
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 29 March 2023
Sec. Cancer Immunity and Immunotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1051431
This article is part of the Research Topic Interplay Between Oncomicrobes, the Microbiota and the Immune System: Impact on Responses to Cancer Immunotherapy View all 7 articles
 Jacob H. Elnaggar1,2
Jacob H. Elnaggar1,2 Victoria O. Huynh1
Victoria O. Huynh1 Daniel Lin2
Daniel Lin2 R. Tyler Hillman3,4,5
R. Tyler Hillman3,4,5 Chike O. Abana2
Chike O. Abana2 Molly B. El Alam2
Molly B. El Alam2 Katarina C. Tomasic2
Katarina C. Tomasic2 Tatiana V. Karpinets4
Tatiana V. Karpinets4 Ramez Kouzy2
Ramez Kouzy2 Jae L. Phan2
Jae L. Phan2 Jennifer Wargo6
Jennifer Wargo6 Emma B. Holliday7
Emma B. Holliday7 Prajnan Das7
Prajnan Das7 Melissa P. Mezzari8
Melissa P. Mezzari8 Nadim J. Ajami4
Nadim J. Ajami4 Erica J. Lynn2
Erica J. Lynn2 Bruce D. Minsky7
Bruce D. Minsky7 Van K. Morris9
Van K. Morris9 Andrea Milbourne3
Andrea Milbourne3 Craig A. Messick6
Craig A. Messick6 Ann H. Klopp2
Ann H. Klopp2 P. Andrew Futreal4
P. Andrew Futreal4 Cullen M. Taniguchi7
Cullen M. Taniguchi7 Kathleen M. Schmeler3
Kathleen M. Schmeler3 Lauren E. Colbert2*
Lauren E. Colbert2*Background: Squamous cell carcinoma of the anus (SCCA) is a rare gastrointestinal cancer. Factors associated with progression of HPV infection to anal dysplasia and cancer are unclear and screening guidelines and approaches for anal dysplasia are less clear than for cervical dysplasia. One potential contributing factor is the anorectal microbiome. In this study, we aimed to identify differences in anal microbiome composition in the settings of HPV infection, anal dysplasia, and anal cancer in this rare disease.
Methods: Patients were enrolled in two prospective studies. Patients with anal dysplasia were part of a cross-sectional cohort that enrolled women with high-grade lower genital tract dysplasia. Anorectal tumor swabs were prospectively collected from patients with biopsy-confirmed locally advanced SCCA prior to receiving standard-of-care chemoradiotherapy (CRT). Patients with high-grade lower genital tract dysplasia without anal dysplasia were considered high-risk (HR Normal). 16S V4 rRNA Microbiome sequencing was performed for anal swabs. Alpha and Beta Diversity and composition were compared for HR Normal, anal dysplasia, and anal cancer.
Results: 60 patients with high-grade lower genital tract dysplasia were initially enrolled. Seven patients had concurrent anal dysplasia and 44 patients were considered HR Normal. Anorectal swabs from 21 patients with localized SCCA were included, sequenced, and analyzed in the study. Analysis of weighted and unweighted UniFrac distances demonstrated significant differences in microbial community composition between anal cancer and HR normal (p=0.018). LEfSe identified that all three groups exhibited differential enrichment of specific taxa. Peptoniphilus (p=0.028), Fusobacteria (p=0.0295), Porphyromonas (p=0.034), and Prevotella (p=0.029) were enriched in anal cancer specimens when compared to HR normal.
Conclusion: Although alpha diversity was similar between HR Normal, dysplasia and cancer patients, composition differed significantly between the three groups. Increased anorectal Peptoniphilus, Fusobacteria, Porphyromonas, and Prevotella abundance were associated with anal cancer. These organisms have been reported in various gastrointestinal cancers with roles in facilitating the proinflammatory microenvironment and neoplasia progression. Future work should investigate a potential role of microbiome analysis in screening for anal dysplasia and investigation into potential mechanisms of how these microbial imbalances influence the immune system and anal carcinogenesis.
Squamous cell carcinoma of the anus (SCCA) is a rare gastrointestinal cancer, affecting 8,000-9,000 patients annually in the US (1). It is linked to prior human papillomavirus (HPV) infections in approximately 90% of cases (2), but factors associated with the cancer development, such as the progression of HPV infection to dysplasia and anal cancer, are unclear. The standard-of-care treatment for patients with localized SCCA is chemoradiotherapy (CRT), which leads to 5-year survival rates of 75-90% (3). However, 30-40% of patients with advanced SCCA experience local recurrence or treatment-related toxicity (4, 5). Despite the use of HPV vaccines, the incidence and morbidity of SCCA continue to rise (6). Anal dysplasia is poorly understood and has been more difficult to detect than lower genital tract dysplasia. Women with HPV-related high-grade dysplasia or cancer of the cervix, vagina, or vulva are at increased risk for the development of anal dysplasia and SCCA (7–9). Prior observational studies have identified anal dysplasia in fewer than 15% of such high-risk women when assessed by high-resolution anoscopy (HRA), which is consistent with epidemiologic data showing that the incidence of anal cancer among women in the United States is much lower than the combined incidence of cervical, vaginal, or vulvar cancer (7, 10). Although some risk factors, such as tobacco use, are shared between both HPV-related diseases, the rarity of anal dysplasia and cancer suggests that additional external factors may modulate this risk. The identification of correlates between host genetics, environmental exposures, and the risk of anal dysplasia or cancer among high-risk patients would have important implications for cancer prevention efforts.
One potential contributing factor is the anal microbiome, which has been implicated in the progression of HPV infection to cancer in other HPV-related carcinogenesis (11). Knowledge about the pervasive influence of commensal bacteria led to the hypothesis that bacterial species found in the cervix and vagina contribute to the development of lower genital tract HPV infection, dysplasia, and cancer (11). Subsequent work using bacterial metagenomic sequencing methods has confirmed these observations by demonstrating a correlation between HPV-related cervical disease and lower levels of Lactobacillus species along with an increased abundance of anaerobic bacteria such as Gardnerella vaginalis, Finegoldia magna, Atopobium vaginae, Dialister invisus, Prevotella buccalis, P. timonensis, and Fusobacterium (12–15). Although the link between the microbiota and cervical carcinogenesis is well established, it is not yet known whether commensal bacteria play a role in the development of HPV-related disease at the anus.
Our previous work demonstrated a role for the anal microbiome in anal cancer response to therapy and toxicity (5). The ability to study the anal microbiome and carcinogenesis is limited by the rarity of the disease and limited patient samples. In this study, we combine anal microbiome samples collected from two prospectively enrolled cohorts, providing a unique opportunity to address this issue. The purpose of this study was to identify differences in anal microbiome composition in the settings of known HPV infection, anal dysplasia, and anal cancer.
