- 1Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Clinical Research Development Unit of Tabriz Valiasr Hospital, Tabriz University of Medical Sciences, Tabriz, Iran
- 3Department of Anatomical Sciences, Faculty of Medicine, Birjand University of Medical Sciences, Birjand, Iran
- 4Department of Pharmacognosy, College of Pharmacy, Hawler Medical University, Erbil, Kurdistan, Iraq
- 5Center of Research and Strategic Studies, Lebanese French University, Erbil, Kurdistan, Iraq
- 6Faculty of Medicine, Birjand University of Medical Sciences, Birjand, Iran
- 7Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 8Institute of Human Genetics, Jena University Hospital, Jena, Germany
- 9Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Programmed death-ligand 1 (PD-L1) is a transmembrane protein with essential roles in the suppression of adaptive immune responses. As an immune checkpoint molecule, PD-L1 can be exploited by cancer cells to evade the anti-tumor attacks initiated by the immune system. Thus, blockade of the PD1/PD-L1 axis can eliminate the suppressive signals and release the antitumor immune responses. Identification of the underlying mechanisms of modulation of the activity of the PD1/PD-L1 axis would facilitate the design of more efficacious therapeutic options and better assignment of patients for each option. Recent studies have confirmed the interactions between miRNAs/lncRNAs/circ-RNAs and the PD1/PD-L1 axis. In the current review, we give a summary of interactions between these transcripts and PD-L1 in the context of cancer. We also overview the consequences of these interactions in the determination of the response of patients to anti-cancer drugs.
Introduction
Programmed death-ligand 1 (PD-L1), alternatively named as CD274 or B7-H1 is a transmembrane protein with essential roles in the suppression of adaptive immune responses. The reaction of the adaptive immune system to external or internal danger signals leads to the expansion of antigen-specific CD8+ and/or CD4+ T cell clones (1). However, when PD-L1 binds to the PD-1 checkpoint, an inhibitory signal is transmitted which decreases the proliferation of antigen-specific T-cells in lymph nodes and at the same time reduces apoptosis of regulatory T cells. PD-1/PD-L1 axis mainly acts at the late stage of induction of T-cell immune responses in peripheral tissues (2).
Immune checkpoint pathways are exploited by cancer cells so as to evade the anti-tumor attacks initiated by the immune system (3). Thus, the blockade of immune checkpoints can eliminate the suppressive signals and release the antitumor immune responses. PD-1 and PD-L1 have been used as key drug targets for the development of immune checkpoint blockade treatment modalities (2). Several PD-1/PD-L1 targeted therapies have been approved for the treatment of several types of malignancies (2). At least three human IgG1 antibodies anti-PD-L1 antibodies have been approved for clinical application (4). While atezolizumab and durvalumab have been designed to eliminate FcγR-binding and effector function, avelumab retains the intact function of Fc (5). BMS-936559 is another PD-L1-targeting antibody that is distinctive from the mentioned approved PD-L1 antibodies since it is an IgG4 mAb with S228P mutation (5). In addition, the fusion protein KN035 contains a distinct domain of the humanized anti-PD-L1 antibody and the Fc of an IgG1 (6). Meanwhile, PD-L1-targeting agents can be prescribed in the form of a prodrug. An example is the agent CX-072 which can be activated by a protease (7). Due to incomplete success and drawbacks of using PD-L-targeting drugs, assessment of expression of PD-L1 is regarded as a marker for prediction of response and identification of patients who gain a favorable clinical response from this type of therapy (4).
The expression of genes can be regulated at the transcriptional and posttranscriptional levels by functional RNA molecules known as non-coding RNAs (8, 9). The expression of ncRNAs, which primarily regulate oncogenes and tumor suppressor genes, is altered in several kinds of human malignancies (10, 11). Recent studies have confirmed the interplay between a variety of non-coding RNAs and the PD1/PD-L1 axis. In fact, several microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) have been shown to modulate the activity of this axis. Identification of molecular mechanisms that modulate the activity of the PD1/PD-L1 axis would facilitate the design of more efficacious therapeutic options and better assignment of patients for each option. In the current review, we give a snapshot of interactions between these transcripts and PD-L1 in the context of cancer. We also overview the consequences of these interactions in the determination of the response of patients to anti-cancer drugs.
Gastrointestinal cancers
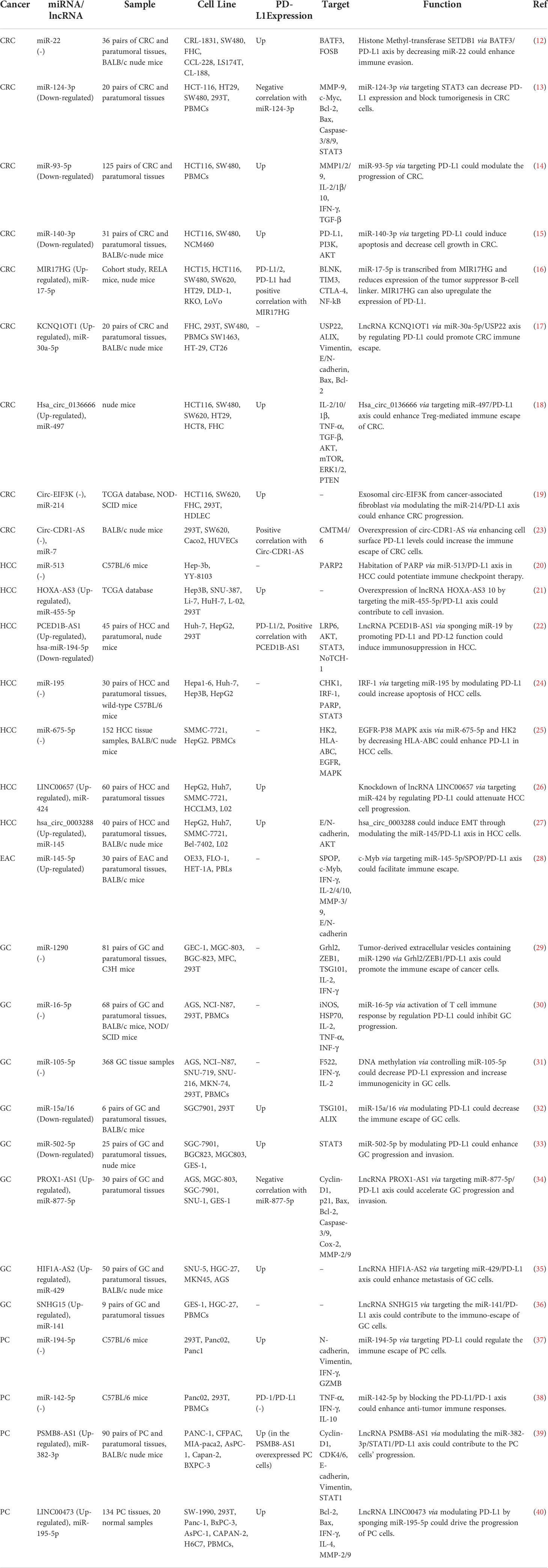
In colorectal cancer (CRC) samples, over-expression of SETDB1 expression has been associated with the expression of PD-L1. Mechanistically, SETDB1 can down-regulate miR-22 levels by decreasing the expression of FOSB. On the other hand, miR-22 down-regulates PD-L1 levels by targeting BATF3. SETDB1 knockdown has enhanced the cytotoxic effects of T cells on tumor cells by influencing the FOSB/miR-22/BATF3/PD-L1 axis, thus hindering the growth of CRC tumors in mice. Cumulatively, the effects of SETDB1 on the activity BATF3/PD-L1 axis leads to immune evasion of CRC tumors (12). miR-124 is another miRNA with a possible regulatory role on PD-L1. Expression of miR-124 is significantly decreased in CRC, and its under-expression has been correlated with the up-regulation of PD-L1 (13). The luciferase assay has validated PD-L1 targeting by miR-124 (13). miR-124 mimics could significantly reduce the expression of PD-L1 at the transcript, protein, and cell surface levels (13). Moreover, this miRNA could inhibit Tregs in co-culture models by influencing levels of IL-10, IL-2, TNF-α, TGF-β, and IFN-γ. Up-regulation of miR-124 could also decrease the proliferation of CRC cells and induce cell cycle arrest at G1 via down-regulating c-Myc. Moreover, this miRNA could induce intrinsic and extrinsic apoptotic pathways, down-regulate CD44 and MMP-9 expression levels, and suppress cell migration and invasion (13). STAT3 signaling has also been suppressed by miR-124 (13). miR‐93‐5p (14) and miR-140-3p (15) are two other miRNAs that can target PD-L1 in CRC cells. Meanwhile, a number of lncRNAs and circRNAs have been found to affect CRC progression by influencing the expression of miRNAs and enhancing the expression of PD-L1. For instance, MIR17HG lncRNA promotes the progression of CRC by influencing the expression of miR-17-5p (16). Besides, KCNQ1OT1 lncRNA released by CRC cells-originated exosomes can mediate immunity through the regulation of PD-L1 ubiquitination via miR-30a-5p/USP22 (17). Hsa_circ_0136666 is also involved in Treg-mediated immune escape through modulation of miR-497/PD-L1 axis (Figure 1A) (18). Cancer-associated fibroblasts have been shown to secrete circEIF3K in their exosomes to enhance progression of CRC through miR-214/PD-L1 axis (19).
