
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 04 October 2022
Sec. Immunological Tolerance and Regulation
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.959729
 Kun Li1
Kun Li1 Ziqiang Wang1,2*
Ziqiang Wang1,2*T cell exhaustion caused by continuous antigen stimulation in chronic viral infections and the tumor microenvironment is a major barrier to successful elimination of viruses and tumor cells. Although immune checkpoint inhibitors should reverse T cell exhaustion, shortcomings, such as off-target effects and single targets, limit their application. Therefore, it is important to identify molecular targets in effector T cells that simultaneously regulate the expression of multiple immune checkpoints. Over the past few years, non-coding RNAs, including microRNAs and long non-coding RNAs, have been shown to participate in the immune response against viral infections and tumors. In this review, we focus on the roles and underlying mechanisms of microRNAs and long non-coding RNAs in the regulation of T cell exhaustion during chronic viral infections and tumorigenesis. We hope that this review will stimulate research to provide more precise and effective immunotherapies against viral infections and tumors.
T cell exhaustion was first observed in murine models of chronic lymphocytic choriomeningitis virus (LCMV) infection. Researchers have found that virus-specific CD8+ T cells can persist indefinitely in chronically infected hosts but are unable to develop antiviral effector functions and thus fail to control viral infection (1, 2). Since then, several types of viruses have been shown to induce T cell exhaustion during viral infections, including herpes simplex virus type 1 (HSV-1), human immunodeficiency virus (HIV), hepatitis B virus (HBV), hepatitis C virus (HCV), Epstein–Barr virus, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a recently identified coronavirus that continuously affects millions of people (3). Growing evidence has revealed that exhausted T cells are also widely distributed in the tumor microenvironment because of persistent exposure to tumor antigens (4–6). During chronic viral infections and tumorigenesis, the exhaustion of virus- or tumor-specific CD4+ and CD8+ T cells is characterized by elevated levels of immune checkpoints, such as programmed death-1 (PD-1), programmed death ligand-1 (PD-L1), cytotoxic T-lymphocyte antigen 4 (CTLA4), T-cell immunoglobulin domain and mucin domain 3 (TIM-3), B- and T-lymphocyte attenuator (BTLA), lymphocyte activation gene 3 (LAG3), and cluster of differentiation 244 (CD244, also known as 2B4). These cells have decreased ability to produce proinflammatory cytokines, such as interleukin-2 (IL-2), tumor necrosis factor-α (TNF-α), and interferon-γ (IFN-γ), and exhibit impaired T cell proliferation (7).
A number of studies have demonstrated that abnormally high expression of immune checkpoints is a hallmark of T cell exhaustion (8–10). By binding to the corresponding ligands, immune checkpoints can induce T cell dysfunction through a variety of mechanisms, including metabolic processes, proliferation, cytokine secretion, recruiting immunosuppressive cells, and impairing T cell memory homeostasis (11, 12) (Figure 1). Although many studies have attempted to reverse T cell exhaustion using immune checkpoint inhibitors, the primary or secondary treatment tolerance, general toxicity, and immune-related adverse events observed in most individuals who receive immune checkpoint blockade therapy. These effects significantly limit their clinical use because of their shortcomings, such as off-target effects and single targets (16). Thus, understanding the regulatory mechanism underlying the expression of these immune checkpoints and identifying molecular targets in effector T cells that can simultaneously regulate the expression of multiple immune checkpoints warrant further research.
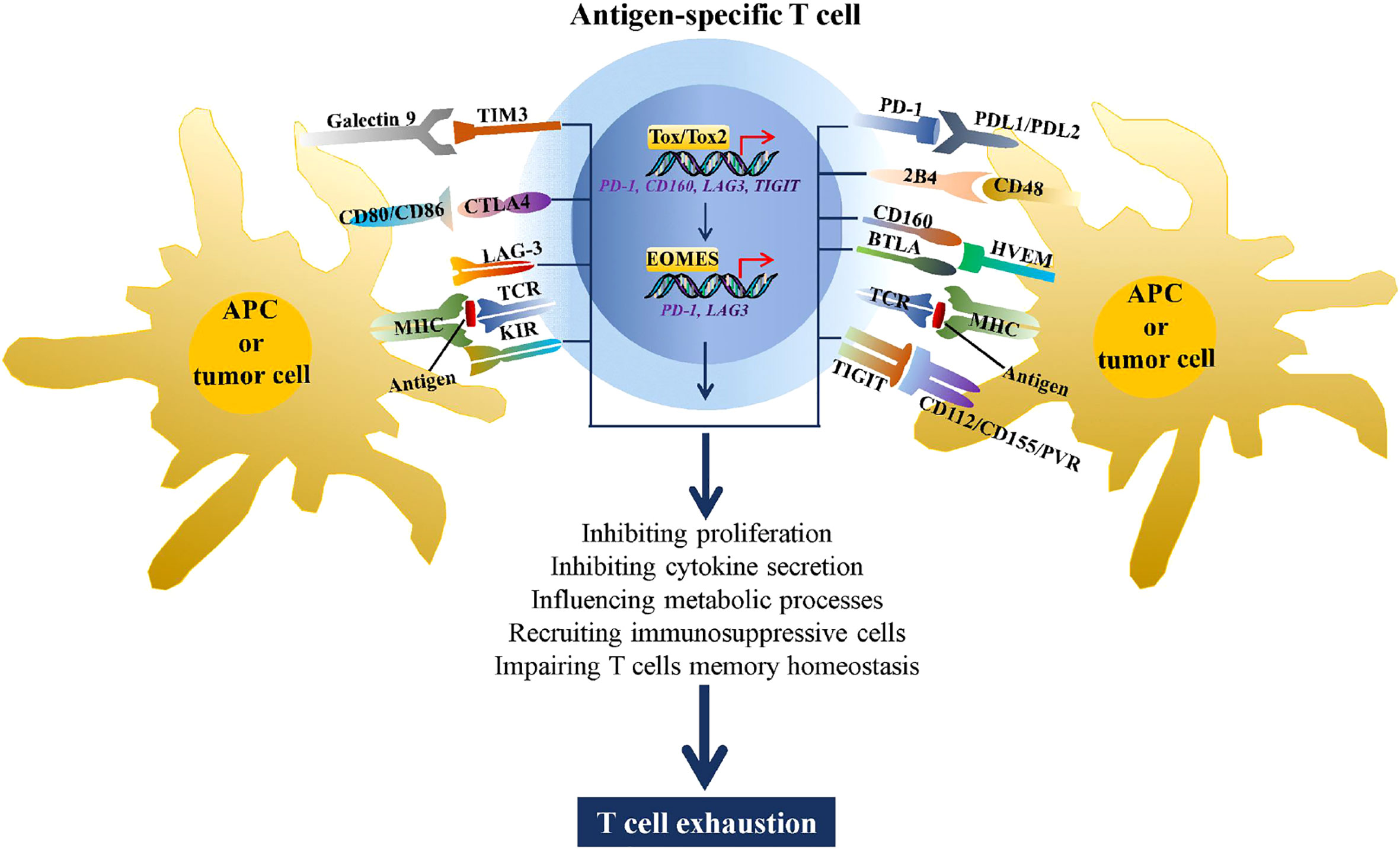
Figure 1 Immune checkpoints increase T cell exhaustion. During chronic viral infections and tumorigenesis, CD4+ and CD8+ T cells recognize antigens presented by MHC complexes on antigen-presenting cells (APCs) or tumor cells through the TCR, stimulating expression of multiple immune checkpoints, such as TIM3, CTLA4 LAG3, killer cell immunoglobulin-like receptor (KIR), PD-1, 2B4, CD160, BTLA, and TIGIT. By binding to the corresponding ligands, these immune checkpoints can drive T cell exhaustion through metabolic processes, inhibiting proliferation and cytokine secretion, recruiting immunosuppressive cells, and impairing T cell memory homeostasis. In addition, the high antigen stimulation of TCR induced expression of TOX/TOX2 and EOMES, two transcriptional factors that are involved in promoting T cell exhaustion by regulating transcription of PD-1, CD160, LAG3, and TIGIT (13–15).
