- 1Translational Cancer Research Facility, National Center for Cancer Care and Research/ Translational Research Institute, Hamad Medical Corporation, Doha, Qatar
- 2National Center for Cancer Care and Research, Hamad Medical Corporation, Doha, Qatar
- 3Qatar University Biomedical Research Center, Qatar University, Doha, Qatar
- 4Translational Research Institute, Academic Health System, Hamad Medical Corporation, Doha, Qatar
- 5Dermatology Institute, Academic Health System, Hamad Medical Corporation, Doha, Qatar
- 6College of Medicine, Qatar University, Doha, Qatar
- 7Communicable Disease Center, Hamad Medical Corporation, Doha, Qatar
- 8Infectious Disease Epidemiology Group, Weill Cornell Medicine–Qatar, Cornell University, Qatar Foundation–Education City, Doha, Qatar
- 9World Health Organization Collaborating Centre for Disease Epidemiology Analytics on HIV/AIDS, Sexually Transmitted Infections, and Viral Hepatitis, Weill Cornell Medicine–Qatar, Cornell University, Qatar Foundation–Education City, Doha, Qatar
- 10Department of Population Health Sciences, Weill Cornell Medicine, Cornell University, New York, NY, United States
- 11Staff Medical Center, Department of Medicine, Hamad Medical Corporation, Doha, Qatar
- 12Medical Intensive Care Unit, Hamad Medical Corporation, Doha, Qatar
Introduction: The BNT162b2 mRNA-based vaccine has shown high efficacy in preventing COVID-19 infection but there are limited data on the types and persistence of the humoral and T cell responses to such a vaccine.
Methods: Here, we dissect the vaccine-induced humoral and cellular responses in a cohort of six healthy recipients of two doses of this vaccine.
Results and discussion: Overall, there was heterogeneity in the spike-specific humoral and cellular responses among vaccinated individuals. Interestingly, we demonstrated that anti-spike antibody levels detected by a novel simple automated assay (Jess) were strongly correlated (r=0.863, P<0.0001) with neutralizing activity; thus, providing a potential surrogate for neutralizing cell-based assays. The spike-specific T cell response was measured with a newly modified T-spot assay in which the high-homology peptide-sequences cross-reactive with other coronaviruses were removed. This response was induced in 4/6 participants after the first dose, and all six participants after the second dose, and remained detectable in 4/6 participants five months post-vaccination. We have also shown for the first time, that BNT162b2 vaccine enhanced T cell responses also against known human common viruses. In addition, we demonstrated the efficacy of a rapid ex-vivo T cell expansion protocol for spike-specific T cell expansion to be potentially used for adoptive-cell therapy in severe COVID-19, immunocompromised individuals, and other high-risk groups. There was a 9 to 13.7-fold increase in the number of expanded T cells with a significant increase of anti-spike specific response showing higher frequencies of both activation and cytotoxic markers. Interestingly, effector memory T cells were dominant in all four participants’ CD8+ expanded memory T cells; CD4+ T cells were dominated by effector memory in 2/4 participants and by central memory in the remaining two participants. Moreover, we found that high frequencies of CD4+ terminally differentiated memory T cells were associated with a greater reduction of spike-specific activated CD4+ T cells. Finally, we showed that participants who had a CD4+ central memory T cell dominance expressed a high CD69 activation marker in the CD4+ activated T cells.
Introduction
In order to limit the rapid spread of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), and its consequences across the globe, many efforts have been focused on developing safe and effective prophylactic vaccines (1). The BNT162b2 vaccine (Pfizer-BioNTech) was the first vaccine to be authorized for emergency use (2). BNT162b2 is a lipid nanoparticle formulated nucleoside-modified messenger RNA (mRNA) encoding the SARS-CoV-2 full-length spike (S) glycoprotein in a prefusion stabilized conformation (2). The vaccine was found safe and demonstrated 95% efficacy for protection against COVID-19 in phase II/III clinical trials (3). Observational data showed that BNT162b2 is highly effective in preventing SARS-CoV-2 infection, related hospitalization, and death (4). However, limited data exist about the persistence of the humoral and T cell responses and the duration of the vaccine-induced protection after the two-dose mRNA vaccination.
Neutralizing antibodies are the best indicators of protective immunity, therefore the quantification of SARS-CoV-2 neutralizing antibody levels induced by vaccination or infection constitutes a critical parameter to determine the protection level against the virus and to assess the potential vaccine effectiveness (5–7). The conventional virus/pseudovirus neutralization assays are considered the reference methods to determine the functional neutralizing ability of antibodies (8, 9). However, these methods require the use of specialized facilities, trained personnel, are time-consuming (2-4 days), and relatively expensive (8, 9). Several surrogates of neutralization cell-based assays have been developed and evaluated to overcome these limitations (10–13). Most of these assays use ELISA or similar platforms requiring multiple time-consuming binding and washing steps (10–13), thus preventing high-throughput screening. Therefore, simple, rapid, and accurate serological tests measuring neutralizing activity are urgently needed to assess the duration of humoral protective immunity in vaccine recipients and in recovered COVID-19 patients. Various new techniques have been employed globally for antibody response monitoring following SARS-CoV-2 infection or immunization (14). Jess Simple Western system is a novel fully automated assay, from Protein Simple, that detects human serum/plasma binding antibodies reactive to five different SARS-CoV-2 viral antigens in a large number of samples in only three hours (15). At the beginning of the pandemic, this assay showed high utility in COVID-19 diagnosis with a sensitivity and specificity of 94% and 93%, respectively (16). Moreover, Jess revealed a substantial agreement of 90% between the results obtained using Jess and ELISA for SARS-CoV-2 Immunoglobulin G (IgG) detection, which substantiates its implementation as a first-line serological test for clinical diagnostics and vaccination monitoring (16). Subsequently, Jess was then used in several studies to characterize SARS-CoV-2 specific humoral response in animal and human systems (17–19).
Alongside the antibody response, recent studies have shown that T cell response plays a dominant role in SARS-CoV-2 viral clearance and protection (20–24). Indeed, several reports indicated that COVID-19 patients with undetectable or impaired humoral responses could recover from the disease, highlighting the importance of the T cell response in virus clearance (20–22). In addition, Hurme et al. demonstrated that T cell memory response in COVID-19 vaccinated, and convalescent individuals could be more persistent than antibody response leading to a more durable source of protection (23, 24). Furthermore, recent studies indicated that T cell response and functionality against SARS-CoV-2 were not affected by the mutations or antigenic variation of the emerging variants of concern as the humoral response (25–27). These findings provide direct evidence that a lack or impairment of the T cell response could be associated with an elevated risk of SARS-CoV-2 infection and severe COVID-19 disease outcome. In the same line, numerous studies indicated that severe outcome in COVID-19 patients was associated with lymphopenia, reduction or disability of the T cell cytotoxic potential, and elevated exhaustion markers (28–30). On the other hand, recent studies demonstrated that immunocompromised patients and the elderly have poor immune responses to the BNT162b2 vaccine, indicating that these patients may not be sufficiently protected against SARS-CoV-2 infection (31–33). Therefore, the development of new therapies that support the cellular response to SARS-CoV-2 by preventing the defect of T cell function may have a significant impact on the outcome of the elderly, immunocompromised, and severe COVID-19 patients after infection or vaccination.
We herein report the dynamics and persistence of antibody and T cell responses in a small cohort of healthy adult recipients of two doses of BNT162b2-mRNA vaccine in the state of Qatar. In addition, we explored the interpersonal variation of the humoral and cellular immune response elicited by BNT162b2 immunization among vaccinated healthy individuals. We further evaluated the Jess technology as a surrogate assay for SARS-CoV-2 antibodies neutralizing activity estimation. Finally, we explored the feasibility and efficacy of a rapid ex-vivo T cell expansion protocol for spike-specific T cell expansion to be potentially used for adoptive-cell therapy in severe COVID-19, immunocompromised patients, and elderly persons.
Material and methods
Study population and sample collection
This study was conducted at the Translational Cancer Facility, National Center for Cancer Care and Research, Hamad Medical Corporation (HMC), Qatar. A total of six healthy participants with no history of SARS-CoV-2 infection, eligible to receive two doses of the BNT162b2 mRNA vaccine (Pfizer-BioNTech) three weeks apart, were enrolled. For each participant, peripheral blood samples were obtained on day 0 (prior vaccination), day 20 (pre-boost), day 34 (14 days post-boost), and day 150 (five months after the first vaccination dose). Peripheral Blood Mononuclear Cells (PBMCs) and serum were isolated and used for serological and T cell responses analysis as reported in supplement 1 (See supplementary material). Demographic characteristics of enrolled participants have been shown in supplement 2 (See supplementary material). This study was approved by the Institutional Review Board (IRB) committee of HMC (Project number MRC-01-21-113), and informed consent was obtained from all study participants.
PBMCs and sera isolation
Peripheral blood samples were collected in EDTA and serum separator tubes at the different time points reported above. Serum was separated by centrifugation at 3200 rpm and stored at -80°C. PBMCs were isolated by density gradient centrifugation using Ficoll Paque Premium (GE Healthcare) and SepMate tubes (STEMCELL Technology) according to the manufacturer’s instructions. Isolated PBMCs were then cryopreserved in a cell recovery medium (Fetal Bovine Serum (FBS, Gibco) supplemented with 10% DMSO (Millipore Sigma) and stored in vapor phase liquid nitrogen until used.
ELISA binding assay
Initially, sera samples were screened for the presence of IgG antibodies against the SARS-CoV-2 recombinant S1 subunit (S1) of the spike protein, using a commercial semi-quantitative ELISA kit (Lionex COVID-19 ELISA-human IgG) as per the manufacturer’s instructions. Briefly, sera samples were diluted at 1:50 in a sample diluent and then added to the microtiter plate (coated with SARS-CoV-2 S1 protein) for 60 mins incubation at room temperature. After a washing step, the conjugate (peroxidase-coupled anti-human antibody) and its substrate (TMB) were added to the wells. The optical density (OD) was measured with an ELISA reader (Epoch Biotek) at 450 nm wavelength. Each sample OD was normalized according to the kit calibrator value, and this normalized value determines the test result. Values below 0.8 were considered negative, values between 0.8 and 1.1 were considered borderline, and values above 1.1 indicate a positive anti-SARS-CoV-2 S1 subunit IgG. All samples were run in duplicates and borderline samples were repeated for confirmation.
Detection of SARS-CoV-2 specific antibodies using jess simple western system
The detection and quantification of anti-SARS-CoV-2 IgG antibodies among vaccinated donors’ sera were assessed using the Jess Simple Western system (Protein Simple). This system enables the detection of human IgG antibodies reactive against five viral antigens simultaneously: S1 Receptor Binding Domain protein (S1-RBD), S1 subunit full length (S1), S2 subunit full length (S2), Spike protein (S), and Nucleocapsid Protein (N) recombinant antigens as reported in supplement 3 (see supplementary material). Samples were run following the manufacturer’s protocol for the 12-230-kDa Jess separation module (Protein Simple). Briefly, the SARS-CoV-2 antigens (Protein Simple) were mixed with 0.1X Sample buffer (Protein Simple) and Fluorescent 5X Master mix (Protein Simple) in the presence of fluorescent molecular weight markers (Protein Simple) and denatured at 95°C for 5 mins. Sera were diluted at 1:10 in the sample buffer. Ladder (12-230-kDa PS-ST02EZ, Protein Simple) and SARS-CoV-2 proteins were run in capillaries. The SARS-CoV-2 specific human antibodies present in the serum samples serve as primary antibodies that were then detected with anti-goat HRP-conjugated anti-human IgG antibody (R&D Systems). The chemiluminescent revelation was established with peroxide/luminol-S (Protein Simple). The digital image of the capillary chemiluminescence was captured with Compass Simple Western software (version 4.1.0, Protein Simple) that automatically calculated the area of the signal (chemiluminescence intensity). Results are represented as the chemiluminescence intensity of each antigen separately.
