
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 05 August 2022
Sec. Comparative Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.981995
This article is part of the Research Topic Pathogenesis and Nutritional Intervention of Intestinal Inflammation in Fish View all 8 articles
 Fufa Qu1
Fufa Qu1 Xuan Zeng1
Xuan Zeng1 Zhenzhen Liu1
Zhenzhen Liu1 Meixing Guo1
Meixing Guo1 Xia Zhang1
Xia Zhang1 Shenping Cao1
Shenping Cao1 Yonghua Zhou1
Yonghua Zhou1 Zhimin He1
Zhimin He1 Jianzhou Tang1
Jianzhou Tang1 Zhuangwen Mao1
Zhuangwen Mao1 Yalin Yang2
Yalin Yang2 Zhigang Zhou2
Zhigang Zhou2 Zhen Liu1*
Zhen Liu1*Mitogen-activated protein kinase kinase kinase 3 (MEKK3) is an evolutionarily conserved Ser/Thr protein kinase of the MEKK family that is essential for the host immune response to pathogen challenges in mammals. However, the immune function of MEKK3s in lower vertebrate species, especially in bony fish, remains largely unknown. In this study, a fish MEKK3 (designated CiMEKK3) gene was cloned and identified from grass carp (Ctenopharyngodon idella). The present CiMEKK3 cDNA encoded a 620 amino acid polypeptide containing a conserved S-TKc domain and a typical PB1 domain. Several potential immune-related transcription factor-binding sites, including activating protein 1 (AP-1), nuclear factor kappa B (NF-κB) and signal transducer and activator of downstream transcription 3 (STAT3), were observed in the 5’ upstream DNA sequence of CiMEKK3. A phylogenetic tree showed that CiMEKK3 exhibits a close evolutionary relationship with MEKK3s from Cyprinus carpio and Carassius auratus. Quantitative real-time PCR analysis revealed that CiMEKK3 transcripts were widely distributed in all selected tissues of healthy grass carp, with a relatively high levels observed in the gill, head kidney and intestine. Upon in vitro challenge with bacterial pathogens (Aeromonas hydrophila and Aeromonas veronii) and pathogen-associated molecular patterns (PAMPs) (lipopolysaccharide (LPS), peptidoglycan (PGN), L-Ala-γ-D-Glu-mDAP (Tri-DAP) and muramyl dipeptide (MDP)), the expression levels of CiMEKK3 in the intestinal cells of grass carp were shown to be significantly upregulated in a time-dependent manner. In vivo injection experiments revealed that CiMEKK3 transcripts were significantly induced by MDP challenge in the intestine; however, these effects could be inhibited by the nutritional dipeptides carnosine and Ala-Gln. Moreover, subcellular localization analysis and luciferase reporter assays indicated that CiMEKK3 could act as a cytoplasmic signal-transducing activator involved in the regulation of NF-κB and MAPK/AP-1 signaling cascades in HEK293T cells. Taken together, these findings strongly suggest that CiMEKK3 plays vital roles in the intestinal immune response to bacterial challenges, which will aid in understanding the pathogenesis of inflammatory bowel disease in bony fish.
Mitogen-activated protein kinases (MAPKs) are a class of conserved serine and threonine protein kinases that are widely present in a variety of organisms and can participate in mediating multiple biological processes in response to extracellular stimuli and cellular stress (1, 2). MAPKs are activated through three-tiered kinase cascades: MAP kinase kinase kinase (MAP3K or MEKK), MAP kinase kinase (MAP2K or MEK) and MAPK (3, 4). In the MAPK signal cascade system, MEKKs are first activated by endogenous and environmental stimuli, which in turn activate downstream dual-specific MKKs that can further phosphorylate Thr and Tyr within the motif Thr-Xaa-Tyr of various MAPKs, including extracellular signal-regulated kinases (ERKs), c-jun N-terminal or stress-activated protein kinases (JNKs/SAPKs) and p38 MAPK, and finally induce the activation of various transcription factors that regulate the expression of effector genes participating in inflammation, apoptosis and development (4–7).
As the initial kinases of three-tiered kinase cascades, MEKKs are essential for signal transduction in the MAPK pathway and mainly consist of MEKK1, MEKK2, MEKK3, MEKK4, transforming growth factor-β-activating kinase 1 (TAK1), apoptosis signal-regulating kinase 1 (ASK1), dual leucine zipper bearing kinase (DLK), and tumor progression locus-2 (Tpl2) (8, 9). Among MEKK family members, MEKK3 has been shown to be highly conserved among eukaryotes and involved in the regulation of cell proliferation, the inflammatory response and tumor development (10–13). In mammals, MEKK3 has been reported to contain an activation loop (A-loop) domain, a Phox and Bem1p (PB1) domain and a serine/threonine kinase catalytic (S-TKc) domain (14). Previously, the A-loop domain was shown to contain some specific serine/threonine phosphorylation sites that are responsible for MEKK3 activation and signal transduction (15–18). The PB1 domain is a secondary structure rich in basic amino acids that could be involved in the transmission of specific intracellular signals in various signaling pathways by the PB1-PB1 interaction with other signal proteins (19–21). The S-TKc domain at the C-terminus of the MEKK3 protein has been shown to be conserved in MEKK family members and was critical for its activation and phosphorylation (22).
In mammals, MEKK3s have been shown to act as crucial regulators of innate immunity against pathogen infections via the involvement of interleukin-1 receptor (IL-1R) and toll-like receptor (TLR) -mediated NF-κB, JNK and p38 cascades, which are essential for inducing the expression of proinflammatory cytokines (12, 23–25). For example, Huang et al. reported that MEKK3 played a decisive role in IL-1-induced and LPS-induced interleukin (IL-6) production by regulating the IKK-NF-κB and JNK-p38 MAPK pathways in mouse embryonic fibroblast (MEF) cell lines (23). In MEKK3-deficient fibroblast cells, MEKK3 is essential for IKK activation and functions downstream of TNF receptor-associated factor 2 (TRAF2) and receptor-interacting protein (RIP) in the TNF-induced NF-κB pathway (24). A study of the macrophage line Raw264.7 showed that the production of LPS-induced IL-6 and granulocyte-macrophage colony-stimulating factor (GM-CSF) was significantly decreased in MEKK3 knockdown cells; however, this decrease was restored by reintroducing human MEKK3 cDNA (12). Recently, Li and colleagues demonstrated that LPS-stimulated proinflammatory cytokine (IL-1β, TNF-α and IL-6) production is significantly regulated by the TAK1-MEKK3 axis in myeloid cells (25). Using knockdown experiments in BV2 cells, it was found that MEKK3 plays a critical role in the development of neuroinflammation in Parkinson’s disease by regulating the NF-κB signaling pathway (26). A recent study in mice found that overexpression of MEKK3 significantly activated IRF7 to trigger strong induction of type I IFNs, while knockdown of MEKK3 in vivo substantially impaired type I IFN induction and increased susceptibility to HSV-1 infection (27).
Due to its important immunological function in mammals, MEKK3 in bony fish has also attracted much attention in recent years. For example, many MEKK3 genes have been cloned and identified in a variety of fish species such as Danio rerio, Cyprinus carpio, Carassius auratus, Pimephales promelas and Triplophysa tibetana. A recent study of a hybrid snakehead (Channa maculate ♀ × Channa argus ♂) showed that fish MEKK3 is involved in the innate immune response to Nocardia seriolae and Aeromonas schubertii challenges (28). However, compared with mammals, the immune function of MEKK3 in fish is still largely unclear. Grass carp (Ctenopharyngodon idella) is one of the most highly produced and economically important freshwater fish species in China. Over the past decades, bacterial enteritis has seriously harmed the healthy breeding of grass carp and caused serious economic losses to aquaculture (29–31). Investigation into the intestinal immune function of C. idella MEKK3, an essential signal transducer of the innate immune response, might facilitate the development of disease control and prevention measures. To this end, a fish MEKK3 (CiMEKK3) gene was identified from C. idella, and its intestinal expression in response to bacterial pathogens (Aeromonas hydrophila and Aeromonas veronii) and PAMPs (lipopolysaccharide (LPS), peptidoglycan (PGN), L-Ala-γ-D-Glu-meso-diaminopimelic acid (Tri-DAP) and muramyl dipeptide (MDP)) challenges was investigated by using quantitative real-time PCR (qRT-PCR). In addition, CiMEKK3 was overexpressed in human embryonic kidney 293T (HEK293T) cells to determine its intracellular localization characteristics and signal transduction function. The data from this study may help to illuminate the function of MEKK3s in the intestinal immunity of bony fish.
Healthy C. idella weighing approximately 30 g were obtained from Hunan Institute of Aquatic Science in Changsha, China, and acclimatized in 30 L tanks with circulating freshwater at 24 ± 1°C for two weeks prior to experimentation. Eight tissues including head kidney, spleen, intestine, liver, gill, blood, muscle and heart were collected from healthy individuals using sterilized scissors and tweezers for further tissue distribution analysis. Tissue samples were ground into powder in liquid nitrogen and stored at -80°C until RNA extraction.
For the in vitro immune challenge experiments, the cultured primary intestinal cells were challenged with bacterial pathogens (A. hydrophila and A. veronii) and PAMPs (LPS, PGN, Tri-DAP and MDP). The experimental protocol was performed according to our previous study (32). Before the challenge experiment, the C. idella intestinal cells were grown in 6-well culture plates with 2 mL of Dulbecco’s modified Eagle’s medium (DMEM, Gibco-BRL, USA) containing 10% fetal bovine serum (FBS, Gibco BRL, USA) per well at 28°C in a humidified incubator provided with 5% CO2. The two bacterial pathogen strains (A. hydrophila and A. veronii) were kindly provided by the Feed Research Institute, Chinese Academy of Agricultural Sciences (33), and cultured in LB medium at 37°C overnight for the challenge experiment. The C. idella intestinal cells were challenged with A. hydrophila (1×107 cfu/mL), A. veronii (1×107 cfu/mL), Tri-DAP (50 μg/mL; In vivoGen), MDP (50 μg/mL; In vivoGen), LPS (10 μg/mL; Sigma-Aldrich) or PGN (10 μg/mL; Sigma-Aldrich) and collected at 0, 3, 6, 12 and 24 h post-challenge. The cells were treated with phosphate-buffered saline (PBS, pH 7.4) used as a control. All the collected cell samples were placed in -80°C immediately and used for gene expression assays.