Non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and long ncRNAs (lncRNAs), are RNA molecules that are not translated into protein products. miRNAs are a group of evolutionarily conserved ncRNAs ranging from 19 to 24 nucleotides in length that play significant roles in post-transcriptional gene regulation through mRNA degradation or translational repression (17). lncRNAs are long functional ncRNAs that are more than 200 nucleotides long (18). Several biological processes are known to be controlled by miRNAs and lncRNAs, including cell differentiation, proliferation, ossification, autophagy, and apoptosis (19–24). miRNAs and lncRNAs also play critical regulatory roles in the progression of various disorders, such as cancers, viral infections, and neurodegenerative diseases (25–30). There is growing evidence that miRNAs and lncRNAs act as regulators of immune escape by directly or indirectly modulating the expression of immune-regulating molecules, especially immune checkpoint proteins, such as members of the CD28, B7, TNF, and TNF receptor families (31, 32). Additionally, miRNAs and lncRNAs play important roles in the differentiation, proliferation, and cytokine secretion of CD4+ and CD8+ T cells (33). Many miRNAs and lncRNAs have been demonstrated to be dysregulated following continuous exposure to antigens in viral infections and tumors, and these miRNAs and lncRNAs play crucial roles in the maintenance of T cell functions (34–37), suggesting that miRNAs and lncRNAs have great potential as therapeutic targets for reversing T cell exhaustion.
In this review, we provide an overview of the roles of miRNAs and lncRNAs in T cell exhaustion during chronic viral infections and tumorigenesis and present their potential implications in reversing T cell exhaustion against human cancers and viral infections, with an emphasis on immune checkpoint regulation. We hope that this review will contribute to the development of non-coding RNA-based therapeutic approaches for the treatment of any type of RNA-related disease, such as SARS-CoV-2.
An increasing number of studies have confirmed that the antigenic load during viral infections directly contributes to CD4+ and CD8+ T cell exhaustion, and several ncRNAs have been identified to be involved. In this section, we summarize the roles of ncRNAs in T cell exhaustion during chronic HSV-1, LCMV, and HIV infections (Table 1).
HSV-1 is a human alpha herpesvirus closely related to orofacial, genital herpes, and encephalitis (49). HSV-1 infection is quite prevalent, with 80–90% of the population infected with HSV-1 during their lifetime. After infection, HSV-1 hijacks host cell factors, including miRNAs and lncRNAs, to facilitate viral replication and gene expression (50–53). A growing body of evidence has revealed that the HSV-1 latency-associated transcript (LAT), a viral lncRNA that is abundantly expressed during latent infection and plays a vital role in the HSV-1 latency reactivation cycle, is an important mediator that promotes viral survival and immune evasion by increasing immune checkpoint expression, thus inducing exhaustion of HSV-1 specific T cells. For example, Homayon et al. found that increased latency in the mouse trigeminal ganglia is associated with higher levels of the exhaustion markers PD-1 and TIM-3, suggesting the functional significance of T cell exhaustion in HSV-1 establishing latency. Further investigation revealed that LAT+ viruses contain more CD8+ T cells expressing PD-1 and TIM-3 than those in LAT- viruses and that CD8α+ dendritic cells are involved in LAT-induced expression of PD-1 and TIM-3 (29, 38, 54). In addition, Lbachir et al. reported that HSV-1 LAT inhibited the cytotoxic function of HSV-1-specific CD8+ T cells and decreased IFN-γ and TNF-α production by directly or indirectly upregulating PD-L1 expression in mouse neuroblastoma cells (40). Moreover, they found that LAT interferes with the phenotypic and functional maturation of HSV-1 antigen-positive dendritic cells and upregulates the expression of PD-1, TIM-3, and CTLA-4 in HSV-specific CD8+ T cells (41, 55).
LCMV infection in mice is a well-established model for investigating the mechanism underlying T cell exhaustion induced by chronic viral infection (56). In studies investigating the role and molecular mechanism of miRNAs during T cell activation and differentiation, miR-31 expression was found to be strongly enhanced by activation of the T cell antigen receptor (TCR) in a calcium- and NFAT-dependent manner (42, 57, 58). Further studies confirmed that miR-31 promotes CD8+ T cell exhaustion and inhibits T cell responses to LCMV infection by increasing expression of c-Maf, a transcription factor, and Ptger2, the receptor for prostaglandin E2 (Ptger2), which contributes to CD8+ T cell dysfunction by upregulating several immune checkpoints, such as PD-1, LAG3, and IL-10 (59–62), and downregulating the expression of several T cell effector molecules, including perforin, granzymes, and osteopontin (42).
miR-155 is another miRNA, the expression of which is affected by TCR activation. Romero et al. reported that miR-155 functions as a key molecule for effector CD8+ T cell accumulation and response to LCMV infection by targeting suppressor of cytokine signaling (SOCS-1), thus promoting cytokine responsiveness and accumulation (63). However, Wherry et al. found increased miR-155 expression in exhausted T cells during LCMV infection and that miR-155 promotes the development, proliferation, accumulation, and long-term durability of terminally differentiated and exhausted T cells through the activator protein 1 (AP-1) transcription factor pathway. Moreover, miR-155 reduced the percentage of CD8+ T cells producing IFN-γ and TNF-α and promoted T cell exhaustion by upregulating the expression of immune checkpoints, including PD-1, CD160, and 2B4 (43).