Generation of SARS-CoV-2 pseudotyped vesicular stomatitis virus and neutralization assay
For the determination of neutralizing antibodies to SARS-CoV-2, we utilized a recombinant ΔG-Vesicular stomatitis virus (VSV) system to generate SARS-CoV-2 pseudovirus as previously described by Whitt (34). Briefly, HEK293T cells were grown in DMEM medium (Gibco) supplemented with 10% Fetal Bovine Serum (Gibco) and 1% Pen/Strep (Gibco) to reach 80-90% of confluence on the day of the experiment. The following day, cell culture media was replaced with Opti-MEM (Gibco) and incubated for 20 mins before transfecting cells with SARS-CoV-2 Spike-TM plasmid (provided by the Viral Pathogenesis Laboratory, Vaccine Research Center, National Institute of Health). After 4 hours, transfection media was replaced with DMEM (Gibco) supplemented with 5% FBS (Gibco), and cells were incubated at 37°C with 5% CO2. After 24 hours, cells were examined for the presence of syncytia due to the expression of the envelope protein. Subsequently, transfected cells were infected with pseudotyped ΔG-luciferase (G*ΔG) (Kerafast, Ref. no. EH1025-PM) at a multiplicity of ~3–5. When most of the cells showed a cytopathic effect (24-30 hours), SARS-CoV-2 VSV pseudovirus was harvested by collecting the supernatant. Supernatants were clarified by centrifugation at 300×g for 10 mins before aliquoting and storing at −80°C. For the titration of pseudotyped viruses, HEK293T cells expressing angiotensin-converting enzyme 2 (ACE2) (BEI) were used. Cells were prepared at 1×106 cell/ml in complete DMEM (Gibco) and added to serially diluted pseudovirus (50 µl of diluted virus added to 50 µl of cells in suspension) in a 96-well cell culture plate and incubated for 2 hours. 100 µl of complete DMEM (Gibco) was then added to the cells and incubated for 48 hours. After incubation, cells were lysed using 30 µl of 1X cell lysis buffer (Promega), and 50 µl of luciferase reagent (Promega) was added. The titer of the pseudovirus was determined by measuring luminescence using a plate reader (Tecan Infinite). To assess the neutralization of SARS-CoV-2 pseudotyped VSV in sera samples, heat-inactivated serum samples (50 to 200-fold) were serially diluted in 60 µl of DMEM media and then incubated with 100 µl pseudovirus (titer 1-2×106 RLU/100 µl) for 30 mins at room temperature. The final volume (160 µl) was then distributed into 3 wells (triplicates) of a 96-cell culture plate. HEK293T-ACE2 cells were then added at 1×106 cells/ml and incubated for 48 hours before reading out luminescence using a plate reader (Infinite 200 PRO). A positive response was defined as a neutralizing activity of 20% or more.
Interferon-γ Enzyme-Linked ImmunoSpot assay
The spike-specific T cell responses to the BNT162b2 vaccine were assessed using the T-spot Discovery SARS-CoV-2 kit (Oxford Immunotec), a modified enzyme-linked immunospot technology. This kit is designed to measure interferon-γ responses to overlapping peptide pools covering peptide sequences of five different SARS-CoV-2 antigens, without HLA restriction. The test specificity to SARS-CoV-2 has been enhanced by removing high homology peptide sequences that are potentially cross-reactive with other coronaviruses. The T-spot discovery SARS-CoV-2 kit was used according to the manufacturer’s protocols. Briefly, 250 000 PBMCs suspended in AIM-V medium (Gibco) were plated into each well of the T-spot plate in duplicates, stimulated with 3 different antigens: S1 spike subunit peptides, peptides coding for sequences with high homology to other coronaviruses, positive control (phytohemagglutinin), and negative control (AIM-V medium) then incubated for 18 hours (37°C, 5% CO2). The interferon-γ secreting T cells were detected using an automated ELISpot reader (Autoimmun Diagnositka GMBH). Results are presented as the mean of the number of spots forming cells (SFC) per 250 000 cells for each panel, subtracting the background (negative control) count. A positive response was defined as an SFC of 10 or more.
Ex-vivo spike-specific T cells expansion
The spike-specific T cells were expanded from vaccinated donors’ PBMCs (collected five months post-vaccination) using a modified protocol for expansion of multivirus-specific T cells targeting cytomegalovirus (CMV), Epstein-Barr virus (EBV), BK virus (BKV), human herpes virus (HHV)-6, respiratory syncytial virus (RSV), adenovirus (Adv) and influenza previously described by Gerdemann et al. (35). Briefly, fresh PBMCs were pulsed with the spike peptide pools at 1 µg of antigen/15 x 106 PBMCs for 30 mins at 37°C. The spike peptide pools (JPT Peptide Technologies) contain a pool of 315 overlapping peptides encompassing the full spike protein. After incubation, cells were resuspended in a virus-specific T cells (VST) medium consisting of 45% Advanced RPMI 1640 (Gibco) supplemented with 45% Click’s medium (Irvine Scientific), 2 mM GlutaMAX (Gibco), 10% FBS (Gibco), 10 ng/ml interleukin 7 (IL-7, Peprotech), and 400 U/ml IL-4 (Peprotech) and transferred to a G-Rex 10 device (Wilson Wolf Manufacturing Corporation). Cells were counted on day six and fresh culture media with cytokines was added. Cells were harvested and evaluated for antigen specificity and functionality on day 11.
Flow cytometry
Expanded spike-specific T cells and PBMCs collected five months post-vaccination were stimulated with the S1 peptide pools (1µg/ml, Oxford Immunotec) for 18 hours. Stimulated PBMCs and T cells were stained with fluorophore-conjugated monoclonal antibodies against CD3 (BD Biosciences), CD4 (BD Biosciences), CD8 (BD Biosciences), CD45RA (BD, Biosciences), CD69 (BD Biosciences), CD107 (BD Biosciences), CD134 (Thermo Fisher), CD137 (Thermo Fisher), and CD197 (BD Biosciences) for phenotypical characterization. All samples were acquired using a Fortessa flow cytometer (BD Biosciences) and the data was analyzed using FlowJo V10 software (BD Biosciences).
Statistical analyses
All statistical analyses and graphs were performed using GraphPad Prism Software (version 9.2.0). The characterization of the humoral and T cell responses dynamics over time was assessed using One-way ANOVA multiple comparison test. The T cell response to S1 antigen before and after expansion was evaluated using the student t test. Correlations between Jess, neutralization, and ELISA immunoassays were analyzed by Pearson correlation and linear regression models. The scatter point represents serum samples (n=24) collected from BNT162b2 vaccinated healthy donors at the baseline, 20-, 34-, and 150-days post-vaccination. The coefficient of correlation (r) represents the strength of the linear relationship between the different immunoassays. The coefficient of determination R squared (R2) represents the percentage of variance in the given data set. The P-value tests whether the regression equation is significant. P-value was considered statistically significant when P ≤ 0.5.
Results
Heterogeneity of the spike-specific antibody response among BNT162b2 vaccinated individuals
The anti-spike (anti-S) binding and neutralizing antibody responses induced by the BNT162b2 vaccine over time were characterized. In this, serum samples were collected from vaccinated participants at four different time points as reported in supplement 1 (See supplementary material). The anti-S and anti-S1 IgG levels were assessed using Jess and ELISA, respectively (Figures 1A, B, 2A, B and supplement 4). The anti-S neutralizing activity was measured using the neutralization assay (Figures 1C and 2C). Overall, our data showed an interpersonal heterogeneity in the vaccine-elicited humoral response among vaccinated individuals. This interpersonal variation was observed at three stages: the induction detected on day 20, the peak response reached on day 34, and the response decline detected 150 days post-vaccination (Figures 1A–C, 2A–C). Jess results showed that the anti-S IgG antibody response (presented by chemiluminescence intensity (CI)) was induced on day 20 in all six participants with different levels, ranging from 502947 to 6719958 CI (Figures 1A and 2A). This response was boosted on day 34 (after the second dose) in all six participants with varying levels, ranging from 5495488 to 12954728 CI (Figures 1A and 2A). However, 150 days post-vaccination a decline in the anti-S IgG levels was observed in 5/6 participants with different magnitudes ranging from 3344080 to 7009985 CI (Figures 1A and 2A). In contrary, VAC-HD1 showed an increase in this response 150 days post-vaccination (Figures 1A and 2A).
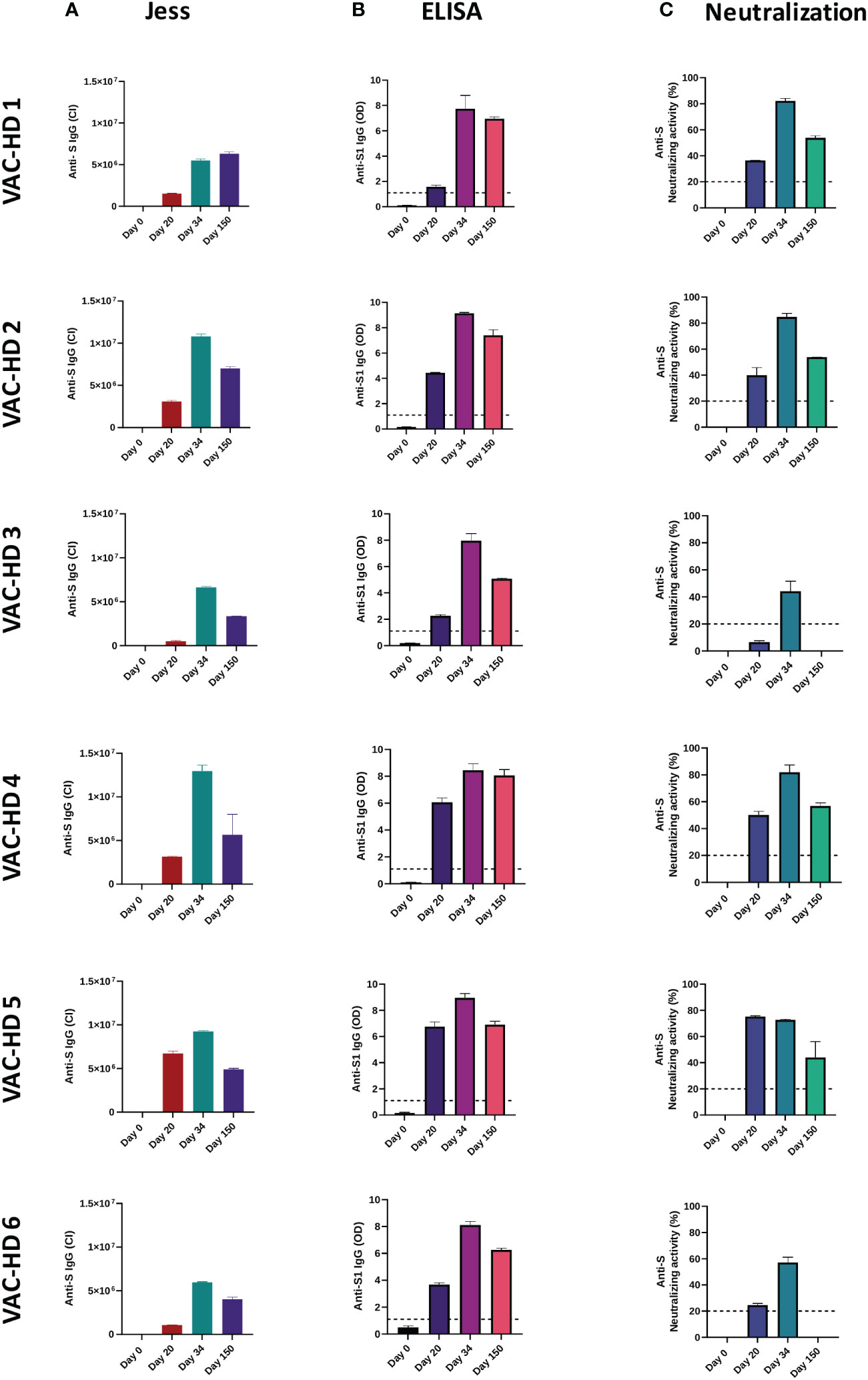
Figure 1 Interpersonal variation of anti-spike binding and neutralizing antibodies responses over time among BNT162b2 vaccinated participants. Serum samples were collected from six BNT162b2 vaccinated participants at the baseline, 20-, 34-, and 150-days post-vaccination. The anti-S and anti-S1 IgG levels were assessed using Jess and ELISA, respectively. The anti-S neutralizing activity was measured using the neutralization assay. (A) Anti-S IgG response in six BNT162b2 vaccinated participant over time (Jess). (B) Anti-S1 IgG response in six BNT162b2 vaccinated participant over time (ELISA). (C) Anti-S neutralizing activity in each BNT162b2 vaccinated participant over time (Neutralization assay). The lines indicate the cut-off value of a positive antibody response.
