- 1Department of Urology, Fudan University Shanghai Cancer Center, Shanghai, China
- 2Department of Interventional Oncology, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 3Affiliated Hospital of Youjiang Medical University for Nationalities, Baise, China
- 4The Medical Department, 3D Medicines Inc., Shanghai, China
- 5Institute for Developmental and Regenerative Cardiovascular Medicine, MOE-Shanghai Key Laboratory of Children’s Environmental Health, Xinhua Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Sarcomatoid differentiation is a highly aggressive pathological characteristic of renal cell carcinoma (RCC) and is characterized by susceptibility to progression and extremely poor prognosis. In this study, we included all genomic alteration events that led to a loss of protein function of MTAP and CDKN2A, and enrolled 5,307 RCC patients with genomic sequencing data from Western and Chinese cohorts. Notably, MTAP/CDKN2AMUT occurred in the Chinese population ~2 times more frequently than in the Western cohort and showed significant co-mutation trends. We found significantly higher proportions of sarcomatoid-positive patients with MTAPMUT or CDKN2AMUT compared with MTAP/CDKN2A wild-type (WT) patients (P < 0.001). Of the 574 RCC samples from the FUSCC cohort and 3,563 RCC samples from 17 independent cohorts, the MTAP/CDKN2AMUT significantly predicted extremely poor outcomes (P < 0.0001). The Western cohort suggested a concordant relationship between MTAP/CDKN2AMUT and sarcomatoid differentiation in RCC. Moreover, although MTAP/CDKN2AMUT RCC may be insensitive to targeted therapy, the high degree of tumor heterogeneity and higher PD-L1 and CXCL13 expression characterizations reflected that MTAP/CDKN2A-deficient features could benefit from immunotherapy for patients with RCC. This study utilized RCC samples from large-scale, global, multicenter sequencing cohorts and first proved that MTAP/CDKN2A deficiency significantly correlates with sarcomatoid differentiation in RCC and predicts aggressive progression, poor prognosis, and primary resistance to targeted therapy and potential favorable responses to immune checkpoint blockade. Unlike conventional targeted therapies, emerging drugs such as immunotherapies or synthetic lethal PRMT5 inhibitors may become novel therapeutic options for patients with MTAP/CDKN2AMUT RCC.
Introduction
Renal cell carcinoma (RCC) is one of the most common genitourinary malignancies (1), accounting for 3% of all malignant tumors, and its incidence is increasing at a rate of 3% every year (2). The incidence rate in major domestic cities of China, such as Beijing, Shanghai, and Hangzhou, has reached more than 8/100,000, which is more than double that of 10 years ago (3, 4). Clear cell renal cell carcinoma (ccRCC) is the predominant pathological type of kidney cancer, which accounts for about 78% of all RCC in adults. Nearly one-third of ccRCC patients encountered lymphatic, bone, or organ metastases at initial diagnosis, and the 5-year survival rate of patients with advanced ccRCC is less than 20% (5, 6). Although classic histological heterogeneity has been widely explored in the research of ccRCC, the latest advances in genomic technologies have demonstrated prominent molecular subtypes, which have assisted in elucidating the precise typing and treatment of ccRCC as well as mechanisms underlying the inevitable occurrence and development essence (7, 8). Therefore, the multi-omics approach from molecular and genomic levels have become important research techniques in the systematic study of tumor occurrence and treatment efficacy improvement for patients with ccRCC.
Sarcomatoid differentiation is a highly aggressive pathological and extremely uncommon characteristic of RCC and is characterized by susceptibility to metastasis and recurrence and extremely poor prognosis. Renal cell carcinoma with sarcomatoid dedifferentiation (sRCC) is insensitive to chemoradiotherapy and targeted therapy, and radical resection is the preferred treatment. Patients who have clear cell histology and a higher percentage of sarcomatoid differentiation may have worse outcomes with VEGF-targeted therapy (9). In addition, the more sarcoma components, the worse the prognosis of the patient. Even with active treatment, the median postoperative survival for patients with sRCC was still not optimistic (10). For example, with over 42 months of median follow-up, RCC patients with sarcomatoid histology who received nivolumab plus ipilimumab still had median overall survival that was not yet reached [(25.2–not estimable); n = 74] versus those who received sunitinib [14.2 months (9.3–22.9); n = 65; P = 0.0004] (11). Meanwhile, the JAVELIN Renal 101 trial enrolled 108 RCC patients with sarcomatoid histology (47 patients in the avelumab plus axitinib arm and 61 in the sunitinib arm), and patients with sRCC in the combination arm had improved efficacy outcomes versus those in the sunitinib arm (12). The median progression-free survival (PFS) was 7.0 months (95% CI, 5.3–13.8 months) versus 4.0 months (95% CI, 2.7–5.7 months), respectively. Although researchers keep on exploring the unique molecular pathogenesis and driver mutation spectrum of sRCC, the need for individual diagnostic or effective treatment options is urgent (13).
Methylthioadenosine (MTA) phosphorylase (MTAP) is a key enzyme of the methionine remediation synthesis pathway. In MTAP-deficient cancers, MTA accumulation selectively inhibits protein arginine methyltransferase 5 (PRMT5) enzyme activity and increases PRMT5 inhibition sensitivity (14). Precisely because of this synthetic lethal mechanism, the development and application of drugs targeting PRMT5 have led to new treatment options for patients with MTAP deletion tumors (15). Since MTAP is located on chromosome 9p21, close to the tumor suppressor CDKN2A, genomic alteration of MTAP/CDKN2A co-occurrence is frequent. At the EAU2021 conference, Necchi et al. compared genomic differences between sRCC and clear cell RCC (ccRCC). The authors found a significant increase in MTAP/CDKN2A loss in sRCC and proposed a potential role of anti-PRMT5 drugs in MTAP-deficient advanced sRCC (16). However, the lack of clinical data and the small sample size were major limitations.
In this study, we included all genomic alteration (GA) events that led to a loss of protein function in the statistical analysis. The study included 574 Chinese patients with RCC from the Fudan University Shanghai Cancer Center (FUSCC); 3,563 RCC samples from 17 independent cohorts were integrated (Western). MTAP/CDKN2A alteration frequency, clinicopathological features, and prognosis were depicted in the FUSCC and Western cohorts. Finally, we included 1,170 Chinese RCC patients from the 3D cohort with panel sequencing data. The relationship of CDKN2A mutations with therapeutic markers was further explored. We hypothesized that MTAP/CDKN2A-deficient features significantly correlate with sarcomatoid differentiation in RCC and predict poor outcomes, inactive targeted treatment responsiveness, and favorable responses to immune checkpoint blockade in patients with RCC.