We performed a retrospective analysis of anorectal microbiome composition using anorectal swab samples obtained from two prospective studies (Figure 1A). In the first study, we obtained anorectal swab samples from study participants enrolled between September 2016 and January 2017 in a cross-sectional study examining the prevalence of anal dysplasia among women with cervical, vaginal, and vulvar dysplasia at the University of Texas MD Anderson Cancer Center and Lyndon B. Johnson General Hospital (NCT02140021). Female patients were eligible for enrollment if they were over the age of 18 and had histologically confirmed cervical, vaginal, or vulvar high-grade dysplasia or invasive squamous cell carcinoma, invasive adenocarcinoma, or adenocarcinoma-in-situ. Pregnant women were excluded, as were any patients with previously documented perianal squamous cell dysplasia or invasive squamous cell carcinoma of the anus. We performed rigorous screening in order to minimize potential GI associated diseases that could alter the microbiome, such as excluding patients with previous abdominal or pelvic radiation therapy. This study was approved by the institutional review board at both the University of Texas MD Anderson Cancer Center (protocol PA2014-0021) and Lyndon B. Johnson General Hospital (eProtocol #14-05-0822).
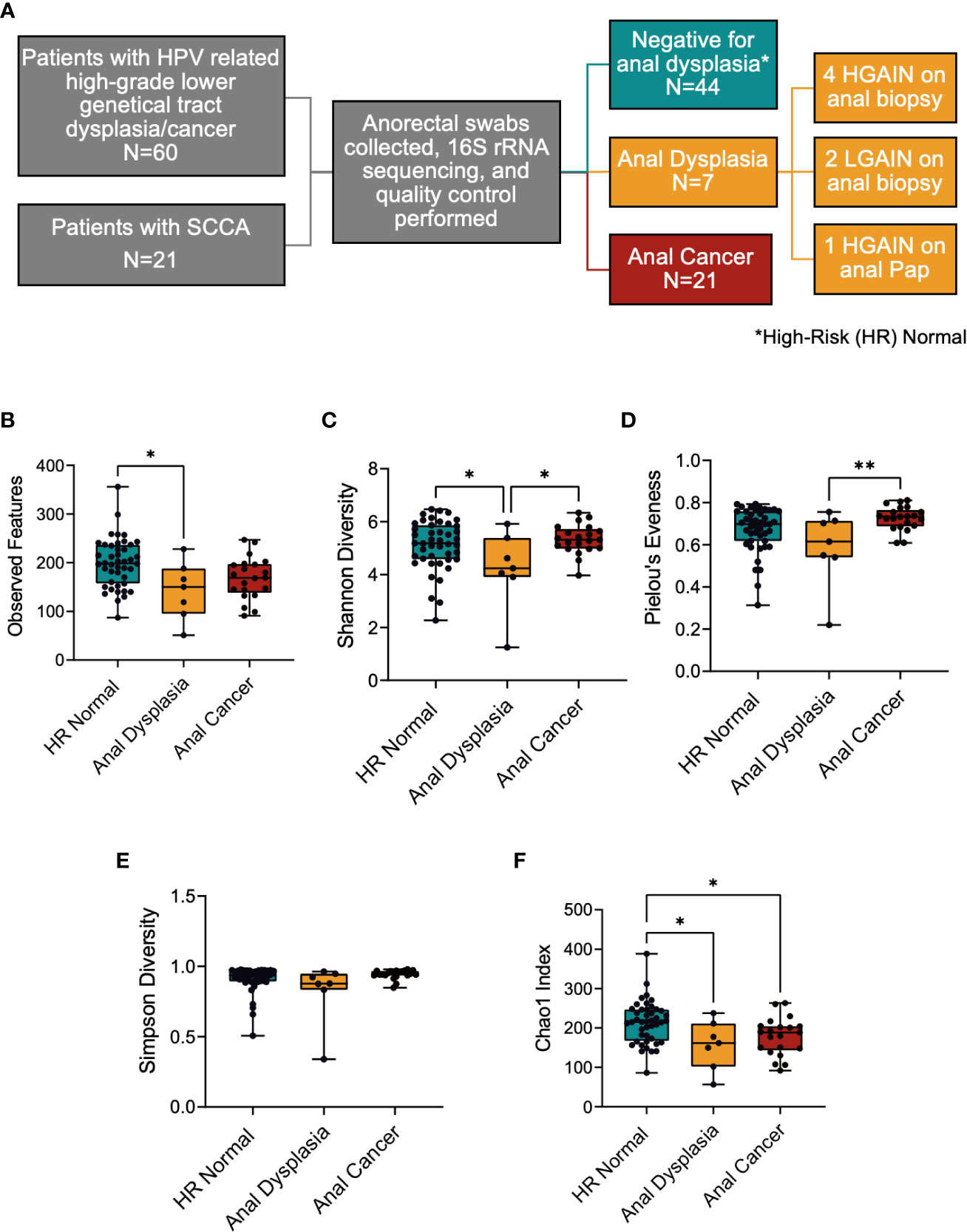
Figure 1 (A) Study overview depicting cohorts used and sample sizes. (B–F) Alpha diversity metrics between the three groups. (B) Richness of species was significantly decreased between HR Normal and Anal Dysplasia groups, (C) Shannon diversity richness index was lower in Anal Dysplasia, (D) Pielou’s Evenness index and (E) Simpson diversity index was greater in Anal Cancer compared to Anal Dysplasia, and (F) Chao1 was greater in HR Normal compared to both Anal Dysplasia and Anal Cancer. Statistical tests were performed using one way ANOVA. *P < 0.05, **P < 0.01. SCCA, Squamous cell carcinoma of the anus; HR Normal, High Risk Normal; H/LGAIN, high-grade or low-grade anal intraepithelial neoplasia.
In the second study, anorectal tumor swabs were prospectively collected from patients with biopsy-confirmed nonmetastatic SCCA receiving standard-of-care treatment as part of an institutional review board–approved study (protocol #2014-0543) at the University of Texas M.D. Anderson Cancer Center from April 2017 to July 2019 as described previously (5, 16). Patients with a previous history of abdominal or pelvic radiation were excluded from the study. Anal Pap smear and high resolution anoscopy were performed on all participants. Anorectal microbial specimens were collected in both studies using a swab biopsy technique.
Following the provision of written, informed consent, clinical and demographic data were collected at enrollment including age, menopausal status, self-reported ethnicity, HPV vaccination status, and human immunodeficiency virus (HIV) status. Information about prior diagnoses of cervical, vaginal, and vulvar dysplasia or cancer was also collected, along with the results of previous cervical or vaginal cytology and HPV testing. Information regarding the use of concurrent systemic antibiotics at the time of enrollment and tobacco use was obtained from the medical record.
Upon enrollment, in addition to standard of care treatment for genital tract disease, all anal dysplasia participants had anal cytology and anal HPV test samples collected, as well as an anorectal swab (Isohelix) prior to treatment for microbiome analysis. High-resolution anoscopy (HRA) was then performed as part of the study protocol. Biopsies were collected of abnormal areas identified by HRA as per standard of care practices. Participants were referred to a colorectal surgeon for anal high-grade anal squamous intraepithelial lesion (HSIL) anal cytology test result (regardless of HRA findings) or if biopsy of abnormal areas revealed high-grade or low-grade anal intraepithelial neoplasia (HGAIN, LGAIN, or AIN2-3). For the present analysis, patients were considered to have “anal dysplasia” if they had either high-grade or low-grade anal intraepithelial neoplasia (AIN1-3) on anal biopsy, or if their anal cytology showed HSIL regardless of HRA findings. Based on the clinical assessments, anal swab specimens were grouped as High-Risk (HR) Normal, Anal Dysplasia, or Anal Cancer (Figure 1A).