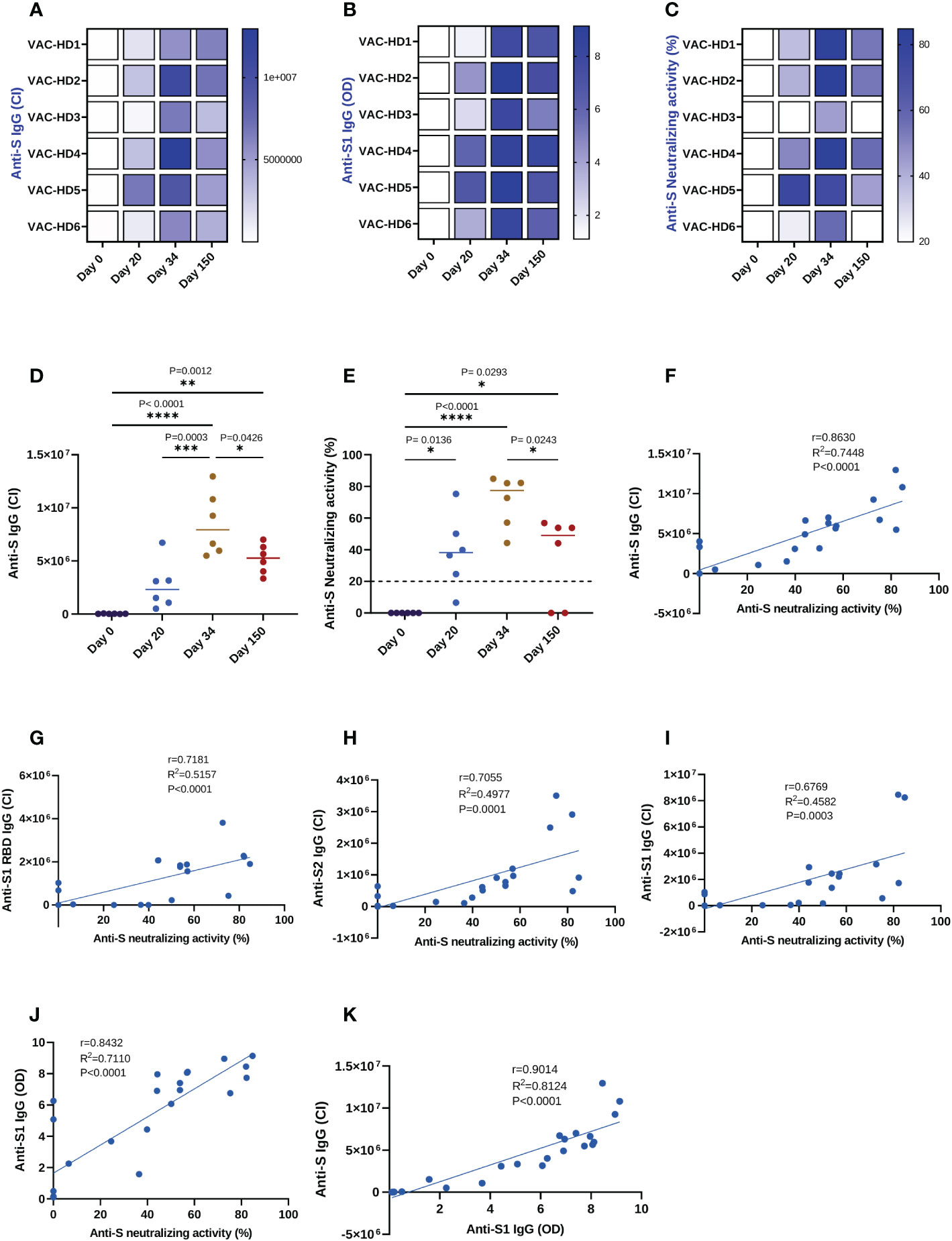
Figure 2 Correlation between the anti-spike binding and neutralizing antibodies responses induced by BNT162b2 vaccination and detected by three different immunoassays. Serum samples were collected from six BNT162b2 vaccinated participants at the baseline, 20-, 34-, and 150-days post-vaccination. The anti-S and anti-S1 IgG levels were assessed using Jess and ELISA, respectively. The anti-S neutralizing activity was measured using the neutralization assay. (A–C) Heat-map of anti-S IgG, anti-S1 IgG, and anti-S neutralizing activity responses in six BNT162b2 vaccinated participants over time. (D) Dynamics of the anti-S IgG levels in six BNT162b2 vaccinated participants over time (Jess). (E) Dynamics of the anti-S neutralizing activity in six BNT162b2 vaccinated participants (Neutralization assay). Each symbol represents an individual participant with a line indicating the median of each time point. One-way ANOVA test was used, P value was considered statistically significant when *P ≤ 0.05. All samples were run in duplicates. (F–I) Correlation between anti-S, S1 RBD, S2, and S1 IgG levels detected by JESS and neutralizing activity, respectively. (J) Correlation between anti-S1 IgG levels detected by semi-quantitative ELISA and neutralizing activity. (K) Correlation between anti-S1 IgG levels detected by ELISA and anti-S IgG levels detected by JESS. All correlations were analyzed by Pearson statistical test and linear regression models. The scatter point represents serum samples (n=24) collected from six BNT162b2 vaccinated participants at the baseline, 20-, 34-, and 150-days post-vaccination, and the blue error band represents the 95% confidence interval. The coefficient of correlation (r) represents the strength of the linear relationship between the different immunoassays. The coefficient of determination (R2) represents the percentage of variance in the given data set. The P-value tests whether the regression equation is significant. P value was considered statistically significant when *P ≤ 0.05. The stars present the level of significance. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
In addition, ELISA results showed that the first vaccination induced the anti-S1 IgG antibody response on day 20 in all six participants with varying degrees of optical density (OD) ranging from 1.58 to 6.75 (Figures 1B and 2B). The second dose increased this response in all six participants with an OD ranging from 7.73 to 9.13 (Figures 1B and 2B). However, five months post-vaccination a decline in the anti-S1 IgG levels was observed in all six participants with an OD ranging from 5.07 to 8.06 (Figures 1B and 2B). We have found also that females (VAC-HD2, 4, 5, and 6) had a stronger anti-S1 IgG antibody response on days 20 and 34 compared to their counterparts in males (VAC-HD1 and 3) (Figures 1B and 2B).
Furthermore, Neutralization results showed that anti-S neutralizing antibodies response was induced on day 20 in all six participants with different levels of neutralizing activity ranging from 6.53 to 75.23% (Figures 1C and 2C). This response was boosted after the second dose in 5/6 participants with a neutralizing activity ranging from 44.27 to 84.76% (Figures 1C and 2C). However, five months post-vaccination a decline in the anti-S neutralizing activity was observed in all six participants with varying magnitudes ranging from 0 to 56.84% (Figures 1C and 2C). Interestingly, we demonstrated that individuals who had low Anti-S neutralizing activity (<25%) on day 20 (VAC-HD3 and 6) tended to have also low response after the second dose and lost this response five months post-vaccination (Figures 1C and 2C). However, individuals who had high anti-S neutralizing activity (>36%) on day 20 (VAC-HD1, 2, 4, and 5) tended to have also high response after the second dose and were able to maintain this response five months post-vaccination (Figures 1C and 2C).
Dynamics of S-specific binding and neutralizing antibody responses following BNT162b2 vaccination
Our data showed that anti-S binding and neutralizing antibodies responses dynamics followed the same trend (Figures 2D, E). Indeed, the anti-S IgG binding antibodies (BAbs) response was induced 20 days after the first dose (26526 CI on day 0 versus 2678053 CI on day 20), significantly boosted with the second dose (2678053 CI on day 20 versus 8519960 CI on day 34, ***P=0.0003), then significantly declined 150 days post-vaccination (8519960 CI on day 34 versus 5205907 CI on day 150, *P=0.0426) (Figure 2D). Identically, we found that the anti-S neutralizing antibodies (NAbs) response was significantly induced 20 days after the priming dose (0% on day 0 versus 38.8% on day 20, *P=0.0136) and was further increased with the booster dose (38.8% on day 20 versus 70.51% on day 34) (Figure 2E). However, the anti-S neutralizing activity had significantly decreased five months post-vaccination as compared to their peak levels at two weeks after the second dose (70.51% on day 34 versus 34.77% on day 150, *P=0.0243) (Figure 2E). Furthermore, we demonstrated that all six participants maintained a detectable anti-S BAbs response five months post-vaccination (Figures 2A and 2D), whereas only four of them maintained the NAbs response (except VAC-HD3 and VAC-HD6) (Figures 2C, E).
Evaluation of Simple automated immunoassay Jess as an alternative to neutralization cell-based assay for SARS-CoV-2 neutralizing activity estimation
We investigated whether the anti-S IgG BAbs levels measured by Jess can substitute the neutralization cell-based assay for the estimation of neutralizing activity in vaccinated individuals. For this purpose, the anti-S BAbs levels and neutralizing activity of anti-S NAbs were measured in serum samples (n=24) collected at the baseline, 20-, 34-, and 150-days post-vaccination using Jess, ELISA, and neutralization assays in order to evaluate the degree of correlation between these 3 immunoassays. We first performed correlation and linear regression analysis on the four different BAbs (anti S, anti-S1 RBD, anti-S2, and anti-S1 IgG) levels detected by Jess and neutralizing activity measured by neutralization assay (Figures 2F–I). Among the four BAbs, anti-S IgG showed a strong positive, statistically significant correlation (r=0.8630, R2 = 0.7448, P <0.0001) with the neutralizing activity (Figure 2F), whereas a moderate correlation (r ranging between 0.6769 and 0.7181) was observed for the remaining IgGs (anti-S1 RBD, anti-S1, and anti-S2) (Figures 2G–I). We then evaluated whether Jess is a better surrogate test for neutralizing activity prediction as compared to another common commercial semi-quantitative ELISA test. Linear regression analysis showed that the linear fit between anti-S IgG detected by Jess and neutralizing activity (R2 = 0.7448, P<0.0001) was substantially higher as compared to the one obtained between anti-S1 IgG levels detected by ELISA and neutralizing activity (R2 = 0.7110, P<0.0001) (Figure 2J). Similarly, a distinguished positive correlation (r=0.9014, R2 = 0.8124, P<0.0001) between anti-S1 IgG and anti-S IgG levels detected by ELISA and Jess respectively was observed (Figure 2K). Overall, these results indicate that the detection of anti-S IgG levels by Jess could be a better surrogate for neutralizing activity estimation compared to ELISA. Moreover, Jess could potentially be a promising alternative that is quicker, cheaper, and easier than the conventional cell-based assays for neutralizing activity estimation.
