- 1Department of Aquatic Animal Health, Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin, China
- 2College of Fisheries and Life Science, Shanghai Ocean University, Shanghai, China
- 3Key Laboratory of Aquatic Animal Diseases and Immune Technology of Heilongjiang Province, Harbin, China
- 4Department of Fisheries and Wildlife, College of Agriculture and Natural Resources, Michigan State University, East Lansing, MI, United States
- 5Department of Pathobiology and Diagnostic Investigation, College of Veterinary Medicine, Michigan State University, East Lansing, MI, United States
- 6College of Veterinary Medicine, Northeast Agricultural University, Harbin, China
Flavobacterium psychrophilum, the etiological agent of bacterial coldwater disease and rainbow trout fry syndrome, causes considerable losses in salmonid aquaculture globally. Systemic F. psychrophilum infections in rainbow trout (Oncorhynchus mykiss) lead to a range of clinical signs, including ulcerative lesions in the skin and muscle and splenitis. Previous studies offered an integrative analysis of the skeletal muscle response to F. psychrophilum infection in rainbow trout. However, little is known about the molecular mechanism of immune response in the spleen, which is an important immune organ of rainbow trout. Here, we investigated the time-course splenic transcriptome profiles in uninfected rainbow trout (CK) and F. psychrophilum–infected rainbow trout at day 3 and day 7 (D3, D7) by RNA-seq analyses. Among the 7,170 differentially expressed genes (DEGs) in the three comparisons (D3 vs. CK, D7 vs. CK, D3 vs. D7), 1,286 DEGs showed consistent upregulation or downregulation at D3 and D7 and were associated with pattern recognition, acute-phase response, complement cascade, chemokine and cytokine signaling, and apoptosis. The Real time quantitative PCR (RT-qPCR) analysis of eight DEGs confirmed the accuracy of the RNA-Sequencing (RNA-seq) data. Our results reflected a general process from pathogen recognition to inflammatory cytokine generation and delineated a putative Toll-like receptor signaling pathway in rainbow trout spleen, following F. psychrophilum infection. Taken together, these results provide new insights into the molecular mechanism of the immune response to F. psychrophilum infection and are a valuable resource for future research on the prevention and control of bacterial coldwater disease during salmon culture.
Introduction
The outbreaks of bacterial coldwater disease (BCWD) and rainbow trout fry syndrome continue to cause considerable losses in freshwater salmonid aquaculture around the globe (1). The etiological agent of this disease, Flavobacterium psychrophilum, is a Gram-negative rod-shaped bacterium (2) that has been reported throughout Asia, Europe, North America, South America, and Australia (3). Although infections have been reported in other fish species, a wide variety of salmonid species are susceptible during freshwater stages (4). Rainbow trout (Oncorhynchus mykiss) infected with F. psychrophilum exhibit a range of clinical signs, including ulcerative lesions and necrotic myositis in skeletal muscle (5). Histologically, extensive damage occurs in many tissues, including the liver, spleen, gill, kidney, and intestine wall (6). Among these, spleen and kidney contain the highest levels of F. psychrophilum, possibly leading to the suppression of the acquired immune system triggered by this pathogen (7). Likewise, the inflammatory infiltrate of spleen and total splenic ellipsoid increased in rainbow trout post-F. psychrophilum infection (8). It was postulated that the spleen size may be an indirect indicator of rainbow trout resistance to F. psychrophilum (9), which has led to an increase in studies into the spleen function in teleost fishes (10). However, there is still lack of a comprehensive understanding on the interaction mechanism in rainbow trout spleen after F. psychrophilum infection.
Next-generation sequencing technologies have deepened the understanding of immune function and immune-related genes in aquatic animals. Splenic transcriptomes obtained using these technologies have provided important information about the immune response in trout (11, 12). In a previous study, the whole-body gene expression between the genetic lines ARS-Fp-R (resistant) and ARS-Fp-S (susceptible) of rainbow trout was analyzed by high-throughput RNA sequencing (RNA-seq), revealing that 1,884 transcripts, including chemokines, complement components, TNF receptor superfamily members, interleukins, Nucleotide-binding Oligomerization Domain (NOD)-like receptor family members, and genes involved in metabolism and wound healing, exhibited a differential expression between F. psychrophilum infection and the control group (13). RNA-Seq has also been applied to uncover the immune response of rainbow trout to several bacterial pathogens like Lactococcus garvieae (12) and Aeromonas salmonicida (14). Taken together, transcriptome analysis can provide a comprehensive understanding of host immunity and help to uncover the underlying immune mechanisms in aquatic animals in response to pathogen infection.
In the present study, we evaluated the rainbow trout spleen transcriptome in response to F. psychrophilum infection using RNA-seq analyses with the aims of: a) providing the transcriptome evidence on the protective effect of acute-phase response (APR), complement systems, Th1-type responses, and caspase-mediated apoptosis in the spleen of rainbow trout against F. psychrophilum invasion and b) generating a valuable resource for future research on the prevention and control of BCWD during salmon culture.
Material and methods
Bacterial strain isolation and culture
The F. psychrophilum isolate CN06, which belongs to ST12 within CC-ST10 (2), originally recovered from the ulcerated muscle of diseased rainbow trout and previously confirmed to be virulent in in vivo challenge experiments, was selected for use in the present study. The CN06 strain was grown for 72 h in the TYES broth medium at 15°C, then collected by centrifuging at 6,000 × g for 5 min and resuspended to a concentration of 108 Colony-Forming Units (CFU)·ml−1 in sterile PBS (pH 7.2). To detect F. psychrophilum in the spleen of infected rainbow trout, a PCR assay was conducted following the procedures described by Li et al. (2).
Bacterial challenge and sampling
Healthy rainbow trout (10–15 g) were obtained from Agrimarine Industrial (Benxi, China), and kept in flow-through tanks for 2 weeks at 14 ± 1°C. During the experiment, 48 rainbow trout (12 fish/m3) were fed commercial trout feed twice a day (1.2% of body weight). After verifying the freedom of F. psychrophilum infection by bacteriological examination, 48 fish were divided randomly and averaged to four groups: two control groups and two infected groups (12 fish per group). Tricaine methane sulfonate (MS222) was used to anesthetize the fish prior to the challenge and prior to tissue sample collection. The infection was induced experimentally by intramuscular injection in the caudal peduncle with 4 × 106 CFU per gram of fish body weight (LD50), whereas fish in the control group were injected with an equivalent volume of phosphate-buffered saline (PBS). Our pre-experiment indicated that rainbow trout showed clinical signs 3 days post-CN06 infection and death occurred 7 days postinfection. Based on the pilot study, the spleen of rainbow trout was euthanized with MS222 and the spleen was collected and frozen immediately in liquid nitrogen at day 3 and day 7 (D3 and D7).
Messenger RNA (mRNA) library construction and sequencing
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Austin, TX, USA), following the manufacturer’s instructions. The RNA concentration was measured by Thermo Scientific™ NanoDrop™ 8000 trace ultraviolet-visible spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
A total of 1-µg RNA per sample was used for RNA sample preparations, and sequencing libraries were generated using an NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA), following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Index-coded samples were clustered on the cBot Cluster Generation System using the TruSeq PE Cluster Kit v4-cBot-HS for Illumina (NEB, USA) according to the manufacturer’s instructions. After cluster generation, library preparations were sequenced on the Illumina platform and double-ended reads were generated.
RNA-seq read mapping and gene functional annotation
The raw reads were firstly filtered using fastp v 0.20.0 by eliminating adaptor sequences and low-quality reads. The clean reads were mapped to the O. mykiss reference genome sequence (genoscope.cns.fr/trout/data/) (15) using Hisat2 (daehwankimlab.github.io/hisat2). Only one mismatch was allowed, and the mapped reads were further analyzed and annotated based on the reference genome data. The Q20, Q30, GC content, and sequence duplication level of the clean data were calculated. The high-quality clean data were used for all the downstream analyses. The mapped reads were annotated by Basic Local Alignment Search Tool (BLAST) searches against the Nr (ncbi.nlm.nih.gov/refseq/release/), Nt (ncbi.nlm.nih.gov/nucleotide/), Pfam (pfam.xfam.org/), Swiss-Prot (expasy.org/resources/uniprotkb-swiss-prot), Cluster of Orthologous Groups of proteins (COG) (ncbi.nlm.nih.gov/research/cog-project/), KO (genome.jp/kegg/), and GO (geneontology.org/) databases. The genes with |log2Ratio| ≥2 and false discovery rate (FDR) ≤0.01 were considered as significantly differentially expressed. The quantification of gene expression levels was estimated by fragments per kilobase of transcript per million fragments mapped (FPKMs). The differential expression analysis of two comparisons (D3 vs. CK and D7 vs. CK) was performed using the DEseq R package. Functional annotation of the DEGs was performed, and the number-annotated DEGs were counted. The GO enrichment analysis was performed using the GOseq R packages, and GO terms with a corrected p-value <0.05 were considered to be enriched. KEGG pathway enrichment was performed using the KOBAS software (16). The TLR signaling pathway was constructed using PathVisio v.3.0.
Construction of the rainbow trout immune gene library
The classification of rainbow trout immune genes was constructed, following a thorough review of teleost fish immunology (10, 17), and the common carp immune gene library (18) was modified to the rainbow trout immune gene library. The modifications were based on gene information obtained by blasting each sequence to databases, including the non-redundant protein sequences (Nr), Swiss-Prot and Pfam databases. According to both classical immunology and new advances in both fish and mammal immunology, nine categories of immune processes were compiled for this study, including “acute phase reactions”, “pattern recognition”, “antigen processing and regulators”, “complement system”, “inflammatory cytokines and receptors”, “adapters, effectors and signal transducers”, “innate immune cells related”, “T/B cell antigen activation”, and “other genes related to immune response”. Additionally, and because some transcripts expressing respiratory burst activity were revealed, respiratory burst activity was added as an additional category of the immune process. Numerous researched categories of immune genes (detailed in Table S4) were applied for the immune process.
