- 1Division of Surgical Oncology, Department of Surgery, University of California Davis Medical Center, Sacramento, CA, United States
- 2School of Veterinary Medicine, University of California Davis, Sacramento, CA, United States
- 3Center for Companion Animal Health Department of Surgical and Radiological Sciences, School of Veterinary Medicine, University of California Davis, Davis, CA, United States
- 4Department of Medical Microbiology and Immunology, University of California Davis, Davis, CA, United States
The microbiome has clearly been established as a cutting-edge field in tumor immunology and immunotherapy. Growing evidence supports the role of the microbiome in immune surveillance, self-tolerance, and response to immune checkpoint inhibitors such as anti PD-L1 and CTLA-4 blockade (1–6). Moreover, recent studies including those using fecal microbial transplantation (FMT) have demonstrated that response to checkpoint immunotherapies may be conferred or eliminated through gut microbiome modulation (7, 8). Consequently, studies evaluating microbiota-host immune and metabolic interactions remain an area of high impact research. While observations in murine models have highlighted the importance of the microbiome in response to therapy, we lack sufficient understanding of the exact mechanisms underlying these interactions. Furthermore, mouse and human gut microbiome composition may be too dissimilar for discovery of all relevant gut microbial biomarkers. Multiple cancers in dogs, including lymphoma, high grade gliomas, melanomas and osteosarcoma (OSA) closely resemble their human analogues, particularly in regard to metastasis, disease recurrence and response to treatment. Importantly, dogs with these spontaneous cancers also have intact immune systems, suggesting that microbiome analyses in these subjects may provide high yield information, especially in the setting of novel immunotherapy regimens which are currently expanding rapidly in canine comparative oncology (9, 10). Additionally, as onco-microbiotic therapies are developed to modify gut microbiomes for maximal responsiveness, large animal models with intact immune systems will be useful for trialing interventions and monitoring adverse events. Together, pre-clinical mechanistic studies and large animal trials can help fully unlock the potential of the microbiome as a diagnostic and therapeutic target in cancer.
Success and limitations of the murine model
Murine models represent the most used model for studying host-microbiome physiology at both functional and mechanistic levels. Due to genetic homogeneity and laboratory environments, mouse models allow for high levels of control and improved experimental reproducibility (11). In cancers with representative murine models, mice have uncovered cancer promoting microbiota, and microbiota associated with improved response to treatment. Murine colon cancer models, for example, have contributed to understanding the influence of the gut microbiome on colorectal tumorigenesis via modulation of inflammation. In these models, transfer of the microbiome from tumor-bearing mice to germ free mice accelerated tumor growth, demonstrating causality (12, 13). Similarly, TLR4 knockout mice have been critical in understanding the oncogenic effect of gut microbiota in liver cancer (14). Sivan et al (15)demonstrated key differences in tumor kinetics and responses to immunotherapy between genetically similar mice bred in two different environments. One cohort showed anti-tumor effects following anti-PD1 checkpoint inhibitor therapy, while the other cohort showed no benefit. However, co-housing the mice together reversed the differences in anti-tumor responses, which were then restored by the administration of oral Bifidobacterium spp (15). Similar observations were made for mice receiving anti-CTLA checkpoint inhibitors with effects modulated by Bacteroides species, (specifically B. fragilis and/or B. thetaiotaomicrom) (16).
Despite the strength of these mechanistic murine studies, there are limitations to mouse models, particularly when considering the complex interactions between microbiome, immune response, and cancer. Laboratory mice demonstrate different microbial compositions to their wild counterparts. Attempts to convert lab-type to wild type mice have successfully altered, but not completely recapitulated, natural microbiome profiles (17). Moreover, animal facilities themselves significantly impact microbiome composition. For example, key differences in dominant taxa have been observed between identical mouse lines from different animal locations (18). Similarly, the use of germ-free mice is both a strength and a weakness of mouse models (19). These sterile conditions allow mice to be born without microbial colonization prior to the introduction of species of interest. However, this germ-free environment also leads to significant impairment of immune system development and responses. For example, mice raised in sterile conditions develop higher levels of IgE and fail to induce the same levels of immune reactivity seen during the “weaning period” of wild-type mice (20–23). This results in susceptibility to certain bacteria and increased risk of immunopathologies, complications which are not resolved with instillation of an “adult” or “wild-type” microbiome (21, 22, 24). Additionally, mouse studies of the microbiome show a large genetic influence which is distinct from humans where <10% of taxa are thought to be heritable (24–28). The high level of genetic homogeneity present in most murine models raises questions of generalizability to the human situation where genetic backgrounds and environmental exposures are vastly more diverse. This concern is reinforced by studies which have demonstrated the impact of environmental influences especially diet and drugs, as the most prominent factors influencing the microbiome in humans (29–31).
Additionally, as laboratory animals, mice do not receive the multi-faceted cancer care that is provided to humans and companion animals such as dogs. While pre-clinical murine models of checkpoint blockade immunotherapy have demonstrated impressive ability to prevent disease progression in various cancer cell lines, translation to human medicine has shown mixed results (32). Clinically meaningful survival benefits of PD-1 & CTLA-4 checkpoint inhibitor therapy have been demonstrated in some cancers, such as melanoma and non-small cell lung cancer, but only in a subset of patients (31–34)While observations from murine studies have demonstrated microbiome-associated factors for responsiveness to treatment, another limitation of murine models is their tendency to be reductionistic in their capacity to explore all potential intersecting variables which influence the cancer-immunity cycle (5–8, 15, 35, 36).
