- 1College of Marine Life Sciences, and Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao, China
- 2Department of Genetics and Cell Biology, Basic Medical College, Qingdao University, Qingdao, China
- 3Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
CD248, also known as endosialin or tumor endothelial marker 1, is a type I single transmembrane glycoprotein. CD248 has been demonstrated to be upregulated in cancers, tumors and many fibrotic diseases in human and mice, such as liver damage, pulmonary fibrosis, renal fibrosis, arthritis and tumor neovascularization. However, no definite CD248 orthologs in fish have been documented so far. In this study, we report the identification of cd248a and cd248b in the zebrafish. Both the phylogenetic analysis and the conserved synteny strongly suggested that zebrafish cd248a and cd248b are orthologs of the human CD248. Both cd248a and cd248b exhibited similar and dynamic expression pattern in early development, both genes had weak maternal expression, the zygotic transcripts were first seen in anterior somites and head mesenchyme, then shifted to eyes and head mesenchyme, later expanded to branchial arches, and gradually declined with development. The expression profiles of cd248a and cd248b were upregulated upon LPS (Lipopolysaccharide) challenge. Both Cd248a protein and Cd248b protein were localized on the cell membrane and cytoplasm, and overexpression of cd248a and cd248b induced the expression of pro-inflammatory cytokines, in vitro and in vivo. Moreover, deficiency of cd248a or cd248b both downregulated the expression of pro-inflammatory cytokines and upregulated anti-inflammatory cytokine. Additionally, loss of cd248a or cd248b both downregulated the expression of pro-inflammatory cytokines after LPS treatment. Taken together, these results indicated that cd248a and cd248b in zebrafish were involved in immune response and would provide further information to understand functions of Cd248 protein in innate immunity of fish.
Highlights
1. Zebrafish cd248a and cd248b are orthologs of human CD248.
2. Zebrafish cd248a and cd248b exhibited similar expression pattern, predominantly on the anterior somites, head mesenchyme, eyes and branchial arches.
3. Zebrafish Cd248 proteins participate in immune response.
Introduction
CD248, also known as endosialin or tumor endothelial marker 1, is a type I single transmembrane glycoprotein (1–3). CD248 was initially identified as an additional tumor stromal antigen (FB5) in human (4). CD248 is a member of C-type lectin domain group 14 family that includes CLEC14A, CD93 and thrombomodulin (also known as TM or THBD), which contain C-type lectin domain (5, 6).
Contrary to the early reports, CD248 is not expressed by endothelial cells, but mainly expressed in stromal fibroblasts, myofibroblasts and a subset of pericytes (1–3). CD248 is expressed in early embryos and is almost undetectable in normal adult mouse tissues except low level in fibroblasts and pericytes (3). CD248 has been shown to be upregulated in cancers, tumours, and many fibrotic diseases, and is considered as a therapeutic target and biomarker (7, 8). It has been shown CD248 participates in angiogenesis, hypoxic regulation, inflammation and reorganization of the extracellular matrix. Vascular smooth muscle cells played a key role in the pathogenesis of atherosclerosis, while CD248 promoted atherosclerosis by remodeling the phenotype of vascular smooth muscle cells (9). Moreover, CD248 expressed in hepatic stellate cells played a key role in regulating the balance between liver fibrosis and hepatocyte proliferation during liver injury, thus endosialin was identified as a therapeutic target in non-neoplastic settings (10). Interestingly, hypoxia, inflammation, and fibrosis in obesity could be eliminated by adipocyte-specific CD248 knockdown, which makes CD248 a potential target for improving metabolic health (11). In addition, CD248 is also involved in renal fibrosis (12), pulmonary fibrosis (13), arthritis (14, 15), and tumor neovascularization (16, 17), and has become a potential target for the treatment of these diseases. Furthermore, CD248 is involved in the immune response. CD248 is expressed on the mesenchymal stromal cells in thymus plays an important role in thymus development and thymus regeneration after infection, further participates in peripheral immunity (18). CD248 also plays an important role in the remodeling of secondary lymphoid organs during adaptive immunity (19). In addition, CD248 expressed by naive human CD8+ T cells inhibits the proliferation of T-cells (20).
CD248 has been extensively studied in humans and mice, but the information regarding cd248 in lower vertebrate is lacking and the role of Cd248 in lower vertebrate is still undefined. In the present study, we identified the orthologs of cd248 in zebrafish, referred as cd248a and cd248b. In brief, we analyzed the expression pattern in early embryos and upon LPS challenge, we also investigated the effect of cd248 on the expression of pro-inflammatory cytokines. Deficiency of cd248a or cd248b downregulated the pro-inflammatory cytokines. In addition, loss of cd248a or cd248b both downregulated the expression of pro-inflammatory cytokines after LPS treatment. Collectively, our data suggested that zebrafish Cd248 proteins were involved in immune response.
Materials and methods
Zebrafish maintenance and cell culture
The AB strain zebrafish (Danio rerio) were maintained at 28 ± 1°C and fed twice daily under 14 hours for light and 10 hours for dark cycle. The embryos were produced by natural mating. The experiments were performed according to the regulations and guidelines issued by Animal Committee of Ocean University of China.
HEK293T cells and RAW264.7 (leukemia cells in mouse macrophage) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) in a 37˚C humidified incubator with 5% CO2 environment. Cell transfections were carried out using Lipofectamine 2000 following the manufacturer’s instructions.
Bioinformatics analysis
The orientation and location of all genes used for gene synteny were referenced on the NCBI website. The protein domains were predicted using SMART website (http://smart.embl.de/).
Phylogenetic tree was constructed by neighbor-joining method and maximum-likelihood method with 1000 bootstrap replicates using MEGA 7.0 (21). All of the amino acid sequences used in bioinformatics analysis were downloaded from NCBI website (https://www.ncbi.nlm.nih.gov/).
Whole-mount in suit hybridization
The fragments of cd248a and cd248b were amplified by PCR using the specific primers P5、P6 and P7、P8, respectively. Both fragments were digested by Nco I (Promega, USA) and then Digoxigenin (DIG)-labeled antisense probe were synthesized in vitro by Sp6 RNA polymerase. The procedures of WISH were following the protocol (22). The staining embryos were observed and photographed under the stereomicroscope (Nikon, Japan).
Microinjection of LPS and quantitative real-time PCR
An aliquot of 6 nL PBS containing 6×10-3 ng LPS (treatment group) or PBS alone (control group) was injected individually into each yolk sac of zebrafish 1-cell stage embryos (23), and two groups were cultured separately in thermostat incubator. At 0 (not injection), 2, 4, 6, 10, 12, 14, 24, 48, 72, 96 and 120 hours post-injection, 40 embryos/larvae were sampled at each stage in each group, and homogenized in Trizol Reagent. Total RNAs extraction and purification using Total RNA Kit I (Omega) according to the manufacturer’s instructions, then were synthesized to cDNA by reverse transcription system (Takara). The expression of cd248a and cd248b mRNA after LPS injection was analyzed by qRT-PCR using ABI 7500 real-time PCR system (Applied Biosystems). The specific primers of cd248a (P9、P10) and cd248b (P11、P12) for qRT-PCR to detect the expression level of the relative mRNA, zebrafish β-actin (P13、P14) gene was chosen as the reference for internal standardization. qRT-PCR reaction conditions were as follows: 95°C for 15s, followed by 40 cycles of 95°C for 5s, 60°C for 15s, and 72°C for 35s. Each experiment was repeated three times. Data were quantified with the comparative Ct method (2-(△△ct)) in order to calculate the relative mRNA expression level (23). The data obtained from qRT-PCR analysis were subjected to two-way analysis of variance (ANOVA) to determine differences in the mean values. Data were shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Subcellular localization
The pcDNA3.1/V5/eGFP vector was formed by cloning eGFP gene into eukaryotic expression vector pcDNA3.1/V5-His A. Subsequently, the complete coding regions of cd248a, cd248b, cd248bΔN(lack of N-terminal transmembrane region), cd248bΔC(lack of C-terminal transmembrane region) and cd248bΔNΔC (lack of N- and C-terminal transmembrane regions) were amplified by PCR using the specific primers (P1、P2, P3、P4, P5、P4, P3、P6 and P5、P6 respectively) containing restriction enzyme cutting sites EcoR I and Xho I, and then the fragments were inserted into the upstream of pcDNA3.1/V5/eGFP to construct the recombinant plasmids. To examine subcellular localization of cd248a and cd248b and its variants in HEK293T, the pcDNA3.1/V5/cd248a/eGFP, pcDNA3.1/V5/cd248b/eGF, pcDNA3.1/V5/cd248bΔN/eGFP, pcDNA3.1/V5/cd248bΔC/eGFP, pcDNA3.1/V5/cd248bΔNΔC/eGFP and pcDNA3.1/V5/eGFP recombinant plasmids were transfected individually into HEK293T cells as described above. At 24h post-transfected, the cells were stained with DAPI. After washing with PBS, the samples were observed from confocal microscope.
Effect of overexpression of zebrafish cd248a and cd248b on pro-inflammatory cytokines expression in mouse macrophages
The open reading frames of cd248a and cd248b were inserted separately into the eukaryotic expression vector pcDNA3.1/V5-His A by EcoR I and Xho I restriction enzyme, and the specific primers used to clone the fragments of cd248a and cd248b were P15、P16 and P17、P18, respectively. Then pcDNA3.1/V5/cd248a, pcDNA3.1/V5/cd248b and pcDNA3.1/V5-His A plasmids were transfected individually into RAW 264.7 cells. At 24h after transfection, the total RNA of the cells was extracted and cDNA was synthesized. Finally, we examined the expression levels of mouse Tnfα (P19 and P20), Il1β (P21 and P22) and Il6 (P23 and P24) by qRT-PCR. Mouse β-actin (P25、P26) gene was chosen as the reference for internal standardization (24). Each experiment was repeated three times.
Effect of overexpression of zebrafish cd248a and cd248b mRNA on pro-inflammatory cytokines expression in vivo
To further verify the effect of overexpression of zebrafish cd248 on pro-inflammatory cytokines in vivo, we synthesized cd248a and cd248b mRNA by SP6 mMESSAGE mMACHINE Kit (ThermoFisher). Diluted mRNAs were injected into 1-cell stage embryos of zebrafish. At 48h after injection, the total RNA of the embryos was extracted and cDNA was synthesized. Finally, we examined the expression levels of tnfα (P27 and P28), il1β (P29 and P30), il6 (P31 and P32), hif1a (P33 and P34), pigf (P35 and P36), vegfr1 (P37 and P38), notch3 (P39 and P40) and mmp9 (P41 and P42) by qRT-PCR. Zebrafish β-actin gene was chosen as the reference for internal standardization (24). Each experiment was repeated three times.