For the in vivo immune challenge experiments, healthy grass carp were injected with the bacterial dipeptide MDP and nutritional dipeptides (carnosine and Ala-Gln) using a 1 mL syringe. In the first immune challenge experiment, C. idella were randomly divided into two groups, the MDP challenge group and the control group, and each group was placed in separate tanks. The fish in the immune challenge group were injected with 100 μl MDP (10 μg/mL, In vivoGen, USA). The control individuals were injected with an equal volume of PBS. After treatment, the grass carp were returned to the tanks and intestines of three individuals in each group were randomly sampled at 0, 3, 6, 12, 24, 48 and 72 h post-injection. In the second immune challenge experiment, grass carp were randomly divided into four groups and were injected intraperitoneally with 100 μl of PBS, MDP (10 μg/mL), MDP (10 μg/mL) + carnosine (5 mmol/L; Sigma-Aldrich) or MDP (10 μg/mL) + Ala-Gln (5 mmol/L; Sigma-Aldrich). Intestines were collected from three individuals in each tank at 12 and 24 h post-injection for gene expression level analysis.
All experiments were performed according to the recommendations of the Guidance of the Care and Use of Laboratory Animals in China. The research presented in this manuscript was approved by the Animal Ethics Committee of Changsha University.
Total RNA was extracted from the harvested intestinal cells and adult tissues of grass carp using RNAiso Plus (Takara, Japan) reagent following the manufacture’s protocol. RNA concentration was measured using the ratio of UV absorbance at 260/280 nm in a NanoDrop 2000 spectrophotometer (Thermo Fisher, USA) and the quality was assessed using 1.5% agarose electrophoresis. The RNA samples were treated with gDNA Eraser (TaKaRa, Japan) to eliminate genomic DNA contamination. Total RNA from each sample was reverse transcribed using the PrimeScript™ 1st Strand cDNA Synthesis Kit (Takara, Japan) and PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Japan) to synthesize the cDNA template for gene cloning and expression analysis, respectively. Finally, the cDNA mix was diluted 10-fold and stored at -80°C for subsequent processing.
Based on the reported C. carpio MEKK3 sequence from the GenBank database, gene-specific primers were designed to amplify the cDNA sequence of CiMEKK3 by reverse transcription PCR (RT-PCR). PCR amplification was performed in a total reaction volume of 50 μl containing 1 μl of cDNA template, 37.75 μl of dH2O, 4 μl of dNTP mixture (2.5 mM each), 5 μl 10×Ex Taq Buffer (Mg2+ plus), 1 μl of each primer (10 μM) and 0.25 μl of Ex Taq DNA Polymerase (TaKaRa, Japan). The PCR conditions were as follows: 94°C for 3 min, 35 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 2 min, and 72°C for 10 min. The PCR products were separated using 1.2% agarose gel/TAE electrophoresis and then purified with a TaKaRa Agarose Gel DNA Purification Kit Ver.2.0 (TaKaRa, Japan). After purification, all of the specific PCR products were cloned into the pMD19-T vector (TaKaRa, Japan). The ligation product was transformed into Escherichia coli DH5α, and three positive colonies were screened and sequenced on a 3730 Applied Biosystems (ABI) DNA sequencer.
The nucleotide and deduced amino acid sequences of the cloned CiMEKK3 gene were analyzed using the BLAST tool available from the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The molecular weight and theoretical isoelectric point were calculated using the pI/Mw tool (https://web.expasy.org/protparam/). The functional domains were deduced with the Simple Modular Architecture Research Tool (SMART) website (http://smart.embl-heidelberg.de/). Potential transcription factor binding sites (TFBSs) in the promoter region of CiMEKK3 were predicted using JASPAR (http://jaspardev.genereg.net/) and AliBaba2 (http://gene-regulation.com/pub/programs/alibaba2/index.html). The exon-intron arrangement of CiMEKK3 based on the DNA sequence from grass carp genome (http://www.ncgr.ac.cn/grasscarp/ ) was determined using the Spidey tool (http://www.ncbi.nlm.nih.gov/spidey/). The identity and similarity of amino acid sequences were calculated with MatGAT2.02 software. The alignment of multiple sequences of MEKK3s from different species was conducted using the MegAlign program with the Clustal W method, and GeneDoc software was employed to visualize the results. A neighbor-joining (NJ) phylogenetic tree of MEKK3 was constructed based on the amino acid sequences of MEKK3s using MEGA 5.0 software, with the bootstrap value set at 1000 replicates. The GenBank accession numbers for the various MEKK3s are as follows: [Homo sapiens] AAB41729.1, [Papio anubis] XP_017806347.1, [Aotus nancymaae] XP_021529429.1, [Cebus imitator] XP_017404666.1, [Mus musculus] NP_036077.1, [Balaenoptera musculus] XP_036692416.1, [Danio rerio] XP_688694.2, [Triplophysa tibetana] KAA0705702.1, [Pimephales promelas] XP_039542275.1, [Cyprinus carpio] XP_018936269.1, [Carassius auratus] XP_026062650.1, [Ctenopharyngodon idella] ON082069.
Quantitative real-time PCR (qRT-PCR) was used to detect the relative mRNA expression levels of CiMEKK3 with β-actin as an internal reference gene. Gene specific primers were designed based on the cDNA sequences of grass carp MEKK3 and β-actin using Primer Premier 5.0 software and are listed in Table 1. qRT-PCR was performed on a Quant-Studio™ 3 Real-Time PCR System (Thermo Fisher, USA) in a total volume of 16 μl containing 8 μl of 2 × SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara, Japan),1 μl of cDNA template, 0.32 μl of ROX, 0.64 μl of each primer, and 5.4 μl of nuclease-free water. The qRT-PCR procedure was as follows: 95°C for 5 min, followed by 45 amplification cycles of 10 s at 95°C, 30 s at 58°C, and 72°C for 10 s, and three biological replicates for each group were conducted. The specificity of each qRT-PCR product was confirmed by melting curve and agarose gel analysis. The relative expression levels of CiMEKK3 were normalized to β-actin expression, and the relative expression values were calculated using the 2-ΔΔCT method (34).
The eukaryotic expression vectors pCMV-N-Flag-CiMEKK3 (CiMEKK3-Flag) and pEGFP-N1-CiMEKK3 (CiMEKK3-GFP) were constructed for mammalian cell transfections using the ClonExpress® II One Step Cloning kit (Vazyme, China) according to the manufacturer’s protocol. The expression plasmids CiMKK4-Flag, CiMKK6-Flag and CiMKK7-Flag were constructed for our previous studies (35, 36). The primer pairs (Table 1) were designed for amplification of the complete open reading frame (ORF) encoding the polypeptide of CiMEKK3. The ORF of CiMEKK3 was cloned into the BamH I/Hind III site of pCMV-N-Flag and Xho I/BamH I site of pEGFP-N1 to generate the plasmids CiMEKK3-Flag and CiMEKK3-GFP, respectively, and then transformed into E. coli, DH5α (TaKaRa, Japan). The colonies were screened on LB plates containing kanamycin at 37°C and sequenced for further verification. All transfection plasmids were prepared from overnight bacterial cultures using the HiPure Plasmid EF Mini Kit (Magen, China) according to the manufacturer’s instructions.
Human embryonic kidney 293T (HEK293T) cells were cultured with DMEM (Gibco-BRL, USA) containing 10% FBS (Gibco BRL, USA) and antibiotics (100 mg/L streptomycin and 105 U/L penicillin, Gibco) at 37°C in a 5% CO2 atmosphere. For plasmid-liposome transfection, cells were seeded overnight and grown to 80–90% confluence at the time of transfection. Then, plasmids were transfected into the cells using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer′s instructions.
To investigate the subcellular localization of CiMEKK3, 1 μg/well of the recombinant plasmid CiMEKK3-GFP or empty plasmid pEGFP-N1 (control) was transfected into HEK293T cells. Prior to transfection, the cells were seeded on sterile coverslips at 1 × 105 cells/well in 6-well plates for overnight growth. Then the cells were transfected using Lipofectamine 2000 with a 2:1 ratio of transfection reagent to endo-free plasmids in serum-free culture medium. At 48 h post-transfection, the transfected cells were fixed with 4% paraformaldehyde, washed with PBS and stained with DAPI to distinguish localization between the cytoplasm and nuclei. The subcellular localization results were observed under a fluorescence microscope.
For the dual-luciferase reporter assays, cells were transiently cotransfected with luciferase reporter vectors NF-κB-Luc (Promega, USA)/AP-1-Luc (Promega, USA), pRL-TK (Promega, USA) (20 ng/well), and expression plasmids CiMEKK3-Flag/CiMKK4-Flag/CiMKK6-Flag/CiMKK7-Flag using Lipofectamine 2000 (Invitrogen, USA). The pRL-TK plasmid was used as the internal control and the plasmid pCMV-N-Flag was used as the negative control. Each experiment was performed in triplicate under similar conditions to obtain biological replicates. After the cells were transfected for 48 h, the firefly and Renilla luciferase activities were measured using a dual-Luciferase reporter assay system (Promega, USA) following the manufacturer’s instructions. The relative luciferase activity is presented as the ratio of firefly luciferase to renilla luciferase.
All the data derived from luciferase assays and qRT-PCR were subjected to one-way analysis of variance (ANOVA) followed by LSD or Duncan’s post-hoc test to determine significant differences among the treatments. The results are shown as the means ± standard error of measurement (SEM). Differences were considered statistically significant at P < 0.05 and extremely significant at P < 0.01.
The cDNA sequence of CiMEKK3 was obtained with RT-PCR and submitted to the GenBank database (No. ON082069). The CiMEKK3 cDNA was 2055 bp in length, which included a 5′-untranslated region (UTR) of 36 bp, a 3′-UTR of 156 bp, and an ORF of 1863 bp encoding a 620 amino acid residue (Figure 1A). The predicted molecular weight of CiMEKK3 was 70.17 kDa, and the theoretical pI was 9.15. Similar to other orthologs, several phosphorylation sites such as Thr294, Thr516, Ser520 and Ser526 were observed in the amino acid residue of CiMEKK3 (Figure 1A). Structural analysis based on the SMART program revealed that MEKK3 contained two conserved domains, including a PB1 domain (positions 47–126 aa) and a typical S_TKc domain (positions 356–616 aa), with the typical features of MEKK3 family proteins (Figure 1B). The genomic organization of CiMEKK3 was analyzed by comparing the genomic DNA and cDNA sequences. As shown in Figure 2A, the DNA sequence of CiMEKK3 possesses a multiexonic gene structure containing sixteen exons separated by fifteen introns, and its mature mRNA sequence was generated by appropriate splicing. Using the JASPAR and AliBaba2 programs, several transcription factor-binding sites, including three nuclear factor kappa B (NF-κB) sites, three activating protein 1 (AP-1) sites, one octamer-binding transcription factor 1 (Oct-1) site, one cAMP response element-binding protein (CREB) site, two signal transducer and activator of downstream transcription 3 (STAT3) sites, one specificity protein 1 (SP1) site and a GATA-binding factor 1 (GATA-1) site were found in the 5’-upstream DNA sequence of CiMEKK3 (Figure 2B).

