
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 23 March 2023
Sec. Cancer Immunity and Immunotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1038651
This article is part of the Research TopicBiomarkers and Immunotherapy for Genitourinary TumorsView all 30 articles
Background: Regarding the global coronavirus disease 2019 (COVID)-19 pandemic, kidney clear cell carcinoma (KIRC) has acquired a higher infection probability and may induce fatal complications and death following COVID-19 infection. However, effective treatment strategies remain unavailable. Berberine exhibits significant antiviral and antitumour effects. Thus, this study aimed to provide a promising and reliable therapeutic strategy for clinical decision-making by exploring the therapeutic mechanism of berberine against KIRC/COVID-19.
Methods: Based on large-scale data analysis, the target genes, clinical risk, and immune and pharmacological mechanisms of berberine against KIRC/COVID-19 were systematically investigated.
Results: In total, 1,038 and 12,992 differentially expressed genes (DEGs) of COVID-19 and KIRC, respectively, were verified from Gene Expression Omnibus and The Cancer Genome Atlas databases, respectively, and 489 berberine target genes were obtained from official websites. After intersecting, 26 genes were considered potential berberine therapeutic targets for KIRC/COVID-19. Berberine mechanism of action against KIRC/COVID-19 was revealed by protein-protein interaction, gene ontology, and Kyoto Encyclopedia of Genes and Genomes with terms including protein interaction, cell proliferation, viral carcinogenesis, and the PI3K/Akt signalling pathway. In COVID-19 patients, ACOX1, LRRK2, MMP8, SLC1A3, CPT1A, H2AC11, H4C8, and SLC1A3 were closely related to disease severity, and the general survival of KIRC patients was closely related to ACOX1, APP, CPT1A, PLK1, and TYMS. Additionally, the risk signature accurately and sensitively depicted the overall survival and patient survival status for KIRC. Numerous neutrophils were enriched in the immune system of COVID-19 patients, and the lives of KIRC patients were endangered due to significant immune cell infiltration. Molecular docking studies indicated that berberine binds strongly to target proteins.
Conclusion: This study demonstrated berberine as a potential treatment option in pharmacological, immunological, and clinical practice. Moreover, its therapeutic effects may provide potential and reliable treatment options for patients with KIRC/COVID-19.
Since its discovery in December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread and levied significant damage to the economies, quality of life, and psychological health of individuals. As of 11 August 2022, the World Health Organization (WHO) (https://covid19.who.int/) reported a 24-hour-emergence of 866,828 new COVID-19 cases, a 580 million increase in the total number of COVID-19 patients worldwide and 6.4 million COVID-19-induced deaths. Despite the rapid development of vaccines and population-wide immunisation campaigns (1, 2), the global epidemiological scenario remains critical owing to significant infectivity, SARS-CoV-2 mutations, and declining neutralising antibody titers (3–6). Given that 5% of COVID-19 patients with acute respiratory distress syndrome (ARDS) require mechanical ventilation and intensive care unit (ICU) admittance (7), effective treatment approaches are urgently required to alleviate symptoms and reduce mortality.
Cancer and COVID-19 may be related as COVID-19 infection is more common in cancer patients (8, 9). In addition, COVID-19 patients with cancer are at a greater risk for serious complications and unfavourable prognosis than those without cancer (10–12), particularly immunocompromised patients (13). Most renal cell carcinoma (kidney clear cell carcinoma, KIRC) cases are malignant with significant mortality (14). Following COVID-19 infection, 28.5% and 19.3% of patients experience immediate renal impairment and die, respectively, indicating that the infection quickly attacks the kidney and endangers the lives of patients (15, 16). This may be linked to the development of sickle cell traits and enhanced ACE2 expression (17, 18). Furthermore, following COVID-19 infection, KIRC patients experience severe complications and elevated mortality risk (19). Similarly, the COVID-19 pandemic delays medical care for KIRC patients (20).KIRC patients have a higher risk of infection and may develop fatal complications and higher mortality from COVID-19.
Therefore, we sought effective treatment methods to reduce the progressive COVID-19-induced deterioration and improve the survival of patients with KIRC. Berberine is a natural compound with strong antibacterial, antiviral, and antitumour activities (21–27). It can resist the inflammatory response of acute lung injury by facilitating nuclear translocation and phosphorylation of nuclear factor erythroid 2–related factor 2 (Nrf2) to reduce the generation of inflammatory factors and reactive oxygen species (ROS) (28); berberine induced apoptosis in virus-infected cells by promoting ROS production (29, 30). This dual effect of resistance to disease damage and viral destruction is specific and important; by directly interacting with the virion, berberine counteracts the infectiousness and inhibits SARS-CoV-2 replication (31). Moreover, berberine decreases circulating inflammatory mediators, including interleukin (IL)-6, tumour necrosis factor (TNF)-α, and C-reactive protein (CRP), in patients with severe COVID-19 (32). Additionally, in combination with photodynamic therapy, berberine accelerates autophagy and apoptosis of KIRC cells by promoting ROS generation, resulting in KIRC cell death (33). It also strengthens apoptosis in KIRC cells by stimulating ROS generation by decreasing c-FLIP and Mcl-1 protein regulation (34). Hence, berberine can inhibit SARS-CoV-2 infection and reproduction, reduce the inflammatory response in COVID-19 patients, and promote apoptosis of KIRC cells by facilitating ROS generation. This study aimed to further explore the berberine therapeutic mechanism in treating KIRC/COVID-19 patients to provide treatment evidence and strategy for these patients (Figure 1).
The COVID-19-related transcriptomic RNA-sequencing datasets, GSE157103 and GSE171110, were acquired from GEO datasets (https://www.ncbi.nlm.nih.gov/geo/). By using the “limma” package in R software (version 4.2) with the the filtering criterion of p < 0.05, and | log fold change | of > 0.585, the COVID-19 differentially expressed genes (DEGs) in two datasets were verified. DEGs of two datasets were intersected reciprocally to screen out the most credible COVID-19- related genes.
The KIRC RNA-seq dataset was downloaded from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/repository). The analogous calculation and filtering criterion were carried out for the identification of KIRC-related genes.
To discover the berberine therapeutic target gene, we deploy a comprehensive retrieval strategy via official websites, including CTD (http://ctdbase.org/), Parm Mapper (http://www.lilab-ecust.cn/pharmmapper/), SEA (https://sea.bkslab.org/), Swiss Target Prediction (http://www.swisstargetprediction.ch/) and TCMSP (http://tcmspw.com/tcmsp.php/). These genes from various websites were merged and considered berberine target genes.
Ultimately, the COVID-19- related genes, KIRC-related genes and berberine target genes were intersected. And these overlapping genes represented therapeutic target genes of berberine against KIRC/COVID-19.
To determine the interactions between target genes of berberine against KIRC/COVID-19, the GeneMANIA (http://genemania.org/), a website predicting the interactions between genes based on the label propagation algorithm and linear regression-based algorithm, was performed to construct a protein-protein interaction (PPI) network. PPI network data was imported into Cytoscape software to build the core network based on crucial topological parameters, including Betweenness-Centrality, Closeness-Centrality and Degree.
In addition, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were executed via the R package “clusterProfiler”, making clear the underlying therapeutic mechanism of berberine against KIRC/COVID-19.
Due to the lack of clinical information in the GSE171110 dataset, the GSE157103 dataset was selected as the primary clinical research objective. Admittedly, mechanical ventilation is an essential life support therapy for patients with respiratory distress, and most critically ill COVID-19 patients desperately need mechanical ventilation (35, 36), which is an important clinical indicator of the severity of COVID-19 patients, especially in the respiratory system. Whether admission in ICU is also a key clinical information to judge the condition of patients with COVID-19, which pays greater attention to holistic life support therapy. Then, the Wilcoxon test was performed to examine which genes are associated with clinical factors of patients with COVID-19. After screening, the target genes were identified the association with each other via the Spearman correlation test.
Based on the “survival”, “survminer” and “glmnet” package in R software, the target genes of KIRC patients were identified by using the univariate COX regression analysis and LASSO Cox regression analysis successively. Genes after being screened would be divided equally into high- and low- expression groups. Kaplan–Meier survival analysis was applied to analyze which genes are closely related to overall survival (OS) between the high- and low- expression groups. These genes would be calculated as risk score according to the computational formula: risk score = sum of coefficients × gene expression level. The KIRC patients were divided into low- and high-risk groups based on the median of risk score. The heatmap could visually describe the difference of gene expression between low- and high-risk groups, and the correlation of each gene detected by the Spearman correlation test.
The relationship between risk signature and OS was proved using Kaplan–Meier survival analysis. The risk signature’s sensitivity and specificity were further evaluated through the area under the curve (AUC) in the time‐dependent receiver operating characteristic (ROC) analyses performed by R packages “survivalROC”. The clinical information, including age, gender, grade, tumour stage, tumour, node and metastasis, were equipped to describe the survival state of KIRC patients in multidimensional and depth. And the differences in the risk signature at different clinical stages would be compared mutually.
Immune response plays an irreplaceable role in the development and treatment of KIRC (37, 38) and COVID-19 infection (39, 40). We estimated 22 immune cell infiltration in KIRC/COVID-19 patients respectively via R package “CIBERSORT”, and analyzed the difference of each immune cell infiltration in genes of berberine against KIRC/COVID-19. Moreover, analysis of immunological function was further investigated through R package “GSVA” and “GSEABase”. The infiltration of stromal cells and immune cells was assessed by R software package (“estimate”), for making clear the tumour microenvironment (TME).
Molecular docking analysis was carried out to predict predominant binding modes of berberine to KIRC/COVID-19-related proteins. We collected structures of target proteins and berberine from RCSB PDB database (https://www.rcsb.org/) and PubChem (https://pubchem.ncbi.nlm.nih.gov/). After removing redundant ligand and hydrone of target proteins via Pymol software (version 2.4), we loaded structures of proteins and berberine into Autodock Vina to perform the molecular docking. When the docking affinity scores < -5.0 kcal/mol, it indicates that berberine has strong binding interaction with target protein.
In total, 19,472 genes were obtained from 10 healthy individuals and 100 COVID-19 patients in the GSE157103 dataset, and 30,183 genes were obtained from 10 healthy individuals and 44 COVID-19 patients in the GSE171110 dataset. Following identification between the healthy and COVID-19 group based on the filtering criterion of p < 0.05, and | log fold change | of > 0.585, 3,505 DEGs (3,399 downregulated and 106 upregulated genes) in the GSE157103 dataset and 6,534 DEGs (3,070 downregulated and 3,464 upregulated genes) in the GSE171110 dataset were obtained (Figures 2A–D). To exclude interference factors and enhance credibility, DEGs in the two datasets were intersected, and 1,038 overlapping genes were considered crucial for COVID-19 infection (Figure 2G).