RNA-seq validation by real-time RT-qPCR
To validate the RNA-seq results, eight DEGs including ccl19, casp8, infgr1, il8, c3, saa, tlr2, and tnf10, which are involved in chemotactic effects, proinflammatory effects, APR, complement cascade, and the Toll-like receptor signaling pathway were further analyzed by RT-qPCR. The RT-qPCR primers are listed in Table S1. Gene expression levels were normalized to that of β-actin, and data were expressed as the mean ± SE. Differences in means between groups were determined using one-way ANOVA, followed by Bonferroni’s post-test. Data with p <0.05 were considered significant. Correlations between the RNA-seq and RT-qPCR data were assessed by multiple linear regression to determine the coefficient of determination (R2) and p. All statistical analyses were performed using GraphPad Prism v.9.00 (GraphPad Software, San Diego, CA, CA, USA).
Results
Transcriptome sequencing, quality control, and gene alignments
Nine cDNA libraries were constructed from rainbow trout spleen to determine changes in the immune system, following F. psychrophilum injection. The raw data generated in each library have been deposited in the NCBI Sequence Read Archive under accession number PRJNA675781. After the quality control of the cDNA libraries, a total of 19,895,398–24,964,841 clean reads were obtained and the percentage of Q30 bases in each sample was more than 92.83% (Table 1). All clean reads were mapped to the O. mykiss reference genome (NCBI O. mykiss Annotation Release 101) (Table S2). The normal distribution of gene expression profiling indicated that the three biological replicates had good consistency (Supplementary Figure S1). Most reads in each library were mapped to the exon regions of the reference genome (Supplementary Figure S2). A total of 8,630 new genes were detected in the three libraries (CK, D3, D7) by RNA-seq analysis, and 74.84% (6,459/8,630) were mapped to the reference genes with homologous sequences in at least one of the multiple databases presented in Table S3.
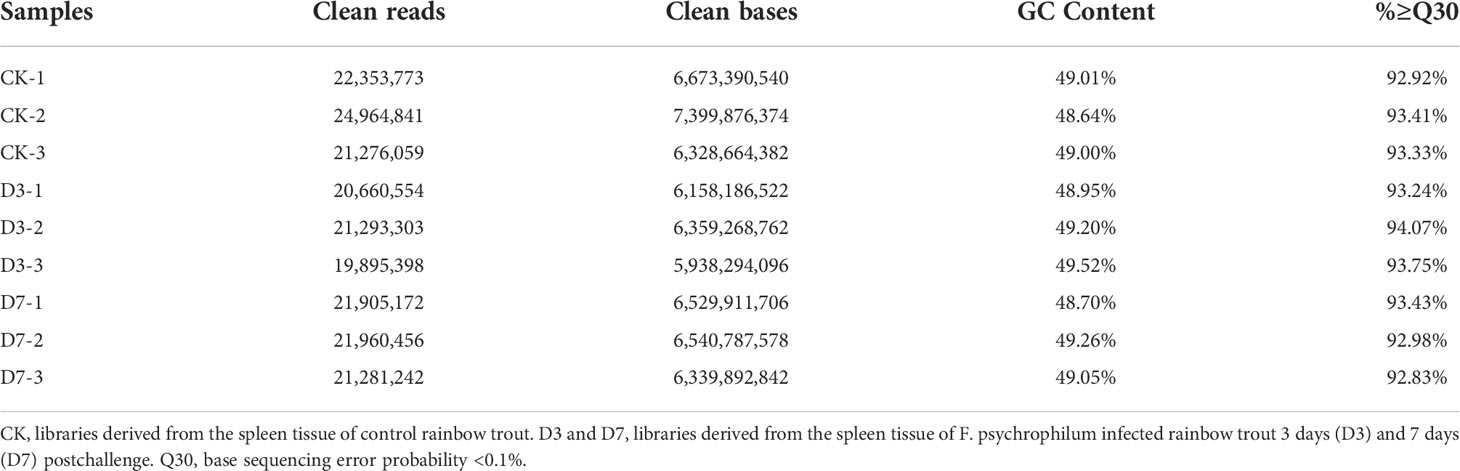
Table 1 Characteristics of the reads from nine sample libraries derived from the spleens of Flavobacterium psychrophilum–challenged and mock-challenged rainbow trout.
Analysis of differentially expressed genes
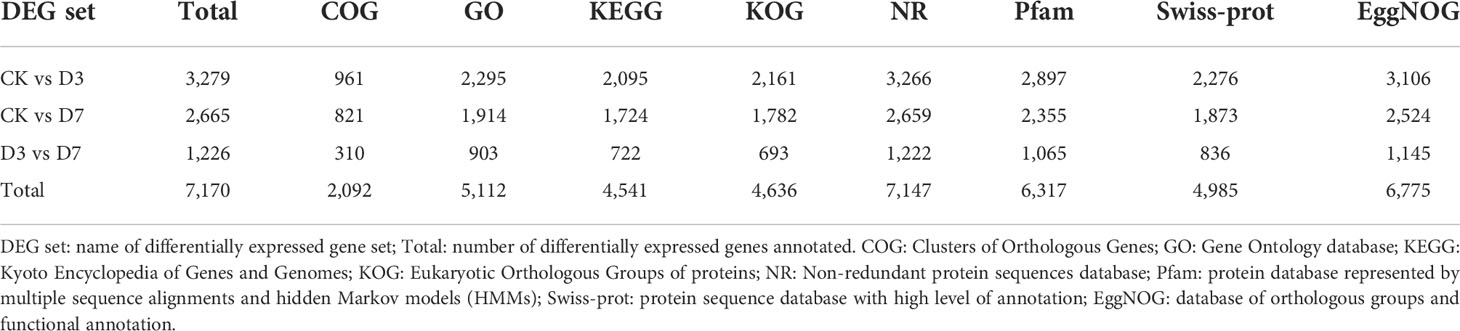
A total of 7,170 DEGs were identified among the three groups (Table 2). Compared with CK, D3 and D7 included 3,376 and 2,737 significant DEGs, respectively. Among them, 1,809 upregulated and 1,567 downregulated genes were identified for CK vs. D3, 1,461 upregulated and 1,276 downregulated genes for CK vs. D7, and 2,601 upregulated and 2,260 downregulated genes for D3 vs. D7 (Supplementary Figure S3). Moreover, 1,286 DEGs were common to the CK vs. D3 and CK vs. D7 comparisons, and 84 DEGs were common to all three comparisons (CK vs. D3, CK vs. D7, D3 vs. D7; Figure 1).
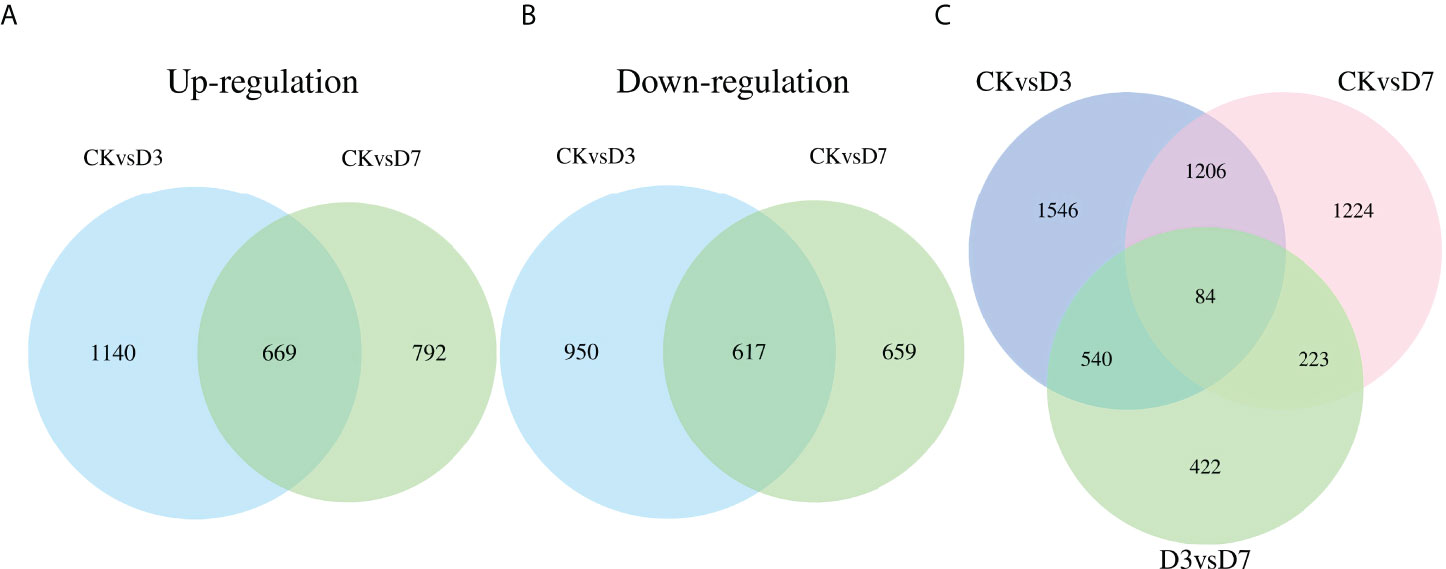
Figure 1 Venn diagrams of differentially expressed genes (DEGs) detected in the spleens of rainbow trout, following Flavobacterium psychrophilum and mock challenge infections. (A, B) Quantities of up- and downregulated DEGs between groups CK (contro l group) vs. D3 (day 3) and CK vs. D7 (day 7). (C) Quantities of DEGs among the three groups.