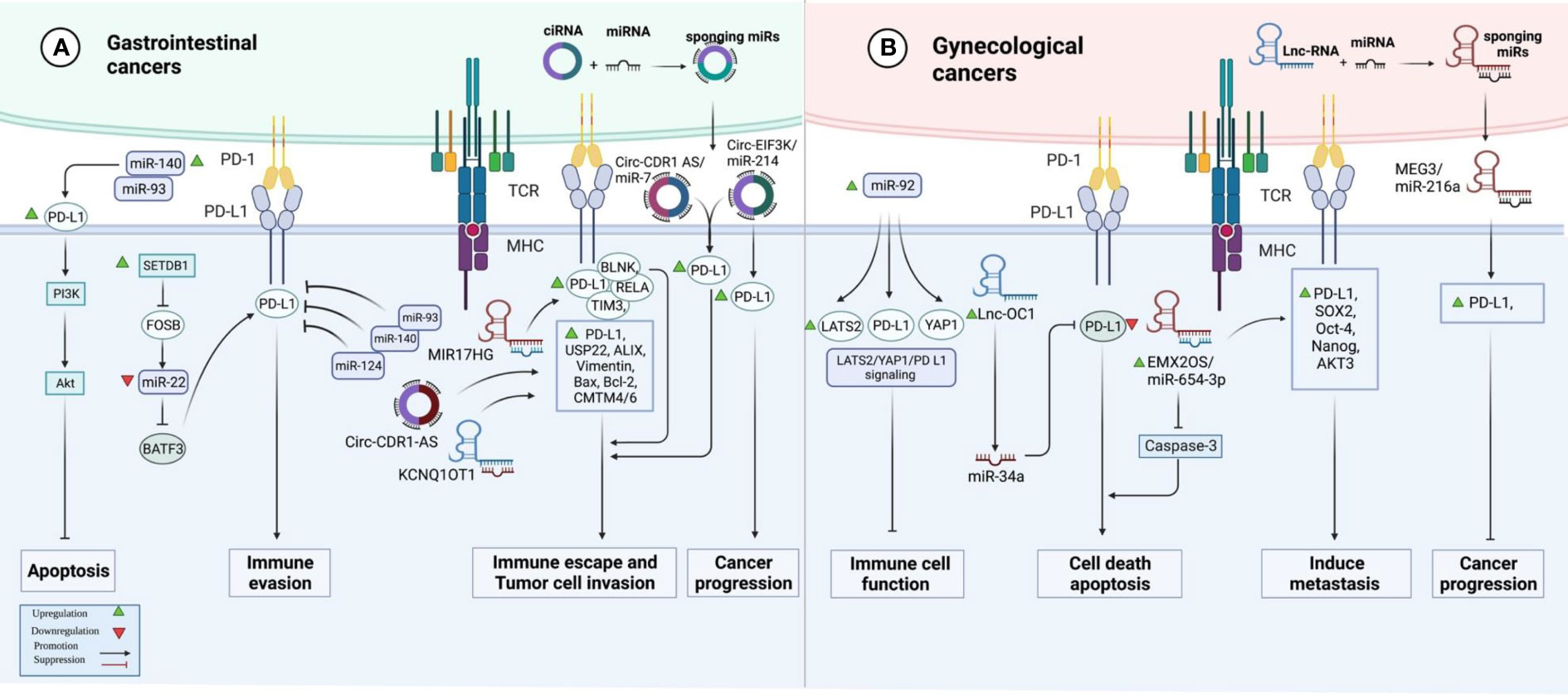
Figure 1 A graphical representation of the ways in which programmed death ligand 1 (PD-L1) and non-coding RNAs interact with one another in gastrointestinal and gynecological cancers.
In hepatocellular carcinoma (HCC), suppression of PARP has enhanced the efficacy of immune checkpoint therapy via influencing the miR-513/PD-L1 axis. Thus, combined administration of the PARP inhibitor olaparib and anti-PD1 has been suggested as a treatment modality in HCC (20). In this type of cancer, HOXA-AS3 lncRNA has been found to promote proliferation and migratory potential through miR-455-5p/PD-L1 axis (21). Another study in HCC has shown a correlation between expressions of PD-L1 and PD-L2. Moreover, this study has demonstrated the up-regulation of PCED1B-AS1. Expression of PCED1B-AS1 has been positively correlated with expression levels of PD-L1 and PDL-2 while being negatively correlated with hsa-miR-194-5p. Mechanistically, PCED1B-AS1 increases expressions of PD-L1 and PD-L2 through sequestering hsa-miR-194-5p, thus inducing PD-L1/2-mediated immunosuppressive effects on T cells (22). Table 1 shows the interactions between non-coding RNAs and PD-L1 in gastrointestinal cancers.
Lung cancer
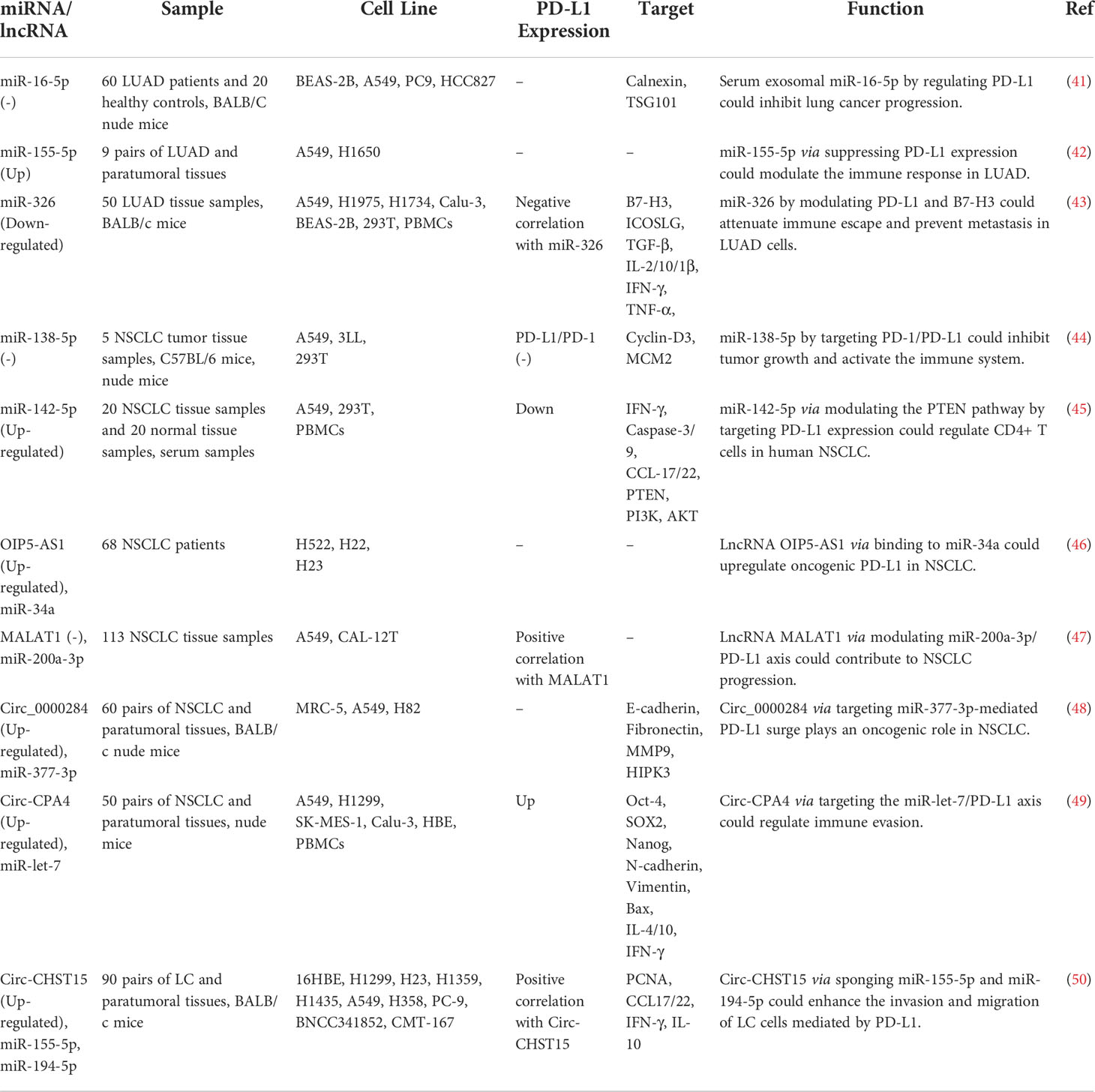
In lung cancer samples, expression of PD-L1 expression has been correlated with the T stage. Treatment with PD-L1 inhibitor has decreased the expression of PD-L1 and diminished T stage in patients suffering from PD-L1-positive lung cancer. In this group of patients, over-expression of PD-L1 or decreased serum exosomal level of miR-16-5p level has been correlated with longer survival upon treatment with a PD-L1 inhibitor. Moreover, this kind of treatment reduced the quantity of exosomes in the sera of PD-L1-positive patients and enhanced serum exosomal levels of miR-16-5p. High exosomal levels of miR-16-5p could depress cell proliferation and migration in cell cultures, and induce apoptosis, particularly in cells treated with a PD-L1 inhibitor. Cumulatively, the miR-16-5p content of serum exosomes possibly inhibits tumor growth and can be used as a marker for PD-L1 inhibitor therapy (41). miR‐155‐5p (42) and miR-326 (43) are two other miRNAs that suppress PD‐L1 expression and attenuate immune escape in lung cancer by targeting PD-L1. In addition, miR-138-5p has been found to affect the activity of the PD-1/PD-L1 axis, inhibit tumor growth and activate the immune system in this type of cancer (44). A number of oncogenic lncRNAs and circRNAs, namely OIP5-AS1, MALAT1, Circ_0000284, Circ-CPA4 and Circ-CHST15 have been shown to act as molecular sponges for PD-L1-targeting miRNAs, thus incresing levels of PD-L1 (Figure 2A) (Table 2).
