- 1Institute for Hepatology, National Clinical Research Center for Infectious Disease, Shenzhen Third People’s Hospital, The Second Affiliated Hospital, School of Medicine, Southern University of Science and Technology, Shenzhen, China
- 2Shenzhen Key Laboratory of Single-Cell Omics Reasearch and Application, Shenzhen, China
- 3Clinical Center for BioTherapy and Institutes of Biomedical Sciences, Zhongshan Hospital, Fudan University, Shanghai, China
- 4Shenzhen Research Center for Communicable Disease Diagnosis and Treatment of Chinese Academy of Medical Science, Shenzhen, China
Amid the ongoing Coronavirus Disease 2019 (COVID-19) pandemic, vaccination and early therapeutic interventions are the most effective means to combat and control the severity of the disease. Host immune responses to SARS-CoV-2 and its variants, particularly adaptive immune responses, should be fully understood to develop improved strategies to implement these measures. Single-cell multi-omic technologies, including flow cytometry, single-cell transcriptomics, and single-cell T-cell receptor (TCR) and B-cell receptor (BCR) profiling, offer a better solution to examine the protective or pathological immune responses and molecular mechanisms associated with SARS-CoV-2 infection, thus providing crucial support for the development of vaccines and therapeutics for COVID-19. Recent reviews have revealed the overall immune landscape of natural SARS-CoV-2 infection, and this review will focus on adaptive immune responses (including T cells and B cells) to SARS-CoV-2 revealed by single-cell multi-omics technologies. In addition, we explore how the single-cell analyses disclose the critical components of immune protection and pathogenesis during SARS-CoV-2 infection through the comparison between the adaptive immune responses induced by natural infection and by vaccination.
Introduction
Single-cell (sc) technologies, including flow cytometry (FACS), mass cytometry (CyTOF), cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq), single-cell RNA sequencing (scRNA-seq), single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq), single-cell T cell receptor sequencing (scTCR-seq), and single-cell B cell receptor sequencing (scBCR-seq), are well reviewed to characterize the heterogeneity of innate or adaptive immune responses to vaccination, infection, and cancer (1, 2). Briefly, they can 1) address immune heterogeneity and identify novel cell subsets; 2) offer more accurate descriptions of cell phenotypes and their responses to vaccination and infection; 3) deduce the transition, differentiation or developmental relationships between cell subsets; 4) explore the function of antigen-specific cells; 5) characterize the immune repertoire such as functional TCRs and BCRs. These single-cell technologies have been extensively adopted to study the protective and pathogenic mechanisms and to develop new strategies to prevent and treat COVID-19 (3–8).
Especially, the antigen-specific adaptive immune responses, which generally play a vital role in controlling most viral infections (9), are revealed by single-cell technologies in SARS-CoV-2 infection and vaccination. This role is reflected in its critical impact on the various clinical outcomes of SARS-CoV-2 infection and on the efficacy of vaccination. B cells, CD4+ T cells, and CD8+ T cells are three critical arms of the adaptive immune system, contributing to the control of SARS-CoV-2 and the development of immune memory (10–12). CD4+ T cells perform effector functions and provide assistance for other immune cells (13), while CD8+ T cells are important tissue-resident lymphocytes which are ready to eradicate virus-infected cells (14). B cells mainly produce antibodies to neutralize viruses or target virus-infected cells in an Fc-mediated manner (15). Both SARS-CoV-2 infection and vaccination effectively elicited T cell and B cell responses against SARS-CoV-2 (3, 16), providing a high level of protection against symptomatic SARS-CoV-2 infection (17), despite the challenges posed to the waned immunity by the immunity-escaping emerging viral variants. Of note, single-cell analysis together with the classic immunological assay, have greatly contributed to deeper understanding of the dynamics of SARS-CoV-2-specific CD4+ and CD8+ T cells, B cells and neutralizing antibodies (18–23) during natural infection and various vaccination (Table 1) (24–36). A comprehensive comparison of the adaptive immune responses with natural infection and vaccination is required to dissect the protective immune properties against SARS-CoV-2 infection. Here, we will review the current knowledge of SARS-CoV-2-specific immune responses during natural infection or vaccination and highlight future directions.
T cell response against SARS-CoV-2
Studies have shown that both SARS-CoV-2 infection and vaccination elicited robust anti-viral T cell responses, showing an increased expression of T-cell relevant cytotoxic signatures (3, 16). A variety of CD4+ and CD8+ T cell subsets were characterized by scRNA-seq analyses. The reported CD4+ T cell subsets by scRNA-seq mainly include naïve (CCR7+, TCF7high, LEF1high), T-helper 1 like cells (TH1, TBX21+ GNLY+), T-helper 2 like cells (TH2, GZMK+ GATA2+), proliferative cells (MKI67+), T follicular helper cells (TFH, ICOS+), T-helper 17 like cells (TH17, RORC+ CCR6+), T-regulatory cells (Treg, CTLA4+ FOXP3+), and interferon-stimulated genes (ISGshigh) cells. The reported CD8+ T cell subsets by scRNA-seq mainly included naïve (CCR7+), effector memory (TEM, GZMK+ LTB+), cytotoxic terminal effector (TTE, GNLY+ GZMB+ PRF1+), exhausted-like (TEX, PDCD1+ LAG3+), tissue-resident memory (TRM, ITGAE+ CXCR6+ ZNF683+), and proliferative (MKI67+) cells (4, 5, 37–53) (Table 2). The antigen specificity, cross-reactivity, composition and transcription features of these T cell subsets have been extensively explored.

Table 2 The T cell and B cell subsets identified by single cell technologies and their canonical markers.
CD4+ T cell responses
SARS-CoV-2 specific CD4+ T cells
Using activation-induced markers (AIM) or MHC-II tetramers, circulating SARS-CoV-2 specific CD4+ T cell responses have been extensively monitored by FACS (54). SARS-CoV-2 infection and vaccination induce robust SARS-CoV-2 specific CD4+ T cell responses that occur prior to the generation of high antibody titers and persist for at least 8-12 months (55). The reactivity of spike-specific CD4+ T cells to various SARS-CoV-2 variants including Omicron is well preserved (56, 57). Both FACS and scRNA-seq studies revealed that infection-induced specific (mainly spike-specific) CD4+ T cell responses showed memory characteristics, including subsets with TH1, TH17 and TFH phenotypes (25, 58, 59). Notably, the vaccination-induced spike-specific CD4+ T cells were predominately TH1-like rather than TFH-like phenotype despite the robust and persistent human TFH cell responses in the draining lymph nodes, whereas these SARS-CoV-2-reactive CD4+ T cells following infection are enriched for both TFH and TH1-like phenotype (60, 61). Moreover, following booster immunization, examined by AIM assay, convalescent individuals mount more robust circulating TFH cell response and generate a distinct spike-specific CD4+ T cell population expressing both IFN-γ and IL-10, compared with those uninfected individuals, which was consistent with better recall responses induced by the hybrid immunity (54, 62) (Figure 1).
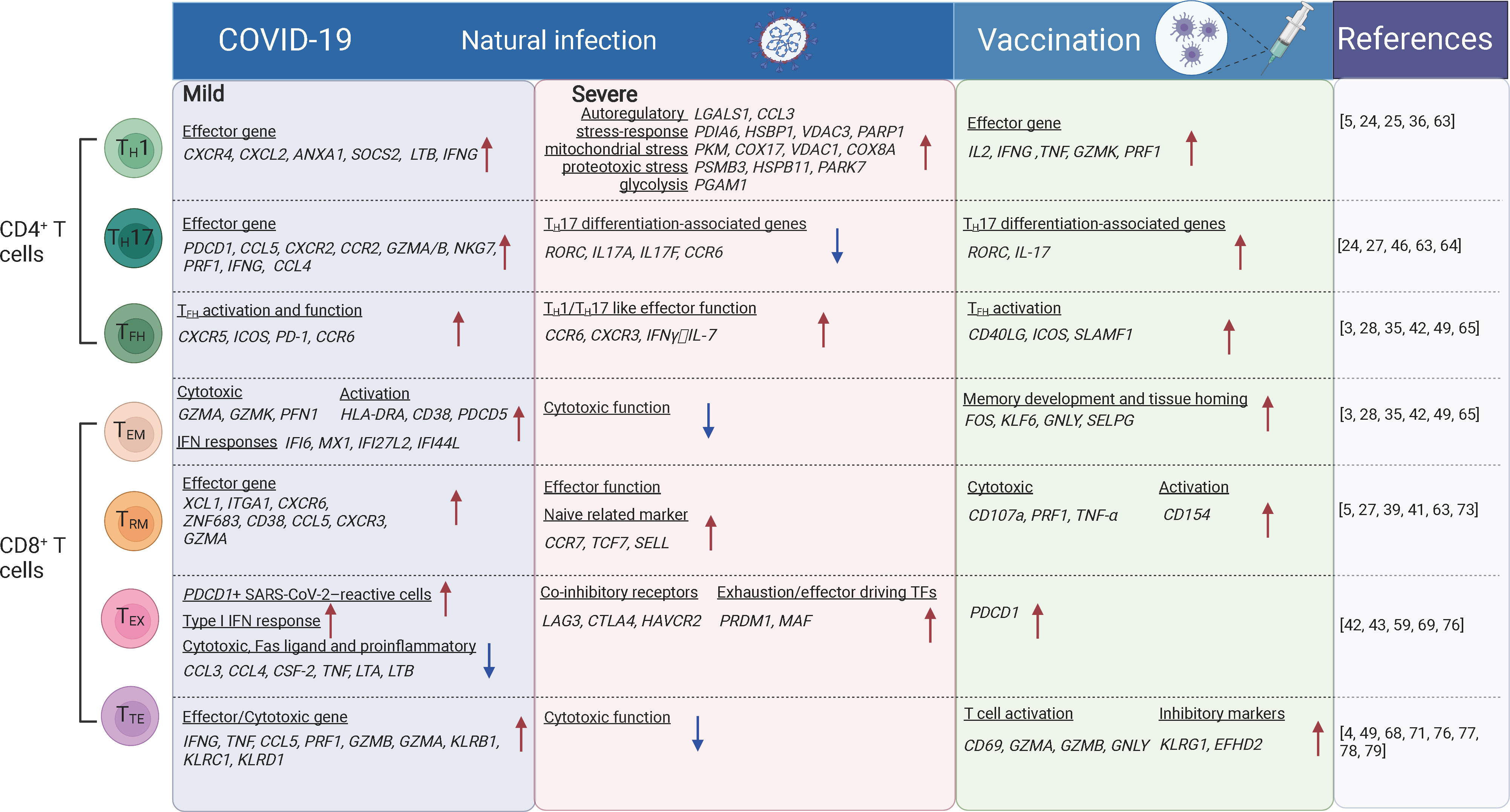
Figure 1 The T cell subsets and their transcriptional changes after SARS-CoV-2 vaccination and infection. The red arrows indicate the genes or pathways that were up-regulated in individuals with COVID-19 or vaccinated compared to the healthy donors, while blue arrows indicate the genes or pathways that were down-regulated. TH1, T-helper 1 like cells; TFH, T follicular helper cells; TH17, T-helper 17 like cells; TEM, effector memory; TTE, cytotoxic terminal effector; TEX, exhausted-like; TRM, tissue-resident memory cells. The figure was created using Biorender.