Classification of differentially expressed genes from different comparisons into immune process and immune gene category
We constructed the rainbow trout immune gene libraries including the immune process; gene categories and annotations are presented in the Table S4. At the level of immune processing, in group CK vs. D3, most immune-related genes were upregulated in the “adapters effectors and signal transducers,” “antigen processing and regulators,” “inflammatory cytokines and receptors,” “patterns recognition,” “other genes related to immune response,” and “T/B cell antigen activation” (Figure 2A, Table 3 and S5). In group CK vs. D7, most immune genes were upregulated in the “adapters effectors and signal transducers”, “inflammatory cytokines and receptors”, “patterns recognition”, “other genes related to immune response,” and “T/B cell antigen activation.” In all groups, “inflammatory cytokines and receptors,” “other genes related to immune response,” and “T/B cell antigen activation” exhibited the greatest DEG number. A Venn diagram was created to demonstrate the relationship between the immune-related DEGs among groups CK vs. D3 and CK vs. D7 (Figure 2B); the comparison results of Venn are presented in Table S6. Comparison results are synthesized on Figure 2B; several DEGs commonly regulated in groups CK vs. D3, and CK vs. D7 within nine immune processes are further depicted (Table 3).
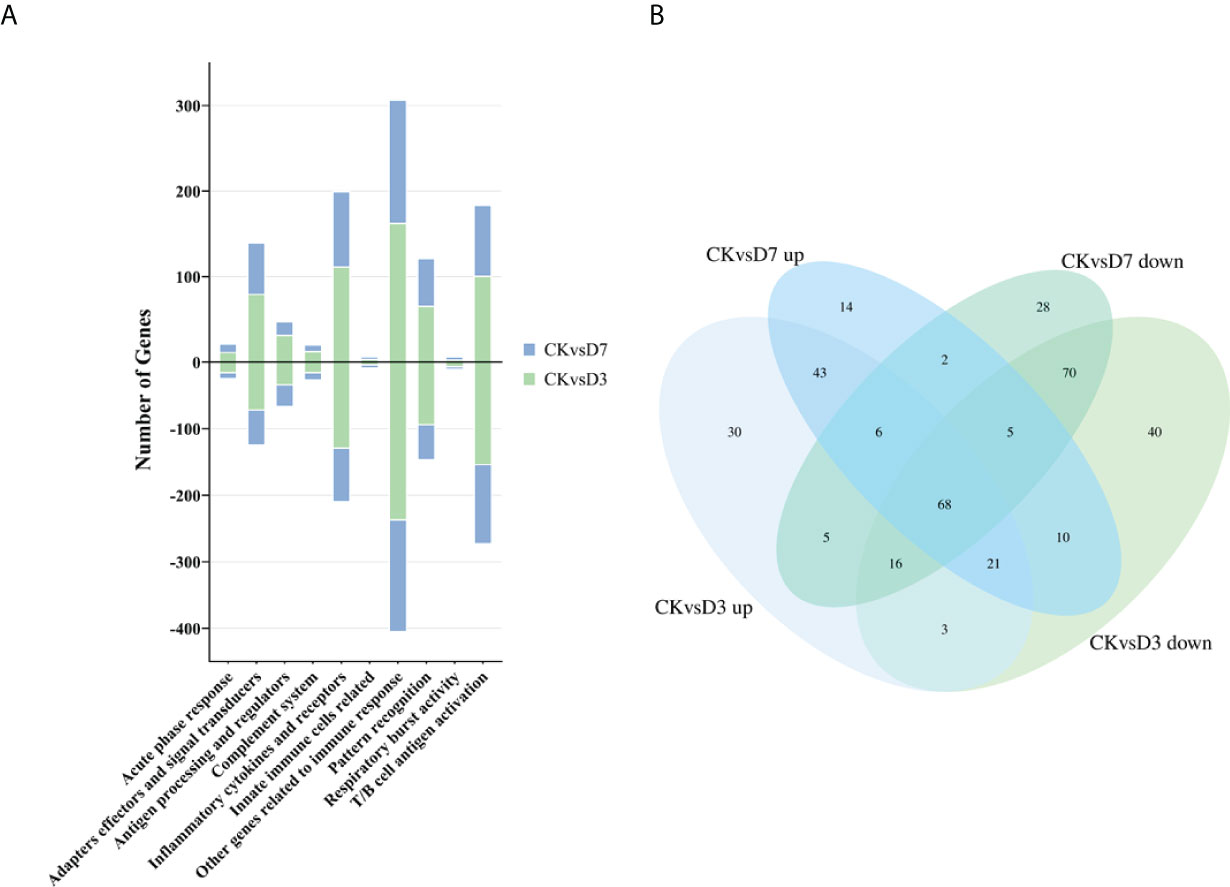
Figure 2 Analysis of the immune-related gene category. (A) Number of DEGs belonging to each of the 10 gene categories. (B) Venn diagram of the immune gene number of up- or downregulated DEGs in groups CK vs. D3 and CK vs. D7.
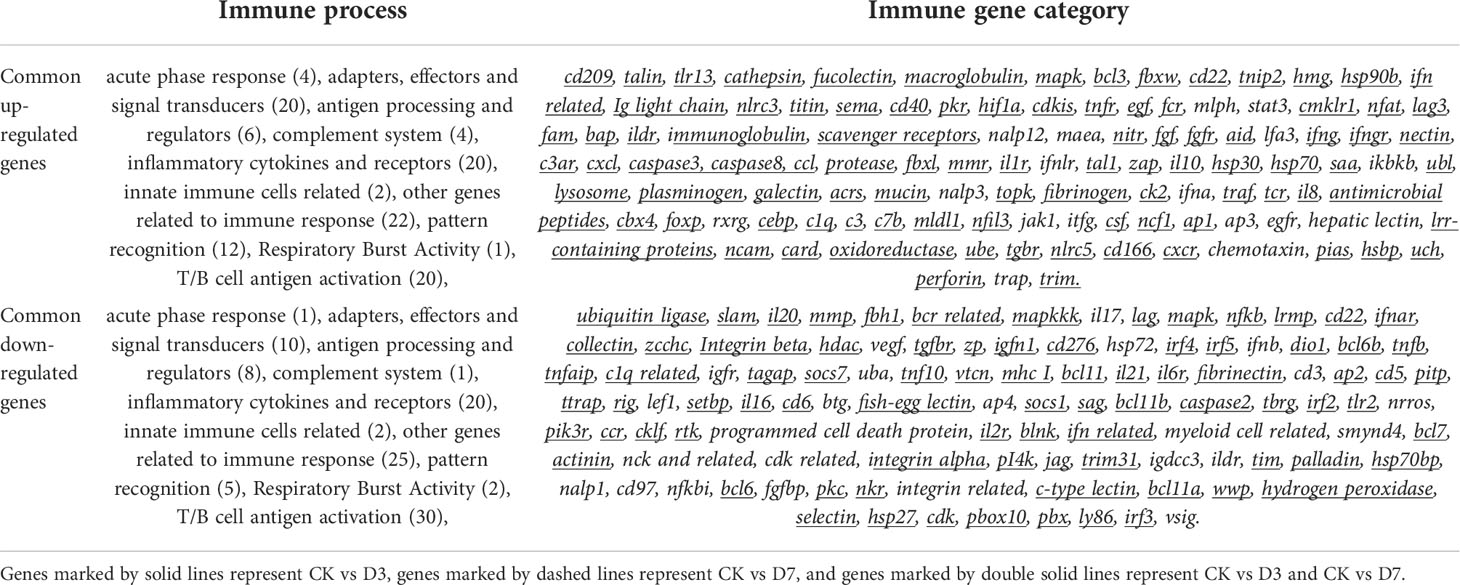
Table 3 The immune process and immune gene category for overlapping genes in the Venn diagram, DEGs with FC>2 is underlined.
Gene ontology analysis of differentially expressed genes
The DEGs were further annotated with terms from the Gene Ontology (GO) database under the three main GO categories: biological process, cellular component, and molecular function. GO terms were assigned to 2,292 and 1,912 DEGs for D3 and D7, respectively (Figure 3; Table S7). Under cellular component, “cell” and “membrane” were the most enriched terms; under molecular function, “catalytic activity” and “binding” were the most enriched terms; and under biological process, “cellular process”, “single-organism process”, “metabolic process”, “response to stimulus”, “biological regulation”, “signaling”, “localization”, “multicellular organismal process”, “developmental process”, “cellular component organization or biogenesis” and “immune system process” were the most enriched terms. The number of DEGs annotated with the term “immune system process” was higher in CK vs. D3 than it was in CK vs. D7.
