AC, Camargo L, Beckedorff FC, Verjovski-Almeida S, Reis EM. Global analysis of biogenesis, stability and sub-cellular localization of lncRNAs mapping to intragenic regions of the human genome. RNA Biol (2015) 12(8):877–92. doi: 10.1080/15476286.2015.1062960
73. Kazimierczyk M, Kasprowicz MK, Kasprzyk ME, Wrzesinski J. Human long noncoding RNA interactome: Detection, characterization and function. Int J Mol Sci (2020) 21(3):1027. doi: 10.3390/ijms21031027
74. Zhang J, Liu M, Liang Y, Zhang M, Huang Z. Correlation between lncRNA H19 rs2839698 polymorphism and susceptibility to NK / T cell lymphoma in Chinese population. J BUON (2021) 26(2):587–91.
75. Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, et al. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol Cancer (2019) 18(1):61. doi: 10.1186/s12943-019-0978-2
76. Gao Y, Wang T, Li Y, Zhang Y, Yang R. Lnc-chop promotes immunosuppressive function of myeloid-derived suppressor cells in tumor and inflammatory environments. J Immunol (2018) 200(8):2603–14. doi: 10.4049/jimmunol.1701721
77. Shang W, Tang Z, Gao Y, Qi H, Su X, Zhang Y, et al. LncRNA RNCR3 promotes chop expression by sponging miR-185-5p during MDSC differentiation. Oncotarget (2017) 8(67):111754–69. doi: 10.18632/oncotarget.22906
78. Shang W, Gao Y, Tang Z, Zhang Y, Yang R. The pseudogene Olfr29-ps1 promotes the suppressive function and differentiation of monocytic MDSCs. Cancer Immunol Res (2019) 7(5):813–27. doi: 10.1158/2326-6066.CIR-18-0443
79. Fujisaka Y, Iwata T, Tamai K, Nakamura M, Mochizuki M, Shibuya R, et al. Long non-coding RNA HOTAIR up-regulates chemokine (C-c motif) ligand 2 and promotes proliferation of macrophages and myeloid-derived suppressor cells in hepatocellular carcinoma cell lines. Oncol Lett (2018) 15(1):509–14. doi: 10.3892/ol.2017.7322
80. Tian X, Ma J, Wang T, Tian J, Zhang Y, Mao L, et al. Long non-coding RNA HOXA transcript antisense RNA myeloid-specific 1-HOXA1 axis downregulates the immunosuppressive activity of myeloid-derived suppressor cells in lung cancer. Front Immunol (2018) 9:473. doi: 10.3389/fimmu.2018.00473
81. Gao Y, Shang W, Zhang D, Zhang S, Zhang X, Zhang Y, et al. Lnc-C/EBPbeta modulates differentiation of MDSCs through downregulating IL4i1 with C/EBPbeta LIP and WDR5. Front Immunol (2019) 10:1661. doi: 10.3389/fimmu.2019.01661
82. Graham M, Adams JM, Cory S. Murine T lymphomas with retroviral inserts in the chromosomal 15 locus for plasmacytoma variant translocations. Nature (1985) 314(6013):740–3. doi: 10.1038/314740a0
83. Ghetti M, Vannini I, Storlazzi CT, Martinelli G, Simonetti G. Linear and circular PVT1 in hematological malignancies and immune response: two faces of the same coin. Mol Cancer (2020) 19(1):69. doi: 10.1186/s12943-020-01187-5
84. Zhong W, Zeng J, Xue J, Du A, Xu Y. Knockdown of lncRNA PVT1 alleviates high glucose-induced proliferation and fibrosis in human mesangial cells by miR-23b-3p/WT1 axis. Diabetol Metab Syndr (2020) 12:33. doi: 10.1186/s13098-020-00539-x
85. Lu J, Xu F, Lu H. LncRNA PVT1 regulates ferroptosis through miR-214-mediated TFR1 and p53. Life Sci (2020) 260:118305. doi: 10.1016/j.lfs.2020.118305
86. Onagoruwa OT, Pal G, Ochu C, Ogunwobi OO. Oncogenic role of PVT1 and therapeutic implications. Front Oncol (2020) 10:17. doi: 10.3389/fonc.2020.00017
87. Wang H, Huang Y, Yang Y. LncRNA PVT1 regulates TRPS1 expression in breast cancer by sponging miR-543. Cancer Manag Res (2020) 12:7993–8004. doi: 10.2147/CMAR.S263383
88. Thakuri BKC, Zhang J, Zhao J, Nguyen LN, Nguyen LNT, Schank M, et al. HCV-associated exosomes upregulate RUNXOR and RUNX1 expressions to promote MDSC expansion and suppressive functions through STAT3-miR124 axis. Cells (2020) 9(12):2715. doi: 10.3390/cells9122715
89. Nie Y, Zhou L, Wang H, Chen N, Jia L, Wang C, et al. Profiling the epigenetic interplay of lncRNA RUNXOR and oncogenic RUNX1 in breast cancer cells by gene in situ cis-activation. Am J Cancer Res (2019) 9(8):1635–49.