Methods
Data collection and preprocessing from the discovery, testing, and validation sets
In the Chinese training cohort, 574 patients with available whole-exome sequencing (WES) data from the FUSCC (Shanghai, China) were included. In the Western testing cohort, the WES sequencing data of 3,563 ccRCC Caucasian patients were obtained from the cBioPortal for Cancer Genomics database (http://www.cbioportal.org/) with gene IDs converted from Ensembl ID to gene symbol matrix. The combined 3D medicine cohort included WES data of DNA extracted from clinically annotated tumor specimens and from whole blood (as the matched germline source) from a total of 1,170 Chinese RCC patients.
Genetic variation analysis
The somatic mutation data were processed using the “maftools” R package to screen and present the genes with the top 20 mutation frequencies. The chi-square test was implemented to analyze the differences in the mutation frequencies of the high-frequency mutant genes among the three cohorts. We counted all genomic alterations (specifically the focal loss of 9p21) to the MTAP and CDKN2A genes that sit next to each other on chromosome 9p21 and referred to the altered status as “MUT.”
Clinicopathological subgroup analysis
We divided patients into different subgroups based on different clinicopathological features, including neoplasm histological grade, cancer metastasis, cancer lymph node stage, neoplasm clinical stage, and race category; different hemogram features, including the levels of hemoglobin, platelet, WBC, and serum calcium; and different clinical therapy data, including individual neoplasm status, sunitinib treatment after operation, new neoplasm event after initial therapy indicator, and response to systematic first-line targeted therapy. The Fisher test was used to compare the differences in different clinical subgroups between the MTAP/CDKN2A-altered and MTAP/CDKN2A-unaltered groups.
Differential gene expression analysis and functional enrichment analysis
To explore the potential biological differences between the MTAP/CDKN2A-altered and MTAP/CDKN2A-unaltered patterns, the “limma” R package was used to identify differentially expressed genes (DEGs), and the threshold value was set as P <0.05, |logFC| ≥3.37. Functional enrichment analyses were carried out to explore the potential functions of the genomic alteration of MTAP/CDKN2A in patients with RCC using the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome databases (17).
Hematoxylin and eosin and immunohistochemistry staining analysis
Hematoxylin and eosin staining was conducted according to routine protocols. Briefly, after deparaffinization and rehydration, tissue sections were stained with hematoxylin solution and eosin solution (ZSGB-BIO, China), followed by dehydration with graded alcohol and clearing in xylene. The mounted slides were then examined and photographed using a LEICA DM3000 LED (Leica DMshare (v3), Germany) following the manufacturer’s protocols and previously described procedure (8). IHC was performed to evaluate the expression level of CXCL13 (1:1,000 dilution; ab246518; Abcam, USA) and PD-L1 (1:300 dilution; No. 19313684, CST, China) in ccRCC samples from the FUSCC according to standard procedures as previously described (8). Staining score and sarcomatoid differentiation features were independently measured by two experienced clinical pathologists.
Statistical analysis
In the statistical analyses, the Wilcoxon test was used to compare the differences between the two groups of samples. The survival curve was analyzed using Kaplan–Meier, and the log-rank test was used to assess the significance for disease-specific survival (DSS), disease-free survival (DFS), overall survival (OS), and PFS. The “survminer” R package was utilized to take the best cutoff value for all survival analyses. A P-value less than 0.05 was considered statistically significant.
Results
Clinical value of MTAP/CDKN2AMUT in sarcomatoid and prognosis of 574 patients with RCC from the FUSCC cohort