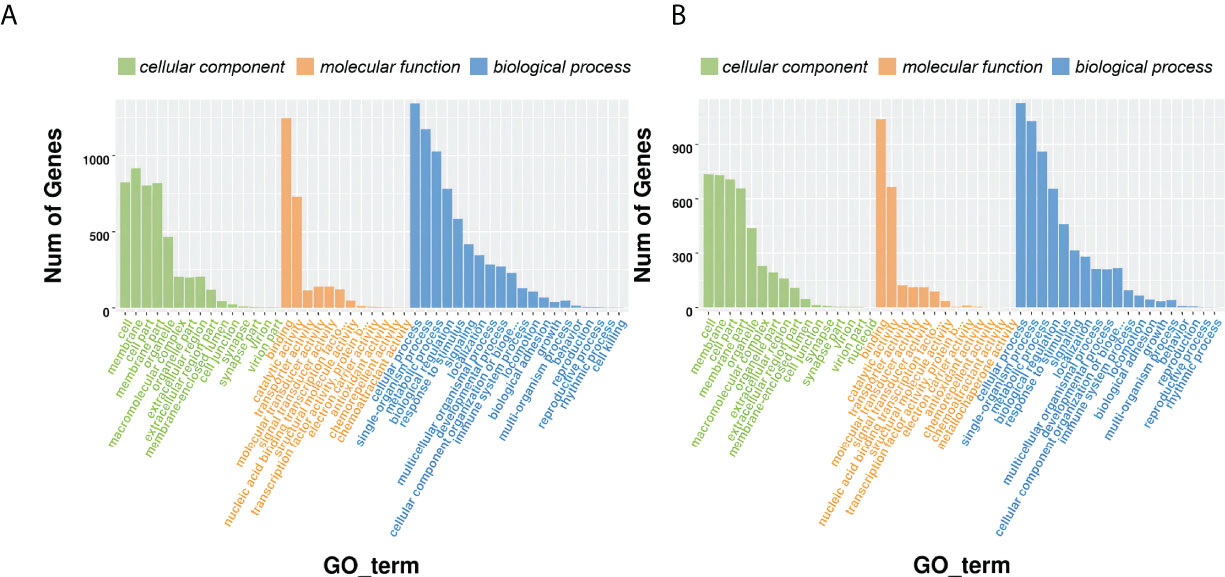
Figure 3 Gene ontology (GO) annotation of the differentially expressed genes. (A) CK vs. D3; (B) CK vs. D7.
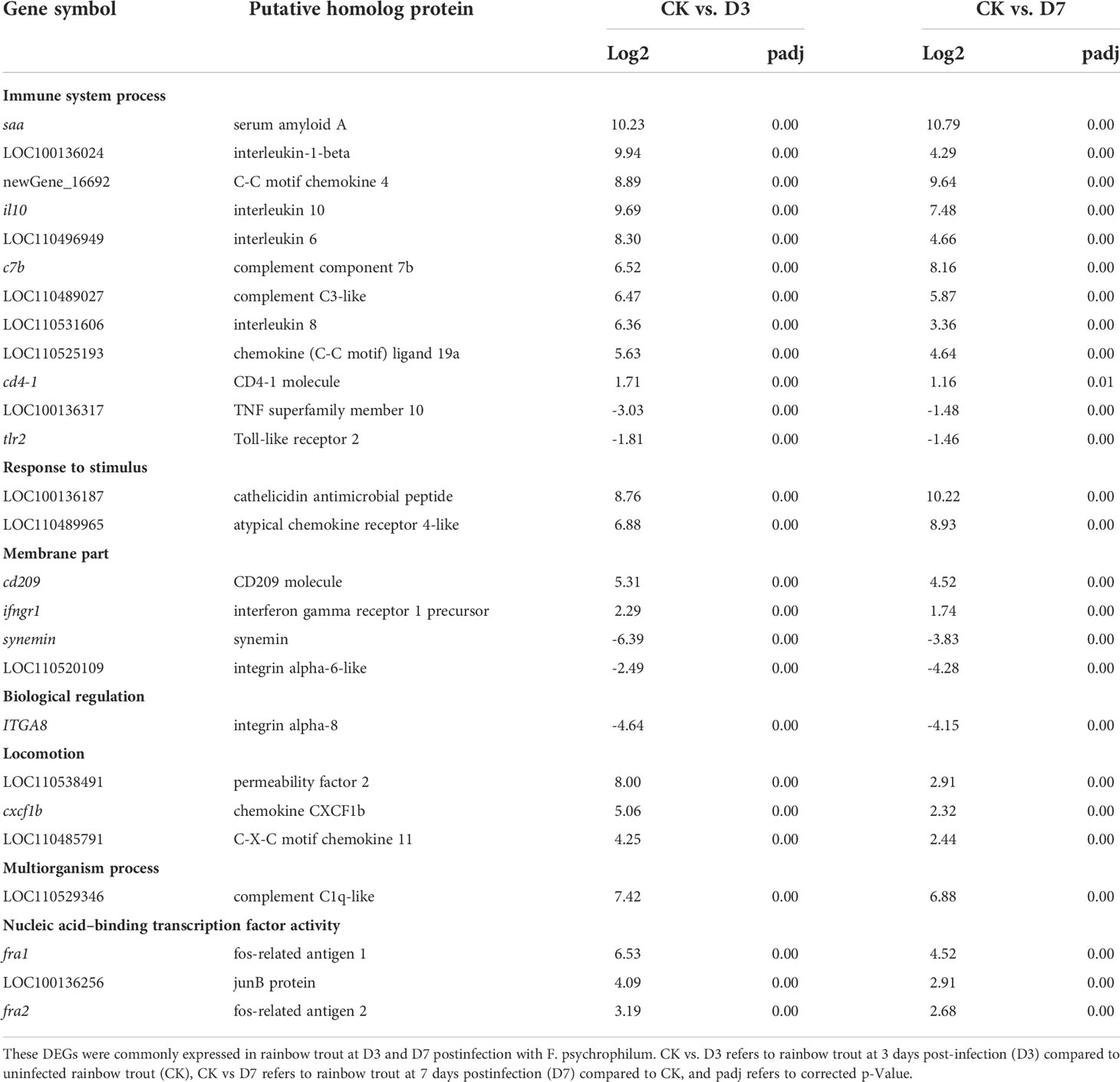
To explore the representative GO terms associated with F. psychrophilum infection in rainbow trout spleen, the details of the main DEGs that may have important immune functions are listed in Table 4. The transcript abundance of cytokine genes il1β, il10, il6, and il8; APR gene saa; chemokine gene cc4, ccl19a, cxcf1b, and cxc11; and complement system genes c7b, c3, and c1q increased in D3 and D7 compared with their abundance in CK, whereas the transcripts associated to the cell membrane including tlr2, itga6, itga8, and the structural molecule activity gene synemin decreased.
Pathway enrichment analysis of the differentially expressed genes
To further explore the immune-related pathways during F. psychrophilum infection in rainbow trout, we performed a KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis of the DEGs. The DEGs from the CK vs. D3 and CK vs. D7 comparisons were mapped to 161 and 186 KEGG pathways, respectively, whereby 27 and 31 of them were significantly enriched (p <0.05). At D3, the DEGs were involved in three immune-related pathways including herpes simplex infection, the NOD-like receptor signaling pathway, and the Toll-like receptor (TLR) signaling pathway. At D7, the DEGs were significantly enriched in the phagosome, oocyte meiosis, cell cycle, and p53 signaling pathways (Figure 4).
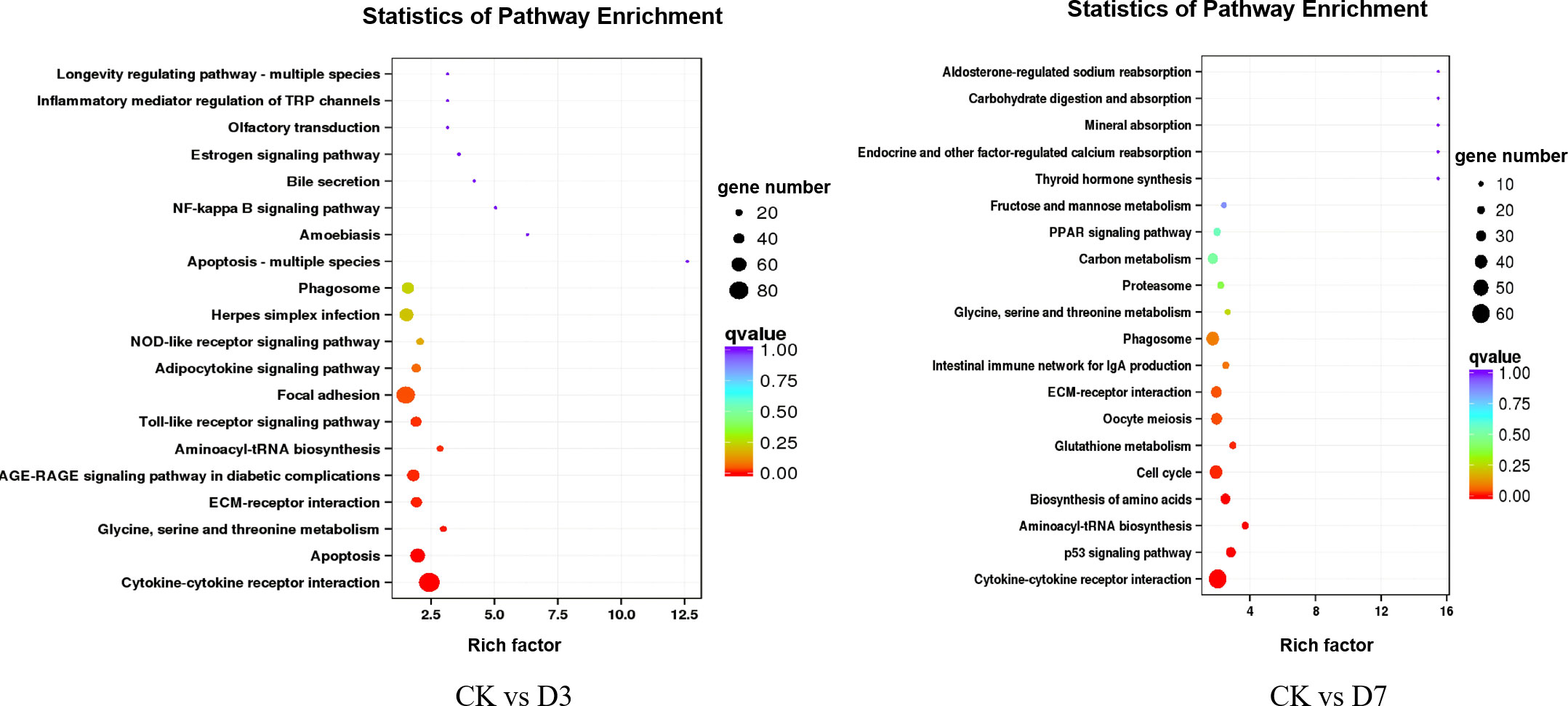
Figure 4 Scatterplot of the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis. Rich factor represents the ratio of the number of DEGs and the number of all genes in the pathways.