Similarly, murine models are able to strictly control diet and environment, both of which are key drivers of microbiome plasticity and composition, but as with germ-free studies, this can be both a strength and a weakness. For example, Matson et al. and Gopalakrishnan et al. (5, 6) used FMT to study whether human commensal microbes would potentiate anti-tumor T cell responses in germ-free mice in two separate studies. Matson et al. demonstrated slower tumor growth in some mice after transferring microbiota from human responders. Gopalakrishnan et al. (6) showed changes in tumor infiltrating cells and upregulation of PD-1 in the tumor microenvironment after FMT. Although these mouse experiments demonstrated significant proof-of-concept results, it is not clear why effects were seen in some, but not all, mice. Given that the mice were evaluated to be highly homogenous and genetically inbred, the mice are expected to respond similarly. It is unclear whether the effects on the treatment would persist when using models with more heterogenous genetic backgrounds and environmental exposures. Additionally, the fact that similar mechanisms were identified in these studies but the precise microorganisms were different highlights potential concerns regarding generalizability and extrapolation as noted above.
Microbiome studies in dogs with spontaneous cancers receiving immunotherapy offer an additional avenue for exploration. As the development of canine immunotherapy advances, including the development of novel caninized anti-PD1, anti-CTLA-4 and anti-PD-L1 monoclonal antibodies, microbiome correlative studies and attempted therapeutic intervention in canines with intact immune systems and a lifetime of commensal gut microbiome symbionts may yield useful observations for human applications (37–39). Human clinical immunotherapy trials are impacted by immune-related adverse effects such as colitis and myocarditis (32, 33, 39, 40). While murine models have been developed to help study these immune-related events, these models are prone to develop auto-immune responses which may limit translational potential (41). Canine models represent an opportunity to bridge mechanistic studies in mice with descriptive studies in humans, and microbiome studies in companion animal dogs can help advance our understanding of how the gut microbiome shapes immunotherapy responses as well as toxicity and adverse events.
Human vs canine microbiome
As in humans, studies of the canine microbiome have evaluated the relationship of the microbiome with inflammatory disease states, development of malignancies, and more recently, response to oncologic therapies (42–47). The “normal” adult microbiota comprises thousands of bacterial species across mucosal and skin surfaces and is determined by environmental and genetic factors. The colonization of both the human and canine gut microbiome, one of the densest bacterial environments on earth, increases in alpha diversity and decreases in beta diversity during the weaning period before stabilizing (0-3 years in humans, 0-9 weeks in canines) (45, 48–50). Exposure to microbes during delivery, early diet, and antibiotic exposure have all been shown to impact gut microbial development (22, 51, 52). Early influences have clinical impact – colonization with C. difficile during the first months of life is associated with increased risk of atopic disorders such as asthma and eczema in children (53). Microbiome compositions in pre-weened puppies display instability of prominent taxa. Puppies that displayed these “immature” microbiome profiles are more suspectilble to diarrheal illness compared to those who have developed stable more “adult” compositions (54). Environmental factors such as diet, drugs, and living conditions also exert key influences on subsequent adult microbiota composition (29, 30). Microbiomes of genetically unrelated co-habitants are consistently demonstrated to be more similar than those of relatives who do not cohabitate, and genetic ancestry does not predict compositional similarities (29, 55). While environment is the primary influencer of composition, there appears to be heritability in human and canine gut microbiomes as well. A study of 400 human twin pairs raised in the same household compared monozygotic vs dizygotic siblings. Interestingly, monozygotic twins demonstrated more similar gut microbiota compared to their dizygotic counterparts (56). Canine comparisons of dog breeds show similar findings, although studies of breed variation indicate that breed does not cause major shifts in microbiome composition or diversity but may influence abundance of specific taxa (46). Although not to the extent of humans or mice, the canine gut microbiota has been surveyed in some detail (50, 57–59). Most large studies agree on the five most prominent phyla in dogs: Firmicutes, Fusobacteria, Bacteroides, Proteobacteria and Actinobacteria, the composition of which is more similar to humans than other commonly studied mammalian species. Coelho et al. (57) compared 129 stool samples from 64 dogs against previously published gut microbial gene catalogs based on similar sequencing methods. The phylum level distribution of genes in the dog was more like the published human data than that of a mouse or pig. When genes were clustered by each species pool, the dog gut gene pool overlapped most with the human gut microbiome (23%) compared to only 4.9% for the murine gene catalog. As in prior human data, the majority of the composition appeared to be influenced by environmental exposures, diet, and disease states. Importantly, certain bacterial species have been found to exert similar effect across species. Obesity, which is a growing problem in both humans and canines, is correlated with a change in Firmicutes/Bacteroides ratios in both humans and canines (60–62). Canine and human inflammatory bowel disorders are associated with a reduction in microbial community diversity and structure characterized by loss of key species or overgrowth of species with genotoxic potential such as Bacteroides fragilis and E. coli (45, 63). In humans, these microbiome shifts have been associated with colorectal cancer (12, 64, 65). While colorectal cancer is less common in dogs, increased E.coli is associated with canine intestinal lymphoma (42). Importantly, human and dog differences in microbiome interactions are also informative. Fusobacterium, for instance, plays a role in energy utilization in the gut as a fermenting species. In humans, F. nucleatum is associated with colorectal cancer and thought to trigger inhibitory T cell receptors that suppress anti-cancer immune response (66, 67). In dogs, however, fusobacteria are associated with maintaining gut health (67).