Effect of cd248 deletion on pro-inflammatory cytokines and anti-inflammatory cytokine
The mutants of cd248a and cd248b were carried out by CRISPR/Cas9 system (25). The sequences for gRNAs were shown in Figures 8A–F. At 48h after fertilization, we extracted the total RNA of WT and mutant embryos and synthesized cDNA. Finally, we examined the expression level of cd248a, cd248b, tnfα, il1β, il6 and il4 (P43 and P44) by qRT-PCR as previously described. These pro-inflammatory cytokines were also examined after LPS treatment. Each experiment was repeated three times.
Supplementary data
All primers used in this paper were shown in Supplemental Table 1. All protein sequences used to construct phylogenetic trees were shown in Supplemental Table 2.
Results
Two orthologs of the human CD248 were present in the zebrafish genome
We performed database searches with the human gene sequences of CD248 and identified two orthologs in the zebrafish genome. Zebrafish cd248a and cd248b contained one single exon similar to that of other species. Zebrafish Cd248 shared similar protein domains as that of human and mouse. The deduced Cd248a had a single peptide, followed by seven conserved functional domains: one CLECT, one CCP domain, five EGF domains and a TM (transmembrane) domain. While the encoded Cd248b consisted a TM domain at the N-terminal, one CLECT domain, five EGF domains and a TM domain in the C-terminal (Figure 1). However, there was no CCP domain in Cd248b, which may be because of its loss during genome duplication. CLECT is C-type lectin domain or carbohydrate-recognition domain. The CLECT is a key structural motif responsible for assisting CD248 to recognize multiple ligands and functions to in defense against pathogens, cell trafficking, and immune regulation. EGF is Epidermal growth factor-like domain. EGF repeats mainly contribute to the interaction of EGF-containing protein with other proteins. CCP is complement control protein domain (also known as short consensus repeats SCRs or SUSHI repeats), CCP domain containing proteins are usually involved in complement pathway (7, 26).
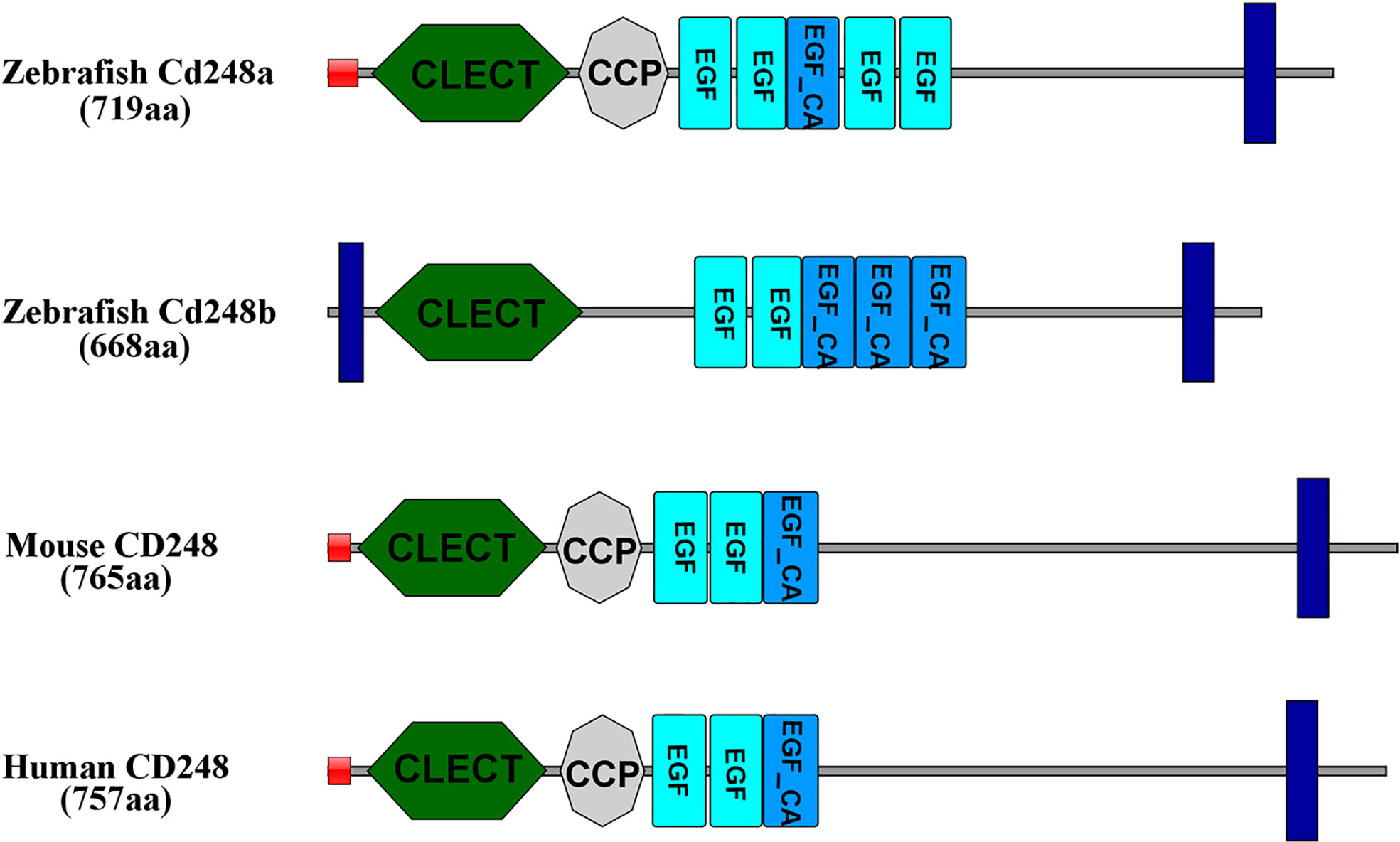
Figure 1 Amino acid functional domains of CD248 protein in human, mouse and zebrafish. Red squares, signal peptide; Blue rectangles, transmembrane region; CLECT, C-type lectin domain; EGF, epidermal growth factor-like domain; EGF_CA, Calcium-binding EGF-like domain; CCP, complement control protein domain.
To explore the possible synteny relationship among the zebrafish cd248 and the human CD248, we analyzed genes surrounding the cd248 loci in the zebrafish and human genomes. While cd248a was located on chromosome 7, cd248b was mapped to chromosome 21. The human CD248 gene resided on chromosome 11. The analysis of the surrounding regions revealed that cd248a and 7 other zebrafish genes (eml3, mta2, taf6l, kcnk7, peli3, rce1a and coro1b) located on chromosome 7 had orthologs on human chromosome 11. Similarly, in the proximity of the cd248b locus, 7 zebrafish genes (ppme1, tmem223, rce1b, esrra, prdx5, nxf1 and bscl2) had their orthologs located near the human CD248 locus on chromosome 11 (Figure 2). This conserved synteny strongly suggested that cd248a and cd248b were co-orthologs of the human CD248 gene. cd248a and cd248b were originated from the presumed teleost-specific whole genome duplication event during evolution (27, 28).
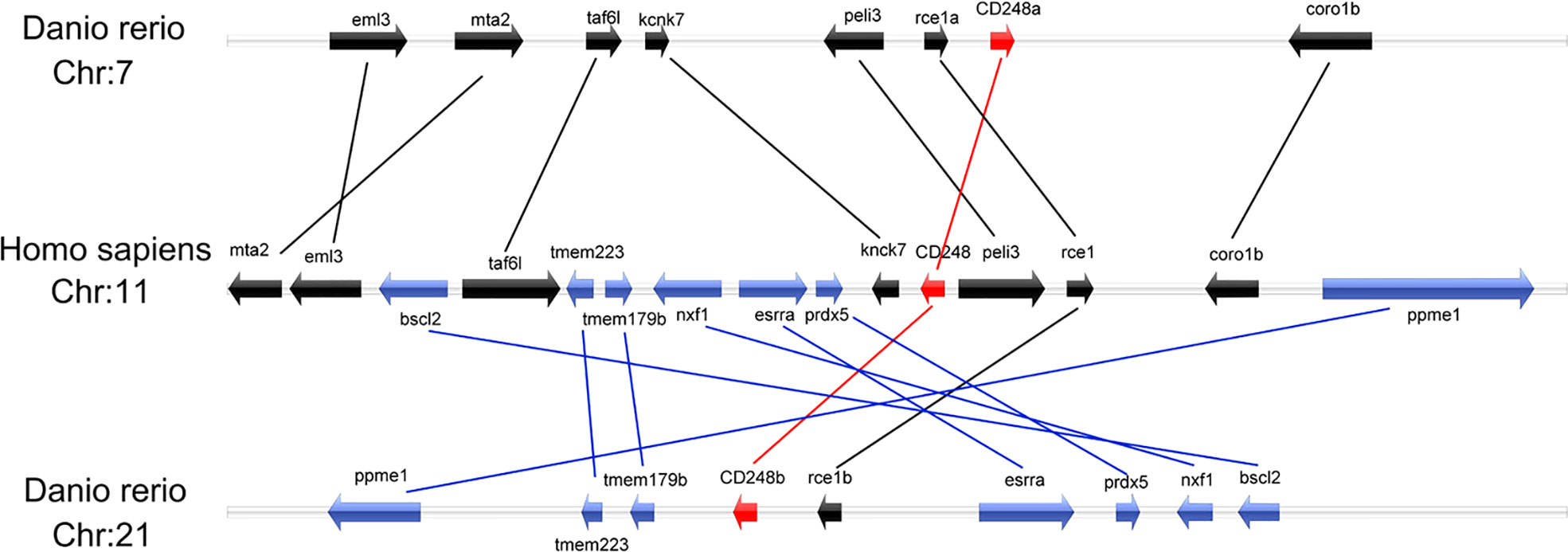
Figure 2 Colinearity analysis between zebrafish cd248a, cd248b and human CD248. All genes were indicated by arrows, and the arrow direction represented the gene direction. Silver strip, chromosome. Chr, chromosome.
Phylogenetic analysis of cd248
CD248 is also a member of C-type lectin domain group 14 family. To determine the relationship between zebrafish Cd248 and corresponding orthologues in different taxa, we conducted a phylogenetic analysis of the predicted protein sequences of C-type lectin domain group 14 family genes across 7 species (human, rat, mouse, frog, chicken, wall lizard and zebrafish) using the neighbor-joining method (Figure 3A) and maximum-likelihood method (Figure 3B). The phylogenetic trees showed that CD248, CD93 and CLEC14A orthologues among various species clustered together and divided into three distinct clades. As expected, zebrafish Cd248a and Cd248b formed a more closely branch, which further supported they were generated by teleost-specific genome duplication.
