
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 11 August 2022
Sec. Inflammation
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.954744
 Yu-Yao He
Yu-Yao He Hai-Feng Zhou
Hai-Feng Zhou Lu Chen
Lu Chen Yan-Ting Wang
Yan-Ting Wang Wan-Li Xie
Wan-Li Xie Zhen-Zhen Xu
Zhen-Zhen Xu Yue Xiong
Yue Xiong Yi-Qi Feng
Yi-Qi Feng Guo-Yang Liu
Guo-Yang Liu Xia Li
Xia Li Jie Liu
Jie Liu Qing-Ping Wu*
Qing-Ping Wu*Fra-1(Fos-related antigen1), a member of transcription factor activator protein (AP-1), plays an important role in cell proliferation, apoptosis, differentiation, inflammation, oncogenesis and tumor metastasis. Accumulating evidence suggest that the malignancy and invasive ability of tumors can be significantly changed by directly targeting Fra-1. Besides, the effects of Fra-1 are gradually revealed in immune and inflammatory settings, such as arthritis, pneumonia, psoriasis and cardiovascular disease. These regulatory mechanisms that orchestrate immune and non-immune cells underlie Fra-1 as a potential therapeutic target for a variety of human diseases. In this review, we focus on the current knowledge of Fra-1 in immune system, highlighting its unique importance in regulating tissue homeostasis. In addition, we also discuss the possible critical intervention strategy in diseases, which also outline future research and development avenues.
In 1988, Cohen DR and Curran T screened rat c-DNA library with FOS DNA probes, and isolated a new cDNA very similar to FOS, which was named FOS Like 1(FOSL1) (1). By the same method, Matsui et al. confirmed in 1990 that FOSL1 existed in human cells, which was 90% similar to that in rats (2). Fra-1, encoded by FOSL1, is one of the members of the FOS family in activator protein(AP-1). The AP-1 complex results from dimerization between members of JUN(c-Jun, JunB, JunD), FOS(c-Fos, FosB, Fra-1, and Fra-2), ATF(Activating Transcription Factor)(ATF2, ATF3/LRF1, B-ATF), and MAF(musculoaponeurotic fibrosarcoma)(c-MAF, MAFA, MAFB, MAFG/F/K, NRL) (3, 4). A large number of studies have shown that the contribution of AP-1 to determining cell fate depends mainly on the relative abundance of AP-1 subunits, dimer composition, quality of stimulation, cell type and cell environment. As a transcription factor, AP-1 controls genes encoding key cellular regulators. Its members can be quickly assembled into dimers by phosphorylation or methylation activation, then bind to the relevant sites of the target genes, leading to the promotion or inhibition of gene expression. At the cellular level, it involves controlling proliferation, differentiation, apoptosis and responding to environmental signals; at the biological level, it plays a vital role in organogenesis, immune response and cognitive function (5–9). The diversity of AP-1 functions makes it very difficult to be studied, and hinders the answers to some basic questions about it. Therefore, a clear understanding of the mechanism of AP-1 members is of great help to further study AP-1.
The immune system plays a key role in most diseases. It is very important to find the key factors regulating immune responses for the pathogenesis and development of various diseases. Several studies have proved the importance of AP-1 signal in immune cell reactions such as macrophages. Meanwhile, the promoter of many cytokines contains AP-1 binding sites, but the specific mechanism is still in the exploratory stage, especially Fra-1. Studies on Fra-1 are mostly focused on the proliferation, apoptosis, differentiation and transformation of cancer cells. The mechanism of its function in bone and tumor biology has been basically established (10, 11), while the properties of immune cell proliferation, activation and differentiation are rarely studied. With the development of technology, the inherent idea that Fra-1 mostly binds to the promoter of target gene to regulate the expression of the target gene has also been questioned (12, 13). Like AP-1, the function of Fra-1 is also affected by many factors. Existing studies have described Fra-1 as a positive regulator of gene transcription involved in innate immunity (14), as well as an inhibitor of gene expression in relevant contexts (15, 16). This may relate to the differences of transcription complex which is consist of Fra-1, as well as the different environment that the cells in or the different stimulations it received, also depending on the type of cells and their initial concentration in the cells. For example, in the case of Th17 cells, Fra-1 has the opposite effect on differentiation in mouse and human cells (17, 18). In addition, in different disease models Fra-1 regulates IL-6 secretion differently even in the same cells (19–21). Interestingly, the role of Fra-1 is related to its cellular localization. Fra-1 trapped in the cytoplasm inhibits Type I interferon responses to Malaria and Viral infections (22), also might be a driving force for IL-8 overexpression (23). Therefore, a comprehensive identification of the role of Fra-1 in immune processes may help to delineate new ways to ameliorate immune-related diseases such as infections, cancer, and cardiovascular diseases.
The earliest and most studied Fos family protein is c-Fos, part of the reason may be that c-Fos knockout does not cause mice to die, while fetuses lacking Fra-1 suffered severe growth retardation due to obstructed placental angiogenesis, resulting in mouse death between E10.0 and E10.5 (24). The importance of Fra-1 in embryonic development made it difficult to study, but the subsequent emergence of specific knockout and “omics” technology made the function of Fra-1 gradually clear. Here, we discuss the structural characteristics and expression regulation mechanism of FOSL1. In addition, we reviewed and summarized some studies on the role of Fra-1 in innate and adaptive immune regulation, as well as its influence on the occurrence and development of some related diseases.
FOSL1 was initially found to be highly expressed in many cancer cells and was defined as a proto-oncogene, which was located at locus 11q13.1 and encoded a mature mRNA with a length of 1.7kb (Figure 1A). FOSL1 encodes a 271 amino acid protein Fra-1 with a relative molecular mass of 29000, and the genome structure consists of four exons and three introns (4, 25). Fra-1 is one of the members of FOS subfamily in the nuclear transcription factor AP-1 family (3), shares the common structural characteristics of AP-1. Like other members of AP-1, Fra-1 is also known as the basic Leucine zipper (bZIP) protein, where the leucine zipper motif (LZ) is used for dimerization and the basic region (BR) is used for binding to specific DNA motifs (Figure 1B). This domain is highly conserved in the AP-1 family (3, 4), and the bZIP domain of Fra-1 is located at 107-161 amino acid sites. The Leucine zipper is composed of extended amino acids, with one leucine occurring every 6 amino acids. The dimer formed by the interaction between Fra-1 and JUN family, forms the “zipper” (Figure 1C) (26). The two subunits constituting the dimer form a continuous α helix, the carboxy-terminal region forms asymmetric helical coils, and the amino terminal region performs base-specific binding to DNA in the main groove (27). Because the amino acid composition in bZIP of FOS family members is slightly different from that of JUN family members, it cannot form homologous dimer with their own members like JUN family, but can only combine with JUN family members to form heterodimers and play a role (3). Moreover, Fra-1 can also heterodimers with other ubiquitous bZIP transcription factors (see Table 1) (13). In addition, related studies have found that since there is a wide electrostatic interaction network between subunits in the αhelix, Fos : Jun heterodimer has stronger affinity to DNA than Jun : Jun homodimer, and it is more stable and shows stronger transcriptional stimulation activity (38, 39). The dimer composed of Fra-1 and Jun preferentially binds to the DNA motif known as the TPA reaction element (TRE; also called AP-1 motif), it can also bind to cAMP-responsive element (CRE), although the latter has a slightly lower affinity than the former. The consensus sequences for TRE/AP-1 and CRE are 5′-TGA(C/G) TCA and 5′- TCACGTCA motifs, respectively (Figure 1C) (3, 26). In addition to the bZIP structure, FOS family members display a second highly homologous at the C-terminal, that is, the region where one of the two instability factors of resides. The C-terminal destabilizing domain (DEST) of Fra-1 protein containing 30-40 residues is necessary for its transformation activity and one of the reasons for its intracellular instability (Figure 1B) (40, 41).
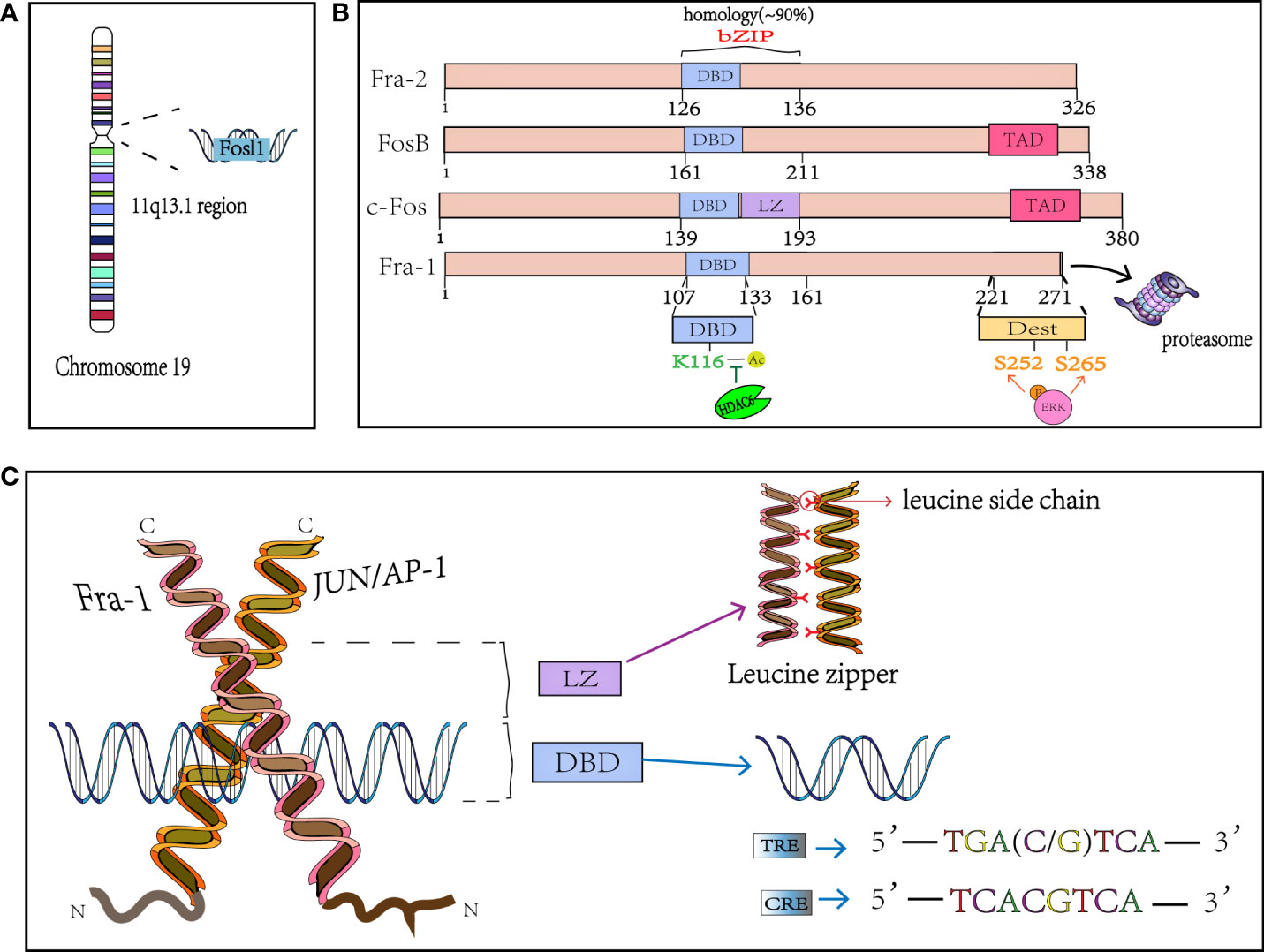
Figure 1 The structure of Fra-1. (A) FOSL1 is on chromosome 19 and located in the 11q13.1. (B) Schematical presentation of the structure of FOS mambers. bZIP, basic Leucine zipper region for dimerisation and DNA-binding and the FOS family has 90% homology in this region; The phosphation of Fra-1 at side S252 and S265 inhibits the COOH-terminal destabilizer increasing Fra-1 half-time and the K116 deacetylation positively controls DNA binding and transactivation. DBD, DNA binding domain; LZ, leucine zipper; TAD, C-terminal transactivating domain. DEST, destabilizer element; HDAC6, Histone deacetylase 6. (C) X-ray structure of the bZIP dimmer AP-1 bound to DNA; Fra-1 and Jun preferentially binds to the DNA motif known as the TRE and CRE. TRE, TPA reaction element; CRE, cAMP-responsive element.
Although these proteins belong to Fos family and share similar structure, the function and expression pattern actually not exactly the same. Skin sections show that Fra-1 is present in all layers other basal layer, and Fra-2 is detected in all layers with an increased level in the upper spinous layer. c-fos is present in the nuclei of the upper spinous and granular layer cells (42). This suggests that c-fos family proteins may exert different function in different types of cells in tissues. Moreover, Fra-1, but not Fra-2, orchestrates the inflammatory state of macrophages by regulating the expression of Arg1 and therefore impedes the resolution of inflammation (15). Another study demonstrated that Fra-1, Fra-2 and c-Fos promote the migration and invasion process of mammary carcinomas with different molecular target (43). In addition, it’s reported that c-Fos and Fra-1 have functional equivalence during vertebrate evolution, and Fra-1 rescues c-Fos-dependent functions such as bone development (44). However, c-Fos cannot totally replace Fra-1 because of the embryonic lethality in Fra-1-defecient mice. These findings show that the interaction and regulatory function among c-fos family proteins are very complicated and highly coordinated, and a detailed understanding of how these proteins determines its propensity to regulate cellular biological events remains further investigations.