While similarities in the development and composition of the gut microbiome present one advantage of the canine model, perhaps the most important is that of a shared environment with humans. As companion animals, dogs inhabit the same environment as humans. Therefore, shifts in composition caused by disease states or therapeutics must be robust enough to compete with the myriad of “real life” influences that patients encounter. Shared environments may also lead to shared taxonomy; canine housemates show similar microbiome profiles, similar to what is observed between human cohabiting with a spouse or partner (68). Interestingly, co-living between species influences microbiome profile as well (69). Companion dogs share more skin flora with their owners than with other dogs and oral microbiomes, which may show relevance in oral melanoma, as there is evidence of oral spread of symbionts between humans and dogs (68, 69). Some have hypothesized that pet ownership may influence the human gut microbiome as well (70). Small differences in abundance for two OTUs have been associated with pet ownership, although additional studies are needed given the risk of type I error (69).
Microbiome in canine therapeutics
Attenuation of immune surveillance and development of immunological tolerance to tumor-derived antigens contribute to cancer development and progression. Novel immunotherapies target one of these two mechanisms to stimulate the subject’s immune system to recognize and target cancer as non-self. Overcoming both intrinsic and acquired resistance to these treatments are critical areas of study, and although several species of interest have been identified, no universal “responder signature” has been identified. Canine analysis that identifies similar bacterial enrichment as in human studies would greatly enhance the chances of replicable and generalizable effects.
Microbiome modulation has demonstrated exciting promise in increasing the number of human patients who may respond to PD-1/PD-L1 checkpoint inhibitor immunotherapy (4–6). Demonstrated first by Gopalakrishnan & Matson et al. (5, 6) in a murine model, and recently by Baruch et al. (7) in a human clinical trial, FMT from a drug-responsive subject to a nonresponsive subject was able to convert a subset of prior non-responders, allowing them to benefit from treatment. Importantly, when considering the human trial of FMT, extensive pre-and post-treatment analysis regarding microbiome composition, local metabolism, and expression of immune-related genes failed to show significant differences between those who became responsive to treatment and those who did not (7). Thus, while successful in proving the concept of FMT to be safe and potentially beneficial, the effects remain correlative. These types of immune-microbiome relationships could benefit from additional studies in dogs with spontaneous cancers in the setting of an intact immune system. FMT is accepted in community veterinary practice making high-powered studies possible once commercially available caninized ICI’s are readily available (59). Repeating FMT trials with canine immunotherapy provides an opportunity to integrate observations of gut microbial alterations with longitudinal changes in metabolism and immune surveillance. Dogs, unlike mice, allow for multiple collections of adequate blood samples that can detect metabolites of short chain fatty acids that can act as ligands for G-protein coupled receptors. Iosine and hypoxanthine have recently been associated with bacteria of interest, for example (71, 72).
While veterinary medicine is waiting for the widespread commercial availability of canine specific PD-1/PD-L1/CTLA-4 mAbs to usher the area of immunotherapy in dogs, other immunotherapy approaches are in use, and comparison of microbiome profiles between responder status in other immune-based therapies should be considered (73, 74). Longitudinal stool and blood sampling of these canine patients for microbiome and metabolic investigation presents a rich area of investigation. Standard treatments for dog cancers include chemotherapy and radiation, which may allow for evaluation of the impacts that traditional therapies have on the microbiome alone or in combination with immunotherapy. Canine lifecycles are also inherently shorter, so rapidly progressing canine cancers allow for collection of samples linked to critical end points such as disease-free interval and time to progression which are highly relevant to the human situation.
Microbiome in the tumor microenvironment
The gut microbiome represents the most extensively classified community in humans and animal models, yet increasing studies have demonstrated that microbial communities in other compartments have clinical implications (74–76). Tumor-associated microbiomes have been identified in multiple human cancers including breast, lung, pancreas, and melanoma tumors (74–79). While tumor infiltrating lymphocytes (TILs) have emerged as a key feature of the immune tumor microenvironment (TME), a deeper molecular and cellular signals responsible for immune-tumor-microbiome communication are not yet understood. While some studies demonstrate a higher number of tumor infiltrating lymphocytes (TILs) in the TME project more positive patient outcomes, others suggest that the upregulation of regulatory T cells in the TME is associated with a more negative outcome (80–82). Although mouse models can provide key insights into the immune-tumor interactions in the TME and are important for mechanistic studies, these models have limitations given the nature of transplanted highly retroviral tumors. Known differences in tumor initiation and promotion are likely to impact the fidelity of the model for microbiome studies (83, 84).