BNT162b2-induced T cells response to SARS-CoV-2 and cross-reactivity with other viral antigens
The cellular immune responses induced by BNT162b2 vaccination were characterized by the measurement of interferon-γ responses to S1 peptide pools (the immunodominant subunit of the S protein) using a relatively novel T-spot assay. This assay is highly specific since the SARS-CoV-2 epitopes having a high degree of homology with other endemic human coronaviruses (huCoVs) were removed from the SARS-CoV-2 antigens panels enabling a specific SARS-CoV-2 response determination. Similar to the antibody response, we observed an immense variation in the T cell responses among the vaccinated participants (Figures 3A, B). T-spot results showed that S1-specific T cell response was significantly induced on day 20 in 5/6 participants with variable levels ranging from 2 to 85 SFC (Figures 3A, B). An increase in this response was observed in all six participants on day 34 with an S1-specific T cell response ranging from 14 to 150 SFC (Figures 3A, B). However, 150 days post-vaccination a decline in the S1-specific response was observed in all six participants with different magnitudes ranging from 6 to 47 SFC (Figures 3A, B). We have found that females (VAC-HD 2, 4, 5, and 6) had a stronger S1-specific T cell response on days 20 and 34 compared to their counterparts in males (VAC-HD1 and 3) (Figures 3A, B).
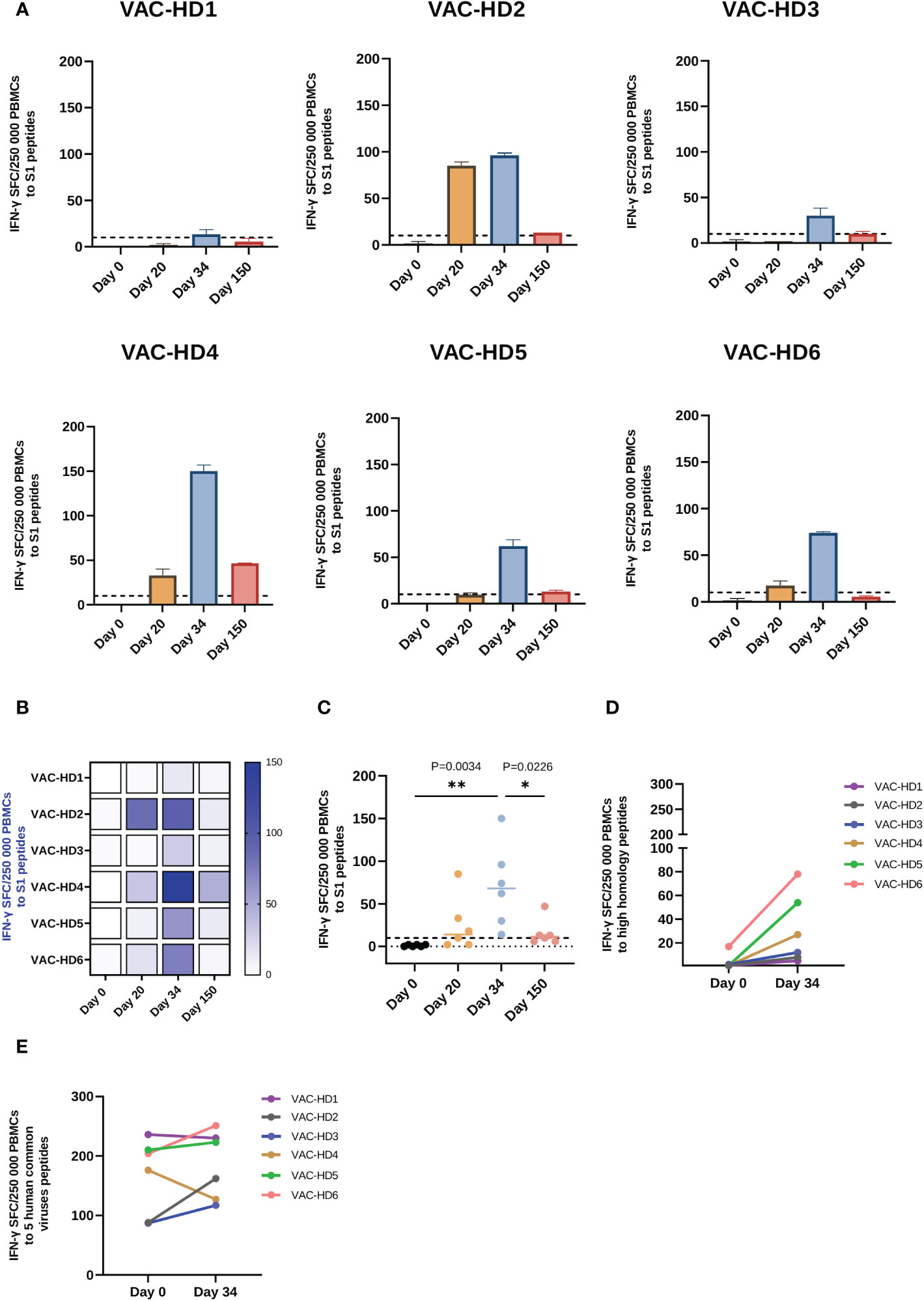
Figure 3 Spike-specific and spike cross-reactive T cell responses in BNT162b2 vaccinated participants. (A) T cell response to S1 peptide pools in six BNT162b2 vaccinated participant at the baseline, 20-, 34-, and 150-days post-vaccination. (B) Heat-map of S1- specific T cell responses in six BNT162b2 vaccinated participants over time (C) Dynamics of S1- specific T cell responses in six BNT162b2 vaccinated participants over time. Each symbol represents an individual participant with a line indicating the median of each time point. (D) T cell response to high homology peptide pools in BNT162b2 vaccinated participants at baseline (Day 0) and on day 34 post-vaccination (after the second dose) (E) T cell response to five human common viruses peptides (CMV, EBV, AdV 3 and 5 and BKV) in BNT162b2 vaccinated participants at baseline (Day 0) and on day 34 post-vaccination (after the second dose). Results are presented as the mean of the number of spots forming cells (SFCs) per 250 000 PBMCs subtracting the background (negative control) count. A positive response was defined as an SFCs of 10 or more. One-way ANOVA test was used, P value was considered statistically significant when *P ≤ 0.05. All samples were run in duplicates. The stars present the level of significance. *P < 0.05; **P < 0.01.
On the other hand, we have demonstrated that the priming dose was able to induce a detectable T cell response (≥ 10 SFCs) against the S1 antigen in only 4/6 participants, whereas after the booster dose, all six participants presented a detectable T cell response ranging from 14 to 150 SFC (Figure 3C). Interestingly, we have shown that T cell response against the S1 antigen was induced 20 days after the priming dose (1 SFC on day 0 versus 25 SFCs on day 20) and significantly increased two weeks after the second dose (1 SFC on day 0 versus 71 SFC on day 34, **P=0.0034) in all the participants (Figure 3C). However, we have observed that T cell response to the S1 antigen was significantly decreased 150 days post-vaccination (71 SFC on day 34 versus 15.83 SFC on day 150, * P=0.0226) (Figure 3C). This decline was observed in all six participants, whereas this T cell response remained detectable 150 days after vaccination in 4/6 participants (Figures 3B, C).
Given the fact that SARS-CoV-2 displays a high level of homology to other human coronaviruses (huCoVs), we evaluated whether the BNT162b2 vaccination could induce a cellular immune response against other huCoVs strains than SARS-CoV-2. Therefore, we compared the T cell response, using high homology peptide pools, on day 0 and day 34 post-vaccination. Interestingly, all six participants showed an increase in T cell response against cross-reactive sequences between SARS-CoV-2 and other huCoVs after the second dose (Figure 3D) with 4/6 participants demonstrated a significant increase in such response (Supplement 5). This result suggests that the booster dose activated and enhanced the T cell responses against other huCoVs strains (priming at day 20 did not enhance this response, data not shown). We further evaluated T cell responses against five different human common viruses peptide pools: CMV, EBV, BKV, Adv 3, and 5 on day 0 and day 34 post-vaccination. As expected, all six participants showed T cell responses to all of these viruses at baseline (day 0 before vaccination), ranging between 87 and 236 SFC due to previous exposure to these viruses (Figure 3E). Importantly, we have demonstrated that this T cell response was increased in 4/6 participants on day 34 post-vaccination (Figure 3E). These results may suggest the presence of cross-reactive epitopes between SARS-CoV-2 and these five viruses.
Rapid ex-vivo expansion of spike-specific T cells from BNT162b2 vaccinated donors
We next investigated whether we could expand the S-specific T cells from BNT162b2 vaccinated donors, five months post-vaccination, using a rapid ex-vivo expansion protocol described in supplement 6 (see supplementary material). Briefly, PBMCs collected from four vaccinated donors were stimulated with the S peptide pools and then cultured in the presence of IL-4 and IL-7 for 11 days in the G-Rex 10 culture device. We first examined the S-specific T cell response in these four participants prior to expansion. T-spot results showed a positive S1-specific T cell response ranging from 10 to 46 SFCs in VAC-HD2, 3, and 4 (Figure 4A). However, VAC-HD 1 lacked detectable S1-specific T cells (SFCs=6, below the positive cutoff threshold) (Figure 4A). Interestingly, the T cells from VAC-HD2, 3, and 4 were expanded up to 9-fold (136x106 cells), 10.4-fold (156x106 cells), and 13.7-fold (206x106 cells), respectively eleven days post-stimulation (Figure 4B). The T cells from VAC-HD1 however failed to adequately expand likely due to the low frequency of the S1-specific T cells before expansion (1.8-fold; 26.5x106 cells) (Figure 4B). These results indicate that the frequency of pre-existing S-specific T cells may play a major role in the expansion of such T cells.
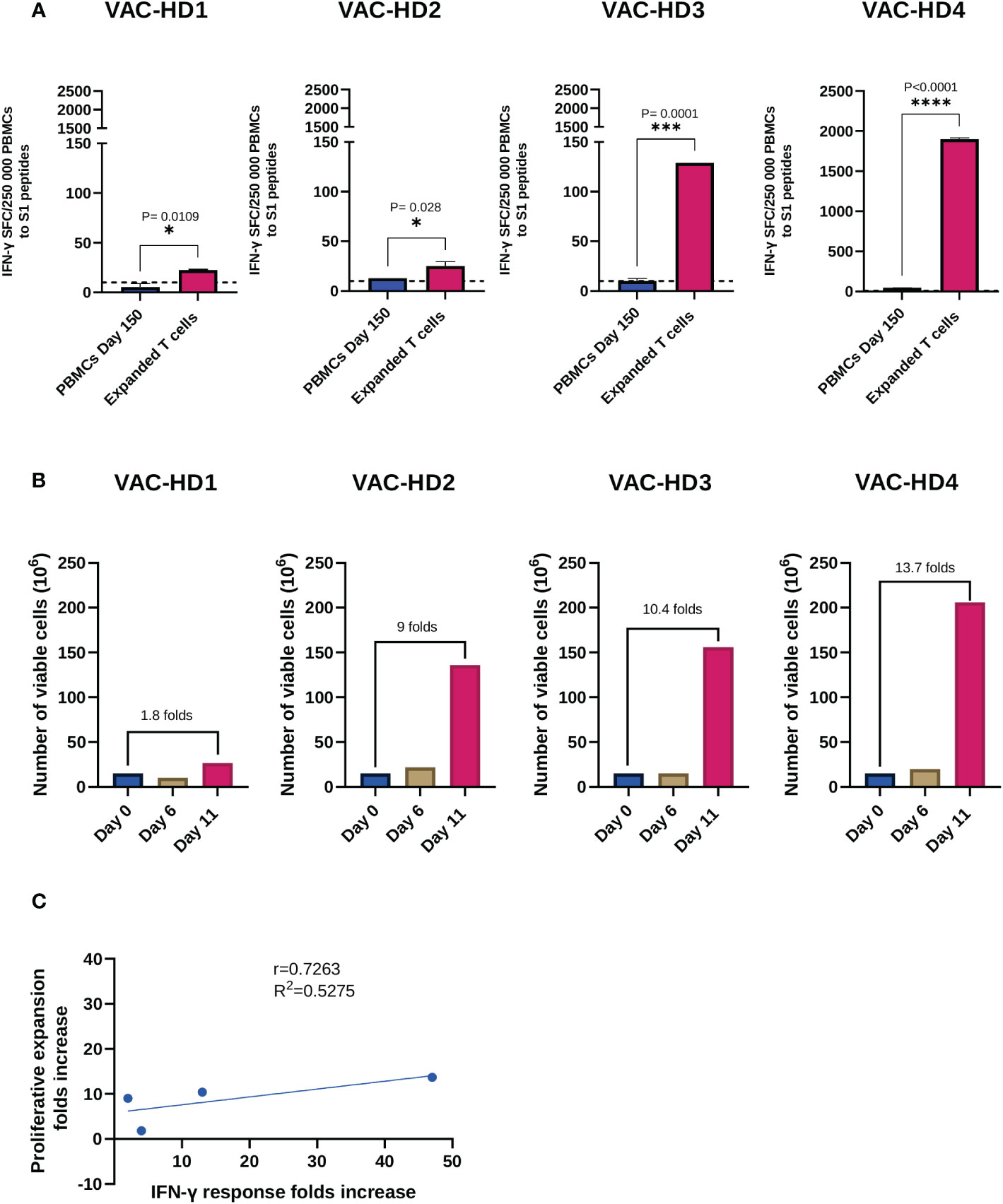
Figure 4 Expansion and functional characterization of expanded spike-specific T cells. (A) IFN-γ secretion by pre-expansion PBMCs collected 150 days post-vaccination and spike-specific expanded T cells following S1 peptide pools overnight stimulation. Results are presented as the mean of the number of spots forming cells (SFCs) per 250 000 PBMCs subtracting the background (negative control) count. A positive response was defined as an SFCs of 10 or more. Student t test was used, P value was considered statistically significant when *P ≤ 0.05. All samples were run in duplicates. (B) Viable cell counts, and fold expansion were assessed on days 0, 6, and 11 of expansion using trypan blue exclusion dye. (C) Correlation between IFN-γ response folds increase and the proliferative expansion folds increase between pre-expansion PBMCs and spike-specific expanded T cells in four BNT162b2 vaccinated participants. The dotted lines represent confidence intervals at 95%. The stars present the level of significance. *P < 0.05; **P < 0.01; ****P < 0.0001.
