- 1Institute of Respiratory Health, Frontiers Science Center for Disease-related Molecular Network, West China Hospital, Sichuan University, Chengdu, China
- 2Department of Respiratory and Critical Care Medicine, Frontiers Science Center for Disease-related Molecular Network, West China Hospital, Sichuan University, Chengdu, China
Tuberculosis (TB) remains a major global health issue, resulting in around 1.5 million people deaths each year. Better diagnostic and therapeutic tools are urgently needed. Circular RNAs (circRNAs) are a new class of noncoding RNAs with a covalently closed structure, and exhibit a tissue-, cell-, and developmental stage-specific expression pattern. Recently, circRNAs were thought to be regulatory molecules implicated in the onset and progression of a series of human diseases including tuberculosis. In tuberculosis, circRNAs have been shown to regulate host anti-TB immune responses, such as decreasing monocyte apoptosis, enhancing autophagy and promoting macrophage polarization. Importantly, circRNAs are physically stable and abundant in several types of body fluids. Therefore they are considered as promising minimally-invasive biomarkers. In this review, we focus on the recent advances in the immune regulatory roles of circRNAs, as well as their potential diagnostic value in TB.
Introduction
Tuberculosis is the second leading infectious disease cause of death globally after COVID-19, caused by Mycobacterium tuberculosis (Mtb), which usually attacks the lung but can affect almost any part of the body. According to the WHO global TB report, approximately 10 million people fell ill with TB and 1.5 million people died from TB globally in 2020 (1). The COVID-19 pandemic largely impacts global TB control because of reduced access to care, leading to around 5% increase of TB deaths compared to 2018 (2).
Although the modern antibiotics and Bacillus Calmette–Guérin (BCG) vaccine dramatically help human beings to fight TB, it has still not been eradicated. The main reasons are that Mtb could develop drug resistance rapidly under the pressure of antibiotics and the BCG vaccine does not work well in adults (3, 4). Another reason is that Mtb is armed with a set of intricate immune escape mechanisms which enable the bacterium to avoid the host immune killing and to survive in host for a long time (5).
Circular RNAs (circRNAs) are a family of recently rediscovered RNA molecules. Originally thought of as mere byproducts of aberrant splicing (6), circRNAs are now appreciated to have important biological roles including regulating gene expression, modulating protein function, encoding proteins and so forth (7). While the roles of immune protein factors during Mtb infection have been extensively studied, the functions of circRNAs in TB remain relatively unclear. In this review, we provide an overview of the current understanding of circRNAs, with a particular focus on their functions in immune regulation. We introduce recent investigations that reveal new roles of host circRNAs in anti-TB immunity, and discuss the potential of circRNAs as novel TB diagnostic biomarkers.
The discovery, biogenesis and function of circRNAs
The discovery of CircRNAs
The first circRNA was discovered in 1976 on viroid particles (8). A few years later, circRNAs were observed by electron microscopy in the cytoplasm of eukaryotic cells (9). Nevertheless, they were mainly considered as “junk RNAs” generated from abnormal splicing events at that time (6). Until 1993, a testis-specific 1.3 kb circRNA from the sex-determining region Y (Sry) gene in mice was molecularly identified. This circRNA was considered to have a potential function in mouse testis (10). Further, next-generation sequencing (NGS) with specific protocols for library preparation ushered in the genome-wide profiling of circRNAs. It is now accepted that circRNAs are the predominant transcript isoforms from thousands of human genes rather than simply accidental byproducts of splicing (11), and their expression is conserved in eukaryotes (12).
The biogenesis of CircRNAs
Based on their composition, circRNAs are currently divided into four categories: circular intronic RNAs (ciRNAs), exon–intron circRNAs (EIciRNAs), exonic circRNAs (ecircRNA) and tRNA intronic circular RNAs (tricRNAs) (Figure 1). The intronic lariat generated from canonical splicing is usually attacked by a debranching enzyme DBR1 and by exonucleases. Thus the cellular lariat is only an intermediate molecule that is usually rapidly degraded, but some lariats appear to be capable of evading this debranching process to form stable ciRNAs (13). ecircRNAs and EIciRNAs come from the back-splicing process wherein a downstream 5′ splice donor site is ligated to an upstream 3′ splice acceptor site (14), unlike canonical splicing that joins an upstream 5′ splice donor site with a downstream 3′ splice acceptor site. The circulation of pre-mRNA during back-splicing is mediated by the base paring between reverse complementary sequences of flanking introns or the dimerization of RNA binding proteins (RBPs) (15, 16). During the pre-mRNA transcription, exon skipping events may occur and forms an excised lariat. Subsequently, the lariat undergoes internal back-splicing leading to the formation of ecircRNAs or EIciRNAs if the flanking intronic sequences are retained in some circumstances (17, 18). EcircRNAs account for over 80% of the identified circRNAs and generally localize in the cytoplasm (9, 15, 19). In contrast, ciRNAs and EIciRNAs are usually localized in the nucleus (13, 18), suggesting that they may regulate gene transcription. TricRNAs are formed by tRNA introns through the cleavage of tRNA introns by tRNA splicing endonuclease (TSEN) complex and the circulation cleaved introns by an unknown ligase (20).