TH1 and TH17 cells
Revealed by CyTOF and scRNA-seq analysis, TH1-like and tissue-resident TH17-like cells were increased and more clonal expanded in patients with mild COVID-19 (5, 43, 63), suggesting virus reactivity of these cells. TH1-like cells from patients with mild COVID-19 exhibited a TH1 polarization state, characterized by upregulation of effector marker genes (PDCD1, CCL5, CXCR2, CCR2, GZMA/B, NKG7, PRF1, IFNG, and CCL4) (5, 63) and genes involved in effector function such as CXCR4, CXCL2, ANXA1, SOCS2 and LTB (5). Whereas in critical COVID-19 cases, TH1-like cells were predominant with the expression of activation markers (HLA-DR, CXCL13), auto-reactive markers (LGALS1, CCL3), stress-response markers (PDIA6, HSBP1, VDAC3, PARP1), mitochondrial stress genes (LDHA, PKM, COX17, VDAC1, COX8A), IL-2 withdrawal-associated stressed genes (MT1E, MT1X), proteotoxic stress genes (PSMB3/B6/D4/A7/C3, HSPB11, PARK7, EIF4EBP1) and glycolysis involved genes (PGAM1), suggesting functional dysregulation of theses TH1-like cells. Tissue-resident TH17-like cells were thought to play protective roles (63, 64), and TH17 response was found suppressed in severe COVID-19 cases, along with the reduced expression of typical TH17-associated genes including RORC, IL17A, IL17F, and CCR6 (46). In vaccination recipients characterized by scRNA-seq and FACS, BNT162b2, mRNA-1273, or CoronaVac primarily induces TH1-like cell responses. These cells express TH1-related cytokines, including IL-2, IFNγ or TNFα, activation markers CD38 and HLA-DRA, cytotoxic molecules GZMK and PRF1, and transcription factor gene TCF7 (24, 25, 36) (Figure 1). Moreover, TH1 cell responses were induced rapidly following infection or vaccination followed by the production of high antibody titers. Robust polyfunctional TH17 responses were also observed within CD4+ T cell compartments following vaccination (24, 27).
TFH cells
Studies of FACS, scRNA-seq, and CITE-seq reported that TFH cells were significantly expanded in asymptomatic and symptomatic cases compared with uninfected people (42, 49), which is indicative of recent antigen encounters and emigration from the germinal center (GC). Spike-specific circulating TFH cells positively correlate with plasma neutralizing activity in COVID-19 patients (65). These cells are enriched for genes involved in type I and type II IFN responses which were notably absent in circulating TFH (cTFH) cells from uninfected people (28). Early TFH cell responses correlate with the neutralizing antibody levels after the first dose of vaccination and with the CD8+ T cell responses after the second dose of vaccination (3). Significant increase in the expression of signature genes for TFH responses (CD40LG, ICOS, SLAMF1, etc.) and the B cell activation (TLR7, CD80/CD86, BCL6) were observed after vaccination (35) (Figure 1). These genes were known to be related to the production of neutralizing antibodies. Moreover, cTFH from vaccinated individuals showed an increased transcriptional signature of TNF-NFκB pathway activation (28), which is linked to improved cTFH survival and robust humoral immune responses in a previous study of influenza vaccination (66).
Pathogenic CD4+ T cells
Some CD4+ T cell subsets are reported to be involved in COVID-19 pathogenesis. Both FACS and scRNA-seq analysis revealed that TH2 cells were increased and more clonally expanded in patients with severe COVID-19 (4, 67, 68) and they expressed higher levels of IL4R and MAF associated with TH2 responses. In contrast, lower levels of TH2 (IL4) responses were observed after vaccination (29). The expression of GATA3 (TH2 transcription factor) was also decreased following SARS-CoV-2 infection and vaccination (24). In addition, ex vivo studies showed an absence of TH2 responses to spike peptides (24). IL-22-expressing CD4+ T cells (TH22-like) were relatively enriched in asymptomatic or mild COVID-19 cases. These cells could be associated with tissue-protective responses which could limit immunopathology (49). Proliferative CD4+ T cells expressing the proliferation marker Ki67 were enriched in cases with greater severity. The negative feedback mechanisms induced by FoxP3 in Tregs are altered in the lung, which may also contribute to immunopathology (46).
In summary, TH1, tissue-resident TH17, and TFH play a protective role in early SARS-CoV-2 infection. These cells could also be elicited by SARS-CoV-2 vaccination. Vaccine-induced spike-specific CD4+ T cell responses peaked more rapidly than antibody responses after the two-doses vaccination. By contrast, the unregulated TH2 and Treg cell responses may contribute to the immunopathology in severe COVID-19 cases.
CD8+ T cell responses
SARS-CoV-2 specific CD8+ T cells
SARS-CoV-2 specific CD8+ T cells have been extensively characterized in recent single-cell analyses by FACS and scRNA-seq. SARS-CoV-2-specific CD8+ T cells show a predominantly effector memory phenotype, in a less terminally differentiated state (69, 70), although terminal effector memory cells and transitional memory cells were also observed for SARS-CoV-2-specific CD8+ T cells. These SARS-CoV-2-specific memory CD8+ T cells are still present at least one year after the infection (71). ScRNA-seq studies revealed that SARS-CoV-2-specific CD8+ T cells during the acute phase of infection expressed genes associated with cytotoxicity (GZMA, GZMK, and PFN1), activation (HLA-DRA, CD38, and PDCD5), proliferation (MKI67, MCM7, and NUDC1), and IFN responses (IFI6, MX1, IFI27L2, and IFI44L), compared with that in the recovery phase (71). More SARS-CoV-2 specific CD8+ TEM cells are accumulated in asymptomatic/mild cases compared with severe/critical cases. In airways, CD8+ TEM cells showed elevated levels of HLA-DR, PD-1 and reduced levels of CD127, suggesting the activated status in situ (Figure 1). SARS-CoV-2 specific CD8+ TEM cells in vaccinees were significantly expanded after the second dose of vaccination examined by scRNA-seq (24). Recently, SARS-CoV-2 specific CD8+ T cells from individuals with vaccination, infection and breakthrough infection were extensively profiled using MHC-I multimers and scRNA-seq. It was reported that breakthrough infection mounted vigorous non-spike-specific responses, while vaccination among previously infected individuals led to the expansion of spike-specific T cells and continued differentiation to CCR7-CD45RA+ phenotype. More importantly, TCR analysis of SARS-CoV-2 reactive CD8+ T cells showed no evidence of repertoire narrowing following repeated exposure (72).
Tissue-resident CD8+ T cells
In addition to SARS-CoV-2-specific CD8+ T cells identified in blood, these cells were also readily detected in tonsils and displayed tissue-resident memory phenotypes with high expression of CD103 and CD69 (73). Analyzed by scRNA-seq, scTCR-seq and CITE-seq, CD8+ TRM cells showed increased proportion and clonal expansion in the airways of mild COVID-19 cases compared with those in severe/critical cases (5, 39, 41, 63). Along the pseudotime differentiation trajectory, CD8+ TRM cells were enriched at the end of the lineage in mild COVID-19 cases, indicating a terminally differentiated phenotype. Moreover, these cells express higher levels of the effector molecules XCL1, ITGAE, CXCR6, and ZNF683 in mild COVID-19 patients than that in severe cases (5, 41) (Figure 1). SARS-CoV-2-specific CD8+ TRM cells persisted at least 2 months after viral clearance in the nasal mucosa (74). Accumulation of both CD8+ TEM cells and CD8+ TRM cells is generally associated with lower disease severity, suggesting that they contribute to better outcomes for COVID-19 cases (49, 75). Interestingly, CD8+ TRM cells in the nasal mucosa were expanded up to 12 days post the first and second doses of SARS-CoV-2 mRNA vaccination (27). Thus, CD8+ TRM cells are important protective cells against the infection of SARS-CoV-2 and other pathogens.
Exhaustion of CD8+ T cells
Studies of FACS, scRNA-seq and CITE-seq also revealed that CD8+ TEX-like cells were phenotypically heterogeneous among patients with different outcomes (42, 43, 76). In addition to the increased expression of the inhibitory checkpoint and cytotoxic markers, CD8+ TEX-like cells were characterized by proliferation with G2M and S gene scores, especially in critical COVID-19 cases (43). Among the co-inhibitory receptors, LAG3, CTLA4, and HAVCR2 were enriched in CD8+ T cells from COVID-19 patients that eventually succumbed to the disease, but PDCD1 and TIGIT were enriched in CD8+ T cells from COVID-19 patients that eventually recovered and discharged (76). The exhaustion/effector driving TFs (PRDM1, MAF) were also upregulated in COVID-19 patients that eventually succumbed to the disease. However, there are evidences that the PD-1-expressing SARS-CoV-2-specific CD8+ TEX-like cells are functional rather than exhausted (69). In addition, SARS-CoV-2 vaccination can also induce PD-1 expression in CD8+ T cells (59) (Figure 1). Hence, whether CD8+ T cell is truly exhausted requires further discussion. Transcriptional signatures alone are not sufficient to indicate whether the cells were more exhausted or merely more activated.
