- 1Institute of Immunology and Molecular Medicine, Jining Medical University, Jining, China
- 2Jining Key Laboratory of Immunology, Jining Medical University, Jining, China
In recent studies, NKG2A is revealed to be a key immune checkpoint for both natural killer (NK) cells and CD8+ T cells. It form heterodimer receptors with CD94, and targets the peptide-presenting human leukocyte antigen-E (HLA-E) molecules. Upon crosslinking, NKG2A/CD94 delivers inhibitory signals for NK cells and CD8+ T cells, while blocking NKG2A can effectively unleash functions of these cytotoxic lymphocytes. The interaction between NKG2A and HLA-E contributes to tumor immune escape, and NKG2A-mediated mechanisms are currently being exploited to develop potential antitumor therapeutic strategies. In addition, growing evidence shows that NKG2A also plays important roles in other immune-related diseases including viral infections, autoimmune diseases, inflammatory diseases, parasite infections and transplant rejection. Therefore, the current work focuses on describing the effect of NKG2A on immune regulation and exploring its potential role in immune-mediated disorders.
Introduction
NKG2A is a member of C-type lectin superfamily (1, 2). Its gene is localized in the natural killer (NK) complex on chromosome 12 and consists of seven exons (3). NKG2A is a single-pass type II integral membrane glycoprotein that contains the cytoplasmic, transmembrane as well as extracellular lectin-like domains (4). The intracellular portion has two ITIMs, which are involved in inhibitory signal transduction (5). NKG2A expression can be detected in cytotoxic lymphocytes, including most NK cells and a subset of CD8+ T cells (6). It is found to be expressed as a heterodimer with CD94, which also belongs to the C-type lectin superfamily (7). The ligands of NKG2A/CD94 heterodimeric receptor are non-classical MHC class I molecules, human leukocyte antigen (HLA)-E in humans and Qa-1 in mice (8). HLA-E is lowly expressed on almost all cell surfaces and displays limited polymorphism (9). Peptides that presented by HLA-E are derived from the leader sequences of the classical MHC class I molecules such as HLAA, HLAB and HLAC (10). Engagement of NKG2A/CD94 receptor with peptide-presenting HLA-E results in the phosphorylation of ITIMs in NKG2A. Phosphorylated ITIMs are responsible for recruiting and activating intracellular phosphatase SHP-1 as well as SHP-2, thus suppressing the activation signals generated by activating receptors such as T cell receptor (TCR) and NKG2D (11). In contrast to classical HLA molecules, which are commonly lost (12), the expression of HLA-E is generally elevated within tumor cells (13). Similar to other immune checkpoint molecules, NKG2A is exploited by tumor cells to achieve immune evasion. In addition, disrupting the interaction of NKG2A with its ligands is shown to be effective in enhancing antitumor immune responses (6, 14, 15). The overexpression of HLA-E is also observed in viral-infected cells, and the NKG2A-HLA-E axis is proved to exert a vital role in viral infection (16). Notably, NKG2A expression is found to be correlated with disease severity in coronavirus disease 2019 (COVID-19) patients (17–20). Apart from that, NKG2A is also involved in the pathological process of other immune-mediated disorders, such as autoimmune diseases, inflammatory diseases, parasite infections and transplant rejection. These findings indicate that NKG2A is a new therapeutic target for managing a variety of immune-mediated disorders. Herein, we review the existing knowledge about NKG2A mediated immune regulation and discuss the implications of NKG2A-targeted immunotherapeutic strategies.
Effects of NKG2A in immunocytes
NKG2A and NK cells
Approximately half of the human peripheral blood NK cells display NKG2A expression (21–23). Its expression is mostly observed in CD56bright immature NK cells and is decreased with stepwise maturation of the NK cells (24). There exits a negative correlation between the expression of NKG2A and killer cell immunoglobulinlike receptors (KIRs), which is implicated in the differentiation process of NK cells (25, 26). Multiple cytokines including interleukin (IL)-21, IL-15, IL-12, IL-10 and transforming growth factor-β (TGF-β) are able to induce the expression of NKG2A in NK cells (27–30). Upon ligands binding, NKG2A/CD94 receptors deliver signals that suppress NK cell functions, while disrupting the interaction of NKG2A/CD94 with Qa-1 or HLA-E activates the cytotoxic activity of NK cells (31–33).
The inhibitory NK receptors for HLA act a pivotal part in the education of NK cells, thereby greatly affecting mature NK cell responsiveness (34). At least one inhibitory NK receptors specific for “self” HLA-I haplotype need to be expressed on mature NK cells for recognizing target cells as well as preventing the activation of NK cells against autologous cells (35). The lack of inhibitory NK receptors for HLA renders NK cells hyporesponsive (36), while educated NK cells that expressing these receptors show higher responsiveness (37). Several studies have shown that NKG2A is required for the education of NK cells (38–40). NKG2A-educated human NK cells were more effective at killing target cells and showed a more dynamic migration behavior (41). NKG2A-educated mouse uterine NK cells were found to be more functionally competent in response to NK1.1 crosslinking (42). Highton et al. reported that NKG2A-educated human NK cells displayed improved responsiveness and metabolic resilience compared to KIRs-educated counterparts (43).
NK cells must maintain an appropriate level of NKG2A expression on the cell surface so as not to destroy normal autologous cells (44). NKG2A can be reused through a relatively rapid recycling process, enabling continuous availability of NKG2A on the cell surface. This recycling process requires energy and the cytoskeleton, but does not require functional ITIMs (44). The interaction of NKG2A/CD94 receptor with its ligands does not affect this recycling process and the expression of NKG2A/CD94 within NK cells (25). According to the fluorescent recovery after photobleaching (FRAP) analysis, most NKG2A/CD94 molecules on the plasma membrane exist in a free-moving form. NKG2A/CD94 is enriched at the contact site after crosslinking, and this enrichment is achieved through lateral diffusion in plasma membrane rather than synthesis of new proteins (45).
NKG2A and T cells
NKG2A is also expressed in T cells, especially CD8+ T cells. NKG2A expression in CD8+ T cells is found to be highly regulated, differing from its expression pattern in NK cells. NKG2A is barely expressed in CD8+ T cells of healthy individuals, but upregulated in tumor lesions and during chronic viral infection (46, 47). The expression of NKG2A in CD8+ T cells can be modulated a number of cytokines such as IL-23, IL-21, IL-15, IL-10, IL-6, IL-4, IL-2 and TGF-β (48–51). In addition, NKG2A expression can be induced by TCR engagement, and is acquired after antigen encounter. Cytotoxic T lymphocyte (CTL) clones sharing the same antigen specificity have the same NKG2A expression pattern, indicating that TCR antigenic specificity dictates the expression of NKG2A (52). NKG2A marks a special CD8+ T cells subset harboring tissue-resident and terminally exhausted features (6, 53–55). Similar to its function in NK cells, NKG2A/CD94 receptor engagement delivers inhibitory signals to CD8+ T cells, thus inhibiting the cytotoxic activity (55). Besides, NKG2A has also been found to be expressed in human CD4+ T cells. The expression of NKG2A/CD94 was observed in anti-CD3 monoclonal antibody (mAb) activated CD4+ T cells under TGF-β and IL-10 treatment. Moreover, NKG2A/CD94 was functional in CD4+ T cells and could inhibit TCR mediated tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ) secretion (56).