Therefore, dogs represent an important translational opportunity for studies of the cancer-immunity cycle, including microbiome studies. Canine immunotherapy, including evaluation of novel therapies in the context of clinical trials, has received significant attention from researchers and funding agencies, including the Cancer Moonshot, and examples of ongoing canine immunotherapy trials can be seen in Table 1 (9, 10). Dogs are diagnosed with naturally occurring cancers which are highly similar to humans, and genetic studies have demonstrated notable overlap in the tumor genetic makeup of human and dog OSA, melanoma, mammary tumors, gliomas and lymphoma (84–90). Canine OSA recapitulates several feature of human OSA including frequent TP53, PI3K, and MAPK pathway mutations with low expression of immuno-associated genes and a trend toward higher mutation burden in metastatic disease (88). Canine mammary tumors and lymphomas share major gene alterations with their human counterparts in addition to having similar tumor mutation burdens (91). Gliomas have been shown to share specific alterations in receptor tyrosine kinases such as TP53 and cell cycle pathways with human pediatric gliomas (89). Overall, the incidence of these canine cancers is believed to be as high or higher than their human counterparts, although precise case numbers are not known since no national database exists for dogs. For example, soft tissue sarcomas make up 10-15% of malignant tumors in dogs with estimates of approximately 7700-31,800 new canine cases per year in the United States compared to soft tissue sarcoma in humans which represents approximately 1% of cancer cases and 10,000 – 12,000 new cases per year in the US (91). Similarly, canine OSA is estimated at approximately 25,000 – 50,000 cases per year, whereas the number of human OSA is approximately 3000 per year in the US (92–94). Data such as these highlight the value of the canine model as an important asset in tumor immunology and immunotherapy studies, especially where an intact immune system is critical for understanding tumor and host biology.
Conclusion
Murine models will always have utility as pre-clinical models, including for microbiome studies. However, due to the complex interactions between the immune system, the TME, and metabolism, utilization of canines for microbiome research is a promising strategy to yield additional data that can bridge our understanding from “proof of concept” to “proof of mechanism”. Further classification of the canine microbiome in cancer, changes in the canine microbiome in response to immunotherapy, and characterization of microbiomes outside the gut will all be important in deriving the most from canine models. This is particularly important now as veterinary medicine has made advances in canine specific immunotherapy agents that mirror human ICI. Canine clinical trials should consider collection of stool samples for microbiome biomarkers, as well as tissue and blood samples as well to correlate microbiome changes with immune infiltration and metabolic changes. As we increase our understanding of the interplay between specific commensal bacteria and the targets of immune therapy, the next step is therapeutic application. Through diet, antibiotics, fecal transfer, or other methods, reliably tailoring gut microbiomes to create maximal responsiveness with minimal side effects is the future of immuno-oncology. Dogs present a step-in bench to bedside science where their developing interventions can be tested on immune intact models exposed to complex environmental influences to ensure they demonstrate durability and utility.
Author contributions
KK and RC conceptualized the manuscript. KK, KI, LP, SC, and AR reviewed current literature. KK and KI wrote the manuscript and generated the associate figure. WC, MK, JE, and RR edited the manuscript and provided context for veterinary literature. RC edited the final paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by National Institutes of Health/National Cancer Institute grants U01 CA224166-01 (RC and RR), R03CA252793 (RC), and T32CA251007 (KK, LP, MK, and RC).
Acknowledgments
We would like to thank the members of the Laboratory for Cancer Immunology at UC Davis for their scientific and editorial input. Figure 1 was created with Biorender.com.
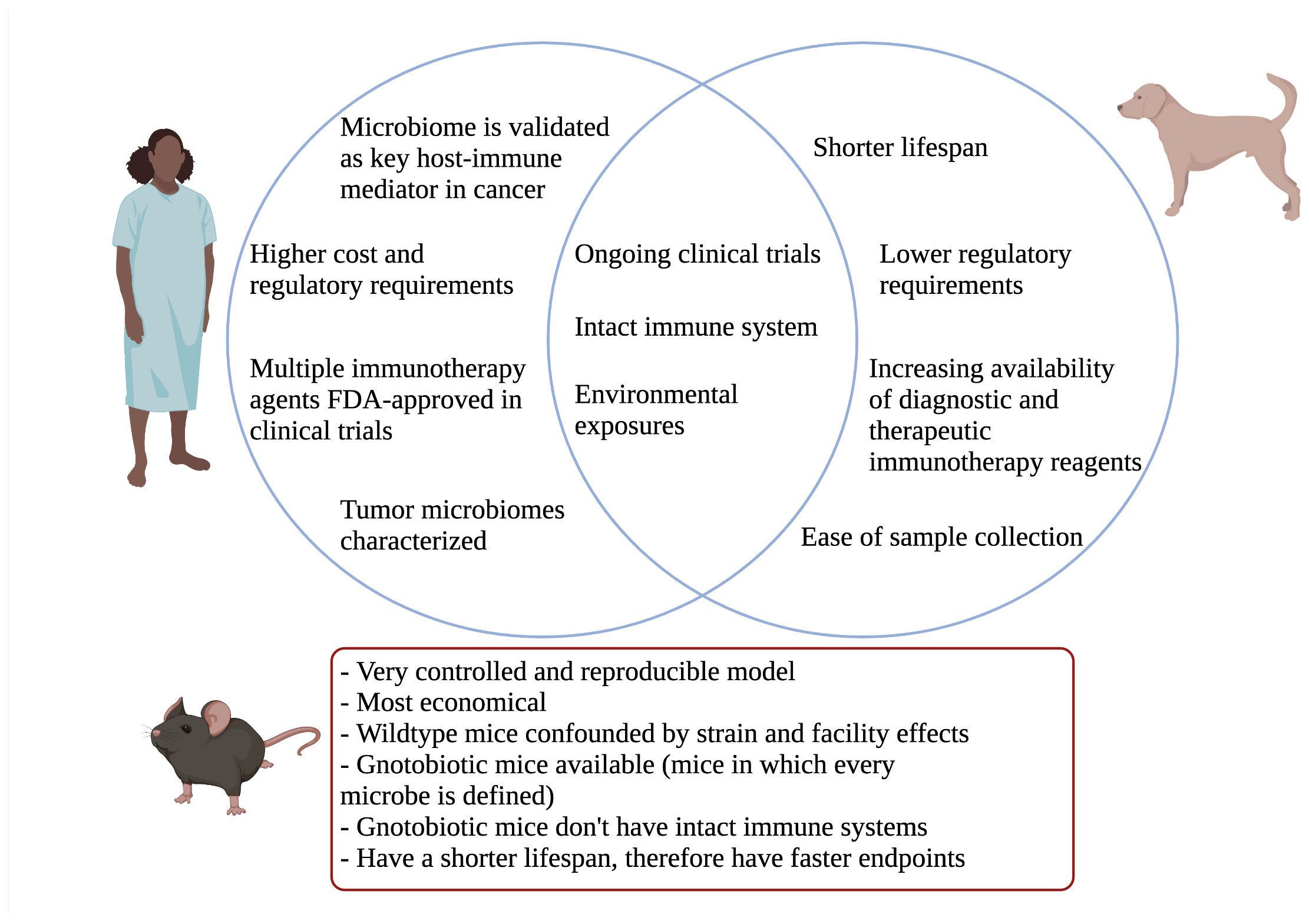
Figure 1 Venn diagram comparing relevant aspects of canine model to humans. Important mouse model differences highlighted below.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA (2007) 104(34):13780–5. doi: 10.1073/pnas.0706625104
