
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 10 August 2022
Sec. T Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.960444
This article is part of the Research Topic Molecular Switches of the Immune System: The E Protein/Id Axis in Hematopoietic Development and Function View all 11 articles
 Aneta Pankow1,2
Aneta Pankow1,2 Xiao-Hong Sun1,2,3*†
Xiao-Hong Sun1,2,3*†T cells develop in the thymus from lymphoid primed multipotent progenitors or common lymphoid progenitors into αβ and γδ subsets. The basic helix-loop-helix transcription factors, E proteins, play pivotal roles at multiple stages from T cell commitment to maturation. Inhibitors of E proteins, Id2 and Id3, also regulate T cell development while promoting ILC differentiation. Recent findings suggest that the thymus can also produce innate lymphoid cells (ILCs). In this review, we present current findings that suggest the balance between E and Id proteins is likely to be critical for controlling the bifurcation of T cell and ILC fates at early stages of T cell development.
The E protein family of transcription factors are crucial molecules engaging in B cell development in the bone marrow and T cells differentiation in the thymus (1, 2). This family consists of proteins encoded by three genes, E2A (also called Tcf3), HEB (Tcf12) and E2-2 (Tcf4) (Figure 1A) (3–5). These proteins share extensive sequence homologies in the activation domains (AD1, LH) and basic helix-loop-helix (bHLH) DNA-binding domain (6–9). E proteins regulate the transcription of their target genes by forming homodimers or heterodimers and bind to E-box sequences (9). The E2A gene gives rise to two proteins, E12 and E47, due to alternative splicing of two adjacent exons, each encoding a basic helix-loop-helix (bHLH) domain (10). While E47 binds DNA avidly as homodimers, E12 does so poorly due to the presence of an inhibitory domain (11). However, both form heterodimers with other bHLH proteins such as MyoD, and bind DNA efficiently. The HEB gene encodes a full-length canonical protein (HEBCan) and a truncated alternate form (HEBAlt), which derives from a transcript initiated in the middle of the gene (12). HEBAlt lacks the AD1 transcription activation domain and has lower transcriptional activities (13). It has an Alt domain at the N-terminus with three tyrosine residues which can be modified by phosphorylation that augments its transcriptional activity (13).
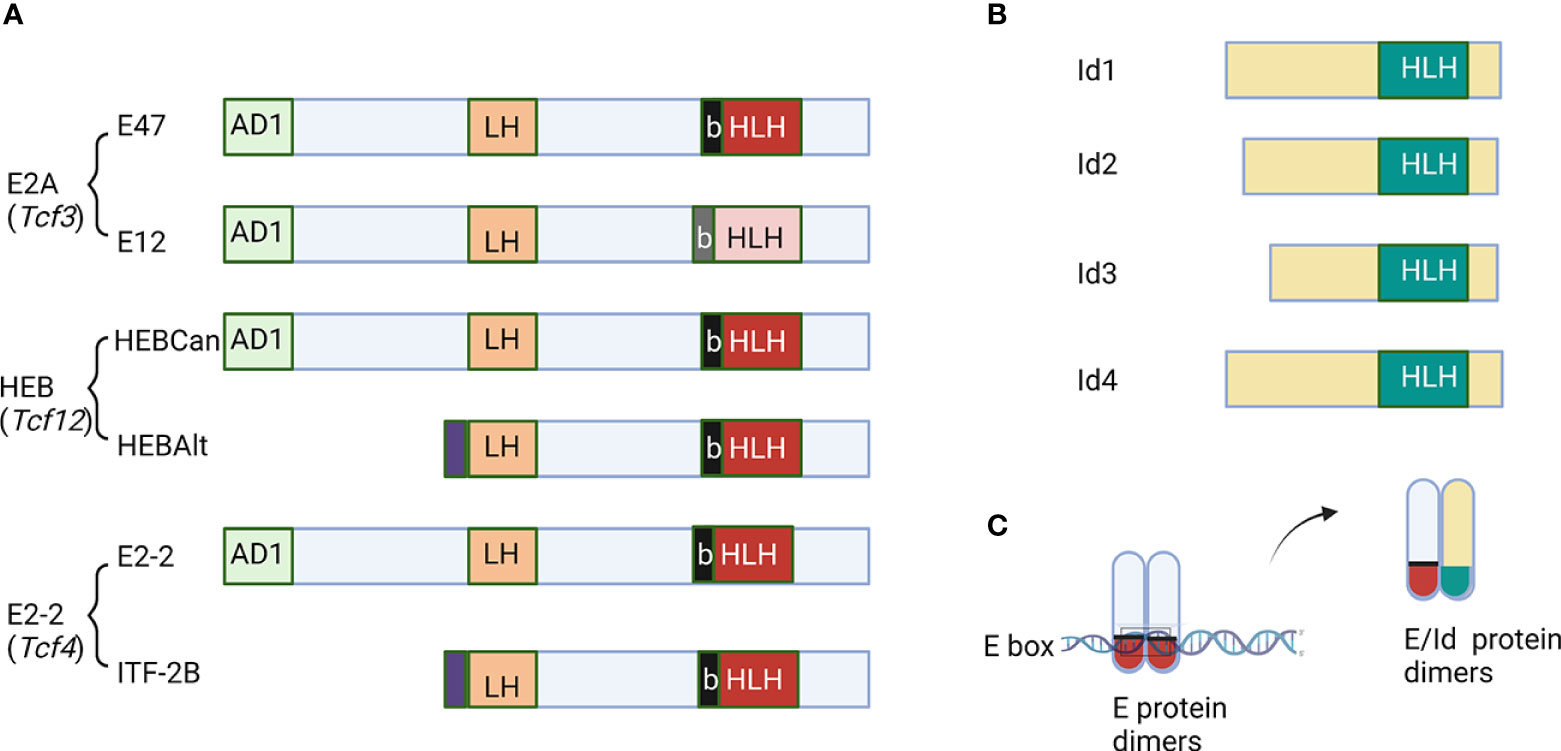
Figure 1 Schematic diagrams of E and Id proteins. (A) The function domains of E proteins are labeled. AD1 and LH are two transcription activation domains. The basic and helix-loop-helix domains are marked as b and HLH, respectively. (B) Id proteins with the HLH domain are shown. (C) The mechanism of action of Id proteins to inhibit DNA binding by E proteins. The figure was created by BioRender.com.
The family of inhibitor of differentiation proteins, Id1-4, antagonize E proteins by dimerizing with them via the helix-loop-helix domain (Figure 1B) (14–19). However, because Id proteins lack the basic amino acids necessary for DNA binding, heterodimers between E and Id proteins cannot bind to E box sequences (Figure 1C). Transcription of the E protein genes is less variable but that of the Id genes is highly dynamic. Therefore, the net E protein activity in a given cell is determined by the levels of both E and Id proteins (16, 17). In this review, we intend to highlight the roles of E and Id proteins in regulating the fate choices between T cells and innate lymphoid cells.
Lymphoid-primed multipotent progenitors (LMPP) and common lymphoid progenitors (CLP) travel from the bone marrow to the thymus and become early T cell progenitors (ETP) (20–23). T cell developmental progression in the thymus can be generally defined by the expression of CD4 and CD8 surface markers: from CD4 and CD8 double negative (DN) to double positive (DP) and then to CD4 or CD8 single positive (SP) (24–27). During the transition from DN to DP stage, an immature CD8 single positive subset (ISP) has been described (28, 29). Within the DN compartment, four subsets (DN1 to DN4) are characterized by the expression of c-kit and CD25 in the order of maturity as c-kit+CD25-, c-kit+CD25+, c-kit-CD25+ and c-kit-CD25- (27). ETPs are at the top of the hierarchy and included in the DN1 subset (23). They give rise to both αβ and γδ T cells, which have distinct T cell receptors (TCRs), different developmental programs and divergent functions. The E2A and HEB genes are both expressed in the thymus. Interestingly, HEBAlt is preferentially produced in the DN and ISP stages. Since E proteins are known to inhibit cell proliferation and HEBAlt acts as a hypomorph (13, 30), whether HEBAlt plays a role in tampering E protein activities during pre-TCR-triggered cell expansion is interesting to be investigated.
The development of αβ T cells is largely driven by αβ TCR signaling events. However, before the formation of pre-TCRs and TCRs, the differentiation of committed T cell precursors is supported by Notch signaling and signaling from cytokine receptors such as that of IL-7 (31–35). Critical transcription factors involved in T cell commitment include TCF1, GATA3 and Bcl11b (36–40). The sequential rearrangements of TCRβ and then TCRα genes catalyzed by the RAG1 and RAG2 recombinases set the milestones of the developmental progression (24, 41–44). The TCRβ locus undergoes recombination between D to J regions and then V to DJ regions to produce functional β chains, which pair with the pre-Tα (45, 46). The pre-TCR complex delivers signals leading to the expansion of DN3 cells and their advancement to the DP stage. The TCRα gene rearrangement occurs at the DP stage, which allows the formation of αβ TCRs, triggers the positive and negative selection and enables the generation of mature SP T cells (47–49). Mature but naive T cells leave the thymus by the upregulation of S1PR1 and CD62L (50–53).