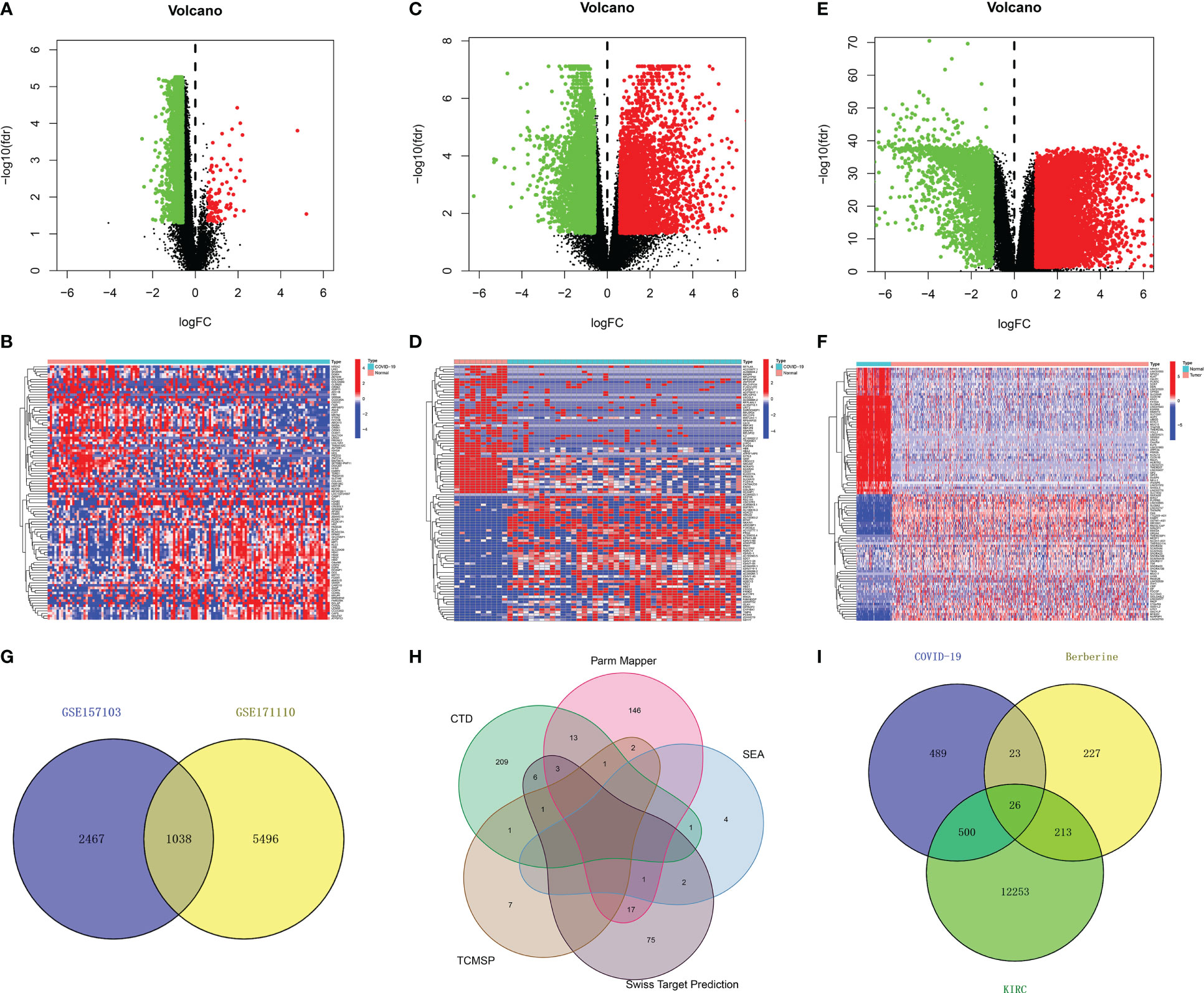
Figure 2 Identification of therapeutic target genes of berberine against KIRC/COVID-19. (A, B) Volcano plot and heatmap of DEGs from GSE157103 data set, and heatmap shown the top 50 DEGs (C, D) DEGs from GSE171110 data set. (E, F) DEGs from TCGA data set. (G) The intersection of GSE157103 and GSE171110 data set. (H) The union set of berberine target genes of each website. (I) The intersection of COVID-19 DEGs, KIRC DEGs and berberine target genes.
In total, 38,125 genes from 72 normal and 539 KIRC tissues were obtained from TCGA. Similarly, 12,992 KIRC DEGs (4,199 down-regulated and 8,793 upregulated genes) (Figures 2E, F) were obtained.
Additionally, 235, 183, 8, 100, and 12 berberine target genes were obtained from CTD, Parm Mapper, SEA, Swiss Target Prediction and TCMSP, respectively. After merging, 489 berberine target genes were obtained (Figure 2H).
In total, 526 DEGs were co-expressed in KIRC/COVID-19 after intersecting, and 26 genes were obtained by crossing 489 berberine target genes with 1,038 COVID-19 and 12,992 KIRC DEGs, indicating that berberine plays a therapeutic role in KIRC/COVID-19 through these genes (Figure 2I).
Proteins, the result of transcription and translation of genes, are elementary substances that directly affect the metabolism, physiology, and pathology of the human body. To verify protein interactions, 26 berberine genes against KIRC/COVID-19 were imported into the GeneMANIA website, and a PPI network was constructed (Figure 3A). Next, the PPI network data were loaded into Cytoscape software, and the topological parameters, including Betweenness-Centrality, Closeness-Centrality and Degree, were calculated via the package “CytoNCA”. Following filtering with a criterion of topological parameters greater than the median, 14 predominant proteins were established in the core network (Figure 3B) (Table 1).
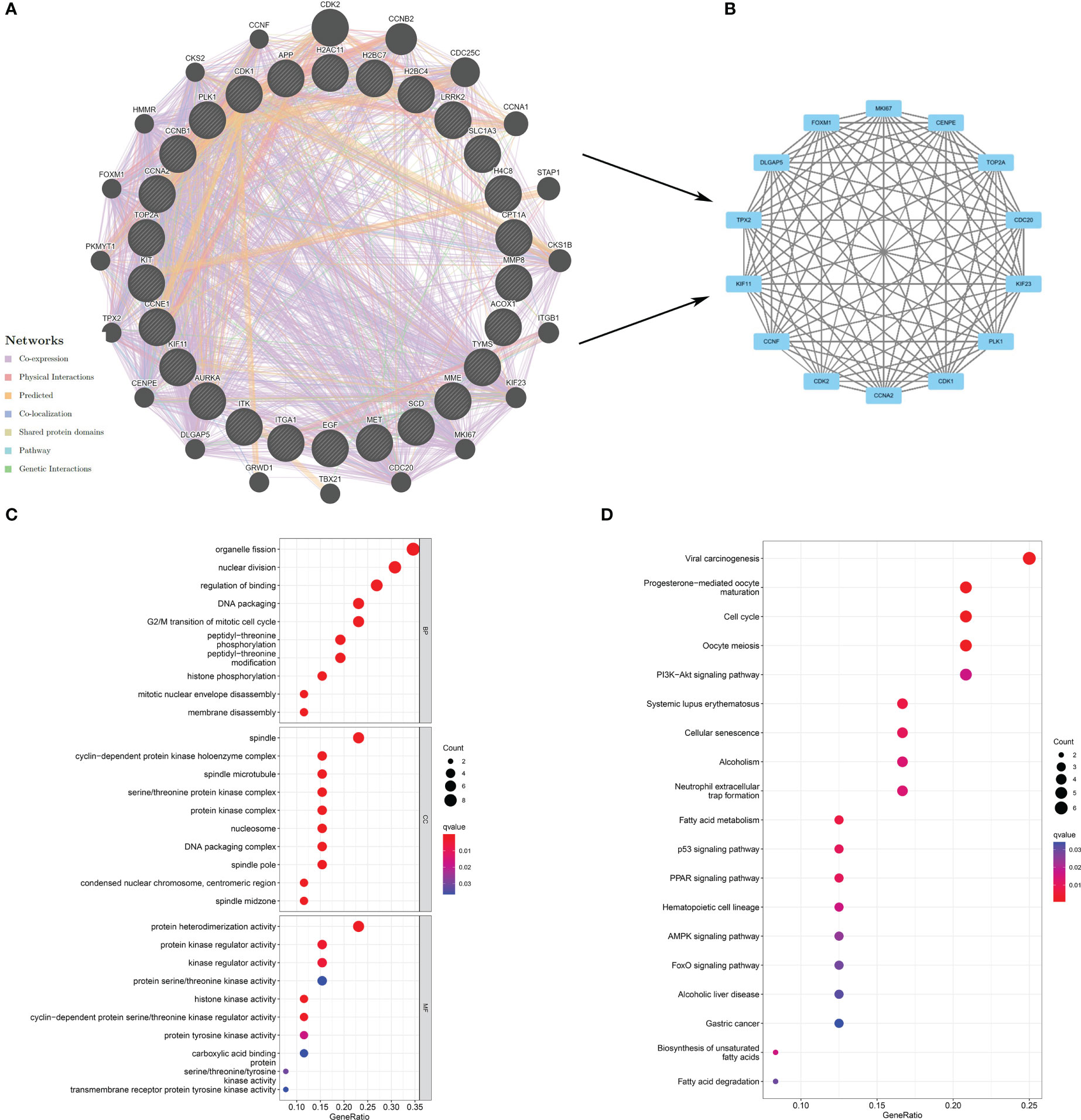
Figure 3 Exploration of the potential mechanism of berberine against KIRC/COVID-19. (A) The PPI network based on therapeutic target genes of berberine against KIRC/COVID-19. (B) the core network of PPI. (C) GO analysis of target genes. (D) KEGG pathway analysis of target genes.
GO analysis revealed that 26 genes were enriched in 771 GO terms, including 669 biological processes (BPs), 85 cellular components (CCs), and 17 molecular functions (MFs). The dominant BPs were organelle fission, nuclear division, binding regulation, DNA packaging, and the G2/M transition of the mitotic cell cycle (Figure 3C). Nineteen KEGG terms were enriched in the KEGG analysis, including viral carcinogenesis, progesterone-mediated oocyte maturation, cell cycle, oocyte meiosis, and PI3K−Akt signalling pathway (Figure 3D).