We next evaluated the specificity of these expanded T cells by measuring IFN-γ secreting T cells in response to S1 stimulation using the T-spot assay, corresponding PBMCs collected 150 days post-vaccination (Pre-expansion PBMCs) were used as a control. Overall, expanded T cells demonstrated an S1-specific IFN-γ production which was significantly higher than the one detected in pre-expanded T cells for all participants (Figure 4A). Our data show that this T cell response varied between the four participants and positively correlated with the proliferative expansion fold (r=0.7263, R2 = 0.5275) (Figure 4C). Significant increase in the number of IFN-γ SFC, after T cell expansion, was observed in the four participants (Figure 4A): VAC-HD1 (4-fold increase from 6-23 SFCs; *P=0.0109), VAC-HD2 (2-fold increase from 13-25 SFCs; *P=0.028), VAC-HD3 (13-fold increase from 10-129 SFCs; ***P=0.0001) and VAC-HD4 (41-fold increase from 47-1900 SFCs; ****P<0.0001) (Figure 4A).
Phenotypic characterization of S-specific expanded T cells
To further analyze the phenotype of the expanded S-specific T cells, the distribution of activated and cytotoxic T cell subsets was assessed using flow cytometry following stimulation with the S1 antigen. We also compared the changes in the distributions of these T cell subsets before and after expansion. The gating strategy and raw data are presented in supplements 7 and 8 (see supplementary material). Overall, the frequencies of both pre-expanded S-specific activated CD4+ (CD4+OX40+CD69+) and CD8+ (CD8+CD137+CD69+) T cells were higher in VAC-HD3 and 4 compared to VAC-HD1 and 2 (Figure 5A). In this, the frequency of S-specific activated CD4+ T cells was 0.93% and 0.73% in VAC-HD1 and 2 versus 5.15% and 5.39% in VAC-HD3 and 4 (Figure 5A). Whereas the frequency of S-specific activated CD8+ T cells was 0.51% and 0.15% for VAC-HD1 and 2 versus 1.39% and 0.7% for VAC-HD3 and 4 (Figure 5A). These results indicate that the frequency of pre-existing S-specific T cells prior to expansion was higher in VAC-HD3 and 4 than in VAC-HD1 and 2 which can explain the fact that the expansion was more efficient for these two cases compared to others (Figure 4B). Interestingly, an increase in the frequencies of expanded CD4+ and/or CD8+ S-activated T cells was recorded in all participants when compared to the pre-expanded population (Figure 5A). We also showed that induction of the T cell activation markers against the S antigen was higher in the CD4+ T cells for VAC-HD1 and VAC-HD3 (4.04% and 8.32% respectively) and in the CD8+ T cells for VAC-HD 2 and VAC-HD4 (1.49% and 13.6% respectively) (Figure 5A).
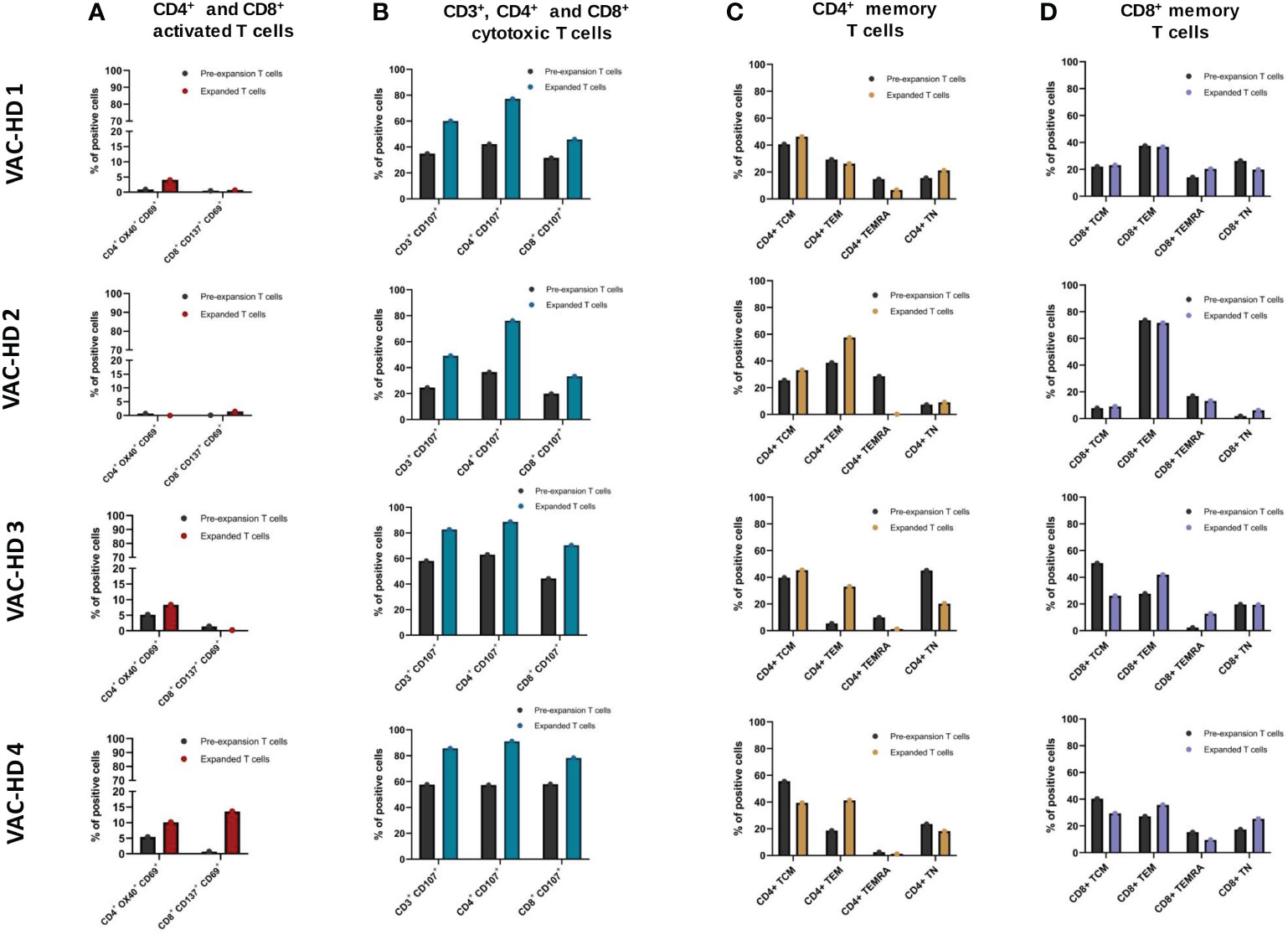
Figure 5 Phenotyping of the spike-specific expanded T cells. (A) Frequency of CD4+ OX40+ CD69+ and CD8+ CD137+ CD69+ activated T cells within pre-expansion and expanded T cells following S1 stimulation in four BNT162b2 vaccinated participants. (B) Frequency of CD3+ CD107+, CD4+ CD107+, and CD8+ CD107+ cytotoxic T cells within pre-expansion and expanded T cells following S1 stimulation in four BNT162b2 vaccinated participants. (C) Frequency of CD4+ naïve (CCR7+CD45RA+), central memory (CCR7+CD45RA−), effector memory (CCR7−CD45RA−), and terminally differentiated memory (CCR7−CD45RA+) within pre-expansion and expanded T cells following S1 stimulation in four BNT162b2 vaccinated participants. (D) Frequency of CD8+ naïve (CCR7+CD45RA+), central memory (CCR7+CD45RA−), effector memory (CCR7−CD45RA−), and terminally differentiated memory (CCR7−CD45RA+) within pre-expansion and expanded T cells following S1 stimulation in four BNT162b2 vaccinated participants.
We next examined the overall changes in CD3+, CD4+, and CD8+ cytotoxic T cell frequencies within the S-specific expanded T cells compared to the pre-expanded T cells. Expanded T cells presented higher frequencies of CD3+CD107+, CD4+CD107+, and CD8+CD107+ cytotoxic T cells compared to the pre-expanded population in all four participants (Figure 5B). In this, the frequencies of CD3+CD107+ increased from 24.6-58.1% (Pre) to 49.1-85.7% (Post), CD4+CD107+ from 36.5-63% (Pre) to 76.1-91.1% (Post), and CD8+CD107+ from 19.9-58% (Pre) to 33.3-78.3% (Post) in all the four participants (Figure 5B). Interestingly, the cytotoxic T cell marker CD107+ was higher in the CD4+ expanded T cell population (76.1% to 91.1%) when compared to the CD8+ counterparts (31.6% to 78.3%) in all four participants (Figure 5B).
In the next step, we examined the distribution of memory T cell subsets in the expanded T cell population. Based on phenotypic markers, T cell subsets can be classified into four subsets: naïve (CCR7+CD45RA+), central memory (CCR7+CD45RA−), effector memory (CCR7−CD45RA−), and terminally differentiated memory (CCR7−CD45RA+). The S-specific expanded CD4+ and CD8+ T cells were found to have different distributions of naïve (TN), effector (TEM), central memory (TCM), and terminally differentiated memory (TEMRA) phenotypes in all four participants (Figures 5C, D). For CD4+ expanded T cells, the TCM subset was dominant in VAC-HD1 and 3 whereas the TEM subset was aberrant in VAC-HD2 and 4 (Figure 5C). However, for CD8+ expanded T cells, the TEM subset was dominated in all four participants with a frequency ranging from 35.7 to 71.6% (Figure 5D). Interestingly, we demonstrated that VAC-HD1 and 2, who showed the highest frequencies of pre-expanded CD4+ TEMRA (Figure 5C), had lower S-specific pre-expanded CD4+ activated T cells (CD4+OX40+CD69+) frequencies in the compared to VAC-HD3 and VAC-HD4 (Figure 5A). Moreover, the frequency of the CD4+ TEMRA subset was greatly reduced after T cell expansion in all 4 participants (Figure 5C). Importantly, we showed that participants who had a CD4+ TCM (CCR7+CD45RA−) dominance (VAC-HD1, 3 and 4) (Figure 5C) expressed high CD69 activation marker in the CD4+ activated T cells among the expanded T cells (Figure 5A).
