
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 23 June 2023
Sec. Autoimmune and Autoinflammatory Disorders : Autoimmune Disorders
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1051247
This article is part of the Research Topic Immune Evasion Mechanisms and Their Role in the Pathogenesis of Autoimmune Disorders View all 6 articles
Objective: Interferon induced with helicase C domain 1 (IFIH1) single-nucleotide polymorphisms (SNP) rs1990760, rs3747517, and rs10930046 have been shown to be closely related to the risk of autoimmune diseases. The aim of this study was firstly to examine the association of the rs1990760 with type 1 diabetes (T1D) in a Chinese population. Secondly, to assess the association of SNP rs1990760, rs3747517, and rs10930046 with autoimmune diseases susceptibility.
Methods: A total of 1,273 T1D patients and 1,010 healthy control subjects in a Chinese population were enrolled in this case-control study. Subsequently, we performed a meta-analysis on the association of the SNP rs1990760, rs3747517, and rs10930046 in the IFIH1 gene with susceptibility to autoimmune diseases. The random and fixed genetic effects models were used to evaluate the association and the effect sizes, including odds ratios (OR) and 95% confidence intervals (CI). Stratification analyses based on ethnicity and the type of autoimmune diseases were performed.
Results: IFIH1 SNP rs1990760 was not associated with a significant risk of T1D in the Chinese population in the case-control study. A total of 35 studies including 70,966 patients and 124,509 controls were identified and included in the meta-analysis. The results displayed significant associations between IFIH1 rs1990760 A allele and rs3747517 C allele and autoimmune diseases risk (OR=1.09, 95% CI: 1.01~1.17; OR=1.24, 95% CI: 1.15~1.25, respectively). Stratified analysis indicated a significant association rs1990760 and rs3747517 with autoimmune diseases risk in the Caucasian population (OR=1.11, 95% CI: 1.02~1.20, OR=1.29, 95% CI: 1.18~1.41, respectively).
Conclusions: This study revealed no association between IFIH1 SNP rs1990760 and T1D in Chinese. Furthermore, the meta-analysis indicated that rs1990760 and rs3747517 polymorphisms, confer susceptibility to autoimmune diseases, especially in the Caucasian population.
Autoimmune diseases (AIDs) are multifactorial diseases. Viral infections, genetic predisposition, and environmental factors contribute to the occurrence of AIDs (1, 2). Targeting different immune organs can lead to different AIDs, and sometimes multiple organs are involved. Common AIDs include type 1 diabetes (T1D), Grave’s disease (GD), Hashimoto’s thyroiditis (HT), systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), multiple sclerosis (MS), and autoimmune Addison’s disease (AAD). Recent research has discovered that some AIDs may share several common mechanisms of pathogenesis or signaling pathways (3, 4), with some also sharing the same genetic background (5, 6). A previous genome-wide association study (GWAS) found that interferon-induced with helicase C domain 1 (IFIH1) rs1990760 was the third most associated single nucleotide polymorphism (SNP) with T1D (7, 8). Researchers also identified an association between several SNPs in the IFIH1 gene with the risk of various AIDs, such as SLE (9), GD (10), and psoriatic arthritis (PsA) (11, 12).
IFIH1, also as known as melanoma differentiation-associated 5 (MDA5), participates in the innate immune system’s recognition of viruses and antiviral processes. The IFIH1 gene, located at 2q24.3, encodes an apoptotic-associated protein activated by viral RNA, which acts as a positive regulator in the virus-induced activation of type I interferon (IFN-I) genes (13, 14). IFIH1 can protects against viral challenges but modestly promotes the risk of autoimmunity (15). Extensive researches in different populations have examined the association of SNPs within the coding region of IFIH1 with T1D (16–27). Previous a meta-analysis have confirmed the relationship between IFIH1 SNP rs1990760 and T1D (28); however, few studies have been able to replicate this finding in T1D populations in China. Therefore, There is a need to revalidate the association between the IFIH1 SNP rs1990760 in Chinese patients with T1D in a larger sample size.
Published articles on the association between common IFIH1 gene SNPs, such as rs1990760, rs3747517, and rs10930046, and AIDs are constrained by various factors such as sample sizes, race, and/or clinical heterogeneity; as a result, contradictory or inconsistent conclusions were made (29). Therefore, to overcome the limitations of individual studies, and resolve inconsistencies, we conducted an all-sided meta-analysis of the association between three SNPs, including rs1990760, rs10930046, and rs3747517, in the IFIH1 gene and multiple AIDs including SLE, T1D, GD, HT, RA, AAD, and MS. We included and retrieved the related case-control articles regarding the above seven autoimmune diseases published in recent years. In particular, the meta-analysis of the association between rs1990760 SNP with T1D included the present Chinese case-control study. The second objective in the present study was to assess the overall effect of the three IFIH1 SNPs in AID susceptibility. To our knowledge, this is the most comprehensive meta-analysis investigating the impact of IFIH1 SNPs on AID susceptibility at present.
This study consisted of two parts. Part one is the research data for the case-control study, and the second part is the literature retrieval data for the meta-analysis. In the case-control study for SNP rs1990760, a total of 1,273 T1D patients of Chinese Han origin (688 males and 585 females; average age at onset: 32.2 ± 17.4 years) were enrolled from October 1999 to December 2012. All participants were from the Department of Metabolism and Endocrinology, The Second Xiangya Hospital of Central South University. T1D was diagnosed based on the World Health Organization classification criteria of diabetes in 1999 (30). T1D patients were considered for inclusion as described previously (31, 32). A group of 1,010 control subjects was recruited from two cross-sectional surveys, including 614 males and 396 females, with an average age of 48.4 ± 14.1 years. The study met the international agreements of the World Medical Association Declaration of Helsinki 2000 regarding the ethical principles for medical research involving human subjects. The study protocol was approved by the ethical committee of The Second Xiangya Hospital of Central South University. Written informed consent was obtained from individual subjects.
DNA was extracted from whole blood by the phenol-chloroform method. Genotyping for rs1990760 was performed using TaqMan Assays in a 7900HT Fast Real-Time PCR system under the conditions recommended by the manufacturer (Array ID: C_2780299_30, Applied Biosystems, USA). An ABI Prism 7900HT Sequence Detection System and the SDS 2.2.2 software (both from Applied Biosystems) were used for allele discrimination. TaqMan typing results were verified by the direct sequencing.
The data retrieved from the literature were described below.