Clinical information in the GSE157103 dataset includes mechanical ventilation and ICU, which are pivotal indicators for evaluating COVID-19 severity. Following the Wilcoxon test, six of the 26 berberine target genes against KIRC/COVID-19 were associated with mechanical ventilation. Compared to the non-mechanical ventilation group, the expression of ACOX1, LRRK2, MMP8, and SLC1A3 was higher, and that of H4C8 and ITK was lower in the mechanical ventilation group (Figure 4A). As MMP8 and SLC1A3 expression increased and CPT1A, H2AC11, H4C8, and ITK expression decreased, the probability of ICU admittance increased (Figure 4B). In addition, the correlation between genes was demonstrated using the Spearman correlation test (Figure 4C). These results revealed that berberine treatment could effectively reduce the utilisation rate of mechanical ventilation and the probability of ICU admittance by targeting these eight genes and inducing a chain reaction among them.
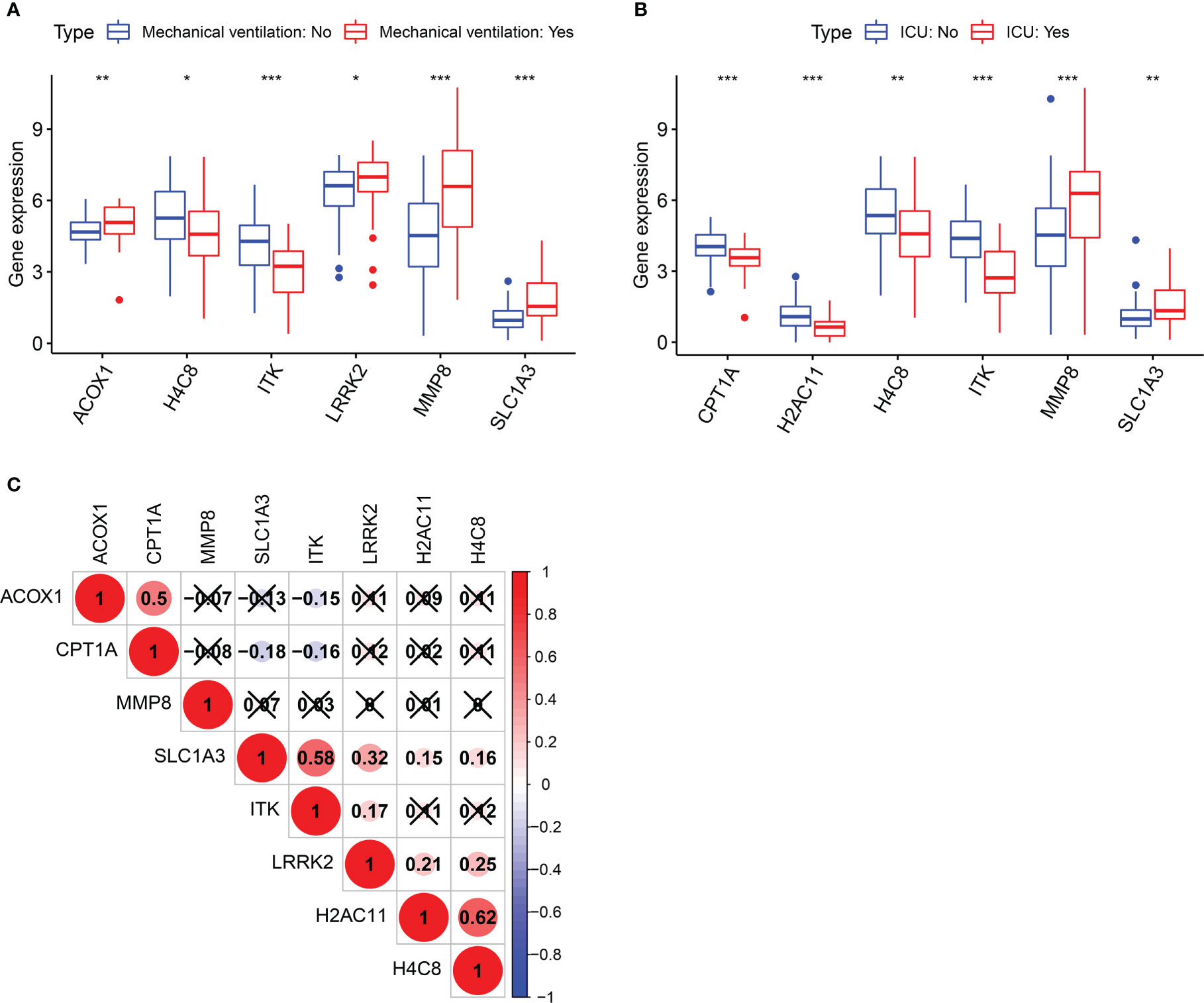
Figure 4 Clinical significance of target genes in COVID-19 patients. (A) Boxplot of target genes associated with mechanical ventilation. (B) Boxplot of target genes associated with ICU admission. (C) The correlation analysis between genes after screening. *p <0.05, **p <0.01, ***p <0.005.
According to univariate Cox regression analysis, 13 of the 26 berberine target genes against KIRC/COVID-19 were closely correlated with OS (Figure 5A). To reduce the influence of multicollinearity among variables and detect the optimal genes, LASSO Cox regression analysis was performed; ACOX1, APP, CPT1A, PLK1 and TYMS were identified as representative signatures of the OS of KIRC patients, and the risk score was forecasted by the computational formula: risk score = (-0.365 × expression ACOX1) + (-0.211 × expression APP) + (-0.077 × expression CPT1A) + (0.663 × expression PLK1) + (-0.036 × expression TYMS) (Figures 5B, C). According to their number, KIRC patients were subdivided into high- and low-risk groups (Figure 6A). The heatmap vividly described the expression of five genes in these groups (Figure 5D). Furthermore, these five genes correlated positively or negatively (Figure 5E). Kaplan-Meier (KM) survival analysis revealed that high-expression genes, including ACOX1, APP, CPT1A, and TYMS, had a higher survival probability over time, whereas PLK1 had the opposite effect (Figures 5F–J).
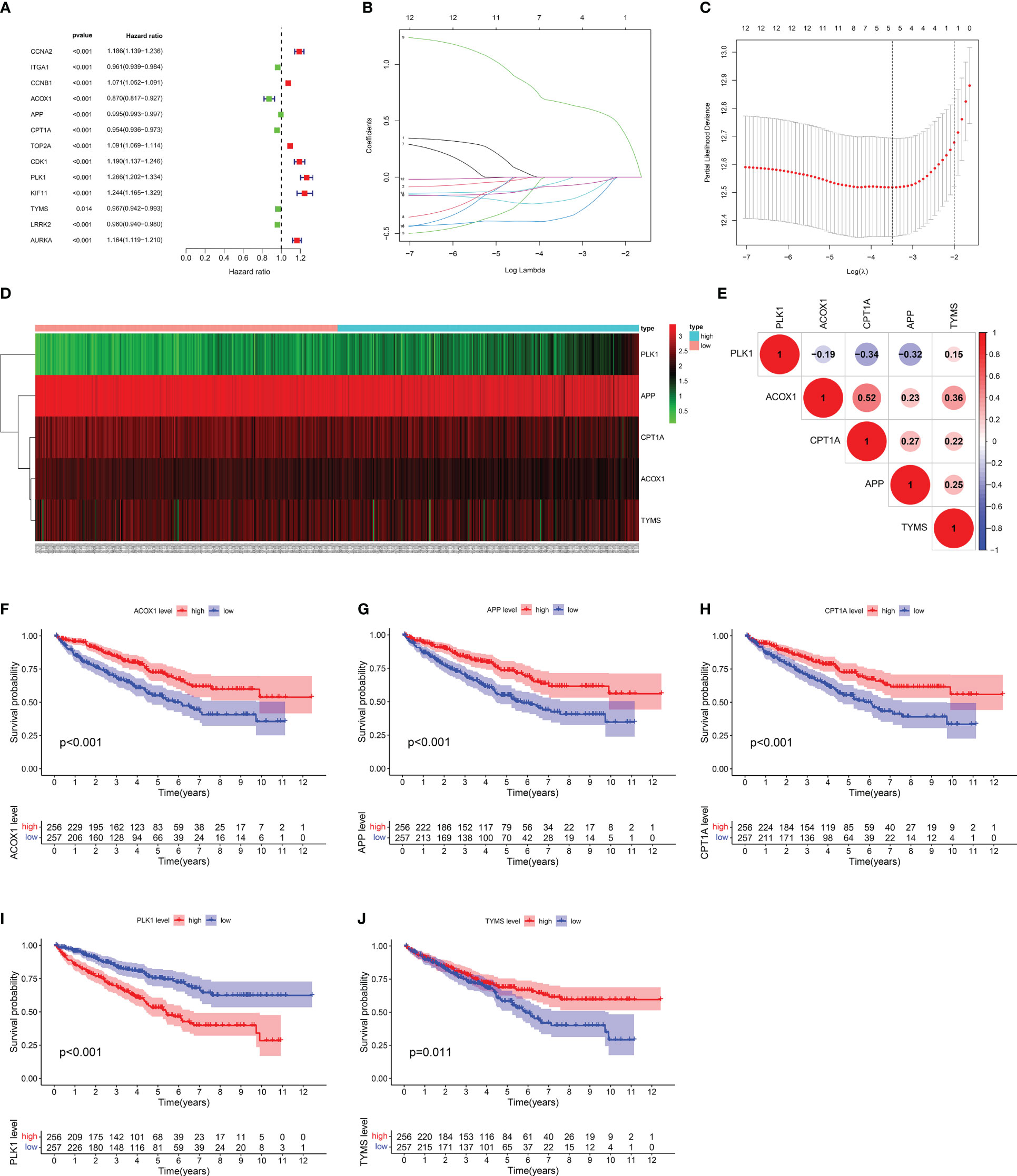
Figure 5 Assessment of target gene signatures for KIRC patients. (A) The forest plot of univariate Cox proportional analysis. (B, C) The LASSO Cox regression analysis for detecting the representative gene. (D) Heatmap of target gene signatures. (E) The correlation analysis between target gene signatures. (F–J) Kaplan-Meier survival analysis of target gene signatures.
