- 1Nephrology Hospital, The First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou, China
- 2Department of Nephrology, Wuhan Fourth Hospital, Puai Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Background: Systemic lupus erythematosus (SLE) is a complex, multisystem autoimmune disease that is characterized by the production of autoantibodies. Although accumulated evidence suggests that the dysregulation of long non-coding RNAs (lncRNAs) is involved in the pathogenesis of SLE, the genetic contributions of lncRNA coding genes to SLE susceptibility remain largely unknown. Here, we aimed to provide more evidence for the role of lncRNA coding genes to SLE susceptibility.
Methods: The genetic association analysis was first adopted from the previous genome-wide association studies (GWAS) and was then validated in an independent cohort. PRDX6-AS1 is located at chr1:173204199-173446294. It spans a region of approximately 240 kb, and 297 single nucleotide polymorphisms (SNPs) were covered by the previous GWAS. Differential expression at the mRNA level was analyzed based on the ArrayExpress Archive database.
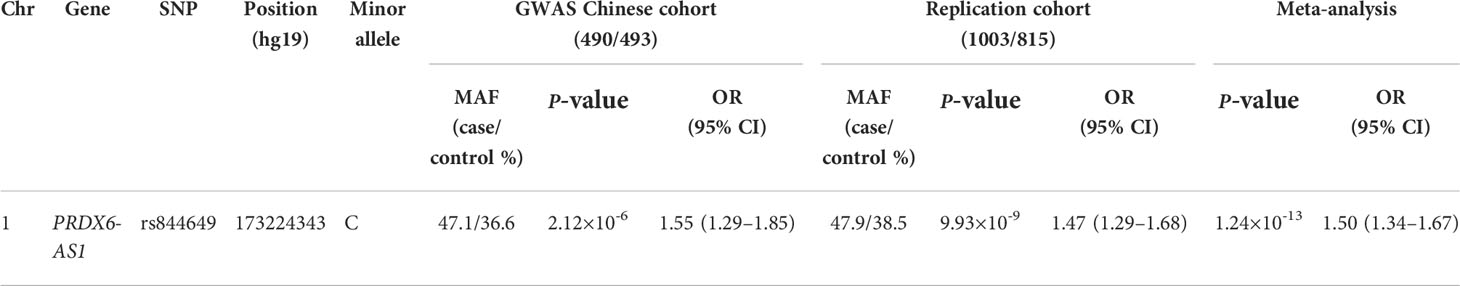
Results: A total of 33 SNPs were associated with SLE susceptibility, with a P<1.68×10-4. The strongest association signal was detected at rs844649 (P=2.12×10-6), according to the previous GWAS. Combining the results from the GWAS Chinese cohort and our replication cohort, we pursued a meta-analysis approach and found a pronounced genetic association between PRDX6-AS1 rs844649 and SLE susceptibility (pmeta=1.24×10-13, OR 1.50, 95% CI: 1.34–1.67). The mRNA expression of PRDX6 was elevated in peripheral blood cells, peripheral blood mononuclear cells (PBMCs), and multiple cell subpopulations, such as B cells, CD4+ T cells, CD3+ cells, and monocytes in patients with SLE. The PRDX6 protein expression level was also increased in patients with SLE compared with healthy donors.
Conclusion: Our study provides new evidence that variants located in lncRNA coding genes are associated with SLE susceptibility.
Introduction
Systemic lupus erythematosus (SLE; OMIM 152700) is a complex, multisystem autoimmune disease that is characterized by the production of autoantibodies with immune complex deposition leading to multiple organ damage (1, 2). The pathogenesis of SLE is still largely unknown, and it is believed that a combination of genetic and environmental factors contributes greatly to the etiology of SLE. Over recent years, increasing evidence has demonstrated that the dysregulation of long non-coding RNAs (lncRNAs) is involved in the pathogenesis of SLE (3, 4).
Long non-coding RNAs (lncRNAs) are a family of non-coding RNAs (ncRNAs) of more than 200 nucleotides in length that do not appear to code functional proteins (5). lncRNAs function both in cis and in trans to regulate gene expression and are involved in regulating various biological processes, such as cell proliferation, apoptosis, inflammation, and immune responses (6). Given the diverse cellular biological processes in which lncRNAs participate, lncRNAs have been implicated in a variety of diseases, including autoimmune diseases such as SLE.
The expression of lncRNAs is dysregulated in SLE and is correlated with clinical manifestations and organ impairments. The lncRNA expression profile in SLE was significantly changed, as there were 3,657 upregulated lncRNAs and 5,211 downregulated lncRNAs in SLE compared with the healthy group (7). Lnc-DC and growth-arrest-specific transcript 5 (GAS5) were downregulated in the plasma of patients with SLE, and their expression levels were negatively correlated with C3 expression and disease activity, respectively (8). nuclear paraspeckle assembly transcript 1 (NEAT1) expression was significantly upregulated in peripheral blood mononuclear cells (PBMCs) in patients with SLE and was positively correlated with disease activity (9). The expression of metastasis associated lung adenocarcinoma transcript 1 (MALAT1) was not only significantly higher in patients with SLE but was also revealed to be associated with interferon (IFN) signatures (10, 11).