Compared with other members of the FOS family, FOSL1 has very similar gene structures both in coding region and noncoding region, especially the size of the third exon and the protein encoded are completely the same (25). Although members of Fos family share similar structural features, there are also some differences. Firstly, in the mouse genome, individual genes of FOS family are not closely located. FOSL1 is on chromosome 19, c-Fos is on chromosome 12, and FosB is located on chromosome 7. In particular, the N-terminal and C-terminal of c-Fos and FosB have a transcriptional activation domain (TAD), while Fra-1 and Fra-2 are not found. At the beginning of discovery, due to the lack of transactivation domain, the entire Fra-1 protein fused with the DNA binding domain of Gal4 shows a lack of any transcriptional activation function (45), so it was once thought to be a transcription suppressor (39, 46, 47). But it was subsequently as a phosphorylation-dependent transcriptional activator (4). In many tumors, these non-transformed FOS proteins, especially Fra-1 and Fra-2, are involved in the progression of many tumors types (4). Thus, the structural characteristics of Fra-1 and the diversity and specificity of the combined dimers lay the foundation for its complex and diverse functions.
Due to the important and diverse functions of FOS family members, precise and complex expression regulation is necessary to avoid the pathological effects of organisms. FOS and JUN are the main existing forms of AP-1 in mammals. The induced expression of ATF and MAF family proteins is more tissue/cell specific than FOS and JUN proteins (3). The expression level of Fra-1 was low in most tissues, but it was found to be quite high in testis and brain (25). In cells, AP-1 family members, including Fra-1, belong to immediate early cell genes (48). When fibroblasts are stimulated by serum, c-Fos and FosB are rapidly and transiently induced, Fra-1 and Fra-2 expressions are delayed but more stable, possibly through the activation of Fra-1 and Fra-2 promoters by Jun/Fos dimer (25, 49, 50). After stimulation, the expression of Fra-1 increased 12 times and lasted longer than that at rest (25). The results showed that Fra-1 was mainly localized in nucleus during indirect immunofluorescence assay (51). The expression characteristics of FOS family proteins may be the basis of their participation in the transition and asynchronous growth of G0-G1 cells (50)
The expression of Fra-1 involves the modification of several histones and recruitment of related proteins, and its transcriptional initiation is started after phosphorylation of S10 on H3 (Figure 2) (52). After receiving stimulation such as serum or growth factor, Myc binds to the corresponding DNA-binding domain and subsequently recruits Mitogen- and stress-activated kinases 1(MSK1/2) or proviral integration site for Moloney murine leukemia virus 1(PIM1) kinase to dephosphorylate H3S10 (52, 53), while TPA activates the extracellular regulated protein kinases(ERK) pathway to mediate Elk1 phosphorylation and TCF-SRE activation, Thus recruiting Aurora Kinase B (AURKB) to phosphorylate the site (54). Phosphorylation at H3S10 forces the 14-3-3 protein to be recruited to chromatin to recognize H3S10ph, which in turn recruits MOF to chromatin, leading to acetylation of H4K16 and H3K9. The P-TEFb/BRD4(positive-elongation factor/Bromodomain Containing 4) complex then recruits to the acetylation site, and catalyzes the phosphorylation of S2 in the carboxyl terminal domain of RNA pol-II(polymerase II), and finally leads to the release of stalled RNSD, resulting in transcriptional extension of FOSL1 (52, 54). In addition, Signal transducers and activators of transcription 3(Stat3) can be phosphorylated and acetylated in response to IL-6 stimulation and then bind to the promoter of FOSL1 to promote its transactivation (55). What’s more, FOSL1 has AP-1 site that allows it to be regulated by Fra-1 itself and other AP-1 family members (25, 56). Studies have found that tumor suppressor gene P53 can recruit to the first promoter region of FOSL1 to positively regulates its transcription, and FOSL1 is the only member of the FOS and JUN family with functional P53 binding sites (57–60).
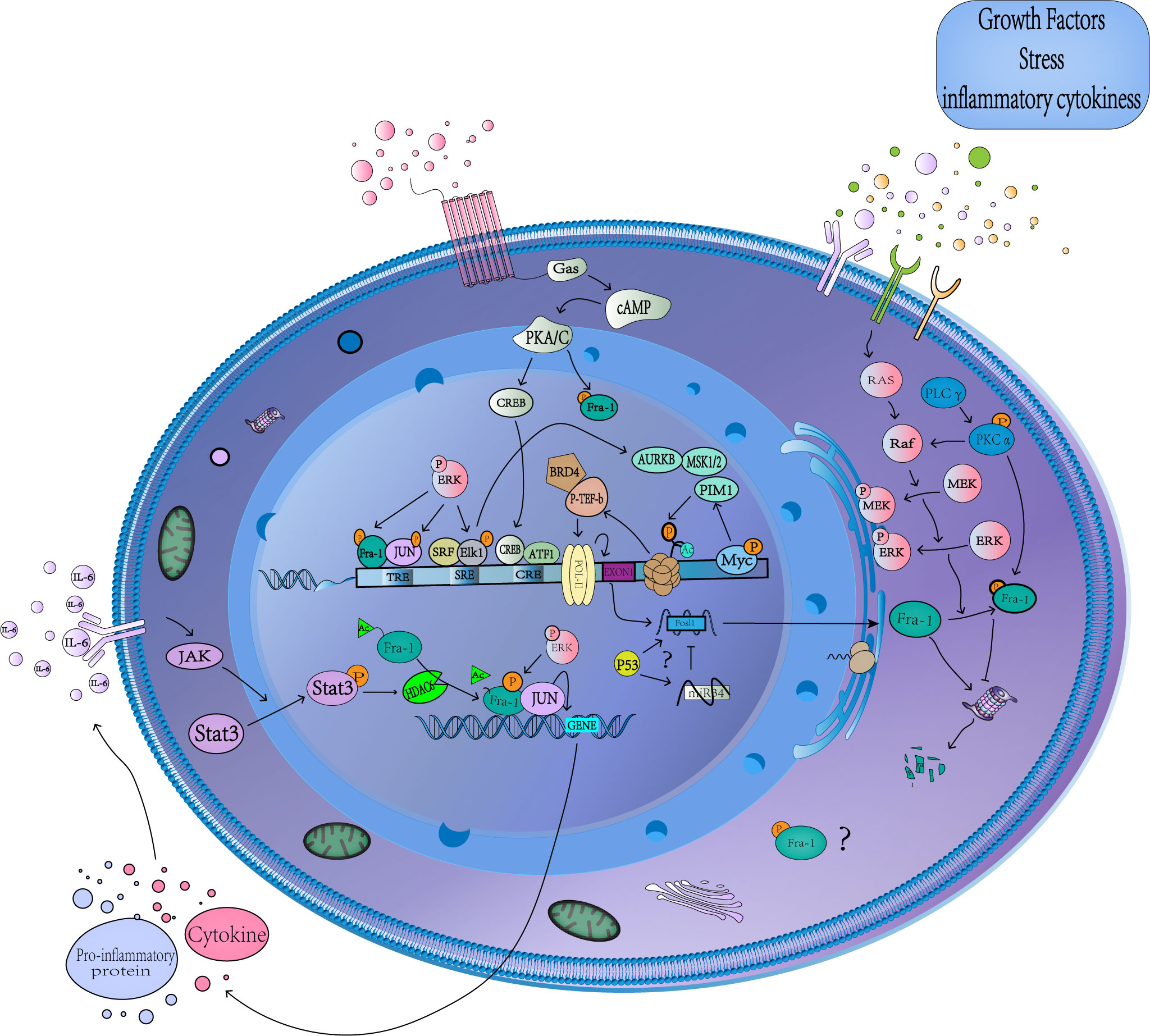
Figure 2 Transcriptional and posttranslational regulation of Fra-1. A schematic representation showing the multiple cellular pathways that converge to regulate Fra-1 function. Stimulation by various factors leads to MAPKs/cAMP/JAK-Stat3 activation, which in turn activates several TFs, causing the expression and phosphorylation of Fra-1, whose production regulation Fra-1 target gene expression. The unphosphorylated Fra-1 in the cytoplasm is easily degraded by proteasome. The question mark represents the unknow role of phosphorylated Fra-1 in the cytoplasm and the sepcific relationship between Fra-1, P53 and miR34. cAMP, Cyclic adenosine monophosphate; CRE, cAMP-responsive element; TRE, TPA reaction element; SRE, serum response element; PKA/C, protein kinase A/C; CREB, cAMP-response element binding protein; MSK1/2, Mitogen- and stress-activated kinases 1 and 2; ERK, extracellular regulated protein kinases; AURKB, Aurora Kinase B; P-TEFb, positive-elongation factor; BRD4, Bromodomain Containing 4; POL-II, polymerase II; ATF1, Activating Transcription Factor 1; Stat3, Signal transducers and activators of transcription 3; MAPK, mitogen-activated protein kinase; HDAC6, Histone Deacetylase 6; IL-6, interleukin-6; SRF, Serum response factor; JAK, janus kinase; RAS, rat sarcoma; Raf, Raf protein kinase; PLC, Phospholipase C.
Fra-1 is essentially an unstable protein, and the regulation of its stability may be the basis of its accumulation (61). Regulation of Fra-1 occurs at multiple levels, not only at the transcription and translation levels, but also under the influence of post-translational modification (such as phosphorylation, ubiquitination, etc.). Many proteins are known to be damaged through covalently linking ubiquitin chains to lysine residues at the n-terminal of the protein, which leads the protein to the proteasome, but there is a class of proteins that do not require prior ubiquitination to degrade (62–64). As mentioned above, Fra-1 is one of these non-ubiquitin-dependent proteasomal degrading proteins, which can be degraded even after all lysine residues are replaced by arginine or its N-terminal is blocked by fusion with Myc epitopes (51, 65).
Numerous studies have found that several kinases can phosphorylate Fra-1 in vitro, including cAMP-dependent kinase (PKA), protein kinase C (PKC), cyclin-dependent kinase 1-cdc2(cdc2) and mitogen-activated protein kinase (MAPK) (8, 41, 66–68). Fra-1 is the target of continuous activation of MAPK cascade, which can increase the transcription of FOSL1 (68), enhance the DNA binding activity of Fra-1 (66), and affect the transcription activity of Fra-1 after being activated (41). The activation of ERK has the greatest impact on Fra-1 content, and there is a linear relationship between the activity of ERK and the Fra-1 protein production rate in the same cell (60). ERK-based activity induces transcription of Fra-1, and its strongly increased activity is required for proteasome-dependent degradation of Fra-1 (20, 69, 70). When two serine residues S252 and S265 at C-terminal of Fra-1 protein are phosphorylated by ERK1/2, the DEST region at C-terminal of Fra-1 is inactivated, thus promoting the stability and accumulation of Fra-1, and the phosphorylation at both sites shows accumulation effect (51, 71). Therefore, under the stimulation of mitogen or in thyroid, colon and breast cancer cells, the high level of Fra-1 expression can be detected, where ERK pathway is highly active due to activation of upstream signal effector factors (51). Fra-1 can also be phosphorylated by protein kinase C (PKC) θ (S265, T223, T230, partially at T217 and T227 residues) and PKCα to prolong the half-time of Fra-1, its Synergistic phosphorylation with ERK-mediated phosphorylation stabilizes Fra-1 from degradation (28, 72, 73). The acetylation at the K116 site in the Fra-1 bZIP region inhibits the activity of Fra-1 and the binding ability to c-Jun independently of the protein stability mechanism. The Histone Deacetylase 6 (HDAC6)-mediated deacetylation at the Fra-1-K116 site can increase its DNA binding and transcriptional activity (74).
Studies have shown that downregulation of many tumor suppressor miRNAs leads to the accumulation of Fra-1 in cells. For example, miR34 can directly target Fra-1 to downregulate its expression, and P53 regulates Fra-1 expression through miR34 dependent manner (75, 76). However, some studies have found that P53 positively regulates the transcription of FOSL1 (57–60), and the specific mechanism remains to be explored. Several members of other tumor suppressor miRNA families (such as miR15/16) also target Fra-1 and affect the disease progression by regulating Fra-1 expression (see Figure 2) (77–79).
The research of Fra-1 was focused on tumors and bone development. Generally, except for cervical cancer (80) and some controversial tumors (81–86), Fra-1 is highly expressed in most tumors and promotes malignant progression of them, control the proliferation and invasion of thyroid (87), bladder (88), lung and pancreatic cells (89). In mice, overexpression of Fra-1 leads to osteosclerosis, and conditional knockout leads to reduced bone mass (90). With the development of technology, there is increasing evidence that AP-1 controls the transcription of multiple inflammation-related genes, such as IL-6, IL-1β, and TNF-α, thereby regulating the inflammation immune response (15, 91, 92).