Systematic literature searches (up to May 2022) were performed by using the online databases PubMed, Web of Science, and Embase, using the following search strategy: (“IFIH1” OR “MDA5”OR “rs1990760”, OR “rs3747517” OR “rs10930046”) AND (“gene” OR “Polymorphism” OR “genetic” OR “allele” OR “variant” OR “mutation”) AND (“autoimmune diseases” OR “autoimmune disease” OR “type 1 diabetes” OR “systemic lupus erythematosus” OR “Grave’s disease” OR “Hashimoto’s thyroiditis” OR “rheumatoid arthritis” OR “multiple sclerosis” OR “autoimmune Addison’s disease”). We also looked for other relevant studies via the references of all confirmed publications. Studies were included if they 1) investigated the association between IFIH1 polymorphisms (rs1990760, rs3747517, or rs10930046) and AIDs; 2) were case-control studies; and 3) provided complete data to proceed with statistical analysis. Family-based association studies, conference abstracts, case reports, reviews, and comments were excluded.
We extracted the following data: the first author, year of publication, country, ethnicity of the participants, type of autoimmune diseases, number of genotypes in both patients and control groups, source of controls, and typing methods. In some studies (9, 17, 33–37), the genotypic data were obtained from the calculation of minor allele frequency (MAF), the total number of cases, and controls accordingly. Data extraction was done independently by Zilin Xiao and Yuemin Zhou. All differences were resolved by discussion. Data from each health cohort were evaluated for compliance with the Hardy-Weinberg equilibrium (HWE) using the Chi-square test. We used the Newcastle-Ottawa Scale (NOS) system to evaluate the quality of included case-control studies (overall NOS > 5) (38).
The case-control association studies were analyzed using the Chi-square test on 2×2 and 2×3 contingency tables for allele and genotype frequencies, respectively. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using Woolf’s method. Significance was defined by P < 0.05.
The whole process of the meta-analysis were conducted by STATA software (version 12.0, Stata Corporation, College Station, TX, USA). For rs1990760, rs3747517, and rs10930046, we performed homozygote (AA vs. GG; CC vs. TT; TT vs CC) and heterozygote (AA vs. AG; CC vs CT; TT vs CT) models, while the allele model (A vs. G; C vs. T; T vs. C), dominant model (AA + AG vs. GG; CC + CT vs. TT; TT + CT vs. TT), and recessive model (AA vs. AG + GG; CC vs. CT + TT; TT vs. CT + CC) were analyzed by the method of Mantel-Haenszel analysis. PHWE < 0.05 was considered statistically significant.
The pooled ORs and 95% CIs were used to assess the association between IFIH1 SNPs and AIDs. The Z-test was used to examine the significance of the pooled ORs. Higgins’ I2 statistic and the Cochran’s Q test were used to evaluate the heterogeneity of included studies. If Pheterogeneity > 0.1 and I2 < 50.0%, heterogeneity among studies were excluded, and a favorable fixed-effects model was applied in the further meta-analysis; if not, the random-effects model was used. Subgroup analyses and sensitivity analyses were used to explore the source of heterogeneity. Regarding publication bias, a funnel plot was utilized for qualitative analysis, and Begg’s test and Egger’s test for quantitative analysis. In cases where PEgger < 0.05, the trim and fill method was used according to previous study. Power tests were performed using the G*Power software v.3.1.9.7. For the calculation of Power in our meta-analysis, the parameter effect size was converted from the corresponding OR value of each study according the previous studies (39).
The clinical characteristics and laboratory data of patients with T1D showed in Supplementary Table 1. The genotype distributions from both T1D patients and controls were in HWE. To evaluate any associations between IFIH1 rs1990760 and T1D, ORs for T1D in A/G allele and genotype, respectively, were calculated together with their corresponding 95% CI with the cohorts indicated in Table 1 (α=0.05, power 100%). The results showed there was no significant association between the rs1990760 and T1D in the Chinese population (P > 0.05). Subsequent meta-analyses included this data. In addition, we divided the patients into two subgroups including the islet autoantibodies positive group and the negative group. However, no statistically significant association was found between these groups.

Table 1 Genotype and allele frequencies for rs1990760 in patients with T1D in Chinese association study.
We primarily searched a total of 284 potential articles from Pubmed (n=97), Web of Science (n=89), and Embase (n=98). We then excluded repeated studies (n=169). After screening the titles and abstracts, 39 articles were reviews, abstracts, comments, or case reports and 73 articles were not about the association between IFIH1 polymorphisms and AIDs. Next, we read the full text and excluded eight studies with incomplete or repeated data. Nine studies were based on family, four studies included non-specific SNPs, and one study showed severe deviation from the HWE. Finally, 10 studies reported rs1990760, one study reported rs3747517, one study reported rs10930046, seven studies reported both rs1990760 and rs3747517, one study reported rs1990760 and rs10930046, and seven studies reported all three SNPs. The flow diagram of the study selection was shown in Figure 1.
Note that among the included studies, there were three multicenter case-control studies and seven studies including more than one autoimmune endocrine disease. One article’s genotype information was from a previous meta-analysis (40). Given the IFIH1 rs3747517 polymorphism sample size, one study was selected (37), though it showed a slight deviation from HWE in controls (0.01 ≤ P < 0.05). One cohort was eliminated for deviating significantly (P < 0.05) from HWE in controls; however, we reserved another cohort in this article (19). People with one AID often had several AIDs simultaneously (21, 41). In an article that included both black and white participants, white participants were in the majority, thus, the data were categorized as white when racially stratified (20). In some studies, where subjects were divided into multiple subgroups, we included data from each subgroup as a separate study. Finally, 35 studies and 66 case-control comparisons were considered to meet the inclusion criteria, including eleven T1D studies (16–24, 29, 40), twelve SLE studies (9, 34–36, 42–49), eight GD studies (10, 24, 25, 50–53), three HT studies (51–53), five MS studies (16, 24, 33, 37, 54), two RA studies (55, 56), and three AAD studies (10, 41, 51), in total including 70,966 cases and 124,509 controls. The characteristics of the selected studies are summarized in Table 2.
The results of the meta-analysis for a possible association of the SNP rs1990760, rs10930046, and rs3747517 in the IFIH1 gene with seven AIDs were summarized in Table 3 and Supplementary Table 2.