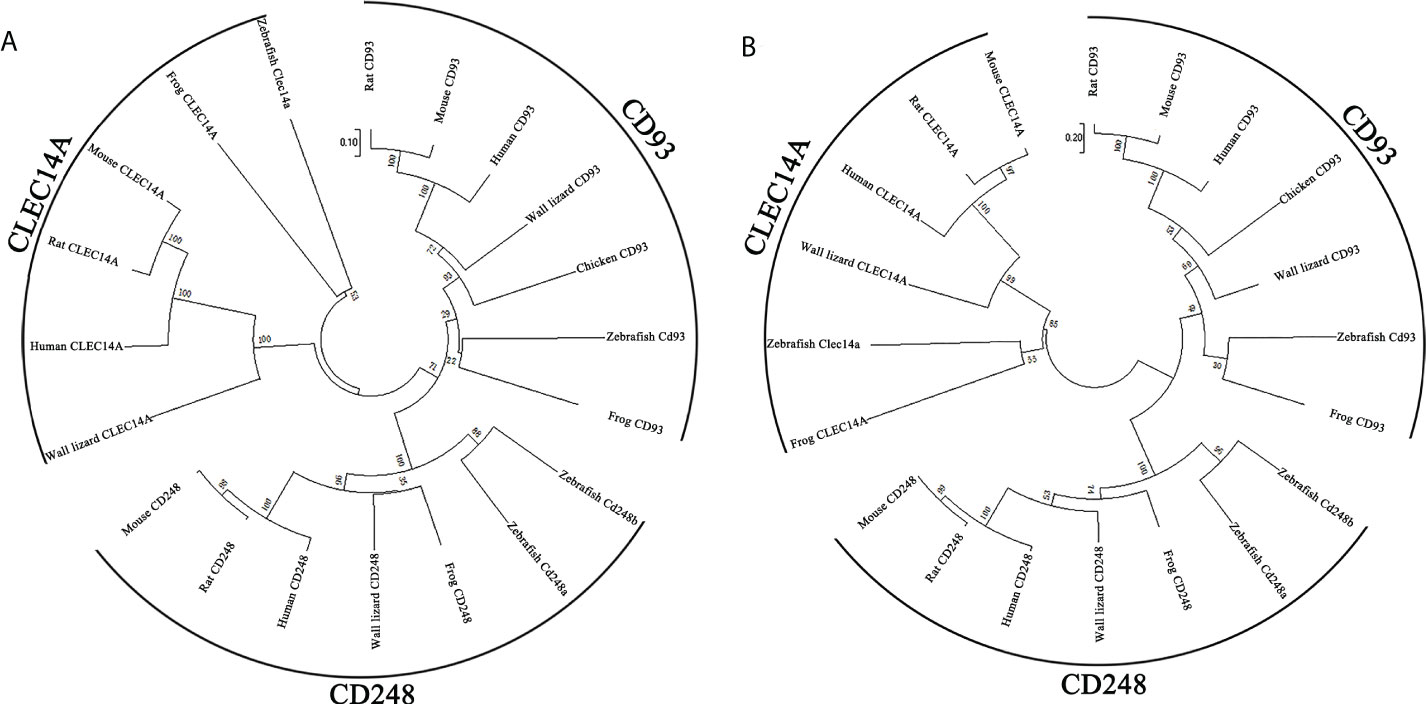
Figure 3 Phylogenetic trees of Cd248a and Cd248b. Phylogenetic trees constructed from the amino acid sequences of homologous genes of various species. CD248, CD93 and CLEC14A are members of the C-type lectin group 14 family. (A) neighbor-joining method; (B) maximum-likelihood method. The reliability of each node was estimated by bootstrapping with 1000 replications. The numbers shown at each node indicate the bootstrap values (%).
Temporospatial expression of cd248
The spatial and temporal expression patterns of cd248a and cd248b were determined by whole-mount in situ hybridization at early stages. cd248a and cd248b exhibited similar expression patterns. cd248a had weak maternal expression (Figures 4A–F). Zygotic transcript of cd248a were first seen in anterior somites and head mesenchyme at 14 hpf (Figures 4G, H); By 24 hpf, the expression was mainly detected in eyes and head mesenchyme (Figure 4I); From 48 to 120 hpf, cd248a showed a gradually declined expression pattern, the signal remained in the eyes, head mesenchyme, Additional expression was also observed in the branchial arches (Figures 4J–M). Weak ubiquitous maternal expression was also seen for cd248b (Figures 5A–E). The expression was observed in anterior somites and head mesenchyme at 10-14 hpf (Figures 5F–H), then shifted to eyes and head mesenchyme from 24 hpf on (Figure 5I); At 48hpf, the expression remained in eyes, head mesenchyme, and also expanded to branchial arches. At later stage, the expression signal gradually declined (Figures 5J–M).
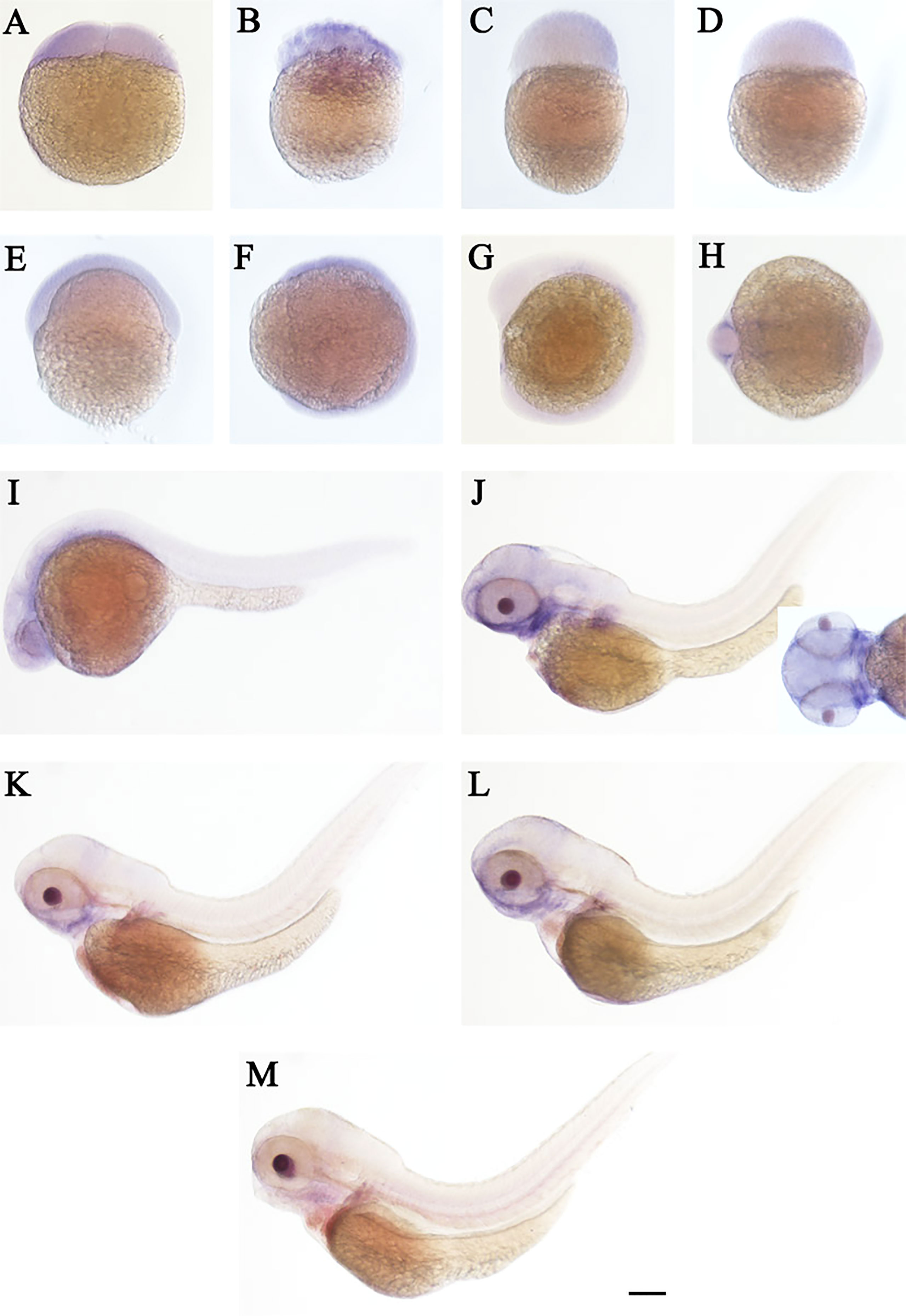
Figure 4 WISH of cd248a in zebrafish early embryos. Stages of embryonic development: (A) 2-cell; (B) 32-cell; (C) 512-cell; (D) sphere; (E) shield; (F) bud; (G, H) 10 somites; (I) 24 hpf; (J) 48 hpf; (K) 72hpf; (L) 96 hpf; (M) 120 hpf. hpf, hours post-fertilization. Scale bar, 100 μm.
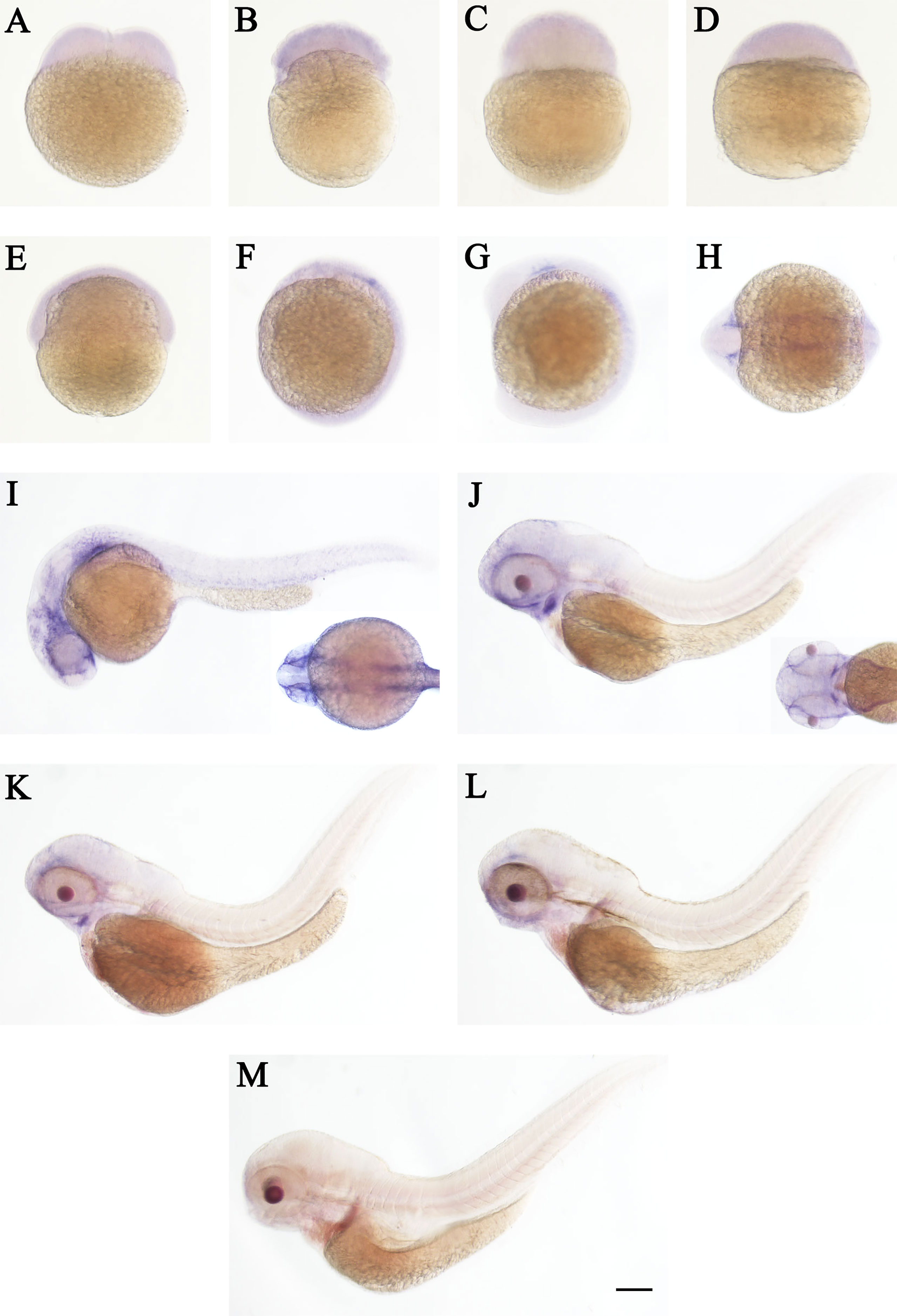
Figure 5 WISH of cd248b in zebrafish early embryos. Stages of embryonic development: (A) 2-cell; (B) 32-cell; (C) 512-cell; (D) sphere; (E) shield; (F) bud; (G, H) 10 somites; (I) 24 hpf; (J) 48 hpf; (K) 72hpf; (L) 96 hpf; (M) 120 hpf. hpf, hours post-fertilization. Scale bar, 100 μm.