The functions of NKG2A in immunopathological settings
NKG2A and tumors
Overexpression of HLA-E is observed in various types of tumors, including solid tumors as well as hematological malignancies (57–63). Meanwhile, HLA-E overexpression predicts a poor prognostic outcome in patients with ovarian, liver, gynecologic, glioblastoma, colorectal, breast, gastric, kidney, esophageal, pancreatic, lung, and head and neck cancer (64–72). HLA-E overexpression may be caused by the interaction of tumor cells with tumor microenvironment (TME), and there was evidence that IFN-γ produced by tumor-reactive immunocytes contributed to the upregulation of HLA-E within tumor cells (73, 74). However, the underlying mechanism is not completely clear so far. Since overexpressed HLA-E functions to inhibit the cytotoxicity of cytotoxic lymphocytes, the blockade of NKG2A-HLA-E axis may possibly enhance the cell-based immunotherapeutic efficacy. In addition, according to multiple studies, NKG2A is also overexpressed in tumor-infiltrating cytotoxic lymphocytes in many types of tumors (55, 66, 75). The increase in the number of NKG2A+ tumor infiltrating lymphocytes (TILs) is correlated with a poor prognosis in patients undergoing colorectal, ovary and liver cancer (65, 72). Moreover, there is a growing body of evidence that NKG2A-HLA-E axis contributes to tumor immune escape. Sheu BC et al. found human cervical cancer cells could upregulate NKG2A expression in CD8+ T cells through an IL-15-dependent mechanism, thus abrogating the antitumor cytotoxicity of TILs (76). The cytotoxic effects of human NK cells and CD8+ T cells on HLA-E expressing B-lymphoblastoid cells were enhanced through RNAi-mediated inhibition of NKG2A expression (77). Kamiya and colleagues knocked out NKG2A protein expression in human peripheral blood NK cells achieved by retroviral transduction of NKG2A blocker, thus generating NKG2Anull NK cells. They further found NKG2Anull NK cells showed higher cytotoxic activity against HLA-E expressing tumor cells in immunodeficient mice (31). According to the results of the in vitro experiment, blocking NKG2A in human NK cells by the humanized anti-NKG2A antibody monalizumab was sufficient for improving the dysfunction of NK cells in chronic lymphocytic leukemia (CLL) (78). Salomé et al. showed that NKG2A was highly expressed in type 1 innate lymphoid cells (ILC1s) of acute myeloid leukemia (AML) patients. Moreover, the cytotoxicity of NKG2A+ ILC1s was impaired when encountering HLA-E-expressing leukemic targets (79). Collectively, the above results indicate that it is worthwhile to develop NKG2A blockade strategies for immunotherapy in cancer patients. However, clinical and preclinical studies showed that blocking NKG2A alone did not appear to be effective for tumor therapy. Monalizumab monotherapy showed very little clinical activity in patients with gynecologic cancers (80). Consistently, data from pre-clinical research suggested that anti-NKG2A mAb alone showed no effect on subcutaneous tumor xenografts in mice (14, 55). Regardless of the above drawbacks, monalizumab is still useful in combination with other immunotherapies. As demonstrated by preliminary data in microsatellite stable colorectal cancer (CRC) patients who typically do not respond to programmed death-ligand 1 (PD-L1)/programmed cell death-1 (PD-1)-based therapy, the combination of monalizumab and durvalumab (an anti-PD-L1 mAb) showed clinical efficacy and safety (81). Andre´ et al. also showed that combined blockade of PD-L1/PD-1 and NKG2A enhanced anticancer immunity in mouse lymphoma tumor models (14). According to the phase II trials interim results, the objective response rate (ORR) in head and neck squamous cell carcinoma (HNSCC) patients receiving monalizumab and cetuximab (an anti-EGFR blocking mAb) combination therapy was 31%, which was superior to previous data obtained from cetuximab monotherapy. The action of the above combinational therapy is probably mediated via NKG2A+ NK cells, rather than NKG2A+ CD8+ T cells (14). Additionally, according to van Hall’s group study based on four mouse models of solid tumors, the antitumor activity of CD8+ T cells responding to peptide vaccination was restored through blocking NKG2A-Qa-1 axis using blocking antibodies or genetic knockout (55). In addition, there are a number of ongoing clinical trials with monalizumab for the treatment of tumors, as shown in Table 1.
NKG2A and viral infections
Altering MHC molecules expression on the infected cell surface is one of the mechanisms that mediate viral immune escape. HLA-E and Qa-1 usually show overexpression on the virus infected cell surfaces and are able to bind peptides derived from viral proteins. HLA-E overexpression was observed in hepatic antigen-presenting cells (APCs) of hepatitis C virus (HCV) infected patients. HLA-E could bind to the viral peptide HCV core aa35–44 and present it on the cell surface, where it interacted with NKG2A/CD94 heterodimers, thereby resulting in immunosuppression (82). HCV infection induced Qa-1 expression in mouse hepatocytes. Abrogation of either NKG2A or Qa-1 signaling was shown to enhance NK function and promote NK cell-dependent HCV clearance (33). Similar findings were obtained during human cytomegalovirus (HCMV) infection. Glycoprotein UL40 encoded by HCMV could bind to HLA-E and interact with NKG2A/CD94 receptors, thereby inhibiting human NK cell activity and leading to immune evasion (83–85). HLA-E expression was enhanced in lymphocytes of human immunodeficiency virus (HIV) infected patients, and viral peptide HIV p24 aa14-22-loaded HLA-E could inhibit NK cell cytotoxic activity by binding to NKG2A (86). However, according to van Stigt Thans et al., HIV-1 downregulated the expression of HLA-E on the surface of infected primary human CD4+ T cells (87). During human papillomavirus (HPV) infection, the decreased expression of classical HLA class I molecules and overexpression of HLA-E were observed. In addition, HLA-E overexpression was associated with the decreased cytotoxicity of NK cells, which was most likely achieved through the interaction with NKG2A/CD94 receptors (88). Interestingly, a recent study showed that not all peptides presented by HLA-E could bind to NKG2A and thus exert inhibitory effects. As discovered by Mbiribindi B and colleagues, human NKG2A+ NK cells was able to recognize and respond to Epstein-Barr virus (EBV) infected autologous B cells. Further studies showed that EBV latent cycle protein-derived peptides impaired the recognition of NKG2A, despite being presented by HLA-E, thereby leading to the absence of inhibition (89).
The expression of NKG2A in NK cells is also generally increased during viral infection. The dysfunction of NK cells and T cells was observed in chronic hepatitis B (CHB) patients, along with the overexpression of inhibitory receptors including PD-1 and NKG2A (90). Hepatitis B e-antigen (HBeAg) was able to induce IL-10 secretion within regulatory T cells (Tregs), thus upregulating NKG2A expression in NK cells of CHB patients (91). The overexpressed NKG2A severely impaired the cytotoxicity of NK cells during HBV infection, which could be restored through the blockade of NKG2A-HLA-E axis (90, 91). Among the EBV reactivation and EBV-chronic graft-versus-host disease (GvHD) patients after hematopoietic stem cell transplantation (HSCT), the frequency of NKG2A+CD56dim NK cells was significantly increased in peripheral blood (92). According to Hendricks et al., the coinfection of cytomegalovirus (CMV) and EBV led to NKG2A+CD56dim NK cell expansion (93). Interestingly, there was a study reveal an innovative viral immune evasion mechanism. According to Wang et al., rodent herpesvirus Peru could counteract mouse NK cell activation by encoding a Qa-1 like protein via RNA splicing (94).
In addition to NK cells, NKG2A also has an impact on CD8+ T cell-mediated antiviral immunity. During polyoma virus infection in mice, NKG2A expression was enhanced in antiviral CD8+ T cells, thereby leading to the decrease of antigen-specific cytotoxicity in the process of virus-mediated oncogenesis and viral clearance (95). During ectromelia virus infection, NKG2A functioned intrinsically within mouse virus-specific CD8+ T cells for limiting excessive activation (96). However, NKG2A does not appear to affect CD8+ T cell-mediated antiviral immunity in all types of viral infections. Miller et al. found NKG2A/CD94 heterodimers showed no inhibitory effect on CD8+ T cell activity during lymphocytic choriomeningitis virus (LCMV) infection in mice (97).
Significantly, recent studies have highlighted a key role of NKG2A in the infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The expression of NKG2A was enhanced in peripheral NK cells and CD8+ T cells of COVID-19 patients and was correlated with disease severity (17–20). Interestingly, the NKG2A+ cytotoxic lymphocytes proportion was reduced among recovered patients (17). Further research has shown that the NKG2A expression is found to be regulated by SARS-CoV-2 spike 1 protein (SP1). The results of in vitro experiments showed that coculture with SP1-transfected lung epithelial cells led to NKG2A overexpression and reduced degranulation in NK cells (98). In addition, NKG2A was also overexpressed in NK cells isolated from bronchoalveolar lavage fluid (BALF) of COVID-19 patients with acute respiratory distress syndrome (ARDS), and the expression level was even higher than that in blood cells (19). In general, the NKG2A-HLA-E axis can be exploited by viruses to limit the activity of cytotoxic lymphocytes, thereby contributing to viral immune escape (Table 2).
NKG2A and autoimmune diseases
NK cells are able to eliminate autoreactive T cells, while NKG2A expressed within NK cells functions to prevent this process (99–101). Hence the blockade of NKG2A-ligand interaction is an efficient approach to treat autoimmune diseases. NKG2A/CD94 receptor has been found to exert a critical influence on experimental autoimmune encephalomyelitis (EAE) by modulating T cell activity. Typically, the interaction of NKG2A with its ligands was essential for immunologic memory development and clonal expansion of autoreactive T cells, as well as contributed to the protection of activated CD4+ T cells from lysis by NKG2A+ NK cell. The blockade of NKG2A-Qa-1 axis could effectively promote the elimination of autoreactive T cells mediated by NK cells, thereby alleviating EAE in mice (102, 103). Consistently, in comparison with CD4+ T cells obtained from Qa-1 wild type mice, NK cells showed higher cytotoxic activity against activated CD4+ T cells isolated from Qa-1 deficient mice (102). As observed from the rheumatoid arthritis (RA) mouse model, blocking NKG2A accelerated NK cell mediated elimination of pathogenic T helper 17 (Th17) cells as well as follicular helper T (Tfh) cells, thus arresting disease progression (104).