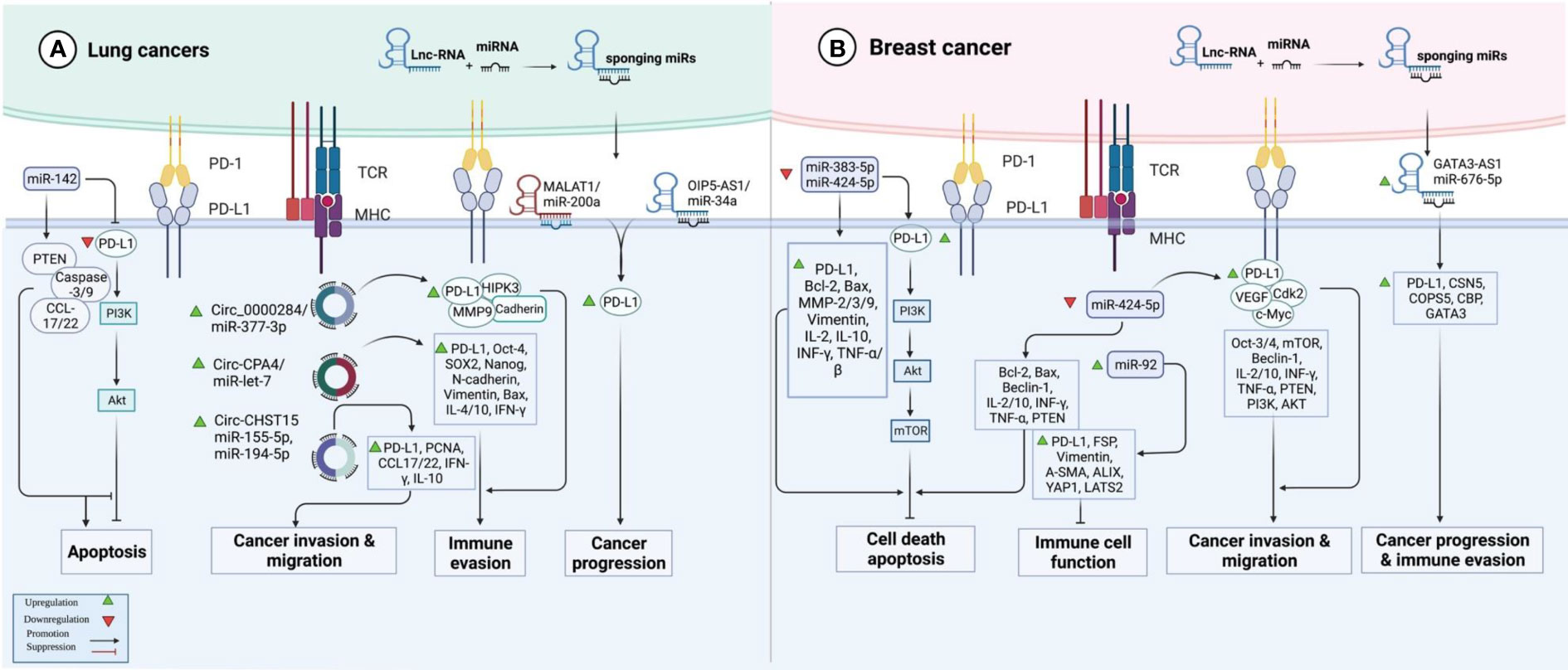
Figure 2 An illustration in graphical form of the methods via which the proteins are known as programmed death ligand 1 (PD-L1) and non-coding RNAs such as miRNAs, lnc-RNAs, and circ-RNAs interact with one another in lung and breast cancer.
Gynecological cancers
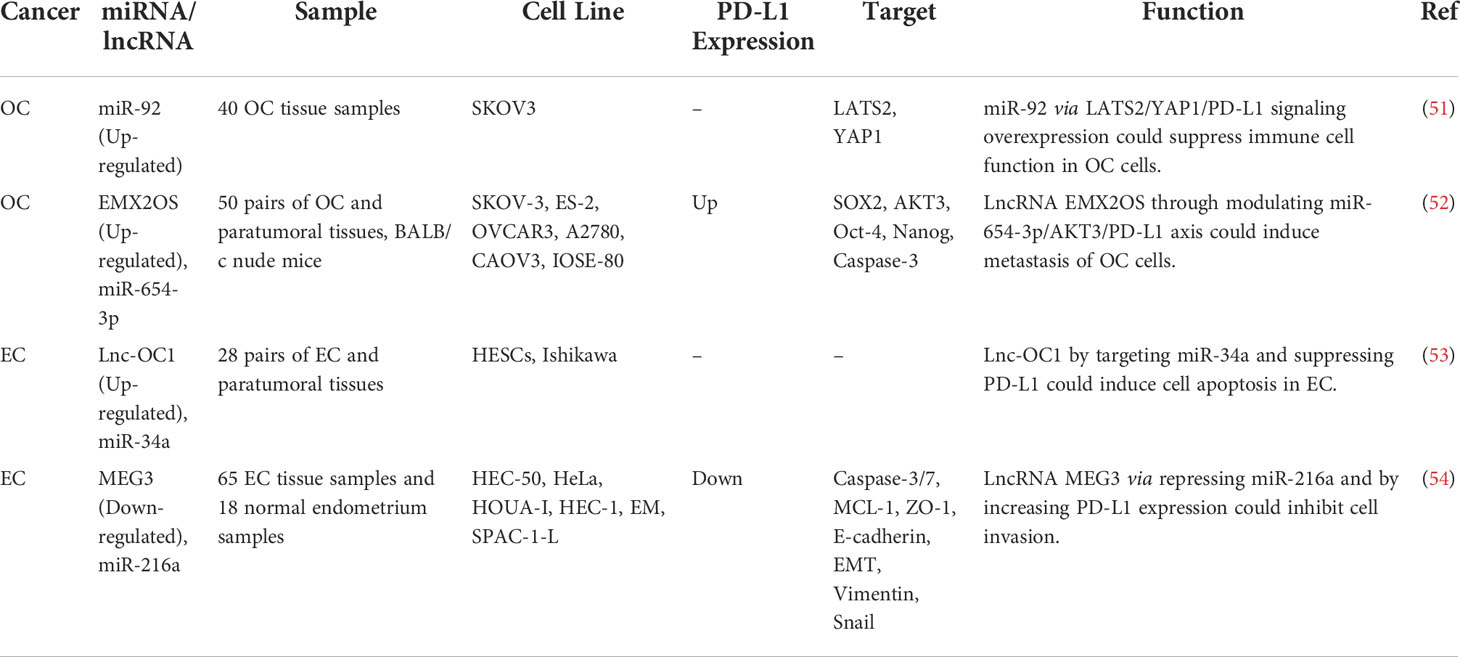
The impact of non-coding RNAs on the expression of PD-L1 has been assessed in ovarian cancer (OC), cervical cancer (CC), and endometrial cancer (EC). In OC, miR-92 has been reported to block the functions of immune cells by modulating the expression of PD-L1 via the LATS2/YAP1 axis (51). Moreover, the sponging effect of EMX2OS on miR-654-3p has been shown to result in the induction of cell proliferation, invasive properties, and sphere formation in OC through regulation of the AKT3/PD-L1 axis (52). In addition, the Lnc-OC1/miR-34a axis has a crucial role in the pathogenesis of EC through the modulation of PD-L1 (53). Conversely, another study has shown that PD-L1 has a tumor suppressor role in aggressive EC and its expression is influenced by MEG3/miR-216a axis (Figure 1B) (54). Table 3 shows the interactions between non-coding RNAs and PD-L1 in gynecological cancers.