Macrophages are cellular components that exist in all tissues and bodies under homeostasis physiological conditions (93–95), which can change their phenotypes in response to many different stimuli. In the early 1990s, two distinct phenotypes of macrophage were described: one called classically activated (or inflammatory) macrophage (M1) and the other called vicariously activated (or wound-healing) macrophage (M2) (96, 97). Macrophages play a dual role in arthritis, myocardial infarction, or other inflammatory diseases, initiating and dispelling inflammation, and a mechanism is needed to program the macrophage phenotype (98). The role of the key transcription factor (TF) family in defining macrophage identity and controlling its function by inducing and maintain specific transcription processes has been fully established (99). Meanwhile, GO analysis of differentially expression genes between WT and Fra-1 deficient macrophage highlighted the important role of Fra-1 in macrophage (15).
First, Fra-1 regulates the secretion of inflammatory cytokines in macrophages (Figure 3). In LPS-induced lung injury mice, Fra-1 is mainly expressed in alveolar macrophages (100), and LPS-induced transcriptional activation of Fra-1 is mediated by NF-kB and ERK1/2 signal transduction. Fra-1 selectively up-regulates LPS-induced NF-kB dependent pro-inflammatory cytokines (IL-1β, MIP-1A) and inhibits the anti-inflammatory response of alveolar macrophage (decreasing IL-10 expression) (20). Fra-1 can also affect the polarization state of macrophages. In myocardial infarction model, Leucine Rich Repeat Containing G Protein-Coupled538 Receptor 4 (Lgr4)-mediated cAMP/PKA/CREB pathway promotes transactivation of Fos in pro-inflammatory macrophage to enhance AP-1 activity, thereby triggering proinflammatory programming in macrophages to coordinate the proinflammatory state of infarcted macrophages (101). In the arthritis model, Fra-1 expression was up-regulated after LPS activated Toll-like receptors (TLR) cascade, and then it was directly bound to the promoter region of Arginase 1 (Arg1) to inhibit its transcription, making macrophages transform into M1 pro-inflammatory phenotype (15). Fra-1 also enhances osteoclast differentiation (102). In addition, it can also affect tumor progression by affecting macrophage activity: Breast cancer cells can overexpress Fra-1 in tumor-associated macrophages, induce macrophages to differentiate from M1 to M2, and then promote tumor immune escape, thereby promoting the invasiveness of breast cancer cells (21, 103). Signaling from CD137, a member of the tumor necrosis factor superfamily, can promote the expression of Fra-1, thus promoting the differentiation and migration of mononuclear macrophage into osteoclasts, then promoting the metastasis of tumor cells (104). Fra-1 also has a certain effect on the activation stage of macrophages. It has been demonstrated that Fra-1 can bind to Lipocalin 2 (Lcn2) promoter to promote neutropil gelatinase-associated lipocalin (NGAL) expression in immune tolerant BMDM(Bone marrow-derived macrophages), which has been described as a marker of inactivated macrophages (14).
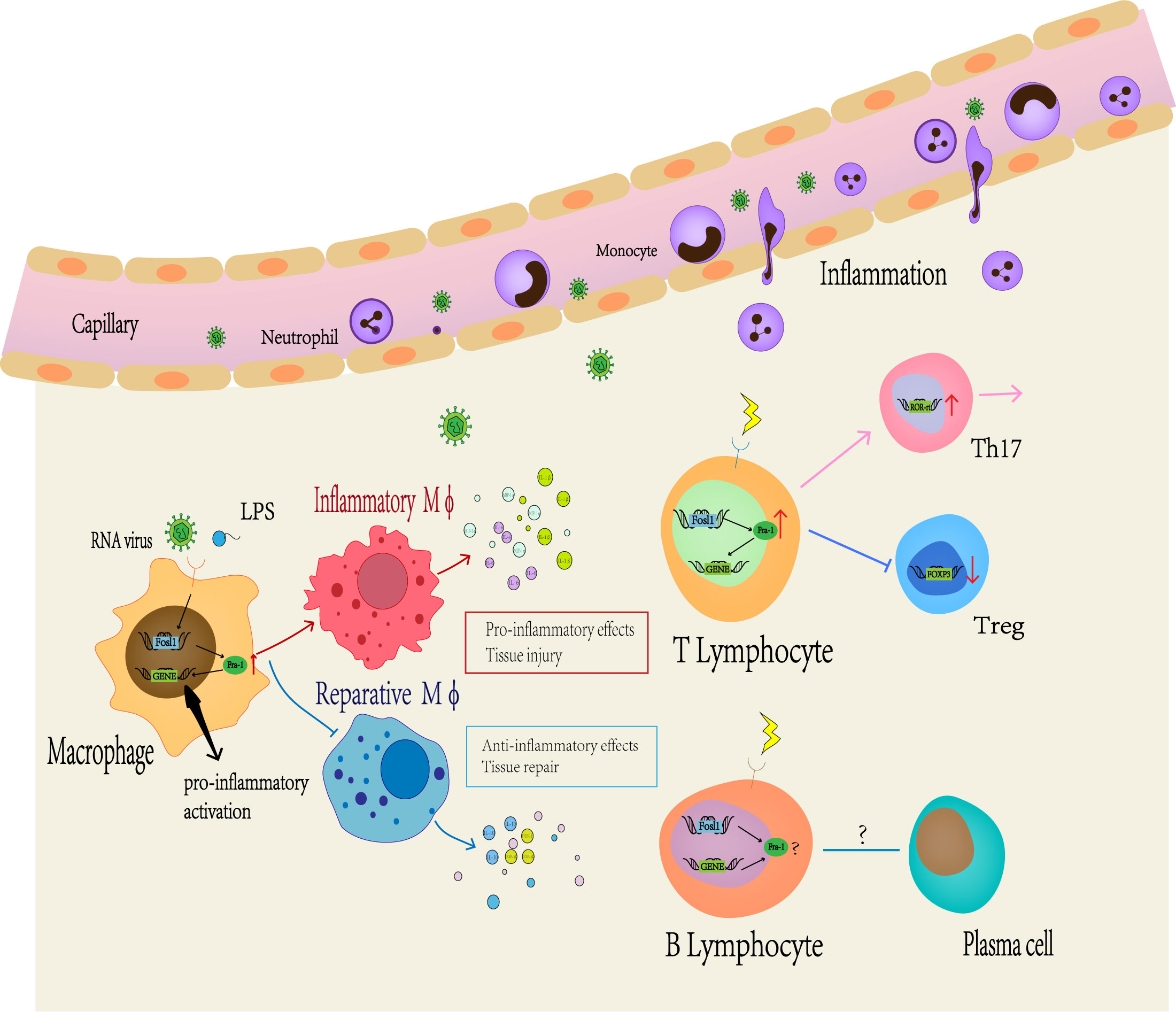
Figure 3 Fra-1 mediated immune regulation. A large number of studies have shown that Fra-1 regulates the function and activity of various immune cells in vivo. At first, the expression level of Fra-1 affects the migration of neutrophils, the differentiation of macrophages and T or B lymphocyte. When the body is infected or stimulated, the expression level of Fra-1 in immune cells increases, prompting more neutrophils to migrate from blood vessels to the site of inflammation, promoting M0 macrophages to differentiate more into M1-type macrophages and secrete more related pro-inflammatory cytokines. In addition, Fra-1 has been reported to be the core of T or B lymphocyte differention in the adaptive immune responses.
Bone marrow precursors give rise to monocytes in blood (105). Together with tissue resident macrophage and dendritic cells (DCs), monocytes are classified as mononuclear macrophages (106), which can serve as a bridge between innate and adaptive immune responses. Some studies have shown that monocytes are highly plastic and can differentiate according to microenvironment changes (107, 108). Most articles use CD14, CCR2 and CD16 to classify monocyte subtypes, which are “classical” monocytes (CD14++CD16-CCR2++), “intermediate” monocytes (CD14++CD16+CCR2+) and “non-classical” monocytes (CD14+CD16++CCR2-) (109–111). Among them, the proportion of “classical” monocytes are the highest, which belongs to inflammatory cells that respond to many stimuli originated from damaged/infected tissue and produce inflammatory cytokines (111, 112); “intermediate” monocytes are highly phagocytic cells that produce high levels of ROS(reactive oxygen species) and inflammatory mediators (113, 114); and non-classical monocytes are reparative/patrolling cells that remove debris from vasculature and produce high levels of anti-inflammatory factors (115, 116). The researchers found that classical monocytes produced high levels of the broadest range of cytokines and chemokines in response to LPS, and this versability of classical monocytes is attributed to AP-1 transcription factors that are most highly expressed at the gene level by classical monocytes (117). Different combinations of the AP-1 transcription factor components are activated by different stimuli and signaling pathways, defining the transcription targets for AP-1 (118–120). What’s more, CD14+ monocytes can up-regulate the expression of Fra-1 to inhibit the production of type I proinflammatory cytokines monocytes and affect the effect of cells after stimulated by RNA virus (121).
Neutrophils, which are a type of polymorphonuclear leukocyte, are well recognized as one of the major players during acute inflammation. They are typically the first leukocytes to be recruited to the inflammatory site and can eliminate pathogens through various mechanisms (122–125). Meanwhile, inappropriate recruitment can lead to tissue damage (126). Therefore, proper control of neutrophil activity and number is important for tissue homeostasis and progression. It has been reported that TGF-βsignal up-regulates the expression of Fra-1, c-Fos and c-Jun in neutrophil nucleus (127). In addition, in most inflammatory diseases, the overexpression of Fra-1 in mice will cause more neutrophils in blood vessels to migrate to tissue, aggravating tissue damage (128).
Traditionally, host immunity is divided into innate and adaptive immune responses. During infection, innate immunity is first stimulated (inflammation) and fully activated within minutes to hours (129). When innate immunity fails to eliminate pathogens, lymphocytes and adaptive immune mechanisms are activated, which can specifically recognize and eliminate pathogens, and it is important for host defense during late infection and secondary infection (130). Modulating adaptive immunity can play an important role in disease progression. T lymphocyte and B lymphocyte are the key cells of adaptive immune response, mainly responsible for the basic functions of cell-mediated immune response and antibody production respectively (131).
CD4+T lymphocytes can differentiate into different subtypes after being stimulated by cytokines, including Th1, Th2, Th9, Th17, Th22, T follicular helper (Tfh), and regulatory T cell (Treg). Under the regulation of intracellular transcription factors, they play different immune roles (132). Studies have shown that Fra-1 and other members of the AP-1 family play multifunctional roles in T cell development (7). In melanoma, Fra-1 is regulated by Ubc13-IKK signaling axis to maintain Treg cell function and inhibit its transformation into effector T cells (133). After overexpression of Fra-1 in CD4+T cells, it can act as a downstream target of Stat3 to induce the differentiation of Th17 cells (17). CD14+ monocytes can induce Fra-1 expression by activating MAPK through TLR7, and preferentially increase the production of Th17 polarizing cytokines to promote the polarization of Th17 cells (121). In the psoriatic model, Sobolev, V., et al. speculated that transcriptional regulation of PPARγ (Peroxisome Proliferator Activated Receptor Gamma) expression by the Fra-1 and Stat3/FOSL1 feedback pathways may be at the core of T cell differentiation (134).
IL-6 can promote the specific differentiation of naive CD4 + T cells, thereby linking congenital and acquired immune responses. The combination of IL-6 and transforming growth factor-β (TGF-β) is essential for the differentiation of Th17 cells (135). It can also inhibit TGF-β-induced Treg differentiation, and induces CD8 + T cells to differentiate into cytotoxic T cells (136). As mentioned above, Fra-1 plays an important role in the process of IL-6 secretion, and it is also the downstream effector molecule. Fra-1 may be the key transcription factor regulating T cell differentiation by these cytokines.
Blimp-1 is a transcription factor that plays an important role in the maturation of plasma cells from B cells to immunoglobulins and in the regulation of T cell homeostasis and tolerance (137). Studies have shown that c-Fos, a member of the Fos family, can positively regulate the expression of Blimp-1 and activate terminal differentiation of B cells (138), and its overexpression can also inhibit germinal center response (139). Increased AP-1 activity leads to early expression of Blimp-1, resulting in premature plasma cell formation and inhibition of germinal center B cell development (140, 141). However, subsequent studies have pointed out that Fra-1 can inhibit the production and development of plasma cells by regulating the direct combination with the Prdm1/Bimp1(PR/SET Domain 1/B lymphocyte induced maturation protein 1) promoter (142). In addition, the activation of B cells by TD antigen can reduce the expression of Fra-1, which can control the production of IgG-RF and inhibit the activation of osteoclasts driven by immune complexes (143). Therefore Fra-1 may be used as a regulator to block the negative effects of autoimmunity by inhibiting the formation of immune complexes. At the same time, in another study, stimulation of CD40 and surface Ig (sIg) receptors on B lymphocytes significantly induced Fra-1 expression by inducing protein kinase C (PKC) (144), which is not consistent with the previously observed phenotype. The regulation mechanism of Fra-1 expression in B cells still needs more research to clarify.
Cytokines are a kind of small molecule proteins with extensive biological activity synthesized and secreted by immune or some non-immune cells. They generally regulate cell growth, differentiation and effect by binding to corresponding receptors, and regulate immune response. Studies have found that Fra-1 plays an important role in the synthesis and secretion of various cytokines as well as the mechanism of action. Promoter regions of inflammatory cytokines and chemokines including TNF-α, IL-1β, IL-11, IL-8 and MCP-1 contain AP-1 binding sites (145, 146), but the reregulate mechanism of Fra-1 on cytokine expression is controversial.