The expression profile of NKG2A within NK cells varies among different autoimmune diseases. Compared with NK cells from healthy individuals, NK cells from systemic lupus erythematosus (SLE) patients showed lower cytotoxicity with enhanced NKG2A expression (105, 106). On the contrary, a decrease in NKG2A expression was observed in NK cells of Graves’ disease (57) and new-onset psoriasis (107) patients. There were reports that the expression of NKG2A in T cells was decreased in SLE patients (108) and rheumatoid arthritis (RA) patients who flared (109), indicating that lack of inhibitory signals might lead to T cell hyperactivation and the immunological disorders. However, this conclusion is not applicable for CD8+ Tregs. CD8+ Tregs function to suppress self-reactive CD4+ T cells activity, thereby alleviating EAE. Upon Binding to Qa-1, NKG2A/CD94 receptor functioned to inhibit CD8+ Tregs activity. Disrupting the interaction of NKG2A/CD94 with Qa-1 unleashed CD8+ Tregs activity and abolished EAE progression in mice (110). Besides, the overexpressed NKG2A in CD8+ Tregs of patients with relapsing multiple sclerosis (MS) may function to limit CD8+ Tregs activity and contribute to disease progression (111).
NKG2A and other immune-related diseases
In addition to tumors, viral infections and autoimmune diseases, NKG2A is also involved in the pathological process of other immune-related diseases including inflammatory diseases, parasite infections and transplant rejection. NKG2A generally exerts immunosuppressive effects in inflammation. As reported by Hall and colleagues, NK cells inhibited the pro-inflammatory function of activated neutrophils through NKG2A-dependent mechanism in a DSS-induced colitis mouse model. Therefore, NKG2A played a protective role by inhibiting inflammation, while blocking NKG2A aggravated neutrophil-induced inflammation and tissue damage (112). In line with this, Zou and colleagues showed that the overexpressed NKG2A in NK cells exerted a vital function in suppressing neutrophil activation, thus alleviating DSS-induced colitis in mice (113). In celiac disease (CD) patients, CD-associated inflammation was marked by a decreased frequency of NKG2A+ natural killer T cells (NKT) and NKG2A+ NK cells, which might be involved in CD-associated tissue damage mediated by cytotoxic lymphocytes (114). Synovial NK cells from arthritis patients exhibited an activated phenotype and were capable of producing TNF-α and IFN-γ. Further, the secretion of these pro-inflammatory cytokines was increased under NKG2A blocking antibody treatment and decreased when NK cells encountered HLA-E expressed target cells, suggesting NKG2A can take part in the regulation of inflammatory response by controlling cytokines secretion in NK cells (115).
The immunosuppressive function of NKG2A has also been demonstrated in parasitic infection studies. In mice with alveolar echinococcosis (AE), which was caused by the infection of Echinococcus multilocularis, the overexpression of NKG2A was observed in NK cells and further led to reduced cytotoxicity by inhibiting IFN-γ secretion (116). Human cystic echinococcosis is caused by the larval stage of Echinococcus granulosus sensu lato. The expression of NKG2A and HLA-E was significantly increased within the hepatic cystic echinococcosis lesion, suggesting an inhibitory microenvironment (117). High expression of NKG2A was observed in human malaria-responsive NK cells (118) and γδ-T cells (119), which provided a homeostatic regulation mechanism for avoiding persistent activation. In addition, the expression of NKG2A was also found to be increased in human NK cells during Toxoplasma gondii (120) and Gnathostoma spinigerum infections (121). In contrast, during Schistosoma japonicum infection, NKG2A expression was significantly decreased in mouse NK cells (122) and NKT cells (123), which might serve as an activation mechanism.
GvHD is one of the major complications and causes of death in HSCT recipients and NKG2A is reportedly involved in GvHD pathogenesis. The percentage of NKG2A+ NK cells was significantly reduced in the peripheral blood of GvHD patients after HSCT in comparison with control subsets. Further, these cells were increased in completely recovered GvHD patients compared with partially recovered or active-stage GvHD patients. Therefore, monitoring the frequency of NKG2A+ NK cells provides clues for GvHD intervention and treatment. In addition, coculture with NKG2A+ NK cells led to decreased IFN-γ secretion as well as proliferation in T cells, indicating that the reduction in NKG2A+ NK cells is most likely the cause, rather than the result, of GvHD. The above evidence highlights the importance of NKG2A+ NK cells in limiting GvHD by suppressing activated self-reactive T cells (124). In line with this, Kordelas et al. also found a reduction of NKG2A+ NK cells in the peripheral blood of GvHD patients after HSCT (125).
Perspectives
A large number of studies have highlighted the critical role of NKG2A in tumors as well as viral infections. It is worth noting that NKG2A is involved in the pathological process of COVID-19 (17–20). The antiviral activity of circulating NK cells and CD8+ T cells is markedly decreased during SARS-CoV-2 infection, which leads to severe impairment of the host immune function (126–128). In COVID-19 patients, NKG2A expression is found to be correlated with the severity of disease (17–20). Therefore, anti-NKG2A mAb monalizumab could represent a possible solution for treating COVID-19 patients. Immune checkpoint blockade is one of the most promising ways to activate antitumor immunity. Unlike other known checkpoint molecules such as cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) and PD-1, NKG2A shows selective expression in cytotoxic lymphocytes including NK cells and CD8+ T cells. This suggests that the NKG2A-HLA-E axis does not appear to affect the initiation or regulation of anti-tumor immunity, but primarily functions in the final stages of tumor killing. Compared with HLA-E, NKG2A is more suitable as the blockade target of NKG2A-HLA-E axis. Apart from NKG2A, HLA-E also binds to NKG2C. NKG2C is expressed in both NK cells and T cells (129–132), and functions as an activating receptor by associating with the DNAX activation protein of 12 kDa (DAP12) signaling adapter (133) (Figure 1). NKG2C and NKG2A recognize mostly overlapping, but partially distinct epitopes on HLA-E (134). Although both NKG2A and NKG2C target HLA-E, the activating receptor shows a much lower affinity for its ligand. Compared with NKG2A, there are some amino acid differences in NKG2C protein, resulting in a 6-fold lower affinity for HLA-E (135, 136). Further, unlike HLA-E, which is expressed on almost all cell surfaces, NKG2A is mainly expressed in tumor lesions. Therefore, blocking NKG2A is more specific. Though NKG2A blockade shows limited effects as a stand-alone therapy, the NKG2A blocking antibody has synergistic effects with other tumor immunotherapies. A central paradigm in current tumor immunotherapy is “combination”, and NKG2A, a modulator of both adaptive and innate immunity, could be an important candidate.