Breast cancer
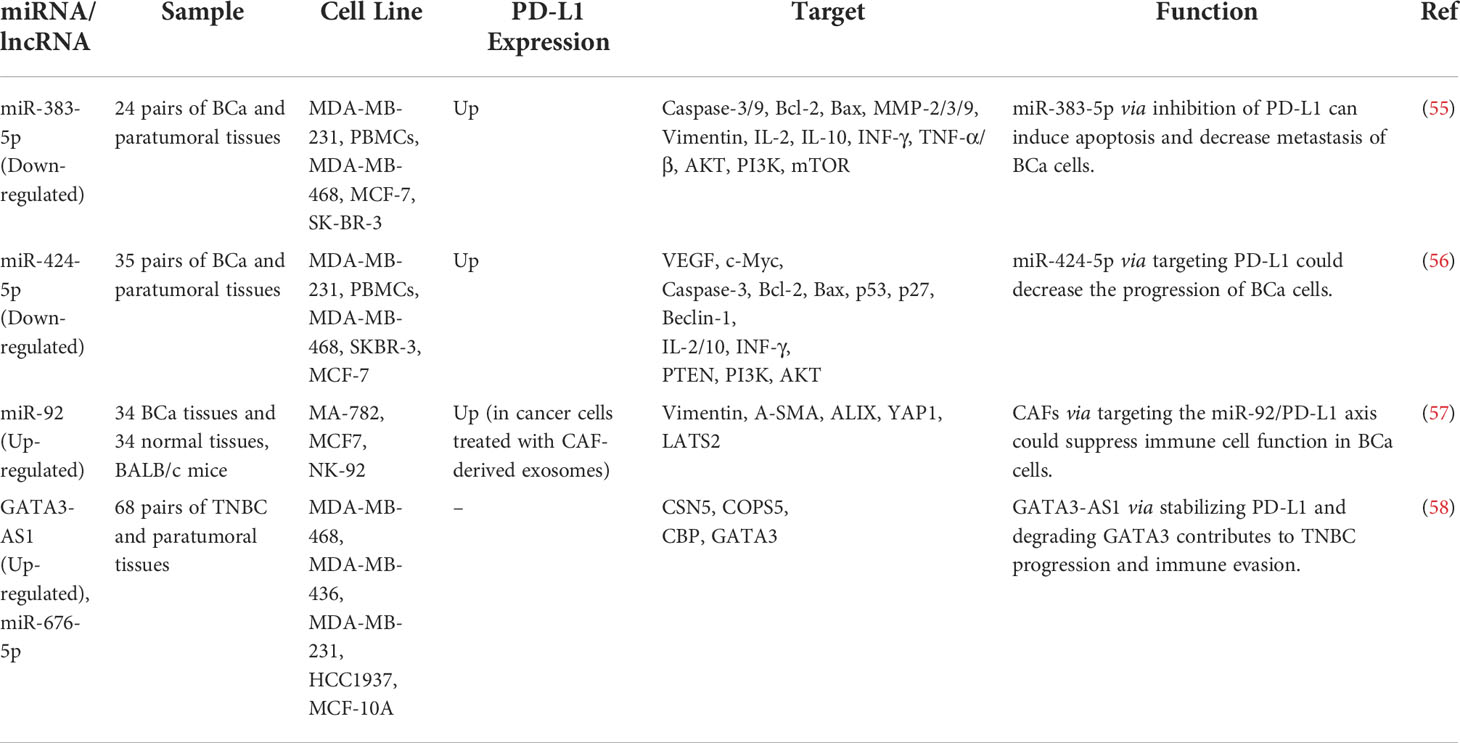
At least four studies have assessed interactions between non-coding RNAs and PD-L1 in the context of breast cancer (BCa) (Table 4). Two down-regulated miRNAs in BCa, namely miR-383-5p (55) and miR-424-5p (56) have been reported to affect the expression of PD-L1 (Figure 2B). Moreover, exosomes secreted by BCa-associated fibroblasts have been shown to assist in the suppression of immune cell function through the transfer of PD-L1 inhibiting miRNA miR-92 (57). Finally, expression of GATA3-AS1 has been found to be markedly increased in triple-negative BCa tissues and cells parallel with a reduction in the proportion of CD8+ T cells. GATA3-AS1 silencing has suppressed the growth of tumor cells and decreased the half-life of the PD-L1 protein. GATA3-AS1 has been shown to induce PD-L1 deubiquitination via miR-676-3p/COPS5 axis. Thus, this lncRNA contributes to the progression of triple-negative BCa and immune evasion via enhancing the stability of PD-L1 protein and inducing GATA3 degradation (58).
Renal cancer
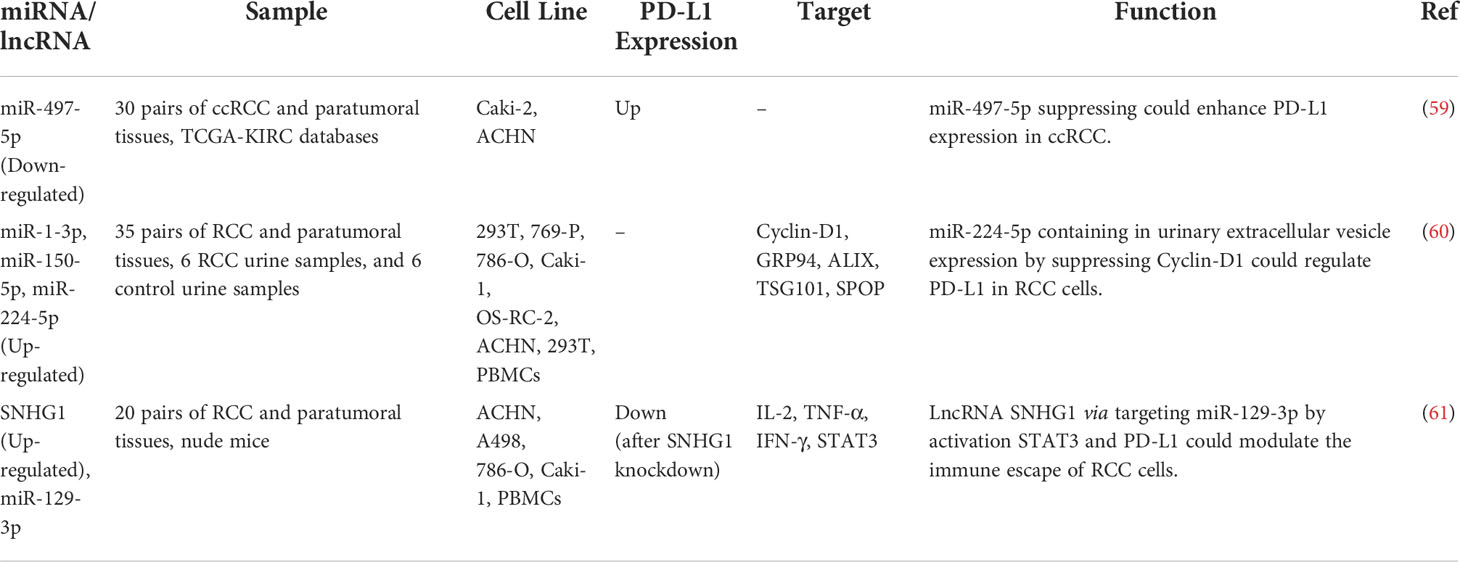
miR-497-5p is a putative tumor suppressor miRNA in renal cancer whose down-regulation leads to the over-expression of PD-L1 in this type of cancer (59). Moreover, urinary extracellular vesicles (EVs) have been found to contain miR-224-5p. Expression of miR-224-5p has been higher in urinary EVs of patients with renal cell carcinoma compared to healthy controls. Over-expressed miR-224-5p could inhibit proliferation and induce cell cycle arrest by targeting CCND1. Besides, miR-224-5p has a role in the induction of invasive and metastatic properties of renal carcinoma cells. Remarkably, miR-224-5p has a role in the enhancement of PD-L1 stability through the suppression of CCND1 (60). Finally, SNHG1 has been found to regulate immune escape in this type of cancer through targeting miR‐129‐3p and subsequently activating STAT3 and PD‐L1 (61). Table 5 shows the interaction between non-coding RNAs and PD-L1 in renal cancer.
Other cancers
The interaction between non-coding RNAs and PD-L1 has also been investigated in laryngeal cancer, anaplastic thyroid carcinoma, osteosarcoma, and diffuse large B-cell lymphoma (DLBCL) (Table 6). In laryngeal cancer, miR-217 by repressing the AEG-1/PD-L1 axis could inhibit metastasis (62). In thyroid carcinoma, UCA1 has been found to affect the miR-148a/PD L1 axis to attenuate the cytotoxic effects of CD8+T cells (63). In osteosarcoma, miR-200a through promoting PTEN-mediated PD-L1 up-regulation could increase immunosuppression (64). Moreover, LINC00657 via targeting miR‐106a could promote metastasis through the activation of PD‐L1 (65). Finally, MALAT1 by sponging miR-195 could enhance tumorigenesis and immune escape of DLBCL (67).