Figure 1 The cDNA sequence and deduced amino acid sequences of CiMEKK3. (A) Nucleotides and amino acids are numbered on the left of the sequences. The start codon (ATG) and stop codon (TGA) are shown in red font. The ORF sequence of CiMEKK3 is indicated in uppercase letters, while the 5′- and 3′-UTR sequences are shown in lowercase. The PB1 domain and S_TKc domain are marked by gray shading and blue underline, respectively. The predicted phosphorylation sites are shown by yellow shading. (B) Functional domains of CiMEKK3 were predicted using the SMART tool.
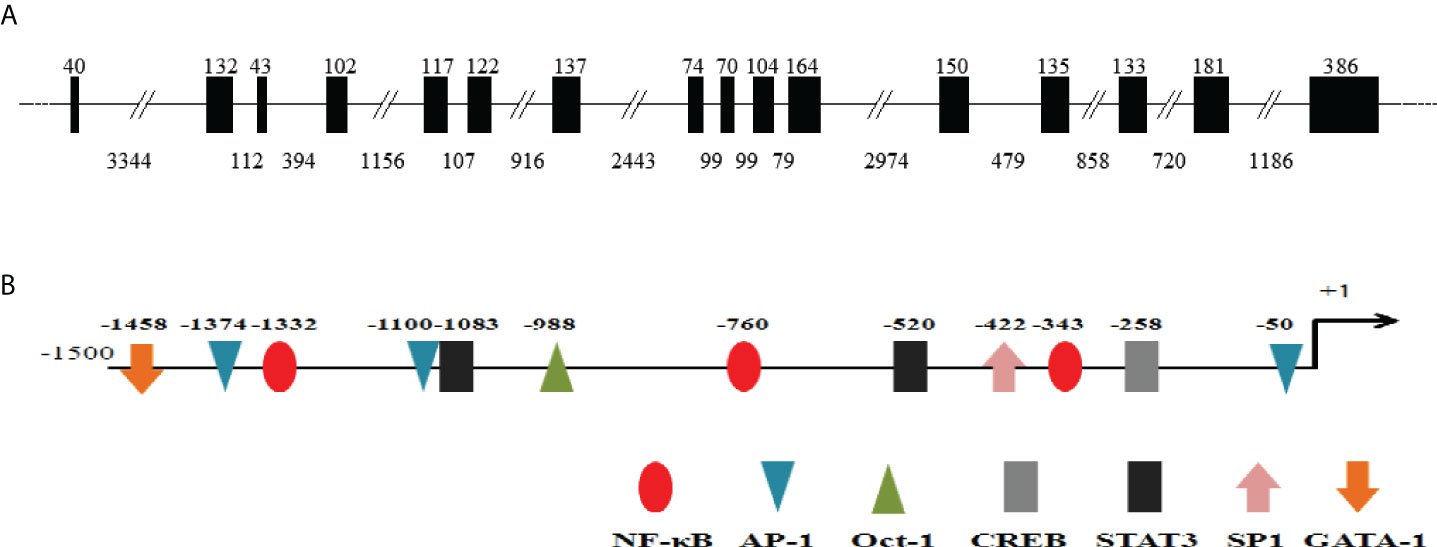
Figure 2 Genomic organization and 5′ flanking regions of CiMEKK3. (A) The intron-exon organization of CiMEKK3. Exons and introns are indicated by black boxes and black lines with the corresponding sizes (bp), respectively. (B) Transcription factor binding sites in the 5′ flanking regions (~1.5 kb) of CiMEKK3. The predicted binding sites are shown by different colors of ellipses, triangles, arrows or rectangles.
Multiple sequence alignment illustrated that the amino acid sequence and functional domains were conserved in vertebrate MEKK3 counterparts and that all contained a typical S-TKc domain and a PB1 domain (Figure 3A). A MatGAT2.01 analysis was conducted to generate a measure of similarity and identity for the CiMEKK3 protein with other homologs. The deduced amino acid sequence of CiMEKK3 shared 76.5–98.1% identity (I) and 86.9–99.0% similarity (S) with MEKK3 sequences from other vertebrate species. Among all the selected MEKK3 sequences, CiMEKK3 was closest to that of Cyprinus carpio (98.1% I, 99.0% S), followed by that of Pimephales promelas (97.7% I, 98.7% S) (Figure 3B). To investigate the evolutionary relationships of MEKK3s, a phylogenetic tree was constructed using the MEKK3 sequences of twelve representative vertebrate species. Overall, the relationships displayed in the cladogram generally agree with those of traditional taxonomy. The MEKK3s from mammals and fish were clustered separately into two branches, and CiMEKK3 was embedded within the fish cluster. In addition, CiMEKK3 exhibited a close evolutionary relationship with MEKK3s from Cyprinus carpio and Carassius auratus (Figure 4).

Figure 3 Multiple alignment of CiMEKK3 with other reported MEKK3 from GenBank database. (A) Identical amino acids are shaded in black and similar amino acids are shaded in gray. The conserved PB1 domain and S_TKc domain are indicated by the red box and blue box, respectively. (B) The similarities (red) and identities (green) of amino acid sequences were analyzed using MatGAT2.02 software.
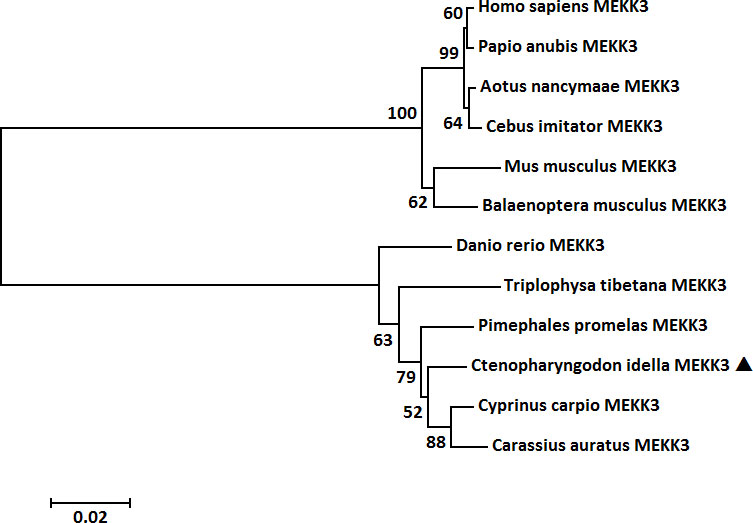
Figure 4 Phylogenetic tree of MEKK3 proteins from representative vertebrate species. The tree was constructed with the maximum likelihood method in MEGA 5.0 software with 1000 bootstrap replications. The bar (0.02) indicates the genetic distance. CiMEKK3 is shown by a black triangle.
The qRT-PCR analysis was employed to determine the tissue expression profiles of CiMEKK3 in healthy grass carp. CiMEKK3 was ubiquitously expressed in all eight examined organs (intestine, liver, blood, muscle, heart, gill, head kidney and spleen), with the highest expression levels in the gill, followed by the head kidney and intestine, and relatively low expression levels in the liver (Figure 5). To obtain the subcellular localization characteristics of CiMEKK3, HEK293T cells were transfected with plasmid pEGFP-N1 or CiMEKK3-GFP using Lipofectamine 2000. Based on the results obtained from fluorescence microscope, CiMEKK3 was distributed mainly in the cytoplasm while the control protein was dispersed throughout the cytoplasm and the nuclear areas, suggesting that the CiMEKK3 protein may be a cytoplasm-localized protein in HEK293T cells (Figure 6).
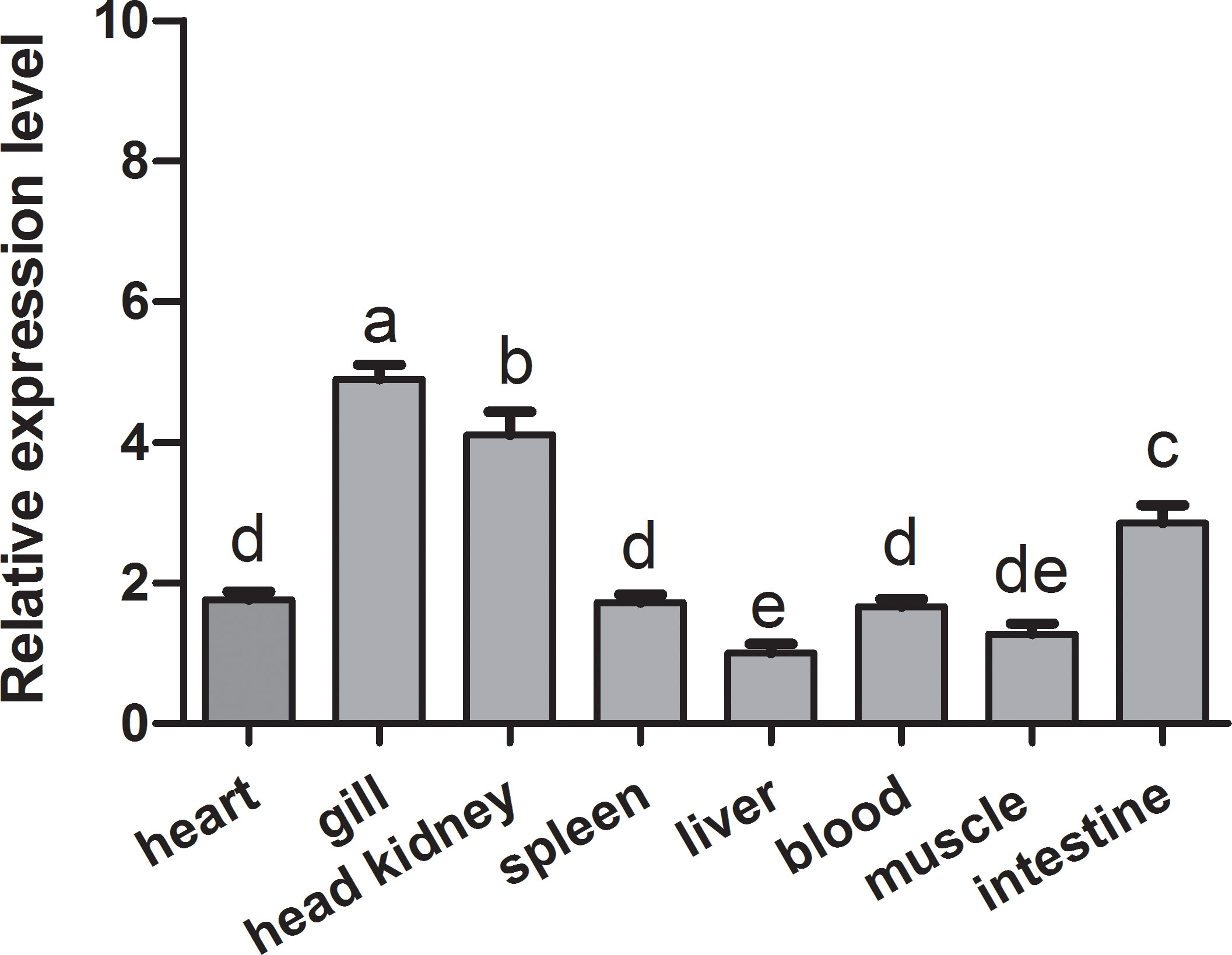
Figure 5 Relative expression levels of CiMEKK3 in various tissues of healthy grass carp. Each bar represents the mean of the normalized expression levels of the replicates (N = 3). Bars marked with different letters indicate significant differences among different tissues (P < 0.05).