First, we summarized the mutation frequencies of MTAP/CDKN2A in the three cohorts, namely, the discovery set (FUSCC cohort, n = 574), the testing set (Western cohort, n = 3,563), and the validation cohort (3D medicine cohort, n = 1,170) (Figure 1A). The results showed the highest frequency of MTAPMUT and CDKN2AMUT (2.96% and 5.75%, respectively) in the FUSCC cohort, and the MTAPMUT frequency was approximately half of CDKN2AMUT frequency. Notably, MTAP/CDKN2AMUT occurred in the Chinese population ~2 times more frequently than in the Western cohort and showed significant co-mutation trends.
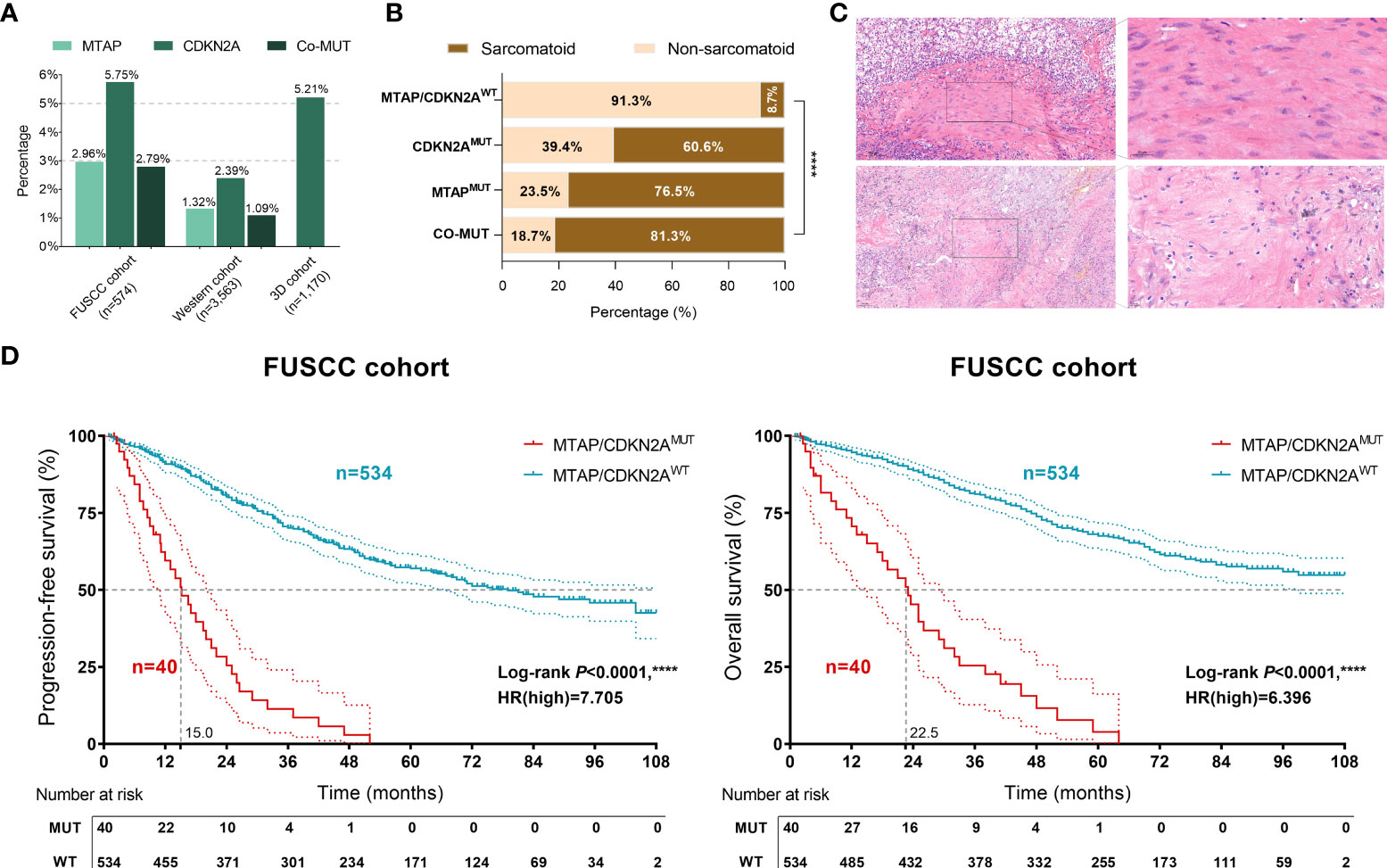
Figure 1 Clinical value of MTAP/CDKN2AMUT in sarcomatoid and the prognosis of 574 patients with renal cell carcinoma (RCC) from the discovery set of the Fudan University Shanghai Cancer Center (FUSCC) cohort. (A) Genomic alteration frequency of MTAP and CDKN2A in 574 Chinese patients with RCC from the FUSCC cohort; 3,563 RCC samples from 17 independent cohorts were integrated (Western cohort), and 1,170 Chinese patients with RCC were included from the 3D medicine cohort with panel sequencing data. (B) The proportion of sarcomatoid pathological features in 574 patients with RCC from the FUSCC cohort in different mutation groups. (C) The representative images of sarcomatoid pathological features in MTAP/CDKN2AMUT samples. (D) Classified by the MTAP/CDKN2AMUT, progression-free survival (PFS) and overall survival (OS) of 574 RCC patients from the FUSCC cohort using Kaplan–Meier curve analysis. ****, P<0.0001.
To verify the relationship between MTAP/CDKN2A mutations and sarcomatoid differentiation, we evaluated the pathological features of 574 samples from the FUSCC cohort and found significantly higher proportions of sarcomatoid-positive patients with MTAPMUT or CDKN2AMUT compared with MTAP/CDKN2A wild-type (WT) patients (P < 0.001; Figure 1B). The majority of MTAP/CDKN2AMUT samples showed sarcomatoid pathological features (Figure 1C). Of the 574 RCC samples, the MTAP/CDKN2AMUT significantly predicted extremely poor PFS [hazard ratio (HR)=7.705, P < 0.0001] and OS (HR = 6.369, P < 0.0001; Figure 1D). Taken together, we found that MTAP/CDKN2AMUT could significantly predict sarcomatoid differentiation and prognosis in RCC.
MTAP/CDKN2AMUT in histopathological subtypes and sarcomatoid differentiation of 3,563 patients with RCC from the Western cohort
To further test our hypothesis, we then explored MTAP/CDKN2AMUT frequencies in 3,563 RCC samples from the Western cohort. As shown in Figure 2A, MTAP/CDKN2AMUT is more common in non-ccRCC, and the frequency of progression and mortality events was significantly increased in the GA group. The frequency of MTAP/CDKN2AMUT in different RCC datasets is shown in Figure 2B. The results revealed that MTAP/CDKN2AMUT is more common in non-ccRCC histopathological subtypes, which is consistent with the consensus that sarcomatoid differentiation occurs more frequently in non-ccRCC. Interestingly, we found remarkable sarcomatoid differentiation in patients with both MTAPMUT and CDKN2AMUT (Figure 2C). Overall, the results from the Western cohort suggest a concordant relationship between MTAP/CDKN2AMUT and sarcomatoid differentiation in RCC.

Figure 2 MTAP/CDKN2AMUT in histopathological subtypes and sarcomatoid differentiation of 3,563 patients with RCC from the Western cohort. (A) Relationship between MTAP/CDKN2AMUT and histopathological subtypes of RCC, disease-free status, and OS status in 3,563 RCC samples from the Western cohort. (B) The frequency of MTAP/CDKN2A genomic alterations in different RCC datasets. (C) Remarkable sarcomatoid differentiation in patients with both MTAPMUT and CDKN2AMUT from The Cancer Genome Atlas. * Ratio is calculated as altered/profiled.
Implications of MTAP/CDKN2AMUT in prognosis, clinicopathological features, and sensitivity to therapy of 3,563 patients with RCC from the Western cohort
Next, we collected RCC samples with available survival and mutation information from the Western cohort. Significantly, the genomic alteration of MTAP prominently predicted shorter DSS (P < 0.0001, HR=5.675), PFS (P < 0.0001, HR = 6.000), DFS (P < 0.0001, HR = 6.008), and OS (P < 0.0001, HR = 3.669) in patients with RCC from the Western testing cohort (Figure 3A). Moreover, the genomic alteration of CDKN2A was also significantly associated with shorter DSS (P < 0.0001, HR = 7.415), PFS (P < 0.0001, HR = 6.441), DFS (P < 0.0001, HR = 5.902), and OS (P < 0.0001, HR = 4.837) in 3,563 patients with RCC from the Western cohort (Figure 3B).