In addition to detection results from clinical samples and experimental animal data, a growing body of evidence suggests that single nucleotide polymorphisms (SNPs) located in lncRNA coding regions are associated with SLE susceptibility. Located in an intronic enhancer, rs13259960 modulates SLEAR expression by impairing signal transducer and activator of transcription 1 (STAT1) recruitment and confers a predisposition to SLE (12). Variants rs205764 and rs547311 contributed to SLE susceptibility by enhancing linc00513 promoter activity, leading to the increased expression of linc00513 in SLE (13).
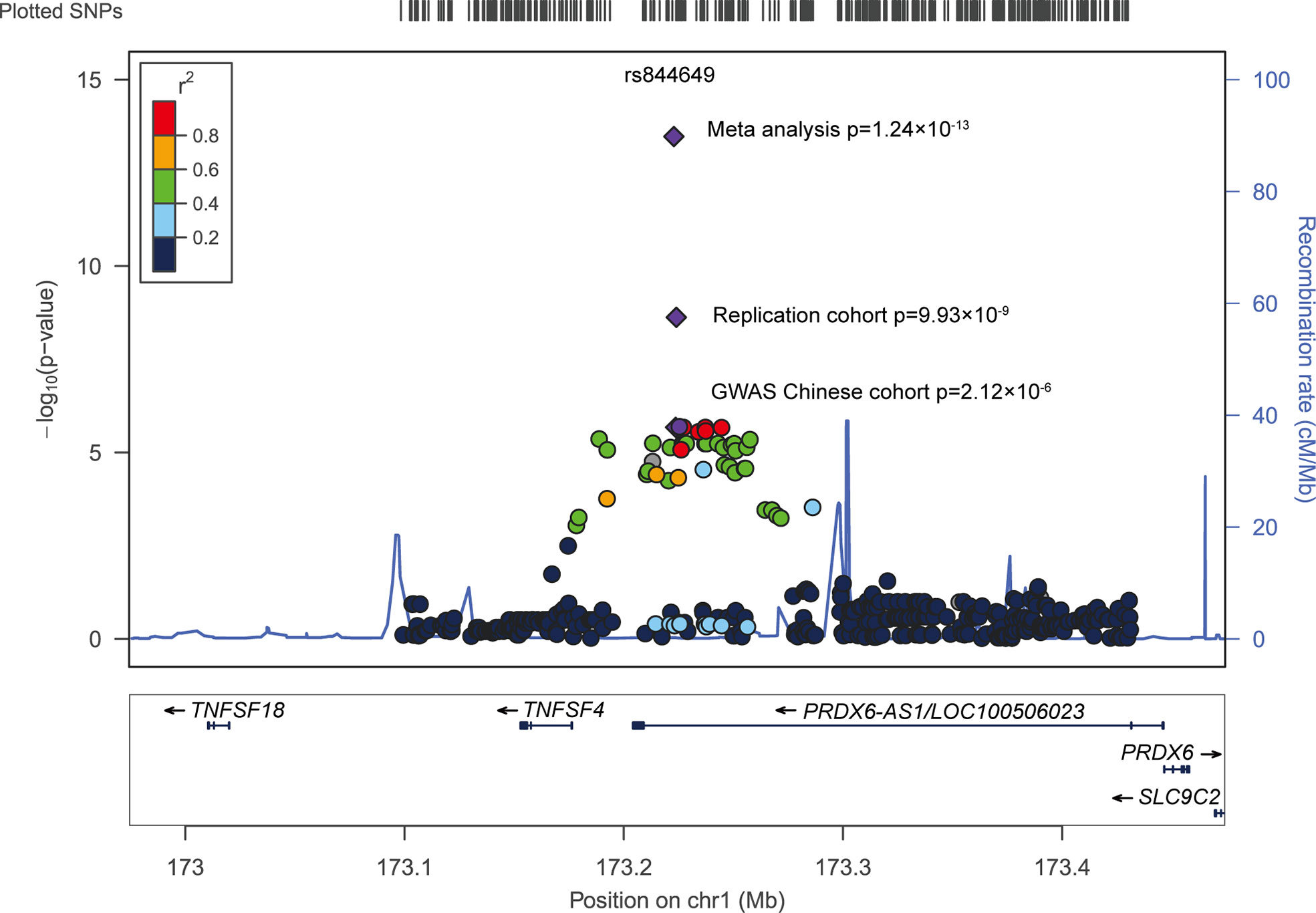
A group of signals concentrated at PRDX6-AS1 was identified when we performed an in-depth data analysis of previous genome-wide association studies (GWAS) (Figure 1). Although these groups of SNPs did not reach statistical significance in the Chinese population, we consider that the limited sample size (490 patients with SLE and 493 healthy donors) might be one of the most important reasons for this. Therefore, in our present study, we replicated the top signal in an independent cohort (1,003 patients with SLE and 815 healthy donors) from the Chinese population. We aimed to explore whether the genetic association signals at PRDX6-AS1 were consistent and aimed to identify its possible function in the pathogenesis of SLE.