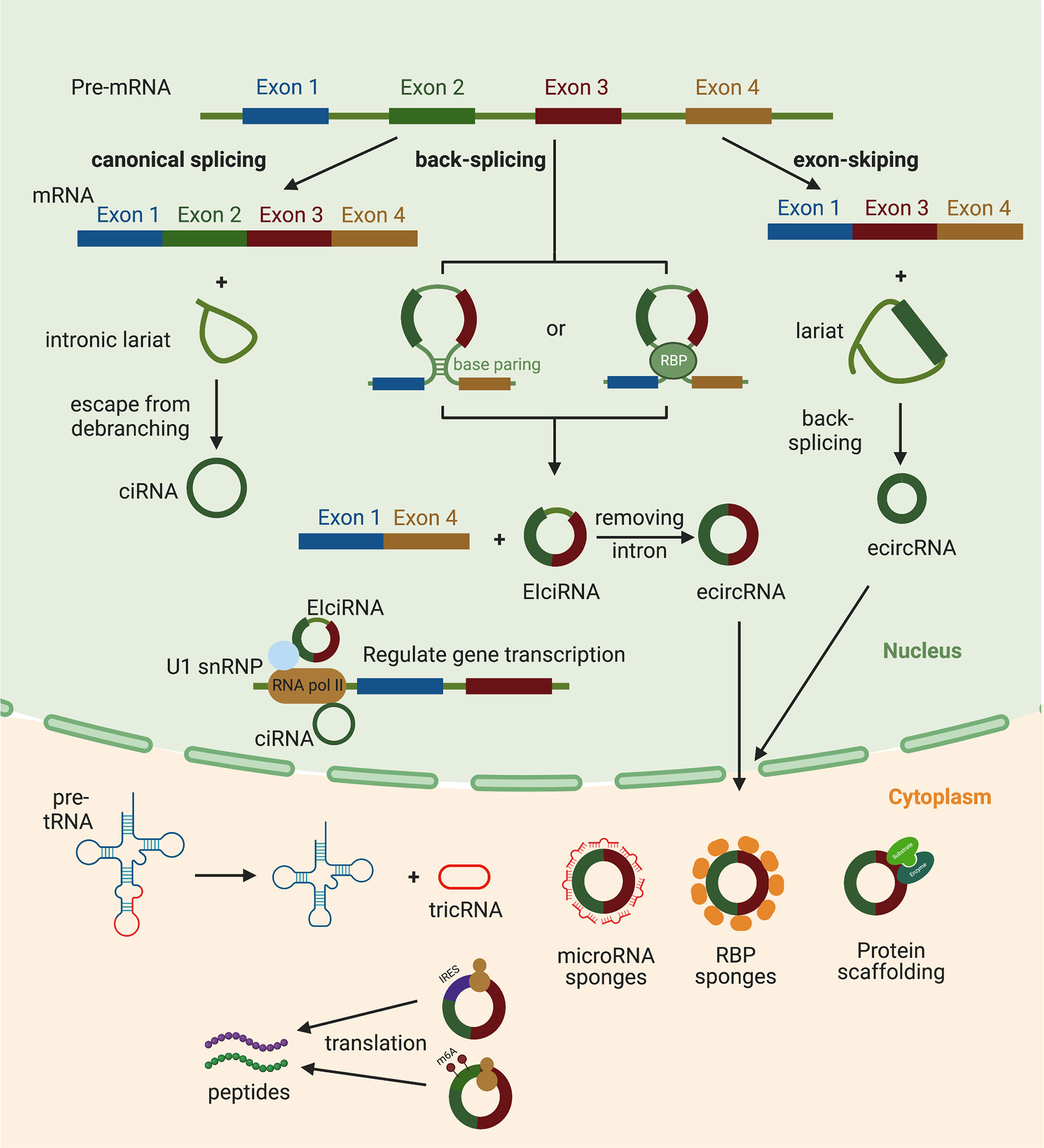
Figure 1 The biogenesis and functions of circRNAs. Exonic circRNAs (EcircRNAs) are generated by a non-canonical back-splicing which is favored by the base paring between reverse complementary sequences (such as Alu repeats) and the dimerization of RNA binding proteins (RBPs). EcircRNAs can also be produced from splicing intermediates called as lariat precursors that are created by an exon-skipping event. Circular intronic RNAs (ciRNAs) are generated from intronic lariats that escape from the debranching step of canonical linear splicing. tRNA intronic circular RNAs (tricRNAs) are formed during the process of pre-tRNA splicing. CiRNAs and EIciRNAs are located in nucleus. CiRNAs can interact with the RNA pol II complex and play a role in regulating parental gene transcription. EIciRNAs can interact with U1 small nuclear ribonucleoproteins and then increase the transcription of their host genes by binding with RNA pol II. EcircRNAs are translocated to cytoplasm after formation, and can act as microRNA sponges, RBP sponges, protein scaffoldings or templates of protein translation.
The biological function of circRNAs
Although circRNAs were discovered decades ago, biological functions have only been studied for a small fraction of the circRNAs identified to date. Most of them were thought to act as miRNA sponges (19, 21–23) (Figure 1). Those circRNAs contain miRNA binding sites, which allow them to sponge miRNAs and restrain miRNAs’ function, thus indirectly regulating the translation of miRNAs’ target mRNAs. CiRS-7, which is highly expressed in the brain, contains more than 70 conserved binding sites for miR-7 (21). It has been reported to regulate the expression of miR-7 target genes and be involved in neuronal function. In addition, some circRNAs have also been demonstrated to act as RBPs’ sponges or protein scaffolds (24–27). The circMbl contains multiple conserved muscleblind (MBL) binding sites and thus is able to be specifically bound by MBL (28). The binding of MBL to circMbl decrease the available cellular MBL proteins levels. Its binding to the flanking introns of circMbl also facilitates the looping of mbl pre-mRNA and promotes the circMbl biogenesis which competes with the linear splicing of pre-mRNA, thus decreasing the production of mbl mRNA (28). By binding to both 3-phosphoinositide-dependent protein kinase 1 (PDK1) and its substrate AKT1, circ-Amotl1 can be as a protein scaffold to facilitate the PDK-1 dependent phosphorylation of AKT1 (25). Furthermore, circRNAs could also be translated into peptides or proteins in a cap-independent manner through internal ribosome entry sites (IRESs) (29–32). For example, circ-FBXW7 contains an ORF that can be translated into a novel 185-amino acids protein called FBXW-185aa, which has been reported to reduce the half-life of c-Myc and inhibit proliferation of cancer cells by interacting with the deubiquitinating enzyme USP28 (33). In addition to IRESs, m6A modification was also reported to be able to drive the translation of circRNAs (31).