αβ T cells possess a large repertoire of TCRs due to a collection of V regions. These TCRs recognize diverse antigens presented by the MHC molecules and elicit subsequent signaling events. CD4+ and CD8+ naïve T cells exit the thymus to be activated and differentiate into helper and cytotoxic effectors in peripheral lymphoid organs, respectively (54, 55). Due to the sheer quantity of thymic output of αβ T cells and their ability to proliferate in response to antigen engagement, αβ T cells are the major players of adaptive T cell immunity.
The development of γδ T cells differs from αβ T cells. Firstly, unlike αβ T cells, γδ T cells do not traverse DP and SP stages during the development. Instead, they undergo γδ lineage commitment and maturation at DN2 and DN3 stages (56–58). Generation of mature γδ T cells depends on the V-J rearrangement of the TCRγ locus and V(D)J recombination in the TCRδ locus, along with Notch signaling. Since the TCRδ gene is embedded in the TCRα locus, TCRα rearrangement, triggered by pre-TCR signaling after an independent rearrangement event of the TCRβ gene, can eliminate the TCRδ gene, thus aborting the γδ T cell fate (59–61). Early precursors of effector γδ T cells in the thymus are identified as CD24+ and then mature to CD24- stage (62, 63). There are three types of γδ T cells classified based on their effector functions, γδ1, γδ17 and innate-like γδ T cells, which secrete interferon γ, IL-17 and interferon γ plus IL-4, respectively (64–66). The development of γδ T cells require stronger TCR signals in the comparison to αβ T cells (67, 68). The gradients of TCR signals determines the development of specific effector subsets. The generation of innate-like γδ T cells depends on the strongest TCR signal as indicated by their higher levels of CD5 compared to other γδ subsets (69). CD5 levels are proportional to TCR signaling strength in the thymus (70, 71). Expression of PLZF transcription factor also depends on ligand ligation with TCR and PLZF is required for the effector function of innate-like γδ T cells (72). Type 1 γδ T cells also require a strong TCR signal and the T-bet transcription factor is critical for γδ1 differentiation (65, 72–75). On the other hand, type 17 γδ T cells rely on a weaker TCR signal for the differentiation (65, 72, 74, 76). In fetal organ culture, addition of activating antibodies against γδ TCR or CD3 impairs the production of γδ17 cells (76). Moreover, RORγt transcription factor is essential for γδ17 development (77). Additionally, CD73 expression marks most of γδ T cells committed to mature into effector cells in the thymus (78).
Distinct subsets of γδ T cells reside in different tissues and develop at different ages in mice (56, 57). The Vγ regions are described by two different nomenclatures. In this review, we will use the one defined by Raulet and colleagues (79). In the early fetal stage, the first wave of γδ T cells is associated with the Vγ3+Vδ1+ subset known as the dendritic epidermal cells, which produce IFNγ (65, 80, 81). The development of the Vγ4+ subset begins at the fetal stage and lasts until birth. The generation of the Vγ2+ subset occurs in the late fetal stage and continues through adulthood. The Vγ2+ subset consists of cells producing IL-17 or IFNγ (82). The IL-17-producing cells become long-lived cells with self-renewal capabilities after birth (83). Vγ1.1+ cells develop at the prenatal stage and this persists through adult life (56). Despite the complicated developmental schemes of γδ T cell differentiation, how γδ TCRs interact with their ligands and elicit signals is less understood. To some extent, γδ T cells are thought to have properties resembling innate cells.
T cell development is a “wasteful’ process”. Every D-J or V-DJ combination only has one third of a chance to create an in-frame joint that result in a full-length TCR chain. It is believed that over 70% of the developing T cells do not reach the mature stage and die because they fail to form pre-TCR (β selection) at the DN3 stage or because they cannot produce a full-length TCRα chain at the DP stage (death by neglect). They can also be eliminated due to excessively strong TCR signaling (negative selection). Are there any alternative fates for these T cell “drop-outs”? Perhaps, innate lymphoid cells are some of the options.
E proteins play pivotal roles in governing the development of αβ T cells. Two of the E protein genes, E2A and EBCan, are expressed in T cells and they have redundant functions. The proteins encoded by these two genes include E12, E47, HEBCan and HEBAlt. Since all knock-out constructs targeted the bHLH domains, E2A or HEB deficient mice lack all of their respective proteins. Germ-line ablation of either E2A or HEB gene partially impairs T cell development by dramatically reducing thymocyte counts (84, 85). The leaky block allows the maturation of small numbers of T cells, which are predisposed to develop T cell lymphoma (84–86). HEB deficiency also reveals a novel role of HEB at the ISP stage (86). In contrast, simultaneous inhibition of all E proteins by expressing Id1 using the proximal promoter of lck in transgenic mice results in a complete block of T cell development, arresting thymocytes at the DN1 stage when the Id1 transgene begins to be expressed (87, 88). Likewise, inducible ablation of both E2A and HEB genes using the plck-Cre transgene results in a developmental arrest at the DN3 stage when the Cre gene is expressed (89).
E protein-mediated control at these early stages of T cell development is multi-dimensional. First, E proteins are known to activate the transcription of Notch1, which encodes the receptor for Notch ligands such as Delta-like 4 in the thymus and ensure the differentiation and survival of T cells (90–92). Second, E proteins are found to activate the transcription of Rag1 and Rag2 (93, 94), which code for the enzymes essential for VDJ recombination of TCR genes. Third, E proteins facilitate TCR gene rearrangement by increasing chromatin accessibility at the TCRβ locus (95). Fourth, the binding of E2A-HEB heterodimers to Ptcra enhancer regulates pre-Tα expression at the DN3 stage (96–98). Finally, the interplay between E proteins and other transcription factors such as TCF1 and LEF1 also contribute to the positive regulation of early T cell development (36, 99).
Following pre-TCR signaling, the Ras-MAP kinase pathway is activated, which leads to the up-regulation of Egr transcription factors and then activation of the Id3 gene (100–103). This suggests that down-regulation of E protein activity is necessary for DN3 cells progress to the DP stage. Indeed, when Rag1 was deleted, T cell development arrested at the DN3 stage (104). However, if E proteins are down-regulated by germline E2A deletion or pLck-Id1 expression, Rag1-/- thymocytes can advance to the DP stage (105, 106). Another mechanism to down-regulate E proteins is to accelerate their ubiquitin-mediated degradation in the presence of Notch signals and MAP kinases activated by pre-TCR signaling (107, 108).
At the DP stage, Id3 expression is transiently triggered by TCR signaling and is involved in the positive selection of developing thymocytes (101, 109). Deleting both Id2 and Id3 genes prevented the progression of positively selected T cells to the SP stage (110). Conversely, low levels of Id1 expression in plck-Id1 heterozygous transgenic mice allows some T cell precursors reach the DP stage but a majority of these cells undergo apoptosis likely due to excessive responses to the normal levels of TCR stimulation (105, 111). This notion was supported by the observation of hyper-activation of NFκB upon ectopic Id1 expression (105, 112). In addition, deleting both E2A and HEB genes also impairs the generation of CD4 SP T cells (110). Collectively, E and Id proteins clearly are the central players in shaping αβ T cell development.
A strong TCR signal triggers the activation of the ERK-Egr-Id3 axis and favors γδ over αβ T cell development (73). Id3 deficiency resulted in an expansion of Vγ1.1+ innate-like γδ T cells, possibly due to the dampening of the strong TCR signaling which normally causes the death of these cells (113, 114). In fetal organ cultures, HEB deficiency impairs the differentiation of Vγ4 and Vγ6-containing γδ17 cells. In et al. postulated two pathways of γδ T cell development (115). Pathway 1, which favors γδ1 cells, depends on strong TCR signaling and up-regulation of Id3. In contrast, pathway 2 mostly occurs in the fetal stage and requires lower levels of TCR signaling and Id3 expression. HEB is necessary for Vγ6+CD73- γδ17 T cells in the fetal stage as well as Vγ4+CD73+ γδ17 T cells in neonates (115). HEB and E2A are thought to activate the transcription of Sox4, Sox13 and Rorc genes necessary for γδ17 differentiation (115, 116). Overall, it appears that Id3 expression plays a critical role in directing γδ T cell development through counterbalancing the function of E proteins.
Innate lymphoid cells (ILCs) are first responders in immune reactions towards environmental insults and microbial infections. ILCs are divided into three groups, ILC1 to ILC3, which play different roles during specific immune responses (117, 118). Even though ILCs share with T cells the transcriptional factors that drive their differentiation and the profiles of cytokine production, they lack T-cell receptors (TCR), thus eliciting innate immunity as opposed to adaptive immunity mediated by T cells (118–120). Each ILC subset has been increasingly recognized to be heterogenous and display different characteristics in different tissues (121). Plasticity between the three ILC subsets also exist, especially under pathophysiological conditions (118, 122). Nevertheless, the general properties and functions of these three subsets of ILCs have been established. The ILC1 group consists of helper-like ILC1s and conventional NK cells (cNK). ILC1s mediate the early immune response upon contact with intracellular pathogens like bacteria and viruses. Their effector function regarding cytokine production is similar to the that of cNK cells, namely secreting IFNγ upon pathogen exposure. However, NK cells but not helper-like ILC1s are cytotoxic and able to produce high levels of cytotoxic granules like perforin and granzymes. The T-bet transcription factor is responsible for ILC1 differentiation and function (123). ILC2s share a transcriptional network and cytokine production profiles with those of type 2 T helper cells (Th2). GATA3 is the signature transcription factor and drives the expression of cytokines including IL-5, IL-13, IL-4, IL-9, and amphiregulin (124–126). RORα is another transcription factor indispensable for ILC2 differentiation (127). ILC2s are crucial for the protection against helminth infection. They are also activated by allergens due to the release of IL-25, IL-33 and TSLP in the tissues, contributing to a number of respiratory diseases such as asthma (128). On the other hand, ILC2s have also been shown to be involved in tissue repair following influenza infection (129). The ILC3 group includes innate immune cells committed to targeting extracellular microbes. They reside mainly in the mucosal tissues and maintain their homeostasis locally. ILC3s express RORγt and produce cytokines such as IL-17A, IL-22, and GM-CSF (118, 123). Lymphoid tissue inducers (LTis) are a subset of ILC3s essential during the fetal stage for supporting the development of lymph nodes and other lymphoid tissues (130).