For the IFIH1 rs1990760 polymorphism, 33 case-control studies containing 47 independent cohorts (39,658 cases and 76,434 controls) were included. The pooled ORs of all qualified case-control studies showed a significant association: allele model (A vs. G, OR=1.09, 95% CI: 1.01~1.17), dominant model (AA+AG vs. GG, OR=1.17, 95% CI: 1.10~1.24), recessive model (AA vs. AG+GG, OR=1.16, 95% CI: 1.10~1.22), homozygote (AA vs. GG, OR=1.26, 95% CI: 1.17~1.36), and heterozygote (AA vs. AG, OR=1.11, 95% CI: 1.06~1.17).
Subsequent subgroup analyses by diseases showed that among T1D, SLE, and GD, a significant association was observed in some genetic models. For example, in the dominant model (T1D: OR=1.23, 95% CI: 1.09~1.38; SLE: OR=1.19, 95% CI: 1.10~1.29; GD: OR=1.28, 95% CI: 1.07~1.53, respectively) (Figure 2). For AAD, no association was detected in any genetic models. But it is worth to mention that AAD association analysis was underpowered in all genetic models tested. RA showed a significant association only in the allele model (OR=1.12, 95% CI: 1.00~1.24). After stratifying by ethnicity, a significant positive association between AIDs and the IFIH1 rs1990760 polymorphism was observed in the Caucasian population. However, for Asians, a significant association was observed only in the dominant model (OR=1.11, 95% CI: 1.02~1.20). The overall analysis suggested the existence of heterogeneity. Subgroup analyses reduced the heterogeneity for many diseases using the dominant and recessive models, but significant heterogeneity remained with the allelic model, even after subgroup analyses. Meanwhile, for HT and MS, the sensitivity analysis indicated a good stability for the estimated OR (Supplementary Figure 1). However, it should be pointed out that HT had a low power to detect a significant association depending on the model and MS was also underpowered for the recessive and allelic models.
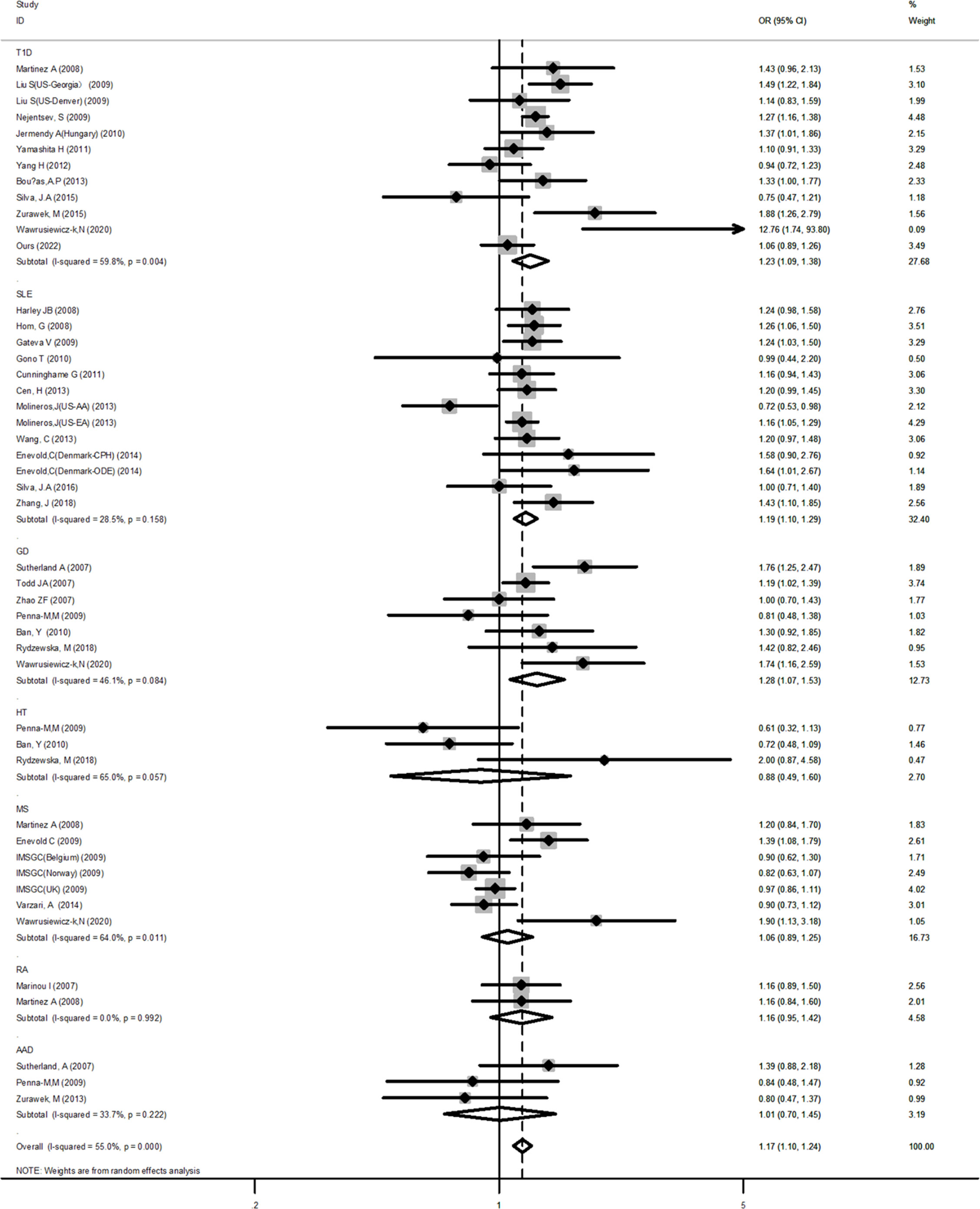
Figure 2 Forest plot for the meta-analysis of the association between the IFIH1 rs1990760 polymorphism and diseases in the subgroup of autoimmune diseases. Synthesized data are based on 33 case-control studies containing 47 independent cohorts that reported IFIH1 rs1990760 genotypic data, by the dominant model (AA+AG vs GG).
For the IFIH1 rs3747517 polymorphism, 13 case-control studies containing 14 independent cohorts (15,440 cases and 20,522 controls) were comprehensively compared. Due to the heterogeneity, all genetic models, except the heterozygote, used a random-effect model in the overall analysis. Results of the meta-analysis on associations between the IFIH1 rs3747517 polymorphism and AIDs showed a significant association in the allele model (C vs. T, OR=1.24, 95% CI: 1.15~1.25), dominant model (CC+CT vs. TT, OR=1.40, 95% CI: 1.22~1.72), recessive model (CC vs. CT+TT, OR=1.23, 95% CI: 1.14~1.32), homozygote (CC vs. TT, OR=1.52, 95% CI: 1.27~1.81), and heterozygote (CC vs. CT, OR=1.16, 95% CI: 1.11~1.22).