Pathogenic CD8+ T cells
CyTOF and scRNA-seq studies reported the early upregulation of effector molecules by CD8+ TTE-cells, typically observed within 7 days from symptoms onset and peaking at 14 days following SARS-CoV-2 infection. CD8+ TTE-cells are associated with effective SARS-CoV-2 clearance and improved clinical outcomes (68, 77). Since CD8+ TTE cells are less enriched in SARS-CoV-2 specific CD8+ T cells (71), they may be bystander-activated and contribute to disease control in other settings (77, 78). In the late stage of infection, CD8+ TTE cells were enriched in cases with greater severity (49, 79), and they are the most proliferative compartments in COVID-19 patients, especially in severe cases (4). The kinetics of TTE-cell responses were prolonged and continued to increase up to day 40 after symptom onset (80). Effector genes expressed by CD8+ TTE cells such as GZMB drive a clear separation between stable and progressive COVID-19 patients (76). In the patients with moderate COVID-19, CD8+ TTE cells showed higher expression of IFNG, TNF, CCL5, PRF1, GZMB, and GZMA, together with genes encoding cytotoxic receptors (KLRB1, KLRC1, and KLRD1) in comparison with severe cases. Besides decreasing cytotoxic function, pro-inflammatory cytokines were also poorly expressed in these cells in patients with severe COVID-19. Hence, peripheral leukocytes are not a major contributor to the putative cytokine storm in COVID-19 cases (81).
Overall, various single-cell technologies revealed that infection elicits more diverse immune phenotypes than vaccination, in which CD4+ and CD8+ T cells activation increased from day 0 to day 14, peaked at day 28, and decreased from day 28 to day 35 (3, 24, 36). Most vaccines elicited TH1-skewed and CD8+ TEM responses (24). Early TFH and TH1 cell responses correlate with effective neutralizing antibody responses after the first dose, whereas CD8+ TEM cell responses were elicited after the second dose (3). Breakthrough infection and booster immunization can induce recall response and continued T cell differentiation.
B cell responses to SARS-CoV-2
B cell subsets
Various B cell subsets were identified using FACS, CyTOF, scRNA-seq, and CITE-seq in COVID-19 patients, including naïve B cells (TCL1A+ SELL+), atypical memory B cells (atMBC, ITGAX+ FCRL5+), activated B cells (ABC, CD21low CD27+ CD10-), memory B cells (MBC, CD21+ CD27+ CD10-), etc. According to their class switching states, memory B cells also comprised class-switched (IgD- CD27+) and non-class-switched (IgD+ CD27+) subsets, where the class-switched memory B cells are thought to have undergone affinity maturation through GC reactions. The identification of antibody-secreting cells (ASCs), like plasmablast (PB, CD79+ MS4A1- MKI67+), ‘active’ plasma cells (XBP1high MS4A1- PRDM1+), and ‘terminal’ plasma cells (XBP1high MS4A1- PRDM1- CCL5high) were also reported following SARS-CoV-2 infection and vaccination (5, 79, 82) (Table 2).
Previous studies have shown that the proportion of CD19+ B cells was increased in severe COVID-19 cases, while transitional B cell subsets were increased in mild/moderate cases. High-dimensional FACS analysis revealed that the proportion of memory B cells was decreased but that of ASCs was increased in severe cases (83). Severe/critical COVID-19 cases also displayed hallmarks of extrafollicular B cell activation which was similar to previously reported in lupus (84) (Figure 2). Expansion of atMBC is also a feature of COVID-19 (85). The increased atMBC proportion was positively correlated with COVID-19 severity and decreased to normal levels after recovery (43, 85). AtMBCs in COVID-19 showed impaired proinflammatory effector functions (86). However, spike-specific atMBCs were abundantly produced during secondary immune responses and SARS-CoV-2 infection induces more atMBC than vaccination (26), suggesting that atMBC are functional in these individuals. Therefore, it is unclear whether atMBC is correlated with impaired humoral immune responses during COVID-19. In contrast to atMBC, vaccine-induced ABCs had a similar capacity to bind to specific viral antigens in both SARS-CoV-2-convalescent individuals and naïve subjects (87).
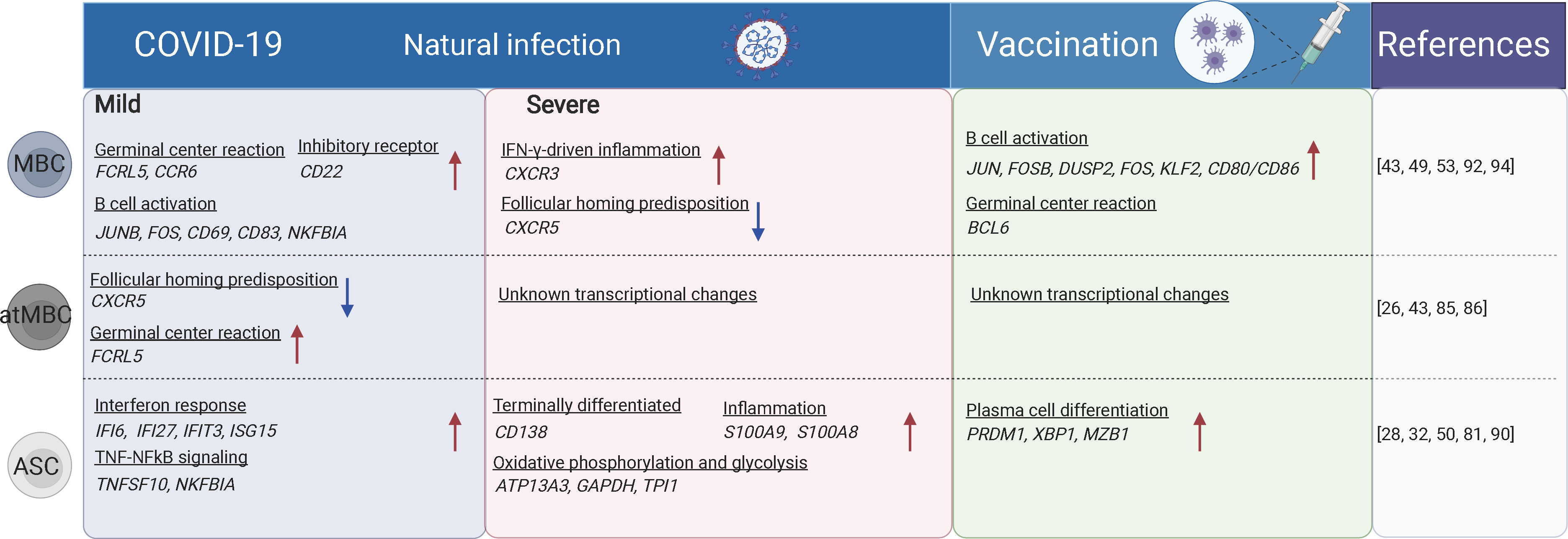
Figure 2 The B cell subsets and their transcriptional changes after SARS-CoV-2 vaccination and infection. The red arrows indicate the genes or pathways that were up-regulated in individuals with COVID-19 or vaccinated compared to the healthy donors, while blue arrows indicate the genes or pathways that were down-regulated. MBC, memory B cells; atMBC, atypical memory B cells; ASC, antibody secreting cells. The figure was created using BioRender.
Plasma cells
Plasma cells produce antibodies against antigens. Both plasma cells and plasmablasts were more expanded in moderate and severe COVID-19 cases compared to mild cases and uninfected controls (50, 81). Antibodies produced shortly after infection are mostly derived from short-lived plasma cells that are developed through an extrafollicular response. Whereas in the later stage, a smaller population of long-lived plasma cells is generated residing in the bone marrow (88). The PRDM1- plasma cells are characterized by sub-optimal differentiation or activation, which may be defective or even inhibit productive antibody responses in COVID-19. In contrast, PRDM1+ plasma cells were supposed to be long-lived populations (89). Plasmablasts are more expanded in hospitalized patients compared to mild/asymptomatic COVID-19 patients (90). The proportions of circulating plasmablasts/plasma cells were positively correlated with the serum levels of TNF, IL-10, and IL-21, which are factors critically involved in B cell differentiation to plasmablasts (42, 90, 91). ScRNA-seq and CITE-seq analyses revealed that these ASCs displayed a high metabolic activity (including oxidative phosphorylation, type I and type II IFN responses, fatty acid metabolism, and mTORC1 signaling), which was reduced following recovery. Meanwhile, memory and naïve B cells showed no significant difference in overall metabolic activity in patients with different disease severity (28). Following vaccination, CD27+CD38bright plasmablasts were substantially increased one week later as examined by FACS (32, 50). Besides, plasmablasts from both COVID-19 patients or individuals following SARS-CoV-2 vaccination were enriched for genes involved in IL-6 receptor signaling (JAK/STAT) and inflammatory response, which is consistent with their roles in promoting plasmablast differentiation (28) (Figure 2).
Germinal center B cells
GC reaction is important and capable of determining the quality of B cell response upon SARS-CoV-2 infection (21). The GC-derived memory B cells and plasma cells are more stable and long-lived (50), and capable of producing high-affinity antibodies (Figure 3). The memory B cells can rapidly differentiate into ASCs upon antigen re-encounter. On the contrary, extrafollicular B cell response leads to the production of low-affinity antibodies and wanes rapidly. ScRNA-seq and multi-color immunofluorescence showed that severe COVID-19 cases showed impaired GC reactions than mild cases (43, 92). Thus, the elevated antibody levels and memory B cell responses in severe COVID-19 could be explained by stronger extrafollicular B cell responses. Recently, using fine-needle sampling of lymph nodes and single-cell analyses, some interesting aspects of GC reactions in SARS-CoV-2 infection and vaccination are revealed (21, 93). ScRNA-seq and FACS analysis showed that SARS-CoV-2 vaccination induced a robust specific GC B cell response in the draining lymph nodes, and these GC B cells were maintained for at least 6 months in some individuals, indicating that they are likely to be undergoing further affinity maturation (21, 53, 94). Consistently, it is shown that SARS-CoV-2 memory B cells undergo continued evolution following vaccination or infection, mainly reflected by memory B cells with the increased somatic hypermutation (SHM) and the encoding of the high-affinity and broadly-reactive antibodies (12, 95–97).