Functional exhaustion of immune cells is also a defining characteristic of HIV-1 chronic infection, and several miRNAs have been identified to be involved in the development of T cell exhaustion observed in HIV-1 infection, including miR-9, miRNA let-7, and miR-146a. Kelleher et al. found that downregulation of miR-9 in progressive HIV-1 infection upregulated the expression of B lymphocyte-induced maturation protein 1 (Blimp-1), a transcriptional repressor of IL-2 expression, and was correlated with PD-1, LAG3, 2B4, IFN-γ, and TNF-α (44, 45, 64). miRNA let-7 is another miRNA that was shown to be significantly downregulated in CD4+ T cells from HIV-1 infected patients, and miRNA let-7 depletion increased the expression of IL-10, an immunosuppressive cytokine that drives the exhaustion of intratumoral CD8+ T cells through a pathway dependent on the transcription factor Blimp-1 (45, 65, 66). miR-146a expression is induced during acute and chronic HIV-1 infections (67, 68). Feng et al. found that chronic HIV-1 infection upregulated the expression of miR-146a and exhaustion markers PD-1 and CTLA-4 following T cell activation, and the depletion of miR-146a improved the antiviral capacity of peripheral blood mononuclear cells from chronic HIV-1 infected patients through the upregulation of antiviral cytokines, such as IFN-γ, IL-2, and TNF-α. Further investigation revealed that miR-146a positively regulates the expression of PD-1 and CTLA-4 (47, 48). These findings suggest that the miR-9/Blimp-1, let-7/IL-10, and miR-146a/PD-1/CTLA-4 axes play important roles in CD4+ T cell dysfunction observed in HIV-1 infection.
In the tumor microenvironment, tumor cells can negatively regulate the functions of tumor-infiltrating T lymphocytes through their association with immunosuppressive cells and the secretion of immunosuppressive cytokines that inhibit tumor-infiltrating T lymphocytes activation by stimulating the expression of multiple inhibitor receptors through binding to cytokine receptors and then triggering signal transducer and activator of transcription (STAT)-dependent signaling pathway (69). In this section, we summarized the roles of ncRNAs in T cell exhaustion during tumorigenesis of melanoma, hepatocellular carcinoma (HCC), breast cancer, nasopharyngeal carcinoma (NPC), gliomas, and lung adenocarcinoma (Table 2).
Melanoma, the most aggressive type of skin cancer, is a common cancer in the western world, increasing in incidence (80), although a rapid decline in death rates has been reported in the United States since the introduction of immune checkpoint inhibitors (81). In a study of the underlying roles and mechanisms of miR-155 in melanoma progression, miR-155 expression was found to be upregulated in effector CD8+ T cells depending on the strength of TCR stimulation and differentiation, and miR-155 knockout promoted tumor growth by enhancing the SOCS-1/STAT5 signaling pathway, thereby impairing cytokine production and resulting in CD8+ T cell exhaustion (63). In addition, miR-155 was found to inhibit T cell senescence and exhaustion to enhance the antitumor response by silencing key transcriptional factors that drive terminal differentiation and exhaustion of T cells via the Ship1/Phf19/Polycomb repressor complex 2 (PRC2) axis (71). Moreover, Li et al. reported that miR-28 was significantly altered in exhausted T cells isolated from a murine melanoma model. Further investigation revealed that miR-28 is involved in T cell exhaustion by directly regulating several immune checkpoints, including PD-1, TIM-3, and BTLA (70).
HCC was the seventh most common cancer and the second leading cause of cancer-related deaths worldwide in 2020 (80). The main risk factors for HCC are chronic HBV or hepatitis C viral infections (82). A study investigating the regulatory network between host factors and T cell exhaustion in HBV-related hepatocellular carcinoma revealed an HBV/pSTAT3/SALL4/miR-200c axis regulated PD-L1 expression and confirmed that miR-200c functions as a therapeutic target to reverse T cell exhaustion during HCC progression (72). In addition, lnc-Tim3 can induce the exhaustion of CD8+ T cells by specifically binding to TIM-3 to upregulate the expression of anti-apoptotic proteins by releasing Bat3 and promoting the activation of p53 and RELA (73).
Breast cancer remains the most diagnosed cancer and the leading cause of cancer-related deaths in women, with an estimated 2.3 million new cases worldwide by 2022 (80). Zhang et al. studied the role of miRNAs in human breast cancer progression and found that the expression of 117 miRNAs was significantly altered in CD8+ PD-1+ and CD8+ PD-1– T cells isolated from 4T1 breast tumor-bearing mice. Among them, miR-149-3p has been shown to promote T cell proliferation, secretion of cytokines IL-2, TNF-α, and IFN-γ, and cytotoxicity of CD8+ T cells against 4T1 breast tumor cells. Further investigation of the molecular mechanism demonstrated that upregulation of miR-149-3p reversed T cell exhaustion by targeting several immune checkpoints, including PD-1, TIM-3, BTLA, and Foxp1 (74). In addition, two downregulated miRNAs, miR-424-5p (75) and miR-138-5p (76), were shown to reduce breast cancer cell viability and restrain T cell exhaustion by upregulating the expression of the effector cytokines IL-2, TNF-α, and IFN-γ, and downregulating the expression of the regulatory cytokine IL-10 by targeting PD-L1.
NPC is a common head and neck cancer of nasopharyngeal epithelium (80). A study to understand the mechanism of T cell development in the tumor microenvironment of NPC found that hypoxia-induced overexpression of miR-24 promoted the transition of activated T cells to exhausted T cells by increasing the expression of CD39, PD-1, and TIM-3. Further investigation revealed that miR-24 reduced ATP production and decreased mitochondrial mass in T cells by binding to and repressing the expression of the proto-oncogene Myc and fibroblast growth factor 11, two essential factors for the reprogramming of mitochondrial dynamics (77).