Discussion
Most of the investigations designed to assess the efficacy, immunogenicity, and protective immunity induced by the BNT162b2 vaccine were based on large cohort studies. However, despite their advantages, these studies are providing only a general observation of the vaccine-induced immune response without dissecting such immune responses that is laborious and expansive to investigate in large cohorts. In the present work, we dissected BNT162b2 vaccine-induced humoral and cellular responses in a small cohort study that includes recipients receiving two doses of this vaccine. Our results showed a broad range of variation in both humoral and cellular responses. This interpersonal variation was observed at three stages: the induction was detected on day 20, the peak response reached on day 34, and the response declined 150 days post-vaccination. The factors involved in the interindividual variation in the human immune response to BNT162b2 vaccine are still largely unknown. Previous reports suggest that the interindividual diversity of the human immune responses to common pathogens and vaccines is determined by intrinsic (age and gender), extrinsic (environment), and genetic factors such as genes coding for human leukocyte antigen alleles, major histocompatibility complex molecules, Toll-like receptors and cytokines (36, 37). Recently, Ward et al. identified age and gender as important determinants of humoral response to BNT162b2 vaccine (38). They showed that antibody positivity was higher in females and the youngest age group (38). Although our study was carried out in a small cohort, we have also shown that females had stronger S1-specific T cells and antibody responses compared to male participants.
Consistent with previous reports (39–41), our results also showed that the dynamics of anti-S binding and neutralizing antibodies responses followed the same trend, where they were induced 20 days post-priming, significantly increased after boosting, then declined five months post-vaccination. The peak level of antibody response detected on day 34 after boosting was associated with an increase of 4-fold in total anti-S binding IgG levels but only a 2-fold increase in the neutralizing activity compared to day 20. Our result is in agreement with a recent study where they showed that the immune response induced at the time of peak response following BNT162b2 vaccination was characterized by a high ratio of non-neutralizing antibodies (42) that may confer protection against SARS-CoV-2 infection (43, 44). Interestingly, we were able to show that all six participants maintained the anti-S IgG binding antibody response five months post-vaccination, whereas only four of them maintained the neutralizing activity response.
Recent studies have demonstrated that neutralizing antibody titers are the most critical parameters for vaccine efficacy evaluation and prediction of SARS-CoV-2 protective immunity (5–7). Many efforts have been made to develop high throughput assays for neutralizing antibody detection that can surrogate the classical neutralizing cell-based assays that require specific laboratory facilities, skilled personnel, and a long (2-4 days) execution time (8, 9). We investigated in this study whether a novel simple automated assay (Jess) could surrogate the neutralization assay for estimation of neutralizing activity. Jess is a robust anti-SARS-CoV-2 binding antibodies surveillance test, which is simple, fully automated, rapid, and can be easily used in laboratories without the need for BSL3 facilities. Using this assay, we have shown that only anti-S IgG (antibodies against the whole spike antigen) had a strong positive significant correlation (r=0.863, R2 = 0.7448, P <0.0001) with the anti-S neutralizing activity compared to other SARS-CoV-2 spike domain-specific IgG (S1-RBD, S1, and S2). Our results are in agreement with two very recent studies that used a chemiluminescent immunoassay for the quantitative determination of SARS-CoV-2 IgG binding antibodies (45, 46). The first study reported a linear correlation between anti-S IgG and surrogate neutralizing antibody levels for wild-type SARS-CoV-2 and variants of concern (VOCs) in BNT162b2 vaccinated and recovered health care workers (45). the second study also showed a strong correlation (R2 = 0.72) between the anti-S antibody IgG titers detected by a chemiluminescent immunoassay and surrogate neutralizing activity (46). Taken together, our results indicate that Jess provides a robust anti-S neutralizing activity surveillance/prediction test.
The characterization of the T cell response in BNT162b2 vaccinated individuals indicated that a single dose of vaccine was not able to induce spike-specific T cell response in 30% of the participants indicating the necessity of a booster dose for efficient and durable protection (47). Similar to this, our results showed that 33% of vaccinated participants lacked a detectable spike-specific T cell response after one dose of this vaccine. However, the second dose was able to induce the spike-specific T cell response in 100% of the participants with a median increase of 3-fold in the T cell response frequencies compared to that observed after priming. Importantly, we demonstrated that the booster dose was able to also activate and enhance T cell responses against other huCoVs suggesting the presence of cross-reactive epitopes between SARS-CoV-2 and other huCoVs. Our findings are supported by a recent study showing the presence of common epitopes between SARS-CoV-2 and huCoVs (48). We have also demonstrated that pre-existing memory T cells, naturally induced during past infections of our participants with the five human common viruses (CMV, EBV, Adv 3 and 5, BKV), were in vivo expanded following the BNT162b2 vaccination. This suggests the presence of cross-reactive epitopes between SARS-CoV-2 and these five viruses derived-antigens. Indeed, the sequence similarity between EBV and SARS-CoV-2 has been well established (49) however, no study related to sequence or epitope similarity for the other viruses have been investigated. Furthermore, our data showed the persistence of spike-specific T cell response five months post-vaccination in 67% of the participants.
BNT162b2 mRNA vaccine clinical trial showed 95% effectiveness in preventing SARS-CoV-2 infection (3). This trial predominantly excluded patients with immunocompromising conditions (50), which present 2% of the global population (51). Indeed, recent studies have demonstrated that a two doses vaccine regimen does not produce sufficient strong immune responses and protection in immunocompromised patients and elderly people (31–33). Considering the ineffectiveness of current SARS-CoV-2 antibody-based immunotherapy due to the development of novel mutations and the immune escape of the VOCs (52), other therapeutic options are warranted. Adoptive SARS-CoV-2 specific T cell therapy represents an attractive therapeutic option in which viral immune escape is likely to be avoided as the recognized T cell epitopes are well conserved among the emerging SARS-CoV-2 variants (25). Moreover, adoptive cellular therapy with ex-vivo expanded specific T cells against other viruses (CMV, EBV, Adv, HHV6, and BKV) has been demonstrated to have efficacy in combating severe viral diseases in patients with immunodeficiency (53). In addition, studies in animal models have also shown that adoptive therapy with CD4+ and/or CD8+ T cells can efficiently control respiratory infections including SARS-CoV-1, MERS, and influenza viruses (54–56). Here, we adapted a rapid ex-vivo T cell expansion protocol for spike-specific T cell expansion to be potentially used for adoptive-cell therapy in severe COVID-19, immunocompromised individuals, and other high-risk groups. In this protocol, we expanded spike-specific T cells from vaccinated donors isolated five months post-vaccination to avoid spike-specific T cells exhaustion during the ex-vivo expansion and to mimic the in vivo stimulation and boosting effect of the booster dose which is usually given between five to six months post-priming. Moreover, the long-term persistence of memory T cells following vaccination or viral infection has been well reported (57, 58). The response mediated by such memory T cells, upon re-exposure to the antigen, is more rapid and effective than the primary response (59). In this rapid protocol (11 days) we were able to expand T cells up to 11-folds in 3/4 participants. A minimum S-specific T cell number ≥10 SFCs was required to support T cell expansion. Moreover, participants who had a higher frequency of pre-expanded S-specific activated CD4+ (CD4+OX40+CD69+) and CD8+ (CD8+CD137+CD69+) T cells tended to have a higher expansion rate of S-specific T cells. The specificity of expanded T cells was measured with a newly modified T-spot assay in which the high-homology peptide sequences cross-reactive with other coronaviruses were removed. Expanded T cells demonstrated a significant increase of S1 spike-specific IFN-γ producing cells compared to the pre-expanded T cells for all participants. Interestingly, these S-specific expanded T cells had higher frequencies of both activation and cytotoxic markers important for viral clearance after re-exposure (60). Adoptively transferring such expanded T cells may be used as an attractive approach to restore and/or boost the cytotoxic T cell response in severe COVID-19, immunocompromised patients and elderly people with impaired cytotoxic T cell response to SARS-CoV-2 (28, 31–33, 61). Interestingly, the cytotoxic T cell marker CD107+ was higher in the CD4+ expanded T cell population when compared to the CD8+ counterparts in all four participants. This result indicates that CD4+ cytotoxic T cells play a major role in the S-specific cell-mediated cytotoxic response following BNT162b2 immunization. In line with this, it has been reported that S-specific T cell response, elicited by SARS-CoV-2 infection, was dominated by the CD4+ subset in COVID-19 patients (62, 63).
It has been well established that central memory (TCM) and effector memory (TEM) T cell subsets have distinct functions and migratory properties (64, 65). Therefore, we examined the distribution of these memory T cell subsets in our spike-specific expanded T cells. There was a variation in the distribution of the CD4+ and CD8+ memory T cell subsets among all four participants. For CD4+ memory T cells, the TCM subset was dominant in 2/4 participants whereas the TEM subset was aberrant in the 2 remaining participants. CD4+ TCM resides within the lymphoid organs and are known for their rapid proliferation and production of IL-2 and IL-10 upon restimulation (66). However, CD4+ TEM reside in peripheral tissues and exhibit immediate cytokine secretion of IFN-γ and IL-4 upon restimulation (66). For CD8+ T cells, the TEM subset was dominated in all four participants. CD8+ TEM also resides in peripheral tissue and provides immediate protection following antigenic stimulation by the secretion of perforin (66). Another subset of memory T cells known as terminally differentiated memory (TEMRA) has been demonstrated to exhibit numerous characteristics of immuno-senescence such as defects in proliferation and effector functions (67). Both CD4+ and CD8+ TEMRA are known to accumulate in the aging process and pathological conditions such as arthritis rheumatoid and persistent viral infection (68–75). Therefore, we investigated this phenomenon in our expanded spike-specific T cells. Interestingly, we found that participants who showed the highest frequencies of CD4+ TEMRA cells prior to expansion tended to have lower S-specific activated CD4+ T cells and a lower fold increase of the S1-specific IFN-γ response of expanded spike-specific T cells compared to the pre-expanded population. These results suggest that this subset of CD4+ TEMRA cells may contribute to the impairment in the S-specific CD4+ T cells development and T cell expansion inefficacy. We have also demonstrated that the frequencies of CD4+ TEMRA were greatly reduced after S-specific T cell expansion. On the other hand, immuno-senescence associated with defects in immune proliferation and effector functions has been shown to correlate with an increased susceptibility to viral infection and a decreased vaccine immunogenicity (76). Indeed, several studies suggested that influenza vaccine inefficacy in aged individuals can be mainly related the immune system immunosenescence (77, 78). Another recent study has also demonstrated that mRNA COVID-19 vaccine immunogenicity was negatively correlated with the accumulation of T cell expressing signs of immunosenescence (79).
CD69 is widely used as an activation marker for T cells and natural killer cells, however, the precise role of this marker in these immune cells is not yet well elucidated (80). Recent evidence suggests that the expression level of CD69 controls the migration and retention of CD4+ memory T cells in their specific niches (81). Similarly, another study suggests that upregulation of CD69, after yellow fever vaccination, can promote T cell migration and retention in the lymph nodes, the home for TCM (80). Therefore, we hypothesized that an increase of the CD69 marker after BNT162b2 vaccination may control the homing and migration of S-specific CD4+ TCM. Interestingly, we showed that participants who had a CD4+ TCM dominance also expressed a high CD69 activation marker in the CD4+ activated T cells among the expanded T cells. Further studies are required to clarify the role of CD69 in vaccine-induced CD4+ memory T cell migration and homing.
In conclusion, this is the first pilot study that highlights the interpersonal heterogeneity of the humoral and cellular responses to the BNT162b2 vaccine. We have demonstrated for the first time a strong correlation between Jess and neutralization cell-based assay. we also validated the feasibility and efficacy of a rapid ex-vivo spike-specific T cell expansion protocol from BNT162b2 vaccinated individuals that can be used in the future to establish a biobank for adoptive transfer of allogeneic HLA-matched spike-specific T cells as therapeutic and/or prevention options in severe COVID-19, immunocompromised patients, and elderly people. The limitation of our study is the use of a small number of participants and further studies with larger sample sizes are needed to confirm our results.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional Review Board (IRB) committee of HMC (Project number MRC-01-21-113). The patients/participants provided their written informed consent to participate in this study.