To further analyze the source of heterogeneity, subgroup analyses and sensitivity analyses were performed. After stratifying by diseases, T1D, SLE, and MS suggested a significant positive correlation (OR=1.30, 95% CI: 1.11~1.52; OR=1.19, 95% CI: 1.09~1.29; OR=1.19, 95% CI: 1.07~1.32; respectively, in the allele model). However, other AIDs did not show a statistically significant difference. Regarding ethnicity, both the Asian and Caucasian populations had significant associations in the above three models (OR=1.94, 95% CI: 1.16~3.24; OR=1.29, 95% CI: 1.18~1.41; respectively, in the dominant model). However, stratification by ethnicity could not explain the source of heterogeneity in the allele model and Asian in the dominant model. We performed a sensitivity analysis to evaluate the influence of the allele model and dominant model, which indicated a good stability for the estimated OR (Figure 3).
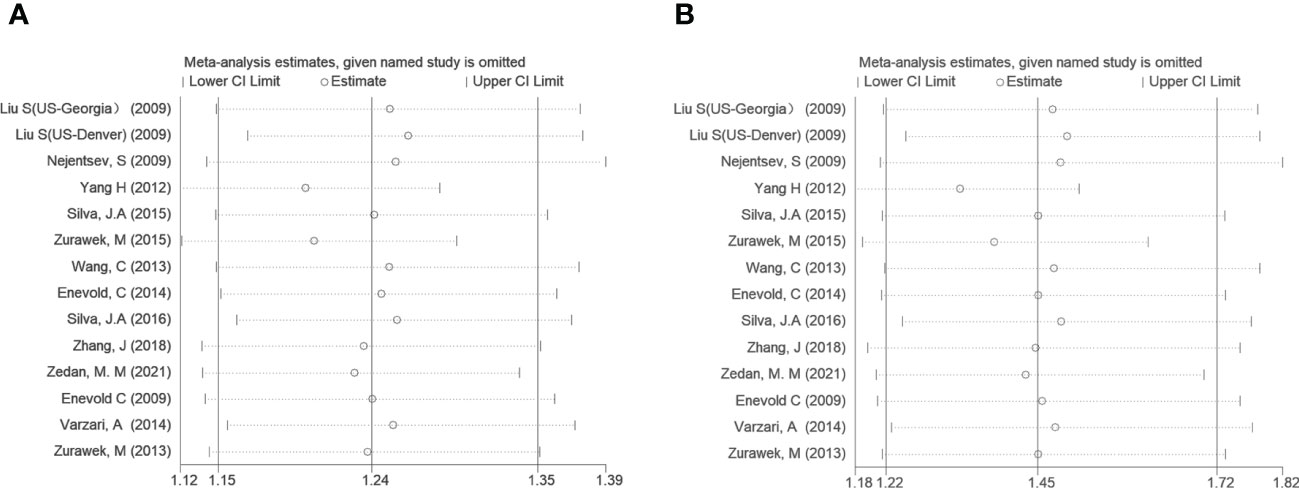
Figure 3 Sensitivity tests of IFIH1 rs3747517 in the allele model (A) and dominant model (B). Spots and horizontal line segments stand for estimated odds ratio (OR) values and 95% confidence intervals (CIs) of pooled data after omitting the labeled study in each line from the entire data pool. All estimated 95% CIs of OR ranges were above one, indicating a stable conclusion in the meta-analysis.
Seven research articles, including eight cohorts (15,868 cases and 27,553 controls), were enrolled. Pooled analysis detected a significant association in the dominant model (TT+TC vs. CC, OR=1.33, 95% CI: 1.15~1.55) and homozygote model (TT vs. CC, OR=1.48, 95% CI: 1.24~1.74) with the fixed-effect model. However, no association was observed among the allele model (T vs. C), heterozygote model (TT vs. TC), and the recessive model (TT vs. TC+CC) (all P>0.05).
There was no significant heterogeneity detected in the dominant model. The ethnicity subgroup analysis suggested inverse outcomes for Caucasians (OR=0.82, 95% CI: 0.72~0.92) and Africans (OR=1.33, 95% CI: 1.18~1.50) in the recessive model. Meanwhile, the problem of heterogeneity was solved. In the dominant model, only the African subgroup showed a statistical difference (OR=1.38, 95% CI: 1.17~1.63), and in the allele model, only the Caucasian subgroup showed statistical difference (OR=0.85, 95% CI: 0.75~0.95). The analysis stratified by the type of AID indicated that the SLE (OR=1.37, 95% CI: 1.17~1.60) subgroup and the T1D (OR=0.84, 95% CI: 0.70~0.99) subgroup had statistical significance. The results of the sensitivity analysis were shown in Supplementary Figure 2, which indicated a slight lack of stability for the estimated OR because of the limited sample sizes.
The funnel plot of the IFIH1 rs1990760 and rs3747517 polymorphisms indicated good symmetry, although rs10930046 did not. Publication bias was assessed using Begg’s test and Egger’s test (Table 4). IFIH1 rs1990760 and rs3747517 showed no statistically significant bias under all models in both tests, while rs10930046 indicated statistically significant bias in different models (in the dominant model PBegg’s = 0.035; in the recessive model PEgger’s = 0.048). Therefore, we used a trim and fill approach to estimate the number of missing studies and incorporated missing hypothetical studies to recalculate the pooled OR values. However, no missing hypothetical studies were shown (Figure 4).