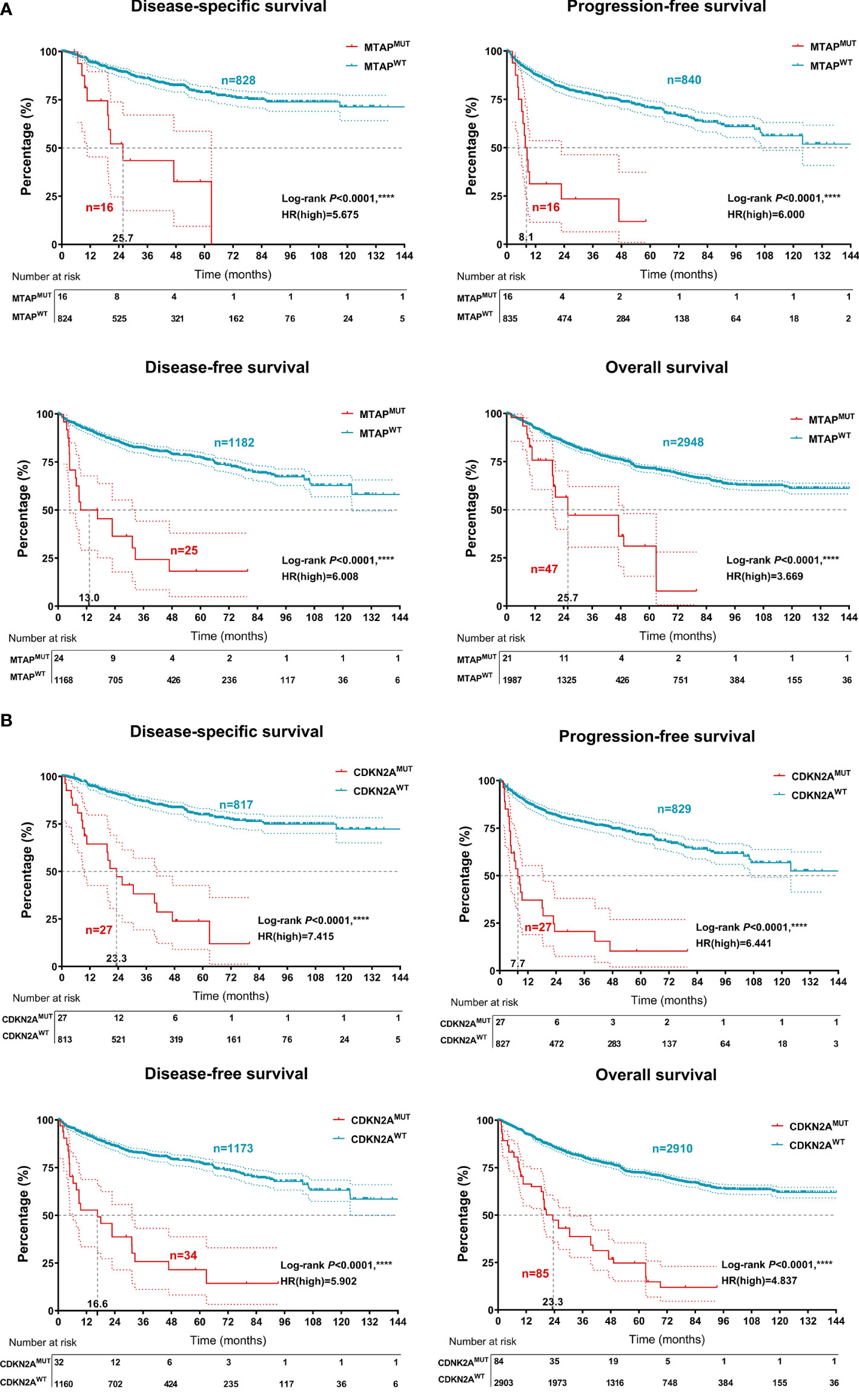
Figure 3 Prognostic implications of MTAPMUT and CDKN2AMUT 3,563 patients with RCC from the testing set of the Western cohort. (A) Classified by the MTAP/CDKN2AMUT, disease-specific survival, PFS, disease-free survival, and OS of RCC patients from the Western cohort using Kaplan–Meier survival analysis. (B) Association between MTAP/CDKN2AMUT and clinicopathological characteristics, association between MTAP/CDKN2AMUT and hemogram, and association between MTAP/CDKN2AMUT and clinical therapeutic efficacy. ****, P<0.0001.
Additionally, the results revealed that MTAP/CDKN2AMUT significantly predicted poor DSS (P < 0.0001, HR = 6.921), PFS (P < 0.0001, HR = 6.295), DFS (P < 0.0001, HR = 5.919), and OS (P < 0.0001, HR = 4.564) in patients with RCC from the testing set (Figure 4A). Moreover, RCC patients with MTAP/CDKN2AMUT showed significantly higher pathology grades (95.8% were G3–G4) and clinical stages (92.5% were III–IV; Figure 4B). Consistently, we found a significantly higher frequency of Asian subjects in the GA group than in the unaltered group. Additionally, patients in the GA group exhibited lower hemoglobin and leukocyte levels and higher platelet and blood calcium levels, reflecting a higher IMDC stage and a higher treatment primary tolerance. Additionally, in the GA group, the proportion of patients with unresectable tumors was 63.5%; 50% of the patients received postoperative sunitinib therapy, and approximately 40% of patients carried new neoplasm events after initial therapy. These findings suggested that patients in the GA group are primarily resistant to conventional targeted therapy (Figures S1A, B). Overall, to our knowledge, this is the first verification of the prominent relationship between MTAP/CDKN2AMUT and sRCC. MTAP/CDKN2AMUT could significantly predict progression, long-term survival, and treatment efficacy in RCC patients.