Author contributions
SD, SM, MM, and VPI: Conceptualized the study, planned, and supervised the experiments. SM: performed experimental work, analyzed data, and generated figures. SM and SD: designed and wrote the manuscript. NT, MM, VPI, MKS, FHA, QF, SH and FS helped in the experimental work. SD, MM, VPI, AR, HMY, FBA, ASO, AAH, MAM, MUA, AWA, SU, LJA, LA, MJ, MG, NA, HMHS, AMJ, MN, AZ, FA and SRA: Critical revision and editing of the scientific contents of the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by Academic Health System, Medical Research Center, Hamad Medical Corporation, Doha, Qatar, grant number MRC-01-21-113, and the Article Processing Charges was funded by Academic Health System, Medical Research Center, Hamad Medical Corporation, Doha, Qatar. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Acknowledgments
We thank all study participants that consented to participate in this project and gratefully acknowledge the Communicable Disease Center staff in Qatar for their support. We acknowledge the Medical Research Center at Hamad Medical Corporation for supporting this work under the approved project MRC-01-21-113.
Conflict of interest
SD, SM, MM, VPI, NT, FA, AR, SH, QF, AWA, FS, LA, MJ, MG, NA, MUA, SU, AMJ, MN, ASO, AAH, MAM, FBA, AZ, SRA, and HMHS were employed by Hamad Medical Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1061255/full#supplementary-material
References
1. Fernandes Q, Inchakalody VP, Merhi M, Mestiri S, Taib N, Moustafa Abo El-Ella D, et al. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann Med (2022) 54(1):524–40. doi: 10.1080/07853890.2022.2031274
2. Lamb YN. BNT162b2 mRNA COVID-19 vaccine: First approval. Drugs (2021) 81(4):495–501. doi: 10.1007/s40265-021-01480-7
3. Skowronski DM, De Serres G. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. New Engl J Med (2021) 384(16):1576–7. doi: 10.1056/NEJMc2036242
4. Dagan N, Barda N, Kepten E, Miron O, Perchik S, Katz MA, et al. BNT162b2 mRNA covid-19 vaccine in a nationwide mass vaccination setting. New Engl J Med (2021) 384(15):1412–23. doi: 10.1056/NEJMoa2101765
5. Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med (2021) 27(7):1205–11. doi: 10.1038/s41591-021-01377-8
6. Cromer D, Steain M, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. Lancet Microbe (2022) 3(1):e52–61. doi: 10.1016/S2666-5247(21)00267-6
7. Chen X, Wang W, Chen X, Wu Q, Sun R, Ge S, et al. Prediction of long-term kinetics of vaccine-elicited neutralizing antibody and time-varying vaccine-specific efficacy against the SARS-CoV-2 delta variant by clinical endpoint. BMC Med (2022) 20(1):36. doi: 10.1186/s12916-022-02249-9
8. Tani H, Kimura M, Tan L, Yoshida Y, Ozawa T, Kishi H, et al. Evaluation of SARS-CoV-2 neutralizing antibodies using a vesicular stomatitis virus possessing SARS-CoV-2 spike protein. Virol J (2021) 18(1):16. doi: 10.1186/s12985-021-01490-7
9. Cantoni D, Mayora-Neto M, Temperton N. The role of pseudotype neutralization assays in understanding SARS CoV-2. Oxford Open Immunol (2021) 2(1):iqab005. doi: 10.1093/oxfimm/iqab005
10. Wisnewski AV, Liu J, Lucas C, Klein J, Iwasaki A, Cantley L, et al. Development and utilization of a surrogate SARS-CoV-2 viral neutralization assay to assess mRNA vaccine responses. PLos One (2022) 17(1):e0262657. doi: 10.1371/journal.pone.0262657
11. Abe KT, Li Z, Samson R, Samavarchi-Tehrani P, Valcourt EJ, Wood H, et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight (2020) 5(19):e142362. doi: 10.1172/jci.insight.142362
12. Byrnes JR, Zhou XX, Lui I, Elledge SK, Glasgow JE, Lim SA, et al. Competitive SARS-CoV-2 serology reveals most antibodies targeting the spike receptor-binding domain compete for ACE2 binding. mSphere (2020) 5(5):e00802-20. doi: 10.1128/mSphere.00802-20
13. Tan CW, Chia WN, Qin X, Liu P, Chen MI, Tiu C, et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol (2020) 38(9):1073–8. doi: 10.1038/s41587-020-0631-z
14. Filchakova O, Dossym D, Ilyas A, Kuanysheva T, Abdizhamil A, Bukasov R. Review of COVID-19 testing and diagnostic methods. Talanta (2022) 244:123409. doi: 10.1016/j.talanta.2022.123409
15. simple B-P. Rapid characterization of human immune response to SARS-CoV-2 antigens using simple Western (2022). Available at: https://www.proteinsimple.com/sars-cov-2-multi-antigen-serology-module.html.
16. Edouard S, Jaafar R, Orain N, Parola P, Colson P, La Scola B, et al. Automated Western immunoblotting detection of anti-SARS-CoV-2 serum antibodies. Eur J Clin Microbiol Infect Dis (2021) 40(6):1309–17. doi: 10.1007/s10096-021-04203-8
17. Davoust B, Guérin P, Orain N, Fligny C, Flirden F, Fenollar F, et al. Evidence of antibodies against SARS-CoV-2 in wild mustelids from Brittany (France). Transboundary emerging Dis (2022) 69(5):e3400-e3407. doi: 10.1101/2022.01.20.477038
18. Castillo-Olivares J, Wells DA, Ferrari M, Chan ACY, Smith P, Nadesalingam A, et al. Analysis of serological biomarkers of SARS-CoV-2 infection in convalescent samples from severe, moderate and mild COVID-19 cases. Front Immunol (2021) 12:748291. doi: 10.3389/fimmu.2021.748291
19. Laidoudi Y, Sereme Y, Medkour H, Watier-Grillot S, Scandola P, Ginesta J, et al. SARS-CoV-2 antibodies seroprevalence in dogs from France using ELISA and an automated western blotting assay. One Health (Amsterdam Netherlands) (2021) 13:100293. doi: 10.1016/j.onehlt.2021.100293
20. Hughes R, Whitley L, Fitovski K, Schneble HM, Muros E, Sauter A, et al. COVID-19 in ocrelizumab-treated people with multiple sclerosis. Multiple sclerosis related Disord (2021) 49:102725. doi: 10.1016/j.msard.2020.102725
21. Asplund Högelin K, Ruffin N, Pin E, Månberg A, Hober S, Gafvelin G, et al. Development of humoral and cellular immunological memory against SARS-CoV-2 despite b cell depleting treatment in multiple sclerosis. iScience (2021) 24(9):103078. doi: 10.1016/j.isci.2021.103078
22. Montero-Escribano P, Matías-Guiu J, Gómez-Iglesias P, Porta-Etessam J, Pytel V, Matias-Guiu JA. Anti-CD20 and COVID-19 in multiple sclerosis and related disorders: A case series of 60 patients from Madrid, Spain. Multiple sclerosis related Disord (2020) 42:102185. doi: 10.1016/j.msard.2020.102185
23. Hurme A, Jalkanen P, Heroum J, Liedes O, Vara S, Melin M, et al. Long-lasting T cell responses in BNT162b2 COVID-19 mRNA vaccinees and COVID-19 convalescent patients. Front Immunol (2022) 13:869990. doi: 10.3389/fimmu.2022.869990
24. Agrati C, Castilletti C, Goletti D, Sacchi A, Bordoni V, Mariotti D, et al. Persistent spike-specific T cell immunity despite antibody reduction after 3 months from SARS-CoV-2 BNT162b2-mRNA vaccine. Sci Rep (2022) 12(1):6687. doi: 10.1038/s41598-022-07741-z
25. Jaiswal V, Lee HJ. Conservation and evolution of antigenic determinants of SARS-CoV-2: An insight for immune escape and vaccine design. Front Immunol (2022) 13:832106. doi: 10.3389/fimmu.2022.832106
26. Choi SJ, Kim DU, Noh JY, Kim S, Park SH, Jeong HW, et al. T Cell epitopes in SARS-CoV-2 proteins are substantially conserved in the omicron variant. Cell Mol Immunol (2022) 19(3):447–8. doi: 10.1038/s41423-022-00838-5
27. Neidleman J, Luo X, McGregor M, Xie G, Murray V, Greene WC, et al. mRNA vaccine-induced T cells respond identically to SARS-CoV-2 variants of concern but differ in longevity and homing properties depending on prior infection status. eLife (2021) 10:e72619. doi: 10.7554/eLife.72619
28. Vigón L, Fuertes D, García-Pérez J, Torres M, Rodríguez-Mora S, Mateos E, et al. Impaired cytotoxic response in PBMCs from patients with COVID-19 admitted to the ICU: Biomarkers to predict disease severity. Front Immunol (2021) 12:665329. doi: 10.3389/fimmu.2021.665329
29. Kusnadi A, Ramírez-Suástegui C, Fajardo V, Chee SJ, Meckiff BJ, Simon H, et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8(+) T cells. Sci Immunol (2021) 6(55):eabe4782. doi: 10.1126/sciimmunol.abe4782
30. Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol (2020) 11:827. doi: 10.3389/fimmu.2020.00827
31. Demaret J, Corroyer-Simovic B, Alidjinou EK, Goffard A, Trauet J, Miczek S, et al. Impaired functional T-cell response to SARS-CoV-2 after two doses of BNT162b2 mRNA vaccine in older people. Front Immunol (2021) 12:778679. doi: 10.3389/fimmu.2021.778679
32. Enßle JC, Campe J, Schwenger A, Wiercinska E, Hellstern H, Dürrwald R, et al. Severe impairment of T-cell responses to BNT162b2 immunization in patients with multiple myeloma. Blood (2022) 139(1):137–42. doi: 10.1182/blood.2021013429
33. Azzolini E, Pozzi C, Germagnoli L, Oresta B, Carriglio N, Calleri M, et al. mRNA COVID-19 vaccine booster fosters b- and T-cell responses in immunocompromised patients. Life Sci alliance (2022) 5(6):137–42. doi: 10.26508/lsa.202201381
34. Whitt MA. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J virological Methods (2010) 169(2):365–74. doi: 10.1016/j.jviromet.2010.08.006
35. Gerdemann U, Keirnan JM, Katari UL, Yanagisawa R, Christin AS, Huye LE, et al. Rapidly generated multivirus-specific cytotoxic T lymphocytes for the prophylaxis and treatment of viral infections. Mol Ther (2012) 20(8):1622–32. doi: 10.1038/mt.2012.130
36. Scepanovic P, Alanio C, Hammer C, Hodel F, Bergstedt J, Patin E, et al. Human genetic variants and age are the strongest predictors of humoral immune responses to common pathogens and vaccines. Genome Med (2018) 10(1):59. doi: 10.1186/s13073-018-0568-8
37. Liston A, Carr EJ, Linterman MA. Shaping variation in the human immune system. Trends Immunol (2016) 37(10):637–46. doi: 10.1016/j.it.2016.08.002
38. Ward H, Whitaker M, Flower B, Tang SN, Atchison C, Darzi A, et al. Population antibody responses following COVID-19 vaccination in 212,102 individuals. Nat Commun (2022) 13(1):907. doi: 10.1038/s41467-022-28527-x
39. Gandolfo C, Anichini G, Mugnaini M, Bocchia M, Terrosi C, Sicuranza A, et al. Overview of anti-SARS-CoV-2 immune response six months after BNT162b2 mRNA vaccine. Vaccines (2022) 10(2):171. doi: 10.3390/vaccines10020171
40. Naaber P, Tserel L, Kangro K, Sepp E, Jürjenson V, Adamson A, et al. Dynamics of antibody response to BNT162b2 vaccine after six months: a longitudinal prospective study. Lancet regional Health Europe (2021) 10:100208. doi: 10.1016/j.lanepe.2021.100208
41. Matusali G, Sberna G, Meschi S, Gramigna G, Colavita F, Lapa D, et al. Differential dynamics of SARS-CoV-2 binding and functional antibodies upon BNT162b2 vaccine: A 6-month follow-up. Viruses (2022) 14(2):312. doi: 10.3390/v14020312
42. Amanat F, Thapa M, Lei T, Ahmed SMS, Adelsberg DC, Carreño JM, et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell (2021) 184(15):3936–48.e10. doi: 10.1016/j.cell.2021.06.005
43. Bahnan W, Wrighton S, Sundwall M, Bläckberg A, Larsson O, Höglund U, et al. Spike-dependent opsonization indicates both dose-dependent inhibition of phagocytosis and that non-neutralizing antibodies can confer protection to SARS-CoV-2. Front Immunol (2021) 12:808932. doi: 10.3389/fimmu.2021.808932
44. Beaudoin-Bussières G, Chen Y, Ullah I, Prévost J, Tolbert WD, Symmes K, et al. A fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection. Cell Rep (2022) 38(7):110368. doi: 10.1016/j.celrep.2022.110368
45. Decru B, Van Elslande J, Steels S, Van Pottelbergh G, Godderis L, Van Holm B, et al. IgG anti-spike antibodies and surrogate neutralizing antibody levels decline faster 3 to 10 months after BNT162b2 vaccination than after SARS-CoV-2 infection in healthcare workers. Front Immunol (2022) 13:909910. doi: 10.3389/fimmu.2022.909910
46. Grandjean L, Saso A, Torres Ortiz A, Lam T, Hatcher J, Thistlethwayte R, et al. Long-term persistence of spike protein antibody and predictive modeling of antibody dynamics after infection with severe acute respiratory syndrome coronavirus 2. Clin Infect Dis an Off Publ Infect Dis Soc America (2022) 74(7):1220–9. doi: 10.1093/cid/ciab607
47. Reynolds CJ, Pade C, Gibbons JM, Butler DK, Otter AD, Menacho K, et al. Prior SARS-CoV-2 infection rescues b and T cell responses to variants after first vaccine dose. Sci (New York NY) (2021) 372(6549):1418–23. doi: 10.1126/science.abh1282
48. Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Sci (New York NY) (2020) 370(6512):89–94. doi: 10.1126/science.abd3871
49. Geraldes CF. SARS-CoV-2 presents amino acid sequences, analogous to those of the Epstein-Barr virus - human gammaherpesvirus 4 (2020). São Paulo, Brazil: Uninove.
