
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 18 August 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.968777
This article is part of the Research Topic Novel Biomarkers for Predicting Response to Cancer Immunotherapy View all 69 articles
Background: The SCF (Skp1-cullin-F-box proteins) complex is the largest family of E3 ubiquitin ligases that mediate multiple specific substrate proteins degradation. Two ring-finger family members RBX1/ROC1 and RBX2/RNF7/SAG are small molecular proteins necessary for ubiquitin ligation activity of the multimeric SCF complex. Accumulating evidence indicated the involvement of RBX proteins in the pathogenesis and development of cancers, but no research using pan-cancer analysis for evaluating their difference has been directed previously.
Methods: We investigated RBX1/2 expression patterns and the association with clinicopathological features, and survivals of cancer patients obtained from the TCGA pan-cancer data. The binding energies of RBX1/2-CUL1 complexes were preliminarily calculated by using molecular dynamics simulations. Meanwhile, we assessed their immune infiltration level across numerous databases, including TISIDB and Timer database.
Results: High expression levels of RBX1/2 were observed in most cancer types and correlated with poor prognosis of patients analyzed. Nonetheless, exceptions were observed: RBX2 expression in KICH was higher than normal renal tissues and played a detrimental role in KICH. The expression of RBX1 was not associated with the prognostic risk of KICH. Moreover, the combination of RBX1 and CUL1 expression is more stable than that of RBX2 and CUL1. RBX1/2 expression showed their own specific characteristics in tumor pathological stages and grades, copy number variation and immune components.
Conclusions: These findings not only indicated that the difference of RBX1/2 might result in varying degrees of tumor progression, but also suggested that they might serve as biomarkers for immune infiltration in cancers, shedding new light on therapeutics of cancers.
The ubiquitin–proteasome system (UPS) is the major proteolytic system that degrades accumulated or misfolded proteins for cellular homeostasis (1, 2). It operates through the presentation of ubiquitin to the substrate proteins using a covalent modification pattern, which involves a series of multienzymes, i.e., Ubiquitin (Ub)-activating enzyme (E1), Ub-conjugating enzyme (E2) and Ub ligase (E3) (3). Among the three enzymes, the E3 ubiquitin ligases play a pivotal role in determining specificity of substrate proteolysis (4, 5). Based on the structural characteristics, E3 enzyme can be divided into four categories: RING E3s, HECT E3s, U-box E3s and RBR E3s (4). The SCF multisubunit complex, the most common RING E3s composing of a scaffold protein cullin1, a Ring protein (RBX1 or RBX2), an adaptor protein and a substrate receptor protein, is the largest family of E3s that promote the degradation of about 20% of UPS-regulated proteins (6, 7).
RBX1/2 usually ubiquitously expressed in human tissues, such as heart, colon, skeletal muscle, and testes (8, 9). RBX proteins can promote ubiquitin transfer from the E2 to the substrates and further enhances cullins activity, therefore, they constitute the catalytic cores of SCF complexes (10). Previous studies have confirmed that RBX proteins were found to be functionally non-redundant. Deletion of RBX1 in mice results in early embryo death (E7.5) due to proliferation failure in a wild-type RBX2 background, whereas inactivation of RBX2 causes late embryo death (E11.5-12.5) associated with cardiovascular defects (11, 12). Although both RBX proteins are highly conservative at protein level, share similar ring finger domain structure, their effect on the regulation of substrate degradation may vary (11). RBX1 mainly mediates proteolysis, including cell cycle regulators (e.g., cell cycle inhibitor p21/p27/p53/p57, and cyclin A/D/E), transcription factors (e.g., E2F1, FOXO1, myc, and c-Jun), DNA replication factor CDT1, and others. RBX2 promotes ubiquitination and degradation of a number of protein substrates, including c-Jun, DEPTOR, HIF-1α, IκBα, NF1, NOXA, p27 and procaspase-3, to degrade different substrates causing various phenotypes (13–16).
To be specific in cancers, RBX1 was shown to be highly expressed in bladder, gastric, prostate and renal cancer (17–19). Notably, RBX2, is rarely expressed in normal tissues, but highly expressed in lung, liver, gastric and renal cancer (20–22). Previous studies on the Ring finger family have focused on the oncogenic function and degradation ability of RBX1 and RBX2 in specific tumors, respectively, which provides a limited understanding of their role in SCF E3s. However, the difference of RBX members in pan-cancer has not been described. To explore the effect of RBX1/2 on the overall picture of SCF complex and in the tumor evolution, we comprehensively analyzed their difference in pan-cancer using the TCGA database in the present study. Their diversities were reflected in the following aspects including mRNA level, protein level, pathological features, prognosis and copy number variation, immune infiltration level.
The CUL1-RBX1 and CUL1-RBX2 complexes were obtained from the Protein Data Bank (RCSB PDB www.rcsb.org) database (23). The molecular dynamic simulation for the CUL1-RBX1/2 complex used PDB ID: 1LDJ and 7 ONI as the templates. A molecular dynamic simulation was performed for the two complexes in a water environment (310 K temperature) with the force field charmm36-feb2021.ff using GROMACS software (24). The binding affinity was calculated using g_mmpbsa and the PyMOL software was used for visualization (25).
We used the UCSC Xena (https://xenabrowser.net/) to download TCGA pan-cancer data, including survival data, clinical data, stemness score (RNA based) and immune subtype (26). RBX1/2 expression was integrated by Perl software. We used the Wilcox test to assess the difference between normal and tumor tissues. P value less than 0.05 is considered as difference. A heatmap and box plot were illustrated by the R-package “ggpubr” and “pheatmap”, respectively. Furthermore, Correlation analysis among Ring finger family genes was performed by R-package “corrplot”.
UALCAN was used to analyze the RBX-proteins expression in several cancers, including BRAC, OV, UCEC and PAAD (27). ∗P< 0.05, ∗∗P< 0.01, and ∗∗∗P< 0.001. Additionally, we obtained box plots of the RBX1/2 expression in different pathological grades and stages via the TISIDB database (28) (http://cis.hku.hk/TISIDB/index.php). Survival analysis of RBX1/2 was used for the “survival” and “survminer” R package. A difference of p less than 0.05 was statistically significant. Meanwhile, we downloaded the TCGA pan-cancer mRNA expression and survival data to conduct the Cox analysis for illustrating the association between RBX1/2 expression and the survival of patients.
Gene Set Cancer Analysis (GSCA) platform is a web server that integrated multiomics data based on TCGA database (29) (http://bioinfo.life.hust.edu.cn/web/GSCA/). Based on CNV module, the proportion of RBX1/2 heterozygous/homozygous and amplification/deletion, Spearman correlation between RBX1/2 mRNA expression and CNV, and the survival difference between their CNV and wild type were displayed in pan-cancer.
The correlation between Ring finger family expression and immune subtypes of different cancer types were explored via the TISIDB database. Furthermore, we selected four types of cancers (COAD, GBM, LIHC, LUAD) to analyze the relationship between RBX1/2 and immune infiltration using Timer database (30)(https://cistrome.shinyapps.io/timer/). Moreover, the associations of RBX1/2 levels with 47 common immune checkpoint genes selected were also evaluated. R software was used to calculate the correlation between RBX1/2 expression and TMB/MSI and the Fmsb R package was used for visualization. Then, we performed the tumor microenvironment analysis for obtaining the estimate score profile by using the “estimate” R package, and the Spearman correlation test for conducting the correlation analysis between RBX1/2 expression and immune score, estimate score, stromal score, DNAss, RNAss and tumor purity in pan-cancer.
All human breast cancer cell lines (MDA-231, BT-474, MCF-7) and normal breast epithelial cell (MCF-10A), lung cancer cell lines (H1975, A549, PC9) and normal lung epithelial cell (BEAS-2B), colorectal cancer cell lines (HCT116, SW480, SW620) and normal colon epithelial cell (HCoEpic), renal cancer cell lines (Caki-1, 786-O, 769-P) and normal renal tubular epithelial cell (HK-2) were purchased from the American Type Tissue Collection (ATCC) and cultured according to the manufacturer’s instructions.
cDNA reverse transcription and fluorescence quantitative PCR amplification were performed using SPARKscript IISYBR Green qRT-PCR Kit (Shandong Sparkjade Biotechnology Co., Ltd.) as previously reported (31). The primers used were as follows: RBX1 forward, 5′-TTGTGGTTGATAACTGTGCCAT -3′,
RBX1 reverse, 5′-GACGCCTGGTTAGCTTGACAT -3′;
RBX2 forward, 5′-TGGAAGACGGAGAGGAAACCT -3′,
RBX2 reverse, 5′-TGAGGGAGAACATCTTGTCGC -3′
β-Actin forward, 5′- CGTGCGTGACATTAAGGAGAAG -3′,
β-Actin reverse, 5′- GGAAGGAAGGCTGGAAGAGTG -3′;.