We know that IL-6 mediates many inflammatory diseases and is considered to be an immune regulator that coordinates innate and adaptive immune responses (19). Morishita, H., et al. reported that overexpression of Fra-1 in RAW264.7 inhibited LPS-stimulated IL-6 production (147). In contrast, the study found that 4T1 breast cancer cells can promote the expression of Fra-1 in RAW264.7 in response to LPS and increase IL-6 production (21). Meanwhile, in LPS-triggered acute lung injury models, there was no significant difference in IL-6 production after knockout of Fra-1 in mouse alveolar macrophages (20). The effect of Fra-1 on cytokines is not absolute, which may be affected by cell types and the initial concentration of Fra-1 in cells, etc. The specific mechanism still needs to be further studied. Interestingly, Stat3 is a major transducer of signals triggered by cytokines and growth factors. However, Stat3 responds to the IL-6 signal on the membrane to phosphorylate downstream Fra-1, resulting in stable and increased intracellular Fra-1 content (55), which can further increase IL-6 expression. The expression of Fra-1 may be a positive feedback mechanism, which is similar to the phenotype that Fra-1 binds to its own promoter to promote expression. Furthermore, Du’s group reported that CCL-5, CCL-19 and CCL-21 could be regulated by Fra1 in medulla thymic epithelial cells (mTECs) and overexpression of Fra1 inhibited the transcriptional level of IL-1β, IL-6, IL-8 and ICAM1. They revealed that Fra1 disrupted inflammatory cytokine secretion by mTECs(medullary thymic epithelial cells) in the MG(myasthenia gravis) thymus (148). Another study conducted by Cho demonstrated that Fra-1-JunB complex directly binds to promoter region of IL-17a and activates its transcription (17). Ishihara et al. found that silencing Fra-1 by siRNA suppressed the expression of TNF-α and IL-1β in BV-2, a microglial cell line (17).
Taken together, these findings indicate that Fra-1 is intimately linked with the transcription of various cytokines in immune cells.
The role of Fra-1 in the immune system enables it to regulate the occurrence and development of some diseases (see Table 2). It has been extensively documented in osteosclerosis and cancer-related diseases, which can be referred to other reviews (60, 71, 164, 165).
Sepsis is a life-threatening disease caused by high morbidity and mortality in hospitals (166). It is characterized by excessive inflammation in the early stage and subsequent abnormal regression of inflammation associated with long-term immunosuppression, which may eventually lead to multiple organ dysfunction. Proper control of inflammation plays an important role in the recovery of sepsis (167), which WHO has identified as a global health priority (168). Studies have proved that Fra-1 plays a certain role in the occurrence and development of sepsis. First, Matrix Metallopeptidase 9 (MMP9) was found to be one of the differentially expressed genes involved in the pathogenesis of sepsis (169). Down-regulation of MMP9 can reduce inflammation, while Fra-1 can occupy the promoter of MMP9 (170, 171). The silencing of Fra-1 destroys the expression of MMP9 (172), thereby reducing sepsis-induced renal inflammation and apoptosis (149). In addition, many studies have found that sepsis in mice overexpressing Fra-1 increases the severity of lung injury (150). In the immunosuppressive phase, specific knockout of Fra-1 in mouse macrophages leads to increased inflammation in immune tolerant mice, and Fra-1 can protect mice against endotoxin-induced sepsis by regulating NGAL (14). The regulation of Fra-1 on inflammation in sepsis is time-dependent, and the specific mechanism of action remains to be studied. Further studies are expected to find that Fra-1 can provide a new direction for sepsis treatment, and finally verify the clinical feasibility.
Knockout of Fra-1 in mice results in embryonic death by blocking the angiogenesis of fetal placenta, indicating the importance of Fra-1 in angiogenesis. Prokineticin 2(PROK2)is an important contributor to tumor angiogenesis (151), Ring Finger Protein 213 (RNF213) is essential for normal vascular development (152), and Leucine Rich Alpha-2-Glycoprotein 1 (LRG1) plays a role in retinal vascularization abnormalities observed in oxygen-induced retinopathy (154). All three have been found to be regulated by Fra-1. It may also serve as a mediator of impaired pulmonary vascular development in neonates (153). In addition, Fra-1 plays a role in atherosclerosis (155), diabetic vascular restenosis (156), myocarditis, myocardial infarction (101) and other diseases. Previous studies suggest that the role of Fra-1 in regulating inflammatory angiogenesis in organs may be a topic for future research.
Expression of Fra-1 is increased in the lungs of people stimulated by LPS or infected with bacteria in vitro (100), and in the adult respiratory distress syndrome (ARDS) (173). Knockout Fra-1 in mice reduced LPS-induced acute lung injury and mortality (128), while overexpression showed opposite results (173). Studies have shown that Fra-1 plays a key role in inflammatory lung injury. Fra-1 plays a protective role in pulmonary fibrosis and regulates early fibro genic response. Fra-1 mediates anti-fibrosis effects by regulating pro-inflammatory, pro-fibrotic and fibrotic gene expression in vivo and in vitro (157, 158). Conversely, Fra-2 has been studied as a causative factor for human fibrosis, and high expression of Fra-2 was observed in idiopathic and autoimmune-mediated pulmonary fibrosis samples (174). The structures of Fra-2 and Fra-1 are very similar, and the opposite effect of the two in the same disease is also worth thinking and studying, and both are considered to be therapeutic targets for pulmonary fibrosis.
Psoriasis is a common inflammatory skin disease that is autoimmune and affects approximately 3% of the world’s population. It is characterized by keratinocyte proliferation, altered differentiation, and increased inflammation and angiogenesis (175). Fra-1 is very important for the stability of psoriasis inflammation, and is overexpressed human pathological tissues (159, 160). PPAR-γ inhibits the transcription of IL-17 by inhibiting its direct transcription factors RORC and STAT3, while FRA-1 can regulate the expression of STATA3, RORC and IL-17 by directly inhibiting PPARγ, thus regulating the development of psoriasis (134). Induction of Fra-1 overexpression in keratinocytes induces increased production of proinflammatory cytokines and chemokines (IL-8 and TNF-a), allowing subsequent immune cells (neutrophils and T cells) to migrate under this proinflammatory background and inducing plaque development in psoriasis (176).
The liver of Fra-1 overexpressed mice was progressively infiltrated by innate and adaptive immune cells. Compared with WT mice, the infiltrated cells in the liver of Fra-1 overexpressed mice were mainly composed of neutrophils and CD3+T cells, while B cells and macrophages rarely appeared in inflammatory sites, showing increased infiltration of activated CD4+CD69+ T cells. B cells, NK cells, and NKT cells decreased dramatically, while neutrophil infiltration increased (161). In DSS-induced colitis model, overexpression of Fra-1 inhibited the activation of NF-kB (162) and reduced the inflammatory response in mice.
The absence of Fra-1 affects the inflammatory stage of arthritis, and the severity of the disease after inducing SIA in Fra-1 deficient mice is reduced. Different from the absence of other AP-1 members, c-Fos deficient mice have more severe arthritis (177). Overexpression of Fra-1 in CD4 + T cells can bind to JunB as a downstream target of Stat3 to induce Th17 cell differentiation and promote autoimmune arthritis (143). Inhibition of AP-1 activity can prevent acute graft-versus-host disease (aGVHD) by altering the differentiation of CD4 + T cells, such as reduced Th17/Th1 population and increased Treg population (163). Other studies have found that the expression of Fra-1 in medullary thymic epithelial cells (mTEC) of myasthenia gravis (MG) patients is increased, and its overexpression may destroy the secretion of inflammatory cytokines in mTEC of MG patients’ thymus. accompanied the increase of CCL5, CCL-19 and CCL-21, and the decrease of ICAM1, IL-6, IL-1β (148).
Numerous studies have confirmed that Fra-1 is crucially involved in human tumor progression and metastasis, thus representing a promising therapeutic target (178, 179). As we all know, inflammation and cancer is intimately linked, the tumor microenvironment (TME) plays a prominent role in the growth of tumor cells. As the major inflammation component of the TME, M2d macrophages are educated by the TME such that they adopt an immunosuppressive role that promotes tumor metastasis and progression (180). Fra-1 can increase the production of the cytokine IL-6 and skew RAW264.7 macrophage cell differentiation into M2d macrophage, then promotes tumor metastasis and progression (21, 103); CD137 promotes the migration of monocytes/macrophages to tumor microenvironment by upregulating the expression of Fra1. It also promoted the differentiation of monocytes/macrophages into osteoclasts at the same time, thus providing a favorable microenvironment for the colonization and growth of breast cancer cells in bone (104); The cytokine IL-6 induces Fra-1 deacetylation, then promoting colorectal cancer stem-like properties (74); In addition, the existence of an aberrant IL-6/STAT3/Fra-1 signaling axis leading to colorectal cancer(CRC) aggressiveness through EMT induction, which suggests novel therapeutic opportunities for the malignant disease (55). A DNA vaccine encoding transcription factor Fra-1 and secretory IL-18 induces a long-lived memory T-cell response which can contribute to tumor regression (181–183). The effect of Fra-1 on immune cells also plays an important role in tumorigenesis.
The complex and diverse functions of AP-1 have made it an actively pursued drug target in past studies, but transcription factors have been considered difficult to target because their activity is triggered by protein-protein or protein-DNA interactions. However, inhibiting key transcription factors such as c-Myc (184, 185), which is previously considered to be drug-free, and then confirmed, this provides confidence and foundation for the suppression of other transcription factors such as AP-1. Selective AP-1 inhibitor T5224 (61) (3-propionic acid (T-5224)) has been investigated in phase II clinical trials for novel c-Fos/AP-1 inhibitors for rheumatoid arthritis (186). Some studies have found that some tumor prevention drugs, such as resveratrol (187), green tea (188) and curcumin (189), work by inhibiting Fra-1 expression in the model system. Retinol like SR11302 inhibits AP-1 activity and FOSL1 expression without activating RARE transcription (12, 190–194). SR11302 inhibits alloreactive T cell response in a dose-dependent manner, showing decreased Th17/Th1 population and increased Treg population in vivo (163). Meanwhile, FOSL1 was found to be the target of anti-epileptic drug levetiracetam (LEV) for inhibiting neuroinflammation. LEV can reduce the expression of FOSL1 in microglia to inhibit its activation and thus inhibit the inflammatory response involved (195). At present, there is no specific small-molecule inhibitor of Fra-1 that is effective both in cells and animals. In fact, the existing selective inhibitors are not only act on Fra-1, which would elicit undesired side effects. As extensive screening of specific inhibitors of Fra-1 is still ongoing, additional improvements may be achieved from future experimentation. Another strategy for targeting Fra-1 is genetic inhibition. Vicent’s group reported that deficiency for Fra-1 in a genetically engineered mouse model of Cholangiocarcinoma using constitutive and inducible short-hairpin RNAs extended mouse survival by decreasing the oncogenic potential of transformed cholangiocytes (179).
Sometimes directly targeting Fra-1 may be difficult at present, but if more targets for Fra-1 regulation can be found, it may be relatively easy to control the target gene and obtain the corresponding phenotype. For example, the expression of the Mevalonate pathway gene HMGCS1 is controlled by direct FOSL1 promoter binding. Genetic and pharmacological inhibition of HMGCS1 and AURKA leads to loss of FOSL1, which can reduce the carcinogenic potential of transformed bile duct cells and prolong mouse survival (179). As previously mentioned, MAP kinase pathway plays a role in the content and stability of Fra-1 in cells. Inhibition of ERK1/2 pathway can block the recruitment of c-Jun and NF-kB transcription factors to endogenous FOSL1 promoter after LPS stimulation to reduce Fra-1 mRNA expression, thereby reducing the expression of inflammatory factors (20). The progression of many diseases is influenced by the immune system, and the role of Fra-1 in the immune system makes it a potential target for disease treatment. The regulation of Fra-1 and its pre- and post-effectors has a great impact on the outcome of the disease. However, the mechanism of Fra-1 in immune cells and immune responses is still unclear, which needs more research to elaborate, and then it is expected to develop effective drugs for clinical use.
As a member of FOS family, Fra-1 plays an important role in cell proliferation, differentiation, tumor transformation, immunity, inflammation and other processes. AP-1 is an important nuclear transcription factor. FOS and JUN families, which mainly exist in mammals, bind to form heterodimers through the leucine zipper domain to regulate the transcription of related target genes. It is involved in MAPK (196), Wnt (197) and other important signaling pathways, which accumulate in cells mainly through transcription and post-translational modification. Since tumor progression is closely related to the intensity of immune response, when most studies focus on the role of Fra-1 in tumors, the other part focuses on the function of Fra-1 in immunity.
Fra-1 affects the differentiation and activation of immune cells, especially macrophages, as well as the secretion of many cytokines (17, 121, 133, 138, 142, 198, 199), but the specific mechanism remains to be further explored. The regulation of the immune response of Fra-1 makes its intracellular expression affect the outcome of many diseases, such as sepsis (149), atherosclerosis (155), myocardial infarction (101), psoriasis (134), colitis (162), etc. This evidence points at the importance of Fra-1 in immune regulation. Thus, it’s necessary to figure out the molecule controls of Fra-1 that maintain the steady-state homeostasis of immune cells and how dysregulation of these processes could precipitate a wide range of immune-associated disorders. Meanwhile, targeting Fra-1 is expected to be an effective intervention strategy. However, there are still many basic questions about Fra-1 that cannot be answered.