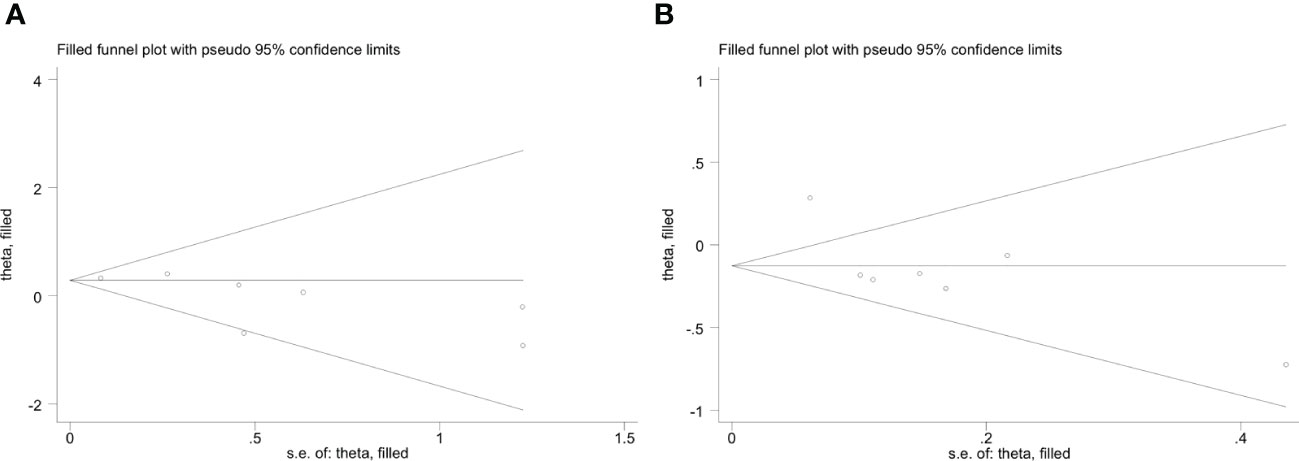
Figure 4 Trim and fill method funnel plots of IFIH1 rs10930046 in the dominant model (A) and recessive model (B). (A) Trim and fill method funnel plots of IFIH1 rs10930046 in the dominant model. Symmetry was verified by Begg’s test with PBegg’s = 0.035. (B) Trim and fill method funnel plots of IFIH1 rs10930046 in the recessive model. Symmetry was verified by Egger’s test with PEgger = 0.048.
As a gene closely related to the pathogenesis of AIDs, IFIH1 has attracted much attention. Herein, we performed a risk association study between the IFIH1 rs1990760 polymorphism and T1D in the Chinese population. Next, we carried out a comprehensive meta-analysis, including seven autoimmune diseases (T1D, SLE, GD, HT, RA, MS, and AAD), three SNPs of IFIH1 (rs1990760, rs3747517, and rs10930046), and three ethnicities (Caucasian, Asian, and African), to investigate whether IFIH1 polymorphisms were associated with AID susceptibility.
HLA, INS, and PTPN22 are commonly demonstrated risk loci in T1D; another seven loci associated with a low polygenic risk score in T1D were identified in the updated GWAS (57). Recent GWAS and expression quantitative trait locus (eQTL) association studies have provided some new insights related to gene expression variation that may help to explain T1D susceptibility and phenotypic diversity (58). The GWAS, in addition to further verification in a case–control and family collection study, has shown evidence of the involvement of the IFIH1 gene SNP rs1990760 in the risk of T1D (7, 59). Meta-analyses have also showed that IFIH1 SNPs are related to T1D (20, 22), SLE (36), and PsA (60). Subsequent associations with IFIH1 have been observed in other AIDs. The meta-analysis by Cen et al., which included literature published before 2013, estimated the association between AID risk and the IFIH1 rs1990760 polymorphism, demonstrating that the IFIH1 rs1990760 confers a risk of SLE, T1D, RA, and MS (45). Previous meta-analyses have mostly focused on one or two AIDs or just one polymorphism, whereas the present study aimed to comprehensively examine the possible association between three common IFIH1 polymorphisms (rs1990760, rs3747517, and rs10930046) and the risk of seven common AIDs. In addition, we included studies published in recent years to ensure that we reported the latest developments.
In the current study, we did not find a significant association between the IFIH1 SNP rs1990760 and T1D in a Chinese population. The results were consistent with two previous Asian studies (29, 40). The reason for this result may be the small population size, therefore, more studies based on Asian populations are needed in the future. The result of the current meta-analysis showed that the IFIH1 rs1990760 allele A and its homozygote and heterozygote significantly increased the overall AID risk. The IFIH1 SNP rs3747517 was also associated with AID susceptibility. Stratification by ethnicity and diseases could explain some of the heterogeneity. Begg’s test and Egger’s test showed no publication bias in rs1990760 and rs3747517. However, for IFIH1 rs10930046, it was found to affect susceptibility to overall AID risk only in the homozygote and dominant model. Unexpectedly, there were some opposite susceptibility results within different disease types and between ethnic groups. This may be partly due to the lack of sufficient research and sample sizes from different sources in the subgroup analysis.
The subgroup analysis result of SNP rs1990760 suggested that it was associated with T1D, SLE, GD, and RA. However, we did not find a correlation between IFIH1 and susceptibility to MS. These results differ from the results of Cen et al., which is probably because we included some newly published articles about GD and MS in our analysis. In addition, we indicated an association between rs1990760 and AID risk in Caucasians, whereas in Asians, only the dominant model showed significant statistical differences. The reason for the ethnic differences may be related to sunlight and other environmental factors, or it may be due to the insufficient sample size in the Asian studies. Rs1990760 and rs3747517 polymorphisms were associated with T1D by GWAS (7, 17, 18, 61); however, GWAS pays more attention to European populations. Therefore, a larger sample size is needed to investigate the association between IFIH1 and AID susceptibility in Asian populations.
We cannot help but wonder if there is a common pathogenesis in these AIDs, and exploration of these mechanisms will help us to understand these diseases more deeply. Several recent studies have discussed this issue. One study analyzed two clinical trials showing the most common shared-targeting molecules and pathways, including the Janus kinase (JAK) signal transducer and transcriptional activator (STAT) pathways (4). The JAK-STAT signaling pathway revealed abnormal STAT signaling in inflammatory conditions and AIDs (62). IFIH1 can augment IFN-α production (13) and the combination of IFN-α and IFNAR can animate the JAK-STAT pathway and induce the transcription of corresponding genes. Thus, IFIH1 may induce the occurrence of AIDs through the JAK-STAT signaling pathway. One study has defined a common gene expression signature for some AIDs (63), and another has shown common genetic characteristics linked to AIDs (3). Meanwhile, IFIH1 can activate the IFN-I response and pro-inflammatory cytokines after recognition of an enterovirus (64). Enterovirus infection may be associated with AIDs such as T1D (65).
There are some strengths and limitations to the current study. A strength is that our meta-analysis included 35 studies and 66 case–control comparisons, and we assessed seven AIDs and three IFIH1 SNPs; therefore, it is the most comprehensive study of its kind to date. This could increase the statistical power of the overall analysis. However, we also note several study limitations. First, Caucasians constitute a large majority of the population in this study, which may lead to unavoidable geographical and ethnic bias. When performing the subgroup analyses, other races had far fewer cases and documents, thus the comparison may not be appropriate. Second, the number of relevant SNP rs10930046 studies was too small. If the meta-analysis included less than 20 original studies, the sensitivities of Begg’s test and Egger’s test were poor. rs1090046 has not shown significant differences in any single case–control study so far; therefore, statistical differences in a single model or stratification are not conclusive. Finally, we found that the original data included in the current meta-analysis were limited, so the influence of factors such as environment and population migration on gene and linkage disequilibrium remains unknown.