CircRNAs in immune regulation
As newly identified macromolecules, their roles in immune regulation attracted great attention. Some evidences supported that circRNAs are directly involved in immune regulation. Y. Grace Chen et al. first showed transfection of exogenous circRNA led to potent induction of innate immunity genes through the nucleic acid sensor RIG-I (34). Another study found that circRNAs can competitively bind and inhibit protein kinase R (PKR), a double-strand RNA activated enzyme in the antiviral signaling pathway, to regulate cellular immune signaling pathways extensively (35). Most cellular cicrRNAs have one or more intra-molecular imperfect RNA duplexes ranging from 16 to 26 bp, which is similar to double-strand RNA structures, to interact with PKR to suppress its activity. When cells are infected with viruses or stimulated by poly(I:C), RNase L degrades cellular circRNAs to release PKR, thus activating the downstream antiviral immunity mechanisms (35).
CircRNAs may also paly roles in the activation and function of immune cells. In response to different environmental stimuli, macrophages could be activated into pro-inflammatory M1-type macrophages or anti-inflammatory M2-type macrophages. One study investigated circRNAs expression profiles in M1- and M2-type macrophages using circRNA microarray. The authors identified 189 differentially expressed circRNAs, indicating that circRNAs may play important roles in regulating or maintaining macrophage polarization (36). Indeed, circRasGEF1B, an LPS-inducible cytoplasmic circRNA, has been reported to regulate the expression of intercellular adhesion molecule 1 (ICAM-1) in macrophages during LPS stimulation (37). ICAM-1 is known to recruit leukocytes to inflamed sites and mediate cell-to-cell interactions during antigen presentation (38, 39). In addition to macrophages, circRNAs’ expression profiles were also examined in neutrophils between healthy subjects and patients with asymptomatic Moyamoya disease (40). In this study, 123 circRNAs were identified differentially expressed between the two groups (40). Another comprehensive circRNA profiling study discovered that circRNA100783 is involved in chronic CD28-associated CD8+ T cell aging (41). It was also found that down-regulation of hsa_circ_0012919 increased the expression of DNMT1, decreased expression of CD70 and CD11a, in CD4+ T cells of patients with systemic lupus erythematous (42).
The role of circRNAs in tuberculosis
Although excise roles of host circRNAs in TB remain to be further investigated, current evidences suggested that circRNAs could act as an important regulator in host anti-TB immune processes. Autophagy is an important cellular defense mechanism against intracellular pathogens including Mtb (43). Several circRNAs have been reported to induce autophagy in host cells. CircTRAPPC6B, one of the downregulated circRNAs in peripheral blood mononuclear cells (PBMCs) from active TB patients, could induce autophagy in Mtb‐infected macrophages by abolishing the repression of miR‐874‐3p on ATG16L1 expression (44). CircAGFG1, an upregulated circRNA in macrophages from TB patients, also has been reported to enhance autophagy whereas reduce apoptosis of Mtb-infected macrophages via the miRNA-1257/Notch axis (45). In contrast, hsa_circ_0045474, which is downregulated in monocytes from patients with TB, negatively regulates the autophagy level in macrophages likely by repressing the expression of miR-582-5p (46). Hsa_circRNA_103571 was also down-regulated in active TB patients. Bioinformatic analysis revealed that it has strong relationship with the biological process of autophagy by analyzing its potential target miRNAs and corresponding target genes of these miRNAs (47). CircRNA_101128 is highly expressed in PBMCs from active TB patients and negatively correlates with the level of its potential target miRNA let-7a, which is a known autophagy regulator, suggesting that it may play a role in regulating autophagy levels (48). Furthermore, some circRNAs, such as circ-Dnmt1, circCDYL, circMUC16, circPAN3, circ-0009910, circ0085131, circRACGAP1, circMOT1, cicr-0023404, circEIF6 and circ-0035483, are known to activate autophagy in cancer cells (49), but their roles in autophagy during Mtb infection remain unknown.
It has been known that macrophage polarization drives granuloma outcome during Mtb infection and Mtb has the potential to modulate macrophage polarization (50). An upregulated circRNA in TB patients, namely hsa_circ_0003528, was found to promote M1 to M2 macrophage polarization by sponging miR-324-5p, miR-224-5p and miR-488-5p and thus upregulating CTLA4 (51). Additionally, hsa-circRNA-100237 was indicated to play a role in TB pathogenesis by regulating macrophage activities (47). The potential mechanism is that the down-regulated hsa-circRNA-100237 in active TB patients could act as an miR-33 sponge, therefore promoting lipid storage by reducing mitochondrial fatty acid oxidation (47, 52). Circ_0001490 expression was down-regulated in the serum of TB patients and M.tb-infected THP-1 macrophages (53). Recently, it has been reported that circ_0001490 repressed M.tb survival and promoted the viability and inflammatory responses of THP-1 macrophages by regulating miR-579-3p/FSTL1 axis (53). CircPWWP2A, which is downregulated in Mtb-infected macrophages, was also reported to be able to protect human macrophages from Mtb-induced cytotoxicity by sponging miR-567 and thus abolishing the suppression of miR-567 on two pro-survival proteins, SIRT1 and PDK1 (54). Furthermore, the circRNA_051239 was reported to be significantly upregulated in drug-resistant TB patients (55). MiR-320a have three binding sites of circRNA_051239 and is significantly down-regulated in the drug-resistant patients (56). Thus, the circRNA_051239 may modulate the development of drug-resistance by targeting miR-320a.