First, why do the same structural molecules play the opposite effect. The structures of most FOS family members are similar, especially Fra-1 and Fra-2, both of which lack the trans-activation domain at their C-terminus (45), but sometimes they show different results in the same disease. For example, Fra-1 has a protective effect on pulmonary fibrosis, but Fra-2 is the pathogenic factor of the disease (157, 158, 174). Secondly, whether the Fra-1/AP-1 transcription factor only binds to the promoter of the target gene plays a role. Prior to the emergence of ‘omics’ research, the molecular transcription research of AP-1 was basically concentrated in the gene promoter region. This view has recently been challenged by genome-wide studies showing that AP-1 frequently binds to enhancer sites far from the gene transcription start site (TSSs), although the nature of this has not been determined (13, 31). It has been found that FOSL1 not only acts on promoters of target genes, but also selectively binds to its enhancers and corresponding Mediators to establish super enhancers (SEs), thus promoting the expression of target genes (12). At present, our understanding of Fra-1 targeted gene and its transcriptional regulation mechanism is still limited. It is not clear whether Fra-1 affects chromatin or whether it affects the expression of target genes in cells by using the preset 3D structural network. Then, whether Fra-1 must be incorporated into the nucleus through transcription factors with other proteins to function. The activation of ERK by Nitrogen mustard (NM) increases Fra-1 levels in mouse epidermis and cultured human keratinocytes, where Fra-1 is mainly expressed in the cytoplasm rather than the nucleus, which contributes to IL-8 expression in NM-damaged skin (23).
In summary, a large number of studies have found that Fra-1 plays a certain role in the immune response process. Intervention at the levels of transcription, translation and post-translational modification can regulate its expression in cells, thereby regulating some cell activities and molecular production. However, the specific mechanism remains unclear, and many problems still need further study.
Y-YH, H-FZ, and LC contributed to the conceptual design, writing, and editing for this manuscript. Y-TW, W-LX, Z-ZX, YX, Y-QF, G-YL, XL, and JL provided valuable discussions and comments. Q-PW revised the manuscript and commented on previous versions of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Grant No. 81901948), the National Natural Science Foundation of China (Grant No. 81873952), and the National Key Research and Development Program of China (Grant No. 2018YFC2001900).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.

1. Cohen DR, Curran T. Fra-1: A serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol Cell Biol (1988) 8(5):2063–9. doi: 10.1128/mcb.8.5.2063-2069.1988
2. Matsui M, Tokuhara M, Konuma Y, Nomura N, Ishizaki R. Isolation of human fos-related genes and their expression during monocyte-macrophage differentiation. Oncogene (1990) 5(3):249–55.
3. Eferl R, Wagner EF. Ap-1: A double-edged sword in tumorigenesis. Nat Rev Cancer (2003) 3(11):859–68. doi: 10.1038/nrc1209
4. Jiang X, Xie H, Dou Y, Yuan J, Zeng D, Xiao S. Expression and function of Fra1 protein in tumors. Mol Biol Rep (2020) 47(1):737–52. doi: 10.1007/s11033-019-05123-9
5. Herdegen T, Waetzig V. Ap-1 proteins in the adult brain: Facts and fiction about effectors of neuroprotection and neurodegeneration. Oncogene (2001) 20(19):2424–37. doi: 10.1038/sj.onc.1204387
6. Hess J, Angel P, Schorpp-Kistner M. Ap-1 subunits: Quarrel and harmony among siblings. J Cell Sci (2004) 117(Pt 25):5965–73. doi: 10.1242/jcs.01589
7. Jochum W, Passegué E, Wagner EF. Ap-1 in mouse development and tumorigenesis. Oncogene (2001) 20(19):2401–12. doi: 10.1038/sj.onc.1204389
8. Shaulian E, Karin M. Ap-1 as a regulator of cell life and death. Nat Cell Biol (2002) 4(5):E131–6. doi: 10.1038/ncb0502-e131
9. Wagner EF, Eferl R. Fos/Ap-1 proteins in bone and the immune system. Immunol Rev (2005) 208:126–40. doi: 10.1111/j.0105-2896.2005.00332.x
10. Jochum W, David JP, Elliott C, Wutz A, Plenk H Jr., Matsuo K, et al. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor fra-1. Nat Med (2000) 6(9):980–4. doi: 10.1038/79676
11. Belguise K, Kersual N, Galtier F, Chalbos D. Fra-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene (2005) 24(8):1434–44. doi: 10.1038/sj.onc.1208312
12. Zhang M, Hoyle RG, Ma Z, Sun B, Cai W, Cai H, et al. Fosl1 promotes metastasis of head and neck squamous cell carcinoma through super-Enhancer-Driven transcription program. Mol Ther J Am Soc Gene Ther (2021) 29(8):2583–600. doi: 10.1016/j.ymthe.2021.03.024
13. Bejjani F, Evanno E, Zibara K, Piechaczyk M, Jariel-Encontre I. The ap-1 transcriptional complex: Local switch or remote command? Biochim Biophys Acta Rev Cancer (2019) 1872(1):11–23. doi: 10.1016/j.bbcan.2019.04.003
14. Cao S, Schnelzer A, Hannemann N, Schett G, Soulat D, Bozec A. The transcription factor fra-1/Ap-1 controls lipocalin-2 expression and inflammation in sepsis model. Front Immunol (2021) 12:701675. doi: 10.3389/fimmu.2021.701675
15. Hannemann N, Cao S, Eriksson D, Schnelzer A, Jordan J, Eberhardt M, et al. Transcription factor fra-1 targets arginase-1 to enhance macrophage-mediated inflammation in arthritis. J Clin Invest (2019) 129(7):2669–84. doi: 10.1172/jci96832
16. Mittelstadt ML, Patel RC. Ap-1 mediated transcriptional repression of matrix metalloproteinase-9 by recruitment of histone deacetylase 1 in response to interferon Β. PLoS One (2012) 7(8):e42152. doi: 10.1371/journal.pone.0042152
17. Moon YM, Lee SY, Kwok SK, Lee SH, Kim D, Kim WK, et al. The fos-related antigen 1-Junb/Activator protein 1 transcription complex, a downstream target of signal transducer and activator of transcription 3, induces T helper 17 differentiation and promotes experimental autoimmune arthritis. Front Immunol (2017) 8:1793. doi: 10.3389/fimmu.2017.01793
18. Shetty A, Tripathi SK, Junttila S, Buchacher T, Biradar R, Bhosale SD, et al. A systematic comparison of Fosl1, Fosl2 and batf-mediated transcriptional regulation during early human Th17 differentiation. Nucleic Acids Res (2022) 50(9):4938–58. doi: 10.1093/nar/gkac256
19. Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta (2011) 1813(5):878–88. doi: 10.1016/j.bbamcr.2011.01.034
20. Mishra RK, Potteti HR, Tamatam CR, Elangovan I, Reddy SP. C-jun is required for nuclear factor-Κb-Dependent, lps-stimulated fos-related antigen-1 transcription in alveolar macrophages. Am J Respir Cell Mol Biol (2016) 55(5):667–74. doi: 10.1165/rcmb.2016-0028OC
21. Wang Q, Ni H, Lan L, Wei X, Xiang R, Wang Y. Fra-1 protooncogene regulates il-6 expression in macrophages and promotes the generation of M2d macrophages. Cell Res (2010) 20(6):701–12. doi: 10.1038/cr.2010.52
22. Cai B, Wu J, Yu X, Su XZ, Wang RF. Fosl1 inhibits type I interferon responses to malaria and viral infections by blocking Tbk1 and Traf3/Trif interactions. mBio (2017) 8(1):e02161–16. doi: 10.1128/mBio.02161-16
23. Ye F, Zeng Q, Dan G, Dong X, Chen M, Sai Y, et al. Nitrogen mustard prevents transport of fra-1 into the nucleus to promote c-fos- and fosb-dependent il-8 induction in injured mouse epidermis. Toxicol Lett (2020) 319:256–63. doi: 10.1016/j.toxlet.2019.10.006
24. Schreiber M, Wang ZQ, Jochum W, Fetka I, Elliott C, Wagner EF. Placental vascularisation requires the ap-1 component Fra1. Dev (Cambridge England) (2000) 127(22):4937–48. doi: 10.1242/dev.127.22.4937
25. Schreiber M, Poirier C, Franchi A, Kurzbauer R, Guenet JL, Carle GF, et al. Structure and chromosomal assignment of the mouse fra-1 gene, and its exclusion as a candidate gene for oc (Osteosclerosis). Oncogene (1997) 15(10):1171–8. doi: 10.1038/sj.onc.1201460
26. Angel P, Karin M. The role of jun, fos and the ap-1 complex in cell-proliferation and transformation. Biochim Biophys Acta (1991) 1072(2-3):129–57. doi: 10.1016/0304-419x(91)90011-9
27. Glover JN, Harrison SC. Crystal structure of the heterodimeric bzip transcription factor c-Fos-C-Jun bound to DNA. Nature (1995) 373(6511):257–61. doi: 10.1038/373257a0
28. Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, et al. Protein kinase c Α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell (2013) 24(3):347–64. doi: 10.1016/j.ccr.2013.08.005
29. Luck K, Kim DK, Lambourne L, Spirohn K, Begg BE, Bian W, et al. A reference map of the human binary protein interactome. Nature (2020) 580(7803):402–8. doi: 10.1038/s41586-020-2188-x
30. Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature (2005) 437(7062):1173–8. doi: 10.1038/nature04209
31. Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between Yap/Taz/Tead and ap-1 at enhancers drives oncogenic growth. Nat Cell Biol (2015) 17(9):1218–27. doi: 10.1038/ncb3216
32. Reinke AW, Baek J, Ashenberg O, Keating AE. Networks of bzip protein-protein interactions diversified over a billion years of evolution. Sci (New York NY) (2013) 340(6133):730–4. doi: 10.1126/science.1233465
33. Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, et al. A proteome-scale map of the human interactome network. Cell (2014) 159(5):1212–26. doi: 10.1016/j.cell.2014.10.050
34. Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, et al. Next-generation sequencing to generate interactome datasets. Nat Methods (2011) 8(6):478–80. doi: 10.1038/nmeth.1597
35. Haenig C, Atias N, Taylor AK, Mazza A, Schaefer MH, Russ J, et al. Interactome mapping provides a network of neurodegenerative disease proteins and uncovers widespread protein aggregation in affected brains. Cell Rep (2020) 32(7):108050. doi: 10.1016/j.celrep.2020.108050
36. Wang J, Huo K, Ma L, Tang L, Li D, Huang X, et al. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol (2011) 7:536. doi: 10.1038/msb.2011.67
37. Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, et al. An empirical framework for binary interactome mapping. Nat Methods (2009) 6(1):83–90. doi: 10.1038/nmeth.1280
38. Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. C-jun dimerizes with itself and with c-fos, forming complexes of different DNA binding affinities. Cell (1988) 55(5):917–24. doi: 10.1016/0092-8674(88)90147-x
39. Ryseck RP, Bravo R. C-jun, jun b, and jun d differ in their binding affinities to ap-1 and cre consensus sequences: Effect of fos proteins. Oncogene (1991) 6(4):533–42.
40. Zoumpourlis V, Papassava P, Linardopoulos S, Gillespie D, Balmain A, Pintzas A. High levels of phosphorylated c-jun, fra-1, fra-2 and atf-2 proteins correlate with malignant phenotypes in the multistage mouse skin carcinogenesis model. Oncogene (2000) 19(35):4011–21. doi: 10.1038/sj.onc.1203732
41. Young MR, Nair R, Bucheimer N, Tulsian P, Brown N, Chapp C, et al. Transactivation of fra-1 and consequent activation of ap-1 occur extracellular signal-regulated kinase dependently. Mol Cell Biol (2002) 22(2):587–98. doi: 10.1128/mcb.22.2.587-598.2002
42. Welter JF, Eckert RL. Differential expression of the fos and jun family members c-fos, fosb, fra-1, fra-2, c-jun, junb and jund during human epidermal keratinocyte differentiation. Oncogene (1995) 11(12):2681–7.
43. Milde-Langosch K, Röder H, Andritzky B, Aslan B, Hemminger G, Brinkmann A, et al. The role of the ap-1 transcription factors c-fos, fosb, fra-1 and fra-2 in the invasion process of mammary carcinomas. Breast Cancer Res Treat (2004) 86(2):139–52. doi: 10.1023/b:Brea.0000032982.49024.71
44. Fleischmann A, Hafezi F, Elliott C, Remé CE, Rüther U, Wagner EF. Fra-1 replaces c-Fos-Dependent functions in mice. Genes Dev (2000) 14(21):2695–700. doi: 10.1101/gad.187900
45. Bergers G, Graninger P, Braselmann S, Wrighton C, Busslinger M. Transcriptional activation of the fra-1 gene by ap-1 is mediated by regulatory sequences in the first intron. Mol Cell Biol (1995) 15(7):3748–58. doi: 10.1128/mcb.15.7.3748
46. Ito E, Sweterlitsch LA, Tran PB, Rauscher FJ 3rd, Narayanan R. Inhibition of pc-12 cell differentiation by the immediate early gene fra-1. Oncogene (1990) 5(12):1755–60.