Isohelix DNA Swabs (Isohelix, SK-2S) were used to collect tissue and fecal material from anal dysplasia and SCCA patients prior to standard of care treatment. Swab specimens were stabilized using BuccalFix DNA Stabilization Solution (Isohelix, BFX-25) within 1 hour of collection and stored at –80°C until DNA isolation. Amplicon sequencing of the 4th hypervariable (V4) region of the bacterial 16S ribosomal RNA (rRNA) gene was performed on anorectal swabs by the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine, as previously described (16, 17). Bacterial genomic DNA was extracted from anorectal swabs using the MoBIO PowerSoil Kit (QIAGEN, 12855-50). The 16S rRNA gene sequencing methods were adapted from the Human Microbiome Project and Earth Microbiome Project (18). The bacterial 16S ribosomal RNA V4 genomic region was PCR amplified and sequenced using 250 bp paired-end reads on a MiSeq sequencer (Illumina, San Diego, CA). The primers used for amplification (515F-806R) contain adapters for MiSeq sequencing and single-index molecular barcodes so that the PCR products may be pooled and sequenced directly.
Sequencing data were processed and analyzed using QIIME2 v2022.2 (19). A QIIME2 provenance diagram is shown in Figure S1A. Sequencing quality control, amplicon sequence variant counts for feature table construction, and representative sequences used for phylogeny tree construction and taxonomic classification were performed using denoising via DADA2 (20) with the pseudo-pooling parameter and identical trim and truncation parameters of 0 and 188, respectively, across all samples. The trim and truncation settings were chosen based on quality scores generated by the QIIME2 platform. A rarefaction sampling depth of 11500 sequences per sample was used when necessary for downstream comparative analysis (Figure S1B). Nonrarefied data tables were used for linear discriminant analysis effect size. A naive Bayes classifier trained on 515/806R V4 ribosomal RNA regions from the SILVA release 132 database was used to assign taxonomy (21). Feature tables constructed using amplicon sequence variant counts were used for downstream comparative diversity analysis. Phylogenetic reference tree construction was performed using a SILVA 128 SATé-enabled phylogenetic placement database (22).
We analyzed the HR Normal, Anal Dysplasia, and Anal Cancer sequencing data using several different alpha diversity metrics (23). Observed Features (richness) provides a count of all identified putative species. Pielou’s evenness index calculates the proportions of individual species in a sample population. The Shannon diversity index accounts for the richness and evenness of taxa within a sample. The Simpson’s diversity index measures the diversity of species in a community. Faith’s phylogenetic diversity (PD) accounts for the phylogenetic differences between species in a sample. The inverse Simpson diversity index measures the relative counts of species that make up the richness, and finally, the Chao1 index estimates richness emphasizing rare species (23).
We generated stacked bar plots of genus level relative counts in ATIMA v3.1.2 (developed by the Center for Metagenomics and Microbiome Research at the Baylor College of Medicine) to observe taxa distribution across groups. Samples in these plots were ordered by relative counts of a specific taxon, e.g., Bacteroides. We also conducted compositional analysis using unweighted and weighted UniFrac and Bray Curtis to generate coordinates for each sample. Principal Coordinates Analysis (PCoA) and biplots were created in ATIMA to visualize group coordinates. Permutational multivariate analysis of variance (PERMANOVA) was used to assess differences in mean and variance between groups. Permutational multivariate analysis of dispersion (PERMDISP) was used to assess differences in dispersion between groups. Linear Discriminant Effect Size (LEfSe) (24) was performed in Miniconda v4.12 (25) was used to identify abundance changes of specific taxa that were enriched between all three groups. The LDA score cutoff was set at 3.5, and the alpha value for the Kruskal-Wallis test among classes was set at 0.05. Cladograms of the LEfSe results were generated to visualize and inspect the nestedness of the differentially enriched taxonomic groupings. Additionally, LEfSe was used with similar parameters to assess pairwise differences between groups. As a confirmatory test, DESeq2 was performed using pairwise comparisons across the groups using the contrast parameter (26), and volcano plots were made using the EnhancedVolcano package (Figure S8) (27). Heatmaps were made using the LEfSe results and colored by the group in which they were enriched. Taxonomic counts were log normalized and plotted using ComplexHeatmap (28). Stacked bar plots based on relative counts were also generated in ATIMA based on overall abundance and the LEfSe results.
For clinical and demographic variables, statistical comparisons were performed in RStudio (v2022.7.0.548) (29) using R v4.2.1 (30). A t-test was used for continuous variables. A Chi-squared test was used to compare categorical variables between HR Normal and Anal Cancer and Wilcoxon signed rank test was used for comparisons between Anal Dysplasia due to its smaller sample size. To compare alpha diversity, statistical tests were performed using pairwise one-way ANOVA to compare between the three groups HR Normal, Anal Dysplasia, and Anal Cancer; adjusted P<0.05 was considered statistically significant. Comparisons between differential abundance of specific taxa was performed on specific taxa using relative counts and compared using a similar one-way ANOVA. GraphPad Prism (GraphPad Software, v9.0) was used to generate graphs for data visualization.
60 patients with high-grade lower genital tract dysplasia were initially enrolled. After excluding patients taking concurrent antibiotics and applying quality filters, anorectal swabs from 51 patients with high-grade lower genital tract dysplasia were sequenced. Of those, 7 patients had concurrent anal dysplasia, 4 HGAIN and 2 LGAIN on anal biopsy and 1 HGAIN on anal Pap. Additionally, this study included anorectal swabs from 21 patients with localized SCCA. In total, 71 swabs (44 HR Normal, 7 Anal Dysplasia, and 21 Anal Cancer) were included, sequenced, and analyzed in the study (Figure 1A). Since this study combines two cohorts and three stages of disease, there are noteworthy differences in patient demographics that are outlined in Table 1. These are grouped by sample type. The age in the HR Normal group (mean=46.1 years) was statically lower than the Anal Cancer group (mean=57 years, P<0.001). However, there was no difference between the Anal Dysplasia group (mean=54 years). Of the participants in the Anal Cancer group, 3 (16.67%) were males. The rest of the participants in the study were females. Additionally, there were differences in other categories including site of recruitment, menopausal status, and ethnicity. Also, one Anal Cancer patient reported a positive HIV diagnosis. Additional clinical and demographic information can be found in Table S1.
In general, the Alpha diversity metrics (Figures 1B–F) displayed a similar pattern of changes among the 3 groups, namely, the greatest was HR Normal, the lowest being Anal Dysplasia, and the intermediate value was Anal Cancer. Specific comparisons revealed that the number of observed features was increased in the HR Normal (mean=119.2) specimens when compared to Anal Dysplasia (mean=142.4, P=0.017, Figure 1B). The Shannon diversity index for Anal Dysplasia (mean=4.169) was significantly lower when compared to both HR Normal (mean= 5.11, P=0.042) and Anal Cancer specimens (mean=5.34, P=0.015, Figure 1C). Additionally, Anal Dysplasia was lower than Anal Cancer for Pielou’s Evenness (Anal Dysplasia mean=0.585, Anal Cancer mean= 0.7255 P=0.019, Figure 1D) and Simpson Diversity indexes (Anal Dysplasia mean=0.8224; Anal Cancer mean=0.9427, P=0.019, Figure 1E). By Chao1 (Figure 1F), HR Normal (mean=213.7) was significantly higher than both Anal Dysplasia (mean=156.7, P=0.026) and Anal Cancer (mean=177.8, P=0.032). Other metrics include Faith PD and Inverse Simpson (Figure S2), and all per sample diversity values and P values can be found in Table S1.