90. Wang H, Li W, Guo R, Sun J, Cui J, Wang G, et al. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int J Cancer (2014) 135(12):2783–94. doi: 10.1002/ijc.28922
91. Sonda N, Simonato F, Peranzoni E, Cali B, Bortoluzzi S, Bisognin A, et al. miR-142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity (2013) 38(6):1236–49. doi: 10.1016/j.immuni.2013.06.004
92. Thevenot PT, Sierra RA, Raber PL, Al-Khami AA, Trillo-Tinoco J, Zarreii P, et al. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity (2014) 41(3):389–401. doi: 10.1016/j.immuni.2014.08.015
93. Xie Q, Ju T, Zhou C, Zhai L. LncRNA RNCR3 promotes the progression of HCC by activating the Akt/GSK3beta signaling pathway. Evid Based Complement Alternat Med (2020) 2020:8367454. doi: 10.1155/2020/8367454
94. Hong Q, Ling L, Huang W, Liu Y, Zhuo Y, Hong Z, et al. LncRNA RNCR3 promotes endothelial cell proliferation and inflammatory cytokine secretion via regulating miR-185-5p/cyclin D2 axis. Environ Sci pollut Res Int (2021) 28(21):27025–32. doi: 10.1007/s11356-020-12117-9
95. Woo CJ, Kingston RE. HOTAIR lifts noncoding RNAs to new levels. Cell (2007) 129(7):1257–9. doi: 10.1016/j.cell.2007.06.014
96. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell (2007) 129:7:1311–23. doi: 10.1016/j.cell.2007.05.022
97. Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol (2014) 7:90. doi: 10.1186/s13045-014-0090-4
98. Yuan C, Ning Y, Pan Y. Emerging roles of HOTAIR in human cancer. J Cell Biochem (2020) 121(5-6):3235–47. doi: 10.1002/jcb.29591
99. Li D, Chai L, Yu X, Song Y, Zhu X, Fan S, et al. The HOTAIRM1/miR-107/TDG axis regulates papillary thyroid cancer cell proliferation and invasion. Cell Death Dis (2020) 11(4):227. doi: 10.1038/s41419-020-2416-1
100. Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood (2009) 113(11):2526–34. doi: 10.1182/blood-2008-06-162164
101. Zhao Y, Wang W, Guan C, Hu Z, Liu L, Li W, et al. Long noncoding RNA HOTAIRM1 in human cancers. Clin Chim Acta (2020) 511:255–9. doi: 10.1016/j.cca.2020.10.011
102. Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci U.S.A. (1993) 90(17):8219–23. doi: 10.1073/pnas.90.17.8219
103. Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity (2010) 32(6):790–802. doi: 10.1016/j.immuni.2010.05.010
104. Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer (2021) 1875(2):188502. doi: 10.1016/j.bbcan.2021.188502
105. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene (2003) 22(39):8031–41. doi: 10.1038/sj.onc.1206928
106. Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: a potential biomarker in cancer. Cancer Manag Res (2018) 10:6757–68. doi: 10.2147/CMAR.S169406
107. Chen G, Zheng Z, Li J, Zhang P, Wang Z, Guo S, et al. Long noncoding RNA PITPNAAS1 silencing suppresses proliferation, metastasis and epithelialmesenchymal transition in nonsmall cell lung cancer cells by targeting microRNA325p. Mol Med Rep (2021) 23(3):212. doi: 10.3892/mmr.2021.11851