Gliomas are the most common and aggressive tumors that arise in the central nervous system and are characterized by poor prognosis (83). Researchers have studied the effects of miR-15a/16 on the antitumor immune response in glioma-infiltrating CD8+ T cells and observed that an miR-15a/16 deficiency in an orthotropic GL261 mouse glioma model attenuated cancer progression by inhibiting tumor growth and prolonged mouse survival. Mechanistic investigations revealed that miR-15a/16 deficiency resulted in tumor-infiltrating CD8+ T cells accumulation and promoted the secretion of IFN-γ, IL-2, and TNF-α. Moreover, miR-15a/16 knockout resulted in lower expression of PD-1, TIM-3, and LAG-3 by targeting mTOR, a crucial regulator of T cell functions (78, 84).
Lung cancer remains the leading cause of cancer-related deaths, with an estimated 2.2 million new cases and 1.8 million deaths worldwide in 2020 (80). Lung adenocarcinoma usually originates from the mucosal glands and represents approximately 40% of all lung cancers. In a study that determined the function and mechanism of circular RNAs (circRNAs) in lung adenocarcinoma, has-circRNA-002178 expression was significantly upregulated in lung adenocarcinoma cells and tissues. This high expression of has-circRNA-002178 induced T cell exhaustion by sponging miR-34 to enhance PD-L1 expression in cancer cells. Moreover, cancer cell-derived circRNA-002178 can be delivered into CD8+ T cells via exosomes, resulting in increased PD-1 expression (79).
Viral infections and tumors are two classes of diseases that pose significant threats to humans (80, 85). During viral infections and tumorigenesis, effector CD4+ and CD8+ T cells act as master players of the adaptive immune system to eradicate the virus and tumor cells from the host. However, T cells chronically exposed to viral and tumor antigens gradually lose their ability to control viral replication and tumor cell growth (86). Thus, T cell-based immunotherapy is a promising approach for the treatment of viral infections and tumors. A growing body of evidence suggests that the ncRNA-mediated upregulation of numerous immune checkpoints is involved in virus- and tumor cell-induced T cell exhaustion (Figure 2). Therefore, ncRNAs are of relevant research interest, and their roles in reversing T cell exhaustion should be considered.
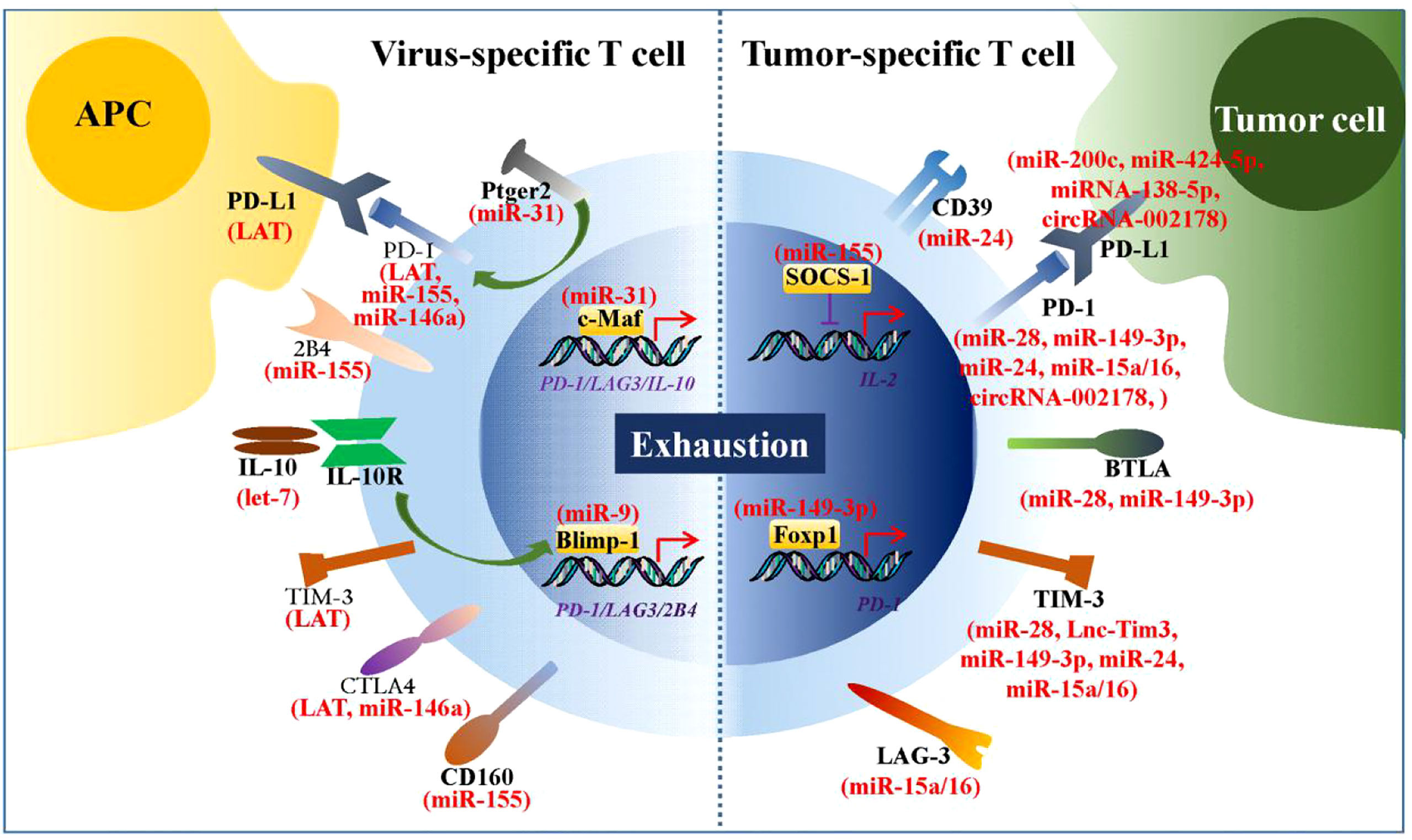
Figure 2 ncRNAs regulate expression of numerous immune checkpoints. During chronic viral infections and tumorigenesis, numbers of ncRNAs are found to be involved in virus- and tumor cell-induced T cell exhaustion by regulating expression of immune checkpoints. These immune checkpoints are mainly inhibitory receptors or the transcriptional factors that regulate their transcription.