Methods
Study subjects
The GWAS Chinese cohort was recruited from Beijing, and it included 490 SLE patients and 493 healthy donors (14). The independent replication cohort, which was enrolled in Henan, consisted of 1,003 patients with SLE (age, 34.5 ± 12.8; female patients, 92.9%) and 815 geographically and ethnically matched healthy donors (age, 45.4 ± 15.7; female donors 49.8%). The individuals diagnosed with SLE fulfilled the revised diagnostic criteria of the American College of Rheumatology (ACR) (15). Written informed consent was obtained from all study subjects, and the study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Zhengzhou University (2019-KY-247).
SNP selection
PRDX6-AS1 is located at chr1:173204199-173446294. It spans a region of approximately 240 kb, and 297 SNPs were covered by previous GWAS (14). Functional annotations were performed among genetically associated SNPs with P<0.05 to explore potential regulatory functions. The top signal was selected for replication.
DNA extraction and genotyping
DNA extraction was performed with a DNA extraction kit (Qiagen, Hilden, Germany). The genotyping for the replication cohort was conducted by the Sequenom MassARRAY platform (Sequenom, Inc., San Diego, CA, USA), and the genotyping yield was higher than 99.5%.
Differential expression analysis
Differential expression at the mRNA level was analyzed based on the ArrayExpress Archive database (http://www.ebi.ac.uk/arrayexpress) (16). It included immune cell subsets (B cells and CD4+ T cells E-GEOD-4588, CD3+ cells E-GEOD-13887, monocyte E-GEOD-46907) (17, 18), PBMCs (E-GEOD-50772) (19), and peripheral blood cells (E-GEOD-20864) (20). The protein level of PRDX6 in sera was also analyzed using data derived from E-MTAB-5900.
Statistical analysis
The distribution of the PRDX6-AS1 rs844649 genotypes was tested for the Hardy–Weinberg equilibrium with the goodness-of-fit χ2 test, which did not violate the Hardy–Weinberg equilibrium (P=0.711 in healthy control individuals in the replication cohort). Genetic association was assessed by a two-tailed χ2 test, and the meta-analysis was used to test Cochran−Mantel−Haenszel statistics. Differential expression analysis was performed with Student’s t-test for continuous variables. Analyses were implemented using the SPSS 19.0 (SPSS, Chicago, IL, USA) and (SAS 9.3; SAS Institute,Cary, NC) software.
Results
PRDX6-AS1 rs844649 polymorphisms associated with SLE susceptibility
A total of 43 SNPs were significantly associated with SLE susceptibility (P<0.05) in the Chinese population (Supplementary Table 1) (14). To reduce the probability of false-positive results caused by the number of tests, Bonferroni correction was applied for multi-testing correction. The level of significance was set at 1.68×10-4, as determined by dividing 0.05 by the number of tests. A total of 33 SNPs were associated with SLE susceptibility with P<1.68×10-4. The strongest association signal was detected at rs844649 (P=2.12×10-6, OR 1.55, 95% CI: 1.29–1.85) (Figure 1 and Table 1).
Based on the results obtained from the above analysis, an independent cohort was enrolled to replicate the association signal at PRDX6-AS1 rs844649. It was observed that the frequency of the risk allele C was 47.9% in patients with SLE and 38.5% in healthy donors, which was similar to the frequency of the GWAS Chinese cohort. The genotype frequencies of PRDX6-AS1 rs844649 in patients and control individuals are presented in Table 2. Carriers of the rs844649 C allele were significantly associated with an increased risk of SLE after adjusting for age and sex.
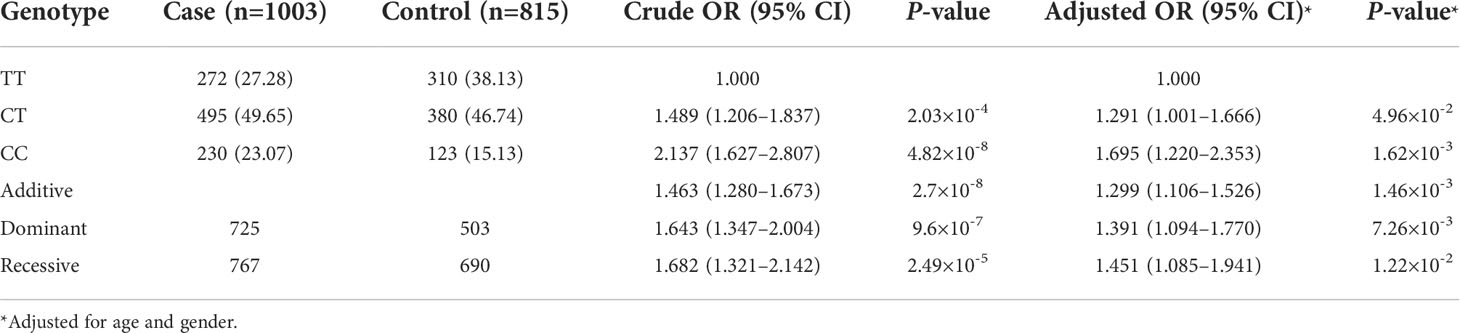
Table 2 Genetic association analysis between rs844649 PRDX6-AS1 with systemic lupus erythematosus (SLE) risk in replication cohort.