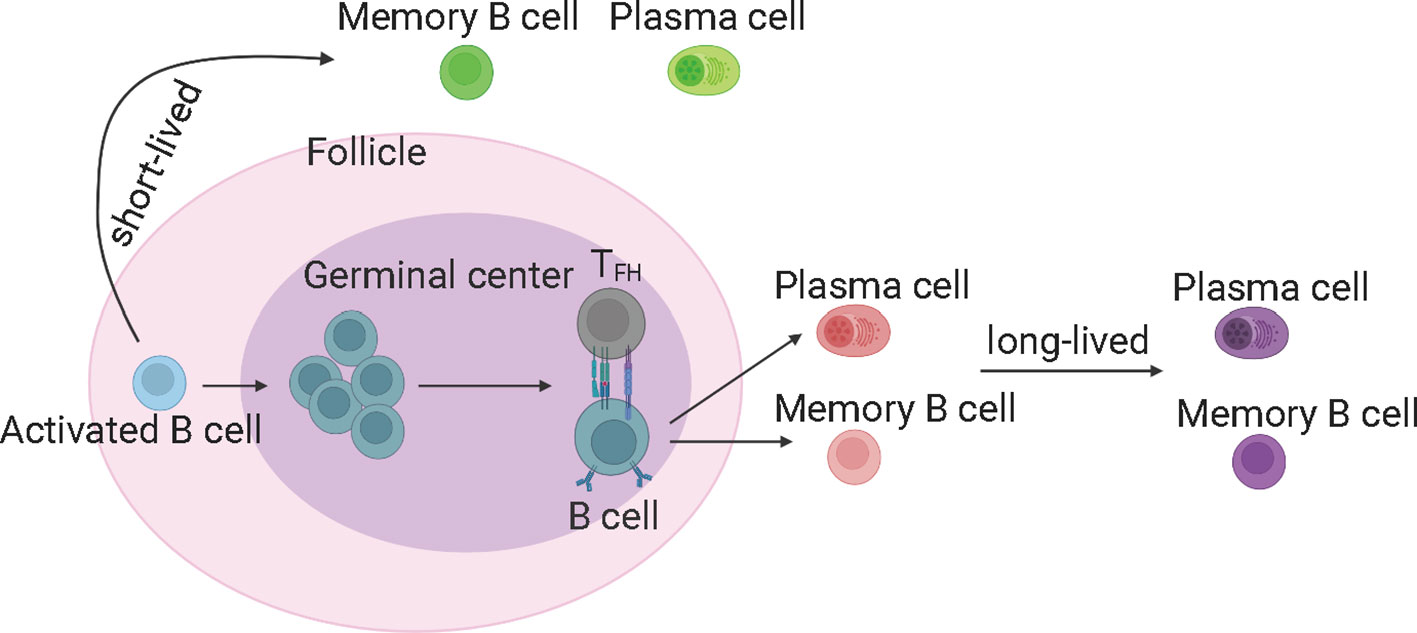
Figure 3 The germinal center reaction and the production of long-lived memory B cells and antibodies. The germinal center derived memory B cells and plasma cells are more stable and long-lived, and able to produce high-affinity antibodies. TFH, T follicular helper cells. The figure was created using BioRender.
SARS-CoV-2 specific B cells
The spike-specific class-switched memory B cells and neutralizing antibodies appeared to be stable up to 6 months after infection in COVID-19 patients (22, 98, 99). Vaccination also elicits robust specific B cell responses (88). Single-cell BCR tracing found that the switched memory B cells increased on day 7, and sustained until day 28 (36). In healthy individuals, SARS-CoV-2-specific antibody responses were enhanced upon the second dose of vaccination. Besides, SHM levels of unswitched memory B cells on day 0 are similar to the clonally expanded switched memory cells on days 7-28, suggesting that vaccine-activated unswitched memory B cells can differentiate into switched memory B cells independent of the GC (36, 50, 100, 101). Recently, SARS-CoV-2 specific B cell responses have been comprehensively probed using oligonucleotide-conjugated SARS-CoV-2 proteins at the single-cell resolution. During the early phase of SARS-CoV-2 infection, the pre-existing seasonal coronavirus cross-reactive memory B cells were activated, whereas the SARS-CoV-2-specific B cell clones were gradually accumulated with time and contributed to the majority of memory B cell pool later. The SARS-CoV-2 specific memory B cells maintained the evolution and affinity-mature over several months, by progressively acquiring SHM in GC (21). Over the course of infection and recovery, the number of activated specific B cells decreases while the resting MBCs increase, irrespective of disease severity.
FACS accompanied with biotinylation labeling analysis revealed that spike-specific MBCs were also robustly induced by SARS-CoV-2 vaccination. The induced specific MBCs persist and increase over time after immunization (102). Moreover, SARS-CoV-2 vaccination in recovered individuals induces a significant increase in the antibody response and B cell response, whereas the second dose of vaccine does not induce any further increase in antibody responses. Spike-specific MBCs were not reduced in individuals who had breakthrough infections. In contrast, 5 to 8 months after breakthrough infections, salivary anti-spike IgA levels declined compared with that before SARS-CoV-2 infection. But these salivary anti-spike IgA levels remained significantly higher than in fully vaccinated individuals who never experienced SARS-CoV-2 infection, possibly due to response to a novel antigen (102).
In summary, both SARS-CoV-2 infection and vaccination induce robust humoral immune responses. Severe COVID-19 is usually accompanied by an elevated level of humoral immune responses, likely due to a more robust extrafollicular B cell response. Although the SARS-CoV-2 antibody levels wane rapidly, the SARS-CoV-2 specific memory B cell responses increase over the first several months following the infection and vaccination. Memory B cells progressively acquire somatic mutations in their immunoglobulin heavy variable genes and continue to undergo clonal evolution due to an ongoing GC response. Finally, The booster immunization was effective in increasing neutralizing antibody titers and breadth encoded by vaccine-induced memory B cells against the SARS-CoV-2 variants.
T cell and B cell immune repertoire
Single-cell based analyses (scTCR-seq and scBCR-seq) have provided key information on the dynamics of TCR and BCR repertoire following SARS-CoV-2 infection or vaccination.
TCR repertoire
TCRs are capable of recognizing fragments of antigen as peptides bound to MHC molecules to target virus-infected cells (103). Identification of SARS-CoV-2 T-cell epitopes, generation of MHC multimers, together with single-cell immune repertoire sequencing will facilitate analyses of SARS-CoV-2-specific TCR repertoires (2, 72, 104, 105). The V(D)J genes usage and immune characteristics of TCRs on SARS-CoV-2-specific T cells have been described by several studies, but their function during SARS-CoV-2 infection needs to be further validated.
SARS-CoV-2 immunodominant epitopes may drive the molecular patterns of T cell responses in COVID-19 patients (105). CD8+ T-cell repertoire to SARS-CoV-2 was heavily skewed by 70% of all TCR mappings to 5.2% (14/269) of candidate antigen pools (106), suggesting that the T cell response is dominated by recognizing a few specific epitopes linked to distinct HLA types. Indeed, TCRs recognizing HLA-A*01:01-restricted TTDPSFLGRY epitope displayed enrichment for the TRBV27 segment (107). YLQPRTFLL-specific TCRs showed a biased usage of TRAV12-1 (71%) and TRBV7-9 (16%), and RLQSLQTYV-specific TCRs use TRAV13-2 (15%) and TRBV6-5 (25%) more frequently as compared with only 3%-4% gene usage in the control TCRs (108). Although TCRs recognizing HLA-A*02:01-restricted YLQPRTFLL and RLQSLQTYV epitopes were observed in convalescent patients, those TCRs were undetectable or at a very low frequency in the total TCR repertoire of the peripheral blood. It is possible that clones specific to the two antigens are both localized in the tissues, and only a limited number of cells are present in the circulation, which was in accordance with our previous findings that CD8+ TRM cells were highly clonally expanded (39, 41). TCRs recognizing HLA-A*11:01-restricted KTFPPTEPK epitope are cross-reactive to various SARS-CoV-2 variants and could confer cytotoxic responses upon encounter with target cells, providing support for developing T-cell epitope incorporating vaccines to prevent continuously emerging SARS-CoV-2 variants (109). By contrast, the antigen-specific TCR repertoire of CD4+ T cells was less studied and understood. It was reported that SARS-CoV-2-specific CD4+ T cells were increased in patients with severe COVID-19, but these cells displayed low functional avidity and clonality in severe cases than those in mild cases (110), which should be investigated to acquire a better understanding of the underlying mechanisms in future studies.
Various RBD- and spike-specific T cell clones were found from different memory subsets of convalescent COVID-19 patients and individuals receiving SARS-CoV-2 mRNA vaccination. However, there is a limited overlap of TCRs between SARS-CoV-2 infection and vaccination. TRAV29/DV5, TRBV5-1, TRAV29/DV5, and TRBV6-5 are biased in convalescent COVID-19 patients. In contrast, TRAV29/DV5, TRBV11-2, TRAV29/DV5, TRBV7-9, TRAV12-2, and TRBV6-2 are frequently used following vaccination (24). These distinct TCR patterns highlight the differences in the breadth of the epitopes recognized in SARS-CoV-2 infections versus vaccinations (111). These immunodominant epitopes and the TCRs confirmed with functional superiority can better inform next-generation vaccine designs.
BCR repertoire
Antibodies, also called immunoglobulins, comprise 5 different classes: IgM, IgD, IgG, IgA, and IgE (112). Different classes of antibodies may form synergy to achieve maximum immune activity against SARS-CoV-2 infection (113). BCR repertoire is the genetic source of neutralizing antibodies. Single-cell analyses have revealed BCR repertoires for total B cells, SARS-CoV-2 specific B cells or plasma cells, facilitating the identification of neutralizing antibodies.