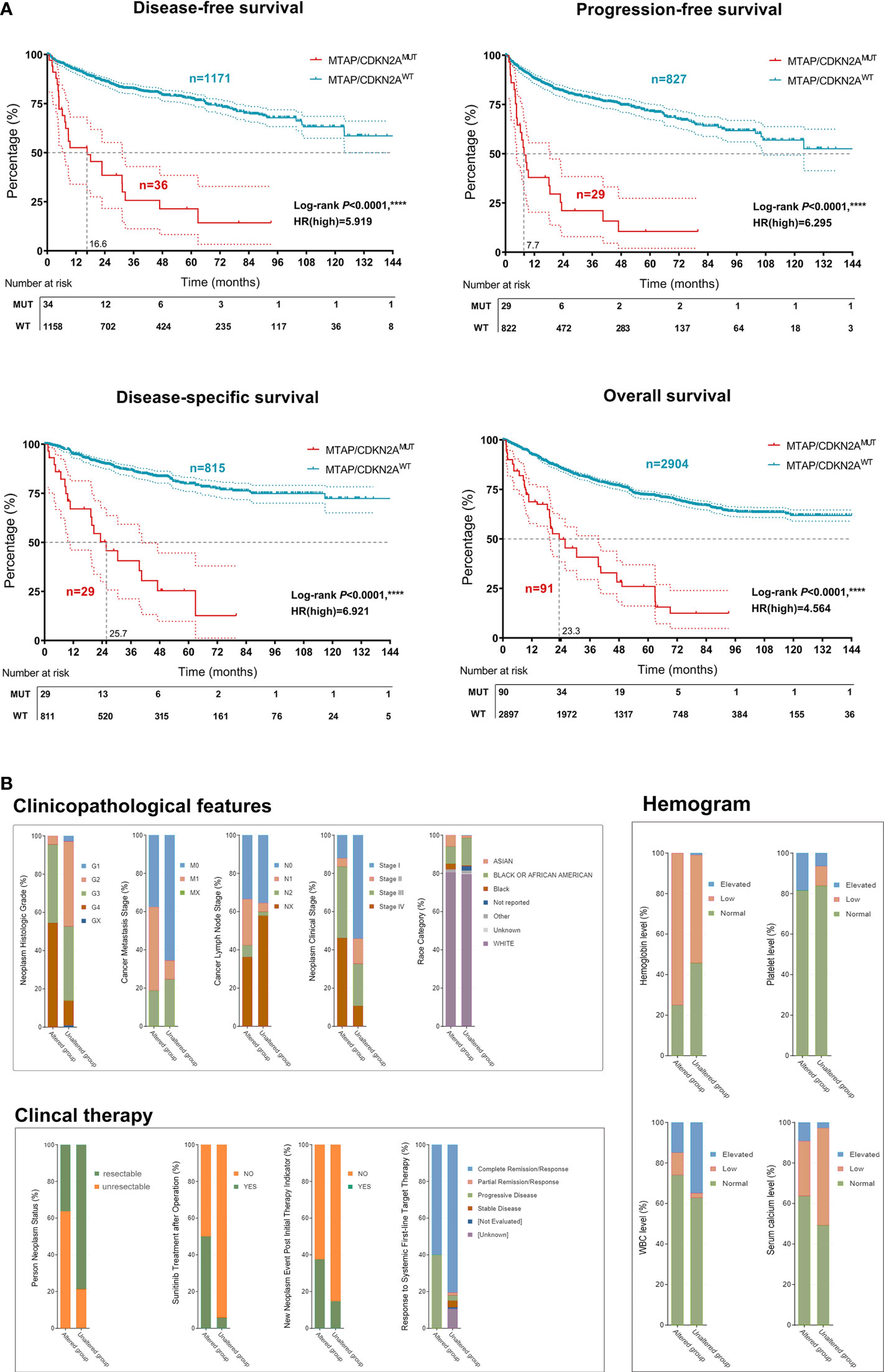
Figure 4 Implications of MTAP/CDKN2AMUT in prognosis, clinicopathological features, and sensitivity to therapy of 3,563 patients with RCC from the Western cohort. (A) Classified by the MTAPMUT, DSS, PFS, DFS, and OS of RCC patients from the Western cohort using Kaplan–Meier survival analysis and log-rank test. (B) Classified by the CDKN2AMUT, DSS, PFS, DFS, and OS of RCC patients from the Western cohort using Kaplan–Meier survival analysis and log-rank test. ****, P<0.0001.
MTAP/CDKN2AMUT predicts higher tumor heterogeneity, tumor microenvironment characterizations, and active responses to immune checkpoint blockade of RCC patients
The above findings aroused our interest regarding the impact of MTAP/CDKN2AMUT on RCC malignant biological functions. Therefore, we analyzed genotype differences in the Western cohort using the “limma” R package with a threshold of Log10(q-value) >2. A total of 363 significantly upregulated genes in the MTAP/CDKN2AMUT group were identified (Figure 5A). Functional enrichment suggested that MTAP/CDKN2AMUT response genes participate in the aggregation and activation of immune cells in the tumor microenvironment (TME). Similarly, pathway enrichment results revealed a marked involvement in the JAK–STAT, interferon, and mTOR signaling pathways, suggesting that MTAP/CDKN2AMUT may activate the anti-tumor immune response in the TME of RCC (Figure 5B). Notably, the 3D cohort showed significantly elevated tumor mutation burden (TMB) and PD-L1 levels in the CDKN2AMUT group (Figure 5C).

Figure 5 MTAP/CDKN2AMUT predicts tumor heterogeneity and TME characterizations of RCC patients from the 3D and FUSCC cohorts. (A) Identification of differential altered genes between MTAP/CDKN2AMUT and unaltered groups in the Western cohort using the “limma” R package with a threshold of Log10(q-value) >2. (B) Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome functional enrichment analyses were performed to speculate on the biological functions and tumor environment changes involved in MTAP/CDKN2AMUT. (C) Tumor mutation burden (TMB) and PD-L1 levels in the CDKN2AMUT group compared with the CDKN2AWT group in 1,170 Chinese patients with RCC from the 3D medicine cohort. (D, E) Protein expression levels of immune checkpoint molecule, PD-L1, and the key lymphokine that recruits tertiary lymphoid structures, CXCL13, were evaluated in ccRCC samples with available genomic data from the discovery set of the FUSCC cohort. The relative expression of PD-L1 and CXCL13 was defined as strong, moderate, and absent/weak staining according to the staining density and intensity of each section. The chi-square test was used to compare differences between groups. *, P<0.05; ****, P<0.0001.
To further portray intratumoral immunophenotypes according to genomic alterations of MTAP and CDKN2A, we evaluated the expression levels of the immune checkpoint molecule, PD-L1, and the key lymphokine that recruits tertiary lymphoid structures, CXCL13, in ccRCC samples with available genomic data from the discovery set of the FUSCC cohort. The relative expression of PD-L1 and CXCL13 was defined as strong, moderate, and absent/weak staining according to the staining density and intensity of each section. Interestingly, significantly higher PD-L1 and CXCL13 expression was found in 30 RCC samples with MTAP/CDKN2AMUT compared with 88 patients in the MTAP/CDKN2AWT group (P < 0.01; Figures 5D, E). In general, these results demonstrated that although MTAP/CDKN2AMUT RCC may be insensitive to targeted therapy, the high degree of tumor heterogeneity and immune-excluded TME reflected that MTAP/CDKN2AMUT patients could benefit from immunotherapies.