Expression responses of cd248 to LPS challenge
The expression profiles of zebrafish cd248 in response to LPS challenge, which mimics infection of pathogenic bacteria, were examined. The dissociation curve of amplified products in all cases showed a single peak, indicating that the amplifications were specific (data not shown). Consistent with the expression pattern in untreated embryos, the mRNA expression level of cd248a in PBS injection group decreased gradually with development, excepted for a slightly higher expression level at 2-4 hours post-injection (hpi) (Figure 6A). By contrast, the mRNA expression of cd248 were upregulated after LPS challenge. The mRNA expression level of cd248a increased soon after injection (2-4 hpi), and decreased at 4-10 hpi, then the expression level increased dramatically at 10-120 hpi (Figure 6A). The expression level of cd248b was highest in 2 hpi, decreased in 2-10 hpi, then remained low in 10-120 hpi (Figure 6B). Upon LPS challenge, the expression level of cd248b decreased in 2-10 hpi and then increased significantly in 10-120 hpi. Taken together, these data demonstrated that zebrafish early embryos/larvae were immunocompetent to LPS challenge via upregulating the expression of cd248.
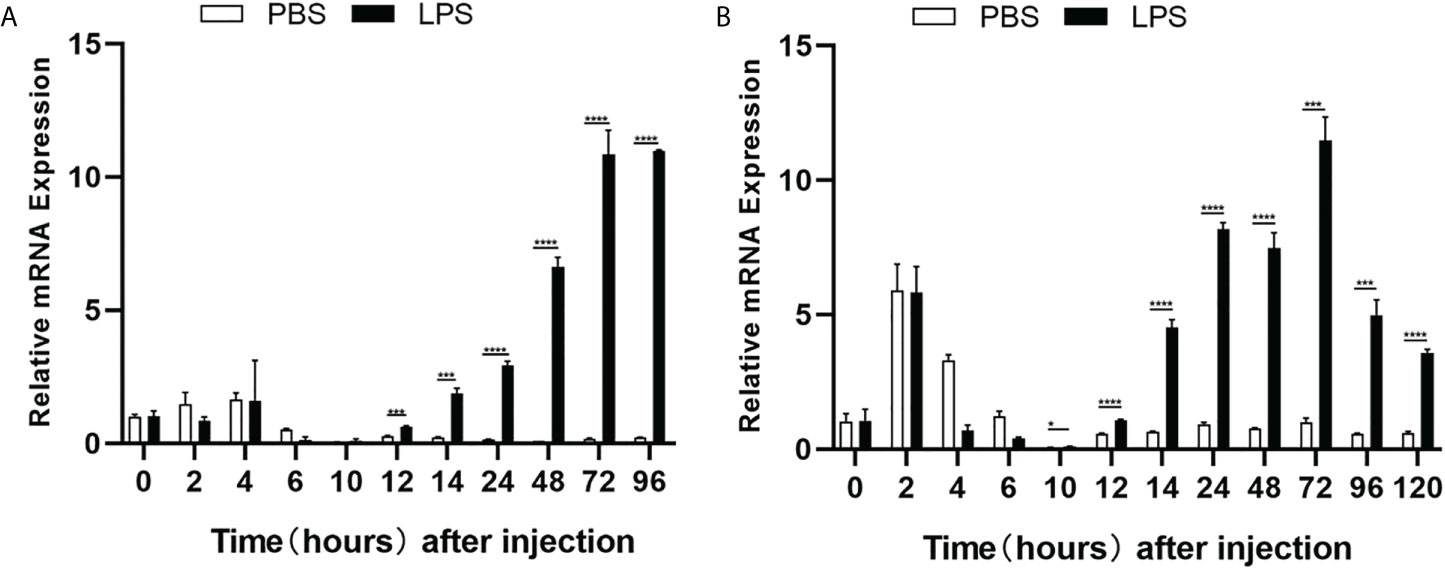
Figure 6 Quantitative analysis of zebrafish cd248a and cd248b genes in response to LPS/PBS treatment. (A) Quantitative analysis of zebrafish cd248a in infected embryos/larvae by microinjecting LPS at 1-cell stage; (B) Quantitative analysis of zebrafish cd248b in infected embryos/larvae by microinjecting LPS at 1-cell stage. PBS, Phosphate Buffered Saline; LPS, lipopolysaccharide. Data were shown as mean ± SD, n = 3. *P < 0.05; ***P < 0.001; ****P < 0.0001.
Cd248 localized on the cell membrane and cytoplasm
To explore subcellular localization of Cd248a and Cd248b, the pcDNA3.1/V5/cd248a/eGFP, pcDNA3.1/V5/cd248b/eGFP, pcDNA3.1/V5/cd248bΔN/eGFP, pcDNA3.1/V5/cd248bΔC/eGFP, pcDNA3.1/V5/cd248bΔNΔC/eGFP and pcDNA3.1/V5/eGFP (control) plasmids were transfected individually into HEK293T cells and stained DAPI at 24h post-transfected. As shown in Figure 7, eGFP was evenly distributed throughout the cell. In contrast, the green fluorescence in both Cd248a-eGFP and Cd248b-eGFP were observed in the periphery of nuclei, indicating they were localized on the cell membrane and cytoplasm. The subcellular localization of Cd248 was consistent with the predicted structures with a transmembrane region. It is striking to note that Cd248b lacking of N- or C-terminal transmembrane region still mainly localized on cell membrane and some in cytoplasm. By contrast, Cd248b lacking both transmembrane regions no longer localized on cell membrane, but in cytoplasm and nuclei as speckles.
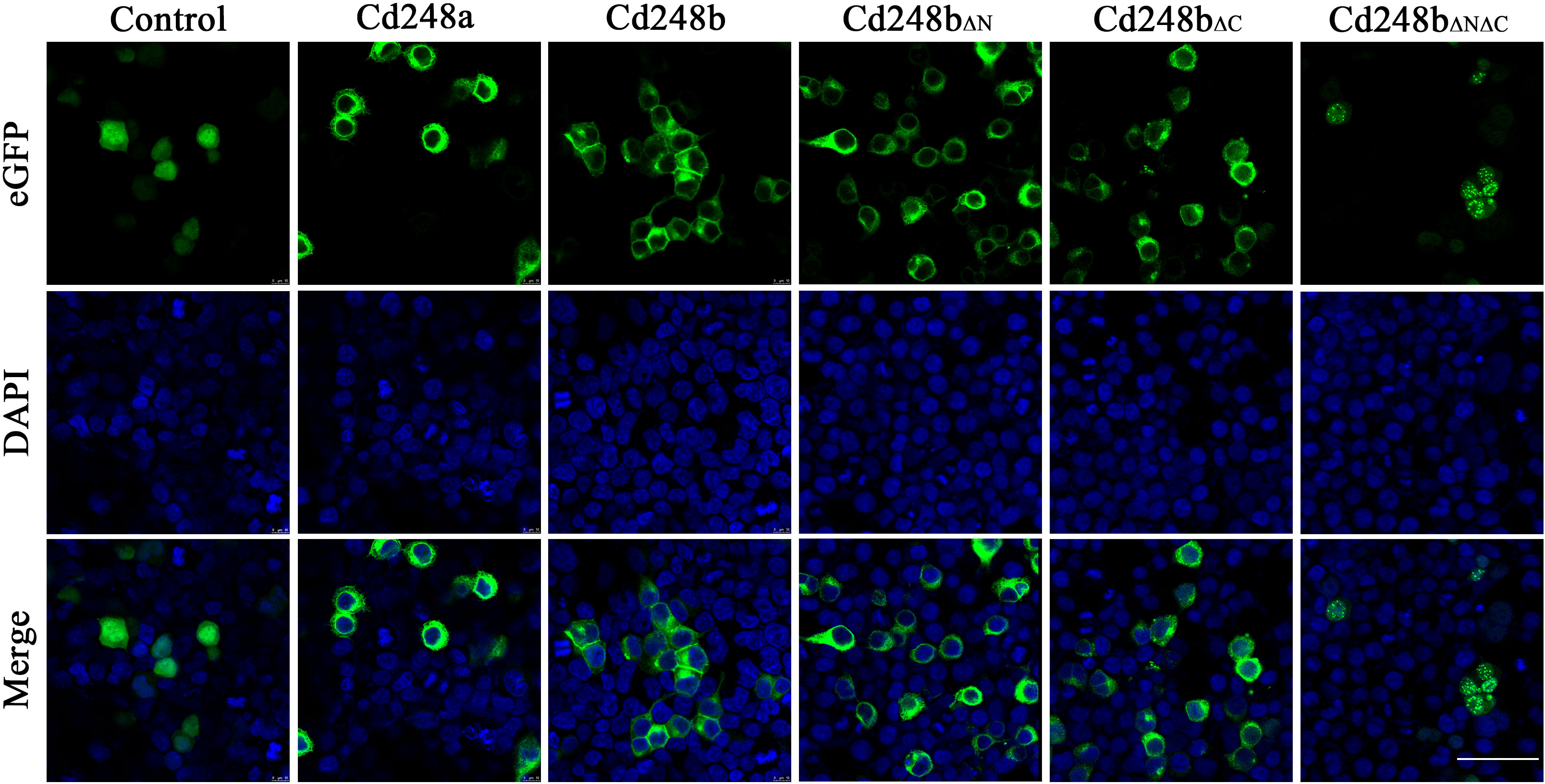
Figure 7 Subcellular localization of Cd248a, Cd248b and truncated Cd248b in HEK293T cells. The pcDNA3.1/V5/Cd248a/eGFP, pcDNA3.1/V5/Cd248b/eGFP and pcDNA3.1/V5/eGFP recombinant plasmids were transfected individually into HET293T cells. The nucleus was stained by DAPI. One representative image for each out of three independent experiments is shown. Scale bar, 50 μm.