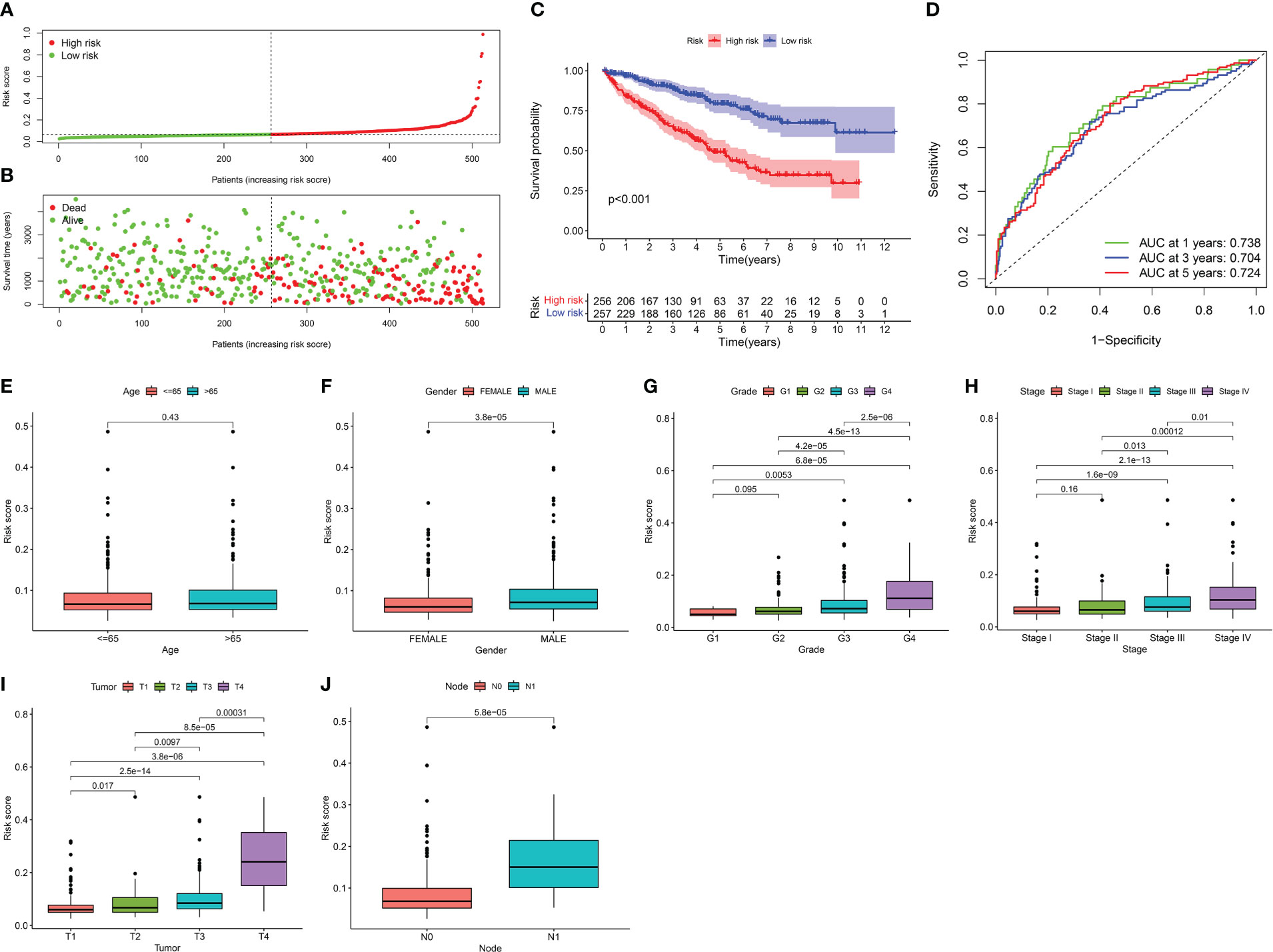
Figure 6 Clinical significance of risk signature for KIRC patients. (A) Distribution of risk scores to be divided into high- and low- risk groups. (B) Distribution of the OS to depict the relationship between OS and risk signature in dead and alive KIRC patients. (C) Kaplan-Meier survival analysis of risk signature. (D) AUC in ROC analysis for risk signature at 1‐, 3‐and 5‐years survival. When AUC is greater than 0.7, the prediction model has reliable accuracy. (E–J) Boxplot reveals the relationship between risk signature and clinical information, containing in age, grade, tumor stage, tumor, lymph node and metastasis deteriorated.
The OS of KIRC patients decreased with an increased risk score, and the survival rate tapered towards death (Figure 6B). KM survival analysis revealed that the prognosis of the high-risk group was substantially worse than that of the low-risk group (Figure 6C). The AUC, a sensitive and specific risk signature indicator, showed reliable predictive accuracy at the 1-, 3-, and 5‐year survival time points (Figure 6D). Except for age, significant differences were observed in the risk signature for each clinical feature. The risk signature increased to varying degrees as the grade, tumour stage, tumour, lymph node, and metastasis deteriorated (Figures 6E–J). These results suggest that the risk signature can effectively reflect the clinical status and prognosis of KIRC patients.
In total, 22 immune cell infiltration was predicted using “CIBERSORT”. We focused on the comparatively large number of neutrophil infiltrations in COVID-19 patients, possibly associated with a sharp neutrophil increase and activation of SARS-CoV-2-induced neutrophil extracellular traps (41–44) (Figures 7A–F, Supplementary File 1). According to the boxplot, high expression of ACOX1, LRRK2, and SLC1A3 was susceptible to neutrophil infiltration, whereas that of CPT1A and ITK was resistant to neutrophil infiltration (Figures 7A–E). The relationship between these genes and neutrophil infiltration was consistent with the relationship between the genes and the utilisation rate of mechanical ventilation or probability of ICU admittance (Figures 4A, B). Moreover, high neutrophil infiltration significantly affected the increased use of mechanical ventilation and ICU stay (Figures 7F, G).
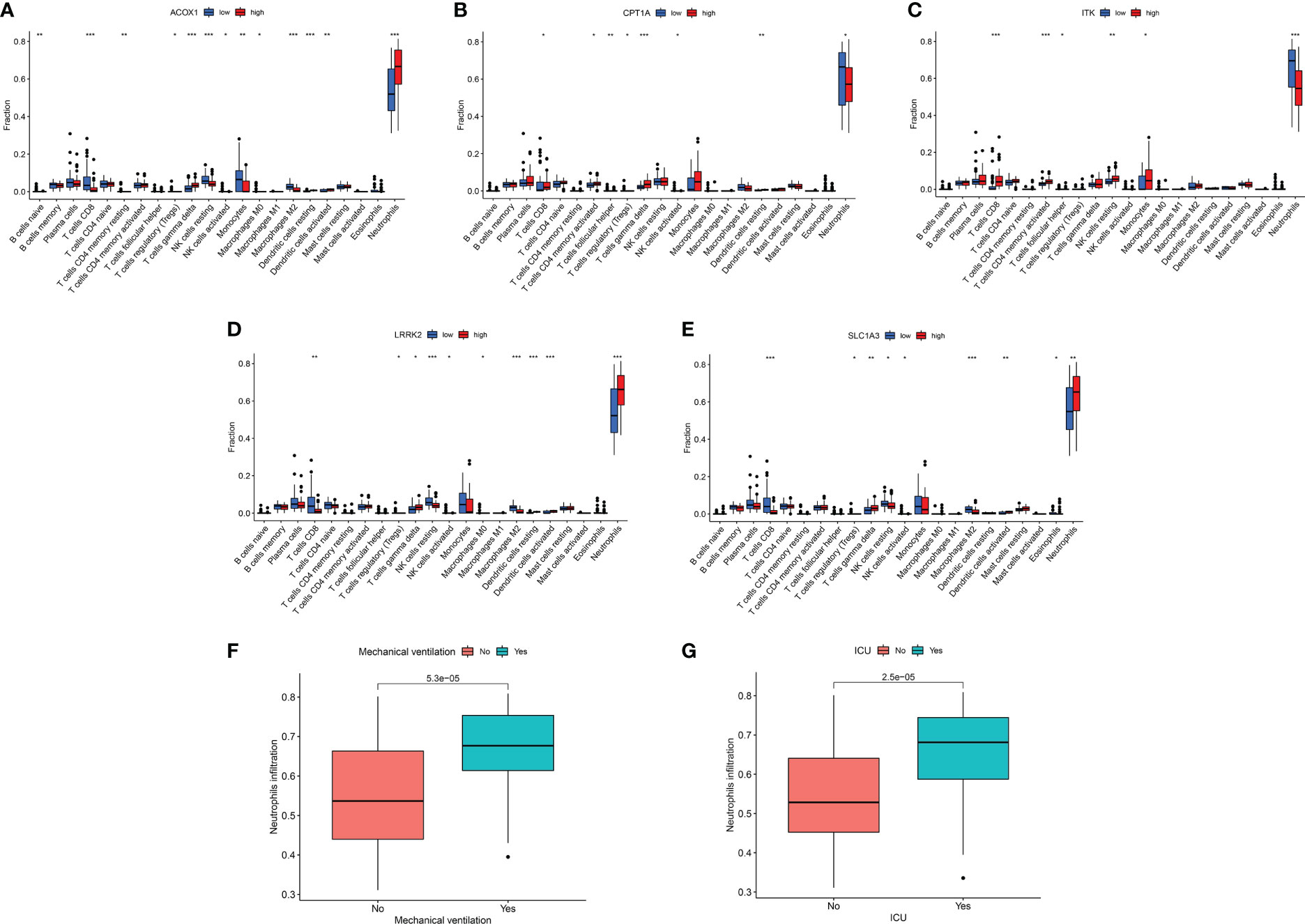
Figure 7 Immune response of target genes for COVID-19 patients. (A–E) Boxplot of target genes associated with 22 immune cell infiltration. (F) Boxplot of neutrophil infiltration associated with mechanical ventilation. (G) Boxplot of neutrophil infiltration associated with ICU admission. *p <0.05, **p <0.01, ***p <0.005.
In KIRC patients, five genes were positively or negatively correlated with 22 immune cells to varying degrees, indicating that berberine may play a therapeutic role by regulating the infiltration of immune cells (Figures 8A–E). Furthermore, seven immune cells, including CD8 T cells, CD4 memory-activated T cells, follicular helper T cells, regulatory T cells (Tregs), macrophages M0, macrophages M1 and activated dendritic cells, were enriched with an increased risk signature; however, CD4 memory resting T cells, macrophages M2, dendritic cells, and resting mast cells, were reduced (Figure 8F). Analysis of immunological function demonstrated that the high-risk group exhibited enhanced APC co-stimulation, CCR, checkpoint, cytolytic activity, inflammation-promotion, parainflammation, T-cell co-inhibition, and T-cell co-stimulation, and inhibited type II IFN response (Figure 8G). The violin plot showed that the high-risk groups correlated with a higher immune score, suggesting that high-risk groups are prone to a hyperimmune response (Figure 8H).