The roles of circRNA in nontuberculous mycobacteria infection was also started to be investigated very recently. One study examined circRNA expression profiles of Mycobacterium avium subsp. paratuberculosis (MAP)-infected bovine monocyte-macrophages and uninfected cells by RNA sequencing (57). Authors identified 39 differentially expressed circRNAs between MAP-infected and uninfected macrophages, including 12 upregulated and 27 downregulated circRNAs in MAP-infected macrophages. Bioinformatic analysis showed that these cicrRNAs might play roles in Th1/Th2/Th17 cell differentiation, necroptosis, and JAK-STAT/chemokine signaling pathways. The other study screened circRNAs’ expression in osteocyte-like cells treated with N-glycosylated muramyl dipeptide (N.g MDP) from Mycobacterium leprae (M. leprae), trying to understand the mechanisms underlying the bone remodeling during M. leprae infection (58). In this study, 724 differentially expressed circRNAs and 724 differentially expressed messenger RNAs were identified, and 58 circRNA–miRNA–mRNA interaction pairs were obtained. The following analysis showed that these 58 genes are uniquely associated with “Circadian Rhythm” including Clock, which was known to be able to regulate bone formation, suggesting that circRNAs may paly roles in bone remodeling during M. leprae infection by regulating Clock genes’ expression.
Anti-tuberculosis drug-induced liver injury (ADLI) often leads to treatment interruptions. To explore ADLI-specific circRNAs, Biao Li et al. assessed the circRNA expression profiles in serums from TB patients with or without ADLI and in hepatocytes treated or untreated with anti-tuberculosis drugs (59). 113 co-differentially expressed circRNAs were identified in this screening. An upregulated circRNA among them, circMARS, was found to play roles in the compensatory repair mechanism of ADLI through the circMARS–miR-6808-5p/-6874-3p/-3157-5p–KMT2C–EGFR function axis. Another study published in this year found a down-regulated circRNA in ADLI patients called has_circ_0093884 could upregulate the expression of an anti-inflammatory protein SIRT1 by binding ribosomal protein S3 and thus regulate the hepatocyte inflammation in ADLI (60).
CircRNAs as diagnostic biomarkers for tuberculosis
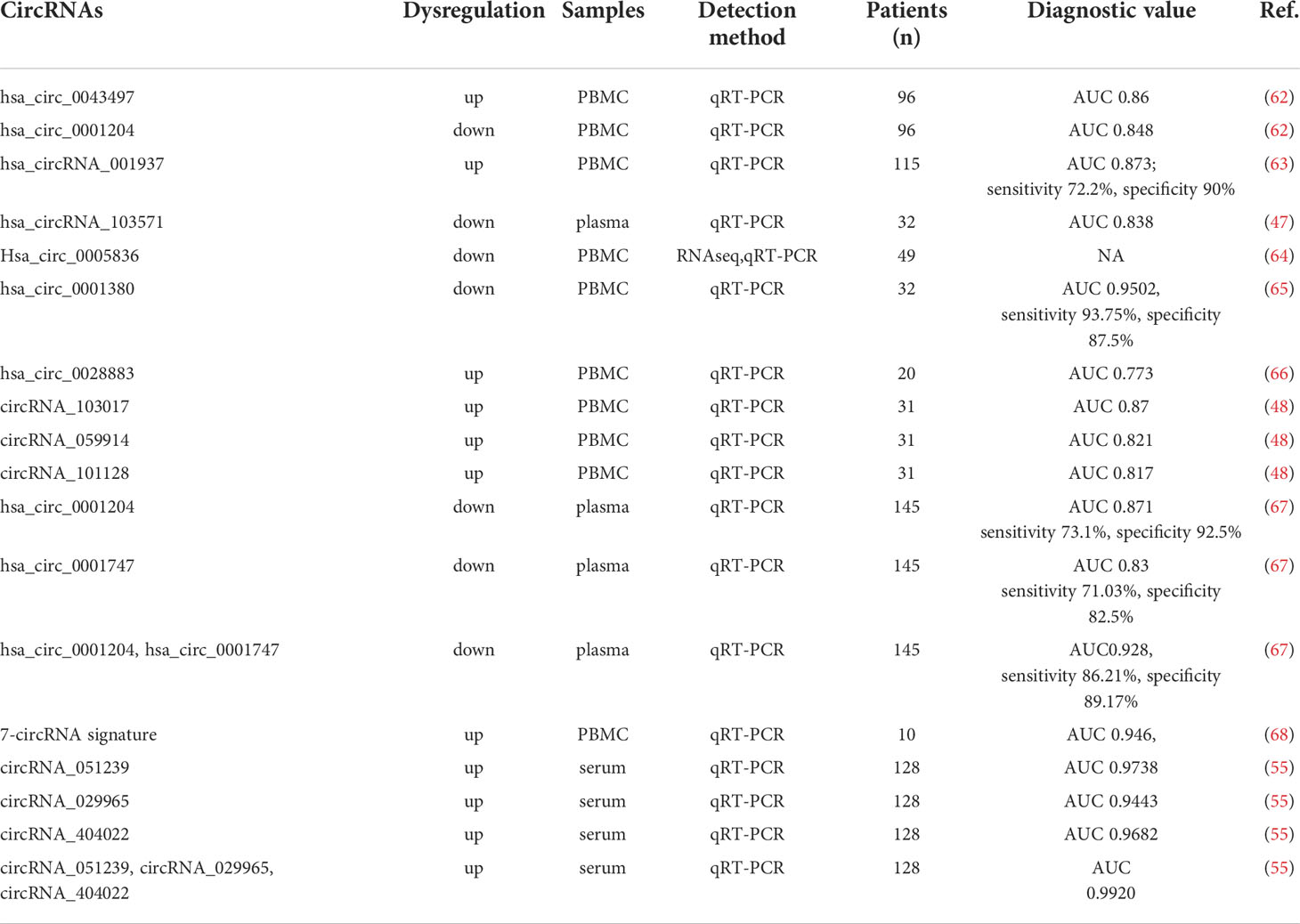
Some circRNAs are known to be expressed in a disease-specific manner. Combined with other features of circRNAs including conservation, stability and high abundance in body fluids, circRNAs are believed to be promising biomarkers for various diseases including TB (61) (Table 1). Using whole transcriptome sequencing, Zhang et al. identified 170 dysregulated circRNAs in whole blood samples from pulmonary TB patients, compared with samples in health individuals (69). Their findings suggested that circRNA-linked competing endogenous RNAs (ceRNA) -mediated regulation of gene is critical for pulmonary TB pathogenesis. Using microarray and quantitative real-time PCR analysis, Huang et al. revealed that a number of circRNAs were differentially expressed in PBMCs from active TB patients. Among them, hsa_ circ_001937 exhibits a substantial diagnostic value for TB with an area-under-curve (AUC) value of 0.873 in the receiver operating characteristic (ROC) curve analysis (63). It was specifically upregulated in patients with TB compared with patients with other lung diseases, and was also correlated with TB severity (63). Yi et al. found that the level of hsa_circRNA_103571 significantly decreased in active TB plasma samples and could be served as a potential biomarker for active TB diagnosis with an AUC value of 0.838 (47). Zhuang et al. reported that hsa_circ_0009128 and hsa_circ_0005836 were significantly down-regulated in the PBMCs of active pulmonary TB (APTB) patients compared with health controls and the hsa_circ_0005836 might act as novel potential biomarkers for APTB (64). Recently, this group reported hsa_circ_0001380 was also significantly downregulated in the PBMCs from TB patients compared with healthy individuals. The ROC