Interactions between non-coding RNAs and PD-L1 in response to chemotherapeutic agents
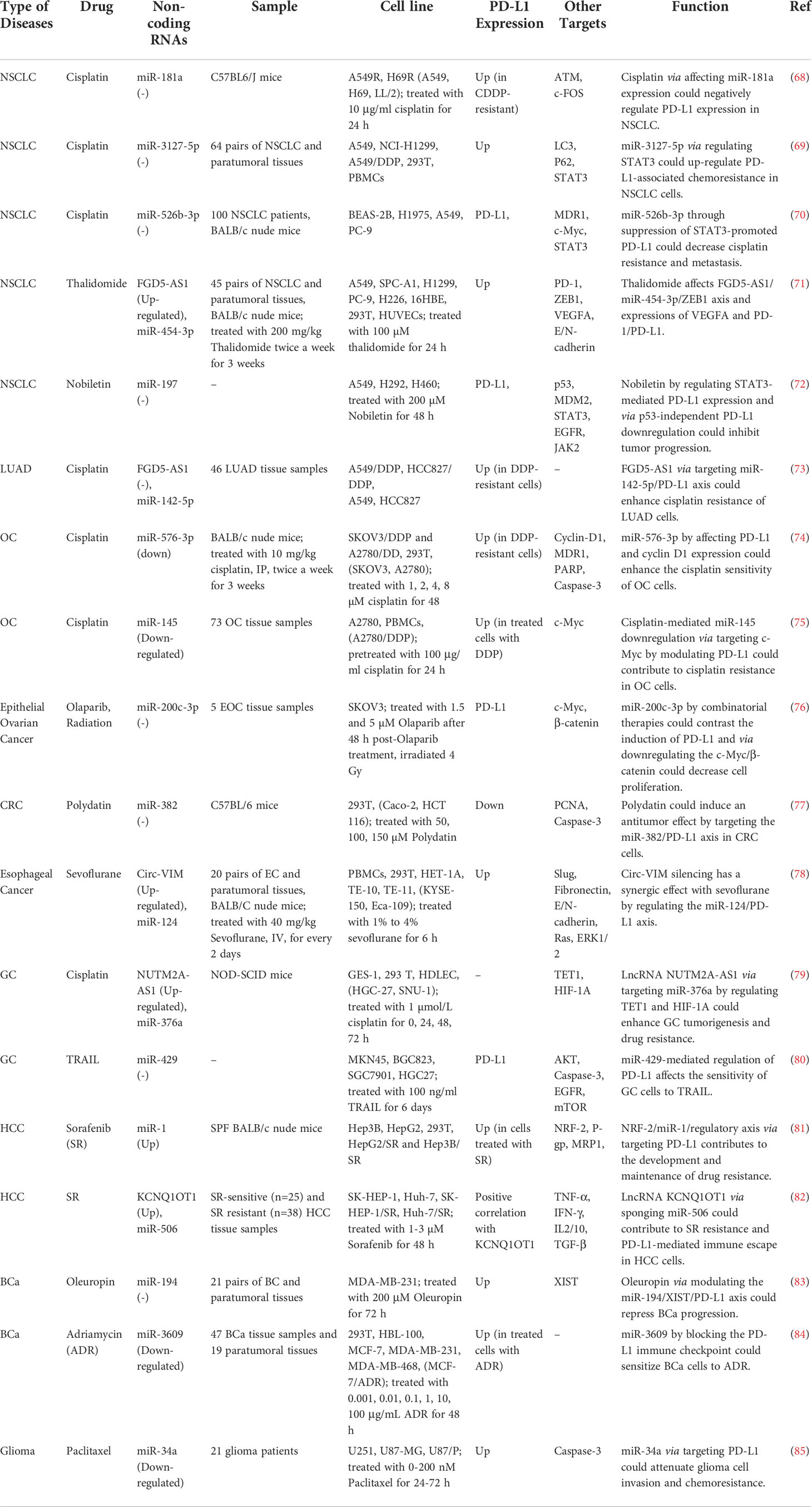
The interaction between non-coding RNAs and PD-L1 can also affect the response of cancer cells to anti-cancer modalities. Moreover, anti-cancer drugs can affect these interactions (Table 7). Cisplatin is the mostly assessed drug in this regard. Cisplatin via modulation of miR-181a expression could negatively regulate PD-L1 expression in NSCLC (68). Meanwhile, miR-3127-5p via regulating STAT3 could up-regulate PD-L1-associated chemoresistance in NSCLC cells (69). Besides, miR-526b-3p through inhibiting STAT3-promoted PD-L1 expression could decrease cisplatin resistance and metastasis (70). miR 576 3p by affecting PD L1 and cyclin D1 expression could enhance the cisplatin sensitivity of OC cells (74).
Thalidomide has been shown to suppress angiogenic process and immune evasion of NSCLC cells by influencing the expression of VEGFA and PD-1/PD-L1 via affecting FGD5-AS1/miR-454–3p/ZEB1 axis (71). Moreover, nobiletin has been shown to regulate PD-L1 expression through the modulation of STAT3, thus inhibiting the progression of NSCLC (72). Meanwhile, miR-34a via targeting PD-L1 could attenuate glioma cell invasion and their chemoresistance (85).
Discussion
PD-L1 has been found to participate in the tumorigenesis process via induction of Tregs and inhibition of antitumor immune responses. Besides, several non-coding RNAs can act as regulators of gene expression, particularly at the posttranscriptional level to affect the expression of PD-L1. In fact, a delicate interactive network exists between non-coding RNAs that influence the expression of PD-L1. Several lncRNA/miRNA or circRNA/miRNA modules have been found in this network. MIR17HG/miR-17-5p, KCNQ1OT1/miR-30a-5p, hsa_circ_0136666/miR-497, circ-EIF3K/miR-214, circ-CDR1-AS/miR-7, HOXA-AS3/miR-455-5p, PCED1B-AS1/hsa-miR-194-5p, LINC00657/miR-424, LINC00657/miR-424, hsa_circ_0003288/miR-145, PROX1-AS1/miR-877-5p, HIF1A-AS2/miR-429, SNHG15/miR-141, PSMB8-AS1/miR-382-3p, LINC00473/miR-195‐5p, OIP5-AS1/miR-34a, MALAT1/miR-200a-3p, circ_0000284/miR-377-3p, circ-CPA4/miR-let-7, circ-CHST15/miR-155-5p, circ-CHST15/miR-194-5p, EMX2OS/miR-654-3p, Lnc-OC1/miR-34a, MEG3/miR-216a, GATA3-AS1/miR-676-5p, LINC00657/miR‐106a and MALAT1/miR-195 are examples of these modules.
In addition to the above-mentioned PD-L1-interacting non-coding RNAs, several non-coding RNAs can indirectly affect the activity of the PD-1/PD-L1 pathway (86). For instance, the up-regulated lncRNA in diffuse large B cell lymphoma SNHG14 adsorbs miR-5590-3p to enhance expression levels of ZEB1, thus activating the PD-1/PD-L1 pathway and inducing immune evasion (87).
The interactions between non-coding RNAs and PD-L1 are also involved in the response of patients to several anti-cancer modalities. These interactions can also be affected by the administration of several treatment modalities. Cisplatin, Adriamycin, Paclitaxel, Thalidomide, Nobiletin, Olaparib, Sevoflurane, Sorafenib, and Oleuropin are examples of drugs that can affect/be affected by such interactions.
Most importantly, several PD-L1-related non-coding RNAs have been found to be secreted in exosomes originated from tumor cells’ microenvironment residing cells. This finding not only shows the extensive impacts of these non-coding RNAs in tumor progression or modulation of the tumor microenvironment but also potentiates these transcripts as biomarkers for the early detection of cancer using biofluids.
The promising results of clinical trials of anti-PD-L1 during recent years have encouraged researchers to find better treatment modalities to enhance the clinical response to these modalities. Non-coding RNAs are putative modulators of PD-L1 expression and activity, and thus can be used as therapeutic targets in this regard. Moreover, expression levels of PD-L1-associated non-coding RNAs can be used as predictive markers for response to PD-L1 inhibitors. In fact, the lack of appropriate response to this type of therapy can be attributed to the levels of expressions of PD-L1-associated non-coding RNAs. Thus, modulation of expression of these transcripts using siRNA or antisense oligonucleotides can enhance clinical response to anti-PD-L1 therapy. Application of these methods needs a prior identification of the pattern of expression of PD-L1-associated non-coding RNAs in patients’ samples and the functional network between these transcripts and PD-L1. This can be achieved using high throughput sequencing methods with a system biology approach. Future studies are needed to propose tissue-specific panels of non-coding RNAs for this purpose.
Data availability statement
The analyzed data sets generated during the study are available from the corresponding author upon reasonable request.