There is a large proportion of the genus Bacteroides among all three groups (blue, Figures 2A, S3A), as well as Prevotella (red, Figure S3B) and the class Clostridia (orange, Figure S3G). Analysis of weighted UniFrac distances demonstrated a significant dispersion in microbial community composition between the three groups (Figure 2B, PERMANOVA P=0.063, PERMDISP P=0.037). Unweighted UniFrac and weighted Bray Curtis comparisons revealed significant differences amongst all three groups (PERMANOVA P=0.001, P=0.020; PERMDISP P=0.557, P=0.902; Figures S4B, C). There was also a significant difference between HR Normal and Anal Cancer (Figure 2C PERMANOVA P=0.018, PERMDISP P=0.019). All PCoA metric comparisons can be observed in Table S1 with corresponding figures in Figure S4. A weighted UniFrac biplot of the three group comparisons (Figure 2B) displayed a pull from the family Enterobacteriaceae (comprising the genera Escherichia and Shigella). There was also a split in the larger cluster between Bacteroides and Prevotella. These findings are also replicated in the pairwise biplot between HR Normal and Anal Cancer (Figure 2C). Additional comparisons using additional metrics and alternative taxonomic levels can be observed in Figure S4.
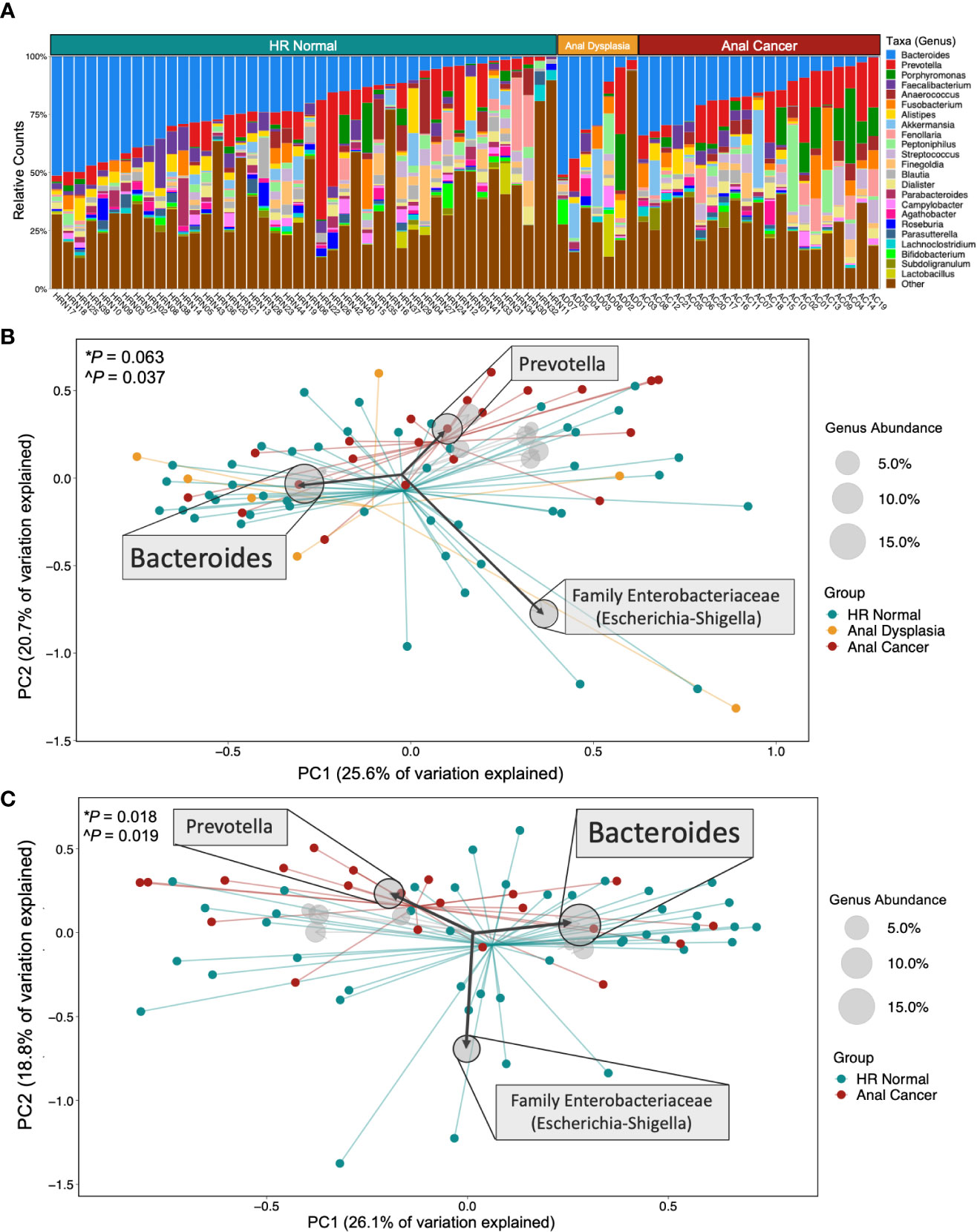
Figure 2 Microbial composition and beta diversity across groups. (A) Stacked bar plot of relative counts for all samples in the study. (B) Comparing all three groups and (C) HR Normal vs Anal Cancer by Weighted UniFrac Principal Coordinate Analysis and biplot showing taxa that pulled samples in a specific direction. Centroids display converging points for each group. The size of the circle represents the abundance of that taxa across all samples displayed, and specific taxa were displayed based on abundance. Statistical tests used *PERMANOVA and ^PERMDISP. HR Normal, High Risk Normal; PC, principal component.
LEfSe identified that all three groups exhibited differential enrichment of specific taxa (Figure 3A, S5A; Table S1), including increased abundance of Peptoniphilus (p=0.025), Firmicutes (p=0.037345458), Anaerococcus (p=0.016), Oscillospiraceae (p=0.043) and Clostridia (p=0.013) among Anal Cancer, Bifidobacterium (p=0.048) among Anal Dysplasia, and Roseburia (p=0.035) and Blautia (p=0.021) among HR Normal. Pairwise comparison between Anal Cancer and HR Normal (Figures 4A, B) showed a similar increased abundance of Peptoniphilus (p=0.025) and Anaerococcus (p=0.023) and additional organisms including Porphyromonas (p=0.027), Prevotella (p=0.023), and Fusobacterium (p=0.028). Anal Cancer also displayed an increased abundance of Clostridia, Firmicutes, Peptoniphilus, Anaerococcus, and Oscillospiraceae when compared to Anal Dysplasia (Figure S5).