47. Yoshioka K, Deng T, Cavigelli M, Karin M. Antitumor promotion by phenolic antioxidants: Inhibition of ap-1 activity through induction of fra expression. Proc Natl Acad Sci USA (1995) 92(11):4972–6. doi: 10.1073/pnas.92.11.4972
48. Bahrami S, Drabløs F. Gene regulation in the immediate-early response process. Adv Biol Regul (2016) 62:37–49. doi: 10.1016/j.jbior.2016.05.001
49. Tulchinsky E. Fos family members: Regulation, structure and role in oncogenic transformation. Histol Histopathol (2000) 15(3):921–8. doi: 10.14670/hh-15.921
50. Kovary K, Bravo R. Existence of different Fos/Jun complexes during the G0-to-G1 transition and during exponential growth in mouse fibroblasts: Differential role of fos proteins. Mol Cell Biol (1992) 12(11):5015–23. doi: 10.1128/mcb.12.11.5015-5023.1992
51. Basbous J, Chalbos D, Hipskind R, Jariel-Encontre I, Piechaczyk M. Ubiquitin-independent proteasomal degradation of fra-1 is antagonized by Erk1/2 pathway-mediated phosphorylation of a unique c-terminal destabilizer. Mol Cell Biol (2007) 27(11):3936–50. doi: 10.1128/mcb.01776-06
52. Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3s10ph and H4k16ac generates a histone code that mediates transcription elongation. Cell (2009) 138(6):1122–36. doi: 10.1016/j.cell.2009.07.031
53. Zippo A, De Robertis A, Serafini R, Oliviero S. Pim1-dependent phosphorylation of histone H3 at serine 10 is required for myc-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol (2007) 9(8):932–44. doi: 10.1038/ncb1618
54. Esnault C, Gualdrini F, Horswell S, Kelly G, Stewart A, East P, et al. Erk-induced activation of tcf family of srf cofactors initiates a chromatin modification cascade associated with transcription. Mol Cell (2017) 65(6):1081–95.e5. doi: 10.1016/j.molcel.2017.02.005
55. Liu H, Ren G, Wang T, Chen Y, Gong C, Bai Y, et al. Aberrantly expressed fra-1 by il-6/Stat3 transactivation promotes colorectal cancer aggressiveness through epithelial-mesenchymal transition. Carcinogenesis (2015) 36(4):459–68. doi: 10.1093/carcin/bgv017
56. Casalino L, De Cesare D, Verde P. Accumulation of fra-1 in ras-transformed cells depends on both transcriptional autoregulation and mek-dependent posttranslational stabilization. Mol Cell Biol (2003) 23(12):4401–15. doi: 10.1128/mcb.23.12.4401-4415.2003
57. Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ Jr. Fluorescent cdna microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene (1999) 18(24):3666–72. doi: 10.1038/sj.onc.1202676
58. Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, et al. Insights into P53 transcriptional function Via genome-wide chromatin occupancy and gene expression analysis. Cell Death differentiation (2012) 19(12):1992–2002. doi: 10.1038/cdd.2012.89
59. Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functions for P53-activated and P53-repressed DNA damage response genes in embryonic stem cells. Mol Cell (2012) 46(1):30–42. doi: 10.1016/j.molcel.2012.01.020
60. Talotta F, Casalino L, Verde P. The nuclear oncoprotein fra-1: A transcription factor knocking on therapeutic applications' door. Oncogene (2020) 39(23):4491–506. doi: 10.1038/s41388-020-1306-4
61. Verde P, Casalino L, Talotta F, Yaniv M, Weitzman JB. Deciphering ap-1 function in tumorigenesis: Fra-ternizing on target promoters. Cell Cycle (Georgetown Tex) (2007) 6(21):2633–9. doi: 10.4161/cc.6.21.4850
62. Hoyt MA, Coffino P. Ubiquitin-free routes into the proteasome. Cell Mol Life Sci CMLS (2004) 61(13):1596–600. doi: 10.1007/s00018-004-4133-9
63. Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta (2008) 1786(2):153–77. doi: 10.1016/j.bbcan.2008.05.004
64. Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J Mol Biol (2009) 386(3):814–27. doi: 10.1016/j.jmb.2008.12.081
65. Basbous J, Jariel-Encontre I, Gomard T, Bossis G, Piechaczyk M. Ubiquitin-independent- versus ubiquitin-dependent proteasomal degradation of the c-fos and fra-1 transcription factors: Is there a unique answer? Biochimie (2008) 90(2):296–305. doi: 10.1016/j.biochi.2007.07.016
66. Gruda MC, Kovary K, Metz R, Bravo R. Regulation of fra-1 and fra-2 phosphorylation differs during the cell cycle of fibroblasts and phosphorylation in vitro by map kinase affects DNA binding activity. Oncogene (1994) 9(9):2537–47.
67. Cook SJ, Aziz N, McMahon M. The repertoire of fos and jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol Cell Biol (1999) 19(1):330–41. doi: 10.1128/mcb.19.1.330
68. Hurd TW, Culbert AA, Webster KJ, Tavaré JM. Dual role for mitogen-activated protein kinase (Erk) in insulin-dependent regulation of fra-1 (Fos-related antigen-1) transcription and phosphorylation. Biochem J (2002) 368(Pt 2):573–80. doi: 10.1042/bj20020579
69. Vial E, Marshall CJ. Elevated erk-map kinase activity protects the fos family member fra-1 against proteasomal degradation in colon carcinoma cells. J Cell Sci (2003) 116(Pt 24):4957–63. doi: 10.1242/jcs.00812
70. Murphy LO, MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol Cell Biol (2004) 24(1):144–53. doi: 10.1128/mcb.24.1.144-153.2004
71. Dhillon AS, Tulchinsky E. Fra-1 as a driver of tumour heterogeneity: A nexus between oncogenes and embryonic signalling pathways in cancer. Oncogene (2015) 34(34):4421–8. doi: 10.1038/onc.2014.374
72. Belguise K, Milord S, Galtier F, Moquet-Torcy G, Piechaczyk M, Chalbos D. The pkcθ pathway participates in the aberrant accumulation of fra-1 protein in invasive er-negative breast cancer cells. Oncogene (2012) 31(47):4889–97. doi: 10.1038/onc.2011.659
73. Belguise K, Cherradi S, Sarr A, Boissière F, Boulle N, Simony-Lafontaine J, et al. Pkcθ-induced phosphorylations control the ability of fra-1 to stimulate gene expression and cancer cell migration. Cancer Lett (2017) 385:97–107. doi: 10.1016/j.canlet.2016.10.038
74. Wang T, Song P, Zhong T, Wang X, Xiang X, Liu Q, et al. The inflammatory cytokine il-6 induces Fra1 deacetylation promoting colorectal cancer stem-like properties. Oncogene (2019) 38(25):4932–47. doi: 10.1038/s41388-019-0763-0
75. Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, et al. Microrna-34 suppresses breast cancer invasion and metastasis by directly targeting fra-1. Oncogene (2013) 32(36):4294–303. doi: 10.1038/onc.2012.432
76. Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue YF, et al. Microrna-34a inhibits migration and invasion of colon cancer cells Via targeting to fra-1. Carcinogenesis (2012) 33(3):519–28. doi: 10.1093/carcin/bgr304
77. Zhang N, Shen Q, Zhang P. Mir-497 suppresses epithelial-mesenchymal transition and metastasis in colorectal cancer cells by targeting fos-related antigen-1. OncoTargets Ther (2016) 9:6597–604. doi: 10.2147/ott.S114609
78. Jin Y, Wang C, Liu X, Mu W, Chen Z, Yu D, et al. Molecular characterization of the microrna-138-Fos-Like antigen 1 (Fosl1) regulatory module in squamous cell carcinoma. J Biol Chem (2011) 286(46):40104–9. doi: 10.1074/jbc.C111.296707
79. Wu J, Ji A, Wang X, Zhu Y, Yu Y, Lin Y, et al. Microrna-195-5p, a new regulator of fra-1, suppresses the migration and invasion of prostate cancer cells. J Trans Med (2015) 13:289. doi: 10.1186/s12967-015-0650-6
80. Xiao S, Zhou Y, Yi W, Luo G, Jiang B, Tian Q, et al. Fra-1 is downregulated in cervical cancer tissues and promotes cervical cancer cell apoptosis by P53 signaling pathway in vitro. Int J Oncol (2015) 46(4):1677–84. doi: 10.3892/ijo.2015.2873
81. Mishra A, Bharti AC, Saluja D, Das BC. Transactivation and expression patterns of jun and Fos/Ap-1 super-family proteins in human oral cancer. Int J Cancer (2010) 126(4):819–29. doi: 10.1002/ijc.24807
82. Gupta S, Kumar P, Kaur H, Sharma N, Saluja D, Bharti AC, et al. Selective participation of c-jun with fra-2/C-Fos promotes aggressive tumor phenotypes and poor prognosis in tongue cancer. Sci Rep (2015) 5:16811. doi: 10.1038/srep16811
83. Robinson CM, Prime SS, Huntley S, Stone AM, Davies M, Eveson JW, et al. Overexpression of junb in undifferentiated malignant rat oral keratinocytes enhances the malignant phenotype in vitro without altering cellular differentiation. Int J Cancer (2001) 91(5):625–30. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1095>3.3.co;2-a
84. Dou Y, Huang D, Zeng X, Zhou Y, Jiang X, Yue C, et al. All-trans retinoic acid enhances the effect of fra-1 to inhibit cell proliferation and metabolism in cervical cancer. Biotechnol Lett (2020) 42(6):1051–60. doi: 10.1007/s10529-020-02847-8
85. Wang C, Li Z, Shao F, Yang X, Feng X, Shi S, et al. High expression of collagen triple helix repeat containing 1 (Cthrc1) facilitates progression of oesophageal squamous cell carcinoma through Mapk/Mek/Erk/Fra-1 activation. J Exp Clin Cancer Res CR (2017) 36(1):84. doi: 10.1186/s13046-017-0555-8
86. Ou L, Wang X, Cheng S, Zhang M, Cui R, Hu C, et al. Verdinexor, a selective inhibitor of nuclear exportin 1, inhibits the proliferation and migration of esophageal cancer Via Xpo1/C-Myc/Fosl1 axis. Int J Biol Sci (2022) 18(1):276–91. doi: 10.7150/ijbs.66612
87. Casalino L, Bakiri L, Talotta F, Weitzman JB, Fusco A, Yaniv M, et al. Fra-1 promotes growth and survival in ras-transformed thyroid cells by controlling cyclin a transcription. EMBO J (2007) 26(7):1878–90. doi: 10.1038/sj.emboj.7601617
88. Sayan AE, Stanford R, Vickery R, Grigorenko E, Diesch J, Kulbicki K, et al. Fra-1 controls motility of bladder cancer cells Via transcriptional upregulation of the receptor tyrosine kinase axl. Oncogene (2012) 31(12):1493–503. doi: 10.1038/onc.2011.336
89. Vallejo A, Perurena N, Guruceaga E, Mazur PK, Martinez-Canarias S, Zandueta C, et al. An integrative approach unveils Fosl1 as an oncogene vulnerability in kras-driven lung and pancreatic cancer. Nat Commun (2017) 8:14294. doi: 10.1038/ncomms14294
90. Eferl R, Hoebertz A, Schilling AF, Rath M, Karreth F, Kenner L, et al. The fos-related antigen fra-1 is an activator of bone matrix formation. EMBO J (2004) 23(14):2789–99. doi: 10.1038/sj.emboj.7600282
91. Srivastava M, Saqib U, Naim A, Roy A, Liu D, Bhatnagar D, et al. The Tlr4-Nos1-Ap1 signaling axis regulates macrophage polarization. Inflammation Res Off J Eur Histamine Res Soc [et al] (2017) 66(4):323–34. doi: 10.1007/s00011-016-1017-z