2. Rodrigues Hoffmann A. The cutaneous ecosystem: the roles of the skin microbiome in health and its association with inflammatory skin conditions in humans and animals. Vet Dermatol (2017) 28(1):60–e15. doi: 10.1111/vde.12408
3. Ni J, Shen T-CD, Chen EZ, Bittinger K, Bailey A, Roggiani M, et al. A role for bacterial urease in gut dysbiosis and crohn’s disease. Sci Transl Med (2017) 9(416):1–11. doi: 10.1126/scitranslmed.aah6888
4. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
5. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science (2018) 359.
6. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
7. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science (2021) 371(6529):602–9. doi: 10.1126/science.abb5920
8. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science (2021) 371(6529):595–602. doi: 10.1126/science.abf3363
9. Park JS, Withers SS, Modiano JF, Kent MS, Chen M, Luna JI, et al. Canine cancer immunotherapy studies: linking mouse and human. J Immunother Cancer (2016) 4:97. doi: 10.1186/s40425-016-0200-7
10. LeBlanc AK, Mazcko CN. Improving human cancer therapy through the evaluation of pet dogs. Nat Rev Cancer (2020) 20(12):727–42. doi: 10.1038/s41568-020-0297-3
11. Nguyen TLA, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech (2015) 8(1):1–16. doi: 10.1242/dmm.017400
12. Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, et al. The gut microbiome modulates colon tumorigenesis. MBio (2013) 4(6):e00692–13. doi: 10.1128/mBio.00692-13
13. Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature (2012) 491(7423):254–8. doi: 10.1038/nature11465
14. Dapito DH, Mencin A, Gwak G-Y, Pradere J-P, Jang M-K, Mederacke I, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell (2012) 21(4):504–16. doi: 10.1016/j.ccr.2012.02.007
15. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
16. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
17. Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science (2019) 365(6452):eaaw4361. doi: 10.1126/science.aaw4361
18. Parker KD, Albeke SE, Gigley JP, Goldstein AM, Ward NL. Microbiome composition in both wild-type and disease model mice is heavily influenced by mouse facility. Front Microbiol (2018) 9:1598. doi: 10.3389/fmicb.2018.01598
19. Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe (2013) 14(5):559–70. doi: 10.1016/j.chom.2013.10.004
20. Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Déjardin F, et al. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity (2019) 50(5):1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014
21. Lubin J-B, Green J, Maddux S, Denu L, Duranova T, Lanza M, et al. Arresting microbiome development limits immune system maturation and resistance to infection. bioRxiv (2022) 1. doi: 10.1101/2022.01.17.476513v1.abstract
22. Bayer F, Ascher S, Pontarollo G, Reinhardt C. Antibiotic treatment protocols and germ-free mouse models in vascular research. Front Immunol (2019) 10:2174. doi: 10.3389/fimmu.2019.02174
23. Labuda JC, Fong KD, McSorley SJ. Cohousing with dirty mice increases the frequency of memory T cells and has variable effects on intracellular bacterial infection. Immunohorizons (2022) 6(2):184–90. doi: 10.4049/immunohorizons.2100069
24. Korach-Rechtman H, Freilich S, Gerassy-Vainberg S, Buhnik-Rosenblau K, Danin-Poleg Y, Bar H, et al. Murine genetic background has a stronger impact on the composition of the gut microbiota than maternal inoculation or exposure to unlike exogenous microbiota. Appl Environ Microbiol (2019) 85(18). doi: 10.1128/AEM.00826-19
25. Bubier JA, Chesler EJ, Weinstock GM. Host genetic control of gut microbiome composition. Mamm Genome (2021) 32(4):263–81. doi: 10.1007/s00335-021-09884-2
26. Campbell JH, Foster CM, Vishnivetskaya T, Campbell AG, Yang ZK, Wymore A, et al. Host genetic and environmental effects on mouse intestinal microbiota. ISME J (2012) 6(11):2033–44. doi: 10.1038/ismej.2012.54
27. Org E, Parks BW, Joo JWJ, Emert B, Schwartzman W, Kang EY, et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res (2015) 25(10):1558–69. doi: 10.1101/gr.194118.115
28. Gacesa R, Kurilshikov A, Vich Vila A, Sinha T, Klaassen MAY, Bolte LA, et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature (2022) 604(7907):732–9. doi: 10.1038/s41586-022-04567-7
29. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature (2014) 505(7484):559–63. doi: 10.1038/nature12820
30. Simões CD, Maukonen J, Kaprio J, Rissanen A, Pietiläinen KH, Saarela M. Habitual dietary intake is associated with stool microbiota composition in monozygotic twins. J Nutr (2013) 143(4):417–23. doi: 10.3945/jn.112.166322