All genes were normalized to β-actin, and the 2−ΔΔCt method was applied to evaluate the relative levels of genes. The comparison between the experimental group and the normal group was performed using the Dunnett’s t test. P less than 0.05 was considered statistically significant.
The SCF complexes are Ring-type E3s that composited of cullin1, SKP1, RBX1/2 and a member of the F-box protein family. Although the abundance of SCF is increased by the variety of F-box proteins, they share the two ring components RBX1 and RBX2 (32–34). RBX1 is constitutively expressed and induced upon mitogen, whereas RBX2 is stress-inducible and induced upon UV, TPA or ROS (14). In this study, we separately calculated the binding affinity of CUL1-RBX1 and CUL1-RBX2 complexes to rough compare stability of SCF complex formed by RBX1/2. The binding energy calculated by the former was -262.59 kJ/mol and the latter was -146.8 kJ/mol (Supplementary Figures 1A, B). The result displayed the combination of RBX1 and CUL1 may be more stable than that of RBX2 and CUL1, suggesting that RBX1 is more likely to form stable SCF complexes to degrade more substrates.
We performed a scale analysis of the expression of RBX1/2 from the TCGA database and found that they are highly expressed in most cancers. However, there were a few apparent exceptions in the 18 types of cancers, a lower RBX1 expression was detected in KICH compare to the matches normal tissues, whereas RBX2 was under expressed in COAD and READ in addition to KICH (Figure 1A). To validate the differences of RBX1/2 expression, we analysed transcriptional expression of these both genes in various tumor cell lines of four common types of cancer (breast, lung, colorectal and renal cancer) and normal cells. Except for the expression of RBX1 in lung cancer and RBX2 in COAD, the experimental results are basically consistent with the bioinformatics analysis (Supplementary Figure 2).
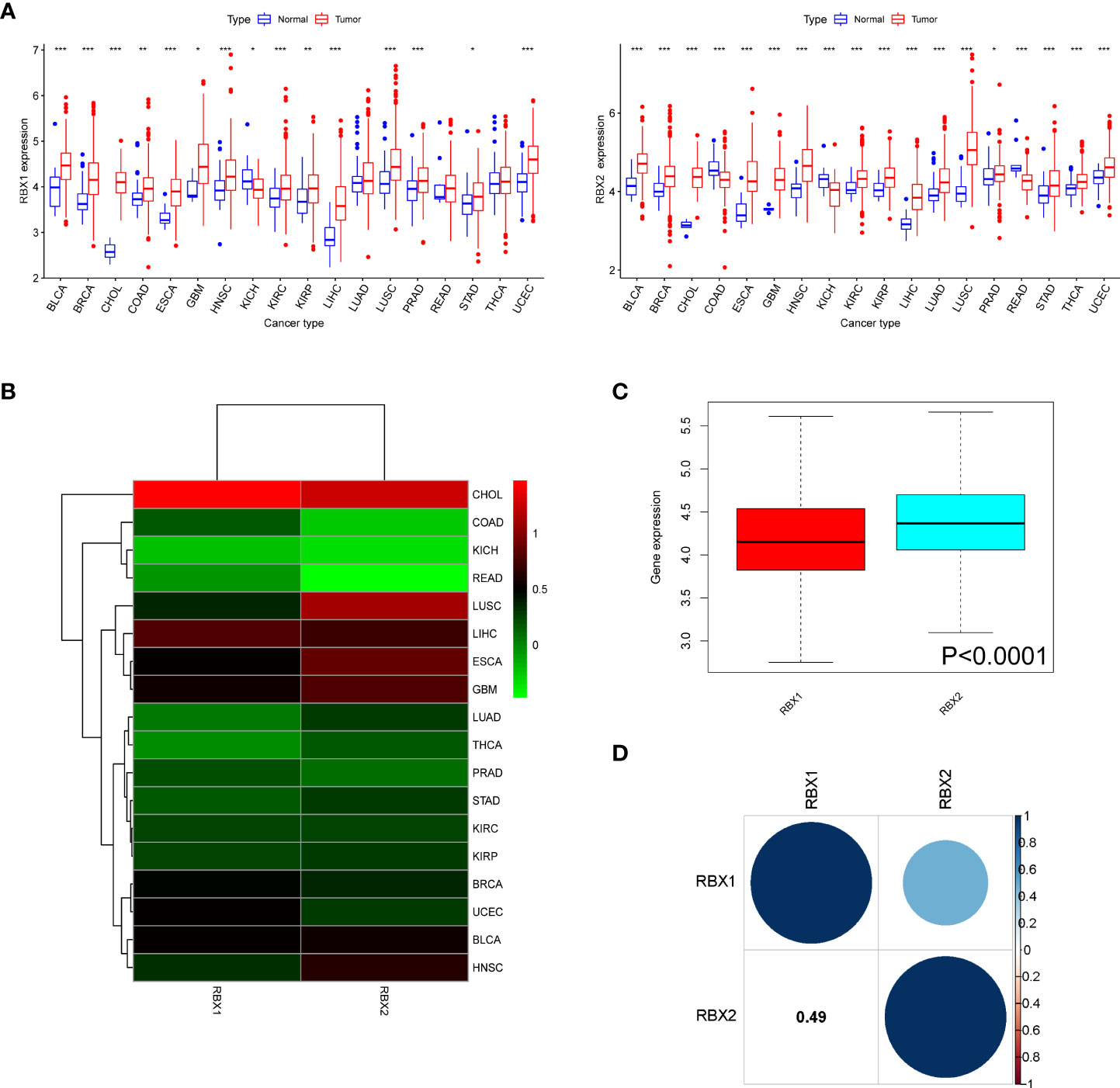
Figure 1 The mRNA expression patterns of RBX1/2 in cancers. (A) Comparison of RBX1/2 expression between tumor and normal samples. (B) Heatmap showing the difference of RBX1/2 gene expression in 18 cancer types from TCGA database. The red and green indicate the high or low expression, respectively. (C) Boxplot illustrating the distribution of RBX1/2 gene expression in various cancer. (D) The correlation between RBX1 and RBX2. The blue dot indicated the positive correlation. *P < 0.05, **P < 0.01, ***P < 0.001.
Further analysis revealed that there were significant difference of Ring finger genes expression comparing primary tumor to adjacent normal tissues, for example, RBX2 expression in COAD tissues was lower than adjacent non-COAD tissues, while RBX1 was in the opposite situation. The difference of RBX2 expression between LUSC and adjacent tissues was much more obvious than that of RBX1 (Figure 1B). Meanwhile, the overall expression level of RBX2 was higher than that of RBX1 in pan-cancer (Figure 1C). We also analyzed that RBX1 and RBX2 are the two genes with significant positive correlation (Correlation coefficient = 0.49, Figure 1D).
We investigated the RBX-proteins expression levels in BRCA, OV, UCEC and PAAD (Figure 2A). The results showed RBX2 expression in BRCA and OV was lower than in normal tissues, while RBX1 expression had no significant difference on the above tumors. Moreover, there was no difference on RBX2 expression in PAAD, however, RBX1 expression was higher in matched normal tissues. Another interesting phenomenon that RBX-proteins expression in UCEC was exact opposite and statistically significant was also illustrated. We showed RBX1/2 expression with pathological grades of KIRC, LIHC, LGG and UCEC using TISIDB database (Figure 2B), revealing that there were no differences in the association between RBX1 expression and clinical grades in LIHC and UCEC, whereas RBX2 expression has statistical significance in association with pathological grade of KIRC, LIHC and UCEC. We also observed the significant correlation between RBX1/2 expression and the pathological stages of several cancers including KIRC, KIRP, LIHC and PAAD (Figure 2C). The expression of RBX1 was not related to the stage of LIHC and PAAD, while RBX2 was in the opposite situation. Moreover, the association with RBX1/2 expression and KIRP stages was completely opposite, RBX1 was significantly correlated with the stages of KIRP. In conclusion, different expression patterns of RBX1/2 in various cancer types may lead to different characterization of tumors.