Figure 3 Taxa-specific changes across three groups. (A) Linear discriminant analysis (LDA) effect size (LEfSe) derived bar graph of enriched taxa between HR Normal, Anal Dysplasia, and Anal Cancer. An LDA of 3.5 or greater was used as a cutoff and an alpha of 0.05 for the Wilcoxon rank-sum test. (B) Heatmap colored horizontally based on the group each taxon was enriched from LEfSe. Values were log normalized. (C–E) Comparison of relative counts between the three groups for specific taxa. Statistical tests were performed using one way ANOVA. *P < 0.05. **P < 0.01. Abv. HR Normal: High Risk Normal.
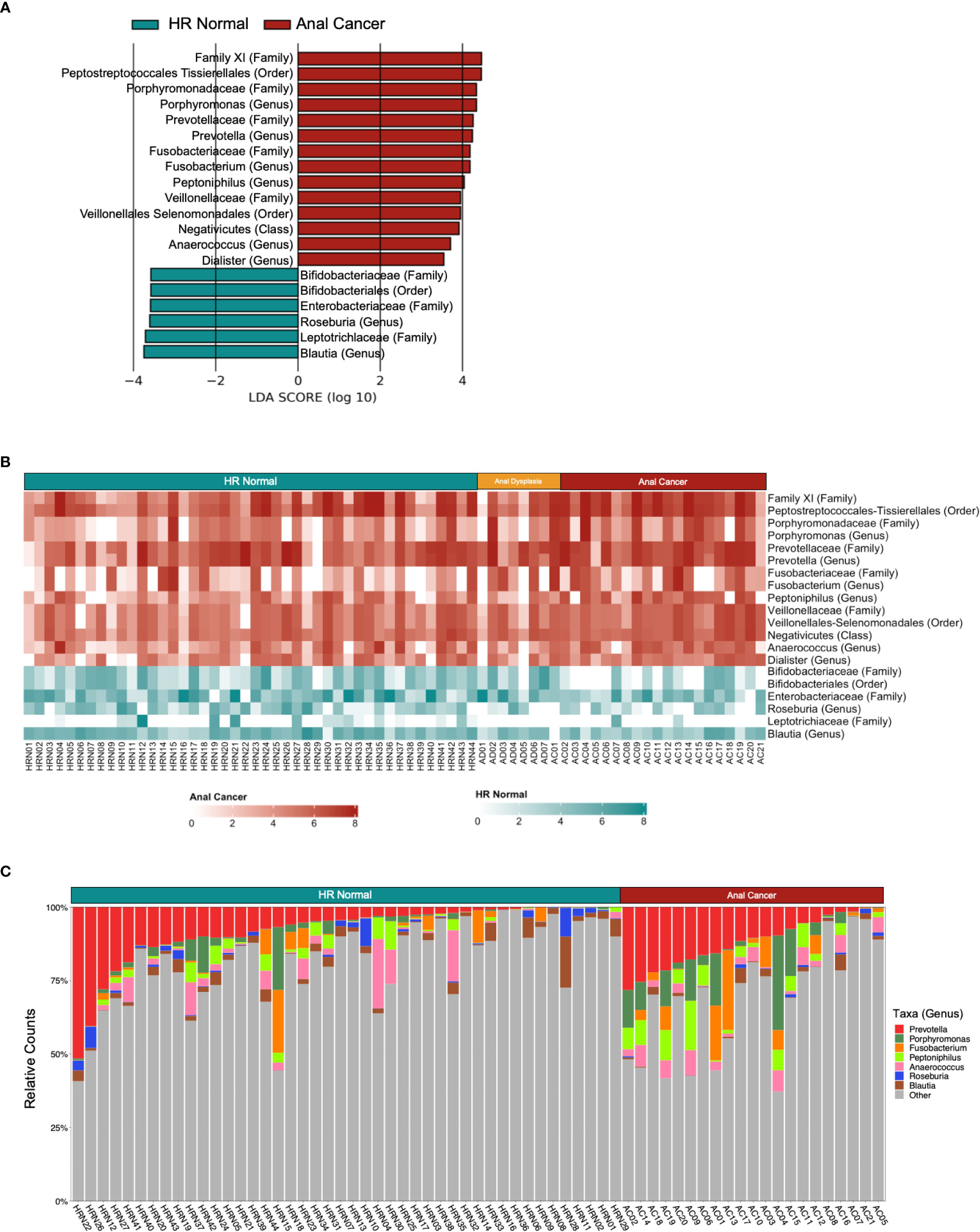
Figure 4 Differently abundant taxa between HR Normal and Anal Cancer. (A) Linear discriminant analysis (LDA) effect size (LEfSe) derived bar graph of enriched taxa between HR Normal and Anal Cancer. (B) Heatmap colored horizontally based on the group each taxon was enriched from LEfSe. Values were log normalized. (C) Stacked bar plot of relative counts for HR Normal and Anal Cancer highlighting the genera enriched from LEfSe. An LDA of 3.5 or greater was used as a cutoff and an alpha of 0.05 for the Wilcoxon rank-sum test.
Using the LEfSe to guide further analyses, we compared taxonomic abundances between the three groups. Heatmaps can be observed comparing the three groups and (Figure 3B) and comparing HR Normal to Anal Cancer (Figure 4B). The groups are colored horizontally based on the group in which they were found to be enriched from LEfSe (Figures 3A, 4A). A stacked bar plot using the genus level enrichments from LEfSe can be observed in Figure 4C, and family level in Figure S6.
When comparing relative counts of specific organisms, there was a significant increase in Porphyromonas between HR Normal (mean=0.0169) and Anal Cancer (mean=0.0629, P=0.015, Figure 3C) which can also be visually observed (green, Figure 4C). There was also significant decrease in Clostridia between Anal Dysplasia (mean=0.1741) and both HR Normal (mean=0.3679, P=0.019, Figure 3D) and Anal Cancer (mean=0.3774, P=0.022). And finally, there was a significant increase in Peptoniphilus in Anal Cancer (mean=0.0423) compared to HR Normal (mean=0.0171, P=0.006) and Anal Dysplasia (mean=0.0077, P=0.027, Figure 3E). Similar comparisons were performed for Anaerococcus, Fusobacterium, Lachnospiraceae, Oscillospirales, Oscillospiraceae, and Prevotella but were not significantly different between groups (Figure S7; Table S1).
Although the microbiome has been well characterized in some HPV-driven cancers, establishing an association between anal cancer and the anorectal microbiome remains difficult due to the rarity of disease and limited patient specimens. We observed primarily compositional and beta diversity differences between our HR Normal, Anal Dysplasia, and Anal Cancer samples with some differences in richness and evenness. We also noted significant enrichment of specific taxa among the Anal Cancer group in comparison to HR Normal, including Peptoniphilus, Prevotella, Porphyromonas, and Fusobacterium (Figures 3, 4).