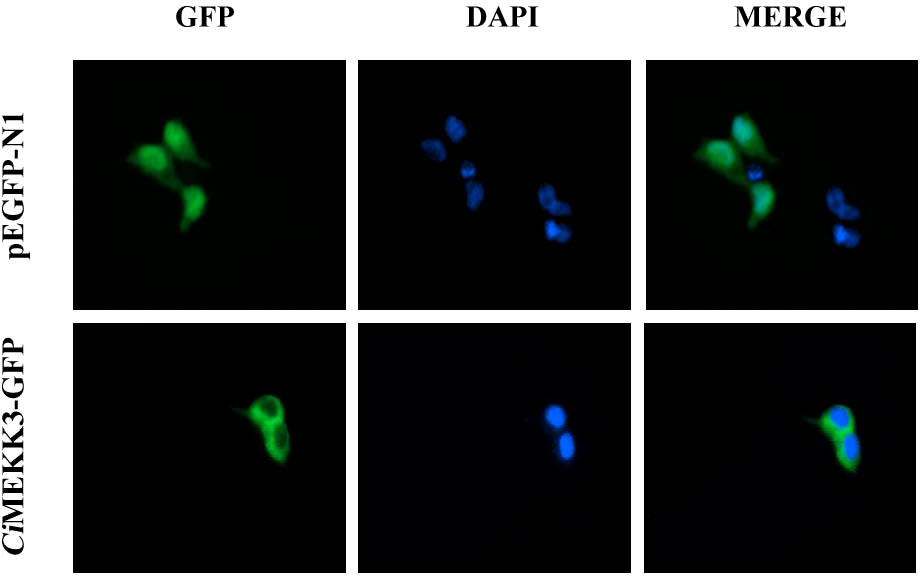
Figure 6 Subcellular localization of CiMEKK3 in HEK293T cells. HEK293T cells were transfected with pEGFP-N1 (upper row) or CiMEKK3-GFP (lower row). At 48 h post-transfection, the cells were fixed with 4% paraformaldehyde, stained with 4′,6-diamidino-2-phenylindole (DAPI), and then observed with fluorescence microscopy.
The transcriptional responses of CiMEKK3 were monitored in intestinal cells after stimulation with A. hydrophila and A. veronii. The qRT-PCR results showed that CiMEKK3 exhibited a strong and elevated response to A. hydrophila and A. veronii infection, and its expression levels in intestinal cells were significantly regulated by these two bacterial pathogen challenges (Figure 7). When challenged with A. hydrophila, the mRNA levels of intestinal CiMEKK3 were upregulated at 3 h post-stimulation (P < 0.01), reaching a peak value at 6 h post-stimulation (P < 0.01), and then returning to control levels at 24 h post-stimulation (Figure 7A). Upon infection with A. veronii, CiMEKK3 transcripts in intestinal cells did not significantly increase until 6 h post-stimulation (P < 0.01), reached the highest level at 12 h post-stimulation (P < 0.01), and then sharply decreased to the original level at 24 h post-stimulation (Figure 7B).
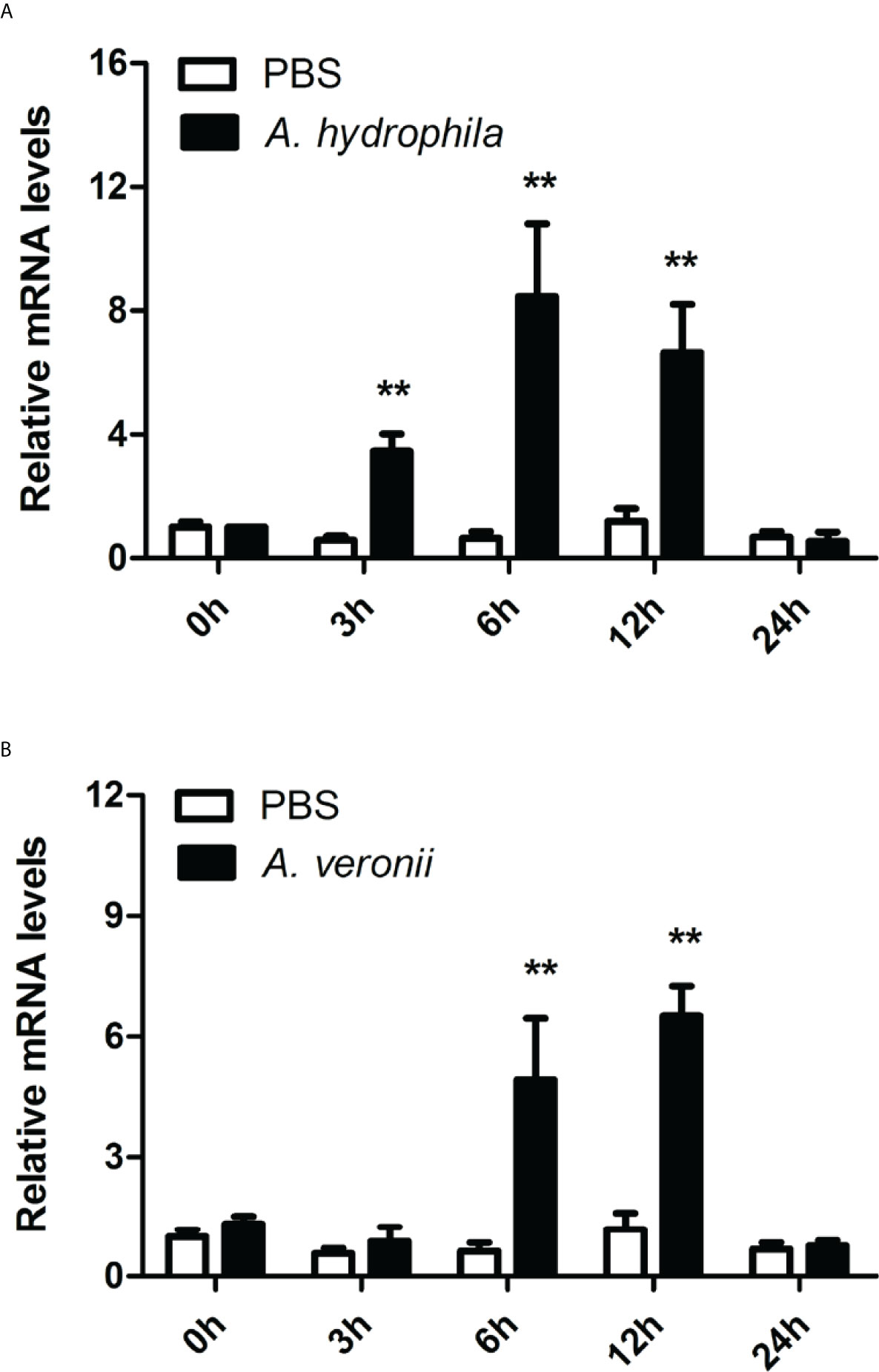
Figure 7 Temporal expression profiles of CiMEKK3 mRNA in intestinal cells after challenge with A. hydrophila (A) or A veronii (B). Each bar represents the mean of the normalized expression levels of replicates (N = 3). Significant differences between the challenge group and the control group are indicated with an asterisk (** represents P < 0.01).
To further investigate the immune function of CiMEKK3 in intestinal cells in vitro, the expression levels of CiMEKK3 were detected after stimulation with typical bacterial PAMPs (MDP, Tri-DAP, PGN and LPS) via qRT-PCR (Figure 8). In the first 3 h of the immune challenge, CiMEKK3 expression only significantly increased in the LPS group compared with that of the PBS control (P < 0.05) (Figure 8A). After 6 h of stimulation, the transcript levels of CiMEKK3 were shown to be significantly induced by MDP, Tri-DAP and LPS (P < 0.05) (Figure 8B). Interestingly, all selected bacterial PAMPs (MDP, Tri-DAP, PGN and LPS) significantly upregulated the expression levels of CiMEKK3 at 12 h post-stimulation (P < 0.05) (Figure 8C). Upon 24 h of PAMP challenge, CiMEKK3 expression was maintained at a relatively high level in the LPS and PGN groups (P < 0.05) but returned to control levels in the MDP and Tri-DAP groups (P > 0.05) (Figure 8D).