In conclusion, this meta-analysis showed that the IFIH1 rs1990760 polymorphism A allele and IFIH1 rs3747517 polymorphism C allele were associated with AIDs. Subsequent stratified analyses found that the rs1990760 polymorphism was associated with T1D, SLE, GD, and RA in different populations. SNP rs3747517 was associated with T1D, SLE, and MS. AIDs risk in both Caucasians and Asians was associated with the IFIH1 rs3747517 polymorphism. Results from IFIH1 rs10930046 did not reach universal statistical significance in our analysis.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by ethical committee of the Second Xiangya Hospital of Central South University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
All authors contributed to the manuscript. ZLX wrote the first draft of the manuscript. SL and ZLX performed the material preparation. YZ and ZLX performed data collection. HP and WY contributed to part of the methods and tables. JQ and ZZ reviewed the manuscript and contributed to the discussion. SL proposed the project and is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors read and approved the final manuscript.
This study was supported by National Key R&D Program of China (2022YFC3603000, 2018YFE0114500), and Hunan Province Health and Wellness High-Level Talent Support Program Funding.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1051247/full#supplementary-material
1. Fujinami RS, von Herrath MG, Christen U, Whitton JL. Molecular mimicry, bystander activation, or viral persistence: infections and autoimmune disease. Clin Microbiol Rev (2006) 19(1):80–94. doi: 10.1128/CMR.19.1.80-94.2006
2. Incorvaia E, Sicouri L, Petersen-Mahrt SK, Schmitz KM. Hormones and AID: balancing immunity and autoimmunity. Autoimmunity (2013) 46(2):128–37. doi: 10.3109/08916934.2012.748752
3. Wang Y, Chen S, Chen J, Xie X, Gao S, Zhang C, et al. Germline genetic patterns underlying familial rheumatoid arthritis, systemic lupus erythematosus and primary sjögren's syndrome highlight T cell-initiated autoimmunity. Ann Rheum Dis (2020) 79(2):268–75. doi: 10.1136/annrheumdis-2019-215533
4. Petitdemange A, Blaess J, Sibilia J, Felten R, Arnaud L. Shared development of targeted therapies among autoimmune and inflammatory diseases: a systematic repurposing analysis. Ther Adv Musculoskelet Dis (2020) 12:1759720x20969261. doi: 10.1177/1759720X20969261
5. Frommer L, Kahaly GJ. Type 1 diabetes and autoimmune thyroid disease-the genetic link. Front Endocrinol (2021) 12:618213. doi: 10.3389/fendo.2021.618213
6. Wu H, Liao J, Li Q, Yang M, Zhao M, Lu Q. Epigenetics as biomarkers in autoimmune diseases. Clin Immunol (Orlando Fla) (2018) 196:34–9. doi: 10.1016/j.clim.2018.03.011
7. Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet (2006) 38(6):617–9. doi: 10.1038/ng1800
8. Robertson CC, Inshaw JRJ, Onengut-Gumuscu S, Chen WM, Santa Cruz DF, Yang H, et al. Fine-mapping, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. Nat Genet (2021) 53(7):962–71. doi: 10.1038/s41588-021-00880-5
9. Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet (2008) 40(2):204–10. doi: 10.1038/ng.81
10. Sutherland A, Davies J, Owen CJ, Vaikkakara S, Walker C, Cheetham TD, et al. Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to graves' disease susceptibility. J Clin Endocrinol Metab (2007) 92(8):3338–41. doi: 10.1210/jc.2007-0173
11. Yin X, Low HQ, Wang L, Li Y, Ellinghaus E, Han J, et al. Genome-wide meta-analysis identifies multiple novel associations and ethnic heterogeneity of psoriasis susceptibility. Nat Commun (2015) 6:6916. doi: 10.1038/ncomms7916
12. Stuart PE, Nair RP, Tsoi LC, Tejasvi T, Das S, Kang HM, et al. Genome-wide association analysis of psoriatic arthritis and cutaneous psoriasis reveals differences in their genetic architecture. Am J Hum Genet (2015) 97(6):816–36. doi: 10.1016/j.ajhg.2015.10.019
13. Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol (Baltimore Md 1950) (2005) 175(5):2851–8. doi: 10.4049/jimmunol.175.5.2851
14. Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature (2006) 441(7089):101–5. doi: 10.1038/nature04734
15. Gorman JA, Hundhausen C, Errett JS, Stone AE, Allenspach EJ, Ge Y, et al. The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat Immunol (2017) 18(7):744–52. doi: 10.1038/ni.3766
16. Martínez A, Santiago JL, Cénit MC, de Las Heras V, de la Calle H, Fernández-Arquero M, et al. IFIH1-GCA-KCNH7 locus: influence on multiple sclerosis risk. Eur J Hum Genet EJHG (2008) 16(7):861–4. doi: 10.1038/ejhg.2008.16
17. Liu S, Wang H, Jin Y, Podolsky R, Reddy MV, Pedersen J, et al. IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Hum Mol Genet (2009) 18(2):358–65. doi: 10.1093/hmg/ddn342
18. Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Sci (New York NY) (2009) 324(5925):387–9. doi: 10.1126/science.1167728
19. Jermendy A, Szatmári I, Laine AP, Lukács K, Horváth KH, Körner A, et al. The interferon-induced helicase IFIH1 Ala946Thr polymorphism is associated with type 1 diabetes in both the high-incidence Finnish and the medium-incidence Hungarian populations. Diabetologia (2010) 53(1):98–102. doi: 10.1007/s00125-009-1561-y
20. Bouças AP, Brondani LA, Souza BM, Lemos NE, de Oliveira FS, Canani LH, et al. The a allele of the rs1990760 polymorphism in the IFIH1 gene is associated with protection for arterial hypertension in type 1 diabetic patients and with expression of this gene in human mononuclear cells. PloS One (2013) 8(12):e83451. doi: 10.1371/journal.pone.0083451