Effects of ectopic overexpression of Cd248 on production of pro-inflammatory cytokines in macrophages
Activated macrophages secrete various pro-inflammatory cytokines, such as Il6, Il1β, Tnfα, these cytokines cause a series of inflammatory reactions (29). Therefore, we sought to determine whether overexpression of cd248a and cd248b regulated the expression of pro-inflammatory cytokines in macrophages. To do this, pcDNA3.1/V5/cd248a, pcDNA3.1/V5/cd248b and pcDNA3.1/V5-HisA (control) plasmids were transfected individually into RAW 264.7 cells, and then the expression levels of Il6, Il1β, Tnfα were detected by qRT-PCR. As shown in Figure 8, the expression levels of Il6, Il1β, Tnfα were significantly upregulated by overexpression of cd248a and cd248b, suggesting that overexpression of cd248a and cd248b increased production of pro-inflammatory mediators in macrophages.
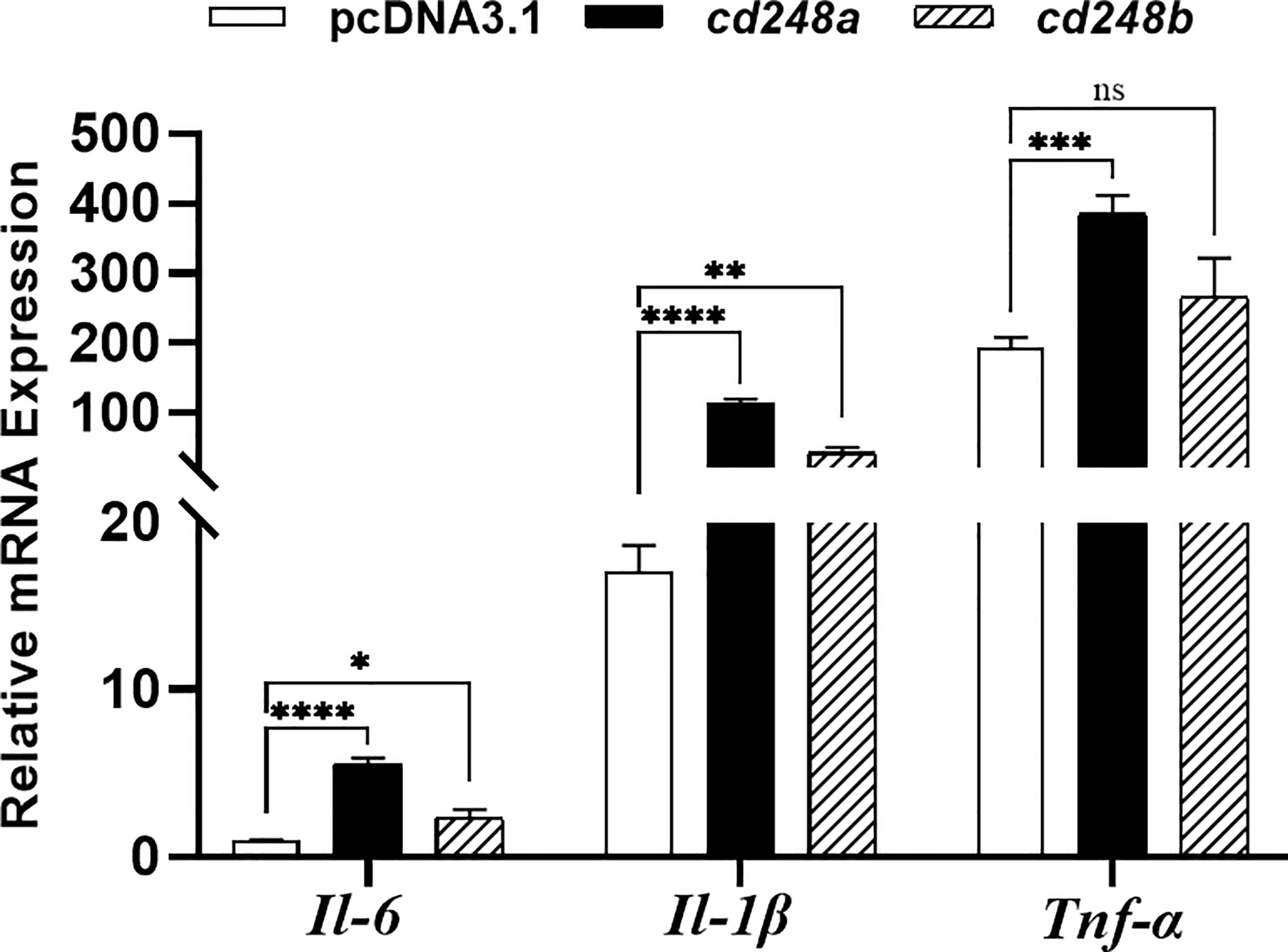
Figure 8 Effect of overexpression of zebrafish cd248a and cd248b on pro-inflammatory cytokines expression in mouse macrophages. The pcDNA3.1/V5/Cd248a, pcDNA3.1/V5/Cd248b and pcDNA3.1/V5-His A recombinant plasmids were transfected individually into RAW264.7 cells. At 24h after transfection, the total RNA of the cells was extracted and cDNA was synthesized. Quantitative analysis of the mRNA expression of mouse Tnfα, Il1β and Il6. Data were shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant.
Effects of ectopic overexpression of cd248 on production of pro-inflammatory cytokines in vivo
Overexpression of cd248a and cd248b could significantly increase the expression of pro-inflammatory cytokines in vitro. To investigate whether overexpression of cd248 has the same effects in vivo, we overexpressed cd248a and cd248b mRNA in zebrafish embryos and verified that they can significantly increase the expression of pro-inflammatory cytokines in vivo (Figures 9A, C). It has been well established that the activation of Notch signal could increase the expression of MMP9 and further increase the expression of inflammatory cytokines (30–33). Our results showed that overexpression of cd248b increased the expression of notch3 and mmp9 (Figure 9D), but that was not the case for cd248a overexpression, which suggested the upregulation of expression pro-inflammatory cytokines upon cd248a overexpression was not regulated by Notch signaling pathway (Figure S1A). We noted VEGF (Vascular endothelial growth factor) signaling pathway is a major regulator of vascular development and lymphatic function, which also plays an important role in tumor growth, retinopathy, tissue inflammation and immunity (34, 35). In this pathway, HIF1A acts as a central link and transmits signals to the downstream PIGF (36–39). PIGF (placental growth factor), is a member of the VEGF family, which can bind VEGFR1 with high affinity, plays a key role in pathological angiogenesis, especially in cancer, cardiovascular, autoimmune and inflammatory diseases (40, 41). Thus, we examined the expression of VEGF signals upon cd248a overexpression, as expected, overexpression of cd248a increased the expression of hif1a and also upregulated the expression of pigf and vegfr1(Figure 9B). However, cd248b overexpression decreased the level of hif1a (Figure S1B). Taken together, the overexpression of cd248a and cd248b upregulated the expression of pro-inflammatory cytokines in vivo by different mechanisms, possibly via VEGF pathway and Notch pathway respectively. These findings suggested that zebrafish cd248a and cd248b participated in the immune response.
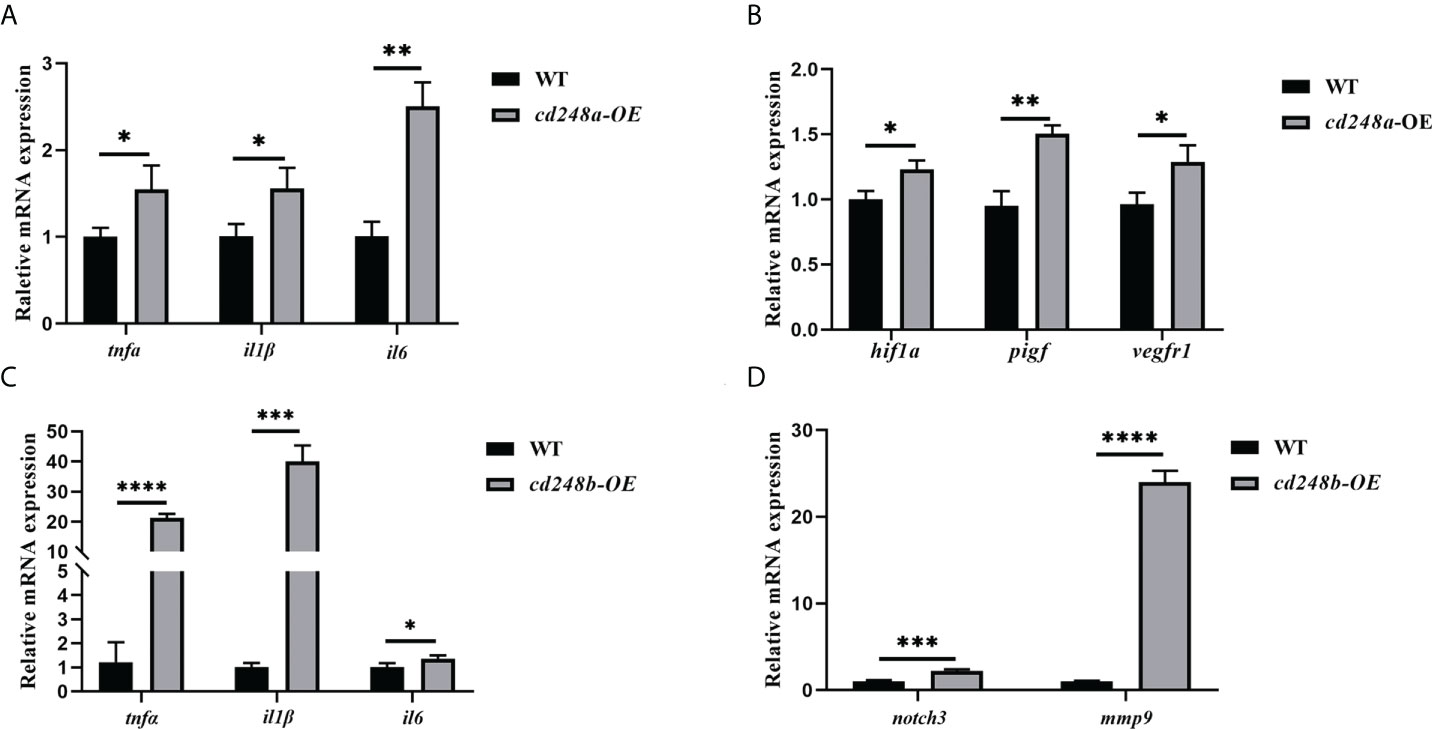
Figure 9 Effect of overexpression of zebrafish cd248a and cd248b on pro-inflammatory cytokines expression in vivo. Diluted mRNAs were injected into one-cell stage embryos of zebrafish. At 48h after injection, the total RNA of the embryos was extracted and cDNA was synthesized. (A, C) Quantitative analysis of the expression of tnfα, il1β and il6 after cd248a and cd248b overexpression in embryos; (B) Quantitative analysis of the expression of hif1a, pigf and vegfr1 after cd248a overexpression in embryos; (D) Quantitative analysis of the expression of notch3 and mmp9 after cd248b overexpression in embryos. cd248a-OE, cd248a overexpression; cd248b-OE, cd248b overexpression. Data were shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Deficiency of cd248 downregulated the expression of pro-inflammatory cytokines and upregulated anti-inflammatory cytokines
To better investigate the role of cd248, we generated cd248a-null mutant and cd248b-null mutant with 23 bp and 10 bp deletions by using the CRISPR/Cas9 system respectively (Figures 10A, B, D, E). The two mutations both caused frameshift in the protein-coding region leading to early termination of translation (Figures 10C, F). Both mRNA level downregulated dramatically in cd248 mutants compared with WT (Figures 10G, H). As described above, overexpression of cd248 could upregulate the expression of pro-inflammatory cytokines, we wondered the effects of impaired cd248 function on the expression of pro-inflammatory cytokines. As expected, both cd248 deletions reduced the expression of pro-inflammatory cytokines (Figure 10I), although no significant difference of the level of tnfα and il6 observed between cd248a mutant and WT (Figure 10I).