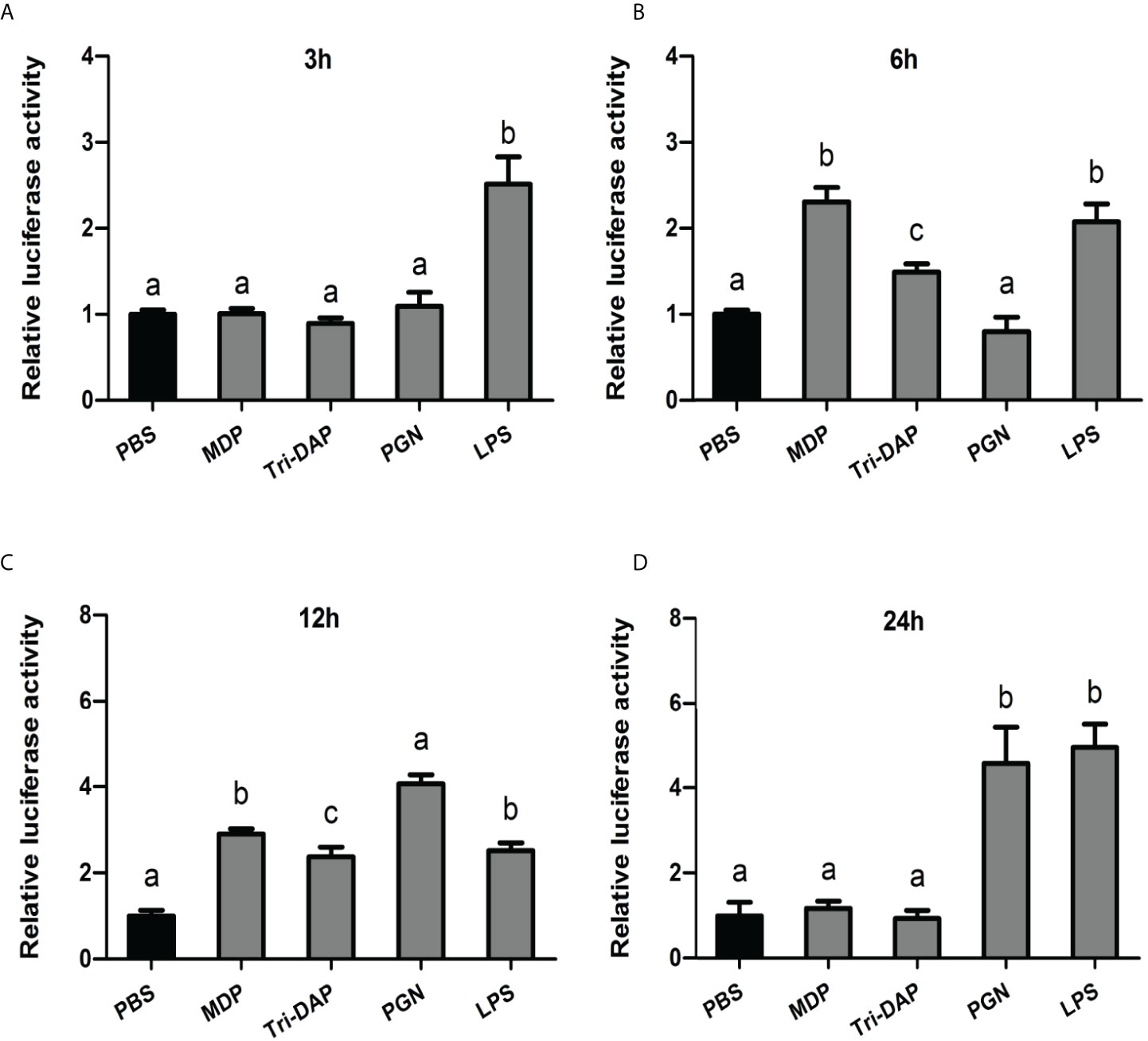
Figure 8 Temporal expression profiles of CiMEKK3 mRNA in intestinal cells at 3 h (A), 6 h (B), 12 h (C) and 24 h (D) post-challenge with MDP, Tri-DAP, PGN or LPS. Data are shown as the mean ± standard error of three individual fish (N = 3). Significant differences between the challenge group and the control group are indicated with different letters (P < 0.05).
The time-course expression levels of the CiMEKK3 transcripts were detected in the MDP-injected intestines of grass carp in vivo. As shown in Figure 9A, the relative expression of CiMEKK3 mRNA was significantly increased at 3 h, 6 h, 12 h, 24 h and 48 h post-injection (P < 0.05) and sharply decreased at 72 h post-injection (P > 0.05) in comparison with the control group. Additionally, a nutritional dipeptide (carnosine or Ala-Gln) and MDP were coinjected into grass carp to study the regulatory mechanism underlying the bacterial MDP-induced expression of CiMEKK3 in intestine. The results from Figure 9B show that the inductive effect of MDP on CiMEKK3 expression was significantly inhibited by carnosine or Ala-Gln treatment in the intestine of grass carp. These data may imply that the nutritional dipeptides carnosine and Ala-Gln may act as effective regulators to alleviate the bacterial MDP-mediated intestinal inflammatory response.
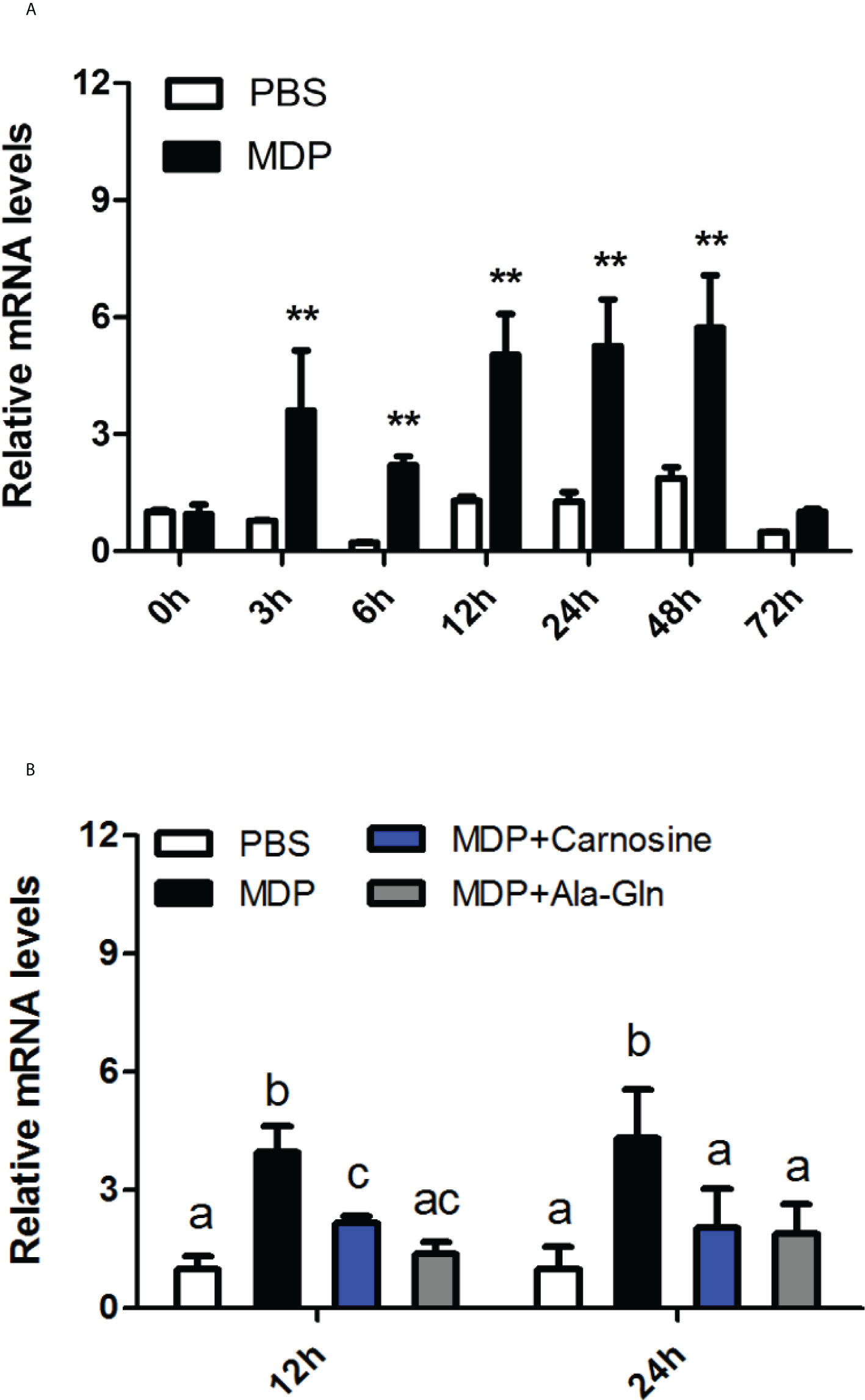
Figure 9 Temporal expression profiles of CiMEKK3 mRNA in the intestine after injection with MDP (A) or MDP + carnosine/Ala-Gln (B). Comparative analysis and statistical tests were performed on the challenge groups and PBS group at the same time point. Each bar represents the mean of the normalized expression levels of replicates (N = 3). Significant differences are indicated with an asterisk (** represents P < 0.01) or different letters (P < 0.05).
Dual-luciferase reporter assays were performed to determine the possible role of CiMEKK3 in the NF-κB and AP-1 signaling pathways. The NF-κB and AP-1 luciferase reporter were significantly activated by overexpression of CiMEKK3 in a dose-dependent manner in HEK293T cells (Figure 10A, B). In particular, it was found that the activating effects of CiMEKK3 overexpression on the AP-1 pathway were stronger than those on NF-κB signaling. Additionally, the luciferase reporter results showed that the activation effects on the AP-1 luciferase reporter of cells cotransfected CiMEKK3-Flag with CiMKK4-Flag, CiMKK6-Flag or CiMKK7-Flag were significantly higher than those of cells transfected with CiMEKK3 or CiMKKs alone (Figure 10C), suggesting that CiMEKK3 could enhance the downstream MKK-induced activation of the AP-1 signaling pathway. Collectively, the present results clearly indicate that CiMEKK3 might serve as an effective activator of the NF-κB and AP-1 signaling pathways.