curve analysis revealed that the AUC value for distinguishing APTB using hsa_circ_0001380 was 0.9502, indicating the high diagnostic value of this circRNA in APTB (65). Another study identified three differentially expressed cicrRNAs by analyzing public GEO datasets and found hsa_circ_0028883 showed a potential diagnostic value in active TB with an AUC value of 0.773 (66). Additionally, Fu et al. characterized the expression profiles of circRNAs in PBMCs of active TB patients and discovered 171 circRNAs were dysregulated in TB samples. Among them, circRNA_059914, circRNA_101128 and circRNA_103017 were significantly upregulated and showed a potential diagnostic power with an AUC value of 0.87, 0.821 and 0.817 respectively (48).
In addition to individual circRNA, the combination of circRNAs has shown a better predictive power in TB diagnosis. One study reported the expression of two circRNAs (hsa_circ_0001204 and hsa_circ_0001747) was significantly decreased in active TB plasma samples compared with health controls. The ROC curve analysis exhibited an AUC value of 0.928 for distinguishing TB patients when hsa_circ_0001747 and hsa_circ_0001204 were used in combination, indicating this circRNA combination could act as a novel biomarker for active TB diagnosis (67). Qian et al. also identified differentially expressed circRNAs in pulmonary TB patients’ PMBCs. Among them, seven circRNAs including hsa_circ_0000414, hsa_circ_0002908, hsa_circ_0000681, hsa_circ_0002362, hsa_circ_0002113, hsa_circ_0008797 and hsa_circ_0063179 were chosen to develop a circRNA-based molecular signature based on pathway analysis. In validated groups, the 7-circRNA-based TB index was significantly higher in TB patients than that in healthy controls with an AUC value of 0.946 (68). Furthermore, Liu et al. identified three circRNAs (circRNA_029965, circRNA_051239 and circRNA_404022) which showed significantly increases in the serum of the active TB patients. The AUC value of the combination of these three circRNAs in ROC curve analysis is as high as 0.992, suggesting these cicrRNAs could serve as ideal potential biomarkers for TB diagnosis (55).
Discussion
The current advanced RNA-sequencing technologies and data analysis algorithms have largely promoted the discovery of host circRNAs. These circRNAs, at least some of them, are now considered to have important functions in the process of Mtb infection. However, current studies on the roles of circRNAs in TB are mainly focused on autophagy and macrophage polarization. Exact mechanisms of circRNAs’ action are still largely unknown. The roles of circRNAs in other immune processes during Mtb infection and their mechanisms need to be comprehensively studied. Also, new technologies, analyses and strategies are still needed to identify key functional circRNAs in TB.
Due to their excellent high abundance and stability in body fluids, circRNAs are regarded as promising diagnostic biomarkers for human diseases including TB. Indeed, as listed in Table 1, dozens of circRNAs or combinations are reported as biomarkers for the diagnosis of TB with varied predictive power. However, several problems/challenges still need to be addressed in the future. Firstly, the samples sizes of current published studies are relatively small. Muti-center large cohort studies are needed to comprehensively evaluate the diagnostic value of these circRNAs. The sensitivity and reliability of using circRNAs as diagnostic biomarkers need further validation. Secondly, as the absolute expression level of circRNAs in samples may vary from person to person, it may be difficult to set up a normal baseline for distinguishing patients from healthy people. Thus, a standardized protocol for circRNAs detection in clinical samples is required. Finally, detection of circRNAs in clinical samples is more expensive and time-consuming currently than protein detection, which may limit the application of circRNAs as biomarkers. It is very important to be considered as the fact that over 95% TB patients are in developing countries.