31. Chae YK, Arya A, Iams W, Cruz MR, Chandra S, Choi J, et al. Current landscape and future of dual anti-CTLA4 and PD-1/PD-L1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non-small cell lung cancer (NSCLC). J Immunother Cancer (2018) 6(1):39. doi: 10.1186/s40425-018-0349-3
32. Lynch TJ, Bondarenko I, Luft A, Serwatowski P, Barlesi F, Chacko R, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line treatment in stage IIIB/IV non–Small-Cell lung cancer: Results from a randomized, double-blind, multicenter phase II study. J Clin Orthod (2012) 30(17):2046–54. doi: 10.1200/JCO.2011.38.4032
33. Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med (2010) 363(8):711–23. doi: 10.1056/NEJMoa1003466
34. Haslam A, Gill J, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for immune checkpoint inhibitor drugs. JAMA Netw Open (2020) 3(3):e200423. doi: 10.1001/jamanetworkopen.2020.0423
35. Choi JW, Withers SS, Chang H, Spanier JA, de la Trinidad VL, Panesar H, et al. Development of canine PD-1/PD-L1 specific monoclonal antibodies and amplification of canine T cell function. PloS One (2020) 15(7):e0235518. doi: 10.1371/journal.pone.0235518
36. Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature (2018) 555(7695):210–5. doi: 10.1038/nature25973
37. Mason NJ, Chester N, Xiong A, Rotolo A, Wu Y, Yoshimoto S, et al. Development of a fully canine anti-canine CTLA4 monoclonal antibody for comparative translational research in dogs with spontaneous tumors. MAbs (2021) 13(1):2004638. doi: 10.1080/19420862.2021.2004638
38. Marable J, Ruiz D, Jaiswal AK, Bhattacharya R, Pantazes R, Agarwal P, et al. Nanobody-based CTLA4 inhibitors for immune checkpoint blockade therapy of canine cancer patients. Sci Rep (2021) 11(1):20763. doi: 10.1038/s41598-021-00325-3
39. Yang JC, Beck KE, Blansfield JA, Tran KQ, Lowy I, Rosenberg SA. Tumor regression in patients with metastatic renal cancer treated with a monoclonal antibody to CTLA4 (MDX-010). J Clin Orthod (2005) 23(16_suppl):2501–1. doi: 10.1200/jco.2005.23.16_suppl.2501
40. Usyk M, Pandey A, Hayes RB, Moran U, Pavlick A, Osman I, et al. Bacteroides vulgatus and bacteroides dorei predict immune-related adverse events in immune checkpoint blockade treatment of metastatic melanoma. Genome Med (2021) 13(1):160. doi: 10.1186/s13073-021-00974-z
41. Omori M, Maeda S, Igarashi H, Ohno K, Sakai K, Yonezawa T, et al. Fecal microbiome in dogs with inflammatory bowel disease and intestinal lymphoma. J Vet Med Sci (2017) 79(11):1840–7. doi: 10.1292/jvms.17-0045
42. Marsella R. Advances in our understanding of canine atopic dermatitis. Vet Dermatol (2021) 32(6):547–e151. doi: 10.1111/vde.12965
43. Gavazza A, Rossi G, Lubas G, Cerquetella M, Minamoto Y, Suchodolski JS. Faecal microbiota in dogs with multicentric lymphoma. Vet Comp Oncol (2018) 16(1):E169–75. doi: 10.1111/vco.12367
44. Pilla R, Suchodolski JS. The role of the canine gut microbiome and metabolome in health and gastrointestinal disease . vol. 6, frontiers in veterinary science. Front Media SA (2020) 11. doi: 10.3389/fvets.2019.00498
45. You I, Kim MJ. Comparison of gut microbiota of 96 healthy dogs by individual traits: Breed, age, and body condition score. Anim (Basel) (2021) 11(8):547–551. doi: 10.3390/ani11082432
46. Mrofchak R, Madden C, Evans MV, Kisseberth WC, Dhawan D, Knapp DW, et al. Urine and fecal microbiota in a canine model of bladder cancer. bioRxiv (2021) 16. doi: 10.1101/2021.12.20.472715.abstract
47. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med (2018) 24(4):392–400. doi: 10.1038/nm.4517
48. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature (2012) 486(7402):222–7. doi: 10.1038/nature11053
49. Huang Z, Pan Z, Yang R, Bi Y, Xiong X. The canine gastrointestinal microbiota: early studies and research frontiers. Gut Microbes (2020) 11(4):635–54. doi: 10.1080/19490976.2019.1704142
50. Peroni DG, Nuzzi G, Trambusti I, Di Cicco ME, Comberiati P. Microbiome composition and its impact on the development of allergic diseases. Front Immunol (2020) 11:700. doi: 10.3389/fimmu.2020.00700
51. Gasparrini AJ, Crofts TS, Gibson MK, Tarr PI, Warner BB, Dantas G. Antibiotic perturbation of the preterm infant gut microbiome and resistome. Gut Microbes (2016) 7(5):443–9. doi: 10.1080/19490976.2016.1218584
52. van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol (2011) 128(5):948–55.e1-3. doi: 10.1016/j.jaci.2011.07.027
53. Burton EN, O’Connor E, Ericsson AC, Franklin CL. Evaluation of fecal microbiota transfer as treatment for postweaning diarrhea in research-colony puppies. J Am Assoc Lab Anim Sci (2016) 55(5):582–7. https://www.ncbi.nlm.nih.gov/pubmed/27657714.
54. Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science. (2016) 352(6285):560–4. doi: 10.1126/science.aad3503
55. Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell (2014) 159(4):789–99. doi: 10.1016/j.cell.2014.09.053
56. Coelho LP, Kultima JR, Costea PI, Fournier C, Pan Y, Czarnecki-Maulden G, et al. Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome (2018) 6(1):72. doi: 10.1186/s40168-018-0450-3
57. Ciaravolo S, Martínez-López LM, Allcock RJN, Woodward AP, Mansfield C. Longitudinal survey of fecal microbiota in healthy dogs administered a commercial probiotic. Front Vet Sci (2021) 8:664318. doi: 10.3389/fvets.2021.664318
58. Schmitz S, Suchodolski J. Understanding the canine intestinal microbiota and its modification by pro-, pre- and synbiotics - what is the evidence? Vet Med Sci (2016) 2(2):71–94. doi: 10.1002/vms3.17
59. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature (2006) 444(7122):1022–3.
60. Tehrani AB, Nezami BG, Gewirtz A, Srinivasan S. Obesity and its associated disease: a role for microbiota? Neurogastroenterol Motil (2012) 24(4):305–11. doi: 10.1111/j.1365-2982.2012.01895.x
61. Salas-Mani A, Jeusette I, Castillo I, Manuelian CL, Lionnet C, Iraculis N, et al. Fecal microbiota composition changes after a BW loss diet in beagle dogs. J Anim Sci (2018) 96(8):3102–11. doi: 10.1093/jas/sky193
62. Vázquez-Baeza Y, Hyde ER, Suchodolski JS, Knight R. Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat Microbiol (2016) 1:16177. doi: 10.1038/nmicrobiol.2016.177
63. O’Keefe SJD. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol (2016) 13(12):691–706. doi: 10.1038/nrgastro.2016.165
64. Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst (2013) 105(24):1907–11. doi: 10.1093/jnci/djt300
65. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer (2017) 17(5):271–85. doi: 10.1038/nrc.2017.13
66. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
67. Chun JL, Ji SY, Lee SD, Lee YK, Kim B, Kim KH. Difference of gut microbiota composition based on the body condition scores in dogs. Hanguk Tongmul Chawon Kwahakhoe Chi (2020) 62(2):239–46. doi: 10.5187/jast.2020.62.2.239
68. Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife (2013) 2:e00458. doi: 10.7554/eLife.00458
69. Oh C, Lee K, Cheong Y, Lee S-W, Park S-Y, Song C-S, et al. Comparison of the oral microbiomes of canines and their owners using next-generation sequencing. PloS One (2015) 10(7):e0131468. doi: 10.1371/journal.pone.0131468
70. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science (2020) 369(6510):1481–9. doi: 10.1126/science.abc3421
71. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol (2016) 16(6):341–52. doi: 10.1038/nri.2016.42
72. Canter RJ, Grossenbacher SK, Foltz JA, Sturgill IR, Park JS, Luna JI, et al. Radiotherapy enhances natural killer cell cytotoxicity and localization in pre-clinical canine sarcomas and first-in-dog clinical trial. J Immunother Cancer (2017) 5(1):98. doi: 10.1186/s40425-017-0305-7
73. Monjazeb AM, Kent MS, Grossenbacher SK, Mall C, Zamora AE, Mirsoian A, et al. Blocking indolamine-2,3-Dioxygenase rebound immune suppression boosts antitumor effects of radio-immunotherapy in murine models and spontaneous canine malignancies. Clin Cancer Res (2016) 22(17):4328–40. doi: 10.1158/1078-0432.CCR-15-3026
74. Tzeng A, Sangwan N, Jia M, Liu C-C, Keslar KS, Downs-Kelly E, et al. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med (2021) 13(1):1–17. doi: 10.1186/s13073-021-00874-2
75. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discovery (2018) 8(4):403–16. doi: 10.1158/2159-8290.CD-17-1134
76. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell (2019) 178(4):795–806.e12. doi: 10.1016/j.cell.2019.07.008
77. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
78. Yust-Katz S, Gigi E, Rosenberg D, Kanner AA, Laviv Y, Benouaich-Amiel A, et al. Tami-40. tumor microbiome and glioblastoma (gbm). Neuro Oncol (2020) 22(Supplement_2):ii221–2.