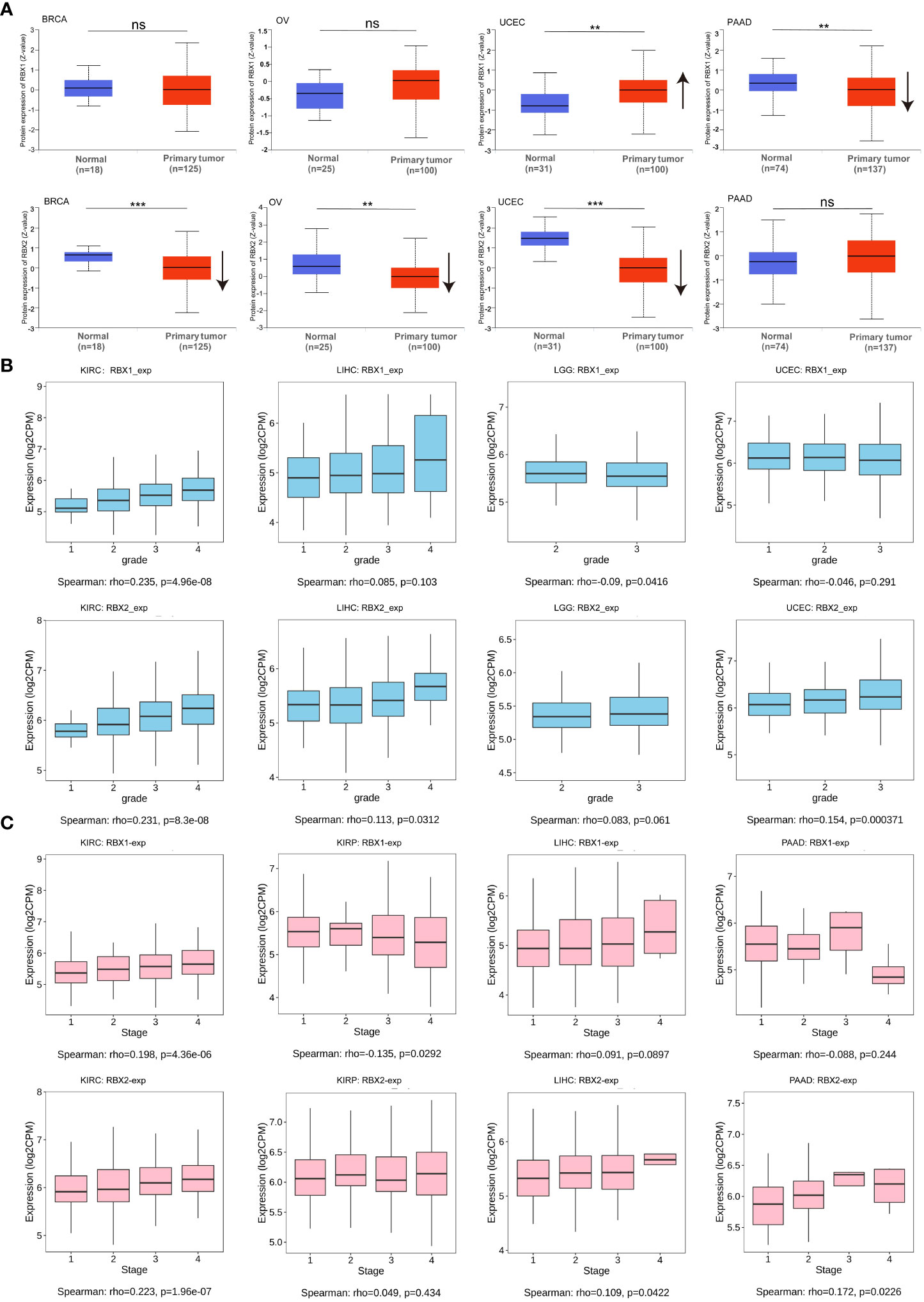
Figure 2 RBX1/2 expression based on tumor types and individual pathological grades and stages. (A) RBX-proteins expression in BRCA, OV, UCEC and PAAD. ns is considered as no statistical difference, **P < 0.01, ***P < 0.001. Up or down arrow represented the expression of tumor samples more or less than the corresponding normal samples, respectively. (B) The expression levels of RBX1/2 were analyzed by tumor pathological grades (grade1, grade2, grade3, grade4) of KIRC, LIHC, LGG and UCEC. P value less than 0.05 is considered as difference. (C) The expression levels of RBX1/2 were analyzed by tumor pathological stages (stage I, stage II, stage III, and stage IV) of KIRC, KIRP, LIHC and PAAD. P value less than 0.05 is considered as difference.
The survival analysis of TCGA database presented a correlation between Ring finger family gene expression and prognosis in several cancers, showing that higher RBX1 expression was associated with poor OS in ACC (P<0.001), KIRC (P=0.011), LIHC (P=0.008), and UVM (P<0.001) (Figure 3A), whereas higher RBX2 expression was linked to poor prognosis in KICH (P=0.025), KIRC (P=0.001), LAML (P=0.026), LGG (P=0.043), LIHC (P=0.005) and PAAD (P=0.038) (Figure 3B). Interestingly, RBX1 had a protective role in OV (P=0.002), PCPG (P=0.014), suggesting RBX1 may exert tumor suppressor effect in OV and PCPG (Figure 3A).
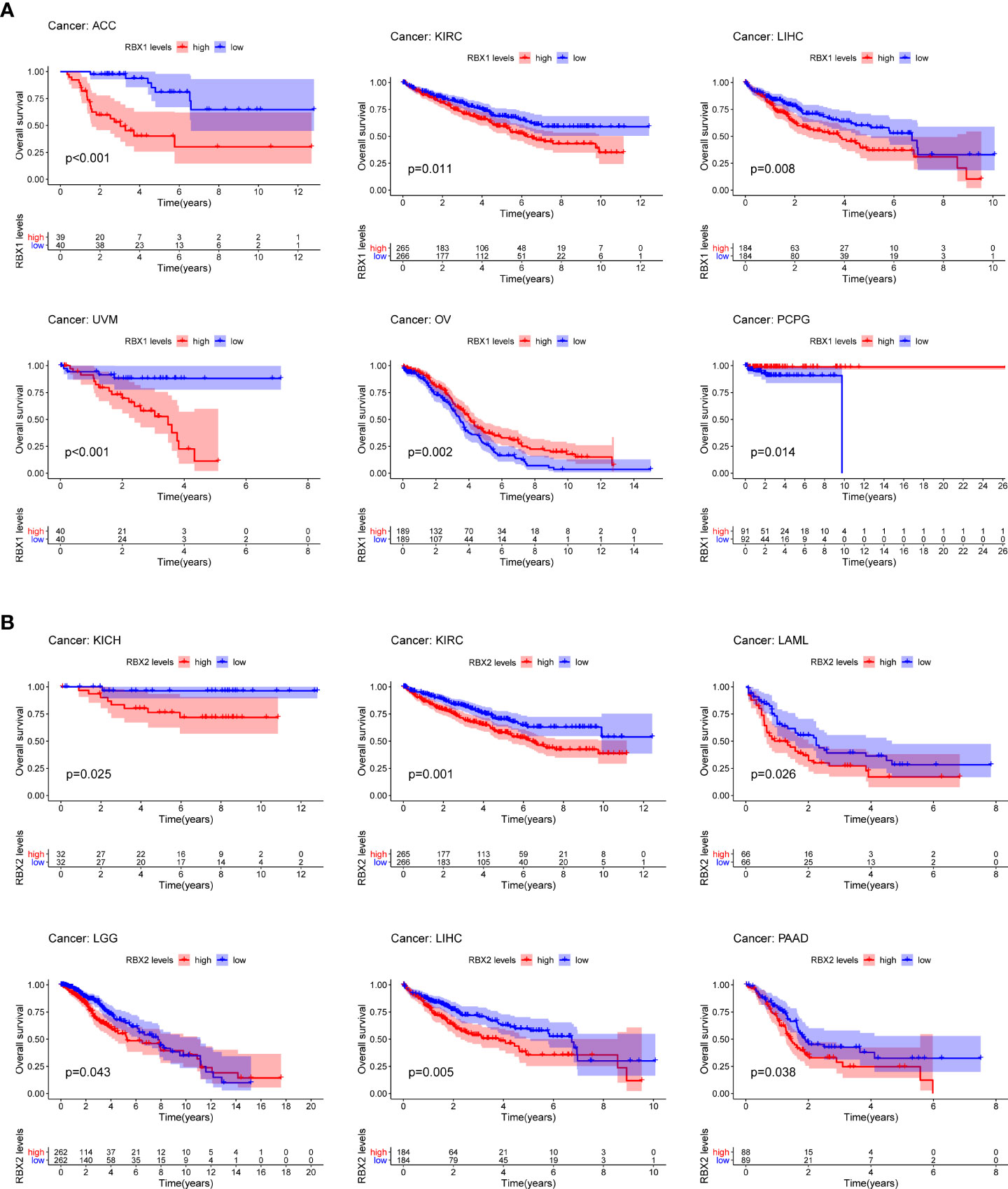
Figure 3 Kaplan-Meier survival curves comparison of high and low expression of Ring finger family gene in pan-cancer. (A) OS survival curves of RBX1 in different cancers: ACC, p<0.001; KIRC, p=0.011; LIHC, p=0.008; UVM, p<0.001; OV, p=0.002; PCPG, p=0.014. (B) OS survival curves of RBX2 in different cancers: KICH, p=0.025; KIRC, p=0.001; LAML, p=0.026; LGG, p=0.043; LIHC, p=0.005; PAAD, p=0.038.