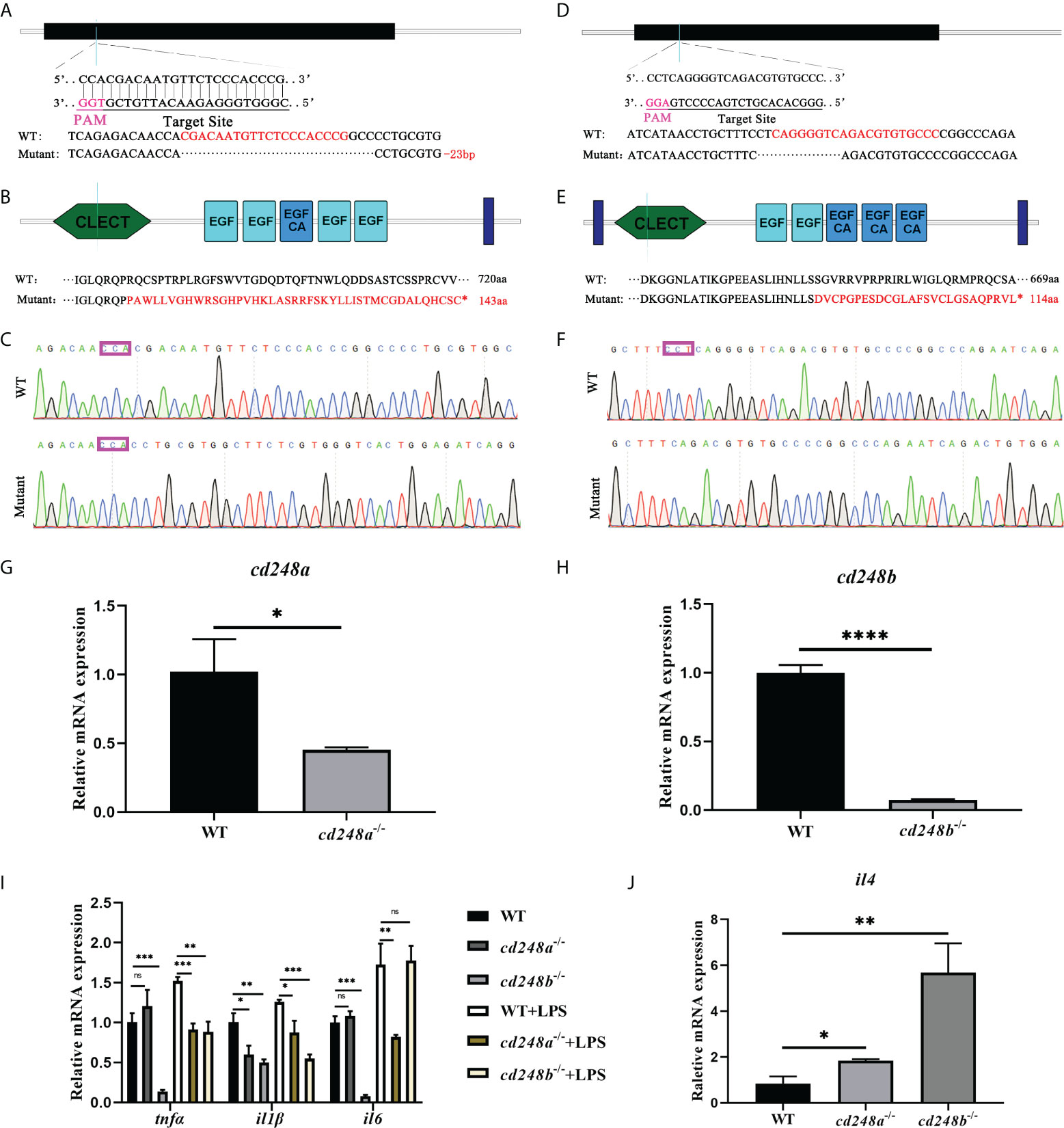
Figure 10 Effect of cd248 deletion on inflammatory cytokines. The mutants of cd248a and cd248b were carried out by CRISPR/Cas9 system. (A–C) CRISPR/Cas9 gRNA design and the genotyping result of the cd248a-mutant allele; (D–F) CRISPR/Cas9 gRNA design and the genotyping result of the cd248b-mutant allele; (G, H) Quantitative analysis of the expression of cd248a or cd248b after cd248a-deletion or cd248b-deletion compared with WT respectively; (I) Quantitative analysis of the expression of zebrafish tnfα, il1β and il6 in the cd248-mutants and LPS treatment. (J) Quantitative analysis of the expression of il4 in the cd248-mutants compared with WT. Data were shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant.
Even after LPS treatment, the expression of pro-inflammatory cytokines in the mutant were lower than that in the WT (Figure 10I). In addition, deficiency of cd248a and cd248b both upregulated the expression of anti-inflammatory factor il4 compared with WT (Figure 10J). Furthermore, the level of VEGF signals in cd248a mutant was comparable with WT (Figure S2A), whereas the expression of Notch signals was reduced in cd248b mutant than WT (Figure S2B). Collectively, these results further confirmed that Cd248 proteins were involved in the innate immune process.
Discussion
CD248 has gained wide attention due to the relevance in human health and disease, particularly in the fields of innate immunity and inflammation (26). However, no definite cd248 orthologs in fish have been documented so far. In this study, we reported the identification of cd248 in zebrafish D. rerio, the first cd248 orthologs in fish.
Zebrafish cd248a and cd248b are orthologs of human CD248 supported by several lines of evidences. Both cd248a and cd248b in zebrafish are intronless, like CD248 homologs in human and mice (15). Zebrafish Cd248 proteins shared similar protein domains as that of other species. No CCP domain was found in human CD248 by SMART domain prediction online tools, although it is reported that a CCP domain is presented in human CD248. The discrepancy may be caused by different tools and different algorithms. In addition, the CCP domain also exists in CD248 of rat, wall lizard, and clawed frog by SMART domain prediction online tools. However, there was no CCP domain in Cd248b, which may be because of its loss during genome duplication. Moreover, there was no CCP domain in Cd248 of medaka, but CCP domain exists in Cd248-like of medaka. This conclusion further confirmed that one of Cd248 lost the CCP domain in the process of fish genome duplication. In humans, the CD248 is located on chromosome 11, while zebrafish cd248a and cd248b are located on chromosome 7 and 21, respectively. Despite the order and direction are different, the genes upstream and downstream of CD248 displayed fine synteny in zebrafish and human. This conserved synteny strongly suggested that cd248a and cd248b are co-orthologs of the human CD248 gene. Alignment with those of other group XIV CTLD-containing proteins revealed that Cd248 shared the highest homology to CD248 (Data not shown). The two independent phylogenetic trees have firmly clustered the zebrafish Cd248a and Cd248b with other vertebrate CD248 homologs. In addition, zebrafish Cd248a and Cd248b shared highly homology, and formed a more closely branch in the phylogenetic tree, which further supported they were generated by teleost-specific genome duplication. We failed to identify authentic CD248 orthologs in invertebrates (such as Amphioxus, Drosophila, Nematodes), CD248 is a member of the group XIV of C-type lectin domain-containing proteins (CTLDcps). CTLDcps appeared with the advent of early vertebrates after a whole genome duplication followed by a sporadic tandem duplication (42). Therefore, we speculated CD248 were arisen only in the vertebrate lineage. It is interesting to note that chicken CD248 (according to NCBI database) clustered together with other species CD93, thus we determined chicken CD248 is actually CD93, not a CD248 ortholog, this is consistent with reported that the losing CD248 in birds was due to chromosomal breakage (42).
Human CD248 is known to be a single transmembrane cell surface glycoprotein (4). The Domain prediction has shown Cd248a possesses a single transmembrane domain while Cd248b contains two transmembrane domains located near the N- and C-termini. The subcellular localization experiment has confirmed both Cd248a and Cd248b expressed on the cell membrane and cytoplasm. Dfi1 protein also contains two transmembrane domains. Similarly, Dfi1 lacking the N-terminal transmembrane region was observed at the cell periphery where full-length Dfi1 normally localizes, whereas the double-truncation mutant was diffusely intracellular (43). Further data suggested the two transmembrane domains contributed to the biogenesis of Dfi1 (43). Cd248b lacking either of transmembrane region still mainly expressed in the cell membrane and cytoplasm, whereas double transmembrane domains deletion mutant was localized in the cytoplasm and nuclei as speckles. These data suggested the two transmembrane domains possibly contributed to the biogenesis of Cd248b and the function of the two transmembrane domains is worthy of further exploration.
CD248 is predominantly expressed on stromal fibroblasts throughout the mesenchyme and on the developing vasculature in mice embryos. The expression of endosialin gradually downregulated during development resulting in great loss of expression in adult tissues (3). In this study, we confirmed that cd248a and cd248b exhibited similar expression patterns in zebrafish. Both genes had weak ubiquitous expression at early stage, the zygotic transcripts were first detected in the head mesenchyme and anterior somite, then mainly in the head mesenchyme, eyes and branchial arches, and gradually declined with development, implying that these two genes may be involved in the formation these tissues.