92. Guha M, Mackman N. Lps induction of gene expression in human monocytes. Cell signalling (2001) 13(2):85–94. doi: 10.1016/s0898-6568(00)00149-2
93. Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol (2005) 5(12):953–64. doi: 10.1038/nri1733
94. Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity (2016) 44(3):439–49. doi: 10.1016/j.immuni.2016.02.024
95. Pollard JW. Trophic macrophages in development and disease. Nat Rev Immunol (2009) 9(4):259–70. doi: 10.1038/nri2528
96. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol (2008) 8(12):958–69. doi: 10.1038/nri2448
97. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity (2014) 41(1):14–20. doi: 10.1016/j.immuni.2014.06.008
98. Tugal D, Liao X, Jain MK. Transcriptional control of macrophage polarization. Arteriosclerosis thrombosis Vasc Biol (2013) 33(6):1135–44. doi: 10.1161/atvbaha.113.301453
99. Molawi K, Sieweke MH. Transcriptional control of macrophage identity, self-renewal, and function. Adv Immunol (2013) 120:269–300. doi: 10.1016/b978-0-12-417028-5.00010-7
100. Rajasekaran S, Tamatam CR, Potteti HR, Raman V, Lee JW, Matthay MA, et al. Visualization of fra-1/Ap-1 activation during lps-induced inflammatory lung injury using fluorescence optical imaging. Am J Physiol Lung Cell Mol Physiol (2015) 309(4):L414–24. doi: 10.1152/ajplung.00315.2014
101. Huang CK, Dai D, Xie H, Zhu Z, Hu J, Su M, et al. Lgr4 governs a pro-inflammatory program in macrophages to antagonize post-infarction cardiac repair. Circ Res (2020) 127(8):953–73. doi: 10.1161/circresaha.119.315807
102. Owens JM, Matsuo K, Nicholson GC, Wagner EF, Chambers TJ. Fra-1 potentiates osteoclastic differentiation in osteoclast-macrophage precursor cell lines. J Cell Physiol (1999) 179(2):170–8. doi: 10.1002/(sici)1097-4652(199905)179:2<170::Aid-jcp7>3.0.Co;2-k
103. Luo YP, Zhou H, Krueger J, Kaplan C, Liao D, Markowitz D, et al. The role of proto-oncogene fra-1 in remodeling the tumor microenvironment in support of breast tumor cell invasion and progression. Oncogene (2010) 29(5):662–73. doi: 10.1038/onc.2009.308
104. Jiang P, Gao W, Ma T, Wang R, Piao Y, Dong X, et al. Cd137 promotes bone metastasis of breast cancer by enhancing the migration and osteoclast differentiation of Monocytes/Macrophages. Theranostics (2019) 9(10):2950–66. doi: 10.7150/thno.29617
105. Hilt ZT, Maurya P, Tesoro L, Pariser DN, Ture SK, Cleary SJ, et al. Β2m signals monocytes through non-canonical tgfβ receptor signal transduction. Circ Res (2021) 128(5):655–69. doi: 10.1161/circresaha.120.317119
106. Ginhoux F, Jung S. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat Rev Immunol (2014) 14(6):392–404. doi: 10.1038/nri3671
107. Yang J, Zhang L, Yu C, Yang XF, Wang H. Monocyte and macrophage differentiation: Circulation inflammatory monocyte as biomarker for inflammatory diseases. biomark Res (2014) 2(1):1. doi: 10.1186/2050-7771-2-1
108. Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, et al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med (2007) 204(12):3037–47. doi: 10.1084/jem.20070885
109. Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, et al. Nomenclature of monocytes and dendritic cells in blood. Blood (2010) 116(16):e74–80. doi: 10.1182/blood-2010-02-258558
110. Sager HB, Hulsmans M, Lavine KJ, Moreira MB, Heidt T, Courties G, et al. Proliferation and recruitment contribute to myocardial macrophage expansion in chronic heart failure. Circ Res (2016) 119(7):853–64. doi: 10.1161/circresaha.116.309001
111. Swirski FK. The spatial and developmental relationships in the macrophage family. Arteriosclerosis thrombosis Vasc Biol (2011) 31(7):1517–22. doi: 10.1161/atvbaha.110.221150
112. Meisner JK, Song J, Price RJ. Arteriolar and venular remodeling are differentially regulated by bone marrow-derived cell-specific Cx3cr1 and Ccr2 expression. PLoS One (2012) 7(9):e46312. doi: 10.1371/journal.pone.0046312
113. Carlin LM, Stamatiades EG, Auffray C, Hanna RN, Glover L, Vizcay-Barrena G, et al. Nr4a1-dependent Ly6c(Low) monocytes monitor endothelial cells and orchestrate their disposal. Cell (2013) 153(2):362–75. doi: 10.1016/j.cell.2013.03.010
114. Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Sci (New York NY) (2007) 317(5838):666–70. doi: 10.1126/science.1142883
115. Zawada AM, Rogacev KS, Rotter B, Winter P, Marell RR, Fliser D, et al. Supersage evidence for Cd14++Cd16+ monocytes as a third monocyte subset. Blood (2011) 118(12):e50–61. doi: 10.1182/blood-2011-01-326827
116. Merino A, Buendia P, Martin-Malo A, Aljama P, Ramirez R, Carracedo J. Senescent Cd14+Cd16+ monocytes exhibit proinflammatory and proatherosclerotic activity. J Immunol (Baltimore Md 1950) (2011) 186(3):1809–15. doi: 10.4049/jimmunol.1001866
117. Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH, et al. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood (2011) 118(5):e16–31. doi: 10.1182/blood-2010-12-326355
118. Echlin DR, Tae HJ, Mitin N, Taparowsky EJ. B-atf functions as a negative regulator of ap-1 mediated transcription and blocks cellular transformation by ras and fos. Oncogene (2000) 19(14):1752–63. doi: 10.1038/sj.onc.1203491
119. Ubeda M, Vallejo M, Habener JF. Chop enhancement of gene transcription by interactions with Jun/Fos ap-1 complex proteins. Mol Cell Biol (1999) 19(11):7589–99. doi: 10.1128/mcb.19.11.7589
120. Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-jun interactions that mediate transcription regulatory specificity. Oncogene (2001) 20(19):2438–52. doi: 10.1038/sj.onc.1204385
121. de Marcken M, Dhaliwal K, Danielsen AC, Gautron AS, Dominguez-Villar M. Tlr7 and Tlr8 activate distinct pathways in monocytes during rna virus infection. Sci Signaling (2019) 12(605):eaaw1347. doi: 10.1126/scisignal.aaw1347
122. Phillipson M, Kubes P. The neutrophil in vascular inflammation. Nat Med (2011) 17(11):1381–90. doi: 10.1038/nm.2514
123. Sadik CD, Kim ND, Luster AD. Neutrophils cascading their way to inflammation. Trends Immunol (2011) 32(10):452–60. doi: 10.1016/j.it.2011.06.008
124. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Sci (New York NY) (2004) 303(5663):1532–5. doi: 10.1126/science.1092385
125. Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol (2013) 13(3):159–75. doi: 10.1038/nri3399
126. Lawrence SM, Corriden R, Nizet V. How neutrophils meet their end. Trends Immunol (2020) 41(6):531–44. doi: 10.1016/j.it.2020.03.008
127. Ratajczak-Wrona W, Wawrusiewicz-Kurylonek N, Garley M, Kretowski AJ, Jablonska E. A proliferation-inducing ligand regulation in polymorphonuclear neutrophils by panax ginseng. Archivum immunologiae therapiae experimentalis (2020) 68(6):32. doi: 10.1007/s00005-020-00597-z
128. Vaz M, Reddy NM, Rajasekaran S, Reddy SP. Genetic disruption of fra-1 decreases susceptibility to endotoxin-induced acute lung injury and mortality in mice. Am J Respir Cell Mol Biol (2012) 46(1):55–62. doi: 10.1165/rcmb.2011-0169OC
129. Netea MG, Balkwill F, Chonchol M, Cominelli F, Donath MY, Giamarellos-Bourboulis EJ, et al. A guiding map for inflammation. Nat Immunol (2017) 18(8):826–31. doi: 10.1038/ni.3790
130. Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. Innate and adaptive immune memory: An evolutionary continuum in the host's response to pathogens. Cell Host Microbe (2019) 25(1):13–26. doi: 10.1016/j.chom.2018.12.006
131. LeBien TW, Tedder TF. B lymphocytes: How they develop and function. Blood (2008) 112(5):1570–80. doi: 10.1182/blood-2008-02-078071
132. Zhu J. T Helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harbor Perspect Biol (2018) 10(10):a030338. doi: 10.1101/cshperspect.a030338
133. Chang JH, Xiao Y, Hu H, Jin J, Yu J, Zhou X, et al. Ubc13 maintains the suppressive function of regulatory T cells and prevents their conversion into effector-like T cells. Nat Immunol (2012) 13(5):481–90. doi: 10.1038/ni.2267
134. Sobolev V, Nesterova A, Soboleva A, Mezentsev A, Dvoriankova E, Piruzyan A, et al. Analysis of pparγ signaling activity in psoriasis. Int J Mol Sci (2021) 22(16):8603. doi: 10.3390/ijms22168603
135. Korn T, Bettelli E, Oukka M, Kuchroo VK. Il-17 and Th17 cells. Annu Rev Immunol (2009) 27:485–517. doi: 10.1146/annurev.immunol.021908.132710
136. Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector Th17 and regulatory T cells. Nature (2006) 441(7090):235–8. doi: 10.1038/nature04753
138. Ohkubo Y, Arima M, Arguni E, Okada S, Yamashita K, Asari S, et al. A role for c-Fos/Activator protein 1 in b lymphocyte terminal differentiation. J Immunol (Baltimore Md 1950) (2005) 174(12):7703–10. doi: 10.4049/jimmunol.174.12.7703
139. Inada K, Okada S, Phuchareon J, Hatano M, Sugimoto T, Moriya H, et al. C-fos induces apoptosis in germinal center b cells. J Immunol (Baltimore Md 1950) (1998) 161(8):3853–61.
140. Kobayashi K, Phuchareon J, Inada K, Tomita Y, Koizumi T, Hatano M, et al. Overexpression of c-fos inhibits down-regulation of a cyclin-dependent kinase-2 inhibitor P27kip1 in splenic b cells activated by surface ig cross-linking. J Immunol (Baltimore Md 1950) (1997) 158(5):2050–6.
141. Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of ap-1 function: A mechanism for the regulation of blimp-1 expression and b lymphocyte differentiation by the b cell lymphoma-6 protooncogene. J Immunol (Baltimore Md 1950) (2002) 169(4):1922–9. doi: 10.4049/jimmunol.169.4.1922
142. Grötsch B, Brachs S, Lang C, Luther J, Derer A, Schlötzer-Schrehardt U, et al. The ap-1 transcription factor Fra1 inhibits follicular b cell differentiation into plasma cells. J Exp Med (2014) 211(11):2199–212. doi: 10.1084/jem.20130795
143. Grötsch B, Lux A, Rombouts Y, Hoffmann AC, Andreev D, Nimmerjahn F, et al. Fra1 controls rheumatoid factor autoantibody production by bone marrow plasma cells and the development of autoimmune bone loss. J Bone mineral Res Off J Am Soc Bone Mineral Res (2019) 34(7):1352–65. doi: 10.1002/jbmr.3705
144. Huo L, Rothstein TL. Isolation and characterization of murine fra-1: Induction mediated by Cd40 and surface ig is protein kinase c dependent. J Immunol (Baltimore Md 1950) (1996) 157(9):3812–8.