BCR clonality was sharply increased at the early stage of infection and then gradually decreased in the convalescent phase to normal levels (79). BCRs in COVID-19 patients exhibited biased VDJ usage compared with that of healthy controls. The reported SARS-CoV-2 neutralizing antibodies are frequently derived from IGHV3 and IGHV1 family, and more than 40 neutralizing antibodies used IGHV3-53 have been identified. Using single-cell BCR sequencing, it was found that the genes of IGHV3 family including IGHV3-7, IGHV3-15, IGHV3-21, IGHV3-23, and IGHV3-30 were over-represented in COVID-19 patients compared with that in the controls. Besides, the preferred IGKVs were IGKV1-17, IGKV2-28, and IGKV3-15, and the preferred IGLVs were IGLV1-44, IGLV2-8, and IGLV3-27 (51). In contrast, after vaccination, IGHV3-33, IGHV3-43, and IGHV3-49 in the IGHV3 family and IGHV1-69D, IGKV1D-39, and IGLV5-45 were preferentially expanded (24). The cause for different V-gene usage between infection- versus vaccine-induced BCRs is unclear, possibly related to different epitope breadth. Infection elicited B cell clones targeting more epitopes on various SARS-CoV-2 proteins, whereas vaccination mainly induces narrowed antibody responses against S1 and RBD domains.
SHM reduction in COVID-19 patients was consistent with an extrafollicular B cell response mentioned above. RBD-specific clones have been shown to display a low level of SHM (below 5%) during the early stages of infection irrespective of disease severity (114). IgG1, IgG3, and IgA1, and to a lesser extent IgA2 and IgE were dominant isotypes that were rarely mutated or unmutated, indicating that they were derived from an early extrafollicular class switching event. This SHM level increases over time, suggesting an ongoing and persistent GC response.
Perspectives on adaptive immune responses to COVID-19 at single-cell solution
We have reviewed the broad applications of single-cell analyses in dissecting adaptive immunity in SARS-CoV-2 infection and vaccination. However, the studies of SARS-CoV-2-specific adaptive immunity are still ongoing and many questions remain unresolved.
Breadth, diversity, magnitude and lifespan of adaptive immunity
These aspects are the most important characteristics of immune defenses against SARS-CoV-2 infection. The SARS-CoV-2 genome encodes six functional open reading frames (ORFs): replicase (ORF1a/ORF1b), spike (S), envelope (E), membrane (M) and nucleocapsid (N), and seven putative ORFs encoding accessory proteins that are interspersed between the structural genes. More than 2, 000 different SARS-CoV-2-derived epitopes have been curated and reported (IEDB; www.iedb.org), exhibiting great epitope diversity. Structural proteins (S, M, E and N) are predominant targets of T cell and B cell responses (115).
Among these reported epitopes, 95% of reported class II and 98% of class I epitopes were fully conserved in different SARS-CoV-2 variants (Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Mu (B.1.621), Delta (B.1.617.2), and Omicron (B.1.1.529)) (30). For S protein, CD8+ T cell epitopes are homogeneously distributed, whereas only a few immunodominant regions were observed for CD4+ T cells (116). While the immunodominant epitopes for CD4+ and CD8+ T cells were noted to be similar in M and N proteins (within 7-101 and 131-213 residues for M protein, and within 31-173 and 201-371 residues for N protein) (105). T cell epitopes in nonstructural proteins show a similar pattern to that of the S protein. CD8+ T cell recognition showed more homogeneous patterns, but CD4+ T cell epitopes were more evident in nsp3 and nsp12 protein (105).
By contrast, B cell epitopes were prone to immune evasion, especially for the key mutations on spike protein which significantly reduced the neutralization activity of antibodies against SARS-CoV-2 variants (56). B cell epitopes have been extensively mapped for the structural proteins including S, N, M, and E proteins, especially the S. Full-length S or S1 domain which contains RBD were considered a good vaccine antigen as they are the main target to induce neutralizing antibody. Considering the broadly reactive T-cell response versus the waning humoral immunity, we need to incorporate these T-cell and B-cell epitopes outside of spike proteins to develop potentially more effective vaccines.
The magnitude of T cell and B cell responses varied when it comes to SARS-CoV-2 infection and vaccination, and is correlated with the disease severity (Figures 4A, B). Within T cells, the memory response is skewed toward more CD4+ T cell responses than that of CD8+ T cells, despite their similar levels immediately after infection (117). Moreover, the S-specific CD4+ T cells showed a limited increase after vaccination compared with CD8+ T cells (59), while the S-specific CD8+ T cell responses after vaccination are weaker. For humoral immunity, vaccination generally induces higher amounts of circulating antibodies compared with infection (Figure 4C).
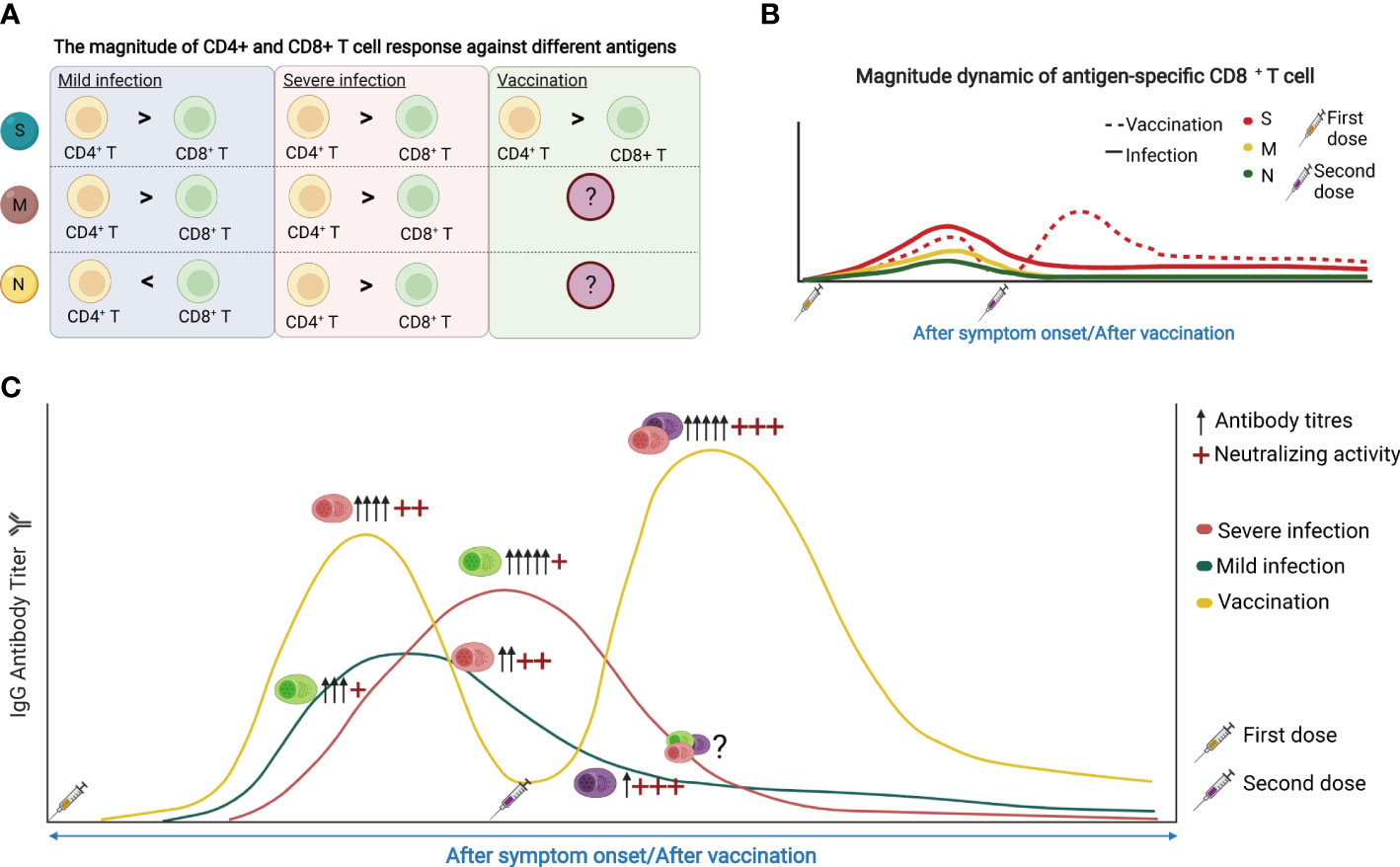
Figure 4 (A) The magnitude of CD4+ and CD8+ T cell response against different antigens. S, spike protein; M, membrane; N, nucleocapsid. (B) The dynamic of SARS-CoV-2 reactive CD8+ T cell response following SARS-CoV-2 infection and vaccination. (C) The dynamic of SARS-CoV-2 reactive antibodies and memory B cells in response to SARS-CoV-2 infection and vaccine. The figure was created using BioRender.
In terms of the duration of memory responses, T cell immunity persists better than antibody responses. However, both SARS-CoV-2 specific memory T cells and SARS-CoV-2 neutralizing antibodies can be detected one year after infection or vaccination (118). Patients with severe COVID-19 showed the delayed engagement of anti-viral CD8+ T cell responses compared with mild cases. CD4+ T cell responses are more robust than that of CD8+ T cells and may even increase in frequency over time, potentially reflecting antigen persistence. However, there was no difference in the magnitude of T-cell responses or neutralizing antibodies in patients with different disease severity one year after infection (118). Vaccines can substantially induce S-specific CD8+ T cell responses which peak in most donors at 9-12 days after immunization (59). These S-specific CD8+ T cells showed effector memory phenotype with expansion capacity, cytokine production, and degranulation capacity, and remained stable irrespective of vaccine booster (Figure 4B). The percentage of B cells increased from day 0-14 but decreased from day 14-28 after immunization. RBD-specific and S1-specific memory B cells may be observed on day 21 and increased gradually over the period of vaccination.