Significantly enriched pathways were mainly classified as immune system and cell growth and death. Thereby, DEGs annotated in these two subcategories were selected and re-enriched for the more explicit functions, as shown in Figure 5. A total of 1,286 DEGs were common to CK vs. D3 and CK vs. D7, and 210 genes in these two subcategories were annotated to 18 KEGG pathways. Among them, apoptosis, Toll-like receptor signaling pathway, cell cycle and p53 signaling pathway were significantly enriched distinguished to other pathways, both in the gene number and the rich factor.
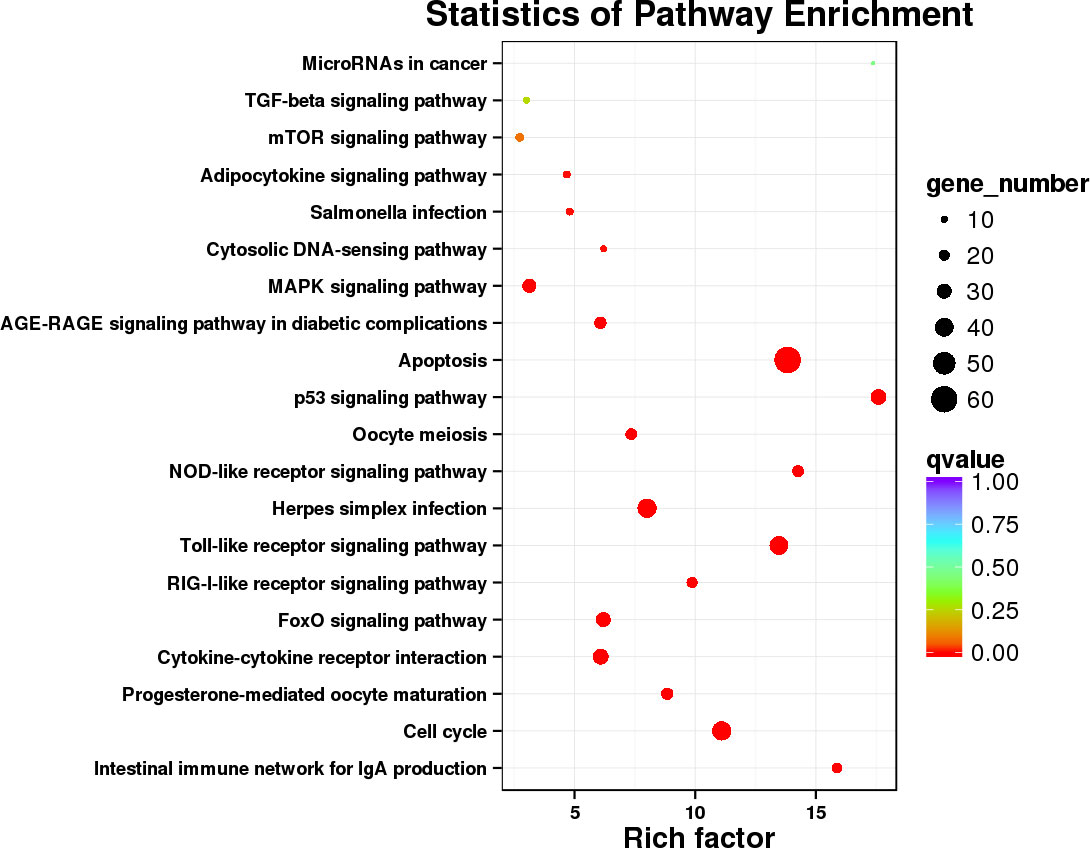
Figure 5 Scatterplot of the KEGG pathways involved in the immune system and cell growth and death. Rich factor represents the ratio of the number of DEGs and the number of all genes in the pathways.
The immune-related DEGs in the TLR signaling pathway mainly contained cytokines, chemokines, and APR genes, such as APR saa; inflammatory cytokines il8, il1β, and il6; interferon receptor ifngr1; apoptosis pathway genes casp8 and casp3; ubiquitin-mediated proteolysis nfkbie and nfkbia, and oncogenic transcription factors jun and fos, which were upregulated expression at D3 and D7 (Figure 6). In contrast, TLRs such as tlr7, tlr9, tlr2, and tril showed downregulated expression at D3 and D7.
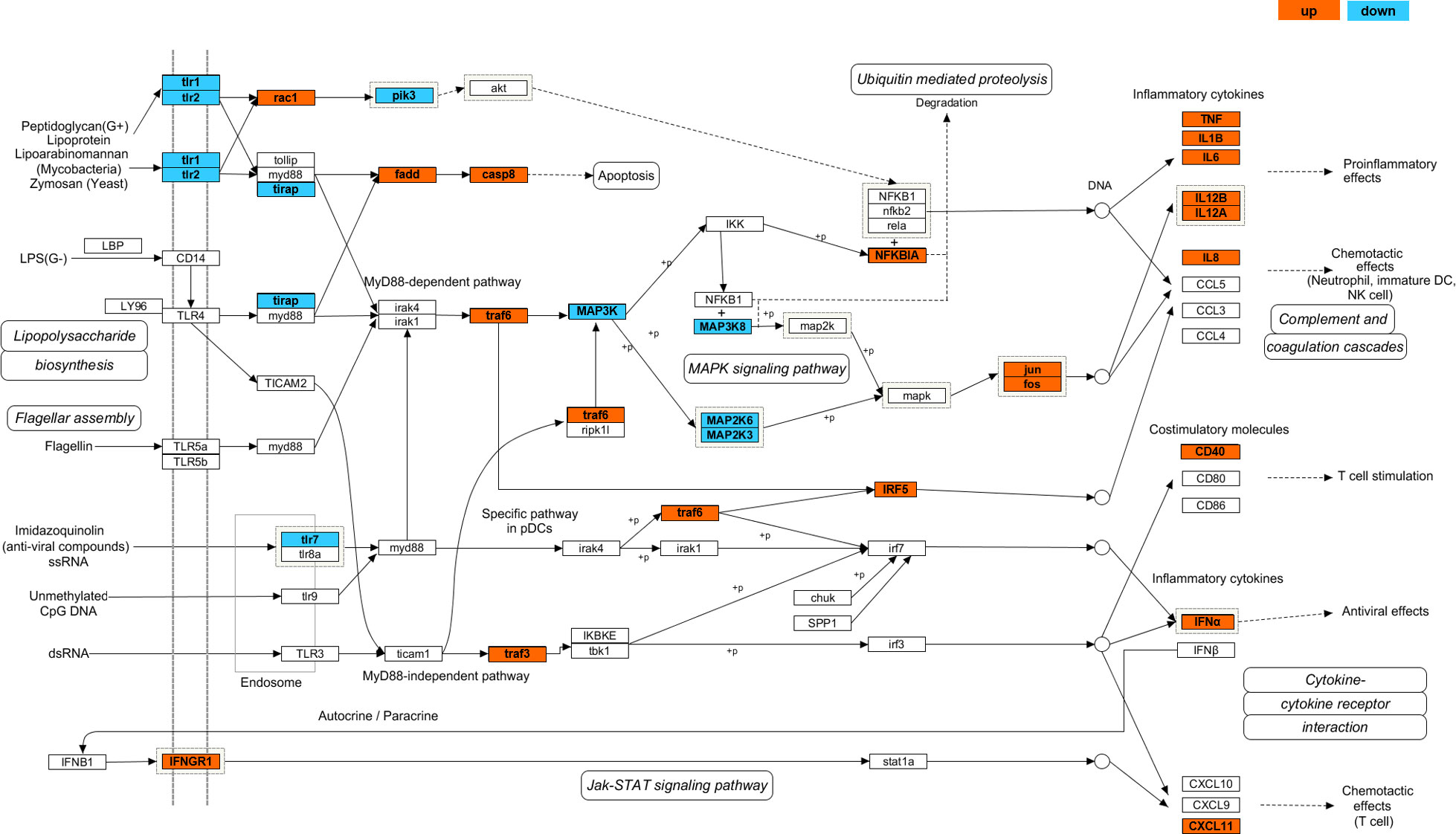
Figure 6 Predicted integrated Toll-like receptor signaling pathway in rainbow trout spleen. The pathway was constructed based on the pathway in Danio rerio (zebrafish) using PathVisio v3. Blue and red indicate a decrease and increase in any of component of the pathways, respectively, which contains D3 and D7 time points postinfection.
Validation of selected differentially expressed genes by RT-qPCR analysis
To validate the RNA-seq analyses, eight immune-related DEGs containing both up- and down-regulated genes were chosen for RT-qPCR analysis (Figure 7). The gene expression fold changes at D3 and D7 measured by RT-qPCR were highly correlated with those obtained by RNA-seq analysis. The significant coefficients between RT-qPCR and RNA-seq for the DEGs in D3 and D7 groups were 0.99 and 0.98, respectively.