Innate lymphoid cells are progenies of hematopoietic stem cells, arising from progenitors destined to become lymphoid cells such as lymphoid-primed multipotent progenitors (LMPPs) or common lymphoid progenitors (CLPs) (20, 21). These progenitors reside in fetal liver or adult bone marrow where ILCs differentiate in addition to B cells. These processes have been extensively studied as summarized below. However, LMPPs and CLPs also travel to the thymus to produce T cells. The capability of the thymus to support ILC differentiation has recently become appreciated (90, 131–133). The divergence of T cell development to ILC fates is an interesting issue to be addressed here. Finally, ILCs are also believed to be derived from tissue-resident progenitors but at what stage these progenitors seed the peripheral tissues and whether all ILC subsets utilizes this mechanism of reproduction are not fully understood.
Innate lymphoid cells develop in the bone marrow from LMPPs or CLPs through a series of intermediate progenitors which progressively lose the potential of giving rise to B cells and then NK cells (120). The progenitors that can generate subsequent progenitors for either ILC or NK cells are called alpha LPs (αLPs), which require the NFIL3 and TOX transcription factors (134). Early innate lymphoid progenitors (EILP) characterized by TCF1 expression, also have a similar differentiation potential (135). Next, common helper ILC progenitors (CHILPs) are regulated by Id2 and responsible for the ILC but not NK subsets (136). ILC progenitors (ILCPs) controlled by PLZF are dedicated to only producing ILCs, and are found in both bone marrow and fetal liver (137). In contrast, NK progenitors (NKPs) which also express Id2 are specialized to become NK cells (120, 137). Although CHILPs or ILCPs have the potential to give rise to all three ILC subsets in vitro when cultured on OP9-DL1 stroma, the predominant subset detected in the bone marrow is ILC2 as well as their precursors called ILC2Ps (138). Moreover, there is also evidence that ILC1s can be generated in adult liver from fetal hematopoietic stem cells (139).
Whether the bone marrow serves as a constant source of ILC2 replenishment has not been well established. Experiments using parabionts suggested tissues such as the lung receive few ILC2s from the blood circulation (140, 141). However, recent single cell RNA sequencing (scRNAseq) data showed a population of ILC2s in the blood of wild type and athymic nude mice, which suggest that these ILC2s may come from the bone marrow or they are the recirculating ILC2s from peripheral tissues (133). IL-18R+ precursors of ILC2s have also been found in the lung and shown to arrive from the blood (142, 143).
In humans, ILC progenitors with biases to different ILC subsets are readily detectable in the blood (144, 145). Likewise, committed ILC1 to ILC3 subsets are also found in the blood (122, 146). These cells are assumed to come from the bone marrow but no direct evidence is available. The frequencies of the ILC subsets are often found to be altered in different disease states, which may potentially serve as biomarkers of these diseases (147–149).
Small numbers of ILCs, particularly ILC2s, have been found in the thymus at pre- and post-natal stages (150–154). This is consistent with the fact T cell progenitors express the transcription factors supporting ILC2 differentiation, namely GATA3, TCF1 and Bcl11b (155).
Whether the thymus is another lymphoid organ capable of exporting ILC precursors or ILCs to peripheral tissues was investigated by using scRNAseq of the lineage negative (Lin-) Thy1+ fraction of the blood of wild type and athymic nude mice (133). Bajana et al. found that about half of the ILC-containing Lin-Thy1+ population, was greatly diminished in the athymic nude mice, which suggest that the production of these cells is thymus-dependent, thus designated td-ILCs. These cells were fractionated into four clusters based on their distinct transcriptomic properties. All td-ILCs express genes commonly expressed in ILCs such as Tcf7 and Il7r but they lack the signature transcription factors that specify ILC1 to ILC3: T-bet, GATA3 and RORγt, suggesting that td-ILCs can be ILC precursors. Indeed, when these cells were isolated as Lin-Thy1+CD127+CD62L+ from the blood and cultured on OP9-DL1 stroma, different subsets of ILCs were generated (133). Whether this population contains disparate progenitors for distinct ILC subsets or progenitors with multiple potentials is to be determined.
Interestingly, td-ILCs express Cd3d, Cd3e and Cd3g but no other T cell specific genes such as Cd4, Cd8a, Rag1, Rag2 and Dntt. Flow cytometry analyses detected CD3ϵ by intracellular staining but not by surface staining (133). Moreover, td-ILCs do not have TCRβ or TCRδ either on the surface or in the cytoplasm, thus indicating that they are not T cells. Using intracellular CD3ϵ (icCD3ϵ) as a marker, Bajana et al. also detected icCD3ϵ+ cells in the lung, small intestine and skin of wild type mice (133). Because these icCD3ϵ + cells are greatly diminished in nude mice, the results were interpreted to mean that icCD3ϵ marks thymus-derived cells. Like in blood td-ILCs, the icCD3ϵ+ cells in the lung and small intestine do not express TCRβ or TCRδ, ruling out the possibility that they are T cells. This suggests that td-ILCs in the blood may home to peripheral tissues where they differentiate into diverse ILC subsets. In the lung, a significant fraction of icCD3ϵ+ ILCs are ST2-RORγt+ ILC3-like cells. In contrast, the lamina propria of small intestine harbors icCD3ϵ+KLRG1-T-bet+ ILC1-like cells. Curiously, the expression levels of GATA3 correlated inversely with those of icCD3ϵ, which suggests that ILC2 differentiation is accompanied by the down-regulation of CD3 expression (133). Although this possibility remains to be investigated, the potential down-regulation of CD3 expression makes it difficult to assess the contribution of thymus-derived ILC2s to the overall ILC2 pool. A lineage-tracing system with a Cre transgene that is specifically and efficiently expressed at the early stages of T cell development would greatly facilitate the estimation of the contribution of thymus-derived ILC2s and further validate the thymic origin of ILC2 subsets.
Additional evidence exist that support the notion that the thymus contributes to the ILC2 pools. Qian et al. showed that not only multipotent progenitors (DN1) but also committed T lineage cells (DN3) from the thymus can differentiate into functional ILC2 on OP9-DL1 stromal cells (132). Consistently, ILC2s isolated from the lung of WT but not nude mice harbor rearranged TCR genes, Tcrb and Tcrg, suggesting that at least some of the ILC2s originated from committed T lineage cells in the thymus (132, 156). While Tcrg rearrangement was readily detectable by electrophoresis, analyses of the D-J and V-DJ recombination in the Tcrb locus required Southern blotting because of the diversity of their rearrangement events. Shin et al. sequenced the rearranged Tcrg segments and found a reduced frequency of in-frame rearrangement in ILC2s compared to that in γδ T cells (156). It was thus concluded that ILC2s are derived from cells which have failed productive γδ TCR rearrangement (156, 157). However, further investigation at the single-cell level could strengthen the conclusion. Despite the rearrangement events detected, ILC2s do not express TCRβ or TCRδ either intracellularly or on the surface.
Likewise, NK cells have also been shown to arise from early T cell precursors in the thymus, suggesting a branch point between T and NK cells (158–160). It remains to be determined if this branch point is similar or different from those giving rise to ILCs.
Id2 is expressed in ILC progenitors and plays an essential role in ILC development, which implicates the involvement of E proteins in regulating ILC differentiation (136, 161). Strikingly, down-regulation of E proteins by the ectopic expression of Id1 in transgenic thymocytes at the DN1 stage or by deletion of the E2A and HEB genes with plck-Cre at the DN3 stage led to dramatic increases in ILC2 production in the thymus (131, 132). As a result, large amounts of ILC2s were exported from the thymus to peripheral tissues throughout the body. The thymus was shown to be responsible for the mass production of ILC2 in Id1 transgenic mice because when the transgene was bred onto the nude background, ILC2 expansion was no longer detectable (132). ILC2s made in the thymus of Id1 transgenic and E protein deficient mice respond to IL-25 or IL-33 stimulation similarly as wild type ILC2s by secreting IL-5 and IL-13 in cultures (131, 132). In vivo, Id1 transgenic mice exhibited greater type 2 responses when treated with papain in the lung or during helminth infection (131). These are likely due to the presence of excessive amounts of ILC2s in Id1 transgenic mice. However, on a per cell basis, Id1 transgenic ILC2s appeared to have a less robust production of IL5 and IL-13 (131). It is not clear if this is due to a cell intrinsic difference or a limitation of stimuli available to all of the extra ILC2s in Id1 transgenic mice. Barshad et al. made a similar observation by treating wild type and Id1 transgenic mice with house dust mites (HDM) (162). By analyzing the chromatin accessibility, they found a reduction in AP-1 and C/EBP binding sites in open chromatins after HDM treatment in Id1 transgenic ILC2s. Whether this is due to a direct or indirect effect of E protein inhibition remained to be determined.