Discussion
The advancement of cancer genome analyses has revealed the detailed genomic landscapes of cancer. Targeted therapies based on the characteristics of the tumor genome are increasingly being offered to patients with cancers (18–20). Our study has essential implications for the design and analysis of molecular epidemiology studies in patients with sarcomatoid differentiation of RCC as well as the somatic characterization of genomes for the Chinese population. Among the Chinese and Caucasian populations, we assessed the mutation landscape and found the highest frequency of MTAPMUT and CDKN2AMUT (2.96% and 5.75%, respectively) in the FUSCC cohort, and the MTAPMUT frequency was approximately half of CDKN2AMUT frequency. Notably, MTAP/CDKN2AMUT occurred in the Chinese population ~2 times more frequently than in the Western cohort and showed significant co-mutation trends.
A previous study identified VHL mutations as the most common gene mutation (72%), followed by PBRM1 (45%), SETD2 (34%), and BAP1 mutations (17%) in 29 Caucasian patients with advanced ccRCC (21). It is also suggested that BAP1, SETD2, and PBRM1 are prevalent co-drivers of tumor grade and invasion of ccRCC and associated with aggressive progression (21–23). However, PBRM1-mutant patients tended to have a higher TMB and might evoke immunotherapy sensitivity (24). Furthermore, Malouf et al. detected genomic profiling, underpinning renal cell carcinoma with sarcomatoid dedifferentiation, and identified TP53 (42.3%), VHL (34.6%), CDKN2A (26.9%), and NF2 (19.2%) as the most frequently altered genes for sRCC (12, 25). Approaches and strategies are needed for other malignancies driven by the loss of CDKN2 and NF2, as revealed by genomic features in the course of clinical management for patients with sRCC. This study speculated that although the initiating mutation in sRCC is similar to other types of RCC, the acquisition of other driver alterations, such as TP53 or NF2, may lead to the generation of the sarcomatoid phenotype. Therefore, understanding the distinct genomic alteration background of the Chinese population with sarcomatoid differentiation of RCC will guide targeted therapies and immunotherapies, paving the way for the clinical practice of precision medicine in this highly lethal cancer.
Previous studies have revealed the unique molecular classification of ccRCC predicting prognosis in Caucasian patients treated with targeted therapies, specifically the TKIs (26). The molecular classification (ccRCC1–4) suggested a high predictive value for favorable outcomes in patients with advanced ccRCC treated with sunitinib (27). This study identified the clusters with MTAP/CDKN2A-altered and MTAP/CDKN2A-unaltered groups with distinct levels of genomic alteration, clinicopathological features, and immune infiltration in patients with RCC. Moreover, RCC patients with MTAP/CDKN2AMUT showed significantly higher pathology grades (95.8% were G3–G4) and clinical stages (92.5% were III–IV). Consistently, we found a significantly higher frequency of Asian subjects in the GA group than in the unaltered group. Additionally, patients in the GA group exhibited lower hemoglobin and leukocyte levels and higher platelet and blood calcium levels, reflecting a higher IMDC stage and a higher treatment primary tolerance. Additionally, in the GA group, the proportion of patients with unresectable tumors was 63.5%; 50% of the patients received postoperative sunitinib therapy, and approximately 40% of patients carried new neoplasm events after initial therapy.
The loss of MTAP expression has been linked to a tumor-promoting effect in multiple cancers, and MTAP may serve as a tumor suppressor (28–30). For example, in 2021, Han et al. found that 9p21 loss, normally CDKN2A (13.5%) and MTAP (9.3%), confers a cold tumor immune microenvironment, poor clinical outcomes, and primary resistance to immune checkpoint therapy for cancers (31). However, this study did not explore the clinicopathological significance of 9p21 deletion in RCC, let alone its impact on the dysregulation of TME. Despite the frequent loss of MTAP expression in high-grade gliomas, MTAP did not appear to be associated with a deteriorating outcome, and in-vitro models demonstrated that MTAP did not affect proliferation, invasion, and migration (32). Unlike in other tumors, non-synonymous mutations, neoantigens, insertions, or deletions caused by chromosomal structural changes and somatic copy number variations were not in significant consistency with the response to immune checkpoint therapies (ICTs) in ccRCC (33). Moreover, the elevated expression of PD-L1 correlated with poor OS, which was also observed in the pro-tumorigenic MTAP/CDKN2AMUT cluster (34, 35).
A previous study pointed out that genetic alteration of CDKN2A was correlated with reduced benefits from ICTs and changes in the TME of urothelial carcinoma (36). Furthermore, CDKN2A deletion and BAP1 mutations, as well as increased expression of MYC transcriptional programs, have been identified as genomic features of aggressive behavior for sarcomatoid and rhabdoid RCC (37). In this study, the relatively immune-infiltrated MTAP/CDKN2AMUT cluster showed a transcriptional signature indicative of pro-tumorigenic immune infiltration in tumors and prominently higher TMB values based on 1,170 Chinese patients with RCC. Interestingly, we identified the mutually exclusive aggressive tumor phenotypes in ccRCC. Through phenotypic analysis, favorable clinical response to ICTs, elevated expression of immune checkpoints, increased abundance of tumor-infiltrated lymphocyte infiltration, and elevated TMB and PD-L1 expression were observed in the MTAP/CDKN2AMUT cluster. However, just as not all solid tumors with high TMB are sensitive to ICTs, patients with high neoantigens are not necessarily accompanied by an elevated level of CD8+ T-cell infiltration in the TME of ccRCC (38). Therefore, under a paradigm of targeted therapies, such as TKIs, two of the clusters that are “immune-infiltrated TME” exerted poorer prognosis but might be uniquely responsive to immune checkpoint blockade, thereby improving treatment outcomes for ccRCC patients. Further tumor type-specific studies are warranted in investigating biomarkers for ICTs.
Conclusion
This study utilized RCC samples from large-scale, global, multicenter sequencing cohorts and first proved that genomic alteration of MTAP/CDKN2A significantly correlates with sarcomatoid differentiation in RCC and predicts aggressive progression, poor prognosis, primary resistance to targeted therapy, and potential favorable responses to immune checkpoint blockade. Unlike conventional targeted therapies, emerging drugs such as immunotherapies or synthetic lethal PRMT5 inhibitors may become novel therapeutic options for patients with MTAP/CDKN2AMUT RCC.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by ethics committee of Fudan University Shanghai Cancer Center (ID: 050432-4-1805C, Shanghai, China). The patients/participants provided their written informed consent to participate in this study.