In summary, due to the aforementioned difference in breadth, activity and duration between the SARS-CoV-2 specific T-cell and B-cell responses, and between infection- and vaccination-induced immunity. We propose to improve current vaccination strategies by adding flavors of T-cell components and using heterologous immunization to mimic hybrid immunity to induce more effective, durable and broadly reactive immune responses.
High-throughput and accurately dissecting antigen-specific immune responses
Single-cell analyses have revealed various aspects of adaptive immune response to SARS-CoV-2 infections and vaccinations, as described above. However, a deep understanding of how these adaptive immune cells are molecularly regulated remains largely unclear. Single-cell multi-omic technologies such as CITE-seq, scATAC-seq, and scBCR/TCR-seq combined with peptide-MHC multimers will help reveal the underlying regulatory mechanisms driving COVID-19 pathology or prompting long-term protective immunity after vaccination (2, 76). After antigen exposure, immune cells are quickly differentiated into different subtypes to eliminate pathogens through various mechanisms (15, 119). Cell state transition or differentiation between different cell subsets is a multistage and multifactorial process, and single-cell multi-omics can facilitate the exploration of the regulatory mechanism underlying these processes, which is now a challenge due to technological limitations (120, 121). For example, combined scRNA-seq and scATAC-seq can simultaneously profile gene expression and open chromatin from the same cell, enabling deeper characterization of cell types/states and uncovering new gene regulatory mechanisms underlying antigen-specific T/B cell activation, differentiation and memory formation (122). While multi-omic single-cell immune profiling can provide full-length V(D)J sequences for TCRs/BCRs, cell surface protein expression, antigen specificity, and gene expression all from a single cell, allowing clonal tracing of antigen-specific T/B response to infection or vaccination (123). Finally, T and B cells respond to antigen stimulation by metabolic remodeling to execute their functions. Recent advances in single-cell metabolomic analyzing techniques will greatly increase our ability to interrogate these metabolic pathways at single-cell level in antigen-specific lymphocytes (124). Together, the integration of these new technologies can accurately dissect the cellular state transition processes and provide detailed temporal resolution of dynamic changes during infection and vaccination.
Furthermore, identifying and/or isolating antigen-specific T cells or B cells is important not only for the understanding of SARS-CoV-2 induced immune response, but also for providing direct solutions for adoptive immunotherapy (125, 126). However, current protocols for characterizing the immune phenotypes of antigen-specific reactions, including activation-induced marker, degranulation, proliferation, ELISA, ELISpot, intracellular cytokine staining, cytotoxicity, and multimer-based assays, are low-throughput, time-consuming and labor-intensive. New high-throughput single-cell based screening technologies are emerging. LIBRA-seq, an emerging multi-omics application, can simultaneously link antigen specificity with BCR sequencing at single-cell solution (82, 127). Other technologies linking antigen specificity with TCR sequencing will be possibly developed in the future (72). These antigen-specific single-cell sequencings provide effective means to comprehensively characterize the B cell or T cell responses and identify neutralizing antibodies or epitope-specific TCRs.
Spatial-temporal immune response
The tissue-resident immunity fights against pathogens at the front line of the host (128–130). Upon SARS-CoV-2 infection, circulating T cells need to be quickly activated and deployed to the site of infection, playing either protective and/or pathogenic roles (131). As the infection resolved, some of these protective effector T cells transformed to long-lived resident T cells, safeguarding the tissues from the potential pathogen re-encounter. In another line of defense, some B cells were initially engaged at the extrafollicular zones where they differentiated into short-lived plasmablasts to provide early antibody responses. Whereas other B cells may undergo GC reactions to produce both long-lived plasma cells and memory B cells (132). Understanding those spatial-temporal processes of SARS-CoV-2 specific T and B cell responses is important but challenging. Although memory T cells and memory B cells were disseminated throughout the body, the source and the distribution of pathogen-specific memory T cell or B cell population have not been systemically elucidated. Single-cell TCR and single-cell BCR sequencing, together with antigen-specific immune cell isolation strategies, can be used to trace the immune cell distribution from the circulation to peripheral tissues, to trace the pathogen-specific memory T (or B) cell turnover, and their differentiation from effector memory phenotype to central memory phenotype over time (4, 28, 72, 123).
In conclusion, the merging multi-omics single-cell technologies will continuously aid the efforts to study adaptive immunity against SARS-CoV-2. The power of these deep immune profiling techniques has already enabled simultaneous analyses of epigenomic, transcriptomic, proteomic, immune repertoire and spatial characteristics of rare populations of SARS-CoV-2 reactive T and B cells. We believe that these advances will greatly prompt both the fundamental and applied studies of SARS-CoV-2 infection and vaccination in the future.
Author contributions
FQ, SZ, and ZZ wrote and edited the manuscript. FQ and YC created the figures and tables. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Science Fund for Distinguished Young Scholars (82025022), the Shenzhen Science and Technology Program (ZDSYS20210623091810030), the Shenzhen Bay Funding (2020B1111340074, 2020B1111340075), and the Central Charity Fund of Chinese Academy of Medical Science (2020-PT310-009). The funders had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wang X, Xu G, Liu X, Liu Y, Zhang S, Zhang Z. Multiomics: unraveling the panoramic landscapes of SARS-CoV-2 infection. Cell Mol Immunol (2021) 18(10):2313–24.
2. Tian Y, Carpp LN, Miller HER, Zager M, Newell EW, Gottardo R. Single-cell immunology of SARS-CoV-2 infection. Nat Biotechnol (2022) 40(1):30–41.
3. Sahin U, Muik A, Vogler I, Derhovanessian E, Kranz LM, Vormehr M, et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature (2021) 595(7868):572–7.
4. Xu G, Qi F, Li H, Yang Q, Wang H, Wang X, et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discovery (2020) 6:73.
5. Wauters E, Van Mol P, Garg AD, Jansen S, Van Herck Y, Vanderbeke L, et al. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res (2021) 31(3):272–90.
6. Ren X, Wen W, Fan X, Hou W, Su B, Cai P, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell (2021) 184(7):1895–913 e19.
7. Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' b cells. Cell (2020) 182(1):73–84 e16.
8. Sinha S, Rosin NL, Arora R, Labit E, Jaffer A, Cao L, et al. Dexamethasone modulates immature neutrophils and interferon programming in severe COVID-19. Nat Med (2022) 28(1):201–11.
12. Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, et al. Evolution of antibody immunity to SARS-CoV-2. Nature (2021) 591(7851):639–44.
13. DuPage M, Bluestone JA. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nat Rev Immunol (2016) 16(3):149–63.
14. Wong P, Pamer EG. CD8 T cell responses to infectious pathogens. Annu Rev Immunol (2003) 21:29–70.
15. Akkaya M, Kwak K, Pierce SK. B cell memory: building two walls of protection against pathogens. Nat Rev Immunol (2020) 20(4):229–38.
16. Combes AJ, Courau T, Kuhn NF, Hu KH, Ray A, Chen WS, et al. Global absence and targeting of protective immune states in severe COVID-19. Nature (2021) 591(7848):124–30.
17. Grigoryan L, Pulendran B. The immunology of SARS-CoV-2 infections and vaccines. Semin Immunol (2020) 50:101422.
18. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol (2020) 21(11):1336–45.
19. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature (2020) 584(7821):457–62.
20. Schulien I, Kemming J, Oberhardt V, Wild K, Seidel LM, Killmer S, et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat Med (2021) 27(1):78–85.
21. Laidlaw BJ, Ellebedy AH. The germinal centre b cell response to SARS-CoV-2. Nat Rev Immunol (2022) 22(1):7–18.
22. Sherina N, Piralla A, Du L, Wan H, Kumagai-Braesch M, Andrell J, et al. Persistence of SARS-CoV-2-specific b and T cell responses in convalescent COVID-19 patients 6-8 months after the infection. Med (N Y) (2021) 2(3):281–95 e4.
23. Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature (2020) 584(7819):115–9.
24. Sureshchandra S, Lewis SA, Doratt BM, Jankeel A, Coimbra Ibraim I, Messaoudi I. Single-cell profiling of T and b cell repertoires following SARS-CoV-2 mRNA vaccine. JCI Insight (2021) 6(24):e153201.
25. Mysore V, Cullere X, Settles ML, Ji X, Kattan MW, Desjardins M, et al. Protective heterologous T cell immunity in COVID-19 induced by the trivalent MMR and tdap vaccine antigens. Med (N Y) (2021) 2(9):1050–71 e7.
26. Pape KA, Dileepan T, Kabage AJ, Kozysa D, Batres R, Evert C, et al. High-affinity memory b cells induced by SARS-CoV-2 infection produce more plasmablasts and atypical memory b cells than those primed by mRNA vaccines. Cell Rep (2021) 37(2):109823.
27. Ssemaganda A, Nguyen HM, Nuhu F, Jahan N, Card CM, Kiazyk S, et al. Expansion of tissue-resident CD8+ T cells and CD4+ Th17 cells in the nasal mucosa following mRNA COVID-19 vaccination. Nat Commun (2022) 13(1):3357.
28. Ivanova EN, Devlin JC, Buus TB, Koide A, Shwetar J, Cornelius A, et al. SARS-CoV-2 mRNA vaccine elicits a potent adaptive immune response in the absence of IFN-mediated inflammation observed in COVID-19. medRxiv (2021). doi: 10.1101/2021.04.20.21255677
29. Arunachalam PS, Scott MKD, Hagan T, Li C, Feng Y, Wimmers F, et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature (2021) 596(7872):410–6.
30. Kramer KJ, Wilfong EM, Voss K, Barone SM, Shiakolas AR, Raju N, et al. Single-cell profiling of the antigen-specific response to BNT162b2 SARS-CoV-2 RNA vaccine. Nat Commun (2022) 13(1):3466.
31. Brewer RC, Ramadoss NS, Lahey LJ, Jahanbani S, Robinson WH, Lanz TV. BNT162b2 vaccine induces divergent b cell responses to SARS-CoV-2 S1 and S2. Nat Immunol (2022) 23(1):33–9.
32. Muller M, Volzke J, Subin B, Muller S, Sombetzki M, Reisinger EC, et al. Single-dose SARS-CoV-2 vaccinations with either BNT162b2 or AZD1222 induce disparate Th1 responses and IgA production. BMC Med (2022) 20(1):29.