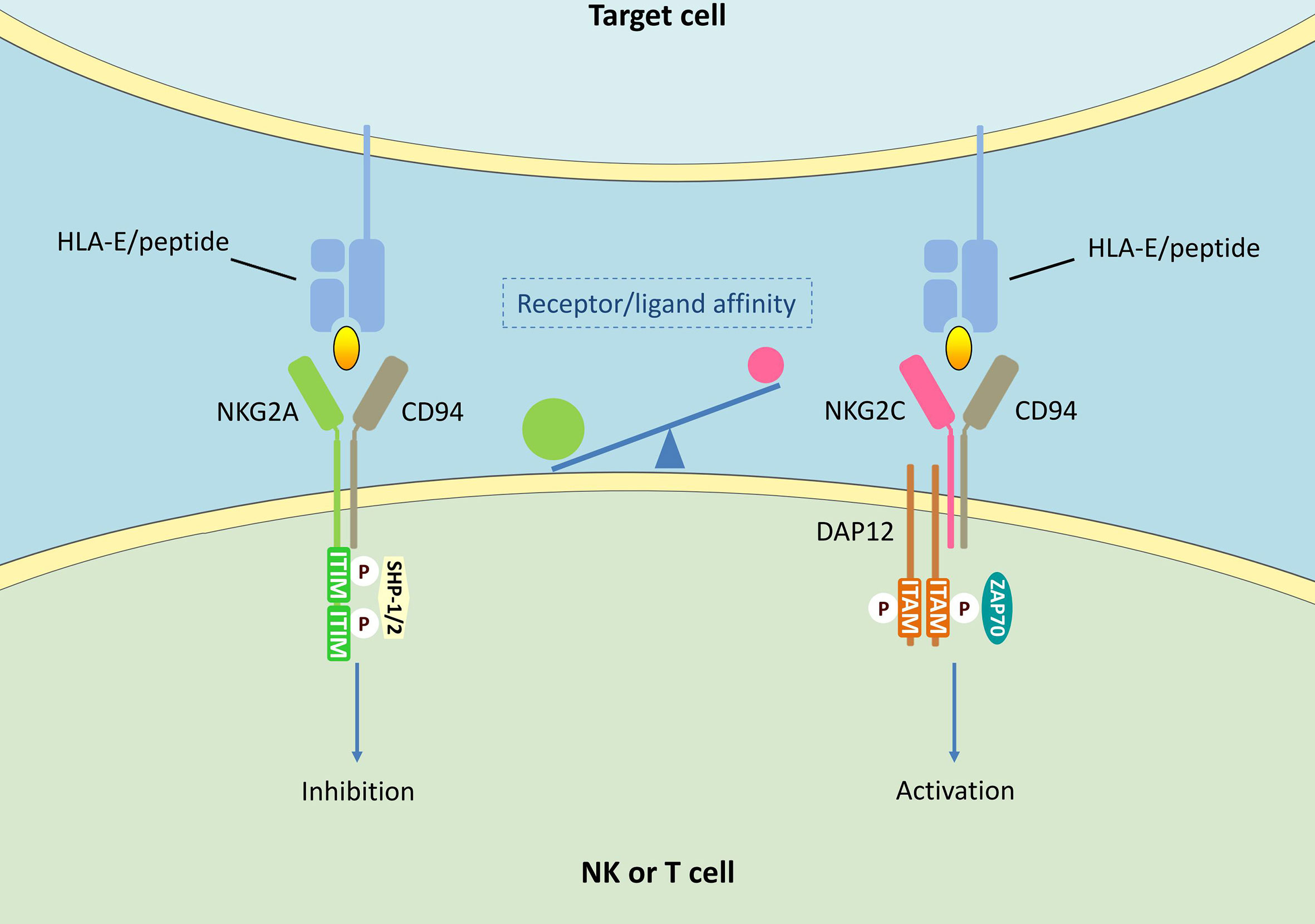
Figure 1 NKG2A/CD94 and NKG2C/CD94 signalings in NK cells and T cells. HLA-E can bind to both NKG2A/CD94 and NKG2C/CD94 receptors. The interaction of NKG2A/CD94 receptor with peptide-presenting HLAE results in the phosphorylation of ITIMs. Phosphorylated ITIMs are responsible for recruiting and activating intracellular phosphatase SHP-1/2, thus delivering negative signals. CD94/NKG2C heterodimers associates with DAP12 containing immune receptor tyrosine activating motifs (ITAMs), thus activating zeta-chain-associated protein kinase 70 kDa (ZAP70) and delivering activating signals.
Author contributions
XW and ZN are responsible for preparation of the published work. HX and ZN are responsible for supervision, review, editing and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
The present study was funded by the National Natural Science Foundation of China (No. 82003027), Doctoral Startup Fund of Jining Medical University (No. 2017JYQD24) and Innovation training program for college students of Jining Medical University (No.cx2021065).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Huang RY, Wang Y, Jhatakia AD, Deng AX, Bee C, Deshpande S, et al. Higher-order structure characterization of nkg2a/cd94 protein complex and anti-nkg2a antibody binding epitopes by mass spectrometry-based protein footprinting strategies. J Am Soc Mass Spectrom (2021) 32(7):1567–74. doi: 10.1021/jasms.0c00399
2. Godal R, Bachanova V, Gleason M, McCullar V, Yun GH, Cooley S, et al. Natural killer cell killing of acute myelogenous leukemia and acute lymphoblastic leukemia blasts by killer cell immunoglobulin-like receptor-negative natural killer cells after NKG2A and LIR-1 blockade. Biol Blood Marrow Transplant (2010) 16(5):612–21. doi: 10.1016/j.bbmt.2010.01.019
3. Plougastel B, Jones T, Trowsdale J. Genomic structure, chromosome location, and alternative splicing of the human NKG2A gene. Immunogenetics (1996) 44(4):286–91. doi: 10.1007/bf02602558
4. Iwaszko M, Bogunia-Kubik K. Clinical significance of the HLA-e and CD94/NKG2 interaction. Arch Immunol Ther Exp (Warsz) (2011) 59(5):353–67. doi: 10.1007/s00005-011-0137-y
5. Mingari MC, Pietra G, Moretta L. Immune checkpoint inhibitors: anti-nkg2a antibodies on board. Trends Immunol (2019) 40(2):83–5. doi: 10.1016/j.it.2018.12.009
6. Ducoin K, Oger R, Bilonda Mutala L, Deleine C, Jouand N, Desfrancois J, et al. Targeting NKG2A to boost anti-tumor CD8 T-cell responses in human colorectal cancer. Oncoimmunology (2022) 11(1):2046931. doi: 10.1080/2162402X.2022.2046931
7. Ram DR, Lucar O, Hueber B, Reeves RK. Simian immunodeficiency virus infection modulates cd94(+) (klrd1(+)) nk cells in rhesus macaques. J Virol (2019) 93(16):e00731-19. doi: 10.1128/JVI.00731-19
8. Guo X, Fang Y, Guo C, Jia Q, Chi Z, Li J, et al. Qa-1b functions as an oncogenic factor in mouse melanoma cells. J Dermatol Sci (2022) 105(3):159–69. doi: 10.1016/j.jdermsci.2022.02.003
9. Li D, Brackenridge S, Walters LC, Swanson O, Harlos K, Rozbesky D, et al. Mouse and human antibodies bind HLA-e-leader peptide complexes and enhance NK cell cytotoxicity. Commun Biol (2022) 5(1):271. doi: 10.1038/s42003-022-03183-5
10. Creelan BC, Antonia SJ. The NKG2A immune checkpoint - a new direction in cancer immunotherapy. Nat Rev Clin Oncol (2019) 16(5):277–8. doi: 10.1038/s41571-019-0182-8
11. Le Dréan E, Vély F, Olcese L, Cambiaggi A, Guia S, Krystal G, et al. Inhibition of antigen-induced T cell response and antibody-induced NK cell cytotoxicity by NKG2A: association of NKG2A with SHP-1 and SHP-2 protein-tyrosine phosphatases. Eur J Immunol (1998) 28(1):264–76. doi: 10.1002/(sici)1521-4141(199801)28:01<264::aid-immu264>3.0.co;2-o
12. Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature (2019) 567(7749):479–85. doi: 10.1038/s41586-019-1032-7
13. Kanevskiy L, Erokhina S, Kobyzeva P, Streltsova M, Sapozhnikov A, Kovalenko E. Dimorphism of hla-e and its disease association. Int J Mol Sci (2019) 20(21):5496. doi: 10.3390/ijms20215496
14. André P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both t and nk cells. Cell (2018) 175(7):1731–43.e13. doi: 10.1016/j.cell.2018.10.014
15. Abd Hamid M, Wang RZ, Yao X, Fan P, Li X, Chang XM, et al. Enriched HLA-e and CD94/NKG2A interaction limits antitumor cd8(+) tumor-infiltrating t lymphocyte responses. Cancer Immunol Res (2019) 7(8):1293–306. doi: 10.1158/2326-6066.CIR-18-0885
16. Grant EJ, Nguyen AT, Lobos CA, Szeto C, Chatzileontiadou DSM, Gras S. The unconventional role of HLA-e: The road less traveled. Mol Immunol (2020) 120:101–12. doi: 10.1016/j.molimm.2020.02.011
17. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol (2020) 17(5):533–5. doi: 10.1038/s41423-020-0402-2
18. Yasin MM, Shehata IH, Elsheikh NG, Elsayed MS. Expression of NKG2A inhibitory receptor on cytotoxic lymphocytes as an indicator of severity in corona virus disease 2019 (COVID-19) patients. Egypt J Immunol (2021) 28(3):157–67.