To date, 11 RNA-based therapeutics, including small interfering RNAs (siRNAs) and antisense oligonucleotides (ASOs), have been approved by the Food and Drug Administration (FDA) and the European Medicines Agency (EMA) for the treatment of liver, muscle, or central nervous system-related diseases (87). In addition, several miRNA-based therapeutics are in phase II or III clinical development for the treatment of keloids, HCV infection, type II diabetes, nonalcoholic fatty liver disease, and Huntington’s disease, but no lncRNA-based therapeutic approaches have entered clinical development. Key challenges facing miRNA- and lncRNA-based therapeutics are the hurdles of low specificity, specific delivery, and immune responses. Therefore, technical advancements in molecular biology, immunology, pharmacology, chemistry, and nanotechnology are needed to improve specificity, tolerance, and delivery.
In addition, it should be investigated how T cell exhaustion-related ncRNAs develop altered expression, along with whether these ncRNAs interact and coordinate to regulate target genes, especially since they are powerful regulators of gene expression. Further studies are needed to elucidate the role of ncRNAs in the immune microenvironment, such as function of ncRNAs as “messenger RNAs” for communication between T cells and tumor/virus-infected cells. More importantly, several groups have reported that the number of T cell exhaustion markers was increased in CD4+ and CD8+ T cells in patients suffering from SARS-CoV-2 infection (88–90). Thus, more attention should be directed towards how to reverse T cell exhaustion during SARS-CoV-2 infection and whether ncRNAs can act as useful therapeutic targets for the treatment.
Overall, this review summarizes and discusses the roles of ncRNAs in T cell exhaustion during chronic viral infections and tumorigenesis and indicates that ncRNAs may be potent targets for enhancing T cell-based immunotherapy against chronic viral infections and tumors.
KL and ZW prepared the manuscript. ZW reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (32000878), Shandong Provincial Natural Science Foundation (ZR2020LZL008, ZR2021LSW017), and the Academic Promotion Programme of Shandong First Medical University (2019LJ001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
LCMV, Lymphocytic choriomeningitis virus; HSV-1, Herpes simplex virus type 1; HIV, Human immunodeficiency virus; HBV, Hepatitis B virus; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; PD-1, Programmed death-1; PD-L1, Programmed death ligand-1; CTLA4, Cytotoxic T-lymphocyte antigen 4; TIM-3, T-cell immunoglobulin domain and mucin domain 3; BTLA, B- and T-lymphocyte attenuator; LAG3, Lymphocyte activation gene 3; CD244, Cluster of differentiation 244; IL-2, Interlekin-2; TNF-α, Tumor necrosis factor-α; IFN-γ, Interferon-γ; NcRNAs, Non-coding RNAs; MiRNAs, MicroRNAs; LncRNAs, Long non-coding RNAs; LAT, Latency-associated transcript; TCR, T cell antigen receptor; Ptger2, Prostaglandin E2; SOCS-1, Suppressor of cytokine signaling; Blimp-1, B lymphocyte-induced maturation protein 1; HCC, Hepatocellular carcinoma; NPC, Nasopharyngeal carcinoma; PRC2, Polycomb repressor complex 2; CircRNAs, Circular RNAs.
1. Gallimore A, Glithero A, Godkin A, Tissot AC, Plückthun A, Elliott T, et al. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J Exp Med (1998) 187(9):1383–93. doi: 10.1084/jem.187.9.1383
2. Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med (1998) 188(12):2205–13. doi: 10.1084/jem.188.12.2205
3. Mishra KP, Singh M, Saraswat D, Ganju L, Varshney R. Dysfunctional state of T cells or exhaustion during chronic viral infections and COVID-19: A review. Viral Immunol (2022) 35(4):284–90. doi: 10.1089/vim.2022.0002
4. Thommen DS, Schumacher TN. T Cell dysfunction in cancer. Cancer Cell (2018) 33(4):547–62. doi: 10.1016/j.ccell.2018.03.012
5. Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol (2016) 16(10):599–611. doi: 10.1038/nri.2016.80
6. Dolina JS, Van Braeckel-Budimir N, Thomas GD, Salek-Ardakani S. CD8+ T cell exhaustion in cancer. Front Immunol (2021) 12:715234. doi: 10.3389/fimmu.2021.715234
8. Budimir N, Thomas GD, Dolina JS, Salek-Ardakani S. Reversing T-cell exhaustion in cancer: Lessons learned from PD-1/PD-L1 immune checkpoint blockade. Cancer Immunol Res (2022) 10(2):146–53. doi: 10.1158/2326-6066.CIR-21-0515
9. ElTanbouly MA, Noelle RJ. Rethinking peripheral T cell tolerance: checkpoints across a T cell’s journey. Nat Rev Immunol (2021) 21(4):257–67. doi: 10.1038/s41577-020-00454-2
10. Wang Z, Li K, Chen W, Wang X, Huang Y, Wang W, et al. Modulation of SRSF2 expression reverses the exhaustion of TILs via the epigenetic regulation of immune checkpoint molecules. Cell Mol Life Sci (2020) 77(17):3441–52. doi: 10.1007/s00018-019-03362-4
11. Okoye IS, Houghton M, Tyrrell L, Barakat K, Elahi S. Coinhibitory receptor expression and immune checkpoint blockade: Maintaining a balance in CD8+ T cell responses to chronic viral infections and cancer. Front Immunol (2017) 8:1215. doi: 10.3389/fimmu.2017.01215
12. Jiang Y, Li Y, Zhu B. T-Cell exhaustion in the tumor microenvironment. Cell Death Dis (2015) 6(6):e1792. doi: 10.1038/cddis.2015.162
13. Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. (2019) 571(7764):265–9. doi: 10.1038/s41586-019-1326-9
14. Llaó-Cid L, Roessner PM, Chapaprieta V, Öztürk S, Roider T, Bordas M, et al. EOMES is essential for antitumor activity of CD8+ T cells in chronic lymphocytic leukemia. Leukemia. (2021) 35(11):3152–62. doi: 10.1038/s41375-021-01198-1
15. Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. (2019) 571(7764):211–8. doi: 10.1038/s41586-019-1325-x
16. Cousin S, Italiano A. Molecular pathways: Immune checkpoint antibodies and their toxicities. Clin Cancer Res (2016) 22(18):4550–5. doi: 10.1158/1078-0432.CCR-15-2569
18. Wang Z, Li K, Huang W. Long non-coding RNA NEAT1-centric gene regulation. Cell Mol Life Sci (2020) 77(19):3769–79. doi: 10.1007/s00018-020-03503-0
19. Wang Z, Zhang S, Li K. LncRNA NEAT1 induces autophagy through epigenetic regulation of autophagy-related gene expression in neuroglial cells. J Cell Physiol (2022) 237(1):824–32. doi: 10.1002/jcp.30556
20. Kundu M, Basu J. The role of microRNAs and long non-coding RNAs in the regulation of the immune response to mycobacterium tuberculosis infection. Front Immunol (2021) 12:687962. doi: 10.3389/fimmu.2021.687962
21. Wang Z, Lu Y, Zhang X, Ren X, Wang Y, Li Z, et al. Serum microRNA is a promising biomarker for osteogenesis imperfecta. Intractable Rare Dis Res (2012) 1 (2):81–5. doi: 10.5582/irdr.2012.v1.2.81
22. Bhat AA, Younes SN, Raza SS, Zarif L, Nisar S, Ahmed I, et al. Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. Mol Cancer. (2020) 19(1):57. doi: 10.1186/s12943-020-01175-9