108. Xu H, Yu B, Shen W, Jin C, Wang L, Xi Y. Over-expression of long non-coding RNA ZEB2-AS1 may predict poor prognosis and promote the migration, invasion, and epithelial-mesenchymal transition of tumor cells in non-small cell lung cancer. Int J Biol Markers (2020) 35:3:29–35. doi: 10.1177/1724600820938385
109. Wu H, Liu A. Long non-coding RNA NEAT1 regulates ferroptosis sensitivity in non-small-cell lung cancer. J Int Med Res (2021) 49(3):300060521996183. doi: 10.1177/0300060521996183
110. Lu X, Yin B, Wang X, Wang F, Li Y, Wang N, et al. Long non-coding RNA-ZNF281 upregulates PTEN expression via downregulation of microRNA-221 in non-small cell lung cancer. Oncol Lett (2020) 20(3):2962–8. doi: 10.3892/ol.2020.11821
111. Xu SH, Bo YH, Ma HC, Zhang HN, Shao MJ. lncRNA LINC00473 promotes proliferation, migration, invasion and inhibition of apoptosis of non-small cell lung cancer cells by acting as a sponge of miR-497-5p. Oncol Lett (2021) 21(6):429. doi: 10.3892/ol.2021.12690
112. Yu W, Sun Z, Yang L, Han Y, Yue L, Deng L, et al. lncRNA PTAR promotes NSCLC cell proliferation, migration and invasion by sponging microRNA101. Mol Med Rep (2019) 20(5):4168–74. doi: 10.3892/mmr.2019.10646
113. Zhong Y, Lin H, Li Q, Liu C, Zhong L. Downregulation of long noncoding RNA GACAT1 suppresses proliferation and induces apoptosis of NSCLC cells by sponging microRNA422a. Int J Mol Med (2021) 47:2:659–67. doi: 10.3892/ijmm.2020.4826
114. Song Z, Du J, Zhou L, Sun B. lncRNA AWPPH promotes proliferation and inhibits apoptosis of nonsmall cell lung cancer cells by activating the wnt/betacatenin signaling pathway. Mol Med Rep (2019) 19(5):4425–32. doi: 10.3892/mmr.2019.10089
115. Zhang X, Wang Q, Xu Y, Wang B, Jia C, Wang L, et al. lncRNA PCAT19 negatively regulates p53 in non-small cell lung cancer. Oncol Lett (2019) 18:6:6795–800. doi: 10.3892/ol.2019.11041
116. Wang T, Tang X, Liu Y. LncRNA-ATB promotes apoptosis of non-small cell lung cancer cells through MiR-200a/beta-Catenin. J BUON (2019) 24(6):2280–6.
117. Qian H, Chen L, Huang J, Wang X, Ma S, Cui F, et al. The lncRNA MIR4435-2HG promotes lung cancer progression by activating beta-catenin signalling. J Mol Med (Berl) (2018) 96(8):753–64. doi: 10.1007/s00109-018-1654-5
118. Tan F, Chen J, Wang B, Du Z, Mou J, Wu Y, et al. LncRNA SBF2-AS1: A budding star in various cancers. Curr Pharm Des (2022) 28(18):1513–22. doi: 10.2174/1381612828666220418131506
119. Ye W, Wu Z, Gao P, Kang J, Xu Y, Wei C, et al. Identified gefitinib metabolism-related lncRNAs can be applied to predict prognosis, tumor microenvironment, and drug sensitivity in non-small cell lung cancer. Front Oncol (2022) 12:939021. doi: 10.3389/fonc.2022.939021
120. Mohamed Gamal El-Din G, Ibrahim FK, Shehata HH, Osman NM, Abdel-Rahman OM, Ali M. Exosomal expression of RAB27A and its related lncRNA lnc-RNA-RP11-510M2 in lung cancer. Arch Physiol Biochem (2020) 128:1–7. doi: 10.1080/13813455.2020.1778036
121. Ke C, Feng X, Li J, Chen S, Hu X. Association between long non-coding RNA HOTAIR polymorphism and lung cancer risk: A systematic review and meta-analysis. Exp Ther Med (2022) 24:2:540. doi: 10.3892/etm.2022.11477