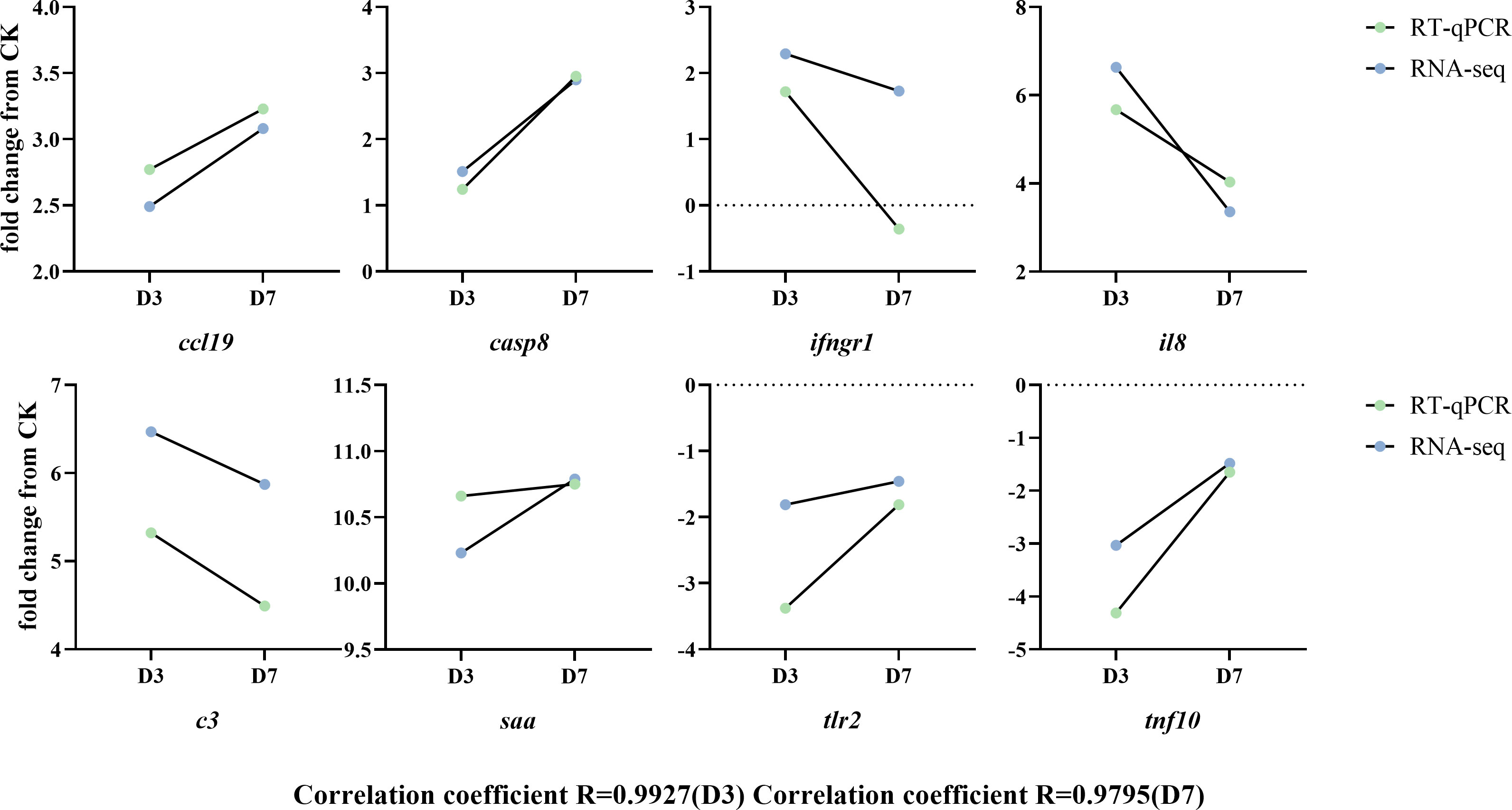
Figure 7 Validation of selected differentially expressed genes by RT-qPCR. Fold changes in rainbow trout at 3 and 7 days postinfection (D3 and D7) with F. psychrophilum are expressed as the ratios of gene expression at D3 and D7 to gene expression in the CK as normalized with the β-actin gene. ccl19, C-C chemokine ligand 19; c3, complement C3; casp8, caspase 8; saa, serum amyloid A; ifngr1, interferon gamma receptor 1 precursor; tlr2, Toll-like receptor 2; il8, interleukin 8/permeability factor 2; tnf10, TNF superfamily member 10.
Discussion
Previous studies have described that the spleen size is an indirect indicator of immunological status in histologically (9). The mechanisms of immune response to F. psychrophilum in spleen are the key events which are vital matters to the interactions occurring in rainbow trout post bacterial infection. But until this study, the transcriptional response in the spleen of rainbow trout against F. psychrophilum infection was not fully understood. Herein and for the first time, attempts to elucidate this mechanism by transcriptome analysis have been performed to explore immunological insensitivity or susceptibility factors to F. psychrophilum infections in the present study. And our results have novel findings and identify the interactions between F. psychrophilum and its susceptible host rainbow trout.
Differentially expressed genes related to the pattern recognition receptors
Pattern recognition receptors (PRRs), including TLRs, C-type lectin receptors (CLRs), scavenger receptors (SRs), and nucleotide oligomerization domain–like receptors (NLRs), are critical for bony fish to develop a higher specialization in innate immunity after recognizing the pathogens. TLRs play an integrated role as a first line of defense against invading pathogens, where they contribute to the inflammation, immune cell regulation, survival, and proliferation of leukocytes. Here, we found that tlr2 expression was downregulated at D3 and D7, suggesting that it may play a negative regulatory role in rainbow trout immune defense, following F. psychrophilum infection. Different to our results, Tang et al. provided the evidence that TLRs in sturgeon showed upregulated expression in head–kidney primary leukoocytes, following LPS and polyI:C stimulation (19). In epithelial or mucosal barriers, tlr2 transcripts in rainbow trout skin were upregulated both at 4 h and 2 d post-immersion vaccination, suggesting that tlr2 was in contact with F. psychrophilum to initiate the subsequent immune response (7). Moreover, tlr7 expression was unchanged at D3 but downregulated at D7, consistent with previous studies (21). Most of the CLRs induce activating or regulatory signal cascades in response to distinct pathogen- or self-derived components. CD209 is shown to be an essential surface factor of the immune defense of rainbow trout, by recognizing high-mannose-containing glycoproteins on pathogens’ outer surfaces (14), and TLR13α1 is responsible for CD209 regulation (24). In the present study, we observed a dramatic increase of the expression level of cd209 and tlr13 in rainbow trout spleen at D3 and D7 post-F. psychrophilum infection (Table S5). Consistent with the findings of Long et al. (14), the tlr13α1 and cd209 genes were both transcriptionally upregulated in the spleens of fish experimentally challenged with another Gram-negative bacterial pathogen of salmonids, A. salmonicida. Of the CLRs, upregulated expression was detected in collectin, fish-egg lectin, and selectin, which could potentially be specific to various pathogens in Japanese eel (Anguilla japonica), European eel (Anguilla Anguilla), and zebrafish (29). The scavenger receptor, also presented in our analysis (Table S5), was dramatically upregulated in rainbow trout spleen post-F. psychrophilum infection at D7. Less studies were reported about SRs in rainbow trout by far. Qiu et al. (20) demonstrated that two variants of rainbow trout SRs with a collagenous structure were identified and found to bind to Gram-negative and -positive bacteria. Our results indicated that SRs may take part in innate immune regulation in rainbow trout against F. psychrophilum infection.
Differentially expressed genes in Toll-like receptor pathway
To better understand the function of TLRs in the immune response of rainbow trout spleen against F. pyschrophilum challenge, we focused on several transcripts involved in the TLR signaling pathway in this research. The TLR signaling pathway both initiates the overactive innate response and activates antigen-producing cells, thus participating in adaptive immune response in fish. Toll-interacting protein (Tollip) can inhibit TLR-mediated cellular responses by suppressing phosphorylation and the kinase activity of IRAK1 (22). Our results showed that the irak1 and irak4 expression levels were unchanged in rainbow trout infected with F. psychrophilum, but one of the components of tollip and traf6 were upregulated at D3 and D7. TRAF6 could mediate downstream signaling cascades in the immune response, but its function in inhibiting the activity of IRAK1 requires further investigation. TRIL (functional component of TLR4) and AP-1 (complex within jun and fos), which is the signal transduction protein in the endpoint of the NF-κB pathway, genes were down- and upregulated postinfection, respectively, which may explain the negative regulation of TRIL in the TLR signaling cascade. Coincidentally, Sepulcre et al. (23) have proven that LPS signaled via a TLR4- and MyD88-independent manner in fish, while the zebrafish TLR4 orthologs negatively regulated the MyD88-dependent signaling pathway. Srivastava et al. (25) demonstrated that the TLR4-TRIF-NFκB axis promotes the rapid secretion of anti-inflammatory cytokines. In addition, ifn-γ (ifng) in the Toll-like pathway not only has anti-viral activity (26) but also assists in the autophagy of intracellular bacteria (27). Our analysis (Table 4) showed that both ifng and ifngr showed upregulated expression, following infection with F. psychrophilum, suggesting that ifng may also play an important role in the immune response of infected rainbow trout. The importance of the TLR signaling pathway–mediated immune response in protection against F. psychrophilum infection needs further investigation.
Differentially expressed genes related to the cytokines
Cytokines, including interferons (IFNs), interleukins (ILs), tumor necrosis factors (TNFs), colony-stimulating factors, and chemokines, could stimulate T and B cells and antigen-presenting cells, mediating the communication between innate and acquired immune in fish. In rainbow trout, higher cytokine abundance in the spleen correlated strongly with lower F. psychrophilum loads in challenged fish (28). In the present study, the transcript levels of il1β1, il1β2, il6, and il8 continuously increased in the spleen of fish postexposure to F. psychrophilum (at D3 and D7), whereas tnf-α was upregulated at D3 and downregulated slightly at D7 (Figure 7, Table S5). Similar results were reported in rainbow trout infected with F. psychrophilum by Kutyrev et al. (30), who found that pro-inflammatory cytokine gene transcripts including il1β1, il1β2, and tnf-α in the spleen were upregulated till 72 h. Hou and Xin et al. (31) evaluated the transcript profiling between symptomatic and asymptomatic trout after Vibrio anguillarum injection and confirmed that cytokines in the spleen including il1β1, il1β2, il8, and tnf-α were dramatically increased (FC>8) in symptomatic trout. Among atypical chemokine receptors, ccl, cxcl, and ck2 transcripts showed a consistent upregulation after F. psychrophilum infection, similar to previous studies (32, 33). The Th1-type cytokine ifn-γ (34), which is induced by il12 in natural killer cells and T cells, was significantly upregulated (FC>4) at D3, but showed no remarkable change at D7. Some interferon regulatory factors (irf1, irf3, irf4b, irf5, irf6, irf8) continued to show upregulation during the later stages of infection (day 6 to day 10) in virus-infected fish spleens (35). Based on our data, irf1, irf7, and irf8 were upregulated at D3. irf2, irf3, and irf4 were downregulated both at D3 and D7, while irf5 was downregulated at D7.