Combining results from the GWAS Chinese cohort and our replication cohort, we pursued a meta-analysis approach and found a pronounced genetic association between PRDX6-AS1 rs844649 and SLE susceptibility (pmeta=1.24×10-13, OR 1.50, 95% CI: 1.34–1.67) (Figure 1 and Table 1).
Functional annotations
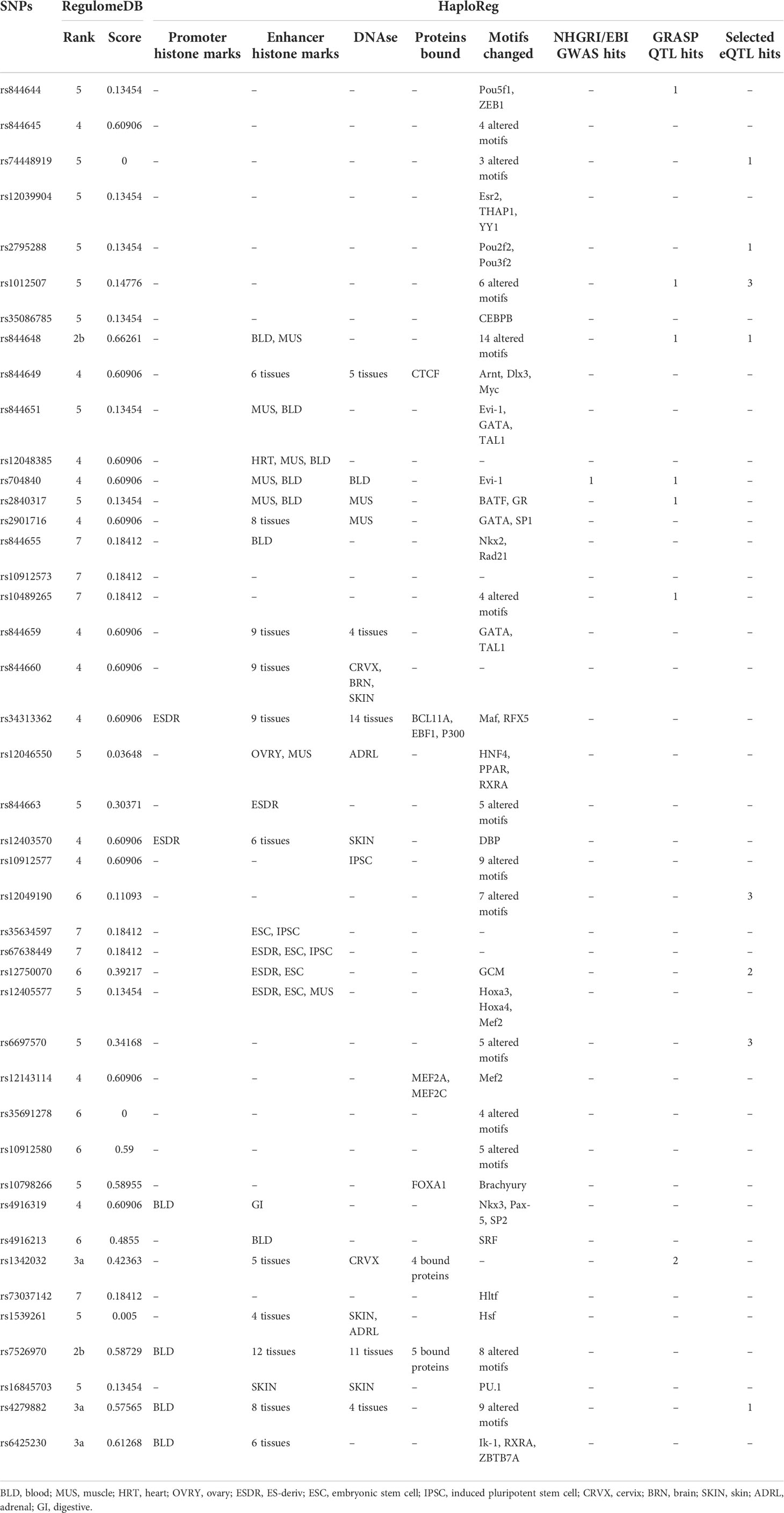
To explore the possible function of rs844649, RegulomeDB and HaploReg were applied to annotate the non-coding genome with known and predicted regulatory elements (21, 22). The RegulomeDB rank was 4, and the score was 0.60906 for rs844649. These findings indicate a potential regulatory function (Table 3). Data from HaploReg showed that rs844649 was located within the region of enhancer histone marks in six tissues, DNAse in five tissues, and in CTCF binding and motif change regions (Arnt, Dlx3, and Myc) (Table 3). Unfortunately, no expression quantitative trait loci (eQTL) effect was predicted to be associated with rs844649.