We further investigated prognosis risk of the Ring finger family genes in pan-cancer by COX analysis (Figure 4). Our results indicated that RBX1 played a detrimental role in ACC, KIRC, LIHC and UVM (HR>1, P<0.05). On the other hand, RBX1 had a protective role in LGG, PCPG and CESC (HR<1, P<0.05). RBX2 acted as a detrimental prognostic factor in ACC, KICH, KIRC, LIHC and PAAD (HR>1, P<0.05). In contrast, RBX2 was a protective prognostic factor in CESC (HR<1, P<0.05). We have enumerated three tumors of the highest incidence (breast, colorectal and lung cancer) to perform comprehensive prognosis analysis with RBX1/2 expression by the PrognoScan database (35) (Table 1). RBX1 and RBX2 were the high-risk genes in breast cancer (RFS). Notably, RBX2 acted as a detrimental prognostic factor in colorectal cancer (OS, DFS) and lung cancer (OS, RFS). However, RBX1 had no significant relation with the prognosis in above cancers. The difference between RBX1 and RBX2 may lead to different tumor outcomes.
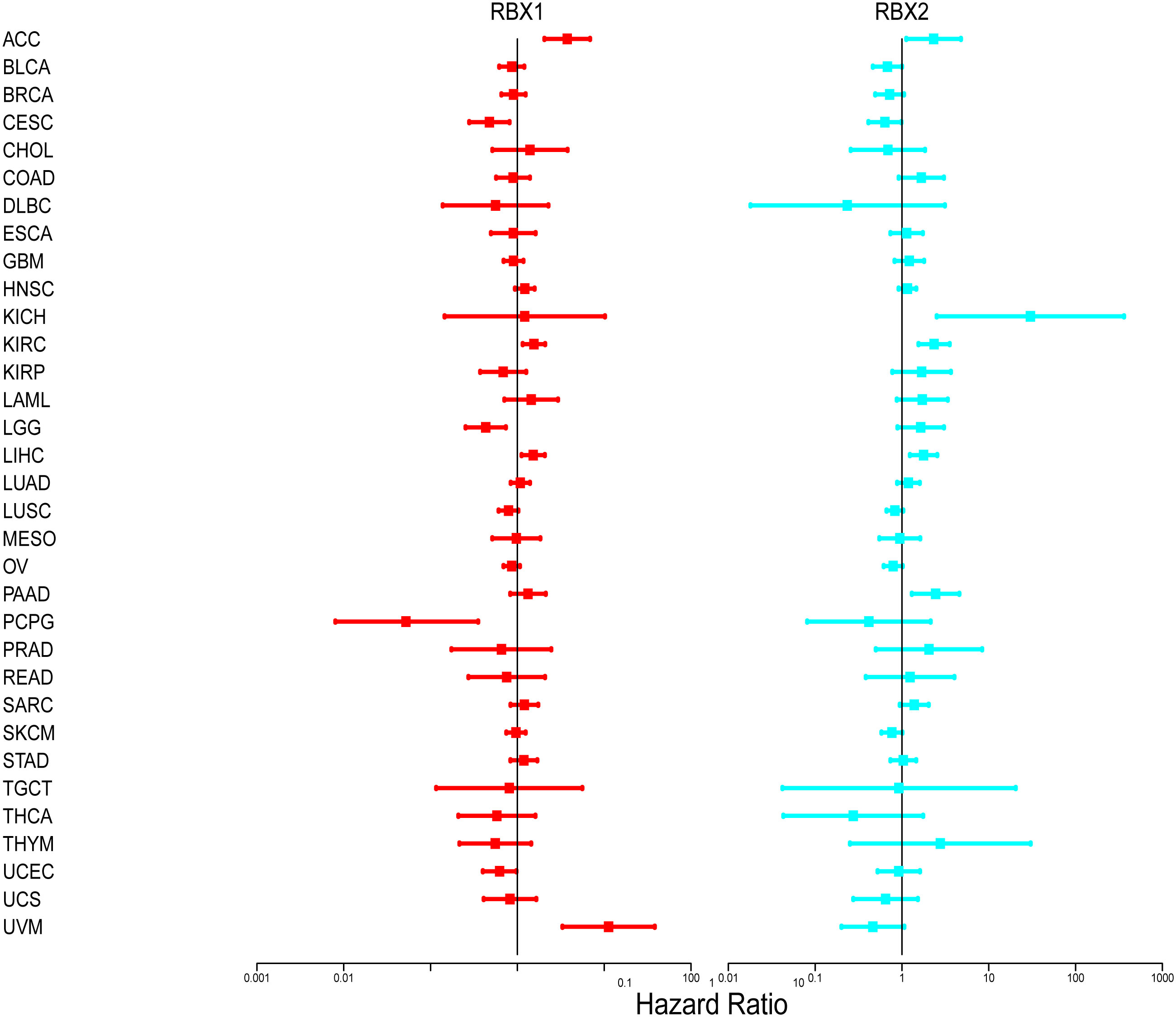
Figure 4 Association of RBX1/2 gene expression with patient’s overall survival for different cancer types. The forest plots with the hazard ratios and 95% confidence intervals for overall survival for different cancer types showing the survival advantage and disadvantage with the increased gene expression of RBX1/2. The univariate Cox proportional hazard regression models were used for the association tests.
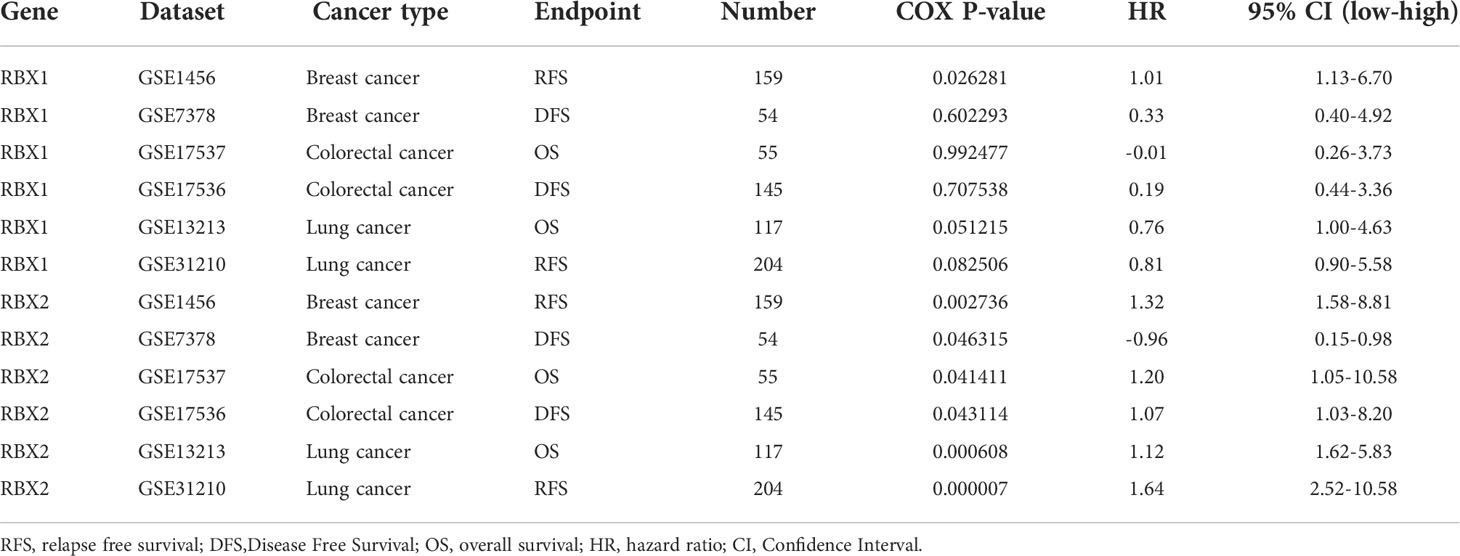
Table 1 Ring finger family gene expression was related to the prognosis of different cancers in PrognoScan.