33. Lee HK, Knabl L, Knabl L, Kapferer S, Pateter B, Walter M, et al. Robust immune response to the BNT162b mRNA vaccine in an elderly population vaccinated 15 months after recovery from COVID-19. medRxiv (2021). doi: 10.1101/2021.09.08.21263284
34. Cao Q, Wu S, Xiao C, Chen S, Chi X, Cui X, et al. Integrated single-cell analysis revealed immune dynamics during Ad5-nCoV immunization. Cell Discovery (2021) 7(1):64.
35. Tong R, Zhong J, Li R, Chen Y, Hu L, Li Z, et al. Characterizing cellular and molecular variabilities of peripheral immune cells in healthy inactivated SARS-CoV-2 vaccine recipients by single-cell RNA sequencing. medRxiv (2021). doi: 10.1101/2021.05.06.21256781
36. Zhang Haoran HY, Zhujun J, Ningning S, Haishuang L, Yudong L, Hui W, et al. Single-cell sequencing and immune function assays of peripheral blood samples demonstrate positive responses of an inactivated SARS-CoV-2 vaccine(2021). Available at: http://dxdoiorg/102139/ssrn3774153.
37. Zheng Y, Liu X, Le W, Xie L, Li H, Wen W, et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell (2020) 11(10):740–70.
38. Bost P, Giladi A, Liu Y, Bendjelal Y, Xu G, David E, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell (2020) 181(7):1475–88 e12.
39. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med (2020) 26(6):842–4.
40. Szabo PA, Dogra P, Gray JI, Wells SB, Connors TJ, Weisberg SP, et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity (2021) 54(4):797–814 e6.
41. Qi F, Xu G, Liao X, Wang F, Yuan J, Wang H, et al. ScRNA-seq revealed the kinetic of nasopharyngeal immune responses in asymptomatic COVID-19 carriers. Cell Discovery (2021) 7(1):56.
42. Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science (2020) 369(6508):eabc8511.
43. Su Y, Chen D, Yuan D, Lausted C, Choi J, Dai CL, et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell (2020) 183(6):1479–95 e20.
44. Shi W, Liu X, Cao Q, Ma P, Le W, Xie L, et al. High-dimensional single-cell analysis reveals the immune characteristics of COVID-19. Am J Physiol Lung Cell Mol Physiol (2021) 320(1):L84–98.
45. Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol (2020) 21(9):1107–18.
46. Kalfaoglu B, Almeida-Santos J, Tye CA, Satou Y, Ono M. T-Cell hyperactivation and paralysis in severe COVID-19 infection revealed by single-cell analysis. Front Immunol (2020) 11:589380.
47. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science (2020) 369(6504):718–24.
48. Zhu L, Yang P, Zhao Y, Zhuang Z, Wang Z, Song R, et al. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity (2020) 53(3):685–96.e3.
49. Stephenson E, Reynolds G, Botting RA, Calero-Nieto FJ, Morgan MD, Tuong ZK, et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med (2021) 27(5):904–16.
50. Sokal A, Chappert P, Barba-Spaeth G, Roeser A, Fourati S, Azzaoui I, et al. Maturation and persistence of the anti-SARS-CoV-2 memory b cell response. Cell (2021) 184(5):1201–13.e14.
51. Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discovery (2020) 6:31.
52. Pusnik J, Richter E, Schulte B, Dolscheid-Pommerich R, Bode C, Putensen C, et al. Memory b cells targeting SARS-CoV-2 spike protein and their dependence on CD4(+) T cell help. Cell Rep (2021) 35(13):109320.
53. Kim W, Zhou JQ, Horvath SC, Schmitz AJ, Sturtz AJ, Lei T, et al. Germinal centre-driven maturation of b cell response to mRNA vaccination. Nature (2022) 604(7904):141–5.
54. Wragg KM, Lee WS, Koutsakos M, Tan HX, Amarasena T, Reynaldi A, et al. Establishment and recall of SARS-CoV-2 spike epitope-specific CD4(+) T cell memory. Nat Immunol (2022) 23(5):768–780.
55. Guerrera G, Picozza M, D'Orso S, Placido R, Pirronello M, Verdiani A, et al. BNT162b2 vaccination induces durable SARS-CoV-2-specific T cells with a stem cell memory phenotype. Sci Immunol (2021) 6(66):eabl5344.
56. Tarke A, Coelho CH, Zhang Z, Dan JM, Yu ED, Methot N, et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from alpha to omicron. Cell (2022) 185(5):847–59 e11.
57. Naranbhai V, Nathan A, Kaseke C, Berrios C, Khatri A, Choi S, et al. T Cell reactivity to the SARS-CoV-2 omicron variant is preserved in most but not all individuals. Cell (2022) 185(7):1259.
58. Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol (2020) 5(48):eabd2071.
59. Oberhardt V, Luxenburger H, Kemming J, Schulien I, Ciminski K, Giese S, et al. Rapid and stable mobilization of CD8(+) T cells by SARS-CoV-2 mRNA vaccine. Nature (2021) 597(7875):268–73.
60. Meckiff BJ, Ramirez-Suastegui C, Fajardo V, Chee SJ, Kusnadi A, Simon H, et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-Reactive CD4(+) T cells in COVID-19. Cell (2020) 183(5):1340–53 e16.
61. Mudd PA, Minervina AA, Pogorelyy MV, Turner JS, Kim W, Kalaidina E, et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell (2022) 185(4):603–13.e15.
62. Rodda LB, Morawski PA, Pruner KB, Fahning ML, Howard CA, Franko N, et al. Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity. Cell (2022) 185(9):1588–601.e14.
63. Zhao Y, Kilian C, Turner JE, Bosurgi L, Roedl K, Bartsch P, et al. Clonal expansion and activation of tissue-resident memory-like Th17 cells expressing GM-CSF in the lungs of severe COVID-19 patients. Sci Immunol (2021) 6(56):eabf6692.
64. Amezcua Vesely MC, Pallis P, Bielecki P, Low JS, Zhao J, Harman CCD, et al. Effector TH17 cells give rise to long-lived TRM cells that are essential for an immediate response against bacterial infection. Cell (2019) 178(5):1176–88 e15.
66. Herati RS, Silva LV, Vella LA, Muselman A, Alanio C, Bengsch B, et al. Vaccine-induced ICOS(+)CD38(+) circulating tfh are sensitive biosensors of age-related changes in inflammatory pathways. Cell Rep Med (2021) 2(5):100262.
67. Graham MB, Braciale VL, Braciale TJ. Influenza virus-specific CD4+ T helper type 2 T lymphocytes do not promote recovery from experimental virus infection. J Exp Med (1994) 180(4):1273–82.
68. Notarbartolo S, Ranzani V, Bandera A, Gruarin P, Bevilacqua V, Putignano AR, et al. Integrated longitudinal immunophenotypic, transcriptional and repertoire analyses delineate immune responses in COVID-19 patients. Sci Immunol (2021) 6(62):eabg5021.
69. Rha MS, Jeong HW, Ko JH, Choi SJ, Seo IH, Lee JS, et al. PD-1-Expressing SARS-CoV-2-Specific CD8(+) T cells are not exhausted, but functional in patients with COVID-19. Immunity (2021) 54(1):44–52 e3.
70. Neidleman J, Luo X, Frouard J, Xie G, Gill G, Stein ES, et al. SARS-CoV-2-Specific T cells exhibit phenotypic features of helper function, lack of terminal differentiation, and high proliferation potential. Cell Rep Med (2020) 1(6):100081.
71. Adamo S, Michler J, Zurbuchen Y, Cervia C, Taeschler P, Raeber ME, et al. Signature of long-lived memory CD8(+) T cells in acute SARS-CoV-2 infection. Nature (2022) 602(7895):148–55.
72. Minervina AA, Pogorelyy MV, Kirk AM, Crawford JC, Allen EK, Chou CH, et al. SARS-CoV-2 antigen exposure history shapes phenotypes and specificity of memory CD8(+) T cells. Nat Immunol (2022) 23(5):781–790.
73. Niessl J, Sekine T, Lange J, Konya V, Forkel M, Maric J, et al. Identification of resident memory CD8(+) T cells with functional specificity for SARS-CoV-2 in unexposed oropharyngeal lymphoid tissue. Sci Immunol (2021) 6(64):eabk0894.
74. Roukens AHE, Pothast CR, Konig M, Huisman W, Dalebout T, Tak T, et al. Prolonged activation of nasal immune cell populations and development of tissue-resident SARS-CoV-2-specific CD8(+) T cell responses following COVID-19. Nat Immunol (2022) 23(1):23–32.
75. Bost P, De Sanctis F, Cane S, Ugel S, Donadello K, Castellucci M, et al. Deciphering the state of immune silence in fatal COVID-19 patients. Nat Commun (2021) 12(1):1428.
76. Unterman A, Sumida TS, Nouri N, Yan X, Zhao AY, Gasque V, et al. Single-cell multi-omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID-19. Nat Commun (2022) 13(1):440.
77. Bergamaschi L, Mescia F, Turner L, Hanson AL, Kotagiri P, Dunmore BJ, et al. Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity (2021) 54(6):1257–75.e8.
78. Maurice NJ, McElrath MJ, Andersen-Nissen E, Frahm N, Prlic M. CXCR3 enables recruitment and site-specific bystander activation of memory CD8(+) T cells. Nat Commun (2019) 10(1):4987.
79. Xu G, Qi F, Wang H, Liu Y, Wang X, Zou R, et al. The transient IFN response and the delay of adaptive immunity feature the severity of COVID-19. Front Immunol (2021) 12:816745.
80. Arunachalam PS, Wimmers F, Mok CKP, Perera R, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science (2020) 369(6508):1210–20.
81. Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martinez-Colon GJ, McKechnie JL, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med (2020) 26(7):1070–6.
82. He B, Liu S, Wang Y, Xu M, Cai W, Liu J, et al. Rapid isolation and immune profiling of SARS-CoV-2 specific memory b cell in convalescent COVID-19 patients via LIBRA-seq. Signal Transduct Target Ther (2021) 6(1):195.