23. Wang ZQ, Lu YQ, Han JX. MicroRNAs: important mediators of ossification. Chin Med J (Engl). (2012) 125(22):4111–6.
24. Wang Z, Lu Y, Han J. Peripheral blood microRNAs: A novel tool for diagnosing disease? Intractable Rare Dis Res (2012) 1(3):98–102. doi: 10.5582/irdr.2012.v1.3.98
25. Li K, Yao T, Zhang Y, Li W, Wang Z. NEAT1 as a competing endogenous RNA in tumorigenesis of various cancers: Role, mechanism and therapeutic potential. Int J Biol Sci (2021) 17(13):3428–40. doi: 10.7150/ijbs.62728
26. Yang T, Wang Y, Liao W, Zhang S, Wang S, Xu N, et al. Down-regulation of EPB41L4A-AS1 mediated the brain aging and neurodegenerative diseases via damaging synthesis of NAD+ and ATP. Cell Biosci (2021) 11(1):192. doi: 10.1186/s13578-021-00705-2
27. Wang Z, Fan P, Zhao Y, Zhang S, Lu J, Xie W, et al. NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell Mol Life Sci (2017) 74(6):1117–31. doi: 10.1007/s00018-016-2398-4
28. Wang Z, Zhao Y, Zhang Y. Viral lncRNA: A regulatory molecule for controlling virus life cycle. Noncoding RNA Res (2017) 2(1):38–44. doi: 10.1016/j.ncrna.2017.03.002
29. Wang Z, Zhao Y, Xu N, Zhang S, Wang S, Mao Y, et al. NEAT1 regulates neuroglial cell mediating aβ clearance via the epigenetic regulation of endocytosis-related genes expression. Cell Mol Life Sci (2019) 76(15):3005–18. doi: 10.1007/s00018-019-03074-9
30. Zhao Y, Wang Z, Mao Y, Li B, Zhu Y, Zhang S, et al. NEAT1 regulates microtubule stabilization via FZD3/GSK3β/P-tau pathway in SH-SY5Y cells and APP/PS1 mice. Aging (Albany NY). (2020) 12(22):23233–50. doi: 10.18632/aging.104098
31. Zhang G, Li N, Li Z, Zhu Q, Li F, Yang C, et al. microRNA-4717 differentially interacts with its polymorphic target in the PD1 3’ untranslated region: A mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget. (2015) 6(22):18933–44. doi: 10.18632/oncotarget.3662
32. Wei J, Nduom EK, Kong LY, Hashimoto Y, Xu S, Gabrusiewicz K, et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol (2016) 18(5):639–48. doi: 10.1093/neuonc/nov292
33. Dooley J, Linterman MA, Liston A. MicroRNA regulation of T-cell development. Immunol Rev (2013) 253(1):53–64. doi: 10.1111/imr.12049
34. Wang X, Shen H, He Q, Tian W, Xia A, Lu XJ. Exosomes derived from exhausted CD8+ T cells impaired the anticancer function of normal CD8+ T cells. J Med Genet (2019) 56(1):29–31. doi: 10.1136/jmedgenet-2018-105439
35. Abdolmohammadi Vahid S, Ghaebi M, Ahmadi M, Nouri M, Danaei S, Aghebati-Maleki L, et al. Altered T-cell subpopulations in recurrent pregnancy loss patients with cellular immune abnormalities. J Cell Physiol (2019) 234(4):4924–33. doi: 10.1002/jcp.27290
36. Möhnle P, Hirschberger S, Hinske LC, Briegel J, Hübner M, Weis S, et al. MicroRNAs 143 and 150 in whole blood enable detection of T-cell immunoparalysis in sepsis. Mol Med (2018) 24(1):54.