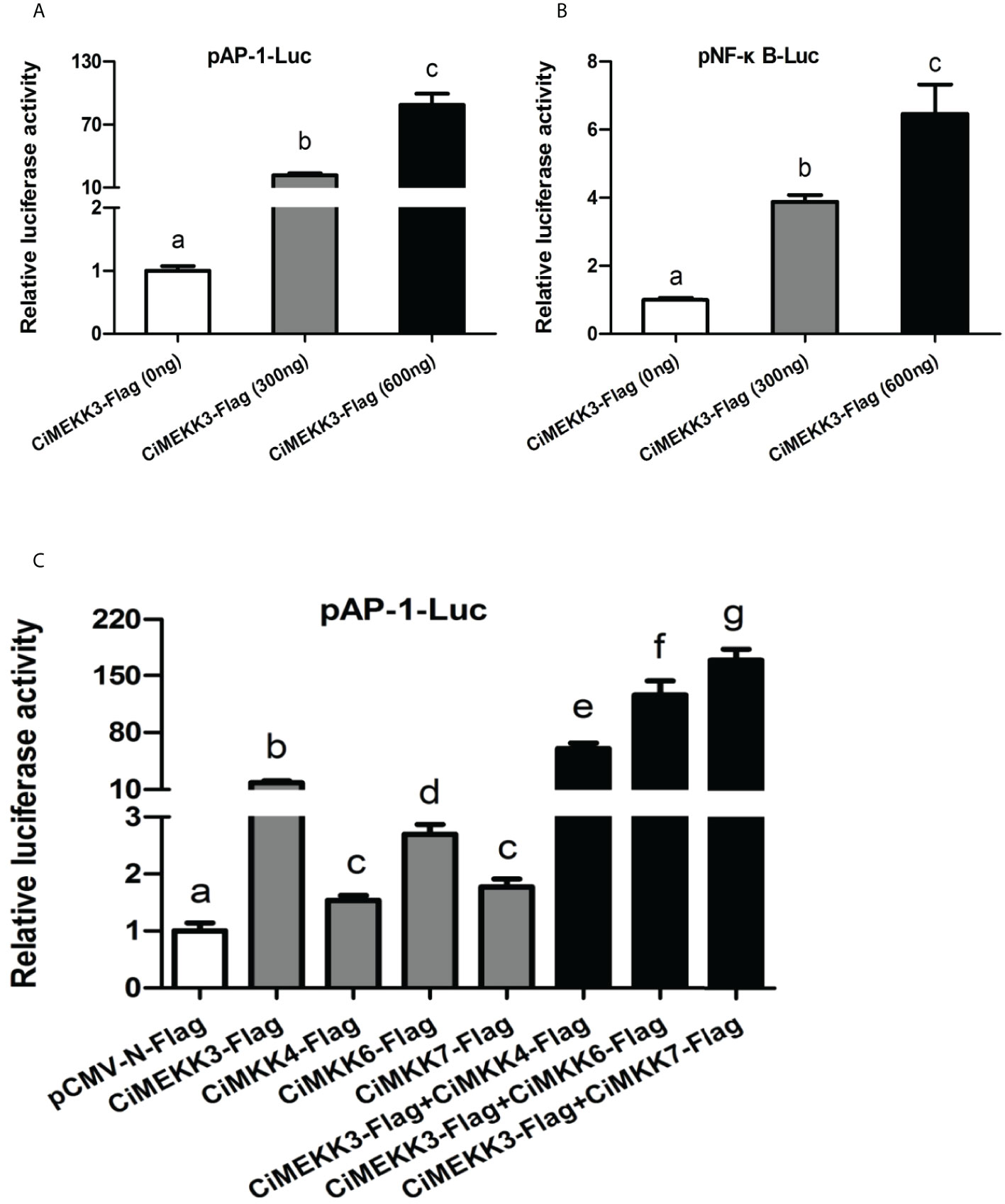
Figure 10 Effects of CiMEKK3 overexpression on the activity of the AP-1 and NF-κB pathways. The CiMEKK3-Flag (0, 300 and 600 ng/well) was cotransfected with 100 ng/well AP-1-Luc (A) or NF-κB-Luc (B) into HEK293T cells. (C) CiMEKK3-Flag (300 ng/well) was cotransfected with 100 ng/well AP-1-Luc, 300 ng/well CiMKK4-Flag, CiMKK6-Flag or CiMKK7-Flag into HEK293T cells. Each bar represents the mean (three replicates) ± standard deviation. Firefly and Renilla luciferase activities were detected in cell lysates 48 h after transfection. Data are the fold changes relative to the empty vector transfected cells. Significant differences are indicated with different letters (P < 0.05).
In vertebrates, MEKKs are essential signaling molecules of the NF-κB and MAPK pathways which play important regulatory roles in the immune response to pathogenic challenges (37, 38). To date, several MEKK family members have been identified from fish such as TAK1 in Oncorhynchus mykiss (39), Paralichthys olivaceus (40) and Megalobrama amblycephala (41), c-Raf in Epinephelus coioides (42) and Oreochromis niloticus (43), and MAP3K4 in C. idella (32). However, information regarding MEKK homologs in bony fish remains limited. In the present study, a member of the fish MEKK family, CiMEKK3, was cloned from C. idella using RT-PCR technology. Similar to other reported MEKK3 proteins, the present CiMEKK3 contains a conserved S-TKc domain and PB1 domain, which were shown to be essential for its activation and interaction with other signaling molecules in the MAPK pathway (19–22). In addition, the kinase catalytic domain of CiMEKK3 contains several phosphorylation sites (Thr294, Thr516, Ser520 and Ser526), which were observed in MEKK3s from mammals. Previous studies have demonstrated that the phosphorylation of specific Ser/Thr sites directly affects MEKK3 activation and its function in intracellular signal transduction under physiological and pathological conditions (15–18). These findings suggested that MEKK3s may possess similar phosphorylation mechanisms and signal transduction functions in mammals and fish. Based on the analysis of the 5′-upstream DNA sequence, several potential immune-related transcription factor-binding sites including NF-κB, AP-1, CREB and STAT3 were observed in the promoter region of CiMEKK3, suggesting that it may be involved in immune-related processes in grass carp. Multiple sequence alignment analysis showed that CiMEKK3 shares higher identity and similarity with other fish MEKK3s than with the reported mammalian homologs. A phylogenetic tree based on the amino acid sequences of MEKK3s revealed that CiMEKK3 shares a close relationship with C. carpio and C. auratus MEKK3. These results indicated that CiMEKK3 is a novel member of the fish MEKK3 family.
Previous studies have reported that MEKK3 is ubiquitously expressed in various tissues and cell types in mammals (12, 14, 25, 44). Recently, fish MEKK3 has been shown to be constitutively expressed in all tissues of healthy hybrid snakehead, including the liver, spleen, head kidney, trunk kidney, skin, gill, muscle, intestine, heart, brain, and blood (28). Similarly, the broad expression patterns of other MEKK family members including TAK1 (39, 40) and MEKK4 (32) have also been observed in fish. In our study, tissue expression analysis revealed that CiMEKK3 is broadly expressed in all selected tissues of healthy grass carp, consistent with the tissue-expression profile of other reported MEKK3s, suggesting the potential roles of CiMEKK3 in various biological processes. In bony fish, the head kidney and intestine are important immune-related tissues that are essential for host defense responses to immune challenges (29, 45). It is well known that fish gills are in direct contact with the aquatic environment, which serves as the first line of immune defense against various pathogen infections (46). Our qRT-PCR results show that CiMEKK3 displays relatively higher expression levels in the gill, head kidney and intestine, indicating that CiMEKK3 may play potential roles in the innate immunity of grass carp. To further explore the distribution and function of CiMEKK3, the subcellular localization of the CiMEKK3 protein was examined using a fluorescence microscope. The results reveal that CiMEKK3 is distributed mainly in the cytoplasm, suggesting that MEKK3 may act as a cytoplasmic localized protein involved in the signal transduction process of the MAPK pathway. Similar results have been observed in hybrid snakehead MEKK3 where CcMEKK3 is exclusively distributed in the cytoplasm of HEK293T cells (28). Moreover, it was noted that other fish MEKK family members, including TAK1 (47), c-Raf (42) and MEKK4 (32), also exist in the cytoplasm of the cells, implying that MEKKs may be mainly involved in biological events in the cytoplasm.
Bacterial enteritis is one of the most frequent and serious infectious diseases that occurs in the intensive cultivation of grass carp (29–31). However, the exact pathogenesis of bacterial enteritis is still not well understood. Similar to other bony fish, innate immunity has been considered a first line of host defense against invading pathogens in the intestines of grass carp (48). Over the past decades, several immune-related signaling pathways, including TLRs (49), IL-1R (50), NF-κB (51) and MAPK/AP-1 (52), have been identified in grass carp and proven to play important roles in the inflammatory response during pathogen infection. In mammals, MEKK3 was shown to act as an essential signal transducer of TLR- and IL-1R-mediated NF-κB, JNK and p38 cascades in response to immune challenge (23). MEKK3 was reported to be involved in regulating the LPS-induced production of proinflammatory cytokines in myeloid cells (25). Recently, a study of fish MEKK3 revealed its potential role in the immune response to pathogens and PAMPs challenge in the hybrid snakehead (28). To determine whether fish MEKK3 is involved in bacterial-induced intestinal inflammation, the expression levels of CiMEKK3 were detected in response to typical aquatic pathogens (A. hydrophila and A. veronii) in intestinal cells of grass carp. Our results show that the transcript levels of CiMEKK3 in intestinal cells are significantly induced by A. hydrophila and A. veronii challenges, suggesting that gram-negative bacterial strains can activate the intestinal MEKK3 pathway in grass carp. To gain more clues about the function of CiMEKK3 in intestinal immune responses, we further analyzed the expression profile of CiMEKK3 after stimulation with LPS and PGN, the important components of gram-negative and gram-positive bacteria, in intestinal cells at different time points. The qRT-PCR data indicated that CiMEKK3 transcripts have a strong responsiveness to LPS and PGN challenges, further implying its potential role in intestinal defense against bacterial infections.
In recent years, much progress has been made with regard to the pathogenesis of bacterial-induced intestinal inflammation in mammals (53). Recent research has found that products of bacterial cell-wall PGN, including MDP and Tri-DAP, could be transported by peptide transporter 1 (PepT1) in epithelial cells of the small intestine, then recognized by the intracellular NBS-LRR proteins (NOD1 and NOD2) and finally result in the transcription of proinflammatory genes to initiate intestinal inflammation through a series of signaling events (54, 55). A large number of studies have shown that bacterial peptide MDP- and Tri-DAP-mediated PepT1/NOD signaling pathways are essential for the intestinal inflammatory response in mammals (56, 57). However, whether MDP and Tri-DAP can induce the intestinal inflammation and their related immune signaling pathways remains poorly understood in bony fish. Over the past few years, our laboratory has conducted work on bacterial peptide-mediated intestinal inflammation in grass carp. Our previous studies showed that the bacterial peptides MDP and Tri-DAP could induce intestinal inflammation and that MAPK pathways participated in the regulation of the intestinal immune response to bacterial peptide challenges in C. idella (52, 58). To better understand the regulatory mechanism of bacterial peptide-induced intestinal inflammation, the expression profile of CiMEKK3 was analyzed after challenge with MDP and Tri-DAP in the intestine of grass carp. The in vitro experiment showed that CiMEKK3 transcript levels in the intestine were significantly increased in a time-dependent manner upon MDP and Tri-DAP challenge. Moreover, we found that the intestinal expression levels of CiMEKK3 induced by MDP challenge could be blocked by the nutritional peptides carnosine and Ala-Gln. Previously, it was reported that PepT1 ligand Lys-Pro-Val (KPV) could inhibit NF-кB signaling and decrease the production of proinflammatory cytokines in Caco2-BBE and Jurkat cells (59). These findings suggested that carnosine and Ala-Gln may exert an anti-inflammatory role similar to that of KPV, which may be useful for the future treatment of intestinal inflammation.