Author contributions
SG-F wrote the manuscript and revised it. MT and GS supervised and designed the study. HS, YP and BH collected the data and designed the figures and tables. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors would like to thank the Clinical Research Development Unit (CRDU) of Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran for their support, cooperation, and assistance throughout the period of. The authors want to acknowledge the clinical research development unit of Tabriz Valiasr Hospital for their support of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CRC, Colorectal Cancer; CC, Cervical cancer; OC, Ovarian Cancer; EC, Endometrial Cancer; HCC, Hepatocellular Carcinoma; EAC, Esophageal Adenocarcinoma; GC, Gastric Cancer; PC, Pancreatic Cancer; LUAD, Lung Adenocarcinoma; NSCLC, Non-Small Cell Lung Cancer; LC, Lung Cancer; PD-L1, Programmed Death-Ligand 1; TNBC, Triple-Negative Breast Cancer; BCa, Breast Cancer; IP, Intraperitoneal; PARP, Polymerase; OIP5-AS1, OPA-Interacting Protein 5 Antisense Transcript 1; TAMs, Tumor-Associated Macrophages; HK2, Hexokinase 2; EGFR, Epidermal Growth Factor Receptor; SPOP, Cullin 3-Speckle Type POZ Protein; IV, Intravenous; FGD5-AS1, FGD5-Antisense 1; Grhl2, Grainy Head-Like 2; HIF1A-AS2, Hypoxia-Inducible Factor-1 Alpha Antisense RNA-2; CHK1, Checkpoint Kinase 1; IRF-1, Interferon Regulatory Factor 1; HUVECs, Human Umbilical Vein Endothelial Cells; CIK, Cytokine-Induced Killer; HUSMC, Human Uterine Smooth Muscle Cells; HDLEC, Human Dermal Lymphatic Endothelial Cells; HESCs, Human Embryonic Stem Cells; PTEN, Chromosome Ten; MMP‐1, Metalloproteinase‐1; NPs, Nanoparticles; CAFs, Cancer-Associated Fibroblasts; NRF-2, Nuclear Factor E2-Related Factor 2; TRAIL, Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand; dMMR, Mismatch Repair Deficiency; BLNK, B-cell linker; PBLs, Peripheral Blood Lymphocytes.
References
1. Spellberg B, Edwards JE Jr. Type 1/Type 2 immunity in infectious diseases. Clin Infect Dis (2001) 32(1):76–102. doi: 10.1086/317537
2. Ghosh C, Luong G, Sun Y. A snapshot of the PD-1/PD-L1 pathway. J Cancer (2021) 12(9):2735–46. doi: 10.7150/jca.57334
3. Marin-Acevedo JA, Dholaria B, Soyano AE, Knutson KL, Chumsri S, Lou Y. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J Hematol Oncol (2018) 11(1):1–20. doi: 10.1186/s13045-018-0582-8
4. Guo L, Wei R, Lin Y, Kwok HF. Clinical and recent patents applications of PD-1/PD-L1 targeting immunotherapy in cancer treatment-current progress, strategy, and future perspective. Front Immunol (2020) 11:1508. doi: 10.3389/fimmu.2020.01508
5. Chen X, Song X, Li K, Zhang T. FcγR-binding is an important functional attribute for immune checkpoint antibodies in cancer immunotherapy. Front Immunol (2019) 10:292. doi: 10.3389/fimmu.2019.00292
6. Zhang F, Wei H, Wang X, Bai Y, Wang P, Wu J, et al. Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade. Cell Discov (2017) 3(1):1–12. doi: 10.1038/celldisc.2017.4
7. Autio KA, Boni V, Humphrey RW, Naing A. Probody therapeutics: An emerging class of therapies designed to enhance on-target effects with reduced off-tumor toxicity for use in immuno-OncologyProbody therapeutics. Clin Cancer Res (2020) 26(5):984–9. doi: 10.1158/1078-0432.CCR-19-1457
8. Hussen BM, Shoorei H, Mohaqiq M, Dinger ME, Hidayat HJ, Taheri M, et al. The impact of non-coding RNAs in the epithelial to mesenchymal transition. Front Mol Biosci (2021) 8. doi: 10.3389/fmolb.2021.665199
9. Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M, Arefian N. Regulatory role of non-coding RNAs on immune responses during sepsis. Front Immunol (2021) 12:798713. doi: 10.3389/fimmu.2021.798713
10. Hussen BM, Azimi T, Abak A, Hidayat HJ, Taheri M, Ghafouri-Fard S. Role of lncRNA BANCR in human cancers: An updated review. Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.689992
11. Ghafouri-Fard S, Hussen BM, Mohaqiq M, Shoorei H, Baniahmad A, Taheri M, et al. Interplay between non-coding RNAs and programmed cell death proteins. Front Oncol (2022) 12. doi: 10.3389/fonc.2022.808475
12. Tian J, Wang W, Zhu J, Zhuang Y, Qi C, Cai Z, et al. Histone methyltransferase SETDB1 promotes immune evasion in colorectal cancer via FOSB-mediated downregulation of MicroRNA-22 through BATF3/PD-L1 pathway. J Immunol Res (2022) 2022. doi: 10.1155/2022/4012920
13. Roshani Asl E, Rasmi Y, Baradaran B. MicroRNA-124-3p suppresses PD-L1 expression and inhibits tumorigenesis of colorectal cancer cells via modulating STAT3 signaling. J Cell Physiol (2021) 236(10):7071–87. doi: 10.1002/jcp.30378
14. Chen YL, Wang GX, Lin BA, Huang JS. MicroRNA-93-5p expression in tumor tissue and its tumor suppressor function via targeting programmed death ligand-1 in colorectal cancer. Cell Biol Int (2020) 44(5):1224–36. doi: 10.1002/cbin.11323
15. Jiang W, Li T, Wang J, Jiao R, Shi X, Huang X, et al. miR-140-3p suppresses cell growth and induces apoptosis in colorectal cancer by targeting PD-L1. OncoTargets Ther (2019) 12:10275. doi: 10.2147/OTT.S226465
16. Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via hippo-YAP signaling. Mol cancer (2019) 18(1):47. doi: 10.1186/s12943-019-1010-6
17. Di Xian LN, Zeng J, Wang L. Lncrna kcnq1ot1 secreted by tumor cell-derived exosomes mediates immune escape in colorectal cancer by regulating pd-l1 ubiquitination via mir-30a-5p/usp22. Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.653808
18. Xu Y-J, Zhao J-M, Gao C, Ni X-F, Wang W, Hu W-W, et al. Hsa_circ_0136666 activates treg-mediated immune escape of colorectal cancer via miR-497/PD-L1 pathway. Cell Signal (2021) 86:110095. doi: 10.1016/j.cellsig.2021.110095
19. Yang K, Zhang J, Bao C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer (2021) 21(1):1–9. doi: 10.1186/s12885-021-08669-9
20. Sun G, Miao G, Li Z, Zheng W, Zhou C, Sun G, et al. Inhibition of PARP potentiates immune checkpoint therapy through miR-513/PD-L1 pathway in hepatocellular carcinoma. J Oncol (2022) 2022. doi: 10.1155/2022/6988923
21. Zeng C, Ye S, Chen Y, Zhang Q, Luo Y, Gai L, et al. HOXA-AS3 promotes proliferation and migration of hepatocellular carcinoma cells via the miR-455-5p/PD-L1 axis. J Immunol Res (2021) 2021. doi: 10.1155/2021/9289719
22. Fan F, Chen K, Lu X, Li A, Liu C, Wu B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol Int (2021) 15(2):444–58. doi: 10.1007/s12072-020-10101-6
23. Tanaka E, Miyakawa Y, Kishikawa T, Seimiya T, Iwata T, Funato K, et al. Expression of circular RNA CDR1−AS in colon cancer cells increases cell surface PD−L1 protein levels. Oncol Rep (2019) 42(4):1459–66. doi: 10.3892/or.2019.7244
24. Yan Y, Zheng L, Du Q, Cui X, Dong K, Guo Y, et al. Interferon regulatory factor 1 (IRF-1) downregulates checkpoint kinase 1 (CHK1) through miR-195 to upregulate apoptosis and PD-L1 expression in hepatocellular carcinoma (HCC) cells. Br J Cancer (2021) 125(1):101–11. doi: 10.1038/s41416-021-01337-6
25. Liu Z, Ning F, Cai Y, Sheng H, Zheng R, Yin X, et al. The EGFR-P38 MAPK axis up-regulates PD-L1 through miR-675-5p and down-regulates HLA-ABC via hexokinase-2 in hepatocellular carcinoma cells. Cancer Commun (2021) 41(1):62–78. doi: 10.1002/cac2.12117
26. Cao X, Zhang G, Li T, Zhou C, Bai L, Zhao J, et al. LINC00657 knockdown suppresses hepatocellular carcinoma progression by sponging miR-424 to regulate PD-L1 expression. Genes Genomics (2020) 42(11):1361–8. doi: 10.1007/s13258-020-01001-y