CD248 is a member of C-type lectin domain group 14 family, which also contains TM (thrombomodulin), CD93 and CLEC14A (6). To date, there are many members of C-type lectin family, served as pattern recognition receptors, play important roles in immune responses (24, 44–52). Loss-of-function studies in mice implied CD248 promoted tumor growth and inflammation (26). Zebrafish Cd248a and Cd248b, members of C-type lectin, were expressed on the cell membrane and cytoplasm, we wondered whether zebrafish Cd248 participated in the immune response. The mRNA expression of cd248a and cd248b were upregulated upon LPS treatment, suggesting cd248a and cd248b were involved in innate immunity of zebrafish. We found that overexpression of cd248a and cd248b both upregulated the expression of pro-inflammatory cytokines in vitro and in vivo. These results suggested that cd248a and cd248b were involved in innate immune responses. It is worth noting that cd248a upregulated the expression of pro-inflammatory cytokines possibly via the VEGF signaling pathway. But cd248b increased the expression of pro-inflammatory cytokines possibly via Notch signaling pathway. Although our study was conducted in zebrafish embryos, and we confirmed overexpression and knockout of cd248a or cd248b influenced on the expression of pro-inflammatory cytokines. We found the link of pro-inflammatory cytokines and VEGF signal or Notch signal in the references, although the data were derived from disease and immunity. Interestingly, in the transcriptomic data of RNA-seq, we noted VEGF signal in cd248a mutant, Notch signal in cd248b mutants changes compared to that in Wild type (Data not shown). Hence, we examined VEGF signal or Notch signal and found that overexpression and knockout of cd248a changed VEGF signal but not Notch signal, overexpression and knockout of cd248b changed Notch signal but not VEGF signal. Therefore, we speculated that cd248a and cd248b promoted pro-inflammatory cytokines possibly via these two signaling pathways respectively. It was uncertain whether cd248a or cd248b directly acts through VEGF signaling pathway or Notch signaling pathway on pro-inflammatory cytokines in zebrafish. It will be interesting to elucidate whether there were other molecules involved and whether other signaling pathways were participated. It was previously reported that human CD248 inhibits the Notch signaling pathway and then activates the expression of pro-inflammatory cytokines (53), which was inconsistent with our conclusion that cd248b activated the Notch signaling pathway and then activated the expression of pro-inflammatory cytokines. In fact, it has been reported that inhibiting Notch signaling pathway promotes the expression of pro-inflammatory cytokines (54), and others reported that activating Notch signaling pathway promotes the expression of pro-inflammatory cytokines (55–60). Different conclusions may be due to different functions of CD248 in different species. In mice, fibronectin and type I and V collagen have been identified as specific ligands of CD248 (1). CD248 promotes inflammation possibly via intracellular signaling via its cytoplasmic tail and/or through extracellular matrix ligands that interact with the ectodomain (15). In zebrafish, to identify the specific ligands of Cd248 protein may help to elucidate how they participated in innate immune responses.
The zebrafish cd248 mutants had no visible phenotype, and we did not observe any cartilage defects by Alcian Blue staining compared with WT (data not shown). Regrettably, we had no specific antibodies against Cd248a protein or Cd248b protein to verify the experimental results at the protein level in the mutants. We only examined the mRNA expression level of the mutants, and the results showed that mRNA levels of cd248a and cd248b were decreased. The absence of cd248a and cd248b both downregulated expression of pro-inflammatory cytokines, but the regulatory level of cd248a was weaker than that of cd248b, which is in part because the more decreased of mRNA level of cd248b in the mutant. Additionally, we will try to obtain the double-mutant of zebrafish cd248 in order to better elucidate the role of Cd248 protein in immune system.
In summary, we identified two orthologs of CD248, cd248a and cd248b, in zebrafish. The duplicated cd248 resemble the mammalian counterpart and exhibited similar expression pattern. Both cd248a and cd248b could be upregulated by LPS challenge. Moreover, overexpression of cd248a or cd248b promoted the production of pro-inflammatory cytokines. Loss-of-function suggested that zebrafish cd248a and cd248b were involved in innate immune process. However, the possible ligands of Cd248 haven’t been elucidated. Additionally, whether the duplicated Cd248 were redundantly involved in early development remains to be further investigated.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
This study was reviewed and approved by Animal Committee of Ocean University of China.
Author contributions
All authors have read and approved the manuscript. XL and RG performed the experiments, wrote and revised the manuscript. SY, XZ, XY, LT, SZ analyzed the data. GJ revised the manuscript. HL designed the experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grants (31872187, 32171139) of Natural Science Foundation of China (NSFC) and grant (2018YFD0900502) of the Ministry of Science and Technology (MOST) of China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.970626/full#supplementary-material
References
1. Tomkowicz B, Rybinski K, Foley B, Ebel W, Kline B, Routhier E, et al. Interaction of endosialin/TEM1 with extracellular matrix proteins mediates cell adhesion and migration. Proc Natl Acad Sci U.S.A. (2007) 104(46):17965–70. doi: 10.1073/pnas.0705647104
2. MacFadyen JR, Haworth O, Roberston D, Hardie D, Webster MT, Morris HR, et al. Endosialin (TEM1, CD248) is a marker of stromal fibroblasts and is not selectively expressed on tumour endothelium. FEBS Lett (2005) 579(12):2569–75. doi: 10.1016/j.febslet.2005.03.071
3. MacFadyen J, Savage K, Wienke D, Isacke CM. Endosialin is expressed on stromal fibroblasts and CNS pericytes in mouse embryos and is downregulated during development. Gene Expr Patterns (2007) 7(3):363–9. doi: 10.1016/j.modgep.2006.07.006
4. Rettig WJ, Garin-Chesa P, Healey JH, Su SL, Jaffe EA, Old LJ. Identification of endosialin, a cell surface glycoprotein of vascular endothelial cells in human cancer. Proc Natl Acad Sci U.S.A. (1992) 89(22):10832–6. doi: 10.1073/pnas.89.22.10832
5. Khan KA, Naylor AJ, Khan A, Noy PJ, Mambretti M, Lodhia P, et al. Multimerin-2 is a ligand for group 14 family c-type lectins CLEC14A, CD93 and CD248 spanning the endothelial pericyte interface. Oncogene (2017) 36(44):6097–108. doi: 10.1038/onc.2017.214
6. Khan KA, McMurray JL, Mohammed F, Bicknell R. C-type lectin domain group 14 proteins in vascular biology, cancer and inflammation. FEBS J (2019) 286(17):3299–332. doi: 10.1111/febs.14985
7. Teicher BA. CD248: A therapeutic target in cancer and fibrotic diseases. Oncotarget (2019) 10(9):993–1009. doi: 10.18632/oncotarget.26590
8. Di Benedetto P, Liakouli V, Ruscitti P, Berardicurti O, Carubbi F, Panzera N, et al. Blocking CD248 molecules in perivascular stromal cells of patients with systemic sclerosis strongly inhibits their differentiation toward myofibroblasts and proliferation: a new potential target for antifibrotic therapy. Arthritis Res Ther (2018) 20(1):223. doi: 10.1186/s13075-018-1719-4
9. Hasanov Z, Ruckdeschel T, Konig C, Mogler C, Kapel SS, Korn C, et al. Endosialin promotes atherosclerosis through phenotypic remodeling of vascular smooth muscle cells. Arterioscler Thromb Vasc Biol (2017) 37(3):495–505. doi: 10.1161/ATVBAHA.116.308455
10. Mogler C, Wieland M, Konig C, Hu J, Runge A, Korn C, et al. Hepatic stellate cell-expressed endosialin balances fibrogenesis and hepatocyte proliferation during liver damage. EMBO Mol Med (2015) 7(3):332–8. doi: 10.15252/emmm.201404246
11. Petrus P, Fernandez TL, Kwon MM, Huang JL, Lei V, Safikhan NS, et al. Specific loss of adipocyte CD248 improves metabolic health via reduced white adipose tissue hypoxia, fibrosis and inflammation. EBioMedicine (2019) 44:489–501. doi: 10.1016/j.ebiom.2019.05.057
12. Smith SW, Croft AP, Morris HL, Naylor AJ, Huso DL, Isacke CM, et al. Genetic deletion of the stromal cell marker CD248 (Endosialin) protects against the development of renal fibrosis. Nephron (2015) 131(4):265–77. doi: 10.1159/000438754
13. Bartis D, Crowley LE, D'Souza VK, Borthwick L, Fisher AJ, Croft AP, et al. Role of CD248 as a potential severity marker in idiopathic pulmonary fibrosis. BMC Pulm Med (2016) 16(1):51. doi: 10.1186/s12890-016-0211-7
14. Choi IY, Karpus ON, Turner JD, Hardie D, Marshall JL, de Hair MJH, et al. Stromal cell markers are differentially expressed in the synovial tissue of patients with early arthritis. PLoS One (2017) 12(8):e0182751. doi: 10.1371/journal.pone.0182751
15. Maia M, de Vriese A, Janssens T, Moons M, van Landuyt K, Tavernier J, et al. CD248 and its cytoplasmic domain: a therapeutic target for arthritis. Arthritis Rheum (2010) 62(12):3595–606. doi: 10.1002/art.27701
16. Rybinski K, Imtiyaz HZ, Mittica B, Drozdowski B, Fulmer J, Furuuchi K, et al. Targeting endosialin/CD248 through antibody-mediated internalization results in impaired pericyte maturation and dysfunctional tumor microvasculature. Oncotarget (2015) 6(28):25429–40. doi: 10.18632/oncotarget.4559
17. Bagley RG, Honma N, Weber W, Boutin P, Rouleau C, Shankara S, et al. Endosialin/TEM 1/CD248 is a pericyte marker of embryonic and tumor neovascularization. Microvasc Res (2008) 76(3):180–8. doi: 10.1016/j.mvr.2008.07.008
18. Lax S, Ross EA, White A, Marshall JL, Jenkinson WE, Isacke CM, et al. CD248 expression on mesenchymal stromal cells is required for post-natal and infection-dependent thymus remodelling and regeneration. FEBS Open Bio (2012) 2:187–90. doi: 10.1016/j.fob.2012.07.003