In the blood of Tcf3fl/flTcf12fl/flplck-Cre mice, an extremely large population of cells (cluster 0) that belong to thymus-dependent ILC precursors was detected using scRNAseq (133). In addition, a subset (cluster 2) with characteristics of NK cells was also markedly enriched (133). These cells can give rise to different ILC/NK subsets when cultured on OP9-DL1 stroma (133). Together, these results suggest that E proteins play multiple roles in suppressing the production of ILC and NK precursors, which may arise at different developmental stages or from different T cell precursors. Whether E proteins suppress the same or different transcriptional programs governing ILC and NK differentiation remains to be investigated.
Ablating E2A and HEB genes starting at the CLP stage using IL7r-Cre increased the production of both ILC2s and LTi-like cells, a subset of ILC3s (90). Conversely, inducible expression of a gain-of-function mutant of E47 by Rag1-Cre impaired the differentiation of ILC2s from ILCP in the bone marrow (163). Furthermore, Id2-/- mice have been shown to be devoid of NK cells and lymph nodes which are initiated by LTi cells (130). Yet, overexpression of Id3 in human hematopoietic stem cells promoted NK differentiation (164). These findings suggest that down-regulating E protein function is instrumental for NK cell differentiation (165). It was further shown that Id2 plays a key role in regulating the production of IL-15 important for NK homeostasis (166, 167).
Inducible deletion of the E2A and HEB genes promoted ILC2 differentiation from CLP, DN1 and DN3 cells on OP9-DL1 stroma by 20-40 folds, which demonstrates a powerful cell-intrinsic suppression by E proteins (132). It is therefore interesting to elucidate the transcriptional programs that underlie the suppression of ILC2 differentiation. Miyazaki et al. performed RNA sequencing and Assay for Transposase-accessible Chromatin Sequencing (ATAC-seq) using DN1 (ETP) cells of fetal thymi of control and Tcf3fl/flTcf12fl/flIl7r-Cre mice. As expected, they found the down-regulation of an array of genes important for T cell development, which include Notch1, Ptcra, Rag1, Rag2 and Cd3d. On the other hand, genes known to be expressed in ILC progenitors or ILC2s were up-regulated. Examples of such genes are Pdcd1, Il18r, Id2, Gata3, Lmo4, Rora, Tox, Est1, Il4, Il1rl1 and Klrg1. The chromatin accessibility assays also showed a shift from the open chromatin patterns of T cells to those of ILCs. While these findings agree with the phenotypes of E protein deficient mice, it is difficult to pinpoint the critical switches that alter the cell fates.
Likewise, Qian et al. conducted RNA sequencing using DN1 or DN3 cells from control and Tcf3fl/flTcf12fl/flRosa26CreERT2 mice cultured on OP9-DL1 stromal cells (132). On day 4 of the culture, tamoxifen was added to the medium and the cells were collected 24 or 72 hours later. Expression of genes important for T cell development decreased whereas those crucial for ILC2 differentiation increased. Even after one day of E-protein ablation, a collection of genes coding for diverse transcription factors became activated. These include Zbtb16, Gata3, Rora, Rxra, Klf6, Ikzf2 and Irf4. While it is possible that E proteins individually repress the transcription of all of these genes, a coordinated program that controls the transcription of critical factors essential for ILC2 differentiation may be at play.
A close-up look at the action of E proteins was carried out by making use of the E47-ER fusion proteins (112), which allowed instant induction of E protein activity upon addition of tamoxifen (168). ILC2s from the thymus of Id1 transgenic mice were transduced with retroviruses expressing E47-ER or empty control viruses. Transduced cells were isolated by sorting for EGFP expressed off the same retroviral vector. After expansion, these cells were then incubated with tamoxifen for 4 or 16 hours and harvested for RNA sequencing or ATAC-seq. Consistent with the function of E proteins as transcription activators, Peng et al. found more genes activated than repressed by E47-ER at both time points (168). Among them are three genes encoding transcriptional repressors, Cbfa2t3, Jdp2 and Bach2 (169–171).
Interestingly, ATAC-seq data showed that a modest increase in chromatin accessibility 4 hours post induction of E47 was followed by a widespread reduction in open chromatin regions 16 hours later. Moreover, the transcription factor motifs enriched in the differential peaks shifted from those bound by bHLH and Ets1 proteins at 4 hours to those recognized by bZip and GATA factors. It is therefore possible that one of the mechanisms whereby E proteins suppress ILC2 differentiation is to control the expression of transcription repressors, which in turn negatively regulate the transcription of genes important for ILC2 differentiation or function. Although this hypothesis has not been validated through genetic complementation studies, the correlation between the alteration of gene expression in Cbfa2t3-/- and E protein deficient mice support this idea (168, 172). Proteins bound to bZip and GATA motifs such as Batf and GATA3 are also known to be crucial for ILC2 function (126, 173, 174).
The RORα transcription factor also plays an important role in ILC2 differentiation (127). Rora-/- mice lack ILC2s but have intact T cell compartments. Recently, Ferreira et al. showed that RORα promotes ILC2 over T cell development by activating the transcription of Id2 and Nfil3, which in turn inhibit the function of E proteins (153). However, in E protein deficient thymocytes, Rora expression is consistently up-regulated (90, 132, 168). Thus, a positive feedback loop may perpetually cause the up-regulation of Rora expression during ILC2 differentiation. There are likely additional transcription factors which act in parallel or in sequence to coordinate the differentiation of ILC2s and possibly ILC progenitors. However, it is clear that E proteins and their inhibitors, Id proteins, play a central role in maintaining the balance between T cell and ILC development.
The major difference between T cells and innate lymphoid cells is the presence and absence of TCRs on their cell surface, respectively. However, there are a number of common features in the differentiation of these two types of cells (175). The thymic environment is conducive to the maturation of both T cells and ILCs (at least ILC2s and ILC3s) by supporting Notch and IL-7 signaling. The thymic progenitors equipped with transcription factors such as TCF1 and GATA3, are able to differentiate into both T cells and ILCs. Obviously, T cell production is the dominating responsibility of the thymus. This is due to the overwhelming effects of TCR-driven T cell expansion and powerful transcriptional programs in place to ensure an adequate T cell output. One of such transcriptional programs is controlled by the balance between E and Id proteins (Figure 2). When E protein activities are high, T cell development proceeds. When Id proteins overcome E proteins, ILCs can develop. Although Id2 has been shown to be expressed in ILC progenitors and play critical roles in ILC differentiation in the bone marrow, expression of Id3 is stimulated by TCR signaling in both αβ and γδ T cells (73, 106). This would create opportunities for developing T cells to divert to the ILC path. However, this possibility needs to be vigorously investigated. It is also interesting to explore whether the large numbers of developing T cells eliminated during the differentiation processes could be recycled into ILCs and used to replenish ILC pools in peripheral tissues. The E/Id axis has clearly been shown to be gate-keepers in the crossroads to T cell and ILC fates but the downstream transcriptional events remain to be further elucidated as the technologies and critical reagents become available.