21. Moura R, Araujo J, Guimarães R, Crovella S, Brandão L. Interferon induced with helicase c domain 1 (IFIH1): trends on helicase domain and type 1 diabetes onset. Gene (2013) 516(1):66–8. doi: 10.1016/j.gene.2012.11.066
22. Silva JA, Tavares NA, Santos MM, Moura R, Guimarães RL, Araújo J, et al. Meta-analysis of STAT4 and IFIH1 polymorphisms in type 1 diabetes mellitus patients with autoimmune polyglandular syndrome type III. Genet Mol Res GMR (2015) 14(4):17730–8. doi: 10.4238/2015.December.21.46
23. Zurawek M, Fichna M, Fichna P, Skowronska B, Dzikiewicz-Krawczyk A, Januszkiewicz D, et al. Cumulative effect of IFIH1 variants and increased gene expression associated with type 1 diabetes. Diabetes Res Clin Pract (2015) 107(2):259–66. doi: 10.1016/j.diabres.2014.11.008
24. Wawrusiewicz-Kurylonek N, Gościk J, Chorąży M, Siewko K, Posmyk R, Zajkowska A, et al. The interferon-induced helicase c domain-containing protein 1 gene variant (rs1990760) as an autoimmune-based pathology susceptibility factor. Immunobiology (2020) 225(1):151864. doi: 10.1016/j.imbio.2019.10.013
25. Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet (2007) 39(7):857–64. doi: 10.1038/ng2068
26. Winkler C, Lauber C, Adler K, Grallert H, Illig T, Ziegler AG, et al. An interferon-induced helicase (IFIH1) gene polymorphism associates with different rates of progression from autoimmunity to type 1 diabetes. Diabetes (2011) 60(2):685–90. doi: 10.2337/db10-1269
27. Klinker MW, Schiller JJ, Magnuson VL, Wang T, Basken J, Veth K, et al. Single-nucleotide polymorphisms in the IL2RA gene are associated with age at diagnosis in late-onset Finnish type 1 diabetes subjects. Immunogenetics. (2010) 62(2):101–7. doi: 10.1007/s00251-009-0417-4
28. Cen H, Wang W, Leng RX, Wang TY, Pan HF, Fan YG, et al. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity (2013) 46(7):455–62. doi: 10.3109/08916934.2013.796937
29. Yang H, Wang Z, Xu K, Gu R, Chen H, Yu D, et al. IFIH1 gene polymorphisms in type 1 diabetes: genetic association analysis and genotype-phenotype correlation in Chinese han population. Autoimmunity (2012) 45(3):226–32. doi: 10.3109/08916934.2011.633134
30. Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabetic Med (1998) 15(7):539–53. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S
31. Luo S, Li X, Huang G, Xie Z, Xiang Y, Dai Z, et al. Distinct two different ages associated with clinical profiles of acute onset type 1 diabetes in Chinese patients. Diabetes/Metab Res Rev (2020) 36(2):e3209. doi: 10.1002/dmrr.3209
32. Niu X, Luo S, Li X, Xie Z, Xiang Y, Huang G, et al. Identification of a distinct phenotype of elderly latent autoimmune diabetes in adults: LADA China study 8. Diabetes/Metab Res Rev (2019) 35(1):e3068. doi: 10.1002/dmrr.3068
33. (IMSGC) IMSGC. The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun (2009) 10(1):11–4. doi: 10.1038/gene.2008.83
34. Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet (2009) 41(11):1228–33. doi: 10.1038/ng.468
35. Molineros JE, Maiti AK, Sun C, Looger LL, Han S, Kim-Howard X, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PloS Genet (2013) 9(2):e1003222. doi: 10.1371/journal.pgen.1003222
36. Silva JA, Lima SC, Addobbati C, Moura R, Brandão LA, Pancoto JA, et al. Association of interferon-induced helicase c domain (IFIH1) gene polymorphisms with systemic lupus erythematosus and a relevant updated meta-analysis. Genet Mol Res (2016) 15(4):gmr15048008. doi: 10.4238/gmr15048008
37. Varzari A, Bruch K, Deyneko IV, Chan A, Epplen JT, Hoffjan S. Analysis of polymorphisms in RIG-i-like receptor genes in German multiple sclerosis patients. J Neuroimmunol (2014) 277(1-2):140–4. doi: 10.1016/j.jneuroim.2014.09.015
38. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol (2010) 25(9):603–5. doi: 10.1007/s10654-010-9491-z
39. Chinn S. A simple method for converting an odds ratio to effect size for use in meta-analysis. Stat Med (2000) 19(22):3127–31. doi: 10.1002/1097-0258(20001130)19:22<3127::AID-SIM784>3.0.CO;2-M
40. Yamashita H, Awata T, Kawasaki E, Ikegami H, Tanaka S, Maruyama T, et al. Analysis of the HLA and non-HLA susceptibility loci in Japanese type 1 diabetes. Diabetes/Metab Res Rev (2011) 27(8):844–8. doi: 10.1002/dmrr.1234
41. Zurawek M, Fichna M, Januszkiewicz D, Fichna P, Nowak J. Polymorphisms in the interferon-induced helicase (IFIH1) locus and susceptibility to addison's disease. Clin Endocrinol (2013) 78(2):191–6. doi: 10.1111/j.1365-2265.2012.04497.x
42. Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. New Engl J Med (2008) 358(9):900–9. doi: 10.1056/NEJMoa0707865
43. Gono T, Kawaguchi Y, Sugiura T, Furuya T, Kawamoto M, Hanaoka M, et al. Interferon-induced helicase (IFIH1) polymorphism with systemic lupus erythematosus and dermatomyositis/polymyositis. Modern Rheumatol (2010) 20(5):466–70. doi: 10.3109/s10165-010-0311-9
44. Cunninghame Graham DS, Morris DL, Bhangale TR, Criswell LA, Syvänen AC, Rönnblom L, et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PloS Genet (2011) 7(10):e1002341. doi: 10.1371/journal.pgen.1002341
45. Cen H, Leng RX, Wang W, Zhou M, Feng CC, Zhu Y, et al. Association study of IFIH1 rs1990760 polymorphism with systemic lupus erythematosus in a Chinese population. Inflammation (2013) 36(2):444–8. doi: 10.1007/s10753-012-9564-0