19. Lax S, Hardie DL, Wilson A, Douglas MR, Anderson G, Huso D, et al. The pericyte and stromal cell marker CD248 (endosialin) is required for efficient lymph node expansion. Eur J Immunol (2010) 40(7):1884–9. doi: 10.1002/eji.200939877
20. Hardie DL, Baldwin MJ, Naylor A, Haworth OJ, Hou TZ, Lax S, et al. The stromal cell antigen CD248 (endosialin) is expressed on naive CD8+ human T cells and regulates proliferation. Immunology (2011) 133(3):288–95. doi: 10.1111/j.1365-2567.2011.03437.x
21. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol (2016) 33(7):1870–4. doi: 10.1093/molbev/msw054
22. Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc (2008) 3(1):59–69. doi: 10.1038/nprot.2007.514
23. Wang M, Li L, Guo Q, Zhang S, Ji D, Li H. Identification and expression of a new Ly6 gene cluster in zebrafish danio rerio, with implications of being involved in embryonic immunity. Fish Shellfish Immunol (2016) 54:230–40. doi: 10.1016/j.fsi.2016.04.005
24. Yang Q, Wang P, Wang S, Wang Y, Feng S, Zhang S, et al. The hepatic lectin of zebrafish binds a wide range of bacteria and participates in immune defense. Fish Shellfish Immunol (2018) 82:267–78. doi: 10.1016/j.fsi.2018.08.012
25. Liu Y, Zhang C, Zhang Y, Lin S, Shi DL, Shao M. Highly efficient genome editing using oocyte-specific zcas9 transgenic zebrafish. J Genet Genomics (2018) 45(9):509–12. doi: 10.1016/j.jgg.2018.05.004
26. Valdez Y, Maia M, Conway EM. CD248: reviewing its role in health and disease. Curr Drug Targets (2012) 13(3):432–9. doi: 10.2174/138945012799424615
27. Glasauer SM, Neuhauss SC. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics (2014) 289(6):1045–60. doi: 10.1007/s00438-014-0889-2
28. Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol (1999) 11(6):699–704. doi: 10.1016/S0955-0674(99)00039-3
29. Youn GS, Lee KW, Choi SY, Park J. Overexpression of HDAC6 induces pro-inflammatory responses by regulating ROS-MAPK-NF-kappaB/AP-1 signaling pathways in macrophages. Free Radic Biol Med (2016) 97:14–23. doi: 10.1016/j.freeradbiomed.2016.05.014
30. Li L, Tan J, Zhang Y, Han N, Di X, Xiao T, et al. DLK1 promotes lung cancer cell invasion through upregulation of MMP9 expression depending on notch signaling. PLoS One (2014) 9(3):e91509. doi: 10.1371/journal.pone.0091509
31. Funahashi Y, Shawber CJ, Sharma A, Kanamaru E, Choi YK, Kitajewski J. Notch modulates VEGF action in endothelial cells by inducing matrix metalloprotease activity. Vasc Cell (2011) 3(1):2. doi: 10.1186/2045-824X-3-2
32. Halade GV, Jin YF, Lindsey ML. Matrix metalloproteinase (MMP)-9: a proximal biomarker for cardiac remodeling and a distal biomarker for inflammation. Pharmacol Ther (2013) 139(1):32–40. doi: 10.1016/j.pharmthera.2013.03.009
33. Medeiros NI, Gomes JAS, Correa-Oliveira R. Synergic and antagonistic relationship between MMP-2 and MMP-9 with fibrosis and inflammation in chagas' cardiomyopathy. Parasite Immunol (2017) 39(8):e12446. doi: 10.1111/pim.12446
34. Shaik-Dasthagirisaheb YB, Varvara G, Murmura G, Saggini A, Potalivo G, Caraffa A, et al. Vascular endothelial growth factor (VEGF), mast cells and inflammation. Int J Immunopathol Pharmacol (2013) 26(2):327–35. doi: 10.1177/039463201302600206
35. Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: Beyond discovery and development. Cell (2019) 176(6):1248–64. doi: 10.1016/j.cell.2019.01.021
36. Syukri A, Budu, Hatta M, Amir M, Rohman MS, Mappangara I, et al. Doxorubicin induced immune abnormalities and inflammatory responses via HMGB1, HIF1-alpha and VEGF pathway in progressive of cardiovascular damage. Ann Med Surg (2022) 76:103501. doi: 10.1016/j.amsu.2022.103501
37. Liu X, Du J, Xi Q. HIF-1alpha ODD polypeptides increased the expression of HIF1 and VEGF in hypoxic rat cortical neuron. Neurol Sci (2011) 32(6):1029–33. doi: 10.1007/s10072-011-0574-8
38. Khromova NV, Kopnin PB, Stepanova EV, Agapova LS, Kopnin BP. p53 hot-spot mutants increase tumor vascularization via ROS-mediated activation of the HIF1/VEGF-a pathway. Cancer Lett (2009) 276(2):143–51. doi: 10.1016/j.canlet.2008.10.049
39. Konstantina A, Lazaris AC, Ioannidis E, Liossi A, Aroni K. Immunohistochemical expression of VEGF, HIF1-a, and PlGF in malignant melanomas and dysplastic nevi. Melanoma Res (2011) 21(5):389–94. doi: 10.1097/CMR.0b013e328347ee33
40. Caporale A, Martin AD, Capasso D, Foca G, Sandomenico A, D'Andrea LD, et al. Short PlGF-derived peptides bind VEGFR-1 and VEGFR-2 in vitro and on the surface of endothelial cells. J Pept Sci (2019) 25(5):e3146. doi: 10.1002/psc.3146
41. Anisimov A, Leppanen VM, Tvorogov D, Zarkada G, Jeltsch M, Holopainen T, et al. The basis for the distinct biological activities of vascular endothelial growth factor receptor-1 ligands. Sci Signal (2013) 6(282):ra52. doi: 10.1126/scisignal.2003905
42. Barbera S, Cucini C. A glimpse into the past: phylogenesis and protein domain analysis of the group XIV of c-type lectins in vertebrates. BMC Genomics (2022) 23(1):420. doi: 10.1186/s12864-022-08659-6
43. Herwald SE, Zucchi PC, Tan S, Kumamoto CA. The two transmembrane regions of candida albicans Dfi1 contribute to its biogenesis. Biochem Biophys Res Commun (2017) 488(1):153–8. doi: 10.1016/j.bbrc.2017.04.158
44. Poisa-Beiro L, Dios S, Ahmed H, Vasta GR, Martinez-Lopez A, Estepa A, et al. Nodavirus infection of sea bass (Dicentrarchus labrax) induces up-regulation of galectin-1 expression with potential anti-inflammatory activity. J Immunol (2009) 183(10):6600–11. doi: 10.4049/jimmunol.0801726
45. He T, Xiao Z, Liu Q, Ma D, Xu S, Xiao Y, et al. Ontogeny of the digestive tract and enzymes in rock bream oplegnathus fasciatus (Temminck et schlegel 1844) larvae. Fish Physiol Biochem (2012) 38(2):297–308. doi: 10.1007/s10695-011-9507-y
46. Zhao ZY, Yin ZX, Weng SP, Guan HJ, Li SD, Xing K, et al. Profiling of differentially expressed genes in hepatopancreas of white spot syndrome virus-resistant shrimp (Litopenaeus vannamei) by suppression subtractive hybridisation. Fish Shellfish Immunol (2007) 22(5):520–34. doi: 10.1016/j.fsi.2006.07.003
47. Willment JA, Brown GD. C-type lectin receptors in antifungal immunity. Trends Microbiol (2008) 16(1):27–32. doi: 10.1016/j.tim.2007.10.012
48. Luo T, Yang H, Li F, Zhang X, Xu X. Purification, characterization and cDNA cloning of a novel lipopolysaccharide-binding lectin from the shrimp penaeus monodon. Dev Comp Immunol (2006) 30(7):607–17. doi: 10.1016/j.dci.2005.10.004
49. Qu B, Yang S, Ma Z, Gao Z, Zhang S. A new LDLa domain-containing c-type lectin with bacterial agglutinating and binding activity in amphioxus. Gene (2016) 594(2):220–8. doi: 10.1016/j.gene.2016.09.009
50. Ling E, Yu XQ. Cellular encapsulation and melanization are enhanced by immulectins, pattern recognition receptors from the tobacco hornworm manduca sexta. Dev Comp Immunol (2006) 30(3):289–99. doi: 10.1016/j.dci.2005.05.005
51. Odintsova NA, Belogortseva NI, Khomenko AV, Chikalovets IV, Luk'yanov PA. Effect of lectin from the ascidian on the growth and the adhesion of HeLa cells. Mol Cell Biochem (2001) 221(1-2):133–8. doi: 10.1023/A:1010936701297
52. Tsutsui S, Iwamoto K, Nakamura O, Watanabe T. Yeast-binding c-type lectin with opsonic activity from conger eel (Conger myriaster) skin mucus. Mol Immunol (2007) 44(5):691–702. doi: 10.1016/j.molimm.2006.04.023
53. Maia M, DeVriese A, Janssens T, Moons M, Lories RJ, Tavernier J, et al. CD248 facilitates tumor growth via its cytoplasmic domain. BMC Cancer (2011) 11:162. doi: 10.1186/1471-2407-11-162
54. Munch J, Grivas D, Gonzalez-Rajal A, Torregrosa-Carrion R, de la Pompa JL. Notch signalling restricts inflammation and serpine1 expression in the dynamic endocardium of the regenerating zebrafish heart. Development (2017) 144(8):1425–40. doi: 10.1242/dev.143362
55. Liu ZJ, Tan Y, Beecham GW, Seo DM, Tian R, Li Y, et al. Notch activation induces endothelial cell senescence and pro-inflammatory response: implication of notch signaling in atherosclerosis. Atherosclerosis (2012) 225(2):296–303. doi: 10.1016/j.atherosclerosis.2012.04.010
56. Qu SY, Lin JJ, Zhang J, Song LQ, Yang XM, Wu CG. Notch signaling pathway regulates the growth and the expression of inflammatory cytokines in mouse basophils. Cell Immunol (2017) 318:29–34. doi: 10.1016/j.cellimm.2017.05.005
57. Wei Z, Chigurupati S, Arumugam TV, Jo DG, Li H, Chan SL. Notch activation enhances the microglia-mediated inflammatory response associated with focal cerebral ischemia. Stroke (2011) 42(9):2589–94. doi: 10.1161/STROKEAHA.111.614834
58. Wang T, Xiang Z, Wang Y, Li X, Fang C, Song S, et al. (-)-Epigallocatechin gallate targets notch to attenuate the inflammatory response in the immediate early stage in human macrophages. Front Immunol (2017) 8:433. doi: 10.3389/fimmu.2017.00433
59. Colombo M, Galletti S, Bulfamante G, Falleni M, Tosi D, Todoerti K, et al. Multiple myeloma-derived jagged ligands increases autocrine and paracrine interleukin-6 expression in bone marrow niche. Oncotarget (2016) 7(35):56013–29. doi: 10.18632/oncotarget.10820
Keywords: zebrafish, cd248a, cd248b, immune, LPS, inflammatory cytokines
Citation: Li X, Guo R, Yang S, Zhang X, Yin X, Teng L, Zhang S, Ji G and Li H (2022) Cd248a and Cd248b in zebrafish participate in innate immune responses. Front. Immunol. 13:970626. doi: 10.3389/fimmu.2022.970626
Received: 17 June 2022; Accepted: 15 August 2022;
Published: 31 August 2022.
Edited by:
Yong-hua Hu, Chinese Academy of Tropical Agricultural Sciences, ChinaReviewed by:
Jiejie Sun, Dalian Ocean University, ChinaGuorong Li, Shandong Normal University, Jinan, China
Copyright © 2022 Li, Guo, Yang, Zhang, Yin, Teng, Zhang, Ji and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyan Li, aG9uZ3lhbmxpQG91Yy5lZHUuY24=; Guangdong Ji, amFtZXNkb25nQG91Yy5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Xianpeng Li1†
Xianpeng Li1† Shicui Zhang
Shicui Zhang Guangdong Ji
Guangdong Ji Hongyan Li
Hongyan Li