In patients with anal cancer, we observed an increase in the abundance of the genera Peptoniphilus (a member of the Clostridia class), Prevotella, Porphyromonas, and Fusobacterium, which are all linked to a range of pro-inflammatory and immune modulating functions. Peptoniphilus is a gram-positive anaerobic commensal that colonizes mucosal surfaces, such as the mouth, gastrointestinal tract, and genitourinary tract. There are reports of a higher abundance of Peptoniphilus in colorectal cancer (CRC) (31), and it is a highly opportunistic pathogenic bacteria that produces calprotectin (32, 33), a protein specific to neutrophil mediated inflammation and neutrophil recruitment (34). Prevotella is an anaerobic gram-negative that inhabits the oral cavity, vagina, and gut and is associated with chronic inflammation (35–37). Prevotella abundance is associated with T helper type 17 (Th17) and T helper type 1 (Th1) mediated mucosal inflammation and immune response through activation of toll-like receptor 2 (38), in addition to stimulating epithelial cells to produce chemokines and cytokines (39, 40). In one study examining the microbiome of men who have sex with men (MSM) patients with anal precancerous lesions, Prevotella was one of the most predictive bacteria associated with high-grade squamous intraepithelial lesions (41). Another study showed an association of Prevotella abundance with persistent cervicovaginal HPV infection (42). Porphyromonas also plays a significant role in CRC (43–46) and is associated with a significant increase in TNF-alpha and IL-6 (47). More specifically, Porphyromonas gingivalis has been well-associated with the occurrence and development of gastrointestinal cancers. In mice orally administered Porphyromonas gingivalis, the mRNA levels of IFN-gamma were elevated in the gut (48). In CRC, Fusobacterium is associated with tumor carcinogenesis, high disease stage, poor tumor differentiation, poor prognosis (49) and metastatic disease (50, 51). Reported mechanisms for these associations in CRC include 1) TLR4-induced initiation of signaling pathways that lead to the secretion of inflammatory cytokines such as TNF-alpha and IL-8 (52, 53), 2) expression of NF-κB, which inhibits apoptosis and stimulates cell proliferation (54), 3) NK T-cell inhibition (55), and 4) recruitment of myeloid-derived suppressor cells that suppress CD4 T-cells (56, 57). We propose that Fusobacterium, along with these other taxa, could affect immune function in anal carcinogenesis and cancer.
This study has several strengths and limitations. A key strength is the inclusion of a relatively homogenous group of patients with a rare disease who share an important risk factor for anal dysplasia, specifically the co-existence of high-grade dysplasia of the lower genital tract. An additional strength of this study is the minimization of ascertainment bias attained through the performance of anal cytology tests and HPV testing as well as HRA on all study participants, consistent with the current diagnostic standard for the detection of anal dysplasia (58). Several limitations exist, however, primarily due to the small sample size given the rarity of anal cancer and anal dysplasia, and sequencing methodology. We can make stronger comparisons between High Risk Normal and Anal Cancer due to their larger sample sizes, while comparisons with Anal Dysplasia are more limited. Nevertheless, these findings are not conclusive. This study is aimed to serve as a pilot for future research in this area. Individual genus composition alone does not fully describe potential interactions between these bacterial genera that could drive the shift from HPV infection to dysplasia and cancer. Although evaluation of co-occurrences of these species in this study is limited by the small sample size, several of these genera are known to interact and function in synergy. For example, Peptoniphilus and Prevotella are found to co-occur in association with recurrence of bacterial vaginosis (29), elevated risk for HIV seroconversion and HIV antiviral treatment resistance (38). In addition, Fusobacterium, Porphyromonas, and Prevotella have been co-associated together in colorectal cancer (59, 60). Another remaining unknown from this study is a description of the function of the individual bacterial species associated with anal cancer. Performing a functional analysis would require shotgun metagenomic sequencing rather than 16S sequencing. However 16S sequencing is superior for compositional analysis (61), which was the primary goal of this study. Future work utilizing whole genome sequencing will describe the functional composition and metabolic analysis of this patient population.
There are significant differences in demographics between HR Normal and Anal Cancer (Table 1). Anal cancer tends to develop later than cervical cancer (62). Therefore, age, and as a result, menopausal status can differ. The Lyndon B. Johnson General Hospital generally cares for a more diverse population which may explain the difference in ethnicity between the two groups. Additionally, there was an Anal Cancer patient with HIV. There an increased risk of HPV-associated cancers, including anal cancer, among individuals with HIV as well as an increased risk with immunosuppression (63). Due to the rarity of anal dysplasia and cancer, a necessary limitation of this study is the small sample size and imbalance between the number of subjects in each group. However, the rate of anal dysplasia in this cohort is consistent with prior reports of incidence in high-risk women (7). In order to mitigate these limitations, we employed rigorous statistical procedures for FDR control and stringent cut-offs for multiple testing. Although this study can provide only exploratory suggestions of correlation due to its design, these results are important for hypothesis generation to guide further studies in this area. Additionally, we plan to perform functional analysis and experimental studies to test these specific hypotheses. Future prospective studies are needed to confirm the generalizability of the observed association between specific bacterial abundance and anal cancer development including mechanisms by which the anorectal microbiota may influence anal dysplasia and cancer risk.
Overall, our findings suggest an association between the anorectal microbiome among anal cancer development. Our work highlights potential roles in an understudied disease that needs to be further explored. Implications of this work could include improved diagnostic tools for this rare and difficult to detect disease, especially among high-risk patients. This could allow us to intervene early and prevent anal dysplasia and the progression to cancer.
The data presented in the study are deposited in the NCBI Sequence Read Archive repository, accession number PRJNA880301.
The studies involving human participants were reviewed and approved by institutional review boards at both the University of Texas MD Anderson Cancer Center (protocol PA2014-0021 & #2014-0543) and Lyndon B. Johnson General Hospital (eProtocol #14-05-0822). The patients/participants provided their written informed consent to participate in this study.
RH, PF, KS, AK, CT, and LC conceived the study. JE, DL, RH, VH, TK, LC, and NA analyzed data. AM, CM, KS, AK, CT, and LC collected samples. AM, CM, AK, JW, PF, KS, and LC jointly supervised the study and provided oversight during the manuscript editing process. JE and VH wrote the first draft and revised the manuscript for submission. All authors contributed to the article and approved the submitted version.
This work was supported in part by the National Institutes of Health (NIH) through MD Anderson’s Cancer Center Support Grant P30CA016672, NIH training grant T32CA101642-11 (RH), the Cancer Prevention & Research Institute of Texas (CPRIT) Scholar in Cancer Research #R1205 (RH), and the University of Texas MD Anderson Cancer Center HPV-related Cancers Moon Shot (LC). CT is a Khalifa Scholar supported by the Khalifa Bin Zayed Al Nahyan Foundation, Childress Family Foundation, and the Dodd Family Foundation. LC received funding from the Radiological Society of North America Resident/Fellow Award.
We thank the patients for their immeasurable contribution and the radiation oncology clinical and laboratory teams at the University of Texas MD Anderson Cancer Center.