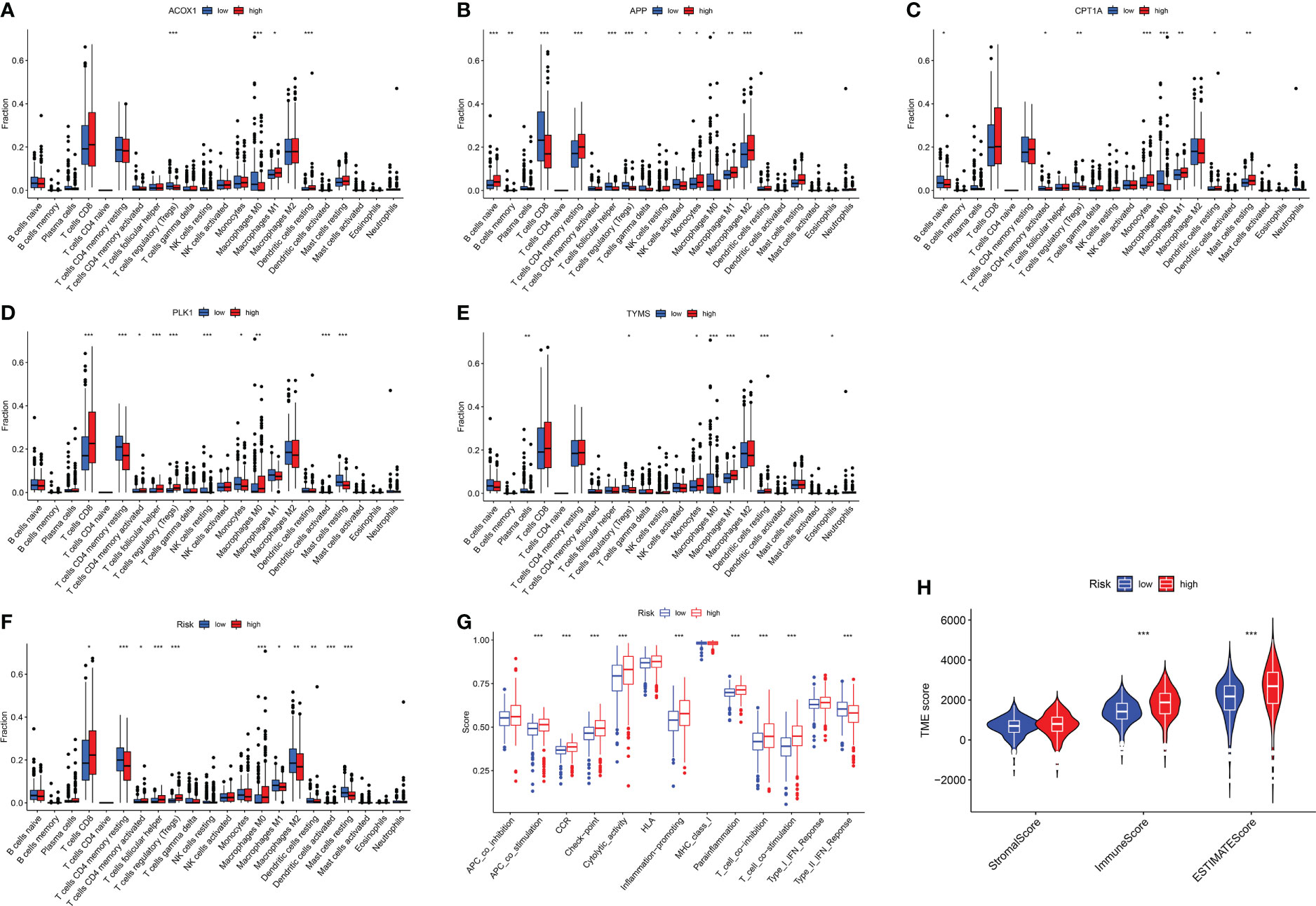
Figure 8 Immune response of target genes for KIRC patients. (A–F) Boxplot of target gene signatures associated with 22 immune cell infiltration. (G) The analysis of immunological functions. (H) the tumor microenvironment score. *p <0.05, **p <0.01, ***p <0.005.
Molecular docking was performed to determine the optimal binding mode of berberine to KIRC/COVID-19 target genes. The docking affinity score output by AutoDock Vina was ACOX1 (-10.4) < PLK1 (-10.1) < H4C8 (-9.7) < H2AC11 (-7.9) < MMP8 (-7.9) < LRRK2 (-7.8) < SLC1A3 (-7.8) < ITK (-7.7) < TYMS (-7.1) < APP (-6.1) < CPT1A (-5.5). All docking affinity score was < -5.0 kcal/mol, indicating that these target genes play an important role in the berberine treatment of KIRC/COVID-19 (Figures 9A–E, Supplementary File 2).
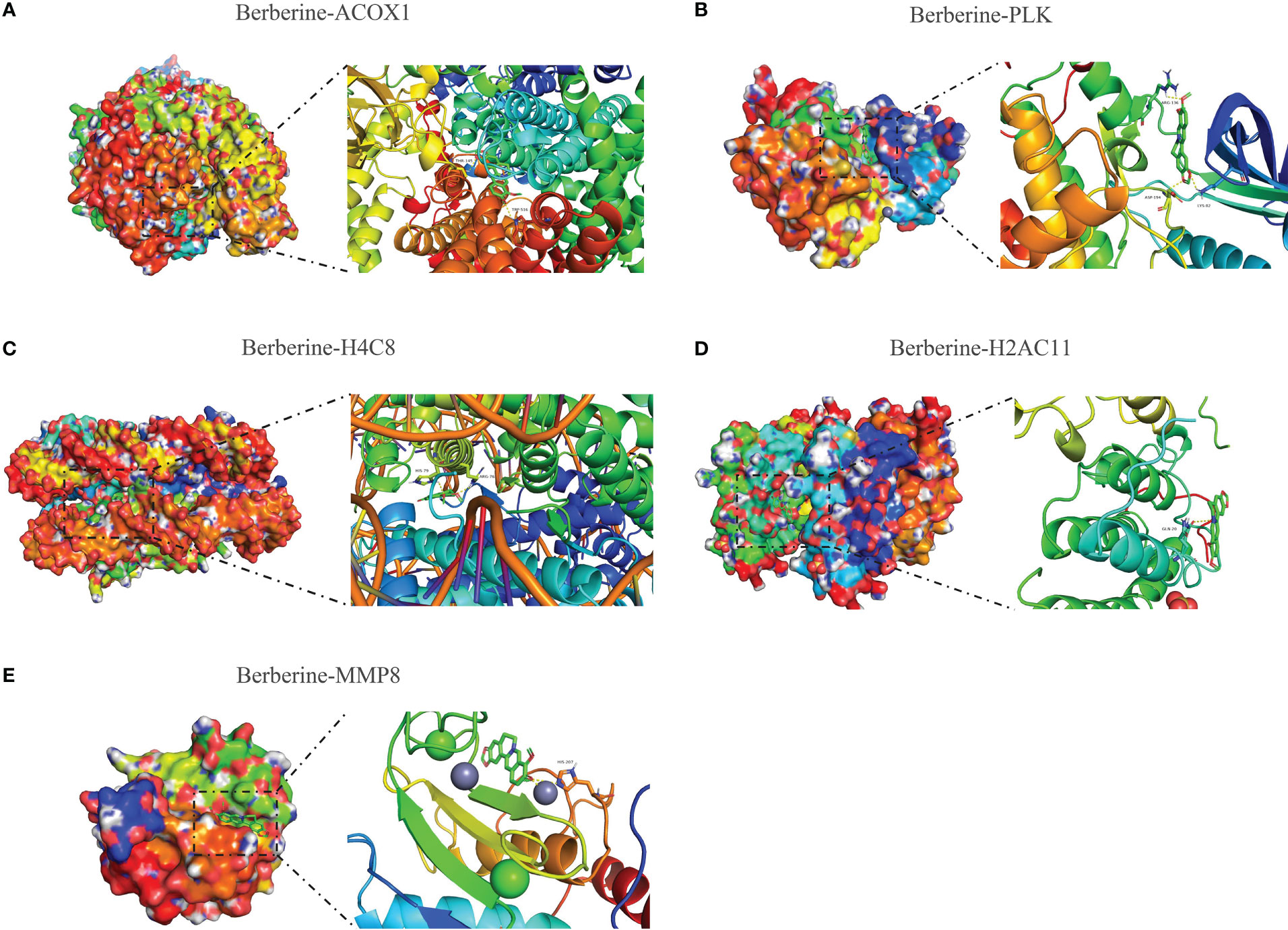
Figure 9 Molecular docking of berberine and target genes. The top 5 docking affinity score. (A) Berberine binding to ACOX1. (B) Berberine binding to PLK. (C) Berberine binding to H4C8. (D) Berberine binding to H2AC11. (E) Berberine binding to MMP8.
The COVID-19 epidemic has significantly affected individuals worldwide and had catastrophic effects on the global economy, health, and psychology (45). Although the vaccine received immediate popularity, the anti-COVID-19 vaccine campaign persisted for years because of the dreadful contagiousness, SARS-CoV-2 mutations, and declining antibody titers (3–6). Although specific medicines have not yet been developed, COVID-19 therapy schedules and management strategies have been continuously investigated (46–48). In China, the mortality rate of COVID-19 patients is 0.41% (24,232 deaths/59,69737 confirmed cases), which is significantly lower than the global average, indicating the superior effectiveness of conventional Chinese medicine in combating COVID-19 (49, 50), such as Lianhua Qingwen capsules, Xuanfei Baidu decoction, and Xuebijing, among others (51). SARS-CoV-2 invades the kidney, harms renal cells, and impairs renal function (52–54), posing a greater threat to KIRC patients. Additionally, KIRC patients are more likely to contract COVID-19 and are more susceptible to dying from its infection (8, 9, 19). We sought an effective therapeutic strategy to decrease possible significant sequelae and increase the survival of KIRC/COVID-19 patients based on the relationship between COVID-19 and KIRC. It has been demonstrated that berberine, an active ingredient in most natural herbs, lowers inflammation in COVID-19 patients and destroys KIRC cells. Using computer-based approaches and algorithms, the possible therapeutic mechanism of berberine in treating KIRC/COVID-19 patients was investigated.
In this study, 26 genes were identified as therapeutic target genes of berberine against KIRC/COVID-19. The PPI network produced by the GeneMANIA website revealed that the 26 therapeutic target genes achieved protein function through a core network composed of 14 genes, such as CDK1, CCNA2, and PLK1, among others. GO analysis revealed that the BPs of 26 target genes were enriched mainly in organelle fission, nuclear division, binding regulation, DNA packaging, and G2/M transition of the mitotic cell cycle. These BPs play an important role in cell proliferation, which is crucial in KIRC/COVID-19 pathogenesis (55, 56). The PI3K-Akt signalling pathway predicted by KEGG analysis was activated by accelerating the phosphorylation of Akt, mTOR, 4E-BP1, and S6K1 during the initial phases of COVID-19 infection (57), resulting in the rapid activation of the translation machinery of viral protein synthesis (58, 59). The MDM2 protein is activated by the aberrant PI3KAkt signalling pathway in KIRC, allowing easier degradation of the tumour suppressor p53 by proteasome machinery (60). According to functional analyses, the berberine mode of action in treating KIRC/COVID-19 may have diverse pathways that interfere with tumour cell proliferation or viral self-replication, particularly the PI3K-Akt signalling pathway.