19. Yaqinuddin A, Kashir J. Innate immunity in COVID-19 patients mediated by NKG2A receptors, and potential treatment using monalizumab, cholroquine, and antiviral agents. Med Hypotheses (2020) 140:109777. doi: 10.1016/j.mehy.2020.109777
20. Shahbaz S, Xu L, Sligl W, Osman M, Bozorgmehr N, Mashhouri S, et al. The quality of SARS-CoV-2-specific t cell functions differs in patients with mild/moderate versus severe disease, and t cells expressing coinhibitory receptors are highly activated. J Immunol (2021) 207(4):1099–111. doi: 10.4049/jimmunol.2100446
21. André P, Brunet C, Guia S, Gallais H, Sampol J, Vivier E, et al. Differential regulation of killer cell ig-like receptors and CD94 lectin-like dimers on NK and T lymphocytes from HIV-1-infected individuals. Eur J Immunol (1999) 29(4):1076–85. doi: 10.1002/(sici)1521-4141(199904)29:04<1076::aid-immu1076>3.0.co;2-z
22. Mahapatra S, Mace EM, Minard CG, Forbes LR, Vargas-Hernandez A, Duryea TK, et al. High-resolution phenotyping identifies NK cell subsets that distinguish healthy children from adults. PloS One (2017) 12(8):e0181134. doi: 10.1371/journal.pone.0181134
23. Manser AR, Uhrberg M. Age-related changes in natural killer cell repertoires: impact on NK cell function and immune surveillance. Cancer Immunol Immunother (2016) 65(4):417–26. doi: 10.1007/s00262-015-1750-0
24. Béziat V, Descours B, Parizot C, Debré P, Vieillard V. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PloS One (2010) 5(8):e11966. doi: 10.1371/journal.pone.0011966
25. Fauriat C, Andersson S, Bjorklund AT, Carlsten M, Schaffer M, Bjorkstrom NK, et al. Estimation of the size of the alloreactive NK cell repertoire: studies in individuals homozygous for the group a KIR haplotype. J Immunol (2008) 181(9):6010–9. doi: 10.4049/jimmunol.181.9.6010
26. Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood (2010) 116(19):3853–64. doi: 10.1182/blood-2010-04-281675
27. Sun G, Zhao X, Li M, Zhang C, Jin H, Li C, et al. CD4 derived double negative T cells prevent the development and progression of nonalcoholic steatohepatitis. Nat Commun (2021) 12(1):650. doi: 10.1038/s41467-021-20941-x
28. Morgan DJ, Davis DM. Distinct effects of dexamethasone on human natural killer cell responses dependent on cytokines. Front Immunol (2017) 8:432. doi: 10.3389/fimmu.2017.00432
29. Brady J, Hayakawa Y, Smyth MJ, Nutt SL. IL-21 induces the functional maturation of murine NK cells. J Immunol (2004) 172(4):2048–58. doi: 10.4049/jimmunol.172.4.2048
30. Mingari MC, Vitale C, Cantoni C, Bellomo R, Ponte M, Schiavetti F, et al. Interleukin-15-induced maturation of human natural killer cells from early thymic precursors: selective expression of CD94/NKG2-a as the only HLA class I-specific inhibitory receptor. Eur J Immunol (1997) 27(6):1374–80. doi: 10.1002/eji.1830270612
31. Kamiya T, Seow SV, Wong D, Robinson M, Campana D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J Clin Invest (2019) 129(5):2094–106. doi: 10.1172/jci123955
32. Wang X, Cui Y, Luo G, Wang Q, Hu J, He W, et al. Activated mouse CD4(+)Foxp3(-) T cells facilitate melanoma metastasis via qa-1-dependent suppression of NK-cell cytotoxicity. Cell Res (2012) 22(12):1696–706. doi: 10.1038/cr.2012.128
33. Zhang C, Wang XM, Li SR, Twelkmeyer T, Wang WH, Zhang SY, et al. NKG2A is a NK cell exhaustion checkpoint for HCV persistence. Nat Commun (2019) 10(1):1507. doi: 10.1038/s41467-019-09212-y
34. Elliott JM, Yokoyama WM. Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol (2011) 32(8):364–72. doi: 10.1016/j.it.2011.06.001
35. Zaghi E, Calvi M, Marcenaro E, Mavilio D, Di Vito C. Targeting NKG2A to elucidate natural killer cell ontogenesis and to develop novel immune-therapeutic strategies in cancer therapy. J Leukoc Biol (2019) 105(6):1243–51. doi: 10.1002/JLB.MR0718-300R
36. Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity (2006) 25(2):331–42. doi: 10.1016/j.immuni.2006.06.013
37. Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature (2005) 436(7051):709–13. doi: 10.1038/nature03847
38. Zhang X, Feng J, Chen S, Yang H, Dong Z. Synergized regulation of NK cell education by NKG2A and specific Ly49 family members. Nat Commun (2019) 10(1):5010. doi: 10.1038/s41467-019-13032-5
39. Lunemann S, Langeneckert AE, Martrus G, Hess LU, Salzberger W, Ziegler AE, et al. Human liver-derived CXCR6(+) NK cells are predominantly educated through NKG2A and show reduced cytokine production. J Leukoc Biol (2019) 105(6):1331–40. doi: 10.1002/JLB.1MA1118-428R
40. Anfossi N, André P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity (2006) 25(2):331–42. doi: 10.1016/j.immuni.2006.06.013
41. Forslund E, Sohlberg E, Enqvist M, Olofsson PE, Malmberg KJ, Önfelt B. Microchip-based single-cell imaging reveals that cd56dimcd57-kir-nkg2a+ nk cells have more dynamic migration associated with increased target cell conjugation and probability of killing compared to cd56dimcd57-kir-nkg2a- nk cells. J Immunol (2015) 195(7):3374–81. doi: 10.4049/jimmunol.1500171
42. Shreeve N, Depierreux D, Hawkes D, Traherne JA, Sovio U, Huhn O, et al. The CD94/NKG2A inhibitory receptor educates uterine NK cells to optimize pregnancy outcomes in humans and mice. Immunity (2021) 54(6):1231–1244 e4. doi: 10.1016/j.immuni.2021.03.021
43. Highton AJ, Diercks BP, Mockl F, Martrus G, Sauter J, Schmidt AH, et al. High metabolic function and resilience of nkg2a-educated nk cells. Front Immunol (2020) 11:559576. doi: 10.3389/fimmu.2020.559576
44. Borrego F, Kabat J, Sanni TB, Coligan JE. NK cell CD94/NKG2A inhibitory receptors are internalized and recycle independently of inhibitory signaling processes. J Immunol (2002) 169(11):6102–11. doi: 10.4049/jimmunol.169.11.6102
45. Sanni TB, Masilamani M, Kabat J, Coligan JE, Borrego F. Exclusion of lipid rafts and decreased mobility of CD94/NKG2A receptors at the inhibitory NK cell synapse. Mol Biol Cell (2004) 15(7):3210–23. doi: 10.1091/mbc.e03-11-0779
46. Borst L, van der Burg SH, van Hall T. The NKG2A-HLA-e axis as a novel checkpoint in the tumor microenvironment. Clin Cancer Res (2020) 26(21):5549–56. doi: 10.1158/1078-0432.CCR-19-2095
47. van Hall T, André P, Horowitz A, Ruan DF, Borst L, Zerbib R, et al. Monalizumab: inhibiting the novel immune checkpoint NKG2A. J Immunother Cancer (2019) 7(1):263. doi: 10.1186/s40425-019-0761-3
48. Cho JH, Kim HO, Webster K, Palendira M, Hahm B, Kim KS, et al. Calcineurin-dependent negative regulation of CD94/NKG2A expression on naive CD8+ T cells. Blood (2011) 118(1):116–28. doi: 10.1182/blood-2010-11-317396
49. Kim HJ, Cantor H. Regulation of self-tolerance by qa-1-restricted CD8(+) regulatory T cells. Semin Immunol (2011) 23(6):446–52. doi: 10.1016/j.smim.2011.06.001
50. Bertone S, Schiavetti F, Bellomo R, Vitale C, Ponte M, Moretta L, et al. Transforming growth factor-beta-induced expression of CD94/NKG2A inhibitory receptors in human T lymphocytes. Eur J Immunol (1999) 29(1):23–9. doi: 10.1002/(sici)1521-4141(199901)29:01<23::aid-immu23>3.0.co;2-y
51. Derre L, Corvaisier M, Pandolfino MC, Diez E, Jotereau F, Gervois N. Expression of CD94/NKG2-a on human T lymphocytes is induced by IL-12: implications for adoptive immunotherapy. J Immunol (2002) 168(10):4864–70. doi: 10.4049/jimmunol.168.10.4864
52. Jabri B, Selby JM, Negulescu H, Lee L, Roberts AI, Beavis A, et al. TCR specificity dictates CD94/NKG2A expression by human CTL. Immunity (2002) 17(4):487–99. doi: 10.1016/s1074-7613(02)00427-2
53. Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med (2018) 24(7):986–93. doi: 10.1038/s41591-018-0078-7
54. Sun H, Sun C, Xiao W, Sun R. Tissue-resident lymphocytes: from adaptive to innate immunity. Cell Mol Immunol (2019) 16(3):205–15. doi: 10.1038/s41423-018-0192-y
55. van Montfoort N, Borst L, Korrer MJ, Sluijter M, Marijt KA, Santegoets SJ, et al. NKG2A blockade potentiates cd8 t cell immunity induced by cancer vaccines. Cell (2018) 175(7):1744–1755.e15. doi: 10.1016/j.cell.2018.10.028
56. Romero P, Ortega C, Palma A, Molina IJ, Peña J, Santamaría M. Expression of CD94 and NKG2 molecules on human CD4(+) T cells in response to CD3-mediated stimulation. J Leukoc Biol (2001) 70(2):219–24.