79. Zhu G, Su H, Johnson CH, Khan SA, Kluger H, Lu L. Intratumour microbiome associated with the infiltration of cytotoxic CD8+ T cells and patient survival in cutaneous melanoma. Eur J Cancer (2021) 151:25–34. doi: 10.1016/j.ejca.2021.03.053
80. Azimi F, Scolyer RA, Rumcheva P, Moncrieff M, Murali R, McCarthy SW, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol (2012) 30(21):2678–83. doi: 10.1200/JCO.2011.37.8539
81. Loi S, Drubay D, Adams S, Pruneri G, Francis PA, Lacroix-Triki M, et al. Tumor-infiltrating lymphocytes and prognosis: A pooled individual patient analysis of early-stage triple-negative breast cancers. J Clin Orthod (2019) 37(7):559–69. doi: 10.1200/JCO.18.01010
82. Kobayashi N, Hiraoka N, Yamagami W, Ojima H, Kanai Y, Kosuge T, et al. FOXP3+ regulatory T cells affect the development and progression of hepatocarcinogenesis. Clin Cancer Res (2007) 13(3):902–11. doi: 10.1158/1078-0432.CCR-06-2363
83. Zhai Y, Haresi AJ, Huang L, Lang D. Differences in tumor initiation and progression of melanoma in the BrafCA ;Tyr-CreERT2;Ptenf/f model between male and female mice. Pigm Cell Melanoma Res (2020) 33(1):119–21. doi: 10.1111/pcmr.12821
84. Simpson S, Dunning MD, de Brot S, Grau-Roma L, Mongan NP, Rutland CS. Comparative review of human and canine osteosarcoma: morphology, epidemiology, prognosis, treatment and genetics. Acta Vet Scand (2017) 59(1):71. doi: 10.1186/s13028-017-0341-9
85. Wunder JS, Gokgoz N, Parkes R, Bull SB, Eskandarian S, Davis AM, et al. TP53 mutations and outcome in osteosarcoma: a prospective, multicenter study. J Clin Oncol (2005) 23(7):1483–90. doi: 10.1200/JCO.2005.04.074
86. Kirpensteijn J, Kik M, Teske E, Rutteman GR. TP53 gene mutations in canine osteosarcoma. Vet Surg (2008) 37(5):454–60. doi: 10.1111/j.1532-950X.2008.00407.x
87. Gardner HL, Sivaprakasam K, Briones N, Zismann V, Perdigones N, Drenner K, et al. Canine osteosarcoma genome sequencing identifies recurrent mutations in DMD and the histone methyltransferase gene SETD2. Commun Biol (2019) 2:266. doi: 10.1038/s42003-019-0487-2
88. Sakthikumar S, Elvers I, Kim J, Arendt ML, Thomas R, Turner-Maier J, et al. SETD2 is recurrently mutated in whole-exome sequenced canine osteosarcoma. Cancer Res (2018) 78(13):3421–31. doi: 10.1158/0008-5472.CAN-17-3558
89. Amin SB, Anderson KJ, Boudreau CE, Martinez-Ledesma E, Kocakavuk E, Johnson KC, et al. Comparative molecular life history of spontaneous canine and human gliomas. Cancer Cell (2020) 37(2):243–57.e7. doi: 10.1016/j.ccell.2020.01.004
90. Alsaihati BA, Ho K-L, Watson J, Feng Y, Wang T, Dobbin KK, et al. Canine tumor mutational burden is correlated with TP53 mutation across tumor types and breeds. Nat Commun (2021) 12(1):4670. doi: 10.1038/s41467-021-24836-9
91. Gustafson DL, Duval DL, Regan DP, Thamm DH. Canine sarcomas as a surrogate for the human disease. Pharmacol Ther (2018) 188:80–96. doi: 10.1016/j.pharmthera.2018.01.012
92. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
93. Dobson JM. Breed-predispositions to cancer in pedigree dogs. ISRN Vet Sci (2013) 2013:941275. doi: 10.1155/2013/941275
Keywords: microbiome, immunotherapy, cancer, comparative oncology, canines
Citation: Kleber KT, Iranpur KR, Perry LM, Cruz SM, Razmara AM, Culp WTN, Kent MS, Eisen JA, Rebhun RB and Canter RJ (2022) Using the canine microbiome to bridge translation of cancer immunotherapy from pre-clinical murine models to human clinical trials. Front. Immunol. 13:983344. doi: 10.3389/fimmu.2022.983344
Received: 30 June 2022; Accepted: 26 July 2022;
Published: 12 August 2022.
Edited by:
Ashutosh K. Mangalam, The University of Iowa, United StatesReviewed by:
Braden C. McFarland, University of Alabama at Birmingham, United StatesCopyright © 2022 Kleber, Iranpur, Perry, Cruz, Razmara, Culp, Kent, Eisen, Rebhun and Canter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert J. Canter, cmpjYW50ZXJAdWNkYXZpcy5lZHU=
 Kara T. Kleber
Kara T. Kleber Khurshid R. Iranpur1
Khurshid R. Iranpur1 Lauren M. Perry
Lauren M. Perry Sylvia M. Cruz
Sylvia M. Cruz Aryana M. Razmara
Aryana M. Razmara William T. N. Culp
William T. N. Culp Michael S. Kent
Michael S. Kent Jonathan A. Eisen
Jonathan A. Eisen Robert B. Rebhun
Robert B. Rebhun Robert J. Canter
Robert J. Canter