145. Roebuck KA, Carpenter LR, Lakshminarayanan V, Page SM, Moy JN, Thomas LL. Stimulus-specific regulation of chemokine expression involves differential activation of the redox-responsive transcription factors ap-1 and nf-kappab. J leukocyte Biol (1999) 65(3):291–8. doi: 10.1002/jlb.65.3.291
146. Zagariya A, Mungre S, Lovis R, Birrer M, Ness S, Thimmapaya B, et al. Tumor necrosis factor alpha gene regulation: Enhancement of C/Ebpbeta-induced activation by c-jun. Mol Cell Biol (1998) 18(5):2815–24. doi: 10.1128/mcb.18.5.2815
147. Morishita H, Saito F, Kayama H, Atarashi K, Kuwata H, Yamamoto M, et al. Fra-1 negatively regulates lipopolysaccharide-mediated inflammatory responses. Int Immunol (2009) 21(4):457–65. doi: 10.1093/intimm/dxp015
148. Li QR, Ni WP, Lei NJ, Yang JY, Xuan XY, Liu PP, et al. The overexpression of Fra1 disorders the inflammatory cytokine secretion by mtec of myasthenia gravis thymus. Scandinavian J Immunol (2018) 88(1):e12676. doi: 10.1111/sji.12676
149. Yao T, Zhang L, Fu Y, Yao L, Zhou C, Chen G. Saikosaponin-d alleviates renal inflammation and cell apoptosis in a mouse model of sepsis Via Tcf7/Fosl1/Matrix metalloproteinase 9 inhibition. Mol Cell Biol (2021) 41(10):e0033221. doi: 10.1128/mcb.00332-21
150. Takada Y, Gresh L, Bozec A, Ikeda E, Kamiya K, Watanabe M, et al. Interstitial lung disease induced by gefitinib and toll-like receptor ligands is mediated by fra-1. Oncogene (2011) 30(36):3821–32. doi: 10.1038/onc.2011.101
151. Lu R, Kujawski M, Pan H, Shively JE. Tumor angiogenesis mediated by myeloid cells is negatively regulated by Ceacam1. Cancer Res (2012) 72(9):2239–50. doi: 10.1158/0008-5472.Can-11-3016
152. Wen J, Sun X, Chen H, Liu H, Lai R, Li J, et al. Mutation of Rnf213a by talen causes abnormal angiogenesis and circulation defects in zebrafish. Brain Res (2016) 1644:70–8. doi: 10.1016/j.brainres.2016.04.051
153. Nitkin CR, Xia S, Menden H, Yu W, Xiong M, Heruth DP, et al. Fosl1 is a novel mediator of Endotoxin/Lipopolysaccharide-induced pulmonary angiogenic signaling. Sci Rep (2020) 10(1):13143. doi: 10.1038/s41598-020-69735-z
154. Wang X, Abraham S, McKenzie JAG, Jeffs N, Swire M, Tripathi VB, et al. Lrg1 promotes angiogenesis by modulating endothelial tgf-Β signalling. Nature (2013) 499(7458):306–11. doi: 10.1038/nature12345
155. Sobolev VV, Zolotarenko AD, Soboleva AG, Sautin ME, Il'ina SA, Sarkisova MK, et al. [Expression of the Fosl1 gene in psoriasis and atherosclerosis]. Genetika (2010) 46(1):104–10. doi: 10.1134/S102279541001014X
156. Zhou C, Wang F, Ma H, Xing N, Hou L, Du Y, et al. Silencing of fos-like antigen 1 represses restenosis Via the Erk/Ap-1 pathway in type 2 diabetic mice. Diabetes Vasc Dis Res (2021) 18(6):14791641211058855. doi: 10.1177/14791641211058855
157. Rajasekaran S, Vaz M, Reddy SP. Fra-1/Ap-1 transcription factor negatively regulates pulmonary fibrosis in vivo. PLoS One (2012) 7(7):e41611. doi: 10.1371/journal.pone.0041611
158. Rajasekaran S, Reddy NM, Zhang W, Reddy SP. Expression profiling of genes regulated by fra-1/Ap-1 transcription factor during bleomycin-induced pulmonary fibrosis. BMC Genomics (2013) 14:381. doi: 10.1186/1471-2164-14-381
159. Zhu W, Li J, Su J, Li J, Li J, Deng B, et al. Fos-like antigen 1 is highly expressed in human psoriasis tissues and promotes the growth of hacat cells in vitro. Mol Med Rep (2014) 10(5):2489–94. doi: 10.3892/mmr.2014.2509
160. Sobolev VV, Zolotorenko AD, Soboleva AG, Elkin AM, Il'ina SA, Serov DN, et al. Effects of expression of transcriptional factor ap-1 Fosl1 gene on psoriatic process. Bull Exp Biol Med (2011) 150(5):632–4. doi: 10.1007/s10517-011-1208-0
161. Kireva T, Erhardt A, Tiegs G, Tilg H, Denk H, Haybaeck J, et al. Transcription factor fra-1 induces cholangitis and liver fibrosis. Hepatol (Baltimore Md) (2011) 53(4):1259–69. doi: 10.1002/hep.24175
162. Takada Y, Ray N, Ikeda E, Kawaguchi T, Kuwahara M, Wagner EF, et al. Fos proteins suppress dextran sulfate sodium-induced colitis through inhibition of nf-kappab. J Immunol (Baltimore Md 1950) (2010) 184(2):1014–21. doi: 10.4049/jimmunol.0901196
163. Park MJ, Moon SJ, Lee SH, Kim EK, Yang EJ, Min JK, et al. Blocking activator protein 1 activity in donor cells reduces severity of acute graft-Versus-Host disease through reciprocal regulation of il-17-Producing T Cells/Regulatory T cells. Biol Blood marrow Transplant J Am Soc Blood Marrow Transplant (2014) 20(8):1112–20. doi: 10.1016/j.bbmt.2014.04.031
164. Sobolev VV, Khashukoeva AZ, Evina OE, Geppe NA, Chebysheva SN, Korsunskaya IM, et al. Role of the transcription factor Fosl1 in organ development and tumorigenesis. Int J Mol Sci (2022) 23(3):1521. doi: 10.3390/ijms23031521
165. Zeng F, He J, Jin X, Liao Q, Chen Z, Peng H, et al. Fra-1: A key factor regulating signal transduction of tumor cells and a potential target molecule for tumor therapy. Biomedicine pharmacotherapy = Biomedecine pharmacotherapie (2022) 150:113037. doi: 10.1016/j.biopha.2022.113037
166. Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: Analysis for the global burden of disease study. Lancet (London England) (2020) 395(10219):200–11. doi: 10.1016/s0140-6736(19)32989-7
167. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA (2016) 315(8):801–10. doi: 10.1001/jama.2016.0287
168. Reinhart K, Daniels R, Kissoon N, Machado FR, Schachter RD, Finfer S. Recognizing sepsis as a global health priority - a who resolution. New Engl J Med (2017) 377(5):414–7. doi: 10.1056/NEJMp1707170
169. He C, Zhang Y, Duan Y, Yu J, Luo B, Jiang N, et al. [Identification of differentially expressed genes and pathways changing in neutrophils of patients with sepsis by bioinformatics analysis]. Xi bao yu fen zi mian yi xue za zhi = Chin J Cell Mol Immunol (2019) 35(6):481–90. doi: 10.13423/j.cnki.cjcmi.008818
170. Huang Y, Wang F, Li H, Xu S, Xu W, Pan X, et al. Inhibition of fibroblast growth factor receptor by Azd4547 protects against inflammation in septic mice. Inflammation (2019) 42(6):1957–67. doi: 10.1007/s10753-019-01056-4
171. Makled MN, El-Kashef DH. Saroglitazar attenuates renal fibrosis induced by unilateral ureteral obstruction Via inhibiting tgf-Β/Smad signaling pathway. Life Sci (2020) 253:117729. doi: 10.1016/j.lfs.2020.117729
172. Kent LN, Rumi MA, Kubota K, Lee DS, Soares MJ. Fosl1 is integral to establishing the maternal-fetal interface. Mol Cell Biol (2011) 31(23):4801–13. doi: 10.1128/mcb.05780-11
173. Fudala R, Allen TC, Krupa A, Cagle PT, Nash S, Gryczynski Z, et al. Increased levels of nuclear factor Κb and fos-related antigen 1 in lung tissues from patients with acute respiratory distress syndrome. Arch Pathol Lab Med (2011) 135(5):647–54. doi: 10.5858/2009-0660-oar1.1
174. Eferl R, Hasselblatt P, Rath M, Popper H, Zenz R, Komnenovic V, et al. Development of pulmonary fibrosis through a pathway involving the transcription factor fra-2/Ap-1. Proc Natl Acad Sci United States America (2008) 105(30):10525–30. doi: 10.1073/pnas.0801414105
175. Langley RG, Krueger GG, Griffiths CE. Psoriasis: Epidemiology, clinical features, and quality of life. Ann rheumatic Dis (2005) 64 Suppl 2(Suppl 2):ii18–23. doi: 10.1136/ard.2004.033217
176. Zolotarenko A, Chekalin E, Piruzian E, Bruskin S. Fra1 mediates the activation of keratinocytes: Implications for the development of psoriatic plaques. Biochim Biophys Acta Mol basis Dis (2018) 1864(12):3726–34. doi: 10.1016/j.bbadis.2018.09.016
177. Redlich K, Hayer S, Ricci R, David JP, Tohidast-Akrad M, Kollias G, et al. Osteoclasts are essential for tnf-Alpha-Mediated joint destruction. J Clin Invest (2002) 110(10):1419–27. doi: 10.1172/jci15582
178. Dai C, Rennhack JP, Arnoff TE, Thaker M, Younger ST, Doench JG, et al. Smad4 represses Fosl1 expression and pancreatic cancer metastatic colonization. Cell Rep (2021) 36(4):109443. doi: 10.1016/j.celrep.2021.109443
179. Vallejo A, Erice O, Entrialgo-Cadierno R, Feliu I, Guruceaga E, Perugorria MJ, et al. Fosl1 promotes cholangiocarcinoma Via transcriptional effectors that could be therapeutically targeted. J Hepatol (2021) 75(2):363–76. doi: 10.1016/j.jhep.2021.03.028
180. Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell (2005) 7(3):211–7. doi: 10.1016/j.ccr.2005.02.013
181. Xiang R, Luo Y, Niethammer AG, Reisfeld RA. Oral DNA vaccines target the tumor vasculature and microenvironment and suppress tumor growth and metastasis. Immunol Rev (2008) 222:117–28. doi: 10.1111/j.1600-065X.2008.00613.x
182. Luo Y, Zhou H, Mizutani M, Mizutani N, Reisfeld RA, Xiang R. Transcription factor fos-related antigen 1 is an effective target for a breast cancer vaccine. Proc Natl Acad Sci USA (2003) 100(15):8850–5. doi: 10.1073/pnas.1033132100
183. Luo Y, Zhou H, Mizutani M, Mizutani N, Liu C, Xiang R, et al. A DNA vaccine targeting fos-related antigen 1 enhanced by il-18 induces long-lived T-cell memory against tumor recurrence. Cancer Res (2005) 65(8):3419–27. doi: 10.1158/0008-5472.Can-04-3120
184. Beaulieu ME, Jauset T, Massó-Vallés D, Martínez-Martín S, Rahl P, Maltais L, et al. Intrinsic cell-penetrating activity propels omomyc from proof of concept to viable anti-myc therapy. Sci Trans Med (2019) 11(484). doi: 10.1126/scitranslmed.aar5012
185. Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, et al. Small-molecule myc inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell (2019) 36(5):483–97.e15. doi: 10.1016/j.ccell.2019.10.001
186. Uchihashi S, Fukumoto H, Onoda M, Hayakawa H, Ikushiro S, Sakaki T. Metabolism of the c-Fos/Activator protein-1 inhibitor T-5224 by multiple human udp-glucuronosyltransferase isoforms. Drug Metab disposition: Biol fate chemicals (2011) 39(5):803–13. doi: 10.1124/dmd.110.037952
187. Yang S, Meyskens FL Jr. Alterations in activating protein 1 composition correlate with phenotypic differentiation changes induced by resveratrol in human melanoma. Mol Pharmacol (2005) 67(1):298–308. doi: 10.1124/mol.104.006023
188. Chung JY, Huang C, Meng X, Dong Z, Yang CS. Inhibition of activator protein 1 activity and cell growth by purified green tea and black tea polyphenols in h-Ras-Transformed cells: Structure-activity relationship and mechanisms involved. Cancer Res (1999) 59(18):4610–7.
189. Prusty BK, Das BC. Constitutive activation of transcription factor ap-1 in cervical cancer and suppression of human papillomavirus (Hpv) transcription and ap-1 activity in hela cells by curcumin. Int J Cancer (2005) 113(6):951–60. doi: 10.1002/ijc.20668
190. Huang C, Ma WY, Dawson MI, Rincon M, Flavell RA, Dong Z. Blocking activator protein-1 activity, but not activating retinoic acid response element, is required for the antitumor promotion effect of retinoic acid. Proc Natl Acad Sci USA (1997) 94(11):5826–30. doi: 10.1073/pnas.94.11.5826
191. Fanjul A, Dawson MI, Hobbs PD, Jong L, Cameron JF, Harlev E, et al. A new class of retinoids with selective inhibition of ap-1 inhibits proliferation. Nature (1994) 372(6501):107–11. doi: 10.1038/372107a0
192. Mishra DK, Kim MP. Sr 11302, an ap-1 inhibitor, reduces metastatic lesion formation in ex vivo 4d lung cancer model. Cancer Microenviron Off J Int Cancer Microenviron Soc (2017) 10(1-3):95–103. doi: 10.1007/s12307-017-0202-0
193. Sun Y, Lin Z, Liu CH, Gong Y, Liegl R, Fredrick TW, et al. Inflammatory signals from photoreceptor modulate pathological retinal angiogenesis Via c-fos. J Exp Med (2017) 214(6):1753–67. doi: 10.1084/jem.20161645
194. Li JJ, Dong Z, Dawson MI, Colburn NH. Inhibition of tumor promoter-induced transformation by retinoids that transrepress ap-1 without transactivating retinoic acid response element. Cancer Res (1996) 56(3):483–9.
195. Niidome K, Taniguchi R, Yamazaki T, Tsuji M, Itoh K, Ishihara Y. Fosl1 is a novel target of levetiracetam for suppressing the microglial inflammatory reaction. Int J Mol Sci (2021) 22(20):10962. doi: 10.3390/ijms222010962
196. Cargnello M, Roux PP. Activation and function of the mapks and their substrates, the mapk-activated protein kinases. Microbiol Mol Biol Rev MMBR (2011) 75(1):50–83. doi: 10.1128/mmbr.00031-10
197. Kikuchi A, Kishida S, Yamamoto H. Regulation of wnt signaling by protein-protein interaction and post-translational modifications. Exp Mol Med (2006) 38(1):1–10. doi: 10.1038/emm.2006.1
198. Vaz M, Rajasekaran S, Potteti HR, Reddy SP. Myeloid-specific fos-related antigen-1 regulates cigarette smoke-induced lung inflammation, not emphysema, in mice. Am J Respir Cell Mol Biol (2015) 53(1):125–34. doi: 10.1165/rcmb.2014-0118OC
Keywords: FRA-1, AP-1, immune system, inflammation, tissue homeostasis
Citation: He Y-Y, Zhou H-F, Chen L, Wang Y-T, Xie W-L, Xu Z-Z, Xiong Y, Feng Y-Q, Liu G-Y, Li X, Liu J and Wu Q-P (2022) The Fra-1: Novel role in regulating extensive immune cell states and affecting inflammatory diseases. Front. Immunol. 13:954744. doi: 10.3389/fimmu.2022.954744
Received: 27 May 2022; Accepted: 22 July 2022;
Published: 11 August 2022.
Edited by:
Eva Reali, University of Milano-Bicocca, ItalyReviewed by:
Jianzhong Shen, Auburn University, United StatesCopyright © 2022 He, Zhou, Chen, Wang, Xie, Xu, Xiong, Feng, Liu, Li, Liu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing-Ping Wu, d3FwMTk2OEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.