We further expanded our annotation to all 43 genetically associated SNPs with P<0.05. The rs844648 and rs7526970 variants had a RegulomeDB rank of 2b (ranks 1 to 7 represent decreasing regulatory evidence), and rs844648 had the highest RegulomeDB score of 0.66261 (Table 3).
Expression analysis of PRDX6
The expression of PRDX6 was elevated in B cells (Figures 2A, B) and CD4+ T cells (Figures 2C, D) in patients with SLE compared with healthy control individuals and approached marginal statistical significance. In CD3+ cells, the expression of PRDX6 was significantly increased in patients with SLE compared with healthy donors (detected by 200844_s_at, P=0.001) (17) (Figures 2E, F). The expression of PRDX6 demonstrated an increasing trend in monocytes in patients with SLE, but this was without statistical significance. This might be due to the limited sample size (18) (Figure 2G).
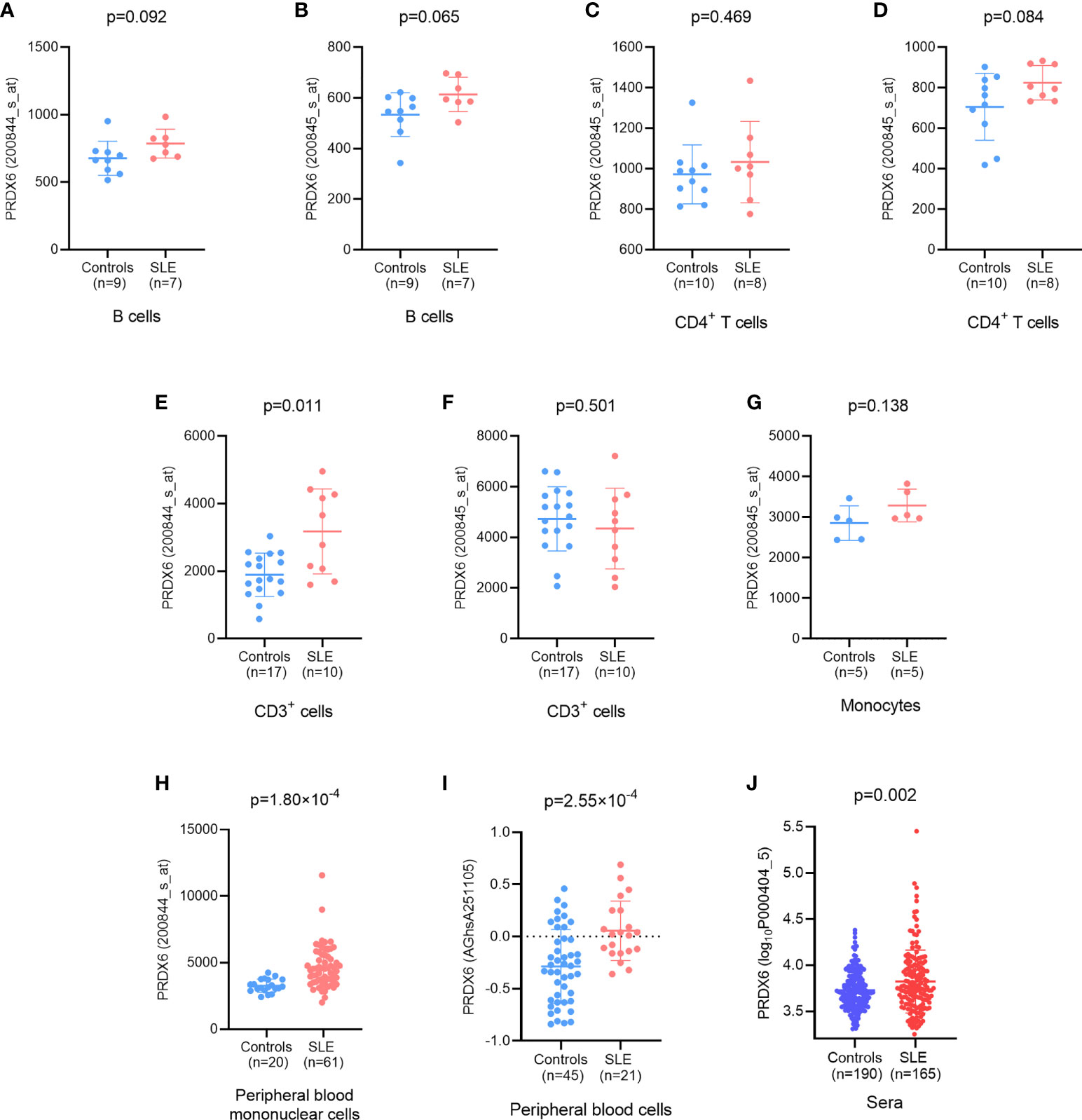
Figure 2 Differential expression of PRDX6 between patients with systemic lupus erythematosus (SLE) and controls. (A, B) At the mRNA level, the expression of PRDX6 was elevated in B cells (E-GEOD-4588); (C, D) CD4+ T cells (E-GEOD-4588); (E, F) CD3+ cells (E-GEOD-13887); (G) monocytes (E-GEOD-46907); (H) Peripheral blood mononuclear cells (PBMCs) (E-GEOD-50772); and (I) peripheral blood cells. (J) The level of PRDX6 was also significantly elevated in patients with SLE compared with healthy donors at protein level (E-MTAB-5900). The analyses were performed based on information available in online databases as referenced.