50. Bergman P, Blennow O, Hansson L, Mielke S, Nowak P, Chen P, et al. Safety and efficacy of the mRNA BNT162b2 vaccine against SARS-CoV-2 in five groups of immunocompromised patients and healthy controls in a prospective open-label clinical trial. EBioMedicine (2021) 74:103705. doi: 10.1016/j.ebiom.2021.103705
51. AstraZeneca. Immunocompromised populations and the risk of viral variants (2022). Available at: https://www.astrazeneca.com/what-science-can-do/topics/covid-19/viral-variants-and-immunocompromised.html.
52. Tian D, Sun Y, Xu H, Ye Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 omicron variant. J Med virology (2022) 94(6):2376–83. doi: 10.1002/jmv.27643
53. Keller MD, Bollard CM. Virus-specific T-cell therapies for patients with primary immune deficiency. Blood (2020) 135(9):620–8. doi: 10.1182/blood.2019000924
54. Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol (Baltimore Md 1950) (2006) 177(5):2888–98. doi: 10.4049/jimmunol.177.5.2888
55. Zhao J, Zhao J, Perlman S. T Cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J virology (2010) 84(18):9318–25. doi: 10.1128/JVI.01049-10
56. Zhao J, Zhao J, Mangalam AK, Channappanavar R, Fett C, Meyerholz DK, et al. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity (2016) 44(6):1379–91. doi: 10.1016/j.immuni.2016.05.006
57. Wang RX, Boland GJ, van Hattum J, de Gast GC. Long-term persistence of T cell memory to HBsAg after hepatitis b vaccination. World J gastroenterology (2004) 10(2):260–3. doi: 10.3748/wjg.v10.i2.260
58. Halwani R, Doroudchi M, Yassine-Diab B, Janbazian L, Shi Y, Said EA, et al. Generation and maintenance of human memory cells during viral infection. Springer Semin immunopathology (2006) 28(3):197–208. doi: 10.1007/s00281-006-0027-2
59. Walker JM, Slifka MK. Longevity of T-cell memory following acute viral infection. Adv Exp Med Biol (2010) 684:96–107. doi: 10.1007/978-1-4419-6451-9_8
60. Rosendahl Huber S, van Beek J, de Jonge J, Luytjes W, van Baarle D. T Cell responses to viral infections - opportunities for peptide vaccination. Front Immunol (2014) 5:171. doi: 10.3389/fimmu.2014.00171
61. Westmeier J, Paniskaki K, Karaköse Z, Werner T, Sutter K, Dolff S, et al. Impaired cytotoxic CD8(+) T cell response in elderly COVID-19 patients. mBio (2020) 11(5):e02243-20. doi: 10.1128/mBio.02243-20
62. Juno JA, Tan HX, Lee WS, Reynaldi A, Kelly HG, Wragg K, et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat Med (2020) 26(9):1428–34. doi: 10.1038/s41591-020-0995-0
63. Stephenson E, Reynolds G, Botting RA, Calero-Nieto FJ, Morgan MD, Tuong ZK, et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med (2021) 27(5):904–16. doi: 10.1038/s41591-021-01329-2
64. Pepper M, Jenkins MK. Origins of CD4(+) effector and central memory T cells. Nat Immunol (2011) 12(6):467–71. doi: 10.1038/ni.2038
65. Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol (2004) 22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702
66. Esser MT, Marchese RD, Kierstead LS, Tussey LG, Wang F, Chirmule N, et al. Memory T cells and vaccines. Vaccine (2003) 21(5-6):419–30. doi: 10.1016/S0264-410X(02)00407-3
67. Zhang J, He T, Xue L, Guo H. Senescent T cells: a potential biomarker and target for cancer therapy. EBioMedicine (2021) 68:103409. doi: 10.1016/j.ebiom.2021.103409
68. Effros RB, Cai Z, Linton PJ. CD8 T cells and aging. Crit Rev Immunol (2003) 23(1-2):45–64. doi: 10.1615/CritRevImmunol.v23.i12.30
69. Kovaiou RD, Grubeck-Loebenstein B. Age-associated changes within CD4+ T cells. Immunol letters (2006) 107(1):8–14. doi: 10.1016/j.imlet.2006.07.006
70. Faint JM, Annels NE, Curnow SJ, Shields P, Pilling D, Hislop AD, et al. Memory T cells constitute a subset of the human CD8+CD45RA+ pool with distinct phenotypic and migratory characteristics. J Immunol (Baltimore Md 1950) (2001) 167(1):212–20. doi: 10.4049/jimmunol.167.1.212
71. Goronzy JJ, Weyand CM. Rheumatoid arthritis. Immunol Rev (2005) 204:55–73. doi: 10.1111/j.0105-2896.2005.00245.x
72. Ponchel F, Verburg RJ, Bingham SJ, Brown AK, Moore J, Protheroe A, et al. Interleukin-7 deficiency in rheumatoid arthritis: consequences for therapy-induced lymphopenia. Arthritis Res Ther (2005) 7(1):R80–92. doi: 10.1186/ar1452
73. van Lier RA, ten Berge IJ, Gamadia LE. Human CD8(+) T-cell differentiation in response to viruses. Nat Rev Immunol (2003) 3(12):931–9. doi: 10.1038/nri1254
74. Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu Rev Immunol (2007) 25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553
75. Moss P, Khan N. CD8(+) T-cell immunity to cytomegalovirus. Hum Immunol (2004) 65(5):456–64. doi: 10.1016/j.humimm.2004.02.014
76. Crooke SN, Ovsyannikova IG, Poland GA, Kennedy RB. Immunosenescence and human vaccine immune responses. Immun Ageing I A (2019) 16:25. doi: 10.1186/s12979-019-0164-9
77. Dugan HL, Henry C, Wilson PC. Aging and influenza vaccine-induced immunity. Cell Immunol (2020) 348:103998. doi: 10.1016/j.cellimm.2019.103998
78. Fulop T, Larbi A, Pawelec G, Cohen AA, Provost G, Khalil A, et al. Immunosenescence and altered vaccine efficiency in older subjects: A myth difficult to change. Vaccines (2022) 10(4):607. doi: 10.3390/vaccines10040607
79. Palacios-Pedrero MÁ, Jansen JM, Blume C, Stanislawski N, Jonczyk R, Molle A, et al. Signs of immunosenescence correlate with poor outcome of mRNA COVID-19 vaccination in older adults. Nat Aging (2022) 2(10):896–905. doi: 10.1038/s43587-022-00292-y
80. Bovay A, Speiser DE, Fuertes Marraco SA. Early drop of circulating T cells negatively correlates with the protective immune response to yellow fever vaccination. Hum Vaccines immunotherapeutics (2020) 16(12):3103–10. doi: 10.1080/21645515.2020.1750249
Keywords: SARS-CoV-2, COVID-19 vaccine, spike-specific immune responses, surrogate neutralization, spike-specific T cells expansion
Citation: Mestiri S, Merhi M, Inchakalody VP, Taib N, Smatti MK, Ahmad F, Raza A, Ali FH, Hydrose S, Fernandes Q, Ansari AW, Sahir F, Al-Zaidan L, Jalis M, Ghoul M, Allahverdi N, Al Homsi MU, Uddin S, Jeremijenko AM, Nimir M, Abu-Raddad LJ, Abid FB, Zaqout A, Alfheid SR, Saqr HMH, Omrani AS, Hssain AA, Al Maslamani M, Yassine HM and Dermime S (2023) Persistence of spike-specific immune responses in BNT162b2-vaccinated donors and generation of rapid ex-vivo T cells expansion protocol for adoptive immunotherapy: A pilot study. Front. Immunol. 14:1061255. doi: 10.3389/fimmu.2023.1061255
Received: 04 October 2022; Accepted: 11 January 2023;
Published: 02 February 2023.
Edited by:
Nargis Khan, University of Calgary, CanadaReviewed by:
Javier Carbone, Gregorio Marañón Hospital, SpainEyad Elkord, University of Nizwa, Oman
Elizabeth Sarmiento, Hospital Central de la Defensa Gómez Ulla, Spain
Copyright © 2023 Mestiri, Merhi, Inchakalody, Taib, Smatti, Ahmad, Raza, Ali, Hydrose, Fernandes, Ansari, Sahir, Al-Zaidan, Jalis, Ghoul, Allahverdi, Al Homsi, Uddin, Jeremijenko, Nimir, Abu-Raddad, Abid, Zaqout, Alfheid, Saqr, Omrani, Hssain, Al Maslamani, Yassine and Dermime. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Said Dermime, c2Rlcm1pbWVAaGFtYWQucWE=; Maysaloun Merhi, bW1lcmhpQGhhbWFkLnFh
 Sarra Mestiri
Sarra Mestiri Maysaloun Merhi
Maysaloun Merhi Varghese P. Inchakalody
Varghese P. Inchakalody Nassiba Taib
Nassiba Taib Maria K. Smatti
Maria K. Smatti Fareed Ahmad4,5
Fareed Ahmad4,5 Afsheen Raza
Afsheen Raza Fatma H. Ali
Fatma H. Ali Queenie Fernandes
Queenie Fernandes Abdul W. Ansari
Abdul W. Ansari Niloofar Allahverdi
Niloofar Allahverdi Shahab Uddin
Shahab Uddin Laith J. Abu-Raddad
Laith J. Abu-Raddad Fatma Ben Abid
Fatma Ben Abid Ahmed Zaqout
Ahmed Zaqout Ali S. Omrani
Ali S. Omrani Hadi M. Yassine
Hadi M. Yassine Said Dermime
Said Dermime