Differentially expressed genes related to complement systems
The activation of a complement system helps the adaptive immune system to more effectively eliminate the foreign pathogens. Specific complement components were found to be regulated under different rearing conditions (36). Marancik et al. (13) found that C3- and C1q-like proteins were upregulated in rainbow trout, following an experimental challenge with F. psychrophilum. In the present study, the complement cascade response genes, such as c7b, c3, c3ar, and c1q-like, were significantly upregulated at D3 and D7 (FC>6). C1q and C3 regulate classical and alternative complement pathways, respectively (37). Our results indicated that complement systems were activated and may constitute an effective response in rainbow trout post-F. psychrophilum infection (13).
Differentially expressed genes related to antigen processing and presentation
In fish, antigen presentation appears to take place locally where the antigen is present. The major histocompatibility complex (MHC II) is involved in the presentation of exogenous antigens by specialists such as macrophages, dendritic cells, and B lymphocytes in fish. Th lymphocytes were activated and secrete cytokines to modulate the adaptive immune response after recognizing this MHC II antigen complex (38). Jirillo et al. (39) demonstrated that IFN-γ and MHC II β chain also enhance MØ respiratory burst, thus enabling the clearance of antigens. It was shown that the ability of rainbow trout gut to mount a T-cell-dependent delayed-type hypersensitivity response based on the release of MIF and of an IFN-γ-like factor (39). mif and mhc-iiβ (Table S5) were upregulated in rainbow trout after the F. psychrophilum challenge, which inferred the efficient cell–cell interactions within the immune system. Therefore, these memory cells can react rapidly to a second antigen encounter and are essential for successful vaccination. Here, we observed that MIF was upregulated at D3 and the MHC II β chain was upregulated at D7. Interestingly, it has been proven that the MHC II β chain could induce the expression of tnf-α and il6 in fish and regulate and coordinate the interaction between cytokines and their receptors (40, 41). It may suggest that the change of MIF and the MHC II b chain on the surface of antigen-presenting cells could stimulate the secretion of cytokines and activation of Th1-type responses in spleen of rainbow trout post F. psychrophilum infection.
Differentially expressed genes related to other immune gene pathways
Serum amyloid A (SAA) is considered a major protein in the APR and is an effector of innate immunity in all vertebrates. We found that saa was upregulated (FC >10) at D3 and D7, indicating the activation of APR after infection, similar to the previous studies (42, 43). Fibrinogen and macroglobulin are involved in the early innate immune system to rapidly isolate and kill invading pathogens (44). As shown in Table 4, both macroglobulin and fibrinogen were upregulated at two time points after infection, suggesting that the pathways activated by the APR were essential antibacterial mechanisms for rainbow trout infected with F. psychrophilum. Cell apoptosis could eliminate microbes and cellular remains and remove inflammatory exudates in an innate immune response. Zeng et al. (45) showed that the apoptosis executioner casp3 and apoptosis initiator casp8 were upregulated in rainbow trout after A. salmonicida and V. anguillarum challenges, which regulated intrinsic and extrinsic apoptosis, respectively. In this study, casp8 and casp3 were upregulated at D3 and D7, while both casp7 and casp14 were significantly upregulated at D3 (FC>7). These results were in accordance with the reports of Aravena (5), which indicated that the F. psychrophilum challenge could induce an increase of caspase-cleaved proteins in the skeletal muscle of rainbow trout. Fu et al. (46) showed that casp7 was involved in apoptosis in puffer fish after a bacterial infection and was dramatically upregulated, consistent with our results.
Conclusion
In the present study, a splenic transcriptome analysis for F. psychrophilum–infected rainbow trout was performed to explore the molecular immune mechanisms for the first time. The DEGs that were induced by F. psychrophilum infection and might be related to immune responses were identified, annotated, and analyzed. PRRs, complement molecules, inflammatory cytokines, chemokines, major histocompatibility complexes, and other immune-related genes might participate in the immune responses of rainbow trout to F. psychrophilum. In conclusion, this work provides a better understanding of the immune system and the defense mechanisms of rainbow trout against F. psychrophilum.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by The Committee of the Ethics on Animal Care and Experiments at Heilongjiang River Fisheries Research Institute of Chinese Academy of Fishery Sciences.
Author contributions
SL and TYL designed the study. FD, DW, and YC carried out the experiments. FD and DW performed data interpretation and drafted the manuscript. FC and DF performed RT-qPCR analysis. TPL and SL provided a critical editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Central Public-interest Scientific Institution Basal Research Fund, HRFRI (NO. HSY202001M) and Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO. 2020TD43).
Acknowledgments
We thank BioMarker Technologies Corporation for providing RNA-seq service and BMKcloud Platform for data analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.965099/full#supplementary-material
References
1. Starliper CE. Bacterial coldwater disease of fishes caused by flavobacterium psychrophilum. J Advanced Res (2011) 2(2):97–108. doi: 10.1016/j.jare.2010.04.001
2. Li S, Chai J, Knupp C, Nicolas P, Wang D, Cao Y, et al. Phenotypic and genetic characterization of flavobacterium psychrophilum recovered from diseased salmonids in China. Microbiol Spectrum (2021) 9(2):e0033021. doi: 10.1128/Spectrum.00330-21
3. Woo PT, Cipriano RC. Fish viruses and bacteria: pathobiology and protection. Boston, MA: CABI (2017). p. 211 P. doi: 10.1079/9781780647784.0211.
4. Nematollahi A, Decostere A, Pasmans F, Haesebrouck F. Flavobacterium psychrophilum infections in salmonid fish. J Fish Diseases (2003) 26:563–74. doi: 10.1046/j.1365-2761.2003.00488.x
5. Rivas-Aravena A, Fuentes-Valenzuela M, Escobar-Aguirre S, Gallardo-Escarate C, Molina A, Valdés JA. Transcriptomic response of rainbow trout (Oncorhynchus mykiss) skeletal muscle to flavobacterium psychrophilum. Comp Biochem Physiol Part D: Genomics Proteomics (2019) 31:100596. doi: 10.1016/j.cbd.2019.100596
6. Cipriano RC, Holt RA. Flavobacterium psychrophilum, cause of bacterial cold-water disease and rainbow trout fry syndrome. Department Interior (2005) 86:44.
7. Ngo TPH. Understanding the fish pathogen flavobacterium psychrophilum diversity for the control of rainbow trout fry syndrome in the united kingdom. Stirling (Scotland: University of Stirling (2016).