Mechanical ventilation and ICU admittance are indispensable life support measures for patients with severe COVID-19 (61–63). The severity of COVID-19 infection in patients intensified as the expression of ACOX1, LRRK2, MMP8, and SLC1A3 increased, and the expression of CPT1A, H2AC11, H4C8, and ITK decreased. These genes play different roles in resisting viral infections and can function as prospective targets for COVID-19 treatment. The enhanced MMP8 production during the early stages of COVID-19 infection is directly linked to the breakdown of the extracellular matrix (ECM), including collagen, laminin, and proteoglycans, which promotes viral multiplication and inflammation (64). ROS production, which is essential for inflammation and lung injury, is enhanced by high levels of ACOX1 protein and function (65, 66). Huang et al. found that IL-2-inducible T-cell kinase (ITK) facilitates γδ T-cell-derived IL-17A production to resist Mycobacterium tuberculosis infection (67). By inhibiting CPT1a expression, increased mitochondrial ROS accelerates the progression of pulmonary fibrosis; however, restoring CPT1a expression impedes pulmonary fibrosis (68). These genes function by controlling ROS production or being controlled by ROS, which may be connected to the interference of berberine with ROS production. Thus, berberine decreases ROS production by controlling the expression levels of these genes, lowering mechanical ventilation and ICU admittance in COVID-19 patients.
Five of the 26 target genes of berberine against KIRC/COVID-19 were closely associated with OS in KIRC patients, as determined by univariate and LASSO Cox regression analyses., Du et al. discovered that lipid deposition plays a significant role in accelerating KIRC progression with hypoxia-inducible factors (HIFs)-induced CPT1A suppression, and with the reversing of this inhibition, CPT1A decreases KIRC progression (69). Increasing fatty acid oxidation by enhancing CPT1A expression may be a promising therapeutic strategy for KIRC (70). Furthermore, HIF-2 increased PLK1 expression, inducing metastasis and drug resistance in KIRC cells (71) and counteracting CPT1A effects. By phosphorylating MCM3, PLK1 promoted KIRC cell growth and prevented apoptosis (72). Regulation of the antagonistic effect between CPT1A and PLK1 may be an advancement to improve the OS of KIRC patients. The survival status of KIRC patients may be more thoroughly and objectively reflected by the risk signature derived from the five gene expression levels in this study and may accurately reveal the change mechanism of tumour grade, stage, lymph nodes, and metastasis. All of the evidence demonstrates the risk signature as a valid indicator of patient prognosis and a tool for evaluating the effectiveness of KIRC.
The immune response significantly contributes to disease development. The fulminant immune response is a body characteristic following SARS-CoV-2 infection (73–75). In this study, abundant neutrophil infiltration was observed in COVID-19 patients compared with the remaining 21 immune cells. This is similar to the results of several studies, and the sharp increase in neutrophils and activation of neutrophil extracellular traps caused further acute injury to the lungs and kidneys (41–44, 76). Cytokine storm-induced inflammation activates ROS, causing severe complications (77). Therefore, it is practical to reduce mechanical ventilation and ICU admittance by decreasing neutrophil infiltration. Abnormal immunological responses in KIRC patients are a significant contributor to disease onset. KIRC patients with a high immune score were at increased risk, suggesting KIRC progressed when a significant proportion of immune cells were concentrated in the tumour microenvironment (TME). In recent years, immune checkpoint inhibitors have gradually become a mainstream therapy and research focus for KIRC (78) and are beneficial for improving the prognosis of patients (79–81). Relative to chemotherapy, treatment with immune checkpoint inhibitors does not appear to significantly increase risk of serious adverse events in COVID-19-positive patients (82). Correcting immune disorders may be a future trend for KIRC treatment, immune checkpoint inhibitors remains an important treatment option even in COVID-19 positive patients.
The treatment mechanism of berberine against KIRC/COVID-19 remains uncertain. In this study, DEGs were screened, and target genes for correlation with clinical information were analysed, revealing immune mechanisms. Finally, molecular docking was used to investigate the best binding mode of berberine to the KIRC/COVID-19 target genes. The results indicate that berberine treatment is an effective strategy for treating KIRC/COVID-19. In a follow-up study, COVID-19-infected KIRC cells will be examined using berberine, and the role of target genes in the treatment process will be analysed to further explore the therapeutic mechanism of berberine against KIRC/COVID-19.
Overall, KIRC patients are at higher risk of contracting COVID-19, and no effective therapies have been developed for both conditions. In this study, berberine was demonstrated as a potential treatment option in pharmacological, immunological, and clinical practice; its therapeutic effect provides a potential and reliable treatment option for patients with KIRC/COVID-19.
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/, https://portal.gdc.cancer.gov/repository, http://ctdbase.org/ http://www.lilab-ecust.cn/pharmmapper/, https://sea.bkslab.org/, http://www.swisstargetprediction.ch/, http://tcmspw.com/tcmsp.php/.
ZZ, KZ and YZ developed the concept of the project. The data was collected by ZZ, WL and FY, and evaluated by XL, KN and XW. KZ and YZ conducted a repeat experiment.All authors reviewed and discussed the results and contributed to the paper preparation. ZZ, KZ and YZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supportive by Natural Science Foundation of Hunan Province (No. 2022JJ40706); Guangdong Provincial High Level Clinical Key Specialties.
We are sincerely grateful to the workers of official websites and the scientists who provided the raw data for official websites.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1038651/full#supplementary-material
Supplementary Figure 1 | Boxplot of H2AC11, H4C8 and MMP8 associated with 22 immune cell infiltration. Without statistical significance in neutrophil infiltration. *p <0.05, **p <0.01, ***p <0.005.
Supplementary Figure 2 | Molecular docking of berberine and target genes.
COVID-19, coronavirus disease 2019; KIRC, kidney clear cell carcinoma; DEGs, differentially expressed genes; GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas databases; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WHO, World Health Organization; ARDS, acute respiratory distress syndrome; Nrf2, nuclear factor erythroid 2–related factor 2; ROS, reactive oxygen species; CRP, C-reactive protein; CCA, Cancer Genome Atlas; CTD, Comparative Toxicogenomics Database; SEA, Spanish Electoral Archive; TCMSP, Traditional Chinese Medicine System Pharmacology; PPI, protein-protein interaction; CCs, cellular components; MFs, molecular functions; ICU, intensive care unit; OS, overall survival; KM, Kaplan-Meier; APC, antigen-presenting cell; CCR, CC chemokine receptors; HIF, hypoxia-inducible factors, TME, tumour microenvironment.
1. Schultz BM, Melo-González F, Duarte LF, Gálvez NMS, Pacheco GA, Soto JA, et al. A booster dose of CoronaVac increases neutralizing antibodies and T cells that recognize delta and omicron variants of concern. mBio (2022) 13(4):e0142322. doi: 10.1128/mbio.01423-22
2. Bellusci L, Grubbs G, Zahra FT, Forgacs D, Golding H, Ross TM, et al. Antibody affinity and cross-variant neutralization of SARS-CoV-2 omicron BA.1, BA.2 and BA.3 following third mRNA vaccination. Nat Commun (2022) 13(1):4617. doi: 10.1038/s41467-022-32298-w
3. Iketani S, Liu L, Guo Y, Liu L, Chan JF, Huang Y, et al. Antibody evasion properties of SARS-CoV-2 omicron sublineages. Nature (2022) 604(7906):553–6. doi: 10.1038/s41586-022-04594-4
4. Leuzinger K, Roloff T, Egli A, Hirsch HH. Impact of SARS-CoV-2 omicron on rapid antigen testing developed for early-pandemic SARS-CoV-2 variants. Microbiol Spectr (2022) 10(4):e0200622. doi: 10.1128/spectrum.02006-22
5. Petersen E, Koopmans M, Go U, Hamer DH, Petrosillo N, Castelli F, et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis (2020) 20(9):e238–44. doi: 10.1016/s1473-3099(20)30484-9
6. Davies NG, Jarvis CI, Edmunds WJ, Jewell NP, Diaz-Ordaz K and Keogh RH. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature (2021) 593(7858):270–4. doi: 10.1038/s41586-021-03426-1
7. Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. Jama (2020) 323(16):1574–81. doi: 10.1001/jama.2020.5394
8. Han HJ, Nwagwu C, Anyim O, Ekweremadu C and Kim S. COVID-19 and cancer: From basic mechanisms to vaccine development using nanotechnology. Int Immunopharmacol (2021) 90:107247. doi: 10.1016/j.intimp.2020.107247
9. Hippisley-Cox J, Coupland CA, Mehta N, Keogh RH, Diaz-Ordaz K, Khunti K, et al. Risk prediction of covid-19 related death and hospital admission in adults after covid-19 vaccination: national prospective cohort study. BMJ (Clinical Res ed.) (2021) 374:n2244. doi: 10.1136/bmj.n2244