In addition to being diagnostic biomarkers, circRNAs could be as potential therapeutic targets or therapeutic strategies for tuberculosis. The disease-promoting circRNAs could be knocked down by RNAi or CRISPR/Cas9-based gene editing. The therapeutic circRNAs could be designed and synthesized artificially according to the clinical need. Liu et al. artificially synthesized a circRNA for the first time and showed this circRNA could inhibit the proliferation of gastric cancer cells in vitro by sponging miR-21 (70). Furthermore, a very recent elegant study found the circRNA vaccine against SARS-CoV-2 enabled higher and more durable antigen production than the modified mRNA vaccine and induced a higher neutralizing antibody titers (71). This study provided a novel idea for the potential application of circRNAs in developing TB vaccine in the future.
Author contributions
QW, DY, YZ, and DW drafted the manuscript. QW and WL supervised and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the startup fund from West China Hospital to Qinglan Wang (137210102).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
2. Pai M, Kasaeva T, Swaminathan S. Covid-19’s devastating effect on tuberculosis care - a path to recovery. N Engl J Med (2022) 386(16):1490–3. doi: 10.1056/NEJMp2118145
3. Jain A, Mondal R. Extensively drug-resistant tuberculosis: Current challenges and threats. FEMS Immunol Med Microbiol (2008) 53(2):145–50. doi: 10.1111/j.1574-695X.2008.00400.x
4. Andersen P, Doherty TM. The success and failure of bcg - implications for a novel tuberculosis vaccine. Nat Rev Microbiol (2005) 3(8):656–62. doi: 10.1038/nrmicro1211
5. Zhai W, Wu F, Zhang Y, Fu Y, Liu Z. The immune escape mechanisms of mycobacterium tuberculosis. Int J Mol Sci (2019) 20:340. doi: 10.3390/ijms20020340
6. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular rna molecules. FASEB J (1993) 7(1):155–60. doi: 10.1096/fasebj.7.1.7678559
7. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular rnas. Mol Cell (2018) 71(3):428–42. doi: 10.1016/j.molcel.2018.06.034
8. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular rna molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
9. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of rna in the cytoplasm of eukaryotic cells. Nature (1979) 280(5720):339–40. doi: 10.1038/280339a0
10. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene sry in adult mouse testis. Cell (1993) 73(5):1019–30. doi: 10.1016/0092-8674(93)90279-y
11. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular rnas are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One (2012) 7(2):e30733. doi: 10.1371/journal.pone.0030733
12. Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular rna is expressed across the eukaryotic tree of life. PloS One (2014) 9(6):e90859. doi: 10.1371/journal.pone.0090859
13. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding rnas. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
14. Vicens Q, Westhof E. Biogenesis of circular rnas. Cell (2014) 159(1):13–4. doi: 10.1016/j.cell.2014.09.005
15. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular rnas are abundant, conserved, and associated with alu repeats. RNA (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
16. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The rna binding protein quaking regulates formation of circrnas. Cell (2015) 160(6):1125–34. doi: 10.1016/j.cell.2015.02.014
17. Barrett SP, Wang PL, Salzman J. Circular rna biogenesis can proceed through an exon-containing lariat precursor. Elife (2015) 4:e07540. doi: 10.7554/eLife.07540
18. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular rnas regulate transcription in the nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
19. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular rnas are a Large class of animal rnas with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
20. Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, et al. Metazoan trna introns generate stable circular rnas in vivo. RNA (2015) 21(9):1554–65. doi: 10.1261/rna.052944.115
21. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural rna circles function as efficient microrna sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
22. Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular rna locus causes mirna deregulation and affects brain function. Science (2017) 357(6357):1254. doi: 10.1126/science.aam8526
23. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular rna profiling reveals an abundant Circhipk3 that regulates cell growth by sponging multiple mirnas. Nat Commun (2016) 7:11215. doi: 10.1038/ncomms11215
24. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of hur target circular rnas uncovers suppression of Pabpn1 translation by Circpabpn1. RNA Biol (2017) 14(3):361–9. doi: 10.1080/15476286.2017.1279788
25. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A circular rna binds to and activates akt phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics (2017) 7(16):3842–55. doi: 10.7150/thno.19764
26. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular rna retards cell cycle progression Via forming ternary complexes with P21 and Cdk2. Nucleic Acids Res (2016) 44(6):2846–58. doi: 10.1093/nar/gkw027
27. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular rna enhancing Foxo3 activity. Cell Death Differ (2017) 24(2):357–70. doi: 10.1038/cdd.2016.133
28. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. Circrna biogenesis competes with pre-mrna splicing. Mol Cell (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
29. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-Znf609 is a circular rna that can be translated and functions in myogenesis. Mol Cell (2017) 66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017
30. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of circrnas. Mol Cell (2017) 66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021
31. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular rnas driven by N(6)-methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
32. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of linc-pint suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun (2018) 9(1):4475. doi: 10.1038/s41467-018-06862-2
33. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of Fbxw7 circular rna in repressing glioma tumorigenesis. J Natl Cancer Inst (2018) 110(3):304–15. doi: 10.1093/jnci/djx166
34. Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, et al. Sensing self and foreign circular rnas by intron identity. Mol Cell (2017) 67(2):228–38.e5. doi: 10.1016/j.molcel.2017.05.022
35. Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. Structure and degradation of circular rnas regulate pkr activation in innate immunity. Cell (2019) 177(4):865–80.e21. doi: 10.1016/j.cell.2019.03.046
36. Zhang Y, Zhang Y, Li X, Zhang M, Lv K. Microarray analysis of circular rna expression patterns in polarized macrophages. Int J Mol Med (2017) 39(2):373–9. doi: 10.3892/ijmm.2017.2852
37. Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK. Inducible Rasgef1b circular rna is a positive regulator of icam-1 in the Tlr4/Lps pathway. RNA Biol (2016) 13(9):861–71. doi: 10.1080/15476286.2016.1207036
38. Long EO. Icam-1: Getting a grip on leukocyte adhesion. J Immunol (2011) 186(9):5021–3. doi: 10.4049/jimmunol.1100646
39. Lebedeva T, Dustin ML, Sykulev Y. Icam-1 Co-stimulates target cells to facilitate antigen presentation. Curr Opin Immunol (2005) 17(3):251–8. doi: 10.1016/j.coi.2005.04.008
40. Ma Q, Li L, Yu B, Jiao L, Han Z, Zhao H, et al. Circular rna profiling of neutrophil transcriptome provides insights into asymptomatic moyamoya disease. Brain Res (2019) 1719:104–12. doi: 10.1016/j.brainres.2019.05.033
41. Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular rna profiling reveals that circular Rna100783 is involved in chronic Cd28-associated Cd8(+)T cell ageing. Immun Ageing (2015) 12:17. doi: 10.1186/s12979-015-0042-z
42. Zhang C, Wang X, Chen Y, Wu Z, Zhang C, Shi W. The down-regulation of Hsa_Circ_0012919, the sponge for mir-125a-3p, contributes to DNA methylation of Cd11a and Cd70 in Cd4(+) T cells of systemic lupus erythematous. Clin Sci (Lond) (2018) 132(21):2285–98. doi: 10.1042/CS20180403
43. Deretic V. Autophagy in tuberculosis. Cold Spring Harb Perspect Med (2014) 4(11):a018481. doi: 10.1101/cshperspect.a018481
44. Luo HL, Pi J, Zhang JA, Yang EZ, Xu H, Luo H, et al. Circular rna Trappc6b inhibits intracellular mycobacterium tuberculosis growth while inducing autophagy in macrophages by targeting microrna-874-3p. Clin Transl Immunol (2021) 10(2):e1254. doi: 10.1002/cti2.1254
45. Shi Q, Wang J, Yang Z, Liu Y. Circagfg1modulates autophagy and apoptosis of macrophages infected by mycobacterium tuberculosis Via the notch signaling pathway. Ann Transl Med (2020) 8(10):645. doi: 10.21037/atm.2020-20-3048
46. Wu M, Liu Z, Zhang S. Down-regulation of Hsa_Circ_0045474 induces macrophage autophagy in tuberculosis Via mir-582-5p/Tnks2 axis. Innate Immun (2022) 28(1):11–8. doi: 10.1177/17534259211064285
47. Yi Z, Gao K, Li R, Fu Y. Dysregulated circrnas in plasma from active tuberculosis patients. J Cell Mol Med (2018) 22(9):4076–84. doi: 10.1111/jcmm.13684
48. Fu Y, Wang J, Qiao J, Yi Z. Signature of circular rnas in peripheral blood mononuclear cells from patients with active tuberculosis. J Cell Mol Med (2019) 23(3):1917–25. doi: 10.1111/jcmm.14093
49. Zhou Z, Zhang Y, Gao J, Hao X, Shan C, Li J, et al. Circular rnas act as regulators of autophagy in cancer. Mol Ther Oncolytics (2021) 21:242–54. doi: 10.1016/j.omto.2021.04.007
50. Marino S, Cilfone NA, Mattila JT, Linderman JJ, Flynn JL, Kirschner DE. Macrophage polarization drives granuloma outcome during mycobacterium tuberculosis infection. Infect Immun (2015) 83(1):324–38. doi: 10.1128/IAI.02494-14
51. Huang Z, Yao F, Liu J, Xu J, Guo Y, Su R, et al. Up-regulation of circrna-0003528 promotes mycobacterium tuberculosis associated macrophage polarization Via down-regulating mir-224-5p, mir-324-5p and mir-488-5p and up-regulating Ctla4. Aging (Albany NY) (2020) 12(24):25658–72. doi: 10.18632/aging.104175
52. Clerbaux LA, Schultz H, Roman-Holba S, Ruan DF, Yu R, Lamb AM, et al. The microrna mir-33 is a pleiotropic regulator of metabolic and developmental processes in drosophila melanogaster. Dev Dyn (2021) 250(11):1634–50. doi: 10.1002/dvdy.344