8. Marancik DP, Leeds TD, Wiens GD. Histopathologic changes in disease-resistant-line and disease-susceptible-line juvenile rainbow trout experimentally infected with flavobacterium psychrophilum. J Aquat Anim Health (2014) 26(3):181–9. doi: 10.1080/08997659.2014.920735
9. Wiens GD, Marancik DP, Zwollo P, Kaattari SL. Reduction of rainbow trout spleen size by splenectomy does not alter resistance against bacterial cold water disease. Dev Comp Immunol (2015) 49(1):31–7. doi: 10.1016/j.dci.2014.11.003
10. Ye J, Kaattari IM, Ma C, Kaattari S. The teleost humoral immune response. Fish Shellfish Immunol (2013) 35(6):1719–28. doi: 10.1016/j.fsi.2013.10.015
11. Cao Y, Wang D, Li S, Zhao J, Xu L, Liu H, et al. A transcriptome analysis focusing on splenic immune-related mcirornas of rainbow trout upon aeromonas salmonicida subsp. salmonicida infection. Fish Shellfish Immunol (2019) 91:350–7. doi: 10.1016/j.fsi.2019.05.048
12. Castro R, Coll J, Blanco MDM, Rodriguez-Bertos A, Jouneau L, Fernández-Garayzábal JF, et al. Spleen and head kidney differential gene expression patterns in trout infected with lactococcus garvieae correlate with spleen granulomas. Veterinary Res (2019) 50(1):32. doi: 10.1186/s13567-019-0649-8
13. Marancik D, Gao G, Paneru B, Ma H, Hernandez AG, Salem M, et al. Whole-body transcriptome of selectively bred, resistant-, control-, and susceptible-line rainbow trout following experimental challenge with flavobacterium psychrophilum. Front Genet (2015) 5:453. doi: 10.3389/fgene.2014.00453
14. Long M, Zhao J, Li T, Tafalla C, Zhang Q, Wang X, et al. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by aeromonas salmonicida subsp. salmonicida. J Proteomics (2015) 122:41–54. doi: 10.1016/j.jprot.2015.03.031
15. Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noël B, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun (2014) 5(1):1–10. doi: 10.1038/ncomms4657
16. Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the kegg orthology (ko) as a controlled vocabulary. Bioinformatics (2005) 21(19):3787–93. doi: 10.1093/bioinformatics/bti430
17. Watts M, Munday B, Burke C. Immune responses of teleost fish. Aust Veterinary J (2001) 79(8):570–4. doi: 10.1111/j.1751-0813.2001.tb10753.x
18. Jia Z, Wu N, Jiang X, Li H, Sun J, Shi M, et al. Integrative transcriptomic analysis reveals the immune mechanism for a cyhv-3-resistant common carp strain. Front Immunol (2021) 12:687151. doi: 10.3389/fimmu.2021.687151
19. Tang R, Wang S, Han P, Zhang Q, Zhang S, Xing X, et al. Toll-like receptor (tlr) 2 and tlr13 from the endangered primitive-ray finned fish dabry’s sturgeon (Acipenser dabryanus) and their expression profiling upon immune stimulation. Aquaculture Rep (2020) 16:100247. doi: 10.1016/j.aqrep.2019.100247
20. Avunje S, Kim W-S, Park C-S, Oh M-J, Jung S-J. Toll-like receptors and interferon associated immune factors in viral haemorrhagic septicaemia virus-infected olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol (2011) 31(3):407–14. doi: 10.1016/j.fsi.2011.06.009
21. Johansson P, Wang T, Collet B, Corripio-Miyar Y, Monte MM, Secombes CJ, et al. Identification and expression modulation of a c-type lectin domain family 4 homologue that is highly expressed in monocytes/macrophages in rainbow trout (Oncorhynchus mykiss). Dev Comp Immunol (2016) 54(1):55–65. doi: 10.1016/j.dci.2015.08.005
22. Vasta GR, Ahmed H, Du S-J, Henrikson D. Galectins in teleost fish: Zebrafish (Danio rerio) as a model species to address their biological roles in development and innate immunity. Glycoconjugate J (2004) 21(8):503–21. doi: 10.1007/s10719-004-5541-7
23. Poynter SJ, Monjo AL, Micheli G, DeWitte-Orr SJ. Scavengers for bacteria: Rainbow trout have two functional variants of MARCO that bind to gram-negative and –positive bacteria. Dev Comp Immunol (2017) 77:95–105. doi: 10.1016/j.dci.2017.07.021
24. Yang YC, Chen SN, Gan Z, Huang L, Nie P. Cloning and functional characterization of IRAK1 from rainbow trout (Oncorhynchus mykiss). Dev Comp Immunol (2021) 114:103780. doi: 10.1016/j.dci.2020.103780
25. Sepulcre MP, Alcaraz-Pérez F, López-Muñoz A, Roca FJ, Meseguer J, Cayuela ML, et al. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol (2009) 182(4):1836–45. doi: 10.4049/jimmunol.0801755
26. Srivastava N, Shelly A, Kumar M, Pant A, Das B, Majumdar T, et al. Aeromonas hydrophila utilizes TLR4 topology for synchronous activation of MyD88 and TRIF to orchestrate anti-inflammatory responses in zebrafish. Cell Death Discovery (2017) 3(1):1–9. doi: 10.1038/cddiscovery.2017.67
27. Li L, Chen SN, Laghari ZA, Huang B, Huo HJ, Li N, et al. Receptor complex and signalling pathway of the two type II IFNs, IFN-γ and IFN-γrel in mandarin fish or the so-called Chinese perch siniperca chuatsi. Dev Comp Immunol (2019) 97:98–112. doi: 10.1016/j.dci.2019.03.016
28. Yin L, Lv M, Qiu X, Wang X, Zhang A, Yang K, et al. IFN-γ manipulates NOD1-mediated interaction of autophagy and edwardsiella piscicida to augment intracellular clearance in fish. J Immunol (2021) 207(4):1087–98. doi: 10.4049/jimmunol.2100151
29. Moore C, Hennessey E, Smith M, Epp L, Zwollo P. Innate immune cell signatures in a BCWD-resistant line of rainbow trout before and after in vivo challenge with flavobacterium psychrophilum. Dev Comp Immunol (2019) 90:47–54. doi: 10.1016/j.dci.2018.08.018
30. Kutyrev I, Cleveland B, Leeds T, Wiens GD. Proinflammatory cytokine and cytokine receptor gene expression kinetics following challenge with flavobacterium psychrophilum in resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol (2016) 58:542–53. doi: 10.1016/j.fsi.2016.09.053
31. Hou Z-S, Xin Y-R, Yang X-D, Zeng C, Zhao H-K, Liu M-Q, et al. Transcriptional profiles of genes related to stress and immune response in rainbow trout (Oncorhynchus mykiss) symptomatically or asymptomatically infected with vibrio anguillarum. Front Immunol (2021) 12:639489. doi: 10.3389/fimmu.2021.639489
32. Cuesta A, Dios S, Figueras A, Novoa B, Esteban M, Meseguer J, et al. Identification of six novel CC chemokines in gilthead seabream (Sparus aurata) implicated in the antiviral immune response. Mol Immunol (2010) 47(6):1235–43. doi: 10.1016/j.molimm.2009.12.014
33. Wang Q, Wang S, Zhang Y, Yu Y, Zhao H, Yang H, et al. The CXC chemokines and CXC chemokine receptors in orange-spotted grouper (Epinephelus coioides) and their expression after Singapore grouper iridovirus infection. Dev Comp Immunol (2019) 90:10–20. doi: 10.1016/j.dci.2018.08.015
34. Hu Y, Alnabulsi A, Alnabulsi A, Scott C, Tafalla C, Secombes CJ, et al. Characterisation and analysis of IFN-gamma producing cells in rainbow trout oncorhynchus mykiss. Fish Shellfish Immunol (2021) 117:328–38. doi: 10.1016/j.fsi.2021.07.022
35. Laghari ZA, Li L, Chen SN, Huo HJ, Huang B, Zhou Y, et al. Composition and transcription of all interferon regulatory factors (IRFs), IRF1-11 in a perciform fish, the mandarin fish, siniperca chuatsi. Dev Comp Immunol (2018) 81:127–40. doi: 10.1016/j.dci.2017.11.015
36. Köbis JM, Rebl A, Kühn C, Korytář T, Köllner B, Goldammer T. Comprehensive and comparative transcription analyses of the complement pathway in rainbow trout. Fish Shellfish Immunol (2015) 42(1):98–107. doi: 10.1016/j.fsi.2014.10.032
37. Sacks SH, Zhou W. The role of complement in the early immune response to transplantation. Nat Rev Immunol (2012) 12(6):431–42. doi: 10.1038/nri3225
38. Castro R, Tafalla C. Overview of fish immunity. Mucosal Health Aquaculture (2015), 3–54. doi: 10.1016/B978-0-12-417186-2.00002-9
39. Jirillo F, Passantino G, Massaro MA, Cianciotta A, Crasto A, Perillo A, et al. In vitro elicitation of intestinal immune responses in teleost fish: evidence for a type IV hypersensitivity reaction in rainbow trout. Immunopharmacol Immunotoxicology (2007) 29(1):69–80. doi: 10.1080/08923970701282544
40. Abós B, Wang T, Castro R, Granja AG, Leal E, Havixbeck J, et al. Distinct differentiation programs triggered by IL-6 and LPS in teleost IgM(+) b cells in the absence of germinal centers. Sci Rep (2016) 6(1):1–16. doi: 10.1038/srep30004
41. Li X, Du H, Liu L, You X, Wu M, Liao Z. MHC class II alpha, beta and MHC class II-associated invariant chains from Chinese sturgeon (Acipenser sinensis) and their response to immune stimulation. Fish Shellfish Immunol (2017) 70:1–12. doi: 10.1016/j.fsi.2017.08.042
42. Raida MK, Buchmann K. Innate immune response in rainbow trout (Oncorhynchus mykiss) against primary and secondary infections with yersinia ruckeri O1. Dev Comp Immunol (2009) 33(1):35–45. doi: 10.1016/j.dci.2008.07.001
43. Chettri JK, Raida MK, Kania PW, Buchmann K. Differential immune response of rainbow trout (Oncorhynchus mykiss) at early developmental stages (larvae and fry) against the bacterial pathogen yersinia ruckeri. Dev Comp Immunol (2012) 36(2):463–74. doi: 10.1016/j.dci.2011.08.014
44. Påhlman LI, Mörgelin M, Kasetty G, Olin AI, Schmidtchen A, Herwald H. Antimicrobial activity of fibrinogen and fibrinogen-derived peptides–a novel link between coagulation and innate immunity. Thromb Haemostasis (2013) 109(05):930–9. doi: 10.1160/TH12-10-0739
45. Zeng C, Hou Z-S, Zhao H-K, Xin Y-R, Liu M-Q, Yang X-D, et al. Identification and characterization of caspases genes in rainbow trout (Oncorhynchus mykiss) and their expression profiles after aeromonas salmonicida and vibrio anguillarum infection. Dev Comp Immunol (2021) 118:103987. doi: 10.1016/j.dci.2020.103987
Keywords: rainbow trout, spleen, transcriptomic analysis, bacterial coldwater disease, Flavobacterium psychrophilum
Citation: Deng F, Wang D, Loch TP, Chen F, Lu T, Cao Y, Fan D and Li S (2022) Time-course transcriptome analyses of spleen in rainbow trout (Oncorhynchus mykiss) post-Flavobacterium psychrophilum infection. Front. Immunol. 13:965099. doi: 10.3389/fimmu.2022.965099
Received: 09 June 2022; Accepted: 11 July 2022;
Published: 09 August 2022.
Edited by:
Alok Agrawal, East Tennessee State University, United StatesReviewed by:
Zhi-Shuai Hou, Ocean University of China, ChinaFang Ma, Tianshui Normal University, China
Copyright © 2022 Deng, Wang, Loch, Chen, Lu, Cao, Fan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaowu Li, bGlzaGFvd3VAaHJmcmkuYWMuY24=
 Furong Deng
Furong Deng Di Wang1,3
Di Wang1,3 Thomas P. Loch
Thomas P. Loch Tongyan Lu
Tongyan Lu Shaowu Li
Shaowu Li