10. Liang W, Guan W, Chen R, Wang W, Li J, Xu K, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol (2020) 21(3):335–7. doi: 10.1016/s1470-2045(20)30096-6
11. Bernard A, Cottenet J, Bonniaud P, Piroth L, Arveux P, Tubert-Bitter P, et al. Comparison of cancer patients to non-cancer patients among COVID-19 inpatients at a national level. Cancers (2021) 13(6):1436–50. doi: 10.3390/cancers13061436
12. Grivas P, Khaki AR, Wise-Draper TM, French B, Hennessy C, Hsu CY, et al. Association of clinical factors and recent anticancer therapy with COVID-19 severity among patients with cancer: a report from the COVID-19 and cancer consortium. Ann Oncol Off J Eur Soc Med Oncol (2021) 32(6):787–800. doi: 10.1016/j.annonc.2021.02.024
13. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese center for disease control and prevention. Jama (2020) 323(13):1239–42. doi: 10.1001/jama.2020.2648
14. Hsieh J, Purdue M, Signoretti S, Swanton C, Albiges L, Schmidinger M, et al. Renal cell carcinoma. Nat Rev Dis Primers (2017) 3:17009. doi: 10.1038/nrdp.2017.9
15. Rostami Z, Mastrangelo G, Einollahi B, Nemati E, Shafiee S, Ebrahimi M, et al. A prospective study on risk factors for acute kidney injury and all-cause mortality in hospitalized COVID-19 patients from Tehran (Iran). Front Immunol (2022) 13:874426. doi: 10.3389/fimmu.2022.874426
16. Chang YC, Lee DJ, Wei CH, Pa CH, Chen CC, Chen HC, et al. SARS-CoV-2 versus other minor viral infection on kidney injury in asymptomatic and mildly symptomatic patients. Virulence (2022) 13(1):1349–57. doi: 10.1080/21505594.2022.2107602
17. Verma A, Huffman JE, Gao L, Minnier J, Wu WC, Cho K, et al. Association of kidney comorbidities and acute kidney failure with unfavorable outcomes after COVID-19 in individuals with the sickle cell trait. JAMA Internal Med (2022) 182(8):796–804. doi: 10.1001/jamainternmed.2022.2141
18. Garreta E, Prado P, Stanifer ML, Monteil V, Marco A, Ullate-Agote A, et al. A diabetic milieu increases ACE2 expression and cellular susceptibility to SARS-CoV-2 infections in human kidney organoids and patient cells. Cell Metab (2022) 34(6):857–873.e9. doi: 10.1016/j.cmet.2022.04.009
19. Tsimafeyeu I, Volkova M, Alekseeva G, Berkut M, Nosov A, Myslevtsev I, et al. Safety and preliminary efficacy of the gam-COVID-Vac vaccine and outcomes of SARS-CoV-2 infection in Russian patients with genitourinary malignancies. J Hematol Oncol (2021) 14(1):192. doi: 10.1186/s13045-021-01205-z
20. Struck J, Schnoor M, Schulze A, Hupe M, Ozimek T, Oppolzer I, et al. Impact of COVID-19 crisis on medical care of patients with metastasized uro-oncologic disease under systemic cancer therapy: a multicenter study in German university hospitals. World J Urol (2022) 40(2):409–18. doi: 10.1007/s00345-021-03868-2
21. Naz F, Kumar M, Koley T, Sharma P, Haque MA, Kapil A, et al. Screening of plant-based natural compounds as an inhibitor of FtsZ from salmonella typhi using the computational, biochemical and in vitro cell-based studies. Int J Biol macromolecules (2022) 219:428–37. doi: 10.1016/j.ijbiomac.2022.07.241
22. Tong J, Hou X, Cui D, Chen W, Yao H, Xiong B, et al. A berberine hydrochloride-carboxymethyl chitosan hydrogel protects against staphylococcus aureus infection in a rat mastitis model. Carbohydr polymers (2022) 278:118910. doi: 10.1016/j.carbpol.2021.118910
23. Sun H, Huang SY, Jeyakkumar P, Cai GX, Fang B and Zhou CH. Natural berberine-derived azolyl ethanols as new structural antibacterial agents against drug-resistant escherichia coli. J medicinal Chem (2022) 65(1):436–59. doi: 10.1021/acs.jmedchem.1c01592
24. Oliva R, Mukherjee S, Manisegaran M, Campanile M, Del Vecchio P, Petraccone L, et al. Binding properties of RNA quadruplex of SARS-CoV-2 to berberine compared to telomeric DNA quadruplex. Int J Mol Sci (2022) 23(10):5690–719. doi: 10.3390/ijms23105690
25. Pizzorno A, Padey B, Dubois J, Julien T, Traversier A, Dulière V, et al. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antiviral Res (2020) 181:104878. doi: 10.1016/j.antiviral.2020.104878
26. Li RT, Chen M, Yang ZC, Chen YJ, Huang NH, Chen WH, et al. AIE-based gold nanostar-berberine dimer nanocomposites for PDT and PTT combination therapy toward breast cancer. Nanoscale (2022) 14(27):9818–31. doi: 10.1039/d2nr03408e
27. Chen Z, Vallega KA, Chen H, Zhou J, Ramalingam SS, Sun SY. The natural product berberine synergizes with osimertinib preferentially against MET-amplified osimertinib-resistant lung cancer via direct MET inhibition. Pharmacol Res (2022) 175:105998. doi: 10.1016/j.phrs.2021.105998
28. Liang Y, Fan C, Yan X, Lu X, Jiang H, Di S, et al. Berberine ameliorates lipopolysaccharide-induced acute lung injury via the PERK-mediated Nrf2/HO-1 signaling axis. Phytotherapy Res PTR (2019) 33(1):130–48. doi: 10.1002/ptr.6206
29. Park GB, Park SH, Kim D, Kim YS, Yoon SH, Hur DY. Berberine induces mitochondrial apoptosis of EBV-transformed b cells through p53-mediated regulation of XAF1 and GADD45α. Int J Oncol (2016) 49(1):411–21. doi: 10.3892/ijo.2016.3502
30. Tai CJ, Jassey A, Liu CH, Tai CJ, Richardson CD, Wong SH, et al. Targeting autophagy augments BBR-mediated cell death in human hepatoma cells harboring hepatitis c virus RNA. Cells (2020) 9(4):908–15. doi: 10.3390/cells9040908
31. Rodriguez-Rodriguez BA, Noval MG, Kaczmarek ME, Jang KK, Thannickal SA, Cifuentes Kottkamp A, et al. Atovaquone and berberine chloride reduce SARS-CoV-2 replication In vitro. Viruses (2021) 13(12):2437–51. doi: 10.3390/v13122437
32. Zhang BY, Chen M, Chen XC, Cao K, You Y, Qian YJ, et al. Berberine reduces circulating inflammatory mediators in patients with severe COVID-19. Br J Surg (2021) 108(1):e9–e11. doi: 10.1093/bjs/znaa021
33. Lopes TZ, de Moraes FR, Tedesco AC, Arni RK, Rahal P, Calmon MF. Berberine associated photodynamic therapy promotes autophagy and apoptosis via ROS generation in renal carcinoma cells. Biomed pharmacother = Biomed pharmacotherapie (2020) 123:109794. doi: 10.1016/j.biopha.2019.109794
34. Lee SJ, Noh HJ, Sung EG, Song IH, Kim JY, Kwon TK, et al. Berberine sensitizes TRAIL-induced apoptosis through proteasome-mediated downregulation of c-FLIP and mcl-1 proteins. Int J Oncol (2011) 38(2):485–92. doi: 10.3892/ijo.2010.878
35. Karagiannidis C, Mostert C, Hentschker C, Voshaar T, Malzahn J, Schillinger G, et al. Case characteristics, resource use, and outcomes of 10 021 patients with COVID-19 admitted to 920 German hospitals: an observational study. Lancet Respir Med (2020) 8(9):853–62. doi: 10.1016/s2213-2600(20)30316-7
36. Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature (2020) 584(7821):463–9. doi: 10.1038/s41586-020-2588-y
37. Denize T, Hou Y, Pignon JC, Walton E, West DJ, Freeman GJ, et al. Transcriptomic correlates of tumor cell PD-L1 expression and response to nivolumab monotherapy in metastatic clear cell renal cell carcinoma. Clin Cancer Res an Off J Am Assoc Cancer Res (2022) 28(18):4045–55. doi: 10.1158/1078-0432.ccr-22-0923
38. Rathmell WK, Rumble RB, Van Veldhuizen PJ, Al-Ahmadie H, Emamekhoo H, Hauke RJ, et al. Management of metastatic clear cell renal cell carcinoma: ASCO guideline. J Clin Oncol Off J Am Soc Clin Oncol (2022) 40(25):2957–95. doi: 10.1200/jco.22.00868
39. Chen L, Guan WJ, Qiu ZE, Xu JB, Bai X, Hou XC, et al. SARS-CoV-2 nucleocapsid protein triggers hyperinflammation via protein-protein interaction-mediated intracellular cl(-) accumulation in respiratory epithelium. Signal transduction targeted Ther (2022) 7(1):255. doi: 10.1038/s41392-022-01048-1
40. de Brabander J, Duijvelaar E, Schippers JR, Smeele PJ, Peters-Sengers H, Duitman JW, et al. Immunomodulation and endothelial barrier protection mediate the association between oral imatinib and mortality in hospitalised COVID-19 patients. Eur Respir J (2022) 60(6):2200780–92. doi: 10.1183/13993003.00780-2022
41. Silva CMS, Wanderley CWS, Veras FP, Gonçalves AV, Lima MHF, Toller-Kawahisa JE, et al. Gasdermin-d activation by SARS-CoV-2 triggers NET and mediate COVID-19 immunopathology. Crit Care (London England) (2022) 26(1):206. doi: 10.1186/s13054-022-04062-5
42. McKenna E, Wubben R, Isaza-Correa JM, Melo AM, Mhaonaigh AU, Conlon N, et al. Neutrophils in COVID-19: Not innocent bystanders. Front Immunol (2022) 13:864387. doi: 10.3389/fimmu.2022.864387
43. Melero I, Villalba-Esparza M, Recalde-Zamacona B, Jiménez-Sánchez D, Teijeira Á, Argueta A, et al. Neutrophil extracellular traps, local IL-8 expression, and cytotoxic T-lymphocyte response in the lungs of patients with fatal COVID-19. Chest (2022) 162(5):1006–16. doi: 10.1016/j.chest.2022.06.007
44. Loyer C, Lapostolle A, Urbina T, Elabbadi A, Lavillegrand JR, Chaigneau T, et al. Impairment of neutrophil functions and homeostasis in COVID-19 patients: association with disease severity. Crit Care (London England) (2022) 26(1):155. doi: 10.1186/s13054-022-04002-3
45. Yang H, Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nature Rev Microbiol (2021) 19(11):685–700. doi: 10.1038/s41579-021-00630-8
46. Agarwal A, Rochwerg B, Lamontagne F, Siemieniuk RA, Agoritsas T, Askie L, et al. A living WHO guideline on drugs for covid-19. BMJ (Clinical Res ed.) (2020) 370:m3379. doi: 10.1136/bmj.m3379