27. Xu G, Zhang P, Liang H, Xu Y, Shen J, Wang W, et al. Circular RNA hsa_circ_0003288 induces EMT and invasion by regulating hsa_circ_0003288/miR-145/PD-L1 axis in hepatocellular carcinoma. Cancer Cell Int (2021) 21(1):1–13. doi: 10.1186/s12935-021-01902-2
28. Zhang L, Wang X, Li Y, Han J, Gao X, Li S, et al. C-myb facilitates immune escape of esophageal adenocarcinoma cells through the miR-145-5p/SPOP/PD-L1 axis. Clin Trans Med (2021) 11(9):e464. doi: 10.1002/ctm2.464
29. Liang Y, Liu Y, Zhang Q, Zhang H, Du J. Tumor-derived extracellular vesicles containing microRNA-1290 promote immune escape of cancer cells through the Grhl2/ZEB1/PD-L1 axis in gastric cancer. Trans Res (2021) 231:102–12. doi: 10.1016/j.trsl.2020.12.003
30. Younesi S, Mahdi Taheri Amin M, Saadati P, Yazdani B, Jamali S, Modarresi M-H, et al. Fine-tuning ofroutine combined first-trimester screening: The ratio of serum-free-beta-human chorionic gonadotropin (fβ-hCG) to pregnancy-associated plasma protein-a (PAPP-a) could improve performance of down syndrome screening program, a retrospective cohort study in Iran. Hum Antibod (2020) 203-210:1–8. doi: 10.3233/HAB-200408
31. Miliotis C, Slack FJ. miR-105-5p regulates PD-L1 expression and tumor immunogenicity in gastric cancer. Cancer Lett (2021) 518:115–26. doi: 10.1016/j.canlet.2021.05.037
32. Yang C, Zhang H, Zhang Q, Deng T, Liu R, Bai M, et al. Exosome-delivered miR-15a/16 target PD-L1 to inhibit immune escape in gastric cancer. (2020). doi: 10.21203/rs.3.rs-29661/v1
33. You W, Liu X, Yu Y, Chen C, Xiong Y, Liu Y, et al. miR-502-5p affects gastric cancer progression by targeting PD-L1. Cancer Cell Int (2020) 20(1):1–13. doi: 10.1186/s12935-020-01479-2
34. Guo T, Wang W, Ji Y, Zhang M, Xu G, Lin S. LncRNA PROX1-AS1 facilitates gastric cancer progression via miR-877-5p/PD-L1 axis. Cancer Manage Res (2021) 13:2669. doi: 10.2147/CMAR.S275352
35. Mu L, Wang Y, Su H, Lin Y, Sui W, Yu X, et al. HIF1A-AS2 promotes the proliferation and metastasis of gastric cancer cells through miR-429/PD-L1 axis. Digest Dis Sci (2021) 66(12):4314–25. doi: 10.1007/s10620-020-06819-w
36. Dang S, Malik A, Chen J, Qu J, Yin K, Cui L, et al. LncRNA SNHG15 contributes to immuno-escape of gastric cancer through targeting miR141/PD-L1. OncoTargets Ther (2020) 13:8547. doi: 10.2147/OTT.S251625
37. Wang C, Li X, Zhang L, Chen Y, Dong R, Zhang J, et al. miR-194-5p down-regulates tumor cell PD-L1 expression and promotes anti-tumor immunity in pancreatic cancer. Int Immunopharmacol (2021) 97:107822. doi: 10.1016/j.intimp.2021.107822
38. Jia L, Xi Q, Wang H, Zhang Z, Liu H, Cheng Y, et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem Biophys Res Commun (2017) 488(2):425–31. doi: 10.1016/j.bbrc.2017.05.074
39. Zhang H, Zhu C, He Z, Chen S, Li L, Sun C. LncRNA PSMB8-AS1 contributes to pancreatic cancer progression via modulating miR-382-3p/STAT1/PD-L1 axis. J Exp Clin Cancer Res (2020) 39(1):1–14. doi: 10.1186/s13046-020-01687-8
40. Zhou WY, Zhang MM, Liu C, Kang Y, Wang JO, Yang XH. Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death-ligand 1 by sponging microRNA-195-5p. J Cell Physiol (2019) 234(12):23176–89. doi: 10.1002/jcp.28884
41. Chen HL, Luo YP, Lin MW, Peng XX, Liu ML, Wang YC, et al. Serum exosomal miR-16-5p functions as a tumor inhibitor and a new biomarker for PD-L1 inhibitor-dependent immunotherapy in lung adenocarcinoma by regulating PD-L1 expression. Cancer Med (2022). doi: 10.1002/cam4.4638
42. Huang J, Weng Q, Shi Y, Mao W, Zhao Z, Wu R, et al. MicroRNA-155-5p suppresses PD-L1 expression in lung adenocarcinoma. FEBS Open Bio (2020) 10(6):1065–71. doi: 10.1002/2211-5463.12853
43. Shao L, He Q, Wang J, He F, Lin S, Wu L, et al. MicroRNA-326 attenuates immune escape and prevents metastasis in lung adenocarcinoma by targeting PD-L1 and B7-H3. Cell Death Discov (2021) 7(1):1–10. doi: 10.1038/s41420-021-00527-8
44. Song N, Li P, Song P, Li Y, Zhou S, Su Q, et al. MicroRNA-138-5p suppresses non-small cell lung cancer cells by targeting PD-L1/PD-1 to regulate tumor microenvironment. Front Cell Dev Biol (2020) 540. doi: 10.3389/fcell.2020.00540
45. Wan J, Ling X, Peng B, Ding G. miR-142-5p regulates CD4+ T cells in human non-small cell lung cancer through PD-L1 expression via the PTEN pathway. Oncol Rep (2018) 40(1):272–82. doi: 10.3892/or.2018.6439
46. Qiao X, Zhao F. Long non-coding RNA opa interacting protein 5-antisense RNA 1 binds to micorRNA-34a to upregulate oncogenic PD-L1 in non-small cell lung cancer. Bioengineered (2022) 13(4):9264–73. doi: 10.1080/21655979.2022.2036904
47. Wei S, Wang K, Huang X, Zhao Z, Zhao Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int J Immunopathol Pharmacol (2019) 33:2058738419859699. doi: 10.1177/2058738419859699
48. Li L, Zhang Q, Lian K. Circular RNA circ_0000284 plays an oncogenic role in the progression of non-small cell lung cancer through the miR-377-3p-mediated PD-L1 promotion. Cancer Cell Int (2020) 20(1):1–11. doi: 10.1186/s12935-020-01310-y
49. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res (2020) 39(1):1–19. doi: 10.1186/s13046-020-01648-1
50. Yang J, Jia Y, Wang B, Yang S, Du K, Luo Y, et al. Circular RNA CHST15 sponges miR-155-5p and miR-194-5p to promote the immune escape of lung cancer cells mediated by PD-L1. Front Oncol (2021) 11:364. doi: 10.3389/fonc.2021.595609
51. Feng S, Sun H, Zhu W. MiR-92 overexpression suppresses immune cell function in ovarian cancer via LATS2/YAP1/PD-L1 pathway. Clin Trans Oncol (2021) 23(3):450–8. doi: 10.1007/s12094-020-02439-y
52. Duan M, Fang M, Wang C, Wang H, Li M. LncRNA EMX2OS induces proliferation, invasion and sphere formation of ovarian cancer cells via regulating the miR-654-3p/AKT3/PD-L1 axis. Cancer Manage Res (2020) 12:2141. doi: 10.2147/CMAR.S229013
53. Liu Y, Chang Y, Cai Y-x. Inhibition of lnc-OC1 induced cell apoptosis and decreased cell viability by releasing miR-34a and inhibiting PD-L1 in endometrial carcinoma. Reprod Sci (2020) 27(10):1848–56. doi: 10.1007/s43032-020-00202-w
54. Xu D, Dong P, Xiong Y, Chen R, Konno Y, Ihira K, et al. PD-L1 is a tumor suppressor in aggressive endometrial cancer cells and its expression is regulated by miR-216a and lncRNA MEG3. Front Cell Dev Biol (2020) 1488. doi: 10.3389/fcell.2020.598205
55. Zhu Q, Li Y, Li L, Guo M, Zou C, Xu Y, et al. MicroRNA-889-3p restrains the proliferation and epithelial-mesenchymal transformation of lung cancer cells via down-regulation of homeodomain-interacting protein kinase 1. Bioengineered (2021) 12(2):10945–58. doi: 10.1080/21655979.2021.2000283
56. Dastmalchi N, Hosseinpourfeizi MA, Khojasteh SMB, Baradaran B, Safaralizadeh R. Tumor suppressive activity of miR-424-5p in breast cancer cells through targeting PD-L1 and modulating PTEN/PI3K/AKT/mTOR signaling pathway. Life Sci (2020) 259:118239. doi: 10.1016/j.lfs.2020.118239
57. Dou D, Ren X, Han M, Xu X, Ge X, Gu Y, et al. Cancer-associated fibroblasts-derived exosomes suppress immune cell function in breast cancer via the miR-92/PD-L1 pathway. Front Immunol (2020) 2026. doi: 10.3389/fimmu.2020.02026
58. Zhang M, Wang N, Song P, Fu Y, Ren Y, Li Z, et al. LncRNA GATA3-AS1 facilitates tumour progression and immune escape in triple-negative breast cancer through destabilization of GATA3 but stabilization of PD-L1. Cell Prolif (2020) 53(9):e12855. doi: 10.1111/cpr.12855