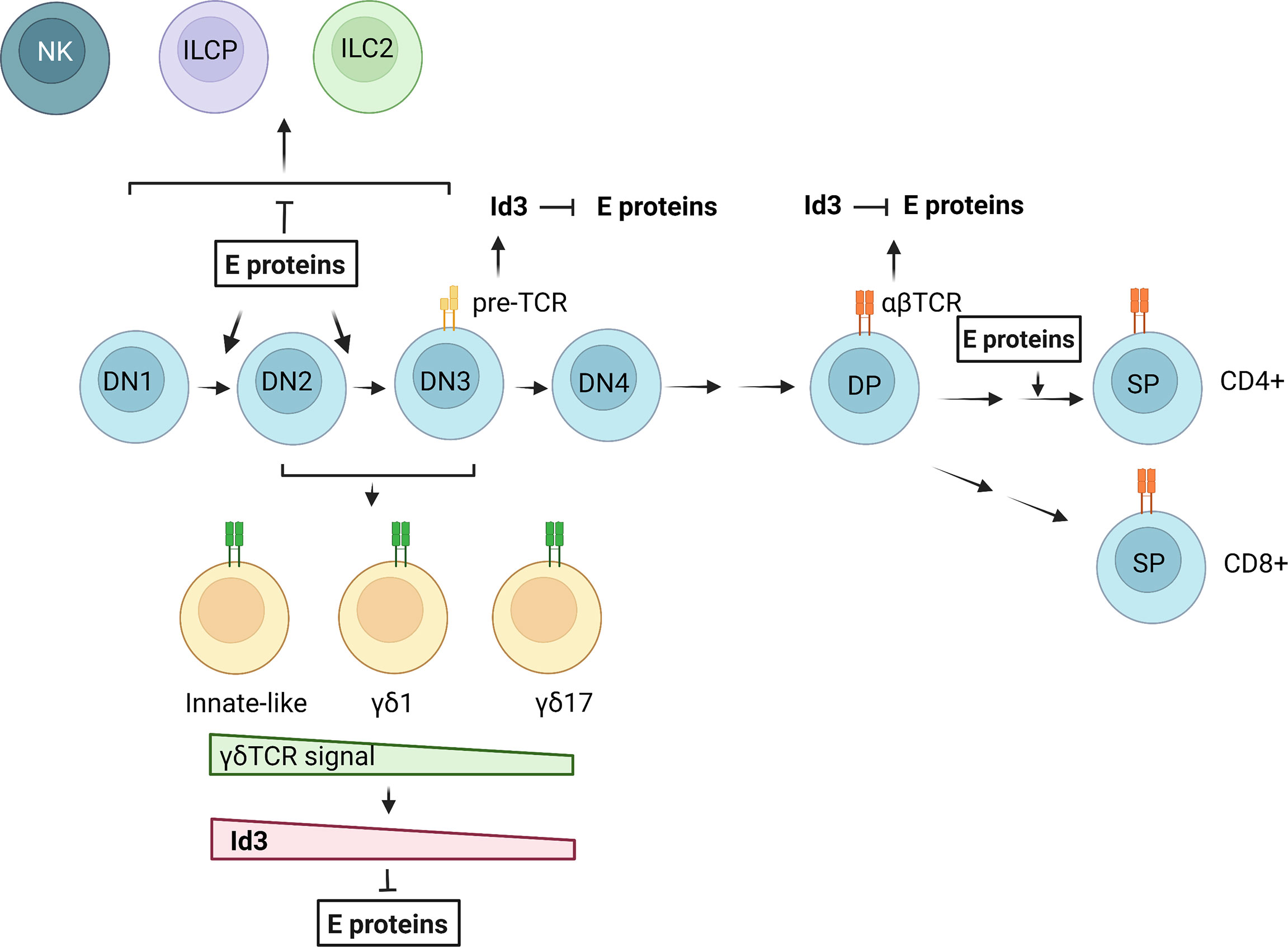
Figure 2 Regulation of T cell and ILC differentiation by E and Id proteins. E proteins promote T cell commitment and differentiation from DN1 to DN3 stages. Pre-TCR and TCR signaling in αβ T cells lead to transient Id3 up-regulation and E protein inhibition. In γδ T cell development, a gradient of γδ TCR signal determines the outcomes of different γδ subsets through regulation of Id3 expression and E protein activities. NKs, ILC2s and ILC precursors (ILCPs) may arise in the DN stages when E protein functions are suppressed. The figure was created by BioRender.com.
AP and X-HS wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work and funds for publication are supported by the National Institute of Allergy and Infectious Diseases (R56A126851).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol (2005) 6(11):1079–86. doi: 10.1038/ni1260
2. Lazorchak A, Jones ME, Zhuang Y. New insights into e-protein function in lymphocyte development. Trends Immunol (2005) 26(6):334–8. doi: 10.1016/j.it.2005.03.011
3. Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobin enhancer binding, daughterless, MyoD and myc proteins. Cell (1989) 56:777–83. doi: 10.1016/0092-8674(89)90682-X
4. Hu JS, Olson EN, Kingston RE. HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding ability of myogenic regulatory factors. Mol Cell Biol (1992) 12(3):1031–42. doi: 10.1128/mcb.12.3.1031-1042.1992
5. Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobin enhancer mE5/kE2 motif. Sci (1990) 247:467–70. doi: 10.1126/science.2105528
6. Aronheim A, Shiran R, Rosen A, Walker MD. The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc Natl Acad Sci USA (1993) 90:8063–7. doi: 10.1073/pnas.90.17.8063
7. Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell (1999) 4(1):63–73. doi: 10.1016/S1097-2765(00)80188-4
8. Quong MW, Massari ME, Zwart R, Murre C. A new transcriptional-activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol (1993) 13:792–800. doi: 10.1128/mcb.13.2.792-800.1993
9. Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell (1989) 58:537–44. doi: 10.1016/0092-8674(89)90434-0
10. Kamps MP, Murre C, Sun X-H, Baltimore D. A new homeobox gene contributes the DNA-binding domain of the t(1:19) translocation protein in pre-b ALL. Cell (1990) 60:547–55. doi: 10.1016/0092-8674(90)90658-2
11. Sun X-H, Baltimore D. An inhibitory domain of E12 prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell (1991) 64:459–70. doi: 10.1016/0092-8674(91)90653-G
12. Wang D, Claus CL, Vaccarelli G, Braunstein M, Schmitt TM, Zuniga-Pflucker JC, et al. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol (2006) 177(1):109–19. doi: 10.4049/jimmunol.177.1.109
13. Yoganathan K, Yan A, Rocha J, Trotman-Grant A, Mohtashami M, Wells L, et al. Regulation of the signal-dependent e protein HEBAlt through a YYY motif is required for progression through T cell development. Front Immunol (2022) 13:848577). doi: 10.3389/fimmu.2022.848577
14. Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein id: A negative regulator of helix-loop-helix DNA binding proteins. Cell (1990) 61:49–59. doi: 10.1016/0092-8674(90)90214-Y
15. Sun X-H, Copeland NG, Jenkins NA, Baltimore D. Id proteins, Id1 and Id2, selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol (1991) 11:5603–11. doi: 10.1128/mcb.11.11.5603-5611.1991
16. Sun XH. Multitasking of helix-loop-helix proteins in lymphopoiesis. Adv Immunol (2004) 84:43–77. doi: 10.1016/S0065-2776(04)84002-1
18. Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D. An id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci USA (1991) 88:1815–9. doi: 10.1073/pnas.88.5.1815
19. Riechmann V, van Cruchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucl Acids Res (1994) 22:749–55. doi: 10.1093/nar/22.5.749
20. Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, et al. Upregulation of Flt3 expression within the bone marrow lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immun (2001) 15(4):659–69. doi: 10.1016/S1074-7613(01)00220-5
21. Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell (1997) 91(5):661–72. doi: 10.1016/S0092-8674(00)80453-5
22. Perry SS, Welner RS, Kouro T, Kincade PW, Sun XH. Primitive lymphoid progenitors in bone marrow with T lineage reconstituting potential. J Immunol (2006) 177(5):2880–7. doi: 10.4049/jimmunol.177.5.2880
23. Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol (2003) 4(2):168–74. doi: 10.1038/ni878
24. Fehling HJ, von Boehmer H. Early alpha beta T cell development in the thymus of normal and genetically altered mice. Curr Opin Immunol (1997) 9(2):263–75. doi: 10.1016/S0952-7915(97)80146-X
25. Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol (1993) 150(10):4244–52.
26. Kisielow P, von Boehmer H. Development and selection of T cells: facts and puzzles. Adv Immunol (1995) 58:87–209. doi: 10.1016/S0065-2776(08)60620-3
27. Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immun (2004) 20(6):735–45. doi: 10.1016/j.immuni.2004.05.004
28. Shortman K, Wilson A, Egerton M, Pearse M, Scollay R. Immature CD4- CD8+ murine thymocytes. Cell Immunol (1988) 113(2):462–79. doi: 10.1016/0008-8749(88)90042-1
29. MacDonald HR, Budd RC, Howe RC. A CD3- subset of CD4-8+ thymocytes: A rapidly cycling intermediate in the generation of CD4+8+ cells. Eur J Immunol (1988) 18(4):519–23. doi: 10.1002/eji.1830180405
30. Prabhu S, Ignatova A, Park ST, Sun X-H. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and ID proteins. Mol Cell Biol (1997) 17(10):5888–96. doi: 10.1128/MCB.17.10.5888
31. Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol (2005) 6(7):663–70. doi: 10.1038/ni1216
32. Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immun (2006) 25(1):105–16. doi: 10.1016/j.immuni.2006.05.010
33. He YW, Nakajima H, Leonard WJ, Adkins B, Malek TR. The common gamma-chain of cytokine receptors regulates intrathymic T cell development at multiple stages. J Immunol (1997) 158(6):2592–9.
34. Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immun (1999) 10(5):547–58. doi: 10.1016/S1074-7613(00)80054-0
35. Haks MC, Oosterwegel MA, Blom B, Spits HM, Kruisbeek AM. Cell-fate decisions in early T cell development: Regulation by cytokine receptors and the pre-TCR. Semin Immunol (1999) 11(1):23–37. doi: 10.1006/smim.1998.0153
36. Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immun (1998) 8(1):11–20. doi: 10.1016/s1074-7613(00)80454-9
37. Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias AM, Chen D, Rothenberg EV. Definition of regulatory network elements for T cell development by perturbation analysis with PU.1 and GATA-3. Dev Biol (2002) 246(1):103–21. doi: 10.1006/dbio.2002.0674
38. Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nat (2011) 476(7358):63–8. doi: 10.1038/nature10279
39. Tydell CC, David-Fung ES, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol (2007) 179(1):421–38. doi: 10.4049/jimmunol.179.1.421
40. Hosokawa H, Romero-Wolf M, Yang Q, Motomura Y, Levanon D, Groner Y, et al. Cell type-specific actions of Bcl11b in early T-lineage and group 2 innate lymphoid cells. J Exp Med (2020) 217(1):e20190972. doi: 10.1084/jem.20190972
41. Mallick CA, Dudley EC, Viney JL, Owen MJ, Hayday AC. Rearrangement and diversity of T cell receptor beta chain genes in thymocytes: A critical role for the beta chain in development. Cell (1993) 73(3):513–9. doi: 10.1016/0092-8674(93)90138-G
42. Hoffman ES, Passoni L, Crompton T, Leu TM, Schatz DG, Koff A, et al. Productive T-cell receptor beta-chain gene rearrangement: Coincident regulation of cell cycle and clonality during development in vivo. Genes Dev (1996) 10(8):948–62. doi: 10.1101/gad.10.8.948
43. Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell (1989) 59(6):1035–48. doi: 10.1016/0092-8674(89)90760-5
44. Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Sci (1990) 248(4962):1517–23. doi: 10.1126/science.2360047
45. von Boehmer H, Aifantis I, Feinberg J, Lechner O, Saint-Ruf C, Walter U, et al. Pleiotropic changes controlled by the pre-t-cell receptor. Curr Opin Immunol (1999) 11(2):135–42. doi: 10.1016/S0952-7915(99)80024-7
46. Born W, Yague J, Palmer E, Kappler J, Marrack P. Rearrangement of T-cell receptor beta-chain genes during T-cell development. Proc Natl Acad Sci U S A (1985) 82(9):2925–9. doi: 10.1073/pnas.82.9.2925
47. Hayday AC, Diamond DJ, Tanigawa G, Heilig JS, Folsom V, Saito H, et al. Unusual organization and diversity of T-cell receptor alpha-chain genes. Nat (1985) 316(6031):828–32. doi: 10.1038/316828a0
48. Yague J, Blackman M, Born W, Marrack P, Kappler J, Palmer E. The structure of V alpha and J alpha segments in the mouse. Nucleic Acids Res (1988) 16(23):11355–64. doi: 10.1093/nar/16.23.11355
49. von Boehmer H. Positive selection of lymphocytes. Cell (1994) 76(2):219–28. doi: 10.1016/0092-8674(94)90330-1
50. McCaughtry TM, Wilken MS, Hogquist KA. Thymic emigration revisited. J Exp Med (2007) 204(11):2513–20. doi: 10.1084/jem.20070601
51. Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nat (2004) 427(6972):355–60. doi: 10.1038/nature02284
52. Allende ML, Dreier JL, Mandala S, Proia RL. Expression of the sphingosine 1-phosphate receptor, S1P1, on T-cells controls thymic emigration. J Biol Chem (2004) 279(15):15396–401. doi: 10.1074/jbc.M314291200
53. Jin R, Wang W, Yao JY, Zhou YB, Qian XP, Zhang J, et al. Characterization of the in vivo dynamics of medullary CD4+CD8- thymocyte development. J Immunol (2008) 180(4):2256–63. doi: 10.4049/jimmunol.180.4.2256
54. Singer A, Bosselut R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: Analysis of the CD4/CD8 lineage decision. Adv Immunol (2004) 83:91–131. doi: 10.1016/S0065-2776(04)83003-7
55. Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol (2002) 2(12):933–44. doi: 10.1038/nri954
56. Xiong N, Raulet DH. Development and selection of gammadelta T cells. Immunol Rev (2007) 215:15–31. doi: 10.1111/j.1600-065X.2006.00478.x
57. Munoz-Ruiz M, Sumaria N, Pennington DJ, Silva-Santos B. Thymic determinants of gammadelta T cell differentiation. Trends Immunol (2017) 38(5):336–44. doi: 10.1016/j.it.2017.01.007
58. Parker ME, Ciofani M. Regulation of gammadelta T cell effector diversification in the thymus. Front Immunol (2020) 11:42. doi: 10.3389/fimmu.2020.00042
59. Kang J, Raulet DH. Events that regulate differentiation of alpha beta TCR+ and gamma delta TCR+ T cells from a common precursor. Semin Immunol (1997) 9(3):171–9. doi: 10.1006/smim.1997.0069
60. Krangel MS, McMurry MT, Hernandez-Munain C, Zhong XP, Carabana J. Accessibility control of T cell receptor gene rearrangement in developing thymocytes. the TCR alpha/delta locus. Immunol Res (2000) 22(2-3):127–35. doi: 10.1385/IR:22:2-3:127
61. Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol (2009) 21(2):133–9. doi: 10.1016/j.coi.2009.03.009
62. Pereira P, Zijlstra M, McMaster J, Loring JM, Jaenisch R, Tonegawa S. Blockade of transgenic gamma delta T cell development in beta 2-microglobulin deficient mice. EMBO J (1992) 11(1):25–31. doi: 10.1002/j.1460-2075.1992.tb05023.x
63. Kreslavsky T, Garbe AI, Krueger A, von Boehmer H. T Cell receptor-instructed alphabeta versus gammadelta lineage commitment revealed by single-cell analysis. J Exp Med (2008) 205(5):1173–86. doi: 10.1084/jem.20072425
64. Ferrick DA, Schrenzel MD, Mulvania T, Hsieh B, Ferlin WG, Lepper H. Differential production of interferon-gamma and interleukin-4 in response to Th1- and Th2-stimulating pathogens by gamma delta T cells in vivo. Nature (1995) 373(6511):255–7. doi: 10.1038/373255a0
65. Jensen KD, Su X, Shin S, Li L, Youssef S, Yamasaki S, et al. Thymic selection determines gammadelta T cell effector fate: Antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immun (2008) 29(1):90–100. doi: 10.1016/j.immuni.2008.04.022
66. Wencker M, Turchinovich G, Di Marco Barros R, Deban L, Jandke A, Cope A, et al. Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nat Immunol (2014) 15(1):80–7. doi: 10.1038/ni.2773
67. Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta lineage fate. Immun (2005) 22(5):583–93. doi: 10.1016/j.immuni.2005.03.014
68. Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, et al. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immun (2005) 22(5):595–606. doi: 10.1016/j.immuni.2005.04.003
69. Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A (2009) 106(30):12453–8. doi: 10.1073/pnas.0903895106
70. Tarakhovsky A, Kanner SB, Hombach J, Ledbetter JA, Muller W, Killeen N, et al. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Sci (1995) 269:535–7. doi: 10.1126/science.7542801
71. Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med (1998) 188(12):2301–11. doi: 10.1084/jem.188.12.2301
72. Fahl SP, Coffey F, Kain L, Zarin P, Dunbrack RL Jr., Teyton L, et al. Role of a selecting ligand in shaping the murine gammadelta-TCR repertoire. Proc Natl Acad Sci U S A (2018) 115(8):1889–94. doi: 10.1073/pnas.1718328115
73. Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, et al. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation notch independent. Immun (2009) 31(4):565–75. doi: 10.1016/j.immuni.2009.07.010
74. Turchinovich G, Hayday AC. Skint-1 identifies a common molecular mechanism for the development of interferon-gamma-secreting versus interleukin-17-secreting gammadelta T cells. Immun (2011) 35(1):59–68. doi: 10.1016/j.immuni.2011.04.018
75. Yin Z, Chen C, Szabo SJ, Glimcher LH, Ray A, Craft J. T-Bet expression and failure of GATA-3 cross-regulation lead to default production of IFN-gamma by gammadelta T cells. J Immunol (2002) 168(4):1566–71. doi: 10.4049/jimmunol.168.4.1566
76. Sumaria N, Grandjean CL, Silva-Santos B, Pennington DJ. Strong TCRgammadelta signaling prohibits thymic development of IL-17A-Secreting gammadelta T cells. Cell Rep (2017) 19(12):2469–76. doi: 10.1016/j.celrep.2017.05.071
77. Barros-Martins J, Schmolka N, Fontinha D, Pires de Miranda M, Simas JP, Brok I, et al. Effector gammadelta T cell differentiation relies on master but not auxiliary Th cell transcription factors. J Immunol (2016) 196(9):3642–52. doi: 10.4049/jimmunol.1501921
78. Coffey F, Lee SY, Buus TB, Lauritsen JP, Wong GW, Joachims ML, et al. The TCR ligand-inducible expression of CD73 marks gammadelta lineage commitment and a metastable intermediate in effector specification. J Exp Med (2014) 211(2):329–43. doi: 10.1084/jem.20131540
79. Garman RD, Doherty PJ, Raulet DH. Diversity, rearrangement, and expression of murine T cell gamma genes. Cell (1986) 45(5):733–42. doi: 10.1016/0092-8674(86)90787-7
80. Xiong N, Kang C, Raulet DH. Positive selection of dendritic epidermal gammadelta T cell precursors in the fetal thymus determines expression of skin-homing receptors. Immun (2004) 21(1):121–31. doi: 10.1016/j.immuni.2004.06.008
81. Havran WL, Chien YH, Allison JP. Recognition of self antigens by skin-derived T cells with invariant gamma delta antigen receptors. Sci (1991) 252(5011):1430–2. doi: 10.1126/science.1828619
82. Narayan K, Sylvia KE, Malhotra N, Yin CC, Martens G, Vallerskog T, et al. Intrathymic programming of effector fates in three molecularly distinct gammadelta T cell subtypes. Nat Immunol (2012) 13(5):511–8. doi: 10.1038/ni.2247
83. Haas JD, Ravens S, Duber S, Sandrock I, Oberdorfer L, Kashani E, et al. Development of interleukin-17-producing gammadelta T cells is restricted to a functional embryonic wave. Immun (2012) 37(1):48–59. doi: 10.1016/j.immuni.2012.06.003
84. Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, et al. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol (1997) 17(8):4782–91. doi: 10.1128/MCB.17.8.4782
85. Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol (1997) 17:7317–27. doi: 10.1128/MCB.17.12.7317