For PBMCs, in comparison to that in healthy subjects, the level of PRDX6 was significantly upregulated in patients with SLE (P=1.8×10-4) (19) (Figure 2H). A markedly increased level of PRDX6 expression was also identified in peripheral blood cells from patients with SLE (P=2.55×10-4) (20) (Figure 2I).
The analysis above was performed at the mRNA level, and the PRDX6 protein expression level was determined using data from E-MTAB-5900 hereafter. According to the data from the protein microarray, the level of PRDX6 was significantly elevated in patients with SLE compared with healthy donors (P=0.002) (Figure 2J).
Discussion
Although accumulated evidence suggests that the dysregulation of lncRNAs is involved in the pathogenesis of SLE, the genetic contributions of lncRNA coding genes to SLE susceptibility remain largely unknown. In our present study, we identified that the rs844649 PRDX6-AS1 variant located on chromosome 1q25.1 was associated with SLE susceptibility, reaching genome-wide significance in two independent cohorts from China. The 1q25.1 region is a hot spot containing potential SLE susceptibility loci. It is well established that tumor necrosis factor ligand superfamily member 4 (TNFSF4) gene polymorphisms are associated with SLE susceptibility in large sample sizes and diverse multiracial and multiethnic populations (23–30). A previous study also revealed that rs10798269, located on 1q25.1, was associated with SLE in women by a genome-wide association scan (31). Despite our data for rs844649 PRDX6-AS1 being from a Chinese population, genetic association results from a Korean population (1,710 patients with SLE vs. 3,167 control individuals) showed a more remarkable result (P=1.67 × 10-12, OR 1.37, 95% CI: 1.25–1.49) (14). However, additional studies are still needed to validate the signal from rs844649 PRDX6-AS1 on chromosome 1q25.1 in more centers with different ethnicities from different centers.
To explore the possible regulatory function of rs844649, we performed functional annotations using a publicly available database. It was observed that rs844649 was located within the regions of enhancer histone marks, DNAse, CTCF binding, and motif changes, but no eQTL effect was identified. Thus, future studies are needed to search for the causal SNP on PRDX6-AS1 from a functional perspective.
Genetic association analysis indicated that the lncRNA PRDX6-AS1 might be involved in the pathogenesis of SLE. Given that one of the first critical functions of a certain lncRNA is to regulate the expression of adjacent protein coding genes, we further examined the expression level of PRDX6 based on an array expression database. Importantly, the mRNA expression of PRDX6 was elevated in peripheral blood cells, PBMCs, and multiple cell subpopulations, including B cells, CD4+ T cells, CD3+ cells, and monocytes, in patients with SLE. The PRDX6 protein expression level was also increased in patients with SLE compared with healthy donors.
Peroxiredoxin (PRDX) 6 belongs to the PRDX family, and three enzymatic activities have been identified for it. These activities are peroxidase, phospholipase A2 (PLA2), and acyl transferase activities (32). Recent advances have highlighted the protective role of PRDX6 against the pathogenesis of SLE through studies using cis-eQTL analysis and Prdx6 knockout (KO) mice (33). SLE risk-associated SNPs downregulate the expression of PRDX6, and Prdx6 deficiency upregulates antibody production (33, 34). Our study revealed that the expression of PRDX6 was upregulated in patients with SLE, as demonstrated by the integrated analysis of multiple studies in different cell subtypes. The increased expression of Prdx6 has been demonstrated in various injury models and patients with peripheral arterial disease (35–37). Whether Prdx6 has protective antioxidant functions or participates in redox imbalance by redox signaling remains controversial. Considering the dual role of PRDX6 in redox imbalance in inflammation models, whether the elevated level of PRDX6 was simply a biomarker of SLE or a driver of disease pathogenesis warrants further investigation.
Conclusion
In this study, we performed a genetic association analysis among a Chinese population and confirmed that the rs844649 PRDX6-AS1 variant on chromosome 1q25.1 was significantly associated with SLE susceptibility. Moreover, PRDX6, the closest gene to PRDX6-AS1, was upregulated in patients with SLE in multiple cell subpopulations. Our study provides new evidence that variants located in lncRNA coding genes are associated with SLE susceptibility.
Data availability statement
The data sets presented in this study can be found in online repositories. The names of the repository/repositories and accession numbers can be found in the article/Supplementary Material.
Ethics statement
The study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Zhengzhou University (2019-KY-247). The patients/participants provided their written informed consent to participate in this study.