59. Qu F, Ye J, Pan X, Wang J, Gan S, Chu C, et al. MicroRNA-497-5p down-regulation increases PD-L1 expression in clear cell renal cell carcinoma. J Drug target (2019) 27(1):67–74. doi: 10.1080/1061186X.2018.1479755
60. Qin Z, Hu H, Sun W, Chen L, Jin S, Xu Q, et al. miR-224-5p contained in urinary extracellular vesicles regulates PD-L1 expression by inhibiting cyclin D1 in renal cell carcinoma cells. Cancers (2021) 13(4):618. doi: 10.3390/cancers13040618
61. Tian P, Wei JX, Li J, Ren JK, Yang JJ. LncRNA SNHG1 regulates immune escape of renal cell carcinoma by targeting miR-129-3p to activate STAT3 and PD-L1. Cell Biol Int (2021) 45(7):1546–60. doi: 10.1002/cbin.11595
62. Miao S, Mao X, Zhao S, Song K, Xiang C, Lv Y, et al. miR-217 inhibits laryngeal cancer metastasis by repressing AEG-1 and PD-L1 expression. Oncotarget (2017) 8(37):62143. doi: 10.18632/oncotarget.19121
63. Wang X, Zhang Y, Zheng J, Yao C, Lu X. LncRNA UCA1 attenuated the killing effect of cytotoxic CD8+ T cells on anaplastic thyroid carcinoma via miR-148a/PD-L1 pathway. Cancer Immunol Immunother (2021) 70(8):2235–45. doi: 10.1007/s00262-020-02753-y
64. Liu Z, Wen J, Wu C, Hu C, Wang J, Bao Q, et al. MicroRNA-200a induces immunosuppression by promoting PTEN-mediated PD-L1 upregulation in osteosarcoma. Aging (Albany NY). (2020) 12(2):1213. doi: 10.18632/aging.102679
65. Zhang J, Chou X, Zhuang M, Zhu C, Hu Y, Cheng D, et al. LINC00657 activates PD-L1 to promote osteosarcoma metastasis via miR-106a. J Cell Biochem (2020) 121(10):4188–95. doi: 10.1002/jcb.29574
66. He B, Yan F, Wu C. Overexpressed miR-195 attenuated immune escape of diffuse large b-cell lymphoma by targeting PD-L1. Biomed Pharmacother (2018) 98:95–101. doi: 10.1016/j.biopha.2017.11.146
67. Wang Q-M, Lian G-Y, Song Y, Huang Y-F, Gong Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large b cell lymphoma by sponging miR-195. Life Sci (2019) 231:116335. doi: 10.1016/j.lfs.2019.03.040
68. Chen Y, Song W, Gao Y, Dong X, Ji X. Increased PD-L1 expression in acquired cisplatin-resistant lung cancer cells via mir-181a. Tohoku J Exp Med (2022). doi: 10.1620/tjem.2022.J013
69. Tang D, Zhao D, Wu Y, Yao R, Zhou L, Lu L, et al. The miR-3127-5p/p-STAT 3 axis up-regulates PD-L1 inducing chemoresistance in non-small-cell lung cancer. J Cell Mol Med (2018) 22(8):3847–56. doi: 10.1111/jcmm.13657
70. Chen K-b, Yang W, Xuan Y, Lin A-j. miR-526b-3p inhibits lung cancer cisplatin-resistance and metastasis by inhibiting STAT3-promoted PD-L1. Cell Death Dis (2021) 12(8):1–10. doi: 10.1038/s41419-021-04033-8
71. Xia Y, Wang W-C, Shen W-H, Xu K, Hu Y-Y, Han G-H, et al. Thalidomide suppresses angiogenesis and immune evasion via lncRNA FGD5-AS1/miR-454–3p/ZEB1 axis-mediated VEGFA expression and PD-1/PD-L1 checkpoint in NSCLC. Chemico-Biol Interact (2021) 349:109652. doi: 10.1016/j.cbi.2021.109652
72. Sp N, Kang DY, Lee J-M, Jang K-J. Mechanistic insights of anti-immune evasion by nobiletin through regulating miR-197/STAT3/PD-L1 signaling in non-small cell lung cancer (NSCLC) cells. Int J Mol Sci (2021) 22(18):9843. doi: 10.3390/ijms22189843
73. Zhu F, Niu R, Shao X, Shao X. FGD5−AS1 promotes cisplatin resistance of human lung adenocarcinoma cell via the miR−142−5p/PD−L1 axis. Int J Mol Med (2021) 47(2):523–32. doi: 10.3390/ijms22189843
74. Zuo Y, Zheng W, Tang Q, Liu J, Wang S, Xin C. miR−576−3p overexpression enhances cisplatin sensitivity of ovarian cancer cells by dysregulating PD−L1 and cyclin D1. Mol Med Rep (2021) 23(1):1–. doi: 10.3892/mmr.2020.11719
75. Sheng Q, Zhang Y, Wang Z, Ding J, Song Y, Zhao W. Cisplatin-mediated down-regulation of miR-145 contributes to up-regulation of PD-L1 via the c-myc transcription factor in cisplatin-resistant ovarian carcinoma cells. Clin Exp Immunol (2020) 200(1):45–52. doi: 10.1111/cei.13406
76. Anastasiadou E, Messina E, Sanavia T, Mundo L, Farinella F, Lazzi S, et al. miR-424-5p -3p contrasts PD-L1 induction by combinatorial therapies and slows proliferation of epithelial ovarian cancer through downregulation of β-catenin and c-myc. Cells (2021) 10(3):519. doi: 10.1111/cei.13406
77. Jin Y, Zhan X, Zhang B, Chen Y, Liu C, Yu L. Polydatin exerts an antitumor effect through regulating the miR-382/PD-L1 axis in colorectal cancer. Cancer Biother Radiopharm (2020) 35(2):83–91. doi: 10.1089/cbr.2019.2999
78. Gao C, Xu Y-J, Qi L, Y-f B, Zhang L, Zheng L. CircRNA VIM silence synergizes with sevoflurane to inhibit immune escape and multiple oncogenic activities of esophageal cancer by simultaneously regulating miR-124/PD-L1 axis. Cell Biol Toxicol (2021) 83-91:1–21. doi: 10.1089/cbr.2019.2999
79. Wang J, Yu Z, Wang J, Shen Y, Qiu J, Zhuang Z. LncRNA NUTM2A-AS1 positively modulates TET1 and HIF-1A to enhance gastric cancer tumorigenesis and drug resistance by sponging miR-376a. Cancer Med (2020) 9(24):9499–510. doi: 10.1002/cam4.3544
80. Lv J, Guo T, Qu X, Che X, Li C, Wang S, et al. PD-L1 under regulation of miR-429 influences the sensitivity of gastric cancer cells to TRAIL by binding of EGFR. Front Oncol (2020) 1067. doi: 10.3389/fonc.2020.01067
81. Li D, F-f S, Wang D, Wang T, Peng J-j, Feng J-Q, et al. Programmed death ligand-1 (PD-L1) regulated by NRF-2/MicroRNA-1 regulatory axis enhances drug resistance and promotes tumorigenic properties in sorafenib-resistant hepatoma cells. Oncol Res (2020) 28(5):467. doi: 10.3727/096504020X15925659763817
82. Zhang J, Zhao X, Ma X, Yuan Z, Hu M. KCNQ1OT1 contributes to sorafenib resistance and programmed death−ligand−1−mediated immune escape via sponging miR−506 in hepatocellular carcinoma cells. Int J Mol Med (2020) 46(5):1794–804. doi: 10.3727/096504020X15925659763817
83. Hamed MM, Handoussa H, Hussein NH, Eissa RA, Abdel-Aal LK, El Tayebi HM. Oleuropin controls miR-194/XIST/PD-L1 loop in triple negative breast cancer: New role of nutri-epigenetics in immune-oncology. Life Sci (2021) 277:119353. doi: 10.1016/j.lfs.2021.119353
84. Li D, Wang X, Yang M, Kan Q, Duan Z. miR3609 sensitizes breast cancer cells to adriamycin by blocking the programmed death-ligand 1 immune checkpoint. Exp Cell Res (2019) 380(1):20–8. doi: 10.1016/j.yexcr.2019.03.025
85. Wang Y, Wang L. miR-34a attenuates glioma cells progression and chemoresistance via targeting PD-L1. Biotechnol Lett (2017) 39(10):1485–92. doi: 10.1007/s10529-017-2397-z
86. Jiang W, Pan S, Chen X, Wang Z-w, Zhu X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol Cancer (2021) 20(1):1–17. doi: 10.1186/s12943-021-01406-7
Keywords: PD-L1, lncRNA, miRNA, expression, cancer
Citation: Ghafouri-Fard S, Shoorei H, Hussen BM, Poornajaf Y, Taheri M and Sharifi G (2022) Interplay between programmed death-ligand 1 and non-coding RNAs. Front. Immunol. 13:982902. doi: 10.3389/fimmu.2022.982902
Received: 30 June 2022; Accepted: 21 October 2022;
Published: 01 November 2022.
Edited by:
Jun Wang, New York University, United StatesReviewed by:
Fangfang Jin, Nanjing University of Chinese Medicine, ChinaYue Sun, Virginia Commonwealth University, United States
Copyright © 2022 Ghafouri-Fard, Shoorei, Hussen, Poornajaf, Taheri and Sharifi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Guive Sharifi, R2libm93QHlhaG9vLmNvbQ==
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Hamed Shoorei
Hamed Shoorei Bashdar Mahmud Hussen
Bashdar Mahmud Hussen Yadollah Poornajaf
Yadollah Poornajaf Mohammad Taheri
Mohammad Taheri Guive Sharifi
Guive Sharifi