CT is on the clinical advisory board of Accuracy, has a patent for oral amifostine as a radioprotectant of the upper gastrointestinal tract issued, licensed, and with royalties paid from Xerient Pharmaceuticals and a pending patent for prolyl hydroxylase inhibitors as a radioprotectant of the gastrointestinal tract, was the lead principal investigator of a multicenter trial testing the effects of high-dose stereotactic body radiation therapy with the radiomodulator, GC4419, and is a paid consultant for Phebra Pty, Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1051431/full#supplementary-material
2. Schiffman M, Doorbar J, Wentzensen N, de Sanjosé S, Fakhry C, Monk BJ, et al. Carcinogenic human papillomavirus infection. Nat Rev Dis Primer (2016) 2:16086. doi: 10.1038/nrdp.2016.86
3. Franklin RA, Giri S, Valasareddy P, Lands LT, Martin MG. Comparative survival of patients with anal adenocarcinoma, squamous cell carcinoma of the anus, and rectal adenocarcinoma. Clin Colorectal Cancer (2016) 15:47–53. doi: 10.1016/j.clcc.2015.07.007
4. Holliday EB, Morris VK, Johnson B, Eng C, Ludmir EB, Das P, et al. Definitive intensity-modulated chemoradiation for anal squamous cell carcinoma: Outcomes and toxicity of 428 patients treated at a single institution. Oncologist (2022) 27:40–7. doi: 10.1093/oncolo/oyab006
5. Lin D, Alam MBE, Jaoude JA, Kouzy R, Phan JL, Elnaggar JH, et al. Microbiome dynamics during chemoradiation therapy for anal cancer. Int J Radiat Oncol Biol Phys (2022) 113:974–84. doi: 10.1016/j.ijrobp.2022.04.037
6. Oliveira CR, Niu YS, Einarsdottir HM, Niccolai LM, Shapiro ED. Disparities in the epidemiology of anal cancer: A cross-sectional time series. Health Equity (2020) 4:382–5. doi: 10.1089/heq.2020.0021
7. Robison K, Cronin B, Bregar A, Luis C, DiSilvestro P, Schechter S, et al. Anal cytology and human papillomavirus genotyping in women with a history of lower genital tract neoplasia compared with low-risk women. Obstet Gynecol (2015) 126:1294–300. doi: 10.1097/AOG.0000000000001135
8. Park IU, Ogilvie JW, Anderson KE, Li Z, Darrah L, Madoff R, et al. Anal human papillomavirus infection and abnormal anal cytology in women with genital neoplasia. Gynecol Oncol (2009) 114:399–403. doi: 10.1016/j.ygyno.2009.05.008
9. Saleem AM, Paulus JK, Shapter AP, Baxter NN, Roberts PL, Ricciardi R. Risk of anal cancer in a cohort with human papillomavirus-related gynecologic neoplasm. Obstet Gynecol (2011) 117:643–9. doi: 10.1097/AOG.0b013e31820bfb16
10. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin (2017) 67:7–30. doi: 10.3322/caac.21387
11. Champer M, Wong A, Champer J, Brito I, Messer P, Hou J, et al. The role of the vaginal microbiome in gynaecological cancer. BJOG Int J Obstet Gynaecol (2018) 125:309–15. doi: 10.1111/1471-0528.14631
12. Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, Téllez-Sosa J, Martínez-Barnetche J, Cortina-Ceballos B, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study. PloS One (2016) 11:e0153274. doi: 10.1371/journal.pone.0153274
13. Piyathilake CJ, Ollberding NJ, Kumar R, Macaluso M, Alvarez RD, Morrow CD. Cervical microbiota associated with higher grade cervical intraepithelial neoplasia in women infected with high-risk human papillomaviruses. Cancer Prev Res (Phila Pa) (2016) 9:357–66. doi: 10.1158/1940-6207.CAPR-15-0350
14. Lee JE, Lee S, Lee H, Song Y-M, Lee K, Han MJ, et al. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PloS One (2013) 8:e63514. doi: 10.1371/journal.pone.0063514
15. So KA, Yang EJ, Kim NR, Hong SR, Lee J-H, Hwang C-S, et al. Changes of vaginal microbiota during cervical carcinogenesis in women with human papillomavirus infection. PloS One (2020) 15:e0238705. doi: 10.1371/journal.pone.0238705
16. Biegert G, El Alam MB, Karpinets T, Wu X, Sims TT, Yoshida-Court K, et al. Diversity and composition of gut microbiome of cervical cancer patients: Do results of 16S rRNA sequencing and whole genome sequencing approaches align? J Microbiol Methods (2021) 185:106213. doi: 10.1016/j.mimet.2021.106213
17. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359:97–103. doi: 10.1126/science.aan4236
18. Methé BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, et al. A framework for human microbiome research. Nature (2012) 486:215–21. doi: 10.1038/nature11209
19. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol (2019) 37:852–7. doi: 10.1038/s41587-019-0209-9
20. Nearing JT, Douglas GM, Comeau AM, Langille MGI. Denoising the denoisers: an independent evaluation of microbiome sequence error-correction approaches. PeerJ (2018) 6:e5364. doi: 10.7717/peerj.5364
21. Robeson MS, O’Rourke DR, Kaehler BD, Ziemski M, Dillon MR, Foster JT, et al. RESCRIPt: Reproducible sequence taxonomy reference database management. PloS Comput Biol (2021) 17:e1009581. doi: 10.1371/journal.pcbi.1009581
22. Janssen S, McDonald D, Gonzalez A, Navas-Molina JA, Jiang L, Xu ZZ, et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. MSystems (2018) 3:e00021-18. doi: 10.1128/mSystems.00021-18
23. Willis AD. Rarefaction, alpha diversity, and statistics. Front Microbiol (2019) 10:2407. doi: 10.3389/fmicb.2019.02407
24. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol (2011) 12:R60. doi: 10.1186/gb-2011-12-6-r60
26. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol (2014) 15:550. doi: 10.1186/s13059-014-0550-8
27. Blighe K, Rana S, Lewis M. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling (2018). Available at: https://bioconductor.org/packages/devel/bioc/vignettes/EnhancedVolcano/inst/doc/EnhancedVolcano.html (Accessed September 16, 2022).
28. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinforma Oxf Engl (2016) 32:2847–9. doi: 10.1093/bioinformatics/btw313
29. RStudio Team. RStudio: Integrated development environment for r. Boston, MA: RStudio, PBC (2022).
30. R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing (2020).
31. Alomair AO, Masoodi I, Alyamani EJ, Allehibi AA, Qutub AN, Alsayari KN, et al. Colonic mucosal microbiota in colorectal cancer: A single-center metagenomic study in Saudi Arabia. Gastroenterol Res Pract (2018) 2018:5284754. doi: 10.1155/2018/5284754
32. Brown K, Church D, Lynch T, Gregson D. Bloodstream infections due to peptoniphilus spp.: report of 15 cases. Clin Microbiol Infect (2014) 20:O857–60. doi: 10.1111/1469-0691.12657
33. Weis S, Schwiertz A, Unger MM, Becker A, Faßbender K, Ratering S, et al. Effect of parkinson’s disease and related medications on the composition of the fecal bacterial microbiota. NPJ Park Dis (2019) 5:1–9. doi: 10.1038/s41531-019-0100-x