Author contributions
Y-YQ and Z-ZZ conceived and designed the experiment. Y-YQ, X-XZ, and X-RL performed the experiments. X-XZ, J-PY, Y-FZ, and YC analyzed the data. Y-YQ, X-XZ, J-PY, and Z-ZZ interpreted the findings. All the authors contributed to writing the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 81900643, 81873611]; the China Postdoctoral Science Foundation [grant number 2019M652592]; the funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We thank all the members of the laboratory for their technical assistance. We also thank the patients and their families, and the healthy donors for their cooperation and for giving consent to participate in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.987385/full#supplementary-material
Abbreviations
SLE, systemic lupus erythematosus; GWAS, genome-wide association study; SNP, single nucleotide polymorphism; lncRNA, long non-coding RNAs; PRDX6, peroxiredoxin 6; PBMCs, peripheral blood mononuclear cells.
References
1. Kiriakidou M, Ching CL. Systemic lupus erythematosus. Ann Intern Med (2020) 172:ITC81–96. doi: 10.7326/AITC202006020
2. Tsokos GC. Autoimmunity and organ damage in systemic lupus erythematosus. Nat Immunol (2020) 21:605–14. doi: 10.1038/s41590-020-0677-6
3. Wu H, Chen S, Li A, Shen K, Wang S, Wang S, et al. LncRNA expression profiles in systemic lupus erythematosus and rheumatoid arthritis: Emerging biomarkers and therapeutic targets. Front Immunol (2021) 12:792884. doi: 10.3389/fimmu.2021.792884
4. Ghafouri-Fard S, Poornajaf Y, Dashti F, Hussen BM, Taheri M, Jamali E. Interaction between non-coding RNAs and interferons: With an especial focus on type I interferons. Front Immunol (2022) 13:877243. doi: 10.3389/fimmu.2022.877243
5. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
6. Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet (2020) 21:102–17. doi: 10.1038/s41576-019-0184-5
7. Luo Q, Li X, Xu C, Zeng L, Ye J, Guo Y, et al. Integrative analysis of long non-coding RNAs and messenger RNA expression profiles in systemic lupus erythematosus. Mol Med Rep (2018) 17:3489–96. doi: 10.3892/mmr.2017.8344
8. Wu GC, Li J, Leng RX, Li XP, Li XM, Wang DG, et al. Identification of long non-coding RNAs GAS5, linc0597 and lnc-DC in plasma as novel biomarkers for systemic lupus erythematosus. Oncotarget (2017) 8:23650–63. doi: 10.18632/oncotarget.15569
9. Zhang F, Wu L, Qian J, Qu B, Xia S, La T, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun (2016) 75:96–104. doi: 10.1016/j.jaut.2016.07.012
10. Yang H, Liang N, Wang M, Fei Y, Sun J, Li Z, et al. Long noncoding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget (2017) 8:77400–6. doi: 10.18632/oncotarget.20490
11. Gao F, Tan Y, Luo H. MALAT1 is involved in type I IFNs-mediated systemic lupus erythematosus by up-regulating OAS2, OAS3, and OASL. Braz J Med Biol Res (2020) 53:e9292. doi: 10.1590/1414-431x20209292
12. Fan Z, Chen X, Liu L, Zhu C, Xu J, Yin X, et al. Association of the polymorphism rs13259960 in SLEAR with predisposition to systemic lupus erythematosus. Arthritis Rheumatol (2020) 72:985–96. doi: 10.1002/art.41200
13. Xue Z, Cui C, Liao Z, Xia S, Zhang P, Qin J, et al. Identification of LncRNA Linc00513 containing lupus-associated genetic variants as a novel regulator of interferon signaling pathway. Front Immunol (2018) 92967. doi: 10.3389/fimmu.2018.02967
14. Sun C, Molineros JE, Looger LL, Zhou XJ, Kim K, Okada Y, et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet (2016) 48:323–30. doi: 10.1038/ng.3496
15. Tan EM, Cohen AS, Fries JF, Masi AT, Mcshane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum (1982) 25:1271–7. doi: 10.1002/art.1780251101
16. Athar A, Fullgrabe A, George N, Iqbal H, Huerta L, Ali A, et al. ArrayExpress update - from bulk to single-cell expression data. Nucleic Acids Res (2019) 47:D711–5. doi: 10.1093/nar/gky964
17. Fernandez DR, Telarico T, Bonilla E, Li Q, Banerjee S, Middleton FA, et al. Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Immunol (2009) 182:2063–73. doi: 10.4049/jimmunol.0803600
18. Rodriguez-Pla A, Patel P, Maecker HT, Rossello-Urgell J, Baldwin N, Bennett L, et al. IFN priming is necessary but not sufficient to turn on a migratory dendritic cell program in lupus monocytes. J Immunol (2014) 192:5586–98. doi: 10.4049/jimmunol.1301319