Nuclear factor-κB (NF-κB) is a group of immune-related transcription factors that play a predominant role in regulating the expression of various immune effectors, including proinflammatory cytokines, antimicrobial peptides and chemokines (60). MEKK3 has been previously shown to participate in the NF-κB signal transduction pathway (23, 24, 26). For example, MEKK3 has been shown to be essential for TNF-induced NF-κB activation in fibroblast cells (24). Additionally, Sun et al. reported that MEKK3 is a central intermediate signaling component in lysophosphatidic acid (LPA) -induced activation of NF-κB. In ovarian epithelial cells, overexpression of MEKK3 has been proven to increase NF-κB activity and the expression of Bcl-2, Bcl-xL and survivin (61). In addition to the NF-κB pathway, MEKK3 is involved in the regulation of MAPK/AP-1 signaling activation (62). Reportedly, MEKK3 is involved in TNFα, IL-1β, and TLR-induced MAPK activation in vivo and in vitro (23). To determine whether fish MEKK3 could activate the NF-κB and AP-1 signaling pathways, CiMEKK3 expression plasmids were cotransfected with the AP-1 or NF-κB luciferase reporter genes into HEK293T cells. Our dual-luciferase reporter assays revealed that overexpression of CiMEKK3 alone could significantly induce the activation of the AP-1 and NF-κB luciferase reporter, which was similar to the results observed in MEKK3 of hybrid snakehead (28). These results suggest that fish MEKK3 may also act as a positive regulator of the NF-κB and AP-1 signaling pathways. Moreover, our results showed that CiMEKK3 may enhance the CiMKK4-, CiMKK6- and CiMKK7-induced activation of the AP-1 luciferase reporter. MKK6 and MKK7 specifically phosphorylate and activate p38 and JNK, respectively, while MKK4 can act as an activator of both the p38 and JNK pathways (63, 64). These findings may suggest that CiMEKK3 regulates the activity of the p38- and JNK-induced AP-1 signaling pathways by interacting with downstream MKKs. Combined with the gene expression profile during immune challenge, it is speculated that CiMEKK3 may act as an important signal transducer of the NF-κB, JNK and p38 MAPK cascades involved in the intestinal immune response of grass carp. However, more experimental evidence is needed to support this speculation.
In conclusion, a functional fish MEKK3 gene (CiMEKK3) was identified and characterized in grass carp, which contained the typical characteristic features of the MEKK3 family. Tissue-specific expression analysis showed that CiMEKK3 mRNA was highly expressed in immune-related tissues of C. idella. The intestinal expression levels of CiMEKK3 mRNA were significantly upregulated after challenge with bacterial pathogens (A. hydrophila and A. veronii) and PAMPs (MDP, Tri-DAP, PGN and LPS). Moreover, overexpression analysis revealed that CiMEKK3 acted as an intracellular signaling molecule involved in the regulation of the NF-κB and AP-1 pathways in HEK293T cells. These results suggested that CiMEKK3 plays essential roles in the intestinal immune response to bacterial challenges, which may provide new insights into the intestinal immunity of bony fish.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, ON082069.
All experiments were performed according to the recommendations of the Guidance of the Care and Use of Laboratory Animals in China. The research presented in this manuscript was approved by the Animal Ethics Committee of Changsha University.
FQ, ZL, and XZe designed the experiments and wrote the manuscript; XZe, ZZL, MG, and XZh conducted the experiments; XZe and ZZL analyzed the data; SC, YZ, ZH, JT, ZM, YY, and ZZ modified the manuscript; all authors reviewed and approved the final manuscript.
This research was supported by the National Natural Science Foundation of China (Grant Nos. U21A20267, 32102813, 31902345 and 31702378), the Hunan Provincial Natural Science Foundation of China (Grant Nos. 2022JJ10053 and 2020JJ4642), the science and technology innovation Program of Hunan Province (2020RC3053), the Project of Scientific Research of the Hunan Provincial Education Department, China (Grant Nos. 20A045 and 20B058), the Training Program for Excellent Young Innovators of Changsha (Grant Nos. kq1707015, kq1802044, kq1905001, kq2009072 and kq2106067).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Seger R, Krebs EG. The mapk signaling cascade. FASEB J Off Publ Fed Am Societies Exp Biol (1995) 9(9):726–35. doi: 10.1096/fasebj.9.9.7601337
2. Cowan KJ, Storey KB. Mitogen-activated protein kinases: New signaling pathways functioning in cellular responses to environmental stress. J Exp Biol (2003) 206(7):1107–15. doi: 10.1242/jeb.00220
3. Cobb MH, Goldsmith EJ. How map kinases are regulated. J Biol Chem (1995) 270(25):14843–6. doi: 10.1074/jbc.270.25.14843
4. Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, et al. Mitogen-activated protein (Map) kinase pathways: Regulation and physiological functions. Endocrine Rev (2001) 22(2):153–83. doi: 10.1210/edrv.22.2.0428
5. Symons A, Beinke S, Ley SC. Map kinase kinase kinases and innate immunity. Trends Immunol (2006) 27(1):40–8. doi: 10.1016/j.it.2005.11.007
6. Yue J, López JM. Understanding mapk signaling pathways in apoptosis. Int J Mol Sci (2020) 21(7):2346. doi: 10.3390/ijms21072346
7. Krens SF, Spaink HP, Snaar-Jagalska BE. Functions of the mapk family in vertebrate-development. FEBS Lett (2006) 580(21):4984–90. doi: 10.1016/j.febslet.2006.08.025
8. Winter-Vann AM, Johnson GL. Integrated activation of Map3ks balances cell fate in response to stress. J Cell Biochem (2007) 102(4):848–58. doi: 10.1002/jcb.21522
9. Craig EA, Stevens MV, Vaillancourt RR, Camenisch TD. Map3ks as central regulators of cell fate during development. Dev dynamics an Off Publ Am Assoc Anatomists (2008) 237(11):3102–14. doi: 10.1002/dvdy.21750
10. Yang J, Boerm M, McCarty M, Bucana C, Fidler IJ, Zhuang Y, et al. Mekk3 is essential for early embryonic cardiovascular development. Nat Genet (2000) 24(3):309–13. doi: 10.1038/73550
11. Wang X, Chang X, Facchinetti V, Zhuang Y, Su B. Mekk3 is essential for lymphopenia-induced T cell proliferation and survival. J Immunol (2009) 182(6):3597–608. doi: 10.4049/jimmunol.0803738
12. Kim K, Duramad O, Qin XF, Su B. Mekk3 is essential for lipopolysaccharide-induced interleukin-6 and granulocyte-macrophage colony-stimulating factor production in macrophages. Immunology (2007) 120(2):242–50. doi: 10.1111/j.1365-2567.2006.02495.x
13. Santoro R, Zanotto M, Carbone C, Piro G, Tortora G, Melisi D. Mekk3 sustains emt and stemness in pancreatic cancer by regulating yap and taz transcriptional activity. Anticancer Res (2018) 38(4):1937–46. doi: 10.21873/anticanres.12431
14. Blank JL, Gerwins P, Elliott EM, Sather S, Johnson GL. Molecular cloning of mitogen-activated Protein/Erk kinase kinases (Mekk) 2 and 3. regulation of sequential phosphorylation pathways involving mitogen-activated protein kinase and c-jun kinase. J Biol Chem (1996) 271(10):5361–8. doi: 10.1074/jbc.271.10.5361
15. Fritz A, Brayer KJ, McCormick N, Adams DG, Wadzinski BE, Vaillancourt RR. Phosphorylation of serine 526 is required for Mekk3 activity, and association with 14-3-3 blocks dephosphorylation. J Biol Chem (2006) 281(10):6236–45. doi: 10.1074/jbc.M509249200
16. Zhang D, Facchinetti V, Wang X, Huang Q, Qin J, Su B. Identification of Mekk2/3 serine phosphorylation site targeted by the toll-like receptor and stress pathways. EMBO J (2006) 25(1):97–107. doi: 10.1038/sj.emboj.7600913
17. Matitau AE, Scheid MP. Phosphorylation of Mekk3 at threonine 294 promotes 14-3-3 association to inhibit nuclear factor kappab activation. J Biol Chem (2008) 283(19):13261–8. doi: 10.1074/jbc.M801474200
18. Sun W, Ge N, Yu Y, Burlingame S, Li X, Zhang M, et al. Phosphorylation of thr-516 and ser-520 in the kinase activation loop of Mekk3 is required for lysophosphatidic acid-mediated optimal ikappab kinase beta (Ikkbeta)/Nuclear factor-kappab (Nf-kappab) activation. J Biol Chem (2010) 285(11):7911–8. doi: 10.1074/jbc.M109.051219
19. Blonska M, You Y, Geleziunas R, Lin X. Restoration of nf-kappab activation by tumor necrosis factor alpha receptor complex-targeted Mekk3 in receptor-interacting protein-deficient cells. Mol Cell Biol (2004) 24(24):10757–65. doi: 10.1128/mcb.24.24.10757-10765.2004
20. Nakamura K, Johnson GL. Pb1 domains of Mekk2 and Mekk3 interact with the Mek5 Pb1 domain for activation of the Erk5 pathway. J Biol Chem (2003) 278(39):36989–92. doi: 10.1074/jbc.C300313200
21. Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. Pb1 domain interaction of P62/Sequestosome 1 and Mekk3 regulates nf-kappab activation. J Biol Chem (2010) 285(3):2077–89. doi: 10.1074/jbc.M109.065102
22. Cheng J, Yu L, Zhang D, Huang Q, Spencer D, Su B. Dimerization through the catalytic domain is essential for Mekk2 activation. J Biol Chem (2005) 280(14):13477–82. doi: 10.1074/jbc.M414258200
23. Huang Q, Yang J, Lin Y, Walker C, Cheng J, Liu Z-g, et al. Differential regulation of interleukin 1 receptor and toll-like receptor signaling by Mekk3. Nat Immunol (2004) 5(1):98–103. doi: 10.1038/ni1014
24. Yang J, Lin Y, Guo Z, Cheng J, Huang J, Deng L, et al. The essential role of Mekk3 in tnf-induced nf-κb activation. Nat Immunol (2001) 2(7):620–4. doi: 10.1038/89769
25. Li K, Wang M, Hu Y, Xu N, Yu Q, Wang Q. Tak1 knockdown enhances lipopolysaccharide-induced secretion of proinflammatory cytokines in myeloid cells Via unleashing Mekk3 activity. Cell Immunol (2016) 310:193–8. doi: 10.1016/j.cellimm.2016.09.011
26. Yao L, Ye Y, Mao H, Lu F, He X, Lu G, et al. Microrna-124 regulates the expression of Mekk3 in the inflammatory pathogenesis of parkinson's disease. J Neuroinflamm (2018) 15(1):13. doi: 10.1186/s12974-018-1053-4
27. Cai M, Huang W, Hu X, Chen A, Zhou X. Mekk3 activates Irf7 to trigger a potent type I interferon induction in response to Tlr7/9 signaling. Mol Immunol (2021) 134:183–91. doi: 10.1016/j.molimm.2021.03.008
28. Li D, Kong L, Cui Z, Zhao F, Deng Y, Tan A, et al. Mekk3 in hybrid snakehead (Channa maculate ♀ ×Channa Argus ♂): Molecular characterization and immune response to infection with nocardia seriolae and aeromonas schubertii. Comp Biochem Physiol Part B Biochem Mol Biol (2021) 256:110643. doi: 10.1016/j.cbpb.2021.110643
29. Song X, Zhao J, Bo Y, Liu Z, Wu K, Gong C. Aeromonas hydrophila induces intestinal inflammation in grass carp (Ctenopharyngodon idella): An experimental model. Aquaculture (2014) 434:171–8. doi: 10.1016/j.aquaculture.2014.08.015
30. Bohai X, Ruifang G, Mulin X. Pathogenetic investigation of the enteritis of the grass carp (Ctenopharyngodon idellus). Acta Hydrobiologica Sin (China) (1988) 12:308–15.