We summarized RBX1/2 CNV landscape in 33 cancer types by using the GSCA database, respectively (Figure 5). The highest heterozygous amplification ratio (45.71%) for RBX1 was found in LUSC, whereas the heterozygous amplification ratio of RBX2 presented a higher level of state in several cancers (>50%) including CESC, HNSC, LUSC and OV. Furthermore, a relatively higher heterozygous deletion ratio (>50%) for RBX1 was found in MESO, OV and UCS. However, RBX2 showed a heterozygous deletion ratio of more than 50% only in PCPG. The homozygous amplification of RBX2, had a significant proportion in some specific cancers containing CESC, ESCA, HNSC, LUSC and OV, for example, RBX2 homozygous amplification in LUSC was accounted for about 20% (Figure 5A). We also explored the association between RBX1/2 CNV and their mRNA expression (Figure 5B). Except for CHOL, DLBC, KICH, KIRC, LAML, PRAD, READ, THYM and UVM, the rest 24 cancer types were statistically significant for the correlation between RBX1 CNV and its mRNA expression. In addition to DLBC, LAML and THCA, RBX2 CNV had also a statistical significance with its mRNA expression in most cancers (Figure 5B). Subsequently, the profile of survival between the two members associated gene set CNV groups in the selected cancers was also summarized. The results suggested that wide type RBX1 had all statistical significance on OS, PFS, DFS and DFI in UCEC and KIRP. However, wide type RBX2 had all statistical significance on above four survival indicators only in UCEC (Figure 5C).
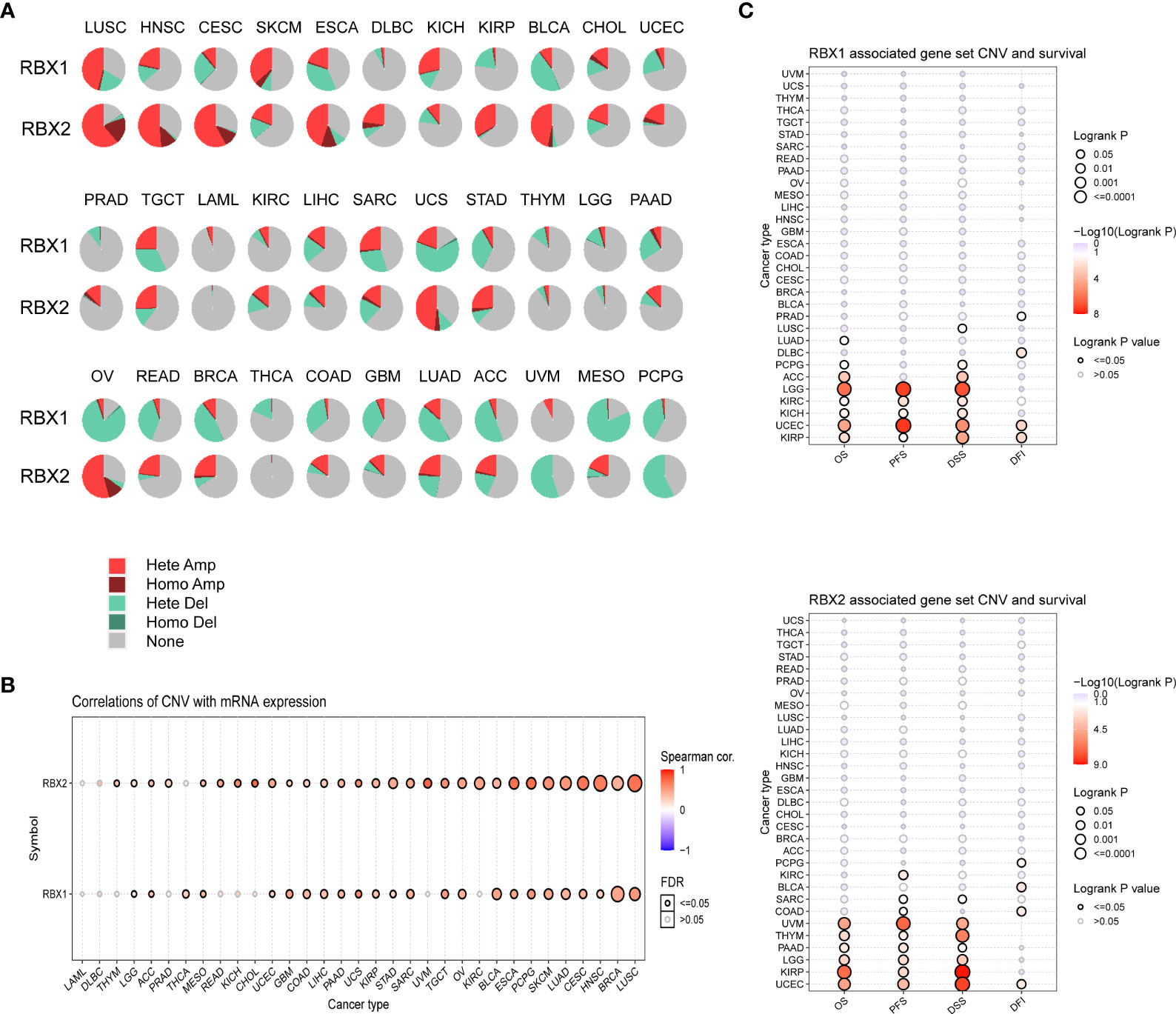
Figure 5 The CNV landscape of RBX1/2 in pan-cancer based on GSCA. (A) The deletion/amplification of heterozygous/homozygous CNV for RBX1/2 in pan-cancer. (B) Correlation between CNV and RBX1/2 mRNA expression in various cancers. (C) The profile of survival between RBX1/2 associated gene set CNV groups in various cancer types. CNV, copy number variation; GSCA, Gene Set Cancer Analysis; Hete Amp, Heterozygous Amplification; Homo Amp, Homozygous Amplification; Hete Del, Heterozygous Deletion; Homo Del, Homozygous Deletion; OS, overall survival; PFS, progression-free survival; DSS, disease specific survival; DFI, disease-free interval.
Previous studies determined that RBX1 and RBX2 were involved in immunomodulatory processes (19, 36), therefore, we compared the relationships between RBX1/2 expression and immune subtypes through the TISIDB database (Figure 6). Immune subtypes were classified into six types, including C1 (wound healing), C2 (IFN-gamma dominant), C3 (inflammatory), C4 (lymphocyte depleted), C5 (immunologically quiet) and C6 (TGF-b dominant). Our analyses showed that RBX1 expression in the immune subtypes of BLCA, UCEC and UVM had no statistical significance, while RBX2 expression in above three cancers was closely related with those immune subtypes. Conversely, RBX2 expression had no correlation with the COAD immune subtypes. Of interest, taking KIRC as the example, RBX1 showed high expression in C2 and C6 types, however, RBX2 expression on C1 immune subtype was the highest in KIRC. Furthermore, we investigated the association with RBX1/2 expression and immune subtypes in the TCGA pan-cancer data, illustrating that the expression of RBX1 was lowest in the C3 immune subtype, while RBX2 was lowest in the C5 immune subtype (Supplementary Figure 3). Based on the above results, we concluded that RBX1/2 expression differs in immune subtypes of various tumor cancers.
Figure 6 The relationship between RBX1/2 expression and pan-cancer immune subtypes. (A) Correlation of RBX1 expression and immune subtypes in BRCA, COAD, LIHC, LUAD, KIRC, STAD, UCEC and UVM. (B) Correlation of RBX2 expression and immune subtypes in BRCA, COAD, LIHC, LUAD, KIRC, STAD, UCEC and UVM. P value less than 0.05 is considered as difference.
Studies indicated that RBX1 expression are associated with the immune suppressive function of Treg cells, and T-cell deficiency, and RBX2 could trigger a series of immune responses, suggesting they may play important roles in regulating immune cells (37, 38). We found a strong correlation between RBX1/2 expression and the levels of immune infiltration in COAD, GBM, LIHC and LUAD by analysis of the TIMER database (Figure 7). The expression of RBX1 was in connection with the infiltration of B cell, CD4+ T cells and neutrophils in above four cancers (Figure 7A). With regard to RBX2, the infiltration of CD8+ T cells and macrophages have a positive correlation with RBX2 in COAD and LIHC (Figure 7B). We also conducted the co-expression analysis to further explore the association between RBX1/2 expression and immune checkpoints in pan-cancer using the TCGA database. As shown in Figure 8A, RBX1 was positively correlated with these immune markers in SARC, TCGT and UVM, whereas the positive association between RBX2 mRNA and immune checkpoints existed in LGG and LIHC (Figure 9A). Interestingly, we found that RBX1 was positively correlated with the expression levels of PD1 (PDCD1) and CTLA-4 in BRCA, KIRP, LIHC, SARC, TCGT, THCA and UVM (Figure 8A). RBX2 had a closely tie with the expression level of PD-L1 (CD274) in BLCA, COAD, HNSC, KIRC, LAML, LIHC, OV, PCPG, PRAD, SKCM, TGCT and THCA (Figure 9A). These results indicated that RBX1/2 might regulate different immune response in various cancer types.