34. Bjarnason I. The use of fecal calprotectin in inflammatory bowel disease. Gastroenterol Hepatol (2017) 13:53–6.
35. Könönen E, Gursoy UK. Oral prevotella species and their connection to events of clinical relevance in gastrointestinal and respiratory tracts. Front Microbiol (2021) 12:798763. doi: 10.3389/fmicb.2021.798763
36. Hammann R. A reassessment of the microbial flora of the female genital tract, with special reference to the occurrence of bacteroides species. J Med Microbiol (1982) 15:293–302. doi: 10.1099/00222615-15-3-293
37. Spiegel CA, Amsel R, Eschenbach D, Schoenknecht F, Holmes KK. Anaerobic bacteria in nonspecific vaginitis. N Engl J Med (1980) 303:601–7. doi: 10.1056/NEJM198009113031102
38. Tett A, Pasolli E, Masetti G, Ercolini D, Segata N. Prevotella diversity, niches and interactions with the human host. Nat Rev Microbiol (2021) 19:585–99. doi: 10.1038/s41579-021-00559-y
39. Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity (2015) 42:965–76. doi: 10.1016/j.immuni.2015.04.019
40. Doerflinger SY, Throop AL, Herbst-Kralovetz MM. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species-specific manner. J Infect Dis (2014) 209:1989–99. doi: 10.1093/infdis/jiu004
41. Ron R, Cabello A, Gosalbes MJ, Sánchez-Conde M, Talavera-Rodríguez A, Zamora J, et al. Exploiting the microbiota for the diagnosis of anal precancerous lesions in men who have sex with men. J Infect Dis (2021) 224:1247–56. doi: 10.1093/infdis/jiab068
42. Dong B, Huang Y, Cai H, Chen Y, Li Y, Zou H, et al. Prevotella as the hub of the cervicovaginal microbiota affects the occurrence of persistent human papillomavirus infection and cervical lesions in women of childbearing age via host NF-κB/C-myc. J Med Virol (2022) 94(11):5519–5534. doi: 10.1002/jmv.28001
43. Liu X-B, Gao Z-Y, Sun C-T, Wen H, Gao B, Li S-B, et al. The potential role of p.gingivalis in gastrointestinal cancer: a mini review. Infect Agent Cancer (2019) 14:23. doi: 10.1186/s13027-019-0239-4
44. Wang X, Jia Y, Wen L, Mu W, Wu X, Liu T, et al. Porphyromonas gingivalis promotes colorectal carcinoma by activating the hematopoietic NLRP3 inflammasome. Cancer Res (2021) 81:2745–59. doi: 10.1158/0008-5472.CAN-20-3827
45. Okumura S, Konishi Y, Narukawa M, Sugiura Y, Yoshimoto S, Arai Y, et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Commun (2021) 12:5674. doi: 10.1038/s41467-021-25965-x
46. Mu W, Jia Y, Chen X, Li H, Wang Z, Cheng B. Intracellular porphyromonas gingivalis promotes the proliferation of colorectal cancer cells via the MAPK/ERK signaling pathway. Front Cell Infect Microbiol (2020) 10:584798. doi: 10.3389/fcimb.2020.584798
47. Mudter J, Neurath MF. Il-6 signaling in inflammatory bowel disease: pathophysiological role and clinical relevance. Inflammation Bowel Dis (2007) 13:1016–23. doi: 10.1002/ibd.20148
48. Liu Y, Huang W, Dai K, Liu N, Wang J, Lu X, et al. Inflammatory response of gut, spleen, and liver in mice induced by orally administered porphyromonas gingivalis. J Oral Microbiol (2022) 14:2088936. doi: 10.1080/20002297.2022.2088936
49. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut (2016) 65:1973–80. doi: 10.1136/gutjnl-2015-310101
50. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. Science (2017) 358:1443–8. doi: 10.1126/science.aal5240
51. Wang S, Liu Y, Li J, Zhao L, Yan W, Lin B, et al. Fusobacterium nucleatum acts as a pro-carcinogenic bacterium in colorectal cancer: From association to causality. Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.710165
52. Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun (2000) 68:3140–6. doi: 10.1128/IAI.68.6.3140-3146.2000
53. Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Inducible expression of human beta-defensin 2 by fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun (2000) 68:2907–15. doi: 10.1128/IAI.68.5.2907-2915.2000
54. DiDonato JA, Mercurio F, Karin M. NF-κB and the link between inflammation and cancer. Immunol Rev (2012) 246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x
55. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity (2015) 42:344–55. doi: 10.1016/j.immuni.2015.01.010
56. Gabrilovich DI, Nagaraj S. Myeloid-derived-suppressor cells as regulators of the immune system. Nat Rev Immunol (2009) 9:162–74. doi: 10.1038/nri2506
57. Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol (2012) 12:253–68. doi: 10.1038/nri3175
58. Leeds IL. Anal cancer and intraepithelial neoplasia screening: A review. World J Gastrointest Surg (2016) 8:41. doi: 10.4240/wjgs.v8.i1.41
59. Rebersek M. Gut microbiome and its role in colorectal cancer. BMC Cancer (2021) 21:1325. doi: 10.1186/s12885-021-09054-2
60. Chen Y, Yang Y, Gu J. Clinical implications of the associations between intestinal microbiome and colorectal cancer progression. Cancer Manag Res (2020) 12:4117–28. doi: 10.2147/CMAR.S240108
61. Mitra A, Grossman Biegert GW, Delgado AY, Karpinets TV, Solley TN, Mezzari MP, et al. Microbial diversity and composition is associated with patient-reported toxicity during chemoradiation therapy for cervical cancer. Int J Radiat Oncol Biol Phys (2020) 107:163–71. doi: 10.1016/j.ijrobp.2019.12.040
62. Darragh TM, Winkler B. Anal cancer and cervical cancer screening: Key differences. Cancer Cytopathol (2011) 119:5–19. doi: 10.1002/cncy.20126
Keywords: anal cancer, anorectal microbiome, HPV-related cancer, anal dysplasia, cancer biology
Citation: Elnaggar JH, Huynh VO, Lin D, Hillman RT, Abana CO, El Alam MB, Tomasic KC, Karpinets TV, Kouzy R, Phan JL, Wargo J, Holliday EB, Das P, Mezzari MP, Ajami NJ, Lynn EJ, Minsky BD, Morris VK, Milbourne A, Messick CA, Klopp AH, Futreal PA, Taniguchi CM, Schmeler KM and Colbert LE (2023) HPV-related anal cancer is associated with changes in the anorectal microbiome during cancer development. Front. Immunol. 14:1051431. doi: 10.3389/fimmu.2023.1051431
Received: 22 September 2022; Accepted: 13 March 2023;
Published: 29 March 2023.
Edited by:
JoAnn M. Sekiguchi, Michigan Medicine, University of Michigan, United StatesReviewed by:
Oscar Medina-Contreras, Mexico Children’s Hospital, MexicoCopyright © 2023 Elnaggar, Huynh, Lin, Hillman, Abana, El Alam, Tomasic, Karpinets, Kouzy, Phan, Wargo, Holliday, Das, Mezzari, Ajami, Lynn, Minsky, Morris, Milbourne, Messick, Klopp, Futreal, Taniguchi, Schmeler and Colbert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lauren E. Colbert, TENvbGJlcnRAbWRhbmRlcnNvbi5vcmc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.