46. Wang C, Ahlford A, Laxman N, Nordmark G, Eloranta ML, Gunnarsson I, et al. Contribution of IKBKE and IFIH1 gene variants to SLE susceptibility. Genes Immun (2013) 14(4):217–22. doi: 10.1038/gene.2013.9
47. Enevold C, Kjær L, Nielsen CH, Voss A, Jacobsen RS, Hermansen ML, et al. Genetic polymorphisms of dsRNA ligating pattern recognition receptors TLR3, MDA5, and RIG-i. association with systemic lupus erythematosus and clinical phenotypes. Rheumatol Int (2014) 34(10):1401–8. doi: 10.1007/s00296-014-3012-4
48. Zhang J, Liu X, Meng Y, Wu H, Wu Y, Yang B, et al. Autoimmune disease associated IFIH1 single nucleotide polymorphism related with IL-18 serum levels in Chinese systemic lupus erythematosus patients. Sci Rep (2018) 8(1):9442. doi: 10.1038/s41598-018-27782-7
49. Zedan MM, Attia ZR, Abd El Azeem RA, Mutawi TM, El Shehawy AS, Bakr A. Genetic polymorphisms in genes involved in the type I interferon system (IFIH1/MDA-5, TNFAIP3/A20, and STAT4): association with SLE risk in Egyptian children and adolescents. J Inflammation Res (2021) 14:3349–58. doi: 10.2147/JIR.S309008
50. Zhao ZF, Cui B, Chen HY, Wang S, Li I, Gu XJ, et al. The A946T polymorphism in the interferon induced helicase gene does not confer susceptibility to graves' disease in Chinese population. Endocrine (2007) 32(2):143–7. doi: 10.1007/s12020-007-9024-z
51. Penna-Martinez M, Ramos-Lopez E, Robbers I, Kahles H, Hahner S, Willenberg H, et al. The rs1990760 polymorphism within the IFIH1 locus is not associated with graves' disease, hashimoto's thyroiditis and addison's disease. BMC Med Genet (2009) 10:126. doi: 10.1186/1471-2350-10-126
52. Ban Y, Tozaki T, Taniyama M, Nakano Y, Ban Y, Hirano T. Genomic polymorphism in the interferon-induced helicase (IFIH1) gene does not confer susceptibility to autoimmune thyroid disease in the Japanese population. Hormone Metab Res = Hormon- und Stoffwechselforschung = Hormones Metabolisme (2010) 42(1):70–2. doi: 10.1055/s-0029-1238293
53. Rydzewska M, Góralczyk A, Gościk J, Wawrusiewicz-Kurylonek N, Bossowska A, Krętowski A, et al. Analysis of chosen polymorphisms rs2476601 a/G - PTPN22, rs1990760 C/T - IFIH1, rs179247 a/G - TSHR in pathogenesis of autoimmune thyroid diseases in children. Autoimmunity (2018) 51(4):183–90. doi: 10.1080/08916934.2018.1486824
54. Enevold C, Oturai AB, Sørensen PS, Ryder LP, Koch-Henriksen N, Bendtzen K. Multiple sclerosis and polymorphisms of innate pattern recognition receptors TLR1-10, NOD1-2, DDX58, and IFIH1. J Neuroimmunol (2009) 212(1-2):125–31. doi: 10.1016/j.jneuroim.2009.04.008
55. Marinou I, Montgomery DS, Dickson MC, Binks MH, Moore DJ, Bax DE, et al. The interferon induced with helicase domain 1 A946T polymorphism is not associated with rheumatoid arthritis. Arthritis Res Ther (2007) 9(2):R40. doi: 10.1186/ar2179
56. Martínez A, Varadé J, Lamas JR, Fernández-Arquero M, Jover JA, de la Concha EG, et al. Association of the IFIH1-GCA-KCNH7 chromosomal region with rheumatoid arthritis. Ann Rheum Dis (2008) 67(1):137–8. doi: 10.1136/ard.2007.073213
57. Qu J, Qu HQ, Bradfield JP, Glessner JT, Chang X, Tian L, et al. Insights into non-autoimmune type 1 diabetes with 13 novel loci in low polygenic risk score patients. Sci Rep (2021) 11(1):16013. doi: 10.1038/s41598-021-94994-9
58. Yu CH, Pal LR, Moult J. Consensus genome-wide expression quantitative trait loci and their relationship with human complex trait disease. Omics (2016) 20(7):400–14. doi: 10.1089/omi.2016.0063
59. Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet (2009) 41(6):703–7. doi: 10.1038/ng.381
60. Budu-Aggrey A, Bowes J, Stuart PE, Zawistowski M, Tsoi LC, Nair R, et al. A rare coding allele in IFIH1 is protective for psoriatic arthritis. Ann Rheum Dis (2017) 76(7):1321–4. doi: 10.1136/annrheumdis-2016-210592
61. Concannon P, Onengut-Gumuscu S, Todd JA, Smyth DJ, Pociot F, Bergholdt R, et al. A human type 1 diabetes susceptibility locus maps to chromosome 21q22.3. Diabetes (2008) 57(10):2858–61. doi: 10.2337/db08-0753
62. Goropevšek A, Holcar M, Avčin T. The role of STAT signaling pathways in the pathogenesis of systemic lupus erythematosus. Clin Rev Allergy Immunol (2017) 52(2):164–81. doi: 10.1007/s12016-016-8550-y
63. Toro-Domínguez D, Carmona-Sáez P, Alarcón-Riquelme ME. Shared signatures between rheumatoid arthritis, systemic lupus erythematosus and sjögren's syndrome uncovered through gene expression meta-analysis. Arthritis Res Ther (2014) 16(6):489. doi: 10.1186/s13075-014-0489-x
64. Chistiakov DA. Interferon induced with helicase c domain 1 (IFIH1) and virus-induced autoimmunity: a review. Viral Immunol (2010) 23(1):3–15. doi: 10.1089/vim.2009.0071
Keywords: IFIH1, polymorphism, autoimmune diseases, type 1 diabetes, meta-analysis
Citation: Xiao Z, Luo S, Zhou Y, Pang H, Yin W, Qin J, Xie Z and Zhou Z (2023) Association of the rs1990760, rs3747517, and rs10930046 polymorphisms in the IFIH1 gene with susceptibility to autoimmune diseases: a meta-analysis. Front. Immunol. 14:1051247. doi: 10.3389/fimmu.2023.1051247
Received: 22 September 2022; Accepted: 08 June 2023;
Published: 23 June 2023.
Edited by:
Fabiana Rizzo, National Institute of Health (ISS), ItalyReviewed by:
Karen Cerosaletti, Benaroya Research Institute, United StatesCopyright © 2023 Xiao, Luo, Zhou, Pang, Yin, Qin, Xie and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuoming Luo, c2h1b21pbmdsdW9AY3N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.