83. Sosa-Hernandez VA, Torres-Ruiz J, Cervantes-Diaz R, Romero-Ramirez S, Paez-Franco JC, Meza-Sanchez DE, et al. B cell subsets as severity-associated signatures in COVID-19 patients. Front Immunol (2020) 11:611004.
84. Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, et al. Extrafollicular b cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol (2020) 21(12):1506–16.
85. Oliviero B, Varchetta S, Mele D, Mantovani S, Cerino A, Perotti CG, et al. Expansion of atypical memory b cells is a prominent feature of COVID-19. Cell Mol Immunol (2020) 17(10):1101–3.
86. Wildner NH, Ahmadi P, Schulte S, Brauneck F, Kohsar M, Lutgehetmann M, et al. B cell analysis in SARS-CoV-2 versus malaria: Increased frequencies of plasmablasts and atypical memory b cells in COVID-19. J Leukoc Biol (2021) 109(1):77–90.
87. Goel RR, Painter MM, Apostolidis SA, Mathew D, Meng W, Rosenfeld AM, et al. mRNA vaccination induces durable immune memory to SARS-CoV-2 with continued evolution to variants of concern. bioRxiv (2021). doi: 10.1101/2021.08.23.457229
88. Turner JS, Kim W, Kalaidina E, Goss CW, Rauseo AM, Schmitz AJ, et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature (2021) 595(7867):421–5.
89. Nutt SL, Fairfax KA, Kallies A. BLIMP1 guides the fate of effector b and T cells. Nat Rev Immunol (2007) 7(12):923–7.
90. Bernardes JP, Mishra N, Tran F, Bahmer T, Best L, Blase JI, et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity (2020) 53(6):1296–314 e9.
91. Berglund LJ, Avery DT, Ma CS, Moens L, Deenick EK, Bustamante J, et al. IL-21 signalling via STAT3 primes human naive b cells to respond to IL-2 to enhance their differentiation into plasmablasts. Blood (2013) 122(24):3940–50.
92. Kaneko N, Kuo HH, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, et al. Loss of bcl-6-Expressing T follicular helper cells and germinal centers in COVID-19. Cell (2020) 183(1):143–57 e13.
93. Tangye SG, Burnett DL, Bull RA. Getting to the (germinal) center of humoral immune responses to SARS-CoV-2. Cell (2022) 185(6):945–8.
94. Turner JS, O'Halloran JA, Kalaidina E, Kim W, Schmitz AJ, Zhou JQ, et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature (2021) 596(7870):109–13.
95. Muecksch F, Wang Z, Cho A, Gaebler C, Ben Tanfous T, DaSilva J, et al. Increased memory b cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature (2022) 607(7917):128–134.
96. Cho A, Muecksch F, Schaefer-Babajew D, Wang Z, Finkin S, Gaebler C, et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature (2021) 600(7889):517–22.
97. Wang Z, Muecksch F, Schaefer-Babajew D, Finkin S, Viant C, Gaebler C, et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature (2021) 595(7867):426–31.
98. Cervia C, Zurbuchen Y, Taeschler P, Ballouz T, Menges D, Hasler S, et al. Immunoglobulin signature predicts risk of post-acute COVID-19 syndrome. Nat Commun (2022) 13(1):446.
99. Pradenas E, Trinite B, Urrea V, Marfil S, Avila-Nieto C, Rodriguez de la Concepcion ML, et al. Stable neutralizing antibody levels 6 months after mild and severe COVID-19 episodes. Med (N Y) (2021) 2(3):313–20 e4.
100. Sokal A, Barba-Spaeth G, Fernandez I, Broketa M, Azzaoui I, de la Selle A, et al. mRNA vaccination of naive and COVID-19-recovered individuals elicits potent memory b cells that recognize SARS-CoV-2 variants. Immunity (2021) 54(12):2893–907 e5.
101. Sokal A, Broketa M, Barba-Spaeth G, Meola A, Fernández I, Fourati S, et al. Analysis of mRNA vaccination-elicited RBD-specific memory b cells reveals strong but incomplete immune escape of the SARS-CoV-2 omicron variant. Immunity (2022).
102. Terreri S, Piano Mortari E, Vinci MR, Russo C, Alteri C, Albano C, et al. Persistent b cell memory after SARS-CoV-2 vaccination is functional during breakthrough infections. Cell Host Microbe (2022) 30(3):400–8 e4.
103. van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol (2011) 11(1):47–55.
104. Chen Z, John Wherry E. T Cell responses in patients with COVID-19. Nat Rev Immunol (2020) 20(9):529–36.
105. Grifoni A, Sidney J, Vita R, Peters B, Crotty S, Weiskopf D, et al. SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe (2021) 29(7):1076–92.
106. Snyder TM, Gittelman RM, Klinger M, May DH, Osborne EJ, Taniguchi R, et al. Magnitude and dynamics of the T-cell response to SARS-CoV-2 infection at both individual and population levels. medRxiv (2020). doi: 10.1101/2020.07.31.20165647
107. Gangaev A, Ketelaars SLC, Isaeva OI, Patiwael S, Dopler A, Hoefakker K, et al. Identification and characterization of a SARS-CoV-2 specific CD8(+) T cell response with immunodominant features. Nat Commun (2021) 12(1):2593.
108. Shomuradova AS, Vagida MS, Sheetikov SA, Zornikova KV, Kiryukhin D, Titov A, et al. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity (2020) 53(6):1245–57 e5.
109. Hu C, Shen M, Han X, Chen Q, Li L, Chen S, et al. Identification of cross-reactive CD8(+) T cell receptors with high functional avidity to a SARS-CoV-2 immunodominant epitope and its natural mutant variants. Genes Dis (2022) 9(1):216–29.
110. Bacher P, Rosati E, Esser D, Martini GR, Saggau C, Schiminsky E, et al. Low-avidity CD4(+) T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity (2020) 53(6):1258–71 e5.
111. Saini SK, Hersby DS, Tamhane T, Povlsen HR, Amaya Hernandez SP, Nielsen M, et al. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8(+) T cell activation in COVID-19 patients. Sci Immunol (2021) 6(58):eabf7550.
112. Taylor PC, Adams AC, Hufford MM, de la Torre I, Winthrop K, Gottlieb RL. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat Rev Immunol (2021) 21(6):382–93.
113. Lv Z, Deng YQ, Ye Q, Cao L, Sun CY, Fan C, et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science (2020) 369(6510):1505–9.
114. Shi D, Weng T, Wu J, Dai C, Luo R, Chen K, et al. Dynamic characteristic analysis of antibodies in patients with COVID-19: A 13-month study. Front Immunol (2021) 12:708184.
115. Tarke A, Sidney J, Kidd CK, Dan JM, Ramirez SI, Yu ED, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med (2021) 2(2):100204.
116. Sarma VR, Olotu FA, Soliman MES. Integrative immunoinformatics paradigm for predicting potential b-cell and T-cell epitopes as viable candidates for subunit vaccine design against COVID-19 virulence. BioMed J (2021) 44(4):447–60.
117. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell (2020) 181(7):1489–501 e15.
118. Guo L, Wang G, Wang Y, Zhang Q, Ren L, Gu X, et al. SARS-CoV-2-specific antibody and T-cell responses 1 year after infection in people recovered from COVID-19: a longitudinal cohort study. Lancet Microbe (2022) 3(5):e348–e356.
119. Kumar BV, Connors TJ, Farber DL. Human T cell development, localization, and function throughout life. Immunity (2018) 48(2):202–13.
120. Andreatta M, Corria-Osorio J, Muller S, Cubas R, Coukos G, Carmona SJ. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat Commun (2021) 12(1):2965.
121. Morgan D, Tergaonkar V. Unraveling b cell trajectories at single cell resolution. Trends Immunol (2022) 43(3):210–29.
122. Ranzoni AM, Tangherloni A, Berest I, Riva SG, Myers B, Strzelecka PM, et al. Integrative single-cell RNA-seq and ATAC-seq analysis of human developmental hematopoiesis. Cell Stem Cell (2021) 28(3):472–87 e7.
123. Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature (2018) 564(7735):268–72.
124. Guo S, Zhang C, Le A. The limitless applications of single-cell metabolomics. Curr Opin Biotechnol (2021) 71:115–22.
125. Parida SK, Poiret T, Zhenjiang L, Meng Q, Heyckendorf J, Lange C, et al. T-Cell therapy: Options for infectious diseases. Clin Infect Dis (2015) 61Suppl 3:S217–24.
126. Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol (2020) 20(11):651–68.
127. Setliff I, Shiakolas AR, Pilewski KA, Murji AA, Mapengo RE, Janowska K, et al. High-throughput mapping of b cell receptor sequences to antigen specificity. Cell (2019) 179(7):1636–46 e15.
128. Gallo O, Locatello LG, Mazzoni A, Novelli L, Annunziato F. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol (2021) 14(2):305–16.
129. Sun H, Sun C, Xiao W, Sun R. Tissue-resident lymphocytes: From adaptive to innate immunity. Cell Mol Immunol (2019) 16(3):205–15.
130. Ardain A, Marakalala MJ, Leslie A. Tissue-resident innate immunity in the lung. Immunology (2020) 159(3):245–56.
131. Woodland DL, Kohlmeier JE. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol (2009) 9(3):153–61.
Keywords: SARS-CoV-2, infection, vaccine, adaptive immune response, antibody production
Citation: Qi F, Cao Y, Zhang S and Zhang Z (2022) Single-cell analysis of the adaptive immune response to SARS-CoV-2 infection and vaccination. Front. Immunol. 13:964976. doi: 10.3389/fimmu.2022.964976
Received: 09 June 2022; Accepted: 10 August 2022;
Published: 02 September 2022.
Edited by:
Yongjun Sui, National Cancer Institute (NIH), United StatesCopyright © 2022 Qi, Cao, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng Zhang, zhangzheng1975@aliyun.com; Shuye Zhang, shuye_zhang@fudan.edu.cn
 Furong Qi
Furong Qi