37. Nguyen LNT, Nguyen LN, Zhao J, Schank M, Dang X, Cao D, et al. Long non-coding RNA GAS5 regulates T cell functions via miR21-mediated signaling in people living with HIV. Front Immunol (2021) 12:601298. doi: 10.3389/fimmu.2021.601298
38. Allen SJ, Hamrah P, Gate D, Mott KR, Mantopoulos D, Zheng L, et al. The role of LAT in increased CD8+ T cell exhaustion in trigeminal ganglia of mice latently infected with herpes simplex virus 1. J Virol (2011) 85(9):4184–97. doi: 10.1128/JVI.02290-10
39. Mott KR, Allen SJ, Zandian M, Ghiasi H. Coregulatory interactions among CD8α dendritic cells, the latency-associated transcript, and programmed death 1 contribute to higher levels of herpes simplex virus 1 latency. J Virol (2014) 88(12):6599–610. doi: 10.1128/JVI.00590-14
40. Chentoufi AA, Kritzer E, Tran MV, Dasgupta G, Lim CH, Yu DC, et al. The herpes simplex virus 1 latency-associated transcript promotes functional exhaustion of virus-specific CD8+ T cells in latently infected trigeminal ganglia: a novel immune evasion mechanism. J Virol (2011) 85(17):9127–38. doi: 10.1128/JVI.00587-11
41. Srivastava R, Dervillez X, Khan AA, Chentoufi AA, Chilukuri S, Shukr N, et al. The herpes simplex virus latency-associated transcript gene is associated with a broader repertoire of virus-specific exhausted CD8+ T cells retained within the trigeminal ganglia of latently infected HLA transgenic rabbits. J Virol (2016) 90(8):3913–28. doi: 10.1128/JVI.02450-15
42. Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. (2007) 27(4):670–84. doi: 10.1016/j.immuni.2007.09.006
43. Stelekati E, Chen Z, Manne S, Kurachi M, Ali MA, Lewy K, et al. Long-term persistence of exhausted CD8 T cells in chronic infection is regulated by MicroRNA-155. Cell Rep (2018) 23(7):2142–56. doi: 10.1016/j.celrep.2018.04.038
44. Seddiki N, Phetsouphanh C, Swaminathan S, Xu Y, Rao S, Li J, et al. The microRNA-9/B-lymphocyte-induced maturation protein-1/IL-2 axis is differentially regulated in progressive HIV infection. Eur J Immunol (2013) 43(2):510–20. doi: 10.1002/eji.201242695
45. Thiele S, Wittmann J, Jäck HM, Pahl A. miR-9 enhances IL-2 production in activated human CD4(+) T cells by repressing blimp-1. Eur J Immunol (2012) 42(8):2100–8. doi: 10.1002/eji.201142203
46. Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, et al. Differential regulation of the let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol (2012) 188(12):6238–46. doi: 10.4049/jimmunol.1101196
47. Teng Y, Luo M, Yu T, Chen L, Huang Q, Chen S, et al. CRISPR/Cas9-mediated deletion of miR-146a enhances antiviral response in HIV-1 infected cells. Genes Immun (2019) 20(4):327–37. doi: 10.1038/s41435-018-0036-x
48. Yu T, Ju Z, Luo M, Hu R, Teng Y, Xie L, et al. Elevated expression of miR-146a correlates with high levels of immune cell exhaustion markers and suppresses cellular immune function in chronic HIV-1-infected patients. Sci Rep (2019) 9(1):18829. doi: 10.1038/s41598-019-55100-2
49. St Leger AJ, Koelle DM, Kinchington PR, Verjans GMGM. Local immune control of latent herpes simplex virus type 1 in ganglia of mice and man. Front Immunol (2021) 12:723809. doi: 10.3389/fimmu.2021.723809
50. Wang Z, Liu Q, Lu J, Fan P, Xie W, Qiu W, et al. Serine/Arginine-rich splicing factor 2 modulates herpes simplex virus type 1 replication via regulating viral gene transcriptional activity and pre-mRNA splicing. J Biol Chem (2016) 291(51):26377–87. doi: 10.1074/jbc.M116.753046
51. Wang Z, Li K, Wang X, Huang W. MiR-155-5p modulates HSV-1 replication via the epigenetic regulation of SRSF2 gene expression. Epigenetics. (2019) 14(5):494–503. doi: 10.1080/15592294.2019.1600388
52. Li K, Wang Z. Splicing factor SRSF2-centric gene regulation. Int J Biol Sci (2021) 17(7):1708–15. doi: 10.7150/ijbs.58888
53. Li K, Wang Z. Speckles and paraspeckles coordinate to regulate HSV-1 genes transcription. Commun Biol (2021) 4(1):1207. doi: 10.1038/s42003-021-02742-6
54. Mott KR, Bresee CJ, Allen SJ, BenMohamed L, Wechsler SL, Ghiasi H. Level of herpes simplex virus type 1 latency correlates with severity of corneal scarring and exhaustion of CD8+ T cells in trigeminal ganglia of latently infected mice. J Virol (2009) 83(5):2246–54. doi: 10.1128/JVI.02234-08
55. Chentoufi AA, Dervillez X, Dasgupta G, Nguyen C, Kabbara KW, Jiang X, et al. The herpes simplex virus type 1 latency-associated transcript inhibits phenotypic and functional maturation of dendritic cells. Viral Immunol (2012) 25(3):204–15. doi: 10.1089/vim.2011.0091
56. Moskophidis D, Lechner F, Pircher H, Zinkernagel RM. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. (1993) 362(6422):758–61. doi: 10.1038/362758a0
57. Zhang L, Ke F, Liu Z, Bai J, Liu J, Yan S, et al. MicroRNA-31 negatively regulates peripherally derived regulatory T-cell generation by repressing retinoic acid-inducible protein 3. Nat Commun (2015) 6:7639. doi: 10.1038/ncomms8639
58. Xue F, Li H, Zhang J, Lu J, Xia Y, Xia Q. miR-31 regulates interleukin 2 and kinase suppressor of ras 2 during T cell activation. Genes Immun (2013) 14(2):127–31. doi: 10.1038/gene.2012.58
59. Chen JH, Perry CJ, Tsui YC, Staron MM, Parish IA, Dominguez CX, et al. Prostaglandin E2 and programmed cell death 1 signaling coordinately impair CTL function and survival during chronic viral infection. Nat Med (2015) 21(4):327–34. doi: 10.1038/nm.3831
60. Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. (2016) 166(6):1500–1511.e9. doi: 10.1016/j.cell.2016.08.052
61. Giordano M, Henin C, Maurizio J, Imbratta C, Bourdely P, Buferne M, et al. Molecular profiling of CD8 T cells in autochthonous melanoma identifies maf as driver of exhaustion. EMBO J (2015) 34(15):2042–58. doi: 10.15252/embj.201490786
62. Moffett HF, Cartwright ANR, Kim HJ, Godec J, Pyrdol J, Äijö T, et al. The microRNA miR-31 inhibits CD8+ T cell function in chronic viral infection. Nat Immunol (2017) 18(7):791–9. doi: 10.1038/ni.3755
63. Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. (2013) 38(4):742–53. doi: 10.1016/j.immuni.2012.12.006
64. Hwang S, Cobb DA, Bhadra R, Youngblood B, Khan IA. Blimp-1-mediated CD4 T cell exhaustion causes CD8 T cell dysfunction during chronic toxoplasmosis. J Exp Med (2016) 213(9):1799–818. doi: 10.1084/jem.20151995
65. Sawant DV, Yano H, Chikina M, Zhang Q, Liao M, Liu C, et al. Adaptive plasticity of IL-10+ and IL-35+ treg cells cooperatively promotes tumor T cell exhaustion. Nat Immunol (2019) 20(6):724–35. doi: 10.1038/s41590-019-0346-9
66. Blackburn SD, Wherry EJ. IL-10, T cell exhaustion and viral persistence. Trends Microbiol (2007) 15(4):143–6. doi: 10.1016/j.tim.2007.02.006
67. Duskova K, Nagilla P, Le HS, Iyer P, Thalamuthu A, Martinson J, et al. MicroRNA regulation and its effects on cellular transcriptome in human immunodeficiency virus-1 (HIV-1) infected individuals with distinct viral load and CD4 cell counts. BMC Infect Dis (2013) 13:250. doi: 10.1186/1471-2334-13-250
68. Rom S, Rom I, Passiatore G, Pacifici M, Radhakrishnan S, Del Valle L, et al. CCL8/MCP-2 is a target for mir-146a in HIV-1-infected human microglial cells. FASEB J (2010) 24(7):2292–300. doi: 10.1096/fj.09-143503
69. Curdy N, Lanvin O, Laurent C, Fournié JJ, Franchini DM. Regulatory mechanisms of inhibitory immune checkpoint receptors expression. Trends Cell Biol (2019) 29(10):777–90. doi: 10.1016/j.tcb.2019.07.002
70. Li Q, Johnston N, Zheng X, Wang H, Zhang X, Gao D, et al. miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget. (2016) 7(33):53735–50. doi: 10.18632/oncotarget.10731
71. Ji Y, Fioravanti J, Zhu W, Wang H, Wu T, Hu J, et al. miR-155 harnesses Phf19 to potentiate cancer immunotherapy through epigenetic reprogramming of CD8+ T cell fate. Nat Commun (2019) 10(1):2157. doi: 10.1038/s41467-019-09882-8
72. Sun C, Lan P, Han Q, Huang M, Zhang Z, Xu G, et al. Oncofetal gene SALL4 reactivation by hepatitis b virus counteracts miR-200c in PD-L1-induced T cell exhaustion. Nat Commun (2018) 9(1):1241. doi: 10.1038/s41467-018-03584-3
73. Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, et al. Long non-coding RNA lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis (2018) 9(5):478. doi: 10.1038/s41419-018-0528-7
74. Zhang M, Gao D, Shi Y, Wang Y, Joshi R, Yu Q, et al. miR-149-3p reverses CD8+ T-cell exhaustion by reducing inhibitory receptors and promoting cytokine secretion in breast cancer cells. Open Biol (2019) 9(10):190061. doi: 10.1098/rsob.190061
75. Dastmalchi N, Hosseinpourfeizi MA, Khojasteh SMB, Baradaran B, Safaralizadeh R. Tumor suppressive activity of miR-424-5p in breast cancer cells through targeting PD-L1 and modulating PTEN/PI3K/AKT/mTOR signaling pathway. Life Sci (2020) 259:118239. doi: 10.1016/j.lfs.2020.118239
76. Rasoolnezhad M, Safaralizadeh R, Hosseinpourfeizi MA, Banan-Khojasteh SM, Baradaran B. MiRNA-138-5p: A strong tumor suppressor targeting PD-L-1 inhibits proliferation and motility of breast cancer cells and induces apoptosis. Eur J Pharmacol (2021) 896:173933. doi: 10.1016/j.ejphar.2021.173933
77. Liu YN, Yang JF, Huang DJ, Ni HH, Zhang CX, Zhang L, et al. Hypoxia induces mitochondrial defect that promotes T cell exhaustion in tumor microenvironment through MYC-regulated pathways. Front Immunol (2020) 11:1906. doi: 10.3389/fimmu.2020.01906
78. Yang J, Liu R, Deng Y, Qian J, Lu Z, Wang Y, et al. MiR-15a/16 deficiency enhances anti-tumor immunity of glioma-infiltrating CD8+ T cells through targeting mTOR. Int J Cancer. (2017) 141(10):2082–92. doi: 10.1002/ijc.30912
79. Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis (2020) 11(1):32. doi: 10.1038/s41419-020-2230-9
80. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
81. Berk-Krauss J, Stein JA, Weber J, Polsky D, Geller AC. New systematic therapies and trends in cutaneous melanoma deaths among US whites, 1986-2016. Am J Public Health (2020) 110(5):731–3. doi: 10.2105/AJPH.2020.305567
82. Thomas London W, Petrick JL, McGlynn KA. Liver cancer. In: Thun M, Linet MS, Cerhan JR, Haiman CA, Schottenfeld D, editors. Cancer epidemiology and prevention, 4th ed. New York: Oxford University Press (2018). p. 635–60.
83. Tan AC, Ashley DM, López GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin (2020) 70(4):299–312. doi: 10.3322/caac.21613
84. Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol (2012) 12(5):325–38. doi: 10.1038/nri3198
85. Alam W, Manzoor F, Farnaz N, Aktar B, Rashid SF. Perception and attitudes towards COVID-19 vaccination among urban slum dwellers in Dhaka, Bangladesh. Lancet Glob Health (2022) 10 Suppl 1:S3. doi: 10.1016/S2214-109X(22)00132-2
86. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol (2015) 15(8):486–99. doi: 10.1038/nri3862
87. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discovery (2021) 20(8):629–51. doi: 10.1038/s41573-021-00219-z
88. Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med (2020) 26(7):1070–6. doi: 10.1038/s41591-020-0944-y
89. Zheng HY, Zhang M, Yang CX, Zhang N, Wang XC, Yang XP, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol (2020) 17(5):541–3. doi: 10.1038/s41423-020-0401-3
Keywords: non-coding RNA, miRNA, lncRNA, T cell exhaustion, immune checkpoint, chronic viral infections, tumorigenesis
Citation: Li K and Wang Z (2022) Non-coding RNAs: Key players in T cell exhaustion. Front. Immunol. 13:959729. doi: 10.3389/fimmu.2022.959729
Received: 02 June 2022; Accepted: 16 September 2022;
Published: 04 October 2022.
Edited by:
Murugaiyan Gopal, Harvard Medical School, United StatesReviewed by:
Qing Rex Lyu, Medical Research Center, Chongqing General Hospital, ChinaCopyright © 2022 Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ziqiang Wang, d2FuZ3ppcWlhbmdAc2RmbXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.