57. Zhang Y, Lv G, Lou X, Peng D, Qu X, Yang X, et al. NKG2A expression and impaired function of NK cells in patients with new onset of graves’ disease. Int Immunopharmacol (2015) 24(1):133–9. doi: 10.1016/j.intimp.2014.09.020
58. Talebian Yazdi M, van Riet S, van Schadewijk A, Fiocco M, van Hall T, Taube C, et al. The positive prognostic effect of stromal CD8+ tumor-infiltrating T cells is restrained by the expression of HLA-e in non-small cell lung carcinoma. Oncotarget (2016) 7(3):3477–88. doi: 10.18632/oncotarget.6506
59. Wu Z, Liang J, Wang Z, Li A, Fan X, Jiang T. HLA-e expression in diffuse glioma: relationship with clinicopathological features and patient survival. BMC Neurol (2020) 20(1):59. doi: 10.1186/s12883-020-01640-4
60. Ferns DM, Heeren AM, Samuels S, Bleeker MCG, de Gruijl TD, Kenter GG, et al. Classical and non-classical HLA class I aberrations in primary cervical squamous- and adenocarcinomas and paired lymph node metastases. J Immunother Cancer (2016) 4:78. doi: 10.1186/s40425-016-0184-3
61. Seliger B, Jasinski-Bergner S, Quandt D, Stoehr C, Bukur J, Wach S, et al. HLA-e expression and its clinical relevance in human renal cell carcinoma. Oncotarget (2016) 7(41):67360–72. doi: 10.18632/oncotarget.11744
62. Zeestraten EC, Reimers MS, Saadatmand S, Goossens-Beumer IJ, Dekker JW, Liefers GJ, et al. Combined analysis of HLA class I, HLA-e and HLA-G predicts prognosis in colon cancer patients. Br J Cancer (2014) 110(2):459–68. doi: 10.1038/bjc.2013.696
63. Eugene J, Jouand N, Ducoin K, Dansette D, Oger R, Deleine C, et al. The inhibitory receptor CD94/NKG2A on CD8(+) tumor-infiltrating lymphocytes in colorectal cancer: a promising new druggable immune checkpoint in the context of HLAE/beta2m overexpression. Mod Pathol (2020) 33(3):468–82. doi: 10.1038/s41379-019-0322-9
64. de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, et al. HLA-e and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol (2010) 185(12):7452–9. doi: 10.4049/jimmunol.1002629
65. Bossard C, Bézieau S, Matysiak-Budnik T, Volteau C, Laboisse CL, Jotereau F, et al. HLA-E/β2 microglobulin overexpression in colorectal cancer is associated with recruitment of inhibitory immune cells and tumor progression. Int J Cancer (2012) 131(4):855–63. doi: 10.1002/ijc.26453
66. Levy EM, Bianchini M, Von Euw EM, Barrio MM, Bravo AI, Furman D, et al. Human leukocyte antigen-e protein is overexpressed in primary human colorectal cancer. Int J Oncol (2008) 32(3):633–41. doi: 10.3892/ijo.32.3.633
67. Kren L, Slaby O, Muckova K, Lzicarova E, Sova M, Vybihal V, et al. Expression of immune-modulatory molecules HLA-G and HLA-e by tumor cells in glioblastomas: an unexpected prognostic significance? Neuropathology (2011) 31(2):129–34. doi: 10.1111/j.1440-1789.2010.01149.x
68. Wolpert F, Roth P, Lamszus K, Tabatabai G, Weller M, Eisele G. HLA-e contributes to an immune-inhibitory phenotype of glioblastoma stem-like cells. J Neuroimmunol (2012) 250(1-2):27–34. doi: 10.1016/j.jneuroim.2012.05.010
69. Gooden M, Lampen M, Jordanova ES, Leffers N, Trimbos JB, van der Burg SH, et al. HLA-e expression by gynecological cancers restrains tumor-infiltrating CD8+ T lymphocytes. Proc Natl Acad Sci U.S.A. (2011) 108(26):10656–61. doi: 10.1073/pnas.1100354108
70. Andersson E, Poschke I, Villabona L, Carlson JW, Lundqvist A, Kiessling R, et al. Non-classical HLA-class I expression in serous ovarian carcinoma: Correlation with the HLA-genotype, tumor infiltrating immune cells and prognosis. Oncoimmunology (2016) 5(1):e1052213. doi: 10.1080/2162402x.2015.1052213
71. Zheng H, Lu R, Xie S, Wen X, Wang H, Gao X, et al. Human leukocyte antigen-e alleles and expression in patients with serous ovarian cancer. Cancer Sci (2015) 106(5):522–8. doi: 10.1111/cas.12641
72. Sun C, Xu J, Huang Q, Huang M, Wen H, Zhang C, et al. High NKG2A expression contributes to NK cell exhaustion and predicts a poor prognosis of patients with liver cancer. Oncoimmunology (2017) 6(1):e1264562. doi: 10.1080/2162402x.2016.1264562
73. Malmberg KJ, Levitsky V, Norell H, de Matos CT, Carlsten M, Schedvins K, et al. IFN-gamma protects short-term ovarian carcinoma cell lines from CTL lysis via a CD94/NKG2A-dependent mechanism. J Clin Invest (2002) 110(10):1515–23. doi: 10.1172/jci15564
74. Gustafson KS, Ginder GD. Interferon-gamma induction of the human leukocyte antigen-e gene is mediated through binding of a complex containing STAT1alpha to a distinct interferon-gamma-responsive element. J Biol Chem (1996) 271(33):20035–46. doi: 10.1074/jbc.271.33.20035
75. Li Q, Cai S, Li M, Zhou X, Wu G, Kang K, et al. Natural killer cell exhaustion in lung cancer. Int Immunopharmacol (2021) 96:107764. doi: 10.1016/j.intimp.2021.107764
76. Sheu BC, Chiou SH, Lin HH, Chow SN, Huang SC, Ho HN, et al. Up-regulation of inhibitory natural killer receptors CD94/NKG2A with suppressed intracellular perforin expression of tumor-infiltrating CD8+ T lymphocytes in human cervical carcinoma. Cancer Res (2005) 65(7):2921–9. doi: 10.1158/0008-5472.can-04-2108
77. Figueiredo C, Seltsam A, Blasczyk R. Permanent silencing of NKG2A expression for cell-based therapeutics. J Mol Med (Berl) (2009) 87(2):199–210. doi: 10.1007/s00109-008-0417-0
78. McWilliams EM, Mele JM, Cheney C, Timmerman EA, Fiazuddin F, Strattan EJ, et al. Therapeutic CD94/NKG2A blockade improves natural killer cell dysfunction in chronic lymphocytic leukemia. Oncoimmunology (2016) 5(10):e1226720. doi: 10.1080/2162402x.2016.1226720
79. Salomé B, Gomez-Cadena A, Loyon R, Suffiotti M, Salvestrini V, Wyss T, et al. CD56 as a marker of an ILC1-like population with NK cell properties that is functionally impaired in AML. Blood Adv (2019) 3(22):3674–87. doi: 10.1182/bloodadvances.2018030478
80. Tinker AV, Hirte HW, Provencher D, Butler M, Ritter H, Tu D, et al. Dose-ranging and cohort-expansion study of monalizumab (iph2201) in patients with advanced gynecologic malignancies: a trial of the canadian cancer trials group (cctg): ind221. Clin Cancer Res (2019) 25(20):6052–60. doi: 10.1158/1078-0432.ccr-19-0298
81. Segal N, Naidoo J, Curigliano G, Patel S, Sahebjam S, Papadopoulos K, et al. First-in-human dose escalation of monalizumab plus durvalumab, with expansion in patients with metastatic microsatellite-stable colorectal cancer. J Clin Oncol (2018) 36:3540–0. doi: 10.1200/JCO.2018.36.15_suppl.3540
82. Nattermann J, Nischalke HD, Hofmeister V, Ahlenstiel G, Zimmermann H, Leifeld L, et al. The HLA-A2 restricted T cell epitope HCV core 35-44 stabilizes HLA-e expression and inhibits cytolysis mediated by natural killer cells. Am J Pathol (2005) 166(2):443–53. doi: 10.1016/s0002-9440(10)62267-5
83. Ulbrecht M, Martinozzi S, Grzeschik M, Hengel H, Ellwart JW, Pla M, et al. Cutting edge: the human cytomegalovirusul40gene product contains a ligand for hla-e and prevents nk cell-mediated lysis. J Immunol (2000) 164(10):5019–22. doi: 10.4049/jimmunol.164.10.5019
84. Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, et al. Surface expression of HLA-e, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science (2000) 287(5455):1031. doi: 10.1126/science.287.5455.1031
85. Wang EC, McSharry B, Retiere C, Tomasec P, Williams S, Borysiewicz LK, et al. UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proc Natl Acad Sci USA (2002) 99(11):7570–5. doi: 10.1073/pnas.112680099
86. Nattermann J, Nischalke HD, Hofmeister V, Kupfer B, Ahlenstiel G, Feldmann G, et al. HIV-1 infection leads to increased HLA-e expression resulting in impaired function of natural killer cells. Antivir Ther (2005) 10(1):95–107. doi: 10.1177/135965350501000107
87. van Stigt Thans T, Akko JI, Niehrs A, Garcia-Beltran WF, Richert L, Sturzel CM, et al. Primary HIV-1 strains use nef to downmodulate hla-e surface expression. J Virol (2019) 93(20):e00719-19. doi: 10.1128/JVI.00719-19
88. Gonçalves MA, Le Discorde M, Simões RT, Rabreau M, Soares EG, Donadi EA, et al. Classical and non-classical HLA molecules and p16(INK4a) expression in precursors lesions and invasive cervical cancer. Eur J Obstet Gynecol Reprod Biol (2008) 141(1):70–4. doi: 10.1016/j.ejogrb.2008.06.010
89. Mbiribindi B, Pena JK, Arvedson MP, Moreno Romero C, McCarthy SR, Hatton OL, et al. Epstein-Barr Virus peptides derived from latent cycle proteins alter NKG2A + NK cell effector function. Sci Rep (2020) 10(1):19973. doi: 10.1038/s41598-020-76344-3
90. Jin X, Yan ZH, Lu L, Lu S, Zhang G, Lin W. Peripheral immune cells exhaustion and functional impairment in patients with chronic hepatitis b. Front Med (Lausanne) (2021) 8:759292. doi: 10.3389/fmed.2021.759292
91. Ma Q, Dong X, Liu S, Zhong T, Sun D, Zong L, et al. Hepatitis b e antigen induces nkg2a(+) natural killer cell dysfunction via regulatory t cell-derived interleukin 10 in chronic hepatitis b virus infection. Front Cell Dev Biol (2020) 8:421. doi: 10.3389/fcell.2020.00421
92. Jaiswal SR, Bhakuni P, Bhagwati G, Aiyar HM, Chakrabarti A, Chakrabarti S. Alterations in nkg2a and nkg2c subsets of natural killer cells following epstein-barr virus reactivation in ctla4ig-based haploidentical transplantation is associated with increased chronic graft-versus-host disease. Transplantation (2020) 104(1):e23–30. doi: 10.1097/tp.0000000000002941
93. Hendricks DW, Balfour HH Jr., Dunmire SK, Schmeling DO, Hogquist KA, Lanier LL. Cutting edge: NKG2C(hi)CD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus. J Immunol (2014) 192(10):4492–6. doi: 10.4049/jimmunol.1303211
94. Wang X, Piersma SJ, Nelson CA, Dai YN, Christensen T, Lazear E, et al. A herpesvirus encoded qa-1 mimic inhibits natural killer cell cytotoxicity through CD94/NKG2A receptor engagement. Elife (2018) 7:e38667. doi: 10.7554/eLife.38667
95. Moser JM, Gibbs J, Jensen PE, Lukacher AE. CD94-NKG2A receptors regulate antiviral CD8(+) T cell responses. Nat Immunol (2002) 3(2):189–95. doi: 10.1038/ni757
96. Rapaport AS, Schriewer J, Gilfillan S, Hembrador E, Crump R, Plougastel BF, et al. The inhibitory receptor NKG2A sustains virus-specific CD8+T cells in response to a lethal poxvirus infection. Immunity (2015) 43(6):1112–24. doi: 10.1016/j.immuni.2015.11.005
97. Miller JD, Peters M, Oran AE, Beresford GW, Harrington L, Boss JM, et al. CD94/NKG2 expression does not inhibit cytotoxic function of lymphocytic choriomeningitis virus-specific CD8+ T cells. J Immunol (2002) 169(2):693–701. doi: 10.4049/jimmunol.169.2.693
98. Bortolotti D, Gentili V, Rizzo S, Rotola A, Rizzo R. SARS-cov-2 spike 1 protein controls natural killer cell activation via the hla-e/nkg2a pathway. Cells (2020) 9(9):1975. doi: 10.3390/cells9091975
99. Gianchecchi E, Delfino DV, Fierabracci A. Natural killer cells: potential biomarkers and therapeutic target in autoimmune diseases? Front Immunol (2021) 12:616853. doi: 10.3389/fimmu.2021.616853
100. Yang Y, Day J, Souza-Fonseca Guimaraes F, Wicks IP, Louis C. Natural killer cells in inflammatory autoimmune diseases. Clin Transl Immunol (2021) 10(2):e1250. doi: 10.1002/cti2.1250
101. Kucuksezer UC, Aktas Cetin E, Esen F, Tahrali I, Akdeniz N, Gelmez MY, et al. The role of natural killer cells in autoimmune diseases. Front Immunol (2021) 12:622306. doi: 10.3389/fimmu.2021.622306
102. Lu L, Ikizawa K, Hu D, Werneck MB, Wucherpfennig KW, Cantor H. Regulation of activated CD4+ T cells by NK cells via the qa-1-NKG2A inhibitory pathway. Immunity (2007) 26(5):593–604. doi: 10.1016/j.immuni.2007.03.017
103. Leavenworth JW, Schellack C, Kim HJ, Lu L, Spee P, Cantor H. Analysis of the cellular mechanism underlying inhibition of EAE after treatment with anti-NKG2A F(ab’)2. Proc Natl Acad Sci USA (2010) 107(6):2562–7. doi: 10.1073/pnas.0914732107
104. Leavenworth JW, Wang X, Wenander CS, Spee P, Cantor H. Mobilization of natural killer cells inhibits development of collagen-induced arthritis. Proc Natl Acad Sci USA (2011) 108(35):14584–9. doi: 10.1073/pnas.1112188108
105. Lin SJ, Kuo ML, Hsiao HS, Lee PT, Chen JY, Huang JL. Activating and inhibitory receptors on natural killer cells in patients with systemic lupus erythematosis-regulation with interleukin-15. PloS One (2017) 12(10):e0186223. doi: 10.1371/journal.pone.0186223
106. Hervier B, Beziat V, Haroche J, Mathian A, Lebon P, Ghillani-Dalbin P, et al. Phenotype and function of natural killer cells in systemic lupus erythematosus: excess interferon-γ production in patients with active disease. Arthritis Rheum (2011) 63(6):1698–706. doi: 10.1002/art.30313
107. Son SW, Kim EO, Ryu ES, Kim TJ, Kim JN, Choi JE, et al. Upregulation of fas and downregulation of CD94/NKG2A inhibitory receptors on circulating natural killer cells in patients with new-onset psoriasis. Br J Dermatol (2009) 161(2):281–8. doi: 10.1111/j.1365-2133.2009.09178.x
108. Wang L, Kang N, Zhou J, Guo Y, Zhang X, Cui L, et al. Downregulation of CD94/NKG2A inhibitory receptor on decreased γδ T cells in patients with systemic lupus erythematosus. Scand J Immunol (2012) 76(1):62–9. doi: 10.1111/j.1365-3083.2012.02705.x
109. Walsh CE, Ryan EJ, O’Farrelly C, Golden-Mason L, FitzGerald O, Veale DJ, et al. Differential expression of NK receptors CD94 and NKG2A by T cells in rheumatoid arthritis patients in remission compared to active disease. PloS One (2011) 6(11):e27182. doi: 10.1371/journal.pone.0027182
110. Lu L, Kim HJ, Werneck MB, Cantor H. Regulation of CD8+ regulatory T cells: Interruption of the NKG2A-Qa-1 interaction allows robust suppressive activity and resolution of autoimmune disease. Proc Natl Acad Sci USA (2008) 105(49):19420–5. doi: 10.1073/pnas.0810383105