Author contributions
Conceptualization: WX, AA, GW, and WL. Data curation and formal analysis: WX, WL, AA, GW, JX, W Shi, JS, and XT. Funding acquisition: WX, YQ, HZ, and DY. Investigation and methodology: WX, AA, WL, GW, JZ, and HZ. Resources and software: JZ, YQ, HZ, and DY. Supervision: JZ, HZ, and DY. Validation and visualization: WX, AA, WL, and JS. Original draft: WX, AA, WL, and GW. Editing: JZ, HZ, and DY. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the “Eagle” Program of Shanghai Anticancer Association (No. SHCY-JC-2021105), the Natural Science Foundation of Shanghai (No. 20ZR1413100). and Shanghai Municipal Health Bureau (No. 2020CXJQ03).
Acknowledgments
We are grateful to all patients for their dedicated participation in the current study. We expressed our sincere gratitude to Ms. Zoo for editing the figures for this study.
Conflict of interest
Author JX was employed by The Medical Department, 3D Medicines Inc., Shanghai, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.953721/full#supplementary-material
Abbreviations
ccRCC, clear cell renal cell carcinoma; DEGs, differentially expressed genes; DFS, disease-free survival; DSS, disease-specific survival; FUSCC, Fudan University Shanghai Cancer Center; GEO, Gene Expression Omnibus; MTAP, methylthioadenosine phosphorylase; PRMT5, protein arginine methyltransferase; OS, overall survival; TIME, tumor immune microenvironment; TME, tumor microenvironment; PFS, progression-free survival; RCC, renal cell carcinoma; sRCC, renal cell carcinoma with sarcomatoid dedifferentiation; TCGA, The Cancer Genome Atlas.
References
1. Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, et al. Renal cell carcinoma. Nat Rev Dis Primers (2017) 3:17009. doi: 10.1038/nrdp.2017.9
2. Siegel R, Miller K, Jemal A. Cancer statistics, 2020. CA: Cancer J Clin (2020) 70:7–30. doi: 10.3322/caac.21590
3. Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) (2021) 134:783–91. doi: 10.1097/CM9.0000000000001474
4. Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R, et al. Cancer incidence and mortality in China, 2016. J Natl Cancer Center (2022) 2:1–9. doi: 10.1016/j.jncc.2022.02.002
5. Miller K, Nogueira L, Mariotto A, Rowland J, Yabroff K, Alfano C, et al. Cancer treatment and survivorship statistics, 2019. CA: Cancer J Clin (2019) 69:363–85. doi: 10.3322/caac.21565
6. Gerlinger M, Rowan A, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New Engl J Med (2012) 366:883–92. doi: 10.1056/NEJMoa1113205
7. Xu W, Anwaier A, Ma C, Liu W, Tian X, Su J, et al. Prognostic immunophenotyping clusters of clear cell renal cell carcinoma defined by the unique tumor immune microenvironment. Front Cell Dev Biol (2021) 9:785410. doi: 10.3389/fcell.2021.785410
8. Xu W, Ma C, Liu W, Anwaier A, Tian X, Shi G, et al. Prognostic value, DNA variation and immunologic features of a tertiary lymphoid structure-related chemokine signature in clear cell renal cell carcinoma. Cancer Immunol Immunother (2022) 71:1923–35. doi: 10.1007/s00262-021-03123-y
9. Golshayan AR, George S, Heng DY, Elson P, Wood LS, Mekhail TM, et al. Metastatic sarcomatoid renal cell carcinoma treated with vascular endothelial growth factor-targeted therapy. J Clin Oncol (2009) 27:235–41. doi: 10.1200/JCO.2008.18.0000
10. Pichler R, Comperat E, Klatte T, Pichler M, Loidl W, Lusuardi L, et al. Renal cell carcinoma with sarcomatoid features: Finally new therapeutic hope? Cancers (Basel) (2019) 11:(422–6). doi: 10.3390/cancers11030422
11. Tannir NM, Signoretti S, Choueiri TK, McDermott DF, Motzer RJ, Flaifel A, et al. Efficacy and safety of nivolumab plus ipilimumab versus sunitinib in first-line treatment of patients with advanced sarcomatoid renal cell carcinoma. Clin Cancer Res (2021) 27:78–86. doi: 10.1158/1078-0432.CCR-20-2063
12. Choueiri TK, Larkin J, Pal S, Motzer RJ, Rini BI, Venugopal B, et al. Efficacy and correlative analyses of avelumab plus axitinib versus sunitinib in sarcomatoid renal cell carcinoma: post hoc analysis of a randomized clinical trial. ESMO Open (2021) 6:100101. doi: 10.1016/j.esmoop.2021.100101
13. Wang Z, Kim TB, Peng B, Karam J, Creighton C, Joon A, et al. Sarcomatoid renal cell carcinoma has a distinct molecular pathogenesis, driver mutation profile, and transcriptional landscape. Clin Cancer Res (2017) 23:6686–96. doi: 10.1158/1078-0432.CCR-17-1057
14. Mavrakis KJ, McDonald ER 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science (2016) 351:1208–13. doi: 10.1126/science.aad5944
15. Mulvaney KM, Blomquist C, Acharya N, Li R, Ranaghan MJ, O'Keefe M, et al. Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol Cell (2021) 81:3481–3495.e3487. doi: 10.1016/j.molcel.2021.07.019
16. Necchi A, Grivas P, Spiess P, Jacob J, Schrock A, Madison R, et al. Methylthioadenosine phosphorylase (MTAP) deletion is more common in sarcomatoid (srcRCC) than in clear cell renal cell carcinoma (ccRCC). Eur Urol (2021) 79:S868–9. doi: 10.1016/S0302-2838(21)01008-3
17. Xu W, Liu WR, Xu Y, Tian X, Anwaier A, Su JQ, et al. Hexokinase 3 dysfunction promotes tumorigenesis and immune escape by upregulating monocyte/macrophage infiltration into the clear cell renal cell carcinoma microenvironment. Int J Biol Sci (2021) 17:2205–22. doi: 10.7150/ijbs.58295
18. Zhou H, Liu J, Zhang Y, Huang Y, Shen J, Yang Y, et al. PBRM1 mutation and preliminary response to immune checkpoint blockade treatment in non-small cell lung cancer. NPJ Precis Oncol (2020) 4:6. doi: 10.1038/s41698-020-0112-3