31. Kong W-G, Li S-S, Chen X-X, Huang Y-Q, Tang Y, Wu Z-X. A study of the damage of the intestinal mucosa barrier structure and function of ctenopharyngodon idella with aeromonas hydrophila. Fish Physiol Biochem (2017) 43(5):1223–35. doi: 10.1007/s10695-017-0366-z
32. Qu F, Li J, Zeng X, She Q, Li Y, Xie W, et al. Grass carp Map3k4 participates in the intestinal immune response to bacterial challenge. Fish Shellfish Immunol (2022) 120:82–91. doi: 10.1016/j.fsi.2021.11.016
33. Ran C, Qin C, Xie M, Zhang J, Li J, Xie Y, et al. Aeromonas veronii and aerolysin are important for the pathogenesis of motile aeromonad septicemia in cyprinid fish. Environ Microbiol (2018) 20(9):3442–56. doi: 10.1111/1462-2920.14390
34. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2– ΔΔCT method. methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
35. Qu F, Tang J, Peng X, Zhang H, Shi L, Huang Z, et al. Two novel mkks (Mkk4 and Mkk7) from ctenopharyngodon idella are involved in the intestinal immune response to bacterial muramyl dipeptide challenge. Dev Comp Immunol (2019) 93:103–14. doi: 10.1016/j.dci.2019.01.001
36. Qu F, Tang J, Liao J, Chen B, Song P, Luo W, et al. Mitogen-activated protein kinase kinase 6 is involved in the immune response to bacterial di-/Tripeptide challenge in grass carp ctenopharyngodon idella. Fish shellfish Immunol (2019) 84:795–801. doi: 10.1016/j.fsi.2018.10.073
37. Clark NM, Marinis JM, Cobb BA, Abbott DW. Mekk4 sequesters Rip2 to dictate Nod2 signal specificity. Curr Biol (2008) 18(18):1402–8. doi: 10.1016/j.cub.2008.07.084
38. Ajibade AA, Wang HY, Wang R-F. Cell type-specific function of Tak1 in innate immune signaling. Trends Immunol (2013) 34(7):307–16. doi: 10.1016/j.it.2013.03.007
39. Jang JH, Kim H, Cho JH. Molecular cloning and functional characterization of Traf6 and Tak1 in rainbow trout, oncorhynchus mykiss. Fish Shellfish Immunol (2019) 84:927–36. doi: 10.1016/j.fsi.2018.11.002
40. Zhao H, Chen Y, Wang Z, Zhang Q, Yu H. Molecular characterization, expression profiles, and immunostimulation responses of Traf6 and Tak1 in Japanese flounder (Paralichthys olivaceus). J Ocean Univ China (2019) 18(1):165–76. doi: 10.1007/s11802-019-3683-2
41. Xu Y, Zhu B, Zhang R, Tang J, Liu Y, Wang W, et al. Tak1 of blunt snout bream promotes nf-kappa b activation Via interaction with Tab1 in response to pathogenic bacteria. Fish Shellfish Immunol (2022) 120:481–96. doi: 10.1016/j.fsi.2021.12.019
42. Mo Z-Q, Lai X-L, Wang W-T, Chen H-P, He Z-C, Han R, et al. Identification and characterization of c-raf from orange-spotted grouper (Epinephelus coioides). Fish shellfish Immunol (2020) 96:311–8. doi: 10.1016/j.fsi.2019.12.017
43. Wei X, Zhao T, Ai K, Zhang Y, Li H, Yang J. C-raf participates in adaptive immune response of Nile tilapia Via regulating lymphocyte activation. Fish shellfish Immunol (2019) 86:507–15. doi: 10.1016/j.fsi.2018.11.072
44. Hernandez ED, Lee SJ, Kim JY, Duran A, Linares JF, Yajima T, et al. A macrophage Nbr1-Mekk3 complex triggers jnk-mediated adipose tissue inflammation in obesity. Cell Metab (2014) 20(3):499–511. doi: 10.1016/j.cmet.2014.06.008
45. Tanekhy M, Matsuda S, Itano T, Kawakami H, Kono T, Sakai M. Expression of cytokine genes in head kidney and spleen cells of Japanese flounder (Paralichthys olivaceus) infected with nocardia seriolae. Veterinary Immunol immunopathology (2010) 134(3-4):178–83. doi: 10.1016/j.vetimm.2009.09.001
46. Koppang EO, Kvellestad A, Fischer U. Fish mucosal immunity: Gill. Mucosal health in aquaculture. (2015) 93–133.
47. Li Y, Li X, Wang Z, Mo Z, Dan X, Luo X, et al. Orange-spotted grouper epinephelus coioides Tak1: Molecular identification, expression analysis and functional study. J Fish Biol (2015) 86(2):417–30. doi: 10.1111/jfb.12550
48. Magnadóttir B. Innate immunity of fish (Overview). Fish shellfish Immunol (2006) 20(2):137–51. doi: 10.1016/j.fsi.2004.09.006
49. Huang W-J, Shen Y, Xu X-Y, Hu M-Y. Identification and characterization of the Tlr18 gene in grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol (2015) 47(2):681–8. doi: 10.1016/j.fsi.2015.09.052
50. Yao F, Yang X, Wang X, Wei H, Zhang A, Zhou H. Molecular and functional characterization of an il-1β receptor antagonist in grass carp (Ctenopharyngodon idella). Dev Comp Immunol (2015) 49(2):207–16. doi: 10.1016/j.dci.2014.11.016
51. Wang H, Zhu Y, Xu X, Wang X, Hou Q, Xu Q, et al. Ctenopharyngodon idella nf-kappa b subunit P65 modulates the transcription of iκbα in cik cells. Fish shellfish Immunol (2016) 54:564–72. doi: 10.1016/j.fsi.2016.04.132
52. Qu F, Xu W, Deng Z, Xie Y, Tang J, Chen Z, et al. Fish c-jun n-terminal kinase (Jnk) pathway is involved in bacterial mdp-induced intestinal inflammation. Front Immunol (2020) 11:459. doi: 10.3389/fimmu.2020.00459
53. Chamaillard M, Girardin SE, Viala J, Philpott DJ. Nods, nalps and naip: Intracellular regulators of bacterial-induced inflammation. Cell Microbiol (2003) 5(9):581–92. doi: 10.1046/j.1462-5822.2003.00304.x
54. Franchi L, Park JH, Shaw MH, Marina-Garcia N, Chen G, Kim YG, et al. Intracellular nod-like receptors in innate immunity, infection and disease. Cell Microbiol (2008) 10(1):1–8. doi: 10.1111/j.1462-5822.2007.01059.x
55. Ingersoll SA, Ayyadurai S, Charania MA, Laroui H, Yan Y, Merlin D. The role and pathophysiological relevance of membrane transporter Pept1 in intestinal inflammation and inflammatory bowel disease. Am J Physiol Gastrointestinal liver Physiol (2012) 302(5):G484–92. doi: 10.1152/ajpgi.00477.2011
56. Ma G, Shi B, Liu J, Zhang H, YinTao Z, Lou X, et al. Nod2-Rip2 signaling contributes to intestinal injury induced by muramyl dipeptide Via oligopeptide transporter in rats. Digestive Dis Sci (2015) 60(11):3264–70. doi: 10.1007/s10620-015-3762-1
57. Dalmasso G, Nguyen HT, Charrier-Hisamuddin L, Yan Y, Laroui H, Demoulin B, et al. Pept1 mediates transport of the proinflammatory bacterial tripeptide l-Ala-{Gamma}-D-Glu-Meso-Dap in intestinal epithelial cells. Am J Physiol Gastrointestinal liver Physiol (2010) 299(3):G687–96. doi: 10.1152/ajpgi.00527.2009
58. Sun Y, Xu W, Li D, Zhou H, Qu F, Cao S, et al. P38 mitogen-activated protein kinases (Mapks) are involved in intestinal immune response to bacterial muramyl dipeptide challenge in ctenopharyngodon idella. Mol Immunol (2020) 118:79–90. doi: 10.1016/j.molimm.2019.12.007
59. Dalmasso G, Charrier-Hisamuddin L, Nguyen HT, Yan Y, Sitaraman S, Merlin D. Pept1-mediated tripeptide kpv uptake reduces intestinal inflammation. Gastroenterology (2008) 134(1):166–78. doi: 10.1053/j.gastro.2007.10.026
60. Hatada EN, Krappmann D, Scheidereit C. Nf-kappab and the innate immune response. Curr Opin Immunol (2000) 12(1):52–8. doi: 10.1016/s0952-7915(99)00050-3
61. Sun W, Wang H, Zhao X, Yu Y, Fan Y, Wang H, et al. Protein phosphatase 2a acts as a mitogen-activated protein kinase kinase kinase 3 (Mekk3) phosphatase to inhibit lysophosphatidic acid-induced ikappab kinase Beta/Nuclear factor-kappab activation. J Biol Chem (2010) 285(28):21341–8. doi: 10.1074/jbc.M110.104224
62. Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by erk, jnk, and P38 protein kinases. Sci (New York NY) (2002) 298(5600):1911–2. doi: 10.1126/science.1072682
63. Schaeffer HJ, Weber MJ. Mitogen-activated protein kinases: Specific messages from ubiquitous messengers. Mol Cell Biol (1999) 19(4):2435–44. doi: 10.1128/MCB.19.4.2435
Keywords: grass carp, MEKK3, molecular characterization, bacterial challenge, intestinal immunity
Citation: Qu F, Zeng X, Liu Z, Guo M, Zhang X, Cao S, Zhou Y, He Z, Tang J, Mao Z, Yang Y, Zhou Z and Liu Z (2022) Functional characterization of MEKK3 in the intestinal immune response to bacterial challenges in grass carp (Ctenopharyngodon idella). Front. Immunol. 13:981995. doi: 10.3389/fimmu.2022.981995
Received: 30 June 2022; Accepted: 18 July 2022;
Published: 05 August 2022.
Edited by:
Hai-peng Liu, Xiamen University, ChinaReviewed by:
Zhendong Qin, Zhongkai University of Agriculture and Engineering, ChinaCopyright © 2022 Qu, Zeng, Liu, Guo, Zhang, Cao, Zhou, He, Tang, Mao, Yang, Zhou and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Liu, MjUzMDAwODVAcXEuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.