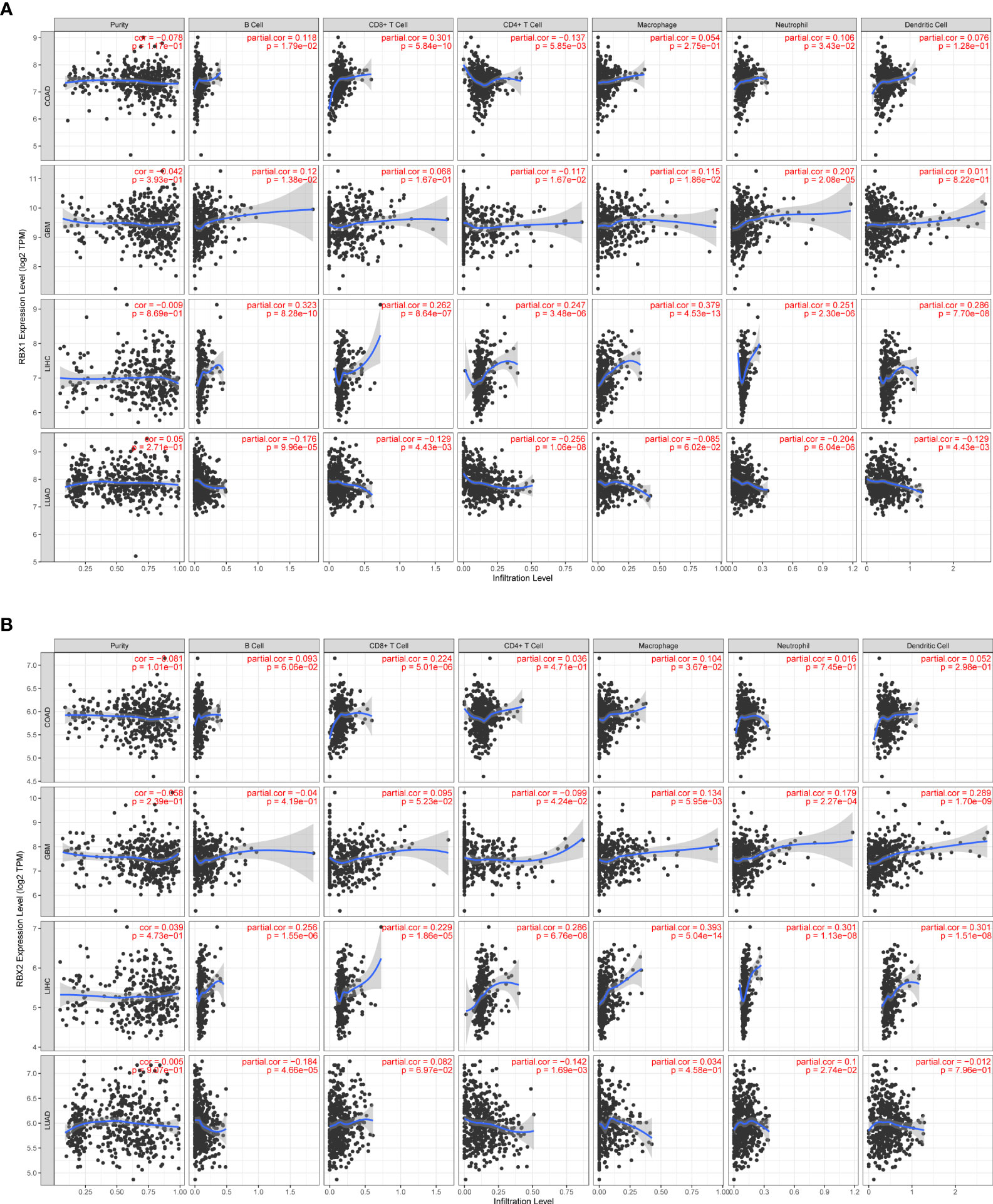
Figure 7 Correlation of RBX1/2 expression with immune infiltration level in COAD, GBM, LIHC and LUAD. (A) RBX1 expression is related with the level of immune infiltration in the above four cancers. (B) RBX2 expression is related with the level of immune infiltration in the above four cancers. P value less than 0.05 is considered as difference.
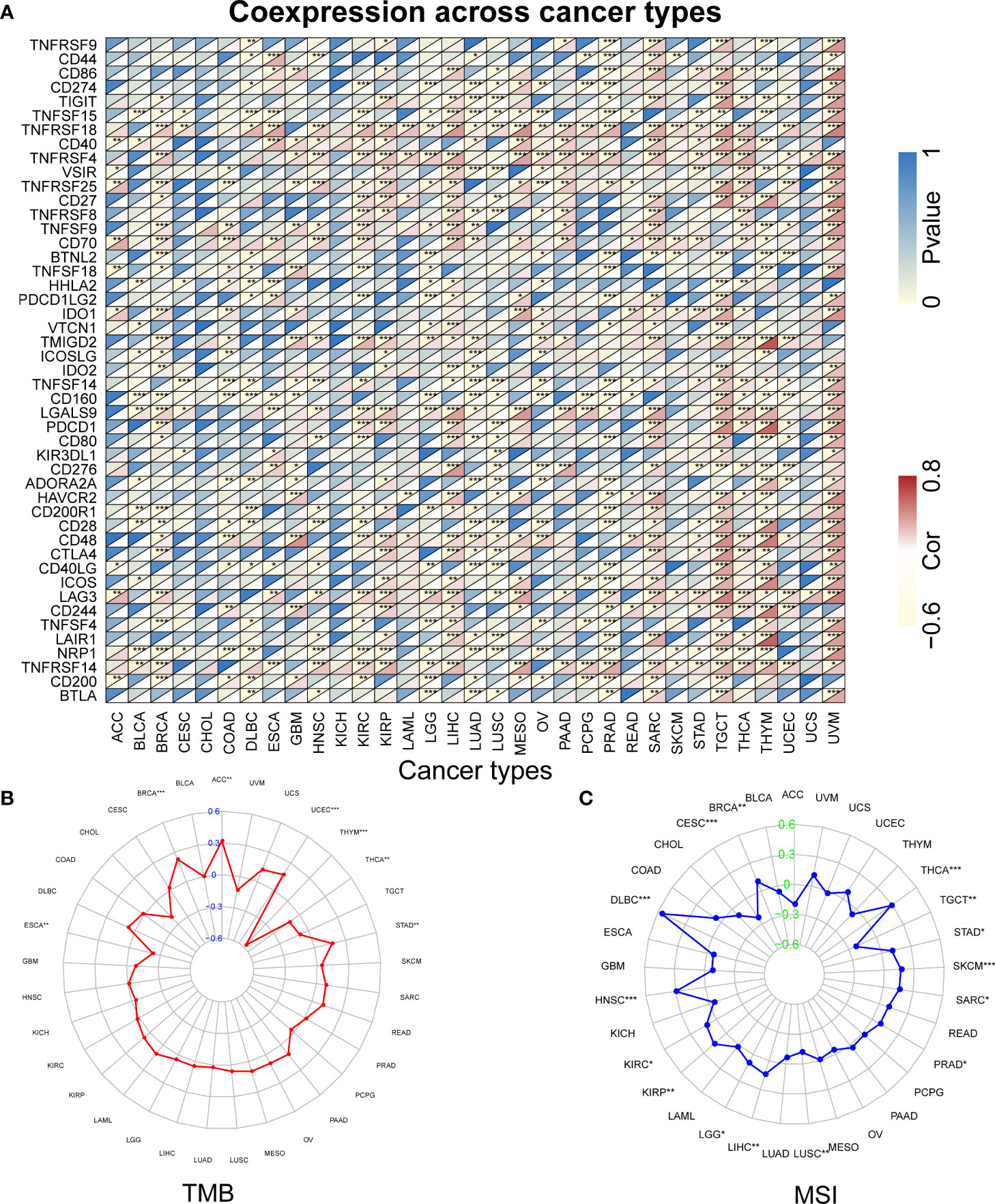
Figure 8 The relationship between RBX1 and immune checkpoints, TMB and MSI based on TCGA database. (A) Heatmap illustrating the relationship between RBX1 and known immune checkpoints. The top left triangle represents the P-value, and the bottom right triangle represents the correlation coefficient. Correlation between RBX1 and TMB (B) and MSI (C). *P < 0.05, **P < 0.01, and ***P < 0.001.

Figure 9 The relationship between RBX2 and immune checkpoints, TMB and MSI based on TCGA database. (A) Heatmap illustrating the relationship between RBX2 and known immune checkpoints. The top left triangle represents the P-value, and the bottom right triangle represents the correlation coefficient. Correlation between RBX2 and TMB (B) and MSI (C). *P < 0.05, **P < 0.01, and ***P < 0.001.