19. Kennedy WP, Maciuca R, Wolslegel K, Tew W, Abbas AR, Chaivorapol C, et al. Association of the interferon signature metric with serological disease manifestations but not global activity scores in multiple cohorts of patients with SLE. Lupus Sci Med (2015) 2:e000080. doi: 10.1136/lupus-2014-000080
20. Lee HM, Sugino H, Aoki C, Nishimoto N. Underexpression of mitochondrial-DNA encoded ATP synthesis-related genes and DNA repair genes in systemic lupus erythematosus. Arthritis Res Ther (2011) 13:R63. doi: 10.1186/ar3317
21. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res (2012) 22:1790–7. doi: 10.1101/gr.137323.112
22. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res (2012) 40:D930–934. doi: 10.1093/nar/gkr917
23. Chang YK, Yang W, Zhao M, Mok CC, Chan TM, Wong RW, et al. Association of BANK1 and TNFSF4 with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun (2009) 10:414–20. doi: 10.1038/gene.2009.16
24. Delgado-Vega AM, Abelson AK, Sanchez E, Witte T, D'alfonso S, Galeazzi M, et al. Replication of the TNFSF4 (OX40L) promoter region association with systemic lupus erythematosus. Genes Immun (2009) 10:248–53. doi: 10.1038/gene.2008.95
25. Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet (2009) 41:1228–33. doi: 10.1038/ng.468
26. Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet (2009) 41:1234–7. doi: 10.1038/ng.472
27. Sanchez E, Webb RD, Rasmussen A, Kelly JA, Riba L, Kaufman KM, et al. Genetically determined Amerindian ancestry correlates with increased frequency of risk alleles for systemic lupus erythematosus. Arthritis Rheum (2010) 62:3722–9. doi: 10.1002/art.27753
28. Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PloS Genet (2010) 6:e1000841. doi: 10.1371/journal.pgen.1000841
29. Sanchez E, Comeau ME, Freedman BI, Kelly JA, Kaufman KM, Langefeld CD, et al. Identification of novel genetic susceptibility loci in African American lupus patients in a candidate gene association study. Arthritis Rheum (2011) 63:3493–501. doi: 10.1002/art.30563
30. Zhou XJ, Lu XL, Nath SK, Lv JC, Zhu SN, Yang HZ, et al. Gene-gene interaction of BLK, TNFSF4, TRAF1, TNFAIP3, and REL in systemic lupus erythematosus. Arthritis Rheum (2012) 64:222–31. doi: 10.1002/art.33318
31. International Consortium for Systemic Lupus Erythematosus, G, Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet (2008) 40:204–10. doi: 10.1038/ng.81
32. Feinstein SI. Mouse models of genetically altered peroxiredoxin 6. Antioxidants (Basel) (2019) 8(4):77. doi: 10.3390/antiox8040077
33. Takeshima Y, Iwasaki Y, Nakano M, Narushima Y, Ota M, Nagafuchi Y, et al. Immune cell multiomics analysis reveals contribution of oxidative phosphorylation to b-cell functions and organ damage of lupus. Ann Rheum Dis (2022) 81:845–53. doi: 10.1136/annrheumdis-2021-221464
34. Marigorta UM, Denson LA, Hyams JS, Mondal K, Prince J, Walters TD, et al. Transcriptional risk scores link GWAS to eQTLs and predict complications in crohn's disease. Nat Genet (2017) 49:1517–21. doi: 10.1038/ng.3936
35. Wang X, Phelan SA, Forsman-Semb K, Taylor EF, Petros C, Brown A, et al. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J Biol Chem (2003) 278:25179–90. doi: 10.1074/jbc.M302706200
36. El Eter E, Al Masri A, Habib S, Al Zamil H, Al Hersi A, Al Hussein F, et al. Novel links among peroxiredoxins, endothelial dysfunction, and severity of atherosclerosis in type 2 diabetic patients with peripheral atherosclerotic disease. Cell Stress Chaperones (2014) 19:173–81. doi: 10.1007/s12192-013-0442-y
Keywords: systemic lupus erythematosus, single nucleotide polymorphisms, PRDX6-AS1, lncRNA, rs844649
Citation: Zhang X-X, You J-P, Liu X-R, Zhao Y-F, Cui Y, Zhao Z-Z and Qi Y-Y (2022) PRDX6AS1 gene polymorphisms and SLE susceptibility in Chinese populations. Front. Immunol. 13:987385. doi: 10.3389/fimmu.2022.987385
Received: 06 July 2022; Accepted: 29 August 2022;
Published: 12 October 2022.
Edited by:
Durga Prasanna Misra, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGI), IndiaReviewed by:
Zhigang Hu, Wuxi Children’s Hospital, ChinaAruna Pal, West Bengal University of Animal and Fishery Sciences, India
Copyright © 2022 Zhang, You, Liu, Zhao, Cui, Zhao and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan-Yuan Qi, cXF5eWlpbGx5eUAxMjYuY29t; Zhan-Zheng Zhao, emhhbnpoZW5nemhhb0B6enUuZWR1LmNu
†These authors contributed equally to this work
 Xiao-Xue Zhang1†
Xiao-Xue Zhang1† Xin-Ran Liu
Xin-Ran Liu Ya-Fei Zhao
Ya-Fei Zhao Yan Cui
Yan Cui Zhan-Zheng Zhao
Zhan-Zheng Zhao Yuan-Yuan Qi
Yuan-Yuan Qi