53. Deng Q, Huang J, Yan J, Mao E, Chen H, Wang C. Circ_0001490/Mir-579-3p/Fstl1 axis modulates the survival of mycobacteria and the viability, apoptosis and inflammatory response in mycobacterium tuberculosis-infected macrophages. Tuberc (Edinb) (2021) 131:102123. doi: 10.1016/j.tube.2021.102123
54. Ma J, Chen XL, Sun Q. Microrna-579 upregulation mediates death of human macrophages with mycobacterium tuberculosis infection. Biochem Biophys Res Commun (2019) 518(2):219–26. doi: 10.1016/j.bbrc.2019.08.035
55. Liu H, Lu G, Wang W, Jiang X, Gu S, Wang J, et al. A panel of circrnas in the serum serves as biomarkers for mycobacterium tuberculosis infection. Front Microbiol (2020) 11:1215. doi: 10.3389/fmicb.2020.01215
56. Cui JY, Liang HW, Pan XL, Li D, Jiao N, Liu YH, et al. Characterization of a novel panel of plasma micrornas that discriminates between mycobacterium tuberculosis infection and healthy individuals. PloS One (2017) 12(9):e0184113. doi: 10.1371/journal.pone.0184113
57. Bao Y, Yao Y, Wang Z, Wu S, Jiang X, Ma H. Analysis of mrna and circrna expression profiles of bovine monocyte-derived macrophages infected with mycobacterium avium subsp. Paratuberculosis Front Microbiol (2021) 12:796922. doi: 10.3389/fmicb.2021.796922
58. Gao ZR, Liu Q, Zhao J, Zhao YQ, Tan L, Zhang SH, et al. A comprehensive analysis of the circrna-Mirna-Mrna network in osteocyte-like cell associated with mycobacterium leprae infection. PloS Negl Trop Dis (2022) 16(5):e0010379. doi: 10.1371/journal.pntd.0010379
59. Li B, Ren Q, Li Y, Tian S, Chong Y, Sun S, et al. Screening differential circular rna expression profiles reveals the regulatory role of circmars in anti-tuberculosis drug-induced liver injury. J Cell Mol Med (2022) 26(4):1050–9. doi: 10.1111/jcmm.17157
60. Yang L, Wang L, Tang Q, Liu Y, Meng C, Sun S, et al. Hsa_Circ_0093884 bound to rna-binding protein Rps3 ameliorates hepatocyte inflammation in anti-tuberculosis drug-induced liver injury by competitively activating Sirt1. Int Immunopharmacol (2022) 110:109018. doi: 10.1016/j.intimp.2022.109018
61. Zhang Z, Yang T, Xiao J. Circular rnas: Promising biomarkers for human diseases. EBioMedicine (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
62. Huang Z, Su R, Deng Z, Xu J, Peng Y, Luo Q, et al. Identification of differentially expressed circular rnas in human monocyte derived macrophages response to mycobacterium tuberculosis infection. Sci Rep (2017) 7(1):13673. doi: 10.1038/s41598-017-13885-0
63. Huang ZK, Yao FY, Xu JQ, Deng Z, Su RG, Peng YP, et al. Microarray expression profile of circular rnas in peripheral blood mononuclear cells from active tuberculosis patients. Cell Physiol Biochem (2018) 45(3):1230–40. doi: 10.1159/000487454
64. Zhuang ZG, Zhang JA, Luo HL, Liu GB, Lu YB, Ge NH, et al. The circular rna of peripheral blood mononuclear cells: Hsa_Circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis. Mol Immunol (2017) 90:264–72. doi: 10.1016/j.molimm.2017.08.008
65. Luo HL, Peng Y, Luo H, Zhang JA, Liu GB, Xu H, et al. Circular rna Hsa_Circ_0001380 in peripheral blood as a potential diagnostic biomarker for active pulmonary tuberculosis. Mol Med Rep (2020) 21(4):1890–6. doi: 10.3892/mmr.2020.10992
66. Zhang X, Zhang Q, Wu Q, Tang H, Ye L, Zhang Q, et al. Integrated analyses reveal Hsa_Circ_0028883 as a diagnostic biomarker in active tuberculosis. Infect Genet Evol (2020) 83:104323. doi: 10.1016/j.meegid.2020.104323
67. Huang Z, Su R, Yao F, Peng Y, Luo Q, Li J. Circulating circular rnas Hsa_Circ_0001204 and Hsa_Circ_0001747 act as diagnostic biomarkers for active tuberculosis detection. Int J Clin Exp Pathol (2018) 11(2):586–94.
68. Qian Z, Liu H, Li M, Shi J, Li N, Zhang Y, et al. Potential diagnostic power of blood circular rna expression in active pulmonary tuberculosis. EBioMedicine (2018) 27:18–26. doi: 10.1016/j.ebiom.2017.12.007
69. Zhang X, Zhu M, Yang R, Zhao W, Hu X, Gan J. Identification and comparison of novel circular rnas with associated Co-expression and competing endogenous rna networks in pulmonary tuberculosis. Oncotarget (2017) 8(69):113571–82. doi: 10.18632/oncotarget.22710
70. Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G, et al. Synthetic circular rna functions as a mir-21 sponge to suppress gastric carcinoma cell proliferation. Mol Ther Nucleic Acids (2018) 13:312–21. doi: 10.1016/j.omtn.2018.09.010
Keywords: circular RNA, circRNA, tuberculosis, mycobacterium tuberculosis, immune response, biomarker
Citation: Wang Q, Yang D, Zuo Y, Wang D and Li W (2022) Emerging roles of circular RNAs in tuberculosis. Front. Immunol. 13:995701. doi: 10.3389/fimmu.2022.995701
Received: 16 July 2022; Accepted: 05 September 2022;
Published: 20 September 2022.
Edited by:
Jiang Pi, Guangdong Medical University, ChinaCopyright © 2022 Wang, Yang, Zuo, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinglan Wang, d2FuZ3FpbmdsYW5Ac2N1LmVkdS5jbg==; Weimin Li, d2VpbWkwMDNAc2N1LmVkdS5jbg==
†These authors have contributed equally to this work
 Qinglan Wang
Qinglan Wang Danni Yang
Danni Yang Yinan Zuo
Yinan Zuo Dan Wang
Dan Wang Weimin Li
Weimin Li