Further analysis found that RBX1 expression was positively correlated with TMB in ACC, BRCA, STAD and UCEC, but negatively correlated with ESCA, THCA and THYM, as seen in Figure 8B. However, RBX2 expression had no relation with TMB in ACC, ESCA, THCA and THYM (Figure 9B). We also found that the RBX1 had a positive association with MSI in BRCA, DLBC, HNSC, KIRC, KIRP, LGG, LIHC, PRAD, SARC, SKCM, STAD and THCA, but had a negative association with CESC, LUSC and TGCT, as seen in Figure 8C. Similarly, correlation analysis between RBX2 expression and MSI was also performed (Figure 9C). In HNSC, KIRC, LIHC, PRAD, READ, SKCM, STAD, THCA and UCEC, RBX2 expression was positively related to MSI, whereas the expression of RBX2 has a negative relationship with GBM (Figure 9C).
To obtain a more comprehensive analysis of the relationship between Ring finger family and immune components, we applied the estimate algorithm to evaluate the stromal and immune scores in 33 cancer types. RBX1/2 existed statistically significance in stromal, immune, and estimate scores (Supplementary Figures 4A-C). Besides, they had a significantly positive or negative correlation with DNAss, RNAss and tumor purity in pan-cancer (Supplementary Figures 4D-F). These results suggested RBX1/2 may be involved in different immune processes in various cancer types.
Previous studies have systematically provided a comprehensive overview on the alterations of SCF E3 ubiquitin ligases in the pathogenesis and development of cancers (39, 40). RBX1/2 were overexpressed in a number of primary cancer tissues, including carcinoma of lung, liver, breast, colon, and renal. Sun Y et al. has demonstrated that inactivation of either RBX1 or RBX2 inhibits carcinogenesis via various mechanisms, including apoptosis and senescence (17, 41, 42). However, two other studies found that only RBX2 overexpression was correlated with the poor prognosis in lung cancer (21); as well as high RBX1 expression was related to poor survival only in KIRC patients and high RBX2 expression had a close relation with poor prognosis in all three types of RCC (22, 43). At present, the comparison of Ring finger family in the same cancer is rare. The underlying mechanisms by which they contribute to different outcomes in cancer patients remains largely unknown. Therefore, we focused on their differences in mRNA level, protein level, pathological parameters, prognosis and etc. by the pan-cancer analysis in this study Supplementary Table 1. Our result showed that RBX1/2 reflected their characteristics respectively in the observation indicators mentioned, for example, RBX2 expression is more differentially expressed than RBX1 in LUSC, which may be one of the reasons that only RBX2 expression is associated with lung cancer prognosis.
Accumulating evidence suggests that the E3s dysfunction can contribute to adverse immune response (44–46). Previously, several studies have observed that RBX1 can promote ubiquitin degradation of HBx-induced PD-L1 protein in HCC cells (47). Meanwhile, RBX2-dependent neddylation played a significant role in the regulation of T-cell responses (38). Thus, there is a dire need for exploring the relationship of RBX1/2 expression and immune components. Using bioinformatics methods, we elucidated the immunological role of the Ring finger family across cancers and provided in first time the gene expression and genetic alteration of RBX1/2 in the regulation of different immune components including their association with PD-L1 expression. This result showed RBX1/2 may be attractive biomarkers of immunotherapy efficacy.
We investigated and integrated information based on bioinformatics and public databases, however, there were still some limitations in the present study. First, whether the Ring finger family is harmful or beneficial remains contradictory because of some conflicting findings from different databases. Second, despite the finding that they were closely associated with immune infiltration and prognosis, we were unable to determine whether these two molecules affected patient survival through immune infiltration. Finally, whether differences in RBX-proteins are a decisive factor in the stability of the SCF complex in pan-cancer needs to be further clarified.
In summary, our results revealed that the important role of Ring finger members in the SCF complex, and the expression profile of RBX1/2 in pan-cancer. Moreover, strong correlations between RBX1/2 and disease prognosis and immune components were proved in the present study. Clinical immune markers, such as PD-1, CTLA-4 and PD-L1, have been confirmed to be closely associated with Ring finger family in a variety of cancers. These findings may provide insights for further investigation of the Ring finger family genes as potential targets in pan-cancer.
The data that support the findings of our study are openly available from the TCGA, UALCAN, TISIDB, PrognoScan, GSCALite and Timer database at (https://tcgadata.nci.nih.gov/tcga/,http://ualcan.path.uab.edu/, http://cis.hku.hk/TISIDB/index.php, http://www.abren.net/PrognoScan/, http://bioinfo.life.hust.edu.cn/web/GSCALite/, https://cistrome.shinyapps.io/timer/).
All authors read and approved the final manuscript. HA designed the overall study and revised the paper. TH and JL drafted the manuscript and performed the data analysis. BS, XL and SL participated in the data collection. All authors contributed to the article and approved the submitted version
This project is supported by the grand “Peking Medical and Health Foundation.” Nr.: F3142C
We thank the databases of the TCGA, UALCAN, TISIDB, PrognoScan, GSCALite, Timer for the availability of the data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.968777/full#supplementary-material
Supplementary Table 1 | The summarization of alterations of RBX1 and RBX2 in all types of cancers. These statistical data are mainly from online databases and R package processed.

1. Park J, Cho J, Song EJ. Ubiquitin-proteasome system (Ups) as a target for anticancer treatment. Arch Pharm Res (2020) 43(11):1144–61. doi: 10.1007/s12272-020-01281-8
2. LaPlante G, Zhang W. Targeting the ubiquitin-proteasome system for cancer therapeutics by small-molecule inhibitors. Cancers (Basel) (2021) 13(12):3079. doi: 10.3390/cancers13123079
3. Li X, Elmira E, Rohondia S, Wang J, Liu J, Dou QP. A patent review of the ubiquitin ligase system: 2015-2018. Expert Opin Ther Pat (2018) 28(12):919–37. doi: 10.1080/13543776.2018.1549229
4. Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem (2005) 41:15–30. doi: 10.1042/eb0410015
5. Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol (2014) 21(4):301–7. doi: 10.1038/nsmb.2780
6. Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1-Rbx1-Skp1-F Boxskp2 scf ubiquitin ligase complex. Nature (2002) 416(6882):703–9. doi: 10.1038/416703a
7. Jackson PK, Eldridge AG. The scf ubiquitin ligase: An extended look. Mol Cell (2002) 9(5):923–5. doi: 10.1016/s1097-2765(02)00538-5
8. Huang Y, Duan H, Sun Y. Elevated expression of Sag/Roc2/Rbx2/Hrt2 in human colon carcinomas: Sag does not induce neoplastic transformation, but antisense sag transfection inhibits tumor cell growth. Mol Carcinog (2001) 30(1):62–70. doi: 10.1002/1098-2744(200101)30:1%3C62::AID-MC1014%3E3.0.CO;2-A
9. Sun Y, Tan M, Duan H, Swaroop M. Sag/Roc/Rbx/Hrt, a zinc ring finger gene family: Molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal (2001) 3(4):635–50. doi: 10.1089/15230860152542989
10. Duda DM, Olszewski JL, Tron AE, Hammel M, Lambert LJ, Waddell MB, et al. Structure of a glomulin-Rbx1-Cul1 complex: Inhibition of a ring E3 ligase through masking of its E2-binding surface. Mol Cell (2012) 47(3):371–82. doi: 10.1016/j.molcel.2012.05.044
11. Wei D, Sun Y. Small ring finger proteins Rbx1 and Rbx2 of scf E3 ubiquitin ligases: The role in cancer and as cancer targets. Genes Cancer (2010) 1(7):700–7. doi: 10.1177/1947601910382776