86. Barndt R, Dai MF, Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. J Immunol (1999) 163(6):3331–43.
87. Kim D, Peng XC, Sun XH. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol Cell Biol (1999) 19(12):8240–53. doi: 10.1128/MCB.19.12.8240
88. Wang HC, Perry SS, Sun XH. Id1 attenuates notch signaling and impairs T-cell commitment by elevating Deltex1 expression. Mol Cell Biol (2009) 29(17):4640–52. doi: 10.1128/MCB.00119-09
89. Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol (2007) 178(9):5717–26. doi: 10.4049/jimmunol.178.9.5717
90. Miyazaki M, Miyazaki K, Chen K, Jin Y, Turner J, Moore AJ, et al. The e-id protein axis specifies adaptive lymphoid cell identity and suppresses thymic innate lymphoid cell development. Immun (2017) 46(5):818–34. doi: 10.1016/j.immuni.2017.04.022
91. Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev (2009) 23(14):1665–76. doi: 10.1101/gad.1793709
92. Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annu Rev Immunol (2010) 28:343–65. doi: 10.1146/annurev.immunol.021908.132719
93. Hsu LY, Lauring J, Liang HE, Greenbaum S, Cado D, Zhuang Y, et al. A conserved transcriptional enhancer regulates RAG gene expression in developing b cells. Immun (2003) 19(1):105–17. doi: 10.1016/S1074-7613(03)00181-X
94. Miyazaki K, Watanabe H, Yoshikawa G, Chen K, Hidaka R, Aitani Y, et al. The transcription factor E2A activates multiple enhancers that drive rag expression in developing T and b cells. Sci Immunol (2020) 5(51):eabb1455. doi: 10.1126/sciimmunol.abb1455
95. Bain G, Romanow WJ, Albers K, Havran WL, Murre C. Positive and negative regulation of V(D)J recombination by the E2A proteins. J Exp Med (1999) 189(2):289–300. doi: 10.1084/jem.189.2.289
96. Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol (2000) 20(18):6677–85. doi: 10.1128/MCB.20.18.6677-6685.2000
97. Takeuchi A, Yamasaki S, Takase K, Nakatsu F, Arase H, Onodera M, et al. E2A and HEB activate the pre-TCR alpha promoter during immature T cell development. J Immunol (2001) 167(4):2157–63. doi: 10.4049/jimmunol.167.4.2157
98. Tremblay M, Herblot S, Lecuyer E, Hoang T. Regulation of pT alpha gene expression by a dosage of E2A, HEB, and SCL. J Biol Chem (2003) 278(15):12680–7. doi: 10.1074/jbc.M209870200
99. Emmanuel AO, Arnovitz S, Haghi L, Mathur PS, Mondal S, Quandt J, et al. TCF-1 and HEB cooperate to establish the epigenetic and transcription profiles of CD4(+)CD8(+) thymocytes. Nat Immunol (2018) 19(12):1366–78. doi: 10.1038/s41590-018-0254-4
100. Alberola-Ila J, Hogquist KA, Swan KA, Bevan MJ, Perlmutter RM. Positive and negative selection invoke distinct signaling pathways. J Exp Med (1996) 184(1):9–18. doi: 10.1084/jem.184.1.9
101. Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the ras- ERK MAPK cascade. Nat Immunol (2001) 2(2):165–71. doi: 10.1038/84273
102. Bettini M, Xi H, Milbrandt J, Kersh GJ. Thymocyte development in early growth response gene 1-deficient mice. J Immunol (2002) 169(4):1713–20. doi: 10.4049/jimmunol.169.4.1713
103. Haks MC, Krimpenfort P, Borst J, Kruisbeek AM. The CD3g chain is essential for development of both the TCRab and TCRgd lineages. EMBO J (1998) 17:1871–82. doi: 10.1093/emboj/17.7.1871
104. Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1 deficient mice have no mature b and T lymphocytes. Cell (1992) 68:869–77. doi: 10.1016/0092-8674(92)90030-G
105. Kim D, Xu M, Nie L, Peng XC, Jimi E, Voll RE, et al. Helix-loop-helix proteins regulate pre-TCR and TCR signaling through modulation of Rel/NF-kappaB activities. Immun (2002) 16(1):9–21. doi: 10.1016/S1074-7613(02)00264-9
106. Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med (2001) 194(6):733–45. doi: 10.1084/jem.194.6.733
107. Nie L, Xu M, Vladimirova A, Sun XH. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J (2003) 22(21):5780–92. doi: 10.1093/emboj/cdg567
108. Nie L, Perry SS, Zhao Y, Huang J, Kincade PW, Farrar MA, et al. Regulation of lymphocyte development by cell-type-specific interpretation of notch signals. Mol Cell Biol (2008) 28(6):2078–90. doi: 10.1128/MCB.00844-07
109. Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immun (2000) 12:17–26. doi: 10.1016/S1074-7613(00)80155-7
110. Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavarone A, Zhuang Y. E protein transcription factors are required for the development of CD4(+) lineage T cells. Immun (2012) 36(3):348–61. doi: 10.1016/j.immuni.2012.02.010
111. Qi Z, Sun XH. Hyperresponse to T-cell receptor signaling and apoptosis of Id1 transgenic thymocytes. Mol Cell Biol (2004) 24(17):7313–23. doi: 10.1128/MCB.24.17.7313-7323.2004
112. Yang Y, Liou HC, Sun XH. Id1 potentiates NF-kappaB activation upon T cell receptor signaling. J Biol Chem (2006) 281(46):34989–96. doi: 10.1074/jbc.M608078200
113. Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J Immunol (2009) 182(9):5306–16. doi: 10.4049/jimmunol.0804249
114. Fahl SP, Kappes DJ, Wiest DL. TCR signaling circuits in alphabeta/gammadelta T lineage choice. In: Soboloff J, Kappes DJ, editors. Signaling mechanisms regulating T cell diversity and function. Boca Raton (FL):CRC Press. (2018). p. 85–104.
115. In TSH, Trotman-Grant A, Fahl S, Chen ELY, Zarin P, Moore AJ, et al. HEB is required for the specification of fetal IL-17-producing gammadelta T cells. Nat Commun (2017) 8(1):2004. doi: 10.1038/s41467-017-02225-5
116. Yang Y, Wang HC, Sun XH. Id1 induces apoptosis through inhibition of RORgammat expression. BMC Immunol (2008) 9:20. doi: 10.1186/1471-2172-9-20
117. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol (2013) 13(2):145–9. doi: 10.1038/nri3365
118. Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. innate lymphoid cells: a new paradigm in immunology. Science (2015) 348(6237):aaa6566. doi: 10.1126/science.aaa6566
119. Zook EC, Kee BL. Development of innate lymphoid cells. Nat Immunol (2016) 17(7):775–82. doi: 10.1038/ni.3481
120. Yang Q, Bhandoola A. The development of adult innate lymphoid cells. Curr Opin Immunol (2016) 39:114–20. doi: 10.1016/j.coi.2016.01.006
121. Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang HE, et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol (2018) 19(10):1093–9. doi: 10.1038/s41590-018-0201-4
122. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell (2018) 174(5):1054–66. doi: 10.1016/j.cell.2018.07.017
123. Artis D, Spits H. The biology of innate lymphoid cells. Nat (2015) 517(7534):293–301. doi: 10.1038/nature14189
124. Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immun (2012) 37(4):634–48. doi: 10.1016/j.immuni.2012.06.020
125. Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immun (2014) 40(3):378–88. doi: 10.1016/j.immuni.2014.01.012
126. Zhu J. GATA3 regulates the development and functions of innate lymphoid cell subsets at multiple stages. Front Immunol (2017) 8:1571. doi: 10.3389/fimmu.2017.01571
127. Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, et al. Transcription factor RORalpha is critical for nuocyte development. Nat Immunol (2012) 13(3):229–36. doi: 10.1038/ni.2208
128. Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol (2015) 15(7):415–28. doi: 10.1038/nri3855
129. Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol (2011) 12(11):1045–54. doi: 10.1038/ni.2131
130. Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nat (1999) 397:702–6. doi: 10.1038/17812
131. Wang HC, Qian L, Zhao Y, Mengarelli J, Adrianto I, Montgomery CG, et al. Downregulation of e protein activity augments an ILC2 differentiation program in the thymus. J Immunol (2017) 198(8):3149–56. doi: 10.4049/jimmunol.1602009
132. Qian L, Bajana S, Georgescu C, Peng V, Wang HC, Adrianto I, et al. Suppression of ILC2 differentiation from committed T cell precursors by e protein transcription factors. J Exp Med (2019) 216(4):884–99. doi: 10.1084/jem.20182100
133. Bajana S, Pankow A, Liu K, Michniowska M, Urban JF Jr., Chen WR, et al. Correlation between circulating innate lymphoid cell precursors and thymic function. iSci (2022) 25(2):103732. doi: 10.1016/j.isci.2022.103732
134. Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, et al. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife (2014) 3:e04406. doi: 10.7554/eLife.04406
135. Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol (2015) 16(10):1044–50. doi: 10.1038/ni.3248
136. Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell (2014) 157(2):340–56. doi: 10.1016/j.cell.2014.03.030
137. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nat (2014) 508(7496):397–401. doi: 10.1038/nature13047
138. Seillet C, Mielke LA, Amann-Zalcenstein DB, Su S, Gao J, Almeida FF, et al. Deciphering the innate lymphoid cell transcriptional program. Cell Rep (2016) 17(2):436–47. doi: 10.1016/j.celrep.2016.09.025
139. Bai L, Vienne M, Tang L, Kerdiles Y, Etiennot M, Escaliere B, et al. Liver type 1 innate lymphoid cells develop locally via an interferon-gamma-dependent loop. Science (2021) 371(6536):aba4177. doi: 10.1126/science.aba4177
140. Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Sci (2015) 350(6263):981–5. doi: 10.1126/science.aac9593
141. Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Sci (2018) 359(6371):114–9. doi: 10.1126/science.aam5809
142. Zeis P, Lian M, Fan X, Herman JS, Hernandez DC, Gentek R, et al. In situ maturation and tissue adaptation of type 2 innate lymphoid cell progenitors. Immun (2020) 53(4):775–92. doi: 10.1016/j.immuni.2020.09.002
143. Ghaedi M, Shen ZY, Orangi M, Martinez-Gonzalez I, Wei L, Lu X, et al. Single-cell analysis of RORalpha tracer mouse lung reveals ILC progenitors and effector ILC2 subsets. J Exp Med (2020) 217(3):20182293. doi: 10.1084/jem.20182293
144. Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell (2017) 168(6):1086–100. doi: 10.1016/j.cell.2017.02.021
145. Nagasawa M, Heesters BA, Kradolfer CMA, Krabbendam L, Martinez-Gonzalez I, de Bruijn MJW, et al. KLRG1 and NKp46 discriminate subpopulations of human CD117(+)CRTH2(-) ILCs biased toward ILC2 or ILC3. J Exp Med (2019) 216(8):1762–76. doi: 10.1084/jem.20190490
146. Mjosberg J, Spits H. Human innate lymphoid cells. J Allergy Clin Immunol (2016) 138(5):1265–76. doi: 10.1016/j.jaci.2016.09.009
147. Cosmi L, Liotta F, Maggi L, Annunziato F. Role of type 2 innate lymphoid cells in allergic diseases. Curr Allergy Asthma Rep (2017) 17(10):66. doi: 10.1007/s11882-017-0735-9
148. Ebbo M, Crinier A, Vely F, Vivier E. Innate lymphoid cells: major players in inflammatory diseases. Nat Rev Immunol (2017) 17(11):665–78. doi: 10.1038/nri.2017.86
149. Xiong T, Turner JE. Innate lymphoid cells in autoimmunity and chronic inflammatory diseases. Semin Immunopathol (2018) 40(4):393–406. doi: 10.1007/s00281-018-0670-4
150. Jones R, Cosway EJ, Willis C, White AJ, Jenkinson WE, Fehling HJ, et al. Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur J Immunol (2018) 48(9):1481–91. doi: 10.1002/eji.201847511
151. Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, Maehr R. A single-cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immun (2018) 48(6):1258–70. doi: 10.1016/j.immuni.2018.04.015
152. Elsaid R, Meunier S, Burlen-Defranoux O, Soares-da-Silva F, Perchet T, Iturri L, et al. A wave of bipotent T/ILC-restricted progenitors shapes the embryonic thymus microenvironment in a time-dependent manner. Blood (2021) 137(8):1024–36. doi: 10.1182/blood.2020006779
153. Ferreira ACF, Szeto ACH, Heycock MWD, Clark PA, Walker JA, Crisp A, et al. RORalpha is a critical checkpoint for T cell and ILC2 commitment in the embryonic thymus. Nat Immunol (2021) 22(2):166–78. doi: 10.1038/s41590-020-00833-w
154. Liu C, Gong Y, Zhang H, Yang H, Zeng Y, Bian Z, et al. Delineating spatiotemporal and hierarchical development of human fetal innate lymphoid cells. Cell Res (2021) 31:1106–22. doi: 10.1038/s41422-021-00529-2
155. De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol (2015) 33:607–42. doi: 10.1146/annurev-immunol-032414-112032
156. Shin SB, Lo BC, Ghaedi M, Scott RW, Li Y, Messing M, et al. Abortive gammadeltaTCR rearrangements suggest ILC2s are derived from T-cell precursors. Blood Adv (2020) 4(21):5362–72. doi: 10.1182/bloodadvances.2020002758
157. Shin SB, McNagny KM. ILC-you in the thymus: A fresh look at innate lymphoid cell development. Front Immunol (2021) 12:681110. doi: 10.3389/fimmu.2021.681110
158. Vargas CL, Poursine-Laurent J, Yang L, Yokoyama WM. Development of thymic NK cells from double negative 1 thymocyte precursors. Blood (2011) 118(13):3570–8. doi: 10.1182/blood-2011-06-359679
159. Veinotte LL, Greenwood CP, Mohammadi N, Parachoniak CA, Takei F. Expression of rearranged TCRgamma genes in natural killer cells suggests a minor thymus-dependent pathway of lineage commitment. Blood (2006) 107(7):2673–9. doi: 10.1182/blood-2005-07-2797
160. Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, Richard-Le Goff O, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol (2006) 7(11):1217–24. doi: 10.1038/ni1395
161. Rankin L, Belz GT. Diverse roles of inhibitor of differentiation 2 in adaptive immunity. Clin Dev Immunol (2011) 2011:281569. doi: 10.1155/2011/281569
162. Barshad G, Webb LM, Ting HA, Oyesola OO, Onyekwere OG, Lewis JJ, et al. E-protein inhibition in ILC2 development shapes the function of mature ILC2s during allergic airway inflammation. J Immunol (2022) 208(5):1007–20. doi: 10.4049/jimmunol.2100414
163. Berrett H, Qian L, Roman O, Cordova A, Simmons A, Sun XH, et al. Development of type 2 innate lymphoid cells is selectively inhibited by sustained e protein activity. Immunohorizons (2019) 3(12):593–605. doi: 10.4049/immunohorizons.1900045
164. Heemskerk MH, Blom B, Oda K, Stegmann AP, Bakker AQ, Weijer K, et al. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med (1997) 186:1597–602. doi: 10.1084/jem.186.9.1597
165. Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of e protein activity. J Exp Med (2007) 204(5):1119–30. doi: 10.1084/jem.20061959
166. Schotte R, Dontje W, Nagasawa M, Yasuda Y, Bakker AQ, Spits H, et al. Synergy between IL-15 and Id2 promotes the expansion of human NK progenitor cells, which can be counteracted by the e protein HEB required to drive T cell development. J Immunol (2010) 184(12):6670–9. doi: 10.4049/jimmunol.0901508
167. Delconte RB, Shi W, Sathe P, Ushiki T, Seillet C, Minnich M, et al. The helix-Loop-Helix protein ID2 governs NK cell fate by tuning their sensitivity to interleukin-15. Immun (2016) 44(1):103–15. doi: 10.1016/j.immuni.2015.12.007
168. Peng V, Georgescu C, Bakowska A, Pankow A, Qian L, Wren JD, et al. E proteins orchestrate dynamic transcriptional cascades implicated in the suppression of the differentiation of group 2 innate lymphoid cells. J Biol Chem (2020) 295(44):14866–77. doi: 10.1074/jbc.RA120.013806
169. Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, et al. ETO, a target of t (8,21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol (2001) 21(19):6470–83. doi: 10.1128/MCB.21.19.6470-6483.2001
170. Jin C, Li H, Murata T, Sun K, Horikoshi M, Chiu R, et al. JDP2, a repressor of AP-1, recruits a histone deacetylase 3 complex to inhibit the retinoic acid-induced differentiation of F9 cells. Mol Cell Biol (2002) 22(13):4815–26. doi: 10.1128/MCB.22.13.4815-4826.2002
171. Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, Karasuyama H, et al. Identification of Bach2 as a b-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3' enhancer. EMBO J (1998) 17(19):5734–43. doi: 10.1093/emboj/17.19.5734
172. Chyla BJ, Moreno-Miralles I, Steapleton MA, Thompson MA, Bhaskara S, Engel M, et al. Deletion of Mtg16, a target of t(16;21), alters hematopoietic progenitor cell proliferation and lineage allocation. Mol Cell Biol (2008) 28(20):6234–47. doi: 10.1128/MCB.00404-08
173. Miller MM, Patel PS, Bao K, Danhorn T, O'Connor BP, Reinhardt RL. BATF acts as an essential regulator of IL-25-responsive migratory ILC2 cell fate and function. Sci Immunol (2020) 5(43):aay3994. doi: 10.1126/sciimmunol.aay3994
174. Xu W, Carr T, Ramirez K, McGregor S, Sigvardsson M, Kee BL. E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood (2013) 121(9):1534–42. doi: 10.1182/blood-2012-08-449447
Keywords: E2A, HEB, innate lymphoid cells, Id2, Id3, T cells
Citation: Pankow A and Sun X-H (2022) The divergence between T cell and innate lymphoid cell fates controlled by E and Id proteins. Front. Immunol. 13:960444. doi: 10.3389/fimmu.2022.960444
Received: 02 June 2022; Accepted: 13 July 2022;
Published: 10 August 2022.
Edited by:
Michele Kay Anderson, University of Toronto, CanadaReviewed by:
Juan Carlos Zuniga-Pflucker, University of Toronto, CanadaCopyright © 2022 Pankow and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Hong Sun, eGlhby1ob25nLXN1bkBvbXJmLm9yZw==
†ORCID: Xiao-Hong Sun, orcid.org/0000-0002-7390-7506
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.