122. Yang L, Xie N, Huang J, Huang H, Xu S, Wang Z, et al. SIK1-LNC represses the proliferative, migrative, and invasive abilities of lung cancer cells. Onco Targets Ther (2018) 11:4197–206. doi: 10.2147/OTT.S165278
123. Cai J, Wang X, Huang H, Wang M, Zhang Z, Hu Y, et al. Down-regulation of long noncoding RNA RP11-713B9.1 contributes to the cell viability in nonsmall cell lung cancer (NSCLC). Mol Med Rep (2017) 16(3):3694–700. doi: 10.3892/mmr.2017.7026
124. Lu W, Cao F, Feng L, Song G, Chang Y, Chu Y, et al. LncRNA Snhg6 regulates the differentiation of MDSCs by regulating the ubiquitination of EZH2. J Hematol Oncol (2021) 14(1):196. doi: 10.1186/s13045-021-01212-0
125. Tian X, Ma J, Wang T, Tian J, Zhang Y, Mao L, et al. Corrigendum: Long non-coding RNA HOXA transcript antisense RNA myeloid-specific 1-HOXA1 axis downregulates the immunosuppressive activity of myeloid-derived suppressor cells in lung cancer. Front Immunol (2019) 10:2929. doi: 10.3389/fimmu.2019.02929
126. Tian X, Zheng Y, Yin K, Ma J, Tian J, Zhang Y, et al. LncRNA AK036396 inhibits maturation and accelerates immunosuppression of polymorphonuclear myeloid-derived suppressor cells by enhancing the stability of ficolin b. Cancer Immunol Res (2020) 8(4):565–77. doi: 10.1158/2326-6066.CIR-19-0595
127. Khodaii Z, Mehrabani Natanzi M, Khalighfard S, Ghandian Zanjan M, Gharghi M, Khori V, et al. Novel targets in rectal cancer by considering lncRNA-miRNA-mRNA network in response to lactobacillus acidophilus consumption: a randomized clinical trial. Sci Rep (2022) 12:1:9168. doi: 10.1038/s41598-022-13297-9
128. Dastmalchi N, Tayefeh-Gholami S, Rajabi A, Safaralizadeh R. PVT1 and ZFAS1 lncRNAs expressions and their biomarker value in gastric cancer tissue sampling among Iranian population. Mol Biol Rep (2021) 48(11):7171–7. doi: 10.1007/s11033-021-06709-y
129. Bayarmaa B, Wu Z, Peng J, Wang Y, Xu S, Yan T, et al. Association of LncRNA MEG3 polymorphisms with efficacy of neoadjuvant chemotherapy in breast cancer. BMC Cancer (2019) 19(1):877. doi: 10.1186/s12885-019-6077-3
130. Guan X, Wen X, Xiao J, An X, Yu J, Guo Y. Lnc-SOX6-1 upregulation correlates with poor risk stratification and worse treatment outcomes, and promotes cell proliferation while inhibits apoptosis in pediatric acute myeloid leukemia. Int J Lab Hematol (2019) 41(2):234–41. doi: 10.1111/ijlh.12952
Keywords: lncRNA, MDSC, lung cancer, targeted therapy, immunotherapy
Citation: Liu Y, Han Y, Zhang Y, Lv T, Peng X and Huang J (2023) LncRNAs has been identified as regulators of Myeloid-derived suppressor cells in lung cancer. Front. Immunol. 14:1067520. doi: 10.3389/fimmu.2023.1067520
Received: 12 October 2022; Accepted: 18 January 2023;
Published: 02 February 2023.
Edited by:
Erika Adriana Eksioglu, Moffitt Cancer Center, United StatesReviewed by:
Chi Ma, National Institutes of Health (NIH), United StatesCopyright © 2023 Liu, Han, Zhang, Lv, Peng and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaochun Peng, cHhjd2Q3ODlAc2luYS5jb20=; Jinbai Huang, amluYmFpaHVhbmdAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.