12. Tan M, Davis SW, Saunders TL, Zhu Y, Sun Y. Rbx1/Roc1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of P27. Proc Natl Acad Sci U.S.A. (2009) 106(15):6203–8. doi: 10.1073/pnas.0812425106
13. Sun Y, Li H. Functional characterization of Sag/Rbx2/Roc2/Rnf7, an antioxidant protein and an E3 ubiquitin ligase. Protein Cell (2013) 4(2):103–16. doi: 10.1007/s13238-012-2105-7
14. Zhou W, Wei W, Sun Y. Genetically engineered mouse models for functional studies of Skp1-Cul1-F-Box-Protein (Scf) E3 ubiquitin ligases. Cell Res (2013) 23(5):599–619. doi: 10.1038/cr.2013.44
15. He H, Gu Q, Zheng M, Normolle D, Sun Y. Sag/Roc2/Rbx2 E3 ligase promotes uvb-induced skin hyperplasia, but not skin tumors, by simultaneously targeting c-Jun/Ap-1 and P27. Carcinogenesis (2008) 29(4):858–65. doi: 10.1093/carcin/bgn021
16. Tan M, Gallegos JR, Gu Q, Huang Y, Li J, Jin Y, et al. Sag/Roc-scf beta-trcp E3 ubiquitin ligase promotes pro-Caspase-3 degradation as a mechanism of apoptosis protection. Neoplasia (2006) 8(12):1042–54. doi: 10.1593/neo.06568
17. Jia L, Soengas MS, Sun Y. Roc1/Rbx1 E3 ubiquitin ligase silencing suppresses tumor cell growth Via sequential induction of G2-m arrest, apoptosis, and senescence. Cancer Res (2009) 69(12):4974–82. doi: 10.1158/0008-5472.Can-08-4671
18. Migita K, Takayama T, Matsumoto S, Wakatsuki K, Tanaka T, Ito M, et al. Prognostic impact of ring box protein-1 (Rbx1) expression in gastric cancer. Gastric Cancer (2014) 17(4):601–9. doi: 10.1007/s10120-013-0318-y
19. Wu Q, Zhou X, Li P, Ding M, You S, Xu Z, et al. Roc1 promotes the malignant progression of bladder cancer by regulating p-Iκbα/Nf-Kb signaling. J Exp Clin Cancer Res (2021) 40(1):158. doi: 10.1186/s13046-021-01935-5
20. Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, et al. Validation of Sag/Rbx2/Roc2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res (2010) 16(3):814–24. doi: 10.1158/1078-0432.Ccr-09-1592
21. Li H, Tan M, Jia L, Wei D, Zhao Y, Chen G, et al. Inactivation of Sag/Rbx2 E3 ubiquitin ligase suppresses Krasg12d-driven lung tumorigenesis. J Clin Invest (2014) 124(2):835–46. doi: 10.1172/jci70297
22. Wang Y, Tan M, Li H, Li H, Sun Y. Inactivation of sag or Roc1 E3 ligase inhibits growth and survival of renal cell carcinoma cells: Effect of bim. Transl Oncol (2019) 12(6):810–8. doi: 10.1016/j.tranon.2019.03.002
23. Jiménez-García B, Teixeira JMC, Trellet M, Rodrigues J, Bonvin A. Pdb-tools web: A user-friendly interface for the manipulation of pdb files. Proteins (2021) 89(3):330–5. doi: 10.1002/prot.26018
24. Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. Gromacs: Fast, flexible, and free. J Comput Chem (2005) 26(16):1701–18. doi: 10.1002/jcc.20291
25. Mooers BHM. Shortcuts for faster image creation in pymol. Protein Sci (2020) 29(1):268–76. doi: 10.1002/pro.3781
26. Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data Via the xena platform. Nat Biotechnol (2020) 38(6):675–8. doi: 10.1038/s41587-020-0546-8
27. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. Ualcan: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
28. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. Tisidb: An integrated repository portal for tumor-immune system interactions. Bioinformatics (2019) 35(20):4200–2. doi: 10.1093/bioinformatics/btz210
29. Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. Gscalite: A web server for gene set cancer analysis. Bioinformatics (2018) 34(21):3771–2. doi: 10.1093/bioinformatics/bty411
30. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. Timer: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res (2017) 77(21):e108–e10. doi: 10.1158/0008-5472.Can-17-0307
31. Dou XW, Liang YK, Lin HY, Wei XL, Zhang YQ, Bai JW, et al. Notch3 maintains luminal phenotype and suppresses tumorigenesis and metastasis of breast cancer Via trans-activating estrogen receptor-A. Theranostics (2017) 7(16):4041–56. doi: 10.7150/thno.19989
32. Ho MS, Tsai PI, Chien CT. F-box proteins: The key to protein degradation. J BioMed Sci (2006) 13(2):181–91. doi: 10.1007/s11373-005-9058-2
33. Kuang P, Tan M, Zhou W, Zhang Q, Sun Y. Sag/Rbx2 E3 ligase complexes with Ubch10 and Ube2s E2s to ubiquitylate B-Trcp1 Via K11-linkage for degradation. Sci Rep (2016) 6:37441. doi: 10.1038/srep37441
34. Tan M, Zhao Y, Kim SJ, Liu M, Jia L, Saunders TL, et al. Sag/Rbx2/Roc2 E3 ubiquitin ligase is essential for vascular and neural development by targeting Nf1 for degradation. Dev Cell (2011) 21(6):1062–76. doi: 10.1016/j.devcel.2011.09.014
35. Mizuno H, Kitada K, Nakai K, Sarai A. Prognoscan: A new database for meta-analysis of the prognostic value of genes. BMC Med Genomics (2009) 2:18. doi: 10.1186/1755-8794-2-18
36. Tan M, Zhu Y, Kovacev J, Zhao Y, Pan ZQ, Spitz DR, et al. Disruption of Sag/Rbx2/Roc2 induces radiosensitization by increasing ros levels and blocking nf-kappab activation in mouse embryonic stem cells. Free Radic Biol Med (2010) 49(6):976–83. doi: 10.1016/j.freeradbiomed.2010.05.030
37. Wu D, Li H, Liu M, Qin J, Sun Y. The Ube2m-Rbx1 neddylation-Cullin-Ring-Ligase proteins are essential for the maintenance of regulatory T cell fitness. Nat Commun (2022) 13(1):3021. doi: 10.1038/s41467-022-30707-8
38. Mathewson ND, Fujiwara H, Wu SR, Toubai T, Oravecz-Wilson K, Sun Y, et al. Sag/Rbx2-dependent neddylation regulates T-cell responses. Am J Pathol (2016) 186(10):2679–91. doi: 10.1016/j.ajpath.2016.06.014
39. Thompson LL, Rutherford KA, Lepage CC, McManus KJ. The scf complex is essential to maintain genome and chromosome stability. Int J Mol Sci (2021) 22(16):8544. doi: 10.3390/ijms22168544
40. Yumimoto K, Yamauchi Y, Nakayama KI. F-box proteins and cancer. Cancers (Basel) (2020) 12(5):1249. doi: 10.3390/cancers12051249
41. Zhang S, Shen Y, Li H, Bi C, Sun Y, Xiong X, et al. The negative cross-talk between Sag/Rbx2/Roc2 and Apc/C E3 ligases in regulation of cell cycle progression and drug resistance. Cell Rep (2020) 32(10):108102. doi: 10.1016/j.celrep.2020.108102
42. Tan M, Li H, Sun Y. Inactivation of Sag/Rbx2/Roc2 E3 ubiquitin ligase triggers senescence and inhibits kras-induced immortalization. Neoplasia (2015) 17(1):114–23. doi: 10.1016/j.neo.2014.11.008
43. Altintas E, Kaynar M, Celik ZE, Celik M, Kilic O, Akand M, et al. Expression of ring box-1 protein and its relationship with fuhrman grade and other clinical-pathological parameters in renal cell cancer. Urol Oncol (2020) 38(1):6.e17–22. doi: 10.1016/j.urolonc.2019.09.019
44. Ye P, Chi X, Cha JH, Luo S, Yang G, Yan X, et al. Potential of E3 ubiquitin ligases in cancer immunity: Opportunities and challenges. Cells (2021) 10(12):3309. doi: 10.3390/cells10123309
45. Zhang Y, Li LF, Munir M, Qiu HJ. Ring-domain E3 ligase-mediated host-virus interactions: Orchestrating immune responses by the host and antagonizing immune defense by viruses. Front Immunol (2018) 9:1083. doi: 10.3389/fimmu.2018.01083
46. Ottina E, Panova V, Doglio L, Kazachenka A, Cornish G, Kirkpatrick J, et al. E3 ubiquitin ligase Hectd2 mediates melanoma progression and immune evasion. Oncogene (2021) 40(37):5567–78. doi: 10.1038/s41388-021-01885-4
Keywords: pan-cancer, ring-finger proteins, SCF complex, prognosis, immune infiltration, difference
Citation: Huang T, Li J, Liu X, Shi B, Li S and An H-X (2022) An integrative pan-cancer analysis revealing the difference in small ring finger family of SCF E3 ubiquitin ligases. Front. Immunol. 13:968777. doi: 10.3389/fimmu.2022.968777
Received: 14 June 2022; Accepted: 01 August 2022;
Published: 18 August 2022.
Edited by:
Jian Song, University Hospital Münster, GermanyReviewed by:
Lele Zhu, University of Texas MD Anderson Cancer Center, United StatesCopyright © 2022 Huang, Li, Liu, Shi, Li and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanxiang An, YW5oYW54aWFuZ0B4bXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.