47. Farge D, Frere C, Connors JM, Khorana AA, Kakkar A, Ay C, et al. 2022 international clinical practice guidelines for the treatment and prophylaxis of venous thromboembolism in patients with cancer, including patients with COVID-19. Lancet Oncol (2022) 23(7):e334–47. doi: 10.1016/s1470-2045(22)00160-7
48. Crook H, Raza S, Nowell J, Young M, Edison P. Long covid-mechanisms, risk factors, and management. BMJ (Clinical Res ed.) (2021) 374:n1648. doi: 10.1136/bmj.n1648
49. Liu M, Gao Y, Yuan Y, Yang K, Shi S, Zhang J, et al. Efficacy and safety of integrated traditional Chinese and Western medicine for corona virus disease 2019 (COVID-19): a systematic review and meta-analysis. Pharmacol Res (2020) 158:104896. doi: 10.1016/j.phrs.2020.104896
50. Lyu M, Fan G, Xiao G, Wang T, Xu D, Gao J, et al. Traditional Chinese medicine in COVID-19. Acta Pharm Sinica. B (2021) 11(11):3337–63. doi: 10.1016/j.apsb.2021.09.008
51. Xia KY, Zhao Z, Shah T, Wang JY, Baloch Z. Composition, clinical efficiency, and mechanism of NHC-approved "Three Chinese medicines and three Chinese recipes" for COVID-19 treatment. Front Pharmacol (2021) 12:781090. doi: 10.3389/fphar.2021.781090
52. Kevadiya BD, Machhi J, Herskovitz J, Oleynikov MD, Blomberg WR, Bajwa N, et al. Diagnostics for SARS-CoV-2 infections. Nat materials (2021) 20(5):593–605. doi: 10.1038/s41563-020-00906-z
53. Hassler L, Reyes F, Sparks MA, Welling P, Batlle D. Evidence for and against direct kidney infection by SARS-CoV-2 in patients with COVID-19. Clin J Am Soc Nephrol CJASN (2021) 16(11):1755–65. doi: 10.2215/cjn.04560421
54. Jansen J, Reimer KC, Nagai JS, Varghese FS, Overheul GJ, de Beer M, et al. SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell (2022) 29(2):217–231.e8. doi: 10.1016/j.stem.2021.12.010
55. Park GJ, Osinski A, Hernandez G, Eitson JL, Majumdar A, Tonelli M, et al. The mechanism of RNA capping by SARS-CoV-2. Nature (2022) 609(7928):793–800. doi: 10.1038/s41586-022-05185-z
56. Xu Z, Chen S, Liu R, Chen H, Xu B, Xu W, et al. Circular RNA circPOLR2A promotes clear cell renal cell carcinoma progression by facilitating the UBE3C-induced ubiquitination of PEBP1 and, thereby, activating the ERK signaling pathway. Mol Cancer (2022) 21(1):146. doi: 10.1186/s12943-022-01607-8
57. Appelberg S, Gupta S, Svensson Akusjärvi S, Ambikan AT, Mikaeloff F, Saccon E, et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerging Microbes infections (2020) 9(1):1748–60. doi: 10.1080/22221751.2020.1799723
58. Kindrachuk J, Ork B, Hart BJ, Mazur S, Holbrook MR, Frieman MB, et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrobial Agents chemotherapy (2015) 59(2):1088–99. doi: 10.1128/aac.03659-14
59. Kuss-Duerkop SK, Wang J, Mena I, White K, Metreveli G, Sakthivel R, et al. Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication. PloS Pathog (2017) 13(9):e1006635. doi: 10.1371/journal.ppat.1006635
60. Dell'Atti L, Bianchi N, Aguiari G. New therapeutic interventions for kidney carcinoma: Looking to the future. Cancers (2022) 14(15):3616–2628. doi: 10.3390/cancers14153616
61. Pérez-Fernández XL, Fuset-Cabanes M, Hernández-Platero L. Noninvasive respiratory strategies and intubation or mortality among patients with acute hypoxemic respiratory failure due to COVID-19. Jama (2022) 327(20):2022–3. doi: 10.1001/jama.2022.5273
62. Greco M, De Corte T, Ercole A, Antonelli M, Azoulay E, Citerio G, et al. Clinical and organizational factors associated with mortality during the peak of first COVID-19 wave: the global UNITE-COVID study. Intensive Care Med (2022) 48(6):690–705. doi: 10.1007/s00134-022-06705-1
63. Sacco C, Del Manso M, Mateo-Urdiales A, Rota MC, Petrone D, Riccardo F, et al. Effectiveness of BNT162b2 vaccine against SARS-CoV-2 infection and severe COVID-19 in children aged 5-11 years in Italy: a retrospective analysis of January-April, 2022. Lancet (London England) (2022) 400(10346):97–103. doi: 10.1016/s0140-6736(22)01185-0
64. Gutman H, Aftalion M, Melamed S, Politi B, Nevo R, Havusha-Laufer S, et al. Matrix metalloproteinases expression is associated with SARS-CoV-2-Induced lung pathology and extracellular-matrix remodeling in K18-hACE2 mice. Viruses (2022) 14(8):1627–39. doi: 10.3390/v14081627
65. Chung HL, Wangler MF, Marcogliese PC, Jo J, Ravenscroft TA, Zuo Z, et al. Loss- or gain-of-Function mutations in ACOX1 cause axonal loss via different mechanisms. Neuron (2020) 106(4):589–606.e6. doi: 10.1016/j.neuron.2020.02.021
66. Lin M, Stewart MT, Zefi S, Mateti KV, Gauthier A, Sharma B, et al. Dual effects of supplemental oxygen on pulmonary infection, inflammatory lung injury, and neuromodulation in aging and COVID-19. Free Radical Biol Med (2022) 190:247–63. doi: 10.1016/j.freeradbiomed.2022.08.004
67. Huang L, Ye K, McGee MC, Nidetz NF, Elmore JP, Limper CB, et al. Interleukin-2-Inducible T-cell kinase deficiency impairs early pulmonary protection against mycobacterium tuberculosis infection. Front Immunol (2019) 10:3103. doi: 10.3389/fimmu.2019.03103
68. Zhang Y, Li T, Pan M, Wang W, Huang W, Yuan Y, et al. SIRT1 prevents cigarette smoking-induced lung fibroblasts activation by regulating mitochondrial oxidative stress and lipid metabolism. J Trans Med (2022) 20(1):222. doi: 10.1186/s12967-022-03408-5
69. Du W, Zhang L, Brett-Morris A, Aguila B, Kerner J, Hoppel CL, et al. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat Commun (2017) 8(1):1769. doi: 10.1038/s41467-017-01965-8
70. Tan SK, Welford SM. Lipid in renal carcinoma: Queen bee to target? Trends Cancer (2020) 6(6):448–50. doi: 10.1016/j.trecan.2020.02.017
71. Dufies M, Verbiest A, Cooley LS, Ndiaye PD, He X, Nottet N, et al. Plk1, upregulated by HIF-2, mediates metastasis and drug resistance of clear cell renal cell carcinoma. Commun Biol (2021) 4(1):166. doi: 10.1038/s42003-021-01653-w
72. Gao Z, Man X, Li Z, Bi J, Liu X, Li Z, et al. PLK1 promotes proliferation and suppresses apoptosis of renal cell carcinoma cells by phosphorylating MCM3. Cancer Gene Ther (2020) 27(6):412–23. doi: 10.1038/s41417-019-0094-x
73. Wheatley AK, Juno JA, Wang JJ, Selva KJ, Reynaldi A, Tan HX, et al. Evolution of immune responses to SARS-CoV-2 in mild-moderate COVID-19. Nat Commun (2021) 12(1):1162. doi: 10.1038/s41467-021-21444-5
74. Li K, Huang B, Wu M, Zhong A, Li L, Cai Y, et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat Commun (2020) 11(1):6044. doi: 10.1038/s41467-020-19943-y
75. Vanshylla K, Di Cristanziano V, Kleipass F, Dewald F, Schommers P, Gieselmann L, et al. Kinetics and correlates of the neutralizing antibody response to SARS-CoV-2 infection in humans. Cell Host Microbe (2021) 29(6):917–929.e4. doi: 10.1016/j.chom.2021.04.015
76. Ma Q, Immler R, Pruenster M, Sellmayr M, Li C, von Brunn A, et al. Soluble uric acid inhibits β2 integrin-mediated neutrophil recruitment in innate immunity. Blood (2022) 139(23):3402–17. doi: 10.1182/blood.2021011234
77. Muhammad W, Zhu J, Zhai Z, Xie J, Zhou J, Feng X, et al. ROS-responsive polymer nanoparticles with enhanced loading of dexamethasone effectively modulate the lung injury microenvironment. Acta biomaterialia (2022) 148:258–70. doi: 10.1016/j.actbio.2022.06.024
78. Liu K, Zhao S, Li J, Zheng Y, Wu H, Kong J, et al. Knowledge mapping and research hotspots of immunotherapy in renal cell carcinoma: A text-mining study from 2002 to 2021. Front Immunol (2022) 13:969217. doi: 10.3389/fimmu.2022.969217
79. Thouvenin J, Alhalabi O, Carlo M, Carril-Ajuria L, Hirsch L, Martinez-Chanza N, et al. Efficacy of cabozantinib in metastatic MiT family translocation renal cell carcinomas. oncologist (2022) 27(12):1041–7. doi: 10.1093/oncolo/oyac158
80. Xu W, Anwaier A, Liu W, Wei G, Su J, Tian X, et al. Genomic alteration of MTAP/CDKN2A predicts sarcomatoid differentiation and poor prognosis and modulates response to immune checkpoint blockade in renal cell carcinoma. Front Immunol (2022) 13:953721. doi: 10.3389/fimmu.2022.953721
81. Chen YW, Tucker MD, Brown LC, Yasin HA, Ancell KK, Armstrong AJ, et al. The association between a decrease in on-treatment neutrophil-to-Eosinophil ratio (NER) at week 6 after ipilimumab plus nivolumab initiation and improved clinical outcomes in metastatic renal cell carcinoma. Cancers (2022) 14(15):3830–43. doi: 10.3390/cancers14153830
82. Mandala M, Lorigan P, De Luca M, Bianchetti A, Merelli B, Bettini A, et al. SARS-CoV-2 infection and adverse events in patients with cancer receiving immune checkpoint inhibitors: an observational prospective study. J immunotherapy Cancer (2021) 9(2):e001694–e001703. doi: 10.1136/jitc-2020-001694
Keywords: coronavirus disease 2019, kidney clear cell carcinoma, berberine, immune mechanism, molecular docking
Citation: Zheng Z, Li X, Nie K, Wang X, Liang W, Yang F, Zheng K and Zheng Y (2023) Identification of berberine as a potential therapeutic strategy for kidney clear cell carcinoma and COVID-19 based on analysis of large-scale datasets. Front. Immunol. 14:1038651. doi: 10.3389/fimmu.2023.1038651
Received: 07 September 2022; Accepted: 14 March 2023;
Published: 23 March 2023.
Edited by:
Jianzhong Ai, Sichuan University, ChinaCopyright © 2023 Zheng, Li, Nie, Wang, Liang, Yang, Zheng and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kairi Zheng, emhlbmdrYWlyaUAxMjYuY29t; Yihou Zheng, MTM4MDk4ODgyMDZAMTM5LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.