111. Correale J, Villa A. Isolation and characterization of CD8+ regulatory T cells in multiple sclerosis. J Neuroimmunol (2008) 195(1-2):121–34. doi: 10.1016/j.jneuroim.2007.12.004
112. Hall LJ, Murphy CT, Quinlan A, Hurley G, Shanahan F, Nally K, et al. Natural killer cells protect mice from DSS-induced colitis by regulating neutrophil function via the NKG2A receptor. Mucosal Immunol (2013) 6(5):1016–26. doi: 10.1038/mi.2012.140
113. Zou Z, Zuo D, Yang J, Fan H. The ANXA1 released from intestinal epithelial cells alleviate DSS-induced colitis by improving NKG2A expression of natural killer cells. Biochem Biophys Res Commun (2016) 478(1):213–20. doi: 10.1016/j.bbrc.2016.07.066
114. Marafini I, Monteleone I, Di Fusco D, Sedda S, Cupi ML, Fina D, et al. Celiac disease-related inflammation is marked by reduction of nkp44/nkp46-double positive natural killer cells. PloS One (2016) 11(5):e0155103. doi: 10.1371/journal.pone.0155103
115. de Matos CT, Berg L, Michaëlsson J, Felländer-Tsai L, Kärre K, Söderström K. Activating and inhibitory receptors on synovial fluid natural killer cells of arthritis patients: role of CD94/NKG2A in control of cytokine secretion. Immunology (2007) 122(2):291–301. doi: 10.1111/j.1365-2567.2007.02638.x
116. Abulizi A, Shao Y, Aji T, Li Z, Zhang C, Aini A, et al. Echinococcus multilocularis inoculation induces NK cell functional decrease through high expression of NKG2A in C57BL/6 mice. BMC Infect Dis (2019) 19(1):792. doi: 10.1186/s12879-019-4417-1
117. Yasen A, Sun W, Aini A, Aji T, Shao Y, Wang H, et al. Single-cell rna sequencing reveals the heterogeneity of infiltrating immune cell profiles in the hepatic cystic echinococcosis microenvironment. Infect Immun (2021) 89(12):e0029721. doi: 10.1128/IAI.00297-21
118. Artavanis-Tsakonas K, Eleme K, McQueen KL, Cheng NW, Parham P, Davis DM, et al. Activation of a subset of human NK cells upon contact with plasmodium falciparum-infected erythrocytes. J Immunol (2003) 171(10):5396–405. doi: 10.4049/jimmunol.171.10.5396
119. D’Ombrain MC, Hansen DS, Simpson KM, Schofield L. Gammadelta-T cells expressing NK receptors predominate over NK cells and conventional T cells in the innate IFN-gamma response to plasmodium falciparum malaria. Eur J Immunol (2007) 37(7):1864–73. doi: 10.1002/eji.200636889
120. Xu X, Zhao M, Liu X, Jiang Y, Zhang H, Zhai X, et al. Toxoplasma gondii infection regulates the balance of activating and inhibitory receptors on decidual natural killer cells. PloS One (2013) 8(2):e55432. doi: 10.1371/journal.pone.0055432
121. Khetsuphan T, Chaisri U, Dechkhajorn W, Benjathummarak S, Dekumyoy P, Ampawong S, et al. Effects of gnathostoma spinigerum infective stage larva excretory-secretory products on NK cells in peripheral blood mononuclear cell culture: focused on expressions of IFN-gamma and killer cell lectin-like receptors. Parasitol Res (2020) 119(3):1011–21. doi: 10.1007/s00436-019-06593-3
122. Li L, Cha H, Yu X, Xie H, Wu C, Dong N, et al. The characteristics of NK cells in schistosoma japonicum-infected mouse spleens. Parasitol Res (2015) 114(12):4371–9. doi: 10.1007/s00436-015-4674-x
123. Cha H, Qin W, Yang Q, Xie H, Qu J, Wang M, et al. Differential pulmonic NK and NKT cell responses in schistosoma japonicum-infected mice. Parasitol Res (2017) 116(2):559–67. doi: 10.1007/s00436-016-5320-y
124. Hu LJ, Zhao XY, Yu XX, Lv M, Han TT, Han W, et al. Quantity and quality reconstitution of nkg2a(+) natural killer cells are associated with graft-versus-host disease after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant (2019) 25(1):1–11. doi: 10.1016/j.bbmt.2018.08.008
125. Kordelas L, Steckel NK, Horn PA, Beelen DW, Rebmann V. The activating nkg2c receptor is significantly reduced in nk cells after allogeneic stem cell transplantation in patients with severe graft-versus-host disease. Int J Mol Sci (2016) 17(11):1797. doi: 10.3390/ijms17111797
126. Qin R, He L, Yang Z, Jia N, Chen R, Xie J, et al. Identification of parameters representative of immune dysfunction in patients with severe and fatal COVID-19 infection: a systematic review and meta-analysis. Clin Rev Allergy Immunol (2022) 1-33. doi: 10.1007/s12016-021-08908-8
127. Zhang S, Gan J, Chen BG, Zheng D, Zhang JG, Lin RH, et al. Dynamics of peripheral immune cells and their HLA-G and receptor expressions in a patient suffering from critical COVID-19 pneumonia to convalescence. Clin Transl Immunol (2020) 9(5):e1128. doi: 10.1002/cti2.1128
128. Zidi I. Puzzling out the COVID-19: Therapy targeting HLA-G and HLA-e. Hum Immunol (2020) 81(12):697–701. doi: 10.1016/j.humimm.2020.10.001
129. Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, López-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood (2004) 104(12):3664–71. doi: 10.1182/blood-2004-05-2058
130. Gumá M, Budt M, Sáez A, Brckalo T, Hengel H, Angulo A, et al. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood (2006) 107(9):3624–31. doi: 10.1182/blood-2005-09-3682
131. Rölle A, Pollmann J, Ewen EM, Le VT, Halenius A, Hengel H, et al. IL-12-producing monocytes and HLA-e control HCMV-driven NKG2C+ NK cell expansion. J Clin Invest (2014) 124(12):5305–16. doi: 10.1172/jci77440
132. Marshall NB, Vong AM, Devarajan P, Brauner MD, Kuang Y, Nayar R, et al. NKG2C/E marks the unique cytotoxic CD4 T cell subset, ThCTL, generated by influenza infection. J Immunol (2017) 198(3):1142–55. doi: 10.4049/jimmunol.1601297
133. Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity (1998) 8(6):693–701. doi: 10.1016/s1074-7613(00)80574-9
134. Wada H, Matsumoto N, Maenaka K, Suzuki K, Yamamoto K. The inhibitory NK cell receptor CD94/NKG2A and the activating receptor CD94/NKG2C bind the top of HLA-e through mostly shared but partly distinct sets of HLA-e residues. Eur J Immunol (2004) 34(1):81–90. doi: 10.1002/eji.200324432
135. Kaiser BK, Barahmand-Pour F, Paulsene W, Medley S, Geraghty DE, Strong RK. Interactions between NKG2x immunoreceptors and HLA-e ligands display overlapping affinities and thermodynamics. J Immunol (2005) 174(5):2878–84. doi: 10.4049/jimmunol.174.5.2878
Keywords: NKG2A, cancer immunotherapy, viral infections, autoimmune diseases, HLA-E
Citation: Wang X, Xiong H and Ning Z (2022) Implications of NKG2A in immunity and immune-mediated diseases. Front. Immunol. 13:960852. doi: 10.3389/fimmu.2022.960852
Received: 03 June 2022; Accepted: 21 July 2022;
Published: 10 August 2022.
Edited by:
Timothy Edward O’Sullivan, University of California, Los Angeles, United StatesReviewed by:
Ana Stojanovic, Heidelberg University, GermanyNadia Guerra, Imperial College London, United Kingdom
Copyright © 2022 Wang, Xiong and Ning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaochen Ning, bmluZ3pjQG1haWwuam5tYy5lZHUuY24=; Huabao Xiong, eGlvbmdodWFiYW9AbWFpbC5qbm1jLmVkdS5jbg==
 Xiaotong Wang1,2
Xiaotong Wang1,2 Zhaochen Ning
Zhaochen Ning