19. Okamura R, Kato S, Lee S, Jimenez RE, Sicklick JK, Kurzrock R, et al. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J Immunother Cancer (2020) 8:e000438(1-6). doi: 10.1136/jitc-2019-000438
20. Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med (2016) 375:819–29. doi: 10.1056/NEJMoa1604958
21. Reig Torras O, Mishra A, Christie A, McKenzie T, Onabolu O, Singla N, et al. Molecular genetic determinants of shorter time on active surveillance in a prospective phase 2 clinical trial in metastatic renal cell carcinoma. Eur Urol (2021) 81:555–8. doi: 10.1016/j.eururo.2021.12.003
22. Espana-Agusti J, Warren A, Chew SK, Adams DJ, Matakidou A. Loss of PBRM1 rescues VHL dependent replication stress to promote renal carcinogenesis. Nat Commun (2017) 8:2026. doi: 10.1038/s41467-017-02245-1
23. Linehan WM, Ricketts CJ. The cancer genome atlas of renal cell carcinoma: findings and clinical implications. Nat Rev Urol (2019) 16:539–52. doi: 10.1038/s41585-019-0211-5
24. Huang Y, Wang J, Jia P, Li X, Pei G, Wang C, et al. Clonal architectures predict clinical outcome in clear cell renal cell carcinoma. Nat Commun (2019) 10:1245. doi: 10.1038/s41467-019-09241-7
25. Malouf GG, Ali SM, Wang K, Balasubramanian S, Ross JS, Miller VA, et al. Genomic characterization of renal cell carcinoma with sarcomatoid dedifferentiation pinpoints recurrent genomic alterations. Eur Urol (2016) 70:348–57. doi: 10.1016/j.eururo.2016.01.051
26. Beuselinck B, Verbiest A, Couchy G, Job S, de Reynies A, Meiller C, et al. Pro-angiogenic gene expression is associated with better outcome on sunitinib in metastatic clear-cell renal cell carcinoma. Acta Oncol (Stockholm Sweden) (2018) 57:498–508. doi: 10.1080/0284186x.2017.1388927
27. Verbiest A, Couchy G, Job S, Zucman-Rossi J, Caruana L, Lerut E, et al. Molecular subtypes of clear cell renal cell carcinoma are associated with outcome during pazopanib therapy in the metastatic setting. Clin Genitourinary Cancer (2018) 16:e605–12. doi: 10.1016/j.clgc.2017.10.017
28. Laera L, Guaragnella N, Giannattasio S, Moro L. 6-thioguanine and its analogs promote apoptosis of castration-resistant prostate cancer cells in a BRCA2-dependent manner. Cancers (Basel) (2019) 11:(945–59). doi: 10.3390/cancers11070945
29. Bertino JR, Waud WR, Parker WB, Lubin M. Targeting tumors that lack methylthioadenosine phosphorylase (MTAP) activity: current strategies. Cancer Biol Ther (2011) 11:627–32. doi: 10.4161/cbt.11.7.14948
30. Christopher SA, Diegelman P, Porter CW, Kruger WD. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res (2002) 62:6639–44.
31. Han G, Yang G, Hao D, Lu Y, Thein K, Simpson BS, et al. 9p21 loss confers a cold tumor immune microenvironment and primary resistance to immune checkpoint therapy. Nat Commun (2021) 12:5606. doi: 10.1038/s41467-021-25894-9
32. Menezes WP, Silva VAO, Gomes INF, Rosa MN, Spina MLC, Carloni AC, et al. Loss of 5'-methylthioadenosine phosphorylase (MTAP) is frequent in high-grade gliomas; nevertheless, it is not associated with higher tumor aggressiveness. Cells (2020) 9:(492–515). doi: 10.3390/cells9020492
33. Braun DA, Hou Y, Bakouny Z, Ficial M, Sant' Angelo M, Forman J, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med (2020) 26:909–18. doi: 10.1038/s41591-020-0839-y
34. Darrow JJ, Avorn J, Kesselheim AS. FDA Approval and regulation of pharmaceuticals, 1983-2018. JAMA (2020) 323:164–76. doi: 10.1001/jama.2019.20288
35. Black JRM, McGranahan N. Genetic and non-genetic clonal diversity in cancer evolution. Nat Rev Cancer (2021) 21:379–92. doi: 10.1038/s41568-021-00336-2
36. Adib E, Nassar AH, Akl EW, Abou Alaiwi S, Nuzzo PV, Mouhieddine TH, et al. CDKN2A alterations and response to immunotherapy in solid tumors. Clin Cancer Res (2021) 27:4025–35. doi: 10.1158/1078-0432.CCR-21-0575
37. Bakouny Z, Braun DA, Shukla SA, Pan W, Gao X, Hou Y, et al. Integrative molecular characterization of sarcomatoid and rhabdoid renal cell carcinoma. Nat Commun (2021) 12:808. doi: 10.1038/s41467-021-21068-9
Keywords: renal cell carcinoma, sarcomatoid differentiation, immunotherapy, genomic alteration, MTAP, CDKN2A, tumor microenvironment
Citation: Xu W, Anwaier A, Liu W, Wei G, Su J, Tian X, Xia J, Qu Y, Zhao J, Zhang H and Ye D (2022) Genomic alteration of MTAP/CDKN2A predicts sarcomatoid differentiation and poor prognosis and modulates response to immune checkpoint blockade in renal cell carcinoma. Front. Immunol. 13:953721. doi: 10.3389/fimmu.2022.953721
Received: 26 May 2022; Accepted: 05 July 2022;
Published: 01 August 2022.
Edited by:
Xiaoran Yin, Second Affiliated Hospital of Xi’an Jiaotong University, ChinaReviewed by:
Jianfeng Yang, Shanghai University of Traditional Chinese Medicine, ChinaGuiming Zhang, The Affiliated Hospital of Qingdao University, China
Jun Wang, Sun Yat-sen University Cancer Center (SYSUCC), China
Copyright © 2022 Xu, Anwaier, Liu, Wei, Su, Tian, Xia, Qu, Zhao, Zhang and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dingwei Ye, ZHd5ZWxpZUAxNjMuY29t; Hailiang Zhang, emhhbmdobDkxOEAxNjMuY29t; Yuanyuan Qu, cXV5eTE5ODdAMTYzLmNvbQ==; Jianyuan Zhao, emhhb2p5QGZ1ZGFuLmVkdS5jbg==
†These authors have contributed equally to this work
 Wenhao Xu
Wenhao Xu Aihetaimujiang Anwaier
Aihetaimujiang Anwaier Wangrui Liu
Wangrui Liu Gaomeng Wei3†
Gaomeng Wei3† Jiaqi Su
Jiaqi Su Xi Tian
Xi Tian Yuanyuan Qu
Yuanyuan Qu Jianyuan Zhao
Jianyuan Zhao Hailiang Zhang
Hailiang Zhang Dingwei Ye
Dingwei Ye